Abstract
Purpose:
To assess the contributions of circulating metabolites for improving upon the performance of the Risk of Ovarian Malignancy Algorithm (ROMA) for risk prediction of ovarian cancer (OvCa) among women with ovarian cysts.
Experimental Design:
Metabolomic profiling was performed on an initial set of sera from 101 serous and non-serous OvCa cases and 134 individuals with benign pelvic masses (BPM). Using a deep learning model, a panel consisting of seven cancer-related metabolites (diacetylspermine, diacetylspermidine, N-(3-acetamidopropyl)pyrrolidin-2-one, N-acetylneuraminate, N-acetyl-mannosamine, N-acetyl-lactosamine, and hydroxyisobutyric acid) was developed for distinguishing early-stage OvCa from BPM. The performance of the metabolite panel was evaluated in an independent set of sera from 118 OvCa cases and 56 subjects with BPM. The contributions of the panel for improving upon the performance of ROMA was further assessed.
Results:
A 7-marker metabolite panel (7MetP) developed in the Training Set yielded an AUC of 0.86 (95% CI: 0.76–0.95) for early-stage OvCa in the independent Test Set. The 7MetP+ROMA model had an AUC of 0.93 (95% CI: 0.84–0.98) for early-stage OvCa in the Test Set, which was improved compared to ROMA alone (0.91 (95% CI: 0.84–0.98); likelihood ratio test p-value:.03). In the entire specimen set, the combined 7MetP+ROMA model yielded a higher positive predictive value (0.68 vs 0.52; 1-sided p<.001) with improved specificity (0.89 vs 0.78; 1-sided p<.001) for early-stage OvCa compared to ROMA alone.
Conclusions:
A blood-based metabolite panel was developed that demonstrates independent predictive ability and complements ROMA for distinguishing early-stage OvCa from benign disease to better inform clinical decision making.
Introduction
Ovarian cysts are found to occur in some 17% of women that undergo transvaginal sonograms (TVS). Most of these cysts are non-cancerous.(1,2) Currently, neither TVS nor cancer antigen 125 (CA125) alone or in combination yield sufficient sensitivity and specificity to distinguish benign form malignant ovarian cysts. (3) The high false positive rates lead to unnecessary surgical procedures, with significant morbidity.(4)
Two risk assessment algorithms, the Risk of Ovarian Malignancy Algorithm (ROMA) and the risk of ovarian cancer algorithm (OVERA), were developed to assess the risk of a mass being cancerous.(5–8) ROMA and OVERA have similar abilities to distinguish malignant from benign pelvic masses.(7,8) Although these algorithms offer high sensitivity for detection of OvCa, specificity is limited.(5–8)
Perturbed cellular metabolism is a hallmark of cancer(9). Several lines of evidence indicate that cellular and systemic metabolic adaptations occur from the earliest phases of cancer development suggesting that metabolites may serve as cancer biomarkers.(10,11) Here, we applied a deep learning approach to metabolic profiles of sera to determine whether a metabolic signature may be uncovered that distinguish early-stage ovarian cancers from benign disease. A model consisting of seven cancer-relevant metabolites, including three polyamines, was developed and tested in an independent set in combination with the ROMA algorithm for OvCa risk prediction among women with ovarian cysts.
Materials and Methods
Specimen Sets
Blood specimens were obtained preoperatively with informed consent under IRB/ethical committees approved protocols (LAB04–0687) at the University of Texas M.D. Anderson Cancer Center (MDACC) and at the Fred Hutchinson Cancer Research Center (FHCRC, IRB 4563) from patients who were admitted for surgery based on a mass found on ultrasound, elevated CA125, or a positive biopsy. This study was approved and monitored by the respective Institutional Review Boards and was conducted in accordance with the Declaration of Helsinki. All human biospecimens were obtained with written informed consent. All patients were fasting at the time of blood collection. Samples were processed on the same day, generally within 4 hours of blood draw, under standardized operating procedures, aliquoted to minimize freeze-thaw cycling effects, and stored in −80°C until use. The specimen set consisted of plasma from 59 patients with stage I-II and 160 patients with stage III-IV invasive epithelial ovarian cancer and from 190 patients with benign pelvic masses. (3,12) Biopsy samples were examined by a certified pathologist for the diagnosis of cancer or benign pelvic condition. Detailed patient and tumor characteristics are provided in Table 1. Information regarding histological ovarian cancer subtypes and benign etiologies are provided in Supplementary Table S1. All participants had provided consent for use of samples in ethically approved secondary studies.
Table 1.
Patient and tumor characteristics
| Patient and Tumor Characteristics | Training Set | Testing Set | ||||
|---|---|---|---|---|---|---|
| Cases | BPM† | P-value‡ | Cases | BPM† | P-value‡ | |
| Number of Subjects, N | 101 | 134 | 118 | 56 | ||
| Age, average (min-max) | 59 (23–85) | 57 (30–83) | 0.14 | 61 (36–86) | 55 (33–79) | 0.009 |
| Menopausal Status, N (%) | ||||||
| Pre | 21 (20.8%) | 39 (29.1%) | 0.17 | 19 (16.1%) | 18 (32.1%) | 0.02 |
| Post | 80 (79.2%) | 95 (70.9%) | 99 (83.8%) | 38 (67.8%) | ||
| Race, N (%) | ||||||
| White | 83 (82.2%) | 104 (77.6%) | 0.18 | 87 (73.7%) | 42 (75%) | 0.52 |
| Black | 5 (5%) | 3 (2.2%) | 9 (7.6%) | 9 (16.1%) | ||
| Asian | 2 (2%) | 3 (2.2%) | 13 (11%) | 1 (1.8%) | ||
| American Indian | - | 1 (0.7%) | - | - | ||
| Pacific Islander | - | - | 9 (7.6%) | 4 (7.1%) | ||
| Unknown | 11 (10.9%) | 23 (17.2%) | - | - | ||
| Stage, N (%) | ||||||
| Stage I and II | 39 (38.6%) | - | 20 (16.9%) | - | ||
| Stage III and IV | 62 (61.4%) | - | 98 (83.1%) | - | ||
| Histological Subtype, N (%) | ||||||
| Serous | 69 (68.3%) | - | 114 (96.6%) | - | ||
| Non-Serous | 32 (31.7%) | - | 1 (0.8%) | - | ||
| Unknown | 3 (2.54%) | |||||
| ROMA, median (25th/75th Percentiles) | 86.85 (36.85–98.03) | 11.14 (6.64–20.66) | <0.0001 | 89.51 (70.55–96.95) | 13.66 (6.42–18.56) | <0.0001 |
individuals with benign pelvic masses (BPM)
Statistical significance was determined by Wilcoxon rank sum tests for continuous variables and Fisher’s exact test or χ2 tests for trend for categorical variables. 2-sided p-values are reported.
Metabolomic analysis
Metabolomic analyses were performed as previously described.(13)
Primary Metabolites and Biogenic Amines
Metabolites were extracted from pre-aliquoted EDTA plasma (10 μL) with 30μL of LCMS grade methanol (ThermoFisher) in a 96-well microplate (Eppendorf). Plates were heat sealed, vortexed for 5 min at 750 rpm, and centrifuged at 2000 × g for 10 minutes at room temperature. The supernatant (10 μL) was carefully transferred to a 96-well plate, leaving behind the precipitated protein. The supernatant was further diluted with 10 μL of 100 mM ammonium formate, pH3. For Hydrophilic Interaction Liquid Chromatography (HILIC) analysis, the samples were diluted with 60 μL LCMS grade acetonitrile (ThermoFisher), whereas samples for C18 analysis were diluted with 60 μL water (GenPure ultrapure water system, Thermofisher). Each sample solution was transferred to 384-well microplate (Eppendorf) for LCMS analysis.
Untargeted Analysis of Primary Metabolites and Biogenic Amines
Untargeted metabolomics analysis was conducted on Waters Acquity™ UPLC system with 2D column regeneration configuration (I-class and H-class) coupled to a Xevo G2-XS quadrupole time-of-flight (qTOF) mass spectrometer. Chromatographic separation was performed using HILIC (Acquity™ UPLC BEH amide, 100 Å, 1.7 μm 2.1× 100mm, Waters Corporation, Milford, U.S.A) and C18 (Acquity™ UPLC HSS T3, 100 Å, 1.8 μm,, 2.1×100mm, Water Corporation, Milford, U.S.A) columns at 45°C.
Quaternary solvent system mobile phases were (A) 0.1% formic acid in water, (B) 0.1% formic acid in acetonitrile and (D) 100mM ammonium formate, pH 3. Samples were separated using the following gradient profile: for the HILIC separation a starting gradient of 95% B and 5% D was increase linearly to 70% A, 25% B and 5% D over a 5min period at 0.4mL/min flow rate, followed by 1 min isocratic gradient at 100 % A at 0.4mL/min flow rate. For C18 separation, a chromatography gradient of was as follows: starting conditions, 100% A, with linear increase to final conditions of 5% A, 95% B followed by isocratic gradient at 95% B, 5% D for 1 min.
Binary pump was used for column regeneration and equilibration. The solvent system mobile phases were (A1) 100mM ammonium formate, pH 3, (A2) 0.1 % formic in 2-propanol and (B1) 0.1 % formic acid in acetonitrile. The HILIC column was stripped using 90% A2 for 5 min followed by 2 min equilibration using 100% B1 at 0.3 mL/min flowrate. Reverse phase C18 column regeneration was performed using 95% A1, 5% B1 for 2 min followed by column equilibration using 5% A1, 95% B1 for 5 min.
Mass Spectrometry Data Acquisition
Mass spectrometry data was acquired using ‘sensitivity’ mode in positive and negative electrospray ionization mode within 50–1200 Da range for primary metabolites and 100–2000 Da for complex lipids. For the electrospray acquisition, the capillary voltage was set at 1.5 kV (positive), 3.0kV (negative), sample cone voltage 30V, source temperature at 120° C, cone gas flow 50 L/h and desolvation gas flow rate of 800 L/h with scan time of 0.5 sec in continuum mode. Leucine Enkephalin; 556.2771 Da (positive) and 554.2615 Da (negative) was used for lockspray correction and scans were performed at 0.5 min. The injection volume for each sample was 3μL, unless otherwise specified. The acquisition was carried out with instrument auto gain control to optimize instrument sensitivity over the samples acquisition time.
Data Processing
Data were processed using Progenesis QI (Nonlinear, Waters). Peak picking and retention time alignment of LC-MS and MSe data were performed using Progenesis QI software (Nonlinear, Waters). Data processing and peak annotations were performed using an in-house automated pipeline as previously described.(13–16) Annotations were determined by matching accurate mass and retention times using customized libraries created from authentic standards and by matching experimental tandem mass spectrometry data against the NIST MSMS, LipidBlast or HMDB v3 theoretical fragmentations; for complex lipids retention time patterns characteristic of lipid subclasses was also considered. To correct for injection order drift, each feature was normalized using data from repeat injections of quality control samples collected every 10 injections throughout the run sequence. Measurement data were smoothed by Locally Weighted Scatterplot Smoothing (LOESS) signal correction (QC-RLSC) as previously described. Values are reported as ratios relative to the median of historical quality control reference samples run with every analytical batch for the given analyte.(13–16)
Assaying of CA125 and HE4
Serum CA125 and HE4 concentrations were measured using the Architect CA125II assay (Abbott Diagnostics, Abbott Park), and the HE4 EIA assay (Fujirebio Diagnostics, Malvern, PA) as previously described.(17) To calculate the ROMA score, a predictive index (PI) was calculated using serum HE4 and CA125 II levels and one of the following equations, depending on the patient’s menopausal status as previously described(7):
Premenopausal: Predictive Index (PI) = −12.0 + 2.38*Natural Log[HE4] + 0.0626*Natural Log[CA 125]
Postmenopausal: Predictive Index (PI) = −8.09 + 1.04*Natural Log[HE4] + 0.732*Natural Log[CA 125]
ROMA percentage were calculated by exp(PI)/(1+exp(PI).
Statistical analysis
An overall schematic workflow of the study is provided in Supplementary Fig. S1. Metabolite selection and model building was performed using metabolic profiles generated from plasma samples from the FHCRC. The method reported by Gedeon was used to prioritize pertinent variables to be included in the model.(18–20) This approach removes irrelevant or noisy variables by analyzing for the relative weight of each variable within the overall data matrix. An importance score is calculated by dividing the absolute value of the weight of an input connecting to an output by the total absolute value of all weights from that input. When applied in the deep learning model, this approach is recursively extended backwards through layers by taking the effect of a neuron on a connected node, then multiplying the derived weight by the effect of the given node on the target output and summing all connecting nodes.
Here, Pjk represents the average contribution of a node j in a layer to a node k in the next layer. w is the weight on the connection and nh is the number of nodes in the next layer.
The contribution of an input neuron to an output is:
Using this approach, 20 iterations with slightly modified hyperparameters were introduced, and the relative variable importance score recalculated for each metabolite. Metabolites that consistently yielded a relative variable importance score >0.7 (corresponding to those metabolites with importance scores in the top 30th percentile) across all 20 iterations were selected to develop an algorithm for distinguishing early-stage OvCa from benign disease. Seven models, including deep learning, random forest, ensemble learning and gradient boosting method algorithms, incorporating the seven metabolites were assessed for distinguishing early-stage OvCa from benign disease. Performance of the models were evaluated using a 5-fold cross validation. To further evaluate model stability, perturbations (e.g. random selection and replacement) were introduced to the Training Set and the performance re-assessed.
A deep learning model (DLM) with 3 hidden layers and 3 nodes in each layer was selected for modeling the 7-marker metabolite panel (7MetP) based on AUC, and the 7MetP using fixed parameters tested for detection of OvCa in the MDACC cohort.
To assess the contributions of the 7MetP and ROMA, we first fitted a logistic regression with the 7MetP and ROMA as two separate predictors (Supplementary Table S2). For ROMA, we used percentage risk as described above.(21) Initial modeling was performed using early-stage OvCa cases and individuals with BPM from the FHCRC and testing of the model performed in the MDACC cohort.
To directly compare the performance of the combined 7MetP+ROMA model with ROMA, we used fixed risk thresholds of 11.4% in premenopausal women and 29.9% for postmenopausal, (21) and calculated positive predictive values (PPV), negative predictive values (NPV) as well as sensitivity and specificity estimates.
The combined score from the logistic regression model were converted to risk by exp(combined score)/(1+exp(combined score)).
Model discrimination was assessed based on receiver operating characteristic curve (ROC), as well as sensitivity and specificity estimates. The 95% confidence intervals (CI) for AUCs were estimated using the Delong method.(22) P-values for specificity and sensitivity were estimated by calculating 2.5 and 97.5 percentiles of 1,000 boot straps on the delta values. All modeling was performed using the h2o package and R statistical program.(18)
Data Availability
Relevant data supporting the findings of this study are available within the Article and Supplementary Information or are available from the authors upon reasonable request.
Results
Feature selection for algorithm training
Untargeted metabolomics was conducted on a Training Set of sera from 101 OvCa cases (39 early stage and 62 late stage) and 134 subjects with BPM from the Fred Hutchinson Cancer Research Center (FHCRC) (Table 1). A total of 475 uniquely annotated metabolites were quantified (Supplementary Table S3). To prioritize metabolites, relative importance scores were calculated using the Gedeon method (19,20) and metabolites selected based on consistently exhibiting an importance score above 0.7. This approach resulted in seven metabolites each of which had prior evidence for cancer relevance (diacetylspermine (DAS)(23), diacetylspermidine (DiAcSpmd)(23), N-(3-acetamidopropyl)pyrrolidin-2-one (N3AP)(23), N-acetylneuraminate (NANA)(24), N-acetyl-mannosamine (NAcMan)(25), N-acetyl-lactosamine (NAcLAC)(26), and hydroxyisobutyric acid (HBA)(27)) that were subsequently used for model building. Individual classifier performance of these metabolites for distinguishing OvCa cases from individuals with BPM ranged from 0.55 to 0.82 (Supplementary Table S4; Supplementary Fig. S2).
Development of a combination rule and validation in an independent test set
We next sought to develop an optimal combination rule that incorporated the seven metabolites for distinguishing early-stage OvCa from benign disease. For model building, we tested seven different machine learning algorithms. Of these, a deep learning model (DLM) with 3 hidden layers and 3 nodes in each layer achieved the highest predictive performance and was used to establish the 7-marker metabolite panel (7MetP), which yielded an AUC of 0.75 (95% CI: 0.66–0.85) for differentiating early-stage OvCa cases from benign disease (Table 2; Supplementary Tables S5–6). When stratifying OvCa cases into serous and non-serous, the 7MetP had respective AUCs of 0.85 (95% CI: 0.79–0.91) and 0.80 (95% CI: 0.71–0.89) (Supplementary Table S7).
Table 2.
Performance of different learning algorithms for differentiating early-stage OvCa cases from BPM in the training set using 5-fold cross validation.
| Model | Hyper parameters | AUC1 | Log Loss | AUCpr2 | Mean per class error | RMSE3 |
|---|---|---|---|---|---|---|
| Deep learning model | Activation: MaxoutWithDropout, hidden layers: [3, 3, 3] | 0.753 | 0.354 | 0.556 | 0.222 | 0.303 |
| Deep learning model | Activation: Tanh, hidden layers: [1,1] | 0.740 | 0.362 | 0.529 | 0.233 | 0.322 |
| StackedEnsemble | Ensemble models: GLM, Deep Learning, Random Forest, Gradient Boost Method | 0.713 | 0.387 | 0.484 | 0.205 | 0.332 |
| Deep learning model | Activation: Tanh, hidden layers: [2,2] | 0.711 | 0.377 | 0.519 | 0.237 | 0.325 |
| Lasso Regression | Lambda =0.2,5 features selected | 0.709 | 0.506 | 0.438 | 0.202 | 0.376 |
| StackedEnsemble | Ensemble models (best of each family): GLM, Deep Learning, Random Forest, Gradient Boost Method | 0.692 | 0.399 | 0.459 | 0.228 | 0.336 |
| GLM | Family: binomial | 0.687 | 0.532 | 0.447 | 0.241 | 0.364 |
| Extremely Randomized Trees (XRT) | - | 0.681 | 0.577 | 0.359 | 0.224 | 0.351 |
| Distributed Random Forest (DRF) | - | 0.679 | 0.746 | 0.355 | 0.216 | 0.354 |
| Gradient Boosting Method | Number of tree: 50, Maximum depth:6 | 0.668 | 0.516 | 0.357 | 0.234 | 0.372 |
AUC: Area under the ROC Curve
AUCpr: Area under the precision recall curve
RMSE: Root-mean-square deviation
Validation of the 7MetP using fixed parameters was performed in an independent Test Set from MD Anderson Cancer Center (MDACC) that consisted of 118 OvCa cases (20 early stage and 98 late stage) and 56 individuals with BPM. The 7MetP yielded an AUC of 0.88 (95% CI: 0.82–0.93) for distinguishing all OvCa cases from individuals with BPM (Supplementary Table S5), and an AUC of 0.86 (95% CI: 0.76–0.95) for early-stage OvCa (Figure 1; Supplementary Table S5).
Figure 1.
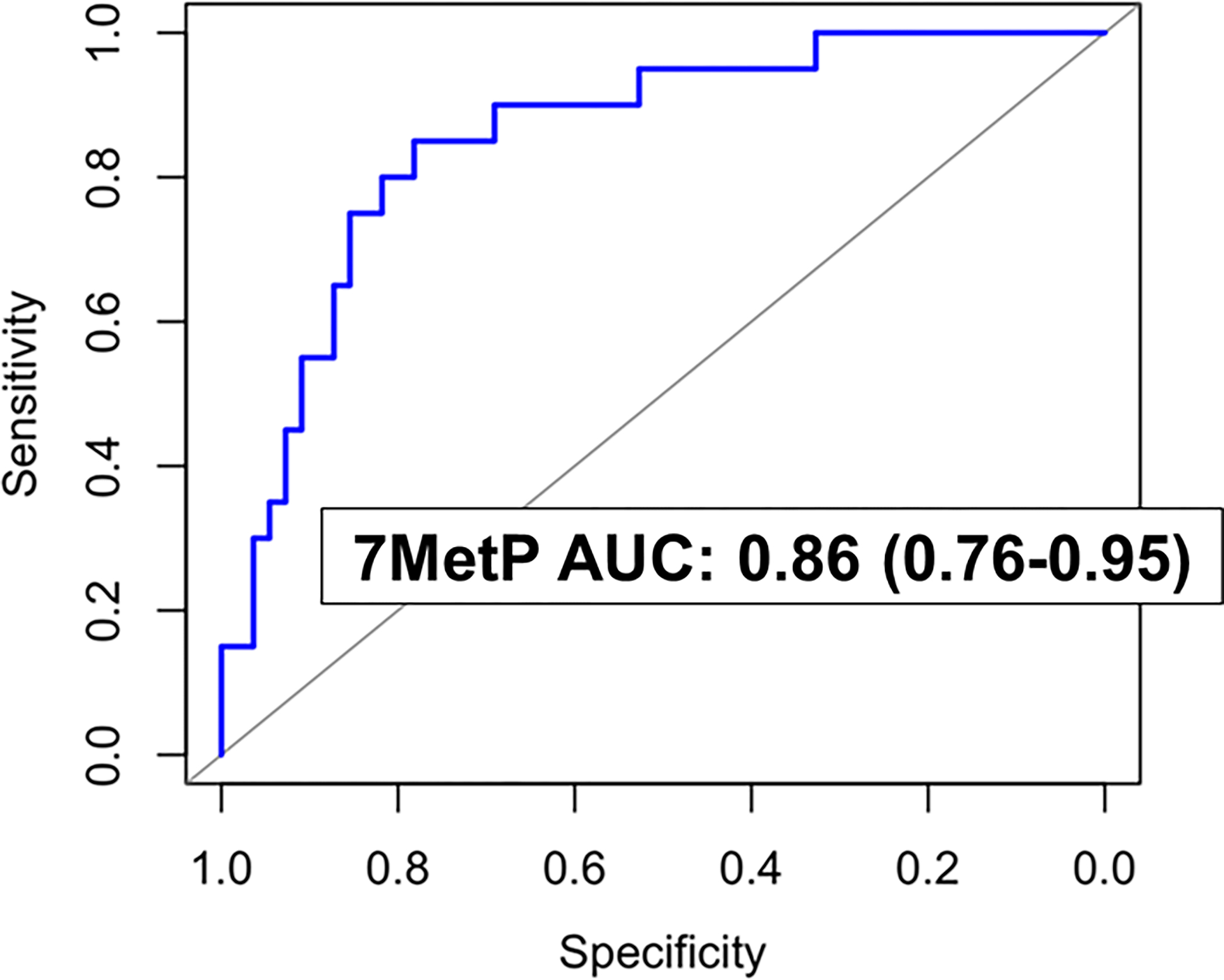
Predictive performance of the 7MetP for distinguishing early-stage OvCa from BPM in the independent Test Set.
Contributions of the metabolite panel with the ROMA algorithm
We next assessed whether the 7MetP would improve upon the predictive performance of the ROMA algorithm. Using model scores derived from the 7MetP and the ROMA algorithm, a logistic regression model for distinguishing early-stage OvCa from BPM was developed in the Training Set and performance evaluated in the Test Set. The combined 7MetP+ROMA yielded an AUC of 0.93 (95% CI: 0.86–1.00) for early-stage OvCa in the Test Set whereas ROMA alone had an AUC of 0.91 (95% CI: 0.84–0.98) (likelihood ratio test p: 0.03) (Table 3). Compared to ROMA, the combined 7MetP+ROMA yielded improvements in the PPV by 21.0% (1-sided p< .001) and specificity by 14.0% (1-sided p< .001) for early stage OvCa (Table 3). When considering all OvCa cases, the combined 7MetP+ROMA model yielded an AUC of 0.97 (95% CI: 0.94–0.99) in the Test Set (Table 4).
Table 3.
Performance estimates of ROMA and the combined 7MetP+ROMA model for early-stage OvCa the Training Set and the independent Testing Set.
| Training Set | |||||
| ROMA | ROMA + 7MetP | Difference | P | ||
| AUC (95% CI) | 0.81 (0.77 – 0.85) | 0.84 (0.80 – 0.88) | 0.03 (0.01 to 0.06) | < .001 | |
| At 11.4% risk threshold for premenopausal and 29.9% for postmenopausal (same risk as ROMA) | Sensitivity | 0.68 (0.60 – 0.77) | 0.68 (0.59 – 0.77) | 0.00 (−0.06 to 0.07) | .49 |
| Specificity | 0.79 (0.75 – 0.83) | 0.88 (0.85 – 0.91) | 0.09 (0.06 to 0.12) | <.001 | |
| PPV | 0.48 (0.40 – 0.55) | 0.62 (0.54 – 0.69) | 0.14 (0.08 to 0.20) | <.001 | |
| NPV | 0.90 (0.87 – 0.93) | 0.91 (0.88 – 0.94) | 0.01 (−0.01 to 0.03) | .14 | |
| Test Set | |||||
| ROMA | ROMA + 7MetP | Difference | P | ||
| AUC (95% CI) | 0.91 (0.84–0.98) | 0.93 (0.86–1.00) | 0.02 (−0.01 to 0.04) | .03 | |
| At 11.4% risk threshold for premenopausal and 29.9% for postmenopausal (same risk as ROMA) | Sensitivity | 0.90 (0.83 – 0.97) | 0.90 (0.81 – 0.97) | 0.00 (−0.09 to 0.09) | .472 |
| Specificity | 0.77 (0.70 – 0.83) | 0.91 (0.86 – 0.95) | 0.14 (0.09 to 0.20) | <.001 | |
| PPV | 0.58 (0.48 – 0.68) | 0.79 (0.67 – 0.87) | 0.21 (0.13 to 0.28) | <.001 | |
| NPV | 0.96 (0.92 – 0.99) | 0.96 (0.93 – 0.99) | 0.01 (−0.03 to 0.04) | .365 | |
Abbreviations: PPV: positive predictive value; NPV: negative predictive value. P-values for comparison of AUCs represent likelihood ratio tests. Risk threshold corresponding to 11.4% in premenopausal women and 29.9% for postmenopausal were chosen based on reported findings from Ortiz-Munoz and colleagues.(21) 1-sided P-values are reported as we expect that the combined 7MetP+ROMA will yield improved performance estimates compared to ROMA alone.
Table 4.
Performance estimates of ROMA and the combined 7MetP+ROMA model for all OvCa in the Training Set and the independent Testing Set.
| Training Set | ||||||
| ROMA | ROMA + 7MetP | Difference | P | |||
| AUC (95% CI) | 0.91 (0.89 – 0.93) | 0.93 (0.91 – 0.94) | 0.01 (0.00 to 0.03) | <.001 | ||
| At 11.4% risk threshold for premenopausal and 29.9% for postmenopausal (same risk as ROMA) | Sensitivity | 0.87 (0.83 – 0.91) | 0.86 (0.82 – 0.90) | −0.01 (−0.04 to 0.02) | .22 | |
| Specificity | 0.79 (0.75 – 0.83) | 0.88 (0.85 – 0.91) | 0.09 (0.06 to 0.12) | <.001 | ||
| PPV | 0.76 (0.71 – 0.80) | 0.84 (0.81 – 0.89) | 0.09 (0.05 to 0.12) | <.001 | ||
| NPV | 0.89 (0.86 – 0.92) | 0.89 (0.86 – 0.92) | 0.00 (−0.02 to 0.02) | .36 | ||
| Test Set | ||||||
| ROMA | ROMA + 7MetP | Difference | P | |||
| AUC (95% CI) | 0.96 (0.94–0.99) | 0.97 (0.94–0.99) | 0.01 (0.00 to 0.01) | .06 | ||
| At 11.4% risk threshold for premenopausal and 29.9% for postmenopausal (same risk as ROMA) | Sensitivity | 0.96 (0.93 – 0.98) | 0.93 (0.90 – 0.96) | −0.03 (−0.05 to −0.01) | .008 | |
| Specificity | 0.76 (0.70 – 0.83) | 0.91 (0.86 – 0.95) | 0.15 (0.10 to 0.20) | < .001 | ||
| PPV | 0.89 (0.86 – 0.93) | 0.96 (0.93 – 0.97) | 0.06 (0.04 to 0.09) | < .001 | ||
| NPV | 0.89 (0.84 – 0.94) | 0.86 (0.81 – 0.91) | −0.03 (−0.08 to 0.01) | .07 | ||
Abbreviations: PPV: positive predictive value; NPV: negative predictive value. P-values for comparison of AUCs represent likelihood ratio tests. Risk threshold corresponding to 11.4% in premenopausal women and 29.9% for postmenopausal were chosen based on reported findings from Ortiz-Munoz and colleagues.(21) 1-sided P-values are reported as we expect that the combined 7MetP+ROMA will yield improved performance estimates compared to ROMA alone.
Performance of the metabolite panel alone and in combination with ROMA in the combined training and test sets
We further evaluated the predictive performance of the 7MetP alone and in combination with ROMA in the entire specimen set (n=219 OvCa cases (59 early stage and 160 late stage and 190 BPM)). The 7MetP had an AUC of 0.85 (95% CI: 0.81–0.88) for distinguishing all OvCa cases from individuals with BPM and an AUC of 0.81 (95% CI: 0.76–0.86) for early-stage OvCa (Supplementary Table S5). The combined 7MetP+ROMA model had a resultant AUC of 0.87 (95% CI: 0.85–0.93) for early-stage OvCa, which was markedly improved compared to ROMA alone (AUC: 0.84 (95% CI: 0.81–0.90); likelihood ratio test p-value: <0.001) (Table 5; Supplementary Table S8). Importantly, compared to ROMA alone, the 7MetP+ROMA model yielded a statistically significantly (1-sided P< .001) higher PPV (0.68 vs 0.52) and specificity (0.89 versus 0.78) for early-stage OvCa (Table 5).
Table 5.
Performance estimates of ROMA and the combined 7MetP+ROMA model for early-stage OvCa in the combined Specimen Set.
| Entire Set | |||||
|---|---|---|---|---|---|
| ROMA | ROMA + 7MetP | Difference | P | ||
| AUC (95% CI) | 0.84 (0.79–0.90) | 0.87 (0.82–0.93) | 0.03 (0.01 to 0.04) | <.001 | |
| At 11.4% risk threshold for premenopausal and 29.9% for postmenopausal (same risk as ROMA) | Sensitivity | 0.76 (0.69 – 0.82) | 0.76 (0.70 – 0.82) | 0.00 (−0.04 to 0.05) | .464 |
| Specificity | 0.78 (0.75 – 0.81) | 0.89 (0.86 – 0.92) | 0.11 (0.08 to 0.14) | <.001 | |
| PPV | 0.52 (0.45 – 0.58) | 0.68 (0.61 – 0.75) | 0.16 (0.11 to 0.21) | <.001 | |
| NPV | 0.91 (0.89 – 0.94) | 0.92 (0.90 – 0.94) | 0.01 (−0.01 to 0.02) | .091 | |
Abbreviations: PPV: positive predictive value; NPV: negative predictive value. P-values for comparison of AUCs represent likelihood ratio tests. Risk threshold corresponding to 11.4% in premenopausal women and 29.9% for postmenopausal were chosen based on reported findings from Ortiz-Munoz and colleagues.(21) 1-sided P-values are reported as we expect that the combined 7MetP+ROMA will yield improved performance estimates compared to ROMA alone.
Discussion
Pelvic masses are relatively common among women of all ages usually necessitating surgery. However, most such masses are benign and only a small percentage of these women will be diagnosed with ovarian cancer.(28)
Algorithms such as OVERA and ROMA were developed to estimate probability of a woman with a pelvic mass harboring a malignancy and to determine whether a patient should be referred to a general gynecologist if the mass is likely to be benign or a gynecologic oncologist if the mass is likely to be malignant.(7,29,30) A gynecologic oncologist has specialized training to dissect nodes, remove the omentum, and to remove as much cancer as possible from the surface of the bowel if extensive disease is found. Although the OVERA and ROMA algorithms offer high sensitivity, they are limited by sub-optimal specificity which can result in high false-positive rates, increased patient anxiety, and unnecessary procedures that are associated with significant morbidity.(31,32) A test that offers high sensitivity and specificity for identifying individuals at high risk of harboring malignant ovarian cysts has potential to better inform clinical decision making and improve patient outcomes. We developed and validated a blood-based metabolite panel that improves prediction of malignancy in combination with ROMA for women presenting with ovarian cysts.
The metabolite panel includes three polyamines, three acetylated carbohydrates, and hydroxyisobutyric acid. We have previously shown utility of circulating polyamines for early detection of ovarian cancer.(15) Polyamines have also been reported to be elevated in urine of individuals with ovarian cancer compared to controls.(33) The acetylated carbohydrates NANA and NAMA are involved in metabolism of sialic acids, which are commonly found in glycans of cell surface glycoproteins and glycolipids. Sialic acids are known to be involved in various aspects of tumorigenesis, including promoting tumor growth and metastasis as well as immune evasion.(34–37) Moreover, sialic acids can accumulate in the circulation due to increased turnover, secretion, and/or shedding.(38) N-acetyllactosamine (LacNAc) is a reported carbohydrate antigen involved in malignant transformation and metastasis.(26,39,40) Hydroxyisobutyric acid, a metabolite derived from valine metabolism, has also been linked to cancer with diagnostic and prognostic applications in ovarian cancer.(27)
On balance, limitations to our study including the unbalanced distribution of histology across the test and validation sets. OvCa cases were largely at advanced stages of disease in the specimen sets with differential representation of non-serous and serous OvCa. The metabolite panel yielded comparable performance for distinguishing serous and non-serous OvCa from BPM. Moreover, our metabolite panel was developed using early-stage OvCa cases and validated in an independent set of early-stage OvCa cases. Contributions of the metabolite panel with the OVERA algorithm was not assessed. Comparison of performance estimates between ROMA and OVERA have shown that they are comparable with tradeoffs in sensitivity and specificity.(7,41,42) Although PPV and NPV estimates are dependent upon the prevalence of disease in the evaluated population, previous investigations have reported ROMA to have a PPV of 42.9% (7), which is consistent with our findings. OVERA has a reported PPV of 40%.(8) In our study, the 7MetP+ROMA model had PPV of 68.0%. Thus, we believe that the metabolite panel has the potential to significantly contribute to the OVERA as well.
In conclusion, we developed and validated a metabolite panel that complements ROMA for improved risk prediction of malignancy among women presenting with ovarian cysts.
Supplementary Material
Translational Relevance.
Two FDA-approved algorithms, the Risk of Ovarian Malignancy Algorithm (ROMA) and the risk of ovarian cancer algorithm (OVERA), have been developed to assess the likelihood of a mass being cancerous. Although these algorithms offer high sensitivity for detection of ovarian cancer, specificity is limited, which can result in high false-positive rates, increased patient anxiety, and unnecessary procedures that is associated with significant morbidity. Here, we developed and independently validated a blood-based metabolite panel for distinguishing early-stage ovarian cancers from benign pelvic masses. We additionally showed that the metabolite panel in combination with ROMA yields a higher positive predictive value with improved specificity for early-stage OvCa compared to ROMA alone. The metabolite panel provides a clinical tool that complements ROMA for improved prediction of malignancy. Such a test would better inform clinical decision making and improve patient outcomes.
Acknowledgements
Supported in part through the Cancer Prevention and Research Institute of Texas grants RP160145 and RP101382, the generous philanthropic contributions to The University of Texas MD Anderson Cancer Center Moon Shots Program™ and a faculty fellowship from The University of Texas MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment (JFF). This work was also supported by grants from the NCI Early Detection Research Network (5 U01 CA200462-02), the MD Anderson Ovarian SPORE (P50 CA217685), National Cancer Institute, Department of Health and Human Services; the Cancer Prevention Research Institute of Texas (RP160145); Golfer’s Against Cancer, Ann and Henry Zarrow Foundation; the Mossy Foundation, the Roberson Endowment, and generous donations from Stuart and Gaye Lynn Zarrow, Barry Elson, and Arthur and Sandra Williams. KAD was partially supported by National Institute of Health SPORE grant P50CA140388, CCTS grant 5UL1TR0003167, CPRIT grant RP160693, and the National Cancer Institute grant CA016672.
Footnotes
Conflict of Interest: Dr. Bast receives royalties from Fujirebio Diagnostics Inc. for the discovery of CA125. An Invention Disclosure Report related to findings reported herein has been submitted to the University of Texas.
References
- 1.Buys SS, Partridge E, Greene MH, Prorok PC, Reding D, Riley TL, et al. Ovarian cancer screening in the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial: findings from the initial screen of a randomized trial. American journal of obstetrics and gynecology 2005;193(5):1630–9. [DOI] [PubMed] [Google Scholar]
- 2.Pavlik EJ, Ueland FR, Miller RW, Ubellacker JM, DeSimone CP, Elder J, et al. Frequency and disposition of ovarian abnormalities followed with serial transvaginal ultrasonography. Obstetrics & Gynecology 2013;122(2 PART 1):210–7. [DOI] [PubMed] [Google Scholar]
- 3.Lu KH, Skates S, Hernandez MA, Bedi D, Bevers T, Leeds L, et al. A 2-stage ovarian cancer screening strategy using the Risk of Ovarian Cancer Algorithm (ROCA) identifies early-stage incident cancers and demonstrates high positive predictive value. Cancer 2013;119(19):3454–61 doi 10.1002/cncr.28183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buys SS, Partridge E, Black A, Johnson CC, Lamerato L, Isaacs C, et al. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening randomized controlled trial. Jama 2011;305(22):2295–303. [DOI] [PubMed] [Google Scholar]
- 5.Dochez V, Caillon H, Vaucel E, Dimet J, Winer N, Ducarme G. Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review. Journal of ovarian research 2019;12(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueland FR, Desimone CP, Seamon LG, Miller RA, Goodrich S, Podzielinski I, et al. Effectiveness of a multivariate index assay in the preoperative assessment of ovarian tumors. Obstetrics & Gynecology 2011;117(6):1289–97. [DOI] [PubMed] [Google Scholar]
- 7.Moore RG, McMeekin DS, Brown AK, DiSilvestro P, Miller MC, Allard WJ, et al. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecologic oncology 2009;112(1):40–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman RL, Herzog TJ, Chan DW, Munroe DG, Pappas TC, Smith A, et al. Validation of a second-generation multivariate index assay for malignancy risk of adnexal masses. American Journal of Obstetrics and Gynecology 2016;215(1):82.e1–.e11 doi 10.1016/j.ajog.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab 2016;23(1):27–47 doi 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science 2020;368(6487) doi 10.1126/science.aaw5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 2015;17(4):351–9 doi 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang WL, Gentry-Maharaj A, Simmons A, Ryan A, Fourkala EO, Lu Z, et al. Elevation of TP53 Autoantibody Before CA125 in Preclinical Invasive Epithelial Ovarian Cancer. Clin Cancer Res 2017;23(19):5912–22 doi 10.1158/1078-0432.Ccr-17-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fahrmann JF, Vykoukal J, Fleury A, Tripathi S, Dennison JB, Murage E, et al. Association Between Plasma Diacetylspermine and Tumor Spermine Synthase With Outcome in Triple-Negative Breast Cancer. J Natl Cancer Inst 2020;112(6):607–16 doi 10.1093/jnci/djz182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahrmann JF, Bantis LE, Capello M, Scelo G, Dennison JB, Patel N, et al. A Plasma-Derived Protein-Metabolite Multiplexed Panel for Early-Stage Pancreatic Cancer. J Natl Cancer Inst 2019;111(4):372–9 doi 10.1093/jnci/djy126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahrmann JF, Irajizad E, Kobayashi M, Vykoukal J, Dennison JB, Murage E, et al. A MYC-Driven Plasma Polyamine Signature for Early Detection of Ovarian Cancer. Cancers (Basel) 2021;13(4) doi 10.3390/cancers13040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vykoukal J, Fahrmann JF, Gregg JR, Tang Z, Basourakos S, Irajizad E, et al. Caveolin-1-mediated sphingolipid oncometabolism underlies a metabolic vulnerability of prostate cancer. Nat Commun 2020;11(1):4279 doi 10.1038/s41467-020-17645-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang WL, Lu Z, Guo J, Fellman BM, Ning J, Lu KH, et al. Human epididymis protein 4 antigen-autoantibody complexes complement cancer antigen 125 for detecting early-stage ovarian cancer. Cancer 2020;126(4):725–36 doi 10.1002/cncr.32582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Candel A, Parmar V, LeDell E, Arora A. Deep learning with H2O. H2O ai Inc 2016:1–21. [Google Scholar]
- 19.Greenwell BM, Boehmke BC, McCarthy AJ. A simple and effective model-based variable importance measure. arXiv preprint arXiv:180504755 2018. [Google Scholar]
- 20.Gedeon TD. Data mining of inputs: analysing magnitude and functional measures. International Journal of Neural Systems 1997;8(02):209–18. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz-Muñoz B, Aznar-Oroval E, García AG, Peris AC, Ballestero PP, Yepes MS, et al. HE4, Ca125 and ROMA algorithm for differential diagnosis between benign gynaecological diseases and ovarian cancer. Tumor Biology 2014;35(7):7249–58. [DOI] [PubMed] [Google Scholar]
- 22.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44(3):837–45. [PubMed] [Google Scholar]
- 23.Fahrmann JF, Irajizad E, Kobayashi M, Vykoukal J, Dennison JB, Murage E, et al. A MYC-Driven Plasma Polyamine Signature for Early Detection of Ovarian Cancer. Cancers 2021;13(4):913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vimr ER, Troy FA. Regulation of sialic acid metabolism in Escherichia coli: role of N-acylneuraminate pyruvate-lyase. Journal of bacteriology 1985;164(2):854–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brigham C, Caughlan R, Gallegos R, Dallas MB, Godoy VG, Malamy MH. Sialic acid (N-acetyl neuraminic acid) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine epimerase. Journal of bacteriology 2009;191(11):3629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadirvelraj R, Yang J-Y, Kim HW, Sanders JH, Moremen KW, Wood ZA. Comparison of human poly-N-acetyl-lactosamine synthase structure with GT-A fold glycosyltransferases supports a modular assembly of catalytic subsites. Journal of Biological Chemistry 2021;296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilvo M, De Santiago I, Gopalacharyulu P, Schmitt WD, Budczies J, Kuhberg M, et al. Accumulated metabolites of hydroxybutyric acid serve as diagnostic and prognostic biomarkers of ovarian high-grade serous carcinomas. Cancer research 2016;76(4):796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curtin JP. Management of the adnexal mass. Gynecologic oncology 1994;55(3):S42–S6. [DOI] [PubMed] [Google Scholar]
- 29.Coulter M, Morell A, Samborski A, Miller M, Moore R. Risk of Ovarian Malignancy Algorithm (ROMA) through time and space: a meta-analysis. Gynecologic Oncology 2021;162:S266 doi 10.1016/S0090-8258(21)01154-9. [DOI] [Google Scholar]
- 30.Carney ME, Lancaster JM, Ford C, Tsodikov A, Wiggins CL. A population-based study of patterns of care for ovarian cancer: who is seen by a gynecologic oncologist and who is not? Gynecol Oncol 2002;84(1):36–42 doi 10.1006/gyno.2001.6460. [DOI] [PubMed] [Google Scholar]
- 31.Bast RC Jr, Klug TL, John ES, Jenison E, Niloff JM, Lazarus H, et al. A radioimmunoassay using a monoclonal antibody to monitor the course of epithelial ovarian cancer. New England journal of medicine 1983;309(15):883–7. [DOI] [PubMed] [Google Scholar]
- 32.Jacobs I, Bast RC Jr. The CA 125 tumour-associated antigen: a review of the literature. Human reproduction 1989;4(1):1–12. [DOI] [PubMed] [Google Scholar]
- 33.Niemi RJ, Roine AN, Hakkinen MR, Kumpulainen PS, Keinanen TA, Vepsalainen JJ, et al. Urinary Polyamines as Biomarkers for Ovarian Cancer. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society 2017;27(7):1360–6 doi 10.1097/igc.0000000000001031. [DOI] [PubMed] [Google Scholar]
- 34.Munkley J, Scott E. Targeting Aberrant Sialylation to Treat Cancer. Medicines (Basel) 2019;6(4) doi 10.3390/medicines6040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Büll C, Boltje TJ, Balneger N, Weischer SM, Wassink M, van Gemst JJ, et al. Sialic Acid Blockade Suppresses Tumor Growth by Enhancing T-cell-Mediated Tumor Immunity. Cancer Res 2018;78(13):3574–88 doi 10.1158/0008-5472.Can-17-3376. [DOI] [PubMed] [Google Scholar]
- 36.Dobie C, Skropeta D. Insights into the role of sialylation in cancer progression and metastasis. British Journal of Cancer 2021;124(1):76–90 doi 10.1038/s41416-020-01126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun H, Zhou Y, Jiang H, Xu Y. Elucidation of Functional Roles of Sialic Acids in Cancer Migration. Front Oncol 2020;10:401 doi 10.3389/fonc.2020.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Wuhrer M, Holst S. Serum sialylation changes in cancer. Glycoconj J 2018;35(2):139–60 doi 10.1007/s10719-018-9820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkitachalam S, Revoredo L, Varadan V, Fecteau RE, Ravi L, Lutterbaugh J, et al. Biochemical and functional characterization of glycosylation-associated mutational landscapes in colon cancer. Scientific reports 2016;6(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu H, Duan W-M, Shu J, Cheng H-X, Wang W-P, Huang X-E, et al. B3GNT2, a polylactosamine synthase, regulates glycosylation of EGFR in H7721 human hepatocellular carcinoma cells. Asian Pacific Journal of Cancer Prevention 2015;15(24):10875–8. [DOI] [PubMed] [Google Scholar]
- 41.Bristow RE, Smith A, Zhang Z, Chan DW, Crutcher G, Fung ET, et al. Ovarian malignancy risk stratification of the adnexal mass using a multivariate index assay. Gynecol Oncol 2013;128(2):252–9 doi 10.1016/j.ygyno.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 42.Ueland FR. A Perspective on Ovarian Cancer Biomarkers: Past, Present and Yet-To-Come. Diagnostics (Basel) 2017;7(1) doi 10.3390/diagnostics7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Relevant data supporting the findings of this study are available within the Article and Supplementary Information or are available from the authors upon reasonable request.


