Abstract
Background: Systemic lupus erythematosus (SLE) is a chronic disease that causes inflammation in cartilage and the lining of blood vessels. Emerging evidence implicates IFN-γ as a major effector molecule in SLE during both active and stable stages. Here, we investigated the effects of IFN-γ on cytokines that play an autoimmune disease-promoting role and Th1-versus-Th2 and B cell dualism in SLE patients and mouse models of SLE. Methods: The levels of pro-inflammatory factors CXCL11, IFN-γ, IL-1β and IL-4, and immune complexes IgG, anti-dsDNA and anti-RNP were assessed through enzyme-linked immunosorbent assays (ELISA). Flow cytometry was performed to measure Th1, Th2 and B cell counts and IFNGR1, IFNGR2, pSTAT1 and TBX21 expression. The pathology of renal tissue from mouse SLE models was investigated through Hematoxylin eosin (H&E) staining. The levels of IgG, anti-dsDNA and anti-RNP were determined through immunofluorescence (IF) assays. Results: Skin damage was observed in SLE patients in both active and stable stages. ELISA analysis showed that SLE patients displayed higher levels of pro-inflammatory factors (CXCL11, IFN-γ, IL-1β and IL-4) and immune complexes (IgG, anti-dsDNA and anti-RNP). The percentage of Th1 and B cells was increased in blood samples from SLE patients with skin lesions (SL) or lupus nephritis (LN). The percentage of Th2 cells among the groups were comparable. Higher levels of IFNGR1, IFNGR2, pSTAT1 and TBX21 were observed in Th1 but not Th2 cells. In SLE mouse models, H&E staining revealed fewer immune complexes in glomerular endothelial cells and decreased hyaline thrombus in the capillary lumen following treatment with anti-IFN-γ antibodies or following IFNGR1 or STAT1 silencing. Conclusion: IFN-γ contributes to the pathogenesis of SLE through the IFNGR1/2-pSTAT1-TBX21 axis and regulates inflammation and immune complex formation in SLE mice.
Keywords: SLE, IFN-γ, inflammation, immune complexes, Th1/2 cells
Introduction
Systemic lupus erythematosus (SLE) is the most common form of lupus, a chronic autoimmune disease that causes severe fatigue and joint pain [1,2]. The etiology of SLE includes environmental factors, genetic predisposition, altered estrogen levels, loss or dysfunction of T lymphocytes, exorbitant proliferation of B cells and the presence of autoantibodies against nuclear antigens (ANA) that target double-stranded DNA (dsDNA) [3,4]. Autoantibodies combine with autoantigens to create immune complexes that deposit in the joints, skin, glomerulus and small blood vessels, resulting in connective tissue damage [5-7].
Cytokines play a pivotal role in the immune response to auto-antigens [8,9]. Interleukin- (IL-)4, IL-1β, IFN-γ and CXCL11 are elevated in SLE [10,11], and immune complexes (IgG, anti-dsDNA and anti-RNP) are key factors during SLE progression [12]. Lupus nephritis (LN) involves immune accumulation at the sites of glomerular and tubular injury [13]. The levels of anti-dsDNA production are also related to disease severity [14]. High levels of proinflammatory cytokines and immune complexes are implicated in SLE progression, but their regulatory mechanisms remain poorly characterized [15,16].
Type I and type II interferons (IFN) are central to the immune response against virus infection. Interferon-γ (IFN-γ) is a type II interferon that is released from Th1 cells, cytotoxic T cells, macrophages, mucosal epithelial cells and NK cells [17]. Its expression is induced by mitogens and cytokines including IL-12, IL-15, IL-18, and type I IFN. IFN-γ is both an important autocrine signal for Antigen-Presenting-Cells (APCs) in the early innate immune response and a key paracrine signal in the adaptive immune system [18,19]. IFN-γ is encoded by the IFNG gene and consists of two antiparallel polypeptide chains. The primary role of IFN-γ is the activation of macrophages to increase phagocytosis and the intracellular destruction of invading pathogens. The IFN-γ receptor is composed of two ligand-binding IFNγR1 chains associated with two signal-transducing IFNγR2 chains that are responsible for signal transduction, also, IFN-γ activates macrophages through a variety of inflammatory, nitrogen and ROS intermediates [20-22]. IFN-γ also downregulates tissue damage associated with inflammation [23]. Furthermore, IFN-γ has been shown to stimulate the pSTAT1-TBX21 axis to facilitate Th1 cell differentiation/activation, the formation of B-cell complexes, and the inhibition of Th2 cell activity. Cellular responses induced by IFN-γ also involve cross-communication with IFN-α/β receptors, which amplify IFN-γ signaling.
In this study, we investigated the role of IFN-γ in the pathogenesis of SLE to verify if IFN modulates the IFNGR1/2-pSTAT1-TBX21 axis during SLE to promote disease progression.
Materials and methods
Samples
This study was approved by the Ethics Committee of the First Affiliated Hospital of China Medical University (AF-SOP-07-1.1-01). All participants signed an informed consent. SLE patients (n=60) were recruited from 2020 to 2022 to provide blood samples. The cohort included 30 SLE patients with skin lesions (SL) and 30 SLE cases with lupus nephritis (LN). Patients were required to meet at least four of the SLE classification criteria of the American College of Rheumatology [24]. Disease activity was evaluated using the Disease Activity Index (SLEDAI) [25]. SLEDAI ≤6 was classified as inactive SLE. SLEDAI ≥6 was classified as active SLE. For inactive patients, oral prednisone (≤0.8 mg/kg/day) with or without immunosuppressive drugs was provided for treatment. For active SLE patients, oral prednisone (≥1 mg/kg/day), intravenous methylprednisolone (40-80 mg/day) and pulsed methylprednisolone (1 g/day, for 3 days), combined with at least one immunosuppressive drug was administered. A total of 30 healthy volunteers with no history of autoimmune disease were randomly selected as a healthy control (HC) group. The inclusion criteria for SLE patients were as follows: no other immune diseases, complete medical records and compliance with examinations. Clinical characteristics of the patients and their classifications of SLE-LP or SLE-LN are shown in Table 1.
Table 1.
Clinical characteristics of SLE patients and healthy controls
| Clinical characteristics | SLE (n=60) | Healthy control (n=30) |
|---|---|---|
| Sex, male/female | 5/60 | 3/30 |
| Age (year) | 32.0 ± 8.10 | 29.0 ± 6.65 |
| Duration (year) | 9.00 ± 4.30 | - |
| SLEDAI scores | 10.6 ± 3.67 | - |
| Disease active, n (%) | 42 (70.0) | - |
| Skin, n (%) | 39 (65.0) | - |
| Kidney, n (%) | 37 (61.7) | - |
| Anti-ds-DNA antibody (IU/ml) | 103.1 ± 20.64 | - |
| CRP (mg/L) | 4.80 ± 1.45 | - |
| ESR (mmH2O) | 31.0 ± 12.8 | - |
| IgG (g/L) | 20.6 ± 4.33 | - |
| IgA (g/L) | 4.81 ± 1.01 | - |
| IgM (g/L) | 1.87 ± 0.46 | - |
| C3 level (g/L) | 0.38 ± 0.07 | - |
| C4 level (g/L) | 0.03 ± 0.01 | - |
| 24 h urine protein (g) | 1.36 ± 0.40 | - |
| Mean prednisone dose, mg/day | 28.1 | - |
| Chloroquine, n (%) | 40 (66.7) | - |
| Immunosuppressantsa, n (%) | 38 (63.3) | - |
AZA, LEF, MMF and cyclophosphamide.
Plasmids and antibodies
Short hairpin RNA (shRNAs) targeting IFNGR1 (sh-IFNGR1), STAT1 (sh-STAT1) and negative controls (sh-NC) were purchased from GenePharma (Shanghai, China). Neutralizing anti-IFN-γ antibodies were purchased from R&D Systems. shRNA sequences were as follows: sh-IFNGR1: 5’-CCGGCCACATAGAATATCAGACTTACTCGAGTAAGTCTGATATTCTATGTGGTTTTTG-3’; sh-NC: 5’-TTCTCCGAACGTGTCACGT-3’.
SLE mouse models
Animal studies were approved by the Ethics Committee of Medical Experimental Animals of the First Affiliated Hospital, China Medical University (AF-SOP-07-1.1-01). Twelve-week-old MRL/lpr female mice (B6.MRL-Faslpr/Nju, n=60) were acquired from the Experiment Animal Center of the Chinese Academy of Medical Sciences (Beijing, China). The mice were divided into the following groups (n=10 per-group): 1) normal saline controls (n=10), 2) intraperitoneal (ip) injection of Lipopolysaccharide (LPS) (75 μg/20 g) to induce SLE, 3) LPS plus injection of anti-IFN-γ antibodies into the tail vein, 4) LPS plus sh-NC, 5) LPS plus sh-IFNGR1, 6) LPS plus sh-STAT1. After 4 weeks, urine samples were collected. The criterion of urine protein >0.5 g was used to confirm successful construction of the model. The mice were anesthetized with isopentane, then abdominal aorta blood, spleen and kidney tissues were collected [26,27].
Hematoxylin & eosin (H&E) staining
Kidney tissues were heated to 60°C, cleared in xylene, and immersed in ethanol at gradient concentrations. Sections were H&E stained (Jrdun Biotechnology Co, Ltd, Shanghai, China) and then imaged under a microscope.
Enzyme-linked immunosorbent assays (ELISA)
Early morning fasting venous blood from SLE patients was collected, and the serum was separated. CXCL11 (ab289695, Abcam, Shanghai, China), IFN-γ (ab282874), IL-1β (ab214025), IL-4 (ab215089), IgG (ab151276), anti-dsDNA (ab287882) and anti-RNP (FS-Ea-04043, R&D Systems, USA) levels were assessed using commercial ELISA kits.
Identification of Th1, Th2 and B cells by Flow cytometry
Early morning fasting venous blood was collected to isolate and quantify PBMCs, Th1, Th2 and B cells by Flow cytometry (Beckman, USA) as previously described [28]. The percentage Th1, Th2, and B cells positive for IFNGR1, IFNGR2, pSTAT1, TBX21 were also measured through Flow cytometry.
Immunofluorescence staining
Mouse kidney tissues were washed in PBS and fixed in 4% paraformaldehyde. Tissues were blocked in 5% BSA and probed with anti-IgG, anti-dsDNA, anti-RNP primary antibodies. Tissues were washed and stained with the appropriate fluorescent-conjugated secondary antibodies and counterstained with 6-Diamidino-2’-phenylindole dihydrochloride (DAPI) (Beyotime Institute of Biotechnology). Cells were imaged on a fluorescent microscope (Olympus Corporation) at 400x magnification.
Statistical analyses
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA). Categorical data were expressed as numbers or percentages and compared using a Chi-squared test or continuity correction as appropriate. Data from triplicate samples were represented as mean ± standard deviation (SD), and differences were compared using a Student’s t-test for pairwise comparisons or a one-way-ANOVA with Tukey post-hoc assessment for multigroup comparisons. P<0.05 was deemed statistically significant.
Results
Patient characteristics
Butterfly rashes and skin damage typically occur in about 52% of SLE patients at the time of diagnosis, which strongly correlates with clinical activity. Figure 1 shows extensive skin damage within our SLE cohort during active disease stages. This damage included erythema around the nails and a vasculitis rash on the hands (Figure 1, P1), bullous lupus erythematosus on the feet (Figure 1, P2), and butterfly erythema on the face (Figure 1, P3-P4).
Figure 1.
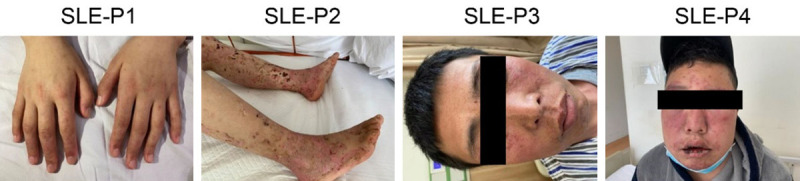
Skin damage in SLE patients. P1 shows erythema around the nails and a vasculitis rash on the hands. P2 shows bullous lupus erythematosus on the feet. P3-4 show butterfly erythema. SLE: systemic lupus erythematosus.
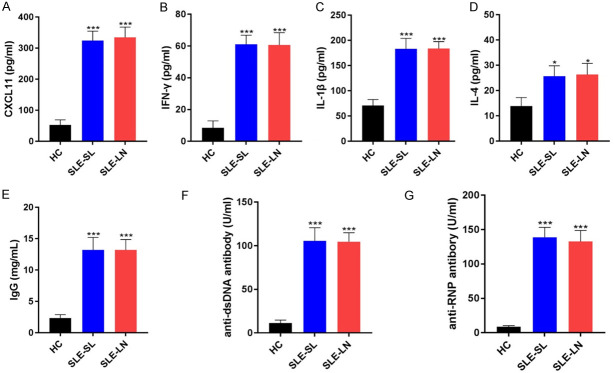
The clinical characteristics of the SLE patients and healthy controls are shown in Table 1. The levels of pro-inflammatory factors CXCL11, IFN-γ, IL-1β and IL-4 were enhanced in SLE-SL and SLE-LN groups compared with those in the HC group (Figure 2A-D). Additionally, the levels of immune complexes IgG, anti-dsDNA and anti-RNP were upregulated in SLE-SL and SLE-LN groups (***P<0.001 vs. HC Figure 2E-G).
Figure 2.
Inflammatory indicators in the blood samples of SLE patients. A-D. Levels of pro-inflammatory CXCL11, IFN-γ, IL-1β and IL-4 measured by ELISA. E-G. Immune complexes IgG, anti-dsDNA and anti-RNP levels. *P<0.05, ***P<0.001 vs. HC group. SLE: systemic lupus erythematosus; HC: healthy controls; SL: skin lesions; LN: lupus nephritis.
Th1, Th2 and B cell percentages in blood samples from SLE patients
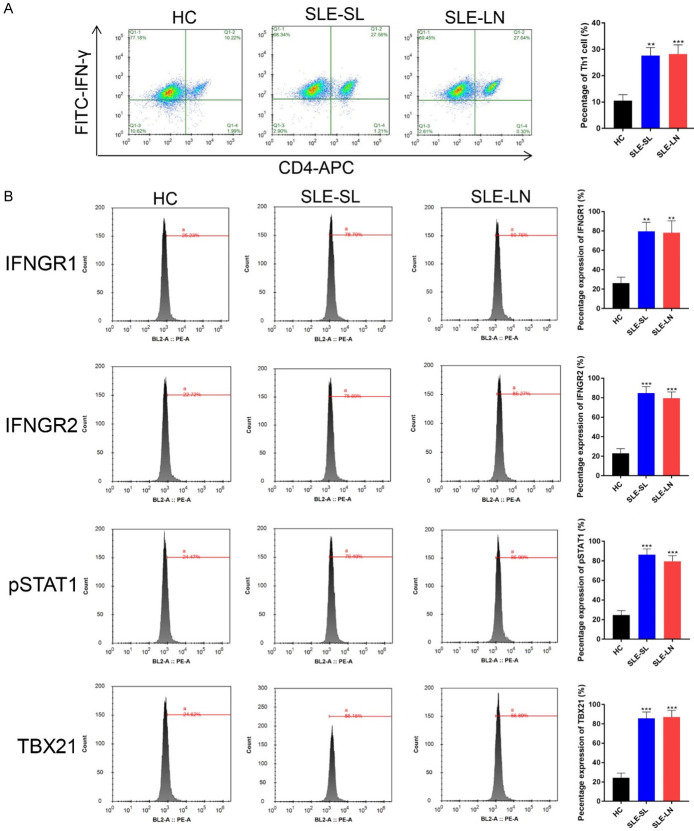
The percentage of Th1 cells increased in the blood samples of SLE patients with SL or LN (Figure 3A). The expression of IFNGR1, IFNGR2, pSTAT1 and TBX21 were upregulated in Th1 cells (Figure 3B). The percentage of Th2 cells showed no differences in SL or LN patients compared to that of healthy volunteers (Supplementary Figure 1A). The expression of IFNGR1, IFNGR2, pSTAT1 and TBX21 also showed no changes in SL or LN Th2 cells (Supplementary Figure 1B). The percentage of B cells was higher in blood samples from all SLE cases (Supplementary Figure 2).
Figure 3.
Th1 cells in the blood samples of SLE patients. A. Th1 cells in the indicated groups were examined by Flow cytometry. B. Expression of IFNGR1, IFNGR2, pSTAT1 and TBX21 assessed by Flow cytometry. **P<0.01, ***P<0.001 vs. HC group. SLE: systemic lupus erythematosus; HC: healthy controls; SL: skin lesion; LN: lupus nephritis.
Correlation of pro-inflammatory factors, immune complexes and cell types in blood samples from SLE patients
A positive correlation between CXCL11, IFN-γ, IL-1β, IL-4 and IgG, or anti-dsDNA, anti-RNP in SLE-SL and SLE-LN cases was confirmed (Figure 4A-C). Moreover, Th1 and B cells were positive for IgG, anti-dsDNA and anti-RNP in SLE cases with SL or LN (Figure 4D and 4F). IL-4 level negatively correlated with IgG, anti-dsDNA and anti-RNP in the blood of SLE patients (Figure 4E). Collectively, these results demonstrate a correlation between pro-inflammatory factors, immune complexes and immune cells in the blood samples of SLE patients.
Figure 4.
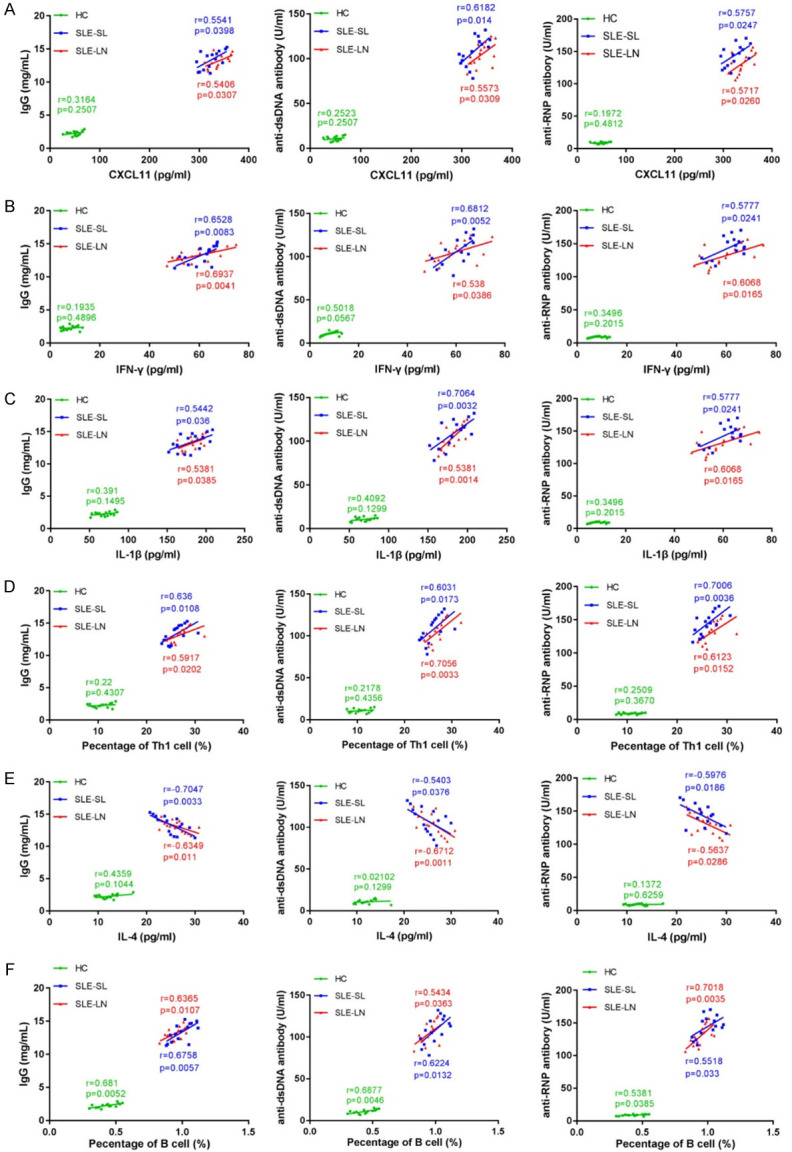
Correlation between pro-inflammatory factors and immune complexes in specific cell types. A-D. Correlation between CXCL11 or IFN-γ, IL-1β, IL-4 and IgG or anti-dsDNA, anti-RNP. E, F. Correlation between Th1 cells, B cells and IgG, or anti-dsDNA and anti-RNP. SLE: systemic lupus erythematosus; HC: healthy controls; SL: skin lesions; LN: lupus nephritis.
Pathology of renal tissue from mouse SLE models
Mouse SLE models were constructed to verify the role of IFN-γ during SLE progression. As shown in Figure 5, H&E staining revealed high deposition of immune complexes in glomerular endothelial cells and decreased hyaline thrombus in the capillary lumen in SLE models. Following the injection of anti-IFN-γ antibodies or IFNGR1/STAT1b silencing, these classic SLE phenotypes were alleviated. This suggested that IFN-γ, IFNGR1 and STAT1 were involved in the progression of SLE.
Figure 5.
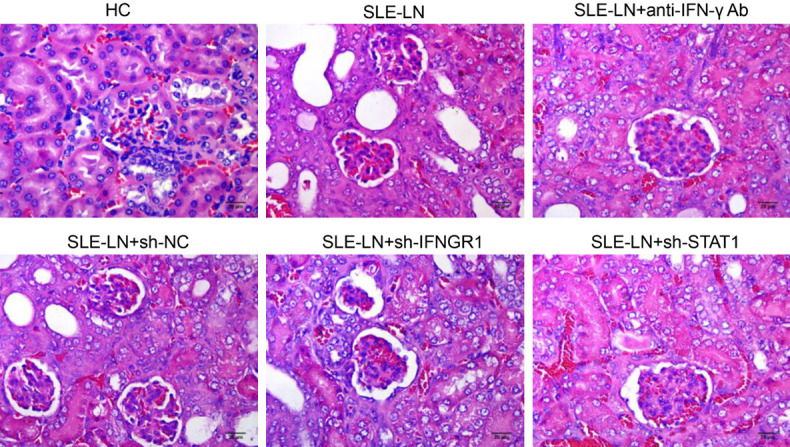
Pathology of mouse renal tissue. Pathology of H&E staining tissue sections from the indicated groups (n=10) (400X). SLE: systemic lupus erythematosus; HC: healthy control; LN: lupus nephritis; NC: negative control. Scale bar: 20 μm.
IFN-γ modulates the IFNGR1/2-pSTAT1-TBX21 axis and regulates inflammation and immune complexes in SLE models
The levels of pro-inflammatory CXCL11, IFN-γ, IL-1β and IL-4 decreased following treatment with anti-IFN-γ antibodies or IFNGR1/STAT1 shRNA (Figure 6A-D). Moreover, the levels of immune complexes (IgG, anti-dsDNA and anti-RNP) were markedly down-regulated following anti-IFN-γ antibody injection or IFNGR1/STAT1 knockdown (Figure 6E-G). Comparable data were observed in renal tissues by IF analysis (Figure 7). These results demonstrate lower levels of inflammation in mouse models following anti-IFN-γ antibody treatment or IFNGR1/STAT1 suppression.
Figure 6.
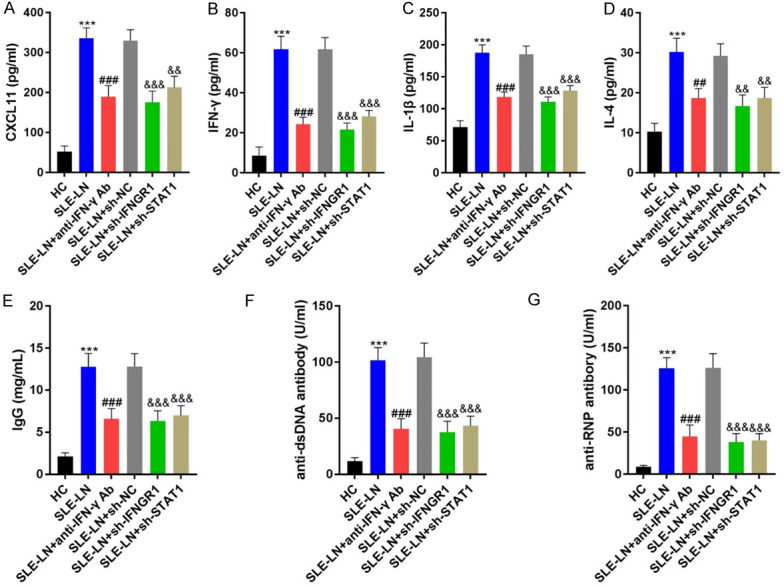
Inflammatory indicators in blood samples from mouse models. A-D. Levels of CXCL11, IFN-γ, IL-1β and IL-4 in the indicated groups were measured by ELISA. E-G. Levels of immune complexes, IgG, anti-dsDNA and anti-RNP. ***P<0.001 vs. HC group; ###P<0.001 vs. SLE-LN group; &&P<0.01, &&&P<0.001 vs. SLE-LN+sh-NC group. SLE: systemic lupus erythematosus; HC: healthy controls; LN: lupus nephritis; NC: negative controls.
Figure 7.
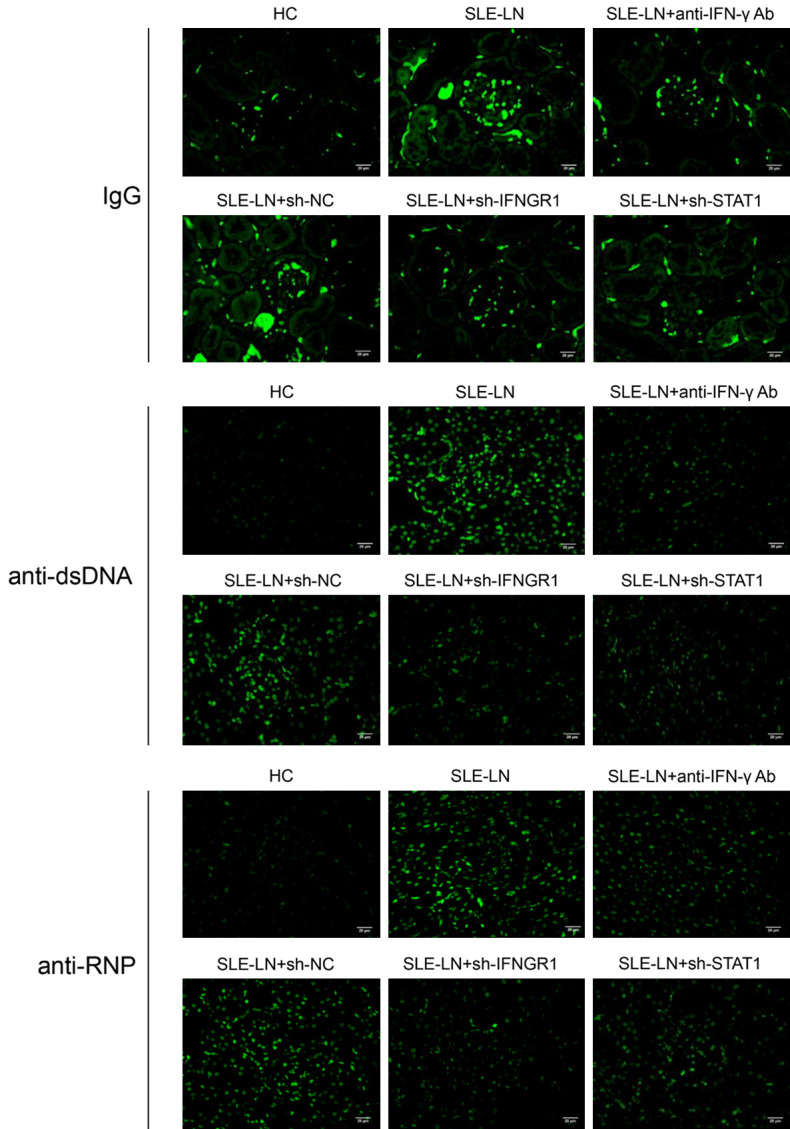
Immunofluorescent analysis of IgG, anti-dsDNA and anti-RNP levels in kidney sections from mouse SLE models. Sections (mouse kidney tissue) from the indicated groups were stained for IgG, anti-dsDNA and anti-RNP. SLE: systemic lupus erythematosus; HC: healthy controls; LN: lupus nephritis; NC: negative controls. (Magnification: 400X, scale bar: 20 μm).
IFN-γ modulates the IFNGR1/2-pSTAT1-TBX21 axis in SLE models
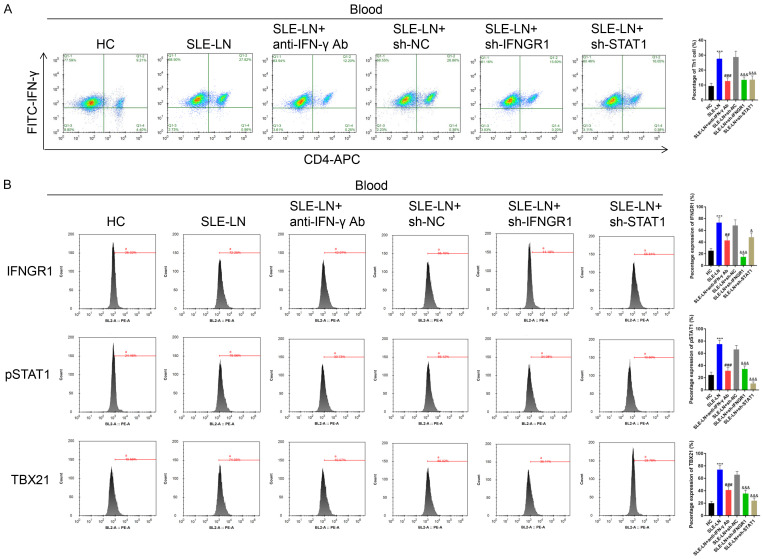
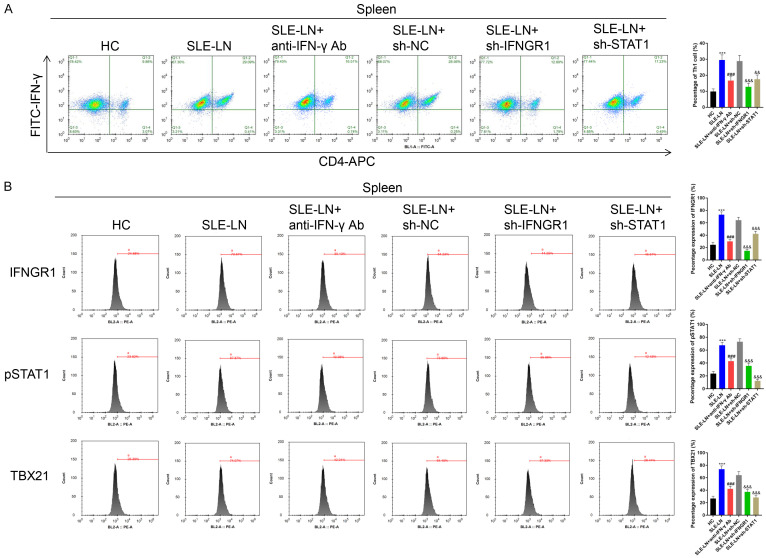
Th1 cell counts in blood and spleen samples decreased following anti-IFN-γ antibody injection or the silencing of IFNGR1/STAT1 in SLE models (Figures 8A and 9A). The levels of IFNGR1, pSTAT1 and TBX21 were also downregulated following treatment with anti-IFN-γ or IFNGR1/STAT1 silencing (Figures 8B and 9B). Th2 cell counts increased following treatment with anti-IFN-γ antibodies or the suppression of IFNGR1/STAT1 (Supplementary Figures 3A and 4A). The level of IFNGR1 was also up-regulated in the SLE-LN+anti-IFN-γ Ab group compared to that of the SLE-LN group. Compared to that of the SLE-LN+sh-NC group, IFNGR1 level was upregulated following STAT1 silencing. The levels of pSTAT1 and TBX21 were unaffected by anti-IFN-γ antibody treatment or IFNGR1 silencing, but decreased following STAT1 silencing in blood and spleen samples (Supplementary Figures 3B and 4B). Moreover, B cell counts in the blood and spleen samples of SLE mice decreased following anti-IFN-γ antibody treatment and IFNGR1/STAT1 silencing (Supplementary Figure 5). Collectively, these data suggest that IFN-γ contributes to the pathogenesis of SLE through the IFNGR1/2-pSTAT1-TBX21 axis through the regulation of inflammation and immune complex formation in SLE mice.
Figure 8.
Th1 cells in blood samples from SLE mouse models. A. Th1 cells in the indicated groups were analyzed by Flow cytometry. B. Expression of IFNGR1, pSTAT1, IFNGR2 and TBX21 determined by Flow cytometry. ***P<0.001 vs. HC group; ###P<0.001 vs. SLE-LN group; &&&P<0.001 vs. SLE-LN+sh-NC group. SLE: systemic lupus erythematosus; HC: healthy controls; LN: lupus nephritis; NC: negative control.
Figure 9.
Th1 cells in spleen samples of SLE mouse models. A. Th1 cells in spleen samples obtained from the indicated groups were examined by Flow cytometry. B. Expression of IFNGR1, pSTAT1, IFNGR2 and TBX21 assessed by Flow cytometry. ***P<0.001 vs. HC group; ###P<0.001 vs. SLE-LN group; &&&P<0.001 vs. SLE-LN+sh-NC group. SLE: systemic lupus erythematosus; HC: healthy controls; LN: lupus nephritis; NC: negative controls.
Discussion
SLE is an autoimmune disease characterized by the generation of autoantibodies [29]. SLE is predominant in women and presents as a range of complex clinical manifestations [30,31]. The discovery of novel biomarkers is of significance to SLE diagnostics and can drive subsequent therapies. NCS 613 is a PDE4 inhibitor that has shown utility in the treatment of SLE through its ability to enhance cAMP levels, suppress systemic inflammation and reduce immune complex deposition [32]. In TLR7-induced SLE, interferon lambda accelerates immune dysregulation and inflammation [33]. The cytomegalovirus protein US31 modulates mono-macrophages in SLE to stimulate inflammation through NFκB2 activation [34]. Let-7f-5p targets NLRP3 to suppress inflammation in SLE patients [35]. In this study, we highlight the occurrence of skin damage during both active and stable SLE stages and confirm the role of the IFNGR1/2-STAT1-TBX21 axis during SLE progression. Our results showed that SLE patients express higher levels of pro-inflammatory CXCL11, IFN-γ, IL-1β and IL-4, and immune complexes IgG, anti-dsDNA and anti-RNP. In SLE mouse models, treatment with anti-IFN-γ antibodies and IFNGR1/STAT1 silencing suppressed immune complex formation in glomerular endothelial cells and decreased inflammation. These data highlight IFN-γ and its downstream effectors as novel therapeutic targets for much-needed anti-SLE therapeutics.
Th cells offer an auxiliary function to cells within the immune system, particularly antigen-presenting cells (APCs) including dendritic cells, macrophages and B cells [36]. CD4+ Th cells are diverse and include Th1, Th2, Th17 and regulatory T cells [37]. Immune cells are sensitized by a specific signature of cytokines and transcription factors [38]. Th1 cells secrete IL-2, IFN-γ, TNF-β and other cytokines [39] and regulate immune responses during organ transplantation rejection, organ specific autoimmune disease and infection [40]. Th2 cells secrete IL-4, IL-6, IL-10, IL-13 and other cytokines, regulate humoral immune responses and induce allergic reactions [41]. Th1, Th2 and B cells have been implicated in SLE progression [10,42,43].
In agreement with previous studies, we found that the percentage of Th1 and B cells increased in the blood samples of SLE patients with SL or LN, whilst Th2 cell percentages decreased. During SLE progression, IRF-8/miR-451a modulates AMPK/mTOR signaling to influence the differentiation of Myeloid-derived suppressor cells (MDSCs) [44]. IL-2 combined with rapamycin in refractory SLE patients improves the long-term balance of Th17/Treg cells [45]. EIF4EBP1 (Eukaryotic Translation Initiation Factor 4E Binding Protein 1) is stabilized by miR-99a-3p and regulates B lymphocyte autophagy to accelerate SLE progression [27]. MiR-301a-3p targets PELI1 (Pellino E3 Ubiquitin Protein Ligase 1) to facilitate the IRAK1 (Interleukin-1 receptor-associated kinase 1) mediated differentiation of Th17 cells, promoting SLE development [46,47].
The IFN-γ receptor (IFNGR) is a heterodimeric receptor composed of IFNGR-1 (α chain) and IFNGR-2 (β chain) chains. IFN-γ binds to the IFNGR-1 subunit with the highest affinity [48]. The interaction between IFNGR1/2 and IFN-γ stimulates Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathways, resulting in the activation, phosphorylation and dimerization of STAT1 [49,50]. STAT1 homodimers then translocate to the nucleus to initiate transcription [51,52]. T-Box Transcription Factor 21 (TBX21, also termed TBET) is a Lineage-defining transcription factor that stimulates Th1 progression from naive Th precursor cells and suppresses Th2 and Th17 through genetic re-programming [53,54].
IFN-γ-STAT1 signaling has been linked to several disease states. The iminosugar derivative WGN-26 inhibits IFN-γ/p-STAT1/T-bet signaling to relieve acute allograft rejection [55]. Suppression of the long non-coding RNA (lncRNA) Sros1 stabilizes Stat1 mRNA to enhance the innate immune response mediated by IFN-γ [56]. The Apelin Receptor (APLNR) modulates IFN-γ signaling in melanoma cells through β-arrestin 1 mediated JAK-STAT1 signaling [57]. The IFN-γ/SOCS1/JAK/STAT1 axis promotes human trophoblast invasion in preeclampsia [58]. SPRY4 modulates IFN-γ-stimulated STAT1 expression in recurrent miscarriages through its effects on trophoblast proliferation and apoptosis [59]. IL-11 inhibits IFN-γ/STAT1 signaling and ROS scavenging to prevent IFN-γ-triggered hepatocyte death [60].
IFN-γ similarly influences SLE development [61]. Increased fatty acid synthesis results in the overproduction of IFN-γ and an imbalance in T helper 1 cells [62]. Elevated level of IFN-γ in patients with SLE activates the B lymphocyte stimulator/B cell-activating factor/TNF ligand superfamily-13B [63]. Splicing factor SRSF1 modulates the G-protein RhoH to reduce IFN-γ production, thereby relieving nephritis [64]. Here, we observed an upregulation of IFNGR1, IFNGR2, pSTAT1 and TBX21 levels in Th1 and B cells, but not in Th2 cells. In the mouse models, H&E staining showed that the deposition of immune complexes in glomerular endothelial cells and the hyaline thrombus in the capillary lumen decreased following treatment with anti-IFN-γ antibodies or IFNGR1 or STAT1 shRNA. This indicated that IFN-γ also modulated the IFNGR1/2-pSTAT1-TBX21 signaling axis.
In summary, we revealed new information regarding the role of the IFN-γ/IFNGR1/2-STAT1-TBX21 signaling axis during SLE progression. Further studies investigating the effects of IFN-γ during inflammation, immune activation and oxidative stress in SLE are now warranted to fully realize the therapeutic potential of targeting this signaling axis for the treatment of SLE.
Disclosure of conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting Information
References
- 1.Fortuna G, Brennan MT. Systemic lupus erythematosus: epidemiology, pathophysiology, manifestations, and management. Dent Clin North Am. 2013;57:631–55. doi: 10.1016/j.cden.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Kiriakidou M, Ching CL. Systemic lupus erythematosus. Ann Intern Med. 2020;172:ITC81–ITC96. doi: 10.7326/AITC202006020. [DOI] [PubMed] [Google Scholar]
- 3.Sharabi A, Tsokos GC. T cell metabolism: new insights in systemic lupus erythematosus pathogenesis and therapy. Nat Rev Rheumatol. 2020;16:100–112. doi: 10.1038/s41584-019-0356-x. [DOI] [PubMed] [Google Scholar]
- 4.Shan J, Jin H, Xu Y. T cell metabolism: a new perspective on Th17/Treg cell imbalance in systemic lupus erythematosus. Front Immunol. 2020;11:1027. doi: 10.3389/fimmu.2020.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truedsson L, Bengtsson AA, Sturfelt G. Complement deficiencies and systemic lupus erythematosus. Autoimmunity. 2007;40:560–6. doi: 10.1080/08916930701510673. [DOI] [PubMed] [Google Scholar]
- 6.Scherlinger M, Guillotin V, Truchetet ME, Contin-Bordes C, Sisirak V, Duffau P, Lazaro E, Richez C, Blanco P. Systemic lupus erythematosus and systemic sclerosis: all roads lead to platelets. Autoimmun Rev. 2018;17:625–635. doi: 10.1016/j.autrev.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Clough JD. Role of autoantibodies and immune complexes in the pathogenesis of systemic lupus erythematosus. J Clin Apher. 1992;7:151–2. doi: 10.1002/jca.2920070313. [DOI] [PubMed] [Google Scholar]
- 8.Voet S, Srinivasan S, Lamkanfi M, van Loo G. Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol Med. 2019;11:e10248. doi: 10.15252/emmm.201810248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramani T, Auletta CS, Weinstock D, Mounho-Zamora B, Ryan PC, Salcedo TW, Bannish G. Cytokines: the good, the bad, and the deadly. Int J Toxicol. 2015;34:355–65. doi: 10.1177/1091581815584918. [DOI] [PubMed] [Google Scholar]
- 10.Guimarães PM, Scavuzzi BM, Stadtlober NP, Franchi Santos LFDR, Lozovoy MAB, Iriyoda TMV, Costa NT, Reiche EMV, Maes M, Dichi I, Simão ANC. Cytokines in systemic lupus erythematosus: far beyond Th1/Th2 dualism lupus: cytokine profiles. Immunol Cell Biol. 2017;95:824–831. doi: 10.1038/icb.2017.53. [DOI] [PubMed] [Google Scholar]
- 11.Raymond WD, Eilertsen GØ, Shanmugakumar S, Nossent JC. The impact of cytokines on the health-related quality of life in patients with systemic lupus erythematosus. J Clin Med. 2019;8:857. doi: 10.3390/jcm8060857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tumurkhuu G, Chen S, Montano EN, Ercan Laguna D, De Los Santos G, Yu JM, Lane M, Yamashita M, Markman JL, Blanco LP, Kaplan MJ, Shimada K, Crother TR, Ishimori M, Wallace DJ, Jefferies CA, Arditi M. Oxidative DNA damage accelerates skin inflammation in pristane-induced lupus model. Front Immunol. 2020;11:554725. doi: 10.3389/fimmu.2020.554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anders HJ, Saxena R, Zhao MH, Parodis I, Salmon JE, Mohan C. Lupus nephritis. Nat Rev Dis Primers. 2020;6:7. doi: 10.1038/s41572-019-0141-9. [DOI] [PubMed] [Google Scholar]
- 14.Rekvig OP. The dsDNA, anti-dsDNA antibody, and lupus nephritis: what we agree on, what must be done, and what the best strategy forward could be. Front Immunol. 2019;10:1104. doi: 10.3389/fimmu.2019.01104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, Li QZ, Yan M, Janke L, Guy C, Linkermann A, Virgin HW, Green DR. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;533:115–9. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Mobarrez F, Fuzzi E, Gunnarsson I, Larsson A, Eketjäll S, Pisetsky DS, Svenungsson E. Microparticles in the blood of patients with SLE: size, content of mitochondria and role in circulating immune complexes. J Autoimmun. 2019;102:142–149. doi: 10.1016/j.jaut.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Kak G, Raza M, Tiwari BK. Interferon-gamma (IFN-γ): exploring its implications in infectious diseases. Biomol Concepts. 2018;9:64–79. doi: 10.1515/bmc-2018-0007. [DOI] [PubMed] [Google Scholar]
- 18.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–89. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 19.Alspach E, Lussier DM, Schreiber RD. Interferon γ and its important roles in promoting and inhibiting spontaneous and therapeutic cancer immunity. Cold Spring Harb Perspect Biol. 2019;11:a028480. doi: 10.1101/cshperspect.a028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arakawa T, Hsu YR, Chang D, Stebbing N, Altrock B. Structure and activity of glycosylated human interferon-gamma. J Interferon Res. 1986;6:687–95. doi: 10.1089/jir.1986.6.687. [DOI] [PubMed] [Google Scholar]
- 21.Ribeiro EB, de Marchi PGF, Honorio-França AC, França EL, Soler MAG. Interferon-gamma carrying nanoemulsion with immunomodulatory and anti-tumor activities. J Biomed Mater Res A. 2020;108:234–245. doi: 10.1002/jbm.a.36808. [DOI] [PubMed] [Google Scholar]
- 22.Jurado A, Carballido J, Griffel H, Hochkeppel HK, Wetzel GD. The immunomodulatory effects of interferon-gamma on mature B-lymphocyte responses. Experientia. 1989;45:521–6. doi: 10.1007/BF01990501. [DOI] [PubMed] [Google Scholar]
- 23.Balce DR, Wang YT, McAllaster MR, Dunlap BF, Orvedahl A, Hykes BL Jr, Droit L, Handley SA, Wilen CB, Doench JG, Orchard RC, Stallings CL, Virgin HW. UFMylation inhibits the proinflammatory capacity of interferon-γ-activated macrophages. Proc Natl Acad Sci U S A. 2021;118:e2011763118. doi: 10.1073/pnas.2011763118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 25.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992;35:630–40. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 26.Conedera FM, Pousa AMQ, Mercader N, Tschopp M, Enzmann V. The TGFβ/Notch axis facilitates Müller cell-to-epithelial transition to ultimately form a chronic glial scar. Mol Neurodegener. 2021;16:69. doi: 10.1186/s13024-021-00482-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang M, Yang B, Deng D. Targeting of EIF4EBP1 by miR-99a-3p affects the functions of B lymphocytes via autophagy and aggravates SLE disease progression. J Cell Mol Med. 2021;25:10291–10305. doi: 10.1111/jcmm.16991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Wu GC, Zhang TP, Yang XK, Chen SS, Li LJ, Xu SZ, Lv TT, Leng RX, Pan HF, Ye DQ. Association of long noncoding RNAs expression levels and their gene polymorphisms with systemic lupus erythematosus. Sci Rep. 2017;7:15119. doi: 10.1038/s41598-017-15156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zucchi D, Elefante E, Calabresi E, Signorini V, Bortoluzzi A, Tani C. One year in review 2019: systemic lupus erythematosus. Clin Exp Rheumatol. 2019;37:715–722. [PubMed] [Google Scholar]
- 30.Sawada T, Fujimori D, Yamamoto Y. Systemic lupus erythematosus and immunodeficiency. Immunol Med. 2019;42:1–9. doi: 10.1080/25785826.2019.1628466. [DOI] [PubMed] [Google Scholar]
- 31.Tsokos GC. Autoimmunity and organ damage in systemic lupus erythematosus. Nat Immunol. 2020;21:605–614. doi: 10.1038/s41590-020-0677-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yougbare I, Keravis T, Lugnier C. NCS 613, a PDE4 inhibitor, by increasing cAMP level suppresses systemic inflammation and immune complexes deposition in kidney of MRL/lpr lupus- prone mice. Biochim Biophys Acta Mol Basis Dis. 2021;1867:166019. doi: 10.1016/j.bbadis.2020.166019. [DOI] [PubMed] [Google Scholar]
- 33.Goel RR, Wang X, O’Neil LJ, Nakabo S, Hasneen K, Gupta S, Wigerblad G, Blanco LP, Kopp JB, Morasso MI, Kotenko SV, Yu ZX, Carmona-Rivera C, Kaplan MJ. Interferon lambda promotes immune dysregulation and tissue inflammation in TLR7-induced lupus. Proc Natl Acad Sci U S A. 2020;117:5409–5419. doi: 10.1073/pnas.1916897117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo G, Ye S, Xie S, Ye L, Lin C, Yang M, Shi X, Wang F, Li B, Li M, Chen C, Zhang L, Zhang H, Xue X. The cytomegalovirus protein US31 induces inflammation through mono-macrophages in systemic lupus erythematosus by promoting NF-κB2 activation. Cell Death Dis. 2018;9:104. doi: 10.1038/s41419-017-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan W, Gu Z, Leng J, Zou X, Chen H, Min F, Zhou W, Zhang L, Li G. Let-7f-5p ameliorates inflammation by targeting NLRP3 in bone marrow-derived mesenchymal stem cells in patients with systemic lupus erythematosus. Biomed Pharmacother. 2019;118:109313. doi: 10.1016/j.biopha.2019.109313. [DOI] [PubMed] [Google Scholar]
- 36.Zhu J. T Helper cell differentiation, heterogeneity, and plasticity. Cold Spring Harb Perspect Biol. 2018;10:a030338. doi: 10.1101/cshperspect.a030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raphael I, Joern RR, Forsthuber TG. Memory CD4(+) T cells in immunity and autoimmune diseases. Cells. 2020;9:531. doi: 10.3390/cells9030531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romagnani S. T-cell subsets (Th1 versus Th2) Ann Allergy Asthma Immunol. 2000;85:9–18. doi: 10.1016/S1081-1206(10)62426-X. quiz 18, 21. [DOI] [PubMed] [Google Scholar]
- 39.Woda A, Picard P, Dutheil F. Dysfunctional stress responses in chronic pain. Psychoneuroendocrinology. 2016;71:127–35. doi: 10.1016/j.psyneuen.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Kidd P. Th1/Th2 balance: the hypothesis, its limitations, and implications for health and disease. Altern Med Rev. 2003;8:223–46. [PubMed] [Google Scholar]
- 41.Dong C, Flavell RA. Th1 and Th2 cells. Curr Opin Hematol. 2001;8:47–51. doi: 10.1097/00062752-200101000-00009. [DOI] [PubMed] [Google Scholar]
- 42.Terrier B, Costedoat-Chalumeau N, Garrido M, Geri G, Rosenzwajg M, Musset L, Klatzmann D, Saadoun D, Cacoub P. Interleukin 21 correlates with T cell and B cell subset alterations in systemic lupus erythematosus. J Rheumatol. 2012;39:1819–28. doi: 10.3899/jrheum.120468. [DOI] [PubMed] [Google Scholar]
- 43.Oster C, Wilde B, Specker C, Sun M, Kribben A, Witzke O, Dolff S. BTLA expression on Th1, Th2 and Th17 effector T-cells of patients with systemic lupus erythematosus is associated with active disease. Int J Mol Sci. 2019;20:4505. doi: 10.3390/ijms20184505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi G, Li D, Zhang D, Xu Y, Pan Y, Lu L, Li J, Xia X, Dou H, Hou Y. IRF-8/miR-451a regulates M-MDSC differentiation via the AMPK/mTOR signal pathway during lupus development. Cell Death Discov. 2021;7:179. doi: 10.1038/s41420-021-00568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao C, Chu Y, Liang Z, Zhang B, Wang X, Jing X, Hao M, Wang Y, An J, Zhang X, Sun L, Chen J. Low dose of IL-2 combined with rapamycin restores and maintains the long-term balance of Th17/Treg cells in refractory SLE patients. BMC Immunol. 2019;20:32. doi: 10.1186/s12865-019-0305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo S, Wu R, Li Q, Zhang G. MiR-301a-3p advances IRAK1-mediated differentiation of Th17 cells to promote the progression of systemic lupus erythematosus via targeting PELI1. J Healthc Eng. 2021;2021:2982924. doi: 10.1155/2021/2982924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li D, Li X, Duan M, Dou Y, Feng Y, Nan N, Zhang W. MiR-153-3p induces immune dysregulation by inhibiting PELI1 expression in umbilical cord-derived mesenchymal stem cells in patients with systemic lupus erythematosus. Autoimmunity. 2020;53:201–209. doi: 10.1080/08916934.2020.1750011. [DOI] [PubMed] [Google Scholar]
- 48.van de Vosse E, van Dissel JT. IFN-γR1 defects: mutation update and description of the IFNGR1 variation database. Hum Mutat. 2017;38:1286–1296. doi: 10.1002/humu.23302. [DOI] [PubMed] [Google Scholar]
- 49.Gao Y, Yang J, Cai Y, Fu S, Zhang N, Fu X, Li L. IFN-γ-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. Int J Cancer. 2018;143:931–943. doi: 10.1002/ijc.31357. [DOI] [PubMed] [Google Scholar]
- 50.Zenke K, Muroi M, Tanamoto KI. IRF1 supports DNA binding of STAT1 by promoting its phosphorylation. Immunol Cell Biol. 2018;96:1095–1103. doi: 10.1111/imcb.12185. [DOI] [PubMed] [Google Scholar]
- 51.Su Q, Wang F, Dong Z, Chen M, Cao R. IFN-γ induces apoptosis in human melanocytes by activating the JAK1/STAT1 signaling pathway. Mol Med Rep. 2020;22:3111–3116. doi: 10.3892/mmr.2020.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strickland MR, Koller EJ, Deng DZ, Ceballos-Diaz C, Golde TE, Chakrabarty P. Ifngr1 and Stat1 mediated canonical Ifn-γ signaling drives nigrostriatal degeneration. Neurobiol Dis. 2018;110:133–141. doi: 10.1016/j.nbd.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu H, Yang J, Jiao S, Li Y, Zhang W, Wang J. T-box transcription factor 21 expression in breast cancer and its relationship with prognosis. Int J Clin Exp Pathol. 2014;7:6906–13. [PMC free article] [PubMed] [Google Scholar]
- 54.Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med. 2011;208:1001–13. doi: 10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao X, Yang H, Xu Y, Xiong Y, Wang G, Ye X, Ye J. Iminosugar derivative WGN-26 suppresses acute allograft rejection via inhibiting the IFN-γ/p-STAT1/T-bet signaling pathway. Int Immunopharmacol. 2014;23:688–95. doi: 10.1016/j.intimp.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 56.Xu H, Jiang Y, Xu X, Su X, Liu Y, Ma Y, Zhao Y, Shen Z, Huang B, Cao X. Inducible degradation of lncRNA Sros1 promotes IFN-γ-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat Immunol. 2019;20:1621–1630. doi: 10.1038/s41590-019-0542-7. [DOI] [PubMed] [Google Scholar]
- 57.Liu Y, Ma X, Yang H, Li X, Ma Y, Ason B, Liu S, Hu LA. APLNR regulates IFN-γ signaling via β-arrestin 1 mediated JAK-STAT1 pathway in melanoma cells. Biochem J. 2022;479:385–399. doi: 10.1042/BCJ20210813. [DOI] [PubMed] [Google Scholar]
- 58.Liu H, Wang W, Liu C. Increased expression of IFN-γ in preeclampsia impairs human trophoblast invasion via a SOCS1/JAK/STAT1 feedback loop. Exp Ther Med. 2021;21:112. doi: 10.3892/etm.2020.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin S, Zhang Y, Zhang J, Tian F, Sun L, He X, Ma X, Zhang J, Liu XR, Zeng W, Lin Y. SPRY4 regulates trophoblast proliferation and apoptosis via regulating IFN-γ-induced STAT1 expression and activation in recurrent miscarriage. Am J Reprod Immunol. 2020;83:e13234. doi: 10.1111/aji.13234. [DOI] [PubMed] [Google Scholar]
- 60.Miyawaki A, Iizuka Y, Sugino H, Watanabe Y. IL-11 prevents IFN-γ-induced hepatocyte death through selective downregulation of IFN-γ/STAT1 signaling and ROS scavenging. PLoS One. 2019;14:e0211123. doi: 10.1371/journal.pone.0211123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu W, Li M, Wang Z, Wang J. IFN-γ mediates the development of systemic lupus erythematosus. Biomed Res Int. 2020;2020:7176515. doi: 10.1155/2020/7176515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwata S, Zhang M, Hao H, Trimova G, Hajime M, Miyazaki Y, Ohkubo N, Satoh Kanda Y, Todoroki Y, Miyata H, Ueno M, Nagayasu A, Nakayamada S, Sakata K, Tanaka Y. Enhanced fatty acid synthesis leads to subset imbalance and IFN-γ overproduction in T helper 1 cells. Front Immunol. 2020;11:593103. doi: 10.3389/fimmu.2020.593103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harigai M, Kawamoto M, Hara M, Kubota T, Kamatani N, Miyasaka N. Excessive production of IFN-gamma in patients with systemic lupus erythematosus and its contribution to induction of B lymphocyte stimulator/B cell-activating factor/TNF ligand superfamily-13B. J Immunol. 2008;181:2211–9. doi: 10.4049/jimmunol.181.3.2211. [DOI] [PubMed] [Google Scholar]
- 64.Katsuyama T, Li H, Krishfield SM, Kyttaris VC, Moulton VR. Splicing factor SRSF1 limits IFN-γ production via RhoH and ameliorates experimental nephritis. Rheumatology (Oxford) 2021;60:420–429. doi: 10.1093/rheumatology/keaa300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.