Abstract
Campylobacter jejuni is a leading cause of infectious diarrhea throughout the world. In addition, there is growing evidence that Guillain-Barré syndrome, an inflammatory demyelinating disease of the peripheral nervous system, is frequently preceded by C. jejuni infection. In the present study, the hrcA-grpE-dnaK gene cluster of C. jejuni was cloned and sequenced. The dnaK gene consists of an open reading frame of 1,869 bp and encodes a protein with a high degree of homology to other bacterial 70-kDa heat shock proteins (HSPs). The overall percentages of identity to the HSP70 proteins of Helicobacter pylori, Borrelia burgdorferi, Chlamydia trachomatis, and Bacillus subtilis were calculated to be 78.1, 60.5, 57.2, and 53.8%, respectively. Regions similar to the Escherichia coli ς70 promoter consensus sequence and to a cis-acting regulatory element (CIRCE) are located upstream of the hrcA gene. Following heat shock, a rapid increase of dnaK mRNA was detectable, which reached its maximum after 20 to 30 min. A 6-His-tagged recombinant DnaK protein (rCjDnaK-His) was generated in E. coli, after cloning of the dnaK coding region into pET-22b(+), and purified by affinity and gel filtration chromatography. Antibody responses to rCjDnaK-His were significantly elevated, compared to those of healthy individuals, in about one-third of the serum specimens obtained from C. jejuni enteritis patients.
Campylobacter jejuni is a bacterial enteric pathogen of increasing medical interest. It is currently regarded as a leading cause of infectious diarrhea throughout the world (35). Particularly, Campylobacter infection is endemic in developing countries, where it contributes considerably to diarrheal disease among young children (38). There is growing evidence that Guillain-Barré syndrome (GBS), a rare but potentially devastating disease of the peripheral nervous system, is frequently preceded by a C. jejuni infection (27). It has been estimated that in the United States about 1 in every 1,000 cases of C. jejuni infection is followed by GBS (23). In spite of the obvious importance of this microorganism, remarkably little is known about the molecular mechanisms involved in virulence, pathogenesis, and immune response during Campylobacter-host interaction (20). Only a few definite protein antigens have been characterized so far. Reconvalescent-phase sera usually react with the 65-kDa flagellin, the 44-kDa major outer membrane protein, and 25- to 29-kDa surface proteins (7, 12, 24, 43). PEB1, which has been proposed to function as an adhesin for adherence of the bacterium to eukaryotic cells, is highly immunogenic (26), and Campylobacter trigger factor has recently been revealed to be a humoral antigen in the human host (16).
GroEL- and GroES-like heat shock proteins (HSPs) of C. jejuni have been shown to elicit a serum immunoglobulin G (IgG) as well as a secretory IgA response in experimentally infected rabbits (46). HSPs are synthesized in virtually all cells under conditions of stress, e.g., as a result of temperature or nutrient change. The best-characterized HSPs belong to the 60-kDa (GroEL) and 70-kDa (DnaK) families and are the most conserved proteins known. Bacterial HSPs have aroused the interest of microbiologists for many years, since they represent major targets of the host’s immune response (19). Although less extensively studied than GroEL, DnaK homologues of many bacterial pathogens have been found to be immunogenic in humans or animals (1, 2, 5, 8, 10, 47). Furthermore, as has been shown by the experimental infection of mice with Borrelia burgdorferi, immunization with proteins containing DnaK-specific sequences may protect against microbial infection (4).
Growing evidence suggests that there are two paradigmatic mechanisms in bacterial heat shock regulation. In the gram-negative species Escherichia coli, heat induction is mediated by the alternative sigma factor ς32, allowing for the coordinate expression of genes that belong to the so-called ς32 regulon (50). After enhancing transcription of heat shock genes, ς32 is sequestered by the DnaK chaperone machine and degraded by ATP-dependent cellular proteases (22, 40). Alternatively, in the gram-positive model organism Bacillus subtilis, a highly conserved palindromic sequence is involved as a cis-acting operator in the regulation of class I (groE and dnaK) heat shock genes. As known so far, this element, designated CIRCE (for controlling inverted repeat of chaperone expression) by Zuber and Schumann (51), is located at the groE and/or dnaK upstream regulatory region of more than 30 bacterial species (for a recent review, see reference 34). CIRCE most probably acts at the DNA level by binding a repressor encoded by the hrcA gene (hrc, for heat shock regulation at CIRCE elements) and, if the inverted repeat is a part of the transcript, by modulating mRNA stability (31, 48, 49). Heat shock regulation in C. jejuni is poorly understood. In front of the C. jejuni dnaJ heat shock gene were observed sequences compatible with E. coli ς32 as well as ς70 consensus sequences (21). We have recently demonstrated that heat-induced transcription of the groESL operon, which is preceded by a CIRCE element, is under the control of a ς70-like promoter (39).
We are interested in the heat shock response of C. jejuni and in the role of this organism’s heat shock proteins as putative immune targets in infectious and autoimmune diseases. As a part of this work, the hrcA-grpE-dnaK gene cluster from C. jejuni was cloned and sequenced. The protein encoded by dnaK was expressed in E. coli, and the humoral response against this protein in C. jejuni-infected patients and in healthy individuals was examined.
MATERIALS AND METHODS
Bacterial strains, vectors, and reagents.
The C. jejuni strain used in this study was isolated in the Department of Hygiene and Microbiology of the University of Würzburg and serotyped as Lior 11. E. coli BL21(DE3) and plasmid pET-22b(+) were purchased from Novagen (Madison, Wis.). pCR-Script SK(+) plasmid and XL-1 blue MRF′ Kanr supercompetent cells were purchased from Stratagene (La Jolla, Calif.). Vent polymerase and restriction enzymes were obtained from New England Biolabs (Beverly, Mass.).
Bacterial cultivation and DNA purification.
C. jejuni was routinely grown on agar plates at 37°C in a microaerophilic environment (5% O2, 10% CO2). To induce heat shock, surface-grown bacterial cells were harvested, subcultured to brain heart broth supplemented with 1% yeast extract (BHIYE) (46), and subsequently shifted to 48°C for 5 to 60 min. E. coli strains were grown at 37°C on Luria-Bertani agar or in Luria-Bertani broth supplemented with ampicillin at a final concentration of 50 μg ml−1, if required. Genomic DNA was isolated as described elsewhere (3). Plasmid DNA was purified by using a QIAprep plasmid kit (Qiagen, Hilden, Germany).
Molecular cloning of the dnaK gene cluster.
Two degenerate oligonucleotide primers, 5′-GG(A/T)AT(A/T)GA(C/T)(C/T)T(A/G/T)GGIACIAC(A/C/T)AA(C/T)TC-3′ and 5′-CC(A/T)GC(A/G/T)ATI(C/T)(G/T)(A/G/T)CCIGC(A/G)TC(C/T)TT-3′, were synthesized according to highly conserved regions of bacterial HSP70 proteins (corresponding to amino acids [aa] 6 to 14 and aa 155 to 162 of E. coli DnaK, respectively). PCR was conducted with a GeneAmp 9600 PCR system (Perkin-Elmer), using the following cycle parameters: 94°C for 30 s, 52°C (42°C during the first 5 cycles) for 30 s, and 72°C for 1 min for 40 cycles. PCR products were ligated with pCR-Script SK(+), cloned into XL1-Blue MRF′ Kanr supercompetent cells, and sequenced. One 480-bp fragment, which exhibited high nucleotide sequence homology to bacterial dnaK sequences, was 32P labeled by random priming (14). A plasmid library was constructed by digesting C. jejuni genomic DNA with BglII and cloning the resulting fragments into pET-22b(+). The library was screened with the radiolabeled 480-bp PCR product in accordance with standard procedures (30). Sequencing revealed that one recovered clone (pdnaK1) with a 1.6-kb insert contained the 3′ end of the putative hrcA gene, the entire grpE gene, and the 5′ end of the dnaK gene. The missing parts of the hrcA and the dnaK genes were obtained by a PCR-based genomic walking strategy as recently described (39). Briefly, seminested PCRs were performed by using two sequence-specific oligonucleotides oriented toward the unknown 3′ end and a random oligonucleotide as the reverse primer, which was used in both the inner and outer PCRs. For each genomic walking step, 10 to 15 arbitrarily chosen primers previously available in our lab were tested. To obtain the missing dnaK 3′ end, two rounds of randomly primed PCR (with random primers 5′-CTCCAAAAACTCATCCTGTACCTT-3′ and 5′-CCTAAATCTCCAGACAAAGCTCAC-3′, respectively) were necessary. A PCR product containing the 5′ end of hrcA was amplified by using random primer 5′-CACGGGAGACTTGGAAAACAC-3′. The resulting amplicons were cloned into pCR-Script SK(+) vectors and sequenced.
DNA sequence analysis.
The DNA sequence was determined on both strands by using a ABI Prism Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, Calif.). DNA sequencing was performed on an ABI model 373A automated sequencer (Applied Biosystems). Nucleic acid and predicted amino acid sequence data were analyzed by using the DNASIS software package (Hitachi, Tokyo, Japan). The Helicobacter pylori DNA sequences were obtained from the Institute for Genomic Research website (18a).
Northern blot analysis.
Total RNA was isolated by using TRIzol reagent (Gibco BRL, Gaithersburg, Md.) as recommended by the manufacturer. To generate a probe, a 539-bp DNA product internal to dnaK was amplified by using oligonucleotide primers 5′-TCACGCAAATCATCAAGAGCT-3′ and 5′-ACTTGATGTTACTCCGCTCTCT-3′ and then 32P labeled. Ten micrograms of total RNA was size separated in a formaldehyde-containing denaturing 1.2% agarose gel. Northern blotting and hybridization were performed according to standard procedures (3). Filters were autoradiographed at −80°C overnight. A 0.24- to 9.5-kb RNA ladder (Gibco BRL) was used as a size marker.
Primer extension analysis.
Transcriptional start sites were determined by primer extension analysis using 5′-IRD800-labeled oligonucleotides (MWG-Biotech, Ebecsberg, Germany). 5′-CATACAC-AGCAACACAAGAATT-3′ is complementary to nucleotides +37 to +58 relative to the dnaK start codon, 5′-TATCTTGCAAATCATCCTGC-3′ is complementary to nucleotides +57 to +76 relative to the grpE translation start site, and 5′-ATAGGCGCATTATCCAAAAG-3′ is complementary to nucleotides +58 to +77 relative to the hrcA start codon. Total RNA was isolated from C. jejuni cells that had been heat shocked for 20 min at 48°C. Contaminating DNA was removed by DNase I (Gibco BRL) treatment. One picomole of each primer was annealed to 10 μg of RNA for 20 min at 52°C and then for 10 min at room temperature. The oligonucleotide primers were extended with 1 U of avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.) at 42°C for 30 min. The samples were precipitated with ethanol and resuspended in a solution containing 2 μl of H2O and 1.5 μl of formamide loading dye (Promega). Nucleotide sequencing was carried out by using a Thermo Sequenase Fluorescent Labelled Primer Cycle Sequencing Kit (Amersham Pharmacia, Little Chalfont, United Kingdom). The extension products and sequencing reactions performed with the same primer were loaded onto a 6% polyacrylamide sequencing gel and analyzed on a model 4000 automated DNA sequencer (LI-COR, Lincoln, Nebr.).
Purification of recombinant C. jejuni DnaK protein.
The full-length coding region of the dnaK gene was amplified by PCR with primers 5′-AAAAGGATAACATATGAGTAAAGTTATAGGTA-3′ and 5′-TCAACTTCAGCGTCGATTAC-3′ (NdeI site is underlined), using 1 U of Vent polymerase per 100-μl reaction volume. The resulting DNA product was digested with NdeI and ligated with pET-22b(+) which had been digested with XhoI, blunt ended with mung bean nuclease, and then digested with NdeI. Thus, the last codon of the dnaK open reading frame (ORF) was fused in frame to the 5′ end of the vector sequence encoding a 6-histidine tail. The nucleotide sequence of the insert was confirmed by sequence analysis to be correct. The plasmid was transformed into E. coli BL21(DE3). Large-scale expression and nondenaturing purification by nickel-nitrilotriacetic acid (Qiagen) metal affinity chromatography were performed as proposed by the manufacturer. Fractions containing the recombinant protein were pooled and applied on a Sephacryl S100-HiPrep 26/69 column (Pharmacia, Freiburg, Germany). The N-terminal sequence of the purified protein was determined by the Edman degradation technique (model 473A amino acid analyzer; Applied Biosystems).
Western blot analysis and ELISA.
For Western blot experiments, the purified recombinant protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Nonspecific binding sites were saturated for 2 h with a solution consisting of 10% fetal calf serum and 1% bovine serum albumin in phosphate-buffered saline (PBS), pH 7.4. The nitrocellulose sheet was cut into strips, which were incubated with either human sera (diluted 1:200) or an anti-histidine tag monoclonal antibody (Dianova, Hamburg, Germany). After the strips were washed with PBS–0.05% Tween 20, bound antibodies were detected with anti-human IgG and anti-mouse IgG peroxidase-conjugated secondary antibodies, respectively. For the enzyme-linked immunosorbent assay (ELISA), 1 μg of recombinant protein ml−1 was adsorbed onto 96-well microtiter plates where were then incubated overnight at 4°C. After the plates were washed with PBS–0.05% Tween 20, remaining binding sites were blocked with a solution consisting of 5% fetal calf serum, 0.1% bovine serum albumin, and 0.03% gelatin in PBS, pH 7.4. Again, the plates were washed and incubated with serially diluted human sera overnight at 4°C. Detection of bound immunoglobulins was achieved by incubation for 30 min at room temperature with peroxidase-coupled anti-human IgG and IgA antibodies, respectively, diluted 1:5,000 in blocking buffer. Antibody binding was visualized with 2 mM ABTS [2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid); Boehringer, Mannheim, Germany] in ABTS substrate buffer (Boehringer). Optical densities (ODs) was read at wavelengths of 405 and 450 nm.
Nucleotide sequence accession number.
The nucleotide sequence of the dnaK locus was assigned GenBank/EMBL accession no. Y17165.
RESULTS
Molecular cloning of the dnaK gene cluster.
PCR on genomic DNA, using two degenerate oligonucleotide primers derived from highly conserved regions of bacterial HSP70 proteins, resulted in the amplification of three DNA fragments of 480 to 500 bp. Nucleotide sequence analysis revealed that one of the isolated fragments (designated dnaK2) was highly homologous to bacterial dnaK genes. A genomic plasmid library was constructed and screened by colony hybridization with the 32P-labeled PCR product. One recombinant plasmid (pdnaK1), carrying a 1.6-kb BglII fragment, was recovered. A database homology search based on nucleotide analysis of the 1.6-kb insert showed that the 5′ end of the putative C. jejuni dnaK gene was present in pdnaK1. The missing 3′ end was obtained by a PCR-based genomic walking approach. After two rounds of randomly primed PCR, two overlapping DNA products of 1.1 and 1.0 kb (dnaK3 and dnaK4, respectively) were amplified, cloned, and sequenced. As was shown by nucleotide sequence analysis, the missing 3′ end of the dnaK gene was contained within the two amplified fragments. To determine the nucleotide sequence of the entire hrcA-grpE-dnaK gene cluster, a 1.2-kb PCR product (hrcA1) was amplified by the genomic walking approach. Cloning and sequencing of the relevant region revealed that the amplicon contained the 5′ end of hrcA and the upstream flanking region.
Nucleotide sequences and analysis of the hrcA, grpE, and dnaK genes.
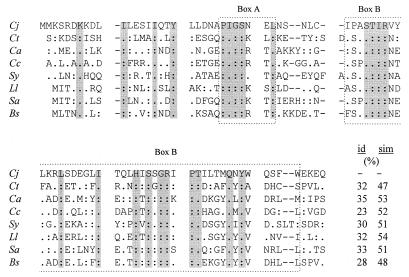
By reconstruction of the genomic DNA sequences from the insert of pdnaK1 and from the three PCR products, the complete nucleotide sequence of the dnaK gene cluster from C. jejuni was determined (Fig. 1B). The dnaK gene consists of an ORF (ORF1) of 1,869 bp and encodes a polypeptide of 623 aa with a predicted molecular mass of 67.3 kDa and a pI of 4.82. The dnaK gene is preceded by a putative ribosome binding site (AAGGAT) 5 nucleotides upstream of the AUG start codon. The average G+C contents at positions 1, 2, and 3 of codons of the putative HSP70 gene are 50.6, 33.2, and 22.0%, respectively, confirming the codon usage commonly found in C. jejuni genes. The predicted protein displayed a high degree of homology to members of the 70-kDa family of HSPs. In particular, the overall degrees of identity to HSP70 proteins of H. pylori (18), Borrelia burgdorferi (2), Chlamydia trachomatis (10), and Bacillus subtilis (44) were calculated to be 78.1, 60.5, 57.2, and 53.8%, respectively. A second ORF (ORF2), which is 525 bp long, was identified 22 bp upstream of dnaK. ORF2 encodes a protein with a calculated molecular mass of 20.0 kDa and a pI of 4.64. This protein exhibited a moderate degree of homology to bacterial GrpE proteins, with overall degrees of identity of 37.8% (H. pylori), 36.6% (Bacillus subtilis), 36.2% (Borrelia burgdorferi), and 34.5% (Chlamydia trachomatis). A third ORF (ORF3) was found upstream of grpE. The stop codon TGA of ORF3 overlaps with the start codon GTG of the grpE gene, and consequently the putative ribosome binding site of grpE (AAGGAG) located 5 bp upstream of the start codon is a part of the preceding ORF. The initiation codon GTG is seldom used in the A+T-rich genome of C. jejuni, and we observed that other strains of C. jejuni used ATG as the grpE start codon. Interestingly, a similar overlapping of grpE with the upstream gene has been reported for the Chlamydia trachomatis hrcA-grpE-dnaK operon (37) and as well as the H. pylori genome (41) (Fig. 1C). ORF3, which is 792 bp long, codes for a protein with a molecular mass of 30.9 kDa and a pI of 5.1. The overall degree of identity of the encoded polypeptide to other bacterial HrcA proteins is low. However, closer inspection revealed a region of greater homology at the N-terminal end of the C. jejuni protein. This region (aa 2 to 83) exhibits identities to HrcA homologues of 35% (Clostridium acetobutylicum), 32% (Chlamydia trachomatis and Lactococcus lactis), 28% (Bacillus subtilis), and 23% (Caulobacter crescentus) (Fig. 2). In some bacterial species (e.g., Bacillus subtilis), dnaJ is a part of the dnaK operon. Therefore, we searched for dnaJ-homologous sequences downstream of dnaK, but no such region could be identified within 600 bp.
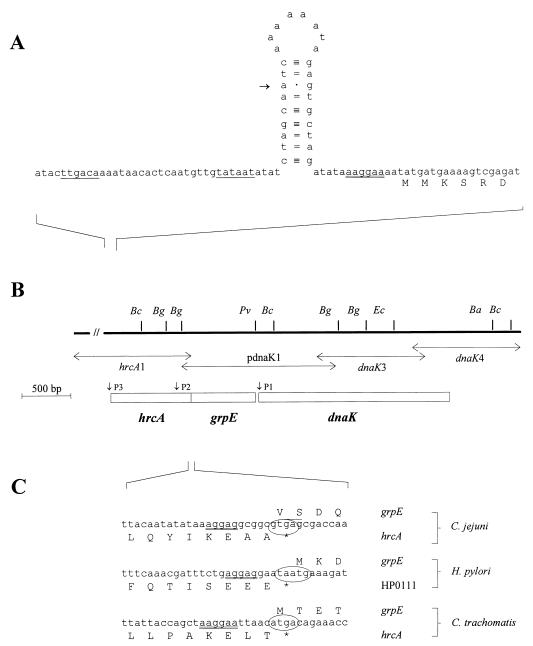
FIG. 1.
(A) The 5′ flanking region and the 5′ end of the hrcA gene. Putative promoter regions are single underlined. A putative ribosome binding site is double underlined. The CIRCE-like inverted repeat is depicted as a stem-loop structure. The putative transcription start site is indicated by an arrow. (B) Restriction map of the dnaK locus. Abbreviations for restriction sites are as follows: Bc, BclI; Bg, BglII; Pv, PvuII; Ec, EcoRI; and Ba, BalI. Vertical arrows indicate the insertion sites of plasmid pdnaK1 and the PCR products, from which the nucleotide sequence was derived. Boxes indicate the hrcA, grpE, and dnaK genes. (C) Nucleotide and amino acid sequences of the hrcA-grpE overlapping region from C. jejuni and corresponding sequences from H. pylori and Chlamydia trachomatis. Putative ribosome binding sites are double underlined. Overlapping start and stop codons are indicated by ovals.
FIG. 2.
Alignment of the deduced N-terminal amino acid sequence of the C. jejuni (Cj) HrcA protein with those from Chlamydia trachomatis (Ct; GenBank accession no. P54306), Clostridium acetobutylicum (Ca; P30727), Caulobacter crescentus (Cc; P54305), Synechococcus sp. (Sy; P72795), L. lactis (Ll; P42370), Staphylococcus aureus (Sa; P45556), and Bacillus subtilis (Bs; P25499). Amino acid residues identical to those of C. jejuni are indicated by colons, while conservative replacements are indicated by dots. Amino acid residues identical or conserved in all species are gray shaded. Gaps, indicated by dashes, have been introduced to maximize similarity. The numbers indicate the percentages of amino acid identity (id) and similarity (sim), respectively, between the N-terminal end of the C. jejuni HrcA protein and the corresponding region of the bacterial homologues.
Features of the noncoding regions.
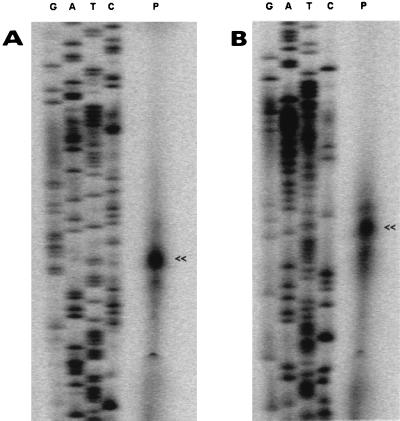
Using oligonucleotides complementary to the 5′ ends of dnaK and grpE, respectively, primer extension analyses revealed a putative transcription start (P1) site 46 bp upstream of the dnaK start codon (Fig. 3A) and a second putative transcription start site (P2) 184 bp upstream of the grpE start codon (data not shown). However, no regions convincingly compatible with ς70 or heat shock promoter consensus sequences (9) were identified upstream of P1 or P2. Surrounding P2, a high-energy (ΔG0 = −24.2 kcal mol−1) putative stem-loop structure was detected within the hrcA coding region, ranging from nucleotides 570 to 616 relative to the hrcA translation start site. Therefore, the possibility that a secondary structure in the RNA stopped the reverse transcriptase during the primer extension reaction cannot be ruled out. A third putative transcription start site (P3) was identified 34 bp upstream of the hrcA translation initiation codon (Fig. 3B). The region in front of P3 exhibited a nucleotide sequence identical to the E. coli ς70 consensus promoter sequence (TTGACA-N16–18-TATAAT). Furthermore, between the putative −10 promoter hexamer and the hrcA translation start site, a putative stem-loop structure similar to the CIRCE inverted repeat was detected. P3 is located at the seventh position of the 5′ branch of the hairpin (Fig. 1A). The C. jejuni sequence differs from the CIRCE consensus sequence (5′-TTAGCACTC-N9-GAGTGCTAA-3′) in that it exhibits three mismatches in the stem and contains 8 nucleotides in the loop of the putative hairpin structure.
FIG. 3.
Mapping of the 5′ end of the dnaK (A) and the hrcA (B) genes by primer extension analysis. IRD800-labeled oligonucleotides complementary to the 5′ ends of dnaK and hrcA, respectively, were hybridized with 10 μg of total RNA (lanes P). The letters G, A, T, and C above the lanes represent products of sequencing reactions using the same oligonucleotide as a primer. Arrowheads indicate the main primer extension products.
A 25-bp structure of dyad symmetry (ΔG0 = −14.2 kcal mol−1) 14 nucleotides downstream of the dnaK stop codon, followed by a stretch of 6 T’s, might function as a rho-independent transcription terminator (11, 28).
Observation of dnaK mRNA levels under conditions of heat stress.
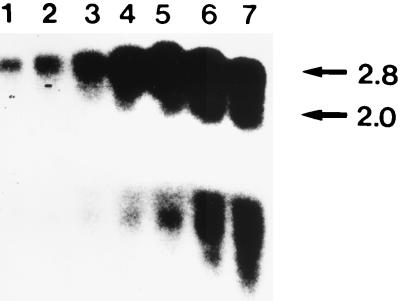
The in vivo transcripts of the dnaK locus were detected by Northern analysis with total RNA obtained from normally grown or heat-stressed bacteria. A 32P-labeled PCR product internal to dnaK hybridized with two mRNA species with molecular sizes of about 2.0 and 2.8 kb (Fig. 4). The 2.8-kb mRNA might correspond lengthwise to a grpE-dnaK transcript, whereas the 2.0-kb mRNA species might correspond to the dnaK gene or constitute a degradation product of the larger transcript. Following heat shock, there was a rapid increase in both mRNA species, with levels reaching a maximum after 20 to 30 min. Even after 60 min, the amount of mRNA was clearly augmented. Under the conditions described in Materials and Methods (growth at 37°C in a microaerobic environment) no dnaK-specific transcripts were detected in C. jejuni cells. However, after longer times of exposure, a signal of low intensity corresponding to a 2.8-kb mRNA species could be detected (data not shown). The possibility of nonspecific degradation of the mRNA was excluded because rehybridization of the blot with a groEL-specific probe yielded a distinct signal (data not shown). Since heat shock experiments were performed in an aerobic-atmosphere environment, C. jejuni cells were cultivated for 60 min at 37°C in BHIYE as a control. Under these conditions, specific mRNA was increased; however, the increase was weaker than that after 5 min of heat shock. This observation is in concordance with published results showing that expression of a putative HSP60 homologue of C. jejuni is induced under aerobic growth conditions (36).
FIG. 4.
Northern blot analysis. RNA was isolated at 5 min (lanes 2), 10 min (lane 3), 15 min (lane 4), 20 min (lane 5), 30 min (lane 6), and 60 min (lane 7) after C. jejuni cells were shifted to 48°C. Lane 1 contains RNA from C. jejuni cells incubated in BHIYE for 60 min at 37°C under aerobic conditions. Filters were hybridized with a 32P-labeled probe internal to dnaK. No signal was detected when using RNA from C. jejuni grown microaerobically at 37°C under the conditions described in Material and Methods (data not shown). The minor mRNA was detected in all RNA samples after longer periods of exposure (data not shown). Transcript sizes are given in kilobases.
Antigenicity of the recombinant DnaK protein.
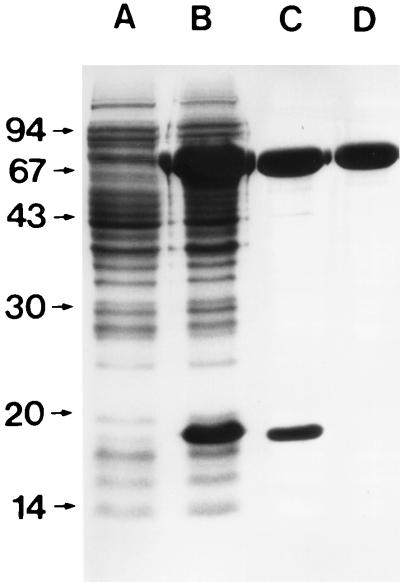
The full-length coding region of the dnaK gene was amplified by PCR and ligated into the prokaryotic expression vector pET-22b(+). Large-scale expression and nondenaturing purification by metal affinity chromatography revealed that in addition to the recombinant DnaK protein (rCjDnaK-His), a polypeptide with an apparent molecular mass of ∼18 kDa, which might be translated from an in-frame ATG codon, was coexpressed and copurified (Fig. 5, lanes B and C). Therefore, fractions containing rCjDnaK-His were pooled and further purified by size exclusion chromatography. The final purified fraction showed a single band with an apparent molecular mass of about 68 kDa (Fig. 5, lane D). The N-terminal sequence of the purified protein was determined by the Edman degradation technique to be SKVIGIDLGT and agreed perfectly with the predicted amino acid sequence.
FIG. 5.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of recombinant C. jejuni DnaK protein. Lanes: A, noninduced bacterial lysate; B, isopropyl-β-d-thiogalactopyranoside (IPTG)-induced E. coli lysate; C, elute from an Ni2+ chelating column; D, rCjDnaK-His after gel filtration. Sizes of protein standards (low molecular weight marker; Pharmacia) are shown to the left (in thousands).
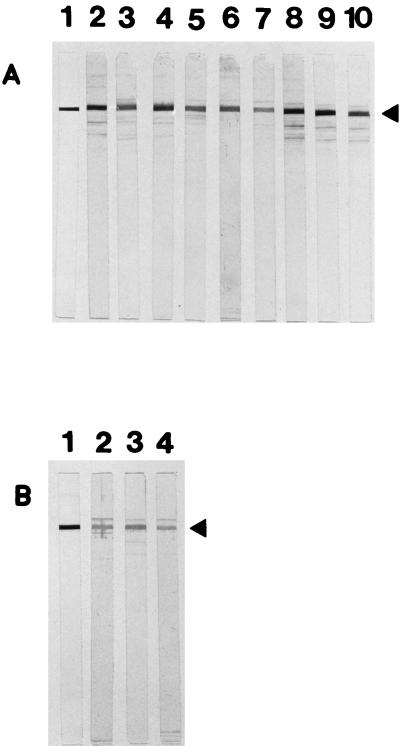
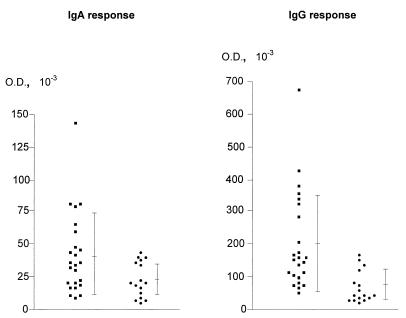
As shown in Fig. 6, the recombinant protein reacted with 9 (of 24) serum specimens from patients with C. jejuni enteritis. Only weak reactivity against rCjDnaK-His was detectable in 3 serum samples (of 16) from healthy volunteers. To quantitatively assess the anti-IgG and anti-IgA antibody responses to rCjDnaK-His, ELISAs were performed. Levels of IgG antibodies directed against rCjDnaK-His in sera from C. jejuni-infected patients (mean OD ± standard deviation [SD], 0.201 ± 0.149) were significantly higher than in sera from healthy controls (0.075 ± 0.042) (P < 0.0005; Mann-Whitney U test) (Fig. 7). In an IgA ELISA, the mean OD ± SD for sera of healthy individuals was 0.041 ± 0.03. Among patients with C. jejuni enteritis, the mean OD ± SD was 0.023 ± 0.013, a difference which was statistically significant as well (P < 0.05) (Fig. 7). By defining positive samples as those with absorbance values equal to, or in excess of, the mean absorbance of the control sera plus 3 SDs, 8 and 5 of 24 serum specimens from C. jejuni-infected patients were found to be anti-IgG and anti-IgA positive, respectively, but all of the controls were negative.
FIG. 6.
Immunoblot analysis of sera from C. jejuni-infected patients (A) (lanes 2 to 10) and from healthy controls (B) (lanes 2 to 4). Blots without reactivities are not shown. Lanes A1 and B1, Western blot analysis using an anti-histidine tag monoclonal antibody. Arrowheads indicate the size of rCjDnaK-His.
FIG. 7.
Serum IgA (A) and IgG (B) responses to rCjDnaK-His of 24 C. jejuni-infected patients (■) and 16 healthy controls (●). Means ± SDs are indicated. ODs were read in an ELISA reader at 405 and 450 nm.
DISCUSSION
In this study, we have cloned, sequenced, and characterized the hrcA-grpE-dnaK gene cluster of C. jejuni. Whereas the polypeptides encoded by the dnaK and grpE genes exhibited distinct homology to the respective proteins of other bacterial species, only a low overall degree of similarity was seen between C. jejuni HrcA and its bacterial homologues. However, at the N-terminal end, a stretch of approximately 85 aa residues exhibited significant homology (∼32% identity and ∼50% similarity) to the corresponding region of other HrcA proteins. This region encompasses two amino acid sequences of extended homology (designated boxes A and B [Fig. 2]), which might be involved in HrcA activity (31). Although the organization of the dnaK gene cluster, unlike the groE operon, differs considerably among bacterial species, the basic structure seems to be hrcA-grpE-dnaK-dnaJ and derivatives thereof (e.g., dnaK-dnaJ in E. coli) (34). In C. jejuni, dnaJ does not appear to be a part of the dnaK operon, a finding that is corroborated by the recently published nucleotide sequence of the dnaJ gene (21). Hence, the organization of the C. jejuni dnaK operon (hrcA-grpE-dnaK without dnaJ) is most similar to those seen in Chlamydia trachomatis (37) and L. lactis (13). Also, in the genome of H. pylori, dnaK and dnaJ are separated (41). In C. jejuni, as well as in Chlamydia trachomatis and H. pylori (between grpE and the preceding gene), but not in the gram-positive species L. lactis, an overlap of translational regulatory signals at the hrcA-grpE intergenic boundary was observed. Therefore, these two genes might be translationally coupled, which would ensure balanced synthesis of HrcA and GrpE proteins.
The hrcA gene is supposed to code for a repressor of the groE and dnaK heat shock genes that acts by binding to a highly conserved inverted repeat designated CIRCE. A putative stem-loop structure very similar to the CIRCE consensus sequence was detected in front of the hrcA start codon. A CIRCE element was found upstream of the C. jejuni groE operon, located as well between the −10 promoter box and the translation initiation site (39). In contrast to L. lactis, in which a CIRCE stem-loop structure is involved in dnaJ expression (42), C. jejuni’s dnaJ gene did not appear to have a CIRCE element in front of it (21).
Northern blot analysis led to the detection of two mRNA species, of 2.8 and 2.0 kb, both of which were strongly heat inducible. The amount of specific mRNA in C. jejuni cells growing microaerobically at 37°C was very small. Similarly, in the closely related species H. pylori, no dnaK transcript was observed in cells grown under nonstressed conditions, although the gene product, the HSP70 protein, was synthesized (18). A possible explanation is that minute amounts of mRNA are sufficient for maintenance of HSP70 protein levels in nonstressed cells. Alternatively, for reasons yet unknown, dnaK mRNA may be prone to specific degradation. Transcript mapping by primer extension identified three putative transcription start points, in front of the hrcA, grpE, and dnaK start codons. The putative −10 and −35 promoter boxes upstream of hrcA perfectly match the E. coli ς70 consensus sequence (17), which is an interesting finding because the consensus sequence for the C. jejuni vegetative promoter in the −35 portion is completely different from that of E. coli (45). Putative promoter regions similar or identical to the E. coli ς70 consensus promoter have been found in front of the dnaJ (21), groE (39), and clpB (38a) heat shock genes from C. jejuni. It can be speculated that a primary hrcA-grpE-dnaK transcript of C. jejuni is initiated from the ς70-like promoter upstream of hrcA. As could be demonstrated for the dnaK operon of Chlamydia trachomatis, methods like reverse transcription-PCR and RNase protection assay, which are considerably more sensitive than standard Northern blot hybridization, were necessary to detect the polycistronic transcript (37).
No regions similar to ς70 or heat shock (ς32) consensus promoters were found corresponding to P1 and P2. Therefore, it may be possible that P1 and/or P2 is an mRNA processing site instead of a transcription start site. Regulation of dnaK expression appears to be complex and may differ among eubacterial species. There has been discussion of posttranscriptional processing of the dnaK operons of Bacillus subtilis and Chlamydia trachomatis giving rise to mRNA species smaller than the primary polycistronic transcript (32, 37, 44). High-energy stem-loop structures, like that surrounding P2, have been described in bacterial polycistronic operons as transcriptional terminators or attenuators or sites crucial for mRNA processing or degradation (15, 29).
Western blot analyses as well as ELISA studies indicate that DnaK is immunoreactive in the human host, though only in a minority of C. jejuni-infected patients. However, when interpreting these data, the fact that serological tests using convalescent-phase sera from C. jejuni-infected patients may produce ambiguous results must be taken into account. First, in some patients, no antibody response to Campylobacter antigens is observed (6), and second, sera from healthy individuals sometimes react with Campylobacter proteins (12). Furthermore, antibodies reacting with epitopes shared by microbial DnaK homologues may contribute to the seropositivity among healthy individuals observed in Western blot experiments. Due to the extensive homology between bacterial HSPs and their mammalian counterparts, the humoral and/or T-cell response against these proteins has been proposed to influence the pathogenesis of autoimmune diseases (19). Cloning of C. jejuni heat shock genes and overexpression of the encoded proteins are first steps in studying their roles, if any, in the pathogenesis of C. jejuni-associated GBS. Since heat shock proteins are currently being discussed as promising candidates for subunit vaccines (25) and a Campylobacter vaccine is urgently needed (33), efforts to rule out the possibility or to demonstrate that C. jejuni HSPs can trigger or support autoimmune mechanisms must be increased.
ACKNOWLEDGMENTS
F.L.T. held a postgraduate training grant from the Deutsche Forschungsgemeinschaft, Graduiertenkolleg “Infektiologie.” This work was supported in part by the Sander-Stiftung (95.038.1).
We thank Uwe Enders for helpful discussions, S. Lukas for technical assistance, and D. Palm for peptide sequencing.
REFERENCES
- 1.Anderson I E, Craggs J K, Dunbar S, Herring A J. Cloning and expression of the 75 kDa DnaK-like protein of Chlamydia psittaci and the evaluation of the recombinant protein by immunoblotting and indirect ELISA. Vet Microbiol. 1997;58:295–307. doi: 10.1016/s0378-1135(97)00166-1. [DOI] [PubMed] [Google Scholar]
- 2.Anzola J, Luft B J, Gorgone G, Dattwyler R J, Soderberg C, Lahesmaa R, Peltz G. Borrelia burgdorferi HSP70 homolog: characterization of an immunoreactive stress protein. Infect Immun. 1992;60:3704–3713. doi: 10.1128/iai.60.9.3704-3713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 4.Bey R F, Larson M E, Lowery D E, Lee B W, Knutson K S, Simonson R R, King V L. Protection of C3H/He mice from experimental Borrelia burgdorferi infection by immunization with a 110-kilodalton fusion protein. Infect Immun. 1995;63:3213–3217. doi: 10.1128/iai.63.8.3213-3217.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birkelund S, Larsen B, Holm A, Lundemose A G, Christiansen G. Characterization of a linear epitope of Chlamydia trachomatis serovar L2 DnaK-like protein. Infect Immun. 1994;62:2051–2057. doi: 10.1128/iai.62.5.2051-2057.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black R E, Perlman D, Clements M L, Levine M M, Blaser M J. Human volunteer studies with Campylobacter jejuni. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C: American Society for Microbiology; 1992. pp. 207–215. [Google Scholar]
- 7.Blaser M J, Hopkins J A, Vasil M L. Campylobacter jejuni outer membrane proteins are antigenic for humans. Infect Immun. 1984;43:986–993. doi: 10.1128/iai.43.3.986-993.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cellier M F M, Teyssier J, Nicolas M, Liautard J P, Marti J, Sri Widada J. Cloning and characterization of the Brucella ovis heat shock protein DnaK functionally expressed in Escherichia coli. J Bacteriol. 1992;174:8036–8042. doi: 10.1128/jb.174.24.8036-8042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowing D W, Bardwell J C A, Craig E A, Woolford C, Hendrix R W, Gross C A. Consensus sequence for Escherichia coli heat shock promoters. Proc Natl Acad Sci USA. 1985;82:2697–2683. doi: 10.1073/pnas.82.9.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danilition S L, Maclean I W, Peeling R, Winston S, Brunham R C. The 75-kilodalton protein of Chlamydia trachomatis: a member of the heat shock protein 70 family? Infect Immun. 1990;58:189–196. doi: 10.1128/iai.58.1.189-196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.d’Aubenton C Y, Brody E, Thermes C. Prediction of a rho-independent Escherichia coli transcription terminator. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 12.Dunn B E, Blaser M J, Snyder E L. Two-dimensional gel electrophoresis and immunoblotting of Campylobacter outer membrane proteins. Infect Immun. 1987;55:1564–1572. doi: 10.21236/ada265461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton T, Shearman C, Gasson M. Cloning and sequence analysis of the dnaK region of Lactococcus lactis subsp. lactis. J Gen Microbiol. 1993;139:3253–3264. doi: 10.1099/00221287-139-12-3253. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specificity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 15.Gagnon G, Vadeboncoeur C, Gauthier L, Frenette M. Regulation of ptsH and ptsI gene expression in Streptococcus salivarius ATCC 25975. Mol Microbiol. 1995;16:1111–1121. doi: 10.1111/j.1365-2958.1995.tb02336.x. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths P L, Park R W A, Connerton I F. The gene for Campylobacter trigger factor: evidence for multiple transcription start sites and protein products. Microbiology. 1995;141:1359–1367. doi: 10.1099/13500872-141-6-1359. [DOI] [PubMed] [Google Scholar]
- 17.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli DNA promoter sequences. Nucleic Acids Res. 1983;11:120–122. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huesca M, Goodwin A, Bhagwansingh A, Hoffman P, Lingwood C A. Characterization of an acidic-pH-inducible stress protein (hsp70), a putative sulfatide binding adhesin, from Helicobacter pylori. Infect Immun. 1998;66:4061–4067. doi: 10.1128/iai.66.9.4061-4067.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Institute for Genomic Research. [Online.] Helicobacter pylori DNA sequences. Gaithersburg, Md: Institute for Genomic Research; 29 September 1998, posting date. http://www.tigr.org/tdb/mdb/hpdb/hpdb.html . [10 September 1998, last access date.] [Google Scholar]
- 19.Kaufmann S H E, Schoel B. Heat shock proteins as antigens in immunity against infection and self. In: Morimoto R, Tissière A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 495–531. [Google Scholar]
- 20.Ketley J M. Pathogenesis of enteric infection by Campylobacter. Microbiology. 1997;143:5–21. doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- 21.Konkel M E, Kim B J, Klena J D, Young C R, Ziprin R. Characterization of the thermal stress response of Campylobacter jejuni. Infect Immun. 1998;66:3666–3672. doi: 10.1128/iai.66.8.3666-3672.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberek K, Wall D, Georgopoulos C. The DnaJ chaperone catalytically activates the DnaK chaperone to preferentially bind the heat shock transcriptional regulator. Proc Natl Acad Sci USA. 1995;92:6224–6228. doi: 10.1073/pnas.92.14.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishu Allos B. Association between Campylobacter infection and Guillain-Barré syndrome. J Infect Dis. 1997;176(Suppl. 2):S125–S128. doi: 10.1086/513783. [DOI] [PubMed] [Google Scholar]
- 24.Nachamkin I, Hart A M. Western blot analysis of the human antibody response to Campylobacter jejuni cellular antigens during gastrointestinal infection. J Clin Microbiol. 1985;21:33–38. doi: 10.1128/jcm.21.1.33-38.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noll A, Bücheler N, Bohn E, Autenrieth I B. Microbial heat shock proteins as vaccines. Behring Inst Mitt. 1997;98:87–98. [PubMed] [Google Scholar]
- 26.Pei Z, Blaser M J. PEB1, the major cell-binding factor of Campylobacter jejuni, is a homolog of the binding component in gram-negative nutrient transport systems. J Biol Chem. 1993;268:18717–18725. [PubMed] [Google Scholar]
- 27.Rees J H, Soudain S E, Gregson N A, Hughes R A. Campylobacter jejuni infection and Guillain-Barré syndrome. N Engl J Med. 1995;333:1374–1379. doi: 10.1056/NEJM199511233332102. [DOI] [PubMed] [Google Scholar]
- 28.Rosenberg M, Court C. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Echevarría M J, de la Cueva G, Diaz-Orejas R. Translational coupling and limited degradation of a polycistronic messenger modulate differential gene expression in the parD stability system of plasmid R1. Mol Gen Genet. 1995;248:599–609. doi: 10.1007/BF02423456. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz A, Tzschaschel B, Schumann W. Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis. Mol Microbiol. 1995;15:421–429. doi: 10.1111/j.1365-2958.1995.tb02256.x. [DOI] [PubMed] [Google Scholar]
- 33.Scott D A. Vaccines against Campylobacter jejuni. J Infect Dis. 1997;176(Suppl. 2):S183–S188. doi: 10.1086/513791. [DOI] [PubMed] [Google Scholar]
- 34.Segal G, Ron E Z. Regulation and organization of the groE and dnaK operons in eubacteria. FEMS Microbiol Lett. 1996;138:1–10. doi: 10.1111/j.1574-6968.1996.tb08126.x. [DOI] [PubMed] [Google Scholar]
- 35.Skirrow M S, Blaser M J. Clinical and epidemiological considerations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C: American Society for Microbiology; 1992. pp. 3–8. [Google Scholar]
- 36.Takata T, Wai S N, Takade A, Sawae Y, Ono J, Amako K. The purification of a GroEL-like stress protein from aerobically adapted Campylobacter jejuni. Microbiol Immunol. 1995;39:639–645. doi: 10.1111/j.1348-0421.1995.tb03245.x. [DOI] [PubMed] [Google Scholar]
- 37.Tan M, Wong B, Engel J N. Transcriptional organization and regulation of the dnaK and groE operons of Chlamydia trachomatis. J Bacteriol. 1996;178:6983–6990. doi: 10.1128/jb.178.23.6983-6990.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor D N. Campylobacter infections in developing countries. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C: American Society for Microbiology; 1992. pp. 20–30. [Google Scholar]
- 38a.Thies, F. L., and G. Giegerich. Submitted for publication.
- 39.Thies, F. L., A. Weishaupt, H. Karch, H.-P. Hartung, and G. Giegerich. Cloning, sequencing, and molecular analysis of the Campylobacter jejuni groE operon. Microbiology, in press. [DOI] [PubMed]
- 40.Tomayasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman A J, Oppenheim A B, Yura T, Yamanaka K, Niki H, Hiraga S, Ogura T. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor ς32. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 42.van Asseldonk M, Simons A, Visser H, de Vos W M, Simons G. Cloning, nucleotide sequence, and regulatory analysis of the Lactococcus lactis dnaJ gene. J Bacteriol. 1993;175:1637–1644. doi: 10.1128/jb.175.6.1637-1644.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wenman W M, Chai J, Louie T J, Goudreau C, Lior H, Newell D G, Pearson A D, Taylor D E. Antigenic analysis of Campylobacter flagellar protein and other proteins. J Clin Microbiol. 1985;21:108–112. doi: 10.1128/jcm.21.1.108-112.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wetzstein M, Völker U, Dedio J, Löbau S, Zuber U, Schiesswohl M, Herget C, Hecker M, Schumann W. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J Bacteriol. 1992;174:3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wösten M M S M, Boeve M, Koot M G A, van Nuenen A C, van der Zeijst B A M. Identification of Campylobacter jejuni promoter sequences. J Bacteriol. 1998;180:594–599. doi: 10.1128/jb.180.3.594-599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Y L, Lee L H, Rollins D M, Ching W M. Heat shock- and alkaline pH-induced proteins of Campylobacter jejuni: characterization and immunological properties. Infect Immun. 1994;62:4256–4260. doi: 10.1128/iai.62.10.4256-4260.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young V D, Lathigra R, Hendrix R, Sweetser D, Young R A. Stress proteins are immune targets in leprosy and tuberculosis. Proc Natl Acad Sci USA. 1988;85:4267–4270. doi: 10.1073/pnas.85.12.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan G, Wong S-L. Regulation of groE expression in Bacillus subtilis: the involvement of the ςA-like promoter and the roles of the inverted repeat sequence (CIRCE) J Bacteriol. 1995;177:5427–5433. doi: 10.1128/jb.177.19.5427-5433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan G, Wong S-L. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J Bacteriol. 1995;177:6462–6468. doi: 10.1128/jb.177.22.6462-6468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yura T, Nagai H, Mori M. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 51.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]