Abstract
Can SARS-CoV-2 hitchhike on the olfactory projection and take a direct and short route from the nose into the brain? We reasoned that the neurotropic or neuroinvasive capacity of the virus, if it exists, should be most easily detectable in individuals who died in an acute phase of the infection. Here, we applied a postmortem bedside surgical procedure for the rapid procurement of tissue, blood, and cerebrospinal fluid samples from deceased COVID-19 patients infected with the Delta, Omicron BA.1, or Omicron BA.2 variants. Confocal imaging of sections stained with fluorescence RNAscope and immunohistochemistry afforded the light-microscopic visualization of extracellular SARS-CoV-2 virions in tissues. We failed to find evidence for viral invasion of the parenchyma of the olfactory bulb and the frontal lobe of the brain. Instead, we identified anatomical barriers at vulnerable interfaces, exemplified by perineurial olfactory nerve fibroblasts enwrapping olfactory axon fascicles in the lamina propria of the olfactory mucosa.
Keywords: COVID-19, SARS-CoV-2, Coronavirus, Delta, Omicron, olfactory, olfactory sensory neuron, perineurial olfactory nerve fibroblast, olfactory bulb, leptomeninges, brain parenchyma, frontal lobe, neurotropism, neuroinvasion, blood-brain barrier, Virchow-Robin space, glia limitans perivascularis, RNAscope
Graphical abstract
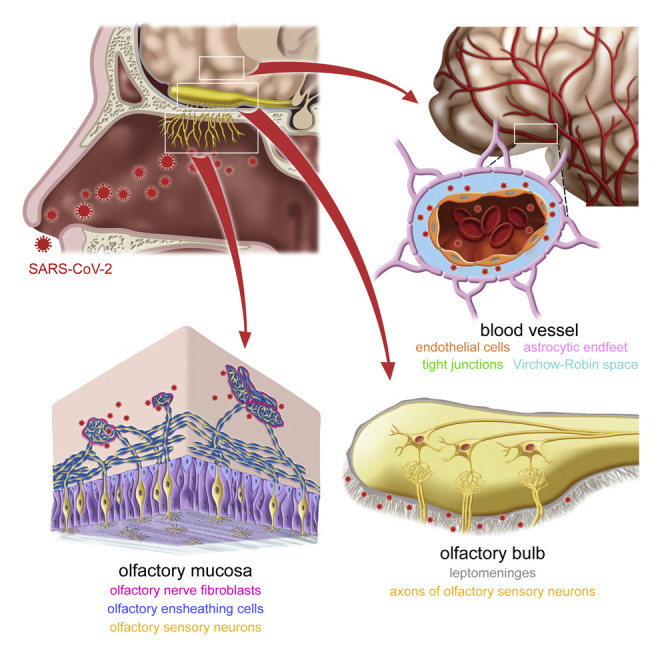
Khan et al. visualized infected cells, ongoing viral replication, and extracellular virions in postmortem tissue samples of COVID-19 patients who died during an acute phase of infection. They identified anatomical barriers protecting against SARS-CoV-2 neuroinvasion at vulnerable interfaces along the olfactory projection and in the frontal lobe of the brain.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the infectious agent responsible for the COVID-19 pandemic.1 Acute respiratory failure is often the cause of death.2 But, despite the name of the viral agent, COVID-19 is not uniquely a respiratory disease.3 , 4 Of relevance here, a plethora of neurological symptoms has become recognized that cannot be linked to the acute respiratory syndrome or be explained by iatrogenic causes.5 Much attention has been given to the hypothesis that SARS-CoV-2 infects neurons or other cell types in the nervous system (neurotropism) or invades the nervous system (neuroinvasion) and thereby causes pathology.6 , 7 , 8 Prominent among the neurological manifestations is olfactory dysfunction.9
Anosmia, the loss of smell, is a common symptom of COVID-19, particularly in the early phases of the pandemic10 but less so with the Omicron variant.11 , 12 , 13 There are only a few millimeters between the olfactory mucosa in the upper part of the nasal cavity and the olfactory bulb at the base and front of the cranial cavity. From the get-go, the olfactory projection has been a prime suspect for offering the virus a direct and short route from the nose into the brain.14 In one scenario, SARS-CoV-2 would infect olfactory sensory neurons (OSNs) at the level of the olfactory epithelium; virions would make it to the olfactory bulb through or along olfactory axon fascicles coursing within the lamina propria of the olfactory mucosa; the virus would spread from the olfactory bulb to the rest of the brain. This hypothetical scenario would explain both the olfactory dysfunction and other neurological manifestations.
A major technical hindrance for histological studies of the olfactory system in humans, living or deceased, is the rapid procurement of tissue samples of suitable quality and unambiguous identity. The olfactory mucosa is an archipelago of islands within the respiratory mucosa. It is not possible to harvest samples of pure olfactory mucosa, let alone pure olfactory epithelium. Furthermore, olfactory bulb biopsies cannot be taken from living patients due to the intracranial position and devastating consequences of the intervention. Adapting an endoscopic technique of skull base surgery,15 we developed a postmortem bedside surgical procedure to rapidly harvest tissue samples of respiratory mucosa, olfactory cleft mucosa, and frontal brain lobe, as well as whole olfactory bulbs.16 We reported on a first cohort of 85 cases, comprised of 70 COVID-19 patients infected with non-variants of concern or the Alpha variant and 15 non-infected control patients. By combining the RNAscope platform of ultrasensitive single-molecule fluorescence RNA in situ hybridization17 with fluorescence immunohistochemistry (IHC), we identified sustentacular cells as the major target cell type for SARS-CoV-2 in the olfactory mucosa. We failed to find evidence for infection of OSNs.
Here, we included a second cohort of 53 cases, comprised of 45 COVID-19 patients who died a few days after diagnosis of infection with the Delta, Omicron BA.1, or Omicron BA.2 variants and 8 non-infected control patients. Arguably, SARS-CoV-2 would have the greatest chance of displaying its hypothetical neurotropic or neuroinvasive capacity in highly vulnerable patients with a colossal failure of the host defense and the gravest outcome of all. But, in this second cohort as well as in the first cohort, we failed to find evidence for neurotropism and neuroinvasion along the olfactory projection and in the frontal lobe of the brain. We discovered that a poorly characterized cell type, the perineurial olfactory nerve fibroblasts (pONFs), forms a hitherto unrecognized anatomical barrier against SARS-CoV-2 virions.
Results
Extended postmortem bedside surgical procedure
The study concept of ANOSMIC-19 (ANnalyzing Olfactory dySfunction Mechanisms In COVID-19) is based on a 24/7 workflow that is initiated by a health care worker on an intensive care unit or a ward placing a call to a team of Ear, Nose, and Throat surgeons shortly after death of a COVID-19 patient. A team is dispatched to the bed and brings along a mobile unit consisting of a monitor, light source, camera, and endoscopic equipment. The surgeons then promptly perform the bedside surgical procedure. Retrospectively, we refer to the cohort described in Khan et al.16 as “cohort-I.”
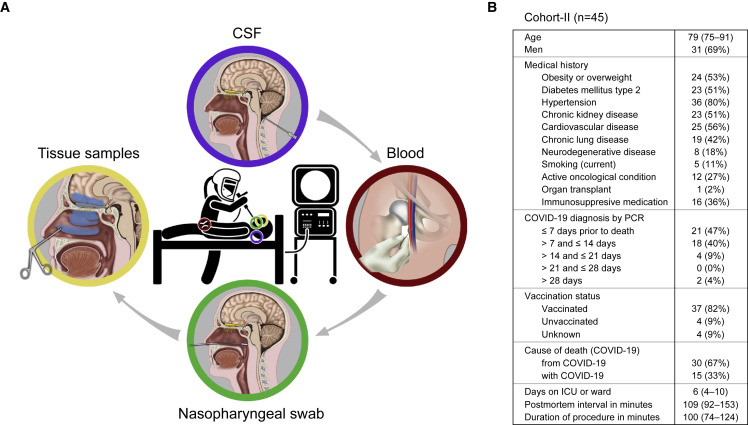
In the new cohort, which we refer to as “cohort-II,” we extended the workflow to include, in most cases, three additional types of samples: a sample of cerebrospinal fluid (CSF) extracted from the cisterna magna, a blood sample drawn from a femoral vein, and several nasopharyngeal swabs taken under endoscopic guidance (Figure 1A). We preferentially included patients who died within ∼14 days after diagnosis of COVID-19 by quantitative reverse-transcription polymerase chain reaction (qRT-PCR), hereafter abbreviated as “days after diagnosis.” Cohort-II is comprised of 45 patients who died from COVID-19 (30 or 67%) or with COVID-19 (15 or 33%) in major hospitals in Leuven (n = 38), Brussels (n = 4), and Bruges (n = 3) between August 2021 and May 2022, and 8 non-infected control patients who died of unrelated causes in Leuven between September 2021 and March 2022 (Figures 1B, S1, and S2). The median of the postmortem interval (PMI) of the COVID cases was 109 min. Cohort-II cases are numbered COVID #71 to #115 and CONTROL #17 to #24. We determined that COVID #71 through #94 were infected with the Delta variant and COVID #95 through #115 with the Omicron BA.1 or Omicron BA.2 variants.
Figure 1.
Extended postmortem bedside surgical procedure
(A) Sequence of four steps to procure bedside samples from deceased COVID-19 patients. Clockwise: in step 1, a CSF sample is extracted from the cisterna magna by a needle puncture below the occiput. In step 2, a blood sample is drawn from a femoral vein. In step 3, several nasopharyngeal swabs are taken under endoscopic guidance. In step 4, further under endoscopic guidance, tissue samples of respiratory mucosa and olfactory cleft mucosa are harvested; using an endoscopic endonasal transcribriform approach, each olfactory bulb is removed separately; finally, tissue samples of the frontal lobe are harvested.
(B) Cohort-II of 45 COVID-19 patients. Time variables are expressed as median and interquartile range Q1–Q3.
Viral loads and serum antibody titers
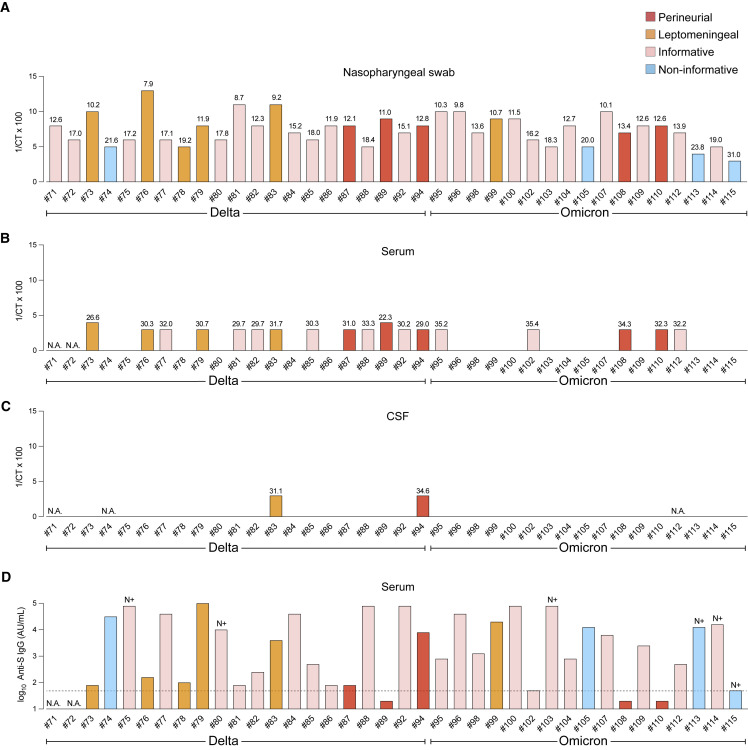
The postmortem nasopharyngeal swabs, serum samples, and CSF samples were analyzed with the TaqPath qRT-PCR assay on the same Quantstudio 7 Flex platform, allowing for direct comparison of the cycle threshold (Ct) values as indications of the viral load. All 38 nasopharyngeal swabs of the Leuven cases tested positive, with Ct values ranging from 7.9 to 31.0 (Figure 2A). Of the 36 available serum samples of the Leuven cases, 18 (50%) tested positive, with Ct values ranging from 22.3 to 35.4 (Figure 2B). Of the 35 available CSF samples of the Leuven cases, 2 (6%) tested positive (Figure 2C); as both cases were RNAemic, we cannot exclude that a traumatic puncture led to contamination with blood. Serum anti-S immunoglobulin G (IgG) antibody titers varied from below the detection limit (<50 arbitrary units [AU]/mL) to above the limit of quantification (>80,000 AU/mL), indicating a broad range of humoral responses against vaccine-derived spike protein and/or virus-derived spike protein (Figures 2D and S3). Seroconversion for anti-N IgG antibodies was observed in six serum samples.
Figure 2.
Postmortem Ct values of qRT-PCR assays and antibody serum titers
(A) Ct values of nasopharyngeal swabs from the 38 Leuven COVID cases. The y axis plots 1/Ct × 100.
(B) Ct values of serum samples.
(C) Ct values of CSF samples.
(D) The y axis plots the log10 values of AU/mL serum for anti-S IgG antibodies. The horizontal stippled line indicates the detection limit of 50 AU/ml for the anti-S IgG measurements. Seroconversion for anti-N IgG is indicated with “N+” above a bar. Samples from COVID #73 and #74 were taken on admission, the others postmortem. N.A., not available. See also Figures S1, S2, and S3.
Taken together, the high viral loads of the nasopharyngeal swabs and the frequent occurrence of an RNAemic state are consistent with an acute phase of infection.
Fatal breakthrough infections
Cohort-I included patients who died between May 2020 and April 2021, an era in which COVID-19 vaccines were not yet authorized or the Belgian vaccination campaign had just been initiated. By contrast, 37 of the 45 COVID cases (82%) in cohort-II were vaccinated. Four (9%) were not vaccinated, and the vaccination status of 4 patients (9%) was unknown. The vaccines most commonly administered were mRNA vaccines tozinameran/Comirnaty and elasomeran/Spikevax, and less frequently administered were vector vaccines ChAdOx1-S/Vaxzevria and Ad26.COV2-S/Janssen Jcovden. An infection with SARS-CoV-2 after vaccination is called a “breakthrough infection,” a term that encompasses a spectrum of outcomes ranging from asymptomatic infection to death.18 , 19 A challenging distinction is that some of the patients who die during a breakthrough infection die from COVID-19 whereas others die with COVID-19. Pragmatically, we coin the term “fatal breakthrough infection” (FBI) for vaccinated patients in our cohorts who died from COVID-19. Of the 37 vaccinated cases in cohort-II, 25 (68%) are, to the best of our clinical judgment, FBI cases. In cohort-I, only COVID #68 was vaccinated, and this patient was judged an FBI case.
Visualizing infected cells, ongoing viral replication, and extracellular virions
SARS-CoV-2 is a positive-sense single-stranded RNA virus. Positive sense means that the viral genomic RNA can be translated directly into protein. A single SARS-CoV-2 virion contains a single positive-sense full-length genomic RNA molecule. We performed confocal imaging of sections stained with a combination of fluorescence RNAscope, which visualizes a single RNA molecule as a dot or “punctum” (plural “puncta”),17 and fluorescence IHC, which visualizes an antigen as an immunoreactive (IR) signal.16 Our panel of viral RNAscope probes consists of SARS-CoV-2-N (nucleocapsid; giving rise to puncta referred to as N puncta), SARS-CoV-2-S (spike; S puncta), SARS-CoV-2-orf1ab (open reading frames 1a and 1b; orf1ab puncta), SARS-CoV-2-N-sense (N-sense puncta), SARS-CoV-2-S-sense (S-sense puncta), and SARS-CoV-2-orf1ab-sense (orf1ab-sense puncta). The sense probes detect negative-sense full-length genomic RNAs and negative-sense subgenomic RNAs, which are produced as intermediates during the viral life cycle but are not incorporated into virions released from cells.20 , 21 RNAscope puncta for sense probes therefore reflect ongoing viral replication, and sense puncta reside in a characteristic perinuclear position. The presence of extracellular N puncta, S puncta, or orf1ab puncta combined with the absence of sense puncta reflects the presence of extracellular virions released from cells. Nucleocapsid-IR signal diffusely fills an infected cell and outlines its contours, thereby facilitating cell type identification.
Delta and Omicron infect ciliated cells in the respiratory epithelium
Ciliated cells are the main target cell type in the respiratory epithelium for non-variants of concern and the Alpha variant.16 Ciliated cells can be identified by puncta for FOXJ1, which encodes a transcription factor involved in ciliogenesis, and by KRT8-IR signal. We detected viral RNA puncta and nucleocapsid-IR signal in respiratory mucosa samples of 41 of the 45 COVID cases of cohort-II (91%). We refer to such cases as “informative” and to the other cases as “non-informative.”16
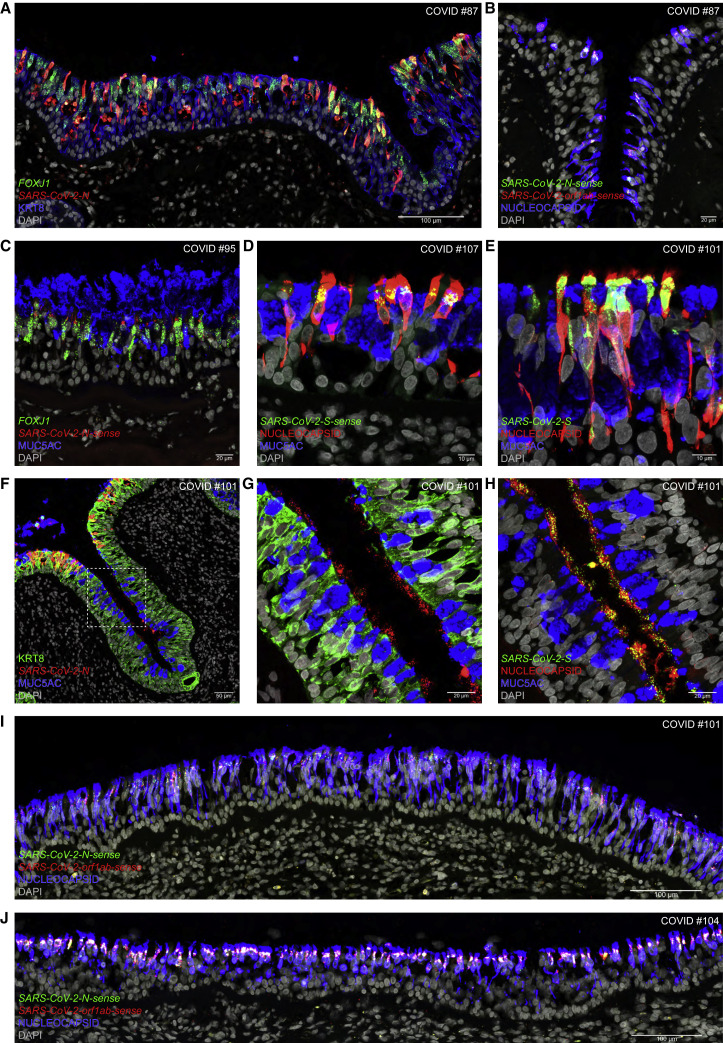
In the respiratory epithelium of COVID #87, a Delta FBI patient who died 3.7 days after diagnosis, many KRT8-IR cells harboring FOXJ1 puncta also harbor N puncta (Figure 3A); nearly all nucleocapsid-IR cells harbor N-sense puncta and orf1ab-sense puncta, reflecting ongoing viral replication (Figure 3B). In COVID #95, an Omicron BA.1 FBI patient who died 1.7 days after diagnosis, cells harboring FOXJ1 puncta harbor N-sense puncta and are situated along a stretch of the respiratory epithelium that is covered by a thick layer of secreted mucus containing IR signal for MUC5AC, a gel-forming glycoprotein secreted by goblet cells (Figure 3C). In COVID #107, an Omicron BA.2 patient who died 1.6 days after diagnosis, many cells with the typical position and shape of ciliated cells are nucleocapsid-IR, and all of these harbor S-sense puncta perinuclearly (Figure 3D). In COVID #101, an Omicron BA.2 patient who died 4.5 days after diagnosis, densely packed S puncta occur in nucleocapsid-IR cells, with IR signal diffusely filling the infected cells and outlining their contours including their cilia (Figure 3E). In this case, some KRT8-IR cells at the apical edge of a crypt or gland duct harbor N puncta and a blob of secreted mucus is MUC5AC-IR (Figure 3F). Deeper in this crypt or gland duct, there are no intracellular N puncta and the surface of the epithelium is covered with clusters of extracellular N puncta (Figure 3G); similar clusters of extracellular S puncta are nucleocapsid-IR (Figure 3H). A massive and diffuse infection across a broad swath of the respiratory epithelium of COVID #101 is revealed in a tiled confocal image: nearly every nucleocapsid-IR cell harbors N-sense puncta and orf1ab-sense puncta (Figure 3I). Likewise, Omicron BA.1 swept across a broad swath of the respiratory epithelium of COVID #104, who died 1.6 days after diagnosis (Figure 3J).
Figure 3.
Infection of the respiratory mucosa with Delta, Omicron BA.1, or Omicron BA.2
Confocal images of sections through the respiratory mucosa of COVID #87 (Delta), #95 (Omicron BA.1), #104 (Omicron BA.1), #101 (Omicron BA.2), and #107 (Omicron BA.2) stained with RNAscope and IHC. Names of genes (italics) and proteins (roman) are at the bottom left, scale bars at the bottom right. DAPI served as nuclear stain.
(A) N puncta are densely packed in cells harboring FOXJ1 puncta or containing KRT8-IR signal.
(B) Nucleocapsid-IR signal is widespread. N-sense puncta and orf1ab-sense puncta reflect ongoing viral replication.
(C) MUC5AC-IR mucus covers a stretch of respiratory epithelium with N-sense puncta in cells harboring FOXJ1 puncta.
(D) S-sense puncta occur at a characteristic perinuclear position in nucleocapsid-IR cells.
(E) Nucleocapsid-IR signal outlines the contours of ciliated cells, which harbor densely packed S puncta.
(F) At the apical edge of a crypt or gland duct, KRT8-IR cells harbor N puncta. Deeper, clusters of extracellular N puncta reflect virions released from cells.
(G) The dashed square in (F) imaged at higher magnification.
(H) In a section near (G), extracellular S puncta colocalize with nucleocapsid-IR signal.
(I and J) Omicron BA.2 (I) or Omicron BA.1 (J) swept through a broad swath of the respiratory epithelium, with nearly every nucleocapsid-IR cell harboring N-sense puncta and orf1ab-sense puncta.
Taken together, Delta, Omicron BA.1, and Omicron BA.2 can mount a massive attack on ciliated cells in the respiratory epithelium. The extracellular N puncta, S puncta, and nucleocapsid-IR signal that we captured in COVID #101 reflect extracellular virions released into the external milieu.
Progressive fasciculation, ensheathment, and enwrapment of olfactory sensory neuron axons in the lamina propria
We found in a subset of cases that SARS-CoV-2 invaded the interstitium of the lamina propria of the olfactory mucosa. Interstitial invasion raises the question of whether virions can infect cells of the olfactory projection at the level of the lamina propria, or hitchhike onto the olfactory projection, and then invade the brain. Before presenting these cases, we describe the anatomical relationships of OSN axons with other cellular components after they exit the olfactory epithelium and enter the lamina propria.
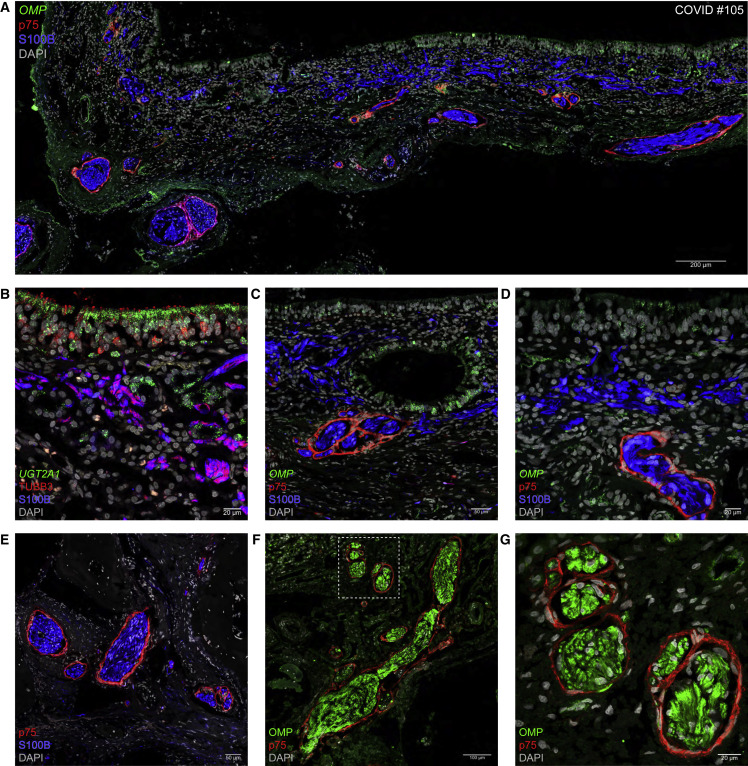
Particularly didactic in this regard is COVID #105, in whom no viral presence was detected. Figure 4A shows a continuous swath of olfactory epithelium rich in cells harboring puncta for OMP, which encodes the olfactory marker protein, a classical marker for mature OSNs. Each mature OSN projects a single axon through a perforation of the basement membrane into the superficial lamina propria, where it is received by olfactory ensheathing cells (OECs), a type of glia cells unique to the olfactory projection.22 , 23 , 24 OECs can be identified with S100B-IR or GFAP-IR signal. In the superficial lamina propria, the OECs run criss-cross and are cut in various planes. Progressively, OSNs and OECs together form olfactory axon fascicles of increasing diameter and become enwrapped by pONFs.25 , 26 , 27 , 28 , 29 , 30 , 31 This cell type can be identified in human tissue sections with p75-IR signal, with p75 as abbreviation for p75NTR, the p75 neurotrophin receptor also known as low-affinity nerve growth factor receptor.32 , 33 Figure 4B shows two major cell types of the olfactory epithelium: sustentacular cells, harboring UGT2A1 puncta, and OSNs, labeled with TUBB3-IR signal34 , 35; in the superficial lamina propria, OSN axons are intimately associated with OECs. Deeper in the lamina propria, transversely cut olfactory axon fascicles are enwrapped individually by pONFs (Figure 4C). The difference in pONF enwrapment of olfactory axon fascicles in superficial vs. deep lamina propria is prominent in Figure 4D. Closer to the cribriform plate, olfactory axon fascicles are thicker, and all have become enwrapped by pONFs (Figure 4E). The OSN provenance of the enwrapped cellular structures can be confirmed with OMP-IR signal (Figures 4F and 4G).
Figure 4.
The lamina propria of the olfactory mucosa
Confocal images of sections through the olfactory mucosa of COVID #105. No viral puncta or nucleocapsid-IR signal were detected in this non-informative case.
(A) OSNs harbor OMP puncta, S100B-IR signal identifies OECs, and p75-IR signal identifies pONFs.
(B) Sustentacular cells in the olfactory epithelium and Bowman’s gland cells in the lamina propria harbor UGT2A1 puncta. OSN axons contain TUBB3-IR signal and OECs contain S100B-IR signal.
(C) Layers of p75-IR pONFs surround olfactory axon fascicles. OMP puncta occur in an intraepithelial cyst or invagination of the olfactory epithelium.
(D) OMP puncta occur in the olfactory epithelium. In the deeper but not in the superficial lamina propria, OECs are enwrapped by pONFs.
(E) Axon fascicles are enwrapped by pONFs.
(F and G) OMP-IR olfactory axon fascicles are enwrapped by pONFs. The dashed square in (F) is imaged at higher magnification in (G).
Taken together, the fuzzy and loose configuration of the progressive fasciculation of OSN axons, ensheathment by OECs, and enwrapment by pONFs poses a vulnerability to the olfactory pathway and the brain, which is only a few millimeters away.
Perineurial olfactory nerve fibroblasts form an anatomical barrier
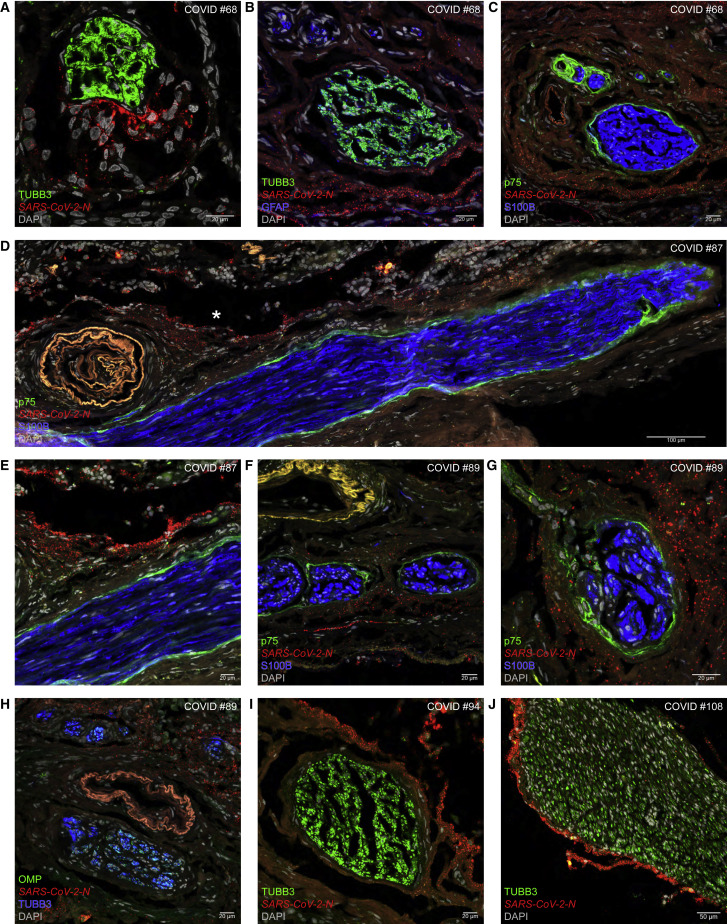
We discovered a pattern of interstitial SARS-CoV-2 spreading by virtue of the RNAscope-based visualization of extracellular virions, consistent with the well-documented hallmark of RNAscope to visualize single RNA molecules as puncta in light microscopy.17 Figure 5 shows that pONFs form an anatomical barrier against such virions. In reference to perineurial ONFs, we refer to seven cases as “perineurial cases": COVID #68 (Alpha, FBI) of cohort-I, and #87 (Delta, FBI), #89 (Delta, FBI), #90 (Delta), #94 (Delta, FBI), #108 (Omicron BA.1, unvaccinated), and #110 (Omicron BA.1) of cohort-II.
Figure 5.
Perineurial olfactory nerve fibroblasts form an anatomical barrier
Confocal images of sections through olfactory mucosa of COVID #68 (Alpha), #87 (Delta), #89 (Delta), #94 (Delta), and #108 (Omicron BA.1).
(A) N puncta do not invade a fascicle of TUBB3-IR OSN axons.
(B) A swarm of N puncta stays separate from fascicles containing OSN axons and GFAP-IR OECs.
(C) In a section close to (B), N puncta remain outside the enwrapment by p75-IR pONFs.
(D) A longitudinally sectioned olfactory axon fascicle containing S100B-IR OECs and p75-IR pONFs is devoid of N puncta. The blood vessel at the left emits autofluorescence.
(E) The area marked with an asterisk in (D) imaged at higher magnification.
(F and G) Arrays of N puncta do not invade axon fascicles containing OECs and enwrapped by pONFs.
(H) Fascicles of OMP-IR and TUBB3-IR OSN axons remain free of N puncta. The blood vessel at the center emits autofluorescence.
(I) N puncta stay clear of a fascicle of OSN axons.
(J) An ant-like column of N puncta escorts a fascicle of OSN axons. See also Figure S4.
In COVID #68, who died 5 days after diagnosis, a collection of N puncta in the interstitium does not intermingle with TUBB3-IR OSN axons (Figure 5A); trains of N puncta remain outside of a fascicle of TUBB3-IR OSN axons tightly associated with GFAP-IR OECs (Figure 5B); and a swarm of N puncta stays clear of the enwrapment by p75-IR pONFs (Figure 5C). In COVID #87, who died 3.7 days after diagnosis, a thick olfactory axon fascicle was cut longitudinally over a distance of ∼1.2 mm and is enwrapped without interruption by a thin layer of p75-IR pONFs; it is accompanied by an array of N puncta in the interstitium (Figure 5D), but the fascicle is devoid of N puncta (Figure 5E). In COVID #89, who died 5.1 days after diagnosis, N puncta are dispersed across the interstitium of the lamina propria but not within olfactory axon fascicles identified with S100B-IR signal of OECs and p75-IR signal of pONFs (Figures 5F and 5G) or with OMP-IR signal and TUBB3-IR signal of OSN axons (Figure 5H). In COVID #94, who died 8.3 days after diagnosis, dense accumulations of N puncta are mutually exclusive with TUBB3-IR OSN axons (Figure 5I). In COVID #108, who died 3.9 days after diagnosis, an ant-like column of N puncta escorts a thick fascicle of TUBB3-IR axons without invading it (Figure 5J).
We confirmed and extended these findings with RNAscope for orf1ab (Figures S4A, S4B, S4D–S4G, and S4I) and S (Figures S4C, S4F, and S4H–S4J): extracellular puncta do not invade TUBB3-IR axon fascicles and stay clear of the enwrapment by p75-IR pONFs. A haze of extracellular nucleocapsid-IR signal colocalizes with viral puncta (Figures S4E, S4F, and S4I). There are no N-sense puncta (Figures S4A, S4B, and S4J) or S-sense puncta (Figures S4D and S4G).
Taken together, pONFs form an anatomical barrier against viral invasion of the olfactory projection at a vulnerable interface in the lamina propria of the olfactory mucosa.
SARS-CoV-2 spares the parenchyma of the olfactory bulb
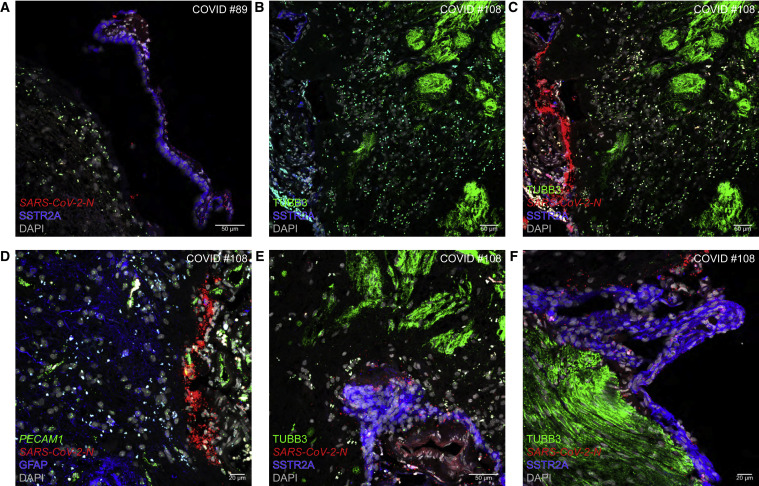
The term “fila olfactoria” is used for olfactory axon fascicles penetrating through foramina of the cribriform plate or coursing within the cranial cavity. These thread-like structures are macroscopically visible (see Video S1 at 1′40″ of Khan et al.16). The external surfaces of the fila olfactoria are contiguous with the leptomeninges (pia and arachnoid) that snugly cover the olfactory bulb. This anatomical contiguity raises the question of whether virions could sneak up from the interstitium of the lamina propria to the leptomeninges of the olfactory bulb by hitchhiking along the olfactory projection and then invade the parenchyma of the olfactory bulb. We reported the presence of viral RNA in the leptomeninges of the olfactory bulb in 11 of the 30 (37%) informative cases of cohort-I.16 We now report a leptomeningeal phenotype in 12 of the 41 informative cases (29%) of cohort-II. Suspiciously, six of the seven perineurial cases are also leptomeningeal.
In COVID #89, N puncta are dispersed across a SSTR2A-IR swath of leptomeninges but do not occur in the parenchyma (Figure 6A). In COVID #108, densely packed N puncta occur in pia mater abutting virus-free parenchyma, in which TUBB3-IR OSN axons terminate in globose structures, the glomeruli (Figures 6B and 6C). Abundant N puncta in the leptomeninges are associated with blood vessels identified with PECAM1 puncta in endothelial cells (Figure 6D). Despite the close proximity to SSTR2A-IR leptomeningeal tissue, N puncta do not occur within TUBB3-IR fila olfactoria or glomeruli (Figure 6E). Sprinkles of N puncta accompany a TUBB3-IR filum olfactorium and are spread across the abutting leptomeninges (Figure 6F).
Figure 6.
The olfactory bulb parenchyma is spared from invasion by SARS-CoV-2
(A) In COVID #89 (Delta), N puncta occur in a partially detached swath of SSTR2A-IR pia mater (to the right) and do not invade the parenchyma (to the left).
(B–F) COVID #108 (Omicron BA.1). The parenchyma including TUBB3-IR glomeruli (to the right) is spared from infection, whereas the abutting SSTR2A-IR pia mater (to the left) contains densely packed N puncta (B and C). An area of pia mater contains densely packed N puncta in the vicinity of PECAM1 puncta identifying endothelial cells, and the parenchyma containing GFAP-IR signal is free of N puncta (D). N puncta are dispersed within leptomeninges but not within TUBB3-IR structures (E). Accompanying, but not invading, a filum olfactorium of TUBB3-IR OSN axons are sprinkles of N puncta, and the abutting leptomeninges contain dispersed N puncta (F).
Taken together, the parenchyma of the olfactory bulb is spared from viral invasion.
Absence of evidence for viral invasion into the parenchyma of the frontal lobe
We harvested brain tissue close to the olfactory bulb, typically the gyrus rectus and sometimes the gyri orbitales of the frontal lobe, in 112 of the 115 cases of cohort-I and cohort-II. We identified the sporadic occurrence of extracellular N or S puncta in 5 of the 112 cases: COVID #60 (non-variant of concern, unvaccinated), #87 (Delta, FBI), #89 (Delta, FBI), #108 (Omicron BA.1, unvaccinated), and #110 (Omicron BA.1). All five cases were RNAemic perimortem. The puncta occurred intravascularly or perivascularly, but not in the parenchyma.
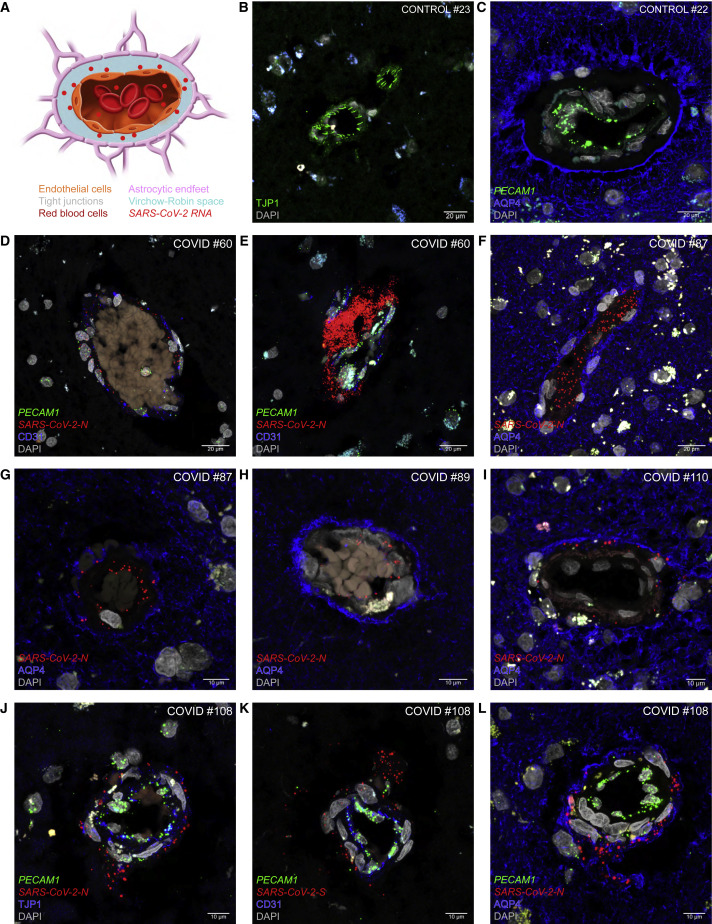
The blood-brain barrier includes the layer of endothelial cells of cerebral blood vessels connected with tight junctions (Figure 7A). The Virchow-Robin space (VRS) is a CSF-filled perivascular space that exists along the larger cerebral blood vessels but not around smaller blood vessels such as capillaries.36 , 37 The glia limitans perivascularis covers the entire cerebral vasculature: all arterioles, capillaries, and venules within the brain parenchyma are surrounded by vascular endfeet of astrocytes.38 The tight junctions among endothelial cells can be visualized as short TJP1-IR stripes, for tight junction protein-1 (Figure 7B). Endothelial cells harbor PECAM1 puncta, and astrocytes, including their endfeet, can be identified with AQP4-IR signal, for aquaporin-4 (Figure 7C).
Figure 7.
Absence of SARS-CoV-2 invasion of the parenchyma of the frontal lobe
(A) Schematic of a cross section through a blood vessel.
(B–L) Confocal images of sections through frontal lobe samples of control cases (B and C) and COVID cases (D–L).
(B) TJP1-IR stripes label tight junctions among endothelial cells of a cerebral blood vessel.
(C) PECAM1 puncta label endothelial cells of a blood vessel and AQP4-IR signal labels the endfeet of astrocytes forming the glia limitans perivascularis. The unstained, dark ring between the area with PECAM1 puncta and the AQP4-IR ring is the VRS.
(D and E) In COVID #60 (non-variant of concern), N puncta occur within the lumen of a blood vessel filled with red blood cells (D). A swarm of N puncta appears to have burst through the endothelial layer of a blood vessel (E). Endothelial cells harbor PECAM1 puncta and contain CD31-IR signal.
(F and G) In COVID #87 (Delta), N puncta are dispersed intravascularly (F) and perivascularly (G). The white dots in the parenchyma represent autofluorescence signal from lipofuscin, emitting in all channels.
(H) In COVID #89 (Delta), N puncta occur within the lumen of a blood vessel filled with red blood cells but not beyond the AQP4-IR ring.
(I) In COVID #110 (Omicron BA.1), N puncta occur perivascularly but not beyond the AQP4-IR ring.
(J–L) In COVID #108 (Omicron BA.1), puffs of N puncta (J and L) or S puncta (K) appear to have leaked through a blood vessel wall identified with PECAM1 puncta (J–L), TJP1-IR signal (J), or CD31-IR signal (K). Viral puncta have not penetrated into the parenchyma (L).
COVID #60 had a malignant brain tumor and died 40 h after diagnosis due to increased intracranial pressure that was treated with high doses of corticosteroids; a serum sample taken a few hours prior to the time of death had a Ct of 33.6. A few N puncta are visible within the lumen of a blood vessel outlined with CD31-IR endothelial cells harboring PECAM1 puncta (Figure 7D), and a compact cluster of N puncta appears to have leaked through the endothelial wall (Figure 7E). In COVID #87, N puncta occur intravascularly and perivascularly but remain confined to the VRS and do not reach beyond the AQP4-IR ring of astrocytic endfeet (Figures 7F and 7G). In COVID #89 (Figure 7H) and in COVID #110 (Figure 7I), N puncta occur within the lumen of a blood vessel but not beyond the AQP4-IR ring. In COVID #108, puffs of N or S puncta appear to have leaked through the endothelial wall identified with PECAM1 puncta (Figures 7J–7L), TJP1-IR signal (Figure 7J), and CD31-IR signal (Figure 7K) but did not proceed into the parenchyma (Figure 7L).
Taken together, we failed to find evidence for viral invasion of the parenchyma of the frontal lobe in 112 of 112 (100%) cases.
The cerebrospinal fluid remains free from infectious SARS-CoV-2 virions
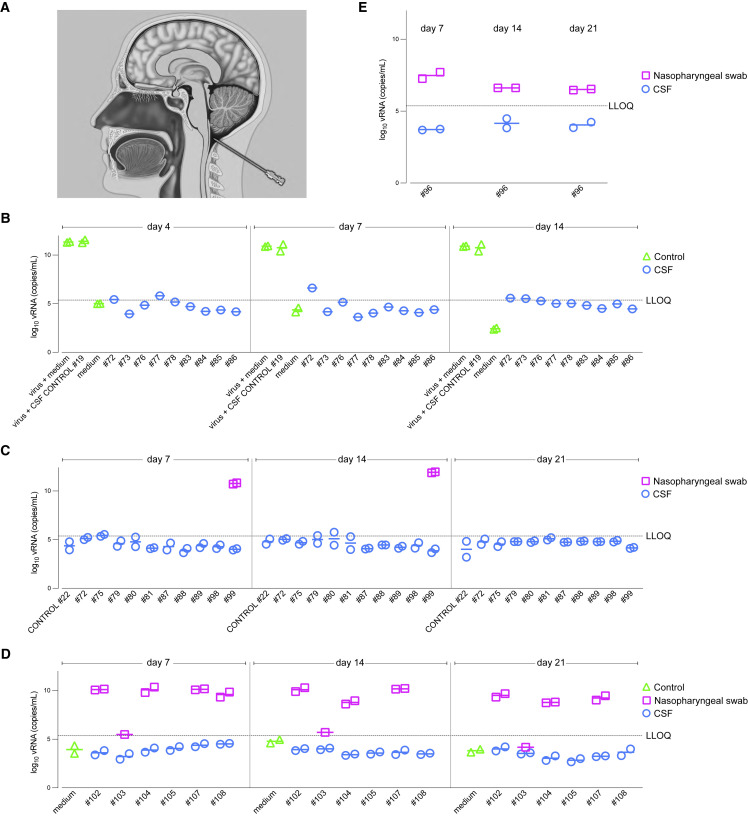
Finally, we asked whether SARS-CoV-2 virus could be grown in culture from CSF samples extracted postmortem from the cisterna magna (Figure 8A). As substrate we used inserts of human airway epithelial cell cultures with an air-liquid interface. These differentiated primary cultures possess the architecture and cellular complexity of the epithelium of the human respiratory tract and serve as assays for profiling antiviral drugs against SARS-CoV-2,39 including Delta and Omicron BA.1.40 An inoculum is applied to the apical side of an insert; the apical side is washed with medium at several time points; RNA is extracted from the wash fluids; and SARS-CoV-2 RNA is quantified by qRT-PCR.
Figure 8.
SARS-CoV-2 cannot be cultured from CSF samples of COVID cases
(A) The cisterna magna is punctured with a needle at the back of the head and a CSF sample is extracted.
(B–E) CSF samples and nasopharyngeal swabs were used to inoculate the apical side of inserts of primary cultures of human airway epithelial cells. CSF does not interfere with replication of a Delta strain; no virus grew from CSF samples of nine COVID cases (B). The positive control was the nasopharyngeal swab of COVID #99 (Omicron BA.1) and the negative control was the CSF sample of CONTROL #22; no virus could be grown from CSF samples of ten COVID cases (C). Paired nasopharyngeal swab and CSF samples of six COVID cases were tested; no virus could be grown from CSF samples (D). Virus could be grown from a freshly taken nasopharyngeal swab, but not from the paired CSF sample, of COVID #96 (Omicron BA.1) (E).
In Figure 8B, we verified that CSF does not inhibit growth of SARS-CoV-2 in this platform by adding CSF from CONTROL #19 to an inoculum of Delta strain hCoV-19/Belgium/rega-7214/2021.41 In parallel, we tested CSF samples of nine Delta cases, including COVID #83, whose CSF sample had a Ct of 31.1; no virus replication was detected. In a second experiment (Figure 8C), virus could be grown from the nasopharyngeal swab, but not from the CSF sample, of COVID #99, and neither could it be grown from CSF samples of nine other Delta and Omicron BA.1 cases, including COVID #87 and #89, whose frontal lobe samples contained N puncta. In a third experiment (Figure 8D), virus could be isolated from nasopharyngeal swabs of four cases, except for COVID #103, who died 14.1 days after diagnosis, but not from paired CSF samples, including COVID #108, whose frontal lobe sample contained N puncta. Finally, virus grew from a freshly taken nasopharyngeal swab, but not from the paired CSF sample of COVID #96 (Figure 8E).
Taken together, in none of 25 COVID cases could we grow SARS-CoV-2 from CSF samples, including from three cases with viral puncta in the frontal lobe sample. By contrast, paired nasopharyngeal swabs routinely gave rise to virus production.
In summary, based on the results of this study and Khan et al.,16 we conclude that SARS-CoV-2 does not infect OSNs, olfactory bulb neurons, or neurons in the frontal lobe; it does not invade olfactory axon fascicles, fila olfactoria, the parenchyma of the olfactory bulb and of the frontal lobe; and virions are not recoverable from CSF samples.
Discussion
In this virocentric view of COVID-19, we visualized infected cells, ongoing viral replication, and extracellular virions in postmortem tissue samples of patients who died during an acute phase of SARS-CoV-2 infection.
Catching SARS-CoV-2 in the act of the attack
Cohort-II of ANOSMIC-19 comprises 45 patients who died from or with COVID-19 shortly after diagnosis in three major hospitals in Belgium over a period of eight months in 2021 and 2022 that spanned the Delta, Omicron BA.1, and Omicron BA.2 waves of the pandemic in Belgium. Our method of procuring postmortem tissue samples takes place at the bedside soon after the death of the patient, with a rapid endoscopic method that allows for precise harvesting of endonasal and intracranial tissue samples. Consistent with the short PMIs, none of the 45 COVID cases and 8 control cases had to be excluded because of poor tissue quality. We report the period until death that elapsed from the time the positive nasopharyngeal swab was taken rather than from the onset of symptoms, which is an unreliable measure, particularly in patients who were already in ill health prior to the infection.
Challenges to find evidence for neuroinvasion by SARS-CoV-2
Our qRT-PCR assays revealed RNAemia in 50% of the postmortem serum samples. RNAemia, the presence of viral RNA in the blood, may reflect viremia, the presence of virus in the blood.42 , 43 This frequent observation of RNAemia and the close association of N and S puncta with blood vessels in the frontal lobe samples of our five COVID cases suggest RNAemia as an explanation for positive qRT-PCR results on homogenized brain samples in some of the literature on autopsies. Blood is present in all tissues, and to various extents. Unless a blood sample from around the time of death is available, caution must be exercised when interpreting claims of neuroinvasion that are largely based on such qRT-PCR analyses.
An early claim of neuroinvasion proposed the olfactory pathway as a port of central nervous system entry.6 But, the sole antibody for a viral antigen that was used in this study6, 3A2, gives similar IR signals in control cases.6 , 44 , 45 , 46 Throughout cohort-I and cohort-II, we have included control cases in parallel, following the same protocols of tissue sample procurement, fixation, processing, staining, and imaging. This two-armed study design safeguards us against false-positive signals during the course of a multi-year project.
The fluorescence RNAscope platform has a high signal-to-noise ratio, yielding a high specificity. The sense probes reflect ongoing viral replication, and sense puncta have a characteristic perinuclear location. The combination of nucleocapsid being the most abundant SARS-CoV-2 protein,47 being a cytosolic protein as opposed to membrane-bound spike, and diffusely filling infected cells and outlining their contours, underlies the strong and distinctive nucleocapsid-IR signals in our confocal images. With the dual visualization of RNAscope puncta and nucleocapsid-IR signal, a given cell type or a particular cell can be unmistakably identified as a target cell type or infected, respectively. Applying these stringent criteria, we failed to find evidence for neurotropism—defined as the ability of a virus to infect and replicate in cells of the nervous system8—in OSNs, olfactory axon fascicles, fila olfactoria, and the parenchyma of the olfactory bulb and of the frontal lobe.
What about neuroinvasiveness, defined as the ability of a virus to enter the nervous system?8 Could virions invade the olfactory projection and the brain without infecting cells? In other words, could SARS-CoV-2 be neuroinvasive but not neurotropic? The pristine condition of our tissue samples, the power of the RNAscope platform, and the precision of confocal imaging enabled us to light-microscopically visualize extracellular virions released from infected cells and spreading interstitially through tissues. The smallest extracellular puncta may well represent single virions, consistent with the extensively documented hallmark of RNAscope visualizing single RNA molecules as puncta.17 By using a battery of RNAscope probes and an anti-nucleocapsid antibody, we failed to find evidence for neuroinvasion of the parenchyma of the olfactory bulb and of the frontal lobe.
But the human brain is a large and anatomically highly structured organ, and we reasoned that we should also look for the presence of infectious virions in the CSF. The brain of an adult human is bathed in ∼150 mL of CSF,48 of which we tapped ∼5 mL from the cisterna magna, which communicates with the other ventricles. We failed to find evidence for infectious virions in CSF samples, including from one COVID case with a positive qRT-PCR result of the CSF sample (#83) and three COVID cases with N puncta in frontal lobe samples (#87, #89, and #108). By contrast, SARS-CoV-2 could be routinely grown from paired nasopharyngeal swabs.
Taken together, we failed to find evidence for neurotropism and neuroinvasion in two successive cohorts comprised of a total of 115 patients infected with non-variants of concern, Alpha, Delta, Omicron BA.1, or Omicron BA.2 and surviving between 0 and 40 days after diagnosis. The colossal failure of host defense would afford the most permissive conditions for viral replication and spreading in these individuals. We submit that our analyses of these cases represent the most stringent test imaginable to date for the neurotropic and neuroinvasive capacity of SARS-CoV-2.
Admittedly, the absence of evidence does not equal the evidence of absence. It is ultimately not possible to prove a negative in science and in medicine. Certain types of neurons may eventually be found to be infected, perhaps in a subset of patients, in certain disease courses or phases, or elsewhere in the brain. A future variant may inflict (collateral) damage to the brain by newly acquired neurotropic or neuroinvasive properties without suffering disadvantage in the incessant evolutionary race to ever-greater transmissibility, fitness, and immune evasion.
Perineurial olfactory nerve fibroblasts form an anatomical barrier
By analogy with peripheral nerves, which have an endoneurium, a perineurium, and an epineurium, pONFs are said to be in a perineurial position with regard to the olfactory nerve. These cells enwrap, as one or a few layers, olfactory axon fascicles consisting of OSN axons ensheathed by OECs. The term “fibroblast” may well reflect the depth of our lack of knowledge about this cell type rather than a cell-biological characteristic. Ultrastructurally, rat pONFs are extremely thin elongated cells with nuclei that reach 15 μm in length.25
In the seven perineurial cases of cohort-I and cohort-II, the lamina propria of the olfactory mucosa contains extracellular N, S, and orf1ab puncta, reflecting virions spreading within the interstitium. Viral invasion of the lamina propria may occur when the olfactory epithelium is structurally damaged due to massive infection of sustentacular cells, ensuing desquamation, and erosion of the basement membrane. Alternatively, and not mutually exclusively, virions may invade the lamina propria via the hematogenous route. Indeed, the five perineurial cases for whom blood samples were available were RNAemic. Collections of extracellular N puncta assume several geometric configurations. They may follow the paths of lowest resistance and be propelled by bulk flow toward the cranial cavity.49 They approach and come so close to olfactory axon fascicles that they seem to trail their contours, but they do not infect pONFs and do not penetrate beyond the p75-IR enwrapment.
The mechanistic basis of the pONF barrier remains to be determined.50 We speculate that this barrier may also be effective against some of the many other pathogens that infect the nasal mucosa and could threaten the brain.51
Absence of evidence for invasion of the parenchyma of the olfactory bulb
We detected extracellular N and S puncta in the leptomeninges (pia mater and arachnoid) that cover the surface of the olfactory bulb. These leptomeninges are anatomically contiguous with the external surfaces of olfactory axon fascicles and fila olfactoria. We reported a leptomeningeal phenotype in 11 of the 30 informative cases of cohort-I in Khan et al.16 and now add 12 of the 41 informative cases of cohort-II for a total of 23 of the 71 informative cases (32%). The absence of sense puncta in the leptomeninges argues against ongoing viral replication at these sites. Suspiciously, six of the seven perineurial cases are also leptomeningeal; the case with the weakest perineurial phenotype (COVID #90) is not leptomeningeal. It is tempting to speculate that perineurial cases and leptomeningeal cases belong to the same spectrum, with virions hitchhiking along the external surface of the pONF enwrapment into the leptomeninges covering the olfactory bulb.
The fate of the leptomeningeal virions is uncertain, but they do not invade the parenchyma of the olfactory bulb. The leptomeninges-parenchyma interface is vulnerable, as the olfactory bulb contains the second-order neurons of the olfactory pathway, the mitral and tufted cells, which project their axons via the olfactory tract to the piriform cortex and many other brain regions. Hence, infection of these neurons would be an effective way for SARS-CoV-2 to spread throughout the brain, but it is a road not taken.
Our findings of leptomeningeal virions in 32% of informative cases are consistent with those of a neuropathological study that examined 16 brain regions in COVID-19 decedents by qRT-PCR for several SARS-CoV-2 gene sequences.52 In 8 out of 21 cases (38%), viral RNA was detected in olfactory bulb samples, and in some of these cases also in amygdala and entorhinal area, brain regions closely connected to the olfactory bulb, but typically not elsewhere in the brain. It would be interesting to determine with RNAscope in cases of the Beach cohort52 whether viral RNA is detectable in the leptomeninges covering the olfactory bulb vs. in the parenchyma.
Absence of evidence for invasion of the parenchyma of the frontal lobe
In a mere 5 of 112 cases of cohort-I and cohort-II for whom frontal lobe tissue samples were available, we detected viral signal—sporadically, as clusters or puffs of extracellular N puncta or S puncta, the proverbial needles in a haystack. Viral puncta did not present within the parenchyma but were confined to the lumen of a blood vessel or appeared to have leaked through the endothelial layer. Viral puncta did make it as far as the VRS, but not beyond the glia limitans perivascularis, which is formed by AQP4-IR vascular endfeet of astrocytes and covers the cerebral vasculature along its entire length of several hundred kilometers. Strikingly, the antibody response to vaccine-derived spike protein and/or virus-derived spike protein was close to non-existent in these five cases: COVID #60, #89, #108, and #110 had anti-S IgG titers below the limit of detection (<50 AU/mL), and COVID #87 had a very low titer of 80.2 AU/mL. Moreover, there was no seroconversion for anti-N IgG in these five cases. Thus, even in the context of an abysmal failure of the humoral response to the vaccine and/or the virus, SARS-CoV-2 failed to invade the parenchyma of the olfactory bulb and of the frontal lobe.
Regular glymphatic cleansing of the brain53 may efficiently dilute virions that made it to the CSF or entered the brain parenchyma. Meningeal lymphatic vessels may help dispose of SARS-CoV-2 virions.54 On the other hand, the integrity of the anatomical barriers may be compromised and breaches resulting in neuroinvasion may be possible. The blood-brain barrier is leaky in neurodegenerative disorders such as Alzheimer’s disease.55 Interestingly, perineurial case #94, whose CSF sample tested positive by qRT-PCR, had Alzheimer’s disease. Microbleeds or fleeting events of microneuroinvasion may go unnoticed in histological studies like ours but might be sufficient to trigger a sequence of pathobiological events culminating in chronic neuroinflammation and lasting dysfunction such as some of the neurological manifestations of long COVID.56
A look ahead
Similar to sustentacular cells in the olfactory epithelium playing the role of unsung heroes of the sense of smell,16 perineurial olfactory nerve fibroblasts are another band of unsung heroes: they form a protective anatomical barrier at a vulnerable interface of the olfactory projection. Through mechanisms that remain to be elucidated, this enigmatic cell type appears to seal olfactory axon fascicles hermetically from invasion by SARS-CoV-2 virions only 100 nm in diameter.
Limitations of the study
The scope of the study was limited to light-microscopically visualizing whether SARS-CoV-2 is capable of neurotropism and neuroinvasion in samples of olfactory cleft mucosa, olfactory bulb, and frontal lobe. We took the viewpoint of the virus and not of the host response for a host of reasons, including the heterogeneity of patients in terms of comorbidities, medical treatments, previous exposures to SARS-CoV-2 (diagnosed or undiagnosed), vaccination regimens, and antibody titers to spike and nucleocapsid antigens. An investigation of neuroinflammation was beyond the scope of the study. Most COVID-19 patients, vaccinated or unvaccinated, recover from their infection, raising the fundamental question as to whether postmortem studies adequately reflect the situation in individuals with a non-fatal disease outcome. This issue will be almost impossible to resolve, as there are no ethically justifiable indications for brain biopsies from living COVID-19 patients. We sampled only the gyrus rectus and gyri orbitales and may have overlooked neurotropism and neuroinvasion elsewhere in the brain. A study of a postmortem cohort cannot be undertaken longitudinally, by its very nature. Instead, we must make inferences from snapshots taken during many singular courses of infection and then attempt to fill in the dots and make associations. It does appear, though, that SARS-CoV-2 is stopped dead in its tracks by several anatomical barriers at vulnerable interfaces, even in extremely weak individuals with an abysmal level of defense who lost the battle.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Aquaporin 4, C-terminus antibody | Millipore | Cat#AB3594; RRID: AB_91530 |
| Anti-CD31 antibody | Abcam | Cat#ab28364; RRID: AB_726362 |
| GFAP antibody | Novus Biologicals | Cat#NB300-141; RRID: AB_10001722 |
| Cytokeratin 8 antibody | Novus Biologicals | Cat#NBP2-67468; RRID: AB_2922444 |
| Mouse monoclonal MUC5AC antibody (45M1) | Thermo Fisher Scientific | Cat#MA5-12178; RRID: AB_10978001 |
| Rabbit monoclonal SARS-CoV/SARS-CoV-2 Nucleocapsid antibody (clone #001) | Sino Biological | Cat#40143-R001; RRID: AB_2827974 |
| Goat Anti-Olfactory Marker Protein Antibody, unconjugated | FUJIFILM Wako Shibayagi | Cat#544-10001-WAKO; RRID: AB_664696 |
| Monoclonal Anti-Nerve Growth Factor Receptor (NGFR p75) antibody | Sigma-Aldrich | Cat#N5408; RRID: AB_477280 |
| S100B Recombinant monoclonal antibody (S100B/1706R) | Enzo Life Sciences | Cat#ENZ-ABS307; RRID: AB_2922445 |
| Rabbit polyclonal antiserum Somatostatin receptor subtype 2A | Biotrend | Cat#NB-49-016-50ul |
| ZO-1 tight junction protein antibody | Novus Biologicals | Cat#NBP1-85046; RRID: AB_11006040 |
| Mouse monoclonal anti-Tubulin beta 3/TUBB3 (TUJ1) | BioLegend | Cat#801202; RRID: AB_10063408 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 | Thermo Fisher Scientific | Cat#A32790; RRID: AB_2762833 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 555 | Thermo Fisher Scientific | Cat#A32794; RRID: AB_2762834 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 555 | Thermo Fisher Scientific | Cat#A32773; RRID: AB_2762848 |
| Donkey anti-Goat IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 555 | Thermo Fisher Scientific | Cat#A32816; RRID: AB_2762839 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 647 | Thermo Fisher Scientific | Cat#A32795; RRID: AB_2762835 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 647 | Thermo Fisher Scientific | Cat#A32787; RRID: AB_2762830 |
| Donkey anti-Goat IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 647 | Thermo Fisher Scientific | Cat#A32849; RRID: AB_2762840 |
| Biological samples | ||
| Tissue, blood, and CSF samples from COVID-19 patients and control patients | Ethical Committee of the University Hospitals Leuven, in Leuven, Belgium (S64042), the General Hospital Sint-Jan Brugge-Oostende AV in Bruges, Belgium (2736), and the Universitair Ziekenhuis Brussel in Brussels, Belgium (EC-2021-360) | clinicaltrials.gov (NCT04445597) |
| Chemicals, peptides, and recombinant proteins | ||
| Mount Solid antifade | abberior | Cat#MM-2011-2X15ML |
| RNAscope Target Retrieval Reagent | Advanced Cell Diagnostics | Cat#322000 |
| RNAscope Protease III | Advanced Cell Diagnostics | Cat#322337 |
| RNAscope Hydrogen Peroxide and Protease Plus | Advanced Cell Diagnostics | Cat# 322330 |
| RNAscope Probe Diluent | Advanced Cell Diagnostics | Cat#300041 |
| 10% neutral buffered formalin | Electron Microscopy Sciences | Cat#15740-04 |
| Tissue-Tek O.C.T. compound | Sakura | Cat#4583 |
| Opal 520 | Akoya Biosciences | Cat#FP1487001KT |
| Opal 570 | Akoya Biosciences | Cat#FP1488001KT |
| Opal 690 | Akoya Biosciences | Cat#FP1497001KT |
| 1x Plus Amplification Diluents | Akoya Biosciences | Cat#FP1498 |
| Donkey serum | Sigma-Aldrich | Cat#S30-100ML |
| Sucrose | Sigma-Aldrich | Cat#S0389-1KG |
| DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher Scientific | Cat#D1306 |
| Critical commercial assays | ||
| RNAscope Multiplex Fluorescent Detection Kit v2 | Advanced Cell Diagnostics | Cat#323110 |
| RNAscope 4-Plex Ancillary Kit | Advanced Cell Diagnostics | Cat#323120 |
| RNAscope Hs-FOXJ1 (C1) | Advanced Cell Diagnostics | Cat#430921 |
| RNAscope Hs-FOXJ1 (C2) | Advanced Cell Diagnostics | Cat#430921-C2 |
| RNAscope Hs-OMP (C1) | Advanced Cell Diagnostics | Cat#824181 |
| RNAscope Hs-PECAM1-O1 (C1) | Advanced Cell Diagnostics | Cat#487381-C1 |
| RNAscope Hs-PECAM1-O1 (C2) | Advanced Cell Diagnostics | Cat#487381-C2 |
| RNAscope Hs-PECAM1-O1 (C4) | Advanced Cell Diagnostics | Cat#487381-C4 |
| RNAscope Hs-UGT2A1 (C1) | Advanced Cell Diagnostics | Cat#814271 |
| RNAscope V-nCoV-N (C1) | Advanced Cell Diagnostics | Cat#846081 |
| RNAscope V-nCoV-N (C2) | Advanced Cell Diagnostics | Cat#846081-C2 |
| RNAscope V-SARS-CoV-2-N-O2-sense (C1) | Advanced Cell Diagnostics | Cat#863841 |
| RNAscope V-SARS-CoV-2-N-O2-sense (C3) | Advanced Cell Diagnostics | Cat#863841-C3 |
| RNAscope V-nCoV2019-orf1ab-sense (C1) | Advanced Cell Diagnostics | Cat#859151 |
| RNAscope V-nCoV2019-S (C1) | Advanced Cell Diagnostics | Cat#848561 |
| RNAscope V-nCoV2019-S-sense (C1) | Advanced Cell Diagnostics | Cat#845701 |
| RNAscope V-nCoV2019-S-sense (C3) | Advanced Cell Diagnostics | Cat#845701-C3 |
| RNAscope V-nCoV-orf1ab-O1 (C1) | Advanced Cell Diagnostics | Cat#859981 |
| Software and algorithms | ||
| Zeiss ZEN 2.6 system | Zeiss | https://www.zeiss.com |
| GraphPad Prism v9.2 | GraphPad | https://graphpad.com |
| Adobe Illustrator 2020 | Adobe | Adobe Creative Cloud |
| Adobe Acrobat Pro DC | Adobe | Adobe Creative Cloud |
| Other | ||
| Zeiss LSM 800 | Zeiss | https://www.zeiss.com |
| Leica CM3050 S cryostat | Leica | https://www.leicabiosystems.com |
Resource availability
Lead contact
Further information and requests should be directed to the lead contact, Peter Mombaerts (peter.mombaerts@gen.mpg.de).
Materials availability
This study did not generate new unique reagents.
Date and code availability
-
•
Clinical data about the patients are confidential, subject to compliance with applicable personal data protection laws, and not publicly available.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Study design and data collection
The foundation of the study protocol ANOSMIC-19 (ANalyzing Olfactory dySfunction Mechanisms in COVID-19) is the bedside procurement of postmortem tissue samples. This national multicenter study was approved by the Ethical Committees of the University Hospitals Leuven in Leuven, Belgium (S64042), of the General Hospital Sint-Jan Brugge-Oostende AV in Bruges, Belgium (2736), and of the Universitair Ziekenhuis Brussel in Brussels, Belgium (EC-2021-360), and registered on clinicaltrials.gov (NCT04445597). The Ethikrat – Kommission des Präsidenten of the Max Planck Society did not require a separate ethics review by a medical ethics committee (Applications No: 2020_14, 2020_30, and 2020_31). Patients were >18 years old at the time of inclusion. Written informed consent from next of kin was obtained prior to tissue harvesting in accordance with the recommendations of the local Ethical Committee.
COVID-19 patients were diagnosed with a SARS-CoV-2 infection by qRT-PCR from a nasopharyngeal swab and died during their subsequent COVID-19 hospitalization. Control patients had a negative qRT-PCR test from a postmortem nasopharyngeal swab taken a few days prior to their time of death and died of other causes than COVID-19. The electronic health records of each patient were retrospectively reviewed and analyzed to obtain information about demographics, comorbidities, disease course, and hospitalization history.
The collection, processing, and disclosure of personal data (such as patient demographic, health, and medical information) are subject to compliance with Regulation (EU) 2016/679, also referred to as the General Data Protection Regulation, and the Belgian Law on the protection of natural persons regarding the processing of personal data. Therefore, combinations of data deemed to be identificatory to specific persons cannot be disclosed. For instance, details of the vaccination record of an individual decedent cannot be reported.
The narrowing of the time between diagnosis and death to ∼14 days led to the inclusion of a greater proportion of patients from a ward (41 or 91%) than from an ICU (4 or 9%) compared to cohort-I (respectively 46% and 54%).
Clinical parameters
Comorbidities were categorized in accordance with international recommendations. Overweight is as a body mass index (BMI) >25 kg/m2, and obesity as a BMI ≥30 kg/m2. Presence of diabetes mellitus type 2 includes previously known and newly diagnosed patients, based on Hb1Ac ≥6.5% or active treatment on admission. Former smokers, defined as having ceased smoking >6 months prior to inclusion, are not considered smokers in Figures 1B and S1. Hypertension is defined as grade 1 hypertension, or treatment with antihypertensive drugs. Chronic kidney disease is defined as the presence of kidney damage or a glomerular filtration rate of <60 mL/min/1.73 m2 for >3 months. Chronic lung disease includes obstructive lung disease (chronic obstructive pulmonary disease, asthma), interstitial lung disease, pulmonary fibrosis, and pulmonary hypertension. Cardiovascular disease comprises heart conditions (such as valvular disease, heart failure, arrhythmias, cardiomyopathies, coronary artery disease), cerebrovascular antecedents, and history of pulmonary embolism. Neurodegenerative disease comprises all forms of dementia and progressive cognitive impairment.
Patients were considered immunocompromised if one of the following criteria was met: (1) an active oncological condition, defined as presence of a solid tumor or hematologic malignancy <6 months prior to inclusion; (2) immunosuppressive drugs as maintenance therapy, including corticosteroids and chemotherapy; (3) recipient of a solid organ transplant.
The cause of death of COVID-19 patients was classified as one out of two categories. (1) Death from COVID-19: hypoxic respiratory failure secondary to SARS-CoV-2 pneumonia or death from COVID-19 sequelae such as acute respiratory distress syndrome, fungal or bacterial superinfection associated with SARS-CoV-2 pneumonia. (2) Death with COVID-19: cause of death not directly related to COVID-19 such as acute cardiac arrest, cerebrovascular accidents, or deterioration of an oncological condition.
Cohort-II patients were included between August 2021 and May 2022. During this period, "vaccinated" was defined as having received, at least two weeks earlier, either two doses of tozinameran/Comirnaty (Pfizer-BioNTech), two doses of elasomeran/Spikevax (Moderna), two doses of ChAdOx1-S/Vaxzevria (AstraZeneca), or a single dose of Ad26.COV2-S/Janssen Jcovden (Johnson & Johnson). Several patients received a booster in the fall of 2021 and some a second booster. The term "vaccinated" in this study encompasses all these vaccination histories. Cohort-I cases died between May 2020 and April 2021, at a time when COVID-19 vaccines did not exist, or the Belgian vaccination campaign had just been rolled out. Only one case of cohort-I, COVID #68, was vaccinated. The definition of "vaccinated" will evolve in the future and the definition of "breakthrough infection" will have to be adapted accordingly. Breakthrough infections occur frequently, and some of the patients who die during the course of a breakthrough infection succumb to the SARS-CoV-2 infection as opposed to passing away with it. The term "fatal breakthrough infection" (FBI) that we here coin is pragmatic and concise, and refers to patients who, to the best of the clinical judgment of the physicians who treated or reviewed the patient files, died from COVID-19 instead of with COVID-19 despite being vaccinated. The term FBI does not imply any underlying cause such as the absence of anti-S IgG in serum on admission or postmortem. In the foreseeable future, unvaccinated patients will no longer represent typical COVID-19 patients.
Method details
CSF sample extraction
The first step of the extended postmortem bedside surgical procedure is to extract a CSF sample from the cerebellomedullary cistern, also known as the cisterna magna. The cisterna magna is located between the medulla oblongata and the cerebellum in the posterior fossa. To achieve optimal exposure for a cisternal puncture, the decedent was positioned in a supine, anti-Trendelenburg position or turned sideways, depending on the physiognomy of the body. The neck of the patient was positioned in flexion, the occiput was palpated downwards until encountering the spine of the axis vertebra and a spinal needle (22 Gauge, 90 mm) was inserted on the midline in the depression between these two points. The needle was directed upwards toward the midpoint of an imaginary line joining the left and right external auditory meatus, until the needle was felt to pierce the atlanto-occipital ligament, thereby entering the cisterna magna. The needle hub was then inspected meticulously to ascertain that the liquid is clear and colorless, before continuing to sample a syringe of 5 mL CSF. In case of a traumatic puncture (revealed as the presence of blood in the needle hub), the procedure was repeated from the beginning.
CSF samples were sent to the National Reference Center for Respiratory Pathogens at the Department of Laboratory Medicine of the University Hospitals Leuven (Leuven, Belgium) for PCR analysis and to the BSL-3 facility of the KU Leuven Rega Institute (Leuven, Belgium) for viral culture.
Blood sample drawing
The second step of the extended postmortem bedside surgical procedure is to draw a blood sample via a femoral puncture or, when available, via an existing central venous line. For the femoral puncture, an 18 Gauge needle was inserted ∼2 cm below the midpoint between the anterior superior iliac spine and the pubic symphysis. Whole blood was collected in 5 mL Serum Separator Tubes (BD Vacutainer). After centrifugation, the serum was stored in sterile tubes and serum samples were sent to the National Reference Center for Respiratory Pathogens at the Department of Laboratory Medicine of the University Hospitals Leuven, (Leuven, Belgium) for qRT-PCR analysis.
Nasopharyngeal swabs
The third step of the extended postmortem bedside surgical procedure entails the initiation of the endoscopic endonasal procedure, using a 4 mm 0° endoscope connected with a camera and monitor and a light source (Karl Storz). Prior to cutting and tissue sample harvesting, three nasopharyngeal swabs were taken under endoscopic guidance. A first nasopharyngeal swab was used immediately for a bedside rapid antigen test, using the Panbio Abbott COVID-19 Rapid Test Device (Abbott, REF#41FK10) against SARS-CoV-2 nucleocapsid antigen. A second nasopharyngeal swab was for TaqPath COVID-19 qRT-PCR and whole-genome sequencing at the Belgian National Reference Center for Respiratory Pathogens at the Department of Laboratory Medicine of the University Hospitals Leuven (Leuven, Belgium). A third nasopharyngeal swab was sent to the BSL-3 facility of the KU Leuven Rega Institute (Leuven, Belgium) for viral culture.
Tissue sample harvesting
The fourth and final step of the extended postmortem bedside surgical procedure consists of two parts: an intranasal and an intracranial part.
Intranasal tissue samples of the respiratory mucosa and olfactory cleft mucosa were collected via a classical endoscopic endonasal procedure with cold instruments. The procedure was described in detail in Khan et al.16 Briefly, to harvest respiratory mucosa, we removed the inferior turbinate and the middle turbinate after cutting their attachments to the lateral nasal wall and nasal roof with Heymann and endoscopic scissors. To harvest olfactory cleft mucosa, an elliptical incision with a sickle or beaver knife was made running over the superior part of the septum, the cribriform plate, and the area of the vertical attachment of the superior turbinate, thus covering the full extent of the olfactory cleft region. A subperiosteal dissection was initiated on the medial side (superior part of the septum) and lateral side (vertical attachment of the turbinates), progressively extending to the center (cribriform plate), where the mucosa was attached only by the remaining fila olfactoria. After transection and tearing of the fila olfactoria, the mucosa was harvested in one or a few pieces.
Intracranial samples of the olfactory bulbs and adjacent brain regions of the frontal lobe were collected via an endoscopic endonasal transcribriform approach. The procedure was described in detail in Khan et al.,16 see also Movie S1 in that paper. Briefly, after opening the cribriform plate with a hammer and chisel to avoid aerosol formation, the dura mater was exposed and incised longitudinally with the sickle knife. The olfactory bulbs were then dissected from the surrounding tissue (such as arachnoidea and blood vessels) and removed in an atraumatic way (blunt dissection with a ball probe) and cut posteriorly from the olfactory tract. Several biopsies of the adjacent brain regions were taken with an upbiting forceps and fixed in 10% formalin for cryopreservation and paraffin embedding. Brain samples were taken from the gyrus rectus or gyri orbitales from the frontal lobe and contained macroscopically gray and white matter.
At each participating hospital, procedures were carried out by a team of two ear, nose, and throat surgeons or trainees specialized in endoscopic sinus surgery, wearing personal protective equipment: gown, gloves, and powered air purifying respirator.
Nasopharyngeal swabs
A dedicated nasopharyngeal swab was taken preprocedurally and stored at −80°C. Later qRT-PCR analysis was performed by the National Reference Center for Respiratory Pathogens at the Department of Laboratory Medicine, University Hospitals Leuven (Leuven, Belgium). Viral RNA extraction was performed with the MagMAX Viral/Pathogen II kit (Thermo Fisher Scientific, Cat#A48383) on a KingFisher Flex System, followed by qRT-PCR with the TaqPath COVID-19 qRT-PCR kit (Thermo Fisher Scientific, Cat#A48067) on a QuantStudio 7 Flex platform (Thermo Fisher Scientific).
Determination of variants of concern
A combination of methods was used to determine the variant of concern based on RNA extracted from postmortem samples or viral cultures. 1) Whole-genome sequencing of RNA extracted from a postmortem nasopharyngeal swab by the Laboratory of Clinical and Epidemiological Virology of the KU Leuven Rega Institute (Leuven, Belgium). 2) Whole-genome sequencing of RNA extracted from a piece of formalin-fixed tissue of the inferior turbinate by GenXPro (Frankfurt, Germany). 3) Whole-genome sequencing of RNA extracted from an apical wash of HAEC-ALI cultures by GenXPro (Frankfurt, Germany). 4) Variant-specific qRT-PCR assays VoXcreen-DO, VoXcreen-BA.1-2, and VoXcreen-BA.2-4-5 on RNA extracted from a piece of formalin-fixed tissue of the inferior turbinate by GenXPro (Frankfurt, Germany). We were unable to determine the variant of concern for COVID #113 and #115. Both patients had a negative rapid antigen test result on a postmortem nasopharyngeal swab. As these cases date from well within the dominance period of Omicron in Belgium, COVID #113 and #115 are grouped in the Omicron subcohort in Figure 2.
Serology
Antibody serum titers were measured in the Department of Laboratory Medicine, University Hospitals Leuven (Leuven, Belgium) on the Abbott Architect platform with the SARS-CoV-2 IgG (anti-N) and IgG II Quant (anti-S) assays using the manufacturer’s cut-offs for positivity of 1.4 S/CO and 50 AU/mL, respectively. Serology for anti-N IgG is scored as positive vs. negative and detection of anti-N IgG is therefore referred to as "seroconversion".
Sample processing
Tissue samples from the 45 COVID-19 cases and 8 control cases were transferred into containers with 10% neutral buffered formalin (Sigma-Aldrich, Cat#HT5011) for >72 h to fix the tissues and inactivate SARS-CoV-2. Samples were treated for cryoprotection by immersing serially in 15%, 25%, and 30% sucrose (Sigma-Aldrich, Cat#S0389-1KG) in 1 x PBS over a period of 9–14 days. The orientation of the samples was recorded before embedding in Tissue-Tek O.C.T. compound (Sakura, Cat#4583) on dry ice. Cryosections of 6–8 μm thickness were cut on a Leica CM3050 S cryostat and collected on SuperFrost Plus Gold slides (Thermo Fisher Scientific/Menzel Gläser, Cat#K5800AMNZ72) or Superfrost Plus Micro slides (VWR, Cat#48311-703). Slides were air-dried at room temperature, and boxes of slides were sealed prior to storage at −80°C. In parallel, tissue samples of olfactory bulb and frontal lobe were processed for paraffin embedding using a fully automated platform, 5 μm paraffin-embedded sections were air-dried and stored at 4°C.
RNAscope in situ hybridization
The fluorescence RNAscope platform was used to visualize viral RNA in the 45 COVID-19 cases and the 8 control cases. Most slides contained multiple sections. Staining was performed with the RNAscope manual assay using the Multiplex Fluorescent Detection Kit v2 (Advanced Cell Diagnostics, Cat#323110) according to manufacturer’s protocols. Briefly, slides were dried at 55°C overnight, then pretreated with hydrogen peroxide, followed by permeabilization in target retrieval reagent (Advanced Cell Diagnostics, Cat#322000) for 3 min in a steamer, and digestion with Protease III (Advanced Cell Diagnostics, Cat#322337) at 40°C for 15 min for cryosections and with Protease Plus (Advanced Cell Diagnostics, Cat#322330) at 40°C for 20 min for paraffin embedded sections. A combination of probes for target RNA detection was hybridized at 40°C for 2 h. Probes in the C4 channel were developed with the RNAscope 4-Plex Ancillary Kit (Advanced Cell Diagnostics, Cat#323120). Signal amplification was followed by development of appropriate HRP channels with dyes Opal 520 (Akoya Biosciences, Cat#FP1487001KT), Opal 570 (Akoya Biosciences, Cat#FP1488001KT), and Opal 690 (Akoya Biosciences, Cat#FP1497001KT). DAPI (Thermo Fisher Scientific, Cat#D1306) served as nuclear stain. Slides were mounted in Mount Solid antifade (abberior, Cat#MM-2011-2X15ML). Confocal images were taken with the Zeiss ZEN 2.6 system on a Zeiss LSM 800.
Immunohistochemistry
For codetection of RNA and protein, IHC was performed after the final step of HRP blocker application in the RNAscope Multiplex Fluorescent Detection protocol. Slides were blocked in 10% donkey serum (Sigma-Aldrich, Cat#S30-100ML) in 0.1% Triton/PBS at room temperature for 1 h. The following primary antibodies were diluted in 2% donkey serum in 0.1% Triton/PBS and incubated at 4°C overnight: Cytokeratin 8 (Novus Biologicals, Cat#NBP2-67468) at 1:300, MUC5AC (Thermo Fisher Scientific, Cat#MA5-12178) at 1:400, SARS-CoV-2 Nucleocapsid (Sino Biological, Cat#40143-R001) at 1:100, Somatostatin receptor subtype 2A/SSTR2A (Biotrend, Cat#NB-49-016-50ul) at 1:4000, TuJ1/TUBB3 (BioLegend, Cat#801202) at 1:100 for olfactory mucosa sections and 1:400 for olfactory bulb sections, GFAP (Novus Biologicals, Cat#NB300-141) at 1:600, NGFR p75 (Sigma-Aldrich, Cat#N5408) at 1:200, S100B (Enzo Life Sciences, Cat#ENZ-ABS307) at 1:300, Aquaporin 4 (Millipore, Cat#AB3594) at 1:100, CD31 (Abcam, Cat#ab28364) at 1:50, ZO-1 tight junction protein (Novus Biologicals, Cat#NBP1-85046) at 1:100 and OMP (FUJIFILM Wako Shibayagi, Cat#544-10001-WAKO) at 1:500. Slides were then washed in 0.1% Triton/PBS 3 × 5 min each followed by incubation with appropriate secondary antibodies at 1:500 in 2% normal donkey serum in 0.1% Triton/PBS at room temperature for 1 h. Secondary antibodies were Alexa Fluor Plus 488 donkey anti-rabbit (Thermo Fisher Scientific, Cat#A32790), Alexa Fluor Plus 555 donkey anti-rabbit (Thermo Fisher Scientific, Cat#A32794), Alexa Fluor Plus 555 donkey anti-mouse (Thermo Fisher Scientific, Cat#A32773), Alexa Fluor Plus 555 donkey anti-goat (Thermo Fisher Scientific, Cat#A32816), Alexa Fluor Plus 647 donkey anti-rabbit (Thermo Fisher Scientific, Cat#A32795), Alexa Fluor Plus 647 donkey anti-mouse (Thermo Fisher Scientific, Cat#A32787), and Alexa Fluor Plus 647 donkey anti-goat (Thermo Fisher Scientific, Cat#A32849). Slides were washed in 0.1% Triton/PBS 3 × 5 min each followed by DAPI (Thermo Fisher Scientific, Cat#D1306) application for nuclei staining. Slides were mounted in Mount Solid antifade (abberior, Cat#MM-2011-2X15ML). For IHC only, slides were pretreated in target retrieval reagent (Advanced Cell Diagnostics, Cat#322000) for 3 min in a steamer. Primary antibody application, secondary antibody detection, DAPI staining, and mounting were performed as above. Confocal images were taken with the Zeiss ZEN 2.6 system on a Zeiss LSM 800.
SARS-CoV-2 growth in culture
Human airway epithelial cells (HAEC) from healthy donors were of bronchial origin in Figures 7B and 7C (Epithelix, Cat#EP01MD) and Figure 8E (developed in-house), and of nasal origin (Epithelix, Cat#EP02MP) in Figure 8D. After arrival in the laboratory, the air-liquid interface (ALI) inserts were washed with and maintained in pre-warmed MucilAir medium (Epithelix, Cat#EP04MM) at 37°C and 5% CO2 for at least four days before use. In case of in-house developed inserts, six-week differentiated inserts in the ALI system were used that showed proper cilia beating and transepithelial electrical resistance measurements. Potential infectious particles from fresh or frozen nasopharyngeal swabs were extracted in 1 mL of MucilAir medium. Viral inocula were prepared in either MucilAir medium or control CSF sample to a final dilution of 5×103 TCID50/mL. On the day of the experiment, the HAEC-ALI cultures were exposed to 200 μL of viral inocula (equal to a multiplicity of infection of 103 TCID50/insert) or 200 μL pure CSF samples or nasopharyngeal swab solutions or fresh media at 35°C for 3 h. The apical sides of the HAEC-ALI cultures were washed with assay medium at indicated days, RNA was extracted from the wash fluids using the NucleoSpin RNA virus kit (Macherey-Nagel, Cat#740956.50), and SARS-CoV-2 RNA was quantified by qRT-PCR using the iTaq universal probes one-step kit (Bio-Rad, Cat#1725141) with a commercial mix of primers for the N gene (IDT Technologies, Cat#10006606) on a LightCycler 96 platform (Roche). The SARS-CoV-2 RNA concentrations are expressed as copies/mL in the wash fluid of the HAEC-ALI cultures by conversion from the Ct values based on a standard curve of DNA in water. The Lower-Limit-of-Quantification (LLOQ) was determined based on a dilution series of a SARS-CoV-2 virus stock treated in the same way as the samples; the LLOQ is the lowest SARS-CoV-2 concentration that was still within the linear range of this serial dilution. In Figure 8D, the inserts inoculated with the nasopharyngeal swab from COVID #108 showed fungal infection on day 10 and were not further analyzed; one insert inoculated with the nasopharyngeal swab from COVID #103 showed fungal infection on day 2 and was not further analyzed. In Figures 8C–8E, samples were tested in duplicate inserts; each replicate and the average are shown.
Acknowledgments
We thank the nurses, doctors, and paramedical staff for their support and cooperation; Piet Maes and Björn Rötter for help with variant determination; Arthur Goby, Peter Leemans, Ling Li, Florian Lindenblatt, and Edisa Turkovic for technical support; Patrick Meeze for drawings; and Thomas Boehm and Simon Feys for comments on a draft. T.N.D.D. received a fellowship from EU Horizon 2020 grant OrganoVIR “Organoids for Virus Research - An innovative training - ITN programme.” C.V. and L.V.G. were supported by FWO Senior Clinical Investigator Fellowships 1803923N and 18B2222N, respectively. J.N. was supported by Covid-19-Fund KU Leuven/UZ Leuven and the COVID-19 call of FWO (G0G4820N). D.R.T. received support from FWO (G0F8516N, G065721N), Stichting Alzheimer Onderzoek (SAO-FRA Belgium, 2020/017), and KU Leuven Internal Funding (C14/17/107; C14/22/132). P. Mombaerts received financial support from the Max Planck Society.
Author contributions
M.K., P. Mombaerts, and L.V.G. conceived, designed, and supervised the project. M. Clijsters, W.B., M. Claerhout, F.C., L.V.B., F.B., K.S., S.K., J.V.L., G.V., L.L., Y.D., P.D.M., J.G., M.J., K.L., P. Meersseman, D.R.T., V.T., C.V., J.W., and L.V.G. collected samples and/or clinical data. M.K., S.C., A.S.D., S.-J.Y., and H.Z. generated RNAscope and/or IHC data. M.K. generated confocal images and made the figures. T.N.D.D., D.J., and J.N. performed HAEC-ALI experiments. P. Mombaerts wrote the original draft of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: November 10, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.neuron.2022.11.007.
Supplemental information
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamers M.M., Haagmans B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022;20:270–284. doi: 10.1038/s41579-022-00713-0. [DOI] [PubMed] [Google Scholar]
- 3.Jonigk D., Werlein C., Acker T., Aepfelbacher M., Amann K.U., Baretton G., Barth P., Bohle R.M., Büttner A., Büttner R., et al. Organ manifestations of COVID-19: what have we learned so far (not only) from autopsies? Virchows Arch. 2022;481:139–159. doi: 10.1007/s00428-022-03319-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merad M., Blish C.A., Sallusto F., Iwasaki A. The immunology and immunopathology of COVID-19. Science. 2022;375:1122–1127. doi: 10.1126/science.abm8108. [DOI] [PubMed] [Google Scholar]
- 5.Dimitriadis K., Meis J., Neugebauer H., Barlinn K., Neumann B., Gahn G., Lochner P., Knier B., Lindemann S., Sühs K.W., et al. Neurologic manifestations of COVID-19 in critically ill patients: results of the prospective multicenter registry PANDEMIC. Crit. Care. 2022;26:217. doi: 10.1186/s13054-022-04080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meinhardt J., Radke J., Dittmayer C., Franz J., Thomas C., Mothes R., Laue M., Schneider J., Brünink S., Greuel S., et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2021;24:168–175. doi: 10.1038/s41593-020-00758-5. [DOI] [PubMed] [Google Scholar]
- 7.Song E., Zhang C., Israelow B., Lu-Culligan A., Prado A.V., Skriabine S., Lu P., Weizman O.E., Liu F., Dai Y., et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021;218:e20202135. doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer L., Laksono B.M., de Vrij F.M.S., Kushner S.A., Harschnitz O., van Riel D. The neuroinvasiveness, neurotropism, and neurovirulence of SARS-CoV-2. Trends Neurosci. 2022;45:358–368. doi: 10.1016/j.tins.2022.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Killingley B., Mann A.J., Kalinova M., Boyers A., Goonawardane N., Zhou J., Lindsell K., Hare S.S., Brown J., Frise R., et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat. Med. 2022;28:1031–1041. doi: 10.1038/s41591-022-01780-9. [DOI] [PubMed] [Google Scholar]
- 10.Cooper K.W., Brann D.H., Farruggia M.C., Bhutani S., Pellegrino R., Tsukahara T., Weinreb C., Joseph P.V., Larson E.D., Parma V., et al. COVID-19 and the chemical senses: supporting players take center stage. Neuron. 2020;107:219–233. doi: 10.1016/j.neuron.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coelho D.H., Reiter E.R., French E., Costanzo R.M. Decreasing incidence of chemosensory changes by COVID-19 variant. Otolaryngol. Head Neck Surg. 2022 doi: 10.1177/01945998221097656. 019459982210976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehgani-Mobaraki P., Patel Z., Zaidi A.K., Giannandrea D., Hopkins C. The Omicron variant of SARS-CoV-2 and its effect on the olfactory system. Int. Forum Allergy Rhinol. 2022 doi: 10.1002/alr.23089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menni C., Valdes A.M., Polidori L., Antonelli M., Penamakuri S., Nogal A., Louca P., May A., Figueiredo J.C., Hu C., et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet. 2022;399:1618–1624. doi: 10.1016/S0140-6736(22)00327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butowt R., Meunier N., Bryche B., von Bartheld C.S. The olfactory nerve is not a likely route to brain infection in COVID-19: a critical review of data from humans and animal models. Acta Neuropathol. 2021;141:809–822. doi: 10.1007/s00401-021-02314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kassam A., Snyderman C.H., Mintz A., Gardner P., Carrau R.L. Expanded endonasal approach: the rostrocaudal axis. Part I. Crista galli to the sella turcica. Neurosurg. Focus. 2005;19:E3. doi: 10.3171/foc.2005.19.1.4. [DOI] [PubMed] [Google Scholar]
- 16.Khan M., Yoo S.J., Clijsters M., Backaert W., Vanstapel A., Speleman K., Lietaer C., Choi S., Hether T.D., Marcelis L., et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell. 2021;184:5932–5949.e15. doi: 10.1016/j.cell.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F., Flanagan J., Su N., Wang L.C., Bui S., Nielson A., Wu X., Vo H.T., Ma X.J., Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Offit P.A. Covid-19 boosters - where from here? N. Engl. J. Med. 2022;386:1661–1662. doi: 10.1056/NEJMe2203329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quandt J., Muik A., Salisch N., Lui B.G., Lutz S., Krüger K., Wallisch A.K., Adams-Quack P., Bacher M., Finlayson A., et al. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci. Immunol. 2022;7:eabq2427. doi: 10.1126/sciimmunol.abq2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fung T.S., Liu D.X. Similarities and dissimilarities of COVID-19 and other coronavirus diseases. Annu. Rev. Microbiol. 2021;75:19–47. doi: 10.1146/annurev-micro-110520-023212. [DOI] [PubMed] [Google Scholar]
- 22.Doucette J.R. The glial cells in the nerve fiber layer of the rat olfactory bulb. Anat. Rec. 1984;210:385–391. doi: 10.1002/ar.1092100214. [DOI] [PubMed] [Google Scholar]
- 23.Barnett S.C., Alexander C.L., Iwashita Y., Gilson J.M., Crowther J., Clark L., Dunn L.T., Papanastassiou V., Kennedy P.G., Franklin R.J. Identification of a human olfactory ensheathing cell that can effect transplant-mediated remyelination of demyelinated CNS axons. Brain. 2000;123:1581–1588. doi: 10.1093/brain/123.8.1581. [DOI] [PubMed] [Google Scholar]
- 24.Choi D., Law S., Raisman G., Li D. Olfactory ensheathing cells in the nasal mucosa of the rat and human. Br. J. Neurosurg. 2008;22:301–302. doi: 10.1080/02688690701883442. [DOI] [PubMed] [Google Scholar]
- 25.Field P.M., Li Y., Raisman G. Ensheathment of the olfactory nerves in the adult rat. J. Neurocytol. 2003;32:317–324. doi: 10.1023/B:NEUR.0000010089.37032.48. [DOI] [PubMed] [Google Scholar]
- 26.Herrera L.P., Casas C.E., Bates M.L., Guest J.D. Ultrastructural study of the primary olfactory pathway in Macaca fascicularis. J. Comp. Neurol. 2005;488:427–441. doi: 10.1002/cne.20588. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Field P.M., Raisman G. Olfactory ensheathing cells and olfactory nerve fibroblasts maintain continuous open channels for regrowth of olfactory nerve fibres. Glia. 2005;52:245–251. doi: 10.1002/glia.20241. [DOI] [PubMed] [Google Scholar]
- 28.Kawaja M.D., Boyd J.G., Smithson L.J., Jahed A., Doucette R. Technical strategies to isolate olfactory ensheathing cells for intraspinal implantation. J. Neurotrauma. 2009;26:155–177. doi: 10.1089/neu.2008.0709. [DOI] [PubMed] [Google Scholar]
- 29.Smithson L.J., Kawaja M.D. A comparative examination of biomarkers for olfactory ensheathing cells in cats and guinea pigs. Brain Res. 2009;1284:41–53. doi: 10.1016/j.brainres.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Smithson L.J., Kawaja M.D. Microglial/macrophage cells in mammalian olfactory nerve fascicles. J. Neurosci. Res. 2010;88:858–865. doi: 10.1002/jnr.22254. [DOI] [PubMed] [Google Scholar]
- 31.Crespo C., Liberia T., Blasco-Ibáñez J.M., Nácher J., Varea E. Cranial pair I: the olfactory nerve. Anat. Rec. 2019;302:405–427. doi: 10.1002/ar.23816. [DOI] [PubMed] [Google Scholar]
- 32.Holbrook E.H., Rebeiz L., Schwob J.E. Office-based olfactory mucosa biopsies. Int. Forum Allergy Rhinol. 2016;6:646–653. doi: 10.1002/alr.21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oprych K., Cotfas D., Choi D. Common olfactory ensheathing glial markers in the developing human olfactory system. Brain Struct. Funct. 2017;222:1877–1895. doi: 10.1007/s00429-016-1313-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holbrook E.H., Leopold D.A., Schwob J.E. Abnormalities of axon growth in human olfactory mucosa. Laryngoscope. 2005;115:2144–2154. doi: 10.1097/01.MLG.0000181493.83661.CE. [DOI] [PubMed] [Google Scholar]
- 35.Holbrook E.H., Wu E., Curry W.T., Lin D.T., Schwob J.E. Immunohistochemical characterization of human olfactory tissue. Laryngoscope. 2011;121:1687–1701. doi: 10.1002/lary.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Virchow R. Ueber die Erweiterung kleinerer Gefässe. Archiv f. pathol. Anat. 1851;3:427–462. doi: 10.1007/BF01960918. [DOI] [Google Scholar]
- 37.Robin C. Recherches sur quelques particularités de la structure des capillaires de l'encéphale. J. Physiol. Homme Anim. 1859;2:537–548. [Google Scholar]
- 38.Ineichen B.V., Okar S.V., Proulx S.T., Engelhardt B., Lassmann H., Reich D.S. Perivascular spaces and their role in neuroinflammation. Neuron. 2022;110:3566–3581. doi: 10.1016/j.neuron.2022.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Do T.N.D., Donckers K., Vangeel L., Chatterjee A.K., Gallay P.A., Bobardt M.D., Bilello J.P., Cihlar T., De Jonghe S., Neyts J., Jochmans D. A robust SARS-CoV-2 replication model in primary human epithelial cells at the air liquid interface to assess antiviral agents. Antiviral Res. 2021;192:105122. doi: 10.1016/j.antiviral.2021.105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Do T.N.D., Claes S., Schols D., Neyts J., Jochmans D. SARS-CoV-2 virion infectivity and cytokine production in primary human airway epithelial cells. Viruses. 2022;14:951. doi: 10.3390/v14050951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdelnabi R., Foo C.S., Jochmans D., Vangeel L., De Jonghe S., Augustijns P., Mols R., Weynand B., Wattanakul T., Hoglund R.M., et al. The oral protease inhibitor (PF-07321332) protects Syrian hamsters against infection with SARS-CoV-2 variants of concern. Nat. Commun. 2022;13:719. doi: 10.1038/s41467-022-28354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Järhult J.D., Hultström M., Bergqvist A., Frithiof R., Lipcsey M. The impact of viremia on organ failure, biomarkers and mortality in a Swedish cohort of critically ill COVID-19 patients. Sci. Rep. 2021;11:7163. doi: 10.1038/s41598-021-86500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Riel D., Embregts C.W.E., Sips G.J., van den Akker J.P.C., Endeman H., van Nood E., Raadsen M., Bauer L., van Kampen J., Molenkamp R., et al. Temporal kinetics of RNAemia and associated systemic cytokines in hospitalized COVID-19 patients. mSphere. 2021;6:e0031121. doi: 10.1128/mSphere.00311-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang A.C., Kern F., Losada P.M., Agam M.R., Maat C.A., Schmartz G.P., Fehlmann T., Stein J.A., Schaum N., Lee D.P., et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature. 2021;595:565–571. doi: 10.1038/s41586-021-03710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aschman T., Mothes R., Heppner F.L., Radbruch H. What SARS-CoV-2 does to our brains. Immunity. 2022;55:1159–1172. doi: 10.1016/j.immuni.2022.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krasemann S., Dittmayer C., von Stillfried S., Meinhardt J., Heinrich F., Hartmann K., Pfefferle S., Thies E., von Manitius R., Aschman T.A.D., et al. Assessing and improving the validity of COVID-19 autopsy studies - A multicentre approach to establish essential standards for immunohistochemical and ultrastructural analyses. eBioMedicine. 2022;83:104193. doi: 10.1016/j.ebiom.2022.104193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., et al. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 48.Rasmussen M.K., Mestre H., Nedergaard M. Fluid transport in the brain. Physiol. Rev. 2022;102:1025–1151. doi: 10.1152/physrev.00031.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lochhead J.J., Thorne R.G. Intranasal delivery of biologics to the central nervous system. Adv. Drug Deliv. Rev. 2012;64:614–628. doi: 10.1016/j.addr.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Ampie L., McGavern D.B. Immunological defense of CNS barriers against infections. Immunity. 2022;55:781–799. doi: 10.1016/j.immuni.2022.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koyuncu O.O., Hogue I.B., Enquist L.W. Virus infections in the nervous system. Cell Host Microbe. 2013;13:379–393. doi: 10.1016/j.chom.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrano G.E., Walker J.E., Tremblay C., Piras I.S., Huentelman M.J., Belden C.M., Goldfarb D., Shprecher D., Atri A., Adler C.H., et al. SARS-CoV-2 brain regional detection, histopathology, gene expression, and immunomodulatory changes in decedents with COVID-19. J. Neuropathol. Exp. Neurol. 2022;81:666–695. doi: 10.1093/jnen/nlac056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hablitz L.M., Nedergaard M. The glymphatic system. Curr. Biol. 2021;31:R1371–R1375. doi: 10.1016/j.cub.2021.08.026. [DOI] [PubMed] [Google Scholar]
- 54.Da Mesquita S., Fu Z., Kipnis J. The meningeal lymphatic system: a new player in neurophysiology. Neuron. 2018;100:375–388. doi: 10.1016/j.neuron.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sweeney M.D., Kisler K., Montagne A., Toga A.W., Zlokovic B.V. The role of brain vasculature in neurodegenerative disorders. Nat. Neurosci. 2018;21:1318–1331. doi: 10.1038/s41593-018-0234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monje M., Iwasaki A. The neurobiology of long COVID. Neuron. 2022;110:3484–3496. doi: 10.1016/j.neuron.2022.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.