Abstract
Context
Despite several reports of familial partial lipodystrophy (FPLD) type 2 (FPLD2) due to heterozygous LMNA variants and FPLD3 due to PPARG variants, the phenotypic differences among them remain unclear.
Objective
To compare the body fat distribution, metabolic parameters, and prevalence of metabolic complications between FPLD3 and FPLD2.
Methods
A retrospective, cross-sectional comparison of patients from 2 tertiary referral centers—UT Southwestern Medical Center and the National Institute of Diabetes and Digestive and Kidney Diseases. A total of 196 females and 59 males with FPLD2 (age 2-86 years) and 28 females and 4 males with FPLD3 (age 9-72 years) were included. The main outcome measures were skinfold thickness, regional body fat by dual-energy X-ray absorptiometry (DXA), metabolic variables, and prevalence of diabetes mellitus and hypertriglyceridemia.
Results
Compared with subjects with FPLD2, subjects with FPLD3 had significantly increased prevalence of hypertriglyceridemia (66% vs 84%) and diabetes (44% vs 72%); and had higher median fasting serum triglycerides (208 vs 255 mg/dL), and mean hemoglobin A1c (6.4% vs 7.5%). Compared with subjects with FPLD2, subjects with FPLD3 also had significantly higher mean upper limb fat (21% vs 27%) and lower limb fat (16% vs 21%) on DXA and increased median skinfold thickness at the anterior thigh (5.8 vs 11.3 mm), calf (4 vs 6 mm), triceps (5.5 vs 7.5 mm), and biceps (4.3 vs 6.8 mm).
Conclusion
Compared with subjects with FPLD2, subjects with FPLD3 have milder lipodystrophy but develop more severe metabolic complications, suggesting that the remaining adipose tissue in subjects with FPLD3 may be dysfunctional or those with mild metabolic disease are underrecognized.
Keywords: PPARG, LMNA, familial partial lipodystrophy, dual-energy X-ray absorptiometry, triglycerides, diabetes mellitus
Familial partial lipodystrophy (FPLD), a rare mostly autosomal dominant disorder, is characterized by marked loss of subcutaneous fat from the extremities, and predisposition to insulin resistance and its metabolic complications, such as diabetes mellitus, dyslipidemia, and hepatic steatosis [1, 2]. The 2 most prevalent types of FPLD are due to disease-causing heterozygous variants in LMNA (FPLD2, or the Dunnigan variety) [3, 4] or in PPARG (FPLD3) genes [5–7]. While FPLD2 has been reported in more than 500 subjects, FPLD3 has been reported in fewer than 100 subjects [2]. Due to extremely rare prevalence of FPLD3 and mostly anecdotal, limited reports in the literature, whether there are phenotypic differences between the 2 types remain unclear. Therefore, we compared body fat distribution using anthropometric measurements and dual-energy X-ray absorptiometry (DXA), metabolic variables, and the prevalence of metabolic disorders between the 2 types of FPLD in patients seen at 2 referral centers in the United States.
Patients and Methods
Subjects
The study protocol was approved by the Institutional Review Board of UT Southwestern Medical Center (UTSW) and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), Bethesda, MD. Adult patients and legal guardians of patients <18 years of age gave a written informed consent; minors provided assent if age appropriate. Patients included in this study presented with familial partial lipodystrophy or were family members of the proband and were found to have heterozygous disease-causing variants either in PPARG (FPLD3) or in LMNA (FPLD2).
Methods
Questionnaire
Demographic data and health history were collected during either physician interview or using a lipodystrophy questionnaire. The presence of metabolic disorders, diabetes mellitus, hypertriglyceridemia, hepatic steatosis, acute pancreatitis, fatty liver, proteinuria, hypertension, coronary heart disease, and polycystic ovarian syndrome were self-reported by the patients.
Mutational analyses
For both the UTSW and the NIDDK cohorts, the LMNA exons, including the splice site regions, were amplified in 11 segments [8] and PPARG exons in 7 segments [5, 9] from 50 ng of genomic DNA using the polymerase chain reaction and exon-specific primers pairs. The purified polymerase chain reaction products were sequenced using dye terminator chemistry and an ABI 3730xl DNA analyzer. Sequence variants were verified by manually inspecting the chromatograms of both the wild-type and mutated products. Pathogenicity of each variant was assessed according to genotype–phenotype segregation, functional studies, and ClinVar predictions (Tables 1 and 2).
Table 1.
Disease-causing heterozygous PPARG variants in subjects with FPLD3
| Patients (n) | Variant cDNA level | Variant protein level | rs number | Clinvar prediction | Functional score [7] | References |
|---|---|---|---|---|---|---|
| 1 | c.282delG | p.K94Nfs*4 | NA | NA | — | — |
| 1 | c.421G>C | p.V141L | NA | NA | −3.5 | — |
| 1 | c.452C>G | p.Y151C | rs1354592503 | Pathogenic | −4.2 | [10, 11] |
| 1 | c.480+120G>C | (?) | NA | NA | — | — |
| 1 | c.580C>T | p.R194W | rs121909146 | Pathogenic | −5.4 | [12] |
| 2 | c.641C>T | p.P214L | NA | NA | −0.48 | — |
| 3 | c.670G>T | p.E224* | NA | NA | — | — |
| 1 | c.971T>C | p.I324T | rs1378972597 | Pathogenic | −2.57 | — |
| 1 | c.1154G>A | p.R385Q | rs140204299 | Uncertain significance | −1.4 | [7] |
| 3 | c.1159C>T | p.P387S | NA | NA | −4.2 | [7, 9] |
| 2 | c.1184A>G | p.K395R | NA | NA | −1.16 | [7, 9] |
| 6 | c.1273C>T | p.R425C | rs72551364 | Pathogenic | −5.37 | [5] |
| 1 | c.1313A>C | p.Q438P | NA | Uncertain significance | −4.9 | [7, 9] |
| 2 | c.1393C>T | p.Q465* | NA | NA | — | — |
| 1 | c.1480+1G>A | (?) | NA | NA | — | — |
| 4 | c.1484C>T | p.P495L | rs121909244 | Pathogenic | −5.58 | [13–15] |
| 1 | c.1495delG | p.E499Rfs*12 | NA | NA | — | — |
Five variants were null or splice site variants. Eleven were missense variants, of which 5 were classified as “pathogenic” and 2 of “uncertain significance” by Clinvar. The 2 variants with “uncertain significance” have been published before. One variant is a novel intronic variant not present in the gnomAD database. For the 4 missense variants that had no information in Clinvar, we looked at the functional score according to Majithia et al [7]. All had negative scores. Two of them had functional scores ≤3.5, which has increased probability of lipodystrophy, and 1 of them, p.P387S, has been previously published by 2 groups. There was evidence of segregation in 3 family members for p.K395R and in 2 family members for p.P214L.
Abbreviation: NA, not available.
Table 2.
Disease-causing heterozygous LMNA variants in subjects with FPLD2
| Patients (n) | Variant cDNA level | Variant protein level | rs number | Clinvar prediction | References |
|---|---|---|---|---|---|
| 2 | c.74G>T | p.R25L | rs61578124 | Uncertain significance | [16] |
| 4 | c.82C>T | p.R28W | rs59914820 | Pathogenic | [9–9] |
| 1 | c.178C>G | p.R60G | rs28928900 | Pathogenic | [20, 21] |
| 8 | c.184C>G | p.R62G | rs56793579 | Pathogenic | [17, 20] |
| 1 | c.575A>T | p.D192V | rs57045855 | Not provided | [20] |
| 2 | c.1045C>T | p.R349W | rs267607555 | Pathogenic/likely pathogenic | [10, 22] |
| 5 | c.1255C>T | p.R419C | rs755686359 | Uncertain significance | [23] |
| 4 | c.1394G>A | p.G465D | rs61282106 | Pathogenic | [17] |
| 5 | c.1445G>T | p.R482L | rs11575937 | Uncertain significance | [17] |
| 92 | c.1444C>T | p.R482W | rs57920071 | Pathogenic | [17, 6–6] |
| 96 | c.1445G>A | p.R482Q | rs11575937 | Pathogenic | [17, 6–6] |
| 1 | c.1458G>T | p.K486N | rs59981161 | Pathogenic | [27] |
| 1 | c.1488+5G>C | NA | rs267607543 | Pathogenic | [28] |
| 2 | c.1567G>C | p.G523R | rs201583907 | Uncertain significance | [29] |
| 2 | c.1580G>C | p.R527P | rs57520892 | Pathogenic | [30] |
| 3 | c.1583C>T | p.T528M | rs57629361 | Uncertain significance | [31] |
| 1 | c.1622G>C | p.R541P | rs61444459 | Pathogenic/likely pathogenic | [32] |
| 1 | c.1744C>T | p.R582C | rs918645468 | Uncertain significance | [22, 33] |
| 12 | c.1745G>A | p.R582H | rs57830985 | Pathogenic | [17, 26, 34] |
| 10 | c.1748C>T | p.S583L | rs59601651 | Uncertain significance | [17] |
| 1 | c.1751G>A | p.R584H | rs56657623 | Uncertain significance | [17, 22] |
| 1 | c.1930C>T | p.R644C | rs142000963 | Benign; uncertain significance | [17, 35] |
All the variants have been associated with lipodystrophy and published before.
Abbreviation: NA, not available.
Biochemical analyses and procedures
Fasting blood samples were collected and were analyzed for biochemical variables. Blood samples for the UTSW patients were sent to Quest Diagnostics (Irving, Texas) for analysis. Serum glucose, lipids, lipoproteins, and liver enzymes of UTSW patients were measured by the photometric method (Beckman Coulter AU clinical analyzer). Blood hemoglobin A1c (HbA1c) was measured by the immunoturbidimetric method (Roche Integra 800 chemistry analyzer). Serum glucose, HbA1c, lipids, and hepatic function tests for the NIDDK samples were conducted in the National Institutes of Health Clinical Center laboratory according to standard methodology [9]. Low-density lipoprotein cholesterol was calculated using Friedewald's equation for those with serum triglycerides values less than 400 mg/dL [36].
Anthropometric measurements
Height and body weight were measured with standard procedures. Skinfold thickness was measured with a Lange caliper (Cambridge Scientific Industries, Cambridge, MD) on the right side of the body. The mean of 3 repeat measurements at each site was calculated.
Dual-energy X-ray absorptiometry
For the UTSW cohort, whole-body DXA scans were acquired on a Discovery W (S/N 80502) model machine according to the procedures recommended by the manufacturer (Hologic, Inc., Bedford, MA) [8, 37]. Subjects changed into a paper gown and were asked to remove all jewelry and other personal effects that could interfere with the DXA scan. Analysis of all scans was performed using Hologic Discovery software version 1.6.5.0 in its default configuration. The analysis of all regional body fat was performed using standard software. The regions of interest were delineated by the following lines: head, pelvis, trunk, hip, spine, leg, and groin as reported by Bazzocchi et al [38]. Regional fat mass values were grouped and analyzed for the following anatomical regions: upper limbs, lower limbs, trunk (chest, axillary, abdomen), and whole body. Mean values of the right and left upper limb fat and right and left lower limb fat (% of regional fat) were calculated. We also calculated the ratio of the lower limb fat to truncal fat. DXA scanning of NIDDK patients was performed on a QDR 4500 (Hologic, Bedford, MA) scanner using Apex 4.0 software and analyzed in the same manner [9].
Statistical Analysis
Data are presented by frequency (percent) for categorical variables, median (minimum and maximum values), or mean ± SD for continuous variables depending on the distribution. The distribution of the continuous variables was assessed by the Shapiro–Wilk normality test and normal probability plots. The comparisons between FPLD3 and FPLD2 were made using the chi square test for categorical variables and the t test for Gaussian distributed variables, and after log transformation for non-Gaussian distributed variables. The comparison of the variables between FPLD3 and FPLD2 were also adjusted by age and sex through linear regression models and the skewed data were log transformed prior to applying in the model.
All analyses were performed using SAS software version 9.4 (SAS Institute, Inc., Cary, NC). A 2-sided P < .05 was the criterion for statistical significance.
Results
Study Population and Demographics
A total of 287 patients with FPLD were included in the study. Of these, 32 (4 males [M], 28 females [F]) had FPLD3 with heterozygous disease-causing PPARG variants (Table 1). The mean ± SD age and body mass index of the subjects with FPLD3 were 40.7 ± 17.2 years and 25.9 ± 3.9 kg/m2, respectively. Seventy-one percent of the subjects were White, 3% were African American, and 25% were in the other category, with no Asian people. A total of 255 subjects (59 M, 196 F) had FPLD2 with heterozygous, disease-causing LMNA variants (Table 2). The mean ± SD age and body mass index of the subjects with FPLD2 were 36.8 ± 16.7 years and 25.5 ± 4.6 kg/m2, respectively. Most subjects (85%) were White, 3% were Asian, 1% were African American, and 11% were in the other category.
Prevalence of Disease-Causing Variants
As shown in Table 1, of the 32 subjects with heterozygous PPARG variants, 6 had the p.R425C variant; 4 had the p.P495L variant; 3 each had p.P387S; 2 each had p.K395R, and 1 each had the p.R194W, p.Y151C, p.I324T, p.R385Q, or p.Q438P variant, all previously reported and considered pathogenic variants according to Clinvar [7, 9, 10–15]. Others had novel heterozygous PPARG variants; of these, 3 had p.E224*; 2 each had p.P214L and p.Q465* variants; and 1 each had p.K94Nfs*4, p.E499Rfs*12, p.V141L, c.1480+1G>A, and c.480+120G>C variants.
As shown in Table 2, of the 255 subjects with heterozygous LMNA variants, 188 had the 2 most common heterozygous pathogenic variants (p.R482Q or p.R482W) associated with FPLD2 [17, 24–26]. The next most prevalent variant, p.R582H [17, 26, 34], was present in 12 patients, followed by p.S583L variant in 10 patients. Eight patients had p.R62G; 5 each had p.R419C and p.R482L; and 4 each had p.R28W and p.G465D [17]; 3 had p.T528M; 2 each had p.R25L, p.R349W, p.G523R, and p.R527P; and 1 each had p.R60G, p.D192V, p.K486N, p.R541P, p.R582C, p.R584H, and p.R644C variants. One subject had an LMNA splice site variant c.1488+5G>C; p (?) [10, 16–35].
Biochemical Parameters and Body Fat Distribution
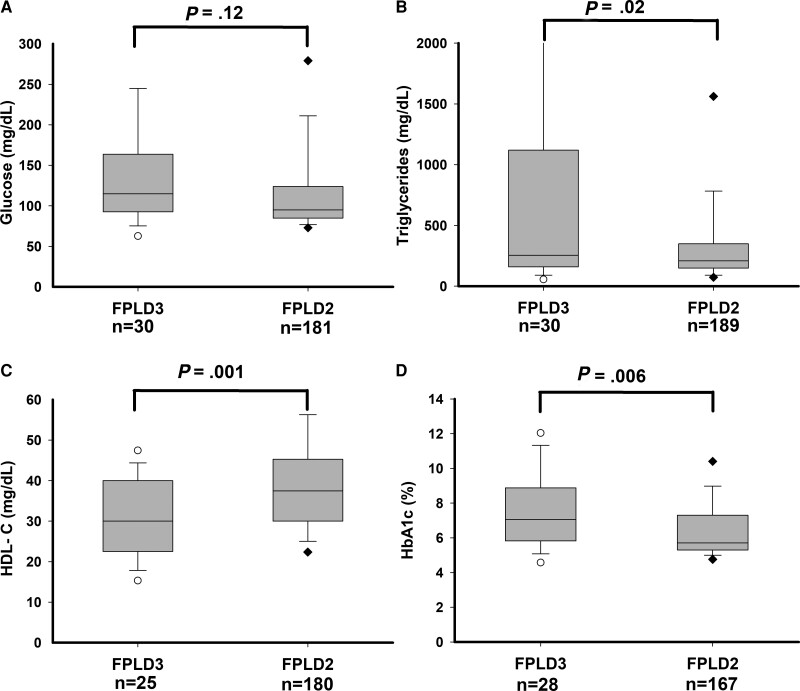
Comparison of biochemical measurements in subjects with FPLD3 and FPLD2 is shown in Table 3 and Fig. 1. Compared with subjects with FPLD2, those with FPLD3 had higher fasting serum triglyceride levels (median 208 vs 255 mg/dL, respectively; P = .018), lower high-density lipoprotein (HDL) cholesterol levels (mean 39.1 vs 30.5 mg/dL, respectively; P = .001), lower low-density lipoproteins cholesterol levels (mean 70 vs 107 mg/dL, respectively; P = .0001), higher blood HbA1c levels (mean 6.4% vs 7.5%, respectively; P = 0.001), but similar fasting serum glucose (median 95 vs 115 mg/dL, respectively; P = .12). There was no significant difference in serum alanine aminotransferase (median 27 vs 22 U/L, respectively; P = .43), aspartate aminotransferase (median 22 vs 22 U/L, respectively; P = .82), or alkaline phosphatase (median 62 vs 55 U/L, respectively; P = .15) levels in the 2 subtypes.
Table 3.
Comparison of biochemical measurements in subjects with familial partial lipodystrophy, FPLD3 and FPLD2
| FPLD3 | FPLD2 | |||||
|---|---|---|---|---|---|---|
| n | Value | n | Value | P value | P value adjusted for sex and age | |
| Sex (M/F) | 4/28 | 59/196 | .16 | NA | ||
| Age (y)a | 32 | 40.7 ± 17.2 | 255 | 36.8 ± 16.7 | .21 | NA |
| Height (cm)a | 28 | 163.4 ± 8.4 | 255 | 164.1 ± 13.8 | .81 | .94 |
| Weight (kg)a | 28 | 70.0 ± 15.7 | 255 | 69.8 ± 18.3 | .96 | .61 |
| BMI (kg/m2)a | 28 | 25.9 ± 3.9 | 255 | 25.5 ± 4.6 | .67 | .56 |
| HDL-Ca (mg/dL) | 25 | 30.5 ± 9.7 | 180 | 39.1 ± 12.4 | .001 | .0006 |
| LDL-Ca (mg/dL) | 19 | 69.6 ± 30.5 | 146 | 106.8 ± 37.2 | .0001 | .0001 |
| HbA1c (%)a | 28 | 7.5 ± 2.2 | 167 | 6.4 ± 1.8 | .006 | .012 |
| Cholesterol (mg/dL)b | 30 | 156 (80-429) | 186 | 191 (60-710) | .04 | .038 |
| Triglycerides (mg/dL)b | 30 | 255 (50-7919) | 189 | 208 (38-11528) | .018 | .04 |
| Glucose (mg/dL)b | 30 | 115 (50-355) | 181 | 95 (68-398) | .12 | .23 |
| ALT (U/L)b | 29 | 22 (8-59) | 55 | 27(10-130) | .43 | .51 |
| AST (U/L)b | 29 | 22 (12-68) | 55 | 22 (9-80) | .82 | .78 |
| ALP (U/L)b | 29 | 55 (29-344) | 51 | 62 (19-159) | .15 | .10 |
Abbreviations: ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; HbA1c, hemoglobin A1c; HDL, high-density lipoprotein; NA, not applicable.
Data shown as mean ± SD.
Data skewed and shown as median (minimum and maximum values).
Figure 1.
Fasting serum glucose, triglycerides, HDL cholesterol, and HbA1c levels in subjects with FPLD3 and FPLD2. (A) Fasting serum glucose, (B) fasting serum triglycerides, (C) HDL-cholesterol, and (D) blood hemoglobin A1c. Numbers of subjects for whom the value of the variable was available are shown below the x-axis. The gray box shows 25th and 75th percentile values with median shown as a horizontal line. The whiskers denote 5th and 95th percentiles. Individual values below the 5th and above the 95th percentile are shown. The 95th percentile of serum triglycerides (5577 mg/dL) for subjects with FPLD3 is not shown. Serum triglycerides exceeding 2000 mg/dL were seen in 5 subjects with FPLD3 (2104, 3115, 4127, 5577, and 7919 mg/dL) and 5 subjects with FPLD2 (2160, 2285, 7740, 9040, and 11 528 mg/dL), which are not shown in the figure. P values are shown above the box plots.
Compared with FPLD2 subjects, those with FPLD3 had higher prevalence of hypertriglyceridemia (66% vs 84%, respectively; P = .05), diabetes (44% vs 72%, respectively; P = .003), history of acute pancreatitis (13% vs 52%, respectively; P < .001), proteinuria (13% vs 26%, respectively; P = .06), and polycystic ovarian syndrome (26% vs 52%, respectively; P = .007), while there was no difference in the prevalence of fatty liver (45% vs 54%, respectively; P = .47), hypertension (41% vs 40%, respectively; P = .88), or coronary heart disease (15% vs 19%, respectively; P = .65) (Table 4).
Table 4.
Prevalence of metabolic disorders in subjects with familial partial lipodystrophy, FPLD3 and FPLD2
| FPLD3 | FPLD2 | |||||
|---|---|---|---|---|---|---|
| N Affected/Total | % Affected | N Affected/Total | % Affected | P value | P value Adjusted for sex and age | |
| Diabetes mellitus | 23/32 | 72.0 | 103/235 | 44.0 | .003 | .02 |
| Hypertriglyceridemia | 26/31 | 84.0 | 140/212 | 66.0 | .05 | .10 |
| Acute Pancreatitis | 15/29 | 52.0 | 32/238 | 13.0 | <.0001 | <.0001 |
| Fatty Liver | 15/28 | 54.0 | 23/51 | 45.0 | .47 | .32 |
| Hypertension | 12/30 | 40.0 | 99/239 | 41.0 | .88 | .41 |
| Proteinuria | 7/27 | 26.0 | 25/200 | 13.0 | .06 | .12 |
| CHD | 5/27 | 19.0 | 36/237 | 15.0 | .65 | .91 |
| PCOS | 14/27 | 52.0 | 42/161 | 26 | .007 | .009 |
Abbreviations: CHD, coronary heart disease; PCOS, polycystic ovarian syndrome.
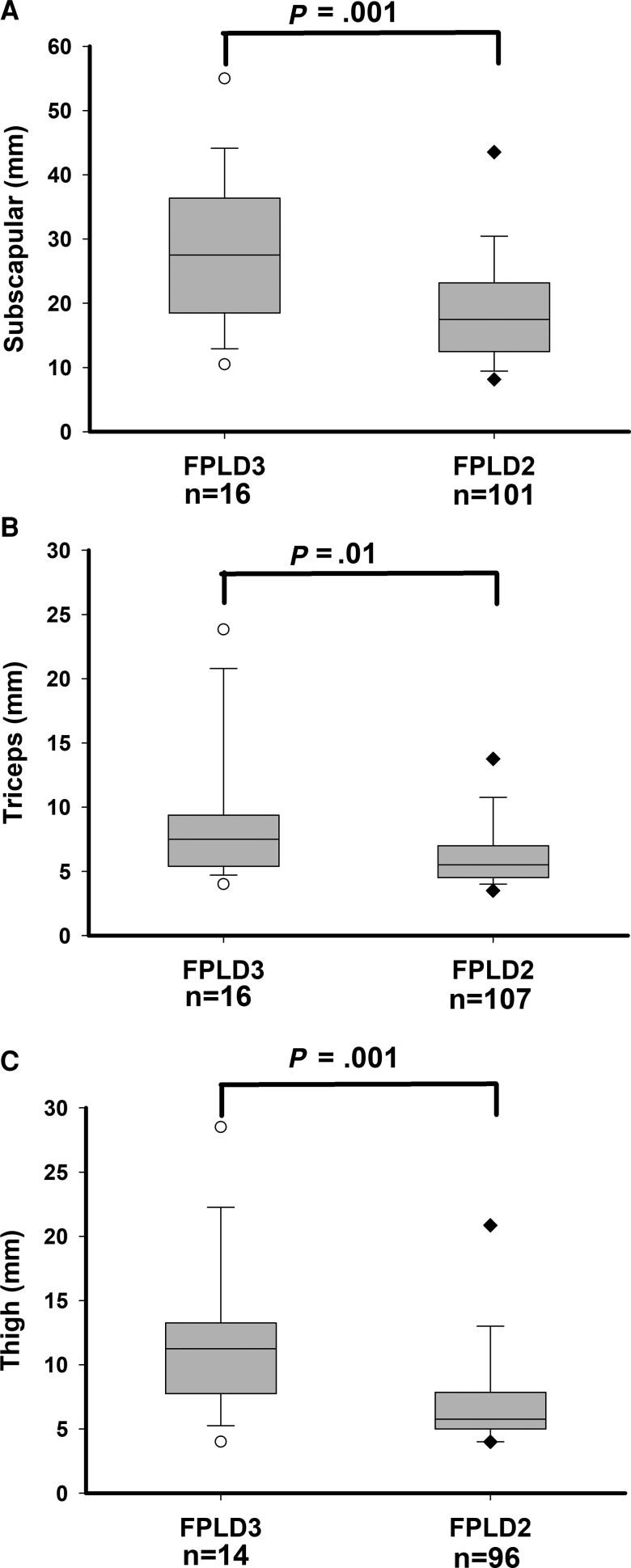
Compared with subjects with FPLD2, subjects with FPLD3 had higher skinfold thickness measurements in the lower and upper limbs: anterior thigh (median 5.8 mm vs 11.3 mm, respectively; P = .001), calf (median 4 mm vs 6 mm, respectively; P = .01), triceps (median 5.5 mm vs 7.5 mm, respectively; P = .01), and biceps (median 4.3 mm vs 6.8 mm, respectively; P = .001) (Table 5 and Fig. 2). Compared with subjects with FPLD2, subjects with FPLD3 also had higher skinfold thickness measurements in the truncal region: subscapular (median 17.5 mm vs 27.5 mm, respectively; P = .001), anterior abdomen (median 15.0 mm vs 24.5 mm, respectively; P = .001), suprailiac (median 9.3 mm vs 16 mm, respectively; P = .001), and axillary (median 12 mm vs 20 mm, respectively; P = .048) but had similar chest skinfold measurements.
Table 5.
Comparison of anthropometric parameters and body fat distribution in subjects with familial partial lipodystrophy, FPLD3 and FPLD2
| FPLD3 | FPLD2 | |||||
|---|---|---|---|---|---|---|
| n | Value | n | Value | P value | P value adjusted for sex and age | |
| Thigh skinfold (mm)a | 14 | 11.3 (4-28.5) | 96 | 5.8 (3-35) | .001 | .0006 |
| Subscapular skinfold (mm)a | 16 | 27.5 (11-55) | 101 | 17.5 (7-48) | .001 | .006 |
| Suprailiac skinfold (mm)a | 14 | 16 (7-45.5) | 103 | 9.3 (4-40) | .001 | .0003 |
| Triceps skinfold (mm)a | 16 | 7.5 (4-23.8) | 107 | 5.5 (3-28.5) | .01 | .004 |
| Biceps skinfold (mm)a | 16 | 6.8 (3-23) | 107 | 4.3 (2-33) | .001 | .002 |
| Calf skinfold (mm)a | 16 | 6 (4-17.5) | 104 | 4 (2-24.5) | .01 | .002 |
| Abdomen skinfold (mm)a | 15 | 24.5 (13-50.0) | 104 | 15.0 (4-60.3) | .001 | .0004 |
| Chest skinfold (mm)a | 9 | 10.5 (5.5-19.5) | 94 | 9.0 (4-46) | .39 | .57 |
| Axillary skinfold (mm)a | 9 | 20.0 (10-29) | 91 | 12 (5-41) | .048 | .11 |
| Upper limb fat (%)b | 18 | 27.3 ± 5.4 | 96 | 21.1 ± 7.8 | .002 | .002 |
| Lower limb fat (%)b | 18 | 20.5 ± 5.6 | 96 | 15.8 ± 6.1 | .003 | .004 |
| Truncal fat (%)b | 18 | 28.9 ± 6.0 | 97 | 26.6 ± 7.0 | .18 | .18 |
| Total fat (%)b | 19 | 26.0 ± 4.6 | 98 | 22.3 ± 6.1 | .013 | .01 |
| Lower limb fat/truncal fat ratiob | 18 | 0.73 ± 0.21 | 96 | 0.60 ± 0.20 | .02 | .013 |
Data skewed and are shown as median (minimum and maximum values).
Data shown as mean ± SD; from dual-energy X-ray absorptiometry.
Figure 2.
Skinfold thickness measurements in subjects with FPLD3 and subjects with FPLD2. (A) Subscapular skinfold thickness (mm) measurements, (B) triceps skinfold thickness (mm) measurements, and (C) thigh skinfold thickness (mm) measurements. Numbers of subjects for whom the value of the variable was available are shown below the x-axis. The gray box shows 25th and 75th percentiles with median shown as a horizontal line. The whiskers denote 5th and 95th percentiles. Individual values below the 5th and above the 95th percentile are shown. P values are shown above the box plots.
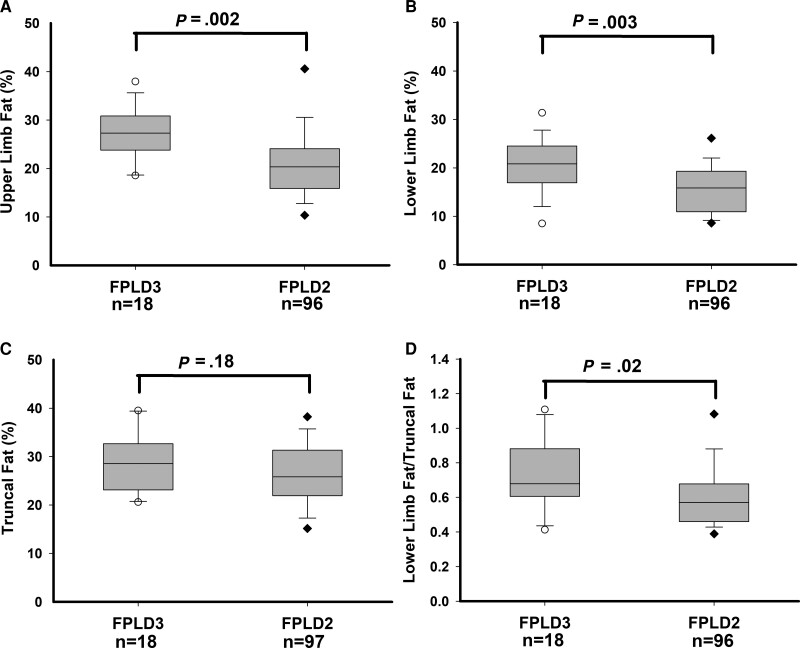
Regional body fat distribution data using DXA were available for 107 subjects with FPLD2 and 19 subjects with FPLD3 (Table 5 and Fig. 3). Compared to FPLD2 subjects, individuals with FPLD3 had higher total body fat (mean 22.3% vs 26.0%, respectively; P = .013), upper limb fat (mean 21.1% vs 27.3%, respectively; P = .002), and lower limb fat (mean 15.8% vs 20.5%, respectively; P = .003), while truncal fat was not different (mean 26.6% vs 28.9%, respectively; P = .18). Compared to FPLD2 subjects, those with FPLD3 also had a higher ratio of lower limb fat/truncal fat (mean 0.60 vs 0.73, respectively; P = .02).
Figure 3.
Regional body fat (% of fat mass/total regional mass) measurements by DXA scan in subjects with FPLD3 and FPLD2. (A) Upper limb fat (%) measurements, (B) lower limb fat (%) measurements, (C) truncal fat (%) measurements, and (D) ratio of lower limb fat/truncal fat. Numbers of subjects for whom the value of the variable was available are shown below the x-axis. The gray box shows 25th and 75th percentiles with median shown as a horizontal line. The whiskers denote 5th and 95th percentiles. Individual values below the 5th and above the 95th percentile are shown. P values are shown above the box plots.
The onset of FPLD2 occurs before the age of puberty [39] and the full phenotype of partial lipodystrophy is only evident during early adulthood. In addition, females with FPLD2 have been reported to have worse metabolic complications than males [40]. Therefore, we also compared all the variables after adjusting for age and sex but observed similar differences as noted when the variables were unadjusted (Tables 3-5). Since some children (<18 years of age) were included in the 2 cohorts (3 in FPLD3 and 41 in FPLD2), we conducted comparisons of all variables after excluding children and we observed similar phenotypic differences, metabolic parameters and prevalence of metabolic complications among adults with FPLD3 and FPLD2 as seen in the entire cohort (Tables S1-S3 [41]).
Discussion
Patients with partial or generalized lipodystrophy syndromes are predisposed to metabolic complications of insulin resistance. The prevailing understanding is that the severity of metabolic complications is correlated with the extent of fat loss [2, 3]. For example, patients with congenital generalized and acquired generalized lipodystrophy syndromes, who have near total absence of body fat, have earlier onset and more severe metabolic complications than those with FPLD or acquired partial lipodystrophy [3]. Compared with partial lipodystrophy syndromes, generalized lipodystrophy syndromes are characterized by near-total lack of adipose tissue and, therefore, adipose tissue depots are not available for storage of circulating triglycerides. These lipids instead “spill over” to ectopic sites such as the liver and skeletal muscles, inducing steatosis and insulin resistance. Our findings that subjects with FPLD3 have more severe metabolic complications of lipodystrophy despite having higher body fat than those with FPLD2 challenge the notion that the extent of anatomic fat loss determines the severity of metabolic derangements in patients with lipodystrophy.
Patients with FPLD3 had more body fat in the central and peripheral body regions (ie, less severe lipodystrophy) than those with FPLD2 based on regional body fat determination by DXA scan, and by measurements of skinfold thickness. However, compared with patients with FPLD2, those with FPLD3 had higher prevalence of diabetes, hypertriglyceridemia, acute pancreatitis, and polycystic ovarian syndrome. They also had higher fasting serum triglycerides and HbA1c. Interestingly, there was no difference in the prevalence of hypertension, hepatic steatosis, and serum levels of liver enzymes, aspartate aminotransferase, or alanine aminotransferase. However, the severity of hepatic steatosis was not assessed in our study by either magnetic resonance spectroscopy or magnetic resonance imaging, which can quantitate the amount of hepatic lipids. Interestingly, similar observations were made by previous investigators in the phenotypic differences among small cohorts of patients with FPLD2 and FPLD3 [42]. It is intriguing to speculate what mechanisms could be responsible for these paradoxical results. Since both patients with FPLD2 and patients with FPLD3 have only partial loss of body fat, mainly from the extremities [10, 43, 44], our data suggest that adipocytes from the nonlipodystrophic regions in patients with FPLD3 may be more dysfunctional than those from patients with FPLD2. Okuno et al [45] reported that troglitazone administration for 15 days normalized mild hyperglycemia and marked hyperinsulinemia in the obese Zucker rats and increased the number of small adipocytes by about 4-fold in both retroperitoneal and subcutaneous adipose tissues, which could be responsible for improved insulin sensitivity. These observations suggest that patients with FPLD3 with PPARG variants may have a reduced number of small, insulin-sensitive adipocytes with preservation of large adipocytes, and this could be another underlying mechanism explaining less severe lipodystrophy despite more insulin resistance in FPLD3 patients compared with patients with FPLD2.
While the precise molecular mechanisms by which LMNA and PPARG variants cause loss of extremity fat are not clearly elucidated, they appear to be strikingly different. For example, LMNA encodes nuclear lamina proteins, lamins A and C, which are widely expressed including in the adipose tissue [46, 47]. Lamins A and C not only interact with the proteins embedded in the inner nuclear membrane but also can directly interact with chromatin [3]. It is believed that LMNA pathogenic variants cause loss of subcutaneous fat from the extremities as a result of nuclear dysfunction resulting in early senescence or apoptosis of adipocytes in these regions [46, 47]. However, why body fat from the face, neck, intra-abdominal region remains well preserved in patients with FPLD2 is not clear. In fact, there is excess fat accumulation in these nonlipodystrophic regions in many patients with FPLD2. For example, patients with FPLD2 have been reported to have increased intra-abdominal fat accumulation on magnetic resonance imaging [43] and autopsy [48, 49]. It is likely that these fat depots are not adversely affected metabolically and participate efficiently in triglyceride storage and release, similar to normal healthy adipose tissue.
On the other hand, PPARᵧ is the master ligand-dependent nuclear transcription factor involved in adipogenesis [50, 51]. PPARG encodes 2 isoforms by alternative splicing, PPARᵧ1 and PPARᵧ2. While PPARᵧ2 is expressed exclusively in adipocytes, PPARᵧ1 is expressed ubiquitously [50, 51]. PPARᵧ also plays a role in lipid storage in adipocytes. We hypothesize that PPARG pathogenic variants causing FPLD3 result in loss of subcutaneous fat from the extremities but also induce a generalized metabolic dysfunction in the remaining, well-preserved, adipose tissue depots, such as those inside the abdomen. This metabolic dysfunction may affect lipogenesis and triglyceride storage as well as lipolysis from the adipocytes. Some of the pathogenic variants in PPARG, but not all, have been documented to be dominant negative [14, 52–54], thus affecting negatively the function of the wild-type expressed protein. Therefore, it is likely that these dominant negative variants may cause worse adipocyte dysfunction but only mild to moderate loss of adipocyte mass. In contrast, LMNA variants may simply affect adipocytes by haploinsufficiency.
Another potential factor contributing to worse metabolic disease despite greater preservation of adipose tissue in FPLD3 vs FPLD2 relates to ease of clinical recognition. Underdiagnosis of FPLD2 has been well documented [55]. As the loss of subcutaneous fat from the extremities in FPLD3 is more subtle than that seen in FPLD2, it is likely that an even a lower proportion of patients with FPLD3 are diagnosed compared with FPLD2. Thus, it is possible that the more severe metabolic disease observed in FPLD3 in this study is due to greater recognition and referral of patients with FPLD3 who have severe metabolic disease to the tertiary referral centers [44].
Thus, future molecular and metabolic studies of nonlipodystrophic adipose tissue from patients with FPLD2 and FPLD3 may provide clues to their dysfunction. Our data suggest that beyond loss of adipose tissue, dysfunction of adipocytes also can contribute to metabolic abnormalities in patients with lipodystrophies.
Acknowledgments
The authors thank the families for participation in the study; Elaine Cochran from NIDDK, NIH, Bethesda, USA, and Claudia Quittner for help with evaluation of patients; Carmel Tovar for LMNA and PPARG sequencing; and UTSW McDermott Center Sequencing Core for sequencing at the UT Southwestern Medical Center, Dallas, TX.
Abbreviations
- DXA
dual-energy X-ray absorptiometry
- FPLD
familial partial lipodystrophy
- HbA1c
hemoglobin A1c
- HDL
high-density lipoprotein
- NIDDK
National Institute of Diabetes and Digestive and Kidney Diseases
- UTSW
UT Southwestern Medical Center
Contributor Information
Chandna Vasandani, Division of Nutrition and Metabolic Diseases and the Center for Human Nutrition, Department of Internal Medicine, UT Southwestern Medical Center, Dallas, TX 75390, USA.
Xilong Li, Department of Population and Data Sciences, UT Southwestern Medical Center, Dallas, TX 75390, USA.
Hilal Sekizkardes, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA.
Rebecca J Brown, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Abhimanyu Garg, Email: abhimanyu.garg@utsouthwestern.edu, Division of Nutrition and Metabolic Diseases and the Center for Human Nutrition, Department of Internal Medicine, UT Southwestern Medical Center, Dallas, TX 75390, USA.
Funding
This work was supported by grants from the National Institutes of Health, R01-DK105448, and UL1TR001105 from the National Center for Advancing Translational Sciences, National Institutes of Health, and by the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases. The funding sources were not involved in study design, analysis and interpretation of data, writing of the paper, and in the decision to submit the article for publication. REDCap was used for data storage.
Author Contributions
C.V. reviewed the medical records, collected and organized the data, performed data interpretation, drafted the initial manuscript, and approved the final manuscript as submitted. X.L. carried out the statistical analysis, reviewed and revised the manuscript, and approved the final manuscript as submitted. H.S. collected and organized the NIH data, reviewed and revised the manuscript, and approved the final manuscript as submitted. R.J.B. performed data interpretation, reviewed and revised the manuscript, and approved the final manuscript as submitted. A.G. conceptualized and designed the study, performed data interpretation, reviewed and revised the manuscript, and approved the final manuscript as submitted.
Conflict of Interest
A.G. consults for Amryt Pharma plc, Regeneron, Kyttaro Limited, Third Rock Ventures, and has received grant support from Amryt Pharma plc, Regeneron, Quintiles, Akcea Pharmaceuticals, and Intercept Pharmaceuticals. R.J.B. has received scientific writing support from Aegerion Pharmaceuticals. The remaining authors have nothing to disclose.
Data Availability
The data sets generated during and/or analyzed during the present study are not publicly available but are available from the corresponding author on reasonable request.
Prior Presentation
An abstract was submitted and presented as a poster at the Endocrine Society Meeting in 2020.
Guarantor Statement
A.G. is the guarantor of this work and as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1. Brown RJ, Araujo-Vilar D, Cheung PT, et al. The diagnosis and management of lipodystrophy syndromes: a multi-society practice guideline. J Clin Endocrinol Metab. 2016;101(12):4500–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hussain I, Patni N, Garg A. Lipodystrophies, dyslipidaemias and atherosclerotic cardiovascular disease. Pathology. 2019;51(2):202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garg A. Acquired and inherited lipodystrophies. N Engl J Med. 2004;350(12):1220–1234. [DOI] [PubMed] [Google Scholar]
- 4. Guenantin AC, Briand N, Bidault G, et al. Nuclear envelope-related lipodystrophies. Semin Cell Dev Biol. 2014;29:148–157. [DOI] [PubMed] [Google Scholar]
- 5. Agarwal AK, Garg A. A novel heterozygous mutation in peroxisome proliferator-activated receptor-gamma gene in a patient with familial partial lipodystrophy. J Clin Endocrinol Metab. 2002;87(1):408–411. [DOI] [PubMed] [Google Scholar]
- 6. Semple RK, Chatterjee VK, O’Rahilly S. PPAR gamma and human metabolic disease. J Clin Invest. 2006;116(3):581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Majithia AR, Tsuda B, Agostini M, et al. Prospective functional classification of all possible missense variants in PPARG. Nat Genet. 2016;48(12):1570–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vasandani C, Li X, Sekizkardes H, Adams-Huet B, Brown RJ, Garg A. Diagnostic value of anthropometric measurements for familial partial lipodystrophy, Dunnigan variety. J Clin Endocrinol Metab. 2020;105(7):2132–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sekizkardes H, Cochran E, Malandrino N, Garg A, Brown RJ. Efficacy of metreleptin treatment in familial partial lipodystrophy due to PPARG vs LMNA pathogenic variants. J Clin Endocrinol Metab. 2019;104(8):3068–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Akinci B, Onay H, Demir T, et al. Clinical presentations, metabolic abnormalities and end-organ complications in patients with familial partial lipodystrophy. Metabolism. 2017;72:109–119. [DOI] [PubMed] [Google Scholar]
- 11. Visser ME, Kropman E, Kranendonk ME, et al. Characterisation of non-obese diabetic patients with marked insulin resistance identifies a novel familial partial lipodystrophy-associated PPARgamma mutation (Y151C). Diabetologia. 2011;54(7):1639–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Monajemi H, Zhang L, Li G, et al. Familial partial lipodystrophy phenotype resulting from a single-base mutation in deoxyribonucleic acid-binding domain of peroxisome proliferator-activated receptor-gamma. J Clin Endocrinol Metab. 2007;92(5):1606–1612. [DOI] [PubMed] [Google Scholar]
- 13. Broekema MF, Savage DB, Monajemi H, Kalkhoven E. Gene-gene and gene-environment interactions in lipodystrophy: lessons learned from natural PPARγ mutants. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864(5):715–732. [DOI] [PubMed] [Google Scholar]
- 14. Barroso I, Gurnell M, Crowley VE, et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402(6764):880–883. [DOI] [PubMed] [Google Scholar]
- 15. Tsai YS, Kim HJ, Takahashi N, et al. Hypertension and abnormal fat distribution but not insulin resistance in mice with P465L PPARgamma. J Clin Invest. 2004;114(2):240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Florwick A, Dharmaraj T, Jurgens J, Valle D, Wilson KL. LMNA Sequences of 60,706 unrelated individuals reveal 132 novel missense variants in A-type lamins and suggest a link between variant p.G602S and type 2 diabetes. Front Genet. 2017;8:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garg A. Lipodystrophies. In: Weiss RE, Refetoff S, eds. Genetic Diagnosis of Endocrine Disorders. 2nd ed. Elsevier; 2016:325–339. [Google Scholar]
- 18. Decaudain A, Vantyghem MC, Guerci B, et al. New metabolic phenotypes in laminopathies: LMNA mutations in patients with severe metabolic syndrome. J Clin Endocrinol Metab. 2007;92(12):4835–4844. [DOI] [PubMed] [Google Scholar]
- 19. Garg A, Speckman RA, Bowcock AM. Multisystem dystrophy syndrome due to novel missense mutations in the amino-terminal head and alpha-helical rod domains of the lamin A/C gene. Am J Med. 2002;112(7):549–555. [DOI] [PubMed] [Google Scholar]
- 20. Subramanyam L, Simha V, Garg A. Overlapping syndrome with familial partial lipodystrophy, Dunnigan variety and cardiomyopathy due to amino-terminal heterozygous missense lamin A/C mutations. Clin Genet. 2010;78(1):66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van der Kooi AJ, Bonne G, Eymard B, et al. Lamin A/C mutations with lipodystrophy, cardiac abnormalities, and muscular dystrophy. Neurology. 2002;59(4):620–623. [DOI] [PubMed] [Google Scholar]
- 22. Mory PB, Crispim F, Freire MB, et al. Phenotypic diversity in patients with lipodystrophy associated with LMNA mutations. Eur J Endocrinol. 2012;167(3):423–431. [DOI] [PubMed] [Google Scholar]
- 23. Haque WA, Oral EA, Dietz K, Bowcock AM, Agarwal AK, Garg A. Risk factors for diabetes in familial partial lipodystrophy, Dunnigan variety. Diabetes Care. 2003;26(5):1350–1355. [DOI] [PubMed] [Google Scholar]
- 24. Cao H, Hegele RA. Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum Mol Genet. 2000;9(1):109–112. [DOI] [PubMed] [Google Scholar]
- 25. Shackleton S, Lloyd DJ, Jackson SN, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet. 2000;24(2):153–156. [DOI] [PubMed] [Google Scholar]
- 26. Speckman RA, Garg A, Du F, et al. Mutational and haplotype analyses of families with familial partial lipodystrophy (Dunnigan variety) reveal recurrent missense mutations in the globular C-terminal domain of lamin A/C. Am J Hum Genet. 2000;66(4):1192–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simon DN, Domaradzki T, Hofmann WA, Wilson KL. Lamin A tail modification by SUMO1 is disrupted by familial partial lipodystrophy-causing mutations. Mol Biol Cell. 2013;24(3):342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morel CF, Thomas MA, Cao H, et al. A LMNA splicing mutation in two sisters with severe Dunnigan-type familial partial lipodystrophy type 2. J Clin Endocrinol Metab. 2006;91(7):2689–2695. [DOI] [PubMed] [Google Scholar]
- 29. Fichna JP, Macias A, Piechota M, et al. Whole-exome sequencing identifies novel pathogenic mutations and putative phenotype-influencing variants in Polish limb-girdle muscular dystrophy patients. Hum Genomics. 2018;12(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bonne G, Di Barletta MR, Varnous S, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet. 1999;21(3):285–288. [DOI] [PubMed] [Google Scholar]
- 31. Savage DB, Soos MA, Powlson A, et al. Familial partial lipodystrophy associated with compound heterozygosity for novel mutations in the LMNA gene. Diabetologia. 2004;47(4):753–756. [DOI] [PubMed] [Google Scholar]
- 32. de Andrade NXS, Adiyaman SC, Yuksel BD, et al. Unusual presentations of LMNA-associated lipodystrophy with complex phenotypes and generalized fat loss: when the genetic diagnosis uncovers novel features. AACE Clin Case Rep. 2020;6(2):e79–e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Montenegro RM J, Costa-Riquetto AD, Fernandes VO, et al. Homozygous and heterozygous nuclear lamin A p.R582C mutation: different lipodystrophic phenotypes in the same kindred. Front Endocrinol (Lausanne). 2018;9:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garg A, Vinaitheerthan M, Weatherall PT, Bowcock AM. Phenotypic heterogeneity in patients with familial partial lipodystrophy (Dunnigan variety) related to the site of missense mutations in lamin a/c gene. J Clin Endocrinol Metab. 2001;86(1):59–65. [DOI] [PubMed] [Google Scholar]
- 35. Mercuri E, Brown SC, Nihoyannopoulos P, et al. Extreme variability of skeletal and cardiac muscle involvement in patients with mutations in exon 11 of the lamin A/C gene. Muscle Nerve. 2005;31(5):602–609. [DOI] [PubMed] [Google Scholar]
- 36. Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18(6):499–502. [PubMed] [Google Scholar]
- 37. Kelly TL, Wilson KE, Heymsfield SB. Dual energy X-ray absorptiometry body composition reference values from NHANES. PLoS One. 2009;4(9):e7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bazzocchi A, Ponti F, Albisinni U, Battista G, Guglielmi G. DXA: technical aspects and application. Eur J Radiol. 2016;85(8):1481–1492. [DOI] [PubMed] [Google Scholar]
- 39. Patni N, Li X, Adams-Huet B, Vasandani C, Gomez-Diaz RA, Garg A. Regional body fat changes and metabolic complications in children with Dunnigan lipodystrophy-causing LMNA variants. J Clin Endocrinol Metab. 2019;104(4):1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garg A. Gender differences in the prevalence of metabolic complications in familial partial lipodystrophy (Dunnigan variety). J Clin Endocrinol Metab. 2000;85(5):1776–1782. [DOI] [PubMed] [Google Scholar]
- 41. Vasandani C, Li X, Sekizkardes H, Brown RJ, Garg A. Supplementary data from: Phenotypic differences among familial partial lipodystrophy due to LMNA or PPARG variants 2022. UT Southwestern Institutional Repository 2022. Deposited 5 August 2022. [DOI] [PMC free article] [PubMed]
- 42. Hegele RA, Joy TR, Al-Attar SA, Rutt BK. Thematic review series: adipocyte biology. Lipodystrophies: windows on adipose biology and metabolism. J Lipid Res. 2007;48(7):1433–1444. [DOI] [PubMed] [Google Scholar]
- 43. Garg A, Peshock RM, Fleckenstein JL. Adipose tissue distribution pattern in patients with familial partial lipodystrophy (Dunnigan variety). J Clin Endocrinol Metab. 1999;84(1):170–174. [DOI] [PubMed] [Google Scholar]
- 44. Malandrino N, Reynolds JC, Brychta RJ, et al. Visceral fat does not contribute to metabolic disease in lipodystrophy. Obes Sci Pract. 2019;5(1):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Okuno A, Tamemoto H, Tobe K, et al. Troglitazone increases the number of small adipocytes without the change of white adipose tissue mass in obese Zucker rats. J Clin Invest. 1998;101(6):1354–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boguslavsky RL, Stewart CL, Worman HJ. Nuclear lamin A inhibits adipocyte differentiation: implications for Dunnigan-type familial partial lipodystrophy. Hum Mol Genet. 2006;15(4):653–663. [DOI] [PubMed] [Google Scholar]
- 47. Worman HJ, Ostlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2(2):a000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haque WA, Vuitch F, Garg A. Post-mortem findings in familial partial lipodystrophy, Dunnigan variety. Diabet Med. 2002;19(12):1022–1025. [DOI] [PubMed] [Google Scholar]
- 49. Ludtke A, Roos GM, van Hettinga M, Horst BA, Worman HJ, Schmidt HH. Post-mortem findings in Dunnigan-type familial partial lipodystrophy. Diabet Med. 2010;27(2):245–246. [DOI] [PubMed] [Google Scholar]
- 50. Aprile M, Cataldi S, Perfetto C, et al. In-vitro-generated hypertrophic-like adipocytes displaying PPARG isoforms unbalance recapitulate adipocyte dysfunctions in vivo. Cells. 2020;9(5):1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. [DOI] [PubMed] [Google Scholar]
- 52. Agostini M, Schoenmakers E, Mitchell C, et al. Non-DNA binding, dominant-negative, human PPARγ mutations cause lipodystrophic insulin resistance. Cell Metab. 2006;4(4):303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gurnell M, Wentworth JM, Agostini M, et al. A dominant-negative peroxisome proliferator-activated receptor gamma (PPARgamma) mutant is a constitutive repressor and inhibits PPARgamma-mediated adipogenesis. J Biol Chem. 2000;275(8):5754–5759. [DOI] [PubMed] [Google Scholar]
- 54. Aprile M, Cataldi S, Ambrosio MR, et al. PPARγΔ5, a naturally occurring dominant-negative splice isoform, impairs PPARγ function and adipocyte differentiation. Cell Rep. 2018;25(6):1577–1592.e6. [DOI] [PubMed] [Google Scholar]
- 55. Gonzaga-Jauregui C, Ge W, Staples J, et al. Clinical and molecular prevalence of lipodystrophy in an unascertained large clinical care cohort. Diabetes. 2020;69(2):249–258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated during and/or analyzed during the present study are not publicly available but are available from the corresponding author on reasonable request.