Abstract
Condensates are biomolecular assemblies that concentrate biomolecules without the help of membranes. They are morphologically highly versatile and may emerge via distinct mechanisms. Nucleic acids–DNA, RNA and poly(ADP-ribose) (PAR) play special roles in the process of condensate organization. These polymeric scaffolds provide multiple specific and nonspecific interactions during nucleation and ‘development’ of macromolecular assemblages. In this review, we focus on condensates formed with PAR. We discuss to what extent the literature supports the phase separation origin of these structures. Special attention is paid to similarities and differences between PAR and RNA in the process of dynamic restructuring of condensates during their functioning.
INTRODUCTION
Even though the phenomenon of phase separation in the self-organization of living matter continues to be one of the most discussed topics in the scientific literature, there are challenges associated with its functional verification in vivo (1). Non-membrane compartments that concentrate specific biomolecules—so-called condensates–may emerge via various mechanisms or their superposition. Understanding how a particular structure is formed is necessary to obtain correct predictions of how it will function, as phase-separated compartments differ significantly in terms of their properties from those generated via binding and bridging.
The key role in condensate architecture belongs to nucleic acids that function as polymeric scaffolds in both stoichiometric and liquid demixing models. In addition to DNA and RNA, nucleic acids include an unusual polymer synthesized in the cells of higher eukaryotes, namely poly(ADP-ribose) [PAR]. Inherent features of this molecule (e.g. high charge density and low complexity) are particularly required in the context of liquid–liquid-phase separation (LLPS). Moreover, PAR can be produced ‘on demand’ at a desired intracellular location and may even function in the extracellular space. In the cell, PAR is concentrated in the biomolecular condensates, whose formation is space- and time-specific (2), for example, the nucleolus (3,4), DNA repair foci (5,6), stress granules (SGs) (7–9), the spindle (10) and transcription hubs (11).
Nonetheless, which observations support that these condensates function as separate phases? In this review, we consider the existing experimental evidence and attempt to understand the functional relevance of the LLPS-driving potential of PAR. We also examine in detail how the similarities and differences between PAR and RNA determine their interplay during the formation and functioning of condensates with a complex architecture.
PHASES VERSUS STRUCTURE
Phase separation
LLPS is a process of demixing of a homogeneous polymer solution into polymer-rich condensates surrounded by a dilute aqueous phase. The fundamental thermodynamic equation used to describe phase behavior of such polymer–water mixtures relates the Gibbs free energy (G) to the enthalpy (H) and entropy (S): ΔG = ΔH – TΔS, where T is the temperature (12). Being intrinsically disadvantageous in terms of entropy, LLPS is possible owing to (i) energetically favorable intermolecular interactions and (ii) the release of water molecules from an entropically unfavorable preorganized hydration shell into the bulk (13,14).
The enthalpic aspect of how LLPS is triggered at the molecular scale has been described in detail in numerous articles (15–18). From this perspective, phase separation in a polymer solution occurs when the graph of a Gibbs free energy change after mixing in a certain range of polymer concentrations acquires a negative curvature due to the contribution of intermolecular interactions’ energy (12,18). Therefore, the key property of molecules that determines their susceptibility to LLPS is their ability to engage in multiple interactions (so-called multivalence). Indeed, multivalent modular and disordered proteins and RNA are key drivers of phase separation events in vitro and in vivo.
Alternative modes of self-organization and condensates of mixed origin
Other mechanisms of molecular self-organization exist besides LLPS. Notably, the term condensates does not imply an obligatory LLPS stage during assembly. Biomolecular condensates were initially defined as intracellular structures that (i) accumulate molecules and (ii) comprise biological molecules, independent of all other characteristics (16).
Condensates that are not formed via LLPS do not have the properties of a separate liquid phase. Two such alternative ways of condensate formation are discussed in the literature on nuclear nonmembrane structures associated with chromatin (19,20). These include binding and bridging (polymer–polymer phase separation). A detailed comparison of non-LLPS and LLPS mechanisms of self-organization is provided by Ref. (19,20). The formation of condensates occurs between two poles: (i) stable complexes with fixed stoichiometry and (ii) thermodynamically driven LLPS in idealized several-component systems (21). A synergy between stoichiometric, structurally specific binding and stochastic interactions in the assembly of biomolecular condensates was recently documented in a combined theoretical-experimental analysis applied to the proteins SynGAP and PSD-95, whose complex coacervate serves as a model for neuronal postsynaptic densities (22). Moreover, a recent paper by Musacchio provides a relevant discussion on the role of site-specific interactions as the primary physicochemical drivers of condensate assembly, even in cases where further stages of its organization involve phase separation (23). A conceptual framework based on synergies between specific internal network structures and phase separation is provided by the work (24).
It has been proposed that condensate functionality is optimized when the interactions that drive the assembly vary widely in affinity (25). Strong-affinity contacts provide structural specificity, while rapidly recycling weak interactions enable time-dependent patterns (25).
Intracellular condensates may have a mixed origin when the LLPS stage is combined with other mechanisms during the assembly. Besides, they may have a complicated architecture, whose individual components act as a separate phase. Finally, due to certain difficulties to specifically manipulate condensate self-organization in vivo, for many structures the role of LLPS in their organization remains controversial.
Methods for studying LLPS and current challenges
According to the ‘gold standard’ of LLPS-derived liquid condensates (19), these droplets must (i) maintain a spherical shape, (ii) merge upon contact and undergo deformation in response to an applied force, (iii) contain mobile molecules that constantly mix inside the droplet and are exchanged with molecules from the environment, (iv) be sensitive to 1,6-hexanediol, (v) be scalable depending on concentration and (vi) pose difficulty for the diffusion of condensate components and inert probes across the interface.
In contrast, in the absence of quantitative measurements, qualitative compliance even with several of these points does not allow to conclude that a given structure is or is not a phase-separated condensate in vivo (1,26). We consider several methods often used to explore different aspects of the ‘gold standard.’ To what extent does a confirmation of the points listed above really demonstrate that a particular structure is LLPS-derived?
Morphological characterization (1,2)
Condensates formed through LLPS in living cells are often smaller than the optical diffraction limit (27). Below this limit, structures of completely different shapes tend to appear spherical due to the scattering of light (1). The development of super-resolution imaging provides opportunities for more detailed research on condensate morphology. In particular, super-resolution imaging and single-molecule tracking have been used to quantitatively assess LLPS within diffraction-limited foci in bacteria (28).
Although the presence of a sub-structure does not negate phase separation, the results obtained using super-resolution microscopy may in some cases cast doubt on whether a structure is phase-separated. For example, stimulation emission depletion microscopy (STED) has been employed to image individual heterochromatin domains (29). Those investigators found that HP1α is enriched within heterogeneously distributed clusters, but its maximal concentration in situ was estimated at 3 mM, that is, significantly less than the critical concentration required for HP1α LLPS in vitro (29). However, these data were obtained with cells fixed with 4 % paraformaldehyde (PFA). It is important to note that PFA fixation used in several super-resolution microscopy techniques, including STED, can both enhance or diminish putative LLPS behaviors, suggesting an advantage of live-cell imaging to study LLPS systems rather than fixed-cell experiments (30).
Detailed information on microscopy methods for morphological characterization of condensates in vitro and in vivo, is provided by (31,32).
The fusion properties of condensates can be quantified in vitro by means of optical trap-induced droplet coalescence. In this approach, individual condensates are optically captured from a solution in two different laser beams and are brought into close proximity to initiate coalescence. The kinetics of droplet fusion and relaxation can be quantitatively determined from fusion videos (33,34). The major experimental techniques for studying physical properties of condensates are described in (34).
Determining biomolecular diffusion (3,6)
Fluorescence recovery after photobleaching (FRAP) is a widely used technique for characterizing dynamic binding interactions of fluorescently labeled molecules in vitro and in vivo (34,35). Tagged molecules are bleached with a strong laser in one spot, and the recovery of the fluorescent signal is analyzed. It is assumed that diffusion of photobleached molecules and their replacement by new ones may reflect rapid reorganization of the ‘liquid’ contents within a compartment (1). However, the recovery is not specific to freely diffusing particles, as even the molecules involved in the formation of stable high-affinity complexes are exchanged (1). In different studies, signal recovery time can vary by up to three orders of magnitude. Nevertheless, it is used to determine whether the investigated compartment is LLPS-derived (1). Indeed, the interpretation of FRAP data often requires caution (34). For example, the FRAP recovery rate depends on molecular identity of the labeled species in heterotypic condensates that correspond to the scaffold–client model (34). Moreover, extracted FRAP data depend on the diffusion model and boundary conditions (34).
Diffusion across a boundary is a criterion for differentiating modes of self-organization. Observing the characteristic motion of tracer particles in cells (passive microrheology) allows characterization of the physical properties of the surrounding material and reveals a boundary constraint (36). Live-cell single molecular tracking (SMT) experiments can be conducted to visualize labeled molecules and categorize their trajectories as either ‘inside’ or ‘outside’ of the compartment (37). This approach illustrates a difference in the behavior of labeled condensate components inside a compartment compared with the rest of cellular volume and helps to quantitatively measure diffusion coefficients (38). In the case of LLPS, the presence of the phase boundary hinders the mobility of inert probes and component molecules (19), while molecular crowding or interactions within a separate phase should affect free diffusion (38). In the case of binding and bridging, inert probes freely diffuse through compartments, and component molecules likewise move freely if they are not bound to the scaffold (19). A theoretical framework to distinguish LLPS and molecular trapping by multiple binding sites from SMT studies is described in Ref. (39). Research on viral replication compartments is a vivid example of how a careful combined use of the above-mentioned approaches allows researchers to avoid erroneous conclusions about the phase-separated nature of a compartment (38). Using SMT, Rad52 motion within DSB repair foci in Saccharomyces cerevisiae cells was found to be consistent with the LLPS model (40). Direct experimental evidence how phase-separated condensates can accelerate the efficiency and specificity of biochemical reactions was likewise obtained by this method. It was shown that transcriptional regulator CBX2 interacts with DNA to assemble condensates on chromatin that facilitate the target-search of CBX2 for its cognate binding sites, thereby enhancing its genomic occupancy (41).
Interaction-disrupting assays (4)
The importance of certain protein domains for LLPS implementation is commonly tested by truncation/modification experiments or by perturbing weak hydrophobic interactions by 1,6-hexanediol treatment (1). These studies make it possible to establish the significance of certain protein–protein or protein–nucleic acid interactions for an analyzed subcellular structure, but cannot prove LLPS as opposed to other possible mechanisms (1). Moreover, it has been found recently that 1,6-hexandiol hyper-condenses chromatin in living cells in a manner distinct from its LLPS-disrupting activity, possibly due to the removal of water molecules around chromatin (42). These data suggest that the results obtained with 1,6-hexandiol treatment must be more carefully reconsidered in the case of chromatin-associated condensates (42).
Concentration-dependent effects (5)
Compartment size scaling in response to changes in component concentration can be a useful indicator for distinguishing between binding, bridging, and LLPS (19). Because LLPS is critically dependent on the concentration of biomolecules, the most important test for liquid demixing is measurement of the critical concentration, above which droplets exist, and below which they do not form (1).
Most studies on LLPS in vivo involve ectopic overexpression (1), although it has been suggested that subcellular systems may be supersaturated and exist on the brink of a two-phase regime, where even the slightest overexpression is able to dramatically influence data interpretation (43). However, there are examples where endogenously labeled proteins behave as overexpressed ones. Thus, liquid-droplet behavior of 53BP1 foci was demonstrated in cells with ectopic 53BP1 overexpression (44,45) and reproduced in live-cell experiments with endogenously tagged 53BP1 (via CRISPR/Cas9) (45). LLPS of heat-shock transcription factor 1 (HSF1) under heat-shock conditions with the formation of liquid-like nuclear stress bodies (nSBs) was observed in HeLa cells transiently expressing enhanced green fluorescent protein (EGFP)-tagged HSF1 and confirmed for endogenous HSF1 using HSF1-knock-in cells (obtained by CRISPR–Cas9-mediated knockout and HaloTag insertion into HSF1 gene locus) (46).
Protein concentrations in cell-free LLPS experiments are often significantly higher (by one to two orders of magnitude) than the intracellular concentrations determined in proteomic studies (47). Nonetheless, numerous LLPS-driver proteins have high local concentrations due to localized translation of their messenger RNA (mRNA) and/or recruitment to larger macromolecules carrying multiple binding sites [e.g., RNA, DNA, membranes (47) or PAR]. Estimated local concentrations for some proteins, for example, synaptic proteins, may by far outweigh their whole-cell concentrations determined proteomically, and therefore justify the use of high concentrations in cell-free experiments (47).
RNA IN BIOMOLECULAR CONDENSATES
Nucleic acids in condensates: the basic principles
As discussed above, the composition of intracellular condensates is highly multicomponent, varied, and time-dependent (48). Therefore, the concept of scaffolds and clients (48,49) has proven to be a useful model for describing compositional control of individual condensates. Scaffolds are the framework components required for the structural integrity of a given condensate. Their deletion or depletion decreases the size and/or number of condensates, whereas their overexpression may have the opposite effect (48). In general, the number of scaffolds is limited. On the contrary, numerous clients are present within a given condensate only under certain conditions, often because of direct interactions with scaffold molecules, whereas they do not significantly affect the condensate structure (48,49). As a rule, scaffolds are multivalent molecules, i.e., polymers containing multiple sites for intra- and intermolecular binding. Nucleic acids (DNA, RNA and PAR) that harbor multiple regions for specific and nonspecific interactions are some of the most important classes of molecules partaking in the architectural organization of condensates.
Nucleic acids with a large negative charge can phase separate consistently with a generic electrostatic mechanism that does not require specific interaction regions and disordered components (50) in a process referred to as complex coacervation (51). In the case of phase-separated droplets, the size, sequence, structure, and rigidity of constituent nucleic acids matters during condensate structuring (52). For example, DNA molecules with more rigid strands form weaker condensates that dissolve at lower ionic strength compared with DNA strands of equal length and charge density, but higher flexibility (53). This is because the force driving LLPS is not strong enough to bend the rigid chains to a conformation optimal for maximal charge neutralization (53). Phase behavior also differs between single-stranded and double-stranded sequences. High LLPS potential of unfolded single-stranded nucleic acids with exposed nucleobases is increased by contributing π-electron–related interactions (54). These include π–π and cation–π interactions of the nucleobase components (54) and sugar–π contacts enabled by deoxyribose moieties (55). In contrast, double-stranded DNA has higher charge density than single-stranded DNA, and binds more strongly to polycations, thereby forming solid aggregates (at low ionic strength) or a liquid–crystalline phase (at high ionic strength), while remaining base-paired (53). The DNA length finely affects the fluidity of DNA-based condensates: short DNAs are reported to drive the formation of liquid-like condensates, whereas longer DNA molecules are prone to forming solid-like structures (56).
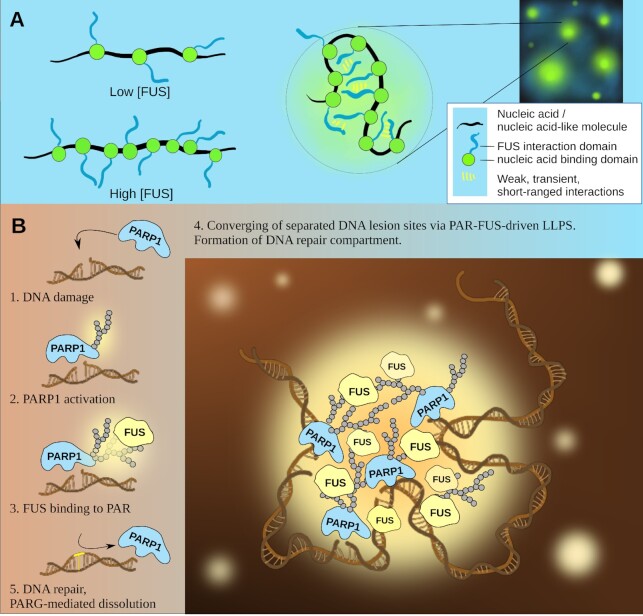
Notably, nucleic-acid–based condensates can be generated by monolayer protein recruitment: a mechanism described by means of the FUS protein is an example (57). FUS consists of a nucleic-acid–binding domain and a disordered low-complexity domain (LCD) that mediates FUS self-attraction. Its adsorption on DNA gives rise to a self-interacting protein–nucleic acid polymer that collapses with the formation of FUS-DNA condensates (57) (see Figure 1A). FUS condensation with RNA is realized in a similar manner: RNA serves as a multivalent platform, whose length controls the valency of FUS (one FUS unit occupies ∼20−25 nts) (58). The results obtained by the authors (58) show that the initial FUS-RNA interaction supports the subsequent FUS-RNA: FUS-RNA interaction mediated by protein–protein contacts and resulting in LLPS. Atomic force microscopy studies revealed a similar phenomenon for PAR: FUS interaction with PAR arising at DNA break sites, which yields compartments that concentrate PAR, FUS, and damaged DNA (see Figure 1B). The contribution of FUS LCD to the compartment formation via the interaction with PAR is clearly detectable, as a deletion of most of this region impairs the formation of large compartments (59). Moreover, the progressive decrease in compartmentalization efficiency is observed with an increase in the number of mutations in FUS LCD, mimicking its phosphorylation (59). FUS can likewise use long PAR polymers (e.g. >16 monomers) as a scaffold for multivalent interactions that drive condensation, as it has been described for FUS and RNA (60).
Figure 1.
A mechanism for the formation of nucleic-acid-based condensates. (A) Monolayer FUS recruitment on DNA (57). (B) FUS interaction with PAR and formation of compartments, concentrating PAR, FUS and damaged DNA (5,6,59).
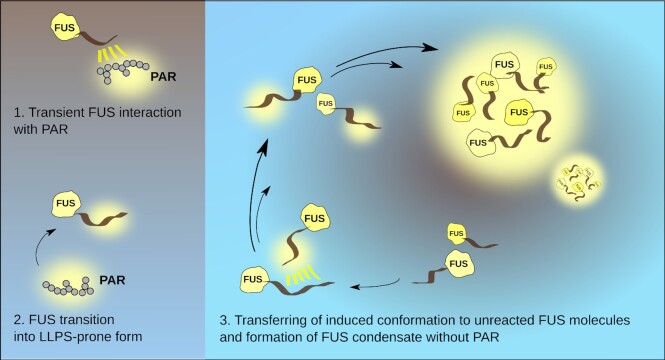
Interestingly, PAR is able to induce FUS condensation in a completely different manner via a trans-acting catalyst-like mechanism (60) (see Figure 2). According to this mechanism, PAR chains of all lengths (>4 monomers) can alter FUS into LLPS-prone form via transient interactions. The primed FUS is able to trigger the formation and maintain FUS condensates on its own (without PAR) (60). Thus, FUS droplets remain intact after treatment of FUS-PAR condensates by PAR-degrading enzyme PARG. Moreover, the PAR-free fraction of PAR-treated FUS recruited unreacted FUS molecules into a condensate (60). A strikingly low concentration of PAR (1 nM) was sufficient to induce FUS condensation; interestingly, this value is three orders of magnitude lower than in the case of RNA (60), and is manifoldly lower than the 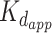 (>200 nM) for PAR–FUS binding. Stimulation of FUS aggregation in vitro by sub-stoichiometric amounts of PAR was also demonstrated previously (5). In contrast to PAR, RNA was demonstrated to stably bind with FUS (
(>200 nM) for PAR–FUS binding. Stimulation of FUS aggregation in vitro by sub-stoichiometric amounts of PAR was also demonstrated previously (5). In contrast to PAR, RNA was demonstrated to stably bind with FUS (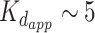 nM) (60), and to be necessary for formation and maintenance of FUS-RNA condensates, as FUS-RNA droplets treated with RNase completely dissolved (60).
nM) (60), and to be necessary for formation and maintenance of FUS-RNA condensates, as FUS-RNA droplets treated with RNase completely dissolved (60).
Figure 2.
A trans-acting catalyst-like mechanism for PAR-triggered FUS condensation.
This sub-stoichiometric mechanism of PAR-mediated LLPS induction partially resembles the sub-stoichiometric action of lncRNAs in the organization of phase-separated condensates (61). The examples are the regulation of Pumilio proteins by lncRNA NORAD and the control of X chromosome inactivation / dosage compensation by the lncRNA Xist (discussed in (61)). In both cases, significant stoichiometric imbalances are bypassed via the two-step mechanism. Initially, the condensate is nucleated by stoichiometric multivalent interactions of the lncRNA with its protein partner, resulting in a local increase in protein concentration. At the following step, homotypic interactions mediated by protein IDRs further amplify the supra-stoichiometric recruitment of additional protein molecules, enlarging the phase-separated compartment (61). Repetitive sequences, which promote multivalent interactions, have proven to be essential for phase transitions seeded by lncRNAs (61,62).
Nucleic acids influence condensate organization in three key ways: they modulate biophysical properties, induce selective nucleation, and recruit additional molecules (52). In this regard, the work based on the ArtiGranule (ArtiG) bottom-up approach is a vivid example. The experiment consists of adding an artificial protein to cells, which forms condensates. Then, an RNA-binding domain is inserted into the protein, and previously biochemically inert condensates of this protein begin to attract RNA within the cell, thus allowing to investigate the contribution of intracellular RNA to the generation of artificial condensates (63). Intracellular RNA has been found to seed the nucleation of condensates, specifying their ultimate composition by recruiting additional RNA-binding proteins (RBPs), and serving as an architectural component that influences condensate morphology (size, number and polydispersity) (63).
RNA in condensates from a microstructural standpoint
As mentioned above, RNA is an essential component of numerous nuclear and cytoplasmic condensates. Properties of scaffold RNA molecules (such as the composition, length, structure, modifications, and expression level) can determine biophysical properties of a nascent condensate: its size, shape, viscosity, fluidity, surface tension, and molecular composition (64). RNA molecules containing tandem repeats (repetitive RNAs, repRNAs) are the most important regulators of dynamic chromatin compartmentalization (62). RepRNAs are generated during the transcription of ribosomal RNA gene clusters in the nucleolus or during transcription of tandem repeats in satellite DNA elements [(peri)centromeres and telomeres], and can originate from repetitive transposon sequences scattered throughout the genome. Local transcription and a high copy number of repRNAs, along with their partial self-complementarity, endow these biomolecules with high potential for the nucleation of chromatin self-organization processes (62).
Short RNAs lacking high valency may influence RBPs phase separation through interaction with their RNA-binding sites, thereby limiting RBPs’ valency or reducing their effective concentration (65). Two RNAs harboring the same number of RBP-binding sites, while having distinct secondary structures, may yield different outcomes during LLPS, as demonstrated for the Whi3 protein from Ashbya gossypii and two RNA transcripts: CLN3 and BNI1 (66,67). It has been suggested that 3D folds of specific RNAs can act as guide scaffolds for spatial positioning of RBPs (65). Proximal location of bound RBPs may favor protein–protein interactions within the complex, whereas more distal positioning may facilitate RBPs’ interactions with the immediate surroundings. Because of these differences, such complexes may nucleate condensates with different compositions and/or material properties (65).
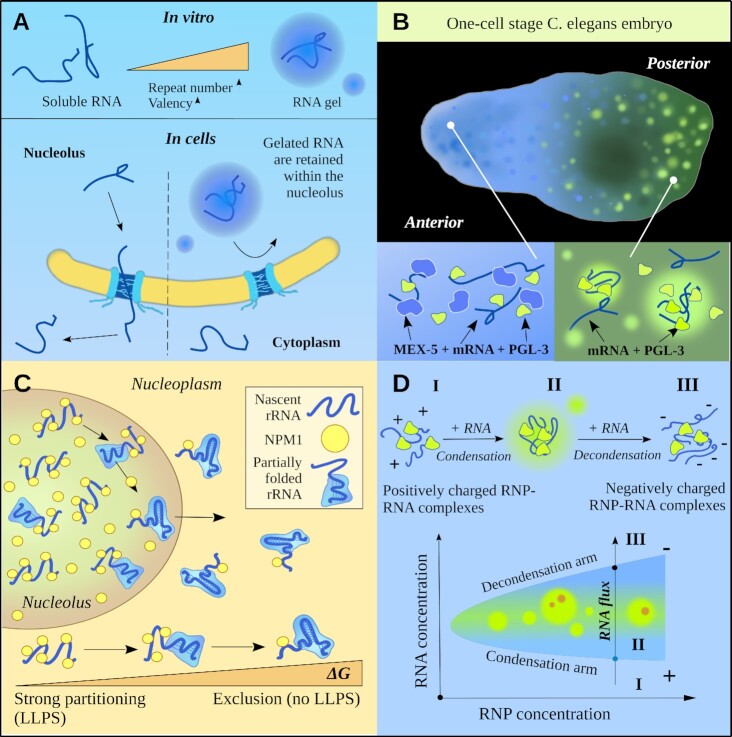
RNA molecules can generate condensates (RNA gels) that do not contain protein components (68). Nucleotide repeat expansion in RNA gives rise to areas of multiple interactions between nitrogenous bases of different RNA molecules. This phenomenon leads to RNA gelation and hinders the release of RNA molecules into the cytoplasm. Consequently, nuclear RNA foci arise, a characteristic of nucleotide repeat expansion disorders (68) (see Figure 3A). RNA gelation takes place when RNA valence rises to a critical level. This may explain why the disease is triggered after nucleotide repeat expansion reaches a threshold repeat number, and why nucleotide repeat expansions located in seemingly unrelated genes can cause similar clinical symptoms (68).
Figure 3.
RNA in biomolecular condensates. (A) A model for the formation of nuclear RNA foci in repeat expansion disorders. Increased RNA valency due to expansion of nucleotide repeat results in RNA gelation and sequestration of RNA molecules into nuclear foci (68). (B) RNA and LLPS in the creation of cellular polar systems. Top: one-cell stage C. elegans embryo. Competition of MEX-5 and PGL-3 proteins for mRNA binding regulates the formation of PGL-3 droplets (81). (C) LLPS-based channeling of ribosomal RNA (rRNA) flux out of the nucleolus. It is proposed that relatively nascent rRNA are available for multiple interactions with scaffolding proteins of the GC-matrix, while rRNA binding with ribosomal proteins decreases their valency, thus disturbing nucleolar LLPS and resulting in the effective emission of fully assembled pre-ribosomal particles (82). (D) Re-entrant liquid condensation. Top: a schematic illustration of three different RNP (ribonucleoprotein complexes):RNA regimes. Bottom: a corresponding phase diagram. The arrow represents the passage of the system from ‘+’ to ‘–’-charged RNPs through a charge-neutral stage and charge inversion when the RNA concentration is increased (RNP concentration is fixed). The shaded area corresponds to the LLPS regime (II). During the condensate dissolution stage, the RNA flux into RNP droplets generates vacuolated coacervates (83).
RNA is the key player in the assembly of liquid condensates with filamentous morphology (although these structures are not spherical, their protein components are as dynamic as those in classic droplets). An example of such a mesh-shaped condensate is the TIS granule network adjoining the rough endoplasmic reticulum, which is important for membrane protein trafficking (69). In vitro and in vivo reconstitution experiments revealed that the minimal set of components of filamentous condensates is a multivalent RBP that concentrates mRNAs with large unstructured regions able to form a pervasive intermolecular mRNA–mRNA interaction network. The underlying RNA matrix acts as a condensate skeleton and prevents full fusion of the spherical liquid-like condensates, thereby generating membraneless organelles with irregular shapes. The large surface area of such structures may promote interactions at the condensate surface and at the interface with other organelles (69).
Composition of RNA–protein systems: assembly manual and a pacemaker
Global phase behavior of RNA–protein systems is determined by the complex interplay between homotypic (RNA–RNA and protein–protein) and heterotypic (RNA–protein) interactions (15). Alterations in the balance of structure-forming contacts may not only shift the phase boundaries, but also govern the formation of complex morphologies and diverse material properties of liquid condensates (70).
Competing protein–protein and protein–RNA interactions regulate the multiphasic structuring and spatial organization of multilayered condensates (71). Different solvation of non-stoichiometric RNP-RNA complexes at compositionally disproportionate mixtures results in a composition-dependent tuning of the condensate surface architecture. Thus, free RNAs in RNA-rich condensates tend to position themselves on the surface due to the larger effective solvation volume of partially bound RNA molecules, as compared to fully complexed ones (71). Indeed, coarse-grained molecular dynamics simulation studies reveal that unequal component stoichiometry in condensates composed of two multivalent associative polymers leads to an enrichment of the majority of species at the interface and a drastic reduction in the surface tension (72). RNA localization on the condensate surface may influence condensate morphogenetic properties, as increasing the RNA surface density implies smaller and more numerous condensates (73). Interestingly, at a high RNA surface density, condensates may even lose sphericity and adopt a clustered morphology akin to TIS granules (73).
Self-organization of heterotypic condensates can be drastically altered by changes to the protein sequence (74). Due to increased propensity for the formation of cation–π and π–π interactions and lower charge density, ssRNA is preferentially associated with the inner phase of model multiphasic condensates formed by polyK, polyR and polyD peptides, while dsRNA is sorted into both the inner and outer phases (75). Moreover, the RNA structure is also influenced by the phase environment: in relatively protein-poor outer phase RNA is well-structured, whereas dsRNA partition within the protein-dense inner phase results in RNA duplexes melting (75). Higher-valency proteins were predicted to concentrate within the core of multicomponent condensates, whereas proteins with lower valency had a tendency to cluster toward the interface (76). Such multilayered architecture was proposed to maximize the density of molecular interactions that stabilize LLPS and reduce the surface tension of condensates (76). Furthermore, using computational methods, the phase behavior of a protein–polynucleotide mixture, as well as co-localization of protein and polynucleotide molecules within condensates could be significantly affected by changes in the protein sequence: increased charge patterning resulted in the formation of structured or patterned condensates (77). Divalent cations were found to have opposed effects on heterotypic (RNA–peptide) and homotypic (RNA–RNA) condensates. As a function of increasing cation concentration, heterotypic phase-separated droplets dissolve to be replaced by homotypic droplets (78). The addition of cations to a solution has opposite effects on the material properties of heterotypic and homotypic condensates: although the relative fluidity increases for RNA–protein droplets, it diminishes for RNA–RNA droplets (78). In accordance, in the presence of Mg2+, RNA molecules with repeat expansion sequences self-assemble into morphologically distinct condensates depending on the cation concentration. As the RNA/Mg2+ ratio increases, the canonical nucleobase-paired structure is substituted by a base-clustered one, especially prominent at the rim, suggesting a hardening process (79).
In a recent study, Laghmach et al. explored how the interplay of the RNA chain length, RNA–protein stoichiometry, and environmental conditions jointly shape the thermodynamic landscape and interfacial properties of RNA–protein condensates (80). The polyelectrolyte charge ratio (RNA:protein charge units) was identified as a key variable influencing the coexistence window of LLPS of protein–RNA mixtures. The RNA chain length was determined as a key parameter governing the surface tension and thermodynamic stability of protein–RNA condensates. Indeed, longer RNAs are more favored to form condensates compared with shorter RNA chains due to reduced entropy cost of binding. Salt and external crowding can likewise tune the LLPS window. Increasing the salt concentration was associated with attenuation of electrostatic interactions between the protein and RNA, resulting in a significant decrease in the LLPS. Under higher pressure, the coexistence window of the RNA–protein mixture expanded considerably, and mixtures containing short RNAs were particularly sensitive to external pressure. External crowding could also counter-balance the destabilizing effect of short RNAs and extend the LLPS window of heterotypic mixtures (80).
Therefore, alteration in the composition of an RNA–protein system in particular is an approach to regulate one- and two-phase regimes for this system. In this article section, we consider several examples of how the composition of an RNA–protein system can act as a ‘pacemaker’ during condensate functioning.
Intracellular polar systems
The competition for RNA between different proteins in conjunction with LLPS can be used to create intracellular polar systems (81). For example, in the Caenorhabditis elegans germ line, a gradient of the polar MEX-5 protein initiates localized phase separation with the formation of RNA–protein P-granules at one of the cell poles, thereby inducing asymmetric division of the embryonic cell. How can the gradient of the MEX-5 protein, which is not a constituent of P-granules, regulate the spatial arrangement of these liquid compartments? The mechanism is based on competition for RNA binding between MEX-5 and a scaffold protein of P-granules, PGL-3 (81). RNA molecules bound to MEX-5 are incapable of LLPS; therefore, the concentration gradient of MEX-5 creates an oppositely directed gradient of RNA molecules accessible to PGL-3: a ‘negative pattern’ of the distribution of P-granules (81) (see Figure 3B).
Channeling of ribosomal subunits
The balance between RNA and protein levels determines the saturation concentration for multicomponent condensates. Heterotypic interactions were reported to promote LLPS and stabilize liquid droplets, whereas homotypic interactions destabilize them (82). An elegant mechanism of directed flow along the nucleolus is based on this principle. The binding of ribosomal RNA to ribosomal proteins reduces the number of binding sites for heterotypic interactions of nucleolar scaffold proteins. Accordingly, in comparison with completely assembled ribosomal subunits, newly synthesized ribosomal RNA transcripts are more accessible to binding by NPM1, SURF6 and other scaffold components of the GC-matrix (82). This situation causes selective exclusion of fully assembled ribosomal subunits, thereby providing a thermodynamic basis for the directed flow of RNA along a nucleolus inside out (82) (see Figure 3C).
Re-entrant phase transitions
A phase transition is called re-entrant if the system undergoing it proceeds from one state to another state–that is macroscopically similar or identical to the original one–through at least two-phase transitions caused by a change in a single parameter (65). Such phase behavior has been discovered for the systems containing RNA (or single-stranded DNA) and cationic intrinsically disordered proteins by varying the concentration of any one of the components (83) (see Figure 3D). Re-entrant LLPS (re-LLPS) may result in multilayered (‘nested’) droplets (65) or condensates containing dynamic vacuoles with a tunable lifetime (83). The formation of hollow structures circumscribed by an ordered RNA–protein envelope (‘bubble condensates’) and micelles has been documented in vitro at a certain RNA: protein ratio under a certain concentration regime (84). Re-entrant phase behavior is ideal for condensates that regulate their lifetime and the duration of biochemical processes facilitated by them (65). Furthermore, this behavior allows for temporal regulation, because metastable condensates may limit their own existence by the synthesis or decay of the second component that influences the LLPS of the first one (65,85).
Re-LLPS has not been investigated in vivo. Nonetheless, several condensates existing in the cell have characteristics reminiscent of re-entrant systems, e.g., a nucleolus (see the section above) and transcription compartments (65), as reviewed below.
AN INTRODUCTION TO THE ROLE OF PAR IN CONDENSATE BIOLOGY
PARPs and PAR
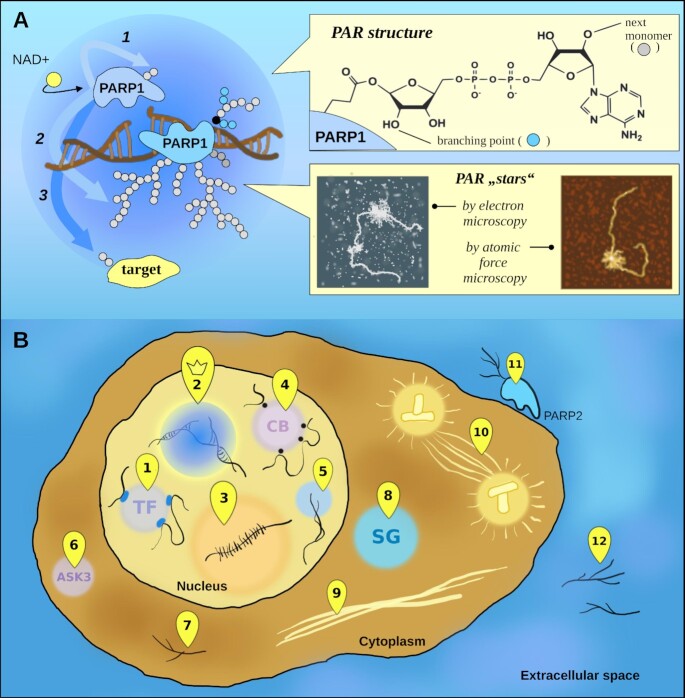
ADP-ribosylation is a reaction involving the transfer of ADP-ribose residues originating from NAD+ to a modification target [a protein, DNA (86–88) or RNA molecule (89)]. In mammals, only four enzymes from the 17-member (ADP-ribosyl)transferase protein family (PARPs 1 and 2 and tankyrases 1 and 2) are able to catalyze the elongation underlying the synthesis of PAR (90,91). Cellular functions of poly(ADP-ribosyl)ation [PARylation] are mainly associated with DNA repair, as DNA breakage is the classic trigger of PAR production, attributed to the folding of catalytically active PARP1 (92) and PARP2 (93). PARP1, the main enzyme for PAR synthesis in the cell, modifies itself by PAR polymers reaching >200 residues in size and up to 100 nm in length (94,95), which look like a highly branched ‘star’ detectable by electron microscopy (95,96) or atomic force microscopy (97) (see Figure 4A). How such a complicated structure is achieved is still not fully understood (98). One suggested explanation is that the branching is catalyzed by PARP2 (99), however, it is well known that PARP1 yields branched PAR as well.
Figure 4.
PAR synthesis and cellular localization. (A) PARPs is performs the PARylation reaction in the form of 1. Intramolecular (in cis-) auto-modification, 2. Intermolecular (in trans-) auto-modification within PARP dimers and 3. trans-modification of non-PARP targets. The points of active PAR production—PAR ‘stars’—are detected by electron microscopy (95,96) or atomic force microscopy (97). (B) PAR is found in cells at several locations: 1.transcription factories; 2. DNA repair foci; 3. the nucleolus; 4. Cajal bodies; 5. telomeres; 6. ASK3 condensates; 7. in the form of protein-free PAR chains; 8. stress granules; 9. pathological aggregates; 10. the spindle; and in extracellular space as 11. a product of PARP2 located on T-cell surface; 12. free PAR chains released in the extracellular matrix due to cell necrosis.
There is evidence that PARP1 and 2 can be activated in other ways: through the interaction with non-B DNA (100), RNA (101–103) [refuted by (104)], including small nucleolar RNAs (105), PAR (99), and phosphorylated kinase ERK2 (106). Moreover, PAR may be translocated within the cell or released into the extracellular space (107). Extracellular PAR can also be produced by the activity of PARP2 located on the surface of T cells (108). PAR is found ubiquitously inside and outside the cell, although DNA damage is the main epicenter of PAR synthesis (see Figure 4B).
Short PAR chains are poor substrates for poly(ADP-ribose) glycohydrolase (PARG) (109). It has been proposed that (ADP-ribosyl)hydrolase 3 (ARH3) activity compensates for the slower processing of short chains by PARG. ARH3 also performs the hydrolysis of terminal seryl-ADP-ribosyl linkages (110), which are inaccessible to PARG (111). PARG, but not ARH3, can resolve branched PAR architecture (112).
The histone PARylation factor 1 (HPF1)—recently reported to form a joint active site with PARP1 and/or 2 (113)–has been shown to limit the PARylation activity of PARPs and stimulate their NAD+ hydrolase activity (114). Further, HPF1 promotes DNA-dependent and DNA-independent autoPARylation of PARP1 and/or 2, as well as the heteroPARylation of histones in the complex with a nucleosome in a defined range of HPF1 and NAD+ concentrations, at which no HPF1-dependent enhancement of the hydrolytic NAD+ consumption occurs (115). The influence that HPF1 has on PAR production and structure in the cell remains to be investigated.
PARPs are implicated in multiple DNA repair pathways and in the maintenance of genomic stability (116). PARylation is believed to be the first wave of DNA damage response signaling. After PAR synthesis, several PAR readers containing different PAR-recognizing modules (117) are recruited to a DNA lesion to remodel chromatin and initiate DNA repair. Thus, in the case of single-strand breaks (SSBs), the rapid recruitment of the core scaffold factor of SSBs repair, XRCC1, to lesion sites is dependent on PARP1 or PARP2 (116). In the case of double-strand breaks (DSBs), PARP1 is important for the early and rapid recruitment of BRCA1 (118), which controls the initial steps of DSB resection and also mediates strand exchange during homologous recombination (HR) (116). During nucleotide excision repair, XPC recruitment to UV lesions is mediated by its binding to PAR (119). PARylation of UV-damaged chromatin through the activity of PARP1 is important for the recruitment of the chromatin-remodeling enzyme ALC1 (120). Initial chromatin relaxation triggered by PARylation and the PAR-binding remodeler activities of ALC1 (121) facilitates the subsequent recruitment of CHD3 and CHD4 for further chromatin remodeling at DNA break sites (122). As a key factor directing cell fate toward apoptosis under the conditions of genotoxic stress, PARP1 is a promising target for treating cancer as well as metabolic, inflammatory, and neurological disorders (123,124).
Interestingly, PAR-dependent chromatin remodeling by ALC1 has been recently identified as a key determinant of PARP inhibitor (PARPi) toxicity in HR-deficient (HRD) cells (125,126). ALC1 loss is synthetic lethal with HRD due to the accumulation of replication-associated DNA damage (125,127) and increased PARP trapping (125,126,128). Nucleosome remodeling by ALC1 is required for the release of trapped PARP2 (128). It was found that the inactivation of ALC1 traps PARP2 but not PARP1 (128). The ubiquitin E3 ligase TRIP12 (129) and the ubiquitin-dependent ATPase p97 (130) are involved in removing of trapped PARP1 and their activities reduce PARPi efficiency. However, PARP trapping is not solely responsible for the ALC1 deficiency/PARPi synthetic phenotype (127). It was proposed that processing of alkylated base damage is a key driver of synthetic lethality with HRD in ALC1–/– cells as well as PARPi sensitivity in HRD (127).
PAR features relevant to condensate microstructuring
Several unique characteristics of PAR, including its low complexity, multivalence, and large charge, may contribute to organization of subcellular condensates, including LLPS-derived ones (2).
Low complexity
It has been proposed that in the case of RNA, the random coiled state of the molecule provides greater accessibility to the binding of short cationic molecules than structured RNA (131). Complex coacervates composed of low-complexity RNA (polyU) and short polyamines (spermine and spermidine) share numerous features with coacervates consisting of intrinsically disordered region (IDR)-containing proteins (131). Indeed, stress-induced low-complexity ribosomal intergenic RNA drives the formation of nucleolar liquid-like foci during the nucleolus conversion to an A-body (132). Consequently, the low complexity of the PAR structure may predispose this polymer to complex coacervation as an LLPS subtype.
Increased flexibility
Molecular dynamics simulations have been employed to determine which geometrical constraints dictate the structural flexibility of PAR chains in solution (133). In that study, PAR was shown to have higher flexibility than single-stranded DNA, likely owing to the presence of two phosphates linking the two deoxyriboses instead of one as in DNA (or one ribose in RNA), and therefore expanding the available conformational space (133). In the same report, molecular dynamics simulations revealed that PAR polymers, especially long ones, adopt numerous different conformations in water, and a stable and defined structure is never reached. A PAR 25-mer adopted a three-globe–shaped conformation, indicating a tendency of monomers to cluster together (133). Therefore, PAR is prone to a dynamic size- and branching-dependent multiglobular conformation, and this intrinsic flexibility allows PAR to recognize multiple proteins and specifically fit their structure (133).
Charge density and distribution
These parameters are of key significance for complex coacervation (134). PAR is the most electronegative natural polymer (135). Compared to DNA and RNA, PAR has a doubled higher negative charge and additional space between ribose residues (2). Notably, polyelectrolyte interactions enable ultrahigh-affinity binding, even though binding partners in this case retain their structural disorder and highly dynamic behavior (136,137), indicating that high-affinity binding and structural disorder are not mutually exclusive (21).
In living organisms, PAR complex coacervation with Ca2+ ions plays a key role during physiological bone biomineralization and pathological vascular calcification (107,138).
Structural heterogeneity and associated metabolic pathways
Different structural elements of a PAR molecule (iso-ADP–ribose, ADP–ribose–ADP–ribose junction and terminal ADP–ribose) can be recognized through a variety of protein domains (117). PAR molecules are highly heterogeneous in terms of chain length and branching. This structural diversity influences the binding characteristics of PAR readers, PAR catabolism and the dynamics of PAR-containing biomolecular condensates (139). In summary, possible outcomes of different PAR structures are as follows: (i) PAR chain length and branching influence the number of PAR readers able to interact with a single PAR molecule and therefore determine their local concentrations. A highly branched polymer may reduce the available space for PAR-binding domains of proteins that recognize linear PAR motifs. (ii) structure-specific binding may result in selective subcellular localization of PAR readers, their regulation, and degradation via PARylation-dependent ubiquitination. (iii) The synthesis of different PAR structures can consume various amounts of NAD+, thus influencing local metabolic pathways. (iv) Given that PAR is highly charged, differences in amounts of building blocks may affect local biophysical properties of the cytoplasm. (v) Structurally distinct PAR molecules behave differently during degradation and can give rise to various sets of catabolites with specific consequences for downstream signaling (139).
The biological relevance of PAR branching is emphasized by the fact that branching frequencies vary considerably during different phases of the DNA damage-induced PARylation reaction and among different mouse tissues. The PAR branching and chain length strongly affect cellular functions, thereby further supporting the notion of a ‘PAR code’ (140).
An additional valence source
In a protein-free state or in the form of a protein PTM, PAR can serve as a source of additional valence mediated by PAR–protein interactions [as is the case for RNA (141)]. The PAR surface, composed of repeated ADP-ribose units, can act as a multivalent platform for protein recruitment. Notably, a larger PAR valence can be achieved in two ways: by increasing the number of modification sites on a given protein, and by increasing PAR length (up to 200 units) and branching (2). In many cases, however, a small polymer such as oligo(ADP-ribose) may be sufficient to achieve desired affinity (8).
Notably, repetitive sequences of lncRNAs are essential for their ability to seed formation of phase-separated condensates, including assemblies organized by the sub-stoichoimetric mechanism (61,62) (see also subsection Nucleic acids in condensates: the basic principles).
Protein PTM
PTMs are the most important regulators of protein–protein and protein–nucleic acid interactions, and therefore represent one of the key approaches to modulate the formation and dynamics of biomolecular condensates (142–144). It is possible that PARylation can alter a conformation and therefore interaction profile of the modified protein similar to (145). Moreover, PAR covalently attached to the protein may be a significantly more specifically organized structure than free PAR (146), and represent a specific platform for initiating the assembly of macromolecules (147).
PAR IN CONDENSATES FROM A MACROSTRUCTURAL POINT OF VIEW. TRULY A sePARate PHASE? PAR-RNA INTERPLAY
The above properties of PAR make it an attractive candidate for an LLPS driver. Indeed, PAR is a necessary component for the assembly of various natural liquid-like biomolecular condensates. In this section, we consider the most relevant examples and attempt to understand supporting evidence of the LLPS mechanism for each structure. We also discuss PAR characteristics that can be important for the organization of these biomolecular assemblies in terms of their macrostructure, that is, 3D shape and material properties.
Despite the similarities, PAR differs from RNA and DNA and has a distinct spectrum of protein partners. In the case of LLPS, this property can cause liquid immiscibility of the PAR-containing phase with other liquid phases and induce condensate subcompartmentalization with the formation of multilayered structures. The reviewed case studies deal with the nucleolus and DNA repair foci.
The nucleolus
Up to 40% of intracellular PARP1 molecules are concentrated in the nucleoli (148), and the disruption of PARP1 enzymatic activity leads to nucleoli disassembly (3).
A series of studies conducted in vitro (149,150) and in vivo (151,152) collectively support a model where the nucleolus behaves as a separate liquid phase in the nucleoplasm (1). Internal architecture of the nucleolus consists of three layers, corresponding to different stages of ribosome biogenesis (153,154). Self-organization of the nucleolar internal structure has been explained by the immiscibility of several liquid phases (149). For example, NPM1 (a scaffold protein of the nucleolus shell) and fibrillarin (a scaffold protein of the nucleolus core) both undergo LLPS in the presence of ribosomal RNA in vitro (150,151).
Nevertheless, NPM1- and fibrillarin-based liquid droplets do not merge with each other and coexist independently. Moreover, fibrillarin-rich droplets are captured by NPM1 droplets, thus giving rise to a core-shell structure characteristic of the nucleolus (149).
Although the exact function of PAR in nucleolus organization is unclear, an interesting analogy has been drawn between the liquid-phase structure of the nucleolus and the presence of PAR in one review (4): in yeast and other lower eukaryotes, nucleoli have only two layers (they lack the fibrillar center, FC) (155), and there is no PARylation in yeast (156). [On the other hand, it is thought that the evolutionary division of a single fibrillar compartment into the FC and DFC was due to significant enlargement of the rDNA intergenic region (155); however, the Saccharomyces cerevisiae rDNA intergenic spacer interrupting 35S RNA coding units is 2.5 kb long (157).] Moreover, nucleolar subcompartment immiscibility is not due to IDRs within fibrillarin and NPM1, which appear to be fully miscible (158). Instead, it is encoded in different RNA-binding domains of these proteins, with distinct substrate specificity (158). Therefore, it is possible that PAR as a specific scaffold played a role in the divergence of FC and DFC subcompartments.
DNA repair foci
DNA damage induces the assembly of temporary compartments concentrating damaged DNA and repair factors. On the one hand, DNA breaks trigger PAR synthesis by activated PARP1, and this PAR is believed to seed the LLPS of FET proteins (FUS, EWSR1 and TAF15) at the lesion site (5). That study involved overexpression of FET proteins, and the conclusions were drawn from in vitro data and indirect in vivo evidence such as roundness, fusion, and alteration of light diffraction (5). Nonetheless, the droplet size changed according to the alteration of protein expression levels (5), indicating a dependence on the critical concentration, which can be considered direct in vivo evidence (1). The positive charges in RGG modules of FET proteins were shown to be necessary for electrostatic PAR binding (5), further indicating that LLPS is possible (the complex coacervation type). PAR ability to nucleate FUS condensates both in vitro and in vivo was also demonstrated by Patel et al. (6). FUS assemblies in FUS-GFP-expressing cells imaged with a digital scanned light-sheet microscope (DSLM) with structured illumination appeared to be spherical, underwent frequent fusion events, and rapidly relaxed into a spherical shape. Their viscosity values, approximated by recovery times of the FRAP were estimated to be around 10- to 100-fold that of water (6).
As demonstrated elsewhere in vitro, PAR production at DNA break sites results in the binding of the FUS protein and subsequent selective concentration of damaged DNA in PAR–FUS compartments (59). It is believed that FUS, by its direct binding to PAR, facilitates DNA repair through the formation of transient compartments enriched with damaged DNA, to which DNA repair proteins are attracted. PAR hydrolysis by PARG enables the dissociation of these compartments, which in turn promotes the turnover of DNA repair in the cell (59,159).
On the other hand, damage-induced long noncoding RNA (dilncRNA) is also synthesized at the sites of DNA double-strand breaks (DSBs), and this dilncRNA recruits the 53BP1 protein, resulting in 53BP1 foci with liquid-droplet properties (44,45). Herein, we consider the experimental evidence pointing to the involvement of the LLPS mechanism in the formation of 53BP1 foci. In the first study, researchers utilized cells expressing near-endogenous levels of GFP-53BP1 (44). Individual GFP-53BP1 foci in these cells manifested a fast homogeneous recovery after photobleaching. Notably, this behavior evolved with time, thereby confirming a progressive increase in internal viscosity (44). In that report, the dynamics (the number, average radius, coalescence, or ripening) and morphology (shape fluctuations) of the 53BP1 condensates were quantified in live cells and supported the viscous-droplet model for 53BP1 foci (44). The average viscosity within foci was estimated from the diffusion coefficient of 53BP1 molecules and appeared to be 500 times higher than that of the nucleoplasm (44). In the second study, live-cell microscopy with ectopic 53BP1 overexpression revealed dynamic spherical 53BP1 compartments that underwent fusion and fission events, and that were highly sensitive to changes in the osmotic pressure, temperature, and ionic strength and to the disruption of hydrophobic interactions by 1,6-hexanediol (45). Moreover, the live-cell experiments were reproduced with endogenously tagged 53BP1 (via CRISPR/Cas9) and confirmed droplet-like behavior of the 53BP1 assemblies (45). The 53BP1 ability to phase separate was validated, and its domains that drive LLPS were identified in vivo by optoDroplet experiments (45). Furthermore, those authors confirmed functional consequences of 53BP1 condensation, because the tumor suppressor protein p53 turned out to be enriched within 53BP1 optoDroplets (45).
3D-SIM indicates that 53BP1 compartments have a hollow architecture: they consist of separate ‘beads’ (53BP1 nanodomains) arranged in a circular fashion around active sites of DSBs, exemplified by XRCC4 or RPA spots (160). In that study, the initial accumulation of 53BP1 was demonstrated to occur immediately after DNA damage induction, and only 10–15 min after 53BP1 nanodomains started to mature to circular microdomains involving the RIF1 factor for shaping chromatin architecture around a DSB (160). The spatiotemporal dynamics of 53BP1 oligomerization during DSB signaling was quantified by Lou et al. (161). They found that preformed 53BP1 dimers are recruited from the nucleoplasm to DSB sites, where the DSB histone code regulates their retention and self-assembly into 53BP1 oligomers.
RIF1 has been recently reported to directly interact with three phosphorylated 53BP1 epitopes, and the simultaneous mutation of these sites abrogated RIF1 accumulation into ionizing radiation-induced foci (162). Interestingly, RIF1 is able to promote 53BP1-dependent DNA repair independently of 53BP1 binding via regulation of shieldin function (162). Notably, the genetic inactivation of RIF1 results in phenotypes similar to 53BP1-null cells (163–165). Indeed, in RIF1-depleted cells, the initial accumulation of 53BP1 was similar to that in wild-type cells, but the resultant 53BP1 nanodomains failed to mature to their circular arrangement, while proteins related to DNA repair (BRCA1 and RPA) lost their focal localization (160). The authors proposed that 53BP1–RIF1 modules safeguard the 3D structure of damaged genomic loci, raise local concentrations of protective factors, and function as a barrier to enzymes whose activity threatens genome stability and epigenetic information encoded in the chromatin architecture (160).
53BP1 foci appear within 5–15 min after genotoxic exposure (166) (or within 30–60 min of DSB induction according to (161)), and can persist in the cell nucleus even for 24 h after DNA repair (167) or up to several days in heavily damaged cells (59,168). Despite rapid PAR degradation (169), PAR foci may have a significantly longer life span due to repeated PARP1 activation on unrepaired DNA breaks. For example, by means of XRCC1 as an indicator of PAR levels, it was demonstrated that PAR foci peak at 1 min after DNA damage, whereas their intensity decreases only to 80% of the maximum in 5 min, and to 40% in 30 min (170). This indicates that both compartments coexist for some period of time.
Given all these data, we hypothesize that 53BP1 nanodomains arise as phase-separated droplets that do not coalesce completely due to rapid maturation and high viscosity [as evidenced by FRAP (44) and 1,6-hexandiol treatment (171)]. Within the 53BP1 microdomain, the emergence of a cavity containing DNA damage and repair factors is ensured by the presence of an incompatible liquid phase, the formation of which precedes the appearance of primary 53BP1 liquid condensates. Interestingly, the formation of 53BP1 foci is delayed in FUS-knockout cells (171). The closure of viscous 53BP1 nanodomains into a circular structure around the ‘core’ is implemented by RIF1 (160). Indeed, according to Altmeyer et al., PAR-mediated assembly of FET proteins at the lesion site is incompatible with the simultaneous accumulation of 53BP1 (5), indicating that the condensates formed by proteins FUS and 53BP1 do not intermix (45). Therefore, at least the intermediate compartment of DSB repair can be assumed to have the structure of a ‘nested’ condensate, with the inner phase generated via PAR-mediated liquid demixing of FET proteins, and the shell organized by phase-separated 53BP1 and dilncRNAs. However, as 53BP1 foci are significantly more long-lived than PAR, it is likely that the layered DNA repair condensate may appear only at early stages and serve for proper assembly of the 53BP1 compartment, which further persists as a hollow structure containing active DSB sites and DNA repair proteins in its center.
Transcription hubs
During transcription, various cis elements (e.g. promoters and enhancers) and trans factors (such as transcription factors [TFs] and coactivators) are recruited to the immediate vicinity of active genes (172). Numerous eukaryotic TFs contain intrinsically disordered low-complexity sequence domains (173), and the LLPS driven by interactions between them may represent an elegant model of transcription coordination and its burst-like dynamics (174,175). A Flory–Huggins free energy model–as applied to the thermodynamic equilibrium of DNA mixed with nonspecific DNA-binding proteins–has uncovered a biphasic regime in solution, and therefore is believed to predict transcription factories (176).
The most vivid illustration of a phase-separated condensate participating in gene expression is the nucleolus. It is thought that smaller transcriptional condensates may be generated by the LLPS mechanism as well, and can promote local biomolecular interactions required for DNA transcription (175,177,178). Nevertheless, the evidence for LLPS taking place during transcription in the cell is rather controversial (1). Transcriptional hubs have a small size and are highly dynamic (173). Moreover, the constituent proteins can interact not only with each other, but also with DNA and RNA (1). Non-specific binding to DNA may produce phenomena similar to, but distinct from, LLPS, as in the case of viral replication compartments (38). Furthermore, accessible DNA sites in the nucleus were recently found to be spatially segregated (179).
Although apparent LLPS is detectable during gross overexpression of TF low-complexity sequence domains, there is no evidence for phase separation of TF hubs (e.g. the EWS/FLI1) formed at endogenous expression levels (173).
Therefore, concerning the topic of LLPS in relation to transcription granules, many authors use cautious terminology, deliberately avoiding the misleading term ‘condensates’ (1,173,178). The regulation of RNA Pol II transcription in terms of both ‘condensates’ (as LLPS-based structures) and hubs is discussed in a recent review (180).
PARPs are key players in transcription. In its inactive state, PARP1 was shown to shape chromatin architecture, where it helps to condense DNA by loop stabilization (181). As a chromatin architectural protein, PARP1 is enriched within heterochromatin (181), while PAR is situated predominantly in actively transcribed genomic regions (182). PARP activity is required for the structure of heat-shock puffs of decondensed transcriptionally activated chromatin in Drosophila (183), and it is reported to be necessary for retaining RNA Pol II within a transcription compartment (11).
In transcription, PARylation modulates DNA-binding activities of specific TFs, and therefore affects their recruitment, triggering chromatin structural rearrangements and modulating the stability and degradation of specific mRNAs (182). PARP1 affects RNA Pol II movement either by reducing or raising the rate of RNA Pol II elongation, depending on the chromatin context (184). In particular, PARylation regulates RNA Pol II silencing during promoter proximal pausing and is involved in the transcriptional response to DNA damage (182). Recently, it has been found that after sensing the lesion, PARP1 interacts with transcription elongation factor P-TEFb, and modifies it at multiple positions (185). PARylation prevents P-TEFb from LLPS that is necessary for Pol II hyperphosphorylation and elongation stimulation (185). Large-scale alterations of the expression of more than 600 genes and several mobile elements in the absence of PARP1 in Drosophila have been documented (186).
Moreover, PAR may serve as a precursor of ATP (187). ATP-Mg affects condensate viscosity, and at a millimolar concentration, it dissolves liquid condensates regardless of its canonical role as an energy source (188). Recently, Mehringer et al. published an study indicating that ATP can prevent fibrillation and enhance protein stability via specific ion effects (salting out) and through interaction of adenosine with delocalized π-systems of amino acid residues in proteins (189). Moreover, ATP chelates most of intracellular Mg2+, the release of which promotes condensation of chromatin by charge neutralization (190). This PAR property is thought to contribute to transcription regulation (191).
Thus, at the moment, it is uncertain to what extent the LLPS mechanism takes part in the regulation of transcription. Therefore, it is not possible to clearly define the role of PAR in the organization of transcriptional condensates. However, one of potential LLPS-concerning models was provided by a recent study, which experimentally confirmed the concept of an RNA-mediated negative feedback mechanism in transcription (192). The study found that small RNAs or low RNA levels at regulatory elements stimulate the transcriptional-condensate assembly, whereas the burst of RNAs (long RNAs or high RNA concentrations) produced during elongation promotes condensate dissolution (192). Accordingly, the inhibition of RNA elongation in vitro and in vivo increases condensate size and prolongs the condensate life span. In contrast, higher levels of local RNA synthesis suppress transcriptional-condensate formation (192). It should be highlighted that the effects of diverse RNAs are sequence independent, suggesting that regulatory potential of RNA is ensured by its inherent high negative-charge density. Furthermore, the re-LLPS model of transcriptional control may account for dynamic burst-like behavior characteristics of transcription (192).
Notably, re-entrant phase behavior during transcription may be more complicated, with condensate nucleation and dissolution being governed by distinct polymers: PAR and RNA (65). PAR is involved in transcription activation, where it mimics RNA and DNA to recruit TFs, and may help with the formation of a transcription compartment, whereas subsequent mRNA production during elongation may dissolve this phase (65).
The spindle
The mitotic spindle is a bipolar dynamic macromolecular structure that arises via self-organized assembly of microtubules (dimers of α- and β-tubulin), microtubule-associated proteins, and motor proteins (193). The spindle is regarded as a condensate with liquid–crystalline properties (194). The LLPS of key proteins involved in spindle function was demonstrated in recent publications (195,196).
Nuclear mitotic apparatus protein (NuMA) regulates spindle polarity (197). In acentrosomal human cells, overexpressed NuMA was reported to form multiple condensates that promote spindle bipolarization by organizing microtubule asters (198). Recently, it became known that at spindle poles, endogenous NuMA also forms numerous condensates, which concentrate tubulins, bind microtubules, and enrich crucial regulators (195). These condensates manifest liquid-like behavior by fusion and fissure, are sensitive to 1,6-hexanediol, and exhibit rapid FRAP recoveries. The phase separation of NuMA is mediated by its C-terminus, whose phosphorylation by Aurora A reduces LLPS and fluidizes NuMA condensates (195). By expressing NuMA mutants in endogenous-NuMA–depleted cells, researchers found that the expression of an LLPS-incapable NuMA mutant results in significantly longer spindle assembly time compared to the wild-type protein, while expression of the unphosphorylatable mutant leads to the assembly of a shorter spindle (195).
In 2004, researchers discovered that PAR is enriched in the spindle and undergoes unusual localization dynamics consistent with a low turnover rate (10). PAR cleavage causes rapid disruption of the spindle structure, and PAR hydrolysis during spindle assembly prevents the formation of the bipolar spindle (10). Moreover, NuMA is a target of covalent modification by PARP5a during mitosis (197), and binds PAR in a noncovalent manner (199). However, PARylation and PAR binding have no influence on NuMA localization to spindle poles (197,199). PAR itself has been detected at spindle poles and is thought to perform a dynamic cross-linking function (199).
Combining these observations with modern data suggestive of NuMA LLPS (195), we speculate that PAR ensures the correct assembly and liquidity of NuMA condensates. We would like to underline the finding that after forming in the plural during nuclear-envelope breakdown, NuMA droplets flow and fuse when transported along microtubules to spindle poles, where they eventually generate large condensates (195). Therefore, it can be theorized that premature solidification of NuMA condensates in the absence of PAR, similar to that observed in the case of unphosphorylatable NuMA, can cause incomplete fusion of droplets and the emergence of extra poles. This question remains open for further research.
Notably, PARP5a PARylates α-tubulin, and this modification recruits the ECT2 factor required for cytokinesis control (200). A comprehensive discussion of PARPs’ participation in mitosis is provided in another review (201).
RNP granules
SGs
SGs are inducible cytoplasmic condensates that concentrate mRNA, RBPs and small ribosomal subunits of eukaryotic cells under stressful conditions (202). SG assembly is triggered by a sudden upsurge of cytoplasmic mRNA levels due to inactivation of translation and release of free mRNAs (202–204).
It has been proposed that SG formation is mediated by LLPS, however, there are several objections to this model (discussed in ref. (202)).
Several proteins that are located within SGs, for example, FUS, hnRNPA1 and TIA-1, have been reported to phase separate in vitro (6,205,206). Nevertheless, only G3BP proteins (G3BP1 and its homolog G3BP2) are essential for SG assembly, because cells with a double knockout of genes G3BP1 and G3BP2 do not form SGs in response to arsenite (207). Light-induced LLPS of G3BP1 in an optogenetic system is sufficient to initiate the formation of SGs in the cell, even in the absence of exogenous stress (208). In contrast, light-induced LLPS of other SG proteins, including TIA1, FUS, and TDP-43, results in liquid-like droplets, whose content and biophysical properties are very different from those of normal SGs (208). The seeding of purified G3BP1 in a mammalian cell lysate induces LLPS with the assembly of condensates, whose RNA and protein composition is highly similar to that of SGs (209). An excellent research study revealed that G3BPs are intracellular sensors of protein-free unfolded RNA and outlines a mechanism underlying G3BP granule formation (202). According to this mechanism, unfolded mRNAs outcompete G3BP autoinhibitory interactions, thereby affording a conformational switch that couples RNA binding to a dramatic increase in multivalence. G3BP1 clusters (210) cross-link RNA molecules into mesh-like inhomogeneous condensates of low protein density (202). Moreover, G3BP1-driven recruitment of RNA into fluid condensates may inhibit RNA entanglement into irreversible structures based on homotypic RNA–RNA interactions (202).
The functions of PAR in SG organization are summarized in a recent review (211). In particular, PAR regulates protein targeting to SGs (8,9,212). Thus, in Drosophila TDP-43 protein binds target substrates modified by Tankyrase/PARP5a, which results in LLPS of TDP-43 and its accumulation in SGs (8). Non-covalent binding of hnRNP A1 with PAR or PARylated targets controls its sorting to SGs, while in vitro addition of PAR to this protein induces its LLPS (9). Moreover, PAR regulates interactions between hnRNP A1 and TDP-43 and promotes co-LLPS of these proteins (9).
In accordance with the forementioned observations, the PAR interaction with G3BP is of particular interest in the context of SG organization. PAR non-covalently binds to the G3BP C-terminal RG-rich domain, allowing G3BP to maintain cytoplasmic localization and enabling subsequent assembly of SGs (213). Notably, it is this RG-rich region of G3BP that must be released—either by high local protein concentrations or via the removal of the inhibitory acidic IDR—to switch G3BP from an autoinhibited state to a multivalent one, which allows for cooperative binding of RNA and for condensate assembly (202).
A recent study indicates that alphavirus ADP-ribosylhydrolase nsP3 suppresses the formation of SGs: its expression gives rise to distinct condensates that lack translation initiation factors, but contain other SG-associated RBPs. The expression of ADP-ribosylhydrolase–deficient nsP3 yields condensates that retain translation initiation factors as well as RBPs, similarly to SGs (214).
Furthermore, it is possible that PAR can finely influence a condensate—s material properties as much as RNA (see also subsection ASK3 condensates). For example, yeast SGs have more viscous solid-like properties than mammalian SGs, which manifest liquid-like behavior (215). It is likely that a reason for this difference is the absence of PARylation in yeast (156).
ASK3 condensates
After osmotic shock, the cell recovers its initial volume under the influence of kinase ASK3 (apoptosis signal-regulating kinase 3). Under hyperosmotic stress, the cell undergoes contraction accompanied by the concentration of cell contents, resulting in ASK3 condensates that transduce an osmosensing signal driving ASK3 inactivation (216). ASK3 condensates are distinct from SGs and P-bodies, which also appear under extreme hyperosmotic conditions, because the markers of these membraneless structures are not colocalized (216).
ASK3 condensation has been suggested to proceed via LLPS. In cells stably expressing ASK3, ASK3 condensates emerge within seconds after hyperosmotic stress initiation, then dynamically move and fuse, showing incomplete but significant FRAP recoveries (216). It is particularly noteworthy that the ASK3 condensation has a ‘spinodal-decomposition-like’ pattern, which can be a good argument in favor of the LLPS mechanism. In vitro, the formation of ASK3 droplets is induced by crowding reagents, but not by alterations of ionic strength. Indeed, in the cell, greater hyperosmolarity increases the number and diminishes the size of ASK3 condensates, thereby pointing to the critical importance of macromolecular crowding in condensate organization.
Notably, ASK3 condensates obtained in vitro are solid-like, as their FRAP recovery is undetectable (216). On the contrary, the addition of either poly(A) RNA or PAR (but not NAD+) suppresses the generation of solid ASK3 condensates. PAR depletion in the cell does not prevent ASK3 condensation under hyperosmotic stress, but significantly inhibits FRAP of the emerging condensates. An ASK3 mutant insensitive to PAR regulation yields ASK3 condensates with significantly reduced FRAP recoveries in vitro and in vivo (216).
Therefore, PAR proves to be important for the maintenance of ASK3 condensate liquidity. The main reason seems to be its ability to serve as a multivalent intermediary coordinating transient interactions between ASK3 molecules (216). However, the report also proposed that flexible and branching PAR may act as a ‘loosening filler’ and promote ASK3 condensate porosity by generating empty spaces within the condensates. It was concluded that this effect significantly influences the viscoelasticity of condensates and enhances ASK3 movement (216).
Pathological aggregates
The overexpression of PARP1 is associated with the pathogenesis of several central-nervous-system disorders, such as strokes, Parkinson’s disease (PD), Alzheimer’s disease, Huntington’s disease, and amyotrophic lateral sclerosis (217). The pathological change common among the last four neurodegenerative diseases is abnormal protein aggregation, for example, when an amyloid protein (such as α-syn, amyloid β, tau, FUS, TDP-43 or hnRNPA1) undergoes conformational alterations giving rise to pathological aggregates associated with disease progression (218). Under physiological conditions, PARylation regulates the assembly/disassembly of SGs containing proteins from this list, whereas dysregulation of PAR levels may induce condensation of irreversible amyloid aggregates and mitigate their cytotoxicity (218). Conversely, amyloid fibrils can stimulate PARP1 activity and elevate PAR levels, which play a major role in cell death (including apoptosis, necroptosis, and parthanatos) as well as activation of PARylation-dependent ubiquitination (218). Consequently, amyloid protein aggregation and PARylation may constitute a feed-forward loop that accelerates the progression of neurodegenerative diseases (218).
In a case study, high levels of PAR were monitored in the cerebrospinal fluid of PD patients (219). PAR has been reported to seed and accelerate α-syn fibrillization in cell-free experiments and in the cell (upon the addition of exogenous PAR or DNA-damaging treatment) and to convert pathological α-syn to a more toxic subtype (219). In a feed-forward loop, preassembled α-syn fibrils activate PARP1 via nitric-oxide–induced DNA damage (219). PAR interactions with hyperphosphorylated α-syn are predominantly observed in PD transgenic murine models of α-syn pathology and in postmortem tissue samples from PD/PDD patients (220). The study confirmed that these interactions involve electrostatic forces between negatively charged PAR and positively charged Lys residues at the α-syn N terminus (220).
A recent study by Rhine et al. elucidated how PAR can trigger FUS transition into LLPS-prone form (60) (discussed also in the subsection nucleic acids in condensates: the basic principles). The FUS primed by short-lived interaction with PAR can recruit unreacted FUS molecules, inducing condensate formation and persistence without PAR (60). This mechanism for transferring an induced conformation from one protein molecule to another resembles prion-like spreading (221), which in the case of FUS was demonstrated for the G156E FUS mutant (222). The influence of nucleic acids on conformational changes and aggregation of amiloidogenic proteins was reviewed in a recent study (223).
Interestingly, the FUS mutation was implicated in aggressive cases of ALS coupled with frontotemporal lobar dementia (FTLD) symptoms, which alters material properties of FUS condensates, and is also connected with increased PAR content that may drive solidification (60).
The ability of PAR to enhance the aggregation of LCD-containing proteins in vitro, including FUS, was also shown by Altmeyer et al. (5). In this study, co-incubation of FET proteins or a model peptide, containing a short prion-like sequence and a triple RGG repeat, with sub-stoichiometric amounts of purified PAR significantly promoted the aggregation process, resulting in larger, more condensed aggregates (5).
A PAR-driven LLPS-like process may play a central part in pathological extracellular-matrix calcification as well, by generating calcified spherical particles reminiscent of those observed during vascular calcification (138).
CONCLUSIONS AND PERSPECTIVES
The emergence of the LLPS paradigm in the context of biomolecular interactions motivated great interest of the scientific community to study the mechanisms of non-membrane cell architecture organization. Critical work by McSwiggen et al. (2019) revealed the limitations of the LLPS concept by demonstrating the insufficiency of descriptive characteristics for conclusions about the phase-separated origin of a particular structure. However, the subsequent development of the topic, in conjunction with the parallel elaboration of methods over the past three years, has led this area to the establishment of significantly more stringent standards for the quality of experiments and the terminology used.
Recently, the question of the validity of the hypothesis that phase separation is the primary driving force behind compartment assembly has been raised in the article by Musacchio (23). The study draws the readers’ attention to the fact that the classical LLPS model as a molecular mechanism for the formation of non-membrane organelles may be considered counterintuitive. In particular, it is not clear how low-affinity and low-specificity interactions can provide selective and localized assembly of macromolecules in place and time under conditions of a large density of competing interactions that takes place in a living cell. How do the compartments formed maintain their identity? And how is the required cellular critical concentration of a particular LLPS-driver adjusted across different tissues and organisms (23)? The author provides several convincing arguments in favor of the fact that the primary drivers of condensate assembly are site-specific interactions (SSIs). This does not negate the fact that the LLPS mechanism can be involved at later stages of compartment organization, investing the structure with the behavior characteristic to LLPS-derived condensates (23). Indeed, even with the theoretical possibility of forming only stoichiometric networks of interactions, the experiment may indicate the occurrence of stochastic binding events that organize preformed stoichiometric clusters into structures of a higher order (22).
Numerous examples of classical droplet-like compartments, even among those considered in our review, confirm the idea of SSIs-induced or restricted phase separation (23). Thus, the sub-stoichiometric action of lncRNAs in the organization of phase-separated condensates (61) is based on a similar principle. Assembly of the nucleolus and transcription hubs requires active transcription, which results in SSIs between protein components and specific sequences on specific RNAs (23). RNA-triggered FUS condensation likewise begins with the interaction of the FUS nucleic-acid-binding domain with RNA molecules (58). G3BP granule formation starts from binding of unfolded mRNAs to specific regions of G3BP, resulting in blocking of autoinhibitory interactions (202). This restricted phase separation can be implemented not only with the participation of RNA, but also in the case of PAR due to SSI with either specific PAR-recognizing domains or high-affinity electrostatic interactions (136).
However, there are two examples discussed above that do not quite fit into the described scheme and deserve closer study. The first one is the ability of PAR to prime FUS into LLPS-prone state (60). Of particular importance is that PAR concentration (1 nM), sufficient for inducing FUS condensation, is manifoldly lower than the 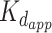 (>200 nM) for PAR–FUS binding (60). FUS aggregation by sub-stoichiometric amounts of PAR is also shown earlier (5). The second example is ASK3 condensation in cells under hyperosmotic stress that has a spinodal-decomposition-like pattern (216) and therefore appears not to have a specific sites of nucleation. Both of these case studies must be further investigated, possibly in accordance with the validation pipeline proposed in (23), to determine whether general phase separation, which is not triggered by SSIs, is the driving force behind their assembly.
(>200 nM) for PAR–FUS binding (60). FUS aggregation by sub-stoichiometric amounts of PAR is also shown earlier (5). The second example is ASK3 condensation in cells under hyperosmotic stress that has a spinodal-decomposition-like pattern (216) and therefore appears not to have a specific sites of nucleation. Both of these case studies must be further investigated, possibly in accordance with the validation pipeline proposed in (23), to determine whether general phase separation, which is not triggered by SSIs, is the driving force behind their assembly.
The hypothesis about the role of PAR in biomolecular condensate formation has been discussed previously (2,21,59,224–226). In the present review, we examined how PAR is involved in condensate organization and attempted to discuss as completely as possible how phase separation–considered a contributing mechanism in the literature–is confirmed for each PAR-containing condensate.
We further paid special attention to the comparison between PAR and RNA, as well as to their interplay during the organization of condensates. The main idea is that PAR and RNA are quite similar, and yet different. Being polymeric scaffolds, PAR and RNA can come into multiple specific and nonspecific contacts. Specific interactions may afford self-organization via binding, bridging, and the formation of stoichiometric complexes, which among other things, may seed LLPS when organizing larger-scale structures. Having different sets of partner proteins, PAR and RNA can ensure the immiscibility of liquid phases and the formation of multilayer condensates.
It was shown that among PAR-binding proteins, more than one third also bind DNA: RNA hybrids, and the most of the last ones contribute to the induction of LLPS. Several proteins exhibited all three of the listed properties. This may suggest that R-loops, dilncRNAs and PAR all contribute to LLPS at DNA lesion sites (227).
Non-specific electrostatic interactions with PAR or RNA may take part in phase separation by complex coacervation. For non-specific interactions, these polyanions are interchangeable. In this regard, they can modulate condensate liquidity in a similar manner, and function as scaffold molecules during droplet assembly/disassembly. PAR can act as a more rapidly synthesized and less site-dependent RNA analog initiating RNA-dependent systems at the early stages of their activity. Similarities between PAR and RNA can be helpful in systems with phase re-entrant behavior.
Furthermore, it must be pointed out that the advantage of PAR for the assembly of transient compartments is evident due to the reversibility of PAR synthesis and degradation (59,159). PAR is important for the turnover of DNA repair, or repair coupled to DNA replication and to other highly dynamic intracellular processes, whereas RNA can contribute to relatively stable condensates.
ACKNOWLEDGEMENTS
The authors are grateful to Dr M.V. Sukhanova for careful reading of the manuscript and useful comments and discussion. We also express our gratitude to Nikolay Shevchuk and Enago Editing Service for language editing. The principles of condensate organization are discussed in the frame of project no. 20-14-00086 from the Russian Science Foundation. The role of PAR in formation of functional structures is discussed within the framework of project no. 21-64-00017 from the Russian Science Foundation.
Contributor Information
Elizaveta E Alemasova, Institute of Chemical Biology and Fundamental Medicine, SB RAS, Novosibirsk 630090, Russia.
Olga I Lavrik, Institute of Chemical Biology and Fundamental Medicine, SB RAS, Novosibirsk 630090, Russia; Novosibirsk State University, Novosibirsk 630090, Russia.
FUNDING
Russian Science Foundation [20-14-00086, 21-64-00017]. Funding for open access charge: Russian Science Foundation [20-14-00086, 21-64-00017].
Conflict of interest statement. None declared.
REFERENCES
- 1. McSwiggen D.T., Mir M., Darzacq X., Tjian R.. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Develop. 2019; 33:1619–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leung A. K.L. Poly(ADP-ribose): a dynamic trigger for biomolecular condensate formation. Trends Cell Biol. 2020; 30:370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boamah E.K., Kotova E., Garabedian M., Jarnik M., Tulin A.V.. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet. 2012; 8:e1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Engbrecht M., Mangerich A.. The nucleolus and PARP1 in cancer biology. Cancers. 2020; 12:1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Altmeyer M., Neelsen K.J., Teloni F., Pozdnyakova I., Pellegrino S., Grøfte M., Rask M.-B.D., Streicher W., Jungmichel S., Nielsen M.L.et al.. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 2015; 6:8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M.et al.. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015; 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- 7. Leung A.K.L., Vyas S., Rood J.E., Bhutkar A., Sharp P.A., Chang P.. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell. 2011; 42:489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McGurk L., Gomes E., Guo L., Mojsilovic-Petrovic J., Tran V., Kalb R.G., Shorter J., Bonini N.M.. Poly(ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell. 2018; 71:703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duan Y., Du A., Gu J., Duan G., Wang C., Gui X., Ma Z., Qian B., Deng X., Zhang K.et al.. PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res. 2019; 29:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang P., Jacobson M.K., Mitchison T.J.. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004; 432:645–649. [DOI] [PubMed] [Google Scholar]
- 11. Zobeck K.L., Buckley M.S., Zipfel W.R., Lis J.T.. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol. Cell. 2010; 40:965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cowie J., Arrighi V.. Polymers: Chemistry and Physics of Modern Materials. 2007; 3rd edn.CRC Press. [Google Scholar]
- 13. Ribeiro S.S., Samanta N., Ebbinghaus S., Marcos J.C.. The synergic effect of water and biomolecules in intracellular phase separation. Nat. Rev. Chem. 2019; 3:552–561. [Google Scholar]
- 14. Ahlers J., Adams E.M., Bader V., Pezzotti S., Winklhofer K.F., Tatzelt J., Havenith M.. The key role of solvent in condensation: Mapping water in liquid-liquid phase-separated FUS. Biophys. J. 2021; 120:1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brangwynne C.P., Tompa P., Pappu R.V.. Polymer physics of intracellular phase transitions. Nat. Phys. 2015; 11:899–904. [Google Scholar]
- 16. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Posey A.E., Holehouse A.S., Pappu R.V.. Phase separation of intrinsically disordered proteins. Methods in Enzymology. 2018; 611:Academic Press Inc; 1–30. [DOI] [PubMed] [Google Scholar]
- 18. Hamley I.W. Introduction to Soft Matter: Synthetic and Biological Self-assembling Materials, Revised Edition. 2007; John Wiley & Sons, Ltd. [Google Scholar]
- 19. Peng A., Weber S.C.. Evidence for and against liquid-liquid phase separation in the nucleus. Non-coding RNA. 2019; 5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Erdel F., Rippe K.. Formation of chromatin subcompartments by phase separation. Biophys. J. 2018; 114:2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spegg V., Altmeyer M.. Biomolecular condensates at sites of DNA damage: More than just a phase. DNA Repair. 2021; 106:103179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin Y.-H., Wu H., Jia B., Zhang M., Chan H.S.. Assembly of model postsynaptic densities involves interactions auxiliary to stoichiometric binding. Biophys. J. 2022; 121:157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Musacchio A. On the role of phase separation in the biogenesis of membraneless compartments. EMBO J. 2022; 41:e109952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mittag T., Pappu R.V.. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell. 2022; 82:2201–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmit J.D., Feric M., Dundr M.. How hierarchical interactions make membraneless organelles tick like clockwork. Trends Biochem. Sci. 2021; 46:525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mir M., Bickmore W., Furlong E.E.M., Narlikar G.. Chromatin topology, condensates and gene regulation: Shifting paradigms or just a phase?. Development (England). 2019; 146:dev182766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hongchen Z., Shipeng S., Yujie S.. Characterization of liquid–liquid phase separation using super-resolution and single-molecule imaging. Biophys. Rep. 2022; 8:2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Azaldegui C.A., Vecchiarelli A.G., Biteen J.S.. The emergence of phase separation as an organizing principle in bacteria. Biophys. J. 2021; 120:1123–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erdel F., Rademacher A., Vlijm R., Tünnermann J., Frank L., Weinmann R., Schweigert E., Yserentant K., Hummert J., Bauer C.et al.. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell. 2020; 78:236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Irgen-Gioro S., Walling V., Chong S.. Fixation can change the appearance of phase separation in living cells. 2022; bioRxiv doi:08 May 2022, preprint: not peer reviewed 10.1101/2022.05.06.490956. [DOI] [PMC free article] [PubMed]
- 31. Shakya A., King J.T.. Modern optical microscopy methods to study biomolecular condensates. Curr. Opin. Coll. Int. Sci. 2021; 52:101421. [Google Scholar]
- 32. Mitrea D.M., Chandra B., Ferrolino M.C., Gibbs E.B., Tolbert M., White M.R., Kriwacki R.W.. Methods for physical characterization of phase-separated bodies and membrane-less organelles. J. Mol. Biol. 2018; 430:4773–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rhine K., Skanchy S., Myong S.. Single-molecule and ensemble methods to probe RNP nucleation and condensate properties. Methods. 2022; 197:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alshareedah I., Kaur T., Banerjee P.R.. Methods for characterizing the material properties of biomolecular condensates. Meth. Enzymol. 2021; 646:143–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sprague B.L., McNally J.G.. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005; 15:84–91. [DOI] [PubMed] [Google Scholar]
- 36. Delarue M., Brittingham G.P., Pfeffer S., Surovtsev I.V., Pinglay S., Kennedy K.J., Schaffer M., Gutierrez J.I., Sang D., Poterewicz G.et al.. mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell. 2018; 174:338–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hansen A.S., Woringer M., Grimm J.B., Lavis L.D., Tjian R., Darzacq X.. Robust model-based analysis of single-particle tracking experiments with Spot-On. eLife. 2018; 7:e33125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McSwiggen D.T., Hansen A.S., Teves S.S., Marie-Nelly H., Hao Y., Heckert A.B., Umemoto K.K., Dugast-Darzacq C., Tjian R., Darzacq X.. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. eLife. 2019; 8:e47098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heltberg M.L., Miné-Hattab J., Taddei A., Walczak A.M., Mora T.. Physical observables to determine the nature of membrane-less cellular sub-compartments. eLife. 2021; 10:e69181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miné-Hattab J., Heltberg M., Villemeur M., Guedj C., Mora T., Walczak A.M., Dahan M., Taddei A.. Single molecule microscopy reveals key physical features of repair foci in living cells. eLife. 2021; 10:e60577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kent S., Brown K., Yang C.-H., Alsaihati N., Tian C., Wang H., Ren X.. Phase-separated transcriptional condensates accelerate target-search process revealed by live-cell single-molecule imaging. Cell Rep. 2020; 33:108248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Itoh Y., Iida S., Tamura S., Nagashima R., Shiraki K., Goto T., Hibino K., Ide S., Maeshima K.. 1,6-hexanediol rapidly immobilizes and condenses chromatin in living human cells. Life Sci. Alliance. 2021; 4:e202001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Narayanan A., Meriin A., Andrews J.O., Spille J.-H., Sherman M.Y., Cisse I.I.. A first order phase transition mechanism underlies protein aggregation in mammalian cells. eLife. 2019; 8:e39695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pessina F., Giavazzi F., Yin Y., Gioia U., Vitelli V., Galbiati A., Barozzi S., Garre M., Oldani A., Flaus A.et al.. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 2019; 21:1286–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kilic S., Lezaja A., Gatti M., Bianco E., Michelena J., Imhof R., Altmeyer M.. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019; 38:e101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang H., Shao S., Zeng Y., Wang X., Qin Y., Ren Q., Xiang S., Wang Y., Xiao J., Sun Y.. Reversible phase separation of HSF1 is required for an acute transcriptional response during heat shock. Nat. Cell Biol. 2022; 24:340–352. [DOI] [PubMed] [Google Scholar]
- 47. Farahi N., Lazar T., Wodak S.J., Tompa P., Pancsa R.. Integration of data from liquid–liquid phase separation databases highlights concentration and dosage sensitivity of llps drivers. Int. J. Mol. Sci. 2021; 22:3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ditlev J.A., Case L.B., Rosen M.K.. Who’s In and Who’s Out-Compositional Control of Biomolecular Condensates. J. Mol. Biol. 2018; 430:4666–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K.. Compositional Control of Phase-Separated Cellular Bodies. Cell. 2016; 166:651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dutagaci B., Nawrocki G., Goodluck J., Ashkarran A.A., Hoogstraten C.G., Lapidus L.J., Feig M.. Charge-driven condensation of RNA and proteins suggests broad role of phase separation in cytoplasmic environments. eLife. 2021; 10:e64004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Overbeek J.T., Voorn M.J.. Phase separation in polyelectrolyte solutions; theory of complex coacervation. J. Cell. Physiol. Supp. 1957; 49:7–26. [PubMed] [Google Scholar]
- 52. André A.A.M., Spruijt E.. Rigidity rules in DNA droplets: Nucleic acid flexibility affects model membraneless organelles. Biophys. J. 2018; 115:1837–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shakya A., King J.T.. DNA local-flexibility-dependent assembly of phase-separated liquid droplets. Biophys. J. 2018; 115:1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mimura M., Tomita S., Sugai H., Shinkai Y., Ishihara S., Kurita R.. Uncharged components of single-stranded DNA modulate liquid-liquid phase separation with cationic linker histone H1. Front. Cell Dev. Biol. 2021; 9:710729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilson K.A., Wells R.A., Abendong M.N., Anderson C.B., Kung R.W., Wetmore S.D.. Landscape of π-π and sugar-π contacts in DNA-protein interactions. J. Biomol. Struct. Dyn. 2016; 34:184–200. [DOI] [PubMed] [Google Scholar]
- 56. Muzzopappa F., Hertzog M., Erdel F.. DNA length tunes the fluidity of DNA-based condensates. Biophys. J. 2021; 120:1288–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Renger R., Morin J.A., Lemaitre R., Ruer-Gruss M., Jülicher F., Hermann A., Grill S.W.. Co-condensation of proteins with single- and double-stranded DNA. Proc. Natl. Acad. Sci. USA. 2022; 119:e2107871119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Niaki A.G., Sarkar J., Cai X., Rhine K., Vidaurre V., Guy B., Hurst M., Lee J.C., Koh H.R., Guo L.et al.. Loss of dynamic RNA interaction and aberrant phase separation induced by two distinct types of ALS/FTD-Linked FUS mutations. Mol. Cell. 2020; 77:82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Singatulina A.S., Hamon L., Sukhanova M.V., Desforges B., Joshi V., Bouhss A., Lavrik O.I., Pastré D.. PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 2019; 27:1809–1821. [DOI] [PubMed] [Google Scholar]
- 60. Rhine K., Dasovich M., Yoniles J., Badiee M., Skanchy S., Ganser L.R., Ge Y., Fare C.M., Shorter J., Leung A.K.L.et al.. Poly(ADP-ribose) drives condensation of FUS via a transient interaction. Mol. Cell. 2022; 82:969–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Unfried J.P., Ulitsky I.. Substoichiometric action of long noncoding RNAs. Nat. Cell Biol. 2022; 24:608–615. [DOI] [PubMed] [Google Scholar]
- 62. Frank L., Rippe K.. Repetitive RNAs as regulators of chromatin-associated subcompartment formation by phase separation. J. Mol. Biol. 2020; 432:4270–4286. [DOI] [PubMed] [Google Scholar]
- 63. Garcia-Jove Navarro M., Kashida S., Chouaib R., Souquere S., Pierron G., Weil D., Gueroui Z.. RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nat. Commun. 2019; 10:3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Roden C., Gladfelter A.S.. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021; 22:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Portz B., Shorter J.. Biochemical timekeeping via reentrant phase transitions. J. Mol. Biol. 2021; 433:166794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang H., Elbaum-Garfinkle S., Langdon E.M., Taylor N., Occhipinti P., Bridges A.A., Brangwynne C.P., Gladfelter A.S.. RNA controls polyq protein phase transitions. Mol. Cell. 2015; 60:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Langdon E.M., Qiu Y., Ghanbari Niaki A., McLaughlin G.A., Weidmann C.A., Gerbich T.M., Smith J.A., Crutchley J.M., Termini C.M., Weeks K.M.et al.. mRNA structure determines specificity of a polyQ-driven phase separation. Science (New York, N.Y.). 2018; 360:922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jain A., Vale R.D.. RNA phase transitions in repeat expansion disorders. Nature. 2017; 546:243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ma W., Zheng G., Xie W., Mayr C.. In vivo reconstitution finds multivalent RNA-RNA interactions as drivers of mesh-like condensates. eLife. 2021; 10:e64252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Boeynaems S., Holehouse A.S., Weinhardt V., Kovacs D., Van Lindt J., Larabell C., Van Den Bosch L., Das R., Tompa P.S., Pappu R.V.et al.. Spontaneous driving forces give rise to protein–RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:7889–7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kaur T., Raju M., Alshareedah I., Davis R.B., Potoyan D.A., Banerjee P.R.. Sequence-encoded and composition-dependent protein–RNA interactions control multiphasic condensate morphologies. Nat. Commun. 2021; 12:872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pyo A. G.T., Zhang Y., Wingreen N.S.. Surface tension and super-stoichiometric surface enrichment in two-component biomolecular condensates. iScience. 2022; 25:103852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cochard A., Garcia-Jove Navarro M., Piroska L., Kashida S., Kress M., Weil D., Gueroui Z.. RNA at the surface of phase-separated condensates impacts their size and number. Biophys. J. 2022; 121:1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fare C.M., Villani A., Drake L.E., Shorter J.. Higher-order organization of biomolecular condensates. Open Biol. 2021; 11:210137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Choi S., Bevilacqua P., Meyer M.O., Keating C.. Phase-specific RNA accumulation and duplex thermodynamics in multiphase coacervate models for membraneless organelles. Nat. Chem. 2022; 14:1110–1117. [DOI] [PubMed] [Google Scholar]
- 76. Sanchez-Burgos I., Espinosa J.R., Joseph J.A., Collepardo-Guevara R.. Valency and binding affinity variations can regulate the multilayered organization of protein condensates with many components. Biomolecules. 2021; 11:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Regy R.M., Dignon G.L., Zheng W., Kim Y.C., Mittal J.. Sequence dependent phase separation of protein–polynucleotide mixtures elucidated using molecular simulations. Nucleic Acids Res. 2020; 48:12593–12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Onuchic P.L., Milin A.N., Alshareedah I., Deniz A.A., Banerjee P.R.. Divalent cations can control a switch-like behavior in heterotypic and homotypic RNA coacervates. Sci. Rep. 2019; 9:12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ma Y., Li H., Gong Z., Yang S., Wang P., Tang C.. Nucleobase Clustering Contributes to the Formation and Hollowing of Repeat-Expansion RNA Condensate. J. Am. Chem. Soc. 2022; 144:4716–4720. [DOI] [PubMed] [Google Scholar]
- 80. Laghmach R., Alshareedah I., Pham M., Raju M., Banerjee P.R., Potoyan D.A.. RNA chain length and stoichiometry govern surface tension and stability of protein–RNA condensates. iScience. 2022; 25:104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Saha S., Weber C.A., Nousch M., Adame-Arana O., Hoege C., Hein M.Y., Osborne-Nishimura E., Mahamid J., Jahnel M., Jawerth L.et al.. Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell. 2016; 166:1572–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Riback J.A., Zhu L., Ferrolino M.C., Tolbert M., Mitrea D.M., Sanders D.W., Wei M.-T., Kriwacki R.W., Brangwynne C.P.. Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020; 581:209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Banerjee P.R., Milin A.N., Moosa M.M., Onuchic P.L., Deniz A.A.. Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew. Chem. Int. Ed. 2017; 56:11354–11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Alshareedah I., Moosa M.M., Raju M., Potoyan D.A., Banerjee P.R.. Phase transition of RNA–protein complexes into ordered hollow condensates. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:15650–15658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Milin A.N., Deniz A.A.. Reentrant phase transitions and non-equilibrium dynamics in membraneless organelles. Biochemistry. 2018; 57:2470–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Talhaoui I., Lebedeva N.A., Zarkovic G., Saint-Pierre C., Kutuzov M.M., Sukhanova M.V., Matkarimov B.T., Gasparutto D., Saparbaev M.K., Lavrik O.I.et al.. Poly(ADP-ribose) polymerases covalently modify strand break termini in DNA fragments in vitro. Nucleic Acids Res. 2016; 44:9279–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zarkovic G., Belousova E.A., Talhaoui I., Saint-Pierre C., Kutuzov M.M., Matkarimov B.T., Biard D., Gasparutto D., Lavrik O.I., Ishchenko A.A.. Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: new insights into DNA ADP-ribosylation. Nucleic Acids Res. 2018; 46:2417–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Matta E., Kiribayeva A., Khassenov B., Matkarimov B.T., Ishchenko A.A.. Insight into DNA substrate specificity of PARP1-catalysed DNA poly(ADP-ribosyl)ation. Sci. Rep. 2020; 10:3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Weixler L., Schäringer K., Momoh J., Lüscher B., Feijs K. L.H., Žaja R.. ADP-ribosylation of RNA and DNA: from in vitro characterization to in vivo function. Nucleic Acids Res. 2021; 49:3634–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Vyas S., Matic I., Uchima L., Rood J., Zaja R., Hay R.T., Ahel I., Chang P.. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014; 5:4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tan E.S., Krukenberg K.A., Mitchison T.J.. Large-scale preparation and characterization of poly(ADP-ribose) and defined length polymers. Anal. Biochem. 2012; 428:126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Eustermann S., Wu W.-F., Langelier M.-F., Yang J.-C., Easton L.E., Riccio A.A., Pascal J.M., Neuhaus D.. Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol. Cell. 2015; 60:742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Obaji E., Maksimainen M.M., Galera-Prat A., Lehtiö L.. Activation of PARP2/ARTD2 by DNA damage induces conformational changes relieving enzyme autoinhibition. Nat. Commun. 2021; 12:3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Alvarez-Gonzalez R., Jacobson M.K.. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987; 26:3218–3224. [DOI] [PubMed] [Google Scholar]
- 95. de Murcia G., Jongstra-Bilen J., Ittel M.E., Mandel P., Delain E.. Poly(ADP-ribose) polymerase auto-modification and interaction with DNA: electron microscopic visualization. EMBO J. 1983; 2:543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hayashi K., Tanaka M., Shimada T., Miwa M., Sugimura T.. Size and shape of poly(ADP-ribose): examination by gel filtration, gel electrophoresis and electron microscopy. Biochem. Biophys. Res. Commun. 1983; 112:102–107. [DOI] [PubMed] [Google Scholar]
- 97. Sukhanova M.V., Abrakhi S., Joshi V., Pastre D., Kutuzov M.M., Anarbaev R.O., Curmi P.A., Hamon L., Lavrik O.I.. Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly(ADP-ribosyl)ation using high-resolution AFM imaging. Nucleic Acids Res. 2016; 44:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Alemasova E.E., Lavrik O.I.. Poly(ADP-ribosyl)ation by PARP1: reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019; 47:3811–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chen Q., Kassab M.A., Dantzer F., Yu X.. PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat. Commun. 2018; 9:3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lonskaya I., Potaman V.N., Shlyakhtenko L.S., Oussatcheva E.A., Lyubchenko Y.L., Soldatenkov V.A.. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J. Biol. Chem. 2005; 280:17076–17083. [DOI] [PubMed] [Google Scholar]
- 101. Léger K., Bär D., Savić N., Santoro R., Hottiger M.O.. ARTD2 activity is stimulated by RNA. Nucleic Acids Res. 2014; 42:5072–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Huambachano O., Herrera F., Rancourt A., Satoh M.S.. Double-stranded DNA binding domain of poly (ADP-ribose) Polymerase-1 and molecular insight into the regulation of its activity. J. Biol. Chem. 2011; 286:7149–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Alemasova E.E., Moor N.A., Naumenko K.N., Kutuzov M.M., Sukhanova M.V., Pestryakov P.E., Lavrik O.I.. Y-box-binding protein 1 as a non-canonical factor of base excision repair. Biochim. Biophys. Acta. 2016; 1864:1631–1640. [DOI] [PubMed] [Google Scholar]
- 104. Nakamoto M.Y., Rudolph J., Wuttke D.S., Luger K.. Non-specific binding of RNA to PARP1 and PARP2 does not lead to catalytic activation. Biochemistry. 2019; 58:5107–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kim D.-S., Camacho C.V., Nagari A., Malladi V.S., Challa S., Kraus W.L.. Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol. Cell. 2019; 75:1270–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cohen-Armon M., Visochek L., Rozensal D., Kalal A., Geistrikh I., Klein R., Bendetz-Nezer S., Yao Z., Seger R.. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol. Cell. 2007; 25:297–308. [DOI] [PubMed] [Google Scholar]
- 107. Chow W.Y., Rajan R., Muller K.H., Reid D.G., Skepper J.N., Wong W.C., Brooks R.A., Green M., Bihan D., Farndale R.W.et al.. NMR spectroscopy of native and in vitro tissues implicates polyADP ribose in biomineralization. Science (New York, N.Y.). 2014; 344:742–746. [DOI] [PubMed] [Google Scholar]
- 108. Morrison A.R., Moss J., Stevens L.A., Evans J.E., Farrell C., Merithew E., Lambright D.G., Greiner D.L., Mordes J.P., Rossini A.A., et al.. ART2, a T cell surface mono-ADP-ribosyltransferase, generates extracellular poly(ADP-ribose). J. Biol. Chem. 2006; 281:33363–33372. [DOI] [PubMed] [Google Scholar]
- 109. Pourfarjam Y., Kasson S., Tran L., Ho C., Lim S., Kim I.-K.. PARG has a robust endo-glycohydrolase activity that releases protein-free poly(ADP-ribose) chains. Biochem. Biophys. Res. Commun. 2020; 527:818–823. [DOI] [PubMed] [Google Scholar]
- 110. Fontana P., Bonfiglio J.J., Palazzo L., Bartlett E., Matic I., Ahel I.. Serine ADP-ribosylation reversal by the hydrolase ARH3. eLife. 2017; 6:e28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Slade D., Dunstan M.S., Barkauskaite E., Weston R., Lafite P., Dixon N., Ahel M., Leys D., Ahel I.. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011; 477:616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Rack J. G.M., Liu Q., Zorzini V., Voorneveld J., Ariza A., Honarmand Ebrahimi K., Reber J.M., Krassnig S.C., Ahel D., van der Marel G.A.et al.. Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal. Nat. Commun. 2021; 12:4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Suskiewicz M.J., Zobel F., Ogden T.E.H., Fontana P., Ariza A., Yang J.-C., Zhu K., Bracken L., Hawthorne W.J., Ahel D.et al.. HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature. 2020; 579:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Rudolph J., Roberts G., Muthurajan U.M., Luger K.. HPF1 and nucleosomes mediate a dramatic switch in activity of PARP1 from polymerase to hydrolase. eLife. 2021; 10:e65773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kurgina T.A., Moor N.A., Kutuzov M.M., Naumenko K.N., Ukraintsev A.A., Lavrik O.I.. Dual function of HPF1 in the modulation of PARP1 and PARP2 activities. Commun. Biol. 2021; 4:1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ray Chaudhuri A., Nussenzweig A.. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017; 18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Teloni F., Altmeyer M.. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016; 44:993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Li M., Yu X.. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013; 23:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Robu M., Shah R.G., Petitclerc N., Brind’Amour J., Kandan-Kulangara F., Shah G.M.. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1658–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Pines A., Vrouwe M.G., Marteijn J.A., Typas D., Luijsterburg M.S., Cansoy M., Hensbergen P., Deelder A., de Groot A., Matsumoto S.et al.. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol. 2012; 199:235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sellou H., Lebeaupin T., Chapuis C., Smith R., Hegele A., Singh H.R., Kozlowski M., Bultmann S., Ladurner A.G., Timinszky G.et al.. The poly(ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage. Mol. Biol. Cell. 2016; 27:3791–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Smith R., Sellou H., Chapuis C., Huet S., Timinszky G.. CHD3 and CHD4 recruitment and chromatin remodeling activity at DNA breaks is promoted by early poly(ADP-ribose)-dependent chromatin relaxation. Nucleic Acids Res. 2018; 46:6087–6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Arruri V.K., Gundu C., Khan I., Khatri D.K., Singh S.B.. PARP overactivation in neurological disorders. Mol. Biol. Rep. 2021; 48:2833–2841. [DOI] [PubMed] [Google Scholar]
- 124. Jeong K.-Y., Lee H.. Inhibition of poly (ADP-Ribose) polymerase: A promising strategy targeting pancreatic cancer with BRCAness phenotype. World J. Gastr. Oncol. 2021; 13:1544–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Verma P., Zhou Y., Cao Z., Deraska P.V., Deb M., Arai E., Li W., Shao Y., Puentes L., Li Y.et al.. ALC1 links chromatin accessibility to PARP inhibitor response in homologous recombination-deficient cells. Nat. Cell Biol. 2021; 23:160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Juhász S., Smith R., Schauer T., Spekhardt D., Mamar H., Zentout S., Chapuis C., Huet S., Timinszky G.. The chromatin remodeler ALC1 underlies resistance to PARP inhibitor treatment. Sci. Adv. 2020; 6:eabb8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Hewitt G., Borel V., Segura-Bayona S., Takaki T., Ruis P., Bellelli R., Lehmann L.C., Sommerova L., Vancevska A., Tomas-Loba A.et al.. Defective ALC1 nucleosome remodeling confers PARPi sensitization and synthetic lethality with HRD. Mol. Cell. 2021; 81:767–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Blessing C., Mandemaker I.K., Gonzalez-Leal C., Preisser J., Schomburg A., Ladurner A.G.. The oncogenic helicase ALC1 regulates PARP inhibitor potency by trapping PARP2 at DNA breaks. Mol. Cell. 2020; 80:862–875. [DOI] [PubMed] [Google Scholar]
- 129. Gatti M., Imhof R., Huang Q., Baudis M., Altmeyer M.. The Ubiquitin Ligase TRIP12 Limits PARP1 trapping and constrains PARP inhibitor efficiency. Cell Rep. 2020; 32:107985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Krastev D.B., Li S., Sun Y., Wicks A.J., Hoslett G., Weekes D., Badder L.M., Knight E.G., Marlow R., Pardo M.C.et al.. The ubiquitin-dependent ATPase p97 removes cytotoxic trapped PARP1 from chromatin. Nat. Cell Biol. 2022; 24:62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Aumiller W.M., Pir Cakmak F., Davis B.W., Keating C.D.. RNA-based coacervates as a model for membraneless organelles: Formation, properties, and interfacial liposome assembly. Langmuir. 2016; 32:10042–10053. [DOI] [PubMed] [Google Scholar]
- 132. Wang M., Tao X., Jacob M.D., Bennett C.A., Ho J.J., Gonzalgo M.L., Audas T.E., Lee S.. Stress-Induced Low Complexity RNA Activates Physiological Amyloidogenesis. Cell Rep. 2018; 24:1713–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. D’Annessa I., Coletta A., Desideri A.. Geometrical constraints limiting the poly(ADP-ribose) conformation investigated by molecular dynamics simulation. Biopolymers. 2014; 101:78–86. [DOI] [PubMed] [Google Scholar]
- 134. Hazra M.K., Levy Y.. Charge pattern affects the structure and dynamics of polyampholyte condensates. Phys. Chem. Chem. Phys. 2020; 22:19368–19375. [DOI] [PubMed] [Google Scholar]
- 135. Drenichev M.S., Mikhailov S.N.. Poly(ADP-ribose): From chemical synthesis to drug design. Bioorg. Med. Chem. Lett. 2016; 26:3395–3403. [DOI] [PubMed] [Google Scholar]
- 136. Borgia A., Borgia M.B., Bugge K., Kissling V.M., Heidarsson P.O., Fernandes C.B., Sottini A., Soranno A., Buholzer K.J., Nettels D.et al.. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018; 555:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Sottini A., Borgia A., Borgia M.B., Bugge K., Nettels D., Chowdhury A., Heidarsson P.O., Zosel F., Best R.B., Kragelund B.B.et al.. Polyelectrolyte interactions enable rapid association and dissociation in high-affinity disordered protein complexes. Nat. Commun. 2020; 11:5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Müller K.H., Hayward R., Rajan R., Whitehead M., Cobb A.M., Ahmad S., Sun M., Goldberga I., Li R., Bashtanova U.et al.. Poly(ADP-ribose) links the DNA damage response and biomineralization. Cell Rep. 2019; 27:3124–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Reber J.M., Mangerich A.. Why structure and chain length matter: on the biological significance underlying the structural heterogeneity of poly(ADP-ribose). Nucleic Acids Res. 2021; 49:8432–8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Aberle L., Krüger A., Reber J.M., Lippmann M., Hufnagel M., Schmalz M., Trussina I.R.E.A., Schlesiger S., Zubel T., Schütz K.et al.. PARP1 catalytic variants reveal branching and chain length-specific functions of poly(ADP-ribose) in cellular physiology and stress response. Nucleic Acids Res. 2020; 48:10015–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Langdon E.M., Gladfelter A.S.. A new lens for RNA localization: liquid-liquid phase separation. Ann. Rev. Microbiol. 2018; 72:255–271. [DOI] [PubMed] [Google Scholar]
- 142. Snead W.T., Gladfelter A.S.. The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol. Cell. 2019; 76:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hofweber M., Dormann D.. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 2019; 294:7137–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Bratek-Skicki A., Pancsa R., Meszaros B., Van Lindt J., Tompa P.. A guide to regulation of the formation of biomolecular condensates. FEBS J. 2020; 287:1924–1935. [DOI] [PubMed] [Google Scholar]
- 145. Mehta S., Algie M., Al-Jabry T., McKinney C., Kannan S., Verma C.S., Ma W., Zhang J., Bartolec T.K., Masamsetti V.P.et al.. Critical role for cold shock protein YB-1 in cytokinesis. Cancers. 2020; 12:E2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Vasil’eva I.A., Anarbaev R.O., Moor N.A., Lavrik O.I.. Dynamic light scattering study of base excision DNA repair proteins and their complexes. Biochim. Biophys. Acta Proteins Proteomics. 2019; 1867:297–305. [DOI] [PubMed] [Google Scholar]
- 147. Vasil’eva I., Moor N., Anarbaev R., Kutuzov M., Lavrik O.. Functional roles of PARP2 in assembling protein-protein complexes involved in base excision DNA repair. Int. J. Mol. Sci. 2021; 22:4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Rancourt A., Satoh M.S.. Delocalization of nucleolar poly(ADP-ribose) polymerase-1 to the nucleoplasm and its novel link to cellular sensitivity to DNA damage. DNA Repair. 2009; 8:286–297. [DOI] [PubMed] [Google Scholar]
- 149. Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P.. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016; 165:1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Mitrea D.M., Cika J.A., Guy C.S., Ban D., Banerjee P.R., Stanley C.B., Nourse A., Deniz A.A., Kriwacki R.W.. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife. 2016; 5:e13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Berry J., Weber S.C., Vaidya N., Haataja M., Brangwynne C.P.. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5237–E5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Weber S.C., Brangwynne C.P.. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr. Biol. 2015; 25:641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Mangan H., Gailin M.O., McStay B.. Integrating the genomic architecture of human nucleolar organizer regions with the biophysical properties of nucleoli. FEBS J. 2017; 284:3977–3985. [DOI] [PubMed] [Google Scholar]
- 154. Farley K.I., Surovtseva Y., Merkel J., Baserga S.J.. Determinants of mammalian nucleolar architecture. Chromosoma. 2015; 124:323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Thiry M., Lafontaine D. L.J.. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 2005; 15:194–199. [DOI] [PubMed] [Google Scholar]
- 156. Citarelli M., Teotia S., Lamb R.S.. Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol. Biol. 2010; 10:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Egidi A., Di Felice F., Camilloni G.. Saccharomyces cerevisiae rDNA as super-hub: the region where replication, transcription and recombination meet. Cell. Mol. Life Sci. 2020; 77:4787–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Lafontaine D. L.J., Riback J.A., Bascetin R., Brangwynne C.P.. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021; 22:165–182. [DOI] [PubMed] [Google Scholar]
- 159. Lavrik O.I. PARPs’ impact on base excision DNA repair. DNA Repair. 2020; 93:102911. [DOI] [PubMed] [Google Scholar]
- 160. Ochs F., Karemore G., Miron E., Brown J., Sedlackova H., Rask M.-B., Lampe M., Buckle V., Schermelleh L., Lukas J.et al.. Stabilization of chromatin topology safeguards genome integrity. Nature. 2019; 574:571–574. [DOI] [PubMed] [Google Scholar]
- 161. Lou J., Priest D.G., Solano A., Kerjouan A., Hinde E.. Spatiotemporal dynamics of 53BP1 dimer recruitment to a DNA double strand break. Nat. Commun. 2020; 11:5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Setiaputra D., Escribano-Díaz C., Reinert J.K., Sadana P., Zong D., Callen E., Sifri C., Seebacher J., Nussenzweig A., Thomä N.H.et al.. RIF1 acts in DNA repair through phosphopeptide recognition of 53BP1. Mol. Cell. 2022; 82:1359–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Chapman J.R., Barral P., Vannier J.-B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D., Boulton S.J.. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 2013; 49:858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Zimmermann M., Lottersberger F., Buonomo S.B., Sfeir A., de Lange T.. 53BP1 regulates DSB repair using Rif1 to control 5’ end resection. Science (New York, N.Y.). 2013; 339:700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T.F., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D.et al.. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013; 49:872–883. [DOI] [PubMed] [Google Scholar]
- 166. Schultz L.B., Chehab N.H., Malikzay A., Halazonetis T.D.. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000; 151:1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Bobkova E., Depes D., Lee J.-H., Jezkova L., Falkova I., Pagacova E., Kopecna O., Zadneprianetc M., Bacikova A., Kulikova E.et al.. Recruitment of 53BP1 proteins for DNA repair and persistence of repair clusters differ for cell types as detected by single molecule localization microscopy. Int. J. Mol. Sci. 2018; 19:3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Shin Y., Chang Y.-C., Lee D. S.W., Berry J., Sanders D.W., Ronceray P., Wingreen N.S., Haataja M., Brangwynne C.P.. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018; 175:1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. D’Amours D., Desnoyers S., D’Silva I., Poirier G.G.. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999; 342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 170. Shao Z., Lee B.J., Rouleau-Turcotte E., Langelier M.-F., Lin X., Estes V.M., Pascal J.M., Zha S.. Clinical PARP inhibitors do not abrogate PARP1 exchange at DNA damage sites in vivo. Nucleic Acids Res. 2020; 48:9694–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Levone B.R., Lenzken S.C., Antonaci M., Maiser A., Rapp A., Conte F., Reber S., Ronchi A.E., Mühlemann O., Leonhardt H.et al.. FUS-dependent liquid-liquid phase separation is an early event in double-strand break repair. 2020; bioRxiv doi:24 August 2020, preprint: not peer reviewed 10.1101/798884. [DOI] [PMC free article] [PubMed]
- 172. Cramer P. Organization and regulation of gene transcription. Nature. 2019; 573:45–54. [DOI] [PubMed] [Google Scholar]
- 173. Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X.et al.. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science (New York, N.Y.). 2018; 361:eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A.. A phase separation model predicts key features of transcriptional control. Cell. 2017; 169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Wei M.-T., Chang Y.-C., Shimobayashi S.F., Shin Y., Strom A.R., Brangwynne C.P.. Nucleated transcriptional condensates amplify gene expression. Nat. Cell Biol. 2020; 22:1187–1196. [DOI] [PubMed] [Google Scholar]
- 176. Le Treut G., Képès F., Orland H.. Phase Behavior of DNA in the Presence of DNA-Binding Proteins. Biophys. J. 2016; 110:51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Cho W.-K., Spille J.-H., Hecht M., Lee C., Li C., Grube V., Cisse I.I.. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science (New York, N.Y.). 2018; 361:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C.et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science (New York, N.Y.). 2018; 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Xie L., Dong P., Chen X., Hsieh T.-H.S., Banala S., Marzio M.D., English B.P., Qi Y., Jung S.K., Kieffer-Kwon K.-R.et al.. 3D ATAC-PALM: super-resolution imaging of the accessible genome. Nat. Methods. 2020; 17:430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Palacio M., Taatjes D.J.. Merging established mechanisms with new insights: Condensates, Hubs, and the regulation of RNA polymerase II transcription. J. Mol. Biol. 2022; 434:167216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Bell N. A.W., Haynes P.J., Brunner K., de Oliveira T.M., Flocco M.M., Hoogenboom B.W., Molloy J.E.. Single-molecule measurements reveal that PARP1 condenses DNA by loop stabilization. Sci. Adv. 2021; 7:eabf3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Páhi Z.G., Borsos B.N., Pantazi V., Ujfaludi Z., Pankotai T.. PARylation during transcription: Insights into the fine-tuning mechanism and regulation. Cancers. 2020; 12:E183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Tulin A., Spradling A.. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science (New York, N.Y.). 2003; 299:560–562. [DOI] [PubMed] [Google Scholar]
- 184. Matveeva E.A., Al-Tinawi Q. M.H., Rouchka E.C., Fondufe-Mittendorf Y.N.. Coupling of PARP1-mediated chromatin structural changes to transcriptional RNA polymerase II elongation and cotranscriptional splicing. Epigenet. Chromatin. 2019; 12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Fu H., Liu R., Jia Z., Li R., Zhu F., Zhu W., Shao Y., Jin Y., Xue Y., Huang J.et al.. Poly(ADP-ribosylation) of P-TEFb by PARP1 disrupts phase separation to inhibit global transcription after DNA damage. Nat. Cell Biol. 2022; 24:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Bordet G., Lodhi N., Guo D., Kossenkov A., Tulin A.V.. Poly(ADP-ribose) polymerase 1 in genome-wide expression control in Drosophila. Sci. Rep. 2020; 10:21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Wright R. H.G., Lioutas A., Le Dily F., Soronellas D., Pohl A., Bonet J., Nacht A.S., Samino S., Font-Mateu J., Vicent G.P.et al.. ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science (New York, N.Y.). 2016; 352:1221–1225. [DOI] [PubMed] [Google Scholar]
- 188. Patel A., Malinovska L., Saha S., Wang J., Alberti S., Krishnan Y., Hyman A.A.. ATP as a biological hydrotrope. Science (New York, N.Y.). 2017; 356:753–756. [DOI] [PubMed] [Google Scholar]
- 189. Mehringer J., Do T.-M., Touraud D., Hohenschutz M., Khoshsima A., Horinek D., Kunz W.. Hofmeister versus Neuberg: is ATP really a biological hydrotrope?. Cell Rep. Phys. Sci. 2021; 2:100343. [Google Scholar]
- 190. Maeshima K., Matsuda T., Shindo Y., Imamura H., Tamura S., Imai R., Kawakami S., Nagashima R., Soga T. Nojiet al.. A transient rise in free Mg2+ ions released from ATP-Mg hydrolysis contributes to mitotic chromosome condensation. Curr. Biol. 2018; 28:444–451. [DOI] [PubMed] [Google Scholar]
- 191. Wright R. H.G., Dily F.L., Beato M.. ATP, Mg2+, nuclear phase separation, and genome accessibility. Trends Biochem. Sci. 2019; 44:565–574. [DOI] [PubMed] [Google Scholar]
- 192. Henninger J.E., Oksuz O., Shrinivas K., Sagi I., LeRoy G., Zheng M.M., Andrews J.O., Zamudio A.V., Lazaris C., Hannett N.M.et al.. RNA-mediated feedback control of transcriptional condensates. Cell. 2021; 184:207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Prosser S.L., Pelletier L.. Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol. 2017; 18:187–201. [DOI] [PubMed] [Google Scholar]
- 194. Oriola D., Jülicher F., Brugués J.. Active forces shape the metaphase spindle through a mechanical instability. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:16154–16159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Sun M., Jia M., Ren H., Yang B., Chi W., Xin G., Jiang Q., Zhang C.. NuMA regulates mitotic spindle assembly, structural dynamics and function via phase separation. Nat. Commun. 2021; 12:7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Pan H., Guan R., Zhao R., Ou G., Chen Z.. Mechanistic insights into central spindle assembly mediated by the centralspindlin complex. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2112039118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Chang P., Coughlin M., Mitchison T.J.. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005; 7:1133–1139. [DOI] [PubMed] [Google Scholar]
- 198. Chinen T., Yamamoto S., Takeda Y., Watanabe K., Kuroki K., Hashimoto K., Takao D., Kitagawa D.. NuMA assemblies organize microtubule asters to establish spindle bipolarity in acentrosomal human cells. EMBO J. 2020; 39:e102378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Chang P., Coughlin M., Mitchison T.J.. Interaction between Poly (ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol. Biol. Cell. 2009; 20:4575–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Li M., Bian C., Yu X.. Poly(ADP-ribosyl)ation is recognized by ECT2 during mitosis. Cell Cycle (Tex.). 2014; 13:2944–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Slade D. Mitotic functions of poly(ADP-ribose) polymerases. Biochem. Pharmacol. 2019; 167:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Guillén-Boixet J., Kopach A., Holehouse A.S., Wittmann S., Jahnel M., Schlüßler R., Kim K., Trussina I.R.E.A., Wang J., Mateju D.et al.. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020; 181:346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Van Treeck B., Protter D. S.W., Matheny T., Khong A., Link C.D., Parker R.. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Begovich K., Wilhelm J.E.. An in vitro assembly system identifies roles for RNA nucleation and ATP in yeast stress granule formation. Mol. Cell. 2020; 79:991–1007. [DOI] [PubMed] [Google Scholar]
- 205. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P.. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015; 163:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Loughlin F.E., West D.L., Gunzburg M.J., Waris S., Crawford S.A., Wilce M.C.J., Wilce J.A.. Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1. Nucleic Acids Res. 2021; 49:2403–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G.M., Ivanov P.et al.. G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016; 212:845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Zhang P., Fan B., Yang P., Temirov J., Messing J., Kim H.J., Taylor J.P.. Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. eLife. 2019; 8:e39578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Freibaum B.D., Messing J., Yang P., Kim H.J., Taylor J.P.. High-fidelity reconstitution of stress granules and nucleoli in mammalian cellular lysate. J. Cell Biol. 2021; 220:e202009079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Niewidok B., Igaev M., Pereira da Graca A., Strassner A., Lenzen C., Richter C.P., Piehler J., Kurre R., Brandt R.. Single-molecule imaging reveals dynamic biphasic partition of RNA-binding proteins in stress granules. J. Cell Biol. 2018; 217:1303–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Jin X., Cao X., Liu S., Liu B.. Functional roles of poly (ADP-ribose) in stress granule formation and dynamics. Front. Cell Dev. Biol. 2021; 9:1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Catara G., Grimaldi G., Schembri L., Spano D., Turacchio G., Lo Monte M., Beccari A.R., Valente C., Corda D.. PARP1-produced poly-ADP-ribose causes the PARP12 translocation to stress granules and impairment of Golgi complex functions. Sci. Rep. 2017; 7:14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Isabelle M., Gagné J.-P., Gallouzi I.-E., Poirier G.G.. Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J. Cell Sci. 2012; 125:4555–4566. [DOI] [PubMed] [Google Scholar]
- 214. Jayabalan A.K., Adivarahan S., Koppula A., Abraham R., Batish M., Zenklusen D., Griffin D.E., Leung A. K.L.. Stress granule formation, disassembly, and composition are regulated by alphavirus ADP-ribosylhydrolase activity. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2021719118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Kroschwald S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I., Richter D., Alberti S.. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 2015; 4:e06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Watanabe K., Morishita K., Zhou X., Shiizaki S., Uchiyama Y., Koike M., Naguro I., Ichijo H.. Cells recognize osmotic stress through liquid–liquid phase separation lubricated with poly(ADP-ribose). Nat. Commun. 2021; 12:1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Thapa K., Khan H., Sharma U., Grewal A.K., Singh T.G.. Poly (ADP-ribose) polymerase-1 as a promising drug target for neurodegenerative diseases. Life Sciences. 2021; 267:118975. [DOI] [PubMed] [Google Scholar]
- 218. Liu C., Fang Y.. New insights of poly(ADP-ribosylation) in neurodegenerative diseases: A focus on protein phase separation and pathologic aggregation. Biochem. Pharmacol. 2019; 167:58–63. [DOI] [PubMed] [Google Scholar]
- 219. Kam T.-I., Mao X., Park H., Chou S.-C., Karuppagounder S.S., Umanah G.E., Yun S.P., Brahmachari S., Panicker N., Chen R.et al.. Poly(ADP-ribose) drives pathologic α-synuclein neurodegeneration in Parkinson’s disease. Science (New York, N.Y.). 2018; 362:eaat8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Puentes L.N., Lengyel-Zhand Z., Lee J.Y., Hsieh C.-J., Schneider M.E., Edwards K.J., Luk K.C., Lee V. M.-Y., Trojanowski J.Q., Mach R.H.. Poly (ADP-ribose) interacts with phosphorylated α-synuclein in post mortem PD samples. Front. Aging Neurosci. 2021; 13:704041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. McAlary L., Plotkin S.S., Yerbury J.J., Cashman N.R.. Prion-like propagation of protein misfolding and aggregation in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 2019; 12:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Nomura T., Watanabe S., Kaneko K., Yamanaka K., Nukina N., Furukawa Y.. Intranuclear aggregation of mutant FUS/TLS as a molecular pathomechanism of amyotrophic lateral sclerosis. J. Biol. Chem. 2014; 289:1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Silva J.L., Vieira T.C., Cordeiro Y., de Oliveira G.A.P.. Nucleic acid actions on abnormal protein aggregation, phase transitions and phase separation. Curr. Opin. Struct. Biol. 2022; 73:102346. [DOI] [PubMed] [Google Scholar]
- 224. Leung A. K.L. Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol. 2014; 205:613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Aguzzi A., Altmeyer M.. Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 2016; 26:547–558. [DOI] [PubMed] [Google Scholar]
- 226. Huang D., Kraus W.L.. The expanding universe of PARP1-mediated molecular and therapeutic mechanisms. Mol. Cell. 2022; 82:2315–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Vågbø C.B., Slupphaug G.. RNA in DNA repair. DNA Repair. 2020; 95:102927. [DOI] [PubMed] [Google Scholar]