Abstract
Wastewater-based surveillance (WBS) for disease monitoring is highly promising but requires consistent methodologies that incorporate predetermined objectives, targets, and metrics. Herein, we describe a comprehensive metagenomics-based approach for global surveillance of antibiotic resistance in sewage that enables assessment of 1) which antibiotic resistance genes (ARGs) are shared across regions/communities; 2) which ARGs are discriminatory; and 3) factors associated with overall trends in ARGs, such as antibiotic concentrations. Across an internationally sourced transect of sewage samples collected using a centralized, standardized protocol, ARG relative abundances (16S rRNA gene-normalized) were highest in Hong Kong and India and lowest in Sweden and Switzerland, reflecting national policy, measured antibiotic concentrations, and metal resistance genes. Asian versus European/US resistomes were distinct, with macrolide-lincosamide-streptogramin, phenicol, quinolone, and tetracycline versus multidrug resistance ARGs being discriminatory, respectively. Regional trends in measured antibiotic concentrations differed from trends expected from public sales data. This could reflect unaccounted uses, captured only by the WBS approach. If properly benchmarked, antibiotic WBS might complement public sales and consumption statistics in the future. The WBS approach defined herein demonstrates multisite comparability and sensitivity to local/regional factors.
Keywords: antibiotic resistance, microbiome, wastewater-based surveillance (WBS), sewage, resistome, metagenomics
Short abstract
This work describes and applies a consistent methodology for interpretation of globally sourced metagenomic data for wastewater-based surveillance of antibiotic resistance.
Wastewater-based surveillance (WBS), which includes the monitoring of sewage for health-related microbes and chemicals,1,2 is an emerging framework for public health surveillance as it quickly and noninvasively provides anonymous population-scale information about human disease and anthropogenic chemical use.3 Activity in this field has focused on xenobiotic and human biomarkers4,5 and the monitoring of infectious diseases such as polio6 and typhoid.7 Most recently, WBS has been broadly deployed for surveillance of COVID-198−10 as well as the spread of antibiotic resistance.11−14 Within any given sewershed, the analysis of temporal or spatial changes in WBS targets (e.g., genes, microbes, chemicals) may provide an early warning of local and regional disease outbreaks,15 while comparisons across sewersheds enable insights into environmental and socioeconomic factors contributing to disease patterns.2 To produce globally actionable information, however, data comparability across disparate systems is crucial and should be considered at the outset of commencing measurements. In this study, we applied a consistent sample collection and analysis protocol16,17 and demonstrated the capability of an array of data analysis approaches18−20 for discriminating internationally sourced sewage samples from Asia (Hong Kong, India, the Philippines), Europe (Sweden, Switzerland), and North America (US) with the aim of advancing a WBS framework for antibiotic resistance surveillance.
Sewage represents a composite of human-associated flora, including pathogens, antibiotic resistant bacteria (ARB), and antibiotic resistance genes (ARGs),21−24 carried across the corresponding community.11,25 The profiles of ARB and ARGs in sewage are expected to be dependent on current and historic antibiotic resistance management practices, including the types and quantities of antibiotics used and local attention to transmission control.21,26 A targeted and well-designed WBS program could help establish baseline levels of antibiotic resistance for a given community, inform effective antibiotic use policy and wastewater treatment practices, and serve as a point of comparison for assessing intervention impacts.2,25,27
Shotgun metagenomic sequencing provides a powerful means to characterize sewage microbiomes28 and viromes,29 producing nucleotide sequences that can be archived and compared to publicly available databases to profile pathogens, pandemic viruses,30,31 and ARG composition (i.e., the “resistome”).32−34 Recent studies have applied metagenomic sequencing to identify compelling sewage resistome trends. Hendriksen et al.11 collected wastewater samples from 79 sites in 60 countries and found ARG abundance correlated with socioeconomic, health, and environmental factors. Using the same data set, Karkman et al. explored to what extent sewage metagenomes (alone or in combination with socioeconomic factors) predict local clinical resistance.13 Pärnänen et al.12 showed that sewage samples collected across a European north–south transect correlated with differences in antibiotic usage and average local temperatures. Going forward, precise surveillance objectives, targets, and metrics must be identified and validated if metagenomics enabled WBS is to be globally recommended and adopted.25,35 In particular, the identification of informative features (e.g., core resistome, discriminatory resistome, antibiotic concentrations) of resistomes and their relationship to other key sewage constituents, including concentrations of metals, metal resistance genes (MRGs), and mobile genetic elements (MGEs), is needed to identify factors that are informative about regional resistance patterns.
Herein, our focus was on the testing of a consistent WBS methodology for sample collection, preservation, processing, and analysis. As such, the number of wastewater treatment plants (WWTPs) that could be sampled was smaller than that employed previously.11,36 The sample sites were specifically chosen to represent known socioeconomic contrasts. We assessed various dimensions of sewage metagenomes, antibiotic concentrations, and the associated metainformation, to identify discriminatory features and factors that might reflect the corresponding resistomes. Taxonomic composition and the diversity of the corresponding microbiomes were evaluated as key constraints on ARG composition.37,38 In addition, plasmids, transposons, and integrons were quantified as plausible carriers of resistance32,33,39 (i.e., indicating the potential for associated ARGs to spread horizontally across species). Genes that could provide coselection opportunities, such as MRGs, were also quantified, because, even if antibiotic use is curbed, metals and other selective agents could still select for ARGs. “Core resistome”37,39,40 and “discriminatory resistome”19 analyses were carried out to account for the ARG “background” of globally distributed or naturally prevalent ARGs (e.g., those found even in 30,000 year old Arctic soils41 and permafrost42) and to differentiate ARGs that result primarily from anthropogenic activity and are of clinical concern. Finally, we investigated how measured antibiotic concentrations relate to country-specific antibiotic sales. Overall, we advance a WBS approach for sewage-based surveillance of antibiotic resistance, focusing monitoring toward targets relevant to risk assessment and antibiotic resistance stewardship.
Materials and Methods
Sample Locations
A total of 14 sewage samples were collected from the influent to 12 WWTPs located in India, Hong Kong, the Philippines, Sweden, Switzerland, and the US, with 2 WWTPs per location and the Hong Kong WWTPs subjected to two separate sampling events in 2016 and 2017 (Supporting Information Table S1). The WWTP capacities ranged from 2.6 to 66 million gallons per day with a majority of the sewage consisting of domestic wastewater, with some industrial and hospital inputs.
Sample Collection and Processing
Sample collection and processing were standardized as described previously.16,17 In brief, 500 mL influent grab samples were collected in sterile polypropylene containers and transported to a local laboratory on ice. Sewage biomass was concentrated in triplicate via vacuum filtration until clogging (15–75 mL) through three separate 0.22-μm mixed cellulose ester membranes (MilliporeSigma Darmstadt, Germany), which were then preserved in 50% ethanol and stored at −20 °C. Ethanol-fixed membrane samples were shipped to Virginia Tech for DNA extraction and biomolecular analyses. Sewage samples (0.5 L) for chemical analysis were acidified and filtered using 0.45-μm glass microfiber filters to remove microorganisms and particulate matter. Na2EDTA (2 mL, 5% v/v in water) and isotopically labeled surrogate standards (50 μL of 1000 μg/L surrogate mix solution) were added to each sample.
DNA Extraction, Shotgun Metagenomic Sequencing, and Analyses
As described previously,16 DNA extraction was carried out using the FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH). While this method was not optimized for plasmid capture, there is experimental evidence (as we43 and others44,45 report) that plasmid DNA can be isolated via this extraction. Additional optimization of the extraction protocol may be necessary to address potential recovery biases. The total mass of extracted DNA was quantified using a Qubit fluorometer. Triplicate DNA extracts were then pooled by equal mass. Composite samples were prepared by pooling equal masses of DNA from triplicate extracts. TrueSeq libraries (Illumina, San Diego, CA) were prepared for shotgun metagenomic sequencing via Illumina HiSeq 2500 with 2 × 100 “bp” paired-end reads. Sequencing was performed at the Virginia Tech Biocomplexity Institute Genomic Sequencing Center (Blacksburg, VA). Library preparation and sequencing were duplicated independently for two samples to verify sequencing reproducibility. With a sequence depth of 20–30 million reads per sample, after joining paired ends and quality trimming, sequencing depth ranged from 10 to 16 million paired-end reads per sample with an average of 13 million reads per sample (Table S2).
Taxonomic, ARG, Plasmid, and MRG Analyses
Sequences were uploaded to the metagenomics RAST server (MG-RAST) and annotated against the RefSeq database to identify bacterial phyla and genera using default parameters.46 Taxonomic beta diversity was examined via nonmetric multidimensional scaling (NMDS)-based ordination according to Euclidian distances between rlog-transformed reads to bacterial genera.47 ARG annotation was carried out via MetaStorm18 with read matching to CARD (protein homolog model) version 1.2.1.48 The pipeline uses the DIAMOND BLASTX71 aligner with the representative hit approach (E-value < 1e-10, identity > 90%, and minimum length of 25 amino acids). The ARGs in CARD version 1.2.1 were assigned into resistance categories based on the updated ARGminer database.20 Manual curation was subsequently performed to fill in missing categories or to update existing categories and to segregate the genes thought to confer resistance via point mutations. Gene names and the ARG category to which they were assigned are found in Supplemental Data 1. If an ARG conferred resistance to two or more antibiotic classes, it was categorized as “multidrug”, except in the case of resistance to macrolide, lincosamide, and streptogramin (MLS) antibiotics, which were assigned as a distinct resistance group. The core resistome is defined as the collection of ARGs that are ubiquitous across the resistomes of all samples analyzed. Core resistome analysis was carried out by subsetting the list of ARGs that were identified in all samples (Supplemental Data 2). Plasmid-associated sequences were annotated using the ACLAME (a CLAssification of Mobile genetic Elements) database, version 0.4.49 MRGs were annotated using the list of experimentally confirmed metal resistance genes from BacMet, antibacterial biocide and metal resistance genes database, version 1.1.50 Gene abundances were normalized within MetaStorm to 16S rRNA gene abundances as previously described.51 NMDS and analysis of similarities (ANOSIM) were conducted considering each individual ARG detected (i.e., not grouped by class) with square root transformation and Bray–Curtis dissimilarities. R-Value cutoffs as defined by Clarke and Warwick52 were used (R > 0.75, well separated; 0.75 > R > 0.25, separated but overlapping; R < 0.25, barely separated). Clinically relevant ARGs were selected from a manually curated list prioritizing human health impacts of ARGs carried by infectious bacteria (Supplemental Data 3). Discriminatory ARGs were identified via the ExtrARG19 machine learning-based approach. ExtrARG is based on the extremely randomized trees classifier that uses a Bayesian strategy to optimize the classifier parameters and identify discriminatory ARGs based on the user-defined categorizing scheme or groups. Differential taxa were explored via Linear discriminant analysis Effect Size (LEfSe) analysis. LEfSe53 enables biomarker discovery among class conditions within metagenomic samples. It first performs a nonparametric Kruskal–Wallis sum-rank test to detect features with differential abundance among the conditions of interest; it then applies an unpaired Wilcoxon rank-sum test to test for biological consistency. LEfSe uses linear discriminant analysis to estimate the effect size of each differentially abundant feature. Reads were assembled de novo in MetaStorm according to default parameters, and scaffolds were mapped against the CARD and ACLAME databases. Network visualization was conducted using Gephi (version 0.8.2). Potential pathogenic ARG hosts (i.e., pathogenic bacteria containing ARGs) were determined using the CARD prevalence tool, which examines the occurrence of each ARG of interest in NCBI archived pathogen chromosomes and assembled whole genome sequences, as well as plasmids.54
Antibiotic Analysis
Sample processing and analysis for antibiotics were conducted as previously described.17 Solid phase extraction was performed on 1 L filtered wastewater samples by conditioning Oasis HLB cartridges with acetonitrile and Nanopure water before the water samples were loaded at a rate of 3–5 mL/min. Cartridges were dried under vacuum and then shipped to the University at Buffalo (NY, USA) for elution and analysis by liquid chromatography with tandem mass spectrometry (LC-MS/MS). LC-MS/MS analysis was carried out using an Agilent 1200 LC system (Palo Alto, CA).
Results and Discussion
Comparison of Resistomes Across a Global Transect of Sewage Samples
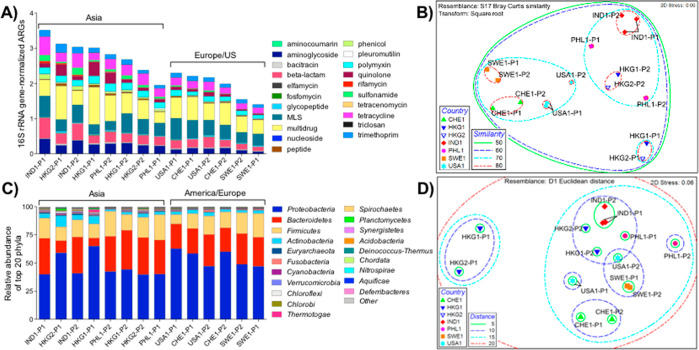
Fourteen influent sewage samples from 12 WWTPs located in six countries were systematically collected by our team using standardized protocols,16,17 profiled, and compared. To identify and quantify ARGs, we performed shotgun metagenomic sequencing and annotated reads using the CARD database.54 We detected an average of 449 ARGs (range: 309–489) at each site, with the Chao index estimating true ARG richness of 501–683 ARGs/sample (average: 577; Figure S1). When considering 16S rRNA gene normalized relative abundances, a clear trend emerged, whereby Asian sewage samples had higher relative abundances of ARGs than European/US samples (Figure 1A, Figures S2 and S3, Supplemental Data 4). Such a pattern is likely driven primarily by the relative inputs of ARGs and ARB from human populations, although other factors could be at play, including differences in relative industrial/hospital inputs or differential selection pressures when sewage travels through the collection network.55,56
Figure 1.
A: ARG distribution and relative abundance by corresponding antibiotic categories. ARGs were annotated via MetaStorm using CARD version 1.2.1. ARG categories were assigned in-house (Supplemental Data 1). Genes corresponding to two or more categories were labeled as “multidrug”. ARG abundances were normalized via MetaStorm to 16S rRNA gene abundances. B: Nonmetric multidimensional scaling (NMDS) ordination of WWTPs according to the ARG-based Bray–Curtis dissimilarities. Ellipses enclose sites of noted similarities, ranging from 0 to 100 (perfect similarity); thus, ellipses with a similarity of 80 represent sites with highest similarity. C: Relative abundance of the top 20 bacterial phyla in WWTP influents. Genus-level annotations were done via MG-RAST using the RefSeq database. D: NMDS ordination of WWTPs according to Euclidian distances between rlog-transformed reads to bacterial genera. Ellipses enclose sites of noted distances, where the shortest distance denotes the highest resemblance, and the longest distance denotes the least resemblance among the sites. Sample names refer to countries (IND: India, PHL: Philippines, USA: United States, CHE: Switzerland, HKG: Hong Kong, SWE: Sweden) along with the visit number (1,2) and WWTP plant number (P).
Hendriksen et al.11 recently reported a global sewage survey, and although the analytical approach was distinct, the trend in ARG relative abundance was strikingly similar to the present study: higher in Africa (median: 2,100 fragments per kilo base per million fragments [FPKM]), Asia (1,200 FPKM), South America (1,900 FPKM), and the Middle East (1,100 FPKM), relative to Europe (750 FPKM), North America (900 FPKM), and Oceania (800 FPKM). Hendriksen et al.11 applied FPKM normalization to address highly variable sequencing library depth (8–398 million reads per sample), while in the present study (20–30 million reads per sample), we normalized to 16S rRNA genes as a biomarker of total bacteria and correspondingly indicate the proportion of bacteria carrying ARGs.51 Hendriksen et al. also applied the ResFinder database57 for ARG annotation, which resulted in detection of an apparently larger number of ARGs. To better evaluate this observed difference, we processed our data with ResFinder using BLASTN58 and consistent annotation parameters (E-value < 1e-10, identity > 90%, and minimum length of 75 nucleotides). We identified a total of 784 distinct ARGs with an average of 320 (range: 234–385). Similarly, we processed the data presented in Hendriksen et al. using CARD and identified 1615 ARGs. Use of either CARD or Resfinder resulted in comparable numbers of total ARGs detected for both scenarios. We attribute the larger number of ARGs detected in Hendriksen et al. to reflect the larger number and diversity of samples examined. Best-hit annotation will inherently result in a higher number of diverse hits if the sample size is larger.
We further compared the ARG profiles (grouped by antibiotic classes) obtained using Resfinder (Figure S4). The general trend was consistent with that obtained using CARD (Figure 1) with Asian samples exhibiting higher relative ARG abundance compared to samples from Europe/US. However, the relative abundance levels were observed to be lower in the case of the Resfinder ARG profiles (0.1–0.4 copies/16S rRNA gene copies) compared to CARD (1.8–4.3 copies/16S rRNA gene copies). Resfinder is a nucleotide database that includes multiple similar variants of the same ARG and primarily incorporates ARGs that are horizontally acquired. Hence, any differences in the ARG profiles could inherently be attributed to the differences in the two databases. The differential number of ARGs included in different databases reflects the rapid evolution of the field of resistome bioinformatics59 and the need to improve comparability across studies for sewage surveillance.60
ARG compositional similarities were viewed via NMDS ordination derived from Bray–Curtis dissimilarity matrices (Figure 1B). Clustering within the NMDS plot was tightest for WWTP influents originating from the same country, with the exception of the two Philippines WWTPs. A second level of clustering was observed between the Asian and European/US samples. One exception was the HKG-P1 influent, which clustered separately from all other samples. This latter result is consistent with the saline nature of this sewage, where ocean water is used for toilet flushing. ANOSIM indicated that sewage samples strongly separated from one another when grouped by location (R = 0.741, p = 0.001). Clusters also strongly separated when grouped as Asia versus Europe/US (R = 0.700; p = 0.002). Three clusters, less strongly separated (R = 0.586; p = 0.001), were apparent when grouped by continent. Importantly, two sets of temporally independent influent samples collected from two Hong Kong WWTPs (HKG1-P1/HKG2-P1 vs HKG1-P2/HKG2-P2) clustered separately, but reproducibly, thus indicating less variation with time relative to geographic location. Ju et al.37 also noted a high level of repeatability when characterizing resistomes in sewage samples from different Swiss WWTPs. Other studies11,61 have similarly shown locational reproducibility, which is key for inferring that observed differences are primarily driven by location rather than the particular time of sample collection.
A number of clinically relevant ARGs (Supplemental Data 3) were detected in the influent of all WWTPs. OXA-type carbapenemase ARGs were ubiquitous and were detected at relatively high abundance (0.03–0.16 copies/16S rRNA gene copies). A variety of other β-lactamase ARG types (GES, CARB, SHV, CTX-M, and TEM) were also broadly detected, although at lower abundances. The qnrS gene (quinolone resistance) was detected in all sewages (0.001–0.24 copies/16S rRNA gene copies) and tended toward higher abundance in Asia (0.085 ± 0.099 copies/16S rRNA gene copies) relative to Europe/US (0.012 ± 0.011 copies/16S rRNA gene copies) but not significantly so (Wilcoxon rank-rum test, p = 0.1649). Others have noted differences in qnr carriage across populations,62,63 but statistical power here may have been insufficient to detect a difference. Reads annotated as mcr-1, mcr-2, and mcr-3 ARGs were found at all of the WWTPs except in HKG2-P1 (mcr-1 not detected) and in HKG1-P2 and IND1-P1 (mcr-2 not detected).
Core and Discriminatory Aspects of Resistomes
We subjected the collected data to core resistome analysis to further delineate similarities among sites. ARGs and resistance classes that were ubiquitous across all sites reflect the “core”. The core resistome of 216 ARGs (Supplemental Data 2) was comprised of ARGs conferring multidrug resistance and resistance to aminoglycosides, β-lactams, MLS, polymyxins, tetracyclines, and trimethoprims. The 25 most abundant ARGs found across all locations included one polymyxin (pmrE), one trimethoprim (dfrE), two tetracycline (tetQ and tetC), seven MLS (msrE, mphD, ermF, mel, macB, mefA, ermB), one sulfonamide (sul1), three multidrug (CRP, msbA, adeJ), two quinolone (qnrS2, qacH), one β-lactam (cfxA6), and two aminoglycoside (aph(3″)-lb and aph(6)-ld) ARGs (Table S3). Notably, among the core ARGs for which information was available, many were “ancient” or “natural background” ARGs identified in prior studies of pristine or less human-impacted environments (e.g., isolated caves, glacial soil, permafrost); thus, these ARGs likely predate the antibiotic era (Supplemental Data 5). However, other core ARGs are known to have become globally distributed more recently (e.g., sul1, sul2, dfrA3, tet(G), aadA8). Based on pairwise comparison, the number of ARGs shared between sites ranged from 296 to 401 (Figure S5) with 349 average shared annotations. The overall proportion of core ARGs averaged 69% (min = 62%, IND1-P2; max = 72%, HKG2-P1). Notably, the least number of reads in common were between one of the Swedish (SWE1-P1) versus one of the Hong Kong sewages (HKG2-P1), which aligns with the Asia versus Europe/US divide in resistome similarity. Consistent with the repeatability of the sampling events, the greatest number of common ARGs was noted for two Hong Kong sewage samples collected six months apart. We note that our definition of the core resistome is operationally defined and as such is dependent on sample sequencing depth. Deeper sequencing may detect additional genes that are shared across all samples.
To focus analysis on the “variable” resistome, we removed core ARGs from the profiles in Figure 1A. This accentuated the continental differences in the sewage resistomes (Figure S6). Specifically, the relative proportions of aminoglycoside, β-lactam, rifamycin, sulfonamide, trimethoprim, and quinolone ARGs tended to increase, while those of MLS, multidrug, polymyxin, and tetracycline ARGs tended to decrease. Interestingly, the proportion of ARGs in the core tended to increase from Asia to Europe/US (Figure S7), which hypothetically could reflect a longer history of antibiotic use and management in Europe/US.
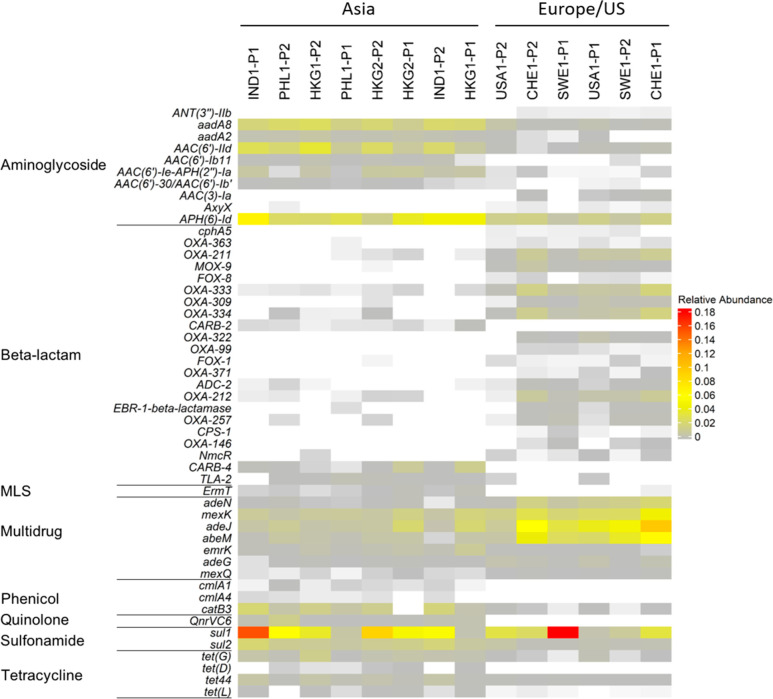
We next characterized the “discriminatory” resistomes (i.e., ARGs that are, in terms of presence or abundance, indicative of local population or region and thus enable locational differentiation). We applied the ExtrARG machine learning-based algorithm19 to identify ARGs that discriminate sewage samples based on a priori selected groups. Sewage samples were grouped at the continent level (i.e., Asia vs Europe/US), and then the relative abundances of the 50 most discriminatory ARGs were visualized using a heatmap (Figure 2). Two aminoglycoside ARGs, aadA8 and aadA2, were prominent in Asia, while ant(3′′)-IIb, aac(3)-Ia, and axyX were more prominent in Europe/US. Various β-lactam ARGs (blaCphA5, blaOXA-363, blaOXA-309, blaOXA-371, blaFOX-8, blaFOX-1, blaCPS-1) were abundant in Europe/US but were found at low concentrations in Asian sewage. Interestingly, blaCARB-2 and blaCARB-4 were not detected in European/US sewage but were found in Asia. The MLS ARG ermT, phenicol ARGs cmlA1 and cmlA4, quinolone ARG qnrVC6, and the tetracycline ARG tet(L) were primarily detected in Asian sewage. The sulfonamide ARG, sul2, was 3× to 5× more abundant in Asian sewage, while a number of the multidrug ARGs (e.g., efflux pumps adeJ, mexK, abeM), were 3× to 15× higher in sewage from Europe/US. Further, we also found that a number of ARGs identified as discriminatory were only discovered within the past decade (e.g., ant(3′′)-IIb, axyX, blaCphA5, blaCPS-1, blaFOX-8, qnrVC6, and qnrVC6; Supplemental Data 6). We suggest that determination of the location-specific distribution of discriminatory ARGs in sewage can be used to evaluate if their global distribution is widening. Prior studies64 indicate that ARG variants within the same ARG class can arise independently from one another prior to becoming common within the human microbiome, and thus, changes in global distribution may suggest potential clinical concern. Our study illustrates the potential of such an approach. However, further validation of its potential will require a greater number of samples per country/region as well as additional historical and epidemiological evidence to ground truth the observations.19,35 Further optimization of metagenomic data generation, such as determination of appropriate sequencing depths, would also be beneficial.
Figure 2.
Heatmap representing the relative abundance of discriminatory ARGs for influent samples grouped according to their sampling location identified using the ExtrARG package.
Comparison of Microbiomes Across the Global Transect of Sewage Samples
Similar to the resistomes, NMDS analysis of the sewage microbiomes (i.e., bacterial taxonomic composition derived from metagenomic data) resulted in clustering of samples as a function of geographic location (Figure 1D, Figure S8). ANOSIM indicated that the sewage microbiomes were most strongly separated when grouped by country (R = 0.701, p = 0.001). When grouped by Asia versus Europe/US, the groups were better separated (R = 0.421, p = 0.001) than when grouped by continent (i.e., Asia vs Europe vs US; R = 0.3; p = 0.003). As was the case for the resistomes, Hong Kong WWTP1 (HKG-P1) clustered separately from the other sewage samples. Similar patterns in the clustering of the resistomes and microbiomes are expected, since phylogenetic constraints to ARG carriage are well-known (e.g., ref (37)); however, differences between the two may signal the broader mobility of the resistome.65
In accordance with the ARG-based class separations for Asia versus Europe/US, LEfSe analysis was performed on the microbiome data (Figure S9). Four and ten classifying genera were identified in European/US versus Asian sewages, respectively. European/US sewages were distinguished by genera belonging to Betaproteobacteria, while Clostridia or Negativicute classes of the Firmicute phylum were discriminatory in Asian sewage. Only two non-Firmicutes genera (Dehalococcoides and Shigella) were found to be discriminatory in Asian sewage. Typically, higher levels of Proteobacteria are thought to reflect environmental conditions within the sewer system, while Firmicutes reflect greater human fecal influence.66,67
Potential parallels between the microbiome and resistome compositions were investigated. First, the Mantel test was applied to either full or continent-specific distance matrices representative of the microbiome (Euclidean distances between log-transformed reads to bacterial genera) and resistome (Bray–Curtis dissimilarity matrix). When comparing all of the collection sites, a strong positive correlation was observed between the ARG and genus-based taxonomy distance matrices (r = 0.74, p < 0.001). When separated by continent, a stronger correlation was observed between the Asian distance matrices (r = 0.80, p < 0.001) than between the European/USA distance matrices (r = 0.48, p = 0.03). For comparative purposes, a Bray–Curtis dissimilarity matrix was generated based on the MetaPhlAn68 output of taxonomic assignments obtained from MetaStorm18 for Mantel-based comparison to the ARG similarity matrix. Again, a strong correlation was observed, both overall (r = 0.76, p < 0.001) and when compared continentally within Asia (r = 0.80, p < 0.001) and Europe/US (r = 0.56, p < 0.01).
Common pathogenic hosts of the discriminatory ARGs identified in Figure 2 were also examined (Table S4). Based on the data reported in the CARD database,54 82 pathogenic bacterial species are known to host these ARGs. Interestingly, discriminatory ARGs most representative of Asian sewages tended to have a broader range of potential host pathogens, whereas discriminatory ARGs associated with European/US sewages tended to have a narrower range.
Genes Providing Opportunities for Coselection and Horizontal Gene Transfer
Genes documented to sometimes be found on plasmids were investigated as indicators of horizontal gene transfer potential. The average relative abundance of such genes identified via alignment to the ACLAME database was 56.7 copies/16S rRNA gene copies, with an observed minimum of 33.3 copies/16S rRNA gene copies in the Philippines (PHL1-P1) and a maximum of 111.6 copies/16S rRNA gene copies in Hong Kong (HKG2-P1; Figure S10). No clear country- or continent-specific patterns were observed in terms of abundance.
Potential associations between ARGs and MRGs known to sometimes occur on plasmids or other MGEs were investigated and visualized via network analysis of assembled scaffolds (Figure S11). While acknowledging that there is uncertainty in the accuracy of scaffolds assembled from metagenomic data,69 we carried out the analysis for empirical comparison assuming a consistent error rate across a sample set generated from the same sequencing platform. A substantial portion of reads were assembled, averaging 34% and ranging from 24% (HKG1-P1) to 50% (PHL1-P2). For each sewage sample, the number of generated scaffolds ranged from 64,576 (USA1-P1) to 243,356 (CHE1-P2), with an average length of 719 bp. All of the major classes of ARGs were detected on the scaffolds, with the most frequently observed belonging to the multidrug, MLS, glycopeptide, and tetracycline categories (Figure S12). The relative distribution of ARGs (based on antibiotic category) among the scaffolds was remarkably similar for all sewage samples. Across all locations, the most common ARGs with plasmid associations were pmrE (peptide ARG), acrB (multidrug efflux), dfrE (trimethoprim ARG), MuxB (multidrug efflux), and rosB (peptide ARG) with 301, 186, 133, 125, and 101 co-occurrences, respectively. pmrE and dfrE were present at high abundance at all of the sites, thus suggesting globalization of these genes. Other notable plasmid ARG co-occurrences include sul1 (sulfonamide ARG, 20 co-occurrences), MCR-3 (peptide, 32 co-occurrences), and the blaCARB, blaOXA, and blaTEM β-lactam ARGs (between 1 and 13 co-occurrences). sul1 is known to be highly associated with both class 1 integrons70 as well as plasmids. Some of these co-occurrences correspond to resistance against carbapenems and colistin, which are critically important antibiotics of last resort. We note that co-occurrence of genes within metagenomes does not necessarily indicate co-occurrence within an individual strain, and thus this result should not be overinterpreted. Nonetheless, this analysis illustrates that clinically important classes of ARGs were readily found on assembled scaffolds that contained genes commonly found on plasmids, an indicator that they are potentially in a mobile form in sewage.
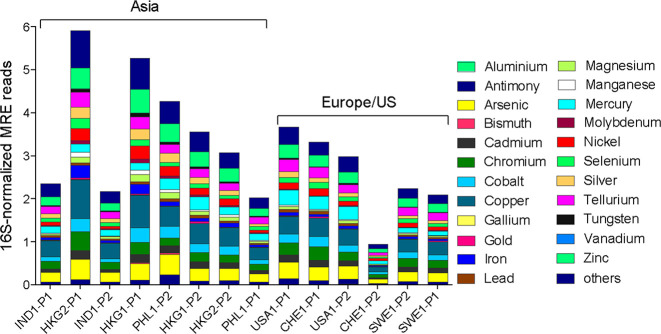
ARGs and MRGs are often subject to coselection pressure when they are both present on a single genetic element, such as a plasmid, or cross-selection if the same gene is both an ARG and an MRG, as is commonly the case for multidrug efflux pumps capable of pumping both metals and antibiotics.71 The ranking of total MRG relative abundances, identified via alignment to the BacMet database,50 was strikingly similar to that of the total ARG relative abundances (Spearman r = 0.66, p = 0.013), except both Indian WWTPs and one Philippines plant ranked much lower than the other Asian WWTPs, while a Swiss WWTP had extremely low MRG relative abundance (Figure 3). This finding is suggestive that there can be common drivers, such as metals, antibiotics, or other agents, either within human gastrointestinal tracts, in industrial inputs, or in sewers themselves, that exert selection pressure for carriage of both ARGs and MRGs.72
Figure 3.
Distribution and relative abundance of metal resistance genes in WWTP influents. Metal resistance genes were annotated via MetaStorm using the BacMet database version 1.1. Metal resistance gene abundances were normalized to 16S rRNA gene abundances. Sample names refer to countries (IND: India, PHL: Philippines, USA: United States, CHE: Switzerland, HKG: Hong Kong, SWE: Sweden) along with the visit number and WWTP plant (P) number. Order is according to the ranked comparison of total ARG relative abundance shown in Figure 1.
Across all locations, multidrug ARGs were the most common type associated with MRGs (Figure S13). The most common co-occurrences within the assembled scaffolds were noted between acrB and MexB ARGs, colocated with copper/zinc MRGs (160 and 83 co-occurrences, respectively), and muxB ARG, colocated with zinc MRGs (96 co-occurrences). These colocations were consistently among the most abundant at all sample sites. A literature review was conducted to examine whether any of these presumed co-occurrences were actually the same gene and thus indicative of cross- rather than coselection. We identified nine genes that were consistently abundant across all the sample sites and confer resistance to both antibiotics and metals, namely, mdtA, mdtB, mdtC, baeS, baeR, cmeB, acrD, mexI, and pmrC. As anticipated, most of these genes are known to be associated with efflux pump systems (Supplemental Data 7).
Sewage Antibiotic Concentrations
Antibiotic, pharmaceutical, and personal care product concentrations measured in these sewage samples were recently reported in detail by Singh et al.,17 and trends relevant to the present study are summarized in Table S5. We note that chemical stability issues precluded quantification of the β-lactams (The penicillins, cephalosporins, and carbapenems all contain a β-lactam ring that readily hydrolyzes in water to form open ring degradation products.73−75) and that, due to logistical constraints, the antibiotic measurements for the Indian sewage were made during a sampling trip that occurred one year later. We measured the highest antibiotic concentrations in Hong Kong sewage, where total concentrations of macrolides, quinolones, sulfonamides, tetracyclines, and trimethoprim reached levels >63,000 ng/L. The antibiotics with the highest concentrations in the sewage were ciprofloxacin (48,100 ng/L, HKG1-P1) and clarithromycin (39,551 ng/L, IND2-P2). In particular, Indian sewage contained very high levels of anhydro-erythromycin and norfloxacin.
To contextualize these measured antibiotic concentrations, we first compared them to predicted no effect concentrations (PNECs; Table S5)76 for resistance selection. All samples, except one from Sweden, were above the PNEC for ciprofloxacin, while norfloxacin was near or above the PNEC in both Swiss sewages and one Indian sewage sample. Clarithromycin was above the PNEC in one US sample. Two Hong Kong sewage samples had concentrations greater than the PNEC for the tetracyclines, while both the Hong Kong and Indian sewages contained the macrolides azithromycin and clarithromycin at levels exceeding the PNEC. It is noted that PNEC values provide a point of reference, but measured concentrations that either exceed or fall short of a PNEC do not necessarily delineate the presence or absence of selection pressure within sewage.76 Resistance metrics quantified in sewage likely reflect both current and historic drivers, including past patterns of antibiotic use and selection pressures that may have occurred in the human gut, that collectively shape the resistome; however, antibiotic measurements only reflect usage at the time of sampling.
We were also interested if measured antibiotic concentrations reflect reported antibiotic sales data. Antibiotic consumption data between 2000 and 2015 for each site was obtained from ResistanceMap.77 As shown in Figure S14, total antibiotic consumption (defined daily doses (DDD) per 1000 population) was reported to be highest in the US, followed by Hong Kong, Sweden, and Switzerland.78 Reported antibiotic use in India and the Philippines has historically been lower than that in Europe/US, but in recent years, reported use in India has increased and as of 2015 is estimated to be similar to that in Sweden and Switzerland. Reported antibiotic use in the Philippines lags relative to other countries and remained fairly stable over the 2000–2015 period.
Based upon sales data alone, while acknowledging geographically and socially variable water usage rates, one might expect that antibiotic concentrations within European/US sewage would be higher than concentrations measured within Asian sewage. In fact, we observe the exact opposite, with much higher levels in Asia (Figure S15). The sum of concentrations of measured antibiotics in both Hong Kong and India was more than 25 times higher than concentrations measured in Sweden and about 10 times higher than in Switzerland. This observation highlights a disconnect between antibiotic sales data and measured sewage concentrations, which cannot be explained by differences in water use. Systems for collecting sales data for antibiotics differ considerably between world regions. In some regions, there may be a substantial portion of antibiotic use that is not easily accounted for through public sales statistics. It may therefore be that trends identified through wastewater surveillance more accurately reflect true usage differences. The measured ARG abundances reflect the trends in the measured antibiotic data (i.e., Hong Kong and Indian sewage ranked highest in total ARG relative abundance; Figure 1A). Given the established link between antibiotic use as a major driver for resistance, this would be in agreement with a higher, not lower, regional use of antibiotics.
While our study only provides a snapshot, as suggested by ref (79) and others, we support the use of WBS of antibiotics as a means for verifying and complementing reported sales and consumption data for antibiotics. The disconnect between antibiotic sales and ARG abundance may reflect the absence of a relationship between antibiotic usage and resistance or it may simply reflect the aforementioned quality of the sales data itself. It may be challenging to accurately estimate absolute levels of use from sewage analyses; still relative differences over time and space could be of great value. It is recognized that some antibiotics (e.g., β-lactams) have short half-lives and/or variable recoveries, and for such antibiotics, it would be particularly challenging to translate sewage concentrations to regional usage. Finally, access to water infrastructure and water usage patterns vary geographically and need to be taken into consideration.
We also explored if prior reports of links between antibiotic concentrations and resistant clinical isolates58,59 held true for sewage resistomes. Overall, few correlations between ARGs and antibiotics were observed (SI Discussion, Supplemental Data 8), which is consistent with other studies.11,22
Surveillance Effectively Reveals Key Sewage Resistome Trends
We observed that the relative abundances of total ARGs were elevated in Asian versus European/US sewages, consistent with general perceptions of rigor in antibiotic use policy across countries22 and the generally higher antibiotic loadings in Asia. The lowest relative abundances of ARGs were found in the sewage from Sweden which has incorporated numerous proactive measures to avoid misuse and overuse of antibiotics.80 Hong Kong, India, and the Philippines, on the other hand, are characterized by dense urban populations and either have problems with illegal antibiotic sales81 or do not require prescriptions for antibiotic use.82
Removal of core ARGs (i.e., those ubiquitously detected in urban sewage) from the resistome profiles accentuated the differences between Asia and Europe/US, while delineation of discriminatory ARGs identified those that uniquely circulate in each corresponding region. In general, a greater variety of ARGs across multiple resistance classes were identified as discriminatory in Asian sewage, while a subset of aminoglycoside and multidrug ARGs was characteristic of European/US sewages. Such knowledge could potentially be used to infer information about the ARGs within local regions in terms of their prevalence, novelty, or their potential carriage by pathogens. While discriminatory analysis could help identify large scale trends, it could be especially valuable when used as a complement to clinical monitoring.
Surveillance is a key component of the global action plan to combat antimicrobial resistance.35,83 Within that context, consistent methods and approaches are required to facilitate robust data comparability. The approach outlined herein provides such consistency and can support analyses requiring high granularity (e.g., comparing local sewersheds, sampling within a sewershed). However, it would be costly to apply everywhere due to its reliance on a dedicated sampling and analysis team. The alternative approach,11 relying upon centralized processing of large numbers of shipped samples, has the capacity to more broadly elucidate global trends. Both approaches are of value moving forward, depending on specific aims, but consistent sample collection and sequencing approaches will be critical to ensuring comparable data are produced to address larger research questions.35 In the future, addition of internal standards and verification that sequencing depths are sufficient to address monitoring objectives would also be a valuable means to enhance quantitative capacity and overall data comparability.84 Implementation of coordinated sewage resistome surveillance efforts will be key to improved understanding of baseline resistome characteristics.59 Such information can be used as a reference point for follow-up sampling to aid in early identification of previously unobserved ARGs that may pose clinical threats, including the emergence of new resistance types.25
Implications for Wastewater-Based Surveillance
WBS is rapidly gaining attention as a highly promising tool for infectious disease monitoring.2,4,11,12,39,85 Here, we demonstrated the potential for metagenomic-based surveillance of ARGs and other key metainformation in sewage to provide a foundation for WBS of antibiotic resistance, and we have illustrated challenges that must be considered as the field develops. In particular, it is critical to define WBS surveillance objectives including the following: what will be monitored, how samples (and appropriate metainformation) will be collected and analyzed, and how the data will ultimately be interrogated prior to investing in extensive sampling campaigns. The hypothesis that the sewage resistome composition differs by geographical location could only be supported through the use of consistent experimental and data analysis protocols, which will be particularly vital in future efforts to further dissect the role of geographical subfactors, such as socioeconomics, population density, antibiotic use, diet, sanitation status, and local climate/temperature.22,23
Comparison of results obtained using widely varying methods (e.g., nucleic acid extraction kits,16 solid-phase extraction protocols17) or metagenomic platforms presents inherent limitations to identifying key factors driving the spread of antibiotic resistance. Data analysis approaches are also important to consider. For example, the CARD database was preferred here over ResFinder, because it is the most comprehensive and is extensively curated. The most recent version of CARD no longer requires manual curation, because SNPs in housekeeping genes are accordingly indicated. However, if the goal is to discover new ARGs, then machine learning or other computational methods may be warranted.86,87 Also, here we normalized ARGs to 16S rRNA genes, because it is the most commonly applied denominator in the literature; but we recognize that normalizing to single copy housekeeping genes88,89 may be a more logical approach to obtaining per cell estimates in the future. Comparative studies leading toward commonly adopted methods would be of value.
The WBS approach defined herein demonstrates a means to delineate specific differences among sewage samples as a function of geography and related covariates. Identification of geographic “hot spots” where attention toward curbing the spread of antibiotic resistance is urgently needed. Incorporating locations in South America and Africa would help determine if there are additional key resistome distinctions among presumed hot spots indicated in prior studies.11 Expanding WBS is especially relevant as we emerge from the new era of pandemic awareness and concern, where multidrug resistant pathogens, ARGs, and infectious viruses have been observed to rapidly spread across international borders.90 Estimates that several thousands of deaths per year globally can be attributed to antibiotic resistance amplifies the need for coordinated, global surveillance. Ultimately, widescale online implementation of metagenomic surveillance of ARGs and other disease determinants within sewage in the context of a WBS framework promises to yield new insights.2,25,35 Such insights will help illuminate the specific factors that drive the dissemination and spread not only of antibiotic resistance but also other diseases of emerging concern.
Acknowledgments
This material is based upon work supported by the U.S. National Science Foundation (NSF) Award OISE: 1545756. Additional funding to D.G.J.L. was provided by the Swedish research council VR (2018-05771) and the Region Västra Götaland under the ALF agreement (grant number ALFGBG-717901). Funding for H.B. was provided by the Swiss National Science Foundation under the NRP72 “Antimicrobial resistance”, grant 167116. Portions of the TOC image were created using BioRender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.1c08673.
Sewage sludge characteristics; sequence metrics; 25 most abundant ARGs; pathogen ARG host listing; antibiotic concentrations; ARG richness; percent relative abundance of ARG classes; heatmap of ARG relative abundance; ARG distribution annotated using Resfinder; shared ARGs across sites; variable resistome composition; proportions of ARGs in core and discriminatory resistomes; bacterial genera across sites; LefSe analysis; plasmid associated reads; co-occurrence of ARGs and plasmid gene markers; heatmap of resistance within assembled scaffolds; co-occurrence of ARGs and MRGs; and antibiotic consumption rates for Asia vs Europe/US (PDF)
Supplemental datasheets: 1) manually curated listing of ARGs and ARG categorization; 2) core ARGs; 3) clinically relevant ARGs; 4) 16S normalized abundances; 5) ancient ARGs; 6) discriminatory ARGs; 7) genes conferring resistance to antibiotics and metals; and 8) correlations between ARGs and antibiotics (XLSX)
Author Contributions
◇ M.V.P.R., E.G., and S.G. contributed equally.
The authors declare no competing financial interest.
Notes
Data Availability: The wastewater influent metagenomic data sets have been deposited in NCBI Short Read Archive (SRA) under bioproject PRJNA527877.
Supplementary Material
References
- Larsen D. A.; Green H.; Collins M. B.; Kmush B. L. Wastewater Monitoring, Surveillance and Epidemiology: A Review of Terminology for a Common Understanding. FEMS Microbes 2021, 2, xtab011. 10.1093/femsmc/xtab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers P. M. C.; Flach C. F.; Larsson D. G. J. A Conceptual Framework for the Environmental Surveillance of Antibiotics and Antibiotic Resistance. Environ. Int. 2019, 130, 104880. 10.1016/j.envint.2019.05.074. [DOI] [PubMed] [Google Scholar]
- Sims N.; Kasprzyk-Hordern B. Future Perspectives of Wastewater-Based Epidemiology: Monitoring Infectious Disease Spread and Resistance to the Community Level. Environ. Int. 2020, 139, 105689. 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P. M.; Tscharke B. J.; Donner E.; O’Brien J. W.; Grant S. C.; Kaserzon S. L.; Mackie R.; O’Malley E.; Crosbie N. D.; Thomas K. V.; Mueller J. F. Wastewater-Based Epidemiology Biomarkers: Past, Present and Future. TrAC-Trends in Analytical Chemistry 2018, 105, 453–469. 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- Zuccato E.; Chiabrando C.; Castiglioni S.; Bagnati R.; Fanelli R. Estimating Community Drug Abuse by Wastewater Analysis. Environ. Health Perspect 2008, 116 (8), 1027–32. 10.1289/ehp.11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovi T.; Shulman L. M.; van der Avoort H.; Deshpande J.; Roivainen M.; EM D. E. G. Role of Environmental Poliovirus Surveillance in Global Polio Eradication and Beyond. Epidemiol Infect 2012, 140 (1), 1–13. 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- Sikorski M. J.; Levine M. M. Reviving the “Moore Swab”: A Classic Environmental Surveillance Tool Involving Filtration of Flowing Surface Water and Sewage Water to Recover Typhoidal Salmonella Bacteria. Appl. Environ. Microbiol. 2020, 86 (13), e00060-20. 10.1128/AEM.00060-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G.; Been F.; Heijnen L.; Petterson S. Implementation of Environmental Surveillance for SARS-Cov-2 Virus to Support Public Health Decisions: Opportunities and Challenges. Curr. Opin Environ. Sci. Health 2020, 17, 49. 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo D.; Quintela-Baluja M.; Corbishley A.; Jones D. L.; Singer A. C.; Graham D. W.; Romalde J. L. Making Waves: Wastewater-Based Epidemiology for Covid-19 - Approaches and Challenges for Surveillance and Prediction. Water Res. 2020, 186, 116404. 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S.; Gwee S. X. W.; Ng J. Q. X.; Lau N.; Koh J.; Pang J. Wastewater Surveillance to Infer Covid-19 Transmission: A Systematic Review. Sci. Total Environ. 2022, 804, 150060. 10.1016/j.scitotenv.2021.150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen R. S.; Munk P.; Njage P.; van Bunnik B.; McNally L.; Lukjancenko O.; Roder T.; Nieuwenhuijse D.; Pedersen S. K.; Kjeldgaard J.; Kaas R. S.; Clausen P.; Vogt J. K.; Leekitcharoenphon P.; van de Schans M. G. M.; Zuidema T.; de Roda Husman A. M.; Rasmussen S.; Petersen B.; Amid C.; Cochrane G.; Sicheritz-Ponten T.; Schmitt H.; Alvarez J. R. M.; Aidara-Kane A.; Pamp S. J.; Lund O.; Hald T.; Woolhouse M.; Koopmans M. P.; Vigre H.; Petersen T. N.; Aarestrup F. M. Global Monitoring of Antimicrobial Resistance Based on Metagenomics Analyses of Urban Sewage. Nat. Commun. 2019, 10 (1), 1124. 10.1038/s41467-019-08853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pärnänen K. M. M.; Narciso-da-Rocha C.; Kneis D.; Berendonk T. U.; Cacace D.; Do T. T.; Elpers C.; Fatta-Kassinos D.; Henriques I.; Jaeger T.; Karkman A.; Martinez J. L.; Michael S. G.; Michael-Kordatou I.; O’Sullivan K.; Rodriguez-Mozaz S.; Schwartz T.; Sheng H.; Sørum H.; Stedtfeld R. D.; Tiedje J. M.; Giustina S. V. D.; Walsh F.; Vaz-Moreira I.; Virta M.; Manaia C. M. Antibiotic Resistance in European Wastewater Treatment Plants Mirrors the Pattern of Clinical Antibiotic Resistance Prevalence. Science Advances 2019, 5 (3), eaau9124. 10.1126/sciadv.aau9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkman A.; Berglund F.; Flach C. F.; Kristiansson E.; Larsson D. G. J. Predicting Clinical Resistance Prevalence Using Sewage Metagenomic Data. Communications Biology 2020, 3 (1), 711. 10.1038/s42003-020-01439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez E. A.; Ramirez D.; Balcázar J. L.; Jiménez J. N. Metagenomic Analysis of Urban Wastewater Resistome and Mobilome: A Support for Antimicrobial Resistance Surveillance in an Endemic Country. Environ. Pollut. 2021, 276, 116736. 10.1016/j.envpol.2021.116736. [DOI] [PubMed] [Google Scholar]
- Daughton C. G. Monitoring Wastewater for Assessing Community Health: Sewage Chemical-Information Mining (SCIM). Sci. Total Environ. 2018, 619–620, 748–764. 10.1016/j.scitotenv.2017.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A. D.; Metch J. W.; Wang Y. L.; Garner E.; Zhang A. N.; Riquelme M. V.; Vikesland P. J.; Pruden A.; Zhang T. Effects of Sample Preservation and DNA Extraction on Enumeration of Antibiotic Resistance Genes in Wastewater. FEMS Microbiology Ecology 2018, 94 (2), fix189. 10.1093/femsec/fix189. [DOI] [PubMed] [Google Scholar]
- Singh R. R.; Angeles L. F.; Butryn D. M.; Metch J. W.; Garner E.; Vikesland P. J.; Aga D. S. Towards a Harmonized Method for the Global Reconnaissance of Multi-Class Antimicrobials and Other Pharmaceuticals in Wastewater and Receiving Surface Waters. Environ. Int. 2019, 124, 361–369. 10.1016/j.envint.2019.01.025. [DOI] [PubMed] [Google Scholar]
- Arango-Argoty G.; Singh G.; Heath L. S.; Pruden A.; Xiao W. D.; Zhang L. Q. Metastorm: A Public Resource for Customizable Metagenomics Annotation. PloS One 2016, 11 (9), e0162442. 10.1371/journal.pone.0162442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.; Arango-Argoty G.; Zhang L.; Pruden A.; Vikesland P. Identification of Discriminatory Antibiotic Resistance Genes among Environmental Resistomes Using Extremely Randomized Tree Algorithm. Microbiome 2019, 7 (1), 123. 10.1186/s40168-019-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango-Argoty G. A.; Guron G. K. P.; Garner E.; Riquelme M. V.; Heath L. S.; Pruden A.; Vikesland P. J.; Zhang L. ARGminer: A Web Platform for Crowdsourcing-Based Curation of Antibiotic Resistance Genes. Bioinformatics 2020, 36 (9), 2966–2973. 10.1093/bioinformatics/btaa095. [DOI] [PubMed] [Google Scholar]
- Bürgmann H.; Frigon D.; Gaze W. H.; Manaia C. M.; Pruden A.; Singer A. C.; Smets B. F.; Zhang T. Water and Sanitation: An Essential Battlefront in the War on Antimicrobial Resistance. FEMS Microbiology Ecology 2018, 94 (9), fiy101. 10.1093/femsec/fiy101. [DOI] [PubMed] [Google Scholar]
- Collignon P.; Beggs J. J.; Walsh T. R.; Gandra S.; Laxminarayan R. Anthropological and Socioeconomic Factors Contributing to Global Antimicrobial Resistance: A Univariate and Multivariable Analysis. Lancet Planet Health 2018, 2 (9), e398–e405. 10.1016/S2542-5196(18)30186-4. [DOI] [PubMed] [Google Scholar]
- Vikesland P.; Garner E.; Gupta S.; Kang S.; Maile-Moskowitz A.; Zhu N. Differential Drivers of Antimicrobial Resistance across the World. Acc. Chem. Res. 2019, 52, 916. 10.1021/acs.accounts.8b00643. [DOI] [PubMed] [Google Scholar]
- Karkman A.; Parnanen K.; Larsson D. G. J. Fecal Pollution Can Explain Antibiotic Resistance Gene Abundances in Anthropogenically Impacted Environments. Nat. Commun. 2019, 10 (1), 80. 10.1038/s41467-018-07992-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarestrup F. M.; Woolhouse M. Using Sewage for Surveillance of Antimicrobial Resistance. Science 2020, 367, 630–632. 10.1126/science.aba3432. [DOI] [PubMed] [Google Scholar]
- Hutinel M.; Huijbers P. M. C.; Fick J.; Ahren C.; Larsson D. G. J.; Flach C. F. Population-Level Surveillance of Antibiotic Resistance in Escherichia Coli through Sewage Analysis. Euro Surveillance 2019, 24 (37), 1800497. 10.2807/1560-7917.ES.2019.24.37.1800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miłobedzka A.; Ferreira C.; Vaz-Moreira I.; Calderón-Franco D.; Gorecki A.; Purkrtova S.; Jan B.; Dziewit L.; Singleton C. M.; Nielsen P. H.; Weissbrodt D. G.; Manaia C. M. Monitoring Antibiotic Resistance Genes in Wastewater Environments: The Challenges of Filling a Gap in the One-Health Cycle. J. Hazard Mater. 2022, 424, 127407. 10.1016/j.jhazmat.2021.127407. [DOI] [PubMed] [Google Scholar]
- Wu L.; Ning D.; Zhang B.; Li Y.; Zhang P.; Shan X.; Zhang Q.; Brown M. R.; Li Z.; Van Nostrand J. D.; Ling F.; Xiao N.; Zhang Y.; Vierheilig J.; Wells G. F.; Yang Y.; Deng Y.; Tu Q.; Wang A.; Global Water Microbiome C.; Zhang T.; He Z.; Keller J.; Nielsen P. H.; Alvarez P. J. J.; Criddle C. S.; Wagner M.; Tiedje J. M.; He Q.; Curtis T. P.; Stahl D. A.; Alvarez-Cohen L.; Rittmann B. E.; Wen X.; Zhou J. Global Diversity and Biogeography of Bacterial Communities in Wastewater Treatment Plants. Nat. Microbiol 2019, 4 (7), 1183–1195. 10.1038/s41564-019-0426-5. [DOI] [PubMed] [Google Scholar]
- Bibby K.; Crank K.; Greaves J.; Li X.; Wu Z.; Hamza I. A.; Stachler E. Metagenomics and the Development of Viral Water Quality Tools. NPJ. Clean Water 2019, 2 (1), 9. 10.1038/s41545-019-0032-3. [DOI] [Google Scholar]
- Izquierdo-Lara R.; Elsinga G.; Heijnen L.; Oude Munnink B. B.; Schapendonk C. M. E.; Nieuwenhuijse D.; Kon M.; Lu L.; Aarestrup F. M.; Lycett S.; Medema G.; Koopmans M.; de Graaf M. Monitoring SARS-CoV-2 Circulation and Diversity through Community Wastewater Sequencing, the Netherlands and Belgium. Emerging Infectious Diseases 2021, 27 (5), 1405–1415. 10.3201/eid2705.204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits-Christoph A.; Kantor R. S.; Olm M. R.; Whitney O. N.; Al-Shayeb B.; Lou Y. C.; Flamholz A.; Kennedy L. C.; Greenwald H.; Hinkle A.; Hetzel J.; Spitzer S.; Koble J.; Tan A.; Hyde F.; Schroth G.; Kuersten S.; Banfield J. F.; Nelson K. L. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 2021, 12 (1), e02703-20. 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Zhang X. X.; Huang K.; Miao Y.; Shi P.; Liu B.; Long C.; Li A. Metagenomic Profiling of Antibiotic Resistance Genes and Mobile Genetic Elements in a Tannery Wastewater Treatment Plant. PloS one 2013, 8 (10), e76079. 10.1371/journal.pone.0076079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. H.; Li J.; Chen H.; Bond P. L.; Yuan Z. G. Metagenomic Analysis Reveals Wastewater Treatment Plants as Hotspots of Antibiotic Resistance Genes and Mobile Genetic Elements. Water Res. 2017, 123, 468–478. 10.1016/j.watres.2017.07.002. [DOI] [PubMed] [Google Scholar]
- D’Costa V. M.; McGrann K. M.; Hughes D. W.; Wright G. D. Sampling the Antibiotic Resistome. Science 2006, 311, 374–377. 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- Pruden A.; Vikesland P. J.; Davis B. C.; de Roda Husman A. M. Seizing the Moment: Now Is the Time for Integrated Global Surveillance of Antimicrobial Resistance in Wastewater Environments. Curr. Opin. Microbiol. 2021, 64, 91–99. 10.1016/j.mib.2021.09.013. [DOI] [PubMed] [Google Scholar]
- Pärnänen K.; Karkman A.; Tamminen M.; Lyra C.; Hultman J.; Paulin L.; Virta M. Evaluating the Mobility Potential of Antibiotic Resistance Genes in Environmental Resistomes without Metagenomics. Sci. Rep 2016, 6, 35790. 10.1038/srep35790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju F.; Beck K.; Yin X.; Maccagnan A.; McArdell C. S.; Singer H. P.; Johnson D. R.; Zhang T.; Burgmann H. Wastewater Treatment Plant Resistomes Are Shaped by Bacterial Composition, Genetic Exchange, and Upregulated Expression in the Effluent Microbiomes. ISME Journal 2019, 13 (2), 346–360. 10.1038/s41396-018-0277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narciso-da-Rocha C.; Rocha J.; Vaz-Moreira I.; Lira F.; Tamames J.; Henriques I.; Martinez J. L.; Manaia C. M. Bacterial Lineages Putatively Associated with the Dissemination of Antibiotic Resistance Genes in a Full-Scale Urban Wastewater Treatment Plant. Environ. Int. 2018, 118, 179–188. 10.1016/j.envint.2018.05.040. [DOI] [PubMed] [Google Scholar]
- Su J. Q.; An X. L.; Li B.; Chen Q. L.; Gillings M. R.; Chen H.; Zhang T.; Zhu Y. G. Metagenomics of Urban Sewage Identifies an Extensively Shared Antibiotic Resistome in China. Microbiome 2017, 5 (1), 84. 10.1186/s40168-017-0298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck C.; Albertsen M.; Telke A.; Ellabaan M.; Nielsen P. H.; Sommer M. O. A. Limited Dissemination of the Wastewater Treatment Plant Core Resistome. Nat. Commun. 2015, 6, 8452. 10.1038/ncomms9452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen H. K.; Moe L. A.; Rodbumrer J.; Gaarder A.; Handelsman J. Functional Metagenomics Reveals Diverse Beta-Lactamases in a Remote Alaskan Soil. Isme Journal 2009, 3 (2), 243–251. 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- D’Costa V. M.; King C. E.; Kalan L.; Morar M.; Sung W. W.; Schwarz C.; Froese D.; Zazula G.; Calmels F.; Debruyne R.; Golding G. B.; Poinar H. N.; Wright G. D. Antibiotic Resistance Is Ancient. Nature 2011, 477 (7365), 457–61. 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- Dai D.; Brown C.; Burgmann H.; Larsson D. G. J.; Nambi I.; Zhang T.; Flach C. F.; Pruden A.; Vikesland P. J. Long-Read Metagenomic Sequencing Reveals Shifts in Associations of Antibiotic Resistance Genes with Mobile Genetic Elements from Sewage to Activated Sludge. Microbiome 2022, 10 (1), 20. 10.1186/s40168-021-01216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumy K. L.; Findlay R. H. Convenient Determination of DNA Extraction Efficiency Using an External DNA Recovery Standard and Quantitative-Competitive PCR. J. Microbiol. Methods 2004, 57 (2), 259–268. 10.1016/j.mimet.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Wang R.; Matsuura N.; Hara-Yamamura H.; Watanabe T.; Honda R. Initial Behaviors and Removal of Extracellular Plasmid Gene in Membrane Bioreactor. Journal of Environmental Management 2021, 298, 113541. 10.1016/j.jenvman.2021.113541. [DOI] [PubMed] [Google Scholar]
- Meyer F.; Paarmann D.; D’Souza M.; Olson R.; Glass E. M.; Kubal M.; Paczian T.; Rodriguez A.; Stevens R.; Wilke A.; Wilkening J.; Edwards R. A. The Metagenomics Rast Server - A Public Resource for the Automatic Phylogenetic and Functional Analysis of Metagenomes. BMC bioinformatics 2008, 9, 386. 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I.; Huber W.; Anders S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with Deseq2. Genome Biology 2014, 15 (12), 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur A. G.; Waglechner N.; Nizam F.; Yan A.; Azad M. A.; Baylay A. J.; Bhullar K.; Canova M. J.; De Pascale G.; Ejim L.; Kalan L.; King A. M.; Koteva K.; Morar M.; Mulvey M. R.; O’Brien J. S.; Pawlowski A. C.; Piddock L. J.; Spanogiannopoulos P.; Sutherland A. D.; Tang I.; Taylor P. L.; Thaker M.; Wang W.; Yan M.; Yu T.; Wright G. D. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57 (7), 3348–57. 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R.; Lima-Mendez G.; Toussaint A. Aclame: A Classification of Mobile Genetic Elements, Update 2010. Nucleic Acids Res. 2010, 38, D57–D61. 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C.; Bengtsson-Palme J.; Rensing C.; Kristiansson E.; Larsson D. G. J. Bacmet: Antibacterial Biocide and Metal Resistance Genes Database. Nucleic Acids Res. 2014, 42 (D1), D737–D743. 10.1093/nar/gkt1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Yang Y.; Ma L.; Ju F.; Guo F.; Tiedje J. M.; Zhang T. Metagenomic and Network Analysis Reveal Wide Distribution and Co-Occurrence of Environmental Antibiotic Resistance Genes. ISME Journal 2015, 9 (11), 2490–2502. 10.1038/ismej.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. R.; Gorley R. N.; Somerfield P. J.; Warwick R. M.. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation; Primer-E: Plymouth, 2014. [Google Scholar]
- Segata N.; Izard J.; Waldron L.; Gevers D.; Miropolsky L.; Garrett W. S.; Huttenhower C. Metagenomic Biomarker Discovery and Explanation. Genome Biology 2011, 12 (6), R60. 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B.; Raphenya A. R.; Alcock B.; Waglechner N.; Guo P.; Tsang K. K.; Lago B. A.; Dave B. M.; Pereira S.; Sharma A. N.; Doshi S.; Courtot M.; Lo R.; Williams L. E.; Frye J. G.; Elsayegh T.; Sardar D.; Westman E. L.; Pawlowski A. C.; Johnson T. A.; Brinkman F. S.; Wright G. D.; McArthur A. G. Card 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45 (D1), D566–D573. 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Yang X.; Qin J.; Lu N.; Cheng G.; Wu N.; Pan Y.; Li J.; Zhu L.; Wang X.; Meng Z.; Zhao F.; Liu D.; Ma J.; Qin N.; Xiang C.; Xiao Y.; Li L.; Yang H.; Wang J.; Yang R.; Gao G. F.; Wang J.; Zhu B. Metagenome-Wide Analysis of Antibiotic Resistance Genes in a Large Cohort of Human Gut Microbiota. Nat. Commun. 2013, 4, 2151. 10.1038/ncomms3151. [DOI] [PubMed] [Google Scholar]
- Pormohammad A.; Nasiri M. J.; Azimi T. Prevalence of Antibiotic Resistance in Escherichia Coli Strains Simultaneously Isolated from Humans, Animals, Food, and the Environment: A Systematic Review and Meta-Analysis. Infect Drug Resist 2019, 12, 1181–1197. 10.2147/IDR.S201324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E.; Hasman H.; Cosentino S.; Vestergaard M.; Rasmussen S.; Lund O.; Aarestrup F. M.; Larsen M. V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67 (11), 2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215 (3), 403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Hendriksen R. S.; Bortolaia V.; Tate H.; Tyson G. H.; Aarestrup F. M.; McDermott P. F. Using Genomics to Track Global Antimicrobial Resistance. Frontiers in Public Health 2019, 7, 242. 10.3389/fpubh.2019.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L.; Coque T. M.; Baquero F. What Is a Resistance Gene? Ranking Risk in Resistomes. Nat. Rev. Microbiol. 2015, 13 (2), 116–123. 10.1038/nrmicro3399. [DOI] [PubMed] [Google Scholar]
- Joseph S. M.; Battaglia T.; Maritz J. M.; Carlton J. M.; Blaser M. J. Longitudinal Comparison of Bacterial Diversity and Antibiotic Resistance Genes in New York City Sewage. Msystems 2019, 4 (4), e00327-19. 10.1128/mSystems.00327-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer-Lluch M.; Calero-Cáceres W.; Jebri S.; Hmaied F.; Muniesa M.; Jofre J. Antibiotic Resistance Genes in Bacterial and Bacteriophage Fractions of Tunisian and Spanish Wastewaters as Markers to Compare the Antibiotic Resistance Patterns in Each Population. Environ. Int. 2014, 73, 167–175. 10.1016/j.envint.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Rutgersson C.; Fick J.; Marathe N.; Kristiansson E.; Janzon A.; Angelin M.; Johansson A.; Shouche Y.; Flach C. F.; Larsson D. G. Fluoroquinolones and Qnr Genes in Sediment, Water, Soil, and Human Fecal Flora in an Environment Polluted by Manufacturing Discharges. Environ. Sci. Technol. 2014, 48 (14), 7825–32. 10.1021/es501452a. [DOI] [PubMed] [Google Scholar]
- Ebmeyer S.; Kristiansson E.; Larsson D. G. J. A Framework for Identifying the Recent Origins of Mobile Antibiotic Resistance Genes. Communications Biology 2021, 4 (1), 8. 10.1038/s42003-020-01545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund D.; Kieffer N.; Parras-Moltó M.; Ebmeyer S.; Berglund F.; Johnning A.; Larsson D. G. J.; Kristiansson E. Large-Scale Characterization of the Macrolide Resistome Reveals High Diversity and Several New Pathogen-Associated Genes. Microbial Genomics 2022, 8 (1), 000770. 10.1099/mgen.0.000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J.; Hammaren R.; Pal C.; Ostman M.; Bjorlenius B.; Flach C. F.; Fick J.; Kristiansson E.; Tysklind M.; Larsson D. G. J. Elucidating Selection Processes for Antibiotic Resistance in Sewage Treatment Plants Using Metagenomics. Sci. Total Environ. 2016, 572, 697–712. 10.1016/j.scitotenv.2016.06.228. [DOI] [PubMed] [Google Scholar]
- McLellan S. L.; Huse S. M.; Mueller-Spitz S. R.; Andreishcheva E. N.; Sogin M. L. Diversity and Population Structure of Sewage-Derived Microorganisms in Wastewater Treatment Plant Influent. Environmental Microbiology 2010, 12 (2), 378–392. 10.1111/j.1462-2920.2009.02075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N.; Waldron L.; Ballarini A.; Narasimhan V.; Jousson O.; Huttenhower C. Metagenomic Microbial Community Profiling Using Unique Clade-Specific Marker Genes. Nat. Methods 2012, 9 (8), 811–814. 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. L.; Keenum I. M.; Dai D.; Zhang L.; Vikesland P. J.; Pruden A. Critical Evaluation of Short, Long, and Hybrid Assembly for Contextual Analysis of Antibiotic Resistance Genes in Complex Environmental Metagenomes. Sci. Rep. 2021, 11 (1), 3753. 10.1038/s41598-021-83081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings M. R.; Gaze W. H.; Pruden A.; Smalla K.; Tiedje J. M.; Zhu Y. G. Using the Class 1 Integron-Integrase Gene as a Proxy for Anthropogenic Pollution. ISME Journal 2015, 9 (6), 1269–79. 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Austin C.; Wright M. S.; Stepanauskas R.; McArthur J. V. Co-Selection of Antibiotic and Metal Resistance. Trends Microbiol. 2006, 14 (4), 176–82. 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Pal C.; Bengtsson-Palme J.; Kristiansson E.; Larsson D. G. J. Co-Occurrence of Resistance Genes to Antibiotics, Biocides and Metals Reveals Novel Insights into Their Co-Selection Potential. BMC Genomics 2015, 16 (1), 964. 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaene E.; Chanteux H.; Servais H.; Mingeot-Leclercq M.-P.; Tulkens Paul M. Comparative Stability Studies of Antipseudomonal B-Lactams for Potential Administration through Portable Elastomeric Pumps (Home Therapy for Cystic Fibrosis Patients) and Motor-Operated Syringes (Intensive Care Units). Antimicrob. Agents Chemother. 2002, 46 (8), 2327–2332. 10.1128/AAC.46.8.2327-2332.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles L. F.; Islam S.; Aldstadt J.; Saqeeb K. N.; Alam M.; Khan M. A.; Johura F.-T.; Ahmed S. I.; Aga D. S. Retrospective Suspect Screening Reveals Previously Ignored Antibiotics, Antifungal Compounds, and Metabolites in Bangladesh Surface Waters. Sci. Total Environ. 2020, 712, 136285. 10.1016/j.scitotenv.2019.136285. [DOI] [PubMed] [Google Scholar]
- Li D.; Yang M.; Hu J.; Zhang Y.; Chang H.; Jin F. Determination of Penicillin G and Its Degradation Products in a Penicillin Production Wastewater Treatment Plant and the Receiving River. Water Res. 2008, 42 (1), 307–317. 10.1016/j.watres.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J.; Larsson D. G. Concentrations of Antibiotics Predicted to Select for Resistant Bacteria: Proposed Limits for Environmental Regulation. Environmental International 2016, 86, 140–149. 10.1016/j.envint.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Joint Programming Initiative on Antimicrobial Resistance. Joint Programming Initiative on Antimicrobial Resistance Mapping Report; 2015.
- Klein E. Y.; Van Boeckel T. P.; Martinez E. M.; Pant S.; Gandra S.; Levin S. A.; Goossens H.; Laxminarayan R. Global Increase and Geographic Convergence in Antibiotic Consumption between 2000 and 2015. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (15), E3463–E3470. 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M.; Kumar R.; Kishor K.; Mlsna T.; Pittman C. U. Jr.; Mohan D. Pharmaceuticals of Emerging Concern in Aquatic Systems: Chemistry, Occurrence, Effects, and Removal Methods. Chem. Rev. 2019, 119 (6), 3510–3673. 10.1021/acs.chemrev.8b00299. [DOI] [PubMed] [Google Scholar]
- Molstad S.; Lofmark S.; Carlin K.; Erntell M.; Aspevall O.; Blad L.; Hanberger H.; Hedin K.; Hellman J.; Norman C.; Skoog G.; Stalsby-Lundborg C.; Tegmark Wisell K.; Ahren C.; Cars O. Lessons Learnt During 20 Years of the Swedish Strategic Programme against Antibiotic Resistance. Bull. World Health Organ 2017, 95 (11), 764–773. 10.2471/BLT.16.184374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes N.Illegal Sale of Antibiotics Still Common in Hong Kong. https://jmsc.hku.hk/2017/03/illegal-sale-of-antibiotics-still-common-in-hong-kong/ (accessed 2021-10-07).
- Morgan D. J.; Okeke I. N.; Laxminarayan R.; Perencevich E. N.; Weisenberg S. Non-Prescription Antimicrobial Use Worldwide: A Systematic Review. Lancet Infect. Dis. 2011, 11 (9), 692–701. 10.1016/S1473-3099(11)70054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (Glass) Report: Early Implementation 2020; 2020.
- Hardwick S. A.; Chen W. Y.; Wong T.; Kanakamedala B. S.; Deveson I. W.; Ongley S. E.; Santini N. S.; Marcellin E.; Smith M. A.; Nielsen L. K.; Lovelock C. E.; Neilan B. A.; Mercer T. R. Synthetic Microbe Communities Provide Internal Reference Standards for Metagenome Sequencing and Analysis. Nat. Commun. 2018, 9 (1), 3096. 10.1038/s41467-018-05555-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenfeld N.; Bisceglia K. J. Emerging Investigators Series: Sewer Surveillance for Monitoring Antibiotic Use and Prevalence of Antibiotic Resistance: Urban Sewer Epidemiology. Environmental Science: Water Research & Technology 2016, 2 (5), 788–799. 10.1039/C6EW00158K. [DOI] [Google Scholar]
- Arango-Argoty G.; Garner E.; Pruden A.; Heath L. S.; Vikesland P.; Zhang L. Deeparg: A Deep Learning Approach for Predicting Antibiotic Resistance Genes from Metagenomic Data. Microbiome 2018, 6 (1), 23. 10.1186/s40168-018-0401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund F.; Österlund T.; Boulund F.; Marathe N. P.; Larsson D. G. J.; Kristiansson E. Identification and Reconstruction of Novel Antibiotic Resistance Genes from Metagenomes. Microbiome 2019, 7 (1), 52. 10.1186/s40168-019-0670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Y.; Tsementzi D.; Hatt J. K.; Bivins A.; Khelurkar N.; Brown J.; Tripathi S. N.; Konstantinidis K. T. Intensive Allochthonous Inputs Along the Ganges River and Their Effect on Microbial Community Composition and Dynamics. Environ. Microbiol 2019, 21 (1), 182–196. 10.1111/1462-2920.14439. [DOI] [PubMed] [Google Scholar]
- Lee K.; Kim D.-W.; Lee D.-H.; Kim Y.-S.; Bu J.-H.; Cha J.-H.; Thawng C. N.; Hwang E.-M.; Seong H. J.; Sul W. J.; Wellington E. M. H.; Quince C.; Cha C.-J. Mobile Resistome of Human Gut and Pathogen Drives Anthropogenic Bloom of Antibiotic Resistance. Microbiome 2020, 8 (1), 2. 10.1186/s40168-019-0774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrazeg M.; Diene S. M.; Medjahed L.; Parola P.; Drissi M.; Raoult D.; Rolain J. M. New Delhi Metallo-Beta-Lactamase around the World: An Ereview Using Google Maps. Eurosurveillance 2014, 19 (20), 2–15. 10.2807/1560-7917.ES2014.19.20.20809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.