Abstract
The host microbiome is polymorphic, compartmentalized, and composed of distinctive tissue microbiomes. While research in the field of cancer immunotherapy has provided an improved understanding of the interaction with the gastrointestinal microbiome, the significance of the tumor-associated microbiome has only recently been grasped. This article provides a state-of-the-art review about the tumor-associated microbiome and sheds light on how local tumor microbiota shapes anticancer immunity and influences checkpoint immunotherapy outcome. The direct route of interaction between cancer cells, immune cells, and microbiota in the tumor microenvironment is emphasized and advocates a focus on the tumor-associated microbiome in addition to the spatially separated gut compartment. Since the mechanisms underlying checkpoint immunotherapy modulation by tumor-associated microbiota remain largely elusive, future research should dissect the pathways involved and outline strategies to therapeutically modulate microbes and their products within the tumor microenvironment. A more detailed knowledge about the mechanisms governing the composition and functional quality of the tumor microbiome will improve cancer immunotherapy and advance precision medicine for solid tumors.
Keywords: Immunotherapy
Linking microbiota with checkpoint immunotherapy
Cancer immunotherapy with immune checkpoint inhibitors (ICIs) has revolutionized the treatment of many advanced solid tumors including several types of carcinoma1–3 and melanoma.4 Biomarkers with a high predictive value for ICI therapy response are needed to (1) save ‘therapeutic time’ in non-responding patients, (2) spare non-responding patients from ICI-related toxicity,5 6 (3) optimize treatment costs in an era of ever-increasing healthcare expenses7 8 and, finally, (4) identify potential novel targets to improve ICI efficacy.9 10 While checkpoint molecule protein expression levels (eg, PD-L1),11 12 tumor mutational burden (TMB),13–15 microsatellite instability (MSI)/DNA mismatch repair deficiency (dMMR),16 and pretreatment infiltration of tumor tissue by immune cells17 can portend the likelihood of response to ICI therapy, their predictive power is limited for various reasons. As an example, high PD-L1 expression may not be therapeutically relevant due to heterogeneity in the signaling circuits associated with response to ICIs,18 and the sum of genetic tumor alterations cannot foretell whether individual mutations result in immunogenic proteins.19 Tumor immune cell infiltration at baseline may also be irrelevant for ICI response prediction if the cells are exhausted/dysfunctional20 21 or if their protective effects are abrogated by immunosuppressive mechanisms in the tumor microenvironment (TME).22–25
The high relevance of microbes for cancer development and growth has recently been acknowledged by the integration of ‘polymorphic microbiomes’ to the latest update of the ‘Hallmarks of Cancer’ concept.26 This is based on the ground-breaking observation that the host microbiome modulates the efficacy of ICI therapies, thereby offering prognostic and therapeutic potential.27–32 The temporally connected use of antibiotics prior to ICI therapy initiation impairs their clinical activity29 33 34 and the fecal microbial composition of ICI-responding versus non-responding melanoma patients differs significantly.30 Early clinical data show that fecal microbiota transplantation (FMT) from responders rescues treatment response in at least a fraction of anti-PD-1-refractory melanoma patients.9 35 While the abundance of certain bacteria such as Akkermansia muciniphila, Bacteroides fragilis, and Bifidobacterium correlates with anticancer immunity and ICI treatment response,28 29 36 37 potentially through immunological and/or metabolic means,38–41 evidence suggests that an overall higher diversity of the host microbiome is associated with prolonged ICI-related survival irrespective of individual microbial species.30 42 43 This indicates that a functionally diverse microbiome less prone to dysbiosis and perturbation, but more likely to generate cancer-resembling T cell epitopes38 as well as a rich metabolic landscape, benefits patients with cancer receiving ICIs.
It is also important to note that the treatment-modulating activity of host microbes in cancer is not limited to immunotherapy but extends to conventional chemotherapeutic drugs, such as nucleoside analogs, antimetabolites, and platinum.44 45 Specifically, Fusobacterium nucleatum orchestrates a molecular program involving innate immune sensing and certain microRNAs to modulate the autophagy pathway and promote cancer cell resistance to capecitabine/oxaliplatin.44 In addition, certain bacteria found in the TME express an isoform of cytidine deaminase that can convert the chemotherapeutic drug gemcitabine to its inactive form (2',2'-difluorodeoxyuridine), thus fostering drug resistance.45
This article not only appraises the role of the gastrointestinal (GI) microbiome in the context of ICIs but also complements the concept of host microbiota as a critical regulator of ICI-induced anticancer immunity by integrating the emerging significance of the local tumor microbiota. A first focus is the concept of microbial compartmentalization and its relevance for the interaction of microbiota with cancer and immune cells. A second focus is the potential translation of this knowledge, that is, how the emerging deconvolution of the local tumor microbiota may be harnessed to design and implement next generation ICI-resensitizing interventions.
Compartmentalization of the host microbiome
The host microbiome refers to the totality of microbes found within an organism. Heterogeneity and complexity characterize the human microbiome whose numerical dimensions are tremendous. As an example, the total number of bacteria in the human body is estimated with 3.8×1013, of which 1012 bacteria are located in extra-GI organs.46 Thus, the total number of bacteria slightly exceeds that of human body cells (~3×1013)46 and the estimated number of non-redundant microbial genes in the gut (~3.3 million) dwarfs the number of human genes (<30,000) by orders of magnitude.47 Viruses, fungi, and other microbes further enrich the human microbiome; however, their relative proportion is small and research in the context of cancer immunotherapy is limited.
Research activities at the intercept of ICIs and the host microbiome have notoriously prioritized the GI microbial habitat possibly for reasons of accessibility and sample availability. In addition, the GI tract, ultimately relating to the whole digestive system from mouth to anus, carries a very high microbial biomass which simplifies microbiome-targeted analyses for technical reasons including a beneficial ‘signal-to-noise’ ratio.48–50 Finally, the GI microbiome appears to be manipulable through antibiotic therapy,51 52 diet,53 oral bacterial supplementation,54 or FMT,35 which all may offer straightforward prospects for ICI-resensitizing interventions.32
Despite this rational emphasis on the GI microbiome, the human microbiome in its totality is much wider and spatially organized. Rich microbial communities have also been identified in the respiratory tract,55 the reproductive tract,56 on skin,57 and in utero,58 among others. The different organ functions and diverse niches relate to both compartmentalized and functionally specialized microbiomes with distinct bacterial communities.59 60 For instance, commensal intestinal microbes support epithelial barrier integrity, digestion of dietary components, and mucosal immunity.61 In contrast, the lung microbiome may provide critical signals to balance local immune responses and promote tolerance to aeroallergens.55 62 63 Dysbiosis in these tissue-specific microbial habitats can cause significant local diseases including inflammatory bowel disease and celiac disease (GI microbiome),64 and pneumonia, allergy and asthma (lung microbiome).55 Despite spatial segregation, different organs and their microbial ecosystems can bidirectionally communicate with each other. Possible mechanisms include the mucosal translocation and systemic circulation of bacteria, bacterial antigens, microbiota-imprinted immune cells, or microbiota-derived metabolites.38 55 65 66
It is important to note that the microbial biomass varies extremely among the different compartments, with a low biomass found in healthy lungs (103–105 bacteria per gram of tissue),55 an intermediate biomass found on skin (104–106 bacteria/micro-organisms per square cm),67 and a very high biomass found in the GI tract (1011–1012 bacteria per gram of luminal content).68 Moreover, in the absence of external triggers such as antibiotic exposure, the structure of organ-specific microbiomes will follow the rules of host coevolution processes,69 70 along with competition for niches and nutrients among the various microbes.71 72
Taken together, the host microbiome extends far beyond the GI compartment and exhibits spatial organization and specialization consistent with organ function. Perturbation of the microbial equilibrium in these compartments can trigger local immune-related diseases, which may sometimes encroach on distant body parts or even become systemic.
Drawing attention to the local tumor microbiome
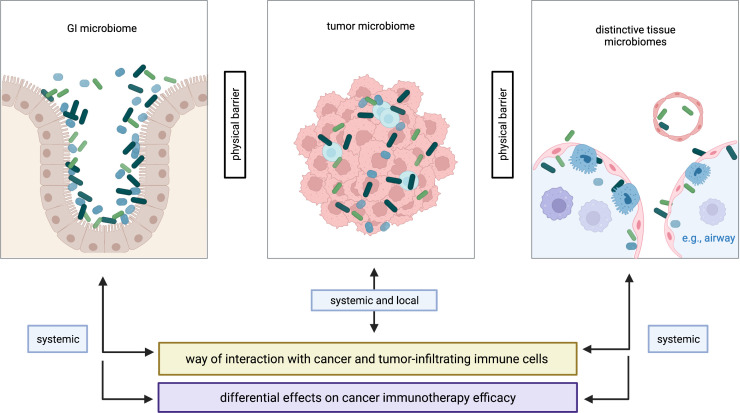
Despite proven involvement in the response to checkpoint immunotherapy,28 29 33 the gut microbiome is spatially separated from the tumor mass in most cancer entities except for GI cancers where immediate tumor–microbiome interactions are evident.73 As previously mentioned, most organs (if not all) harbor distinct microbial ecosystems and, thus, bacteria may coevolve alongside the growing tumors, shape local anticancer immunity and influence tumor progression.74 An interesting finding is that matched primary and metastatic colorectal tumors harbor nearly identical Fusobacterium strains, suggesting that microbial tumor habitats are conserved during the metastatic process (potentially through comigration of cancer cells and bacteria) and that niche-dependent coevolution processes may subsequently occur.75 Fundamentally, interactions of tumor-associated microbiota with cancer cells and tumor-infiltrating immune cells have a local and a systemic aspect, with juxtacrine and paracrine signaling circuits involved, respectively, while cross-compartment microbial interactions require systemic circulation (figure 1).
Figure 1.
Compartmentalization of the microbiome in cancer. Distinctive microbiomes are found in various anatomical compartments of patients with cancer, here categorized as GI microbiome, tumor microbiome and distinctive tissue microbiomes. These microbiomes are physically separated yet functionally interact with each other. The tumor microbiota plays an outstanding role as interactions with cancer cells and tumor-infiltrating immune cells may be more direct as compared with the GI microbiome and other tissue microbiomes. Specifically, interactions with tumor and immune cells can occur via the systemic route (eg, circulation) in all three scenarios, while local (‘physical’) interactions are exclusive to the tumor microbiome. This suggests distinct effects of different microbial habitats on cancer immunotherapy efficacy. Figure is created with BioRender.com. GI, gastrointestinal.
In lung cancer, research has shown that lower airway dysbiosis following enrichment of oral commensals (eg, Veillonella) likely after microaspiration and/or reduced airway clearance leads to an unfavorable immune landscape that fosters cancer progression.76 77 Mechanistically, the authors showed that dysbiosis activates the IL-17 pathway as well as other signaling cascades to generate an inflammatory TME and induce host transcriptomic changes that exacerbate malignancy.76 77 Of note, pulmonary dysbiosis through induced bacterial pneumonia leads to TLR4 activation on bronchoepithelial cells and IL-6 secretion, facilitating hepatic sinusoidal adhesion of non-small cell lung cancer (NSCLC) cells thereby augmenting liver metastasis.78 Another important study discovered that local lung microbiota can induce cancer-promoting inflammation by activating tissue-resident γδ T cells via IL-1β/IL-23 produced from myeloid cells, leading to upregulation of IL-17 and tumor cell proliferation.79 Thus, evidence suggests that lung cancer development is linked to early interactions between lung microbiota and local immune cells which may be therapeutically harnessed by neutralizing the key mediators of this inflammatory cascade.
In murine models, it was recently demonstrated that intratumoral accumulation of specific microbes (ie, Bifidobacterium) is critical for response to CD47-targeted immunotherapy in subcutaneously growing tumors.80 Tumor site-associated Bifidobacterium facilitates treatment response in a STING-dependent and interferon (IFN)-dependent fashion and augments cross-priming by dendritic cells (DCs).80 Moreover, a large study of pan-cancer microbial profiling showed the frequent intratumoral presence of bacterial components including DNA, RNA, and outer membrane/cell wall constituents.81 Interestingly, this study found that bacterial components were also detectable in the tumor tissue of cancer entities without direct connection to the external environment (eg, glioblastoma, bone cancer, and ovarian cancer) and that breast cancers harbored a particularly rich microbial flora.81 Other important lessons from this study are the observations that (1) intratumor bacteria are dominated by intracellular species which are found both within tumor and immune cells and that (2) signatures of intratumor bacteria correlate with specific clinicopathological tumor characteristics as well as the response to ICI therapy.81 Along similar lines, a recent study revealed an association of tumor-resident intracellular bacteria with metastatic activity and cytoskeletal rearrangements in circulating tumor cells,82 thus suggesting the tumor-associated microbiota as an antimetastasis target. As discussed in more detail below, the origin of tumor-populating microbiota might be diverse and various local, distant, and systemic sources are potentially relevant. In addition, there might be crosstalk of microbiota and their products between organ systems as well as between organs and tumors.65 75 83 84
A comprehensive study based on treatment-naïve The Cancer Genome Atlas (TCGA) samples found tumor-discriminatory and stage-discriminatory power of the cancer-associated microbiome.85 Importantly, the microbial signature of the blood alone could predict the cancer type and discriminate between healthy individuals and patients with cancer with great accuracy, thus holding great potential for screening and diagnostic applications.85 In pancreatic cancer, the tumor microbiome is considerably more abundant as compared with the healthy pancreas45 and fosters tumor growth by suppressing anticancer immunity at least in part through a mechanism involving differential Toll-like receptor activation in monocytic cells.86 Interestingly, long-term survivors of pancreatic cancer show a higher diversity of their tumor microbiome and exhibit a distinct intratumoral microbiota signature highly predictive of outcome and correlated to tumor immune cell infiltration in murine models.87
These data collectively indicate an important role of the local tumor microbiome in malignant growth and the anticancer immune response. Drawing attention to this poorly researched aspect of tumor biology is warranted and may ultimately open new avenues for microbiome-centered interventions to optimize cancer immunotherapy.
Techniques and challenges in profiling the tumor-associated microbiome
Inner organs, even those that are in constant exchange with the environment (such as the lungs), have been historically considered as ‘sterile’ mostly due to technical limitations in culturing microbes from corresponding tissue samples.55 However, this view has changed dramatically owing to the advent and widespread adoption of culture-independent methods for microbial profiling roughly 15 years ago.55 Culture-independent methods for microbial detection and identification include molecular, immunological, microscopic, spectrometric, and spectroscopic approaches.88
Tumor-associated and other tissue microbiomes typically entail a low to very low microbial biomass which can only be investigated using highly sensitive and specific methods.43 50 55 In the absence of prior knowledge allowing for more targeted investigations, researchers typically aim at dissecting the bulk tumor-associated microbiome (or bacteriome), thus making ‘universal species coverage’ a top priority. All these criteria are met by modern sequencing-based technologies which have therefore prevailed in the systematic profiling of microbes from clinical samples such as tumor tissue.
16S rRNA gene amplicon sequencing,89 90 that is, the sequencing of the RNA component of the 30S subunit of the prokaryotic ribosome (on DNA level), depicts a powerful method for profiling bacteria from tumor specimens.43 81 91–93 PCR amplification of the genomic 16S region prior to sequencing ensures high assay sensitivity. Since this method is specific for bacteria (more precisely, prokaryotes), the stoichiometric predominance of host cell/tumor DNA is less disturbing for analysis. Therefore, 16S rRNA gene amplicon sequencing represents a straightforward and cost-efficient approach for profiling tumor-associated bacteria without producing huge amounts of data. A clear downside of this method is the fact that it does not cover non-bacterial microbes such as viruses and fungi. However, novel strategies have been developed to increase the accuracy and coverage of 16S rRNA sequencing of tumor-resident bacteria including 5R multiplexed sequencing81 and the use of biotinylated 16S primers.82
In contrast, shotgun metagenomic sequencing represents an unselective approach detecting all DNAs found within a complex sample based on short, overlapping DNA fragments and bioinformatic sequence assembly using large reference databases.94 Shotgun metagenomic sequencing thus covers microbial species from all biological kingdoms (including viruses, which are strictly spoken not living organisms) as well as host normal/tumor DNA.94 The species coverage of shotgun sequencing is therefore theoretically complete, even though massive enrichment of host cell DNA in tumor tissue (sometimes accounting for >99% of total DNA in the sample) typically results in a very low coverage of most bacteria, potentially posing significant analytical challenges especially with regard to downstream taxonomic ranks. The rare fraction of the microbial target signal also explains the huge amounts of data produced as well as the associated high costs of this sequencing modality. Nonetheless, shotgun metagenomic sequencing has important advantages especially when the microbial biomass is high, including enhanced detection of bacteria and a more granular analysis of microbial diversity.90 Shotgun sequencing has been successfully applied to the metagenomic analysis of tumor tissue, including formalin-fixed, paraffin-embedded samples.85 95 96
Further sequencing-based technologies for deep microbial profiling and/or expression profiling are currently being developed, one such example being microbial single-cell RNA-sequencing using a combinatorial barcoding strategy,97 and another one being the scDual-Seq method simultaneously capturing pathogen and host transcriptomes.98 Thus, further optimization of culture-independent methods for microbial analysis is ongoing and additional analytical options will be available in the future.
Contamination and low sensitivity represent major challenges in characterizing the tumor-associated microbiome. As a general rule, contamination gets more problematic/confounding the lower the microbial biomass and the smaller the analyzed tissue specimens. Cross-contamination with other microbes can occur during the sampling procedure itself or when the retrieved sample is handled in non-sterile environments.43 50 In addition, it is important to note that laboratory-grade reagents will contain remnants of environmental microbial DNA which can critically affect sequence-based microbiome studies.48 49 It is therefore proposed that non-template and paraffin controls as well as other specific quality checks (eg, addressing batch-specific and center-specific effects) are performed to mitigate potential interference from contamination.48 49 81
In summary, sensitive sequence-based methods are available to profile the tumor-associated microbiome but further technical developments are expected in the near future to overcome current limitations. Researchers should be aware of the challenges and pitfalls of sequence-based microbiome studies in tumor tissue and interpret their results accordingly.
Clinical relevance of the tumor-associated microbiome
The accumulating evidence on the fundamental role of the tumor-associated microbiome in cancer development is also suggestive of prognostic and therapeutic potential in terms of microbiome-based patient assessment and microbiome-targeted interventions. Unsurprisingly, the most comprehensive data come from colorectal cancer (CRC),73 99 100 a tumor entity in direct and steady contact with large amounts of bacteria and comparatively easy to access. As an example, studies have shown that the long-term use of antibiotics in earlier life is associated with later colorectal adenoma formation,101 and that certain bacterial species (eg, F. nucleatum) are enriched in CRC tissue, aggravating intestinal tumorigenesis and associating with metastatic lymph node involvement.102–104 In line, 16S rRNA gene amplicon sequencing has revealed an association of CRC and reduced microbial diversity in feces.105 Other clinically interesting observations include the overabundance of certain bacteria in CRC tissue and their association with an MSI-high phenotype, chemo-resistance and recurrence, and poor prognosis.44 106 107 Of note, while a majority of CRC cancer cases are treated locally or with chemotherapy, the use of ICIs is approved for patients with metastatic MSI-high/dMMR CRC, a patient population making up roughly 5% of metastatic CRC cases.108 For this subset, the significance of the tumor-associated microbiome for ICI treatment response is potentially high and warrants further investigation in the future.
In the absence of actionable driver mutations, cancer immunotherapy with ICIs has become the main pillar of treatment for advanced NSCLC, either in combination with antiproliferative chemotherapy or as monotherapy. Among carcinomas, NSCLC is at the forefront of clinical experience with ICIs, including data on the interaction with the distant GI microbiome and antibiotics. To take into account more proximal microbial habitats, we recently analyzed the microbial composition of bronchoscopic tumor biopsies from ICI-treated NSCLC patients using 16S rRNA gene amplicon sequencing. These analyses revealed the consistent presence of bacterial signatures in the samples and suggested high intratumoral abundance especially of Firmicutes, Bacteroidetes, and Proteobacteria.43 Interestingly, a higher microbial tumor diversity was associated with improved overall survival irrespective of ICI treatment response, indicating treatment-unrelated beneficial effects of tumor microbial diversity in NSCLC patients.43 Our analyses further showed that high tumorous abundance of Gammaproteobacteria was associated with low PD-L1 expression and poor progression-free survival (PFS), suggesting that Gammaproteobacteria are preferentially found in PD-L1 low-expressing tumors which typically do not respond well to ICI therapy.43 However, it currently remains elusive whether Gammaproteobacteria mechanistically participate in the regulation of PD-L1 levels in the tumor bed.
In melanoma, another tumor entity successfully treated with ICIs, scarce data are available regarding the significance of the local skin microbiome for treatment response and patient outcome. Nevertheless, preclinical evidence suggests that melanoma progression is associated with dysbiosis of the skin microbiome, which might be exploited for future therapeutic development.109 In hepatocellular carcinoma (HCC), various bacterial phyla are detectable in tumor tissue possibly after translocation from the gut compartment following disruption of epithelial barrier integrity.110 Consistent with observations in NSCLC,43 the bacterial diversity is higher in HCC tissue as compared with adjacent liver tissue, suggesting that intratumoral microbes might contribute to HCC pathology.110 Retrospective clinical data suggested that HCC of non-viral etiology (particularly, non-alcoholic steatohepatitis (NASH)-associated HCC) is less sensitive to ICI treatment potentially through NASH-related changes in T cell activation,111 suggesting that the local liver microbiota may shape—or even foster—the response to ICI therapy. Finally, a large study on various carcinoma types, melanoma, and brain cancer revealed an association of intratumoral bacteria, or their predicted metabolic functions, with clinicopathological features (eg, molecular tumor subtypes) and response to ICIs.81 It is worth mentioning that the precise origin of intratumoral bacteria remains frequently unknown. Possible sources include surrounding tissue (ie, local tissue-specific microbiomes),109 translocation from a different anatomical compartment (eg, the gut),110 and comigration of cancer cells and bacteria from the primary tumor in the case of metastatic lesions.75 Of note, evidence also suggests that most bacteria found in tumors are opportunistic inhabitants and that only few examples exist where the relationship with cancer development is causal (one such example being Helicobacter pylori in certain gastric cancer types).112 Irrespective, particular bacterial species, genera, families, etc and/or the totality of the tumor-associated microbiome may modulate the behavior of established tumors and influence both natural and treatment-reinforced anticancer immunity. Finally, it is also important to note that some bacteria (eg, Bifidobacterium)80 113 and oncolytic viruses (eg, vesicular stomatitis virus)114 have a natural tropism to colonize/infect tumors after systemic administration.
Taken together, emerging evidence suggests that the local tumor microbial composition plays an important role in cancer development, progression, and the response to immune-mediated treatment. The tumor-associated microbiome hence depicts a rational target for enhancing cancer immunotherapy efficacy and boosting response rates.
Putative underlying mechanisms
Few data are available deciphering the mechanisms governing the modulatory function of the local tumor microbiome for cancer growth and ICI outcome. One publication based on subcutaneously growing tumors found that tumor-associated Bifidobacterium facilitates CD47-based immunotherapy dependent on STING signaling in tumor DCs as well as intratumoral type I IFN and T cells.80 In line, concomitant CD47 blockade and STING targeting improves immunotherapy efficacy through enhanced tumor cell phagocytosis, stimulating anticancer immunity.115
In CRC, F. nucleatum binds to intestinal epithelial cells via adhesins and activates β-catenin to promote cancer cell stemness,73 a principal tumor cell characteristic generally associated with metastatic progression, treatment failure, and relapse.116–121 In addition, F. nucleatum has proinflammatory effects on the TME through mechanisms involving TLR4 and MyD88 signaling.73 Most interestingly, viable Fusobacterium is not only found in primary CRC tissue but retained in distal metastases, suggesting that Fusobacterium may be able to migrate to metastatic sites alongside the disseminating cancer cells.75 Other mechanisms of cancer promotion by CRC-associated bacteria include impaired DNA repair and genotoxicity (eg, via colibactin, reactive oxygen species and UshA)73 122, both of which may be relevant for cancer immunotherapy through increasing the TMB.
It is also well established that gut microbes can help boost anticancer immunity through various mechanisms including the formation of protective tertiary lymphoid structures and immune cell recruitment into the TME.28 29 123 An interesting observation is that microbiome-specific/reactive T cells are associated with favorable anticancer immunity and response to ICI therapy.28 39 Although the mechanisms behind this association are yet to be elucidated, immunological mimicry of tumor neoantigens by microbial species and corresponding T cell cross-reactivity might at least in part explain why microbes are functionally relevant for cancer immunotherapy.28 31 39 43 124 125 In support, preliminary data suggest that a higher diversity of the microbiome either in the gut compartment or at the tumor site may be associated with favorable treatment response and/or outcome in patients receiving ICIs.30 38 42 43 Another—non-mutually exclusive—explanation for the functional involvement of microbes in cancer immunotherapy is their metabolic landscape which comprises factors with known immune-modulatory activity, such as short-chain fatty acids (SCFAs) and inosine.40 41 126 As an example, SCFAs impact T cell differentiation into both immunosuppressive T regulatory cells and effector T cells, whose presence negatively and positively associates with ICI efficacy, respectively.127 Inosine has been shown to foster Th1 cell differentiation and activation acting through T cell specific A2A receptor signaling, promoting anticancer immunity and immunotherapy response.126 In a study on triple-negative breast cancer, the Clostridia-associated metabolite trimethylamine N-oxide (TMAO) was shown to activate CD8+ T cells and antitumor-polarized M1 macrophages, and high plasma TMAO concentrations were also correlated to better response to immunotherapy as well as prolonged PFS.128 Finally, L-arginine produced from ammonia by engineered, tumor-homing bacteria was shown to increase intratumoral L-arginine concentrations and improve the response to ICI therapy dependent on T cells.129
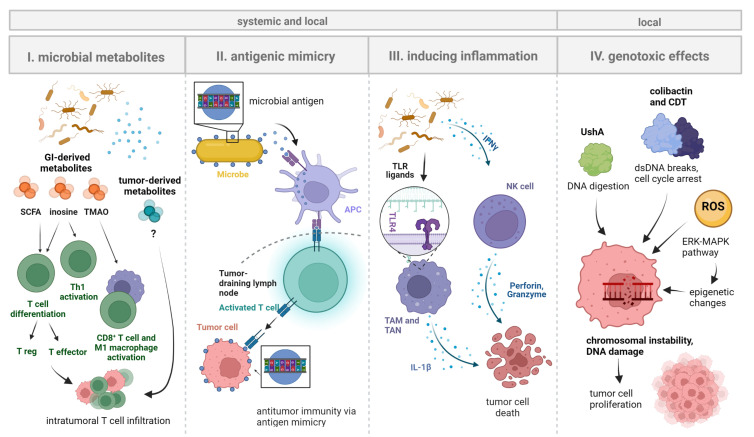
In sum, mechanistic data on the role of the local tumor microbiome in cancer immunotherapy are sparse at this stage and respective research is warranted. Extrapolating principal findings from the gut compartment would infer that tumor-associated microbes influence cancer immunotherapy efficacy through (1) direct effects on cancer cells and (2) modulation of anticancer immunity. Here, it is important to separate local from systemic effects of microbes, and to highlight that especially immune-related effects will often have a local and a systemic dimension whereas cancer cell-specific effects may be more spatially confined to the TME (figure 2).
Figure 2.
Systemic and local effects of microbes on tumorigenesis and immune cells. Shown are principal mechanisms by which microbiota may influence tumorigenesis and local immune cells that take effect on cancer immunotherapy efficacy. Microbiota can have direct effects on the immunotherapy target cells (ie, cancer cells) or regulate the amplitude and quality of the anticancer immune response both on and off treatment: (1) Microbial metabolites (of GI microbiota, likely also of local microbiota) may promote T cell activation and differentiation, resulting in the accumulation of both proinflammatory (eg, effector T cells) and immunosuppressive (eg, T regulatory cells) immune cell populations in the TME. (2) MHC class II-expressing cells (eg, DCs) presenting microbial antigens may activate T cells in tumor-draining lymph nodes. Molecular mimicry of tumor antigens by microbial peptides would result in tumor-directed T cell activity, thereby influencing cancer immunotherapy efficacy. (3) Bacterial molecules (pathogen-associated molecular patterns) such as LPS may bind TLRs (eg, TLR4) on TAMs/TANs or directly activate tumor-associated NK cells (eg, via IFNγ signaling). This may activate downstream pro-inflammatory pathways and/or promote tumor cell death. (4) Genotoxic microbial products can induce DNA damage in cancer cells to reshape the tumor genome and promote cancer cell proliferation. Examples include colibactin and CDT that both induce transient cell cycle arrest (promoting the occurrence of genomic errors during the repair process) and double-strand breaks leading to chromosomal instability139 140as well as the novel genotoxin ushA (acting via DNA digestion) and ROS that directly damage cancer cells and/or induce epigenetic changes via ERK-MAPK signaling. Figure is created with BioRender.com.73 122 (IMPORTANT: move this citation to the sentence before right after "ERK-MAPK signaling".) APC, antigen-presenting cell; CDT, cytolethal distending toxin; DC, dendritic cell; LPS, lipopolysaccharide, NK, natural killer (cell); ROS, reactive oxygen species; SCFA, short-chain fatty acid; TLR, Toll-like receptor; TAM, tumor-associated macrophage; TAN, tumor-associated neutrophil; TMAO, trimethylamine N-oxide; TME, tumor microenvironment.
Therapeutic and dietary perspectives
Harnessing the microbiome for enhancing ICI performance and/or increasing response rates has been a goal since the initial discovery of microbiome-dependent modulation of ICI efficacy28 and is being actively pursued.31 32 Inspired by both mechanistic studies28 29 and clinical observations,33 34 the concept of FMT from ICI top-responding patients has been clinically tested in phase I and II trials.9 35 These studies showed the feasibility of the approach and provided preliminary evidence for the efficacy of ICI reinduction following FMT in otherwise treatment-refractory melanoma.9 35 Of note, FMT was associated with distinct proteomic and metabolic signatures as well as favorable immunological changes both within the TME and in the periphery.9 35 More recently, this concept was refined and a randomized phase I study was conducted where patients received an ICI combination (nivolumab and ipilimumab) with or without CBM588, a bifidogenic live bacterial product orally supplemented.130 Although this study failed to demonstrate longitudinal changes in the relative abundance of Bifidobacterium, the addition of CBM588 to combined ICIs was safe and prolonged the progression-free interval.130 Another recent study addressed the role of dietary fiber intake and probiotics on ICI response.10 Interestingly, a high-fiber diet was associated with improved PFS especially when no probiotics were supplemented.10 Parallel preclinical studies suggested that the beneficial effect of dietary fibers may have been mediated by a higher frequency of IFN-producing cytotoxic T cells in the TME,10 and a mechanistic basis for this could be reprogramming of tumor-associated monocytes by bacterial-derived STING agonists and regulation of intratumoral natural killer (NK) cell/DC crosstalk.131 Adopting a fasting-mimicking diet has also been shown to positively modulate anticancer immunity by regulating immunosuppressive cell populations and enhancing cytotoxic T cell responses in a clinical trial.132 These data set the stage for larger clinical trials to investigate potential synergy with anticancer treatments including ICIs.132 Of note, future technology will also allow the targeted manipulation of individual genes within a complex microbiome to better understand host–microbiome interactions and potentially optimize cancer therapies through reverse microbial genetics.133 It will be of importance to identify key metabolic TME changes that are likely to elicit vulnerabilities, hence contributing to the development of precision nutrition approaches that augment the efficacy of cancer immunotherapy together with targeted microbial modulation.
Here, again, we raise the question of how findings from the GI microbiota, including data on dietary intervention and FMT, can be extrapolated and translated to the local tumor microbiome? It is important to stress that the microbial fingerprint is quite well conserved among paired primary and metastatic tumors, suggesting interdependence of both microbial habitats and, potentially, common therapeutic vulnerabilities/opportunities.75 One possible explanation for this is comigration of cancer cell clusters and bacteria from the primary tumor during the formation of metastases.75 The tumor-associated microbiome may also be subject to influence from translocated GI microbiota following spontaneous events such as disruption of gut epithelial barrier function, thus delineating direct gut–tumor microbial crosstalk.110 Furthermore, there is growing evidence for microbial crosstalk between organ systems and typical examples include the gut–lung axis55 65 and the gut–brain axis.134 135 Among others, GI microbiota influence systemic immune tones by affecting myelopoiesis in bone marrow, with the complexity of the GI microbiota correlating with the size of the myeloid cell pool.65
An interesting perspective for microbiome-targeted intervention is the natural tumor tropism of certain bacteria (and viruses). As an example, intratumoral accumulation of the gut commensal Bifidobacterium has been observed after systemic administration (ie, intravenous injection) and was found to facilitate local immunotherapy via STING signaling.80 113 The tumor tropism of Bifidobacterium is not completely understood but one may hypothesize that leaky blood vessels and hypoxic conditions within the TME create a permissive niche for the preferential accumulation of this anaerobic gut commensal potentially further supported by local immune privilege.80 136 The response to immunotherapy was also rescued when Bifidobacterium was administered directly into tumor tissue,80 suggesting that tumor-specific delivery of microbes might be a feasible option to enhance cancer immunotherapy at least when the tumor mass is physically accessible. Other bacteria with tumor-homing/colonizing capacity include Escherichia coli129 and Salmonella typhimurium,136 both of which warrant investigation as therapeutic vehicles for targeted genetic TME modulation and (re)sensitization to cancer immunotherapy.
Finally, a large study showed that human tumors harbor a rich microbiome whose composition is biased towards intracellular bacteria present inside both tumor and immune cells.81 As intracellular bacteria—just like viruses—are subject to endogenous antigen presentation via MHC class I (and II), they are principal targets for adaptive immunity that could also be treated with tumor-specific, microbiota-targeted vaccination.91 In other words, intracellular tumor bacteria represent a potential source of tumor-associated/specific antigens, thus qualifying for therapeutic protocols meeting the principles of personalized cancer vaccination,137 138 even though tumor coevolution and immunoediting processes need to be considered.
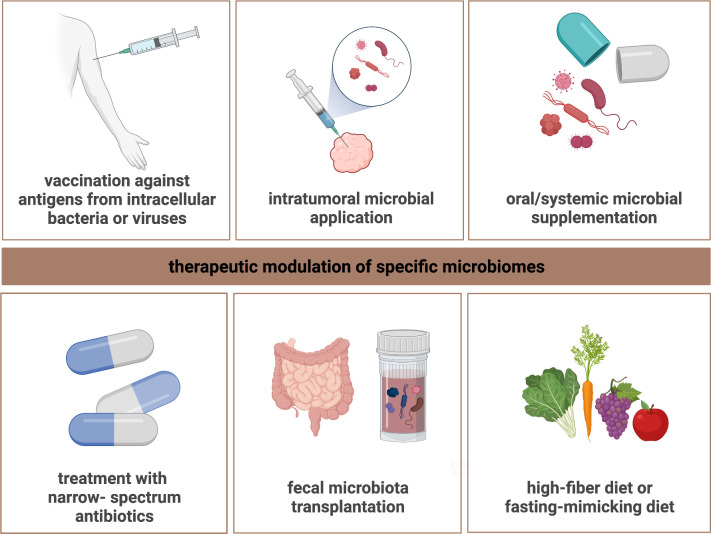
Collectively, various strategies are conceivable to modulate the microbiome for higher response rates and greater efficacy of cancer immunotherapy (figure 3). Future research is warranted to delineate clinically actionable therapeutic options and advance microbiome-centered interventions beyond the gut compartment.
Figure 3.
Concepts for microbiome-centered interventions. Specific microbial compartments may be amenable for therapeutic intervention to resensitize to ICI treatment. Targeted modulation of specific microbiomes may be achieved based on different conceptual approaches. Supplementing particular microbial species (eg, Bifidobacterium) using different routes of administration could modulate the tumor microbiome, the GI microbiome, and the lung microbiome, respectively, to increase the diversity and/or make the functional composition more beneficial. On the other hand, the use of narrow-spectrum antibiotics against harmful bacteria or FMT from ICI-responding patients could restore ICI efficacy in otherwise non-responding/refractory patients. Personalized vaccination against tumor-associated antigens from intracellular bacteria or viruses represents another strategy by which microbial species might be harnessed for ICI-resensitizing intervention. Finally, adopting specific dietary behaviors such as high-fiber- or fasting-mimicking diet may also facilitate ICI performance by inducing favorable immunological changes in the TME. Figure is created with BioRender.com. FMT, fecal microbiota transplantation; GI, gastrointestinal; ICI, immune checkpoint inhibitor; TME, tumor microenvironment.
Concluding remarks
The clinical relevance of the tumor-associated microbiome for cancer immunotherapy efficacy is increasingly recognized. Microbes within the TME are associated with clinicopathological tumor characteristics and shape the anticancer immune response through immunological and metabolic means. In addition, tumor-associated microbes have direct effects on cancer cells including the regulation of tumor cell stemness and genotoxicity. Preliminary data suggest that there is at least some degree of interdependence between the tumor and the gut microbiome, and that the composition of the tumor microbiome may be amenable for therapeutic modulation to re-sensitize to ICI therapy. An increase in the overall tumor microbial diversity as well as targeted supplementation/elimination of beneficial/harmful microbes and genetic microbial engineering all represent rational therapeutic concepts. However, it is important to note that research on the tumor-associated microbiome is still in its infancy, with many current unknowns and only few mechanisms resolved at this stage. Challenges for developing the field include tissue sample/biopsy availability (especially in advanced-stage/inoperable cancer), low microbiota abundance in tumor tissue, cross-contamination during sample retrieval, intrapatient and interpatient variation of tumor-associated microbiomes, the identification of ‘driver’ (causal) versus ‘passenger’ (associated) microbes, and the elucidation of feasible therapeutic strategies. Notwithstanding, the tumor-associated microbiome offers great potential and the clinical perspective is to use knowledge about the tumor microbiota to deliver informed, microbiome-tailored precision ‘immune-microbioloncology’. To reach this goal, future studies should aim at systematically characterizing different tumor microbiomes and identify therapeutically relevant phyla, classes, orders, families, genera, and/or species. In a second step, strategies for the therapeutic modulation of the tumor microbiome should be delineated, considering both systemic and local supplementation approaches, dietary interventions, and the use of narrow-spectrum antibiotics. Higher response rates and enhanced efficacy of cancer immunotherapy are the overarching goals of microbiome-centered interventions.
Acknowledgments
Figures were created with BioRender.com.
Footnotes
Contributors: Wrote the first draft of the paper: MB and MHB. Wrote the final version of the paper: MB, LH, FB, AP, DW, SS, RS, HT, and MHB. Generated the figures: LH. Approved the paper for publication: MB, LH, FB, AP, DW, SS, RS, HT, and MHB.
Funding: Research leading to this work was kindly supported by the Lungenliga St.Gallen-Appenzell based in St.Gallen, Switzerland (grant to MB, no grant number available), and the Stiftung Propter Homines based in Vaduz, Principality of Liechtenstein (grant to MB, no grant number available). DW was supported by the Deutsche Krebshilfe (grant No. DKH 70112994). DW and AP were supported by the In Memoriam Gabriel Salzner Stiftung. LH was supported by the OeGHO.
Competing interests: MB serves as an advisor for Pantec Biosolutions AG. The other authors have no potential conflicts of interest to declare. No medical writer was involved in the preparation of the manuscript.
Provenance and peer review: Commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1.Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 2018;378:2078–92. 10.1056/NEJMoa1801005 [DOI] [PubMed] [Google Scholar]
- 2.Galsky MD, Arija José Ángel Arranz, Bamias A, et al. Atezolizumab with or without chemotherapy in metastatic urothelial cancer (IMvigor130): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet 2020;395:1547–57. 10.1016/S0140-6736(20)30230-0 [DOI] [PubMed] [Google Scholar]
- 3.Motzer RJ, Rini BI, McDermott DF, et al. Nivolumab plus ipilimumab versus sunitinib in first-line treatment for advanced renal cell carcinoma: extended follow-up of efficacy and safety results from a randomised, controlled, phase 3 trial. Lancet Oncol 2019;20:1370–85. 10.1016/S1470-2045(19)30413-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robert C, Ribas A, Schachter J, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol 2019;20:1239–51. 10.1016/S1470-2045(19)30388-2 [DOI] [PubMed] [Google Scholar]
- 5.Postow MA, Sidlow R, Hellmann MD. Immune-Related adverse events associated with immune checkpoint blockade. N Engl J Med 2018;378:158–68. 10.1056/NEJMra1703481 [DOI] [PubMed] [Google Scholar]
- 6.Johnson DB, Reynolds KL, Sullivan RJ, et al. Immune checkpoint inhibitor toxicities: systems-based approaches to improve patient care and research. Lancet Oncol 2020;21:e398–404. 10.1016/S1470-2045(20)30107-8 [DOI] [PubMed] [Google Scholar]
- 7.Nesline MK, Knight T, Colman S, et al. Economic burden of checkpoint inhibitor immunotherapy for the treatment of non-small cell lung cancer in US clinical practice. Clin Ther 2020;42:1682–98. 10.1016/j.clinthera.2020.06.018 [DOI] [PubMed] [Google Scholar]
- 8.Verma V, Sprave T, Haque W, et al. A systematic review of the cost and cost-effectiveness studies of immune checkpoint inhibitors. J Immunother Cancer 2018;6:128. 10.1186/s40425-018-0442-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davar D, Dzutsev AK, McCulloch JA, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021;371:595–602. 10.1126/science.abf3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spencer CN, McQuade JL, Gopalakrishnan V, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 2021;374:1632–40. 10.1126/science.aaz7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doroshow DB, Bhalla S, Beasley MB, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol 2021;18:345–62. 10.1038/s41571-021-00473-5 [DOI] [PubMed] [Google Scholar]
- 12.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther 2015;14:847–56. 10.1158/1535-7163.MCT-14-0983 [DOI] [PubMed] [Google Scholar]
- 13.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samstein RM, Lee C-H, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202–6. 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim ES, Velcheti V, Mekhail T, et al. Blood-based tumor mutational burden as a biomarker for atezolizumab in non-small cell lung cancer: the phase 2 B-F1RST trial. Nat Med 2022;28:939–45. 10.1038/s41591-022-01754-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch Repair-Deficient/Microsatellite Instability-High metastatic colorectal cancer. J Clin Oncol 2018;36:773–9. 10.1200/JCO.2017.76.9901 [DOI] [PubMed] [Google Scholar]
- 17.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banchereau R, Leng N, Zill O, et al. Molecular determinants of response to PD-L1 blockade across tumor types. Nat Commun 2021;12:3969. 10.1038/s41467-021-24112-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumacher TN, Scheper W, Kvistborg P. Cancer neoantigens. Annu Rev Immunol 2019;37:173–200. 10.1146/annurev-immunol-042617-053402 [DOI] [PubMed] [Google Scholar]
- 20.Thommen DS, Schumacher TN. T cell dysfunction in cancer. Cancer Cell 2018;33:547–62. 10.1016/j.ccell.2018.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Li Y, Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis 2015;6:e1792. 10.1038/cddis.2015.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariathasan S, Turley SJ, Nickles D, et al. Tgfβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiss M, Vande Walle L, Saavedra PHV, et al. IL1β promotes immune suppression in the tumor microenvironment independent of the inflammasome and gasdermin D. Cancer Immunol Res 2021;9:309–23. 10.1158/2326-6066.CIR-20-0431 [DOI] [PubMed] [Google Scholar]
- 24.Gurusamy D, Clever D, Eil R, et al. Novel "Elements" of Immune Suppression within the Tumor Microenvironment. Cancer Immunol Res 2017;5:426–33. 10.1158/2326-6066.CIR-17-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf D, Fiegl H, Zeimet AG, et al. High RIG‐I expression in ovarian cancer associates with an immune‐escape signature and poor clinical outcome. Int J Cancer 2020;146:2007–18. 10.1002/ijc.32818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov 2022;12:31–46. 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- 27.Zitvogel L, Ma Y, Raoult D, et al. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science 2018;359:1366–70. 10.1126/science.aar6918 [DOI] [PubMed] [Google Scholar]
- 28.Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079–84. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018;359:91–7. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 30.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finlay BB, Goldszmid R, Honda K. Can we harness the microbiota to enhance the efficacy of cancer immunotherapy? Nat Rev Immunol 2020. [DOI] [PubMed] [Google Scholar]
- 32.Derosa L, Routy B, Desilets A, et al. Microbiota-Centered interventions: the next breakthrough in Immuno-Oncology? Cancer Discov 2021;11:2396–412. 10.1158/2159-8290.CD-21-0236 [DOI] [PubMed] [Google Scholar]
- 33.Derosa L, Hellmann MD, Spaziano M, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Annals of Oncology 2018;29:1437–44. 10.1093/annonc/mdy103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schett A, Rothschild SI, Curioni-Fontecedro A, et al. Predictive impact of antibiotics in patients with advanced non small-cell lung cancer receiving immune checkpoint inhibitors : Antibiotics immune checkpoint inhibitors in advanced NSCLC. Cancer Chemother Pharmacol 2020;85:121–31. 10.1007/s00280-019-03993-1 [DOI] [PubMed] [Google Scholar]
- 35.Baruch EN, Youngster I, Ben-Betzalel G, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021;371:602–9. 10.1126/science.abb5920 [DOI] [PubMed] [Google Scholar]
- 36.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science 2015;350:1084–9. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derosa L, Routy B, Thomas AM, et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat Med 2022;28:315–24. 10.1038/s41591-021-01655-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boesch M, Baty F, Rothschild SI, et al. Tumour neoantigen mimicry by microbial species in cancer immunotherapy. Br J Cancer 2021;125:313–23. 10.1038/s41416-021-01365-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balachandran VP, Łuksza M, Zhao JN, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017;551:512–6. 10.1038/nature24462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomura M, Nagatomo R, Doi K, et al. Association of short-chain fatty acids in the gut microbiome with clinical response to treatment with nivolumab or pembrolizumab in patients with solid cancer tumors. JAMA Netw Open 2020;3:e202895. 10.1001/jamanetworkopen.2020.2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malczewski AB, Navarro S, Coward JI, et al. Microbiome-derived metabolome as a potential predictor of response to cancer immunotherapy. J Immunother Cancer 2020;8:e001383. 10.1136/jitc-2020-001383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin Y, Dong H, Xia L, et al. The diversity of gut microbiome is associated with favorable responses to Anti–Programmed death 1 immunotherapy in Chinese patients with NSCLC. J Thorac Oncol 2019;14:1378–89. 10.1016/j.jtho.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 43.Boesch M, Baty F, Albrich WC, et al. Local tumor microbial signatures and response to checkpoint blockade in non-small cell lung cancer. Oncoimmunology 2021;10:1988403. 10.1080/2162402X.2021.1988403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017;170:548–63. 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geller LT, Barzily-Rokni M, Danino T, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017;357:1156–60. 10.1126/science.aah5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016;14:e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014;12:87. 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Goffau MC, Lager S, Salter SJ, et al. Recognizing the reagent microbiome. Nat Microbiol 2018;3:851–3. 10.1038/s41564-018-0202-y [DOI] [PubMed] [Google Scholar]
- 50.Marsh RL, Nelson MT, Pope CE, et al. How low can we go? the implications of low bacterial load in respiratory microbiota studies. Pneumonia 2018;10:7. 10.1186/s41479-018-0051-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chou S, Zhang S, Guo H, et al. Targeted antimicrobial agents as potential tools for modulating the gut microbiome. Front Microbiol 2022;13:879207. 10.3389/fmicb.2022.879207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene 2020;39:4925–43. 10.1038/s41388-020-1341-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wastyk HC, Fragiadakis GK, Perelman D, et al. Gut-microbiota-targeted diets modulate human immune status. Cell 2021;184:4137–53. 10.1016/j.cell.2021.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montalban-Arques A, Katkeviciute E, Busenhart P, et al. Commensal Clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe 2021;29:1573–88. 10.1016/j.chom.2021.08.001 [DOI] [PubMed] [Google Scholar]
- 55.Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol 2019;20:1279–90. 10.1038/s41590-019-0451-9 [DOI] [PubMed] [Google Scholar]
- 56.Rowe M, Veerus L, Trosvik P, et al. The reproductive microbiome: an emerging driver of sexual selection, sexual conflict, mating systems, and reproductive isolation. Trends Ecol Evol 2020;35:220–34. 10.1016/j.tree.2019.11.004 [DOI] [PubMed] [Google Scholar]
- 57.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol 2018;16:143–55. 10.1038/nrmicro.2017.157 [DOI] [PubMed] [Google Scholar]
- 58.Mishra A, Lai GC, Yao LJ, et al. Microbial exposure during early human development primes fetal immune cells. Cell 2021;184:3394–409. 10.1016/j.cell.2021.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol 2013;14:646–53. 10.1038/ni.2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tropini C, Earle KA, Huang KC, et al. The gut microbiome: connecting spatial organization to function. Cell Host Microbe 2017;21:433–42. 10.1016/j.chom.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol 2020;38:23–48. 10.1146/annurev-immunol-070119-115104 [DOI] [PubMed] [Google Scholar]
- 62.Gollwitzer ES, Saglani S, Trompette A, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med 2014;20:642–7. 10.1038/nm.3568 [DOI] [PubMed] [Google Scholar]
- 63.Gollwitzer ES, Marsland BJ. Impact of early-life exposures on immune maturation and susceptibility to disease. Trends Immunol 2015;36:684–96. 10.1016/j.it.2015.09.009 [DOI] [PubMed] [Google Scholar]
- 64.Bäckhed F, Fraser CM, Ringel Y, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe 2012;12:611–22. 10.1016/j.chom.2012.10.012 [DOI] [PubMed] [Google Scholar]
- 65.Dang AT, Marsland BJ. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol 2019;12:843–50. 10.1038/s41385-019-0160-6 [DOI] [PubMed] [Google Scholar]
- 66.Sharon G, Garg N, Debelius J, et al. Specialized metabolites from the microbiome in health and disease. Cell Metab 2014;20:719–30. 10.1016/j.cmet.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loomis KH, Wu SK, Ernlund A, et al. A mixed community of skin microbiome representatives influences cutaneous processes more than individual members. Microbiome 2021;9:22. 10.1186/s40168-020-00963-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet 2003;361:512–9. 10.1016/S0140-6736(03)12489-0 [DOI] [PubMed] [Google Scholar]
- 69.Mallott EK, Amato KR. Host specificity of the gut microbiome. Nat Rev Microbiol 2021;19:639–53. 10.1038/s41579-021-00562-3 [DOI] [PubMed] [Google Scholar]
- 70.Groussin M, Mazel F, Alm EJ. Co-Evolution and Co-speciation of Host-Gut bacteria systems. Cell Host Microbe 2020;28:12–22. 10.1016/j.chom.2020.06.013 [DOI] [PubMed] [Google Scholar]
- 71.Sorbara MT, Pamer EG. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol 2019;12:1–9. 10.1038/s41385-018-0053-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hardy BL, Merrell DS. Friend or foe: interbacterial competition in the nasal cavity. J Bacteriol 2021;203. 10.1128/JB.00480-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tilg H, Adolph TE, Gerner RR, et al. The intestinal microbiota in colorectal cancer. Cancer Cell 2018;33:954–64. 10.1016/j.ccell.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 74.Whisner CM, Athena Aktipis C. The role of the microbiome in cancer initiation and progression: how microbes and cancer cells utilize excess energy and promote one another's growth. Curr Nutr Rep 2019;8:42–51. 10.1007/s13668-019-0257-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017;358:1443–8. 10.1126/science.aal5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsay J-CJ, Wu BG, Sulaiman I, et al. Lower airway dysbiosis affects lung cancer progression. Cancer Discov 2021;11:293–307. 10.1158/2159-8290.CD-20-0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zitvogel L, Kroemer G. Lower airway dysbiosis exacerbates lung cancer. Cancer Discov 2021;11:224–6. 10.1158/2159-8290.CD-20-1641 [DOI] [PubMed] [Google Scholar]
- 78.Gowing SD, Chow SC, Cools-Lartigue JJ, et al. Gram-Negative pneumonia augments non-small cell lung cancer metastasis through host Toll-like receptor 4 activation. J Thorac Oncol 2019;14:2097–108. 10.1016/j.jtho.2019.07.023 [DOI] [PubMed] [Google Scholar]
- 79.Jin C, Lagoudas GK, Zhao C, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell 2019;176:998–1013. 10.1016/j.cell.2018.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi Y, Zheng W, Yang K, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via sting signaling. J Exp Med 2020;217. 10.1084/jem.20192282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nejman D, Livyatan I, Fuks G, et al. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science 2020;368:973–80. 10.1126/science.aay9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fu A, Yao B, Dong T, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022;185:1356–72. 10.1016/j.cell.2022.02.027 [DOI] [PubMed] [Google Scholar]
- 83.Wan X, Song M, Wang A, et al. Microbiome crosstalk in immunotherapy and antiangiogenesis therapy. Front Immunol 2021;12:747914. 10.3389/fimmu.2021.747914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pal S, Perrien DS, Yumoto T, et al. The microbiome restrains melanoma bone growth by promoting intestinal NK and Th1 cell homing to bone. J Clin Invest 2022;132. 10.1172/JCI157340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poore GD, Kopylova E, Zhu Q, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020;579:567–74. 10.1038/s41586-020-2095-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pushalkar S, Hundeyin M, Daley D, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov 2018;8:403–16. 10.1158/2159-8290.CD-17-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Riquelme E, Zhang Y, Zhang L, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 2019;178:e12:795–806. 10.1016/j.cell.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ferone M, Gowen A, Fanning S, et al. Microbial detection and identification methods: bench top assays to omics approaches. Compr Rev Food Sci Food Saf 2020;19:3106–29. 10.1111/1541-4337.12618 [DOI] [PubMed] [Google Scholar]
- 89.Sanschagrin S, Yergeau E. Next-Generation sequencing of 16S ribosomal RNA gene amplicons. JoVE 2014;90. 10.3791/51709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ranjan R, Rani A, Metwally A, et al. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun 2016;469:967–77. 10.1016/j.bbrc.2015.12.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kalaora S, Nagler A, Nejman D, et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 2021;592:138–43. 10.1038/s41586-021-03368-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loke MF, Chua EG, Gan HM, et al. Metabolomics and 16S rRNA sequencing of human colorectal cancers and adjacent mucosa. PLoS One 2018;13:e0208584. 10.1371/journal.pone.0208584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choi S, Chung J, Cho M-L, et al. Analysis of changes in microbiome compositions related to the prognosis of colorectal cancer patients based on tissue-derived 16S rRNA sequences. J Transl Med 2021;19:485. 10.1186/s12967-021-03154-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Quince C, Walker AW, Simpson JT, et al. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol 2017;35:833–44. 10.1038/nbt.3935 [DOI] [PubMed] [Google Scholar]
- 95.Debesa-Tur G, Pérez-Brocal V, Ruiz-Ruiz S, et al. Metagenomic analysis of formalin-fixed paraffin-embedded tumor and normal mucosa reveals differences in the microbiome of colorectal cancer patients. Sci Rep 2021;11:391. 10.1038/s41598-020-79874-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kwon M, Seo S-S, Kim M, et al. Compositional and functional differences between microbiota and cervical carcinogenesis as identified by shotgun metagenomic sequencing. Cancers 2019;11:309. 10.3390/cancers11030309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuchina A, Brettner LM, Paleologu L, et al. Microbial single-cell RNA sequencing by split-pool barcoding. Science 2021;371. 10.1126/science.aba5257. [Epub ahead of print: 19 02 2021]. 10.1126/science.aba5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Avital G, Avraham R, Fan A, et al. scDual-Seq: mapping the gene regulatory program of Salmonella infection by host and pathogen single-cell RNA-sequencing. Genome Biol 2017;18:200. 10.1186/s13059-017-1340-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014;15:317–28. 10.1016/j.chom.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat Commun 2015;6:6528. 10.1038/ncomms7528 [DOI] [PubMed] [Google Scholar]
- 101.Cao Y, Wu K, Mehta R, et al. Long-term use of antibiotics and risk of colorectal adenoma. Gut 2018;67:672–8. 10.1136/gutjnl-2016-313413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012;22:299–306. 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013;14:207–15. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012;22:292–8. 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 2013;105:1907–11. 10.1093/jnci/djt300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 2014;33:1381–90. 10.1007/s10096-014-2081-3 [DOI] [PubMed] [Google Scholar]
- 107.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016;65:1973–80. 10.1136/gutjnl-2015-310101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.André T, Shiu K-K, Kim TW, et al. Pembrolizumab in Microsatellite-Instability-High advanced colorectal cancer. N Engl J Med 2020;383:2207–18. 10.1056/NEJMoa2017699 [DOI] [PubMed] [Google Scholar]
- 109.Mekadim C, Skalnikova HK, Cizkova J, et al. Dysbiosis of skin microbiome and gut microbiome in melanoma progression. BMC Microbiol 2022;22:63. 10.1186/s12866-022-02458-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Komiyama S, Yamada T, Takemura N, et al. Profiling of tumour-associated microbiota in human hepatocellular carcinoma. Sci Rep 2021;11:10589. 10.1038/s41598-021-89963-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pfister D, Núñez NG, Pinyol R, et al. Nash limits anti-tumour surveillance in immunotherapy-treated HCC. Nature 2021;592:450–6. 10.1038/s41586-021-03362-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cummins J, Tangney M. Bacteria and tumours: causative agents or opportunistic inhabitants? Infect Agent Cancer 2013;8:11. 10.1186/1750-9378-8-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kimura NT, Taniguchi S, Aoki K, et al. Selective localization and growth of Bifidobacterium bifidum in mouse tumors following intravenous administration. Cancer Res 1980;40:2061–8. [PubMed] [Google Scholar]
- 114.Wang L, Chard Dunmall LS, Cheng Z, et al. Remodeling the tumor microenvironment by oncolytic viruses: beyond oncolysis of tumor cells for cancer treatment. J Immunother Cancer 2022;10:e004167. 10.1136/jitc-2021-004167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kosaka A, Ishibashi K, Nagato T, et al. Cd47 blockade enhances the efficacy of intratumoral STING-targeting therapy by activating phagocytes. J Exp Med 2021;218. 10.1084/jem.20200792. [Epub ahead of print: 01 11 2021]. 10.1084/jem.20200792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boesch M, Onder L, Cheng H-W, et al. Interleukin 7-expressing fibroblasts promote breast cancer growth through sustenance of tumor cell stemness. Oncoimmunology 2018;7:e1414129. 10.1080/2162402X.2017.1414129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boesch M, Zeimet AG, Reimer D, et al. The side population of ovarian cancer cells defines a heterogeneous compartment exhibiting stem cell characteristics. Oncotarget 2014;5:7027–39. 10.18632/oncotarget.2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boesch M, Spizzo G, Seeber A. Concise review: aggressive colorectal cancer: role of epithelial cell adhesion molecule in cancer stem cells and epithelial-to-mesenchymal transition. Stem Cells Transl Med 2018;7:495–501. 10.1002/sctm.17-0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zeimet AG, Reimer D, Sopper S, et al. Ovarian cancer stem cells. Neoplasma 2012;59:747–55. 10.4149/neo_2012_094 [DOI] [PubMed] [Google Scholar]
- 120.Hatina J, Boesch M, Sopper S, et al. Ovarian cancer stem cell heterogeneity. Adv Exp Med Biol 2019;1139:201–21. 10.1007/978-3-030-14366-4_12 [DOI] [PubMed] [Google Scholar]
- 121.Boesch M, Sopper S, Zeimet AG. Heterogeneity of cancer stem cells: rationale for targeting the stem cell niche. Biochim Biophys Acta 1866;2016:276–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu Y, Fu K, Wier EM, et al. Bacterial genotoxin accelerates transient Infection-Driven murine colon tumorigenesis. Cancer Discov 2022;12:236–49. 10.1158/2159-8290.CD-21-0912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Overacre-Delgoffe AE, Bumgarner HJ, Cillo AR, et al. Microbiota-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. Immunity 2021;54:2812–24. 10.1016/j.immuni.2021.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sioud M. T-cell cross-reactivity may explain the large variation in how cancer patients respond to checkpoint inhibitors. Scand J Immunol 2018;87:e12643. 10.1111/sji.12643 [DOI] [PubMed] [Google Scholar]
- 125.Leng Q, Tarbe M, Long Q, et al. Pre‐existing heterologous T‐cell immunity and neoantigen immunogenicity. Clin Transl Immunol 2020;9:e01111. 10.1002/cti2.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mager LF, Burkhard R, Pett N, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020;369:1481–9. 10.1126/science.abc3421 [DOI] [PubMed] [Google Scholar]
- 127.Park J, Kim M, Kang SG, et al. Short-Chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-s6K pathway. Mucosal Immunol 2015;8:80–93. 10.1038/mi.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang H, Rong X, Zhao G, et al. The microbial metabolite trimethylamine N-oxide promotes antitumor immunity in triple-negative breast cancer. Cell Metab 2022;34:581–94. 10.1016/j.cmet.2022.02.010 [DOI] [PubMed] [Google Scholar]
- 129.Canale FP, Basso C, Antonini G, et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature 2021;598:662–6. 10.1038/s41586-021-04003-2 [DOI] [PubMed] [Google Scholar]
- 130.Dizman N, Meza L, Bergerot P, et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: a randomized phase 1 trial. Nat Med 2022;28:704–12. 10.1038/s41591-022-01694-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lam KC, Araya RE, Huang A, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell 2021;184:5338–56. 10.1016/j.cell.2021.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vernieri C, Fucà G, Ligorio F, et al. Fasting-Mimicking diet is safe and reshapes metabolism and antitumor immunity in patients with cancer. Cancer Discov 2022;12:90–107. 10.1158/2159-8290.CD-21-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jin W-B, Li T-T, Huo D, et al. Genetic manipulation of gut microbes enables single-gene interrogation in a complex microbiome. Cell 2022;185:e22:547–62. 10.1016/j.cell.2021.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.WL W, Adame MD, Liou CW. Microbiota regulate social behaviour via stress response neurons in the brain. Nature 2021;595:409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Perry RJ, Peng L, Barry NA, et al. Acetate mediates a microbiome–brain–β-cell axis to promote metabolic syndrome. Nature 2016;534:213–7. 10.1038/nature18309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zheng JH, Nguyen VH, Jiang S-N, et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci Transl Med 2017;9. 10.1126/scitranslmed.aak9537 [DOI] [PubMed] [Google Scholar]
- 137.Sahin U, Türeci Özlem. Personalized vaccines for cancer immunotherapy. Science 2018;359:1355–60. 10.1126/science.aar7112 [DOI] [PubMed] [Google Scholar]
- 138.Lang F, Schrörs B, Löwer M, et al. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat Rev Drug Discov 2022;21:261–82. 10.1038/s41573-021-00387-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Thakur BK, Malaisé Y, Martin A. Unveiling the mutational mechanism of the bacterial genotoxin colibactin in colorectal cancer. Mol Cell 2019;74:227–9. 10.1016/j.molcel.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 140.Gagnaire A, Nadel B, Raoult D, et al. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol 2017;15:109–28. 10.1038/nrmicro.2016.171 [DOI] [PubMed] [Google Scholar]