Abstract
BACKGROUND & AIMS:
Epidemiologic and murine studies suggest that dietary emulsifiers promote development of diseases associated with microbiota dysbiosis. Although the detrimental impact of these compounds on the intestinal microbiota and intestinal health have been demonstrated in animal and in vitro models, impact of these food additives in healthy humans remains poorly characterized.
METHODS:
To examine this notion in humans, we performed a double-blind controlled-feeding study of the ubiquitous synthetic emulsifier carboxymethylcellulose (CMC) in which healthy adults consumed only emulsifier-free diets (n = 9) or an identical diet enriched with 15 g per day of CMC (n = 7) for 11 days.
RESULTS:
Relative to control subjects, CMC consumption modestly increased postprandial abdominal discomfort and perturbed gut microbiota composition in a way that reduced its diversity. Moreover, CMC-fed subjects exhibited changes in the fecal metabolome, particularly reductions in short-chain fatty acids and free amino acids. Furthermore, we identified 2 subjects consuming CMC who exhibited increased microbiota encroachment into the normally sterile inner mucus layer, a central feature of gut inflammation, as well as stark alterations in microbiota composition.
CONCLUSIONS:
These results support the notion that the broad use of CMC in processed foods may be contributing to increased prevalence of an array of chronic inflammatory diseases by altering the gut microbiome and metabolome (ClinicalTrials.gov, number NCT03440229).
Keywords: Emulsifier, Metabolism, Microbiota, Metabolome
Graphical Abstract
Consumption of highly processed foods has increased dramatically since the mid-20th century, and is associated with increased incidence of several chronic inflammatory diseases. Among these are inflammatory bowel disease (IBD)1 and metabolic syndrome,2 both of which are associated with, and thought to be promoted by, alterations in gut microbiota.3–5 A common feature of highly processed foods is the use of 1 or more emulsifiers or thickeners (referred hereafter as emulsifiers) that are added to enhance texture and extend shelf life. Some of the emulsifiers that are commonly added to foods, such as lecithin, are a natural component of unprocessed foods, whereas others, such as carboxymethylcellulose (CMC), are synthetic. Despite lack of extensive safety testing, CMC was approved in the 1960s for use in foods at concentrations up to 2% (wt/wt) by regulatory agencies, including the US Food and Drug Administration and European Commission based on the GRAS (generally regarded as safe) designation developed by these agencies. Part of the basis for presuming that CMC, and some other emulsifiers, are safe is that they are not well absorbed and thus mostly eliminated in feces. However, such passage through the intestine allows these products to directly interact with gut microbiota and the intestinal mucosa. For example, CMC has been shown to impact gut transit time6 and alter fecal bile acid profiles.7 More recent studies show that CMC impacts human microbiota composition and gene expression in vitro, and in mice, wherein its impacts on gut microbiota promote the development of colitis or metabolic syndrome.8–12 These findings compelled us to investigate the extent to which CMC impacts intestinal-microbiota interactions in humans.
Examination of how an individual food component affects human microbiota is complicated by interindividual heterogeneity in factors such as quantity of the food consumed, background diet quality and composition, and gut microbiota composition. To minimize the potential confounding impact of these factors, we performed an inpatient (domiciled) study that ensured protocol adherence, identical background diets, and enabled daily monitoring and specimen collections before, during, and after CMC consumption, or lack thereof.
Methods
Study Design
General information.
This randomized, controlled-feeding study took place in the University of Pennsylvania’s Center for Human Phenomic Science (CHPS), and was registered at https://ClinicalTrials.gov as trial no. NCT03440229. The first 3 days of the study were as an outpatient followed by 11 days as an inpatient, as presented Figure 1A. Once admitted to the CHPS unit, participants were not allowed to leave the unit unless accompanied by study staff. The study included 16 healthy volunteers between the ages of 18 and 60 years.
Figure 1.
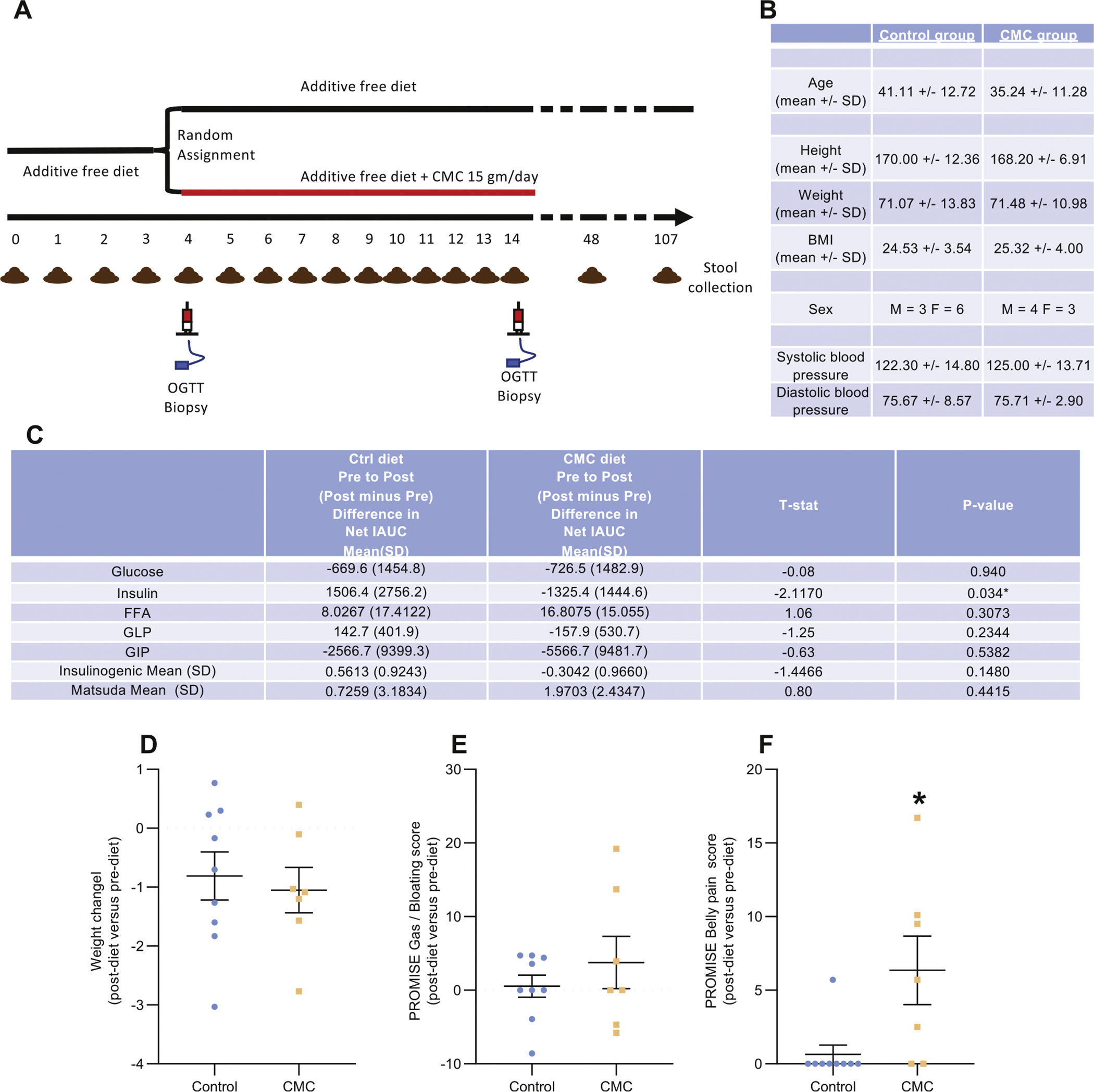
Effect of CMC consumption on metabolic parameters. (A) Schematic representation of the inpatient study that enabled daily monitoring and specimen collections before, during, and after CMC consumption, or lack thereof, and presenting timing of oral glucose tolerance tests, intestinal biopsies, and feces collection. (B) Biomorphometric characterization of the study’s participants at the beginning of the study. (C) Effect of dietary emulsifier CMC consumption on various metabolic parameters, measured both pre- and postintervention. (D–F) Effect of dietary emulsifier CMC consumption on weight (D), PROMISE gas/bloating (E), and belly pain (F) scores, measured both pre- and postintervention. OGTT, oral glucose tolerance test. Significance was determined using Mann-Whitney test; *P < 0.05 compared with control group.
Study endpoint and objectives.
There were no pre-specified efficacy or safety endpoints for this study. The objectives were to (1) establish a tractable and physiologic means of measuring CMC consumption and its metabolic impact in healthy volunteers; (2) examine the extent to which CMC consumption affects human gut microbiota composition, gene expression, and/or localization; and (3) explore effects of CMC consumption on a range of inflammatory and metabolic parameters that characterize metabolic syndrome. These included concentration of lipocalin in feces and interferon-γ, interleukin-17, interleukin-8, and inducible protein-10 in serum. In addition, insulin sensitivity was assessed with a 2.5-hour oral glucose tolerance testing performed after an overnight fast on inpatient days 1 and 11. Insulin sensitivity was measured as change in insulin divided by change in glucose from time 0 to 30 minutes.
Sample size calculation.
Power calculation was based on measure of bacterial-epithelial distance, which provides a quantitative parameter whose diminution is associated with disease (colitis and metabolic syndrome) in both mice and humans.12,13 Specifically, the difference in mean distance of the nearest bacteria to the epithelium between patients with and without diabetes was 19.13 μm. The within-group standard deviation (SD) for patients without diabetes was 7.17 μm. The within-group SD for those with diabetes was even smaller. With a sample size of 8 subjects per group and assuming a within-group SD of 7.17 μm, we projected to have 90% and 80% power to detect a difference in the distance of the nearest bacteria to the epithelium between the treatment groups (CMC vs no CMC) that is 35% and 44% smaller than the difference observed between patients with and without diabetes, respectively.
Changes to methods after trial commencement.
As fully detailed Supplementary Table 1, the study design was modified to improve participant recruitment. More specifically, although the first 3 participants stayed at CHPS for the washout period, the remaining 13 participants were allowed to go back home with provided in-house cooked food for the washout period. Moreover, although the CMC treatment duration was 14 days for the 3 first participants, it was 11 days for the remaining 13 participants. Importantly, the only data from days 12, 13, and 14 for the 3 first participants used in the analysis were the mucosal biopsies that were collected on day 14.
Recruitment.
Participants were recruited via advertising the study on an online system at the University of Pennsylvania from April 12, 2018, to January 16, 2019.
Early withdrawal of participants.
No participant was withdrawn from the study.
Eligibility criteria for participants.
Inclusion criteria were ability to give informed consent and age 18 to 60 years. Exclusion criteria were diagnosis with IBD, celiac disease, or other chronic intestinal disorders; baseline bowel frequency less than every 2 days or more than 3 times daily; current smoker; body mass index <18.5 or >40 at screening; more than 2 of the criteria for metabolic syndrome (waist circumference >89 cm for women or 102 cm for men, diagnosis of diabetes mellitus or baseline HbA1c >6.4% or a fasting glucose level of greater than 100 mg/dL; systolic blood pressure >130 mm Hg or diastolic blood pressure >85 mm Hg or treated with medications for hypertension; fasting triglycerides >149 mg/dL or treated with medications for hypertriglyceridemia; fasting high-density lipoprotein cholesterol <40 mg/dL in men or <50 mg/dL in women or treated with medications for hypercholesterolemia); known substance abuse disorder or consumption of illicit drugs or alcohol in the 24 hours before admission; prior bowel resection surgery other than appendectomy; white blood cell count less than 3500 per μL or an absolute neutrophil count of less than 1000 per μL; platelet count of less than 100,000 per μL or an international normalized ratio greater than 1.2; estimated glomerular filtration rate <60 mL/min per 1.73 m2; pregnant or lactating women; use of antibiotics in the 6 months before screening; use of laxatives or antidiarrhea medications in the 2 weeks before screening; use of anticholinergic medications, narcotics, antacids, nonsteroidal anti-inflammatory drugs, or dietary supplements in the week before screening; human immunodeficiency virus infection, AIDS, or other known conditions resulting in immunosuppression; allergies or intolerance to the components of the study diets; following a vegan or vegetarian diet; and experienced diarrhea within the 2 weeks before screening.
Blinding.
The study used concealed allocation with neither the participants nor the research team being aware of the treatment assignment during the screening phase and until all data were collected. The research team remained blinded to treatment assignment until all biopsies had been reviewed to assess for bacteria distance from the epithelium, and data were analyzed for this outcome together with the oral glucose tolerance tests and inflammatory markers.
Intervention.
All food was prepared within the CHPS metabolic kitchen without emulsifiers (unless specifically added). All participants followed the same Western-style diet (the only difference being portion size). The macronutrient percentages of calories for the study diet were 55% carbohydrate, 30% fat, and 15% protein. The diet provided is considered healthy with a Healthy Eating Index score of 75.14–16 The diet was composed of 2 menus that were consumed on alternating days. Water, black coffee, and plain tea were provided as desired. Participants had access to additional servings of food beyond the meals provided; however, the entire serving of the previous meal must have been consumed to receive additional servings.
For the 3 days before admission, participants ate an emulsifier-free diet at home with food provided by the CHPS metabolic kitchen. After admission to CHPS, all participants consumed the same emulsifier-free diet until dinner on the first day of the inpatient stay. Thus, all participants had approximately 80 hours of emulsifier-free washout time before administration of the food containing CMC (source: Modernist Pantry, Eliot, ME) or matched CMC-free food.
Participants were randomly assigned to receive 0 or 15 g per day of CMC (9 and 7 participants in each arm of the study) using concealed allocation by Dr Hongzhe Li. Because of the small sample size, we used block randomization with a block size of 4 participants. Beginning with the dinner meal on inpatient day 4, all participants consumed 3 servings of brownie and 3 servings of sorbet per day, each containing 0 or 2.5 g CMC per serving. The brownie and sorbet servings were provided at 3 scheduled meals and 3 scheduled snacks. Before eating any other food on the study menu, participants were required to consume the brownie and sorbet servings. Neither the participant nor the investigators were aware of which diet participants were assigned until the analyses of metabolic parameters, inflammatory parameters, microbiome composition, and bacteria-mucosa distance assessment had been performed.
Physical activity was monitored during the 3 days before admission to CHPS through the use of a FitBit Flex. During the inpatient portion of the study, participants were required to attain within 10% of the average number of daily steps that they took in the 3 days before admission.
Sample collection.
Urine was collected before starting the outpatient study diet and each morning of the inpatient stay after an overnight fast and aliquoted and frozen at −80°C. Blood was collected after an overnight fast before breakfast at the screening visit, at a postscreening visit before admission, and on days 1 to 4, 8, 10, and 11 of the inpatient study, and 1 month after discharge. Plasma was separated from the blood samples and stored frozen at −80°C for use in metabolomic studies. Stool samples were collected without preservatives or stabilizers before starting the outpatient diet, daily during the inpatient stay, and at 1 and 3 months after discharge. The first stool sample of the day was aliquoted and frozen at −80°C. All other stool samples during the inpatient stay were weighed and then discarded. On inpatient days 1 and 11 (or 14 for the first 3 participants), each participant underwent a sigmoidoscopy to obtain biopsies from the area of approximately 15 cm from the anal verge, which correlates with approximately the rectosigmoid junction. No bowel preparation was used before the sigmoidoscopy. Biopsy samples were placed in Carnoy solution for nondenaturing confocal microscopy.
Additional data collection.
We collected information on the participant’s usual diet using the Diet History Questionnaire II, a food frequency questionnaire developed by the National Cancer Institute. On inpatient days 2, 3, 5, 6, 9, and 10, following lunch, participants completed a standard food satiety questionnaire using a 150-mm visual analog scale to measure satiety and hunger as per Doucet et al.17,18 Diet quality was assessed using the Healthy Eating Index 2015.14 On days 1 and 11, participants completed the Patient-Reported Outcomes Measurement Information System (PROMIS) scales for belly pain (version 1.0 – 5a) and gas/bloating (version 1.0 – 13a).
Measurements of Circulating Metabolic Parameters and Cytokines
Serum cytokines were assayed using the Luminex 100 Multi-analyte System by the University of Maryland’s Cytokine Core Laboratory.
Serum Lipopolysaccharide- and Flagellin-specific Immunoglobulins
See the supplemental methods section.
Microbiota Analysis by 16S Ribosomal RNA Gene Sequencing Using Illumina Technology
See the supplemental methods section.
16S Ribosomal RNA Gene Sequence Analysis
See the supplemental methods section.
Microbiota Analysis by Shotgun Sequencing Using Illumina Technology
See the supplemental methods section.
Bacterial Density Quantification by 16S Ribosomal RNA qPCR
See the supplemental methods section.
Quantification of Fecal Lipocalin-2 by Enzyme-linked Immunosorbent Assay
For quantification of fecal lipocalin-2 (Lcn-2) by enzyme-linked immunosorbent assay (ELISA), frozen fecal samples were reconstituted in phosphate-buffered saline containing 0.1% Tween 20 to a final concentration of 100 mg/mL and vortexed for 20 minutes to get a homogeneous fecal suspension.19 These samples were then centrifuged for 10 minutes at 14,000g and 4°C. Clear supernatants were collected and stored at −20°C until analysis. Lcn-2 levels were estimated in the supernatants using Duoset Human Lcn-2 ELISA kit (R&D Systems, Minneapolis, MN) using the colorimetric peroxidase substrate tetramethylbenzidine, and optical density was read at 450 nm (Molecular Devices, San Jose, CA).
Fecal Flagellin and Lipopolysaccharide Load Quantification
See the supplemental methods section.
Immunostaining of Mucins and Localization of Bacteria by Fluorescence In Situ Hybridization
See the supplemental methods section.
Metabolomic Analysis of Stool and Urine Samples
Stool and urine sample preparation for nuclear magnetic resonance (NMR) were performed as previously described.20 1H NMR spectra were acquired on a Bruker Avance NEO 600-MHz spectrometer equipped with an inverse cryogenic probe (Bruker Biospin, Rheinstetten, Germany) at 298 K. A typical 1D NMR spectrum named NOESYPR1D was acquired for each sample. The metabolites were assigned based on published results21 and confirmed with a series of 2D NMR spectra. All 1H NMR spectra were adjusted for phase and baseline using Chenomx (Chenomx Inc, Edmonton, Alberta, Canada). The chemical shift of 1H NMR spectra were referenced to sodium 3-trimethylsilyl [2,2,3,3-d4] propionate at δ 0.00. Supplementary Table 2 lists all the quantitated metabolites and their characteristics (Moieties, δ 1H [ppm] and δ 13C [ppm]). The relative contents of metabolites were calculated by normalizing to the total sum of the spectral integrals. The quantification of metabolites, including CMC, in stool was calculated by NMR peak area against trimethylsilylpropanoic acid using Chenomx. The lower limit of CMC detection using the NMR approach is approximately >1 μM for pure CMC and 1 to 10 μM for CMC in stool and urine samples. For CMC absolute quantification, 5 concentrations were used in triplicate, with a lower limit of detection of 0.5 mg/mL, as presented in Supplementary Figure 12.
AccQ•Tag Amino Acid Analysis of Stool Samples
Amino acids were extracted from stool samples with 1 mL of ice-cold methanol/water (2:1) solution (containing 2.5 μM of Norvaline), followed by homogenization (Precellys; Bertin Technologies, Rockville, MD) with 1.0-mm-diameter zirconia/silica beads (BioSpec, Bartlesville, OK), 3 freeze-thaw cycles, and centrifugation (Eppendorf, Hamburg, Germany). Supernatant was collected, evaporated to dryness (Thermo Scientific, Waltham, MA) and then resuspend in 50 μL 0.1N HCl solution. Amino acid derivation with AccQ•Tag reagents (Waters, Milford, MA) was conducted according to the manufacturer’s protocol. Briefly, 10 μL of stool extract was mixed with 70 μL of AccQ•Tag Ultra borate buffer and 20 μL of AccQ•Tag Ultra reagent in a Total Recovery Vial. The vials were capped and vortexed for several seconds and proceed for 10 minutes at 55 °C. Amino acids were detected by Waters Xevo TQS coupled with PDA, and AccQ•Tag Ultra Column (C18 1.7 μm 2.1 × 100 mm) with in-line filter (Waters, Milford, MA) were used for separation.22 Results were quantified by comparing integrated peak areas against a standard curve.
Statistical Analysis
Significance was determined using t tests, Mann-Whitney test, 1-way analysis of variance (ANOVA) corrected for multiple comparisons with a Bonferroni posttest, 2-way ANOVA corrected for multiple comparisons with a Bonferroni posttest (or mixed-effect analysis when some values were missing), or repeated t tests corrected with the false discovery rate approach where appropriate (GraphPad Prism software, version 6.01; La Jolla, CA). Differences were noted as significant at P ≤ .05.
Results
We enrolled 16 subjects, deemed healthy based on lack of disease history or current evidence of metabolic syndrome (see Methods), who were randomly assigned with concealed allocation to the CMC-containing (n = 7) or control (n = 9) diets, with both investigators and subjects blinded to assignments (Figure 1A and Supplementary Table 1). The groups were similar in terms of age, gender, body mass index, and blood pressure (Figure 1B). At the time of screening, subjects in both groups were consuming similar diets as indicated using principal coordinate analysis (PCoA) to visualize the varied food recall responses provided by subjects on study enrollment (Supplementary Figure 1). On study days 4 to 14, all subjects consumed 3 servings of brownies and 3 servings of sorbet that lacked or contained 2.5 g CMC per serving. Both groups of subjects exhibited reductions in body weight of about 1 kg and had modest improvements in glycemic control over the course of the study, the extent of which did not vary significantly between the 2 groups except that a modest decrease in serum insulin levels was seen in the CMC-fed group (Figure 1C and D). CMC consumption was not associated with severe adverse events or alterations in serum levels of inflammatory cytokines, nor did it have an appreciable impact on appetite, food consumption, or bloating (Figure 1E and Supplementary Figure 2). Moreover, levels of anti-lipopolysaccharide and anti-flagellin immunoglobulin G antibodies, which have been used as an indirect measure of gut permeability,23,24 did not change over the course of the study in control or CMC-fed subjects (Supplementary Figure 3). CMC consumption did associate with a modestly significant increase in postprandial abdominal pain (Figure 1F, P = .019).
Microbiota Composition
Microbiota composition of daily-collected fecal specimens was characterized by 16S ribosomal RNA (rRNA) gene sequencing. In accord with previous studies,25 PCoA of the pairwise distances (unweighted UniFrac) between samples revealed strong clustering within subjects, indicating that extent of interindividual variations in gut microbiota composition exceeds impacts of short-term alterations in diet (Figure 2A, permutational multivariate ANOVA [Permanova] P = .001). Consequently, as a means of focusing on the potential impact of CMC on each individual subject, we used samples collected the morning of day 4, the day on which the subjects began consuming CMC in the study, to normalize all microbiota composition data. This approach revealed that subjects fed CMC displayed greater changes in microbiota composition during the intervention period, resulting in PCoA plots showing clear treatment-based clustering after 10 days of CMC consumption (Figure 2B, Permanova day 0 P = .928, day 9 P = .228, day 14 P = .002). Moreover, analysis of Bray-Curtis distance changes from the morning of day 4 revealed a trend toward greater microbiota alterations during the intervention period in the CMC group compared with the control group (Figure 2C, P = .102). These relative shifts in microbiota composition occurred without significant alterations in daily fecal weight (Supplementary Figure 4) or fecal bacterial density between the control and CMC groups (Figure 2D, diet effect P = .503). Phyla and order-level analysis did not reveal significant differences in the CMC and control groups during the intervention period, Supplementary Figure 5). Investigation of the most significantly differentially abundant sequence variants (SVs) between CMC and control groups revealed SVs that were generally stably represented in control subjects on day 14 compared with day 4, with relative values being very close to 1 (Supplementary Figure 6), whereas the relative abundance of these SVs were markedly affected by CMC consumption, including decreases in Faecalibacterium prausnitzii and Ruminococcus sp., and increases in Roseburia sp. and Lachnospiraceae (Supplementary Figure 6). Although it is difficult to reliably ascribe functional consequences to these alterations, we note that CMC consumption induced loss of F. prausnitzii, which is associated with health and known to mediate production of beneficial metabolites such as short-chain fatty acids.26–28
Figure 2.
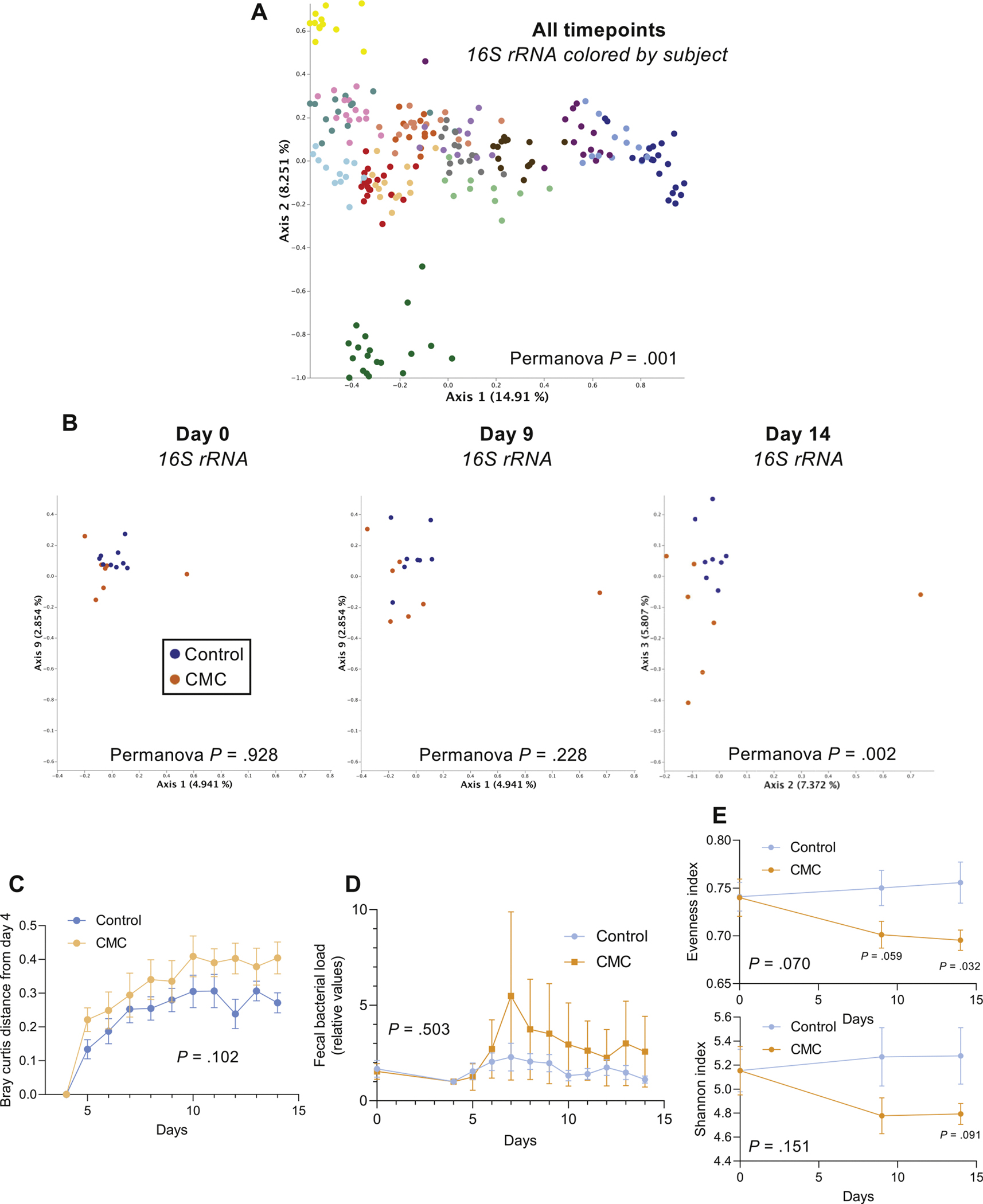
Effect of CMC consumption on microbiota composition. (A) PCoA of the unweighted UniFrac distance matrix of the study participants’ microbiota assessed by 16S rRNA gene sequencing. All time points are included in the representation, and samples are colored by participants. (B) PCoA of the Bray-Curtis distance matrix at days 0, 9, and 14 of study participants’ microbiota composition after normalization of every SV based on day 4 value, with samples colored by group. (C) Changes in the microbial community structure over time, as measured by Bray-Curtis distance from day 4 to subsequent days, for each group. (D) Fecal bacterial load assessed by 16S quantitative polymerase chain reaction (qPCR). (E) Changes in Evenness and Shannon alpha diversity measures for CMC intervention versus control groups, at days 0, 9, and 14. Significance was determined using 2-way ANOVA corrected for multiple comparisons with a Bonferroni posttest (E), multiple t tests (E) or Permanova analysis (A, B).
CMC also reduced microbiota richness, which is a hallmark of various diseases states,29 as revealed by decrease in the evenness (Figure 2E, diet effect P = .070, with P = .059 at day 9 and P = .032 at day 14) and Shannon indices (diet effect P = .151 with P = .091 at day 14). To further investigate impacts of CMC on microbiota composition, we next performed shotgun metagenomic sequencing on fecal samples collected shortly before or after 10 days of CMC consumption (days 4 and 14, respectively). Quality filtered reads were assigned to taxa and function. Use of PCoA analysis of the Bray-Curtis distances to compare all of the samples (ie, pre- and post-CMC) showed within subject clustering both taxonomically and functionally (Figure 3A and Supplementary Figure 7A), reflecting patterns observed using 16S rRNA gene sequence data. Nonetheless, there was clear post-treatment clustering of samples from control and CMC-fed subjects based on taxonomic (Supplementary Figure 7B and C), and, especially, function-based analysis (Figure 3B and C, PCoA analysis of the Bray-Curtis distances, see the Method section for details). The significantly altered functional categories that drove such clustering, identified via Maaslin2, comprised a variety of microbial metabolic pathways, suggesting that CMC-induced alteration in microbiota composition might have broad impacts on microbiota function (Figure 3D).
Figure 3.
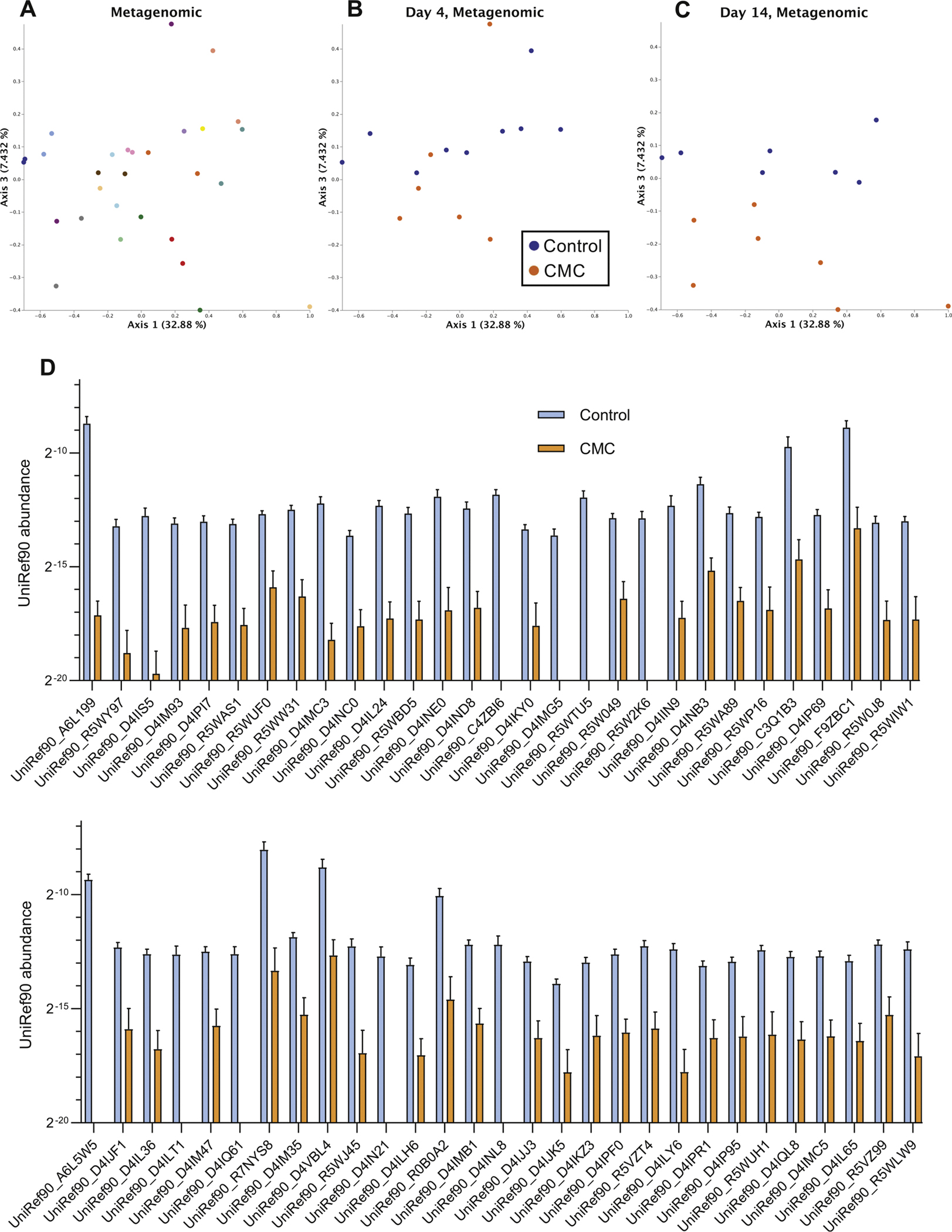
Effect of CMC consumption on fecal metagenome. (A) PCoA of the Bray-Curtis dissimilarity of study’s participants metagenome (uniref90 categories) assessed by shotgun sequencing. Days 4 and 14 are included in the representation, and samples are colored by participants. (B, C) PCoA of the Bray-Curtis distance matrix at day 4 (B) and 14 (C) of study’s participants metagenome assessed by shotgun sequencing, with samples colored by group.
Changes in Fecal Metabolome
To investigate the functional consequences of CMC’s impacts on microbiota, we first measured fecal levels of molecules known to mediate host-microbiota interactions. Use of TLR4 and TLR5 reporter cells revealed, respectively, that fecal levels of lipopolysaccharide and flagellin were not affected by CMC consumption (Figure 4A and B, mixed-effects analysis with Bonferroni multiple comparisons tests, diet effect P = .413 for Figure 4A and P = .220 for Figure 4B, Bonferroni corrected P > .1 for all days). There was no significant change in levels of fecal lipocalin-2, an inflammatory marker (Figure 4C, mixed-effects analysis with Bonferroni multiple comparisons tests, diet effect P = .258, Bonferroni corrected P > .1 for all days). Next, we sought to broadly examine the extent to which CMC altered the fecal metabolome, which is both shaped by gut microbiota and mediates many of its impacts on the host. We used a 1H NMR-based targeted assay capable of quantitating about 40 metabolites that are reliably detected in stools of a healthy person, many of which can be influenced by the gut microbiota. In accord with the notion that, in general, there is far less interperson heterogeneity in microbiota metabolic function than in species composition,30 we compared fecal metabolomes between control and CMC-fed subjects, without normalization to correct for basal variation among subjects. Accordingly, before CMC consumption (day 4), no significant clustering by study group was evident for the fecal metabolome (Figure 4D, Permanova day 0 P = .573). In contrast, following CMC consumption, this approach showed a clear ability to distinguish fecal metabolomes of control vs CMC-fed subjects (Figure 4D, Permanova day 9 P = .001, day 14 P = .001). Concomitantly, display of individual values of each metabolite for each subject on day 14 (Supplementary Figure 8), as well as viewing mean values for each group over time via a heat map (Figure 4E), demonstrated that fecal metabolomes of CMC-fed subjects were, on average, depleted in an array of microbiota-related metabolites, including short-chain fatty acids and essential amino acids. Such changes were clearly evident by 3 days after initiating CMC consumption and remained throughout the period of CMC consumption and had resolved when subjects were resampled approximately 1 month later (day 48) (Figure 4E). Moreover, NMR-based detection of fecal amino acid concentration demonstrated a decrease in the fecal amounts of numerous amino acids, as presented in Supplementary Figure 9. The depletion of metabolites in feces of CMC subjects occurred despite lack of significant difference in fecal bacterial density (Figure 2D, adjusted P = .503) or change in total stool mass produced per subject (Supplementary Figure 4, within-group change in stool weight P = .903 for control group and P = .990 for CMC group), arguing against it reflecting loss of bacteria or stool dilution. Nor did CMC directly inhibit NMR-based detection of amino acids (Supplementary Figure 10), indicating that the reductions in these metabolites did not reflect a technical artifact but rather that CMC feeding depleted an array of microbiota-related metabolites.
Figure 4.
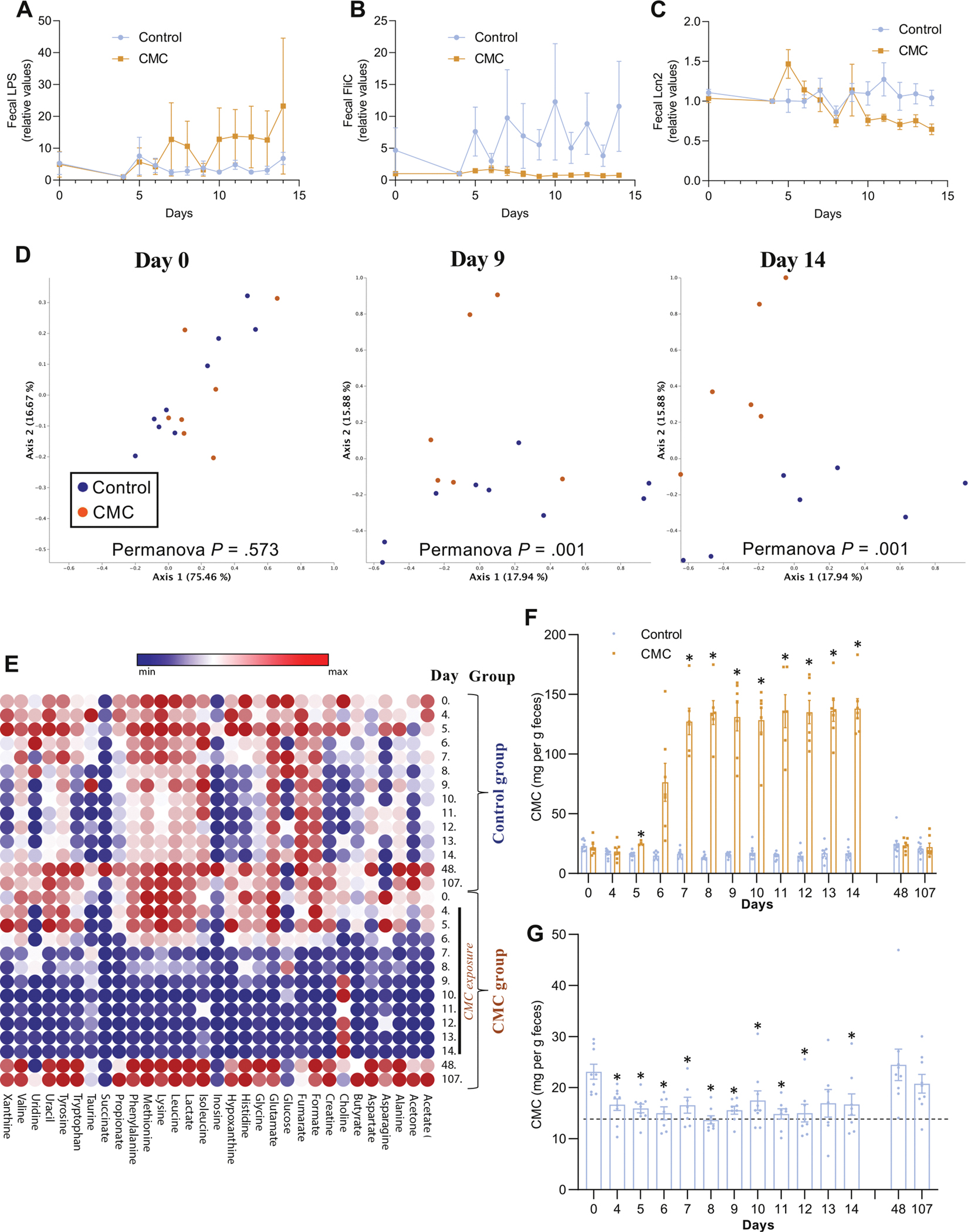
Effect of CMC consumption on the fecal metabolome. (A) Changes of the fecal level of bioactive lipopolysaccharide from day 0 to subsequent days measured with HEK-TLR4 reporter cells. (B) Changes of the fecal level of bioactive flagellin from day 0 to subsequent days measured with HEK-TLR5 reporter cells. (C) Changes of the fecal level of the inflammatory marker Lipocalin-2 from day 0 to subsequent days. (D) PCoA of the Euclidean distance at days 0, 9, and 14 of study participants’ fecal metabolome, with samples colored by group. (E) Heatmap presenting participants fecal metabolome over the course of the study. (F) Changes of the fecal level of CMC from day 0 to subsequent days in both control and CMC-treated groups. (G) Changes of the fecal level of CMC from day 0 to subsequent days in control group. Significance was determined using 2-way ANOVA corrected for multiple comparisons with a Bonferroni posttest or repeated t tests corrected with the false discovery rate approach for (G); *P < .05 compared with control group for (F), *P < .05 compared with day 0 for (G).
A New Assay for CMC Quantification
Animal studies using radiolabeled CMC indicate that most of the label is eliminated in feces, suggesting that this compound is poorly absorbed.31 Hence, we developed a new 1H NMR-based assay that detected copious amounts of seemingly intact CMC in feces of subjects receiving the CMC-containing diet compared with participants consuming the control diet and compared with their usual diet (Figure 4F and Supplementary Figure 11). Although the nonzero levels of CMC measured by this assay may reflect background (ie, another fecal metabolite with spectral properties similar to CMC), the significant decreased level in the participants consuming an additive-free diet (P < .05 for all time points except day 13) and subsequent increase at day 48 and 107 after the study suggests that the readout is capturing CMC contained in processed foods that were consumed before or after participation in our study (Figure 4G). In further accord with the notion that CMC is not absorbed, it was undetectable in urine, nor were alterations in the urinary metabolome associated with CMC consumption (Supplementary Figure 12). Thus, our results comport with the notion that CMC is nonabsorbed but significantly altered the host-microbiota relationship.
Distance Between the Intestinal Mucosa and the Microbiota and Identification of CMC-sensitive Subjects
A characteristic of altered host-microbiota interactions in a range of chronic inflammatory diseases, including IBD, metabolic syndrome, and cancer, is encroachment of gut microbiota into the normally near-sterile inner mucus layer. Hence, we hypothesized that CMC consumption might result in microbiota reduced bacterial-epithelial distance as measured via confocal microscopy in distal colonic biopsies preserved in Carnoy’s solution collected before or after the intervention period. On average, bacterial-epithelial distance did not change over the course of the study in the control or CMC group. However, 2 individual subjects within the CMC group showed a marked reduction in this parameter, such that their biopsies showed bacteria in very close proximity to the epithelium following CMC exposure (Figure 5A and Supplementary Figure 13), reminiscent of observations made in patients with IBD.32 Application of Fisher’s exact test to the observation that 2 of 7 CMC-fed subjects and 0 of 9 control subjects displayed this phenotypic change over the course of the study yielded a 2-tailed P value of 0.175, which does not meet common standards of being statistically significant but nonetheless suggests a reasonable likelihood it was a consequence of CMC treatment. Accordingly, we examined if any of the clinical and/or microbiota parameters might give insight into these seemingly CMC-sensitive subjects. Although these subjects did not respond differently in terms of clinical parameters or inflammatory markers, they had significantly greater relative changes in microbiota composition in response to CMC consumption relative to other participants in the CMC group (Figure 5B and C, group effect P = .004). Moreover, these subjects displayed significantly increased levels of fecal lipopolysaccharide (Figure 5D, group effect P = .005). Analysis of the metagenomic data at the functional level using beta diversity measurement of the Bray-Curtis distance revealed that these 2 participants had strikingly greater relative changes in microbiota function in response to CMC consumption relative to the other participants of the CMC group (Figure 5G, P = .0002). Analysis of morphometric characteristics taken at the beginning of the clinical trial revealed that CMC-sensitive subjects are both male and are older compared with other members of the CMC group, without any other significant differences (weight, height, body mass index, systolic blood pressure, diastolic blood pressure; Figure 5H). Collectively, these results suggest that some individuals may be prone to develop alterations in the host-microbiota interactions in response to CMC consumption, and future studies are warranted to investigate the long-term consequences on intestinal health.
Figure 5.
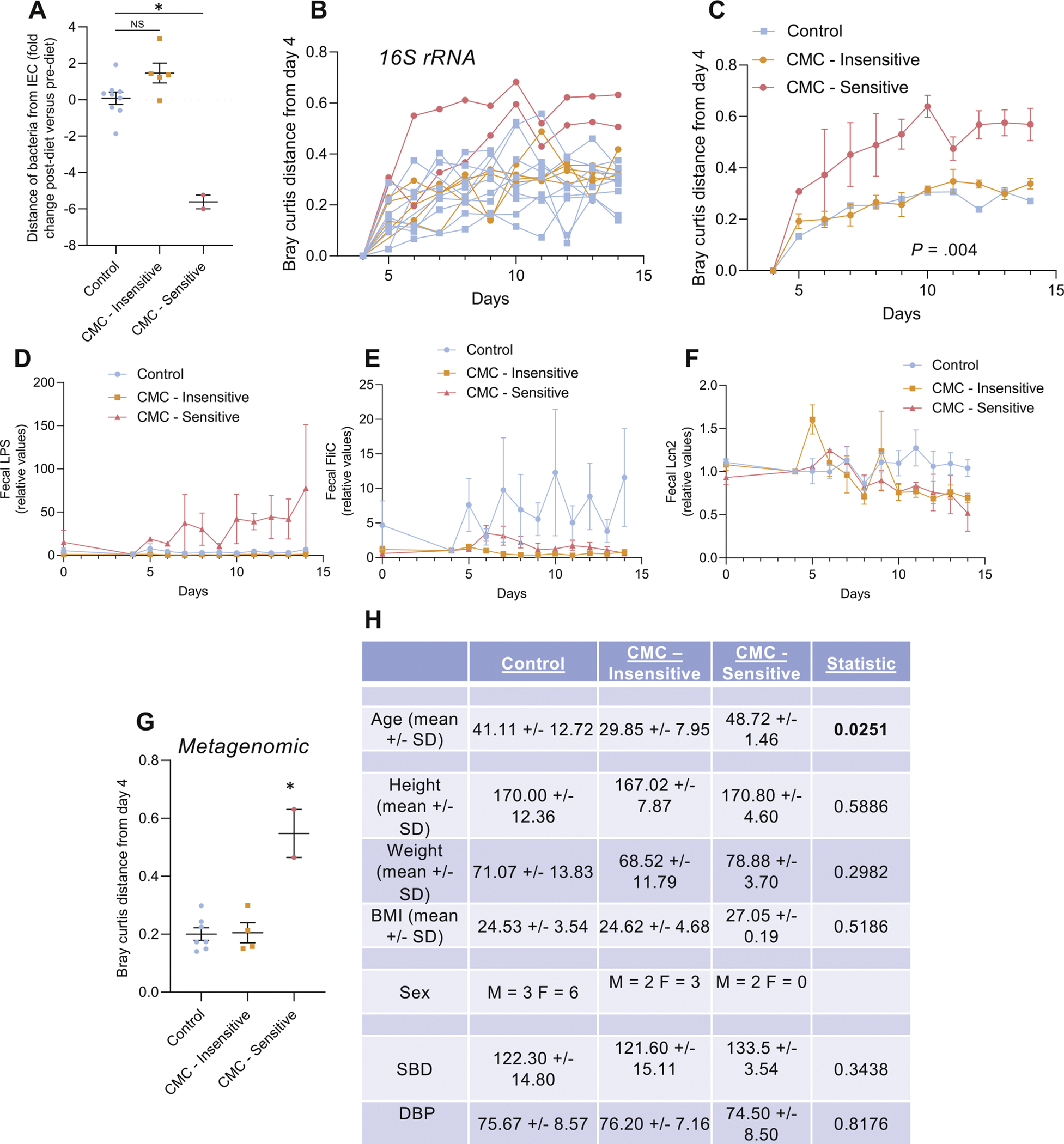
Intersubject variability in the response to CMC consumption. (A) Effect of dietary emulsifier CMC consumption on microbiota localization (distance of the closest bacteria from the surface of the epithelium), measured both pre- and postintervention. (B) Changes of the Bray-Curtis distance matrix, for each study’s participant from the CMC-treated group, from day 4 to subsequent days. (C) Changes of the Bray-Curtis distance matrix, for the CMC-Insensitive and the CMC-Sensitive groups, from day 4 to subsequent days. (D) Changes of the fecal level of bioactive LPS from day 0 to subsequent days measured with HEK-TLR4 reporter cells. (E) Changes of the fecal level of bioactive flagellin from day 0 to subsequent days measured with HEK-TLR5 reporter cells. (F) Changes of the fecal level of the inflammatory marker Lipocalin-2 from day 0 to subsequent days. (G) Effect of dietary emulsifier CMC consumption on fecal metagenome measured through Bray-Curtis distance. (H) Biomorphometric characterization of study’s participants at the beginning of the study and according to CMC sensitivity status. Significance was determined using 1-way ANOVA corrected for multiple comparisons with a Bonferroni posttest (A and G) or 2-way ANOVA corrected for multiple comparisons with a Bonferroni posttest (C, D, E, and F). BMI, body mass index; DBP, diastolic blood pressure; IEC, intestinal epithelial cell; LPS, lipopolysaccharide; NS, not statistically significant; SBP, systolic blood pressure.
Discussion
That the post-mid-20th century increased incidence of chronic inflammatory diseases has been roughly paralleled by increased consumption of highly processed foods has long suggested the possibility that some components of such foods promote inflammation. Appreciation of the role of the intestinal microbiota in driving inflammation led to interest in food additives capable of perturbing the host-microbiota relationship. Our previous findings that some dietary emulsifiers can affect microbiota in vitro and in animal models, whereby they promote inflammatory diseases, suggest that these compounds might be one specific example of this notion.9–12 However, the extent to which such substances actually increase risk of disease in the doses and frequency in which they are consumed by humans remains far less clear. Our findings reported herein that consumption of one widely used food additive, namely the synthetic emulsifier CMC, affected microbiota in humans in a seemingly detrimental manner are a step toward filling this knowledge gap.
Epidemiologic-based studies of food additives have limited power to assess consequences of specific food additives for numerous reasons. For one, concentrations of these components in commercially prepared foods are not widely reported, making extremely challenging to quantitatively estimate food additive consumption in humans.33 Furthermore, processed foods often contain multiple potentially detrimental ingredients, making the driver of associations difficult to identify. Randomized controlled trials to assess the impact of food additives on disease incidence are very challenging because of the long period of follow-up required. Nonetheless, they remain the gold standard means to identify specific ingredients, for example, artificial sweeteners.34 Indeed, controlled-feeding studies, such as ours, are ideal to study the physiologic response of humans to short-term dietary exposures in a tightly controlled setting. Our design allowed us to focus on microbiota changes that are associated with chronic diseases, where a role in causation has been proposed. We observed stark changes in gut microbiota, fecal metabolome, and, in a subset of the participants, encroachment of microbiota on the gut epithelium. The predominant changes in the fecal metabolome on CMC feeding was loss of purportedly beneficial metabolites. We envision this change likely reflected loss of key taxa and/or general disruption of microbial community homeostasis. We also demonstrate that CMC consumption can be assayed by quantitating its level in feces, thus providing a tool to facilitate longer-term studies that could address the extent to which CMC exposure promotes chronic diseases that are increasingly prevalent in developed countries.
The dose of CMC (15 g per person per day) used in this study likely exceeds CMC intake of most individuals but might approximate the total amount of emulsifier consumption by persons whose diets are largely comprised of highly processed foods that contain numerous emulsifiers, many of which appear to detrimentally affect human microbiotas in vitro.8 Although this study focused on one specific food additive, CMC, the results obtained support the need to apply this paradigm to other dietary emulsifiers, and mixtures thereof, at lower concentration, thus better mimicking their use in processed foods. Further, we view it as important to discern the extent to which the highly heterogeneous impact of emulsifier on human microbiota in vitro is recapitulated in vivo.8 Finally, although our study was not powered to discover CMC-sensitive/CMC-insensitive participants, our results nonetheless suggest that microbiota responsiveness to this food additive may be highly personalized. Although follow-up studies are needed to better understand such interindividual variability and assess its role in driving microbiota-mediated disease states, our observations argue that a particular food additive might perturb the host-microbiota relationship to promote disease in a subpopulation of individuals. If our results are confirmed in larger studies with longer-term follow-up, the identified mechanism(s) may inform healthy food choices and enable the development of healthier processed foods.
Supplementary Material
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT
Some widely used food additives, including dietary emulsifiers, alter gut microbiota and promote inflammation in in vitro and animal models, but applicability of such observations to humans remains poorly characterized. To begin to fill this knowledge gap, we investigated the impact of the synthetic dietary emulsifier carboxymethylcellulose on healthy human volunteers.
NEW FINDINGS
Addition of carboxymethylcellulose to a healthy additive-free diet increased postprandial abdominal discomfort and altered intestinal microbiota composition. Moreover, carboxymethylcellulose consumption starkly impacted the fecal metabolome, including depletion of health-promoting metabolites such as short-chain fatty acids and free amino acids. Furthermore, some individuals displayed microbiota encroachment into the normally sterile inner mucus layer following carboxymethylcellulose consumption.
LIMITATIONS
This study was focused on the short-term impacts of carboxymethylcellulose, particularly on gut microbiome. Assessing the extent to which these changes would persist in states of long-term consumption of carboxymethylcellulose and/or other emulsifiers, and determining their phenotypic consequences, would require additional studies.
IMPACT
That carboxymethylcellulose consumption by humans impacted the microbiome supports the notion that wide use of this compound, and perhaps other dietary emulsifiers, in processed foods may have contributed to increased incidence of chronic inflammatory diseases.
Acknowledgments
The authors thank the Penn Center for Nutritional Science and Medicine and the Host-Microbial Analytic and Repository Core of the Center for Molecular Studies in Digestive and Liver Diseases (National Institutes of Health P30 DK050306). The authors thank Dr. Philip Smith from the Penn State Metabolomics Facility, and Nicholas Youngblut from the MPI. All authors had access to the study data and reviewed and approved the final manuscript.
Funding
This work was supported by National Institutes of Health grants DK115180, 5UL1TR001878, and P30-DK050306, and by the Max Planck Society. Benoit Chassaing’s laboratory is supported by a Starting Grant from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. ERC-2018-StG- 804135), a Chaire d’Excellence from IdEx Université de Paris (ANR-18-IDEX-0001), an Innovator Award from the Kenneth Rainin Foundation, a grant from the French Research Agency (Emulbiont, ANR-21-CE15–0042–01), and the national program “Microbiote” from INSERM. Andrew D. Patterson’s laboratory is supported by the USDA National Institute of Food and Federal Appropriations under Project PEN04607 and Accession number 1009993. The funders had no role in the design of the study and data collection, analysis and interpretation, or in manuscript writing.
Abbreviations used in this paper:
- ANOVA
analysis of variance
- CHPS
Center for Human Phenomic Science
- CMC
carboxymethylcellulose
- IBD
inflammatory bowel disease
- NMR
nuclear magnetic resonance
- PCoA
principal coordinate analysis
- Permanova
permutational multivariate ANOVA
- rRNA
ribosomal RNA
- SV
sequence variant
Footnotes
Conflict of interest
Dr Lewis consulted or served on an advisory board for Eli Lilly and Company, Samsung Bioepis, UCB, Bristol-Myers Squibb, Nestle Health Science, Merck, Celgene, Janssen Pharmaceuticals, Bridge Biotherapeutics, Entasis Therapeutics, AbbVie, Pfizer, Gilead, Arena Pharmaceuticals, Protagonist Therapeutics, Amgen, and Scipher Medicine. He has had research funding from Nestle Health Science, Takeda, Janssen Pharmaceuticals, and AbbVie. The remaining authors disclose no conflicts.
CRediT Authorship Contributions
Order of Authors (with Contributor Roles):
Benoit Chassaing, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – original draft: Supporting)
Charlene Compher, PhD (Conceptualization: Supporting; Formal analysis: Supporting; Investigation: Supporting; Project administration: Supporting; Writing – original draft: Supporting)
Brittaney Bonhomme, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Writing – original draft: Supporting)
Qing Liu, PhD (Data curation: Supporting; Formal analysis: Supporting; Writing – original draft: Supporting)
Yuan Tian, PhD (Data curation: Supporting; Formal analysis: Supporting; Writing – original draft: Supporting)
William Walters, PhD (Data curation: Supporting; Formal analysis: Supporting; Writing – original draft: Supporting)
Lisa Nessel, PhD (Data curation: Supporting; Writing – review & editing: Supporting)
Clara Delaroque, PhD student (Data curation: Supporting; Formal analysis: Supporting)
Fuhua Hao, PhD (Data curation: Supporting; Formal analysis: Supporting)
Victoria Gershuni, PhD (Writing – review & editing: Supporting)
Lillian Chau, PhD (Data curation: Supporting; Writing – review & editing: Supporting)
Josephine Ni, PhD (Writing – review & editing: Supporting)
Meenakshi Bewtra, PhD (Writing – review & editing: Supporting)
Lindsey Albenberg, PhD (Writing – review & editing: Supporting)
Alexis Bretin, PhD (Data curation: Supporting; Writing – original draft: Supporting)
Liam McKeever, PhD (Conceptualization: Supporting; Data curation: Supporting; Writing – review & editing: Supporting)
Ruth Ley, PhD (Data curation: Supporting; Writing – original draft: Supporting)
Andrew Patterson, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Writing – original draft: Supporting)
Gary Wu, MD, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Supervision: Supporting; Writing – original draft: Supporting)
Andrew Gewirtz, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Resources: Supporting; Supervision: Supporting; Validation: Supporting; Writing – original draft: Supporting)
James Lewis, MD, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Resources: Supporting; Writing – original draft: Supporting)
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2021.11.006.
References
- 1.Marion-Letellier R, Amamou A, Savoye G, et al. Inflammatory bowel diseases and food additives: to add fuel on the flames. Nutrients 2019;11:1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez Steele E, Juul F, Neri D, et al. Dietary share of ultra-processed foods and metabolic syndrome in the US adult population. Prev Med 2019;125:40–48. [DOI] [PubMed] [Google Scholar]
- 3.Caruso R, Lo BC, Nunez G. Host-microbiota interactions in inflammatory bowel disease. Nat Rev Immunol 2020;20:411–426. [DOI] [PubMed] [Google Scholar]
- 4.Neurath MF. Host-microbiota interactions in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17:76–77. [DOI] [PubMed] [Google Scholar]
- 5.Dabke K, Hendrick G, Devkota S. The gut microbiome and metabolic syndrome. J Clin Invest 2019;129:4050–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultz J. Carboxymethylcellulose as a colloid laxative. Am J Dig Dis 1949;16:319–322. [DOI] [PubMed] [Google Scholar]
- 7.Smits CH, Veldman A, Verkade HJ, et al. The inhibitory effect of carboxymethylcellulose with high viscosity on lipid absorption in broiler chickens coincides with reduced bile salt concentration and raised microbial numbers in the small intestine. Poult Sci 1998;77:1534–1539. [DOI] [PubMed] [Google Scholar]
- 8.Naimi S, Viennois E, Gewirtz AT, et al. Direct impact of commonly used dietary emulsifiers on human gut microbiota. Microbiome 2021;9:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viennois E, Bretin A, Dube PE, et al. Dietary emulsifiers directly impact adherent-invasive E. coli gene expression to drive chronic intestinal inflammation. Cell Rep 2020;33:108229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viennois E, Merlin D, Gewirtz AT, et al. Dietary emulsifier-induced low-grade inflammation promotes colon carcinogenesis. Cancer Res 2017;77:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chassaing B, Van de Wiele T, De Bodt J, et al. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut 2017;66:1414–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chassaing B, Koren O, Goodrich JK, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015;519:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chassaing B, Raja SM, Lewis JD, et al. Colonic microbiota encroachment correlates with dysglycemia in humans. Cell Mol Gastroenterol Hepatol 2017;4:205–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krebs-Smith SM, Pannucci TE, Subar AF, et al. Update of the Healthy Eating Index: HEI-2015. J Acad Nutr Diet 2018;118:1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panizza CE, Shvetsov YB, Harmon BE, et al. Testing the predictive validity of the Healthy Eating Index-2015 in the multiethnic cohort: is the score associated with a reduced risk of all-cause and cause-specific mortality? Nutrients 2018;10:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petimar J, Smith-Warner SA, Fung TT, et al. Recommendation-based dietary indexes and risk of colorectal cancer in the Nurses’ Health Study and Health Professionals Follow-up Study. Am J Clin Nutr 2018;108:1092–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doucet E, Laviolette M, Imbeault P, et al. Total peptide YY is a correlate of postprandial energy expenditure but not of appetite or energy intake in healthy women. Metabolism 2008;57:1458–1464. [DOI] [PubMed] [Google Scholar]
- 18.Doucet E, Pomerleau M, Harper ME. Fasting and postprandial total ghrelin remain unchanged after short-term energy restriction. J Clin Endocrinol Metab 2004;89:1727–1732. [DOI] [PubMed] [Google Scholar]
- 19.Chassaing B, Srinivasan G, Delgado MA, et al. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One 2012;7:e44328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang L, Huang J, Wang Y, et al. Eliminating the dication-induced intersample chemical-shift variations for NMR-based biofluid metabonomic analysis. Analyst 2012;137:4209–4219. [DOI] [PubMed] [Google Scholar]
- 21.Tian Y, Zhang L, Wang Y, et al. Age-related topographical metabolic signatures for the rat gastrointestinal contents. J Proteome Res 2012;11:1397–1411. [DOI] [PubMed] [Google Scholar]
- 22.Armenta JM, Cortes DF, Pisciotta JM, et al. Sensitive and rapid method for amino acid quantitation in malaria biological samples using AccQ.Tag ultra performance liquid chromatography-electrospray ionization-MS/MS with multiple reaction monitoring. Anal Chem 2010;82:548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole CR, Frem JC, Schmotzer B, et al. The rate of bloodstream infection is high in infants with short bowel syndrome: relationship with small bowel bacterial over-growth, enteral feeding, and inflammatory and immune responses. J Pediatr 2010;156:941–947.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedirko V, Tran HQ, Gewirtz AT, et al. Exposure to bacterial products lipopolysaccharide and flagellin and hepatocellular carcinoma: a nested case-control study. BMC Med 2017;15:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quevrain E, Maubert MA, Michon C, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016;65:415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenoir M, Martin R, Torres-Maravilla E, et al. Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. Gut Microbes 2020;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen OFA, Claassen E. The mechanistic link between health and gut microbiota diversity. Sci Rep 2018;8:2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bar A, Van Ommen B, Timonen M. Metabolic disposition in rats of regular and enzymatically depolymerized sodium carboxymethylcellulose. Food Chem Toxicol 1995;33:901–907. [DOI] [PubMed] [Google Scholar]
- 32.Johansson ME, Gustafsson JK, Holmen-Larsson J, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 2014;63:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chazelas E, Druesne-Pecollo N, Esseddik Y, et al. Exposure to food additive mixtures in 106,000 French adults from the NutriNet-Sante cohort. Sci Rep 2021;11:19680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suez J, Korem T, Zeevi D, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014;514:181–186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


