ABSTRACT
The initiation of a spatially and temporally limited inflammation is essential for tissue and bone remodelling by the periodontal ligament (PdL) located between teeth and alveolar bone. Nutritional components may cause alterations in the inflammatory response of PdL fibroblasts to mechanical stress such as those occurring during orthodontic tooth movement (OTM). Recently, we reported an attenuated pro-inflammatory response of human PdL fibroblasts (HPdLFs) to compressive forces when stimulated with oleic acid (OA), a monounsaturated fatty acid particularly prominent in the Mediterranean diet. Fatty acids could serve as alternative source of acetyl-CoA, thereby affecting epigenetic histone marks, such as histone 3 lysine acetylation (H3Kac) in a lipid metabolism-dependent manner. In this study, we aimed to investigate the extent to which OA exerts its anti-inflammatory effect in compressed HPdLFs via changes in H3Kac. Six-hour compressed HPdLFs showed increased H3Kac when cultured with OA. Inhibition of histone deacetylases resulted in a comparable IL10-increase as observed in compressed OA-cultures. In contrast, inhibition of histone acetyltransferases, particularly p300/CBP, in compressed HPdLFs exposed to OA normalized the inflammatory response to control levels. OA-dependent increased association of H3Kac to IL10 promoter regions in compressed HPdLFs further strengthened the assumption that OA exhibits its anti-inflammatory properties via modulation of this epigenetic mark. In conclusion, our study strongly suggests that nutritional components can directly affect PdL cells via changes in their epigenetic code. Since epigenetic inhibitors are already widely used clinically, they may hold promise for novel approaches for personalized orthodontic treatment that incorporates nutritional and metabolism-related changes.
KEYWORDS: Histone acetylation, periodontal ligament, tooth movement, oleic acid, IL10
Introduction
Oral health and the functions of the various cell types in the oral cavity can be affected by dietary components, including specific micro-nutrients such as vitamins and minerals, as well as macro-nutrients including carbohydrates, proteins, and fatty acids [1]. Thus, a bidirectional relationship exists as nutritional components may influence oral health, but conversely, compromised oral cavity integrity may also impair an individual`s eating ability [2]. In addition to oral infectious diseases, particularly both acute and chronic systemic diseases with an impact on oral health such as obesity-linked disorders may compromise the patient’s nutritional status [3]. Similarly, inadequate nutrition due to an unbalanced diet can affect the development and integrity of the oral cavity and the progression of oral diseases [1].
Under healthy conditions, fatty acids (FA) are important macro-nutrients required for normal cellular functions [4,5]. However, excessive or imbalanced supply can impair these processes and thus alter cell-specific gene expression [6–13]. While saturated fatty acids (SFAs) have been shown to stimulate the expression of genes encoding pro-inflammatory cytokines such as tumour necrosis factor α (TNFα), interleukin 6 (IL6), IL8, IL1α, and IL1β [6–11,14,15], monounsaturated fatty acids (MUFAs) are mostly considered to have anti-inflammatory effects [9,12,13,16,17]. Among those MUFAs, oleic acid (OA; omega-9 fatty acid; 18:1), which is predominantly present in the Mediterranean diet, has been shown to counterbalance pro-inflammatory effects of SFAs [9,13,16,17]. In this context, OA-related increased anti-inflammatory IL10 secretion was reported [18], contributing to the down-regulation of pro-inflammatory cytokines [19,20]. Moreover, there is evidence that an OA-enriched diet contributes to the treatment and prevention of obesity, as it can lead to a reduction in abdominal fat and thus positively influence body composition [21].
We recently reported that OA in contrast to the SFA palmitic acid (PA) attenuates the force-induced pro-inflammatory response of human periodontal ligament fibroblasts (HPdLFs) [22], which are the main cell type of the periodontal ligament (PdL). As connective tissue between the teeth and the alveolar bone, the PdL has important functions in modulating the response of soft and hard tissues to mechanical stimuli, such as those that occur during trauma and mastication, but also in orthodontic treatments [23]. During tissue remodelling caused by mechanical stimulation, HPdLFs promote a transient aseptic tissue inflammation, which is essential for tissue and bone remodelling processes [24]. Interfering with inflammatory signalling may affect oral tissue and bone remodelling during orthodontic treatment, which could be critical in regard to risks, such as attachment loss, root resorption, and tooth loss [25,26]. However, the reasons explaining how OA exerts anti-inflammatory effects on the meachanoreactive inflammatory response of HPdLFs that may influence bone remodelling processes remained unclear.
Interestingly, studies indicate that fatty acids can serve as an alternative source of acteyl-coenzyme A (acetyl-CoA) [4,5], which is a key metabolic intermediate that particularly affects the activity and specificity of various enzymes as well as the acetylation state of a wide variety of proteins, including histones [27]. This highly dynamic histone modification alters chromatin architecture and regulates the accessibility of genes without direct changes to the DNA, therefore being assigned to epigenetic regulatory mechanisms [28]. There are various histone acetyltransferases (HATs), known as epigenetic writers, which perform target-specific acetylation of lysines of histone tail proteins, for which acetyl-CoA is required [27]. Among these are the E1A binding protein p300 and the CREB-binding protein (CREBBP or CBP), which in combination with their HAT function additionally act as adaptor proteins and recruit the basal transcriptional machinery, thereby promoting gene expression of corresponding genes [29]. Epigenetic erasers in this process are histone deacetylases (HDACs), which specifically remove acetylation marks on histone lysines and reversing site-specific transcriptional activation [28]. Overactive or dysfunctional HATs or HDACs could cause widespread changes in the cellular transcriptional profile, which however, might also dependent on their functional interplay [30]. While substrate specificity of HATs is mainly determined by multi-protein complexes, the availability of acetyl-CoA may also influence both HAT specificity and activity [31,32]. In this context, it has been demonstrated that increased availability of various fatty acids and thus of FA-derived acetyl-CoA rapidly induces the acetylation of histones [5].
In view of the potential effect of increased OA exposure on acetyl-CoA levels and thus on histone acetylation, our aim was to analyse to what extent this contributes to its anti-inflammatory effect in compressed HPdLFs. Thus, the results of our study should contribute to the understanding of how metabolic changes due to dietary composition affect the epigenetic regulation of inflammatory markers in the PdL and thus may have implications for the mechanoreactive response of PdL fibroblasts.
Materials and methods
Cell culture
Commercially acquired human periodontal ligament fibroblasts (HPdLFs, Lonza, Basel, Switzerland) are pooled from several donors. They were grown in Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific, Carlsbad, CA, USA) containing 4.5 g/L glucose, 10% heat-inactivated foetal bovine serum (Thermo Fisher Scientific, Carlsbad, CA, USA), 100 U/ml penicillin, 100 µg/ml streptomycin and 50 mg/L L-ascorbic acid at 37°C, 5% CO2, and 95% humidity. When reaching 75% confluency, HPdLFs were passaged with 0.05% Trypsin/EDTA (Thermo Fisher Scientific, Carlsbad, CA, USA). Passage four to eight were used for experimental setups. For transcriptional analysis, HDAC and HAT activity assay as well as CHIP, 2.5 × 104 HPdLFs were seeded into each well of a 6-well plate. For TPH1 adhesion assay, 5 × 103 HPdLFs were seeded on glass coverslips into each well of a 24-well-plate.
THP1 cells (DMSZ, Braunschweig, Germany) were cultured in RPMI 1640 medium (Thermo Fisher Scientific, Carlsbad, CA, USA) that contains 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C, 5% CO2 and 95% humidity. Passages were performed weekly and 1 × 106 cells were reseeded into a T175 culture flask (Thermo Fisher Scientific, Carlsbad, CA, USA).
Fatty acid exposure
Prior treatment with 200 µM oleic acid (OA) for six days, freshly seeded HPdLFs were subsequently cultured for 24 h in culture medium. The stimulation with OA was performed as described previously [33]. Briefly, OA was dissolved at 70°C in sterile water containing 50 mM NaOH, complexed with 37°C preheated bovine serum albumin (BSA, Seqens IVD, Limoges, France) and diluted in culture medium. As control, 0.66% BSA was applied.
Inhibitor treatment
Different concentrations of FK228 between 10 nM and 100 nM were used to inhibit histone deacetylases in 75% confluent HPdLFs. For inhibition of histone acetyltransferases, 1 nM to 10 nM anacardic acid (AA) or 1 nM to 10 nM C646 were applied. For final experiments, 10 nM FK228, 10 nM AA or 1 nM C646 were used. Since the inhibitors were dissolved in DMSO, this was used as control.
Mechanical compression
HPdLFs were stimulated by compressive force as previously described [22]. Briefly, sterile glass plates with 2 g/cm2 were placed in 6-well plates for six hours at 37°C, 5% CO2, and 95% humidity. In 24-well plates, the application of compressive force was performed by centrifugation for six hours at 37°C and a force of 7.13 g/cm2, which was the minimal condition of the centrifuge.
THP1 cell adherence assay
For visualization of the inflammatory response of specifically stimulated HPdLFs, analysis of THP1 cell adhesion was performed as previously described [22]. Briefly, after staining of non-adherent cells with 15 µM Celltracker CMFDA (Thermo Fisher Scientific, Carlsbad, CA, USA) for 30 min at 37°C, 50 × 103 cells were added to each well of cultured HPdLFs in a 24-well plate. After cell adhesion for 30 min at 37°C, 5% CO2, and 95% humidity, non-adhered THP1 cells were removed with prewarmed PBS and remaining cells were fixated in 4% paraformaldehyde for 10 min. Cell nuclei were stained for 5 min with DAPI (1:10,000 in PBS) and coverslips were embedded with Mowiol® 4–88 (Carl Roth, Karlsruhe, Germany) on glass object slides for microscopic imaging.
RNA extraction and quantitative PCR
TRIzol Reagent (Thermo Fisher Scientific, Carlsbad, CA, USA)/1-bromo-3-chloropropane was used to isolate RNA of specifically treated HPdLFs for expression analysis. After purification with the RNA Clean and Concentrator-5 kit (Zymo Research, Freiburg, Germany), quantity and quality was tested with Nanodrop 2000 (Avantor, Radnor, PA, USA). cDNA synthesis was performed with SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, Carlsbad, CA, USA) and Oligo(dt)18 primers (Thermo Fisher Scientific, Carlsbad, CA, USA), according to the manufacturer's protocol. Luminaris Colour HiGreen qPCR Master Mix (Thermo Fisher Scientific, Carlsbad, CA, USA) and the qTOWER3 (Analytik Jena, Jena, Germany) was used for performing quantitative PCR. All analysed genes are displayed in Table 1 with their respective primer sequences. RPL22 and TBP were used as reference genes. To evaluate primer quality and specificity, melting curve analysis and agarose gel electrophoresis were performed. Primer efficiency was analysed with a cDNA dilution series. The efficiency corrected ΔΔCT method was used for data analysis [34]. Each condition was analysed at least in biological triplicates with technical duplicates.
Table 1.
qPCR primer sequences of human genes indicated in 5`-3` direction. bp, base pairs; fw, forward; Length, amplicon length; rev, reverse.
| Gene | Gene Symbol | NCBI Gene ID | Primer Sequence | Length |
|---|---|---|---|---|
| CREB binding protein | CREBBP | 1387 | fw: GCCAACAAGAAGAAGCCCAGrev: GGATCACGAAGAAGACCTCCT | 100 bp |
| E1A binding protein p300 | EP300 | 2033 | fw: TGACCAAGGGAGACAGCAAArev: GAGGCGGATCACAAAGAAGAC | 182 bp |
| Histone deacetylase 1 | HDAC1 | 3065 | fw: AGCTCCACATCAGTCCTTCCArev: TTCGTCCTCATCGCCACTCT | 170 bp |
| Histone deacetylase 2 | HDAC2 | 3066 | fw: ACTGATGCTTGGAGGAGGTGrev: CTGGAGTGTTCTGGTTTGTCA | 185 bp |
| Histone deacetylase 3 | HDAC3 | 8841 | fw: GCTGGGTGGTGGTGGTTATArev: TTCTGATTCTCGATGCGGGT | 174 bp |
| Interleukin 10 | IL10 | 3586 | fw: AGCCATGAGTGAGTTTGACArev: AGAGCCCCAGATCCGATTTT | 141 bp |
| Ribosomal protein L22 | RPL22 | 6146 | fw: TGATTGCACCCACCCTGTAGrev: GGTTCCCAGCTTTTCCGTTC | 98 bp |
| TATA-box binding protein associated factor 1 | TAF1 | 6872 | fw: CCAGTGCCACTCCAGAAAAGrev: TCCCATCCTCCTCCTCATCA | 114 bp |
| TATA-box binding protein | TBP | 6908 | fw: CGGCTGTTTAACTTCGCTTCCrev: TGGGTTATCTTCACACGCCAAG | 86 bp |
Immunofluorescent staining
After specific treatment, HPdLFs cultured on coverslips were fixed in 4% PFA for 10 minutes and washed with PBS before incubation with the primary and secondary antibodies for 45 minutes each. Nuclei were stained with DAPI (Thermo Fisher Scientific, Carlsbad, CA, USA; 1:10,000 in PBS). The pan-antibody mouse-anti-human H3K9/14/18/23/27ac (ab47915, Abcam, Cambridge, UK; 1:500) was used to detect this histone modifications in HPdLFs and goat-anti-Mouse-Cy5 (Jackson ImmunoResearch, Baltimore Pike, PA, USA; 1:1000) was used for fluorescently labelling. Each condition was tested in independent biological triplicates with technical duplicates.
MTT cell vitality test
Cell vitality of HPdLFs was analysed with MTT colorimetric assay (Sigma Aldrich, St. Louis, Missouri, USA) according to manufacturer's protocol. Colorimetric analysis was performed with the plate reader Infinite ® M Nano (TECAN, Männedorf, Swiss). Each condition was analysed in biological triplicates with technical duplicates.
Nuclear extraction and HDAC/HAT activity assay
Nuclear extraction was performed with the EpiQuik Nuclear Extraction Kit (EpiGentek, Farmingdale, New York, USA) according to manufacturer's protocol. The activities of histone deacetylases (HDAC) and histone acetyltransferases (HAT) were analysed with EpiQuik HDAC Activity/Inhibition Assay Kit (EpiGentek, Farmingdale, New York, USA) and EpiQuik HAT Activity/Inhibition Assay Kit (EpiGentek, Farmingdale, New York, USA) according to manufacturer's guidelines, respectively. Assay read out was done with the plate reader Infinite ® M Nano (TECAN, Männedorf, Swiss). Each condition was analysed in biological triplicates with technical duplicates.
Chromatin Immunoprecipitation (ChIP)-qPCR
After treatment, crosslinking of DNA and protein was performed with 1% formaldehyde in PBS for 10 min and neutralized with 120 mM glycine. Cells were harvested in ice-cold PBS, aliquoted with an approximate cell number of 1 × 106 cells per tube and centrifuged for 5 min at 1000 × g at 4°C prior storage at −80°C. Chromatin immunoprecipitation was performed with the Zymo-Spin ChIP Kit (Zymo Research, Freiburg, Germany) according to manufacturer's protocol Following ChIP-validated antibodies were used for precipitation: mouse-anti-human H3K9/14/18/23/27ac (Thermo Fisher Scientific, Carlsbad, CA, USA) and mouse-anti-IgG (Thermo Fisher Scientific, Carlsbad, CA, USA). One percent input controls were stored throughout the ChIP process. The determination of the amount of specific DNA fragments bound to H3K9/14/18/23/27ac was performed as previously described [35]. Briefly, a 20-cycle primer-specific pre-amplification was performed prior quantitative analysis with the qTOWER3 (Analytik Jena, Jena, Germany) according to the manufacturer's protocol using Luminaris Colour HiGreen qPCR Master Mix (Thermo Fisher Scientific, Carlsbad, CA, USA). Primer sequences for three promoter regions (#1, #2 and #3) as well as one non-promoter region (#4) are displayed in Table 2. Normalization of DNA content was performed according to the per cent-input method in relation to the analysed input probes [36]. Each condition was analysed in biological triplicates with technical duplicates.
Table 2.
qPCR primer sequences of human IL10 in promoter regions (#1, #2, #3) and a non-promoter region (#4) indicated in 5`-3` direction. bp, base pairs; fw, forward; Length, amplicon length; rev, reverse.
| Location | Label | Primer Sequence | Length |
|---|---|---|---|
| promoter region | #1 | fw: TGAAGAAGTCCTGATGTCACrev: TTACCTATCCCTACTTCCCC | 187 bp |
| promoter region | #2 | fw: AGCACTACCTGACTAGCATArev: AGAGACTGGCTTCCTACAG | 192 bp |
| promoter region | #3 | fw: GGGGACCCAATTATTTCTCArev: TGGGCTACCTCTCTTAGAAT | 188 bp |
| non-promoter region | #4 | fw: GCTTAGAGCGTTTCCAGACCrev: CTCCCCACTGTAGACATCCA | 131 bp |
Microscopy, image analysis and statistics
The THP1 adhesion assay and the H3Kac immunofluorescent staining was imaged with the inverted confocal laser scanning microscope TCS SP5 (Leica, Wetzlar, Germany). Analysis of the THP1 cell number and the fluorescent intensity of H3K9/14/18/23/27ac staining was performed with Fiji software (https://imagej.net/Fiji, accessed on 01.04.2017). Fluorescence intensity was asses as previously reported [37]. Briefly, microscopic scanning of each experiment was carried out on the same day with pre-warmed lasers and under identical settings for each treatment condition. To avoid overexposure due to the prominent H3Kac staining, the presumably most intense condition was determined in each experiment prior to scanning, and the scanning settings were calibrated to this condition. Mean grey values (MGVs) of H3Kac stainings were measured in the nuclei of 270 cells per condition. The background of each image was subtracted from the MGVs of each cell, and the intensity of the MGVs was visualized as a thermal LUT. In agreement with our microscopic imaging protocol, MGVs were normalized to control conditions and presented as percent changes.
For statistical analysis, Graph Pad Prism (https://www.graphpad.com, accessed on 01.02.2021). Adobe Photoshop CS5 (https://adobe.com, accessed on 01.02.2013) was used for figure illustration. One-way ANOVA and post hoc test (Tukey) were used as statistical tests. Significance levels: p value < 0.05 */#/§; p value < 0.01 **/##/§§; p value < 0.001 ***/###/§§§.
Results
Oleic acid exposure enhanced the force-related up-regulation of H3 lysine acetylation
To determine the influence of oleic acid (OA) on force-induced changes in H3 lysine acetylation (H3Kac), we first examined the expression of key epigenetic regulators of histone acetylation and deacetylation in periodontal cells by quantitative PCR (Figure 1(a,b)). Yet, we could not detect any changes in genes encoding key histone acetylases (TAF1, EP300, CREBBP) and ubiquitously expressed histone deacetylases (HDAC1-3), neither due to fatty acid exposure nor to compressive stress. However, those HATs and HDACs act in large protein complexes that determine their activity and also specificity [30]. Thus, the formation of those epigenetic modifications may indeed change according to the corresponding conditions.
Figure 1.
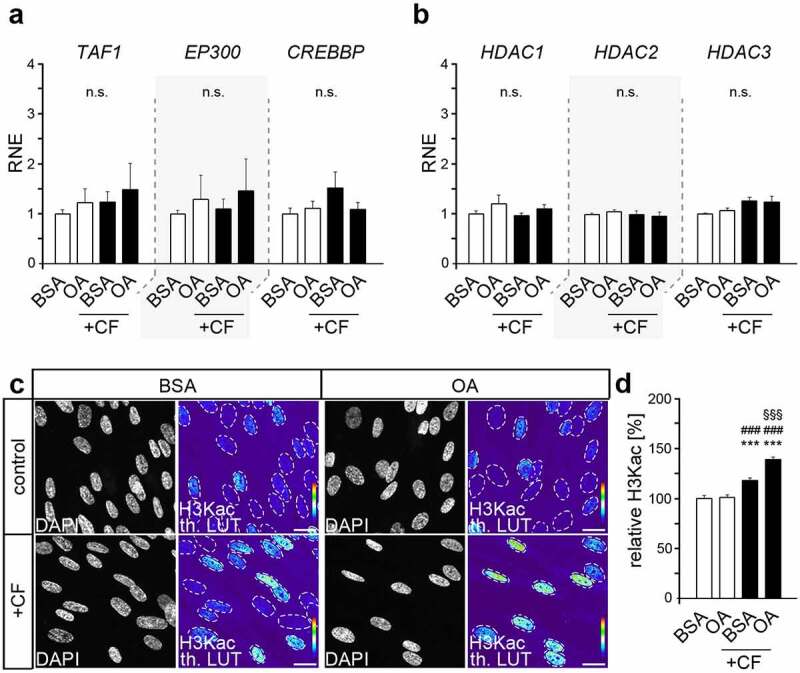
Compressive force increases global histone lysine acetylation further augmented by oleic acid exposure. (a, b) Quantitative expression analysis of genes encoding histone acetyl transferases TAF1, EP300 and CREBBP (a) and encoding histone deacetylases HDAC1, HDAC2 and HDAC3 (b) in human periodontal ligament fibroblasts (HPdLFs) exposed to oleic acid (OA) and stimulated with compressive force (CF) compared to BSA controls. (c, d) Analysis of global H3 lysine acetylation level (H3Kac) presented in representative microphotographs of HPdLFs treated with OA and stimulated with CF in comparison to BSA controls (c) and analysed in (d) in relation to BSA control as percent changes. DAPI labels the cell nuclei and H3Kac staining is shown in thermal LUT (H3Kac th. LUT), indicating staining intensity. The dashed lines illustrate the positions of the cell nuclei. *** p < 0.001 in relation to BSA, ### p < 0.001 in relation to PA, §§§ p < 0.001 in relation to BSA + CF; One-Way ANOVA and post hoc test (Tukey). Scale bars: 10 μm in (c). n.s., not significant; RNE, relative normalized expression.
Semi-quantitative analysis of acetylated H3 lysines (K9/14/18/23/27) was performed by immunofluorescent labelling to reveal differences in the formation of these epigenetic modifications due to OA as well as compressive force (Figure 1(c,d)). While baseline levels remained comparable, force-related increase in H3K acetylation was higher in OA-cultures (38.61% ± 2.59) than in BSA control (21.80% ± 2.70; p value = 0.0085 x 105, ***). Thus, these data suggest that changes in the acetylation of H3 lysines may be involved in the OA-induced changes of the inflammatory response of HPdLFs to compressive forces.
Histone lysine acetylation promote IL10 expression in HPdLFs
To evaluate the role of the highly dynamic histone acetylation and deacetylation processes for the mechanoreactive inflammatory response of HPdLFs, we first inhibited histone deacetylation activity in unstressed control cells with FK228, which is a classic HDAC inhibitor with wide application in research and clinical settings [38].
To avoid possible toxic effects, the influence of different inhibitor concentrations within the usual range of application on the total cell number after one day of application was determined by MTT assay (Figure 2a). At all concentrations used, no toxic effects were observed in one-day treatment. However, even at the lowest concentration of 10 nM, a drastic reduction in metabolic activity was observed after more than 2 days of cultivation (Figure 2b).
Figure 2.
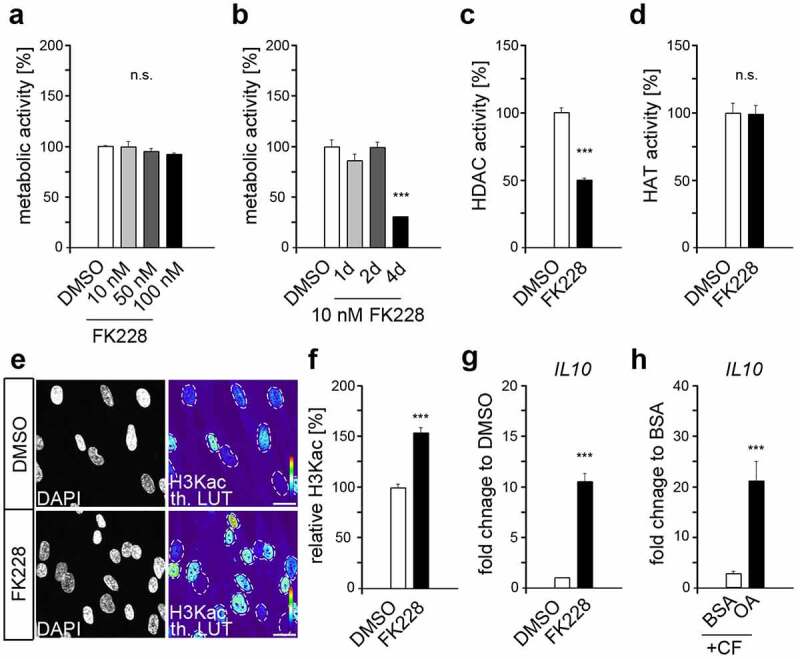
Histone lysine deacetylation revealed increased IL10 expression comparable to that of compressed HPdLFs exposed to oleic acid. (a) Analysis of the metabolic activity of human periodontal ligament fibroblasts (HPdLFs) treated one day with different concentrations of FK228, a histone deacetylase (HDAC) inhibitor. DMSO was used as control. (b) Analysis of metabolic activity of HPdLFs treated for 1d, 2d or 4d with 10 nM FK228 compared to DMSO control. (c, d) Analysis of the activity of HDACs (c) and histone acetyltransferases (HATs; d) in HPdLFs treated one day with 10 nM FK228 in comparison to DMSO control. (e, f) Analysis of global H3K acetylation level (H3Kac) visualized in representative microphotographs of HPdLFs treated one day with 10 nM FK228 compared to DMSO control (e) and analysed in (f) in relation to DMSO control as percent change. DAPI labels the cell nuclei and H3Kac staining is shown in thermal LUT (H3Kac th. LUT), indicating intensity. The dashed lines illustrate the positions of the cell nuclei. (g, h) Quantitative expression analysis of IL10 in HPdLFs treated one day with 10 nM FK228 (g) as well as in HPdLFs exposed to oleic acid (OA) and stimulated with compressive force (CF) in comparison to BSA control. Expression data in (f) were normalized to the non-forced BSA control (not shown). *** p < 0.001 in relation to DMSO in (b), (c), (f) and (g), and in relation to BSA in (h); One-Way ANOVA and post hoc test (Tukey). Scale bars: 10 μm in (e). n.s., not significant.
The efficacy of FK228 in blocking HDAC activity was with 49.09% ± 3.45 inhibition rate significant even at 10 nM (Figure 2c), which was used for further experiments for these reasons. FK228 had no direct effect on the activity of histone acetyltransferases (Figure 2d), but resulted in increased H3Kac levels (Figure 2(e,f)), presumably due to decrease deacetylation activity of blocked HDACs in FK288-treated fibroblasts, whereas HATs remained comparably active. However, in this way, an OA-related increase in H3K acetylation was simulated, but without a variety of other changes due to fatty acid treatment. The pro-inflammatory response of HPdLF to several stimuli can be limited by anti-inflammatory cytokines including interleukin 10 (IL-10) [39]. Quantitative expression analysis revealed a significant increase of IL10 transcription levels in HPdLFs exposed to FK228 (Figure 2g) indicating an important role of H3K acetylation in regulating IL10 gene activity. This hypothesis was further strengthened by the increased expression of IL10 in compressed OA-cultures compared to the respective BSA control (Figure 2h) suggesting IL-10 as a promising candidate for the anti-inflammatory effect of OA.
OA-related anti-inflammatory effects in force-stressed HPdLFs depends on H3K acetylation of IL10 promoter regions
To determine whether the increase in H3K acetylation induced by OA treatment has an anti-inflammatory effect, we reduced H3K acetylation by specific HAT inhibitors. For this purpose, we used anacardic acid (AA) with a broad specificity and C646 as a specific p300/CBP inhibitor [40,41]. Both compounds had a positive effect on HPdLFs growth even at low concentrations (Figure 3a), reliably decreased HAT activity (Figure 3b), and reduced acetylation of H3 lysines (Figure 3(c,d)).
Figure 3.
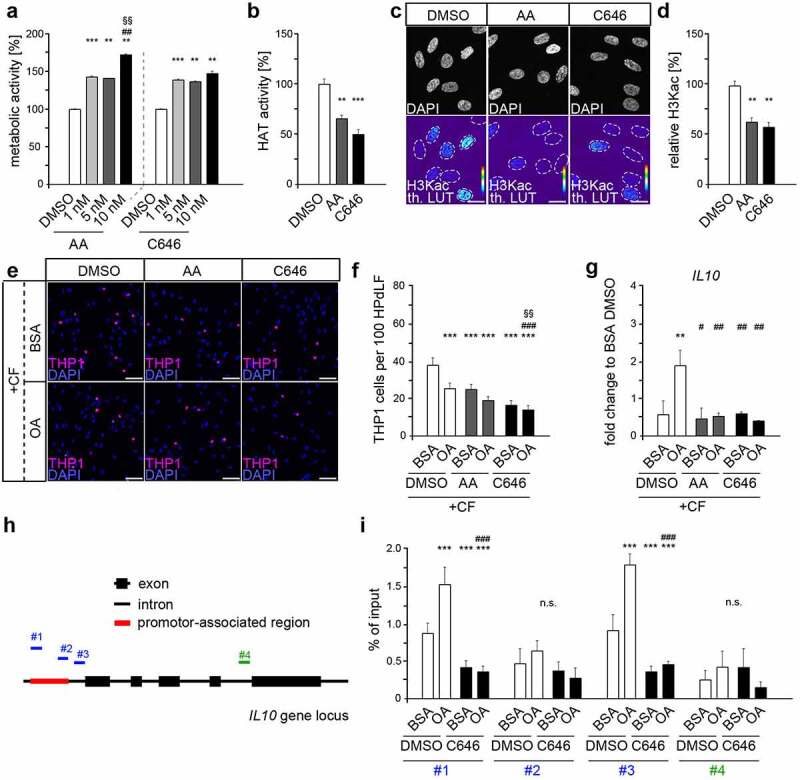
Oleic acid exerts its anti-inflammatory effect in force-stressed HPdLFs via increased histone acetylation potentially on the IL10 gene promoter (a) Analysis of the metabolic activity of human periodontal ligament fibroblasts (HPdLFs) treated one day with different concentrations of anacardic acid (AA) or C646, both are histone acetyltransferase (HAT) inhibitors. DMSO was used as control. (b) Analysis of HAT activity in HPdLFs treated one day with 10 nM AA or 1 nM C646 in relation to DMSO control. (c, d) Analysis of global H3K acetylation level (H3Kac) visualized in representative microphotographs of HPdLFs treated one day with 10 nM AA or 1 nM C646 compared to DMSO control (c) and analysed in (d) in relation to DMSO control as percent change. DAPI labels the cell nuclei and H3Kac staining is shown in thermal LUT (H3Kac th. LUT), indicating intensity. The dashed lines illustrate the positions of the cell nuclei. (e, f) Analysis showing representative microscopic images of fluorescently labelled, adherent THP1 monocytic cells (violet) on compressed HPdLFs exposed to oleic acid (OA) after one-day treatment with 10 nM AA or 1 nM C646 in relation to BSA and DMSO controls (e) analysed as relative number of THP1 cells per 102 HPdLFs in (f). Cell nuclei were labelled with DAPI. (g) Quantitative expression analysis of IL10 in compressed HPdLFs exposed to OA after one-day treatment with 10 nM AA or 1 nM C646 in comparison to BSA and DMSO controls. Expression data were normalized to the non-forced BSA DMSO control (not shown). (h) Locations of DNA primer amplified regions in the human IL10 gene locus indicating promoter-associated pairs (blue, #1, #2 and #3) and a non-promoter-associated pair (green, #4). (i) Quantitative analysis of the association of specific IL10 gene regions shown in (e) with acetylated H3K in compressed HPdLFs exposed to OA and treated for one day with 1 nM C646 in comparison to the respective controls. Data were normalized to the sample input controls. ** p < 0.01, *** p < 0.001 in relation to DMSO in (a), (b) and (c) and in relation to BSA DMSO + CF in (f), (g) and (i); ## p < 0.01 in relation to 1 nM AA in (a); # p < 0.05, ### p < 0.001 in relation to OA DMSO + CF in (f), (g) and (i). §§ p < 0.01 in relation to 5 nM AA in (A) and in relation to BSA C646 + CF in (f); One-Way ANOVA and post hoc test (Tukey). Scale bars: 10 μm in (c) and (e). n.s., not significant.
We functionally analysed the pro-inflammatory response of compressed HPdLFs cultured in the presence of AA and C646, and detected reduced numbers of adherent THP1 cells compared to the respective BSA control (Figure 3(e,f)) suggesting general anti-inflammatory properties of these HAT inhibitors. Nevertheless, further OA-related anti-inflammatory properties in compressed HPdLFs appeared to be prevented by both AA and C646, as THP1 cell adhesion was comparable to BSA control. Quantitative PCR revealed down-regulated IL10 expression in force-stressed OA-cultures treated with both AA and C646 compared to respective DMSO controls (Figure 3g). Thus, the detected expression levels of the inhibitor-treated OA-cultures were comparable to those of the corresponding controls. Overall, these data strongly implicate that IL-10 may be modulated by H3K acetylation and that p300/CBP has a major role in this regulation in HPdLFs.
Acetylated H3K promote transcriptional activation of nearby genes [28]. To determine whether the IL10 gene could be a direct target of OA-modulated H3Kac, we performed CHIP-qPCR to assess its association to acetylated H3 lysines (Figure 3(h,i)). We detected enhanced association of this permissive histone mark in the IL10 promoter and promoter-associated regions of positions #1 and #3 of the force-stressed OA-cultures compared with the respective BSA control, whereas region #2 and the intron control #4 were not affected. Treatment with C646 significantly reduced this association resulting in comparable values to those for the BSA controls. This further strengthen a major role of H3K acetylation in the anti-inflammatory effects of OA, particularly via regulation of IL10 expression.
Discussion
This study examined potential epigenetic mechanisms for the previously reported attenuated mechanoreactive immune response of HPdLF upon stimulation with the fatty acid OA [22]. Compressive force increased the global level of H3K acetylation, and exposure to OA further enhanced this force-related increase in global H3Kac. Epigenetic inhibitor studies and the increase in the association of acetylated H3 lysines with IL10 promoter-specific regions strongly implied a role of this epigenetic modification in compressed HPdLF treated with OA. In summary, our data indicate that the anti-inflammatory effects of OA in stressed HPdLF appear to be regulated, at least to some extent, by p300/CBP-modulated H3K acetylation of IL10, providing an intriguing link between nutrition, oral tissue function, and epigenetic regulatory mechanisms.
In addition to direct binding and post-translational modifications of transcription factors, fatty acids can regulate gene expression by altering epigenetic modification such as histone marks [42,43]. As recently shown, FA-related changes in the epigenetic code may thereby condition hyperinflammatory responses of HPdLFs to combined mechanical and peripathogenic stimuli [39]. In the context of the MUFA oleic acid, this has been specifically shown in glucose-starved C2C12 mouse myoblast cells, which showed up-regulated H3K23 acetylation when supplemented with 200 µM OA [44]. In this study, we found increased acetylation of H3 lysines in HPdLFs only when they were additionally stimulated with mechanical stress but not without. In contrast to Jo et al. [44], who used medium with low glucose content, we continuously cultured in a medium with higher glucose levels necessary for proper HPdLF proliferation and function. Starvation also makes HPdLFs more sensitive to additional stimuli [45]. However, we can only speculate on what impact additional prior starvation may have had on global H3K acetylation levels. Since glucose is one of the major sources of acetyl-CoA [46], additional starvation could cause more drastic effects as fatty acid-related sources of this key factor become much more important [5]. However, considering the general oversupply of sugar in today’s western society, we therefore consider our experimental set-up to be well suited.
Independent of fatty acid exposure, compressive force alone also triggered an increase in H3K acetylation in HPdLFs, supporting comparable results in bone marrow mesenchymal stem cells (MSC) [47]. In this study, perpendicular compressive forces on parallel-aligned cells triggered decreased HDAC activity leading to increased histone acetylation. This pathway, presumably mediated by the nuclear matrix protein lamin A/C [47], may be of interest because PdL cells also exhibit robust lamin A/C expression that is even increased in a force-dependent manner for lamin A [48]. In this aspect, in several cell types OA treatment triggered the formation of lamin A/C-positive nuclear tubules [49], which are important for calcium homoeostasis that can considerably affect histone acetylation [50]. Thus, changes in lamin A/C expression or function could be accountable for the increased H3K acetylation that we detected in compressed HPdLFs treated with OA. Future studies could focus on the specific intracellular and intranuclear alterations of specific signal transduction pathways and transducers caused by simultaneous exposure to fatty acids and mechanical forces.
Although we did not measure HDAC activity in compressed OA-cultured HPdLFs, the comparable IL10 expression increase triggered by FK228-mediated HDAC inhibition suggests that it could be sufficient for an anti-inflammatory response to restrict the activity of those epigenetic erasers. This seems to be in accordance with HDAC inhibition studies in several cell types, such as bone marrow–derived antigen-presenting cells [51], RAW264.7 macrophages [52] as well as KB31, C2C12, and 3T3-J2 cell lines [53]. Although we did not detect global changes in HAT activity after FK228-induced inhibition of HDAC that could directly explain the changes in IL10 expression, it is not unlikely that specific HATs were altered in activity or specificity because of the interconnectivity of HAT and HDAC [30]. In addition, other transcriptional regulatory mechanisms could be altered by OA that modulate the expression of IL10 [54]. However, a wide variety of HAT inhibitor studies suggests that IL-10 is also directly regulated by HAT activity [55–59]. This was also evident here, as mechanically stressed HPdLFs cultured in OA no longer exhibited elevated IL10 levels after HAT inhibition with anacardic acid and p300/CBP-specific C646. Although both inhibitors have anti-inflammatory properties [60], which we also demonstrated, they have previously been shown to negatively regulate IL-10 expression and secretion [56,58,59]. Our data now suggest that the anti-inflammatory effects of OA in force-loaded HPdLFs are mediated by p300/CBP since its inhibition was sufficient to prevent OA-related reduced THP1 cell adhesion and enhanced lysine acetylation at the IL10 promoter. This seems not unlikely in view of the fact that C646 competes with acetyl-CoA for the p300 Lys-CoA binding pocket [41]. Since alterations in acetyl-CoA concentrations could affect p300 histone acetylation activity [31,32], increased availability of acetyl-CoA by OA exposure should thereby no longer have an impact. The general and OA-specific reduction in histone acetylation at IL10 promoter sites further supports this.
The results of our study are limited by several reasons. Because of the complex metabolic influences that individual fatty acids may have, in vivo it is not only the excess of one fatty acid that is decisive for metabolic alterations, but in particular, the ratio to other fatty acids as well as other micro- and macro-nutrients. This complicates in vitro studies, such as ours. Additionally, we performed our analyses only on a certain concentration of OA and mechanical stimulation. However, studies show that this concentration is achieved by increased intake through OA-rich diet [61], as well as in the range also detected in the serum of obese patients [62–66]. After six hours of mechanical stimulation, the early phase of the inflammatory response can be seen. Moreover, examination of specific H3 lysine acetylation rather than an average of H3K9, K14, K18, K23, and K27 acetylation due to pan-antibody could provide further insight into the specific changes that OA might induce. In addition, further analysis of other anti-inflammatory cytokines could provide a more comprehensive view of the changes associated with OA, which was, however, beyond the scope of the study.
In summary, our data highlight an important influence of oleic acid on the acetylation profile of histone lysines in human PdL fibroblasts under mechanical compression. Thus, also in PdL tissue, epigenetic modifications, and associated transcriptional changes are associated with metabolic shifts induced by dietary components, as well as also seen in obese patients, which may influence tooth movement. As personalized treatment in orthodontics offers great potential in terms of nutritional modifications, insights into the effects of fatty acids on oral functionality are of great importance. In addition, considering that epigenetic inhibitors already used in a variety of diseases and some of them could be applied locally, this might be a possibility to reduce metabolic disease-related orthodontic impairments and associated risks for root resorption and bone loss.
Funding Statement
This work was supported by the Interdisciplinary Centre of Clinical Research of the Medical Faculty Jena under Grant MSP-08 and the Program for the Support of Third-Party Funding for Young Scientists 2018 Program Line B (Basic) of the Friedrich-Schiller University Jena under Grant DRM/2018-10Friedrich-Schiller-Universität Jena.
Authors contributions
LS performed experiments and analysis as well as wrote the original manuscript draft; KvB performed experiments; CJ revised the manuscript draft and contributed to project supervision; JS designed the project, acquired funding, supervised the project, performed experiments, figure illustration, revised the manuscript draft and performed revision of the manuscript. All authors have read and agreed to the published version of the manuscript.
Data availability statement
The datasets of this study are available from the corresponding author, JS, on reasonable request. The data are not publicly available due to very large size of microscopy images https://figshare.com/projects/Oleic_acid-related_anti-inflammatory_effects_in_force-stressed_PdL_fibroblasts_are_mediated_by_H3_lysine_acetylation_associated_with_altered_IL10_expression/130172
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Gondivkar SM, Gadbail AR, Gondivkar RS, et al. Nutrition and oral health. Dis Mon. 2019;65(6):147–154 [DOI] [PubMed] [Google Scholar]
- [2].Touger-Decker R;, Mobley C.. Academy of,N.; dietetics, position of the academy of nutrition and dietetics: oral health and nutrition. J Acad Nutr Diet. 2013;113(5):693–701. [DOI] [PubMed] [Google Scholar]
- [3].Deshpande NC, Amrutiya MR. Obesity and oral health - is there a link? An observational study. J Indian Soc Periodontol. 2017;21(3):229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ferreri C, Masi A, Sansone A, et al. Fatty acids in membranes as homeostatic, metabolic and nutritional biomarkers: recent advancements in analytics and diagnostics. Diagnostics. 2016;7(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].McDonnell E, Crown SB, Fox DB, et al. Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep. 2016;17(6):1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sokolova M, Vinge LE, Alfsnes K, et al. Palmitate promotes inflammatory responses and cellular senescence in cardiac fibroblasts. Biochim Biophys Acta. Mol Cell Biol Lipids. 2017;1862(2):234–245. [DOI] [PubMed] [Google Scholar]
- [7].Pillon NJ, Azizi PM, Li YE, et al. Palmitate-induced inflammatory pathways in human adipose microvascular endothelial cells promote monocyte adhesion and impair insulin transcytosis. Am J Physiol Endocrinol Metab. 2015;309(1):E35–44. [DOI] [PubMed] [Google Scholar]
- [8].Nemecz M, Constantin A, Dumitrescu M, et al. The distinct effects of palmitic and oleic acid on pancreatic beta cell function: the elucidation of associated mechanisms and effector molecules. Front Pharmacol. 2019;9:1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gillet C, Spruyt D, Rigutto S, et al. Oleate abrogates palmitate-induced lipotoxicity and proinflammatory response in human bone marrow-derived mesenchymal stem cells and osteoblastic cells. Endocrinology. 2015;156(11):4081–4093. [DOI] [PubMed] [Google Scholar]
- [10].de Souza CO, Valenzuela CA, Baker EJ, et al. Palmitoleic acid has stronger anti-inflammatory potential in human endothelial cells compared to oleic and palmitic acids. Mol Nutr Food Res. 2018;62(20):e1800322. [DOI] [PubMed] [Google Scholar]
- [11].de Lima-Salgado TM, Alba-Loureiro TC, Do Nascimento CS, et al. Molecular mechanisms by which saturated fatty acids modulate tnf-alpha expression in mouse macrophage lineage. Cell Biochem Biophys. 2011;59(2):89–97. [DOI] [PubMed] [Google Scholar]
- [12].Carrillo C, Cavia Mdel M, Alonso-Torre S. Role of oleic acid in immune system; mechanism of action; a review. Nutricion hospitalaria. 2012;27(4):978–990. [DOI] [PubMed] [Google Scholar]
- [13].Al-Shudiefat AA, Sharma AK, Bagchi AK, et al. Oleic acid mitigates tnf-alpha-induced oxidative stress in rat cardiomyocytes. Mol Cell Biochem. 2013;372(1–2):75–82. [DOI] [PubMed] [Google Scholar]
- [14].Korbecki J, Bajdak-Rusinek K, et al. The effect of palmitic acid on inflammatory response in macrophages: an overview of molecular mechanisms. Inflamm Res. 2019;68(11):915–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhou BR, Zhang JA, Zhang Q, et al. Palmitic acid induces production of proinflammatory cytokines interleukin-6, interleukin-1beta, and tumor necrosis factor-alpha via a nf-kappab-dependent mechanism in hacat keratinocytes. Mediators Inflamm. 2013;2013:530429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Drosatos-Tampakaki Z, Drosatos K, Siegelin Y, et al. Palmitic acid and dgat1 deficiency enhance osteoclastogenesis, while oleic acid-induced triglyceride formation prevents it. J Bone Miner Res. 2014;29(5):1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ravaut G, Legiot A, Bergeron KF, et al. Monounsaturated fatty acids in obesity-related inflammation. Int J Mol Sci. 2020;22(1):330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Milanski M, Degasperi G, Coope A, et al. Saturated fatty acids produce an inflammatory response predominantly through the activation of tlr4 signaling in hypothalamus: implications for the pathogenesis of obesity. J Neurosci. 2009;29(2):359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fuchs AC, Granowitz EV, Shapiro L, et al. Clinical, hematologic, and immunologic effects of interleukin-10 in humans. J Clin Immunol. 1996;16(5):291–303. [DOI] [PubMed] [Google Scholar]
- [20].Niho Y, Niiro H, Tanaka Y, et al. Role of il-10 in the cross regulation of prostaglandins and cytokines in monocytes. Acta Haematol. 1998;99(3):165–170. [DOI] [PubMed] [Google Scholar]
- [21].Tutunchi H, Ostadrahimi A, Saghafi-Asl M. The effects of diets enriched in monounsaturated oleic acid on the management and prevention of obesity: a systematic review of human intervention studies. Adv Nutr. 2020;11(4):864–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Symmank J, Appel S, Bastian JA, et al. Hyperlipidemic conditions impact force-induced inflammatory response of human periodontal ligament fibroblasts concomitantly challenged with p. Gingivalis-lps. Int J Mol Sci. 2021;22(11):6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jonsson D, Nebel D, Bratthall G, et al. The human periodontal ligament cell: a fibroblast-like cell acting as an immune cell. J Periodontal Res. 2011;46(2):153–157. [DOI] [PubMed] [Google Scholar]
- [24].Yamaguchi M, Fukasawa S. Is inflammation a friend or foe for orthodontic treatment?: inflammation in orthodontically induced inflammatory root resorption and accelerating tooth movement. Int J Mol Sci. 2021;22(5):2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Weltman B, Vig KW, Fields HW, et al. Root resorption associated with orthodontic tooth movement: a systematic review. Am J Orthodont Dentofac Orthoped. 2010;137(4):462–476. discussion 412A. [DOI] [PubMed] [Google Scholar]
- [26].Wishney M. Potential risks of orthodontic therapy: a critical review and conceptual framework. Aust Dent J. 2017;62(1):86–96. [DOI] [PubMed] [Google Scholar]
- [27].Pietrocola F, Galluzzi L, Bravo-San Pedro JM, et al. Acetyl coenzyme a: a central metabolite and second messenger. Cell Metab. 2015;21(6):805–821. [DOI] [PubMed] [Google Scholar]
- [28].Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76(1):75–100. [DOI] [PubMed] [Google Scholar]
- [29].Dancy BM, Cole PA. Protein lysine acetylation by p300/cbp. Chem Rev. 2015;115(6):2419–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Peserico A, Simone C. Physical and functional hat/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. 2011;2011:371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Henry RA, Kuo YM, Andrews AJ. Differences in specificity and selectivity between CBP and p300 acetylation of histone h3 and h3/h4. Biochemistry. 2013;52(34):5746–5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Henry RA, Kuo YM, Bhattacharjee V, et al. Changing the selectivity of p300 by acetyl-coa modulation of histone acetylation. ACS Chem Biol. 2015;10(1):146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Symmank J, Chorus M, Appel S, et al. Distinguish fatty acids impact survival, differentiation and cellular function of periodontal ligament fibroblasts. Sci Rep. 2020;10(1):15706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- [35].Symmank J, Bayer C, Reichard J, et al. Neuronal Lhx1 expression is regulated by DNMT1-dependent modulation of histone marks. Epigenetics. 2020;15(11):1259–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Haring M, Offermann S, Danker T, et al. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;3(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Symmank J, Bayer C, Schmidt C, et al. DNMT1 modulates interneuron morphology by regulating Pak6 expression through crosstalk with histone modifications. Epigenetics. 2018;13(5):536–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yoon S, Eom GH. HDAC and HDAC inhibitor: from cancer to cardiovascular diseases. Chonnam Med J. 2016;52(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schuldt L, Reimann M, von Brandenstein K, et al. Palmitate-triggered cox2/pge2-related hyperinflammation in dual-stressed pdl fibroblasts is mediated by repressive h3k27 trimethylation. Cells. 2022;11(6):955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hemshekhar M, Sebastin Santhosh M, Kemparaju K, et al. Emerging roles of anacardic acid and its derivatives: a pharmacological overview. Basic Clin Pharmacol Toxicol. 2012;110(2):122–132. [DOI] [PubMed] [Google Scholar]
- [41].Bowers EM, Yan G, Mukherjee C, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol. 2010;17(5):471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Choi SW, Claycombe KJ, Martinez JA, et al. Nutritional epigenomics: a portal to disease prevention. Adv Nutr. 2013;4(5):530–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang Y, Kutateladze TG. Diet and the epigenome. Nat Commun. 2018;9(1):3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jo C, Park S, Oh S, et al. Histone acylation marks respond to metabolic perturbations and enable cellular adaptation. Exp Mol Med. 2020;52(12):2005–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Al-Rekabi Z, Fura AM, Juhlin I, et al. Hyaluronan-cd44 interactions mediate contractility and migration in periodontal ligament cells. Cell Adh Migr. 2019;13(1):138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hsieh WC, Sutter BM, Ruess H, et al. Glucose starvation induces a switch in the histone acetylome for activation of gluconeogenic and fat metabolism genes. Mol Cell. 2022;82(1):60–74 e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li Y, Chu JS, Kurpinski K, et al. Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys J. 2011;100(8):1902–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Denes BJ, Bolton C, Illsley CS, et al. Notch coordinates periodontal ligament maturation through regulating lamin a. J Dent Res. 2019;98(12):1357–1366. [DOI] [PubMed] [Google Scholar]
- [49].Lagace TA, Ridgway ND. The rate-limiting enzyme in phosphatidylcholine synthesis regulates proliferation of the nucleoplasmic reticulum. Mol Biol Cell. 2005;16(3):1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Plackic J, Preissl S, Nikonova Y, et al. Enhanced nucleoplasmic ca(2+) signaling in ventricular myocytes from young hypertensive rats. J Mol Cell Cardiol. 2016;101:58–68. [DOI] [PubMed] [Google Scholar]
- [51].Villagra A, Cheng F, Wang HW, et al. The histone deacetylase hdac11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10(1):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Leus NG, van den Bosch T, van der Wouden PE, et al. Hdac1-3 inhibitor ms-275 enhances il10 expression in raw264.7 macrophages and reduces cigarette smoke-induced airway inflammation in mice. Sci Rep. 2017;7(1):45047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Di Liddo R, Valente S, Taurone S, et al. Histone deacetylase inhibitors restore il-10 expression in lipopolysaccharide-induced cell inflammation and reduce il-1beta and il-6 production in breast silicone implant in c57bl/6j wild-type murine model. Autoimmunity. 2016;1–11. doi: 10.3109/08916934.2015.1134510. [DOI] [PubMed] [Google Scholar]
- [54].Rutz S, Ouyang W. Regulation of interleukin-10 expression. Adv Exp Med Biol. 2016;941:89–116. [DOI] [PubMed] [Google Scholar]
- [55].Hammitzsch A, Tallant C, Fedorov O, et al., Cbp30, a selective CBP/p300 bromodomain inhibitor, suppresses human th17 responses. Proceedings of the National Academy of Sciences of the United States of America; 2015. 112, p. 10768–10773. doi: 10.1073/pnas.1501956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bai AH, Wu WK, Xu L, et al. Dysregulated lysine acetyltransferase 2b promotes inflammatory bowel disease pathogenesis through transcriptional repression of interleukin-10. J Crohn’s Colitis. 2016;10(6):726–734. [DOI] [PubMed] [Google Scholar]
- [57].Zheng Z, Huang G, Gao T, et al. Epigenetic changes associated with interleukin-10. Front Immunol. 2020;11:1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hedrich CM, Rauen T, Apostolidis SA, et al., Stat3 promotes il-10 expression in lupus t cells through trans-activation and chromatin remodeling. Proceedings of the National Academy of Sciences of the United States of America; 2014. 111, p. 13457–13462. 10.1073/pnas.140802311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sondergaard JN, Poghosyan S, Hontelez S, et al. Dc-script regulates il-10 production in human dendritic cells by modulating nf-kappa bp65 activation. J Iimmunol. 2015;195(4):1498–1505. [DOI] [PubMed] [Google Scholar]
- [60].Gomes Junior AL, Islam MT, Nicolau LAD, et al. Anti-inflammatory, antinociceptive, and antioxidant properties of anacardic acid in experimental models. ACS omega. 2020;5(31):19506–19515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Temme EH, Mensink RP, Hornstra G. Comparison of the effects of diets enriched in lauric, palmitic, or oleic acids on serum lipids and lipoproteins in healthy women and men. Am J Clin Nutr. 1996;63(6):897–903. [DOI] [PubMed] [Google Scholar]
- [62].Bierman EL, Dole VP, Roberts TN. An abnormality of nonesterified fatty acid metabolism in diabetes mellitus. Diabetes. 1957;6(6):475–479. [DOI] [PubMed] [Google Scholar]
- [63].Taskinen MR, Bogardus C, Kennedy A, et al. Multiple disturbances of free fatty acid metabolism in non insulin-dependent diabetes. Effect of oral hypoglycemic therapy. J Clin Invest. 1985;76(2):637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Soares EA, Nakagaki WR, Garcia JA, et al. Effect of hyperlipidemia on femoral biomechanics and morphology in low-density lipoprotein receptor gene knockout mice. J Bone Miner Metab. 2012;30(4):419–425. [DOI] [PubMed] [Google Scholar]
- [65].Cistola DP, Small DM. Fatty acid distribution in systems modeling the normal and diabetic human circulation. A 13c nuclear magnetic resonance study. J Clin Invest. 1991;87(4):1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kissebah AH, Alfarsi S, Adams PW, et al. Role of insulin resistance in adipose tissue and liver in the pathogenesis of endogenous hypertriglyceridaemia in man. Diabetologia. 1976;12(6):563–571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets of this study are available from the corresponding author, JS, on reasonable request. The data are not publicly available due to very large size of microscopy images https://figshare.com/projects/Oleic_acid-related_anti-inflammatory_effects_in_force-stressed_PdL_fibroblasts_are_mediated_by_H3_lysine_acetylation_associated_with_altered_IL10_expression/130172


