Abstract
Hippocampal subfields exhibit differential vulnerabilities to Alzheimer’s disease-associated pathology including abnormal accumulation of amyloid-β deposition and neurofibrillary tangles. These pathological processes extensively impact on the structural and functional interconnectivities of the subfields and may explain the association between hippocampal dysfunction and cognitive deficits. In this study, we investigated the degree of alterations in the microstructure of hippocampal subfields across the clinical continuum of Alzheimer’s disease.
We applied a grey matter-specific multi-compartment diffusion model (Cortical-Neurite orientation dispersion and density imaging) to understand the differential effects of Alzheimer’s disease pathology on the hippocampal subfield microstructure. A total of 119 participants were included in this cross-sectional study. Participants were stratified into three categories, cognitively normal (n = 47), mild cognitive impairment (n = 52), and Alzheimer’s disease (n = 19). Diffusion MRI, plasma biomarkers and neuropsychological test scores were used to determine the association between the microstructural integrity and Alzheimer’s disease-associated molecular indicators and cognition. For Alzheimer’s disease-related plasma biomarkers, we studied amyloid-β, total tau and neurofilament light; for Alzheimer’s disease-related neuropsychological tests, we included the Trail Making Test, Rey Auditory Verbal Learning Test, Digit Span and Montreal Cognitive Assessment.
Comparisons between cognitively normal subjects and those with mild cognitive impairment showed significant microstructural alterations in the hippocampal cornu ammonis (CA) 4 and dentate gyrus region, whereas CA 1–3 was the most sensitive region for the later stages in the Alzheimer’s disease clinical continuum. Among imaging metrics for microstructures, the volume fraction of isotropic diffusion for interstitial free water demonstrated the largest effect size in between-group comparisons. Regarding the plasma biomarkers, neurofilament light appeared to be the most sensitive biomarker for associations with microstructural imaging findings in CA4-dentate gyrus. CA 1–3 was the subfield which had stronger correlations between cognitive performance and microstructural metrics. Particularly, poor performance on the Rey Auditory Verbal Learning Test and Montreal Cognitive Assessment was associated with decreased intracellular volume fraction.
Overall, our findings support the value of tissue-specific microstructural imaging for providing pathologically relevant information manifesting in the plasma biomarkers and neuropsychological outcomes across various stages of Alzheimer’s disease.
Keywords: diffusion magnetic resonance imaging, microstructure, Alzheimer’s disease, hippocampal subfields, plasma biomarkers
Shahid et al. report that microstructural imaging can detect subtle changes in individual hippocampal subfields across the clinical diagnostic continuum of Alzheimer’s disease. The imaging metrics are associated with plasma biomarkers and cognitive outcomes in both a group and region-specific manner.
See Toniolo (https://doi.org/10.1093/brain/awac190) for a scientific commentary on this article.
See Toniolo (https://doi.org/10.1093/brain/awac190) for a scientific commentary on this article.
Introduction
Sporadic Alzheimer’s disease is a neurodegenerative disease that is one of the most common causes of dementia. Symptomatic phases of Alzheimer’s disease progression, starting with the prodromal phase of mild cognitive impairment (MCI) and advancing with dementia severity, are well characterized by pathological alterations in cortical structures and white matter degeneration.1 These pathological alterations are also shown to correlate with cognitive decline.1 The neuropathological hallmarks of Alzheimer’s disease include abnormal accumulation of amyloid-β (Aβ) in extracellular neuritic plaques and intra-cellular neurofibrillary tangles, which are composed of hyperphosphorylated microtubule-associated tau protein.2,3 The accumulation of these pathological proteins begins years, even decades, before the onset of clinical symptoms.4
A number of studies have reported abnormal deposition of Aβ and neurofibrillary tangles in the medial temporal lobe, including the hippocampus, during the early stages of Alzheimer’s disease.5 The hippocampus along with its parahippocampal network connections is considered to be one of the most important regions supporting episodic memory.6 Since episodic memory is one of the earliest and most severely affected cognitive functions in Alzheimer’s disease,7 the involvement of the hippocampus in Alzheimer’s disease is of primary research interest.
The hippocampus is a heterogeneous and complex region consisting of functionally and anatomically interconnecting, yet distinct subfields.8 These subfields include the subiculum complex (anterior hippocampus), the cornu ammonis (CA) subregions comprising CA1–4 (posterior regions), the dentate gyrus (DG) and the hippocampal fissure. A number of histopathological studies suggest that there are differential Alzheimer’s disease-associated pathological changes among various hippocampal subfields.9 The Alzheimer’s disease-associated differential changes among the subfields are also observed in structural MRI studies using volumetric,9 shape-based10 and diffusion MRI.11,12
The accumulation of neurofibrillary tangles and Aβ-aggregates starts at the very early stages of the Alzheimer’s disease in the hippocampal regions. These pathological proteins disrupt the tissue microstructural organization, resulting in deterioration of the tissue cytoarchitecture and myeloarchitecture, causing sclerosis and partial breakdown of intracellular organelles. Alterations in these microstructural barriers likely influence the water diffusivity profile of the underlying tissue in the hippocampus.13 These microstructural changes are often a precursor of macroscopic volumetric changes and some studies have reported that these changes are independent of macroscopic volume loss.14–17 On this basis, several studies have employed diffusion tensor imaging (DTI) to detect Alzheimer’s disease-associated pathological alterations in the microstructural organization of hippocampal tissue.11,12,18–21 Recent studies, however, have cast doubt on the sensitivity of DTI-derived microstructural indices in detecting Alzheimer’s disease-associated changes in the hippocampus, compared with macroscopic volumetric changes.12,22,23
Owing to the heterogeneous microstructural organization of grey matter, we believe that some of the discrepancies in the literature maybe due to methodological limitation of DTI in mapping Alzheimer’s disease-associated pathological alterations in the hippocampus. Since DTI assumes the diffusion process to be Gaussian, it can only provide average measurements of water diffusion from multiple compartments (e.g. intracellular space, extracellular matrix, compartments with isotropic diffusivity). These compartments would likely exhibit different diffusivities, shapes and orientations; thus, DTI cannot properly capture diffusivity profile in regions of complex white matter fibre configurations, highly heterogeneous regions such as the grey matter and voxels contaminated by partial volume effect24–28 To address some of the limitations of DTI, a recent study employed a white matter neurite orientation dispersion and density imaging (NODDI) model29 to estimate the sensitivity of the multi-compartment model over the single tensor scheme in the hippocampus of healthy ageing participants30 The authors reported higher sensitivity of the NODDI model to age-related differences in grey matter compared with DTI.
The aim of the present cross-sectional study was to elucidate the microstructural alterations in hippocampal subfields during the prodromal and dementia stages of Alzheimer’s disease. We examined the differential effects of Alzheimer’s disease-related pathology on the hippocampal subfield microstructure using a tissue-specific multi-compartment diffusion model. We hypothesize that the grey matter-specific multi-compartment model (Cortical-NODDI)31 would be sensitive to Alzheimer’s disease-associated pathological alterations in hippocampal subfield microstructures across various stages of the disease. To the end, we investigate the relationship of the NODDI-derived grey matter microstructural metrics in the hippocampal subfields with Alzheimer’s disease plasma biomarkers, namely, the ratio of Aβ42 to Aβ40, total tau and neurofilament light chain (NfL). We further hypothesize that these grey matter microstructural metrics will be significantly associated with critical cognitive performance in Alzheimer’s disease.
Materials and methods
Study participants
A total of 119 participants from the Indiana Memory and Aging Study (IMAS) at the Indiana Alzheimer’s Disease Research Center (IADRC) were included in this cross-sectional study. The participants included cognitively normal controls (n = 47), individuals with mild cognitive impairment (MCI; n = 52) and patients with Alzheimer disease (n = 19). Demographic distribution of the participants is provided in Table 1. All IADRC participants received the Uniform Data Set (UDS3)32 battery (used in National Institute on Aging Alzheimer’s Disease Research Centers) and additional neuropsychological tests used at the IADRC with special emphasis on memory and executive function (see details in the neuropsychological assessment section). Exclusion criteria were significant cerebrovascular disease or malformations; history of systemic chemotherapy or radiation therapy to the head; current major depression; history of schizophrenia, bipolar disorder, developmental disability, Parkinson’s disease or other neurological disorders, brain surgery, brain infection, or significant head injury; and alcohol or illicit drug dependency. All participants provided written informed consent according to procedures approved by the Institutional Committee for the Protection of Human Subjects at Indiana University School of Medicine.
Table 1.
Demographic, plasma biomarkers and cognitive profiles of the participants
| Group | P-value | |||||
|---|---|---|---|---|---|---|
| CN (n = 47) | MCI (n = 52) | AD (n = 19) | CN versus MCI | CN versus AD | MCI versus AD | |
| Age (years) | 70.75 ± 4.78 | 72.98 ± 6.61 | 72.84 ± 8.41 | 0.06 | 0.32 | 0.95 |
| Education (years) | 16.57 ± 2.34 | 15.67 ± 2.85 | 15.42 ± 3.17 | 0.08 | 0.16 | 0.76 |
| Sex (female/male) | 36/11 | 30/22 | 12/8 | 0.04 | 0.26 | 0.68 |
| Race (Caucasian/African American/Asian/others) | 38/9/0/0 | 45/7/0/0 | 14/5/1/0 | 0.44 | 0.25 | 0.09 |
| APOE ε4 carrier status: sample size (0:1)a | n = 42 (27/15) | n = 45 (17/28) | n = 17 (4/13) | 0.01 | 0.005 | 0.29 |
| Aβ40 (pg/ml) | 272.74 ± 62.82 | 291.54 ± 58.49 | 278.76 ± 69.40 | 0.23 | 0.82 | 0.63 |
| Aβ42 (pg/ml) | 14.54 ± 3.86 | 13.78 ± 3.20 | 12.61 ± 3.74 | 0.40 | 0.19 | 0.41 |
| Aβ42/Aβ40 | 0.053 ± 0.009 | 0.047 ± 0.006 | 0.045 ± 0.005 | 0.002 | 0.001 | 0.29 |
| T-tau (pg/ml) | 3.96 ± 1.54 | 3.83 ± 0.93 | 4.32 ± 0.76 | 0.67 | 0.33 | 0.12 |
| NfL (pg/ml) | 19.12 ± 7.78 | 25.41 ± 10.75 | 36.34 ± 16.31 | 0.01 | 0.009 | 0.07 |
| TMT-A | 30.46 ± 9.08 | 43.40 ± 28.17 | 80.00 ± 42.96 | 0.003 | <0.001 | 0.008 |
| TMT-B | 71.06 ± 24.76 | 130.63 ± 74.92 | 221.87 ± 70.84 | <0.001 | <0.001 | <0.001 |
| TMT-(B−A) | 40.59 ± 20.59 | 90.83 ± 73.78 | 145.25 ± 40.99 | <0.001 | <0.001 | 0.008 |
| RAVLT-IR | 46.83 ± 8.04 | 29.88 ± 6.86 | 23.85 ± 6.67 | <0.001 | <0.001 | 0.06 |
| RAVLT-DR | 10.02 ± 2.75 | 2.78 ± 2.74 | 1.28 ± 2.56 | <0.001 | <0.001 | 0.19 |
| Digit span forward | 8.04 ± 2.14 | 7.33 ± 2.51 | 6.26 ± 2.25 | 0.13 | 0.013 | 0.13 |
| Digit span backward | 7.21 ± 2.15 | 5.58 ± 2.09 | 4.53 ± 3.33 | <0.001 | 0.009 | 0.26 |
| MoCA | 26.27 ± 2.01 | 20.76 ± 3.78 | 12.58 ± 5.73 | <0.001 | <0.001 | <0.001 |
AD = Alzheimer’s disease; CN = cognitively normal; DR = delayed recall; IR = immediate recall; MoCA = Montreal Cognitive Assessment; RAVLT-IR = sum score of initial five learning trials. P-values were derived from the Welch’s t-test except for sex, race and APOE where P-values were obtained using chi-squared test (χ2 test).
0 = non-carrier for alleles ε2 ε3 and ε3 ε3; 1 = carrier for alleles ε2 ε4, ε3 ε4, and ε4 ε4.
Clinical and neuropsychological assessment
Participants were evaluated using a detailed neuropsychological protocol, including measures of memory, attention, executive function, language, spatial ability, general intellectual ability and psychomotor speed, as well as other tests in standard dementia screens. Tests included, but were not limited to, the Trail Making Test [TMT, Part A, Part B and the difference (B − A)], Rey Auditory Verbal Learning Test (RAVLT) immediate recall and delayed recall, digit span (forward and backward), MoCA, clinical dementia rating scale, the 15-item geriatric depression scale and the cognitive change index to evaluate self-reported subjective cognitive decline.33,34
Diagnoses were made based on a multidisciplinary clinical consensus panel review aligning with the criteria by National Institute on Aging-Alzheimer’s Association workgroups (NIH-AA)35 at the time of study initiation. The consensus panel included neurologists (M.R.F., L.G.A.), clinical neuropsychologists (F.W.U., A.J.S.), geriatric psychiatrists and other disciplines and trainees. Supplementary Table 1 lists selected primary tests and diagnostic criteria. Participants without measurable cognitive deficits in all tests36–38 and without significant memory concerns (Total < 20 on the first 12-items of the self-report form of the cognitive change index33) were considered cognitively normal. The MCI individuals had significant complaints about their cognition (reported by themselves, an informant or as assessed by a clinician). They also demonstrated significant deficits (>1.5 SD below normal) in either memory or other cognitive domains without significant impairment in daily functioning.35 Alzheimer’s disease dementia participants exhibited a significant decline in cognition and daily functioning (assessed by the UDS functional assessment scale39) and met the criteria for Alzheimer’s disease diagnosis as recommended by NIH-AA.40 Neuropsychological performance across all groups is provided in Table 1.
Plasma fluid analysis
Blood samples were collected by venipuncture. A 10 ml EDTA (purple-top) tube was used to collect whole blood which was centrifuged at 4°C, 1962 × g for 15 min. Plasma was aliquoted into cryovials, frozen upright and stored in a −80°C freezer within 2 h of collection until analysis. Plasma Aβ40, Aβ42, NfL and total tau concentrations were measured using singleplex single molecule array (Simoa) assays on an HD-1 analyser according to instructions provided by the manufacturer (Quanterix). The measurements were performed in one round of experiments using one batch of reagents by board-certified laboratory technicians who were blinded to clinical data. Coefficients of variation were 6.3–10%.
MRI
Image acquisition
Imaging was performed on a single Siemens Prisma 3 T scanner with a 64-channel RF receiver head coil. All participants underwent anatomical T1-weighted, high-resolution T2-weighted and multi-shell diffusion MRI. T1-weighted imaging was acquired using a 3D magnetization-prepared rapid-gradient echo (MPRAGE) sequence with imaging parameters matching the Alzheimer’s Disease Neuroimaging Initiative-2 protocol (http://adni.loni.usc.edu/methods/documents/mri-protocols/). The high-resolution turbo-spin-echo T2-weighted images were acquired using a high in-plane resolution of 0.4 × 0.4 mm2 in an oblique plane perpendicular to the main axis of the hippocampus. The other acquisition parameters were as follows: slice thickness = 2.0 mm, repetition time/echo time = 8310/50 ms, flip angle = 122o, field of view = 175 mm2, number of slices = 32 and a GRAPPA acceleration factor of 2.41 The diffusion MRI was carried out using single-shot spin-echo echo-planar imaging with a hybrid diffusion imaging (HYDI)-encoding scheme.42 The HYDI-encoding scheme contained three zero-diffusion-weighting (b-value = 0 s/mm2) and five concentric-diffusion-weighting shells for a total of 142 diffusion-sensitizing gradient directions (six directions at b-values = 250 s/mm2, 21 at 1000 s/mm2, 24 at 2000 s/mm2, 30 at 3250 s/mm2 and 61 at 5000 s/mm2).43,44 The remaining acquisition parameters were as follows: multi-band acceleration factor = 3, repetition time/echo time = 2690/83.6 ms, field of view = 240 mm2, acquisition matrix = 120 × 120, voxel resolution = 2 × 2 × 2 mm3, 86 slices, diffusion duration (δ) = 20.50 ms and diffusion time (Δ) = 39.69 ms. An additional b = 0 s/mm2 image with reverse phase encoding polarity was acquired for susceptibility-induced geometric distortion correction.
Image processing
The hippocampal subfields were first delineated on the T1-weighted and high-resolution T2-weighted images using FreeSurfer software. The individual subfields were then combined into three distinct regions of interest before transforming to the diffusion space, where the diffusion microstructural indices were extracted. The following are step by step details. Hippocampal subfield segmentation was carried out with Freesurfer 7.1.1 (https://surfer.nmr.mgh.harvard.edu) using T1-weighted and high-resolution T2-weighted image data. To minimize partial volume effect, the subfields were combined as follows: CA1 and CA2/3 were combined as CA1–3; CA4 and GC-ML-DG were combined as CA4-DG; and parasubiculum, presubiculum and subiculum were combined as subiculum. Left and right regions were also combined. Bilateral hippocampal volumes and total intracranial volumes were also obtained as covariates in the statistical analyses described below. For each subject, the subfield segmentation quality was visually inspected.
The raw diffusion-weighted images (DWI) were preprocessed to reduce signal noise,45,46 effects from Gibbs ringing,47 subject motion,48 susceptibility induced geometric distortions48 and B1 field inhomogeneity.49 Multi-compartment microstructural imaging was performed using NODDI, which has been shown to be more specific to the underlying microstructure.29 To achieve a more physiologically plausible representation of the grey matter microstructure, the grey matter-optimized intracellular intrinsic parallel diffusivity of 1.1 mm2/s was used instead of a white matter-specific value of 1.7 mm2/s.31,50 Orientation dispersion index (ODI), volume fraction of isotropic water diffusivity (VISO), intracellular volume fraction (VFIC) and extracellular volume fraction (VFEC) parametric maps were obtained using the Dmipy toolbox, which is a python-based open-source application programming interface based on the DIPY framework.51
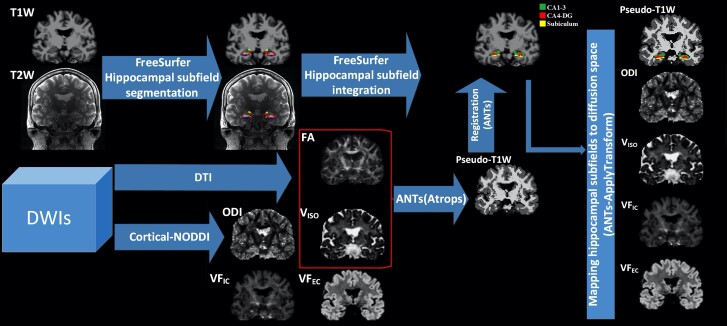
To extract NODDI-derived microstructural parameters from bilateral hippocampal subfields, the pipeline proposed by Nazeri et al.52 was adopted. Briefly, for each subject, a white matter fraction map was generated from a fractional anisotropy map. Using the ‘Atropos’ function in the advanced normalization tools,53 a grey matter fraction map was obtained by subtracting the white matter fraction map and VISO (CSF) map from 1.0. The three binarized tissue fractional maps were weighted according to the tissue type (grey matter = 2, white matter = 1, CSF = 0). Using these weighted images, a pseudo-T1-weighted image was obtained by adding all three weightings.26 A high quality pseudo-T1-weighted image increases the accuracy of transformation between T1-weighted space and diffusion space. The pseudo-T1-weighted image in diffusion space was then non-linearly registered to the T1-weighted image using the advanced normalization registration tool.53 FreeSurfer generated bilateral hippocampal subfields were then transformed to subject diffusion space using inverse transform matrix, which was generated during the forward registration. The regional (subfield) mean values of the ODI, VISO, VFIC and VFEC were calculated using the FSL ‘fslstats’ tool. To obtain robust regional mean values on diffusion space, individual grey matter fraction maps were scaled at 0.85 and then binarized to generate a robust grey matter mask. Each regional mean value was then extracted within the confines of the robust grey matter mask. The robust mean values were winsorized by excluding ±5% of the regional extreme values. The general schematic of the workflow is shown in Fig. 1.
Figure 1.
The schematics of the image processing framework. To generate bilateral hippocampal subfields masks in subject’s T1-weighted space, subject-specific T1-weighted and high-resolution T2-weighted images were used in FreeSurfer.Subject-specific DWIs were used to generate DTI-derived fractional anisotropy (FA) and cortical-NODDI-derived parametric maps of microstructure (i.e. ODI, VISO, VFIC, and VFEC). For each subject, FA and VISO maps were used in ANTs to generate the pseudo-T1-weighted map. The pseudo-T1-weighted map was linearly registered to the subject’s T1-weighted image. Bilateral hippocampal subfield masks were mapped to the subject’s diffusion space. For each subject, regional (i.e. subfield) mean values of parametric maps were calculated for further analyses.
Statistical analysis
Table 1 shows the demographic profile, plasma biomarker characteristics and neuropsychological performance test scores of the participants. For categorical variables (i.e. sex, APOE ε4 status and race), the between-group differences were compared using χ2 tests. For non-categorical data, the between-group attributes were compared using Welch’s unequal variance t-test. To investigate general group differences in the hippocampal subfield microstructural metrics, multivariate analysis of covariance (MANCOVA) with general linear models was used. Post hoc tests were then conducted to further understand individual group-wise comparisons. Across the study cohorts and within each group, we further investigated associations of diffusion microstructural metrics in the hippocampal subfields with the plasma biomarkers of Alzheimer’s disease pathology using partial correlation. To evaluate the relationship between the hippocampal subfield microstructure and cognitive performance, partial correlations models were used to test the association between the microstructural metrics and neuropsychological scores across the cohorts. The above analyses accounted for the effects of age, sex, education, APOE ε4 status and total intracranial volume using wild bootstrap with 5000 samples in SPSS (IBM SPSS version 27). To account for multiple comparisons across the three regions of interest, false discovery rate correction using Benjamini-Hochberg criterion (α = 0.05) was used. PFDR < 0.05 was deemed significant.
Data availability
The data used in this study were acquired via NIH-NIA funded R01 projects collected through the Indiana Alzheimer’s Disease Research Center (IADRC; NIH P30). Therefore, we will comply with the NIH data sharing policy and guidance (http://grants.nih.gov/grants/policy/data_sharing/data_sharing_guidance.htm) as well as the data sharing plan outlined by the IADRC.
Briefly, we will make the data available as early as feasible to qualified researchers who have obtained institutional review board approval from their institution and who are willing to sign a data-sharing agreement. Requestors must agree to NIH policies regarding privacy, data security and ethical practices, including the requirement that no attempt will be made to determine the identities of participants or their relatives. The principal investigators will review requests for anonymized human imaging data. Requestors will be encouraged to develop collaborative analyses with the project investigators, but this will not be required for data access. The data processing and analysis codes used in this study are from open-source software tools can be freely downloaded (see the ‘Materials and methods’ section). The code developed in-house will be available upon request and follow aforementioned data sharing policy.
Results
Participant characteristics
The demographic, plasma biomarker and neuropsychological profiles of the participants are summarized in Table 1. There were no significant differences among the cognitively normal, MCI and Alzheimer’s disease participants in terms of age, race or years of education. Overall, there were more female participants in all groups. The sex distribution was significantly different between the cognitively normal and MCI participants, with more males in the MCI group compared with the cognitively normal group. The APOE ε4 status was significantly different between groups with more APOE ε4 carriers in the MCI and Alzheimer’s disease groups than the cognitively normal group. In terms of the plasma biomarkers, although no group differences were observed in individual Aβ40 and Aβ42, the Aβ42/Aβ40 ratio was significantly different between cognitively normal and MCI participants and between cognitively normal and Alzheimer’s disease participants, with Alzheimer’s disease and MCI patients exhibiting lower Aβ42/Aβ40 ratio values relative to cognitively normal participants. NfL was significantly lower in cognitively normal compared to MCI and Alzheimer’s disease participants. In terms of neuropsychological test scores, TMT-A and TMT-B were significantly different among all three groups, with poorer performance in MCI and Alzheimer’s disease participants. The RAVLT scores were significantly higher in cognitively normal relative to both MCI and Alzheimer’s disease participants. MoCA scores were significantly different across all three groups, with MCI participants showing lower performance than cognitively normal subjects and Alzheimer’s disease participants showing lower performance than both MCI and cognitively normal subjects.
Sensitivity of diffusion microstructural metrics across the hippocampal subfields
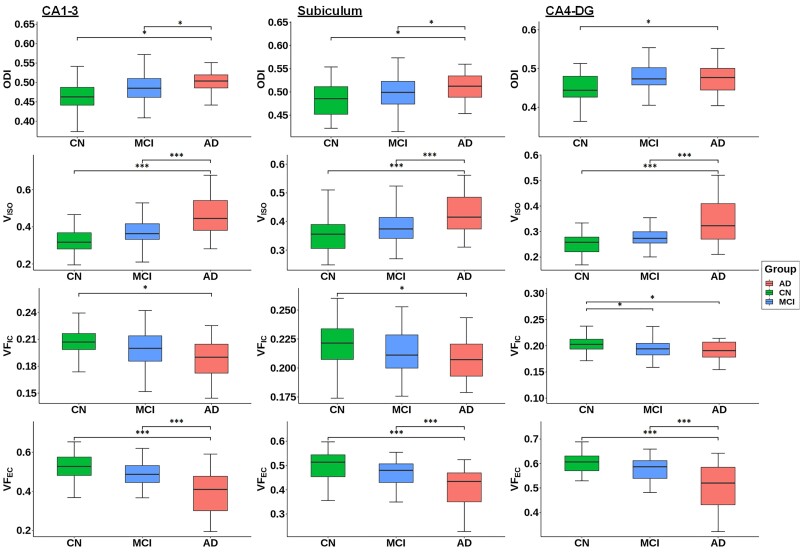
The NODDI derived microstructural indices in the hippocampal subfields were compared across the three clinical groups. The overall group-difference analysis showed statistical significance across multiple hippocampal subfields, except for VFIC in the subiculum (Table 2). Large effect sizes in group differences were observed in VISO in CA1–3 (ηp2 = 0.24) as well as VFEC in CA1–3 (ηp2 = 0.21) and the subiculum (ηp2 = 0.22). To further explore the pair-wise group differences in the microstructural indices across the hippocampal subfields, FDR-corrected post-hoc analyses were performed (Fig. 2 and Table 3). In the early stage of the Alzheimer’s disease clinical continuum, VFIC exhibited significant alterations in CA4-DG, where the MCI participants had significantly lower VFIC compared to the cognitively normal group (PFDR < 0.05; Hedge’s g = 0.53). At the later stage of the Alzheimer’s disease clinical continuum, significant group differences between cognitively normal and Alzheimer’s disease were observed in all the microstructural indices across all the subfields. The largest effect sizes were observed in VISO and VFEC (PFDR < 0.001, Hedge’s g > 1). When comparing between the MCI and Alzheimer’s disease participants, VISO exhibited the largest effect size in CA1–3 (PFDR < 0.001; Hedge’s g = −0.95) and in CA4-DG (PFDR < 0.001; Hedge’s g = −0.98). Overall, in the later Alzheimer’s disease clinical continuum, both CA1–3 and CA4-DG showed great sensitivity in group differences with high number of significant findings and large averaged effect sizes (averaged Hedge’s g = 1.02 and 1.03, respectively). Furthermore, VISO was the most sensitive microstructural metric with the largest averaged effect size (averaged Hedge’s g = 1.11).
Table 2.
Comparisons of diffusion microstructural metrics in the hippocampal subfields across groups
| Subfield region | df | df error | F | P-value | Partial Eta squared (ηp2) |
|---|---|---|---|---|---|
| ODI | |||||
| CA1–3 | 2 | 103 | 6.438 | 0.002 | 0.118 |
| Subiculum | 2 | 103 | 3.222 | 0.044 | 0.063 |
| CA4-DG | 2 | 103 | 4.181 | 0.018 | 0.080 |
| VISO | |||||
| CA1–3 | 2 | 103 | 14.946 | <0.001 | 0.237 |
| Subiculum | 2 | 103 | 8.541 | <0.001 | 0.151 |
| CA4-DG | 2 | 103 | 10.731 | <0.001 | 0.183 |
| VFIC | |||||
| CA1–3 | 2 | 103 | 3.950 | 0.022 | 0.076 |
| Subiculum | 2 | 103 | 2.869 | 0.062 | 0.056 |
| CA4-DG | 2 | 103 | 4.518 | 0.013 | 0.086 |
| VFEC | |||||
| CA1–3 | 2 | 103 | 13.076 | <0.001 | 0.214 |
| Subiculum | 2 | 103 | 13.862 | <0.001 | 0.224 |
| CA4-DG | 2 | 103 | 10.037 | <0.001 | 0.173 |
MANCOVA (df = 2) controlling for age, sex, education, APOE ε4 status and total intracranial volume.
Figure 2.
Group differences of Cortical-NODDI-derived ODI, VISO, VFIC and VFEC in the hippocampal subfields among the cognitively normal, MCI and Alzheimer’s disease participants. The comparison was conducted using general linear model with age, sex, level of education, APOE ε4 status and total intracranial volume as covariates. Multiple comparisons across three regions of interest (i.e. subfields) were adjusted by FDR using the Benjamini-Hochberg criterion (α = 0.05). *PFDR < 0.05; **PFDR < 0.01; ***PFDR < 0.001. AD = Alzheimer disease; CN = cognitively normal.
Table 3.
Pair-wise group differences with the post hoc analyses
| Subfields | Diffusion indices | CN versus MCI | CN versus AD | MCI versus AD | |||
|---|---|---|---|---|---|---|---|
| Mean difference (CN-MCI) | Effect size (Hedge’s g) | Mean difference (CN-AD) | Effect size (Hedge’s g) | Mean difference (MCI-AD) | Effect size (Hedge’s g) | ||
| CA1–3 | ODI | −0.010 | −0.548 | −0.037** | −1.025 | −0.028** | −0.602 |
| VISO | −0.015 | −0.554 | −0.107*** | −1.412 | −0.092*** | −0.950 | |
| VFIC | 0.007 | 0.435 | 0.017** | 0.901 | 0.011 | 0.440 | |
| VFEC | 0.017 | 0.56 | 0.109*** | 1.359 | 0.092*** | 0.856 | |
| Subiculum | ODI | −0.001 | −0.246 | −0.025* | −0.757 | −0.024* | −0.461 |
| VISO | −0.011 | −0.439 | −0.070*** | −1.191 | −0.060*** | −0.782 | |
| VFIC | 0.006 | 0.301 | 0.014* | 0.572 | 0.008 | 0.275 | |
| VFEC | 0.022 | 0.673 | 0.088*** | 1.427 | 0.067*** | 0.797 | |
| CA4-DG | ODI | −0.016 | −0.633 | −0.032** | −0.766 | −0.016 | −0.172 |
| VISO | −0.005 | −0.498 | −0.069*** | −1.313 | −0.064*** | −0.984 | |
| VFIC | 0.010* | 0.528 | 0.015** | 0.788 | 0.005 | 0.187 | |
| VFEC | 0.004 | 0.49 | 0.073*** | 1.380 | 0.068*** | 0.918 | |
AD = Alzheimer disease; CN = cognitively normal. Analysis corrected for age, sex, education, APoE ε4 status and total intracranial volume. Bootstrap results are based on 5000 bootstrap samples. Multiple comparisons across three regions of interest (i.e. subfields) were adjusted by FDR using Benjamini-Hochberg criterion (α = 0.05).
*PFDR < 0.05; **PFDR < 0.01; ***PFDR < 0.001.
Association between the diffusion microstructural indices and the plasma biomarkers
To understand the relation of the Alzheimer’s disease plasma biomarkers to hippocampal subfield microstructures, partial correlation of the diffusion MRI metrics in the subfields with the plasma biomarkers was carried out first across all groups. Since the rate of accumulation of these pathological proteins may differ across various clinically defined stages of the disease, the analysis was further carried out separately for each group. Across all the participants (Table 4), only NfL had significant associations with the diffusion microstructural metrics. The significant associations were in CA4-DG with VISO (r = 0.36; PFDR < 0.05) and with VFEC (r = −0.30; PFDR < 0.05). In the cognitively normal participants (Supplementary Table 2), similarly, NfL was positively associated with VISO in CA4-DG (r = 0.47; PFDR < 0.05). In the Alzheimer’s disease participants (Supplementary Table 4), on the other hand, the total tau level showed significant negative associations with the ODI in the subiculum (r = −0.98; PFDR < 0.05) and CA4-DG (r = −0.98; PFDR < 0.05). The associations were not significant in the MCI participants (Supplementary Table 3). Within all three groups, no significant associations were observed for any microstructural metrics with the Aβ42/Aβ40 ratio.
Table 4.
Partial correlation between the diffusion microstructural metrics and the plasma biomarkers in all participants
| Plasma biomarkers | CA1–3 | Subiculum | CA4-DG | |||
|---|---|---|---|---|---|---|
| r | P-value | r | P-value | r | P-value | |
| ODI | ||||||
| Aβ42/Aβ40 | −0.246 | 0.047a | −0.060 | 0.635 | −0.249 | 0.044a |
| T-tau | 0.009 | 0.940 | 0.094 | 0.453 | 0.014 | 0.908 |
| NfL | 0.246 | 0.047a | 0.011 | 0.933 | 0.251 | 0.042a |
| VISO | ||||||
| Aβ42/Aβ40 | −0.088 | 0.482 | −0.191 | 0.125 | −0.021 | 0.865 |
| Total tau | 0.150 | 0.228 | −0.018 | 0.887 | −0.014 | 0.911 |
| NfL | 0.258 | 0.037a | 0.154 | 0.217 | 0.357 | 0.003* |
| VFIC | ||||||
| Aβ42/Aβ40 | −0.067 | 0.593 | −0.010 | 0.939 | 0.013 | 0.916 |
| Total tau | 0.069 | 0.583 | −0.067 | 0.595 | 0.218 | 0.078 |
| NfL | −0.019 | 0.883 | −0.142 | 0.256 | −0.149 | 0.231 |
| VFEC | ||||||
| Aβ42/Aβ40 | 0.124 | 0.320 | 0.207 | 0.095 | 0.072 | 0.566 |
| Total tau | −0.170 | 0.173 | −0.031 | 0.807 | −0.070 | 0.577 |
| NfL | −0.249 | 0.043a | −0.193 | 0.120 | −0.299 | 0.015* |
r = correlation coefficient. Analysis adjusted for age, sex, education, APoE ε4 status and total intracranial volume. Bootstrap results are based on 5000 bootstrap samples. P-values in bold survived FDR correction for multiple comparison at PFDR < 0.05 using Benjamini-Hochberg criterion (α = 0.05).
*PFDR < 0.05.
Uncorrected P < 0.05.
Association between the diffusion microstructural metrics and the neuropsychological performance scores
To understand the association between hippocampal subfield microstructural alterations and cognitive performance, a partial-correlation analysis was performed between the diffusion metrics and neuropsychological test scores (Table 5). After FDR correction (α = 0.05), VFIC exhibited the most associations with the neuropsychological scores across all the subfields (0.20 < r < 0.33). High VFIC was associated with better cognitive performance in the MoCA and RAVLT-immediate recall. Particularly, VFIC had the highest correlation coefficient with the MoCA in CA1–3 (r = 0.33; PFDR < 0.05). Among the neuropsychological tests, TMT was the least sensitive instrument for correlating with the imaging microstructural biomarkers. Across the subfields, CA1–3 demonstrated as the most important subfield with most significant associations and highest effect sizes (r > 0.45) between cognitive performance and the microstructural metrics. In CA1–3, high cognitive performance (i.e. MoCA and RAVLT) was associated with decreased microstructural dispersion (ODI) and interstitial free-water diffusion (VISO), and with increased intracellular and extracellular volume fractions for restricted (VFIC) and hindered (VFEC) diffusion, respectively.
Table 5.
Partial correlation between the neuropsychological test scores and diffusion MRI metrics
| Neuropsychological tests | CA1–3 | Subiculum | CA4-DG | |||
|---|---|---|---|---|---|---|
| r | P-value | r | P-value | r | P-value | |
| ODI | ||||||
| MoCA | −0.272 | 0.007* | −0.175 | 0.089 | −0.166 | 0.106 |
| RAVLT-IR | −0.261 | 0.023a | −0.061 | 0.601 | −0.166 | 0.152 |
| RAVLT-DR | −0.220 | 0.052 | −0.133 | 0.243 | −0.148 | 0.193 |
| TMT (B−A) | 0.079 | 0.470 | −0.077 | 0.484 | 0.192 | 0.078 |
| VISO | ||||||
| MoCA | −0.532 | <0.001* | −0.333 | 0.001* | −0.403 | <0.001* |
| RAVLT-IR | −0.370 | 0.001* | −0.089 | 0.445 | −0.077 | 0.511 |
| RAVLT-DR | −0.327 | 0.003* | −0.155 | 0.174 | −0.109 | 0.338 |
| TMT (B−A) | 0.019 | 0.860 | 0.022 | 0.839 | −0.054 | 0.624 |
| VFIC | ||||||
| MoCA | 0.330 | 0.001* | 0.236 | 0.020* | 0.201 | 0.049* |
| RAVLT-IR | 0.257 | 0.025* | 0.257 | 0.025* | 0.268 | 0.019* |
| RAVLT-DR | 0.136 | 0.232 | 0.213 | 0.059 | 0.178 | 0.117 |
| TMT (B−A) | −0.183 | 0.094 | −0.118 | 0.282 | −0.293 | 0.006* |
| VFEC | ||||||
| MoCA | 0.494 | <0.001* | 0.411 | <0.001* | 0.423 | <0.001* |
| RAVLT-IR | 0.289 | 0.011* | 0.199 | 0.085 | 0.048 | 0.678 |
| RAVLT-DR | 0.312 | 0.005* | 0.203 | 0.073 | 0.114 | 0.316 |
| TMT (B−A) | 0.023 | 0.838 | −0.089 | 0.417 | 0.116 | 0.291 |
DR = delayed recall; IR = immediate recall; r = correlation coefficient. Analysis adjusted for age, sex, education, APoE ε4 status and total intracranial volume. Bootstrap results are based on 5000 bootstrap samples. P-values in bold survived FDR correction for multiple comparison at PFDR < 0.05 using Benjamini-Hochberg criterion (α = 0.05).
*PFDR < 0.05.
Uncorrected P < 0.05.
Discussion
The present study investigates Alzheimer’s disease-associated alterations in the microstructural organization of the hippocampal subfields using multi-shell diffusion MRI. While single-shell DTI-based measurements have been reported previously in the whole hippocampus18–21 and the subfields,11,12 there have been very limited diffusion studies with a multi-compartment model focusing on the hippocampal subfields in Alzheimer’s disease. Advanced compartment modelling may offer more specific pathophysiological explanation by decomposing the diffusion signals into several biologically meaningful components. Using this novel neuroimaging technique, we observed graded changes across the clinical continuum of Alzheimer’s disease and differential changes among the hippocampal subfields.
This study showed that the regional diffusion microstructural indices had differential effects along the clinical continuum of Alzheimer’s disease. An early sign of microstructural changes from normal cognition to MCI is indicated by one significant group-difference finding (i.e. VFIC in CA4-DG). At the later stage, approximately 70% of the microstructural indices across the subfields differed between MCI and Alzheimer’s disease, and all the comparisons between cognitively normal subjects and those with Alzheimer’s disease were significant. Upon examining individual diffusion metrics in the prodromal and clinical stages of Alzheimer’s disease, we found reduced intra- and extra-cellular volume fractions and increased tissue dispersion and isotropic fast diffusion.
Our observation of increased isotropic diffusion may explain previous DTI findings in MCI and Alzheimer’s disease. Elevated DTI-derived mean diffusivity was previously reported in the hippocampus of Alzheimer’s disease and to a lesser extend in MCI participants.13,19–21,54,55 This increase of mean diffusivity may be attributed to the increased fast isotropic diffusion arising from the oedematous changes due to Alzheimer’s disease-associated dendritic loss, neuronal shrinkage, axonal degeneration, or disruption of cellular membrane integrity.
In the present study, differential changes among the hippocampal subfields were also observed and microstructures of the CA regions appeared to be highly susceptible to pathology. While CA4-DG showed emerging signs of early decrease in the intracellular volume fraction, in the later stages of the Alzheimer’s disease continuum, both CA1–3 and CA4-DG had large effect sizes in group differences and associations with the Alzheimer’s disease blood biomarkers. For the associations with neuropsychological outcomes, CA1–3 demonstrated most sensitivity and the largest effect sizes. The CA1 subfield has been the focus of previous pathophysiological studies, showing significant loss in neurons and synapses in Alzheimer’s disease.56–61 Such neuronal loss is also related to cognition on the Mini Mental State Examination.59 On the other hand, the DG is thought to be the neurogenesis center of the hippocampus,62–64 which has been reported to be dysfunctional in animal models of Alzheimer’s disease.65,66
Molecular biomarkers of Alzheimer’s disease pathology are most useful during early disease stages prior to dementia.13,14,67 Microstructural alterations have been reported to exhibit a varying degree of association with CSF derived biomarkers of Alzheimer’s disease pathology.68,69 Recent studies have demonstrated that the blood-based biomarkers can achieve similar performances to CSF biomarkers in detecting CNS amyloid and tau deposition in Alzheimer’s disease70,71 This study provides supporting evidence to the utility of the plasma biomarkers in detecting hippocampal regional microstructural alterations. Our inter- and intra-group analyses demonstrated significant associations between plasma biomarkers levels and microstructural metrics in the hippocampal subfields. The direction of associations suggests elevated NfL is corresponding to neurodegenerative changes with increased interstitial free water and the extracellular hindered water components. Across the subfields, the results also highlight the differential effects of pathology on the hippocampal subfield microstructure. CA4-DG and the subiculum had elevated NfL and total tau associated with microstructural degeneration. On the other hand, CA1–3 showed some emerging relationships between the blood markers and diffusion metrics, though they did not survive multiple comparison corrections.
The present study demonstrated significant associations between neuropsychological test scores and regional hippocampal microstructural alterations. Consistent with the group comparison results, where the VFIC showed the earliest detectable difference (cognitively normal versus MCI), VFIC was one of the most sensitive microstructural metrics in the association with clinical outcomes. VFIC demonstrated strong associations with most neuropsychological scores across all hippocampal subfields. Overall, poor performance in neuropsychological tests was associated with decreased intracellular volume fraction, likely from decreased neurite density. These results support the hypothesis that neuronal loss and synaptic impairment are strongly associated with cognitive deficits.72,73 Furthermore, a recent study on young onset Alzheimer’s disease demonstrated an association between the NODDI derived neurite density index in the cortical grey matter and Mini-Mental State Examination.25
This study has a few limitations that need to be acknowledged. First, our sample in general was highly educated (years of education ≥ 15 years), which may limit generalizability of findings to populations with lower educational levels. Second, due to the cross-sectional nature of the study, the reported differences are group effects rather than intra-individual changes as a result of disease progression. Future longitudinal studies are warranted to confirm the current findings. Third, the study used standard resolution (2 × 2 × 2 mm3) diffusion MRI data to estimate microstructural changes in relatively small structures as the hippocampal subfields. Thus, we chose not to further divide CA1–3 or CA4-DG. In addition, we focused on the primary grey-matter part of the hippocampal subfields. Thus, the fimbria and parahippocampal cortices were not included in this analysis. To mitigate the partial volume effect due to a finite resolution, we used grey matter specific multi-compartment modelling to (i) achieve a physiologically plausible representation of grey matter microstructure; and (ii) to isolate CSF and white matter partial volume contaminations. In addition, we have taken upmost care in quality assurance/quality control of the co-registration between high-resolution anatomical images and diffusion images. As the observed changes in diffusion MRI parametric maps may reflect different physiological/pathophysiological processes, caution must be used when interpreting and generalizing these results. Lastly, while our results may contribute to the collated evidence relating to the biological definition of Alzheimer’s disease [i.e. the A/T (N) system], the study focuses on the Alzheimer’s disease clinical continuum and the groups were stratified based on the clinical criteria rather than A/T (N) biomarker criteria.74 The relation of the A/T (N) system and microstructural imaging will be our future research focus by including phosphorylated tau representing ‘T’ to complement the existing blood biomarker data, Aβ42/Aβ40 ratio for ‘A’ and NfL for ‘N’.
Despite the limitations, the results of this study demonstrate the efficacy of microstructural imaging in detecting subtle changes in the hippocampal subfields across the clinical diagnostic continuum of Alzheimer’s disease. We found the association of the microstructural imaging indices with the molecular biomarkers of Alzheimer’s disease pathology in a group-specific manner as well as a region-specific manner. In addition, the changes in the microstructural indices of the hippocampal subfields may explain the participants’ neuropsychological outcomes.
Supplementary Material
Acknowledgements
We are grateful for the participation of the individuals in this research study without whom this research would not be possible. The MRI scans were performed at the In-Vivo Imaging Core, Indiana University School of Medicine (IUSM). We also thank the staff of the Indiana ADRC, Center for Neuroimaging and In-Vivo Imaging Core. We thank Brooke M. Patz, MA, CCRP, CPM for assistance with sample processing and storage.
Abbreviations
- Aβ
amyloid-β
- CA
cornu ammonis
- DG
dentate gyrus
- DTI
diffusion tensor imaging
- DWI
diffusion weighted image
- MCI
mild cognitive impairment
- MoCA
Montreal Cognitive Assessment
- NfL
neurofilament light chain
- NODDI
neurite orientation dispersion and density imaging
- ODI =
orientation dispersion index
- RAVLT
Rey Auditory Verbal Learning Test
- TMT
Trail Making Test
- VFEC
extracellular volume fraction
- VFIC
intracellular volume fraction
- VISO
volume fraction of isotropic water diffusivity
Contributor Information
Syed Salman Shahid, Center for Neuroimaging, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA; Indiana Alzheimer’s Disease Research Center, Indiana University School of Medicine, Indianapolis, IN, USA.
Qiuting Wen, Center for Neuroimaging, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA; Indiana Alzheimer’s Disease Research Center, Indiana University School of Medicine, Indianapolis, IN, USA.
Shannon L Risacher, Center for Neuroimaging, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA; Indiana Alzheimer’s Disease Research Center, Indiana University School of Medicine, Indianapolis, IN, USA; Stark Neuroscience Research Institute, Indiana University School of Medicine, Indianapolis, IN, USA.
Martin R Farlow, Indiana Alzheimer’s Disease Research Center, Indiana University School of Medicine, Indianapolis, IN, USA; Stark Neuroscience Research Institute, Indiana University School of Medicine, Indianapolis, IN, USA; Department of Neurology, Indiana University School of Medicine, Indianapolis, IN, USA.
Frederick W Unverzagt, Indiana Alzheimer’s Disease Research Center, Indiana University School of Medicine, Indianapolis, IN, USA; Department of Psychiatry, Indiana University School of Medicine, Indianapolis, IN, USA.
Liana G Apostolova, Center for Neuroimaging, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA; Indiana Alzheimer’s Disease Research Center, Indiana University School of Medicine, Indianapolis, IN, USA; Stark Neuroscience Research Institute, Indiana University School of Medicine, Indianapolis, IN, USA; Department of Neurology, Indiana University School of Medicine, Indianapolis, IN, USA; Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN, USA.
Tatiana M Foroud, Indiana Alzheimer’s Disease Research Center, Indiana University School of Medicine, Indianapolis, IN, USA; Stark Neuroscience Research Institute, Indiana University School of Medicine, Indianapolis, IN, USA; Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN, USA.
Henrik Zetterberg, Department of Psychiatry and Neurochemistry, The Sahlgrenska Academy, University of Gothenburg, Mölndal, Sweden; Clinical Neurochemistry Laboratory, Sahlgrenska University Hospital, Mölndal, Sweden; Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK; UK Dementia Research Institute at UCL, London, UK.
Kaj Blennow, Department of Psychiatry and Neurochemistry, The Sahlgrenska Academy, University of Gothenburg, Mölndal, Sweden; Clinical Neurochemistry Laboratory, Sahlgrenska University Hospital, Mölndal, Sweden.
Andrew J Saykin, Center for Neuroimaging, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA; Indiana Alzheimer’s Disease Research Center, Indiana University School of Medicine, Indianapolis, IN, USA; Stark Neuroscience Research Institute, Indiana University School of Medicine, Indianapolis, IN, USA; Department of Neurology, Indiana University School of Medicine, Indianapolis, IN, USA; Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN, USA.
Yu Chien Wu, Center for Neuroimaging, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA; Indiana Alzheimer’s Disease Research Center, Indiana University School of Medicine, Indianapolis, IN, USA; Stark Neuroscience Research Institute, Indiana University School of Medicine, Indianapolis, IN, USA.
Funding
This work was supported by the National Institutes of Health R01 AG053993 (Y.C.W.), R01 NS112303 (Y.C.W.), P30 AG010133 (A.J.S.), R01 AG019771 (A.J.S.), R01 AG061788 (S.L.R.) and K01 AG049050 (S.L.R.). The MRI scans were funded, in part, by the Department of Radiology and Imaging Sciences at IUSM via the Advanced Imaging Research Technology Development (AirTD) program. H.Z. is a Wallenberg Scholar supported by grants from the Swedish Research Council (#2018-02532), the European Research Council (#681712), Swedish State Support for Clinical Research (#ALFGBG-720931), the Alzheimer Drug Discovery Foundation (ADDF), USA (#201809-2016862), the Alzheimer’s Disease Strategic Fund and the Alzheimer's Association (#ADSF-21-831376-C, #ADSF-21-831381-C and #ADSF-21-831377-C), the Olav Thon Foundation, the Erling-Persson Family Foundation, Stiftelsen för Gamla Tjänarinnor, Hjärnfonden, Sweden (#FO2019-0228), the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 860197 (MIRIADE) and the UK Dementia Research Institute at UCL. K.B. is supported by the Swedish Research Council (#2017-00915), the Alzheimer Drug Discovery Foundation (ADDF), USA (#RDAPB-201809-2016615), the Swedish Alzheimer Foundation (#AF-742881), Hjärnfonden, Sweden (#FO2017-0243), the Swedish state under the agreement between the Swedish government and the County Councils, the ALF-agreement (#ALFGBG-715986), the European Union Joint Program for Neurodegenerative Disorders (JPND2019-466-236), and the National Institute of Health (NIH), USA, (grant #1R01AG068398-01).
Competing interests
H.Z. has served at scientific advisory boards and/or as a consultant for Alector, Eisai, Denali, Roche Diagnostics, Wave, Samumed, Siemens Healthineers, Pinteon Therapeutics, Nervgen, AZTherapies, CogRx and Red Abbey Labs, has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure and Biogen, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work). K.B. has served as a consultant, at advisory boards, or at data monitoring committees for Abcam, Axon, Biogen, JOMDD/Shimadzu. Julius Clinical, Lilly, MagQu, Novartis, Prothena, Roche Diagnostics and Siemens Healthineers, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program. A.J.S. receives support from multiple NIH grants (P30 AG010133, P30 AG072976, R01 AG019771, R01 AG057739, U01 AG024904, R01 LM013463, R01 AG068193, T32 AG071444, and U01 AG068057 and U01 AG072177). He has also received support from Avid Radiopharmaceuticals, a subsidiary of Eli Lilly (in kind contribution of PET tracer precursor); Bayer Oncology (Scientific Advisory Board); Eisai (Scientific Advisory Board); Siemens Medical Solutions USA, Inc. (Dementia Advisory Board); Springer-Nature Publishing (Editorial Office Support as Editor-in-Chief, Brain Imaging and Behavior). The other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Márquez F, Yassa MA. Neuroimaging biomarkers for Alzheimer's disease. Mol Neurodegener. 2019;14:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002:297:353–356. [DOI] [PubMed] [Google Scholar]
- 3. Tsai J, Grutzendler J, Duff K, Gan WB. Fibrillar amyloid deposition leads to local synaptic abnormalities and breakage of neuronal branches. Nat Neurosci. 2004;7:1181–1183. [DOI] [PubMed] [Google Scholar]
- 4. Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. [DOI] [PubMed] [Google Scholar]
- 6. Dickerson BC, Eichenbaum H. The episodic memory system: neurocircuitry and disorders. Neuropsychopharmacology. 2010;35:86–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soininen HS, Scheltens P. Early diagnostic indices for the prevention of alzheimer's disease. Ann Med. 1998;30:553–559. [DOI] [PubMed] [Google Scholar]
- 8. Small SA, Schobel SA, Buxton RB, Witter MP, Barnes CA. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat Rev Neurosci. 2011;12:585–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De Flores R, La Joie R, Landeau B, et al. Effects of age and Alzheimer's disease on hippocampal subfields: comparison between manual and FreeSurfer volumetry. Hum Brain Mapp. 2015;36:463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blanken AE, Hurtz S, Zarow C, et al. Associations between hippocampal morphometry and neuropathologic markers of Alzheimer's disease using 7 T MRI. Neuroimage Clin. 2017;15:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li YD, Dong HB, Xie GM, Zhang LJ. Discriminative analysis of mild Alzheimer's disease and normal aging using volume of hippocampal subfields and hippocampal mean diffusivity: an in vivo magnetic resonance imaging study. Am J Alzheimers Dis Other Demen. 2013;28:627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mak E, Gabel S, Su L, et al. Multi-modal MRI investigation of volumetric and microstructural changes in the hippocampus and its subfields in mild cognitive impairment, Alzheimer's disease, and dementia with Lewy bodies. Int Psychogeriatr. 2017;29:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weston PS, Simpson IJ, Ryan NS, Ourselin S, Fox NC. Diffusion imaging changes in grey matter in Alzheimer's disease: a potential marker of early neurodegeneration. Alzheimers Res Ther. 2015;7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Montal V, Vilaplana E, Alcolea D, et al. Cortical microstructural changes along the Alzheimer’s disease continuum. Alzheimers Dement. 2018;14:340–351. [DOI] [PubMed] [Google Scholar]
- 15. Ringman JM, O'neill J, Geschwind D, et al. Diffusion tensor imaging in preclinical and presymptomatic carriers of familial Alzheimer's disease mutations. Brain. 2007;130:1767–1776. [DOI] [PubMed] [Google Scholar]
- 16. Canu E, Mclaren DG, Fitzgerald ME, et al. Microstructural diffusion changes are independent of macrostructural volume loss in moderate to severe Alzheimer's disease. J Alzheimers Dis. 2010;19:963–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ridha BH, Barnes J, Bartlett JW, et al. Tracking atrophy progression in familial Alzheimer's disease: a serial MRI study. Lancet Neurol. 2006;5:828–834. [DOI] [PubMed] [Google Scholar]
- 18. Tang X, Qin Y, Wu J, Zhang M, Zhu W, Miller M. Shape and diffusion tensor imaging based integrative analysis of the hippocampus and the amygdala in Alzheimer's disease. Magn Reson Imaging. 2016;34:1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scola E, Bozzali M, Agosta F, et al. A diffusion tensor MRI study of patients with MCI and AD with a 2-year clinical follow-up. J Neurol Neurosurg Psychiatry. 2010;81:798–805. [DOI] [PubMed] [Google Scholar]
- 20. Muller MJ, Greverus D, Dellani PR, et al. Functional implications of hippocampal volume and diffusivity in mild cognitive impairment. Neuroimage. 2005;28:1033–1042. [DOI] [PubMed] [Google Scholar]
- 21. Kantarci K, Petersen RC, Boeve BF, et al. DWI Predicts future progression to Alzheimer disease in amnestic mild cognitive impairment. Neurology. 2005;64:902–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brueggen K, Dyrba M, Barkhof F, et al. Basal forebrain and hippocampus as predictors of conversion to Alzheimer's disease in patients with mild cognitive impairment—a multicenter DTI and volumetry study. J Alzheimers Dis. 2015;48:197–204. [DOI] [PubMed] [Google Scholar]
- 23. Clerx L, Visser PJ, Verhey F, Aalten P. New MRI markers for Alzheimer's disease: a meta-analysis of diffusion tensor imaging and a comparison with medial temporal lobe measurements. J Alzheimers Dis. 2012;29:405–429. [DOI] [PubMed] [Google Scholar]
- 24. Gong NJ, Wong CS, Chan CC, Leung LM, Chu YC. Aging in deep gray matter and white matter revealed by diffusional kurtosis imaging. Neurobiol Aging. 2014;35:2203–2216. [DOI] [PubMed] [Google Scholar]
- 25. Parker TD, Slattery CF, Zhang J, et al. Cortical microstructure in young onset Alzheimer's disease using neurite orientation dispersion and density imaging. Hum Brain Mapp. 2018;39:3005–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nazeri A, Chakravarty MM, Rotenberg DJ, et al. Functional consequences of neurite orientation dispersion and density in humans across the adult lifespan. J Neurosci. 2015;35:1753–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rathi Y, Pasternak O, Savadjiev P, et al. Gray matter alterations in early aging: a diffusion magnetic resonance imaging study. Hum Brain Mapp. 2014;35:3841–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henf J, Grothe MJ, Brueggen K, Teipel S, Dyrba M. Mean diffusivity in cortical gray matter in Alzheimer's disease: the importance of partial volume correction. Neuroimage Clin. 2018;17:579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61:1000–1016. [DOI] [PubMed] [Google Scholar]
- 30. Venkatesh A, Stark SM, Stark CEL, Bennett IJ. Age- and memory- related differences in hippocampal gray matter integrity are better captured by NODDI compared to single-tensor diffusion imaging. Neurobiol Aging. 2020;96:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fukutomi H, Glasser MF, Zhang H, et al. Neurite imaging reveals microstructural variations in human cerebral cortical gray matter. Neuroimage. 2018;182:488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weintraub S, Besser L, Dodge HH, et al. Version 3 of the Alzheimer disease centers’ neuropsychological test battery in the uniform data set (UDS). Alzheimer Dis Assoc Disord. 2018;32:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rattanabannakit C, Risacher SL, Gao S, et al. The cognitive change index as a measure of self and informant perception of cognitive decline: relation to neuropsychological tests. J Alzheimers Dis. 2016;51:1145–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jessen F, Amariglio RE, Van Boxtel M, et al. A conceptual framework for research on subjective cognitive decline in preclinical Alzheimer's disease. Alzheimers Dement. 2014;10:844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Albert MS, Dekosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berg L. Clinical Dementia Rating (CDR). Psychopharmacol Bull. 1988;24:637–639. [PubMed] [Google Scholar]
- 37. Wechsler D; Manual for the Wechsler Adult Intelligence Scale (rev.ed.) . The Pscyhological Corporation, Harcourt Brace Janvanovich, Inc; 1987. [Google Scholar]
- 38. Nasreddine ZS, Phillips NA, Bédirian V, et al. The Montreal cognitive assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 2005;53:695–699. [DOI] [PubMed] [Google Scholar]
- 39. Pfeffer RI, Kurosaki TT, Harrah CH Jr, Chance JM, Filos S. Measurement of functional activities in older adults in the community. J Gerontol. 1982;37:323–329. [DOI] [PubMed] [Google Scholar]
- 40. Mckhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cong S, Risacher SL, West JD, et al. Volumetric comparison of hippocampal subfields extracted from 4-minute accelerated vs. 8-minute high-resolution T2-weighted 3 T MRI scans. Brain Imaging Behav. 2018;12:1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu YC, Alexander AL. Hybrid diffusion imaging. Neuroimage. 2007;36:617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wen Q, Mustafi SM, Li J, et al. White matter alterations in early-stage Alzheimer's disease: A tract-specific study. Alzheimers Dement (Amst). 2019;11:576–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wen Q, Risacher SL, Xie L, et al. Tau-related white-matter alterations along spatially selective pathways. Neuroimage. 2021;226:117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Veraart J, Fieremans E, Jelescu IO, Knoll F, Novikov DS. Gibbs ringing in diffusion MRI. Magn Reson Med. 2016;76:301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Veraart J, Novikov DS, Christiaens D, Ades-Aron B, Sijbers J, Fieremans E. Denoising of diffusion MRI using random matrix theory. Neuroimage. 2016;142:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kellner E, Dhital B, Kiselev VG, Reisert M. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn Reson Med. 2016;76:1574–1581. [DOI] [PubMed] [Google Scholar]
- 48. Andersson JL, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. [DOI] [PubMed] [Google Scholar]
- 50. Guerrero JM, Adluru N, Bendlin BB, et al. Optimizing the intrinsic parallel diffusivity in NODDI: an extensive empirical evaluation. PLoS One. 2019;14:e0217118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alimi A, Fick R, Wassermann D, Deriche R. Dmipy: A diffusion microstructure imaging toolbox in python to improve research reproducibility. In: Computational Imaging of the Central Nervous System. Athena. 2019. [Google Scholar]
- 52. Nazeri A, Mulsant BH, Rajji TK, et al. Gray matter neuritic microstructure deficits in schizophrenia and bipolar disorder. Biol Psychiatry. 2017;82:726–736. [DOI] [PubMed] [Google Scholar]
- 53. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jacobs HI, Van Boxtel MP, Gronenschild EH, et al. Decreased gray matter diffusivity: a potential early Alzheimer's disease biomarker? Alzheimers Dement. 2013;9:93–97. [DOI] [PubMed] [Google Scholar]
- 55. Fellgiebel A, Yakushev I. Diffusion tensor imaging of the hippocampus in MCI and early Alzheimer's disease. J Alzheimers Dis. 2011;26:257–262. [DOI] [PubMed] [Google Scholar]
- 56. West MJ, Coleman PD, Flood DG, Troncoso JC. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer's disease. Lancet. 1994:344:769–772. [DOI] [PubMed] [Google Scholar]
- 57. Price JL, Ko AI, Wade MJ, Tsou SK, McKeel DW, Morris JC. Neuron number in the entorhinal cortex and CA1 in preclinical Alzheimer disease. Arch Neurol. 2001;58:1395–1402. [DOI] [PubMed] [Google Scholar]
- 58. Rössler M, Zarski R, Bohl J, Ohm TG. Stage-dependent and sector-specific neuronal loss in hippocampus during Alzheimer's disease. Acta Neuropathol. 2002;103:363–369. [DOI] [PubMed] [Google Scholar]
- 59. Akram A, Christoffel D, Rocher AB, et al. Stereologic estimates of total spinophilin-immunoreactive spine number in area 9 and the CA1 field: relationship with the progression of Alzheimer's disease. Neurobiol Aging. 2008;29:1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Scheff SW, Price DA, Schmitt FA, Dekosky ST, Mufson EJ. Synaptic alterations in CA1 in mild Alzheimer disease and mild cognitive impairment. Neurology. 2007;68:1501–1508. [DOI] [PubMed] [Google Scholar]
- 61. Simic G, Kostovic I, Winblad B, Bogdanovic N. Volume and number of neurons of the human hippocampal formation in normal aging and Alzheimer's disease. J Comp Neurol. 1997;379:482–494. [DOI] [PubMed] [Google Scholar]
- 62. Disouky A, Lazarov O. Adult hippocampal neurogenesis in Alzheimer's disease. Prog Mol Biol Transl Sci. 2021;177:137–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ohm TG. The dentate gyrus in Alzheimer's disease. Prog Brain Res. 2007;163:723–740. [DOI] [PubMed] [Google Scholar]
- 64. Horgusluoglu E, Nudelman K, Nho K, Saykin AJ. Adult neurogenesis and neurodegenerative diseases: A systems biology perspective. Am J Med Genet B Neuropsychiatr Genet. 2017;174:93–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Moreno-Jimenez EP, Flor-Garcia M, Terreros-Roncal J, et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer's disease. Nat Med. 2019;25:554–560. [DOI] [PubMed] [Google Scholar]
- 66. Richetin K, Steullet P, Pachoud M, et al. Tau accumulation in astrocytes of the dentate gyrus induces neuronal dysfunction and memory deficits in Alzheimer's disease. Nat Neurosci. 2020;23:1567–1579. [DOI] [PubMed] [Google Scholar]
- 67. Jack CR J, Bernstein MA, Borowski BJ, et al. Update on the magnetic resonance imaging core of the Alzheimer's disease neuroimaging initiative. Alzheimers Dement. 2010;6:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Q, Wang Y, Liu J, et al. Quantification of white matter cellularity and damage in preclinical and early symptomatic Alzheimer's disease. Neuroimage Clin. 2019;22:101767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Molinuevo JL, Ripolles P, Simo M, et al. White matter changes in preclinical Alzheimer's disease: a magnetic resonance imaging-diffusion tensor imaging study on cognitively normal older people with positive amyloid beta protein 42 levels. Neurobiol Aging. 2014;35:2671–2680. [DOI] [PubMed] [Google Scholar]
- 70. Palmqvist S, Insel PS, Stomrud E, et al. Cerebrospinal fluid and plasma biomarker trajectories with increasing amyloid deposition in Alzheimer’s disease. EMBO Mol Med. 2019;11:e11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Palmqvist S, Tideman P, Cullen N, et al. Prediction of future Alzheimer's disease dementia using plasma phospho-tau combined with other accessible measures. Nat Med. 2021;27:1034–1042. [DOI] [PubMed] [Google Scholar]
- 72. Heneka MT, Carson MJ, El Khoury J, et al. Neuroinflammation in Alzheimer's disease. Lancet Neurol. 2015;14:388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shankar GM, Walsh DM. Alzheimer's disease: synaptic dysfunction and Abeta. Mol Neurodegener. 2009;4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jack CR J, Bennett DA, Blennow K, et al. NIA-AA research framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study were acquired via NIH-NIA funded R01 projects collected through the Indiana Alzheimer’s Disease Research Center (IADRC; NIH P30). Therefore, we will comply with the NIH data sharing policy and guidance (http://grants.nih.gov/grants/policy/data_sharing/data_sharing_guidance.htm) as well as the data sharing plan outlined by the IADRC.
Briefly, we will make the data available as early as feasible to qualified researchers who have obtained institutional review board approval from their institution and who are willing to sign a data-sharing agreement. Requestors must agree to NIH policies regarding privacy, data security and ethical practices, including the requirement that no attempt will be made to determine the identities of participants or their relatives. The principal investigators will review requests for anonymized human imaging data. Requestors will be encouraged to develop collaborative analyses with the project investigators, but this will not be required for data access. The data processing and analysis codes used in this study are from open-source software tools can be freely downloaded (see the ‘Materials and methods’ section). The code developed in-house will be available upon request and follow aforementioned data sharing policy.