Abstract
The pathogeneses of the 2 major forms of diabetes, type 1 and type 2, differ with respect to their major molecular insults (loss of immune tolerance and onset of tissue insulin resistance, respectively). However, evidence suggests that dysfunction and/or death of insulin-producing β-cells is common to virtually all forms of diabetes. Although the mechanisms underlying β-cell dysfunction remain incompletely characterized, recent years have witnessed major advances in our understanding of the molecular pathways that contribute to the demise of the β-cell. Cellular and environmental factors contribute to β-cell dysfunction/loss through the activation of molecular pathways that exacerbate endoplasmic reticulum stress, the integrated stress response, oxidative stress, and impaired autophagy. Whereas many of these stress responsive pathways are interconnected, their individual contributions to glucose homeostasis and β-cell health have been elucidated through the development and interrogation of animal models. In these studies, genetic models and pharmacological compounds have enabled the identification of genes and proteins specifically involved in β-cell dysfunction during diabetes pathogenesis. Here, we review the critical stress response pathways that are activated in β cells in the context of the animal models.
Keywords: islet, endoplasmic reticulum stress, autophagy, integrated stress response, diabetes
Diabetes is a syndromic disorder characterized by hyperglycemia that includes manifestations of dyslipidemia, neuropathy, and microvascular and macrovascular complications. Worldwide, diabetes has increased to epidemic proportions, afflicting hundreds of millions of individuals (1). The traditional classification of the disorder as either type 1 (T1D) (β-cell autoimmunity-mediated) or type 2 (T2D) (insulin resistance and β-cell dysfunction-mediated) belies a more heterogeneous pathophysiology that, in some individuals, exhibit features of more than one type. For example, a recent study showed that up to 41% of individuals with T2D exhibit evidence of autoimmunity (islet β-cell-specific T-cell reactivity) with higher frequency of β-cell dysfunction (2). Conversely, in individuals with T1D, postmortem studies show that less than 24% of T1D individuals have any detectable pathologic evidence of insulitis (3). Other studies show a striking persistence of proinsulin secretion even in long-standing T1D (4), reflecting preservation of β cells in a disorder thought to be defined by their complete absence (5). These and other studies describe an overall picture wherein “endotypes” of T1D, T2D, and other, less common forms (eg, secondary diabetes, monogenic diabetes, gestational diabetes, ketosis-prone diabetes) represent disorders along a phenotypic and pathologic spectrum with an underlying feature being the loss or dysfunction of insulin-producing β cells.
As a central focus of disease, β cells must maintain regulated insulin release despite the prevalence of “stressors” (proinflammatory cytokines, free fatty acids, elevated glucose) that are characteristic of the disease process. The nature of the β-cell response to these diabetogenic stressors is arguably the single most crucial factor in the progression to disease since it dictates the robustness of insulin release as well as production of antigens that trigger the immune response. In this minireview, we discuss key molecular pathways and processes triggered by diabetogenic stressors—endoplasmic reticulum (ER) stress, the integrated stress response (ISR), the oxidative stress response, autophagy, and cellular senescence—and how these processes or their dysregulation contribute to β-cell dysfunction. Particular attention is paid to the animal models that are used to study these pathways and processes.
Endoplasmic Reticulum Stress in β-Cell Dysfunction
ER stress occurs when the demand for protein folding exceeds the ER folding capacity (6). As specialized secretory cells, β cells have a well-developed ER and several mechanisms in place to handle the enormous demand for insulin protein folding (7). Normally, β cells respond to alterations in blood glucose levels with a proportional synthesis and secretion of insulin. However, increased insulin demands during insulin resistance or the loss of β-cell mass from autoimmune attack creates an increased synthetic demand on residual β cells, thereby triggering ER stress. Studies both in mice and humans have explored the role of ER stress in diabetes (8, 9). ER stress may exacerbate or trigger T1D in mice, as evidenced by both the activation of ER stress-related genes and the expansion of ER in β cells (10, 11). Likewise, ER stress markers are elevated in β cells from pancreatic tissues of donors with T1D (12). Similar evidence has been presented in the context of T2D and has been well-reviewed previously (13). In the setting of ER stress, misfolded proteins are often the target of new posttranslational modifications, which serve as neoantigens that activate the immune response in T1D (14).
Unfolded Protein Response Is a Homeostatic Mechanism to Mitigate Endoplasmic Reticulum Stress
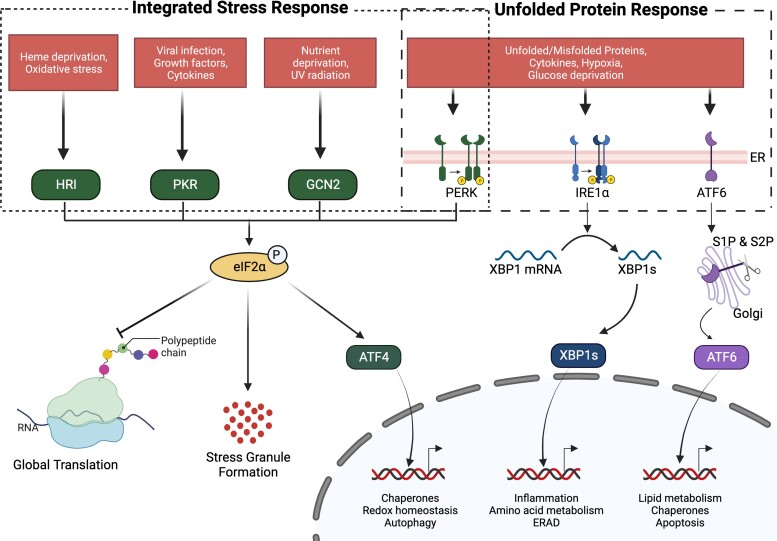
To alleviate ER stress and regain ER homeostasis, cells activate the unfolded protein response (UPR) pathway that (a) slows global protein synthesis; (b) upregulates chaperones that aid in protein folding; and (c) activates ER-associated degradation to degrade irreparably misfolded proteins (15). This response is activated by 3 UPR stress sensors: inositol requiring enzyme1α (IRE1α), activating transcription factor 6 (ATF6), and PKR-like endoplasmic reticulum kinase (PERK) (Fig. 1). Under normal conditions, the UPR sensors bind to a chaperone, binding immunoglobulin protein (BiP or GRP78) (16). Under ER stress conditions, BiP changes conformation and dissociates from the UPR sensors. BiP then binds to misfolded proteins and shuttles them from the ER lumen to the cytoplasm for proteasomal degradation (17). Deletion of any of the UPR sensors (IRE-1α, ATF6, or PERK) in mice induces diabetes, decreases β-cell mass, and impairs glucose metabolism (18–21).
Figure 1.
Integrated stress response (ISR) and unfolded protein response (UPR): The ISR (left side of figure) is a cellular response to extracellular stress signals that leads to activation of distinct kinases (PERK, HRI, GCN2, PKR). All 4 kinases phosphorylate eIF2α, which inhibits global protein translation and rescues the cell from stress. The UPR (right side of figure) is triggered by the presence of unfolded proteins in the endoplasmic reticulum (ER) and leads to the activation of 3 separate arms (PERK, IRE1α, ATF6). The PERK arm is shared with the ISR. The collective effect of the UPR is to delimit protein synthesis and activate chaperones to aid in protein folding.
Three “Arms” of the Unfolded Protein Response Are Essential for Endoplasmic Reticulum Homeostasis in β Cells
When activated by ER stress, IRE1α uses its endoribonuclease domain to splice Xbp1 messenger RNA (mRNA) to ensure encoding of mature X-box-binding protein 1 (XBP1) (22). XBP1 is a transcription factor that promotes expression of chaperones and ER-associated degradation proteins (eg, the Sel1L-Hrd1 complex) (23). β-cell–specific deletion of Sel1L, which is a cofactor of Hrd1, impairs ER-associated degradation and results in loss of β-cell identity (13). Conditional, β-cell–specific deletion of the gene encoding IRE1α (Ern1) in mice impairs glycemic control, decreases insulin secretion, and decreases folding of proinsulin (19). Similarly, β-cell–specific deletion of Xbp1 in mice decreases insulin secretion and results in hyperglycemia (24). The IRE1α pathway could help delay T1D development.
The second arm of the UPR, ATF6, has an autocatalytic site that is cleaved by site 1/2 proteases (S1P and S2P) in the Golgi (25, 26). Cleaved ATF6 acts as a transcription factor to activate genes encoding chaperones and ER-associated degradation proteins (27). ATF6 protects against viral infection-induced T1D in mice on administration of tauroursodeoxycholic acid (TUDCA) to enhance protein folding. These effects were not seen in animals with β-cell–specific Atf6 deletion, suggesting that TUDCA exerts its effects through the ATF6 arm of the UPR pathway (10). However, more studies need to be conducted to assess the role of ATF6 in T1D.
The third arm of the UPR, PERK, phosphorylates eukaryotic translation initiation factor 2 subunit 1 (EIF2S1 also known as eIF2α), which inhibits global mRNA translation (28). As expected, PERK-deficient mice show decreased eIF2α phosphorylation, enhanced global mRNA translation, and increased ER stress, which is accompanied by loss of β cells (21, 29). Germline deletion of the gene encoding PERK (Eif2ak3) results in neonatal diabetes (30), and its postnatal deletion results in a pleiotropic response in β cells with increased proliferation, and accumulation of proinsulin preceding increased cell death (31). With chronic ER stress, PERK promotes translation of the gene encoding activating transcription factor 4 (ATF4), which activates the gene encoding C/EBP-homologous protein (CHOP) to promote apoptosis (32). Thus, deletion of CHOP in C57BL/6 mice expands β-cell mass, alleviates ER stress, and improves glycemic control (33, 34). Although whole-body deletion of CHOP in nonobese diabetic (NOD) mice (a mouse model of T1D) has no effect on diabetes incidence, apoptosis, or insulitis, it reduces the early production of insulin autoantibodies (35). Studies suggest that the UPR and PERK pathway also play a role in human T1D. These studies report a strikingly similar trend of increased CHOP and BiP expression in islets isolated both from mice and human donors with T1D (11, 12). In addition, islets from donors with T1D have decreased expression of genes involved in the adaptation to ER stress (36). Collectively, these studies demonstrate that β-cell ER stress is activated during the pathogenesis of T1D and may contribute to β-cell dysfunction.
It should be noted that other proteins can affect the ER stress response indirectly, either by altering ER calcium homeostasis or altering transcription of genes that are required for ER homeostasis. For example, sarcoendoplasmic reticulum Ca2+ ATPase 2 (SERCA2) maintains a 3-fold higher concentration of Ca2+ within the ER, relative to the cytosol, to ensure proper functioning of ER-resident chaperones and protein folding machinery (37). Mice haploinsufficient for the gene encoding SERCA2 show elevated ER stress as well as impaired glucose tolerance in β cells after high-fat diet feeding (38). Similarly, deletion or deficiency of the gene encoding Wolframin (Wfs1), another protein required for ER calcium loading, results in ER stress, impaired glucose tolerance, and β-cell death (39). In humans, mutations in the WFS1 gene result in Wolfram syndrome, which includes β-cell loss (with juvenile-onset diabetes), sensorineural hearing loss, and optic atrophy (40). Thus, both direct and indirect causes of ER stress can lead to T1D. Collectively, the literature on β-cell ER stress supports the notion that the UPR and ER stress function as homeostatic mechanisms that allow the cell to recover from stresses that enhance protein demands, but that this adaptive mechanism can become maladaptive in the setting of prolonged stress and lead to eventual impairment and death of β cells.
Integrated Stress Response
The PERK pathway of the UPR is also a branch of the ISR. The ISR is a cytoprotective response to stresses such as viral infections, inflammation, and changes in gut microbiota and nutrient availability. These stresses are sensed by at least 1 of 4 specialized protein kinases: PERK, general control nonderepressible 2 (GCN2), protein kinase R (PKR), or heme-regulated inhibitor (HRI) (41). Each activated kinase phosphorylates eIF2α, which represses global translation and simultaneously promotes translation of mRNAs that mitigate stress (28) (see Fig. 1). The ISR, like the UPR, is an adaptive response whose activation is designed to relieve cellular stress and promote survival. However, when ISR activation is chronic and stress persists, the ISR can promote cellular apoptosis (42). In pancreatic β cells, this chronic activation of the ISR is now being investigated, as it may contribute to diabetes pathogenesis. Each of the 4 eIF2α kinases is activated on its dimerization and autophosphorylation. Although their catalytic domains share homology, their regulatory domains differ substantially so that each kinase can respond to distinct stress signals (43, 44). As noted previously, PERK is activated by ER stress. We next discuss the remaining 3 eIF2α kinases in greater depth. We also review the existing studies on β-cell function and diabetes pathogenesis that display hallmarks of ISR activation.
eIF2α Kinases of the Integrated Stress Response
The ISR kinase GCN2 senses cellular amino acid levels (21). During amino acid starvation, there is an increase in the levels of uncharged transfer RNAs (tRNAs) (45). These uncharged tRNAs bind to the histidyl-tRNA synthetase-related domain of GCN2, which activates the GCN2 kinase domain through autophosphorylation. Once activated, GCN2 phosphorylates eIF2α (46). Global deletion of the gene encoding GCN2 (Eif2ak4) in mice results in no overt phenotype. However, on high-fat diet feeding, Eif2ak4−/− mice exhibit impaired glucose tolerance and reduced β-cell mass (47). A role for GCN2 in the context of T1D pathogenesis has not yet been reported.
PKR is activated predominantly by double-stranded RNA in response to viral infection (48, 49). When its C-terminal kinase domains dimerize, PKR becomes autophosphorylated at T446 and subsequently activated (50). Activated PKR phosphorylates eIF2α to inhibit viral and host protein synthesis. In addition to double-stranded RNA, PKR can be activated by oxidative and ER stress, growth factor deprivation, cytokines, bacterial infection, ribotoxic stress, stress granules, and heparin (as reviewed in Pakos-Zebrucka et al) (41). Viral infections, which activate PKR, have been implicated as early triggers of T1D, and islets from T1D donors show higher expression of PKR (51). PKR has also recently been demonstrated to be activated in human islets following infection by the SARS-CoV-2 virus, and its subsequent signaling cascade may lead to β-cell dysfunction in the setting of COVID-19 (52). In a mouse study, inhibiting PKR protected against streptozotocin (STZ)-induced diabetes by downregulating the c-Jun N-terminal kinase (JNK) pathway (53). Hence, PKR may play a role in T1D development after viral infections as well as intracellular stresses.
HRI kinase is also activated on dimerization and autophosphorylation of its kinase domain. Its kinase activity is regulated by 2 heme-binding domains at the N-terminus and the kinase insertion domain. Heme inhibits HRI kinase activity by promoting disulfide bonds that lock HRI in an inactive dimer (54). Conversely, when heme is absent, noncovalent interactions form between HRI molecules, resulting in an active HRI dimer. Besides heme absence, other known activators of HRI include oxidative stress, nitric oxide, and 26S proteasome inhibition (55–57). The mechanisms of HRI activation by these stressors are unknown. To date, most studies of HRI have been performed in erythroid cells owing to the higher expression of HRI in erythrocytes. However, heme-independent HRI activators, such as oxidative stress and nitric oxide (synthesized in inflammation), are active in β cells during the initiation and progression of diabetes. Currently, no studies link prolonged activation of HRI to β-cell dysfunction.
As mentioned, all 4 kinases phosphorylate eIF2α (41). EIF2α is the critical regulatory subunit of the eIF2 complex (also consisting of eIF2β and eIF2γ), which plays a central role in initiating mRNA translation. When eIF2α is phosphorylated, it blocks eIF2β-mediated exchange of GDP for GTP, which is needed to deliver initiator methionyl-tRNA to the preinitiation complex. As a result, phosphorylated eIF2α globally reduces mRNA translation. However, a select set of mRNAs become preferentially translated that relieve cell stress and reinitiate translation (58).
Another outcome of ISR activation is the formation of nonmembranous stress granules that consist of translationally repressed mRNA and associated RNA-binding proteins (59). Because stress granules can also be released as extracellular vesicle contents (60–62), their contents could serve as a means of β-cell-to-immune cell communication in T1D (63). Under normal conditions, stress granule formation is reversible. However, under chronic stress or pathological conditions, stress granules accumulate, either due to increased stress granule formation or decreased stress granule clearance (64). In human islets, inhibiting the ISR reduced stress granule formation after inflammatory cytokine treatment and increased production of the immunosuppressive protein PD-L1 (65). A key player in stress granule assembly is Ras GTPase-activating protein-binding protein 1 (G3BP1); deleting G3BP1 in neurons resulted in fewer stress granules (66). Another player in stress granule assembly is deoxyhypusine synthase (DHPS), an enzyme required for the posttranslational modification (hypusination) of eukaryotic translation initiation factor 5A (eIF5A). Knockdown of DHPS results in significant inhibition of oxidative-stress induced stress granule formation (67–69), and systemic DHPS inhibition in NOD mice reduces ER stress and incidence of autoimmune diabetes (70, 71). Interestingly, another study suggests that the hypusine circuit can also promote ER stress and the IRE-1α pathway of the UPR (72). Together, these studies suggest that inhibition of stress granule assembly, perhaps through the manipulation of the ISR, could play a role in potentially delaying autoimmune diabetes incidence.
Additional studies suggest that a prolonged ISR contributes to T1D pathogenesis. Recent evidence points toward dysregulation of the ISR in pancreata of pre-T1D autoantibody-positive (AAb+) donors owing to repression of ISR kinase gene expression (PERK, GCN2, PKR) (73). However, there was elevated expression of the ISR genes in T1D donor pancreata, suggesting a chronic activation of ISR. Notably, there is an increase in EIF2AK3 (PERK) in T1D donor pancreata, consistent with activation of the ER stress arm of the ISR (73). As such, these data suggest that attenuating the ISR pathway may protect against diabetes. Indeed, during SARS-CoV2 infection of β cells, which can suppress insulin secretion and worsen diabetes, inhibiting the ISR allowed for recovery of β-cell function and reversed markers of cellular dedifferentiation (52). Further studies are needed to clarify the timing, magnitude, and duration of the ISR during diabetes pathogenesis as new ISR-targeting therapies are investigated. Collectively, emerging data on the ISR indicate that phosphorylation of eIF2α may be a mechanism to attenuate mRNA translation that is not restricted simply to the PERK arm of the ER stress cascade, and that other factors not directly related to ER stress (eg, viral infection or nutrient depravation) may be contributing factors in the eventual decline of β cells in diabetes.
Oxidative Stress in β-Cell Dysfunction
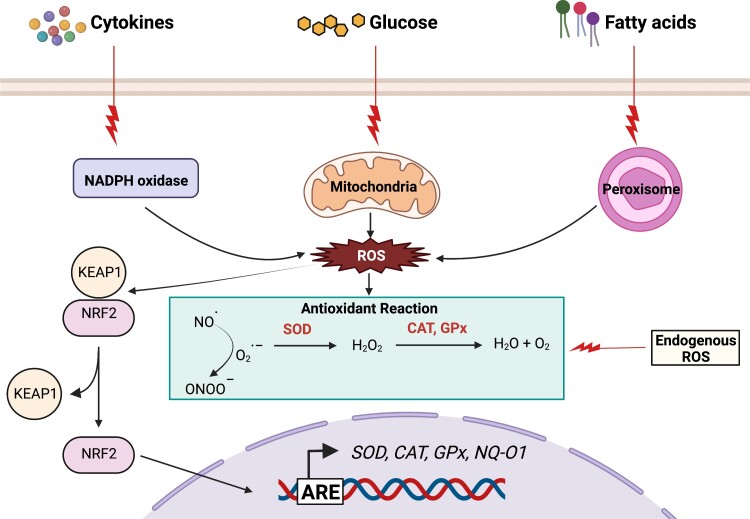
Oxidative stress and ER stress pathways are closely intertwined, as oxidative stress can lead to protein misfolding and ER stress, and ER stress can affect redox homeostasis (see Cao and Kaufman for a review) (74). Oxidative stress occurs when the accumulation of reactive oxygen species (ROS) exceeds the antioxidant scavenging capacity of the cell. Failure to resolve ROS accumulation leads to oxidative stress, damage to cellular components, and ultimately cell death (75). These highly reactive radicals are produced by mechanisms that are both endogenous and exogenous with respect to the β cells (Fig. 2) (76). Endogenous ROS is released as a byproduct of metabolic activity in mitochondria (77) and peroxisomes (78, 79). Other endogenous producers of free radicals include NADPH oxidases, nitric oxide synthases, and lipoxygenases (76, 80, 81). The exogenous factors that induce ROS generation in β cells include cytokines, glucose, and free fatty acids, as well as environmental factors such as radiation, chemicals, carcinogens, cigarette smoke, and alcohol consumption (76, 82). Hyperglycemia increases intracellular levels of advanced glycation end products, glucose oxidation, and lipid peroxidation (83), leading to ROS generation (84) and ultimately diminishing insulin release. Studies have also shown that hyperinsulinemia promotes the generation of free radicals by an NADPH-dependent mechanism (85). In addition, islets may have lower levels of antioxidant enzymes than other tissues, making them more vulnerable to oxidative damage (86, 87). Because β cells have an exceptionally high protein biosynthetic load, excessive NADPH is generated leading to elevated superoxide anion radical (O2−) production by promoting NADPH oxidase activity, rendering β cells susceptible to oxidative damage (88).
Figure 2.
Oxidative stress promotes an antioxidant response. Exposure to different stressors generates reactive oxygen species (ROS) through the NADPH oxidase system, mitochondria, peroxisomes, and through endogenous enzymatic reactions. Abnormal accumulation of ROS triggers the dissociation of the master antioxidant response transcription factor NRF2 from its inhibitor KEAP1. Free NRF2 translocates to the nucleus, binds to antioxidant response elements (ARE), and promotes expression of anti-oxidant response genes.
Oxidative stress damages various cellular targets, including nucleic acids, lipids, and proteins (89). DNA oxidation results in single-stranded breaks and formation of 8-hydroxyguanosine, a relatively stable oxidation product used to measure oxidative DNA damage (90). DNA damage is associated with cellular senescence, apoptosis, and cancer (91). Lipid oxidation via lipoxygenases and cyclooxygenases can disrupt membrane integrity and fluidity and increase cell permeability. Moreover, lipid oxidation generates more radicals that can then damage DNA and proteins (92–94). Protein oxidation can lead to alteration of enzyme activity, loss of protein function, protein aggregation, and increased immunogenicity (95–97). Thus, ROS can inflict widespread damage to cells.
At low levels, ROS can act as important second messengers (98). However, when there is an accumulation of ROS, these signaling pathways may promote diabetes pathogenesis. For example, ROS activate mammalian target of rapamycin complex 1 (mTORC1), a protein complex downstream of AKT signaling that is growth-promoting but at chronic levels of activation can also promote apoptosis (99). ROS can also activate mitogen-activated protein kinase, extracellularly regulated kinase, and JNK pathways, which promote cellular senescence and trigger apoptosis in β cells (100). Importantly, ROS can induce inflammation through nuclear factor-κB (NF-κB), thereby promoting the production of proinflammatory cytokines tumor necrosis factor-α and interleukin-6 (101, 102), suggesting an active role for ROS in the inflammatory conditions of T1D.
The cellular response to ROS is controlled by the master antioxidant transcription factor nuclear factor-erythroid factor 2-related factor 2 (NRF2) (Fig. 2). NRF2 activity is tightly regulated by Kelch-like ECH-associated protein 1 (Keap1), which binds to NRF2 in the cytosol and targets it for ubiquitin-mediated degradation. However, under oxidative stress, binding of Keap1 to NRF2 is disrupted, so NRF2 can translocate to the nucleus and activate transcription of a host of antioxidant genes (103). In mice, deletion of the NRF2 gene leads to impaired glucose tolerance and exacerbated hyperglycemia in chemically induced STZ or alloxan models of T1D (104, 105). Notably, the plasma of individuals with early T1D exhibits an increase in oxidative stress but no increase in antioxidant activity, implying a failed antioxidant response system (106). In support of this finding, induction of NRF2 in the NOD murine model of autoimmune diabetes represses the onset of hyperglycemia (107). In further support, hypomorphs or deletions of Keap1 in NOD mice activate NRF2, reduce T-cell infiltrates into the islet, and protect against hyperglycemia (107). Collectively, these studies confirm the importance of the antioxidant system in maintaining β-cell health.
Some of the antioxidant genes that NRF2 upregulates include those encoding catalase, superoxide dismutase (SOD), glutathione peroxidase (GPX), NAD(P)G: quinone oxidoreductase 1 (NQ-O1), and glutathione-S-transferase (GST) (108). These enzymes convert ROS to nontoxic metabolites, thereby protecting the cells from damage. Catalase is a peroxisomal enzyme that reduces hydrogen peroxide and helps remove islet amyloid fibrils, which are formed from misfolded islet amyloid polypeptide and impair β-cell function (109). Cat−/− mice have impaired glucose tolerance (110). SODs are superoxide scavengers located on the mitochondrial and extracellular matrix (111). Deficiency of extracellular SOD exacerbates β-cell damage on alloxan-induced diabetes (112). Similarly, deficiency of Cu, Zn superoxide dismutase (SOD1) in mice increases blood glucose, impairs glucose-stimulated insulin secretion, and downregulates PDX1 signaling (113). GPXs are located in the cytoplasm and mitochondria and reduce hydrogen peroxide and peroxide radicals (114). GPX1-deficient mice have a similar but less pronounced phenotype as SOD1-deficient mice (113). However, mouse deficiency of both SOD1 and GPX1 does not exacerbate either individual phenotype, suggesting that superoxide-mediated oxidative stress is primarily responsible for diabetes outcomes (113). NQ-O1 detoxifies quinones (115). Deficiency of NQ-O1 worsens STZ-induced β-cell death and accelerates hyperglycemia (116). Last, GSTs inactivate secondary metabolites, such as unsaturated aldehydes, epoxides, and hydroperoxides (117). In mice, deficiency of GSTP (pi isoform) does not affect fasting blood insulin or glucose levels but impairs glucose tolerance, which was reversed by inhibiting JNK (118). This suggests that the glucose intolerance with GSTP deletion is mediated through activation of the JNK pathway.
In addition to the major antioxidant enzymes, other proteins regulate oxidative stress through NRF2 signaling, including thioredoxin (119), thioredoxin interacting protein (TXNIP) (120, 121), thioredoxin reductase 1, and 12-lipoxygenase (12-LOX) (122–126). Inhibition of 12-LOX or thioredoxin reductase 1 in islets results in upregulation of the antioxidant response through NRF2 (124, 127). Also, 12-LOX plays a role both in myeloid cells (macrophages) and β cells, as tissue-specific removal of 12-LOX in either cell type in NOD mice (65, 128) protects against autoimmune diabetes. Loss of 12-LOX in β cells in a model of T2D protects against obesity-induced dysglycemia (126). Similarly, overexpression of thioredoxin in β cells of NOD mice delays the development of autoimmune diabetes (119). By contrast, loss of TXNIP (an inhibitor of thioredoxin) in mice leads to increased β-cell mass and resistance to STZ-induced diabetes development (129). Finally, it was demonstrated that TXNIP induces the production of islet amyloid polypeptide in β cells, suggesting that activity of TXNIP may promote production of islet amyloid polypeptide-induced β-cell dysfunction in T2D (130). Together, these studies suggest that identification of targets in the antioxidant response pathway would yield more therapeutic possibilities both for T1D and T2D.
Mouse and human studies suggest that oxidative stress plays a role in T1D pathogenesis. In prediabetic NOD mice, oxidative stress–induced damage is elevated in islets and vascular tissues (131). This oxidative stress in β cells is amplified by the aberrant cytokine production of infiltrating immune cells (132). In humans with T1D, markers of oxidative stress such as lipid peroxides are elevated, whereas levels of antioxidants like SOD and GPX are reduced (133). Recently, a study found that serum levels of derivatives of reactive oxygen metabolites were significantly increased in youth with T1D, consistent with an increase in systemic oxidative stress in T1D (134). Longitudinal profiling of plasma proteins during T1D progression consistently shows increased expression of oxidative stress markers and a spike in antioxidant enzymes that immediately precedes conversion to islet autoimmunity (135). In concordance, another study found that SOD antioxidant activity is elevated in the erythrocytes of children at the onset of T1D, followed by a decline in antioxidant defense over time (136). Predictably, individuals with hereditary catalase deficiency (known as acatalasemia) have higher prevalence of diabetes (137).
Taken together, the aforementioned studies implicate a critical role for oxidative species in altering protein structure and function in the β cell. Given their relatively low native antioxidant response, β cells function in a precarious environment where prolonged alterations in oxidative species or redox states can have a profound effect on function and survival.
Autophagy in Diabetes
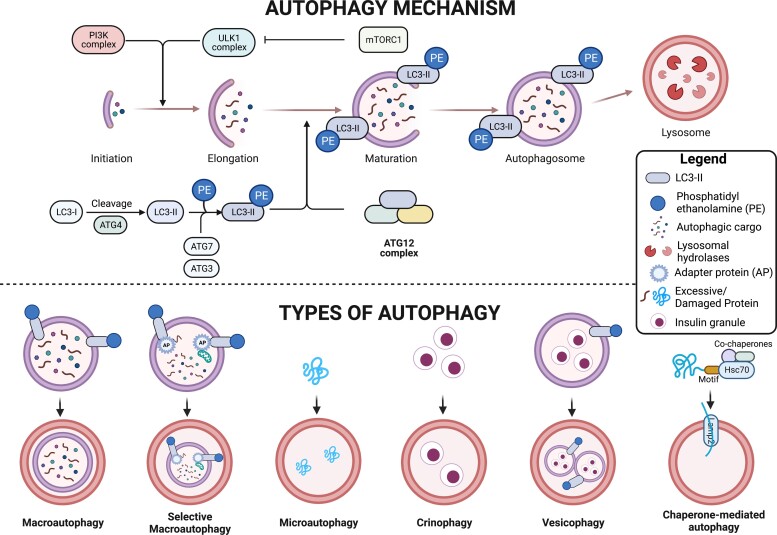
Autophagy is an evolutionarily conserved degradation and recycling process that is triggered by viral infections, ER stress, oxidative stress, viral infections, or nutrient starvation (138). Different forms of autophagy are activated under different contexts. In macroautophagy, a host of autophagy-related genes (Atg) (139) work through a series of well-orchestrated steps to sequester substrates in double-membraned vesicles called phagophores. Eventually, the microtubule-associated protein 1 light chain 3 (LC3) conjugation system (made up of LC3, ATG7, and ATG3) causes LC3 to become lipidated, which helps expand phagophores into autophagosomes. The autophagosome cargo is obtained directly from the cytoplasm or brought to the phagophore by adapter proteins such as p62 (selective autophagy). Autophagosomes then fuse with the lysosomes, and enzymes in the acidic lysosomal lumen degrade and recycle cargo into building blocks that the cell can use to regain homeostasis (140). In chaperone-mediated autophagy, proteins containing a “KFERQ” motif are targeted for degradation and chaperoned to the lysosomes where they are ultimately degraded (141). In microautophagy, proteins and organelles directly invaginate the lysosomes for degradation and recycling (142). A schematic of the autophagy process and the different types of autophagy is presented in Fig. 3.
Figure 3.
Autophagy promotes β-cell survival. Autophagy is a cellular nutrient sensing mechanism that is also activated under different stress conditions. The top panel shows the stages of autophagy and key proteins involved in the process. Phosphatidylethanolamine (PE) is a lipid moiety that is added to the protein LC3 during the autophagosome elongation and maturation stage and is used as a marker for autophagosomes. The bottom panel shows the types of autophagy and how the cargo is carried to the lysosomes for degradation. Macroautophagy: autophagosome and its cargo are degraded in lysosomes. In selective macroautophagy, cargo is tethered by adapter proteins (AP) and brought to autophagosomes for degradation in lysosomes. Microautophagy: Proteins directly fuse with lysosomes. Crinophagy: Insulin granules directly fuse with lysosomes. Vesicophagy: Insulin granules are engulfed in autophagosomes, which then fuse with lysosomes. Chaperone-mediated autophagy: Proteins are translocated to lysosomes with the help of cytosolic and lysosomal chaperones.
Many genes involved in selective autophagy contain T1D susceptibility loci (143). Thus, there has been a lot of interest in understanding the role of autophagy in T1D. Dysfunctional autophagy is detrimental to β-cell health and reduces insulin secretion (144, 145). Additionally, inhibiting autophagy results in chronic ER stress and β-cell apoptosis (146). Organelle-specific autophagy—such as mitophagy (degrades mitochondria), crinophagy (degrades insulin granules), and lipophagy (degrades lipid droplets)—also regulates β-cell function (145). The roles of different types of autophagy in β cells have been reviewed in depth (147). Here, we focus on models that have been used to study β-cell autophagy in diabetes, as well as other autophagy models that could be applied to β cells.
Molecular Mechanisms of Autophagy and Their Role in β-Cell Health and Disease
The first step in autophagosome membrane formation involves a complex of proteins, one of which is Beclin 1. Constitutively active mutant Beclin 1 mice (and therefore constitutively active autophagy) have improved insulin sensitivity but impaired insulin secretion resulting in glucose intolerance on a high-fat diet (148). In selective autophagy, the adapter proteins have LC3 interacting regions, which help tether substrates to LC3 on the autophagosomal membrane. One of these adapter proteins is p62. LC3 and p62, as autophagosome markers, are used to detect impaired autophagy when they accumulate. LC3 has been shown to accumulate in insulin-positive cells in autoantibody-positive donors (149) and both LC3 and p62 in T2D human islets (150, 151). However, deletion of the gene encoding p62 in β cells displays no overt phenotype, suggesting that p62 is dispensable during normal β-cell function (152). Global and β-cell–specific deletions of Parkin, a protein involved in mitochondrial degradation, show contrasting effects. Whereas global deletion impairs glucose tolerance (153), β-cell–specific deletion has no apparent effect on glucose tolerance and insulin secretion (154). These studies highlight the importance of focusing on β-cell–specific deletions to elucidate the functional roles for autophagy-related genes in β-cell health. Genetic models of autophagy in the context of diabetes have been previously reviewed (145). Briefly, pancreas-specific deletion of Clec16a—encoding an E3 ubiquitin ligase and T1D risk gene that functions in mitophagy (155)—impairs glucose tolerance, glucose-stimulated insulin secretion, and mitochondrial respiration. Similarly, β-cell–specific deletion of Miro1, a mitophagy-related gene whose encoded protein prevents translocation of Parkin to mitochondria, impairs insulin secretion, and disrupts mitochondrial function (156).
All types of autophagy end with lysosomal degradation. Lysosomes must maintain a low pH to activate proteolytic cathepsin enzymes. The lysosomal lumen is acidified by a large multimeric vacuolar H + ATPase complex consisting of 2 functional domains (v1 and v0), each containing about 6 to 8 subunits. In mice, β-cell–specific deletion of a gene encoding one of the subunits, Atp6ap2, leads to impaired insulin secretion and accumulation of vacuolar structures (157). Global deletion of the T1D-risk gene Ctsh, a lysosomal cysteine protease, disrupts insulin secretion and raises fasting blood glucose, further emphasizing the role of autophagy in maintaining β-cell homeostasis (158).
Although early autophagy has been studied in diabetes, there is little information on the role of lysosomes. Notably, both in T1D and T2D, there is possible dysfunction at the final clearance step by the lysosomes. Therefore, studies on lysosome biogenesis (eg, the role of lysosome transcription factor EB), acidification, and function (eg, the role of different cathepsins) will inform us on how to target this process in β cells to prevent diabetes progression. Additionally, the idea that microautophagy and chaperone-mediated autophagy maintain β-cell homeostasis has been understudied. Therefore, studies targeting each type of autophagy will reveal new targets that promote β-cell survival under stress conditions, both in T1D and T2D.
Senescence in Diabetes
When cellular stress is unmitigated, it leads to irreparable cell damage (eg, DNA damage) that forces cells to stop dividing and undergo senescence. Senescence causes cell cycle arrest, apoptosis resistance, and secretion of various senescence-associated factors that are collectively termed as senescence-associated secretory phenotype (159). The initial cell cycle exit is mediated by cyclin-dependent kinase inhibitors in the p53/p21CIPI and/or p16INKa/Rb pathways. This is followed by an increase in senescence-associated β-galactosidase (SA-βgal) activity and release of senescence-associated secretory phenotypes that potentiate tissue remodeling by recruiting immune cells (160). In β cells, senescence-associated cyclin-dependent kinase inhibitors, such as p21 and p16, are activated under stress conditions (161, 162). Mice with a transgene that facilitates the targeted deletion of cells containing p16INKa (INK-ATTAC mice) have improved β-cell function (162), suggesting that modulation of the markers of cell cycle improves β-cell function.
Recent evidence shows that some remnant β cells in NOD mice and β cells in recent onset and long-standing T1D donors have DNA damage-associated senescence features (163, 164). These studies suggest that DNA damage–associated senescence is an early feature in the pathogenesis of T1D. Senescent β cells of NOD mice exhibit increased expression of p21, p16INKa, and the cell death regulator Bcl-2 and increased SA-βgal activity (163). Notably, treatment of NOD mice with Bcl-2 inhibitors (senolytics) eliminates senescent cells and halts autoimmune destruction of β cells (163). Senolytics such as ABT263, which targets the Bcl-2 pathway, also restore β-cell identity and glucose mechanism in the context of T2D (162). To date, many studies have used p16INKa genetic mouse lines to model senescence in β cells during T2D (162). Because p21 is also elevated in senescent cells, p21 genetic models (165) could help uncover new therapeutic targets. Whether senescence is a driver or aftermath of ER stress or oxidative stress is still debatable. However, recent evidence points toward the UPR being a major contributor that controls senescence hallmarks (166), suggesting that ER stress and oxidative stress might precede a senescence phenotype.
Alternative Models to Study Endoplasmic Reticulum Stress, Oxidative Stress, Autophagy, and Senescence in Diabetes
Although mutant mouse models are ideal to study specific genes in diabetes pathogenesis, they are often time-consuming and laborious to develop. Additionally, because many stress response genes are essential housekeeping genes, many are embryonically lethal when deleted. Nevertheless, this issue can be circumvented by using conditional deletion of genes post maturity. This strategy can be especially challenging with difficult-to-breed mice, such as NOD mice. Gene delivery or knockdown techniques that use antisense oligos (167, 168), morpholinos (169, 170), and adeno-associated virus vectors (171) allow for targeted gene deletion or insertion without elaborate breeding schemes. These targeted gene delivery systems can also be combined with genetic models to further validate the role of the gene of interest.
Pharmacological inhibitors and activators provide additional insight into the role of pathways under different stress conditions. For example, the UPR and ER stress in β cells and islets have been studied by exposing cells to various chemicals (172), such as thapsigargin (SERCA inhibitor), tunicamycin (inhibits glycosylation of nascent proteins), TUDCA (chemical chaperone), and imatinib (tyrosine kinase inhibitor). Similarly, oxidative stress in β cells has been studied with diabetogenic agents, such as STZ (173) and alloxan (88). Verapamil, a calcium channel blocker that lowers the antioxidant TXNIP levels, preserves β-cell function by modulating both oxidative stress and ER stress pathways (174).
To study autophagy, chemical manipulators that target different steps of the degradation pathway have been used successfully. For example, metformin targets adenosine 5′-monophosphate-activated protein kinase (AMPK), which inhibits mTORC1 and therefore promotes autophagy. Rapamycin, torin1, and resveratrol also inhibit mTORC1 to promote autophagy (175–177). BH3 mimetics induce autophagy by disrupting the interaction between Beclin 1 (a part of the PI3K complex) and Bcl-2. On the other hand, PI3K inhibitors, such as wortmannin, 3-methyladenine, and spautin-1, inhibit autophagy initiation. Similarly, ULK and ATG4B inhibitors prevent elongation of the phagophore (175–177). Autophagy can also be inhibited by blocking adapter proteins, such as p62 and optineurin, using LC3 interacting region inhibitors. The last stage of degradation, where autophagosomes fuse to lysosomes and components are degraded in the acidic lysosomal lumen, can be blocked by chloroquine, bafilomycin A, or cystatin B (175–177).
It is difficult to replicate senescence in vitro. However, a recent study using sublethal doses of etoposide showed a DNA damage–induced senescence phenotype in β cells, thereby paving the way for more relevant studies in vitro using human islets (178). Although these pharmacological strategies do not exactly mimic physiological stress responses, they can improve our understanding of the pathways involved in β-cell homeostasis (179).
In addition to chemical modulators, zebrafish represent tractable models for monitoring stress response systems in vivo, considering the ease of handling and optical transparency. For example, ER stress (180–185), oxidative stress (186–188), inflammation (189, 190), and autophagy (191) have been extensively studied using zebrafish models.
Stress Response Pathways Converge on Activation of Inflammation
The aforementioned stress pathways are capable of damaging β cells themselves; however, studies show that they primarily mediate their effects by activating local inflammation that amplifies the damage to the cells (192–194). Inflammation is a critical immune response that clears pathogens and damaged cells, repairs tissue, and releases molecular mediators that amplify or resolve inflammation (195). The inflammatory response involves a variety of cytokines, chemokines, and proteins that directly or indirectly induce IκB kinase-β (IκKβ), JNK, or signal transducer and activator of transcription (STAT) signaling cascades. IκKβ is involved in the activation of NF-κB–mediated inflammation, wherein NF-κB upregulates proinflammatory cytokines expression (196). Similarly, JNK signaling upregulates proinflammatory and profibrotic factors (197). Activation of different STATs by cytokines also promotes inflammation (198). When inflammation is limited and appropriate, it prevents disease. However, maladaptive inflammation often triggers the development and progression of diseases, including diabetes (199). T1D is an autoimmune disorder in which the islets are inflamed by substantial numbers of infiltrating immune cells, a condition known as insulitis (200). Accumulating evidence suggests that the immune system is not only critical in T1D, but also has a substantial role in T2D pathogenesis. T2D is characterized by insulin insufficiency and insulin resistance. Among the factors contributing to T2D are oxidative stress, ER stress, glucotoxicity, lipotoxicity, and amyloid deposition in the pancreas. Interestingly, each of these factors is associated with inflammation (201–204). As a result, T2D is increasingly being viewed as an inflammatory condition, and immune system dysfunction is a focus of research in T2D pathogenesis. Because inflammation is common to both forms of diabetes, therapies that seek to dampen the stress response pathways could be critical for halting diabetes pathogenesis.
Conclusions
The UPR, antioxidant response, and autophagy are all activated by stress. Each of these stress response systems intersects with one another to promote cell survival and to maintain cellular homeostasis. However, prolonged activation redirects the cellular machinery into apoptosis and senescence (205), which both contribute to diabetes pathogenesis. In T1D and T2D, there is evidence of increased ER stress (11, 206), oxidative stress (131, 207), and autophagy (149, 151, 208). Although β cells can overcome acute ER stress, when overburdened, they fail to put the brakes on their stress mitigation systems and redirect themselves to destruction (36). Genetic mouse models have been key in identifying how these stress pathways contribute to β-cell dysfunction and death. We list these models in Table 1. Although global gene deletions provide valuable information, they do not specifically elucidate the role of the genes in β-cell health and function. Therefore, β-cell–specific genetic models and targeted chemical manipulations are critical for identifying potential therapeutic targets for T1D and T2D. Altogether, these studies highlight the need to harness strategies that reduce stress and restore β-cell homeostasis.
Table 1.
Mouse models for studying stress-related and stress-response pathways in β cells in the context of diabetes pathogenesis
| Pathway | Gene | Models | Key findings | References |
|---|---|---|---|---|
| ER stress | Grp78 | Whole body Heterozygotes; overproduction in β cells | Improved glycemic levels; resistant to high-fat diet–induced hyperinsulinemia | (209, 210) |
| Eif2ak3 | Germline deletion | Neonatal diabetes | (30, 31) | |
| Atf6 | β-cell–specific deletion | Increased β-cell apoptosis | (10) | |
| Ern1 | β-cell–specific deletion | Impaired glycemic control, decreased proinsulin folding, decreased insulin secretion | (19) | |
| Xbp1 | β-cell–specific deletion | Reduced insulin secretion and hyperglycemia | (24) | |
| Ddit3 | Whole-body deletion; β-cell–specific deletion | Improved glycemic control; increased β-cell mass; decreased ER stress; reduced production of insulin autoantibodies | (33–35, 211) | |
| ISR | Eif2ak4 | Whole-body deletion | Impaired glucose tolerance; reduced β-cell mass following high-fat diet feeding | (47) |
| Eif2s1 | Whole-body heterozygous and homozygous mutation (S51A) | Severe hypoglycemia; early lethality | (212, 213) | |
| Oxidative stress | Nfe2l2 | Whole-body deletion | Impaired glucose tolerance; hyperglycemia | (104, 105) |
| Keap1 | Whole-body deletion | Delay in T1D incidence; reduction in T-cell infiltrates into islet | (107) | |
| Sod | Whole-body deletion | Increased β-cell damage on alloxan exposure; hyperglycemia and impaired glucose-stimulated insulin secretion | (112, 113) | |
| Gpx1 | Whole-body deletion | Reduced insulin secretion | (113) | |
| Nqo1 | Whole-body deletion | Increased β-cell death and hyperglycemia | (116) | |
| Gstp1 | Whole-body deletion | Impaired glucose tolerance | (118) | |
| Autophagy | Atg7 | β-cell–specific deletion (embryonic); induced β-cell–specific deletion (adult) | Reduced β-cell mass; reduced insulin levels; impaired glucose tolerance; improved β-cell function on short-term deletion and impaired β-cell function on long-term deletion during high-fat diet | (144, 146, 214–217) |
| Rptor | β-cell–specific deletion (adult) | Reduced β-cell mass; hyperglycemia | (218) | |
| Becn1 | Mutant (F121A) | Improved insulin sensitivity with impaired insulin secretion | (148) | |
| Sqstm1 | Whole-body deletion; β-cell–specific deletion | Insulin resistance and obesity (whole-body deletion); no apparent phenotype (β-cell–specific deletion) | (152, 219) | |
| Prkn | Whole-body deletion; β-cell–specific deletion | Impaired glucose tolerance (whole-body deletion); normal glucose tolerance (β-cell–specific deletion) | (153, 154) | |
| Clec16a | Pancreas-specific deletion | Impaired glucose tolerance; impaired glucose-stimulated insulin secretion | (155) | |
| Miro1 | β-cell–specific deletion | Impaired insulin secretion and disrupted mitochondrial function | (156) | |
| Atp6ap2 | β-cell–specific deletion | Impaired insulin secretion | (157) | |
| Ctsh | β-cell–specific deletion | Impaired insulin secretion; hyperglycemia | (158) |
Abbreviations: ER, endoplasmic reticulum; T1D, type 1 diabetes.
Acknowledgments
The following search terms were used either alone or in combination on PubMed to identify references for this review: islet stress, β-cell stress, oxidative stress, integrated stress response, autophagy, diabetes, type 1 diabetes, type 2 diabetes, neoantigen, obesity, autoimmunity, insulin resistance, senescence, de-differentiation, virus, cytokines, lipotoxicity, hyperglycemia, glucolipotoxicity, free fatty acids, insulin, ER stress, and unfolded protein response.
Abbreviations
- 12-LOX
12-lipoxygenase
- ATF6
activating transcription factor 6
- CHOP
C/EBP-homologous protein
- DHPS
deoxyhypusine synthase
- ER
endoplasmic reticulum
- GCN2
general control nonderepressible 2
- GPX
glutathione peroxidase
- GST
glutathione-S-transferase
- HRI
heme-regulated inhibitor
- IRE1α
inositol requiring enzyme1α
- ISR
integrated stress response
- JNK
c-Jun N-terminal kinase
- Keap1
Kelch-like ECH-associated protein 1
- LC3
light chain 3
- mRNA
messenger RNA
- mTORC1
mammalian target of rapamycin complex 1
- NF-κB
nuclear factor-κB
- NOD
nonobese diabetic
- NQ-O1
NAD(P)G: quinone oxidoreductase 1
- NRF2
nuclear factor-erythroid factor 2-related factor 2
- PERK
PKR-like endoplasmic reticulum kinase
- PKR
protein kinase R
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- STAT
signal transducer and activator of transcription
- STZ
streptozotocin
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- tRNA
transfer RNA
- TUDCA
tauroursodeoxycholic acid
- TXNIP
thioredoxin interacting protein
- UPR
unfolded protein response
- XBP1
X-box-binding protein 1
Contributor Information
Abhishek Kulkarni, Kovler Diabetes Center and Department of Medicine, The University of Chicago, Chicago, Illinois 60637, USA.
Charanya Muralidharan, Kovler Diabetes Center and Department of Medicine, The University of Chicago, Chicago, Illinois 60637, USA.
Sarah C May, Kovler Diabetes Center and Department of Medicine, The University of Chicago, Chicago, Illinois 60637, USA.
Sarah A Tersey, Kovler Diabetes Center and Department of Medicine, The University of Chicago, Chicago, Illinois 60637, USA.
Raghavendra G Mirmira, Kovler Diabetes Center and Department of Medicine, The University of Chicago, Chicago, Illinois 60637, USA.
Financial Support
Research in the Mirmira laboratory is supported by the National Institutes of Health (grant Nos. U01 DK127786, R01 DK124906, R01 DK060581, and R01 DK105588, all to R.G.M.; and P30 DK020595 to the University of Chicago), and Diabetes Research Connection awards (to C.M. and A.K.).
Author Contributions
Conceptualization, A.K., C.M. and R.G.M.; writing—original draft preparation, C.M. and A.K.; writing—review and editing, C.M., A.K., S.C.M., S.A.T, and R.G.M.; supervision, R.G.M. All authors have read and agreed to the final version of the manuscript.
Disclosures
The authors have nothing to disclose.
Data Availability
Data sharing is not applicable to this article as no data sets were generated or analyzed during the present study.
References
- 1. Saeedi P, Petersohn I, Salpea P, et al. ; IDF Diabetes Atlas Committee . Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract. 2019;157:107843. [DOI] [PubMed] [Google Scholar]
- 2. Brooks-Worrell B, Hampe CS, Hattery EG, et al. ; GRADE Beta-cell Ancillary Study Network . Islet autoimmunity is highly prevalent and associated with diminished β-cell function in patients with type 2 diabetes in the GRADE study. Diabetes. 2022;71(6):1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. In’t Veld P. Insulitis in human type 1 diabetes: a comparison between patients and animal models. Semin Immunopathol. 2014;36(5):569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sims EK, Bahnson HT, Nyalwidhe J, et al. . Proinsulin secretion is a persistent feature of type 1 diabetes. Diabetes Care. 2019;42(2):258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campbell-Thompson M, Fu A, Kaddis JS, et al. . Insulitis and β-cell mass in the natural history of type 1 diabetes. Diabetes. 2016;65(3):719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schröder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569(1-2):29–63. [DOI] [PubMed] [Google Scholar]
- 7. Papa FR. Endoplasmic reticulum stress, pancreatic β-cell degeneration, and diabetes. Cold Spring Harb Perspect Med. 2012;2(9):a007666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marchetti P, Bugliani M, Lupi R, et al. . The endoplasmic reticulum in pancreatic beta cells of type 2 diabetes patients. Diabetologia. 2007;50(12):2486–2494. [DOI] [PubMed] [Google Scholar]
- 9. Cnop M, Toivonen S, Igoillo-Esteve M, Salpea P. Endoplasmic reticulum stress and eIF2α phosphorylation: the Achilles heel of pancreatic β cells. Mol Metab. 2017;6(9):1024–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Engin F, Yermalovich A, Nguyen T, et al. . Restoration of the unfolded protein response in pancreatic β cells protects mice against type 1 diabetes. Sci Transl Med. 2013;5(211):211ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tersey SA, Nishiki Y, Templin AT, et al. . Islet β-cell endoplasmic reticulum stress precedes the onset of type 1 diabetes in the nonobese diabetic mouse model. Diabetes. 2012;61(4):818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marhfour I, Lopez XM, Lefkaditis D, et al. . Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia. 2012;55(9):2417–2420. [DOI] [PubMed] [Google Scholar]
- 13. Shrestha N, De Franco E, Arvan P, Cnop M. Pathological β-cell endoplasmic reticulum stress in type 2 diabetes: current evidence. Front Endocrinol (Lausanne). 2021;12:650158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marre ML, McGinty JW, Chow IT, et al. . Modifying enzymes are elicited by ER stress, generating epitopes that are selectively recognized by CD4+ T cells in patients with type 1 diabetes. Diabetes. 2018;67(7):1356–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pandey VK, Mathur A, Kakkar P. Emerging role of unfolded protein response (UPR) mediated proteotoxic apoptosis in diabetes. Life Sci. 2019;216:246–258. [DOI] [PubMed] [Google Scholar]
- 16. Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5(3):a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rao RV, Bredesen DE. Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr Opin Cell Biol. 2004;16(6):653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hassler JR, Scheuner DL, Wang S, et al. . The IRE1α/XBP1s pathway is essential for the glucose response and protection of β cells. PLoS Biol. 2015;13(10):e1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsuchiya Y, Saito M, Kadokura H, et al. . IRE1-XBP1 pathway regulates oxidative proinsulin folding in pancreatic β cells. J Cell Biol. 2018;217(4):1287–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Usui M, Yamaguchi S, Tanji Y, et al. . Atf6α-null mice are glucose intolerant due to pancreatic β-cell failure on a high-fat diet but partially resistant to diet-induced insulin resistance. Metabolism. 2012;61(8):1118–1128. [DOI] [PubMed] [Google Scholar]
- 21. Zhang P, McGrath BC, Reinert J, et al. . The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22(19):6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee K, Tirasophon W, Shen X, et al. . IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16(4):452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park SM, Kang TI, So JS. Roles of XBP1s in transcriptional regulation of target genes. Biomedicines. 2021;9(7):791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee AH, Heidtman K, Hotamisligil GS, Glimcher LH. Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proc Natl Acad Sci U S A. 2011;108(21):8885–8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen J, Prywes R. Dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6. J Biol Chem. 2004;279(41):43046–43051. [DOI] [PubMed] [Google Scholar]
- 26. Ye J, Rawson RB, Komuro R, et al. . ER Stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–1364. [DOI] [PubMed] [Google Scholar]
- 27. Bommiasamy H, Back SH, Fagone P, et al. . ATF6α induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci. 2009;122(10):1626–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism12. Adv Nutr. 2012;3(3):307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feng D, Wei J, Gupta S, McGrath BC, Cavener DR. Acute ablation of PERK results in ER dysfunctions followed by reduced insulin secretion and cell proliferation. BMC Cell Biol. 2009;10(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harding HP, Zeng H, Zhang Y, et al. . Diabetes mellitus and exocrine pancreatic dysfunction in Perk–/– mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7(6):1153–1163. [DOI] [PubMed] [Google Scholar]
- 31. Gao Y, Sartori DJ, Li C, et al. . PERK is required in the adult pancreas and is essential for maintenance of glucose homeostasis. Mol Cell Biol. 2012;32(24):5129–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rozpędek W, Pytel D, Mucha B, Leszczyńska H, Diehl JA, Majsterek I. The role of the PERK/eIF2α/ATF4/CHOP signaling pathway in tumor progression during endoplasmic reticulum stress. Curr Mol Med. 2016;16(6):533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song B, Scheuner D, Ron D, Pennathur S, Kaufman RJ. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J Clin Invest. 2008;118(10):3378–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yong J, Parekh VS, Reilly SM, et al. . Chop/Ddit3 depletion in β cells alleviates ER stress and corrects hepatic steatosis in mice. Sci Transl Med. 2021;13(604):eaba9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Satoh T, Abiru N, Kobayashi M, et al. . CHOP deletion does not impact the development of diabetes but suppresses the early production of insulin autoantibody in the NOD mouse. Apoptosis. 2011;16(4):438–448. [DOI] [PubMed] [Google Scholar]
- 36. Chen CW, Guan BJ, Alzahrani MR, et al. . Adaptation to chronic ER stress enforces pancreatic β-cell plasticity. Nat Commun. 2022;13(1):4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lytton J, Westlin M, Burk SE, Shull GE, MacLennan DH. Functional comparisons between isoforms of the sarcoplasmic or endoplasmic reticulum family of calcium pumps. J Biol Chem. 1992;267(20):14483–14489. [PubMed] [Google Scholar]
- 38. Tong X, Kono T, Anderson-Baucum EK, et al. . SERCA2 deficiency impairs pancreatic β-cell function in response to diet-induced obesity. Diabetes. 2016;65(10):3039–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fonseca SG, Ishigaki S, Oslowski CM, et al. . Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest. 2010;120(3):744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Urano F. Wolfram syndrome: diagnosis, management, and treatment. Curr Diab Rep. 2016;16(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17(10):1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Costa-Mattioli M, Walter P. The integrated stress response: from mechanism to disease. Science. 2020;368(6489):eaat5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34(Pt 1):7–11. [DOI] [PubMed] [Google Scholar]
- 44. Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: their structures and functions. Cell Mol Life Sci. 2013;70(19):3493–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Aldana CRV, Wek RC, Segundo PS, Truesdell AG, Hinnebusch AG. Multicopy tRNA genes functionally suppress mutations in yeast eIF-2 alpha kinase GCN2: evidence for separate pathways coupling GCN4 expression to unchanged tRNA. Mol Cell Biol. 1994;14(12):7920–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6(2):269–279. [DOI] [PubMed] [Google Scholar]
- 47. Kanno A, Asahara S, Furubayashi A, et al. . GCN2 regulates pancreatic β cell mass by sensing intracellular amino acid levels. JCI Insight. 2020;5(9):e128820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR activation by dsRNA. J Mol Biol. 2008;381(2):351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Clemens MJ, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17(9):503–524. [DOI] [PubMed] [Google Scholar]
- 50. Vattem KM, Staschke KA, Wek RC. Mechanism of activation of the double-stranded-RNA-dependent protein kinase, PKR: role of dimerization and cellular localization in the stimulation of PKR phosphorylation of eukaryotic initiation factor-2 (eIF2). Eur J Biochem. 2001;268(13):3674–3684. [DOI] [PubMed] [Google Scholar]
- 51. Apaolaza PS, Balcacean D, Zapardiel-Gonzalo J, et al. ; nPOD-Virus Group . Islet expression of type I interferon response sensors is associated with immune infiltration and viral infection in type 1 diabetes. Sci Adv. 2021;7(9):eabd6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tang X, Uhl S, Zhang T, et al. . SARS-CoV-2 infection induces beta cell transdifferentiation. Cell Metab. 2021;33(8):1577–1591.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Udumula MP, Mangali S, Kalra J, et al. . High fructose and streptozotocin induced diabetic impairments are mitigated by indirubin-3-hydrazone via downregulation of PKR pathway in Wistar rats. Sci Rep. 2021;11(1):12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Han AP, Yu C, Lu L, et al. . Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 2001;20(23):6909–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ill-Raga G, Tajes M, Busquets-García A, et al. . Physiological control of nitric oxide in neuronal BACE1 translation by heme-regulated eIF2α kinase HRI induces synaptogenesis. Antioxid Redox Signal. 2015;22(15):1295–1307. [DOI] [PubMed] [Google Scholar]
- 56. McEwen E, Kedersha N, Song B, et al. . Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem. 2005;280(17):16925–16933. [DOI] [PubMed] [Google Scholar]
- 57. Yerlikaya A, Kimball SR, Stanley BA. Phosphorylation of eIF2alpha in response to 26S proteasome inhibition is mediated by the haem-regulated inhibitor (HRI) kinase. Biochem J. 2008;412(3):579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Palam LR, Baird TD, Wek RC. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem. 2011;286(13):10939–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu Y, Zhang Z, Li Y, Li Y. The regulation of integrated stress response signaling pathway on viral infection and viral antagonism. Front Microbiol. 2022;12:814635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. de Jong OG, Verhaar MC, Chen Y, et al. . Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012;1(1):18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jimenez L, Yu H, McKenzie AJ, et al. . Quantitative proteomic analysis of small and large extracellular vesicles (EVs) reveals enrichment of adhesion proteins in small EVs. J Proteome Res. 2019;18(3):947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Statello L, Maugeri M, Garre E, et al. . Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS One. 2018;13(4):e0195969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Aguirre RS, Kulkarni A, Becker MW, et al. . Extracellular vesicles in β cell biology: role of lipids in vesicle biogenesis, cargo, and intercellular signaling. Mol Metab. 2022;63:101545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. Elife. 2016;5:e18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Piñeros AR, Kulkarni A, Gao H, et al. . Proinflammatory signaling in islet β cells propagates invasion of pathogenic immune cells in autoimmune diabetes. Cell Rep. 2022;39(13):111011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Martin S, Zekri L, Metz A, et al. . Deficiency of G3BP1, the stress granules assembly factor, results in abnormal synaptic plasticity and calcium homeostasis in neurons. J Neurochem. 2013;125(2):175–184. [DOI] [PubMed] [Google Scholar]
- 67. Anderson-Baucum E, Piñeros AR, Kulkarni A, et al. . Deoxyhypusine synthase promotes a pro-inflammatory macrophage phenotype. Cell Metab. 2021;33(9):1883–1893.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kulkarni A, Anderson CM, Mirmira RG, Tersey SA. Role of polyamines and hypusine in β cells and diabetes pathogenesis. Metabolites. 2022;12(4):344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li CH, Ohn T, Ivanov P, Tisdale S, Anderson P. eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS One. 2010;5(4):e9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Colvin SC, Maier B, Morris DL, Tersey SA, Mirmira RG. Deoxyhypusine synthase promotes differentiation and proliferation of T helper type 1 (Th1) cells in autoimmune diabetes. J Biol Chem. 2013;288(51):36226–36235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Imam S, Prathibha R, Dar P, et al. . eIF5A inhibition influences T cell dynamics in the pancreatic microenvironment of the humanized mouse model of type 1 diabetes. Sci Rep. 2019;9(1):1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Anderson A, Kulkarni A, Mirmira RG. 259-LB: inhibition of the polyamine-hypusine circuit delimits islet ß-cell stress in the context of T1D via suppression of IRE-1a signaling. Diabetes. 2022;71(Suppl 1):259-LB. [Google Scholar]
- 73. Hiller H, Beachy DE, Lebowitz JJ, et al. . Monogenic diabetes and integrated stress response genes display altered gene expression in type 1 diabetes. Diabetes. 2021;70(8):1885–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cao SS, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid Redox Signal. 2014;21(3):396–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014;94(3):909–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pham-Huy LA, He H, Pham-Huy C. Free radicals, antioxidants in disease and health. Int J Biomed Sci. 2008;4(2):89–96. [PMC free article] [PubMed] [Google Scholar]
- 77. Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–247. [DOI] [PubMed] [Google Scholar]
- 78. Del Río LA, López-Huertas E. ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol. 2016;57(7):1364–1376. [DOI] [PubMed] [Google Scholar]
- 79. Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763(12):1755–1766. [DOI] [PubMed] [Google Scholar]
- 80. Phaniendra A, Jestadi DB, Periyasamy L. Free radicals: properties, sources, targets, and their implication in various diseases. Indian J Clin Biochem. 2015;30(1):11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Holmström KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15(6):411–421. [DOI] [PubMed] [Google Scholar]
- 82. Lobo V, Patil A, Phatak A, Chandra N. Free radicals, antioxidants and functional foods: impact on human health. Pharmacogn Rev. 2010;4(8):118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Maritim AC, Sanders RA, Watkins JB. Diabetes, oxidative stress, and antioxidants: a review. J Biochem Mol Toxicol. 2003;17(1):24–38. [DOI] [PubMed] [Google Scholar]
- 84. Di Naso FC, Simões Dias A, Porawski M, Marroni NAP. Exogenous superoxide dismutase: action on liver oxidative stress in animals with streptozotocin-induced diabetes. Exp Diabetes Res. 2011;2011:754132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ceolotto G, Bevilacqua M, Papparella I, et al. . Insulin generates free radicals by an NAD(P)H, phosphatidylinositol 3′-kinase-dependent mechanism in human skin fibroblasts ex vivo. Diabetes. 2004;53(5):1344–1351. [DOI] [PubMed] [Google Scholar]
- 86. Delmastro MM, Piganelli JD. Oxidative stress and redox modulation potential in type 1 diabetes. Clin Dev Immunol. 2011;2011:593863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lenzen S, Drinkgern J, Tiedge M. Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radic Biol Med. 1996;20(3):463–466. [DOI] [PubMed] [Google Scholar]
- 88. Lenzen S. The mechanisms of alloxan- and streptozotocin-induced diabetes. Diabetologia. 2008;51(2):216–226. [DOI] [PubMed] [Google Scholar]
- 89. Bokov A, Chaudhuri A, Richardson A. The role of oxidative damage and stress in aging. Mech Ageing Dev. 2004;125(10-11):811–826. [DOI] [PubMed] [Google Scholar]
- 90. Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-T and A-C substitutions. J Biol Chem. 1992;267(1):166–172. [PubMed] [Google Scholar]
- 91. Wyld L, Bellantuono I, Tchkonia T, et al. . Senescence and cancer: a review of clinical implications of senescence and senotherapies. Cancers (Basel). 2020;12(8):2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kwiecien S, Jasnos K, Magierowski M, et al. . Lipid peroxidation, reactive oxygen species and antioxidative factors in the pathogenesis of gastric mucosal lesions and mechanism of protection against oxidative stress-induced gastric injury. J Physiol Pharmacol. 2014;65(5):613–622. [PubMed] [Google Scholar]
- 93. Landar A, Zmijewski JW, Dickinson DA, et al. . Interaction of electrophilic lipid oxidation products with mitochondria in endothelial cells and formation of reactive oxygen species. Am J Physiol Heart Circ Physiol. 2006;290(5):H1777–H1787. [DOI] [PubMed] [Google Scholar]
- 94. Winczura A, Zdżalik D, Tudek B. Damage of DNA and proteins by major lipid peroxidation products in genome stability. Free Radic Res. 2012;46(4):442–459. [DOI] [PubMed] [Google Scholar]
- 95. Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272(33):20313–20316. [DOI] [PubMed] [Google Scholar]
- 96. Celi P, Gabai G. Oxidant/antioxidant balance in animal nutrition and health: the role of protein oxidation. Front Vet Sci. 2015;2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wall SB, Oh JY, Diers AR, Landar A. Oxidative modification of proteins: an emerging mechanism of cell signaling. Front Physiol. 2012;3:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pizzino G, Irrera N, Cucinotta M, et al. . Oxidative stress: harms and benefits for human health. Oxid Med Cell Longev. 2017;2017:8416763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Krakauer T. mTORC1 activation, and metabolic derangement contribute to the susceptibility of diabetics to infections. Med Hypotheses. 2015;85(6):997–1001. [DOI] [PubMed] [Google Scholar]
- 100. Son Y, Cheong YK, Kim NH, Chung HT, Kang DG, Pae HO. Mitogen-activated protein kinases and reactive oxygen species: how can ROS activate MAPK pathways? J Signal Transduct. 2011;2011:792639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chelombitko MA. Role of reactive oxygen species in inflammation: a minireview. Moscow Univ Biol Sci Bull. 2018;73(4):199–202. [Google Scholar]
- 102. Mittal M, Siddiqui MR, Tran K, Reddy SP, Malik AB. Reactive oxygen species in inflammation and tissue injury. Antioxid Redox Signal. 2014;20(7):1126–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284(20):13291–13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Aleksunes LM, Reisman SA, Yeager RL, Goedken MJ, Klaassen CD. Nuclear factor erythroid 2-related factor 2 deletion impairs glucose tolerance and exacerbates hyperglycemia in type 1 diabetic mice. J Pharmacol Exp Ther. 2010;333(1):140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lou Y, Kong M, Li L, et al. . Inhibition of the Keap1/Nrf2 signaling pathway significantly promotes the progression of type 1 diabetes mellitus. Oxid Med Cell Longev. 2021;2021:7866720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Marra G, Cotroneo P, Pitocco D, et al. . Early increase of oxidative stress and reduced antioxidant defenses in patients with uncomplicated type 1 diabetes: a case for gender difference. Diabetes Care. 2002;25(2):370–375. [DOI] [PubMed] [Google Scholar]
- 107. Yagishita Y, Uruno A, Chartoumpekis DV, Kensler TW, Yamamoto M. Nrf2 represses the onset of type 1 diabetes in non-obese diabetic mice. J Endocrinol 2019; 240(3):403–416. doi: 10.1530/JOE-18-0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. Oxidative stress and antioxidant defense. World Allergy Organ J. 2012;5(1):9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Glorieux C, Calderon PB. Catalase, a remarkable enzyme: targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol Chem. 2017;398(10):1095–1108. [DOI] [PubMed] [Google Scholar]
- 110. Heit C, Marshall S, Singh S, et al. . Catalase deletion promotes prediabetic phenotype in mice. Free Radic Biol Med. 2017;103:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Fukai T, Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid Redox Signal. 2011;15(6):1583–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sentman ML, Jonsson LM, Marklund SL. Enhanced alloxan-induced beta-cell damage and delayed recovery from hyperglycemia in mice lacking extracellular-superoxide dismutase. Free Radic Biol Med. 1999;27(7-8):790–796. [DOI] [PubMed] [Google Scholar]
- 113. Wang X, Vatamaniuk MZ, Roneker CA, et al. . Knockouts of SOD1 and GPX1 exert different impacts on murine islet function and pancreatic integrity. Antioxid Redox Signal. 2011;14(3):391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lubos E, Loscalzo J, Handy DE. Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2011;15(7):1957–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ross D, Siegel D. The diverse functionality of NQO1 and its roles in redox control. Redox Biol. 2021;41:101950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yeo SH, Noh JR, Kim YH, et al. . Increased vulnerability to β-cell destruction and diabetes in mice lacking NAD(P)H:quinone oxidoreductase 1. Toxicol Lett. 2013;219(1):35–41. [DOI] [PubMed] [Google Scholar]
- 117. Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45(1):51–88. [DOI] [PubMed] [Google Scholar]
- 118. Dastidar S G, Jagatheesan G, Haberzettl P, et al. . Glutathione S-transferase P deficiency induces glucose intolerance via JNK-dependent enhancement of hepatic gluconeogenesis. Am J Physiol Endocrinol Metab. 2018;315(5):E1005–E1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Hotta M, Tashiro F, Ikegami H, et al. . Pancreatic beta cell-specific expression of thioredoxin, an antioxidative and antiapoptotic protein, prevents autoimmune and streptozotocin-induced diabetes. J Exp Med. 1998;188(8):1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lerner AG, Upton JP, Praveen PVK, et al. . IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012;16(2):250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Chen J, Saxena G, Mungrue IN, Lusis AJ, Shalev A. Thioredoxin-interacting protein: a critical link between glucose toxicity and beta-cell apoptosis. Diabetes. 2008;57(4):938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Conteh AM, Reissaus CA, Hernandez-Perez M, et al. . Platelet-type 12-lipoxygenase deletion provokes a compensatory 12/15-lipoxygenase increase that exacerbates oxidative stress in mouse islet β cells. J Biol Chem. 2019;294(16):6612–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hernandez-Perez M, Kulkarni A, Samala N, et al. . A 12-lipoxygenase-Gpr31 signaling axis is required for pancreatic organogenesis in the zebrafish. FASEB J. 2020;34(11):14850–14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hernandez-Perez M, Chopra G, Fine J, et al. . Inhibition of 12/15-lipoxygenase protects against β-cell oxidative stress and glycemic deterioration in mouse models of type 1 diabetes. Diabetes. 2017;66(11):2875–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kulkarni A, Nadler JL, Mirmira RG, Casimiro I. Regulation of tissue inflammation by 12-lipoxygenases. Biomolecules. 2021;11(5):717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tersey SA, Maier B, Nishiki Y, Maganti AV, Nadler JL, Mirmira RG. 12-Lipoxygenase promotes obesity-induced oxidative stress in pancreatic islets. Mol Cell Biol. 2014;34(19):3735–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Stancill JS, Hansen PA, Mathison AJ, Schmidt EE, Corbett JA. Deletion of thioredoxin reductase disrupts redox homeostasis and impairs β-cell function. Function (Oxf). 2022;3(4):zqac034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kulkarni A, Pineros AR, Walsh MA, et al. . 12-Lipoxygenase governs the innate immune pathogenesis of islet inflammation and autoimmune diabetes. JCI Insight. 2021;6(14):147812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Masson E, Koren S, Razik F, et al. . High beta-cell mass prevents streptozotocin-induced diabetes in thioredoxin-interacting protein-deficient mice. Am J Physiol Endocrinol Metab. 2009;296(6):E1251–E1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Jing G, Westwell-Roper C, Chen J, Xu G, Verchere CB, Shalev A. Thioredoxin-interacting protein promotes islet amyloid polypeptide expression through miR-124a and FoxA2. J Biol Chem. 2014;289(17):11807–11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Haskins K, Bradley B, Powers K, et al. . Oxidative stress in type 1 diabetes. Ann N Y Acad Sci. 2003;1005(1):43–54. [DOI] [PubMed] [Google Scholar]
- 132. Chou ST, Tseng ST. Oxidative stress markers in type 2 diabetes patients with diabetic nephropathy. Clin Exp Nephrol. 2017;21(2):283–292. [DOI] [PubMed] [Google Scholar]
- 133. Francescato MP, Stel G, Geat M, Cauci S. Oxidative stress in patients with type 1 diabetes mellitus: is it affected by a single bout of prolonged exercise? PLoS One. 2014;9(6):e99062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Morandi A, Corradi M, Orsi S, et al. . Oxidative stress in youth with type 1 diabetes: not only a matter of gender, age, and glycemic control. Diabetes Res Clin Pract. 2021;179:109007. [DOI] [PubMed] [Google Scholar]
- 135. Liu CW, Bramer L, Webb-Robertson BJ, Waugh K, Rewers MJ, Zhang Q. Temporal expression profiling of plasma proteins reveals oxidative stress in early stages of type 1 diabetes progression. J Proteomics. 2018;172:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Domínguez C, Ruiz E, Gussinye M, Carrascosa A. Oxidative stress at onset and in early stages of type 1 diabetes in children and adolescents. Diabetes Care. 1998;21(10):1736–1742. [DOI] [PubMed] [Google Scholar]
- 137. Góth L, Eaton JW. Hereditary catalase deficiencies and increased risk of diabetes. Lancet 2000;356(9244):1820-1821. [DOI] [PubMed] [Google Scholar]
- 138. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43(1):67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hansen M, Rubinsztein DC, Walker DW. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol. 2018;19(9):579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. [DOI] [PubMed] [Google Scholar]
- 141. Cuervo AM. Chaperone-mediated autophagy: Dice's “Wild” idea about lysosomal selectivity. Nat Rev Mol Cell Biol. 2011;12(8):535–541. [DOI] [PubMed] [Google Scholar]
- 142. Li W, Li J, Bao J. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69(7):1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ramos-Rodríguez M, Pérez-González B, Pasquali L. The β-cell genomic landscape in T1D: implications for disease pathogenesis. Curr Diab Rep. 2021;21(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Jung HS, Chung KW, Won Kim J, et al. . Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8(4):318–324. [DOI] [PubMed] [Google Scholar]
- 145. Pearson GL, Gingerich MA, Walker EM, Biden TJ, Soleimanpour SA. A selective look at autophagy in pancreatic β-cells. Diabetes. 2021;70(6):1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bartolome A, Guillen C, Benito M. Autophagy plays a protective role in endoplasmic reticulum stress-mediated pancreatic β cell death. Autophagy. 2012;8(12):1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Muralidharan C, Linnemann AK. β-Cell autophagy in the pathogenesis of type 1 diabetes. Am J Physiol Endocrinol Metab. 2021;321(3):E410–E416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Yamamoto S, Kuramoto K, Wang N, et al. . Autophagy differentially regulates insulin production and insulin sensitivity. Cell Rep. 2018;23(11):3286–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Muralidharan C, Conteh AM, Marasco MR, et al. . Pancreatic beta cell autophagy is impaired in type 1 diabetes. Diabetologia. 2021;64(4):865–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Abe H, Uchida T, Hara A, et al. . Exendin-4 improves β-cell function in autophagy-deficient β-cells. Endocrinology. 2013;154(12):4512–4524. [DOI] [PubMed] [Google Scholar]
- 151. Masini M, Bugliani M, Lupi R, et al. . Autophagy in human type 2 diabetes pancreatic beta cells. Diabetologia. 2009;52(6):1083–1086. [DOI] [PubMed] [Google Scholar]
- 152. Honda A, Komiya K, Hara A, et al. . Normal pancreatic β-cell function in mice with RIP-Cre-mediated inactivation of p62/SQSTM1. Endocr J. 2018;65(1):83–89. [DOI] [PubMed] [Google Scholar]
- 153. Hoshino A, Ariyoshi M, Okawa Y, et al. . Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic β-cell function in diabetes. Proc Natl Acad Sci U S A. 2014;111(8):3116–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Corsa CAS, Pearson GL, Renberg A, et al. . The E3 ubiquitin ligase parkin is dispensable for metabolic homeostasis in murine pancreatic β cells and adipocytes. J Biol Chem. 2019;294(18):7296–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Soleimanpour SA, Gupta A, Bakay M, et al. . The diabetes susceptibility gene Clec16a regulates mitophagy. Cell. 2014;157(7):1577–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Chen L, Liu C, Gao J, et al. . Inhibition of Miro1 disturbs mitophagy and pancreatic β-cell function interfering insulin release via IRS-Akt-Foxo1 in diabetes. Oncotarget. 2017;8(53):90693–90705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Binger KJ, Neukam M, Tattikota SG, et al. . Atp6ap2 deletion causes extensive vacuolation that consumes the insulin content of pancreatic β cells. Proc Natl Acad Sci U S A. 2019;116(40):19983–19988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Fløyel T, Brorsson C, Nielsen LB, et al. . CTSH regulates β-cell function and disease progression in newly diagnosed type 1 diabetes patients. Proc Natl Acad Sci U S A. 2014;111(28):10305–10310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Herranz N, Gil J. Mechanisms and functions of cellular senescence. J Clin Invest. 2018;128(4):1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Muñoz-Espín D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15(7):482–496. [DOI] [PubMed] [Google Scholar]
- 161. Brawerman G, Thompson PJ. Beta cell therapies for preventing type 1 diabetes: from bench to bedside. Biomolecules. 2020;10(12):E1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Aguayo-Mazzucato C, Andle J, Lee TB, et al. . Acceleration of β cell aging determines diabetes and senolysis improves disease outcomes. Cell Metab. 2019;30(1):129–142.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Thompson PJ, Shah A, Ntranos V, Van Gool F, Atkinson M, Bhushan A. Targeted elimination of senescent beta cells prevents type 1 diabetes. Cell Metab. 2019;29(5):1045–1060.e10. [DOI] [PubMed] [Google Scholar]
- 164. Horwitz E, Krogvold L, Zhitomirsky S, et al. . β-Cell DNA damage response promotes islet inflammation in type 1 diabetes. Diabetes. 2018;67(11):2305–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Wang B, Wang L, Gasek NS, et al. . An inducible p21-Cre mouse model to monitor and manipulate p21-highly-expressing senescent cells in vivo. Nat Aging. 2021;1(10):962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Abbadie C, Pluquet O. Unfolded protein response (UPR) controls major senescence hallmarks. Trends Biochem Sci. 2020;45(5):371–374. [DOI] [PubMed] [Google Scholar]
- 167. Ämmälä C, Drury WJ, Knerr L, et al. . Targeted delivery of antisense oligonucleotides to pancreatic β-cells. Sci Adv. 2018;4(10):eaat3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Knerr L, Prakash TP, Lee R, et al. . Glucagon like peptide 1 receptor agonists for targeted delivery of antisense oligonucleotides to pancreatic beta cell. J Am Chem Soc. 2021;143(9):3416–3429. [DOI] [PubMed] [Google Scholar]
- 169. Moulton JD. Using morpholinos to control gene expression. Curr Protoc Nucleic Acid Chem. 2006;27(1):4301–43024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Chen S, Sbuh N, Veedu RN. Antisense oligonucleotides as potential therapeutics for type 2 diabetes. Nucleic Acid Ther. 2021;31(1):39–57. [DOI] [PubMed] [Google Scholar]
- 171. Rehman KK, Trucco M, Wang Z, Xiao X, Robbins PD. AAV8-mediated gene transfer of interleukin-4 to endogenous β-cells prevents the onset of diabetes in NOD mice. Mol Ther. 2008;16(8):1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Sahin GS, Lee H, Engin F. An accomplice more than a mere victim: the impact of β-cell ER stress on type 1 diabetes pathogenesis. Mol Metab. 2021;54:101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Pandey S, Dvorakova MC. Future perspective of diabetic animal models. Endocr Metab Immune Disord Drug Targets. 2020;20(1):25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Ovalle F, Grimes T, Xu G, et al. . Verapamil and beta cell function in adults with recent-onset type 1 diabetes. Nat Med. 2018;24(8):1108–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11(9):709–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Towers CG, Thorburn A. Therapeutic targeting of autophagy. EBioMedicine. 2016;14:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov. 2017;16(7):487–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Brawerman G, Pipella J, Thompson PJ. DNA damage to β cells in culture recapitulates features of senescent β cells that accumulate in type 1 diabetes. Mol Metab. 2022;62:101524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Bergmann TJ, Fregno I, Fumagalli F, et al. . Chemical stresses fail to mimic the unfolded protein response resulting from luminal load with unfolded polypeptides. J Biol Chem. 2018;293(15):5600–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Seth A, Stemple DL, Barroso I. The emerging use of zebrafish to model metabolic disease. Dis Model Mech. 2013;6(5):1080–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Benchoula K, Khatib A, Jaffar A, et al. . The promise of zebrafish as a model of metabolic syndrome. Exp Anim. 2019;68(4):407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Kulkarni A, Ibrahim S, Haider I, et al. . A novel 2-hit zebrafish model to study early pathogenesis of non-alcoholic fatty liver disease. Biomedicines. 2022;10(2):479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Schlegel A, Gut P. Metabolic insights from zebrafish genetics, physiology, and chemical biology. Cell Mol Life Sci. 2015;72(12):2249–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Zang L, Maddison LA, Chen W. Zebrafish as a model for obesity and diabetes. Front Cell Dev Biol. 2018;6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Zang L, Shimada Y, Nishimura N. Development of a novel zebrafish model for type 2 diabetes mellitus. Sci Rep. 2017;7(1):1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Kulkarni AA, Conteh AM, Sorrell CA, et al. . An in vivo zebrafish model for interrogating ROS-mediated pancreatic β-cell injury, response, and prevention. Oxidative Med Cell Longevity. 2018;2018:e1324739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Willcox A, Richardson SJ, Bone AJ, Foulis AK, Morgan NG. Analysis of islet inflammation in human type 1 diabetes. Clin Exp Immunol. 2009;155(2):173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Ehses JA, Perren A, Eppler E, et al. . Increased number of islet-associated macrophages in type 2 diabetes. Diabetes. 2007;56(9):2356–2370. [DOI] [PubMed] [Google Scholar]
- 189. Ibrahim S, Harris-Kawano A, Haider I, Mirmira RG, Sims EK, Anderson RM. A novel Cre-enabled tetracycline-inducible transgenic system for tissue-specific cytokine expression in the zebrafish: CETI-PIC3. Dis Model Mech. 2020;13(6):dmm042556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Kulkarni A, Anderson R, Mirmira RG. A zebrafish tailfin injury assay protocol for quantifying immune cell migration and infiltration. STAR Protoc. 2022;3(1):101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Varga M, Fodor E, Vellai T. Autophagy in zebrafish. Methods. 2015;75:172–180. [DOI] [PubMed] [Google Scholar]
- 192. Hasnain SZ. Endoplasmic reticulum and oxidative stress in immunopathology: understanding the crosstalk between cellular stress and inflammation. Clin Transl Immunology. 2018;7(7):e1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Maamoun H, Benameur T, Pintus G, Munusamy S, Agouni A. Crosstalk between oxidative stress and endoplasmic reticulum (ER) stress in endothelial dysfunction and aberrant angiogenesis associated with diabetes: a focus on the protective roles of heme oxygenase (HO)-1. Front Physiol. 2019;10:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Zhang K. Integration of ER stress, oxidative stress and the inflammatory response in health and disease. Int J Clin Exp Med. 2010;3(1):33–40. [PMC free article] [PubMed] [Google Scholar]
- 195. Chen L, Deng H, Cui H, et al. . Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2018;9(6):7204–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Yung JHM, Giacca A. Role of c-Jun N-terminal kinase (JNK) in obesity and type 2 diabetes. Cells. 2020;9(3):E706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Banerjee S, Biehl A, Gadina M, Hasni S, Schwartz DM. JAK-STAT signaling as a target for inflammatory and autoimmune diseases: current and future prospects. Drugs. 2017;77(5):521–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Bennett JM, Reeves G, Billman GE, Sturmberg JP. Inflammation—nature's way to efficiently respond to all types of challenges: implications for understanding and managing “the epidemic” of chronic diseases. Front Med (Lausanne). 2018;5:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Tsalamandris S, Antonopoulos AS, Oikonomou E, et al. . The role of inflammation in diabetes: current concepts and future perspectives. Eur Cardiol. 2019;14(1):50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Donath MY, Størling J, Maedler K, Mandrup-Poulsen T. Inflammatory mediators and islet beta-cell failure: a link between type 1 and type 2 diabetes. J Mol Med (Berl). 2003;81(8):455–470. [DOI] [PubMed] [Google Scholar]
- 202. Ehses JA, Ellingsgaard H, Böni-Schnetzler M, Donath MY. Pancreatic islet inflammation in type 2 diabetes: from alpha and beta cell compensation to dysfunction. Arch Physiol Biochem. 2009;115(4):240–247. [DOI] [PubMed] [Google Scholar]
- 203. Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol. 2008;8(12):923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Masters SL, Dunne A, Subramanian SL, et al. . Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat Immunol. 2010;11(10):897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Nam HY, Han MW, Chang HW, Kim SY, Kim SW. Prolonged autophagy by MTOR inhibitor leads radioresistant cancer cells into senescence. Autophagy. 2013;9(10):1631–1632. [DOI] [PubMed] [Google Scholar]
- 206. Lee JH, Lee J. Endoplasmic reticulum (ER) stress and its role in pancreatic β-cell dysfunction and senescence in type 2 diabetes. Int J Mol Sci. 2022;23(9):4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Ježek P, Jabůrek M, Plecitá-Hlavatá L. Contribution of oxidative stress and impaired biogenesis of pancreatic β-cells to type 2 diabetes. Antioxid Redox Signal. 2019;31(10):722–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Bugliani M, Mossuto S, Grano F, et al. . Modulation of autophagy influences the function and survival of human pancreatic beta cells under endoplasmic reticulum stress conditions and in type 2 diabetes. Front Endocrinol (Lausanne). 2019;10:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Teodoro-Morrison T, Schuiki I, Zhang L, Belsham DD, Volchuk A. GRP78 overproduction in pancreatic beta cells protects against high-fat-diet-induced diabetes in mice. Diabetologia. 2013;56(5):1057–1067. [DOI] [PubMed] [Google Scholar]
- 210. Ye R, Jung DY, Jun JY, et al. . Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010;59(1):6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Maris M, Overbergh L, Gysemans C, et al. . Deletion of C/EBP homologous protein (Chop) in C57Bl/6 mice dissociates obesity from insulin resistance. Diabetologia. 2012;55(4):1167–1178. [DOI] [PubMed] [Google Scholar]
- 212. Scheuner D, Vander Mierde D, Song B, et al. . Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11(7):757–764. [DOI] [PubMed] [Google Scholar]
- 213. Scheuner D, Song B, McEwen E, et al. . Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001;7(6):1165–1176. [DOI] [PubMed] [Google Scholar]
- 214. Chen C, Ricks D, Gittes GK, et al. . Autophagy protects pancreatic beta cell mass and function in the setting of a high-fat and high-glucose diet. Sci Rep. 2017;7(1):16348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Chu KY, Mellet N, Thai LM, Meikle PJ, Biden TJ. Short-term inhibition of autophagy benefits pancreatic β-cells by augmenting ether lipids and peroxisomal function, and by countering depletion of n-3 polyunsaturated fatty acids after fat-feeding. Mol Metab. 2020;40:101023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Ebato C, Uchida T, Arakawa M, et al. . Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8(4):325–332. [DOI] [PubMed] [Google Scholar]
- 217. Quan W, Hur KY, Lim Y, et al. . Autophagy deficiency in beta cells leads to compromised unfolded protein response and progression from obesity to diabetes in mice. Diabetologia. 2012;55(2):392–403. [DOI] [PubMed] [Google Scholar]
- 218. Blandino-Rosano M, Barbaresso R, Jimenez-Palomares M, et al. . Loss of mTORC1 signalling impairs β-cell homeostasis and insulin processing. Nat Commun. 2017;8(1):16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Rodriguez A, Durán A, Selloum M, et al. . Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3(3):211–222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no data sets were generated or analyzed during the present study.