Abstract
Purpose:
Chemokines are essential for immune cell trafficking and are considered to have a major impact on the composition of the tumor microenvironment. CX-chemokine receptor 4 (CXCR4) is associated with poor differentiation, metastasis, and prognosis in pancreatic ductal adenocarcinoma (PDAC). This study provides a comprehensive molecular portrait of PDAC according to CXCR4 mRNA expression levels.
Experimental Design:
The Cancer Genome Atlas database was used to explore molecular and immunologic features associated with CXCR4 mRNA expression in PDAC. A large real-word dataset (n = 3,647) served for validation and further exploratory analyses. Single-cell RNA analyses on a publicly available dataset and in-house multiplex immunofluorescence (mIF) experiments were performed to elaborate cellular localization of CXCR4.
Results:
High CXCR4 mRNA expression (CXCR4high) was associated with increased infiltration of regulatory T cells, CD8+ T cells, and macrophages, and upregulation of several immune-related genes, including immune checkpoint transcripts (e.g., TIGIT, CD274, PDCD1). Analysis of the validation cohort confirmed the CXCR4-dependent immunologic TME composition in PDAC irrespective of microsatellite instability–high/mismatch repair–deficient or tumor mutational burden. Single-cell RNA analysis and mIF revealed that CXCR4 was mainly expressed by macrophages and T-cell subsets. Clinical relevance of our finding is supported by an improved survival of CXCR4high PDAC.
Conclusions:
High intratumoral CXCR4 mRNA expression is linked to a T cell– and macrophage-rich PDAC phenotype with high expression of inhibitory immune checkpoints. Thus, our findings might serve as a rationale to investigate CXCR4 as a predictive biomarker in patients with PDAC undergoing immune checkpoint inhibition.
Translational Relevance.
Chemokines are essential for immune cell trafficking and are considered to have a major impact on the composition of the tumor microenvironment (TME). CX-chemokine receptor 4 (CXCR4) is associated with poor differentiation, metastasis, and prognosis in pancreatic ductal adenocarcinoma (PDAC). Therefore, we aimed to provide novel insights of the molecular portrait and the TME of PDAC according to CXCR4 mRNA expression. Our in-depth characterization of two different cohorts revealed that PDAC samples harboring a high CXCR4 mRNA expression are associated with an upregulation of several immune-related genes, immune checkpoints, tumor-infiltrating cytotoxic CD8+ T cells, and macrophages. Single-cell RNA and immunofluorescence analyses indicated that CXCR4 is expressed on CD8+ T cells and correlated with cytotoxicity markers and exhaustion genes. High CXCR4 expression was linked to improved survival in PDAC. Collectively, our findings unravel novel molecular and immunologic insights, and might serve as a first impulse to investigate CXCR4 as a potential predictive biomarker in future clinical trials.
Introduction
The prognosis of pancreatic ductal adenocarcinoma (PDAC) remains poor with a 5-year overall survival (OS) of only 9% (1). Despite impressive advances in the field of immuno-oncology, checkpoint inhibition seems to be ineffective in the majority of patients with PDAC (2–4). Use of immune checkpoint inhibitors (ICI) as a therapeutic strategy is currently limited to the small fraction of patients with microsatellite instability–high (MSI-H) or mismatch repair–deficient (dMMR) chemo-refractory metastatic PDAC (5–7). Thus, chemotherapy remains the mainstay of treatment in metastatic PDAC (8). To improve outcomes in patients with metastatic PDAC, deeper insights on molecular mechanisms and the composition of the tumor microenvironment (TME) might unravel novel promising targets.
The process of pancreatic carcinogenesis is orchestrated by various cell types, including cancer, stromal, healthy pancreatic, and local as well as newly recruited immune cells, that all contribute to a delicate network of cellular communication and interaction regulated in part by chemokine ligands and receptors (9–12). Local chemokines gradients coordinate directed cellular movement, and are therefore considered to have a major impact on the cellular composition of the TME (13). The CX-chemokine receptor 4 (CXCR4) is a seven-transmembrane-spanning G-protein coupled cell-surface receptor, which binds the alpha chemokine CX-chemokine ligand 12 (CXCL12, also called stromal cell-derived factor 1 or SDF-1). The CXCR4/CXCL12 axis is active in multiple organs, especially in the bone marrow, lungs, liver, and spleen (14), and plays an essential role in physiologic processes and immune cell trafficking (15). It promotes chemotaxis, cell adhesion and migration, as well as cell proliferation and survival (16). In cancer, CXCR4 is frequently overexpressed in a variety of entities and has been associated with poor outcome (17). In PDAC, CXCR4 is overexpressed on cancerous when compared with normal nonneoplastic pancreatic tissue, and was linked to poor differentiation, metastasis, and prognosis (18–21).
Taking the current evidence into consideration, there is strong rationale to investigate CXCR4 as a potential therapeutic target in PDAC. For this reason, we aimed to explore the transcriptomic/genetic landscape, the TME, molecular features and the prognostic value according to CXCR4 mRNA expression in PDAC. We employed the publicly available The Cancer Genome Atlas (TCGA) dataset to explore the association of distinct molecular features with CXCR4 mRNA expression. In a second step, we evaluated a large cohort of 3,647 patients with PDAC to corroborate our findings. Finally, a publicly available single-cell dataset and multiplex immunofluorescence (mIF) were used to elaborate cellular distribution of CXCR4 expression in PDACs.
Materials and Methods
Analysis of the TCGA cohort
The analysis was done with R, version 4.2.0. A total of 147 primary PDAC samples from the TCGA Pancreatic adenocarcinoma (PAAD) cohort were analyzed (22). The normalized (level 3) RNA sequencing (RNA-seq) read count data were extracted and transformed with the log2 (FKPM + 1) function as described previously (23). Immune cell infiltration was estimated using the QuanTIseq method (24). The QuanTIseq algorithm is a computational method developed to analyze bulk RNA-seq data that quantifies ten different immune-cell types relevant for cancer immunology (25). Genes associated with T-cell exhaustion (26), cytotoxic responses (27), IFNγ, expanded immune (28), and tumor inflammation responses (29) were extracted from the respective published signatures. Gene signature scores were calculated with the gene set variation analysis (GSVA) algorithm (30).
Clustering of the TCGA cancer samples in respect to the QuanTIseq infiltration estimates was done with a combined self-organizing map (som() function, package kohonen, grid size for samples: 7 × 7, estimates: 4 × 4, sum-of-squares distance) and k-means algorithm (Manhattan distance, k = 3 clusters; refs. 31–33). The home-developed wrappers for clustering quality control and visualization are available at https://github.com/PiotrTymoszuk/clustTools.
The TCGA samples were stratified in CXCR4low and CXCR4high tumors based on the optimal expression cutoff displaying the largest difference in tumor-related OS determined by Mantel–Henszel test (23). Differences in GSVA gene signature scores and gene expression levels between the CXCR4 strata were assessed by Benjamini–Hochberg–corrected two-tailed t test (FDR; ref. 34). In the whole transcriptome differential gene expression analysis, significant differences were identified by pFDR < 0.05 and 1.5-fold regulation cutoffs. Signaling pathway modulation was investigated with spia() tool from SPIA package (35). Significantly modulated pathways were defined by the aggregated, Benjamini–Hochberg–corrected enrichment/perturbation P value (pGFDR) < 0.05 (34). The complete TCGA cohort analysis pipeline is available from GitHub (https://github.com/PiotrTymoszuk/CXCR4-TCGA).
Real-world patient cohort
A total of 3,647 PDAC samples were centrally submitted to a CLIA/CAP-certified laboratory (Caris Life Sciences; Phoenix, AZ). This study was conducted in accordance with the guidelines of the Declaration of Helsinki, Belmont report, and U.S. Common rule. In keeping with 45 CFR 46.101(b)(4), this study was performed using retrospective, de-identified clinical data. Therefore, this study is considered institutional review board—exempt and no patient consent was necessary from the subjects.
Next-generation sequencing
Next-generation sequencing (NGS) was performed on genomic DNA isolated from formalin-fixed, paraffin-embedded (FFPE) tumor samples using the NextSeq platform (Illumina, Inc., San Diego, CA). A custom-designed SureSelect XT assay was used to enrich 592 whole-gene targets (Agilent Technologies). All variants were detected with >99% confidence based on allele frequency and amplicon coverage, with an average sequencing depth of >500 coverage and an analytic sensitivity of 5%. Prior to molecular testing, manual microdissection techniques were used to insure that the specimen contained as much tumor tissue as possible.
mRNA expression (whole transcriptome sequencing)
CXCR4 expression was evaluated on mRNA isolated from FFPE tumor samples using the Illumina NovaSeq platform (Illumina, Inc., San Diego, CA) and Agilent SureSelect Human All Exon V7 bait panel (Agilent Technologies); transcripts per million (TPM) were reported. Gene fusions were detected using Illumina Novaseq platform as previously described (36). In addition, immune cell fractions were calculated by QuanTIseq (24) using transcriptomic data (25).
IHC
IHC was performed on FFPE tissue sections of glass slides. Slides were stained using automated staining techniques, according to the manufacturer's instructions, and were optimized and validated per CLIA/CAP and ISO requirements. The primary antibody clone used against programmed death-ligand 1 (PD-L1) was SP142 (Ventana Medical Systems, Inc., Tucson, AZ). The staining was regarded as positive if its intensity on the membrane of the tumor cells was ≥2+ and the percentage of positively stained cells was >5%.
Tumor mutational burden
Tumor mutational burden (TMB) was measured by counting all non-synonymous missense, nonsense, inframe insertion/deletion and frameshift mutations found per tumor that had not been previously described as germline alterations in dbSNP151, Genome Aggregation Database databases or benign variants identified by geneticists from Caris Life Sciences. A cut-off point of ≥10 mutations per MB (mt/MB) was used based on the KEYNOTE-158 pembrolizumab trial (7), which showed that patients with a TMB of ≥10 mt/MB across several tumor types had higher response rates than patients with a TMB of <10 mt/MB. Caris Life Sciences is a participant in the Friends of Cancer Research TMB Harmonization Project (37).
MSI and MMR genes
A combination of multiple test platforms was used to determine MSI-H/dMMR status of the tumors profiled, including fragment analysis (FA, Promega), IHC [MLH1, M1 antibody; MSH2, G2191129 antibody; MSH6, 44 antibody; and PMS2, EPR3947 antibody (Ventana Medical Systems, Inc.)] and NGS (for tumors tested with NextSeq platform, 7,000 target microsatellite loci were examined and compared with the reference genome hg19 from the University of California).
Single-cell RNA-seq analysis
The respective 10x Genomics matrix files from (Cellranger processed GSE154778 Dataset; ref. 38) of 10 patients with primary PDAC were imported in Scanpy version 1.8.0 (39) running with Python version 3.8. Each sample was QC-filtered for the occurrence of the gene in a minimum of 3 cells. To exclude low-quality cells (droplets) and duplets cell codes with less than 200 or more than 8,000 were filtered out. Moreover, cells with less than 2,000 total transcripts were filtered out as well as those having and more than 30% mitochondrial transcripts. Finally, cells passing quality control were stored as AnnData object (h5ad).
We used Combat (40) for batch correction, integration, and common embedding after dimension reduction for each patient sample. Cell types were identified and annotated by a set of cell type–specific markers such as, CD3E, NKG7, CD163, CDH5, EPCAM, KRT19, CD79A, ACTA2, COL1A1, and MKI67. Data analysis and graphical visualization was performed with scanpyv.1.8.0, anndata v.0.7.5, umap v.0.5.1, numpyv.1.19.2, scipy v.1.5.2, pandas v.1.1.3, scikit-learn v.0.24.2, statsmodels v.0.12.0, and python-igraph v.0.9.1.
mIF
Human PDAC samples (n = 6) were fixed in 4% paraformaldehyde and embedded in paraffin. Five-micrometer sections were used for the immunofluorescence staining. Immunofluorescence staining on FFPE tissue was performed using the Opal 7-Color Automated Immunohistochemistry Kit (cat: NEL821001KT, Akoya Biosciences). A multiplex panel of immune markers was developed with antibodies against: CXCR4 (clone EPUMBR3, cat: ab181020, dilution 1:300, Abcam), CD8 (clone C8/144B, cat: M710301–2, dilution 1:200, Dako/Agilent), CD68 (clone PG-M1, cat: M087601–2, dilution 1:400, Dako/Agilent), Cytokeratin (Cytokeratin 7, clone OV-TL, cat: M701801–2, dilution 1:500, Dako/Agilent + Cytokeratin 19, clone A53-B/A2.26, cat: 760–4281, RUI, Cell Marque/Roche). The staining procedure was performed using an automated staining system (BOND-RX; Leica Biosystems). All markers were sequentially applied and paired with respective Opal fluorophores. To visualize cell nuclei, the tissue was stained with 4‘,6-diamidino-2-phenylindole (spectral DAPI, Akoya Biosciences). Stained slides were scanned using Mantra 2 Quantitative Pathology Workstation (Akoya Biosciences) and representative images from each tissue were acquired with the Mantra Snap software version 1.0.4. Spectral unmixing, multispectral image acquisition was carried out using the InForm Tissue Analysis Software version 2.4.10 (Akoya Biosciences).
Statistics
Differences in immune cell levels between the immune infiltration clusters of the TCGA cohort were assessed by Kruskal–Wallis test, CXCR4 expression level differences between the clusters were investigated by one-way ANOVA. Association of CXCR4 transcript levels and QuanTIseq estimates was investigated with Spearman's correlation. Prevalence of molecular alterations among CXCR4 mRNA expression quartiles of the real-world cohort were analyzed using χ2 or Fisher exact tests. CXCR4 TPM distribution was analyzed using non-parametric Kruskal–Wallis testing. Similarly, TME cell fractions were analyzed among mRNA expression quartiles as described previously (24). A value of < 0.05 was considered a trending difference; P values were further corrected for multiple comparison using the Benjamini–Hochberg method to avoid type I error and an adjusted P value (q-value) of < 0.05 was considered a significant difference.
Real-world overall survival (rwOS) information was obtained from insurance claims data and calculated from first specimen collection to last contact. Kaplan–Meier estimates were calculated for molecularly defined patient cohorts. Kaplan–Meier plots were automatically generated by the CODEai software provided by CARIS Life Science. Significance was defined as a P value < 0.05.
Data availability
The deidentified sequencing data are owned by Caris Life Sciences. The datasets generated and analyzed during the current study are available from the authors upon reasonable request and with permission of Caris Life Sciences. Qualified researchers may contact the corresponding author with their request.
Results
CXCR4 expression is linked to the immunologic TME composition in the TCGA cohort
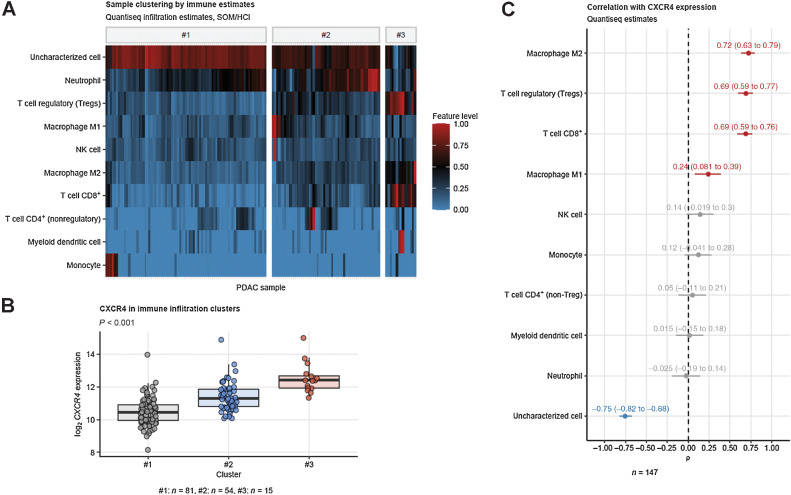
To investigate heterogeneity of CXCR4 mRNA expression in respect to quantity and quality of tumor stromal immune cell infiltration, we subjected the PDAC samples of the TCGA PAAD cohort (n = 147) to unsupervised clustering in respect to immune cell abundance estimates obtained with the QuanTIseq algorithm (24, 36). By means of combined self-organizing map and hierarchical clustering (Supplementary Fig. S1; ref. 31), three clusters of PDAC samples were identified differing significantly in the content of M1 and M2 tumor-associated macrophages (TAM), nonregulatory and regulatory (Treg) CD4+ T cells and cytotoxic CD8+ T cells as well as uncharacterized, bona fide malignant cells (Fig. 1A; Supplementary Fig. S2).
Figure 1.
Predominant mRNA expression of CXCR4 in CD8+ and TAM-rich PDACs. Tumor samples were clustered in respect to QuanTIseq immune infiltration estimates with a two-step self-organizing map/hierarchical clustering procedure (Supplementary Fig. S1). Differences in immune infiltration estimates and log2 CXCR4 expression between the three subsets were determined by one-way ANOVA and P values corrected for multiple testing with Benjamini–Hochberg method. A, Values of immune infiltration estimates in the #1, #2, and #3 tumor subsets presented as a heat map. B, log2 CXCR4 expression in the tumor subsets. P value for the expression difference is shown in the plot heading. N numbers of samples assigned to the tumor subsets are provided under the plot. C, Spearman correlation of the QuaTIseq immune infiltration estimates with CXCR4 mRNA expression. ρ correlation coefficients with 95% CIs are shown for the significant immune feature estimates. Red, positive correlation; blue, negative correlation. N, number of observations is indicated below the plot.
CXCR4 expression was the highest in the immune infiltration cluster #3 with the highest levels of CD8+ T cells, Treg and M2 TAMs (Fig. 1; Supplementary Fig. S2). The clustering results were corroborated by correlation analysis showing strong positive association of CXCR4 expression with M2 TAM, Treg, and CD8+ T cell estimates and strong negative association with levels of uncharacterized bona fide malignant cells (Fig. 1C).
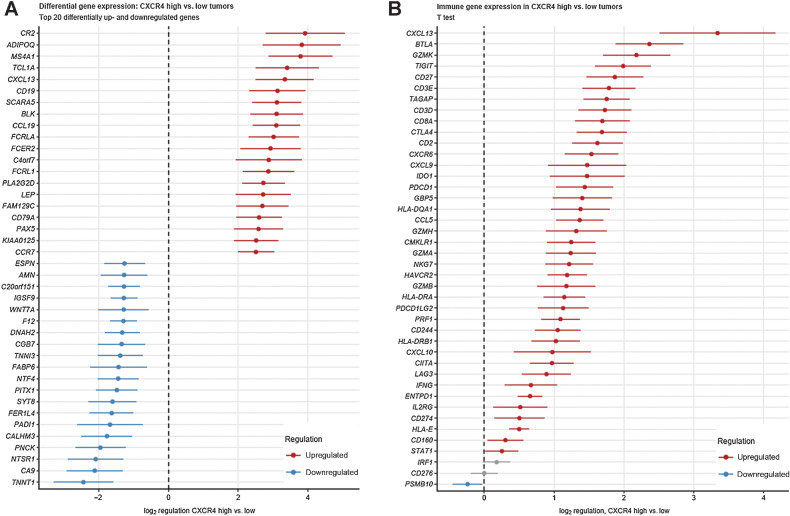
Next, we stratified the TCGA samples into CXCR4low and CXCR4high tumors by automatically determined CXCR4 mRNA expression cutoffs providing the largest split in cancer-specific OS. A trend toward improved OS was observed for the CXCR4high strata (P = 0.074). Analysis of whole transcriptome differential gene expression in the CXCR4 subsets revealed significant upregulation of 2,332 and downregulation of 605 transcripts in CXCR4high tumors when compared with CXCR4low samples (Supplementary Fig. S3). The twenty most down- or upregulated genes are provided in Fig. 2A. Regarding genes involved in immune processes it revealed that genes encoding for chemokines (CXCL9, CXCL10, and CXCL13), granzymes, T-cell lineage markers (CD3D, CD3E, CD2, CD8A) along with T-cell exhaustion markers (TIGIT, PDCD1, HAVCR2, CTLA4, LAG3) represented a prominent group of genes expressed at significantly higher levels in CXCR4high compared with CXCR4low samples (Fig. 2B). Amongst others, a higher activity of natural killer (NK) cell–mediated cytoxicity, chemokine signaling, leukocyte transendothelial migration, cytokine–cytokine receptor interaction, MAPK and NF-κB signaling pathways was predicted for CXCR4high tumors by signaling pathway perturbation analysis based on the differentially regulated gene sets (Supplementary Fig. S4).
Figure 2.
A, Differences in gene expression between CXCR4high and CXCR4low tumors. Specimens were stratified as CXCR4high versus CXCR4low expressors. Log2 differences of the Top 20 differentially up- and downregulated genes between the CXCR4high versus CXCR4low samples with 95% CIs are presented. B, Differences in expression of unique gene members of immune signatures between CXCR4high and CXCR4low tumors. Differences in expression of unique gene members of immune signatures between CXCR4high and CXCR4low tumors. Samples were stratified as CXCR4high versus CXCR4low expressors. Log2 differences in expression between the CXCR4high versus CXCR4low samples with 95% CIs are presented.
Clinical characteristics of the validation cohort according to CXCR4 mRNA expression
For further exploration and validation of CXCR4 mRNA expression in PDAC we investigated a large cohort of patients with PDAC. In total, tumor samples from 3,647 patients with PDAC were centrally analyzed in a CLIA-certified laboratory of which 53.7% (n = 1,960) were male. Median age was 66 and 67 years for males and females, respectively. Advanced (metastatic) disease was present in 57.4% of cases (n = 2,092).
Using whole transcriptome sequencing we quantified CXCR4 mRNA expression levels in the whole cohort. CXCR4 mRNA values ranged from 0 to 793 TPM with a median value of 32 TPM. CXCR4 mRNA expression was higher in primary tumors compared with distant metastasis (38 vs. 28 TPM, P < 0.0001).
For further detailed comparison we stratified the cohort into four subsets according to quartiles (Q) of CXCR4 mRNA expression: Q1 ranged from 0 to 17 TPM, Q2 from 17 TPM to 32 TPM, Q3 from 32 to 59 TPM, and Q4 from 59 to 793 TPM. No differences regarding gender or age in the four subsets were observed (Table 1).
Table 1.
Characteristics of the evaluated real-world cohort according to CXCR4 mRNA expression status.
| Characteristics of the evaluated cohort | ||||
|---|---|---|---|---|
| CXCR4 quartiles | Q1 N (%) | Q2 N (%) | Q3 N (%) | Q4 N (%) |
| Numbers (n) | 912 | 912 | 911 | 912 |
| CXCR4 mRNA expression ranges (TPM) | 0–17 | 17–32 | 32–59 | 59–793 |
| Median age (years) | 67 | 66 | 67 | 67 |
| Gender | ||||
| Female | 416 (45.6) | 408 (44.7) | 425 (46.7) | 438 (48.0) |
| Male | 496 (54.4) | 504 (55.3) | 486 (53.3) | 474 (52.0) |
| Tissue site | ||||
| Primary | 346 (37.9) | 316 (34.6) | 385 (42.3) | 508 (55.7) |
| Metastasis | 566 (62.1) | 596 (65.4) | 526 (57.7) | 404 (44.3) |
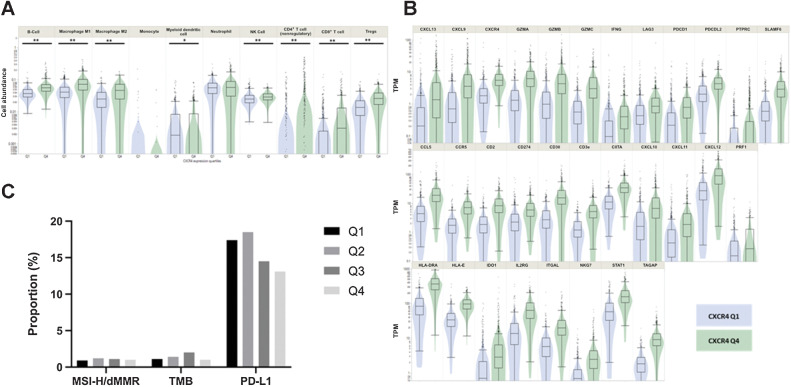
High CXCR4 mRNA expression is linked to increased peri-tumoral infiltration of several immune cells and respective immune-related genes
Next, we sought to confirm alterations of the immune infiltration profile in respect to CXCR4 mRNA expression suggested by the observations from the TCGA cohort, by comparing levels of immune cell abundance estimates calculated by the QuanTIseq algorithm (24) according to the Q1 versus Q4 CXCR4 strata. The CXCR4high (Q4) group was characterized by a significant increase of B cells, macrophages (M1 and M2), NK cells, Tregs, as well as CD4+ and CD8+ T cells (all, P < 0.0005). Conversely, lower levels of dendritic cell infiltration were detected in the CXCR4high versus the CXCR4low subcohort (Fig. 3A). At the level of specific transcripts, CXCR4high PDACs were characterized by an increase of HLA-E, HLA-DRA, chemokines [CXCL9, CXCL10, C-C chemokine receptor 5 (CCR5), CCL5], granzymes (GZMA, GZMB, GZMK), LAG3, IDO, and INFγ RNA levels (Fig. 3B). Differential gene expression, with subsequent pathway modulation and enrichment analysis (35, 41), suggests a higher activity of NK-cell cytotoxicity, chemokine and focal adhesion pathways together with increased MAPK signaling in the CXCR4high compared with the CXCR4low cohort.
Figure 3.
Correlation of immune-related genes and immune cell estimates in CXCR4high and CXCR4low PDACs. A, High CXCR4 mRNA expression (green; Q4) is linked to an increase of several immune-related genes using whole transcriptomic analyses (all, P < 0.05). B, QuanTiseq analysis suggests an increase of immune cell infiltration in CXCR4high (Q4; green) compared with CXCR4low (Q1; blue) PDACs. C,CXCR4 mRNA expression is independent of MSI-H/dMMR and TMB. PD-L1 IHC expression shows a decrease from Q1 to Q4. **, P < 0.001; *, P < 0.01.
To determine the relationship between CXCR4 mRNA expression and biomarkers predicting response toward ICI we next analyzed MSI-H/dMMR status, TMB and PD-L1 expression in the validation cohort. While PD-L1 expression status decreased from Q1 to Q4 (17.4% vs. 13.1%, P = 0.017), MSI-H/dMMR prevalence and TMB levels were comparable between the subcohorts (Fig. 3C).
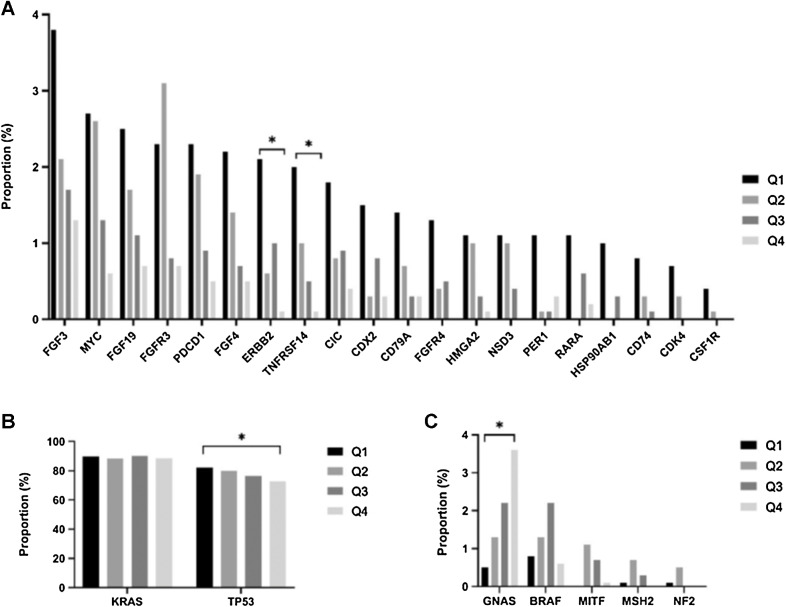
The genetic landscape significantly differs between the CXCR4 subgroups
Using NGS we investigated genetic alterations and copy-number alterations (CNA) in the four subsets. TP53 mutations were more frequently observed in Q1 compared with Q4 (82.2% vs. 72.7%, P < 0.0005, q = 0.0043), whereas a higher frequency of GNAS mutations were detected in Q4 compared with Q1 (3.6% vs. 0.5%, P < 0.0005, q = 0.0054; Fig. 4B and C; Supplementary Table S1). CNAs were more frequently observed in CXCR4low compared with CXCR4high tumors. For example, CNAs in ERBB2 (2.1% vs. 0.1%, P < 0.0005, q = 0.028) and TNFRSF14 (2.0% vs. 0.1%, P < 0.0005, q = 0.028) were more frequently detected in Q1 than in Q4 (Fig. 4A).
Figure 4.
Significant differences of genetic alterations according to CXCR4 mRNA expression. A, Copy-number alterations between CXCR4 subgroups. B and C, Significant point mutations between the CXCR4high and CXCR4low tumors. *, q < 0.05.
Clinical impact of high CXCR4 mRNA expression levels in PDAC
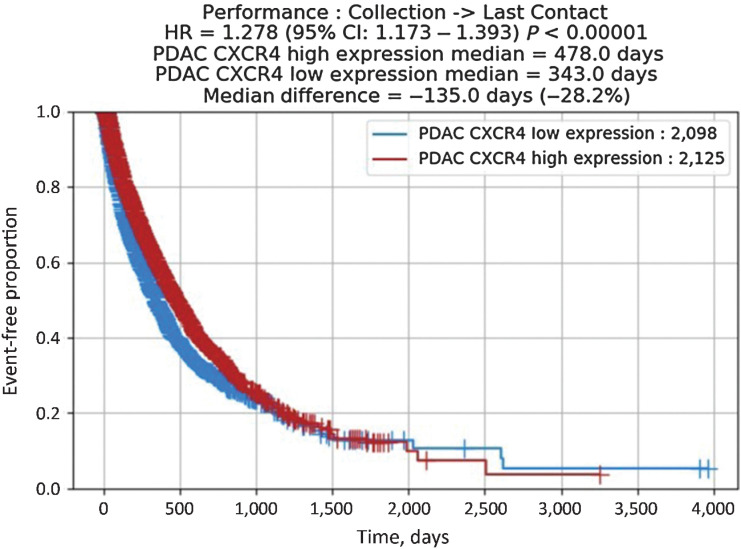
To explore the prognostic value of CXCR4 mRNA expression we compared the survival of patients with CXCR4high and CXCR4low tumor stratified by the median expression value (32 TPM). For this kind of analysis, real-world data for 1,758 patients were available. Univariate analysis of rwOS was significantly better in the CXCR4high compared with the CXCR4low subset [HR, 1.278; 95% confidence interval (CI), 1.173–1.393; P < 0.0001; Fig. 5]. A multivariable Cox proportional hazard modeling, considering various standard of care chemotherapeutic agents (e.g., gemcitabine, nab-paclitaxel, 5-fluorouracil), however, revealed that CXCR4high versus CXCR4low mRNA expression is not an independent predictive variable for rwOS.
Figure 5.
rwOS according to CXCR4 mRNA expression. Median OS in patients with CXCR4high expression was 478 days, whereas mOS in CXCR4low expressers was 343 days (HR, 1.278; 95% CI, 1.173–1.393; P < 0.0001).
CXCR4 expression is predominantly detected in immune cells according to single-cell RNA-seq and immunofluorescence
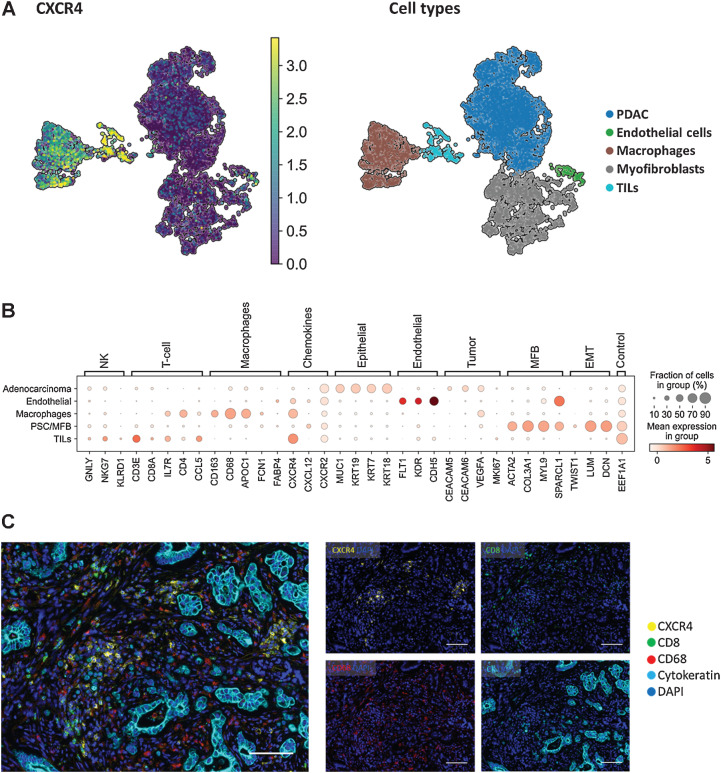
To further delineate CXCR4 expression on the single-cell level we reanalyzed a publicly available PDAC dataset (GSE154778), comprising single-cell RNA-seq data from 10 primary PDAC samples. Following cell type annotation, reanalysis revealed CXCR4 mRNA expression mainly in macrophages (CD68+), and tumor-infiltrating lymphocytes (Fig. 6A and B). In-depth analysis of CXCR4 expression in different immune cell subclusters could not be performed due to the small number of cells available in the dataset. Immunofluorescence experiments of in-house PDAC samples again revealed that CXCR4 is mainly expressed on macrophages and CD8+ T cells (Fig. 6C).
Figure 6.
Single-cell analysis reveals CXCR4 mRNA expression on macrophages and tumor-infiltrating lymphocytes in a publicly available PDAC dataset. A, Integrated UMAP for CXCR4, CD3E, CD68, and the respective annotated cell types from dissociated primary tumors obtained from patients with PDAC (n = 10). Strong CXCR4 gene expression was found in the macrophages and tumor-infiltrating lymphocytes cell clusters. B, Dotblot expression analysis of annotated cell types in the dataset with cell type–specific markers and targets CXCR4, CXCL12. C, Representative immunofluorescence images of a PDAC sample with CXCR4 expression on macrophages and CD8+ T-cells. mIF staining of CXCR4 (yellow), CD8 (green), CD68 (red), Cytokeratin (cyan), and DAPI (blue) in PDAC. Scale bar, 100 μmol/L.
Discussion
To the best of our knowledge, this study represents the largest and most comprehensive genetic and transcriptomic analysis of CXCR4 mRNA expression in PDAC due to the investigation of a large cohort of patients and two independent publicly available datasets.
Our comprehensive profiling approach unravelled distinct immunologic features associated with CXCR4 mRNA expression. In the TCGA dataset, CXCR4high samples showed a significant positive correlation with immune cell estimates of CD8+ T cells, Tregs and macrophages. These observations were validated in a large cohort of patients with PDAC, where the QuanTIseq analysis revealed a significant increase of CD8+ T cells, CD4+ T cells, Tregs, and macrophages in CXCR4high tumors. Of note, microsatellite status and TMB were comparable between the investigated quartiles. Thus, the immunologic properties of CXCR4 mRNA expression in PDAC are irrespective of these distinct features.
Besides the increased immune cell infiltration surrogates, an upregulation of several chemokines was noted in the TCGA dataset and in the validation cohort in PDACs with the CXCR4high phenotype including CXCL13, CXCL9, CXCL10, and CCR5. Of those, CXCL13 was the most upregulated gene in samples harboring high CXCR4 mRNA expression. Recently, Lin and colleagues reported that CXCL13 is secreted by follicular T helper cells leading to recruitment of CD8+ T and B cells in PDAC (42). CXCL9 is a chemokine physiologically binding to CXCR3 on T cells facilitating recruitment of cytotoxic CD8+ T cells to the TME (43). Of note, CXCL9 expression was found to be a potential biomarker for ICI response. According to a meta-analysis including >1,000 patients receiving ICI therapy, high CXCL9 expression outperformed the CD8+ effector and T cell–inflamed signature with regard to ICI response (44). A study conducted by Romero and colleagues provided additional evidence that a chemokine signature consisting of CCL4, CCL5, CXCL9, and CXCL10 is associated with a CD8+ T cell–inflamed phenotype in PDAC (45). In line with these findings, we also noticed a significant upregulation of CCL5, CXCL9, CXCL10—but not of CCL4—in patients with high CXCR4 mRNA expression levels. The CCR5 which physiologically binds to the three chemokine ligands CCL3, CCL4, and CCL5 was also found to be significantly upregulated in CXCR4high patients (46–48). In general, CCR5 has been linked to stimulation of tumor-cell proliferation and contributes to an immune-suppressive TME (49). In PDAC, high CCR5 expression was associated with poor differentiation (50). In a human PDAC xenograft model, the CCR5 antagonist maraviroc induced remission of liver metastasis via inhibition of cell-cycle kinase complexes resulting in G1 phase arrest (51). Again, a high CCR5 expression was found in our CXCR4high subset. In T-cell activation, CCR5 and CXCR4 are considered pivotal players as they deliver co-stimulatory signals and modulate T-cell response. It is believed that close interaction between CXCR4 and CCR5 regulates chemokine-receptor signaling versatility (52).
In parallel to the features that are associated with CD8+ T-cell infiltration, an upregulation of immune checkpoints and genes associated with immune evasion was observed including CD274 (PD-L1), PDCD1 (PD1), PDCD1LG2 (PD-L2), LAG3, IDO, TIGIT, CTLA4, and SLAMF6, suggesting T-cell exhaustion by continuous T-cell stimulation (53), which limit the endogenous anticancer T-cell reactivity (54). Restoration of deficient antitumor immunity by inhibiting immune checkpoints, such as anti—programmed cell death protein 1 (PD-1) or anti–PD-L1 inhibitors, have revolutionized anticancer treatment (2, 43). However, so far efficacy of ICI in PDAC is mainly restricted to patients harboring a MSI-H/dMMR status (5–7). Efforts have been made to increase efficacy of ICI in PDAC by combinational approaches. For example, in the randomized phase II PRINCE trial the addition of sotigalimab, a CD40 agonistic antibody, and/or nivolumab plus chemotherapy was evaluated in the first line setting. The combination of sotigalimab/nivolumab and chemotherapy failed to improve survival. Despite extensive immunologic work-up, no specific subset of patients benefitting from this combination could be identified (55). However, immunologic analyses of the nivolumab/chemotherapy and sotigalimab/chemotherapy arm underlined the potential of thorough immunologic work-up to improve patient selection for trials investigating ICI in PDAC. Freed-Pastor and colleagues (56) highlighted the immune evasive features of tumor-infiltrating leukocytes with a terminally exhausted T-cell phenotype in the majority of PDACs. Most importantly, they delineated the CD155/TIGIT axis as a crucial player in inducing and maintaining immune evasion. Of note, TIGIT was one of the most upregulated genes in CXCR4high patients in our study. We therefore speculate that CXCR4high PDAC are characterized by an increased abundance of intra-tumor exhausted T cells. Our hypothesis is supported by findings obtained from the single-cell RNA dataset, which reveals substantial CXCR4 gene expression in T-cell clusters, and—to a significant lower extent—also in in PDAC cells.
When focusing on patterns of cancer-associated genetic changes with respect to the CXCR4 expression pattern in PDAC, we observed a lower frequency of CNAs in CXCR4high patients. This might reflect a distinct and different tumor biology, since the burden of CNA correlates with oncogenic driver alterations (57). An induction of CNAs due to TP53 mutations in the course of carcinogenesis has already been proposed (57). In line with this hypothesis, we observed a higher prevalence of TP53 mutations in CXCR4low compared with CXCR4high tumors.
According to this observation, patients with CXCR4high PDAC have a superior OS, as shown in our large validation cohort. A previous study comprising early-stage PDACs of the TCGA dataset was in line with our findings (58). Our analysis of the TCGA cohort revealed an association of high CXCR4 expression with a trend toward improved survival. These observations are in contrast to a meta-analysis encompassing 1,183 patients (21). This discrepancy might arise from different methodologies used in the evaluation of the CXCR4 expression. While high CXCR4 expression established by IHC was reported to be a poor prognostic factor, the study investigating the TCGA dataset and our study applied mRNA sequencing for CXCR4 mRNA quantification. In addition, our cohort included a substantially higher number of patients as compared with the meta-analysis conducted by Krieg and colleagues (21). Moreover, our analysis represents the first study investigating the predictive value of CXCR4 in patients with PDAC. Our exploratory analysis revealed no significant survival differences in patients with PDAC that were treated with various standard-of-care chemotherapeutic agents, such as gemcitabine and taxanes. The retrospective design prohibited deeper analyses of the clinical data (e.g., PFS or response rates) according to CXCR4 mRNA expression levels. Nevertheless, more evidence is necessary to establish the prognostic and predictive value of CXCR4 mRNA expression in PDAC.
Besides the retrospective study design, which prohibited additional clinical correlation, further limitations apply to our study: even though our findings are suggestive that CXCR4 mRNA expression might be a valuable biomarker for patients with PDAC undergoing ICI we were not able to test this hypothesis in a suitable cohort.
Of note, the phase II COMBAT/KEYNOTE-202 trial assessed the efficacy of the CXCR4 antagonist BL-8040 (motixafortide) in combination with the anti–PD-1 antibody pembrolizumab ± chemotherapy in patients with pretreated metastatic PDAC (59). This combination therapy led to changes in the composition of the TME as depicted by an increase of intratumor CD8+ T cells while immunosuppressive cells decreased. While the combination of BL-8040 and pembrolizumab (= cohort 1) only showed a low activity with an objective response rate (ORR) of 3.4%, the combination of BL-8040, pembrolizumab and the NAPOLI-1 regimen (liposomal irinotecan, 5-fluorouracil and leucovorin;=cohort 2) yielded an ORR of 13.2% (59, 60). In the light of the response rates of the NAPOLI-1 approval study (61), which reported an ORR of 16%, the efficacy of the combinational approach of anti-CXCR4, anti–PD-1, and the NAPOLI-1 regimen appears to have only modest synergistic activity. However, to the best of our knowledge, intratumor CXCR4 mRNA expression has not been assessed within the COMBAT trial. On the basis of our findings, the correlation of CXCR4 mRNA expression and patients´ outcome might be helpful to improve patient stratification for this innovative treatment approach. Future prospective studies or at least post hoc analyses of ICI trials enlightening the predictive role of CXCR4 in PDAC are clearly desirable.
Even though, we observed an association of CXCR4 with immune cell infiltration and that CXCR4 is mainly expressed on CD8+ T cells and macrophages, we are not able confirm mechanistic features of this finding. To gain a deeper understanding between a potential CXCR4-dependent immune cell recruitment and PDAC additional in vitro and in vivo studies are required. Finally, the single-cell dataset only included a small number of cells which prohibited detailed correlation of CXCR4 with subsets of tumor-infiltrating lymphocytes and macrophages. In summary, our study indicates that comprehensive molecular and immunologic characterization might pave the way toward patient selection for future clinical trials. On the basis of our results it seems reasonable to investigate the potential of CXCR4 mRNA expression as a predictive biomarker in trials evaluating ICI and/or CXCR4 antagonist combinations. Thus, post hoc exploratory analyses of already completed clinical trials are desirable to further delineate the role of CXCR4 mRNA expression in PDAC.
Supplementary Material
Acknowledgments
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
Y. Baca is an employee of Caris Life Sciences. J. Xiu is an employee of Caris Life Sciences. P. Tymoszuk reports owning a data science enterprise ‘Data Analytics as a Service Tirol' and works as a freelance data scientist and biostatistician. A. Petrillo reports personal fees from Eli Lilly, Merck Serono, Servier, and BMS; grants from Bayer and Roche; and grants and personal fees from MSD and Amgen outside the submitted work. A.F. Shields reports research support provided by Caris Life Sciences for genomic analysis. M.E. Salem reports consulting and speaking for Taiho Oncology, AstraZeneca, Daiichi Sankyo, Bristol-Myers Squibb, Merck, Pfizer, QED Therapeutics, Novartis, and Exelixis. J.L. Marshall reports personal fees from Caris Life Science, Indivumed, Bayer, Merck, Taiho, and Pfizer outside the submitted work. M. Hall reports other support from Caris during the conduct of the study; and personal fees from Eisai and Natera outside the submitted work; in addition, M. Hall has a patent for hereditary risk detection issued; and collaborative research only (no financial support or grants or fees) with the following: Myriad, Invitae. W.M. Korn reports Caris Life Sciences employment and stock ownership. C. Nabhan reports other support from Caris Life Sciences during the conduct of the study. E. Lou reports research grants from the American Association for Cancer Research (AACR-Novocure Tumor-Treating Fields Research Award, Grant Number 19-60-62-LOU); American Cancer Society Research Scholar Grant RSG-22-022-01-CDP; and the Minnesota Ovarian Cancer Alliance in 2019, 2021, and 2022; honorarium and travel expenses for a research talk at GlaxoSmithKline in 2016; honoraria and travel expenses for lab-based research talks, and equipment for laboratory-based research, Novocure, Ltd, 2018-2021; honorarium for panel discussion organized by Antidote Education for a CME module on diagnostics and treatment of HER2+ gastric and colorectal cancers, funded by Daiichi Sankyo, 2021 (honorarium donated to lab); consultant, Nomocan Pharmaceuticals (unpaid); Scientific Advisory Board Member, Minnetronix, LLC, 2018-present (unpaid); consultant and speaker honorarium, Boston Scientific US, 2019; institutional principal investigator for clinical trials sponsored by Celgene, Novocure, Intima Biosciences, and the NCI, and University of Minnesota membership in the Caris Life Sciences Precision Oncology Alliance (unpaid); we acknowledge and thank the following groups for donations in support of cancer research: Friends and family of Gayle Huntington; the Mu Sigma Chapter of the Phi Gamma Delta Fraternity, University of Minnesota (FIJI); the Litman Family Fund for Cancer Research; Dick and Lynnae Koats; and Love Like Laurie Legacy. S. Choo reports personal fees from BMS, Ipsen, MSD, Merck Serono, Eisai, Bayer, and AstraZeneca outside the submitted work. A. Seeber reports other support from Caris Life Sciences during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
F. Kocher: Conceptualization. A. Puccini: Conceptualization. G. Untergasser: Formal analysis. A. Martowicz: Data curation, methodology. K. Zimmer: Data curation. A. Pircher: Conceptualization. Y. Baca: Data curation. J. Xiu: Formal analysis. J. Haybaeck: Conceptualization. P. Tymoszuk: Formal analysis. R.M. Goldberg: Methodology. A. Petrillo: Writing–original draft. A.F. Shields: Validation, visualization. M.E. Salem: Data curation. J.L. Marshall: Project administration. M. Hall: Investigation. W.M. Korn: Software. C. Nabhan: Supervision. F. Battaglin: Visualization. H.-J. Lenz: Project administration. E. Lou: Project administration, writing–review and editing. S.-P. Choo: Investigation. C.-K. Toh: Formal analysis. S. Gasteiger: Data curation, methodology. R. Pichler: Formal analysis, writing–review and editing. D. Wolf: Writing–original draft. A. Seeber: Conceptualization, resources, data curation, formal analysis, supervision, validation, methodology, project administration, writing–review and editing.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 2. Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti–PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Reilly EM, Oh DY, Dhani N, Renouf DJ, Lee MA, Sun W, et al. Durvalumab with or without tremelimumab for patients with metastatic pancreatic ductal adenocarcinoma: a phase II randomized clinical trial. JAMA Oncol 2019;5:1431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henriksen A, Dyhl-Polk A, Chen I, Nielsen D. Checkpoint inhibitors in pancreatic cancer. Cancer Treat Rev 2019;78:17–30. [DOI] [PubMed] [Google Scholar]
- 5. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch repair deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair–deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol 2020;38:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sohal DP, Mangu PB, Khorana AA, Shah MA, Philip PA, O'Reilly EM, et al. Metastatic pancreatic cancer: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol 2016;34:2784–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balkwill F. Cancer and the chemokine network. Nat Rev Cancer 2004;4:540–50. [DOI] [PubMed] [Google Scholar]
- 10. Chow MT, Luster AD. Chemokines in cancer. Cancer Immunol Res 2014;2:1125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol 2017;17:559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ozga AJ, Chow MT, Luster AD. Chemokines and the immune response to cancer. Immunity 2021;54:859–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol 2014;32:659–702. [DOI] [PubMed] [Google Scholar]
- 14. Zhou W, Guo S, Liu M, Burow ME, Wang G. Targeting CXCL12/CXCR4 axis in tumor immunotherapy. Curr Med Chem 2019;26:3026–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Busillo JM, Benovic JL. Regulation of CXCR4 signaling. Biochim Biophys Acta 2007;1768:952–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dewan MZ, Ahmed S, Iwasaki Y, Ohba K, Toi M, Yamamoto N. Stromal cell–derived factor-1 and CXCR4 receptor interaction in tumor growth and metastasis of breast cancer. Biomed Pharmacother 2006;60:273–6. [DOI] [PubMed] [Google Scholar]
- 17. Zhao H, Guo L, Zhao J, Weng H, Zhao B. CXCR4 over-expression and survival in cancer: a system review and meta-analysis. Oncotarget 2015;6:5022–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang J, Liu C, Mo X, Shi H, Li S. Mechanisms by which CXCR4/CXCL12 cause metastatic behavior in pancreatic cancer. Oncol Lett 2018;15:1771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cui K, Zhao W, Wang C, Wang A, Zhang B, Zhou W, et al. The CXCR4-CXCL12 pathway facilitates the progression of pancreatic cancer via induction of angiogenesis and lymphangiogenesis. J Surg Res 2011;171:143–50. [DOI] [PubMed] [Google Scholar]
- 20. Wehler T, Wolfert F, Schimanski CC, Gockel I, Herr W, Biesterfeld S, et al. Strong expression of chemokine receptor CXCR4 by pancreatic cancer correlates with advanced disease. Oncol Rep 2006;16:1159–64. [PubMed] [Google Scholar]
- 21. Krieg A, Riemer JC, Telan LA, Gabbert HE, Knoefel WT. CXCR4–A prognostic and clinicopathological biomarker for pancreatic ductal adenocarcinoma: a meta-analysis. PLoS One 2015;10:e0130192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Network CGAR. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017;32:185–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kocher F, Tymoszuk P, Amann A, Sprung S, Salcher S, Daum S, et al. Deregulated glutamate to pro-collagen conversion is associated with adverse outcome in lung cancer and may be targeted by renin-angiotensin-aldosterone system (RAS) inhibition. Lung Cancer 2021;159:84–95. [DOI] [PubMed] [Google Scholar]
- 24. Finotello F, Mayer C, Plattner C, Laschober G, Rieder D, Hackl H, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med 2019;11:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Plattner C, Finotello F, Rieder D. Deconvoluting tumor-infiltrating immune cells from RNA-seq data using quanTIseq. Methods Enzymol 2020;636:261–85. [DOI] [PubMed] [Google Scholar]
- 26. Woroniecka K, Chongsathidkiet P, Rhodin K, Kemeny H, Dechant C, Farber SH, et al. T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin Cancer Res 2018;24:4175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan K, Lu Y, Yan Z, Wang Y. 9-Gene Signature Correlated With CD8. Front Immunol 2021;12:622563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFNγ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Danaher P, Warren S, Lu R, Samayoa J, Sullivan A, Pekker I, et al. Pan-cancer adaptive immune resistance as defined by the tumor inflammation signature (TIS): results from The Cancer Genome Atlas (TCGA). J Immunother Cancer 2018;6:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vesanto J, Alhoniemi E. Clustering of the self-organizing map. IEEE Trans Neural Netw 2000;11:586–600. [DOI] [PubMed] [Google Scholar]
- 32. Kohonen T. Self-organizing maps. Springer Series in Information Sciences (Vol. 30). Springer Berlin Heidelberg; 1995. [Google Scholar]
- 33. Wehrens R, Kruisselbrink J. Flexible self-organizing maps in kohonen 3.0. J Stat Softw 2018;87:1–18. [Google Scholar]
- 34. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289–300. [Google Scholar]
- 35. Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS, et al. A novel signaling pathway impact analysis. Bioinformatics 2009;25:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abraham J, Heimberger AB, Marshall J, Heath E, Drabick J, Helmstetter A, et al. Machine learning analysis using 77,044 genomic and transcriptomic profiles to accurately predict tumor type. Transl Oncol 2021;14:101016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Merino DM, McShane LM, Fabrizio D, Funari V, Chen SJ, White JR, et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the friends of Cancer research TMB harmonization project. J Immunother Cancer 2020;8:e000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin W, Noel P, Borazanci EH, Lee J, Amini A, Han IW, et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med 2020;12:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Büttner M, Miao Z, Wolf FA, Teichmann SA, Theis FJ. A test metric for assessing single-cell RNA-seq batch correction. Nat Methods 2019;16:43–9. [DOI] [PubMed] [Google Scholar]
- 41. Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010;11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin X, Ye L, Wang X, Liao Z, Dong J, Yang Y, et al. Follicular helper T cells remodel the immune microenvironment of pancreatic cancer via secreting CXCL13 and IL21. Cancers (Basel) 2021;13:3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Litchfield K, Reading JL, Puttick C, Thakkar K, Abbosh C, Bentham R, et al. Meta-analysis of tumor- and T cell–intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 2021;184:596–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Romero JM, Grünwald B, Jang GH, Bavi PP, Jhaveri A, Masoomian M, et al. A four-chemokine signature is associated with a T cell–inflamed phenotype in primary and metastatic pancreatic cancer. Clin Cancer Res 2020;26:1997–2010. [DOI] [PubMed] [Google Scholar]
- 46. Lederman MM, Sieg SF. CCR5 and its ligands: a new axis of evil? Nat Immunol 2007;8:1283–5. [DOI] [PubMed] [Google Scholar]
- 47. Blanpain C, Migeotte I, Lee B, Vakili J, Doranz BJ, Govaerts C, et al. CCR5 binds multiple CC-chemokines: MCP-3 acts as a natural antagonist. Blood 1999;94:1899–905. [PubMed] [Google Scholar]
- 48. Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry 1996;35:3362–7. [DOI] [PubMed] [Google Scholar]
- 49. Hemmatazad H, Berger MD. CCR5 is a potential therapeutic target for cancer. Expert Opin Ther Targets 2021;25:311–27. [DOI] [PubMed] [Google Scholar]
- 50. Singh SK, Mishra MK, Eltoum IA, Bae S, Lillard JW, Singh R. CCR5/CCL5 axis interaction promotes migratory and invasiveness of pancreatic cancer cells. Sci Rep 2018;8:1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang H, Zepp M, Georges RB, Jarahian M, Kazemi M, Eyol E, et al. The CCR5 antagonist maraviroc causes remission of pancreatic cancer liver metastasis in nude rats based on cell-cycle inhibition and apoptosis induction. Cancer Lett 2020;474:82–93. [DOI] [PubMed] [Google Scholar]
- 52. Contento RL, Molon B, Boularan C, Pozzan T, Manes S, Marullo S, et al. CXCR4-CCR5: a couple modulating T-cell functions. Proc Natl Acad Sci USA 2008;105:10101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wherry EJ, Kurachi M. Molecular and cellular insights into T-cell exhaustion. Nat Rev Immunol 2015;15:486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jiang Y, Li Y, Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis 2015;6:e1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Padrón LJ, Maurer DM, O'Hara MH, O'Reilly EM, Wolff RA, Wainberg ZA, et al. Sotigalimab and/or nivolumab with chemotherapy in first-line metastatic pancreatic cancer: clinical and immunologic analyses from the randomized phase II PRINCE trial. Nat Med 2022;28:1167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Freed-Pastor WA, Lambert LJ, Ely ZA, Pattada NB, Bhutkar A, Eng G, et al. The CD155/TIGIT axis promotes and maintains immune evasion in neoantigen-expressing pancreatic cancer. Cancer Cell 2021;39:1342–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hieronymus H, Murali R, Tin A, Yadav K, Abida W, Moller H, et al. Tumor copy-number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. Elife 2018;7:e37294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu QY, Yang CK, Rong LJ, Li JC, Lei LM. Investigation of the association between C-X-C motif chemokine receptor subunits and tumor infiltration levels and prognosis in patients with early-stage pancreatic ductal adenocarcinoma. Oncol Lett 2020;20:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bockorny B, Semenisty V, Macarulla T, Borazanci E, Wolpin BM, Stemmer SM, et al. BL-8040, a CXCR4 antagonist, in combination with pembrolizumab and chemotherapy for pancreatic cancer: the COMBAT trial. Nat Med 2020;26:878–85. [DOI] [PubMed] [Google Scholar]
- 60. Bockorny B, Macarulla T, Semenisty V, Borazanci E, Feliu J, Ponz-Sarvise M, et al. Motixafortide and pembrolizumab combined to nanoliposomal irinotecan, fluorouracil, and folinic acid in metastatic pancreatic cancer: the COMBAT/KEYNOTE-202 trial. Clin Cancer Res 2021;27:5020–7. [DOI] [PubMed] [Google Scholar]
- 61. Wang-Gillam A, Li CP, Bodoky G, Dean A, Shan YS, Jameson G, et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): a global, randomized, open-label, phase III trial. Lancet 2016;387:545–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The deidentified sequencing data are owned by Caris Life Sciences. The datasets generated and analyzed during the current study are available from the authors upon reasonable request and with permission of Caris Life Sciences. Qualified researchers may contact the corresponding author with their request.