Abstract
Depilatory creams are widely used to remove unwanted body hair, but people with “sensitive skin” are subject to depilatory-induced skin burn/inflammation. It remains unknown what makes their skin more sensitive than others. Herein, we demonstrate that epidermal fatty acid binding protein (E-FABP) expressed in the skin plays a critical role in promoting depilatory-induced acute skin inflammation in mouse models. While Nair, a common depilatory cream, removed hair by breaking down keratin disulfide bonds, it activated cytosolic phospholipase A2 (cPLA2), leading to activation of the arachidonic acid (AA)/E-FABP/PPARβ signaling pathway in keratinocytes. Specifically, PPARβ activation induced downstream targets (e.g. COX2) and chemokine (e.g. CXCL1) production, which systemically mobilized neutrophils and recruited them to localize in the skin for acute inflammatory responses. Importantly, E-FABP deletion by CRISPR-Cas9 reduced cPLA2/PPARβ activation in keratinocytes, and genetic deletion of E-FABP protected mice from Nair-induced neutrophil recruitment and skin inflammation. Our findings suggest E-FABP as a molecular sensor for “sensitive skin” by triggering depilatory-induced, lipid-mediated skin inflammatory responses.
Introduction
Depilatory creams provide a highly effective, convenient and painless way to remove unwanted body hair. However, there are many complaints that depilatories can cause significant skin burn and/or topical irritant reactions (Kiene et al., 2020). Most depilatory creams contain strong alkaline-based chemicals, like sodium thioglycolate and calcium thioglycolate, which break down disulfide bonds in hair keratin, leading to hair loss (Lee et al., 2008). While FDA-approved depilatories are generally skin safe, skin reactions, including redness, pain, burning, etc., occur in some people even though they strictly follow the instructions (Prigot et al., 1962). Those who develop skin irritation are determined to have “sensitive skin”, but what makes their skin more sensitive to depilatories remains largely unknown.
As the largest organ in the body, the skin consists of two main layers: an outer layer of epidermis and an inner layer of dermis (Swann, 2010). Different cell populations reside in each layer. Keratinocytes make up over 90% of the cells in the epidermis. Melanocytes and immune cells, including skin γδ T cells and macrophages (known as Langerhans cells), also reside in the epidermis (Yanez et al., 2017). The dermis mainly consisting of collagen and elastic tissue is less cellular than the epidermis, but multiple immune cell populations occupy the dermis, such as macrophages, dendritic cells, CD4+ and CD8+ T cells, γδ T cells, NK cells and mast cells (Nestle et al., 2009). Besides providing nutrients to the skin, dermal blood vessels help maintaining local immune cell balance under a variety of physiological and pathological settings. It is of great interest to understand which cellular and molecular sensor (s) in subjects with sensitive skin trigger depilatory cream-induced inflammatory skin responses.
Skin inflammation usually occurs when skin cells are activated by external/internal stimuli, such as chemicals, UV, pathogen-associated molecular patterns (PAMPs), damage-associated molecular patterns (DAMPs), etc. Depending on the stimuli, different signaling pathways (e.g. peroxisome proliferator-activated receptor PPAR signaling, chemokine signaling, and cytokine signaling pathways) are activated to induce innate/adaptive immune responses (Schwingen et al., 2020). Neutrophils are the first immune cells to infiltrate the skin, thereby instigating an inflammatory response (Marzano et al., 2019).
Lipid metabolites such as arachidonic acid (AA)-derived eicosanoids (e.g. leukotriene B4, LTB4, prostaglandins, PGEs) are strong chemoattractants for neutrophils (Petri and Sanz, 2018). Fatty acid binding proteins (FABPs) play a central role in coordinating lipid responses (Li et al., 2020; Storch and Corsico, 2008; Zimmer et al., 2004). Epidermal FABP (E-FABP, also known as FABP5), the predominant FABP isoform in skin, promotes epidermal proliferation through activating PPARβ/δ signaling (Kannan-Thulasiraman et al., 2010). Our studies also demonstrate that E-FABP is critical in mediating high fat diet-induced skin inflammation (Zhang and Li, 2014; Zhang et al., 2015). Thus, we proposed that E-FABP might function as a molecular sensor triggering lipid-mediated signaling for neutrophil-mediated skin inflammation.
Nair, a common commercially available depilatory lotion, is widely used for hair removal in different mouse models (Qin et al., 2015). In the current study, using Nair-induced skin inflammation in mouse models, we found that E-FABP was critical in enhancing the expression of cytoplasmic phospholipase A2 (cPLA2), a rate limiting enzyme hydrolyzing AA from membrane phospholipids in keratinocytes, thus promoting lipid-mediated chemokine signaling and neutrophil recruitment. In addition, mice deficient in E-FABP exhibited reduced neutrophil recruitment and Nair-induced skin reactions.
Results
Nair-induced acute skin inflammation is associated with systemic neutrophil mobilization
Although depilatory hair removal creams are generally safe for use, some people are subject to depilatory cream-induced acute inflammatory skin reactions. The cause for this is unknown. In our mouse studies using Nair to remove hair, we noticed that some mice exhibited strong acute skin reactions (Supplementary Figure S1A). To determine the underlying cellular and molecular mechanisms by which Nair induced skin inflammation, we collected skin and other immune tissues from shaved mice treated with or without Nair and compared their immunophenotypic and functional alterations (Figure 1A). In the epidermis of Nair-treated mice, there were elevated CD45+ cells compared to untreated mice (Figure 1B,1C), suggesting increased infiltration of immune cells. Neutrophils were rare in the epidermis of untreated mice, but significantly increased in response to Nair treatment (Figure 1D, 1E). Similar as the epidermis, there were increased CD45+ cells in the dermis (Figure 1F, 1G), and neutrophil recruitment to the dermis was more obvious as determined by flow cytometric analyses (Figure 1H, 1I) and immunofluorescence staining (Figure 1J). Macrophages, but not other immune cells, were also increased in the epidermal and dermal layers (Supplementary Figure S1B, S1C). Normal skin contains resident macrophages but rarely neutrophils. To determine the origin of the increased neutrophil infiltration in the skin, we analyzed immune cell populations in the blood, spleen and bone marrow (BM) of mice treated with or without Nair. Interestingly, Nair treatment remarkably increased neutrophils in the blood (Figure 1K,1L) and the spleen (Figure 1M, 1N), but decreased their prevalence in the BM (Figure 1O, 1P), suggesting the mobilization of neutrophils from the BM to the periphery. By contrast, Nair treatment did not induce systemic alterations in macrophages or other immune cells (Supplementary Figure S1D–S1F). These data suggest that Nair treatment induces systemic neutrophil mobilization and local recruitment to the skin.
Figure 1. Nair-induced acute skin inflammation is associated with neutrophil recruitment.
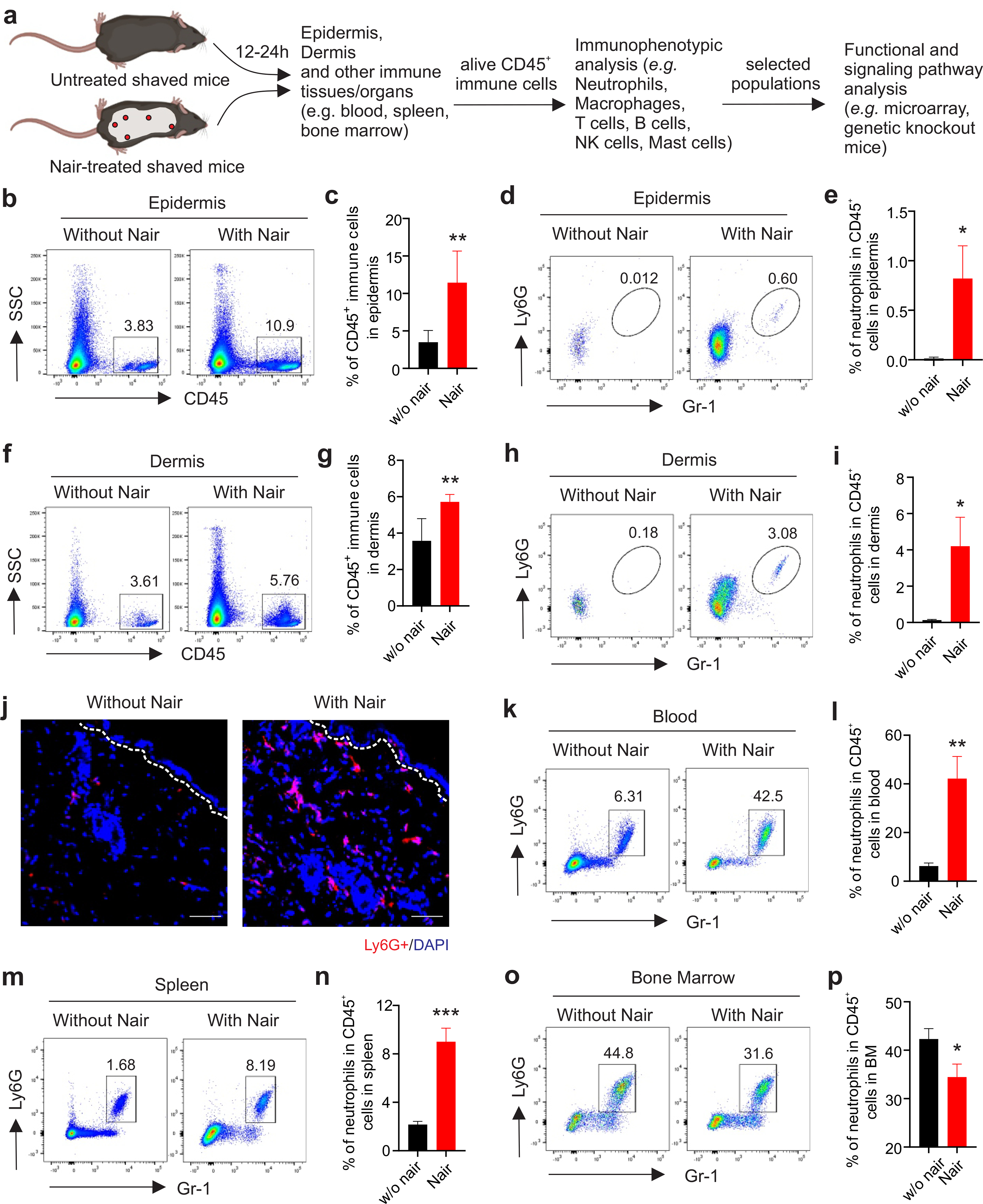
(a) Experimental procedure of dissecting Nair-induced skin inflammation.
(b,c) flow cytometric analysis of CD45+ immune cells in the epidermis of mice treated with or without Nair (b) and average of CD45+ cells in the epidermis is shown (c).
(d,e) neutrophil infiltration in the epidermis of mice treated with or without Nair (d), and average of neutrophil percentage in CD45+ cells is shown (e).
(f, g) flow cytometric analysis of CD45+ immune cells in the dermis of mice treated with or without Nair (f) and average of CD45+ cells in the dermis is shown (g).
(h,i) neutrophil infiltration in the dermis of mice treated with or without Nair (h) and average of neutrophil percentage in CD45+ cells is shown (i).
(j) Immunofluorescence staining of Ly6G+ neutrophils (red) in mice treated with or without Nair (Bar=50μM).
(k,l) flow cytometric analysis of neutrophil percentage in the peripheral blood from mice treated with or without Nair (k), and average percentage of neutrophils in total CD45+ cells is shown (l).
(m,n) flow cytometric analysis of neutrophil percentage in the spleen from mice treated with or without Nair (m) and average percentage of neutrophils in total CD45+ cells is shown (n).
(o,p) flow cytometric analysis of neutrophil percentage in the bone marrow from mice treated with or without Nair (o) and average percentage of neutrophils in total CD45+ cells is shown (p).
Data are shown as mean ± SEM (*p<0.05, **p<0.01, ***p<0.001).
To confirm if neutrophil skin infiltration was specifically induced by Nair treatment, but not by clipper-related physical skin damages or other effects (e.g. grooming or fighting), we further analyzed neutrophil infiltration in unshaved vs shaved skin without Nair treatment (Supplementary Figure S2A). As compared to unshaved mice, shaved mice did not exhibit increased infiltrations of CD45+ immune cells and neutrophils in both epidermis (Supplementary Figure S2B–S2E) and dermis (Supplementary Figure S2F–S2I), which demonstrate no/few physical damage-associated neutrophil infiltrations in shaved mice. As no hair growing out of mouse ears, we also compared neutrophil infiltrations in mouse ears treated with or without Nair. CD45+ immune cells (Supplementary Figure S3A) and neutrophils (Supplementary Figure S3D) in the Nair-treated ear tissue were significantly increased when compared to untreated ears. Of note, both percentage and absolute numbers of CD45+ cells (Supplementary Figure S3B, S3C) and neutrophils (Supplementary Figure S3E, S3F) were increased in response to the Nair treatment. Altogether, these data demonstrate that Nair treatment specifically induces neutrophil-mediated skin inflammation.
Nair induces neutrophil infiltration in the skin through the cPLA2/CXCL1 axis
Neutrophils are the primary innate immune cells that are rapidly recruited to inflamed tissues. Depending on the inflammatory insult, neutrophils are specifically regulated by multiple chemoattractants (Petri and Sanz, 2018). Lipids derived from AA, such as LBT4, are strong chemoattractants for neutrophil recruitment (Chou et al., 2010; Sadik and Luster, 2012). AA can also be metabolized through other enzymatic pathways, such as cyclooxygenase 2 (COX2)/prostaglandin E2 (PGE2), to promote neutrophil chemotaxis by chemokines, such as CXCL1 and CXCL2 (Wang et al., 2014) (Figure 2A). To dissect the molecular mechanisms underlying Nair-induced neutrophil skin recruitment, we measured the expression of key enzymes/chemokines responsible for neutrophil recruitment in the skin treated with or without Nair. We found that cytosolic phospholipase A2 (cPLA2), the rate-limiting enzyme responsible for AA release from membrane phospholipids, was significantly upregulated in response to Nair treatment (Figure 2B). However, enzymes for LTB4 synthesis, including Alox5 and Lta4H (Figure 2C, 2D), did not respond to Nair treatment, suggesting a minimal role of LTB4 in mediating Nair-induced neutrophil recruitment. Instead, Nair treatment significantly enhanced the COX2/PGE2/CXCL1/CXCR2 signaling axis (Figure 2E–2I). Inflammation-associated cytokines (e.g. IL-1β, IL-6) (Figure 2J, 2K) were also upregulated in Nair-treated skin. Other chemokines/receptors associated with neutrophil/macrophage recruitment, including IL-8, CCL17, CCL22, CCL2, CCR2 (Supplementary Figure S4A–S4E), lipid synthesis enzymes (e.g. Alox12, PGI2, TXA2) (Supplementary Figure S4F–S4H) and cytokines (e.g. IL-36γ, TNFα) (Supplementary Figure S4I, S4J) were not significantly altered in response to Nair treatment. These data suggest that the cPLA2/CXCL1 axis, but not the cPLA2/LTB4 axis, was mainly responsible for neutrophil recruitment. To further validate our observations, we measured skin cPLA2 activity (Figure 2L) and chemoattractant levels of LTB4 and CXCL1 in the skin and in serum of mice treated with or without Nair by ELISA (Figure 2M–2O). Our results confirmed that Nair-induced neutrophil recruitment was associated with activated cPLA2/CXCL1 signaling.
Figure 2. Nair treatment induces activation of the cPLA2/COX2/PGE2/CXCL1 axis.
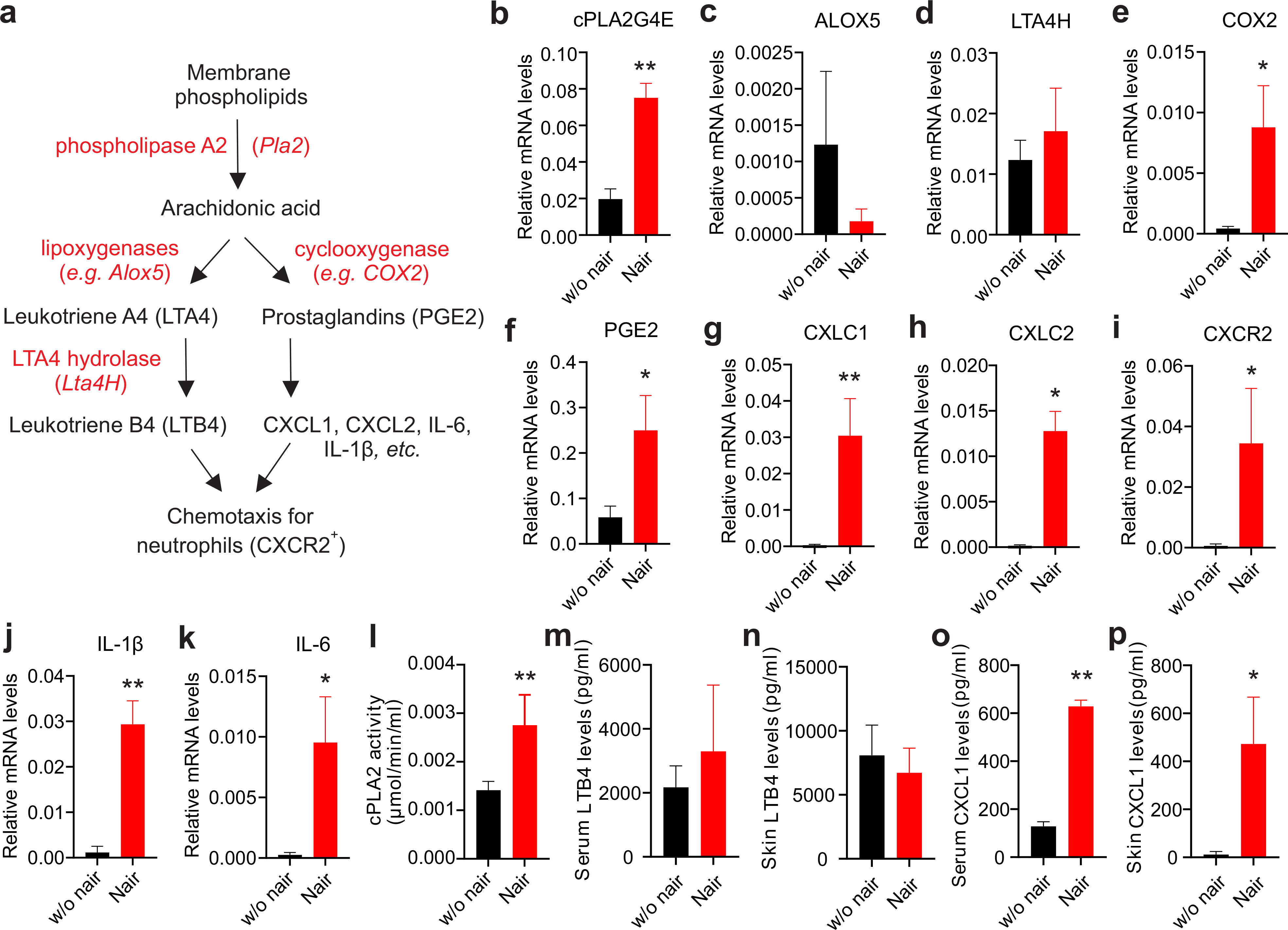
(a) cPLA2-induced neutrophil recruitment pathways and related molecules.
(b-k) real-time PCR analysis of expression of cPLA2G4E (b), Alox5 (c), LTA4H(d), COX2 (e), PGE2 (f), CXCL1 (g), CXCL2 (h), CXCR2 (i), IL-1 (j) and IL-6 (k) in the skin treated with or without Nair.
(l) Measurement of skin cPLA2 activity from mice treated with or without Nair
(m,n) ELSIA analysis of LTB4 levels in serum (l) and in the skin (m) from mice treated with or without Nair.
(o,p) ELSIA analysis of CXCL1 levels in serum (n) and in the skin (o) from mice treated with or without Nair.
Data are shown as mean ± SEM (*p<0.05, **p<0.01).
Nair-activated E-FABP/PPARβ signaling promotes cPLA2 expression in keratinocytes
Fatty acid binding proteins (FABPs) play a central role in facilitating lipid transportation and responses inside cells (Hotamisligil and Bernlohr, 2015; Li et al., 2020). E-FABP preferentially binds AA released by cPLA2 and clusters with COX2 for the synthesis of PGE2 (Bogdan et al., 2018; Zimmer et al., 2004). As expected, we observed that E-FABP was the predominant isoform expressed in the skin (Figure 3A), and Nair treatment increased the expression of both E-FABP and cPLA2 (Figure 3B). As E-FABP binds AA which is released by cPLA2 hydrolysis, we hypothesized a positive regulatory mechanism between E-FABP and cPLA2 expression. Since immunofluorescent staining showed that Nair treatment mainly increased E-FABP expression in epidermal keratinocytes (Figure 3C), we used keratinocytes to address our hypothesis. Deletion of E-FABP with CRISPR-Cas9 decreased the expression of cPLA2G4E, cPLA2G4A, the main isoforms of cPLA2 in keratinocytes (Figure 3D), suggesting an important role for E-FABP in regulating cPLA2 expression. It is known that E-FABP binding to AA facilitates its nuclear translocation and activates nuclear transcription factor PPARβ (Armstrong et al., 2014). We observed that PPARβ, which was predominantly expressed in the skin, was upregulated by Nair treatment (Figure 3E). Moreover, treatment of keratinocytes with thioglycolic acid (TGA), the main active ingredient in Nair, induced E-FABP nuclear translocation (Figure 3F), suggesting a role of E-FABP/PPARβ signaling in regulating cPLA2 expression. Using the well-known PPARβ agonist GW101516, we confirmed that E-FABP was critical in controlling PPARβ-induced cPLA2 expression in keratinocytes (Figure 3G). Taken together, our results suggest a novel observation that E-FABP plays a critical role in regulating Nair-induced cPLA2 expression in a PPARβ-mediated pathway.
Figure 3. Nair treatment induces activation of the E-FABP/PPARβ signaling.
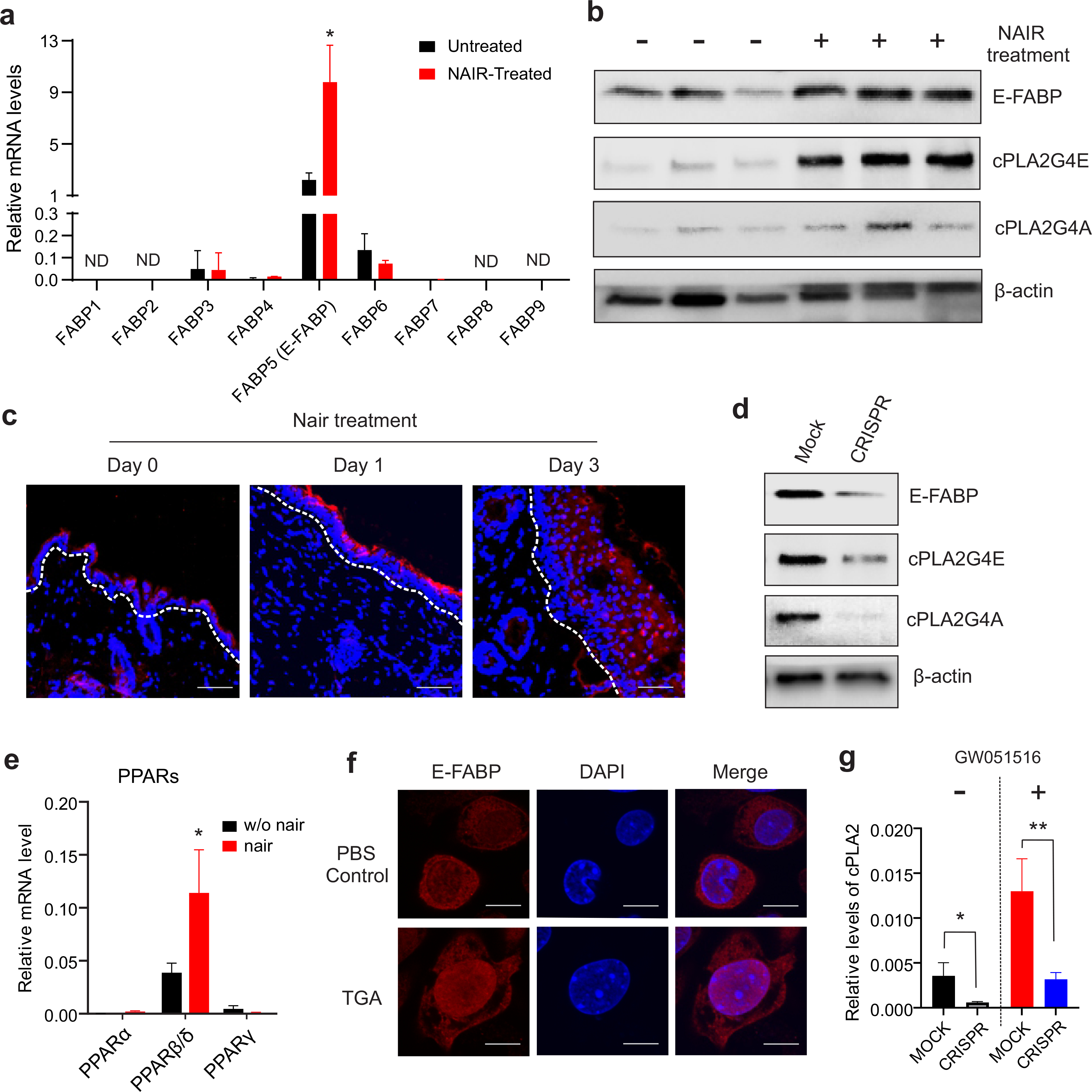
(a) Realtime PCR analysis of FABP family gene expression profile in the skin treated with or without Nair.
(b) Western blotting analysis of E-FABP, cPLA2GE, cPLA2G4A expression in the skin.
(c) Immunofluorescence staining of E-FABP expression (red) in the skin treated with or without Nair at the indicated time points (DAPI for nuclei) (Bar=50μM).
(d) Western blotting analysis of expression of E-FABP, cPLA2G4A, cPLA2G4E in keratinocytes stably transfected with MOCK or CRISPR for E-FABP deletion.
(e) Realtime PCR analysis of expression levels of PPAR family gene in the skin treated with or without Nair
(f) Immunofluorescence analysis of E-FABP nuclear translocation in keratinocytes treated with PBS or thioglycolate acid (TGA) (Bar=5μM).
(g) Real-time PCR analysis of cPLA2 expression in E-FABP knockdown or control keratinocytes treated with or without PPARβ agonist GW051516.
Data are shown as mean ± SEM (*p<0.05, **p<0.01).
Mice deficient in E-FABP exhibit reduced cPLA2 expression in the skin
We next employed E-FABP-deficient mice to validate the E-FABP/cPLA2 regulation that we observed. We collected skin from Nair-treated WT and E-FABP−/− mice and performed Affymetrix microarray analysis. Comparison of the differentially expressed genes in the skin of WT and E-FABP−/− mice suggested that E-FABP was mainly involved in cellular differentiation, growth and proliferation, connective tissue development, lipid metabolism, cancer and cardiovascular diseases (Supplementary Table S1). Gene network analyses showed that expression of cPLA2 was involved in molecular transport as well as lipid and small molecule metabolism (Supplementary Figure S5). Heatmap analysis of lipid chemoattractants, chemokines and the PPAR family demonstrated that E-FABP deficiency in the skin downregulated the cPLA2 family, particularly cPLA2G4E and cPLA2G4F. E-FABP deficiency also significantly reduced PPARβ expression (Figure 4A). Using real-time PCR, we confirmed that E-FABP deficient mice exhibited reduced expression of PLA2G4E, PLA2G4F, PPARβ (Figure 4B–4E). Of note, E-FABP deficiency did not affect expression of COX2 (Figure 4F) nor ALOX5 (Figure 4G) in the skin, suggesting a specific effect of E-FABP on cPLA2 expression. Western blotting and immunofluorescent staining further confirmed the reduced expression of cPLA2G4E in the skin of E-FABP−/− mice when compared to WT mice (Figure 4H, 4I). Altogether, our results demonstrated a role for E-FABP in regulating cPLA2 expression, thus contributing to Nair-induced lipid signaling and inflammatory responses in the skin.
Figure 4. E-FABP deficiency decreases cPLA2 expression in mouse skin.
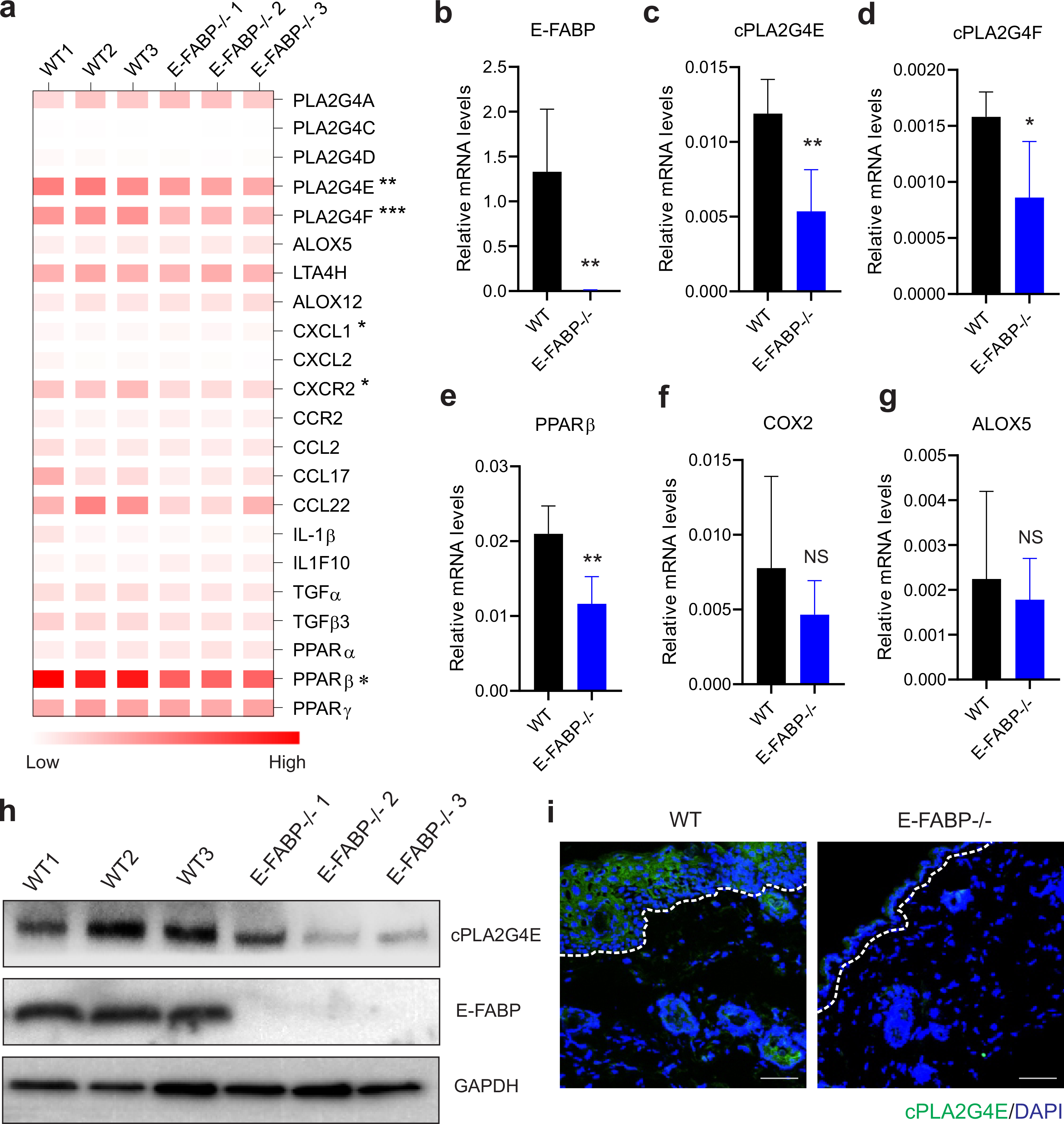
(a) Heatmap of cPLA2 related genes in the skin of Nair-treated WT and E-FABP−/− mice.
(b-g) Realtime PCR validation of expression of E-FABP (b), cPLA2G4E (c), cPLA2G4F (d), PPARβ (e), COX2 (f) and ALOX5 (g) in the skin of Nair-treated WT and E-FABP−/− mice.
(h) Western blotting analysis of cPLA2G4E and E-FABP expression in Nair-treated WT and E-FABP−/− mice
(i) Immunofluorescence staining of cPLA2G4E expression (green) in the skin epidermis of Nair-treated WT and E-FABP−/− mice (Bar=50μM).
Data are shown as mean ± SEM (*p<0.05, **p<0.01).
E-FABP deficiency protects mice from development of Nair-induced skin inflammation
As E-FABP was critical in regulating the expression of cPLA2 which was involved in neutrophil infiltration in skin (Figure 2), we reasoned that E-FABP deficiency inhibited Nair-induced skin inflammation. To this end, we compared Nair-induced acute skin inflammation between WT and E-FABP−/− mice. While Nair treatment induced acute skin inflammation starting on day 1 and peaking on day 3, E-FABP deficiency dramatically reduced Nair-induced skin inflammation (Figure 5A). Analysis of neutrophil infiltration in the dermis on day 1 after Nair treatment demonstrated that E-FABP deficiency dramatically reduced neutrophil skin recruitment as shown by flow cytometric analysis (Figure 5B, 5C) and immunofluorescent staining (Figure 5D). Further analyses of neutrophil mobilization in the blood and in the spleen also demonstrated that E-FABP deficiency reduced systemic neutrophil mobilization from bone marrow (Figure 5E–5I). We also compared neutrophil recruitment in WT and E-FABP−/− mice on day 3 after Nair treatment. Similar results of impaired neutrophil recruitment from bone marrow in E-FABP−/− mice were observed in the skin, blood and spleen (Supplementary Figure S6). Taken together, these data demonstrate that E-FABP expression is critical in mediating Nair-induced neutrophil recruitment and skin inflammation.
Figure 5. E-FABP deficiency protects mice from Nair-induced neutrophil-mediated skin inflammation.
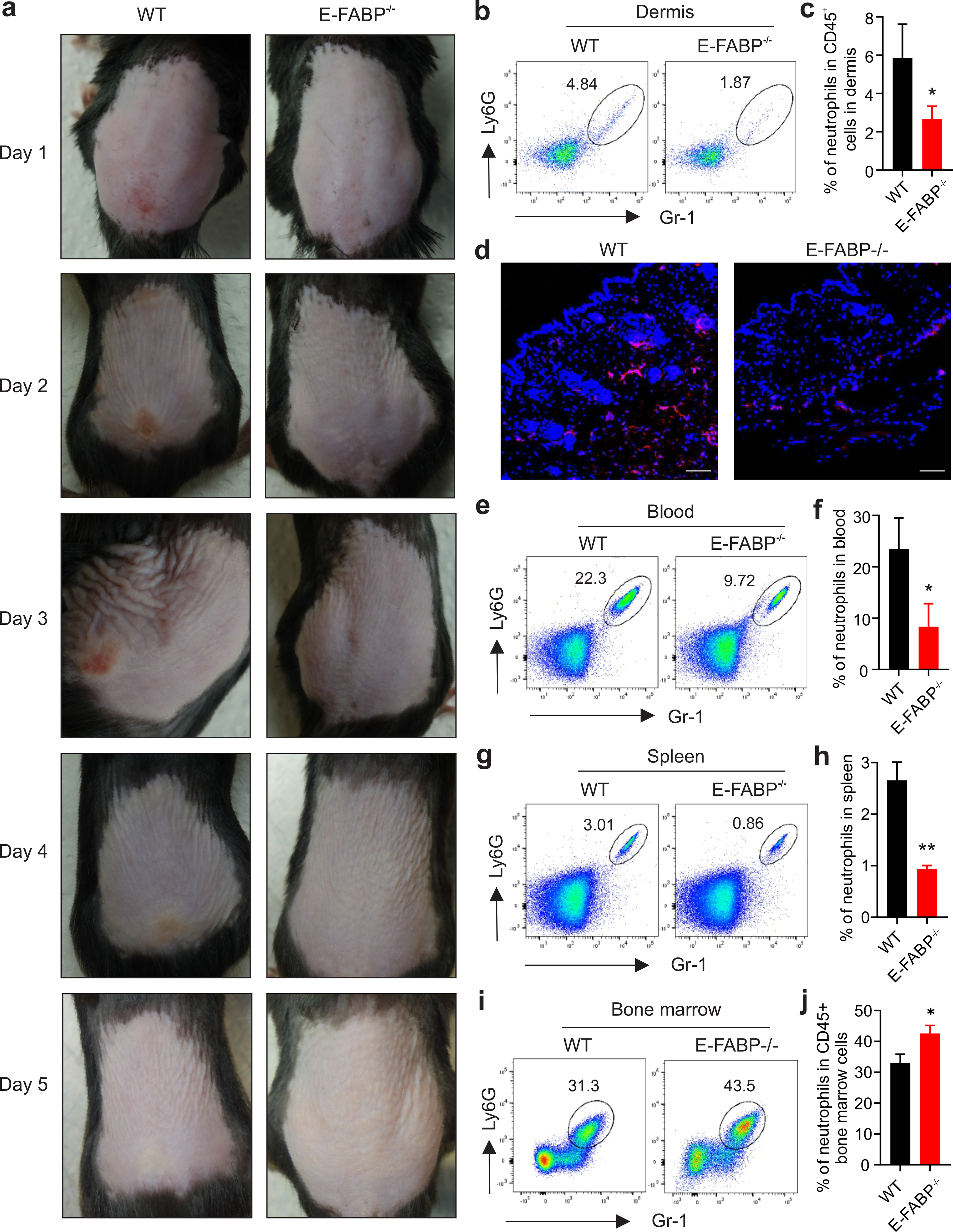
(a) Representative pictures of Nair-induced skin inflammation in WT and E-FABP−/− mice treated at the indicated time points.
(b,c) Flow cytometric analysis of neutrophil percentage in the dermis of Nair-treated WT and E-FABP−/− mice (b) and average percentage of dermal neutrophils in CD45+ cells is shown (c).
(d) Immunofluorescence staining of neutrophils (red) in the skin of Nair-treated WT and E-FABP−/− mice (Bar=50μM).
(e,f) Flow cytometric analysis of neutrophil percentage in the blood of Nair-treated WT and E-FABP−/− mice (e) and average percentage of dermal neutrophils in CD45+ cells is shown (f).
(g,h) Flow cytometric analysis of neutrophil percentage in the spleen of Nair-treated WT and E-FABP−/− mice (g) and average percentage of dermal neutrophils in CD45+ cells is shown (h).
(i,j) Flow cytometric analysis of neutrophil percentage in the bone marrow of Nair-treated WT and E-FABP−/− mice (i) and average percentage of neutrophils in CD45+ bone marrow cells is shown (j).
Data are shown as mean ± SEM (*p<0.05, **p<0.01).
Discussion
Depilatory cream-induced skin burn/inflammation may occur in people with “sensitive skin”. However, the molecular and cellular mechanisms that lead to skin hypersensitivity remain unknown. Our studies demonstrated that the expression of E-FABP in skin keratinocytes functions as a molecular sensor to promote neutrophil recruitment through activation of the PPARβ/cPLA2/CXCL1 signaling axis.
The main ingredients in most depilatory creams are salts of thioglycolic acid (e.g. calcium thioglycolate). Hair is made of keratins, fibrous proteins cross-linked by disulfide bonds. When combined with bases, the proton in thioglycolate’s thiol group (-SH) is released and the free sulfur atom is able to attack keratin disulfide bonds, leading to skin hair degradation. However, proteins in the skin other than keratins also contain disulfide bonds, which are natural targets of thioglycolate. For example, PLA2 is rich in disulfide bonds, which regulate protein structure and catalytic activity (Burke and Dennis, 2009; Janssen et al., 1999). Therefore, depilatory cream application is likely to influence PLA2 activity during hair removal.
Depending on cellular location, calcium-dependence and protein sequence and structure, the PLA2 superfamily consists of four main types: secreted PLA2 (sPLA2), cytosolic (cPLA2), calcium-independent PLA2 (iPLA2) and platelet activating factor (PAF) acetyl hydrolases (Burke and Dennis, 2009). In each main type, there are multiple subtypes of family members. For instance, there are at least 11 isoforms in sPLA2 family, 6 isoforms in cPLA2 family and 9 isoforms in iPLA2 family (Murakami et al., 2018). In general, PLA2 hydrolyzes the release of fatty acids from the sn-2 position of membrane phospholipids. The released free fatty acid (e.g. AA, linoleic acid) is further metabolized by different enzymes (e.g. LOX5, COX2) to generate numerous lipid metabolites, including leukotrienes and prostaglandins, which are involved in multiple physiological and inflammatory processes. As the largest organ in the human body, the skin contains multiple PLA2 isoforms, which are involved in maintaining pathophysiologic activity in the skin (Vlachojannis et al., 2005; Yamamoto et al., 2011). However, it remains unclear as to which PLA2 isoform is critical in skin tissue/cells, which fatty acids on the sn-2 position are sensitive to the pertinent PLA2 isoform, and how PLA2-released fatty acids and their metabolites influence different types of skin inflammation.
In our Nair-induced acute skin inflammation models we identified that cPLA2G4E was highly expressed in Nair-treated skin. Compared to the skin of untreated mice, Nair treatment further increased the expression of cPLA2G4E. Interestingly, the COX2/PGE2 pathway was also upregulated, accompanied by elevated levels of chemokines (e.g. CXCL1/CXCL2) that target neutrophils. By contrast, we did not observe activation of the ALOX5/LTB4 pathway, suggesting that LTB4 is not the main chemoattractant in the setting of Nair-induced inflammation. In delineating the mechanistic basis of preferential activation of cPLA2/COX2/PGE2/CXCL1 signaling in this setting, we observed that E-FABP, the predominant isoform of FABPs expressed in the skin, was significantly upregulated. AA and other U-shape fatty acids (e.g. linoleic acid) bind E-FABP for nuclear translocation and PPARβ activation (Armstrong et al., 2014). It is thus likely that AA released from membrane phospholipids by cPLA2 binds E-FABP and activates PPARβ signaling, which further induces downstream targets (e.g.COX2) and subsequent proinflammatory mediators (e.g. E-FABP, CXCL1, IL-1β) (Morgan et al., 2010; Wang et al., 2014). Our results demonstrate the cPLA2/E-FABP/PPARβ axis involved in Nair-induced skin inflammation.
As E-FABP is mainly expressed in keratinocytes, but not in neutrophils (Ogawa et al., 2011; Rolph et al., 2006; Zhang et al., 2014), we further validated our findings by in vitro cellular studies using E-FABP depletion in keratinocytes and in vivo mouse studies with E-FABP−/− mice. Deletion of E-FABP with CRISPR dramatically inhibited cPLA2G4E expression in keratinocytes. Mice lacking E-FABP also exhibited reduced PLA2G4E expression in the skin epidermis. More importantly, E-FABP deficiency significantly alleviated Nair-induced acute skin inflammation in mouse models. When we analyzed cellular components in inflamed skin, we observed that neutrophils were rapidly recruited from the bone marrow to the epidermis and dermis in an E-FABP-dependent manner. We also observed macrophage activation in the skin (data not shown), suggesting that macrophages might be involved in Nair-induced skin inflammation too. In response to the COX2-derived PGE2 signaling, macrophages have been shown to express proinflammatory mediators such as CXCL1, IL-1β, etc. (Castoldi et al., 2020; Wang et al., 2006). It is likely that Nair-induced cPLA2/E-FABP activation promotes COX2/PGE2 production in keratinocytes, leading to proinflammatory mediator expression in macrophages, thus amplifying neutrophil recruitment and skin inflammation.
Skin inflammation induced by chemical depilatory creams is often ascribed to patients with “sensitive skin”. However, what drives skin hypersensitivity remains largely unknown. Our studies demonstrate that E-FABP expression in skin is critical in Nair-triggered, lipid-mediated, neutrophil-infiltrated skin inflammation. Thus, people with high skin E-FABP expression are likely to be at risk to depilatory cream-induced skin inflammation. Like other FABP members, E-FABP expression is modulated by genetic, hormonal, dietary and pharmacological factors (Bu et al., 2011; Furuhashi and Hotamisligil, 2008). Recent studies demonstrated that while consumption of high fat diets (HFD) can lead to E-FABP-mediated skin inflammation (Zhang et al., 2015), the composition of dietary fatty acids is also critical to regulation of E-FABP expression and HFD-induced skin inflammation (Zeng et al., 2018). As U-shape fatty acids (e.g. AA, linoleic acid) have been shown to facilitate E-FABP nucleic translocation, diets rich in these fatty acids may enhance skin sensitivity to depilatory cream-induced side effects.
In summary, we characterized the cellular and molecular features of Nair-induced acute skin inflammation using mouse models. Upon Nair treatment, we observed that cPLA2 in keratinocytes was activated to release AA and other fatty acids, which were transported by E-FABP to the nucleus for PPARβ activation and downstream chemokine/cytokine expression, leading to neutrophil mobilization and acute skin inflammation. These findings have both mechanistic and clinical implications as they not only uncover E-FABP as a molecular sensor for “sensitive skin”, but also suggest that dietary or pharmacological inhibition of E-FABP activity may offer a strategy for prevention/treatment of depilatory cream-induced inflammatory skin diseases.
Materials and Methods
Mice
8–12 week-old E-FABP deficient (E-FABP−/−) and their wild type (WT) littermates (all C57BL/6 background) were bred and housed in the animal facility of the University of Louisville. All animal manipulations were carried out according to the protocol approved by the Institutional Animal Care and Use Committee at the University of Louisville.
Nair-induced skin inflammation mouse models
To characterize Nair-induced skin inflammation, both male and female mice were studied in the experiments. Approximately 15–18 cm2 of mouse dorsal fur was removed under anesthesia using an electric animal clipper. Nair, a popular hair removal lotion, was applied to the clipped area (about 400mg/mice) for 10 mins to fully remove hair. Mice were euthanized on day 1, 3, or 5 after Nair treatment to observe immune cell phenotype and function in different tissues, including skin, peripheral blood, spleen, lymph nodes and bone marrow.
Epidermal and dermal cell separation and analyses
Skin cell preparation followed previous studies (Morris et al., 2019; Zhang et al., 2018). Briefly, clipped dorsal skin from WT or E-FABP−/− mice treated with or without Nair was removed using thumb forceps and scissors. With hairy side down onto a thin Petri dish the removed skin was scraped off all subcutaneous tissue until semi-translucent. The skin was sliced into about 0.5 cm × 1cm strips and put into a 0.25% trypsin digesting solution.
After digestion at 32°C for 1.5 hours, the epidermis was scraped off into PBS with a scalpel blade, and was dispersed with a pipette for at least 10 min. After being filtered through a 40μM cell strainer, epidermal cells were suspended in PBS for future experiments.
To obtain single cells from the dermis, the epidermis-removed skin was further cut into small pieces and digested in an enzymatic solution containing 0.2mg/ml hyaluronidase (SIGMA), 0.02mg/ml DNase I (Roche, Mannheim, Germany) and 0.5 mg/ml collagenase Type II (Worthington Biochemical, Lakewood, NJ) in 5% FBS RPMI-1640 medium for 45 mins at 37°C. The single cell suspension of the dermis was filtered with a 40μM cell strainer for following analyses.
Deletion of E-FABP in keratinocytes by CRISPR-Cas9
Mouse keratinocyte cell line MPEK was purchases from ZenBio (Research Triangle Park, NC) and cultured in CnT-PR medium (ZenBio). To establish stable E-FABP knockout keratinocytes in vitro, MPEK cells were constructed with the CRISPR-Cas9 genome editing technology from ORIGENE (Fabp5 mouse gene knockout kit, Cat# KN505491) according to the manufacturer protocol. Briefly, when MPEK cells reached 50–70% confluence in culture, they were co-transfected with guide RNA targeting to E-FABP genome sequence including GFP/puromycin sequence using Oligofectamin (Life Technologies). After 48 hours upon transfection, cells were split 1:10 and were cultured for another 3 days. The procedure was repeated until 3 weeks post transfection, then GFP+ or GFP− cells were purified with a FACS Aria II sorter. The efficiency of E-FABP deletion in MPEK cell clones were identified with the Western blotting.
Western blotting
Western blotting was used to determine the levels of E-FABP and cPLA2 in CRISPR-transfected MPEK cells. The levels of cPLA2 and E-FABP in the skin of WT and E-FABP−/− mice treated with or without Nair were also determined by the Western blotting as previously described (Zeng et al., 2018). In brief, 20 μg proteins (quantified by BCA assay) in each sample were loaded for SDS-PAGE, transferred to PVDF membrane and blotted with respective antibodies (anti-mFABP5,cat#: AF1476; anti-PLA2G4A, Cat #:5479, anti-PLA2G4E, Cat#:18088–1-AP). β-actin was probed as a loading control. Image Quant TL system was applied to calculate the relative protein quantification.
Analysis of immune cell phenotype by flow cytometric staining
For phenotypic analysis, immune cells in the epidermis, dermis, peripheral blood, spleen, draining lymph nodes and bone marrow were surface-stained with different antibodies, including anti-CD45 (Cat#103116,Biolegend), anti-CD11b(Cat#612800, Biolegend), anti-F4/80 (Cat#, 123120, Biolegend), anti-Gr-1 (Cat# 108442, Biolegend), anti-Ly6G (Cat# 127639, Biolegend), anti-CD3 (Cat# 612771, Biolegend), anti-γδ T (Cat# 118124, Biolegend), anti-CD4 (Cat#100449, Biolegend), anti-CD8 (Cat# 100742, Biolegend), anti-B220 (Cat#, 103232, Biolegend), anti-NK1.1 (Cat#108710, Biolegend), anti-cKit (Cat#105805, Biolegend), anti-FcεRIα (Cat# 134307, Biolegend), anti-CD11c (Cat# 117310, Biolegend), anti-CD207 (Cat# 144204, Biolegend), for 30 mins at 4°C in PBS containing 1% FBS. Cells were washed with 1× PBS once, and acquired using BD FACS Fortessa and BD FACS Aria II Cell Sorter. Dead cells were excluded using the Zombie UV™ Fixable Viability Kit (Cat# 423108, Biolegend). All the data were analyzed with FlowJo 10.1 software.
Confocal analysis
For the analysis of immune cell infiltration, skin tissues obtained from mice treated with or without Nair were snap-frozen with cryo-embedding OCT media (4583, Sakura Finetek USA) at −80°C. Frozen samples cut into 7μm sections were mounted on gelatin-coated slides, and immediately incubated in ice with blocking buffer (5% BSA) for 30 mins. Samples were stained with anti-Ly6G (Cat# 127607, Biolegend), anti-Gr-1 (Cat#,108442 Biolegend), E-FABP (Cat# AF1476, R&D system), cPLA24E (Cat # 18088–1-AP, Thermo) for 30 mins on ice, then stained with 0.2μM DAPI (Cat# 4038S, Cell Signaling Technology) for 15 mins. Slides were analyzed with a Nikon A1 laser scanning confocal microscope, and images captured by Olympus confocal microscopy and analyzed by FV10-ASW Viewer software (Ver 3.1).
Quantitative RT-PCR
For real-time PCR analysis, RNA was extracted from the skin tissues using PureLink™ RNeasy Mini Kit (Cat# 12183025, Invitrogen). Complementary DNA synthesis was performed with a QuantiTect Reverse Transcription Kit (Cat# 205314, Qiagen) as previously described (Hao et al., 2018a; Hao et al., 2018b; Liu et al., 2020). Quantitative PCR was performed with Power SYBR Green PCR Master Mix (Cat# 4368708, Applied biosystems) using StepOnePlus™ Real-Time PCR Systems (Applied Biosystems) to analyze the expression of cPLA2 family members, ALOX5, LTA4H, COX2, PGE2, CXLC1, CXCL2, CXCR2, IL-1β, IL-6, FABP family members, PPARα, PPARβ/δ, PPARγ, CCL17, CCL2, CCR2, ALOX12, TNF-α, TGFβ, IL1F10 (see primer sequences in Supplementary Table 2). Relative mRNA levels were determined using the housekeeping HPRT1 gene as a reference.
Enzyme-linked immunosorbent assay (ELISA)
Skin tissues were homogenized in cell lysis buffer (Cat# 9803S, Cell signaling) containing protease inhibitor (Cat# 78429, Thermo). The homogenates were centrifuged at 12,000 rpm for 10 minutes. Protein levels were measured by Pierce BCA protein assay kit (Cat#23225, Thermo) according to the manufacturer’s protocol. Skin cPLA2 activity was measured by cPLA2 assay kit (No. 765021, Cayman Chemical). Quantitative analysis of LTB4 (No.520111, Cayman Chemical) and CXCL1(Cat# 447507, Biolegend) were measured by ELISA according to the manufacturer’s instructions.
Statistical Analysis
A two-tailed Student t-test was performed for comparison, p <0.05 was considered defined significant difference.
Supplementary Material
Acknowledgements
The authors thank Dr. Rebecca Morris for help with skin cell separation protocol. This study was supported by R01AI137324 (to B. Li) and NIH R01CA180986 (to B. Li). The opinions expressed in this paper are the authors’ own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
Footnotes
Conflicts of Interest
The authors state no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability statement
Datasets related to this article can be found at http://DOI:10.17632/6bmzzf4rcv.2, an open-source online data repository hosted at Mendeley Data.
References
- Armstrong EH, Goswami D, Griffin PR, Noy N, and Ortlund EA (2014). Structural basis for ligand regulation of the fatty acid-binding protein 5, peroxisome proliferator-activated receptor beta/delta (FABP5-PPARbeta/delta) signaling pathway. J Biol Chem 289, 14941–14954. 10.1074/jbc.M113.514646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan D, Falcone J, Kanjiya MP, Park SH, Carbonetti G, Studholme K, et al.(2018). Fatty acid-binding protein 5 controls microsomal prostaglandin E synthase 1 (mPGES-1) induction during inflammation. J Biol Chem 293, 5295–5306. 10.1074/jbc.RA118.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L, Salto LM, De Leon KJ, and De Leon M (2011). Polymorphisms in fatty acid binding protein 5 show association with type 2 diabetes. Diabetes Res Clin Pract 92, 82–91. 10.1016/j.diabres.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JE, and Dennis EA (2009). Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res 50 Suppl, S237–242. 10.1194/jlr.R800033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi A, Monteiro LB, van Teijlingen Bakker N, Sanin DE, Rana N, et al. (2020). Triacylglycerol synthesis enhances macrophage inflammatory function. Nat Commun 11, 4107. 10.1038/s41467-020-17881-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou RC, Kim ND, Sadik CD, Seung E, Lan Y, Byrne MH, et al. (2010). Lipid-cytokine-chemokine cascade drives neutrophil recruitment in a murine model of inflammatory arthritis. Immunity 33, 266–278. 10.1016/j.immuni.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M, and Hotamisligil GS (2008). Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov 7, 489–503. 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Yan F, Zhang Y, Triplett A, Zhang Y, Schultz DA, et al. (2018a). Expression of Adipocyte/Macrophage Fatty Acid-Binding Protein in Tumor-Associated Macrophages Promotes Breast Cancer Progression. Cancer Res 78, 2343–2355. 10.1158/0008-5472.CAN-17-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Zhang Y, Yan X, Yan F, Sun Y, Zeng J, et al. (2018b). Circulating Adipose Fatty Acid Binding Protein Is a New Link Underlying Obesity-Associated Breast/Mammary Tumor Development. Cell Metab 28, 689–705 e685. 10.1016/j.cmet.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS, and Bernlohr DA (2015). Metabolic functions of FABPs--mechanisms and therapeutic implications. Nat Rev Endocrinol 11, 592–605. 10.1038/nrendo.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen MJ, Verheij HM, Slotboom AJ, and Egmond MR (1999). Engineering the disulphide bond patterns of secretory phospholipases A2 into porcine pancreatic isozyme. The effects on folding, stability and enzymatic properties. Eur J Biochem 261, 197–207. 10.1046/j.1432-1327.1999.00256.x. [DOI] [PubMed] [Google Scholar]
- Kannan-Thulasiraman P, Seachrist DD, Mahabeleshwar GH, Jain MK, and Noy N (2010). Fatty acid-binding protein 5 and PPARbeta/delta are critical mediators of epidermal growth factor receptor-induced carcinoma cell growth. J Biol Chem 285, 19106–19115. 10.1074/jbc.M109.099770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiene J, Maiman RE, and DeKlotz CMC (2020). Severe irritant reaction following sequential waxing and use of a chemical depilatory cream in an adolescent. Pediatr Dermatol 37, 190–191. 10.1111/pde.14060. [DOI] [PubMed] [Google Scholar]
- Lee JN, Jee SH, Chan CC, Lo W, Dong CY, and Lin SJ (2008). The effects of depilatory agents as penetration enhancers on human stratum corneum structures. J Invest Dermatol 128, 2240–2247. 10.1038/jid.2008.82. [DOI] [PubMed] [Google Scholar]
- Li B, Hao J, Zeng J, and Sauter ER (2020). SnapShot: FABP Functions. Cell 182, 1066–1066 e1061. 10.1016/j.cell.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Jin R, Hao J, Zeng J, Yin D, Yi Y, et al. (2020). Consumption of the Fish Oil High-Fat Diet Uncouples Obesity and Mammary Tumor Growth through Induction of Reactive Oxygen Species in Protumor Macrophages. Cancer Res 80, 2564–2574. 10.1158/0008-5472.CAN-19-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzano AV, Ortega-Loayza AG, Heath M, Morse D, Genovese G, and Cugno M (2019). Mechanisms of Inflammation in Neutrophil-Mediated Skin Diseases. Front Immunol 10, 1059. 10.3389/fimmu.2019.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E, Kannan-Thulasiraman P, and Noy N (2010). Involvement of Fatty Acid Binding Protein 5 and PPARbeta/delta in Prostate Cancer Cell Growth. PPAR Res 2010. 10.1155/2010/234629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Readio N, Boland K, Johnson K, Lad S, Singh A, et al. (2019). Isolation of Mouse Epidermal Keratinocytes and Their In Vitro Clonogenic Culture. J Vis Exp. 10.3791/58701. [DOI] [PubMed] [Google Scholar]
- Murakami M, Yamamoto K, and Taketomi Y (2018). Phospholipase A2 in skin biology: new insights from gene-manipulated mice and lipidomics. Inflamm Regen 38, 31. 10.1186/s41232-018-0089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle FO, Di Meglio P, Qin JZ, and Nickoloff BJ (2009). Skin immune sentinels in health and disease. Nat Rev Immunol 9, 679–691. 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa E, Owada Y, Ikawa S, Adachi Y, Egawa T, Nemoto K, et al. (2011). Epidermal FABP (FABP5) regulates keratinocyte differentiation by 13(S)-HODE-mediated activation of the NF-kappaB signaling pathway. J Invest Dermatol 131, 604–612. 10.1038/jid.2010.342. [DOI] [PubMed] [Google Scholar]
- Petri B, and Sanz MJ (2018). Neutrophil chemotaxis. Cell Tissue Res 371, 425–436. 10.1007/s00441-017-2776-8. [DOI] [PubMed] [Google Scholar]
- Prigot A, Garnes AL, and Nwagbo U (1962). Evaluation of a chemical depilatory for preoperative preparation of five hundred fifteen surgical patients. Am J Surg 104, 900–906. 10.1016/0002-9610(62)90466-x. [DOI] [PubMed] [Google Scholar]
- Qin W, Baran U, and Wang R (2015). Lymphatic response to depilation-induced inflammation in mouse ear assessed with label-free optical lymphangiography. Lasers Surg Med 47, 669–676. 10.1002/lsm.22387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolph MS, Young TR, Shum BO, Gorgun CZ, Schmitz-Peiffer C, Ramshaw IA,et al. (2006). Regulation of dendritic cell function and T cell priming by the fatty acid-binding protein AP2. J Immunol 177, 7794–7801. 10.4049/jimmunol.177.11.7794. [DOI] [PubMed] [Google Scholar]
- Sadik CD, and Luster AD (2012). Lipid-cytokine-chemokine cascades orchestrate leukocyte recruitment in inflammation. J Leukoc Biol 91, 207–215. 10.1189/jlb.0811402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwingen J, Kaplan M, and Kurschus FC (2020). Review-Current Concepts in Inflammatory Skin Diseases Evolved by Transcriptome Analysis: In-Depth Analysis of Atopic Dermatitis and Psoriasis. Int J Mol Sci 21. 10.3390/ijms21030699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch J, and Corsico B (2008). The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr 28, 73–95. 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- Swann G (2010). The skin is the body’s largest organ. J Vis Commun Med 33, 148–149. 10.3109/17453054.2010.525439. [DOI] [PubMed] [Google Scholar]
- Vlachojannis GJ, Scholz-Pedretti K, Fierlbeck W, Geiger H, Pfeilschifter J, and Kaszkin M (2005). Enhanced expression of group IIA secreted phospholipase A2 by elevated glucose levels in cytokine-stimulated rat mesangial cells and in kidneys of diabetic rats. Clin Nephrol 63, 356–367. 10.5414/cnp63356. [DOI] [PubMed] [Google Scholar]
- Wang D, Fu L, Ning W, Guo L, Sun X, Dey SK, et al. (2014). Peroxisome proliferator-activated receptor delta promotes colonic inflammation and tumor growth. Proc Natl Acad Sci U S A 111, 7084–7089. 10.1073/pnas.1324233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Wang H, Brown J, Daikoku T, Ning W, Shi Q, et al. (2006). CXCL1 induced by prostaglandin E2 promotes angiogenesis in colorectal cancer. J Exp Med 203, 941–951. 10.1084/jem.20052124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Taketomi Y, Isogai Y, Miki Y, Sato H, Masuda S, et al. (2011). Hair follicular expression and function of group X secreted phospholipase A2 in mouse skin. J Biol Chem 286, 11616–11631. 10.1074/jbc.M110.206714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez DA, Lacher RK, Vidyarthi A, and Colegio OR (2017). The role of macrophages in skin homeostasis. Pflugers Arch 469, 455–463. 10.1007/s00424-017-1953-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Zhang Y, Hao J, Sun Y, Liu S, Bernlohr DA, et al. (2018). Stearic Acid Induces CD11c Expression in Proinflammatory Macrophages via Epidermal Fatty Acid Binding Protein. J Immunol 200, 3407–3419. 10.4049/jimmunol.1701416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hao J, Zeng J, Li Q, Rao E, Sun Y, et al. (2018). Epidermal FABP Prevents Chemical-Induced Skin Tumorigenesis by Regulation of TPA-Induced IFN/p53/SOX2 Pathway in Keratinocytes. J Invest Dermatol 138, 1925–1934. 10.1016/j.jid.2018.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, and Li B (2014). E-FABP: regulator of immune function. Oncoscience 1, 398–399. 10.18632/oncoscience.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li Q, Rao E, Sun Y, Grossmann ME, Morris RJ, et al. (2015). Epidermal Fatty Acid binding protein promotes skin inflammation induced by high-fat diet. Immunity 42, 953–964. 10.1016/j.immuni.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun Y, Rao E, Yan F, Li Q, Zhang Y, et al. (2014). Fatty acid-binding protein E-FABP restricts tumor growth by promoting IFN-beta responses in tumor-associated macrophages. Cancer Res 74, 2986–2998. 10.1158/0008-5472.CAN-13-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer JS, Dyckes DF, Bernlohr DA, and Murphy RC (2004). Fatty acid binding proteins stabilize leukotriene A4: competition with arachidonic acid but not other lipoxygenase products. J Lipid Res 45, 2138–2144. 10.1194/jlr.M400240-JLR200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets related to this article can be found at http://DOI:10.17632/6bmzzf4rcv.2, an open-source online data repository hosted at Mendeley Data.


