Abstract
The NuA4 lysine acetyltransferase complex acetylates histone and nonhistone proteins and functions in transcription regulation, cell cycle progression, and DNA repair. NuA4 harbors an interesting duality in that its catalytic module can function independently and distinctly as picNuA4. At the molecular level, picNuA4 anchors to its bigger brother via physical interactions between the C-terminus of Epl1 and the HSA domain of Eaf1, the NuA4 central scaffolding subunit. This is reflected at the regulatory level, as picNuA4 can be liberated genetically from NuA4 by disrupting the Epl1−Eaf1 interaction. As such, removal of either Eaf1 or the Epl1 C-terminus offers a unique opportunity to elucidate the contributions of Eaf1 and Epl1 to NuA4 biology and in turn their roles in balancing picNuA4 and NuA4 activities. Using high-throughput genetic and gene expression profiling, and targeted functional assays to compare eaf1Δ and epl1-CΔ mutants, we found that EAF1 and EPL1 had both overlapping and distinct roles. Strikingly, loss of EAF1 or its HSA domain led to a significant decrease in the amount of picNuA4, while loss of the Epl1 C-terminus increased picNuA4 levels, suggesting starkly opposing effects on picNuA4 regulation. The eaf1Δ epl1-CΔ double mutants resembled the epl1-CΔ single mutants, indicating that Eaf1’s role in picNuA4 regulation depended on the Epl1 C-terminus. Key aspects of this regulation were evolutionarily conserved, as truncating an Epl1 homolog in human cells increased the levels of other picNuA4 subunits. Our findings suggested a model in which distinct aspects of the Epl1−Eaf1 interaction regulated picNuA4 amount and activity.
Keywords: histone acetylase, histone acetylation, chromatin, yeast genetics, gene regulation
Introduction
Chromatin structure facilitates DNA compaction inside the cell nucleus and contributes to the regulation of nuclear processes like transcription, DNA repair, and DNA replication (Ehrenhofer-Murray 2004; Rando and Chang 2009). The basic building block of chromatin is the nucleosome, which consists of 146 bp of DNA, wrapped around a histone protein octamer (Luger et al. 1997). Chromatin structure is dynamic, modulated by a variety of mechanisms including histone variants, ATP-dependent chromatin remodeling, and the posttranslational addition of chemical groups on to the histones. An astounding number of diverse chemical modifications have been detected on histone proteins to date, of which acetylation, methylation, and phosphorylation are among the best characterized (Bannister and Kouzarides 2011; Huang et al. 2014).
Histone acetylation is most commonly associated with active transcription and is regulated by the combined activity of lysine acetyltransferases (KATs) that deposit the modification, and lysine deacetylases that remove it (Shahbazian and Grunstein 2007; Steunou et al. 2014). Several of these KAT complexes exist in the budding yeast Saccharomyces cerevisiae, but only one is required for viability under normal growth conditions, the NuA4 (nucleosome acetyltransferase of H4) complex (Smith et al. 1998; Clarke et al. 1999). Among histones, NuA4 acetylates the N-terminal tails of H4, H2A, and the H2A variant H2A.Z (Doyon and Côté 2004; Babiarz et al. 2006; Keogh et al. 2006). NuA4 can also acetylate nonhistone proteins, including, but not limited to, several of its resident subunits (Lin et al. 2008; 2009; Lu et al. 2011; Mitchell et al. 2011, 2013; Downey et al. 2015). Functionally, while NuA4 has a major role in regulating the expression of ribosomal protein (RP) genes (Reid et al. 2000; Rossetto et al. 2014), it is also closely linked to cell cycle progression, DNA damage tolerance and repair, and the establishment of heterochromatin–euchromatin boundaries at subtelomeric regions (Doyon and Côté 2004; Zhang et al. 2004; Downs et al. 2004; Babiarz et al. 2006; Zhou et al. 2011; Renaud-Young et al. 2015; Cheng et al. 2018).
In its entirety, the NuA4 complex consists of 13 subunits, encoded by a mixture of essential and nonessential genes, and structurally arranged in a modular fashion (Auger et al. 2008). Several subunits are key to NuA4’s core biochemical function and composition. Chief among them is Esa1/Kat5, the catalytic subunit encoded by an essential gene (Smith et al. 1998). Esa1/Kat5 partners with Epl1, Yng2, and Eaf6 to form the catalytic submodule of NuA4. An important feature of the catalytic submodule is the evolutionarily conserved EpcA (Enhancer of Polycomb A) domain found at the N-terminus of Epl1, which interacts with Esa1/Kat5 and the other subunits to coordinate submodule assembly (Boudreault et al. 2003; Selleck et al. 2005; Xu et al. 2016). More broadly, the components of the NuA4 catalytic submodule collectively support Esa1/Kat5 activity, providing structural stability, orienting Esa1/Kat5, and mediating its interaction with the nucleosome substrate (Boudreault et al. 2003; Selleck et al. 2005; Chittuluru et al. 2011; Xu et al. 2016; Steunou et al. 2016; Searle et al. 2017). In addition to being integral to NuA4 catalytic submodule composition, Epl1 anchors this entity to the rest of NuA4. Specifically, through residues located in its C-terminal half, Epl1 interacts with Eaf1, a protein that serves as the scaffold for NuA4 assembly. This interaction involves regions located in the Eaf1 N-terminus, including its HSA domain and a highly charged region adjacent to it (Boudreault et al. 2003; Auger et al. 2008; Setiaputra et al. 2018; Wang et al. 2018). Beyond the catalytic module, the remaining NuA4 subunits form 3 additional submodules that are also anchored to the Eaf1 scaffold (Auger et al. 2008; Szerlong et al. 2008; Rossetto et al. 2014; Setiaputra et al. 2018; Wang et al. 2018). The function of each of these submodules in the context of NuA4 biology is less well understood but include auxiliary roles in chromatin binding and recruitment of NuA4 to specific genomic locations (Brown et al. 2001; Lu et al. 2009; Zhou et al. 2010; Rossetto et al. 2014; Sathianathan et al. 2016; Steunou et al. 2016; Klein et al. 2018; Gómez-Zambrano et al. 2018).
The established role of Eaf1 as a central scaffold for NuA4 submodules presents an interesting conundrum. Specifically, contrary to Esa1/Kat5, Eaf1 is encoded by a nonessential gene (Auger et al. 2008). At the same time, cells lacking EAF1 have reduced tetra-acetylated H4 levels and are sensitive to a wide range of genotoxic agents, much like strains defective in Esa1/Kat5 function (Kobor et al. 2004; Krogan et al. 2004). In its most basic interpretation, this suggests that some level of Esa1/Kat5 activity must remain in eaf1Δ mutants despite the integrity of the entire NuA4 complex being greatly compromised (Auger et al. 2008; Mitchell et al. 2008). One explanation for this seemingly counterintuitive finding is the fact that the NuA4 catalytic submodule can actually exist and function on its own as a smaller complex called piccolo NuA4 (picNuA4) (Boudreault et al. 2003). It is intriguing that at the subunit composition level, picNuA4 is identical to the catalytic submodule of NuA4. At the same time, these complexes have distinct roles within the cell. For instance, picNuA4 catalyzes nontargeted global histone acetylation in vivo, while NuA4 performs locus-specific histone acetylation. Furthermore, picNuA4 preferentially acetylates nucleosomes over free histones in vitro, while NuA4 acetylates both to the same extent (Reid et al. 2000; Boudreault et al. 2003; Nourani et al. 2004; Selleck et al. 2005; Friis et al. 2009; Uprety et al. 2015).
Adding to the complexity of the relationship between NuA4 and its smaller cousin, removal of either Eaf1 or the Epl1 C-terminus via genetic means liberates picNuA4 from the rest of the NuA4 complex (Boudreault et al. 2003; Auger et al. 2008; Mitchell et al. 2008). While this has been clearly established, a remaining key question is whether these perturbations result in similar levels of picNuA4 activity and NuA4-related defects, respectively. Here, we address this question by directly comparing the effects of losing the entire Eaf1 protein vs. the Epl1 C-terminus using a variety of approaches. While large-scale genetic and gene expression analyses revealed commonalities between the eaf1Δ and epl1-CΔ mutants, these also highlighted clear differences, indicating unique contributions to picNuA4 and NuA4 function. In addition, both the eaf1Δ and eaf1 HSAΔ mutant had more severe defects than the epl1-CΔ mutants across a spectrum of analyses. Perhaps most strikingly, several Eaf1 associated defects were partially rescued by removal of the Epl1 C-terminus. These included growth phenotypes and bulk H4 and H2A.Z acetylation, as well as substantial effects relating to the abundance of picNuA4, which were decreased in the eaf1Δ mutant but robustly increased upon truncation of the Epl1 C-terminus, regardless of EAF1 status. This activity was likely evolutionarily conserved in humans, as truncating a known Epl1 homolog resulted in similar increases to the levels of other picNuA4 subunits. Taken together, this work suggested distinct roles for Eaf1 and Epl1 in regulating the assembly, function, and most likely the balance of NuA4 and picNuA4 levels in the cell, an effect mediated at least in part through an intricate interplay between the Epl1 C-terminus and the HSA domain of Eaf1.
Materials and methods
Yeast strains, plasmids, and yeast techniques
All strains used in this study were generated using standard genetic techniques (Ausubel 1987) and are listed in Table 1. Strains for Epistatic Mini-Array Profiling (E-MAP) and mRNA expression profiling were derived from BY4741 as per standard procedure for those methods, while all other experiments used strains created in the W303 background. Complete gene deletions, EPL1 truncations, and 3′ end integration of in-frame epitope tags [HA, tandem affinity purification (TAP), 3xFLAG, or eGFP] were achieved using the 1-step gene integration of PCR-amplified modules method (Longtine et al. 1998; Gelbart et al. 2001). Integrated modules were generated using the KAPA HiFi HotStart DNA Polymerase following manufacturer protocol (Roche). All double mutant strains were generated by mating, sporulation, and tetrad dissection.
Table 1.
Yeast strains used in this study.
| Strain number | Relevant genotype |
|---|---|
| MKY583 | BY4741, MATα, his3Δ1 leu2Δ0 LYS2 met15Δ0 ura3Δ0 Δcan1::MATαPr-HIS3 Δlyp1::MATαPr-LEU2 |
| MKY1419 | MKY583, eaf1 1-982-3xHA::NAT |
| MKY1428 | MKY583, eaf1Δ-3xHA::NAT |
| MKY1429 | MKY583, EPL1-3xFLAG::KAN |
| MKY1430 | MKY583, epl1(1-485)-3xFLAG::KAN |
| MKY1431 | MKY583, epl1(1-380)-3xFLAG::KAN |
| MKY1432 | BY4742, MATa his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0 |
| MKY1433 | MKY1432, eaf1 1-982-3xHA::NAT |
| MKY1442 | MKY1432, eaf1Δ-3xHA::NAT |
| MKY1443 | MKY1432, EPL1-3xFLAG::KAN |
| MKY1444 | MKY1432, epl1(1-485)-3xFLAG::KAN |
| MKY1445 | MKY1432, epl1(1-380)-3xFLAG::KAN |
| MKY6 | W303, MATA, ADE2, can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 lys2Δ |
| MKY1762 | MKY6, EPL1-TAP::TRP |
| MKY1763 | MKY6, EPL1-TAP::TRP, eaf1::HIS |
| MKY1764 | MKY6, epl1(1-485)-TAP::TRP |
| MKY1765 | MKY6, epl1(1-485)-TAP::TRP, eaf1::HIS |
| MKY1766 | MKY6, epl1(1-380)-TAP::TRP |
| MKY1767 | MKY6, epl1(1-380)-TAP::TRP, eaf1::HIS |
| MKY1768 | MKY6, pRS316 |
| MKY1769 | MKY6, EPL1-TAP::TRP, [pRS316] |
| MKY1770 | MKY6, EPL1-TAP::TRP, eaf1::HIS [pRS316] |
| MKY1771 | MKY6, EPL1-TAP::TRP, eaf1::HIS [pRS316, EAF1] |
| MKY1772 | MKY6, EPL1-TAP::TRP, eaf1::HIS [pRS316, eaf1-HSAΔ] |
| MKY1773 | MKY6, EPL1-TAP::TRP, eaf1::HIS [pRS316, eaf1-SANTΔ] |
| MKY1774 | MKY6, EPL1-TAP::TRP, eaf1::HIS [pRS316, eaf1-HSAΔSANTΔ] |
| MKY1775 | MKY6, epl1(1-485)-TAP::TRP, eaf1::HIS [pRS316] |
| MKY1776 | MKY6, epl1(1-485)-TAP::TRP, eaf1::HIS [pRS316, EAF1] |
| MKY1777 | MKY6, epl1(1-485)-TAP::TRP, eaf1::HIS [pRS316, eaf1-HSAΔ] |
| MKY1778 | MKY6, epl1(1-485)-TAP::TRP, eaf1::HIS [pRS316, eaf1-SANTΔ] |
| MKY1779 | MKY6, epl1(1-485)-TAP::TRP, eaf1::HIS [pRS316, eaf1-HSAΔSANTΔ] |
| MKY2048 | MKY6, EPL1-3xFLAG::KAN |
| MKY2049 | MKY6, EPL1-3xFLAG::KAN, eaf1::HIS |
| MKY2050 | MKY6, epl1(1-485)-3xFLAG::KAN |
| MKY2051 | MKY6, epl1(1-380)-3xFLAG::KAN |
| MKY7 | W303, MATA, ade2-1, can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 |
| DSY191 | MKY7, epl1(1-380)-GFP::URA |
| DSY192 | MKY7, epl1(1-485)-GFP::URA |
| DSY197 | MKY7, EPL1-GFP::URA |
| MKY399 | W303, MATα, ADE2, can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 |
| MKY2097 | MKY399, epl1::HIS [pRS315, 3xFLAG-EPL1] |
| MKY2098 | MKY399, epl1::HIS [pRS315, 3xFLAG-epl1K648R] |
| MKY2099 | MKY399, epl1::HIS, eaf1::KAN [pRS315, 3xFLAG-EPL1] |
| MKY2100 | MKY399, epl1::HIS, eaf1::KAN [pRS315, 3xFLAG-epl1K648R] |
The EPL1 and EAF1 genes were PCR amplified from genomic DNA using the KAPA HiFi HotStart DNA Polymerase (Roche) and cloned into the pRS315 (LEU2) and pRS316 (URA3) centromeric vectors, respectively. The epl1-K648R point mutant and internal deletions of EAF1 were made by adapting the QuikChange site-directed mutagenesis method (Agilent). For recombinant expression in Escherichia coli, pET15-HIS-eaf1-(1-538) was subcloned into pET28a using NdeI and HindIII restriction sites to generate pET28a-HIS-eaf1-(1-538). All plasmids were confirmed by DNA sequencing and are listed in Table 2, and primer sequences used for cloning can be found in Supplementary Table 1.
Table 2.
Yeast plasmids used in this study.
| Name | Description | Source |
|---|---|---|
| pRS316 | URA3 ARS-CEN | |
| pMK570 | URA3 ARS-CEN EAF1, expressed using endogenous promoter | This study |
| pMK571 | URA3 ARS-CEN eaf1-HSAΔ (Δ1042–1254) | This study |
| pMK572 | URA3 ARS-CEN eaf1-SANTΔ (Δ1936–2121) | This study |
| pMK573 | URA3 ARS-CEN eaf1-HSAΔSANTΔ (Δ1042–1254, 1936–2121) | This study |
| pMK664 | pET28a HIS-eaf1-(1-538) | This study |
| pRS315 | LEU2 ARS-CEN | |
| pMK701 | LEU2 ARS-CEN 3xFLAG-EPL1 | This study |
| pMK702 | LEU2 ARS-CEN 3xFLAG-epl1-K648R | This study |
Immunoblotting
To determine bulk protein and histone acetylation levels, whole cell extracts were prepared using glass bead lysis in the presence of trichloroacetic acid (Foiani et al. 1994) and analyzed by immunoblotting. Antibodies against H4 (Abcam), tetra-acetylated H4 (Upstate), H4K5ac (Millipore), H4K8ac (Abcam), H4K12ac (Active Motif), H4K16ac (Millipore), H2A.Z (Active Motif), H2A.Z K14ac (Upstate), rabbit IgG (Millipore), FLAG (Sigma), and Pgk1 (Life Technologies) were purchased from the indicated vendors while antibodies against Esa1/Kat5 and Eaf1 were described previously (Allard et al. 1999; Auger et al. 2008). Immunoblots were scanned with the Odyssey Infrared Imaging System (Licor) and quantified using Image Studio (Licor).
E-MAP
E-MAP assays were performed as described previously (Schuldiner et al. 2006). Using a Singer robot, EAF1 and EPL1 alleles were crossed to a library of 1,536 mutants representing genes involved in transcription, RNA processing, and chromatin biology. All strains were screened in triplicate and scores were calculated as previously described (Collins et al. 2006; Schuldiner et al. 2006). The full E-MAP profiles generated and used for our specific analyses can be found in Supplementary File 1. Genetic interactions with S scores greater than 2 or less than −2.5 were deemed significant.
mRNA expression profiling
Gene expression profiling was performed as described previously (van de Peppel et al. 2003). EAF1 and EPL1 alleles were processed 4 times from 2 independently inoculated cultures. Dual-channel 70-mer oligonucleotide arrays were used with a common reference wild-type RNA. After RNA isolation, all steps were operated using robotic liquid handlers. Scores were calculated as previously described (van de Peppel et al. 2003; van Bakel and Holstege 2004). Differentially expressed genes were determined using a P-value of <0.01 and a minimum fold change of >1.7 compared to wild type (van Wageningen et al. 2010). Database for Annotation, Visualization, and Integrated Discovery (DAVID) was used for Gene Ontology (GO) enrichment analysis using the list of genes present in the array as the background, and the Benjamini–Hochberg method for multiple testing correction (Huang et al. 2009a,b).
Growth and genotoxic sensitivity assays
Overnight cultures grown in YPD were diluted to OD600 0.5. Cells were 10-fold serially diluted and spotted onto solid YPD plates or plates with 0.005% methyl-methanesulfonate (MMS), 50 mM hydroxyurea (HU), or 1% formamide. For strains containing URA3 plasmids, the cultures were grown in SC-URA media overnight and spotted onto SC-URA plates with or without the indicated genotoxic agents. Plates were incubated at the indicated temperature for 3 days.
RT-quantitative PCR
Yeast cultures were grown in YPD to an OD600 of 0.5. RNA was extracted and purified using the Qiagen Rneasy Mini Kit as per manufacturer protocol. cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen). cDNA was analyzed using a Rotor-Gene 6000 (Qiagen) and PerfeCTa SYBR green FastMix (Quanta Biosciences). mRNA levels were normalized to TUB1 mRNA levels. Samples were analyzed in triplicates from 3 independent RNA preparations. Primer sequences are listed in Supplementary Table 1.
Chromatin immunoprecipitation-quantitative PCR
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously (Schulze et al. 2009). Briefly, yeast cells (500 ml for histone ChIP and 250 ml for Epl1-FLAG ChIP) were grown in YPD to an OD600 of 0.5–0.6 and crosslinked with 1% formaldehyde for 20 min before chromatin extraction. Chromatin was sonicated (Bioruptor, Diagenode; 10 cycles, 30 s on/off, high setting) to yield an average DNA fragment of 500 bp. Anti-tetra-acetylated H4 (Upstate), or anti-FLAG (Sigma) antibodies coupled to 60 μl of protein A magnetic beads (Invitrogen) were incubated with each sample. After reversal of crosslinking and DNA purification, the immunoprecipitated and input DNA were analyzed by quantitative PCR (qPCR). Samples were analyzed in triplicate from 3 independent ChIP experiments. H4 acetylation was normalized to % input (0.7%), while Epl1-FLAG occupancy was normalized to an intergenic region of Chromosome V (Keogh and Buratowski 2004). Primers used are listed in Supplementary Table 1.
Chromatin association assay
Chromatin association assays were performed as previously described (Wang et al. 2009). Cells were diluted to OD600 0.15 and collected at logarithmic phase. Cells were resuspended in prespheroplast buffer [100 mM PIPES/KOH (pH 9.4), 10 mM DTT, 0.1% sodium azide] and incubated with rotation for 10 min at room temperature, followed by incubation with 20 mg/ml Zymoylase-100T (Seikagaku Corporation) for 20 min at 37°C. The resulting spheroplasts were washed with wash buffer [50 mM HEPES/KOH (pH 7.5), 100 mM KCl, 2.5 mM MgCl2, 0.4 M sorbitol] and resuspended in an equal volume of EB [50 mM HEPES/KOH (pH 7.5), 100 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 1 mM PMSF, and Complete Protease Inhibitor cocktail; Roche]. Spheroplasts were lysed with 1% Triton X-100. Whole cell extract samples were saved, and the remaining lysate was centrifuged through a sucrose gradient (EB + 0.25% Triton X-100 and 30% sucrose) separating the chromatin bound proteins from the rest (supernatant fraction). All 3 fractions were analyzed by SDS–PAGE and immunoblotting. Rabbit IgG (Millipore) was used to detect Epl1-TAP, and antibodies against Pgk1 (Life Technologies) and H4 (Abcam) were used as controls for the supernatant and chromatin fractions, respectively.
Fluorescence microscopy
eGFP-tagged strains were grown to mid-log phase in SC-URA media. 2.5 μg/ml of DAPI was added to the culture and incubated for 30 min. Live cells were applied to glass slides and visualized using an Olympus Fluoview FV1000 laser scanning confocal microscope.
TAP of yeast NuA4 and picNuA4 complexes
Immunoprecipitation of native protein complexes was performed from 1 l of cultures that were harvested at an OD600 of 1.00. Large-scale purification was adapted from a previously described protocol with minor modifications (Mitchell et al. 2008). Cells were lysed using a KRUPS coffee grinder with dry ice pellets and subsequently resuspended in TAP buffer [20 mM HEPES (pH 8), 350 mM NaCl, 10% glycerol, 0.1% Tween-20, 1× phosphatase inhibitor mix, and Complete Protease Inhibitor cocktail]. NP-40 was added to a final concentration of 1% prior to centrifugation at 3,000 × g for 10 min at 4°C. Crude extracts were incubated for 3 h at 4°C with 200 µl of IgG (Millipore) crosslinked to M-270 Epoxy beads (Invitrogen). Beads were washed 3 times with 4 ml of TAP buffer. Protein complexes were eluted in 30 µl of 0.1 M citrate (pH 3.1), loaded into a 4–20% gradient gel (Bio-Rad), and silver stained or used for immunoblotting as described above.
In vitro Epl1 and Eaf1 small-scale interaction assays
Binary interaction assays using recombinant proteins were done using 0.7 g of T7 Express E. coli (NEB) coexpressing His-Eaf1(1-538) and either GST, GST-Epl1(1-485), or GST-Epl1(486-833). Cells were lysed by sonication in lysis buffer [50 mM Tris (pH 7.0), 150 mM NaCl, 0.1% Triton X-100, 5% glycerol, 0.5 mM DTT, and 2 mM PMSF]. Lysates were cleared by centrifugation at 23,600 × g for 30 min and the supernatant was incubated with 150 μl of glutathione agarose resin (Thermo Fisher) for 1 h. The resin was washed with 1 ml of wash buffer (Lysis buffer without PMSF and DTT). Protein was eluted from the beads through boiling in SDS sample buffer.
Purification of human picNuA4 complexes
pRevCMV-3FLAG and pCDNA3-HA-based vectors for expression in mammalian cells were constructed using standard procedures. Purification of human picNuA4 complexes from transient transfections was performed in HEK293T cells. Cells were transfected near confluency by the calcium phosphate method with 5 µg of each plasmid (pRevCMV for 3FLAG-EPC1 constructs/truncations and pCDNA3 for HA-Tip60/Kat5, HA-ING3, and HA-MEAF6) per 150-mm plate. Transfection efficiency was monitored with a GFP-expressing vector. Cells were harvested 48 h posttransfection, and whole cell extracts were prepared followed by anti-FLAG immunoprecipitation and elution as previously described (Avvakumov et al. 2012; Lalonde et al. 2013). Proteins representing various NuA4 subunits were detected via immunoblotting using the following antibodies: anti-Tip60 (Santa Cruz), anti-ING3 (Abcam), anti-MEAF6 (Abcam), anti-FLAG M2-HRP (Sigma), and anti-Brd8 (Bethyl).
Results
High-throughput genetic interaction and gene expression analysis of eaf1Δ and epl1-CΔ mutants revealed functional differences
The interaction between Eaf1 and the Epl1 C-terminus anchors picNuA4 to the rest of the NuA4 complex (Boudreault et al. 2003; Auger et al. 2008; Mitchell et al. 2008). As such, genetic manipulations that remove either Eaf1 or the Epl1 C-terminus and thus decouple picNuA4 from NuA4 have been widely used to examine the function and regulation of picNuA4. However, whether these manipulations result in similar consequences to NuA4 and picNuA4 activity remains unclear. Thus, to understand how Eaf1 and Epl1 contribute to cellular function and the biology of NuA4, we first analyzed eaf1Δ and EPL1 C-terminal truncation mutants using a combination of E-MAP and gene expression microarrays. Here, we included 2 previously described epl1-CΔ mutants (Boudreault et al. 2003): epl1(1-485) and epl1(1-380), which remove various portions of the Epl1 C-terminus without disrupting the N-terminal region and the conserved EpcA domain important for picNuA4 assembly (Boudreault et al. 2003; Selleck et al. 2005; Fig. 1a). Reassuringly, both Epl1 truncations also recapitulated the cellular localization of the full length Epl1 protein, being found primarily in the cell nucleus (Supplementary Fig. 1).
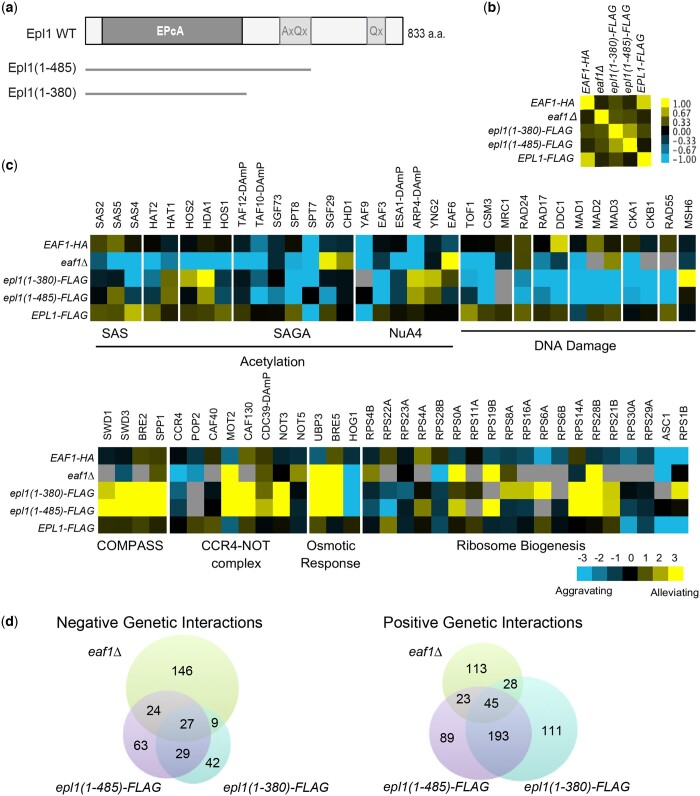
Fig. 1.
Genetic interaction analysis revealed similarities and distinctions between eaf1 and epl1 mutants. a) Schematic representation of the epl1-CΔ mutations. The conserved EpcA domain is shown, along with the alanine and glutamine rich regions. b) Spearman’s rho correlation of genetic interaction profiles of wild type and the indicated mutant strains. Yellow and blue indicate positive and negative correlations respectively. c) Representative genetic interactions for the eaf1Δ and epl1-CΔ mutants with genes encoding subunits of the indicated complexes. Blue indicates aggravating interactions, yellow represents alleviating interactions, and gray denotes missing data. d) Venn diagrams showing significant negative and positive genetic interactions between eaf1Δ, epl1(1-485), and epl1(1-380).
We used E-MAP to generate high-throughput genetic interaction profiles for the eaf1Δ and epl1-CΔ mutants to compare the roles of EAF1 and EPL1 in cellular function. Consistent with both Eaf1 and Epl1 being components of the NuA4 complex, their overall genetic interaction profiles were positively correlated, although we note that the 2 epl1-CΔ mutants had stronger correlations to one another than to the eaf1Δ mutant (Fig. 1b). At a more granular level, focusing on genes and complexes broadly related to NuA4 biology, at least 3 different categories of genetic interactions could be clearly visualized (Fig. 1c). First, we identified genes that had the same general patterns of genetic interactions between the eaf1Δ and the 2 epl1-CΔ mutants, whether they were positive or negative. This category included the osmotic stress response genes BRE5, UBP3, and HOG1 (Solé et al. 2011), genes encoding subunits of the CCR4-NOT transcription regulatory complex, and genes involved in the DNA damage response. A second set of genes had distinct patterns between the mutants, especially when we compared eaf1Δ to the 2 epl1-CΔ mutants. The most obvious examples of this set included genes encoding subunits of the COMPASS histone methyltransferase complex and subunits of various histone deacetylases (HOS2, HDA1), while genes encoding the SAS, HAT2 and SAGA complexes showed a similar, but more nuanced trend. The third, and perhaps most ambiguous category were complexes where the type of interaction with our specific mutants varied depending on individual genes of a complex or process. This latter category included NuA4 itself, as well genes involved in ribosome biogenesis. A more quantitative analysis of our genetic interaction profiles confirmed these general gene and complex-focused trends (Fig. 1d). Specifically, while there were a substantial number of shared positive and negative interactions between eaf1Δ, and the 2 epl1-CΔ mutants, there were also at least as many unique interactions especially when we contrasted eaf1Δ to the 2 epl1-CΔ mutants combined. We do note that despite encoding different versions of the same gene, the 2 epl1-CΔ mutants each had a fair number of allele-specific genetic interactions, both when compared to the eaf1Δ mutant but also among themselves (Fig. 1d).
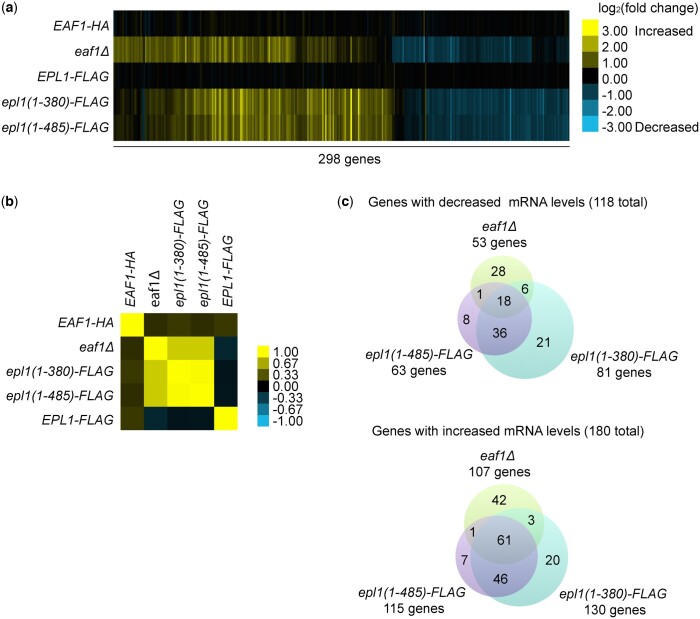
Consistent with the genetic interaction findings, the gene expression profiles of the eaf1Δ and epl1-CΔ mutants were also highly correlated, while at the same time displaying clear differences. Again, we found that the 2 epl1-CΔ mutants were more similar to one another than to the eaf1Δ mutant (Fig. 2, a and b) even though the shorter epl1(1-380) mutant had more significant (fold change ≥1.7 and P ≤ 0.01) gene expression alterations compared to the longer epl1(1-485) mutant (Fig. 2c). Overall, a total of 118 genes showed significantly decreased mRNA levels in the eaf1Δ and epl1-CΔ mutants compared to wild type, an effect consistent with NuA4 being involved in gene activation (Doyon and Côté 2004) (Fig. 2c). However, we note that a total of 180 genes showed significantly increased mRNA levels in the mutants, indicating possible repressive or indirect functions for NuA4. Collectively, 18 downregulated and 61 upregulated genes changed significantly in all 3 mutants (Fig. 2c). The latter were involved in “response to temperature stimulus” (P-value = 3.00E−19) and metabolic processes like “carbohydrate catabolic process” (P-value = 7.30E−05), “glycoside metabolic process” (P-value = 1.60E−04), and “protein catabolic process” (P-value = 2.7E−04), while the former had no significantly enriched terms via GO enrichment analysis. However, we also identified unique enrichments for the eaf1Δ and epl1-CΔ mutants; genes upregulated in the eaf1Δ mutant only were enriched for pathways involved in “transposition” (P-value = 4.40E−03) and “cell death” (P-value = 3.50E−03), while genes uniquely downregulated had no significantly enriched pathways. Although several genes were uniquely affected in the epl1-CΔ mutants, these were not enriched for any GO categories. Taken together, the high-throughput genetic and gene expression profiling data suggested both shared and unique roles for EAF1 and EPL1 in cellular processes and in the biology of NuA4.
Fig. 2.
Gene expression profiles supported shared and unique roles for EAF1 and EPL1. a) Heat map showing the log2 fold changes for 298 genes with significant changes in expression in at least one of the eaf1Δ or epl1-CΔ mutants. Absolute fold changes greater than 1.7 (corresponding to values greater than 0.77 or less than −0.77 on the heatmap) were considered significant. Yellow indicates upregulated genes, and blue represents downregulated genes. b) Spearman’s rho correlation of the gene expression profile of wild type and mutant strains. c) Venn diagrams showing significantly upregulated and downregulated genes in eaf1Δ, epl1(1-485), and epl1(1-380).
The eaf1Δ and epl1-CΔ mutants differed in their impact on cell growth and bulk histone H4 and H2A.Z acetylation
Given the differences observed at the level of genetic interaction and gene expression profiles, we next sought to determine how the eaf1Δ and epl1-CΔ mutants related to one another in the context of previously described NuA4-associated phenotypes. To this end, we generated eaf1Δ epl1(1-380) and eaf1Δ epl1(1-485) double mutants via mating and tetrad dissection and examined the effect of the eaf1Δ and epl1-CΔ single and double mutants on cell growth, bulk H4 and H2A.Z acetylation, and promoter H4 acetylation and expression of representative RP genes. Consistent with previous reports (Boudreault et al. 2003; Auger et al. 2008), the eaf1Δ and epl1-CΔ mutants had severe growth defects when exposed to MMS, HU, and formamide (Fig. 3a). While the eaf1Δ and epl1(1-380) mutants had similar growth defects, the longer epl1(1-485) mutant had less severe phenotypes compared to both the eaf1Δ and epl1(1-380) mutants, most clearly seen upon MMS exposure. We note that under these conditions, the eaf1Δ epl1(1-485) double mutant mirrored the epl1(1-485) mutant phenotype, suggesting that the effect of EAF1 was dependent on the presence of the Epl1 C-terminus.
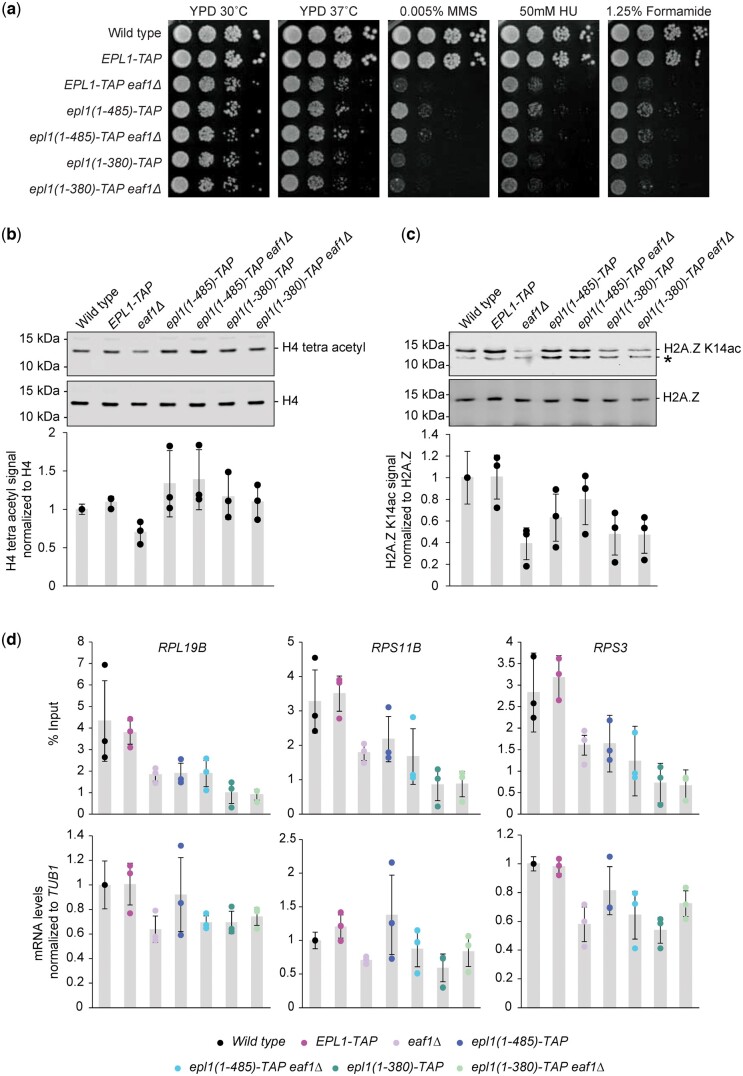
Fig. 3.
Epl1 C-terminus truncation masked some of the effects of deleting EAF1. a) Logarithmic serial dilutions of the indicated strains were plated on YPD media, with or without genotoxic agents, and incubated for 3 days at the indicated temperature. The eaf1Δ and epl1-CΔ mutants showed decreased growth in the presence of the genotoxic agents. The eaf1Δ epl1(1-485) mutant showed milder growth defects under MMS compared to the eaf1Δ single mutant. Bulk histone acetylation was examined from whole cell extracts of the indicated strains, analyzed by protein blotting with (b) anti-tetra-acetylated H4 and (c) anti-H2A.Z K14ac antibodies. Antibodies against H4 and H2A.Z were used as loading controls. * denotes a background band in the H2A.Z K14ac immunoblot; we note that it fluctuates for unknown reasons in the epl1-CΔ mutants. The tetra-acetylated H4 and H2A.Z K14ac signals were quantified relative to the H4 and H2A.Z signals, respectively, and normalized to the wild-type strain. Averages were taken across 3 replicates, with error bars indicating the standard deviation. d) Disruption of NuA4 function in the eaf1Δ and epl1-CΔ single and double mutants resulted in both decreased mRNA levels and H4 promoter acetylation at RP candidate genes: RPL19B, RPS11B, and RPS3. Enrichment of tetra-acetylated H4 was normalized to % input. mRNA levels were measured by RT-qPCR and normalized to TUB1 mRNA levels. Error bars represent standard deviation of the means for 3 independent experiments.
We next examined the effects of the eaf1Δ and epl1-CΔ single and double mutants on bulk H4 and H2A.Z acetylation. As reported previously (Kobor et al. 2004; Krogan et al. 2004; Babiarz et al. 2006), the eaf1Δ mutant had decreased tetra-acetylated H4 and H2A.Z K14ac levels compared to wild type (Fig. 3, b and c). In contrast, the epl1(1-380) mutant had no appreciable effect on tetra-acetylated H4 but did have decreased H2A.Z K14ac levels, while the longer epl1(1-485) mutant had no effect on either H4 acetylation or H2A.Z K14ac levels. These data thus revealed not only differences to the eaf1Δ mutant but also some nuanced ones between the 2 epl1-CΔ mutants. Perhaps more interestingly, the H2A.Z and H4 acetylation levels in the eaf1Δ epl1-CΔ double mutants resembled those observed in the respective epl1-CΔ single mutants. Specifically, the H4 acetylation defect present in the eaf1Δ single mutant was normalized in both double mutants, while the epl1(1-485) mutant also normalized the H2A.Z acetylation defects. Since previous reports also examined lysine-specific H4 acetylation in eaf1Δ mutants and arrived at contrasting findings (Kobor et al. 2004; Auger et al. 2008), we also measured the levels of H4K5ac, K8ac, K12ac, and K16ac individually (Supplementary Fig. 2). Compared to wild type, the eaf1Δ mutant had very modest decreases in H4K5ac and K12ac and no decrease in K16ac, while wild type levels of acetylation were observed in the epl1-CΔ single and corresponding double mutants across all 3 lysine sites. In the case of H4K8ac, however, we observed a surprising increase in acetylation in the epl1-CΔ mutants, while the eaf1Δ single mutant had wild-type levels.
Given that NuA4 recruitment and subsequent H4 acetylation plays a key role in RP gene regulation (Reid et al. 2000), we next explored whether the differences in bulk H4 tetra-acetylation observed between the eaf1Δ and epl1-CΔ mutants were also associated with specific RP gene promoter acetylation and gene expression patterns. In contrast to the bulk findings, the eaf1Δ and epl1-CΔ mutants had decreased promoter H4 acetylation at 3 representative RP genes: RPL19B, RPS11B, and RPS3 (Fig. 3d), an effect also seen in the eaf1Δ epl1-CΔ double mutants. In addition, the mRNA levels of all 3 genes decreased for the eaf1Δ, epl1(1-380), and eaf1Δ epl1-CΔ mutants in comparison to wild type. In contrast, the epl1(1-485) mutant showed no decrease in mRNA levels. As such, our results indicated that, while loss of the Epl1 C-terminus was sufficient to overcome the global H4 tetra-acetylation defects in an eaf1Δ background, the suppression did not extend to gene loci specifically targeted by the full NuA4 complex.
In addition to the RP genes, we examined promoter acetylation and mRNA levels from 4 additional genes (BIO5, AQR1, PES4, INH1) which showed altered expression in the microarray data in either the eaf1Δ or epl1-CΔ mutants (Fig. 2a). In comparison to wild type, the mRNA levels increased for both PES4 in the epl1-CΔ and eaf1Δ epl1-CΔ mutants, and for INH1 in the epl1(1-380) and eaf1Δ epl1-CΔ mutants (Supplementary Fig. 3a). In contrast to the RP genes, none of the mutants showed differences in H4 acetylation at the promoters of tested loci (BIO5, AQR1, PES4, INH1) (Supplementary Fig. 3b).
The Epl1 C-terminus was required for its association with chromatin
To understand the molecular underpinnings of the functional differences between the eaf1Δ and epl1-CΔ mutants on NuA4 phenotypes, we first tested whether the genetic manipulations affected the association of Epl1 with chromatin. This was a particularly attractive possibility, given that Eaf1 is the nexus between Epl1 and the various NuA4 submodules that contribute to chromatin recruitment and binding (Auger et al. 2008). Consistent with previous studies (Searle et al. 2017), chromatin association of Epl1 was strong in wild-type cells but weak in the epl1-CΔ mutants, an effect that perplexedly was somewhat more pronounced in the epl1(1-485) mutant compared to the epl1(1-380) mutant (Fig. 4a). Unexpectedly, Epl1 chromatin association was unaffected by loss of Eaf1, suggesting that the Epl1 C-terminus likely contributed to Epl1’s anchoring to chromatin, either directly by itself or indirectly through interactions with other NuA4 subunits.
Fig. 4.
The Epl1 C-terminus was required for its interaction with chromatin, independent of EAF1 status. a) Epl1’s association with chromatin was assessed by western blot analysis of cellular fractions generated by centrifugation of whole cell extract over a sucrose gradient. W, whole cell extract; S, soluble nonchromatin fraction; C, chromatin pellet. In each case, the fractionation efficiency was judged by the levels of H4 and Pgk1, found in the chromatin and the supernatant fraction, respectively. b) Epl1 enrichment at the promoters of RP genes was lost in the epl1-CΔ mutants, based on ChIP-qPCR analysis of the representative genes RPL21A, RPL39, RPL17B, RPL19B, RPS11B, and RPS3. Results are depicted as box plots overlaid with the individual measurements from each replicate.
Given that we observed decreased promoter acetylation and mRNA expression defects at RP genes in both the eaf1Δ and epl1-CΔ single mutants, we next tested whether these bulk chromatin association findings were reflective of localization at specific NuA4-regulated loci. Since NuA4 was previously shown to be recruited to the promoters of RP genes (Reid et al. 2000; Rossetto et al. 2014), we performed chromatin immunoprecipitation followed by qPCR (ChIP-qPCR) at several representative RP gene promoters, including the 3 that were examined above. As expected, wild-type Epl1 was enriched at the promoters of all 6 tested RP genes (Fig. 4b). However, and in contrast to the bulk chromatin association assay, Epl1 enrichment at the RP genes was dependent upon Eaf1, as it was lost in the eaf1Δ mutant. Epl1 enrichment was also lost in both epl1-CΔ mutants, findings that collectively suggested that the locus-specific recruitment of picNuA4, as well as RP gene expression and promoter H4 acetylation, required the entire NuA4 complex.
The eaf1Δ and epl1-CΔ mutants had differential effects on the amount of picNuA4 in the cell
Careful examination of the chromatin association assays described above hinted that the eaf1Δ and epl1-CΔ mutants might result in different amounts of Epl1 levels in the cell [compare whole cell extract fractions (W) in Fig. 4a]. To examine this further, Epl1 protein levels were measured from total protein extracts prepared from eaf1Δ and epl1-CΔ single and double mutants (Fig. 5a). Confirming the results described above, the eaf1Δ mutant indeed had decreased levels of Epl1, while the 2 epl1-CΔ mutants resulted in robustly increased Epl1 levels relative to wild type. Again, the eaf1Δ epl1-CΔ double mutants mirrored the respective epl1-CΔ mutant phenotype, suggesting that the effect of EAF1 on Epl1 protein levels was dependent on the Epl1 C-terminus. We note that the changes in Epl1 protein levels likely resulted from posttranscriptional events, as RT-qPCR analysis of EPL1 mRNA levels revealed little difference between the strains (Fig. 5b). In fact, the eaf1Δ mutant showed a slight increase in EPL1 mRNA levels relative to wild type, a somewhat unexpected finding given that the eaf1Δ mutant on its own showed a decrease in Epl1 protein levels.
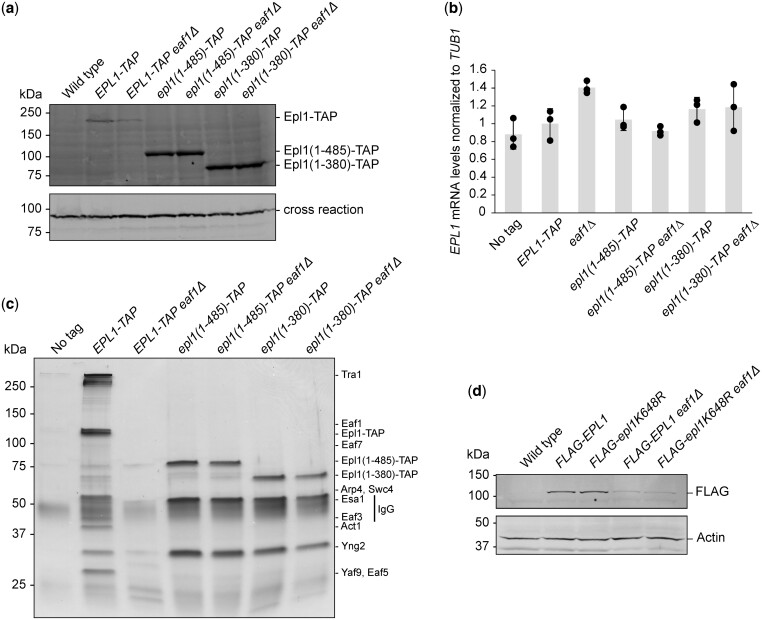
Fig. 5.
Truncating Epl1 stabilized picNuA4 and increased Epl1 protein levels independent of Eaf1 status. a) Epl1 protein levels were strongly decreased in the eaf1Δ mutant but increased in the epl1-CΔ mutants. A cross-reactive band was used as a loading control. b) EPL1 expression was similar across all strains. The mRNA levels of EPL1 was measured by RT-qPCR and normalized to levels of TUB1 mRNA. Error bars represent the standard error of 3 independent biological replicates. c) Immunopurification of NuA4 using Epl1-TAP demonstrated a decrease in the amount of Epl1-associated NuA4 subunits in the eaf1Δ mutant, while only subunits found in picNuA4 were observed in the epl1-CΔ mutants. Purified fractions from indicated strains were loaded onto a 4–20% gradient SDS-PAGE gel and visualized by silver staining. Bands corresponding to NuA4 subunits are indicated on the left. Untagged Epl1 was used as a negative control. d) The Epl1 K648R point mutation did not restore Epl1 protein levels in the eaf1Δ mutant, as measured in whole cell extracts from strains expressing the indicated plasmid-derived FLAG-Epl1 constructs. PGK1 was used as a loading control.
Having documented differences in Epl1 protein levels, we next tested if they resulted in changes to the amount of NuA4 or picNuA4 in the cell. To this end, TAP-tagged versions of Epl1, Epl1(1-485) or Epl1(1-380) were purified from strains with or without deletion of EAF1 and analyzed using SDS-PAGE followed by silver staining. As expected, purification of wild-type Epl1 captured most known subunits of NuA4 (Fig. 5c). Furthermore, and in agreement with previous reports (Auger et al. 2008; Mitchell et al. 2008), loss of EAF1 resulted in a decreased ability to purify NuA4 subunits via Epl1, with many subunits being absent or below the visual detection level employed here. However, we note that the other picNuA4 subunits including Yng2 and Esa1/Kat5 were present in the purification, albeit at much lower levels. In contrast, but still consistent with previous findings (Auger et al. 2008), purification of Epl1(1-485) or Epl1(1-380) resulted in high amounts of copurifying Yng2 and Esa1/Kat5 without significant levels of other NuA4 subunits, indicating the presence of high levels of picNuA4 in the epl1-CΔ mutants. Perhaps most surprisingly given the eaf1Δ mutant phenotypes, removal of Eaf1 had no effect on the biochemical purification of picNuA4 liberated through shortened versions of Epl1, as similar amounts of picNuA4 were obtained from both eaf1Δ epl1-CΔ double mutants as the respective epl1-CΔ single mutants.
These data suggested a role for the Epl1 C-terminus in the regulation of picNuA4, likely coordinated through its interaction with Eaf1. Building upon previously published findings which indicate that Eaf1 binds to Epl1 somewhere between amino acids 381–832 (Setiaputra et al. 2018), we generated recombinant Epl1 proteins based on the epl1(1-485) mutant for directed small scale in vitro binding assays. Using this binary and somewhat reductionist approach, we found that Epl1(1-485) interacted with Eaf1 in vitro, while Epl1(486-832) did not (Supplementary Fig. 4).
We were intrigued by the possibility that Eaf1 might contribute to picNuA4 stability through an indirect mechanism, perhaps by preventing other factors from interacting with the Epl1 C-terminus. As our E-MAP findings suggested a functional connection to the Ubp3/Bre5 ubiquitin protease, we hypothesized that a ubiquitin-mediated degradation pathway could be involved. Consistent with this idea, high-throughput mass spectrometry data indicate that K648 of Epl1, which was removed in both of our epl1-CΔ mutants, might be a site of ubiquitination (Swaney et al. 2013) and is found in close proximity to Eaf1 (Setiaputra et al. 2018). To test whether the ubiquitination of Epl1 K648 was a factor in regulating Epl1 protein abundance, we generated plasmids carrying N-terminally FLAG tagged wild type or Epl1 K648R and introduced them into epl1Δ and epl1Δ eaf1Δ backgrounds. Perhaps not surprising given the exploratory nature of high-throughput mass spectrometry data and the current lack of existing functional data about this potential ubiquitination site of Epl1, the Epl1 K648R mutant did not change the amount of Epl1 in the cell compared to wild type in the absence of Eaf1 (Fig. 5d).
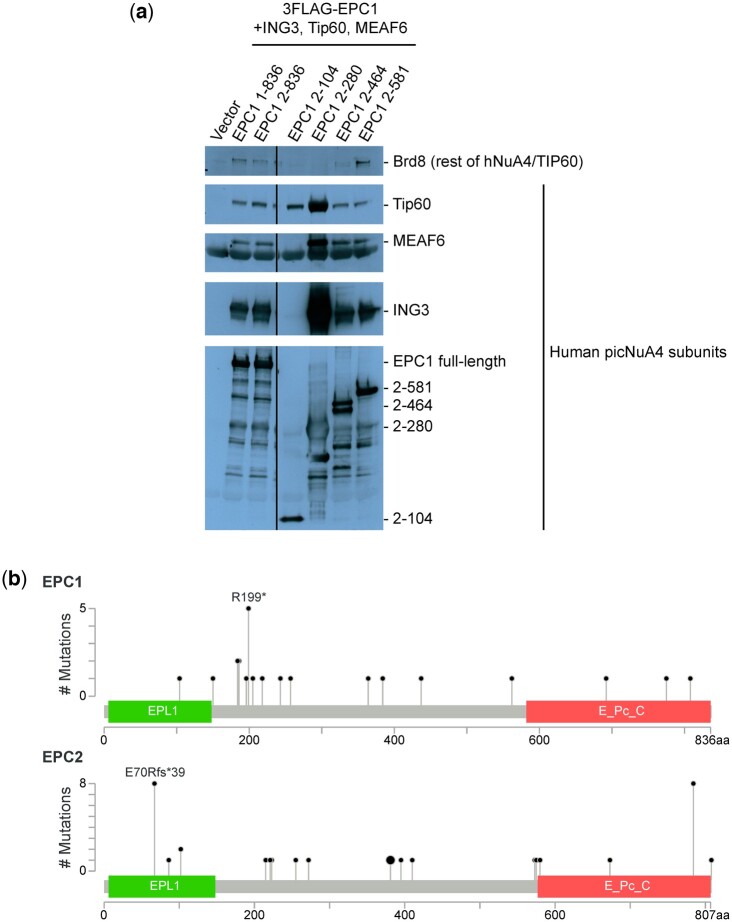
Moving beyond budding yeast, we hypothesized that the evolutionary conservation and functional importance of the Epl1 C-terminus might be reflective of a broader role in NuA4 biology. To begin testing this hypothesis, we expressed truncated forms of EPC1, one of the 2 human homologs of Epl1, in HEK293T cells and measured their effect on NuA4 composition and subunit levels. Immunopurification of EPC1(2-280), which is analogous to Epl1(1-380) (Supplementary Fig. 5), resulted in the separation of human picNuA4 from the rest of the hNuA4/TIP60 complex as well as higher levels of all 4 picNuA4 subunits compared to when wild type or shorter versions of EPC1 were present, effects reminiscent of our findings in yeast (Fig. 6a). Given that the effects of truncating Epl1 appeared conserved across species, we wondered if the functional importance of this interaction might be reflected in human disease. Examining existing large-scale genomic and disease-related data of both human homologs of EPL1, EPC1 and EPC2, we found that truncating mutations in EPC1 and EPC2 have been observed in cancer patients (Fig. 6b) (Cerami et al. 2012; Gao et al. 2013).
Fig. 6.
The effect of truncating Epl1 on picNuA4 levels was conserved from yeast to humans. a) The EPC1(2-280) mutant that is homologous to Epl1(1-380) led to increased picNuA4 levels. A series of C-terminal truncation mutants of 3FLAG-EPC1 were immunopurified after cotransfection with HA-Tip60/Kat5, HA-ING3, and HA-MEAF6 and analyzed by SDS-PAGE followed by immunoblotting using antibodies against FLAG and the indicated hNuA4/TIP60 subunits. b) The human homologs of EPL1, EPC1, and EPC2 were truncated in cancer patients. Lollipop plots of the EPC1 and EPC2 proteins show the location of previously identified truncating mutations, along with annotated protein domains. Analysis and images were generated using the cBio Cancer Genomics Portal (Cerami et al. 2012; Gao et al. 2013).
The Eaf1 HSA domain was required for NuA4 stability and function
Given the profound impact of losing Eaf1 on NuA4 and picNuA4 integrity when the Epl1 C-terminus was intact, we attempted to determine which specific domains of Eaf1 mediated these effects. Internal deletions of untagged EAF1 lacking the HSA and/or SANT domains were created and tested for their ability to recapitulate the eaf1Δ mutant phenotypes across a range of the assays used previously to assess NuA4 function (Fig. 7a). Overall, we found that loss of the SANT domain had no effect on H4 and H2A.Z acetylation levels, cell growth under genotoxic stress, Epl1 protein levels, and the ability to purify NuA4 and picNuA4 subunits via Epl1 affinity purification (Fig. 7, b–e and Supplementary Fig. 6a). In contrast, loss of the HSA domain closely mirrored the complete loss of EAF1, as previously hinted at in an earlier study (Wang et al. 2018). We note, however, that the levels of Eaf1 appeared to decrease in the eaf1 HSAΔ and eaf1 HSAΔSANTΔ mutants (Supplementary Fig. 6a), suggesting that our findings could in part be due decreased Eaf1 protein levels in addition to the loss of the HSA domain specifically. Combining the EAF1 domain mutants with the epl1(1-485) or epl1(1-380) alleles led to growth phenotypes that mirrored the epl1(1-485) or epl1(1-380) single mutants (Supplementary Fig. 6b), further suggesting that eaf1 HSAΔ phenocopied the eaf1Δ mutant. Collectively, these results pointed specifically to the HSA domain of Eaf1 as having a key role in regulating NuA4 and picNuA4 function.
Fig. 7.
The Eaf1 HSA domain was required for NuA4 stability and function. a) Schematic representation of Eaf1 internal deletion alleles: HSAΔ, SANTΔ, and HSAΔ/SANTΔ. b) The loss of the HSA domain led to a similar defect in bulk histone acetylation as the complete loss of EAF1. Whole cell extracts of the indicated strains were analyzed by protein blotting with anti-tetra-acetylated H4 or anti-H2A.Z K14ac antibodies. Antibodies against H4 and H2A.Z were used as a loading controls. * indicates a background band. c) Logarithmic serial dilutions of the indicated strains were plated onto SC-URA media containing the indicated genotoxic agents and incubated for 3 days at the indicated temperature. Strains lacking the Eaf1 HSA domain recapitulated the eaf1Δ mutant phenotype. d) Purification of Epl1 revealed decreased association of NuA4 subunits in the eaf1 HSAΔ strain. Purified fractions from the indicated strains were loaded onto a 4–20% gradient SDS–PAGE gel and visualized by silver staining. Bands corresponding to NuA4 subunits are indicated on the left. Untagged Epl1 was used as a negative control. e) Epl1 purifications of the indicated strains were visualized by immunoblotting to verify Epl1-immunoprecipitation and Esa1/Kat5 association. IgG and anti-Esa1/Kat5 antibodies were used to detect Epl1-TAP and Esa1/Kat5, respectively.
Discussion
This work explored the functional and structural connection between 2 key regulatory subunits of the NuA4 and picNuA4 HAT complexes in S. cerevisiae. As such, our experiments illuminated the circuitry between of the Eaf1 protein and the Epl1 C-terminus in regulating NuA4 and picNuA4 function and composition. Genome-wide expression and genetic interaction data from eaf1Δ and epl1-CΔ mutants revealed not only an expected set of common requirements, but more surprisingly, remarkably different phenotypic and functional consequences. Detailed biochemical examination revealed that EAF1 was needed for detectable picNuA4 formation, a requirement that could be overcome when the Epl1 C-terminus was removed. Despite being sufficient to restore global H4 acetylation levels in an eaf1Δ background, the picNuA4 released in epl1-CΔ mutants displayed reduced interaction with chromatin and occupancy at NuA4-targeted loci, highlighting its distinct function from NuA4. Collectively, our work revealed unexpected differences between epl1-CΔ and eaf1Δ in NuA4 and picNuA4 activity and suggested that the interaction between the Epl1 C-terminus and Eaf1, either directly or indirectly, was necessary for picNuA4 complex stability and the balance between NuA4 and its small cousin in cellular function. In support of an evolutionary conserved function, key biochemical features of this interplay were recapitulated in human cells and in turn might be associated with cancer.
The data presented here provide functional genomics and biochemical data to clarify 2 existing and somewhat conflicting models of NuA4 structural integrity (Auger et al. 2008; Mitchell et al. 2008). The first model suggests that upon deletion of the scaffolding Eaf1 protein, the various submodules of the NuA4 complex remain intact, leaving behind a functional and stoichiometric picNuA4 complex sufficient for un-targeted chromatin acetylation (Auger et al. 2008). The alternative model suggests that Eaf1 is essential for NuA4 complex integrity and that upon deletion of EAF1, there is a significant reduction in the level of NuA4 in the cell (Mitchell et al. 2008). Both models place Eaf1 at the center of NuA4 function through a combination of biochemical and genetic means. However, the defining difference between the 2 scenarios centers on the importance of Eaf1 in regulating the quantitative balance between the various submodules, and in particular picNuA4. In principle, the data presented here supported the coexistence of both models. The strong decrease of H4 acetylation in the eaf1Δ mutant suggested that the released picNuA4 had compromised HAT activity. As evidenced by the NuA4 purification experiments, we indeed observed a reduction in the levels of picNuA4 subunits that copurified with Epl1-TAP when EAF1 was deleted. However, and perhaps most importantly, in contrast to the eaf1Δ mutant, the eaf1Δ epl1-CΔ mutants had a fully active minimal picNuA4 capable of maintaining basal H4 acetylation levels, suggesting that the catalytic module of NuA4 existed as a functional and independent entity. In addition, the differences in promoter H4 acetylation and mRNA levels at representative RP genes we found in the epl1-CΔ mutants, combined with the loss of Epl1 enrichment at several of these RP promoters, was consistent with this version of picNuA4 having lost the targeting functions present in NuA4. Furthermore, these RP locus-specific effects in both the epl1-CΔ and eaf1Δ single mutants were in agreement with previously proposed models suggesting picNuA4 is diffused over the genome and is responsible for basal, untargeted chromatin acetylation (Boudreault et al. 2003; Auger et al. 2008; Friis et al. 2009). This was also supported by our findings at non-RP loci, which did not show altered promoter H4 acetylation, suggesting picNuA4 was sufficient to appropriately acetylate other loci in the cell.
Our comprehensive analysis of the functional consequences of loss of Eaf1 or the Epl1 C-terminus, either alone or in combination, might further consolidate these 2 prevailing models. While both models suggest that loss of Eaf1 or loss of the Epl1 C-terminus liberate picNuA4, the biochemical data presented here suggested that they result in dramatically different levels of this moiety, at least as judged by our analytical methods. At the same time, our functional genomics data identified both overlapping and divergent gene expression clusters between the mutants, as well as common but also unique genes required to support growth of eaf1Δ and epl1-CΔ mutants, respectively. Taken together with the biochemical examination, our data suggested that these 2 genetic manipulations, while sharing important functional consequences, also led to some unexpected and distinct outcomes for NuA4 and picNuA4. The double mutant analysis of cells lacking Eaf1 and the Epl1 C-terminus further pointed toward a complex relationship between these 2 NuA4 HAT complex subunits. In line with previous reports, cells lacking EAF1 displayed decreased fitness under genotoxic stress and reduced global H4 acetylation, most likely as a result of the dissociation of the NuA4 complex into its submodules (Boudreault et al. 2003; Kobor et al. 2004; Babiarz et al. 2006; Auger et al. 2008; Mitchell et al. 2008). However, this effect was in large part due to the presence of the Epl1 C-terminus, given that removal of the C-terminal domain was sufficient to restore the bulk H4 acetylation defect and to a lesser degree, the growth defects of the eaf1Δ mutant. It is tempting to speculate that the mechanism underlying the suppression of eaf1Δ mutant phenotypes by introduction of the epl1-CΔ mutation was due to the increased Epl1 protein levels and the stable assembly of picNuA4 seen in our analytical-scale purification experiments.
The decreased cellular levels of at least 2 picNuA4 subunits, Epl1 and Esa1/Kat5, observed in the absence of wild-type Eaf1 strongly complemented the notion that this scaffold protein had a central role in NuA4 assembly and stability, as established previously by both functional and cryo-EM structural studies (Auger et al. 2008; Mitchell et al. 2008; Setiaputra et al. 2018; Wang et al. 2018). However, adding a surprising—and we believe very important—mechanistic twist to the existing model, we found that Eaf1 was only required for picNuA4 submodule function and stability in the presence of full length Epl1 protein. Specifically, our functional and biochemical findings established a key regulatory role for the Epl1 C-terminus in overcoming the defects caused by the loss of Eaf1. In the broader context of the role of Eaf1 in NuA4 biology, it is interesting to note that the Eaf1 protein might also play an as of yet poorly defined role in regulating the levels of the Eaf3/5/7 (TINTIN) NuA4 submodule, as these are slightly decreased in the absence of EAF1 (Rossetto et al. 2014; Setiaputra et al. 2018; Wang et al. 2018). More specifically related to our work, detailed domain deletion analysis of Eaf1 identified its HSA domain as an important factor in regulating the stability of picNuA4, as eaf1 HSAΔ cells exhibited reduced picNuA4 levels akin to the eaf1Δ mutant. Taken together, our data suggested that the interface between the Eaf1 HSA domain and the Epl1 C-terminus was required for maintaining the equilibrium of NuA4 and picNuA4 in the cell, either directly or indirectly. However, we acknowledge an apparent conflict between our NuA4 protein complex purifications from cells and the in vitro protein–protein interactions presented here as to the exact delineation of the Eaf1 interaction region on Epl1. This is perhaps reflective of the reduced complexity of binary in vitro protein–protein interaction assays compared to the more native protein complex purifications from cells. Here, the latter likely reflects the role of other NuA4 subunits present in vivo that may affect the Eaf1–Epl1 interaction. Specifically, Arp4 and Act1 have been shown to interact with the same region of Eaf1 and to stabilize its interaction with Epl1 (Szerlong et al. 2008; Wang et al. 2018).
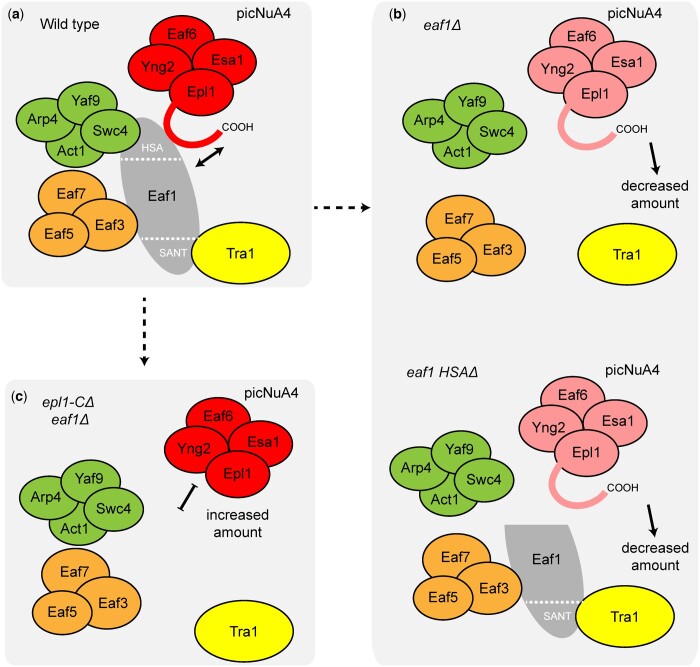
Consolidating these results, our findings expand upon previous work suggesting that picNuA4 docks onto Eaf1 through the Epl1 C-terminus (Auger et al. 2008; Szerlong et al. 2008; Setiaputra et al. 2018; Wang et al. 2018) (Fig. 8a) and show that beyond this physical interaction, the Eaf1 HSA domain and Epl1 C-terminus are key regulators of the protein levels and perhaps the balance of NuA4 and picNuA4 in the cell. We found that Eaf1, particularly its HSA domain, was required for Epl1 and picNuA4 stability (Fig. 8b). Our data also demonstrated that upon truncation of the Epl1 C-terminus, Epl1 protein levels were increased and picNuA4 function and stability were restored in an eaf1Δ background (Fig. 8c). Our identification of ubiquitin-mediated degradation pathway genes being genetically linked to NuA4, in combination with high-throughput mass spectrometry data suggesting ubiquitination of K648 of Epl1, raised the possibility of ubiquitin dependent degradation of Epl1 being, in part, responsible for its decrease in the eaf1Δ background. However, our data suggested that under conditions tested, mutation of K648 to a nonmodifiable arginine had no effect on Epl1 protein levels in the absence of Eaf1. We do not know whether this was reflective of the documented redundancy in the biology of ubiquitin modifications (Trulsson and Vertegaal 2021), or perhaps hinted at nonubiquitination pathways playing a role in regulating Epl1 protein levels. Regardless of the exact mechanisms underlying this circuitry, our work illuminated the importance of the relationship between Eaf1 and the Epl1 C-terminus in picNuA4 formation and supported a model where the stability and levels of picNuA4 were regulated by the Epl1 C-terminus in concert with the central scaffold protein, Eaf1. This provided a means by which the loss of Eaf1 could indeed have a significant impact on NuA4 and picNuA4 levels, while also allowing mechanisms for picNuA4 to stably exist independently of NuA4, unifying the 2 previous models (Auger et al. 2008; Mitchell et al. 2008).
Fig. 8.
Model for the regulatory mechanisms of Eaf1 and the Epl1 C-terminus in picNuA4 stability. a) The catalytic submodule (picNuA4) associates with NuA4 through the Epl1 C-terminus and the Eaf1 HSA domain. b) Deletion of Eaf1 or its HSA domain leads to reduced picNuA4 levels, perhaps indicative of decreased protein stability. c) Truncation of the Epl1 C-terminus in an eaf1Δ background increases picNuA4 levels and restores H4 acetylation.
While we feel this model is compelling and inclusive of the existing data, we note that there are several questions that remain to be explored. First while overall similar, the 2 epl1-CΔ mutants had important differences that are hard to reconcile with this model and the existing data. For instance, while the epl1(1-380) mutant had more severe growth phenotypes than the epl1(1-485) mutant, both mutants led to similar levels and composition of picNuA4. As the 105 amino acid region that comprises the differences between the 2 epl1 truncations lacks any annotated protein domain and is predicted to be unstructured (Xu et al. 2016), it is unclear what caused the nuanced variance in phenotype between the epl1(1-380) and epl1(1-485) mutants. Second, we note that while our model is consistent with the observed levels of H4 tetra-acetylation, it does not easily explain the levels of acetylation of H2A.Z, an important target of NuA4, which actually differed between the epl1-CΔ mutants. This difference suggested H2A.Z acetylation did not simply require adequate levels of picNuA4. However, the epl1-CΔ mutants did alleviate the more severe defect seen in the eaf1Δ mutant, indicating that picNuA4 at least partially contributed to H2A.Z acetylation. Finally, it remains to be understood why the robust increase in picNuA4 levels observed in the epl1-CΔ mutants did not result in an increase in bulk histone acetylation levels. Related, it is unclear whether any of our manipulations affected the acetylation of any nonchromatin substrates that NuA4 acetylates (Lin et al. 2008, 2009; Lu et al. 2011; Mitchell et al. 2011, 2013; Downey et al. 2015).
Putting our findings on the role of the Epl1 C-terminus in regulating NuA4 and picNuA4 in the context of the human EPL1 homologs, EPC1 and EPC2, might also provide some intriguing insight into the biology of NuA4/TIP60 across species. In support of a possible evolutionary conservation for the role of Epl1 and Eaf1 in NuA4/TIP60 biology identified here, we found that truncating one of the 2 human EPL1 homologs (EPC1) also increased the levels of picNuA4 subunits in a human cell line. It is also interesting to note that the C-terminus of EPC1 interacts with the methyl-histone binding protein MBTD1, which is a nonconserved peripheral subunit of hNuA4/TIP60 (Jacquet et al. 2016). This is consistent with our finding that Epl1 had an Eaf1-independent chromatin association mechanism that depended on its own C-terminus, although the exact nature of this anchoring, and the molecular interactions involved, has yet to be determined. Moving forward, the evolutionarily conserved role of EPC1 truncations on picNuA4 subunit amounts and composition in human cells raises the intriguing possibility that additional functional consequences of the circuitry between Epl1 and Eaf1 are also conserved, including the regulation of H4 and H2A.Z acetylation levels. To that end, the identification of truncating mutations in cancer patients in both EPC1 and EPC2 might point to some relevant processes, as Tip60/Kat5 can act as either a tumor suppressor or an oncogene depending on the type of cancer (Gorrini et al. 2007; Coffey et al. 2012).
Supplementary Material
Acknowledgments
The authors wish to thank Kristy Dever for critical reading of the article and Dr. Gian Luca Negri for help with accessing data from the cBio Cancer Genomics Portal.
Funding
This work was supported by the Canadian Institutes of Health Research (CIHR) through operating grants to MSK (MOP-119383) and JC (FDN-143314), Canada graduate scholarships to PYTL and NL, and a PDF fellowship to NA and by the Natural Sciences and Engineering Research Council (NSERC) through an operating grant to CKY (418157-2012), Canada graduate scholarships to ACK, HTB, and DTS, and a PDF fellowship to MJA. JC holds the Canada Research Chair in Chromatin Biology and Molecular Epigenetics.
Conflicts of interest
None declared.
Contributor Information
Phoebe Y T Lu, Centre for Molecular Medicine and Therapeutics, British Columbia Children’s Hospital Research Institute, Department of Medical Genetics, University of British Columbia, Vancouver, BC V5Z 4H4, Canada.
Alyssa C Kirlin, Centre for Molecular Medicine and Therapeutics, British Columbia Children’s Hospital Research Institute, Department of Medical Genetics, University of British Columbia, Vancouver, BC V5Z 4H4, Canada.
Maria J Aristizabal, Centre for Molecular Medicine and Therapeutics, British Columbia Children’s Hospital Research Institute, Department of Medical Genetics, University of British Columbia, Vancouver, BC V5Z 4H4, Canada.
Hilary T Brewis, Centre for Molecular Medicine and Therapeutics, British Columbia Children’s Hospital Research Institute, Department of Medical Genetics, University of British Columbia, Vancouver, BC V5Z 4H4, Canada.
Nancy Lévesque, Centre for Molecular Medicine and Therapeutics, British Columbia Children’s Hospital Research Institute, Department of Medical Genetics, University of British Columbia, Vancouver, BC V5Z 4H4, Canada.
Dheva T Setiaputra, Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC V6T 1Z3, Canada.
Nikita Avvakumov, Department of Molecular Biology, Medical Biochemistry, and Pathology, Laval University Cancer Research Center, CHU de Québec-Université Laval Research Center-Oncology Division, Quebec City, QC G1R 3S3, Canada.
Joris J Benschop, Center for Molecular Medicine, Molecular Cancer Research, University Medical Center Utrecht, Utrecht 3584 CX, The Netherlands.
Marian Groot Koerkamp, Princess Máxima Center for Pediatric Oncology, Utrecht 3584 CS, The Netherlands.
Frank C P Holstege, Princess Máxima Center for Pediatric Oncology, Utrecht 3584 CS, The Netherlands.
Nevan J Krogan, Department of Cellular and Molecular Pharmacology, University of California, San Francisco, San Francisco, CA 94158, USA.
Calvin K Yip, Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC V6T 1Z3, Canada.
Jacques Côté, Department of Molecular Biology, Medical Biochemistry, and Pathology, Laval University Cancer Research Center, CHU de Québec-Université Laval Research Center-Oncology Division, Quebec City, QC G1R 3S3, Canada.
Michael S Kobor, Centre for Molecular Medicine and Therapeutics, British Columbia Children’s Hospital Research Institute, Department of Medical Genetics, University of British Columbia, Vancouver, BC V5Z 4H4, Canada.
Data Availability
Microarray data were submitted to the Gene Expression Omnibus under accession number GSE137261. This publication is accompanied by Supplementary File 1, which contains the E-MAP data used in this study, Supplementary Table 1, and Supplementary Figs 1–6.
Supplemental material is available at GENETICS online.
Literature cited
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Côté J.. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18(18):5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger A, Galarneau L, Altaf M, Nourani A, Doyon Y, Utley RT, Cronier D, Allard S, Côté J.. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol. 2008;28(7):2257–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel M, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. PA: John Wiley & Sons, Inc., Media; 1987. [Google Scholar]
- Avvakumov N, Lalonde M-E, Saksouk N, Paquet E, Glass KC, Landry A-J, Doyon Y, Cayrou C, Robitaille GA, Richard DE, et al. Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol Cell Biol. 2012;32(3):689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Halley JE, Rine J.. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20(6):700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Côté J.. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17(11):1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL.. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292(5525):2333–2337. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Jobin-Robitaille O, Billon P, Buisson R, Niu H, Lacoste N, Abshiru N, Côté V, Thibault P, Kron SJ, et al. Phospho-dependent recruitment of the yeast NuA4 acetyltransferase complex by MRX at DNA breaks regulates RPA dynamics during resection. Proc Natl Acad Sci U S A. 2018;115(40):10028–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittuluru JR, Chaban Y, Monnet-Saksouk J, Carrozza MJ, Sapountzi V, Selleck W, Huang J, Utley RT, Cramet M, Allard S, et al. Structure and nucleosome interaction of the yeast NuA4 and Piccolo-NuA4 histone acetyltransferase complexes. Nat Struct Mol Biol. 2011;18(11):1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L.. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19(4):2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey K, Blackburn TJ, Cook S, Golding BT, Griffin RJ, Hardcastle IR, Hewitt L, Huberman K, McNeill HV, Newell DR, et al. Characterisation of a Tip60 specific inhibitor, NU9056, in prostate cancer. PLoS One. 2012;7(10):e45539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS.. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7(7):R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Johnson JR, Davey NE, Newton BW, Johnson TL, Galaang S, Seller CA, Krogan N, Toczyski DP.. Acetylome profiling reveals overlap in the regulation of diverse processes by sirtuins, gcn5, and esa1. Mol Cell Proteomics. 2015;14(1):162–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Côté J.. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16(6):979–990. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Côté J.. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14(2):147–154. [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem. 2004;271(12):2335–2349. [DOI] [PubMed] [Google Scholar]
- Foiani M, Marini F, Gamba D, Lucchini G, Plevani P.. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol. 1994;14(2):923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis RMN, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC.. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 2009;37(12):3969–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T.. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol Cell Biol. 2001;21(6):2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Zambrano Á, Crevillén P, Franco-Zorrilla JM, López JA, Moreno-Romero J, Roszak P, Santos-González J, Jurado S, Vázquez J, Köhler C, et al. Arabidopsis SWC4 binds DNA and recruits the SWR1 complex to modulate histone H2A.Z deposition at key regulatory genes. Mol Plant. 2018;11(6):815–832. [DOI] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448(7157):1063–1067. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009a;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y.. SnapShot: histone modifications. Cell. 2014;159(2):458–458.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert J-P, Roques C, Pandita RK, Paquet E, Herst P, Gingras A-C, Pandita TK, et al. The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol Cell. 2016;62(3):409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M-C, Buratowski S.. Using chromatin immunoprecipitation to map cotranscriptional mRNA processing in Saccharomyces cerevisiae. Methods Mol Biol. 2004;257:1–16. [DOI] [PubMed] [Google Scholar]
- Keogh M-C, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20(6):660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BJ, Ahmad S, Vann KR, Andrews FH, Mayo ZA, Bourriquen G, Bridgers JB, Zhang J, Strahl BD, Côté J, et al. Yaf9 subunit of the NuA4 and SWR1 complexes targets histone H3K27ac through its YEATS domain. Nucleic Acids Res. 2018;46(1):421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J.. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2(5):E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Baetz K, Keogh M-C, Datta N, Sawa C, Kwok TCY, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci U S A. 2004;101(37):13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde M-E, Avvakumov N, Glass KC, Joncas F-H, Saksouk N, Holliday M, Paquet E, Yan K, Tong Q, Klein BJ, et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013;27(18):2009–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-Y, Lu J-Y, Zhang J, Walter W, Dang W, Wan J, Tao S-C, Qian J, Zhao Y, Boeke JD, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136(6):1073–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-Y, Qi Y, Lu J-Y, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD.. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 2008;22(15):2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR.. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. [DOI] [PubMed] [Google Scholar]
- Lu J-Y, Lin Y-Y, Sheu J-C, Wu J-T, Lee F-J, Chen Y, Lin M-I, Chiang F-T, Tai T-Y, Berger SL, et al. Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell. 2011;146(6):969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PYT, Lévesque N, Kobor MS.. NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components. Biochem Cell Biol. 2009;87(5):799–815. [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ.. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. [DOI] [PubMed] [Google Scholar]
- Mitchell L, Huard S, Cotrut M, Pourhanifeh-Lemeri R, Steunou A-L, Hamza A, Lambert J-P, Zhou H, Ning Z, Basu A, et al. mChIP-KAT-MS, a method to map protein interactions and acetylation sites for lysine acetyltransferases. Proc Natl Acad Sci U S A. 2013;110(17):E1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell L, Lambert J-P, Gerdes M, Al-Madhoun AS, Skerjanc IS, Figeys D, Baetz K.. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol Cell Biol. 2008;28(7):2244–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell L, Lau A, Lambert J-P, Zhou H, Fong Y, Couture J-F, Figeys D, Baetz K.. Regulation of septin dynamics by the Saccharomyces cerevisiae lysine acetyltransferase NuA4. PLoS One. 2011;6(10):e25336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani A, Utley RT, Allard S, Côté J.. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 2004;23(13):2597–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Chang HY.. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JL, Iyer VR, Brown PO, Struhl K.. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6(6):1297–1307. [DOI] [PubMed] [Google Scholar]
- Renaud-Young M, Lloyd DC, Chatfield-Reed K, George I, Chua G, Cobb J.. The NuA4 complex promotes translesion synthesis (TLS)-mediated DNA damage tolerance. Genetics. 2015;199(4):1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto D, Cramet M, Wang AY, Steunou A-L, Lacoste N, Schulze JM, Côté V, Monnet-Saksouk J, Piquet S, Nourani A, et al. Eaf5/7/3 form a functionally independent NuA4 submodule linked to RNA polymerase II-coupled nucleosome recycling. EMBO J. 2014;33(12):1397–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathianathan A, Ravichandran P, Lippi JM, Cohen L, Messina A, Shaju S, Swede MJ, Ginsburg DS.. The Eaf3/5/7 subcomplex stimulates NuA4 interaction with methylated histone H3 Lys-36 and RNA polymerase II. J Biol Chem. 2016;291(40):21195–21207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Weissman JS, Krogan NJ.. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods. 2006;40(4):344–352. [DOI] [PubMed] [Google Scholar]
- Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A.. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35(5):626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle NE, Torres-Machorro AL, Pillus L.. Chromatin regulation by the NuA4 acetyltransferase complex is mediated by essential interactions between enhancer of polycomb (Epl1) and Esa1. Genetics. 2017;205(3):1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck W, Fortin I, Sermwittayawong D, Côté J, Tan S.. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol Cell Biol. 2005;25(13):5535–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiaputra D, Ahmad S, Dalwadi U, Steunou A-L, Lu S, Ross JD, Dong M-Q, Côté J, Yip CK.. Molecular architecture of the essential yeast histone acetyltransferase complex NuA4 redefines its multi-modularity. Mol Cell Biol. 2018;38(9): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M.. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. [DOI] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD.. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A. 1998;95(7):3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé C, Nadal-Ribelles M, Kraft C, Peter M, Posas F, de Nadal E.. Control of Ubp3 ubiquitin protease activity by the Hog1 SAPK modulates transcription upon osmostress. EMBO J. 2011;30(16):3274–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steunou A-L, Cramet M, Rossetto D, Aristizabal MJ, Lacoste N, Drouin S, Côté V, Paquet E, Utley RT, Krogan N, et al. Combined action of histone reader modules regulates NuA4 local acetyltransferase function but not its recruitment on the genome. Mol Cell Biol. 2016;36(22):2768–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steunou A-L, Rossetto D, Côté J.. Regulating chromatin by histone acetylation. In: Workman JL, Abmayr SM, editors. Fundamentals of Chromatin, Fundamentals of Chromatin. New York, NY: Springer New York; 2014. p. 147–212. [Google Scholar]
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villén J.. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR.. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15(5):469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trulsson F, Vertegaal ACO.. Site-specific proteomic strategies to identify ubiquitin and SUMO modifications: challenges and opportunities. Semin Cell Dev Biol. 2021. 10.1016/j.semcdb.2021.11.006 [DOI] [PubMed] [Google Scholar]
- Uprety B, Sen R, Bhaumik SR.. Eaf1p is required for recruitment of NuA4 in targeting TFIID to the promoters of the ribosomal protein genes for transcriptional initiation in vivo. Mol Cell Biol. 2015;35(17):2947–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Holstege FCP.. In control: systematic assessment of microarray performance. EMBO Rep. 2004;5(10):964–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Peppel J, Kemmeren P, van Bakel H, Radonjic M, van Leenen D, Holstege FCP.. Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep. 2003;4(4):387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wageningen S, Kemmeren P, Lijnzaad P, Margaritis T, Benschop JJ, de Castro IJ, van Leenen D, Groot Koerkamp MJA, Ko CW, Miles AJ, et al. Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell. 2010;143(6):991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AY, Schulze JM, Skordalakes E, Gin JW, Berger JM, Rine J, Kobor MS.. Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation. Proc Natl Acad Sci U S A. 2009;106(51):21573–21578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ahmad S, Zhang Z, Côté J, Cai G.. Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex. Nat Commun. 2018;9(1):1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Li C, Chen Z, Jiang S, Fan S, Wang J, Dai J, Zhu P, Chen Z.. The NuA4 core complex acetylates nucleosomal histone H4 through a double recognition mechanism. Mol Cell. 2016;63(6):965–975. [DOI] [PubMed] [Google Scholar]
- Zhang H, Richardson DO, Roberts DN, Utley R, Erdjument-Bromage H, Tempst P, Côté J, Cairns BR.. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol Cell Biol. 2004;24(21):9424–9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BO, Wang S-S, Xu L-X, Meng F-L, Xuan Y-J, Duan Y-M, Wang J-Y, Hu H, Dong X, Ding J, et al. SWR1 complex poises heterochromatin boundaries for antisilencing activity propagation. Mol Cell Biol. 2010;30(10):2391–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BO, Wang S-S, Zhang Y, Fu X-H, Dang W, Lenzmeier BA, Zhou J-Q.. Histone H4 lysine 12 acetylation regulates telomeric heterochromatin plasticity in Saccharomyces cerevisiae. PLoS Genet. 2011;7(1):e1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data were submitted to the Gene Expression Omnibus under accession number GSE137261. This publication is accompanied by Supplementary File 1, which contains the E-MAP data used in this study, Supplementary Table 1, and Supplementary Figs 1–6.
Supplemental material is available at GENETICS online.