Abstract
Enzymatic catalysis is critically dependent on selectivity, active site architecture, and dynamics. To contribute insights into the interplay of these properties, we established an approach with NMR, crystallography, and MD simulations focused on the ubiquitous phosphotransferase adenylate kinase (AK) isolated from Odinarchaeota (OdinAK). Odinarchaeota belongs to the Asgard archaeal phylum that is believed to be the closest known ancestor to eukaryotes. We show that OdinAK is a hyperthermophilic trimer that, contrary to other AK family members, can use all NTPs for its phosphorylation reaction. Crystallographic structures of OdinAK-NTP complexes revealed a universal NTP-binding motif, while 19F NMR experiments uncovered a conserved and rate-limiting dynamic signature. As a consequence of trimerization, the active site of OdinAK was found to be lacking a critical catalytic residue and is therefore considered to be “atypical.” On the basis of discovered relationships with human monomeric homologs, our findings are discussed in terms of evolution of enzymatic substrate specificity and cold adaptation.
A universal NTP-binding motif and trimerization-dependent active site geometry was discovered in an enzyme from Odinarchaeota.
INTRODUCTION
The “hot spring” hypothesis suggests that life may have originated in geothermal sites (1) that can be found both on land and in deep sea vents (2). Enzymes isolated from microorganisms that colonize such high-temperature habitats are of general interest to protein science because they may be considered as ancestral proteins. Comparison of their structure and function with modern variants can provide mechanistic insight into various functional, structural, and biophysical aspects such as relaxed versus constricted substrate specificity and cold adaptation. A phylum of archaea named Asgard archaea was reported 2015 and was isolated close to the black smoker vent field called Loki’s castle situated on the arctic mid-ocean ridge (3). The Asgard archaea phylum consists of Loki-, Thor-, Heimdall-, and Odinarchaeota (4) and is proposed to represent the closest known ancestors to eukaryotic cells (5). An argument in favor of the evolutionary linkage between archaea and eukaryotic cells is the presence of so called eukaryotic signature proteins such as the Ras superfamily of small guanosine triphosphatases (3).
The essential enzyme adenylate kinase (AK) (6) is a member of the larger nucleoside monophosphate (NMP) kinase family of enzymes and is ubiquitously present in all known organisms. A comparative approach of structure, function, and dynamics of AK isolated from different organisms can provide new insights into molecular properties that are important for catalytic rate enhancement. To this end, we have undertaken an integrated structural biology approach centered on an AK isolated from Odinarchaeota (OdinAK) and compared its structural and functional properties with mesophilic AKs of both human and bacterial origin. We have previously discovered that AKs that are destined to different compartments in human cells have different substrate specificities. The mitochondria-specific enzyme AK3 is specific for guanosine 5′-triphosphate (GTP) as phosphoryl donor, whereas the cytosolic enzyme AK1 is specific for adenosine 5′-triphosphate (ATP) (7). The structural basis underlying the GTP versus ATP selectivity lies in the orientation of a short loop, denoted as the selectivity loop, that presents either a backbone hydrogen bond donor or acceptor for the specific recognition of GTP and ATP, respectively (7). Here, we show that, contrary to all known AKs, OdinAK can use all nucleoside triphosphates (NTPs) available in the cell [ATP, GTP, cytidine 5′-triphosphate (CTP), uridine 5′-triphosphate (UTP), and 3′-deoxythymidine 5′-triphosphate (dTTP)] as phosphoryl donor and therefore exhibits a broad substrate preference. The key structural feature of the universal NTP-binding motif is a unique selectivity loop that harbors a glutamine that through its side chain can both accept and donate hydrogen bonds to direct NTP binding. Further, comparative structural analysis of OdinAK with mesophilic homologs provides a potential evolutionary mechanism for enzymatic cold adaptation that is rooted in the oligomeric state of the enzyme.
RESULTS
OdinAK has a unique selectivity loop
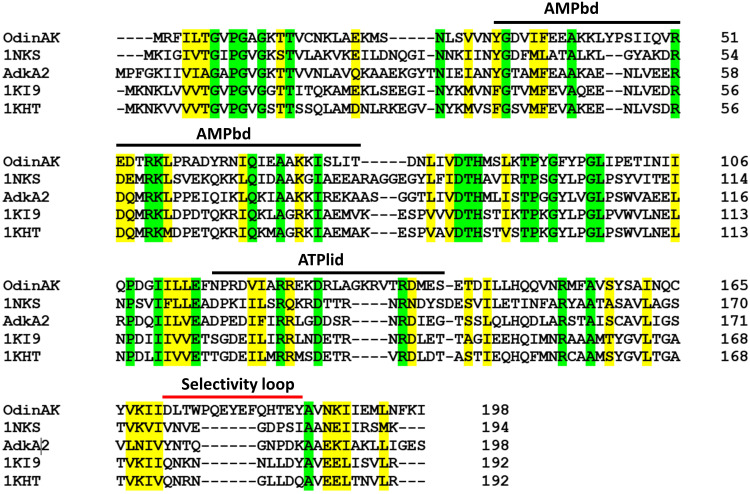
The primary sequences of AKs from Odin-, Thor, Heimdall-, and Lokiarchaeota were identified from the annotated genomic sequences of the respective organisms (3, 4). Because Odinarchaeota appears to be the oldest Asgard lineage based on 16S ribosomal sequencing (8), we targeted AK from this lineage. There exist two AK enzymes that have been annotated to Odinarchaeota with the gene names adkA1 (UniProt accession code: A0A1Q9N9I8) and adkA2 (A0A1Q9N7J7). To select the most interesting enzyme for our studies, we generated a multiple sequence alignment between AdkA1 (hereafter named OdinAK), AdkA2, and three archaeal AKs for which the structures have been determined (Fig. 1). The alignment revealed the distinct feature that OdinAK has a significantly longer selectivity loop compared to the other enzymes including AdkA2. Hence, the substrate recognition mechanism of OdinAK may differ from the established recognition of ATP or GTP with the selectivity loop (7), and therefore, we pursued a deeper analysis into OdinAK.
Fig. 1. Multiple sequence alignment of archaeal AK enzymes.
Shown is a multiple sequence alignment between the two AK enzymes from Odinarchaeota (i.e., AdkA1, hereafter named OdinAK, and AdkA2) and three other archaeal AKs for which the crystallographic structure has been solved. The three other AK sequences are from Sulfolobus acidocaldarius (PDB ID: 1NKS), M. thermolithotrophicus (PDB ID: 1KI9), and Methanococcus voltae (PDB ID: 1KHT). The location of the ATP- and AMP-binding domains (ATPlid and AMPbd) is indicated, together with the location of the selectivity loop (7). The alignment was generated with Clustal Omega. Identical residues are colored green, while similar residues are colored yellow.
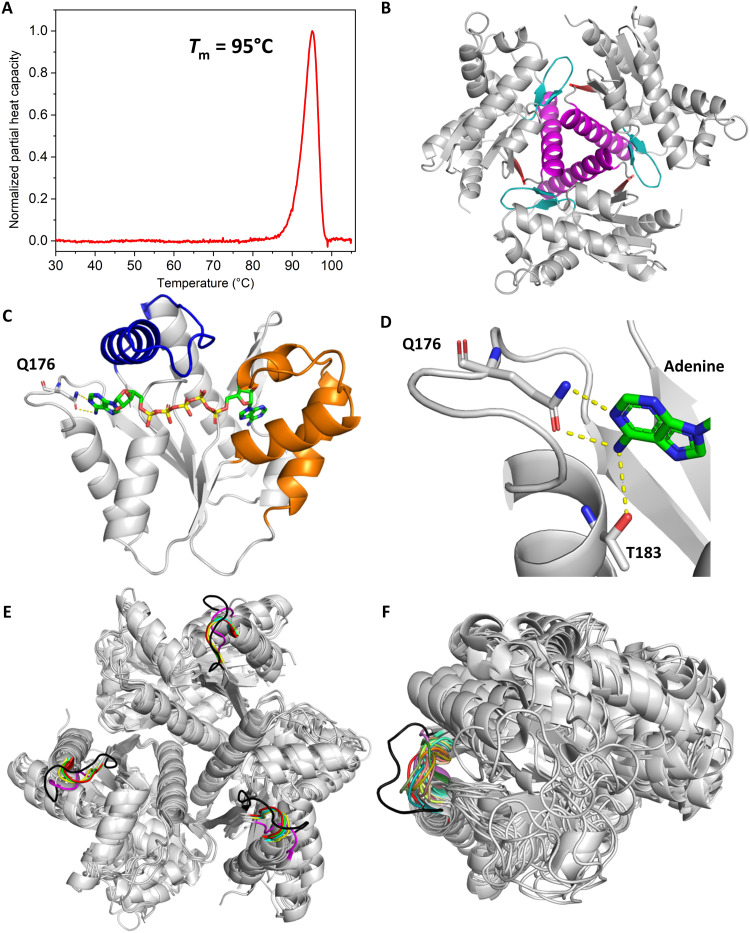
The vast majority of AKs that have been studied at the level of protein structure preferentially uses ATP to phosphorylate adenosine 5′-monophosphate (AMP) (9), and only a few prefer GTP (7). We have recently discovered the importance of the selectivity loop for NTP selectivity by its ability to adopt unique conformations for the respective NTP (fig. S1) (7), and if a structure is available, then it is straightforward to predict if an NMP kinase is specific for ATP or GTP. In the absence of a structural model for OdinAK, an informed guess suggests that the most probable substrate is ATP. Most Odinarchaeota lineages are of hyperthermophilic origin and have been isolated from high temperature habitats (4). The melting point of an enzyme serves as an indicator to the habitat temperature of an organism and this is, for instance, reflected in a range of melting points in Escherichia coli proteins of 37° to 67°C and a corresponding range of 70° to 99°C for proteins from the hyperthermophilic bacterium Thermus thermophilus (10). Here, we find that the hyperthermophilic nature of Odinarchaeota is reflected in the molecular properties of OdinAK by establishing that substrate-free OdinAK has a melting point (Tm) of 95°C (Fig. 2A) as quantified with differential scanning calorimetry (DSC). No difference in the melting point was observed for OdinAK supplemented with 630 μM ATP and 5 mM MgCl2 (fig. S2). In any case, the extreme thermostability of OdinAK demonstrates that the enzyme has evolved to tolerate the extreme temperatures that are present in hydrothermal vents. AKs of archaeal origin are trimeric as inferred from the cases where structures have been reported (11–13). Consistent with these findings, we found that the biological oligomerization state of OdinAK is trimeric as determined from quantitative light scattering experiments (fig. S3). The structure of apo OdinAK (Fig. 2B) was solved by x-ray crystallography using molecular replacement with the structure of Methanococcus thermolithotrophicus AK (1KI9.pdb) (11) as the search model. The overall structure has the trimeric NMP kinase fold where trimerization is mediated through interactions between trimerization helices (amino acid residues E142 to N163) and between β-hairpin loops (residues M86 to P96) and β strands (residues Y166 to I169) of adjacent monomers (Fig. 2B and table S1).
Fig. 2. Structure of hyperthermophilic OdinAK.
(A) The thermal stability of OdinAK was followed with DSC. The Tm was determined from the inflection point of the thermogram and was found to be 95°C. The data have been corrected with the solvent background, and the normalized partial heat capacity is displayed. (B) Crystal structure of trimeric apo OdinAK showing trimer contacts between trimerization helices (magenta), β-hairpins (teal), and β strands (red) of adjacent monomers. (C) Structure of a single subunit of closed OdinAK in complex with Ap5A. The specific recognition of the adenosine base of ATP by the side chain of Q176 is highlighted. (D) Zoom-in on the selectivity loop showing recognition of the ATP base by the side chains of Q176 and T183. (E and F) Superimposition of 5 known trimeric and archaeal (E) and 30 monomeric (F) AK structures from the PDB illustrating that OdinAK has a unique and significantly longer selectivity loop (shown in black) relative to all other AKs.
Because ATP was predicted to be the most likely substrate for phosphorylation of AMP by OdinAK, we determined the crystal structure of the enzyme in complex with the tight binding ATP/AMP-mimicking inhibitor, P1,P5-di(adenosine-5′) pentaphosphate (Ap5A) (Fig. 2C) (9). The apo and Ap5A structures provide snapshots of the extreme stages during catalysis and reveal that there exist “induced fit” like conformational changes to both the ATPlid and the AMP-binding domain (AMPbd). Although the overall structure of OdinAK displays the signature AK interaction principles for binding of ATP and AMP (fig. S4), the enzyme contains a unique mode for recognizing the base of ATP. In general, AKs contain a short selectivity loop that resides between the last β strand and the C-terminal α helix, and substrate recognition is accomplished through a hydrogen bond formed between the nucleobase and a backbone atom in the selectivity loop (fig. S1) (7). In contrast, the selectivity loop of OdinAK is significantly longer consistent with the multiple sequence alignment in Fig. 1 and provides a platform for optimal positioning of the Q176 side chain to form a hydrogen bond with ATP (Fig. 2D). In addition, there exists a hydrogen bond donated from the NH2 group on the C6 carbon of the adenosine base to the side-chain oxygen of T183. The extended length and unique nature of the selectivity loop in OdinAK are illustrated by comparing its structure with other known AK structures from archaea and other organisms that are available in Protein Data Bank (PDB) (Fig. 2, E and F). In short, the ATP recognition mode is defined by interactions with Q176 and T183. Because a glutamine side chain contains both a hydrogen bond acceptor and donor, we hypothesized that OdinAK can use both ATP and GTP as a substrate for AMP phosphorylation.
Binding of nucleotides to OdinAK, kinetics, and conformational dynamics during catalysis
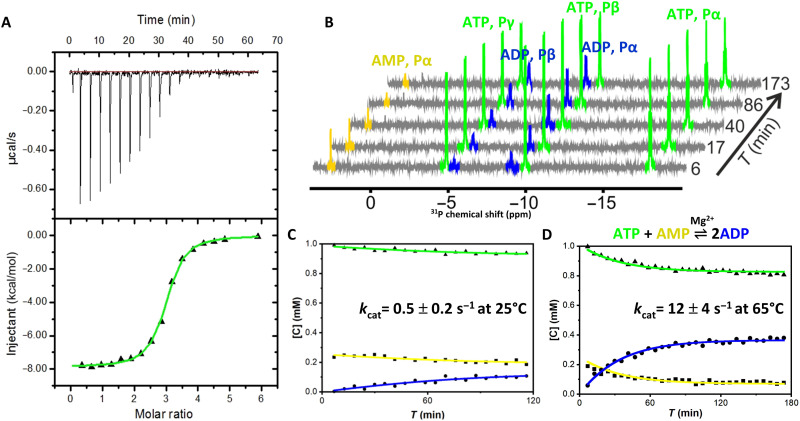
To elucidate the substrate selectivity of OdinAK, the dissociation constant (Kd) and catalytic activity (kcat) were quantified for ATP.Although the endogenous temperature of OdinAK is expected to be above 70°C (given its hyperthermophilic nature), for practical reasons and for the possibility to compare values to other AK enzymes, the Kd value for ATP binding to OdinAK was determined at 25°C with isothermal titration calorimetry (ITC). The Kd was determined to be 0.3 ± 0.1 μM with a molar ratio of 3:1 (ATP to OdinAK) as anticipated for a trimer (Fig. 3A and Table 1). Because the physiologically active state of ATP inside cells is in a complex with Mg2+, we also determined the Kd for ATP in the presence of Mg2+. It was found to be 0.2 ± 0.1 μM (fig. S5) and is within experimental uncertainty equal to the case without Mg2+. This similarity in ATP-binding affinities in the presence and absence of Mg2+ also fits with the previous finding that the Kd for ATP binding (in the absence of Mg2+) to E. coli AK (AKeco) (14) matches the Michaelis constant (Km) value obtained from an enzymatic assay (performed with Mg2+) (15). ATP binds to each monomer of OdinAK independently because the ITC data could be fitted with one collective Kd of 0.3 ± 0.1 μM without observable signs of cooperativity. Comparing with the Kd of 53 μM for ATP binding to AKeco (14), it is evident that OdinAK binds ATP with high affinity. The catalytic activity of OdinAK was quantified using a real-time 31P nuclear magnetic resonance (NMR) assay, especially developed for quantitative determination of kcat values for phosphoryl transfer (16). 31P NMR is favorable for following the AK reaction because it can be used to accurately distinguish between the relevant nucleoside phosphates [i.e., AMP, adenosine 5′-diphosphate (ADP), and ATP]. In short, the assay is initiated by mixing ATP, AMP, and Mg2+ with a catalytic concentration of OdinAK, and the kcat value is extracted from quantitative fits of the rate equations to the buildup of ADP and the simultaneous depletion of ATP and AMP (16). Because of the inherently low sensitivity of NMR, the assay is performed at saturating nucleoside phosphate concentrations and therefore only kcat (and not Km) can be extracted from the experiment. This limitation can however be compensated for by the Kd values determined using ITC. Representative NMR spectra from the 31P activity assay are illustrated in Fig. 3B. Enzymes that catalyze the AK reaction have evolved by suppressing unwanted water mediated hydrolysis of ATP (17), and we could demonstrate the same suppression for OdinAK as deduced from the 31P NMR assay (fig. S6).
Fig. 3. OdinAK substrate-binding affinity and enzyme kinetics.
(A) ITC binding isotherm between OdinAK and ATP at 25°C. The best fitted isotherm is shown as a green line, and fitted parameters are displayed in Table 1. (B) Representative 31P NMR spectra showing the real-time evolution of mono-, di-, and trinucleoside phosphates from AMP, ADP, and ATP, respectively, over time. The experiment was initiated by mixing ATP, AMP, and Mg2+ together with catalytic amounts of OdinAK. (C and D). Quantification of enzymatic activity by a real-time 31P NMR assay at 25°C (C) and 65°C (D). The rate equations (16) were fitted to the evolution of nucleotide concentrations to obtain kcat (Table 2).
Table 1. Thermodynamic parameters of binding between OdinAK and its substrates determined using ITC at 25°C.
| Ligand | N | Kd (μM) | ΔH0 (kcal/mol) | −TΔS0 (kcal/mol) | ΔG0 (kcal/mol) |
| ATP | 2.9 ± 0.1 | 0.3 ± 0.1 | −7.4 ± 0.6 | −1.5 ± 0.8 | −8.9 ± 0.2 |
| ATP + Mg2+ | 2.9 ± 0.3 | 0.2 ± 0.1 | −9.7 ± 2.2 | 0.55 ± 2.51 | −9.1 ± 0.4 |
| GTP | 3.1 ± 0.1 | 7.2 ± 0.6 | −3.8 ± 0.1 | −3.3 ± 0.1 | −7 ± 0.1 |
| CTP | 2.8 ± 0.2 | 28.7 ± 7 | −4.0 ± 0.6 | −2.2 ± 0.7 | −6.2 ± 0.1 |
| UTP | 2.7 ± 0.1 | 4.6 ± 0.5 | −4.5 ± 0.1 | −2.7 ± 0.1 | −7.3 ± 0.1 |
| dTTP | 2.8 ± 0.1 | 24.2 ± 8 | −1.8 ± 0.1 | −4.5 ± 0.1 | −6.3 ± 0.1 |
| ADP | 3.1 ± 0.1 | 0.8 ± 0.1 | −8.3 ± 0.5 | −0.1 ± 0.5 | −8.2 ± 1 |
| ADP + Mg2+ | 2.8 ± 0.1 | 0.06 ± 0.01 | −11.3 ± 0.4 | 1.5 ± 0.5 | −9.8 ± 0.1 |
The catalytic activity of OdinAK was quantified at the two temperatures: 25° and 65°C. The rationale for selecting these temperatures is that 25°C allows a comparison of activities to other AK enzymes (particularly AKeco), while 65°C was selected as a reasonable proxy of the expected hyperthermophilic habitat of Odinarchaeota. By using the 31P NMR assay (16), we found that kcat of OdinAK with ATP as phosphoryl donor was 0.5 ± 0.2 s−1 at 25°C and 12 ± 4 s−1 at 65°C (Fig. 3, C and D, and Table 2). Again, benchmarking toward AKeco that has a kcat value of 360 s−1 at 25°C (15), it is evident that the absolute magnitude of kcat of OdinAK is low. The free energy differences for activities and ATP-binding affinities of OdinAK versus AKeco at 25°C are hence −16 and −13 kJ mol−1, respectively. The near proportional energetic differences indicate that the low catalytic activity of OdinAK is predominantly dictated by a high binding affinity. Combining our data supports that OdinAK can be safely classified as a member of the AK family.
Table 2. OdinAK enzyme kinetics and dynamics obtained using 31P real-time NMR and 19F NMR relaxation dispersion (RD), respectively, at 65°C.
| Kinetics | Dynamics | |||||
| Substrates (+ Mg2+) | kcat (s−1) | kex (s−1) | P closed | P open | kclose (s−1) | kopen (s−1) |
| ADP | 12 ± 4 | 1476 ± 99 | 0.9919 | 0.0081 | 1463 ± 98 | 13 ± 2 |
| ATP + AMP | 12 ± 4 | 1360 ± 109 | 0.9915 | 0.0085 | 1357 ± 114 | 11 ± 2 |
| GTP + AMP | 11 ± 3 | 1324 ± 70 | 0.9855 | 0.0145 | 1294 ± 56 | 18 ± 1 |
| CTP + AMP | 10 ± 2 | 1495 ± 76 | 0.9852 | 0.0148 | 1473 ± 104 | 21 ± 2 |
| UTP + AMP | 14 ± 2 | 1403 ± 106 | 0.9873 | 0.0127 | 1384 ± 99 | 18 ± 2 |
| dTTP + AMP | 13 ± 3 | 1409 ± 93 | 0.9865 | 0.0135 | 1387 ± 89 | 19 ± 3 |
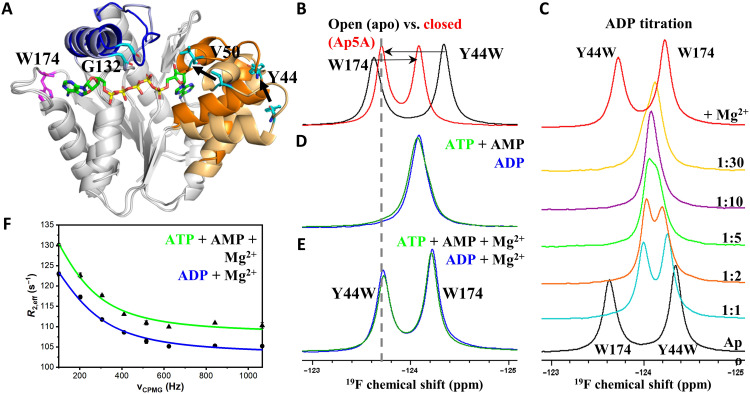
The core of our catalytic model for AK catalysis is a rate-limiting substrate release step that mechanistically is dependent on slow opening of the substrate-binding domains as demonstrated for both bacterial (AKeco) (18) and human (AK3) (7) enzymes. To probe whether this dynamic signature is also a feature of OdinAK, we turned to NMR and spin relaxation experiments. Characterization of dynamic exchange processes can be obtained from relaxation dispersion (RD) experiments (19, 20) providing (i) kinetics (rate constants), (ii) thermodynamics (populations), and (iii) structural information (chemical shifts). These parameters can be quantified if the dynamic process occurs on the micro- to millisecond time scale (21) and if the chemical shift for a particular nucleus is different for the states in between which exchange occurs. An RD experiment from which data can be fitted will display a decrease in signal intensity as function of an applied so called CPMG (Carr-Purcell-Meiboom-Gill) field (21). Because the molecular weight of trimeric OdinAK (69 kDa) is too large for standard NMR approaches, we sought to use the favorable properties of the 19F nuclei (22). To enable fluorine experiments, we incorporated 19F in tryptophan residues by supplementing a minimal growth medium with 5-fluoroindole (23). The approach was to acquire 19F RD experiments (24) under turnover conditions, i.e., with saturating concentrations of substrates and Mg2+. OdinAK contains one tryptophan (W174) that is located in the selectivity loop (Fig. 4A). This position, however, was silent to conformational dynamics on the CPMG time scale (i.e., micro- to milliseconds) (21) and consequently yielded flat dispersion profiles (fig. S7). Therefore, we introduced single-tryptophan replacements at several positions (Y44, V50, and G132) in the enzyme in search for a sensitive probe of conformational dynamics (Fig. 4A). The replacement of the tyrosine at position 44 with 5-fluorotryptophan yielded 19F dispersion profiles with substantial amplitude for quantitative analysis. The enzymatic activity of the Y44W variant at 65°C is virtually identical to the wild type as deduced from 31P NMR (fig. S8).
Fig. 4. OdinAK conformational dynamics during catalysis.
(A) Selection of suitable probe sites for 19F NMR by comparing the structures of OdinAK in open (substrate-free) and closed (Ap5A-bound) states. Wild-type W174 (magenta) located in the selectivity loop yielded flat dispersion profiles and therefore Y44, V50, and G132 (cyan) were tested for insertion of 19F probe. (B) One-dimensional (1D) 19F NMR spectra of apo (black) and Ap5A-bound (red) OdinAK Y44W at 65°C, showing chemical shifts of mutant Y44W and wild-type W174 peaks as fingerprints for open and closed states of the enzyme. (C) Titration of OdinAK Y44W with ADP at 65°C showing that the enzyme adopts the closed conformation only in the presence of both binding site substrates and Mg2+. (D) 19F NMR spectra of substrate-bound OdinAK Y44W at 65°C. The green and blue spectra correspond to saturating concentrations [ATP (1:10) + AMP (1:10)] and ADP (1:20), respectively. Under these conditions, the signals from both tryptophans (44 and 174) are overlapping. (E) 19F NMR spectra of OdinAK Y44W under turnover conditions, i.e., substrate-bound states supplemented with 5 mM Mg2+ at 65°C. Addition of Mg2+ shifts the enzyme toward the closed state as deduced from the chemical shift of Y44W and indicated with the dashed line. (F) Quantification of dynamics during catalysis from 19F RD experiments under turnover conditions at 65°C. The observed R2 relaxation rate (R2,eff) obtained from Y44W is plotted against the applied CPMG field for samples generated from ATP, AMP, and Mg2+ (green) or ADP and Mg2+ (blue). The fitted parameters are shown in table S2, and rate constants for enzyme closing (kclose) and opening (kopen) are shown in Table 2.
To quantify the internal dynamics of OdinAK that is relevant for enzymatic turnover, it is necessary to have both substrates and magnesium ions present. 19F chemical shifts of the OdinAK Y44W variant demonstrate that the enzyme:substrate complex adopts its fully closed conformation only upon the addition of Mg2+ (Fig. 4, B to E). Therefore, magnesium has at least two important roles for OdinAK catalysis. First, as generally established for processes involving ATP, magnesium is an essential cofactor that increases the rate of phosphoryl transfer (25, 26). Second, magnesium modulates the open/closed equilibrium in the presence of substrates and shuttles the enzyme:substrate complex to the closed and active conformation. Consistent with the latter, ITC revealed a 13-fold increase in binding affinity with ADP in the presence of Mg2+ (Table 1 and fig. S9). Magnesium thus has a significantly stronger effect on a catalytic state (i.e., ATP and AMP present) compared to the scenario with only ATP. RD curves of Y44W were indicative of internal dynamics and yielded a sizeable modulation of peak intensity as function of applied CPMG field (Fig. 4F). In line with a fast exchange process, the dispersion profiles displayed featureless and monotonic decays. Fast exchanges only allow fitting of a global exchange rate constant (kex) that, for a two-state process (here closing and opening of substrate-binding domains), is equal to the sum of the microscopic rate constants (i.e., kex = kclose + kopen). The microscopic rate constants can, however, be extracted if the chemical shift difference for the exchanging states can be measured in a separate experiment. Assuming that the exchange dynamics of Y44W is reporting on closing and opening of the substrate-binding domains (Supplementary Text), it is possible to measure the difference in 19F chemical shift from NMR spectra of open (substrate-free) and closed (Ap5A-bound) states (refer to Materials and Methods for a more detailed description). By applying this assumption and introducing the relevant difference in 19F chemical shift (table S2), we found that the microscopic rate constants kclose and kopen correspond to 1357 ± 114 s−1 and 11 ± 2 s−1 at 65°C, respectively (Fig. 4F and Table 2). The absolute magnitudes of kopen (11 ± 2 s−1) and kcat (12 ± 4 s−1) overlap within their experimental uncertainties and suggest that enzymatic turnover by OdinAK is rate-limited by kopen and that the slowest step in the enzymatic cycle is opening of the substrate-binding domains. Overall, the signature dynamics found for both bacterial and human AKs can therefore be extended also to Odinarchaeota. The presence of rate-limiting conformational dynamics in AK enzymes across vastly different species suggests that this feature may be evolutionarily conserved in all AK enzymes and potentially also in the larger NMP kinase family of enzymes.
OdinAK contains a universal NTP-binding motif
Intrigued by the finding that the selectivity loop of OdinAK presents a glutamine side chain for its recognition of the adenosine base of ATP, we hypothesized that the enzyme can also use GTP and potentially other NTPs (CTP, UTP, and dTTP) as phosphoryl donor. The underlying physiochemical argument is that specific recognition of ATP or GTP is dependent on the ability of an enzyme to provide a hydrogen bond acceptor (ATP case) or hydrogen bond donor (GTP case) (fig. S1) (7). These boundary conditions are satisfied by a glutamine side chain with its ability to both accept and donate hydrogen bonds. From the 31P NMR assay, we found that OdinAK can use GTP and all other NTPs as substrate for phosphorylation of AMP. As deduced from 31P NMR, the kcat values for all NTPs are, within experimental uncertainty, comparable to the value observed for ATP (Table 2 and fig. S10), and on the basis of ITC, the binding affinities (Kd) for the remaining NTPs range between 4 and 30 μM, with UTP being the highest among them (Table 1 and fig. S11). The finding that OdinAK can use all naturally occurring NTPs implies that the selectivity loop of OdinAK harbors a universal NTP-binding motif.
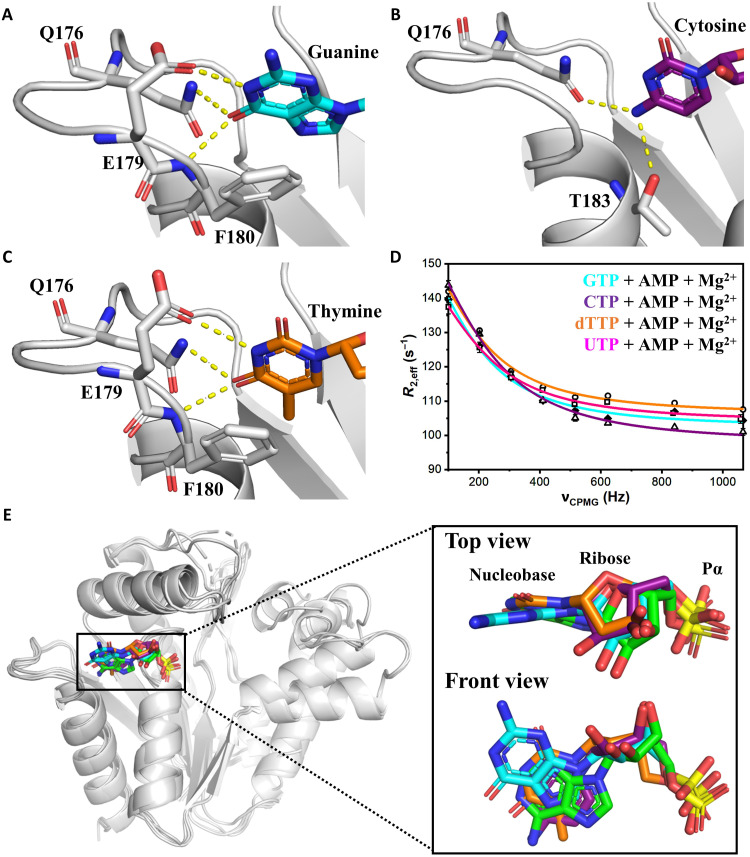
To decipher the molecular basis for the broad substrate specificity profile of OdinAK, we determined the crystal structures of the enzyme in complexes with GTP, CTP, and dTTP (Fig. 5, A to C, and fig. S12). As predicted from the Ap5A complex, the dual hydrogen bonding capacity of the Q176 side chain plays a vital role in all complexes. In the GTP complex, the key aspect in recognition is that the side chain of Q176 donates a hydrogen bond to the carbonyl oxygen at C6 position of the guanosine base, contrasting the ATP case where the side chain accepts a hydrogen bond from the substituent on C6 carbon. In addition, there are hydrogen bonds between the backbone nitrogen of F180 and the carbonyl oxygen on C6 position of the guanosine base and between the side chain of E179 and N1 position of the guanosine base (Fig. 5A). The shift in hydrogen bonding pattern is accomplished with only minor conformational changes in the selectivity loop and invariant positioning of Q176.
Fig. 5. Structural basis for universal NTP binding by OdinAK.
(A to C) Crystal structures of OdinAK in complex with NTPs demonstrate that the key determinant of selective binding of the nucleobases is the interactions formed by the side chain of Q176. In addition, the interactions formed by other important side chains are indicated in the different complexes. (A) Structure of OdinAK in complex with GTP. (B) Structure of OdinAK in complex with CTP. (C) Structure of OdinAK in complex with dTTP. (D) 19F CPMG RD profiles obtained from fluorinated Y44W for different substrates under turnover conditions (compare to Fig. 4F). The resulting fits from the data (table S2 and Table 2) show that OdinAK is rate-limited by slow conformational dynamics for all NTPs. (E) A key factor for the broad substrate specificity of OdinAK is harnessing of the endogenous flexibility of NTP molecules. Shown is a superimposition of NTP substrates bound to OdinAK. The color coding is as follows: ATP (green), GTP (cyan), CTP (purple), and dTTP (orange). The overlay was generated by superimposing the carbon α atoms of OdinAK, and for simplicity, only the base, ribose, and α phosphate are shown for the NTPs.
By solving the structures of complexes with the pyrimidines, CTP (Fig. 5B) and dTTP (Fig. 5C), a general pattern emerges with two distinct modes of NTP recognition used by OdinAK. The two modes are dependent on the substituent at C6 carbon of the nucleobase, and either of the modes is recruited depending on the base. The first mode is used in the cases of ATP and CTP and is defined by interactions with residues Q176 and T183. Conversely, the second mode is recruited in GTP and dTTP binding and comprises interactions with residues Q176, E179, and F180. Thus, the common denominator in recognition of all NTPs is the hydrogen bonding to the side chain of Q176, acting either as a hydrogen bond donor or acceptor. In addition, the malleability of torsion angles in the nucleosides allows the one-ringed pyrimidines to extend all the way between the fixed position of the α phosphate and the side chain of Q176. While the crystal structures reveal the structural basis for the universal NTP binding capacity of OdinAK, 19F RD experiments demonstrate that the dynamic signature with rate-limiting and slow conformational dynamics during enzyme opening is also preserved for all NTPs (Fig. 5D, Table 2, and fig. S13).
Together, the key physiochemical and structural principles underlying the universal NTP selectivity of OdinAK are (i) an extended selectivity loop providing a platform for optimal positioning of the Q176 side chain for NTP recognition, (ii) the ability of Q176 side chain to act both as hydrogen bond donor and acceptor, and (iii) by exploiting the endogenous flexibility of the NTP substrates (Fig. 5E). Furthermore, in performing the catalytic function, the dynamic signature of the enzyme, i.e., slow conformational and rate-limiting dynamics, is preserved for all NTP substrates.
Extreme thermostability of OdinAK is dependent on trimerization
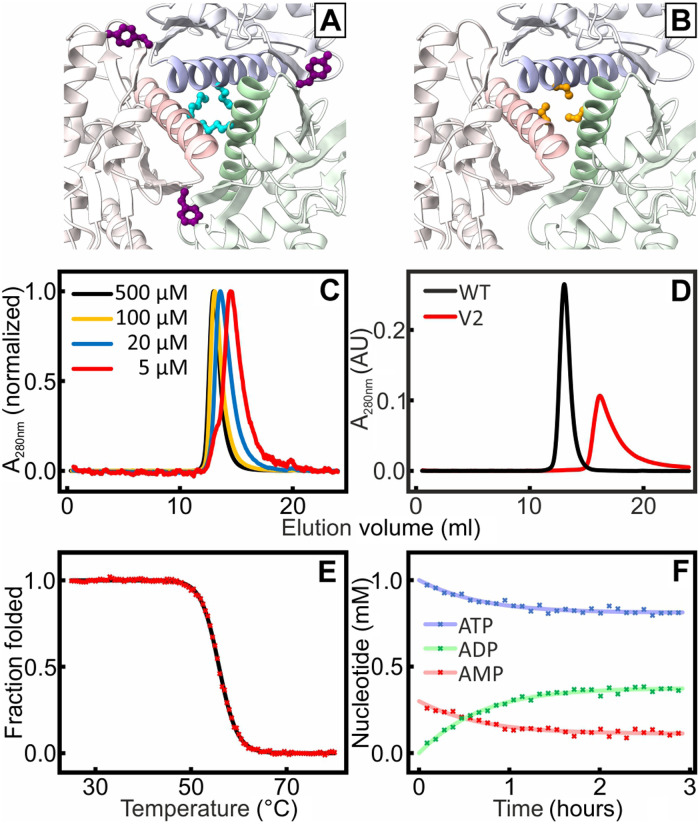
The trimerization interfaces of OdinAK are extensive with, on average, 824 ± 8 Å2 buried surface area per interface and monomer as calculated with the interactive tool PDBePISA. This is equal to a total burial of ~1600 Å2 per interaction interface, which is in good agreement with expectations for protein-protein interactions (i.e., 500 to 2000 Å2) (27). To test the contribution of the trimerization interfaces to the extreme thermal stability of OdinAK, we designed two OdinAK variants denoted as V1 and V2 (Fig. 6, A and B), both of which are designed to destabilize the trimerization interfaces significantly. In V1, M154 and Y166 were mutated to glutamate, both replacing a hydrophobic residue with a negatively charged residue (i.e., the M154E/Y166E double mutant; Fig. 6A). The assembly status of the variants was then quantified with size exclusion chromatography (SEC). For V1, we did not observe a fully monomeric variant but rather a concentration-dependent partitioning between monomer, dimer, and trimer as inferred from concentration-dependent elution profiles (Fig. 6C). Because the perturbation of the interfaces in V1 was not sufficient to fully shuttle the enzyme to a monomer, we designed the triple-mutant V2 (building upon V1) that contains the replacements M154R, S158R, and Y166R. Serine-58 is included because it forms a plug in the trimerization helices by participating in a ring like structure (Fig. 6B). With these replacements, the resulting enzyme is fully monomeric as assessed from the SEC elution volume (Fig. 6D). The Tm of the substrate-free monomeric OdinAK variant V2 was found to be 56°C (Fig. 6E), which is almost 40°C lower compared to the wild-type enzyme and is very similar to that of substrate-free monomeric AKeco [~57°C (28)]. The catalytic activity of the monomeric triple mutant was quantified with 31P NMR assay and found to be virtually identical (0.5 s−1) to the wild-type trimer at 25°C (Fig. 6F). Assuming that the solvent exposed positions in V2 does not significantly affect the thermal stability of the monomer, it appears that trimerization of the wild-type enzyme contributes significantly toward its thermal stability.
Fig. 6. Formation of monomeric OdinAK by disruption of trimerization interfaces.
The trimerization interfaces were destabilized by replacing key amino acid residues identified from structural analysis. In V1, M154 (cyan) and Y166 (purple) (A) were mutated into glutamic acid. In V2, Met154, Tyr166, and Ser158 (orange) (B) were mutated into arginines. (C) SEC of different concentration samples of V1 showed that the elution for samples with decreasing concentration samples was shifted toward higher volumes, indicating the presence of an equilibrium between different oligomeric states. (D) The SEC profile for V2 (red) indicated a fully monomeric state that elutes at a volume that is significantly larger than that of a wild-type (WT) reference trimer (black). (E) Thermal unfolding of V2 probed with CD spectroscopy at 222 nm; Tm was determined to 56°C. (F) The kcat value of monomeric V2 at 25°C is 0.5 s−1 as quantified from the 31P NMR activity assay.
To address the mechanism of assembly of the trimeric state, we performed molecular dynamics (MD) simulations of the wild type and the V1 variant starting from both the substrate-free and Ap5A-bound crystal structures (fig. S14). Because both simulations provided very similar results, we focus the discussion here on the simplest system that is the substrate-free state. As expected from the stable interfaces, we observed small conformational fluctuations with very low root mean square deviation (RMSD) of the trimerization helices during 500-ns MD simulations of the wild type (fig. S14A). In contrast, distinct changes are observed in the V1 variant during the MD simulations, showing titling of trimerization helices relative to the wild-type enzyme (fig. S14B). This led to an opening and enhanced RMSD of the trimerization interfaces in the V1 variant. By applying an equilibrium argument, these changes can be viewed as the final adjustments that occur during assembly of the native trimer and can be described as a relaxation of the trimer interfaces. Last, we note that the AMPbd fluctuated between the open and closed orientations in the wild type while it remained open in the V1 variant (fig. S14C).
Together, the data illustrate four aspects: (i) trimerization is required for the extreme thermostability of OdinAK, (ii) monomeric variants can be stabilized with amino acid replacements at solvent exposed interface positions, (iii) the final stage during assembly of the native trimer is corresponding to a relaxation of trimer interfaces, and (iv) the catalytic activity of the engineered monomer is comparable to that of the trimer. Therefore, we conclude that an important and functional role of OdinAK trimerization is to support its extreme thermostability. It should, however, be noted that there also exist monomeric AK enzymes with exceptional thermal stability (18), whose thermostability mechanism differs from that of OdinAK.
Active site architecture in OdinAK and linkages to human AK6
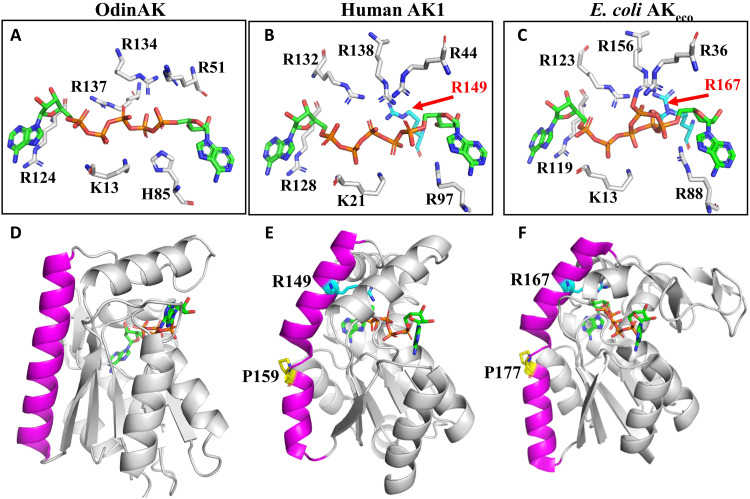
Having established that the catalytic activity of OdinAK is low at 25°C, with a kcat value of 0.5 s−1 compared to the corresponding values of 360 s−1 (15) and 140 s−1 (fig. S15) for AKeco and AK1, respectively, we analyzed crystallographic structures of AK:Ap5A complexes in search for a structural underpinning for this difference in activity. The OdinAK active site (Fig. 7A) is composed of six residues: K13, R51, H85, R124, R134, and R137. The “typical” AK active site was discovered through the structure determination of AKeco in complex with Ap5A (9), and this active site architecture is fully conserved in human AK1. A comparison of the OdinAK active site with those of the typical monomeric human AK1 (Fig. 7B) and AKeco (Fig. 7C) reveals an overall strong structural similarity and two notable differences. First, OdinAK contains a histidine residue (H85) where the corresponding residue in AK1 and AKeco is arginine (R97 and R88). The histidine residue in the active site is a conserved feature of archaeal AKs as observed from the archaeal AK structures deposited in the PDB (PDB IDs: 6PSP, 3H86, 1KHT, 6HF7, 6LN3, and 1NKS). Second, the active sites of both AK1 and AKeco contain one additional arginine residue (R149 and R167, respectively), which is absent in OdinAK. These arginines are relevant for catalysis because the replacement of R167 with an alanine in AKeco results in a 150-fold reduction of kcat to 2.4 s−1 at 25°C (26), and the kcat value of the R167A AKeco variant is comparable to that of OdinAK at 25°C (0.5 s−1). In summary, the active site of OdinAK is “atypical” in relation to the well described typical AK active sites.
Fig. 7. Active site architecture in trimeric and monomeric AK enzymes.
(A to C) Annotated active sites in AK enzymes. (A) The active site of trimeric OdinAK in complex with Ap5A (PDB ID: 7OWE) is atypical because it is lacking a critical arginine and has one of the otherwise conserved arginine residues replaced with histidine (His85). (B) The typical AK active site of monomeric human AK1 in complex with Ap5A (PDB ID: 1Z83) with the critical arginine-149 present. (C) The active site of AKeco in complex with two ADP molecules (PDB ID: 7APU) is a representative of a bacterial and monomeric enzyme that has the critical arginine-167 present. (D) The straight and close to ideal conformation of the trimerization helix (magenta) in OdinAK occludes the possibility of a typical active site because the N terminus of the trimerization helix projects away from the active site. (E) The corresponding α helix in monomeric AK1 is kinked due to a conserved proline residue at position 159. The kink places the N terminus of the α helix in an optimal position for complementation of the active site by R149. (F) Analogous to AK1, the α helix of AKeco is kinked by the presence of P177, which results in complementation of the active site by R167.
What are then the underlying topological and structural features responsible for typical versus atypical AK active sites? Analysis of our cocrystal structure of OdinAK in complex with Ap5A and previously solved structures revealed a direct connection between trimeric/monomeric states and active site architecture (Fig. 7, D to F). In Fig. 7D, the structure of one monomer of trimeric OdinAK is shown in complex with Ap5A. The trimerization helix (magenta) has a straight conformation that adopts a close to ideal helical geometry. Turning to monomeric AKs (Fig. 7, E and F), the corresponding helix is kinked due to a conserved proline residue present in the monomeric enzymes (P159 in AK1 and P177 in AKeco). This kink positions the N terminus of the helix in close proximity to the substrate and allows the additional arginine (R149 in AK1 and R167 in AKeco) to complement the typical active site, thereby increasing the catalytic activity to these enzymes. Because the trimerization of OdinAK is dependent on straight trimerization helices, the N terminus of these helices is positioned too far away from the active site to enable complementation with the critical arginine. Thus, trimerization of OdinAK results in an atypical active site with low catalytic activity but provides a compensation in thermal stability of the enzyme.
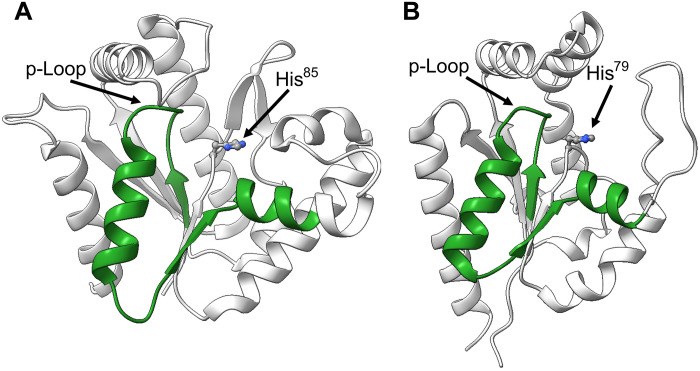
The evolutionary linkage and argument that Asgard archaea are the closest known relatives to eukaryotic organisms are based on both 16S ribosomal RNA sequences (8) and phylogenetic analysis of proteins (such as profilins) (29). To identify possible evolutionary linkages between OdinAK and human enzymes, we interrogated human sequences in UniProt with a basic local alignment search tool (BLAST) search using OdinAK as query. The top hit was found to be nuclear human AK6 (hAK6), also known as human coilin-interacting protein (30). Although hAK6 is the closest human homolog to OdinAK, the sequence similarity is limited to 19 identical residues in a 36–amino acid segment that is equivalent to the conserved Walker-A p-loop segment (fig. S16) (31). The sequence similarity is displayed on both OdinAK and hAK6 structures in Fig. 8 (A and B). A notable aspect of hAK6, as inferred from its structure (PDB ID: 1RKB) (30), is that, similar to archaeal AKs, it contains the conserved histidine in the active site (Fig. 8B). This feature is distinct to the remaining human AKs for which the structures have been determined. At present, nine human AKs have been identified, and these enzymes are expressed in different cellular compartments (32). Of these enzymes, the structures have been solved for cytosolic AK1 (PDB ID: 1Z83), mitochondrial AK2 (PDB ID: 2C9Y), mitochondrial matrix AK3 (PDB ID: 6ZJB) (7), mitochondrial AK4 (PDB ID: 2AR7), cytosolic/nuclear AK5 (PDB ID: 2BWJ), and nuclear hAK6 (PDB ID: 1RKB) (33). It should be noted that it is not clear whether hAK6 works exclusively as an AK inside human cells and that it has been suggested to have roles in gene transcription, ribosome synthesis, cellular metabolism, cell proliferation, apoptosis, DNA damage responses, and genome stability (30). In addition, the catalytic turnover for the AK reaction is very slow (kcat is equal to 0.0063 s−1) for hAK6 (34), and it also displays low adenosine triphosphatase (ATPase) activity (kcat is equal to 0.00048 s−1) (34). Irrespective of its function inside human cells, both the active site histidine and the similarities between hAK6 and OdinAK suggest that hAK6 may be of archaeal origin.
Fig. 8. Sequence similarities between OdinAK and nuclear hAK6 displayed on crystallographic structures.
The figure illustrates the sequence similarities obtained by interrogating the human proteome with a BLAST search using OdinAK as queries. The top hit for both OdinAK was hAK6. (A) The sequence similarity between OdinAK and hAK6 is confined to residues 4 to 39 in OdinAK, and this segment is colored in green on molecule A of the apo OdinAK structure (PDB ID: 7OWH). The p-loop and H85 are indicated. (B) The sequence similarity between OdinAK and hAK6 illustrated on the hAK6 structure (PDB ID: 1RKB). For hAK6, the segment is consists of amino acid residues 7 to 41. The p-loop together with His79 is indicated. The details of the sequence similarities obtained from the BLAST search are shown in fig. S16.
DISCUSSION
Our current and previous studies on AK enzymes from all domains of life allow us to discuss our findings for selectivity and catalysis in an evolutionary context. Over the course of evolution and with increasing complexity of life, enzymes typically become highly specific toward their substrates. Hence, it has been proposed that, while modern enzymes may be selective, primitive enzymes might display a broader substrate specificity, tolerating different but chemically related substrates (35, 36). This view is supported by experiments (36) but has also been challenged (37). Here, we present data providing molecular and structural insight into an ancient enzyme (OdinAK) that displays such broad substrate selectivity. Enzymes that use NTPs for their biological function have generally evolved a strict specificity that is governed by specific interactions with the purine or pyrimidine bases. Well-known examples are G proteins and protein kinases that display strong selectivity toward GTP (38) or ATP (39), respectively. On the other hand, ribonucleotide reductase has evolved to bind all NTPs to reduce them into deoxynucleotide triphosphates (40). Here, we have discovered that the AK from the Asgard archaea Odinarchaeota can use all NTPs for the phosphorylation of AMP. The broad selectivity of this enzyme is rooted in two components: the specific geometry of the selectivity loop containing a hydrogen bond donor and acceptor (Q176) that renders two distinct binding modes and the exploitation of the endogenous flexibility of torsion angles in the NTP substrates. We have previously discovered the structural basis for the feature that AKs are strictly selective for one “designated” NTP and that this selectivity is dependent on the orientation of the selectivity loop (7). As mentioned, there exist at least nine human AK isoforms (32), and it has been shown that the organelle-specific cytoplasmic AK1 (this study) and mitochondrial AK3 (7) are specific for ATP and GTP, respectively. In light of the strict selectivity of the human enzymes and the broad selectivity profile of OdinAK, our study provides experimental support to the idea that more primitive enzymes may display a broader substrate preference, while modern counterparts have evolved to be more selective (35, 36). From a BLAST search, we discovered that hAK6 and OdinAK are evolutionary related through the p-loop segment and the presence of a conserved histidine in the active site. The conservation of the p-loop segment is intriguing because it has been proposed to be one of a set of defined and ancestral peptides that is proposed to have served as cofactors in an early RNA world (41). These similarities open the possibility that hAK6, from an evolutionary aspect, may be of archaeal origin. The evolution of eukaryotic organelles is assumed to be the result of different events where the classic example is the eukaryotic mitochondria that is believed to have originated from an endosymbiotic event (42). There exists a substantial controversy on the evolution of the eukaryotic nucleus, and different models have been proposed including a prokaryotic origin (43). Our finding that hAK6 contains conserved archaeal sequence and that structural features suggest a relationship between the human cell nucleus and archaeal predecessors. In contrast to the difference in selectivity, the dynamic signature of AK appears to be conserved through evolution because it has been shown that members from the three domains of life all have a catalytic mechanism that is rate-limited by slow conformational dynamics and the comparable catalytic rates between the wild-type OdinAK and the R167A variant of E. coli AK (26). This feature has now been shown for Archaeal (OdinAK; this study), bacterial (E. coli AK) (18), and eukaryotic AKs (human AK3) (7).
There exists an evidence supporting the idea that the ancient environment of the early Earth was hot and the first seeds of life originated before it cooled down to present temperatures (37). Following this idea, contemporary enzymes isolated from mesophilic and psychrophilic organisms have adapted their catalytic activity to cooler temperatures, and this generally translates to an enzymatic activity (kcat) or catalytic efficiency (kcat /Km) that scales inversely with habitat temperature (44). We have shown that yeast cells require a minimum threshold in the enzymatic activity of AK to support viability of the cells (6), and this corresponds to around 5 to 10% of the wild-type kcat value. In light of this threshold activity that is substantially higher than the activity of OdinAK reported here, cold adaptation of enzymatic activity is necessary to support cellular viability when the environmental temperature changes over the course of evolution. Analogous to the many possible routes for thermal stability of enzymes (45), different mechanisms underlying cold adaptation of enzymes have been proposed. These mechanisms include active site dynamics (46), conformational flexibility (47), modulation of product release through conformational dynamics (48), and nonzero heat capacities for the rate-limiting step of catalysis (49). A common denominator for these models is that the mechanisms are inferred from correlations of temperature dependencies of catalytic rates with quantification of dynamics. In contrast to these conclusions based on correlations, we discuss a model for enzymatic cold adaptation that is based on atomic coordinates. As illustrated in Fig. 7, the active site of trimeric OdinAK is atypical, whereas typical active sites are so far only observed in monomeric AKs where a critical arginine is complementing the active site. Hence, together with the role of the enzyme trimerization in the extreme thermostability of OdinAK, we propose a mechanism for cold adaptation where the evolution of a monomeric enzyme from a trimer results in a substantial boost in activity at cooler temperatures as a result of complementation of the active site by the critical arginine. The model contains two key evolutionary events, corresponding to the formation of a monomeric species from the trimer and thereafter, or in concert, formation of a bent “trimerization” helix, resulting in positioning of the arginine to complete the active site. Our results demonstrate that the first step corresponding to transformation of the trimer to functional monomers can be accomplished with only a few amino acid replacements. This “monomerization” step also accompanies a significant loss of thermal stability, which then would be tolerated due to the reduction of the surrounding temperature during cold adaptation. The second step requires larger changes to the structural topology of the enzyme and could be approached with, for instance, experiments based on direct evolution. As a concluding remark, we note that further molecular and functional studies on proteins from Odinarchaeota may uncover additional novel insights in protein function.
MATERIALS AND METHODS
Reagents
The genes for OdinAK and human AK1 (UniProt accession IDs: A0A1Q9N9I8 and P00568) were synthesized by GenScript. Substrates (AMP, ADP, ATP, GTP, CTP, UTP, and dTTP), Ap5A, and 5-fluoroindole were purchased from Sigma-Aldrich. 15NH4Cl and D2O were purchased from Cambridge Isotope Laboratories.
Protein production
OdinAK and AK1 coding sequences were subcloned into the pET24d vector. OdinAK variants were generated using site-directed mutagenesis. Both constructs were transformed into E. coli BL21 (DE3) cells and grown to an optical density at 600 nm around 0.7 in LB medium containing kanamycin (50 μg/ml) at 37°C. Protein expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside, and the cultures were grown at 20°C overnight. For 19F-labeled OdinAK samples, cultures were grown in M9 minimal medium where 5-fluoroindole (60 mg/liter; dissolved in dimethyl sulfoxide) (23) was added during induction. Cells were harvested by centrifugation at 5000g for 30 min and lysed by sonication in buffer containing 50 mM tris-HCl (pH 7.5) and 1% Triton X-100. The lysates were centrifuged at 60,000g for 30 min, and the supernatants were loaded on to a Blue Sepharose (GE Healthcare) affinity chromatography column equilibrated with a 50 mM tris-HCl buffer at pH 7.5. The proteins were eluted using a salt gradient up to 1 M NaCl in the same buffer as used for equilibration. In the case of OdinAK, the lysate was heat-shocked at 65°C for 30 min, before Blue Sepharose. The eluted proteins were further purified using SEC (HiLoad 16/600 Superdex 75) in buffer containing 30 mM Mops (pH 7.0) and 50 mM NaCl. The purity and quality of purified proteins were assessed using SDS–polyacrylamide gel electrophoresis and NMR.
X-ray crystallography
Purified OdinAK was concentrated to ~20 mg/ml. Crystals of the native protein were obtained in drop F12 in a Morpheous crystallization screen at 18°C using the vapor diffusion hanging drop method. Cocrystals of OdinAK complexes contained threefold excess of Ap5A, CTP, GTP, or dTTP. Crystallization drops of all four OdinAK complexes contained between 0.1 and 1 μl of protein solution mixed with equal amounts of precipitant and equilibrated against 1 ml of precipitant solution containing the following: (i) for OdinAK-Ap5A, drop E7 from the PACT screen premium (Molecular Dimensions); (ii) for OdinAK-GTP, 0.1 M bis-tris propane (pH 6.5) and 15 to 18% polyethylene glycol (PEG) 3350; and (iii) for OdinAK-TTP and OdinAK-CTP, 0.1 M bis-tris propane (pH 7.0) and 15 to 18% PEG 3350. Crystals were cryoprotected in 30% PEG 3350 or 6000 before vitrification in N2 stream maintained at 100 K using a Cryostream Cooler (Oxford Cryosystems). High-resolution synchrotron diffraction data at 100 K were collected at beamlines P13 at the PETRA laboratory (Hamburg, Germany) and BioMAX at MAX IV (Lund, Sweden). Data were processed and scaled using the routines XDS, Pointless, and Aimless from the CCP4 suite (50). Data collection statistics are listed in table S1. The phases for structure determination were obtained by molecular replacement using PHASER from the PHENIX package and the AK structure from M. thermolithotrophicus [PDB ID: 1KI9 (11)] as search model. All structures were built and refined using Coot (51) and PHENIX REFINE. The atomic coordinates and structure factors [PDB IDs: 7OWH (OdinAK apo), 7OWE (OdinAK-Ap5A), 7OWJ (OdinAK-GTP), 7OWL (OdinAK-CTP), and 7OWK (OdinAK-dTTP)] were deposited in the PDB, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ.
Isothermal titration calorimetry
OdinAK and substrates were prepared in buffer containing 30 mM Mops (pH 7.0) and 50 mM NaCl. Protein concentrations varying from 40 to 100 μM were titrated against substrate concentrations ranging from 0.4 to 1.5 mM at 25°C. In the case of OdinAK-ADP-Mg2+, 5 mM MgCl2 was included in both cell and syringe buffers. ITC experiments were performed using a MicroCal ITC200 calorimeter (MicroCal-Malvern). The binding isotherms were fitted to the “one set of sites” model implemented in Origin 7 (OriginLab). Errors were estimated from experiments performed in triplicate (i.e., technical replicates).
Differential scanning calorimetry
The thermostability was analyzed using a MicroCal VP-Capillary DSC (MicroCal Inc., Northampton, MA). Before measurement, solutions were degassed on MicroCal’s Thermovac unit. The sample concentration was 21 μM based on trimer concentration in a buffer consisting of 30 mM Mops (pH 7.0) and 50 mM NaCl.
SEC-MALS
OdinAK elution peak was further analyzed by SEC–multiangle light scattering (MALS) using an ÄKTApure system (GE Healthcare) coupled to a miniDAWN TREOS II detector and an OptiLab T-rEX online refractive index detector (Wyatt Technology). The absolute molar mass was calculated by analyzing the scattering data using ASTRA analysis software package, version 7.2.2.10 (Wyatt Technology). Bovine serum albumin (BSA) was used for calibration, and proteins were separated on a Superdex 200 10/300 analytical column (GE Healthcare) with a flow rate of 0.4 ml/min. Sample (200 μl) at 2 mg/ml was injected and eluted in 30 mM Mops (pH 7) and 150 mM NaCl buffer. The refractive index increment of Odin was set at 0.185 ml/g, and the extinction coefficient for ultraviolet detection at 280 nm was calculated from the primary sequence. The calculated molecular weight is the average from three runs, with the error being the SD.
Analytical SEC
Analytical SEC experiments were conducted using a Superose 12 10/300 GL column (Cytiva) equilibrated with a 30 mM Mops buffer at pH 7.0 supplemented with 50 mM NaCl. In each run, 200 μl of the sample was injected and then eluted with 1.1 column volumes (24.9 ml) of equilibration buffer, running at 0.8 ml/min. The protein was detected by absorption at 280 nm.
Enzymatic activity
Enzymatic activities were quantified with either a coupled ATPase assay (15) or a 31P real-time NMR assay (16). The ATPase assay was used for AK1 following the procedures described previously (7). To obtain the Km and the reaction velocity at saturating substrate conditions (kcat), the Michaelis-Menten equation (Eq. 1) was fitted to the determined reactions velocities (V) for the various substrate concentrations ([S]). The reaction velocities were scaled with the total enzyme concentration [E]tot according to
| (1) |
For OdinAK, 31P NMR assay was used because it is suitable for enzymes with low absolute values of kcat and because the NMR spectrometer allows accurate control of temperature over a wide range. The reaction was initiated by adding OdinAK to a final concentration of 10 nM to a reaction mixture containing either 1 mM ATP, GTP, CTP, UTP, or dTTP, in addition to 300 μM AMP, 5 mM MgCl2, BSA (0.2 mg/ml), and 7% D2O (v/v) in buffer containing 30 mM Mops (pH 7.0) and 50 mM NaCl at 65°C. The buildup of nucleoside diphosphate (NDP) and depletion of NTP and AMP were monitored by acquiring one-dimensional (1D) 31P NMR spectra at constant time intervals using a 600-MHz Bruker Avance III HD spectrometer equipped with a broadband observe (BBO) cryoprobe. The peaks were integrated, using TopSpin 3.6 (Bruker Biospin), to obtain the concentrations of mono-, di-, and triphosphates and plotted against the reaction times. The data were fitted as described previously (16) to quantify the kcat using OriginPro 2020 (OriginLab). The error estimates were obtained from three or more technical replicates.
19F NMR spectroscopy
19F-labeled OdinAK was prepared in a buffer containing 30 mM Mops (pH 7.0), 50 mM NaCl, and 15% D2O (v/v). Experiments were performed at 65°C on a 600-MHz Bruker Avance III HD spectrometer (19F Larmor frequency of 564 MHz) equipped with a 5-mm 19F cryoprobe. The typical π/2 pulse length was 13.8 μs, the acquisition time was 0.14 s, and the relaxation delay was 3 s. The CPMG RD experiments were run as pseudo-2D in interleaved mode with eight scans to complete the phase cycle and typically 96 numbers of averages yielding in total 768 numbers of scans per increment in the CPMG loop counter. The loop counter list contained 18 entries between 2 and 20 echoes including repeats for error estimation that, together with the total CPMG period of 10 ms, yielded CMPG frequencies in the interval of approximately 100 to 1000 Hz. A two-site model valid in the fast exchange regime (52) was fitted to the RD data
| (2) |
where R2,eff is the observed transverse relaxation rate; is the transverse relaxation rate at infinite CPMG field; pA and pB are the populations of major and minor exchanging states; δω is the difference in chemical shifts between exchanging states in rads per second; νCPMG is the applied CPMG field strength per second and νCPMG = 1/4τ where 2τ is the delay between the centers of two successive 180° pulses; and kex is the observed exchange kinetics defined as kex = kf + kr or alternatively as
In the fast exchange regime, the parameters that can be fitted are defined as “C,” , and kex (table S2). The parameters pA, pB, and kex can be extracted from the exchange product C under the condition that δω can be measured in a separate experiment. The microscopic rate constants are then kf = pBkex and kr = pAkex. In our analysis, we assume that exchange occurs between open and closed states and therefore kf = kclose and kr = kopen. The chemical shift difference (δω) between open and closed states can be measured in 19F NMR spectra with the assumption that the apo enzymes correspond to the open state and that the Ap5A-bound enzyme corresponds to the closed state. Using this procedure, the microscopic rate constants in the forward (kclose) and reverse directions (kopen) were computed. Errors were estimated from the fits of the data to Eq. 2. The error estimates were obtained from three replicate experiments.
MD simulations
The systems for MD simulations were prepared on the basis of both the substrate-free (7OWH.pdb) and Ap5A-bound (7OWE.pdb) OdinAK structures, using CHARMM-GUI (53). In each system, the protonation state of each residue was deduced on the basis of the possible hydrogen bonding interactions with neighboring residues at pH 7.5, and all three monomers were included in the system preparation. The N terminus and C terminus were truncated with the CHARMM’s NTER and CTER patches, respectively. For the V1 system (i.e., M154E/T166E double mutant), the mutated residues were model-built on the basis of the CHARMM 36m force field parameters (54). All missing hydrogen atoms were built on the basis of the H-Build facility of CHARMM. Then, each system was solvated with 100-Å cubic box of transferable intermolecular potential with 3 points (TIP3P) (55) water models and neutralized at 150 mM NaCl, followed by the removal of any TIP3P water molecules that overlapped with any protein heavy atom and Na+ and Cl− ions. This resulted in a total of 93,999 atoms for the wild type and 93,992 atoms for the V1 variant, respectively, for the substrate-free systems; the total numbers of atoms of the Ap5A-bound systems were 94,242 for the wild type and 94,194 for the V1 mutant.
The systems were energy-minimized with different positional restraints/constraints: first, 500 minimization steps with fixed heavy atoms, 1000 steps with fixed protein and water heavy atoms and Na+ and Cl− ions, 2000 steps with harmonic restraints on the protein heavy atoms, and, lastly, 1500 steps without constraints and restraints. The resulting systems were then heated to 300.0 K and equilibrated for 300 ps at constant temperature. For each system, we then performed the 500-ns MD simulations at 300 K and 1 bar. The temperature and pressure of the simulated systems were maintained at their target values using the Langevin thermostat (56) and the Monte Carlo barostate (57) available in the CHARMM/BlaDE module (58). The leapfrog Verlet was used with the 2-fs time integration using SHAKE (59) applied to all bonds with hydrogen atoms. Electrostatic interactions were evaluated using the particle mesh Ewald method (60), and the real-space interactions were evaluated with the 12-Å cutoff. The same cutoff was used in the evaluation of the van der Waals interactions with force switching between 10 and 12 Å. During the MD simulations, the coordinates were saved at every 10-ps intervals for postprocessing analysis.
Acknowledgments
The Protein Expertise Platform (PEP) at the Umeå University is acknowledged for providing reagents for protein production, and M. Lindberg at PEP is appreciated for preparation of plasmids. We acknowledge MAX IV Laboratory (Lund, Sweden) for time on BioMAX and DESY (Hamburg, Germany) for time on PETRA-3. All NMR experiments were performed at the Swedish NMR Center at Umeå University. We also acknowledge the Swedish National Infrastructure for Computing (SNIC) at the High Performance Computing Center North (HPC2N) and the National Energy Research Scientific Computing Center (NERSC) for computational resources.
Funding: This work was financially supported by the Swedish Research Council (2017-04203 to M.W.-W., 2019-03771 to E.S.-E., and 2016-03599 to R.B.), the Knut and Alice Wallenberg Foundation (to R.B.), the Kempe foundation (SMK-1869 to R.B.), the Carl Trygger Foundation (to L.B.), a postdoctoral fellowship from the Carl Tryggers Foundation (17.504 to A.V.), and the National Institutes of Health (R01GM132481 to M.W.-W. and K.N).
Author contributions: A.V. performed ITC, 19F NMR, and 31P NMR and performed data analysis. E.Å.-Z. and C.G. performed crystallization of OdinAK in its different states. U.H.S. and E.S.-E. were responsible for collection of diffraction data and structure determinations. T.S. and A.U.M. performed 19F NMR and 31P NMR experiments. T.S. contributed with quantitative analysis of RD experiments. L.B. performed and analyzed DSC and ITC experiments. K.N. was responsible for MD simulations. P.R. performed the analytical SEC experiments and 31P NMR experiments and evaluated the hydrolysis experiment. R.B. performed and analyzed the SEC-MALS experiment. A.V., M.W.-W., and E.S.-E. performed analysis of protein structures. M.W.-W. guided the project and A.V., E.S.-E., and M.W.-W. drafted the manuscript. The manuscript was written through contributions of all authors, and all authors reviewed and revised the manuscript. All authors have given approval to the final version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The plasmids used in this study can be provided by M.W.-W.’s pending scientific review and a completed material transfer agreement. Requests for the plasmids should be submitted to M.W.-W. (magnus.wolf-watz@umu.se). The crystallographic structures have been deposited at the PDB with accession codes 7OWH (OdinAK-native), 7OWE (OdinAK-Ap5A), 7OWJ (OdinAK-GTP), 7OWK (OdinAK-dTTP), and 7OWL (OdinAK-CTP).
Supplementary Materials
This PDF file includes:
Supplementary Text
Tables S1 and S2
Figs. S1 to S16
REFERENCES AND NOTES
- 1.Deamer D., Singaram S., Rajamani S., Kompanichenko V., Guggenheim S., Self-assembly processes in the prebiotic environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1809–1818 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rona P. A., Klinkhammer G., Nelsen T. A., Trefry J. H., Elderfield H., Black smokers, massive sulfides and vent biota at the Mid-Atlantic Ridge. Nature 321, 33–37 (1986). [Google Scholar]
- 3.Spang A., Saw J. H., Jørgensen S. L., Zaremba-Niedzwiedzka K., Martijn J., Lind A. E., van Eijk R., Schleper C., Guy L., Ettema T. J. G., Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaremba-Niedzwiedzka K., Caceres E. F., Saw J. H., Bäckström D., Juzokaite L., Vancaester E., Seitz K. W., Anantharaman K., Starnawski P., Kjeldsen K. U., Stott M. B., Nunoura T., Banfield J. F., Schramm A., Baker B. J., Spang A., Ettema T. J. G., Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Fournier G. P., Poole A. M., A briefly argued case that Asgard archaea are part of the eukaryote tree. Front. Microbiol. 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tükenmez H., Magnussen H. M., Kovermann M., Bystrom A., Wolf-Watz M., Linkage between fitness of yeast cells and adenylate kinase catalysis. PLOS ONE 11, e0163115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogne P., Dulko-Smith B., Goodman J., Rosselin M., Grundström C., Hedberg C., Nam K., Sauer-Eriksson A. E., Wolf-Watz M., Structural basis for GTP versus ATP selectivity in the NMP kinase AK3. Biochemistry 59, 3570–3581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eme L., Spang A., Lombard J., Stairs C. W., Ettema T. J. G., Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 15, 711–723 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Müller C. W., Schulz G. E., Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 Å resolution. A model for a catalytic transition state. J. Mol. Biol. 224, 159–177 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Jarzab A., Kurzawa N., Hopf T., Moerch M., Zecha J., Leijten N., Bian Y., Musiol E., Maschberger M., Stoehr G., Becher I., Daly C., Samaras P., Mergner J., Spanier B., Angelov A., Werner T., Bantscheff M., Wilhelm M., Klingenspor M., Lemeer S., Liebl W., Hahne H., Savitski M. M., Kuster B., Meltome atlas-thermal proteome stability across the tree of life. Nat. Methods 17, 495–503 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Criswell A. R., Bae E., Stec B., Konisky J., Phillips G. N. Jr., Structures of thermophilic and mesophilic adenylate kinases from the genus Methanococcus. J. Mol. Biol. 330, 1087–1099 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Vonrhein C., Bonisch H., Shafer G., Schulz G. E., The structure of a trimeric archaeal adenylate kinase. J. Mol. Biol. 282, 167–179 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Shibanuma Y., Nemoto N., Yamamoto N., Sampei G. I., Kawai G., Crystal structure of adenylate kinase from an extremophilic archaeon Aeropyrum pernix with ATP and AMP. J. Biochem. 168, 223–229 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Ådén J., Wolf-Watz M., NMR identification of transient complexes critical to adenylate kinase catalysis. J. Am. Chem. Soc. 129, 14003–14012 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Åden J., Verma A., Schug A., Wolf-Watz M., Modulation of a pre-existing conformational equilibrium tunes adenylate kinase activity. J. Am. Chem. Soc. 134, 16562–16570 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Rogne P., Sparrman T., Anugwom I., Mikkola J. P., Wolf-Watz M., Realtime (31)P NMR investigation on the catalytic behavior of the enzyme adenylate kinase in the matrix of a switchable ionic liquid. ChemSusChem 8, 3764–3768 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Åden J., Weise C. F., Brännström K., Olofsson A., Wolf-Watz M., Structural topology and activation of an initial adenylate kinase-substrate complex. Biochemistry 52, 1055–1061 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Wolf-Watz M., Thai V., Henzler-Wildman K., Hadjipavlou G., Eisenmesser E. Z., Kern D., Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat. Struct. Mol. Biol. 11, 945–949 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Mulder F. A., Mittermaier A., Hon B., Dahlquist F. W., Kay L. E., Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 8, 932–935 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Loria J. P., Rance M., Palmer A. G., A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332 (1999). [Google Scholar]
- 21.Palmer A. G. III, NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 104, 3623–3640 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Gonti S., Westler W. M., Miyagi M., Bann J. G., Site-specific labeling and19F NMR provide direct evidence for dynamic behavior of the anthrax toxin pore ϕ-clamp structure. Biochemistry 60, 643–647 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crowley P. B., Kyne C., Monteith W. B., Simple and inexpensive incorporation of 19F-tryptophan for protein NMR spectroscopy. Chem. Commun. 48, 10681–10683 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Liebau J., Tersa M., Trastoy B., Patrick J., Rodrigo-Unzueta A., Corzana F., Sparrman T., Guerin M. E., Mäler L., Unveiling the activation dynamics of a fold-switch bacterial glycosyltransferase by 19F NMR. J. Biol. Chem. 295, 9868–9878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf F. I., Cittadini A., Chemistry and biochemistry of magnesium. Mol. Aspects Med. 24, 3–9 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Ojeda-May P., Mushtaq A. U. I., Rogne P., Verma A., Ovchinnikov V., Grundström C., Dulko-Smith B., Sauer U. H., Wolf-Watz M., Nam K., Dynamic connection between enzymatic catalysis and collective protein motions. Biochemistry 60, 2246–2258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., Sawyer N., Regan L., Protein-protein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 22, 510–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsson U., Wolf-Watz M., Overlap between folding and functional energy landscapes for adenylate kinase conformational change. Nat. Commun. 1, 111 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Akil C., Robinson R. C., Genomes of Asgard archaea encode profilins that regulate actin. Nature 562, 439–443 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Xu R. D., Yang Y. F., Zheng X. F., Unique structural features of the adenylate kinase hCINAP/AK6 and its multifaceted functions in carcinogenesis and tumor progression. FEBS Lett. 595, 2071–2084 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Walker J. E., Saraste M., Runswick M. J., Gay N. J., Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panayiotou C., Solaroli N., Karlsson A., The many isoforms of human adenylate kinases. Int. J. Biochem. Cell Biol. 49, 75–83 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Ren H., Wang L., Bennett M., Liang Y., Zheng X., Lu F., Li L., Nan J., Luo M., Eriksson S., Zhang C., Su X. D., The crystal structure of human adenylate kinase 6: An adenylate kinase localized to the cell nucleus. Proc. Natl. Acad. Sci. U.S.A. 102, 303–308 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drakou C. E., Malekkou A., Hayes J. M., Lederer C. W., Leonidas D. D., Oikonomakos N. G., Lamond A. I., Santama N., Zographos S. E., hCINAP is an atypical mammalian nuclear adenylate kinase with an ATPase motif: Structural and functional studies. Proteins 80, 206–220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen R. A., Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30, 409–425 (1976). [DOI] [PubMed] [Google Scholar]
- 36.O’Brien P. J., Herschlag D., Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 6, R91–R105 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Wheeler L. C., Lim S. A., Marqusee S., Harms M. J., The thermostability and specificity of ancient proteins. Curr. Opin. Struct. Biol. 38, 37–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pai E. F., Kabsch W., Krengel U., Holmes K. C., John J., Wittinghofer A., Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature 341, 209–214 (1989). [DOI] [PubMed] [Google Scholar]
- 39.Knighton D. R., Zheng J., ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., Sowadski J. M., Crystal-structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 (1991). [DOI] [PubMed] [Google Scholar]
- 40.Fairman J. W., Wijerathna S. R., Ahmad M. F., Xu H., Nakano R., Jha S., Prendergast J., Welin R. M., Flodin S., Roos A., Nordlund P., Li Z., Walz T., Dealwis C. G., Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat. Struct. Mol. Biol. 18, 316–322 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alva V., Soding J., Lupas A. N., A vocabulary of ancient peptides at the origin of folded proteins. eLife 4, e09410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagan L., On the origin of mitosing cells. J. Theor. Biol. 14, 255–274 (1967). [DOI] [PubMed] [Google Scholar]
- 43.Devos D. P., Graf R., Field M. C., Evolution of the nucleus. Curr. Opin. Cell Biol. 28, 8–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feller G., Molecular adaptations to cold in psychrophilic enzymes. Cell. Mol. Life Sci. 60, 648–662 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petsko G. A., Structural basis of thermostability in hyperthermophilic proteins, or “there’s more than one way to skin a cat”. Methods Enzymol. 334, 469–478 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Kohen A., Cannio R., Bartolucci S., Klinman J. P., Enzyme dynamics and hydrogen tunnelling in a thermophilic alcohol dehydrogenase. Nature 399, 496–499 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Zavodszky P., Kardos J., Svingor, Petsko G. A., Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 7406–7411 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovermann M., Ådén J., Grundström C., Elisabeth Sauer-Eriksson A., Sauer U. H., Wolf-Watz M., Structural basis for catalytically restrictive dynamics of a high-energy enzyme state. Nat. Commun. 6, 7644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hobbs J. K., Jiao W., Easter A. D., Parker E. J., Schipper L. A., Arcus V. L., Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates. ACS Chem. Biol. 8, 2388–2393 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luz Z., Meiboom S., Nuclear magnetic resonance study of the protolysis of trimethylammonium ion in aqueous solution—Order of the reaction with respect to solvent. J. Chem. Phys. 39, 366 (1963). [Google Scholar]
- 53.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Best R. B., Vendruscolo M., Structural interpretation of hydrogen exchange protection factors in proteins: Characterization of the native state fluctuations of CI2. Structure 14, 97–106 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 56.Brünger A., Brooks C. L., Karplus M., Stochastic boundary conditions for molecular dynamics simulations of ST2 water. Chem. Phys. Lett. 105, 495–500 (1984). [Google Scholar]
- 57.Ferguson N., Capaldi A. P., James R., Kleanthous C., Radford S. E., Rapid folding with and without populated intermediates in the homologous four-helix proteins Im7 and Im9. J. Mol. Biol. 286, 1597–1608 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Hayes R. L., Buckner J., Brooks III C. L., BLaDE: A basic lambda dynamics engine for GPU-accelerated molecular dynamics free energy calculations. J. Chem. Theory Comput. 17, 6799–6807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryckaert J. P., Ciccotti G., Berendsen H. J. C., Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of N-alkanes. J. Comput. Phys. 23, 327–341 (1977). [Google Scholar]
- 60.Darden T., York D., Pedersen L., Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Tables S1 and S2
Figs. S1 to S16