ABSTRACT
To date, several studies have reported an alarming increase in pathogen resistance to current antibiotic therapies and treatments. Therefore, the search for effective alternatives to counter their spread and the onset of infections is becoming increasingly important. In this regard, microorganisms of the former Lactobacillus genus have demonstrated the ability to reduce the virulence of pathogens. In addition to the production of bioactive substances, self- and coaggregation, and substrate competition, lactobacilli influence gene expression by downregulating genes associated with the virulence of pathogens. As demonstrated in many in vivo and in vitro trials, lactobacilli counteract and inhibit various virulence factors that favor pathogens, including the production of toxins, biofilm formation, host cell adhesion and invasion, and downregulation of virulence genes linked to quorum sensing. The aim of this review is to summarize current studies on the inhibition of pathogen virulence by lactobacilli, an important microbial group well known in the industrial and medical fields for their technological and probiotic properties that benefit human hosts with the potential to provide an important aid in the fight against pathogens besides use of the current therapies. Further research could lead to the identification of new strains that, in addition to alleviating adverse effects, could improve the efficacy of antibiotic therapies or play an important preventive role by reducing the onset of pathogen infections if regularly taken.
KEYWORDS: lactobacilli, virulence, probiotics, pathogen suppression
INTRODUCTION
Lactobacilli, the term used in this work to refer to the former Lactobacillus genus (1), are lactic acid bacteria with fundamental roles in modern society and economies and are essential in the production and conservation of many food and feed products. Owing to their long history of safe use and their fermentative and bioprotective abilities, which ensure the quality and safety of products, they have received the designations of generally recognized as safe by the Food and Drug Administration and qualified presumption of safety by the European Food Safety Authority (EFSA) (2, 3). Due to their properties, several strains of this group have been identified as probiotics, defined by FAO and WHO as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (4, 5), and their inactivated cells or their cell-free supernatants (CFS) hosting numerous beneficial components are also considered postbiotics, defined as “preparation of inanimate microorganisms and/or their components that confers a health benefit on the host” (6). They are also part of the human natural bacterial flora, in which they have a regulatory role in protecting hosts against colonization bypathogens and exert beneficial effects, such as increasing and improving nutrient assimilation during digestion or stimulating host tissues (7). Prolonged consumption of these bacteria leads to modification of the human gastrointestinal microbial flora, thus stimulating the immune system and decreasing pathogen adhesion (8). Owing to the interconnection between the gastrointestinal tract and the central nervous system, known as the gut-brain axis, these effects also arise from the production of signaling molecules with brain modulation abilities (9, 10). Lactobacilli are also effective in the prevention and treatment of gastrointestinal and urogenital tract diseases because of their antimicrobial properties (11, 12) and confer numerous beneficial effects, such as alleviating lactose intolerance, reducing blood cholesterol and incidence and progression of cancer, stimulating immunity, and preventing and treating diarrheal diseases, stomach ulcers, and infectious diseases (13, 14). Furthermore, lactobacilli inhibit pathogen growth through nutrient subtraction, competition for substrate, and the production of molecules such as bacteriocins, enzymes, organic acids, and hydrogen peroxide (15). Other important mechanisms include the ability to self-aggregate and coaggregate, which allow lactobacilli to adhere to each other or other microbial species. These adhesive properties provide lactobacilli with the ability to adhere to the mucosa, thereby limiting pathogen adhesion and creating a microenvironment in which their strict proximity allows the increase of inhibitory effects of the secreted substances (16).
In addition to these well-known properties, lactobacilli inhibit various virulence genes encoding transacting proteins associated with infective mechanisms, which are fundamental in bacterial virulence, as reviewed in Table 1. Among these mechanisms, one of the most important is the quorum sensing (QS) system, which leads to the production of different chemical molecules, named autoinducers, which alter gene expression. Through these signal-response systems, different bacteria coordinate their behaviors on a population scale, acting as multicellular organisms (17). QS systems regulate many microbial pathways, including biofilm formation, sporulation, antibiotic synthesis, induction of virulence factors, host infection, and bacteriocin synthesis. Autoinducer 2 (AI-2), produced by the LuxS enzyme (luxS gene), is of particular interest because it is associated with the expression of genes involved in pathogen motility, adhesion, and internalization. AI-2 also plays a fundamental role in biofilm formation, a common feature among pathogenic species that increases their adhesion to surfaces, provides them with nutrients, and confers resistance to external factors, thus making bacteria more virulent and resistant to antibiotic treatments (18–20). Moreover, antiviral activity, a property of particular interest in medical applications, has been observed in specific strains of lactobacilli and might be used to prevent viral adhesion and propagation (21).
TABLE 1.
Summary of virulence genes affected by lactobacilli
| Bacteria | Gene | Protein | Function | Reference |
|---|---|---|---|---|
| Listeria monocytogenes | fbp | Fibronectin-binding protein | Adhesion to epithelial cells | 114 |
| flaA | Flagellin | Motility | 28 | |
| hly | Hemolysin listeriolysin O (LLO) | Survival inside macrophages | 27 | |
| iap | Invasion-associated protein | Invasion of epithelial cells | 114 | |
| plcA, plcB | PlcA and PlcB phospholipases | Survival inside macrophages | 27 | |
| prfA | Transcriptional activator of hly and plc genes | 29 | ||
| sigB | Stress response regulon | 31 | ||
| Autolysin amidase (Ami) | Bacterial adhesion on enterocytes | 29 | ||
| Actin-polymerizing protein (ActA) | Required for actin polymerization allowing intracytoplasmic movement | 30 | ||
| Internalin A (InlA) Internalin B (IlnB) | Adhesion and internalization inside enterocytes | 27 | ||
| Listeria adhesion protein (LAP) | Bacterial adhesion on enterocytes | 29 | ||
| Salmonella spp. | avrA | AvrA | Inhibition of innate immunity | 56 |
| hilA | HilA | Regulation of Salmonella pathogenicity island 1 gene expression | 48 | |
| hilC hilD | HilC, HilD | Transcriptional regulators of hilA | 48 | |
| invH | Outer membrane lipoprotein InvH | Facilitates the translocation of proteins, including SipC, from the cytoplasm to the membrane | 53 | |
| nmp | Outer membrane-associated protein | Bacterial porin formation | 114 | |
| prgK | PrgK periplasmatic protein | Type III secretion system | 49 | |
| sip | Sip effector protein | Induction of inflammation response | 52 | |
| sop | Salmonella outer Protein B | Lipid phosphatase critical in enteropathogenicity | 50 | |
| sptP | SptP effector protein | Recovery of the host cytoskeleton after the infection | 55 | |
| spv | Promoter of the virulence genes of nontyphoid Salmonella serovars | 51 | ||
| ssrB | SsrB | Activation of genes needed for intracellular survival | 57 | |
| Campylobacter jejuni | cadF | Outer membrane protein CadF | Adhesion to intestinal epithelial cells | 76 |
| cdt | Cytolethal distending toxin | Toxin composed by three subunits, involved in cell adhesion and inhibition of cell division | 76 | |
| cia | Campylobacter invasion antigen B | Invasion potential | 76 | |
| fla | Flagellin | Motility and colonization | 76 | |
| flh | Flagellin | Motility and colonization | 76 | |
| luxS | LuxS enzyme | Production of autoinducer 2 (AI-2) | 79 | |
| Escherichia coli | eaeA | Intimin | Attachment to cell surface | 86 |
| fliC | Flagellin | Motility | 96 | |
| hly | Enterohemolysin and α-hemolysin | Toxins with hemolytic activity | 87 | |
| ler | LEE1-encoded regulator | Transcriptional activator of LEE genes | 94 | |
| luxS | LuxS enzyme | Production of autoinducer 2 (AI-2) | 97, 98 | |
| qseA | QseA effector protein | LEE1 gene activator | 95 | |
| stx | Shiga-like toxin Stx | Toxin causing diarrhea and other disorders | 89 | |
| tir | Translocated intimin protein | Adhesion to epithelial cells | 93 | |
| Adhesins | Adhesion on both abiotic and cell surfaces | 91 | ||
| Intimin receptor EspE | Type III secretion system that allows attaching and effacing (A/E) lesions | 92 | ||
| Clostridium spp. | luxS | LuxS enzyme | Production of autoinducer 2 (AI-2) | 125 |
| tcdA | Enterotoxin A | Toxin that causes diarrhea and intestinal damage | 119, 120 | |
| tcdB | Toxin B | Toxin with strong cytotoxic effect | 119, 120 | |
| txeR | σ factor | Induces RNA polymerase to recognize the promoters of tdc genes | 121 | |
| Staphylococcus aureus | agr | QS system that regulates virulence factors | 130 | |
| ica | Biofilm formation | 137 | ||
| mecA | Methicillin resistance | 136 | ||
| sae | Regulatory locus that activates the production of different exoproteins | 131 | ||
| sbi | Ig-binding protein | Binding to IgG and blood coagulation | 135 | |
| sea | Enterotoxin A | Food poisoning | 132 | |
| spa | Protein A | Inhibition of phagocytosis | 135 | |
| ssl1 | Staphylococcus superantigen-like protein (SSL-1) | Inhibition of metalloproteases | 134 | |
| tst | Toxic shock syndrome toxin 1 (TSST-1) | Superantigen that causes organ dysfunctions associated with high mortality rate | 133 | |
| Helicobacter spp. | cagA | CagA cytotoxin | Alteration of intracellular signal transduction | 148 |
| fla | Flagellin | Motility | 149 | |
| vacA | VacA cytotoxin | Fusion between endosomes and lysosomes in eukaryotic cells | 148 | |
| Pseudomonas spp. | exo | Cytotoxins belonging to the type III effector proteins family | Toxins that cause different damage to the host | 157 |
| fleSR | Flagellin | Flagella necessary for swimming/swarming motility | 158 | |
| lasI/R | LasI/R protein | QS system that regulates virulence factors | 162 | |
| ndvB | Biofilm formation | 157 | ||
| pil | Pilin | Type IV pili necessary for twitching motility | 158 | |
| rhI/R | RhI/R protein | QS system that regulates virulence factors | 162 | |
| Klebsiella pneumoniae | sugE | Biofilm formation | 163 | |
| treC | 163 | |||
| Streptococcus spp. | ftf | Fructosyltransferase | Adhesion | 168 |
| gtf | Glucotransferase | Production of exopolysaccharides | 167 | |
| luxS | LuxS enzyme | Production of autoinducer 2 (AI-2) | 171 | |
| sag | Streptolysin S | Toxin that causes erythrocyte lysis | 177 | |
| tft | Glucosyltransferase (GTF) | Production of exopolysaccharides | 167 | |
| Neisseria gonorrhoeae | Major outer protein porin PorB | Suppression of neutrophil oxidative burst and apoptosis | 187 | |
| N. gonorrhoeae lipooligosaccharide (LOS) | Adhesion and invasion of the host cells | 187 | ||
| Opacity proteins (Opa) | Colonization of the mucosal epithelium | 187 | ||
| Pilin | Type IV pili for twitching motility, immune evasion, and colonization | 187 | ||
| Trichomonas vaginalis | Lipophosphoglycan | Adherence factor | 186 | |
| Gardnerella vaginalis | sld | Sialidase | Adhesion to cells and surfaces | 188 |
| vly | Vaginolysin | Inhibition of immune response | ||
| Candida albicans | ALS3 | Adhesins | Adhesion properties | 195 |
| BCR1 | Biofilm formation | 195 | ||
| CPH1 | Biofilm formation | 195 | ||
| ECE1 | Yeast-to-hyphal morphogenesis | 196 | ||
| EFG1 | Biofilm formation | 195 | ||
| HWP1 | Adhesins | Adhesion properties | 195 | |
| MspI | Major peptidoglycan hydrolase | Chitin hydrolysis | 204 | |
| Saps | Hydrolytic enzymes | 196 | ||
| TEC1 | Biofilm formation | 195 | ||
| CDR1, CDR2, and MDR1 proteins | Resistance to drugs and immune system | 195 | ||
| Aggregatibacter actinomycetemcomitans | ltxA | Leukotoxins | Induces the death of leukocytes | 220 |
| cdtB | Cytolethal distending toxin | Diarrheal disease-causing toxin |
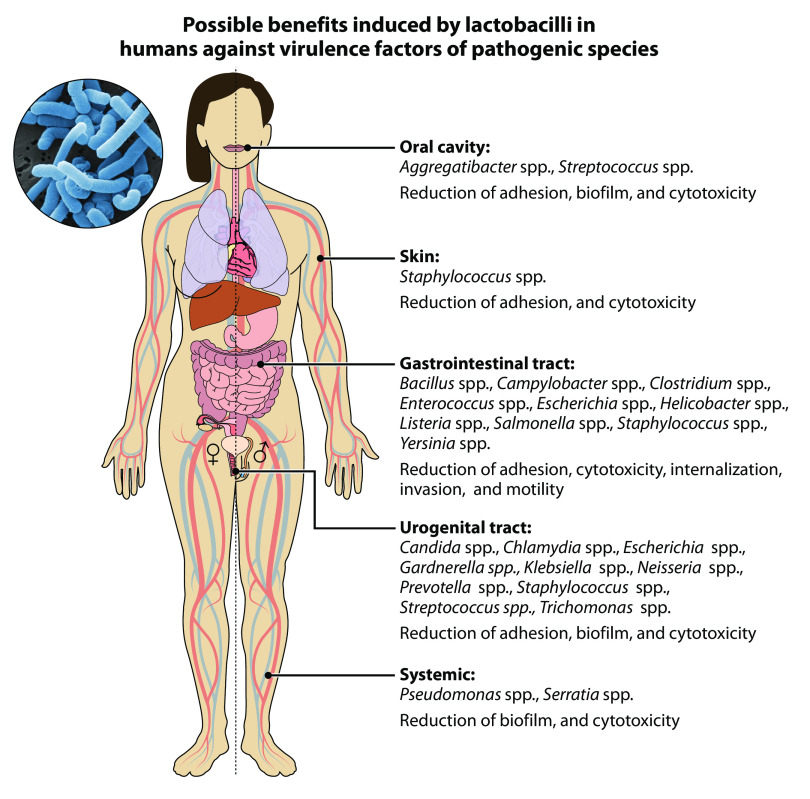
Pathogenic bacteria are an important threat to human health, as they represent 4 of the top 10 causes of death worldwide (22). Currently, infections are treated mainly with antibiotics, whose discovery dates to the first half of the 20th century. However, the extensive and prolonged use of these substances has led to a natural evolutionary phenomenon of adaptation that has contributed to the spread of antibiotic resistance (23). Consequently, infections have become more difficult because antibiotics have become less effective in counteracting pathogens, thus enabling their survival and even replication in the presence of therapeutic levels of drugs. If no action is taken, multidrug-resistant pathogens have been expected to cause 10 million deaths by the year 2050. Therefore, identifying new effective methods will be critical to counteract the spread of pathogens and simultaneously decrease the use of antibiotics (24) in medical and zootechnical fields (25). The present review summarizes available data from original studies reporting the effectiveness of lactobacilli in counteracting the virulence of pathogenic species such as Aggregatibacter actinomycetemcomitans, Bacillus cereus, Campylobacter jejuni (Cj), Candida albicans, Chlamydia trachomatis, Clostridium spp., Enterococcus faecalis, Escherichia coli (Ec), Gardnerella vaginalis, Helicobacter spp., Klebsiella spp., Listeria monocytogenes (Lm), Neisseria gonorrhoeae, Pseudomonas spp., Prevotella bivia, Salmonella spp., Serratia marcescens, Staphylococcus aureus (Sa), Streptococcus spp., Trichomonas vaginalis, and Yersinia enterocolitica, as summarized in Fig. 1.
FIG 1.
Possible benefits induced by lactobacilli in humans against virulence factors of pathogenic species.
LISTERIA MONOCYTOGENES
Listeria monocytogenes (Lm) is the etiological agent of listeriosis, a severe foodborne disease with a low incidence rate but a high mortality rate that poses a serious public health concern (26). Internalization of this pathogen occurs via invasion of macrophages and nonphagocytic cells, a capability conferred by the internalin proteins InlA and InlB, while the production of hemolysin listeriolysin O (LLO) and PlcA and PlcB phospholipases, encoded by the hly and plc genes, respectively, enables macrophage survival (27). The presence of Listeria adhesion protein (LAP) and autolysin amidase Ami, which enhance bacterial adhesion, prfA transcriptional activator, ActA actin polymerization protein, sigB stress response factor, and flagellin, encoded by flaA gene, all contribute to Lm virulence (28–31). Several studies have reported the reduction of all of these virulence factors (Table S1 in the supplemental material). In vitro trials have revealed that lactobacilli, through the production of organic acids and proteinaceous molecules and their interaction with mucosal epithelial cells, significantly decreased inflammation during the invasion of Lm (32). Coculture with Lactiplantibacillus plantarum significantly decreased Lm virulence toward HT-29 cells (33). On Caco-2 cells, Lpb. plantarum and Lacticaseibacillus rhamnosus coinoculation significantly reduced the Lm survival ratio under simulated digestion, thus inhibiting cell adhesion and invasion and downregulating the sigB, hly, inlA, inlB, and prfA genes (34, 35). This property was also observed for Limosilactobacillus reuteri, Limosilactobacillus fermentum, and Lpb. plantarum with lower LLO production, epithelial E-cadherin-binding ability, and expression of virulence genes, while in an in vivo trial, these strains increased survival of Galleria mellonella inoculated with lethal doses of Lm (36). In addition, preexposure to bioengineered Lacticaseibacillus casei and Lacticaseibacillus paracasei preserved tight barrier junction integrity and decreased Lm-mediated cytotoxicity and adhesion, whereas these effects were not observed on Lm already attached to Caco-2 cells (37, 38). Other in vivo studies confirmed the antilisterial activities of lactobacilli. In murine models, the administration of Lcb. paracasei and Lcb. casei systematically decreased the dissemination of Lm (39), whereas Latilactobacillus sakei 2a lowered lesions and edema of the intestinal villi (40). Levilactobacillus brevis reduced the propagation and dispersion of Lm in the intestines, spleen, and liver without affecting neutrophils and lymphocyte values (41). In infected chickens, supplementation with Lactobacillus acidophilus and Lpb. plantarum attenuated Lm adhesion, pore formation, and invasion, downregulating the expression of LLO, InlA, InlB, Ami, and flagellin. Moreover, a decreased load of Lm in the cecum, skin, liver, and spleen, a decrease in serum cytokines, and an upregulation of anti-inflammatory-related genes were observed (42). In addition, Lm cocultured with bacteriocin-producing Llb. sakei 1 resulted in diminished hemolytic activity (43, 44), thus indicating the effectiveness of lactobacilli in preventing Lm adhesion to abiotic surfaces (45, 46).
SALMONELLA SPP.
Salmonella enterica (Slm) is a pathogen that affects both humans and animals. Septicemia and enteric fever are common clinical manifestations of serovars Typhi and Paratyphi, whereas bacteremia is typical of nontyphoidal Salmonellae, such as S. enterica serovar Typhimurium (SlmT), Enteritidis (SlmE), Heidelberg (SlmH), and Javiana (SlmJ) (47). Salmonella pathogenicity islands (SPI) group hilA, hilC, and hilD invasion genes (48) and prgK, which are associated with type III secretion system 1 (T3SS1) and T3SS2 systems (49), as well as sop genes, which are important in enteropathogenesis (50). The virulence traits of nontyphoid Salmonella serovars are also enhanced by the spv plasmidic gene (51). The invH gene promotes tissue invasion both in vivo and in vitro and is related to the expression of the sip gene, which is involved in host translocation (52, 53). During infection, Slm invades macrophages and dendritic and epithelial cells (54), thus promoting survival and replication thanks to avrA, sptP, and ssrB genes (48, 55–57). Several studies have demonstrated that lactobacilli and their metabolites downregulate genes associated with Slm virulence (Table S2 in the supplemental material). Lactobacillus bulgaricus, Lcb. paracasei, and Lcb. rhanosus, for example, downregulate the sipA, sipB, sopB, spvB, hilA, hilD, and invH genes in SlmE, SlmT, and SlmH (50), whereas hilA and hilD along with hilC and sipC are also downregulated by other probiotic lactobacilli (58). In SlmT-infected chickens administered lactobacilli, almost all SPI virulence genes (hilA, hilC, hilD, sopB, sopD, sopE2, sipA, avrA, and sptP, but not sipC) were downregulated, thus decreasing infection in the liver and spleen (59, 60). In addition, Lbc. acidophilus and Lpb. plantarum reduced the expression of the invA, avrA, hilA, ssrB, and sopD genes and the invasiveness of SlmT, thus altering the function of the type III secretion system (61, 62). A Lbc. acidophilus strain was also able to delay the internalization of SlmT, also altering its swimming motility (63). Other lactobacilli and their metabolites showed substantial antivirulence properties toward Slm in in vivo studies; for example, different Lpb. plantarum strains interfered with the growth and virulence of SlmT on Vero cells. These lactobacilli, which had higher ciprofloxacin resistance than the pathogen, significantly reduced its adherence, invasion, and cytotoxicity (64). Preexposure of HT29 cells to live Lbc. acidophilus, Lcb. rhanosus, and Lcb. casei decreased the induced cytotoxicity and the expression of virulence genes, particularly those related to the invasiveness of SlmJ (65). Also, on thermally stressed Caco-2 cells, Lcb. rhanosus reduced the severity of Slm infection (66). The adhesion of SlmT to the same cell line was inhibited by molecules secreted by lactobacilli, in particular lactic acid produced from Lcb. casei Shirota, Lbc. acidophilus, Lcb. rhanosus, and Lbc. amylovorus, whereas Lactobacillus johnsonii and Lpb. plantarum produced unknown inhibitory substances with anti-Salmonella activity (67). A bioengineered Lcb. casei strain overproducing conjugated linoleic acids (CLA) competitively excluded SlmT in a mixed culture and altered biofilm formation, adherence, and invasive activity toward INT-407 host cells, thus downregulating expression of the invG, invH, prgK, hilA, hilC, hilD, and invF genes (68, 69).
Live lactobacilli cells and their CFSs show antivirulence effects against Slm. Lcb. paracasei CFS lowered SlmE adhesion to Caco-2 cells (70), whereas the CFS produced by Lbc. acidophilus induced the release of lipopolysaccharide in SlmT, a decrease in intracellular ATP correlated with bacterial death, bacterial membrane permeabilization, and increased sensitivity to sodium dodecyl sulfate (71). In a trial evaluating the expression of the SlmE hilA-lacZY transcriptional fusion, 24 h of incubation with spent medium from a Lactobacillus species strain isolated from poultry resulted in an absence of β-galactosidase activity. In comparison, SlmE, grown in Slm-spent medium, showed a 4-fold higher expression of hilA (72). Other properties of lactobacilli have been demonstrated in vivo. Lcb. casei inhibited the invasion and decreased the survival of SlmT in Caco-2 cells and mice, thus lowering the cecal colonization levels and the bacterial translocation rate to the spleen, liver, and mesenteric lymph nodes. In addition, administration of Lcb. casei to infected mice significantly delayed the occurrence of 100% animal mortality from 9 to 15 days (73). Pretreatment with washed cells and CFS of Ligilactobacillus salivarius, Lactobacillus delbrueckii subsp. delbrueckii, and Lpb. plantarum inhibited SlmT attachment to the cecal mucus of infected chickens (74). The immune system modulation ability of lactobacilli was observed in Slm-infected mice, in which Lactobacillus zeae, Lpb. plantarum, and Lmb. reuteri increased the proinflammatory cytokine response. This induced response was more effective with a combination of lactobacilli isolates than with a single strain (75).
CAMPYLOBACTER JEJUNI
Campylobacter jejuni (Cj) is a commensal microorganism that is found in both domestic and wild animals and is responsible for campylobacteriosis, a severe foodborne diarrheal disease. Its virulence and survival in humans are linked to a variety of factors, including flagellum motility conferred by fla and flh genes, adhesion capacity conferred by cia and cadF genes, and cytolethal distending toxin encoded by cdtA, cdtB, and cdtC genes, interfering with cell division (76). Lactobacilli, already recognized for their ability to relieve gastrointestinal symptoms caused by pathogenic infections, have been found to decrease Cj invasiveness (Table S3 in the supplemental material) (77). In vitro experiments revealed that the prolonged colonization of E12 cells with different lactobacilli attenuated Cj association, internalization, and translocation to the basolateral medium in transwells (78). On Caco-2 cells, various lactobacilli exhibited antagonistic effects against this pathogen, lowering the expression of genes involved in invasion (ciaB), motility (flaA, flaB, and flhA), and AI-2 production (luxS). These strains increased Cj macrophage phagocytosis and the expression of interferon-γ (IFN-γ), interleukin-1β (IL-1β), IL-12p40, IL-10, and chemokines in macrophages (79). Similarly, the CFS of a genetically engineered Lcb. casei overexpressing the mcrA gene decreased Cj adhesion to, and invasion of, HD-11 and HeLa cells and altered the expression of cadF, cdtB, ciaB, and flaB genes (80). The expression of ciaB and flaA virulence genes in C. jejuni was downregulated by Lbc. acidophilus CFS, according to real-time PCR (RT-PCR) analysis. The effect of the same strain has been tested on luxS-mutant Cj and downregulated only the ciaB gene, thereby suggesting an active role of luxS in the modulation of Cj virulence even when lactobacilli strains were added (81).
ESCHERICHIA COLI
Although Escherichia coli (Ec) is commonly part of the commensal intestinal microbiota in both human and animal intestines, some opportunistic strains transmitted via the fecal-oral route can cause disease in humans. Pathogenic Ec can be classified as extraintestinal or diarrhoeagenic and can be further subdivided into different pathovars: enteropathogenic (EPEC), enterohemorrhagic (EHEC), enterotoxigenic (ETEC), enteroinvasive (EIEC), enteroaggregative (EAEC), Shiga toxin-producing (STEC), adherent invasive (AIEC), and diffusively adherent (DAEC) (82, 83). Whereas EIEC is an intracellular pathogen that invades and replicates within epithelial cells and macrophages, other pathogenic Ec strains interact with the epithelium through the expression of specific genes such as the eaeA gene, which regulates attachment to intestinal cells (84–86). An important virulence factor is the production of toxins, such as cell-associated enterohemolysin and α-hemolysin, encoded by hlyA, hlyB, hlyC, and hlyD genes in STEC (87). ETEC and EHEC are the main causes of enteric diseases in humans each year (88) owing to the ability of EHEC to produce verotoxin and Shiga-like toxins (Stx1 and Stx2) (89) and the ability of ETEC to produce toxins and adhesins (90, 91). EHEC has a pathogenicity island called locus of enterocyte effacement (LEE), which encodes gene regulators, adhesin, the type III secretion system, and proteins, including the translocated intimin receptor (tir) and Esp proteins that enhance adhesion to epithelial cells (92, 93). LEE1-encoded regulator (ler) activity is controlled by QS autoinducer 3 (AI-3) and by epinephrine and norepinephrine hormones (94), whereas the qseA gene encodes the QseA effector protein, which directly activates the LEE1 gene (95). EHEC is further characterized by the presence of a flagellum encoded by the fliC gene (96). Different lactobacilli and their metabolites alter the gene expression and consequently the virulence of Ec (Table S4 in the supplemental material). For example, Lmb. reuteri downregulated the epinephrine-mediated induction of ler in EHEC (94). CFS from Lbc. acidophilus supplementation in yogurt reduced the severity of infection and the attachment and colonization of EHEC and downregulated tumor necrosis factor-α (TNF-α) in infected mice. These effects were supported by RT-PCR, which detected a decrease in the expression of the stxB2, qseA, luxS, tir, ler, eaeA, and hlyB genes (97). Another study found that CFS of the same strain reduced extracellular AI-2 concentrations and downregulated other virulence-associated genes (tir, espA, fliC, espD, luxS, eaeA, ler, hylB, and qseA), but no modification in Shiga toxin production has been observed (98). CFS and lactic acid produced by Lmb. reuteri significantly inhibited uropathogenic Ec (UPEC), thus reducing the production of virulence factors involved in the adhesion process, such as adhesion outer membrane proteins A and X, urogenital tract adherence promoter factor type 1, and P fimbriae subunits (99). Furthermore, studies conducted on different cell lines have confirmed the anti-Ec activity of several Lactobacillus strains. The adhesion ability of two Ec strains on Hep-2 and T84 cells was reduced after pretreatment with Lbc. acidophilus and Lcb. rhanosus (100). Whereas Lbc. jensenii and Lbc. gasseri inhibited adhesion of DAEC to HeLa cells, Lmb. reuteri also reduced Ec internalization in the same cell line (101). Also, Lpb. plantarum and Lcb. rhanosus inhibited Ec adherence to HT-29 cells by increasing the expression of intestinal mucins MUC2 and MUC3 (102). Also, an interference of induced cell signaling against DAEC caused by Lbc. acidophilus abolished the structural and functional microvilli alteration in human enterocyte-like cells (103, 104). As also reported for Slm, CLA overproducer Lcb. casei strain altered biofilm formation and modified Ec adhesion and invasion in INT407 cells (68). The combination of Lcb. rhanosus with oligosaccharides resulted in an effective antidiarrheal formulation, owing to the increased autoaggregation and coaggregation properties of this strain. The inhibition of adherence to HT-29 cells was maximal with a Lcb. rhanosus and inulin combination and significantly decreased the production of cyclic AMP, cyclic GMP, and related toxins (105). In an in vitro EHEC infection model, Lcb. rhanosus, Lbc. gasseri, Lcb. casei, and Lpb. plantarum have been studied on C2BBe1 human colon epithelial cells. Among the tested strains, live Lcb. rhanosus cells significantly reduced pathogen internalization, whereas this effect has not been observed with dead Lcb. rhanosus cells or conditioned medium, thus implying that lactobacilli modulate the intracellular mechanism responsible for EHEC internalization (106). Multiple lactobacilli were also effective in inhibiting the Ec quorum sensing system, such as Llb. sakei and Lbc. acidophilus cell extract, which significantly inhibited AI-2-like activity without affecting EHEC growth. Moreover, Lbc. acidophilus cell extracts inhibited biofilm formation on abiotic surfaces and HT-29 cell adhesion and downregulated the expression of several virulence factors associated with AI-2-like activity, particularly proteins involved in sulfur metabolism and membrane-associated functions (107, 108). In vivo experiments have shown similar results, including a significant decrease in adhesion and improvements in the immune system of infected animals. In a murine model, Lactobacillus kefiranofaciens treatment prevented EHEC infection-induced symptoms, Shiga toxin penetration, bacterial translocation, renal and intestinal damage, and increased mucosal EHEC-specific IgA responses. Lactobacilli also had protective effects in Caco-2 cells, reducing cell death and epithelial integrity loss induced by the pathogen (109). The ability of Ec to adhere to pig intestine brush borders decreased in a dose-dependent manner after administration of recombinant engineered fimbriae-producing Lbc. acidophilus (110). In an in vivo trial, the ability of Ec to disrupt the intestinal barrier and increase permeability was significantly reduced by administering Lpb. plantarum to rats, indicating a beneficial effect on the intestinal tract (111). Lcb. casei Shirota treatment of Ec in a murine urinary tract infection model inhibited growth and reduced inflammatory responses (112). In addition, exopolysaccharides produced during fermentation demonstrated in vivo anti-Ec activity, as reuterin and levan from Lmb. reuteri contained in weanling pig feed that reduced the number of Ec and the amount of heat-stable enterotoxin in colonic digesta (113). In addition, Lcb. casei strains decreased virulence gene expression in EHEC, SlmT, and Lm, particularly downregulating the Ec eaeA, SlmT nmpC, and Lm fbp and iap genes (114). Also, pretreatment of Caco-2 cells with live and heat-killed Lbc. acidophilus dose-dependently inhibited the adhesion and invasive properties of EPEC, Lm, SlmT, and Yersinia pseudotuberculosis (115, 116). Another study investigating the effect of pretreatment of Caco-2 and HT-29 cells with lactobacilli reported that one Lvb. brevis, two Lpb. plantarum, and two Lcb. paracasei strains inhibited EPEC and SlmE adhesion to both cell lines (117).
CLOSTRIDIUM SPP.
Hospital-acquired infections have severe consequences for already debilitated patients, and several studies have shown the effectiveness of lactobacilli in preventing the onset of such complications, as in the case of Clostridium difficile (Cd). This nosocomial bacterium infects the human gastrointestinal tract (118) and is characterized by two major virulence factors: enterotoxin A, expressed by the tdcA gene and causes diarrhea and intestinal mucosa damage, and toxin B, expressed by the tcdB gene and has strong cytotoxic effects (119, 120). Another important virulence factor is the txeR gene, which encodes a sigma factor that directs RNA polymerase to recognize the promoters of the tcdA and tcdB genes (121). Several lactobacilli have inhibitory effects on Cd virulence factors (Table S5 in the supplemental material), particularly on the production of toxins, as demonstrated by various in vitro studies. Coculture of lactobacilli with Cd on Vero cells significantly decreased TcdA and TcdB toxins in spent supernatants and increased their intracellular concentrations, thereby suggesting a possible antagonistic mechanism that could reduce the synthesis and/or secretion of toxins (122). S-layer proteins extracted from Lentilactobacillus kefiri strains inhibited the damage caused by Cd-spent culture supernatants in Vero cells, and this activity was higher in aggregating strains than in nonaggregating strains, thus indicating a direct interaction between S-layer proteins and clostridial toxins. The same results were not obtained with live Lbc. kefiri cells, thereby indicating a different interaction between the soluble S-layer proteins and those located on the surface of the bacterium (123). Lbc. acidophilus CFS significantly reduced the cytotoxic and cytopathic effects of a hypervirulent Cd strain culture filtrate on human epithelial cells by decreasing pathogen attachment on HT-29 and Caco-2 cells (124). Inhibition of Cd virulence factors has also been observed in vivo. The administration of Lbc. acidophilus in Cd-inoculated mice altered QS molecule production, lowering the transcriptional levels of luxS, tcdA, tcdB, and txeR genes and increasing mouse survival ratios by as much as 80% (125). Furthermore, the administration of Lmb. reuteri significantly decreased Cd colonization and concentrations of toxins in the cecum and decreased the numbers of rotavirus, a human virus that causes gastroenteritis in infants and children, after both pretreatment and coincubation of the pathogen and the probiotic with HT-29 cells (126). In a protection model, an engineered Lactobacillus strain expressing TcdB-neutralizing antibody fragments delayed the death of infected hamsters (127), whereas in mice, an engineered Lcb. casei expressing Clostridium perfringens alpha-toxin toxoid induced the production of antibodies capable of neutralizing C. perfringens alpha-toxin and increasing levels of cytokines and interferon-γ in the serum and spleen lymphocytes (128).
STAPHYLOCOCCUS AUREUS
Staphylococcus aureus (Sa) is an opportunistic pathogen accounting for 76% of all skin and soft tissue infections in humans (129) due to the expression of several virulence factors regulated by the agr QS system and the sae gene (130, 131). Sa produces a variety of toxins, including sea enterotoxins, which cause food poisoning (132), toxic shock syndrome toxin 1 (TSST-1) expressed by the tst gene, a superantigen that causes multiple organ dysfunctions and is associated with a high mortality rate (133), and Staphylococcus superantigen-like protein 1 (SSL-1), which inhibits the activity of matrix metalloproteases (134). The ability to evade the host immune system is promoted by the production of protein A (spa), a surface protein that prevents phagocytosis, and immunoglobulin-binding protein (sbi), which binds IgG and is involved in blood coagulation (135). Furthermore, the mecA gene confers methicillin resistance to Sa (136), and the expression of the ica operon promotes biofilm formation (137). Several studies demonstrated that lactobacilli can effectively counteract the virulence factors of this pathogen (Table S6 in the supplemental material). Either cocultivation or CFS from different lactobacilli strains inhibited Sa biofilm formation, as in the case of the cocultivation with Lcb. rhanosus (138) and acid CFS from Lbc. acidophilus that also inhibited lipase from biofilm and planktonic cells with a significant effect on methicillin-resistant Sa (139). In a study conducted on CFS produced by Lpb. plantarum, inhibition of the growth of Sa was observed, whereas CFS produced by Lmb. fermentum inhibited the expression of the icaA and icaR operons, thus limiting biofilm formation (140). CFS obtained from Lpb. plantarum, Lmb. fermentum, and Lmb. reuteri strains dependently decreased the expression of the sea, sae, agrA, tst, spa, and spi genes (141), and, in particular, the production of SSL-1 was significantly reduced when Sa was grown in Lmb. reuteri supernatant (142). Furthermore, Lbc. acidophilus and Lmb. fermentum have demonstrated a significant reduction of Sa adherence even on abiotic surfaces, most notably catheters and surgical implants (143, 144), thus suggesting a potential for the application of lactobacilli in the medical field to prevent the spread of nosocomial infections. The inhibitory effect of lactobacilli on Sa has also been confirmed in vitro. For example, Lbc. crispatus and Lactobacillus jensenii coaggregated with Sa, preventing pathogen adhesion to vaginal cells (145), whereas live Lcb. casei cells affected Sa internalization, and both live and heat-killed Lcb. casei cells reduced Sa adhesion in bMEC cells (146). Depending on their growth phase, concentration, competition, and the presence of surface layer proteins, Lgb. salivarius and Lpb. plantarum significantly inhibited Sa adherence to Caco-2 cells (147).
HELICOBACTER SPP.
Helicobacter is an important genus involved in food-borne illness. The clinical manifestations are determined by the genetics and behaviors of the human hosts (i.e., diet or smoking status) as well as bacterial virulence. cagA and vacA cytotoxin-associated genes are important in this regard; cagA alters intracellular signal transduction, and vacA induces the fusion between endosomes and lysosomes (148). Another important virulence factor is the production of flagellin, which is induced by the expression of flaA and flab genes and provides the motility necessary for stomach colonization (149). Several studies have provided clear evidence that lactobacilli and their metabolites could decrease virulence factors of this species (Table S7 in the supplemental material). For example, the compounds produced by a Lcb. casei strain reduced the expression of genes codifying for flagellins in Helicobacter pylori (flaA and flaB) and SlmT (flaC), decreasing the motility and related internalization abilities (150). Similar results were obtained from a Lmb. reuteri strain, which significantly reduced the expression of flaA and vacA genes (151), whereas Lactiplantibacillus paraplantarum CFS reduced the adherence of H. pylori on AGS cells (152). Pretreatment with live and UV-killed Lgb. salivarius strains promoted the modification of the interleukin and chemokine response in the same cell line, in addition to downregulating 8 of 12 genes belonging to the H. pylori Cag pathogenicity island. This immunomodulatory effect was not dependent on adhesion or bacteriocin production, but after Lgb. salivarius exposure, CagA protein accumulated inside H. pylori cells, probably because of the loss of CagA secretion functionality (153). In vivo tests on Helicobacter hepaticus-stimulated macrophages from IL-10-deficient mice have been performed to investigate TNF-α-inhibitory Lmb. reuteri and Lcb. paracasei. These lactobacilli effectively decreased intestinal inflammation by lowering the levels of the proinflammatory colonic cytokines TNF-α and IL-12 but had no effects on H. hepaticus vitality (154). Lbc. acidophilus eradicated H. pylori from colonized children in 6.5% of subjects, while no spontaneous clearance was observed in untreated children, demonstrating the efficacy of lactobacilli administration in humans (155).
PSEUDOMONAS SPP., STREPTOCOCCUS SPP., AND KLEBSIELLA SPP.
Biofilms are microorganism aggregations within an extracellular matrix composed of proteins, exopolysaccharides, water, nutrients (such as polysaccharides and amino acids), and ions. The ability to form biofilms is an important common property that increases pathogen virulence, conferring adhesiveness and resistance to the host immune system and antibiotics (156). Biofilm formation is a characteristic trait of Pseudomonas spp., Streptococcus spp., and Klebsiella spp., all of which can establish ecological niches in which they replicate and become infectious to humans. Also in this case, lactobacilli and their metabolites have proven to be effective in inhibiting specific virulence factors of these pathogens (Table S8 in the supplemental material).
Pseudomonas aeruginosa, one of the most common pathogens in the hospital setting, owes its pathogenicity to various virulence factors (besides biofilm formation), such as the secretion of toxins (157) and the presence of flagella and pili (158). P. aeruginosa biofilm formation and elastase production were effectively inhibited by Lmb. fermentum, Lbc. zeae, and Lcb. paracasei (159), whereas Apilactobacillus kunkeei exhibited in vitro antibiofilm properties and attenuated P. aeruginosa infection in a G. mellonella model (160). Other in vivo tests were performed to evaluate the effects of Lpb. plantarum on P. aeruginosa acyl-homoserine-lactones, elastases, and biofilm virulence factors. In a burned mouse model, lactobacilli inhibited P. aeruginosa colonization, thus improving tissue repair and enhancing pathogen phagocytosis (161). Crude extract from Companilactobacillus crustorum degraded N-homoserine lactone and significantly enhanced biofilm sensitivity to azithromycin, thereby inhibiting biofilm formation and reducing the thickness of already formed biofilms. Real-time quantitative PCR (RT-qPCR) analysis revealed downregulation of lasI/R and rhlI/R QS virulence genes as well as inhibition of chitinase, protease, rhamnolipid, alginate, pyocyanin, and exopolysaccharide synthesis (162).
Klebsiella pneumoniae, a pathogenic bacterium associated with urinary infections that occur primarily in hospitalized patients and are frequently connected with the use of medical devices, is another microorganism whose pathogenicity relies on the ability to form biofilms (163). In this regard, Lmb. fermentum cells and their acid supernatants exerted antibiofilm properties against K. pneumoniae on catheters (164). In addition, Lbc. acidophilus and Lmb. fermentum or their supernatants hindered pathogen spread within biofilms, since no K. pneumoniae live cells were found after treatment (165).
Streptococcus mutans is the main etiological agent of human dental caries, owing to its virulence factors such as the aforementioned ability to form biofilms (166) as well as glucosyltransferases encoded by gtf and tft genes, which enable the production of exopolysaccharides and thus the formation of plaque (167), and fructosyltransferase (ftf), which is essential in adhesion (168). Different lactobacilli produce biosurfactants that downregulate the expression of S. mutans biofilm-forming genes, for example, Lmb. fermentum and Lbc. acidophilus, which reduced gtfB and gtfC gene expression modifying the surface and adhesion properties of the pathogen (169, 170), Lmb. reuteri, which reduced gftB, gtfC, and fft gene expression (168), and Lbc. acidophilus, which downregulated gtf and luxS (171). Similar results were obtained with the coculture of S. mutans with Lcb. casei, which downregulated luxS and gftB, spaP, and gbpB adhesion genes (172). Likewise, Lcb. casei, Lmb. reuteri, Lpb. plantarum, Lgb. salivarius, Lcb. rhanosus, and Lmb. reuteri decreased biofilm formation and downregulated the gtf genes, significantly decreasing bacterial attachment to surfaces (173–175).
Lactobacilli were also effective against Streptococcus pyogenes, a pathogen that affects humans exclusively and causes a variety of disorders ranging from asymptomatic transport to mild and superficial infections of the skin and mucous membranes to systemic diseases (176). Its virulence depends on the production of toxins, in particular streptolysin S encoded by the sag operon, which causes erythrocytes lysis (177). The combination of Lcb. rhanosus and Lmb. reuteri and their spent media were the most effective in reducing S. pyogenes adherence in FaDu and Detroit 562 host cells, inhibiting hemolytic activity through the downregulation of sag operon expression with a consequent decrease in streptolysin S production (178). In addition, a Lpb. plantarum strain decreased the levels of IL-17 and IL-23 in Hep-2 and A549 cells exposed to S. pyogenes by inducing the Toll-like receptor 2 (TLR2)/TLR4 surface receptors involved in the immune response (179).
UROGENITAL-CORRELATED PATHOGENS
Urogenital tract infections are major causes of disease in women. Several pathogenic species, including Candida albicans, Chlamydia trachomatis, Ec, Gardnerella vaginalis, Neisseria gonorrhoeae, Prevotella bivia, Streptococcus agalactiae, and Trichomonas vaginalis, are involved in the onset of disorders that, if untreated, can cause serious irreversible complications (180). In healthy individuals, the vaginal microbiota is dominated by lactobacilli (181), which protect against infections by inhibiting pathogen colonization via several mechanisms (Table S9 in the supplemental material), such as increasing microbiota adhesion through the production of biosurfactants, competition for host cell receptors, or direct killing through the production of hydrogen peroxide and bacteriocins (182). Inhibition of pathogen adhesion has been observed both in cell lines and on abiotic surfaces. Lbc. acidophilus, Lbc. gasseri, and Lbc. jensenii isolated from the human vagina were able to autoaggregate and strongly adhere to vaginal cell surfaces (183), whereas Lpb. plantarum coaggregated with pathogens such as S. agalactiae, G. vaginalis, and Ec (184). Moreover, a Lbc. acidophilus strain was able to inhibit Staphylococcus epidermidis and UPEC attachment on abiotic surfaces (185). Other urogenital tract pathogens include Trichomonas vaginalis, which causes trichomoniasis, Neisseria gonorrhoeae, which causes gonorrhea, and Gardnerella vaginalis, which is responsible for the initiation of bacterial vaginosis due to its ability to form biofilm. The most important virulence factor of T. vaginalis and N. gonorrhoeae is vaginal cell adhesion ability (186, 187), whereas G. vaginalis produces vaginolysin (vly), which inhibits the immune response, and sialidase (sld), an enzyme that releases salicylic acid, which improves adherence to cells and surfaces. Lactobacilli isolated from the human vagina showed significant inhibitory activities toward T. vaginalis, N. gonorrhoeae, and G. vaginalis. In particular, pretreatment with Lbc. crispatus competitively excluded G. vaginalis adhesion to HeLa cells, reducing the expression of vly and sld virulence genes (188), whereas Lbc. gasseri and Lbc. jensenii inhibited adhesion of T. vaginalis and N. gonorrhoeae to VEC and Hec-1-B cell lines, respectively (189, 190). Furthermore, a recombinant Lbc. jensenii secreting two domain CD4 proteins prevented the entrance of human immunodeficiency virus (HIV) into HeLa cells (191). Different trials observed the ability of Lbc. gasseri, Lbc. crispatus, and Lbc. helveticus to counteract vaginal-associated pathogens, specifically protecting cervix epithelial cells against the effects of P. bivia, toxin-producing G. vaginalis, and UPEC, inhibiting their adhesion to HeLa cells (192, 193). Similar results were obtained from Lbc. helveticus, which was able to inhibit the adhesion of G. vaginalis and UPEC to HeLa cells and internalization of UPEC and SlmT on HeLa and Caco2 cells, respectively (194).
Candida albicans is an opportunistic pathogenic yeast that resides in the oral cavity and gastrointestinal and urogenital tracts and is responsible for oral and vulvovaginal candidiasis. Its pathogenicity arises from multiple factors, including adherence promoted by various types of adhesins (Als3 and Hwp1), biofilm formation (Ece1, Als3, Bcr1, Efg1, Tec1, and Cph1), resistance to drugs, and the immune system through overexpression of Cdr1, Cdr2, and Mrd1 proteins (195), yeast-to-hyphal morphogenesis (Ece1), and hydrolytic enzymes (Saps) (196). Probiotic lactobacilli are effectively used in medical treatments to limit the spread of C. albicans by maintaining the balance of microbiota and producing inhibitory substances active against the pathogen (197–199). Lactobacilli isolated from women produced biosurfactants that significantly reduced C. albicans adhesion and prevented the formation of biofilms, and maximal results were obtained with Lbc. gasseri, Lmb. reuteri, Lbc. acidophilus, and Lcb. paracasei (200). Similar effects were obtained by coinoculating Lpb. plantarum, Lmb. fermentum, Lbc. gasseri, and Lmb. reuteri with C. albicans. Their autoaggregative properties, enhanced by low pH values and biofilm-forming ability, resulted in vaginal tract colonization, whereas coaggregation with C. albicans prevented yeast adhesion (201). Lbc. gasseri and Lactobacillus crispatus CFS coincubation with C. albicans significantly reduced the expression of Hwp1 and Ece1, Als3, Bcr1, Efg1, Tec1, and Cph1 genes, lowering biofilm formation, whereas CFS from Lbc. crispatus inhibited C. albicans adhesion to HeLa cells (202). Another important mechanism of virulence inhibition is the modification of the hyphal structure. Several studies found that Lcb. rhanosus reduced hyphal elongation (203), and Lcb. rhanosus, Lcb. paracasei, and Lcb. casei were effective against C. albicans hyphal morphogenesis because they expressed the MspI gene, encoding a major peptidoglycan hydrolase that hydrolyzes chitin (204). Proteinase and hemolysin activities were reduced in C. albicans grown with Lcb. rhanosus, with alterations to antifungal susceptibility (205). In addition, Lcb. rhanosus affected adhesion, invasion, and hyphal extension, preventing oral epithelial tissue damage. This effect was correlated with glucose depletion and repression of ergosterol synthesis (206). Several lactobacilli had different effects on C. albicans-induced interleukin in VK2/E6E7 cells; for example, Lcb. rhanosus alone or in combination with Lmb. reuteri inhibited the increase in IL-1α and IL-8, whereas their supernatants increased IL-8 and IP-10 levels (207). In addition, Lbc. crispatus lowered C. albicans adhesion to VK2/E6E7 cells, thus upregulating IL-2, IL-6, and IL-17 while downregulating IL-8 (208), and to HeLa cells, lowering IL-8 and increasing β-defensin 2 and 3 (209). In the same cell line, a reduction in adhesion was attributed to antifungal activity arising from the inhibition of histone deacetylase by Lbc. crispatus, Limosilactobacillus vaginalis, and Lbc. gasseri (210). Several studies have investigated the effects of lactobacilli on gene expression of this pathogen. An extract from a Lactobacillus species strain, owing to high levels of oleic and myristic acid, affected C. albicans virulence (hyphal formation, proteinase, and phospholipase secretion), thus decreasing also Hwp1, Plb2, and Sap1 virulence gene expression (211). Moreover, CFSs of Lbc. crispatus, Lbc. gasseri, Lbc. acidophilus, and Lbc. jensenii effectively decreased the yeast-to-hyphal transition and the expression of hyphae-specific genes Als3, Hwp1, and Ece1, whereas Nrg1, a negative transcriptional regulator, was upregulated (212). Lcb. rhanosus and its supernatant reduced C. albicans filamentation and biofilm formation in vitro, altering the expression of Bcr1, Hwp1, and Als3 adhesion genes and Cph1 transcriptional regulatory genes. The same strain was tested on G. mellonella infected with C. albicans, and this treatment increased larval survival up to 80% (213). Lcb. paracasei, Lmb. fermentum, and Lcb. rhanosus also attenuated candidiasis in G. mellonella by increasing hemocyte quantity, upregulating galiomicin and gallerymicin antifungal peptide genes, slowing hyphal formation, and lowering biofilm development by downregulating the Als3, Hwp1, Efg1, and Cph1 genes (214). In other studies, Lbc. acidophilus and its filtrate inhibited C. albicans filamentation and biofilm formation, increasing the G. mellonella survival rate (215).
OTHER PATHOGENS
Multiple studies have been conducted on other pathogens and have shown encouraging results (Table S10 in the supplemental material). The modulating effect of lactobacilli on the immune system had positive effects in both mice inoculated with Yersinia enterocolitica and children infected with Enterococcus faecalis. In the first case, Lpb. plantarum had an immunomodulatory effect on infected BALB/c mice, resulting in a decrease in the anti-inflammatory cytokine IL-10 and an increase in IgA production (216). The administration of Lcb. rhanosus to children colonized with vancomycin-resistant En. faecalis led to immune system modulation, preventing the onset of infection (217). Lpb. plantarum also increased the virulence of Serratia marcescens, which causes hospital-acquired infections and whose antibiotic resistance poses a severe risk to patients, and of Bacillus cereus, which causes food poisoning. In relation to inoculum concentration and temperature, Lpb. plantarum reduced the hemolytic activity and protease and lecithinase expression of B. cereus (218), whereas CFS from Lbc. acidophilus and Lpb. plantarum affected the resistance of Se. marcescens to ceftriaxone and completely inhibited swarming motility (219). In addition, the CFS of Lgb. salivarius and Lbc. gasseri significantly reduced the virulence gene expression of Aggregatibacter actinomycetemcomitans, an oral pathogen that causes localized periodontitis by producing leukotoxins (LtzA) and cytolethal distending toxin (CdtB) (220).
CONCLUSIONS
Despite the development of various effective therapies, bacterial infections continue to pose a major threat to public health. In this regard, as described herein, lactobacilli capable of counteracting the virulence abilities of pathogenic microorganisms could be used to support existing treatments.
Some of these mechanisms include the reduction of the adhesive and invasive properties, the ability to self-aggregate and coaggregate with the pathogens, direct downregulation of virulence genes, and the production of metabolites with specific activities that can affect and modulate the host immune response. In addition, their presence has a bioprotective effect on both abiotic surfaces and cellular tissues. Lactobacilli, through competition for substrate and their steric hindrance, can inhibit pathogen activity and reduce their ability to adhere to epithelial cells, hence preventing the onset of diseases.
Although from review of the literature, many authors have demonstrated the ability to reduce virulence factors in pathogens by lactobacilli (our sincere apologies go to colleagues whose work was involuntarily not cited); however, there are still few studies conducted directly on humans validating all these capabilities observed in in vitro and in vivo tests on animals. Further research on this topic would thus help understand and advance the real applications of this microbial group to counteract pathogen virulence.
Lactobacilli, which have always been used by mankind and have a long history of safe use by humans in food preservation and processing, are currently also used as probiotics thanks to their proven beneficial properties. In addition to this, current whole-genome sequencing techniques provide additional assurance of safety, as evidenced by the recent EFSA statement, which recommends genetic characterization of all microbial strains before their use in food applications (221). Knowledge of the whole genome enables the identification of all potential risk factors present in lactobacilli (222), thus increasing the safety of use even in debilitated patients in hospital settings, where complete safety of the bacterial strains used must be ensured. In fact, beyond the current use as probiotics to alleviate the adverse effects of antibiotic therapies, lactobacilli could be used also as adjuvants for antibiotics, owing to their ability to counteract pathogens and their virulence properties. Infectious disease prevention is a fundamental achievement to limit the widespread use of drugs to strictly necessary cases, thus hindering the spread of antibiotic resistance. This issue has made treatment of infection more difficult in recent years; therefore, identifying alternative treatments is increasingly important to decrease the use of antibiotics while also improving host health. Given that the average age of the world population is rising, the consequences of demographic aging are expected to have severe repercussions on numerous social dynamics in the future, including an increase in the cost of public health. To reduce the number of hospitalizations and consequently the costs of health care, the condition of older and fragile people must be improved. The identification and study of strains with probiotic and antivirulence activity against pathogens may lead to the development of therapies that can be combined with current antibiotic treatments, thus reducing their adverse effects on patients while increasing their effectiveness. Furthermore, consistent intake of strains capable of reducing the likelihood of pathological manifestations in hosts, such as through the consumption of food formulations, could also be used to prevent infections, thereby reducing antibiotic use.
ACKNOWLEDGMENTS
The authors would like to thank Patrick Lane (ScEYEnce Studios) for the prompt willingness and competence in helping us to improve the presentation of the work creating Figure 1.
Footnotes
Supplemental material is available online only.
Contributor Information
Lucilla Iacumin, Email: lucilla.iacumin@uniud.it.
Laurie E. Comstock, University of Chicago
REFERENCES
- 1.Zheng J, Wittouck S, Salvetti E, Franz CMAP, Harris HMB, Mattarelli P, O'Toole PW, Pot B, Vandamme P, Walter J, Watanabe K, Wuyts S, Felis GE, Gänzle MG, Lebeer S. 2020. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 70:2782–2858. 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 2.Plavec TV, Berlec A. 2020. Safety aspects of genetically modified lactic acid bacteria. Microorganisms 8:297. 10.3390/microorganisms8020297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EFSA BIOHAZ Panel (Panel on Biological Hazards). 2016. Update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 4: suitability of taxonomic units notified to EFSA until March 2016. EFSA J 14:e04522. 10.2903/j.efsa.2016.4522. [DOI] [Google Scholar]
- 4.FAO/WHO. 2001. Report of a joint FAO/WHO expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria- Report of a Joint FAO/WHO Working Group on drafting guidelines for the evaluation of probiotics in food. https://www.fao.org/3/a0512e/a0512e.pdf.
- 5.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. 2014. Expert consensus document: the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11:506–514. 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 6.Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EMM, Sanders ME, Shamir R, Swann JR, Szajewska H, Vinderola G. 2021. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol 18:649–667. 10.1038/s41575-021-00440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liévin-Le Moal V, Servin AL. 2014. Anti-infective activities of Lactobacillus strains in the human intestinal microbiota: from probiotics to gastrointestinal anti- infectious biotherapeutic agents. Clin Microbiol Rev 27:167–199. 10.1128/CMR.00080-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Cerbo A, Palmieri B, Aponte M, Morales-Medina JC, Iannitti T. 2016. Mechanisms and therapeutic effectiveness of lactobacilli. J Clin Pathol 69:187–203. 10.1136/jclinpath-2015-202976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao K, Mu CL, Farzi A, Zhu WY. 2020. Tryptophan metabolism: a link between the gut microbiota and brain. Adv Nutr 11:709–723. 10.1093/advances/nmz127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turroni F, Ventura M, Buttó LF, Duranti S, O’Toole PW, Motherway MOC, Van Sinderen D. 2014. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell Mol Life Sci 71:183–203. 10.1007/s00018-013-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slover CM, Danziger L. 2008. Lactobacillus: a review. Clin Microbiol Newsl 30:23–27. 10.1016/j.clinmicnews.2008.01.006. [DOI] [Google Scholar]
- 12.de Melo Pereira GV, de Oliveira Coelho B, Magalhães Júnior AI, Thomaz-Soccol V, Soccol CR. 2018. How to select a probiotic? A review and update of methods and criteria. Biotechnol Adv 36:2060–2076. 10.1016/j.biotechadv.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Masood MI, Qadir MI, Shirazi JH, Khan IU. 2011. Beneficial effects of lactic acid bacteria on human beings. Crit Rev Microbiol 37:91–98. 10.3109/1040841X.2010.536522. [DOI] [PubMed] [Google Scholar]
- 14.Ohashi Y, Ushida K. 2009. Health-beneficial effects of probiotics: its mode of action. Anim Sci J 80:361–371. 10.1111/j.1740-0929.2009.00645.x. [DOI] [PubMed] [Google Scholar]
- 15.Hilmi HTA. 2010. Lactic acid bacteria and their antimicrobial peptides: induction, detection, partial characterization, and their potential applications. Academic dissertation. University of Helsinki, Helsinki, Finland. [Google Scholar]
- 16.Collado MC, Surono I, Meriluoto J, Salminen S. 2007. Indigenous dadih lactic acid bacteria: cell-surface properties and interactions with pathogens. J Food Sci 72:M89–M93. 10.1111/j.1750-3841.2007.00294.x. [DOI] [PubMed] [Google Scholar]
- 17.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 18.Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. 2005. Making “sense” of metabolism: autoinducer-2, LUXS and pathogenic bacteria. Nat Rev Microbiol 3:383–396. 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- 19.Verderosa AD, Totsika M, Fairfull-Smith KE. 2019. Bacterial biofilm eradication agents: a current review. Front Chem 7:824. 10.3389/fchem.2019.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carpentier B, Chassaing D. 2004. Interactions in biofilms between Listeria monocytogenes and resident microorganisms from food industry premises. Int J Food Microbiol 97:111–122. 10.1016/j.ijfoodmicro.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 21.Dicks LMT, Grobbelaar MJ. 2021. Double-barrel shotgun: probiotic lactic acid bacteria with antiviral properties modified to serve as vaccines. Microorganisms 9:1565. 10.3390/microorganisms9081565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO. 2017. World health statistics 2017: Monitoring health for the SDGs, sustainable development goals. World Health Organization, Geneva. [Google Scholar]
- 23.Hutchings MI, Truman AW, Wilkinson B. 2019. Antibiotics: past, present and future. Curr Opin Microbiol 51:72–80. 10.1016/j.mib.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Rello J, Parisella FR, Perez A. 2019. Alternatives to antibiotics in an era of difficult-to-treat resistance: new insights. Expert Rev Clin Pharmacol 12:635–642. 10.1080/17512433.2019.1619454. [DOI] [PubMed] [Google Scholar]
- 25.Syed B, Wein S, Ruangapanit Y. 2020. The efficacy of synbiotic application in broiler chicken diets, alone or in combination with antibiotic growth promoters on zootechnical parameters. JWPR 10:469–479. 10.36380/jwpr.2020.54. [DOI] [Google Scholar]
- 26.Disson O, Moura A, Lecuit M. 2021. Making sense of the biodiversity and virulence of Listeria monocytogenes. Trends Microbiol 29:811–822. 10.1016/j.tim.2021.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Camejo A, Carvalho F, Reis O, Leitão E, Sousa S, Cabanes D. 2011. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2:379–394. 10.4161/viru.2.5.17703. [DOI] [PubMed] [Google Scholar]
- 28.Piercey MJ, Hingston PA, Truelstrup Hansen L. 2016. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15°C. Int J Food Microbiol 223:63–74. 10.1016/j.ijfoodmicro.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Quereda JJ, Andersson C, Cossart P, Johansson J, Pizarro-Cerdá J. 2018. Role in virulence of phospholipases, listeriolysin O and listeriolysin S from epidemic Listeria monocytogenes using the chicken embryo infection model. Vet Res 49:13. 10.1186/s13567-017-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacquet C, Gouin E, Jeannel D, Cossart P, Rocourt J. 2002. Expression of ActA, Ami, InlB, and Listeriolysin O in Listeria monocytogenes of human and food origin. Appl Environ Microbiol 68:616–622. 10.1128/AEM.68.2.616-622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sibanda T, Buys EM. 2022. Listeria monocytogenes pathogenesis: the role of stress adaptation. Microorganisms 10:1522. 10.3390/microorganisms10081522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corr S, Hill C, Gahan CGM. 2006. An in vitro cell-culture model demonstrates internalin- and hemolysin-independent translocation of Listeria monocytogenes across M cells. Microb Pathog 41:241–250. 10.1016/j.micpath.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Dutra V, Silva AC, Cabrita P, Peres C, Malcata X, Brito L. 2016. Lactobacillus plantarum LB95 impairs the virulence potential of Gram-positive and Gram-negative food-borne pathogens in HT-29 and vero cell cultures. J Med Microbiol 65:28–35. 10.1099/jmm.0.000196. [DOI] [PubMed] [Google Scholar]
- 34.Iglesias MB, Viñas I, Colás-Medà P, Collazo C, Serrano JCE, Abadias M. 2017. Adhesion and invasion of Listeria monocytogenes and interaction with Lactobacillus rhamnosus GG after habituation on fresh-cut pear. J Funct Foods 34:453–460. 10.1016/j.jff.2017.05.011. [DOI] [Google Scholar]
- 35.Dong Q, Zhang W, Guo L, Niu H, Liu Q, Wang X. 2020. Influence of Lactobacillus plantarum individually and in combination with low O2-MAP on the pathogenic potential of Listeria monocytogenes in cabbage. Food Control 107:106765. 10.1016/j.foodcont.2019.106765. [DOI] [Google Scholar]
- 36.Upadhyay A, Upadhyaya I, Mooyottu S, Venkitanarayanan K. 2016. Eugenol in combination with lactic acid bacteria attenuates Listeria monocytogenes virulence in vitro and in invertebrate model Galleria mellonella. J Med Microbiol 65:443–455. 10.1099/jmm.0.000251. [DOI] [PubMed] [Google Scholar]
- 37.Koo OK, Amalaradjou MAR, Bhunia AK. 2012. Recombinant probiotic expressing Listeria adhesion protein attenuates Listeria monocytogenes virulence in vitro. PLoS One 7:e29277. 10.1371/journal.pone.0029277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathipa MG, Bhunia AK, Thantsha MS. 2019. Internalin AB-expressing recombinant Lactobacillus casei protects Caco-2 cells from Listeria monocytogenes-induced damages under simulated intestinal conditions. PLoS One 14:e0220321. 10.1371/journal.pone.0220321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Archambaud C, Nahori M-A, Soubigou G, Bećavin C, Laval L, Lechat P, Smokvina T, Langella P, Lecuit M, Cossart P. 2012. Impact of lactobacilli on orally acquired listeriosis. Proc Natl Acad Sci USA 109:16684–16689. 10.1073/pnas.1212809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bambirra FHS, Lima KGC, Franco BDGM, Cara DC, Nardi RMD, Barbosa FHF, Nicoli JR. 2007. Protective effect of Lactobacillus sakei 2a against experimental challenge with Listeria monocytogenes in gnotobiotic mice. Lett Appl Microbiol 45:663–667. 10.1111/j.1472-765X.2007.02250.x. [DOI] [PubMed] [Google Scholar]
- 41.Riaz A, Noureen S, Liqat I, Arshad M, Arshad N. 2019. Antilisterial efficacy of Lactobacillus brevis MF179529 from cow: an in vivo evidence. BMC Complement Altern Med 19:37. 10.1186/s12906-019-2444-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng Q, Shi H, Luo Y, Zhao H, Liu N. 2020. Effect of dietary lactobacilli mixture on Listeria monocytogenes infection and virulence property in broilers. Poult Sci 99:3655–3662. 10.1016/j.psj.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alves VF, Lavrador MAS, De Martinis ECP. 2003. Bacteriocin exposure and food ingredients influence on growth and virulence of Listeria monocytogenes in a model meat gravy system. J Food Saf 23:201–217. 10.1111/j.1745-4565.2003.tb00363.x. [DOI] [Google Scholar]
- 44.Martinez RCR, De Martinis ECP. 2005. Evaluation of bacteriocin-producing Lactobacillus sakei 1 against Listeria monocytogenes 1/2a growth and haemolytic activity. Brazilian J Microbiol 36:83–87. 10.1590/S1517-83822005000100016. [DOI] [Google Scholar]
- 45.Winkelströter LK, Gomes BC, Thomaz MRS, Souza VM, De Martinis ECP. 2011. Lactobacillus sakei 1 and its bacteriocin influence adhesion of Listeria monocytogenes on stainless steel surface. Food Control 22:1404–1407. 10.1016/j.foodcont.2011.02.021. [DOI] [Google Scholar]
- 46.Martinez RCR, De Martinis ECP. 2005. Antilisterial activity of a crude preparation of Lactobacillus sakei 1 bacteriocin and its lack of influence on Listeria monocytogenes haemolytic activity. Food Control 16:429–433. 10.1016/j.foodcont.2004.05.002. [DOI] [Google Scholar]
- 47.Andino A, Hanning I. 2015. Salmonella enterica: survival, colonization, and virulence differences among serovars. ScientificWorldJournal 2015:520179. 10.1155/2015/520179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lucas RL, Lee CA. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J Bacteriol 183:2733–2745. 10.1128/JB.183.9.2733-2745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergeron JRC, Brockerman JA, Vuckovic M, Deng W, Okon M, Finlay BB, McIntosh LP, Strynadka NCJ. 2018. Characterization of the two conformations adopted by the T3SS inner-membrane protein PrgK. Protein Sci 27:1680–1691. 10.1002/pro.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muyyarikkandy MS, Amalaradjou MA. 2017. Lactobacillus bulgaricus, Lactobacillus rhamnosus and Lactobacillus paracasei attenuate Salmonella Enteritidis, Salmonella Heidelberg and Salmonella Typhimurium colonization and virulence gene expression in vitro. Int J Mol Sci 18:2381. 10.3390/ijms18112381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guiney DG, Fierer J. 2011. The role of the spv genes in Salmonella pathogenesis. Front Microbiol 2:129. 10.3389/fmicb.2011.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pati NB, Vishwakarma V, Jaiswal S, Periaswamy B, Hardt WD, Suar M. 2013. Deletion of invH gene in Salmonella enterica serovar Typhimurium limits the secretion of Sip effector proteins. Microbes Infect 15:66–73. 10.1016/j.micinf.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 53.Lou L, Zhang P, Piao R, Wang Y. 2019. Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front Cell Infect Microbiol 9:270. 10.3389/fcimb.2019.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang L, Wang P, Song X, Zhang H, Ma S, Wang J, Li W, Lv R, Liu X, Ma S, Yan J, Zhou H, Huang D, Cheng Z, Yang C, Feng L, Wang L. 2021. Salmonella Typhimurium reprograms macrophage metabolism via T3SS effector SopE2 to promote intracellular replication and virulence. Nat Commun 12:879. 10.1038/s41467-021-21186-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haraga A, Miller S. 2003. A Salmonella enterica serovar Typhimurium translocated leucine-rich repeat effector protein inhibits NF-κB-dependent gene expression. Infect Immun 71:4052–4058. 10.1128/IAI.71.7.4052-4058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao AP, Petrof EO, Kuppireddi S, Zhao Y, Xia Y, Claud EC, Sun J. 2008. Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PLoS One 3:e2369. 10.1371/journal.pone.0002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pérez-Morales D, Banda MM, Chau NYE, Salgado H, Martínez-Flores I, Ibarra JA, Ilyas B, Coombes BK, Bustamante VH. 2017. The transcriptional regulator SsrB is involved in a molecular switch controlling virulence lifestyles of Salmonella. PLoS Pathog 13:e1006497. 10.1371/journal.ppat.1006497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abdelhafez SM, Abdelwahab AMO, Ammar AA, Eldemerdash AS. 2016. Molecular studies on the prophylactic effect of probiotics on Salmonella Typhimurium infected chicks. Benha Vet Med J 31:73–82. 10.21608/bvmj.2016.31264. [DOI] [Google Scholar]
- 59.Wang C, Wang J, Gong J, Yu H, Pacan JC, Niu Z, Si W, Sabour PM. 2011. Use of Caenorhabditis elegans for preselecting Lactobacillus isolates to control Salmonella Typhimurium. J Food Prot 74:86–93. 10.4315/0362-028X.JFP-10-155. [DOI] [PubMed] [Google Scholar]
- 60.Yang X, Brisbin J, Yu H, Wang Q, Yin F, Zhang Y, Sabour P, Sharif S, Gong J. 2014. Selected lactic acid-producing bacterial isolates with the capacity to reduce salmonella translocation and virulence gene expression in chickens. PLoS One 9:e93022. 10.1371/journal.pone.0093022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andino A, Zhang N, Diaz-Sanchez S, Yard C, Pendleton S, Hanning I. 2014. Characterization and specificity of probiotics to prevent Salmonella infection in mice. FFHD 4:370–380. 10.31989/ffhd.v4i8.148. [DOI] [Google Scholar]
- 62.Song F, Liu J, Zhao W, Huang H, Hu D, Chen H, Zhang H, Chen W, Gu Z. 2020. Synergistic effect of eugenol and probiotic Lactobacillus plantarum ZS2058 against Salmonella infection in C57BL/6 mice. Nutrients 12:1611. 10.3390/nu12061611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liévin-Le Moal V, Amsellem R, Servin AL. 2011. Impairment of swimming motility by antidiarrheic Lactobacillus acidophilus strain LB retards internalization of Salmonella enterica serovar Typhimurium within human enterocyte-like cells. Antimicrob Agents Chemother 55:4810–4820. 10.1128/AAC.00418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abdel-Daim A, Hassouna N, Hafez M, Ashor MSA, Aboulwafa MM. 2013. Antagonistic activity of Lactobacillus isolates against Salmonella Typhi in vitro. Biomed Res Int 2013:680605. 10.1155/2013/680605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burkholder KM, Fletcher DH, Gileau L, Kandolo A. 2019. Lactic acid bacteria decrease Salmonella enterica Javiana virulence and modulate host inflammation during infection of an intestinal epithelial cell line. Pathog Dis 77:ftz025. 10.1093/femspd/ftz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burkholder KM, Bhunia AK. 2009. Salmonella enterica serovar Typhimurium adhesion and cytotoxicity during epithelial cell stress is reduced by Lactobacillus rhamnosus GG. Gut Pathog 1:14. 10.1186/1757-4749-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Makras L, Triantafyllou V, Fayol-Messaoudi D, Adriany T, Zoumpopoulou G, Tsakalidou E, Servin A, De Vuyst L. 2006. Kinetic analysis of the antibacterial activity of probiotic lactobacilli towards Salmonella enterica serovar Typhimurium reveals a role for lactic acid and other inhibitory compounds. Res Microbiol 157:241–247. 10.1016/j.resmic.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 68.Peng M, Tabashsum Z, Patel P, Bernhardt C, Biswas D. 2018. Linoleic acids overproducing Lactobacillus casei limits growth, survival, and virulence of Salmonella Typhimurium and enterohaemorrhagic Escherichia coli. Front Microbiol 9:2663. 10.3389/fmicb.2018.02663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tabashsum Z, Peng M, Bernhardt C, Patel P, Carrion M, Rahaman SO, Biswas D. 2020. Limiting the pathogenesis of Salmonella Typhimurium with berry phenolic extracts and linoleic acid overproducing Lactobacillus casei. J Microbiol 58:489–498. 10.1007/s12275-020-9545-1. [DOI] [PubMed] [Google Scholar]
- 70.Jankowska A, Laubitz D, Antushevich H, Zabielski R, Grzesiuk E. 2008. Competition of Lactobacillus paracasei with Salmonella enterica for adhesion to Caco-2 cells. J Biomed Biotechnol 2008:357964. 10.1155/2008/357964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coconnier-Polter M-H, Liévin-Le Moal V, Servin AL. 2005. A Lactobacillus acidophilus strain of human gastrointestinal microbiota origin elicits killing of enterovirulent Salmonella enterica serovar Typhimurium by triggering lethal bacterial membrane damage. Appl Environ Microbiol 71:6115–6120. 10.1128/AEM.71.10.6115-6120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durant JA, Corrier DE, Stanker LH, Ricke SC. 2000. Salmonella Enteritidis hilA gene fusion response after incubation in spent media from either S. Enteritidis or a poultry Lactobacillus strain. J Environ Sci Health B 35:599–610. 10.1080/03601230009373295. [DOI] [PubMed] [Google Scholar]
- 73.Hudault S, Liévin V, Bernet-Camard MF, Servin AL. 1997. Antagonistic activity exerted in vitro and in vivo by Lactobacillus casei (strain GG) against Salmonella Typhimurium C5 infection. Appl Environ Microbiol 63:513–518. 10.1128/aem.63.2.513-518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Craven SE, Williams DD. 1998. In vitro attachment of Salmonella Typhimurium to chicken cecal mucus: effect of cations and pretreatment with Lactobacillus spp. isolated from the intestinal tracts of chickens. J Food Prot 61:265–271. 10.4315/0362-028x-61.3.265. [DOI] [PubMed] [Google Scholar]
- 75.Hu J-L, Yu H, Kulkarni RR, Sharif S, Cui SW, Xie M-Y, Nie S-P, Gong J. 2015. Modulation of cytokine gene expression by selected Lactobacillus isolates in the ileum, caecal tonsils and spleen of Salmonella-challenged broilers. Avian Pathol 44:463–469. 10.1080/03079457.2015.1086725. [DOI] [PubMed] [Google Scholar]
- 76.Eryildiz C, Tabakcioglu K, Kuyucuklu G, Sakru N. 2020. Investigation of antimicrobial resistance and virulence genes of Campylobacter isolates from patients in tertiary hospital in Edirne, Turkey. Indian J Med Microbiol 38:157–161. 10.4103/ijmm.IJMM_20_78. [DOI] [PubMed] [Google Scholar]
- 77.Mohan V. 2015. The role of probiotics in the inhibition of Campylobacter jejuni colonization and virulence attenuation. Eur J Clin Microbiol Infect Dis 34:1503–1513. 10.1007/s10096-015-2392-z. [DOI] [PubMed] [Google Scholar]
- 78.Alemka A, Clyne M, Shanahan F, Tompkins T, Corcionivoschi N, Bourke B. 2010. Probiotic colonization of the adherent mucus layer of HT29MTXE12 cells attenuates Campylobacter jejuni virulence properties. Infect Immun 78:2812–2822. 10.1128/IAI.01249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taha-Abdelaziz K, Astill J, Kulkarni RR, Read LR, Najarian A, Farber JM, Sharif S. 2019. In vitro assessment of immunomodulatory and anti-Campylobacter activities of probiotic lactobacilli. Sci Rep 9:17903. 10.1038/s41598-019-54494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tabashsum Z, Peng M, Salaheen S, Comis C, Biswas D. 2018. Competitive elimination and virulence property alteration of Campylobacter jejuni by genetically engineered Lactobacillus casei. Food Control 85:283–291. 10.1016/j.foodcont.2017.10.010. [DOI] [Google Scholar]
- 81.Mundi A, Delcenserie V, Amiri-Jami M, Moorhead S, Griffiths MW. 2013. Cell-free preparations of Lactobacillus acidophilus strain La-5 and Bifidobacterium longum strain NCC2705 affect virulence gene expression in Campylobacter jejuni. J Food Prot 76:1740–1746. 10.4315/0362-028X.JFP-13-084. [DOI] [PubMed] [Google Scholar]
- 82.Chapman TA, Wu X-Y, Barchia I, Bettelheim KA, Driesen S, Trott D, Wilson M, Chin JJ-C. 2006. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl Environ Microbiol 72:4782–4795. 10.1128/AEM.02885-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fratamico PM, DebRoy C, Liu Y, Needleman DS, Baranzoni GM, Feng P. 2016. Advances in molecular serotyping and subtyping of Escherichia coli. Front Microbiol 7:644. 10.3389/fmicb.2016.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaper JB, Nataro JP, Mobley HLT. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 85.Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 8:26–38. 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 86.Donnenberg MS, Tacket CO, James SP, Losonsky G, Nataro JP, Wasserman SS, Kaper JB, Levine MM. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Invest 92:1412–1417. 10.1172/JCI116717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taneike I, Zhang H-M, Wakisaka-Saito N, Yamamoto T. 2002. Enterohemolysin operon of Shiga toxin-producing Escherichia coli: a virulence function of inlammatory cytokine production from human monocytes. FEBS Lett 524:219–224. 10.1016/S0014-5793(02)03027-2. [DOI] [PubMed] [Google Scholar]
- 88.Beraldo LG, Borges CA, Maluta RP, Cardozo MV, Rigobelo EC, de Ávila FA. 2014. Detection of Shiga toxigenic (STEC) and enteropathogenic (EPEC) Escherichia coli in dairy buffalo. Vet Microbiol 170:162–166. 10.1016/j.vetmic.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 89.Amin MA, Hashem HR, El-Mahallawy HS, Abdelrahman AA, Zaki HM, Azab MM. 2022. Characterization of enterohemorrhagic Escherichia coli from diarrhoeic patients with particular reference to production of Shiga-like toxin. Microb Pathog 166:105538. 10.1016/j.micpath.2022.105538. [DOI] [PubMed] [Google Scholar]
- 90.Umpiérrez A, Ernst D, Fernández M, Oliver M, Casaux ML, Caffarena RD, Schild C, Giannitti F, Fraga M, Zunino P. 2021. Virulence genes of Escherichia coli in diarrheic and healthy calves. Rev Argent Microbiol 53:34–38. 10.1016/j.ram.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 91.McWilliams BD, Torres AG. 2014. Enterohemorrhagic Escherichia coli adhesins. Microbiol Spectr 2. 10.1128/microbiolspec.EHEC-0003-2013. [DOI] [PubMed] [Google Scholar]
- 92.Beltrametti F, Kresse AU, Guzmán CA. 1999. Transcriptional regulation of the pas gene of enterohemorrhagic Escherichia coli. J Bacteriol 181:3409–3418. 10.1128/JB.181.11.3409-3418.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Navarro-Garcia F, Serapio-Palacios A, Ugalde-Silva P, Tapia-Pastrana G, Chavez-Dueñas L. 2013. Actin cytoskeleton manipulation by effector proteins secreted by diarrheagenic Escherichia coli pathotypes. Biomed Res Int 2013:374395. 10.1155/2013/374395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jelčić I, Hüfner E, Schmidt H, Hertel C. 2008. Repression of the locus of the enterocyte effacement-encoded regulator of gene transcription of Escherichia coli O157:H7 by Lactobacillus reuteri culture supernatants is LuxS and strain dependent. Appl Environ Microbiol 74:3310–3314. 10.1128/AEM.00072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sharp FC, Sperandio V. 2007. QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect Immun 75:2432–2440. 10.1128/IAI.02003-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beutin L, Strauch E, Zimmermann S, Kaulfuss S, Schaudinn C, Männel A, Gelderblom HR. 2005. Genetical and functional investigation of fliC genes encoding flagellar serotype H4 in wildtype strains of Escherichia coli and in a laboratory E. coli K-12 strain expressing flagellar antigen type H48. BMC Microbiol 5:4. 10.1186/1471-2180-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zeinhom M, Tellez AM, Delcenserie V, El-Kholy AM, El-Shinawy SH, Griffiths MW. 2012. Yogurt containing bioactive molecules produced by Lactobacillus acidophilus la-5 exerts a protective effect against enterohemorrhagic Escherichia coli in mice. J Food Prot 75:1796–1805. 10.4315/0362-028X.JFP-11-508. [DOI] [PubMed] [Google Scholar]
- 98.Medellin-Peña MJ, Wang H, Johnson R, Anand S, Griffiths MW. 2007. Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl Environ Microbiol 73:4259–4267. 10.1128/AEM.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cadieux PA, Burton JP, Devillard E, Reid G. 2009. Lactobacillus by-products inhibit the growth and virulence of uropathogenic Escherichia coli. J Physiol Pharmacol 60:13–18. [PubMed] [Google Scholar]
- 100.Sherman PM, Johnson-Henry KC, Yeung HP, Ngo PSC, Goulet J, Tompkins TA. 2005. Probiotics reduce enterohemorrhagic Escherichia coli O157:H7- and enteropathogenic E. coli O127:H6-induced changes in polarized T84 epithelial cell monolayers by reducing bacterial adhesion and cytoskeletal rearrangements. Infect Immun 73:5183–5188. 10.1128/IAI.73.8.5183-5188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leccese Terraf MC, Juarez Tomás MS, Rault L, Le Loir Y, Even S, Nader-Macías MEF. 2017. In vitro effect of vaginal lactobacilli on the growth and adhesion abilities of uropathogenic Escherichia coli. Arch Microbiol 199:767–774. 10.1007/s00203-016-1336-z. [DOI] [PubMed] [Google Scholar]
- 102.Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. 1999. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol 276:941–949. 10.1152/ajpgi.1999.276.4.G941. [DOI] [PubMed] [Google Scholar]
- 103.Liévin-Le Moal V, Amsellem R, Servin AL, Coconnier M-H. 2002. Lactobacillus acidophilus (strain LB) from the resident adult human gastrointestinal microflora exerts activity against brush border damage promoted by a diarrhoeagenic Escherichia coli in human enterocyte-like cells. Gut 50:803–811. 10.1136/gut.50.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Atassi F, Brassart D, Grob P, Graf F, Servin AL. 2006. Vaginal Lactobacillus isolates inhibit uropathogenic Escherichia coli. FEMS Microbiol Lett 257:132–138. 10.1111/j.1574-6968.2006.00163.x. [DOI] [PubMed] [Google Scholar]
- 105.Anand S, Mandal S, Singh KS, Patil P, Tomar SK. 2018. Synbiotic combination of Lactobacillus rhamnosus NCDC 298 and short chain fructooligosaccharides prevents enterotoxigenic Escherichia coli infection. LWT 98:329–334. 10.1016/j.lwt.2018.08.061. [DOI] [Google Scholar]
- 106.Hirano J, Yoshida T, Sugiyama T, Koide N, Mori I, Yokochi T. 2003. The effect of Lactobacillus rhamnosus on enterohemorrhagic Escherichia coli infection of human intestinal cells in vitro. Microbiol Immunol 47:405–409. 10.1111/j.1348-0421.2003.tb03377.x. [DOI] [PubMed] [Google Scholar]
- 107.Kim Y, Oh S, Park S, Seo JB, Kim S-H. 2008. Lactobacillus acidophilus reduces expression of enterohemorrhagic Escherichia coli O157:H7 virulence factors by inhibiting autoinducer-2-like activity. Food Control 19:1042–1050. 10.1016/j.foodcont.2007.10.014. [DOI] [Google Scholar]
- 108.Park H, Yeo S, Ji Y, Lee J, Yang J, Park S, Shin H, Holzapfel W. 2014. Autoinducer-2 associated inhibition by Lactobacillus sakei NR28 reduces virulence of enterohaemorrhagic Escherichia coli O157: H7. Food Control 45:62–69. 10.1016/j.foodcont.2014.04.024. [DOI] [Google Scholar]
- 109.Chen YP, Lee TY, Hong WS, Hsieh HH, Chen MJ. 2013. Effects of Lactobacillus kefiranofaciens M1 isolated from kefir grains on enterohemorrhagic Escherichia coli infection using mouse and intestinal cell models. J Dairy Sci 96:7467–7477. 10.3168/jds.2013-7015. [DOI] [PubMed] [Google Scholar]
- 110.Chu H, Kang S, Ha S, Cho K, Park S-M, Han K-H, Sang KK, Lee HG, Seung HH, Yun CH, Choi Y. 2005. Lactobacillus acidophilus expressing recombinant K99 adhesive fimbriae has an inhibitory effect on adhesion of enterotoxigenic Escherichia coli. Microbiol Immunol 49:941–948. 10.1111/j.1348-0421.2005.tb03687.x. [DOI] [PubMed] [Google Scholar]
- 111.Mangell P, Nejdfors P, Wang M, Ahrné S, Weström B, Thorlacius H, Jeppsson B. 2002. Lactobacillus plantarum 299v inhibits intestinal permeability. Dig Dis Sci 47:511–516. 10.1023/A:1017947531536. [DOI] [PubMed] [Google Scholar]
- 112.Asahara T, Nomoto K, Watanuki M, Yokokura T. 2001. Antimicrobial activity of intraurethrally administered probiotic Lactobacillus casei in a murine model of Escherichia coli urinary tract infection. Antimicrob Agents Chemother 45:1751–1760. 10.1128/AAC.45.6.1751-1760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang Y, Galle S, Le MHA, Zijlstra RT, Gänzle MG. 2015. Feed fermentation with reuteran- and levan-producing Lactobacillus reuteri reduces colonization of weanling pigs by enterotoxigenic Escherichia coli. Appl Environ Microbiol 81:5743–5752. 10.1128/AEM.01525-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Peng M, Reichmann G, Biswas D. 2015. Lactobacillus casei and its byproducts alter the virulence factors of foodborne bacterial pathogens. J Funct Foods 15:418–428. 10.1016/j.jff.2015.03.055. [DOI] [Google Scholar]
- 115.Coconnier MH, Bernet MF, Kernéis S, Chauvière G, Fourniat J, Servin AL. 1993. Inhibition of adhesion of enteroinvasive pathogens to human intestinal Caco-2 cells by Lactobacillus acidophilus strain LB decreases bacterial invasion. FEMS Microbiol Lett 110:299–305. 10.1111/j.1574-6968.1993.tb06339.x. [DOI] [PubMed] [Google Scholar]
- 116.Bernet MF, Brassart D, Neeser JR, Servin AL. 1994. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut 35:483–489. 10.1136/gut.35.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fonseca HC, de Sousa Melo D, Ramos CL, Dias DR, Schwan RF. 2021. Probiotic properties of lactobacilli and their ability to inhibit the adhesion of enteropathogenic bacteria to Caco-2 and HT-29 cells. Probiotics Antimicrob Proteins 13:102–112. 10.1007/s12602-020-09659-2. [DOI] [PubMed] [Google Scholar]
- 118.Binyamin D, Nitzan O, Azrad M, Hamo Z, Koren O, Peretz A. 2021. The microbial diversity following antibiotic treatment of Clostridioides difficile infection. BMC Gastroenterol 21:166. 10.1186/s12876-021-01754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lyerly DM, Krivan HC, Wilkins TD. 1988. Clostridium difficile: its disease and toxins. Clin Microbiol Rev 1:1–18. 10.1128/CMR.1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Awad MM, Johanesen PA, Carter GP, Rose E, Lyras D. 2014. Clostridium difficile virulence factors: insights into an anaerobic spore-forming pathogen. Gut Microbes 5:579–593. 10.4161/19490976.2014.969632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, Rood JI, Sonenshein AL, Dupuy B. 2002. Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J Bacteriol 184:5971–5978. 10.1128/JB.184.21.5971-5978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Trejo FM, Pérez PF, De Antoni GL. 2010. Co-culture with potentially probiotic microorganisms antagonises virulence factors of Clostridium difficile in vitro. Antonie Van Leeuwenhoek 98:19–29. 10.1007/s10482-010-9424-6. [DOI] [PubMed] [Google Scholar]
- 123.Carasi P, Trejo FM, Pérez PF, De Antoni GL, de los Angeles Serradell M. 2012. Surface proteins from Lactobacillus kefir antagonize in vitro cytotoxic effect of Clostridium difficile toxins. Anaerobe 18:135–142. 10.1016/j.anaerobe.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 124.Najarian A, Sharif S, Griffiths MW. 2019. Evaluation of protective effect of Lactobacillus acidophilus La-5 on toxicity and colonization of Clostridium difficile in human epithelial cells in vitro. Anaerobe 55:142–151. 10.1016/j.anaerobe.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 125.Yun B, Oh S, Griffiths MW. 2014. Lactobacillus acidophilus modulates the virulence of Clostridium difficile. J Dairy Sci 97:4745–4758. 10.3168/jds.2014-7921. [DOI] [PubMed] [Google Scholar]
- 126.Sagheddu V, Uggeri F, Belogi L, Remollino L, Brun P, Bernabè G, Moretti G, Porzionato A, Morelli L, Castagliuolo I, Elli M. 2020. The biotherapeutic potential of Lactobacillus reuteri characterized using a target-specific selection process. Front Microbiol 11:532. 10.3389/fmicb.2020.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Andersen KK, Strokappe NM, Hultberg A, Truusalu K, Smidt I, Mikelsaar RH, Mikelsaar M, Verrips T, Hammarström L, Marcotte H. 2016. Neutralization of Clostridium difficile toxin B mediated by engineered lactobacilli that produce single-domain antibodies. Infect Immun 84:395–406. 10.1128/IAI.00870-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gao X, Ma Y, Wang Z, Bai J, Jia S, Feng B, Jiang Y, Cui W, Tang L, Li Y, Wang L, Xu Y. 2019. Oral immunization of mice with a probiotic Lactobacillus casei constitutively expressing the α-toxoid induces protective immunity against Clostridium perfringens α-toxin. Virulence 10:166–179. 10.1080/21505594.2019.1582975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Parlet CP, Brown MM, Horswill AR. 2019. Commensal staphylococci influence Staphylococcus aureus skin colonization and disease. Trends Microbiol 27:497–507. 10.1016/j.tim.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oogai Y, Matsuo M, Hashimoto M, Kato F, Sugai M, Komatsuzawa H. 2011. Expression of virulence factors by Staphylococcus aureus grown in serum. Appl Environ Microbiol 77:8097–8105. 10.1128/AEM.05316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Giraudo AT, Calzolari A, Cataldi AA, Bogni C, Nagel R. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett 177:15–22. 10.1111/j.1574-6968.1999.tb13707.x. [DOI] [PubMed] [Google Scholar]
- 132.Argudín MÁ, Mendoza MC, Rodicio MR. 2010. Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel) 2:1751–1773. 10.3390/toxins2071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zheng Y, Qin C, Zhang X, Zhu Y, Li A, Wang M, Tang Y, Kreiswirth BN, Chen L, Zhang H, Du H. 2020. The tst gene associated Staphylococcus aureus pathogenicity island facilitates its pathogenesis by promoting the secretion of inflammatory cytokines and inducing immune suppression. Microb Pathog 138:103797. 10.1016/j.micpath.2019.103797. [DOI] [PubMed] [Google Scholar]
- 134.Tang A, Caballero AR, Bierdeman MA, Marquart ME, Foster TJ, Monk IR, O’Callaghan RJ. 2019. Staphylococcus aureus superantigen-like protein SSL1: a toxic protease. Pathogens 8:2. 10.3390/pathogens8010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Atkins KL, Burman JD, Chamberlain ES, Cooper JE, Poutrel B, Bagby S, Jenkins ATA, Feil EJ, van den Elsen JMH. 2008. S. aureus IgG-binding proteins SpA and Sbi: host specificity and mechanisms of immune complex formation. Mol Immunol 45:1600–1611. 10.1016/j.molimm.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 136.Lakhundi S, Zhang K. 2018. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31:e00020-18. 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Omidi M, Firoozeh F, Saffari M, Sedaghat H, Zibaei M, Khaledi A. 2020. Ability of biofilm production and molecular analysis of spa and ica genes among clinical isolates of methicillin-resistant Staphylococcus aureus. BMC Res Notes 13:19. 10.1186/s13104-020-4885-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fornitano ALP, Amendola I, Santos SSF, Silva CRG, Leao MVP. 2019. Lactobacillus rhamnosus versus Staphylococcus aureus: influence on growth and expression of virulence factors. J Dent Maxillofac Res 2:29–33. https://ologyjournals.com/jdsomr/jdsomr_00025.php. [Google Scholar]
- 139.Jabbar H, Hala F, Radeef M. 2011. Capability of Lactobacillus acidophilus supernatant to inhibit production of lipase from methicillin-resistant Staphylococcus aureus. J Univ Anbar Pure Sci 5:1–5. [Google Scholar]
- 140.Melo TA, Dos Santos TF, De Almeida ME, Junior LAGF, Andrade EF, Rezende RP, Marques LM, Romano CC. 2016. Inhibition of Staphylococcus aureus biofilm by Lactobacillus isolated from fine cocoa. BMC Microbiol 16:250. 10.1186/s12866-016-0871-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ramezani M, Zainodini N, Hakimi H, Zarandi ER, Bagheri V, Bahramabadi R, Zare-Bidaki M. 2020. Cell-free culture supernatants of lactobacilli modify the expression of virulence factors genes in Staphylococcus aureus. Jundishapur J Microbiol 12 10.5812/jjm.96806. [DOI] [Google Scholar]
- 142.Laughton JM, Devillard E, Heinrichs DE, Reid G, Mccormick JK. 2006. Inhibition of expression of a staphylococcal superantigen-like protein by a soluble factor from Lactobacillus reuteri. Microbiology (Reading) 152:1155–1167. 10.1099/mic.0.28654-0. [DOI] [PubMed] [Google Scholar]
- 143.Reid G, Tieszer C. 1994. Use of lactobacilli to reduce the adhesion of Staphylococcus aureus to catheters. Int Biodeterior Biodegradation 34:73–83. 10.1016/0964-8305(95)00011-9. [DOI] [Google Scholar]
- 144.Gan BS, Kim J, Reid G, Cadieux P, Howard JC. 2002. Lactobacillus fermentum RC‐14 inhibits Staphylococcus aureus infection of surgical implants in rats. J Infect Dis 185:1369–1372. 10.1086/340126. [DOI] [PubMed] [Google Scholar]
- 145.Younes JA, van der Mei HC, van den Heuvel E, Busscher HJ, Reid G. 2012. Adhesion forces and coaggregation between vaginal staphylococci and lactobacilli. PLoS One 7:e36917. 10.1371/journal.pone.0036917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bouchard DS, Rault L, Berkova N, Le Loir Y, Even S. 2013. Inhibition of Staphylococcus aureus invasion into bovine mammary epithelial cells by contact with live Lactobacillus casei. Appl Environ Microbiol 79:877–885. 10.1128/AEM.03323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ren D, Li C, Qin Y, Yin R, Li X, Tian M, Du S, Guo H, Liu C, Zhu N, Sun D, Li Y, Jin N. 2012. Inhibition of Staphylococcus aureus adherence to Caco-2 cells by lactobacilli and cell surface properties that influence attachment. Anaerobe 18:508–515. 10.1016/j.anaerobe.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 148.Chang W-L, Yeh Y-C, Sheu B-S. 2018. The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J Biomed Sci 25:68. 10.1186/s12929-018-0466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yan J, Liang S-H, Mao Y-F, Li li W, Li S-P. 2003. Construction of expression system for flaA and flaB genes of Helicobacter pylori and determination of immunoreactivity and antigenicity of recombinant proteins. World J Gastroenterol 9:2240–2250. 10.3748/wjg.v9.i10.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liévin Le Moal V, Fayol-Messaoudi D, Servin AL. 2013. Compound(s) secreted by Lactobacillus casei strain Shirota YIT9029 irreversibly and reversibly impair the swimming motility of Helicobacter pylori and Salmonella enterica serovar Typhimurium, respectively. Microbiology (Reading) 159:1956–1971. 10.1099/mic.0.067678-0. [DOI] [PubMed] [Google Scholar]
- 151.Urrutia-Baca VH, Escamilla-García E, de la Garza-Ramos MA, Tamez-Guerra P, Gomez-Flores R, Urbina-Ríos CS. 2018. In vitro antimicrobial activity and downregulation of virulence gene expression on Helicobacter pylori by reuterin. Probiotics Antimicrob Proteins 10:168–175. 10.1007/s12602-017-9342-2. [DOI] [PubMed] [Google Scholar]
- 152.Ki M-R, Ghim S-Y, Hong I-H, Park J-K, Hong K-S, Ji A-R, Jeong K-S. 2010. In vitro inhibition of Helicobacter pylori growth and of adherence of cagA-positive strains to gastric epithelial cells by Lactobacillus paraplantarum KNUC25 isolated from kimchi. J Med Food 13:629–634. 10.1089/jmf.2009.1265. [DOI] [PubMed] [Google Scholar]
- 153.Ryan KA, O'Hara AM, van Pijkeren J-P, Douillard FP, O'Toole PW. 2009. Lactobacillus salivarius modulates cytokine induction and virulence factor gene expression in Helicobacter pylori. J Med Microbiol 58:996–1005. 10.1099/jmm.0.009407-0. [DOI] [PubMed] [Google Scholar]
- 154.Pena JA, Rogers AB, Ge Z, Ng V, Li SY, Fox JG, Versalovic J. 2005. Probiotic Lactobacillus spp. diminish Helicobacter hepaticus-induced inflammatory bowel disease in interleukin-10-deficient mice. Infect Immun 73:912–920. 10.1128/IAI.73.2.912-920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gotteland M, Poliak L, Cruchet S, Brunser O. 2005. Effect of regular ingestion of Saccharomyces boulardii plus inulin or Lactobacillus acidophilus LB in children colonized by Helicobacter pylori. Acta Paediatr 94:1747–1751. 10.1111/j.1651-2227.2005.tb01848.x. [DOI] [PubMed] [Google Scholar]
- 156.Jamal M, Ahmad W, Andleeb S, Jalil F, Imran M, Nawaz MA, Hussain T, Ali M, Rafiq M, Kamil MA. 2018. Bacterial biofilm and associated infections. J Chinese Med Assoc 81:7–11. 10.1016/j.jcma.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 157.Veesenmeyer JL, Hauser AR, Lisboa T, Rello J. 2009. Pseudomonas aeruginosa virulence and therapy: evolving translational strategies. Crit Care Med 37:1777–1786. 10.1097/CCM.0b013e31819ff137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sultan M, Arya R, Kim KK. 2021. Roles of two-component systems in Pseudomonas aeruginosa virulence. Int J Mol Sci 22:12152. 10.3390/ijms222212152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Alexandre Y, Le Berre R, Barbier G, Le Blay G. 2014. Screening of Lactobacillus spp. for the prevention of Pseudomonas aeruginosa pulmonary infections. BMC Microbiol 14:107. 10.1186/1471-2180-14-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Berríos P, Fuentes JA, Salas D, Carreño A, Aldea P, Fernández F, Trombert AN. 2018. Inhibitory effect of biofilm-forming Lactobacillus kunkeei strains against virulent Pseudomonas aeruginosa in vitro and in honeycomb moth (Galleria mellonella) infection model. Benef Microbes 9:257–268. 10.3920/BM2017.0048. [DOI] [PubMed] [Google Scholar]
- 161.Valdéz JC, Peral MC, Rachid M, Santana M, Perdigón G. 2005. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: the potential use of probiotics in wound treatment. Clin Microbiol Infect 11:472–479. 10.1111/j.1469-0691.2005.01142.x. [DOI] [PubMed] [Google Scholar]
- 162.Cui T, Bai F, Sun M, Lv X, Li X, Zhang D, Du H. 2020. Lactobacillus crustorum ZHG 2–1 as novel quorum-quenching bacteria reducing virulence factors and biofilms formation of Pseudomonas aeruginosa. LWT 117:108696. 10.1016/j.lwt.2019.108696. [DOI] [Google Scholar]
- 163.Wu M-C, Lin T-L, Hsieh P-F, Yang H-C, Wang J-T. 2011. Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS One 6:e23500. 10.1371/journal.pone.0023500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Maldonado NC, Silva De Ruiz C, Cecilia M, Nader-Macias ME. 2007. A simple technique to detect Klebsiella biofilm-forming-strains. Inhibitory potential of Lactobacillus fermentum CRL 1058 whole cells and products, p 52–59. In Vilas AM (ed), Communicating Current Research Educcational Topics and Trends in Applied Microbiolgy. FORMATEX, Guadalajara, Mexico. [Google Scholar]
- 165.Al-Mathkhury HJF, Aded Assa SD. 2012. Inhibitory effect of lactobacilli filtrate on Klebsiella pneumoniae biofilm. Iraqui Postgrad Med J 11:168–179. [Google Scholar]
- 166.Lemos JA, Palmer SR, Zeng L, Wen ZT, Kajfasz JK, Freires IA, Abranches J, Brady LJ. 2019. The biology of Streptococcus mutans. Microbiol Spectr 7. 10.1128/microbiolspec.GPP3-0051-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Krzyściak W, Jurczak A, Kościelniak D, Bystrowska B, Skalniak A. 2014. The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis 33:499–515. 10.1007/s10096-013-1993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Salehi R, Salehi A, Savabi O, Tahmourespour A, Eslami G, Kamali S, Kazemi M. 2014. Effects of Lactobacillus reuteri-derived biosurfactant on the gene expression profile of essential adhesion genes (gtfB, gtfC and ftf) of Streptococcus mutans. Adv Biomed Res 3:169. 10.4103/2277-9175.139134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tahmourespour A, Salehi R, Kasra Kermanshahi R. 2011. Lactobacillus acidophilus-derived biosurfactant effect on GTFB and GTFC expression level in Streptococcus mutans biofilm cells. Braz J Microbiol 42:330–339. 10.1590/S1517-83822011000100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tahmourespour A, Salehi R, Kermanshahi RK, Eslami G. 2011. The anti-biofouling effect of Lactobacillus fermentum-derived biosurfactant against Streptococcus mutans. Biofouling 27:385–392. 10.1080/08927014.2011.575458. [DOI] [PubMed] [Google Scholar]
- 171.Ahmed A, Dachang W, Lei Z, Jianjun L, Juanjuan Q, Yi X. 2014. Effect of Lactobacillus species on Streptococcus mutans biofilm formation. Pak J Pharm Sci 27:1523–1528. [PubMed] [Google Scholar]
- 172.Wen ZT, Yates D, Ahn SJ, Burne RA. 2010. Biofilm formation and virulence expression by Streptococcus mutans are altered when grown in dual-species model. BMC Microbiol 10:111. 10.1186/1471-2180-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wasfi R, Abd El-Rahman OA, Zafer MM, Ashour HM. 2018. Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J Cell Mol Med 22:1972–1983. 10.1111/jcmm.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Söderling EM, Marttinen AM, Haukioja AL. 2011. Probiotic lactobacilli interfere with Streptococcus mutans biofilm formation in vitro. Curr Microbiol 62:618–622. 10.1007/s00284-010-9752-9. [DOI] [PubMed] [Google Scholar]
- 175.Wu C-C, Lin C-T, Wu C-Y, Peng W-S, Lee M-J, Tsai Y-C. 2015. Inhibitory effect of Lactobacillus salivarius on Streptococcus mutans biofilm formation. Mol Oral Microbiol 30:16–26. 10.1111/omi.12063. [DOI] [PubMed] [Google Scholar]
- 176.Fiedler T, Koller T, Kreikemeyer B. 2015. Streptococcus pyogenes biofilms—formation, biology, and clinical relevance. Front Cell Infect Microbiol 5:15. 10.3389/fcimb.2015.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Molloy EM, Cotter PD, Hill C, Mitchell DA, Ross RP. 2011. Streptolysin S-like virulence factors: the continuing sagA. Nat Rev Microbiol 9:670–681. 10.1038/nrmicro2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Saroj SD, Maudsdotter L, Tavares R, Jonsson A-B. 2016. Lactobacilli interfere with Streptococcus pyogenes hemolytic activity and adherence to host epithelial cells. Front Microbiol 7:1176. 10.3389/fmicb.2016.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rizzo A, Losacco A, Carratelli CR, Di Domenico M, Bevilacqua N. 2013. Lactobacillus plantarum reduces Streptococcus pyogenes virulence by modulating the IL-17, IL-23 and Toll-like receptor 2/4 expressions in human epithelial cells. Int Immunopharmacol 17:453–461. 10.1016/j.intimp.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 180.Spurbeck RR, Arvidson CG. 2011. Lactobacilli at the front line of defense against vaginally acquired infections. Future Microbiol 6:567–582. 10.2217/fmb.11.36. [DOI] [PubMed] [Google Scholar]
- 181.Redondo-Lopez V, Cook RL, Sobel JD. 1990. Emerging role of lactobacilli in the control and maintenance of the vaginal bacterial microflora. Rev Infect Dis 12:856–872. 10.1093/clinids/12.5.856. [DOI] [PubMed] [Google Scholar]
- 182.Reid G, Cook RL, Bruce AW. 1987. Examination of strains of lactobacilli for properties that may influence bacterial interference in the urinary tract. J Urol 138:330–335. 10.1016/s0022-5347(17)43137-5. [DOI] [PubMed] [Google Scholar]
- 183.Boris S, Suárez JE, Vázquez F, Barbés C. 1998. Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect Immun 66:1985–1989. 10.1128/IAI.66.5.1985-1989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kmet V, Lucchini F. 1997. Aggregationpromoting factor in human vaginal Lactobacillus strains. FEMS Immunol Med Microbiol 19:111–114. 10.1111/j.1574-695X.1997.tb01079.x. [DOI] [PubMed] [Google Scholar]
- 185.Hawthorn LA, Reid G. 1990. Exclusion of uropathogen adhesion to polymer surfaces by Lactobacillus acidophilus. J Biomed Mater Res 24:39–46. 10.1002/jbm.820240105. [DOI] [PubMed] [Google Scholar]
- 186.Mielczarek E, Blaszkowska J. 2016. Trichomonas vaginalis: pathogenicity and potential role in human reproductive failure. Infection 44:447–458. 10.1007/s15010-015-0860-0. [DOI] [PubMed] [Google Scholar]
- 187.Quillin SJ, Seifert HS. 2018. Neisseria gonorrhoeae host adaptation and pathogenesis. Nat Rev Microbiol 16:226–240. 10.1038/nrmicro.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Castro J, Martins AP, Rodrigues ME, Cerca N, Onderdonk AB. 2018. Lactobacillus crispatus represses vaginolysin expression by BV associated Gardnerella vaginalis and reduces cell cytotoxicity. Anaerobe 50:60–63. 10.1016/j.anaerobe.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 189.Phukan N, Parsamand T, Brooks AES, Nguyen TNM, Simoes-Barbosa A. 2013. The adherence of Trichomonas vaginalis to host ectocervical cells is influenced by lactobacilli. Sex Transm Infect 89:455–459. 10.1136/sextrans-2013-051039. [DOI] [PubMed] [Google Scholar]
- 190.Spurbeck RR, Arvidson CG. 2010. Lactobacillus jensenii surface-associated proteins inhibit Neisseria gonorrhoeae adherence to epithelial cells. Infect Immun 78:3103–3111. 10.1128/IAI.01200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chang TL-Y, Chang C-H, Simpson DA, Xu Q, Martin PK, Lagenaur LA, Schoolnik GK, Ho DD, Hillier SL, Holodniy M, Lewicki JA, Lee PP. 2003. Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proc Natl Acad Sci USA 100:11672–11677. 10.1073/pnas.1934747100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Atassi F, Brassart D, Grob P, Graf F, Servin AL. 2006. Lactobacillus strains isolated from the vaginal microbiota of healthy women inhibit Prevotella bivia and Gardnerella vaginalis in coculture and cell culture. FEMS Immunol Med Microbiol 48:424–432. 10.1111/j.1574-695X.2006.00162.x. [DOI] [PubMed] [Google Scholar]
- 193.Atassi F, Pho Viet Ahn DL, Lievin-Le Moal V. 2019. Diverse expression of antimicrobial activities against bacterial vaginosis and urinary tract infection pathogens by cervicovaginal microbiota strains of Lactobacillus gasseri and Lactobacillus crispatus. Front Microbiol 10:2900. 10.3389/fmicb.2019.02900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Atassi F, Brassart D, Grob P, Graf F, Servin AL. 2006. In vitro antibacterial activity of Lactobacillus helveticus strain KS300 against diarrhoeagenic, uropathogenic and vaginosis-associated bacteria. J Appl Microbiol 101:647–654. 10.1111/j.1365-2672.2006.02933.x. [DOI] [PubMed] [Google Scholar]
- 195.Maras B, Maggiore A, Mignogna G, D’Erme M, Angiolella L. 2021. Hyperexpression of CDRs and HWP1 genes negatively impacts on Candida albicans virulence. PLoS One 16:e0252555. 10.1371/journal.pone.0252555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mayer FL, Wilson C, Hube B. 2013. Candida albicans pathogenicity mechanisms. Virulence 4:119–128. 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.De Seta F, Parazzini F, De Leo R, Banco R, Maso GP, De Santo D, Sartore A, Stabile G, Inglese S, Tonon M, Restaino S. 2014. Lactobacillus plantarum P17630 for preventing Candida vaginitis recurrence: a retrospective comparative study. Eur J Obstet Gynecol Reprod Biol 182:136–139. 10.1016/j.ejogrb.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 198.da Silva Dantas A, Lee KK, Raziunaite I, Schaefer K, Wagener J, Yadav B, Gow NA. 2016. Cell biology of Candida albicans–host interactions. Curr Opin Microbiol 34:111–118. 10.1016/j.mib.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Grin PM, Kowalewska PM, Alhazzan W, Fox-Robichaud AE. 2013. Lactobacillus for preventing recurrent urinary tract infections in women: meta-analysis. Can J Urol 20:6607–6614. [PubMed] [Google Scholar]
- 200.Itapary dos Santos C, Ramos França Y, Duarte Lima Campos C, Quaresma Bomfim MR, Oliveira Melo B, Assunção Holanda R, Santos VL, Gomes Monteiro S, Buozzi Moffa E, Souza Monteiro A, Andrade Monteiro C, Monteiro-Neto V. 2019. Antifungal and antivirulence activity of vaginal Lactobacillus spp. products against Candida vaginal isolates. Pathogens 8:150. 10.3390/pathogens8030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Santos CMA, Pires MCV, Leão TL, Hernández ZP, Rodriguez ML, Martins AKS, Miranda LS, Martins FS, Nicoli JR. 2016. Selection of Lactobacillus strains as potential probiotics for vaginitis treatment. Microbiology (Reading) 162:1195–1207. 10.1099/mic.0.000302. [DOI] [PubMed] [Google Scholar]
- 202.Matsuda Y, Cho O, Sugita T, Ogishima D, Takeda S. 2018. Culture supernatants of Lactobacillus gasseri and Lbc. crispatus inhibit Candida albicans biofilm formation and adhesion to HeLa cells. Mycopathologia 183:691–700. 10.1007/s11046-018-0259-4. [DOI] [PubMed] [Google Scholar]
- 203.Graf K, Last A, Gratz R, Allert S, Linde S, Westermann M, Gröger M, Mosig AS, Gresnigt MS, Hube B. 2019. Keeping Candida commensal: how lactobacilli antagonize pathogenicity of Candida albicans in an in vitro gut model. Dis Model Mech 12:dmm039719. 10.1242/dmm.039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Allonsius CN, Vandenheuvel D, Oerlemans EFM, Petrova MI, Donders GGG, Cos P, Delputte P, Lebeer S. 2019. Inhibition of Candida albicans morphogenesis by chitinase from Lactobacillus rhamnosus GG. Sci Rep 9:2900. 10.1038/s41598-019-39625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Oliveira VMC, Santos SSF, Silva CRG, Jorge AOC, Leão MVP. 2016. Lactobacillus is able to alter the virulence and the sensitivity profile of Candida albicans. J Appl Microbiol 121:1737–1744. 10.1111/jam.13289. [DOI] [PubMed] [Google Scholar]
- 206.Mailänder-Sánchez D, Braunsdorf C, Grumaz C, Müller C, Lorenz S, Stevens P, Wagener J, Hebecker B, Hube B, Bracher F, Sohn K, Schaller M. 2017. Antifungal defense of probiotic Lactobacillus rhamnosus GG is mediated by blocking adhesion and nutrient depletion. PLoS One 12:e0184438-19. 10.1371/journal.pone.0184438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Martinez RCR, Seney SL, Summers KL, Nomizo A, De Martinis ECP, Reid G. 2009. Effect of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 on the ability of Candida albicans to infect cells and induce inflammation. Microbiol Immunol 53:487–495. 10.1111/j.1348-0421.2009.00154.x. [DOI] [PubMed] [Google Scholar]
- 208.Niu X-X, Li T, Zhang X, Wang S-X, Liu Z-H. 2017. Lactobacillus crispatus modulates vaginal epithelial cell innate response to Candida albicans. Chin Med J (Engl) 130:273–279. 10.4103/0366-6999.198927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Rizzo A, Losacco A, Carratelli CR. 2013. Lactobacillus crispatus modulates epithelial cell defense against Candida albicans through Toll-like receptors 2 and 4, interleukin 8 and human β-defensins 2 and 3. Immunol Lett 156:102–109. 10.1016/j.imlet.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 210.Parolin C, Marangoni A, Laghi L, Foschi C, Palomino RAÑ, Calonghi N, Cevenini R, Vitali B. 2015. Isolation of vaginal lactobacilli and characterization of anti-Candida activity. PLoS One 10:e0131220-17. 10.1371/journal.pone.0131220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ahmad A. 2013. Probiotic Lactobacillus affects the expression of virulence in Candida albicans. J Microb Biochem Technol 5:5948. 10.4172/1948-5948.S1.011. [DOI] [Google Scholar]
- 212.Wang S, Wang Q, Yang E, Yan L, Li T, Zhuang H. 2017. Antimicrobial compounds produced by vaginal Lactobacillus crispatus are able to strongly inhibit Candida albicans growth, hyphal formation and regulate virulence-related gene expressions. Front Microbiol 8:564. 10.3389/fmicb.2017.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ribeiro FC, de Barros PP, Rossoni RD, Junqueira JC, Jorge AOC. 2017. Lactobacillus rhamnosus inhibits Candida albicans virulence factors in vitro and modulates immune system in Galleria mellonella. J Appl Microbiol 122:201–211. 10.1111/jam.13324. [DOI] [PubMed] [Google Scholar]
- 214.Rossoni RD, Fuchs BB, de Barros PP, dos Santos Velloso M, Jorge AOC, Junqueira JC, Mylonakis E. 2017. Lactobacillus paracasei modulates the immune system of Galleria mellonella and protects against Candida albicans infection. PLoS One 12:e0173332-17. 10.1371/journal.pone.0173332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vilela SFG, Barbosa JO, Rossoni RD, Santos JD, Prata MCA, Anbinder AL, Jorge AOC, Junqueira JC. 2015. Lactobacillus acidophilus ATCC 4356 inhibits biofilm formation by C. albicans and attenuates the experimental candidiasis in Galleria mellonella. Virulence 6:29–39. 10.4161/21505594.2014.981486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.De Montijo-Prieto S, Moreno E, Bergillos-Meca T, Lasserrot A, Ruiz-López MD, Ruiz-Bravo A, Jiménez-Valera M. 2015. A Lactobacillus plantarum strain isolated from kefir protects against intestinal infection with Yersinia enterocolitica O9 and modulates immunity in mice. Res Microbiol 166:626–632. 10.1016/j.resmic.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 217.Felipe EMM, Fanny MJ, Isabela A, Celia RG, Mariella VPL, Silvana SFDS. 2016. Relationship between the probiotic Lactobacillus rhamnosus and Enterococcus faecalis during the biofilm formation. Afr J Microbiol Res 10:1182–1186. 10.5897/AJMR2016.7990. [DOI] [Google Scholar]
- 218.Reda FM. 2014. Detoxification of enterotoxigenic Bacillus cereus (JX455159) isolated from meat by a local strain of Lactobacillus plantarum (JX282192). Ann Microbiol 64:287–296. 10.1007/s13213-013-0662-5. [DOI] [Google Scholar]
- 219.Vahedi-Shahandashti R, Kasra-Kermanshahi R, Shokouhfard M, Ghadam P, Feizabadi MM, Teimourian S. 2017. Antagonistic activities of some probiotic lactobacilli culture supernatant on Serratia marcescens swarming motility and antibiotic resistance. Iran J Microbiol 9:348–355. [PMC free article] [PubMed] [Google Scholar]
- 220.Nissen L, Sgorbati B, Biavati B, Belibasakis GN. 2014. Lactobacillus salivarius and Lbc. gasseri down-regulate Aggregatibacter actinomycetemcomitans exotoxins expression. Ann Microbiol 64:611–617. 10.1007/s13213-013-0694-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.EFSA. 2007. Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA—opinion of the scientific committee. The EFSA Journal 587:1–16. 10.2903/j.efsa.2007.587. [DOI] [Google Scholar]
- 222.Colautti A, Arnoldi M, Comi G, Iacumin L. 2022. Antibiotic resistance and virulence factors in lactobacilli: something to carefully consider. Food Microbiol 103:103934. 10.1016/j.fm.2021.103934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S10. Download jb.00272-22-s0001.pdf, PDF file, 0.3 MB (316KB, pdf)