Significance
Group 3 innate lymphoid cells (ILC3s) mediate immune responses in bacterial infections and autoimmunity. Intestinal ILC3s encounter metabolic signals derived from the diet and microbiota, yet the impact of these metabolites on ILC3s is unclear. Here we demonstrate that polyamines and their associated metabolic pathways are enriched in ILC3s. Treatment with putrescine, a key polyamine species, enhanced ILC3 production of IL-22. Conditional deletion of ornithine decarboxylase 1 (ODC1), the rate-limiting enzyme in polyamine biosynthesis, in ILC3s reduced IL-22 expression and increased mice susceptibility to infection by Citrobacter rodentium. In an autoimmune colitis model, ODC1 deficiency in ILC3s protected mice from colitis by reducing IL-22 production. We conclude that ILC3 polyamine biosynthesis is a promising therapeutic target for intestinal infections and autoinflammatory disorders.
Keywords: innate lymphoid cells, polyamines, IL-22, ornithine decarboxylase, enteritis
Abstract
Group 3 innate lymphoid cells (ILC3s) are RORγT+ lymphocytes that are predominately enriched in mucosal tissues and produce IL-22 and IL-17A. They are the innate counterparts of Th17 cells. While Th17 lymphocytes utilize unique metabolic pathways in their differentiation program, it is unknown whether ILC3s make similar metabolic adaptations. We employed single-cell RNA sequencing and metabolomic profiling of intestinal ILC subsets to identify an enrichment of polyamine biosynthesis in ILC3s, converging on the rate-limiting enzyme ornithine decarboxylase (ODC1). In vitro and in vivo studies demonstrated that exogenous supplementation with the polyamine putrescine or its biosynthetic substrate, ornithine, enhanced ILC3 production of IL-22. Conditional deletion of ODC1 in ILC3s impaired mouse antibacterial defense against Citrobacter rodentium infection, which was associated with a decrease in anti-microbial peptide production by the intestinal epithelium. Furthermore, in a model of anti-CD40 colitis, deficiency of ODC1 in ILC3s markedly reduced the production of IL-22 and severity of inflammatory colitis. We conclude that ILC3-intrinsic polyamine biosynthesis facilitates efficient defense against enteric pathogens as well as exacerbates autoimmune colitis, thus representing an attractive target to modulate ILC3 function in intestinal disease.
Innate lymphoid cells (ILCs) are lymphocytes which lack T or B cell receptors and constitutively reside at barrier tissues (1–3). Functional diversification of ILC subsets parallels the current paradigm for T helper cell differentiation with group 1 ILCs (ILC1s), group 2 ILCs (ILC2s), and group 3 ILCs (ILC3s) mirroring Th1, Th2, and Th17, respectively. The murine small intestine lamina propria (siLP) contains major populations of ILC3s and ILC2s with minor populations of ILC1s and natural killer cells. ILC3s originate from progenitors derived from fetal liver and bone marrow which populate the intestine during embryonic development and early after birth (4–6). ILC3s are characterized by the expression of the lineage-determining transcription factor, retinoic acid (RA)-related orphan receptor γ isoform t (RORγT), and the capacity to produce IL-22, IL-17A/F, and granulocyte-machrophage colony stimulating factor (GM-CSF). In the intestine, ILC3s provide two major effector functions: the rapid production of immunoregulatory cytokines and the formation of tertiary lymphoid tissue (7). These roles are delegated among three functionally distinct subsets based on the expression of NKp46 and CCR6. NKp46+ ILC3s are abundantly distributed throughout the siLP but are markedly reduced in the colon and mesenteric lymph nodes (8). This subset is dependent on microbiota-derived signals for differentiation and cytokine production (9). They also exhibit functional plasticity; they express T-BET, produce interferon-γ (IFN-γ), and are capable of transdifferentiation to RORγT− ILC1s (10–12). NKp46−CCR6− double-negative (DN) ILC3s are proposed to contain intestinal precursors which acquire T-BET expression to differentiate into NKp46+ ILC3s (13). Last, CCR6+ ILC3s are proposed to induce formation of tertiary lymphoid tissue such as cryptopatches (CPs) and isolated lymphoid follicles (14–17). As opposed to NKp46+ ILC3s, these cells are localized to the base of intestinal crypts in CPs and constitutively produce IL-22 independent of microbiota-derived signals (9).
The differentiation and function of ILC3s are significantly influenced by metabolism. We have previously shown that intrinsic metabolic control mediated by mTORC1-HIF1α and the production of mitochondrial reactive oxygen species are necessary for ILC3 proliferation and production of IL-22 and IL-17 (18). These studies demonstrated a significant role for metabolic adaptations in regulating ILC3 differentiation and function. However, the contribution of other forms of metabolism in intestinal ILC3s remains unknown. In order to comprehensively identify metabolic pathways of biological significance in ILCs, we mined a previously published single-cell RNA-seq (scRNA-seq) dataset of small intestinal ILCs (19). Our analysis identified a strong enrichment for polyamine biosynthetic genes in ILC3s as compared to other ILCs.
Polyamines are polycationic alkylamines that are mainly derived from arginine via the concerted action of arginase-1 and ornithine decarboxylase (ODC1) (20, 21). Intracellular polyamines are highly abundant, with concentrations in the millimolar range (20, 21). The levels of these metabolites are kept under tight control by de novo synthesis, intracellular recycling, and import/export. Aside from host-synthesized polyamines, microbiota are a significant source of polyamines (22). De novo synthesis of polyamines is regulated by the activity and expression of ODC1, which catalyzes the decarboxylation of ornithine to putrescine. Putrescine is used as a substrate for the synthesis of spermidine and then spermine by spermidine synthase (SRM) and spermine synthases (SMS), respectively. While multiple metabolic enzymes participate in intracellular recycling and export of polyamines, acetylation by the enzyme spermidine/spermine-N1-acetyltranferase (SSAT/SAT1) is the initiating step in all these reactions. As polyamines are protonated at physiological pH, they are incapable of passive diffusion across the cell membrane. While it is understood that bidirectional transport of polyamines is energy-dependent and saturable, the molecular players in polyamine transport are not yet characterized (23). Broadly, polyamines promote cell proliferation, regulate gene transcription, and control translation elongation and termination (20, 24, 25). Polyamines have previously been described to play important roles in adaptive lymphocytes and myeloid cells. Early work has demonstrated a strong association between polyamine biosynthesis via ODC1 and activation-dependent proliferation of lymphocytes (26, 27). Later work demonstrated that Odc1 is a transcriptional target of c-MYC, which is a well-known downstream target of antigen receptor and IL-2 signaling (28–32). ODC1 facilitates lymphocyte proliferation through multiple mechanisms, but the most well-described has been the hypusination of eukaryotic initiation factor 5A (eIF5A) (24). Hypusine is a rare posttranslational modification which has only been reported to occur on a single protein, eIF5A, and which uses spermidine as its main substrate. Hypusination of eIF5A stimulates translation of transcripts containing polyproline motifs and is necessary for life. It is thus postulated that polyamine biosynthesis is up-regulated during cellular proliferation to accommodate the extensive bioenergetic and translational demands of this process. Conditional deletion of Odc1 in T cells demonstrated widespread dysfunction in TH specification during in vitro polarization, and ODC1-deficient T cells were more pathogenic in a model of T cell transfer colitis (33). Similar work showed a defect in the proliferation and viability of CD4+ T cells in vitro but this defect was not observed in vivo in a model of experimental autoimmune encephalitis (34). Aside from mediating hypusination, polyamines have also been described to act as epigenetic regulators due to their polycationic nature. While the exact mechanisms remain undefined, deficiencies in Odc1 have been associated with changes in histone modifications and chromatin accessibility (35, 36). Given the enrichment of polyamine biosynthetic genes in ILC3s and the documented effects of polyamines on immune cell function, we hypothesized that polyamines may play important roles in regulating ILC3 biology.
Results
Integration of Single Cell Transcriptomics and Untargeted Metabolomics Identifies Polyamine Biosynthetic Gene Enrichment in ILC3s.
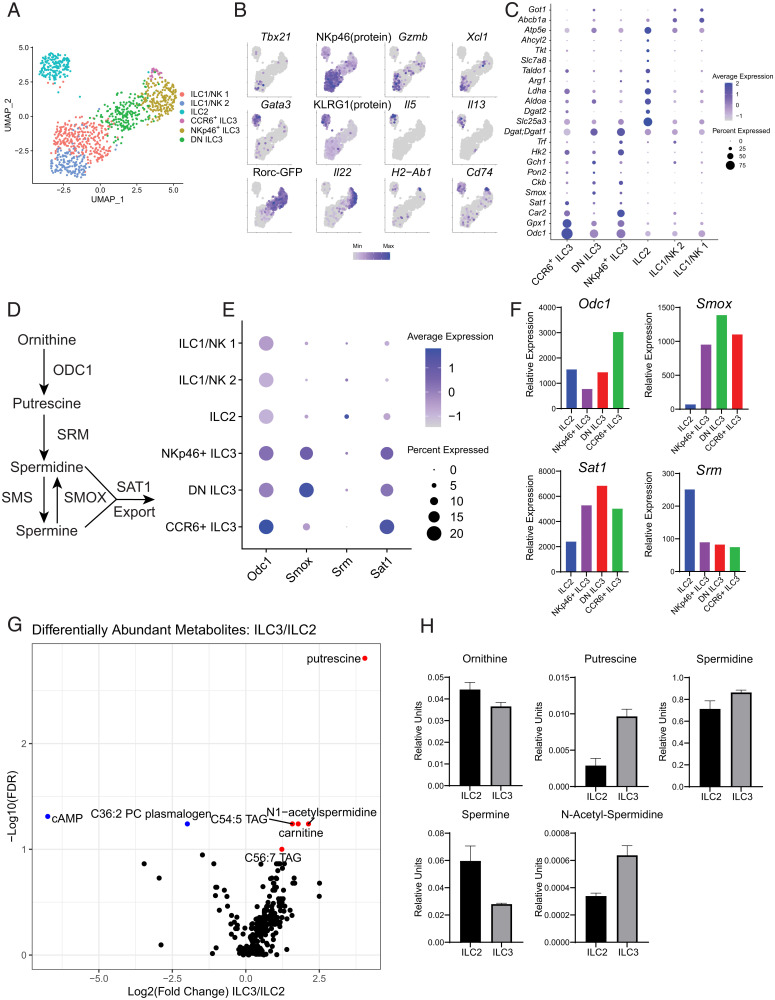
We analyzed a previously published single-cell RNA-seq (scRNA-seq) dataset of small intestinal ILCs to identify differentially enriched metabolic pathways (19). Our reanalysis of these data resolved all intestinal ILC populations, as well as three phenotypically distinct ILC3 subsets. Annotation of these clusters was performed by manual examination of marker genes, Rorc-eGFP reporter expression, and antibody staining for KLRG1 and NKp46, as these cells were index-sorted (Fig. 1 A and B). ILC3 marker genes were notable for genes related to polyamine metabolism (Odc1, Sat1, Smox) and ROS metabolism (Gpx1 and Pon2) (Fig. 1C). The enrichment of ROS metabolic genes supports previous findings that activated ILC3s accumulate mitochondrial ROS (18). ILC2-enriched genes were involved in lipid metabolism (Dgat1, Dgat2), amino acid import (Slc7a8), and arginine metabolism (Arg1). Both Dgat1 and Arg1 have previously been shown to be important for the proper homeostasis and activation of ILC2s (37, 38). These findings provided support that our transcriptomic approach could reliably identify metabolic pathways of functional significance. Out of all the metabolic genes, ornithine decarboxylase 1 (Odc1) stood out as the most strongly expressed marker of ILC3 clusters, especially in CCR6+ ILC3s (Fig. 1C). ODC1 is the rate-limiting enzyme in polyamine biosynthesis, facilitating the conversion of ornithine into putrescine (Fig. 1D). Downstream polyamine metabolic genes, Smox and Sat1, were also enriched in ILC3 subsets (Fig. 1E). This was recapitulated in bulk RNA-seq profiles of ILCs provided by the ImmGen Consortium (Fig. 1F) (39). Furthermore, these data show that Odc1 is most highly expressed in CCR6+ ILC3s compared with all ILC subsets, demonstrating that ILC3s strongly express several components of the polyamine metabolic pathway.
Fig. 1.
scRNA-seq and metabolomics of intestinal ILCs identifies enrichment of polyamine biosynthesis in ILC3s. (A) Uniform manifold approximation and projection (UMAP) of intestinal ILC subsets. (B) Feature plots showing enrichment of marker genes and selected protein/reporter molecules specific to ILC subsets. (C) Dot plot showing enrichment of metabolic enzymes in ILC subsets. (D) Schema showing polyamine biosynthetic and catabolic pathways. (E) Dot plot showing enrichment of enzymes involved in polyamine biosynthesis and catabolism in ILC3 subsets. (F) Bulk RNA-seq profiles of polyamine metabolic genes for ILC2 and ILC3 subsets. Obtained from ImmGen. (G) Volcano plot showing differentially abundant metabolites between ILC2s and ILC3s (n = 4). (H) Targeted validation of polyamine biosynthetic intermediates in ILC2s and ILC3s (n = 2). Results are shown as mean ± SEM.
In parallel with our transcriptomic approach, we additionally sought to characterize the metabolome of the major intestinal ILC subsets. We reasoned that these data, when used together, would synergize to power the discovery of true biological differences in metabolism. We performed untargeted metabolomics profiling of intestinal ILC2s and ILC3s from steady-state mice using liquid chromatography-mass spectrometry (LC-MS). We detected 18,036 metabolic features distributed among lipids, amines, and cationic metabolites (Fig. 1G). By running known chemical standards in parallel, we were able to reliably annotate 336 of these features. Using this method, comparison of metabolites in ILC3s and ILC2s demonstrated an enrichment for two polyamines, putrescine and N-acetyl-spermidine, in ILC3s as well as an enrichment of cyclic adenosine monophosphate (cAMP) in ILC2s (Fig. 1G). The latter finding is in accordance with published findings that ILC2s are under homeostatic regulation by calcitonin gene-related peptide (CGRP) which acts through a Gαs-protein-coupled receptor to promote accumulation of intracellular cAMP (40, 41). This result provided evidence that our assay could detect biologically significant metabolites. We validated the enrichment of polyamines in ILC3s using targeted high-performance LC-MS (Fig. 1H). The enrichment of putrescine was especially significant as it is the direct product of ODC1, providing a strong link between our transcriptomic and metabolomic data.
Polyamines Support ILC3 Production of IL-22.
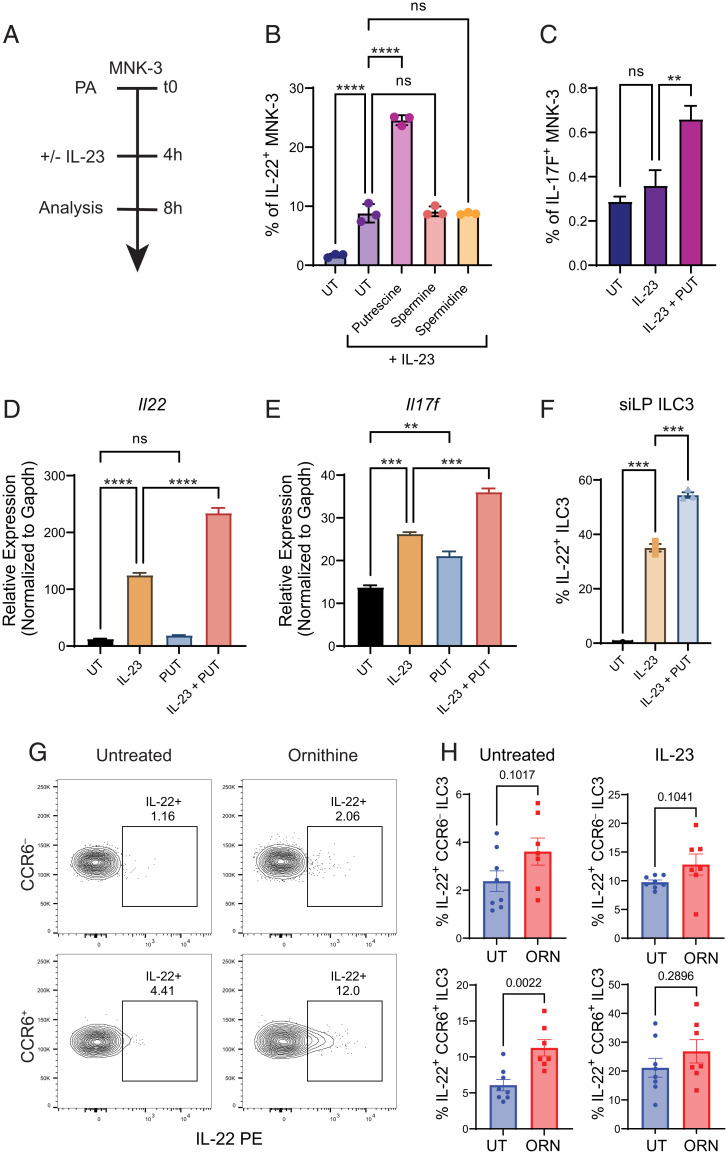
In order to understand the role of polyamines on ILC3 function, we required an in vitro system of ILC3 function where we could tightly control polyamine levels. For this purpose, we utilized MNK-3 cells, an ILC3-like cell line. MNK-3 cells express RORγT, lack T cell lineage markers, express classical ILC3 surface markers (CD117, CD127, NKp46, CCR6, CXCR5, and CD90.2), and model ILC3 expression of IL-22 and IL-17A/F (42). Using MNK-3 cells, we performed a chemical screen with multiple polyamine species (Fig. 2A). When stimulated with IL-23, MNK-3 cells pretreated with putrescine produced significantly more IL-22 (Fig. 2B). However, other polyamine species such as spermidine and spermine did not significantly affect IL-22 production. While IL-23 stimulation alone is not sufficient to induce IL-17F production from MNK-3 cells, we found that IL-23 and putrescine together induced significantly higher IL-17F production (Fig. 2C). We then asked whether the enhancement of IL-22 protein production occurred via transcriptional regulation or posttranscriptional regulation. We performed quantitative RT-PCR on MNK-3 cells treated with IL-23 or IL-23 and putrescine. We found that although putrescine was unable to induce Il22 transcription alone, it significantly enhanced Il22 transcription when combined with IL-23 (Fig. 2D). Surprisingly, we found that putrescine was sufficient to induce Il17f transcription by itself and that this induction was further enhanced with IL-23 cotreatment (Fig. 2E). We also found that putrescine enhanced IL-22 production in primary ILC3s treated with IL-23 (Fig. 2F). In order to investigate the roles of polyamine function on ILC3 function in vivo, we fed wild-type (WT) mice with ornithine, the biosynthetic substrate for putrescine, for two weeks via drinking water. After two weeks of ornithine supplementation, siLP CCR6± ILC3s produced significantly more IL-22 at baseline, compared with untreated controls, consistent with their high expression of Odc1 (Fig. 2 G and H). After stimulation with IL-23, IL-22 production was still higher in the ornithine-fed group but was no longer statistically significant. In contrast, ornithine supplementation did not significantly alter IL-22 production by CCR6− ILC3s at baseline or after stimulation ex vivo. These data demonstrate that the polyamine putrescine positively regulates transcription of Il22 in MNK-3 cells and ILC3s, and that supplementation with its metabolic precursor, ornithine, enhances IL-22 production by CCR6+ ILC3s in vivo.
Fig. 2.
Putrescine enhances Il22 transcription by MNK-3 cells and primary ILC3s. (A) Diagram of experimental timeline for in vitro studies. (B) Frequency of IL-22+ MNK-3 cells stimulated with IL-23 in the presence of indicated polyamines (n = 3). (C) Frequency of IL-17F+ MNK-3 cells after stimulation with IL-23 ± putrescine (n = 3). (D, E) Relative expression of Il22 and Il17f in MNK-3 cells under the indicated conditions. (n = 3). (F) Frequency of IL-22+ sorted primary ILC3s after stimulation with IL-23 ± putrescine (n = 3). (G) Representative FACS plots of IL-22+ CCR6– and CCR6+ ILC3s from untreated and ornithine-fed mice. (H) Frequency of IL-22+ CCR6– and CCR6+ ILC3s without and with IL-23 stimulation from untreated and ornithine-fed mice (n = 7 to 8). Data from B, C are representative of 3 independent experiments. Data from D–F are representative of two independent experiments. Data from G and H are pooled from two independent experiments. Results are shown as mean ± SEM. P values were calculated using the two-tailed Student’s t test or one-way ANOVA and the Tukey's multiple comparisons test. ns, not significant. **P < 0.01; ***P < 0.001; ****P < 0.0001. PA = polyamines; UT = untreated; PUT = putrescine; ORN = ornithine.
ILC3-Intrinsic Polyamine Biosynthesis Supports Control of Enteric Pathogens.
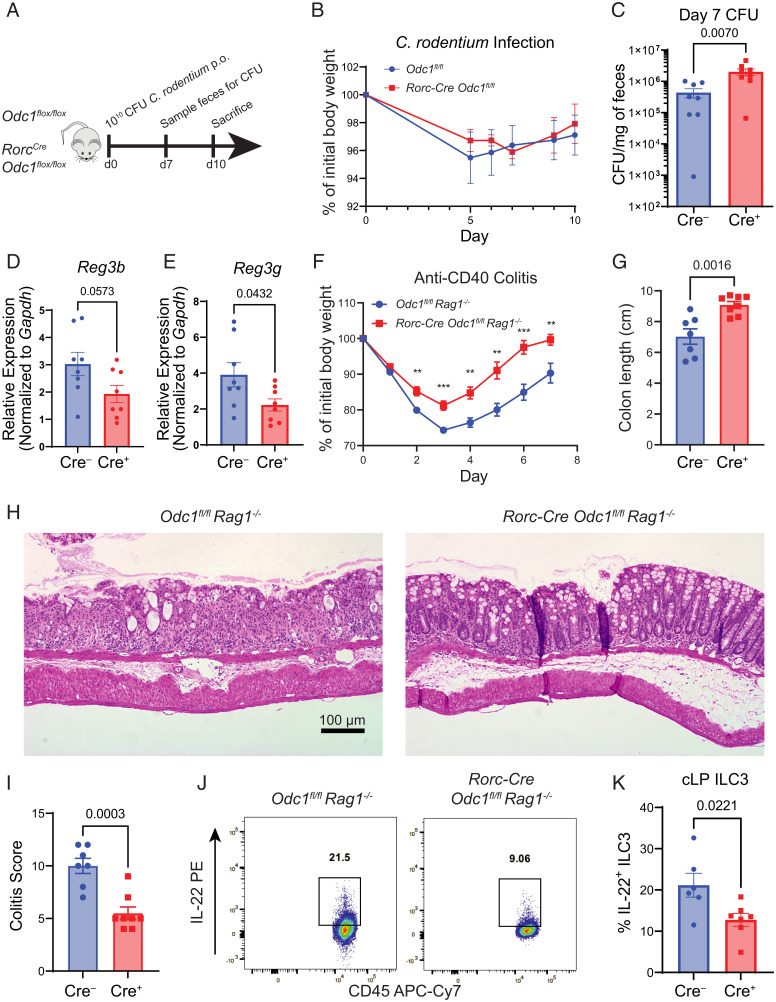
We then asked whether ILC3-intrinsic polyamine metabolism was necessary for ILC3 functions in vivo. To address this, we generated RorcCre Odc1flox/flox mice (Odc1ΔILC3,T) to ablate polyamine biosynthesis in ILC3s. It is important to note that RorcCre is also active in all conventional T cells. Odc1ΔILC3,T mice developed normally and displayed no signs of pathology or sickness. Quantification of siLP immune cell subsets revealed no significant differences in T cell or ILC3 subsets (SI Appendix, Figs. S1 and S2). Functional profiling of ODC1-deficient ILC3s at steady-state demonstrated moderate but nonsignificant reductions in IL-22 and IL-17A production (SI Appendix, Figs. S1 and S2). ILC3s and IL-22 play a critical role in resistance to Citrobacter rodentium by augmenting intestinal epithelial barrier function (43, 44, 45). Given the enhancement of Il22 expression by polyamines, we hypothesized that Odc1ΔILC3,T mice would be less protected against infection by enteric pathogens. Thus, we infected Odc1ΔILC3,T and WT controls with C. rodentium and monitored them for infection severity. As the RorcCre recombines in both ILC3s and all conventional T cells, we only examined mice during the innate phase of the infection (Fig. 3A). Consistent with previous data showing partial redundancy of ILC3s in preventing weight loss during C. rodentium infection (46), we did not observe any significant changes in weight (Fig. 3B). However, assessment of CFU on day 7 before the adaptive immune system responds to infection showed that Odc1ΔILC3,T mice contained significantly more bacterial burden compared to Cre-negative controls (Fig. 3C). IL-22 protects against C. rodentium infection through the induction of antimicrobial peptides by the intestinal epithelium. To test whether this mechanism may be involved in the impaired protection observed in Odc1ΔILC3,T mice, we analyzed bulk colonic tissue of infected mice for antimicrobial peptide gene expression by quantitative RT-PCR (Fig. 3 D and E). Indeed, we detected reductions in both the expression of Reg3b and Reg3g. Importantly, we did not detect any quantitative differences in ILC or TH subsets during C. rodentium infection (SI Appendix, Figs. S1 and S3). Altogether, our data demonstrate that ILC3-intrinsinc polyamine biosynthesis is important for control of enteric pathogen colonization, partially through the induction of antimicrobial peptides by the intestinal epithelium. This occurs independently of T cells, as ODC1-deficient T cells are not affected at this early time point of infection.
Fig. 3.
ILC3-specific deficiency of ODC1 renders mice susceptible to infection with C. rodentium but is protective during anti-CD40–induced colitis. (A) Diagram of experimental timeline for C. rodentium infection. (B) Percentage weight loss during C. rodentium infection (n = 4). (C) Colony-forming unit (CFU) counted from fecal pellets from control and Odc1ΔILC3,T mice on day 7 of C. rodentium infection (n = 8). (D, E) Relative expression of Reg3b and Reg3g from whole distal colon tissue of control and Odc1ΔILC3,T mice on day 10 of C. rodentium infection (n = 8). (F) Percentage weight loss during anti-CD40-induced colitis (n = 7–8). (G) Colon length of control and Odc1ΔILC3 mice on day 7 of anti-CD40-induced colitis (n = 7–8). (H) Representative hematoxylin and eosin (H&E) sections of distal colon on day 7 of anti-CD40–induced colitis. (I) Histologic colitis score of H&E slides of distal colon from control and Odc1ΔILC3 mice on day 7 of anti-CD40–induced colitis (n = 7 to 8). (J) Representative FACS plot of IL-22+ ILC3s from colon lamina propria (cLP) of control and Odc1ΔILC3 mice on day 4 of anti-CD40–induced colitis. (K) Frequency of IL-22+ colonic ILC3s from control and Odc1ΔILC3 mice following stimulation with PMA/Ionomycin for 4 h (n = 6). Data from B is representative of two independent experiments. Data from C–K are pooled from two independent experiments. Results are shown as mean ± SEM. P values for C were calculated using the Mann–Whitney test. All other P values were calculated using two-tailed Student’s t test. **P < 0.01; ***P < 0.001.
Polyamine Biosynthesis Enhances ILC3-Driven Inflammatory Colitis.
We then asked whether polyamine biosynthesis may be active in a model of ILC3-driven autoimmunity. We chose the model of anti-CD40–induced colitis as ILC3s play an essential and nonredundant pathogenic role in this model that is dependent on production of IL-22 and GM-CSF (46, 47). Since this model requires the injection of anti-CD40 in RAG-deficient mice, we crossed Odc1ΔILC3,T mice to a RAG-deficient background (Odc1ΔILC3). Because polyamines positively regulate ILC3 production of IL-22, we hypothesized that Odc1ΔILC3 mice would be protected against anti-CD40 colitis. Indeed, following injection of anti-CD40, Odc1ΔILC3 mice demonstrated attenuated weight loss and greater recovery as compared to Cre-negative controls (Fig. 3F). Measurement of colon length also demonstrated less severe colonic shortening in Odc1ΔILC3 mice, indicative of decreased inflammation (Fig. 3G). Histologic analysis of colonic sections from injected mice demonstrated reduced histopathology in specimens from Odc1ΔILC3 mice (Fig. 3 H and I). We observed that Odc1-deficient ILC3s produced significantly less IL-22 on day 4 of induced colitis, consistent with the pathogenic role for this cytokine in the disease process (Fig. 3 J and K). In summary, these data demonstrate that ILC3-intrinsic polyamine biosynthesis drives autoinflammatory pathology during anti-CD40 colitis.
Transcriptome of Polyamine-Enriched and Polyamine-Deprived Conditions Implicates the Transcriptional Regulator NR4A1.
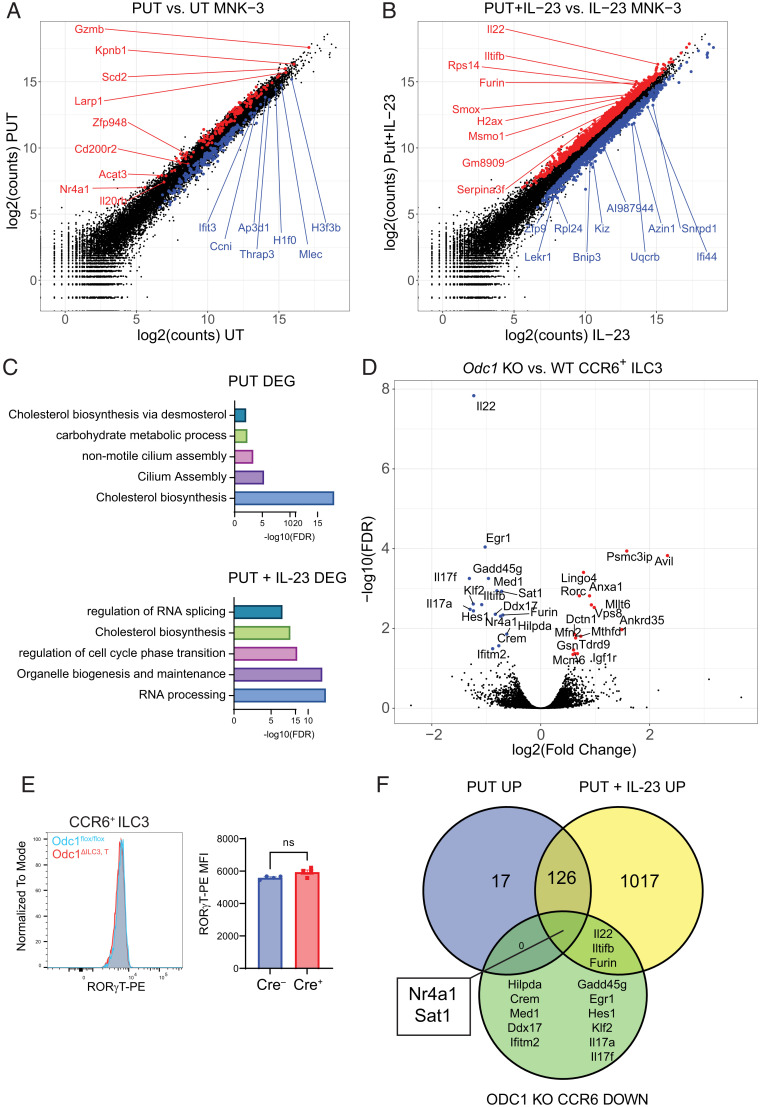
In order to investigate the mechanism by which polyamines promote the production of IL-22, we performed bulk RNA-seq of polyamine-treated MNK-3 cells with or without stimulation with IL-23 (Fig. 4A). Compared to untreated cells, treatment with putrescine produced 362 unique differentially expressed genes (DEGs; 145 up-regulated, 217 down-regulated). In IL-23 stimulated cells, pretreatment with putrescine produced 2415 DEGs (1164 up-regulated, 1251 down-regulated) (Fig. 4B). Gene ontology enrichment of DEGs from both conditions revealed terms related to cholesterol biosynthesis, RNA processing, and cell cycle transition (Fig. 4C). In order to validate these findings in vivo, we performed RNA-sequencing on ODC1 knockout (KO) CCR6+ ILC3s and WT controls (Fig. 4D). CCR6+ ILC3s were selected on the basis of their high Odc1 expression relative to other ILC3 subsets and previous data showing that ornithine selectively increased IL-22 production by this subset. In contrast to studies with MNK-3, ODC1 KO ILC3s had fewer DEGs (31, 15 up-regulated, 16 down-regulated) when compared to ODC1 WT ILC3s. However, manual inspection of DEGs revealed genes of known functional significance in ILC3s (Rorc, Il17a, Il7f, Il22). We also observed significant down-regulation of the polyamine catabolic enzyme Sat1, indicating some degree of metabolic compensation in the absence of ODC1. Notably, ODC1 KO ILC3s had significantly reduced expression of Il22. While ODC1 KO ILC3s displayed increased expression of Rorc, measurement of RORγT protein by intracellular staining revealed equivalent amounts between ODC1 KO and WT ILC3s (Fig. 4E). As Lingo4, which is directly adjacent to Rorc, was also significantly elevated in ODC1 KO ILC3s, we attribute the increase of these two transcripts to the presence of the Rorc-Cre transgene. We then overlapped down-regulated genes from ODC1 KO ILC3s with up-regulated genes from putrescine-treated MNK-3 cells in order to identify consistent polyamine-dependent genes (Fig. 4F). As expected, Il22 was down-regulated in ODC1 KO ILC3s and up-regulated in stimulated MNK-3 cells pretreated with putrescine. Surprisingly, only two genes were shared by all three conditions: Sat1 and Nr4a1. As discussed above, changes in Sat1 are likely a form of metabolic auto-regulation due to increased or decreased polyamine flux. NR4A1 has previously been shown to positively regulate ILC3 function as Nr4a1 KO ILC3s have defective production of IL-22 (48). Importantly, Nr4a1 was up-regulated by putrescine treatment in the absence of stimulation. Thus, polyamines may enhance IL-22 transcription through induction of Nr4a1 transcription prior to stimulation.
Fig. 4.
Transcriptomic profile of polyamine-stimulated MNK-3 cells and ODC1-deficient ILC3s implicates polyamine regulation of the transcription factor NR4A1. (A) DEGs between putrescine-treated and untreated MNK-3 cells. (B) DEGs between putrescine and IL-23–treated MNK-3 and IL-23–treated MNK-3 cells. (C) Gene ontology enrichment of DEGs from putrescine vs. untreated MNK-3 and putrescine and IL-23–treated vs. IL-23–treated MNK-3 cells. (D) Volcano plot showing DEGs between ODC1-deficient vs. WT CCR6+ ILC3s. (E) Representative histogram and quantification of RORγT MFI in CCR6+ ILC3s from control and Odc1ΔILC3,T mice (n = 4). Data are representative of two experiments. (F) Venn diagram showing overlap between up-regulated genes from putrescine-treated vs. untreated MNK-3 cells, up-regulated genes from putrescine and IL-23-treated vs. IL-23–treated MNK-3 cells, and down-regulated genes from ODC1-deficient vs. WT CCR6+ ILC3s. Results are shown as mean ± SEM. P values in E calculated by two-tailed Student’s t test. ns = not significant. PUT = putrescine; UT = untreated; KO = knockout; DEG = differentially expressed genes.
Discussion
In this study, we utilized single-cell transcriptomics to identify a novel association between polyamine metabolism and ILC3s which was validated using metabolomics. Using a novel mouse model of ILC3-intrinsic deletion of ODC1, the rate-limiting enzyme of polyamine biosynthesis, we demonstrated that polyamine biosynthesis was required for both optimal antibacterial defense and ILC3-driven autoimmune colitis.
Using an ILC3-like cell line and primary ILC3s, we demonstrate that polyamines positively regulate transcription of Il22, and to a lesser extent Il17f. Concordantly, we find decreased expression of Il22, Il17a, and Il17f in ODC1-deficient CCR6+ ILC3s, but production of these cytokines are only modestly decreased ex vivo. Notwithstanding, we show that ILC3-intrinsic deletion of ODC1 is sufficient to impair host control of the enteric pathogen C. rodentium with no detectable involvement from adaptive immune cells. This impaired pathogen control is partially due to decreased induction of antimicrobial peptides from the intestinal epithelium, which may be a result of decreased ILC3-derived IL-22. Consistent with these findings, we also show that ODC1 is required for the pathogenesis of ILC3-driven autoimmune colitis. Deficiency of ODC1 substantially reduces ILC3-derived IL-22 and colonic pathology during colitis. Transcriptomic analysis of MNK-3 cells and ILC3s in polyamine-enriched and deprived conditions highlight the transcription factor NR4A1 as a possible mediator in polyamine-driven support of Il22 transcription. These results establish a novel role for polyamines in regulating the function of ILC3s and demonstrate a mechanism by which polyamine metabolism can regulate mucosal immune responses.
Both dietary and microbial sources contribute to the levels of intestinal polyamines and their metabolic precursors. Among dietary sources, soy-based foods or fermented products, such as cheese, are rich in polyamines, especially spermidine. Microbial production of polyamines in the intestine remains incompletely understood. While certain human intestinal microflora have been shown to synthesize polyamines, it is appreciated that microbial polyamines arise from collective biosynthetic pathways shared between communities of bacteria (49). Indeed, many individual intestinal species do not possess a complete biosynthetic pathway for the generation of polyamines from arginine. Thus, therapeutic manipulation of intestinal polyamine abundance through diet, probiotics, or chemical modulators is a promising avenue for the modulation of ILC3 activity in intestinal disease (50, 51, 52, 53, 54).
One notable aspect of our study is the divergent roles that polyamines play in ILC3s compared to adaptive lymphocytes. Polyamines are strongly associated with lymphocyte proliferation and induction of Odc1 is driven by c-MYC (29). However, we detect strong Odc1 expression by ILC3s, which are relatively nonproliferative in comparison to adaptive lymphocytes. Consistent with this nonproliferative feature, ILC3s express low levels of c-MYC. An important question remains as to what drives Odc1 expression in ILC3s. Recent work utilizing inducible deletion of RORγT and RORα in ILCs demonstrated a down-regulation of Arg1, Odc1, and Sat1, suggesting that Odc1 may be induced by retinoic acid signaling, though it is hard to interpret given the fundamental role of these transcription factors in shaping the whole ILC3 identity (55).
In addition to differences in upstream regulators, polyamines appear to play a fundamentally different function in ILC3s compared to T cells. ODC1-deficient T cells display overt defects in proliferation and lineage fidelity and present with massive changes in transcription and chromatin accessibility (33, 36). These changes are attributed to defects in hypusination of EIF5a, which interfere with the translation of chromatin factors required for epigenomic stability. In contrast, we observe a more nuanced role for polyamines in ILC3s. The role of putrescine in enhancing IL-22 transcription occurs on a relatively short timeframe, with 4-h treatment being sufficient to observe robust changes. Compared to the widespread epigenetic dysregulation in ODC1-deficient T cells, we observe a small, but biologically important, set of differentially regulated genes in ODC1-deficient ILC3s. One possible explanation for these discrepancies is that ODC1-deficient ILC3s can partially compensate for the loss of polyamine biosynthesis through alternative mechanisms such as polyamine salvage from the extracellular environment. Given the multifaceted roles of polyamines in biology, it is possible that changes due to hypusination defects occur during conditions of extreme polyamine deprivation whereas other roles for polyamines manifest during periods of relative deficiency. Thus, the mechanism by which ODC1 supports Il22 transcription may represent a previously unappreciated role for polyamines in immune cells. Our transcriptional analyses implicate the transcription factor NR4A1 as a potential polyamine-dependent regulator of IL-22, consistent with a previous report showing that Nr4a1 KO ILC3s display defective IL-22 production (48). Polyamines have been previously described to regulate transcription through modulation of histone modifications such as H3K9me1/2/3 and H3K9ac (56, 57). Further studies examining how polyamines modulate NR4A1 are warranted.
Materials and Methods
Experimental details on animals, reagents and cell lines, cell extraction from tissues, antibody staining for flow cytometry and sorting, sequencing and metabolomic analyses, infection and colitis models, histology, and statistical analyses for this study are described in detail in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Tom Burris, Cyrielle Billon, Patrick Woster, Manoocher Soleimani, Mark R. Burns, the Genome Technology Access Center at McDonnell Genome Institute, and the members of the metabolomics and FACS core facilities at Washington University School of Medicine. Funding information: This work was supported by NIH 5F30DK127540, 5T32DK077653 (V.P.), T32DK00713045 (S.C.), DK043351, DK097485, DK117263 (R.J.X), R01DK124699, and R21AI159210 (M.C.).
Footnotes
Reviewers: A.C., Heidelberg University; and G.S., Universita degli Studi di Roma La Sapienza.
M.C. receives research support from Pfizer. R.J.X. is a cofounder of Celsius Therapeutics. All the other authors have nothing to disclose.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2214900119/-/DCSupplemental.
Data, Materials, and Software Availability
RNAseq data generated in this study are deposited in the Gene Expression Omnibus (GSE214152) (58).
References
- 1.Vivier E., et al. , Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Bando J. K., Colonna M., Innate lymphoid cell function in the context of adaptive immunity. Nat. Immunol. 17, 783–789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klose C. S. N., Artis D., Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 17, 765–774 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Bando J. K., Liang H.-E., Locksley R. M., Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat. Immunol. 16, 153–160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Constantinides M. G., McDonald B. D., Verhoef P. A., Bendelac A., A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klose C. S. N., et al. , Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Withers D. R., Hepworth M. R., Group 3 innate lymphoid cells: Communications hubs of the intestinal immune system. Front. Immunol. 8, 1298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willinger T., Metabolic control of innate lymphoid cell migration. Front. Immunol. 10, 2010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savage A. K., Liang H.-E., Locksley R. M., The development of steady-state activation hubs between adult LTi ILC3s and primed macrophages in small intestine. J. Immunol. 199, 1912–1922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vonarbourg C., et al. , Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt(+) innate lymphocytes. Immunity 33, 736–751 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernink J. H., et al. , Interleukin-12 and -23 control plasticity of CD127(+) Group 1 and Group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 43, 146–160 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Cella M., et al. , Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 20, 980–991 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klose C. S. N., et al. , A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature 494, 261–265 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Kiss E. A., et al. , Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334, 1561–1565 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Eberl G., Littman D. R., Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science 305, 248–251 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Kanamori Y., et al. , Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+ IL-7R+ Thy1+ lympho-hemopoietic progenitors develop. J. Exp. Med. 184, 1449–1459 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Randall T. D., Mebius R. E., The development and function of mucosal lymphoid tissues: A balancing act with micro-organisms. Mucosal Immunol. 7, 455–466 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Di Luccia B., Gilfillan S., Cella M., Colonna M., Huang S. C.-C., ILC3s integrate glycolysis and mitochondrial production of reactive oxygen species to fulfill activation demands. J. Exp. Med. 216, 2231–2241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gury-BenAri M., et al. , The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–1246.e13 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Pegg A. E., Functions of polyamines in mammals. J. Biol. Chem. 291, 14904–14912 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R. A. Casero, Jr, Murray Stewart T., Pegg A. E., Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 18, 681–695 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto M., Benno Y., The relationship between microbiota and polyamine concentration in the human intestine: A pilot study. Microbiol. Immunol. 51, 25–35 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Poulin R., Casero R. A., Soulet D., Recent advances in the molecular biology of metazoan polyamine transport. Amino Acids 42, 711–723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutierrez E., et al. , eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasini A., Caldarera C. M., Giordano E., Chromatin remodeling by polyamines and polyamine analogs. Amino Acids 46, 595–603 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Bowlin T. L., McKown B. J., Babcock G. F., Sunkara P. S., Intracellular polyamine biosynthesis is required for interleukin 2 responsiveness during lymphocyte mitogenesis. Cell. Immunol. 106, 420–427 (1987). [DOI] [PubMed] [Google Scholar]
- 27.Kay J. E., Pegg A. E., Effect of inhibition of spermidine formation on protein and nucleic acid synthesis during lymphocyte activation. FEBS Lett. 29, 301–304 (1973). [DOI] [PubMed] [Google Scholar]
- 28.Bello-Fernandez C., Packham G., Cleveland J. L., The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. U.S.A. 90, 7804–7808 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R., et al. , The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowlin T. L., McKown B. J., Sunkara P. S., Increased ornithine decarboxylase activity and polyamine biosynthesis are required for optimal cytolytic T lymphocyte induction. Cell. Immunol. 105, 110–117 (1987). [DOI] [PubMed] [Google Scholar]
- 31.Schall R. P., Sekar J., Tandon P. M., Susskind B. M., Difluoromethylornithine (DFMO) arrests murine CTL development in the late, pre-effector stage. Immunopharmacology 21, 129–143 (1991). [DOI] [PubMed] [Google Scholar]
- 32.Nitta T., Igarashi K., Yamashita A., Yamamoto M., Yamamoto N., Involvement of polyamines in B cell receptor-mediated apoptosis: Spermine functions as a negative modulator. Exp. Cell Res. 265, 174–183 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Puleston D. J., et al. , Polyamine metabolism is a central determinant of helper T cell lineage fidelity. Cell 184, 4186–4202.e20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu R., et al. , De novo synthesis and salvage pathway coordinately regulate polyamine homeostasis and determine T cell proliferation and function. Sci. Adv. 6, eabc4275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardbower D. M., et al. , Ornithine decarboxylase regulates M1 macrophage activation and mucosal inflammation via histone modifications. Proc. Natl. Acad. Sci. U.S.A. 114, E751–E760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner A., et al. , Metabolic modeling of single Th17 cells reveals regulators of autoimmunity. Cell 184, 4168–4185.e21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karagiannis F., et al. , Lipid-droplet formation drives pathogenic Group 2 innate lymphoid cells in airway inflammation. Immunity 52, 620–634.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Monticelli L. A., et al. , Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat. Immunol. 17, 656–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida H., et al. ; Immunological Genome Project, The cis-regulatory atlas of the mouse immune system. Cell 176, 897–912.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H., et al. , Transcriptional atlas of intestinal immune cells reveals that neuropeptide α-CGRP modulates Group 2 innate lymphoid cell responses. Immunity 51, 696–708.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagashima H., et al. , Neuropeptide CGRP limits Group 2 innate lymphoid cell responses and constrains Type 2 inflammation. Immunity 51, 682–695.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allan D. S., et al. , An in vitro model of innate lymphoid cell function and differentiation. Mucosal Immunol. 8, 340–351 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Satoh-Takayama N., et al. , Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Zheng Y., et al. , Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Goto Y., et al. , Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345, 1254009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song C., et al. , Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med. 212, 1869–1882 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buonocore S., et al. , Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 464, 1371–1375 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B., et al. , Circular RNA circZbtb20 maintains ILC3 homeostasis and function via Alkbh5-dependent m6A demethylation of Nr4a1 mRNA. Cell. Mol. Immunol. 18, 1412–1424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tofalo R., Cocchi S., Suzzi G., Polyamines and gut microbiota. Front. Nutr. 6, 16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss T. S., et al. , Intracellular polyamine levels of intestinal epithelial cells in inflammatory bowel disease. Inflamm. Bowel Dis. 10, 529–535 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Hong S.-K. S., et al. , Increased expression and cellular localization of spermine oxidase in ulcerative colitis and relationship to disease activity. Inflamm. Bowel Dis. 16, 1557–1566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakamura A., et al. , Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat. Commun. 12, 2105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gobert A. P., et al. , Protective role of spermidine in colitis and colon carcinogenesis. Gastroenterology 162, 813–827.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carriche G. M., et al. , Regulating T-cell differentiation through the polyamine spermidine. J. Allergy Clin. Immunol. 147, 335–348.e11 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Fiancette R., et al. , Reciprocal transcription factor networks govern tissue-resident ILC3 subset function and identity. Nat. Immunol. 22, 1245–1255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Latour Y. L., Gobert A. P., Wilson K. T., The role of polyamines in the regulation of macrophage polarization and function. Amino Acids 52, 151–160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.A. Yurdagul, Jr, et al. , ODC (ornithine decarboxylase)-dependent putrescine synthesis maintains MerTK (MER tyrosine-protein kinase) expression to drive resolution. Arterioscler. Thromb. Vasc. Biol. 41, e144–e159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.V. Peng, M. Colonna, Ornithine decarboxylase supports ILC3 responses in infectious and autoimmune colitis through positive regulation of IL-22 transcription. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214152. Deposited 25 September 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data generated in this study are deposited in the Gene Expression Omnibus (GSE214152) (58).