Summary
Microtubules have spatiotemporally complex posttranslational modification patterns. How cells interpret this tubulin modification code is largely unknown. We show that C. elegans katanin, a microtubule severing AAA ATPase mutated in microcephaly and critical for cell division, axonal elongation, and cilia biogenesis, responds precisely, differentially, and combinatorially to three chemically distinct tubulin modifications: glycylation, glutamylation, and tyrosination, but is insensitive to acetylation. Glutamylation and glycylation are antagonistic rheostats with glycylation protecting microtubules from severing. Katanin exhibits graded and divergent responses to glutamylation on the α- and β-tubulin tails, and these act combinatorially. The katanin hexamer central pore constrains the polyglutamate chain patterns on β-tails recognized productively. Elements distal to the katanin AAA core sense α-tubulin tyrosination, and detyrosination downregulates severing. The multivalent microtubule recognition that enables katanin to read multiple tubulin modification inputs explains in vivo observations and illustrates how effectors can integrate tubulin code signals to produce diverse functional outcomes.
eToc
Microtubules have spatiotemporally controlled posttranslational modification patterns. How this “tubulin code” is read by cells is largely unknown. Szczesna et al. show that distinct tubulin posttranslational modification patterns on α- and β-tubulin tails can act combinatorially and have divergent outcomes on the activity of the microtubule severing enzyme katanin.
Graphical Abstract
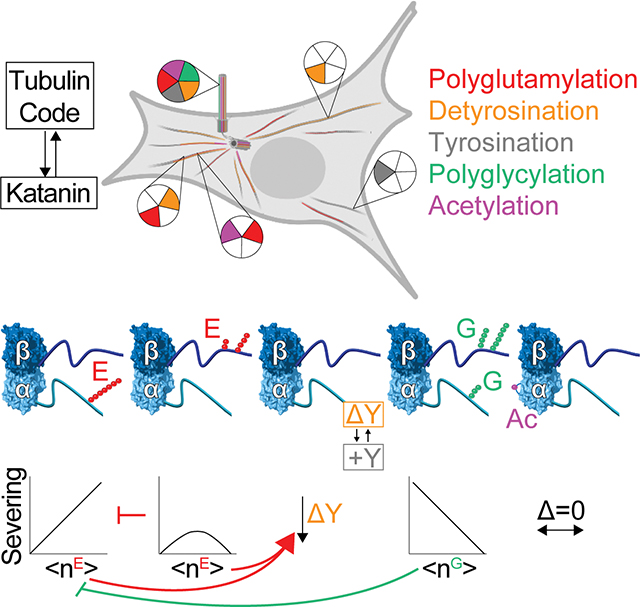
Introduction
Microtubules, polymers of αβ-tubulin heterodimers, are essential cytoskeletal components. They give structure to cells, build morphologically varied structures such as spindles, cilia, and flagella, and serve as tracks for intracellular transport. To support these diverse functions, microtubule networks are constantly remodeled by motors and microtubule-associated proteins that aid their nucleation and growth, disassembly, and organization. How the action of these effectors is precisely coordinated spatially and temporally is an active frontier in microtubule research.
Microtubules have chemically diverse posttranslational modifications, including acetylation, methylation, detyrosination, glutamylation and glycylation (reviewed in MacTaggart et al.1; Roll-Mecak2). Recent discoveries firmly point to microtubule posttranslational modifications acting as part of a “tubulin code” that together with tubulin isotypes regulates the recruitment and activity of microtubule effectors (reviewed in Roll-Mecak2). The levels, types and spatial distribution of microtubule posttranslational modifications are stereotyped in cells and dysregulation of modification patterns cause human pathologies, including neurodegeneration and cancers (reviewed in Bodakuntla et al.3; Wattanathamsan et al.4). Axonemes of cilia and flagella are especially enriched with tubulin modifications. Axonemal tubulin can have as many as 22 glutamates5, 6 and 40 glycines7. These modifications display precise and stereotyped patterns such that even adjacent microtubules have distinct chemical signatures. For example, the A tubule in the microtubule doublet is mostly tyrosinated while the attached B tubule is detyrosinated, glutamylated and glycylated. Specific outer doublet microtubules also have different glutamylation levels8, 9. The microtubules in the central doublet are mostly detyrosinated and have low glutamylation levels10–12. Dynamic microtubules at cilia tips have no detectable polyglycylation13. Most tubulin in the mammalian brain is glutamylated and detyrosinated, and these modifications can occur on the same tubulin dimer14. Brain tubulin has a preponderance of 3–6 glutamates on α- and β-tubulin and as many as 11 and 7 glutamates, respectively15, and tubulin modifying enzymes are upregulated during neuronal differentiation16. The spatial segregation of modifications in neurons is striking. Stable microtubules in axons are enriched in acetylation, detyrosination and glutamylation and dynamic microtubules in growth cones are mostly unmodified/tyrosinated17, 18. Modifications are spatially segregated even within the dense microtubule bundles in dendrites with stable, acetylated microtubules in the core of the bundle and the dynamic, tyrosinated microtubules arranged concentrically at the periphery19. These precise, stereotyped organizations indicate regulatory pathways for the patterning of modifications and their subsequent readout by effectors. Indeed, modification-dependent interactions regulate microtubule dynamics20, organelle distribution21, axonal trafficking22, 23 and ciliary beating12, 24. Despite knowing about this tubulin diversity for decades, we are still in the early stages of understanding how modifications affect microtubule regulators, individually and in combination, largely due to the lack of in vitro reconstitution systems until very recently2.
Modifications concentrate on the intrinsically disordered C-terminal tails of tubulin which form a dense lawn on the microtubule surface. Katanins are a subfamily of microtubule-severing enzymes25, 26 important in cellular processes ranging from cilia biogenesis and axonal elongation to phototropism. They require tubulin C-terminal tails for activity (reviewed in Lynn et al.27; McNally et al.28). Katanins belong to the superfamily of AAA ATPases (ATPases Associated with diverse cellular Activities) and consist of a catalytic subunit, p60, and a regulatory subunit, p8027. They perform their functions by extracting tubulin subunits from microtubules, thus both remodeling the microtubule through healing with soluble GTP-tubulin as well as severing it along its length29. Katanin severing releases microtubules nucleated by the γ-TURC complex30–33 and katanin loss leads to disorganized spindles and defects in chromosome segregation34–38. Katanin participates in cilia biogenesis39–43 and has been implicated in deciliation44. In neurons, katanin family members are important for axonal outgrowth30, 45 and branch formation46, programmed dendrite pruning47 and dendrite branching48. Consistent with their broad roles, katanin mutations cause cerebral malformations such as microcephaly and autism49–51 as well as cancers52.
Not surprisingly, dysregulated, or excessive microtubule severing by katanin is deleterious (reviewed in McNally et al.28), as exemplified by the tight regulation of katanin at the transcriptional, translational, and posttranslational levels28. Katanin disassembles microtubules by forming a hexamer that engages the β-tubulin tail and dislodges the tubulin subunit out of the microtubule29, 53, a mechanism common among AAA ATPases54, 55. Given the centrality of the tubulin tails to katanin-mediated microtubule severing, posttranslational modifications, overwhelmingly concentrated on tubulin tails, are prime candidates to regulate katanin. Indeed, mutation of glutamylation sites on Tetrahymena tubulin phenocopies katanin loss42, 56 and katanin overexpression leads to selective loss of the ciliary B-tubule42 which is highly glutamylated10, 57. Moreover, defects caused by partial katanin depletion can be rescued by overexpressing the glutamylase TTLL658. However, we do not know whether and how katanin responds directly to tubulin modifications. Spastin, a closely related severing enzyme, is regulated by glutamylation59, 60. It is not clear whether katanin responds the same to modifications since, unlike spastin, katanin functions as part of a complex with its regulatory subunit61 and these enzymes are not functionally redundant in vivo.
Here, we use recombinantly engineered and differentially modified microtubules to establish how the complex chemical landscape of the microtubule cytoskeleton regulates katanin. We show that katanin serves as a hub that integrates multiple chemically diverse tubulin modifications. We find that katanin responds differently to glutamylation on the α- and β-tubulin tails, with α-tail glutamylation being strongly stimulatory and β-tail glutamylation eliciting a biphasic response. Glutamylation on these two tails acts combinatorially, with glutamylation on the β-tail depressing the stimulation elicited by α-tubulin glutamylation. Structural modeling of katanin bound to glutamylated tubulin tails sheds light on the molecular mechanism for this differential regulation. Unlike spastin, we find that katanin senses the tyrosinated α-tubulin tail through elements distal to its motor core and is inhibited by detyrosination. This inhibition can be overcome by glutamylation demonstrating the combinatorial regulation of different tubulin modifications. Lastly, by generating differentially glycylated microtubules, we show that katanin binding and severing are proportionally inhibited by glycylation. This inhibition is operational also on glutamylated microtubules. Thus, glutamylation and glycylation act as antagonistic rheostats for katanin recruitment, with glycylation establishing exclusionary zones protected against severing. Our work shows that different glutamylation patterns introduced by distinct TTLL enzymes are interpreted differentially, providing a molecular explanation for the diversification of TTLL enzymes in organisms with complex microtubule arrays. Our work also provides a molecular mechanism for in vivo observations in many model systems and shows how katanin uses its multivalent microtubule recognition to read multiple tubulin modification inputs for precise spatial and temporal control in cells.
Results
α-tail polyglutamylation proportionally enhances katanin
Tubulin glutamylation involves the addition of glutamates to internal glutamates in tubulin tails62–65. It is catalyzed by tubulin tyrosine ligase like (TTLL) enzymes, specialized in adding either mono- or polyglutamate chains of variable lengths to either α- or β-tubulin tails66, 67. Glutamylation is prevalent in neurons15, 68, as well as in cilia5, 6. Katanin localizes to glutamylated microtubules42, 59, 69, 70. However, whether katanin responds directly and quantitatively to glutamylation or to the pattern of glutamylation on the α- versus β-tubulin tails remains unknown. This is a central question because the α- and β- tails contribute differently to katanin activity53, and α- and β-tubulin glutamylation levels are regulated differently during development22.
To generate microtubules with defined glutamylation levels, we purified unmodified human tubulin71 and used recombinant glutamylases to modify it in vitro. We first modified taxol-stabilized microtubules with TTLL6 which prefers to modify α-tails66, 67 (Figure S1A, STAR Methods). TTLL6 localizes to cilia12, 72 and is highly expressed in ciliated tissues where it is important for cilia biogenesis and function73. Glutamylation was quantified using reverse-phase liquid chromatography mass spectrometry (LC-MS; STAR Methods). Using microscopy-based assays (STAR Methods) we found that katanin recruitment to microtubules increases gradually and monotonically with α-tail glutamylation (Figures 1A, B, C, S1B). This increase in microtubule binding is accompanied by an increase in severing (Figures 1D, E, F, S1C). At a mean glutamate number (<nE>) of ~ 3 on α-tubulin severing is enhanced ~5-fold. At <nE>α ~ 7 severing is enhanced ~ 9-fold compared to unmodified microtubules. Katanin undergoes microtubule templated hexamerization that activates it for severing74. To distinguish between the effects of glutamylation on katanin associated with the microtubule versus its activation once on the microtubule, we performed assays with differentially glutamylated microtubules in which we established the minimum katanin concentration needed to observe a severing event. This experiment was performed over a long observation period, an order of magnitude longer than the time needed to reach steady-state binding. These measurements revealed that the minimum katanin concentration at which a severing event is observed decreases with TTLL6-mediated glutamylation (Figure S1E) from ~ 4.5 nM for unmodified microtubules to ~1 nM for microtubules with <nE>α ~ 5, suggesting that the assembly of a severing active species increases concomitant with α-tubulin glutamylation. Consistent with this, binding assays at 4 nM katanin and with the slow-hydrolysable ATP analog ATPγS, which promotes katanin hexamerization, show a less pronounced dependence of katanin binding with glutamylation than with ATP (Figure S1F), and binding in the presence of ATPγS has a shallower dependence on glutamylation (Figure S1G) than severing at the same enzyme concentration of 20 nM (Figure 1E, F). We cannot measure binding affinity at 20 nM katanin and ATP because the enzyme severs the microtubules before measurements are complete. Thus, TTLL6 glutamylation increases katanin microtubule association and lowers the activation energy for severing as a function of glutamylation level.
Figure 1. Proportional stimulation of microtubule severing by polyglutamylation on the α-tubulin tail by TTLL6.
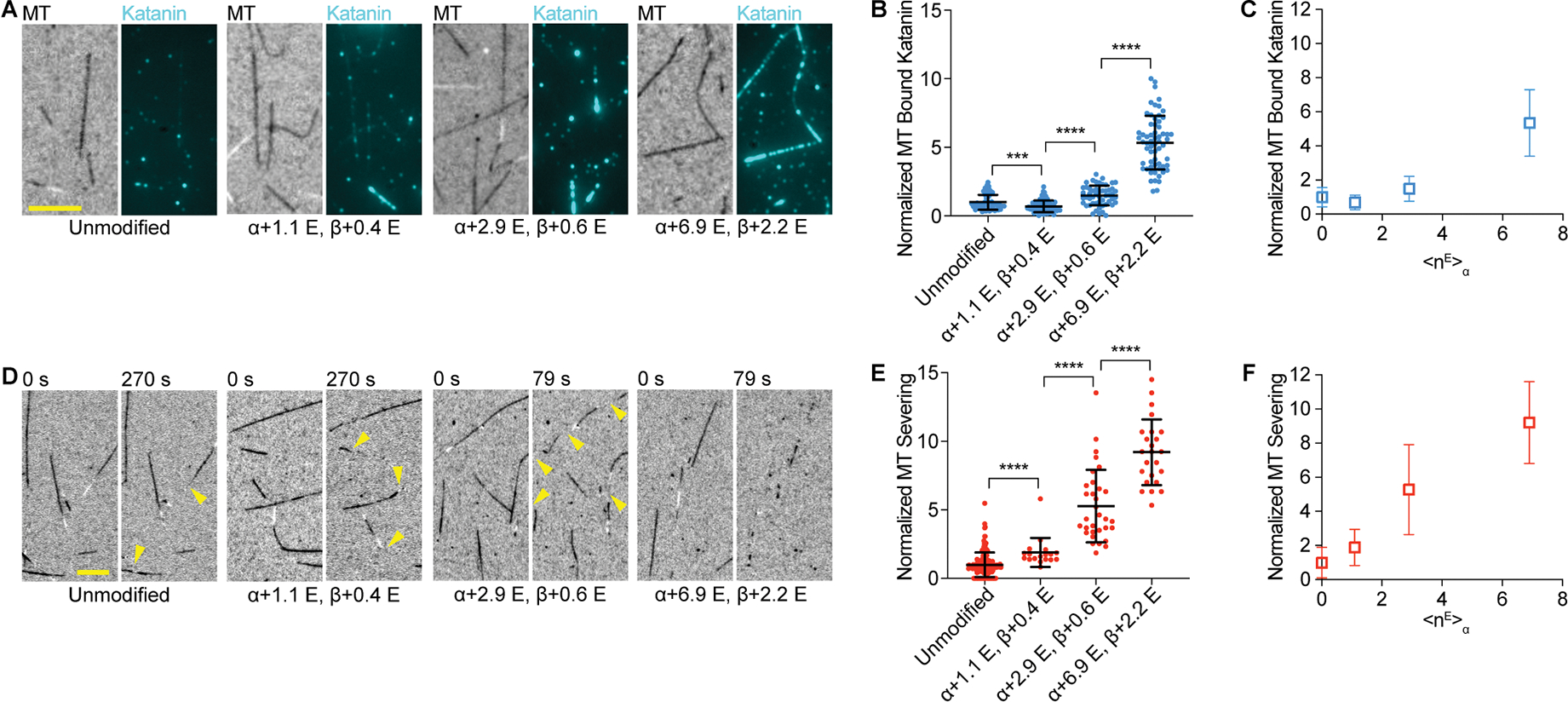
(A) Katanin association with unmodified and glutamylated microtubules with increasing glutamylation levels added by TTLL6. The weighted mean of the number of glutamates (<nE>) added to α- and β-tubulin are denoted α + <nE>; β + <nE>. Assays at 4 nM katanin, 1 mM ATP; Scale bar, 5 μm.
(B) Katanin recruitment to microtubules increases with TTLL6-catalyzed glutamylation levels. Katanin levels normalized to those on unmodified microtubules; n=52, 75, 52 and 62 microtubules from multiple chambers for unmodified and <nE>α ~ 1.1, 2.9 and 6.9, respectively.
(C) Normalized katanin levels bound to microtubules as a function of glutamates added on α-tubulin by TTLL6.
(D) Severing of unmodified and glutamylated microtubules with increasing glutamylation levels added by TTLL6. Time after end of katanin perfusion shown above. Reactions at 20 nM katanin, 1 mM ATP; cellular katanin concentrations ~ 3–28 nM130,131,132. Yellow arrows, severing. Scale bar, 5 μm.
(E) Severing increases with TTLL6-catalyzed glutamylation levels. Severing rates normalized to that of unmodified microtubules (STAR Methods); n = 152, 18, 32 and 24 microtubules from multiple chambers for unmodified and <nE>α ~ 1.1, 2.9 and 6.9, respectively.
(F) Normalized severing as a function of glutamates added on α-tubulin by TTLL6; Mean and S.D., p-value ≤ 0.001 (***), 0.0001 (****) by Mann-Whitney or t-test. LC-MS of microtubules and additional data in Figure S1.
β-tail polyglutamylation biphasically enhances and inhibits katanin
Next, we modified microtubules preferentially on β-tubulin tails using TTLL760, 75, 76 (STAR Methods, Figure S2A). TTLL7 is critical for neurite outgrowth and is upregulated during neuronal differentiation77. It also localizes to cilia67. As for TTLL6, we find that katanin microtubule association increases monotonically with glutamylation (Figures 2A, B, C). However, unlike for TTLL6-modified microtubules, severing gradually increases with glutamylation, but then decreases above a threshold (Figures 2D, E, F). The transition between stimulation and inhibition occurs at <nE> ~7.2 on β-tubulin where severing is enhanced ~ 3-fold over unmodified microtubules (Figures 2E, F). Spastin has a similar biphasic response60. Higher glutamylation levels are inhibitory even though they do not decrease katanin microtubule binding (Figure S2B). Interestingly, β-tubulin glutamylation by TTLL7 stimulates severing to a lesser extent than α-tubulin glutamylation by TTLL6: at <nE>α of 2.9, severing is enhanced ~ 5-fold, but only ~ 3-fold at <nE> β of 3.5. Similarly, at <nE> α of 6.9, katanin severing is enhanced ~ 9-fold, but only ~ 3-fold at <nE> β of 7.2 (Figure 2F).
Figure 2. Biphasic regulation of severing by polyglutamylation on the β-tubulin tail by TTLL7.
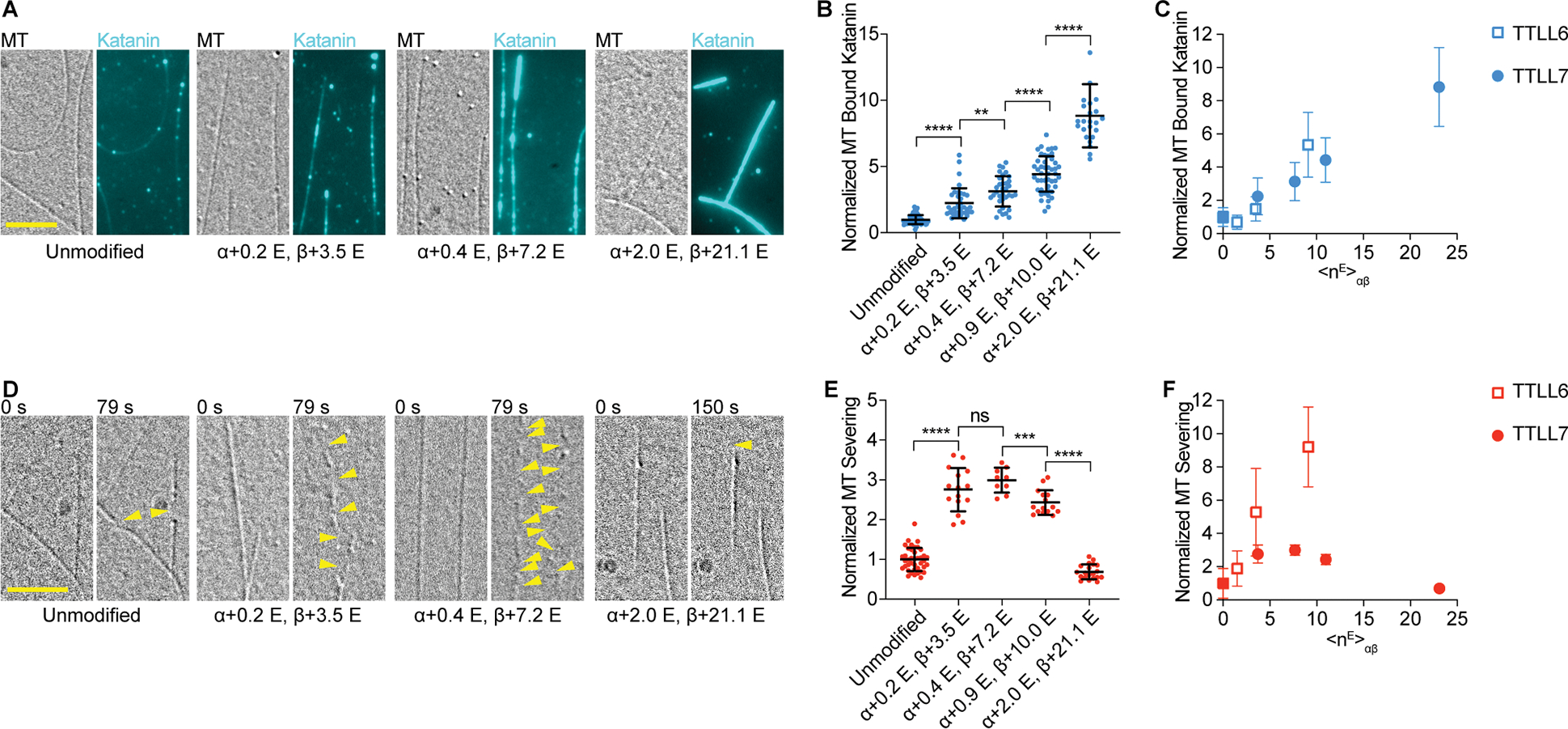
(A) Katanin association with unmodified and glutamylated microtubules with increasing glutamylation levels added by TTLL7. Assays at 4 nM katanin, 1 mM ATP; Scale bar, 5 μm.
(B) Katanin recruitment to microtubules increases with TTLL7-catalyzed glutamylation levels. Katanin levels normalized to those on unmodified microtubules; n= 67, 37, 34, 47, 24 microtubules from multiple chambers for unmodified and <nE>β ~ 3.5, 7.2, 10.0 and 21.1, respectively. Note: one data point of 16.51 for binding to <nE>β ~ 21.1 is above the y axis limit.
(C) Normalized katanin levels bound to microtubules as a function of glutamates added on tubulin by TTLL6 (open squares) and TTLL7 (filled circles); <nE>αβ, weighted average of glutamates on tubulin.
(D) Severing of unmodified microtubules and microtubules with increasing glutamylation levels introduced by TTLL7. Reactions at 20 nM katanin, 1 mM ATP; Yellow arrows, severing. Scale bar, 5 μm.
(E) Severing varies biphasically with TTLL7-catalyzed glutamylation levels. Severing rates normalized to that of unmodified microtubules; n= 40, 15, 9, 15, 20 microtubules from multiple chambers for unmodified and <nE>β ~ 3.5, 7.2, 10.0 and 21.1, respectively.
(F) Normalized severing as a function of glutamates added on tubulin by TTLL6 (open squares) and TTLL7 (filled circles). Means and S.D., p-value > 0.05 (ns), ≤ 0.01 (**), 0.001 (***), 0.0001 (****) by 2-tailed t-test. LC-MS of microtubules and additional data in Figure S2.
Readout of β-tail glutamylation by the katanin pore
The observed response to TTLL6 and 7-mediated glutamylation on microtubule severing could be due to the position and length of the glutamate chains added by these enzymes and the different functions the α- and β-tubulin tails have in katanin-mediated severing. The β-tail is threaded through the katanin central pore and required for severing while the α-tail contributes to binding but is not essential53. TTLL6 adds long glutamate chains preferentially on α-tubulin at E44366 while TTLL7 can add chains at multiple positions in the β-tail76, 78. MS/MS mapping of the TTLL7-introduced glutamylation sites on the βI-tail show preferential glutamylation at E441 followed by E438 and to a lesser extent at E439, E442 and E443 (Figure 3A, Figure S3A, B, STAR Methods). Extracted-ion chromatograms (XIC) for βI-tail peptide species with increasing glutamate numbers show the gradual decrease in the dominant peak for glutamylation at E441 and the gradual appearance of additional peaks for tails modified at additional sites (Figure S3A). Katanin uses two electropositive interconnected spirals in its central pore (pore dimensions in Figure S4A) to coordinate the glutamate-rich β-tail essential for severing53. We wanted to gain an atomic-level understanding of whether and how polyglutamate branches can be accommodated in the katanin pore. We modeled katanin in complex with tubulin tails having one or two glutamate branches at positions determined from our MS/MS data. Energy minimization of these structures showed all models have excellent geometry with no clashes in the substrate binding pore (Figure 3B, C, Table S1, Figures S4B–H; STAR Methods). These models revealed that a single poly-Glu branch can be accommodated at any position and can run in either of the two directions parallel to the tubulin tail (Figures 3E, F, S4 C, D; Models 1 and 2 in Table S1 and Data S1), providing additional binding energy and enhancing severing. Addition of a second branch, one or more residues long (Figure 3A, G, H, Models 3, 4, 5, 6 in Table S1, Figures S4E–H, and Data S1) can be accommodated, but with restrictions. Specifically, the second branch can be accommodated in either polarity at any position if the second branch is short and does not overlap with the first one (Figures 3G, S4E, Model 3 in Table S1). A longer branch can be accommodated only in the opposite polarity from the first one (Figures 3H, S4F–H; Models 4, 5 and 6, Data S1), otherwise there are steric clashes between the second branch and pore loops [20.12 clash score versus zero for the other branch configurations (Table S1 and Figure 3H)]. Once the second longer poly-Glu branch is added there is an entropic penalty because only a subset of configurations is compatible to bind in the pore. If two branches overlap, the three polypeptide chains (tubulin tail and the two poly-Glu branches) do not fit in the pore (Figures 3G, H, S4I, Model 7 in Table S1 and Data S1). Thus, second poly-Glu branches can be added in any configuration if short. They become inhibitory when juxtaposed with the primary branch at any position along their length as they thread through the pore. At higher glutamylation levels with two glutamate chains initiated by TTLL7, most configurations that the poly-Glu branches sample are not compatible with translocation. The increase in binding with increased β-tail glutamylation (Figures 2A, B, C) indicates that katanin is still able to bind some of these chain structures which provide additional negative charges that the electropositive pore loops recognize; however, these branch configurations do not allow the enzyme to assemble productively around the modified tail to extract tubulin, thus resulting in gradual inhibition beyond <nE> ~7 (Figures 2E, F). Consistent with this, the minimum katanin concentration at which TTLL7-glutamylated microtubules are severed initially decreases from 4.5 to 3.5 nM, but then slowly increases with glutamylation levels (Figure S2C), unlike for TTLL6-glutamylated microtubules where this concentration shows a monotonic decrease (Figure S1E).
Figure 3. Digital regulation of katanin severing by polyglutamate branch structure.
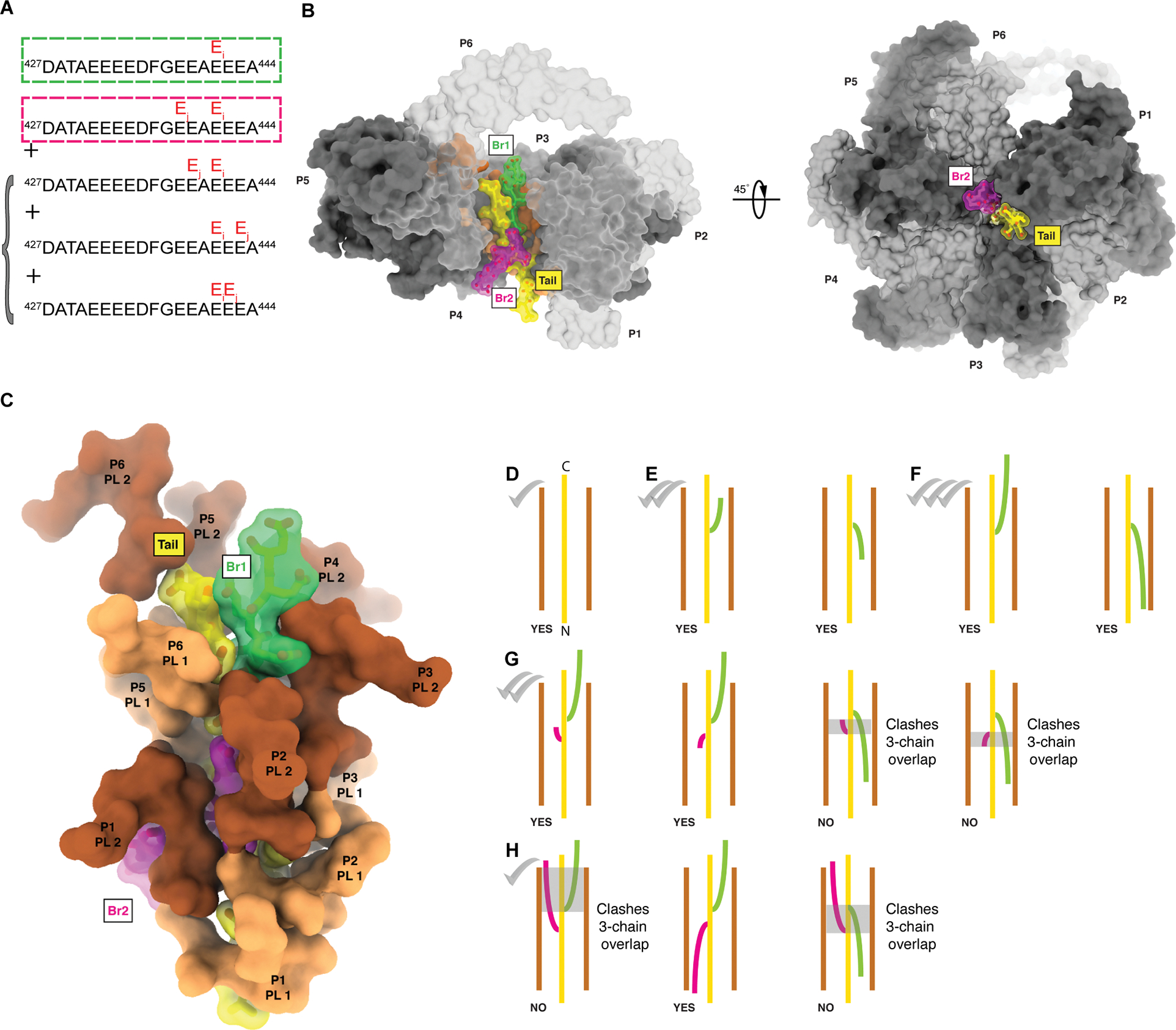
(A) Positions of glutamate chains added by TTLL7 on the β-tail. Main poly-Glu chain, green box, secondary poly-Glu chain, magenta box. Additional minor species detected shown in bracket. For MS/MS spectra and XIC see Figure S3. β-tail residues beyond DATA are intrinsically disordered and not visible in microtubule cryo-EM reconstructions133,100.
(B) Left, side-view, right, top-view through the pore of the modeled katanin hexamer (grey and light grey for alternating protomers, labeled P1 through P6), bound to a β-tubulin tail (yellow) with two poly-Glu branches (branch 1, green, six glutamates; branch 2, magenta, seven glutamates, branch geometry as in Model 5 in Table S1).
(C) Coordination of the polyglutamylated tail by katanin pore loops 1 (PL1, orange) and 2 (PL2, brown).
(D-H) Schematics of tubulin tails (yellow) with different poly-Glu branch structures (green and magenta) in the katanin pore. Branched peptide geometries that can be accommodated in the pore are labeled “YES”; geometries that cannot are labeled “NO”. Severing activity level indicated with checkmarks, (D); A single poly-Glu branch of any length can be accommodated in the katanin pore in either polarity, Models 1 and 2 in Table S1 and Data S1, (E, F); A second poly-Glu branch can be accommodated in either polarity if it is short so that it is not juxtaposed with the first branch, Model 3 in Table S1 and Data S1, (G); Two long poly-Glu branches can be accommodated only if of opposite polarities, models 4, 5, 6 in Table S1 and Data S1, (H). No more than two polypeptide chains (tail + one poly-Glu branch) can be accommodated at the same pore height – overlap highlighted with grey transparent boxes. The substrate is too wide to fit without clashes in the katanin pore when the two branches are juxtaposed (Model 7 in Table S1, Figure S4I, Data S1).
Combinatorial regulation by glutamylation on α- and β-tubulin tails
In cells, microtubules are modified combinatorially by multiple TTLLs. We thus sought to understand how the combined glutamylation on α- and β-tubulin by TTLL6 and 7 regulates katanin. We identified a series of conditions that result in a wide range of glutamylation levels on α- and β-tubulin (Figures 4, S5A, B, STAR Methods) that are within ranges found in vivo. Severing assays with these differentially modified microtubules show that glutamylation on α-and β-tubulin synergize for higher activity when the glutamate number on the β-tail is in the stimulatory regime (<nE> β < ~ 7; Figure 4A). Once β-tubulin glutamylation is in the inhibitory regime (<nE> β > ~ 7), the increase in glutamates on the α-tail cannot relieve this inhibition, even though increased α-tubulin glutamylation increases katanin microtubule binding (Figure 4B, S5B, S5E). Thus, the addition of multiple poly-Glu branches on the β-tail acts as a dominant negative for severing despite contributing to increased microtubule binding, consistent with our structural analyses which show that multi-branch configurations on the β-tail lead to unproductive binding to the microtubule (Figure 3). Lastly, when examining microtubules with similar overall glutamate numbers on αβ-tubulin (<nE>αβ), severing is higher for microtubules with higher numbers of glutamates on α-tubulin. Taken together, these data indicate that severing is stimulated most on microtubules with the highest fraction of glutamylation on α-tubulin (Figure 4A), while microtubule recruitment increases with increasing glutamylation of both α-and β-tubulin (Figure 4B).
Figure 4. Combinatorial regulation by polyglutamylation on α- and β-tubulin tails.
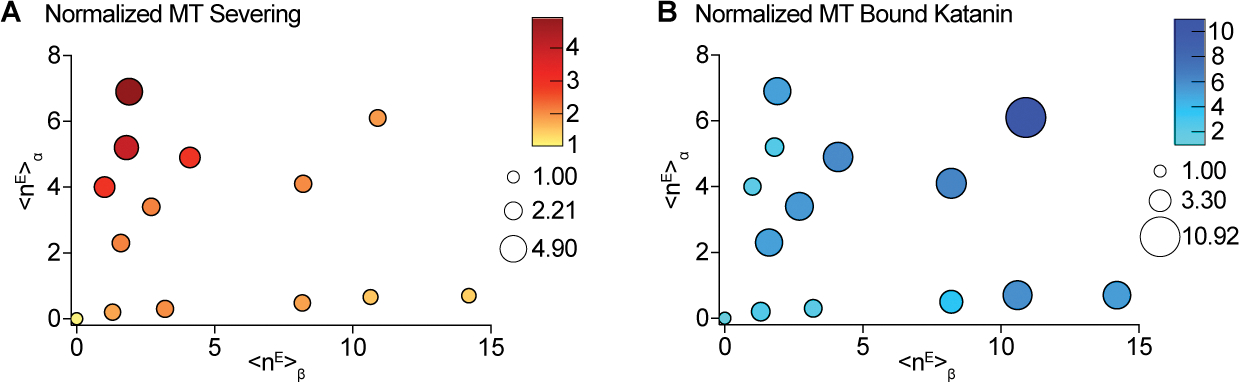
(A, B) Bubble chart of normalized microtubule severing (A) and binding (B) as a function of glutamylation on α- (y-axis) and β-tubulin (x-axis). <nE>α and <nE>β, weighted average of glutamates on α- or β-tubulin, respectively. Scatter plots of these data and LC-MS of microtubules in Figure S5.
Binary regulation of katanin by the detyrosination/tyrosination cycle
Given the contribution of the α-tail to katanin microtubule binding and the stimulation of microtubule binding and severing by its glutamylation, we examined whether α-tubulin detyrosination also regulates katanin. Detyrosination involves the reversible removal of the α-tubulin terminal tyrosine. It regulates the activity of microtubule effectors, including dynein/dynactin79,23, MCAK80,81 and CLIP-17020,82. We thus generated detyrosinated microtubules through enzymatic modification by vasohibin-1 complexed with the small vasohibin binding protein (VASH1/SVBP)83,84 (Figure S6A, STAR Methods). Experiments with these microtubules showed that detyrosination decreases katanin microtubule binding (Figures 5A, B) while also decreasing severing (Figures 5C, D). The stimulatory effect of the terminal tyrosine is also supported by experiments with recombinant engineered tyrosinated and detyrosinated microtubules (Figures S6B, C). The closely related severing enzyme spastin does not distinguish between tyrosinated and detyrosinated α-tubulin60. Unlike spastin, katanin functions in complex with a regulatory subunit, p80. The microtubule interacting and trafficking (MIT) domain and linker in the catalytic p60 subunit together with the α-helical C-terminal domain of p80 (p80C) form a high-affinity microtubule binding module85,86,87,88 (Figure 5E). TIRF-based microtubule binding assays with recombinant, isotypically pure, tyrosinated and detyrosinated α1A/βIII microtubules show that this p60/p80 microtubule binding module (ΔAAA katanin) is sufficient to sense tyrosination. The terminal tyrosine makes a dominant contribution to the binding energy because removal of the entire α-tubulin tail has no further significant effect (Figure 5F). Previous work showed that katanin p60 is more potently inhibited by tyrosinated α-tubulin tail peptides fused to BSA than detyrosinated ones89. Our ATPase assays show very weak ATPase stimulation of the p60 subunit by the α-tail, regardless of tyrosination status (Figure S6D). This contrasts with the β-tail which binds in the katanin central pore and stimulates the ATPase53 and indicates that the α-tail, in isolation, does not promote hexamerization by binding to the AAA central pore. Thus, the tyrosinated α-tail is sensed by elements outside the catalytic AAA core, consistent with a model where the AAA core engages the β-tubulin tail while additional interactions with the α-tail are made through the flexible katanin arms which harbor an additional microtubule binding module sensitive to tyrosination.
Figure 5. Microtubule detyrosination inhibits katanin-mediated severing and polyglutamylation can overcome this inhibition.
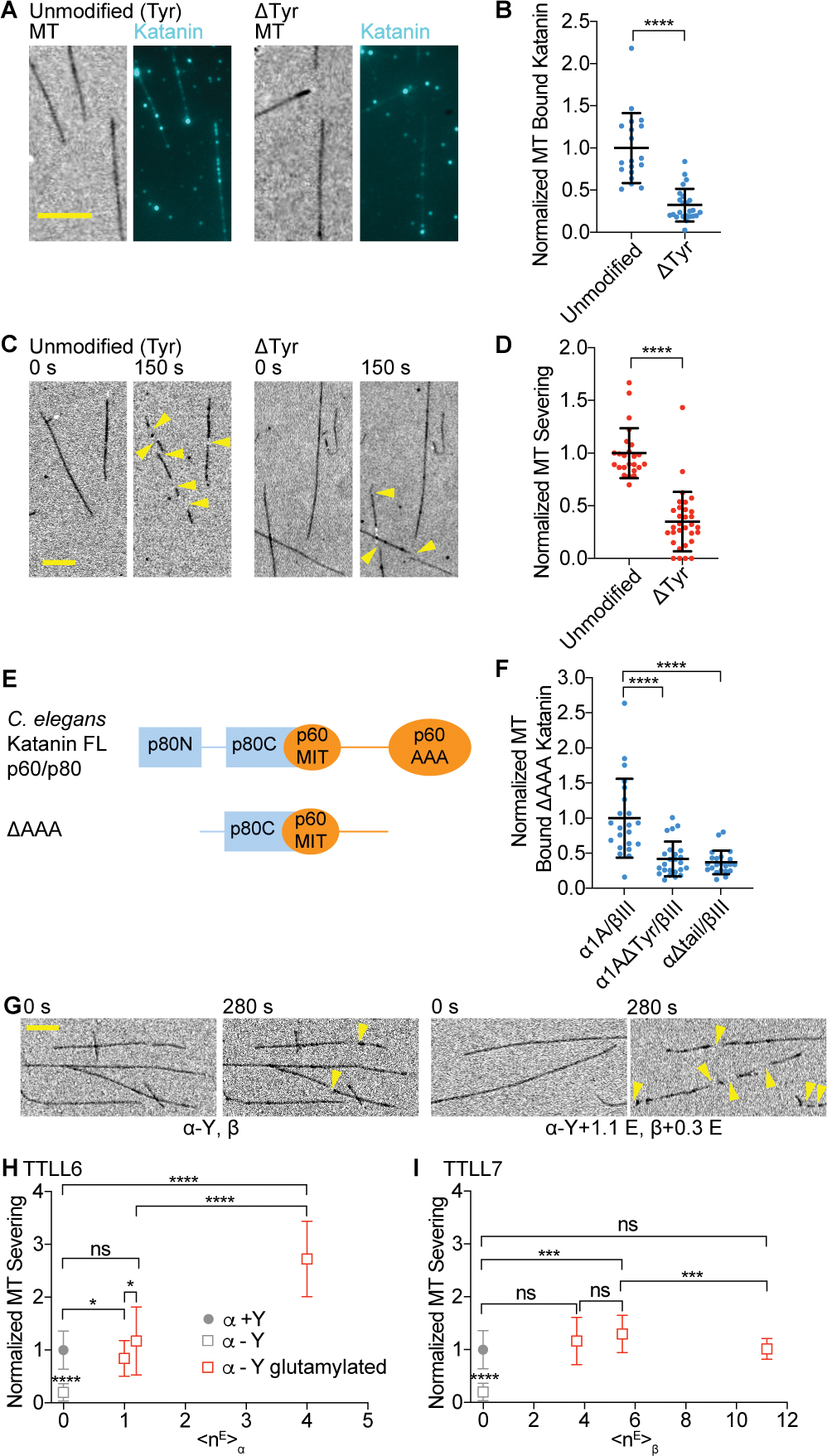
(A) Katanin association to unmodified tyrosinated and detyrosinated microtubules. Scale bar, 5 μm.
(B) Normalized katanin bound to tyrosinated and detyrosinated microtubules. Assays at 4 nM katanin, 1 mM ATP; n = 19 tyrosinated, 24 detyrosinated microtubules from multiple chambers.
(C) Severing of unmodified tyrosinated and detyrosinated microtubules. Yellow arrows, severing. Scale bar, 5 μm.
(D) Normalized severing of tyrosinated and detyrosinated microtubules. Reactions at 20 nM katanin, 1 mM ATP; n = 24, 31 tyrosinated and detyrosinated microtubules, respectively, from multiple chambers. LC-MS of microtubules and additional data in Figure S6.
(E) Domain organization of C. elegans katanin showing interactions between p60 and p80 subunits.
(F) Microtubule binding of p60ΔAAA/p80Cterm to recombinant human tyrosinated (α1A/βIII), detyrosinated (α1AΔY/βIII), and microtubules missing their α-tubulin tails (α1AΔtail/βIII); n = 22, 24 and 22 tyrosinated, detyrosinated and α-tailless microtubules, respectively from multiple chambers.
(G) Severing of detyrosinated microtubules (left) and detyrosinated microtubules glutamylated by TTLL6 (right). Yellow arrows, severing. Scale bar, 5 μm.
(H, I) Severing of detyrosinated microtubules with progressively higher glutamylation levels added by TTLL6 (H) or TTLL7 (I), normalized to severing of unmodified microtubules. Filled circle, tyrosinated, open square, detyrosinated microtubules with different glutamylation levels; Severing at 20 nM katanin, 1 mM ATP; n ≥ 29 microtubules from multiple chambers. All error bars, S.D. p-value > 0.05 (ns), p ≤ 0.05 (*), 0.001 (***), 0.0001 (****) by Mann-Whitney test.
Glutamylation overrides katanin inhibition by detyrosination
In cells, glutamylation and detyrosination are frequently found on the same microtubule. Both modifications accumulate on microtubules with long lifetimes (reviewed in Yu et al.90). Since α-tail glutamylation is strongly stimulatory and detyrosination is inhibitory we investigated the combinatorial effect of these two modifications. We found that polyglutamylation on either the α- or β-tail overcame the inhibitory effect of detyrosination, with α-tubulin glutamylation having the strongest effect (Figures 5G–I). At <nE>α ~ 1, detyrosinated microtubules are severed as effectively as tyrosinated microtubules. At higher <nE>α, glutamylation was strongly stimulatory (Figures 5G, H). Similarly, glutamylation on β-tubulin can overcome the inhibitory effects of detyrosination such that at <nE>β ~ 4, severing activity is comparable to that on tyrosinated microtubules (Figure 5I). This enhancement over the detyrosinated substrate persists at higher glutamylation levels. Thus, katanin can effectively remodel long-lived microtubules that have accumulated both detyrosination and glutamylation marks.
Lys40 acetylation has no effect on katanin
Glutamylated and detyrosinated microtubules are also frequently acetylated91,92. This overlap has made it difficult to establish causation in cellular experiments. Previous studies in fibroblasts reported a higher sensitivity to severing by katanin of microtubules acetylated on α-tubulin Lys4093. Unlike detyrosination and glutamylation, which alter tubulin tails on the outside microtubule surface, Lys40 α-tubulin acetylation occurs in the microtubule lumen94,95,96 where it cannot be directly sensed by katanin. Acetylation alters the conformation of a loop close to lateral polymerization interfaces94 and allows microtubules to bend without breaking97,98.
Given the connection between acetylation and the strength of microtubule lattice interactions, as well as cellular studies pointing to acetylation as a possible regulator of severing93, we examined katanin activity on acetylated microtubules generated by in vitro enzymatic modification using tubulin acetyltransferase71 (Figure S7A). Contrary to in vivo observations, acetylation does not affect katanin microtubule association or severing (Figures 6, S7B). Because acetylation at Lys40 affects lateral interactions between protofilaments, and taxol binds between protofilaments and changes microtubule rigidity as well as lattice parameters99,100, we also assayed GMPCPP microtubules, to exclude any confounding effects from taxol. These experiments also showed no effect of acetylation (Figures S7C, D, E). Therefore, the correlation between microtubule acetylation and higher severing in cells is due to other factors, likely glutamylation which we now show is stimulatory for severing, and frequently also found on stable microtubules that are acetylated. Our results are consistent with experiments in Tetrahymena that showed no increased katanin binding to acetylated microtubules70. Katanin is inhibited by microtubule associated proteins such as MAP4, tau and MAP2101,102,103 and the higher sensitivity to severing of acetylated microtubules in vivo could also reflect an exclusion of protective microtubule associated proteins.
Figure 6. Katanin is not regulated by α-tubulin Lys40 acetylation.
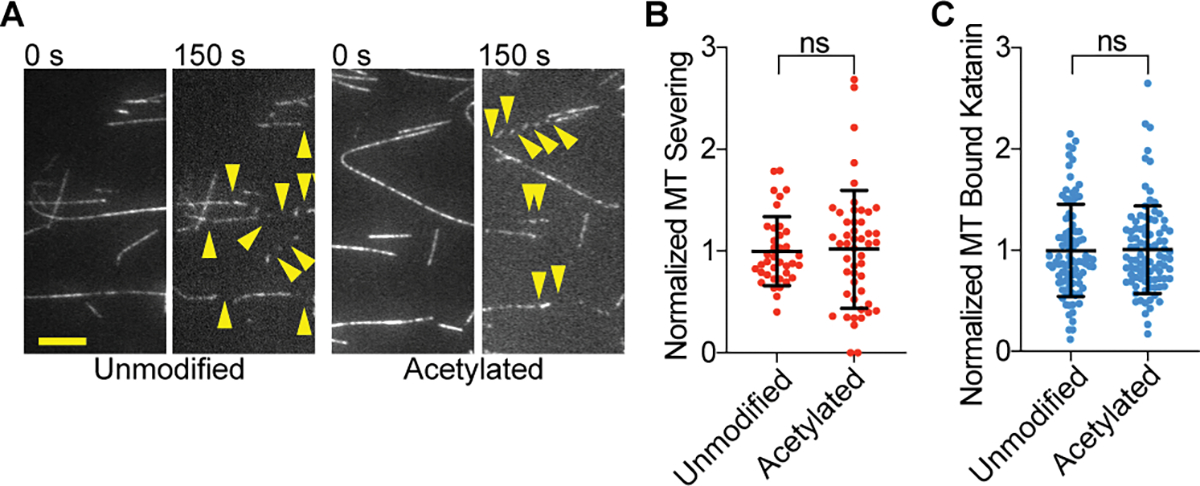
(A) Severing of unmodified and acetylated microtubules. Yellow arrows, severing events. Scale bar, 5 μm.
(B) Normalized severing of unmodified and acetylated microtubules. Reactions at 20 nM katanin, 1 mM ATP; n = 89, 92 microtubules from multiple chambers for unmodified and acetylated, respectively.
(C) Normalized katanin levels bound to unmodified and acetylated microtubules. Binding performed at 4 nM katanin, 1 mM ATP; n = 38, 49 unmodified and acetylated microtubules, respectively, from multiple chambers. All error bars, S. D., p-value > 0.05 (ns) by Mann-Whitney test. LC-MS of microtubules and additional data in Figure S7.
Microtubule glycylation inhibits katanin binding and severing
Because populations of pure glycylated microtubules have hitherto been unavailable, this is the biochemically least well understood among tubulin modifications characterized by covalent addition of amino acids. Glycylation is highly abundant in the axonemes of cilia and flagella where it is important for their stability73,104. Katanin is important for cilia biogenesis42 as well as deciliation105,44. Axonemal tubulin can have as many as 40 and 26 glycines on α- and β-tubulin respectively7. In mice, loss of glycylation affects axonemal dynein conformation resulting in sperm motility24. Thus, it is important to uncover the effectors through which glycylation performs its function and the effect of this modification on axonemal proteins.
Unlike glutamylation, where TTLLs can both initiate and elongate glutamate chains, glycine chains are added through the action of two distinct enzyme classes106,104 (reviewed in Garnham et al.107): one that strictly initiates the chain by adding one glycine to an internal glutamate (monoglycylation) and one that catalyzes elongation from this glycine to form a polyglycine chain (polyglycylation). We reconstituted tubulin glycylation in vitro by using recombinant TTLL3, a monoglycylase78 and TTLL10, an elongase (Figure S8; STAR Methods). Addition of only monoglycines to α- and β-tubulin by TTLL3 reduced severing (Figures 7A, B). Elongation of polyglycine chains from these monoglycines (<nG>α ~5.8 and <nG>β ~ 7.7) further decreased severing by 79% compared to unmodified microtubules, with almost complete inhibition at <nG> αβ ~ 25 (Figure 7B–C). Monoglycylation and polyglycylation both inhibit katanin recruitment to microtubules proportional to glycylation levels (Figures 7D, E, S9A, B). This decrease in binding is in stark contrast with the stimulation elicited by poly-Glu of comparable length (Figures 7F, 1C, 2C). A katanin construct missing the AAA domain also shows a reduction in microtubule binding as a function of glycylation (Figure 7G, S9C), indicating that the inhibition is not mediated exclusively through interactions between the AAA pore and the glycylated tubulin tail, which is less electronegative and bulkier because of the added glycines, but also through microtubule binding interfaces outside the AAA core by direct masking of interfaces or indirectly through conformational changes.
Figure 7. Katanin binding and severing is inhibited by glycylation.
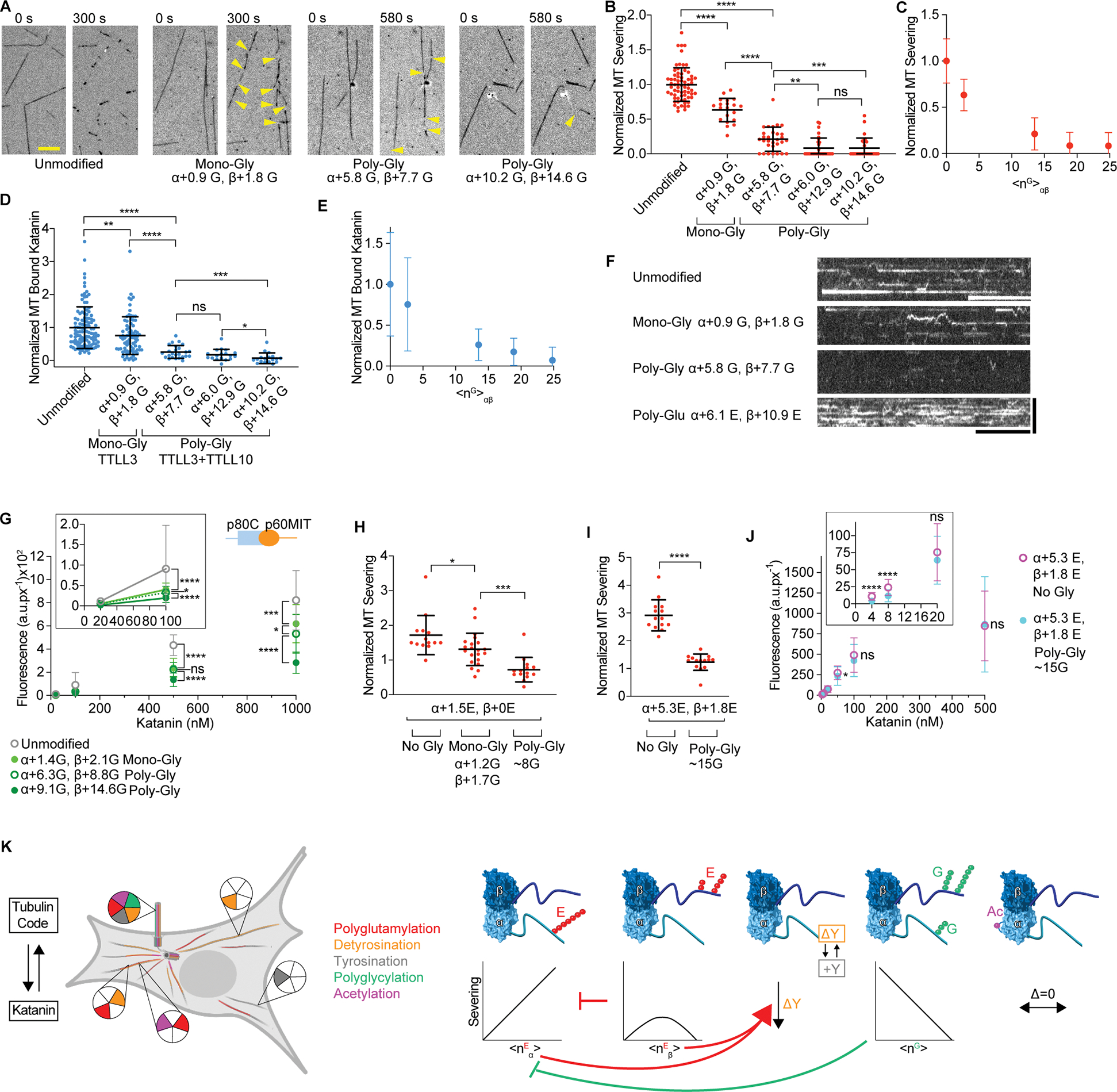
(A) Severing of unmodified, monoglycylated and polyglycylated microtubules. The weighted mean of the number of glycines (<nG>) added to α- and β-tubulin are denoted α + <nG>; β + <nG>. Yellow arrows, severing events. Scale bar, 5 μm.
(B) Severing decreases with glycylation levels; rates normalized to those on unmodified microtubules; n = 68, 20, 30, 30 and 30 microtubules from multiple chambers for unmodified, α+0.9 G β+1.8 G, α+5.8 G β+7.7 G, α+6.0 G β+12.9 G and α+10.2 G β+14.6 G microtubules, respectively. Reactions at 20 nM katanin, 1 mM ATP. LC-MS of microtubules and additional data, Figure S8.
(C) Normalized severing as a function of total glycines on the tubulin dimer <nG>αβ.
(D) Katanin association with microtubules decrease with glcylation; Katanin levels normalized to those on unmodified microtubules; n = 112, 77, 25, 18 and 18 microtubules from multiple chambers for unmodified, α+0.9 G β+1.8 G, α+5.8 G β+7.7 G, α+6.0 G β+12.9 G and α+10.2 G β+14.6 G microtubules, respectively. LC-MS of microtubules in Figure S8.
(E) Normalized katanin levels bound to microtubules as a function of total glycines on the tubulin dimer, <nG>αβ.
(F) Kymographs showing katanin binding to unmodified, monoglycylated, polyglycylated and polyglutamylated microtubules. Katanin concentration 8 nM for all conditions in D-F except glutamylation where 4 nM was used due to the higher binding affinity. Scale bar, 10 s (horizontal), 5 μm (vertical).
(G) Background-corrected fluorescence of katanin p60ΔAAA/p80Cterm bound to microtubules at various concentrations. Grey, unmodified, light green, monoglycylated α+1.4 G β+2.1 G, dark green open circles, polyglycylated α+6.3 G β+8.8 G, dark green filled circles, polyglycylated α+9.1 G β+14.6 G. n ≥ 22 microtubules for each condition. Scatter plots of this data, Figure S9C.
(H, I) Mono- and polyglycylation inhibit severing of TTLL6-glutamylated microtubules with low (H) or high (I) glutamylation levels (STAR Methods); n ≥ 14 microtubules for each condition from two chambers.
(J) Background-corrected fluorescence of katanin bound to the microtubules in (G) at various concentrations and in the presence of ATPγS; n≥19 microtubules for each condition from two chambers. All error bars, S. D.; p > 0.05 (ns), ≤ 0.05 (*), 0.01 (**), 0.001 (***), 0.0001 (****) by Mann-Whitney (in B, D, H, I) and 2-tailed t-test (in G, J).
(K) Differential and combinatorial regulation of katanin by the tubulin code.
We also investigated the combined effects of glycylation and glutamylation, a situation that could be relevant to cilia. We first glutamylated microtubules with TTLL6 and then monoglycylated with TTLL3. Glutamylation by TTLL6 and glycylation by TTLL3 are carefully choreographed during cilia biogenesis, with TTLL6 glutamylation appearing early, and required for their biogenesis, and monoglycylation later, and required for maintenance73. As cilia mature, polyglutamylation gradually decreases while polyglycylation increases42. LC-MS experiments showed that TTLL3 glycylated the glutamylated tubulin species (Figure S9D). We then extended the polyglycyl chains using TTLL10 (STAR Methods). The mass spectra of the polyglycylated species were too complex to assign exact glycine numbers for each species; however, we could estimate polyglycylation levels by performing Western blot analyses with antibodies that recognize poly-Gly chains104,108 and calibrating these against samples with known poly-Gly levels (Figure S9E). These experiments revealed that severing of glutamylated microtubules is progressively inhibited by mono- and polyglycylation (Figures 7H, I). Polyglycylation can inhibit even severing of microtubules with high glutamylation levels which are on their own highly stimulatory, returning severing close to that of unmodified microtubules (Figure 7I). This reduction in severing occurs even though similar katanin levels are recruited to the microtubule under these conditions (Figure 7J), suggesting that polyglycylation cannot overcome the stimulatory effects on binding elicited by polyglutamylation, but still inhibits severing due to an impairment in the activation of katanin, likely due to defective assembly of the enzyme around the polyglycylated tubulin tails. Taken together, our experiments demonstrate that glycylation is a potent inhibitor of katanin.
Discussion
Our work demonstrates that katanin is a hub that responds quantitatively to three chemically distinct tubulin posttranslational modifications, modulating its microtubule binding and severing activities. As polymers that explore most of the cellular space, microtubules are the largest signaling platforms, able to integrate inputs from distant sites. As has been prominently shown e.g., in mitogenic signaling, posttranslational modifications, most notably phosphorylation, are key to controlling and organizing the crosstalk between molecules recruited to signaling platforms. Analogously, recent studies revealed that chemically diverse posttranslational modifications pattern the microtubule network, controlling the specificity and timing of recruitment of microtubule effectors (reviewed in Roll-Mecak2). We show that tubulin modifications elicit complex and divergent functional outputs from katanin. Polyglutamylation of α-tubulin by TTLL6 increases both microtubule binding and severing, proportional with the number of added glutamates (Figures 1, 7K). This direct regulation by TTLL6 offers a mechanistic explanation for recent in vivo work that shows TTLL6 overexpression rescues axonal pathfinding errors due to partial katanin depletion58. It also explains why katanin preferentially severs the axonemal B-tubules42 which are glutamylated12. Katanin also participates in the formation of the central pair in motile cilia by severing microtubules close to the cilium tip109. As katanin severs the elongating outer doublet microtubules, the resulting seeds are stabilized and organized for elongation into the central pair. Interestingly, mutations in the tubulin tail that abolish glutamylation and glycylation also result in loss of the central pair in Tetrahymena42,56, as do TTLL6 mutations, raising the possibility that glutamylation may help specify severing during ciliogenesis.
In contrast to the stimulatory effects of glutamylation, by reconstituting glycylated microtubules in vitro, we show that glycylation by TTLL3 and TTLL10, also highly abundant in cilia, decreases both katanin microtubule binding and severing (Figure 7). Inhibition is proportional with glycine number. Furthermore, the inhibitory effect of glycylation is also operational on microtubules that are already glutamylated by TTLL6, a situation relevant to function in cilia. Glutamylation is required for cilia biogenesis73, while glycylation accumulates as cilia mature, controls their length110,111 and is required for their stability73,104,111. In the absence of glycylation, cilia disassemble. The inhibition of katanin severing by glycylation could be one component that participates in modulating cilia stability.
Glutamylation on the β-tail by TTLL7, a glutamylase whose expression increases with, and is important for neuronal differentiation77 regulates katanin severing biphasically. This biphasic response is due to the ability of katanin hexamer to thread through its pore two polypeptide chains simultaneously, the β-tubulin tail and one polyglutamate chain of any length, and its inability to accommodate two polyglutamate chains on the β-tail if they are juxtaposed, regardless of polarity (Figure 3). This constraint on molecular recognition of the modified tubulin tail explains why spastin shows a similar response to β-tail glutamylation60 since the two enzymes have similar pore sequences and architectures112,113,53. The closely related AAA ATPase VPS4 can accommodate a circular peptide in its pore114 further supporting our finding that the pore for this subclass of AAA ATPases is wide enough to accommodate two polypeptide chains. Other AAA ATPases such as ClpXP and the proteosome can translocate two polypeptide chains through their pores115,116,117,118.
α- and β-tubulin glutamylation affect axonal transport differently22, suggesting that the differential effect of glutamylation on α- and β-tubulin that we now demonstrate in vitro for severing enzymes may have parallels more broadly in other effectors. Our work demonstrates in an in vitro reconstituted system that there is regioselectivity to the readout of tubulin glutamylation and that this modification does not act simply by adding global negative charge to tubulin, providing a mechanistic explanation for the functional diversification of TTLL glutamylases. Moreover, the different effects on severing of hyperglutamylation of α- versus β-tubulin and the combinatorial mechanism with which they can act illustrates that phenotypes that result from loss of CCP1 deglutamylase activity on both α- and β-tubulin119 cannot be easily interpreted in the absence of more quantitative mechanistic investigations. For example, hyperglutamylation on α-tubulin enhances severing, but concomitant hyperglutamylation on β-tubulin acts as a dominant negative and will inhibit severing (Figure 4).
It was previously proposed that polyglycylation does not only change the overall charge on the tubulin dimer, but also leads to masking of the microtubule surface by the tubulin tails120 as polyglycine peptides favor compact, collapsed conformations unlike polyglutamate peptides which prefer extended conformations121. Indeed, recent molecular dynamics simulations showed that polyglutamylated tubulin tails are extended while polyglycylated ones collapse close to the microtubule surface122. We now show that poly-Gly chains block the productive interaction between the katanin pore and tubulin tail, while also masking additional interfaces that interact with katanin outside its AAA core (Figure 7G). Thus, glutamylation and glycylation not only change the net charge on tubulin tails, which are recognized by electrostatic interactions with katanin, both in the pore and accessory microtubule binding domains53, but also control access to the microtubule surface differently: the extended polyglutamate chains increase the odds of productive collision, with the tails acting as a lasso to recruit katanin, while polyglycine chains block interactions.
By modulating two polymodifications, glutamylation and glycylation, cells can elicit a wide range of functional outputs. Glycylation and glutamylation are not evenly distributed along the axoneme110,9 and they anti-correlate in their abundance104,111, at least partly due to overlapping modification sites78,123. Consequently, they can provide localized areas of protection against severing or concentrate severing activity in a small region, potentially during deciliation. Many proteins bind microtubules through interactions between their positively charged motifs and the negatively charged tubulin tails124. We hypothesize that glutamylation and glycylation can create gradients of attachment and detachment of proteins along the axoneme. Future work using in vitro glycylated microtubule generated with the method we describe here will establish whether glycylation functions as a more general inhibitor of microtubule interactions.
Unlike spastin60, katanin activity is higher on tyrosinated microtubules (Figures 5A–F, S6). The α-tail terminal tyrosine is recognized by a microtubule binding platform outside the AAA core which comprises the MIT domain, poorly conserved with spastin, and the p80 regulatory subunit, not shared with spastin. This tyrosination recognition module shows no homology to known tyrosination sensors such as CLIP-170 Cap-Gly domains or MCAK80,125. Interestingly, katanins can assemble with multiple regulatory subunits126,37. These are not as well-conserved as the catalytic subunit, and are thought to target katanins for distinct functions in the cell27,28. Part of this specialization could also stem from different responses to microtubule modifications. The downregulation by detyrosination that we observe can be rescued by glutamylation on either the α- or β-tails (Figures 5G–I). The experiments in this study were conducted with C. elegans katanin and human microtubules. Given the high degree of sequence conservation among species, especially in the central pore that coordinates the modified tubulin tail, we anticipate qualitatively similar responses for mammalian katanins, especially for glutamylation and glycylation. An interesting area of investigation will be how the various proteins that target and regulate specific katanin isoforms will synergize with the regulation by tubulin modifications that we report here.
Katanin assembles into a hexamer with the AAA ring at the center and flexible arms emanating from it that also bind microtubules127. Because of the multivalent microtubule recognition whereby distinct structural units, i.e., the AAA core and the flexible microtubule binding modules recognize different tubulin posttranslational modifications, katanin isn’t just regulated differentially by chemically distinct modifications, but is also able to integrate them for quantitatively different outputs. We anticipate that other modular microtubule associated proteins contain multiple tubulin modification recognition motifs that allow them to integrate different tubulin code inputs for diverse functional outcomes. The divergent effects of tubulin modifications that we show here for katanin make it imperative to know the tubulin code milieu in which all cytoskeletal effectors operate in cells. Finally, the different effects of distinct glutamylation patterns and the inhibitory effect of polyglycine chains we uncovered have broader implications for non-cytoskeletal proteins such as nucleosome assembly factors (NAPs) and cyclic GMP-AMP synthase (cGAS) that are regulated by different TTLL enzymes128,129.
Limitations of the study
We performed our severing assays at katanin concentrations close to those reported in vivo in various cell types130,131,132; however it is possible that the exact magnitude of the effects that we see in our in vitro assays is different in cells, depending on the local concentration of katanin, its posttranslational modification (such as phosphorylation35) as well as the presence of other regulatory factors such as MAPs which have been shown to control katanin access to the microtubule. Our in vitro reconstitution work will serve as a reference for future studies of katanin regulation in vivo.
STAR Methods
RESOURCES AVAILABILITY
LEAD CONTACT
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Antonina Roll-Mecak (Antonina@nih.gov).
MATERIALS AVAILABILITY
All plasmids used in this study are available upon request from the authors.
DATA AND CODE AVAILABILITY
Microscopy and mass spectrometry data reported in this paper will be shared by the lead contact upon request. This paper does not use any original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
EXPRESSION IN BACULOVIRUS SYSTEM
Recombinant human tubulin, VASH1/SVBP, TTLL3 and TTLL10 were expressed in Sf9 insect cells. Cells were cultured in Sf-900™ III SFM media (Life Technologies) at 27°C, 125 rpm.
E. COLI EXPRESSION STRAINS
Katanin was expressed in BL21 (DE3) Competent E.coli (NEB, C2527H). TTLL6 was expressed in E. coli Arctic Express DE3 (Agilent). TTLL7 and TAT1 were expressed in E. coli Rosetta2(DE3)pLysS (Novagen).
HUMAN CELL LINES
Unmodified human tubulin was purified from tsA201 cells. tsA201 cells were cultured in Freestyle293 supplemented with 2% FBS and 1xPenicillin/Streptomycin at 37°C, 125 rpm, 8% CO2, and 70% humidity. For large scale growth, medium contained additional 2 mM glutamine.
METHOD DETAILS
PROTEIN EXPRESSION, PURIFICATION AND LABELING
The katanin p60/p80 heterodimer from C. elegans134 was expressed in E. coli and purified as described previously127. The ΔAAA construct was also co-expressed and purified as a complex (Key Resource Table). Katanin p80 was fused to an Atto488-labeled peptide using Sortase A135 as previously described53. The microtubule severing activity of the labeled katanin construct is similar to that of the unlabeled protein53. TTLL7 and 6 were purified as described previously75,66. The gene for Xenopus tropicalis TTLL10 (residues 105–570) was cloned into pCoofy28136 (Addgene plasmid # 44004; http://n2t.net/addgene:44004; RRID:Addgene_44004) with an N-terminal GST tag and bacmid was produced by transformation of DH10EMBacY (Geneva Biotech). Baculovirus was produced by transfection of and subsequent amplification with ExpiSf9 cells (Thermo Fisher Scientific). Large scale expression was done in Sf9 cells. Cells were infected at a density of 2.5×106 cells/mL with an MOI of 2 and then grown for 48 hrs before harvest. Cells were lysed using a microfluidizer, and cell lysate was clarified by centrifugation at 450,000 rcf for 60 minutes. The fusion protein was captured using GST affinity chromatography. TTLL10 was liberated using 3C protease at a 1:500 molar ratio. TTLL10 was further purified on a heparin column, followed by size-exclusion chromatography, and flash-frozen in 20 mM Tris pH 8.0, 10 mM MgCl2, 100 mM NaCl, 2 mM TCEP, 10% Glycerol.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-polyGly rabbit polyclonal | (Rogowski et al.104; Xia et al.139) | N/A |
| Anti-α-tubulin DM1α mouse monoclonal | Sigma | Cat# T9026-.2ML |
| LICOR IRDye 680LT Goat anti-Mouse IgG (H + L) | LI-COR BioScience | Cat# 926-68020 |
| LICOR IRDye 800CW Goat anti-Rabbit IgG (H + L) | LI-COR BioScience | Cat# 926-32211 |
| Bacterial and virus strains | ||
| E.coli BL21 (DE3) | NEB | Cat# C2527H |
| E. coli Arctic Express (DE3) | Agilent | Cat# 230192 |
| E. coli Rosetta2(DE3)pLysS | Novagen | Cat# 71401 |
| Chemicals, peptides, and recombinant proteins | ||
| GMPCPP | Jena Bioscience | Cat# NU-405L |
| Catalase from bovine liver | Sigma Aldrich | Cat#C40 |
| Glucose Oxidase from Aspergillus niger | Sigma Aldrich | Cat#G7141 |
| Pluronic F-127 | Life Technologies | Cat#P6866 |
| Casein | Sigma Aldrich | Cat#C5890 |
| Neutravidin | Life Technologies | Cat#31000 |
| C.elegans Katanin MEI-1/MEI-2 | This study (Zehr et al.127) | N/A |
| Human Unmodified Tubulin from tSA201 Cells | This study (Vemu et al.71) | N/A |
| H.sapiens α1A-Tubulin (NP_001257328) H.sapiens βIII-Tubulin (NM_006077) | (Chen et al.20, Vemu et al.133) | N/A |
| H.sapiens α1A-Tubulin detyrosinated H.sapiens βIII-Tubulin (NM_006077) | (Chen et al.20) | N/A |
| H.sapiens α1A-Tubulin tailless (aa 1-439) H.sapiens βIII-Tubulin (NM_006077) | (Chen et al.20) | N/A |
| Porcine Brain Tubulin | Cytoskeleton, Inc. | Cat# T238P |
| HiLyte647-Labeled Porcine Tubulin | Cytoskeleton, Inc. | Cat# TL670M |
| HiLyte488-Labeled Porcine Tubulin | Cytoskeleton, Inc. | Cat# TL488M |
| Biotin-Labeled Porcine Tubulin | Cytoskeleton, Inc. | Cat# T333P-B |
| Atto488-Labeled C.elegans Katanin | This study (Zehr et al.53) | N/A |
| Atto488-Labeled C. elegans Katanin MEI-1 (aa 1-150)/MEI-2 (aa 68-280) | This study | N/A |
| M. musculus TTLL6 aa 51-502 | (Mahalingan et al.66) | NP_766387.2 |
| X. tropicalis TTLL7 aa1-520 | (Valenstein et al.60) | NP_001136132.1 |
| X. tropicalis TTLL3 aa 6-569 | (Garnham et al.78) | N/A |
| X. tropicalis TTLL10 aa 105-570 | This study | N/A |
| Human VASH1/SVBP | T.R. Brummelkamp (Netherlands Cancer Institute) (Nieuwenhuis et al. 84) | N/A |
| D. rerio TAT1 aa 1-196 | Szyk et al.96, Vemu et al.71 | N/A |
| Peptide: VDSVEGEGEEEGEEY | Bio-Synthesis Inc/ (Zehr et al.53) | N/A |
| Peptide: VDSVEGEGEEEGEE | Bio-Synthesis Inc | N/A |
| Critical commercial assays | ||
| EnzCheck™ Phosphate Assay Kit | Thermo Fisher | Cat# E6646 |
| Deposited data | ||
| Katanin Hexamer In The Spiral Conformation Bound To Substrate | (Zehr et al.53) | PDBID: 6UGD |
| Experimental models: Cell lines | ||
| Human: tsA201 Cells Derived From HEK293 | Millipore Sigma | Cat# 96121229-1VL |
| Sf9 insect cells | Gibco | Cat#12659-017 |
| Oligonucleotides | ||
| Primers for C.elegans Katanin Mei-1 aa 1–150 in pCDF-Duet: introducing the stop codon after aa 150: GGCTGATGTGAAACCGGCGAATtaaACCCAGGGCATT, AATGCCCTGGGTTTAATTCGCCGGTTTCACATCAGCC | Eurofins Genomics LLC | N/A |
| Primers for C.elegans Katanin Mei-1 aa 1–150 in pCDF-Duet: insertion of 3C protease site and StrepTagII: GCTGATGTGAAACCGGCGAATctggaagttctgttccaggggccctcagcgtggagccacccgcagttcgagaaataaACCCAGGGCATTCTGCC, GGCAGAATGCCCTGGGTttatttctcgaactgcgggtggctccacgctgagggcccctggaacagaacttccagATTCGCCGGTTTCACATCAGC | Eurofins Genomics LLC | N/A |
| Primers for C.elegans Katanin Mei-2 aa 68–280 with sortase tag in pMAL C5X: cloning of Mei-2 aa 68–280 to pMAL C5X: GAATTCcatatgGAAAACCTATATTTCCAATCAAGACGTCCATTGCATTCCGAG, GTCGACggatccTTACTTATGGCTGGAAACTTTTTTCAATCGC | Eurofins Genomics LLC | N/A |
| Primers for C.elegans Katanin Mei-2 aa 68–280 with sortase tag in pMAL C5X: insertion of sortase tag: gaaaaaagtttccagccataagGGCGGTGGCGGTTCTCTGCCGGAAACCGGCGGTtaaAAGCTTgcggccgc, gcggccgcAAGCTTttaACCGCCGGTTTCCGGCAGAGAACCGCCACCGCCcttatggctggaaacttttttc | Eurofins Genomics LLC | N/A |
| Recombinant DNA | ||
| Human VASH1 in pFastBac™-based vector | T.R. Brummelkamp (Netherlands Cancer Institute) (Nieuwenhuis et al.84) |
N/A |
| Human SVBP in pFastBac™-based vector | T.R. Brummelkamp (Netherlands Cancer Institute) (Nieuwenhuis et al. 84) |
N/A |
| H.sapiens α1A-Tubulin and βIII-Tubulin in pFastBac™-Dual, Codon Optimized for Baculovirus Expression | (Vemu et al.71) | N/A |
| C.elegans Katanin Mei-1 in pCDF-Duet | Francis McNally (UC Davis) (McNally et al.134) |
NP_492257.1 |
| C.elegans Katanin Mei-2 in pMAL-CRI | Francis McNally (UC Davis) (McNally et al.134) |
NP_491894.1 |
| C.elegans Katanin Mei-2 with sortase tag in pMAL-CRI | This study (Zehr et al.53) | NP_491894.1 |
| C.elegans Katanin Mei-1 aa 1–150 in pCDF-Duet | This study | NP_492257.1 |
| C.elegans Katanin Mei-2 aa 68–280 with sortase tag in pMAL C5X | This study | NP_491894.1 |
| Software and algorithms | ||
| ImageJ Fiji | (Schindelin et al.141) | https://fiji.sc |
| Prism | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| Software for modeling: Coot | (Emsley et al.143) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Software for modeling: Phenix | (Moriarty et al.142) | https://phenix-online.org/download/ |
| Software for protein structure image rendering: ChimeraX, | (Goddard et al.147) | https://www.cgl.ucsf.edu/chimerax/ |
| Software for protein structure image rendering: UCSF Chimera | (Pettersen et al.146) | https://www.cgl.ucsf.edu/chimera/ |
| MassHunter | Agilent | https://www.agilent.com/en/product/software-informatics/mass-spectrometry-software |
| PyMOL | (Schrodinger148) | https://pymol.org/2/#page-top |
| Biorender (part of Fig. 7K and graphical abstract) | Biorender | https://biorender.com |
GENERATION OF MODIFIED MICROTUBULES
Generation of glutamylated microtubules. Tubulin was purified from tsA201 cells using TOG1 domain-affinity chromatography71,137 and was subject to a polymerization/depolymerization cycle to remove any remaining microtubule-associated proteins14,138. Mass spectrometry did not detect posttranslational modifications on this purified tubulin14, 71. Unmodified microtubules used in experiments were taxol-stabilized and contained 1–1.5% biotinylated tubulin (Cytoskeleton Inc.)60 and no fluorescent tubulin unless specified. The choice of taxol-stabilized microtubules eliminates the possible confounding effects of the modifications themselves on microtubule dynamics. Differentially glutamylated microtubules were obtained by enzymatic treatment with TTLL666 or TTLL775,60 at 1:10 molar ratio of enzyme to tubulin in 20 mM HEPES pH 7.0, 50 mM NaCl, 10 mM MgCl2, 1 mM Glu, 1 mM TCEP, 1 mM ATP, 10 μM taxol at room temperature. Incubation times for TTLL6 were 1 hr. for <nE>α ~ 1.1, 7.5 hr. for <nE>α ~ 2.9, 22 hr. with TTLL6 added twice at 1:20 molar ratio each for <nE>α ~ 6.9. Incubation times for TTLL7 were 30 min for <nE>β ~ 3.5, 1 hr. for <nE>β ~ 7.2, 1.5 hr. for <nE>β ~ 10, 3 hr. for <nE>β ~21. The enzyme was removed by a 300 mM KCl salt wash followed by passage through a 60% glycerol cushion60. Control microtubules were prepared the same way by either omitting glutamate (TTLL7) or adding aspartate (TTLL6), which is not a substrate for TTLL glutamylases. To generate microtubules with differentially modified α- and β-tubulin, unmodified microtubules were first incubated with TTLL6 followed by modification with TTLL7. For microtubules with long polyglutamate chains on α- and β-tubulin (8 or more Glu on β-tubulin), TTLL6 was first removed by salt wash before addition of TTLL7. Control microtubules were prepared identically, with aspartate substituting for glutamate. The numbers of glutamates added to α- and β-tubulin was determined by LC-MS60,71. The spectra were deconvoluted in MassHunter software (Agilent) and display the characteristic distribution of masses with peaks separated by 129 Da corresponding to one glutamate (Figures S1, S2, S5). The extent of tubulin glutamylation on α- or β-tubulin was determined by calculating the weighted average of peak intensities for each tubulin species present.
Generation of detyrosinated microtubules. Taxol-stabilized unmodified microtubules were first detyrosinated with 1:50 molar ratio of human VASH1/SVBP84 for 3 h at 37°C followed by a 300 mM KCl salt wash and centrifugation through glycerol cushion to remove the enzyme. Completion of the reaction was confirmed by mass spectrometry. Mass spectra displayed a 163 Da shift in the α-tubulin mass, corresponding to the loss of a tyrosine (Figure S6). Recombinant human tyrosinated (α1A/βIII), detyrosinated tubulin (α1A-Y/βIII) and tubulin missing the α-tubulin tail (αΔtail/βIII) were purified as described in Chen et al.20 and Vemu et al.133. Microtubules were assembled with 1.5% biotin brain tubulin (Cytoskeleton).
Generation of detyrosinated glutamylated microtubules. Taxol-stabilized unmodified microtubules were glutamylated with either TTLL6 or TTLL7 as described above. TTLL6 was then inhibited by addition of 1 mM ATPγS followed by addition of human VASH1/SVBP at 1:20 molar ratio to tubulin for 5h at 37°C. Modification enzymes were removed by a 300 mM KCl salt wash and centrifugation through a glycerol cushion. TTLL7 was removed by a 300 mM KCl salt wash as described above followed by treatment with VASH1/SVBP at 1:20 molar ratio to tubulin for 3h at 37°C followed by a second 300 mM KCl salt wash to remove VASH1/SVBP. Control tyrosinated or detyrosinated microtubules were treated the same way with aspartate instead of glutamate which does not serve as a substrate for TTLL6 and 7 enzymes under these conditions. The numbers of glutamates added to α- and β-tubulin were determined by LC-MS. The spectra displayed the characteristic distribution of masses with peaks separated by 129 Da corresponding to one glutamate.
Generation of acetylated microtubules. Taxol-stabilized unmodified microtubules with 4% fluorescent tubulin (HiLyte647 for binding assays, HiLyte488 for severing assays, both from Cytoskeleton Inc.) and 1.5% biotinylated tubulin, or GMPCPP-stabilized unmodified microtubules with 1.5% biotinylated tubulin were acetylated with tubulin acetyltransferase as previously described60,71 at 1:1 molar ratio for 24 h at room temperature. Control microtubules were incubated with coenzyme A instead of acetyl-coenzyme A. The acetyltransferase was removed by high salt wash as previously described71, but in the case of GMPCPP-stabilized microtubules, no glycerol cushion was used for centrifugation. Completion of the reaction was confirmed by mass spectrometry. Mass spectra displayed a 42 Da shift in the α-tubulin mass, corresponding to the addition of a single acetate group. No unacetylated α-tubulin was detected (Figure S7).
Generation of glycylated microtubules. Taxol-stabilized unmodified microtubules with 1.5% biotinylated tubulin were incubated with Xenopus tropicalis TTLL378, at 1:10 molar ratio of enzyme to tubulin at room temperature for 16 h in 20 mM HEPES pH 7.0, 10 mM KCl, 5 mM MgCl2, 1 mM TCEP, 1 mM ATP, 1 mM [13C]-glycine and 20 μM Taxol. To obtain polyglycylated microtubules, microtubules were incubated first with TTLL3 as above, followed by addition of KCl to 50 mM and 1:20 molar ratio of Xenopus tropicalis TTLL10 and additional 30 min incubation for <nG>α ~ 5.8, <nG>β ~ 7.7, and 2 hr. for <nG>α ~ 6.0, <nG>β ~ 12.9. To obtain the longest polyglycine chains with <nG>α ~ 10.2, <nG>β ~ 14.6, microtubules were first incubated with 1:5 molar ratio of TTLL3 for 24 h followed by addition of 1:20 TTLL10 for 3 h. The enzymes were removed by 300 mM KCl salt wash followed by centrifugation through a 60% glycerol cushion in Brb80 (80 mM K-PIPES pH 6.8, 1 mM EGTA, 1 mM MgCl2), 20 μM taxol for 12 min at 109,000 × g at 37°C. The pellet was washed with Brb80, 14 mM β-mercaptoethanol and 20 μM taxol and resuspended in the same buffer. Control microtubules were prepared identically but without glycine. The numbers of glycines added to α- and β-tubulin was determined by LC-MS. The spectra display the characteristic distribution of masses with peaks separated by 58 Da corresponding to one [13C]-glycine (Figure S8). The extent of tubulin glycylation on α- or β-tubulin was determined by calculating the weighted average of peak intensities for each tubulin species present.
Generation of glycylated glutamylated microtubules. Taxol-stabilized unmodified microtubules with 1.5% biotinylated tubulin were glutamylated with TTLL6. The modification enzyme was removed by a 300 mM KCl salt wash as described above. Then, they were glycylated with [13C]-glycine by addition of glycylation enzymes: 1:10 TTLL3 for the mono-glycylated samples, or 1:10 TTLL3 followed by 1:20 TTLL10 for polyglycylated samples in the conditions described above for the glycylation only reactions. Each sample was split in half after the glutamylation reaction: half was glycylated and the other half was incubated in the same conditions without glycine to generate the glutamylated only control. The number of glutamate residues added to tubulin was determined by LC-MS. The number of glycines added by TTLL3 in the monoglycylated sample was also determined by LC-MS. The spectra of the polyglycylated polyglutamylated samples were too complex to allow peak assignment for glycine number determination. The glycine numbers shown in Figure 7 H–J are based on Western blot analysis calibrated against polyglycylated microtubules with known glycine numbers as determined by LC/MS. Briefly, microtubules with <nG>αβ of 9.4, 15.1, 18.9 and 23.7 as well as polyglutamylated microtubules with unknown amount of polyglycylation were separated by SDS-PAGE and transferred onto a nitrocellulose membrane. α-tubulin and polyglycylated tubulin were detected with mouse DM1α (Sigma Aldrich) and polyclonal anti-poly-Gly antibodies104,139,108, respectively. Goat secondary antibodies conjugated with LiCor dyes were used for fluorescence detection. Fluorescent signal was measured, and background corrected. The polyglycylation signal was normalized to the tubulin signal and plotted as a function of <nG>αβ (Figure S9E). The polyglycylation level of polyglutamylated microtubules was approximated from the linear regression.
MS/MS ANALYSES
Taxol-stabilized unmodified microtubules were glutamylated with TTLL7 as above but using [13C]-glutamate as substrate to facilitate the differentiation of the posttranslationally added glutamates from the many glutamates present in the tubulin tails. Reactions were stopped by addition of 50 mM EDTA. Tubulin was digested in solution with AspN at 1:15 ratio of AspN to tubulin for 8 h at 37°C as previously described78. Peptides were desalted and injected into a nano-LC-MS/MS system where an Ultimate 3000 HPLC (Thermo-Dionex) was coupled to an Orbitrap Lumos mass spectrometer (Thermo Scientific) via an Easy-Spray ion source (Thermo Scientific). The composition of mobile phases A and B was 0.1% formic acid in HPLC water, and 0.1% formic acid in HPLC acetonitrile, respectively. The Thermo Scientific Orbitrap Lumos mass spectrometer was operated in data-dependent mode. The MS1 scans were performed in orbitrap with a resolution of 120K and a mass range of 400–1600 m/z, and an automatic gain control (AGC) value of 2 × 1e5. The MS2 scans were conducted in ion trap with an AGC target of 3 × 1e4. Two LC-MS/MS runs were performed for each sample. In the first run, peptides were separated on an ES800 Easy-Spray column (75-μm inner diameter, 15 cm length, 3 μm C18 beads; Thermo Scientific). The LC-MS/MS data acquisition was performed without a priority list and normal mass range was used for MS2 scan. In the 2nd run, an ES902 column (75-μm inner diameter, 25 cm length, 3 μm C18 beads; Thermo Scientific) was used. The species of interest were included in the priority list. High mass range was used for MS2 scan. For the database search, raw data were converted to peak list files in mgf format using Mascot Distiller (version 2.7.1.0). Database search was performed using Mascot Daemon (2.6.0) against the Sprot Human database. The following parameters were used for the database search. Enzyme: AspN; Variable modifications: 1 to 8 [13C]-glutamate on Glu residue; Mass values: Monoisotopic; Peptide mass tolerance: ± 10 ppm; Fragment mass tolerance: ± 0.6 Da; Max missed cleavages: 4. Peptides with heavy Glu modification matched by database search were manually curated.
MICROTUBULE SEVERING ASSAYS
Microtubule severing assays were performed as described previously60,140. Flow chambers of ~7 μl volume were constructed of silanized coverslips and slides. Flow chamber construction and silanization protocol is covered in detail in a previously published methods chapter140. Microtubules prepared as described in the section above were immobilized in the flow chamber with 0.02 mg/ml NeutrAvidin (Thermo Scientific) and washed with BRB80 with 2 mg/ml casein and 20 μM Taxol. The chambers were equilibrated with 15 μl of severing mixture without katanin (47 mM PIPES pH 6.8, 3.3 mM HEPES pH 7.0, 110 mM KCl, 2.2 mM MgCl2, 1.3 mg/ml casein, 0.6 mM EGTA, 2.5% glycerol, 9.1 mM 2-mercaptoethanol, 0.2 mM TCEP, 1 mM ATP, 1% Pluronic F127, 8.7 μM Taxol) supplemented with an oxygen scavenger mix to remove free oxygen from solution (7.5 U/μL catalase, 0.15 U/μL glucose oxidase, 20 mM glucose). Assays with GMPCPP-stabilized microtubules were performed the same way, but without the addition of Taxol. The chamber was placed on the microscope stage, field of view was chosen and 15 μl of freshly prepared severing mixture containing 20 nM katanin was perfused while recording. The 20 nM katanin concentration was chosen because it is close to the cellular katanin concentration of 3–28 nM130, 131,132. Microtubule images were acquired by differential interference contrast (DIC) or interference reflection microscopy (IRM)20 on a Nikon Eclipse Ti-E equipped with a CoolSnap camera (Photometrics) or an ORCA Flash4.0 V2 sCMOS camera (Hamamatsu), respectively, at 1 or 0.5 Hz for 10 min total. Taxol-stabilized acetylated microtubules were imaged at 1 Hz by TIRF on a Nikon Ti-E equipped with an iXON3–897 EMCCD camera (Andor). Microtubule severing progress was monitored by counting manually the number of observed severing sites after perfusion of katanin60. Microtubule severing rates were determined from the time required to observe one severing event per 10 μm of microtubule. Assays were performed at 21°C. To determine the minimal katanin concentration to observe a severing event, assays were performed as above, and microtubules were visualized by IRM. Katanin was perfused and images were acquired for 10 minutes at 0.5 Hz. The concentration of katanin was decreased each time, until there was only one or no microtubule severing events in the field of view containing at least 20 microtubules for 10 minutes after katanin perfusion.
MICROTUBULE BINDING ASSAYS
Microtubules were immobilized in the chamber the same way as for severing assays. Atto488-labeled katanin was perfused into the chamber in the same buffer as the one used for severing assays (47 mM PIPES pH 6.8, 3.3 mM HEPES pH 7.0, 110 mM KCl, 2.2 mM MgCl2, 1.3 mg/ml casein, 0.6 mM EGTA, 2.5% glycerol, 9.1 mM 2-mercaptoethanol, 0.2 mM TCEP, 1 mM ATP, 1% Pluronic F127) supplemented with an oxygen scavenger mix to remove free oxygen from solution (7.5 U/μL catalase, 0.15 U/μL glucose oxidase, 20 mM glucose). Binding assays were performed at 4 nM katanin except for glycylated microtubules and detyrosinated microtubules for which the katanin concentration was 8 nM because of the weaker binding for these microtubules. Binding assays for the truncated katanin construct (Figure 5F) were conducted at 50 nM katanin due to weaker binding of this construct compared to full length katanin which makes multivalent interactions with the microtubule. Assays were performed at 21°C. Images were acquired using an inverted total internal reflection fluorescence (TIRF) microscope (Nikon Ti-E with TIRF attachment) equipped with an iXON3–897 EMCCD camera (Andor). The excitation light was provided by a 488 nM diode laser at 20 mW. Light was delivered to the sample through a 100× 1.49 NA TIRF objective (Nikon CFI Apo TIRF 100x) and the emission filter FF01–550/88 or FF03–525/50 (Semrock) was used. Images were acquired continuously at 100 msec exposure with 488 laser and ND2 filter. An image of the microtubules in DIC or IRM was taken before starting image acquisition. The average background-corrected intensity along microtubules was measured on a sum of frames from 25 to 60 s in ImageJ Fiji141 and normalized for microtubule length.
To measure the binding of katanin to modified microtubules as a function of katanin concentration, Atto488-labeled katanin was perfused as above but 1 mM ATPγS was used instead of ATP to prevent severing. After 2 min incubation, microtubules were imaged by IRM and katanin by TIRF with 488 nm laser excitation and 100 ms exposure. The truncated katanin construct without AAA domain was imaged the same way but with no nucleotide in solution and at 50 mM KCl. Multiple fields of view were imaged. Background corrected line scan intensities were measured using Fiji141 and normalized to microtubule length.
MODELING OF GLUTAMYLATED TAILS BOUND TO KATANIN
The model for the katanin hexamer in the spiral conformation bound to tubulin tail substrate (ID: 6UGD)53 was used as a starting model for building katanin models bound to a 14-residue peptide with one or two polyglutamate branches in various configurations. The branch point structure was generated using https://chemdrawdirect.perkinelmer.cloud/js/sample/index.html#. Phenix eLBOW142 was used to obtain the pdb and crystallographic information (cif) file for the branch point. VPS4 bound to cyclic peptide (PDB ID 6OO2) was used as a guide to position the backbone of a branch within the pore114. Multiple tubulin tail substrates with one poly-glutamate chain at position 11, two poly-glutamate chains at positions 8 and 11, 9 and 11 or 11 and 12 were generated using Coot143. These positions are the most frequently modified positions within the βI and βIVb tubulin tails based on the MS/MS data. Using Coot, we modeled a link between the C05 atom of the branch’s Cγ and the N-terminal nitrogen atom of a poly-Glu branch. The remaining glutamates within a branch are α-linked. The atomic models were subjected to energy minimization in Phenix144,145. The atomic models were refined in real-space against the cryo-EM map for the katanin hexamer in the spiral conformation with substrate (EMD-20761)53 using Phenix145. To preserve the modelled link of the branch glutamate during refinement it was added as a bond in the custom geometry restrains window of the Phenix real space refinement GUI. Model statistics are listed in Table S1. Figures were prepared in UCSF Chimera146 and ChimeraX147. A Pymol148 session file for all the models is provided as Data S1. Pore dimensions shown in Figure S4 were measured manually or with MOLEonline149 and gave similar results.
ATPase ASSAYS
Peptides were purchased from Bio-Synthesis. ATPase assays with α-tubulin peptides were performed as described previously using an EnzChek Phosphate Assay (Life Technologies)53. Initial rates were calculated from the linear portion of the reaction profiles after addition of 1 mM ATP. ATPase rates were adjusted by subtraction of the measured release of phosphate in the absence of ATP.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were plotted and analyzed in Prism (Graphpad Inc). If the distribution of data was normal, a 2-tailed t-test was used. If the data were not normally distributed, a Mann-Whitney test was used. Each condition for binding and severing was measured in multiple chambers. The number of analyzed microtubules is indicated in the figure legends.
Supplementary Material
Data S1 (related to Figure 3 and Table S1).
Pymol session file with Models 1 through 7 shown in Table S1.
Tubulin tail, ball-and-stick, yellow; Branch 1, ball-and-stick, green; Branch 2, ball-and-stick, magenta; Branch positions labeled. Class in Model 7 indicated. Subset of pore loop 1 and 2 residues shown in cpk representation.
Highlights.
Katanin responds differentially to distinct glutamylation patterns on α- and β-tails
Glycylation and glutamylation have antagonistic effects on katanin activity
Katanin has a novel tyrosination recognition element outside its AAA core
Katanin integrates different tubulin modification inputs for divergent outcomes
Acknowledgements
We thank Duck-Yeon Lee at the National Heart, Lung, and Blood Institute Biochemistry Core for mass spectrometer access, T.R. Brummelkamp (Netherlands Cancer Institute) for VASH1/SVBP plasmids, F. J. McNally (U.C. Davis) for katanin plasmids. A.R.M. thanks A. Ferre-D’Amare for support during the pandemic. A.R.M is supported by the intramural programs of the National Institute of Neurological Disorders and Stroke (NINDS) and the National Heart, Heart, and Blood Institute (NHLBI).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.MacTaggart B, and Kashina A (2021). Posttranslational Modifications of the Cytoskeleton. Cytoskeleton 78, 142–173. 10.1002/cm.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roll-Mecak A (2020). The tubulin code in microtubule dynamics and information encoding. Developmental cell 54, 7–20. 10.1016/j.devcel.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodakuntla S, Janke C, and Magiera MM (2021). Tubulin polyglutamylation, a regulator of microtubule functions, can cause neurodegeneration. Neuroscience Letters 746, 135656. 10.1016/j.neulet.2021.135656. [DOI] [PubMed] [Google Scholar]
- 4.Wattanathamsan O, and Pongrakhananon V (2021). Post-translational modifications of tubulin: their role in cancers and the regulation of signaling molecules. Cancer Gene Therapy, 1–8. 10.1038/s41417-021-00396-4. [DOI] [PubMed] [Google Scholar]
- 5.Geimer S, Teltenkötter A, Plessmann U, Weber K, and Lechtreck KF (1997). Purification and characterization of basal apparatuses from a flagellate green alga. Cell motility and the cytoskeleton 37, 72–85. . [DOI] [PubMed] [Google Scholar]
- 6.Schneider A, Plessmann U, Felleisen R, and Weber K (1998). Posttranslational modifications of trichomonad tubulins; identification of multiple glutamylation sites. FEBS letters 429, 399–402. 10.1016/s0014-5793(98)00644-9. [DOI] [PubMed] [Google Scholar]
- 7.Wall KP, Pagratis M, Armstrong G, Balsbaugh JL, Verbeke E, Pearson CG, and Hough LE (2016). Molecular determinants of tubulin’s C-terminal tail conformational ensemble. ACS chemical biology 11, 2981–2990. 10.1021/acschembio.6b00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kann ML, Prigent Y, Levilliers N, Bré MH, and Fouquet JP (1998). Expression of glycylated tubulin during the differentiation of spermatozoa in mammals. Cell motility and the cytoskeleton 41, 341–352. . [DOI] [PubMed] [Google Scholar]
- 9.Péchart I, Kann ML, Levilliers N, Bre MH, and Fouquet JP (1999). Composition and organization of tubulin isoforms reveals a variety of axonemal models. Biology of the Cell 91, 685–697. 10.1111/j.1768-322x.1999.tb01113.x. [DOI] [PubMed] [Google Scholar]
- 10.Lechtreck KF, and Geimer S (2000). Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell motility and the cytoskeleton 47, 219–235. . [DOI] [PubMed] [Google Scholar]
- 11.Multigner L, Pignot-Paintrand I, Saoudi Y, Job D, Plessmann U, Rüdiger M, and Weber K (1996). The A and B tubules of the outer doublets of sea urchin sperm axonemes are composed of different tubulin variants. Biochemistry 35, 10862–10871. 10.1021/bi961057u. [DOI] [PubMed] [Google Scholar]
- 12.Suryavanshi S, Eddé B, Fox LA, Guerrero S, Hard R, Hennessey T, Kabi A, Malison D, Pennock D, and Sale WS (2010). Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Current biology 20, 435–440. 10.1016/j.cub.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wloga D, Joachimiak E, Louka P, and Gaertig J (2017). Posttranslational modifications of tubulin and cilia. Cold Spring Harbor perspectives in biology 9, a028159. 10.1101/cshperspect.a028159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vemu A, Atherton J, Spector JO, Moores CA, and Roll-Mecak A (2017). Tubulin isoform composition tunes microtubule dynamics. Molecular biology of the cell 28, 3564–3572. 10.1091/mbc.E17-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redeker V (2010). Mass spectrometry analysis of C-terminal posttranslational modifications of tubulins. Methods in cell biology 95, 77–103. 10.1016/S0091-679X(10)95006-1. [DOI] [PubMed] [Google Scholar]
- 16.Moutin MJ, Bosc C, Peris L, and Andrieux A (2021). Tubulin post-translational modifications control neuronal development and functions. Developmental neurobiology 81, 253–272. 10.1002/dneu.22774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond JW, Huang C-F, Kaech S, Jacobson C, Banker G, and Verhey KJ (2010). Posttranslational modifications of tubulin and the polarized transport of kinesin-1 in neurons. Molecular biology of the cell 21, 572–583. 10.1091/mbc.E09-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robson SJ, and Burgoyne RD (1989). Differential localisation of tyrosinated, detyrosinated, and acetylated α-tubulins in neurites and growth cones of dorsal root ganglion neurons. Cell motility and the cytoskeleton 12, 273–282. 10.1002/cm.970120408. [DOI] [PubMed] [Google Scholar]
- 19.Katrukha EA, Jurriens D, Salas Pastene DM, and Kapitein LC (2021). Quantitative mapping of dense microtubule arrays in mammalian neurons. Elife 10. 10.7554/eLife.67925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Kholina E, Szyk A, Fedorov VA, Kovalenko I, Gudimchuk N, and Roll-Mecak A (2021). α-tubulin tail modifications regulate microtubule stability through selective effector recruitment, not changes in intrinsic polymer dynamics. Developmental Cell. 10.1016/j.devcel.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng P, Obara CJ, Szczesna E, Nixon-Abell J, Mahalingan KK, Roll-Mecak A, Lippincott-Schwartz J, and Blackstone C (2022). ER proteins decipher the tubulin code to regulate organelle distribution. Nature 601, 132–138. 10.1038/s41586-021-04204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodakuntla S, Yuan X, Genova M, Gadadhar S, Leboucher S, Birling M.c., Klein D, Martini R, Janke C, and Magiera MM (2021). Distinct roles of α-and β-tubulin polyglutamylation in controlling axonal transport and in neurodegeneration. The EMBO Journal 40, e108498. 10.15252/embj.2021108498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nirschl JJ, Magiera MM, Lazarus JE, Janke C, and Holzbaur EL (2016). α-Tubulin tyrosination and CLIP-170 phosphorylation regulate the initiation of dynein-driven transport in neurons. Cell reports 14, 2637–2652. 10.1016/j.celrep.2016.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gadadhar S, Viar GA, Hansen JN, Gong A, Kostarev A, Ialy-Radio C, Leboucher S, Whitfield M, Ziyyat A, and Touré A (2021). Tubulin glycylation controls axonemal dynein activity, flagellar beat, and male fertility. Science 371. 10.1126/science.abd4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNally FJ, and Vale RD (1993). Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75, 419–429. Doi 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 26.Vale RD (1991). Severing of stable microtubules by a mitotically activated protein in Xenopus egg extracts. Cell 64, 827–839. 10.1016/0092-8674(91)90511-v. [DOI] [PubMed] [Google Scholar]
- 27.Lynn NA, Martinez E, Nguyen H, and Torres JZ (2021). The Mammalian Family of Katanin Microtubule-Severing Enzymes. Frontiers in Cell and Developmental Biology 9, 692040. 10.3389/fcell.2021.692040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNally FJ, and Roll-Mecak A (2018). Microtubule-severing enzymes: From cellular functions to molecular mechanism. Journal of Cell Biology 217, 4057–4069. 10.1083/jcb.201612104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vemu A, Szczesna E, Zehr EA, Spector JO, Grigorieff N, Deaconescu AM, and Roll-Mecak A (2018). Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science 361. 10.1126/science.aau1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmad FJ, Yu W, McNally FJ, and Baas PW (1999). An essential role for katanin in severing microtubules in the neuron. The Journal of cell biology 145, 305–315. 10.1083/jcb.145.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong C, Xu H, Zhang R, Tanaka N, Takeichi M, and Meng W (2017). CAMSAP3 accumulates in the pericentrosomal area and accompanies microtubule release from the centrosome via katanin. Journal of cell science 130, 1709–1715. 10.1242/jcs.198010. [DOI] [PubMed] [Google Scholar]
- 32.Keating T, Peloquin J, Rodionov V, Momcilovic D, and Borisy G (1997). Microtubule release from the centrosome. Proceedings of the National Academy of Sciences 94, 5078–5083. 10.1073/pnas.94.10.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura M, Ehrhardt DW, and Hashimoto T (2010). Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nature cell biology 12, 1064–1070. 10.1038/ncb2110. [DOI] [PubMed] [Google Scholar]
- 34.Huang J, Liang Z, Guan C, Hua S, and Jiang K (2021). WDR62 regulates spindle dynamics as an adaptor protein between TPX2/Aurora A and katanin. Journal of Cell Biology 220, e202007167. 10.1083/jcb.202007167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loughlin R, Wilbur JD, McNally FJ, Nédélec FJ, and Heald R (2011). Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147, 1397–1407. 10.1016/j.cell.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNally K, Audhya A, Oegema K, and McNally FJ (2006). Katanin controls mitotic and meiotic spindle length. The Journal of cell biology 175, 881–891. 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srayko M, Buster DW, Bazirgan OA, McNally FJ, and Mains PE (2000). MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes & development 14, 1072–1084. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D, Rogers GC, Buster DW, and Sharp DJ (2007). Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. The Journal of cell biology 177, 231–242. 10.1083/jcb.200612011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dymek EE, Lefebvre PA, and Smith EF (2004). PF15p is the chlamydomonas homologue of the Katanin p80 subunit and is required for assembly of flagellar central microtubules. Eukaryotic Cell 3, 870–879. 10.1128/Ec.3.4.870-879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dymek EE, and Smith EF (2012). PF19 encodes the p60 catalytic subunit of katanin and is required for assembly of the flagellar central apparatus in Chlamydomonas. Journal of cell science 125, 3357–3366. 10.1242/jcs.096941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu WF, Pomp O, Ben-Omran T, Kodani A, Henke K, Mochida GH, Timothy WY, Woodworth MB, Bonnard C, and Raj GS (2014). Katanin p80 regulates human cortical development by limiting centriole and cilia number. Neuron 84, 1240–1257. 10.1016/j.neuron.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma N, Bryant J, Wloga D, Donaldson R, Davis RC, Jerka-Dziadosz M, and Gaertig J (2007). Katanin regulates dynamics of microtubules and biogenesis of motile cilia. The Journal of cell biology 178, 1065–1079. 10.1083/jcb.200704021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun L, Cui L, Liu Z, Wang Q, Xue Z, Wu M, Sun T, Mao D, Ni J, and Pastor-Pareja JC (2021). Katanin p60-like 1 sculpts the cytoskeleton in mechanosensory cilia. Journal of Cell Biology 220. 10.1083/jcb.202004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirvis M, Siemers KA, Nelson WJ, and Stearns TP (2019). Primary cilium loss in mammalian cells occurs predominantly by whole-cilium shedding. PLoS biology 17, e3000381. 10.1371/journal.pbio.3000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin SC, Im S-K, Jang E-H, Jin KS, Hur E-M, and Kim EE (2019). Structural and molecular basis for katanin-mediated severing of glutamylated microtubules. Cell Reports 26, 1357–1367. e1355. 10.1016/j.celrep.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 46.Qiang L, Yu W, Liu M, Solowska JM, and Baas PW (2010). Basic fibroblast growth factor elicits formation of interstitial axonal branches via enhanced severing of microtubules. Molecular biology of the cell 21, 334–344. 10.1091/mbc.E09-09-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee H-H, Jan LY, and Jan Y-N (2009). Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proceedings of the National Academy of Sciences 106, 6363–6368. 10.1073/pnas.0902051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart A, Tsubouchi A, Rolls MM, Tracey WD, and Sherwood NT (2012). Katanin p60-like1 promotes microtubule growth and terminal dendrite stability in the larval class IV sensory neurons of Drosophila. Journal of Neuroscience 32, 11631–11642. 10.1523/JNEUROSCI.0729-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartholdi D, Stray-Pedersen A, Azzarello-Burri S, Kibaek M, Kirchhoff M, Oneda B, Rodningen O, Schmitt-Mechelke T, Rauch A, and Kjaergaard S (2014). A newly recognized 13q12.3 microdeletion syndrome characterized by intellectual disability, microcephaly, and eczema/atopic dermatitis encompassing the HMGB1 and KATNAL1 genes. Am J Med Genet A 164A, 1277–1283. 10.1002/ajmg.a.36439. [DOI] [PubMed] [Google Scholar]
- 50.Mishra-Gorur K, Caglayan AO, Schaffer AE, Chabu C, Henegariu O, Vonhoff F, Akgumus GT, Nishimura S, Han W, Tu S, et al. (2015). Mutations in KATNB1 Cause Complex Cerebral Malformations by Disrupting Asymmetrically Dividing Neural Progenitors. Neuron 85, 228. 10.1016/j.neuron.2014.12.046. [DOI] [PubMed] [Google Scholar]
- 51.Stessman HA, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, Kvarnung M, Gerdts J, Trinh S, Cosemans N, et al. (2017). Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet 49, 515–526. 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu W, Wu H, Cheng Z, Huang S, and Rao H (2018). The role of katanin p60 in breast cancer bone metastasis. Oncol Lett 15, 4963–4969. 10.3892/ol.2018.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zehr EA, Szyk A, Szczesna E, and Roll-Mecak A (2020). Katanin Grips the beta-Tubulin Tail through an Electropositive Double Spiral to Sever Microtubules. Dev Cell 52, 118–131 e116. 10.1016/j.devcel.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, and Levchenko I (2004). Sculpting the proteome with AAA+ proteases and disassembly machines. Cell 119, 9–18. 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shorter J, and Southworth DR (2019). Spiraling in Control: Structures and Mechanisms of the Hsp104 Disaggregase. Cold Spring Harb Perspect Biol 11. 10.1101/cshperspect.a034033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thazhath R, Liu C, and Gaertig J (2002). Polyglycylation domain of beta-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat Cell Biol 4, 256–259. 10.1038/ncb764. [DOI] [PubMed] [Google Scholar]
- 57.Fouquet JP, Prigent Y, and Kann ML (1996). Comparative immunogold analysis of tubulin isoforms in the mouse sperm flagellum: unique distribution of glutamylated tubulin. Molecular Reproduction and Development: Incorporating Gamete Research 43, 358–365. . [DOI] [PubMed] [Google Scholar]
- 58.Ten Martin D, Jardin N, Giudicelli F, Gasmi L, Vougny J, Haumaître C, Nicol X, Janke C, Fassier C, and Hazan J (2022). A key role for p60-Katanin in axon navigation is conditioned by the tubulin polyglutamylase TTLL6. bioRxiv. [Google Scholar]
- 59.Lacroix B, Van Dijk J, Gold ND, Guizetti J, Aldrian-Herrada G, Rogowski K, Gerlich DW, and Janke C (2010). Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. Journal of Cell Biology 189, 945–954. 10.1083/jcb.201001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valenstein ML, and Roll-Mecak A (2016). Graded control of microtubule severing by tubulin glutamylation. Cell 164, 911–921. 10.1016/j.cell.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartman JJ, Mahr J, McNally K, Okawa K, Iwamatsu A, Thomas S, Cheesman S, Heuser J, Vale RD, and McNally FJ (1998). Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell 93, 277–287. Doi 10.1016/S0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- 62.Alexander JE, Hunt DF, Lee MK, Shabanowitz J, Michel H, Berlin SC, Macdonald TL, Sundberg RJ, Rebhun LI, and Frankfurter A (1991). Characterization of posttranslational modifications in neuron-specific class III beta-tubulin by mass spectrometry. Proceedings of the National Academy of Sciences 88, 4685–4689. 10.1073/pnas.88.11.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edde B, Rossier J, Le Caer J-P, Desbruyeres E, Gros F, and Denoulet P (1990). Posttranslational glutamylation of alpha-tubulin. Science 247, 83–85. 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- 64.Redeker V, Melki R, Promé D, Le Caer J-P, and Rossier J (1992). Structure of tubulin C-terminal domain obtained by subtilisin treatment The major α and β tubulin isotypes from pig brain are glutamylated. FEBS letters 313, 185–192. 10.1016/0014-5793(92)81441-n. [DOI] [PubMed] [Google Scholar]
- 65.Rüdiger M, Plessman U, Klöppel K-D, Wehland J, and Weber K (1992). Class II tubulin, the major brain β tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS letters 308, 101–105. 10.1016/0014-5793(92)81061-p. [DOI] [PubMed] [Google Scholar]
- 66.Mahalingan KK, Keenan EK, Strickland M, Li Y, Liu Y, Ball HL, Tanner ME, Tjandra N, and Roll-Mecak A (2020). Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes. Nature Structural & Molecular Biology 27, 802–813. 10.1038/s41594-020-0462-0. [DOI] [PubMed] [Google Scholar]
- 67.Van Dijk J, Rogowski K, Miro J, Lacroix B, Eddé B, and Janke C (2007). A targeted multienzyme mechanism for selective microtubule polyglutamylation. Molecular cell 26, 437–448. 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 68.Audebert S, Koulakoff A, Berwald-Netter Y, Gros F, Denoulet P, and Eddé B (1994). Developmental regulation of polyglutamylated alpha-and beta-tubulin in mouse brain neurons. Journal of cell science 107, 2313–2322. 10.1242/jcs.107.8.2313. [DOI] [PubMed] [Google Scholar]
- 69.Joachimiak E, Waclawek E, Niziolek M, Osinka A, Fabczak H, Gaertig J, and Wloga D (2020). The LisH Domain-Containing N-Terminal Fragment is Important for the Localization, Dimerization, and Stability of Katnal2 in Tetrahymena. Cells 9, 292. 10.3390/cells9020292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waclawek E, Joachimiak E, Hall MH, Fabczak H, and Wloga D (2017). Regulation of katanin activity in the ciliate Tetrahymena thermophila. Molecular Microbiology 103, 134–150. 10.1111/mmi.13547. [DOI] [PubMed] [Google Scholar]
- 71.Vemu A, Garnham CP, Lee D-Y, and Roll-Mecak A (2014). Generation of differentially modified microtubules using in vitro enzymatic approaches. Methods in enzymology 540, 149–166. 10.1016/B978-0-12-397924-7.00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JE, Silhavy JL, Zaki MS, Schroth J, Bielas SL, Marsh SE, Olvera J, Brancati F, Iannicelli M, and Ikegami K (2012). CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nature genetics 44, 193–199. 10.1038/ng.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bosch Grau M, Gonzalez Curto G, Rocha C, Magiera MM, Marques Sousa P, Giordano T, Spassky N, and Janke C (2013). Tubulin glycylases and glutamylases have distinct functions in stabilization and motility of ependymal cilia. Journal of Cell Biology 202, 441–451. 10.1083/jcb.201305041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hartman JJ, and Vale RD (1999). Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science 286, 782–785. 10.1126/science.286.5440.782. [DOI] [PubMed] [Google Scholar]
- 75.Garnham CP, Vemu A, Wilson-Kubalek EM, Yu I, Szyk A, Lander GC, Milligan RA, and Roll-Mecak A (2015). Multivalent microtubule recognition by tubulin tyrosine ligase-like family glutamylases. Cell 161, 1112–1123. 10.1016/j.cell.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mukai M, Ikegami K, Sugiura Y, Takeshita K, Nakagawa A, and Setou M (2009). Recombinant mammalian tubulin polyglutamylase TTLL7 performs both initiation and elongation of polyglutamylation on β-tubulin through a random sequential pathway. Biochemistry 48, 1084–1093. 10.1021/bi802047y. [DOI] [PubMed] [Google Scholar]
- 77.Ikegami K, Mukai M, Tsuchida J. i., Heier RL, MacGregor GR, and Setou M (2006). TTLL7 is a mammalian β-tubulin polyglutamylase required for growth of MAP2-positive neurites. Journal of Biological Chemistry 281, 30707–30716. 10.1074/jbc.M603984200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garnham CP, Yu I, Li Y, and Roll-Mecak A (2017). Crystal structure of tubulin tyrosine ligase-like 3 reveals essential architectural elements unique to tubulin monoglycylases. Proceedings of the National Academy of Sciences 114, 6545–6550. 10.1073/pnas.1617286114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McKenney RJ, Huynh W, Vale RD, and Sirajuddin M (2016). Tyrosination of α-tubulin controls the initiation of processive dynein–dynactin motility. The EMBO journal 35, 1175–1185. 10.15252/embj.201593071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peris L, Wagenbach M, Lafanechère L, Brocard J, Moore AT, Kozielski F, Job D, Wordeman L, and Andrieux A (2009). Motor-dependent microtubule disassembly driven by tubulin tyrosination. Journal of Cell Biology 185, 1159–1166. 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sirajuddin M, Rice LM, and Vale RD (2014). Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nature cell biology 16, 335–344. 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peris L, Thery M, Fauré J, Saoudi Y, Lafanechère L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M, and Wordeman L (2006). Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. The Journal of cell biology 174, 839–849. DOI 10.1083/jcb.200512058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aillaud C, Bosc C, Peris L, Bosson A, Heemeryck P, Van Dijk J, Le Friec J, Boulan B, Vossier F, and Sanman LE (2017). Vasohibins/SVBP are tubulin carboxypeptidases (TCPs) that regulate neuron differentiation. Science 358, 1448–1453. 10.1126/science.aao4165. [DOI] [PubMed] [Google Scholar]
- 84.Nieuwenhuis J, Adamopoulos A, Bleijerveld OB, Mazouzi A, Stickel E, Celie P, Altelaar M, Knipscheer P, Perrakis A, and Blomen VA (2017). Vasohibins encode tubulin detyrosinating activity. Science 358, 1453–1456. 10.1126/science.aao5676. [DOI] [PubMed] [Google Scholar]
- 85.Faltova L, Jiang K, Frey D, Wu Y, Capitani G, Prota AE, Akhmanova A, Steinmetz MO, and Kammerer RA (2019). Crystal structure of a heterotetrameric katanin p60: p80 complex. Structure 27, 1375–1383. e1373. 10.1016/j.str.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 86.Jiang K, Rezabkova L, Hua S, Liu Q, Capitani G, Altelaar AM, Heck AJ, Kammerer RA, Steinmetz MO, and Akhmanova A (2017). Microtubule minus-end regulation at spindle poles by an ASPM–katanin complex. Nature cell biology 19, 480–492. 10.1038/ncb3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McNally KP, Bazirgan OA, and McNally FJ (2000). Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. Journal of cell science 113, 1623–1633. 10.1242/jcs.113.9.1623. [DOI] [PubMed] [Google Scholar]
- 88.Rezabkova L, Jiang K, Capitani G, Prota AE, Akhmanova A, Steinmetz MO, and Kammerer RA (2017). Structural basis of katanin p60: p80 complex formation. Scientific reports 7, 1–8. 10.1038/s41598-017-14194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bailey ME, Sackett DL, and Ross JL (2015). Katanin severing and binding microtubules are inhibited by tubulin carboxy tails. Biophysical journal 109, 2546–2561. 10.1016/j.bpj.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu I, Garnham CP, and Roll-Mecak A (2015). Writing and reading the tubulin code. Journal of Biological Chemistry 290, 17163–17172. 10.1074/jbc.R115.637447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fouquet JP, Kann ML, Edde B, Wolff A, Desbruyeres E, and Denoulet P (1994). Differential distribution of glutamylated tubulin during spermatogenesis in mammalian testis. Cell motility and the cytoskeleton 27, 49–58. 10.1002/cm.970270106. [DOI] [PubMed] [Google Scholar]
- 92.Schulze E, Asai DJ, Bulinski JC, and Kirschner M (1987). Posttranslational modification and microtubule stability. The Journal of cell biology 105, 2167–2177. 10.1083/jcb.105.5.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sudo H, and Baas PW (2010). Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. Journal of Neuroscience 30, 7215–7226. 10.1523/Jneurosci.0048-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eshun-Wilson L, Zhang R, Portran D, Nachury MV, Toso DB, Löhr T, Vendruscolo M, Bonomi M, Fraser JS, and Nogales E (2019). Effects of α-tubulin acetylation on microtubule structure and stability. Proceedings of the National Academy of Sciences 116, 10366–10371. 10.1073/pnas.1900441116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soppina V, Herbstman JF, Skiniotis G, and Verhey KJ (2012). Luminal localization of α-tubulin K40 acetylation by cryo-EM analysis of fab-labeled microtubules. PloS one 7, e48204. 10.1371/journal.pone.0048204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Szyk A, Deaconescu AM, Spector J, Goodman B, Valenstein ML, Ziolkowska NE, Kormendi V, Grigorieff N, and Roll-Mecak A (2014). Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell 157, 1405–1415. 10.1016/j.cell.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Portran D, Schaedel L, Xu Z, Théry M, and Nachury MV (2017). Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nature cell biology 19, 391–398. 10.1038/ncb3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu Z, Schaedel L, Portran D, Aguilar A, Gaillard J, Marinkovich MP, Théry M, and Nachury MV (2017). Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356, 328–332. 10.1126/science.aai8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kellogg EH, Hejab NM, Howes S, Northcote P, Miller JH, Díaz JF, Downing KH, and Nogales E (2017). Insights into the distinct mechanisms of action of taxane and non-taxane microtubule stabilizers from cryo-EM structures. Journal of molecular biology 429, 633–646. 10.1016/j.jmb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang R, LaFrance B, and Nogales E (2018). Separating the effects of nucleotide and EB binding on microtubule structure. Proceedings of the National Academy of Sciences 115, E6191–E6200. 10.1073/pnas.1802637115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McNally KP, Buster D, and McNally FJ (2002). Katanin-mediated microtubule severing can be regulated by multiple mechanisms. Cell motility and the cytoskeleton 53, 337–349. 10.1002/cm.10080. [DOI] [PubMed] [Google Scholar]
- 102.Qiang L, Yu W, Andreadis A, Luo M, and Baas PW (2006). Tau protects microtubules in the axon from severing by katanin. Journal of Neuroscience 26, 3120–3129. 10.1523/JNEUROSCI.5392-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Siahaan V, Krattenmacher J, Hyman AA, Diez S, Hernández-Vega A, Lansky Z, and Braun M (2019). Kinetically distinct phases of tau on microtubules regulate kinesin motors and severing enzymes. Nature cell biology 21, 1086–1092. 10.1038/s41556-019-0374-6. [DOI] [PubMed] [Google Scholar]
- 104.Rogowski K, Juge F, Van Dijk J, Wloga D, Strub J-M, Levilliers N, Thomas D, Bré M-H, Van Dorsselaer A, and Gaertig J (2009). Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell 137, 1076–1087. 10.1016/j.cell.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 105.Lohret TA, McNally FJ, and Quarmby LM (1998). A role for katanin-mediated axonemal severing during Chlamydomonas deflagellation. Molecular biology of the cell 9, 1195–1207. 10.1091/mbc.9.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ikegami K, and Setou M (2009). TTLL10 can perform tubulin glycylation when co-expressed with TTLL8. FEBS letters 583, 1957–1963. 10.1016/j.febslet.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 107.Garnham CP, and Roll-Mecak A (2012). The chemical complexity of cellular microtubules: tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton 69, 442–463. 0.1002/cm.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhuang Z, Cummings SW, Roll-Mecak A, and Tanner ME (2022). Phosphinic acid-based inhibitors of tubulin polyglycylation. Chemical Communications 58, 6530–6533. 10.1039/d2cc01783k. [DOI] [PubMed] [Google Scholar]
- 109.Liu H, Zheng J, Zhu L, Xie L, Chen Y, Zhang Y, Zhang W, Yin Y, Peng C, and Zhou J (2021). Wdr47, Camsaps, and Katanin cooperate to generate ciliary central microtubules. Nature communications 12, 1–15. 10.1038/s41467-021-26058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gadadhar S, Dadi H, Bodakuntla S, Schnitzler A, Bièche I, Rusconi F, and Janke C (2017). Tubulin glycylation controls primary cilia length. Journal of Cell Biology 216, 2701–2713. 10.1083/jcb.201612050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wloga D, Webster DM, Rogowski K, Bré M-H, Levilliers N, Jerka-Dziadosz M, Janke C, Dougan ST, and Gaertig J (2009). TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Developmental cell 16, 867–876. 10.1016/j.devcel.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 112.Han H, Schubert HL, McCullough J, Monroe N, Purdy MD, Yeager M, Sundquist WI, and Hill CP (2020). Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation. Journal of Biological Chemistry 295, 435–443. 10.1074/jbc.AC119.009890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sandate CR, Szyk A, Zehr EA, Lander GC, and Roll-Mecak A (2019). An allosteric network in spastin couples multiple activities required for microtubule severing. Nature structural & molecular biology 26, 671–678. 10.1038/s41594-019-0257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han H, Fulcher JM, Dandey VP, Iwasa JH, Sundquist WI, Kay MS, Shen PS, and Hill CP (2019). Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases. Elife 8, e44071. 10.7554/eLife.44071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bodnar NO, and Rapoport TA (2017). Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell 169, 722–735. e729. 10.1016/j.cell.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Burton RE, Siddiqui SM, Kim YI, Baker TA, and Sauer RT (2001). Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. The EMBO journal 20, 3092–3100. 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kraut DA, and Matouschek A (2011). Proteasomal degradation from internal sites favors partial proteolysis via remote domain stabilization. ACS chemical biology 6, 1087–1095. 10.1021/cb2002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee C, Prakash S, and Matouschek A (2002). Concurrent translocation of multiple polypeptide chains through the proteasomal degradation channel. Journal of Biological Chemistry 277, 34760–34765. 10.1074/jbc.M204750200. [DOI] [PubMed] [Google Scholar]
- 119.Magiera MM, Bodakuntla S, Žiak J, Lacomme S, Marques Sousa P, Leboucher S, Hausrat TJ, Bosc C, Andrieux A, and Kneussel M (2018). Excessive tubulin polyglutamylation causes neurodegeneration and perturbs neuronal transport. The EMBO journal 37, e100440. 10.15252/embj.2018100440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roll-Mecak A (2015). Intrinsically disordered tubulin tails: complex tuners of microtubule functions? Paper presented at: Seminars in cell & developmental biology (Elsevier). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tran HT, Mao A, and Pappu RV (2008). Role of backbone– solvent interactions in determining conformational equilibria of intrinsically disordered proteins. Journal of the American Chemical Society 130, 7380–7392. 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- 122.Bigman LS, and Levy Y (2021). Modulating Microtubules: A Molecular Perspective on the Effects of Tail Modifications. Journal of molecular biology 433, 166988. 10.1016/j.jmb.2021.166988. [DOI] [PubMed] [Google Scholar]
- 123.Redeker V, Levilliers N, Vinolo E, Rossier J, Jaillard D, Burnette D, Gaertig J, and Bré M-H (2005). Mutations of tubulin glycylation sites reveal cross-talk between the C termini of α-and β-tubulin and affect the ciliary matrix in Tetrahymena. Journal of Biological Chemistry 280, 596–606. 10.1074/jbc.M408324200. [DOI] [PubMed] [Google Scholar]
- 124.Ramkumar A, Jong BY, and Ori-McKenney KM (2018). ReMAPping the microtubule landscape: How phosphorylation dictates the activities of microtubule-associated proteins. Developmental Dynamics 247, 138–155. 10.1002/dvdy.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Steinmetz MO, and Akhmanova A (2008). Capturing protein tails by CAP-Gly domains. Trends in biochemical sciences 33, 535–545. 10.1016/j.tibs.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 126.Cheung K, Senese S, Kuang J, Bui N, Ongpipattanakul C, Gholkar A, Cohn W, Capri J, Whitelegge JP, and Torres JZ (2016). Proteomic analysis of the mammalian katanin family of microtubule-severing enzymes defines katanin p80 subunit B-like 1 (KATNBL1) as a regulator of mammalian katanin microtubule-severing. Molecular & Cellular Proteomics 15, 1658–1669. 10.1074/mcp.M115.056465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zehr E, Szyk A, Piszczek G, Szczesna E, Zuo X, and Roll-Mecak A (2017). Katanin spiral and ring structures shed light on power stroke for microtubule severing. Nature structural & molecular biology 24, 717–725. 10.1038/nsmb.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miller KE, and Heald R (2015). Glutamylation of Nap1 modulates histone H1 dynamics and chromosome condensation in Xenopus. Journal of Cell Biology 209, 211–220. 10.1083/jcb.201412097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xia P, Ye B, Wang S, Zhu X, Du Y, Xiong Z, Tian Y, and Fan Z (2016). Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nature immunology 17, 369–378. 10.1038/ni.3356. [DOI] [PubMed] [Google Scholar]
- 130.Itzhak DN, Davies C, Tyanova S, Mishra A, Williamson J, Antrobus R, Cox J, Weekes MP, and Borner GH (2017). A mass spectrometry-based approach for mapping protein subcellular localization reveals the spatial proteome of mouse primary neurons. Cell reports 20, 2706–2718. 10.1016/j.celrep.2017.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Itzhak DN, Tyanova S, Cox J, and Borner GH (2016). Global, quantitative and dynamic mapping of protein subcellular localization. elife 5, e16950. 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McNally FJ, and Thomas S (1998). Katanin is responsible for the M-phase microtubule-severing activity in Xenopus eggs. Molecular biology of the cell 9, 1847–1861. DOI 10.1091/mbc.9.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vemu A, Atherton J, Spector JO, Szyk A, Moores CA, and Roll-Mecak A (2016). Structure and Dynamics of Single-isoform Recombinant Neuronal Human Tubulin. Journal of Biological Chemistry 291, 12907–12915. 10.1074/jbc.C116.731133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McNally K, Berg E, Cortes DB, Hernandez V, Mains PE, and McNally FJ (2014). Katanin maintains meiotic metaphase chromosome alignment and spindle structure in vivo and has multiple effects on microtubules in vitro. Molecular biology of the cell 25, 1037–1049. 10.1091/mbc.E13-12-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Theile CS, Witte MD, Blom AE, Kundrat L, Ploegh HL, and Guimaraes CP (2013). Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nature protocols 8, 1800–1807. 10.1038/nprot.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Scholz J, Besir H, Strasser C, and Suppmann S (2013). A new method to customize protein expression vectors for fast, efficient and background free parallel cloning. BMC biotechnology 13, 1–11. 10.1186/1472-6750-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Widlund PO, Podolski M, Reber S, Alper J, Storch M, Hyman AA, Howard J, and Drechsel DN (2012). One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Molecular biology of the cell 23, 4393–4401. 10.1091/mbc.E12-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Castoldi M, and Popov AV (2003). Purification of brain tubulin through two cycles of polymerization–depolymerization in a high-molarity buffer. Protein expression and purification 32, 83–88. 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- 139.Xia L, Hai B, Gao Y, Burnette D, Thazhath R, Duan J, Bré M-H, Levilliers N, Gorovsky MA, and Gaertig J (2000). Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. The Journal of cell biology 149, 1097–1106. DOI 10.1083/jcb.149.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ziółkowska NE, and Roll-Mecak A (2013). In vitro microtubule severing assays. In Adhesion Protein Protocols (Springer; ), pp. 323–334. [DOI] [PubMed] [Google Scholar]
- 141.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Moriarty NW, Grosse-Kunstleve RW, and Adams PD (2009). electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallographica Section D: Biological Crystallography 65, 1074–1080. 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta crystallographica section D: biological crystallography 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 144.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, and Grosse-Kunstleve RW (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D: Biological Crystallography 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallographica Section D: Structural Biology 74, 531–544. 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 147.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science 27, 14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schrodinger, LLC (2015). The PyMOL Molecular Graphics System, Version 1.8.
- 149.Pravda L, Sehnal D, Toušek D, Navrátilová V, Bazgier V, Berka K, Svobodová Vařeková R, Koča J, Otyepka M (2018) MOLEonline: a web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Research 46, W368–W373. 10.1093/nar/gky309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 (related to Figure 3 and Table S1).
Pymol session file with Models 1 through 7 shown in Table S1.
Tubulin tail, ball-and-stick, yellow; Branch 1, ball-and-stick, green; Branch 2, ball-and-stick, magenta; Branch positions labeled. Class in Model 7 indicated. Subset of pore loop 1 and 2 residues shown in cpk representation.
Data Availability Statement
Microscopy and mass spectrometry data reported in this paper will be shared by the lead contact upon request. This paper does not use any original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


