Abstract
In the development of colon cancer, the intestinal dysbiosis and disruption of barrier function are common manifestations. In the current study, we hypothesized that host factors, e.g., vitamin D receptor deficiency or adenomatous polyposis coli (APC) mutation, contribute to the enhanced dysbiosis and disrupted barrier in the pathogenesis of colorectal cancer (CRC). Using the human CRC database, we found enhanced tumor-invading bacteria and reduced colonic VDR expression, which was correlated with a reduction of Claudin-10 mRNA and protein. In the colon of VDRΔIEC mice, deletion of intestinal epithelial VDR led to lower protein of tight junction protein Claudin-10. Lacking VDR and a reduction of Claudin-10 are associated with an increased number of tumors in the mice without myeloid VDR. Intestinal permeability was significantly increased in the mice with myeloid VDR conditional deletion. Further, mice with conditional colonic APC mutation showed reduced mucus layer, enhanced bacteria in tumors, and loss of Claudin-10. Our data from human samples and colon cancer models provided solid evidence- on the host factor regulation of bacterial translocation and dysfunction on barriers in colonic tumorigenesis. Studies on the host factor regulation of microbiome and barriers could be potentially applied to risk assessment, early detection, and prevention of colon cancer.
Keywords: Adenomatous polyposis coli (APC), Claudin, Barrier function, Inflammation, Colon cancer, Myeloid cells, Microbiome, Mucin, Tight junction, Vitamin D, Vitamin D receptor
Introduction
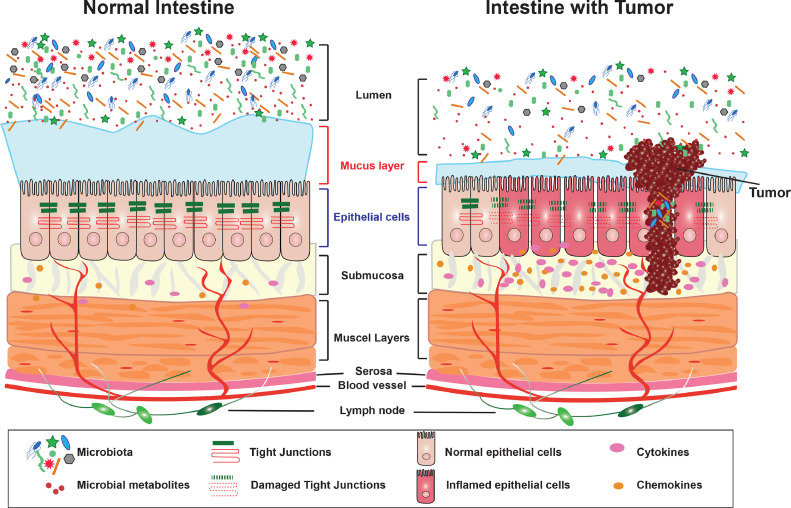
In the healthy colon, several barrier layers, including the balanced microbiota, strong mucus layer, epithelial barriers, and local immunity, protect the intestine against inflammation and cancer. However, the tumor development in the colon is related to the microbial dysbiosis, narrowed mucus layer, and damaged barrier functions. Moreover, these dysfunctional changes could further induce the dislocation of intestinal microbiome from the lumen to the intestinal epithelium and tumors.
Multiple factors (e.g., risk genes, environmental triggers/lifestyle, immunity, and microbiome) contribute to the pathogenesis of colorectal cancer (CRC) [1], [2], [3], [4], [5], [6]. The intestinal dysbiosis and disruption of barrier function are common manifestations of CRC. Tight junction (TJ) structures are essential in intestinal innate immunity and barrier functions. Changes in expression and distribution of TJ proteins, such as Claudin -2, -5, and -8 lead to discontinuous TJs and barrier dysfunction in active Crohn's disease [7,8]. Vitamin D/Vitamin D receptor (VDR) is known to protect against dysbiosis and tumorigenesis [9]. Vitamin D is an environmental factor related to exposure to sunlight, dietary, and lifestyle, whereas VDR acts as a host factor in maintaining healthy innate immunity. Considering the multiple functional roles of VDR in the development of cancer, it is important to dissect the mechanisms by which VDR contributes to barrier function in protecting the host from tumorigenesis.
In the current study, we tested the hypothesis that a host factor, e.g., VDR deficiency or adenomatous polyposis coli (APC) mutation [10], leads to the reduced thickness of mucus layer and enhanced penetration of microbiota into the epithelial cells, thus enhancing colonic tumorigenesis in vivo. The positively correlated status of VDR and Claudins (e.g., Claudin-10) contribute to the altered intestinal permeability in tumorigenesis. In the intestinal epithelial VDR knockout (VDR∆IEC) mice, myeloid cell VDR deletion (VDRlyz) mice, and an APC colonic mutated mouse model, bacterial translocation and inflammation in the genetic deficient context further altered TJ proteins, e.g., Claudin-10, and manipulated the barrier functions in the development of colon cancer. Studies on the host factor regulation of intestinal microbiome and barriers could be potentially applied to risk assessment, early detection, and prevention of colon cancer. These studies will provide an additional avenue to restore microbiome and barrier functions for risk assessment and prevention of CRC.
Materials and methods
Human tissue samples
This study was performed in accordance with approval from the UIC Ethics Committee (Institutional Review Board: 2017-0384). Colorectal tissue samples were obtained from 10 CRC patients with neoplasia and 10 patients without neoplasia (49–74 years old).
Gene expression datasets
For expression analyses, we used microarray data reported in the NCBI Gene Expression Omnibus database [11]. We gathered data by searching the Gene Expression Omnibus [11] (https://www.ncbi.nlm.nih.gov/geo/) for expression profiling studies using colonic biopsy samples from normal controls as well as colon cancer biopsy samples from colon cancer subjects. We randomly selected the GEO database reference GSE 21510 [12], 44 normal controls and 104 colon cancer patients were analyzed. Both were subjected to further analyses.
Animals
VDRΔIEC mice were obtained by crossing the VDRLoxp mice, originally provided by Dr. Geert Carmeliet [13] with villin-cre mice (Jackson Laboratory, 004586), as we previously reported [14], [15], [16]. We further backcrossed this strain with C57BL/6 mice for more than 10 generations after arriving our animal facility [17]. VDRΔlyz mice were obtained by crossing the VDRLoxP mice with Lyz-cre mice (Jackson Laboratory, 004781). VDRΔIEC mice were obtained by crossing the VDRLoxp mice with villin-cre mice (Jackson Laboratory, 004586), as we previously reported [14,16]. Experiments were performed on 2-3 months old mice including male and female.
Intestinal-specific VDR-overexpressing (O-VDR) mice were generated in C57BL/6 mice strain background, as reported in our recent study [18]. VDR expression in O-VDR mice is Cre driven [18]. This O-VDRloxp mouse line is labeled as O-VDRloxp in our gain of function study, distinct from the VDRloxP/loxP mouse made for VDRΔIEC mice. Experiments were performed on 2–3 months old mice, including male and female.
The colon-specific APC knockout (APCΔCEC) mice were generated by breeding APC15loxp mice (Jackson Laboratory, 029275) and CDX2P-NLS Cre mice (Jackson Laboratory, 009350). The 6-8 week-old female and male APC15loxp and APCΔCEC mice were randomly chosen for experimental groups. APC15ΔCEC mice spontaneously developed colonic tumors at >21-week old at the UIC animal facility.
Mice were provided with water ad libitum and maintained in a 12 h dark/light cycle. The animal work was approved by the UIC Office of Animal Care. The animal work was approved by the UIC Office of Animal Care (ACC 15-231,17-218, 20-201, 21-120, and 21-177).
Induction of colon cancer by AOM-DSS in mice
Mice were treated with 10mg/kg of AOM (Sigma-Aldrich, Milwaukee, WI, USA) by intraperitoneal injection as previously described [9,19,20]. After a 7-day recovery period, mice received three cycles of 2% DSS in the drinking water. Tumor counts and measurements were performed in a blinded fashion under a stereo-dissecting microscope (Nikon SMZ1000, Melville, NY, USA). Microscopic analysis was performed for the severity of inflammation and dysplasia on hematoxylin and eosin-stained ‘Swiss rolled’ colons by a gastrointestinal pathologist blinded to treatment conditions.
Western blot analysis and antibodies
Mouse colonic epithelial cells were collected by scraping the tissue from the colon of the mouse, including the proximal and distal regions. The cells were sonicated in lysis buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, pH 8.0, 1% Triton X-100) with 0.2 mM sodium ortho-vanadate, and protease inhibitor cocktail. The protein concentration was measured using the BioRad Reagent (BioRad, Hercules, CA, USA). Equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with primary antibodies. Villin was used as an internal control for intestinal epithelial cells. Beta-actin was also used as a control for equal loading of proteins. The following antibodies were used: anti-Claudin-10 (Invitrogen, 38-8400, Carlsbad, CA, USA), anti-Claudin-7 (Invitrogen, 34-9100, Carlsbad, CA, USA), anti-Claudin-3 (Invitrogen, 34-1700, Carlsbad, CA, USA), anti-Claudin-4 (Invitrogen, 36-4800, Carlsbad, CA, USA), anti-Claudin-7 (Invitrogen, 34-9100, Carlsbad, CA, USA), anti-villin (Santa Cruz Biotechnology, SC-58897, Dallas, TX, USA) or anti-β-actin (Sigma-Aldrich, A5316, St. Louis, MO, USA) antibodies and were visualized by ECL (Thermo Fisher Scientific, 32106, Waltham, MA, USA). Membranes that were probed with more than one antibody were stripped before re-probing.
Intestinal permeability
Fluorescein Dextran (Molecular weight 3 kDa, diluted in HBSS) was gavaged (50 mg/g mouse). Four hours later, mouse blood samples were collected for fluorescence intensity measurement, as previously reported [21].
Immunofluorescence
Colonic tissues were freshly isolated and embedded in paraffin wax after fixation with 10% neutral buffered formalin. Immunofluorescence was performed on paraffin-embedded sections (4 μm), after preparation of the slides as described previously [22] followed by incubation for 1 hour in blocking solution (2% bovine serum albumin, 1% goat serum in HBSS) to reduce nonspecific background, as we previously reported [23]. The tissue samples were incubated overnight with primary antibodies at 4°C. The anti-Claudin-10 antibody was used. Slides were washed 3 times for 5 minutes each at room temperature in wash buffer. Samples were then incubated with secondary antibodies (goat anti-rabbit, Alexa Fluor 488, Molecular Probes, CA; 1:200) for 1 hour at room temperature. Tissues were mounted with SlowFade Antifade Kit (Life technologies, s2828, Grand Island, NY, USA), followed by a coverslip, and the edges were sealed to prevent drying. Specimens were examined with a Zeiss laser scanning microscope LSM 710 (Carl Zeiss Inc., Oberkochen, Germany).
Immunohistochemistry (IHC)
After preparation of the slides, antigen retrieval was achieved by incubating the slides for 15 min in hot preheated sodium citrate (pH 6.0) buffer and 30 min of cooling at room temperature. Endogenous peroxidases were quenched by incubating the slides in 3% hydrogen peroxide for 10 min, followed by three rinses with HBSS, and incubation for 1 hour in 3% BSA + 1% goat serum in HBSS to reduce nonspecific background. Primary antibodies VDR (1:300) were applied for overnight in a cold room. The secondary antibody (1:100, Jackson ImmunoResearch Laboratories, Cat.No.115-065-174, West Grove, PA, USA). The slides were incubated with vectastain ABC reagent (Vector Laboratories, Cat.No. PK-6100, Burlingame, CA 94010, USA) for 1 hour. After washing with HBSS, color development was achieved by applying a peroxidase substrate kit (Vector Laboratories, Cat.No. SK-4800, Burlingame, CA, USA). The duration of peroxidase substrate incubation was determined through pilot experiments and was then held constant for all of the slides. After washing in distilled water, the sections were counterstained with haematoxylin (Leica, Cat.No.3801570, Wetzlar, Germany), dehydrated through ethanol and xylene, and cover‐slipped using a permount (Fisher Scientific, Cat.No. SP15-100, Waltham, MA, USA).
Fluorescence In Situ Hybridization
FISH was performed using antisense single-stranded DNA probes targeting the bacterial 16S ribosomal RNA [24]. The EUB338 (5’-GCTGCCTCCCGTAGGAGT-3’) conjugated to Alexa Fluor488 probe was used for universal bacteria [25]. FISH was performed on paraffin sections of mucosal biopsies fixed with Carnoy's fixative or with 10% formalin solution. Before performing FISH assay, 5 μm tissue sections were baked overnight at 55°C. Tissue sections were deparaffinized in xylene, dehydrated with 100% ethanol, air dried, incubated in 0.2 mol/L HCl for 20 minutes, and heated in 1 mmol/L sodium thiocyanate at 80°C for 10 minutes. Samples were pepsin-digested (4% pepsin in 0.01 N HCl) for 20 minutes at 37°C), slides were washed in wash buffer (0.3 mol/L NaCl, 0.03 mol/L sodium citrate, pH 7, and 0.1% SDS), fixed in 10% buffered formalin for 15 minutes, washed and dried, and hybridized with the probes at 5 ng/μL concentration each for 5 minutes at 96°C in hybridization buffer (0.9 mol/L NaCl, 30% formamide, 20 mmol/L Tris-HCl, pH 7.4, and 0.01% SDS) and incubated at 37°C overnight. Slides were washed 4 times for 5 minutes each at 45°C in wash buffer. For visualization of the epithelial cell nuclei, the slides were counterstained with 4′, 6′-diamidino-2-phenylindole/antifade solution. Slides were examined with a Zeiss laser scanning microscope LSM 710 (Carl Zeiss, Inc). Fluorescence staining intensity was determined by using ImageJ software.
Alcian Blue /PAS staining
Alcian blue-PAS staining was performed on paraffin sections of colon tissue fixed with Carnoy's fixative. Before performing Alcian blue-PAS assay, 5 μm colon tissue sections were baked overnight at 55°C. Colon tissue sections were deparaffinized in xylene, dehydrated with 100% ethanol. The air-dry slides were stained with Alcian blue solution (1 g of Alcian blue, pH 2.5, 3 mL/L of acetic acid, and 97 mL of distilled water) for 30 min for goblet cells. This was followed by rinsing in tap water for 10 min, oxidizing in periodic acid (5 g/L) for 5 min, rinsing in tap water for 10 min, and staining in Schiff's reagent as a counter stain (Electron Microscopy Sciences, Cat. NO. 26853-01, Hatfield, PA, USA) for 10 min. After washing in distilled water for 10 minutes, dehydrated through ethanol and xylene, and cover‐slipped using a permount (Fisher Scientific, Cat.No. SP15-100, Waltham, MA, USA).
Goblet cell- number-
Alcian blue-PAS-stained slides were used for counting goblet cells in the colon. Acidic (blue), neutral (pink), and mixed (purple) goblet cells were counted in each crypt. Goblet cell number was calculated by counting the average number of goblet cells in 3 crypts as one point. Three points were randomly selected for each mouse. Mean values for the number of goblet cells were calculated and analyzed using Welch's t test.
Multiplex ELISA assay
Mouse blood samples were collected by cardiac puncture and placed in tubes containing EDTA (10 mg/mL). Mouse cytokines were measured using a Cytokine & Chemokine Convenience 26-Plex Mouse ProcartaPlex™ Panel 1 (Invitrogen, EPXR260-26088-90, Carlsbad, CA, USA) according to the manufacturer's instructions. Briefly, beads of defined spectral properties were conjugated to protein-specific capture antibodies and added along with samples (including standards of known protein concentration, control samples, and test samples) into the wells of a filter-bottom microplate, where proteins bound to the capture antibodies over the course of a 2-hour incubation. After washing the beads, protein-specific biotinylated detector antibodies were added and incubated with the beads for 1 hour. After removal of excess biotinylated detector antibodies, the streptavidin-conjugated fluorescent protein R-phycoerythrin was added and allowed to incubate for 30 minutes. After washing to remove unbound streptavidin–R-phycoerythrin, the beads were analyzed with the Luminex detection system (Bio-rad, Bio-Plex 200 Systems, Hercules, CA).
Statistical analysis
All data were expressed as the mean ± SD. All statistical tests were 2-sided. All p-values < 0.05 were considered statistically significant. After the Shapiro-Wilk test confirmed that the data are normally distributed, the differences between samples were analyzed using unpaired Student's t-test for two groups and one-way ANOVA for more than two groups as appropriate, respectively. The p-values in ANOVA analysis were adjusted using the Tukey method to ensure accurate results. Pearson correlation analyses and scatter plots were conducted for staining intensity changes between VDR protein and Claudin-5, using SAS version 9.4 (SAS Institute, Inc., Cary, NC, USA) and R software (R Core Team (2021), R Foundation for Statistical Computing, Vienna, Austria). Other statistical analyses were performed using GraphPad Prism 6 (GraphPad, Inc., San Diego, CA., USA).
Results
Enhanced tumor-invading bacteria, reduced VDR, and decreased Claudin-10 in tumor samples from human CRC
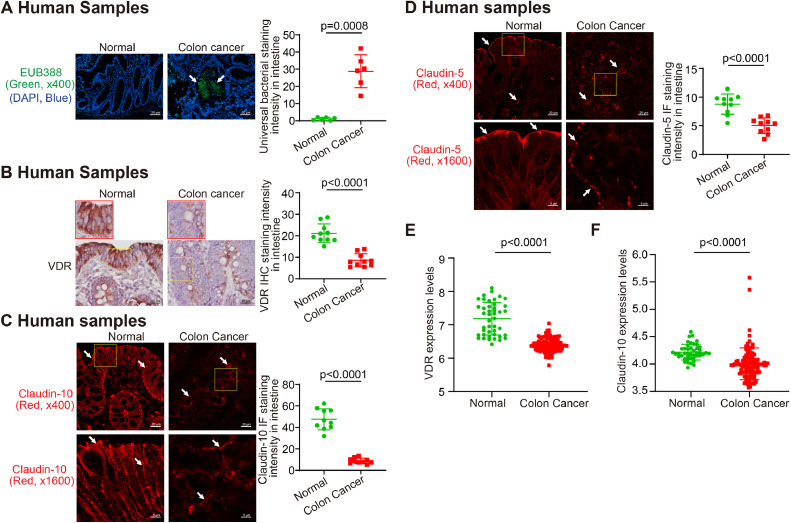
In the human tumor samples, we identified enhanced bacteria by fluorescent in situ hybridization (Fig. 1A). We speculated that enhanced bacteria in tumors were associated with the status of VDR. We then tested intestinal VDR by IHC staining in normal colon controls and patients with colon cancer. Compared with the normal intestine, the intestine of patients had significantly lower VDR expression (n=10; Welch's t test) (Fig. 1B). At the mRNA level, we found reduced VDR expression in patients with colon cancer (GEO database GSE21510, Normal, n=44; colon cancer, n=104) (Fig. 1E). We believe the bacterial translocation to the tumors is related to the disrupted intestinal barrier functions through tight junctions (TJs). By IF staining, we showed the reduced TJ protein Claudin-10 and -5 in patients’ samples, compared to the normal colon (Fig. 1C and 1D). Compared with normal intestines, colon cancer patients’ intestines had significantly lower Claudin-10 mRNA expression (GEO database GSE21510, data were expressed as mean ± SD; Normal, n=44; colon cancer, n=104) colon (Fig. 1F). These data indicate the reduced VDR in the colon of CRC patients is associated with the reduced intestinal tight junctions for barrier and enhanced bacterial translocation to tumors.
Fig. 1.
Enhanced bacterial translocation, reduced VDR, and decreased Claudin-10 expression in CRC patients. (A) Bacteria in tumors from CRC patient by Fluorescence in situ hybridization [39]. (Images are representative of experiments in triplicate; normal, n = 6; colon cancer, n = 6; Welch's t test). (B) Significantly lowered VDR expression in the colon of CRC patients, compared with the normal intestines (Images are representative of three experiments; normal, n = 10; colon cancer, n = 10; Welch's t test). (C) Significantly reduced Claudin-10 in the colon of CRC patients, compared with the normal intestines. (Images are representative of experiments in triplicate; normal, n = 10; colon cancer, n = 10; Welch's t test). (D) Significantly reduced Claudin-5 in the colon of CRC patients, compared with the normal intestines. (Images are representative of three experiments; normal, n = 10; colon cancer, n = 10; Welch's t test). (E) Reduced VDR expression in patients with CRC (GEO database GSE21510, data were expressed as mean ± SD; normal, n = 44; colon cancer, n = 104; Welch's t test). (F) Reduced Claudin-10 expression in patients with CRC (GEO database GSE21510, data were expressed as mean ± SD; normal, n = 44; colon cancer, n = 104; Welch's t test). All p values are shown in the figure.
Intestinal VDR-specific knockout led to reduced mucus thickness in colonic tumors
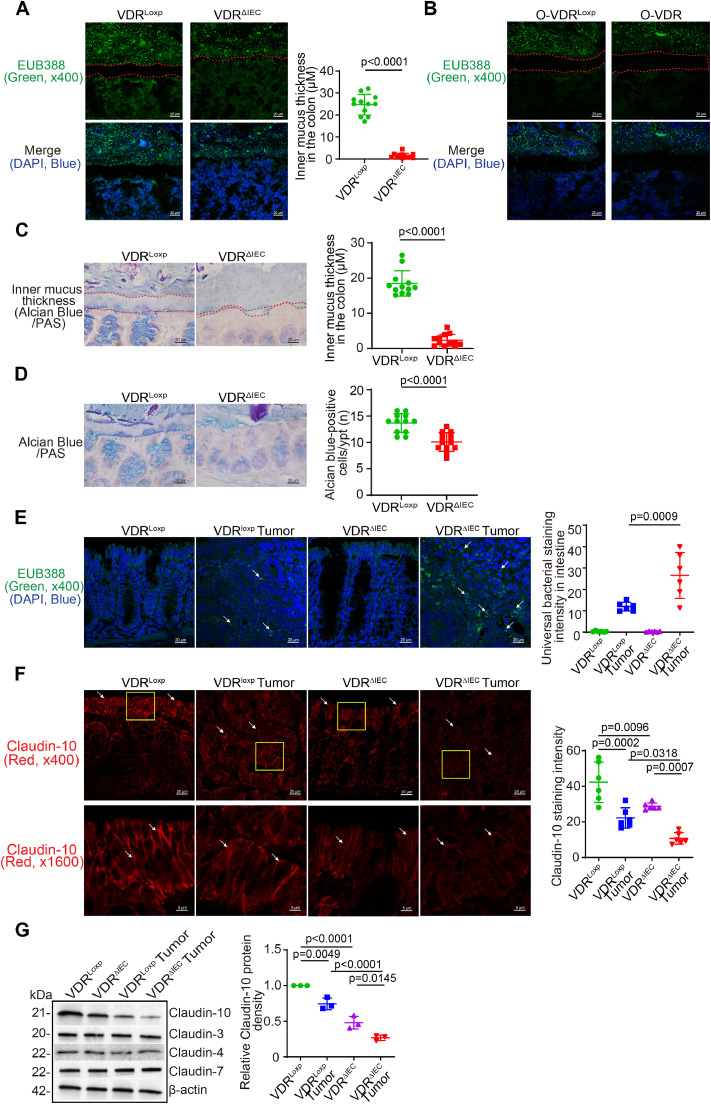
To investigate the mechanism of host factor regulation of microbiome and intestinal barrier function, we then examined the animal models with intestinal VDR deletion in the intestinal epithelial cells (VDRΔIEC) or myeloid cells (VDRΔLyz) [26], [27], [28]. The VDRΔIEC mice were known to be susceptible to the chemical-induced tumorigenesis in the colon, based on our recent reports [9,26]. Here, we found that mucus thickness declined in the colon of VDRΔIEC mice, compared to the VDRLoxp mice, without any treatment (Fig. 2A). In addition to the narrowed mucus layer, there was dislocation of intestinal microbiota from the lumen to the gut epitheliums and tumor tissue. However, in the VDR overexpressed mice and their loxp controls, we did not find the changes in the mucus thickness or bacterial penetrations (Fig. 2B). We further examine the mucus layer by Alcian Blue/PAS staining in the colon of VDRLoxp and VDRΔIEC mice. The narrowed mucus layer was significant in the colon of VDRΔIEC mice (Fig. 2C). Goblet cells were significantly reduced on the the colon of VDRΔIEC mice, compaed to the VDRLoxp mice (Fig. 2D). Bacteria were increased in the tumor tissue of VDRΔIEC mice, compared to the tumor tissue of VDRLoxp mice (Fig. 2E). Moreover, TJ Claudin-10 distribution in the colonic epithelial cells was decreased in VDRΔIEC mice compared to VDRloxp mice at basal level. Claudin-10 was further decreased in tumors of VDRΔIEC mice, compared to those of VDRLoxp mice by immunofluorescence staining (Fig. 2F). The reduced Claudin-10 protein in VDRΔIEC mice was further confirmed by Western blots (Fig. 2G), whereas TJ Claudin-3, 4, and 7 did not change with the VDR status. Please note that the level of colonic Claudin-10 protein was significantly reduced in the VDRΔIEC mice compared to VDRloxp mice without any treatment. It is known that tumorigenesis is related to the microbial dysbiosis, narrowed mucus layer, and damaged barrier functions. These data suggest that the host regulation of barrier dysfunction was preexisting pre-cancer and got worse in the tumor development.
Fig. 2.
Intestinal epithelial VDR-specific knockout reduced mucus thickness, enhanced bacterial penetration, and reduced Claudin-10 expression in the VDRΔIEC mice. (A) Colonic mucus thickness declined in the VDRΔIEC mice, compared to the VDRLoxp mice. Representative images of FISH staining of the colon of VDRLoxp and VDRΔIEC mice, using the universal bacterial DNA probe EUB338-Alexa Fluor 488 [40], and nuclear staining DAPI (blue). The space between apical membranes of the epithelial cells and gut microbiome was indicated by dashed red lines. Images are from a single experiment and are representative of 4 mice per group. Three points were randomly selected for each mouse (data are expressed as mean ± SD. n = 12, Welch's t test). (B) Colonic mucus thickness was not changed in the O-VDR mice, compared to O-VDRLoxp mice. Images are from a single experiment and are representative of 4 mice per group. (C) Mucus thickness declined in the colon of VDRΔIEC mice compared to VDRLoxp mice. Representative images of Alcian Blue/PAS staining of the colon of VDRLoxp and VDRΔIEC mice. The space between apical membranes of the epithelial cells and gut microbiome was indicated by dashed red lines. Images are from a single experiment and are representative of 4 mice per group. Three points were randomly selected for each mouse (data are expressed as mean ± SD. n = 12, Welch's t test). (D) Reduced Goblet cells in the colon of VDRΔIEC mice, compared to VDRLoxp mice. Images are from a single experiment and are representative of 4 mice per group. Three points were randomly selected for each mouse (data are expressed as mean ± SD. n = 12, Welch's t test). (E) Enhanced bacteria in the tumors of the VDRΔIEC mice, as indicated in FISH staining. Images are from a single experiment and are representative of 6 mice per group (data are expressed as mean ± SD. n= 6, one-way ANOVA test) [41]. Claudin-10 was decreased in VDRΔIEC mice, compared to the VDRLoxp mice at the basal level. Claudin-10 was further decreased in tumors of the VDRΔIEC mice by immunofluorescence staining. Images are from a single experiment and are representative of 6 mice per group (data are expressed as mean ± SD. n=6, one-way ANOVA test). (G) Claudin-10 was decreased in VDRΔIEC mice compared to VDRloxp mice at the basal level and also in tumors by Western blots. (data are expressed as mean ± SD. n=3, one-way ANOVA test). All p values are shown in the figure.
Mice with myeloid VDR deletion developed more and larger tumors, compared to the VDRLoxp mice
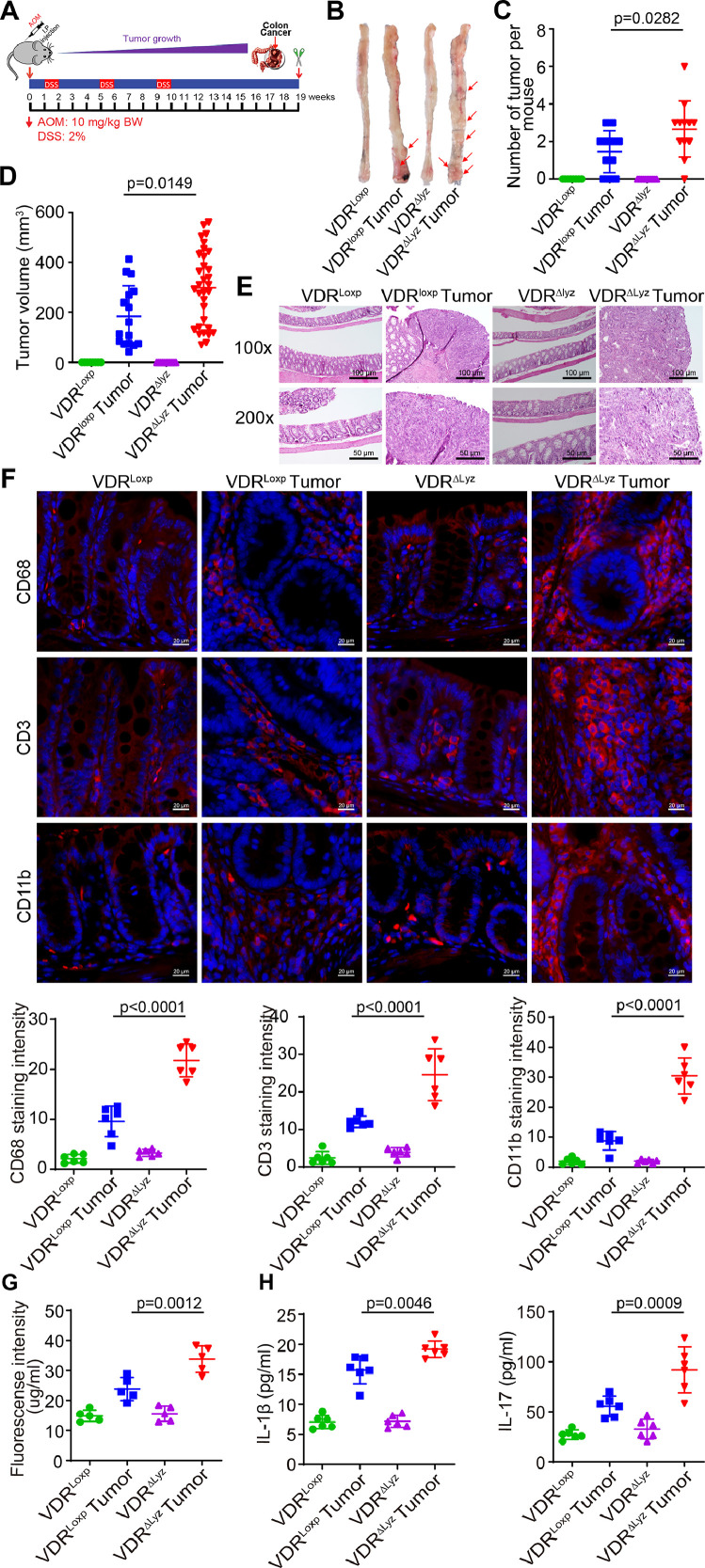
We then studied the VDRΔLyz mice with myeloid VDR deletion to understand the role of immunity in the interplays among the host factor, microbial dysbiosis, and barrier functions. VDRΔLyz mice and their VDRLoxp controls were treated with the AOM/DSS to induce colon cancer (Fig. 3A). Representative colons from different groups and colonic tumors in situ were shown in Fig. 3B. Tumor numbers and volumes in the VDRΔLyz mice were significantly greater than those in the VDRLoxp-AOM/DSS group (Fig. 3C and 3D). Representative H&E staining of “Swiss rolls” of representative colons from the indicated groups (Fig. 3E). Levels of CD68, CD3, and CD11b for immune cells (monocyte/macrophages, granulocytes, and natural killer cells) were increased significantly in the tumor tissues of VDRΔLyz mice, compared to those in VDRLoxp mice (Fig. 3F). Permeability of the VDRΔLyz mice was significantly increased measured by fluorescence intensity in serum (Fig. 3G). Serum cytokines, such as IL-1β and IL-17, were also significantly increased in the AOM-DSS-treated VDRΔLyz mice (Fig. 3H), suggesting increased inflammation in the mice.
Fig. 3.
VDRΔLyz mice with Myeloid VDR deletion developed more and larger tumors, compared with the VDRloxp mice. (A) A schematic overview of the AOM/DSS-induced colon cancer model. AOM (10 mg/kg) was injected on day 0. Starting at Day 7, 2% DSS solution was administered to mice in drinking water with a duration of 7 days. Then, the mice were given DSS-free drinking water for 3 weeks. After that, an additional two cycles of DSS drinking water were administered prior to scarification at Week 19. (B) Colonic tumors in situ. Representative colons from different groups. Tumors were indicated with red arrows. (C) Tumor numbers in AOM-DSS induced colon cancer model: VDRLoxp and VDRΔLyz mice (data are expressed as mean ± SD. n = 8, 11, 10, and 12 for the VDRLoxp control, VDRLoxp tumor, VDRΔLyz control, and VDRΔLyz tumor groups, one-way ANOVA test). (D) The tumor volume in AOM-DSS induced colon cancer model: VDR+/+ and VDR−/− mice (data are expressed as mean ± SD. n = 8, 11, 10, and 12 for the VDRLoxp control, VDRLoxp tumor, VDRΔLyz control, and VDRΔLyz tumor groups, respectively; one-way ANOVA test). (E) Representative H&E staining of “Swiss rolls” of representative colons from the indicated groups. Images are from a single experiment and are representative of 6 mice per group. (F) Levels of CD68, CD3, and CD11b were increased significantly in the tumor tissues of VDRΔLyz mice, compared to the tumor tissues of VDRLoxp mice by IF staining. Images are from a single experiment and are representative of 6 mice per group (data are expressed as mean ± SD. n = 6, one-way ANOVA test). All p values are shown in the figure. (G)Intestine permeability increased in the VDRΔLyz mice with tumors. Experiments were assayed in triplicates (data are expressed as mean ± SD; n = 5, one-way ANOVA test). (H) Serum cytokines IL-1β and IL-17 were significantly increased in the AOM-DSS-treated VDRΔLyz mice. Experiments were assayed in triplicates (data are expressed as mean ± SD. n= 6, one-way ANOVA test). All p values are shown in the figure.
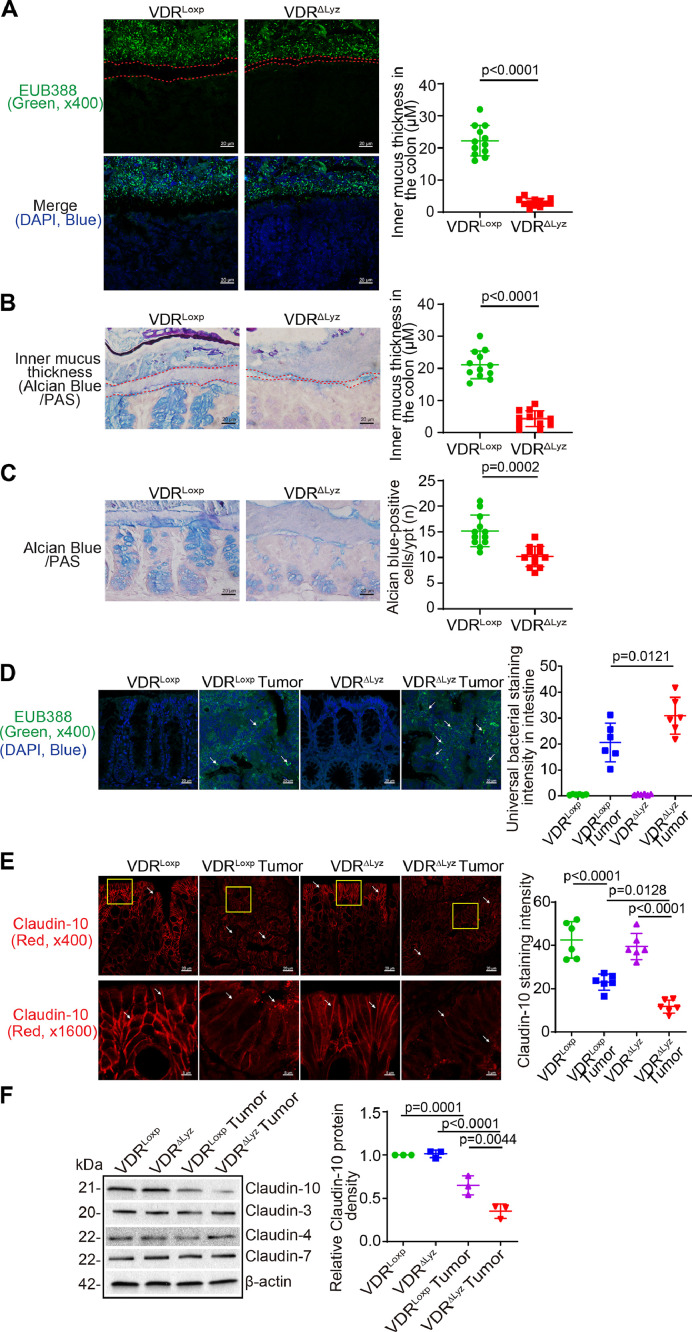
Myeloid cells VDR-specific knockout induced mucus thickness decline, bacterial translocation, and reduced Claudin-10 in the colon
As expected, colonic mucus thickness declined in the VDRΔLyz mice colon, compared to VDRLoxp mice (Fig. 4A). The status of mucus layer was further confirmed by Alcian Blue/PAS staining. The narrowed mucus layer was significant in the colon of VDRΔLyz mice (Fig. 4B). Goblet cells were significantly reduced on the the colon of VDRΔLyz mice, compared to the VDRLoxp mice (Fig. 4C). Bacteria were increased in the tumors of VDRΔLyz mice, compared to those of VDRLoxp mice (Fig. 4D). TJ Claudin-10 was significantly decreased in VDRΔLyz mice with tumors (Fig. 4E). The reduced Claudin-10 at the protein level in VDRΔIEC mice was further confirmed by Western blots, whereas TJ Claudins 3, 4, and 7 did not change with the VDR status in the myeloid cells (Fig. 4F).
Fig. 4.
Myeloid cells VDR-specific knockout led to reduced mucus thickness, bacterial dislocation, and reduced Claudin-10 expression tumor tissues of VDRΔLyz mice. (A) Mucus thickness decline in the colon of the VDRΔLyz mice in FISH staining. Images are from a single experiment and are representative of 4 mice per group. Three points were randomly selected for each mouse. (Data are expressed as mean ± SD. n = 12, Welch's t test). (B) Mucus thickness decline in the VDRΔLyz mice colon compared to VDRLoxp mice. Representative images of Alcian Blue/PAS staining of the colon of VDRLoxp and VDRΔLyz mice. The space between apical membranes of the epithelial cells and gut microbiome is indicated by a dashed red line. Images are from a single experiment and are representative of 4 mice per group. Three points were randomly selected for each mouse (data are expressed as mean ± SD. n = 12, Welch's t test). (C) Goblet cells number decreased in the VDRΔLyz mice colon compared to VDRLoxp mice. Images are from a single experiment and are representative of 4 mice per group. Three points were randomly selected for each mouse (data are expressed as mean ± SD. n = 12, Welch's t test). (D) Bacteria were increased in the tumors of VDRΔLyz mice as indicated. Images are from a single experiment and are representative of 6 mice per group (data are expressed as mean ± SD. n = 6, one-way ANOVA test). (E) Claudin-10 was decreased in the tumors of VDRΔLyz mice, compared to VDRLoxp mice by immunofluorescence staining. Images are from a single experiment and are representative of 6 mice per group (data are expressed as mean ± SD. n = 6, one-way ANOVA test) [41]. Claudin-10 was decreased in tumor tissue of VDRΔLyz mice by Western blots (data are expressed as mean ± SD. n = 3, one-way ANOVA test). Claudin-3, Claudin-4 and Claudin-7 were unchanged in the colon of VDRΔLyz mice compared to VDRLoxp mice treated with AOM/DSS. All p values are shown in the figure.
Reduced mucus thickness, enhanced bacteria, and decreased Claudin-10 in tumors spontaneously developed in the APC15ΔCEC mice.
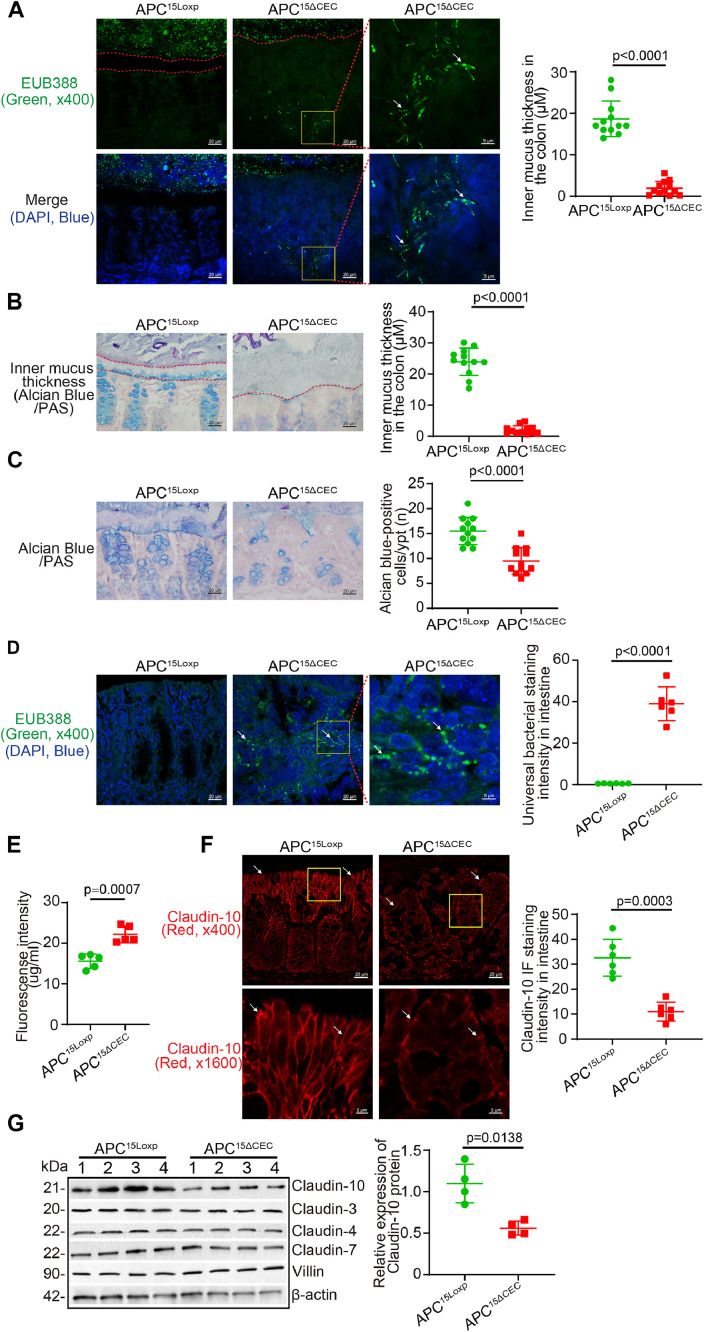
To study the host factor regulation of altered mucus layer and bacterial translocation to tumors we then use another colon cancer model: APC15ΔCEC CRC model [29,30]. Colonic APC knockout (APC15ΔCEC) mice were generated by breeding APC15Loxp mice and CDX2P-NLS Cre mice. APC15ΔCEC mice spontaneously developed colonic tumors at >21- week old at the UIC animal facility. We investigated the changes of barrier and microbiome in the spontaneous tumor formation process in the APC15ΔCEC mice. Colonic mucus thickness was reduced in the APC115ΔCEC mice, compared to that in the APC15Loxp mice (Fig. 5A). Bacteria were signifcantly increased in tumors of APC115ΔCEC mice (Fig. 5B). Claudin-10 was decreased in APC15ΔCEC mice, compared to APC15Loxp mice by IF (Fig. 5C) and Western blots (Fig. 5D). However, Claudins- 3, 4 and 7 did not change in the colon of APC15ΔCEC mice compared to APC15Loxp mice (Fig. 5D). These data suggest that the genetic factor of APC mutation in the colon could lead to altered microbiome distribution and disrupted barriers.
Fig. 6.
The working model for the host factor in regulating protective microbiota, mucus layer, and epithelial barriers. In the healthy intestine, the balanced microbiota, strong mucus layer and protective epithelial barriers keep the intestine from inflammation and tumorigenesis. However, in the susceptible host (e.g., VDR deficiency, APC mutation), there is preexisting barrier dysfunction and dysbiosis. The tumor development worsens the microbial dysbiosis, narrows mucus layer, and damages barrier functions by modifying tight junctions (TJs). Moreover, these dysfunctional changes could further induce the dislocation of microbiota from the lumen to the intestinal epithelium and tumors.
Fig. 5.
Reduced mucus thickness and decreased Claudin-10 expression in the colon of APC15ΔCEC mice. (A) Mucus thickness in the colon were reduced in the APC15ΔCEC mice, compared to APC15Loxp mice. Bacteria were enhanced in the tumors of APC15ΔCEC mice. Images are from a single experiment and are representative of 4 mice per group. Three points were randomly selected for measurement per mouse (data are expressed as mean ± SD. n = 12, Welch's t test). (B) Mucus thickness decline in the APC15ΔCEC mice colon by Alcian Blue/PAS staining. Representative images of Alcian Blue/PAS staining of the colon of APC15Loxp and APC15ΔCEC mice. The apical membranes of the epithelial cells and gut microbiome are indicated by dashed red lines. Images are from a single experiment and are representative of 4 mice per group. Three points were randomly selected for each mouse (data are expressed as mean ± SD. n = 12, Welch's t test). (C) The numbers of goblet cells were reduced in the APC15ΔCEC mice colon compared to APC15Loxp mice. Images are from a single experiment and are representative of 4 mice per group. Three points were randomly selected for measurement per mouse (data are expressed as mean ± SD. n = 12, Welch's t test). All p values are shown in the figure. (D) Bacteria were increased in 10% formalin-fixed tumor tissues of APC15ΔCEC mice. Images are from a single experiment and are representative of 6 mice per group (data are expressed as mean ± SD. n = 6, one-way ANOVA test). (E) Intestine permeability increased in the APC15ΔCEC mice compared to APC15Loxp mice (data are expressed as mean ± SD. n = 5, Welch's t test). (F) Claudin-10 was decreased in APC15ΔCEC mice compared to APC15Loxp mice. Images are from a single experiment and are representative of 6 mice per group (data are expressed as mean ± SD. n = 6, one-way ANOVA test). (G) Claudin-10 was decreased in in APC15ΔCEC mice, compared to the APC15Loxp mice by Western blots (data are expressed as mean ± SD. n = 4, Welch's t test). Claudin-3, Claudin-4 and Claudin-7 were unchanged in the colon of APC15ΔCEC mice compared to the APC15Loxp mice. All p values are shown in the figure.
Discussion
In the current study, we reported that a host factor, conditional VDR deficiency or colonic APC mutation, led to the reduced thickness of mucus layer and enhanced penetration of microbiota into the epithelial cells, thus enhancing colonic tumorigenesis. In the healthy colon, the microbiota, mucus layer, epithelial tight junctions, and local immune cells keep the host from inflammation and cancer. However, the microbial dysbiosis, narrowed mucus layer, and damaged barrier functions by modifying regulators enhance the tumorigenesis. Moreover, these dysfunctional changes further induce the dislocation of microbiota from the lumen to the epitheliums and tumor tissue. Previous studies have demonstrated that conditional VDR deletion in intestinal epithelial cells led to dysbiosis [28,31,32]. Here, our data from human samples and various cancer models provided solid evidences on the host factor regulation of local dysbiosis, bacterial penetration to the epithelial cells, and dysfunction on barriers in the colon pre-cancer or in the development of cancer.
TJ protein Claudins have a big family with at least 27 members in mouse and human [33]. A previous study showed expression profiles of some CLDN genes and prognostic value in CRC samples classified by their molecular subtypes. It suggested that CRC heterogeneity must be taken into account when assessing CLDN potential value as prognostic markers or therapeutic targets [34]. We explored the human datasets and reported that the mRNA expression of Claudin -1, -2, -12, -19 were increased in colon cancer patients; the mRNA expression of Claudin -5, -8, -15, -17, -20, -23 were decreased in patients with colon cancer. Claudin -3, -4, -6, -7, -9, -10, -11, -14, -16, -18 did not show distinct changes at the mRNA level between patients with colon cancer and normal controls. However, these observations are only at the mRNA levels. The biological functions of junction proteins rely on their protein distributions and interactions with other junctional proteins. The level of mRNA could not completely reflect the biological and pathological functions of junction proteins in the development of cancer. Currently, there are limited cellular studies on the individual Claudin protein and its alteration by the upstream regulator from the host. Our data on the Claudin-10 distribution and protein level in the human CRC tumors and chemical-induced and genetic colon cancer models start to fill the gap in the field.
VDR was a transcriptional factor regulating the genes of several TJ Claudins [35,36]. The positively correlated status of VDR and Claudins (e.g., Claudin-5, and 15) contributes to the altered intestinal permeability in chronic inflammation and tumorigenesis [21,26,37]. We have demonstrated that conditional VDR deletion in a tissue specific manner led to dysbiosis [27,28,31,32]. In the intestinal epithelial VDR knockout VDR∆IEC mice and VDR∆lyz mice, dysbiosis and inflammatory cytokines in the VDR deficient context altered TJs and manipulated the barrier functions. Our current studies showed narrowed mucus layer and bacterial translocation, which weakens the multiple layers of the intestinal barriers in absence of VDR. Moreover, the narrow mucus layer and bacterial translocation in the APC15∆CEC mice validate the role of host factor in a genetic cancer model. These studies provide strong rationale and an additional avenue to restore microbiome and barrier functions for risk assessment and prevention of CRC in the future [38].
In the current study, we have demonstrated the importance of myeloid VDR in barrier function and protecting the host against tumorigenesis. Without any treatment, VDRΔLyz mice already showed less mucus layer and bacterial penetration to the epithelial cells, compared to the VDRLoxp mice. Tumor numbers and volumes were significantly greater in the AOM/DSS treated VDRΔLyz mice than those in the VDRLoxp with the AOM/DSS. VDR deficiency in the immune cells leads to less epithelial Claudin 10, more infiltration, increased inflammation, suggesting the cross talks between the local immune cells and epithelial cells in the colon. VDRΔLyz mice are known with reduced beneficial bacterial profile and altered functions of metabolites based on our previous studies [27,28]. All together, these results provide us with insights into the innate immunity in the interplays among the host factor, microbial dysbiosis, and intestinal barrier functions. It is known that balanced microbiota, strong mucus layer, epithelial barriers, and local immunity together protect the intestine against inflammation and cancer. Further studies are needed to understand of the mechanisms and impacts of the dynamic interactions of microbiota, immune cells, and epithelial cells in health and cancer.
In summary, our studies from human samples and various colon cancer models have provided evidence on the host factor regulation of dysbiosis and barrier dysfunction in the pre-cancer or in the development of cancer. For the long-run, insights into the host - microbiome interactions and barriers could be potentially applied to risk assessment, early detection, and prevention of colon cancer and other cancers, as well.
Author contributions
YZ: acquisition, analysis, and interpretation of data; drafting the manuscript; and statistical analysis. JZ: assistance with animal models, Western blots, and manuscript and figure drafting. YX: statistical analysis, and manuscript drafting. JS: study concept and design, analysis and interpretation of data, writing the manuscript for important intellectual content, obtained funding, and study supervision.
Acknowledgments
We would like to thank Jason S. Xia for proofreading.
Footnotes
Author Contributions: YZ: acquisition, analysis, and interpretation of data; drafting the figures and manuscript; and statistical analysis. JZ: assistance with animal models, western blots, and figure drafting. YX: statistical analysis, and manuscript drafting. JS: study concept and design, analysis and interpretation of data, writing the manuscript for important intellectual content, obtained funding, and study supervision.
Funding: This research was funded by the UIC Cancer Center, the NIDDK/National Institutes of Health grant R01 DK105118 and R01DK114126, VA Merit Award VA 1 I01 BX004824-01, and DOD BC160450P1 to Jun Sun. The study sponsors played no role in the study design, data collection, analysis, and interpretation of data. The contents do not represent the views of the United States Department of Veterans Affairs or the United States Government.
Conflicts of Interest: The authors declare no conflict of interest. The funders played no role in the study design, the collection, analyses, or interpretation of data, the writing of the manuscript, or the decision to publish the results.
References
- 1.Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158:322–340. doi: 10.1053/j.gastro.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun J, Kato I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016;3:130–143. doi: 10.1016/j.gendis.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun J. Impact of bacterial infection and intestinal microbiome on colorectal cancer development. Chin Med J. 2021 doi: 10.1097/CM9.0000000000001979. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fletcher R, Wang YJ, Schoen RE, Finn OJ, Yu J, Zhang L. Colorectal cancer prevention: Immune modulation taking the stage. Biochim Biophys Acta Rev Cancer. 2018;1869:138–148. doi: 10.1016/j.bbcan.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Wei H, Zhou Y, Szeto CH, Li C, Lin Y, Coker OO, Lau HCH, Chan AWH, Sung JJY, et al. High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterology. 2022;162:135–149. doi: 10.1053/j.gastro.2021.08.041. e132. [DOI] [PubMed] [Google Scholar]
- 6.Kwong TNY, Wang X, Nakatsu G, Chow TC, Tipoe T, Dai RZW, Tsoi KKK, Wong MCS, Tse G, Chan MTV, et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology. 2018;155:383–390. doi: 10.1053/j.gastro.2018.04.028. e388. [DOI] [PubMed] [Google Scholar]
- 7.Horas K, Zheng Y, Fong-Yee C, Macfarlane E, Manibo J, Chen YZ, Qiao J, Gao MX, Haydar N, McDonald MM, et al. Loss of the Vitamin D receptor in human breast cancer cells promotes epithelial to mesenchymal cell transition and skeletal colonization. J Bone Miner Res. 2019;34:1721–1732. doi: 10.1002/jbmr.3744. [DOI] [PubMed] [Google Scholar]
- 8.Zeissig S, Burgel N, Gunzel D, Richter J, Mankertz J, Wahnschaffe U, Kroesen AJ, Zeitz M, Fromm M, Schulzke JD. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut. 2007;56:61–72. doi: 10.1136/gut.2006.094375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YG, Lu R, Wu S, Chatterjee I, Zhou D, Xia Y, Sun J. Vitamin D receptor protects against dysbiosis and tumorigenesis via the JAK/STAT pathway in intestine. Cell Mol Gastroenterol Hepatol. 2020;10:729–746. doi: 10.1016/j.jcmgh.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dove WF, Cormier RT, Gould KA, Halberg RB, Merritt AJ, Newton MA, Shoemaker AR. The intestinal epithelium and its neoplasms: genetic, cellular and tissue interactions. Philos Trans R Soc Lond B Biol Sci. 1998;353:915–923. doi: 10.1098/rstb.1998.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukamoto S, Ishikawa T, Iida S, Ishiguro M, Mogushi K, Mizushima H, Uetake H, Tanaka H, Sugihara K. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clin Cancer Res. 2011;17:2444–2450. doi: 10.1158/1078-0432.CCR-10-2884. [DOI] [PubMed] [Google Scholar]
- 13.Van Cromphaut SJ, Dewerchin M, Hoenderop JG, Stockmans I, Van Herck E, Kato S, Bindels RJ, Collen D, Carmeliet P, Bouillon R, et al. Duodenal calcium absorption in vitamin D receptor-knockout mice: functional and molecular aspects. Proc Natl Acad Sci U S A. 2001;98:13324–13329. doi: 10.1073/pnas.231474698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Y-g Zhang, R Lu, Xia Y, Zhou D, Petrof E, Claud EC, Sun J. Lack of vitamin D receptor leads to hyperfunction of claudin-2 in intestinal inflammatory responses. Inflammatory Bowel Diseases. 2018;25:97–110. doi: 10.1093/ibd/izy292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y-G, Lu R, Wu S, Chatterjee I, Zhou D, Xia Y, Sun J (2020). Vitamin D receptor protects against dysbiosis and tumorigenesis via the JAK/STAT pathway in intestine Biorxiv02. [DOI] [PMC free article] [PubMed]
- 16.Wu S, Zhang YG, Lu R, Xia Y, Zhou D, Petrof EO, Claud EC, Chen D, Chang EB, Carmeliet G, et al. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2015;64:1082–1094. doi: 10.1136/gutjnl-2014-307436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu R, Zhang Y, Xia Y, Zhang J, Kaser A, Blumberg R, Sun J. Paneth cell alertness to pathogens maintained by vitamin D receptors. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatterjee I, Zhang Y, Zhang J, Lu R, Xia Y, Sun J. Overexpression of vitamin D receptor in intestinal epithelia protects against colitis via upregulating tight junction protein claudin 15. J Crohn's Colitis. 05 March, 2021 doi: 10.1093/ecco-jcc/jjab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu R, Wu S, Zhang YG, Xia Y, Liu X, Zheng Y, Chen H, Schaefer KL, Zhou Z, Bissonnette M, et al. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis. 2014;3:e105. doi: 10.1038/oncsis.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu R, Wu S, Zhang YG, Xia Y, Zhou Z, Kato I, Dong H, Bissonnette M, Sun J. Salmonella protein AvrA activates the STAT3 signaling pathway in colon cancer. Neoplasia. 2016;18:307–316. doi: 10.1016/j.neo.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang YG, Lu R, Xia Y, Zhou D, Petrof E, Claud EC, Sun J. Lack of Vitamin D receptor leads to hyperfunction of claudin-2 in intestinal inflammatory responses. Inflamm Bowel Dis. 2019;25:97–110. doi: 10.1093/ibd/izy292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu R, Wu S, Liu X, Xia Y, Zhang YG, Sun J. Chronic effects of a Salmonella type III secretion effector protein AvrA in vivo. PLoS One. 2010;5:e10505. doi: 10.1371/journal.pone.0010505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YG, Zhu X, Lu R, Messer JS, Xia Y, Chang EB, Sun J. Intestinal epithelial HMGB1 inhibits bacterial infection via STAT3 regulation of autophagy. Autophagy. 2019 doi: 10.1080/15548627.2019.1596485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLong EF, Wickham GS, Pace NR. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 25.Tan SM, Yung PYM, Hutchinson PE, Xie C, Teo GH, Ismail MH, Drautz-Moses DI, Little PFR, Williams RBH, Cohen Y. Primer-free FISH probes from metagenomics/metatranscriptomics data permit the study of uncharacterised taxa in complex microbial communities. NPJ Biofilms Microbiomes. 2019;5:17. doi: 10.1038/s41522-019-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Garrett S, Carroll RE, Xia Y, Sun J. Vitamin D receptor upregulates tight junction protein claudin-5 against colitis-associated tumorigenesis. Mucosal Immunol. 2022 doi: 10.1038/s41385-022-00502-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chatterjee I, Lu R, Zhang Y, Zhang J, Dai Y, Xia Y, Sun J. Vitamin D receptor promotes healthy microbial metabolites and microbiome. Sci Rep. 2020;10:7340. doi: 10.1038/s41598-020-64226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Zhang Y, Xia Y, Sun J. Imbalance of the intestinal virome and altered viral-bacterial interactions caused by a conditional deletion of the vitamin D receptor. Gut Microbes. 2021;13 doi: 10.1080/19490976.2021.1957408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robanus-Maandag EC, Koelink PJ, Breukel C, Salvatori DC, Jagmohan-Changur SC, Bosch CA, Verspaget HW, Devilee P, Fodde R, Smits R. A new conditional Apc-mutant mouse model for colorectal cancer. Carcinogenesis. 2010;31:946–952. doi: 10.1093/carcin/bgq046. [DOI] [PubMed] [Google Scholar]
- 30.Flisikowska T, Merkl C, Landmann M, Eser S, Rezaei N, Cui X, Kurome M, Zakhartchenko V, Kessler B, Wieland H, et al. A porcine model of familial adenomatous polyposis. Gastroenterology. 2012;143 doi: 10.1053/j.gastro.2012.07.110. 1173-1175 e1177. [DOI] [PubMed] [Google Scholar]
- 31.Lu R, Zhang YG, Xia Y, Zhang J, Kaser A, Blumberg R, Sun J. Paneth cell alertness to pathogens maintained by vitamin D receptors. Gastroenterology. 2021;160:1269–1283. doi: 10.1053/j.gastro.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu S, Yoon S, Zhang YG, Lu R, Xia Y, Wan J, Petrof EO, Claud EC, Chen D, Sun J. Vitamin D receptor pathway is required for probiotic protection in colitis. Am J Physiol Gastrointest Liver Physiol. 2015;309:G341–G349. doi: 10.1152/ajpgi.00105.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K, et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011;585:606–612. doi: 10.1016/j.febslet.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 34.Cherradi S, Martineau P, Gongora C, Del Rio M. Claudin gene expression profiles and clinical value in colorectal tumors classified according to their molecular subtype. Cancer Manag Res. 2019;11:1337–1348. doi: 10.2147/CMAR.S188192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yong-guo Zhang SW, Lu Rong, Zhou David, Wan Jiandi, Zhou Jingsong, Carmeliet Geert, Petrof Elaine, Claud Erika C., Sun Jun. Tight junction CLDN2 gene is a direct target of the vitamin D receptor. Scientific Report. 2015 doi: 10.1038/srep10642. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang YG, Wu S, Sun J. Vitamin D, Vitamin D receptor, and tissue barriers. Tissue Barriers. 2013;1 doi: 10.4161/tisb.23118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatterjee I, Zhang Y, Zhang J, Lu R, Xia Y, Sun J. Overexpression of Vitamin D receptor in intestinal epithelia protects against colitis via upregulating tight junction protein Claudin 15. J Crohns Colitis. 2021;15:1720–1736. doi: 10.1093/ecco-jcc/jjab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J, Zhang YG. Vitamin D receptor influences intestinal barriers in health and disease. Cells. 2022;11 doi: 10.3390/cells11071129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li XY, Boudjelal M, Xiao JH, Peng ZH, Asuru A, Kang S, Fisher GJ, Voorhees JJ. 1,25-Dihydroxyvitamin D3 increases nuclear vitamin D3 receptors by blocking ubiquitin/proteasome-mediated degradation in human skin. Mol Endocrinol. 1999;13:1686–1694. doi: 10.1210/mend.13.10.0362. [DOI] [PubMed] [Google Scholar]
- 40.Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, Cho KR, Fearon ER. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 2007;67:9721–9730. doi: 10.1158/0008-5472.CAN-07-2735. [DOI] [PubMed] [Google Scholar]
- 41.Boivin GP, Washington K, Yang K, Ward JM, Pretlow TP, Russell R, Besselsen DG, Godfrey VL, Doetschman T, Dove WF, et al. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology. 2003;124:762–777. doi: 10.1053/gast.2003.50094. [DOI] [PubMed] [Google Scholar]