Abstract
Concerns regarding inappropriate leakage of sensitive personal information as well as unauthorized data use are increasing with the growth of genomic data repositories. Therefore, privacy and security of genomic data have become increasingly important and need to be studied. With many proposed protection techniques, their applicability in support of biomedical research should be well understood. For this purpose, we have organized a community effort in the past 8 years through the integrating data for analysis, anonymization and sharing consortium to address this practical challenge. In this article, we summarize our experience from these competitions, report lessons learned from the events in 2020/2021 as examples, and discuss potential future research directions in this emerging field.
Keywords: genome privacy, genome security, genomic data analysis, genomic data sharing, community effort
BACKGROUND
As sequencing technology advances, the cost of short-read sequencing at greater depth and higher sensitivity has been significantly reduced, and personalized whole genome sequencing analysis is becoming increasingly affordable.1 As human genome data are currently available to a limited group of researchers, sharing these data with the broader scientific community may help accelerate discoveries and decrease disparities in access. At the same time, privacy and security concerns regarding inappropriate leakage of sensitive personal information or unauthorized data access will increase. For example, recent incidents such as the SolarWinds flaw2 allow attackers to bypass authentication and obtain sensitive data such as patients’ genomes. The impact of such attacks would be (1) deep: for example, attackers may be able to find a person’s ancestors and may try to link to additional data and predict an individual’s health issues; (2) wide: for example, hackers can link the information to the person’s family members; and (3) permanent: the leaked data will be indelible and cannot be retracted.
It is natural that most genomics researchers focus on genome data analysis methods, with only a much smaller community of computer scientists and informaticians working on the preservation of privacy. Given the rapid growth of genomic data and related analysis techniques, genome privacy (ie, information leakage)3 and security (ie, unauthorized data access)4 have become increasingly important,5 not only for protecting patients’ sensitive biometric data and complying with regulations (eg, Health Insurance Portability and Accountability Act,6 General Data Protection Regulation,7 and others) but also for supporting biomedical research.
Both genome privacy and security have been attracting great attention in the past decade, across multiple disciplines such as Genetics/Heredity, Biotechnology, Microbiology, and Medical Informatics, as shown in Figure 1 [data collected from the Web of Science (WOS)8] These categories are predefined by the WOS, and the counts indicate the number of papers in each category. The statistics are presented as a Tree Map Chart.
Figure 1.
Publication categories for genome privacy (top panel) and security (bottom panel), using statistics from Web of Science8 on December 14, 2021.
The rest of this article is organized as follows: we first summarize our prior conference results and impact on the community in the “The Integrating Data for Analysis, Anonymization and Sharing Community Effort for Practical Privacy, and Security Protection” section, followed by a competition topic introduction and analysis in the “Topics and Methods” section. We then use the competitions in 2020 (“Lessons Learned from the 2020 iDASH Competition” section) and 2021 (“Lessons Learned from the 2021 iDASH Competition” section) as examples to demonstrate in detail what scientific results were produced. Finally, we discuss potential future trends in the “Anticipated Future Research Trends” section and conclusions in the “Conclusion” section.
THE INTEGRATING DATA FOR ANALYSIS, ANONYMIZATION AND SHARING COMMUNITY EFFORT FOR PRACTICAL PRIVACY AND SECURITY PROTECTION
Computer scientists and informaticians strive to develop practical and rigorous privacy and security methods to help human genome researchers protect sensitive data. In an ideal setting, we would be equipping researchers with tools that tune the amount of data protection according to consent, trust in the data recipient, as well as intended use. However, such tools are not yet ready and much needs to be done to develop, implement, and test systems that rely on specific privacy protection techniques. A thorough evaluation of the usefulness of existing privacy and security techniques that are appropriate for the biomedical context becomes critical. Although there have been surveys9–11 on the protection of privacy and security for genomic data analysis and sharing, most of them focus on theory. The research community needs practical benchmarking datasets that can be used for comprehensive evaluation of privacy and security techniques in real-world applications. Without direct comparisons of different methods in real-world scenarios, we cannot effectively evaluate their capabilities and understand their limitations. Both methods and technology are evolving fast, so what could be considered not feasible just a few years ago may now be ready for real-world applications. To narrow the gap between theory and practice, we initiated in 2012 the integrating data for analysis, anonymization and sharing (iDASH) consortium,12 which has become a premier biomedical privacy and security annual workshop where teams present their solutions to carefully selected problems in genome privacy and security. Specifically, we built a community focusing on the connection of both theoretical and practical aspects of genome privacy and security. Our goal is to promote the development of novel and practical protection methods to deal with the critical and emerging privacy and security challenges in human genomic research. Our competitions evaluate creative privacy and security methods with real genomic analysis tasks.
TOPICS AND METHODS
The first step to initiate the community efforts is to determine a set of highly relevant and critically needed gnomic privacy/security research topics. During the process of data analysis across multiple institutions, there are several possible ways to share information, within which our topics lies (a glossary is shown in Table 1):
Table 1.
Glossary of topics for iDASH competitions
| # | Topic | Description and references |
|---|---|---|
| 1 | Privacy-preserving data sharing | Allow differentially private federated data analysis with fragmented data from distributed sources21 |
| 2 | Secure release | Support differentially private data release with mitigated risks of information leakage22 |
| 3 | Secure outsourcing | Delegate data storage and analysis on untrusted third party servers23,24 |
| 4 | Homomorphic encryption | Support encrypted operations to match the plaintext operation with advanced cryptographic techniques, without leaking information25,26 |
| 5 | Secure collaboration | Collaboration among two or more parties to perform a computation jointly, without sharing their own raw data27 |
| 6 | Secure multiparty computation | Cryptographic techniques to perform computation jointly by two or more parties on encrypted data28 |
| 7 | Beacon service | Evaluation of a human genomic data sharing service developed by the GA4GH to check whether a human genomic dataset contains a genome with a specific variant (nucleotide) at a specific chromosomal location29 |
| 8 | Privacy-preserving search | Support for the calculation of distances between two genome sequences, without revealing variants30 |
| 9 | Encryption testing | Allowing genetic testing on encrypted data and results that can only be decrypted by data owners who have the secret key |
| 10 | Deduplication | Removal of duplicate records in a database31 |
| 11 | Software guard extensions | Application of isolation techniques developed by Intel hardware to protect data in use32 |
| 12 | Secure search | Identification of a query record in an encrypted database33 |
| 13 | Blockchain and smart contract | Distributed ledger technology that allows both decentralized sharing of data (blockchain34–36) and code (smart contracts37–39) |
| 14 | Secure machine learning | Building of machine learning models from encrypted data40–42 |
| 15 | Privacy-preserving machine learning | Execution of plaintext models on encrypted data to preserve data privacy43–47 |
iDASH: integrating data for analysis, anonymization and sharing.
Sharing raw data. The most straightforward way is to share the raw data across institutions. However, patient data are too sensitive to be shared directly without any protection due to privacy concerns and associated institutional data sharing policies. Therefore, possible methods to enhance data protection during the data sharing process include data perturbation (eg, adding noise to the data) to avoid sensitive information leakage (privacy-preserving data sharing13) encrypting and outsourcing the computation to a trusted third party [secure outsourcing,14 homomorphic encryption (HE),14–20 and encryption testing15] linking patients across different institutions without using sensitive data (deduplication16) hardware-supported secured analysis (software guard extensions16,18,19 and privacy-preserving machine learning (ML)18–20) encrypting queries and databases for genomic data (secure search17) and adopting a decentralized architecture to avoid central-server risks such as single point of failure (blockchain and smart contract17,18,20)
Sharing intermediate analysis results but not the raw data. Another possible way is to share partially summarized data (ie, intermediate results) among institutions, to allow joint analysis without sharing the raw (ie, observational level) data directly. However, designing the computational algorithms to allow intermediate result sharing without leaking patient-level data can be challenging. Therefore, we focused on topics related to algorithm developing, such as secure collaboration,14,18–20 secure multiparty computation,14–18 privacy-preserving search,15 and secure ML.18
Sharing only the final analysis results. Yet another way is to only share analytical results. However, there might still be privacy concerns (eg, exposing more information than expected by the differential privacy (DP) criterion with a small privacy budget), which occurs in particular when the sample size is small that the patients’ information can be “reversed engineered” from the shared final results. Plausible methods to mitigate the risk include anonymizing genome-wide association studies (GWAS) and genome sequence comparison results (secure release13) and randomly flipping query results to avoid patients’ information being inferred from repeated queries (eg, through the beacon service15)
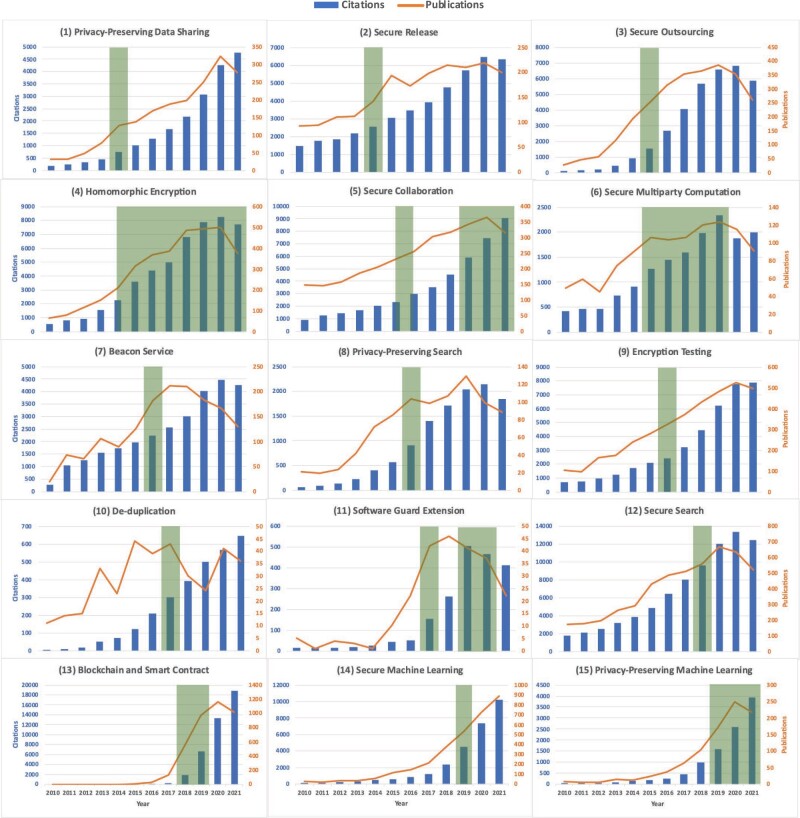
We summarize topics associated with privacy and security techniques in each track of the iDASH competitions in Table 2. Most of these 15 topics have only been emerging at the time of competition, but most of them are now recognized to be important by the scientific community. This can be shown in our publication and citation analysis (Figure 2). We observe an upward trend, with the top 5 (in terms of publications) being blockchain, smart contracts, secure ML, secure search, and secure outsourcing through HE. We also present the years in which the iDASH competition selected a particular topic, showing that our community efforts were timely and in line with current research and development directions. Also, our competition was organized while many papers in these topics were being published, thereby allowing us to take advantage of the growing interest in genome privacy and security as emphasized by iDASH. To provide more details about the outcomes and lessons learned from our competition, we use the most recent competitions, organized in 2020 and 2021, as our examples.
Table 2.
Topics for iDASH competitions, by year13–20
| # | Topic | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | 2020 | 2021 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Privacy-preserving data sharing | X | |||||||
| 2 | Secure release | X | |||||||
| 3 | Secure outsourcing | X | |||||||
| 4 | Homomorphic encryption | X | X | X | X | X | X | X | |
| 5 | Secure collaboration | X | X | X | X | ||||
| 6 | Secure multiparty computation | X | X | X | X | X | |||
| 7 | Beacon service | X | |||||||
| 8 | Privacy-preserving search | X | |||||||
| 9 | Encryption testing | X | |||||||
| 10 | Deduplication | X | |||||||
| 11 | Software guard extensions | X | X | X | |||||
| 12 | Secure search | X | |||||||
| 13 | Blockchain and smart contract | X | X | X | |||||
| 14 | Secure machine learning | X | |||||||
| 15 | Privacy-preserving machine learning | X | X | X |
iDASH: integrating data for analysis, anonymization and sharing.
Figure 2.
General trends in scientific publications and citations for iDASH competition topics. The data source is WOS8 as of December 14, 2021. We also show the years (shaded boxes) in which the iDASH competition focused on a particular topic, showing that our community efforts are timely and in line with these research topics.13–17 In general, the trends are upwards in both publications and citations, with the largest numbers for blockchain and smart contracts (topic # 13), secure machine learning (topic # 14), secure search (topic # 12), homomorphic encryption (topic # 4), and encryption testing (topic # 9). Citations and publications for Software Guard Extension seem to be trending down. iDASH: integrating data for analysis, anonymization and sharing; WOS: Web of Science.
To benchmark and evaluate these important topics, we organized, with the participation of community members from all over the world, eight annual iDASH competitions (2014–2021), aimed at tackling state-of-the-art privacy and security challenges. Each competition contained two to four different tracks (as shown in Supplementary Table ST1), and the iDASH consortium generated 40 publications13–17,21–26,30,40–67 from 23 tracks. These papers have been cited 1491 times (max = 137, min = 3, median = 29.5, average = 37.3) as of April 2022,68 demonstrating the impact of the competition on the field (a diagram of the total citations, as well as the citations per year published, is shown in Supplementary Figure SF1). Two meetings were virtual, while the others were scheduled right before or after a relevant conference in a particular city (so we referred to them as being “colocated” with a conference). Participants were mainly from North America in 2014, while in 2021 the community had expanded to multiple continents, representing an ever-growing, world-wide group of researchers whose focus is on tackling practical genome privacy and security issues. Particularly, the following three regions have demonstrated strong interest in this field: North America, Europe, and Asia.
LESSONS LEARNED FROM THE 2020 iDASH COMPETITION
The iDASH community effort has promoted pragmatic privacy research in key biomedical areas; it has been producing new and promising results that enable practical biomedical data protection at rest or in use. For example, in 2020:
In track 1 (Secure multilabel tumor classification using HE), we observed that most teams were utilizing linear/logistic regression models to implement cancer classifiers. These models had been improved significantly over the years through the HE competition, and HE is quite scalable and efficient now. The top solutions achieved a micro-Area Under the receiver operating characteristic Curve (micro-AUC, a measure for multilabel classification69) of ≥0.97 to classify 11 cancer types from encrypted genetic variants of 909 samples, within 5 minutes. These results show the feasibility of applying plaintext ML models to encrypted data for secure classification within acceptable time.
In track 2 [Privacy-preserving clustering of single-cell transcriptomics data in Software Guard eXtension (SGX)], we observed that two submission teams achieved comparable accuracy for Clustering through Imputation and Dimensionality Reduction algorithms70 when running on up to 10 000 single-cell sequences. However, the computing overhead of the best-performing solution increased 5 times for the input of 3000 cells up to over 20 times for the input of 10 000 cells, indicating that there is still plenty of room for further improvement to reduce the computation overhead of the SGX-based algorithms. These results suggest that the implementation of clustering algorithms for single-cell RNA-seq data on SGX is efficient on a moderate single-cell dataset but is still not efficient enough for large datasets.
In track 3 (Differentially private federated learning for a cancer prediction model), we were impressed by the innovative solutions, which achieved almost perfect model accuracy while enforcing a high DP standard (ie, DP with a privacy budget of 3.0 or lower). The training process of the best-performing solution was very fast, comparable with the efficiency of training an ML model on all data, unprotected, by a single party. These results suggest that the federated learning methods have advanced significantly in the past few years and could be ready for practical applications in biomedical research today.
LESSONS LEARNED FROM THE 2021 iDASH COMPETITION
Another set of examples comes from our competition in 2021:
In track 1 (Data sharing consent for health-related data using contracts on blockchain), we found that it was feasible to store patients’ willingness to share their digital health records in seven categories (demographics, mental health, biospecimen, family history, genetic, general clinical information, and sexual/reproductive health) for a given clinical/genomic study on blockchain, at up to ∼6800 records per hour (or ∼1.889 records per second). These results show that this emerging blockchain and smart contract technology has improved over past years and could become increasingly feasible in supporting real-world applications (eg, recording patients’ data sharing consents), without requiring high-throughput storage.
In track 2 (HE-based secure viral strain classification), the performance of the solutions was highly impressive. Almost all teams did very well in classification performance (many reported micro-AUC >0.99), indicating that secure viral strain classification was a highly practical task. There was large variability for the time cost in the secure computation, ranging from a few seconds to hours. The best solutions balanced the computation involved in all steps (preprocessing, key generation, encryption, classification, and decryption), and optimized computational costs to classify four SARS-CoV2 viral strains from 2000 homomorphically encrypted genomes within a few seconds. These results are highly encouraging for the practical use of HE to safeguard data privacy in high-performance classification models (eg, deep learning) for viral strain identification.
In track 3 (Confidential computing), we observed that federated learning algorithms submitted by participating teams were very efficient (ie, produced results within a minute) in training an ML model jointly by two parties (with each holding their individual training datasets). The task was to predict the potential risk of wild-type transthyretin amyloid cardiomyopathy from thousands of features extracted from electronic health records (EHRs). These solutions achieved comparable accuracy, and the ML model trained directly on the joint datasets under DP with a required protection level, ensuring that no private information in the EHR held by one party was leaked to the other party during the learning process. These evaluation results suggest that efficient DP-based algorithms could be used to build ML models from distributed training sets with satisfactory accuracy.
ANTICIPATED FUTURE RESEARCH TRENDS
We identify the following five directions of future genomic privacy research, which represent the emerging challenges that we plan to explore in the future competitions:
Combining federated learning and secure computing. There are some recent trends in this direction to combine the strength of both techniques to achieve better performance and a stronger privacy guarantee. Multikey HE71,72 is an example in which HE and secure multiparty computation can crossfertilize to improve efficiency and reduce the memory footprint in federated learning. Another example is the combination of DP and HE to enable a “refreshed” calculation of gradient with mitigated privacy and the development of a DP global ML model.73 A challenge for these hybrid solutions is the unification of security standards so that the overall security will not be lowered by the least secure component in the combined architecture. This is a very active area of research, and we expect highly innovative models to be developed.
Efficient training and evaluation of deep learning models on encrypted genomic data. We observe that many secure operations on encrypted genomic data, which were originally considered to be purely theoretical, have become more practical for real deployment.74 For example, recent work on secure genome imputation67 demonstrates that well-optimized HE-based regression models can meet the time and memory requirements that are comparable to or lower than those of nonsecure methods. We believe that this is just the beginning of a new era of secure deep learning on encrypted genomic data and that the community will witness the emergence of new models that are highly secure and efficient. Despite exciting progress, there are still many challenges in making encrypted genomics data analysis practical and scalable. HE algorithms are not friendly to high-order polynomials, and efficient implementation requires a deep understanding of parallelization. We will focus on closing the technology gap in future competitions by designing challenges related to these issues to push the front of encrypted genomic data analysis with state-of-the-art deep learning models.
Trusted hardware/software combinations. Recent studies show that the hybrid approaches that combine hardware (eg, SGX) and software (eg, HE and secure multiparty computation) offer efficient solutions to genomic data analyses. For example, SAFETY75 and DyPS76 are hybrid computational frameworks to perform secure GWAS on distributed genomic datasets using HE and SGX techniques. Kockan et al77 developed an approximation algorithm to accelerate a secure GWAS algorithm running in SGX that achieves comparable accuracy and efficiency to those of nonsecure counterparts. Bomai et al78 developed another hybrid approach combining multikey HE and SGX for GWAS and human genome computing. Widanage et al79 developed an SGX-based big-data analytics workflow HySec-Flow, which showcases privacy-preserving genomic computing tasks such as reads’ alignment. The future challenges along the direction include the extension of the approaches to emerging hardware architectures for confidential computing, such as Intel’s Trust Domain eXtension80 and AMD’s Secure Encrypted Virtualization,81 and the development of novel approaches that combine the hardware and software solutions to achieve stronger data protection and better performance for privacy-preserving genomic data analyses.
Distributed database and secure computing using smart contracts. Recent studies proposed to adopt smart contracts for consent management in genomic data sharing,82 COVID-19 data tracking,83 clinical X-ray image storing,84 and biomedical training certificate recording.85 As blockchain technology becomes more mature, we anticipate more genomic/biomedical applications to be proposed and developed. That said, the scalability of blockchain is still considered a bottleneck for large-scale data storage. Therefore, we plan to focus on performance improvement when designing future competition tasks on this topic.
Use of genome privacy technologies to support Ethical, Legal and Social Implications (ELSI) research. Novel genome privacy technologies can serve as enablers to circumvent ELSI barriers to support data sharing and federated learning. For example, researchers are implementing HE and DP within Informatics for Integrating Biology and the Bedside framework86 to enable an efficient privacy-preserving explorer for genetic cohorts. Secure multiparty computing models have been developed to enable privacy-preserving drug-target interaction protocols87 and large-scale GWAS analysis.3 We expect that future research in genomics privacy will be more tightly connected to ELSI requirements (specifically, to understand the emerging ELSI issues) and provide novel technology solutions to support scientific discoveries.
CONCLUSION
Our efforts to organize competitions and workshops to address practical privacy and security topics for genomic data analysis have created a solid global community, attracted interest from interdisciplinary teams around the world, and pushed the frontier of safeguarding patient data while advancing genomic research. Although the biomedical and healthcare privacy community is still small and iDASH competitions have started less than a decade ago, the impacts of our competitions/workshops start to become prominent with the citations generated by the 40 papers related to our community efforts in the past 8 years. From these experiences, we learned that such a community-driven approach could attract more researchers to devote themselves to genomic privacy and security research. We plan to continue this endeavor to grow the international community and facilitate biomedical privacy and security studies. In the 2022 iDASH competition, for example, we are focusing on four emerging topics:88 (1) blockchain-based recording of human subjects’ compliance training certificates, (2) secure model evaluation on homomorphically encrypted genotype data, (3) confidential computing for clustering single-cell transcriptomics data, and (4) secure record linkage. Using cutting-edge technology, theoretical developments and practical implementations can be integrated to provide highly deployable solutions that improve privacy protection and security for genomic data analysis and sharing. Specifically, we suggest that the following mature technologies can readily be implemented and even deployed today by entities stewarding genomic data: secure genome imputation, homomorphic encrypted GWAS, secure ancestry inference for admixed populations, ML-based confidential-computing for disease prognosis, secure single-cell data analyses, and polygenic risk score.89
FUNDING
The competitions are funded by the U.S. National Institutes of Health (NIH) (R13HG009072). T-TK is partly funded by the U.S. NIH (R00HG009680, R01HL136835, and R01GM118609). LO-M and XJ are funded by the U.S. NIH (R01LM013712 and RHL136835A). XFW and HT are funded by the U.S. NIH (R01HG010798). LO-M is funded by the U.S. NIH (RM1HG011558, R01LM013712, R01HL136835, and R01HG011066). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
AUTHOR CONTRIBUTORS
T-TK contributed to conceptualization, data curation, formal analysis, investigation, methodology, software, validation, visualization, and writing—original draft. XJ, HT, XFW, AH, MK, KP, DB, TB, JK, WL, and HC contributed to data curation, formal analysis, investigation, methodology, visualization, and writing—review and editing. LO-M contributed to conceptualization, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, visualization, and writing—review and editing.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGEMENTS
We want to extend our special thanks to evaluators and meeting moderators from the three institutions, including UTHealth (Luyao Chen), Indiana University (Weijie Liu, Tianhao Mao, Diyue Bu, Lei Wang), and UCSD administration (Morgan Von Ebke). We also thank contributors who provide fruitful discussions from UCI (Kai Zheng and the team) and Cedar Sinai (Spencer Soohoo and the team). We thank all teams that participated in the competitions, as well as Dr. Heidi Sofia.
CONFLICT OF INTEREST STATEMENT
None declared.
Contributor Information
Tsung-Ting Kuo, UCSD Health Department of Biomedical Informatics, University of California San Diego, La Jolla, California, USA.
Xiaoqian Jiang, School of Biomedical Informatics, The University of Texas Health Science Center at Houston, Houston, Texas, USA.
Haixu Tang, Luddy School of Informatics, Computing, and Engineering, Indiana University Bloomington, Bloomington, Indiana, USA.
XiaoFeng Wang, Luddy School of Informatics, Computing, and Engineering, Indiana University Bloomington, Bloomington, Indiana, USA.
Arif Harmanci, School of Biomedical Informatics, The University of Texas Health Science Center at Houston, Houston, Texas, USA.
Miran Kim, Department of Mathematics, Hanyang University, Seoul, Republic of Korea; Department of Computer Science, Hanyang University, Seoul, Republic of Korea.
Kai Post, UCSD Health Department of Biomedical Informatics, University of California San Diego, La Jolla, California, USA.
Diyue Bu, Luddy School of Informatics, Computing, and Engineering, Indiana University Bloomington, Bloomington, Indiana, USA.
Tyler Bath, UCSD Health Department of Biomedical Informatics, University of California San Diego, La Jolla, California, USA.
Jihoon Kim, UCSD Health Department of Biomedical Informatics, University of California San Diego, La Jolla, California, USA.
Weijie Liu, Luddy School of Informatics, Computing, and Engineering, Indiana University Bloomington, Bloomington, Indiana, USA.
Hongbo Chen, Luddy School of Informatics, Computing, and Engineering, Indiana University Bloomington, Bloomington, Indiana, USA.
Lucila Ohno-Machado, UCSD Health Department of Biomedical Informatics, University of California San Diego, La Jolla, California, USA; Division of Health Services Research & Development, Veteran Affairs San Diego Healthcare System, San Diego, California, USA.
Data Availability
The data underlying this article are available in the article.
REFERENCES
- 1. NHGRI. Genome Sequencing Program (GSP). www.genome.gov/sequencingcostsdata Accessed August 25, 2022
- 2. Lakshmanan R. A New SolarWinds Flaw Likely Had Let Hackers Install SUPERNOVA Malware; 2020. https://thehackernews.com/2020/12/a-new-solarwinds-flaw-likely-had-let.html Accessed December 28, 2020
- 3. Cho H, Wu DJ, Berger B.. Secure genome-wide association analysis using multiparty computation. Nat Biotechnol 2018; 36 (6): 547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fiume M, Cupak M, Keenan S, et al. Federated discovery and sharing of genomic data using Beacons. Nat Biotechnol 2019; 37 (3): 220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang S, Jiang X, Singh S, et al. Genome privacy: challenges, technical approaches to mitigate risk, and ethical considerations in the United States. Ann NY Acad Sci 2017; 1387 (1): 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The104thUnitedStatesCongress. Health Insurance Portability and Accountability Act (HIPAA) of 1996; 1996. https://uslaw.link/citation/us-law/public/104/191 Accessed September 15, 2022.
- 7. European Parliament, Council of the European Union. General Data Protection Regulation (GDPR); 2016. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32016R0679 Accessed September 15, 2022.
- 8. Clarivate Analytics. Web of Science; 2020. https://clarivate.com/webofsciencegroup/solutions/web-of-science/ Accessed September 15, 2022.
- 9. Al Aziz MM, Sadat MN, Alhadidi D, et al. Privacy-preserving techniques of genomic data—a survey. Brief Bioinform 2019; 20 (3): 887–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonomi L, Huang Y, Ohno-Machado L.. Privacy challenges and research opportunities for genomic data sharing. Nat Genet 2020; 52 (7): 646–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wan Z, Hazel JW, Clayton EW, Vorobeychik Y, Kantarcioglu M, Malin BA.. Sociotechnical safeguards for genomic data privacy. Nat Rev Genet 2022; 23 (7): 429–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohno-Machado L, Bafna V, Boxwala A. et al. ; iDASH team. iDASH: integrating data for analysis, anonymization, and sharing. J Am Med Inform Assoc 2012; 19 (2): 196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang X, Zhao Y, Wang X, et al. A community assessment of privacy preserving techniques for human genomes. BMC Med Inform Decis Mak 2014; 14 (S1): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang H, Jiang X, Wang X, et al. Protecting genomic data analytics in the cloud: state of the art and opportunities. BMC Med Genomics 2016; 9 (1): 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang S, Jiang X, Tang H, et al. A community effort to protect genomic data sharing, collaboration and outsourcing. npj Genomic Med 2017; 2 (1): 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Tang H, Wang S, et al. iDASH secure genome analysis competition 2017. BMC Med Genomics 2018; 11 (Suppl 4): 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuo T-T, Jiang X, Tang H, et al. iDASH secure genome analysis competition 2018: Blockchain genomic data access logging, homomorphic encryption on GWAS, and DNA segment searching. BMC Med Genomics 2020; 13 (Suppl 7): 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. iDASH Privacy & Security Workshop. Secure Genome Analysis Competition 2019; 2019. http://www.humangenomeprivacy.org/2019/ Accessed September 15, 2022.
- 19. iDASH Privacy & Security Workshop. Secure Genome Analysis Competition 2020;2020. http://www.humangenomeprivacy.org/2020/ Accessed September 15, 2022.
- 20. iDASH Privacy & Security Workshop. Secure Genome Analysis Competition 2021;2021. http://www.humangenomeprivacy.org/2021/ Accessed September 15, 2022.
- 21. Yu F, Ji Z.. Scalable privacy-preserving data sharing methodology for genome-wide association studies: an application to iDASH healthcare privacy protection challenge. BMC Med Inform Decis Mak 2014; 14 (S1): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang S, Mohammed N, Chen R.. Differentially private genome data dissemination through top-down specialization. BMC Med Inform Decis Mak 2014; 14 (S1): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y, Dai W, Jiang X, Xiong H, Wang S.. Foresee: fully outsourced secure genome study based on homomorphic encryption. BMC Med Inform Decis Mak 2015; 15 (5): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sousa JS, Lefebvre C, Huang Z, et al. Efficient and secure outsourcing of genomic data storage. BMC Med Genomics 2017; 10 (Suppl 2): 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu W-J, Yamada Y, Sakuma J.. Privacy-preserving genome-wide association studies on cloud environment using fully homomorphic encryption. BMC Med Inform Decis Mak 2015; 15 (5): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim M, Lauter K.. Private genome analysis through homomorphic encryption. BMC Med Inform Decis Mak 2015; 15 (5): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cahill V, Gray E, Seigneur J-M, et al. Using trust for secure collaboration in uncertain environments. IEEE Pervasive Comput 2003; 2 (3): 52–61. [Google Scholar]
- 28. Cramer R, Damgård IB.. Secure Multiparty Computation. Cambridge, United Kingdom: Cambridge University Press; 2015. [Google Scholar]
- 29. Global Alliance for Genomics and Health. Genomics. A federated ecosystem for sharing genomic, clinical data. Science 2016; 352 (6291): 1278–80. [DOI] [PubMed] [Google Scholar]
- 30. Carpov S, Tortech T.. Secure top most significant genome variants search: iDASH 2017 competition. BMC Med Genomics 2018; 11 (Suppl 4): 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyer DT, Bolosky WJ.. A study of practical deduplication. ACM Trans Storage 2012; 7 (4): 1–20. [Google Scholar]
- 32. Costan V, Devadas S.. Intel SGX Explained. Lyon, France: Cryptology ePrint Archive; 2016. [Google Scholar]
- 33. Pham H, Woodworth J, Amini Salehi M.. Survey on secure search over encrypted data on the cloud. Concurr Comput Pract Exp 2019; 31 (17): e5284. [Google Scholar]
- 34. Kuo T-T, Kim H-E, Ohno-Machado L.. Blockchain distributed ledger technologies for biomedical and health care applications. J Am Med Inform Assoc 2017; 24 (6): 1211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakamoto S. Bitcoin: a peer-to-peer electronic cash system. Decentralized Bus Rev 2008: 21260. [Google Scholar]
- 36. Greenspan G. MultiChain Private Blockchain—White Paper. London, United Kingdom: Coin Sciences Ltd; 2015. [Google Scholar]
- 37. Kuo T-T, Zavaleta Rojas H, Ohno-Machado L.. Comparison of blockchain platforms: a systematic review and healthcare examples. J Am Med Inform Assoc 2019; 26 (5): 462–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu H, Sun H, Wu D, Kuo T-T. Comparison of smart contract blockchains for healthcare applications. In: AMIA Annual Symposium: American Medical Informatics Association, Bethesda, MD; 2019. [PMC free article] [PubMed]
- 39. Buterin V. A next-generation smart contract and decentralized application platform. White Paper 2014; 3 (27): 2-1. [Google Scholar]
- 40. Kim A, Song Y, Kim M, Lee K, Cheon JH.. Logistic regression model training based on the approximate homomorphic encryption. BMC Med Genomics 2018; 11 (Suppl 4): 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen H, Gilad-Bachrach R, Han K, et al. Logistic regression over encrypted data from fully homomorphic encryption. BMC Med Genomics 2018; 11 (Suppl 4): 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bonte C, Vercauteren F.. Privacy-preserving logistic regression training. BMC Med Genomics 2018; 11 (Suppl 4): 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim M, Song Y, Li B, Micciancio D.. Semi-parallel logistic regression for GWAS on encrypted data. BMC Med Genomics 2020; 13 (7): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carpov S, Gama N, Georgieva M, Troncoso-Pastoriza JR.. Privacy-preserving semi-parallel logistic regression training with fully homomorphic encryption. BMC Med Genomics 2020; 13 (7): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blatt M, Gusev A, Polyakov Y, Rohloff K, Vaikuntanathan V.. Optimized homomorphic encryption solution for secure genome-wide association studies. BMC Med Genomics 2020; 13 (7): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim D, Son Y, Kim D, Kim A, Hong S, Cheon JH.. Privacy-preserving approximate GWAS computation based on homomorphic encryption. BMC Med Genomics 2020; 13 (7): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sim JJ, Chan FM, Chen S, Tan BHM, Aung KMM.. Achieving GWAS with homomorphic encryption. BMC Med Genomics 2020; 13 (7): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Constable SD, Tang Y, Wang S, Jiang X, Chapin S.. Privacy-preserving GWAS analysis on federated genomic datasets. BMC Med Inform Decis Mak 2015; 15 (5): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y, Blanton M, Almashaqbeh G.. Secure distributed genome analysis for GWAS and sequence comparison computation. BMC Med Inform Decis Mak 2015; 15 (5): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wan Z, Vorobeychik Y, Kantarcioglu M, Malin B.. Controlling the signal: practical privacy protection of genomic data sharing through Beacon services. BMC Med Genomics 2017; 10 (Suppl 2): 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Al Aziz MM, Ghasemi R, Waliullah M, Mohammed N.. Aftermath of bustamante attack on genomic beacon service. BMC Med Genomics 2017; 10 (Suppl 2): 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang XS, Huang Y, Zhao Y, Tang H, Wang X, Bu D. Efficient genome-wide, privacy-preserving similar patient query based on private edit distance. In: Proceedings of the 22nd ACM SIGSAC Conference on Computer and Communications Security; 2015.
- 53. Al Aziz MM, Alhadidi D, Mohammed N.. Secure approximation of edit distance on genomic data. BMC Med Genomics 2017; 10 (Suppl 2): 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Çetin GS, Chen H, Laine K, Lauter K, Rindal P, Xia Y.. Private queries on encrypted genomic data. BMC Med Genomics 2017; 10 (Suppl 2): 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ziegeldorf JH, Pennekamp J, Hellmanns D, et al. BLOOM: BLoom filter based oblivious outsourced matchings. BMC Med Genomics 2017; 10 (Suppl 2): 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim M, Song Y, Cheon JH.. Secure searching of biomarkers through hybrid homomorphic encryption scheme. BMC Med Genomics 2017; 10 (Suppl 2): 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Laud P, Pankova A.. Privacy-preserving record linkage in large databases using secure multiparty computation. BMC Med Genomics 2018; 11 (Suppl 4): 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen F, Wang C, Dai W, et al. PRESAGE: PRivacy-preserving gEnetic testing via SoftwAre guard extension. BMC Med Genomics 2017; 10 (Suppl 2): 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gursoy G, Bjornson R, Green ME, Gerstein M.. Using blockchain to log genome dataset access: efficient storage and query. BMC Med Genomics 2020; 13 (7): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pattengale ND, Hudson CM.. Decentralized genomics audit logging via permissioned blockchain ledgering. BMC Med Genomics 2020; 13 (7): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ma S, Cao Y, Xiong L.. Efficient logging and querying for blockchain-based cross-site genomic dataset access audit. BMC Med Genomics 2020; 13 (7): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ozdayi MS, Kantarcioglu M, Malin B.. Leveraging blockchain for immutable logging and querying across multiple sites. BMC Med Genomics 2020; 13 (7): 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sotiraki K, Ghosh E, Chen H.. Privately computing set-maximal matches in genomic data. BMC Med Genomics 2020; 13 (7): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hasan MZ, Mahdi MSR, Sadat MN, Mohammed N.. Secure count query on encrypted genomic data. J Biomed Inform 2018; 81 (2018): 41–52. [DOI] [PubMed] [Google Scholar]
- 65. Kuo T-T, Bath T, Ma S, et al. Benchmarking blockchain-based gene-drug interaction data sharing methods: a case study from the iDASH 2019 secure genome analysis competition blockchain track. Int J Med Inform 2021; 154: 104559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gürsoy G, Brannon CM, Gerstein M.. Using Ethereum blockchain to store and query pharmacogenomics data via smart contracts. BMC Med Genomics 2020; 13 (1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim M, Harmanci AO, Bossuat J-P, et al. Ultrafast homomorphic encryption models enable secure outsourcing of genotype imputation. Cell Syst 2021; 12 (11): 1108–20.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Google. Google Scholar. http://scholar.google.com/ Accessed September 15, 2022.
- 69. Wu X-Z, Zhou Z-H. A unified view of multi-label performance measures. In: International Conference on Machine Learning. PMLR; 2017.
- 70. Lin P, Troup M, Ho JW.. CIDR: ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biol 2017; 18 (1): 59–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen H, Chillotti I, Song Y. Multi-key homomorphic encryption from TFHE. In: International Conference on the Theory and Application of Cryptology and Information Security. Springer; 2019.
- 72. Chen H, Dai W, Kim M, Song Y. Efficient multi-key homomorphic encryption with packed ciphertexts with application to oblivious neural network inference. In: Proceedings of the 2019 ACM SIGSAC Conference on Computer and Communications Security; 2019.
- 73. Kim M, Lee J, Ohno-Machado L, Jiang X.. Secure and differentially private logistic regression for horizontally distributed data. IEEE Trans Inf Forensics Secur 2020; 15: 695–710. [Google Scholar]
- 74. Jiang X, Kim M, Lauter K, Song Y. Secure outsourced matrix computation and application to neural networks. In: Proceedings of the 2018 ACM SIGSAC Conference on Computer and Communications Security; 2018. [DOI] [PMC free article] [PubMed]
- 75. Sadat MN, Al Aziz MM, Mohammed N, Chen F, Jiang X, Wang S.. Safety: secure gwAs in federated environment through a hybrid solution. IEEE/ACM Trans Comput Biol Bioinform 2019; 16 (1): 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pascoal T, Decouchant J, Boutet A, Esteves-Verissimo P.. Dyps: dynamic, private and secure GWAS. Proc Priv Enh Technol 2021; 2021 (2): 214–34. [Google Scholar]
- 77. Kockan C, Zhu K, Dokmai N, et al. Sketching algorithms for genomic data analysis and querying in a secure enclave. Nat Methods 2020; 17 (3): 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bomai A, Aldeen MS, Zhao C. Privacy-preserving GWAS computation on outsourced data encrypted under multiple keys through hybrid system. In: 2020 IEEE 7th International Conference on Data Science and Advanced Analytics (DSAA). IEEE; 2020.
- 79. Widanage C, Liu W, Li J, et al. HySec-Flow: privacy-preserving genomic computing with SGX-based big-data analytics framework. In: 2021 IEEE 14th International Conference on Cloud Computing (CLOUD). IEEE; 2021. [DOI] [PMC free article] [PubMed]
- 80. Intel. Trust Domain Extensions White Paper; 2020. https://www.intel.com/content/dam/develop/external/us/en/documents/tdx-whitepaper-v4.pdf Accessed September 15, 2022.
- 81. AMD. Secure Encrypted Virtualization White Paper; 2020. https://www.amd.com/en/processors/amd-secure-encrypted-virtualization Accessed August 14, 2022
- 82. Albalwy F, Brass A, Davies A.. A blockchain-based dynamic consent architecture to support clinical genomic data sharing (ConsentChain): Proof-of-concept study. JMIR Med Inform 2021; 9 (11): e27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Marbouh D, Abbasi T, Maasmi F, et al. Blockchain for COVID-19: review, opportunities, and a trusted tracking system. Arab J Sci Eng 2020; 45 (12): 9895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mun Li M, Kuo T-T.. Previewable contract-based on-chain X-ray image sharing framework for clinical research. Int J Med Inform 2021; 156: 104599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tellew J, Kuo T-T.. CertificateChain: decentralized healthcare training certificate management system using blockchain and smart contracts. JAMIA Open 2022; 5 (1): ooac019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Raisaro JL, Pradervand S, Colsenet R, et al. Protecting privacy and security of genomic data in i2b2 with homomorphic encryption and differential privacy. IEEE/ACM Trans Comput Biol Bioinform 2018; 15 (5): 1413–26. [DOI] [PubMed] [Google Scholar]
- 87. Hie B, Cho H, Berger B.. Realizing private and practical pharmacological collaboration. Science 2018; 362 (6412): 347–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. iDASH Privacy & Security Workshop. Secure Genome Analysis Competition 2022; 2022. http://www.humangenomeprivacy.org/2022/ Accessed August 8, 2022
- 89. Li R, Chen Y, Ritchie MD, Moore JH.. Electronic health records and polygenic risk scores for predicting disease risk. Nat Rev Genet 2020; 21 (8): 493–502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article.