Zhou et al. establish murine models of anti-CTLA-4–mediated intestinal irAEs. These reveal common immune signatures and the importance of fecal microbiome dysbiosis as irAE-driving mechanisms, which enable preclinical therapeutic interventions. Key immune features are validated in a cohort of melanoma patients with ICB-associated intestinal irAEs.
Abstract
Immune checkpoint blockade (ICB) has revolutionized cancer treatment, yet quality of life and continuation of therapy can be constrained by immune-related adverse events (irAEs). Limited understanding of irAE mechanisms hampers development of approaches to mitigate their damage. To address this, we examined whether mice gained sensitivity to anti-CTLA-4 (αCTLA-4)–mediated toxicity upon disruption of gut homeostatic immunity. We found αCTLA-4 drove increased inflammation and colonic tissue damage in mice with genetic predisposition to intestinal inflammation, acute gastrointestinal infection, transplantation with a dysbiotic fecal microbiome, or dextran sodium sulfate administration. We identified an immune signature of αCTLA-4–mediated irAEs, including colonic neutrophil accumulation and systemic interleukin-6 (IL-6) release. IL-6 blockade combined with antibiotic treatment reduced intestinal damage and improved αCTLA-4 therapeutic efficacy in inflammation-prone mice. Intestinal immune signatures were validated in biopsies from patients with ICB colitis. Our work provides new preclinical models of αCTLA-4 intestinal irAEs, mechanistic insights into irAE development, and potential approaches to enhance ICB efficacy while mitigating irAEs.
Introduction
Therapeutic blockade of immune checkpoint mechanisms has provided the most significant breakthrough in cancer treatment in the past decade with demonstrated clinical success in controlling a variety of aggressive tumors (Curran et al., 2010; Hellmann et al., 2018; Hodi et al., 2018; Leach et al., 1996; Motzer et al., 2018). These treatments are unique in that they target the host immune system and not tumors and operate by unleashing or reinvigorating antitumor immunity. A common strategy to suppress immune checkpoint molecules involves antibody-mediated blockade of regulatory surface proteins on T cells (Wei et al., 2018). For instance, anti-cytotoxic T lymphocyte antigen-4 (αCTLA-4) prolongs T cell costimulation, T cell activation, and T cell–mediated antitumor immunity (Joosse et al., 2019; Leach et al., 1996; Pedicord et al., 2011; Wei et al., 2018). Clinically, αCTLA-4 has demonstrated potent ability to control highly lethal tumors such as metastatic melanoma and significantly prolong the lifespan of cancer patients who previously had few therapeutic options (Hellmann et al., 2018; Hodi et al., 2018; Motzer et al., 2018).
As immune checkpoint blockades (ICBs) such as αCTLA-4 have been advanced in the cancer clinic, it is apparent that these therapies can also lead to off-target tissue toxicities or immune-related adverse events (irAEs; Das and Johnson, 2019; Dougan et al., 2021; Postow et al., 2018). Approximately, 50% of patients treated with ICB experience one or more irAEs (Esfahani et al., 2020). These events cause additional morbidities and may lead to discontinuation of therapy or, in rare instances, death. ICB-associated irAEs arise unpredictably during therapy or following completion, yet little is understood about the mechanisms that drive their development. Nonetheless, expanding use of αCTLA-4 in cancer patients is desirable. This is due to the ability of αCTLA-4 to promote long-lasting T cell–mediated antitumor immunity that persists after completion of therapy, the distinct mechanism of αCTLA-4 alone or in combination with anti-programmed cell death protein 1 (αPD-1), and the increased use of αCTLA-4/αPD-1 combination therapy, which demonstrates improved tumor control versus single agent treatments (Larkin et al., 2015; Pedicord et al., 2011; Wei et al., 2017; Wei et al., 2019). Therefore, delineating the underlying mechanisms of αCTLA-4–mediated irAEs and developing approaches to mitigate tissue toxicity are crucial needs.
While multiple immunological mechanisms have been proposed for irAEs (Dougan et al., 2021), roles for T cells have been studied most extensively. For example, αCTLA-4 treatment is associated with a rapid diversification of CD4+ and CD8+ T cell repertoires and the generation of self-reactive clonotypes in cancer patients (Oh et al., 2017), consistent with CTLA-4 inhibitory function during T cell priming. Moreover, CD8+ T cells with enhanced cytotoxic and proliferative states appeared to originate from tissue-resident populations in ICB-associated colitis (Luoma et al., 2020). By contrast, there is little insight into whether the innate immune system contributes to irAEs despite the fact that innate subsets regulate T cell tolerance and have key roles in inducing and shaping T cell responses.
Many common irAEs occur at barrier sites between the environment and host tissues (Dougan, 2017; Dougan and Pietropaolo, 2020). For instance, αCTLA-4 therapy is associated with a high risk of colitis, including debilitating grade 3–4 intestinal irAE. Homeostasis of barrier sites is regulated by a complex network of immune responses, including principle activities of myeloid and dendritic cell (DC) subsets that mediate tolerance to the host microbiome. These findings collectively suggest disruptions in immune homeostasis predispose or contribute to ICB-driven irAEs, yet the toxicity of αCTLA-4 at barrier sites has been difficult to replicate in mice. To address this gap, we utilized mice genetically predisposed to intestinal inflammation, infected with an acute gastrointestinal pathogen, transplanted with a dysbiotic fecal microbiome, or treated with dextran sodium sulfate (DSS) to evaluate αCTLA-4–driven intestinal toxicity. These models revealed irAE-driving mechanisms, including neutrophil accrual and systemic IL-6 release, which allowed us to develop therapeutic interventions to mitigate intestinal toxicity and improve αCTLA-4–mediated tumor control. We validated our models by comparison to immune signatures in human ICB-associated colitis, underscoring their utility. Collectively, our work provides mechanistic insight into irAE development and novel preclinical models for further identification of strategies to mitigate αCTLA-4–associated irAEs.
Results
Disruption of homeostatic immunity reveals αCTLA-4–mediated intestinal toxicity
We used an inflammation-prone mouse model with a defined genetic lesion to evaluate whether disruption of homeostatic immunity predisposes to αCTLA-4–mediated intestinal toxicity. The CD11c-Cre+ Stat3f/f mice (Stat3Δ/Δ) have a deletion of the transcription factor STAT3 from DCs, which activate naive T lymphocytes. In DCs, STAT3 restrains expression of costimulatory molecules and cytokines upon encounter with TLR agonists (Chrisikos et al., 2020; Chrisikos et al., 2022; Hillmer et al., 2016; Melillo et al., 2010; Zhang et al., 2014). Thus, Stat3Δ/Δ mice are prone to autoinflammatory colitis due to loss of STAT3 anti-inflammatory function, hyperactive responses of DCs to the intestinal microbiome, and increased T cell activity.
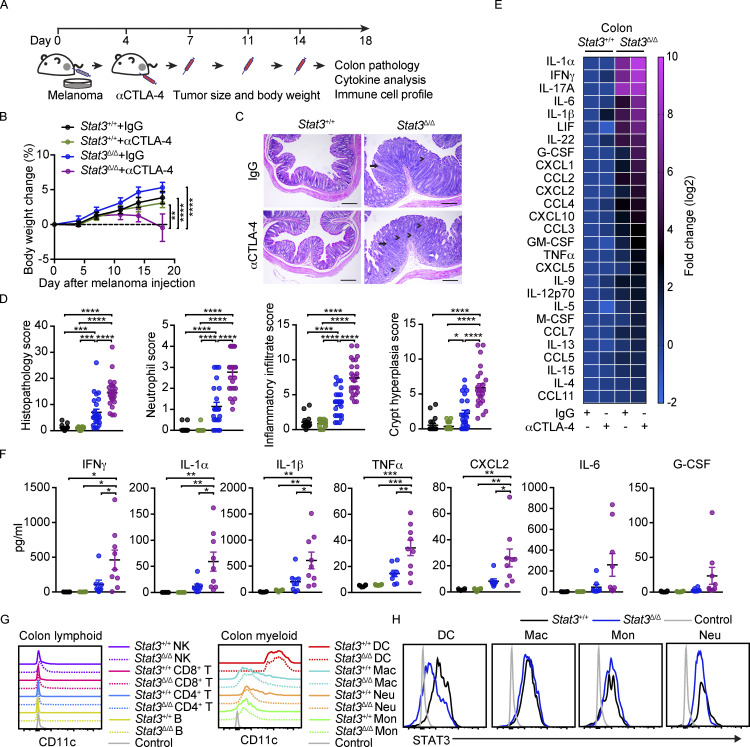
Adult Stat3Δ/Δ mice were challenged with B16-OVA melanoma tumors and treated with αCTLA-4 or control IgG. Stat3Δ/Δ mice on αCTLA-4 therapy failed to gain body weight over time, in contrast to control groups (Fig. 1, A and B). Histological examination revealed exacerbated colon damage in this cohort, accompanied by crypt hyperplasia and increased inflammatory and neutrophilic infiltrates (Fig. 1, C and D; and Table S1). αCTLA-4–treated Stat3Δ/Δ mice also showed significant accumulation of proinflammatory factors in colon tissue, including IFN-γ, IL-1α, IL-1β, TNF-α, and CXCL2 (Fig. 1, E and F). A number of other factors such as IL-6, G-CSF, IL-17A, and CXCL1 followed a similar trend of enrichment in Stat3Δ/Δ mice on αCTLA-4 (Fig. 1, E and F). By contrast, none of the measured cytokines or chemokines were induced significantly upon αCTLA-4 treatment in Stat3+/+ mice (Fig. 1, E and F). Collectively, our data suggest a discrete group of proinflammatory factors as candidates mediating intestinal tissue toxicity in αCTLA-4–treated Stat3Δ/Δ mice.
Figure 1.
αCTLA-4–mediated intestinal toxicity in inflammation-prone mice. Stat3Δ/Δ and Stat3+/+ mice bearing B16-OVA tumors were treated biweekly for 2 wk with IgG or αCTLA-4 beginning 4 d after tumor establishment, as indicated. Body weight was measured over time. Colon pathology was evaluated 18–19 d following tumor establishment. (A) Schematic diagram of the experimental approach. (B) Body weight over time; n = 21 per group. (C) Representative photomicrographs of colonic mucosa; intense inflammatory infiltrate in the LP (arrowheads) and crypt hyperplasia (arrow) are indicated; scale bar = 100 microns; H&E. (D) Summed scores for histopathology, neutrophil infiltrate, mixed inflammatory cell infiltrate, and crypt hyperplasia are shown; n = 18–24 per group. (E) Mean concentration of differentially expressed cytokines and chemokines in colon tissue (fold change absolute log2 > 1), determined by multiplex assays. Results were normalized to Stat3+/+ + IgG group and transformed to log2; n = 6–9 per group. (F) Cytokine concentration in colon tissues from individual mice (each mouse represented by one dot) determined by multiplex assays; n = 6–9 per group. (G) CD11c expression on colonic LP immune cells. (H) Intracellular staining of STAT3 in colonic LP myeloid cells. Data shown as mean ± SEM. Results from two to five independent experiments. Data were analyzed by two-way ANOVA (B), one-way ANOVA (D and F). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
To confirm the specificity of our Stat3Δ/Δ model, we evaluated CD11c and STAT3 expression within colonic lamina propria (LP) immune subsets. Lymphoid cells express low to undetectable amounts of CD11c; myeloid populations such as macrophages, monocytes, and neutrophils express intermediate amounts of CD11c; and DCs express the highest amounts of CD11c (Fig. 1 G and Fig. S1, A and B). STAT3 was efficiently depleted from colonic LP DCs but not other myeloid cells in Stat3Δ/Δ mice (Fig. 1 H), thus validating the specificity of Stat3 targeting.
Figure S1.
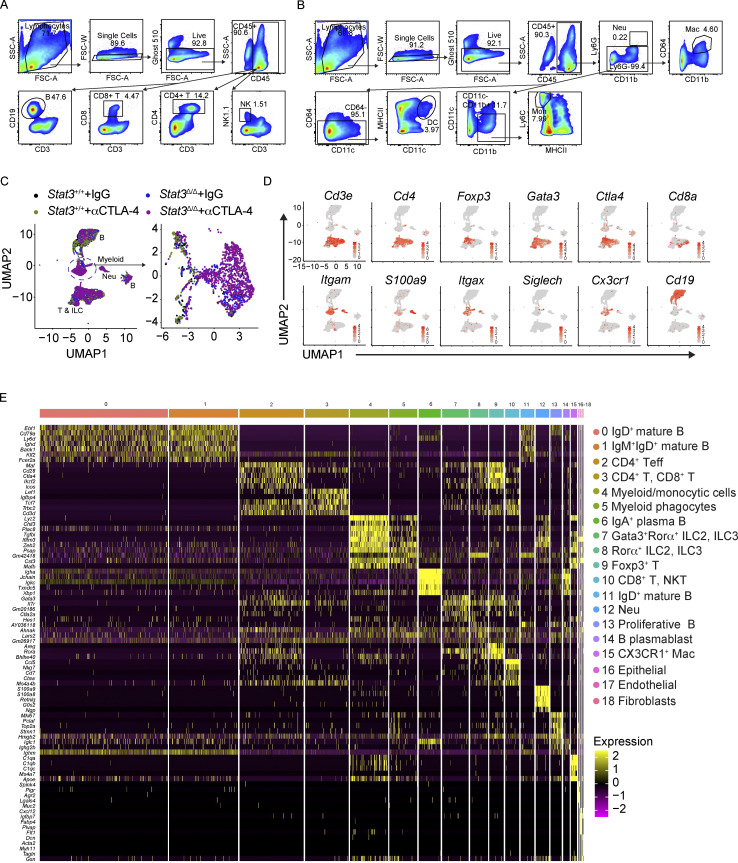
Flow cytometry gating strategy for colonic LP immune cells and scRNAseq analysis of colonic LP immune cells following αCTLA-4 therapy. (A) Gating strategy of colonic LP lymphoid cell subsets. (B) Gating strategy of colonic LP myeloid cell subsets. (C–E) Stat3Δ/Δ and Stat3+/+ mice bearing B16-OVA tumors were treated biweekly for 2 wk with IgG or αCTLA-4. Colonic LP immune cells were subjected to scRNAseq as indicated in Fig. 2; n = 7–8 per group. (C) Distinct composition of colonic LP immune cell populations in mice of indicated genotypes and treatments (left), with expanded view of myeloid clusters (right). (D) Feature plots of selected cluster-defining genes. (E) Heatmap of the top five differentially expressed genes in each cluster.
αCTLA-4 remodels the intestinal immune repertoire in inflammation-prone conditions
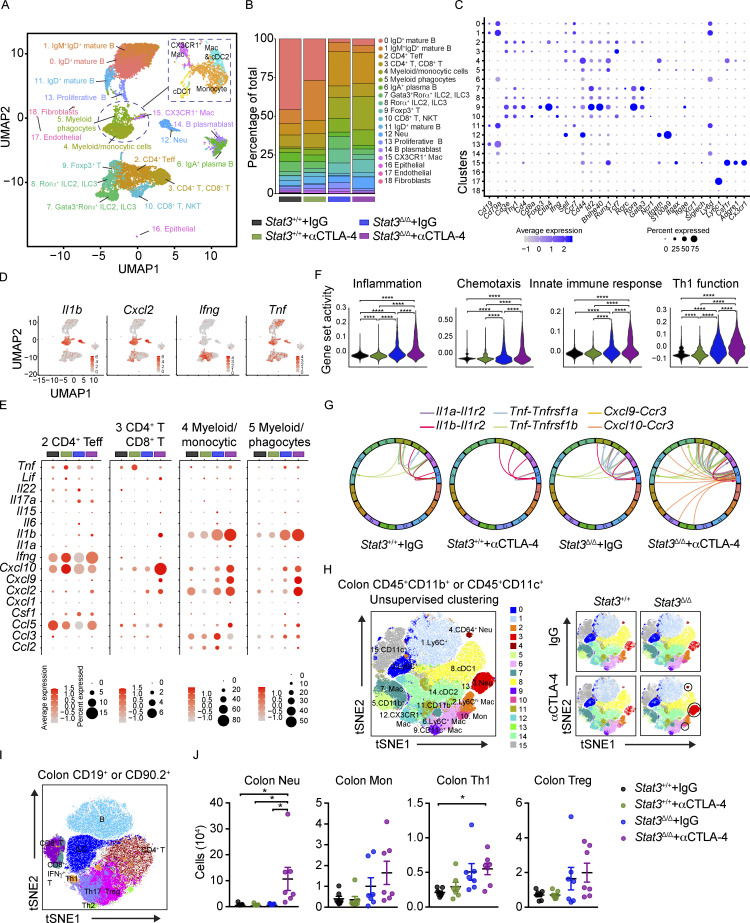
We next evaluated immunological mechanisms of intestinal toxicity using unbiased high-dimensional immune profiling. We conducted single-cell RNA sequencing (scRNAseq) of CD45+ colonic LP immune cells purified from melanoma-bearing Stat3+/+ and Stat3Δ/Δ mice treated with αCTLA-4 or control IgG. The scRNAseq data, representing 14,039 cells from our four experimental groups, were concatenated and graphed using unsupervised clustering to identify transcriptionally related populations. This resulted in elaboration of 19 discrete clusters, including 16 immune and 3 nonimmune clusters (Fig. 2 A). Importantly, each immune cluster (clusters 0–15) contained cells from all experimental groups (Fig. 2 B and Fig. S1 C). To estimate the identity of clusters, we assessed expression of canonical lineage-associated genes and compared differentially expressed genes to their representation in immune lineages according to the ImmGen database (Fig. 2 C, Fig. S1, D and E, and Table S2). The lymphoid compartment consisted of two RORα+ innate lymphoid cell clusters (clusters 7 and 8), six distinct B cell clusters (clusters 0, 1, 6, 11, 13, 14), and single clusters of CD4+ T effector (Teff) cells (cluster 2) and Foxp3+ T cells (cluster 9). Interestingly, two additional lymphoid subpopulations appeared to cluster on the basis of their functional markers rather than their known cellular subtype. Cluster 3 contained a mixture of both CD4+ T and CD8+ T cells lacking or expressing low levels of Ifng and Cd44, while expressing higher Ccr7 and Sell (CD62L) as compared with other lymphocyte clusters, suggesting a less activated phenotype (Fig. 2 C and Table S2). This cluster was defined as CD4+ T, CD8+ T cells. Cluster 10 contained activated CD8+ T cells as well as natural killer T (NKT) cells and was defined as CD8+ T, NKT (Fig. 2 C and Table S2). The myeloid compartment included discrete clusters of neutrophils (cluster 12) and CX3CR1+ macrophages (cluster 15). In addition, we identified a cluster representing cells with myeloid and monocyte features (cluster 4, defined as myeloid/monocytic cells) and a separate cluster enriched for macrophage and DC genes (cluster 5, defined as myeloid phagocytes; Fig. 2 C, Fig. S1, D and E, and Table S2). Cross-genotype assessments showed increases in the relative proportions of neutrophils, myeloid/monocytic cells, myeloid phagocytes, Foxp3+ T cells, and CD4+ Teff cells in Stat3Δ/Δ mice, while the frequencies of mature B cell clusters were decreased (Fig. 2 B). Moreover, αCTLA-4 therapy drove substantial increases in neutrophils and myeloid/monocytic cells in Stat3Δ/Δ mice relative to other groups including the Stat3Δ/Δ IgG controls (Fig. 2 B), suggesting myeloid infiltration is associated with intestinal toxicity.
Figure 2.
αCTLA-4 therapy remodels the intestinal immune landscape in inflammation-prone conditions. Stat3Δ/Δ and Stat3+/+ mice bearing B16-OVA tumors were treated biweekly for 2 wk with IgG or αCTLA-4. Colonic LP immune cells (live CD45+ cells) were isolated by FACS 18–19 d after tumor establishment and completion of therapy. (A–G) Colonic LP immune cells were subjected to scRNAseq. (A) UMAP plot showing distinct clusters generated from a merged dataset of the four experimental groups, based on transcriptomic analysis of 14,039 individual cells. Results represent 4,461 cells from the Stat3+/+ + IgG group, 4,734 cells from the Stat3+/+ + αCTLA-4 group, 1,341 cells from the Stat3Δ/Δ + IgG group, and 3,503 cells from the Stat3Δ/Δ + αCTLA-4 group; from seven to eight mice per group. Dimensionality reduction analysis identified 16 major immune clusters and three minor nonimmune clusters. (B) Proportion of individual clusters in each experimental group. (C) Dot plots of selected cluster-defining genes. (D) Feature plots of combined groups depicting single-cell mRNA expression of proinflammatory factors. (E) Dot plots showing differentially expressed cytokines and chemokines in T cell and myeloid clusters among experimental groups. (F) Expression module scores of Gene Ontology terms (inflammation, chemotaxis, innate immune response, and Th1 function) computed for the aggregated dataset of individual experimental groups. (G) Analysis of cytokine and chemokine receptor–ligand pairs across clusters of each experimental group. All shown interactions were statistically significant based on a permutation test. Arrows denote directionality from ligand to receptor. (H–J) Colonic LP immune cells were analyzed by multiparameter flow cytometry. (H) tSNE plots showing unsupervised analyses of merged live myeloid cells (CD11b+ or CD11c+) from the four experimental groups (left); tSNE plots of aggregated myeloid cells of individual experimental groups (right); n = 7 in each group. (I) tSNE plot showing prospective analyses of merged live lymphoid cells (CD90.2+ or CD19+) from colonic LP from the four experimental groups; n = 7 per group. (J) Absolute counts of neutrophils, monocytes, Th1, and Treg cells in colonic LP; n = 7 per group. Data shown as mean ± SEM. (H–J) Results from two independent experiments. Data were analyzed by one-way ANOVA (F and J). * P < 0.05, **** P < 0.0001.
To address the source(s) of candidate toxicity-driving soluble factors, we mapped the expression of cytokines and chemokines to discrete immune clusters (Fig. 2 D and Fig. S2 A). This process identified neutrophils, myeloid/monocytic cells, and myeloid phagocytes as major producers of Il1a, Il1b, and Cxcl2. Innate lymphoid cell and T cell populations were enriched for Ifng, while both myeloid and lymphoid subsets produced Tnfa (Fig. 2 D). Further evaluation revealed that myeloid and T cell populations were responsible for expression of the majority of cytokine and chemokines analyzed (Fig. S2 B). The percentage of cells expressing inflammatory cytokine and chemokine transcripts or the intensity of their expression on a per-cell basis was increased in these immune clusters in αCTLA-4–treated Stat3Δ/Δ mice (Fig. 2 E and Fig. S2 B). Of note, we observed increases in the IFN-γ–responsive genes Cxcl9 and Cxcl10, as well as IL-1α/β–responsive Cxcl2, consistent with elevated IFN-γ, IL-1α, and IL-1β in colon tissues of αCTLA-4–treated Stat3Δ/Δ mice (Fig. 2 E and Fig. 1 F). In addition, the CD8+ T, NKT and CD4+ T, CD8+ T clusters showed enhanced expression of cytotoxic factors, such as Gzma, Gzmb, or Gzmk, in αCTLA-4–treated Stat3Δ/Δ mice (Fig. S2 C). By contrast, αCTLA-4 therapy had minimal effect in Stat3+/+ mice on the expression of cytokine, chemokine, or cytotoxic genes (Fig. 2 E and Fig. S2, B and C). These data indicate αCTLA-4 treatment activates inflammatory and cytotoxic genes in myeloid, CD4+ Teff, and CD8+ T, NKT immune subsets in the colonic LP of inflammation-susceptible mice.
Figure S2.
Expression of pro-inflammatory factor mRNAs in colonic LP immune cells from mice on αCTLA-4 therapy. Colonic LP immune cells were subjected to scRNAseq as indicated in Fig. 2; n = 7–8 per group. (A) Feature plots of combined groups depicting single-cell mRNA expression of pro-inflammatory factors. (B) Dot plots showing differentially expressed cytokines across clusters and treatment groups. (C) Cytotoxic gene signature in T cells across different treatment groups.
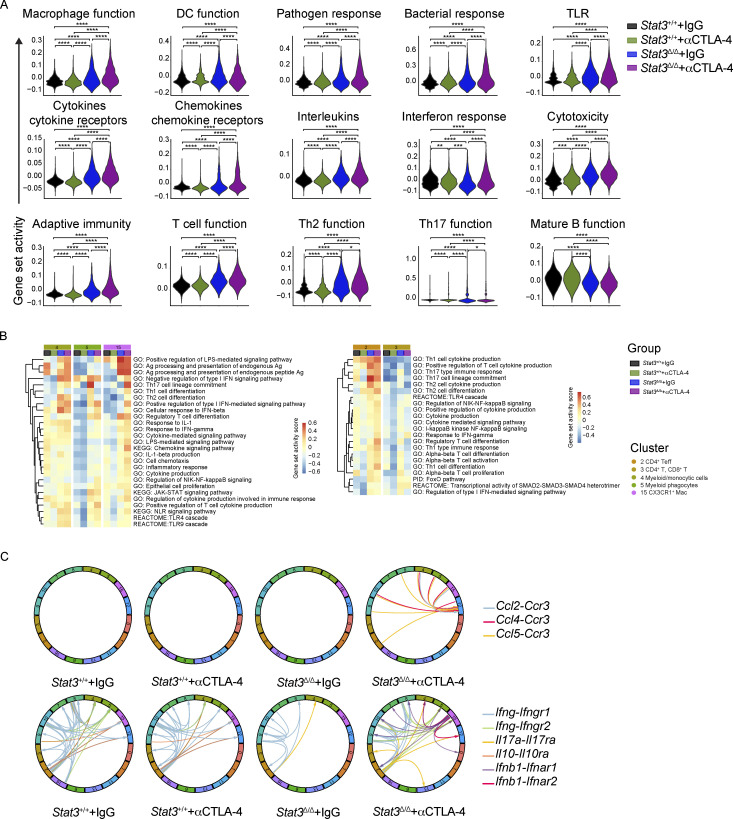
We next evaluated the gene set activity scores at both experimental group and cluster levels to assess the collective influence of αCTLA-4 therapy. The group level scores indicated that colonic LP cells from αCTLA-4–treated Stat3Δ/Δ mice had increased expression of genes associated with immune activation, including inflammation, chemotaxis, innate immune response, and T helper 1 (Th1) function (Fig. 2 F and Fig. S3 A). The cluster level scores within specific experimental groups showed immune pathway activation in the majority if not all immune populations in Stat3Δ/Δ mice treated with αCTLA-4 (Fig. S3 B). In addition, a number of cytokine and receptor pairs were significantly upregulated in αCTLA-4–treated Stat3Δ/Δ mice, including Cxcl9/Cxcl10/Ccl2/Ccl4/Ccl5 and Ccr3, Il1a/Il1b and Il1r2, Tnf and Tnfrsf1a/Tnfrsflb, Ifnb1 and Ifnar1/Ifnar2, as well as Il17a and Il17ra (Fig. 2 G, Fig. S3 C, and Table S3). Collectively, our data suggest αCTLA-4 treatment in inflammation-prone conditions drives immune-activating transcriptional responses and cellular functions of colon-infiltrating immune cells, which may contribute to αCTLA-4–mediated toxicity.
Figure S3.
Pathway analysis of colonic LP immune cells from mice on αCTLA-4 therapy. Colonic LP immune cells were subjected to scRNAseq as indicated in Fig. 2; n = 7–8 per group. (A) Expression module scores of Gene Ontology (GO) terms, computed for the aggregated dataset of individual groups. (B) Gene set activation score of myeloid (left) and T (right) cells in each experimental group computed by GSVA. (C) Analysis of cytokine and chemokine receptor–ligand pairs across clusters of each experimental group. All shown interactions were statistically significant based on a permutation test (Table S3). Arrows denote directionality from ligand to receptor.
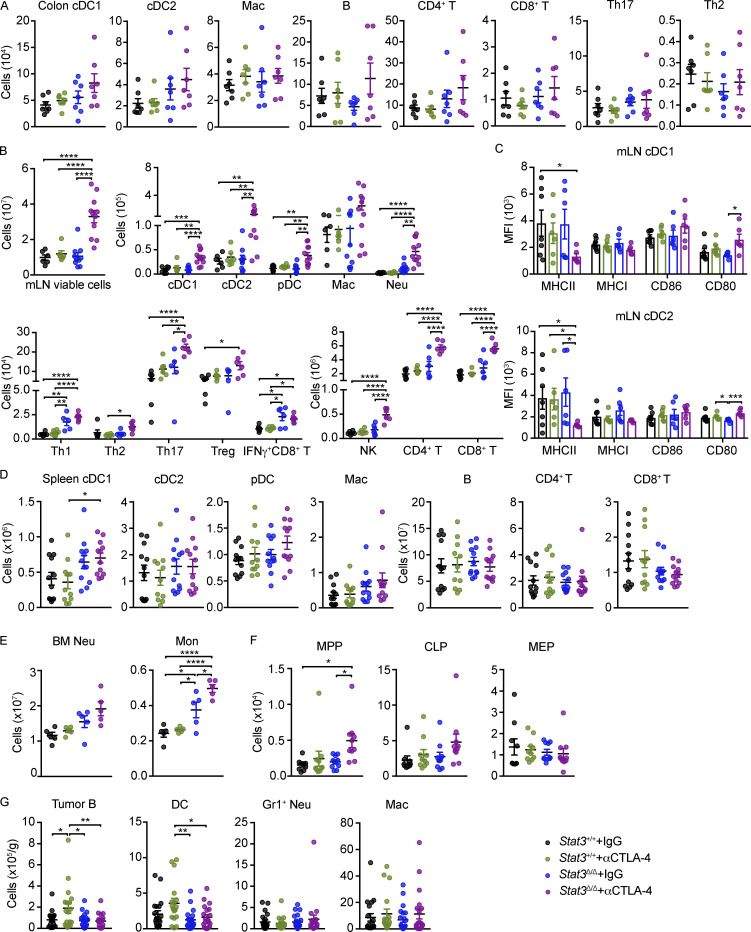
To further validate intestinal immune responses in melanoma-bearing mice during αCTLA-4 treatment, we performed multiparameter flow cytometry of colonic LP and intestinal draining lymph node (mesenteric LN [mLN]) subsets (Fig. 2, H–J; and Fig. S4, A–C). Evaluation of colonic LP data by prospective and unsupervised clustering analyses confirmed enrichment of neutrophils and IFN-γ+ CD4+ Th1 cells in Stat3Δ/Δ mice during αCTLA-4 therapy (Fig. 2, H–J), consistent with our scRNAseq results. Moreover, Stat3Δ/Δ mice treated with αCTLA-4 showed a trend toward elevated monocytes, Foxp3+ T cells (Treg), type 1 conventional DCs (cDC1s), and cDC2s in the colonic LP (Fig. 2 J and Fig. S4 A). By contrast, αCTLA-4 therapy did not significantly affect the abundance of immune populations infiltrating the colonic LP in Stat3+/+ mice (Fig. 2 J and Fig. S4 A). Total mLN cellularity was increased by αCTLA-4 therapy in Stat3Δ/Δ mice, as well as the abundance of other immune populations analyzed (Fig. S4 B). In addition, expression of the costimulatory molecule CD80 was increased on mLN-associated cDC1s and cDC2s in αCTLA-4–treated Stat3Δ/Δ mice, while MHC-II expression was reduced (Fig. S4 C), suggesting an altered activation state. These data confirm specific increases in infiltrating neutrophils and Th1 subsets in colonic LP associated with intestinal irAE and imply general immune activation in mLNs.
Figure S4.
Characterization of immune responses in mice on αCTLA-4 therapy. Stat3Δ/Δ and Stat3+/+ mice with B16-OVA tumors were treated biweekly for 2 wk with IgG or αCTLA-4. (A) Absolute numbers of colonic LP immune subsets; n = 7 per group. (B) Absolute numbers of mLN immune cell populations. For plots of viable cells and myeloid cells, n = 5–11 per group. For plots of lymphoid cells, n = 5–6 per group. (C) Mean fluorescence intensity (MFI) of MHC and co-stimulatory molecules on mLN DCs; n = 6–7 per group. (D) Absolute numbers of splenic immune cells; n = 11–12 per group. (E) Absolute numbers of neutrophils and monocytes in BM; n = 5 per group. (F) Absolute numbers of MPPs (multipotent progenitors), CLPs (common lymphoid progenitors) and MEPs (megakaryocyte-erythroid progenitors) in BM; n = 9–10 per group. (G) Absolute number of tumor-infiltrating B cells, DCs, neutrophils, and macrophages; n = 18–21 per group. Data shown as mean ± SEM. Results from two to five independent experiments. Data were analyzed by one-way ANOVA (D, F, and G). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
αCTLA-4 drives IL-6 release and myelopoiesis in inflammation-prone mice
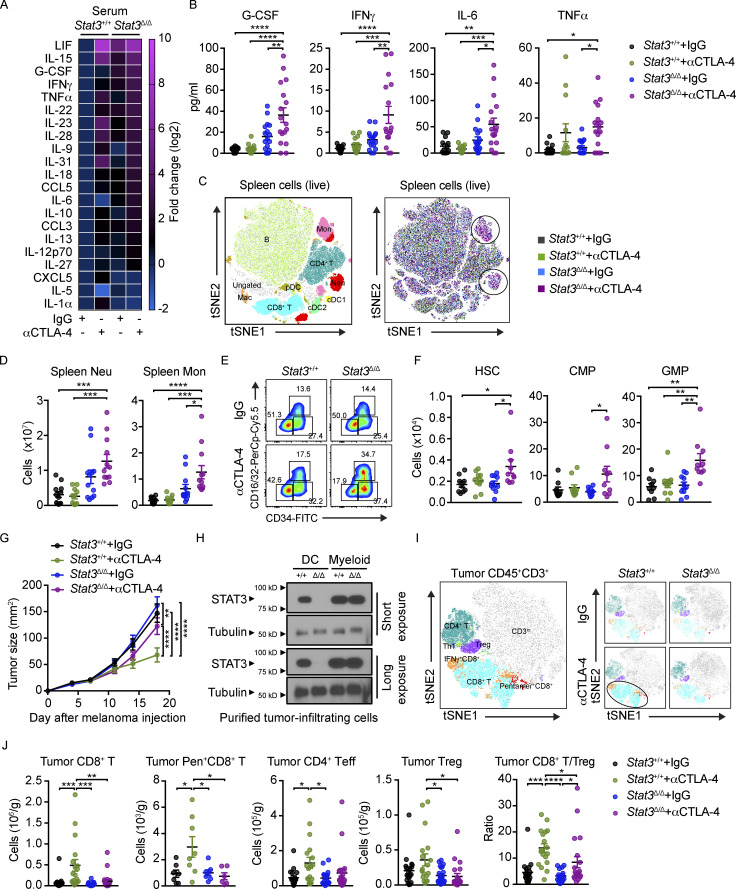
We next evaluated whether systemic factors or cellular responses were associated with intestinal tissue toxicity in melanoma-bearing mice. Treatment with αCTLA-4 stimulated specific proinflammatory factors in the circulation of Stat3Δ/Δ mice, including G-CSF, IFN-γ, IL-6, and TNF-α (Fig. 3, A and B). Additional cytokines were induced by αCTLA-4 therapy in both genotypes, and several factors showed upregulation in serum but not colon (e.g., LIF, IL-15, IL-23, IL-28, and IL-31; Fig. 3 A), suggesting generalized immune activation distinct from irAE. Nonetheless, three cytokines (i.e., IFN-γ, IL-6, and G-CSF) were upregulated in both serum and colon tissue of αCTLA-4–treated Stat3Δ/Δ mice (Fig. 3 B and Fig. 1 F), correlating with intestinal toxicity. Elevated IL-6 is associated with ICB-mediated colitis in humans, and increases in circulating IFN-γ and IL-6 have been linked previously with CTLA-4 blockade (Bamias et al., 2017; Connolly et al., 2019; Esfahani et al., 2020). Thus, increased amounts of IFN-γ, IL-6, and potentially G-CSF in circulation may serve as a “cytokine signature” of intestinal toxicity associated with αCTLA-4 ICB.
Figure 3.
Systemic cytokine release and myelopoiesis with αCTLA-4 therapy in inflammation-prone conditions. Stat3Δ/Δ and Stat3+/+ mice with B16-OVA tumors were treated biweekly for 2 wk with IgG or αCTLA-4. Serum cytokines and spleen and BM immune cells were analyzed 18–19 d after tumor establishment and completion of therapy. (A) Mean concentration of differently expressed cytokines in serum, determined by multiplex assays, fold change absolute log2 > 1; n = 13–17 per group. (B) Serum cytokines from individual mice analyzed by multiplex assays; n = 13–17 per group. (C) tSNE plot showing prospective clustering of live spleen cells merged from the four experimental groups (left); distribution of spleen immune cells, colored based on experimental groups (right); n = 6–7 in each group. (D) Neutrophil and monocyte amounts in spleen; n = 11–12 per group. (E) Representative flow plots showing CD34+ CD16/32− CMPs, CD34+ CD16/32+ GMPs, and CD34− CD16/32− MEPs (megakaryocyte-erythroid progenitors) gated from the lin− Sca-1− CD117+ population. (F) Absolute amounts of HSCs, CMPs, GMPs in each experimental group; n = 9–10 per group. (G) Tumor size over time; n = 24–30 per group. (H) STAT3 expression in tumor-infiltrating DCs (CD11c+) or myeloid cells (CD11b+ CD11c−) determined by immunoblotting. The filter was cut horizontally to separate differentially sized proteins and probed with antibodies to STAT3 or tubulin. The filters were reassembled according to the original gel orientation for each exposure time. (I) tSNE plots showing merged CD45+ CD3+ cells from the four experimental groups (left), and tSNE plots of aggregated CD45+ CD3+ cells of individual experimental groups (right); n = 6–8 per group. (J) Number of tumor-infiltrating CD8+ T cells, OVA-specific SIINFEKL/H-2Kb pentamer+ CD8+ T cells, CD4+ Foxp3− Teff cells, CD4+ Foxp3+ Treg, and CD8+/Treg ratios. For SIINFEKL/H-2Kb pentamer+ CD8+ T cells, n = 6–8 per group; for other plots, n = 18–21 per group. Data shown as mean ± SEM. Results from two to five independent experiments. Data were analyzed by one-way ANOVA (B, D, F, and J) or two-way ANOVA (G). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. Source data are available for this figure: SourceData F3.
IFN-γ, IL-6, and G-CSF are well-established mediators of myelopoiesis due to their ability to enhance proliferation and differentiation of bone marrow (BM) myeloid progenitor cells (Mirantes et al., 2014; Morales-Mantilla and King, 2018; Panopoulos and Watowich, 2008; Walker et al., 2008). Consistently, we detected increased amounts of neutrophils and monocytes in spleen and BM of melanoma-bearing Stat3Δ/Δ mice treated with αCTLA-4 (Fig. 3, C and D; and Fig. S4, D and E). Investigation of BM hematopoietic stem cells (HSCs) and progenitor subsets revealed specific increases in HSCs, multipotent progenitors, and myeloid progenitors, including common myeloid (CMPs) and granulocyte-monocyte progenitors (GMPs), in αCTLA-4–treated Stat3Δ/Δ mice (Fig. 3, E and F; and Fig. S4 F). These data indicate αCTLA-4 drives BM myelopoiesis in mice prone to intestinal inflammation. Taken together, our results suggest αCTLA-4 therapy in inflammation-susceptible conditions leads to systemic cytokine release, sustained myeloid production, immune cell transcriptional reprogramming, and intestinal myeloid infiltration and toxicity.
Melanoma tumors are nonresponsive to αCTLA-4 in inflammation-prone conditions
Growth of B16-OVA melanoma tumors was refractory to αCTLA-4 therapy in Stat3Δ/Δ animals, while Stat3+/+ mice responded to treatment (Fig. 3 G). Importantly, we confirmed that STAT3 depletion was restricted to tumor-infiltrating DCs, while tumor-infiltrating myeloid cells, which constitute the majority of tumor immune cells, were STAT3-sufficient (Fig. 3 H). Tumor nonresponsiveness in Stat3Δ/Δ mice correlated with fewer tumor-infiltrating CD8+ T cells, less antigen-specific (SIINFEKL/H-2Kb pentamer+) CD8+ T cells, a lower ratio of CD8+ T cells to Foxp3+ Tregs, and overall less tumor immune activation, relative to Stat3+/+ animals treated with αCTLA-4 (Fig. 3, I and J; and Fig. S4 G). We did not observe effects on tumor-associated neutrophils or macrophages, or Treg depletion with αCTLA-4 (Fig. 3 J and Fig. S4 G). Thus, Stat3Δ/Δ mice demonstrate poor lymphocyte infiltration and tumor nonresponsiveness upon αCTLA-4 treatment. Taken together, our results support the use of Stat3Δ/Δ mice as an irAE model system mimicking therapeutic nonresponsiveness and tissue toxicity.
Single-agent therapeutic interventions to enhance αCTLA-4 efficacy and suppress irAE
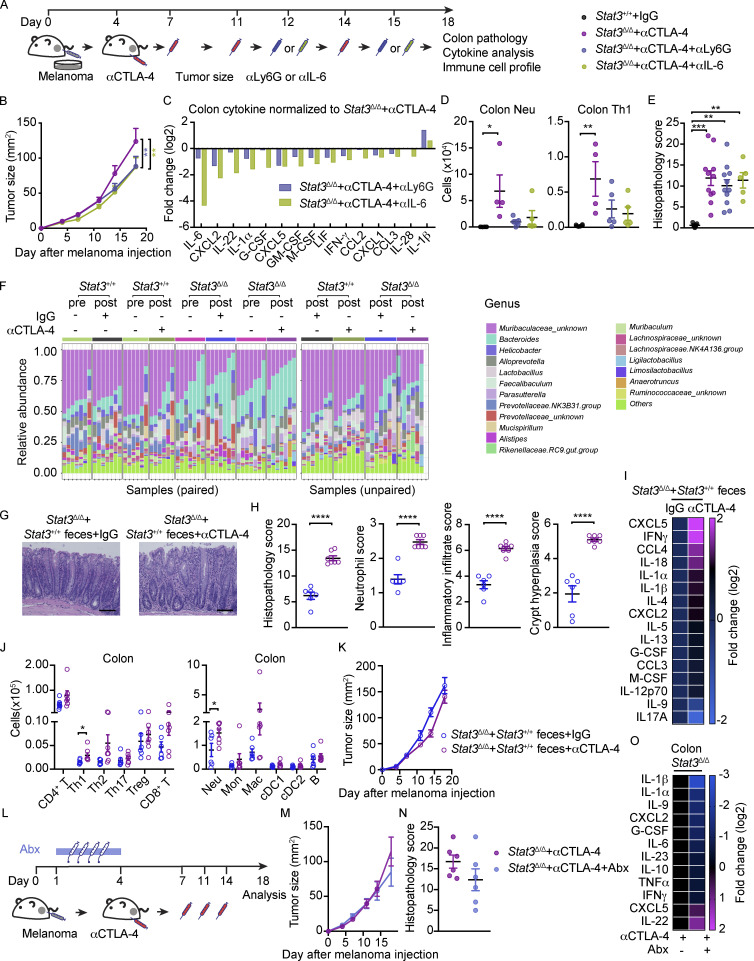
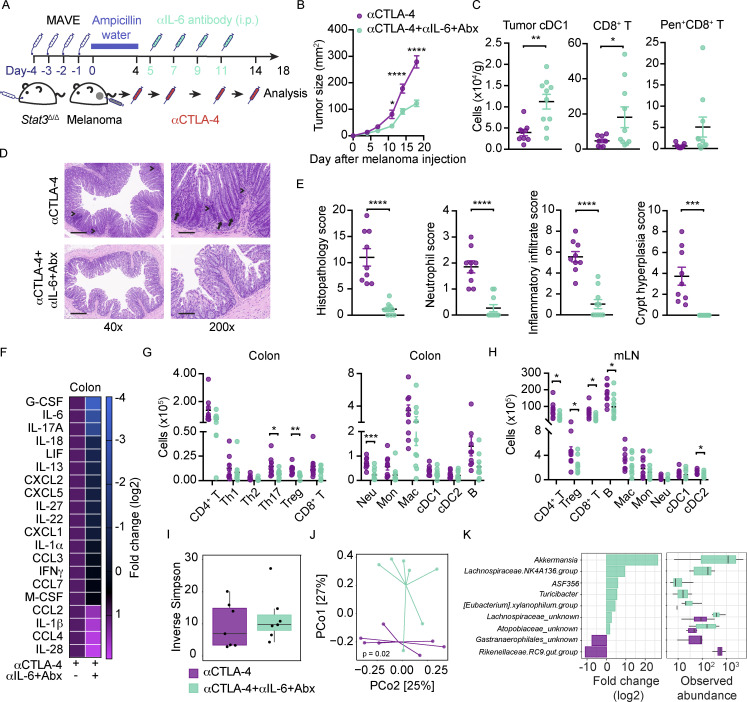
A major goal of irAE research is to identify approaches to mitigate tissue toxicity while maintaining or improving therapeutic efficacy to ICB. In our inflammation-prone model, myeloid cells, Th1 cells, and, to some extent, CD8+ T cell function correlate with αCTLA-4–mediated irAE. Of these populations, myeloid cells are most likely to impede ICB-mediated control of tumors; therefore, we employed strategies to block their function (Fig. S5 A). We found that inhibition of neutrophils by Ly6G antibody (αLy6G) treatment or blockade of IL-6 activity via anti-IL-6 (αIL-6) enhanced the antitumor efficacy of αCTLA-4 in Stat3Δ/Δ mice (Fig. S5 B). Moreover, immune signatures of irAE in the colonic LP, including proinflammatory cytokine amounts, neutrophils, and Th1 cells were diminished upon αLy6G or αIL-6 treatment (Fig. S5, C and D). Analyses of colon histopathology, however, did not indicate an effect of αLy6G or αIL-6 therapy on colitis (Fig. S5 E). While these results indicate neutrophils and IL-6 are key drivers of αCTLA-4–mediated intestinal toxicity and tumor nonresponsiveness, the inability to reverse intestinal damage suggests additional irAE-driving factors.
Figure S5.
Effects of neutrophil depletion, IL-6 blockade, antibiotics, or FMT from WT mice on αCTLA-4–associated toxicity. (A–E) Stat3Δ/Δ mice with B16-OVA tumors were treated biweekly for 2 wk with IgG or αCTLA-4; Ly6G or IL-6 antibodies (αLy6G or αIL-6, respectively) were injected 12 and 15 d after tumor establishment. Colon cytokines and immune subsets were analyzed 18 d after tumor establishment and completion of therapy. (A) Schematic diagram of experiment. (B) Tumor growth over time; n = 11–17 per group. (C) Differentially expressed cytokines in colon (i.e., cytokines with fold change <0.7 or >1.4); expression normalized to mean concentration in αCTLA-4–treated Stat3Δ/Δ mice; n = 8–14 per group. (D) Neutrophil and Th1 cell numbers in colonic LP; n = 4–6 per group. (E) Histopathology score; n = 5–11 per group. (F) Stat3Δ/Δ and Stat3+/+ mice with B16-OVA tumors were treated biweekly for 2 wk with IgG or αCTLA-4. Fecal samples were collected prior to (3 d) and following αCTLA-4 treatment (18 d); microbiome composition was characterized by 16S rRNA gene profiling, as described in Fig. 4. Composition plots representing relative abundance of microbial taxa (obtained from 16S rRNA gene profiling of the fecal samples) at the genus level. (G–K) Recipient Stat3Δ/Δ mice were given fecal transplantation from donor Stat3+/+ mice. Recipient Stat3Δ/Δ mice were injected with B16-OVA cells and treated with αCTLA-4 or IgG i.p. biweekly for 2 wk. Organ samples were collected 18 d after melanoma injection. (G) Representative photomicrographs of colonic mucosa, scale bar = 100 microns; H&E. (H) Summed scores for histopathology, neutrophil infiltrate, mixed inflammatory cell infiltrate, and crypt hyperplasia are shown; n = 6–7 per group. (I) Differentially expressed cytokines in colon (i.e., cytokines with fold change <0.7 or >1.4); n = 6–7 per group. (J) Absolute amounts of immune cells in colonic LP; n = 6–7 per group. (K) Tumor size over time; n = 6–7 per group. (L–O) Stat3Δ/Δ mice with B16-OVA tumors were treated biweekly for 2 wk with αCTLA-4. Mice received broad-spectrum antibiotics (metronidazole, ampicillin, vancomycin, and enrofloxacin; Abx) daily for 4 d prior to first αCTLA-4 injection. (L) Schematic diagram of experiment. (M) Tumor size over time; n = 11–16 per group. (N) Histopathology scores; n = 6 for each group. (O) Differentially expressed cytokines from colon (i.e., cytokines with fold change <0.7 or >1.4), normalized to mean concentration in αCTLA-4–treated Stat3Δ/Δ mice; n = 6 for each group. Data shown as mean ± SEM. Results from two to three independent experiments. Data were analyzed by two-way ANOVA (B), one-way ANOVA (D and E), statistical method implemented in the R package DESeq2 (F), two-tailed unpaired Student’s t test (H and J). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Dysbiosis and contribution of the intestinal microbiome to αCTLA-4–mediated colitis
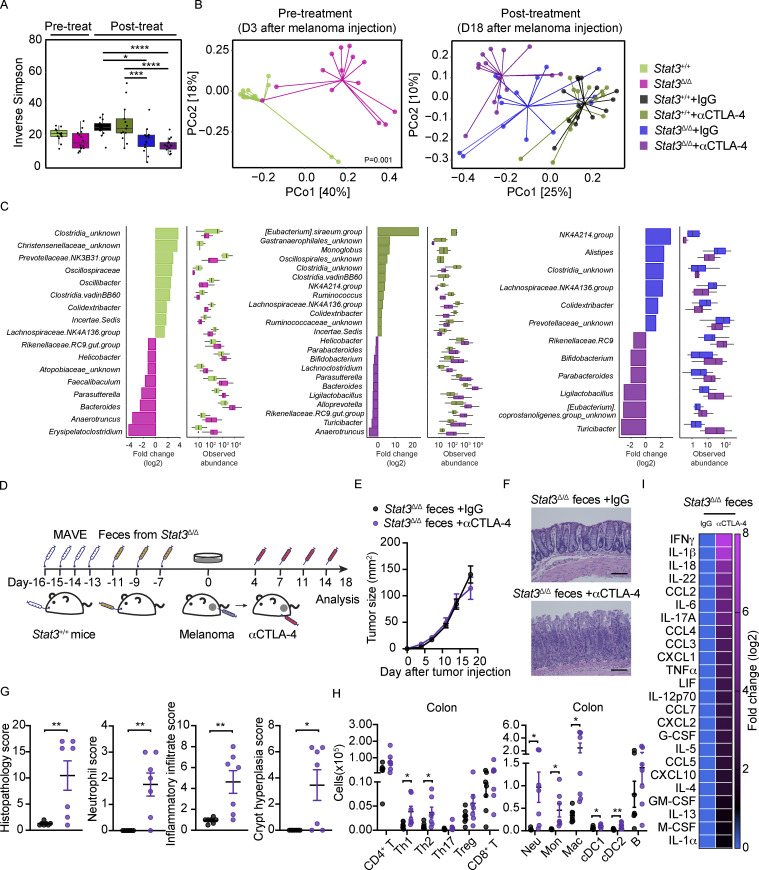
The intestinal microbiome is linked with antitumor response to ICB, irAEs, and non-ICB-associated intestinal inflammation (Cheng et al., 2020; Dubin et al., 2016; Gopalakrishnan et al., 2018a; Gopalakrishnan et al., 2018b; McCulloch et al., 2022; Vétizou et al., 2015; Wang et al., 2018). Accordingly, we evaluated the diversity and composition of the fecal microbiome in cohoused tumor-bearing Stat3Δ/Δ mice and controls, collected prior to or following αCTLA-4 therapy, by targeted 16S ribosomal RNA (rRNA) gene profiling. These studies revealed Stat3Δ/Δ mice have a trend toward reduced fecal bacterial community diversity at baseline compared with Stat3+/+ mice (Fig. 4 A). Alpha diversity was reduced in Stat3Δ/Δ mice following tumor growth and αCTLA-4 therapy relative to Stat3+/+ mice (Fig. 4 A). We also detected differences in microbial beta diversity (Fig. 4 B), indicating the community structure of the fecal microbiome is substantially distinct in Stat3+/+ versus Stat3Δ/Δ mice. Moreover, the fecal microbial beta diversity of Stat3Δ/Δ mice was significantly changed upon αCTLA-4 treatment (P = 0.004), while these effects were not observed in Stat3+/+ mice (P = 0.9; Fig. 4 B).
Figure 4.
Dysbiosis and FMT induces sensitivity to αCTLA-4–mediated irAE. (A–C) Stat3Δ/Δ and Stat3+/+ mice with B16-OVA tumors were treated biweekly for 2 wk with IgG or αCTLA-4. Feces were collected prior to (day 3) or following αCTLA-4 treatment (day 18); fecal microbiome composition was determined by 16S rRNA gene profiling. (A) Box-and-whisker plot represents within-sample diversity using Inverse Simpson diversity scores of indicated mice and treatments groups; n = 13–14 per group. (B) Beta diversity analysis using Bray–Curtis dissimilarity compares between-sample diversity of the indicated groups. Based on PCA each of the fecal microbiome profiles are represented in terms of the top-two principal components; n = 13–14 per group. (C) Using microbial abundance data aggregated at the genus level, each of the three plot-pair shows significant differentially abundant taxa (P value <0.05) are associated with the indicated treatment groups. The bar plot shows the log-fold change, and the box-and-whisker plot shows the normalized abundance of the differentially abundant taxa identified using DESeq2; n = 13–14 per group. (D–I) Recipient Stat3+/+ mice were given fecal transplantation by feces suspension from donor Stat3Δ/Δ mice following microbiome depletion by broad spectrum antibiotics (metronidazole, ampicillin, vancomycin, and enrofloxacin; MAVE), then Stat3+/+ mice were injected with B16-OVA cells and treated with αCTLA-4 or IgG i.p. biweekly for 2 wk. Organ samples were collected 18 d after melanoma injection. (D) Schematic diagram of the experimental approach. (E) Tumor size over time; n = 7 per group. (F) Representative photomicrographs of colonic mucosa; scale bar = 100 microns; H&E. (G) Summed scores for histopathology, neutrophil infiltrate, mixed inflammatory cell infiltrate, and crypt hyperplasia are shown; n = 7 per group. (H) Absolute amounts of immune cells in colonic LP; n = 7 per group. (I) Differential cytokine amounts in colon tissue (n = 7 per group), fold change <0.7 or >1.4. Data shown as mean ± SEM. Results from two independent experiments. Data were analyzed by Wilcoxon Rank-Sum test (A), Bray–Curtis dissimilarity (B), statistical test implemented in the R package DESeq2 (C), two-tailed unpaired Student’s t test (G and H). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Differential abundance analysis revealed significant enrichment of several taxa including Anaerotruncus, Bacteroides, Parasutterella, Helicobacter, and Rikenellaceae genera in Stat3Δ/Δ mice compared with Stat3+/+ mice, as well as Stat3Δ/Δ mice treated with αCTLA-4, while others such as Clostridia and Lachnospiraceae were significantly enriched in Stat3+/+ mice (Fig. 4 C and Fig. S5 F). Therefore, our data indicate Stat3Δ/Δ mice have a distinct fecal microbiome composition relative to Stat3+/+ controls. Strikingly, these differences were maintained in the Stat3Δ/Δ and Stat3+/+ mice despite being cohoused, indicating a profound influence of the genetic lesion in Stat3Δ/Δ mice on the intestinal microbiome. Moreover, αCTLA-4 therapy appeared to promote further alterations in the microbiome of Stat3Δ/Δ animals, suggesting an interplay with intestinal inflammation and tissue toxicity.
To investigate roles for the dysbiotic gut microbiome in αCTLA-4–mediated responses, we performed fecal microbiome transplant (FMT) experiments (Fig. 4 D). These studies demonstrated that transplantation of fecal material from Stat3Δ/Δ mice into Stat3+/+ (WT) mice rendered tumor nonresponsiveness to therapy and sensitivity to αCTLA-4 intestinal irAEs (Fig. 4, E–G). This was accompanied by significant increases in colonic myeloid cells, CD4+ T subsets, cDC1s, and cDC2s upon αCTLA-4 therapy (Fig. 4 H). Colon inflammatory cytokines were also increased following αCTLA-4 treatment (Fig. 4 I). By contrast, reciprocal FMT using feces from WT mice transplanted into Stat3Δ/Δ mice did not confer tumor responsiveness or prevent intestinal irAEs (Fig. S5, G–K). Taken together, our data indicate intestinal microbiome dysbiosis directly confers susceptibility to αCTLA-4–driven intestinal irAEs and tumor nonresponsiveness to therapy. Our results also imply inflammation-prone conditions in the gut contribute to these unfavorable responses to ICB.
Combination treatment with αIL-6 and antibiotics enhances αCTLA-4 efficacy and reduces irAEs in the preclinical setting
To further evaluate roles for the dysbiotic microbiome in αCTLA-4–mediated intestinal toxicity and tumor nonresponsiveness, we treated Stat3Δ/Δ mice with broad-spectrum antibiotics (Abx) to ablate gut bacterial communities. We performed Abx treatment alone or in combination with IL-6 blockade (αIL-6) as our data indicate this proinflammatory factor contributes to irAE (Fig. 5 A and Fig. S5, C, D, and L). While Abx alone did not enhance αCTLA-4 efficacy or improve intestinal pathology significantly, this approach led to reduced proinflammatory cytokines in the colon (Fig. S5, M–O). By contrast, combination treatment with Abx and αIL-6 significantly enhanced the antitumor efficacy of αCTLA-4 in Stat3Δ/Δ mice (Fig. 5 B). This was associated with increases in tumor-infiltrating cDC1s and CD8+ T cells, as well as tumor antigen-specific CD8+ T cells (SIINFEKL/H-2Kb pentamer+ CD8+; Fig. 5 C). Furthermore, Abx and αIL-6 combination therapy significantly reduced intestinal tissue pathology, colon inflammatory cytokine amounts, colonic LP neutrophil and CD4+ T subset infiltration, and immune accrual in mLNs (Fig. 5, D–H). These data underscore key roles for the dysbiotic microbiome in Stat3Δ/Δ mice in mediating tumor nonresponsiveness and intestinal tissue toxicity during αCTLA-4 therapy. Moreover, our results point to interventions that improve αCTLA-4–mediated tumor control and inhibit intestinal irAE in our preclinical model using combination Abx and IL-6 blockade.
Figure 5.
Combination treatment with αIL-6 and antibiotics enhances αCTLA-4 efficacy and reduces irAEs. Stat3Δ/Δ mice were given broad-spectrum Abx (metronidazole, ampicillin, vancomycin, and enrofloxacin; MAVE) by oral gavage for 4 d. B16-OVA tumors were established, and mice were maintained on ampicillin-containing drinking water until initiation of αCTLA-4 therapy. Mice were treated biweekly for 2 wk with αCTLA-4. αIL-6 was injected (i.p.) on day 5, 7, 9, and 11. (A) Schematic diagram of the experiment. (B) Tumor growth over time; n = 9–10 per group. (C) Numbers of tumor-infiltrating cDC1s, CD8+, and OVA-specific SIINFEKL/H-2Kb pentamer+ CD8+ T cells; n = 9–10 per group. (D) Representative photomicrographs of colonic mucosa; intense inflammatory (arrowheads) and neutrophil (arrow) infiltrate in the LP are indicated; scale bar = 500 μm for 40× magnification, 100 μm for 200× magnification; H&E. (E) Histopathology scores; n = 9–10 for each group. (F) Differentially expressed cytokines from colon (i.e., cytokines with fold change <0.7 or >1.4), normalized to mean concentration in αCTLA-4–treated Stat3Δ/Δ mice; n = 5 for each group. (G and H) Absolute amounts of immune cells in colonic LP (G) and mLN (H); n = 9–10 per group. (I) Box-and-whisker plot of within-sample diversity using Inverse Simpson diversity measure of indicated treatments groups. (J) Beta diversity analysis using Bray–Curtis dissimilarity compares between-sample diversity of the indicated groups. The ordination plot represents the microbiome profile of the samples in terms of the top-two PCs obtained from the principal coordinate; n = 7 per group. (K) Using microbial abundance data aggregated at the family level, the analysis (using R package DESeq2) identifies significant differentially abundant taxa (P < 0.05) associated with the indicated treatment groups. For the identified taxa, the bar plot (left) reports log-fold change and the box-and-whisker plot (right) compares the indicated two-treatment groups in terms of normalized abundance data; n = 7 per group. Data shown as mean ± SEM from two independent experiments. Data were analyzed by two-way ANOVA (B), two-tailed unpaired Student’s t test (C, E, G, and H), Bray–Curtis dissimilarity (J), statistical test implemented in the R package DESeq2 (K). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
To study the effect of αIL-6 and Abx on the intestinal microbiome of Stat3Δ/Δ mice treated with αCTLA-4, we performed 16S rRNA gene profiling. While alpha diversity was unchanged by αIL-6 and Abx, we found effects on the community structure (Fig. 5, I and J). Identification of differentially abundant taxa revealed enrichment of Akkermansia and Lachnospiraceae by αIL-6 and Abx intervention, while Rikenellaceae were relatively enhanced in Stat3Δ/Δ mice treated with αCTLA-4 alone (Fig. 5 K). These results reveal an intestinal microbiome composition associated with a favorable antitumor response and suppression of ICB-mediated inflammation. Our data align with recent results from human ICB-treated melanoma patients (McCulloch et al., 2022), further supporting the utility of our animal models in evaluating therapeutic interventions to alleviate intestinal irAEs and improve ICB efficacy.
Acute intestinal infection or DSS administration in WT mice drives αCTLA-4–induced toxicity
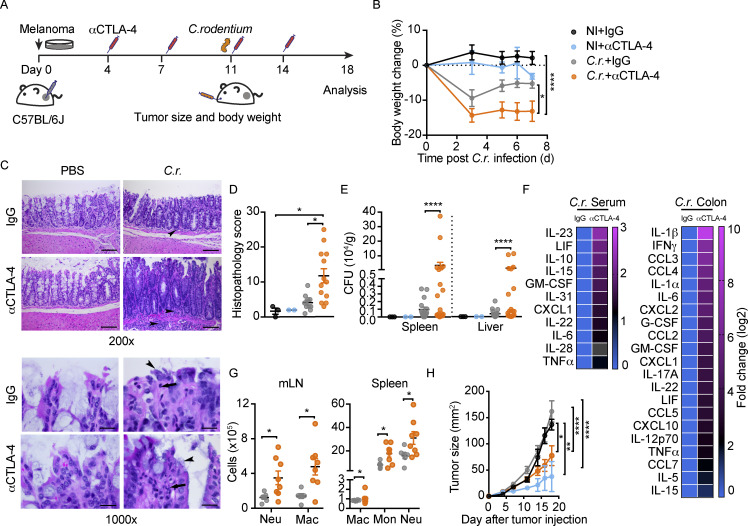
To evaluate whether distinct approaches affecting intestinal immune homeostasis render sensitivity to αCTLA-4–mediated irAEs, we used acute gastrointestinal infection in WT mice. Melanoma-bearing WT mice were infected orally with Citrobacter rodentium, which models infection with enteropathogenic and enterohaemorrhagic Escherichia coli strains in humans (Bouladoux et al., 2017). Animals were infected during the course of αCTLA-4 therapy to mimic an acquired gastrointestinal infection in humans undergoing ICB treatment. These experiments indicated C. rodentium infection enhanced body weight loss as well as intestinal tissue damage in mice treated with αCTLA-4 (Fig. 6, A–D). We detected elevated bacterial burden in spleen and liver following C. rodentium infection in the αCTLA-4–treatment group (Fig. 6 E), consistent with a loss of intestinal barrier function. Furthermore, C. rodentium infection promoted an irAE immune signature in mice on αCTLA-4 therapy, including elevated IL-1β, IFN-γ, IL-1α, IL-6, G-CSF, and TNF-α in colon, increased IL-6 in circulation, and systemic myeloid accrual (Fig. 6, F and G). Melanoma tumors in WT mice infected with C. rodentium were responsive to αCTLA-4, yet showed a trend for decreased responsiveness relative to uninfected mice treated with αCTLA-4 (Fig. 6 H). These results indicate acute inflammation resulting from intestinal infection may have a modest but nonsignificant effect on therapeutic responsiveness to αCTLA-4, but can contribute to intestinal irAE. Moreover, this irAE response is accompanied by IL-6 elevation and myeloid accrual, suggesting common irAE-driving mechanisms in inflammation-prone and acute infection conditions.
Figure 6.
Disruption of gut homeostasis by acute intestinal infection associates with αCTLA-4–induced toxicity. C57BL/6J mice with B16-OVA tumors were treated biweekly for 2 wk with IgG or αCTLA-4. Mice were infected with 4–6 × 109 C. rodentium on day 11 by oral gavage (C.r.) or remained uninfected (NI). Animals were euthanized 18 d after tumor establishment and completion of therapy. (A) Schematic diagram of experiment. (B) Body weight change over time; n = 2–5 per NI group, n = 11–12 per C.r. group. (C) Representative photomicrographs of proximal colonic mucosa. Note the intense inflammatory infiltrate in the LP (arrowheads), scale bar = 100 microns (upper panels); and neutrophil infiltrate (arrow) and C. rodentium load on colonic epithelium (arrowheads), scale bar = 20 microns (lower panels); H&E. (D) Histopathological scores of proximal colon at the experimental endpoint; n = 2–3 per NI group, n = 11–13 per C.r. group. (E) CFUs in spleen and liver as indicated; n = 2–3 per NI group, n = 16–20 per C.r. group. (F) Differential cytokine amounts in colon tissue (n = 2 per group) and serum (n = 10 for C.r. + IgG group, n = 11 for C.r. + αCTLA-4 group) as indicated, fold change absolute log 2 > 1. (G) Absolute amounts of immune cells in mLN and spleen as indicated; n = 6–8 per group. (H) Tumor size over time; n = 2–3 per NI group, n = 9–11 per C.r. group. Data shown as mean ± SEM, from four independent experiments. Data were analyzed by two-way ANOVA (B and H), one-way ANOVA (D), F test to compare variations (E), and two-tailed unpaired Student’s t test (G). * P < 0.05, ** P < 0.01, **** P < 0.0001.
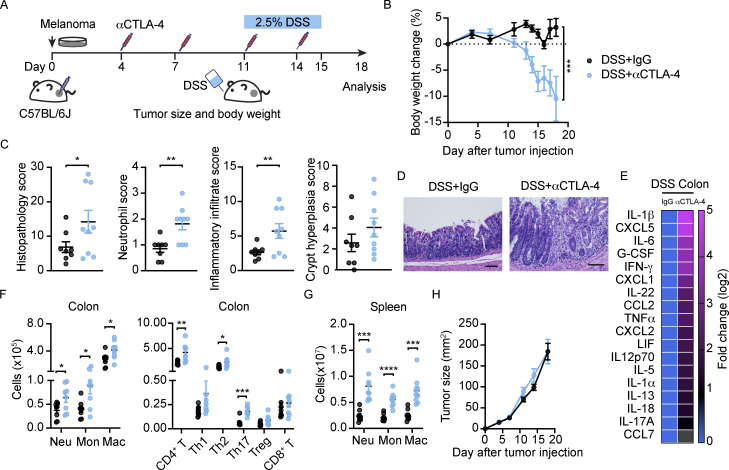
To assess whether the intestinal irAEs observed in Stat3Δ/Δ mice and WT mice infected with C. rodentium were more broadly reflective of αCTLA-4–mediated toxicity, we evaluated the effect of DSS treatment, a common approach to induce experimental colitis (Fig. 7 A). DSS administration in WT tumor-bearing mice on αCTLA-4 led to significant reduction in body weight and evidence of increased colon histopathology, analogous to our findings in αCTLA-4–treated Stat3Δ/Δ mice and WT mice infected with C. rodentium (Fig. 7, B–D). We also detected similar αCTLA-4–mediated immune signatures. For instance, colon myeloid cells and CD4+ T cells, as well as colon inflammatory cytokines and chemokines, including IL-1β, IL-6, G-CSF, IFN-γ, TNF-α, CXCL2, and IL-1α, were elevated in αCTLA-4 DSS-treated WT mice (Fig. 7, E and F). Moreover, we observed systemic increases in splenic neutrophils, monocytes, and macrophages, paralleling responses in αCTLA-4–treated Stat3Δ/Δ and C. rodentium–infected WT mice (Fig. 7 G). In addition, the antitumor effect of αCTLA-4 was disrupted by DSS-induced colitis (Fig. 7 H). These data suggest common immune mechanisms are elicited by αCTLA-4 therapy in conditions of deregulated intestinal immune homeostasis.
Figure 7.
Disruption of gut homeostasis by DSS associates with αCTLA-4–induced toxicity. C57BL/6J mice with B16-OVA tumors were treated biweekly for 2 wk with IgG or αCTLA-4. Mice were given 2.5% DSS in drinking water on day 11–15 after tumor establishment. Animals were euthanized 18 d after tumor establishment. (A) Schematic diagram of experiment. (B) Body weight change over time; n = 8 per group. (C) Histopathological scores of colon at the experimental endpoint; n = 8–9 per group. (D) Representative photomicrographs of colonic mucosa, scale bar = 100 microns; H&E. (E) Differential cytokine amounts in colon tissue, fold change absolute log 2 > 1; n = 8–9 per group. (F and G) Absolute amounts of immune cells in colon (F) and spleen (G) as indicated; n = 8–9 per group. (H) Tumor size over time; n = 8 per group. Data are shown as mean ± SEM, from two independent experiments. Data were analyzed by two-way ANOVA (B) and two-tailed unpaired Student’s t test (C, F and G). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Myeloid and lymphoid signatures in human ICB-associated colitis
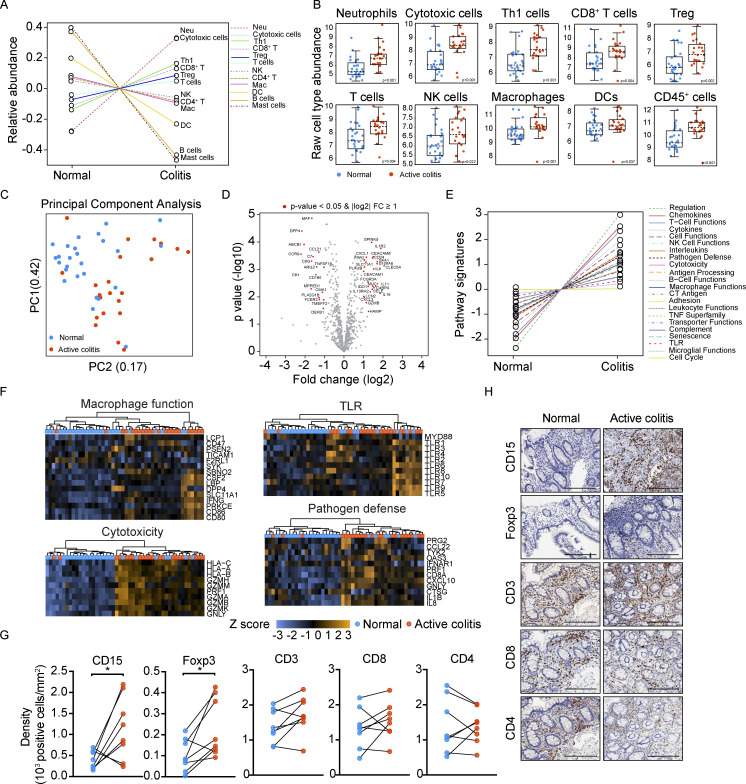
To address the clinical relevance of our murine models, we analyzed immune infiltrates in inflamed intestinal tissue biopsies from individuals with advanced melanoma undergoing ICB therapy at MD Anderson Cancer Center who had developed clinical grade 3–4 colitis (patients, n = 15; biopsy samples, n = 22; Table S4). Matched biopsies of noninflamed intestinal tissue were collected from a subset of these individuals (patients, n = 7; biopsy samples, n = 7); normal colon biopsies were also obtained from unmatched individuals who underwent surgery for colorectal carcinoma without immunotherapy (patients, n = 19; biopsy samples, n = 19; Table S4). The ICB-treated patient cohort included individuals treated with αCTLA-4, αPD-1, combination ICB, and with or without steroid therapy for ICB-associated colitis (Table S4). Gene expression analyses indicated elevated neutrophil, cytotoxic, Th1, and CD8+ T cell signatures in colitis samples, with enrichment of myeloid chemoattractants and inflammatory mediators (e.g., CXCL1, CXCL3, IL8, IL19), and factors associated with T and NK cell effector function (FCGR3A, GZMB; Fig. 8, A and B; and Table S5). Moreover, comprehensive immune response pathway analysis revealed hallmarks of immune activation in the colitis group, including elevated cytokine, chemokine, T cell function, and cytotoxicity gene signatures (Fig. 8, C–F; and Table S5). By evaluating matched tissue samples (patients, n = 8) using immunohistochemistry (IHC), we found significant accumulation of CD15+ neutrophils and Foxp3+ T cells in inflamed areas versus healthy tissue (Fig. 8 G and Table S6). Inflamed regions consisted of crypt abscesses, evident by accumulation of CD15+ positive neutrophils; furthermore, cells that transmigrated through the epithelium into the lumen also appeared to be neutrophils (Fig. 8 H). These results indicate key similarities in the intestinal immune response between our murine irAE models and melanoma patients with ICB-associated colitis. Additionally, the data agree with recent findings from three distinct human intestinal irAE cohorts, which noted increased cytotoxic T cells, neutrophils, and neutrophil chemoattractants in colon tissue (Hailemichael et al., 2022; Luoma et al., 2020). Thus, the collective results suggest our irAE models reflect immune responses in human ICB-associated colitis, with ICB-mediated toxicity driven by neutrophils and activated T cells infiltrating intestinal tissue.
Figure 8.
Immune signatures and association with αCTLA-4 response of human intestinal irAE. (A–F) Colitis regions (n = 22, from 15 individuals with ICB-associated colitis) and normal intestinal biopsies (n = 26, from 19 individuals with colorectal carcinoma and 7 with ICB-associated colitis) were evaluated by NanoString using the human nCounter PanCancer Immune Profiling Panel of 770 cancer-related and immune response genes. (A) Estimation of immune subset abundance using expression of cell type–specific marker genes. Data represent relative abundance scores. (B) Data shown indicate raw cell type abundance scores of individual immune subsets. (C) PCA of all biopsies. (D) Volcano plot showing differential gene expression. Results are normalized to healthy tissue biopsy group. (E) Plot of pathway signature scores for colitis and normal biopsy samples. (F) Heatmaps showing gene expression in individual pathways and samples. (G and H) Staining density (G) and representative IHC images (H) from CD15, Foxp3, CD3, CD8, or CD4 detection in intestinal biopsies from melanoma patients with ICB-associated colitis; scale bar = 200 μm. Matched biopsies diagnosed as active colitis and without diagnostic abnormality from a total of eight patients were analyzed. (B and D) Two-tailed unpaired Student’s t test. * P < 0.05.
Discussion
We found αCTLA-4 stimulates intestinal tissue damage in mice with genetic predisposition to intestinal inflammation, microbiome dysbiosis, acute gastrointestinal infection, or DSS administration, indicating that perturbation of intestinal homeostasis is important to drive murine ICB-associated toxicity. The immune responses observed in our models mirror key immune signatures in human ICB-associated colitis, including accumulation of neutrophils, cytotoxic and IFN-γ+ CD8+ and CD4+ T cells, and inflammatory cytokines such as IFN-γ and IL-6. Moreover, our results suggest systemic release of myeloid-activating cytokines and hematopoietic remodeling to favor myelopoiesis induces a feed-forward mechanism that sustains myeloid-mediated intestinal tissue destruction and inhibits antitumor immunity. Identification of these mechanisms was crucial to intervene therapeutically in the preclinical setting; we found that combination treatment with IL-6 blockade and Abx improved tumor control by αCTLA-4 and reduced intestinal tissue damage. Importantly, our models provide additional opportunities for preclinical investigation of irAE-suppressive therapeutics.
Immune mechanisms of αCTLA-4–driven intestinal irAE
The immunological mechanisms driving ICB-associated irAEs, particularly roles for myeloid subsets, are poorly characterized (Esfahani et al., 2020). We found myeloid populations, predominantly neutrophils in the colonic LP of murine and human irAE lesions, were tightly linked with ICB-mediated intestinal toxicity. Although a limitation of our work is the varied ICB and steroid treatments within our patient cohort, our results agree with a recent study that reported strong neutrophil signatures in colitis biopsies from two independent human cohorts (Hailemichael et al., 2022). Furthermore, our scRNAseq showed immune-activating transcriptional signatures were enhanced in murine intestinal myeloid populations upon αCTLA-4 treatment, suggesting myeloid functional activation is a contributing factor in intestinal irAE. These results warrant further investigation into mechanisms by which ICB targeting adaptive T lymphocytes can also unleash innate, myeloid-mediated responses.
Our model with a defined genetic lesion, Stat3 deficiency in DCs, shows hyperresponsiveness in the intestinal DC compartment can promote experimental ICB-associated colitis, further supporting the concept of myeloid-driving irAE mechanisms in the gut. Through the use of additional immune perturbations in WT mice, including infection, FMT with a dysbiotic microbiome, or DSS, our work indicates that a shift away from immune homeostasis, toward an activated immune state, enhances irAE likelihood. Animal models have key differences in immune status with humans; moreover, our Stat3Δ/Δ mice are unlikely to reflect the basal immune status of the majority of patients treated with ICB, posing limitations to our study. For instance, Stat3Δ/Δ mice present with features of elevated intestinal and systemic immune responses, compared with WT mice, in the absence of αCTLA-4 therapy. While the majority of these responses were further exaggerated in Stat3Δ/Δ mice treated with αCTLA-4, colonic neutrophil accumulation and BM myelopoiesis appeared to be uniquely driven by αCTLA-4 therapy, suggesting interplay between inflammation-prone conditions and ICB. Collectively, these data suggest that pre-existing subacute or acute gut inflammation from genetic or environmental factors in cancer patients may predispose them to ICB-associated intestinal irAEs. Indeed, prior studies demonstrate that inflammatory bowel disease increases the risk of severe gastrointestinal adverse events in patients treated with ICB and can lead to discontinuation of therapy (Abu-Sbeih et al., 2020; Grover et al., 2020; Johnson et al., 2016). Thus, our models may serve as useful tools to identify additional irAE-driving mechanisms and mitigative strategies for cancer patients with inflammatory bowel disease or other conditions predisposed to inflammation.
We observed BM hematopoietic remodeling and emergency myelopoiesis in inflammation-prone mice treated with αCTLA-4; similarly, αCTLA-4 treatment drove systemic myeloid increases with C. rodentium infection or DSS administration. These findings align with studies that identify correlations between increased circulating neutrophil:lymphocyte ratios and poor response to ICB (McCulloch et al., 2022). While we found neutrophil depletion reduced the intestinal irAE immune signature and increased antitumor efficacy of αCTLA-4, neutrophil ablation is not optimal for immunocompromised cancer patients. Rather, specific interventions to decrease localized neutrophil accrual or myeloid-mediated inflammation and consequent tissue damage may provide improved opportunities to suppress irAEs in a clinical setting.
Our comprehensive immune profiling also showed IFN-γ–producing and cytotoxic T cell subsets are associated with murine and human intestinal irAE. In addition, we identified a trend for increased Treg in our inflammation-prone irAE model and significant Treg accrual in human ICB-associated colitis biopsies, suggesting compensatory anti-inflammatory mechanisms were activated. Importantly, these immune features align with studies of independent human intestinal irAE cohorts (Hailemichael et al., 2022; Luoma et al., 2020). While αCTLA-4 is reported to have antagonistic or depletion effects on Tregs in mice, this has not been observed in humans (Wei et al., 2018) and, critically, we did not detect Treg depletion in tumors or peripheral tissues of our αCTLA-4–treated mice. Moreover, our scRNAseq experiments suggest crosstalk mechanisms between myeloid and T cell compartments were enhanced in irAE conditions, particularly via specific cytokine–receptor pairs, including type I and II IFNs, IFN-responsive CXCL9/CXCL10, TNF, and IL-1 pathways. These data are consistent with the concept that myeloid hyperreactivity cooperates with off-target activation of T cell responses in intestinal irAE.
Cytokine drivers of αCTLA-4–associated intestinal irAE
We identified a suite of inflammatory factors that correlate with intestinal irAE, including systemic increases in IL-6, IFN-γ, and G-CSF. This cytokine signature aligns closely with reports from others that have evaluated human ICB-associated colitis (Andrews et al., 2021; Hailemichael et al., 2022; Lim et al., 2019; Luoma et al., 2020). Moreover, these factors are well-known drivers of myeloid cell production, immune activation, and inflammation (Mirantes et al., 2014; Morales-Mantilla and King, 2018; Panopoulos and Watowich, 2008; Walker et al., 2008), consistent with enhanced myelopoiesis and transcriptional reprogramming of colonic LP immune cells in our inflammation-prone irAE model. While IFN-γ can disrupt epithelial barrier junctions (Bruewer et al., 2003) and potentially contribute to intestinal irAE, IFN-γ–producing T cells are beneficial for tumor control, thus targeting this cytokine is not ideal. By contrast, IL-6 and G-CSF have established immunosuppressive effects in the tumor environment and are produced by numerous populations including activated myeloid cells, stromal subsets, and tumors (Gabrilovich and Nagaraj, 2009; Meyer et al., 2018). We found that therapeutic blockade of IL-6 improved αCTLA-4 anti-tumor efficacy in inflammation-prone mice, while combination of IL-6 blockade and Abx effectively mitigated intestinal irAE. These results align well with a recent report that demonstrated IL-6 blockade reduced the severity of experimental autoimmune encephalomyelitis in tumor-bearing mice treated with αCTLA-4 ICB (Hailemichael et al., 2022). Thus, IL-6 inhibition may be a novel strategy to enhance tumor immunity and reduce irAE risk in the clinic (Uemura et al., 2016).
Contribution of the intestinal microbiome to irAE and tumor control by αCTLA-4
The gut microbiome has key roles in mediating the therapeutic response to ICB, yet also contributes to irAEs. Recent work has generated important correlatives between specific taxa and response or toxicity with ICB treatment (Andrews et al., 2021; Chaput et al., 2017; Gopalakrishnan et al., 2018b; McCulloch et al., 2022; Routy et al., 2018). For instance, enrichment of bacterial taxa including Bacteroides genera, Rikenellaceae genera, and Sutterellaceae spp. correlates with poor response to PD-1 blockade and intestinal irAEs (Andrews et al., 2021; McCulloch et al., 2022). Moreover, these taxa were linked with a microbial gene signature of enhanced LPS synthesis, as well as enriched inflammatory gene expression in shed luminal cells, consistent with concomitant systemic and localized inflammation (McCulloch et al., 2022). We identified similar microbiome characteristics in fecal samples from Stat3Δ/Δ mice, including enrichment of Bacteroides, Parasutterella, and Rikenellaceae genera, along with enhanced LPS signaling responses in colonic myeloid populations evaluated by scRNAseq. By contrast, Lachnospiraceae was significantly enriched in Stat3+/+ mice, which aligns with enrichment of this taxa in humans responsive to ICB (McCulloch et al., 2022; Routy et al., 2018).
Critically, using FMT, we demonstrated a direct mechanistic link between the dysbiotic gut microbiome of Stat3Δ/Δ mice and αCTLA-4–mediated intestinal inflammation, tissue toxicity, and poor therapeutic efficacy. By contrast, the reciprocal FMT approach, providing feces from WT to Stat3Δ/Δ mice, failed to improve tumor response to therapy or mitigate αCTLA-4–mediated intestinal irAEs. This may be due to the inherent hypersensitivity of Stat3Δ/Δ DCs to microbiome-derived TLR agonists and consequent elevation in inflammatory responses (Chrisikos et al., 2020; Chrisikos et al., 2022; Melillo et al., 2010). Moreover, by microbiome profiling following intervention with Abx and IL-6 blockade in Stat3Δ/Δ mice, we identified enrichment of Akkermansia and Lachnospiraceae genera, which aligns with enrichment of these taxa in humans responsive to αPD-1 (McCulloch et al., 2022; Routy et al., 2018). Thus, our Stat3Δ/Δ model recapitulates important findings in human ICB-treated individuals. In addition, our FMT studies reveal that specific microbiome components render sensitivity to intestinal irAEs and reduce tumor response to αCTLA-4. FMT of a colitogenic or dysbiotic microbiome provides a potential translational platform for evaluating irAE-driving taxa in human colitis fecal samples. Collectively, our data lay groundwork for further exploration of targeted microbiome interventions to reduce irAE risk and maintain or improve ICB antitumor efficacy.
In summary, the models we describe have key immune correlates with human ICB-associated colitis and provide new preclinical approaches to test therapies to mitigate intestinal irAEs, including targeted manipulation of the gut microbiome. Moreover, our results reveal that dysbiosis or a shift from immune homeostasis toward an immune-activated state predisposes to intestinal irAE in mice. These findings suggest irAEs may arise after homeostatic or tolerance thresholds in particular organ sites are breached by combined effects from localized immune-stimulatory signals and systemic ICB-mediated immune activation, consistent with a recent study in a preclinical skin irAE model (Hu et al., 2022). Our work provides a basis to develop an integrated assessment utilizing multiple platforms to translate preclinical studies into predictive models for ICB-associated irAEs.
Materials and methods
Mice
C57BL/6J mice were acquired from The Jackson Laboratory. B6.Cg-Tg (Itgax-cre)1-1Reiz/J mice (CD11c-Cre; 008068; The Jackson Laboratory) were bred with Stat3f/f mice (Caton et al., 2007; Takeda et al., 1998) to generate CD11c-Cre+ Stat3f/f mice (Stat3Δ/Δ) and Cre-negative Stat3f/f littermate controls (termed herein, Stat3+/+). All mice were maintained in a specific pathogen–free animal facility at MD Anderson Cancer Center. Mice of both sexes were analyzed, ranging in age from 7 through 13 wk. Age- and sex-matched littermates were chosen randomly for inclusion in treatment groups and were used in accordance with MD Anderson Cancer Center Institutional Animal Care and Use Committee–approved protocols.
Murine melanoma and αCTLA-4 therapy
Murine B16-F10 melanoma cells stably expressing OVA (B16-OVA cells) were cultured in DMEM medium (Gibco) containing 10% FBS and 1% penicillin–streptomycin (Zhou et al., 2020). Stat3Δ/Δ and Stat3+/+ mice were shaved on the abdomen and injected s.c.) with 2 × 105 melanoma cells on one side. InVivoPlus anti-mouse CTLA-4 (9H10; BioXcell) or IgG (polyclonal Syrian hamster; BioXcell) were diluted in 200 μl InVivoPure pH 7.0 dilution buffer (BioXcell) and injected i.p. biweekly for 2 wk. Antibodies were delivered at a dose of 200 μg/mouse for the first treatment and 100 μg/mouse for subsequent treatments. Body weight and tumor size (length × width) were measured every 3–4 d. Peripheral blood and organ samples were collected 18–19 d after melanoma injection. Peripheral blood was obtained from anesthetized mice; animals were euthanized, and colon, tumor, spleen, BM, tumor-draining lymph node, and mLN samples were harvested. Fecal samples were collected from mice prior to αCTLA-4 or IgG therapy and on the day of tissue harvest. Mice were euthanized if tumor sizes reached a maximum of 20 mm in any direction or if tumor ulceration occurred; these animals were excluded from further analysis.
Treatment with αIL-6 or αLy6G
Melanoma-bearing Stat3Δ/Δ and Stat3+/+ mice on αCTLA-4 therapy were injected i.p. with anti-mouse Ly6G (1A8; BioXcell) at 12 and 15 d after tumor establishment using 75–250 μg/mouse. A separate group of mice was treated with anti-mouse IL-6 (MP5-20F3; BioXcell) two to four times between 4 and 15 d after tumor establishment using 200 μg/mouse. Anti-trinitrophenol rat IgG2a (2A3; BioXcell) or anti-HRP rat IgG1 isotype control (HRPN; BioXcell) were used as isotype controls.
αIL-6 and Abx combination therapy
Mice received 100 μl of broad-spectrum antibiotic cocktail by oral gavage daily for a total of 4 d prior to tumor injection. The antibiotic cocktail contained ampicillin (1 g/liter; Sigma-Aldrich), vancomycin (0.5 g/liter; Gold Biotechnology), metronidazole (1 g/liter; Sigma-Aldrich), and enrofloxacin (1 g/liter; Sigma-Aldrich). After oral gavage, the mice were maintained on ampicillin-containing (1 g/liter) drinking water for another 5 d until initiation of αCTLA-4 therapy. αIL-6 (200 μg/mouse) was injected (i.p.) every other day for a total of four treatments as indicated in the figure legends.
FMT
Stat3Δ/Δ and Stat3+/+ mice (4 wk old) were separated into different cages according to genotype. Mice were treated with 100 μl of a broad-spectrum antibiotic cocktail by oral gavage daily for a total of 5 d prior to tumor injection. The antibiotic cocktail contained ampicillin (1 g/liter), vancomycin (0.5 g/liter), metronidazole (1 g/liter), and enrofloxacin (1 g/liter). After oral gavage, the mice were allowed to recover for 2 d. FMTs were performed by oral gavage with 50 μl of fecal suspension delivered every other day for a total of three times. Fecal suspensions were prepared by collecting fresh feces from donor Stat3Δ/Δ and Stat3+/+ mice (8 wk old). Fecal material was weighed and smashed in sterile PBS (2 mg feces/50 μl PBS) and then filtered through a 40-μm cell strainer. Recipient Stat3+/+ mice received FMT from donor Stat3Δ/Δ mice and recipient Stat3Δ/Δ mice received FMT from donor Stat3+/+ mice. After FMT, mice were allowed to recover for a week before tumor injection. FMT recipients were then injected s.c. with B16-OVA cells and treated with αCTLA-4 or IgG i.p. biweekly for 2 wk. Antibodies were delivered at a dose of 200 μg/mouse for the first treatment and 100 μg/mouse for subsequent treatments. Mice of different treatment groups were maintained in separate cages throughout the duration of the experiment. Organ samples were collected 18 d after melanoma injection.
C. rodentium infection
C. rodentium (strain DBS100; 51459; ATCC) cultures were established using an inoculation of 10 ml LB medium with 5 μl of concentrated bacterial stock, followed by overnight incubation at 37°C in a bacterial shaker incubator. The following day, 3 ml of the bacterial culture was transferred to 300 ml LB medium and incubated for an additional 2.5 h until OD600 = 0.8–1.0. The bacterial concentration (OD600 of 1.0 = 5 × 108 bacteria/ml) was confirmed by plating serially diluted bacterial cultures on LB plates and determining CFU activity (Bouladoux et al., 2017).
C57BL/6J mice were injected s.c. with B16-OVA cells and treated with αCTLA-4 or IgG as indicated. Mice were infected with C. rodentium 11 d after tumor establishment, using oral gavage with 4–6 × 109 C. rodentium in a total volume of 200 μl PBS per mouse. Body weight and tumor sizes were measured over time following infection, and mice were euthanized 18 d after tumor injection (7 d after infection) or when weight loss was >20%. To evaluate tissue CFU, spleen and liver were collected following euthanasia; organs were weighed, homogenized using a sterile syringe plunger, and homogenates were diluted in 0.5–1 ml sterile PBS. Serial dilutions of 100 μl were transferred to LB plates, spread, and incubated at 37°C for 24 h. CFU was enumerated after 24 h.
DSS administration in melanoma-bearing mice
C57BL/6J mice were injected s.c. with B16-OVA cells and treated with αCTLA-4 or IgG i.p. biweekly for 2 wk. Antibodies were delivered at a dose of 200 μg/mouse for the first treatment and 100 μg/mouse for subsequent treatments. Mice were given 2.5% DSS (MP Biomedicals) in drinking water 11–15 d after melanoma injection for a total of 5 d. Organ samples were collected 18 d after melanoma injection.
Immune cell isolation from colonic LP
Following dissection, colon tissue was placed in ice-cold PBS immediately and maintained on ice for further processing. Mesenteric material, fat, and Peyer’s patches were removed. Colon sections were cut open, feces removed, and colon tissue was cut into 1–2 cm pieces. In some experiments, 1 cm of the proximal colon and 1 cm of the distal colon was harvested for protein lysate preparation or histological assessments, leaving the remaining colon tissue for immune cell isolation. Colon tissue pieces were placed in 8-ml ice-cold PBS and shaken vigorously by hand at room temperature for 2 min. Tissue pieces were transferred to 20 ml PBS containing 1 mM dithiothreitol (Sigma-Aldrich), 30 mM EDTA (Sigma-Aldrich), and 10 mM Hepes (Sigma-Aldrich) at room temperature. The samples were placed in a bacterial shaker incubator horizontally, rotated at 37°C for 10 min at 200 rpm, and then shaken by hand at room temperature for 1 min (Kim et al., 2018). Tissue pieces were transferred to new tubes containing a fresh solution of 20 ml PBS, 30 mM EDTA, and 10 mM Hepes, and were shaken for a second 10 min as indicated. The tissue pieces were rinsed gently with RPMI medium containing 2% heat-inactivated FBS (RPMI/2% FBS) at room temperature. Tissue pieces were transferred to tubes containing 5 ml of RPMI medium, 10% FBS, 200 U/ml collagenase VIII (Sigma-Aldrich), and 150 μg/ml DNase I (Sigma-Aldrich). Tissues were digested by shaking in a bacterial shaker incubator at 100 rpm at 37°C for 1 h and then shaken by hand at room temperature for 1 min. The digested tissues were placed on a 100 μm cell strainer; single cells in the flow-through were collected. Cells were centrifuged at 600 g for 5 min at 4°C and resuspended in 4 ml 40% Percoll (Sigma-Aldrich) in RPMI/2% FBS. The Percoll-cell solution was layered on top of 4 ml 80% Percoll in RPMI/2% FBS and centrifuged at 1,300 g at room temperature for 20 min; spins were concluded without using the centrifuge brake. The cells at the interface of the 40 and 80% Percoll solutions were collected; cells were washed once with ice-cold PBS/2% FBS prior to antibody incubations.
Immune cell isolation from BM
BM cells were flushed and red blood cells were removed using RBC Lysis Buffer (Tonbo Bioscience). Lineage− (lin−) BM progenitors were enriched by magnetic bead separation using biotinylated rat anti-mouse antibodies against CD3 (145-2C11; eBioscience), CD45R (RA36B2; eBioscience), CD11b (M1/70; eBioscience), Ter-119 (Ter-119; eBioscience), and Ly6G/Ly6C (RB6-8C5; eBioscience) followed by negative selection with anti-rat Ig conjugated Biomag beads (Miltenyi Biotec). The lin− cells were washed once with ice-cold PBS prior to antibody incubations.
Immune cell isolation from tumors, spleen, and LNs
Single-cell suspensions were generated from tumors, spleen, and LNs by smashing tissues with a sterile syringe plunger onto a 100-μm cell strainer; cells were rinsed through the strainer with RPMI/2% FBS at room temperature. For spleen cell isolations, red blood cells were first removed using RBC Lysis Buffer (Tonbo Bioscience) following the manufacturer’s recommendations. Cells were centrifuged at 600 g for 5 min at 4°C and resuspended in ice-cold PBS/2% FBS prior to antibody incubations.
Antibody staining and flow cytometry
Single-cell suspensions were plated in 96-well V-bottom plates at 1 × 105–3 × 106 cells per sample. For T cell subset analysis, samples were incubated with 0.5 μg/ml ionomycin (Sigma-Aldrich), 50 ng/ml PMA (Sigma-Aldrich), and GolgiStop (BD Biosciences) for 4–6 h for intracellular cytokine accumulation prior to Fc receptor blocking. For all samples except lin− BM progenitors, cell surface Fc receptors were blocked by incubation with rat anti-mouse CD16/32 antibody (1:100; Tonbo Bioscience) for 15 min at 4°C. Cells were incubated with fluorescently conjugated antibodies for surface makers for 30 min at 4°C; subsequently, T cells were fixed and permeabilized using commercial reagents (Intracellular Fixation and Permeabilization Buffer Set; eBioscience) and stained with antibodies against intracellular proteins for 30 min at 4°C (Liu et al., 2019). All antibodies were purchased from BioLegend, eBioscience, BD Biosciences, or Tonbo Biosciences (Table S7). Dead cells were discriminated in all experiments using Ghost Dye Violet 510 (Tonbo Biosciences). OVA-specific CD8+ T cells were analyzed by staining with SIINFEKL/H-2Kb-Pentamer (F093-4A; ProImmune). Precision Count Beads (BioLegend) were added into samples for cell number calculation.
Flow cytometry analysis was performed on LSRII and LSRFortessa flow cytometers (BD). Data was acquired using FACSDiva 8.0 software (BD). Flow cytometry files (.fcs) were analyzed using FlowJo 10.6.2 software (BD) through traditional manual gating or unsupervised gating strategies. Dimension reduction was performed using the tSNE algorithm, and clustering was performed using the FlowSOM algorithm in FlowJo 10.6.2.
For generating tSNE plots, the flow cytometry data from three tissue types, colon (n = 28), tumor (n = 29), and spleen (n = 27) were analyzed with dimension reduction and unbiased clustering algorithms along with traditional, manual gating methods. For both the colon myeloid (maximum 150,000 events per mouse) and tumor T cell (maximum 10,000 events per mouse) panels, an equal number of viable single CD45+ cells were evaluated from each biological murine sample and concatenated into one file, unless the flow cytometry file contained a low number of events, in which case all events in the described population were used. For the spleen panel, the concatenated file was generated using viable single cells, with a maximum of 50,000 events per mouse. For the colon T cell panel, a total of 37,625 viable single CD90.2+ or CD19+ cells were evaluated from each treatment group. Next, the dimension reduction algorithm tSNE (software embedded plugin) and the unbiased clustering algorithm FlowSOM (version 2.6) were used to identify cell populations, using all surface markers in the panel for algorithm computation except viability dye and CD45. The algorithm input population for the colon myeloid panel was single, live, CD45+ myeloid cells (CD11b+ CD11c−, CD11b−CD11c+, and CD11b+ CD11c+ cells); the colon T cell panel was single, live lymphoid cells (CD90.2+ CD19−, CD90.2−CD19+, and CD90.2+ CD19+ cells); the tumor T cell panel was single, live, CD45+ CD3+ cells; and the spleen panel was single, live cells. Manually gated populations, FlowSOM clustered populations, mouse genotype and treatment groups, and surface marker expression were overlaid on tSNE maps to confirm immune subset populations in an unbiased manner and to visually represent the changes in immune subset populations with treatment groups. Manually gated and FlowSOM cluster population frequencies of single, live, CD45+ or single, live cells were calculated for all mice and for individual mice for statistical analysis.
Cytokine detection by multiplex assays
To prepare protein lysates from colon tissue, 1 cm of the proximal colon was harvested, washed with PBS, and minced into small pieces (∼1 mm). Colon pieces were resuspended in 500 μl nonenzymatic cell dissociation buffer (Sigma-Aldrich) and treated by sonication on ice for 1 min for 20 timed cycles using the Ultrasonic Processor W-385, Heat Systems-Ultrasonics, Inc. Cycle time was set to 2 s, 60% duty cycle at maximum output control. Samples were centrifuged at 2,000 g for 10 min at 4°C to remove cell debris. Supernatants were collected and stored at −80°C until analysis.
For serum preparation, whole blood was collected by cardiopuncture and allowed to clot undisturbed at room temperature for 30 min. Clots were removed by centrifugation at 2,000 g for 10 min at 4°C. Serum samples were collected and stored at −80°C until analysis.
Cytokines in colon lysates and serum were measured by the ProcartaPlex assay (mouse ProcartaPlex panel 1A; Invitrogen) performed on a Luminex 200 machine (Luminex). Cytokines and chemokines analyzed were as follows: CXCL1 (C-X-C motif chemokine 1), CXCL2, CXCL5, CXCL10, CCL2 (C-C motif ligand 2), CCL3, CCL4, CCL5, CCL7, Eotaxin, IFN-α, IFN-γ, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12p70, IL-13, IL-15/IL-15R, IL-17A, IL-18, IL-22, IL-23, IL-27, IL-28, IL-31, G-CSF, GM-CSF, M-CSF, leukemia inhibitory factor, and TNF-α.
Histopathology and scoring of murine samples
Colon samples were opened longitudinally and contents were removed gently by flushing with PBS. The colon was wrapped in a “Swiss roll” and fixed in 10% neutral-buffered formalin. In some experiments, 1.0–1.5 cm of the distal colon was collected, gently flushed with PBS, separated into two to three small pieces, and fixed in formalin. Tissue samples were routinely processed into paraffin blocks, sectioned at 5 microns, and stained with H&E. Architectural and cytomorphologic features were evaluated and scored by a board-certified veterinary pathologist using a semiquantitative scale generated for this study (Table S1), with 0 = no change/lesion and 4 = severe change/tissue destruction. Histologic features scored included inflammatory infiltrates (estimate of populations of neutrophils or mixed leukocytes in the LP or submucosa) and crypt hyperplasia (mitotic figures above proliferation zone, pseudostratification of colonic epithelial nuclei, altered crypt architecture, and loss of goblet cells). The sum of all scored features is reported as “total histopathology.” Data from individual animals were reported as the mean scores from two to three tissue sections or regions per mouse (scores were determined separately from the proximal and distal colon of Swiss roll sections).
Immunoblot assays
Single-cell suspensions were generated from tumors by dissociating tissues with a sterile syringe plunger onto a 100-μm cell strainer. DCs (CD11c+) and myeloid cells (CD11c− CD11b+) were purified by FACS from tumors by flow cytometry. Total cell lysates and immunoblots were performed as described previously (Liu et al., 2021) using rabbit polyclonal antibodies against STAT3 (79D7; Cell Signaling Technology) and Tubulin (B-5-1-2; Abcam).
scRNAseq
Library generation
Colonic LP immune cells were isolated and sorted for live CD45+ cells by FACS. Cells were resuspended at a concentration of 700–1,200 cells/μl for microfluidics (Chromium Single-Cell Controller; 10x Genomics). Cells were loaded into the chip and run using the Chromium Single Cell 3′ Reagent Kit v3 (10x Genomics) according to the manufacturer’s instructions. Resuspended single cells were partitioned in gel beads in emulsion and lysed. Lysis was followed by RNA barcoding, reverse transcription, PCR amplification (12–14 cycles), fragmentation, ligation, and sample index PCR. cDNAs were quantified by D5000 ScreenTape (Agilent Technologies) on an Agilent 4200 TapeStation system (Agilent Technologies). Sequencing-ready scRNAseq libraries were quantified by 2100 Bioanalyzer (Agilent Genomics) instrument. Sequencing was performed on an Illumina NextSeq 500 machine (San Diego), and four indexed samples were multiplexed into one output flow cell using NextSeq 500/550 High Output Kit v2.5 in paired-end sequencing (R1, 26nt; R2, 98nt, and i7 index 8nt) at the MD Anderson Cancer Center South Campus RNAseq core facility.
Quantification of scRNAseq reads
Sequencing data were analyzed using the Cellranger pipeline (10x Genomics, version 3.0, https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger) to generate gene count matrices. Cellranger mkfastq (10x Genomics) was used to demultiplex raw base call files from the Illumina NextSeq 500 machine into sample-specific fastq files. Next, fastq files for each sample were processed with Cellranger count (10x Genomics), which takes fastq files and performs alignment, filtering, barcode counting, and unique molecular identifier counting. The expected cell numbers for each sample were 104. Samples were aligned to the murine genome (mm10), filtered, and quantified. The resulting analysis files for each sample were aggregated using the cellranger aggr pipeline, which performed a between-sample normalization step and merged four samples into one.
Preprocessing of scRNAseq data
Quality control (QC), normalization, and analysis were done using Seurat 3.0 (Butler et al., 2018; Stuart et al., 2019). Genes that were expressed in <3 cells were excluded. Cells with unique feature counts <200 or >2,500 genes were removed, which are potentially poor- and high-PCR artifact cells. Finally, cells that had over 25% mitochondrial counts were excluded, as these are low-quality cells, dying cells, or doublets. Cells were normalized using regression to remove the percent mitochondrial DNA variable via the scTransform command. After QC and normalization, anchors were identified and integrated colonic CD45+ immune cell datasets from the four groups were compiled into one, enabling them to be jointly analyzed as an integrated expression matrix (FindIntegrationAnchors and IntegrateData command). Next, PCA was performed, and dimensionality of the dataset was determined by the heuristic method by generating an “Elbow plot” (RunPCA and ElbowPlot command). We observed an “elbow” around PC 15–16, suggesting that the majority of true signals are captured in the first 16 PCs. For clustering and visualization, a Uniform Manifold Approximation and Projection (UMAP) dimensional reduction was performed on a matrix using the first 16 PCAs to obtain a two-dimensional representation of the cell states with resolution 0.1–1.2, leading to 7–22 clusters (RunUMAP, FindNeighbors, and FindClusters command). A resolution of 0.6 was chosen for the analysis.
Doublet removal analysis
Droplet-based scRNAseq introduces multiplets where multiple cells are encapsulated into a single drop. In our analysis, we evaluated doublet scores for all analyzed cells with the R package DoubletFinder v2.0 (McGinnis et al., 2019). We calculated the doublet scores for the four groups separately with the same pN = 0.25 for all four groups, with pK = 0.01 for IgG-treated Stat3+/+ mice, pK = 0.01 for αCTLA-4–treated Stat3+/+ mice, pK = 0.02 for IgG-treated Stat3Δ/Δ mice, pK = 0.01 for αCTLA-4–treated Stat3Δ/Δ mice, and with estimated doublet rates = 0.045 for IgG or αCTLA-4–treated Stat3+/+ mice, 0.015 for IgG-treated Stat3Δ/Δ mice, and 0.035 for αCTLA-4–treated Stat3Δ/Δ mice. The DoubletFinder package automatically determined thresholds for the four groups based on the given estimated doublet rates. Cells with doublet scores higher than thresholds were identified as doublets by the algorithm initially. To further identify doublets that are falsely reported as negative, we performed outlier analysis based on the clustering information. The principle is clustering in subgroups captures subtle variations that distinguish doublets from singlets. We normalized and scaled the expression data again within each subgroup (obtained in the preprocessing step, resolution = 0.6) and identified new clusters again within the subgroup. For some subgroups, we observed new clusters isolated from the major population with higher level of doublet scores. We considered these clusters as outliers and classified them as multiplets.
Identification of cluster-specific genes and marker-based classification
To identify marker genes, the FindAllMarkers command was used with likelihood-ratio test for single-cell gene expression. For each cluster, only genes that were expressed in >25% of cells with at least 0.25-fold difference and with a P value <0.01 were considered. Plots were generated with either the DimPlot, FeaturePlot, or VlnPlot commands. To characterize clusters, we compared the expression of the top 50 differentially expressed genes in each cluster to gene expression in purified immune subsets analyzed by RNAseq on the ImmGen platform (Table S2; Gubin et al., 2018; Heng et al., 2008).
Evaluation of gene set activities
We selected gene sets of interests (metabolic pathways and bioprocess gene sets) from MSigDB (C2 and C5) and quantified their activation levels with gene set variation analysis (GSVA) and Seurat (Hanzelmann et al., 2013; Subramanian et al., 2005). For group-level comparisons (Fig. 2 F), we used the AddModuleScore function in Seurat to calculate the average expression values of the given genes that occurred in the gene set for each cell. The scores were then summarized with the violin plots in Fig. 2 F among four treatment groups regardless of cell types. For cluster-level comparisons (Fig. S3 B), we calculated the average normalized expression (the data slot of the RNA assay in a Seurat object) and used them to evaluate the gene set activity scores with GSVA.
Cell crosstalk analysis
We performed the crosstalk analysis for the major cell populations (clusters 0–15) based on the average expression levels of ligand–receptor pairs. We excluded the minor populations (clusters 16–18) because the gene expression levels might be inaccurate due to the low abundance of each cluster. The ligand–receptor pairs analyzed were extracted from a mouse ortholog dataset derived from a human dataset (Ramilowski et al., 2015; Skelly et al., 2018). We used a similar strategy to evaluate the intensity of cell–cell crosstalk (Luoma et al., 2020). First, we calculated the average normalized expression levels (the data slot of the RNA assay in a Seurat object) of all ligand/receptor genes across all cell populations of interest (clusters 0–15) and kept ligand/receptor genes with average expression levels >1 as candidate genes. Second, among the candidate genes, we calculated the ligand–receptor scores between cluster i and cluster j as SL,R = EL,i·ER,j, where EL,i is the average expression level of the ligand L in cluster i, ER,j is the average expression level of the receptor in cluster j, EL,i > 1 and ER,j > 1. Third, we used the permutation tests to filter ubiquitous pairs (Table S3). For each ligand–receptor pair, we randomly shuffled the cell type identities N times (N = 10,000) and calculated the ligand–receptor scores S′L,R according to the shuffled identities. The P value of the pair is evaluated as the count of the event S′L,R > SL,R divided by N. We visualized the ligand–receptor pairs with P value <0.001 in Fig. 2 G with the R package Circlize (v0.4.8; Gu et al., 2014).
Targeted 16S rRNA gene profiling
To approximate the gut microbiome, fecal material was subjected to 16S rRNA sequencing. For data shown in Fig. 4, A–C, feces were collected from melanoma-bearing Stat3Δ/Δ and Stat3+/+ mice 3 d after tumor establishment (prior to αCTLA-4 treatment) and 18 d after tumor injection (following four doses of αCTLA-4). Sample preparation and 16S rRNA sequencing was performed in collaboration with the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine. For data shown in Fig. 5, I–K, feces were collected from melanoma-bearing Stat3Δ/Δ mice 18 d after tumor injection, following four doses of αCTLA-4 with or without αIL-6 and Abx therapy. Sample preparation and 16S rRNA sequencing was performed in collaboration with the MD Anderson Cancer Center Microbiome Core. Library preparation and sequencing followed our previously published methodology (Gopalakrishnan et al., 2018b). In brief, bacterial genomic DNA was extracted using the PowerSoil DNA Isolation Kit (MO BIO Laboratories). The 16S ribosomal gene V4 region was amplified by PCR and sequenced on the MiSeq platform (Illumina) using the 2 × 250 bp paired-end protocol, yielding paired-end reads with a near-complete overlap. The primers used for amplification contain adapters for MiSeq sequencing, and single-end barcodes allow pooling and direct sequencing of PCR products. Quality-filtered sequences with >97% identity were processed with the Divisive Amplicon Denoising Algorithm 2 generating amplicon sequence variants that were classified with the SILVA v138 (Callahan et al., 2016; Quast et al., 2013). The relative abundance of each amplicon sequence variant was determined for all samples. Sequencing depths ranged from 13,007 to 38,117, with a median of 25,464 reads per sample. A rarefaction cutoff of 13,077 reads was applied to the dataset for analyses. Fecal microbiome abundance data from the samples were analyzed in the statistical software R using phyloseq, DESeq2, and ggplot2 packages. Within-sample diversity of the microbiome samples was reported using Inverse Simpson alpha diversity and then compared using Wilcoxon Rank-Sum test. Between-sample diversity was compared using beta diversity analysis with Bray–Curtis dissimilarity. Then the ordination plot represents samples in terms of the top two principal components obtained from principal coordinate analysis. PERMANOVA analyses (with 999 permutations) were used to compare microbial diversity between any two treatment groups. Using the statistical method implemented in the R package DESeq2 (Love et al., 2014), we identified significant differentially abundant taxa that are associated with the treatment groups.
Patient cohorts for RNA and IHC studies
We evaluated samples from a cohort of patients with advanced melanoma (stage III/IV) who received ICB therapy at MD Anderson Cancer Center between 12/28/2013 and 01/17/2019 and had developed clinical grade 3–4 colitis (n = 15; Table S4) for RNA expression by NanoString. ICB treatment includes αCTLA-4 (ipilimumab), αPD-1 (pembrolizumab or nivolumab), or both (ipilimumab plus pembrolizumab or nivolumab). Regions of colitis (n = 22) and normal, noninflamed tissue (n = 7) were biopsied at the time of clinical grade 3–4 colitis (Table S4). Biopsy samples were immediately formalin-fixed and paraffin-embedded (FFPE) upon collection. Colitis-associated pathology or normal tissue architecture was confirmed independently by two pathologists using H&E-stained samples. An additional set of biopsies from noninflamed intestine (n = 19) was obtained from 19 colorectal carcinoma patients who underwent surgery without ICB treatment. These samples were included as controls in RNA analyses. For IHC studies, we analyzed samples from a cohort of patients with advanced melanoma (stage III/IV) who received ICB therapy at MD Anderson Cancer Center and had developed clinical grade 3–4 colitis (n = 8; Table S6). Regions of colitis (n = 12) and normal, noninflamed tissue (n = 8) were biopsied at the time of clinical grade 3–4 colitis (Table S6). Biopsy samples were obtained following informed consent. These studies were conducted under the MD Anderson Cancer Center Institutional Review Board–approved protocols following the Declaration of Helsinki.
Gene expression profiling by NanoString
NanoString was performed using the nCounter PanCancer Immune Profiling Panel Human V1.1 (NanoString Technologies) representing 770 cancer-related and immune response genes. RNA was extracted from FFPE blocks using the Highpure RNA Isolation Kit (Roche). For gene expression studies, 1 μg of RNA was used per sample. Hybridization was performed for 16–18 h at 65°C and then loaded onto the nCounter Prep Station for binding and washing prior to scanning and capture of 600 fields using the nCounter (NanoString Technologies).
Gene expression data analysis was performed using nSolver 4.0 analysis software (NanoString Technologies; Table S5). For QC, the overall performance of the nCounter assay was evaluated by imaging and binding density QC metrics. Next, the performance of the positive control was assessed using the positive control linearity and limit of detection parameters. In addition, we performed an overall visual inspection of the data and assessed the severity of any QC flags. For background correction, we used two methods, background thresholding and background subtraction. For standard normalization, we used a combination of positive control normalization, which uses synthetic positive control targets, and codeSet content normalization, which uses housekeeping genes, to apply a sample-specific correction factor to all target probes within that sample lane. For generating normalized NanoString gene expression data, ratios and fold change were calculated in the log2 scale. The relative abundance scores were determined by subtracting the total immune score from a single cell type score (all except CD4+ T cells) or the T cell score (CD4+ T cells). The total immune cell score is defined as the average of B, T, CD45+, macrophage, and cytotoxic cell scores. Cell type profiling, pathway score, and differential gene expression were analyzed by nSolver Advanced Analysis software plugin 2.0.
IHC of colitis biopsies
For analysis of intestinal immune cell infiltration in melanoma patient biopsies, colitis and normal intestinal FFPE tissue blocks were serially sectioned for IHC staining. IHC studies were performed on a BOND automated system in combination with the BOND Polymer Refine Detection reagents (Leica Biosystems). IHC sections were stained with mouse or rabbit anti-human antibodies against CD3 (1:100; polyclonal; Agilent), CD4 (1:80; 4B12; Leica Biosystems), CD8 (1:100; C8/144B; Thermo Fisher Scientific), Foxp3 (1:50; 206D; Biolegend), or CD15 (1:40; MMA; BD Biosciences) at the indicated dilutions. Rabbit anti-mouse IgG and polymer anti-rabbit poly-HRP-IgG were used as secondary antibodies (BOND Polymer Refine Detection kit, Leica Biosystems). The substrate chromogen, 3,3′-diaminobenzidine tetrahydrochloride hydrate, was used to visualize immune complexes via a brown precipitate (BOND Polymer Refine Detection kit, Leica Biosystems). Hematoxylin (blue) counterstaining allowed visualization of cell nuclei. Slides were scanned and digitized using the Aperio Scanscope AT Turbo scanner (Leica Biosystems).
Quantitative analysis of IHC staining was conducted using the software Aperio ImageScope (Leica Biosystems). Five random areas were selected using a customized algorithm for each marker to determine the number of positive cells at a high-power field. The data were expressed as the number of positive cells per mm2. IHC staining was interpreted in conjunction with H&E-stained sections.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 8 or R. Statistical analyses of immune cell amounts, cytokine and chemokine amounts, histopathological scores, tissue CFU, and human IHC scores were determined by two-tailed Student’s t test or ordinary one-way ANOVA with Tukey’s multiple comparisons test using Prism. Body weight change and tumor size measurements were evaluated by ordinary two-way ANOVA with Sidak multiple comparisons using Prism. A 95% confidence interval was calculated. For 16S rRNA gene profiling, alpha diversity was analyzed by the Wilcoxon Rank-Sum test, beta diversity was analyzed by Bray–Curtis dissimilarity, and differentially abundant taxa were identified by statistical method implemented in the R package DESeq2. Data are from distinct samples or animals, presented as mean ± SEM. Differences were considered significant when P < 0.05.
Online supplemental material
Fig. S1 shows the flow cytometry gating strategy for colonic LP immune cells and scRNAseq analysis of murine colonic LP immune cells following αCTLA-4 therapy. Fig. S2 shows expression of proinflammatory factor and granzyme mRNAs in colonic LP immune cells from mice on αCTLA-4 therapy. Fig. S3 shows pathway analysis and crosstalk between colonic LP immune cells from mice on αCTLA-4 therapy. Fig. S4 shows characterization of the immune response in various tissues from mice on αCTLA-4 therapy. Fig. S5 shows the effects of neutrophil depletion, IL-6 blockade, or antibiotics on αCTLA-4–associated intestinal toxicity. Table S1 shows the scoring system for colonic inflammation. Table S2 shows differential gene expression of scRNAseq clusters. Table S3 shows ligand–receptor pairs across scRNAseq clusters. Table S4 provides data for the patient cohort used in NanoString analyses. Table S5 shows normalized gene expression of samples analyzed by NanoString. Table S6 provides data for the patient cohort used for IHC studies. Table S7 includes the antibodies used for flow cytometry analyses.
Supplementary Material
shows the scoring system for colonic inflammation.
shows differential gene expression of clusters.
shows ligand–receptor pairs across clusters.
shows the patient cohort for NanoString.
shows normalized gene expression of NanoString samples.
shows the patient cohort for IHC.
is the flow cytometry antibody list.
contains original blots for Fig. 3.
Acknowledgments
We thank Kathryn Newton, Dr. Xianxiu Wan, Xiuping Liu, and Dr. Changgong Liu for technical assistance; Dr. Yongwoo David Seo for review of clinical data; Dr. James Mancuso for review of the manuscript; and the MD Anderson Research Histology Core Laboratory and South Campus Flow Core for advice and assistance. We acknowledge the support of the High Performance Computing for research facility at the University of Texas MD Anderson Cancer Center for providing computational resources that have contributed to the research results reported in this paper.
This work was supported by grants from National Institutes of Health (NIH) to S.S. Watowich (R01AI109294, R01AI133822, and a developmental research project grant from P50CA221703); a joint NIH R01 supplement to S.S. Watowich and P. Hwu (R01AI109294-04S1 and 3R01CA187076-05S1); the Wilks Family Fund to P. Hwu and A. Diab; the Cancer Prevention and Research Institute of Texas (CPRIT) Research Training award to T.T. Chrisikos, R.L. Babcock, and A.P. Cogdill (RP170067); the CPRIT Research Training award to E.M. Park and L.M. Kahn (RP210028); the Parker Institute for Cancer Immunotherapy Bridge Scholar Award, the University of Texas MD Anderson Cancer Center Support Grant New Faculty Award supported by the NIH/National Cancer Institute (NCI; P30CA016672), the CPRIT Scholar in Cancer Research Grant to M.M. Gubin (RR190017); NIH T32 to S.H. Cass (CA009599); NIH/NCI Cancer Center Support Grant P30 (CA016672) and NIH/National Heart, Lung, and Blood Institute R01 (HL158796) to C.B. Peterson. J.P. Allison is a CPRIT Scholar in Cancer Research and Director of the Parker Institute for Cancer Immunotherapy at MD Anderson. The Research Histology Core Laboratory and The Advanced Cytometry & Sorting Core Facility are supported by NCI P30CA0166722 to MD Anderson.
Author contributions: Y. Zhou conceptualized the studies, designed and performed experiments, collected and analyzed data, and wrote the manuscript. Y.B. Medik, B. Patel, E.M. Park, R.L. Babcock, T.T. Chrisikos, L.M. Kahn, A.M. Dyevoich, J.E. Pineda, and H.S. Li assisted with mouse sample collection and data analysis. D.B. Zamler and S. Chen analyzed scRNAseq data. T. Chapman, M.C. Wong, A.K. Mishra, N.J. Ajami, S.B. Johnson, J. Wang, and M.A. Wadud Khan assisted with the 16s profiling and data analysis. S. Schneider and K. Clise-Dwyer assisted with flow cytometry data analysis. S.H. Cass collected and analyzed patient cohort information. K. Wani and R. Arora identified samples for Nanostring and IHC. M.T. Tetzlaff, W.C. Foo, D.A. Ledesma, and C.W. Hudgens performed IHC staining and analyzed data. A.Y. Joon analyzed NanoString data. E.M. Whitley, M.G. Raso, and X. Tang performed histopathology analyses of mouse colon samples. A.P. Cogdill, D.H. Johnson, C.B. Peterson, W. Peng, X. Zhang, M.M. Gubin, G.E. Diehl, J.P. Allison, P. Hwu, N.J. Ajami, A. Diab, and J.A. Wargo provided advice on experimental design, data analyses, and data presentation. S.S. Watowich conceptualized the studies, designed experiments, wrote the manuscript, contributed to funding acquisition, administered the project, and supervised the study. All authors read, edited, and approved the final manuscript.
Data availability
The scRNAseq data (Fig. 2, A–G) are available at the Gene Expression Omnibus under accession code GSE159892. 16s RNA gene profiling data (Fig. 4, A–C; and Fig. 5, I–K) are available at NCBI BioProject under project code PRJNA670730.
The code generated for this current study is available from the corresponding author upon request. The code is stored at https://github.com/chansigit/irae_reproduction.
References
- Abu-Sbeih, H., Faleck D.M., Ricciuti B., Mendelsohn R.B., Naqash A.R., Cohen J.V., Sellers M.C., Balaji A., Ben-Betzalel G., Hajir I., et al. 2020. Immune checkpoint inhibitor therapy in patients with preexisting Inflammatory Bowel Disease. J. Clin. Oncol. 38:576–583. 10.1200/JCO.19.01674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, M.C., Duong C.P.M., Gopalakrishnan V., Iebba V., Chen W.S., Derosa L., Khan M.A.W., Cogdill A.P., White M.G., Wong M.C., et al. 2021. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat. Med. 27:1432–1441. 10.1038/s41591-021-01406-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamias, G., Delladetsima I., Perdiki M., Siakavellas S.I., Goukos D., Papatheodoridis G.V., Daikos G.L., and Gogas H.. 2017. Immunological characteristics of colitis associated with anti-CTLA-4 antibody therapy. Cancer Invest. 35:443–455. 10.1080/07357907.2017.1324032 [DOI] [PubMed] [Google Scholar]
- Bouladoux, N., Harrison O.J., and Belkaid Y.. 2017. The Mouse Model of infection with citrobacter rodentium. Curr. Protoc. Immunol. 119:19.15.11–19.15.25. 10.1002/cpim.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruewer, M., Luegering A., Kucharzik T., Parkos C.A., Madara J.L., Hopkins A.M., and Nusrat A.. 2003. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J. Immunol. 171:6164–6172. 10.4049/jimmunol.171.11.6164 [DOI] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., and Holmes S.P.. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 13:581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton, M.L., Smith-Raska M.R., and Reizis B.. 2007. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 204:1653–1664. 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput, N., Lepage P., Coutzac C., Soularue E., Le Roux K., Monot C., Boselli L., Routier E., Cassard L., Collins M., et al. 2017. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 28:1368–1379. 10.1093/annonc/mdx108 [DOI] [PubMed] [Google Scholar]
- Cheng, W.Y., Wu C.Y., and Yu J.. 2020. The role of gut microbiota in cancer treatment: Friend or foe? Gut. 69:1867–1876. 10.1136/gutjnl-2020-321153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrisikos, T.T., Zhou Y., Kahn L.M., Patel B., Denne N.L., Brooks A., Shen L., Wang J., and Watowich S.S.. 2022. STAT3 inhibits autocrine IFN signaling in type I conventional dendritic cells. J. Immunol. 209:1286–1299. 10.4049/jimmunol.2101104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrisikos, T.T., Zhou Y., Li H.S., Babcock R.L., Wan X., Patel B., Newton K., Mancuso J.J., and Watowich S.S.. 2020. STAT3 inhibits CD103(+) cDC1 vaccine efficacy in murine breast cancer. Cancers. 12:E128. 10.3390/cancers12010128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, C., Bambhania K., and Naidoo J.. 2019. Immune-related adverse events: A case-based approach. Front. Oncol. 9:530. 10.3389/fonc.2019.00530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, M.A., Montalvo W., Yagita H., and Allison J.P.. 2010. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc. Natl. Acad. Sci. USA. 107:4275–4280. 10.1073/pnas.0915174107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S., and Johnson D.B.. 2019. Immune-related adverse events and anti-tumor efficacy of immune checkpoint inhibitors. J. Immunother. Cancer. 7:306. 10.1186/s40425-019-0805-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan, M.. 2017. Checkpoint blockade toxicity and immune homeostasis in the gastrointestinal tract. Front. Immunol. 8:1547. 10.3389/fimmu.2017.01547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan, M., Luoma A.M., Dougan S.K., and Wucherpfennig K.W.. 2021. Understanding and treating the inflammatory adverse events of cancer immunotherapy. Cell. 184:1575–1588. 10.1016/j.cell.2021.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan, M., and Pietropaolo M.. 2020. Time to dissect the autoimmune etiology of cancer antibody immunotherapy. J. Clin. Invest. 130:51–61. 10.1172/JCI131194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin, K., Callahan M.K., Ren B., Khanin R., Viale A., Ling L., No D., Gobourne A., Littmann E., Huttenhower C., et al. 2016. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 7:10391. 10.1038/ncomms10391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfahani, K., Elkrief A., Calabrese C., Lapointe R., Hudson M., Routy B., Miller W.H. Jr., and Calabrese L.. 2020. Moving towards personalized treatments of immune-related adverse events. Nat. Rev. Clin. Oncol. 17:504–515. 10.1038/s41571-020-0352-8 [DOI] [PubMed] [Google Scholar]
- Gabrilovich, D.I., and Nagaraj S.. 2009. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 9:162–174. 10.1038/nri2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan, V., Helmink B.A., Spencer C.N., Reuben A., and Wargo J.A.. 2018a. The Influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 33:570–580. 10.1016/j.ccell.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan, V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., et al. 2018b. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 359:97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover, S., Ruan A.B., Srivoleti P., Giobbie-Hurder A., Braschi-Amirfarzan M., Srivastava A., Buchbinder E.I., Ott P.A., Kehl K.L., Awad M.M., et al. 2020. Safety of immune checkpoint inhibitors in patients with pre-existing inflammatory bowel disease and microscopic colitis. JCO Oncol. Pract. 16:e933–e942. 10.1200/JOP.19.00672 [DOI] [PubMed] [Google Scholar]
- Gu, Z., Gu L., Eils R., Schlesner M., and Brors B.. 2014. Circlize implements and enhances circular visualization in R. Bioinformatics. 30:2811–2812. 10.1093/bioinformatics/btu393 [DOI] [PubMed] [Google Scholar]
- Gubin, M.M., Esaulova E., Ward J.P., Malkova O.N., Runci D., Wong P., Noguchi T., Arthur C.D., Meng W., Alspach E., et al. 2018. High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell. 175:1443. 10.1016/j.cell.2018.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailemichael, Y., Johnson D.H., Abdel-Wahab N., Foo W.C., Bentebibel S.E., Daher M., Haymaker C., Wani K., Saberian C., Ogata D., et al. 2022. Interleukin-6 blockade abrogates immunotherapy toxicity and promotes tumor immunity. Cancer Cell. 40:509–523.e6. 10.1016/j.ccell.2022.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann, S., Castelo R., and Guinney J.. 2013. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, M.D., Ciuleanu T.E., Pluzanski A., Lee J.S., Otterson G.A., Audigier-Valette C., Minenza E., Linardou H., Burgers S., Salman P., et al. 2018. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 378:2093–2104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng, T.S.P., Painter M.W., and Immunological Genome Project C.. 2008. The immunological genome project: Networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Hillmer, E.J., Zhang H., Li H.S., and Watowich S.S.. 2016. STAT3 signaling in immunity. Cytokine Growth Factor Rev. 31:1–15. 10.1016/j.cytogfr.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi, F.S., Chiarion-Sileni V., Gonzalez R., Grob J.J., Rutkowski P., Cowey C.L., Lao C.D., Schadendorf D., Wagstaff J., Dummer R., et al. 2018. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 19:1480–1492. 10.1016/S1470-2045(18)30700-9 [DOI] [PubMed] [Google Scholar]
- Hu, Z.I., Link V.M., Lima-Junior D.S., Delaleu J., Bouladoux N., Han S.J., Collins N., and Belkaid Y.. 2022. Immune checkpoint inhibitors unleash pathogenic immune responses against the microbiota. Proc. Natl. Acad. Sci. USA. 119:e2200348119. 10.1073/pnas.2200348119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D.B., Sullivan R.J., Ott P.A., Carlino M.S., Khushalani N.I., Ye F., Guminski A., Puzanov I., Lawrence D.P., Buchbinder E.I., et al. 2016. Ipilimumab therapy in patients with advanced melanoma and preexisting autoimmune disorders. JAMA Oncol. 2:234–240. 10.1001/jamaoncol.2015.4368 [DOI] [PubMed] [Google Scholar]
- Joosse, M.E., Nederlof I., Walker L.S.K., and Samsom J.N.. 2019. Tipping the balance: Inhibitory checkpoints in intestinal homeostasis. Mucosal Immunol. 12:21–35. 10.1038/s41385-018-0113-5 [DOI] [PubMed] [Google Scholar]
- Kim, M., Galan C., Hill A.A., Wu W.J., Fehlner-Peach H., Song H.W., Schady D., Bettini M.L., Simpson K.W., Longman R.S., et al. 2018. Critical role for the microbiota in CX3CR1(+) intestinal mononuclear phagocyte regulation of intestinal T Cell responses. Immunity. 49:151–163.e5. 10.1016/j.immuni.2018.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J., Chiarion-Sileni V., Gonzalez R., Grob J.J., Cowey C.L., Lao C.D., Schadendorf D., Dummer R., Smylie M., Rutkowski P., et al. 2015. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373:23–34. 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, D.R., Krummel M.F., and Allison J.P.. 1996. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 271:1734–1736. 10.1126/science.271.5256.1734 [DOI] [PubMed] [Google Scholar]
- Lim, S.Y., Lee J.H., Gide T.N., Menzies A.M., Guminski A., Carlino M.S., Breen E.J., Yang J.Y.H., Ghazanfar S., Kefford R.F., et al. 2019. Circulating cytokines predict immune-related toxicity in melanoma patients receiving anti-PD-1-based immunotherapy. Clin. Cancer Res. 25:1557–1563. 10.1158/1078-0432.CCR-18-2795 [DOI] [PubMed] [Google Scholar]
- Liu, C., Zha Z., Zhou C., Chen Y., Xia W., Wang Y.N., Lee H.H., Yin Y., Yan M., Chang C.W., et al. 2021. Ribonuclease 7-driven activation of ROS1 is a potential therapeutic target in hepatocellular carcinoma. J. Hepatol. 74:907–918. 10.1016/j.jhep.2020.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Zhou Y., Li M., Wang Y., Yang L., Yang S., Feng Y., Wang Y., Wang Y., Ren F., et al. 2019. Absence of GdX/UBL4A protects against inflammatory diseases by regulating NF-кB signaling in macrophages and dendritic cells.. Theranostics. 9:1369–1384. 10.7150/thno.32451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma, A.M., Suo S., Williams H.L., Sharova T., Sullivan K., Manos M., Bowling P., Hodi F.S., Rahma O., Sullivan R.J., et al. 2020. Molecular pathways of colon inflammation induced by cancer immunotherapy. Cell. 182:655–671.e22. 10.1016/j.cell.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch, J.A., Davar D., Rodrigues R.R., Badger J.H., Fang J.R., Cole A.M., Balaji A.K., Vetizou M., Prescott S.M., Fernandes M.R., et al. 2022. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat. Med. 28:545–556. 10.1038/s41591-022-01698-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis, C.S., Murrow L.M., and Gartner Z.J.. 2019. DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8:329–337.e4. 10.1016/j.cels.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo, J.A., Song L., Bhagat G., Blazquez A.B., Plumlee C.R., Lee C., Berin C., Reizis B., and Schindler C.. 2010. Dendritic cell (DC)-specific targeting reveals Stat3 as a negative regulator of DC function. J. Immunol. 184:2638–2645. 10.4049/jimmunol.0902960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, M.A., Baer J.M., Knolhoff B.L., Nywening T.M., Panni R.Z., Su X., Weilbaecher K.N., Hawkins W.G., Ma C., Fields R.C., et al. 2018. Breast and pancreatic cancer interrupt IRF8-dependent dendritic cell development to overcome immune surveillance. Nat. Commun. 9:1250. 10.1038/s41467-018-03600-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirantes, C., Passegue E., and Pietras E.M.. 2014. Pro-inflammatory cytokines: Emerging players regulating HSC function in normal and diseased hematopoiesis. Exp. Cell Res. 329:248–254. 10.1016/j.yexcr.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Mantilla, D.E., and King K.Y.. 2018. The role of interferon-gamma in hematopoietic stem cell development, homeostasis, and disease. Curr. Stem Cell Rep. 4:264–271. 10.1007/s40778-018-0139-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer, R.J., Tannir N.M., McDermott D.F., Aren Frontera O., Melichar B., Choueiri T.K., Plimack E.R., Barthelemy P., Porta C., George S., et al. 2018. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med. 378:1277–1290. 10.1056/NEJMoa1712126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, D.Y., Cham J., Zhang L., Fong G., Kwek S.S., Klinger M., Faham M., and Fong L.. 2017. Immune Toxicities Elicted by CTLA-4 blockade in cancer patients Are associated with early diversification of the T-cell repertoire. Cancer Res. 77:1322–1330. 10.1158/0008-5472.CAN-16-2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos, A.D., and Watowich S.S.. 2008. Granulocyte colony-stimulating factor: Molecular mechanisms of action during steady state and “emergency” hematopoiesis. Cytokine. 42:277–288. 10.1016/j.cyto.2008.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedicord, V.A., Montalvo W., Leiner I.M., and Allison J.P.. 2011. Single dose of anti-CTLA-4 enhances CD8+ T-cell memory formation, function, and maintenance. Proc. Natl. Acad. Sci. USA. 108:266–271. 10.1073/pnas.1016791108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow, M.A., Sidlow R., and Hellmann M.D.. 2018. Immune-related adverse events associated with immune checkpoint blockade. N. Engl. J. Med. 378:158–168. 10.1056/NEJMra1703481 [DOI] [PubMed] [Google Scholar]
- Quast, C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., and Glockner F.O.. 2013. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski, J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B., and Forrest A.R.R.. 2016. Corrigendum: A draft network of ligand-receptor-mediated multicellular signalling in human.. Nat. Commun. 7:10706. 10.1038/ncomms10706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy, B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillere R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., et al. 2018. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 359:91–97. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- Skelly, D.A., Squiers G.T., McLellan M.A., Bolisetty M.T., Robson P., Rosenthal N.A., and Pinto A.R.. 2018. Single-Cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 22:600–610. 10.1016/j.celrep.2017.12.072 [DOI] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. 3rd, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, K., Kaisho T., Yoshida N., Takeda J., Kishimoto T., and Akira S.. 1998. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: Generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol. 161:4652–4660 [PubMed] [Google Scholar]
- Uemura, M., Trinh V.A., Haymaker C., Jackson N., Kim D.W., Allison J.P., Sharma P., Vence L., Bernatchez C., Hwu P., and Diab A.. 2016. Selective inhibition of autoimmune exacerbation while preserving the anti-tumor clinical benefit using IL-6 blockade in a patient with advanced melanoma and Crohn's disease: A case report. J. Hematol. Oncol. 9:81. 10.1186/s13045-016-0309-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vétizou, M., Pitt J.M., Daillere R., Lepage P., Waldschmitt N., Flament C., Rusakiewicz S., Routy B., Roberti M.P., Duong C.P.M., et al. 2015. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 350:1079–1084. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, F., Zhang H.H., Matthews V., Weinstock J., Nice E.C., Ernst M., Rose-John S., and Burgess A.W.. 2008. IL6/sIL6R complex contributes to emergency granulopoietic responses in G-CSF- and GM-CSF-deficient mice. Blood. 111:3978–3985. 10.1182/blood-2007-10-119636 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Wiesnoski D.H., Helmink B.A., Gopalakrishnan V., Choi K., DuPont H.L., Jiang Z.D., Abu-Sbeih H., Sanchez C.A., Chang C.C., et al. 2018. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 24:1804–1808. 10.1038/s41591-018-0238-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S.C., Anang N.A.A.S., Sharma R., Andrews M.C., Reuben A., Levine J.H., Cogdill A.P., Mancuso J.J., Wargo J.A., Pe'er D., and Allison J.P.. 2019. Combination anti-CTLA-4 plus anti-PD-1 checkpoint blockade utilizes cellular mechanisms partially distinct from monotherapies. Proc. Natl. Acad. Sci. USA. 116:22699–22709. 10.1073/pnas.1821218116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S.C., Duffy C.R., and Allison J.P.. 2018. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8:1069–1086. 10.1158/2159-8290.CD-18-0367 [DOI] [PubMed] [Google Scholar]
- Wei, S.C., Levine J.H., Cogdill A.P., Zhao Y., Anang N.A.A.S., Andrews M.C., Sharma P., Wang J., Wargo J.A., Pe'er D., and Allison J.P.. 2017. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell. 170:1120–1133.e17. 10.1016/j.cell.2017.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Hu H., Greeley N., Jin J., Matthews A.J., Ohashi E., Caetano M.S., Li H.S., Wu X., Mandal P.K., et al. 2014. STAT3 restrains RANK- and TLR4-mediated signalling by suppressing expression of the E2 ubiquitin-conjugating enzyme Ubc13. Nat. Commun. 5:5798. 10.1038/ncomms6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Slone N., Chrisikos T.T., Kyrysyuk O., Babcock R.L., Medik Y.B., Li H.S., Kleinerman E.S., and Watowich S.S.. 2020. Vaccine efficacy against primary and metastatic cancer with in vitro-generated CD103(+) conventional dendritic cells. J. Immunother. Cancer. 8:e000474. 10.1136/jitc-2019-000474 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the scoring system for colonic inflammation.
shows differential gene expression of clusters.
shows ligand–receptor pairs across clusters.
shows the patient cohort for NanoString.
shows normalized gene expression of NanoString samples.
shows the patient cohort for IHC.
is the flow cytometry antibody list.
contains original blots for Fig. 3.
Data Availability Statement
The scRNAseq data (Fig. 2, A–G) are available at the Gene Expression Omnibus under accession code GSE159892. 16s RNA gene profiling data (Fig. 4, A–C; and Fig. 5, I–K) are available at NCBI BioProject under project code PRJNA670730.
The code generated for this current study is available from the corresponding author upon request. The code is stored at https://github.com/chansigit/irae_reproduction.