Significance
TP53 mutations drive tumor development but require additional cooperating lesions to do so. Our omics analyses in a sporadic breast-cancer model driven by a somatic Trp53R245W mutation catalogue a list of potential cooperative driver mutations. In particular, we identify a Pip5k1cG67E mutation in the majority of tumors that stabilizes the Pip5k1c protein and activates Pi3k/Akt/mTOR signaling. A homolog of PIP5K1C, PIP5K1A is frequently coamplified with PI4KB, activating PI3K/AKT/mTOR in breast cancer. Our findings support the cooperation of somatic TP53 mutations with PI3K/AKT/mTOR signaling in driving breast tumorigenesis and suggest possible combination therapies to target these cancers.
Keywords: p53, breast cancer, Pip5k1c, genomics
Abstract
Alterations of the tumor suppressor TP53, one of the most common events in cancer, alone are insufficient for tumor development but serve as drivers of transformation. We sought to identify cooperating events through genomic analyses of a somatic Trp53R245W mouse model (equivalent to the TP53R248W hot spot mutation in human cancers) that recapitulates metastatic breast–cancer development. We identified cooperating lesions similar to those found in human breast cancers. Moreover, we identified activation of the Pi3k/Akt/mTOR pathway in most tumors via mutations in Pten, Erbb2, Kras, and/or a recurrent Pip5k1c mutation that stabilizes the Pip5k1c protein and activates Pi3k/Akt/mTOR signaling. Another PIP5K1C family member, PIP5K1A, is coamplified with PI4KB in 18% of human breast cancer patients; both encode kinases that are responsible for production of the PI3K substrate, phosphatidylinositol 4,5-bisphosphate. Thus, the TP53R248W mutation and PI3K/AKT/mTOR signaling are major cooperative events driving breast-cancer development. Additionally, a combination of two US Food and Drug Administration (FDA)-approved drugs, tigecycline and metformin, which target oxidative phosphorylation downstream of PI3K signaling, inhibited tumor cell growth and may be repurposed for breast-cancer treatment. These findings advance our understanding of how mutant p53 drives breast-tumor development and pinpoint the importance of PI3K/AKT/mTOR signaling, expanding combination therapies for breast-cancer treatment.
Breast cancer is the most commonly diagnosed cancer in women (1). Genomic alterations are generally recognized as contributing to breast malignancy (2). Somatic missense mutations in the TP53 gene are among the most-frequent genomic alterations in human breast cancer (3, 4). Specifically, arginine-to-tryptophan/glutamine mutations at codon 248 (TP53R248W/Q) and arginine-to-histidine mutation at codon 175 (TP53R175H) are “hotspot” mutations that occur more frequently than other TP53 mutations in breast cancer (5, 6). Recently, our laboratory generated the first sporadic breast cancer mouse models that allow mutating p53 specifically in mammary epithelia while maintaining a wild-type (WT) stroma and immune system (7), thereby faithfully mimicking the somatic TP53 mutations in human breast cancer. Using these models, we demonstrated that somatic Trp53R245W/+ mice (corresponding to TP53R248W/+ in humans) required long latency (∼20 mo) to develop spontaneous breast tumors, while the somatic Trp53R172H/+ mice (corresponding to TP53R175H/+ in human) required additional insults, e.g., radiation or loss of the WT Trp53 allele through Cre/loxP-mediated deletion (Trp53R172H/flx) for tumor formation (7). Regional whole-exome sequencing of two Trp53R245W/+ primary breast tumors and their corresponding lung metastases not only identified genomic heterogeneity of these tumors but also demonstrated early dissemination of metastases, providing experimental validation of human studies (7–9). Collectively, these data support our mouse models as a valuable tool for human breast cancer studies and suggest that additional cooperative events are needed for somatic Trp53 mutations to facilitate tumorigenesis.
The PI3K/AKT/mTOR pathway is a critical signaling transduction system that regulates multiple aspects of cellular metabolism and growth and is one of the most-activated pathways in human cancers (10, 11). The major genomic alternations activating PI3K/AKT/mTOR pathway in human breast cancer are PIK3CA mutations or PTEN mutations/deletions (12). PIK3CA, mutated in >30% of breast cancer, encodes p110α protein, which is the catalytic subunit of PI3K that phosphorylates phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) to produce phosphatidylinositol-3,4,5-trisphosphate (PIP3), which further activates the downstream AKT/mTOR signaling (10). PTEN dephosphorylates PIP3 to prevent the activation of this pathway, and PTEN is lost in ∼10% of breast cancer (13). Pik3ca mutations or Pten deletions have been shown to cooperate with p53 alterations to drive breast tumorigenesis in mouse models (14, 15). Although, we also identified a Pten mutation in one of the two somatic Trp53R245W/+ breast tumors sequenced (7), whether and how Pi3k/Akt/mTOR signaling is activated in our somatic mutant Trp53-driven breast cancer model is not clearly understood.
Metabolic reprogramming, featured as upregulated glycolysis, represents a potential vulnerability for targeted therapies (16, 17). Metabolic heterogeneity, however, poses a significant barrier to developing such therapies (18, 19). Oxidative phosphorylation (OXPHOS), which has been falsely assumed to be downregulated in cancers, is still active and serves as the major energy source for most cancer cells (19, 20). Consistent with this paradox, PI3K/AKT/mTOR signaling has diverse effects on cellular energetics via regulating nutrient sensing, metabolic enzymes, and mitochondrial dynamics (21, 22). PI3K/AKT activates glycolysis to supply energy and metabolites to fuel the growth and expansion of cancer cells (16, 17), whereas mTOR coordinates mitochondrial activity and biogenesis to promote tumorigenesis (21, 23–25). As the main function of mitochondria, OXPHOS has also been shown to augment the tumor-initiating activities of cancer stem cells and to confer cell survival advantage with drug treatment or oxidative stress (26–28). Targeting OXPHOS is therefore another promising strategy for cancer treatment (29, 30). However, like other cancer therapies, the stratification of patients that can benefit from treatment remains challenging.
In this study, we took advantage of the breast-tumor samples generated in our sporadic breast-cancer mouse models (7) and performed whole genomic and transcriptome analyses of tumors. We identified a Pip5k1c mutation activating Pi3k signaling in mouse breast tumors and subsequently classify coamplification of PIP5K1A and PI4KB on chromosome 1q21 as a frequent event activating PI3K signaling in human breast tumors with TP53 mutations. These findings reveal a potential anticancer strategy by targeting both PI3K and mutant p53 in breast cancer.
Results
Identification of Cooperating Mutations in Mutant p53-Driven Breast Tumors.
In order to explore the mutational landscape and molecular pathways altered in breast tumors driven by somatic Trp53 mutations, we performed whole-genome sequencing (WGS) of nine Trp53R245W/+ tumors as listed in Table 1. All tumors were from mice with an identical C57BL/6J×BALB/c F1 hybrid background. Lack of expression of Esr1, Pgr, and Erbb2 in these tumors suggested all were triple-negative breast cancer (TNBC) (SI Appendix, Fig. S1). The survival of mice bearing these tumors ranged from 14.7 to 23.1 mo. Six tumors were pathologically diagnosed as breast adenocarcinomas, two were anaplastic carcinomas, and one was a poorly differentiated carcinoma. Additionally, six tumors were accompanied by visible metastases in lung and/or liver. Thus, somatic Trp53R245W as a driver mutation in mammary epithelial cells led to multiple types of breast cancers, indicative of different evolutionary trajectories in these tumors. The WT Trp53 allele was completely lost in five tumors, partially lost in two tumors, and retained in another two tumors. Thus, it was not necessary to lose the WT Trp53 allele in these mice for tumor development.
Table 1.
Summary of mouse breast tumors driven by somatic Trp53R245W included in this study
| Tumor | Tumor genotype | Diagnosis | LOH | WT Trp53 retention | Metastasis site | Age at endpoint (mo) |
|---|---|---|---|---|---|---|
| 245–1 | p53R245W/+ | Anaplastic carcinoma | Partial | No | ND | 17.1 |
| 245–4 | p53R245W/+ | Adenocarcinoma | Yes | No | Lung | 19.4 |
| 245–5 | p53R245W/+ | Anaplastic carcinoma | Yes | No | Lung, liver | 16.9 |
| 245–14 | p53R245W/+ | Poorly differentiated carcinoma | No | Yes | Lung | 14.7 |
| 245–17 | p53R245W/+ | Adenocarcinoma | Yes | No | Lung | 21.7 |
| 245–20 | p53R245W/+ | Adenocarcinoma | Yes | No | ND | 22.1 |
| 245–21 | p53R245W/+ | Adenocarcinoma | Partial | No | Lung | 23.1 |
| 245–24 | p53R245W/+ | Adenocarcinoma | Yes | No | ND | 22.1 |
| 245–27 | p53R245W/+ | Adenocarcinoma | No | Yes | Lung | 20.9 |
ND, not detectable; LOH, loss of heterozygosity.
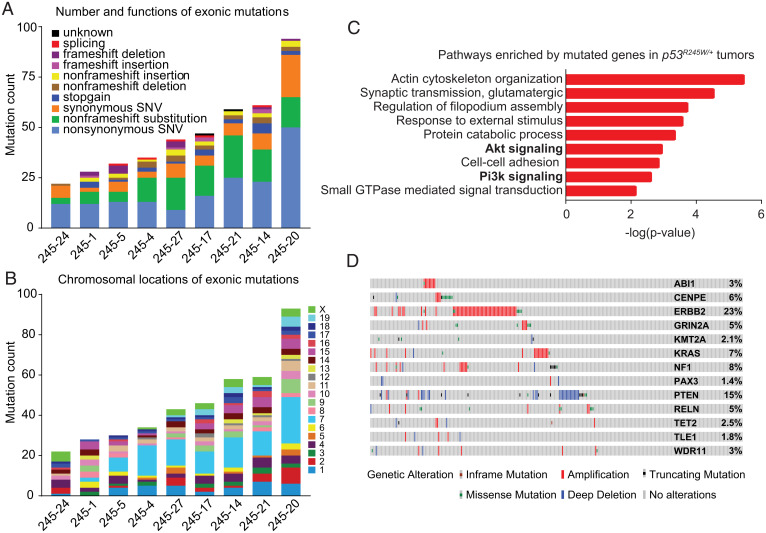
WGS analysis of nine tumors using two mouse tails as controls revealed a large No. of somatic mutations across individual tumors, ranging from ∼6,000 to ∼14,000 mutation counts per tumor (SI Appendix, Fig. S2A). The average mutation frequency of the whole genome was 3.74 (range 2.26–5.55) mutations/Mb. These mutations were mainly intronic or intergenic and the majority spanned chromosomes 1, 3, 7, 15, 16, and X (SI Appendix, Fig. S2 A and B). A small No. of mutations were exonic, with most being nonsynonymous mutations or nonframeshift substitutions (SI Appendix, Fig. S2A and Fig. 1A). Specifically, 22–94 exonic mutations were identified per tumor sample. The distribution of exonic mutations across chromosomes followed the similar pattern of total mutations (Fig. 1B). Gene Oncology (GO) enrichment analysis (31) of all exonic mutations identified pathways related to cell migration, such as actin cytoskeleton organization, regulation of filopodium assembly, and cell–cell adhesion (Fig. 1C). GO analysis also identified the enrichment of Pi3k signaling and Akt signaling, highlighting a role of the Pi3k/Akt/mTOR signaling in these breast tumors (Fig. 1C). GO analyses of breast-tumor samples with metastasis (6) separated from those without obvious metastases (3) also indicated that Pi3k/Akt/mTOR signaling was enriched (SI Appendix, Fig. S3). Analysis of the exonic mutations also documented a list of genes identified as cancer genes by OncoKB, a database that provides up-to-date information about the biological and clinical implications of potential cancer gene alterations (32), or previous publications (33–40) (SI Appendix, Table S1). These genes included some common driver mutations, such as ERBB2, PTEN, NF1, and KRAS, in human cancers, including breast cancer, as well as driver mutations implicated in cancer types other than breast cancer, such as BIRC6 (34) and WDR11 (39, 40). All of these cancer genes are mutated in TP53-mutant human breast tumors as demonstrated in cBioPortal (https://www.cbioportal.org/) (41, 42) (Fig. 1D), indicating that our mouse models recapitulated at least some of the driver mutations in human breast tumors. The occurrence of these driver mutations in the context of Trp53 mutation suggested their contribution to mutant p53-driven breast tumorigenesis.
Fig. 1.
WGS identifies disruption of Pi3k/Akt/mTOR signaling in Trp53R245W/+ breast tumors. (A) The graph showing exonic mutation count for each tumor. Types of mutations were also shown. A total of nine tumors were sequenced. (B) A graph depicting distribution of exonic mutations on different chromosomes for each tumor. (C) GO enrichment analysis of the mutated genes identified in Trp53R245W/+ tumors. GTPase, guanosine triphosphatase. (D) The identified cancer genes in our mouse tumors were input into cBioPortal to generate OncoPrint for these genes in TP53-mutant human breast tumors (n = 280) in The Cancer Genome Atlas data. All identified cancer genes in mouse tumors were altered in TP53-mutant human breast tumors to various extents.
Identification of a Recurrent Pip5k1c Mutation as a Potential Driver for Breast Cancer.
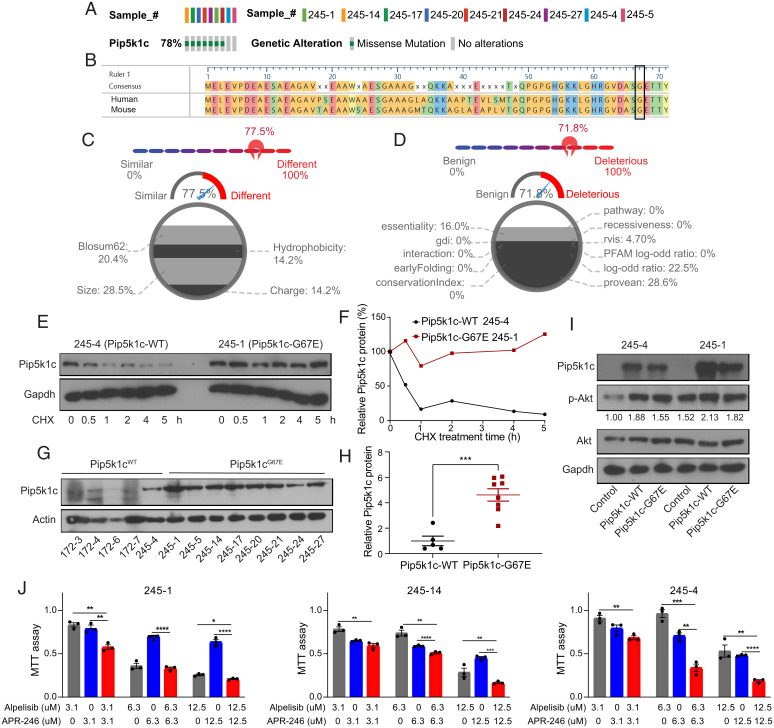
Recurrent mutations in tumors can be highly informative of the biological processes driving tumor evolution and are usually predicted to be cancer driver mutations (43, 44). To identify such driver mutations in our mouse breast tumors, we compiled the mutations that occurred in at least two tumor samples (SI Appendix, Fig. S4). Surprisingly, a large No. of genes were mutated at high frequency in these tumors: 23 genes were mutated in more than 50% of the tumors. In particular, Pip5k1c was found to be mutated in 78% (7/9) of the Trp53R245W/+ tumors (SI Appendix, Fig. S4 and Fig. 2A). This mutation was of interest because Pi3k signaling was found to be dysregulated in these tumors (Fig. 1C) and Pip5k1c is a member of the phosphatidylinositol 4-phosphate 5-kinase (Pip5k) family that catalyzes the production of PI(4,5)P2 and is an upstream activator of Pi3k signaling (45, 46).
Fig. 2.
WGS identifies a recurrent Pip5k1cG67E mutation activating Pi3k/Akt/mTOR signaling as a potential driver for p53R245W/+ breast tumors. (A) OncoPrint representation of the Pip5k1c mutation identified in Trp53R245W/+ tumors. (B) Alignment of the first 71 amino acids of human PIP5K1C amino acid sequence with mouse Pip5k1c amino acid sequence. Glycine (G) at codon 67 is conserved between human and mouse. (C) Comparison of glycine and glutamic acid in DEOGEN2 generates a score of 0.775, showing the difference between the two amino acids. (D) Human PIP5K1CG67E is predicted to be deleterious and disease causing, with a score of 0.718 by DEOGEN2. (E) Western blot showing the stability of Pip5k1c protein in tumor-derived cell lines treated with 33 μg/mL cycloheximide (CHX) and harvested at different time points. (F) Quantification of the western blot results in E. (G) Western blot showing the protein levels of Pip5k1c in Pip5k1cWT tumors and Pip5k1cG67E tumors. 172–3, 172–4, 172–6, and 172–7 are mouse mammary tumors driven by somatic p53R172H that maintain WT Pip5k1c. (H) Quantification of the western blot results in G. (I) Western blot showing the protein levels of Pip5k1c and p-Akt in Pip5k1cWT and Pip5k1cG67E overexpressing cells with quantification of p-Akt normalized to Akt. (J) MTT assays in 245–1, 245–14, and 245–4 cells treated with alpelisib or APR-246 for 3 d. Results were shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Strikingly, a guanine to adenine mutation occurring at the same codon of Pip5k1c was consistently observed in these tumors, resulting in a glycine (G) to glutamic acid (E) mutation at the amino acid position 67 (Pip5k1cG67E) (SI Appendix, Fig. S5A). We performed Sanger sequencing using DNAs from Trp53R245W/+ tumors and control tails and validated this mutation in the seven Trp53R245W/+ tumor samples as identified by WGS (SI Appendix, Fig. S5B). Sanger sequencing results also indicated that the Pip5k1cG67E mutation was in a heterozygous state. In addition, we identified the mutation in an additional tumor sample, 245–5, which had a low rate of the G to A mutation and thus was missed by WGS (SI Appendix, Fig. S5B). We confirmed that the Pip5k1cG67E mutation was a somatic mutation because this mutation was found in tumor 245–1, but not found in the tail of mice bearing tumor 245–1 (SI Appendix, Fig. S5B). The observation of identical mutations occurring at high frequency in mouse breast tumors is not unique to our tumors. A recent publication reported that an identical cytosine to thymine mutation in Ptprh was observed in 81% MMTV-PyMT breast tumors and the mutation increased the EGFR signaling dependence of tumor cells and enhanced their sensitivity to erlotinib treatment (47).
Human and mouse Pip5k1c protein sequences exhibit >90% similarity, with conservation of the glycine at the amino acid position 67 (Fig. 2B). The change from a neutral glycine to an acidic glutamic acid may have an evident impact on protein structure and function. To predict how the Pip5k1cG67E mutation may affect the function of protein, we used DEOGEN2, which analyzes and predicts the deleterious nature of a single amino acid variant in human proteins (48). DEOGEN2 showed that amino acids glutamic acid and glycine are relatively different, with a score of 0.775, and that the variant is likely deleterious, with an overall score of 0.718 (0 is benign and 1 is deleterious, with the prediction cutoff point at 0.5) (Fig. 2 C and D). As reference, p53R175H and p53R248W were also predicted to be deleterious, with an overall score close to 1 (SI Appendix, Fig. S6). Therefore, Pip5k1cG67E variant was predicted to be disease causing and likely oncogenic in mouse breast tumors driven by somatic Trp53R245W. Analysis of human breast cancer data showed PIP5K1C was amplified in 12 breast cancer patients, six of which had concurrent TP53 mutations (SI Appendix, Fig. S7A). Although the No. of patients with PIP5K1C amplification is low, amplification correlates with higher mRNA expression and worse overall survival (SI Appendix, Fig. S7 B–D). We therefore speculated that the Pip5k1cG67E mutation increased the stability of Pip5k1c protein. Indeed, the half-life of Pip5k1c protein in a cycloheximide-treated cell line generated from Pip5k1c-mutant tumor 245–1 was much longer than that generated from a Pip5k1c-WT tumor 245–4 (Fig. 2 E and F). Similarly, the Pip5k1c protein was quite stable in the Pip5k1c-mutant 245–14 cell line, but not in Pip5k1c-WT 172–7 cell line, which is from a mouse breast tumor initiated by Trp53R172H (SI Appendix, Fig. S8 A and B). Moreover, Pip5k1c protein levels were relatively lower in Pip5k1c-WT tumors than in Pip5k1c-mutant tumors (Fig. 2 G and H). Additionally, overexpression of both WT and mutant Pip5k1c activated the Pi3k/Akt/mTOR signaling, as demonstrated by increased phospho-Akt (p-Akt) (Fig. 2I). Therefore, the Pip5k1cG67E mutation stabilizes the Pip5k1c protein and activates Pi3k/Akt/mTOR signaling.
We next treated Trp53R245W/+ tumor-derived cell lines with a US Food and Drug Administration (FDA)-approved PI3K inhibitor, alpelisib (49), and demonstrated that cancer cells responded to Pi3k inhibition (Fig. 2J). Additionally, we combined alpelisib with APR-246, which targets mutant p53 and reactivates WT p53 functions, albeit with off-target effects (50), and showed that their combination achieved greater repression of cancer cell growth (Fig. 2J). These findings not only underscored the cooperation of Pi3k signaling with mutant p53 in driving breast cancer but also supported the application of dual repression of Pi3k and mutant p53 in treating breast cancer.
Identification of OXPHOS as a Vulnerability in p53-Mutant TNBC.
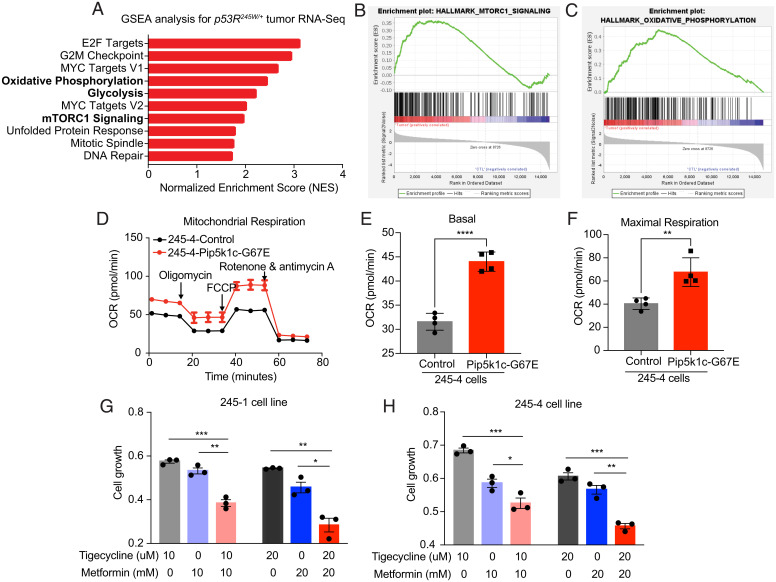
To gain insight into the signaling pathways disrupted by the mutations identified in tumors driven by somatic Trp53R245W, we performed RNA sequencing (RNA-seq) on the same tumors and compared with isolated mammary epithelia from three mice of the same background. Gene Set Enrichment Analysis (GSEA) (51) of the RNA-seq data re-emphasized the role of Pi3k/Akt/mTOR signaling by identifying the activation of mTORC1 in these tumors (Fig. 3 A and B). Additionally, we identified the activation of both glycolysis and OXPHOS in tumors (Fig. 3 A and C), consistent with the role of Pi3k/Akt in regulating glycolysis (16, 17) and mTOR in regulating mitochondrial kinetics (21, 23–25). Focusing on OXPHOS, we performed Seahorse Mito Stress analysis on 245–4-control cells and 245–4-Pip5k1cG67E cells. We demonstrated that expression of Pip5k1cG67E promoted mitochondrial respiration (Fig. 3 D–F), supporting the activation of OXPHOS in mouse breast tumors and also consistent with previous reports in human breast cancer (52, 53). To test the essentiality of OXPHOS in these tumors, we treated Trp53R245W/+ tumor-derived cell lines with two FDA-approved drugs that have been reported to target mitochondria—tigecycline repressing mitochondrial translation (54) and metformin inhibiting mitochondrial complex I (55). We showed that both tigecycline and metformin repressed the growth of tumor-derived cell lines and that combination of the two drugs achieved better efficacy in repressing cell growth (Fig. 3 G and H). Collectively, these results highlighted the role of OXPHOS in somatic mutant p53-driven mouse TNBC and supported the application of therapies targeting OXPHOS in treating these tumors.
Fig. 3.
Oxidative phosphorylation is a vulnerability in Trp53R245W/+ breast tumors. (A) GSEA of RNA-seq data of murine Trp53R245W/+ tumors sequenced in Fig. 1. Graph representing the top 10 significantly upregulated pathways in tumors. (B) mTORC1 signaling was activated in Trp53R245W/+ tumors as demonstrated by GSEA. (C) Oxidative phosphorylation was activated in Trp53R245W/+ tumors as demonstrated by GSEA. (D) Mito Stress assay results in control 245–4 cells and Pip5k1cG67E-overexpressing 245–4 cells. (E) Basal respiration as measured by OCR (oxygen consumption rate) in 245–4-control cells and Pip5k1cG67E-overexpressing 245–4 cells. (F) Maximal respiration in 245–4-control cells and Pip5k1cG67E-overexpressing 245–4 cells. (G and H) MTT assays in (G) 245–1 and (H) 245–4 cells treated with tigecycline and metformin for 3 d. Results were shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
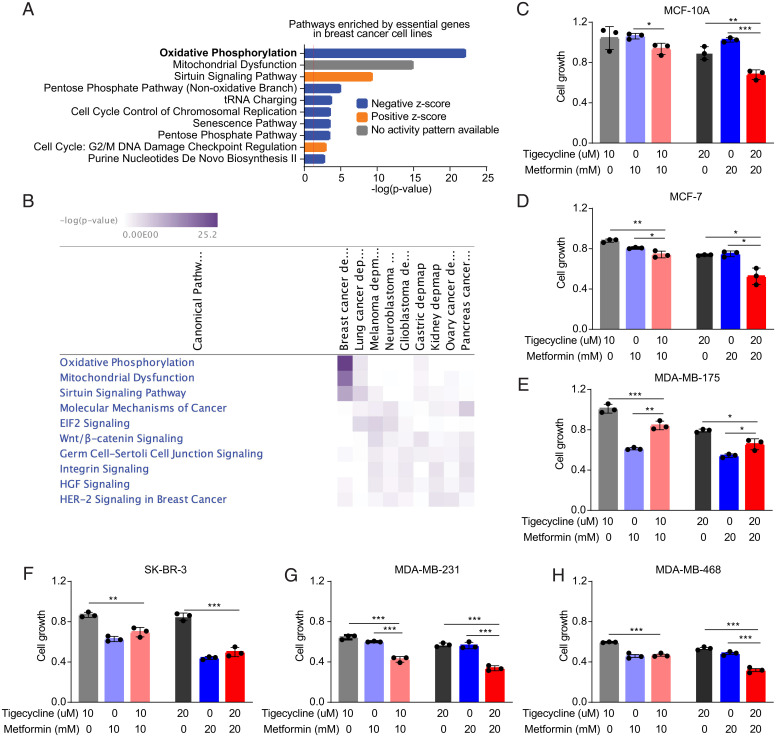
To comprehensively evaluate the essentiality of OXPHOS in human breast cancer, we took advantage of the Dependency Map (DepMap) (https://depmap.org/portal/) portal, which compiles results of large-scale genetic screens performed in human cancer cell lines. Detailed information of the breast cell lines studied are shown in SI Appendix, Table S2. Ingenuity Pathway Analysis (IPA) with the essential genes identified in breast cancer cells identified OXPHOS and mitochondrial dysfunction as top essential pathways (Fig. 4A), supporting that OXPHOS is a vulnerability in human breast cancer. Intriguingly, OXPHOS and mitochondrial dysfunction did not show up as top essential pathways in other eight types of solid cancers that have relatively large No. of cell lines included in Depmap (SI Appendix, Fig. S9). These two pathways were the most significantly enriched essential pathways in breast cancer compared with other cancers (Fig. 4B).
Fig. 4.
Oxidative phosphorylation is a vulnerability in human TNBC. (A) Top 10 pathways by IPA of the essential genes identified by genetic screens in human breast cancer cell lines. (B) Comparison of essential pathways in breast cancer and other types of cancers. (C–H) MTT assays in (C) MCF-10A (normal cell line, TP53WT), (D) MCF-7 (ER+ HER2−, TP53WT), (E) MDA-MB-175 (ER+ HER2−, TP53WT), (F) SK-BR-3 (ER− HER2+, TP53R175H), (G) MDA-MB-231 (TNBC, TP53R280K), and (H) MDA-MB-468 (TNBC, TP53R273H) cells treated with tigecycline and metformin for 3 d. Results were shown as mean ± SE (SEM). *P < 0.05; **P < 0.01; ***P < 0.001.
Analysis of human breast cancer sequencing data identified PTEN signaling was the most significantly downregulated pathway in patients with TP53 missense mutations compared with patients with WT TP53 (SI Appendix, Fig. S10). Downregulation of PTEN signaling is an indicator of activation of PI3K signaling and its downstream OXPHOS. To further validate the dependency of human breast cancer cells on OXPHOS, we treated a panel of cell lines: MCF-10A (normal cell line, TP53WT), MCF-7 (ER+ HER2−, TP53WT), MDA-MB-175 (ER+ HER2−, TP53WT), SK-BR-3 (ER− HER2+, TP53R175H), MDA-MB-231 (TNBC, TP53R280K), and MDA-MB-468 (TNBC, TP53R273H) with tigecycline and metformin. Our results demonstrated that MCF-10A responded poorly to tigecycline or metformin treatment; MCF-7, MDA-MB-175, and SK-BR-3 showed minimal response to tigecycline and some response to metformin; and the two TNBC cell lines, MDA-MB-231 and MDA-MB-468, responded to both tigecycline and metformin (Fig. 4 C–H). Additionally, combination of tigecycline and metformin achieved slightly better efficacy in MCF-10A and MCF-7 but achieved much better efficacy in MDA-MB-231 and MDA-MB-468, suggesting that TP53-mutant TNBC cells respond better to the combination therapy.
Identification of Coamplification of PI4KB and PIP5K1A as a Frequent Event Activating PI3K Signaling to Drive Human TP53-Mutant Breast Cancer.
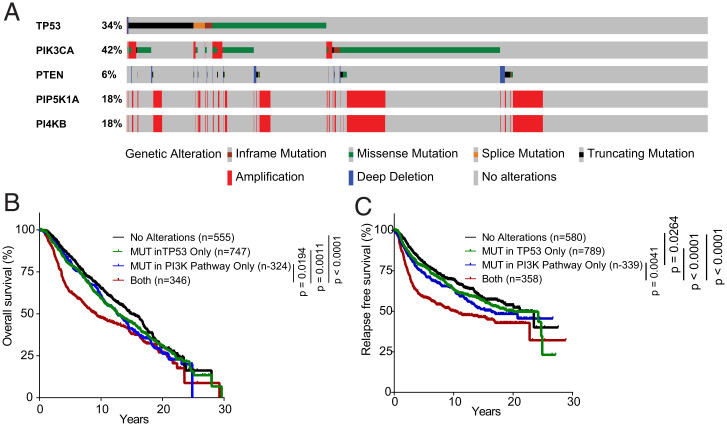
The identification of Pip5k1cG67E mutant as an activator of Pi3k/Akt/mTOR signaling suggests that genomic alterations in the genes encoding phosphatidylinositol kinases for the production of PI(4,5)P2 represent additional mechanisms for PI3K activation in breast cancer. The production of PI(4,5)P2 is regulated step by step by two kinase families: (1) the PIP5K family, including PIP5K1A, PIP5K1B, and PIP5K1C, catalyzes the production of PI(4,5)P2 from PI4P and (2) the PI4K (phosphatidylinositol 4-kinase) family, including PI4KA, PI4KB, PI4K2A, and PI4K2B, catalyzes the production of PI4P. We examined the METABRIC data (56, 57) in the cBioPortal analysis tool (www.cBioPortal.org) and found that PI4KB and PIP5K1A were coamplified in 18% of human breast cancer (Fig. 5A). Intriguingly, both PI4KB and PIP5K1A are located on chromosome 1q21, the amplification of which is frequently found in breast cancer and is associated with tumor-initiating characteristics (58). PI4KB and PIP5K1A have been shown individually to regulate PI3K/AKT/mTOR signaling (59, 60); however, the role of coamplification of PI4KB and PIP5K1A in cancer has not been studied. Since that PI4KB catalyzes the production of PI4P and that PIP5K1A catalyzes the production of PI(4,5)P2 from PI4P, it is very likely that coamplification of PI4KB and PIP5K1A augments the production of PI(4,5)P2 to hyperactivate PI3K signaling. Sixty-five percent (258 out of 357) of the PI4KB/PIP5K1A coamplified patients are ER+HER2− (SI Appendix, Tables S3 and S4), indicating that coamplification of PI4KB and PIP5K1A may preferentially occur in this subtype. Progesterone receptor status, however, is not available for these patients. Approximately thirty percent of PI4KB/PIP5K1A coamplification co-occurs with TP53 mutations (Fig. 5A), suggesting the possibility of PI4KB and PIP5K1A coamplification cooperating with mutant TP53 to drive breast tumorigenesis. Additionally, coamplification of PI4KB and PIP5K1A is concurrent with PIK3CA mutations in 8.6% of patients, indicative of hyperactivation of PI3K signaling in these patients. We next stratified patients with PI4KB/PIP5K1A coamplification, PIK3CA mutations, and PTEN mutations as those with alterations in PI3K pathway and found that patients with mutations in both the PI3K pathway and TP53 gene had worse overall survival and relapse-free survival than those with mutations in PI3K pathway only or those with TP53 mutations only (Fig. 5 B and C). These data re-emphasize the cooperation of PI3K pathway and TP53 mutations in tumor initiation and progression.
Fig. 5.
Coamplification of PI4KB and PIP5K1A as a frequent event to activate PI3K signaling in TP53-mutant breast cancer. (A) OncoPrint of PI4KB, PIP5K1A together with TP53, PIK3CA, and PTEN in human breast cancer (n = 2,173). (B) Overall survival of the breast-cancer patients with different mutation status of TP53 and PI3K-related genes (PI4KB, PIP5K1A, PIK3CA, and PTEN). (C) Relapse-free survival of the breast-cancer patients with different mutation status of TP53 and PI3K-related genes. Patients that had mutations in TP53, but not in other genes, were designated as MUT in TP53 only; patients that had mutations in PI3K-related genes, but not in TP53, were designated as MUT in PI3K pathway only; patients that had mutations in both TP53 and one of the PI3K-related genes were designated as both; and all other patients were designated as no alterations.
Discussion
TP53 mutations are the most frequent genomic alterations in human breast cancer (3, 4). Hotspot mutations in TP53, such as TP53R248W, occur more frequently than other mutations and correlate with more-aggressive disease and worse prognosis (61–63). Our laboratory generated the first sporadic breast cancer mouse models that faithfully recapitulate human breast tumor etiology (7), laying the foundation to identify cooperative mutations required for somatic mutant Trp53-driven breast cancer. We performed WGS on tumors from these models and identified a list of mutated cancer genes that may function as cooperative lesions for somatic Trp53 mutations in breast cancer. The identified cancer genes include Erbb2, Kras, and Pten, as well as other genes, the role of which have not been elucidated in breast cancer. GO analysis of all mutated genes also recognizes Pi3k signaling as a cooperative event for mutant p53, consistent with previous reports (14, 15). Additionally, we identify recurrent mutations that may facilitate the tumorigenesis initiated by somatic Trp53R245W. Particularly, we identify and validate a recurrent Pip5k1cG67E mutation that stabilizes the Pip5k1c protein to amplify Pi3k/Akt/mTOR signaling.
Although we did not identify the same PIP5K1CG67E mutation in human breast cancer, and PIP5K1C is only amplified in a small percentage of human breast tumors, we identify an analog to PIP5K1C—PIP5K1A—amplified in 18% of human breast cancer, suggesting that genomic alterations in the genes responsible for PI(4,5)P2 production represent a frequent event for PI3K activation in breast cancer. Intriguingly, PI4KB, which functions upstream of PIP5K1A, is located on the same chromosomal region as PIP5K1A and is coamplified with PIP5K1A in the 18% of human breast cancer. Therefore, coamplification of PI4KB and PIP5K1A enhances the production of PI(4,5)P2 to hyperactivate PI3K signaling. Additionally, the co-occurrence of PI4KB/PIP5K1A amplification with TP53 mutations and the worse prognosis in patients harboring both PI3K activation and TP53 mutations further support their cooperation in driving breast-cancer progression. Consistent with the essential role of Pi3k signaling in these somatic Trp53R245W-driven breast tumors, inhibition of Pi3k represses the growth of tumor-derived cell lines. Importantly, the combination of drugs targeting Pi3k and mutant p53 achieves better efficacy than either drug alone. These data not only stress the cooperation of PI3K signaling with mutant TP53 in driving breast tumorigenesis but also shed light on therapeutic strategies targeting both PI3K and mutant p53.
Transcriptomic analysis in these somatic Trp53R245W-driven breast tumors identified activation of Pi3k/Akt/mTOR signaling and its downstream metabolic pathways, including glycolysis and OXPHOS. Disrupting different steps of OXPHOS using FDA-approved drugs tigecycline or metformin inhibits the growth of mouse TNBC cell lines initiated by somatic Trp53R245W, and combination of the two drugs achieves better efficacy than just one. Intriguingly, OXPHOS is the top essential pathway in human breast cancer cell lines, but not in other types of cancer cell lines. Tigecycline, metformin, or their combination potently represses the growth of human TP53-mutant TNBC cell lines. Therefore, our findings potentially open therapeutic opportunities for breast-cancer patients and especially TNBC patients. In addition, these results point to a different direction for repurposing the two FDA-approved drugs, tigecycline and metformin, which have been used for bacterial infections and diabetes, respectively, for cancer treatment.
Finally, we propose one model in which the frequent amplification of chromosome 1q21 containing PI4KB and PIP5K1A leads to overexpression of PI4KB and PIP5K1A, which subsequently boosts the production of PI(4,5)P2 to overactivate PI3K/AKT/mTOR signaling (SI Appendix, Fig. S11). The hyperactivation of PI3K signaling rewires cellular energetics (to cooperate with mutant p53) to promote breast tumorigenesis. In summary, our findings uncover potential cooperative driver mutations and pathways for somatic Trp53 mutations, reveal mutations activating Pi3k/Akt/mTOR pathway, and shed light on therapeutic strategies for breast-cancer patients.
Materials and Methods
Cell Lines.
Human breast cancer cell lines were obtained and authenticated from the Cytogenetics and Cell Authentication Core at Department of Genetics at MD Anderson Cancer Center. All cancer cell lines were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
WGS.
DNA was isolated using the Qiagen DNeasy Blood & Tissue Kits according to manufacturer’s instructions. WGS for tumor DNAs was performed at MD Anderson Advanced Technology Genomics Core (ATGC) with NovaSeq6000 150-nt PE flow cells. DNA from two tails of mice from the same cohorts was included as normal control. The quality of raw, 150-bp paired-end reads in FASTQ format was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). After quality check, the reads were aligned to the mouse reference genome (GRCm38 from the Wellcome Sanger Institute) using BWA (64). Summary statistics of read alignments were obtained using SAMtools flagstat (65). Then the Picard tool of MarkDuplicates was employed to mark and remove duplicate reads (broadinstitute.github.io/picard/). Local realignment around insertions and deletions was then performed to correct mapping-related artifacts using the genome analysis toolkit (GATK) (66–68) RealignerTargetCreator and IndelRealigner tools. Next, base quality scores were recalibrated using the GATK tools BaseRecalibrator and PrintReads in order to correct sequencing errors and other experimental artifacts. After the above preprocessing steps, somatic mutations were called using GATK MuTect2 v4.1.0.0 for tumors and their corresponding normal controls (mouse tails). We filtered out likely false-positive variants using the default parameters of the GATK tools FilterMutectCalls, CollectSequencingArtifactMetrics, and FilterByOrientationBias. The mutations, including single nucleotide variants and short insertions/deletions were functionally annotated using ANNOVAR with the corresponding databases for mouse GRCm38/mm10 (69). Further filtering steps were applied: (1) we removed all variants occurring in simple repeat regions (simpleRepeat.txt, downloaded from hgdownload.cse.ucsc.edu/goldenpath/mm10/database) (70), (2) we filtered out known variation using the Sanger variant dbSNP142 databases, (3) we removed variants replicated in all nine Trp53R245W/+ tumors, (4) we kept mutations with approximate read depth ≥20 in both tumor and normal samples, (5) we kept mutations with allele fraction of alternate alleles >0.1 in the tumor and <0.05 in the normal, and (6) we kept the overlapping variants for the tumor sample called against the two normal samples.
Western Blots.
Tissues were homogenized and protein lysates were prepared using radioimmunoprecipitation assay buffer. Equal amounts of lysates were resolved by SDS-PAGE and transferred to ImmunBlot polyvinylidene difluoride membranes (Bio-Rad). Membranes were blocked and probed with specific primary antibodies. Anti-P53 antibody was purchased from Santa Cruz (#sc-6243). Anti-PIP5K1C antibody was purchased from Milipore Sigma (#K1894 or #K2019). Anti-Phospho-Akt (#4060), anti-Actin (#4970) and anti-GAPDH (#2118S) antibodies were purchased from Cell Signaling Technology. Bound antibodies were detected with secondary antibodies conjugated with horseradish peroxidase and visualized by enhanced chemiluminescent substrate (Pierce/Thermo Fisher Scientific). Quantification was performed with ImageJ normalizing to internal control GAPDH or Actin.
Generation of Pip5k1cG67E Expressing Vector.
Pip5k1c cDNA was synthesized by Twist Bioscience. Pip5k1cG67E cDNA was generated by site-directed mutagenesis using WT Pip5k1c cDNA as template and cloned into pBabe-puro (Addgene plasmid No. 1764, a gift from Hartmut Land & Jay Morgenstern & Bob Weinberg).
Mammary Epithelia Isolation.
Isolation of murine mammary epithelia followed published protocol (71).
RNA Extraction.
Total RNA was isolated from homogenized tissues using TRIzol and Qiagen RNeasy Kits. Briefly, appropriate volume of TRIzol was added to homogenized tissues and incubated at room temperature for 5 min. Chloroform was then added to the tissue/TRIzol mixture and mixed by vortex (chloroform:trizol, 1:5 in volume). After incubating at room temperature for 3 min, the chloroform/tissue/TRIzol mixture was centrifuged at 12,000g at 4 °C for 30 min. The upper phase was transferred to a new tube without disturbing the interphase. Then 1.5 volumes of 100% ethanol was added to the upper phase, mixed thoroughly by inverting several times, loaded to RNeasy spin column, and centrifuged for 15 s at >8,000g. The flow-through was discarded. The column was washed with buffer RW1, treated with DNaseI, and then washed with RW1 and RPE buffers. Thirty to one hundred microliters nuclease-free water was added to the column to elute RNA.
RNA-Seq.
RNA-seq was performed at MD Anderson ATGC with NextSeq500 High. The sample library was prepared using Illumina TruSeq stranded protocol. Raw, 75-bp paired-end reads in FASTQ format were initially checked for read quality using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and then aligned to the mouse reference genome (Gencode GRCm38 (72)) using TopHat2 (73). Alignment quality was evaluated from the output of TopHat2. The BAM file with mapped reads for each sample was sorted using SAMtools (65) and then served as an input for HTSEq (74) to count reads that mapped to genes. The read counts for all samples were then normalized using the trimmed mean of M method implemented in the R Bioconductor package edgeR to generate the abundance for each gene (75, 76). Based on the count data, edgeR was also used to perform differential gene expression analysis between groups of samples using the generalized linear model likelihood ratio test. For the comparisons involving tumor samples, we applied the R limma package voom transformation to the RNA-seq data and then fit the mixed model by treating the groups of interest as a fixed effect and sample identification as a random effect as we estimated the correlation between the duplicated samples that were sequenced more than once (77, 78). Benjamini–Hochberg correction was applied to the resulting P values for multiple testing adjustment.
Metabolic Assays.
Agilent Seahorse XF Cell Mito Stress Test Kit was performed according to manufacturer’s protocol. Briefly, 5,000 cells were plated in Seahorse XF cell culture plates the day prior to analysis. On the day of experiment, media was replaced with XF Base Medium containing required supplements. Cartridge was incubated in calibrant prior to drug addition to injection ports. Oligomycin, fluoro-carbonyl cyanide phenylhydrazone, and rotenone and antimycin were added to final concentrations of 1.5 μM, 1 μM, 0.5 μM, and 0.5 μM, respectively. Data were collected in the Seahorse XFe96 Analyzer.
Statistics.
Unpaired Student's t test was used for comparing the means of two groups. Log-rank test was used for comparing Kaplan–Meier survival curves. P values less than 0.05 were recognized as statistically significant. Statistical analysis was performed in Graphpad Prism 9.
Supplementary Material
Acknowledgments
This research was supported by the Cancer Prevention and Research Institute of Texas grant RP180313 and the NIH grant CA82577 to G.L. Sequencing experiments were performed with The University of Texas MD Anderson Cancer Center Advanced Technology Genomics Core, which was supported, in part, by a Cancer Center Support Grant from the National Cancer Institute (CA016672). Authors thank members of the Lozano lab for helpful discussions.
Footnotes
Reviewers: C.E., Thomas Jefferson University; and J.J., Tulane School of Medicine.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2210618119/-/DCSupplemental.
Data, Materials, and Software Availability
WGS data reported in this paper have been deposited in NCBI BioProject database with ID PRJNA858446 (79). RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus database with accession No. GSE207604 (80).
All other data are included in the manuscript and/or SI Appendix.
References
- 1.DeSantis C., Ma J., Bryan L., Jemal A., Breast cancer statistics, 2013. CA Cancer J. Clin. 64, 52–62 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Weir B., Zhao X., Meyerson M., Somatic alterations in the human cancer genome. Cancer Cell 6, 433–438 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Anonymous; Cancer Genome Atlas Network, Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosh R., Rotter V., When mutants gain new powers: News from the mutant p53 field. Nat. Rev. Cancer 9, 701–713 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Silwal-Pandit L., et al. , TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin. Cancer Res. 20, 3569–3580 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Walerych D., Napoli M., Collavin L., Del Sal G., The rebel angel: Mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 33, 2007–2017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., et al. , Somatic Trp53 mutations differentially drive breast cancer and evolution of metastases. Nat. Commun. 9, 3953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosseini H., et al. , Early dissemination seeds metastasis in breast cancer. Nature 540, 552–558 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harper K. L., et al. , Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 540, 588–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samuels Y., et al. , High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Liu P., Cheng H., Roberts T. M., Zhao J. J., Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 8, 627–644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukohara T., PI3K mutations in breast cancer: Prognostic and therapeutic implications. Breast Cancer (Dove Med. Press) 7, 111–123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stemke-Hale K., et al. , An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 68, 6084–6091 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S., Liu J. C., Kim D., Datti A., Zacksenhaus E., Targeted Pten deletion plus p53-R270H mutation in mouse mammary epithelium induces aggressive claudin-low and basal-like breast cancer. Breast Cancer Res. 18, 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams J. R., et al. , Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res. 71, 2706–2717 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Ward P. S., Thompson C. B., Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297–308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faubert B., Solmonson A., DeBerardinis R. J., Metabolic reprogramming and cancer progression. Science 368, 152–161 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensley C. T., et al. , Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashton T. M., McKenna W. G., Kunz-Schughart L. A., Higgins G. S., Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. 24, 2482–2490 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Morita M., et al. , mTOR controls mitochondrial dynamics and cell survival via MTFP1. Mol. Cell 67, 922–935.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Hoxhaj G., Manning B. D., The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 20, 74–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunningham J. T., et al. , mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736–740 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Morita M., et al. , mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 14, 473–480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita M., et al. , mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Haq R., et al. , Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 23, 302–315 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vazquez F., et al. , PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 23, 287–301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valle S., et al. , Exploiting oxidative phosphorylation to promote the stem and immunoevasive properties of pancreatic cancer stem cells. Nat. Commun. 11, 5265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollak M., Targeting oxidative phosphorylation: Why, when, and how. Cancer Cell 23, 263–264 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Jia D., et al. , Elucidating cancer metabolic plasticity by coupling gene regulation with metabolic pathways. Proc. Natl. Acad. Sci. U.S.A. 116, 3909–3918 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y., et al. , Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakravarty D., et al. , OncoKB: A precision oncology knowledge base. JCO Precis. Oncol. 1, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao X., et al. , ACADL plays a tumor-suppressor role by targeting Hippo/YAP signaling in hepatocellular carcinoma. NPJ Precis. Oncol. 4, 7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luk I. S., et al. , BIRC6 targeting as potential therapy for advanced, enzalutamide-resistant prostate cancer. Clin. Cancer Res. 23, 1542–1551 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Kung P. P., et al. , Chemogenetic evaluation of the mitotic kinesin CENP-E reveals a critical role in triple-negative breast cancer. Mol. Cancer Ther. 13, 2104–2115 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Liang Y., et al. , LSD1-mediated epigenetic reprogramming drives CENPE expression and prostate cancer progression. Cancer Res. 77, 5479–5490 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Rai D., Kim S. W., McKeller M. R., Dahia P. L., Aguiar R. C., Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 3111–3116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zavadil J., Brezinová J., Svoboda P., Zemanová Z., Michalová K., Smad5, a tumor suppressor candidate at 5q31.1, is hemizygously lost and not mutated in the retained allele in human leukemia cell line HL60. Leukemia 11, 1187–1192 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Wei L., et al. , Exome sequencing analysis of murine medulloblastoma models identifies WDR11 as a potential tumor suppressor in Group 3 tumors. Oncotarget 8, 64685–64697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chernova O. B., et al. , A novel member of the WD-repeat gene family, WDR11, maps to the 10q26 region and is disrupted by a chromosome translocation in human glioblastoma cells. Oncogene 20, 5378–5392 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostrow S. L., Barshir R., DeGregori J., Yeger-Lotem E., Hershberg R., Cancer evolution is associated with pervasive positive selection on globally expressed genes. PLoS Genet. 10, e1004239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stobbe M. D., et al. , Recurrent somatic mutations reveal new insights into consequences of mutagenic processes in cancer. PLOS Comput. Biol. 15, e1007496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Bout I., Divecha N., PIP5K-driven PtdIns(4,5)P2 synthesis: Regulation and cellular functions. J. Cell Sci. 122, 3837–3850 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Ishihara H., et al. , Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J. Biol. Chem. 273, 8741–8748 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Rennhack J. P., et al. , Integrated analyses of murine breast cancer models reveal critical parallels with human disease. Nat. Commun. 10, 3261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raimondi D., et al. , DEOGEN2: Prediction and interactive visualization of single amino acid variant deleteriousness in human proteins. Nucleic Acids Res. 45 (W1), W201–W206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.André F., et al. ; SOLAR-1 Study Group, Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 380, 1929–1940 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Zandi R., et al. , PRIMA-1Met/APR-246 induces apoptosis and tumor growth delay in small cell lung cancer expressing mutant p53. Clin. Cancer Res. 17, 2830–2841 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitaker-Menezes D., et al. , Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: Visualizing the therapeutic effects of metformin in tumor tissue. Cell Cycle 10, 4047–4064 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones R. A., et al. , RB1 deficiency in triple-negative breast cancer induces mitochondrial protein translation. J. Clin. Invest. 126, 3739–3757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skrtić M., et al. , Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 20, 674–688 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wheaton W. W., et al. , Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife 3, e02242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pereira B., et al. , The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Curtis C., et al. ; METABRIC Group, The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goh J. Y., et al. , Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med. 23, 1319–1330 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Semenas J., et al. , The role of PI3K/AKT-related PIP5K1α and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 111, E3689–E3698 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morrow A. A., et al. , The lipid kinase PI4KIIIβ is highly expressed in breast tumors and activates Akt in cooperation with Rab11a. Mol. Cancer Res. 12, 1492–1508 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Olivier M., et al. , The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin. Cancer Res. 12, 1157–1167 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Liu D. P., Song H., Xu Y., A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene 29, 949–956 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou G., et al. , Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol. Cell 54, 960–974 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKenna A., et al. , The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DePristo M. A., et al. , A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van der Auwera G. A., et al. , From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 43, 11.10.11–11.10.33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang K., Li M., Hakonarson H., ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuhn R. M., Haussler D., Kent W. J., The UCSC genome browser and associated tools. Brief. Bioinform. 14, 144–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen-Ngoc K. V., et al. , 3D culture assays of murine mammary branching morphogenesis and epithelial invasion. Methods Mol. Biol. 1189, 135–162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frankish A., et al. , GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47 (D1), D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim D., et al. , TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Law C. W., Chen Y., Shi W., Smyth G. K., voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smyth G. K., Michaud J., Scott H. S., Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067–2075 (2005). [DOI] [PubMed] [Google Scholar]
- 79.X. Yu et al., Omics analyses of a somatic Trp53 R245W/+ breast cancer model identify cooperating driver events activating PI3K/Akt/mTOR signaling. BioProject. https://www.ncbi.nlm.nih.gov/bioproject/858446. Deposited 13 July 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.X. Yu et al., Omics analyses of a somatic Trp53R245W/+ breast cancer model identify cooperating driver events activating PI3K/Akt/mTOR signaling. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207604. Deposited 6 July 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WGS data reported in this paper have been deposited in NCBI BioProject database with ID PRJNA858446 (79). RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus database with accession No. GSE207604 (80).
All other data are included in the manuscript and/or SI Appendix.