Significance
Cell-free DNA (cfDNA) is nonrandomly fragmented and contains a wealth of molecular information useful for noninvasive prenatal testing and cancer detection. cfDNA fragmentomics contains information beyond genetics, such as gene expression inference. However, the feasibility of using cfDNA fragmentomics for deducing cfDNA methylomics remains unexplored. This study demonstrated the possibility of using cfDNA fragmentation patterns to deduce the methylation patterns of cfDNA molecules, breaking free from the limitation of bisulfite sequencing. By using cfDNA cleavage profiles surrounding a cytosine-phosphate-guanine (CpG) site, we determined the methylation status ranging from a particular region down to a single CpG assisted by a deep learning algorithm. Both genetic and epigenetic information of cfDNA can therefore be obtained in a single nondestructive assay.
Keywords: epigenetics, fragmentomics, cancer detection, noninvasive prenatal testing, liquid biopsy
Abstract
Cell-free DNA (cfDNA) fragmentation patterns contain important molecular information linked to tissues of origin. We explored the possibility of using fragmentation patterns to predict cytosine-phosphate-guanine (CpG) methylation of cfDNA, obviating the use of bisulfite treatment and associated risks of DNA degradation. This study investigated the cfDNA cleavage profile surrounding a CpG (i.e., within an 11-nucleotide [nt] window) to analyze cfDNA methylation. The cfDNA cleavage proportion across positions within the window appeared nonrandom and exhibited correlation with methylation status. The mean cleavage proportion was ∼twofold higher at the cytosine of methylated CpGs than unmethylated ones in healthy controls. In contrast, the mean cleavage proportion rapidly decreased at the 1-nt position immediately preceding methylated CpGs. Such differential cleavages resulted in a characteristic change in relative presentations of CGN and NCG motifs at 5′ ends, where N represented any nucleotide. CGN/NCG motif ratios were correlated with methylation levels at tissue-specific methylated CpGs (e.g., placenta or liver) (Pearson’s absolute r > 0.86). cfDNA cleavage profiles were thus informative for cfDNA methylation and tissue-of-origin analyses. Using CG-containing end motifs, we achieved an area under a receiver operating characteristic curve (AUC) of 0.98 in differentiating patients with and without hepatocellular carcinoma and enhanced the positive predictive value of nasopharyngeal carcinoma screening (from 19.6 to 26.8%). Furthermore, we elucidated the feasibility of using cfDNA cleavage patterns to deduce CpG methylation at single CpG resolution using a deep learning algorithm and achieved an AUC of 0.93. FRAGmentomics-based Methylation Analysis (FRAGMA) presents many possibilities for noninvasive prenatal, cancer, and organ transplantation assessment.
Fragmentation patterns of cell-free DNA (cfDNA) molecules contain a wealth of molecular information related to their tissues of origin (1). For instance, compared with the background DNA molecules that are mainly derived from the hematopoietic system (2, 3), size shortening of fetal and tumoral DNA molecules occurs in the plasma DNA of pregnant women and cancer patients, respectively (4–6). In addition, a series of 10-bp periodicities were present in fetal and tumoral DNA molecules below 146 bp, with a relative reduction in the major peak at 166 bp (1). Such characteristic size profiles suggest that the fragmentation of cfDNA may be associated with nucleosome structures (5, 7). Many important characteristics pertaining to cfDNA fragmentation have been unveiled recently, such as nucleosome footprints (8, 9), fragment end motifs (10), preferred ends (7, 11), and jagged ends (12), which are examples of fragmentomic markers (1).
cfDNA fragmentomics is an emergent and actively pursued area, with wide-ranging biological and clinical implications. It has been reported that the use of fragmentation patterns of cfDNA could inform the expression status of genes (13, 14). Using mouse models, DNA nucleases (e.g., DNASE1L3) were found to play important roles in the generation of plasma DNA molecules (15, 16). Fragmentomic features, such as cfDNA end motifs and jagged ends, were further demonstrated to be useful for monitoring DNA nuclease activities, providing biomarkers for autoimmune diseases (e.g., systemic lupus erythematosus) (17, 18). In addition, the deficiencies of nuclease activities in a mouse model resulted in altered DNA methylation profiles of plasma DNA molecules (19). However, how cfDNA fragmentation patterns interplay with DNA methylation in human individuals under different pathophysiological conditions, such as pregnancy and oncogenesis, and in healthy patients without nuclease deficiency, is unknown. It is also not known whether fragmentomic features can be used to deduce cfDNA methylation status.
A widely employed way to assess DNA methylation is through bisulfite sequencing (20). A key limitation of this approach is the severe degradation of DNA molecules caused by the bisulfite treatment (21), which greatly increases the sampling variation when analyzing rare target molecules (e.g., tumoral cfDNA at early stages of cancer). Many efforts have been made toward overcoming this issue. For example, Vaisvila et al. developed enzymatic methyl sequencing for which DNA molecules were treated using tet methylcytosine dioxygenase 2 and T4 phage β-glucosyltransferase, followed by the apolipoprotein B mRNA editing enzyme catalytic subunit 3A (APOBEC3A) treatment. Cytosine conversion based on enzymatic processes was reported to be much less destructive (22). Recently, researchers developed approaches making use of third-generation sequencing technologies such as single-molecule real-time sequencing (Pacific Biosciences) (23) and nanopore sequencing (24) to analyze cytosine-phosphate-guanine (CpG) methylation patterns in native DNA molecules, theoretically overcoming the above-mentioned limitation. However, compared with second-generation sequencing (also called next-generation sequencing [NGS]) technologies, the throughput of third-generation sequencing technologies is generally lower and the sequencing cost per nucleotide (nt) is much higher, thus restricting its immediate application in clinical settings. Here, we explore the feasibility of enabling the assessment of DNA methylation using fragmentomic characteristics of cfDNA molecules deduced from NGS results without the use of bisulfite or enzymatic treatment. If successful, such an approach could leverage the high throughput of NGS while obviating the use of chemical/enzymatic conversion and could potentially be readily integrated into currently used NGS-based platforms for cfDNA analysis.
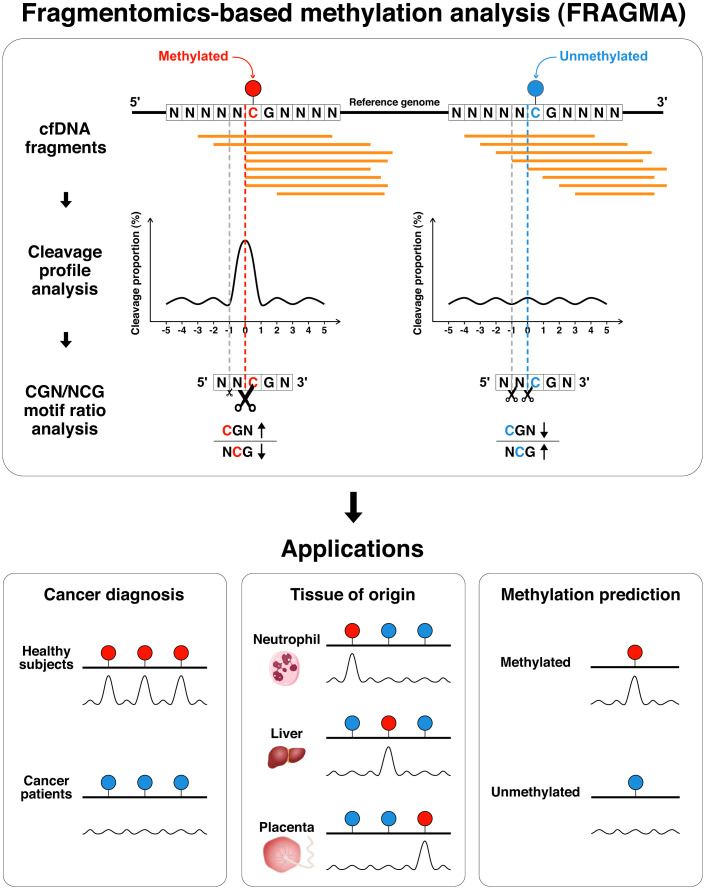
In this study, we utilize the fragmentation patterns proximal to a CpG site for deducing its methylation status. The fragmentation pattern is depicted by the frequency of cfDNA fragment ends at each position within a certain nt range relative to a CpG of interest, termed a cleavage profile (Fig. 1). Such a cleavage profile varies according to the methylation status of the CpG site of interest, providing the basis for methylation analysis by using fragmentomic features. We further correlated two types of end motifs (CGN and NCG; N represents any nucleotide of A, C, G, or T) resulting from differential cutting in the measurement window related to DNA methylation, attempting to construct a simplified approach for methylation analysis. Modeling CpG methylation using cfDNA fragmentation may facilitate noninvasive prenatal testing, cancer detection, and tissue-of-origin analysis (Fig. 1). Furthermore, we explore the feasibility of using deep learning to deduce the methylation status at single CpG resolution through the cleavage profile (Fig. 1). We refer to this FRAGmentomics-based Methylation Analysis as FRAGMA in this study.
Fig. 1.
Schematic for FRAGMA of cfDNA molecules. cfDNA molecules were sequenced by massively parallel sequencing and aligned to the human reference genome. The cleavage proportion within an 11-nt window (the cleavage measurement window) was used to measure the cutting preference of cfDNA molecules. The patterns of cleavage proportion within a window (the cleavage profile) depended on the methylation status of one or more CpG sites associated with that window. For example, a methylated CpG site might confer a higher probability of cfDNA cutting at the cytosine in the CpG context, but an unmethylated site might not. Such methylation-dependent differential fragmentation within a cleavage measurement window resulted in the change in CGN/NCG motif ratio. Thus, the CGN/NCG motif ratio provided a simplified version for reflecting CpG methylation, allowing cfDNA tissue-of-origin analysis of cfDNA and cancer detection. Furthermore, the great number of cleavage profiles derived from cfDNA molecules might provide an opportunity to train a deep learning model for methylation prediction at the single CpG resolution.
Results
DNA Methylation Directly Affects the Cleavage Profile of cfDNA.
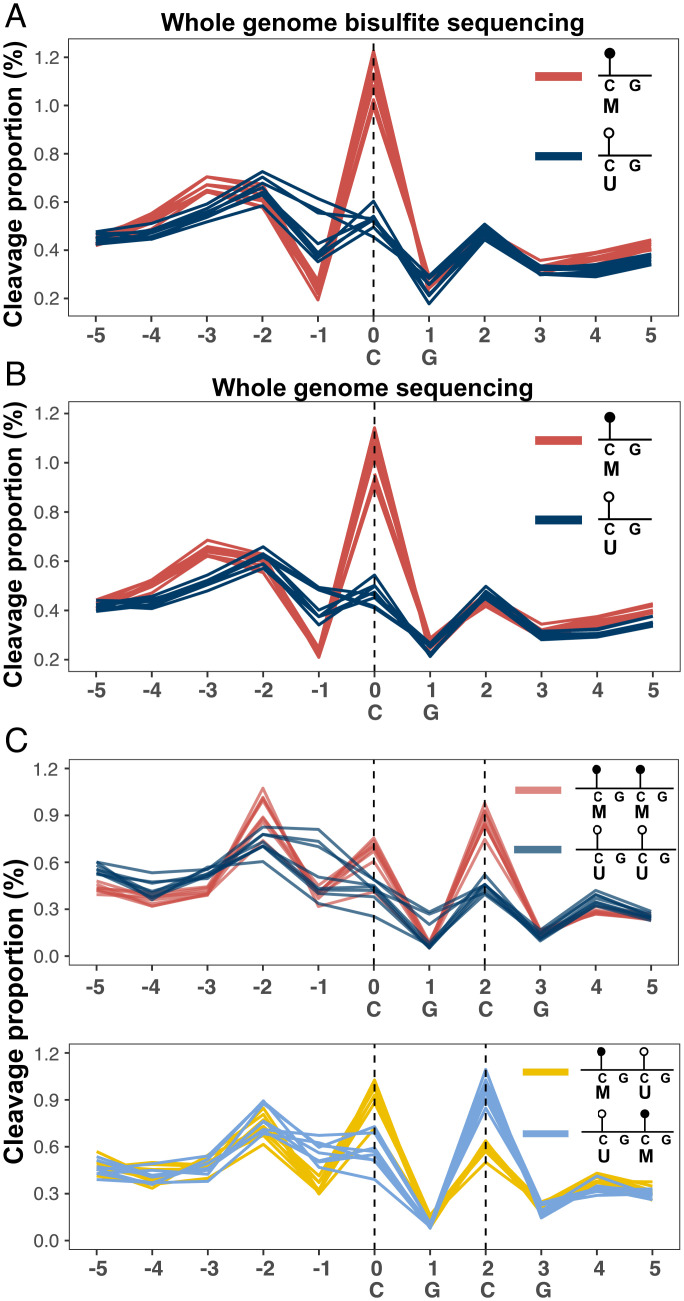
To investigate the relationship between DNA methylation and cfDNA fragmentation, we examined the cleavage profile related to hypermethylated and hypomethylated CpG sites based on bisulfite sequencing results of plasma DNA from eight healthy controls in a previous study (10). The hypermethylated and hypomethylated CpG sites were defined as CpG sites with a methylation index (i.e., percentage of methylated cytosines) > 70% and < 30%, respectively. The cleavage profile was constructed according to the cleavage proportion (i.e., the percentage of fragment ends with respect to the sequencing depth [%]) across each nt within an 11-nt genomic window centered on a CpG site. For simplicity, we termed such an 11-nt genomic window as a cleavage measurement window. The mean cleavage proportion in a plasma DNA sample was calculated for each nt position relative to the CpG site of interest across 4,631,823 cleavage measurement windows associated with a hypermethylated CpG site. The mean cleavage proportion across 307,831 cleavage measurement windows associated with a hypomethylated CpG site was determined similarly. As shown in Fig. 2A, an approximately twofold higher cleavage proportion was observed at hypermethylated CpG sites (position 0) (median: 1.13; range: 0.99–1.23) than at hypomethylated CpG sites (median: 0.53; range: 0.45–0.60) (P < 0.001, Wilcoxon rank-sum test). In contrast, a lower cleavage proportion at position −1 was observed in hypermethylated CpG sites compared with hypomethylated CpG sites (median: 0.24 versus 0.41; range: 0.19–0.27 versus 0.35–0.61) (P < 0.001, Wilcoxon rank-sum test). Notably, this relationship between cfDNA cleavage profile and predefined methylation patterns could be reproduced in paired nonbisulfite sequencing data (Fig. 2B).
Fig. 2.
Cleavage proportion depending on CpG methylation status. (A) Cleavage profiles related to hypermethylated (red lines) and hypomethylated (blue lines) CpGs in plasma DNA of eight healthy controls based on whole-genome bisulfite sequencing data. Each line represents one sample. (B) Cleavage profiles related to hypermethylated (red lines) and hypomethylated (blue lines) CpGs in plasma DNA of eight healthy controls based on whole-genome nonbisulfite sequencing data. Each line represents one sample. (C) Cleavage profiles in windows each containing two tandem CpG dinucleotides spanning positions 0, 1, 2, and 3 (i.e., CGCG subsequence) in plasma DNA of eight healthy controls. Red, dark blue, yellow, and light blue lines correspond to the cleavage profiles with different methylation configurations of two immediately adjacent CpG sites: “MM,” “UU,” “MU,” and “UM,” where M and U represent hypermethylated and hypomethylated states, respectively.
We further studied how cleavage profiles were affected by the methylation configuration of two tandem CpG dinucleotides spanning positions of 0, 1, 2, and 3 in a window (i.e., CGCG subsequence). A significantly higher cleavage proportion was observed at positions 0 and 2 when both CpG sites were hypermethylated, compared with those where both CpG sites were hypomethylated (P < 0.001, Wilcoxon rank-sum test) (Fig. 2C and SI Appendix, Fig. S1A). When the methylation state of the tandem CpG sites was opposite, the increased cleavage proportion occurred at the hypermethylated cytosine (Fig. 2C and SI Appendix, Fig. S1B). These results suggested that DNA methylation was associated with the cfDNA cleavage pattern, wherein a higher cleavage proportion at a CpG site was associated with higher methylation.
CGN/NCG Motif Ratio of cfDNA Reflects its Methylation Level.
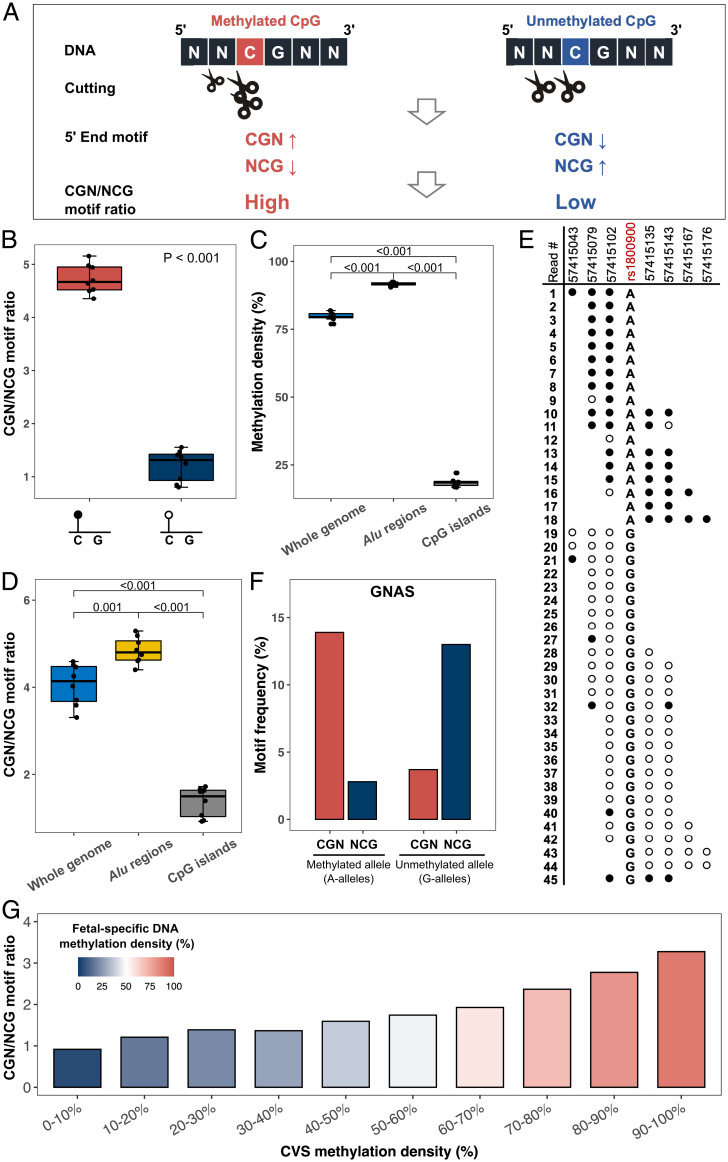
The differential cleavage of cfDNA at positions 0 and −1 relative to a CpG site, depending on methylation status, would lead to the differential presentation of end motifs. Methylated CpG sites tended to have more endpoints at position 0, enriching 5′ CGN motifs (N represents any nucleotide of A, C, G, or T), but less at the position −1, depleting 5′ NCG motifs (Fig. 3A). In contrast, the unmethylated CpG site attenuated such a cutting preference (Fig. 3A). Indeed, for plasma DNA samples of healthy controls, the ratio of 5′ CGN to NCG end motifs (i.e., CGN/NCG motif ratio) was found to be significantly higher at hypermethylated CpG sites (median: 4.70; range: 4.35–5.16) than at hypomethylated sites (median: 1.31; range: 0.80–1.55) (P < 0.001, Wilcoxon rank-sum test) (Fig. 3B and SI Appendix, Fig. S2A). We hypothesized that the CGN/NCG motif ratio of cfDNA molecules originating from a genomic region could be used to inform the methylation level of that region. As shown in Fig. 3C, Alu regions showed higher methylation levels while CpG islands showed lower methylation levels, compared to the overall methylation level of the whole human genome. We observed that the CGN/NCG motif ratios across Alu regions, CpG islands, and the whole genome were concordant with the methylation levels determined by bisulfite sequencing (Fig. 3D), which were further confirmed in the matched nonbisulfite sequencing data (SI Appendix, Fig. S2 B and C).
Fig. 3.
CGN/NCG motif ratio analysis. (A) Illustration of the biological principle for CGN/NCG motif ratio. A methylated CpG confers a higher cleavage probability at the cytosine of the CpG context but a lower cleavage probability at one base before the CpG context, compared with an unmethylated CpG. Such differential cutting leads to an increase of CGN motifs but a decrease of NCG motifs. Therefore, we expected to observe higher CGN/NCG motif ratios on hypermethylated CpG sites compared to those on hypomethylated CpGs. (B) Box plot of CGN/NCG motif ratio between hypermethylated and hypomethylated CpGs from plasma DNA of eight healthy control samples. (C) Methylation density of cfDNA molecules measured by bisulfite sequencing across the whole genome, Alu regions, and CpG islands from eight healthy control samples, respectively. (D) CGN/NCG motif ratios of whole genome, Alu regions, and CpG islands, respectively. (E) Methylation status of sequenced fragments mapped to an imprinting region (GNAS gene, located at chr20:57,415,043–57,415,176). Each row with the back (methylated) and white (unmethylated) dots represents one plasma DNA molecule. Each dot represents one CpG site. Two groups of sequenced fragments carried A alleles and G alleles, respectively, at an SNP (rs1800900). cfDNA molecules carrying A-alleles are methylated while those with G-alleles are unmethylated. (F) The frequencies of CGN and NCG motifs related to the imprinting region. (G) The CGN/NCG motif ratios from fetal-specific cfDNA in maternal plasma DNA (first trimester) correlated with the methylation levels in the paired chorionic villus sampling (CVS) biopsy. CpGs were grouped into 10 groups according to the methylation levels from the paired CVS biopsy. The y axis represents the CGN/NCG motif ratio of fetal-specific cfDNA, and the graded colors in the bars represent the different methylation densities of fetal-specific cfDNA.
To further investigate the resolution that CGN/NCG motif ratio–based methylation analysis could achieve, we analyzed plasma DNA molecules from a region involving genomic imprinting that conferred differential DNA methylation depending on parental origin (e.g., the GNAS gene, located at chr20:57,415,043–57,415,176). Those sequenced reads were obtained from a first-trimester pregnancy sample in a previous study (25). As shown in Fig. 3E, DNA fragments carrying alleles of A or G at an single-nucleotide polymorphism (SNP) site (the single nucleotide polymorphism database [dbSNP] ID: rs1800900) were inherited from different parents. Those DNA fragments carrying G alleles were unmethylated, while those carrying A alleles were methylated. Intriguingly, cfDNA fragments carrying A alleles (methylated) showed a higher frequency of 5′-CGN end motifs (13.89% versus 3.70%) but a lower frequency of 5′-NCG end motifs (2.78% versus 12.96%), compared with those cfDNA fragments carrying G alleles (unmethylated) (Fig. 3F). A similar correlation between 5′-CGN and NCG end motifs and allele-specific methylation could be observed in another region exhibiting genomic imprinting (the MEST gene, located at chr7:130,132,754–130,132,884; SI Appendix, Fig. S2D). Taken together, these results suggest that in addition to reflecting regional methylation levels, the CGN/NCG motif ratio could inform allele-specific methylation patterns.
We further explored the relationship between the cfDNA cleavage patterns and the methylation states within stretches of DNA with several adjacent CpGs by analyzing the CGN/NCG motif ratios across different combinations of methylation states for cfDNA fragments with two and three adjacent CpGs, respectively. As shown in SI Appendix, Fig. S2 E and F, the CGN/NCG motif ratio was significantly higher in those cfDNA molecules starting with a methylated CpG at the 5′ end, compared with molecules starting with an unmethylated CpG at the 5′ end. Interestingly, the cleavage of molecules starting with the unmethylated CpG at the 5′ end seemed to be relatively enhanced by the presence of methylation of the adjacent CpGs (SI Appendix, Fig. S2 E and F). These data suggested that the methylation status of the CpG that was immediately adjacent to the cleavage site of interest showed a more pronounced impact on the cfDNA cleavages than those CpG sites farther away from the cleavage site. In addition, the data also suggested that the cleavage of the CpG at the 5′ end might at least in part be affected by the methylation status of adjacent CpGs.
Furthermore, in a first-trimester pregnant woman we observed a correlation between CGN/NCG motif ratios and methylation densities of fetal-specific DNA molecules from different genomic regions (Fig. 3G). A similar correlation between the fetal CGN/NCG motif ratio and the methylation density of the placenta tissues could be seen in a third-trimester sample (SI Appendix, Fig. S2E). These results implied that using the CGN/NCG motif ratio might help construct the fetal methylome in maternal plasma.
DNASE1L3 Plays a Role in Methylation-Aware Fragmentation.
DNASE1L3 carries nuclear localization signals, reported to be present in the nucleus of cells and bound to chromatin (26). Recently, Chen et al. reported that DNASE1L3 protein could be detectable in human blood plasma based on Western blot (27). DNASE1L3 can function extracellularly and intracellularly. DNASE1L3 activities seem to influence cfDNA fragmentation patterns according to mouse model data (19), thus potentially confounding the apparent measurement of cfDNA methylation using the CGN/NCG motif ratio. For instance, the knockout of the Dnase1l3 gene has resulted in a lower overall methylation level compared with wild-type mice (19). Hence, we studied how the DNASE1L3 activity would affect the cfDNA fragmentomics-based methylation measurement in the plasma of human patients.
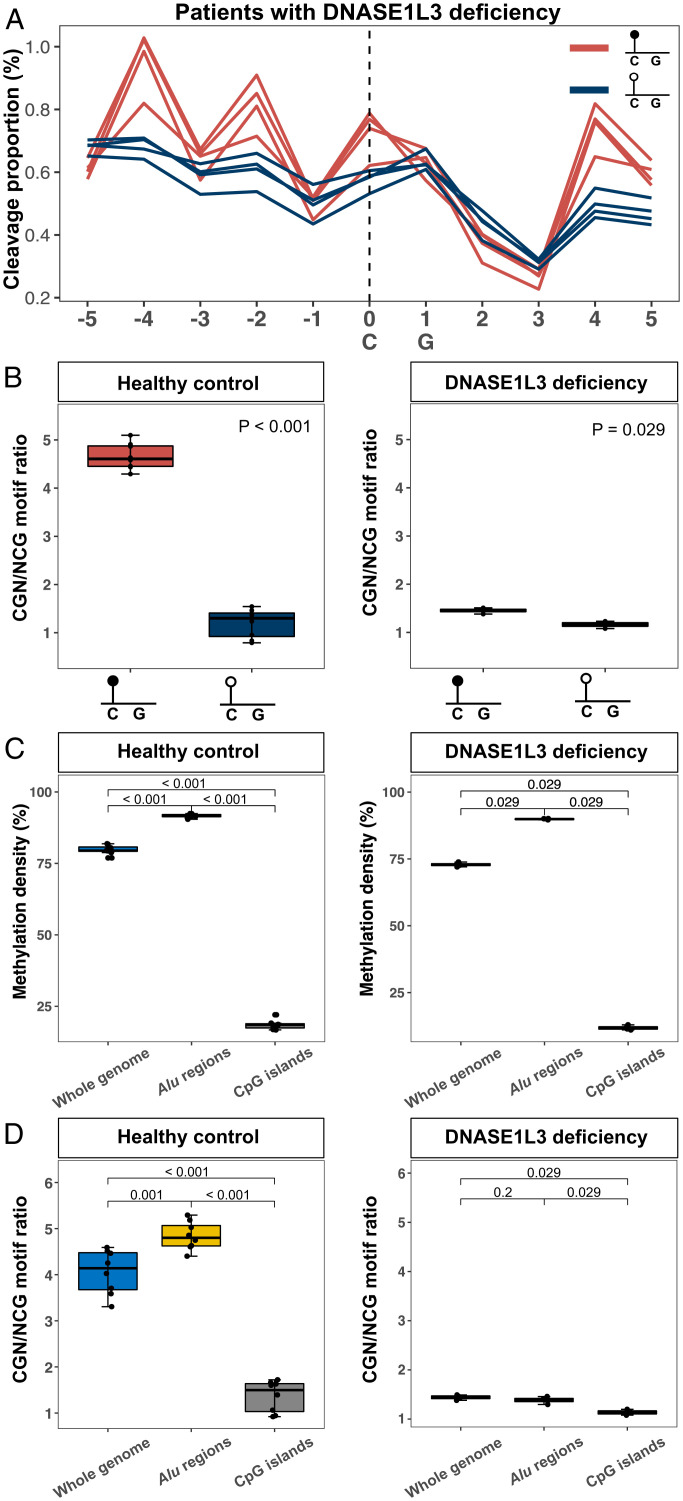
We analyzed and compared the cleavage profile of plasma DNA from four individuals with DNASE1L3 deficiency based on bisulfite sequencing in a previous study (18). In contrast to cleavage patterns in healthy individuals for which a higher cleavage proportion at position 0 and a lower cleavage proportion at position −1 was observed in hypermethylated CpG sites compared with hypomethylated ones (Fig. 2A), such position-specific cleavage patterns were drastically diminished in the plasma DNA of patients with DNASE1L3 deficiency (Fig. 4A). Due to the alteration in cleavages mediated by DNASE1L3 activities, the difference in the CGN/NCG motif ratio between hypermethylated and hypomethylated CpG sites was substantially reduced in DNASE1L3-deficient patients (i.e., approximately from a fourfold difference down to a 1.3-fold difference) (Fig. 4B). These results suggested that in addition to methylation patterns, DNASE1L3 activity was another factor acting on methylation-aware cleavages. The influence of DNASE1L3 activities on the CGN/NCG motif ratio appeared in a genome-wide manner, as the deficiency of DNASE1L3 could obscure the correlation between the CGN/NCG motif ratio and DNA methylation across the whole genome, Alu regions, and CpG islands (Fig. 4 C and D).
Fig. 4.
DNASE1L3 activity affecting cfDNA cleavage profile. (A) Cleavage profiles associated with hypermethylated (red lines) and hypomethylated (blue lines) CpGs for four patients with DNASE1L3 deficiency. (B) CGN/NCG motif ratios between hypermethylated and hypomethylated CpGs in plasma DNA of healthy controls (Left) and patients with DNASE1L3 deficiency (Right). (C) Methylation density of the whole genome, Alu regions, and CpG islands for plasma DNA samples from healthy controls (Left) and patients with DNASE1L3 deficiency (Right). (D) CGN/NCG motif ratios across the whole genome, Alu regions, and CpG islands in healthy controls (Left) and patients with DNASE1L3 deficiency (Right).
Methylation-Aware Cleavage Patterns Inform Tissues of Origin of cfDNA Molecules.
We demonstrated that DNA methylation was correlated with cfDNA cleavage profiles and that the CGN/NCG motif ratio of cfDNA could be used to inform cfDNA methylation levels of genomic regions of interest. Plasma DNA is a mixture comprising cfDNA molecules originating from different tissues (28, 29). The tissues of origin of cfDNA molecules can be determined using tissue-specific methylation patterns (3, 28–31). Thus, we tested whether the cfDNA cleavage profile might be reflective of tissue-specific hypermethylated and hypomethylated CpG sites and whether its associated CGN/NCG motif ratio could be used as a surrogate for tissue contribution to the plasma DNA pool.
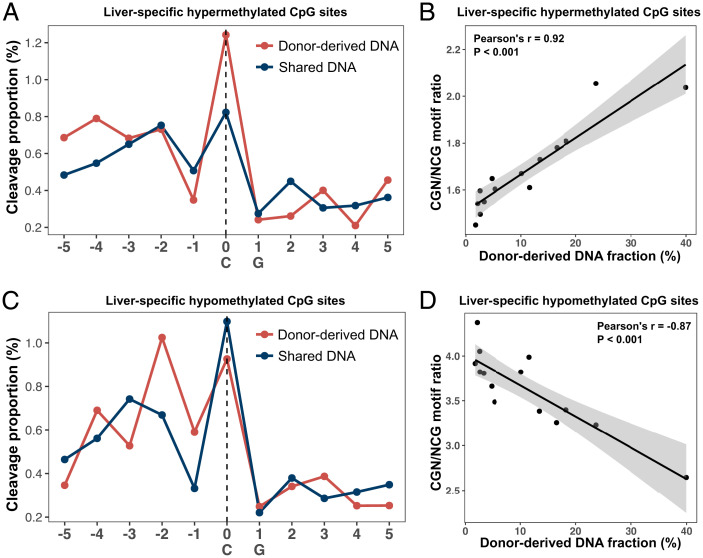
To this end, we first used liver transplantation as a model to explore the feasibility of tracing the tissue-specific cleavage profile surrounding a CpG. We analyzed plasma DNA samples from 14 liver transplant recipients previously reported (32). We identified liver-specific hypermethylated (n = 258,630) and hypomethylated (n = 226,417) CpG sites by comparing bisulfite sequencing results between the liver tissues and buffy coat samples (see details in SI Appendix, Methods and Materials). Fig. 5A shows that the donor-derived DNA molecules gave rise to a 51.0% increase in cleavage proportion at position 0 of liver-specific hypermethylated CpG sites, compared with shared DNA molecules mainly of hematopoietic origin. In contrast, the corresponding cleavage proportion decreased at position −1 by 31.3%. Thus, the presentation of CGN and NCG motifs would be expected to proportionally change depending on the specific cutting preference linked to differentially methylated CpG sites. Indeed, the CGN/NCG motif ratio from the liver-specific hypermethylated CpGs showed a strong positive correlation with the donor-derived DNA fraction (i.e., liver DNA fraction) deduced by the SNP-based approach (Pearson’s r = 0.92; P < 0.001; Fig. 5B). On the other hand, the cleavage profile at liver-specific hypomethylated CpG sites tended to show patterns opposite of those at hypermethylated CpG sites (Fig. 5C). For the hypomethylated CpG sites, the CGN/NCG motif ratio strongly correlated with the donor-derived DNA fraction in an inverse manner (Pearson’s r = −0.87; P < 0.001; Fig. 5D).
Fig. 5.
Liver-specific cleavage profile readily used for deducing liver DNA contribution in plasma DNA of liver transplant patients. (A) Cleavage profiles associated with liver-specific hypermethylated CpGs deduced from donor-derived DNA (red line) and shared DNA (blue line). Donor-derived DNA was defined as cfDNA carrying donor-specific alleles that were absent in recipient genomes, while the shared DNA was defined as cfDNA molecules carrying the alleles existing in both the donor and recipient genomes. For the cleavage profile analysis, donor-derived and shared DNA were pooled together from 14 liver transplant samples. (B) The CGN/NCG motif ratio associated with liver-specific hypermethylated CpGs was positively correlated with the donor-derived DNA fraction. (C) Cleavage profiles associated with liver-specific hypomethylated CpGs were analyzed in a similar way as liver-specific hypermethylated CpGs. (D) The CGN/NCG motif ratio associated with liver-specific hypomethylated CpGs was negatively correlated with the donor-derived DNA fraction.
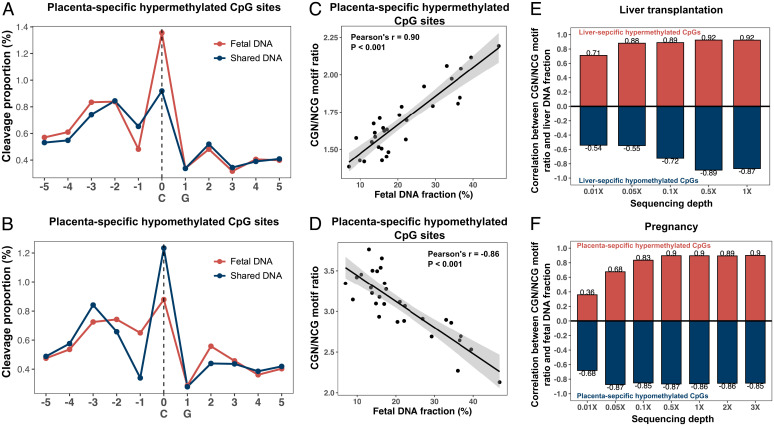
In addition, we validated the hypothesis that cfDNA cleavage patterns on CpG sites with tissue-specific methylation were informative for tissue-of-origin analysis using a pregnancy model. We obtained bisulfite sequencing results of maternal plasma DNA from 30 pregnant women in a previous study (10). Compared with buffy coat DNA samples, we identified 184,430 and 1,922,990 placenta-specific hypermethylated and hypomethylated CpG sites, respectively (see details in SI Appendix, Methods and Materials). The cleavage pattern depending on methylation states observed in liver-specific methylation was well generalized to plasma DNA molecules associated with placenta-specific methylation in pregnant women (Fig. 6 A and B). A high positive correlation was observed between the CGN/NCG motif ratio of placenta-specific hypermethylated CpGs and the fetal DNA fraction deduced by the SNP-based approach (Pearson’s r = 0.90; P < 0.001; Fig. 6C), whereas a high negative correlation was observed in placenta-specific hypomethylated CpG sites (Pearson’s r = −0.86; P < 0.001; Fig. 6D). These data provided further evidence that the cfDNA cleavage profile was useful for deducing contributions of plasma DNA molecules from different tissues.
Fig. 6.
Tissue-specific cleavage profiled used for tissue-of-origin analysis. Cleavage profiles of placenta-specific hypermethylated (A) and hypomethylated (B) CpGs in fetal-specific DNA (red line) and shared DNA (blue line) were determined, respectively. Fetal-specific and shared DNA molecules in maternal plasma were pooled together from 30 pregnant women. The CGN/NCG motif ratios associated with placenta-specific hypermethylated (C) and hypomethylated CpGs (D) were positively and negatively correlated with fetal DNA fractions, respectively. (E) and (F) Impact of the sequencing depth on the performance of tissue-of-origin analysis. Pearson’s correlation coefficient between the CGN/NCG motif ratio from liver-specific hypermethylated (red) and hypomethylated (blue) CpGs and liver DNA fraction (E). Pearson’s correlation coefficient between the CGN/NCG motif ratio from placenta-specific hypermethylated (red) and hypomethylated (blue) CpGs and fetal DNA fraction. X-axis represents different sequencing depths (F).
To explore the feasibility of using shallow sequencing depth for FRAGMA, we performed a down-sampling analysis on the correlation between the targeted tissue-specific DNA fraction and the CGN/NCG motif ratio derived from a set of tissue-specific differentially methylated CpGs. The Pearson’s correlation coefficient (r) reached > 0.8 at sequencing depths of 0.05× and 0.5× for liver-specific hypermethylated and hypomethylated CpGs, respectively (Fig. 6E). For pregnant women, sequencing depths of 0.1× and 0.05× allowed a Pearson’s r > 0.80 for placenta-specific hypermethylated and hypomethylated CpGs, respectively (Fig. 6F). These data suggested that it was feasible to use the CGN/NCG motif ratio to reflect the tissue-specific methylation levels based on a shallow sequencing depth. In addition, we compared the placental contributions in the plasma samples from 30 pregnant women, which were respectively deduced using FRAGMA and bisulfite-based methylation analysis (31) through varying sequencing depths. The overall performance between these two approaches seemed to be comparable (SI Appendix, Fig. S3A).
Moreover, we performed computer simulation analysis (details in SI Appendix, Methods and Materials) to study how the sequencing depth and the fractional DNA concentration of the target tissue would affect the detection of target molecules. As a result, a higher sequencing depth would be required to achieve the same level of area under the receiver operating characteristic curve (AUC) as the fractional concentration of the target tissue DNA decreased in the plasma DNA pool. To obtain an AUC of 0.95, the desired sequence depths were deduced to be 100×, 200×, 700×, and 1400× for a plasma DNA sample with the fractional concentration of the target tissue DNA of 100%, 50%, 20%, and 10%, respectively (SI Appendix, Fig. S3B). Of note, this simulation was based on the analysis of single CpG sites. The sequencing depth per locus could be greatly reduced when focusing on a set of informative CpG sites. For example, if one analyzes 1,000 tissue-specific CpG sites in a plasma DNA sample with 10% target tissue DNA, then the desired sequence depths would theoretically be 1.4× (i.e., 1,400/1,000).
Clinical Implications of Aberrations in CGN/NCG Motif Ratios.
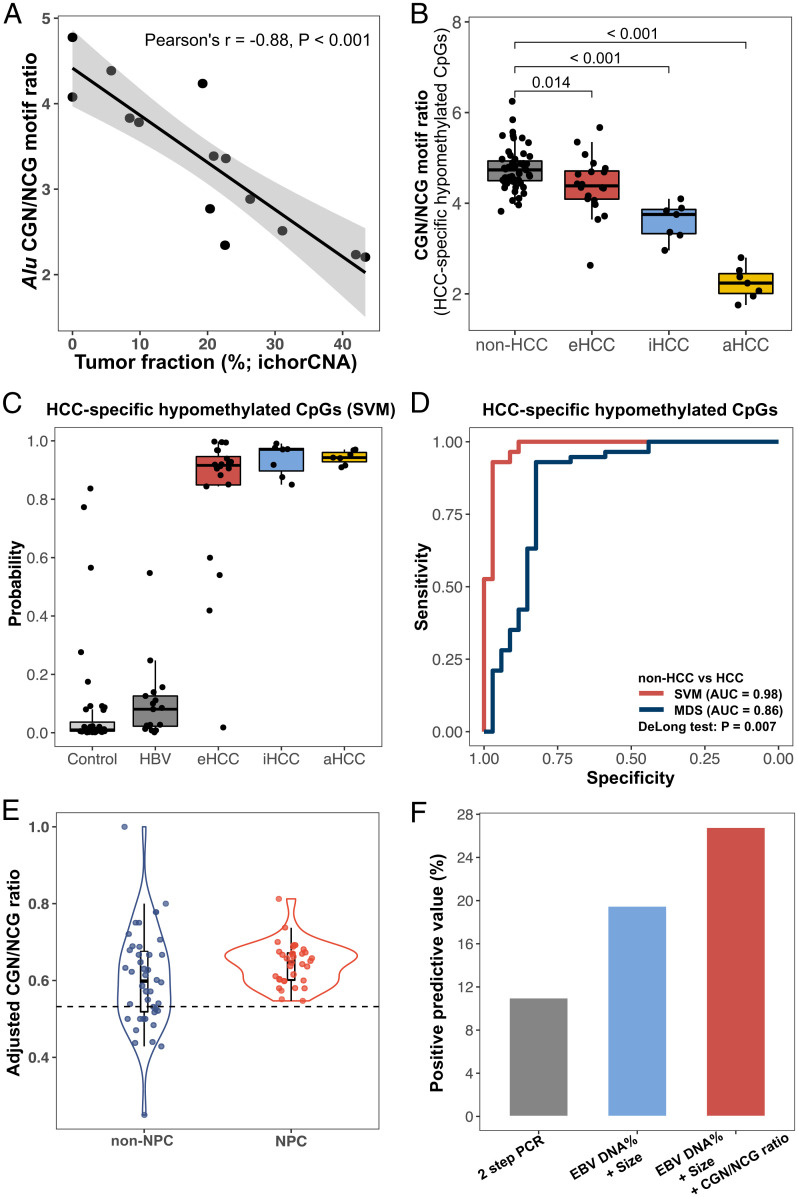
Genome-wide hypomethylation, typically in repetitive elements (e.g., Alu), frequently occurs in various cancers (33) and can be detected in the plasma DNA of cancer patients (34). Hence, we attempted to employ the CGN/NCG motif ratio as an indicator of plasma DNA methylation changes caused by cfDNA molecules released from tumor cells. We indeed observed a negative correlation between the CGN/NCG motif ratio from Alu regions with the tumor DNA fraction estimated by copy number aberrations (ichorCNA) (35) in hepatocellular carcinoma (HCC) patients (Pearson’s r = −0.88, P < 0.001; Fig. 7A), suggesting that more hypomethylated fragments shed from the tumor cells were cut to the 5′ NCG position, resulting in a lower CGN/NCG motif ratio. Interestingly, such a correlation was higher than a previously reported metric, the motif diversity score (MDS) (Pearson’s r = 0.59; P = 0.026), which reflected the evenness of 256 5′ 4-mer end motifs (SI Appendix, Fig. S4A) (10). These results indicated that the CGN/NCG motif ratio from the Alu region might be useful for estimating the tumor DNA contribution.
Fig. 7.
The use of end motifs resulting from the differential cutting within the cleavage measurement window for cancer detection. (A) The correlation between CGN/NCG motif ratio originating from Alu regions and tumor DNA fraction determined by copy number aberrations in patients with HCC. (B) The CGN/NCG motif ratio concerning HCC-specific hypomethylated CpGs in plasma DNA among non-HCC patients (healthy controls and HBV carriers) and HCC patients with early (eHCC), intermediate (iHCC), and advanced (aHCC) stages. (C) HCC probability determined by SVM models using CG-containing motifs (i.e., CGA, CGT, CGC, CGG, ACG, TCG, CCG, and GCG). (D) ROC (receiver operating characteristic curve) analysis between CG-containing motifs and motif diversity scores. (E) The adjusted CGN/NCG motif ratios of informative CpGs between non-NPC and NPC patients. (F) PPVs archived by PCR-based assay, the approach based on EBV DNA proportion and size ratio, and the approach based on the combined EBV DNA proportion, size ratio, and cleavage motifs.
Compared with non-HCC individuals including healthy controls and individuals with chronic hepatitis B virus (HBV) infection but without HCC (i.e., HBV carriers), the CGN/NCG motif ratio derived from HCC-specific hypomethylated CpG sites seemed to be significantly lower in the HCC group, with a gradual decline over tumor stages (Fig. 7B). Such a decrease in CGN/NCG motif ratios coincided with an increase in tumor DNA fractions across tumor stages (mean tumor DNA fractions: 2.4%, 9.2%, and 29.8% in patients with early-stage HCC (eHCC; [range: 0.00–13.9%]), intermediate-stage HCC (iHCC; [range: 0.00–20.9%]), and advanced-stage HCC (aHCC; [range: 20.4–43.4%]), respectively (SI Appendix, Table S1). To take full advantage of CGN and NCG motifs, we adopted a support vector machine (SVM) model using all CG-containing end motifs (i.e., ACG, CCG, GCG, TCG, CGA, CGC, CGG, and CGT) from HCC-specific hypomethylated CpG sites based on a leave-one-out analysis. SVM model based on CG-containing motifs achieved an AUC of 0.98 (Fig. 7 C and D), leading to a significantly better performance than the MDS (AUC: 0.86; Fig. 7D) (P = 0.007, DeLong test). With a specificity of 96%, the sensitivities were 80%, 100%, and 100% for the eHCC, iHCC, and aHCC detection, respectively. Of note, CGN/NCG motif ratios related to CpG sites with HCC-specific hypermethylation seemed to lose power in distinguishing HCC patients from non-HCC individuals (SI Appendix, Fig. S4B), possibly because of the effect of the reduced DNASE1L3 activity (10).
In addition to the methylation of the nuclear genome, differential methylation signals were reported to be present in viral cfDNA molecules such as Epstein-Barr virus (EBV) DNA between individuals with and without nasopharyngeal carcinoma (NPC); these were useful for NPC screening (36). Hence, we reasoned that the CGN/NCG motif ratio of plasma EBV DNA could be another dimension to detect NPC. We reanalyzed the previous nonbisulfite sequencing dataset comprising 272 individuals positive for EBV but without NPC and 65 EBV-positive individuals with NPC (37). We identified 1,425 informative CpG sites in the EBV genome showing an up-regulation of the adjusted CGN/NCG motif ratio (ratio of CGN motif with respect to the total of CGN and NCG motifs) in patients with NPC (n = 31) compared with non-NPC individuals (n = 230) (SI Appendix, Fig. S5C). Those sites were further confirmed with a higher methylation index in patients with NPC than in non-NPC individuals in a previous bisulfite sequencing dataset (SI Appendix, Fig. S5D) (36). In the remaining samples comprising 42 non-NPC individuals and 34 patients with NPC, the adjusted CGN/NCG motif ratio from plasma EBV DNA molecules related to informative CpG sites showed a significant difference between the NPC and non-NPC groups (P = 0.041, Wilcoxon rank-sum test; Fig. 7E). If we adopted an adjusted CGN/NCG motif ratio cutoff of 0.532, combined with previously published metrics (EBV DNA proportion and EBV DNA fragment size ratio), then the positive predictive value (PPV) reached 26.8%, which was higher than the qPCR assay (PPV: 11.0%) (38) and the approach based on EBV DNA proportion and EBV DNA fragment size ratio (PPV: 19.6%) (Fig. 7F) (37). Thus, the data implied that the cleavage profile related to viral cfDNA might be another important molecular feature for developing novel diagnostic tools for virus-driven cancers.
Methylation Status Prediction at Single CpG Resolution Using a Deep Learning Model Trained from Cleavage Profile.
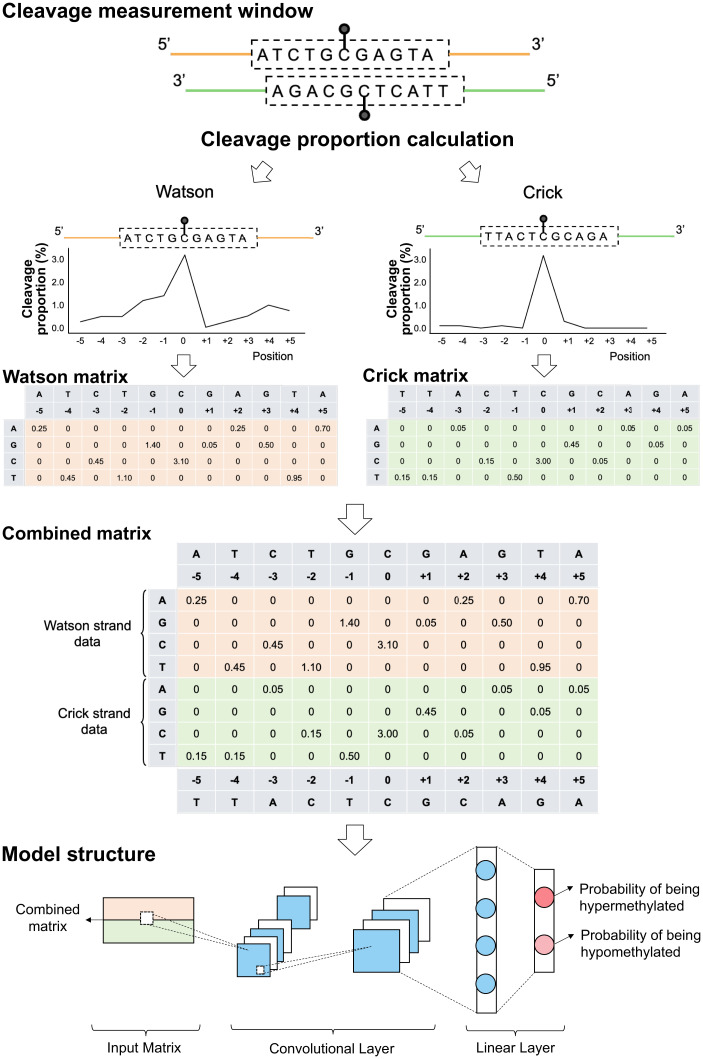
We had determined that the use cfDNA cleavage patterns and the resultant CGN/NCG motif enabled the deduction of cfDNA methylation states across regions. One important further goal would be to accurately discern the methylation status of individual CpG sites by taking advantage of the cfDNA cleavage profile surrounding a CpG site. To test the feasibility of this aim, we explored one of the deep learning algorithms, the convolutional neural network (CNN), to predict a CpG methylation index above 70% or below 30% by analyzing the cfDNA cleavage patterns around a CpG site. Fig. 8 shows the workflow for constructing a CNN model using cleavage patterns. The 5-nt upstream (e.g., ATCTG) and 5-nt downstream (e.g., GAGTA) of the cytosine at a CpG site being analyzed were presented as 5′-[ATCTG]C[GAGTA]-3′ for the Watson strand—the cleavage measurement window of the Watson strand. The relative positions of this sequence corresponded to −5, −4, −3, −2, −1, 0, +1, +2, +3, +4, and +5, respectively. The center position (i.e., position 0) corresponded to the cytosine at the CpG site subjected to the methylation analysis. The cfDNA cleavage proportion for each position was constructed into a 2-dimensional (2-D) matrix according to the sequence context (Fig. 8). For example, for position −1, corresponding to guanine (G), the cleavage proportion (1.40) was filled in the corresponding cell between a column of −1 and a row of G. The remaining rows corresponding to A, C, and T in the Watson strand were filled by 0. The cleavage profile and sequence context originating from the Crick strand (′5-[TTACT]C[GCAGA]-3′) were processed similarly (Fig. 8). The data matrices from the Watson and Crick strands were combined to make a combined cleavage measurement window for downstream analysis.
Fig. 8.
Schematic for methylation status prediction at single CpG resolution using a CNN model based on cleavage profiles. For illustration purposes, the 5 nt upstream (e.g., ATCTG) and 5 nt downstream (e.g., GAGTA) of the cytosine at a CpG site (i.e., the cleavage measurement window) being analyzed were presented as 5′-[ATCTG]C[GAGTA]-3′ for the Watson strand. The relative positions of this sequence corresponded to −5, −4, −3, −2, −1, 0, +1, +2, +3, +4, and +5, respectively. The central position 0 corresponded to the cytosine at the CpG site that was subjected to the methylation analysis. The cleavage proportion for each position was constructed into a 2-D matrix according to the sequence context. For instance, for a position of −1 corresponding to the base of guanine (G), the cleavage proportion associated with G (1.40) was filled in the corresponding cell between a column of −1 and a row of G. The remaining rows corresponding to A, C, and T in the Watson strand were filled by 0. The cleavage profiles and sequence context originating from the Crick strand (‘5-[TTACT]C[GCAGA]-3′) were processed similarly. The data matrices from the Watson and Crick strand were put together into a combined matrix to train and test a CNN model.
To obtain sufficient sequencing depth for profiling the cleavage proportion, we pooled bisulfite sequencing data from eight healthy controls and 13 HBV carriers. A set of hypermethylated and hypomethylated CpG sites with a sequence depth of >50× and at least 10 3-mer end motifs containing a CG dinucleotide within the cleavage measurement window were used to train and test the above-mentioned CNN model. To train a CNN, we utilized a number of combined cleavage measurement windows originating from hypermethylated and hypomethylated cytosines. The model parameters learned from the training datasets were used to analyze the testing dataset to output a probabilistic score (referred to as the methylation score in this study), indicating the likelihood of a CpG site being hypermethylated (see details in SI Appendix, Methods and Materials).
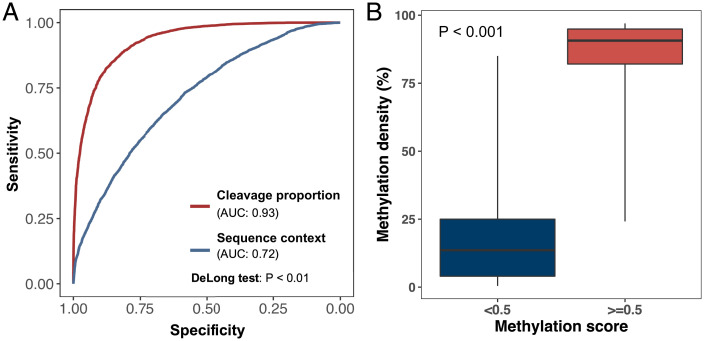
We achieved an AUC of 0.93 to classify whether one CpG site was hypermethylated or hypomethylated (Fig. 9A). Moreover, CpG sites with a methylation score of < 0.5 showed significantly lower methylation indices, compared with CpG sites with a methylation score of ≥ 0.5 (Fig. 9B) (P < 0.001, Wilcoxon rank-sum test). Of note, if we trained the CNN model only using sequence context surrounding CpG sites without cfDNA cleavage patterns, then the performance significantly declined to an AUC of 0.72 (P < 0.001, DeLong test), highlighting that the cleavage patterns contributed significantly to the accuracy of methylation analysis. These results demonstrated the feasibility of exploiting the cfDNA fragmentomic patterns to deduce methylation status at a single CpG level.
Fig. 9.
Evaluation for CNN model for methylation analysis using cleavage measurement windows. (A) ROC analysis for the performance of the CNN model by using cleavage measurement windows (red line) and sequence context (blue line) in a testing dataset. (B) The box plot illustrated the CpG methylation density detected by bisulfite sequencing between two CpG groups with a methylation score < 0.5 or ≥ 0.5 in a testing dataset.
Discussion
In this study, we demonstrated a link between the cleavage profile of a CpG site and its methylation status. The use of the cleavage profile enabled the deduction of the methylation status in a genomic region and even at a single CpG resolution. The resultant CGN/NGC motif ratio derived from cleavage patterns could serve as a potential biomarker for noninvasive prenatal, organ transplantation, and cancer assessment. These data form the basis of the FRAGMA technology. One advantage of this approach for methylation analysis could be directly applied to widely practiced, massively parallel sequencing of plasma DNA, obviating special methylation-aware treatments, such as bisulfite-based or enzymatic cytosine conversion–based treatments. Such cfDNA cleavage-based methylation deduction enabled an extra dimension to the data mining of general massively parallel sequencing of cfDNA molecules, bringing together genetic and epigenetic analyses in one simplified assay.
cfDNA cleavage patterns appeared to highly correlate with methylation configurations. The methylated CpG sites conferred a higher chance of cutting to the cytosine (i.e., generated ends at position 0) but a lower chance of cutting to the position 1-nt immediately upstream of that CpG dinucleotide (i.e., generated ends at position −1). The unmethylated CpG sites switched to produce the opposite cleavage patterns at these two positions. Such methylation-aware cutting preferences generated characteristic 5′ 3-mer end motifs, i.e., CGN and NCG motifs. The CGN/NCG motif ratio metric was positively correlated with methylation levels of plasma DNA, reflective of methylation patterns of cellular genomes that contributed DNA to plasma. Of note, the use of such cleavage patterns across a set of CpG sites with tissue-specific methylation (hypomethylation or hypermethylation) could inform the contributions from various tissues (e.g., the placenta, liver, and blood cells) to plasma DNA. These data could also provide insight into the future development of PCR-based cfDNA assays. For example, the preferential cfDNA cleavages, depending on methylation status, could result in a biased measurement in PCR-based assays such as droplet digital PCR (ddPCR) and amplicon-based target sequencing. For instance, one could develop a ddPCR assay to quantify the liver DNA contribution to the plasma DNA pool, utilizing a genomic region that was specifically methylated in the liver tissue but unmethylated in other cell types such as blood cells. Because the methylated molecules would be preferentially cleaved compared to unmethylated molecules, a certain amount of liver-derived cfDNA molecules from such a region would not be able to form amplifiable signals, resulting in an underestimation of liver DNA contribution. If these kinds of methodologies could take into account the presence of differential cleavages of cfDNA, then the assay performance and the accuracy of the data interpretation could be further enhanced.
The impact of nuclease activity on fragmentation should be considered when interpreting cleavage profiles for cfDNA methylation analysis in a clinical scenario, such as cancer detection. Our data showed that the DNASE1L3 deficiency largely diminished the difference in cleavage profiles between methylated and unmethylated CpG sites (Fig. 4B), perhaps influencing the cfDNA cleavage–based methylation analysis. For example, for patients with HCC, CGN/NCG motif ratios related to CpG sites with HCC-specific hypomethylated sites showed good differentiating power of HCC patients from non-HCC individuals, whereas the motif ratio related to the CpG sites with HCC-specific hypermethylation exhibited much poorer differentiating power. One likely reason is that the decrease of DNASE1L3 activity in HCC may partially cancel out the expected increase of the CGN/NCG motif ratio conferred by the hypermethylation. On the other hand, if we focused on the CGN/NCG motif ratio on HCC-specific hypomethylation CpGs, then the decreased signal of the CGN/NCG motif ratio conferred by reduced methylation could be enhanced by the down-regulated DNASE1L3 activity (i.e., caused more cuts at position −1). Therefore, a detailed understanding of the interplay between methylation and nuclease activity would facilitate a synergistic combination of these two types of signals for cancer detection. Moreover, using CG-containing 3-mer motifs based on SVM, we achieved an AUC of 0.98 for differentiating between patients with and without HCC, suggesting that the appropriate analysis of end motifs as a result of DNA nucleases would improve the diagnostic performance. Interestingly, by combining EBV quantity and size properties (37) with cleavage patterns (i.e., the adjusted CGN/NCG ratio) related to EBV DNA in plasma, the performance of NPC screening could be further improved with a PPV of 26.8%, up from 19.6% based on EBV DNA proportion and size parameters.
Furthermore, this study has demonstrated that we could exploit the spectrum of cleavage patterns across an 11-nt window centered on a CpG to predict its methylation state using a deep learning algorithm. Deep learning is widely used in image pattern recognition (39). Cleavage signals were organized into a matrix depending on sequencing context and positions relative to the CpG site in a way that the patterns could be used to train a CNN model. The trained CNN was able to differentiate between hypomethylated and hypermethylated CpG sites, with an AUC of 0.93. The results suggested that deep learning is a feasible method to recognize CpG methylation based on cleavage patterns. The analytical workflow provided an exemplar of translating fragmentomic signals into signals that can be harnessed by artificial intelligence.
The mechanistic basis whereby DNASE1L3 prefers to cut methylated CpG sites remains to be explored. Nevertheless, one might gain mechanistic insights from the DNASE1 enzyme, which exhibits homology to DNASE1L3 (40). DNA cleavage by DNase I was reported to be enhanced at least eightfold at methylated CpGs compared with unmethylated CpGs, based on bisulfite sequencing of naked DNA digested by DNase I (41). Methylation-induced narrowing of the minor groove might be one of the factors that enhances the contact between DNase I and the DNA substrate (41). Moreover, one report demonstrated that DNA methylation might induce the conformation change of DNA wrapping around its accompanying histones; CpG methylation might cause the internal regions of DNA to be “overwrapped” around a histone octamer (42). Hence, we speculated that the underlying mechanism governing the methylation-aware cleavage profile might in part be related to the accessibility of nuclease to the DNA substrate, perhaps depending on chromatin structures and DNA conformation. Future studies could provide further biophysical and biochemical insights into the methylation-aware cleavages of plasma DNA by examining the conformational changes and structural properties in relation to the interactions of DNA molecules, histones, and various nucleases.
In summary, we developed the FRAGMA methodology, utilizing the cleavage profile to reflect the CpG methylation. Prediction of the methylation status of individual CpG sites can be achieved by exploiting a deep learning algorithm to process the cleavage patterns. FRAGMA provides relatively easy access to the signals hidden in cleavage patterns, providing a potential biomarker for noninvasive prenatal testing, organ transplantation, and cancer assessment. More cost-effective versions of FRAGMA may be developed by targeting CGN and NCG motifs. The unraveling of linkages between cfDNA cleavage patterns and methylation opens many possibilities for maximizing the value of plasma DNA sequencing, through integrating genetic and epigenetic analyses in a single assay.
Materials and Methods
Sample Collection and Processing.
All patients involved in this study gave written informed consent, and the study was approved by The Joint Chinese University of Hong Kong—Hospital Authority New Territories East Cluster Clinical Research Ethics Committee under the Declaration of Helsinki.
The datasets used in this study are summarized in detail in SI Appendix, Table S2.
Sequencing Alignment.
After base calling, the sequencing reads were preprocessed by removing the adaptor sequences and low-quality bases (i.e., a quality score of < 20). The trimmed reads in a FASTQ format were analyzed for the nonbisulfite and bisulfite sequencing data, respectively, as described previously (10). The paired-end reads, each with a proper alignment and a spanning insert size of < 600 bp, were used for downstream analysis.
Cleavage Proportion.
To analyze the preference of cfDNA cutting at nucleotides surrounding a CpG, we used the cleavage proportion to measure the relative cutting frequency as in the formula below:
where sequencing depth was defined as the number of sequence fragments covering at a site i, and fragment ends refer to 5′ ends in this study. As an example, if one genomic site was covered by 100 sequenced fragments and five ends terminated at that site, then the cleavage proportion was 5%. A higher cleavage proportion indicated a higher cutting preference.
Cleavage profile was defined as the cleavage proportions across positions within a cleavage measurement window centered on a CpG. The window was defined as 5-nt upstream and downstream of a CpG. When we analyzed the cleavage profile of a number of cleavage measurement windows, the mean cleavage proportion of each relative position was used. As the CpG methylation at the Watson and Crick strands was often symmetrical, cleavage profiles of the Watson and Crick strands were merged in the 5′ to 3′ direction for downstream analysis.
CGN/NCG Motif Ratio.
The CGN/NCG motif ratio was defined as follows:
The CGN/NCG motif ratio referred to the number of cfDNA fragments carrying 5′ CGN end motifs (i.e., 5′- CGA, CGT, CGG, and CGC motifs) divided by the number of 5′ NCG end motifs (i.e., 5′- ACG, TCG, GCG, and CCG motifs). The 5′ end motif was determined as previously described (10).
Supplementary Material
Acknowledgments
This work was supported by the Research Grants Council of the Hong Kong Special Administrative Region Government under the theme-based research scheme (T12-403/15-N and T12-401/16-W) and the Innovation and Technology Commission (InnoHK initiative). Y.M.D.L. is supported by an endowed chair from the Li Ka Shing Foundation. We thank Professor Peter Pak-Hang Cheung for his technical assistance.
Footnotes
Reviewers: I.D.V., Cornell University; and Y.D., Hebrew University of Jerusalem.
Competing interest statement: A patent application on the described technology has been filed by Q.Z., G.K., P.J., R.Q., L.J., R.W.K.C., K.C.A.C., and Y.M.D.L.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209852119/-/DCSupplemental.
Data, Materials, and Software Availability
Sequencing raw data examined were obtained from previous studies (10, 18, 23, 25, 31, 32, 34, 36, 37) and are summarized in SI Appendix, Table S2. The corresponding data accession numbers in the European Genome-Phenome Archive (EGA) (https://www.ebi.ac.uk/ega/) include EGAS00001000566 (43), EGAS00001002707 (44), EGAS00001003408 (45), EGAS00001003409 (46), EGAS00001004642 (47), and EGAS00001005562 (48).
References
- 1.Lo Y. M. D., Han D. S. C., Jiang P., Chiu R. W. K., Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 372, eaaw3616 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Lui Y. Y., et al. , Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 48, 421–427 (2002). [PubMed] [Google Scholar]
- 3.Moss J., et al. , Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang P., et al. , Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. U.S.A. 112, E1317–E1325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo Y. M., et al. , Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2, 61ra91 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Underhill H. R., et al. , Fragment length of circulating tumor DNA. PLoS Genet. 12, e1006162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang P., et al. , Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 115, E10925–E10933 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snyder M. W., Kircher M., Hill A. J., Daza R. M., Shendure J., Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun K., et al. , Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 29, 418–427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang P., et al. , Plasma DNA end-motif profiling as a fragmentomic marker in cancer, pregnancy, and transplantation. Cancer Discov. 10, 664–673 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Chan K. C., et al. , Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends. Proc. Natl. Acad. Sci. U.S.A. 113, E8159–E8168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang P., et al. , Detection and characterization of jagged ends of double-stranded DNA in plasma. Genome Res. 30, 1144–1153 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esfahani M. S., et al. , Inferring gene expression from cell-free DNA fragmentation profiles. Nat. Biotechnol. 40, 585–597 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulz P., et al. , Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet. 48, 1273–1278 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Han D. S. C., et al. , The biology of cell-free DNA fragmentation and the roles of DNASE1, DNASE1L3, and DFFB. Am. J. Hum. Genet. 106, 202–214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serpas L., et al. , Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc. Natl. Acad. Sci. U.S.A. 116, 641–649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan R. W. Y., et al. , Plasma DNA profile associated with DNASE1L3 gene mutations: Clinical observations, relationships to nuclease substrate preference, and in vivo correction. Am. J. Hum. Genet. 107, 882–894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding S. C., et al. , Jagged ends on multinucleosomal cell-free DNA serve as a biomarker for nuclease activity and systemic lupus erythematosus. Clin. Chem. 68, 917–926 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Han D. S. C., et al. , Nuclease deficiencies alter plasma cell-free DNA methylation profiles. Genome Res. 31, 2008–2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frommer M., et al. , A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 89, 1827–1831 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grunau C., Clark S. J., Rosenthal A., Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res. 29, E65 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaisvila R., et al. , Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA. Genome Res. 31, 1280–1289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tse O. Y. O., et al. , Genome-wide detection of cytosine methylation by single molecule real-time sequencing. Proc. Natl. Acad. Sci. U.S.A. 118, e2019768118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson J. T., et al. , Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Lun F. M., et al. , Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin. Chem. 59, 1583–1594 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Napirei M., Wulf S., Eulitz D., Mannherz H. G., Kloeckl T., Comparative characterization of rat deoxyribonuclease 1 (Dnase1) and murine deoxyribonuclease 1-like 3 (Dnase1l3). Biochem. J. 389, 355–364 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M., et al. , Fragmentomics of urinary cell-free DNA in nuclease knockout mouse models. PLoS Genet. 18, e1010262 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng A. P., et al. , Cell-free DNA tissues of origin by methylation profiling reveals significant cell, tissue, and organ-specific injury related to COVID-19 severity. Med (N Y) 2, 411–422.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng A. P., et al. , Cell-free DNA profiling informs all major complications of hematopoietic cell transplantation. Proc. Natl. Acad. Sci. U.S.A. 119, e2113476118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehmann-Werman R., et al. , Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. U.S.A. 113, E1826–E1834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun K., et al. , Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. U.S.A. 112, E5503–E5512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gai W., et al. , Liver- and colon-specific DNA methylation markers in plasma for investigation of colorectal cancers with or without liver metastases. Clin. Chem. 64, 1239–1249 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Feinberg A. P., Tycko B., The history of cancer epigenetics. Nat. Rev. Cancer 4, 143–153 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Chan K. C., et al. , Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 18761–18768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adalsteinsson V. A., et al. , Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam W. K. J., et al. , Methylation analysis of plasma DNA informs etiologies of Epstein-Barr virus-associated diseases. Nat. Commun. 10, 3256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam W. K. J., et al. , Sequencing-based counting and size profiling of plasma Epstein-Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. U.S.A. 115, E5115–E5124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan K. C. A., et al. , Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med. 377, 513–522 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Hosny A., Parmar C., Quackenbush J., Schwartz L. H., Aerts H. J. W. L., Artificial intelligence in radiology. Nat. Rev. Cancer 18, 500–510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keyel P. A., Dnases in health and disease. Dev. Biol. 429, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazarovici A., et al. , Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. U.S.A. 110, 6376–6381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J. Y., Lee T. H., Effects of DNA methylation on the structure of nucleosomes. J. Am. Chem. Soc. 134, 173–175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang K. C., et al. , Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. European Genome-Phenome Archive. https://ega-archive.org/studies/EGAS00001000566. Deposited 11 April 2013. [DOI] [PMC free article] [PubMed]
- 44.Lam W. K. J., et al. , Sequencing-based counting and size profiling of plasma Epstein-Barr virus DNA enhance population screening of nasopharyngeal carcinoma. European Genome-Phenome Archive. https://ega-archive.org/studies/EGAS00001002707. Deposited 14 April 2018. [DOI] [PMC free article] [PubMed]
- 45.Lam W. K. J., et al. , Methylation analysis of plasma DNA informs etiologies of Epstein-Barr virus-associated diseases. European Genome-Phenome Archive. https://ega-archive.org/studies/EGAS00001003408. Deposited 22 July 2019. [DOI] [PMC free article] [PubMed]
- 46.Zhou Q., et al. , Plasma DNA motif analysis. European Genome-Phenome Archive. https://ega-archive.org/studies/EGAS00001003409. Deposited 5 January 2020.
- 47.Tse O. Y. O., et al. , Genomewide detection of cytosine methylation by single molecule real-time sequencing. European Genome-Phenome Archive. https://ega-archive.org/studies/EGAS00001004642. Deposited 25 January 2021.
- 48.Zhou Q., et al. , Jagged ends of plasma DNA (human). European Genome-Phenome Archive. https://ega-archive.org/studies/EGAS00001005562. Deposited 19 May 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing raw data examined were obtained from previous studies (10, 18, 23, 25, 31, 32, 34, 36, 37) and are summarized in SI Appendix, Table S2. The corresponding data accession numbers in the European Genome-Phenome Archive (EGA) (https://www.ebi.ac.uk/ega/) include EGAS00001000566 (43), EGAS00001002707 (44), EGAS00001003408 (45), EGAS00001003409 (46), EGAS00001004642 (47), and EGAS00001005562 (48).