Abstract
Aedes aegypti is a major vector of arboviruses that cause dengue, chikungunya, yellow fever, and Zika. Although recent success in reverse genetics has facilitated rapid progress in basic and applied research, integration of forward genetics with modern technologies remains challenging in this important species, as up to 47% of its chromosome is refractory to genetic mapping due to extremely low rate of recombination. Here, we report the development of a marker-assisted mapping strategy to readily screen for and genotype only the rare but informative recombinants, drastically increasing both the resolution and signal-to-noise ratio. Using marker-assisted mapping, we mapped a transgene that was inserted in a >100-Mb recombination desert and a sex-linked spontaneous red-eye (re) mutation just outside the region. We subsequently determined, by CRISPR/Cas9-mediated knockout, that cardinal is the causal gene of re, which is the first forward genetic identification of a causal gene in Ae. aegypti. The identification of the causal gene of the sex-linked re mutation provides the molecular foundation for using gene editing to develop versatile and stable genetic sexing methods. To facilitate genome-wide forward genetics in Ae. aegypti, we generated and compiled a number of lines with markers throughout the genome. Thus, by overcoming the challenges presented by the vast recombination deserts and the scarcity of markers, we have shown that effective forward genetic analysis is increasingly feasible in this important arboviral vector species.
Keywords: sex locus, homomorphic sex chromosome, sex separation, cardinal, eye color, causal gene, dengue, Zika
Introduction
Aedes aegypti is a major vector of a number of arboviruses and filarial worms. Effective control of this 1 species could help reduce or prevent a number of vector-borne infectious diseases including dengue, chikungunya, yellow fever, and Zika. Current strategies to reduce the incidence and burden of these diseases depend heavily on effective vector control, which is hindered by increasing insecticide resistance. Novel control strategies, informed by improved understanding of mosquito biology, are urgently needed. Recent years witnessed rapid accumulation of genomic resources (e.g. Matthews et al. 2018) and effective applications of CRISPR/Cas9-mediated reverse genetic analysis of gene function in Ae. aegypti (e.g. Hall et al. 2015; Li et al. 2017; Aryan et al. 2020). However, successful integration of forward genetics with modern technologies to identify causal genes of important traits or phenotypes, as championed by many plant and animal geneticists (e.g. Schneeberger 2014; Navarro-Escalante et al. 2020; Feng et al. 2021a), still awaits in this important vector species.
Recombination is fundamental to forward genetic analysis and quantitative trait loci (QTL) mapping. Mapping a locus that contains the causal gene is possible when recombination separates the causal gene locus from its neighboring sequences, resulting in an enrichment of markers tightly linked to the causal gene in individuals manifesting the phenotype. However, Ae. aegypti has a lower per megabase recombination rate (∼0.3 cM/Mb) than Anopheles gambiae (0.8 cM/Mb) and Drosophila melanogaster (1.6 cM/Mb) (Wilfert et al. 2007). In addition, up to 47% of its chromosome surrounding the centromere belongs to so-called low recombination regions (LRRs), with rates much lower than the 0.3 cM/Mb average (Juneja et al. 2014; Dudchenko et al. 2017; Fontaine et al. 2017). Although it is not unique to Ae. aegypti to have LRR near centromeres, having nearly half of the genome in LRR further complicates genetic efforts to map genes that determine particular traits. We are not aware of any report of the successful determination of a causal gene using forward genetics in Ae. aegypti, despite the intense interest in identifying genes that underly insecticide and pathogen resistance or genes that could be used as selectable markers for genetic sexing (Koskinioti et al. 2021; Ward et al. 2021). The presence of extensive regions with very low recombination rates, or recombination deserts, has also been reported in many plant and animal species (Stapley et al. 2017a,b). Therefore, forward genetic mapping in these recombination deserts is a significant and broadly important challenge.
Markers flanking a genetic locus of interest have been used to isolate recombinants to increase the effectiveness of mapping efforts in Drosophila (Chen et al. 2008; Ding et al. 2016) and other insects (Loehlin et al. 2010). We hypothesize that a marker-assisted mapping (MAM) strategy will overcome the bottleneck imposed by the vast recombination deserts and enable effective forward genetic studies in Ae. aegypti. Using MAM, we rapidly mapped a transgene insertion in a region of suppressed recombination near the sex locus in Ae. aegypti. Using the same method in conjunction with introgression analysis, we also identified the causal gene for a spontaneous red-eye (re) mutation in Ae. aegypti, which was first reported nearly 60 years ago (McClelland 1962, 1966). The identification of the causal gene of the sex-linked re mutation provides the molecular foundation for future improvements and expansion of genetic sexing strains. Although MAM does not require densely populated markers, we generated and compiled a number of lines with markers interspersed in the genome to facilitate forward genetic analysis in this important vector species.
Materials and methods
Mosquito rearing
The Liverpool (LVP) strain (obtained from www.BEI.org) and the RED strain (obtained from the Severson laboratory at the University of Notre Dame) of Ae. aegypti were maintained at Virginia Tech at 26–28C and 60–70% relative humidity with a 14/10-h day/night light cycle. Adult mosquitoes were fed 10% sucrose and fed blood using artificial membrane feeders and defibrinated sheep’s blood (Colorado Serum Company, Denver, CO, USA).
Generating P10 and other transgenic marker lines
A piggyBac donor plasmid (500 ng/μl, Supplementary Fig. 1) that contains an EGFP transformation marker driven by the Ae. aegypti polyubiquitin promoter and was coinjected with an in vitro transcribed piggyBac mRNA (300 ng/μl) into less than 1-h-old embryos of the LVP strain of Ae. aegypti (Coates et al. 1998). The piggyBac-hsp70-transposase (Handler et al. 1998) was used as a template for in vitro transcription using the mMessage mMachine T7 Ultra kit (Thermofisher), followed by MEGAclear (Thermofisher) column purification. Approximately 1,200 embryos were injected, resulting in 150 G0 females and 160 G0 males. Surviving G0 females were mated to LVP males in pools of 20–25. Each G0 male was mated individually with 5 LVP females in individual cages and mosquitoes from 15 to 20 of these cages were merged into 1 large pool. G1 larvae were screened for green fluorescence using a Leica M165 FC fluorescence microscope. Positive G1 individuals were out-crossed to LVP mosquitoes to ensure that all transgene cassettes were stably inherited to the G2 generation. The same method was used to generate the P10, R4M, and Pub_DsRed_P1 transgenic lines and all relevant donor constructs were described in Supplementary Fig. 1.
Identification of the transgene insertion sites for P10 and 4 other strains
Oxford Nanopore sequencing was performed using genomic DNA isolated from 4 transgene-positive individuals. The Qiagen Genomic Tip DNA Isolation kit (Cat. No. 10223, Qiagen, Hilden, Germany) was used. These 4 males were homogenized using a hand held pestle motor mixer (Cole Palmer) with a sterile disposable plastic pestle (USA Scientific) for approximately 10 s in the lysis buffer. The homogenate was incubated with 300 mAU Proteinase K (Qiagen, Hilden, Germany) overnight at 55°C and then transferred into a 15-ml conical tube for centrifugation at 5,000 × g for 15 min at 4°C to remove debris. The lysate was loaded on a 20-G column for purification following the protocol. The purity, approximate size, and concentration of the DNA were measured using a nanodrop spectrophotometer, 0.5% agarose gel electrophoresis, and Qubit dsDNA assay, respectively. Approximately 2,000 ng of gDNA was used to prepare the library using the 1D ligation sequencing kit (SQK-LSK108; Oxford Nanopore Technologies, UK). Adapter ligation was performed at room temperature for 45 min. Approximately 500 ng of the prepared library was loaded on the flow cell. Approximately 7.6 Gb were obtained and submitted to NCBI (PRJNA718905). Four long reads that contain the piggyBac insertion were identified and used to map the location of the P10 insertion (Supplementary Method 1 and Supplementary Figs. 1 and 2). Oxford Nanopore sequencing was also performed for the P14, R4M, Pub_DsRed_P1 (this study), and J2 (Hall et al. 2014) transgenic lines and the transgene insertion sites were identified using the same method (Supplementary Method 1 and Supplementary Fig. 1). Oxford Nanopore reads that contain these transgenes were provided in the Supplementary Data.
Sample collections for MAM
The positions of the sex locus and P10 are shown in Fig. 1. The crossing scheme is shown in Fig. 2a. GFP-positive, black-eyed P10 males (n = 66) were mated with GFP-negative, red-eyed females (n = 37) to generate the F1 progeny. The GFP-positive, black-eyed F1 males (n = 26) were backcrossed with GFP-negative, red-eyed females (n = 46) to generate F2 progeny. The F2 progenies were screened and counted. The following numbers of individuals were pooled for genomic DNA isolation and Illumina sequencing: male positive black eyed (n = 60), male positive red eyed (n = 12), female positive black eyed (n = 20), female negative red eyed (n = 60), male negative red eyed (n = 11), and female positive black eyed (n = 7). Not all of the informative recombinant progeny shown in Fig. 2b are sequenced. Illumina sequencing coverage and other statistics are provided in Supplementary Table 1.
Fig. 1.
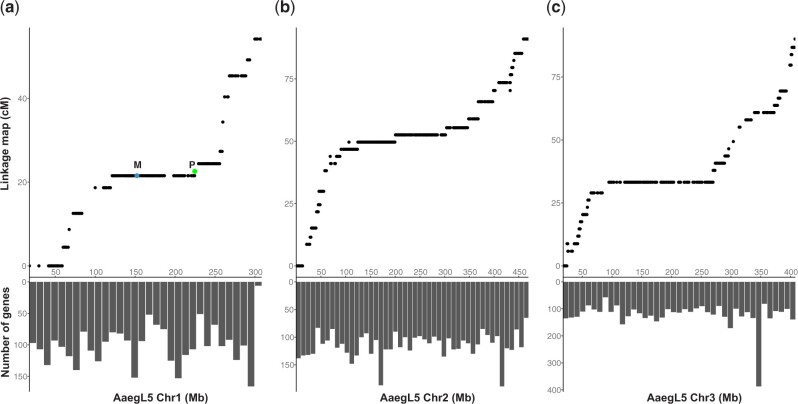
Vast recombination deserts in Ae. aegypti contain thousands of genes. a–c) Chromosomes 1, 2, and 3, respectively. The X-axis is the chromosome position. The top panels are genetic distances measured in centimorgan. The lower panels are the number of protein-coding genes in the corresponding 10-Mb window. LRRs in Ae. aegypti were previously shown in all 3 chromosomes by mapping the genetic cross data (Juneja et al. 2014) to an earlier version of the assembly (Dudchenko et al. 2017; Fontaine et al. 2017). The top panels remapped the same genetic cross data to the most recent AaegL5 PacBio-based assembly (Matthews et al. 2018). On chromosome 1, the positions of the male-determining locus (M) and a GFP-expressing transgene named P10 (P, green dot, 224 Mb, Supplementary Figs. 1 and 2) are indicated. M and P showed a genetic distance of 1.02–1.55 cM according to 2 screens of 1,353 and 3,145 progenies, respectively (Table 1).
Fig. 2.
MAM identified the location of the P10 transgene. a) The parental and backcrosses that yield the F2 progeny (b) used for MAM. In this example, P (the P10 GFP transgene insertion) is the gene to be mapped. Two flanking markers, the dominant male-determining locus M and a wild-type black-eye allele B (or +), are used to assist the mapping of P. The corresponding alleles m and re on the homologous chromosome are not shown. M/m individuals are males and m/m individuals are females. B/re (or +/re) individuals are black eyed and re/re individuals are red eyed. The genotype of the females in both the parental and the backcross is m/m, N/N (negative for the transgene insertion), and re/re. The genotype of the F1 male is M/m, P/N, and B/re (or +/re) and only the dominant alleles are shown for simplicity. b) Identifying the informative recombinants in the progenies of the backcross: the phenotype of the F2 progeny, which is solely determined by the genotype of the gamete of the F1 male, is used for MAM. The presence of the easily scorable markers (sex and eye color) flanking the gene of interest P enables the identification of the vast number of noninformative F2 progenies, which maintain the parental linkage: MPB, males (M) that are positive for GFP (P) and black eyed (B); or FNR, females (m) that are negative for GFP (N) and red eyed (re). Thus, we are able to exclude the noninformative progeny and focus on the informative recombinants MPR, FBP, MNR, and FNB. The numbers below each genotype/phenotype are the actual numbers of individuals identified from the backcross. More than 3,000 (1,556 + 1,511 = 3,067) noninformative individuals are excluded from the MAM analysis. The number of individuals sequenced is indicated in materials and methods. c) Expected Δ(SNP index) values differ under 3 sampling scenarios: (1) MAM, only the informative F2 progeny (P and N progeny showing recombination between markers M and B) are genotyped; (2) extreme genotyping (EG), thousands of P and N progeny are genotyped; and (3) limited genotyping (LG), a limited sample of the P and N progeny is genotyped. As shown in (d), Δ(SNP index) is the signal used for mapping. The recombination rate between M and B is approximately 2.5% in the current example. Thus, genotyping 1,000 P and 1,000 N progeny will likely only include 25 informative P and 25 informative N progeny having recombination between the flanking markers, severely limiting both the signal-to-noise ratio and the resolution of the mapping result. The position of the “peak” in LG could also be misleading due to limited sampling. d) MAM identifies the location of the P10 transgene insertion. The tricube‐smoothed Δ(SNP index) is shown in 1-Mb sliding windows. The Δ(SNP index) value is calculated by comparing all informative P10 positive recombinants (MPR and FPB) with all informative P10 negative recombinants (FNB and MNR). The corresponding 2-sided confidence intervals are shown as red (90%) or blue (95%) lines. X-axis is the genomic position on the homomorphic sex chromosome 1. The confidence intervals were estimated using 10,000 replicate simulations for all SNP positions with the given read depths. Positions from 223,288,153 to 226,114,389 exceed 95% confidence and the left and right inflection points of this peak are 223,412,798–225,833,455, respectively.
MAM data analysis
Adapters and low-quality Illumina reads were trimmed by Trimmomatic v0.38 (Bolger et al. 2014) and quality control was performed using FastQC v0.11.8. (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Variants were called using GATK v4.1 (McKenna et al. 2010) following the “Best Practices workflow” (DePristo et al. 2011; Van der Auwera et al. 2013). First, paired reads of each sample were aligned to the Ae. aegypti genome L5 from VectorBase (www.vectorbase.org) using BWA-MEM v0.7.17 (Li and Durbin 2009) and sam files were converted to bam files and sorted by samtools (Li et al. 2009). PCR duplicates were marked by GATK. SNPs of each sample were called by GATK using the Haplotype caller mode. GVCF files were combined by GATK CombineGVCFs. Combined GVCF files were converted to VCF files by GATK GenotypeGVCFs. Variants are selected from VCF to a table by GATK VariantsToTable for the subsequent Bulk Segregant Analysis by QTLseqr (Takagi et al. 2013; Mansfeld and Grumet 2018). Variants were filtered using GATK. The number of SNPs, minimum read depth, and tricube-smoothed Δ(SNP index) within a 1-Mb sliding window were calculated by QTLseqr. The slope of Δ(SNP index) was used to calculate the inflection point. SNP index and Δ(SNP index) are defined as follows.
To map the P10 insertion (Fig. 2d), male negative red eye (MNR) and female negative black eye (FNB) were used as the low bulk while the female positive black eye (FPB) and male positive red eye (MPR) were used as the high bulk. Variants were filtered based on 10 < total read depth < 400, 0.2 ≤ sum of reference allele frequency of 2 bulks ≤ 0.8, per sample read depth ≥ 5 and GATK GQ score ≥ 99. Approximately 2.7 million SNPs remained after variant filtering. To map the re locus (Fig. 3a), FPB and FNB were used as the low bulk while MNR and MPR were used as the high bulk. Variants were filtered based on the same parameters as mentioned above. Approximately 2.5 million SNPs remained after variant filtering.
Fig. 3.
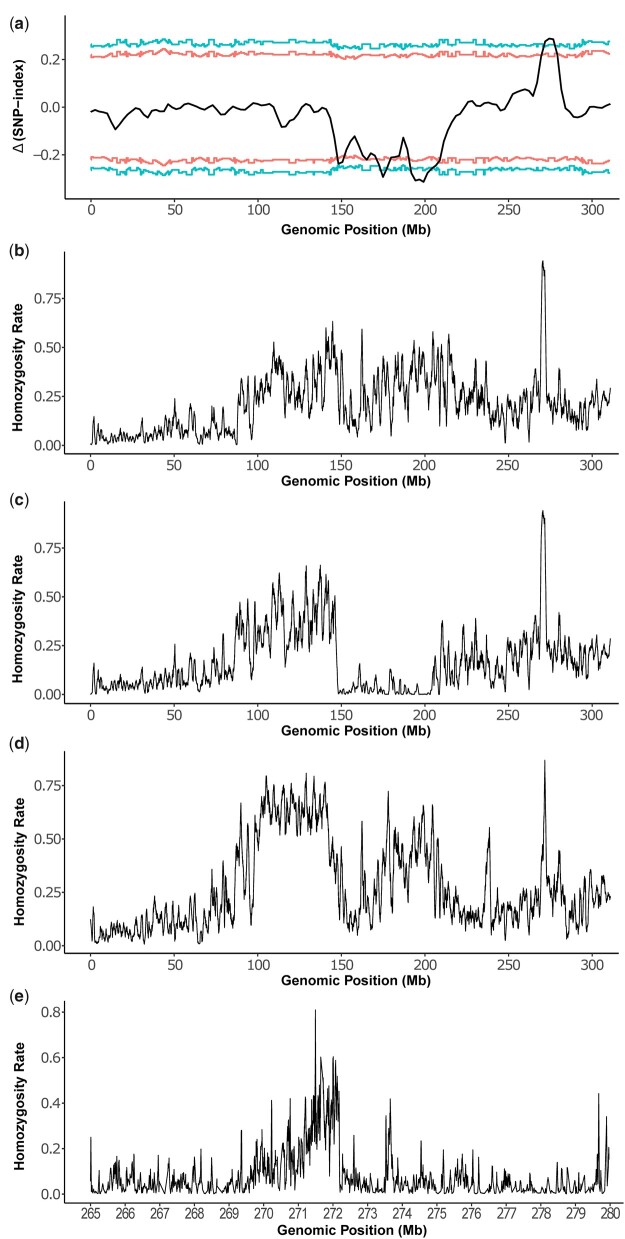
Mapping of re. a) MAM of re by plotting the Δ(SNP index) values in 1-Mb sliding windows. The corresponding 2-sided confidence intervals are shown as red (90%) or blue (95%) lines. X-axis is the genomic position on the homomorphic sex chromosome 1. The confidence intervals were estimated using 10,000 replicate simulations for all the SNP positions with the given read depths. The broad depression between 150 and 210 Mb likely reflects the sex-linked differences while the peak around 275 Mb is likely the location of re. Positions from 271,120,117 to 277,843,549 exceed 95% confidence and the left and right inflection points of this peak are 271,970,355 and 276,823,052, respectively. Homozygosity plots of red-eyed hybrids from F8THAF9MEX_F (b), F8THAF9MEX_M (c), and F7PAKF3BRA_F (d). X-axis is the genomic location on chromosome 1. Y-axis is the homozygosity of the alternative or nonreference alleles, which is calculated as (the number of unique alternative SNPs showing 100% frequency)/(the number of all unique alternative SNPs) in a 1-Mb window. We focused on unique alternative SNPs as we are only interested in SNPs showing 100% frequency. The peak for (b)–(d) is at 270–271, 270–271, and 271–272 Mb, respectively. e) Homozygosity analysis between 265 and 280 Mbp on chromosome 1 by pooling all 3 hybrid red-eyed samples. Y-axis is the homozygosity rate of the alternative or nonreference alleles, which is calculated as described above but in a 20-kb window. The highest level of homozygosity is at position 271,505,401.
Simulations with 10,000 bootstrap replicates were used to calculate the 2-sided confidence intervals. The simulation was performed based on data derived from read depths and the type and size of the population following the protocol described in QTLseqr (Mansfeld and Grumet 2018). Briefly, alternative allele frequency was sampled 100,000 times per individual according to a 25% probability for it to be 0 or 1 and a 50% probability for it to be 0.5. FH, the alternative allele frequency of the high bulk, was calculated by averaging the sampled alternative allele frequency from all individuals of the high bulk. For each read depth (D), which reflects the true range in the data, an SNP-index was obtained using the following formula in R: SNP-indexHighBulk = rbinom (n = 1, size = D, prob = FH)/D. rbinom (n = 1, size = D, prob = FH) refers to randomly selecting from a binomial distribution with FH as the probability of success and D as the number of trials. This simulation was repeated 10,000 times (n = 10,000) in R with rbinom (n = 10,000, size = D, prob = FH)/D. Similar simulations were performed for the low bulk. By subtracting the simulated SNP-indexHighBulk from the simulated SNP-indexLowBulk, values for the Δ(SNP index) were obtained from these 10,000 replicates. The extreme quantiles of these Δ(SNP index) were used to estimate confidence intervals.
Ae. aegypti strains used for homozygosity analysis
All strains described in this section were maintained in the insectary of the Insect Pest Control Laboratory (Joint FAO/IAEA Center of Nuclear Techniques in Food and Agriculture, Seibersdorf, Austria) at 27 ± 1°C, 80% relative humidity and a 12/12-h day/night photoperiod. Adult mosquitoes were provided a 10% sucrose solution and females were fed with porcine blood twice per week. The blood used was collected in Vienna, Austria, during routine slaughtering of pigs in a nationally authorized abattoir, conducted at the highest possible standards strictly following EU laws and regulations. Egg collections were initiated 72 h after the last blood feeding using moistened oviposition papers (white germination paper, Sartorius Stedium Biotech, Austria). The Rexvillle strain was provided by Dr. Margareth Capurro at the Department of Parasitology, University of Sao Paulo, Brazil. All Rexvillle individuals have red eye color which is evident throughout all developmental stages and darkens as adults age. The Rexvillle strain failed to complement the red-eye mutation (re) in the RED strain (Supplementary Fig. 3). The Rexville red-eye phenotype was introgressed into the BRA, MEX, THA, and PAK strains to create red-eye GSSs with different genomic backgrounds in a separate study (Augustinos et al. 2020, 2022; Koskinioti et al. 2021). The 11-generation introgression protocol is described in Supplementary Method 2.
Crosses and sample preparation for the homozygosity analysis
The above-mentioned introgressed GSS contains the re mutation in divergent genetic backgrounds, which could be used to narrow down the location of re, by identifying regions of enriched homozygosity. Thirty F8 red-eye females from the THA Red-Eye GSS and 15 F9 red-eye males from the MEX Red-Eye GSS were mass-crossed. All progeny were red eyed and a pool of 20 freshly emerged adult females (F8THAF9MEX_F) and a pool of 20 males (F8THAF9MEX_M) were used to extract DNA using the QIAGEN Genomic tip 20/G kit (Cat. No.: 10223, Qiagen, Hilden, Germany). In another sample, 1 wild-type BRA male was crossed with 5 virgin wild-type BRA females to create an isomale BRA line. Males from this isomale line were crossed with red-eyed Rexville females and the F1 was sib mated. F2 red-eye females were then backcrossed with the wild-type black-eyed males from the isomale line. F3 black-eyed males were crossed with the red-eyed females originated from the F7 PAK Red-eye GSS. Twenty red-eyed female progeny (F7PAKF3BRA_F) were used for HMW DNA extraction, as described above. Genomic DNA from F8THAF9MEX_F, F8THAF9MEX_M, and F7PAKF3BRA_F were sequenced by Illumina (Supplementary Table 1).
Homozygosity analysis
Mapping and variant calling were performed as described for MAM except that samples are the hybrid sequencing samples F8THAF9MEX_F, F8THAF9MEX_M, and F7PAKF3BRA_F. As the coverage is lower than those in the MAM analysis, variant filtering was adjusted based on 5 < total read depth < 500, only 1 alternative in the variant, and GATK GQ score ≥50. As shown in Fig. 3, b–e, within a given sliding window size (e.g. 1 Mb or 20 kb), the alternative homozygosity rate is calculated as [the number of unique alternative variants showing 100% frequency (SNP-index = 1)] divided by [the number of all unique alternative variants (SNP-index >0)]. We focused on unique alternative SNPs as we are only interested in SNPs showing 100% frequency.
Mosquito imaging
Imaging of larvae, pupae, and adult specimens was performed using an AMSCOPE LED Trinocular Zoom Stereo Microscope 3.5X-180X and 10 MP USB3 Camera (https://www.amscope.com/applications/veterinary-zoology/entomology/led-trinocular-zoom-stereo-microscope-3-5x-180x-and-10mp-usb3-camera.html). Imaging of pupae shown in Supplementary Fig. 3 was performed using an AMSCOPE 3.5X-180X Manufacturing 144-LED Zoom Stereo Microscope with 8MP Digital Camera (https://www.amscope.com/3-5x-180x-manufacturing-144-led-zoom-stereo-microscope-with-8mp-digital-camera.html) and the software AMSCOPE version x64, 3.7.7303 (built: April 29, 2016).
sgRNA design and sgRNA synthesis
sgRNAs were designed to target 2 regions within the predicted coding sequence of the cardinal gene (Fig. 4a). The first sgRNA target region is located in exon 3 at 664–737 bp from the predicted start codon and the second in exon 5 at 1,698–1,776 bp and is near the region targeted for the recent knockout of cardinal in Culex quinquefaciatus (Feng et al. 2021b). All 5 sgRNAs (Supplementary Table 2) were designed using CHOPCHOP (Montague et al. 2014; Labun et al. 2016, 2019) and CRISPOR (Concordet and Haeussler 2018) web tools. sgRNA DNA templates were synthesized by PCR using previously described protocols (Bassett et al. 2013) using primers listed in Supplementary Table 3. sgRNA was synthesized by T7 in vitro transcription using the MEGAscript T7 kit, purified using the MEGAclear kit (Thermo Fisher Scientific), and then aliquoted for embryonic microinjections (Basu et al. 2015).
Fig. 4.
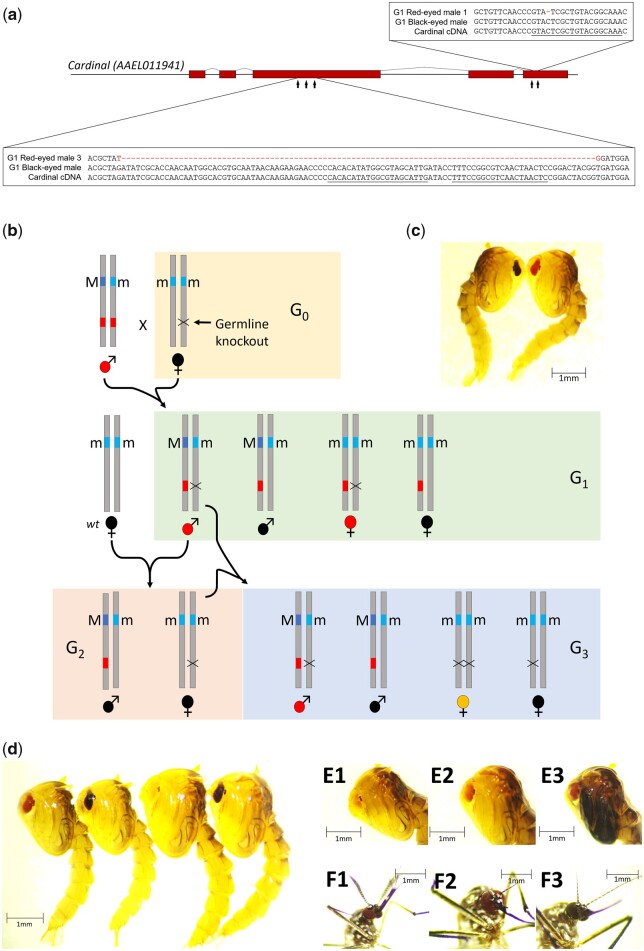
Cardinal is the causal gene that underlies the spontaneous re mutation. a) Illustration of the cardinal gene and the relative positions of the 5 sgRNAs. A 1-bp deletion (in red) in red-eyed G1 male #1 and a 99-bp deletion (in red) in red-eyed G1 male #3 are also shown. The 1-bp deletion was found in the other sequenced red-eyed G1 individuals (male #1, 5, 6, and 7). None of the deletions was found in black-eyed G1 individuals. The sgRNA sequences near the deletion sites are underlined. b) Design for germline knockout of cardinal (G1) and the crossing scheme used to generate homozygous cardinal knockout (G3). The wild-type (+) ere color allele is not shown and the re allele is indicated as a red box. c) G1 black-eye (left) and red-eye (right) phenotypes. Four red-eyed G1 females and 7 red-eyed G1 males were identified among approximately 500 black-eyed G1 progeny. Sequence confirmation of the knockout is shown in (a). d) Four phenotypes observed in the G3 progeny shown in (b). From left to right: red-eyed male, black-eyed male, yellow-eyed female, black-eyed female. The G1 red-eye male has the 99-bp deletion (G1 red-eye male #3). (e) The eyes in the G3 yellow-eyed female pupa turn red over the course of pupal development. E1–3: 1-, 2-, and 3-day-old pupae. To highlight the eye phenotype, the contrast was adjusted to 20% and the brightness was adjusted to 20% (for E1–2) and 40% (E3). F) One-day-old G3 adult eye phenotypes of the red-eyed male (F1), yellow-turn-red-eyed female (F2), and black-eyed female (F3). To highlight the eye phenotypes, the contrast of images was adjusted to 25% and the brightness to 25% (F1–2) and 50% (F3). All unmodified photos are found in the Supplementary File.
CRISPR/Cas9-mediated knockout of cardinal
For somatic knockout of cardinal, LVP males were mated with RED female mosquitoes in a G−1 cross. The resulting G0 embryos were injected with a mixture containing 300 ng/μl Cas9 mRNA and 100 ng/μl each of sgRNAs 1, 2, and 3. Somatic knockout of cardinal was evaluated visually at the larval pupal stages. To produce a germline knockout of cardinal (Fig. 4, b–f), an injection mixture containing 300 ng/μl Cas9 mRNA and 100 ng/μl each of sgRNAs 1, 2, 4, and 5 was injected into LVP embryos. Male and female G0 adult survivors were separated and mated with RED adults of the opposite sex. Specific cardinal mutations were validated by sequencing PCR amplicons from each sgRNA target region. Briefly, genomic DNA extracted using the QiaAMP gDNA-micro kit (Qiagen) or Zymo Quick-gDNA (Zymo Research) was used as the template for PCR using primers listed in Supplementary Table 3. PCR amplicons were purified using the Nucleospin PCR and gel clean-up kit (Macherey-Nagel) and sequenced using Sanger (Genomics Sequencing Center, Virginia Tech). Trace files used to validate germline knockouts of cardinal are in the Supplementary File.
Results
The vast recombination deserts in Ae. aegypti contain thousands of genes
LRRs, which show little or no recombination at the resolution of the available genetic crosses (Juneja et al. 2014), encompass up to 47% of a chromosome in Ae. aegypti (Dudchenko et al. 2017). To enable analysis using the most recent assembly, we mapped data from the same genetic crosses (Juneja et al. 2014) to the most recent AaegL5 assembly (Matthews et al. 2018). As shown in Fig. 1a, little or no recombination was observed in a ∼110-Mb region surrounding the sex locus, which is at ∼152 Mb on the homomorphic sex chromosome 1 in Ae. aegypti. Similar recombination deserts exist in all 3 chromosomes (Fig. 1, upper panels). Large LRRs are not necessarily problematic if they are devoid of genes. Unfortunately, these recombination deserts are not gene poor (Fig. 1, lower panels), making forward genetic mapping of genes in these regions a challenging problem.
The markers and the chromosomal region under investigation
We seek to map a recently obtained transgene insertion in the sex-linked LRR using 2 flanking markers to establish the MAM strategy. The transgenic insertion line is named P10 (or P for P10 positive, Supplementary Fig. 1 and Table 1) and the transgene expresses a polyUb-driven GFP. The transgene is inserted at position 224.7 Mb of the q arm of chromosome 1 (Fig. 1a), as shown by Oxford Nanopore sequencing (Supplementary Fig. 2). As shown in Fig. 2a, the 2 flanking markers are the sex locus (M/m being male and m/m being female) and an eye-color locus (B/re or +/re being black eyed or wild type and re/re being red eyed). The genetic distance between P and the sex locus (M locus, 151.68–152.95 Mb on chromosome 1) is approximately 1.02–1.55 cM, according to 2 screens of 1,353 and 3,145 progenies, respectively (Table 1). Thus, the per megabase recombination rate in the ∼72 Mb (224.68–152.95 = 71.73 Mb) region is 0.014–0.022 cM/Mbp, approximately 73–114-fold lower than that of the D. melanogaster (1.6 cM/Mb) (Wilfert et al. 2007). The eye color locus (re) is also on the q arm of chromosome 1, 2–3 cM away from the M locus, with an unknown chromosomal location (Koskinioti et al. 2021).
Table 1.
Sex linkage of the P10 (GFP) transgene insertion.
| GFP-positive male | GFP-negative male | GFP-positive female | GFP-negative female | |
|---|---|---|---|---|
| G2 | 86 | 0 | 1 | 84 |
| G8 | 743 | 12 | 9 | 589 |
| (P10/N; +/re) × (N/N; re/re) | 1,573 | 22 | 10 | 1,540 |
N stands for negative for the P10 transgene insertion; + stands for the wild-type black-eye allele. Genetic distance between the sex locus (M/m) and the P10 insertion is calculated as follows: (12 + 9)/(12 + 9 + 743 + 589) = 1.55% = 1.55 cM, according to the G8 data, and (22 + 10)/(22 + 10 + 1,573 + 1,540) = 1.02% = 1.02 cM, according to the cross shown in the last row. The numbers are too low in G2 for an accurate estimation.
Rapid and effective mapping of the P10 transgene in a region of low recombination using MAM
We seek to map the P10 transgene using 2 flanking markers sex and eye color. The relative location of M, P, and B (or +) is shown in Fig. 2 while the corresponding recessive alleles m, N (N for negative), and re are not shown. The general strategy of MAM and its advantages are depicted in Fig. 2, a–c. Only informative recombinants (Fig. 2b) are selected for genotyping analysis based on the breakage of linkage between the selectable markers (M and B) flanking the target locus (P), thus greatly increasing both the resolution and the accuracy of genetic mapping (Fig. 2c). In the example shown in Fig. 2, approximately 97.5% of the noninformative progeny (MPB: male positive black eyed; FNR: female negative red eyed) are ignored, thus both the resolution and signal-to-noise ratio are increased by 4000%. The accuracy of mapping is also improved as MAM ensures sufficient sampling of informative recombinants by ignoring the vast number of noninformative progeny. The much-reduced number of samples make it feasible to genotype the positive and control groups either as individuals or in bulk, as in bulk-segregant analysis (Schneeberger 2014), to identify causal genes in the previously inaccessible regions of suppressed recombination. To map P10 (P) by MAM and to also map re in a later analysis (see below), we grouped the informative recombinants into 4 categories for Illumina sequencing (Supplementary Table 1). The 4 groups include MPR; FPB; FNB; and MNR. As shown in Fig. 2d, when MPR and FPB (all informative P10-positive recombinants) are compared with FNB and MNR (all informative P10-negative recombinants), MAM successfully mapped the P10 location to be between 223.3 and 226.1 Mb with 95% confidence. The midpoint of the peak is 224.7 Mb, precisely the location of the P10 insertion.
MAM of re
The above-mentioned sequencing data are also used to map re, as we can compare the recombinant male red eyed and the recombinant female black eyed samples. A potential caveat is the lack of resolution from the telomeric side of re, as the cross was originally intended for mapping P10 and a marker to the telomeric side of re was not used. However, given the interest in developing re as a marker for GSSs (Koskinioti et al. 2021), the fact that re is one of the earliest reported spontaneous mutations isolated in Ae. aegypti (McClelland 1962, 1966), and the lack of any successful examples of forward genetic identification of causal genes in Ae. aegypti, we attempted to map re using the existing data. As shown in Fig. 3a, the re locus is mapped to the region between 271 and 278 Mbp with 95% confidence.
Mapping re using the introgression lines
The above mapping result for re is unexpected as the previously published data indicate that the genetic distance between the sex locus and the 271–278-Mb region is at least 10 cM (Dudchenko et al. 2017), much higher than the 2.48 cM observed between the sex locus and re (Koskinioti et al. 2021). We sought independent evidence for the location of re. As a part of an ongoing effort to develop GSSs, the Rexville red-eye strain (Koskinioti et al. 2021) was introgressed with Ae. aegypti from various regions for a minimum of 11 generations to establish GSSs of various local genetic backgrounds (Supplementary Methods) (Augustinos et al. 2022). The re-containing strain used in the above-mentioned MAM experiment is the RED strain, which is homozygous for the re locus on chromosome 1, the s (spot abdomen) locus on chromosome 2, and the b/t (black tarsus) on chromosome 3 (Craig and Hickey 1967). Genetic complementation crosses confirmed that the re in RED and the red-eye locus in the Rexville-derived introgression lines share the same mutant gene (Supplementary Fig. 3). These introgressed GSSs contain the re mutation in divergent genetic backgrounds, which could be used to narrow down the location of re, by identifying regions of enriched homozygosity. To increase the cost-effectiveness, we performed Illumina sequencing of red-eyed hybrids (Supplementary Table 1) from crosses between different GSSs to further highlight the enrichment of homozygosity near the re locus. As shown in Fig. 3, b–d, homozygosity is the highest in a region around 271–272 Mb. We note that the homozygosity peak of the 3 red-eyed introgression samples falls on the centromeric side of the MAM peak (271–278 Mb), which is consistent with expectation as the P10 marker is to the centromeric side of re and there is no marker to the telomeric side of re.
Identification of a candidate gene cardinal
To further narrow down the candidate region, we focused the homozygosity analysis on the 265–280 Mb region using combined SNP data from all 3 hybrid red-eyed samples. As shown in Fig. 3e, the region with the highest homozygosity is around 271.5 Mb. Interestingly, a gene encoding a chorion peroxidase, which is an ortholog of the Drosophila eye pigment gene cardinal, is located between 271.574 and 271.583 Mb and this gene is expressed throughout development after 20 h postegg deposition and its transcripts are enriched in the brain/head of both sexes in Ae. aegypti (Matthews et al. 2018). Thus, cardinal is the top candidate for the causal gene for the spontaneous re mutation. Indeed, the cardinal gene sequences in red-eyed samples show a number of nonsynonymous mutations leading to changes of amino acid residues that are conserved across divergent mosquitoes (Supplementary Fig. 4).
CRISPR/Cas9-mediated knockout of cardinal produced somatic knockouts showing mosaic red-eyed G0 individuals
To increase the likelihood to observe a somatic cardinal knockout phenotype in the G0, we first performed CRISPR/Cas9-mediated knockout in heterozygous +/re individuals. The sgRNAs used to target cardinal are shown in Fig. 4a and provided in Supplementary Table 2. Mosaic red eyes were indeed observed in +/re G0 individuals (Supplementary Fig. 5). Such somatic mosaicism may reflect knockout of the wild-type cardinal allele (cd−) in some cells, resulting in the red-eye phenotype in the cd−/re genotyped cells. Thus, the result is consistent with cardinal being the causal gene of the re mutation. However, we cannot rule out the possibility that the mosaic phenotype may have nothing to do with re and it simply resulted from somatic knockout of both copies of the cardinal gene. Regardless, this result confirmed that the sgRNAs are effective and the cardinal gene is indeed involved in eye color in Ae. Aegypti, as recently demonstrated in Culex quinquefasciatus and An. gambiae (Carballar-Lejarazú et al. 2020; Feng et al. 2021b).
Germline knockout of cardinal showed that it is the causal gene of the re mutant
To isolate germline knockout of cardinal (Fig. 4b), sgRNAs and Cas9 mRNAs were injected in embryos collected from wild-type Lvp females (+/+) mated with wild-type Lvp males (+/+). The resulting G0 females were black-eyed and selected to mate with red-eyed males (re/re). Four female and 7 male red-eyed G1 individuals were identified among approximately 500 black-eyed G1 individuals (counts shown in Supplementary Table 4; genotypes shown in Fig. 4b, G1 box; G1 phenotype shown in Fig. 4c). PCR and subsequent Sanger sequencing of 5 red-eyed G1 males using primers flanking the sgRNAs (Supplementary Table 3) identified 2 types of mutations (Fig. 4a). A single base deletion that results in a frameshift was found in 4 of the G1 males (#1, 5, 6, and 7) and a 99-base deletion in the other (G1 male #3). However, none of these deletions were found in the black-eyed G1 individuals (Fig. 4a). These red-eyed G1 individuals can only have 1 re allele from the father and 1 cd− allele from the mother. Therefore, cardinal knockouts (cd−) resulting from either 1- or 99-bp deletion fail to complement re, suggesting that cardinal is indeed the causal gene of the re mutant.
Establishing and characterizing a homozygous cardinal knockout
To obtain homozygous knockout individuals, black-eye G2 females (+/cd−) were backcrossed to their father, red-eye G1 male #3 (re/cd−; Fig. 4b). Their G3 offspring were evaluated, 4 genotypes were expected, and 4 phenotypes were observed: red-eyed males, black-eyed males, yellow-eyed females, and black-eyed females (Fig. 4b, G3 box, and Supplementary Table 5). The yellow-eyed females are homozygous for the 99-bp cardinal deletion. The yellow-eyed phenotype is observed in the prepupal compound eye (Fig. 4d and Supplementary Fig. 6). However, the yellow eyes gradually turned into red eyes over the course of pupae development and adult emergence (Fig. 4, e and f). Therefore, the yellow-eye phenotype is in fact yellow-turn-red. No yellow-eyed L2/L3 instars were found and the ocelli of the yellow-eyed prepupa are red, indicating that the yellow-turn-red phenotype is associated with the remodeling of the compound eye during metamorphosis. The sibling red-eyed males inherited 1 copy of the re spontaneous mutation and 1 copy of the cardinal knockout (Fig. 4b, G3 box), again indicating that cd− does not complement re. As expected, when the G2 females (+/cd−) were crossed to RED males (re/re), approximately half of the offspring are red eyed in each sex (Supplementary Fig. 7 and Supplementary Table 6).
Towards genome-wide MAM in Ae. Aegypti
P10 was mapped using sex and re as the flanking markers, approximately 72 and 47 Mb away on either side. It was possible to map re using only the P10 marker. Therefore, it is not necessary to have densely populated markers for MAM to succeed. However, to extend MAM to the entire genome and to provide additional resources for fine mapping, we began a systematic effort to generate and compile an expanding list of marker lines throughout the genome. Initially, we focused on the homomorphic sex-determining chromosome 1 by performing Oxford Nanopore Sequencing of 4 newly generated and 1 existing sex-linked transgene-insertion lines to identify their insertion sites (Table 2 and Supplementary Method 1). We also compiled published transgenic lines with known genomic insertions as well as knockout or knock-in lines that provide screenable markers throughout the genome (Table 2). In total, we identified lines with 44 morphological or transgenic markers with 21, 9, and 14 on chromosomes 1, 2, and 3, respectively. Some are independent lines that mark the same gene. Importantly, multiple markers are available in the recombination deserts on all 3 chromosomes (Table 2). Therefore, genetic resources are available for genome-wide MAM in and outside the recombination deserts in Ae. aegypti.
Table 2.
Morphological and transgenic markers available for marker-assisted mapping in Ae. aegypti.
| Chr | Position | Gene or transgene | Marker | Reference |
|---|---|---|---|---|
| 1 | 17,701,465 | AAEL014686 (Ir7f) a | 3xP3_DsRed | (Jové et al. 2020) |
| 1 | 20,424,072 | N1 (nix transgene)b | Pub_GFP | (Aryan et al. 2020) |
| 1 | 37,734,383 | Ir7a (manual annotation) a | 3xP3_DsRed | (Jové et al. 2020) |
| 1 | 65,092,968 | 15xQUAS-CaMPARI2c | 3xP3_ECFP | (Shankar et al. 2020) |
| 1 | 83,180,924 | AAEL006830 (yellow) d | yellow body | (Li et al. 2017) |
| 1 | 85,989,081 | AAEL002922 (IR8a) a | Pub_DsRed | (Raji et al. 2019) |
| 1 | 85,989,081 | AAEL002922 (IR8a) a | 3xP3_DsRed | (Shankar et al. 2020) |
| 1 | 107,942,728 | AAEL016999 (white) d | white eye | (Li et al. 2017) |
| 1 | 114,522,328 | J2b | Pub_DsRed | (Hall et al. 2014); this work |
| 1 | 152,616,641 | Nix | maleness | (Hall et al. 2015) |
| 1 | 161,792,846 | AAEL019619 a | Pub_GFP | (Lutrat et al. 2022) |
| 1 | 180,261,137 | P14c | Pub_GFP | this work |
| 1 | 201,036,504 | AAEL005793 (ebony) d | dark body | (Li et al. 2017) |
| 1 | ∼205.6 Mb | Sensorb | 3xP3-DsRED | (Adelman et al. 2008) |
| 1 | ∼205.6 Mb | C42 (Vg-CecA)b | 3xP3_GFP | (Krzywinska et al. 2016) |
| 1 | 223,544,654 | R4Mc | 3xP3_DsRed | this work |
| 1 | 224,678,384 | P10c | Pub_GFP | this work |
| 1 | 243,246,615 | AAEL002167 (Gr2) a | Pub_ECFP | (Duvall et al. 2017) |
| 1 | ∼266.2 Mb | Pub_DsRed_P1c | Pub_DsRed | this work |
| 1 | 271,574,080 | AAEL011941 (cardinal) d | red eye | this work |
| 1 | 271,574,080 | AAEL011941 (cardinal) d | yellow-turn-red eye | this work |
| 2 | 7,643,838 | QUAS-Syt1: tdTomatoc | 3xP3_ECFP | (Zhao et al. 2021) |
| 2 | 67,565,706 | N2 (nix transgene)b | Pub_GFP | (Aryan et al. 2020) |
| 2 | 127,230,326 | AAEL008879 (kmo) d | white eye | (Aryan et al. 2013) |
| 2 | 234,641,436 | AAEL018153 (brp) a | 3XP3-dsRed | (Zhao et al. 2021) |
| 2 | 302,311,047 | AAEL002380 (Gr1) a | 3xP3_DsRed2 | (Shankar et al. 2020) |
| 2 | 305,065,798 | AAEL010779 (pickpocket 204) a | Pub_DsRed | (Kistler et al. 2015) |
| 2 | 324,350,134 | exu_crec | Pub-EYFP | (Shankar et al. 2020) |
| 2 | 329,084,910 | 15XQUAS-mCD8::GFPc | 3xP3_ECFP | (Shankar et al. 2020) |
| 2 | 458,973,530 | AAEL000704 (synaptotagmin) a | 3XP3-dsRed | (Zhao et al. 2021) |
| 3 | 11,155,320 | AAEL010058 (Gr3) a | Pub_ECFP | (McMeniman et al. 2014) |
| 3 | 11,821,895 | AAEL000582 (pickpocket 301) a | Pub_ECFP | (Kistler et al. 2015) |
| 3 | 15,095,953 | AAEL006498 (GPRop1) a | 3xP3_DsRed | (Zhan et al. 2021) |
| 3 | 15,095,953 | AAEL006498 (GPRop1) a | 3xP3_GFP | (Zhan et al. 2021) |
| 3 | 63,253,679 | AAEL006259 (GPRop2) a | 3xP3_DsRed | (Zhan et al. 2021) |
| 3 | 63,253,679 | AAEL006259 (GPRop2) a | 3xP3_GFP | (Zhan et al. 2021) |
| 3 | 104,935,424 | AAEL000048 (GR4) a | 3xP3_DsRed | (Jové et al. 2020) |
| 3 | 114,494,813 | AAEL005776 (orco) a | 3xP3_DsRed2 | (Shankar et al. 2020) |
| 3 | 114,494,813 | AAEL005776 (orco) a | 3xP3_DsRed | (Herre et al. 2022) |
| 3 | 114,494,813 | AAEL005776 (orco) a | 3xP3_DsRed | (Zhao et al. 2022) |
| 3 | 203,135,372 | AAEL009813 (IR25a) a | 3xP3_DsRed | (Herre et al. 2022) |
| 3 | 212,799,454 | exu_Cas9c | Opie2_DsRed | (Li et al. 2017) |
| 3 | 386,240,391 | AAEL029058 (GCTL-3) a | Pub_GFP | (Li et al. 2020) |
| 3 | 396,626,080 | AAEL006360 (IR76b) a | 3xP3_DsRed | (Herre et al. 2022) |
The positions shown are either gene positions in the case of gene knockout/knock-in or the first nucleotide of the transposon insertion target site. In some cases, precise transposon insertion sites are not determined due to the tandem repeats immediate flanking sequences while the approximate locations are identified according to sequences further out. Strains with unknown marker sites or strains that are not amenable to genetic screening are not included.
Knocked in transformation marker is the screenable marker. Some genes such as IR8a, GPRop1, GPRop2, and orco have multiple independent knock-in strains. In these cases, the precise knock-in sites within the gene may vary.
Transposon-mediated insertion that contains a marker and a cargo that is functional in at least 1 tissue of 1 sex.
Transposon-mediated insertion that either contains only the transformation marker or a marker plus a cargo that is not functional by itself.
Gene knockout or mutation confers the screenable phenotype.
Discussion
This study addresses two major bottlenecks to forward genetic studies in Ae. aegypti. We demonstrate that MAM can overcome the challenge of mapping genes in the vast recombination deserts by isolating and genotyping only the small number of informative recombinants. We also begin a systematic effort to provide and compile genetic strains with markers spanning the entire genome. Therefore, effective forward genetic analysis is increasingly feasible for this important arboviral vector species. We discuss practical considerations and future perspectives when applying MAM to identify the genetic determinants of various traits and describe the implications of identifying the causal gene of the spontaneous re mutation to improving genetic sexing strains.
MAM enables effective forward genetic analysis in and outside the recombination desert in Ae. aegypti
We designed the MAM strategy to overcome the low recombination bottleneck by selecting and genotyping only the rare but informative recombinants between easy-to-screen markers flanking the target locus, thus, drastically improving the resolution, signal-to-noise ratio, and cost-effectiveness for genetic mapping (Fig. 2). Using MAM, we successfully mapped P10, a dominant transgene insertion that shows an extremely low rate of recombination relative to the sex locus (0.014–0.022 cM/Mbp) and the red-eye (re) marker (1.46 cM/46 Mb = 0.032 cM/Mb). The MAM peak spans from 223.3 Mb to 226.1 Mb with the midpoint being 224.7 Mb, precisely the location of the P10 insertion. Therefore, we have demonstrated the power of MAM in making forward genetics accessible in the LRRs which were previously inaccessible or challenging.
Despite the lack of a marker to the telomeric side of re, we were able to map re to a broader peak between 271.1 and 277.8 Mb at a 95% confidence interval. The location of the re causal gene cardinal is at 271.5 Mb, much closer to the centromeric side (271.1 Mb) of the peak than to the telomeric side. The lack of resolution at the telomeric side, as indicated by the 6.3 Mb distance between 271.5 and 277.8 Mb, likely reflects the lack of a marker to “assist” in selecting informative recombinants. We showed that homozygosity analysis using hybrids between repeatedly introgressed strains can also facilitate mapping studies. The introgressed GSSs are highly valuable but time consuming to create as a minimum of 11 generations were used in this study, thus may not be available to other mapping studies. However, even in the absence of one of the two flanking markers and without the use of additional resources such as these introgression lines, MAM can still identify the candidate locus with the causal gene being proximal to the marker side of the peak. The identification of cardinal as the causal gene for the spontaneous re mutant is the first report of a successful identification of the causal gene using forward genetics in Ae. aegypti. The cardinal gene is outside the previously defined LRRs (Juneja et al. 2014; Dudchenko et al. 2017; Fontaine et al. 2017). Thus, we have shown that MAM enabled forward genetics both in and outside the LRRs.
As mentioned earlier, the location of re is somewhat unexpected as the previously published data indicate that the genetic distance between the sex locus and the 271.5 Mb region is at least 10 cM (Dudchenko et al. 2017), much higher than the 2–3 cM between the sex locus and re (Koskinioti et al. 2021). Although such a discrepancy may result from different genetic backgrounds or other conditions that may affect the rate of recombination (Craig and Hickey 1967), it may also reflect a limitation of the previous mapping studies in which a small number of progenies were genotyped. The ease of screening for the P10 and re markers affords a high-resolution estimate of the recombination rate based on rapid screening of a large number of progenies. This is especially important when working with loci in the LRRs.
Further considerations of genome-wide MAM in Ae. aegypti
As discussed earlier, a single marker can successfully assist with mapping the causal gene from dozens of megabases away. Therefore, the published marker lines and the lines we generated in this study (Table 2) will enable genome-wide MAM in and outside the recombination deserts in Ae. aegypti. As we demonstrated by mapping the causal gene for re, MAM can be used to fine map the large list of spontaneous mutations, which are mostly single-locus traits that are apparently abundant in Ae. aegypti populations (e.g. Craig and Hickey 1967). Mapping multi-locus quantitative traits is intrinsically more challenging than what we described here especially if the traits are not easy to score. However, with the help of flanking markers MAM can assist with fine mapping a large-effect QTL just as it can with fine mapping a single-locus qualitative trait by mitigating the impediment imposed by low recombination (Supplementary Fig. 8). When mapping traits that are time or labor intensive to score, such as insecticide or viral resistance, MAM provides another advantage as assays will only need to be performed on a small number of informative recombinants. The small number of informative samples also make it feasible to genotype the positive and control groups as barcoded individuals. It may also be cost-effective to only sequence or genotype the genomic region between the assisting markers.
When performing MAM, the genetic distance between the flanking markers determines the level of enrichment and the number of progeny that need to be screened to obtain a sufficient number of informative recombinants for a target resolution (Fig. 2). Markers flanking a recombination desert will confer high enrichment but require screening a large number of progeny. We anticipate that additional marker lines will rapidly accumulate as CRISPR/Cas9-mediated editing is reported at a fast pace. Some knockouts (e.g. Li et al. 2017; this study) will produce screenable phenotypes and most knock-ins (e.g. Zhao et al. 2021) will contain a screenable transformation marker. Although some of the knock-in would also result in an interruption of the gene, this normally will not prevent them from being a useful marker line as MAM will involve crossing the marker line with a strain of interest followed by a backcross with the same strain. Therefore, the allele interrupted by the transformation marker will not be homozygous during the process. In addition, transposon-mediated random insertion is also producing a rapidly increasing number of Ae. aegypti strains with easy to screen transformation markers and high-throughput strategies (e.g. Stern 2017) and long-read sequencing described in this study (Supplementary Method 1) make it easy to identify transgene insertion sites. These transposon insertions may contain a cargo as they are often designed to express a gene of interest, which may or may not affect their use as a marker for MAM. As an alternative, transformation-marker-only strains can be readily generated and characterized for use in MAM (Table 2; Supplementary Fig. 1). As densely populated markers are not necessary, a small number of strains, each contains multiple markers on a different chromosome arm, may be sufficient for genome-wide MAM in Ae. aegypti. A stock center that maintains and distributes even just a small number of useful Ae. aegypti strains will be an invaluable resource to the community and will significantly facilitate the genetic research of this important arboviral vector. Ae. aegypti embryos can be kept dormant for 6 months or more, which reduces the cost and simplifies the operation of such a resource center.
Insights into the function of cardinal
We did not observe any phenotypic differences between re/re and re/cd− individuals. However, the homozygous progeny of a 99-bp deletion in cardinal showed a phenotype of eyes gradually turning from yellow to red, which is associated with the remodeling of the compound eye during metamorphosis. The cardinal gene encodes a peroxidase, which catalyzes the formation of the eye pigment xanthommatin from 3-hydroxykynurenine in the ommochrome pathway and is believed to also participate in the synthesis of ommin (Howells et al. 1977; Harris et al. 2011; Osanai-Futahashi et al. 2016; Zhang et al. 2017; Xu et al. 2020). Indeed, knockout of cardinal near its C terminus in C. quinquefasciatus resulted in a red-eye mutation visible from the larval to the early adult (Feng et al. 2021b), while an amorphic cardinal knockout in the diamond back moth resulted in yellow-eyed adults, which turned into red eyed as they aged (Xu et al. 2020). The gradual change was explained by the slow autooxidation of the yellow pigment 3-hydroxykynurenine leading to the eventual accumulation of the red pigment xanthommatin. Similarly, the 99-bp cardinal deletion in Ae. aegypti likely resulted in either an amorphic mutation or a more severe hypomorphic mutation than the spontaneous re.
Implications to the development of GSSs
The development of GSSs has been a key challenge to SIT programs from the start. Indeed, it took more than 20 years to develop one of the most successful GSSs for the Mediterranean fruit fly, the Ceratitis capitata VIENNA 8 GSS (Franz et al. 2021). Only recently, the causal gene of the white-pupae (wp) mutation used in the VIENNA 8 GSS was identified (Ward et al. 2021). The necessity to develop a mosquito GSS does not only stem from economic concerns but also from the behavioral traits of the species since only females feed on vertebrate hosts and transmit pathogens. Several mosquito GSSs were developed for Anopheles species since the 1970s (Curtis 1978; Kaiser et al. 1978; Lines and Curtis 1985; Robinson 1986; Yamada et al. 2012, 2015). However, the high genetic instability of these strains and in some cases the reliance on insecticide resistance as a selection marker are problematic. Recent field releases of transgenic, irradiated and/or Wobachia-infected males showed promising results for suppressing Aedes populations in areas of moderate size (reviewed in Papathanos et al. 2018; Kittayapong et al. 2019; Zheng et al. 2019; Crawford et al. 2020; Balatsos et al. 2021). However, robust, precise, and economical sex separation remains a significant bottleneck to large-scale implementations of genetic strategies to control mosquito-borne infectious diseases.
The current red-eye GSS has shown excellent performance and genetic stability under laboratory conditions (Koskinioti et al. 2021) and an induced chromosomal inversion further increased the precision and effectiveness of these GSS lines (Augustinos et al. 2020). In this study, we identified the causal gene of the red-eye phenotype. The identification of the red-eye causal gene will facilitate the development of GSS lines, not only for Ae. aegypti but also for other medically important mosquitoes such as Ae. albopictus and Anopheles species. For example, the red-eye phenotype can be created by knocking out cardinal in these species using CRISPR/Cas-mediated genome editing approaches. Tight sex linkage could be achieved either by irradiation-induced or CRISPR/Cas9-induced chromosomal translocation, or by CRISPR/Cas9-mediated knock-in of a wild-type cardinal in the male sex locus. This would significantly accelerate the process of the GSS development thus saving time and resources.
Concluding remarks
We anticipate that MAM in conjunction with the rapidly accumulating genetic resources described in this study will significantly improve forward genetic studies in Ae. aegypti and facilitate the mapping of the genetic determinants for single gene trait as well as large-effect loci identified from QTL analyses, including genes responsible for insecticide and viral resistance. Similar applications can be extended to other plant and animal species with large regions of suppressed recombination including those of economic significance (Stapley et al. 2017a,b).
Supplementary Material
Acknowledgments
This study benefited from discussions during meetings in the frame of the IAEA Coordinated Research Project “Generic approach for the development of genetic sexing strains for SIT applications.” We thank Song Li for discussions on data analysis and Lindy McBride for providing a list of publications that report knock-in transgenic marker lines. We thank the Chloé Lahondère laboratory for use of the Amscope microscope imaging system and for advice on imaging. We thank Margareth Capurro, Jorge Aurelio Torres Monzon, Pat Kittayapong, and Muhammad Misbah-ul-Haq for kindly providing the BRA, MEX, THA, and PAK strains used in the present study.
Funding
This work is supported by NIH grants R01AI123338 and 1R01AI157491 and the Virginia Agriculture Experimental Station. AC is supported by a fellowship from the Robert Wood Johnson Foundation. This study is also financially supported by the Joint FAO/IAEA Insect Pest Control Subprogramme of the Joint FAO/IAEA Centre of Nuclear Techniques in Food and Agriculture and the U.S. State Department in the frame of the “Surge Expansion of the Sterile Insect Technique (SIT) to Control Mosquito Populations that Transmit the Zika Virus” project.
Conflicts of interest
None declared.
Contributor Information
Chujia Chen, Genetics Bioinformatics and Computational Biology Program, Virginia Tech, Blacksburg, VA 24061, USA; Fralin Life Sciences Institute, Virginia Tech, Blacksburg, VA 24061, USA.
Austin Compton, Fralin Life Sciences Institute, Virginia Tech, Blacksburg, VA 24061, USA; Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Katerina Nikolouli, Insect Pest Control Laboratory, Joint FAO/IAEA Centre of Nuclear Techniques in Food and Agriculture, Department of Nuclear Sciences and Applications, IAEA Laboratories, 2444 Seibersdorf, Austria.
Aihua Wang, Fralin Life Sciences Institute, Virginia Tech, Blacksburg, VA 24061, USA; Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Azadeh Aryan, Fralin Life Sciences Institute, Virginia Tech, Blacksburg, VA 24061, USA; Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Atashi Sharma, Fralin Life Sciences Institute, Virginia Tech, Blacksburg, VA 24061, USA; Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Yumin Qi, Fralin Life Sciences Institute, Virginia Tech, Blacksburg, VA 24061, USA; Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Camden Dellinger, Fralin Life Sciences Institute, Virginia Tech, Blacksburg, VA 24061, USA; Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Melanie Hempel, Fralin Life Sciences Institute, Virginia Tech, Blacksburg, VA 24061, USA; Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Mark Potters, Fralin Life Sciences Institute, Virginia Tech, Blacksburg, VA 24061, USA; Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Antonios Augustinos, Insect Pest Control Laboratory, Joint FAO/IAEA Centre of Nuclear Techniques in Food and Agriculture, Department of Nuclear Sciences and Applications, IAEA Laboratories, 2444 Seibersdorf, Austria.
David W Severson, Department of Biological Sciences, University of Notre Dame, Notre Dame, IN 46556, USA.
Kostas Bourtzis, Insect Pest Control Laboratory, Joint FAO/IAEA Centre of Nuclear Techniques in Food and Agriculture, Department of Nuclear Sciences and Applications, IAEA Laboratories, 2444 Seibersdorf, Austria.
Zhijian Tu, Genetics Bioinformatics and Computational Biology Program, Virginia Tech, Blacksburg, VA 24061, USA; Fralin Life Sciences Institute, Virginia Tech, Blacksburg, VA 24061, USA; Department of Biochemistry, Virginia Tech, Blacksburg, VA 24061, USA.
Data Availability
Strains and plasmids are available upon request. Original image files are available at FigShare: https://doi.org/10.25386/genetics.20098100. Illumina and Oxford Nanopore sequencing data are deposited and available at https://www.ncbi.nlm.nih.gov/sra/PRJNA718905.
Supplemental material available at GENETICS online.
Literature cited
- Adelman ZN, , AndersonMAE, , MorazzaniEM, , Myles KM.. A transgenic sensor strain for monitoring the RNAi pathway in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2008;38(7):705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryan A, Anderson MA, Myles KM, Adelman ZN.. Talen-based gene disruption in the dengue vector Aedes aegypti. PLoS One. 2013;8(3):e60082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryan A, Anderson MAE, Biedler JK, Qi Y, Overcash JM, Naumenko AN, Sharakhova MV, Mao C, Adelman ZN, Tu Z.. Nix alone is sufficient to convert female Aedes aegypti into fertile males and myo-sex is needed for male flight. Proc Natl Acad Sci U S A. 2020;117(30):17702–17709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustinos AA, Misbah-Ul-Haq M, Carvalho DO, de la Fuente LD, Koskinioti P, Bourtzis K.. Irradiation induced inversions suppress recombination between the m locus and morphological markers in Aedes aegypti. BMC Genet. 2020;21(Suppl 2):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustinos AA, Nikolouli K, de la Fuente LD, Misbah-ul-Haq M, Carvalho DO, Bourtzis K.. Introgression of the Aedes aegypti red-eye genetic sexing strains into different genomic backgrounds for sterile insect technique applications. Front Bioeng Biotechnol. 2022;10:821428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balatsos G, Puggioli A, Karras V, Lytra I, Mastronikolos G, Carrieri M, Papachristos DP, Malfacini M, Stefopoulou A, Ioannou CS, et al. Reduction in egg fertility of Aedes albopictus mosquitoes in Greece following releases of imported sterile males. Insects. 2021;12(2):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL.. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4(1):220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Aryan A, Overcash JM, Samuel GH, Anderson MA, Dahlem TJ, Myles KM, Adelman ZN.. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc Natl Acad Sci U S A. 2015;112(13):4038–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballar-Lejarazú R, Ogaugwu C, Tushar T, Kelsey A, Pham TB, Murphy J, Schmidt H, Lee Y, Lanzaro GC, James AA.. Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae. Proc Natl Acad Sci USA. 2020;117(37):22805–22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ahlford A, Schnorrer F, Kalchhauser I, Fellner M, Viragh E, Kiss I, Syvänen A-C, Dickson BJ.. High-resolution, high-throughput SNP mapping in Drosophila melanogaster. Nat Methods. 2008;5(4):323–329. [DOI] [PubMed] [Google Scholar]
- Coates CJ, Jasinskiene N, Miyashiro L, James AA.. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A. 1998;95(7):3748–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet JP, Haeussler M.. Crispor: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46(W1):W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig GB Jr, Hickey WA.. Current status of the formal genetics of Aedes aegypti. Bull World Health Organ. 1967;36(4):559–562. [PMC free article] [PubMed] [Google Scholar]
- Crawford JE, Clarke DW, Criswell V, Desnoyer M, Cornel D, Deegan B, Gong K, Hopkins KC, Howell P, Hyde JS, et al. Efficient production of male Wolbachia-infected Aedes aegypti mosquitoes enables large-scale suppression of wild populations. Nat Biotechnol. 2020;38(4):482–492. [DOI] [PubMed] [Google Scholar]
- Curtis CF. Genetic sex separation in Anopheles arabiensis and the production of sterile hybrids. Bull World Health Organ. 1978;56(3):453–454. [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Berrocal A, Morita T, Longden KD, Stern DL.. Natural courtship song variation caused by an intronic retroelement in an ion channel gene. Nature. 2016;536(7616):329–332. [DOI] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. De novo assembly of the Aedes aegypti genome using hi-c yields chromosome-length scaffolds. Science. 2017;356(6333):92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall LB, Basrur NS, Molina H, McMeniman CJ, Vosshall LB. Apeptide signaling system that rapidly enforces paternity in the Aedes aegypti mosquito. Curr Biol. 2017;27(23):3734–3742.e5. [DOI] [PMC free article] [PubMed]
- Feng J, Zhang X, Zhang M, Guo L, Qi T, Tang H, Zhu H, Wang H, Qiao X, Xing C, et al. Physical mapping and InDel marker development for the restorer gene Rf2 in cytoplasmic male sterile CMS-D8 cotton. BMC Genomics. 2021a;22(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Kambic L, Nishimoto JHK, Reed FA, Denton JA, Sutton JT, Gantz VM.. Evaluation of Gene Knockouts by CRISPR as Potential Targets for the Genetic Engineering of the Mosquito Culex quinquefasciatus. CRISPR J. 2021b;4(4):595–608. 10.1089/crispr.2021.0028 34280034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine A, Filipovic I, Fansiri T, Hoffmann AA, Cheng C, Kirkpatrick M, Rasic G, Lambrechts L.. Extensive genetic differentiation between homomorphic sex chromosomes in the mosquito vector, Aedes aegypti. Genome Biol Evol. 2017;9(9):2322–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz G, Bourtzis K, Cáceres C.. Practical and operational genetic sexing systems based on classical genetic approaches in fruit flies, an example for other species amenable to large-scale rearing for the sterile insect technique. In: Dyck VA, Hendrichs J, Robinson AS, editors. Sterile Insect Technique. Boca Raton, FL (USA): CRC Press; 2021. p. 575–604. [Google Scholar]
- Hall AB, Basu S, Jiang X, Qi Y, Timoshevskiy VA, Biedler JK, Sharakhova MV, Elahi R, Anderson MAE, Chen X-G, et al. A male-determining factor in the mosquito Aedes aegypti. Science. 2015;348(6240):1268–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AB, Timoshevskiy VA, Sharakhova MV, Jiang X, Basu S, Anderson MA, Hu W, Sharakhov IV, Adelman ZN, Tu Z.. Insights into the preservation of the homomorphic sex-determining chromosome of Aedes aegypti from the discovery of a male-biased gene tightly linked to the M-locus. Genome Biol Evol. 2014;6(1):179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AM, McCombs SD, Fraser MJ, Saul SH.. The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. Proc Natl Acad Sci U S A. 1998;95(13):7520–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DA, Kim K, Nakahara K, Vasquez-Doorman C, Carthew RW.. Cargo sorting to lysosome-related organelles regulates siRNA-mediated gene silencing. J Cell Biol. 2011;194(1):77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herre M, Goldman OV, Lu T-C, Caballero-Vidal G, Qi Y, Gilbert ZN, Gong Z, Morita T, Rahiel S, Ghaninia M,. et al. Non-canonical odor coding in the mosquito. Cell. 2022;185(17):3104–3123.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howells AJ, Summers KM, Ryall RL.. Developmental patterns of 3-hydroxykynurenine accumulation in white and various other eye color mutants of Drosophila melanogaster. Biochem Genet. 1977;15(11–12):1049–1059. [DOI] [PubMed] [Google Scholar]
- Jové V, Gong Z, Hol FJ, Zhao Z, Sorrells TR, Carroll TS, Prakash M, McBride CS, Vosshall LB.. Sensory discrimination of blood and floral nectar by Aedes aegypti mosquitoes. Neuron. 2020;108(6):1163–1180.e1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja P, Osei-Poku J, Ho YS, Ariani CV, Palmer WJ, Pain A, Jiggins FM.. Assembly of the genome of the disease vector Aedes aegypti onto a genetic linkage map allows mapping of genes affecting disease transmission. PLoS Negl Trop Dis. 2014;8(1):e2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P, Seawright J, Dame D, Joslyn D.. Development of a genetic sexing system for Anopheles albimanus. J Econ Entomol. 1978;71(5):766–771. [Google Scholar]
- Kistler KE, Vosshall LB, Matthews BJ.. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 2015;11(1):51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittayapong P, Ninphanomchai S, Limohpasmanee W, Chansang C, Chansang U, Mongkalangoon P.. Combined sterile insect technique and incompatible insect technique: the first proof-of-concept to suppress Aedes aegypti vector populations in semi-rural settings in Thailand. PLoS Negl Trop Dis. 2019;13(10):e0007771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinioti P, Augustinos AA, Carvalho DO, Misbah-Ul-Haq M, Pillwax G, de la Fuente LD, Salvador-Herranz G, Herrero RA, Bourtzis K.. Genetic sexing strains for the population suppression of the mosquito vector Aedes aegypti. Philos Trans R Soc Lond B: Biol Sci. 2021;376(1818):20190808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinska E, , KokozaV, , MorrisM, , De La Casa-EsperonE, , RaikhelAS, , Krzywinski J.. The sex locus is tightly linked to factors conferring sex-specific lethal effects in the mosquito Aedes aegypti. Heredity. 2016;117(6):408–416. 10.1038/hdy.2016.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E.. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44(W1):W272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E.. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47(W1):W171–W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-H, Cai Y, Li J-C, Su MP, Liu W-L, Cheng L, Chou S-J, Yu G-Y, Wang H-D, Chen C-H.. C-type lectins link immunological and reproductive processes in Aedes aegypti. Iscience. 2020;23(9):101486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R., . 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Bui M, Yang T, Bowman CS, White BJ, Akbari OS.. Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc Natl Acad Sci U S A. 2017;114(49):E10540–E10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lines JD, Curtis CF.. Genetic sexing systems in Anopheles arabiensis Patton (Diptera: Culicidae). J Econ Entomol. 1985;78(4):848–851. [DOI] [PubMed] [Google Scholar]
- Loehlin DW, Oliveira DCSG, Edwards R, Giebel JD, Clark ME, Cattani MV, van de Zande L, Verhulst EC, Beukeboom LW, Muñoz-Torres M, et al. Non-coding changes cause sex-specific wing size differences between closely related species of Nasonia. PLoS Genet. 2010;6(1):e1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutrat C, Burckbuchler M, Olmo RP, Beugnon R, Fontaine A, Baldet T, Bouyer J, Marois E. Combining two genetic sexing strains allows sorting of non-transgenic males for aedes genetic control. bioRxiv, 2022. 10.1101/2022.03.11.483912. [DOI] [PMC free article] [PubMed]
- Mansfeld BN, Grumet R.. QTLseqr: an R package for bulk segregant analysis with next-generation sequencing. Plant Genome. 2018;11(2):180006. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, Glassford WJ, Herre M, Redmond SN, Rose NH, et al. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature. 2018;563(7732):501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland G. Sex-linkage in Aedes aegypti. Trans Roy Soc Trop Med Hyg. 1962;56(4). [Google Scholar]
- McClelland GA. Sex-linkage at two loci affecting eye pigment in the mosquito Aedes aegypti (Diptera: Culicidae). Can J Genet Cytol. 1966;8(2):192–198. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman CJ, , CorfasRA, , MatthewsBJ, , RitchieSA, , Vosshall LB.. Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell. 2014;156(5):1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E.. CHOPCHOP: a CRISPR/Cas9 and talen web tool for genome editing. Nucleic Acids Res. 2014;42(Web Server issue):W401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Escalante L, Zhao C, Shukle R, Stuart J.. BSA-Seq discovery and functional analysis of candidate hessian fly (Mayetiola destructor) avirulence genes. Front Plant Sci. 2020;11:956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osanai-Futahashi M, Tatematsu KI, Futahashi R, Narukawa J, Takasu Y, Kayukawa T, Shinoda T, Ishige T, Yajima S, Tamura T, et al. Positional cloning of a Bombyx pink-eyed white egg locus reveals the major role of cardinal in ommochrome synthesis. Heredity (Edinb). 2016;116(2):135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papathanos PA, Bourtzis K, Tripet F, Bossin H, Virginio JF, Capurro ML, Pedrosa MC, Guindo A, Sylla L, Coulibaly MB, et al. A perspective on the need and current status of efficient sex separation methods for mosquito genetic control. Parasit Vectors. 2018;11(Suppl 2):654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raji JI, Melo N, Castillo JS, Gonzalez S, Saldana V, Stensmyr MC, DeGennaro M.. Aedes aegypti mosquitoes detect acidic volatiles found in human odor using the IR8a pathway. Curr Biol. 2019;29(8):1253–1262. e1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AS. Genetic sexing in Anopheles stephensi using dieldrin resistance. J Am Mosq Control Assoc. 1986;2(1):93–95. [PubMed] [Google Scholar]
- Schneeberger K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat Rev Genet. 2014;15(10):662–676. [DOI] [PubMed] [Google Scholar]
- Shankar S, Tauxe GM, Spikol ED, Li M, Akbari OS, Giraldo D, McMeniman CJ. Synergistic coding of human odorants in the mosquito brain. bioRxiv, 2020.
- Stapley J, Feulner PGD, Johnston SE, Santure AW, Smadja CM.. Recombination: the good, the bad and the variable. Philos Trans R Soc Lond B: Biol Sci. 2017a;372(1736):20170279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Feulner PGD, Johnston SE, Santure AW, Smadja CM.. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philos Trans R Soc Lond B: Biol Sci. 2017b;372(1736):20160455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. Tagmentation-based mapping (tagmap) of mobile DNA genomic insertion sites. bioRxiv 037762, 2017. https://doi.org/10.1101/037762.
- Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, et al. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013;74(1):174–183. [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 10 11–11 10 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CM, Aumann RA, Whitehead MA, Nikolouli K, Leveque G, Gouvi G, Fung E, Reiling SJ, Djambazian H, Hughes MA, et al. White pupae phenotype of tephritids is caused by parallel mutations of a MFS transporter. Nat Commun. 2021;12(1):491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfert L, Gadau J, Schmid-Hempel P.. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity (Edinb). 2007;98(4):189–197. [DOI] [PubMed] [Google Scholar]
- Xu X, Harvey-Samuel T, Yang J, Alphey L, You M.. Ommochrome pathway genes kynurenine 3-hydroxylase and cardinal participate in eye pigmentation in Plutella xylostella. BMC Mol Cell Biol. 2020;21(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Benedict MQ, Malcolm CA, Oliva CF, Soliban SM, Gilles JR.. Genetic sex separation of the malaria vector, Anopheles arabiensis, by exposing eggs to dieldrin. Malar J. 2012;11:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Vreysen MJ, Bourtzis K, Tschirk W, Chadee DD, Gilles JR.. The Anopheles arabiensis genetic sexing strain ANO IPCL1 and its application potential for the sterile insect technique in integrated vector management programmes. Acta Trop. 2015;142:138–144. [DOI] [PubMed] [Google Scholar]
- Zhan Y, San Alberto DA, Rusch C, Riffell JA, Montell C.. Elimination of vision-guided target attraction in Aedes aegypti using CRISPR. Curr Biol. 2021;31(18):4180–4187.e4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lin Y, Shen G, Tan X, Lei C, Long W, Liu H, Zhang Y, Xu Y, Wu J, et al. Pigmentary analysis of eggs of the silkworm Bombyx mori. J Insect Physiol. 2017;101:142–150. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tian D, McBride CS.. Development of a pan-neuronal genetic driver in Aedes aegypti mosquitoes. Cell Rep Methods. 2021;1(3):100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zung JL, Hinze A, Kriete AL, Iqbal A, Younger MA, Matthews BJ, Merhof D, Thiberge S, Ignell R, et al. Mosquito brains encode unique features of human odour to drive host seeking. Nature. 2022;605(7911):706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhang D, Li Y, Yang C, Wu Y, Liang X, Liang Y, Pan X, Hu L, Sun Q, et al. Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature. 2019;572(7767):56–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. Original image files are available at FigShare: https://doi.org/10.25386/genetics.20098100. Illumina and Oxford Nanopore sequencing data are deposited and available at https://www.ncbi.nlm.nih.gov/sra/PRJNA718905.
Supplemental material available at GENETICS online.