Abstract
Klebsiella pneumoniae is an opportunistic pathogen causing nosocomial and community-acquired infections. Klebsiella has developed resistance against antimicrobials including the last resort class; carbapenem. Currently, treatment options for carbapenem-resistant-Klebsiella (CRK) are very limited. This study aims to restore carbapenem effectiveness against CRK using celastrol and thymol. Clinical Klebsiella isolates were identified using biochemical and molecular methods. Antimicrobial susceptibility was determined using disk-diffusion method. Carbapenemase-production was tested phenotypically and genotypically. Celastrol and thymol-MICs were determined and the carbapenemase-inhibitory effect of sub-MICs was investigated. Among 85 clinical Klebsiella isolates, 72 were multi-drug-resistant and 43 were meropenem-resistant. Phenotypically, 39 isolates were carbapenemase-producer. Genotypically, blaNDM1 was detected in 35 isolates, blaVIM in 17 isolates, blaOXA in 18 isolates, and blaKPC was detected only in 6 isolates. Celastrol showed significant inhibitory effect against carbapenemase-hydrolytic activity. Meropenem-MIC did not decrease in presence of celastrol, only 2-fold decrease was observed with thymol, while 4–64 fold decrease was observed when meropenem was combined with both celastrol and thymol. Furthermore, thymol increased CRK cell wall-permeability. Molecular docking revealed that celastrol is superior to thymol for binding to KPC and VIM-carbapenemase. Our study showed that celastrol is a promising inhibitor of multiple carbapenemases. While meropenem-MIC were not affected by celastrol alone and decreased by only 2-folds with thymol, it decreased by 4–64 folds in presence of both celastrol and thymol. Thymol increases the permeability of CRK-envelope to celastrol. The triple combination (meropenem/celastrol/thymol) could be useful for developing more safe and effective analogues to restore the activity of meropenem and other β-lactams.
Subject terms: Antimicrobials, Infection
Introduction
Klebsiella is a Gram-negative bacterium that belongs to the family Enterobacteriaceae. Klebsiella pneumoniae is the most medically important Klebsiella species and one of the major opportunistic pathogens associated with hospital outbreaks [1]. K. pneumoniae can colonize the intestinal tract, skin, nose and throat of healthy individuals. However, K. pneumoniae could cause various infections in hospitalized patients, most commonly pneumonia, wound, soft tissue, and urinary tract infections [2]. K. pneumoniae infections are particularly a problem among neonates, the elderly, and immunocompromised patients. In addition K. pneumoniae can cause significant number of serious community-acquired infections including pyogenic liver abscess, pneumonia, and meningitis [3].
Klebsiella pneumoniae has developed different resistance mechanisms to antimicrobials. These mechanisms include: production of inactivating enzymes, target site modification, reducing the intracellular concentration of antimicrobials either through active expulsion of drug by efflux pump or by changing membrane permeability, or through development of an alternative metabolic pathways [4]. β–lactam is the largest and most important class of antibiotics that have different sub-classes including penicillins, cephalosporins, carbapenems and monobactam. β–lactam antibiotics are widely used in treatment of K. pneumonia infections. Bacterial resistance to β–lactams has increased dramatically with the production of β-lactamases [5]. Carbapenems are among the last-line antibiotics that are used against resistant Gram-negative bacteria. The rapid dissemination of carbapenem-resistant Enterobacteriaceae (CRE) especially Carbapenem resistant-Klebsiella (CRK) represents a global public health threat [6].
Carbapenem-resistance occurs mainly due to production of various types of carbapenemases. Based on the molecular structure and amino acid sequences (ambler classes), carbapenemases belong to class A and D enzymes depend on serine for β-lactams hydrolysis, while class B metallo-betalactamase contains divalent zinc ions important for substrate hydrolysis [7]. In Enterobacteriaceae, the most prevalent class A-carbapenemases is the Klebsiella pneumoniae carbapenemase (KPC). The class B Metallo-β-lactamases (MBLs) mainly include New Delhi MBL (NDM), Verona Integron-encoded MBL (VIM), and IMP-type carbapenemases. While the most frequently detected class D is the OXA-type β-lactamases [8]. Other carbapenem resistance mechanisms include loss of expression or mutation of porin-encoding genes and overexpression of genes encoding efflux pumps [7].
Carbapenem resistance in Gram-negative pathogens is dramatically limiting treatment options, it is obvious that novel therapies are urgently needed. Recently, new carbapenemase inhibitors with activity against CRE have been approved for clinical use including avibactam, relebactam, and vaborbactam [8]. The development of these inhibitors represents a step in the fight against antimicrobial resistance that needs to be followed by further steps.
Celastrol is a pentacyclic-triterpenoid that is extracted from the root Pulp of the Tripterygium wilfordii plant [9]. Celastrol is a component of Chinese medicine that is used to treat cancer, inflammatory, autoimmune, and neurodegenerative diseases [10]. Celastrol has been recently exploited to treat obesity and type 2 diabetes mellitus [11]. In addition, celastrol has been found to exhibit a growth inhibitory activity against Gram-positive bacteria [12]. Furthermore, celastrol was found to reduce staphyloxanthin biosynthesis and biofilm formation in Staphylococcus aureus [13, 14]. Besides, celastrol shows antiviral activity against hepatitis viruses [15].
Although celastrol exhibits many biological activities, no previous study has characterized the β-lactamase inhibitory potential of celastrol [16]. However, pentacyclic triterpenoids (e.g., corosolic acid) and polycyclic terpene (e.g., oleanolic acid) were reported as β-lactamase inhibitors. Based on similarities in chemical structure and biological activities [17], celastrol is thought to show a similar activity as a potential β-lactamase inhibitor. This study aims to evaluate the antibacterial activity of celastrol against CRK and to investigate its carbapenemase-inhibitory potential.
Materials and methods
Bacterial isolates and chemicals
A total of 85 clinical isolates of Klebsiella were recovered from specimens sent to clinical laboratory of Zagazig University hospital, Zagazig, Egypt. Clinical specimens were from different sources including blood, urine, sputum, pus, and endotracheal tube aspirate. Celastrol, Dimethyl sulphoxide (DMSO), Ethylene-diamine tetra-acetic acid (EDTA), Resazurin, and thymol were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Bacterial media and antibiotics disks were purchased from Oxoid (UK).
Phenotypic and molecular identification of Klebsiella species by 16S-rRNA gene sequencing
Klebsiella isolates were identified as being Gram-negative rods with lactose fermenting activity on MacConkey agar. Confirmatory tests include: IMViC tests, growth on triple sugar iron and motility tests [18].
Molecular identification was based on sequencing of 16S rRNA gene, gDNA was extracted as described previously [19]. Briefly, a colony of isolate was suspended in 50 µl of nuclease-free water, heated to 100 °C for 10 min using Biometra T-GRADIENT thermocycler (Rudolf-Wissell-Str. Göttingen, Germany). The bacterial debris were removed by centrifugation at 21,000 × g for 10 min, the supernatant was used as template in a polymerase chain reaction (PCR). The universal primers 341 F and R806 were used for amplification of bacterial 16S rRNA [20]. These primers were supplied by Sigma Aldrich (Petaluma, USA).
The COSMO PCR RED 2x Master Mix (Willowfort UK) was used. The PCR cycling conditions were as follows: an initial denaturation for 5 min at 94 °C, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 2 min, and then a final extension for 5 min at 72 °C. The amplified PCR products were electrophoresed on 2% agarose gel, and photographed with gel documentation system (Cleaver Scientific Ltd, UK). The PCR products were purified using Thermo scientific GeneJET PCR purification kits (Vilnius, Lithuania), according to the manufacturer’s instructions. Purified PCR samples were used for sequencing according to service requirements, where 5 μl of template DNA (20–80 ng) were mixed with 5 μl of 341 F primer (5 pmol μl−1). PCR samples were sequenced using the Illumina HiSeq platform using 300 PE chemistry (GATC-Biotech, Konstanz, Germany). The obtained sequences were used to draw the phylogenetic tree and to perform multiple sequence alignment. Phylogenetic analyses were conducted in MEGA11 [21].
Antimicrobial susceptibility testing
Antimicrobial susceptibility of Klebsiella isolates was performed by disk diffusion method according to the clinical and laboratory standard institute (CLSI) guidelines [22]. Briefly, a sterile cotton swab was dipped in Mueller Hinton broth (MHB) with a turbidity equivalent to 0.5 McFarland standards, and used to inoculate the surface of dried Mueller Hinton agar (MHA) plate. The inoculated plates were left to dry for 3–5 min, and the antibiotic disks were placed on them. Plates were incubated at 37 °C for 18 h. The diameter of inhibition zone was measured, recorded, and interpreted according to CLSI (2). The used antibiotic disks include meropenem (MEM, 10 μg), piperacillin-tazobactam (TZP, 100/10 μg), ceftriaxone (CRO, 30 μg), cefepime (FEP, 30 μg), cefoperazone (CFP, 75 μg), aztreonam (ATM, 30 μg), gentamicin (GN, 10 μg), amikacin (AK, 30 μg), azithromycin (AZM, 15 μg), tetracycline (TE, 30 μg), tigecycline (TGC, 15 μg), levofloxacin (LEV, 5 μg), ofloxacin (OFX, 5 μg), trimethoprim-sulfamethoxazole (SXT, 1.25/23.75 μg), and chloramphenicol (C, 30 μg).
Phenotypic detection of carbapenemase-producing Klebsiella by Carba NP test
The Carba NP colorimetric assay for detection of carbapenemase production is based on detection of acidic products due to hydrolysis of imipenem using phenol red indicator [22]. A modified protocol that uses colonies (instead of bacterial extracts) and using 0.1% Triton X-100 as cells lytic agent was used [23]. Briefly, a colony of pure bacterial culture was suspended in tube containing 100 µl of solution A (phenol red 0.05% + ZnSO410 mM + 0.1% (vol/vol) Triton X-100 solution, adjusted to pH= 7.8), or solution B (solution A + 12 mg ml−1 imipenem) then vortexed for 10 s and incubated at 35 °C for up to 2 h. The tube containing solution A was used as control and tube containing solution B as test. Carbapenemase-production was indicated by the appearance of orange or yellow color in the test tube while the control tube remained red [23].
Detection of carbapenemase encoding genes by PCR
Conventional PCR was conducted for the detection of presence of Metallo-β-lactamase genes (blaNDM and blaVIM) or carbapenemases genes (blaOXA-9 -and blaKPC-1) in Klebsiella isolates. The primers were supplied by IDT (Integrated DNA Technologies, Coralville, Iowa, USA). The sequences of the primers: blaNDM, blaOXA, blaKPC-1 [24], and blaVIM [25] are listed in Supplementary Table 1. PCR mixture contained 25 μl of MasterMix, 2 μl of each primer, 2 μl of DNA template, and nuclease free water to 50 μl. The ampilification conditions were: initial denaturation at 95 °C for 3 min followed by 30 cycles of denaturation at 95 °C for 5 s, annealing for 30 s at temperature indicated in supplementary Table 1, and extension at 72 °C for 1 min, followed by final extension at 72 °C for 5 min.
Determination of minimum inhibitory concentration (MIC) of meropenem, celastrol, and thymol against CRK isolates
Meropenem-MIC was determined by broth microdilution method for CRK isolates that were positive for carbapenemase production. Briefly, 3 colonies were used of each isolate (from overnight culture) to inoculate 5 ml of MHB. Broth cultures were incubated for 18 h at 37 °C. Cultures were then diluted in sterile saline and the turbidity was adjusted to 0.5 McFarland’s standard. Then 1/100 dilution of this suspension was made in sterile MHB. In sterile 96 wells-microplate, serial dilutions of tested chemicals were prepared in sterile MHB including: meropenem (0.5–1024 µg ml−1), celastrol (0.5–1024 µg ml−1) and thymol (5–2400 µg ml−1). 50 µl of the bacterial suspension was added to each well containing the serially diluted chemicals. The plates were incubated for 18 h at 37 °C and examined for bacterial growth. The lowest concentration that inhibited visible bacterial growth was reported as MIC [22]. After determination of meropenem-MIC, isolates with the highest MIC were used in the next analysis, where the MICs of celastrol and thymol were determined by the broth microdilution method.
Cell viability measurement with Alamar Blue assay
Alamar Blue (Resazurin) assay was performed to assess the effect of sub-MIC of celastrol and thymol on the metabolic activity of CRK as described previously [26]. Briefly, bacterial isolates were incubated alone, with 128 µg ml−1 of celastrol, with 300 µg ml−1 of thymol, or with a combination of celastrol (128 µg ml−1) and thymol (300 µg ml−1) for 24 h at 37 °C. Cells were collected by centrifugation at 8000 rpm for 10 min and resuspended in freshly prepared Phosphate-buffered saline (PBS). 0.1 ml of 6.5 mg ml−1 resazurin stock solution (prepared in PBS) was added to 0.9 ml of cell suspension. The reaction solutions were incubated in dark for 4 h at 37 °C. Sterile PBS with resazurin was used as blank. The fluorescent intensity of the reduced resazurin (resorufin) was observed at 590 nm emission and 560 nm excitation wavelengths.
Carbapenemase inhibition assay of crude periplasmic extract
This technique measured the hydrolytic activity of meropenem at a final concentration of (100 μM) using ultraviolet-visible (UV-Vis) spectrophotometry in the presence and absence of enzyme inhibitor. EDTA (reported inhibitor of MBLs) was used to validate the test [27]. Briefly, bacteria were cultured overnight on MHA supplemented with meropenem (4 μg ml−1). A standard number of bacterial cells [~5 × 108 cells ml−1; optical density (OD) at 600 nm (OD 600) = 0.65] were suspended in 1 ml of 20 mM Tris–HCl buffer containing 2% v/v Triton X. After vigorous mixing, the suspension was left at room temperature for 10 min and then centrifuged at 4000 rpm for 15 min. The supernatant was collected, and 180 µl of the supernatant was added to 96 sterile microtiter-plate. The first well was used to estimate the background absorbance (blank). The second well contained 100 μM of meropenem, the third well contained 100 μM of meropenem and 64 µg ml−1 of celastrol, the fourth well contained 100 μM meropenem and 128 µg ml−1 of celastrol and the fifth well contained 100 μM meropenem and 300 µg ml−1 of thymol. Finally, ddH2O was added to each well to make the final volume 200 μl. Hydrolysis of meropenem was measured using a microplate reader (synergy HT BioTek) at 297 nm. Meropenem initial absorbance was measured immediately after the inclusion of the antibiotic. and the reaction plate was incubated at 37 °C for 60 min [28]. The hydrolysis of meropenem was measured after the incubation period. And The hydrolysis index was calculated as follows:
hydrolysis index = [Initial Absorbance – Absorbance after 60 min]/Initial Absorbance.
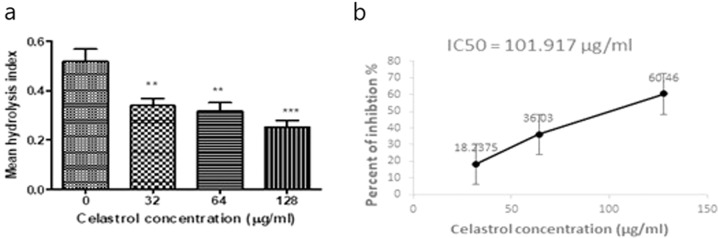
The half maximal inhibitory concentration (IC50) was calculated using Microsoft Excel. IC50 is defined as the minimum concentration of celastrol required for inhibiting the activity of the extracted carbapenemases by 50% Four concentrations of celastrol (0, 32, 64, and 128 µg ml−1) were used to determine the percentage of carbapenemases inhibition, then the IC50 was calculated.
Determination of meropenem MIC in presence of sub-MIC of celastrol and/or thymol
The MIC of meropenem was determined in presence of sub-MIC (64 µg ml−1 and 128 µg ml−1) of celastrol. Furthermore, meropenem MIC was determined in presence of ¼ (300 µg ml−1) and 1/8 (150 µg ml−1) MIC of thymol. Finally, a triple combination of meropenem, celastrol (64, 128 µg ml−1), and thymol (150, 300 µg ml−1) was evaluated. The change of meropenem MIC in presence of tested inhibitors was determined using the broth microdilution method as described previously [22].
Effect of sub-MIC concentrations of thymol on the lytic activities of SDS and Triton X 100 on K. pneumoniae outer membrane
This experiment was used to investigate thymol ability to increase the permeability of Klebsiella outer membrane to boost the activity of less permeable celastrol [29]. This technique is based on the sensitization of cells to lytic action of the detergents sodium dodecyl sulfate (SDS) and Triton X-100 by thymol. Briefly, a standard inoculum of bacteria (OD630 of 0.5) was treated with 300 µg ml−1 of thymol for 10 min at room temperature and added to microplate wells that already contained either SDS (0.1 and 1%), Triton X-100 (0.1 and 1%) or buffer solution. Turbidity of the cell suspensions was then monitored with Synergy microplate reader (Agilent, Santa Clara, USA) as described previously [30]. Cell death caused by the sudden influx of these lytic agents was determined by measuring the decrease in OD (Relative turbidity %).
In silico analysis by molecular docking
The crystal structures of the proteins were retrieved from the Protein Data Bank (https://www.rcsb.org/). These proteins include: the class A carbapenemase KPC-2 (PDB-ID: 3DW0) [31], NDM-1 (PDB-ID: 3SPU) [32], β-lactamase OXA-181 (PDB-ID: 5OE0) [33] and VIM-2 MBL (PDB-ID: 5YD7) [34]. Both celastrol and thymol were drawn into Marvin Sketch of Marvin suite (http://www.chemaxon.com) and lowest energy three-dimensional conformer for each was generated. Dock module of MOE (Molecular Operating Environment) version MOE 2019.0102,2 [35] on a computer having Pentium 1.6 GHz workstation, 512 MB memory using windows operating system, was utilized in docking studies. Tested compounds were docked into the rigid binding pocket of the protein using flexible ligand mode. The free energy of binding of the ligand is estimated using the GBVI/WSA ΔG as a force field-based scoring function [36]. Molecular docking was performed to investigate how ligand binds to protein target [37].
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 5.0.1 for Windows, GraphPad Software (San Diego, California USA). Carbapenemase inhibition assay and cell viability measurement with Alamar Blue data were analyzed using one-way ANOVA followed by Dunnett’s post hoc test. The Paired t-test was used for analysis of the permeability assay of the outer membrane data. The probability value (P < 0.05) was considered as the level of significance.
Results
Identification and taxonomic classification of isolated strains
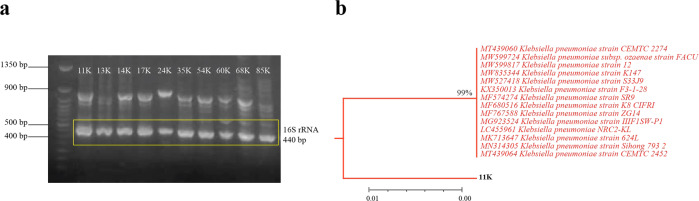
A total of 85 clinical Klebsiella isolates were identified using biochemical tests, isolates with high resistance to carbapenm were confirmed by molecular analysis. Molecular identification was based on 16S rRNA gene sequence, the 16S rRNA gene was amplified by PCR giving a fragment of 440 bp (Fig. 1a). After sequencing, sequences were submitted to the Basic Local Alignment Search Tool of Nucleotides (BLASTN) service in NCBI to align and compare the sequences in relation to reference sequences in the Genebank. BLASTN analysis confirmed that the queried sequences represent a partial sequence of the 16S rRNA of the genus Klebsiella. The constructed phylogenetic tree (Fig. 1 b) reveals that all the inspected clinical isolates were completely related to K. pneumoniae reference strains.
Fig. 1.
Molecular identification and phylogenetic analysis of bacterial isolates. a Gel electrophoresis of PCR products (440 bp) of genes encoding 16S rRNA of suspected K. pneumoniae, b representative phylogenetic tree of the partial 16S rRNA gene sequence from isolate 11 K compared to sequences of the most related K. pneumoniae strains recognized by BLASTN
Susceptibility of Klebsiella isolates to different classes of antimicrobials
Among the 85 clinical Klebsiella isolates, 84.70% were multi-drug resistant (MDR) as they were resistant to three or more antimicrobial classes. High percentage of isolates were resistant to ceftriaxone (83.52%), cefoperazone (82.35%), azithromycin (75.3%), tetracycline (72.9%), cefepime (71.76%), trimethoprim /sulfamethoxazole (71.76%), aztreonam (70.85%), piperacillin /tazobactam (64.7%), ofloxacin and levofloxacin (63.5% each). 43 isolates (50.58%) were found to be resistant to meropenem. Less that 50% of isolates were resistant to tigecycline (45.9%) and chloramphenicol (31.8%). The susceptibility profile of Klebsiella isolates is shown in Supplementary Fig. 1.
Phenotypic and genotypic detection of carbapenemase-producing Klebsiella isolates
Carba NP test was performed on the forty-three isolates that showed resistance to meropenem in susceptibility test. This test reflected that 39 isolates of 43 were carbapenemases producers as shown in Fig. 2a. Furthermore, PCR was performed on the 43 meropenem-resistant isolates to detect genes encoding for carbapenemases. Out of the 43 carbapenem-resistant isolates, the blaNDM-1 was detected in 35 isolates (81%), the blaVIM was detected in 17 (39.5%) isolates, the blaOXA was detected in 18 (41.8%) isolates, and blaKPC was detected in 6 (13.9%) isolates (Fig. 2b). Only three isolates did not have any of the tested genes.
Fig. 2.
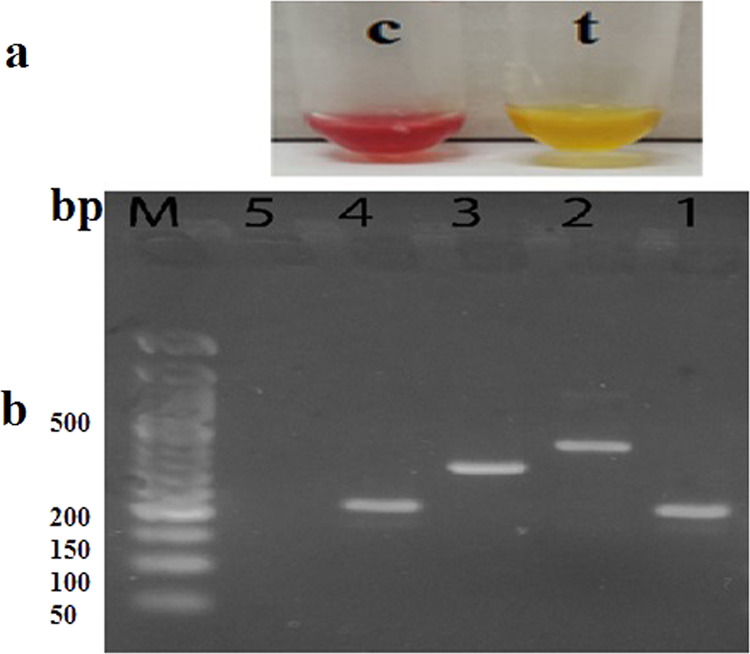
Phenotypic and genotypic detection of carbapenemase activity in Klebsiella isolates. a The Carba NP test was used for phenotypic analysis, positive results were indicated by the change of color of inoculated tube (t) to yellow: b Genotypic detection of carbapenemase genes by PCR in CRK isolates: lane 1: bla NDM-1 gene (209 bp), lane 2: bla VIM gene (382 bp), lane 3: bla OXA gene (315 bp), lane 4: bla KPC gene (209 bp), lane 5: negative control, M: 50 bp DNA-ladder
Cell viability measurement with Alamar Blue
This experiment was conducted to determine the effect of celastrol and thymol (alone or in combination) in the used concentrations on the viability of the bacterial cells. As outlined in the materials and methods section, viable cells reduce the non-fluorescing form of this chromogenic indicator dye by cellular dehydrogenases to a pink, fluorescent form, that can be monitored spectrophotometrically. Cell viability is expressed as fluorescence intensity. The experiment revealed that Klebsiella isolates cultured in the presence of celastrol (128 µg ml−1), thymol (300 µg ml−1), or in combination, showed no significant decrease in fluorescence intensity from that of the control culture (Fig. 3).
Fig. 3.
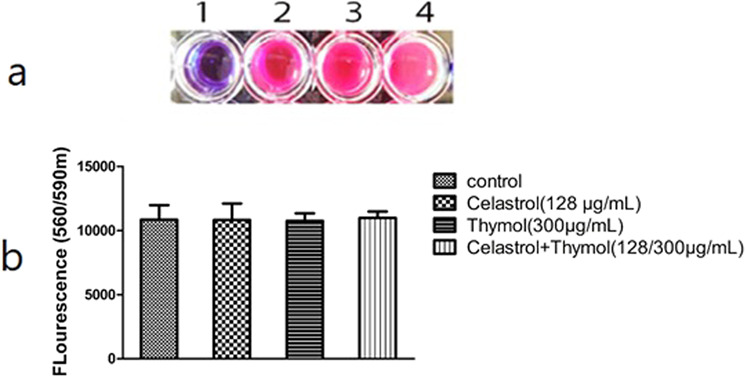
Cell viability measurement with Alamar Blue. a Color change of resazurin dye 1: blank, 2: Celastrol, 3: Thymol, 4: Control. b fluorescence intensity of the treated culture was not significantly decreased from that of the control culture
Evaluation of the effect of celastrol and thymol on meropenem-MIC in carbapenemase-producing isolates
Eight isolates with the highest MIC were used to study the effect of both celastrol and thymol on meropenem MIC (Table 1). MIC of celastrol for all 8 isolates was higher than 1 mg ml−1. MIC of thymol was 1200 µg ml−1 for the 8 selected isolates except for isolates 35k and 68k, where the MIC was 600 µg ml−1. Table 1 shows the change in MIC of meropenem alone, meropenem in a combination of 128 μg ml−1 of celastrol, meropenem in combination with 300 μg ml−1 of thymol, and MIC of meropenem in a triple combination.
Table 1.
The effect of sub-MIC of celastrol, thymol or combination of them on the MIC of meropenem against K.pneumoniae isolates
| Isolate’s No. | MEM | MEM + Celastrol | MEM + Thymol | MEM + Celastrol + Thymol* |
|---|---|---|---|---|
| 11k | 256 | 256 | 128(2) | 32(8) |
| 13k | 256 | 256 | 128(2) | 8(32) |
| 14k | 32 | 32 | 16(2) | <1(>32) |
| 17k | 512 | 512 | 256(2) | 128(4) |
| 35k | 64 | 64 | 32(2) | <1(>64) |
| 60k | 128 | 128 | 128(0) | 32(4) |
| 68k | 256 | 256 | 256(0) | 64(4) |
| 85k | 128 | 128 | 128(0) | 32(4) |
MEM meropenem, (fold change in MEM-MIC was listed between brackets)
At sub-MIC of celastrol (128 µg ml−1) the eight Klebsiella isolates showed no decrease in the MIC of meropenem. At sub-MIC of thymol (150, 300 μg ml−1) showed either no change or only 2 fold reduction in MIC of meropenem.The combination of sub- MIC of celastrol (128 μg ml−1), and thymol (300 μg ml−1) exerted a significant reduction in meropenem MIC (4 to 64 fold reduction).
Celastrol significantly inhibited the hydrolytic activity of carbapenemases in the crude periplasmic extract
Enzyme inhibition assays were performed to detect the activities of carbapenemases in bacterial culture supernatants when co-incubated with different concentrations of celastrol (32, 64, 128 µg ml−1). Celastrol showed a significant inhibitory effect against the activity of carbapenemases when co-incubated in culture supernatants in concentration dependant matter (P < 0.05). The celastrol IC50 for inhibition of carbapenemases hydrolytic activities was 101.9 μg ml−1 (Fig. 4).
Fig. 4.
Celastrol inhibitory effect on the hydrolytic activity of the carbapenemases. a significant inhibitory effect of different celastrol concentrations on the activities carbapenemases detected by enzyme inhibition assays following co-incubation, b Percentage of carbapenemase activities inhibited by celastrol in a concentration-dependent manner. **indicates P < 0.01; ***indicates P < 0.001
Effect of sub-MIC of thymol on lytic activities of SDS and Triton-X 100 on K. pneumoniae outer membrane
The lytic agents (SDS and Triton-X) showed higher lytic activity in presence of sub-MIC (300 µg ml−1) of thymol. The lytic activity expressed as relative turbidity percentage of bacterial suspension measured at 630 nm. The result showed that sub-MIC of thymol significantly (P-value = 0.0084) increased the permeability of lytic agents through the outer membrane of tested Klebsiella isolates (Supplementary Table 2).
Molecular docking results
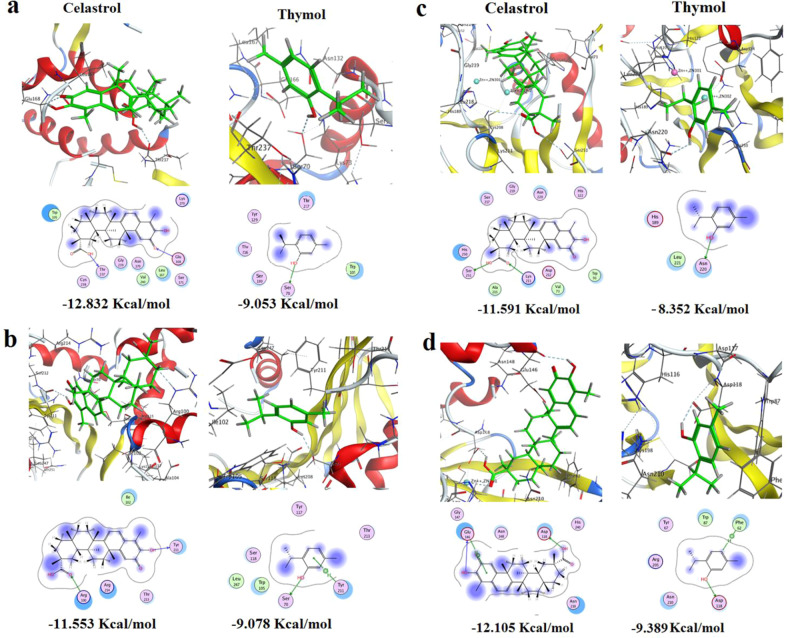
The docking results of celastrol against the crystal structures of the class A carbapenemase KPC revealed that the electronegative oxygen atom doubly bonded to carbon atom of position −11, has constructed an H-bond with the amino acid Glu168. The other terminal of the ligand was firmly stabilized at the core of the pocket via another H-bond between the OH of the carboxyl group linked to a carbon atom of position-2 and the conserved H-acceptor amino acid Thr237. Moreover, the hydrophobic/hydrophilic interactions improved binding affinity and total recognition of the ligand inside the core of the pocket ending up with a ligand/receptor complex of −12.8318186 Kcal mol−1 stability. While, with thymol, the hydroxyl group of its phenolic structure has formed H-bond with the conserved H-acceptor amino acid Ser70 with a free binding energy of −9.05285263 Kcal/mol (Fig. 5a).
Fig. 5.
The putative binding modes (2D & 3D) of celastrol and thymol and their free binding energies expressed in Kcal/mol in the active site of the predicted 3D structure of K. pneumoniae carbapenemases a Class A KPC (KPC-2-3DW0), b Class D OXA (OXA-181 5OE0) c Class-B NDM (NDM-1 3SPU), d Class-B Verona Integron-encoded MBL (VIM-2 5YD7). The blue and cyan shadow of the ligand and active site amino acids respectively indicated strong hydrophobic/hydrophilic interactions
For protein target OXA-181, the H-donor hydroxyl group at position-10 of celastrol formed an H-bond with the conserved amino acid Tyr211, whereas the carboxyl carbonyl (H-acceptor fragment) constructed H-bond with the conserved H-donor amino acid Arg100. Added to hydrophobic/hydrophilic interactions the ligand scored a free binding energy of −11.5534611 Kcal/mol. On the other hand, the phenyl ring of thymol formed an arene-H bond with Tyr211, while its phenolic OH assembled an H-bond with Ser70 at the core of the active site to afford ligand/receptor complex with −9.07757473 Kcal/mol stability (Fig. 5b).
On the other hand, docking of celastrol against the New Delhi MBL (NDM) protein target (PDB: 3SPU) showed the role of the carboxyl group at position-2 as its H-acceptor carbonyl and H-donor hydroxyl groups have formed two H-bonds with H-donor Lys211 and H-acceptor Ser251, respectively, with highest free binding energy −11.5906353 Kcal/mol. For thymol, H-bond was constructed between its hydroxyl group and the conserved H-acceptor amino acid Asn220. Besides, the phenyl ring with its hydrocarbon substituents in positions-2, and 5 exhibited conspicuous hydrophobic/hydrophilic interactions giving rise to the highest free binding energy −8.35244465 Kcal mol−1 (Fig. 5c).
Furthermore, the docking was extended to VIM-2 Verona Integron-MBL (PDB ID: 5YD7), and the two OH groups of celastrol, at position-10 and of carboxyl group at position-2, displayed two H-bonds with Glu146 and Asp118, respectively. Furthermore, the terminal non-classical phenyl ring formed arene-H-bond with Glu146 leading to the highest free binding energy −12.1051254 Kcal mol−1. In addition, the aromatic essential α-amino acid Phe62 established an arene-H-bond with the aromatic hydrogen at position-4 of thymol ligand whose active phenolic OH formed an H-bond with the conserved amino acid Asp118 leading to free binding energy of −9.3893404 Kcal mol−1 (Fig. 5d). Overall, celastrol is persistently dominant over thymol in all investigated protein targets and showed the best activity on KPC-2 with a free binding energy of about −12.832 Kcal mol−1 followed by VIM-2 with a free binding energy of about −12.105 Kcal mol−1.
Discussion
Klebsiella pneumoniae is a major health-care pathogen that is responsible for various infections including urinary tract, pneumonia, meningitis, and pyogenic liver abscess [3]. A total of 85 Klebsiella isolates were recovered from various clinical sources, identified by biochemical testing, and confirmed by 16S rRNA sequencing. Antibiotic susceptibility testing showed a higher frequency of MDR (84.70%) than recently reported in other study (50%) [38], the highest resistance rates among tested antibiotics were against cefoperazone and ceftriaxone followed by azithromycin. The lowest resistance rates were against chloramphenicol followed by tigecycline. The low resistance rates to chloramphenicol can be attributed to the limited use of chloramphenicol in clinical practice (topical eye drops) due to its wide range of adverse effects [39]. Tigecycline is a relatively new antibiotic with a low resistance rate [40], in addition, its use is limited to cases of bacteremia and severe pneumonia due to its large volume of distribution resulting in low blood concentration [41].
Approximately 51% of our isolates were CRK which is consistent with another Egyptian study that reported 53.7% resistance to carbapenem in Klebsiella [42]. This estimate is slightly lower than the 55% reported from the Europian countries [43] among carbapenemase-producing Enterobacteriaceae (EuSCAPE). Higher resistance percentages were reported from Thailand (97%), and United States (63%) [44, 45]. Unfortunately, some local studies reveals that carbapenem resistance rate in Egypt is on the rise [46].
CRK isolates were tested for carbapenemases production and for the presence of four genes representing 3 classes of carbapenemases. Out of the 43 CRK, 40 isolates (93.02%) carried one or more carbapenemase genes. The gene with the highest prevalence was bla-NDM-1 (81%) followed by bla-OXA (41.8%), and bla-VIM (39.5%) while bla-KPC gene has the lowest prevalence (13.9%). Similarly, other studies in Egypt showed high prevalence of bla-NDM-1 (80.5 and 94.1%) and reported 36.4 and 47% for bla VIM [25, 46]. While another study reported lower rate of bla-NDM-1 gene (33.3%), bla-OXA gene (30.7%), and only 2.5% for bla-KPC gene [38]. Most of our isolates carry multiple carbapenemase-encoding genes which is consistent with previous report [46]. This can be explained by the fact that genes encoding β-lactamases are mostly carried on mobile genetic elements and thus facilitate their transmition and accumulation within hospital isolates. Enterobacteriaceae acquire resistance to carbapenems via different mechanisms [47], the most common mechanism is the production of different classes of carbapenemases [8, 46].
CPK-infections are difficult to treat and have limited therapeutic options as seen from our antibiotic susceptibility results. Hence, it is very critical to restore the activity of carbapenems by developing drugs that can be combined with carbapenems to inhibit the action of carbapenemases. Recently two combinations were approved for treatment of carbapenemase-producing bacteria; vaborbactam in combination with meropenem was approved by food and drug administration (FDA) for treatment of complicated urinary tract infections and pneumonia [8]. Vaborbactam only inhibits the class-A carbapenemase KPC with no activity against class-B or D [48]. Relebactam is another FDA-approved carbapenemase inhibitor marketed in combination with imipenem. It does not show in vitro activity against class-D OXA-48 β-lactamases but has potent activity against class-A and class-C β-lactamases [49]. It was reported that triterpenoids such as corosolic acid and polycyclic terpene acids such as oleanolic acid had β-lactamase inhibitory activity [17]. Celastrol has similar chemical structure and biological activity to corosolic and oleanolic acid. Hence, in this study, celastrol was evaluated as potential inhibitor of carbapenemases to restore the activity of meropenem on CRK. Our results showed that celastrol had activity on all tested classes of carbapenemases (KPC, NDM, OXA and VIM-2), while Zhou et al. [17] reported activity against KPC enzyme only. Moreover, the study of Zhou and coworkers used only a standard E. coli strain to test the activity of corosolic acid, and have reported a maximum MIC of only 14 µg ml−1. In our study we tested the effect of celastrol on MDR-clinical CRK isolates (which is more reliable and realistic). It is expected that if corosolic acid is tested on clinical isolates to have much higher MICs.
The antibacterial activity of celastrol was tested against CRK isolates with no inhibitory activity observed for concentrations up to 1024 µg ml−1. This is consistent with previous study reported that celastrol showed low MIC against Gram-positive bacteria and no growth inhibitory activity against Gram-negative bacteria in the used concentration [12]. Our study presumed that celastrol possibly acts by the same mechanism on both Gram-positive and Gram-negative bacteria. However, the outer membranes of Gram-negative bacteria act as a permeability barrier, holding celastrol physically distant from its target [50].
Triton-X was used to extract the crude periplasmic enzymes. After incubation with sub-MIC of celastrol (128, 64, and 32 µg ml−1), celastrol showed significant inhibition of meropenem hydrolysis in concentration-dependent manner with the highest activity contributed to 128 µg ml−1. After this meropenem-MIC was determined in combination with the same concentrations of celastrol. There was no reduction in meropenem-MIC combined with any of the used concentrations. This confirmed the theory that was suggested previously [12] stating that celastrol cannot pass the Gram-negative outer membrane.
The challenge here was to deliver celastrol to the periplasmic space where it can inhibit carbapenemase activity. Disrupting the outer membrane could increase permeability and give celastrol a chance to pass through it. Thymol is a phytochemical that belongs to phenolic monoterpenes [51]. Thymol was described as a natural outer membrane permeabilizer that can increase the intracellular concentration of hydrophobic antibiotics such as erythromycin, azithromycin, sulphamethoxazole/trimethoprim, and novobiocin [29, 51, 52]. One of the intrinsic resistance mechanisms in Gram-negative bacteria is the inner membrane that protects the cell against extracellular toxic compounds. In this membrane, porins regulate the internal accumulation of hydrophilic molecules including antibiotics [53]. Thymol disrupts this membrane by lipophilic action, as it integrates within the polar head groups of the lipid bilayer, inducing alterations of the cell-membrane permeability [54].
To confirm that the sub-MIC of celastrol (128 µg ml−1) and thymol (300 µg ml−1) do not affect the viability of bacteria, cell viability assay with Alamar-Blue was conducted. It was found that neither celastrol nor thymol or their combination showed any significant decrease in bacteria viability.
Thymol outer membrane disrupting effects were evaluated in this study by testing their effects on the lytic activity of triton X-100 and SDS. The presence of lipopolysaccharide molecules in the outer membrane gives Gram-negative an intrinsic resistance against hydrophilic antibiotics and detergents such as SDS and Triton X-100. It was found that thymol in a concentration 300 µg ml−1 (1/4 MIC) sensitized bacterial cells to the cell lytic activity of SDS and Triton X-100 by 15–29% and with a significant difference from untreated culture (P-value = 0.0084), indicating a weakening of the outer membrane barrier. Our findings are consistent with that reported previously [29] despite using lower concentration. We used 150 µg ml−1 of thymol, but it showed no significant difference.
After establishing the thymol disrupting effect on the protective outer membrane of Klebsiella isolates, we evaluated a triple combination of celastrol (128, 64, or 32 µg ml−1), and thymol (300 µg ml−1) and meropenem. We found that celastrol (128 µg ml−1) in presence of thymol reduced meropenem-MIC by (4–64) fold change and rendered two of the test isolates sensitive to meropenem again. This is explained by that thymol increased the permeability of celastrol into the bacterial cell in a concentration high enough to inhibit the hydrolytic activity of carbapenemases thus protecting meropenem and decreasing its MIC. Celastrol at concentrations of 64 or 32 µg ml−1 showed no decrease in meropenem-MIC. It was observed that meropenem-MIC was decreased by a maximum of 2-fold when combined with thymol alone.
One of the main issues about celastrol is its toxicity which can strictly limit its clinical applications [55]. Some useful approaches to reduce celastrol toxicity include: combination therapy (which is proposed in our study), synthesis of structural derivatives, development of new formulations and use of more safe routs of administration [56]. For example, it was reported that the use of celastrol-nanoparticles successfully reduced systemic toxicity and also solve the problem of its poor solubility [57]. Furthermore, a recent study suggested a relative safety of celastrol when administered through an oral route [58]. In addition, celastrol can be applied topically for treatment of burn and wound infections caused by CRK. [59]. However the best solution for the toxicity problem is to use celastrol as a lead compound to develop structural analogues with more selectivity to bacterial carbapenemases
The docking results of celastrol and thymol against different carbapenemases revealed the size and bulky structure of celastrol and the distribution of H-donor/ acceptor polar substituents COOH, carbonyl, and OH groups of celastrol were found to be necessary for stabilization of ligand/ receptor complex. While the small size of thymol was found to be less convenient as its binding to protein targets was mainly attributed to its phenolic OH. Of note, the interaction energy expressed in Kcal/mol, reflects how stable is the ligand-protein complex, the lower the interaction energy the more stable the ligand/receptor complex, and the higher the affinity of the compound into the target-receptor.
Since NDM-MβL is one of the most prevalent β-lactamases in Klebsiella, a recent in silico study screened natural compounds as potential inhibitors of NDM. The study identified critical residues for NDM inhibition including Ser251, Asn220, Asp124, Lys211, and His122 residues [60]. Another molecular docking study on NDM inhibitors identified six potent inhibitors with free binding energy ranging from −11 to −12 Kcal mol−1 and reported that H-bonding was mainly through Lys211 and Asn220 residues [61]. Consistent with these studies, our study showed that celastrol and thymol could bind NDM via Lys211, Ser251, and Asn220 amino acid residues.
In addition, a recent study used molecular docking to evaluate the effect of celastrol on some targets for treating thyroid carcinoma [62]. It is worth mentioning that free binding energy scores of celastrol on the selected targets in their study ranged from −7.0 to −9.9 Kcal mol−1 [62], which is much lower than the free binding energy scores in our study (ranged from −11.55 to −12.8 Kcal mol−1) which means superior activity of celastrol on bacterial carbapenemases compared to human targets.
In conclusion, our investigation revealed that celastrol is a promising inhibitor of carbapenemases. Thymol increases the permeability of the Klebsiella envelope to celastrol. Celastrol can be considered as lead compound that could help in the design of new molecules or formulations with more solubility and less toxicity that can be used to restore the activity of carbapenem antibiotics.
Supplementary information
Acknowledgements
The authors would like to acknowledge the medical staff at clinical laboratories in Zagazig University hospitals and El-Ahrar educational hospital for providing clinical specimens. The authors also thank Dr. Fathy Serry (Professor of Microbiology and Immunology) for manuscript proofreading. Celastrol was a kind gift from Dr. Fatma Al-zahraa Yehia, Microbiology and Immunology Department, Zagazig University.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Data availability
The datasets used /or analyzed in the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Ethical approval
Not applicable as the specimens are provided by the hospital clinical laboratory and there was no direct contact with patients.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41429-022-00566-y.
References
- 1.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112:E3574–81. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reyes J, Aguilar AC. Carbapenem-resistant Klebsiella pneumoniae: Microbiology key points for clinical practice. Int J Gen Med. 2019;12:437–36. doi: 10.2147/IJGM.S214305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, et al. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis. 2002;8:160–6. [DOI] [PMC free article] [PubMed]
- 4.Moya C, Maicas S. Antimicrobial resistance in Klebsiella pneumoniae strains: mechanisms and outbreaks. Proceedings. 2020;66:11. doi: 10.3390/proceedings2020066011. [DOI] [Google Scholar]
- 5.Worthington RJ, Melander C. Overcoming resistance to β-lactam antibiotics. J Org Chem. 2013;78:4207–13. doi: 10.1021/jo400236f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han Y, Huang L, Liu C, Huang X, Zheng R, Lu Y, et al. Characterization of carbapenem-resistant Klebsiella pneumoniae ST15 clone coproducing KPC-2, CTX-M-15, and SHV-28 spread in an intensive care unit of a tertiary hospital. Infect Drug Resist. 2021;14:767–73. doi: 10.2147/IDR.S298515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin Infect Dis. 2019;69:S521–8. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vázquez-Ucha JC, Arca-Suárez J, Bou G, Beceiro A. New carbapenemase inhibitors: clearing the way for the β-lactams. Int J Mol Sci. 2020;21:9308. doi: 10.3390/ijms21239308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascão R, Fonseca JE, Moita LF. Celastrol: A spectrum of treatment opportunities in chronic diseases. Front Med. 2017;4:69.. doi: 10.3389/fmed.2017.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo L, Zhang Y, Al-Jamal KT. Recent progress in nanotechnology-based drug carriers for celastrol delivery. Biomater Sci. 2021;9:6355–80. doi: 10.1039/D1BM00639H. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Lee J, Salazar Hernandez MA, Mazitschek R, Ozcan U. Treatment of obesity with celastrol. Cell. 2015;161:999–1011. doi: 10.1016/j.cell.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padilla-Montaño N, de León Guerra L, Moujir L. Antimicrobial activity and mode of action of celastrol, a nortriterpen quinone isolated from natural sources. Foods. 2021;10:591. doi: 10.3390/foods10030591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo S-G, Lee S-Y, Lee S-M, Lim K-H, Ha E-J, Eom Y-B. Activity of novel inhibitors of Staphylococcus aureus biofilms. Folia Microbiologica. 2017;62:157–67. doi: 10.1007/s12223-016-0485-4. [DOI] [PubMed] [Google Scholar]
- 14.Yehia FA, Yousef N, Askoura M. Celastrol mitigates staphyloxanthin biosynthesis and biofilm formation in Staphylococcus aureus via targeting key regulators of virulence; in vitro and in vivo approach. BMC Microbiol. 2022;22:106. doi: 10.1186/s12866-022-02515-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tseng C-K, Hsu S-P, Lin C-K, Wu Y-H, Lee J-C, Young K-C. Celastrol inhibits hepatitis C virus replication by upregulating heme oxygenase-1 via the JNK MAPK/Nrf2 pathway in human hepatoma cells. Antivir Res. 2017;146:191–200. doi: 10.1016/j.antiviral.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma H, Kumar P, Deshmukh RR, Bishayee A, Kumar S. Pentacyclic triterpenes: New tools to fight metabolic syndrome. Phytomedicine. 2018;50:166–77. doi: 10.1016/j.phymed.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Lv X, Chen M, Guo Y, Ding R, Liu B, et al. Characterization of corosolic acid as a KPC-2 Inhibitor that increases the susceptibility of KPC-2-positive bacteria to carbapenems. Front Pharm. 2020;11:1047. doi: 10.3389/fphar.2020.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen DS, Aucken HM, Abiola T, Podschun R. Recommended test panel for differentiation of Klebsiella species on the basis of a trilateral interlaboratory evaluation of 18 biochemical tests. J Clin Microbiol. 2004;42:3665–9. doi: 10.1128/JCM.42.8.3665-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed O, Dablool A. Quality improvement of the DNA extracted by boiling method in Gram negative bacteria. Int J Bioassays. 2017;6:5347–9. doi: 10.21746/ijbio.2017.04.004. [DOI] [Google Scholar]
- 20.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis Version 11. Mol Biol Evol. 2021;38:3022–7. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. Performance standards for antimicrobial susceptibility testing. 28th ed. CLSI supplement M100. 2018. https://clsi.org/media/1930/m100ed28_sample.pdf.
- 23.Pasteran F, Tijet N, Melano RG, Corso A. Simplified protocol for Carba NP test for enhanced detection of carbapenemase producers directly from bacterial cultures. J Clin Microbiol. 2015;53:3908–11. doi: 10.1128/JCM.02032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Ganiny AM, El-mahdy AM, Abd El Latif HK, Ibrahem RH, Abdelsabour HI. Phenotypic and genotypic detection of beta-lactams resistance in Klebsiella pneumonia from Egyptian hospitals revealed carbapenem resistance by OXA and NDM genes. AfrJ Microbiol Res. 2016;10:339–47. doi: 10.5897/AJMR2015.7871. [DOI] [Google Scholar]
- 25.Abbas HA, Kadry AA, Shaker GH, Goda RM. Impact of specific inhibitors on metallo-beta-carbapenemases detected in Escherichia coli and Klebsiella pneumoniae isolates. Micro Pathog. 2019;132:266–74. doi: 10.1016/j.micpath.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Gargotti M, Lopez-Gonzalez U, Byrne HJ, Casey A. Comparative studies of cellular viability levels on 2D and 3D in vitro culture matrices. Cytotechnology. 2018;70:261–73. doi: 10.1007/s10616-017-0139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Routsias JG, Tsakogiannis D, Katsiki M, Marinou D, Mavrouli M, Vrioni G, et al. Development of a new spectrophotometric assay for rapid detection and differentiation of KPC, MBL and OXA-48 carbapenemase-producing Klebsiella pneumoniae clinical isolates. Int J Antimicrob Agents. 2020;56:106211. doi: 10.1016/j.ijantimicag.2020.106211. [DOI] [PubMed] [Google Scholar]
- 28.Pal A, Tripathi A. Quercetin inhibits carbapenemase and efflux pump activities among carbapenem-resistant Gram-negative bacteria. Acta Pathologica, Microbiologica, et Immunologica Scandinavica. 2020;128:251–9. doi: 10.1111/apm.13015. [DOI] [PubMed] [Google Scholar]
- 29.Farrag HA, Abdallah N, Shehata MMK, Awad EM. Natural outer membrane permeabilizers boost antibiotic action against irradiated resistant bacteria. J Biomed Sci. 2019;26:69. doi: 10.1186/s12929-019-0561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alakomi HL, Saarela M, Helander IM. Effect of EDTA on Salmonella enterica serovar Typhimurium involves a component not assignable to lipopolysaccharide release. Microbiology. 2003;149:2015–21. doi: 10.1099/mic.0.26312-0. [DOI] [PubMed] [Google Scholar]
- 31.Petrella S, Ziental-Gelus N, Mayer C, Renard M, Jarlier V, Sougakoff W. Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A β-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrob Agents Chemother. 2008;52:3725–36. doi: 10.1128/aac.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King D, Strynadka N. Crystal structure of New Delhi metallo‐β‐lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011;20:1484–91. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lund BA, Thomassen AM, Carlsen TJO, Leiros HK. Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis. Acta Crystallogr F: Struct Biol Commun. 2017;73:579–87. doi: 10.1107/S2053230X17013838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen JZ, Fowler DM, Tokuriki N. Comprehensive exploration of the translocation, stability and substrate recognition requirements in VIM-2 lactamase. Elife. 2020;9:e56707. doi: 10.7554/eLife.56707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inc CCG. Molecular operating environment (MOE) version 2019.0102. 1010 Sherbooke St. West, Suite# 910, Montreal: Chemical Computing Group Inc; 2019. [Google Scholar]
- 36.Labute P. The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J Comput Chem. 2008;29:1693–8. doi: 10.1002/jcc.20933. [DOI] [PubMed] [Google Scholar]
- 37.Jacquemard C, Drwal MN, Desaphy J, Kellenberger E. Binding mode information improves fragment docking. J Cheminform. 2019;11:24. doi: 10.1186/s13321-019-0346-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherif M, Palmieri M, Mirande C, El-Mahallawy H, Rashed HG, Abd-El-Reheem F, et al. Whole-genome sequencing of Egyptian multidrug-resistant Klebsiella pneumoniae isolates: a multi-center pilot study. Eur J Clin Microbiol Infect Dis. 2021;40:1451–60. doi: 10.1007/s10096-021-04177-7. [DOI] [PubMed] [Google Scholar]
- 39.Flach AJ. Fatal aplastic anemia following topical administration of ophthalmic chloramphenicol. Am J Ophthalmol. 1982;94:420–2. doi: 10.1016/0002-9394(82)90540-2. [DOI] [PubMed] [Google Scholar]
- 40.Pournaras S, Koumaki V, Spanakis N, Gennimata V, Tsakris A. Current perspectives on tigecycline resistance in Enterobacteriaceae: susceptibility testing issues and mechanisms of resistance. Int J Antimicrob Agents. 2016;48:11–18. doi: 10.1016/j.ijantimicag.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 41.Barbour A, Schmidt S, Ma B, Schiefelbein L, Rand KH, Burkhardt O, et al. Clinical pharmacokinetics and pharmacodynamics of tigecycline. Clin Pharmacokinet. 2009;48:575–84. doi: 10.2165/11317100-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 42.Kotb S, Lyman M, Ismail G, Abd El Fattah M, Girgis SA, Etman A, et al. Epidemiology of Carbapenem-resistant Enterobacteriaceae in Egyptian intensive care units using National healthcare-associated infections surveillance data, 2011-2017. Antimicrob Resist Infect Control. 2020;9:2. doi: 10.1186/s13756-019-0639-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–29. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paveenkittiporn W, Lyman M, Biedron C, Chea N, Bunthi C, Kolwaite A, et al. Molecular epidemiology of carbapenem-resistant Enterobacterales in Thailand, 2016-2018. Antimicrob Resist Infect Control. 2021;10:88. doi: 10.1186/s13756-021-00950-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlsson M, Lutgring JD, Ansari U, Lawsin A, Albrecht V, McAllister G, et al. Molecular characterization of carbapenem-resistant Enterobacterales collected in the United States. Micro Drug Resist. 2022;28:389–97. doi: 10.1089/mdr.2021.0106. [DOI] [PubMed] [Google Scholar]
- 46.Abdelaziz NA. Phenotype-genotype correlations among carbapenem-resistant Enterobacterales recovered from four Egyptian hospitals with the report of SPM carbapenemase. Antimicrob Resist Infect Control. 2022;11:13. doi: 10.1186/s13756-022-01061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernández L, Hancock REW. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012;25:661–81. doi: 10.1128/cmr.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lomovskaya O, Sun D, Rubio-Aparicio D, Nelson K, Tsivkovski R, Griffith DC, et al. Vaborbactam: spectrum of beta-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob Agents Chemother. 2017;61:e01443–17. doi: 10.1128/aac.01443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papp-Wallace KM, Barnes MD, Alsop J, Taracila MA, Bethel CR, Becka SA, et al. Relebactam is a potent inhibitor of the KPC-2 β-lactamase and restores imipenem susceptibility in KPC-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2018;62:e00174–18. doi: 10.1128/aac.00174-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zgurskaya HI, López CA, Gnanakaran S. Permeability barrier of Gram-negative cell envelopes and approaches to bypass It. ACS Infect Dis. 2015;1:512–22. doi: 10.1021/acsinfecdis.5b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowalczyk A, Przychodna M, Sopata S, Bodalska A, Fecka I. Thymol and Thyme essential oil new insights into selected therapeutic applications. Molecules. 2020;25:4125. doi: 10.3390/molecules25184125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veras HNH, Rodrigues FFG, Botelho MA, Menezes IRA, Coutinho HDM, Costa JGM. Enhancement of aminoglycosides and β-lactams antibiotic activity by essential oil of Lippia sidoides Cham. and the Thymol. Arab J Chem. 2017;10:S2790–55. doi: 10.1016/j.arabjc.2013.10.030. [DOI] [Google Scholar]
- 53.Yap PSX, Yiap BC, Ping HC, Lim SHE. Essential oils, a new horizon in combating bacterial antibiotic resistance. Open Microbiol J. 2014;8:6–14. doi: 10.2174/1874285801408010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu J, Zhou F, Ji BP, Pei RS, Xu N. The antibacterial mechanism of carvacrol and thymol against Escherichia coli. Lett Appl Microbiol. 2008;47:174–9. doi: 10.1111/j.1472-765x.2008.02407.x. [DOI] [PubMed] [Google Scholar]
- 55.Li Z, Guo Z, Chu D, Feng H, Zhang J, Zhu L. Effectively suppressed angiogenesis-mediated retinoblastoma growth using celastrol nanomicelles. Drug Deliv. 2020;27:358–66. doi: 10.1080/10717544.2020.1730522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi J, Li J, Xu Z, Chen L, Luo R, Zhang C, et al. Celastrol: a review of useful strategies overcoming its limitation in anticancer application. Front Pharmacol. 2020;11:558741. doi: 10.3389/fphar.2020.558741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo L, Luo S, Du ZW, Zhou ML, Li PW, Fu Y, et al. Targeted delivery of celastrol to mesangial cells is effective against mesangio-proliferative glomerulonephritis. Nat Commun. 2017;8:878. doi: 10.1038/s41467-017-00834-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu W, Wang L, Du G, Guan Q, Dong T, Song L, et al. Effects of microbiota on the treatment of obesity with the natural product Celastrol in rats. Diabetes Metab J. 2020;44:747–63. doi: 10.4093/dmj.2019.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chhibber S, Gondil VS, Singla L, Kumar M, Chhibber T, Sharma G, et al. Effective topical delivery of H-AgNPs for eradication of Klebsiella pneumoniae–induced burn wound infection. AAPS PharmSciTech. 2019;20:169. doi: 10.1208/s12249-019-1350-y. [DOI] [PubMed] [Google Scholar]
- 60.Salari-jazi A, Mahnam K, Sadeghi P, Damavandi MS, Faghri J. Discovery of potential inhibitors against New Delhi metallo-β-lactamase-1 from natural compounds: in silico-based methods. Sci Rep. 2021;11:2390. doi: 10.1038/s41598-021-82009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thiyagarajan CTP. Virtual screening and molecular docking of Ndm1 inhibitor for treatment of Klebsiella pneumonia infection. Int J Life Sci Pharma Res. 2016;6:L50–57. [Google Scholar]
- 62.Yi J, Tian M, Hu L, Kang N, Ma W, Zhi J, et al. The mechanisms of celastrol in treating papillary thyroid carcinoma based on network pharmacology and experiment verification. Ann Transl Med. 2021;9:866. doi: 10.21037/atm-21-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used /or analyzed in the current study are available from the corresponding author on reasonable request.