Abstract
The malp gene of Mycoplasma fermentans is shown to occur in single copy but to encode two discrete translated forms of lipid-modified surface protein that can be differentially expressed on isolates within this species: MALP-2, a 14-amino-acid (2-kDa) lipopeptide with potent macrophage-stimulatory activity (P. F. Mühlradt, M. Kiess, H. Meyer, R. Süssmuth, and G. Jung, J. Exp. Med. 185:1951–1958, 1997), and MALP-404, an abundant, full-length (404-amino-acid) surface lipoprotein of 41 kDa, previously designated P41 (K. S. Wise, M. F. Kim, P. M. Theiss, and S.-C. Lo, Infect. Immun. 61:3327–3333, 1993). The sequences, transcripts, and translation products of malp were compared between clonal isolates of strains PG18 (known to express P41) and II-29/1 (known to express high levels of MALP-2). Despite conserved malp DNA sequences containing full-length open reading frames and expression of full-length monocistronic transcripts in both isolates, Western blotting using a monoclonal antibody (MAb) to the N-terminal MALP-2 peptide revealed marked differences in the protein products expressed. Whereas PG18 expressed abundant MALP-404 with detectable MALP-2, II-29/1 revealed no MALP-404 even in samples containing a large comparative excess of MALP-2. Colony immunoblots with the MAb showed uniform surface expression of MALP-2 in II-29/1 populations. A second MAb to an epitope of MALP-404 outside the MALP-2 sequence predictably failed to stain II-29/1 colonies but uniformly stained PG18 populations. Collectively, these results provide evidence for novel posttranscriptional (probably posttranslational) processing pathways leading to differential intraspecies expression of a major lipoprotein, and a potent macrophage-activating lipopeptide, on the surface of M. fermentans. In the course of this study, a striking conserved motif (consensus, TD-G--DDKSFNQSAWE--), designated SLA, was identified in MALP-404; this motif is also distributed among selected lipoproteins and species from diverse bacterial genera, including Bacillus, Borrelia, Listeria, Mycoplasma, and Treponema. In addition, malp was shown to flank a chromosomal polymorphism. In eight isolates of M. fermentans examined, malp occurred upstream of an operon encoding the phase-variable P78 ABC transporter; but, in three of these isolates, a newly discovered insertion sequence, IS1630 (of the IS30 class), was located between these genes.
Lipid-modified translation products expressed on the surface of mycoplasmas are increasingly recognized as an important class of proteins contributing to fundamental biological and pathogenic processes of these organisms, including adaptive surface variation now widely observed in this group of wall-less procaryotes (3, 4, 28, 61) and modification of assorted host cell functions, characteristic of bacterial modulins (20). First, surface lipoproteins representing abundant mycoplasma coat proteins (4, 27, 50, 66), adhesins (5, 16, 55, 57, 65), and transporters (52) have been shown to undergo rapid, heritable alteration in expression (phase variation) or structure, due to underlying mutational instabilities directly associated with corresponding genes. Genes encoding products with these diverse functions can occur in single copy or as families of related sequences distributed in the limited genome of these organisms. To date, mutations associated with genes encoding primary gene products are the most prevalent mechanisms known to determine population diversity involving mycoplasma surface lipoprotein expression or structural variation. However, evidence has also been presented for unidentified factors that indirectly affect other surface properties in a phase-variable manner, such as the accessibility of specific surface epitopes on proteins that are continually expressed (51, 53). It is likely that additional and novel mechanisms may be employed by mycoplasmas to generate surface diversity in populations, which appears to be a common theme in the survival of these obligate parasitic pathogens. One type of variation not previously reported in mycoplasmas is the use of posttranslational pathways to generate alternative forms of primary lipoprotein gene products.
A second major impact of mycoplasmal lipoproteins lies in the potent immunomodulatory activities associated with the lipid-modified N-terminal region of these processed membrane proteins (33, 35, 36, 38). Due to the relatively large number (relative to the total gene content) and diversity of lipoproteins in mycoplasmas, documented by genomic sequencing (17, 21) and experimental evidence (11, 56), and the lack of other components such as lipopolysaccharide or cell wall constituents in these organisms, lipid-modified proteins are likely to be a primary element responsible for the immunomodulatory role of mycoplasmas that has been extensively documented in their respective animal hosts or in vitro (12, 29, 44). In this regard, the activities, biogenesis, and intraspecies variation of lipoproteins and corresponding lipopeptides are important features to be understood in the analysis of mycoplasmal pathogenesis.
Past studies of the lipoproteins and related products in Mycoplasma fermentans in our laboratories (11, 33, 35, 36, 38, 43, 51–53, 59) and by others (15, 18, 26, 40, 41) have contributed to the understanding of mycoplasmal surface variation and immunomodulatory activities associated with these surface membrane components. In the present report, we document a novel form of surface variation that involves a previously reported major lipoprotein (P41) and a potent macrophage-activating lipopeptide (MALP-2) of M. fermentans. P41 is an abundant amphiphilic lipoprotein, first defined (59) in the type strain PG18 by detergent fractionation and metabolic labeling with [3H]palmitate and [35S]cysteine and shown by monospecific antibody (Ab) to be present also in the strain Incognitus (sb51 isolate). Subsequently, this protein has been identified as a common target of the Ab response to M. fermentans in the human host (22). MALP-2 is a lipopeptide isolated from M. fermentans. It has been rigorously purified, shown to have an exceptionally high macrophage-activating activity (in the pmolar range), and structurally solved (33). Significantly, the MALP-2 lipopeptide contains only two fatty acid chains. This feature was shown to be compatible with a characteristic S-(2,3-di-O-acyloxypropyl) cysteinyl group at the Cys residue in the +1 position (+1 Cys residue) generated after cleavage of a typical procaryotic prolipoprotein (6); but, it is highly distinctive in that a third fatty acid normally linked in other eubacteria by an amide bond to the free amino group of Cys is absent. This is particularly intriguing in light of recent genomic analysis (17, 21) that has failed to identify an ortholog of the bacterial apolipoprotein N-acyl transferase (47) that is responsible for this final acylation reaction, in the two mycoplasma genome sequences reported to date.
This report defines the single gene, malp, that encodes both the P41 and MALP-2 products of M. fermentans and reveals a novel mechanism operating in mycoplasmas that determines the relative expression of these two components on variants within the species, and possibly within clonal populations. The potential role of this variation in determining antigenic and immunomodulatory properties of propagating populations is discussed. In the course of this study, a highly distinctive protein sequence motif distributed selectively among individual lipoproteins of diverse bacterial genera is defined, and a marked chromosomal polymorphism associated with the malp gene is shown to occur in M. fermentans isolates, due to the presence or absence of a newly defined insertion-like element, IS1630.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
M. fermentans PG18 (clone 39) was grown in modified Hayflick medium as described elsewhere (59). The cloned M. fermentans isolate II-29/1 was propagated in GBF-3 medium as described previously (33). Mycoplasma colonies were grown by plating liquid cultures on respective solid medium containing 1% (for PG18) or 0.7% (for II-29/1) Noble agar (Difco Laboratories, Detroit, Mich.). Escherichia coli DH10B (Gibco BRL, Grand Island, N.Y.) and strains containing plasmid pZero2.1 (Invitrogen, Carlsbad, Calif.) and its derivatives were grown at 37°C in Luria-Bertani medium, supplemented with 50 μg of kanamycin per ml when appropriate. The species Mycoplasma hyorhinis SK76 (42), Mycoplasma hominis 1620 (64), and Mycoplasma arginini G230 (provided by Richard DelGiudice) were propagated in Hayflick medium supplemented as described in the respective references.
Synthesis of a MALP-2-specific probe.
With the exception of oligonucleotides MALP-F1 and MALP-R1, purchased from Gibco BRL, the oligonucleotides used in this study were synthesized on a model 3948 nucleic acid synthesis and purification system (Applied Biosystems, Inc. [ABI], Foster City, Calif.) by the University of Missouri Molecular Biology Program DNA Core Facility. Inosine (I)-containing degenerate oligonucleotides MALP-F1 [5′ TG(A/G) GGI AA(C/T) AA(C/T) GA(C/T) GA(A/G) 3′] and MALP-R1 [5′ (C/T)TT (C/T)TC (C/T)TT (A/G)AA IGA IAT (A/G)TT 3′] were synthesized based on the MALP-2 peptide sequence (33). These two primers (25 pmol each) were used in a standard PCR to generate a MALP-2-derived 42-bp product from genomic DNA (50 ng) from M. fermentans II-29/1. The thermocycle parameters included 30 cycles of 94°C for 1 min, 40°C for 1 min, and 72° for 1 min. A gel slice containing this amplicon was excised from a 3% (wt/vol) low-melting-point agarose gel (FMC BioProducts, Rockland, Maine) and melted at 68°C for 10 min, and a fraction was used as the template in a second PCR amplification using the same primers and conditions, but in the presence of digoxigenin-11-labeled dUTP (DIG-dUTP). The resulting DIG-labeled PCR product was used as a probe for Southern analysis and E. coli colony hybridization at 42°C under standard conditions, as outlined by the product supplier (Boehringer Mannheim, Indianapolis, Ind.).
PCR amplification and chromosomal linkage of the malp gene in M. fermentans strains.
The oligonucleotides MALP-F2 (5′ TTG AGA TAT TTA AGC AAA ATA TCT A 3′) and MALP-R2 (5′ ATT TTC CAG CAT TTT TTT GAT 3′) were used to amplify the entire MALP-404 protein coding sequence from genomic DNA from various M. fermentans strains, kindly provided by S.-C. Lo, Armed Forces Institute of Pathology, Washington, D.C. These included DNA preparations from strains K7, MT-2, M39A, M70B, SK5, and Incognitus. DNA template (10 ng) was used in a standard PCR consisting of 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1.5 min. The resulting PCR amplicons were purified by using QiaQuick spin columns (Qiagen, Santa Clarita, Calif.) and sequenced directly with primers MALP-F2 and MALP-R2 and with two additional internal primers.
To determine the linkage of malp to the downstream operon p63-p58-p35-p78 encoding the P78 ABC transporter (52) and to characterize the intervening region, PCR was performed under standard conditions with oligonucleotides MALP-F3 (5′ AAT TTA CCA CAC ATC ACC TGT T 3′) and P63-R (5′ GGT CGG GTT CAT ATA GTC CAA 3′) on genomic DNA samples from various M. fermentans strains.
Inverse PCR methods.
Inverse PCR was used to isolate DNA sequences upstream of the malp gene from M. fermentans PG18. Briefly, approximately 500 ng of genomic DNA was digested with either BglII or EcoRI, purified by the QiaQuick (Qiagen) spin column procedure, and concentrated in vacuo. Dilutions of the DNA sample were self-ligated in a 20-μl volume with 1 U of T4 DNA ligase for 3 to 4 h at room temperature. Samples from the ligation reaction were then amplified under standard PCR conditions using primers MALP-F3 and ORF3-R1 (5′ TAA TAG CAT GAC GAG CTT CAT 3′), for the 1.6-kb fragment from the ligation of EcoRI-generated DNA fragments, and primers MALP-F4 (5′ CTC AGA CCA AGG TAT GAT TCA A 3′) and ORF2-R1 (5′ CTC CAT TAT TTG AGT TGA TTG GAA 3′), for amplifying the 1.6-kb fragment from the ligation of BglII fragments (shown in Fig. 2). The resulting amplicons were purified and sequenced directly by primer walking.
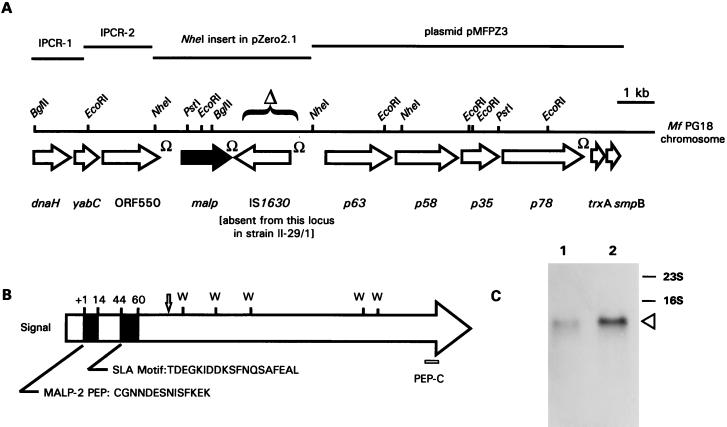
FIG. 2.
Cloning and characterization of the malp-containing region of the M. fermentans PG18 chromosome. (A). A restriction map of the chromosomal region containing the malp gene is shown as a solid horizontal line. The location and direction of each ORF are indicated by a large open arrow, except those for malp, which are indicated by a solid arrow. DNA fragments that were cloned or generated by inverse PCR are indicated by solid lines above the restriction map. Stem-loop structures that may function as transcription terminators are indicated by Ω. IS1630, the 1.4-kb insertion sequence that is absent from the malp-p63 intergenic region in strain II-29/1, is identified by a bracket and Δ. IPCR-1 and IPCR-2 are regions sequenced by inverse PCR as described in Materials and Methods. Plasmid pMFPZ3 was described elsewhere (52). (B) Schematic representation of the MALP-404 prolipoprotein, showing locations of the signal peptide, the MALP-2 lipopeptide sequence and the SLA motif (solid blocks), and a peptide sequence near the C terminus (PEP-C), used to generate a specific PAb. The positions of five Trp residues, all encoded by UGA codons, are indicated by W’s. The vertical arrow indicates the position after which an alternative C-terminal portion of this gene product has been predicted (19). (C) Northern analysis showing expression of a full-length malp transcript in M. fermentans PG18 and II-29/1. Total RNA from liquid cultures of strain PG18 (lane 1) or strain II-29/1 (lane 2) was separated by denaturing agarose gel electrophoresis, transferred to a nylon membrane, and probed with a 536-nt DIG-labeled probe encompassing the MALP-2 peptide sequence. The positions of the 16S and 23S rRNA bands are indicated on the right. The arrowhead indicates the position of the hybridizing ∼1,400-nt transcript.
DNA preparation and hybridization methods.
Genomic DNA from M. fermentans PG18, M. hyorhinis SK76, and M. arginini G230 was prepared as described previously (62). Small-scale preparations of genomic DNA from M. fermentans II-29/1 were prepared by using a Wizard genomic DNA isolation kit (Promega, Madison, Wis.) in accordance with the supplied instructions. For Southern hybridization, genomic DNA was digested to completion with either NheI, PstI, or EcoRI, resolved by electrophoresis in 0.7% (wt/vol) agarose gels, transferred to positively charged nylon membranes (Boehringer Mannheim), and hybridized with a denatured 42-nucleotide (nt) DIG-labeled probe. Hybridization and washing steps were carried out at 42°C (because of the small size and degenerate nature of the probe) in standard buffer solutions described in the guide provided by the manufacturer (Boehringer Mannheim). Hybridizing bands were detected by using nonradioactive detection methods, as described previously (64). Colony hybridization of transformants with DIG-labeled probes was carried out by following the recommendations of the manufacturer of the nonradioactive detection system (Boehringer Mannheim).
Cloning of the DNA sequence encoding MALP-2.
NheI-digested chromosomal DNA from M. fermentans PG18 and II-29/1 was ligated to XbaI-digested pZero2.1 cloning vector under conditions recommended by the supplier. Following transformation into competent E. coli DH10B cells (Gibco BRL), kanamycin-resistant transformants were screened by standard colony hybridization methods with the DIG-labeled 42-bp MALP-2 probe, as described for Southern analysis. Multiple hybridizing clones were isolated for both the 4.1-kb NheI fragment from strain PG18 and the 2.7-kb fragment from strain II-29/1. The nucleotide sequence for each of these NheI fragments was determined by using a combination of specific deletion subclones (generated from restriction sites present within the cloned region and the vector polylinker) and sequence-generated oligonucleotide primers.
DNA sequencing and computer analysis.
DNA sequencing reactions were performed by the University of Missouri Molecular Biology Program DNA Core Facility by using Taq dye terminators and a Prism 377 automated DNA sequencer (ABI). DNA and protein sequences were analyzed by using the GCG software package (Genetics Computer Group, Madison, Wis.).
RNA extraction and analysis.
Total RNA was extracted from mycoplasmas harvested by centrifugation from 15-ml late-logarithmic-phase cultures of M. fermentans, using an RNeasy kit (Qiagen). The RNA (5 μg) was denatured and separated by electrophoresis through formaldehyde-containing agarose gels, by standard procedures (46). The RNA was then transferred to a positively charged nylon membrane (Boehringer Mannheim) by capillary blotting, immobilized by UV cross-linking, and incubated with a denatured DIG-labeled DNA probe derived from the MALP-404 coding sequence (using high-stringency hybridization conditions recommended by the membrane supplier [Boehringer Mannheim]). This 536-nt probe was prepared by PCR using oligonucleotides MALP-2 F5 (5′ TAT TAG GAT TGA GTC CTA TTG CT 3′) and MALP-2 R3 (5′ TTA ATC AAC TTG CAA TTG CAT A 3′) and corresponds to the region between amino acid residues −16 to +160 of MALP-404. Following hybridization, the membrane was washed and developed by using standard procedures for chemiluminescent detection of DIG-labeled molecules (Boehringer Mannheim).
Peptide synthesis and antipeptide antibodies.
Synthetic peptides were prepared as previously described (11, 63) on a model 432A peptide synthesizer (ABI) by using standard Fmoc (9-fluorenylmethylcarbonyl) chemistries. Two synthetic peptides, CGNNDESNISFKEK (the MALP-2 peptide sequence located at the N terminus of the predicted MALP-404 lipoprotein) and CKEAIKMFKELPEDFVKYINSDKALKDGNK (an internal sequence near the C terminus of MALP-404) were synthesized and coupled through an appended N-terminal Cys residue to keyhole limpet hemocyanin (KLH), using a hapten-carrier conjugation kit (Pierce Chemical Co., Rockford, Ill.). Abs to the MALP-2 peptide were generated in BALB/c mice that were injected intraperitoneally three times at weekly intervals with 20 to 50 μg of KLH-peptide emulsified in incomplete Freund adjuvant. Preimmune serum and serum samples taken at least 1 week after the last injection were used at a dilution of 1:200 for immunostaining.
Hybridoma construction and screening.
Monoclonal Ab (MAb) 4444H7.A {immunoglobulin G1(κ) [IgG1(κ)]} to MALP-404 and MAb 4443H2.C [IgG2b(κ)] to P76 were generated by fusion of the myeloma cell line SP2/0 with splenocytes from BALB/c mice immunized with freeze-thaw-disrupted whole organisms of M. fermentans PG18 (51). MAb F208C2B1 [IgG2b(κ)] to the MALP-2 peptide was generated by methods described previously (10). Briefly, 9 months after injection with the KLH–MALP-2 peptide conjugate (described above), a mouse was injected intraperitoneally with 25 μg of the same conjugate dissolved in phosphate-buffered saline. Three days later, splenocytes were fused with the murine myeloma cell line NS-1 (ATCC TIB 18) and hybridomas were selected. Fusions and selection were performed at the University of Missouri—Columbia Molecular Biology Program Cell and Immunobiology Core Facility.
An enzyme-linked immunosorbent assay using immobilized synthetic peptides was used to screen MAbs, as described previously (10). Briefly, a saturating amount of biotinylated peptide was immobilized in 96-well plates (Immulon 2; Dynatech Laboratories, Inc., Chantilly, Va.) that had been previously coated with 0.5 μg of neutral avidin (NeutrAvidin; Pierce Chemical Co.) per well and blocked with phosphate-buffered saline containing 3% bovine serum albumin (fraction V; Fisher Scientific, Fairlawn, N.J.). A biotinylated peptide comprising an irrelevant sequence in VlpA (10) was used as a negative control to ensure Ab specificity. Peroxidase-conjugated, goat anti-mouse IgG (whole molecule; Cappel Organon Teknika, Durham, N.C.) was used as the secondary Ab to select for bound MAb. The MAb isotype was determined by Western immunostaining with secondary Abs specific for murine heavy- and light-chain subclasses (Southern Biotechnology Associates, Inc., Birmingham, Ala.).
Detergent fractionation, PAGE, immunoblotting, and immunoprecipitation.
Triton X-114 (TX-114) phase fractionation was performed as previously described (60), using 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (Pefabloc SC; Boehringer Mannheim) as a protease inhibitor. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blotting of proteins were performed as described previously (42, 53). Mycoplasma colony immunoblotting was performed as described before (60).
To resolve low-molecular-weight proteins and the MALP-2 lipopeptide, a Tris-Tricine buffer system (48) was used with modifications (2), employing a 10% resolving gel and a 4% stacking gel. The bromophenol blue dye front was run to within 1 cm of the end of the 11-cm gel. Prestained protein molecular size standards (2.8- to 43-kDa range; Gibco BRL) were used. Western blotting of these gels was performed by using polyvinylidene difluoride (PVDF)-type membranes with a pore size of 0.1 to 0.2 μm (ABI). Electrophoretic transfer was performed in transfer buffer (48 mM Tris, 39 mM glycine, 0.04% SDS, 20% [vol/vol] methanol [pH 8.6]) at 4°C for 1 h at a current of 0.7 to 0.8 A in a Transphor electrophoresis unit (TE52X; Hoeffer Scientific Instruments, San Francisco, Calif.). The membrane was blocked overnight at 4°C with TS buffer (42) containing 0.05% Tween 20 detergent and 10% newborn calf serum. Membranes were immunostained as described above.
Immunoprecipitation was carried out as described previously (58) by using a detergent phase preparation from M. fermentans PG18 adjusted to 0.05% TX-114 and the precipitating MAb 4444H7.A immobilized on immunosorbent Sepharose 4B beads conjugated to affinity-purified goat antibody to mouse IgG (Cappel Organon Teknika, Durham, N.C.).
Nucleotide sequence accession numbers.
The sequences reported in this paper are deposited in GenBank under accession number AF100324 for the malp genomic region from strain PG18 and accession numbers AF099209 (II-29/1), AF099210 (Incognitus), AF099211 (SK5), AF099212 (MT-2), AF099213 (K7), AF099214 (M39A), and AF099215 (M70B) for the malp genes from the M. fermentans isolates indicated in parentheses.
RESULTS
Identification and cloning of MALP-2-encoding DNA sequences from M. fermentans.
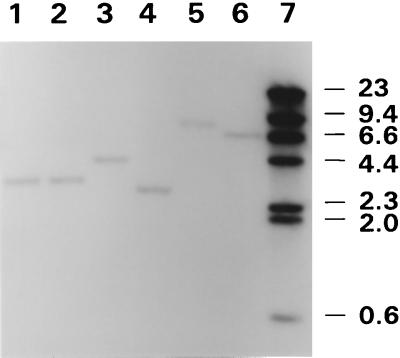
A reverse genetics approach was used to identify and clone the MALP-2-encoding gene from M. fermentans since the 14-amino-acid MALP-2 lipopeptide sequence from strain II-29/1 was known (33). Two opposing, nonoverlapping degenerate oligonucleotides (one 18mer and one 21mer) were used to amplify a 42-bp product, which encompassed the entire 14-amino-acid coding sequence, by using M. fermentans II-29/1 DNA as the template. This product was reamplified with incorporation of DIG-dUTP to generate a probe for Southern hybridization analysis, in order to identify corresponding sequences in genomic restriction digests from M. fermentans PG18 and II-29/1. A single hybridizing band was observed in each strain (Fig. 1). The probe did not identify hybridizing bands in restriction digests of genomic DNA from M. hominis 1620, M. hyorhinis SK76, or M. arginini G230 DNA (data not shown), indicating that closely related sequences were absent from these assorted mycoplasmal species. Since a single hybridizing band was detected in each of several restriction enzyme digests of the M. fermentans genomic DNA analyzed, the MALP-2-encoding sequence most likely occurs at a single location in the genomes of both strains PG18 and II-29/1.
FIG. 1.
Identification of the malp gene sequence in M. fermentans strains. Genomic DNA from M. fermentans PG18 (lanes 1, 3, and 5) or strain II-29/1 (lanes 2, 4, and 6) was digested with EcoRI (lanes 1 and 2), NheI (lanes 3 and 4), or PstI (lanes 5 and 6), transferred to a nylon membrane, and hybridized with a denatured, 42-nt DIG-labeled malp probe. The probe was generated by PCR using degenerate oligonucleotides, based on the MALP-2 lipopeptide sequence, as described in Materials and Methods. Lane 7 contains DIG-labeled λ HindIII markers (Boehringer Mannheim), the sizes of which are indicated in kilobase pairs. The sizes of the single hybridizing fragments in each lane are 3 kb (lanes 1 and 2), 4.1 kb (lane 3), 2.7 kb (lane 4), 8 kb (lane 5), and 6.5 kb (lane 6).
Interestingly, several restriction enzymes yielded hybridizing bands that differed in size between PG18 and II-29/1 (for example, NheI and PstI shown in Fig. 1), whereas other restriction enzymes produced indistinguishable hybridizing fragments in each strain (e.g., EcoRI shown in Fig. 1). These restriction fragment length polymorphisms (RFLP) raised the possibility that variation between these genomes occurred in the local DNA sequence or in the organization of flanking genes, or possibly through size variation within a single gene. This RFLP is addressed further in a later section. To characterize the sequences encoding and flanking MALP-2 (and later to analyze the RFLP), the 4.1-kb NheI fragment from PG18 and the 2.7-kb NheI fragment from II-29/1 were cloned in E. coli, as described in Materials and Methods, and the nucleotide sequence of each fragment was determined.
Features of the malp gene and its predicted product.
The MALP-2 coding sequence occurred once in each of the cloned NheI fragments from M. fermentans PG18 and II-29/1 (Fig. 2A). In each clone, a translated sequence corresponding to the known MALP-2 sequence was found near the 5′ end of a long open reading frame (ORF) (Fig. 2B) that begins with an AUG start codon preceded by a consensus ribosome binding site (AAGGAG) 6 nt upstream. The DNA sequence spanning the ORF is rich in A+T residues (31% G+C) and the ORF contains five UGA (Trp) codons, both features characteristic of mycoplasma genes. The ORF encodes a predicted 48-kDa polypeptide of 428 amino acid residues. The gene containing this long ORF, which includes the sequence coding for the MALP-2 peptide, is designated malp. The predicted protein sequence possesses a typical 25-amino-acid signal sequence containing a motif for lipoprotein cleavage (AVSC) which is predicted to occur on the N-terminal side of the single Cys residue in the ORF. Previous structural characterization of MALP-2 (33) revealed a corresponding 14-amino-acid product with a lipid modification on the +1 Cys, confirming the modification and functional signal cleavage site. The predicted mass of the 404-amino-acid lipoprotein (after signal peptide cleavage) is 45 kDa. This product is here designated MALP-404 on the basis of its encoded sequence. Since there is no stop codon immediately following Lys14 (the C-terminal residue of the MALP-2 lipopeptide), and because Southern hybridization analysis indicated that the DNA sequence encoding MALP-2 is present as a single copy in the chromosome, the MALP-2 lipopeptide is most likely derived from this ORF. Hydropathy analysis indicated that MALP-404 is hydrophilic, with no putative transmembrane domains. Like several other mycoplasma lipoproteins (11), it is predicted to be anchored in the membrane by the hydrophobic Cys-linked lipid modification and has been shown to be oriented on the external side of the single plasma membrane of the organism (59).
A comparison of the malp gene sequences in strains PG18 and II-29/1 revealed a near identity, with only seven nucleotide differences, of which five result in amino acid substitutions at various positions in the product. Therefore, each strain contains an uninterrupted ORF of identical length, with identical sites of translation initiation, termination, and signal processing.
Characterization of the MALP-404 lipoprotein in M. fermentans PG18.
To examine the expression of the MALP-404 lipoprotein, and to define specific reagents for its subsequent analysis, several Abs were used. A previous study from this laboratory (59) identified an abundant lipoprotein of approximately 41 kDa, designated P41. This amphiphilic protein partitioned into the TX-114 phase during detergent fractionation of M. fermentans PG18 organisms and corresponded to a major [3H]palmitate-labeled protein in this fraction. This product was originally identified by hyperimmune antiserum raised against the gel-excised protein (59). Subsequently, MAb 4444H7.A was shown to bind an indistinguishable product in Western blots (unpublished data and Fig. 3A). Using these and other reagents, two strategies were used to establish the identity of the MALP-404 product and its relationship to the previously reported P41 lipoprotein. First (Fig. 3A), a hyperimmune polyclonal antiserum (PAb) raised against the synthetic 14-amino-acid peptide corresponding to the MALP-2 lipopeptide sequence and a subsequent MAb, F208C2B1, derived from this regimen, were used to immunostain Western blots of TX-114-fractionated proteins from M. fermentans PG18. These Abs, which had been shown to specifically bind the synthetic peptide immobilized in an enzyme-linked immunosorbent assay (data not shown), identified an abundant 41-kDa amphiphilic protein of PG18. The mobility of this protein was indistinguishable from that immunostained with MAb 4444H7.A or with a PAb raised against a C-terminal 30-amino-acid synthetic peptide of the predicted MALP-404 lipoprotein (Fig. 3A). Importantly, this latter reagent established that the predicted full-length MALP-404 is expressed and formally defined the reading frame of this malp gene product in M. fermentans PG18.
FIG. 3.
Identification of MALP-404 in TX-114 phase-fractionated proteins from M. fermentans PG18. (A) Total proteins from the TX-114 phase were separated by SDS-PAGE, transferred to nitrocellulose, and immunostained with preimmune mouse serum (lane 2), hyperimmune PAb generated to the MALP-2 synthetic peptide (lane 3) or to a C-terminal synthetic peptide within the MALP-404 ORF (lane 4), hyperimmune PAb to gel-excised 41-kDa protein (lane 5), MAb F208C2B1 to the MALP-2 synthetic peptide (lane 6), or MAb 4444H7.A to a previously described (59) 41-kDa protein (lane 7). As a control to indicate sizes of other proteins in this fraction, a blot strip was also stained with a combination of MAbs to amphiphilic proteins P78, P76, P61, and P38 (lane 1) described previously (59) or in this report (protein P76). (B) Immunoprecipitation of MALP-404 and localization of multiple epitopes on the product. The abundant 41-kDa amphiphilic protein recognized by MAb 4444H7.A was immunoprecipitated with this MAb from TX-114 phase proteins, as described in Materials and Methods. Immunoprecipitated protein (along with precipitating MAb) was separated by SDS-PAGE, transferred to nitrocellulose, and immunostained with a series of Abs, including MAb 4444H7.A used for immunoprecipitation (lane 1), PAb to gel-excised P41 (lane 2), MAb F208C2B1 to the MALP-2 synthetic peptide (lane 3), or PAb to the C-terminal synthetic peptide of the MALP-404 ORF product (lane 4). A negative control, showing the absence of staining by preimmune mouse antisera is also shown (lane 5). The positions of the immunoprecipitating immunoglobulin heavy (H) and light (L) chains (which are lightly stained by the secondary Ab reagent) are indicated on the left.
A second strategy confirmed that each of these Ab reagents recognized MALP-404 rather than possible comigrating products. This was particularly important to determine for the reagents that were not directed against known peptide sequences encoded by malp. Therefore, MAb 4444H7.A was used to immunoprecipitate the 41-kDa product from TX-114-extracted proteins from strain PG18. The immunoprecipitated material was subjected to standard SDS-PAGE and to Western analysis by using a MAb to the MALP-2 synthetic peptide, a PAb to the C-terminal MALP-404 peptide sequence, and a PAb to gel-excised P41 (Fig. 3B). All of these Abs (but not preimmune Ab) specifically recognized the 41-kDa protein immunoprecipitated by MAb 4444H7.A, confirming that the MALP-404 and the abundant P41 lipoprotein detected by this MAb were in fact the same translated product. MAbs to other lipoproteins of M. fermentans revealed no corresponding products coprecipitating with MALP-404 (data not shown). These results therefore established that strain PG18 expresses MALP-404 as a major membrane protein constituent and defined multiple Abs that bind to different regions of this translated product. Notably, MAb 4444H7.A recognizes an epitope on MALP-404 distinct from the N-terminal MALP-2 lipopeptide sequence recognized by MAb F208C2B1.
Intraspecies distribution of the malp gene.
To investigate the presence and possible intraspecies variation of the primary sequence of MALP-2 or MALP-404, the entire malp coding sequence was amplified by PCR from chromosomal DNA from six additional M. fermentans isolates and the amplicons were directly sequenced. The six isolates included the Incognitus strain of M. fermentans, clonal isolates from individuals with AIDS (SK5) or acute respiratory disease (M39A and M70B), and two clonal isolates derived (MT-2) or potentially derived (K7) from contaminated tissue culture. Comparison of the deduced MALP-404 amino acid sequences from these six isolates, together with those determined for PG18 and II-29/1, revealed no interruption or frameshift in the coding sequence, and only minor differences in the primary sequence, with none occurring in the MALP-2 peptide region. The ORFs from II-29/1 and PG18 were the most dissimilar (differences in seven nucleotides and in five amino acids in the encoded product), whereas sequences from M39A, M70B, and PG18 were identical at the DNA level. Sequences of the malp gene from strains MT-2 and Incognitus differed from each other by only one silent nucleotide substitution, and their deduced translation products differed from that encoded in the PG18 genome by four amino acids. The deduced MALP-404 sequences from strains MT-2 and Incognitus are distinctive in that each contains an additional Ala residue, following Ala261 (due to a directly repeated GCA sequence), that is absent from the six other MALP-404 sequences analyzed.
Definition of a lipoprotein motif selectively distributed among diverse bacteria.
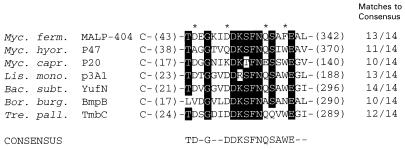
A FastA search of GenBank with the MALP-404 sequence revealed similarity to products encoded by ORFs from different bacteria. The most extensive similarity (32% identity over 175 residues; P = 2 × 10−21) was found between MALP-404 and the Ag 243-5 lipoprotein, originally described (54) as a product of M. arginini but recently shown in studies from our laboratory (13) to be the P47 lipoprotein of M. hyorhinis, to which the originally described feature of tumor metastasis promotion in a nude mouse model (54) has now been ascribed. The lipoprotein P39 (also designated BmpB) of Borrelia burgdorferi also contains an extended region of similarity to MALP-404 (27% identity over 180 amino acids; P = 2 × 10−8). In addition to the modest similarity exhibited over extended regions of these two lipoproteins, a 19-amino-acid region of striking local sequence conservation was shared by MALP-404 and, surprisingly, several additional lipoproteins from diverse bacteria (Fig. 4). This sequence, designated the SLA (selective lipoprotein-associated) motif, was found in MALP-404, P47, and BmpB ORFs, as well as ORFs encoding putative lipoproteins of unknown function from Bacillus subtilis, Treponema pallidum, Mycoplasma capricolum, and Listeria monocytogenes. Interestingly, the conserved motif is similarly positioned in all of these lipoproteins, occurring from 17 to 43 residues downstream of the predicted N-terminal lipid-modified +1 Cys residue of each ORF. The significance of this motif is currently unknown, but the conservation of its sequence and relative position argues for some hitherto-unknown function associated with the specific lipoproteins containing it. These selected lipoproteins are not ubiquitous in mycoplasmas or other bacteria, since the motif could not be identified in the complete genome sequences from the mycoplasmas M. genitalium or M. pneumoniae, the gram-negative bacteria, E. coli, Haemophilus influenzae, or Helicobacter pylori, or any of the sequenced archeons.
FIG. 4.
Amino acid sequence alignment of the SLA motif of various lipoproteins. SLA motif-containing lipoproteins were identified and aligned by the FastA search program (GCG). The sequences of MALP-404 from M. fermentans, P47 from M. hyorhinis (13) (GenBank accession no. D16674), P20 deduced lipoprotein from M. capricolum (Z33368), p3A1 protein from L. monocytogenes (S80336), YufN from B. subtilis (Z93937), P39 or BmpB from Borrelia burgdorferi (L24194 and U49938, respectively), and TmbC lipoprotein from T. pallidum (X57836) are shown. Amino acid residues present in at least six of the seven lipoproteins are highlighted with a black background, and residues conserved among five of the lipoproteins are indicated with an asterisk. A consensus SLA motif sequence was derived from the seven sequences and is shown below the sequence alignment. The numbers in parentheses to the left indicate the number of residues between the putative lipid-modified +1 Cys residue and the first amino acid of the motif. Those in parentheses to the right indicate the number of residues between the last residue of the motif and those encoded by the stop codon of the ORF.
Organization and IS1630-associated polymorphism of the malp region of the M. fermentans chromosome.
Sequence analysis of the 4.1- and 2.7-kb NheI fragments containing the malp genes of strains PG18 and II-29/1, respectively, revealed that the RFLP observed in Southern analysis (Fig. 1) is not due to differences in the malp DNA sequences but instead results from the presence of a newly discovered insertion sequence (IS)-like element immediately downstream of the malp gene in PG18 (Fig. 2A), which is absent from this corresponding region of the genome in strain II-29/1. Interestingly, this element, designated IS1630, is oriented in the opposite direction from all other genes in this region of the M. fermentans PG18 chromosome (Fig. 2A). IS1630 is distinct from the IS-like element described previously in M. fermentans by Lo and coworkers (23) and is related to a characteristic group typified by IS30 of E. coli (8). Since IS1630 is approximately 1.4 kb in size, its absence from the NheI fragment from II-29/1 accounts for the observed 1.4-kb size difference between the cloned 2.7- and 4.1-kb NheI fragments from II-29/1 and PG18, respectively, as well as the 1.4-kb size difference between the respective PstI fragments from these strains that hybridize with a MALP-2-specific probe (Fig. 1 and 2A). IS1630 is not restricted to the malp region of the M. fermentans chromosome, as Southern analysis revealed that the IS element is present in multiple copies in the genomes of both strains PG18 and II-29/1 (data not shown).
Analysis of the sequence downstream of malp indicated that this NheI fragment overlaps a genomic region containing four genes, p63-p58-p35-p78, that comprise an ABC transporter operon (52). Further PCR analysis using forward and reversed primers in the malp and p63 gene regions, respectively (Fig. 2A), confirmed the presence of this gene organization in both strains PG18 and II-29/1. Application of this PCR analysis to the additional six isolates of M. fermentans revealed (data not shown) that in all cases, malp was linked to p63 and that IS1630 was present in the intergenic region in two of the six strains tested, M39A and M70B. In contrast, IS1630, although detected by PCR in the genome of all M. fermentans isolates used in this study (data not shown), was absent from this intergenic region in the remaining four strains, Incognitus, MT-2, SK5, and K7. The presence of identical malp DNA sequences and equivalent malp-IS1630 linkages between strains M39A, M70B, and PG18 suggests that these three strains may be very closely related. This notion is consistent with the results of Campo et al. (9) from Southern analysis of these three strains, using restriction polymorphisms associated with an unrelated IS element.
To determine further the genomic context in the region of malp in strain PG18, inverse PCR and DNA sequencing were used to characterize three ORFs 5′ to the malp gene. In addition, sequencing of a previously isolated p78-containing plasmid clone (52) was carried out to identify two ORFs 3′ to p78. Each of the five additional ORFs identified in the contiguous 15-kb region shown in Fig. 2A had identifiable orthologs in other mycoplasmas, but none was predicted to be a lipoprotein or other translocated surface protein. The genes were dnaH, yabC, and orf550 upstream of malp and trxA and smpB downstream of p78. The presence of flanking genes encoding the delta subunit of DNA polymerase III (dnaH) and thioredoxin (trxA) indicated that the genomic region surrounding malp is chromosomal. The closest homologs and other features of the ORFs from this chromosomal region are summarized in Table 1. Interestingly, the malp gene and IS1630 are the only sequences present in this chromosomal region that do not have orthologous counterparts in the complete published genome sequences of Mycoplasma genitalium (17) and Mycoplasma pneumoniae (21).
TABLE 1.
Products of ORFsa in chromosomal region of malp in M. fermentans PG18
| ORF product designation | Size (amino acids) | Closest homolog (identity)b | MG homologc | MP homologd | Motifse |
|---|---|---|---|---|---|
| DnaH | 305 | DnaH, B. subtilis (21/256) | MG007 | D12_orf253 | NDf |
| YabC | 236 | HI1654, H. influenzae (42/223) | MG056 | D09_orf276 | UPF0011 |
| ORF 550 | 550 | MG423, M. genitalium (28/550) | MG423/139 | C12_orf561 | ND |
| MALP-404 | 428 | P47, M. hyorhinis (32/175) | ND | ND | LP + SLA |
| TnpA IS1630 | 387 | Transposase IS30, E. coli (27/349) | ND | ND | ND |
| P63g | 601 | YufO, B. subtilis (38/597) | MG119 | A65_orf572 | ATP |
| P58g | 537 | MG120, M. genitalium (27/434) | MG120 | A65_orf517 | ND |
| P35g | 327 | MG121, M. genitalium (35/321) | MG121 | A65_orf311 | ND |
| P78g | 681 | ND | ND | ND | LP |
| TrxA | 104 | TrxA, Cyanidium sp. (32/96) | MG124 | A65_orf102 | ND |
| SmpB | 147 | SmpB, E. faecalis (40/147) | MG059 | D09_orf147 | ND |
Relative position and orientation of each ORF are indicated in Fig. 2.
Basis for homology is given as number of identical amino acid residues/total number of amino acid residues in region (FastA).
MG homolog is from the M. genitalium database; designations refer to a system described previously (17).
MP homolog is from the M. pneumoniae database; designations refer to a system described previously (21).
Motif abbreviations: UPF0011, uncharacterized protein family 0011 (as defined in the GCG Motif program); LP, lipoprotein signal peptide and cleavage motif; SLA, selective lipoprotein-associated motif (this work); ATP, ATP binding site.
ND, none detected.
ORF products previously characterized (52).
Immunological detection of the MALP-2 lipopeptide and intraspecies variation in the levels of malp-encoded products.
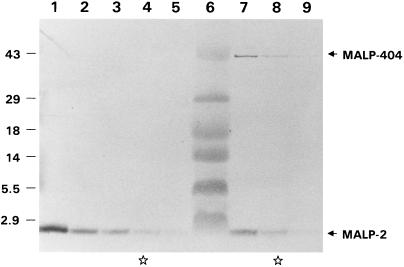
The data presented above indicate that the MALP-2 lipopeptide is expressed from a larger ORF that encodes MALP-404, an abundant TX-114 phase membrane surface lipoprotein of M. fermentans PG18 (59). In a previous report (33), the potent macrophage-stimulating activity attributed to the purified MALP-2 lipopeptide was shown to vary between PG18 and the high producer, isolate II-29/1. To further examine this variability between isolates, we sought to directly monitor the expression of the MALP-2 lipopeptide and MALP-404 lipoprotein, using specific MAb reagents. By employing Tris-Tricine gels for resolution of the lipopeptide, PVDF membranes to bind the lipopeptide after electrotransfer, and F208C2B1, the potent MAb to the MALP-2 synthetic peptide, a strong immunostaining band, which migrated close to the established (33) 2-kDa mass of MALP-2, could be readily detected in total protein extracted from either strain PG18 or strain II-29/1, (Fig. 5). As expected, the MAb to the MALP-2 peptide also immunostained the 41-kDa polypeptide in strain PG18, representing MALP-404. Surprisingly, no MALP-404 could be detected in total cellular protein (Fig. 5, lanes 1 to 5) or in the TX-114 phase protein fraction (data not shown) of II-29/1. The lack of staining of MALP-404 in II-29/1 is not due to differences in the epitope recognized by the MAb, since the MALP-2 lipopeptide sequence deduced from the DNA sequence is identical in each strain. In additional experiments, MALP-404 could not be detected in total protein extracts of II-29/1 with MAb 4444H7.A, polyclonal anti-P41, or the PAb raised against the C-terminal synthetic peptide of MALP-404 (data not shown). These data suggest that under the conditions employed, the only product of the malp gene detectable in II-29/1 is the MALP-2 lipopeptide.
FIG. 5.
Strain variation in the relative levels of MALP-2 and MALP-404. Total proteins from late-logarithmic-phase cultures of M. fermentans II-29/1 (lanes 1 to 5) or PG18 (lanes 7 to 9) were separated through a Tris-Tricine gel, transferred to a PVDF membrane, and immunostained with MAb F208C2B to the MALP-2 peptide sequence as described in Materials and Methods. Twofold dilutions of each protein sample were analyzed over a range that included equivalent immunostaining of the MALP-2 lipopeptide. The most concentrated sample from PG18 (lane 7) contained 10 μg of protein. Lane 6 contains low-molecular-weight markers (Gibco BRL), the sizes of which (in kilodaltons) are indicated on the left. The positions of the MALP-2 lipopeptide and the 41-kDa MALP-404 are indicated on the right. Lanes indicated by stars contain samples of the two strains with approximately the same amount of immunostained MALP-2.
To compare the relative ratios of MALP-2 to MALP-404 between strains PG18 and II-29/1, a series of twofold dilutions of total protein from each strain was subjected to SDS-PAGE and Western analysis with the MAb to MALP-2. The results (Fig. 5) show that whereas MALP-404 can be detected in the most-dilute protein sample from strain PG18 shown (2.5 μg of total protein), MALP-404 is not detected in extracts from II-29/1, even in samples containing eightfold-greater amounts of MALP-2. Therefore, the relative steady-state ratio of MALP-404 to MALP-2 is dramatically lower in II-29/1 (if MALP-404 is expressed at all) than in PG18. Furthermore, this analysis revealed no additional polypeptide species containing the MALP-2 peptide in either strain, indicating that alternative forms or possible intermediates of processing are either absent or occur at undetectable levels.
Distribution of malp products in clonal populations.
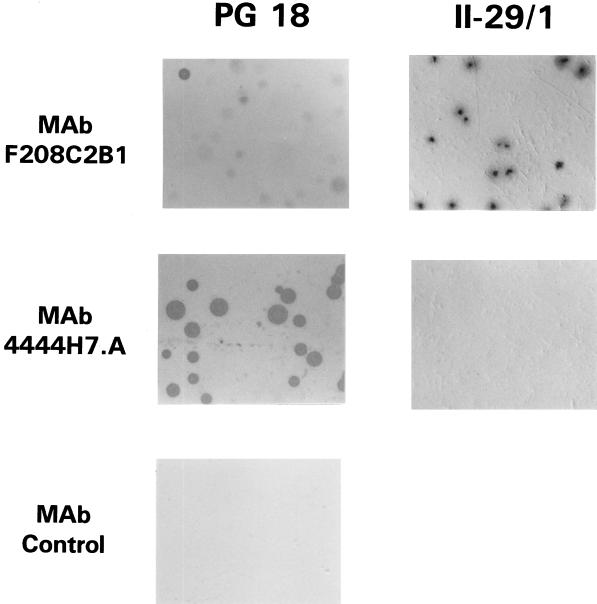
Although the mycoplasmal isolates analyzed in this study had been cloned during isolation, the distribution of malp translation products was examined further to assess the possibility of differential expression of MALP-2 and MALP-404 within propagating clonal populations. In particular, Western blot analysis of the two strains was uninformative in resolving whether all of the cells in the broth culture from PG18 express MALP-2 and MALP-404 or whether variation in the expression pattern within the population occurred during growth in liquid medium. Similarly, the actual proportion of cells in the II-29/1 population that are producing MALP-2 could not be determined by such analysis. To address these questions, a portion of the broth cultures used for Western analysis was plated onto solid medium and the resulting colonies were analyzed by colony immunoblotting with the MAb to the N-terminal MALP-2 peptide, F208C2B1, or MAb 4444H7.A, which recognizes a distinct epitope on MALP-404 outside the region of this N-terminal peptide and is therefore operationally specific for the full-length product identified previously (Fig. 3 and 5). As shown in Fig. 6, colonies derived from the PG18 culture all strongly immunostained with MAb 4444H7.A but stained with variable intensity when the MAb to the MALP-2 peptide, F208C2B1, was used. In contrast, all colonies derived from the II-29/1 culture strongly immunostained with the MAb to the MALP-2 peptide, but none showed any staining with MAb 4444H7.A. These data indicate that in strain II-29/1, each colony (i.e., the great majority of cells within a colony) expresses surface-associated MALP-2 lipopeptide but no (or undetectable levels of) MALP-404, whereas in strain PG18, each CFU gives rise to a colony expressing the longer form of the malp gene product bearing the MAb 4444H7.A epitope. The strong, uniform staining of PG18 by this MAb, in contrast to the variable staining by the MAb to the MALP-2 peptide, is also noteworthy. Specifically, while the expression of MALP-404 appears to occur throughout the PG18 population, the variable staining with the MAb to the N-terminal MALP-2 sequence could reflect various means of surface masking of this epitope within the MALP-404 product and/or the differential expression of the shorter, exposed MALP-2 lipopeptide by some organisms in the population. Masking mechanisms have been documented in strain PG18 (51, 53). In any case, the results presented in Fig. 5 and 6 indicate that whereas MALP-404 is a significant component of the surface architecture of M. fermentans PG18, it is not present in detectable quantities either in liquid cultures or on colonies of strain II-29/1 taken at early passage after initial cloning (33). Moreover, the ability to immunostain MALP-2 lipopeptide on II-29/1 colonies suggests that this abundant macrophage-activating component is accessible on the surface of these mycoplasmas, apparently as a membrane-anchored lipopeptide.
FIG. 6.
Population analysis of the expression profile of surface-associated MALP-2 and MALP-404. Colony blots were prepared from clonal populations of M. fermentans PG18 (left panels) or II-29/1 (right panels) and immunostained with MAb F208C2B1 to the MALP-2 peptide (upper panels) or MAb 4444H7.A to a distinct epitope on MALP-404 (middle panels). Each panel represents an equivalent area taken from the same blot of a single plate of colonies from the strain indicated. The size of colonies ranges from 0.2 to 0.5 mm in diameter. As a negative control to rule out nonspecific staining, a third panel from the strain PG18 immunoblot was incubated with an irrelevant isotype-matched MAb to P70 of M. hyorhinis (7) (MAb control).
Transcriptional analysis of the malp gene suggests a posttranscriptional mechanism determining differential expression of MALP-2 and MALP-404.
Downstream of the MALP-404 coding sequence is a DNA sequence that is predicted to form a stem-loop structure (ΔG = −11.3 kcal) that may function as a transcription terminator. This feature is present both in strain PG18 (containing IS1630 in this region) and in II-29/1 (lacking IS1630 in this region). This structure, together with the absence of closely linked and similarly oriented ORFs in the vicinity of malp, suggests that the MALP-404 ORF may be translated from a monocistronic mRNA. Further examination of the coding sequence and the upstream putative promoter region failed to suggest any mechanism to explain the differential expression (between PG18 and II-29/1) of the MALP-2 lipopeptide and MALP-404, either at the DNA or transcriptional levels. Nevertheless, to directly characterize the malp transcript in each strain, total RNA was extracted from cultures of strain PG18 or early-passage II-29/1 and subjected to Northern analysis using a 536-nt probe encompassing the portion of MALP-404 between amino acid residues −16 to +160. The results of this analysis (Fig. 2C) revealed a single hybridizing band of approximately 1.4 kb in each strain. This is consistent with the translation of MALP-404 from a monocistronic mRNA and suggests that the IS1630 does not affect the overall size of the malp transcript. In conjunction with the full-length ORFs present in PG18 and II-29/1, these results further support a posttranscriptional and probably posttranslational mechanism that dictates the strain differences in expression of the MALP-2 versus MALP-404 products from this gene.
DISCUSSION
This study identifies a single gene encoding the immunomodulatory MALP-2 lipopeptide and the 41-kDa MALP-404 surface lipoprotein of M. fermentans and documents key features of translation and processing of these products, as well as marked differences in their resultant patterns of expression on different isolates of this species. The structure, topology, and variable posttranslational processing of malp-encoded products, and the polymorphic nature of the malp chromosomal locus, all underscore a potentially novel role for this mycoplasmal system in adaptation and pathogenesis in the human host.
Based on previous evidence (33, 59) and the results of the present study, the two malp-encoded translation products are predicted and demonstrated to be surface associated and lipid modified, based on colony immunoblot analysis, detergent phase partitioning, direct structural determination (for MALP-2), and [3H]palmitate labeling and accessibility to surface proteolysis (for MALP-404). The malp gene sequence revealed a signal peptide and a lipoprotein cleavage site, AVSC, very similar to those targeting membrane translocation and acylation of other mycoplasma lipoproteins (11, 56). Matrix-associated laser desorption/ionization time of flight mass spectrometry of MALP-2 indicated that this signal was cleaved and that the acylation of MALP-2 was atypical for procaryotes in that the N-linked acylation was absent. The ability to directly sequence a product likely to be related to MALP-404 (discussed below) implies that the full-length lipoprotein may have these same N-terminal features (19). A further example of macrophage-stimulatory lipoproteins and lipopeptides with two O-ester-linked fatty acids and a free N terminus on the +1 Cys comes from recent studies with M. hyorhinis (34). It is conceivable that lipoproteins from mollicutes of the genus Mycoplasma may generally contain only two acyl groups on the +1 Cys. This could have general ramifications in affecting properties of lipoproteins that determine their ability to interact with and partition into the single mycoplasmal lipid bilayer membrane, as well as in their solubility and micellar formation, all possibly related to the potency of their immunomodulatory activity.
Colony immunoblot analysis of PG18 indicated that virtually all or a great majority of cells express MALP-404. Since Western blot analysis confirmed that MALP-2 is present in PG18, at least some cells must produce both products. The pathway for the biogenesis of MALP-2 in PG18 is not known, but since only full-length mRNA species could be detected from malp, the mechanism must operate co- or posttranslationally, although mechanisms hitherto unknown in procaryotes, such as RNA editing, cannot be formally discounted. Two quite distinct pathways can be envisioned. In one model, the MALP-2 lipopeptide is derived by processing of the larger MALP-404 product. In this case, the full-length 41-kDa polypeptide is translated, lipid modified, and then cleaved by signal peptidase II to remove the signal peptide, prior to the separate processing event(s) that generate MALP-2. A slight variation on this model entails processing of the lipid-modified 41-kDa molecule to MALP-2, prior to signal peptide cleavage. In either scenario, processing to MALP-2 must occur after translocation through the membrane. An alternative cotranslational model invokes a cytoplasmic processing apparatus that acts upon the translation product (perhaps as the nascent peptide) prior to membrane translocation, lipid modification, and signal peptide cleavage. In neither model is it clear what determines the size of the final surface-associated MALP-2 product or whether a precise site of processing is recognized. The simplest prospective candidate for the processing step is an endoprotease, specifically cleaving at a single site C-terminal of Lys14 (the terminal residue in MALP-2). This is consistent with a lack of detectable intermediates, which might otherwise be anticipated if nonspecific proteolytic cleavage occurred.
In contrast to the two translation products seen in M. fermentans PG18, only the MALP-2 lipopeptide was detected in extracts of the clonal isolate II-29/1 despite the presence of a full-length mRNA. This result defines an important, novel form of variation, wherein the absence of a major antigen, but prevalence of an immunomodulatory lipopeptide, occurs on the mycoplasma surface as a result of posttranslational modification. Although the precise mechanism and steps of biosynthesis are not known, it is useful to speculate that strain variability in the levels of MALP-404 and MALP-2 may reflect different extents of processing, possibly due to (i) differential access to, or presence of, a specific protease; (ii) different levels of components that specifically inhibit such a protease, or that interact with the substrate in a way to render it resistant to processing to the MALP-2 form; or (iii) differences in the efficiency of processing this specific lipoprotein through the secretory apparatus of the cell, thereby leading to differences in degradation of the larger product during transit. These represent likely and testable hypotheses.
Posttranslational processing adds a novel level of complexity to our understanding of mycoplasma lipoproteins and surface antigenic variation within a species; however, the extent to which this mechanism causes intraspecies variation has yet to be explored. Reports suggest that a full-length MALP-404 protein may be expressed in strain Incognitus (59) and in two M. fermentans-contaminated human cell lines (19, 31) (see below), but the presence of the shorter MALP-2 product was not assessed in these studies. That variation in posttranslational processing underlies further complexities in propagating populations is suggested by our recent observation (25) that upon increased passage of II-29/1 clones in liquid culture, variants that express MALP-404 arise, as measured both by Western and colony immunoblotting with MAb 4444H7.A, the reagent specific for the longer form. It is not yet known whether these clonal variants are capable of reversing this phenotypic switch (the hallmark of phase variation), but if so, this would confer a phase-variable phenotype based on differential processing of a primary gene product. Although the precise mechanism that results in the emergence of the MALP-404-expressing variants is not known, our initial PCR analysis indicates that the malp gene organization is maintained in such populations, without an insertion of IS1630. This result rules out any correlation between the presence or absence of a malp-linked copy of IS1630 and the predominant translation product that is expressed. While intraclonal variation of MALP-404 expression is yet to be fully characterized, two important technical points are addressed by the emergence of MALP-404 in propagating populations of II-29/1. First, the immunostaining of these mycoplasmas with MAb 4444H7.A demonstrates that the cognate epitope is present in the MALP-404 product encoded by the malp gene of this strain and that the lack of positive staining of the colony immunoblot shown in Fig. 5 is not due to amino acid substitutions affecting the MAb 4444H7.A epitope. Second, such populations formally demonstrate that the strain and the conditions used for cultivation do not lead to any generalized proteolytic degradation of surface proteins, including MALP-404. Thus, the biogenesis of MALP-2 is unlikely to be a result of generalized proteolytic activity. It may be of interest to note that in at least one clone of another mycoplasma species, M. hyorhinis, truncated forms of the variable lipoprotein VlpA exist in addition to the longer forms of this Vlp (34).
Two recent studies reporting genes with sequences similar to malp are noteworthy, in the context of assigning immunological roles to MALP-404 or its various regions and as a cautionary note regarding the attribution of well-known immunological properties of whole mycoplasmas in tissue culture to this specific translation product. In one study (19, 24), a protein associated with (and initially thought to be derived from) the human HL60 promyelocytic cell line was subsequently recognized as a gene product of M. fermentans (19) to which the property of promoting proliferation and activation of HL60 cells was attributed. Interestingly, the gene sequence, derived from a cDNA library of infected cell cultures, predicted a product that resembled the N-terminal region of MALP-404 but reflected another reading frame product following amino acid position 90 (see Fig. 2), due to the absence of a base in the DNA sequence corresponding to this position. Our study revealed no such alternative form of a malp gene product in eight strains examined, and the bona fide reading frame for MALP-404 was confirmed by its immunostaining with an Ab to our predicted C-terminal peptide sequence. In a second series of studies, Matsumoto et al. incorrectly identified the MALP-404 gene sequence as a human cell product in which UGA codons were suggested to encode selenocysteine and proposed that a complement-activating property found in cell culture supernatants was associated with this soluble protein product (31). These authors recently reassigned the origin of the gene as M. fermentans (30, 49), based on its high sequence similarity with the sequence reported by Hall et al. (19). Although not formally identifying their tissue culture contaminants as M. fermentans, Matsumoto et al. also suggested that the gene and corresponding antigen were represented in several cultures displaying complement-activating properties (30). Finally, these authors showed Southern hybridization of DNA from contaminated lines that generate multiple sizes of restriction fragments (30). Whether this represents malp-linked polymorphisms associated with multiple contaminants of M. fermentans, or possibly other mycoplasmal species, is not clear. Overall, these reports underscore the need for extremely rigorous definition of (i) the activities measured, (ii) the source of the components with the activity, (iii) the physical nature and state of the moiety endowing the property, and (iv) high levels of purity (for authentic or recombinant products) to avoid confounding modulatory compounds present in most eubacteria, including multiple mycoplasmal species (20). With these strict requirements in mind, we suggest that the lipid moiety of MALP-2 is the critical element in macrophage activation and that the solubility and differential expression of MALP-2 and MALP-404 may provide critically discrete physical forms of this product that could affect the antigenic properties and the immunomodulatory properties of M. fermentans. We have not ruled out other activities associated with the protein sequence of MALP-404, but currently see no definitive evidence for such activities.
One of the most intriguing aspects of the MALP-404 sequence is the SLA motif. A cursory examination of the known binding motifs of human major histocompatibility complex molecules (39) reveals that the SLA sequence contains the nonapeptide motif -G------F, which could be presented to T cells by the human class I major histocompatibility complex molecule HLA-B*5101, provided that this peptide can be generated from SLA after appropriate processing by the antigen-presenting cells. Incidentally, there exists a rare multifaceted autoimmune disease, Behcet’s disease, which is strongly HLA-B51 associated in many ethnic groups (14, 32). The etiology of Behcet’s disease is unknown, but infectious agents have been discussed as a possible cause from early on, as recently reviewed (45). It is tempting to speculate that this rheumatic autoimmune disease may be triggered in susceptible B51-positive patients by protein antigens from quite different genera of eubacteria expressing the conserved SLA sequence. Whether this motif is associated with an immunological activity per se or might serve other functions in the organism, such as a recognition motif for proteolytic processing to MALP-2, analogous to other motifs involved in regulatory proteolysis (1, 37), is currently under investigation.
ACKNOWLEDGMENTS
We thank Shyh-Ching Lo, Joseph Tully, and Richard DelGiudice for providing mycoplasmal isolates and DNA preparations and Jeong Im for preparation of peptides and conjugates.
This work was supported in part by U.S. Public Health Service grant AI32219 (to K.S.W.) from the National Institute of Allergy and Infectious Diseases and by grant Mu 672/2-2 (to P.F.M.) from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Alley M R, Maddock J R, Shapiro L. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- 2.Applied Biosystems. User bulletin no. 42. Foster City, Calif: Applied Biosystems, Inc.; 1991. ProBlott™ applications in SDS-PAGE, electroblotting and protein sequencing; pp. 1–12. [Google Scholar]
- 3.Behrens A, Heller M, Kirchhoff H, Yogev D, Rosengarten R. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect Immun. 1994;62:5075–5084. doi: 10.1128/iai.62.11.5075-5084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhugra B, Voelker L R, Zou N, Yu H, Dybvig K. Mechanism of antigenic variation in Mycoplasma pulmonis: interwoven, site-specific DNA inversions. Mol Microbiol. 1996;18:703–714. doi: 10.1111/j.1365-2958.1995.mmi_18040703.x. [DOI] [PubMed] [Google Scholar]
- 5.Boesen T, Emmersen J, Jensen L T, Ladefoged S A, Thorsen P, Birkelund S, Christiansen G. The Mycoplasma hominis vaa gene displays a mosaic gene structure. Mol Microbiol. 1998;29:97–110. doi: 10.1046/j.1365-2958.1998.00906.x. [DOI] [PubMed] [Google Scholar]
- 6.Braun V, Wu H C. Lipoproteins, structure, function, biosynthesis and model for protein export. In: Ghuysen J-M, Hakenbeck R, editors. New comprehensive biochemistry. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 319–341. [Google Scholar]
- 7.Bricker T M, Boyer M J, Keith J, Watson-McKown R, Wise K S. Association of lipids with integral membrane surface proteins of Mycoplasma hyorhinis. Infect Immun. 1988;56:295–301. doi: 10.1128/iai.56.2.295-301.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calcutt, M. J., and K. S. Wise. Unpublished results.
- 9.Campo L, Larocque P, La Malfa T, Blackburn W D, Watson H L. Genotypic and phenotypic analysis of Mycoplasma fermentans strains isolated from different host tissues. J Clin Microbiol. 1998;36:1371–1377. doi: 10.1128/jcm.36.5.1371-1377.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citti C, Kim M F, Wise K S. Elongated versions of Vlp surface lipoproteins protect Mycoplasma hyorhinis escape variants from growth-inhibiting host antibodies. Infect Immun. 1997;65:1773–1785. doi: 10.1128/iai.65.5.1773-1785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleavinger C M, Kim M F, Im J H, Wise K S. Identification of mycoplasma membrane proteins by systematic TnphoA mutagenesis of a recombinant library. Mol Microbiol. 1995;18:283–293. doi: 10.1111/j.1365-2958.1995.mmi_18020283.x. [DOI] [PubMed] [Google Scholar]
- 12.Cole B C, Naot Y, Stanbridge E J, Wise K S. Interactions of mycoplasmas and their products with lymphoid cells in vitro. In: Razin S, Barile M F, editors. The mycoplasmas. 4. Mycoplasma pathogenicity. New York, N.Y: Academic Press, Inc.; 1985. pp. 203–257. [Google Scholar]
- 13.Droesse, M., and K. S. Wise. Unpublished results.
- 14.Falk K, Rotzschke O, Takiguchi M, Gnau V, Stevanovic S, Jung G, Rammensee H G. Peptide motifs of HLA-B51, -B52 and -B78 molecules, and implications for Behcet’s disease. Int Immunol. 1995;7:223–228. doi: 10.1093/intimm/7.2.223. [DOI] [PubMed] [Google Scholar]
- 15.Feng S-H, Lo S-C. Induced mouse spleen B-cell proliferation and secretion of immunoglobulin by lipid-associated membrane proteins of Mycoplasma fermentans incognitus and Mycoplasma penetrans. Infect Immun. 1994;62:3916–3921. doi: 10.1128/iai.62.9.3916-3921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsyth M H, Tourtellotte M E, Geary S J. Localization of an immunodominant 64 kDa lipoprotein (LP 64) in the membrane of Mycoplasma gallisepticum and its role in cytadherence. Mol Microbiol. 1992;6:2099–2106. doi: 10.1111/j.1365-2958.1992.tb01383.x. [DOI] [PubMed] [Google Scholar]
- 17.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrmann J, Nguyen D, Utterback T R, Saudek D M, Phillips C A, Merrick J M, Tomb J F, Dougherty B A, Bott K F, Hu P C, Lucier T S, Peterson S N, Smith H O, Hutchison III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 18.Garcia J, Lemercier B, Roman-Roman S, Rawadi G. A Mycoplasma fermentans-derived synthetic lipopeptide induces AP-1 and NF-κB activity and cytokine secretion in macrophages via the activation of MAPK pathways. J Biol Chem. 1998;273:34391–34398. doi: 10.1074/jbc.273.51.34391. [DOI] [PubMed] [Google Scholar]
- 19.Hall R E, Agarwal S, Kestler D P, Cobb J A, Goldstein K M, Chang N S. cDNA and genomic cloning and expression of the P48 monocytic differentiation/activation factor, a Mycoplasma fermentans gene product. Biochem J. 1996;319:919–927. doi: 10.1042/bj3190919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson B, Poole S, Wilson M. Bacterial modulins: a novel class of virulence factors which cause host tissue pathology by inducing cytokine synthesis. Microbiol Rev. 1996;60:316–341. doi: 10.1128/mr.60.2.316-341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman R W, O’Sullivan F X, Schafermeyer K R, Moore T L, Roussell D, Watson-McKown R, Kim M F, Wise K S. Mycoplasma infection and rheumatoid arthritis: analysis of their relationship using immunoblotting and an ultrasensitive polymerase chain reaction detection method. Arthritis Rheum. 1997;40:1219–1228. doi: 10.1002/1529-0131(199707)40:7<1219::AID-ART5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 23.Hu W S, Wang R Y, Liou R S, Shih J W, Lo S C. Identification of an insertion-sequence-like genetic element in the newly recognized human pathogen Mycoplasma incognitus. Gene. 1990;93:67–72. doi: 10.1016/0378-1119(90)90137-g. [DOI] [PubMed] [Google Scholar]
- 24.Kestler D P, Agarwal S, Hall R E. Up-regulation of cytokine mRNA in human monocytes and myeloid cell lines by the differentiation/activation factor p48. Immunology. 1995;86:463–468. [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, M. F., and K. S. Wise. Unpublished results.
- 26.Kostyal D A, Butler G H, Beezhold D H. A 48-kilodalton Mycoplasma fermentans membrane protein induces cytokine secretion by human monocytes. Infect Immun. 1994;62:3793–3800. doi: 10.1128/iai.62.9.3793-3800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lysnyansky I, Rosengarten R, Yogev D. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J Bacteriol. 1996;178:5395–5401. doi: 10.1128/jb.178.18.5395-5401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markham P F, Glew M D, Whithear K G, Walker I D. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect Immun. 1993;61:903–909. doi: 10.1128/iai.61.3.903-909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall A J, Miles R J, Richards L. The phagocytosis of mycoplasmas. J Med Microbiol. 1995;43:239–250. doi: 10.1099/00222615-43-4-239. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto M, Nishiguchi M, Kikkawa S, Nishimura H, Nagasawa S, Seya T. Structural and functional properties of complement-activating protein M161Ag, a Mycoplasma fermentans gene product that induces cytokine production by human monocytes. J Biol Chem. 1998;273:12407–12414. doi: 10.1074/jbc.273.20.12407. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto M, Takeda J, Inoue N, Hara T, Hatanaka M, Takahashi K, Nagasawa S, Akedo H, Seya T. A novel protein that participates in nonself discrimination of malignant cells by homologous complement. Nat Med. 1997;3:1266–1270. doi: 10.1038/nm1197-1266. [DOI] [PubMed] [Google Scholar]
- 32.Mizuki N, Ohno S, Ando H, Chen L, Palimeris G D, Stavropoulos-Ghiokas E, Ishihara M, Goto K, Nakamura S, Shindo Y, Isobe K, Ito N, Inoko H. A strong association between HLA-B*5101 and Behcet’s disease in Greek patients. Tissue Antigens. 1997;50:57–60. doi: 10.1111/j.1399-0039.1997.tb02835.x. [DOI] [PubMed] [Google Scholar]
- 33.Mühlradt P F, Kiess M, Meyer H, Süssmuth R, Jung G. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J Exp Med. 1997;185:1951–1958. doi: 10.1084/jem.185.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mühlradt P F, Kiess M, Meyer H, Süssmuth R, Jung G. Structure and specific activity of macrophage-stimulating lipopeptides from Mycoplasma hyorhinis. Infect Immun. 1998;66:4804–4810. doi: 10.1128/iai.66.10.4804-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mühlradt P F, Meyer H, Jansen R. Identification of S-(2,3-dihydroxypropyl)cystein in a macrophage-activating lipopeptide from Mycoplasma fermentans. Biochemistry. 1996;35:7781–7786. doi: 10.1021/bi9602831. [DOI] [PubMed] [Google Scholar]
- 36.Mühlradt P F, Schade U. MDHM, a macrophage-stimulatory product of Mycoplasma fermentans, leads to in vitro interleukin-1 (IL-1), IL-6, tumor necrosis factor, and prostaglandin production and is pyrogenic in rabbits. Infect Immun. 1991;59:3969–3974. doi: 10.1128/iai.59.11.3969-3974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popham P L, Hahn T W, Krebes K A, Krause D C. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs post-translationally. Proc Natl Acad Sci USA. 1997;94:13979–13984. doi: 10.1073/pnas.94.25.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quentmeier H, Schmitt E, Kirchhoff H, Grote W, Mühlradt P F. Mycoplasma fermentans-derived high-molecular-weight material induces interleukin-6 release in cultures of murine macrophages and human monocytes. Infect Immun. 1990;58:1273–1280. doi: 10.1128/iai.58.5.1273-1280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rammensee H G, Friede T, Stevanovic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 40.Rawadi G, Ramez V, Lemercier B, Roman-Roman S. Activation of mitogen-activated protein kinase pathways by Mycoplasma fermentans membrane lipoproteins in murine macrophages: involvement in cytokine synthesis. J Immunol. 1998;160:1330–1339. [PubMed] [Google Scholar]
- 41.Rawadi G, Roman-Roman S, Castedo M, Dutilleul V, Susin S, Marchetti P, Geuskens M, Kroemer G. Effects of Mycoplasma fermentans on the myelomonocytic lineage. Different molecular entities with cytokine-inducing and cytocidal potential. J Immunol. 1996;156:670–678. [PubMed] [Google Scholar]
- 42.Rosengarten R, Wise K S. The Vlp system of Mycoplasma hyorhinis: combinatorial expression of distinct size variant lipoproteins generating high-frequency surface antigenic variation. J Bacteriol. 1991;173:4782–4793. doi: 10.1128/jb.173.15.4782-4793.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruschmeyer D, Thude H, Mühlradt P F. MDHM, a macrophage-activating product of Mycoplasma fermentans, stimulates murine macrophages to synthesize nitric oxide and become tumoricidal. FEMS Immunol Med Microbiol. 1993;7:223–230. doi: 10.1111/j.1574-695X.1993.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 44.Ruuth E, Praz F. Interactions between mycoplasmas and the immune system. Immunol Rev. 1989;112:133–160. doi: 10.1111/j.1600-065x.1989.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 45.Sakane T. New perspective on Behcet’s disease. Int Rev Immunol. 1997;14:89–96. doi: 10.3109/08830189709116847. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 47.Sankaran K, Wu H C. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem. 1994;269:19701–19706. [PubMed] [Google Scholar]
- 48.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 49.Seya T, Begum N A, Matsumoto M. Mycoplasma origin of tumor cell protein. Nat Med. 1998;4:133. doi: 10.1038/nm0298-133a. [DOI] [PubMed] [Google Scholar]
- 50.Simmons W L, Zuhua C, Glass J I, Simecka J W, Cassell G H, Watson H L. Sequence analysis of the chromosomal region around and within the V-1-encoding gene of Mycoplasma pulmonis: evidence for DNA inversion as a mechanism for V-1 variation. Infect Immun. 1996;64:472–479. doi: 10.1128/iai.64.2.472-479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theiss P, Karpas A, Wise K S. Antigenic topology of the P29 surface lipoprotein of Mycoplasma fermentans: differential display of epitopes results in high-frequency phase variation. Infect Immun. 1996;64:1800–1809. doi: 10.1128/iai.64.5.1800-1809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Theiss P, Wise K S. Localized frameshift mutation generates selective, high-frequency phase variation in a surface lipoprotein encoded by a mycoplasma ABC transporter operon. J Bacteriol. 1997;179:4013–4022. doi: 10.1128/jb.179.12.4013-4022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Theiss P M, Kim M F, Wise K S. Differential protein expression and surface presentation generates high-frequency antigenic variation in Mycoplasma fermentans. Infect Immun. 1993;61:5123–5128. doi: 10.1128/iai.61.12.5123-5128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ushio S, Iwaki K, Taniai M, Ohta T, Fukuda S, Sugimura K, Kurimoto M. Metastasis-promoting activity of a novel molecule, Ag 243-5, derived from mycoplasma, and the complete nucleotide sequence. Microbiol Immunol. 1995;39:393–400. doi: 10.1111/j.1348-0421.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 55.Washburn L R, Weaver K E, Weaver E J, Donelan W, Al-Sheboul S. Molecular characterization of Mycoplasma arthritidis variable surface protein MAA2. Infect Immun. 1998;66:2576–2586. doi: 10.1128/iai.66.6.2576-2586.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wieslander Å, Boyer M J, Wroblewski H. Membrane and protein structure. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 93–112. [Google Scholar]
- 57.Wilton J L, Scarman A L, Walker M J, Djordjevic S P. Reiterated repeat region variability in the ciliary adhesin gene of Mycoplasma hyopneumoniae. Microbiology. 1998;144:1931–1943. doi: 10.1099/00221287-144-7-1931. [DOI] [PubMed] [Google Scholar]
- 58.Wise K S, Kim M F. Major membrane surface proteins of Mycoplasma hyopneumoniae selectively modified by covalently bound lipid. J Bacteriol. 1987;169:5546–5555. doi: 10.1128/jb.169.12.5546-5555.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wise K S, Kim M F, Theiss P M, Lo S-C. A family of strain-variant surface lipoproteins of Mycoplasma fermentans. Infect Immun. 1993;61:3327–3333. doi: 10.1128/iai.61.8.3327-3333.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wise K S, Kim M F, Watson-McKown R. Variant membrane proteins. In: Razin S, Tully J G, editors. Molecular and diagnostic procedures in mycoplasmology. New York, N.Y: Academic Press; 1995. pp. 227–241. [Google Scholar]
- 61.Wise K S, Yogev D, Rosengarten R. Antigenic variation. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 473–490. [Google Scholar]
- 62.Yogev D, Watson-McKown R, McIntosh M A, Wise K S. Sequence and TnphoA analysis of a Mycoplasma hyorhinis protein with membrane export function. J Bacteriol. 1991;173:2035–2044. doi: 10.1128/jb.173.6.2035-2044.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yogev D, Watson-McKown R, Rosengarten R, Im J H, Wise K S. Increased structural and combinatorial diversity in an extended family of genes encoding Vlp surface proteins of Mycoplasma hyorhinis. J Bacteriol. 1995;177:5636–5643. doi: 10.1128/jb.177.19.5636-5643.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Q, Wise K S. Molecular basis of size and antigenic variation of a Mycoplasma hominis adhesin encoded by divergent vaa genes. Infect Immun. 1996;64:2737–2744. doi: 10.1128/iai.64.7.2737-2744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Q, Wise K S. Localized reversible frameshift mutation in an adhesin gene confers a phase-variable adherence phenotype in mycoplasma. Mol Microbiol. 1997;25:859–869. doi: 10.1111/j.1365-2958.1997.mmi509.x. [DOI] [PubMed] [Google Scholar]
- 66.Zheng X, Teng L J, Watson H L, Glass J I, Blanchard A, Cassell G H. Small repeating units within the Ureaplasma urealyticum MB antigen gene encode serovar specificity and are associated with antigen size variation. Infect Immun. 1995;63:891–898. doi: 10.1128/iai.63.3.891-898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]