Summary
Skeletal muscle generation of ammonia, an endogenous cytotoxin, is increased during exercise. Perturbations in ammonia metabolism consistently occur in chronic diseases, and may blunt beneficial skeletal muscle molecular responses and protein homeostasis with exercise. Phosphorylation of skeletal muscle proteins mediates cellular signaling responses to hyperammonemia and exercise. Comparative bioinformatics and machine learning-based analyses of published and experimentally derived phosphoproteomics data identified differentially expressed phosphoproteins that were unique and shared between hyperammonemic murine myotubes and skeletal muscle from exercise models. Enriched processes identified in both hyperammonemic myotubes and muscle from exercise models with selected experimental validation included protein kinase A (PKA), calcium signaling, mitogen-activated protein kinase (MAPK) signaling, and protein homeostasis. Our approach of feature extraction from comparative untargeted “omics” data allows for selection of preclinical models that recapitulate specific human exercise responses and potentially optimize functional capacity and skeletal muscle protein homeostasis with exercise in chronic diseases.
Subject areas: Biological sciences, Cell biology, Functional aspects of cell biology, Omics, Proteomics
Graphical abstract
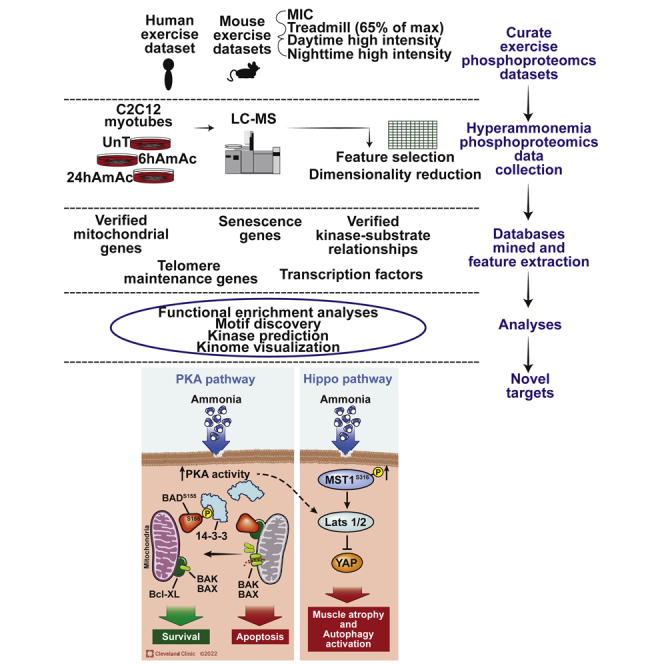
Highlights
-
•
Hyperammonemia occurs in a number of chronic diseases and physical exercise
-
•
Beneficial responses to exercise may be blunted by increased muscle ammoniagenesis
-
•
Comparative phosphoproteomics show potential modifiable shared molecular responses
-
•
Exercise capacity in chronic disease may be improved by targeting hyperammonemia
Biological sciences; Cell biology; Functional aspects of cell biology; Omics; Proteomics.
Introduction
Ammonia, an endogenous cytotoxin generated during amino acid and purine nucleotide catabolism and the gut microbiome, is metabolized to urea in hepatocytes (Adeva et al., 2012; Dasarathy and Hatzoglou, 2018). Dysregulated ammonia metabolism occurs in a number of chronic diseases, including liver cirrhosis, heart failure, and chronic obstructive pulmonary disease (Dasarathy and Hatzoglou, 2018; Medeiros et al., 2014; Valero et al., 1974). During the consequent hyperammonemia, skeletal muscle becomes a major organ for ammonia uptake (Ganda and Ruderman, 1976; Lockwood et al., 1979; Qiu et al., 2013) with complex molecular and metabolic perturbations(Dasarathy and Hatzoglou, 2018; Davuluri et al., 2016a, 2016b; Kumar et al., 2021; Medeiros et al., 2014; Valero et al., 1974; Welch et al., 2021). However, circulating and skeletal muscle ammonia concentrations do not parallel each other, which may be because of an increased expression of inducible skeletal muscle ammonia transporter, RhBG (Kant et al., 2019; McDaniel et al., 2016; Qiu et al., 2013). Studies were performed in our previously reported myotube model using ammonium acetate in the medium at concentrations higher than those in circulation that, however, achieved ammonia levels similar to those in the skeletal muscle of a preclinical rat model and human patients with cirrhosis (McDaniel et al., 2016; Qiu et al., 2013). The biological and translational relevance of hyperammonemia is because of tissue responses to this endogenous cytotoxin (Chen and Dunn, 2016; Dasarathy et al., 2017). Public datasets exist for ammonia-induced transcriptional and phosphoproteomic responses in mammals (Sequence Read Archive: SRP 313829) (Welch et al., 2021); however, despite the presence of multiple public datasets of protein modifications such as phosphorylation (a major post-translational regulatory modification) (Huang et al., 2018; Huang and Fraenkel, 2009; Lee et al., 2006), including the effects of hyperammonemia in the cerebellum (Brunelli et al., 2012), there are no published data on hyperammonemia-induced skeletal muscle protein phosphorylation in mammals.
Increased skeletal muscle ammoniagenesis with elevated circulating ammonia concentration that occurs during exercise and muscle contraction is believed to be a causal factor for fatigue (Banister and Cameron, 1990; Chen et al., 2020; Dudley et al., 1983; Eriksson et al., 1985; Gorostiaga et al., 2014; Graham et al., 1987, 1990, 1993, 1995, 1990, 1987, 1995; Graham and MacLean, 1998; Katz et al., 1986a, 1986b; MacLean et al., 1991; Mutch and Banister, 1983). However, the impact of hyperammonemia on other exercise responses including functional capacity, skeletal muscle organelle function, and molecular alterations and protein homeostasis is not known. Exercise results in adaptive cellular signaling responses in the skeletal muscle, but the mediators of these global alterations have not been specifically evaluated (Amar et al., 2021; Maier et al., 2022; McGee and Hargreaves, 2020; Pillon et al., 2020; Srisawat et al., 2017). Regulatory layers of cellular responses include chromosomal conformation, transcription, translation and post-translational modifications including phosphorylation. Because skeletal muscle contraction is critical for exercise, a number of studies have evaluated transcriptomics and proteomics responses during exercise and, recently, meta-analyses of public databases have been published (Amar et al., 2021; Pillon et al., 2020; Srisawat et al., 2017). Despite phosphorylation being a critical mediator of cellular function, there are few phosphoproteomics data from disparate mouse models and human exercise studies published to date (Hoffman et al., 2015; Needham et al., 2019; Nelson et al., 2019; Steinert et al., 2021). A goal of developing animal models is to perform mechanistic studies that can be translated to human interventions (Thu et al., 2017; Timson, 1990). However, similar to the diversity of human responses, there are significant differences between mouse models of exercise. Given that no animal model recapitulates all human exercise responses, we developed an approach to identify models to evaluate specific hypotheses based on shared pathways or molecules in response to exercise. Such a strategy has the potential to be applied to study other interventions also. This is of particular interest because despite extensive data on the benefits and responses of exercise training in healthy subjects, in disease states, phenotype and molecular responses are not consistent, and mediators of differential responses have not been well studied (Bellar et al., 2020; McGee and Hargreaves, 2020). Therefore, a strategy to determine optimal animal models of exercise that most closely recapitulate the desired study outcomes of exercise in humans is needed (Feng et al., 2019).
To identify skeletal muscle molecular responses with exercise that may be shared with those because of ammonia, a comparative overlay of skeletal muscle phosphoproteomics in response to exercise and hyperammonemia was performed. Comprehensive analyses of phosphoproteomics data in an established in vitro model of skeletal muscle hyperammonemia were performed. Using a number of supervised and unsupervised approaches, these data were then compared with published, public domain skeletal muscle phosphoproteomics data from human subjects and mice in response to exercise. Even though public datasets are freely accessible and can be reused for data-driven studies, comparative meta-analyses are only recently being reported (Bono and Hirota, 2020; Srisawat et al., 2017; van Wijk et al., 2014). Integrating multiple unbiased datasets from skeletal muscle during hyperammonemia across molecular layers of chromatin access, proteomics and transcriptomics in cellular, murine and human skeletal muscle, we identified global changes in the differential expression of molecules in a number of diverse pathways including protein synthesis, mitochondrial oxidative function, and senescence (Davuluri et al., 2019; Welch et al., 2021). These multiomics analyses revealed changes in a number of pathways, but post-translational modifications of proteins that are critical to mediate functional responses were not reported. Phosphorylation of molecules has been among the most studied modifications and the regulatory role of kinases has been reported extensively (Ramazi and Zahiri, 2021). The global phosphoproteomics landscape in skeletal muscle during hyperammonemia has not been evaluated and can provide insights into regulatory responses and its relevance to human physiology and homeostatic responses.
Our analyses of published exercise-induced skeletal muscle phosphoproteomics showed protein kinase A (PKA), calcium, and mitogen-activated protein kinase (MAPK) signaling, and protein homeostasis were among the most enriched pathways. In our experimental model of hyperammonemia in myotubes, we noted distinct and shared temporal patterns of protein phosphorylation compared with untreated controls. Changes in PKA, matrix metalloproteases, and integrin signaling occurred early, whereas later, cell cycle control signaling, DNA damage signaling, and PKA signaling were among the most enriched pathways. Experimentally, we validated a number of highly differentially expressed phosphoproteins during hyperammonemia including phosphorylation of inhibitor of nuclear factor kappa B kinase subunit β (pIKKβ), decreased ribosomal protein S6 (Rps6) phosphorylation (as reported earlier) (Davuluri et al., 2016a, 2016b, 2019), increased phosphorylation of the HIPPO signaling core kinase Mst2 (also known as Stk3/4), and lower phosphorylation of minichromosome maintenance complex component 2 (Mcm2). These molecules were chosen because of their high relevance in skeletal muscle functional responses (Knight and Kothary, 2011). Our complementary bioinformatics analyses of published data overlaid on experimental results allow for selection of appropriate models and may provide insights into potential mediators of global/specific responses in exercise and hyperammonemia including changes in functional capacity, skeletal muscle protein homeostasis and organelle function.
Results
Phosphoproteomic landscape during hyperammonemia in myotubes
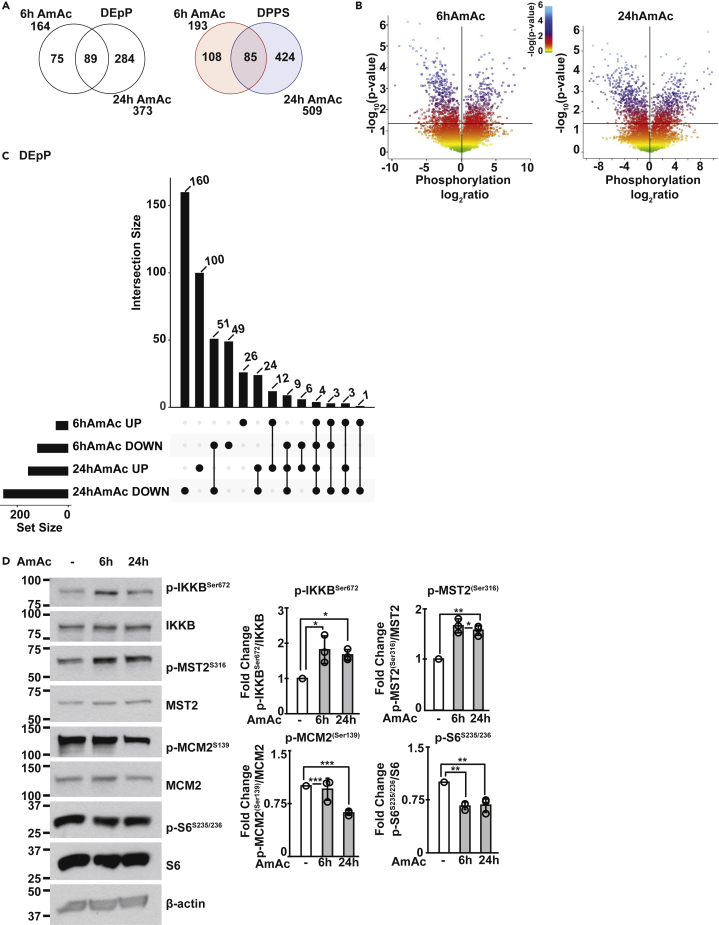
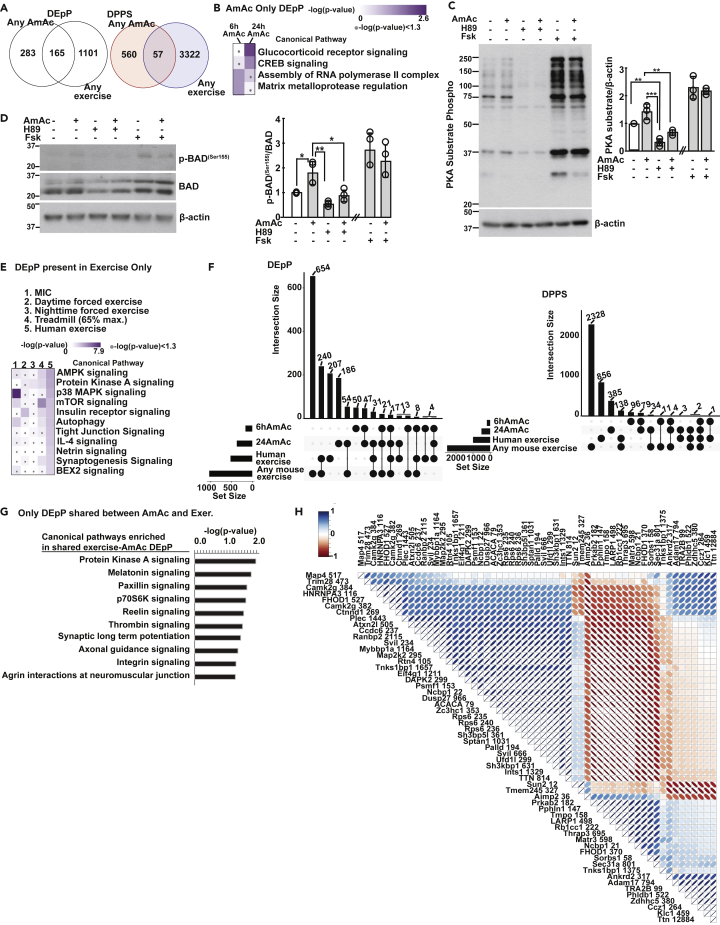
Because exercise increases muscle ammoniagenesis (Bellar et al., 2020; Calvert et al., 2010; Chen et al., 2020; Graham and MacLean, 1998) and hyperammonemia results in perturbed skeletal muscle proteostasis, we first examined the phosphoproteomics landscape during hyperammonemia in myotubes to determine whether there are post-translational changes that may mediate decreased protein synthesis. In the 6 and 24h hyperammonemic myotube datasets, there were 448 DEpP that were identified in hyperammonemic myotubes with 164 total (75 unique) DEpP in the 6hAmAc treatment group and 373 total (284 unique) DEpP in the 24hAmAc treatment group with 89 shared DEpP (Figure 1A). We next identified unique and shared DPPS within the hyperammonemia datasets. Of the 617 DPPS identified in the ammonia-treated C2C12 myotubes, there were 193 total (108 unique) DPPS in the 6hAmAc treatment group and 509 total (424 unique) DPPS in the 24hAmAc treatment group with 85 shared DPPS (Figure 1A). In hyperammonemic myotubes, there were more DEpP with DPPS that had decreased phosphorylation than increased phosphorylation (DOWN or UP as compared to controls, respectively) in each treatment group (6hAmAc: 122(74.3%) total DOWN vs. 49(29.9%) UP, with 7 DEpP that had both increased and decreased phosphorylation at two or more DPPS); 24hAmAc: 255(68.4%) DOWN and 158(42.4%) UP (with 40 DEpP that had both increased and decreased phosphorylation at two or more DPPS)) as seen on volcano and UpSet plots (Figures 1B and 1C). Immunoblots were performed to experimentally validate critical phosphoproteomics data in myotubes. Consistent with the untargeted phosphoproteomics data, increased phosphorylation of inhibitor of nuclear factor kappa B kinase subunit β (IKKβSer672) and Mammalian Sterile 20-like kinase 2 (MST2Ser316) were observed in response to hyperammonemia compared to no treatment in myotubes (Figure 1D). Phosphorylation of IKKβ results in nuclear translocation of p65NF-kB and transcriptional responses during hyperammonemia (Qiu et al., 2013). Phosphorylation of MST2 mediates muscle atrophy (Chen, 2005; Wei et al., 2013). In contrast, MCM2Ser139 and S6 ribosomal proteinSer235/236 exhibited decreased phosphorylation in hyperammonemic (versus untreated) myotubes in our phosphoproteomics datasets and in validation experiments. Inactivation of MCM2 and S6 ribosomal protein leads to decreases in cell proliferation and protein synthesis (Fei and Xu, 2018; Rosario et al., 2020; Zhou et al., 2021).
Figure 1.
Temporal course of differentially expressed phosphoproteins and phosphorylated phosphosites in myotubes during hyperammonemia
Differentially expressed phosphoproteins (DEpP) and phosphorylated phosphosites (DPPS) were identified in murine C2C12 myotubes treated with10mM ammonium acetate (AmAc) for 6 and 24h compared to untreated controls.
(A) Venn diagrams showing the number of shared and unique DEpP/DPPS.
(B) Volcano plots comparing significance to expression of DPPS.
(C) UpSet plot showing the number of DEpP in each group based on the direction of expression change (UP = increased expression, DOWN = decreased expression vs. untreated controls).
(D) Representative immunoblots and densitometries for p-IKKBS672, total IKKB, p-MST2S316, total MST2, p-MCM2S139, total MCM2, p-S6S235/236, and total S6 in untreated and 6hAmAc and 24hAmAc myotubes. All myotube experiments were done in n = 3 biological replicates (one 24hAmAc phosphoproteomics data replicate was removed from downstream analyses because of outlier status). For densitometry, data shown as mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 on ANOVA with Bonferroni post-hoc analysis. Statistical significance cutoff for DEpP/DPPS was p-adj <0.05 (Student’s t test with Benjamini Hotchberg correction). IKKB = Inhibitor of nuclear factor kappa B kinase subunit beta, MST2 = Mammalian sterile 20-like kinase 2; MCM2 = mini chromosome maintenance protein 2.
Extending our analyses, we generated networks of known protein-protein interactions for the most connected proteins that were differentially phosphorylated in each hyperammonemic dataset (Figure S1) and can potentially be used to identify novel phosphorylation signaling cascades. We then determined the hyperammonemic DEpP that overlapped with our previously published quantitative proteomics data in hyperammonemic myotubes (Welch et al., 2021) to determine whether any pair of DEpP and differentially expressed total protein (DEP) had a shared direction of expression (Figure S2). There was no significant correlation between the expression levels of all the 24hAmAc DEP and DEpP (Figure S2A). However, of the 19 shared DEP and DEpP, the expression patterns of 10 molecules were concordant whereas the remaining had a discordant pattern (Figure S2B). Functional enrichment analysis of all overlapping DEpP/DEP showed enrichment of apoptosis-induced DNA fragmentation and formation of senescence-associated heterochromatin foci that are consistent with reports that ammonia promotes senescence in multiple cell types (Gorg et al., 2015; Jo et al., 2021; Kumar et al., 2021).
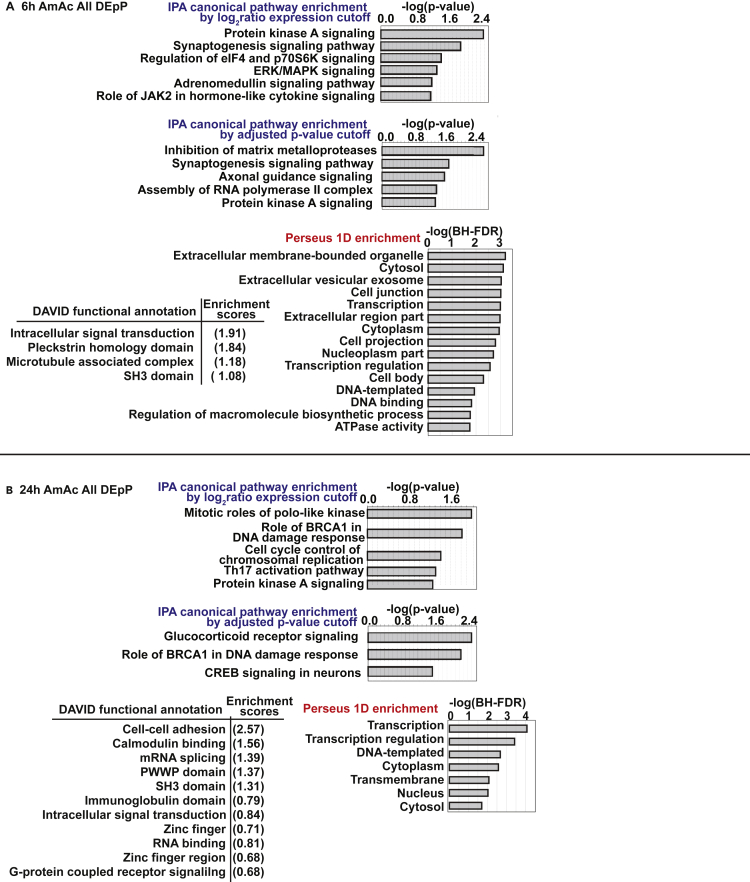
Next, to determine the global and temporal hyperammonemic phosphoproteomic landscape, functional enrichment analyses were performed on the phosphoproteomics datasets from the hyperammonemic myotubes at 6 and 24h compared to untreated controls (Figures 2A, 2B, and S3A–S3C). Datasets were evaluated separately for pathway enrichment including all DEpP in the 6hAmAc set, all DEpP in the 24hAmAc set, and smaller subsets of DEpP that were unique to only the 6hAmAc or the 24hAmAc datasets when compared to each other. Furthermore, a subset of those DEpP that were shared between both 6hAmAc and 24hAmAc datasets was also interrogated for functional enrichment. The curated pathway that was enriched in both the early (6h) and late (24h) hyperammonemia dataset was PKA signaling (Figures 2A and 2B). Enrichment scores of pathways differed between 6hAmAc and 24hAmAc datasets (Figures 2A and 2B), suggesting temporal changes in adaptive/maladaptive responses during hyperammonemia that are consistent with our previous reports (Welch et al., 2021). Components of senescence including DNA damage and cell cycle regulation were also enriched during hyperammonemia (Figure 2B). A number of other processes were also enriched in the unique and shared 6hAmAc and 24hAmAc datasets (Figures S3A–S3C).
Figure 2.
Functional enrichment analysis of proteins that are differentially phosphorylated in myotubes during hyperammonemia
Analyses of shared and unique differentially expressed phosphoproteins (DEpP) in murine C2C12 myotubes treated for 6 and 24h with 10mM ammonium acetate (AmAc) compared to untreated controls.
(A and B) Physiologically relevant pathways enriched in the phosphoproteomics datasets from hyperammonemic myotubes curated using IPA, DAVID, and Perseus. All experiments were done in n = 3 biological replicates (one 24hAmAc replicate was removed from downstream analyses because of outlier status). Statistical significance cutoff for full datasets using IPA was performed using both log2ratio>|2.5| and padj<0.05 (Student’s t test with Benjamini-Hotchberg false discovery rate correction (BH-FDR)). Foreground DEpP in DAVID analyses was padj<0.05. IPA pathway significance cutoff was the default-log(p value) ≥ 1.3. Perseus 1D analysis significance cutoff was the default BH-FDR>0.02. Green color = DEpP identified in the data subset, Black color = DEpP not identified in the data subset.
Because the PKA signaling pathway was enriched in both hyperammonemic datasets (and was among the most enriched in the 6hAmAc dataset), we then evaluated if regulation occurred at one or more molecular levels upstream of phosphorylation. We compared the DEpP from our 6hAmAc and 24h AmAc datasets contained within the PKA signaling pathway to our previously published differentially expressed molecules in ATACseq, RNAseq, and proteomics datasets from hyperammonemic myotubes; RNAseq and proteomics datasets from skeletal muscle from mice following ammonia treatment; and RNAseq from human skeletal muscle from patients with cirrhosis (Welch et al., 2021) (Figures S4A–S4C). Within the 24hAmAc dataset, Polo-like kinase (PLK) signaling, which regulates cell cycle and is involved in cellular senescence(Kim et al., 2013; Lee et al., 2014), was one of the most enriched pathways (Figure 2A). Phosphorylation of other cell cycle regulatory serine/threonine kinases, including cyclin dependent kinases (CDK) that are involved in senescence (Kim et al., 2013; Sadaie et al., 2015), was also significantly altered by ammonia (Figure S5A) as reported earlier (Gorg et al., 2015; Jo et al., 2021; Welch et al., 2021). We also observed that PKA and PLK signaling components (Figure S5B) were both altered during hyperammonemia (6hAmAc, 24hAmAc). These data demonstrate that cellular functions including PKA signaling are regulated at multiple molecular levels. To dissect potential interactions between the PKA and PLK components, we generated a network map and a correlation matrix between known PKA and PLK targets in our UnT and 24hAmAc datasets only as PLK was not among the highly enriched pathways in the 6hAmAc dataset (Figure S5C). We observed multiple molecules in these pathways that showed positive and negative correlations, suggesting that such critical regulatory molecules (PKA, PLK) may have indirect regulatory relationships even if the molecules themselves do not interact.
Our phosphoproteomics studies in hyperammonemic myotubes with experimental validation also showed altered expression of a critical member of the HIPPO signaling pathway, which is altered in a context dependent manner in muscle fibers and myogenically committed satellite cells (Gnimassou et al., 2017; Watt et al., 2018). The core elements of the HIPPO pathway converge to activate or inhibit mammalian ste-20 like kinase 1 and 2 (Mst1/2) or mitogen-activated protein kinase kinase kinases (MAP4K) (Plouffe et al., 2016; Watt et al., 2018). We dissected the HIPPO signaling and target responses in our previously published proteomics dataset (Welch et al., 2021) and the phosphoproteomics dataset in hyperammonemic myotubes, which showed changes in HIPPO signaling components during hyperammonemia (Figure S5D). We also noted that using our list of non-differentially phosphorylated phosphoproteomics as the background and queried at the level of DPPS, HIF1α signaling was also identified as significant (p < 0.05) in the 24hAmAc dataset (Figure S5E) and consistent with our previous report of increased HIF1α during hyperammonemia(Welch et al., 2021). The supervised heatmaps generated for PKA, CDK, PLK, HIPPO and HIF1α were then complemented by unsupervised analyses that showed similar clustering of samples in the datasets (Figures S4C, S5A, S5B, S5D, and S5E).
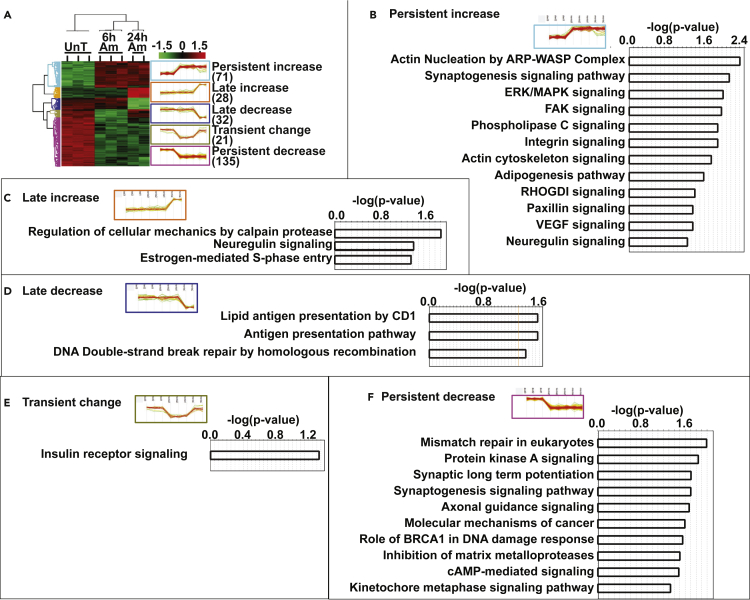
Temporal dynamics of phosphorylation during hyperammonemia were further evaluated by hierarchical clustering with dimensionality reduction and feature selection of DPPS (Figures 3 and S6). These analyses allowed for identification of differential responses with supervised analyses of the 287 DPPS that segregated into 5 distinct patterns or clusters of differentially regulated protein phosphorylation at early (6h) and late (24h) exposure to ammonia (Figure 3A). Persistent increase/decrease clusters included those DPPS with an increase (n = 71) and decrease (n = 135) in phosphorylation that remained high/low in both 6hAmAc and 24hAmAc. Late increase/decrease clusters included DPPS with a delayed (at 24h) increase (n = 28) or decrease (n = 32) in phosphorylation. Transient change cluster included DPPS that were altered at 6hAmAc but reversed to untreated levels at 24h (n = 21). Even though other clusters could be defined, they were not analyzed as part of our dimensionality reduction/feature selection approach to ensure sufficient DPPS in each cluster. The Persistent increase and Persistent decrease clusters yielded the most pathway enrichment. In the Persistent increase cluster, we identified actin nucleation by the ARP-WASP complex (central role in assembly of actin networks required for diverse cellular processes including cell motility and morphogenesis (Goode et al., 2001)), ERK/MAK signaling, and synaptogenesis (shares molecules with PKA signaling pathway) (Figure 3B). In the Late increase cluster, regulation of cellular mechanics by calpain protease was enriched (Figure 3C). In the Late decrease cluster, DNA damage repair pathway (Chen et al., 2007) was enriched (Figure 3D). The Transient change cluster had enrichment of insulin receptor signaling (Figure 3E). Mismatch repair, PKA signaling pathway, synaptic potentiation and synaptogenesis were the most enriched pathways in the Persistent decrease cluster (Figure 3F). These data show that skeletal muscle development, protein homeostasis, contractile and metabolic functions were among the most enriched pathways. Even though our analyses suggested enrichment of different pathways, we noted a number of shared molecules between multiple pathways (on IPA database) including PKA and synaptogenesis (Figure S6).
Figure 3.
Hierarchical clustering of phosphorylated proteins in hyperammonemic myotubes reveals distinct temporal differences
Hierarchical clustering, heatmap, and cluster analysis of differentially phosphorylated phosphosites (DPPS) and differentially expressed phosphoproteins (DEpP) in untreated (UnT) myotubes or those treated with 6h or 24h of 10mM ammonium acetate (AmAc).
(A) Heatmap of DPPS with hierarchical clustering into 5 groups based on the direction of temporal change: Persistent increase (DPPS that have increased phosphorylation at both 6hAmAc and 24hAmAc compared to UnT), Late increase (DPPS that have no change at 6hAmAc but have increased phosphorylation at 24hAmAc compared to UnT), Late decrease (DPPS that have no change at 6hAmAc but have decreased phosphorylation at 24hAmAc compared to UnT), Transient change (DPPS that have increased phosphorylation at 6hAmAc but have decreased phosphorylation at 24hAmAc compared to UnT), and Persistent decrease (DPPS that have decreased phosphorylation at both 6hAmAc and 24hAmAc compared to UnT).
(B–F) Pathway enrichment of DEpP in each of the 5 identified clusters: Persistent increase, Late increase, Late decrease, Transient change, and Persistent decrease. All experiments were done in n = 3 biological replicates (one 24hAmAc replicate was removed from downstream analyses because of outlier status). Statistical significance cutoff for DEpP/DPPS was padj<0.05 (Student’s t test with Benjamini-Hotchberg correction). Significance for canonical pathways was the default-log(p value) ≥1.3 using a right-sided Fisher exact test.
Analyses of the clusters of DEpP using KEGG and GO allowed us to link the regulation of phosphorylation patterns to biological functions (Figures S7A–S7E). Supervised analyses showed clustering of DEpP involved in regulation of transcription and cell adhesion (Persistent increase cluster; Figure S7A) and were consistent with our overall pathway enrichment findings in hyperammonemia at 6 and 24h (Figures 2A–2D). Synapse and synaptosome are regulated by PKA (Hoover et al., 2001; Munno et al., 2003), a pathway consistently enriched during both 6hAmAc and 24hAmAc. A number of other regulatory responses that were identified are consistent with previous reports of RNAseq and proteomics responses and pathway enrichments during skeletal muscle hyperammonemia reported previously (Kumar et al., 2021; Welch et al., 2021).
Exercise-induced skeletal muscle phosphoproteomics responses
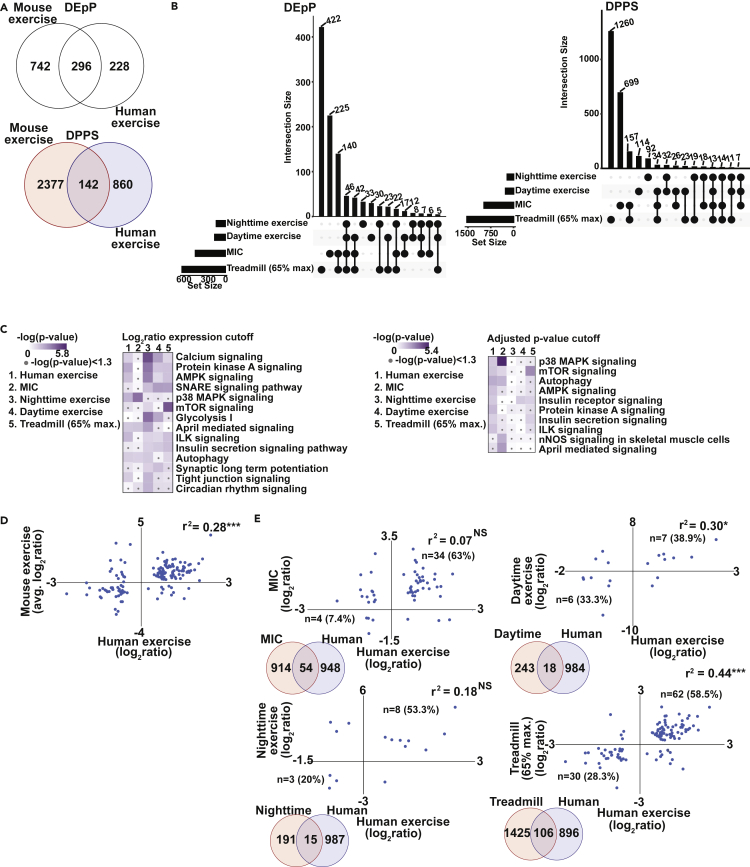
Because we are interested in developing a framework to identify modifiable skeletal muscle targets that can potentially improve functional capacity and protein homeostasis during exercise, we next performed integration of previously published phosphoproteomics datasets from skeletal muscle in mouse and human models of exercise and identified the mouse models of exercise that best recapitulate human exercise responses. The 4 mouse muscle phosphoproteomics datasets used for these analyses (electrically evoked maximal-intensity contraction (MIC), single bout treadmill running at 65% of maximal (max.) running speed in mice, daytime high-intensity treadmill running, nighttime high-intensity treadmill running) (Maier et al., 2022; Nelson et al., 2019; Steinert et al., 2021) were compared to the phosphoproteomic responses to a single bout of exercise in humans (Hoffman et al., 2015) (Figure 4). There were 296 shared differentially expressed phosphoproteins (DEpP) and 142 shared differentially phosphorylated phosphosites (DPPS) between all of the mouse exercise datasets and the human exercise dataset (Figure 4A). To determine responses that are related to individual mouse models of exercise or muscle contraction, we identified the shared and unique DEpP and DPPS from the 4 mouse exercise datasets (Figure 4B) and correlated each of the mouse datasets against each other (Figure S8). There were 14 DPPS shared across all mouse exercise models. There were 3 datasets that included only male mice (Maier et al., 2022; Steinert et al., 2021) and 1 dataset (Treadmill exercise at 65% of max. running speed) that included only female mice (Nelson et al., 2019). Among all 3 male mouse phosphoproteomics datasets, there were 7 shared DPPS/DEpP that were unique to the male models, perhaps related to reported sex differences in exercise responses (Hunter, 2016). Within the 7 shared DPPS/DEpP, the greatest decrease in differential expression was noted in phosphorylation of Ankyrin Repeat Domain 2 (Ankrd2), a member of the muscle ankyrin repeat protein (MARP) family of proteins that is highly expressed in skeletal muscle. Ankrd2 regulates sarcomeric activity and mechanosignaling pathways (Cenni et al., 2019; Lun et al., 2014), both of which are of relevance during exercise. The greatest increase in differential expression occurred in phosphorylation of Nascent Polypeptide Associated Complex Subunit Alpha (Naca) which prevents newly synthesized peptides from incorrect translocation and is involved in muscle growth, regeneration and myofibril organization (Li et al., 2009; Park et al., 2010b). Adding the female mouse dataset (treadmill at 65% max. running speed), showed that Naca phosphorylation was shared across models but on different phosphorylation sites in different models (Figure S9A). We also observed the greatest overlap in number of DPPS in the mouse exercise datasets between the MIC male mouse model and the treadmill exercise at 65% max. running speed female mouse model (n = 218), which also had a significant correlation between DPPS expression (r2 = 0.26, p < 0.001). The most enriched pathways (curated for relevance to skeletal muscle) included calcium signaling, PKA and synaptic transmission including vesicle fusion (SNARE), protein homeostasis (mTOR, AMPK), and insulin secretion signaling pathways (Figure 4C). To further dissect the direction of change in each dataset, heatmaps for these pathways were created (Figure S9B). Changes in mTORC1 and insulin signaling are known responses to exercise (Hawley, 2009; Hawley et al., 2018), but the relation between exercise and PKA is not as well studied (Azevedo Voltarelli et al., 2021; Berdeaux and Stewart, 2012; Hostrup et al., 2018; Jessen et al., 2021). Other new pathways identified included the TNF superfamily member 13 (APRIL) involvement during exercise responses. Of intererst, expression of components of each enriched pathway was not consistent in the significance or direction of change providing an explanation for differences in responses among models.
Figure 4.
Differentially phosphorylated phosphoproteins and phosphosites in murine vs human skeletal muscle following exercise or maximal intensity contractions
(A) Venn diagram of differentially expressed phosphoproteins (DEpP) and differentially phosphorylated phosphosites (DPPS) that are unique and shared in mouse and human exercise datasets.
(B) UpSet plot comparing 4 mouse models of exercise and their DEpP and DPPS.
(C) Canonical pathways enriched in each exercise dataset.
(D) Scatterplot comparing the averaged mouse DPPS expression to the human exercise DPPS expression.
(E) Expression of DPPS in each mouse exercise dataset was compared to that of the human exercise dataset using Pearson’s correlation analysis. Significance cutoff for DEpP/DPPS was padj<0.05 for mouse data and padj<0.05 and expression fold change >|1.5| for human data. Significance for canonical pathways was-log(p value) ≥1.3 using a right-sided Fisher exact test. MIC = maximal intensity contraction, Treadmill (65% max.) exercise = mice exercised at 65% of their maximal running speed on a treadmill, Daytime exercise = mice that underwent high-intensity treadmill running during the zeitgeber time (ZT)0 period of “lights on,” Nighttime exercise = mice that underwent high-intensity treadmill running during the ZT12 period of “lights off.”
Comparison of shared mouse and human exercise DPPS expression using Pearson’s correlation analysis showed that the phosphorylation pattern (i.e., whether the protein had increased or decreased phosphorylation in the mouse and human datasets) was significantly correlated (r2 = 0.28, p < 0.001) (Figures 4A and 4D). We next compared the expression of the shared DPPS between each individual exercise model. We used this approach to determine the mouse model that best recapitulates human muscle phosphoproteomics responses. Even though the MIC dataset had the greatest percentage of positively correlated phosphorylated proteins (63%) with the human dataset, the correlation coefficient was not significant (r2 = 0.07). The DPPS expression from the skeletal muscle of mice that underwent free treadmill running at 65% of max.intensity was the most correlated with those in the human exercise model (r2 = 0.44, p < 0.001), whereas the daytime high intensity treadmill running was the next most correlated with human responses (r2 = 0.30, p < 0.05) (Figure 4E). These data are consistent with our initial observations that molecular changes are not necessarily consistent across exercise models and may be context dependent because of differences in potential mediator(s) of these post-translational modifications.
Integrated analyses of exercise related skeletal muscle phosphoproteomics public datasets
Exercise-induced skeletal muscle hyperammonemia is believed to contribute to fatigue and potentially limit benefits of exercise, especially in chronic diseases (Banister and Cameron, 1990; Mutch and Banister, 1983). We therefore integrated our hyperammonemic myotube phosphoproteomics datasets with the published exercise datasets in mice and human to identify responses unique to either hyperammonemia, exercise, or those that were shared between the DEpP/DPPS under the two conditions in skeletal muscle and myotubes (Figure 5). Across any exercise (mouse and/or human) and any hyperammonemia (6hAmAc and/or 24hAmAc) dataset, there were 57 shared DPPS (165 DEpP), 560 DPPS (283 DEpP) unique to hyperammonemia, and 3322 DPPS (1101 DEpP) unique to exercise (Figure 5A). In the DEpP unique to the AmAc datasets, the most enriched pathways included glucocorticoid receptor signaling, CREB signaling, assembly of RNA polymerase II, and matrix metalloprotease regulation, all of which are potentially regulated by PKA (Felinski et al., 2001; Park et al., 2010a; Rangarajan et al., 1992) (Figure 5B). These findings are consistent with our experimental studies that the total (overall) PKA substrate phosphorylation was not significantly altered in response to hyperammonemia (Figure 5C) but that specific targets including phosphorylation of BCL2 associated agonist of cell death (BADSer155) were significantly increased with hyperammonemia (Figure 5D). In the DEpP unique to exercise datasets (as compared to hyperammonemia datasets), AMPK signaling, PKA signaling, MAPK signaling and mTOR signaling were among the most enriched pathways (Figure 5E). Regulatory interaction of PKA with these pathways have been reported including via the A-kinase anchoring proteins (AKAP) (Djouder et al., 2010; Waltereit and Weller, 2003; Zhang et al., 2017). We identified differential phosphorylation of AKAP12 and 13 (Figures S10A and S10B), but these regulatory interactomes between PKA and the pathways/mediators of ammonia and/or exercise responses need to be experimentally evaluated. We found that there were 2 DPPS (17 DEpP) that were found in the 6hAmAc, 24hAmAc, any of the mouse exercise datasets, and in the human exercise dataset (Figure 5F). The DEpP that were shared between any exercise dataset and any hyperammonemia dataset (n = 165) were analyzed for functional enrichment (Figure 5G), PKA was identified to be consistently enriched in many of the full datasets and subsets including those shared between hyperammonemic myotubes and human/mouse exercise models (Figure S10C). To further dissect the responses in exercise and hyperammonemia, we identified both PKA signaling and target molecules on our previously published proteomics datasets (Welch et al., 2021) and compared them with the DEpP/DPPS in the different exercise models and ammonia datasets. These analyses showed that even though the PKA pathway was enriched in the unbiased datasets, the specific signaling molecules and targets were different, helping explain the discord between in vivo exercise and in vitro hyperammonemia (Figures 5B, 5E, and 5G). In addition, RNA binding, processing, splicing, and transport were among the pathways enriched in DEpP shared between exercise and 6hAmAc and exercise and 24hAmAc DEpP (Figure S10D). We then generated a correlation matrix between all exercise (mice and human datasets) and all hyperammonemic DPPS (6hAmAc, 24hAmAc) and found that the majority of the DPPS across these models were positively correlated (Figure 5H) showing that certain clusters of molecules are highly correlated during hyperammonemia that show known and potentially new interactomes or regulomes (Figure S11) and, interestingly, PKA pathway components were among those highly correlated.
Figure 5.
Differentially phosphorylated proteins in hyperammonemia are shared with those in skeletal muscle from exercised mice and humans
(A) Venn diagrams of differentially expressed phosphoproteins (DEpP) and differentially phosphorylated phosphosites (DPPS) present in at least one of the 6h/24h ammonium acetate (AmAc) treated C2C12 myotube datasets compared to DEpP and DPPS, respectively, from at least one of the exercise (mouse or human) datasets.
(B) Canonical pathways enriched in DEpP unique to the hyperammonemia datasets compared to the exercise datasets.
(C) Representative immunoblots and densitometry of a protein kinase A (PKA) substrate motif (RRXS∗/T∗) phosphorylation in murine C2C12 myotubes treated with 10mM AmAc, 50uM H89 (PKA activator), and 20uM forskolin (Fsk; PKA inhibitor). Separate membranes with the same samples were used to generate loading controls for this panel.
(D) Representative immunoblots and densitometry of p-BAD(Ser155) in myotubes treated with 10mM AmAc, 50uM H89, and 20uM Fsk.
(E) Heatmap of enriched canonical pathways in DEpP unique to the exercise datasets and not found in the hyperammonemic datasets.
(F) UpSet plot showing unique and shared DEpP and DPPS among the 6hAmAc and 24hAmAc, mouse exercise, and human exercise datasets.
(G) Bar graph of enriched canonical pathways from the subset of DEpP that are shared between at least one hyperammonemia and at least one exercise dataset.
(H) Correlation matrix of shared DPPS between any exercise dataset and any hyperammonemic dataset (Blue = positive correlation and Red = negative correlation).Myotube experiments were done in n = 3 biological replicates (one 24hAmAc replicate was removed from downstream analyses because of outlier status). Densitometry data are mean ± SD. ∗p<0.05; ∗∗p<0.01; ∗∗∗p<0.001 ANOVA with Bonferroni post-hoc analysis. Statistical significance cutoff for DEpP/DPPS was padj<0.05 for ammonia and mouse data and padj<0.05 and expression fold change >|1.5| for human data. Significance for canonical pathways was-log(p value) ≥1.3 using a right-sided Fisher exact test. Daytime exercise = mice that underwent high-intensity treadmill running during the zeitgeber time (ZT)0 period of “lights on”; MIC = maximal intensity contraction; Nighttime exercise = mice that underwent high-intensity treadmill running during the ZT12 period of “lights off”, Treadmill (65% max.) exercise = mice exercised at 65% of their maximal running speed on a treadmill.
To identify the potential contribution of ammonia to post-translational modification of skeletal muscle proteins after exercise and to determine the exercise model(s) with the most shared molecules within each ammonia dataset, we compared these individual datasets with each other (Figures S12A–12F). Shared DEpP and DPPS among the 7 separate datasets (Figure S12F) showed only 2 DEpP shared among all the data (Titin, Plectin). Titin is a PKA target and one of the largest muscle proteins (Fukuda et al., 2005). Plectin regulates Wnt signaling (Yin et al., 2021) and consequently ribosome biogenesis via the β-catenin-cMYC axis during hyperammonemia (Davuluri et al., 2019). We also identified all of the shared DPPS on DEpP across all exercise and hyperammonemia datasets and created a score that allows for unbiased clustering based on number of phosphosites and absolute value of expression log2ratio of DPPS in each dataset (Figure S13). The shared DEpP with the highest scores, i.e., most phosphorylation sites with greatest absolute value of differential expression, had enrichment of structural proteins and molecular processes whereas the shared DEpP with the lowest scores, i.e., fewer phosphorylation sites with lower absolute value of differential expression, had enrichment of signaling (PKA, AMPK and mTORC1), structural proteins, and longevity pathways. Generating such a score has the potential to identify the murine model that has the greatest change in differential phosphorylation of each DEpP that were shared with human exercise and will allow for identification of preclinical models based on biological relevance and evaluation of the regulatory role of individual molecules (using genetic or pharmacologic modulation).
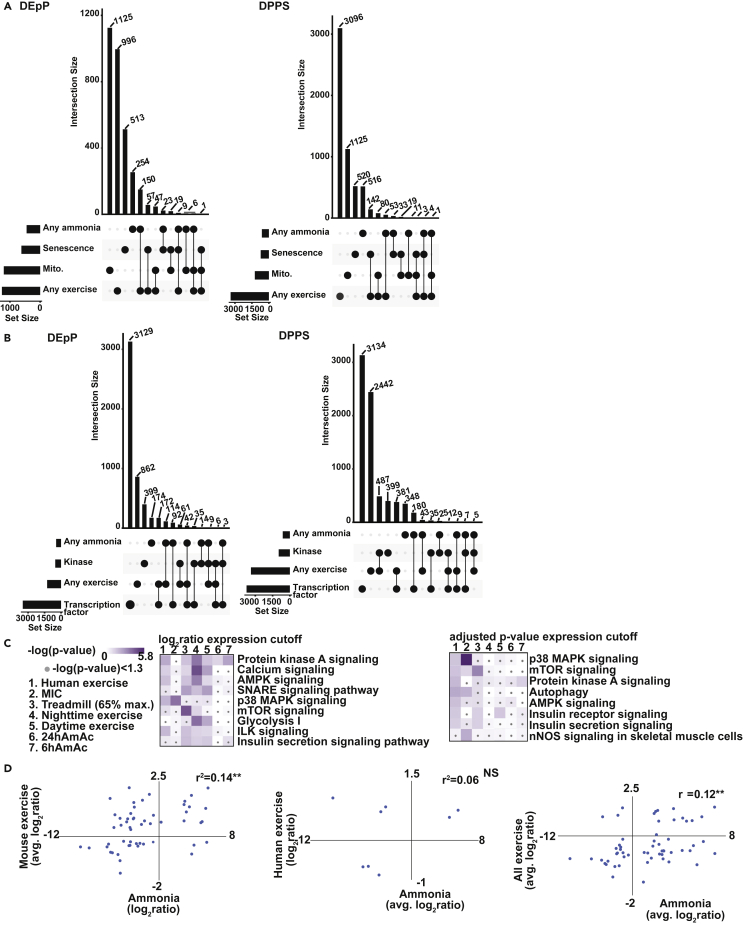
We and others have also reported that hyperammonemia causes skeletal muscle mitochondrial dysfunction and senescence whereas exercise promotes mitochondrial biogenesis and oxidative function and mitigates aging related effects via effects on fibroadipogenic precursors and stem cells (Cartee et al., 2016; Fiorenza et al., 2019; Kumar et al., 2021; Saito and Chikenji, 2021; Saito et al., 2020; Silva et al., 2009). We therefore performed a comparative phosphoproteomics analysis of the unique and overlapping DEpP in verified mitochondrial (Rath et al., 2021) and senescence genes (Avelar et al., 2020; Zhao et al., 2016) in the hyperammonemic myotube and exercise datasets (Figures 6 and S14A–S14D). Analysis of senescence related proteins showed 36 DPPS on 32 proteins that were shared between hyperammonemia and the senescence databases (Avelar et al., 2020; Zhao et al., 2016), but only 3 DPPS that were also shared with the exercise datasets (Figure 6A). Because telomere signaling is involved in DNA damage, and cell cycle regulation (Venturelli et al., 2014; Wan et al., 2021), processes which were significantly enriched in several datasets and clusters, we compared the ammonia and exercise datasets to a database of genes involved in telomere maintenance (TelNet)(Braun et al., 2018) and found there were 158 DPPS in either ammonia dataset (34% of the ammonia DPPS) and 519 (15%) of the exercise DPPS present in the Telnet dataset (Figure S14B). To identify change in phosphorylation status of kinases and transcription factors shared between exercise and hyperammonemia, we compared the exercise and hyperammonemia datasets to databases of known transcription factors and kinases (Figures 6B and S14C). Heatmaps of the mitochondrial and senescence-related DPPS during hyperammonemia in myotubes or in response to exercise in skeletal muscle showed differential alteration in components of the oxidative phosphorylation, TCA cycle regulatory and senescence-related proteins (Figures S15A–S15D).
Figure 6.
Comparative analyses of phosphoproteomics during hyperammonemia and exercise
(A) UpSet plots of unique and shared differentially expressed phosphoproteins (DEpP) present in at least one of the 6h or the 24h ammonium acetate (AmAc)-treated myotube datasets (Any ammonia) and those DEpP present in at least one of the mouse or human exercise datasets (Any exercise) compared to genes in a verified mitochondrial-localized protein database (MitoCarta3.0), and genes in senescence databases (CSgene and CellAge).
(B) UpSet plot of unique and shared DEpP present in at least one of the 6hAmAc or the 24hAmAc-treated myotube datasets (Any ammonia) and those DEpP present in at least one of the mouse or human exercise datasets (Any exercise) compared to transcription factor and kinase databases.
(C) Heatmap of canonical pathways enriched in the DEpP from the 6hAmAc and 24hAmAc datasets, mouse exercise datasets, and the human exercise dataset.
(D) Scatterplots of expression levels for DPPS in the mouse exercise and hyperammonemic myotube datasets. All myotube experiments were done in n = 3 biological replicates (one 24hAmAc replicate was removed from downstream analyses because of outlier status). Statistical significance cutoff for UpSet plot and scatterplot DEpP/DPPS was padj<0.05 for ammonia and mouse data and padj<0.05 and expression fold change >|1.5| for human data. Functional enrichment analyses were performed with two cutoffs: 1)log2ratio cutoffs were used to identify a similar proportion of foreground molecules in each dataset (500-800), 2) q<0.05 (as defined by each dataset) was used uniformly. The foregrounds were analyzed against each respective dataset as the background. Significance for canonical pathway enrichment was-log(p value) ≥1.3 using a right-sided Fisher exact test. ∗p<0.05, ∗∗p<0.01, Daytime exercise = mice that underwent high-intensity treadmill running during the zeitgeber time (ZT)0 period of “lights on”; MIC = maximal intensity contraction; Mito. = verified mitochondrial-localized molecules; Nighttime exercise = mice that underwent high-intensity treadmill running during the ZT12 period of “lights off”; NS = non-significant; Treadmill (65% max.) exercise = mice exercised at 65% of their maximal running speed on a treadmill.
Using functional enrichment analyses, we identified PKA, mTOR, AMPK signaling and cell cycle regulation/senescence/longevity to be overrepresented pathways in the human exercise dataset, at least one of the mouse exercise datasets, and at least one of the hyperammonemia datasets (Figure 6C). We also observed that SNARE signaling pathway which is involved in vesicle transport and synaptic transmission (Mukund and Subramaniam, 2020) was enriched in some of these models. A number of molecules in the SNARE pathway are also shared with the PKA signaling pathway, consistent with reports of PKA regulation of the SNARE complex (Chheda et al., 2001). The synaptogenesis pathway that was enriched during hyperammonemia shared a number of components with the PKA pathway. Similar to the observations with the PKA pathway, we also observed shared molecules between ERK/MAPK. APRIL and p38 MAPK signaling pathways were also enriched during hyperammonemia (Figure S15E). We then generated scatterplots to compare the DPPS expression levels across the hyperammonemic and exercise datasets (Figure 6D) that showed that the mouse exercise and hyperammonemia models are significantly positively correlated (r2 = 0.12, p < 0.01).
Phosphoproteomic responses during hyperammonemia and exercise revealed shared and unique motifs
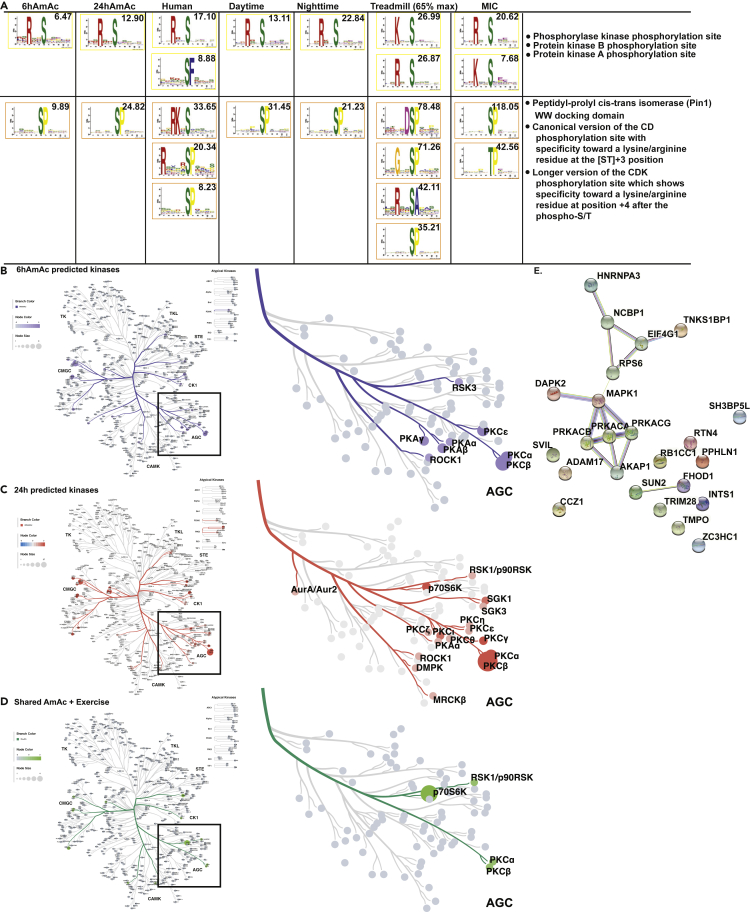
To determine temporal changes in the motifs contained in the hyperammonemia dataset, we analyzed the 6hAmAc and 24hAmAc datasets separately for motif enrichment and compared them with the motifs in the exercise datasets (Figure 7). In the 6hAmAc total DPPS dataset, there were 2 significant phosphorylation motifs (all p-S) identified and in the 24hAmAc total DPPS dataset there were 5 significant phosphorylation motifs (all p-S) identified. Phosphorylation motifs in the various exercise datasets also showed 2 phosphorylation motifs (xxRxxSxxxxx; xxxxxSPxxxx) shared between the hyperammonemia (6hAmAc, 24hAmAc) and exercise (mouse and human) datasets (Figure 7A). These phosphorylation motifs were predicted targets for phosphorylase kinase (regulates glycogen metabolism), protein kinase B (mTOR1 kinase that regulates protein homeostasis) and PKA (multiple targets involved in skeletal muscle (Knight and Kothary, 2011)) for motif 1 (xxRxxSxxxxx). Targets for motif 2 (xxxxxSPxxxx) included proline directed kinases, peptidyl-prolyl cis-trans isomerase (Pin1) WW docking domain and the canonical and long version of the cyclin-dependent kinase (CDK) phosphorylation site (Figure 7A). Of interest, PKA also has kinase interactions with phosphorylase kinase and PKB (Brushia and Walsh, 1999; Filippa et al., 1999) that can explain the high enrichment of PKA motifs in DEpP in our datasets. We identified the proteins that contained PKA target motifs and generated a connectivity network that showed the multiple levels of interactions and regulation by PKA during exercise and hyperammonemia (Figure S16). Pin1 is a type of foldase and belongs to the parvulin family that regulates mitotic activity. Pin1 interacts with mitotic phosphoproteins and regulates cell cycle and senescence (Shen et al., 1998; Toko et al., 2014), consistent with previous reports on ammonia-induced cell senescence (Gorg et al., 2015; Jo et al., 2021; Welch et al., 2021). The CDK molecules have regulatory roles in senescence and differentiation of myoblasts that can explain their relevance in muscle senescence (Etienne et al., 2020).
Figure 7.
Motifs and kinome analyses in the phosphoproteomics datasets
(A) Motifs and predicted kinases identified in differentially phosphorylated phosphosites (DPPS) from myotubes treated with 6 and 24h of 0mM ammonium acetate (AmAc) and skeletal muscle from mice and human exercise models. Scores shown in each panel indicates best fit with a known motif from the Eukaryotic Linear Motif database.
(B–D) Predicted kinases using NetworKIN, NetPhorest, and weighted CORAL kinome trees for DPPS in the 6hAmAc and 24hAmAc datasets and DPPS shared between any hyperammonemic (6hAmAc, 24hAmAc) dataset and any mouse or human exercise datasets. Enlarged kinome tree subsets show predicted protein kinases from the A, G and C (AGC) family in the respective datasets. Weighting is based on NetworKIN enrichment score.
(E) STRING protein-protein interaction network shows PKA, MAPK1, and their known interactions with DEpP shared between the hyperammonemia and exercise datasets. All myotube experiments were done in n = 3 biological replicates (one 24hAmAc replicate was removed from downstream analyses because of outlier status).
We performed complementary analyses using NetPhorest (Horn et al., 2014) and NetworKIN (Horn et al., 2014) to predict kinase-substrate interactions for the DPPS present in the AmAc and exercise phosphoproteomics datasets and CORAL (Metz et al., 2018) to visualize the frequency of the predicted kinases. We also found that the most frequently predicted kinases in either the 6hAmAc or 24hAmAc DPPS were casein kinase 2 (CK2), PKC, and Cdc-like kinases (CLK) and PKA (Figures 7B and 7C). Changes in CK2 were consistent with our previous report that the Wnt-β-catenin pathway which promotes ribosomal biogenesis and is regulated by casein kinase is impaired during hyperammonemia (Davuluri et al., 2019). On the kinome tree, the evolutionarily conserved serine-threonine protein kinase A,G,C (AGC) family of enzymes, which mediate signaling responses in a number of pathways, was enriched in the datasets. Given the relevance of these molecules in senescence, these observations are consistent with ammonia-induced post-mitotic senescence in skeletal muscle and modulation of muscle senescence by exercise (Cartee et al., 2016; Kumar et al., 2021). The subset of DPPS shared between any exercise dataset (human or mouse) and any hyperammonemia set (6hAmAc or 24hAmAc) generated a smaller number of predicted kinases, but the AGC family was again represented in this comparative analysis (Figure 7D). Given the consistent enrichment of our DEpP/DPPS datasets with PKA, an STRING network of the shared molecules between ammonia and exercise with a PKA node identifies potential interactions between these DEpP and PKA via MAPK1 (Figure 7E). Sequential analyses to identify regulatory interactions can be used to identify additional biological processes that are involved in exercise.
To identify protein kinase-substrate relationships within the clusters (Jamal et al., 2021), we performed substrate motif analysis for each temporal cluster of DEpP (Figure S17). In the P. increase and decrease clusters, the recognition motifs for phosphorylation by PKA, PKC, casein kinase I, GSK3 and Ca2+/calmodulin-dependent protein kinase 2 (CAMK2) family members were noted. Interestingly, this motif was overrepresented in both the P. increase and P. decrease cluster DPPS, which suggests that they are regulated by distinct and potentially common kinases. In the Late increase cluster, Casein kinase II and PAK2 kinase, which are important for muscle homeostasis, and in the Late decrease cluster, motifs for Casein kinase I and β-adrenergic receptor kinase were enriched. T. change cluster DPPS were recognized by cyclin-dependent kinase (CDK) and MAPK families. These analyses revealed that phosphorylation targets during hyperammonemia induced signaling are regulated by distinct sets of kinases.
Our integrated muscle phosphoproteomics dataset can also be interrogated in future comparative analyses with the approaches we have used in these studies. An index of Tables (Table S1) that allows for ease for determining the supporting data for our bioinformatics figures are provided as individual excel files for rigor and ease of reproducibility (Tables S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12, S13, S14, S15, S16, S17, S18, S19, S20, S21, S22, S23, S24, S25, S26, S27, and S28).
Discussion
We observed that ammonia, a cytotoxin consistently generated during exercise (Bellar et al., 2020; Chen et al., 2020; Gorostiaga et al., 2014; Graham et al., 1990, 1993, 1995; Graham and MacLean, 1998), results in differential phosphorylation events and enrichment of pathways in myotubes. A number of unique and shared phosphoproteomics responses between myotube hyperammonemia and skeletal muscle from exercise models were identified. Among the phosphorylated proteins shared between one or more hyperammonemic and skeletal muscle from exercise model dataset, PKA pathway had the highest enrichment. Regulatory interactions of PKA targets including AMPK, protein homeostasis and senescence mediators were also identified. Experimental studies in myotubes validated these phosphoproteomic responses during hyperammonemia and suggest that targeting either PKA or its substrates may be of relevance in chronic diseases with perturbed ammonia metabolism.
Our detailed phosphoproteomics analyses in hyperammonemic myotubes are consistent with and expand published data on the global molecular responses (Welch et al., 2021). In addition, because of our previous investigation of ammonia-induced changes in myotubes from the chromosomal to the translational level, we were able to identify those proteins regulated at only the phosphorylation level, only the protein level, or both. A number of molecules including those involved in cell cycle regulation and senescence, mitochondrial function, and protein synthesis that are known responses to hyperammonemia were regulated at both the proteomic and phosphoproteomic levels. Individual molecules that were shared between proteomics and phosphoproteomics in myotubes and whose expression levels changed in the same direction included metabolism regulatory molecule pyruvate dehydrogenase (Pdha1); Serine and Arginine Rich Splicing Factor 6 (Srsf6), an RNA splicing factor; Syntaxin (Stx7), a vesicle trafficking molecule; and Neurite outgrowth inhibitor (Rtn4), another membrane trafficking molecule that also interacts with apoptosis regulatory protein Bcl2 like-1 gene. Pathways that were enriched in the subsets of DE-Ps only regulated at the protein level and DEpP only regulated at the phosphoprotein level contained proteins regulating both known and previously unknown responses to hyperammonemia. The functional and regulatory relevance of these proteins in the skeletal muscle is still not completely clear and require experimental studies to identify their biological role during hyperammonemic stress (Davuluri et al., 2016b).
Hyperammonemia blunts protein synthesis via multiple mechanisms as reported by us earlier (Davuluri et al., 2016a, 2016b, 2019; McDaniel et al., 2016; Welch et al., 2021) and the phosphoproteomics analyses in the present studies also suggest reduced signaling in the protein synthesis pathway and impaired mitochondrial oxidative function, a critical requirement for ATP synthesis during contractile function. The present studies are consistent with recent data that muscle hyperammonemia promotes a senescence associated molecular phenotype that adversely affects multiple signaling and metabolic pathways (Kumar et al., 2021). Our phosphoproteomics analyses in myotubes show that, in addition to the alterations in mitochondrial proteins, telomere regulatory pathways are also altered and may contribute to muscle senescence during abnormal ammonia metabolism in chronic disease. Although exercise in healthy subjects promotes muscle plasticity via improved mitochondrial function and multiple signaling responses (Cartee et al., 2016), hyperammonemia worsens muscle plasticity. These observations suggest that dysregulated ammonia metabolism in chronic diseases (Dasarathy and Hatzoglou, 2018) may perturb skeletal muscle functional capacity, contractile function, and signaling responses to exercise.
Recently, two metanalyses of skeletal muscle transcriptomics related to exercise in humans have been published (Amar et al., 2021; Pillon et al., 2020). A number of factors including the duration and type of exercise, sex of the subject, location of the muscle biopsied and the time interval between termination of exercise and biopsy influenced the transcriptomic responses. In one study, 66 datasets were analyzed for exercises (n = 59) or inactivity (n = 6) and related differentially expressed skeletal muscle mRNA (Pillon et al., 2020). Distinct and shared patterns of transcriptomics were observed between acute endurance and resistance exercise whereas the expression of a number of genes including NR4A3 and GADD45 were differentially expressed between acute exercise and inactivity. In a subsequent metanalyses, the temporal course of skeletal muscle transcriptomics showed SMAD3, as a central hub of the regulatory responses to exercise (Amar et al., 2021). Both studies identified PGC1α, a regulator of mitochondrial biogenesis as an exercise responsive gene. However, our studies on muscle phosphoproteomics responses to exercise and ammonia in myotubes revealed enrichment of mitochondrial dysfunction pathways but not PGC1α targets, reiterating the lack of consistent concordance across layers of regulation in cells. We have previously shown that hyperammonemia causes reversible skeletal muscle mitochondrial dysfunction in addition to reversible post-mitotic senescence. Many of the molecules identified in these analyses were regulated by or altered the phosphorylation status of downstream signaling molecules.
In our integrated analyses of skeletal muscle phosphoproteomics responses in different models of exercise, we identified that there is enrichment of the PKA pathway including RNA processing, splicing, cell cycle regulation, as well as well recognized exercise responses including AMPK signaling, mTORC1 signaling and insulin responses (Hawley, 2009; Hawley et al., 2018; Hoffman et al., 2015). However, previous studies on skeletal muscle transcriptomics or proteomics do not report changes in PKA expression. The MetaMEx database (Pillon et al., 2020) showed that in acute aerobic exercise, PKA transcripts did not show significant changes, suggesting that PKA regulation of downstream molecules may be dependent on phosphorylation rather than on transcript expression. In the skeletal muscle, PKA regulates the neuromuscular junction and decreases mitochondrial oxidative function (Antipenko et al., 1999; Rudolf et al., 2013). The mechanism by which PKA is modulated during exercise is currently believed to be because of sympathetic activation (Bruno et al., 2014; Cairns and Borrani, 2015). However, our previous studies on the global landscape during hyperammonemia, a consistent metabolic consequence of exercise, showed enrichment of the PKA pathway in myotube transcriptomics and proteomics (Welch et al., 2021), and the present studies showed that PKA kinase activity is enriched during hyperammonemia, but the canonical target, CREB, is not phosphorylated at reported phosphorylation sites. Instead, during hyperammonemia, PKA regulates apoptosis via BAD with decreased apoptosis, consistent with previous data on preferential senescence over apoptosis (Kumar et al., 2021). Our analyses also suggest that PKA may regulate senescence via interactions between PKA signaling and CDKs as have been reported in other systems (Arsenijevic et al., 2006; Makarevich et al., 2010). Thus post-mitotic senescence in myotubes may be the consequence of convergence of multiple regulatory pathways whose molecular components may not necessarily directly interact. This is similar to the lack of direct correlation of their expressions of PKA and PLK in our bioinformatics analyses or in published data. However, both kinases have been reported to target the anaphase promoting complex/cyclosome (APC) complex, a ubiquitin ligase that targets cyclin B and factors that regulate metaphase-anaphase transition and mitotic exit and transcription (Kotani et al., 1998; Martin and Strebhardt, 2006). In addition to senescence, a number of CDK molecules regulate skeletal muscle morphogenesis that is also controlled by the HIPPO pathway (Watt et al., 2018). Consistently, our analyses suggested enrichment of HIPPO signaling pathways. Experimentally, we identified increased phosphorylation of MST2, a HIPPO signaling inhibitor. This is of relevance given that HIPPO signaling controls organ growth and regeneration via regulating cell proliferation and cell fate across species and multiple organs (Meng et al., 2016; Watt et al., 2018). Our studies showed that even though expression of HIPPO targets were not significantly altered in quantitative proteomics in hyperammonemic myotubes, phosphorylation of a number of HIPPO target proteins was altered. These data suggest that ammonia differentially regulates HIPPO responses in a time and context dependent manner and could be a potential target, along with PKA signaling, to optimize exercise induced skeletal muscle protein homeostasis and functional capacity, especially in chronic diseases, by mitigating perturbations related to skeletal muscle ammoniagenesis.
Our analyses merging all data in each dataset complement a recently described “personalized phosphoproteomics” — linking biological function using phenotype differences in response to exercise in human subjects (Needham et al., 2022). By taking into consideration duplicate annotations that may be secondary to transcript variants as well as orthologous phosphosites across species, these analyses provide a unique resource for additional analyses to address novel questions or relate these observations to future unbiased datasets that may be published in response to either hyperammonemia or exercise. However, challenges for such “across dataset analyses” include the complexity of the data, heterogeneity in models, and non-uniform experimental protocols that contribute to differences in responses. With the increasing use of machine learning and artificial intelligence-based approaches (including supervised and unsupervised analyses), comparative overlays, and functional enrichment analyses, unique and shared responses can be identified for mechanistic studies and therapeutic targeting. Additional challenges include the increasing availability of computational tools for such analyses. A large number of algorithms and post sample analyses bioinformatics tools have been developed to analyze unbiased data. It can be, therefore, challenging for scientists to translate developer tools to biological systems to express and interpret data in a meaningful manner (Lee et al., 2015; Mangul et al., 2019; Savage and Zhang, 2020). Our use of standard and widely used comparative molecular and pathway analyses allows for similar approaches to be applied rapidly and easily across models by an interdisciplinary group of investigators who may not be proficient in advanced bioinformatics or pipeline development and provides rigor and reproducibility.
Our systems biology approach to evaluate specific phosphoproteomic signatures will help identify preclinical models based on the scientific hypothesis and biological relevance. Specifically, our analyses suggest that ammonia lowering, especially during chronic diseases with perturbed ammonia metabolism including liver cirrhosis, heart failure, chronic obstructive lung disease and renal failure(Dasarathy and Hatzoglou, 2018; Medeiros et al., 2014; Valero et al., 1974), may improve exercise capacity and functional responses. This is especially relevant given the availability of ammonia lowering agents for human use and recent data that ammonia lowering can be beneficial in chronic diseases (Kumar et al., 2017, 2021). Future studies on the effects of ammonia-lowering interventions on exercise induced improvement in functional capacity and skeletal muscle protein homeostasis will be of high translational relevance. Our studies also lay the foundation for novel strategies to improve exercise capacity and beneficial responses by modulating other potential mediators of adverse responses to exercise(Bellar et al., 2020) while maintaining tissue and whole organism benefits.
Limitations of the study
Despite the physiological and translational relevance of our studies to identify the potential ammonia-related responses during exercise, it is possible that our myotube model may not reproduce skeletal muscle concentrations during exercise. However, blood concentrations of ammonia with exercise are similar to those reported in human cirrhosis and ammoniagenesis occurs within the skeletal muscle during exercise. Therefore, exercise-induced ammoniagenesis is likely to result in tissue concentrations that are similar to those observed in patients with cirrhosis and potentially other chronic diseases with hyperammonemia (Chen et al., 2020; Dasarathy and Hatzoglou, 2018; Medeiros et al., 2014; Qiu et al., 2013; Valero et al., 1974). Plasma ammonia concentrations decrease rapidly because of hepatic disposal in healthy subjects, but not necessarily in patients with disease because of impaired ureagenesis (Shangraw and Jahoor, 1999; Zheng et al., 2018). Another potential difference between exercise-induced ammoniagenesis in healthy subjects and subsequent responses and those in our experimental models is the temporal course of skeletal muscle phosphoproteomics responses during exercise. We have used the existing exercise phosphoproteomics datasets generated from healthy human subjects or wild-type mice, but the temporal course and severity of muscle hyperammonemia in chronic disease and exercise are currently unknown. Our myotube model recapitulates the tissue concentrations of ammonia in the resting state in cirrhosis, but with exercise, blood ammonia increases further (Dietrich et al., 1990) with muscle concentrations being even higher than in the basal state. Such a response during exercise in chronic disease may result in greater or different alterations in muscle molecular responses than those observed in our myotube model of hyperammonemia, but there are no published data on muscle ammonia in response to exercise in chronic diseases. Our analyses are therefore likely to be more conservative than the responses with exercise in chronic disease. Exercise related tissue ammonia concentrations will allow for direct comparisons of tissue levels, but even though such data are currently not available, future studies on ammonia lowering during exercise will be helpful for clinical translation.
Another potential confounder is that myotube cultures do not reproduce the fiber type differences in vivo, which can explain some of the differences observed between the cellular, animal and human studies observed in these and reported in other studies (Welch et al., 2021). However, differentiated C2C12 murine myotubes have been shown to recapitulate a number of skeletal muscle responses and in vitro studies allow for determining responses to stressors without the whole body adaptive responses compensating for the cell-specific responses (Abdelmoez et al., 2020; Davuluri et al., 2016b; Kumar et al., 2021; McMahon et al., 1994; Nedachi et al., 2008). Also, some of the differences in physiological, kinase and signaling and responses between myotubes and in vivo models may be related to the sex of the mice or gender of human subjects, muscle group used and exercise protocols. Hence, the numbers of DPPS on DEpP are not always concordant because even if the DEpP are shared, the DPPS on those proteins may not necessarily match, suggesting that phosphorylation at different sites can have variable functional responses depending on the confounders that influence data integration across models. Despite these limitations, our analyses, including experimental data, lay the foundation for future mechanistic studies including ammonia measurements and responses to ammonia lowering in human subjects with chronic diseases in response to exercise. Even though our analyses across models helped identify unique and shared exercise-induced responses and perturbations during hyperammonemia across molecular layers (that some call “omics layers”), pathways including PKA signaling may be potential targets for modulating skeletal muscle protein homeostasis (proteostasis) in response to exercise. Our analyses or experiments do not provide direct evidence of a mechanistic evidence that exercise induced ammoniagenesis lessens the beneficial responses including skeletal muscle functional capacity and protein homeostasis. By querying public datasets of untargeted phosphoproteomics from skeletal muscle from mouse and human exercise models, we identified that the treadmill at 65% of max. running speed and MIC mouse models had the most concordant phosphoproteins with exercise responses in human muscle and therefore may be optimal models to use for future exercise-based studies with hyperammonemia. Studies in preclinical models with loss/gain of function with or without ammonia lowering are needed to establish such a link given the challenges of mechanistic studies in humans.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-Bad antibody (Ser155) (rabbit polyclonal) | Cell Signaling | Cat# 9297; RRID:AB_2062131 |
| Phospho-PKA Substrate antibody (RRXS∗/T∗) (clone 1000G7E) (rabbit monoclonal antibody) | Cell Signaling | Cat# 9624; RRID:AB_331817 |
| Phospho-IKKB antibody (Ser672) (rabbit polyclonal) | Aviva Systems Biology | Cat# OAAB16292 |
| IKK-beta antibody (clone 10AG2) (mouse monoclonal) | Millipore | Cat# 05-535; RRID:AB_2122161 |
| Phospho-MST2 antibody (Ser316) (rabbit polyclonal) | Invitrogen | PA-105065 |
| MST2 antibody (rabbit polyclonal) | Cell Signaling | Cat# 3952; RRID:AB_2196471 |
| Phospho-MCM2 antibody (Ser139) (clone D1Z8X) (rabbit monoclonal) | Cell Signaling | Cat#12958; RRID:AB_2798069 |
| MCM2 antibody (clone D7G11) XP(R) (rabbit monoclonal) | Cell Signaling | Cat# 3619; RRID:AB_2142137 |
| Phospho-S6 antibody (Ser235/326) (rabbit polyclonal) | Cell Signaling | Cat# 2211; RRID:AB_331679 |
| S6 ribosomal protein antibody (clone 54D2) (mouse monoclonal) | Cell Signaling | Cat# 2317; RRID:AB_2238583 |
| β-actin antibody (clone C4) (mouse monoclonal) | Santa Cruz | Cat#: sc-47778; RRID:AB_626632 |
| Anti-rabbit IgG, HRP-linked antibody | Cell Signaling | Cat#7074; RRID:AB_2099233 |
| Anti-mouse IgG, HRP-linked antibody | Cell Signaling | Cat#: 7076; RRID:AB_330924 |
| Chemicals, peptides, and recombinant proteins | ||
| Ammonium Acetate | Sigma Aldrich | Cat# A7330 |
| RIPA buffer | Thermo Fisher | Cat# 89901 |
| Forskolin, Coleus forskohlii in DMSO | EMD Millipore | Cat# 344282 |
| H-89, Dihydrochloride | EMD Millipore | Cat# 371962 |
| Deposited data | ||
| Hyperammonemia-dependent and independent skeletal muscle phosphoproteomic responses during exercise | This paper | ProteomeXchange: PXD031372 |
| Mitochondrial responses during hyperammonemia | ProteomeXchange | ProteomeXchange: PXD026955 |
| Transcriptomic, proteomic and phosphoproteomic underpinnings of daily exercise performance and Zeitgeber activity of endurance training in mouse skeletal muscle | ProteomeXchange | ProteomeXchange: PXD026461 |
| The exercise-regulated skeletal muscle phosphoproteome | ProteomeXchange | ProteomeXchange: PXD001543 |
| Phosphoproteomic screening of exercise-like treatments reveals drug interactions regulating protein secretion | ProteomeXchange | ProteomeXchange: PXD014322 |
| Phosphoproteomics of rodent exercise models | ProteomeXchange | ProteomeXchange: PXD010452 |
| Impact of maximal-intensity contractions and rapamycin on the proteome and phosphoproteome of mouse skeletal muscle | University of California San Diego (UCSD) MassIVE Database | MassIVE database: MSV000086732 |
| Integrated molecular landscape perturbations underlie cellular responses during hyperammonemia [ATAC-seq] | GEO | GSE171642 |
| Integrated molecular landscape perturbations underlie cellular responses during hyperammonemia [human RNA-seq] | GEO | GSE171643 |
| Integrated molecular landscape perturbations underlie cellular responses during hyperammonemia [mouse RNA-seq] | GEO | GSE171644 |
| Integrated molecular landscape perturbations underlie cellular responses during hyperammonemia | GEO | GSE171645 |
| Experimental models: Cell lines | ||
| C2C12 myotubes | ATCC | CRL-1772 |
| Software and algorithms | ||
| IPA | QIAGEN, Inc. | https://digitalinsights.qiagen.com/IPA |
| g:Profiler | N/A | https://biit.cs.ut.ee/gprofiler/gost |
| RStudio | N/A | Rstudio.com |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Perseus | MaxQuant | https://maxquant.net/perseus/ |
| Phomics | N/A | http://phomics.jensenlab.org/phospho_enrichment |
| STRING | String Consortium 2022 | https://string-db.org/ |
| Other | ||
| Phosphoproteomics Scatterplot code | Zenodo and GitHub | v0.1SitesPerProtein dasaraslab/Unbiased: Phosphoproteomics Scatterplot https://zenodo.org/badge/latestdoi/392334492 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Srinivasan Dasarathy (dasaras@ccf.org).
Materials availability
This study did not generate any new reagents.
Experimental model and subject details
Cell lines
Murine C2C12 myoblasts (ATCC, Manassas, VA) were differentiated as previously described (Qiu et al., 2012, 2013). In brief, myoblasts were grown at 37°C in Dulbecco’s modified Eagle Medium (DMEM) with 10% fetal bovine serum (proliferation medium) to near confluence. Proliferation medium was replaced with differentiation medium (DMEM with 2% horse serum) for 48h. Differentiated myotubes were then treated with 10mM ammonium acetate, a concentration that reproduces tissue concentrations of ammonia in a rat model and human patients with cirrhosis and (McDaniel et al., 2016; Qiu et al., 2013). Even though it is not known if these concentrations reproduce skeletal muscle concentrations during exercise, we have chosen this model because we expect tissue concentrations to increase even further with exercise based on consistent increases induced blood ammonia with exercise (Calvert et al., 2010; Dietrich et al., 1990).
Method details
Sample preparation and phosphoproteomics assays
Label free proteomics were performed in samples as previously described (Welch et al., 2021). In brief, cells were lysed in ∼500μl of Urea Lysis Buffer (20mM HEPES, 9M urea, 1X HALT protease and phosphatase inhibitor cocktail, pH 8.0 and protein concentrations measured using bicinchoninic acid assay as previously reported. Protein extracts were reduced by alkylated dithiothreitol and digested with trypsin overnight at room temperature. After digestion, peptides were purified (desalted), lyophilized and spiked with phosphopeptide standard I (Protea Biosciences Group, Inc. #PS-180-1) with 6 pool each of three synthetic phosphopeptides [DRVpYIHPF (Angiotensin II), IKNLQpSLDPSH (Cholecystokinin 10-20) and DFNKFHpTFPQTAIGV (Calcitonin 15-29)]. These peptides were spiked in after digestion and prior to enrichment to serve as a measure of enrichment efficiency. Two of these phosphopeptides were identified in the LCMS/MS experiments. The data from these samples were searched against the full mouse UniProtKB database considering S, T, and Y phosphorylation as a dynamic modification. Equal amounts of peptide from each sample were phospho-enriched using Thermo Scientific™ High-Select™ TiO2 Phosphopeptide Enrichment Kit, and the eluted peptides were dried immediately. Each sample was reconstituted in 30μl 1% acetic acid and spiked in 10μl 50 fmole/μl Pierce Peptide Retention Time Calibration Mixture (12.5 fmole/μl final concentration) as reference. The LC-MS system was a ThermoScientific Fusion Lumos mass spectrometer system. The HPLC column was a Dionex 15 cm × 75 μm internal diameter Acclaim Pepmap C18, 2μm, 100 Å reversed-phase capillary chromatography column. Peptides from 5μL of the extract were eluted from the column by an acetonitrile/0.1% formic acid gradient at a flow rate of 0.25 μl/min introduced into the source of the mass spectrometer on-line. The digest was analyzed using the data dependent multitask capability of the instrument acquiring full scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequence in successive instrument scans.
Raw mass spectra were searched against the mouse UniProt protein database with 16,996 entries (downloaded on July 9th, 2019) using Sequest algorithm of Proteome Discoverer software (Orsburn, 2021) (V2.3 (https://www.thermofisher.com/order/catalog/product/IQLAAEGABSFAKJMAUH). Carbamidomethyl C was selected as a fixed modification, whereas oxidation (M), acetylation (protein N-term), and phosphorylation (STY) were selected as variable modifications. Perseus software 1.5.8.5 (74) was used for quantitative analysis of the results from Proteome Discoverer V2.3. The raw intensity of each phosphorylation site was normalized on the basis of starting amount of proteins. Analysis of variance (ANOVA) with permutation-based false discovery rate (control at 0.05) was used to detect statistically significant differences in phospho-peptide levels between developmental stages. Hierarchical clustering of the z-score transformed abundance of the statistically significant phosphorylation sites was performed using Euclidean distance and the average linkage method. Sequence logos around phosphorylated residues were created (PhosphoLogo) for subsets of statistically significant sites based on profile plots (increasing or decreasing). The total number of peptides identified in all eighteen samples was 14453 with 9232 phosphopeptides. The experimental approach is shown in the graphical abstract.
Quantitative analysis
The relative abundance of the phosphopeptides in these samples was determined using a label free approach. This method involves aligning chromatograms and determining the normalized abundance for each peptide. The LFQ value was either derived from an identified peptide precursor or a precursor peak whose peptide was identified in another sample. If the precursor peak was not found in a sample, the data point would be missing. These data were normally not missing at random but were either due to their abundances below the detection limit of the instrument or not presented in the samples. The censoring missing values were imputed using a quantile regression approach that imputes missing data using random draws from a truncated distribution with parameters estimated using quantile regression in Perseus (Tyanova et al., 2016). The data matrix was log2 transformed and the missing values were imputed from a truncated normal distribution. The imputed data were taken randomly from the distribution of the data in the column down shift 1.8 σ and the width of selection was set at 0.3 σ. The LFQ intensities and imputed values for each protein across all 9 samples are provided in Figure S18A). These normalized abundances were used to calculate the LFQ ratios. The counts plots for all 9 samples indicates that the 24hr Ammonia-1 sample is an outlier due to the identification of only 1026 phosphopeptides (Figure S18B). The low number of identified phosphopeptides may be due to poor digestion of this sample. PCA plots were plotted without this sample (Figure S18C). Sample comparisons for phosphosites in the hyperammonemic datasets are shown in Figures S18D and S18E.
Experimental validation
Immunoblots of lysates from differentiated myotubes were performed as previously described (Qiu et al., 2012). In brief, after denaturation using Laemmli buffer, proteins were subjected to electrophoresis in a 10% tris-glycine gel, electrotransferred to PVDF membranes, transfer and equal loading ascertained by Ponceau staining, incubated with primary (1:1000) and secondary antibodies (1:10,000), and developed using enhanced chemiluminescence assay. Densitometry of blots were quantified by ImageJ (Schneider et al., 2012). For the protein kinase A regulation, cells were treated with forskolin 20μM for 6h to activate and H89 (50μM for 6h) to inhibit PKA. Classical PKA target cyclic AMP response binding (CREB) protein phosphorylation at the canonical serine133 site, and Bcl2 associated agonist of cell death (BAD) at serine 155 was also quantified by immunoblots. Other phosphorylated proteins that were experimentally validated included MST2Ser316, MCM2Ser319, and ribosomal S6Ser235/236. These specific phosphoproteins were chosen because they are critical regulators of skeletal muscle protein homeostasis (proteostasis); published literature supports changes in one or more of these molecules across tissues during hyperammonemia (IKKB and ribosomal S6 protein) and validated antibodies are commercially available to test molecules of high relevance in maintenance of muscle mass/proteostasis that were highly enriched in our pathway analyses/DEpP.
Curation of published exercise datasets
A search of ProteomeXchange, a consortium of unified submissions in the proteomics field (Vizcaino et al., 2014) followed by a manual search of publications identified 4 mouse phosphoproteomics datasets in which either exercise or electrical contraction were used (Maier et al., 2022; Nelson et al., 2019; Steinert et al., 2021) and one human exercise phosphoproteomics dataset (Hoffman et al., 2015) that were compared with controls in skeletal muscle. We compared these public datasets of DEpP/DPPS from mice and humans to our data in myotubes during hyperammonemia. We recognize the heterogeneity of the exercise protocols and used overlay approaches to determine unique and shared responses.
Mouse exercise datasets
In the MIC dataset, mouse muscle was obtained following max. intensity contractions to electrical stimulation were elicited in anesthetized male mice using an electrode placed on the sciatic nerve which was stimulated at 100Hz with 0.5 ms pulses at 4-7 V for 10 sets of 6 contractions (Steinert et al., 2021). Each contraction was elicited for 3 seconds and was followed by a short (10 second) rest period with 1 min rest between sets. Mice were sacrificed 1h after this regimen and the tibialis anterior was collected for the phosphoproteomics analyses. In our analysis, we compared the group of mice that underwent MIC plus vehicle (n = 4) to the group of mice that did not undergo MIC plus vehicle (n = 4).
In the dataset from the daytime exercise mice (Maier et al., 2022), skeletal muscle from male mice who underwent high intensity treadmill exercise at Zeitgeber time 0 (e.g. early morning exercise, at the time of light onset) was compared to the skeletal muscle of sedentary mice (n = 3 in each group). The exercise regimen was performed using a treadmill with a shock grid on which a max. exercise capacity test was performed with the treadmill speed increasing by 2 meters per minute every 2 minutes at a 15 degree slope until exhaustion. In the nighttime exercise dataset from the same publication (Maier et al., 2022), a similar high intensity treadmill exercise was performed at Zeitgeber time 12 (e.g. early nighttime exercise at the time of light offset) with a mild electrical shock as for the daytime exercise mice above compared to sedentary mice analyzed at the same Zeitgeber time 12 (n = 3 in each group). Mice in all groups compared in this analysis were sacrificed immediately after exhaustion and the quadriceps and gastrocnemius muscles were analyzed for phosphoproteomics.
In the mouse dataset from the treadmill (65% max.) mice (Nelson et al., 2019), data were obtained from skeletal muscle from 3-month-old female mice that underwent a single bout of treadmill running and compared to that from sedentary mice (n = 5 in each group). Max. running speed per mouse was determined using a 10 degree incline with a 5-minute warm up at 10 meters/minute and speed increased by 1.2 meters per minute every minute until exhaustion (defined as the speed at which the mouse was unable to keep up with the treadmill). The experiment was performed at 65% of each mouse’s max. running speed (approximately 20 meters/minute) for 30 minutes at a 10 degree incline. Control mice were rested on a still treadmill. Mice were sacrificed immediately after the exercise episode and the gastrocnemius muscle was analyzed for phosphoproteomics studies.
Human exercise dataset
We compared hyperammonemic cellular and mouse exercise phosphoproteomics data to human exercise data from skeletal muscle biopsies from untrained healthy males prior to and after undergoing one bout of high-intensity exercise on a cycle (n = 4 at each timepoint). Exercise was performed for 6 minutes at 85% of maximum work capacity (Wmax) and then to exhaustion at 92% of Wmax, which occurred after 9–11 minutes of exercise time (Hoffman et al., 2015). Muscle biopsies were taken from the vastus lateralis before and after exercise completion and phosphoproteomics analyses were performed.
Phosphoproteomics datasets from hyperammonemic myotubes and those from skeletal muscle in exercise models were aligned and merged to generate a searchable table.
Bioinformatics approaches
Comparative phosphoproteomics analyses were performed across myotubes, mouse and human muscles using machine learning approaches including feature selection and supervised and unsupervised analyses of different datasets. Supervised analyses used labeled datasets (group to which the data belongs is identified) for differential expression analyses. This allowed for pattern identification of the groups (UnT, 6h AmAc and 24hAmAc). Subsequently, an unsupervised analysis was done where the labels were removed and the data were analyzed for hierarchical clustering to determine if our pattern identification was valid. Even though this is a limited use of machine learning, it did allow us to demonstrate that hyperammonemia did result in distinct signatures that had unique and overlapping features with exercise responses in vivo.
Feature visualization
R studio (version 4.1.2) was used for the visualizations. The following R packages were used for the Venn diagrams-Venn (https://github.com/dusadrian/venn), ggplot2(Wickham, 2016) (https://ggplot2.tidyverse.org), and ggpolypath (https://CRAN.R-project.org/package=ggpolypath). Venn diagrams of DEpP were created by removing duplicated proteins with multiple phosphosites and counting the unique phosphoproteins in a dataset. To generate the correlation matrix, we used corrplot(Wei and Simko, 2021), https://github.com/taiyun/corrplot] and magrittr [https://magrittr.tidyverse.org/articles/magrittr.html] R packages. Excel (Microsoft Corporation. (2021). Version 16.56.Microsoft Excel. Retrieved from https://www.microsoft.com/en-us/microsoft-365/excel) was used to create scatterplots of shared DPPS. UpSet plots were generated using R package UpSetR (version 1.4.0) (Lex et al., 2014).
Dataset comparisons
In order to compare the mouse phosphosites in the hyperammonemic myotube and mouse exercise datasets to the human exercise dataset and to use the downstream motif prediction tools, we used PhosphoSitePlus to identify orthologous phosphosites between mouse and human(Hornbeck et al., 2015). When we merged the human/mouse datasets, we only included those mouse proteins that had a human ortholog to capture the most overlap with exercise and hyperammonemia possible. We next determined whether there was any overlap between the hyperammonemic and exercise datasets with the senescence databases CSgene (Zhao et al., 2016) and CellAge (Avelar et al., 2020) which we combined to create a curated senescence database, verified mitochondrial genes from Mitocarta3.0 (Rath et al., 2021), transcription factors (Chawla et al., 2013), kinases (UniProt Consortium, 2021), and telomere maintenance(Braun et al., 2018) databases. For rigor and reproducibility of our analyses, we chose those bioinformatics tools that were consistently functional across operating systems, produced high quality of results, had an ability to adapt to additional data, and allowed for efficiency and audit trails to evaluate the logic of the analyses. We therefore chose the PhosphoSitePlus databases over others to determine orthologous phosphorylation sites across mouse and human proteomes for these analyses as well as multiple functional enrichment tools including both open-source and proprietary software.
Motif identification
Two motif-based sequence analysis tools from Meme-suite.org (MEME version 5.3.3) were used, MoMo (Cheng et al., 2019), and Tomtom (Gupta et al., 2007). First, MoMo was used to discover phosphorylation motifs within the datasets and subsets using the following parameters: algorithm--motif x, motif width—11, Eliminate duplicate peptides identical for width—11, yes, Minimum number of occurrences—5, Motif-x P-value Threshold—0.000001. The motif-x algorithm score, which identifies the sum over the significant position/residue pairs of-log(pbinomial)). Fg_size is the total number of foreground peptides with the given central modification and fg_match is the number of foreground peptides that match the motif. This score was used to identify motifs enriched in our data. Next, the Tomtom tool was used to search these motifs against the Eukaryotic Linear Motif (ELM 2018) database (Kumar et al., 2020). Parameters used in Tomtom were as follows: Motiftype—Protein, Target motifs—Protein Motifs, Eukaryotic Linear Motif (ELM 2018), Search with just one motif—yes, Motif column comparison function—Pearson correlation coefficient, Significance threshold—E-value<10, Activate complete scoring—yes. Output motifs from Tomtom were investigated using the ELM resource for Functional Sites in Proteins (Kumar et al., 2020), a manually curated database from over 3000 publications identifying of experimentally validated short linear motifs (n = 3523) within 289 motif classes.
Identifying known and predicted kinase-substrate relationships
The kinase-substrate database from PhosphoSitePlus (Hornbeck et al., 2015) was used to identify experimentally validated kinase-substrate relationships for the phosphosites identified in the analyzed datasets. Prediction for kinase-substrate relationships were made using NetPhorest (v2.1) and NetworKIN (v3.0) that are components of kinomeXplorer, which incorporate artificial intelligence methods including neural networks (Horn et al., 2014). The NetworKIN score is a NetPhorest probability score combined with the STRING-derived proximity score using the naïve Bayes method.
Weighted CORAL (Clear and customizable visualization of human kinome data) (Metz et al., 2018) (http://phanstiel-lab.med.unc.edu/CORAL/) kinome trees were used to visualize the most frequent predicted kinase families enriched in the studied datasets. The kinome trees were weighted by quantifying the number of times a kinase was predicted to interact with a DPPS in a dataset (determined by NetworKIN score) for a kinase-substrate relationship (Hoffman et al., 2015; Needham et al., 2019; Nelson et al., 2019; Steinert et al., 2021).
Quantification and statistical analysis
Differential expression for DEpP/DPPS was determined by comparing the expression of the treated (e.g. exercise or ammonia) sample to its respective control (e.g. resting or untreated) sample. Differential expression of the phosphoprotein (DEpP) was then defined as an adjusted p-value <0.05 irrespective of the phosphorylation site (serine/threonine). DPPS was defined as an adjusted p value of p<0.05 but accounting for the phosphorylation site to allow for comparisons between the same proteins but on different phosphorylation sites which not be identified on DPPS analyses. Similarly, shared and unique DPpP and DPPS were defined as follows: if at least one phosphoproteins/phosphosite was common between mouse and human datasets, it was considered to be shared and if there were no common phosphorylated proteins/phosphosites across dataset, it was considered unique to the model in which this was differentially expressed.
Feature selection was performed by filtering DEpP/DPPS based on an adjusted p-value (false discovery rate or q-value which is obtained by calculating the fraction of accepted hits from the permuted data divided by the accepted hits from the measured data and normalized by the total number of randomizations) of <0.05 (Tyanova et al., 2016).
When we paired the ammonia and exercise datasets, the average expression ratio of the phosphoprotein pairs was calculated. This was a modification of a method previously described by others (Bono and Hirota, 2020) to quantify the log2ratio expression of DPPS that are identified across similar models. For example, if a DPPS is found in both the 6hAmAc and 24hAmAc dataset, then the average log2ratio of the two scores is calculated. If a DPPS is shared in 3 mouse models, the DPPS for exercise is the average log2ratio across the 3 datasets. If the averaged ratio was over the threshold for upregulation, it was considered to be upregulated and vice versa for downregulation. Since we only considered DPPS, there were no unchanged molecules considered in this analysis. The relation between models was evaluated using the averaged expression scores plotted for each pair and the correlations were either positive or negative and were located on the top right and bottom left quadrants respectively. Pairs that did not correlate between models were defined in the top left and bottom right quadrants. The percentage of correlated and non-correlated molecules were not calculated because they depend on the total number of shared and unique genes. Hence, to provide a perspective of the total number of molecules being evaluated for the AE score correlations, Venn diagrams of shared and unique molecules were provided with each model pair being compared.
Frequency-expression plots were created from the DPPS for each gene across datasets (log2ratio of exercise/control). Specifically, the protocol used was:
-
1.
For each gene, count the number of total sites up-regulated across all datasets.
-
2.
For each gene, count the number of total sites down-regulated across all datasets.
-
3.Store the maximum value between step 1 & step 2
-
a.Ex: TTN
-
i.TTN Position 10-> 6h: 1.0, 24h: 2.0, MIC: −4.0, Human: 1.5
-
ii.TTN Position 20-> 6h: −1.5, 24h: −2.4, MIC: 6.0, Human: 2.0
-
iii.Step 1 results in 5 up-regulated sites for TTN, Step 2 results in 3 down-regulated sites for TTN.
-
iv.Store a value of 5 for the maximum value of shared directional sites
-
i.
-
a.
If there is a tie between the maximum values between genes, the tie is broken by ordering by the average of the absolute value of the fold changes for each gene’s sites.
Functional enrichment analyses
To account for differences in statistical analyses across the mouse and human exercise phosphoproteomics datasets, comparative functional enrichment analyses were performed using two methods of feature selection and dimensionality reduction: first, using greatest expression differences in phosphorylation as compared to controls and, next, using the p-value cutoffs specified in each published dataset. To avoid reliance only on one pathway algorithm or gene list (Liberzon et al., 2011, 2015), we used GO (Harris et al., 2004), KEGG (Kanehisa et al., 2017; Kanehisa and Goto, 2000), and Reactome (Fabregat et al., 2018) databases.
IPA (QIAGEN Inc., https://www.qiagenbio-informatics.com/products/ingenuity-pathway-analysis), DAVID (Huang da et al., 2009a, 2009b), Perseus (Cox and Mann, 2012; Tyanova et al., 2016), g:Profiler (https://biit.cs.ut.ee/gprofiler/), and STRING were used for functional enrichment analyses. Given the differences in algorithms and gene lists for different approaches, we used a combination of tools including IPA (Kramer et al., 2014) that provided pathway enrichments, DAVID (Huang da et al., 2009b) to obtain an annotated gene list and functional enrichment using gene datasets, and Perseus (Tyanova et al., 2016) to determine if expression values of individual phosphoproteins have a preference to be systematically larger or smaller than the global distribution of expression values (Cox and Mann, 2012). These varied approaches allowed for a broad exploration of pathway and biological process enrichment discovery of these integrated datasets.
DAVID
DAVID functional enrichment analysis was performed for the complete hyperammonemia phosphoproteomics datasets using a foreground of regulated (FDR <0.05) sites and using a background of unregulated sites (FDR≥0.05). DAVID functional enrichment analysis was performed for the phosphoproteomics data subsets of the ‘6hAmAc Only’ sites, the ‘24hAmAc Only’ sites, and the hyperammonemic clusters and a background using the entire phosphoproteomics data (Huang da et al., 2009a; 2009b).
Ingenuity pathway analysis (IPA)
Canonical pathways shown in figures were filtered for relevance and ordered based on a-log(p-value) ≥ 1.3. Exercise and hyperammonemia data in IPA were analyzed using the dataset phosphorylated proteins as the background and filtered by log fold change for the foreground proteins in order to understand what pathways were enriched using the greatest change in expression in each group. Since the statistical approaches and number of samples in each of the published datasets were variable, we performed functional enrichments in IPA on the full datasets by defining the foreground of differentially expressed proteins using 2 approaches: 1) Absolute value of log2ratio change cutoffs was adjusted per dataset (6hAmAc and 24hAmAc>|2.5|; Nighttime, Daytime>|1|; MIC, Treadmill (65% max.), Human >|0.5|) to achieve 500-800 proteins to normalize for differences in machines and batch effect, 2) A uniform significance cutoff at the DEpP level of q<0.05 in each dataset. The background against which enrichment was identified for each hyperammonemia and exercise datasets was the full dataset of phosphoproteins identified in each project. For the data subsets, i.e. ‘AmAc’ only, ‘Exercise only’, and ‘Shared AmAc and Exercise,’ the foreground data used were the DEpP which were analyzed against the background of all phosphoproteins within the hyperammonemia datasets (‘AmAc only’), the exercise datasets (‘Exercise only’), or both (‘Shared AmAc and Exercise’ sites).
Perseus
Perseus 1D analysis was performed for the hyperammonemia datasets using all phosphoproteins without any significance cutoff for protein at 6hAmAc and 24hAmAc and the default settings for pathway significance (Benjamini-Hotchberg method, FDR<0.02, removal of duplicate phosphoproteins if more than one site was phosphorylated on the protein) in the hyperammonemia datasets.
g:Profiler
g:Profiler analysis was only performed for subsets of shared hyperammonemia and exercise DEpP and were performed using the gene lists of interest against a homo sapiens genome background.
All canonical pathways that had representation within a dataset or subset are listed in supplementary tables that correspond to each figure that contains functional enrichment analyses.
Phomics
(http://phomics.jensenlab.org/phospho_enrichment) analysis (Munk et al., 2016) was used to determine the functional enrichment of phospho-proteomics datasets as compared to phospho-proteomics background molecules that were not differentially expressed in the datasets of interest.
STRING
The STRING database was queried to showprotein-protein network interactions between proteins of interest. Connections between DEpP subsets were identified using the “multiple proteins by names/identifiers” search tab (https://stringdb.org/cgi/input?sessionId=bI6QSAkvXKc1&input_page_active_form=multiple_identifiers).
Acknowledgments
Funded in part by NIH RO1 GM119174; RO1 DK113196; P50 AA024333; RO1 AA021890; 3U01AA026976 - 03S1; UO1 AA 026976; R56HL141744; UO1 DK061732; 5U01DK062470-17S2; R21 AR 071046. P50 AA024333; K12 HL141952 (AA) and the American College of Gastroenterology Clinical Research Award and NIH KO8 AA028794 (NW).
Author contributions
N.W., conceptualization, data curation, formal analysis, investigation, resources, software, validation, visualization, writing original draft, review and editing. S.S.S., data curation, formal analysis, visualization, investigation, writing original draft. R.M., data curation, formal analysis, software, visualization, writing original draft, review and editing. M.S.M., data curation, formal analysis, visualization, review and editing. A.B., investigation, writing original draft, review and editing.S.M., investigation, writing original draft, review and editing. A.K.C., review and editing. J.S., investigation, writing original draft, review and editing. A.H.A., investigation, writing original draft, review and editing. L.L., data curation, formal analysis, investigation, writing original draft, review and editing. B.W., investigation, writing original draft, review and editing. T.H., data curation, formal analysis, investigation, methodology, resources, supervision, visualization, writing original draft, review and editing. S.D., conceptualization, data curation, formal analysis, funding acquisition, investigation, resources, software, supervision, validation, visualization, writing original draft, review and editing.
Declaration of interests
The authors declare no competing interests.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105325.
Supplemental information
Data and code availability
-
•
The hyperammonemia phosphoproteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier ProteomeXchange: PXD031372 and https://doi.org/10.6019/PXD031372. Previously published hyperammonemia datasets for cellular RNAseq are publicly available at NCBI Sequence Read Archives database NCBI BioProject: PRJNA495054. Previously published proteomics, cellular RNA-Seq datasets, cellular ATAC-Seq dataset and human and mouse RNA-Seq datasets, are available at NCBI Gene Expression Omnibus as a superseries with accession number GSE171645 can be directly located at the following URLs:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171642.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171643.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171644.
The previously published hyperammonemia MS proteomics data are available at ProteomeXchange Consortium via the PRIDE partner repository (http://www.proteomexchange.org) with dataset identifier ProteomeXchange: PXD026955 and https://doi.org/10.6019/PXD026955. Other published data were curated from the manuscripts: Maier et al. proteomics datasets available at ProteomeXchange, accession code ProteomeXchange: PXD026461 (Maier et al., 2022), ProteomeXchange: PXD001543 (Hoffman et al., 2015), ProteomeXchange: PXD014322 (Needham et al., 2019), ProteomeXchange: PXD010452 (Nelson et al., 2019), the University of California San Diego (UCSD) MassIVE Database: MSV000086732 (Steinert et al., 2021).
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI can be found in the key resources table.
-
•
All other data are contained within the article. Any additional information needed to reanalyze the data reported within the article will be provided by the lead contact upon request.
References
- Abdelmoez A.M., Sardón Puig L., Smith J.A.B., Gabriel B.M., Savikj M., Dollet L., Chibalin A.V., Krook A., Zierath J.R., Pillon N.J. Comparative profiling of skeletal muscle models reveals heterogeneity of transcriptome and metabolism. Am. J. Physiol. Cell Physiol. 2020;318:C615–C626. doi: 10.1152/ajpcell.00540.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeva M.M., Souto G., Blanco N., Donapetry C. Ammonium metabolism in humans. Metabolism. 2012;61:1495–1511. doi: 10.1016/j.metabol.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Amar D., Lindholm M.E., Norrbom J., Wheeler M.T., Rivas M.A., Ashley E.A. Time trajectories in the transcriptomic response to exercise - a meta-analysis. Nat. Commun. 2021;12:3471. doi: 10.1038/s41467-021-23579-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antipenko A., Frías J.A., Parra J., Cadefau J.A., Cussó R. Effect of chronic electrostimulation of rabbit skeletal muscle on calmodulin level and protein kinase activity. Int. J. Biochem. Cell Biol. 1999;31:303–310. doi: 10.1016/s1357-2725(98)00112-5. [DOI] [PubMed] [Google Scholar]
- Arsenijevic T., Degraef C., Dumont J.E., Roger P.P., Pirson I. G1/S Cyclins interact with regulatory subunit of PKA via A-kinase anchoring protein. Cell Cycle. 2006;5:1217–1222. doi: 10.4161/cc.5.11.2802. [DOI] [PubMed] [Google Scholar]
- Avelar R.A., Ortega J.G., Tacutu R., Tyler E.J., Bennett D., Binetti P., Budovsky A., Chatsirisupachai K., Johnson E., Murray A., et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 2020;21:91. doi: 10.1186/s13059-020-01990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo Voltarelli V., Coronado M., Goncalves Fernandes L., Cruz Campos J., Jannig P.R., Batista Ferreira J.C., Fajardo G., Chakur Brum P., Bernstein D. Beta2-Adrenergic signaling modulates mitochondrial function and morphology in skeletal muscle in response to aerobic exercise. Cells. 2021;10 doi: 10.3390/cells10010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banister E.W., Cameron B.J. Exercise-induced hyperammonemia: peripheral and central effects. Int. J. Sports Med. 1990;11:S129–S142. doi: 10.1055/s-2007-1024864. [DOI] [PubMed] [Google Scholar]
- Bellar A., Welch N., Dasarathy S. Exercise and physical activity in cirrhosis: opportunities or perils. J. Appl. Physiol. 2020;128:1547–1567. doi: 10.1152/japplphysiol.00798.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdeaux R., Stewart R. cAMP signaling in skeletal muscle adaptation: hypertrophy, metabolism, and regeneration. Am. J. Physiol. Endocrinol. Metab. 2012;303:E1–E17. doi: 10.1152/ajpendo.00555.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono H., Hirota K. Meta-analysis of hypoxic transcriptomes from public databases. Biomedicines. 2020;8:E10. doi: 10.3390/biomedicines8010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun D.M., Chung I., Kepper N., Deeg K.I., Rippe K. TelNet - a database for human and yeast genes involved in telomere maintenance. BMC Genet. 2018;19:32. doi: 10.1186/s12863-018-0617-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelli L., Campagna R., Airoldi L., Cauli O., Llansola M., Boix J., Felipo V., Pastorelli R. Exploratory investigation on nitro- and phospho-proteome cerebellum changes in hyperammonemia and hepatic encephalopathy rat models. Metab. Brain Dis. 2012;27:37–49. doi: 10.1007/s11011-011-9268-4. [DOI] [PubMed] [Google Scholar]
- Bruno N.E., Kelly K.A., Hawkins R., Bramah-Lawani M., Amelio A.L., Nwachukwu J.C., Nettles K.W., Conkright M.D. Creb coactivators direct anabolic responses and enhance performance of skeletal muscle. EMBO J. 2014;33:1027–1043. doi: 10.1002/embj.201386145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brushia R.J., Walsh D.A. Phosphorylase kinase: the complexity of its regulation is reflected in the complexity of its structure. Front. Biosci. 1999;4:D618–D641. doi: 10.2741/brushia. [DOI] [PubMed] [Google Scholar]
- Cairns S.P., Borrani F. beta-Adrenergic modulation of skeletal muscle contraction: key role of excitation-contraction coupling. J. Physiol. 2015;593:4713–4727. doi: 10.1113/JP270909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert L.D., Steiner M.C., Morgan M.D., Singh S.J. Plasma ammonia response to incremental cycling and walking tests in COPD. Respir. Med. 2010;104:675–681. doi: 10.1016/j.rmed.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Cartee G.D., Hepple R.T., Bamman M.M., Zierath J.R. Exercise promotes healthy aging of skeletal muscle. Cell Metabol. 2016;23:1034–1047. doi: 10.1016/j.cmet.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenni V., Kojic S., Capanni C., Faulkner G., Lattanzi G. Ankrd2 in mechanotransduction and oxidative stress response in skeletal muscle: new cues for the pathogenesis of muscular laminopathies. Oxid. Med. Cell. Longev. 2019;2019:7318796. doi: 10.1155/2019/7318796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla K., Tripathi S., Thommesen L., Lægreid A., Kuiper M. TFcheckpoint: a curated compendium of specific DNA-binding RNA polymerase II transcription factors. Bioinformatics. 2013;29:2519–2520. doi: 10.1093/bioinformatics/btt432. [DOI] [PubMed] [Google Scholar]
- Chen H.W., Dunn M.A. Muscle at risk: the multiple impacts of ammonia on sarcopenia and frailty in cirrhosis. Clin. Transl. Gastroenterol. 2016;7:e170. doi: 10.1038/ctg.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.H., Hales C.N., Ozanne S.E. DNA damage, cellular senescence and organismal ageing: causal or correlative? Nucleic Acids Res. 2007;35:7417–7428. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Minegishi Y., Hasumura T., Shimotoyodome A., Ota N. Involvement of ammonia metabolism in the improvement of endurance performance by tea catechins in mice. Sci. Rep. 2020;10:6065. doi: 10.1038/s41598-020-63139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Grant C.E., Noble W.S., Bailey T.L. MoMo: discovery of statistically significant post-translational modification motifs. Bioinformatics. 2019;35:2774–2782. doi: 10.1093/bioinformatics/bty1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chheda M.G., Ashery U., Thakur P., Rettig J., Sheng Z.H. Phosphorylation of Snapin by PKA modulates its interaction with the SNARE complex. Nat. Cell Biol. 2001;3:331–338. doi: 10.1038/35070000. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinf. 2012;13(Suppl 16):S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasarathy S., Hatzoglou M. Hyperammonemia and proteostasis in cirrhosis. Curr. Opin. Clin. Nutr. Metab. Care. 2018;21:30–36. doi: 10.1097/MCO.0000000000000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasarathy S., Mookerjee R.P., Rackayova V., Rangroo Thrane V., Vairappan B., Ott P., Rose C.F. Ammonia toxicity: from head to toe? Metab. Brain Dis. 2017;32:529–538. doi: 10.1007/s11011-016-9938-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri G., Allawy A., Thapaliya S., Rennison J.H., Singh D., Kumar A., Sandlers Y., Van Wagoner D.R., Flask C.A., Hoppel C., et al. Hyperammonaemia-induced skeletal muscle mitochondrial dysfunction results in cataplerosis and oxidative stress. J. Physiol. 2016;594:7341–7360. doi: 10.1113/JP272796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri G., Giusto M., Chandel R., Welch N., Alsabbagh K., Kant S., Kumar A., Kim A., Gangadhariah M., Ghosh P.K., et al. Impaired ribosomal biogenesis by noncanonical degradation of beta-catenin during hyperammonemia. Mol. Cell Biol. 2019;39:004511-18. doi: 10.1128/MCB.00451-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri G., Krokowski D., Guan B.J., Kumar A., Thapaliya S., Singh D., Hatzoglou M., Dasarathy S. Metabolic adaptation of skeletal muscle to hyperammonemia drives the beneficial effects of l-leucine in cirrhosis. J. Hepatol. 2016;65:929–937. doi: 10.1016/j.jhep.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich R., Bachmann C., Lauterburg B.H. Exercise-induced hyperammonemia in patients with compensated chronic liver disease. Scand. J. Gastroenterol. 1990;25:329–334. doi: 10.3109/00365529009095494. [DOI] [PubMed] [Google Scholar]
- Djouder N., Tuerk R.D., Suter M., Salvioni P., Thali R.F., Scholz R., Vaahtomeri K., Auchli Y., Rechsteiner H., Brunisholz R.A., et al. PKA phosphorylates and inactivates AMPKalpha to promote efficient lipolysis. EMBO J. 2010;29:469–481. doi: 10.1038/emboj.2009.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley G.A., Staron R.S., Murray T.F., Hagerman F.C., Luginbuhl A. Muscle fiber composition and blood ammonia levels after intense exercise in humans. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 1983;54:582–586. doi: 10.1152/jappl.1983.54.2.582. [DOI] [PubMed] [Google Scholar]
- Eriksson L.S., Broberg S., Björkman O., Wahren J. Ammonia metabolism during exercise in man. Clin. Physiol. 1985;5:325–336. doi: 10.1111/j.1475-097x.1985.tb00753.x. [DOI] [PubMed] [Google Scholar]
- Etienne J., Liu C., Skinner C.M., Conboy M.J., Conboy I.M. Skeletal muscle as an experimental model of choice to study tissue aging and rejuvenation. Skeletal Muscle. 2020;10:4. doi: 10.1186/s13395-020-0222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei L., Xu H. Role of MCM2-7 protein phosphorylation in human cancer cells. Cell Biosci. 2018;8:43. doi: 10.1186/s13578-018-0242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felinski E.A., Kim J., Lu J., Quinn P.G. Recruitment of an RNA polymerase II complex is mediated by the constitutive activation domain in CREB, independently of CREB phosphorylation. Mol. Cell Biol. 2001;21:1001–1010. doi: 10.1128/MCB.21.4.1001-1010.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R., Wang L., Li Z., Yang R., Liang Y., Sun Y., Yu Q., Ghartey-Kwansah G., Sun Y., Wu Y., et al. A systematic comparison of exercise training protocols on animal models of cardiovascular capacity. Life Sci. 2019;217:128–140. doi: 10.1016/j.lfs.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippa N., Sable C.L., Filloux C., Hemmings B., Van Obberghen E. Mechanism of protein kinase B activation by cyclic AMP-dependent protein kinase. Mol. Cell Biol. 1999;19:4989–5000. doi: 10.1128/MCB.19.7.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorenza M., Lemminger A.K., Marker M., Eibye K., Iaia F.M., Bangsbo J., Hostrup M. High-intensity exercise training enhances mitochondrial oxidative phosphorylation efficiency in a temperature-dependent manner in human skeletal muscle: implications for exercise performance. Faseb. J. 2019;33:8976–8989. doi: 10.1096/fj.201900106RRR. [DOI] [PubMed] [Google Scholar]
- Fukuda N., Wu Y., Nair P., Granzier H.L. Phosphorylation of titin modulates passive stiffness of cardiac muscle in a titin isoform-dependent manner. J. Gen. Physiol. 2005;125:257–271. doi: 10.1085/jgp.200409177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganda O.P., Ruderman N.B. Muscle nitrogen metabolism in chronic hepatic insufficiency. Metabolism. 1976;25:427–435. doi: 10.1016/0026-0495(76)90075-5. [DOI] [PubMed] [Google Scholar]
- Gnimassou O., Francaux M., Deldicque L. Hippo pathway and skeletal muscle mass regulation in mammals: a controversial relationship. Front. Physiol. 2017;8:190. doi: 10.3389/fphys.2017.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B.L., Rodal A.A., Barnes G., Drubin D.G. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J. Cell Biol. 2001;153:627–634. doi: 10.1083/jcb.153.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görg B., Karababa A., Shafigullina A., Bidmon H.J., Häussinger D. Ammonia-induced senescence in cultured rat astrocytes and in human cerebral cortex in hepatic encephalopathy. Glia. 2015;63:37–50. doi: 10.1002/glia.22731. [DOI] [PubMed] [Google Scholar]
- Gorostiaga E.M., Navarro-Amézqueta I., Calbet J.A.L., Sánchez-Medina L., Cusso R., Guerrero M., Granados C., González-Izal M., Ibáñez J., Izquierdo M. Blood ammonia and lactate as markers of muscle metabolites during leg press exercise. J. Strength Condit Res. 2014;28:2775–2785. doi: 10.1519/JSC.0000000000000496. [DOI] [PubMed] [Google Scholar]
- Graham T., Bangsbo J., Saltin B. Skeletal muscle ammonia production and repeated, intense exercise in humans. Can. J. Physiol. Pharmacol. 1993;71:484–490. doi: 10.1139/y93-070. [DOI] [PubMed] [Google Scholar]
- Graham T.E., Bangsbo J., Gollnick P.D., Juel C., Saltin B. Ammonia metabolism during intense dynamic exercise and recovery in humans. Am. J. Physiol. 1990;259:E170–E176. doi: 10.1152/ajpendo.1990.259.2.E170. [DOI] [PubMed] [Google Scholar]
- Graham T.E., MacLean D.A. Ammonia and amino acid metabolism in skeletal muscle: human, rodent and canine models. Med. Sci. Sports Exerc. 1998;30:34–46. doi: 10.1097/00005768-199801000-00006. [DOI] [PubMed] [Google Scholar]
- Graham T.E., Pedersen P.K., Saltin B. Muscle and blood ammonia and lactate responses to prolonged exercise with hyperoxia. J. Appl. Physiol. 1987;63:1457–1462. doi: 10.1152/jappl.1987.63.4.1457. [DOI] [PubMed] [Google Scholar]
- Graham T.E., Turcotte L.P., Kiens B., Richter E.A. Training and muscle ammonia and amino acid metabolism in humans during prolonged exercise. J. Appl. Physiol. 1995;78:725–735. doi: 10.1152/jappl.1995.78.2.725. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley J.A. Molecular responses to strength and endurance training: are they incompatible? Appl. Physiol. Nutr. Metabol. 2009;34:355–361. doi: 10.1139/H09-023. [DOI] [PubMed] [Google Scholar]
- Hawley J.A., Lundby C., Cotter J.D., Burke L.M. Maximizing cellular adaptation to endurance exercise in skeletal muscle. Cell Metabol. 2018;27:962–976. doi: 10.1016/j.cmet.2018.04.014. [DOI] [PubMed] [Google Scholar]
- Hoffman N.J., Parker B.L., Chaudhuri R., Fisher-Wellman K.H., Kleinert M., Humphrey S.J., Yang P., Holliday M., Trefely S., Fazakerley D.J., et al. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metabol. 2015;22:922–935. doi: 10.1016/j.cmet.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover F., Mathiesen I., Skålhegg B.S., Lømo T., Taskén K. Differential expression and regulation of the PKA signalling pathway in fast and slow skeletal muscle. Anat. Embryol. 2001;203:193–201. doi: 10.1007/s004290000155. [DOI] [PubMed] [Google Scholar]
- Horn H., Schoof E.M., Kim J., Robin X., Miller M.L., Diella F., Palma A., Cesareni G., Jensen L.J., Linding R. KinomeXplorer: an integrated platform for kinome biology studies. Nat. Methods. 2014;11:603–604. doi: 10.1038/nmeth.2968. [DOI] [PubMed] [Google Scholar]
- Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostrup M., Reitelseder S., Jessen S., Kalsen A., Nyberg M., Egelund J., Kreiberg M., Kristensen C.M., Thomassen M., Pilegaard H., et al. Beta2 -adrenoceptor agonist salbutamol increases protein turnover rates and alters signalling in skeletal muscle after resistance exercise in young men. J. Physiol. 2018;596:4121–4139. doi: 10.1113/JP275560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang H., Arighi C.N., Ross K.E., Ren J., Li G., Chen S.C., Wang Q., Cowart J., Vijay-Shanker K., Wu C.H. iPTMnet: an integrated resource for protein post-translational modification network discovery. Nucleic Acids Res. 2018;46:D542–D550. doi: 10.1093/nar/gkx1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.S.C., Fraenkel E. Integrating proteomic, transcriptional, and interactome data reveals hidden components of signaling and regulatory networks. Sci. Signal. 2009;2:ra40. doi: 10.1126/scisignal.2000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S.K. The relevance of sex differences in performance fatigability. Med. Sci. Sports Exerc. 2016;48:2247–2256. doi: 10.1249/MSS.0000000000000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal S., Ali W., Nagpal P., Grover A., Grover S. Predicting phosphorylation sites using machine learning by integrating the sequence, structure, and functional information of proteins. J. Transl. Med. 2021;19:218. doi: 10.1186/s12967-021-02851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen S., Eibye K., Christensen P.M., Hostrup M., Bangsbo J. No additive effect of acetaminophen when co-ingested with caffeine on cycling performance in well-trained young men. J. Appl. Physiol. 2021;131:238–249. doi: 10.1152/japplphysiol.00108.2021. [DOI] [PubMed] [Google Scholar]
- Jo D., Kim B.C., Cho K.A., Song J. The cerebral effect of ammonia in brain aging: blood-brain barrier breakdown, mitochondrial dysfunction, and neuroinflammation. J. Clin. Med. 2021;10:2773. doi: 10.3390/jcm10132773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S., Davuluri G., Alchirazi K.A., Welch N., Heit C., Kumar A., Gangadhariah M., Kim A., McMullen M.R., Willard B., et al. Ethanol sensitizes skeletal muscle to ammonia-induced molecular perturbations. J. Biol. Chem. 2019;294:7231–7244. doi: 10.1074/jbc.RA118.005411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A., Broberg S., Sahlin K., Wahren J. Muscle ammonia and amino acid metabolism during dynamic exercise in man. Clin. Physiol. 1986;6:365–379. doi: 10.1111/j.1475-097x.1986.tb00242.x. [DOI] [PubMed] [Google Scholar]
- Katz A., Sahlin K., Henriksson J. Muscle ammonia metabolism during isometric contraction in humans. Am. J. Physiol. 1986;250:C834–C840. doi: 10.1152/ajpcell.1986.250.6.C834. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Cho J.H., Kim J.R. Downregulation of Polo-like kinase 1 induces cellular senescence in human primary cells through a p53-dependent pathway. J. Gerontol. A Biol. Sci. Med. Sci. 2013;68:1145–1156. doi: 10.1093/gerona/glt017. [DOI] [PubMed] [Google Scholar]
- Knight J.D., Kothary R. The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis. Skeletal Muscle. 2011;1:29. doi: 10.1186/2044-5040-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani S., Tugendreich S., Fujii M., Jorgensen P.M., Watanabe N., Hoog C., Hieter P., Todokoro K. PKA and MPF-activated polo-like kinase regulate anaphase-promoting complex activity and mitosis progression. Mol. Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Davuluri G., Silva R.N.E., Engelen M.P.K.J., Ten Have G.A.M., Prayson R., Deutz N.E.P., Dasarathy S. Ammonia lowering reverses sarcopenia of cirrhosis by restoring skeletal muscle proteostasis. Hepatology. 2017;65:2045–2058. doi: 10.1002/hep.29107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Welch N., Mishra S., Bellar A., Silva R.N., Li L., Singh S.S., Sharkoff M., Kerr A., Chelluboyina A.K., et al. Metabolic reprogramming during hyperammonemia targets mitochondrial function and post-mitotic senescence. JCI Insights. 2021;6 doi: 10.1172/jci.insight.154089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Gouw M., Michael S., Sámano-Sánchez H., Pancsa R., Glavina J., Diakogianni A., Valverde J.A., Bukirova D., Čalyševa J., et al. ELM-the eukaryotic linear motif resource in 2020. Nucleic Acids Res. 2020;48:D296–D306. doi: 10.1093/nar/gkz1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.C.H., Jones A.R., Hubbard S.J. Computational phosphoproteomics: from identification to localization. Proteomics. 2015;15:950–963. doi: 10.1002/pmic.201400372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Jang C., Lee K.A. Polo-like kinases (plks), a key regulator of cell cycle and new potential target for cancer therapy. Dev. Reprod. 2014;18:65–71. doi: 10.12717/DR.2014.18.1.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.Y., Huang H.D., Hung J.H., Huang H.Y., Yang Y.S., Wang T.H. dbPTM: an information repository of protein post-translational modification. Nucleic Acids Res. 2006;34:D622–D627. doi: 10.1093/nar/gkj083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A., Gehlenborg N., Strobelt H., Vuillemot R., Pfister H. UpSet: visualization of intersecting sets. IEEE Trans. Visual Comput. Graph. 2014;20:1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Randall W.R., Du S.J. skNAC (skeletal Naca), a muscle-specific isoform of Naca (nascent polypeptide-associated complex alpha), is required for myofibril organization. Faseb. J. 2009;23:1988–2000. doi: 10.1096/fj.08-125542. [DOI] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood A.H., McDonald J.M., Reiman R.E., Gelbard A.S., Laughlin J.S., Duffy T.E., Plum F. The dynamics of ammonia metabolism in man.Effects of liver disease and hyperammonemia. J. Clin. Invest. 1979;63:449–460. doi: 10.1172/JCI109322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.S., Chen J., Lange S. Probing muscle ankyrin-repeat protein (MARP) structure and function. Anat. Rec. 2014;297:1615–1629. doi: 10.1002/ar.22968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean D.A., Spriet L.L., Hultman E., Graham T.E. Plasma and muscle amino acid and ammonia responses during prolonged exercise in humans. J. Appl. Physiol. 1991;70:2095–2103. doi: 10.1152/jappl.1991.70.5.2095. [DOI] [PubMed] [Google Scholar]
- Maier G., Delezie J., Westermark P.O., Santos G., Ritz D., Handschin C. Transcriptomic, proteomic and phosphoproteomic underpinnings of daily exercise performance and zeitgeber activity of training in mouse muscle. J. Physiol. 2022;600:769–796. doi: 10.1113/JP281535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich A.V., Sirotkin A.V., Rafay J. Comparison of effects of protein kinase A, mitogen-activated protein kinase, and cyclin-dependent kinase blockers on rabbit ovarian granulosa cell functions. Horm. Metab. Res. 2010;42:936–943. doi: 10.1055/s-0030-1267226. [DOI] [PubMed] [Google Scholar]
- Mangul S., Martin L.S., Hill B.L., Lam A.K.M., Distler M.G., Zelikovsky A., Eskin E., Flint J. Systematic benchmarking of omics computational tools. Nat. Commun. 2019;10:1393. doi: 10.1038/s41467-019-09406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B.T., Strebhardt K. Polo-like kinase 1: target and regulator of transcriptional control. Cell Cycle. 2006;5:2881–2885. doi: 10.4161/cc.5.24.3538. [DOI] [PubMed] [Google Scholar]
- McDaniel J., Davuluri G., Hill E.A., Moyer M., Runkana A., Prayson R., van Lunteren E., Dasarathy S. Hyperammonemia results in reduced muscle function independent of muscle mass. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;310:G163–G170. doi: 10.1152/ajpgi.00322.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee S.L., Hargreaves M. Exercise adaptations: molecular mechanisms and potential targets for therapeutic benefit. Nat. Rev. Endocrinol. 2020;16:495–505. doi: 10.1038/s41574-020-0377-1. [DOI] [PubMed] [Google Scholar]
- McMahon D.K., Anderson P.A., Nassar R., Bunting J.B., Saba Z., Oakeley A.E., Malouf N.N. C2C12 cells: biophysical, biochemical, and immunocytochemical properties. Am. J. Physiol. 1994;266:C1795–C1802. doi: 10.1152/ajpcell.1994.266.6.C1795. [DOI] [PubMed] [Google Scholar]
- Medeiros W.M., Carvalho A.C., Peres P., De Luca F.A., Gun C. The dysfunction of ammonia in heart failure increases with an increase in the intensity of resistance exercise, even with the use of appropriate drug therapy. Eur. J. Prev. Cardiol. 2014;21:135–144. doi: 10.1177/2047487312460520. [DOI] [PubMed] [Google Scholar]
- Meng Z., Moroishi T., Guan K.L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz K.S., Deoudes E.M., Berginski M.E., Jimenez-Ruiz I., Aksoy B.A., Hammerbacher J., Gomez S.M., Phanstiel D.H. Coral: clear and customizable visualization of human kinome data. Cell Syst. 2018;7:347–350.e1. doi: 10.1016/j.cels.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukund K., Subramaniam S. Skeletal muscle: a review of molecular structure and function, in health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020;12:e1462. doi: 10.1002/wsbm.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk S., Refsgaard J.C., Olsen J.V., Jensen L.J. From phosphosites to kinases. Methods Mol. Biol. 2016;1355:307–321. doi: 10.1007/978-1-4939-3049-4_21. [DOI] [PubMed] [Google Scholar]
- Munno D.W., Prince D.J., Syed N.I. Synapse number and synaptic efficacy are regulated by presynaptic cAMP and protein kinase A. J. Neurosci. 2003;23:4146–4155. doi: 10.1523/JNEUROSCI.23-10-04146.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutch B.J., Banister E.W. Ammonia metabolism in exercise and fatigue: a review. Med. Sci. Sports Exerc. 1983;15:41–50. [PubMed] [Google Scholar]
- Nedachi T., Fujita H., Kanzaki M. Contractile C2C12 myotube model for studying exercise-inducible responses in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1191–E1204. doi: 10.1152/ajpendo.90280.2008. [DOI] [PubMed] [Google Scholar]
- Needham E.J., Hingst J.R., Parker B.L., Morrison K.R., Yang G., Onslev J., Kristensen J.M., Højlund K., Ling N.X.Y., Oakhill J.S., et al. Personalized phosphoproteomics identifies functional signaling. Nat. Biotechnol. 2022;40:576–584. doi: 10.1038/s41587-021-01099-9. [DOI] [PubMed] [Google Scholar]
- Needham E.J., Humphrey S.J., Cooke K.C., Fazakerley D.J., Duan X., Parker B.L., James D.E. Phosphoproteomics of acute cell stressors targeting exercise signaling networks reveal drug interactions regulating protein secretion. Cell Rep. 2019;29:1524–1538.e6. doi: 10.1016/j.celrep.2019.10.001. [DOI] [PubMed] [Google Scholar]
- Nelson M.E., Parker B.L., Burchfield J.G., Hoffman N.J., Needham E.J., Cooke K.C., Naim T., Sylow L., Ling N.X., Francis D., et al. Phosphoproteomics reveals conserved exercise-stimulated signaling and AMPK regulation of store-operated calcium entry. EMBO J. 2019;38:e102578. doi: 10.15252/embj.2019102578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsburn B.C. Proteome discoverer-A community enhanced data processing suite for protein informatics. Proteomes. 2021;9:15. doi: 10.3390/proteomes9010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.H., Moon Y., Shin C.M., Chung J.H. Cyclic AMP suppresses matrix metalloproteinase-1 expression through inhibition of MAPK and GSK-3beta. J. Invest. Dermatol. 2010;130:2049–2056. doi: 10.1038/jid.2010.62. [DOI] [PubMed] [Google Scholar]
- Park C.Y., Pierce S.A., von Drehle M., Ivey K.N., Morgan J.A., Blau H.M., Srivastava D. skNAC, a Smyd1-interacting transcription factor, is involved in cardiac development and skeletal muscle growth and regeneration. Proc. Natl. Acad. Sci. USA. 2010;107:20750–20755. doi: 10.1073/pnas.1013493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon N.J., Gabriel B.M., Dollet L., Smith J.A.B., Sardón Puig L., Botella J., Bishop D.J., Krook A., Zierath J.R. Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat. Commun. 2020;11:470. doi: 10.1038/s41467-019-13869-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe S.W., Meng Z., Lin K.C., Lin B., Hong A.W., Chun J.V., Guan K.L. Characterization of hippo pathway components by gene inactivation. Mol. Cell. 2016;64:993–1008. doi: 10.1016/j.molcel.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Thapaliya S., Runkana A., Yang Y., Tsien C., Mohan M.L., Narayanan A., Eghtesad B., Mozdziak P.E., McDonald C., et al. Hyperammonemia in cirrhosis induces transcriptional regulation of myostatin by an NF-kappaB-mediated mechanism. Proc. Natl. Acad. Sci. USA. 2013;110:18162–18167. doi: 10.1073/pnas.1317049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Tsien C., Thapalaya S., Narayanan A., Weihl C.C., Ching J.K., Eghtesad B., Singh K., Fu X., Dubyak G., et al. Hyperammonemia-mediated autophagy in skeletal muscle contributes to sarcopenia of cirrhosis. Am. J. Physiol. Endocrinol. Metab. 2012;303:E983–E993. doi: 10.1152/ajpendo.00183.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazi S., Zahiri J. Database; 2021. Posttranslational Modifications in Proteins: Resources, Tools and Prediction Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan P.N., Umesono K., Evans R.M. Modulation of glucocorticoid receptor function by protein kinase A. Mol. Endocrinol. 1992;6:1451–1457. doi: 10.1210/mend.6.9.1435789. [DOI] [PubMed] [Google Scholar]
- Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T.J., Goodman R.P., Grabarek Z., Haas M.E., Hung W.H.W., et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49:D1541–D1547. doi: 10.1093/nar/gkaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario F.J., Powell T.L., Gupta M.B., Cox L., Jansson T. mTORC1 transcriptional regulation of ribosome subunits, protein synthesis, and molecular transport in primary human trophoblast cells. Front. Cell Dev. Biol. 2020;8:583801. doi: 10.3389/fcell.2020.583801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf R., Khan M.M., Lustrino D., Labeit S., Kettelhut I.C., Navegantes L.C.C. Alterations of cAMP-dependent signaling in dystrophic skeletal muscle. Front. Physiol. 2013;4:290. doi: 10.3389/fphys.2013.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M., Dillon C., Keith W.N., Narita M., Young A.R.J., Cairney C.J., Godwin L.S., Torrance C.J., Bennett D.C., Narita M., Narita M. Cell-based screen for altered nuclear phenotypes reveals senescence progression in polyploid cells after Aurora kinase B inhibition. Mol. Biol. Cell. 2015;26:2971–2985. doi: 10.1091/mbc.E15-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Chikenji T.S. Diverse roles of cellular senescence in skeletal muscle inflammation, regeneration, and therapeutics. Front. Pharmacol. 2021;12:739510. doi: 10.3389/fphar.2021.739510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Chikenji T.S., Matsumura T., Nakano M., Fujimiya M. Exercise enhances skeletal muscle regeneration by promoting senescence in fibro-adipogenic progenitors. Nat. Commun. 2020;11:889. doi: 10.1038/s41467-020-14734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage S.R., Zhang B. Using phosphoproteomics data to understand cellular signaling: a comprehensive guide to bioinformatics resources. Clin. Proteomics. 2020;17:27. doi: 10.1186/s12014-020-09290-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangraw R.E., Jahoor F. Effect of liver disease and transplantation on urea synthesis in humans: relationship to acid-base status. Am. J. Physiol. 1999;276:G1145–G1152. doi: 10.1152/ajpgi.1999.276.5.G1145. [DOI] [PubMed] [Google Scholar]
- Shen M., Stukenberg P.T., Kirschner M.W., Lu K.P. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 1998;12:706–720. doi: 10.1101/gad.12.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva L.A., Pinho C.A., Scarabelot K.S., Fraga D.B., Volpato A.M.J., Boeck C.R., De Souza C.T., Streck E.L., Pinho R.A. Physical exercise increases mitochondrial function and reduces oxidative damage in skeletal muscle. Eur. J. Appl. Physiol. 2009;105:861–867. doi: 10.1007/s00421-008-0971-8. [DOI] [PubMed] [Google Scholar]
- Srisawat K., Shepherd S.O., Lisboa P.J., Burniston J.G. A systematic review and meta-analysis of proteomics literature on the response of human skeletal muscle to obesity/type 2 diabetes mellitus (T2DM) versus exercise training. Proteomes. 2017;5:E30. doi: 10.3390/proteomes5040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert N.D., Potts G.K., Wilson G.M., Klamen A.M., Lin K.H., Hermanson J.B., McNally R.M., Coon J.J., Hornberger T.A. Mapping of the contraction-induced phosphoproteome identifies TRIM28 as a significant regulator of skeletal muscle size and function. Cell Rep. 2021;34:108796. doi: 10.1016/j.celrep.2021.108796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu V.T., Kim H.K., Han J. Acute and chronic exercise in animal models. Adv. Exp. Med. Biol. 2017;999:55–71. doi: 10.1007/978-981-10-4307-9_4. [DOI] [PubMed] [Google Scholar]
- Timson B.F. Evaluation of animal models for the study of exercise-induced muscle enlargement. J. Appl. Physiol. 1990;69:1935–1945. doi: 10.1152/jappl.1990.69.6.1935. [DOI] [PubMed] [Google Scholar]
- Toko H., Hariharan N., Konstandin M.H., Ormachea L., McGregor M., Gude N.A., Sundararaman B., Joyo E., Joyo A.Y., Collins B., et al. Differential regulation of cellular senescence and differentiation by prolyl isomerase Pin1 in cardiac progenitor cells. J. Biol. Chem. 2014;289:5348–5356. doi: 10.1074/jbc.M113.526442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero A., Alroy G., Eisenkraft B., Itskovitch J. Ammonia metabolism in chronic obstructive pulmonary disease with special reference to congestive right ventricular failure. Thorax. 1974;29:703–709. doi: 10.1136/thx.29.6.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk K.J., Friso G., Walther D., Schulze W.X. Meta-analysis of Arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell. 2014;26:2367–2389. doi: 10.1105/tpc.114.125815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli M., Morgan G.R., Donato A.J., Reese V., Bottura R., Tarperi C., Milanese C., Schena F., Reggiani C., Naro F., et al. Cellular aging of skeletal muscle: telomeric and free radical evidence that physical inactivity is responsible and not age. Clin. Sci. 2014;127:415–421. doi: 10.1042/CS20140051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J.A., Sun Z., Farrah T., Bandeira N., et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltereit R., Weller M. Signaling from cAMP/PKA to MAPK and synaptic plasticity. Mol. Neurobiol. 2003;27:99–106. doi: 10.1385/MN:27:1:99. [DOI] [PubMed] [Google Scholar]
- Wan M., Gray-Gaillard E.F., Elisseeff J.H. Cellular senescence in musculoskeletal homeostasis, diseases, and regeneration. Bone Res. 2021;9:41. doi: 10.1038/s41413-021-00164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt K.I., Goodman C.A., Hornberger T.A., Gregorevic P. The hippo signaling pathway in the regulation of skeletal muscle mass and function. Exerc. Sport Sci. Rev. 2018;46:92–96. doi: 10.1249/JES.0000000000000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B., Dui W., Liu D., Xing Y., Yuan Z., Ji G. MST1, a key player, in enhancing fast skeletal muscle atrophy. BMC Biol. 2013;11:12. doi: 10.1186/1741-7007-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Simko V. R Package 'corrplot': visualization of a correlation matrix. 2021. https://github.com/taiyun/corrplot Version 0.92.
- Welch N., Singh S.S., Kumar A., Dhruba S.R., Mishra S., Sekar J., Bellar A., Attaway A.H., Chelluboyina A., Willard B.B., et al. Integrated multiomics analysis identifies molecular landscape perturbations during hyperammonemia in skeletal muscle and myotubes. J. Biol. Chem. 2021;297:101023. doi: 10.1016/j.jbc.2021.101023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Yin H., Han S., Cui C., Wang Y., Li D., Zhu Q. Plectin regulates Wnt signaling mediated-skeletal muscle development by interacting with Dishevelled-2 and antagonizing autophagy. Gene. 2021;783:145562. doi: 10.1016/j.gene.2021.145562. [DOI] [PubMed] [Google Scholar]
- Zhang J., Wang Y., Liu X., Dagda R.K., Zhang Y. How AMPK and PKA interplay to regulate mitochondrial function and survival in models of ischemia and diabetes. Oxid. Med. Cell. Longev. 2017;2017:4353510. doi: 10.1155/2017/4353510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Chen L., Qu H. CSGene: a literature-based database for cell senescence genes and its application to identify critical cell aging pathways and associated diseases. Cell Death Dis. 2016;7:e2053. doi: 10.1038/cddis.2015.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.K., Zhao J.H., Yan Y., Lian T.Y., Ye J., Wang X.J., Wang Z., Jing Z.C., He Y.Y., Yang P. Metabolic reprogramming of the urea cycle pathway in experimental pulmonary arterial hypertension rats induced by monocrotaline. Respir. Res. 2018;19:94. doi: 10.1186/s12931-018-0800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang M., Zhou Z., Wang W., Duan J., Wu G. Expression and prognostic value of MCM family genes in osteosarcoma. Front. Mol. Biosci. 2021;8:668402. doi: 10.3389/fmolb.2021.668402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The hyperammonemia phosphoproteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier ProteomeXchange: PXD031372 and https://doi.org/10.6019/PXD031372. Previously published hyperammonemia datasets for cellular RNAseq are publicly available at NCBI Sequence Read Archives database NCBI BioProject: PRJNA495054. Previously published proteomics, cellular RNA-Seq datasets, cellular ATAC-Seq dataset and human and mouse RNA-Seq datasets, are available at NCBI Gene Expression Omnibus as a superseries with accession number GSE171645 can be directly located at the following URLs:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171642.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171643.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171644.
The previously published hyperammonemia MS proteomics data are available at ProteomeXchange Consortium via the PRIDE partner repository (http://www.proteomexchange.org) with dataset identifier ProteomeXchange: PXD026955 and https://doi.org/10.6019/PXD026955. Other published data were curated from the manuscripts: Maier et al. proteomics datasets available at ProteomeXchange, accession code ProteomeXchange: PXD026461 (Maier et al., 2022), ProteomeXchange: PXD001543 (Hoffman et al., 2015), ProteomeXchange: PXD014322 (Needham et al., 2019), ProteomeXchange: PXD010452 (Nelson et al., 2019), the University of California San Diego (UCSD) MassIVE Database: MSV000086732 (Steinert et al., 2021).
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI can be found in the key resources table.
-
•
All other data are contained within the article. Any additional information needed to reanalyze the data reported within the article will be provided by the lead contact upon request.