To the Editor
Advances in highly multiplexed imaging have enabled the comprehensive profiling of complex tissues in healthy and diseased states, facilitating the study of fundamental biology and human disease at spatially resolved, subcellular resolution1,2. Although the rapid innovation of biological imaging brings significant scientific value, the proliferation of technologies without unification of interoperable standards has created challenges that limit the analysis and sharing of results. The adoption of community-designed next-generation file formats (NGFF) is a proposed solution to promote bioimaging interoperability at scale3. Here we introduce Viv (https://github.com/hms-dbmi/viv), an open-source bioimaging visualization library that supports OME-TIFF4 and OME-NGFF3 directly on the web. Viv addresses a critical limitation of most web-based bioimaging viewers by removing a dependency on server-side rendering, offering a flexible toolkit for browsing multi-terabyte datasets on both mobile and desktop devices—without software installation.
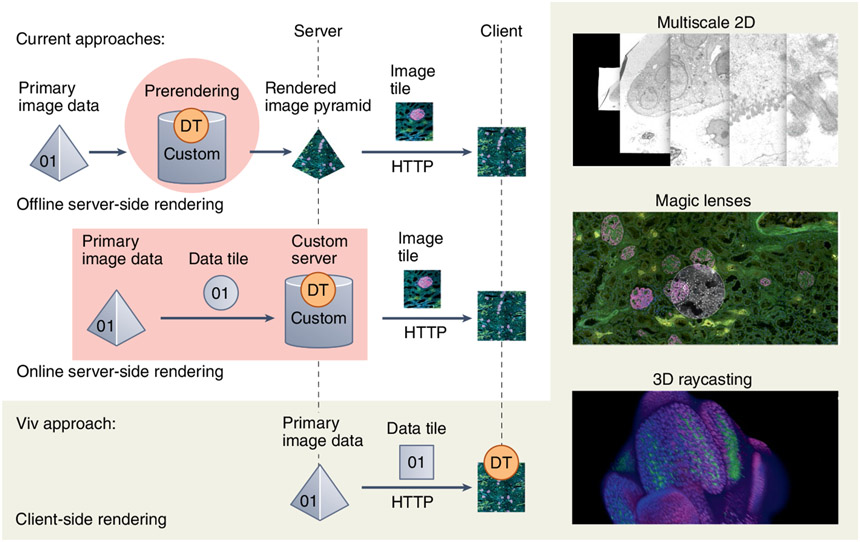
Viv functions more similarly to popular desktop bioimaging applications than to corresponding web alternatives (Supplementary Note 1). Most web viewers require the pre-translation of large binary data files into rendered images (PNG or JPEG) for display in a browser client. Two existing approaches perform this step (server-side rendering) but differ with regard to when rendering occurs and how much of the binary data is transformed at one time (Fig. 1). The offline option performs all rendering before an application is deployed to users, meaning that all channel groupings and data transformations are fixed and cannot be adjusted via the user interface. The online option supports on-demand, user-defined rendering but introduces latency when exploring data transformations and requires active maintenance of complex server infrastructure. Neither approach provides a flexible solution to directly view datasets saved in open formats from large-scale public data repositories, and the transient data representation introduced by server-side rendering inhibits interoperability with other visualization and analysis software.
Fig. 1 ∣. Overview of data flow and rendering approaches for web-based bioimage data visualization and Viv features.
DT indicates the location where data is transformed into an image. The right column displays a subset of Viv’s flexible client-side rendering, including multiscale 2D pyramids, magic lenses and 3D volumes via raycasting.
Viv implements purely client-side rendering to decouple the browser from the server and still offer the flexibility of on-demand multichannel rendering. Existing web viewers also leverage graphical processing unit (GPU)-accelerated rendering but are generally tailored toward single-channel volumetric datasets and, crucially, lack the ability to reuse and compose features for existing or novel applications5,6. In contrast, Viv’s modularity allows core functionality to be repurposed and extended. The library consists of two primary components: (i) data-loading modules for OME-TIFF and OME-NGFF and (ii) configurable GPU programs that render full-bit-depth primary data on user devices.
The data-loading component of Viv is responsible for fetching and decoding atomic ‘chunks’ of compressed channel data from binary files via HTTP. We targeted both OME-TIFF and OME-NGFF to suit the diverse needs of the bioimaging community and promote the use of interoperable open standards. Although OME-TIFF is more ubiquitous, its binary layout and metadata model are too restrictive to efficiently represent large volumes and datasets with high dimensionality. OME-NGFF is designed to address these limitations and is natively accessible from the cloud due to the underlying multidimensional Zarr format (https://doi.org/10.5281/zenodo.3773450). Zarr provides direct access to individual chunks, whereas TIFF requires seeking to access chunks from different planes, introducing long latencies when reading datasets with large c, t or z dimensions3. Similar to approaches popularized in genomics7, we propose indexing OME-TIFF to improve random chunk retrieval times. Our method, Indexed OME-TIFF, improves read efficiency and is implemented as an optional extension to OME-TIFF (https://doi.org/10.6084/m9.figshare.19416344).
Viv’s image layers coordinate fetching data chunks and GPU-accelerated rendering. All rendering occurs on the client GPU, allowing continuous and immediate updates of properties such as color mapping, opacity, channel visibility and affine coordinate transformation without additional data transfer. Library users can modify rendering with custom WebGL shaders, a feature we used to implement ‘magic lenses’8 that apply local data transformations: for example, rescaling brightness or filtering specific channels.
Because all rendering is moved to the browser, Viv does not rely on a server like previous web viewers for highly multiplex and multiscale images, making it flexible and capable of being embedded within a variety of applications (https://doi.org/10.6084/m9.figshare.19416401). We integrated Viv into Jupyter Notebooks (https://github.com/hms-dbmi/vizarr) via Imjoy9 to enable a remote human-in-the-loop multimodal image registration workflow and direct browsing of OME-NGFF images. Viv also underlies the imaging component of the Vitessce single-cell visualization framework10, demonstrating its ability to integrate with and power other modular tools that include imaging modalities. Finally, we developed Avivator (http://avivator.gehlenborglab.org), a stand-alone image viewer that showcases Viv’s rich feature set.
With its extensibility and minimal deployment requirements, Viv presents a novel toolkit with which to build a wide range of reusable bioimaging applications. It is best suited for displaying 2D visualizations of highly multiplex and multiscale datasets, although 3D visualizations via raycasting are also supported. We do not expect Viv to replace existing web-based viewers but instead to shepherd a new generation of web-based visualization and analysis tools to complement desktop software also built around interoperable standards.
Code availability
Viv is open source and available under an MIT license at https://github.com/hms-dbmi/viv and at https://www.npmjs.com/package/@hms-dbmi/viv.
Supplementary Material
Acknowledgements
We would like to thank members of the OME community for their guidance, as well as M. deCaestecker, E. Neumann and M. Brewer from Vanderbilt University and Vanderbilt University Medical Center for their efforts generating the imaging mass spectrometry and microscopy data highlighted in the referenced the use cases. Viv was developed with funding from the US National Institutes of Health (NIH; OT2OD026677, T15LM007092, T32HG002295) and National Science Foundation (NSF; DGE1745303) and the Harvard Stem Cell Institute (CF-0014-17-03). The use cases in this manuscript were supported with additional funding from the NIH (U54DK120058, 2P41GM103391, OT2OD026671) and NSF (CBET1828299).
Footnotes
Competing interests
N.G. is a co-founder and equity owner of Datavisyn. All other authors declare no competing interests
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41592-022-01482-7.
References
- 1.HuBMAP Consortium. Nature 574, 187–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rozenblatt-Rosen O et al. Cell 181, 236–249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore J et al. Nat. Methods 18, 1496–1498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg IG et al. Genome Biol. 6, R47 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saalfeld S, Cardona A, Hartenstein V & Tomancak P Bioinformatics 25, 1984–1986 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boergens KM et al. Nat. Methods 14, 691–694 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Li H Bioinformatics 27, 718–719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bier EA, Stone MC, Pier K, Buxton W & DeRose TD in Proceedings of the 20th Annual Conference on Computer Graphics and Interactive Techniques 73–80 (Association for Computing Machinery, 1993). [Google Scholar]
- 9.Ouyang W, Mueller F, Hjelmare M, Lundberg E & Zimmer C Nat. Methods 16, 1199–1200 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Keller MS, Gold I, McCallum C, Manz T, Kharchenko V, & Gehlenborg N Preprint at OSF Preprints 10.31219/osf.io/y8thv (2021). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.