Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, is a highly transmittable and pathogenic human coronavirus that first emerged in China in December 2019. The unprecedented outbreak of SARS-CoV-2 devastated human health within a short time leading to a global public health emergency. A detailed understanding of the viral proteins including their structural characteristics and virulence mechanism on human health is very crucial for developing vaccines and therapeutics. To date, over 1800 structures of non-structural, structural, and accessory proteins of SARS-CoV-2 are determined by cryo-electron microscopy, X-ray crystallography, and NMR spectroscopy. Designing therapeutics to target the viral proteins has several benefits since they could be highly specific against the virus while maintaining minimal detrimental effects on humans. However, for ongoing and future research on SARS-CoV-2, summarizing all the viral proteins and their detailed structural information is crucial. In this review, we compile comprehensive information on viral structural, non-structural, and accessory proteins structures with their binding and catalytic sites, different domain and motifs, and potential drug target sites to assist chemists, biologists, and clinicians finding necessary details for fundamental and therapeutic research.
1. Introduction
Infections with the newly emerged beta coronavirus (CoV) named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) are now widespread, affecting more than 210 countries and regions on earth (Tiwari et al., 2020). As of June 8, 2022, 537 million cases have been confirmed, with over 6.32 million deaths worldwide (World Health Organization, 2021) (“WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard with Vaccination Data,” 2021). During the covid-19 pandemic, biomedical research received unprecedented attention. Chemists, biologists, and clinicians from all fields of biomedical science worked together to accelerate diagnostic testing and enable the development of vaccines and therapeutics for covid. Initial milestones were the publication of the viral genome sequence, solving the structure of SARS-CoV- 2 main protease, and spike (S) glycoprotein. Preliminary analyses were suggested that SARS-CoV-2 has a close evolutionary association (96 % nucleotide sequence identity) with the SARS-like bat coronaviruses (Lam et al., 2020a). The early information that the SARS-CoV-2 receptor binding domain (RBD) has a higher hACE2 binding affinity than SARS-CoV RBD, and the interactions between RBD and hACE2 were the key step for the viral life cycle (Fan et al., 2020, Wrapp et al., 2020). These were crucial for understanding the molecular basis of the viral life cycle, and atomic-scale resolution of structures provided the target for designing structure-based vaccines or drugs.
Up to now, over 1800 structures of various viral proteins of SARS-CoV-2 have been resolved and reported (https://www.rcsb.org). It is important to compile the structural information together to design and develop new structure-based therapeutics. This review summarizes the current knowledge on the genome constitution and structure–function relationships of different viral proteins, and a short overview of the repurposed drugs and vaccines with therapeutic potential and ongoing trials.
2. Genome organization and overview of SARS-CoV-2 proteins
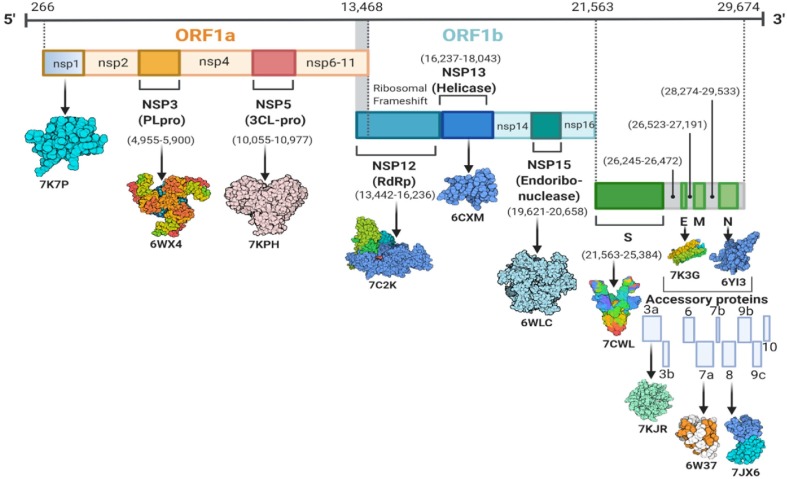
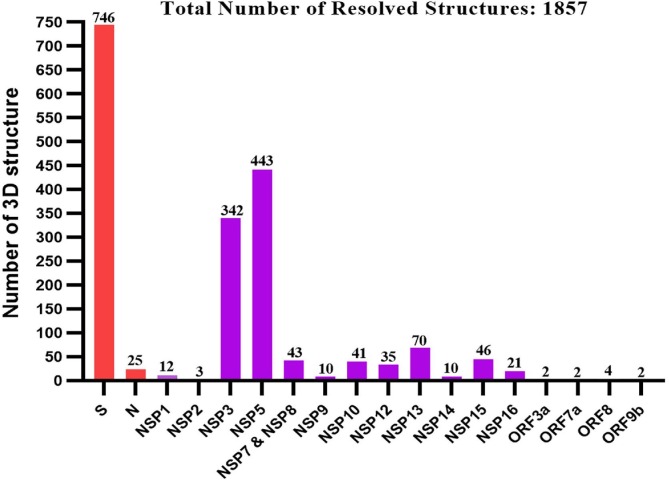
SARS-CoV-2 is a positive-sense single-stranded RNA genome of 29.8–29.9 kb nucleotides that encodes a lengthy polyprotein of 9860 amino acids (Saxena et al., 2020). The NCBI viral database has about 485,141 completed nucleotide sequences for SARS-CoV-2 as of November 5, 2021 (Hatcher et al., 2017). The genome contains four structural proteins such as spike (S) protein, Envelope (E) protein, Membrane (M) protein, and Nucleocapsid (N) protein, along with 14 ORFs that encode 27 proteins and is about 80 % identical to the human coronavirus (Wu et al., 2020a). SARS-CoV-2 has fourteen open reading frames (ORFs) in its genome, separated into two portions. Cellular ribosomes directly translate ORF1a and ORF1ab into two polyproteins (pp1a and pp1ab), found in the first two-thirds of the viral genome from the 5′ end. Two viral proteases, papain-like protease (PLpro) and main protease (Mpro or CLpro), then process the polyproteins and generate sixteen nonstructural proteins, nsp1–nsp16 (Y. Chen et al., 2020a), (Lu et al., 2020a). Furthermore, eight accessory proteins such as ORF3a, ORF3b, p6, ORF7a, ORF7b, ORF8b, ORF9b, and ORF14 are located at the 3′ end (Wu et al., 2020a). The genome organization and corresponding proteins have been depicted in Fig. 1 . A summarized function and structural characteristics of structural, non-structural, and accessory proteins have been listed in Table 1 . More than 1500 structures of SARS-CoV-2 proteins have been submitted to the Protein Data Bank (PDB) database (https://rcsb.org/covid19) since January 2020, as presented in Fig. 2 and Supplementary Table S1. In the case of SARS-CoV-2 structural proteins, ∼747 spike proteins, an envelope protein, and ∼25 nucleocapsid protein structures have been deposited into the PDB database. However, the membrane protein structure of SARS-CoV-2 has not been reported yet. Among the non-structural proteins of SARS-CoV-2, a higher number of structures have been resolved for nsp5 (443), followed by nsp3, nsp13, nsp15, nsp7 & 8, nsp10, nsp12, nsp16, nsp9, and nsp14, respectively. In addition, several structures of SARS-CoV-2 accessory proteins were deposited into the PDB database, whereas only four structures for ORF8 and two structures for each of the ORF3a, ORF7a, and ORF9b proteins.
Fig. 1.
Genomic representation of SARS-CoV-2 consisting of open reading frames that encode structural, non-structural, and accessory proteins.
Table 1.
Brief description of various structural, non-structural, and accessory proteins of SARS-CoV-2.
| Protein name | Length (aa) | Function | Binding site/ catalytic residues | Different domain and motifs | Drug binding sites | Ref. |
|---|---|---|---|---|---|---|
| S (Spike) | 1273 | Mediates binding to ACE2 | K417, E484, N487, F486, N501 | NTD (14–306), RBD (331–528), CTD1 (529–591), CTD2 (592–686), HR1 (910–985), HR2 (1163–1211), TM (1212–1234), CT (1235–1273) |
CTD of S1: V382 L390, C391, T393, T430, L517, A520, A522, L527, N544, L546, N564, F565, F782, A1056 S2 Domain: I870, D867, A1056, P1057, G1059, H1058, S730, M 730 M731, Y733, V860, L861, P863 |
(Chowdhury et al., 2020, Zhang et al., 2021) |
| E (Envelope) | 75 | Involved in virus morphogenesis and assembly | E8, N15, L18, L21, V25, L28, A32, T35 | NTD (1–8), TM (9–38), CTD (39–75) |
T9, G10, T11, I13, A36, L37, S16, N15, I33, E8, N15 | (Bhowmik et al., 2020, Mandala et al., 2020) |
| M (Membrane) | 222 | Important for the budding process of coronaviruses | NTD (1–19), Triple-TM (20–100), CTD (101–222), Motifaromatic-XX-aromatic motif (91-WXXY-94), Di-leucine motif (219-LL-220) |
Y50, L51, L54, L93, A98 | (Bhowmik et al., 2020, Yan et al., 2022) | |
| N (Nucleocapsid) | 419 | Promotes genome packaging, RNA chaperoning, intracellular protein transport, DNA degradation, interference in host translation |
A50, T57, H59, R89, R92, I94, S105, R107, R149, Y172 |
NTD (1–50), RBD (51–174), Linker (175–246), Dimerization domain (247–365), CTD (366–419) |
N48, N49, T50, A51, R89, Y112, Y110 | (Bhowmik et al., 2020, Cubuk et al., 2021, Khan et al., 2021b) |
| NSP1 | 180 | Recommended as leader protein which inhibit host translation and degrade host mRNAs | P153-N160, S166-N178 |
NTD (1–128), CTD (148–180), Motif KH (164–165) |
V35, E36, L39, V89, Y97, F143, F157, Q158 | (Schubert et al., 2020, Singh et al., 2021) |
| NSP2 | 638 | Binds to prohibitin 1 (PHB1) and 2 (PHB2) | V126, A127, C132, V157, L169, C240, Y242, W243, T256, G257 |
NTD (1–345), CTD (438–638) | P15, D16, N94, V96, A227 | (Ma et al., 2021, Maiti et al., 2020) |
| NSP3 (PLpro) | 1945 | Responsible for cleaving of NSP1, NSP2, and NSP3 from the N-terminal region of pp1a and 1ab | C111, H272, D286 | UbI1 (1–108), HVR (109–206), Mac1 or X (207–386), SUD (387–745), UbI2 (746–805), PLPro (806–1058), NBD (1059–1200), MD (1201–1340), TM (1341–1567), Y domain (1568–1945) |
L162, G163, D164, E167, P247, P248, Y264, Y268, Q269, Y273 | (Fu et al., 2021, Osipiuk et al., 2021, Yan et al., 2022) |
| NSP4 | 500 | Potential transmembrane scaffold protein which helps modify ER membranes | NA | TM1 (10–30), TM2 (280–300). TM3 (305–330), TM4 (355–380), CTD (381–500) |
NA | (Santerre et al., 2021, Yan et al., 2022) |
| NSP5 (3CLpro) | 306 | Cleaves viral polyprotein | C145, H41 |
N-finger (1–9), Domain-I (10–99), Domain-II (100–182), Domain-III (198–303) |
T24, T25, T26, H41, F140, L141, N142, G143, C145, H163, E166, P168, H172, Q189, T190, A191, Q192 | (Jin et al., 2020, Khan et al., 2021b, Rahman et al., 2020) |
| NSP6 | 290 | Induction of autophagosomes from host ER (Cottam et al., 2014, Cottam et al., 2014) | NA | NA | NA | |
| NSP7 | 83 | Forms hexadecameric complex with nsp8 for viral replication and participate as a cofactor for nsp12 | S4, D5, K7, C8, H36, L40, N37, V33 S15, L14, V11, A30, W29, E23 | Replicase domain (1–83) | R21, K43, D44 | (Wilamowski et al., 2021) |
| NSP8 | 198 | Makes heterodimer with nsp7 and nsp12 | P183, Y149, V131, M129, P133, A125, K127, V130, P121, L122, A110, L128, N118, I119, T123, K79, L117, I106, N109, P116, V115, M94, D112, C114, D99, L95, N104, L91, V83, L98, F92, A162, T84, R80, Q88, M90, I185, M87, A86 | Shaft domain (6–104), Head domain (105–196) |
A102, A150, R190, A194 | (Wilamowski et al., 2020) |
| NSP9 | 198 | RNA-binding protein which may participate in viral replication | N33, G100, M101, V102, L103, G104, S105 |
Single domain protein (1–109) Motif GxxxG (100–104) |
M12, S13, N33, T35, F40, L42, L94, N98 | (Khan et al., 2021b, Littler et al., 2021a) |
| NSP10 | 139 | Forms heterodimer complex with nsp14 and nsp16, acting as a cofactor for both and stimulates ExoN (viral exoribonuclease) and 2-O-methyltransferase activity | N3, V4, T5, F8, K9, D10, P20, T21, Q22, P24, T25, H26, L27, L38, C39, D41, F60, K61, M62, N63, Y64, V66, Y69, T127, N129, N130, T131, K196, K200, I201 | Single domain protein (1–139) | V21, D22, A26, G35, Q36, P37, I38, GLY52, Q65, R78, P107, V108 | (Halder, 2021, Lin et al., 2021) |
| NSP11 | 13 | Unknown | NA | NA | NA | NA |
| NSP12 (RdRp) | 932 | Replication and methylation | Y420, F415, F441, F440, F442, N552, A443, P412, G413, D445, Q444, T409, N447, R392, D390, L391, L389, N403, V405, L388, T402, L387, N386, A379, P323, L270, F396, F326, V398, L271, L514, P328, M666, V330, Y273, T324, T344, L329, P339, M380, R331, V338, K332, Y374, F340, A383, D336, S384, V341, S518, F407, L371, F368, D523, W509, S759, D760, D761 |
NiRAN (1–250), C-terminal RdRp (398–932), Motif G (499–511), Motif F (544–560), Motif A (612–626), Motif B (678–710), Motif C (753–767), Motif D (771–796), Motif E (810–820) |
M542, K545, S549, K551, R553, R555, V557, D618, C622, ASP623, S682, S759, D760, D761, R836 | (Ahmed et al., 2020, Khan et al., 2021b, Zhang et al., 2020c) |
| NSP13 (Helicase) | 596 | A helicase core domain participates in binding interaction with ATP. Zn-binding domain is involving in replication and transcription | R178, H230, N361, S468, T532, D534 | ZBD (1–100), SD (101–150), 1B domain (151–261), 1A domain (262–442), 2A domain (443–601) |
V45, Y70, F90, P283, G285, T286, G287, K288, H290, R443, E540 | (Chen et al., 2020, Malone et al., 2021, Yan et al., 2021) |
| NSP14 (ExoN) | 527 | Acting on both ssRNA and dsRNA in a 3′ to 5′ direction and a N7-guanine methyltransferase activity | D90, E92, E191, H268, D273 | Flanking region (1–50), ExoN (51–287), N7-MTase (288–527), DEDD motif |
W385, N386, Y420, F426, F506 |
(Devkota et al., 2021, Tahir, 2021) |
| NSP15 | 346 | Uridine-specific endoribonuclease activity | H235, H250, K290, T341, Y343, S294 |
N-domain (1–64), Middle domain (65–182), endoU (207–347) |
F44, E45, D92, H250, Y290, V292, C293, S294, Y343 | (Khan et al., 2021b, Kim et al., 2021) |
| NSP16 | 298 | RNA-cap methyltransferase |
K46, D130, K170, E203 | NTD (1–29), Mtase domain (30–210), CTD (211–298) |
A80, T83, A84, L86, T94, L95, L96, V97, D98, S99, D100 | (Rosas-Lemus et al., 2020, Vithani et al., 2021) |
| ORF3a | 275 | Infection, inducing apoptosis | NA | NTD (1–34) TM1 (35–56), TM2 (76–99), TM3 (103–125), CR domain (127–133), CTD (208–264), TRAF3-binding motif (36–40), CBM (141–149), Motif YXXΦ (160–163), Motif EXD (171–173) |
Y61, I62, I63, T64, I118, V121, R122, Y206 | (Kern et al., 2021) |
| ORF6 | 61 | Type 1 IFN antagonist | D53, E55, M58, E59, D61 | Interaction motif (56–61) | NA | (Gordon et al., 2020, Li et al., 2022) |
| ORF7a | 121 | Triggers an immune response in host cells | NA | Signal peptide (1–15), Ig-like ectodomain (16–96), TM region (97–116), ER retention motif (117–121) |
E33, C35, S36, S37, T39, Y40, E41, G42, S44, P45, F46, P48, F65 | (Gorgulla et al., 2021) |
| ORF8 | 121 | Disrupts IFN-I signaling when exogenously overexpressed in cells | P85, F86, T87, I88, N89, C90, Q91, E92 | D1 domain (1–15), D2 domain (16–121), Catalytic core motif (85–92) |
I47-L60, V62, D63, Y73–I76, Y79, T80, Q91, K94, L95 | (Cavasotto et al., 2021, Hassan et al., 2021) |
NTD = N-terminal Domain, RBD = Receptor Binding Domain, CTD = C-terminal Domain, TM = Transmembrane Domain, UbI1 = ubiquitin-like domain 1, SUD = SARS-unique domain, HVR = hypervariable region, PL2pro = papain-like protease, MD = Marker domain, NBD = nucleic acid-binding domain, ZBD = zinc-binding domain, SD = stalk domain.
Fig. 2.
Histogram of numbers of PDB entries of each of the SARS-CoV-2 proteins (Last update: March 15,2022) (www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html). Among of the deposited structures, 40 % S protein structures are in complex with antibodies and human ACE2, 19 % are nsp3, and 24 % are nsp5, while the rest of the other protein structures comprises 22 % of the total structures. Interestingly, some of the proteins have just a few copies of structure or no structure at all, i.e., membrane protein (M).
3. Structural proteins
3.1. Spike protein (S)
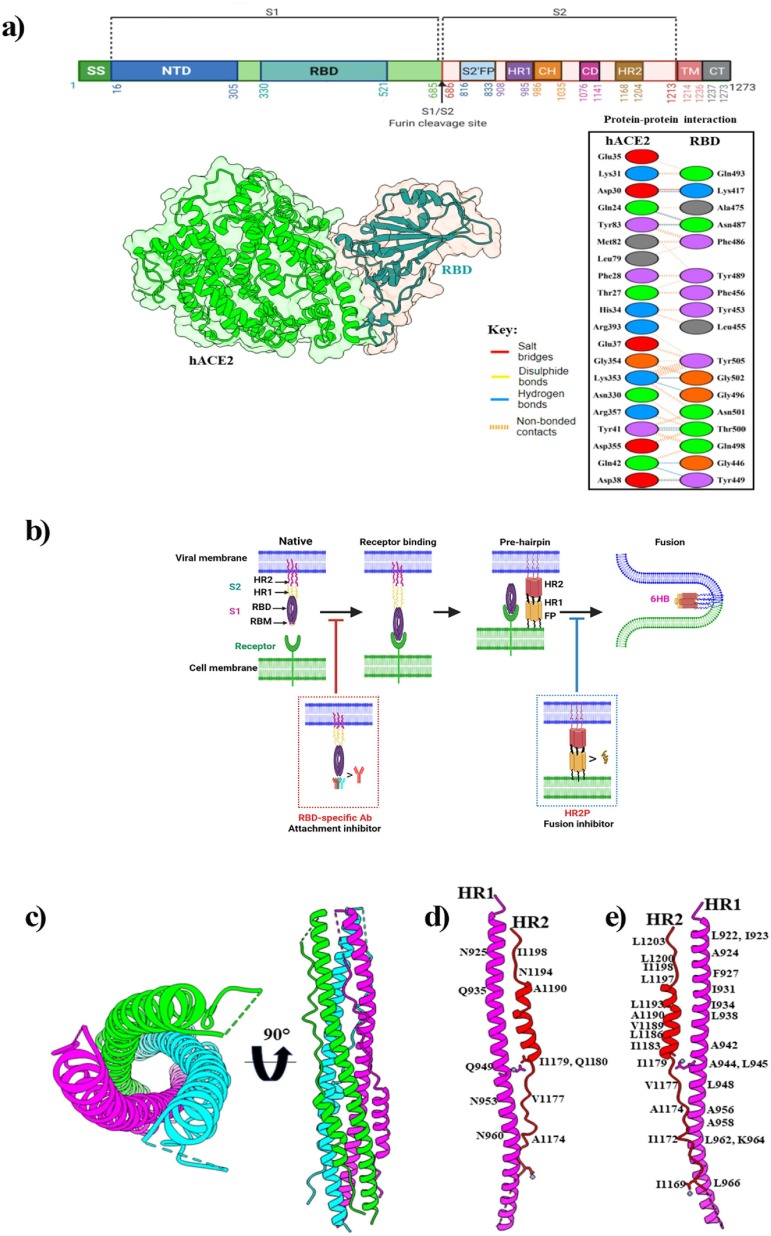
Among the structural proteins, the spike glycoprotein (S) is the most important. The S protein mediates the virus entry to the host cell, enhances virulence, and determines the life cycle of the virus (Walls et al., 2020). Structurally S protein is a homotrimer; three polypeptide chains of S protein are assembled to form a functional protein (Benton et al., 2020). Each monomer of the spike protein has two subunits S1 and S2, which mediate receptor-binding and membrane fusion (Fig. 3 a) (Wrapp et al., 2020). Furthermore, S1 is subdivided into an N-terminal domain (NTD) and the receptor-binding domain (RBD), which directly binds to the extracellular peptidase domain (PD) of host cell surface receptor called hACE2 of human respiratory epithelial cells (Lam et al., 2020b, Walls et al., 2020). The highly mutable RBD domain consists of a five-stranded antiparallel β-sheet (β1- β3- β5- β4- β2) core, which is flanked by a short helix. The receptor-binding motif, RBM (437–508), forms a cradle-like conformation for receptor binding (Xia et al., 2020).
Fig. 3.
(a) Schematic representation of SARS-CoV-2 spike protein primary structure and hACE2-RBD complex. Here, the top panel shows different colors domains. SS, single sequence; NTD, N-terminal domain; RBD, receptor-binding domain; S1, subdomain 1; S2, subdomain 2; S1/S2, S1/S2 protease cleavage site; S2′, S2′ protease cleavage site; FP, fusion peptide; HR1, heptad repeat 1; CH, central helix; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. Arrows indicate the protease cleavage site. The lower-left panel shows the cartoon and surface representations of the overall structure of the SARS-CoV-2 RBD bound to hACE2 (PDB: 6M0J) and protein–protein interactions are presented in the lower-right panel. (b) The mechanism of human ACE2 and SARS-CoV-2 S protein-mediated virus attachment and fusion. In the native state, the S2 subunit is encapsulated by the S1 subunit. Several conformational changes occur in the S2 subunit after viral RBD engagement with the receptor. The HR1-trimer core structure is formed from three HR1 molecules, and three HR2 molecules bind to the HR1-trimer to form 6-HB, which mediates membrane fusion. A neutralizing antibody that targets RBD blocks viral infection by blocking RBD’s interaction with cellular receptors. The membrane fusion process is inhibited by the fusion inhibitor that blocks 6-HB formation. (c) Six-helix bundle fusion core is comprised of three HR2-helices packed in the HR1 side grooves (PDB: 6M1V). Structures in the top view (left panel) and side view (right panel) are presented respectively. Here, three HR1/HR2 chains are colored light green, magenta, and cyan. (d) The detailed interactions between HR1 and HR2, and residues involved in the H-bond interactions are labelled. (e) Residues involved in the hydrophobic interactions are labeled.
Basically, the interaction with the hACE2 occurs through the α1 helix of RBD which is augmented by the engagement of two polar residues of the middle segment of the α1 helix as well as the linker between β3 and β4 loops and the α2 helix (Wrapp et al., 2020). The interacting residues of RBD with the host cell receptors have been reported in several studies, shedding light on the structural basis of receptor recognition (Othman et al., 2020, Yan et al., 2020). The RBM remains buried inside the protein in the closed state, and in the open conformation, it interacts with ACE2. In the S-ACE2 complex, four disulfide bonds (C336–C361, C379–C432, C391–C525, and C480–C488) stabilize the RBD structure, and the RBM forms a concave outer surface to accommodate the N-terminal helix of ACE2. Ten H-bonds, a salt bridge, and several hydrophobic interactions contribute to ACE2 engagement (Fig. 3 a) (Lan et al., 2020). The S2 subunit comprises an N-terminal fusion peptide (FP); two heptad repeats (HR1 and HR2) separated by a central helix (CH) and a connector domain (CD); a transmembrane domain (TM), and a cytoplasmic tail (CT) (Wrapp et al., 2020, Xia et al., 2020). The S protein contains three conformational states during the membrane fusion process: a native state (prefusion), an intermediate state (pre-hairpin), and a post-fusion hairpin state (stable). During the fusion process, the FP is inserted into the host cell membrane, which leads S2 to the pre-hairpin intermediate state, forming an α-helical anti-parallel complex between HR1 and HR2, where the loop region acts as a hinge and builds a six-helix bundle (6HB) that brings the cellular and viral lipid bilayers in close proximity (Ling et al., 2020, Wang et al., 2021), as shown in Fig. 3 b. In post-fusion state, HR1 and HR2 of the S2 subunit are associated with each other forming a six-helix bundle (6HB) fusion core where three HR2 helices surround the HR1 helices in an anti-parallel arrangement (Fig. 3 c). This complex structure is highly stable and plays a significant role in membrane fusion (Schütz et al., 2020). Protein-protein interaction between HR1 and HR2 revealed that multiple H-bond contacts were established between amino acids from HR1 (N925, Q935, Q949, N953, N960) and amino acids from HR2 (A1174, V1177, I1179, Q1180, A1190, N1194, I1198) (Fig. 3 d). HR hairpin trimerization is mediated by a collection of tandemly ordered seven-residue repeats, with the repeatedly presented hydrophobic amino acids forming a strong hydrophobic face (Fig. 3 e). Notably, the HR2 domains of SARS-CoV-2 and SARS-CoV-1 are identical, while the HR1 domains show variations; considering HR2 a good target site for developing potential fusion inhibitor (Schütz et al., 2020, Xia et al., 2020, Yan and Gao, 2021). A range of antiviral agents has been developed to target the S protein, including antibodies, inhibitors, and vaccines. In general, inhibitors of S glycoproteins prevent virus-membrane fusion by competitively blocking RBD-ACE2 interaction (Chowdhury et al., 2020). Such inhibitors include arbidol (umifenovir) (Padhi et al., 2021) and ivermectin (Caly et al., 2020). Haste et al. reported that RBD-ACE2 interactions are blocked by three kinds of antibodies (RBD1, RBD2, RBD-3 mAbs) through both steric hindrance and direct competition for interface residues. Here, RBD-1mAbs largely overlap with the RBM; the RBD-2mAbs move to the “Peak” of the RBM from the center of ACE binding; and the RMD-3mAbs bind to the “Mesa” of the RBM from the center of ACE2 binding. (Hastie et al., 2021). Furthermore, a pan-CoV fusion inhibitor, such as EK1 has been used to target HR1 of the S2 domain to inhibit membrane fusion (Efaz et al., 2021, Wang et al., 2021).
3.2. Membrane protein (M)
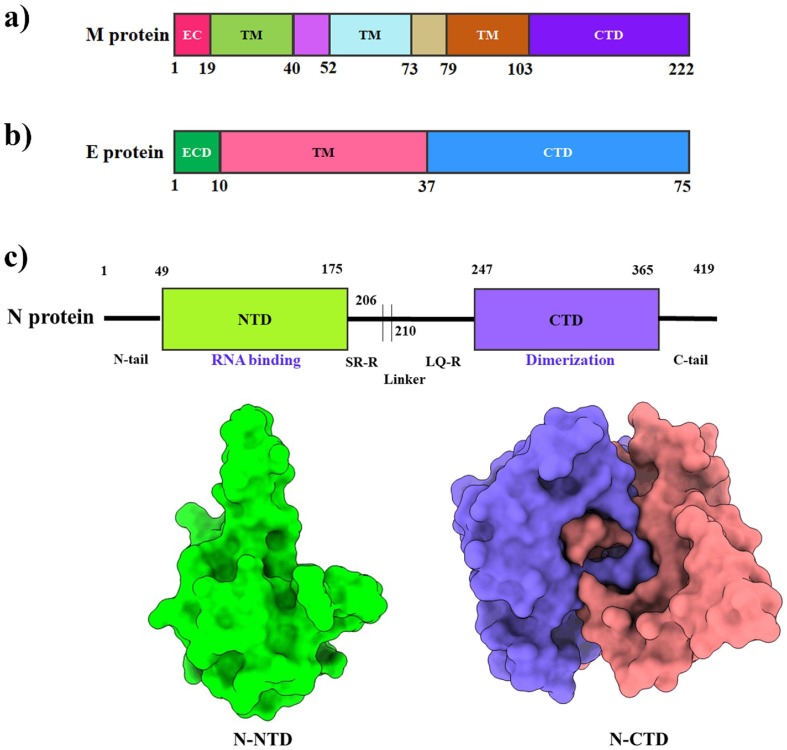
The M protein is the most abundant structural protein and the major component of the viral envelope (Tseng et al., 2013). Moreover, M protein is crucial for viral assembly, morphogenesis (Hu et al., 2003), budding (Voβ et al., 2009), and recruitment of S protein to the virus assembly and budding site. The M protein is also required for genome packing (Hu et al., 2003), and nucleocapsid inclusion into the virion (Voβ et al., 2009). The M protein consists of three primary domains: an ectodomain at the N-terminus, three transmembrane helices (TMH1-TMH3), and an endodomain at the C-terminus (Fig. 4 a) (Mahtarin et al., 2020) TM1-TM2 intersegment is thought to be in the interior, while the TM2-TM3 intersegment is in the exterior (Hu et al., 2003). The C-terminal region is predicted to have at least two casein kinase II phosphorylation sites (TSR at codon 171, SQR at codon 183) related to S, E, and N protein interaction (Hu et al., 2003). These interactions are required for membrane bending (budding) and operate as a checkpoint to form new virions (Ujike and Taguchi, 2015). Furthermore, SARS-CoV M protein residues L218 and L219 are needed for nucleocapsid packing (Liu et al., 2010, Tseng et al., 2013). Antigenic epitopes have been found in the TM1 and TM2 regions of the SARS-CoV M protein, leading to the designing of peptide inhibitors or vaccines against this protein. Along with the experimental study, researchers have recently employed computational techniques such as molecular dynamics (MD) simulations to uncover several potential drugs (such as remdesivir) that have a higher affinity for M protein (Khan et al., 2021a), but further studies are required to confirm these interactions.
Fig. 4.
Structural features of the membrane (M), envelope (E), and nucleocapsid (N) protein of SARS-CoV-2. (a, b) Schematic representation of the domains of the M and E proteins. (c) The upper panel shows a schematic representation of the SARS-CoV-2 N protein, which consist of two domains, i.e. the N-terminal domain (NTD) and the C-terminal domain (CTD). The lower-left panel shows a surface of the amino-terminal (N-terminal) domain (PDB: 6M3M). The lower-right panel shows the surface representation of the dimeric carboxy-terminal (C-terminal) domain (PDB: 6YUN).
3.3. Envelope protein (E)
The SARS-CoV-2 envelope (E) protein is the smallest of all structural proteins and is found primarily in the host cell's endoplasmic reticulum (ER) and Golgi complex, where it is involved in viral assembly, pathogenesis, and release (Westerbeck and Machamer, 2019). The topology of E protein consists of a five-helix bundle, surrounded by a dehydrated narrow pore and bipartite channel. In terms of amino acid composition, E proteins are highly divergent but structurally highly conserved in various genera of β-coronaviruses with a hydrophilic ectodomain at the N-terminus, a hydrophobic transmembrane domain (TMD), and a lengthy hydrophilic C-terminal endodomain (Fig. 4 b) (Schoeman and Fielding, 2019). Amantadine (AMT) and hexamethylene amiloride (HMA) are two ion-channel drugs having guanidinium group to interact with polar residues at the entrance of ion channel and engage the amino-terminal lumen, blocking ion channel activity of the E protein (Mandala et al., 2020, Pervushin et al., 2009). Moreover, peptide inhibitors derived from Ec18 can be employed to inhibit the interactions between the E protein and PLAS1 (the human cell junction protein) (Chai et al., 2021). Furthermore, Bacillus Calmette-Guerin (BCG) vaccination is used as an alternative approach for treating COVID-19 that induces specific host immunity targeting the SARS-CoV-2 E protein (Nuovo et al., 2020).
3.4. Nucleocapsid protein (N)
SARS-CoV-2 N protein is encoded in the structural ORF situated at the 3′ end. In the domain architecture of SARS-CoV-2 N protein, it possesses three highly conserved domains: an N-terminal domain (NTD), a linker region or an RNA-binding domain, and a C-terminal domain (CTD) (Fig. 4 c). The core region of the NTD constitutes β-sheets situated in a five-stranded anti-parallel position. Two α-helices occur inside the β-sheet core, forming an overall conformation of β1-α1-β2-β2′-β3′-β3-β4-α2-β5 in which β2′ and β3′ forms a long basic β-hairpin structure (Dinesh et al., 2020).The N-CTD monomer comprises a structure of η1-α1-α2-η2-α3-α4-β1-β2-α5-η3 orientation which contains five α-helices, three 310 (η) helices, and two anti-parallel β-strands that forms a β-hairpin structure. The CTD forms a compact homodimeric structure, and the monomer of CTD is assumed to be unstable (Zhou et al., 2020). The N-CTD dimeric structure is stabilized by 40H-bond and 389 hydrophobic interactions (Zinzula et al., 2021). The CTD region of SARS-CoV is essential for RNA binding (residue 248–280) (Chen et al., 2007, Takeda et al., 2008), and it remains almost conserved in SARS-CoV-2 (corresponding residue 247–279) with one amino acid replacement (SARS-CoV Gln268 → Ala267 SARS-CoV-2) (Zhou et al., 2020). The C-terminal tail mediates higher order self-assembly forming tetramer, hexamer, and possible higher oligomeric forms (Chang et al., 2013). The N protein has been observed to inhibit interferon β production for modulating the innate immune response of host cells, although the mechanism of this process is quite unclear (Kopecky-Bromberg et al., 2007, Lu et al., 2011). The N protein is a target for diagnosis due to higher sequence conservancy, less prone to mutation, and induces a strong protective immune response in the host compared to other drug target proteins (3CLpro, PLpro, and S protein) (Kannan et al., 2020). The nCoV396 monoclonal antibody, isolated from the blood of convalescent COVID-19 patients, forms H-bonds and hydrophobic interactions with several residues (Q163, L167, and K169) of the N-NTD to stabilize the protein complexes. These interactions work together to help the antibody neutralize the N protein's antigenicity (Kang et al., 2021). The N protein has also been inhibited by small molecules (such as PJ34 and rapamycin) that interfere with the RNA binding of N-NTD and dimerization of N-CTD (Matsuo, 2021, Peng et al., 2020).
4. Non-Structural proteins (NSP)
4.1. nsp1
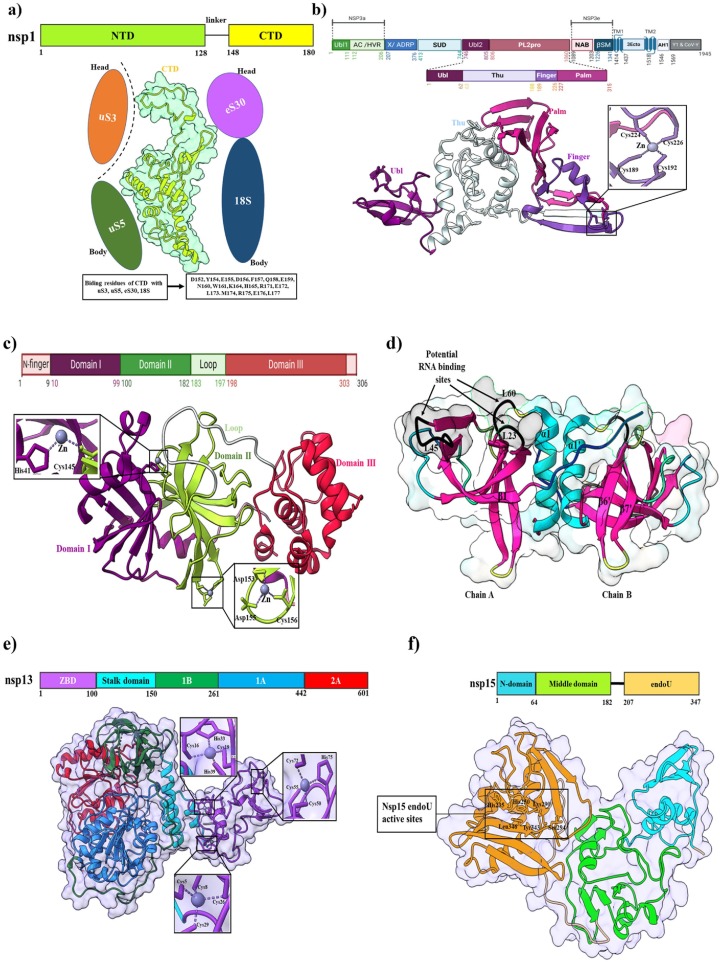
Nsp1 is the N-terminal cleavage product released from polyprotein precursors pp1a and pp1ab by viral papain-like protease (PLpro) through proteolysis (Clark et al., 2021). The structure of the nsp1-40S ribosomal subunit complex was resolved by the cryo-EM which revealed the mechanism of translation inhibition (Fig. 5 a) (Schubert et al., 2020). The hydrophobic core of the β-barrel comprises three layers, where the first layer is formed by side chains of residues L16, L18, V69, L88, L107, and L123, but the opening of the β-barrel at this layer is obstructed by α1 helix side-chain residue L46. The middle layer consists of residues V20, L53, I71, V86, and V121, while the bottom layer features residues V84 and L104. Moreover, the two 310 helices in the globular domain form an H-bond interaction between R24 and Q63 which stabilize the position of two of the largest loops in the globular domain, the β1-α1 (L21 – S34) loop and the β2-β3 (E54 – P67) loop (Semper et al., 2021). Several surface residues, such as E36, E37, E41, K47, K58, R124, and K125 are significant for mRNA binding; these are highly conserved in SARS-CoV-2 (Almeida et al., 2007). Guardeño et al., reported that two C-terminal regions (aa 122–130 and aa 155–165) of nsp1 are important to inhibit IFN responses and/or antiviral signaling (Jimenez-Guardeño et al., 2015). Recently, Vankadari et al. reported that several natural product molecules including garinolic acid, glycyrrhizic acid, tirilazad, and lobaric acid were considered as potential nsp1 inhibitors. These molecules were also screened by preliminary computational studies, and it is reported that they can possibly block the nsp1/SL1 complex formation (Vankadari et al., 2020).
Fig. 5.
Structures of SARS-CoV-2 non-structural proteins. (a) The upper panel shows the domains organization of SARS-CoV-2 nsp1; NTD (N-terminal domain) and CTD (C-terminal domain). The middle panel shows the cartoon and surface representation of nsp1 CTD in complex with the host ribosomal 40S subunit (PDB: 7K7P). The lower panel shows binding residues of nsp1 CTD with the 40S ribosomal subunit. (b) The upper panel shows the domain organization of SARS-CoV-2 nsp3. The lower-panel shows the cartoon representation of functional domains. The conserved zinc-binding motifs are highlighted in the SARS-CoV-2 nsp3 structure (PDB: 6WRH). The coordinate details of the zinc-binding residues are shown in stick representation. (c) Schematic representation of domain features of SARS-CoV-2 nsp5 (Mpro) in the upper panel. The lower panel shows the cartoon representation of Mpro (PDB: 6YB7) with magnified views on the two zinc-finger motifs. The domain I, domain II, and domain III are colored with violet, lemon, and red, respectively. Residues involved in zinc-coordination are shown in sticks. (d) The cartoon and surface representation of crystal structure of dimeric SARS-CoV-2 nsp9 (PDB: 6WXD). Predicted RNA binding sites on the surface of one face of the SARS-CoV-2 nsp9 dimer are identified with black arrows: between β7 and α1 (L60), β2 and β3 (L23), and β4 and β5 (L45). (e) The upper panel shows the domain organization of SARS-CoV-2 nsp13. Here, the zinc-binding domain (ZBD), stalk domain, 1B, 1A, and 2A are colored with violet, cyan, green, blue, and red, respectively. The lower panel shows a cartoon and surface representation of the crystal structure of SARS-CoV-2 nsp13 (PDB: 7NIO). Magnified views on the three zinc-finger motifs in SARS-CoV-2 helicase apo form. Residues involved in zinc-coordination are shown in sticks. (f) The upper panel shows the primary structure of SARS-CoV-2 nsp15. Here, N-domain, N-terminal domain, middle domain, and endoU domain are colored with blue, lemon, and brown, respectively. The lower panel shows a cartoon and surface representation of the crystal structure of SARS-CoV-2 nsp15 (PDB: 6WLC). The rectangular box shows active sites of the endoU domain.
4.2. nsp2
SARS-CoV-2 nsp2 is the second protein of pp1, which contains two domains, i.e., the N-terminal domain (1–345) and the C-terminal domain (438–638) (Heo and Feig, 2020). Although SARS-CoV-2 nsp2 has been involved in viral processes, its exact functions and the structural basis remain unknown (Y. Chen et al., 2020b). Gupta et al. reported that a highly conserved cysteine residue coordinating a Zn2+ ion in a zinc ribbon-like motif is structurally highly similar to RNA binding proteins. Probably, this motif is important for nsp2 interactions with nucleic acids (Gupta et al., 2021). Currently, there is no known inhibitor of nsp2. From molecular docking, some candidates for nsp2 inhibitors have been proposed. For instance, nigellidine is an indazole-alkaloid that binds to nsp2′s entry pocket. It establishes an H-bond with nsp2 Cys240, allowing it to occupy the nsp2 entrance channel, formed by many residues (L169, V126, W243, A127, C132, T256, G257, Y242, and V157) (Maiti et al., 2022). The immunogenicity of nsp2 may be exploited to develop inactivated or live attenuated virus vaccines.
4.3. nsp3
SARS-CoV-2 nsp3 is the largest membrane-bound protein (1945 aa) with several domains (Báez-Santos and st. John, S.E., Mesecar, A.D., , 2015, Wu et al., 2020b). It acts as a membrane-anchored scaffold that associates with the host proteins and other nsps to form the viral replication-transcription complex (Angelini et al., 2013). Nsp3 consists of the N-terminal Nsp3a domain (includes ubiquitin-like domain 1 (Ubl1) and acidic domain (Ac) or hypervariable region (HVR), macrodomain-X, SARS unique domains (SUDs), ubiquitin-like domain 2 (Ubl2), papain-like protease domain (PL2pro), nucleic acid-binding (NAB) domain, beta coronavirus-specific marker (βSM) domain, transmembrane domains (TM), nsp3 ectodomain (3Ecto), amphipathic helix 1 (AH1), Y1 and CoV-Y domain (Fig. 5 b) (Lei et al., 2018). The known functional role of the Ubl1 domain in CoVs is linked to ssRNA binding and interaction with the N protein (Hurst et al., 2013, Hurst et al., 2010, Serrano et al., 2009). In the case of SARS-CoV, the Ubl1 domain binds ssRNA containing AUA patterns (Serrano et al., 2009). Following the Ubl1, the second subdomain, the Glu-rich acidic region resides at the N-terminus of nsp3. Both domains together are also called “Nsp3a” (Neuman et al., 2008). Currently, the function of the Glu-rich acidic domain in CoVs is unknown. Although Glu-or Asp-rich proteins are often engaged in many biological roles, such as metal-ion binding, DNA/RNA mimicry, and protein–protein interactions (Chou and Wang, 2015). A conserved X domain or macrodomain (also called Nsp3b) follows the hypervariable region in all CoVs (Gorbalenya et al., 1991, Neuman, 2016, Neuman et al., 2008). Recently, several studies reported that the macrodomain plays a role in revoking the innate immune response of host cells (Eriksson et al., 2008, Fehr et al., 2016, Fehr et al., 2015, Kuri et al., 2011). Imbert et al. reported that the macrodomain has binding interaction with the RNA-dependent RNA polymerase (Imbert et al., 2008). If this interaction exists in the virus life cycle, two proteins can impact each other's enzymatic activity (Lei et al., 2018). The exact functional role of the Ubl2 domain is not clear. Frieman et al. reported that the Ubl2 domain is essential for antagonizing the host innate immune response by blocking IRF3 or the NF-κB pathway (Frieman et al., 2009). Recently, a study reported that SARS-CoV-2 PLpro recognizes LXGG tetrapeptide motif between nsp1 and nsp2, nsp2 and nsp3, and nsp3 and nsp (Rut et al., 2020a). The catalytically active PLpro domain of SARS-CoV cleaves PPla at three cleavage sites at the N- terminus (176ELNGG↓AV182, 814RLKGG↓AP820, and 2736SLKGG↓KI2742) to release nsp1, nsp2, and nsp3 through proteolytical processes (Lei et al., 2018). According to the UniprotKB database (UniProtKB: P0DTD1), these three cleavage sites have been found for SARS-CoV-2 at the N-terminus (176ELNGG↓AV182, 814RLKGG↓AP820, and 2759ALKGG↓KI2765) of ppla; this process is essential for viral replication (Harcourt et al., 2004). The PLpro monomer comprises four distinct domains, three of which adopt an extended right-hand fold with a distinct thumb, finger, and palm subdomains (Lei et al., 2018). The first 62 residues fold into a ubiquitin-like domain (Ubl). This domain is well separated from the other three domains that interact with each other and form a compact globular conformation (Alfuwaires et al., 2017). The Ubl domain adopts a β-grasp fold similar to ubiquitin and is highly conserved in most β-CoVs, including SARS-CoV and MERS-CoV (Lei et al., 2014, Yang et al., 2014). The central thumb subdomain of SARS-CoV-2 PLpro is predominantly helical, comprised of six α-helices and one β-strand, and a catalytic Cys111 residue contributes to the active site. Amino acids from 189 to 314-fold into the finger and palm domains. The C-terminal region of the PLpro domain is made up of mostly β-strands. The finger domain consists of one α-helices (α8), one long (β7) and two short (β8 and β9) β-strands, and the palm domain is made up of eight β-strands. A Zn-ion is coordinated by four cysteine residues (Cys189, Cys192, Cys224, and Cys226) inducing from two β-hairpins is located between β7 and β9 of the finger domain. Although the conformations of the zinc finger are variable between different CoV PL2pro (Lei et al., 2014), the motif is significant for proteolytic activity and structural stability (Barretto et al., 2005). The active sites are located at the interface of the thumb domain and middle of the palm domain and comprise the typical Cys111, His272, and Asp286 triad, adjacent to the flexible “blocking loop” (BL2) containing Trp106 (Bagherzadeh et al., 2020). This loop comprises six amino acid residues (GNYQCG) in the enzyme of SARS-CoV PLpro (Lei et al., 2018). The enzyme has four substrate recognition subsites (S1-S4) (Arya et al., 2020). Residues Gly271, Trp106, Cys111, and Tyr112 are included in the S1-subsite, whereas residues Leu162, Asp164, Gly271, and Tyr273 are engaged in shaping the conserved S2 subsite. S3 subsite had residue Gly271 that is partially solvent-exposed, and S4 subsite includes residues Asp302, Pro228, Tyr264, Tyr268, Tyr273, and Thr301 that are buried and structured (Kong et al., 2015). The competitive inhibitors of SARS-CoV PLpro are bound at the S2 and S4 subsites (Baez-Santos et al., 2014, Kong et al., 2015). The deubiquitinase and deISGylating activity of CoV PLpro are well installed, but the elaborated mechanism of the PLpro antagonism of the host innate immune response is still uncertain (Lei and Hilgenfeld, 2017). Various cytokines such as TNFs and IFNs are induced to inhibit virus replication by the IRF3 and the NF-κB pathway (Hiscott et al., 2006). However, the protease activity of SARS-CoV PLpro is significant to block the TNF-α or NF-κB signaling pathway (Frieman et al., 2009). So, it is a valuable target protease for treating of SARS-CoV-2 infections (Ansori et al., 2021). Several PLpro inhibitors such as VIR251, GRL-0617, and YM155 have been used to block the active site of PLpro. The VIR251 inhibitor binds with the active site of PLpro, several H-bonds and hydrophobic interactions are involved in stabilizing the complex (Rut et al., 2020b). GRL-0617 inhibitor targets the USP domain; it can block the C-terminus binding of ISG15 to PLpro by strong interaction between GRL-0617 and PLpro (Fu et al., 2021). The YM155 inhibits the activity of the PLpro protease and blocks ISG15 from binding to PLpro at the C-terminus (Zhao et al., 2021).
4.4. nsp4
SARS-CoV-2 nsp4 is predicted to be a membrane-spanning protein that is released by the combined activity of the nsp3 and nsp5 proteases (Graham et al., 2008). To date, limited structural information on this protein is available (Almazán et al., 2006). This protein contains four transmembrane domains: N-terminal, lumenal, TM3, and C-terminal domain (Bonilla et al., 1994). TMs 1 to 3 and a specific charged residue are essential for productive virus infection and the C-terminal domain is exposed at the cytoplasmic face of the membrane (Manolaridis et al., 2009, Xu et al., 2009). It is speculated that the nsp4 helps anchoring of viral RTC (replication-transcription) complex in association with other integral viral membrane proteins such as nsp3 and nsp6 (Almazán et al., 2006, Hagemeijer et al., 2014, Hagemeijer et al., 2011). Induction of concentrated foci in the perinuclear region and redistribution of proteins from ER to the foci results from the co-expression of nsp4 (mainly the large luminal loop) and nsp3C (Angelini et al., 2013, Hagemeijer et al., 2011). Several studies reported that mutation in the lumenal domain causes loss of nsp4 glycosylation, membrane rearrangement, and RNA replication (Angelini et al., 2013, Gadlage et al., 2010).
4.5. nsp5
The SARS-CoV-2 nsp5 or main protease (Mpro), also known as 3C-like protease (3CLpro), leads to the processing of viral polyprotein and is also a promising target for antiviral therapy (Anand et al., 2003, Ziebuhr et al., 2000). The SARS-CoV-2 Mpro monomer consists of N-terminal domains (domain-I and domain-II) and C-terminal domain-III (Fig. 5 c) (L. Zhang et al., 2020b). The domain-I and II are anti-parallel β-barrels that form the active site containing a catalytic dyad with Cys145 and His41 at their interface and the last C-terminal helix in domain-III is involved in dimerization through a salt-bridge interaction between Glu290 of a protomer and Arg4 of the other (Zhang et al., 2020b). The N-terminal tail, termed the “N-finger” of molecule B, forms an intercommunicating attachment between domain-II of molecule A and domains-II and III of the parent monomers. This peculiar arrangement is stabilized by Ser1 and Glu166 residues, forming some key H-bonds (Cannalire et al., 2020). The active site at the interface of domain-I and domain-II contains five substrate binding subsite pockets (S1, S2, S3, S4, and S5), where S2, S4, and S5 are flexible by binding diverse chemical groups (Kneller et al., 2020). The S1 site (Phe140, Ser144, Asn142, His163, Glu166, and His172) shaped by Ser1 of promoter B, which interacts with Glu166 of promoter A (Lee et al., 2020); S2 is a cleft formed by Met48 and the backbone of Asp187-Gln189; S3 and S4 subsites are extended toward the solvent, and included residues are significant for conformational shifts upon interactions with ligands (Cannalire et al., 2020, Kneller et al., 2020). Several antiviral inhibitors have been utilized to treat COVID-19, including N3, Calpain inhibitor II (UAW241), α-ketoamide, and Oral antiviral PF-07321332. In 3CLpro, the N3 inhibitor forms non-covalent interactions with H163, H164, E166, Q189, and T190 as well as covalent interactions with C145 (Xiong et al., 2021). Calpain inhibitor II interacts with His163 through a weak H-bond and multiple hydrophobic interactions with C145, H162, and E166 (Sacco et al., 2020). The α-keto (L. Zhang et al., 2020a) and PF-07321332 (Ledford et al., 2021) form covalent interactions with the Cys145-His41 catalytic dyad.
4.6. nsp6
SARS-CoV-2 nsp6 is a multiple-spanning transmembrane protein located to the endoplasmic reticulum (ER) (Benvenuto et al., 2020, Cottam et al., 2011). It is associated with the generation of autophagosomes to release viral components to lysosomes for degradation (Cottam et al., 2014, Cottam et al., 2014). Oostra et al. reported that SARS-CoV nsp6 and mouse hepatitis virus (MHV) nsp6 contain six TM domains (Oostra et al., 2008). In TM2 and TM3 domains, the highly conserved lysine and histidine residues are designated as KH loop, but the function of this cytosolic loop is unknown. Another point is that both N- and C-terminus are exposed to the cytosol (Baliji et al., 2009, Oostra et al., 2008). Notably, the C-terminal domain possesses a palmitoylation site (Hagemeijer et al., 2012) and predicted it to be cysteine residue within the conserved G (X)C (X)G motif (Baliji et al., 2009).
4.7. nsp7, nsp8, and nsp12
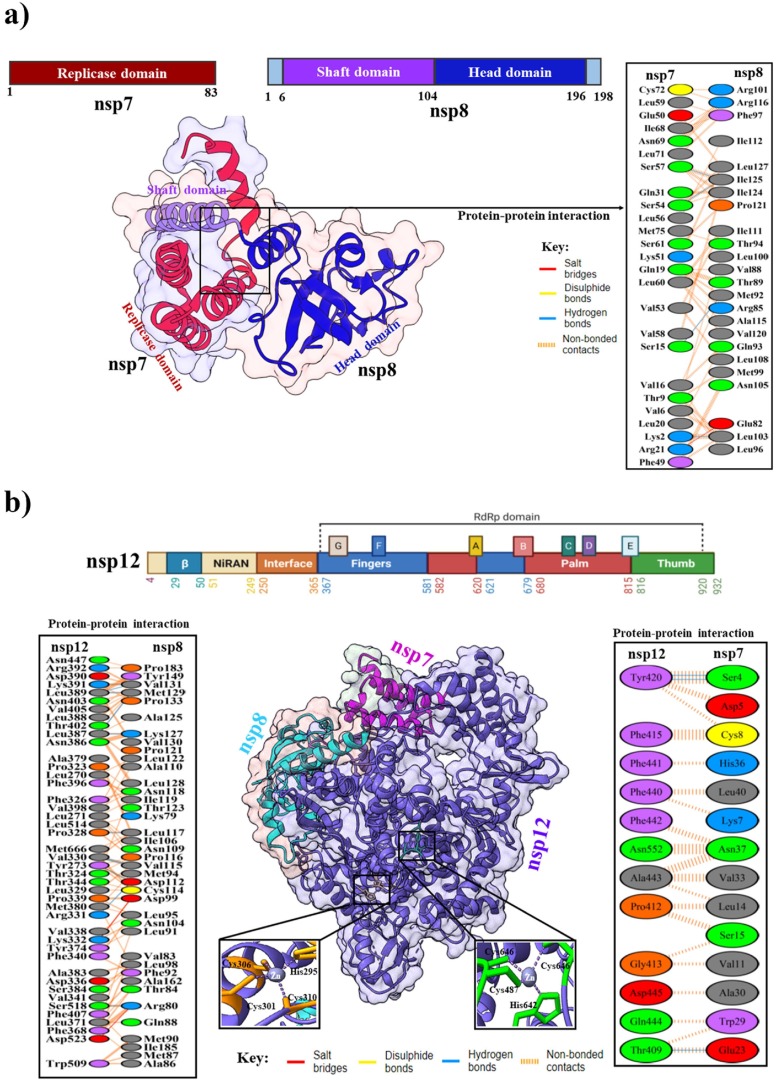
The SARS-CoV-2 nsp7 is composed of α-helical structure with three helical bundle folds, whereas nsp8 has two subdomains: An N-terminal ‘shaft’ domain (6–104 residues) and C-terminal ‘head’ domain comprised of four anti-parallel β-strands118 ( Fig. 6 a) (Konkolova et al., 2020). The crystal structure of SARS-CoV-2 nsp7 with nsp8 is a hollow cylindrical hexadecameric complex that forms a dimer conformation with negatively charged outer skin and positive charged inner core channel. The channel is mainly formed by attaching the four N-terminus helices of nsp8, of which the structure resembles the “shaft” of a “golf-club” (Zhai et al., 2005). This charge distribution helps the phosphate backbone of nucleic acid to pass the cylindrical channel without any electrostatic repulsion (Krishna et al., 1994). The cylindrical nsp7-nsp8 complex is stabilized by a salt bridge, four H-bond, and 90 hydrophobic interactions. The structural details of the nsp7-nsp8 complex suggest that the development of a potential allosteric inhibitor can block RdRp activity. Distinct from nucleotide analogs that directly target RdRp, allosteric inhibitors disrupt the assembly of nsp7-nsp8-nsp12, which inhibits the activity of RdRp machinery (Biswal et al., 2021).
Fig. 6.
(a) Domain organization of nsp7 and nsp8. Cartoon representation of SARS-CoV-2 nsp7 bound to the C-terminal of nsp8 in the lower-left panel (PDB: 6M5I). The lower-right panel shows protein–protein interactions between SARS-CoV-2 nsp7 and nsp8. (b) The overall topology of SARS-CoV-2 nsp12 with different colors domains. Cartoon representation in the below (left panel) shows SARS-CoV-2 nsp12-nsp7-nsp8 complex (PDB: 7BV2). The conserved zinc-binding motifs are highlighted in the SARS-CoV-2 nsp12 structure. The coordinate details of the zinc-binding residues are shown in stick representation. Protein-protein interactions between SARS-CoV-2 nsp12, nsp7, and nsp8 are shown in the left and right panels.
Nsp12 is a multi-subunit RNAdependent RNA polymerase (RdRp) (van Hemert et al., 2008). Structurally, nsp12 contains two main functional domains, i.e., N-terminal (1–379) and a polymerase domain (398–919) (Yin et al., 2020). The polymerase domain at the C-terminus part resembles a “right hand” cupped shaped conformation with finger subdomain (398–581, 628–687), palm subdomain (582–627, 688–815), and a thumb subdomain (816–919) (Fig. 6 b) (de Clercq, 2006). The N-terminus of RdRp contains a norovirus RdRp-associated nucleotidyltransferase domain or NiRAN domain (115–250), and an extended N-terminal β-hairpin domain (31–50) (Yin et al., 2020). The NiRAN domain is followed by an interface domain (251–365), connected to the RdRp domain, and the β-hairpin domain inserts into a groove clamped by the palm domain and NiRAN domain (Gao et al., 2020). However, the active site of RdRp is situated at the finger and thumb sub-domain interface, which are the center of the substrate domain where RNA synthesis takes place (Cheng et al., 2005). In the NiRAN domain and fingers domain, two Zn2+ ions are situated distally from the RdRp catalytic site, coordinated with highly conserved residues that may be essential for structural stability. Two Mg2+ ions are coordinated to D618, D760, D761, and NTP in the palm subdomain, which forms the polymerase catalytic core (Yin et al., 2020). The complex of SARS-CoV-2 nsp12 is bound to nsp7 and nsp8 (PDB: 7BV2), forming two salt bridges, 11H-bonds, and 166 hydrophobic interactions with nsp8 and three H-bond, and 40 hydrophobic interactions with nsp7. The nsp7-nsp8 heterodimer's attachment to the finger loop stabilizes the polymerase domain, allowing for greater affinity for template RNA. The second subunit of nsp8 is thought to play a key role in polymerase activity, potentially by binding to the template RNA and providing an expanded interaction surface, which keeps the RNA strand in place. In the presence of both nsp7 and nsp8, nsp12's binding affinity is increased significantly to template-primer RNA and the polymerase activity is also enhanced (Gao et al., 2020). Currently, numerous nucleoside analog drugs such as remdesivir, and galidesivir (adenosine analogs) (Elfiky, 2020), ribavirin and favipiravir (guanine analogs) (de Clercq, 2019), molnupiravir (Sheahan et al., 2020), sofosbuvir (uridine analog) (Gane et al., 2013) have been used to block the catalytic active site of RdRp. Moreover, RdRp is also targeted by the drug Suramin, a poly-sulfonated trypan blue derivative that effectively suppresses a range of viruses, including SARS-CoV-2 (Zoltner et al., 2020).
4.8. nsp9
SARS-CoV-2 nsp9 is an ssRNA-binding protein involved in viral replication (Littler et al., 2020). Structurally, nsp9 is homologous to a subdomain of serine protease, particularly the first domain of picornaviral 3CLpro (PDB ID: 1L1N) and the second domain of the SARS-CoV 3CLpro (PDB ID: 1P9U, 1Q2W, and 1P9S). Like other nsp9 homologs it exhibits an unusual fold that is yet to be observed outside coronaviruses (Littler et al., 2020, Sutton et al., 2004). The folding core consists of a 6-stranded enclosed β-barrel, from which most of the extended loops are observed outward (Fig. 5 d) (Biswas et al., 2021). Two glycine-rich loops, such as β2-β3 and β3-β4 are positively charged and involved in RNA-binding. The N-terminal β-strand in SARS-CoV-2 nsp9 forms a dimer interface with the C-terminal α1-helix. The GXXXG motif is highly conserved at the dimer interface, allowing the helixes in a close pack at the residues G100 and G104 (Miknis et al., 2009). Littler et al. reported that hydrophobic residues of the α1-helix form a funnel-like hydrophobic cavity on either side of the dimer interface. In the structure of SARS-CoV-2 nsp9, the N-terminal tag is incorporated together with a rhinoviral 3C protease sequence (LEVL). This 3C sequence entered into the other side cavities of the dimer interface and proximal to the GXXXG motif. Furthermore, additional β-sheet interactions are formed by the 3C sequence with the N-terminus from the neighboring protomer (Biswas et al., 2021). The uracil-analog FR6, which has a weak backbone affinity with nsp9 (Littler et al., 2021b). This compound induces a hexameric form and modifies the oligomerization state, altering RNA entry channels and thus affect RNA binding. Furthermore, FR6 disrupts the dimer interface of the nsp9 GXXXG, impacting RNA binding and viral proliferation (Hu et al., 2017).
4.9. nsp10, nsp14, and nsp16
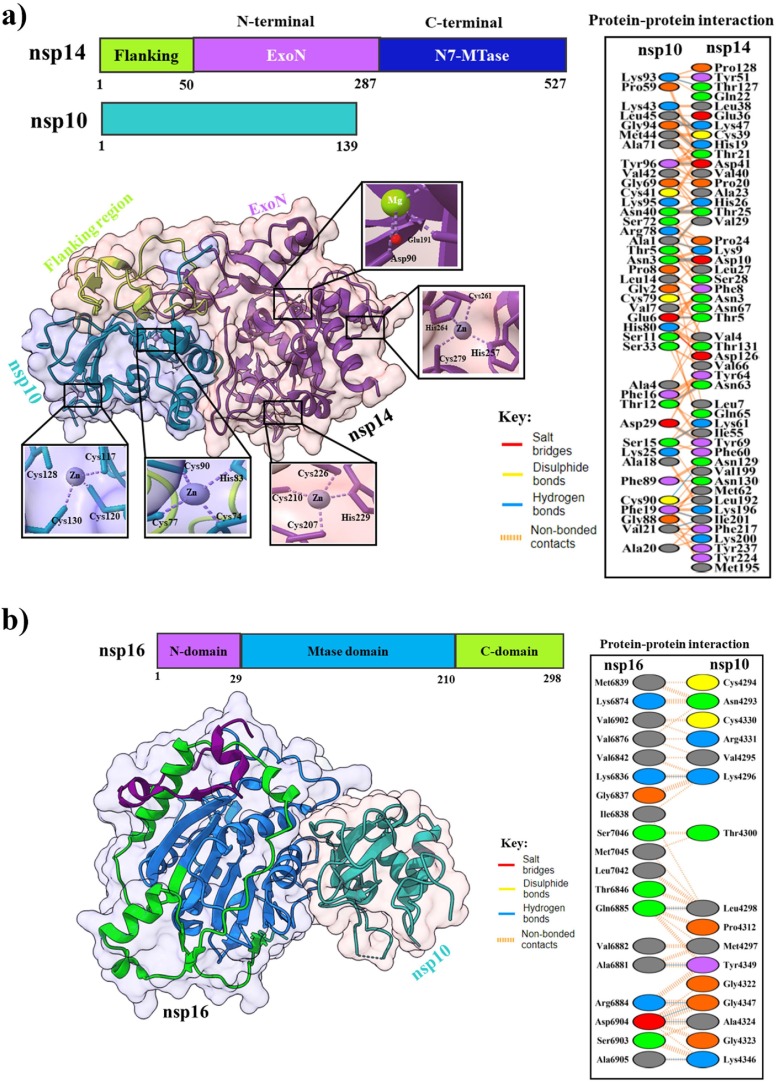
The nsp10 protein is a small, single-domain protein with 139 amino acids that acts as a scaffold to bind with the nsp14 (exonuclease and N7-methyltransferase) and nsp16 (20-Omethyltransferase) for forming the mRNA cap methylation complex (Chen et al., 2013). The crystal structure of nsp10 (PDB: 7DIY) shows that it comprises a helical domain, an anti-parallel β-sheet, and two zinc-binding sites (Fig. 7 a). A short peptide, K29 is found in SARS-CoV nsp10 (resides 68–96) that can inhibit the 2′-O-methyltransferase activity of nsp16 (Ke et al., 2012).
Fig. 7.
(a) Overall topology of SARS-CoV-2 nsp10 and nsp14. The lower-left panel shows the cartoon and surface representations of the nsp10 and nsp14 complex (PDB: 7DIY). Zinc and magnesium ions are shown as spheres and are colored magenta and lemon. Magnified views on the two zinc-finger motifs in nsp10 and nsp14, and one magnesium-finger motif in nsp14. Residues involved in zinc and magnesium-coordination are shown in sticks. The lower-right panel shows protein–protein interactions between nsp10 and nsp14 proteins. (b) Domain organization of SARS-CoV-2 nsp16 with different colors. The lower-left panel shows the cartoon and surface representations of the nsp16 and nsp10 complex (PDB: 7L6R). The lower right-panel shows protein–protein interactions between nsp16 and nsp10.
The nsp14 is a bifunctional protein, which contains the N-terminal exonuclease (ExoN) domain and N7 methyltransferase (N7-MTase) domain for RNA cap formation (Fig. 7 a) (Konkolova et al., 2020), and is also critical for proofreading, repair, and safeguarding the viral genome as a part of the replication-transcription complex (RTC) throughout the viral life cycle (Bouvet et al., 2012). The ExoN domain contains two zinc-finger motifs (C207-C210-C226-H229 and H257-C261-H264-C279) are sterically located close to the MTase domain, connecting the β11/ β12, β13/ β14, and α4/ α5 intervening loops to helix α5. In the nsp10-nsp14 complex structure, nsp10 helices α1, α2, α3, η1, strands β1, β2 and most of their intervening loops are involved in the binding including nsp14 helix η1, strands β2, β8, β11, a long N-terminal loop and multiple intervening loops (β2/ β3, β3/α1, β7/ β8, α3/ β11, and β11/ β12) (Lin et al., 2021). Several studies revealed that the relationship between nsp14-nsp10 is explicitly responsible for establishing the ExoN activity, i.e., 35-fold enhanced activity (Bouvet et al., 2012, Ma et al., 2015). For SARS-CoV-2, nsp10 in complex with nsp14 stabilizes the ExonN activity by binding of a salt bridge, 20H-bonds, and 272 hydrophobic interactions. Therefore, the ExoN domain's catalytic pocket visibility collapses in the absence of nsp10 (Ziebuhr et al., 2004). Nsp14 is an exoribonuclease that removes both nucleotide analogs and mis-incorporated nucleotides from the nascent RNA, which can lead to the development of nucleotide analog-based antiviral resistance (Ferron et al., 2017, Robson et al., 2020). This issue can be resolved using a combination of nsp14 inhibitors and nucleotide analogs (such as sofosbuvir, remdesivir, and ribavirin) (Eastman et al., 2020, Jockusch et al., 2020, Khater et al., 2021). For example, the activity of ExoN is inhibited by 3′-deoxy nucleotide analogs (Liu et al., 2021); Ebselen and Disulfiram, zinc-ejecting agents, inhibit the activity of nsp14 via their three Zn-binding sites (Chen et al., 2021, Sargsyan et al., 2020); Several SAM analogs and competitive inhibitors such as sinefungin (SFG), aurintricarboxylic acid (ATA), and S-adenosylhomocysteine (SAH) impede N7-Mase activity (Ahmed-Belkacem et al., 2020, He et al., 2004) ultimately preventing 5′-end cap formation (Devkota et al., 2021). Additionally, to these inhibitors, mutations in nsp14 can impair virus replication and induce a higher level of the interferon response. Therefore, the development of live attenuated virus vaccines and antibodies are alternative options for this target (Graham et al., 2012, Lu et al., 2020b).
The nsp16 is an S-adenosyl methionine (SAM) dependent and m7GpppA (Cap-1) specific protein which catalyzes the 5′-methyl capping of viral mRNA (Bouvet et al., 2010). It contains a canonical SAM-MTase fold with eight stranded cores twisted β-sheet flanked by three helices on one side and two α-helices on the other side (Fig. 7 b) (Viswanathan et al., 2020). Nsp16 is inactive by itself and requires nsp10 for the activity. Nsp10 interacts with nsp16 through seven H-bonds and 90 hydrophobic interactions to stabilize the SAM binding site (Lin et al., 2020, Rosas-Lemus et al., 2020). The genetic disruption of SARS-CoV nsp16 causes a tenfold reduction in the viral RNA synthesis. Disrupting the nsp10/nsp14 or nsp10/nsp16 interface might be an excellent therapeutic target method since the interaction of nsp10 with nsp14 and nsp16 is required for their optimum activity (Bouvet et al., 2014). RNA-cap methyltransferase (nsp16) might be a crucial target in developing antiviral drugs against SARS-CoV-2, although no effective inhibitors or licensed drugs are currently available (Rohaim et al., 2021).
4.10. nsp13 (Helicase)
SARS-CoV-2 nsp13 is an RNA helicase that unwinds double-stranded RNA (dsRNA) or DNA (dsDNA) into single strands in an ATP-dependent manner, and its helicase activity can be enhanced by binding of polymerase protein (Jia et al., 2019). Guo et al. reported that nsp13 has an inhibitory role in regulating type I interferon production, and overexpression of nsp13 suppressed IFN-β levels in the host cell (Guo et al., 2021). The structural studies of SARS-CoV-2 nsp13 contain an N-terminal zinc-binding domain (ZBD), two helicase subdomains RecA1 (1A) and RecA2 (2A), a β-barrel 1B domain being connected to ZBD via a helical “stalk” region (Fig. 5 e) (Yan et al., 2020).Two copies of nsp13 can interact with the core nsp7-nsp8-nsp12 complex on opposing sides of the RNA binding cleft. In both cases, the interactions are mediated from the ZBD (residues V45, N46, M68, I79, S80, F81, F90, G91, L92, Y93, K94, N95) and 1B domain (residues V193, Q194. H230, R248, Y253, L256), which interacts with the “shaft” and “head” domains of nsp8 and the thumb domain of nsp12. The structure of nsp13 in a complex with nsp8 and nsp12, might have potential implications for helicase activity and regulation (Chen et al., 2020c). Zeng et al. identified novel inhibitors such as FPA-124 and suramin-like compounds that can inhibit viral helicase activities (Zeng et al., 2021).
4.11. nsp15
SARS-CoV-2 nsp15 is a uridine-specific endoribonuclease and is highly conserved across coronaviruses. It is responsible for interference with the innate immune response (Pillon et al., 2021). An earlier study reported that overexpression of nsp15 inhibits the interferon (IFN) response and the mitochondrial antiviral signaling protein (MAVS) mediated apoptosis in SARS-CoV (Frieman et al., 2009). Nsp15 contains three discrete domains: N-terminal “wing” domain, middle “body” domain and C-terminal catalytic NendoU “wing” domain (Fig. 5 f) (Gordon et al., 2020). The N-terminal domain consists of two α-helices with three anti-parallel β-sheets: uridine-specific endoribonuclease (Ricagno et al., 2006). The middle domain comprises ten β-strands, a major mixed β-sheet, three small and two short α-helices, which form a hexamer resulting in the concave surfaces that may act as an interaction hub (Joseph et al., 2007). The C-terminal catalytic NendoU domain contains two anti-parallel β-sheets that play a catalytic function in SARS-CoV-2 and likely many others coronaviruses family (Ulferts and Ziebuhr, 2011). However, the active site in the C-terminal is formed by six major residues (His235, His250, Lys290, Thr341, Tyr343, and Ser294), which is significant for replication in the host cell (Xu et al., 2006). The inhibitors that target nsp15, are modified oligonucleotides containing derivatives or uracil derivatives, such as 2′-fluorine-modified RNA and tipiracil on the uridine ribose (Guo et al., 2017, Kim et al., 2021). As part of the nsp15/ tipiracil complex, tipiracil inhibits the activity of EndoU and analogously binds to uridine. EndoU is inhibited by compounds that perturb the stability of the hexamer conformation (Kim et al., 2021, Kish and Uppal, 2016). Additionally, SARS-CoV-2 with nsp15 defects has been proposed as a live attenuated virus vaccine (Hackbart et al., 2020).
5. Accessory factors
5.1. ORF3a
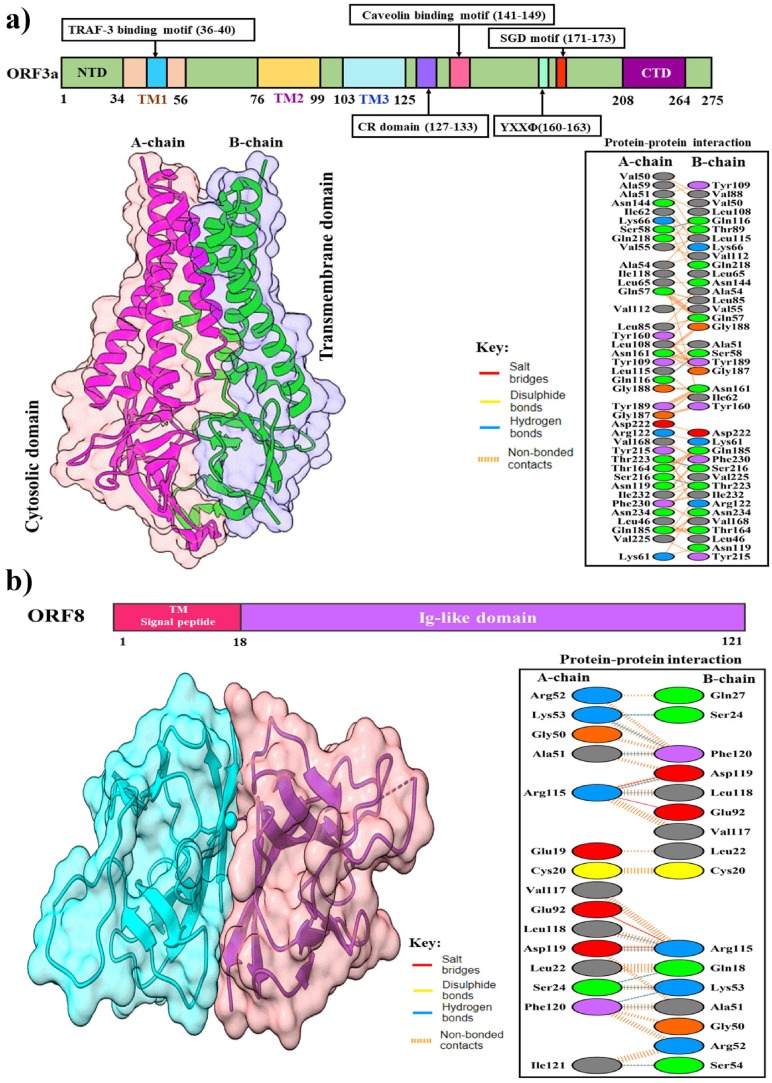
ORF3a is a membrane-associated protein located between the gene of the spike and envelope protein (Yu et al., 2004), which is associated with apoptosis, pathogenicity, and virus release (Ren et al., 2020). The recent cryo-EM structure of SARS-CoV-2 ORF3a (PDB: 6XDC) has N-terminal ectodomain, three transmembrane regions, a cysteine-rich domain, YxxΦ domain, diacidic domain, and C-terminal endodomain (Fig. 8 a) (Kern et al., 2021). Cysteine-rich domain forms homo and hetero-tetramers, which cluster together with S protein by forming inter-chain disulfide bonds and a potassium-permeable ion channel (McBride and Fielding, 2012). Potassium–permeable ion channel is significant for integrating viral particles (Issa et al., 2020). It also interacts with the M and E proteins and helps viral assembly. It downregulates the type 1 interferon receptor by inducing serine phosphorylation within the IFN alpha-receptor subunit 1 (IFNAR1) degradation motif. By promoting TRAF3-dependent ubiquitination of caspase recruitment domain (ASC), ORF3a protein activates the NLR family pyrin domain containing 3 (NLRP3) inflammasome and inflammation plays an important role in viral infection (Siu et al., 2019). Apoptosis is a cell-death program that occurs in response to several extrinsic and intrinsic signals such as cellular stress, including virus infection. The ORF3a protein also triggers the mitochondrial death pathway by activating p38 MAP kinase (Padhan et al., 2008). The tyrosine-based sorting motif, YXXΦ, is essential for its trafficking to the plasma membrane from Golgi. ORF3a interacts with Caveolin-1, a protein that is part of lipid-rich regions of the membrane and plays an important role in cell signaling, cell cycle, and virus uptake (Minakshi and Padhan, 2014). Many signaling pathways are regulated by caveolin-1, including the extracellularly regulated kinase (ERK) and inducible nitric oxide synthase (iNOS) pathways. These two critical pathways are involved in cell survival, proliferation, and response to viral infection (Padhan et al., 2007). The ORF3a-HMOX1 (Gordon et al., 2020) complex plays an important role in anti-inflammatory effects through the NLRPS pathway (Lee and Chau, 2002, Lv et al., 2018). Treatment of COVID-19 with drugs that block the interaction between ORF3a and HMOX1 is an effective approach. In addition, anti-ORF3a antibodies have been detected in the plasma of patients convalescing from COVID-19 (Grifoni et al., 2020, Oja et al., 2020). Suppressing the expression of ORF3a in SARS-CoV resulted in less virus release and less morbidity (Castaño-Rodriguez et al., 2018, Lu et al., 2006).
Fig. 8.
Structures of SARS-CoV-2 accessory proteins. (a) The upper panel shows a schematic representation of the domains of SARS-CoV-2 ORF3a protein; N-terminal ectodomain, three transmembrane regions, a cysteine-rich domain, YxxΦ domain, diacidic domain, and C-terminal endodomain. The lower-left panel shows a cartoon and surface representation of cryo-EM structure of dimeric ORF3a (PDB: 6XDC). The lower-right panel shows protein–protein interactions between the A-chain and B-chain of the dimeric structure. (b) The upper panel shows the overall topology of SARS-CoV-2 ORF8. The lower-left panel shows the cartoon and surface representation of the crystal structure of dimeric ORF8 (PDB: 7JTL). The lower-right panel shows detailed protein–protein interactions between the A-chain and B-chain of the dimeric structure.
5.2. ORF6
The ORF6 of SARS-CoV-2 is a 61 amino acid long protein found in the endoplasmic reticulum and membranes of vesicles such as lysosomes and autophagosomes (Lee et al., 2021). It is a potent IFN antagonist, a role reported in SARS-CoV previously reported by Kopecky-Bromberg and coworkers (Kopecky-Bromberg et al., 2007). In SARS-CoV, ORF6 was found to co-localize and interact with nsp8. It is still unknown whether ORF6 has a role in RNA synthesis or whether its association with nsp8 influences the replication-transcription complex's activity (Mariano et al., 2020).
5.3. ORF7a
SARS-CoV-2 ORF7a is a type-1 transmembrane protein that contains a signal peptide, luminal domain or ectodomain, transmembrane segment, and cytoplasmic tail (Liu et al., 2014). Huang et al. reported that SARS-CoV ORF7a physically interacts with SARS-CoV S protein and ORF3a protein for incorporation into mature virions (Huang et al., 2006). An earlier study reported no significant impact on viral RNA replication and synthesis in cell culture due to the elimination of ORF7a from the SARS-CoV genome (Yount et al., 2005). Schaecher et al. reported that both ORF7a and ORF7b deletion do not significantly impact on SARS-CoV replication (Schaecher et al., 2007).
5.4. ORF8
The ORF8 is a unique accessory protein of SARS-CoV-2 having an N-terminal signal peptide (1–17) for transport to the endoplasmic reticulum, and an Ig-like domain (18–121) consisting of a beta-strand core (Fig. 8 b) (Hassan et al., 2021). A study reported a novel immunoglobulin (Ig) domain that may play a role as a potential immune modulator to reduce the host immune response against the virus and it contains highly conserved cysteine residues that formed disulfide-bond might enhance to dimerization (Grifoni et al., 2020). Although ORF8 does not participate in viral replication, it possibly plays a direct role in viral pathogenesis by interaction with host molecules and possibly played a significant role in viral trafficking into the cells (Gordon et al., 2020). However, the protein is fast-evolving in SARS-CoV-2 viruses that might be considered a mutational hotspot of the recent pandemic (Su et al., 2020). Drugs can be developed based on the crystal structure of SARS-CoV-2 ORF8 to perturb the interaction between ORF8 and a variety of host proteins, including IL17RA (interleukin 17 receptor), LOX (lysyl oxidase), and GDF15 (growth/differentiation factor 15) (Zinzula, 2021). A substantial antibody response is also generated in the host by ORF8 protein, which has become the major serological marker for SARS-CoV-2 infection (Gordon et al., 2020, Hachim et al., 2020).
6. Potential vaccines for COVID-19
There has been an increased effort around the world to develop vaccines in response to the SARS-CoV-2 outbreak and emerging variants. In order to fight infection, vaccines are administered to people of all ages to build and strengthen humoral and cellular immunity (Fahmi et al., 2021). Globally, 37 authorized/ approved vaccines have been developed and 97 vaccines are in various phases of trial (https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker; accessed on July 24, 2022). The majority of these are aimed at inducing neutralizing antibodies against the spike protein (S). As a result, human ACE-2 receptors are prevented from being occupied by these antibodies, preventing viral entry (Thanh Le et al., 2020).
Currently, several vaccine candidates are being tested in Phase 3 clinical trials (Table 2 ) The mRNA-1273 and mRNA-BNT162b2 vaccines both are encapsulated in a lipid nanoparticle (LNP) that encodes full-length sequence of SARS-CoV-2 S protein and stabilized prefusion conformation, including a transmembrane anchor and an entire S1-S2 cleavage site (Jackson et al., 2020, Sahin et al., 2021). A similar mechanism of action is expressed by both Pfizer and Moderna vaccines. The purpose of these vaccines is to induce both B- and T-cell responses against the spike protein (Patel et al., 2022). The Ad5-nCoV is a replication-defective adenovirus type-5 vectored vaccine that encodes the full-length of SARS-CoV-2 S protein. Although this vaccine's efficacy is relatively high, but a drawback is that it may not be effective for people with recessive infectious diseases (Han et al., 2021). The AZD1222 is a recombinant adenovirus vaccine that was developed using codon-optimized S glycoprotein. In the shuttle (plasmid) vector, amino acid (2 to 1273) and tissue plasminogen activator (tPA) leader sequences at 5′end are encapsulated (Kaur and Gupta, 2020). In the human body, these modified adenoviruses cannot replicate as the gene which facilitates virion assembly has been removed and introducing the SARS-CoV-2 antigenic component into the host cell in a safe way (Li et al., 2021). This approach results in the expression of the S protein by host cells, which activates a potent humoral and cell-mediated immune response (Ewer et al., 2020, van Doremalen et al., 2020).
Table 2.
Current vaccine candidates are in Phase 3 clinical evaluation against SARS-CoV-2.
| Candidate | Vaccine type | Manufacturer | Mechanism of action | Efficacy |
|---|---|---|---|---|
| AZD1222 | Viral Vector | Oxford/AstraZeneca | The ChAdOx1 vector has been engineered to contain genetic information that encodes the S protein of the wild-type SARS-CoV-2 | 62–90 % |
| mRNA-1273 | mRNA | Moderna/US NIAID | Activates T cells to assist in the development of B cells that produce antibodies; initiating an adaptive immune response to the virus' S protein | 95 % |
| BNT162b2 | mRNA | Pfizer/BioNTech | Lipid nanoparticles encapsulate the mRNA of SARS-CoV-2 S protein. Immunological responses are triggered when these proteins are released from the injected cells | 95 % |
| Convidicea Ad5-nCoV | Viral vector | CanSino | Enables to recognize the SARS-CoV-2 S protein and triggers an immune response | 65.7 % in moderate cases, and 90.98 % in severe cases |
| Sputnik V/Gam-COVID Vac | Viral vector | Gamaleya Research Institute of Epidemiology and Microbiology | Enables to recognize the SARS-CoV-2 S protein and triggers an immune response | 92 % |
| JNJ-78436735/ Ad26.COV2.S | Viral vector | Johnson and Johnson/ Janssen | A recombinant, non-replicative human adenovirus vector that recognize the antigen of S protein without the virus to propagate once inside human cells | 66 % |
| CoronaVac | Inactivated | Sinovac Biotech | Well tolerated and β-propiolactone-activation virus provides an immune response in the host cell | 50 % |
| BBIBP-CorV | Inactivated | Sinopharm (Beijing) | It induced robust humoral responses in a short period of time | 79 % |
| Covaxin | Inactivated | Bharat Biotech | Well tolerated and β-propiolactone-activation virus provides an immune response in the host cell | 100 % in severe cases and 70 % in asymptomatic cases |
| NVX-CoV2373 | Recombinant nanoparticle | Novavax | S protein-containing nanoparticles are injected into the arm muscle to activate the antigen-presenting cells | 89 % |
| ZF2001 | Protein subunit | Anhui Zhifei Longcom Bio | Full immunity still under investigation | NA |
| Unknown | Inactivated | Sinopharm (Wuhan) | After 14 days, an immunogenic, significant neutralizing antibody response is observed | 73 % |
| EpiVacCorona | Protein subunit | Federal Budgetary Research Institution State Research Center of Virology and Biotechnology | Induces virus-specific and neutralizing antibodies | 82 % |
| Data source: (Covid-19: China approves Sinopharm, xxxx, Kaur and Gupta, 2020, Kyriakidis et al., 2021, Sanyaolu et al., 2022) | ||||
Sputnik V is an adenoviral vaccine containing two vectors which carry the SARS-CoV-2 gene for S protein (Chugh et al., 2021). The SARS-CoV-2 S protein is transferred into the cells through the gene-containing vector (rAd26) during the first vaccination, which then triggers an immune response. During a second vaccination, an additional vector (rAd5) is introduced to the body for boosting immunity, and ensuring long-term protection (Zahid et al., 2021). The JNJ-78436735 is a recombinant vector vaccine that expresses the SARS-CoV-2 spike protein within cells using a human adenoviral vector. By introducing a segment of DNA from SARS-CoV-2 into the adenovirus, which has been genetically modified so that it cannot replicate in the body. CoronaVac is an inactivated vaccine. A dead version of SARS-CoV-2 is used to prevent replication, however, the surface spike protein is preserved to trigger the immune system to form antibodies against the live virus (Wong et al., 2022).
7. Conclusion
In response to the SARS-CoV-2 pandemic, considerable progress has been made in revealing the structural features of the non-structural, structural, and accessory proteins of SARS-CoV-2 and their functionalities in the virus's life cycle. The viral protein’s structures have assisted the development of various antiviral drugs and antibody treatments. Particularly, antiviral drug Veklury (also known as remdesivir) was designed targeting the RNA-dependent polymerase. In addition, Paxlovid and Lagevrio (molnupiravir) were developed inhibiting the main protease, and several monoclonal antibody treatments are offered for treatment targeting the spike protein. However, for emerging variants, continuous research and development initiatives are expected to improve the strategy of the structural-based rational drug design. Drugs and biological products targeting the various conserved binding sites or multiple key sites of viral and host proteins in the life cycle of SARS-CoV-2 can assist developing more potent therapeutics for covid treatment. In summary, this review provided details structural insights of the representative viral proteins of SARS-CoV-2 to advance the development of antiviral drugs, peptides, antibody, and vaccine preventing and treating the COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We would like to acknowledge Md. Nazmus Samdani, Niaz Morshed, Rumman Reza, Department of Pharmacy, University of Dhaka, Dhaka-1000, Bangladesh; Golam Md. Adil, Himadree Sarkar, Department of Genetic Engineering and Biotechnology, Shahjalal University of Science and Technology, Sylhet-3114, Bangladesh; Shafiqul Islam, Shaila Akter, Sadia Afrose Esha, Md Ackas Ali, Division of Infectious Diseases and Division of Computer Aided Drug Design, The Red-Green Research Centre, BICCB, 16 Tejkunipara, Tejgaon, Dhaka, 1215, Bangladesh; Nazma Sultana Lupin, Department of Microbiology, Friedrich Schiller University of Jena, Germany; Kaniz Fatema, Global Centre for Environmental Remediation, University of Newcastle, Australia for their contribution in the data collection.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imbio.2022.152302.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Ahmed S., Mahtarin R., Ahmed S.S., Akter S., Islam M.S., Mamun A.A., Islam R., Hossain M.N., Ali M.A., Sultana M.U.C., Parves M.D.R., Ullah M.O., Halim M.A. Investigating the binding affinity, interaction, and structure-activity-relationship of 76 prescription antiviral drugs targeting RdRp and Mpro of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020;39:1. doi: 10.1080/07391102.2020.1796804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed-Belkacem R., Sutto-Ortiz P., Guiraud M., Canard B., Vasseur J.J., Decroly E., Debart F. Synthesis of adenine dinucleosides SAM analogs as specific inhibitors of SARS-CoV nsp14 RNA cap guanine-N7-methyltransferase. Eur. J. Med. Chem. 2020;201 doi: 10.1016/J.EJMECH.2020.112557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfuwaires M., Altaher A., Kandeel M. Molecular dynamic studies of interferon and innate immunity resistance in MERS CoV non-structural protein 3. Biol. Pharm. Bull. 2017;40:345–351. doi: 10.1248/bpb.b16-00870. [DOI] [PubMed] [Google Scholar]
- Almazán F., DeDiego M.L., Galán C., Escors D., Álvarez E., Ortego J., Sola I., Zuñiga S., Alonso S., Moreno J.L., Nogales A., Capiscol C., Enjuanes L. Construction of a Severe Acute Respiratory Syndrome Coronavirus Infectious cDNA Clone and a Replicon To Study Coronavirus RNA Synthesis. J. Virol. 2006;80:10900–10906. doi: 10.1128/jvi.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M.S., Johnson M.A., Herrmann T., Geralt M., Wüthrich K. Novel β-Barrel Fold in the Nuclear Magnetic Resonance Structure of the Replicase Nonstructural Protein 1 from the Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2007;81:3151–3161. doi: 10.1128/jvi.01939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) Structure: Basis for design of anti-SARS drugs. Science. 2003;1979(300):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J., et al. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4(e00524-13) doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansori A.N.M., Kharisma V.D., Fadholly A., Tacharina M.R., Antonius Y., Parikesit A.A. Severe acute respiratory syndrome Coronavirus-2 emergence and its treatment with alternative medicines: a review. Res J Pharm Technol. 2021;14:5551–5557. doi: 10.52711/0974-360X.2021.00967. [DOI] [Google Scholar]
- Arya, R., Das, A., Prashar, V., Kumar, M., 2020. Potential inhibitors against papain-like protease of novel coronavirus (SARS-CoV-2) from FDA approved drugs. Chemrxiv.Org. https://doi.org/10.26434/chemrxiv.11860011.v2.
- Baez-Santos Y.M., Mielech A.M., Deng X., Baker S., Mesecar A.D. Catalytic Function and Substrate Specificity of the Papain-Like Protease Domain of nsp3 from the Middle East Respiratory Syndrome Coronavirus. J. Virol. 2014;88:12511–12527. doi: 10.1128/jvi.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y.M., st. John, S.E., Mesecar, A.D., The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015 doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagherzadeh, K., Daneshvarnejad, K., Abbasinazari, M., Azizian, H., 2020. In silico Repositioning for Dual Inhibitor Discovery of SARS-CoV-2 (COVID- 19) 3C-like Protease and Papain-like Peptidase. https://doi.org/10.20944/preprints202004.0084.v1.
- Baliji S., Cammer S.A., Sobral B., Baker S.C. Detection of Nonstructural Protein 6 in Murine Coronavirus-Infected Cells and Analysis of the Transmembrane Topology by Using Bioinformatics and Molecular Approaches. J. Virol. 2009;83:6957–6962. doi: 10.1128/jvi.00254-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The Papain-Like Protease of Severe Acute Respiratory Syndrome Coronavirus Has Deubiquitinating Activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/jvi.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D.J., Wrobel A.G., Xu P., et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/S41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto D., Angeletti S., Giovanetti M., Bianchi M., Pascarella S., Cauda R., Ciccozzi M., Cassone A. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020;81:e24–e27. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmik D., Nandi R., Jagadeesan R., Kumar N., Prakash A., Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infection, Genetics and Evolution. 2020;84 doi: 10.1016/J.MEEGID.2020.104451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal M., Diggs S., Xu D., Khudaverdyan N., Lu J., Fang J., Blaha G., Hai R., Song J. Two conserved oligomer interfaces of NSP7 and NSP8 underpin the dynamic assembly of SARS-CoV-2 RdRP. Nucleic Acids Res. 2021;49:5956–5966. doi: 10.1093/NAR/GKAB370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas N., Kumar K., Mallick P., Das S., Kamal I.M., Bose S., Choudhury A., Chakrabarti S. Structural and Drug Screening Analysis of the Non-structural Proteins of Severe Acute Respiratory Syndrome Coronavirus 2 Virus Extracted From Indian Coronavirus Disease 2019 Patients. Front. Genet. 2021;12 doi: 10.3389/FGENE.2021.626642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla P.J., Gorbalenya A.E., Weiss S.R. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: Heterogeneity among MHV strains. Virology. 1994;198:736–740. doi: 10.1006/viro.1994.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In Vitro Reconstitution of SARS-Coronavirus mRNA Cap Methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci U S A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Lugari A., Posthuma C.C., Zevenhoven J.C., Bernard S., Betzi S., Imbert I., Canard B., Guillemot J.C., Lécine P., Pfefferle S., Drosten C., Snijder E.J., Decroly E., Morelli X. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannalire R., Cerchia C., Beccari A.R., di Leva F.S., Summa V. Targeting SARS-CoV-2 Proteases and Polymerase for COVID-19 Treatment: State of the Art and Future Opportunities. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Rodriguez, C., Honrubia, J.M., Gutiérrez-Álvarez, J., DeDiego, M.L., Nieto-Torres, J.L., Jimenez-Guardeño, J.M., Regla-Nava, J.A., Fernandez-Delgado, R., Verdia-Báguena, C., Queralt-Martín, M., Kochan, G., Perlman, S., Aguilella, V.M., Sola, I., Enjuanes, L., 2018. Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. mBio 9. https://doi.org/10.1128/MBIO.02325-17. [DOI] [PMC free article] [PubMed]
- Cavasotto C.N., Lamas M.S., Maggini J. Functional and druggability analysis of the SARS-CoV-2 proteome. Eur. J. Pharmacol. 2021;890 doi: 10.1016/J.EJPHAR.2020.173705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J., Cai Y., Pang C., Wang L., McSweeney S., Shanklin J., Liu Q. Structural basis for SARS-CoV-2 envelope protein recognition of human cell junction protein PALS1. Nat. Commun. 2021;12 doi: 10.1038/S41467-021-23533-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Chen C.-M.-M., Chiang M., Hsu Y., Huang T. Transient Oligomerization of the SARS-CoV N Protein – Implication for Virus Ribonucleoprotein Packaging. PLoS ONE. 2013;8:e65045. doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-Y., Chang C., Chang Y.-W., Sue S.-C., Bai H.-I., Riang L., Hsiao C.-D., Huang T. Structure of the SARS Coronavirus Nucleocapsid Protein RNA-binding Dimerization Domain Suggests a Mechanism for Helical Packaging of Viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Fei C.Y., Chen Y.P., Sargsyan K., Chang C.P., Yuan H.S., Lim C. Synergistic Inhibition of SARS-CoV-2 Replication Using Disulfiram/Ebselen and Remdesivir. ACS Pharmacol Transl Sci. 2021;4:898–907. doi: 10.1021/ACSPTSCI.1C00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020 doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A., et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/J.CELL.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tao J., Sun Y., Wu A., Su C., Gao G., Cai H., Qiu S., Wu Y., Ahola T., Guo D. Structure-Function Analysis of Severe Acute Respiratory Syndrome Coronavirus RNA Cap Guanine-N7-Methyltransferase. J. Virol. 2013;87:6296–6305. doi: 10.1128/jvi.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Zhang W., Xie Y., Jiang W., Arnold E., Sarafianos S.G., Ding J. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology. 2005;335:165–176. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.C., Wang A.H.J. Structural D/E-rich repeats play multiple roles especially in gene regulation through DNA/RNA mimicry. Mol. BioSyst. 2015;11:2144–2151. doi: 10.1039/c5mb00206k. [DOI] [PubMed] [Google Scholar]
- Chowdhury S.M., Talukder S.A., Khan A.M., Afrin N., Ali M.A., Islam R., Parves R., Mamun A., al, Sufian, Md.A., Hossain, M.N., Hossain, M.A., Halim, M.A., Antiviral Peptides as Promising Therapeutics against SARS-CoV-2. J. Phys. Chem. B. 2020;124:9785–9792. doi: 10.1021/ACS.JPCB.0C05621. [DOI] [PubMed] [Google Scholar]
- Chugh H., Awasthi A., Agarwal Y., Gaur R.K., Dhawan G., Chandra R. A comprehensive review on potential therapeutics interventions for COVID-19. Eur. J. Pharmacol. 2021;890 doi: 10.1016/J.EJPHAR.2020.173741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L.K., Green T.J., Petit C.M. Structure of Nonstructural Protein 1 from SARS-CoV-2. J. Virol. 2021;95 doi: 10.1128/JVI.02019-20/ASSET/CA14D612-0C60-4620-A1E6-D2A037BDE51E/ASSETS/GRAPHIC/JVI.02019-20-F0007.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335. doi: 10.4161/AUTO.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10(8):1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covid-19: China approves Sinopharm vaccine for general use - BBC News [WWW Document], n.d. URL https://www.bbc.com/news/world-asia-china-55498197 (accessed 8.21.22).
- Cubuk, J., Alston, J.J., Incicco, J.J., Singh, S., Stuchell-Brereton, M.D., Ward, M.D., Zimmerman, M.I., Vithani, N., Griffith, D., Wagoner, J.A., Bowman, G.R., Hall, K.B., Soranno, A., Holehouse, A.S., 2021. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nature Communications 2021 12:1 12, 1–17. https://doi.org/10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed]
- de Clercq E. Potential antivirals and antiviral strategies against SARS coronavirus infections. Expert Rev Anti Infect Ther. 2006 doi: 10.1586/14787210.4.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Clercq E. New Nucleoside Analogues for the Treatment of Hemorrhagic Fever Virus Infections. Chem. Asian J. 2019;14:3962–3968. doi: 10.1002/ASIA.201900841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota K., Schapira M., Perveen S., Yazdi A.K., Li F., Chau I., Ghiabi P., Hajian T., Loppnau P., Bolotokova A., Satchell K.J.F., Wang K., Li D., Liu J., Smil D., Luo M., Jin J., Fish P., v., Brown, P.J., Vedadi, M., Probing the SAM Binding Site of SARS-CoV-2 Nsp14 In Vitro Using SAM Competitive Inhibitors Guides Developing Selective Bisubstrate Inhibitors. Slas Discovery. 2021;26:1200. doi: 10.1177/24725552211026261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh, D.C., Chalupska, D., Silhan, J., Veverka, V., Boura, E., 2020. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. bioRxiv 2020.04.02.022194. https://doi.org/10.1101/2020.04.02.022194. [DOI] [PMC free article] [PubMed]
- Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 2020;6:672. doi: 10.1021/ACSCENTSCI.0C00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efaz F.M., Islam S., Talukder S.A., Akter S., Tashrif M.Z., Ali M.A., Sufian M.A., Parves M.R., Islam M.J., Halim M.A. Repurposing fusion inhibitor peptide against SARS-CoV-2. J. Comput. Chem. 2021;42:2283–2293. doi: 10.1002/JCC.26758. [DOI] [PubMed] [Google Scholar]
- Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020;253 doi: 10.1016/J.LFS.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson K.K., Cervantes-Barragán L., Ludewig B., Thiel V. Mouse Hepatitis Virus Liver Pathology Is Dependent on ADP-Ribose-1″-Phosphatase, a Viral Function Conserved in the Alpha-Like Supergroup. J. Virol. 2008;82:12325–12334. doi: 10.1128/jvi.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer, K.J., Barrett, J.R., Belij-Rammerstorfer, S., Sharpe, H., Makinson, R., Morter, R., Flaxman, A., Wright, D., Bellamy, D., Bittaye, M., Dold, C., Provine, N.M., Aboagye, J., Fowler, J., Silk, S.E., Alderson, J., Aley, P.K., Angus, B., Berrie, E., Bibi, S., Cicconi, P., Clutterbuck, E.A., Chelysheva, I., Folegatti, P.M., Fuskova, M., Green, C.M., Jenkin, D., Kerridge, S., Lawrie, A., Minassian, A.M., Moore, M., Mujadidi, Y., Plested, E., Poulton, I., Ramasamy, M.N., Robinson, H., Song, R., Snape, M.D., Tarrant, R., Voysey, M., Watson, M.E.E., Douglas, A.D., Hill, A.V.S., Gilbert, S.C., Pollard, A.J., Lambe, T., Ali, A., Allen, E., Baker, M., Barnes, E., Borthwick, N., Boyd, A., Brown-O’Sullivan, C., Burgoyne, J., Byard, N., Puig, I.C., Cappuccini, F., Cho, J.S., Cicconi, P., Clark, E., Crocker, W.E.M., Datoo, M.S., Davies, H., Dunachie, S.J., Edwards, N.J., Elias, S.C., Furze, J., Gilbride, C., Harris, S.A., Hodgson, S.H.C., Hou, M.M., Jackson, S., Jones, K., Kailath, R., King, L., Larkworthy, C.W., Li, Y., Lias, A.M., Linder, A., Lipworth, S., Ramon, R.L., Madhavan, M., Marlow, E., Marshall, J.L., Mentzer, A.J., Morrison, H., Noé, A., Pipini, D., Pulido-Gomez, D., Lopez, F.R., Ritchie, A.J., Rudiansyah, I., Sanders, H., Shea, A., Silk, S., Spencer, A.J., Tanner, R., Themistocleous, Y., Thomas, M., Tran, N., Truby, A., Turner, C., Turner, N., Ulaszewska, M., Worth, A.T., Kingham-Page, L., Alvarez, M.P.P., Anslow, R., Bates, L., Beadon, K., Beckley, R., Beveridge, A., Bijker, E.M., Blackwell, L., Burbage, J., Camara, S., Carr, M., Colin-Jones, R., Cooper, R., Cunningham, C.J., Demissie, T., Maso, C. di, Douglas, N., Drake-Brockman, R., Drury, R.E., Emary, K.R.W., Felle, S., Feng, S., Ford, K.J., Francis, E., Gracie, L., Hamlyn, J., Hanumunthadu, B., Harrison, D., Hart, T.C., Hawkins, S., Hill, J., Howe, E., Howell, N., Jones, E., Keen, J., Kelly, S., Kerr, D., Khan, L., Kinch, J., Koleva, S., Lees, E.A., Lelliott, A., Liu, X., Marinou, S., McEwan, J., Morey, E., Morshead, G., Muller, J., Munro, C., Murphy, S., Mweu, P., Nuthall, E., O’Brien, K., O’Connor, D., O’Reilly, P.J., Oguti, B., Osborne, P., Owino, N., Parker, K., Pfafferott, K., Provstgaard-Morys, S., Ratcliffe, H., Rawlinson, T., Rhead, S., Roberts, H., Sanders, K., Silva-Reyes, L., Smith, C.C., Smith, D.J., Szigeti, A., Thomas, T.M., Thompson, A., Tonks, S., Varughes, R., Vichos, I., Walker, L., White, C., White, R., Yao, X.L., Conlon, C.P., Frater, J., Cifuentes, L., Baleanu, I., Bolam, E., Boland, E., Brenner, T., Damratoski, B.E., Datta, C., Muhanna, O. el, Fisher, R., Galian-Rubio, P., Hodges, G., Jackson, F., Liu, S., Loew, L., Morgans, R., Morris, S.J., Olchawski, V., Oliveria, C., Parracho, H., Pabon, E.R., Tahiri-Alaoui, A., Taylor, K., Williams, P., Zizi, D., Arbe-Barnes, E.H., Baker, P., Batten, A., Downing, C., Drake, J., English, M.R., Henry, J.A., Iveson, P., Killen, A., King, T.B., Larwood, J.P.J., Mallett, G., Mansatta, K., Mirtorabi, N., Patrick-Smith, M., Perring, J., Radia, K., Roche, S., Schofield, E., Naude, R. te W., Towner, J., Baker, N., Bewley, K.R., Brunt, E., Buttigieg, K.R., Charlton, S., Coombes, N.S., Elmore, M.J., Godwin, K., Hallis, B., Knott, D., McInroy, L., Shaik, I., Thomas, K., Tree, J.A., Blundell, C.L., Cao, M., Kelly, D., Skelly, D.T., Themistocleous, A., Dong, T., Field, S., Hamilton, E., Kelly, E., Klenerman, P., Knight, J.C., Lie, Y., Petropoulos, C., Sedik, C., Wrin, T., Meddaugh, G., Peng, Y., Screaton, G., Stafford, E., 2020. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nature Medicine 2020 27:2 27, 270–278. https://doi.org/10.1038/s41591-020-01194-5.
- Fahmi M., Kharisma V.D., Ansori A.N.M., Ito M. Retrieval and Investigation of Data on SARS-CoV-2 and COVID-19 Using Bioinformatics Approach. Adv. Exp. Med. Biol. 2021;1318:839–857. doi: 10.1007/978-3-030-63761-3_47/COVER. [DOI] [PubMed] [Google Scholar]
- Fan, X., Cao, D., Kong, L., Zhang, X., 2020. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nature Communications 2020 11:1 11, 1–10. https://doi.org/10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed]
- Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 Macrodomain Promotes Virulence in Mice with Coronavirus-Induced Encephalitis. J. Virol. 2015;89:1523–1536. doi: 10.1128/jvi.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7:1–12. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F., Subissi L., de Morais A.T.S., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci U S A. 2017;115:E162–E171. doi: 10.1073/PNAS.1718806115/SUPPL_FILE/PNAS.201718806SI.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe Acute Respiratory Syndrome Coronavirus Papain-Like Protease Ubiquitin-Like Domain and Catalytic Domain Regulate Antagonism of IRF3 and NF-κB Signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/jvi.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E, Mesecar A.D., Baric R.S., et al. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z., Huang B., Tang J., Liu S., Liu M., Ye Y., Liu Z., Xiong Y., Zhu W., Cao D., Li J., Niu X., Zhou H., Zhao Y.J., Zhang G., Huang H. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021;12 doi: 10.1038/S41467-020-20718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadlage M.J., Sparks J.S., Beachboard D.C., Cox R.G., Doyle J.D., Stobart C.C., Denison M.R. Murine Hepatitis Virus Nonstructural Protein 4 Regulates Virus-Induced Membrane Modifications and Replication Complex Function. J. Virol. 2010;84:280–290. doi: 10.1128/jvi.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gane E.J., Stedman C.A., Hyland R.H., Ding X., Svarovskaia E., Symonds W.T., Hindes R.G., Berrey M.M. Nucleotide Polymerase Inhibitor Sofosbuvir plus Ribavirin for Hepatitis C. N. Engl. J. Med. 2013;368:34–44. doi: 10.1056/NEJMOA1208953/SUPPL_FILE/NEJMOA1208953_DISCLOSURES.PDF. [DOI] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;1979(368):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E., v., Lai, M.M.C., Putative papain-related thiol proteases of positive-strand RNA viruses Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, α- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D.E., Jang, G.M., Bouhaddou, M., Xu, J., Obernier, K., White, K.M., O’Meara, M.J., Rezelj, V. v., Guo, J.Z., Swaney, D.L., Tummino, T.A., Hüttenhain, R., Kaake, R.M., Richards, A.L., Tutuncuoglu, B., Foussard, H., Batra, J., Haas, K., Modak, M., Kim, M., Haas, P., Polacco, B.J., Braberg, H., Fabius, J.M., Eckhardt, M., Soucheray, M., Bennett, M.J., Cakir, M., McGregor, M.J., Li, Q., Meyer, B., Roesch, F., Vallet, T., mac Kain, A., Miorin, L., Moreno, E., Naing, Z.Z.C., Zhou, Y., Peng, S., Shi, Y., Zhang, Z., Shen, W., Kirby, I.T., Melnyk, J.E., Chorba, J.S., Lou, K., Dai, S.A., Barrio-Hernandez, I., Memon, D., Hernandez-Armenta, C., Lyu, J., Mathy, C.J.P., Perica, T., Pilla, K.B., Ganesan, S.J., Saltzberg, D.J., Rakesh, R., Liu, X., Rosenthal, S.B., Calviello, L., Venkataramanan, S., Liboy-Lugo, J., Lin, Y., Huang, X.P., Liu, Y.F., Wankowicz, S.A., Bohn, M., Safari, M., Ugur, F.S., Koh, C., Savar, N.S., Tran, Q.D., Shengjuler, D., Fletcher, S.J., O’Neal, M.C., Cai, Y., Chang, J.C.J., Broadhurst, D.J., Klippsten, S., Sharp, P.P., Wenzell, N.A., Kuzuoglu-Ozturk, D., Wang, H.Y., Trenker, R., Young, J.M., Cavero, D.A., Hiatt, J., Roth, T.L., Rathore, U., Subramanian, A., Noack, J., Hubert, M., Stroud, R.M., Frankel, A.D., Rosenberg, O.S., Verba, K.A., Agard, D.A., Ott, M., Emerman, M., Jura, N., von Zastrow, M., Verdin, E., Ashworth, A., Schwartz, O., d’Enfert, C., Mukherjee, S., Jacobson, M., Malik, H.S., Fujimori, D.G., Ideker, T., Craik, C.S., Floor, S.N., Fraser, J.S., Gross, J.D., Sali, A., Roth, B.L., Ruggero, D., Taunton, J., Kortemme, T., Beltrao, P., Vignuzzi, M., García-Sastre, A., Shokat, K.M., Shoichet, B.K., Krogan, N.J., 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020 583:7816 583, 459–468. https://doi.org/10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed]
- Gorgulla, C., Das, K.M.P., Leigh, K.E., Cespugli, M., Fischer, P.D., Wang, Z.-F., Tesseyre, G., Pandita, S., Shnapir, A., Calderaio, A., Gechev, M., Rose, A., Lewis, N., Hutcheson, C., Yaffe, E., Luxenburg, R., Herce, H.D., Durmaz, V., Halazonetis, T.D., Fackeldey, K., Patten, J.J., Chuprina, A., Dziuba, I., Plekhova, A., Moroz, Y., Radchenko, D., Tarkhanova, O., Yavnyuk, I., Gruber, C., Yust, R., Payne, D., Näär, A.M., Namchuk, M.N., Davey, R.A., Wagner, G., Kinney, J., Arthanari, H., 2021. A multi-pronged approach targeting SARS-CoV-2 proteins using ultra-large virtual screening. iScience 24. https://doi.org/10.1016/J.ISCI.2020.102021. [DOI] [PMC free article] [PubMed]
- Graham R.L., Sparks J.S., Eckerle L.D., Sims A.C., Denison M.R. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133:88–100. doi: 10.1016/j.virusres.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Becker M.M., Eckerle L.D., Bolles M., Denison M.R., Baric R.S. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 2012;18:1820–1826. doi: 10.1038/NM.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/J.CELL.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, G., Gao, M., Gao, X., Zhu, B., Huang, J., Luo, K., Zhang, Y., Sun, J., Deng, M., Lou, Z., 2021. SARS-CoV-2 non-structural protein 13 (nsp13) hijacks host deubiquitinase USP13 and counteracts host antiviral immune response. Signal Transduction and Targeted Therapy 2021 6:1 6, 1–3. https://doi.org/10.1038/s41392-021-00509-3. [DOI] [PMC free article] [PubMed]
- Guo F., Li Q., Zhou C. Synthesis and biological applications of fluoro-modified nucleic acids. Org. Biomol. Chem. 2017;15:9552–9565. doi: 10.1039/C7OB02094E. [DOI] [PubMed] [Google Scholar]
- Gupta, M., Azumaya, C.M., Moritz, M., Pourmal, S., Diallo, A., Merz, G.E., Jang, G., Bouhaddou, M., Fossati, A., Brilot, A.F., Diwanji, D., Hernandez, E., Herrera, N., Kratochvil, H.T., Lam, V.L., Li, F., Li, Y., Nguyen, H.C., Nowotny, C., Owens, T.W., Peters, J.K., Rizo, A.N., Schulze-Gahmen, U., Smith, A.M., Young, I.D., Yu, Z., Asarnow, D., Billesbølle, C., Campbell, M.G., Chen, J., Chen, K.-H., Chio, U.S., Dickinson, M.S., Doan, L., Jin, M., Kim, K., Li, J., Li, Y.-L., Linossi, E., Liu, Y., Lo, M., Lopez, J., Lopez, K.E., Mancino, A., Moss, F.R., Paul, M.D., Pawar, K.I., Pelin, A., Pospiech, T.H., Puchades, C., Remesh, S.G., Safari, M., Schaefer, K., Sun, M., Tabios, M.C., Thwin, A.C., Titus, E.W., Trenker, R., Tse, E., Tsui, T.K.M., Wang, F., Zhang, K., Zhang, Y., Zhao, J., Zhou, F., Zhou, Y., Zuliani-Alvarez, L., QCRG Structural Biology Consortium, Agard, D.A., Cheng, Y., Fraser, J.S., Jura, N., Kortemme, T., Manglik, A., Southworth, D.R., Stroud, R.M., Swaney, D.L., Krogan, N.J., Frost, A., Rosenberg, O.S., Verba, K.A., 2021. CryoEM and AI reveal a structure of SARS-CoV-2 Nsp2, a multifunctional protein involved in key host processes. bioRxiv. https://doi.org/10.1101/2021.05.10.443524.
- Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R.A.P.M., Poon L.L.M., Peiris J.S.M., Valkenburg S.A. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- Hackbart M., Deng X., Baker S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci U S A. 2020;117:8094–8103. doi: 10.1073/PNAS.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Ulasli M., Vonk A.M., Reggiori F., Rottier P.J.M., de Haan C.A.M. Mobility and Interactions of Coronavirus Nonstructural Protein 4. J. Virol. 2011;85:4572–4577. doi: 10.1128/jvi.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Rottier P.J.M., de Haan C.A.M. Biogenesis and Dynamics of the Coronavirus Replicative Structures. Viruses. 2012;4:3245. doi: 10.3390/V4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Monastyrska I., Griffith J., van der Sluijs P., Voortman J., van Bergen en Henegouwen P.M., Vonk A.M., Rottier P.J.M., Reggiori F., de Haan C.A.M. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder U.C. Predicted antiviral drugs Darunavir, Amprenavir, Rimantadine and Saquinavir can potentially bind to neutralize SARS-CoV-2 conserved proteins. J. Biol. Res. Thessaloniki. 2021;28:1–58. doi: 10.1186/S40709-021-00149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Xu P., Ye Q. Analysis of COVID-19 vaccines: Types, thoughts, and application. J. Clin. Lab. Anal. 2021;35 doi: 10.1002/JCLA.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of Severe Acute Respiratory Syndrome Coronavirus Replicase Products and Characterization of Papain-Like Protease Activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S.S., Aljabali A.A.A., Panda P.K., Ghosh S., Attrish D., Choudhury P.P., Seyran M., Pizzol D., Adadi P., Abd El-Aziz T.M., Soares A., Kandimalla R., et al. A unique view of SARS-CoV-2 through the lens of ORF8 protein. Comput. Biol. Med. 2021;133 doi: 10.1016/J.COMPBIOMED.2021.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S.S., Aljabali A.A.A., Panda P.K., Ghosh S., Attrish D., Choudhury P.P., Seyran M., Pizzol D., Adadi P., Abd El-Aziz T.M., Soares A., Kandimalla R., Lundstrom K., Lal A., Azad G.K., Uversky V.N., Sherchan S.P., Baetas-da-Cruz W., Uhal B.D., Rezaei N., Chauhan G., Barh D., Redwan E.M., Dayhoff G.W., Bazan N.G., Serrano-Aroca Á., El-Demerdash A., Mishra Y.K., Palu G., Takayama K., Brufsky A.M., Tambuwala M.M. A unique view of SARS-CoV-2 through the lens of ORF8 protein. Comput. Biol. Med. 2021;133 doi: 10.1016/J.COMPBIOMED.2021.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M., Li H., Bedinger D., Schendel S.L., Moses Dennison S., Li K., Rayaprolu V., Yu X., Mann C., Zandonatti M., Avalos R.D., Zyla D., Buck T., Hui S., Shaffer K., Hariharan C., Yin J., Olmedillas E., Enriquez A., Parekh D., Abraha M., Feeney E., Horn G.Q., Aldon Y., Ali H., Aracic S., Cobb R.R., Federman R.S., Fernandez J.M., Glanville J., Green R., Grigoryan G., Lujan Hernandez A.G., Ho D.D., Huang K.Y.A., Ingraham J., Jiang W., Kellam P., Kim C., Kim M., Kim H.M., Kong C., Krebs S.J., Lan F., Lang G., Lee S., Leung C.L., Liu J., Lu Y., MacCamy A., McGuire A.T., Palser A.L., Rabbitts T.H., Tehrani Z.R., Sajadi M.M., Sanders R.W., Sato A.K., Schweizer L., Seo J., Shen B., Snitselaar J.L., Stamatatos L., Tan Y., Tomic M.T., van Gils M.J., Youssef S., Yu J., Yuan T.Z., Zhang Q., Peters B., Tomaras G.D., Germann T., Saphire E.O. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study. Science. 2021;374:472–478. doi: 10.1126/SCIENCE.ABH2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., Schaffer A.A., Rodney Brister J. Virus Variation Resource - improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45:D482–D490. doi: 10.1093/NAR/GKW1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Adonov A., Traykova-Adonova M., Cao J., Cutts T., Grudesky E., Deschambaul Y., Berry J., Drebot M., Li X. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem. Biophys. Res. Commun. 2004;320:1199–1203. doi: 10.1016/J.BBRC.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, L., Feig, M., 2020. Modeling of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Proteins by Machine Learning and Physics-Based Refinement. bioRxiv. https://doi.org/10.1101/2020.03.25.008904.
- Hiscott J., Nguyen T.L.A., Arguello M., Nakhaei P., Paz S. Manipulation of the nuclear factor-κB pathway and the innate immune response by viruses. Oncogene. 2006 doi: 10.1038/sj.onc.1209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Chen C., Li H., Dou Y., Zhou M., Lu D., Zong Q., Li Y., Yang C., Zhong Z., Singh N., Hu H., Zhang R., Yang H., Su D. Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9. Protein Sci. 2017;26:1037–1048. doi: 10.1002/PRO.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Wen J., Tang L., Zhang H., Zhang X., Li Y., Wang J., Han Y., Li G., Shi J., Tian X., Jiang F., Zhao X., Wang J., Liu S., Zeng C., Wang J., Yang H. The M protein of SARS-CoV: basic structural and immunological properties. Genomics Proteom. Bioinform. Beijing Genomics Institute. 2003;1:118–130. doi: 10.1016/S1672-0229(03)01016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Ito N., Tseng C.-T.-K., Makino S. Severe Acute Respiratory Syndrome Coronavirus 7a Accessory Protein Is a Viral Structural Protein. J. Virol. 2006;80:7287–7294. doi: 10.1128/JVI.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Ye R., Goebel S.J., Jayaraman P., Masters P.S. An Interaction between the Nucleocapsid Protein and a Component of the Replicase-Transcriptase Complex Is Crucial for the Infectivity of Coronavirus Genomic RNA. J. Virol. 2010;84:10276–10288. doi: 10.1128/jvi.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Characterization of a Critical Interaction between the Coronavirus Nucleocapsid Protein and Nonstructural Protein 3 of the Viral Replicase-Transcriptase Complex. J. Virol. 2013;87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Snijder E.J., Dimitrova M., Guillemot J.C., Lécine P., Canard B. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa, E., Merhi, G., Panossian, B., Salloum, T., Tokajian, S., 2020. SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis. mSystems 5. https://doi.org/10.1128/msystems.00266-20. [DOI] [PMC free article] [PubMed]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMOA2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/NAR/GKZ409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guardeño J.M., Regla-Nava J.A., Nieto-Torres J.L., DeDiego M.L., Castaño-Rodriguez C., Fernandez-Delgado R., Perlman S., Enjuanes L. Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jockusch S., Tao C., Li X., Chien M., Kumar S., Morozova I., Kalachikov S., Russo J.J., Ju J. Sofosbuvir terminated RNA is more resistant to SARS-CoV-2 proofreader than RNA terminated by Remdesivir. Sci. Rep. 2020;10 doi: 10.1038/S41598-020-73641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal Structure of a Monomeric Form of Severe Acute Respiratory Syndrome Coronavirus Endonuclease nsp15 Suggests a Role for Hexamerization as an Allosteric Switch. J. Virol. 2007;81:6700–6708. doi: 10.1128/jvi.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Yang M., He S., Wang Y., Chen X., Chen Y.Q., Hong Z., Liu J., Jiang G., Chen Q., Zhou Z., Zhou Z., Huang Z., Huang X., He H., Zheng W., Liao H.X., Xiao F., Shan H., Chen S. A SARS-CoV-2 antibody curbs viral nucleocapsid protein-induced complement hyperactivation. Nat. Commun. 2021;12 doi: 10.1038/S41467-021-23036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S., Subbaram K., Ali S., Kannan H. Molecular Characterization and Amino Acid Homology of Nucleocapsid (N) Protein in SARS-CoV-1, SARSCoV-2, MERS-CoV, and Bat Coronavirus. J Pure Appl Microbiol. 2020;14:757–763. doi: 10.22207/JPAM.14.SPL1.13. [DOI] [Google Scholar]
- Kaur S.P., Gupta V. COVID-19 Vaccine: A comprehensive status report. Virus Res. 2020;288 doi: 10.1016/J.VIRUSRES.2020.198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M., Chen Y., Wu A., Sun Y., Su C., Wu H., Jin X., Tao J., Wang Y., Ma X., Pan J.A., Guo D. Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2’-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 2012;167:322–328. doi: 10.1016/J.VIRUSRES.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern, D.M., Sorum, B., Mali, S.S., Hoel, C.M., Sridharan, S., Remis, J.P., Toso, D.B., Kotecha, A., Bautista, D.M., Brohawn, S.G., 2021. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nature Structural & Molecular Biology 2021 28:7 28, 573–582. https://doi.org/10.1038/s41594-021-00619-0. [DOI] [PMC free article] [PubMed]
- Khan M.T., Irfan M., Ahsan H., Ahmed A., Kaushik A.C., Khan A.S., Chinnasamy S., Ali A., Wei D.Q. Structures of SARS-CoV-2 RNA-Binding Proteins and Therapeutic Targets. Intervirology. 2021;64:55–68. doi: 10.1159/000513686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F.I., Kang T., Ali H., Lai D. Remdesivir Strongly Binds to RNA-Dependent RNA Polymerase, Membrane Protein, and Main Protease of SARS-CoV-2: Indication From Molecular Modeling and Simulations. Front. Pharmacol. 2021;12 doi: 10.3389/FPHAR.2021.710778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khater S., Kumar P., Dasgupta N., Das G., Ray S., Prakash A. Combining SARS-CoV-2 Proofreading Exonuclease and RNA-Dependent RNA Polymerase Inhibitors as a Strategy to Combat COVID-19: A High-Throughput in silico Screening. Front. Microbiol. 2021;12 doi: 10.3389/FMICB.2021.647693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Wower, Maltseva N., et al. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun Biol. 2021;4(1):193. doi: 10.1038/S42003-021-01735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Wower J., Maltseva N., Chang C., Jedrzejczak R., Wilamowski M., Kang S., Nicolaescu V., Randall G., Michalska K., Joachimiak A. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun Biol. 2021;4 doi: 10.1038/S42003-021-01735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kish T., Uppal P. Trifluridine/Tipiracil (Lonsurf) for the Treatment of Metastatic Colorectal Cancer. Pharmacy and Therapeutics. 2016;41:314. [PMC free article] [PubMed] [Google Scholar]
- Kneller D.W., Galanie S., Phillips G., O’Neill H.M., Coates L., Kovalevsky A. Malleability of the SARS-CoV-2 3CL Mpro Active-Site Cavity Facilitates Binding of Clinical Antivirals. Structure. 2020;28:1313–1320.e3. doi: 10.1016/j.str.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Shaw N., Yan L., Lou Z., Rao Z. Structural view and substrate specificity of papain-like protease from avian infectious bronchitis virus. J. Biol. Chem. 2015;290:7160–7168. doi: 10.1074/jbc.M114.628636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkolova E., Klima M., Nencka R., Boura E. Structural analysis of the putative SARS-CoV-2 primase complex. J. Struct. Biol. 2020;211 doi: 10.1016/j.jsb.2020.107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. J. Virol. 2007;81:548. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna T.S.R., Kong X.P., Gary S., Burgers P.M., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Kuri T., Eriksson K.K., Putics A., Züst R., Snijder E.J., Davidson A.D., Siddell S.G., Thiel V., Ziebuhr J., Weber F. The ADP-ribose-1″-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J. Gen. Virol. 2011;92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- Kyriakidis N.C., López-Cortés A., González E.V., Grimaldos A.B., Prado E.O. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. npj Vaccines. 2021;6 doi: 10.1038/S41541-021-00292-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, S.D., Bordin, N., Waman, V.P., Scholes, H.M., Ashford, P., Sen, N., Dorp, L. van, Rauer, C., Dawson, N.L., Pang, C.S.M., Abbasian, M., Sillitoe, I., Edwards, S.J.L., Fraternali, F., Lees, J.G., Santini, J.M., Orengo, C.A., 2020. SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Scientific Reports 2020 10:1 10, 1–14. https://doi.org/10.1038/s41598-020-71936-5. [DOI] [PMC free article] [PubMed]
- Lam, T.T.-Y., Jia, N., Zhang, Y.-W., Shum, M.H.-H., Jiang, J.-F., Zhu, H.-C., Tong, Y.-G., Shi, Y.-X., Ni, X.-B., Liao, Y.-S., Li, W.-J., Jiang, B.-G., Wei, W., Yuan, T.-T., Zheng, K., Cui, X.-M., Li, J., Pei, G.-Q., Qiang, X., Cheung, W.Y.-M., Li, L.-F., Sun, F.-F., Qin, S., Huang, J.-C., Leung, G.M., Holmes, E.C., Hu, Y.-L., Guan, Y., Cao, W.-C., 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020 583:7815 583, 282–285. https://doi.org/10.1038/s41586-020-2169-0. [DOI] [PubMed]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Ledford H., et al. COVID antiviral pills: what scientists still want to know. Nature. 2021;599:358–359. doi: 10.1038/D41586-021-03074-5. [DOI] [PubMed] [Google Scholar]
- Lee T.S., Chau L.Y. Heme oxygenase-1 mediates the anti-inflammatory effect of interleukin-10 in mice. Nat. Med. 2002;8:240–246. doi: 10.1038/NM0302-240. [DOI] [PubMed] [Google Scholar]
- Lee, J.-G., Huang, W., Lee, H., van de Leemput, J., Kane, M.A., Han, Z., 2021. Characterization of SARS-CoV-2 proteins reveals Orf6 pathogenicity, subcellular localization, host interactions and attenuation by Selinexor. Cell & Bioscience 2021 11:1 11, 1–12. https://doi.org/10.1186/S13578-021-00568-7. [DOI] [PMC free article] [PubMed]
- Lee J., Worrall L.J., Vuckovic M., Rosell F.I., Gentile F., Ton A.T., Caveney N.A., Ban F., Cherkasov A., Paetzel M., Strynadka N.C.J. Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-19662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Hilgenfeld R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 2017;591:3190–3210. doi: 10.1002/1873-3468.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Mesters J.R., Drosten C., Anemüller S., Ma Q., Hilgenfeld R. Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features. Antiviral Res. 2014;109:72–82. doi: 10.1016/j.antiviral.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018 doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tenchov R., Smoot J., Liu C., Watkins S., Zhou Q. A Comprehensive Review of the Global Efforts on COVID-19 Vaccine Development. ACS Cent. Sci. 2021;7:512–533. doi: 10.1021/ACSCENTSCI.1C00120/ASSET/IMAGES/LARGE/OC1C00120_0012.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Wen Y., Guo H., Yang T., Yang H., Ji X. Molecular Mechanism of SARS-CoVs Orf6 Targeting the Rae1–Nup98 Complex to Compete With mRNA Nuclear Export. Front Mol Biosci. 2022;8:1345. doi: 10.3389/FMOLB.2021.813248/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Chen H., Ye F., Chen Z., Yang F., Zheng Y., Cao Y., Qiao J., Yang S., Lu G. Crystal structure of SARS-CoV-2 nsp10/nsp16 2′-O-methylase and its implication on antiviral drug design. Signal Transduct Target Ther. 2020;5 doi: 10.1038/S41392-020-00241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Chen H., Chen Z., Yang F., Ye F., Zheng Y., Yang J., Lin X., Sun H., Wang L., Wen A., Dong H., Xiao Q., Deng D., Cao Y., Lu G. Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity. Nucleic Acids Res. 2021;49:5382–5392. doi: 10.1093/NAR/GKAB320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling R., Dai Y., Huang B., Huang W., Yu J., Lu X., Jiang Y. In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. Peptides. 2020;N.Y.) 130 doi: 10.1016/j.peptides.2020.170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littler D.R., Gully B.S., Colson R.N., Rossjohn J. Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9. 2020;iScience 23 doi: 10.1016/J.ISCI.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littler D.R., Mohanty B., Lowery S.A., Colson R.N., Gully B.S., Perlman S., Scanlon M.J., Rossjohn J. Binding of a pyrimidine RNA base-mimic to SARS-CoV-2 nonstructural protein 9. J. Biol. Chem. 2021;297 doi: 10.1016/J.JBC.2021.101018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littler D.R., Mohanty B., Lowery S.A., Colson R.N., Gully B.S., Perlman S., Scanlon M.J., Rossjohn J. Binding of a pyrimidine RNA base-mimic to SARS-CoV-2 nonstructural protein 9. J. Biol. Chem. 2021;297 doi: 10.1016/J.JBC.2021.101018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K., Shukla A., et al. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/J.ANTIVIRAL.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Shi W., Becker S.T., Schatz D.G., Liu B., Yang Y. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Science. 2021;373:1142–1146. doi: 10.1126/SCIENCE.ABI9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sun Y., Qi J., Chu F., Wu H., Gao F., Li T., Yan J., Gao G.F. The Membrane Protein of Severe Acute Respiratory Syndrome Coronavirus Acts as a Dominant Immunogen Revealed by a Clustering Region of Novel Functionally and Structurally Defined Cytotoxic T-Lymphocyte Epitopes. J. Infect. Dis. 2010;202:1171–1180. doi: 10.1086/656315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Cai H., Lu M., Ma Y., Li A., Gao Y., Zhou J., Gu H., Li J., Gu J. Porcine Epidemic Diarrhea Virus Deficient in RNA Cap Guanine-N-7 Methylation Is Attenuated and Induces Higher Type I and III Interferon Responses. J. Virol. 2020;94 doi: 10.1128/JVI.00447-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Pan J., Tao J., Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K.L., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc Natl Acad Sci U S A. 2006;103:12540–12545. doi: 10.1073/PNAS.0605402103/ASSET/F2252789-9397-46B2-88A7-D09F6F161BC6/ASSETS/GRAPHIC/ZPQ0330631550005.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., Su W., Yu Q., Zhang M., Di C., Lin X., Wu M., Xia Z. Heme oxygenase-1 protects airway epithelium against apoptosis by targeting the proinflammatory NLRP3-RXR axis in asthma. J. Biol. Chem. 2018;293:18454–18465. doi: 10.1074/JBC.RA118.004950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Chen Y., Wu W., Chen Z., et al. Structure and Function of N-Terminal Zinc Finger Domain of SARS-CoV-2 NSP2. Virol Sin. 2021;36(5):1104–1112. doi: 10.1007/S12250-021-00431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl. Acad. Sci. 2015;112:9436–9441. doi: 10.1073/PNAS.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtarin R., Islam M.J., Ullah M.O., Ali M.A., Halim M.A., et al. Structure and dynamics of membrane protein in SARS-CoV-2. J Biomol Struct Dyn. 2020;40(10):4725–4738. doi: 10.1080/07391102.2020.1861983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S., Banerjee A., Nazmeen A., Kanwar M., Das S. Active-site Molecular docking of Nigellidine with nucleocapsid- NSP2-MPro of COVID-19 and to human IL1R-IL6R and strong antioxidant role of Nigella-sativa in experimental rats. J. Drug Target. 2020;1–23 doi: 10.1080/1061186X.2020.1817040. [DOI] [PubMed] [Google Scholar]
- Maiti S., Banerjee A., Nazmeen A., Kanwar M., Das S. Active-site molecular docking of nigellidine with nucleocapsid-NSP2-MPro of COVID-19 and to human IL1R-IL6R and strong antioxidant role of Nigella sativa in experimental rats. J. Drug Target. 2022;1–11 doi: 10.1080/1061186X.2020.1817040. [DOI] [PubMed] [Google Scholar]
- Malone B., Chen J., Wang Q., Llewellyn E., Choi Y.J., Olinares P.D.B., Cao X., Hernandez C., Eng E.T., Chait B.T., Shaw D.E., Landick R., Darst S.A., Campbell E.A. Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/PNAS.2102516118/-/DCSUPPLEMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala V.S., McKay M.J., Shcherbakov A.A., Dregni A.J., Kolocouris A., Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020;27:1202–1208. doi: 10.1038/S41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolaridis I., Wojdyla J.A., Panjikar S., Snijder E.J., Gorbalenya A.E., Berglind H., Nordlund P., Coutard B., Tucker P.A. Structure of the C-terminal domain of nsp4 from feline coronavirus. Acta Crystallogr. D Biol. Crystallogr. 2009;65:839–846. doi: 10.1107/S0907444909018253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariano G., Farthing R.J., Lale-Farjat S.L.M., Bergeron J.R.C. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front Mol Biosci. 2020:344. doi: 10.3389/FMOLB.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T. Viewing SARS-CoV-2 Nucleocapsid Protein in Terms of Molecular Flexibility. Biology (Basel) 2021;10 doi: 10.3390/BIOLOGY10060454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., Fielding B. The Role of Severe Acute Respiratory Syndrome (SARS)-Coronavirus Accessory Proteins in Virus Pathogenesis. Viruses. 2012;4:2902–2923. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miknis Z.J., Donaldson E.F., Umland T.C., Rimmer R.A., Baric R.S., Schultz L.W. Severe Acute Respiratory Syndrome Coronavirus nsp9 Dimerization Is Essential for Efficient Viral Growth. J. Virol. 2009;83:3007–3018. doi: 10.1128/jvi.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K. The YXXΦ motif within the severe acute respiratory syndrome coronavirus (SARS-CoV) 3a protein is crucial for its intracellular transport. Virol J. 2014;11:75. doi: 10.1186/1743-422X-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res. 2016 doi: 10.1016/j.antiviral.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., Wüthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics Analysis Unravels the Functional Repertoire of Coronavirus Nonstructural Protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/jvi.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuovo, G., Tili, E., Suster, D., Matys, E., Hupp, L., Magro, C., 2020. Strong homology between SARS-CoV-2 envelope protein and a Mycobacterium sp. antigen allows rapid diagnosis of Mycobacterial infections and may provide specific anti-SARS-CoV-2 immunity via the BCG vaccine. Ann Diagn Pathol 48. https://doi.org/10.1016/J.ANNDIAGPATH.2020.151600. [DOI] [PMC free article] [PubMed]
- Oja A.E., Saris A., Ghandour C.A., Kragten N.A.M., Hogema B.M., Nossent E.J., Heunks L.M.A., Cuvalay S., Slot E., Linty F., Swaneveld F.H., Vrielink H., Vidarsson G., Rispens T., van der Schoot E., van Lier R.A.W., ten Brinke A., Hombrink P. Divergent SARS-CoV-2-specific T- and B-cell responses in severe but not mild COVID-19 patients. Eur. J. Immunol. 2020;50:1998–2012. doi: 10.1002/EJI.202048908. [DOI] [PubMed] [Google Scholar]
- Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P.J., te Lintelo E.G., Rottier P.J.M., de Haan C.A.M. Topology and Membrane Anchoring of the Coronavirus Replication Complex: Not All Hydrophobic Domains of nsp3 and nsp6 Are Membrane Spanning. J. Virol. 2008;82:12392–12405. doi: 10.1128/jvi.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk, J., Azizi, S.A., Dvorkin, S., Endres, M., Jedrzejczak, R., Jones, K.A., Kang, S., Kathayat, R.S., Kim, Y., Lisnyak, V.G., Maki, S.L., Nicolaescu, V., Taylor, C.A., Tesar, C., Zhang, Y.A., Zhou, Z., Randall, G., Michalska, K., Snyder, S.A., Dickinson, B.C., Joachimiak, A., 2021. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nature Communications 2021 12:1 12, 1–9. https://doi.org/10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed]
- Othman H., Bouslama Z., Brandenburg J.T., da Rocha J., Hamdi Y., Ghedira K., Srairi-Abid N., Hazelhurst S. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: Similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem. Biophys. Res. Commun. 2020;527:702–708. doi: 10.1016/j.bbrc.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhan K., Tanwar C., Hussain A., Hui P.Y., Lee M.Y., Cheung C.Y., Peiris J.S.M., Jameel S. Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin. J. Gen. Virol. 2007;88:3067–3077. doi: 10.1099/vir.0.82856-0. [DOI] [PubMed] [Google Scholar]
- Padhan K., Minakshi R., Towheed M.A., bin, Jameel, S., Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J. Gen. Virol. 2008;89:1960–1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- Padhi A.K., Seal A., Khan J.M., Ahamed M., Tripathi T. Unraveling the mechanism of arbidol binding and inhibition of SARS-CoV-2: Insights from atomistic simulations. Eur. J. Pharmacol. 2021;894 doi: 10.1016/J.EJPHAR.2020.173836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R., Kaki M., Potluri V.S., Kahar P., Khanna D. A comprehensive review of SARS-CoV-2 vaccines: Pfizer. Moderna & Johnson & Johnson. 2022 doi: 10.1080/21645515.2021.2002083. 10.1080/21645515.2021.2002083 18. https://doi.org/10.1080/21645515.2021.2002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Du N., Lei Y., Dorje S., Qi J., Luo T., Gao G.F., Song H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39 doi: 10.15252/EMBJ.2020105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K., Tan E., Parthasarathy K., Lin X., Jiang F.L., Yu D., Vararattanavech A., Tuck W.S., Ding X.L., Torres J. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5 doi: 10.1371/JOURNAL.PPAT.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon, M.C., Frazier, M.N., Dillard, L.B., Williams, J.G., Kocaman, S., Krahn, J.M., Perera, L., Hayne, C.K., Gordon, J., Stewart, Z.D., Sobhany, M., Deterding, L.J., Hsu, A.L., Dandey, V.P., Borgnia, M.J., Stanley, R.E., 2021. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nature Communications 2021 12:1 12, 1–12. https://doi.org/10.1038/s41467-020-20608-z. [DOI] [PMC free article] [PubMed]
- Rahman, Md.M., Saha, T., Islam, K.J., Suman, R.H., Biswas, S., Rahat, E.U., Hossen, Md.R., Islam, R., Hossain, M.N., Mamun, A. al, Khan, M., Ali, M.A., Halim, M.A., 2020. Virtual screening, molecular dynamics and structure–activity relationship studies to identify potent approved drugs for Covid-19 treatment. https://doi.org/10.1080/07391102.2020.1794974 1–11. https://doi.org/10.1080/07391102.2020.1794974. [DOI] [PMC free article] [PubMed]
- Ren, Y., Shu, T., Wu, D., Mu, J., Wang, C., Huang, M., Han, Y., Zhang, X.-Y., Zhou, W., Qiu, Y., Zhou, X., 2020. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cellular & Molecular Immunology 2020 17:8 17, 881–883. https://doi.org/10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed]
- Ricagno S., Coutard B., Grisel S., Brémond N., Dalle K., Tocque F., Campanacci V., Lichière J., Lantez V., Debarnot C., Cambillau C., Canard B., Egloff M.P. Crystallization and preliminary X-ray diffraction analysis of Nsp15 from SARS coronavirus. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2006;62:409–411. doi: 10.1107/S1744309106009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., Rocchi P., Ng W.L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell. 2020;79:710–727. doi: 10.1016/J.MOLCEL.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohaim M.A., el Naggar R.F., Clayton E., Munir M. Structural and functional insights into non-structural proteins of coronaviruses. Microb. Pathog. 2021;150 doi: 10.1016/J.MICPATH.2020.104641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Lemus, M., Minasov, G., Shuvalova, L., Inniss, N.L., Kiryukhina, O., Wiersum, G., Kim, Y., Jedrzejczak, R., Maltseva, N.I., Endres, M., Jaroszewski, L., Godzik, A., Joachimiak, A., Satchell, K.J.F., 2020. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. bioRxiv. https://doi.org/10.1101/2020.04.17.047498.
- Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., el Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti-COVID-19 drug design. Sci. Adv. 2020;6 doi: 10.1126/SCIADV.ABD4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L. Sci. Adv. 2020;6 doi: 10.1126/SCIADV.ABE0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin, U., Muik, A., Vogler, I., Derhovanessian, E., Kranz, L.M., Vormehr, M., Quandt, J., Bidmon, N., Ulges, A., Baum, A., Pascal, K.E., Maurus, D., Brachtendorf, S., Lörks, V., Sikorski, J., Koch, P., Hilker, R., Becker, D., Eller, A.K., Grützner, J., Tonigold, M., Boesler, C., Rosenbaum, C., Heesen, L., Kühnle, M.C., Poran, A., Dong, J.Z., Luxemburger, U., Kemmer-Brück, A., Langer, D., Bexon, M., Bolte, S., Palanche, T., Schultz, A., Baumann, S., Mahiny, A.J., Boros, G., Reinholz, J., Szabó, G.T., Karikó, K., Shi, P.Y., Fontes-Garfias, C., Perez, J.L., Cutler, M., Cooper, D., Kyratsous, C.A., Dormitzer, P.R., Jansen, K.U., Türeci, Ö., 2021. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature 2021 595:7868 595, 572–577. https://doi.org/10.1038/s41586-021-03653-6. [DOI] [PubMed]
- Santerre M., Arjona S.P., Allen C.N., Shcherbik N., Sawaya B.E. Why do SARS-CoV-2 NSPs rush to the ER? J. Neurol. 2021;268:2013. doi: 10.1007/S00415-020-10197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyaolu A., Okorie C., Marinkovic A., Prakash S., Williams M., Haider N., Mangat J., Hosein Z., Balendra V., Abbasi A.F., Desai P., Jain I., Utulor S., Abioye A. Current advancements and future prospects of COVID-19 vaccines and therapeutics: a narrative review. 2022;10 doi: 10.1177/25151355221097559. doi: 10.1177/25151355221097559. 251513552210975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargsyan K., Lin C.C., Chen T., Grauffel C., Chen Y.P., Yang W.Z., Yuan H.S., Lim C. Multi-targeting of functional cysteines in multiple conserved SARS-CoV-2 domains by clinically safe Zn-ejectors. Chem. Sci. 2020;11:9904–9909. doi: 10.1039/D0SC02646H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, S.K., Kumar, S., Baxi, P., Srivastava, N., Puri, B., Ratho, R.K., 2020. Chasing COVID-19 through SARS-CoV-2 spike glycoprotein. VirusDisease 2020 31:4 31, 399–407. https://doi.org/10.1007/S13337-020-00642-7. [DOI] [PMC free article] [PubMed]
- Schaecher S.R., Touchette E., Schriewer J., Buller R.M., Pekosz A. Severe Acute Respiratory Syndrome Coronavirus Gene 7 Products Contribute to Virus-Induced Apoptosis. J. Virol. 2007;81:11054–11068. doi: 10.1128/jvi.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:1–22. doi: 10.1186/S12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, K., Karousis, E.D., Jomaa, A., Scaiola, A., Echeverria, B., Gurzeler, L.A., Leibundgut, M., Thiel, V., Mühlemann, O., Ban, N., 2020. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nature Structural & Molecular Biology 2020 27:10 27, 959–966. https://doi.org/10.1038/s41594-020-0511-8. [DOI] [PubMed]
- Schütz D., Ruiz-Blanco Y.B., Münch J., Kirchhoff F., Sanchez-Garcia E., Müller J.A. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv. Drug Deliv. Rev. 2020 doi: 10.1016/j.addr.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semper C., Watanabe N., Savchenko A. Structural characterization of nonstructural protein 1 from SARS-CoV-2. iScience. 2021;24 doi: 10.1016/j.isci.2020.101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Johnson M.A., Chatterjee A., Neuman B.W., Joseph J.S., Buchmeier M.J., Kuhn P., Wüthrich K. Nuclear Magnetic Resonance Structure of the Nucleic Acid-Binding Domain of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein 3. J. Virol. 2009;83:12998–13008. doi: 10.1128/jvi.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/SCITRANSLMED.ABB5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Bhardwaj V.K., Das P., Purohit R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2. Comput. Biol. Med. 2021;135 doi: 10.1016/J.COMPBIOMED.2021.104555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K., Yuen K.-S., Castano-Rodriguez C., Ye Z., Yeung M., Fung S., Yuan S., Chan C., Yuen K.-Y., Enjuanes L., Jin D. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y.C.F., Anderson, D.E., Young, B.E., Linster, M., Zhu, F., Jayakumar, J., Zhuang, Y., Kalimuddin, S., Low, J.G.H., Tan, C.W., Chia, W.N., Mak, T.M., Octavia, S., Chavatte, J.M., Lee, R.T.C., Pada, S., Tan, S.Y., Sun, L., Yan, G.Z., Maurer-Stroh, S., Mendenhall, I.H., Leo, Y.S., Lye, D.C., Wang, L.F., Smith, G.J.D., 2020. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7B and orf8 during the early evolution of SARS-CoV-2. mBio 11, 1–9. https://doi.org/10.1128/mBio.01610-20. [DOI] [PMC free article] [PubMed]
- Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L.M., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–353. doi: 10.1016/J.STR.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir M. Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target. J. Med. Virol. 2021;93:4258. doi: 10.1002/JMV.27009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M., Chang C., Ikeya T., Güntert P., Chang Y., Hsu Y., Huang T., Kainosho M. Solution Structure of the C-terminal Dimerization Domain of SARS Coronavirus Nucleocapsid Protein Solved by the SAIL-NMR Method. J. Mol. Biol. 2008;380:608–622. doi: 10.1016/j.jmb.2007.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020 doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Tiwari, R., Dhama, K., Sharun, K., Iqbal Yatoo, M., Singh Malik, Y., Singh, R., Michalak, I., Sah, R., Katterine Bonilla-Aldana, D., Rodriguez-Morales, A.J., 2020. Veterinary Quarterly COVID-19: animals, veterinary and zoonotic links COVID-19: animals, veterinary and zoonotic links. https://doi.org/10.1080/01652176.2020.1766725. [DOI] [PMC free article] [PubMed]
- Tseng Y.-T., Chang C.-H., Wang S.-M., Huang K.-J., Wang C.-T. Identifying SARS-CoV Membrane Protein Amino Acid Residues Linked to Virus-Like Particle Assembly. PLoS ONE. 2013;8:e64013. doi: 10.1371/journal.pone.0064013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike, M., Taguchi, F., 2015. Incorporation of Spike and Membrane Glycoproteins into Coronavirus Virions. Viruses 2015, Vol. 7, Pages 1700-1725 7, 1700–1725. https://doi.org/10.3390/V7041700. [DOI] [PMC free article] [PubMed]
- Ulferts R., Ziebuhr J., et al. Nidovirus ribonucleases: Structures and functions in viral replication. RNA Biol. 2011;8(2):295–304. doi: 10.4161/RNA.8.2.15196. [DOI] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., Feldmann F., Allen E.R., Sharpe H., Schulz J., Holbrook M., Okumura A., Meade-White K., Pérez-Pérez L., Edwards N.J., Wright D., Bissett C., Gilbride C., Williamson B.N., Rosenke R., Long D., Ishwarbhai A., Kailath R., Rose L., Morris S., Powers C., Lovaglio J., Hanley P.W., Scott D., Saturday G., de Wit E., Gilbert S.C., Munster V.J. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/S41586-020-2608-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., van den Worm S.H.E., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-Coronavirus Replication/Transcription Complexes Are Membrane-Protected and Need a Host Factor for Activity In Vitro. PLoS Pathog. 2008;4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankadari N., Jeyasankar N.N., Lopes W.J. Structure of the SARS-CoV-2 Nsp1/5’-Untranslated Region Complex and Implications for Potential Therapeutic Targets, a Vaccine, and Virulence. J. Phys. Chem. Lett. 2020;11:9659–9668. doi: 10.1021/ACS.JPCLETT.0C02818. [DOI] [PubMed] [Google Scholar]
- Viswanathan, T., Arya, S., Chan, S.-H., Qi, S., Dai, N., Misra, A., Park, J.-G., Oladunni, F., Kovalskyy, D., Hromas, R.A., Martinez-Sobrido, L., Gupta, Y.K., 2020. Structural basis of RNA cap modification by SARS-CoV-2. Nature Communications 2020 11:1 11, 1–7. https://doi.org/10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed]
- Vithani N., Ward M.D., Zimmerman M.I., Novak B., Borowsky J.H., Singh S., Bowman G.R. SARS-CoV-2 Nsp16 activation mechanism and a cryptic pocket with pan-coronavirus antiviral potential. Biophys. J . 2021;120:2880–2889. doi: 10.1016/J.BPJ.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voß D., Pfefferle S., Drosten C., Stevermann L., Traggiai E., Lanzavecchia A., Becker S. Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein. Virol J. 2009;6:1–13. doi: 10.1186/1743-422X-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/J.CELL.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xia S., Zhu Y., Lu L., Jiang S. Pan-coronavirus fusion inhibitors as the hope for today and tomorrow. Protein Cell. 2021 doi: 10.1007/s13238-020-00806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck J.W., Machamer C.E. The Infectious Bronchitis Coronavirus Envelope Protein Alters Golgi pH To Protect the Spike Protein and Promote the Release of Infectious Virus. J. Virol. 2019;93 doi: 10.1128/JVI.00015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data [WWW Document], n.d. URL https://covid19.who.int/ (accessed 9.20.21).
- Wilamowski, M., Kim, Y., Jedrzejczak, R., Maltseva, N., Endres, M., Godzik, A., Michalska, K., Joachimiak, A., 2020. Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 [WWW Document]. To be published. URL https://www.rcsb.org/structure/6WTC (accessed 10.2.21).
- Wilamowski M., Hammel M., Leite W., Zhang Q., Kim Y., Weiss K.L., Jedrzejczak R., Rosenberg D.J., Fan Y., Wower J., Bierma J.C., Sarker A.H., Tsutakawa S.E., Pingali S.V., O’Neill H.M., Joachimiak A., Hura G.L. Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication. Biophys. J . 2021;120:3152–3165. doi: 10.1016/J.BPJ.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.-A., Yap C.G., Jahan N.K., Pillai N., Wong L.-A., Yap C.G., Jahan N.K., Pillai N. COVID-19 Vaccine: Review of the Mechanism of Action of Different Types of Vaccine. Open Access Library Journal. 2022;9:1–20. doi: 10.4236/OALIB.1108624. [DOI] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;1979(367):1260–1263. doi: 10.1126/SCIENCE.ABB2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S., Liu, M., Wang, C., Xu, W., Lan, Q., Feng, S., Qi, F., Bao, L., Du, L., Liu, S., Qin, C., Sun, F., Shi, Z., Zhu, Y., Jiang, S., Lu, L., 2020. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Research 2020 30:4 30, 343–355. https://doi.org/10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed]
- Xiong M., Su H., Zhao W., Xie H., Shao Q., Xu Y. What coronavirus 3C-like protease tells us: From structure, substrate selectivity, to inhibitor design. Med. Res. Rev. 2021;41:1965–1998. doi: 10.1002/MED.21783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhai Y., Sun F., Lou Z., Su D., Xu Y., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. New Antiviral Target Revealed by the Hexameric Structure of Mouse Hepatitis Virus Nonstructural Protein nsp15. J. Virol. 2006;80:7909–7917. doi: 10.1128/jvi.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Lou Z., Ma Y., Chen X., Yang Z., Tong X., Zhao Q., Xu Y., Deng H., Bartlam M., Rao Z. Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0006217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., Zhang, Y., Ge, J., Zheng, L., Gao, Y., Wang, T., Jia, Z., Wang, H., Huang, Y., Li, M., Wang, Q., Rao, Z., Lou, Z., 2020. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nature Communications 2020 11:1 11, 1–6. https://doi.org/10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed]
- Yan, W., Zheng, Y., Zeng, X., He, B., Cheng, W., 2022. Structural biology of SARS-CoV-2: open the door for novel therapies. Signal Transduction and Targeted Therapy 2022 7:1 7, 1–28. https://doi.org/10.1038/s41392-022-00884-5. [DOI] [PMC free article] [PubMed]
- Yan F.F., Gao F. Comparison of the binding characteristics of SARS-CoV and SARS-CoV-2 RBDs to ACE2 at different temperatures by MD simulations. Brief Bioinform. 2021;22:1122–1136. doi: 10.1093/BIB/BBAB044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., Liu Z., Wang H., Li Y., Chen Y., Guddat L.W., Wang Q., Rao Z., Lou Z. Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis. Cell. 2021;184:184. doi: 10.1016/J.CELL.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Chen X., Bian G., Tu J., Xing Y., Wang Y., Chen Z. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J. Gen. Virol. 2014;95:614–626. doi: 10.1099/vir.0.059014-0. [DOI] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;1979(368):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe Acute Respiratory Syndrome Coronavirus Group-Specific Open Reading Frames Encode Nonessential Functions for Replication in Cell Cultures and Mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/jvi.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-J., Chen Y.-C., Hsiao C.-H., Kuo T.-C., Chang S.C., Lu C.-Y., Wei W.-C., Lee C.-H., Huang L.-M., Chang M.-F., Ho H.-N., Lee F.-J.-S. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 2004;565:111. doi: 10.1016/J.FEBSLET.2004.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahid M.N., Moosa M.S., Perna S., Buti E., bin, A review on COVID-19 vaccines: stages of clinical trials, mode of actions and efficacy. 2021;28:225–233. doi: 10.1080/25765299.2021.1903144. doi: 10.1080/25765299.2021.1903144. [DOI] [Google Scholar]
- Zeng J., Weissmann F., Bertolin A.P., Posse V., Canal B., Ulferts R., Wu M., Harvey R., Hussain S., Milligan J.C., Roustan C., Borg A., McCoy L., Drury L.S., Kjaer S., McCauley J., Howell M., Beale R., Diffley J.F.X. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp13 helicase. Biochem. J. 2021;478:2405–2423. doi: 10.1042/BCJ20210201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Sun F., Li X., et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12(11):980–986. doi: 10.1038/NSMB999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. α-Ketoamides as Broad-Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, Synthesis, and Activity Assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/ACS.JMEDCHEM.9B01828. [DOI] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;1979(368):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.F., Stephen P., Stephen P., Thériault J.F., Wang R., Lin S.X. Novel Coronavirus Polymerase and Nucleotidyl-Transferase Structures: Potential to Target New Outbreaks. J. Phys. Chem. Lett. 2020;11:4430–4435. doi: 10.1021/ACS.JPCLETT.0C00571. [DOI] [PubMed] [Google Scholar]
- Zhang J., Xiao T., Cai Y., Chen B. Structure of SARS-CoV-2 spike protein. Curr Opin Virol. 2021;50:173–182. doi: 10.1016/J.COVIRO.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Du X., Duan Y., Pan X., Sun Y., You T., Han L., Jin Z., Shang W., Yu J., Guo H., Liu Q., Wu Y., Peng C., Wang J., Zhu C., Yang X., Yang K., Lei Y., Guddat L.W., Xu W., Xiao G., Sun L., Zhang L., Rao Z., Yang H. High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors. Protein Cell. 2021;12:877. doi: 10.1007/S13238-021-00836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Zeng R., von Brunn A., Lei J. Structural characterization of the C-terminal domain of SARS-CoV-2 nucleocapsid protein. Molecular Biomedicine. 2020;1:2. doi: 10.1186/s43556-020-00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004 doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000 doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Zinzula L. Lost in deletion: The enigmatic ORF8 protein of SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021;538:116–124. doi: 10.1016/J.BBRC.2020.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzula L., Basquin J., Bohn S., Beck F., Klumpe S., Pfeifer G., Nagy I., Bracher A., Hartl FU., Baumeister W., et al. High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2. Biochem. Biophys. Res. Commun. 2021;538:54–62. doi: 10.1016/J.BBRC.2020.09.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltner M., Campagnaro G.D., Taleva G., Burrell A., Cerone M., Leung K.F., Achcar F., Horn D., Vaughan S., Gadelha C., Zíková A., Barrett M.P., de Koning H.P., Field M.C. Suramin exposure alters cellular metabolism and mitochondrial energy production in African trypanosomes. J. Biol. Chem. 2020;295:8331–8347. doi: 10.1074/JBC.RA120.012355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.