Abstract
A total of 617 fecal specimens were collected on 18 Hotan Black chicken farms in Southern Xinjiang, China, and tested for the presence of Cryptosporidium spp. by PCR of the small subunit ribosomal RNA (SSU rRNA) gene. The overall infection rate by Cryptosporidium spp. was 11.5% (71/617), and ten of the 18 farms were positive. The infection rate by Cryptosporidium spp. was 14.5% (48/331) in the 30–60 d group, higher than chickens in the <30 d (12.0%, 15/125), 60–90 d (6.9%, 5/72), and >90 d (3.4%, 3/89) groups. Cryptosporidium meleagridis (n = 38) and C. baileyi (n = 33) were confirmed by sequencing analysis. A total of 25 of the 38 C. meleagridis-positive specimens were subtyped successfully at the gp60 gene, including one known subtype (IIIbA23G1R1, n = 1) and two novel subtypes, named IIIbA25G1R1 (n = 20) and IIIbA31G1R1 (n = 4). The results showed that infection by Cryptosporidium spp. in Hotan Black Chickens was common in this area and the distribution of C. meleagridis subtypes had regional characteristics.
Keywords: Cryptosporidium, Genetic, Subtype, Chicken, China
Abstract
Un total de 617 échantillons fécaux ont été prélevés dans 18 élevages de poulets noirs du Hotan dans le sud du Xinjiang, en Chine, et testés pour la présence de Cryptosporidium spp. par PCR du gène de la petite sous-unité de l’ARN ribosomique (ARNr SSU). Le taux d’infection global par Cryptosporidium spp. était de 11,5 % (71/617) et dix des 18 élevages étaient positifs. Le taux d’infection de Cryptosporidium spp. était de 14,5 % (48/331) dans le groupe 30–60 jours, supérieur à celui des poulets dans les groupes <30 jours (12,0 %, 15/125), 60–90 jours (6,9 %, 5/72) et >90 jours (3,4 %, 3/89). Cryptosporidium meleagridis (n = 38) et C. baileyi (n = 33) ont été confirmés par analyse de séquençage. Vingt-cinq des 38 spécimens positifs pour C. meleagridis ont été sous-typés avec succès au niveau du gène gp60, dont un sous-type connu (IIIbA23G1R1, n = 1) et deux nouveaux sous-types, nommés IIIbA25G1R1 (n = 20) et IIIbA31G1R1 (n = 4). Les résultats ont montré que l’infection par Cryptosporidium spp. chez les poulets noirs du Hotan était commune dans cette zone et que la distribution des sous-types de C. meleagridis avait des caractéristiques régionales.
Introduction
Cryptosporidium is considered an important zoonotic protozoan parasite of various animals, including mammals, birds, reptiles, fishes, and amphibians [10]. Morphological, biological, and molecular data have validated 46 recognized Cryptosporidium species [15, 24]. Among these, 23 Cryptosporidium species have been identified in humans [23]. Epidemiological data have revealed that most human cryptosporidiosis cases are caused by C. hominis, C. parvum, and C. meleagridis [23]. DNA sequence analysis of the 60-kDa glycoprotein (gp60) gene is the most common subtyping tool [34]. In previous subtyping studies of C. meleagridis based on the gp60 gene, ten subtype families (IIIa to IIIj) have been established, and eight (IIIa to IIIh) of which were found in birds and humans [23]. Based on the principle of “One Health”, it is a key step to prevent and control cryptosporidiosis by investigating infection by Cryptosporidium in animal hosts at the subtype level.
Chickens are among the most widespread domestic animals. Currently, seven species (C. meleagridis, C. baileyi, C. parvum, C. galli, C. andersoni, C. ubiquitum and C. ornithophilus) and five genotypes (C. avian genotypes VII, VIII, IX, C. genotype BrPR1 and C. xiaoi-like genotype) of Cryptosporidium have been recognized in chickens [26]. In China, the infection rate by Cryptosporidium in chickens was reported to be 1.3% to 10.2% in Shandong, Henan, Hebei, Hubei and Zhejiang Provinces, with C. baileyi being the dominant Cryptosporidium species [4, 11, 21, 32, 33]. Hotan Black chicken is an excellent local breed, mainly distributed in the Minfeng, Lop, and Hotan counties of the Xinjiang Uygur Autonomous Region (herein referred to as Xinjiang), China. However, the prevalence and species/subtypes of Cryptosporidium in Hotan Black chicken remain unclear. This study aimed to examine the occurrence and genetic characterization of Cryptosporidium in Hotan Black chickens in Xinjiang.
Materials and methods
Ethics statement
The Animal Ethical Committee of Wenzhou Medical University (WMU-2020-12) reviewed and approved the protocol of this study. Before beginning the study, we contacted the owners of these animals and obtained their permission to profile their chickens. Fecal specimens were collected without harming the animals.
Specimens collection
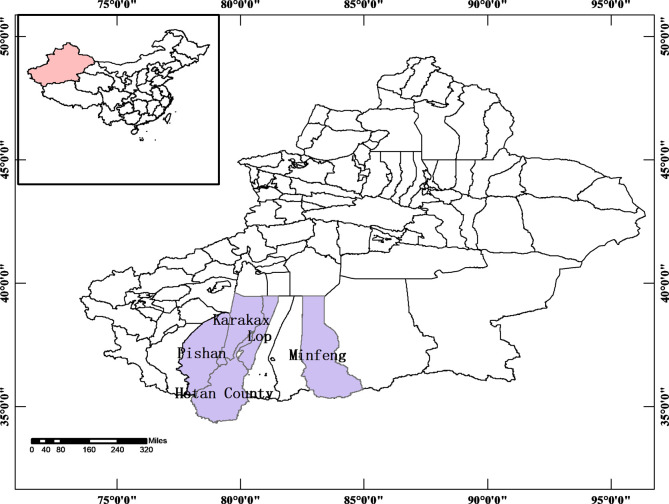
Between June 2020 and July 2021, 617 chickens were collected from 18 private farms in villages of Hotan, Lop, Minfeng, Karakax, and Pishan counties of Xinjiang (Fig. 1). The farm owners randomly selected the chickens that were subsequently housed in individual flat-surfaced cages away from contact with other animals. Then, one fecal specimen (2–5 g) for each animal was collected from the tray under the individual cage using a sterile disposable latex glove and placed into an individual sterile plastic container. All the specimens were kept in iceboxes, transported to the laboratory and stored at 4 °C. All chickens were healthy looking without any apparent infection at sampling.
Figure 1.
Specific locations where specimens were collected in this study.
DNA extraction
Approximately 200 mg of stool specimens were picked using autoclaved disposable bamboo sticks to extract genomic DNA of Cryptosporidium using an E.Z.N.A. Stool DNA kit (Omega Biotek Inc., Norcross, GA, USA), following the manufacturer’s procedure. The eluted DNA (200 μL) was kept at −20 °C until PCR analysis.
Genotyping and subtyping of Cryptosporidium spp.
All extracted DNA were examined by nested PCR at the SSU rRNA gene of Cryptosporidium [35]. Cryptosporidium-positive specimens were subtyped by nested PCR at the gp60 gene [31]. EasyTaq PCR SuperMix (TransGene Biotech Co. Ltd., Beijing, China) was used for all PCR amplifications. Double distilled water and cattle derived C. andersoni DNA were used as the negative and positive control, respectively. Each specimen was PCR amplified twice. All the secondary PCR products were resolved on 1.5% agarose gels stained with GelRed (Biotium Inc., Fremont, CA, USA).
Nucleotide sequence analysis
All positive secondary PCR products were sent for bidirectional sequencing at GENEWIZ (Suzhou, China). The obtained sequences were aligned against each other and reference sequences from GenBank using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Clustal X 2.1 (http://www.clustal.org/).
Phylogenetic analyses
A phylogenetic tree was constructed in MEGA7 using the maximum likelihood method based on the Tamura-Nei model to evaluate the genetic relationship between the gp60 subtypes of C. meleagridis. The representative nucleotide sequences obtained in this study were deposited in the GenBank database under accession numbers: ON209450 to ON209452 for the SSU rRNA gene and ON210984 to ON210986 for the gp60 gene.
Statistical analysis
A chi-square analysis determined the correlation between the Cryptosporidium prevalence in farm regions and age groups using SPSS version 19.0. Results with p < 0.05 were considered statistically significant.
Results
Occurrence of Cryptosporidium in Hotan Black chickens
The overall infection rate by Cryptosporidium in Hotan Black chickens was 11.5% (71/617). Ten out of the 18 farms had Cryptosporidium positive specimens, with the highest infection rate in Farm Hotan5 (46.2%, 6/13), and the lowest in farm Pishan18 (2.4%, 1/41) (Table 1). The infection rates by Cryptosporidium in chickens on each farm were significantly different (χ2 = 142.2, df = 17, p < 0.01). The Cryptosporidium infection rate in the 30–60 d Hotan Black chickens (14.5%, 48/331) was higher than in the <30 d (12.0%, 15/125), 60–90 d (6.9%, 5/72), and >90 d (3.4%, 3/89) groups (Table 2), with significant differences among different age groups (χ2 = 10.202, df = 3, p < 0.05).
Table 1.
Prevalence and distribution of C. baileyi and C. meleagridis as well as subtypes of C. meleagridis in chickens in this study.
| Sampling locations | No. positive/No. examined (%) | OR (95 CI) | No. positive C. baileyi (%) | No. positive C. meleagridis (%)/subtype (n) |
|---|---|---|---|---|
| Hotan1 | 5/22 (22.7) | 11.8 (3.7–41.7) | 5 (22.7) | 0 |
| Hotan2 | 0/16 (0) | 0 | 0 | |
| Hotan3 | 0/45 (0) | 0 | 0 | |
| Hotan4 | 0/14 (0) | 0 | 0 | |
| Hotan5 | 6/13 (46.2) | 34.3 (14.8–77.5) | 5 (38.5) | 1 (7.7)/IIIbA23G1R1 (1) |
| Hotan6 | 0/72 (0) | 0 | 0 | |
| Karakax15 | 11/32 (34.4) | 21.0 (17.0–51.8) | 3 (9.4) | 8 (25.0)/IIIbA25G1R1 (6) |
| Karakax16 | 9/29 (31.0) | 18.0 (13.1–48.9) | 3 (10.3) | 6 (20.7)/IIIbA25G1R1 (4) |
| Lop7 | 5/38 (13.2) | 6.0 (1.9–24.4) | 0 | 5 (13.2) |
| Lop8 | 3/58 (5.2) | 2.2 (0–11.0) | 1 (1.7) | 2 (3.4) |
| Lop9 | 0/22 (0) | 0 | 0 | |
| Lop10 | 5/20 (25.0) | 13.3 (4.2–45.8) | 0 | 5 (25.0)/IIIbA25G1R1 (3) |
| Lop11 | 0/23 (0) | 0 | 0 | |
| Minfeng12 | 21/46 (45.7) | 33.6 (30.7–60.6) | 11 (24.0) | 10 (21.7)/IIIbA31G1R (3), IIIbA25G1R1 (7) |
| Minfeng13 | 0/20 (0) | 0 | 0 | |
| Minfeng14 | 5/72 (6.9) | 2.8 (0.9–13.0) | 4 (5.6) | 1 (1.4)/IIIbA31G1R (1) |
| Pishan17 | 0/34 (0) | 0 | 0 | |
| Pishan18 | 1/41 (2.4) | 1.0 (0–7.4) | 1 (2.4) | 0 |
| Total | 71/617 (11.5) | 5.2 (0.9–14) | 33 (5.3) | 38 (6.2)/IIIbA23G1R1 (1), IIIbA25G1R1 (20), IIIbA31G1R (4) |
Table 2.
Infection and subtype distribution of Cryptosporidium species by age.
| Age | No. positive/No. examined (%) | OR (95 CI) | No. positive C. baileyi (%) | No. positive C. meleagridis (%)/subtype (n) |
|---|---|---|---|---|
| <30 d | 15/125 (12.0) | 3.9 (6.2–17.8) | 8 (6.4) | 7 (5.6)/IIIbA23G1R1 (1), IIIbA25G1R1 (4) |
| 30–60 d | 48/331 (14.5) | 4.9 (10.7–18.3) | 20 (6.0) | 28 (8.5)/IIIbA25G1R1 (16), IIIbA31G1R (4) |
| 60–90 d | 5/72 (6.9) | 2.1 (0.9–13) | 4 (5.6) | 1 (1.4) |
| >90 d | 3/89 (3.4) | 1.0 (0–7.2) | 1 (1.1) | 2 (2.2) |
Genetic characterizations of Cryptosporidium at the SSU rRNA gene
From the 71 sequences of Cryptosporidium at the SSU rRNA gene, 38 (ON209452) were identical to C. meleagridis isolated from Parrot (MN410718), while 23 (ON209450) and 10 (ON209451) sequences were identical to C. baileyi isolated sequences from Chinese grosbeak (MN410723) and from silky fowl in China (KU744847), respectively. On the ten Cryptosporidium positive farms, C. baileyi was found on farms Hotan1 and Pishan18, and C. meleagridis was found on farms Lop7 and Lop10, while the remaining six farms were positive for both C. meleagridis and C. baileyi (Table 1). No mixed infections were observed in any examined chickens.
The infection rates by C. meleagridis and C. baileyi were 7.7% (35/456) and 6.1% (28/456) in the <60 d group of Hotan black chickens, and 1.9% (3/161) and 3.1% (5/161) in the >60 d group, respectively. The infection rate by C. meleagridis was higher in the <60 d chickens than in the >60 d groups (Table 2). In contrast, the two age groups had no statistical difference in C. baileyi infection.
Subtyping of C. meleagridis at the gp60 gene
A total of 25 of the 38 C. meleagridis isolates were successfully PCR amplified at the gp60 gene. Three subtypes of C. meleagridis were identified, including one known subtype (IIIbA23G1R1; n = 1) and two novel subtypes, named IIIbA25G1R1 (n = 20) and IIIbA31G1R1 (n = 4). Subtype IIIbA25G1R1 was only found on farms Lop10, Karakax15, and Karakax16, while subtypes IIIbA23G1R1 (n = 1) and IIIbA31G1R1 (n = 1) were only present on farms Hotan5 and Minfeng14, respectively. Farm Minfeng12 had both IIIbA31G1R and IIIbA25G1R1 subtypes (Table 1). All three subtypes of C. meleagridis were found only in <60 d groups of Hotan Black chickens (Table 2).
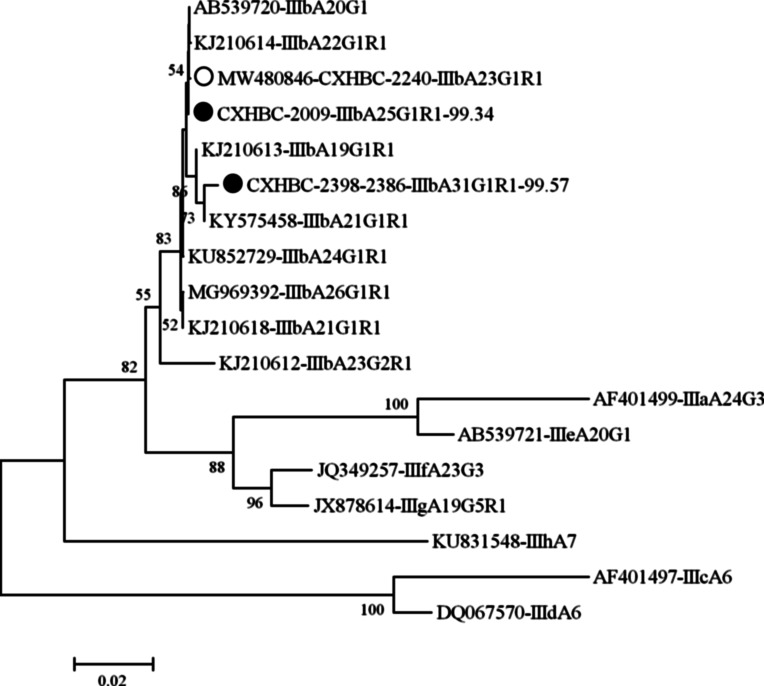
Phylogenetic analysis of C. meleagridis
All three subtypes of C. meleagridis obtained in this study clustered into subtype family IIIb, as expected (Fig. 2). The known subtype IIIbA23G1R1 and the novel one IIIbA25G1R1 clustered into one sub-branch together with IIIbA20G1 and IIIbA22G1R1, while subtype IIIbA31G1R1 clustered into a sub-branch together with IIIbA19G1R1 and IIIbA21G1R1.
Figure 2.
Phylogenetic tree based on the N-J analysis of gp60 sequences of C. meleagridis. Phylogenetic relationships of C. meleagridis subtypes identified in chicken here and parts of other known subtypes deposited in GenBank were inferred by a neighbor-joining analysis of gp60 sequences of C. meleagridis based on genetic distances by the Kimura two-parameter model. The numbers on the branches are percent bootstrapping values from 1000 replicates. The triangles filled in black and white indicate novel and known subtypes identified in this study, respectively.
Discussion
Molecular epidemiological surveys of Cryptosporidium in chickens have been conducted in 12 countries with infection rates from 0.7% to 75.0% (Table 3) [1, 2, 4, 5, 7, 8, 11–14, 16, 17, 20, 21, 27–30, 32, 33]. This is the first study that revealed the presence of Cryptosporidium in chickens in Xinjiang, with an average infection rate of 11.5% which is higher than other studies from China (1.3–10.0%), Germany (7.0%), Iran (0.7–8.0%), Jordan (4.8%), Kenya (7.8%), Syria (9.9%), and Tunisia (4.5%) (Table 3). However, chickens from Algeria (15.3–34.0%), Bangladesh (15.7%), Brazil (12.6–25.6%), Côte d’Ivoire (16.1%), and Sweden (75.0%) had higher Cryptosporidium infection rates than those observed in the present study (Table 3). The differences in prevalence may be related to the sensitivity and specificity of the detection methods, the health status of hosts, territorial environment, overall sample size, feeding conditions, and drinking water sources, amongst others.
Table 3.
Prevalence and species/genotype distribution of Cryptosporidium in chickens worldwide.
| Country | No. positive/No. examined (%) | Species/genotypes of Cryptosporidium (n) | References |
|---|---|---|---|
| Algeria | 31/90 (34.0) | C. meleagridis (26); C. baileyi (5) | [16] |
| 23/150 (15.3) | C. baileyi (12); C. meleagridis (11) | [17] | |
| Bangladesh | 31/197 (15.7) | C. baileyi (17); C. meleagridis (12); C. parvum (2). | [18] |
| Brazil | 24/190 (12.6) | C. baileyi (18); C. meleagridis (1); C. parvum (4); Cryptosporidium sp. (1) | [19] |
| 20/130 (15.4) | C. meleagridis (15); C. baileyi (4); Mix (1) | [20] | |
| 91/351 (25.6)a | C. meleagridis (57); C. baileyi (15); C. parvum (3); C. genotype BrPR1 (3) | [21] | |
| China | 5/206 (2.4) | C. baileyi (5) | [10] |
| 179/2015 (8.9) | C. meleagridis (3); C. baileyi (176) | [11] | |
| 38/385 (9.9) | C. baileyi (33); C. meleagridis (2); Avian genotype II (3) | [12] | |
| 11/829 (1.3) | C. baileyi (5); C. meleagridis (2); C. galli (2); C. xiaoi-like genotype (2) | [13] | |
| 48/471 (10.2) | C. meleagridis (15); C. baileyi (33) | [14] | |
| 132/1001 (13.2) | C. meleagridis (78); C. baileyi (48) | [22] | |
| Côte d’Ivoire | 5/31 (16.1) | C. meleagridis (4); C. parvum (1) | [23] |
| Germany | 18/256 (7.0) | C. parvum (13); C. baileyi (2); C. avian genotypes VII, VIII, IX (one each) | [24] |
| Iran | 7/1000 (0.7) | C. baileyi (7) | [25] |
| 8/100 (8.0) | C. baileyi (6); C. parvum (2) | [26] | |
| Jordan | 1/21 (4.8) | C. baileyi (1) | [27] |
| Kenya | 25 (7.8)a | C. andersoni (2); C. meleagridis (4); C. parvum (3); C. ubiquitum (1) | [28] |
| Sweden | 12/16 (75.0)a | C. meleagridis (6); C. baileyi (1); C. galli (3) | [9] |
| Syria | 11/111 (9.9) | C. baileyi (9); Unknown genotype (2)b | [30] |
| Tunisia | 9/200 (4.5)a | C. meleagridis (1) | [31] |
The numbers of species are not consistent with the numbers of positives in some countries because not all isolates were genotyped successfully for Brazil, Kenya, Sweden and Tunisia.
Two samples showed unknown profiles in comparison to species.
In this study, the Hotan Black chickens aged 30–60 days had the highest infection rate by Cryptosporidium (14.5%), followed by those aged <30 days (12.0%), 60–90 days (6.9%) and >90 days (3.4%). In one previous study, chickens aged 61–90 days (17.8%) showed a significantly higher infection rate than chickens <30 d (8.3%), 31–60 days (12.0%) and >90 days (6.8%) of age, which was partially in agreement with the results of our study [22]. Usually, warming of chickens can be completely stopped at the age of >30 days, and they then leave the hotbed for flat-surfaced cages, living at their natural room temperature. At this age, the chickens’ immune system is not mature enough. Once the rearing environment is contaminated with Cryptosporidium oocysts, these chickens are easily infected with Cryptosporidium.
In this study, 38 C. meleagridis and 33 C. baileyi were identified, which was similar to a study where 26 C. meleagridis and five C. baileyi were identified in chickens in Algeria [1], and a study where 15 C. meleagridis and four C. baileyi were identified in chickens in Brazil [5]. Previously, 281 Cryptosporidium isolates were identified in chickens in China, with C. baileyi (n = 252) being the primary, widely distributed species, followed by C. meleagridis (n = 22), C. ornithophilus (synonym: C. avian genotype II) (n = 3), C. galli (n = 2), and C. xiaoi-like genotype (n = 2) [4, 11, 21, 32, 33]. In fact, all the studies from China except from Ezhou City, Hubei Province showed that both C. meleagridis and C. baileyi were present in chickens [4, 11, 21, 32, 33]. This shows that C. baileyi and C. meleagridis are two common species in chickens. However, the high proportion of C. meleagridis in chickens detected in the present study indicates that the constituent proportions of C. baileyi and C. meleagridis in the chickens were significantly different compared with other studies conducted in China, which may represent the geographical genetic distribution characteristics of Cryptosporidium in chickens in this area. This phenomenon may be caused by the different transmission routes of Cryptosporidium in southern Xinjiang due to its unique natural environment and geographical conditions.
Cryptosporidium baileyi is the most common species detected in chickens and has been reported in over 20 avian hosts [24]. This species was also detected in the stools of two immunodeficient patients, probably due to the disrupted immune system [6, 19]. In some birds, experimental cross-transmission of C. baileyi from naturally infected broilers to other hosts has been successful, but mice and goats did not become infected [24]. Cryptosporidium meleagridis has also been reported in a wide range of avian species, as well as in humans, dogs, deer, minks, cattle, and rodents [18, 24, 26]. In Peru and Thailand, C. meleagridis accounts for 10–20% of human Cryptosporidium infections, with a frequency similar to C. parvum infections [3, 25]. In China, C. meleagridis has been isolated from humans, broiler chickens, laying hens, quails, cattle, minks, and wastewater samples, suggesting C. meleagridis as a zoonotic disease agent with significance to human health [9]. Thus, it is necessary to avoid or reduce the occurrence, transmission, and re-infection of this pathogen among individuals within each farm by improving husbandry practices related to feeding and water.
To date, 28 subtypes have been identified in the subtype family IIIb, and 23 of these can infect humans, while the other five subtypes (IIIbA15G1, IIIbA21G1R1d, IIIbA22G1R1, IIIbA22G1R1a and IIIbA27G1) were found only in birds or water (Table S1). Therefore, the subtypes in family IIIb have potential zoonotic transmission. In this study, three subtypes (IIIbA23G1R1, IIIbA25G1R1, and IIIbA31G1R) were identified. However, subtypes IIIbA25G1R1 and IIIbA31G1R found here were not described previously, which may indicate geographical segregation characteristics in the distribution of C. meleagridis subtypes derived from chickens. More investigations will need to be done for source attribution of infection of the two subtypes and to understand their transmission dynamics in the future.
In conclusion, this study reports the first detection of Cryptosporidium in Hotan Black chicken with an 11.5% overall infection rate. Cryptosporidium meleagridis and C. baileyi were identified, with C. meleagridis being the predominated one, and two novel C. meleagridis subtypes were identified, named IIIbA25G1R1and IIIbA31G1R1. Therefore, future investigations on the genetic characteristics of regional distribution of avian Cryptosporidium should be conducted.
Conflict of interest
The authors declare that they have no competing interests.
Supplementary materials
The supplementary material is available at https://www.parasite-journal.org/10.1051/parasite/2022051/olm
Acknowledgments
This study was supported by the Program for Young and Middle-aged Leading Science, Technology, and Innovation of the Xinjiang Production & Construction Corps (2018CB034).
Cite this article as: Feng X, Xin L, Yu F, Song X, Zhang J, Deng J, Qi M & Zhao W. 2022. Genetic characterization of Cryptosporidium spp. in Hotan Black Chickens in China reveals two novel subtypes of Cryptosporidium meleagridis. Parasite 29, 50.
Footnotes
Edited by Jean-Lou Justine
References
- 1.Baroudi D, Khelef D, Goucem R, Adjou KT, Adamu H, Zhang H, Xiao L. 2013. Common occurrence of zoonotic pathogen Cryptosporidium meleagridis in broiler chickens and turkeys in Algeria. Veterinary Parasitology, 196(3–4), 334–340. [DOI] [PubMed] [Google Scholar]
- 2.Berrilli F, D’Alfonso R, Giangaspero A, Marangi M, Brandonisio O, Kaboré Y, Glé C, Cianfanelli C, Lauro R, Di Cave D. 2012. Giardia duodenalis genotypes and Cryptosporidium species in humans and domestic animals in Côte d’Ivoire: occurrence and evidence for environmental contamination. Transactions of the Royal Society of Tropical Medicine and Hygiene, 106(3), 191–195. [DOI] [PubMed] [Google Scholar]
- 3.Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, Gilman RH, Xiao L. 2008. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerging Infectious Diseases, 14(10), 1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao S, Xu M, Jiang Y, Liu H, Yuan Z, Sun L, Cao J, Shen Y. 2020. Prevalence and genetic characterization of Cryptosporidium, Giardia and Enterocytozoon in chickens from Ezhou, Hubei, China. Frontiers in Veterinary Science, 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Cunha MJR, Cury MC, Santin M. 2018. Molecular characterization of Cryptosporidium spp. in poultry from Brazil. Research in Veterinary Science, 118, 331–335. [DOI] [PubMed] [Google Scholar]
- 6.Ditrich O, Palkovic L, Sterba J, Prokopic J, Loudova J, Giboda M. 1991. The first finding of Cryptosporidium baileyi in man. Parasitology Research, 77(1), 44–47. [DOI] [PubMed] [Google Scholar]
- 7.Essendi WM, Muleke C, Miheso M, Otachi E. 2022. Genetic diversity of Cryptosporidium species in Njoro Sub County, Nakuru, Kenya. Journal of Parasitic Diseases, 46(1), 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewald MPC, Martins FDC, Caldart ET, Vieira FEG, Yamamura MH, Sasse JP, Barros LD, Freire RL, Navarro IT, Garcia JL. 2017. The first study of molecular prevalence and species characterization of Cryptosporidium in free-range chicken (Gallus gallus domesticus) from Brazil. Brazilian Journal of Veterinary Parasitology, 26(4), 472–478. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Xiao L. 2017. Molecular Epidemiology of Cryptosporidiosis in China. Frontiers in Microbiology, 8, 1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia RJ, Hayman DT. 2016. Origin of a major infectious disease in vertebrates: The timing of Cryptosporidium evolution and its hosts. Parasitology, 143(13), 1683–1690. [DOI] [PubMed] [Google Scholar]
- 11.Gong Z, Kan Z-Z, Huang J-M, Fang Z, Liu X-C, Gu Y-F, Li W-C. 2021. Molecular prevalence and characterization of Cryptosporidium in domestic free-range poultry in Anhui Province, China. Parasitology Research, 120(10), 3519–3527. [DOI] [PubMed] [Google Scholar]
- 12.Hamidinejat H, Jalali MH, Jafari RA, Nourmohammadi K. 2014. Molecular determination and genotyping of Cryptosporidium spp. in fecal and respiratory samples of industrial poultry in Iran. Asian Pacific Journal of Tropical Medicine, 7(7), 517–520. [DOI] [PubMed] [Google Scholar]
- 13.Helmy YA, Krucken J, Abdelwhab EM, von Samson-Himmelstjerna G, Hafez HM. 2017. Molecular diagnosis and characterization of Cryptosporidium spp. in turkeys and chickens in Germany reveals evidence for previously undetected parasite species. PLoS One, 12(6), e0177150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hijjawi N, Mukbel R, Yang R, Ryan U. 2016. Genetic characterization of Cryptosporidium in animal and human isolates from Jordan. Veterinary Parasitology, 228, 116–120. [DOI] [PubMed] [Google Scholar]
- 15.Ježková J, Limpouchová Z, Prediger J, Holubová N, Sak B, Konečný R, Kvĕtoňová D, Hlásková L, Rost M, McEvoý J, Rajsky D, Feng Y, Kváč M. 2021. Cryptosporidium myocastoris n. sp. (Apicomplexa: Cryptosporidiidae), the species adapted to the nutria (Myocastor coypus). Microorganisms, 9(4), 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabir MHB, Han Y, Lee SH, Nugraha AB, Recuenco F, Murakoshi F, Xuan X, Kato K. 2020. Prevalence and molecular characterization of Cryptosporidium species in poultry in Bangladesh. One Health, 9, 100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kassouha M, Soukkarieh C, Alkhaled A. 2016. First genotyping of Cryptosporidium spp. in pre-weaned calves, broiler chickens and children in Syria by PCR-RFLP analysis. Veterinary Parasitology, 225, 86–90. [DOI] [PubMed] [Google Scholar]
- 18.Khine NO, Chimnoi W, Kamyingkird K, Kengradomkij C, Saetiew N, Simking P, Saengow S, Jittapalapong S, Inpankaew T. 2021. Molecular detection of Giardia duodenalis and Cryptosporidium spp. from stray dogs residing in monasteries in Bangkok, Thailand. Parasitology International, 83, 102337. [DOI] [PubMed] [Google Scholar]
- 19.Kopacz Z, Kvac M, Piesiak P, Szydlowicz M, Hendrich AB, Sak B, McEvoy J, Kicia M. 2020. Cryptosporidium baileyi pulmonary infection in immunocompetent woman with benign neoplasm. Emerging Infectious Diseases, 26(8), 1958–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laatamna AK, Belkessa S, Khalil A, Afidi A, Benmahdjouba K, Belalmi R, Benkrour M, Ghazel Z, Hakem A, Aissi M. 2018. Prevalence of Cryptosporidium spp. in farmed animals from steppe and high plateau regions in Algeria. Tropical Biomedicine, 35(3), 724–735. [PubMed] [Google Scholar]
- 21.Liao C, Wang T, Koehler AV, Fan Y, Hu M, Gasser RB. 2018. Molecular investigation of Cryptosporidium in farmed chickens in Hubei Province, China, identifies “zoonotic” subtypes of C. meleagridis. Parasites & Vectors, 11(1), 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X, Xin L, Qi M, Hou M, Liao S, Qi N, Li J, Lv M, Cai H, Hu J, Zhang J, Ji X, Sun M. 2022. Dominance of the zoonotic pathogen Cryptosporidium meleagridis in broiler chickens in Guangdong, China, reveals evidence of cross-transmission. Parasites & Vectors, 15(1), 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan U, Zahedi A, Feng Y, Xiao L. 2021. An update on zoonotic Cryptosporidium species and genotypes in humans. Animals (Basel), 11(11), 3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan UM, Feng Y, Fayer R, Xiao L. 2021. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia – a 50 year perspective (1971–2021). International Journal for Parasitology, 51(13–14), 1099–1119. [DOI] [PubMed] [Google Scholar]
- 25.Sannella AR, Suputtamongkol Y, Wongsawat E, Cacciò SM. 2019. A retrospective molecular study of Cryptosporidium species and genotypes in HIV-infected patients from Thailand. Parasites & Vectors, 12(1), 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santana BN, Ferrari ED, Nakamura AA, Silva GSD, Meireles MV. 2022. Validation of a one-tube nested real-time PCR assay for the detection of Cryptosporidium spp. in avian fecal samples. Brazilian Journal of Veterinary Parasitology, 31(1), e000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santana BN, Kurahara B, Nakamura AA, da Silva Camargo V, Ferrari ED, da Silva GS, Nagata WB, Meireles MV. 2018. Detection and characterization of Cryptosporidium species and genotypes in three chicken production systems in Brazil using different molecular diagnosis protocols. Preventive Veterinary Medicine, 151, 73–78. [DOI] [PubMed] [Google Scholar]
- 28.Shahbazi P, Aligolzadeh A, Khordadmehr M, Hashemzadeh Farhang H, Katiraee F. 2020. Molecular study and genotyping of Cryptosporidium baileyi and Cryptosporidium parvum from free-range and commercial broiler chickens in Guilan province, Iran. Comparative Immunology, Microbiology and Infectious Diseases, 69, 101411. [DOI] [PubMed] [Google Scholar]
- 29.Silverlas C, Mattsson JG, Insulander M, Lebbad M. 2012. Zoonotic transmission of Cryptosporidium meleagridis on an organic Swedish farm. International Journal for Parasitology, 42(11), 963–967. [DOI] [PubMed] [Google Scholar]
- 30.Soltane R, Guyot K, Dei-Cas E, Ayadi A. 2007. Prevalence of Cryptosporidium spp. (Eucoccidiorida: Cryptosporiidae) in seven species of farm animals in Tunisia. Parasite, 14(4), 335–338. [DOI] [PubMed] [Google Scholar]
- 31.Stensvold CR, Beser J, Axén C, Lebbad M. 2014. High applicability of a novel method for gp60-based subtyping of Cryptosporidium meleagridis. Journal of Clinical Microbiology, 52(7), 2311–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Xue X, Li J, Zhou Q, Yu Y, Du A. 2014. Cryptosporidiosis in broiler chickens in Zhejiang Province, China: molecular characterization of oocysts detected in fecal samples. Parasite, 21, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, Jian F, Sun Y, Hu Q, Zhu J, Wang F, Ning C, Zhang L, Xiao L. 2010. Large-scale survey of Cryptosporidium spp. in chickens and Pekin ducks (Anas platyrhynchos) in Henan, China: prevalence and molecular characterization. Avian Pathology, 39(6), 447–451. [DOI] [PubMed] [Google Scholar]
- 34.Xiao L, Feng Y. 2017. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food and Waterborne Parasitology, 8–9, 14–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson RCA, Fayer R, Lal AA. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Applied and Environmental Microbiology, 65(8), 3386–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplementary material is available at https://www.parasite-journal.org/10.1051/parasite/2022051/olm