Abstract
Abstract
The field of pediatric critical care has been hampered in the era of precision medicine by our inability to accurately define and subclassify disease phenotypes. This has been caused by heterogeneity across age groups that further challenges the ability to perform randomized controlled trials in pediatrics. One approach to overcome these inherent challenges include the use of machine learning algorithms that can assist in generating more meaningful interpretations from clinical data. This review summarizes machine learning and artificial intelligence techniques that are currently in use for clinical data modeling with relevance to pediatric critical care. Focus has been placed on the differences between techniques and the role of each in the clinical arena. The various forms of clinical decision support that utilize machine learning are also described. We review the applications and limitations of machine learning techniques to empower clinicians to make informed decisions at the bedside.
Impact
Critical care units generate large amounts of under-utilized data that can be processed through artificial intelligence.
This review summarizes the machine learning and artificial intelligence techniques currently being used to process clinical data.
The review highlights the applications and limitations of these techniques within a clinical context to aid providers in making more informed decisions at the bedside.
Introduction
Critical illness in children leads to millions of hospital admissions to a Pediatric Intensive Care Unit (PICU).1,2 Since the inception of the field more than nearly six decades ago, outcomes for these patients have steadily improved, with PICU mortality rates as low as 1–2%.3–5 While clinical research has played a role in improving outcomes, there are surprisingly few therapies in pediatric critical care supported by high levels of evidence. For example, in recent guidelines for the care of children with sepsis6 and traumatic brain injury,7 the vast majority of recommendations were supported by “low” quality of evidence. Reasons for this paucity of evidence-based therapies include heterogeneity within the age spectrum seen in the specialty, limitations of extrapolation of adult studies, low rate of mortality necessitating other outcomes of possibly lower interest, patient volumes lower than adult critical care, and heterogeneity within clinical diagnoses (e.g., sepsis). A reliance on traditional ways to collect and analyze data has also limited the field of pediatric critical care research.
New paradigm-shifting approaches in machine learning, predictive modeling, functional immunophenotyping, and artificial intelligence (AI) have been developed to improve understanding and specificity in refining definitions of disease. There has been rapid growth both in computing power and data storage, enabling a wide range of applications for machine learning and AI within medicine. The term AI refers to the domain of tasks that historically required human input, while machine learning is the subset of AI where learning from data exists without explicit programming.8 Both have impacted drug discovery, personalized diagnostics, therapeutics, and medical imaging.9,10 Within the realm of pediatric critical care, the use of these techniques has the potential to significantly improve our understanding of disease and of therapeutic efficacy. In this review, we will outline different machine learning techniques, provide an overview of current AI applications and specific machine learning/AI limitations, and discuss how these technologies will further the field of pediatric critical care.
Machine learning
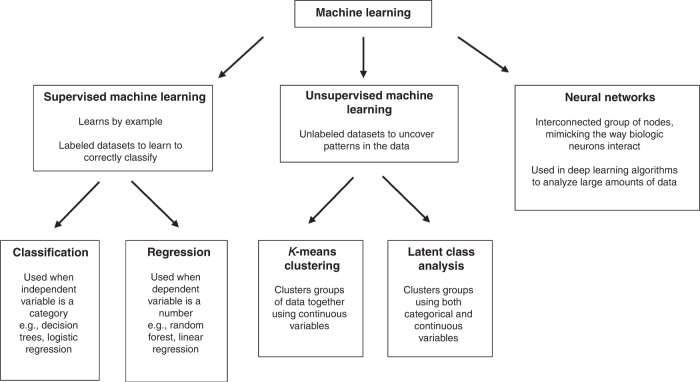
In machine learning, algorithms are used to correctly classify a piece of data or make correct predictions by examining other data provided. There are three broad stratifications: supervised machine learning, unsupervised machine learning, and neural networks (Fig. 1).
Fig. 1. Types of machine learning.
Examples of only a few types of machine learning, including subcategories of supervised and unsupervised machine learning.
Supervised machine learning
Supervised machine learning is the most prevalent in medicine.11–19 In supervised learning, labeled datasets are used to train an algorithm to correctly classify data.11–19 To train an algorithm, the labeled data is used to deduce any association between independent and dependent variables. The respective weights of independent variables are adjusted within the algorithm until it arrives at the best fit that has the least error in predicting the dependent variable. The trained model derived from this process is then validated on additional datasets to assess the generalizability of the algorithm. Algorithm performance can be assessed in several ways including sensitivity, accuracy, and area under the curve of a receiver operating characteristic curve20 (Table 1).
Table 1.
Common Measures of Model Performance.
| Description | Advantages | Limitations | |
|---|---|---|---|
| Accuracy | Ratio of correct predictions to total number of predictions made. (TP + TN/Total) | Easy to understand, works well if there are an equal number of samples in each class | Does not represent a clinically meaningful number if the classes are unbalanced, or if there is a high cost of misclassification (rare but fatal disease) |
| Precision (positive predictive value) |
Number of correct positives divided by number of positive test results TP/(TP + FP) |
Gives information about performance with respect to false positives. Goal is to minimize false positives | No information about false negatives |
| Sensitivity/recall |
Number of correct positive results divided by all that are actually positive TP/(TP + FN) |
Gives information about performance with respect to false negatives. Goal is to minimize false negatives | No information about false positives |
| Specificity | Number of correct negative results divided by all that are actually negative TN/(FP + TN) | Useful to characterize the rate of true negatives compared to predicted negatives | No information about true positives. |
| F1 score |
Measure of the accuracy of a test– represents a harmonic mean between precision and recall. F1 = 2 × ((Precision × Recall)/(Precision + Recall)) |
Balances both precision and recall, for instance a high precision with low recall may have a high accuracy but would have a lower F1 score | Harder to calculate than an arithmetic mean, may be difficult to interpret unless familiar with the concept |
| Mean absolute error | Average of difference between original values and predicted values | Measure of the distance between prediction and actual input | Does not give any insight into direction of error (under or over prediction) |
| Mean squared error | Similar to mean absolute error but takes square of the difference | Easier to compute gradient differences | Can emphasize the effect of larger errors over smaller errors |
| Logarithmic loss | Classifier must assign probabilities for each prediction. Penalizes false classifications. Values closer to 0 indicate higher accuracy | Very strong if many observations | Weak if few observations. Maximizing log loss may lead to better probability estimation but at cost of accuracy |
| Area under receiver operator curve (AUROC) | Probability a true positive will have a higher predicted probability than true negative across all thresholds | Useful for discrimination. Helpful to visually assess performance over range of thresholds, useful to compare across models. Higher is better (1.0 = perfect) | Not clinically relevant, can be biased if classes are unbalanced |
| Area under precision-recall curve (AUPRC) | Average probability that a positive prediction will be true across all sensitivities | Useful for discrimination. Reflects overall probability that a positive prediction is a true positive. Higher is better (1.0 = perfect). Better positioned in rare events than AUROC, and helpful to visually assess performance | May be difficult to interpret, some performance is also graphed at clinically irrelevant regions |
| Hosmer–Lemeshow | Observed probability vs predicted probability across varying ranges of prediction | Useful for assessing model calibration. Visually represents the data, allows easy observation of areas where the model may have poor performance | Groupings of ranges are arbitrary. May struggle with smaller datasets |
| Scaled Brier’s Score | Squared difference between observations and prediction, scaled to account for the event rate | Explains variance so useful for both discrimination and calibration. Higher is better (1.0 = perfect). Good measure of overall predictive performance | Does not give information about individual predictions and may represent performance at clinically irrelevant regions |
Supervised machine learning methods are often task-driven and can complete classification, and regression tasks (Fig. 1). Classification is used when the outcome of interest is a categorical variable (alive/dead, high risk/low risk, etc.). The model uses the independent variables in the labeled dataset to determine the category of the dependent variable. A commonly used example of supervised machine learning is training a model to relate a patient’s demographics and smoking history to a certain outcome such as lung cancer.21 Commonly used algorithms for classification include logistic regression, k-nearest neighbors, decision trees, gradient boosting, support vector machines, and naive Bayes algorithms.9 Regression is used to predict a numerical value for the dependent variable. These models produce continuous outcomes and have been used outside medicine to predict house prices, stock prices, or sales. Commonly used algorithms for regression include linear regression, lasso regression, polynomial regression, support vector regression, random forest algorithms, and boosted as well as ensemble methods.22
In pediatrics, supervised machine learning is commonly used for prognostic predictions. In prognostic models, the algorithm is used for risk stratification for outcomes of interest.23–26 Examples of this include using machine learning to determine the risk of serious bacterial infection in a cohort of children in the emergency department,24 to determine if a subgroup of critically ill patients would be more likely to benefit from corticosteroids,23 and to determine the risk of developing childhood asthma.26
Predictive modeling can be used to predict whether a patient responds to treatment or is at risk for clinical deterioration.27–30 In the hospital setting, real-time or recent clinical data can be used as a decision-support tool to alert clinicians to subtle signs of clinical deterioration that can be acted on prior to decompensation. Examples of this include using algorithms to detect the need for transfer to an intensive care unit (ICU)29,30 and to detect deterioration of children in the cardiac ICU.13
Unsupervised machine learning
Unsupervised machine learning is used to find previously undetected patterns and clusters in unlabeled data (Fig. 1).11–15,31–35 Unsupervised machine learning may serve as a data exploration tool as it requires less manual intervention as it involves unlabeled data. While these techniques can yield previously undiscovered patterns, the groupings may not necessarily be clinically meaningful without clinician insight. In addition to data exploration, unsupervised machine learning can also be used for classification tasks.
Examples of unsupervised machine learning include cluster analysis; where data is grouped based on similarities, differences, and associations. Dimensionality reduction can be used in large datasets in the preproduction phase and reduces the number of variables while preserving the integrity of the dataset, making it more manageable for analysis. Common clustering algorithms include latent class or profile analysis, k-means clustering, and hierarchical clustering.
Latent class or profile analysis is the most used unsupervised machine learning technique in pediatric research.32–35 Latent class or profile analysis allows the detection of a possible unmeasured group within a population by inferring patterns or indicators from the observed variables.36 This differs from cluster analysis which uses a distance from a specific measure to assign grouping, while latent class or profile analysis estimates the probability of each unit belonging to a class.36 Recent reanalysis using latent class analysis of the RESTORE (Randomized Evaluation of Sedation Titration for Respiratory Failure) and BALI (Biomarkers in Children with Acute Lung Injury) studies have shown that while patients may fit under a unifying definition of pediatric acute respiratory distress syndrome (ARDS) within these groups there may be hypoinflammatory and hyperinflammatory phenotypes.33 Adult literature has shown that similar phenotypes in ARDS have varying responses to targeted therapies (e.g., Positive End Expiratory Pressure (PEEP), fluid overload, statins).37–39 Researchers have also used it to identify phenotypes in critically ill children with sepsis34 and near-fatal asthma.35
Neural networks
Artificial neural networks are another type of machine learning modeling and take inspiration from biological neural networks; they may be supervised or unsupervised. They are most comparable to gradient boosting methods and are a popular classifier algorithm.9 They consist of layers of neurons, with an input layer, one or more hidden layers, and an output layer. Input layers typically consist of input variables such as physiologic or lab markers, and hidden layers have a function applied (a series of calculations including weighing or combining input variables) to predict the output layer.40 Two common types of neural networks include recurrent neural networks—which can process large amounts of data and “learn” from missed predictions and convolutional neural networks which specialize in transforming imaging data.40 While several applications have been published,41–43 historic limitations include their “black box” nature and difficulty in determining clinical importance. Recent advances such as detector randomized input sampling or generative adversarial networks have substantially reduced the “black box” nature of neural networks, these techniques have allowed researchers to even determine which portions of an x-ray were important to an algorithm in predicting if an image belonged to a COVID-19 positive or negative patient.44,45
Predictive modeling techniques
There are several widely used illness severity scores in pediatric critical care that were developed over the past four decades using traditional approaches. The first widespread physiology-based scoring system to assess the risk of mortality in critically ill children was the Physiologic Stability Index (PSI) which was published in 1984.46 The same group of investigators simplified the PSI into the Pediatric Risk of Mortality (PRISM) score several years later, which improved usability by reducing the number of variables from 34 to 14.47 Another group developed the Pediatric Index of Mortality (PIM) in 1996.48 Also based on the PSI, the PIM score only required eight variables present within the first hour of PICU care. These scores have undergone serial refinement to the PRISM IV and PIM3 scores by adjusting what variables are included, their cut-offs, and their weights.49,50 Because mortality in the PICU is uncommon, other illness severity scores like the Pediatric Logistic Organ Dysfunction (PELOD) score, PELOD-2 score, and Pediatric Sequential Organ Failure Assessment (pSOFA) score are intended to quantify organ dysfunction. The pediatric organ dysfunction information update mandate (PODIUM) developed contemporary criteria to define pediatric single- and multi-organ dysfunction.51 The panel of 88 content experts from 47 institutions appraised the body of present-day peer-reviewed evidence defining pediatric organ failure for 11 organ systems. The goals of this endeavor are to promote early recognition and appropriate treatment of pediatric organ dysfunction to create a globally accepted platform for universal nomenclature, promoting enhanced multi-institutional collaborative research.
There are inherent limitations to these scores and the methods used to develop them. While PRISM and PIM relied on variables already established as predictive in the PSI score, the PSI variables themselves were selected subjectively via the consensus of “a group of pediatric intensivists”. Similarly, variables for PELOD were chosen by the Delphi method and in PODIUM the final variables were voted on by the panel of content experts after being selected through a rigorous examination of the literature. While expert consensus does identify variables associated with outcomes of interest, it is inherently limited in scope and prone to bias. Many of the variables themselves are single values of continuous variables (e.g., heart rate), with the “worst” value in a specified time range being used for scoring. Improved methods to identify and weigh variables could enable predictive scores to be improved for use on cohorts and refined sufficiently for use on individual patients. Many of these scores have also been developed to describe outcomes and stratify the severity of illness across the population or individual intensive care unit level and may be limited in their ability for individual patient prediction.
A large reason for the paucity of widely recognized and validated pediatric predictive tools includes the low mortality rate and substantially lower numbers of patients compared with critically ill adults. Many in the field are moving away from developing additional tools to predict mortality or define the severity of specific dysfunctional organ systems. There is now momentum targeting more nuanced outcomes such as disease trajectory, clinical deterioration during hospitalization, and the development of new cognitive or physical disability. Utilizing modern monitoring systems through machine learning and AI, the field is rapidly advancing towards higher-level predictive modeling that will likely soon become standard to the care of critically ill pediatric patients. Several single-center prospective and retrospective studies have recently been published underscoring the importance of advancing this field in addition to highlighting the significant momentum building worldwide. Several groups have started to define machine learning algorithms to predict the development of sepsis or septic shock in pediatric inpatients.52–54 This is in addition to the use of machine learning in pediatrics for predicting 30-day readmissions,55 need for massive transfusion following blunt trauma,56 risk of cerebral hemorrhage in preterm infants,57 and early prediction of AKI.58,59 Recent publications also include utilizing machine learning to predict the absence of serious bacterial infection at the time of pediatric intensive care unit admission, with a goal to reduce antibiotic days per patient.60 Finally, machine learning is being utilized to predict long-term neurologic outcomes in pediatric traumatic brain injury patients.61–63 Ultimately, the utilization of dynamic trends in physiologic data and changes in laboratory values over time, together with high fidelity machine learning algorithms, will provide a more robust and fertile landscape for outcome prediction in the critically ill pediatric patient.
Clinical decision support
The widespread adoption of electronic health records (EHR) has been followed by the increased development of clinical decision support systems (CDSS). These systems range from medication interaction alerts to patient safety reminders.64–67 CDSS have been demonstrated to improve process measures and clinical outcomes.68,69 The utilization of machine learning algorithms for CDSS is more recent and rapidly expanding.
Prediction models have been the most implemented modality of machine learning in clinical medicine.70–72 These models use machine learning techniques to synthesize large amounts of patient data into simplified scores that providers can use to assess each patient’s risk.73 The clinical application of these models includes the prediction of kidney injury,74 significant clinical deterioration,75 and mortality.76,77 These models are frequently derived using single-center data with validation performed on a separate cohort of patients admitted to the same center.74–77 The rise in popularity of similar models has led to calls for greater rigor in their derivation to ensure true clinical utility.78 Another group of models includes those created by EHR vendors that are available to use in hospitals that are paying for a particular EHR. While these models tend to be derived from larger datasets, their proprietary nature leads to limited information being published regarding their validation. Attempts at external validation have raised concerns about the validity of these models.79,80
A different approach to CDSS is the augmentation of data visualization to assist physicians in better understanding trends in real time. One system that utilizes this approach is the Etiometry (Etiometry Inc., Boston, MA) risk analytics algorithm81 that includes a data aggregation and visualization system in addition to a risk analytics engine. The T3 (tracking, trajectory, and triggering) data visualization system continuously aggregates real-time patient data including vital signs and select lab data. Similarly, Sickbay (Medical Informatics Corp., Houston, TX) is a vendor-neutral platform that aggregates data to improve data visualization.82 This contrasts with conventional data monitoring in the ICU which is limited to nurse-validated recordings at fixed time intervals. Current EHRs usually store and present data at hourly intervals. These models aggregate all data points continuously and therefore attempt to provide a more holistic picture.83
Some platforms use algorithms to provide additional functionality beyond the aggregation of patient data. Sickbay allows the use of continuous physiologic data to develop real-time risk calculators for outcomes. Published examples of this include predicting deterioration in children with congenital heart disease28 and predicting the need for extracorporeal membrane oxygenation (ECMO) in neonates with congenital diaphragmatic hernias.84 In contrast, Etiometry leverages continuous physiologic data through a proprietary machine learning algorithm to provide real-time risk-based analysis of patient deterioration. The algorithm uses patient data to continuously calculate the risk of inadequate delivery of oxygen (IDO2) and inadequate ventilation of carbon dioxide (IVCO2) that can be used as proxies to predict clinical deterioration. To their credit, the development of these metrics has been described in detail, providing users with an in-depth understanding of their workings.85 Publications testing the accuracy and utility of Etiometry models have had mixed results. One study demonstrated that the IDO2 index was significantly elevated in patients who failed to be weaned off vasoactive infusions compared to those who were weaned successfully.86 Another study found that the IDO2 index was outperformed by a conventional scoring system in predicting adverse events in children after cardiac bypass.87
Other examples include an algorithm developed by Better Care (Better Care Inc., Barcelona, Spain) that is currently being used in Spain88 and the Continuous Monitoring of Event Trajectories (CoMET) developed at the University of Virginia.89 In addition to using patient vital signs, the Better Care algorithm also incorporates ventilator metrics and analytics such as asynchrony in its features. This is built on previous efforts by the same company to utilize machine learning to categorize patient-ventilator interactions.90 CoMET combines continuous physiologic data and lab values in its algorithm to predict the risk of urgent intubation and assist in the early detection of sepsis.91 However, there are currently no published reports describing the performance of this system in an external pediatric cohort.
While the development of CDSS has been promising, there are challenges that have limited widespread adoption.92 Limitations include the knowledge and time required to deploy and maintain an update-to-date and clinically relevant CDSS. A good CDSS must further be well-integrated into the clinician’s existing workflow. Another area of concern is transparency and understanding. The workings of a CDSS must be transparent to garner trust. Providers are answerable to their patients and are unlikely to utilize a metric in their decision-making that they do not fully understand, and therefore cannot explain to their patients. Finally, the CDSS must continue to adapt to changes in both clinical guidelines and practice patterns. A stagnant CDSS that is created based on a historical dataset and not updated will lose accuracy and eventually become redundant. This concept is known as data drift, and updates to a CDSS are required if the data distribution passes a prespecified threshold.93 These systems must therefore be reviewed and updated regularly, either at fixed intervals or with significant changes in practice or the target population to remain relevant.94 Additional limitations that are specific to a pediatric population include the smaller patient population and fewer large datasets available to train algorithms.95 Within that smaller population, there is also greater heterogeneity due to the differences in normal ranges and at times treatment strategies by age group. Furthermore, adverse outcomes in pediatric patients tend to be more infrequent compared to adults. This limits the accuracy of a model derived to predict those rare outcomes. The financial consideration of implementing CDSS can be significant, limiting the widespread adoption of tools that are yet unproven in clinical efficacy. These limitations were highlighted in a recent study in which most pediatric critical care providers were neutral or disagreed that current predictive algorithms provided useful information.96 Providers agreed that important goals included evidence-based CDSS with a proven impact on patient safety, that were well-placed and delivered at the right time.96 Providers expressed concern about the accuracy of CDSS, the effect on practitioners’ critical reasoning, and the burden of increased time spent on the computer.96
Limitations
Several machine learning and AI-derived tools have failed to live up to their promise when deployed clinically. It is crucial to understand the pitfalls in their development and implementation, and why so many have struggled to make an impact at the bedside. Specific examples range from sepsis prediction to imaging classification.79,97,98
During development particular focus needs to be paid to the definitions and scope of the model, and the selection of predictor variables. Several important characteristics must be true, the predictors must not have collinearity with the predicted outcome and be known prior to the outcome.78 This is vital to ensure that the information provided by the model is clinically actionable. Predictors that become known either immediately before or after the outcome event occurs do not provide an opportunity for clinical intervention. Observable data including blood pressure or heart rate may be perturbed in sepsis only after the condition has developed, limiting any time to make actionable predictions.99 The model must also be able to retain accuracy when applied to new data and thus be generalizable to be useful to the bedside clinician.
Care should be further taken during data preprocessing, first to ensure data accuracy, and further to not disregard potentially useful information by binning continuous variables. Doing so often introduces assumptions into the model that are not biologically plausible (e.g., a model may treat a hemoglobin of 3 the same as 6.7 if a dichotomous predictor of hemoglobin <7 exists). While grouping variables may be useful for easy bedside prediction, we recommend close consideration of these tradeoffs when developing a model. Particular attention must also be paid to how specific models handle missing data. Extensive data preprocessing may yield better results but may dramatically limit real-world applications if the preprocessing is not able to be done in real time.
When developing models to evaluate binary outcomes, a key concept is the number of events per variable (EPV)—which represents the number of events/outcomes divided by the number of predictor variables.100 EPV may provide guidance on sample size requirements. Further attention is required in regression models to avoid overfitting. Overfitting occurs when the model begins to describe the random error in the data rather than the true relationship between variables—this often occurs as the model becomes more complex and reduces generalizability outside the original dataset.
When evaluating models, it is important to determine how a model has been evaluated and validated. Standard evaluation frequently includes internal validation, which is determining if model performance is reproducible in the same population it was derived from. This frequently means the same dataset and can be performed by either holding out a particular set of patients for model validation, or k-fold cross-validation. K-fold cross-validation generally includes partitioning the data into subsamples, the model is then trained on all subsamples except one and validated on the remaining subsample (Fig. 2).78 The data are then shuffled, and this is process is repeated until a stable model is derived. A rare outcome limits model performance since most mathematical models are designed to distinguish between two outcomes (i.e., event vs. no event) of equal likelihood. This is particularly important when considering applying machine learning to predict rare outcomes in pediatric patients. External validation determines if the model is reproducible in a distinct population from the one it was trained on. Models trained on single-center data often demonstrate high accuracy but fail to perform similarly in new populations, again limiting their generalizability.
Fig. 2. K-fold cross-validation.
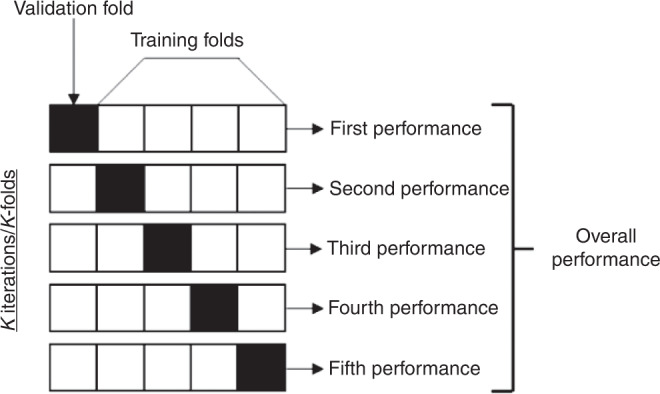
Example of K-fold cross validation. A different subset of the data is used for training and validation in each fold, and performance is based off combined performance in the validation folds.
Performance metrics for machine learning models also differ from common statistical models (Table 1). It is crucial to separate a model’s discrimination – its ability to separate events/outcomes from non-events/outcomes (e.g., Fig. 3), from its calibration—which is its ability to specify the probability of the outcome. Common measures such as area under the receiver operating characteristic curve (AUROC) are mainly a measure of discrimination and may be falsely high when predicting rare events. For rare events, it is also important to consider a series of metrics, including the area under the precision-recall curve (AUPRC), which reflects positive predictive value and sensitivity (the average probability that a positive prediction is true across all sensitivities). Other metrics may also provide further clarity, especially in rare events on a model’s performance, including its specificity, sensitivity, and F1 score which combines the precision and recall into a single metric.101 Understanding the calibration of a model is also crucial in knowing if the model will be useful clinically (e.g., there is a large difference between predicting an outcome will occur 51% of the time, or 90% of the time). Ultimately while AUROC and AUPRC curves are important, to the bedside clinician the important factors are the positive and negative predictive values for any algorithm, which will depend on the prevalence of the outcome in the local population.
Fig. 3. Confusion Matrix.
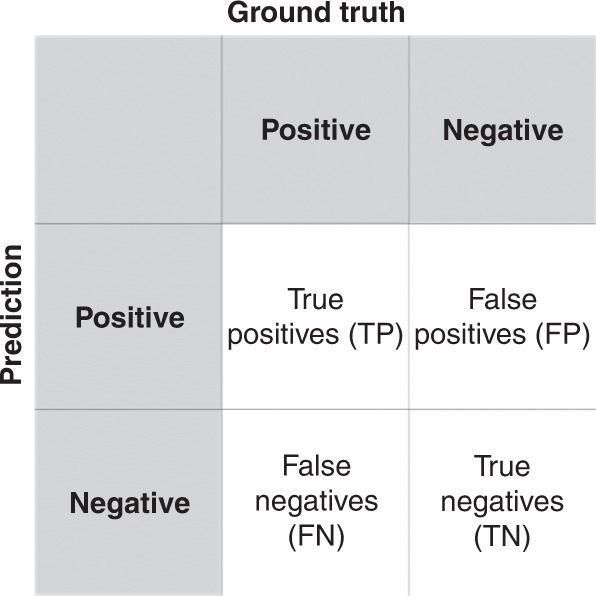
Example of a 2x2 confusion matrix, comparing prediction to ground truth.
A recent concern has been how the incorporation of any systematic bias in the underlying dataset may perpetuate the bias in the algorithm, especially if the algorithm is utilized for triage or treatment decisions. These risks can be understood in models where there is higher clinical correlation or plausibility but a recent paper has shown that a deep learning model was able to identify self-reported race from radiological images even when the data was corrupted or cropped, and this capability was incredibly difficult to isolate.102 Further work is necessary before the broad deployment of these types of models.
Along the same lines, it is important to also understand how provider beliefs and actions affect machine learning models.103 Actions performed by clinicians such as obtaining certain lab tests are based on prior knowledge of disease patterns or clinical intuition. Patients whom a clinician is more concerned about expectedly may have more labs or diagnostic tests performed. Machine learning models that incorporate the results of these actions may in fact be predicting provider behavior and not disease patterns. Conversely, models trained exclusively on patient data that is independent of provider actions (e.g., vital signs on admission) may provide a more distilled approximation of the disease process but not reflect the realities of patient care within the hospital system. Overall, machine learning models that are trained on both clinician-initiated and clinician-independent variables are likely to encompass both physician intuition and patient factors in their predictions. Pediatric critical care generates swaths of clinician-independent data that can be harnessed to train more models that are agnostic of provider behavior patterns and home in on the disease process.
In evaluating model performance then, it is important to consider the variables used in model training, how the model was validated (retrospective vs prospective), how performance was reported, and if performance included measures of discrimination, calibration, and clinically relevant measures such as positive or negative predictive values in a representative population. Educating clinicians on understanding these metrics and critically appraising machine learning models will be essential in ensuring successful adoption at the bedside.
For these reasons, the addition of machine learning and artificial intelligence algorithms to pediatric critical care is not meant to replace decision-making by bedside staff. Rather, it should augment the knowledge employed to develop individualized care plans for increasingly complex patients and will ultimately lead to improved nuance and discrimination of diverse phenotypes within organ system failures.
Conclusions
Common machine learning and artificial intelligence techniques hold promise in their applications in predictive modeling and clinical decision support however to be fully impactful to the field, common pitfalls that may explain why current tools have failed to live up to their promise must be considered for the field to mature. As these tools and techniques become ubiquitous, understanding how they were developed and how to evaluate them will be vital for pediatric intensivists.
Author contributions
All authors made substantial contributions to the conception and design, drafting and revision, and provided final approval of this manuscript.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jung M, et al. Age-specific distribution of diagnosis and outcomes of children admitted to ICUs: a population-based cohort study. Pediatr. Crit. Care Med. 2019;20:e301–e310. doi: 10.1097/PCC.0000000000001978. [DOI] [PubMed] [Google Scholar]
- 2.Crow SS, et al. Epidemiology of pediatric critical illness in a population-based birth cohort in Olmsted County, MN. Pediatr. Crit. Care Med. 2017;18:e137–e145. doi: 10.1097/PCC.0000000000001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epstein D, Brill JE. A history of pediatric critical care medicine. Pediatr. Res. 2005;58:987–996. doi: 10.1203/01.PDR.0000182822.16263.3D. [DOI] [PubMed] [Google Scholar]
- 4.Gupta P, Gossett J, Rao Rettiganti M. 60: Trends in mortality rates in pediatric intensive care units in the United States from 2004 to 2015. Crit. Care Med. 2018;46:30. doi: 10.1097/01.ccm.0000528115.33510.1b. [DOI] [Google Scholar]
- 5.Markovitz BP, Kukuyeva I, Soto-Campos G, Khemani RG. PICU volume and outcome: a severity-adjusted analysis. Pediatr. Crit. Care Med. 2016;17:483–489. doi: 10.1097/PCC.0000000000000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss SL, et al. Surviving sepsis campaign international guidelines for the management of septic shock and sepsis-associated organ dysfunction in children. Pediatr. Crit. Care Med. 2020;21:e52–e106. doi: 10.1097/PCC.0000000000002198. [DOI] [PubMed] [Google Scholar]
- 7.Kochanek PM, et al. Management of pediatric severe traumatic brain injury: 2019 consensus and guidelines-based algorithm for first and second tier therapies. Pediatr. Crit. Care Med. 2019;20:269–279. doi: 10.1097/PCC.0000000000001737. [DOI] [PubMed] [Google Scholar]
- 8.Helm JM, et al. Machine learning and artificial intelligence: definitions, applications, and future directions. Curr. Rev. Musculoskelet. Med. 2020;13:69–76. doi: 10.1007/s12178-020-09600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez G. Artificial intelligence in the intensive care unit. Crit. Care. 2020;24:101. doi: 10.1186/s13054-020-2785-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovejoy CA, Buch V, Maruthappu M. Artificial intelligence in the intensive care unit. Crit. Care. 2019;23:7. doi: 10.1186/s13054-018-2301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N. Engl. J. Med. 2019;380:1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez-Pinto LN, Luo Y, Churpek MM. Big data and data science in critical care. Chest. 2018;154:1239–1248. doi: 10.1016/j.chest.2018.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams JB, Ghosh D, Wetzel RC. Applying machine learning to pediatric critical care data. Pediatr. Crit. Care Med. 2018;19:599–608. doi: 10.1097/PCC.0000000000001567. [DOI] [PubMed] [Google Scholar]
- 14.Alanazi HO, Abdullah AH, Qureshi KN. A critical review for developing accurate and dynamic predictive models using machine learning methods in medicine and health care. J. Med. Syst. 2017;41:69. doi: 10.1007/s10916-017-0715-6. [DOI] [PubMed] [Google Scholar]
- 15.Lonsdale H, Jalali A, Ahumada L, Matava C. Machine learning and artificial intelligence in pediatric research: current state, future prospects, and examples in perioperative and critical care. J. Pediatr. 2020;221S:S3–S10. doi: 10.1016/j.jpeds.2020.02.039. [DOI] [PubMed] [Google Scholar]
- 16.Choudhary, R. & Gianey, H. K. Comprehensive review on supervised machine learning algorithms. In 2017 International Conference on Machine Learning and Data Science (MLDS) 37–43 (2017).
- 17.Shafaf N, Malek H. Applications of machine learning approaches in emergency medicine; a review article. Arch. Acad. Emerg. Med. 2019;7:34. [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhury A, Rosenthal J, Waring J, Umeton R. Applying self-supervised learning to medicine: review of the state of the art and medical implementations. Informatics. 2021;8:59. doi: 10.3390/informatics8030059. [DOI] [Google Scholar]
- 19.Grogan KL, et al. A narrative review of analytics in pediatric cardiac anesthesia and critical care medicine. J. Cardiothorac. Vasc. Anesth. 2020;34:479–482. doi: 10.1053/j.jvca.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Casp. J. Intern. Med. 2013;4:627–635. [PMC free article] [PubMed] [Google Scholar]
- 21.Sidey-Gibbons JAM, Sidey-Gibbons CJ. Machine learning in medicine: a practical introduction. BMC Med. Res. Methodol. 2019;19:64. doi: 10.1186/s12874-019-0681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhai Q, et al. Using machine learning tools to predict outcomes for emergency department intensive care unit patients. Sci. Rep. 2020;10:20919. doi: 10.1038/s41598-020-77548-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong HR, et al. Combining prognostic and predictive enrichment strategies to identify children with septic shock responsive to corticosteroids. Crit. Care Med. 2016;44:e1000–e1003. doi: 10.1097/CCM.0000000000001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramgopal, S., Horvat, C. M., Yanamala, N. & Alpern, E. R. Machine learning to predict serious bacterial infections in young febrile infants. Pediatrics10.1542/peds.2019-4096 (2020). [DOI] [PMC free article] [PubMed]
- 25.Berger RP, et al. Derivation and validation of a serum biomarker panel to identify infants with acute intracranial hemorrhage. JAMA Pediatr. 2017;171:e170429. doi: 10.1001/jamapediatrics.2017.0429. [DOI] [PubMed] [Google Scholar]
- 26.Kothalawala DM, et al. Prediction models for childhood asthma: a systematic review. Pediatr. Allergy Immunol. 2020;31:616–627. doi: 10.1111/pai.13247. [DOI] [PubMed] [Google Scholar]
- 27.Kwon JM, et al. Deep learning algorithm to predict need for critical care in pediatric emergency departments. Pediatr. Emerg. Care. 2021;37:e988–e994. doi: 10.1097/PEC.0000000000001858. [DOI] [PubMed] [Google Scholar]
- 28.Rusin CG, et al. Prediction of imminent, severe deterioration of children with parallel circulations using real-time processing of physiologic data. J. Thorac. Cardiovasc. Surg. 2016;152:171–177. doi: 10.1016/j.jtcvs.2016.03.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, S. J. et al. Development and validation of a deep-learning-based pediatric early warning system: a single-center study. Biomed. J.10.1016/j.bj.2021.01.003 (2021). [DOI] [PMC free article] [PubMed]
- 30.Zhai H, et al. Developing and evaluating a machine learning based algorithm to predict the need of pediatric intensive care unit transfer for newly hospitalized children. Resuscitation. 2014;85:1065–1071. doi: 10.1016/j.resuscitation.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen B, et al. Mining tasks and task characteristics from electronic health record audit logs with unsupervised machine learning. J. Am. Med. Inf. Assoc. 2021;28:1168–1177. doi: 10.1093/jamia/ocaa338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy K, et al. Subphenotypes in critical care: translation into clinical practice. Lancet Respir. Med. 2020;8:631–643. doi: 10.1016/S2213-2600(20)30124-7. [DOI] [PubMed] [Google Scholar]
- 33.Dahmer, M. K. et al. Identification of phenotypes in paediatric patients with acute respiratory distress syndrome: a latent class analysis. Lancet Respir. Med.10.1016/S2213-2600(21)00382-9 (2021). [DOI] [PMC free article] [PubMed]
- 34.Zhang Z, Zhang G, Goyal H, Mo L, Hong Y. Identification of subclasses of sepsis that showed different clinical outcomes and responses to amount of fluid resuscitation: a latent profile analysis. Crit. Care. 2018;22:347. doi: 10.1186/s13054-018-2279-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolli S, et al. 973: latent class analysis of pediatric patients with near-fatal asthma. Crit. Care Med. 2021;49:484. doi: 10.1097/01.ccm.0000729780.23521.f5. [DOI] [Google Scholar]
- 36.Sinha P, Calfee CS, Delucchi KL. Practitioner’s guide to latent class analysis: methodological considerations and common pitfalls. Crit. Care Med. 2021;49:e63–e79. doi: 10.1097/CCM.0000000000004710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calfee CS, et al. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir. Med. 2018;6:691–698. doi: 10.1016/S2213-2600(18)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Famous KR, et al. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am. J. Respir. Crit. Care Med. 2017;195:331–338. doi: 10.1164/rccm.201603-0645OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calfee CS, et al. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir. Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A, F., Shah, N., Z, W. & Raman, L. Machine learning: Brief overview for biomedical researchers. J. Transl. Sci.10.15761/JTS.1000343 (2020).
- 41.Meyer A, et al. Machine learning for real-time prediction of complications in critical care: a retrospective study. Lancet Respir. Med. 2018;6:905–914. doi: 10.1016/S2213-2600(18)30300-X. [DOI] [PubMed] [Google Scholar]
- 42.Kamaleswaran R, et al. Applying artificial intelligence to identify physiomarkers predicting severe sepsis in the PICU. Pediatr. Crit. Care Med. 2018;19:e495–e503. doi: 10.1097/PCC.0000000000001666. [DOI] [PubMed] [Google Scholar]
- 43.Shah, N. et al. Neural networks to predict radiographic brain injury in pediatric patients treated with extracorporeal membrane oxygenation. J. Clin. Med.10.3390/jcm9092718 (2020). [DOI] [PMC free article] [PubMed]
- 44.DeGrave, A. J., Janizek, J. D. & Lee, S. I. AI for radiographic COVID-19 detection selects shortcuts over signal. Nat. Mach. Intell. 3, 610–619 (2021).
- 45.Savage, N. Breaking into the black box of artificial intelligence. Nature10.1038/d41586-022-00858-1 (2022). [DOI] [PubMed]
- 46.Yeh TS, Pollack MM, Ruttimann UE, Holbrook PR, Fields AI. Validation of a physiologic stability index for use in critically ill infants and children. Pediatr. Res. 1984;18:445–451. doi: 10.1203/00006450-198405000-00011. [DOI] [PubMed] [Google Scholar]
- 47.Pollack MM, Ruttimann UE, Getson PR. Pediatric risk of mortality (PRISM) score. Crit. Care Med. 1988;16:1110–1116. doi: 10.1097/00003246-198811000-00006. [DOI] [PubMed] [Google Scholar]
- 48.Shann F, Pearson G, Slater A, Wilkinson K. Paediatric index of mortality (PIM): a mortality prediction model for children in intensive care. Intensive Care Med. 1997;23:201–207. doi: 10.1007/s001340050317. [DOI] [PubMed] [Google Scholar]
- 49.Straney L, et al. Paediatric index of mortality 3: an updated model for predicting mortality in pediatric intensive care. Pediatr. Crit. Care Med. 2013;14:673–681. doi: 10.1097/PCC.0b013e31829760cf. [DOI] [PubMed] [Google Scholar]
- 50.Pollack MM, et al. The Pediatric Risk of Mortality Score: update 2015. Pediatr. Crit. Care Med. 2016;17:2–9. doi: 10.1097/PCC.0000000000000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bembea MM, et al. Pediatric Organ Dysfunction Information Update Mandate (PODIUM) contemporary organ dysfunction criteria: executive summary. Pediatrics. 2022;149:S1–S12. doi: 10.1542/peds.2021-052888B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spaeder MC, et al. Predictive analytics in the pediatric intensive care unit for early identification of sepsis: capturing the context of age. Pediatr. Res. 2019;86:655–661. doi: 10.1038/s41390-019-0518-1. [DOI] [PubMed] [Google Scholar]
- 53.Liu R, et al. Prediction of impending septic shock in children with sepsis. Crit. Care Explor. 2021;3:e0442. doi: 10.1097/CCE.0000000000000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott HF, et al. Development and validation of a predictive model of the risk of pediatric septic shock using data known at the time of hospital arrival. J. Pediatr. 2020;217:145.e6–151.e6. doi: 10.1016/j.jpeds.2019.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou H, Albrecht MA, Roberts PA, Porter P, Della PR. Using machine learning to predict paediatric 30-day unplanned hospital readmissions: a case-control retrospective analysis of medical records, including written discharge documentation. Aust. Health Rev. 2021;45:328–337. doi: 10.1071/AH20062. [DOI] [PubMed] [Google Scholar]
- 56.Shahi N, et al. Decision-making in pediatric blunt solid organ injury: a deep learning approach to predict massive transfusion, need for operative management, and mortality risk. J. Pediatr. Surg. 2021;56:379–384. doi: 10.1016/j.jpedsurg.2020.10.021. [DOI] [PubMed] [Google Scholar]
- 57.Turova V, et al. Machine learning models for identifying preterm infants at risk of cerebral hemorrhage. PLoS ONE. 2020;15:e0227419. doi: 10.1371/journal.pone.0227419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandokji I, et al. A time-updated, parsimonious model to predict AKI in hospitalized children. J. Am. Soc. Nephrol. 2020;31:1348–1357. doi: 10.1681/ASN.2019070745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong J, et al. Machine learning model for early prediction of acute kidney injury (AKI) in pediatric critical care. Crit. Care. 2021;25:288. doi: 10.1186/s13054-021-03724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin, B., DeWitt, P. E., Scott, H. F., Parker, S. & Bennett, T. D. Machine learning approach to predicting absence of serious bacterial infection at PICU admission. Hosp. Pediatr.10.1542/hpeds.2021-005998 (2022). [DOI] [PMC free article] [PubMed]
- 61.Kayhanian S, et al. Modelling outcomes after paediatric brain injury with admission laboratory values: a machine-learning approach. Pediatr. Res. 2019;86:641–645. doi: 10.1038/s41390-019-0510-9. [DOI] [PubMed] [Google Scholar]
- 62.Tunthanathip T, Oearsakul T. Application of machine learning to predict the outcome of pediatric traumatic brain injury. Chin. J. Traumatol. 2021;24:350–355. doi: 10.1016/j.cjtee.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daley M, et al. Pediatric severe traumatic brain injury mortality prediction determined with machine learning-based modeling. Injury. 2022;53:992–998. doi: 10.1016/j.injury.2022.01.008. [DOI] [PubMed] [Google Scholar]
- 64.The Office of the National Coordinator for Health Information Technology (ONC). Clinical decision support. https://www.healthit.gov/topic/safety/clinical-decision-support (2018).
- 65.Muylle KM, Gentens K, Dupont AG, Cornu P. Evaluation of an optimized context-aware clinical decision support system for drug-drug interaction screening. Int. J. Med. Inf. 2021;148:104393. doi: 10.1016/j.ijmedinf.2021.104393. [DOI] [PubMed] [Google Scholar]
- 66.Lytle KS, Short NM, Richesson RL, Horvath MM. Clinical decision support for nurses: a fall risk and prevention example. Comput. Inf. Nurs. 2015;33:530–537. doi: 10.1097/CIN.0000000000000192. [DOI] [PubMed] [Google Scholar]
- 67.Haroz EE, et al. Designing a clinical decision support tool that leverages machine learning for suicide risk prediction: development study in partnership with Native American care providers. JMIR Public Health Surveill. 2021;7:e24377. doi: 10.2196/24377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwan JL, et al. Computerised clinical decision support systems and absolute improvements in care: meta-analysis of controlled clinical trials. BMJ. 2020;370:m3216. doi: 10.1136/bmj.m3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bright TJ, et al. Effect of clinical decision-support systems: a systematic review. Ann. Intern. Med. 2012;157:29–43. doi: 10.7326/0003-4819-157-1-201207030-00450. [DOI] [PubMed] [Google Scholar]
- 70.Peiffer-Smadja N, et al. Machine learning for clinical decision support in infectious diseases: a narrative review of current applications. Clin. Microbiol. Infect. 2020;26:584–595. doi: 10.1016/j.cmi.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 71.Buchlak QD, et al. Machine learning applications to clinical decision support in neurosurgery: an artificial intelligence augmented systematic review. Neurosurg. Rev. 2020;43:1235–1253. doi: 10.1007/s10143-019-01163-8. [DOI] [PubMed] [Google Scholar]
- 72.Fernandes M, et al. Clinical decision support systems for triage in the emergency department using intelligent systems: a review. Artif. Intell. Med. 2020;102:101762. doi: 10.1016/j.artmed.2019.101762. [DOI] [PubMed] [Google Scholar]
- 73.Handelman GS, et al. eDoctor: machine learning and the future of medicine. J. Intern. Med. 2018;284:603–619. doi: 10.1111/joim.12822. [DOI] [PubMed] [Google Scholar]
- 74.Sanchez-Pinto LN, Khemani RG. Development of a prediction model of early acute kidney injury in critically ill children using electronic health record data. Pediatr. Crit. Care Med. 2016;17:508–515. doi: 10.1097/PCC.0000000000000750. [DOI] [PubMed] [Google Scholar]
- 75.Mayampurath A, et al. A vital sign-based model to predict clinical deterioration in hospitalized children. Pediatr. Crit. Care Med. 2020;21:820–826. doi: 10.1097/PCC.0000000000002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aczon MD, Ledbetter DR, Laksana E, Ho LV, Wetzel RC. Continuous prediction of mortality in the PICU: a recurrent neural network model in a single-center dataset. Pediatr. Crit. Care Med. 2021;22:519–529. doi: 10.1097/PCC.0000000000002682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwizera A, et al. A machine learning-based triage tool for children with acute infection in a low resource setting. Pediatr. Crit. Care Med. 2019;20:e524–e530. doi: 10.1097/PCC.0000000000002121. [DOI] [PubMed] [Google Scholar]
- 78.Leisman DE, et al. Development and reporting of prediction models: guidance for authors from editors of Respiratory, Sleep, and Critical Care journals. Crit. Care Med. 2020;48:623–633. doi: 10.1097/CCM.0000000000004246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wong A, et al. External validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern. Med. 2021;181:1065–1070. doi: 10.1001/jamainternmed.2021.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hwang AB, Schuepfer G, Pietrini M, Boes S. External validation of EPIC’s Risk of Unplanned Readmission model, the LACE+ index and SQLape as predictors of unplanned hospital readmissions: a monocentric, retrospective, diagnostic cohort study in Switzerland. PLoS ONE. 2021;16:e0258338. doi: 10.1371/journal.pone.0258338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Etiometry Inc. Etiometry, T3. https://www.etiometry.com/ (2022).
- 82.Medical Informatics Corp. Sickbay. https://michealthcare.com/sickbay/ (2022).
- 83.Sanchez Cordero, A. Wired. https://www.wired.co.uk/article/autodoctor-artificial-intelligence-healthcare (2017).
- 84.Cruz, S. M. et al. A novel multimodal computational system using near-infrared spectroscopy predicts the need for ECMO initiation in neonates with congenital diaphragmatic hernia. J. Pediatr. Surg.10.1016/j.jpedsurg.2017.10.031 (2017). [DOI] [PubMed]
- 85.Baronov D, McManus M, Butler E, Chung D, Almodovar MC. Next generation patient monitor powered by in-silico physiology. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2015;2015:4447–4453. doi: 10.1109/EMBC.2015.7319382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goldsmith MP, et al. Use of a risk analytic algorithm to inform weaning from vasoactive medication in patients following pediatric cardiac surgery. Crit. Care Explor. 2021;3:e0563. doi: 10.1097/CCE.0000000000000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rogers L, et al. The inadequate oxygen delivery index and low cardiac output syndrome score as predictors of adverse events associated with low cardiac output syndrome early after cardiac bypass. Pediatr. Crit. Care Med. 2019;20:737–743. doi: 10.1097/PCC.0000000000001960. [DOI] [PubMed] [Google Scholar]
- 88.BetterCare. Data processing. https://bettercare.es/#data-processing (2022).
- 89.Nihon Kohden Digital Health Solutions Inc. Continuous monitoring of event trajectories. https://amp3d.biz/comet/ (2021).
- 90.Blanch L, et al. Validation of the Better Care® system to detect ineffective efforts during expiration in mechanically ventilated patients: a pilot study. Intensive Care Med. 2012;38:772–780. doi: 10.1007/s00134-012-2493-4. [DOI] [PubMed] [Google Scholar]
- 91.UVAHealth Physician Resource. UVA Children’s at forefront of technologies that signal early illness, prevent death. https://www.uvaphysicianresource.com/predictive-monitoring-technology/ (2021).
- 92.Shortliffe EH, Sepúlveda MJ. Clinical decision support in the era of artificial intelligence. JAMA. 2018;320:2199–2200. doi: 10.1001/jama.2018.17163. [DOI] [PubMed] [Google Scholar]
- 93.Duckworth C, et al. Using explainable machine learning to characterise data drift and detect emergent health risks for emergency department admissions during COVID-19. Sci. Rep. 2021;11:23017. doi: 10.1038/s41598-021-02481-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Futoma J, Simons M, Panch T, Doshi-Velez F, Celi LA. The myth of generalisability in clinical research and machine learning in health care. Lancet Digit. Health. 2020;2:e489–e492. doi: 10.1016/S2589-7500(20)30186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moore MM, Slonimsky E, Long AD, Sze RW, Iyer RS. Machine learning concepts, concerns and opportunities for a pediatric radiologist. Pediatr. Radiol. 2019;49:509–516. doi: 10.1007/s00247-018-4277-7. [DOI] [PubMed] [Google Scholar]
- 96.Dziorny, A. C. et al. Clinical decision support in the PICU: implications for design and evaluation. Pediatr. Crit. Care Med.10.1097/PCC.0000000000002973 (2022). [DOI] [PubMed]
- 97.Roberts M, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021;3:199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- 98.Fleuren LM, et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020;46:383–400. doi: 10.1007/s00134-019-05872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goh KH, et al. Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare. Nat. Commun. 2021;12:711. doi: 10.1038/s41467-021-20910-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Austin PC, Steyerberg EW. Events per variable (EPV) and the relative performance of different strategies for estimating the out-of-sample validity of logistic regression models. Stat. Methods Med. Res. 2017;26:796–808. doi: 10.1177/0962280214558972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tharwat A. Classification assessment methods. Appl. Comput. Inform. 2021;17:168–192. doi: 10.1016/j.aci.2018.08.003. [DOI] [Google Scholar]
- 102.Gichoya JW, et al. AI recognition of patient race in medical imaging: a modelling study. Lancet Digit. Health. 2022;4:e406–e414. doi: 10.1016/S2589-7500(22)00063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Beaulieu-Jones BK, et al. Machine learning for patient risk stratification: standing on, or looking over, the shoulders of clinicians? NPJ Digit. Med. 2021;4:62. doi: 10.1038/s41746-021-00426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.