Abstract
The human Na+/H+ antiporter NHA2 (SLC9B2) transports Na+ or Li+ across the plasma membrane in exchange for protons, and is implicated in various pathologies. It is a 537 amino acids protein with an 82 residues long hydrophilic cytoplasmic N‐terminus followed by a transmembrane part comprising 14 transmembrane helices. We optimized the functional expression of HsNHA2 in the plasma membrane of a salt‐sensitive Saccharomyces cerevisiae strain and characterized in vivo a set of mutated or truncated versions of HsNHA2 in terms of their substrate specificity, transport activity, localization, and protein stability. We identified a highly conserved proline 246, located in the core of the protein, as being crucial for ion selectivity. The replacement of P246 with serine or threonine resulted in antiporters with altered substrate specificity that were not only highly active at acidic pH 4.0 (like the native antiporter), but also at neutral pH. P246T/S versions also exhibited increased resistance to the HsNHA2‐specific inhibitor phloretin. We experimentally proved that a putative salt bridge between E215 and R432 is important for antiporter function, but also structural integrity. Truncations of the first 50–70 residues of the N‐terminus doubled the transport activity of HsNHA2, while changes in the charge at positions E47, E56, K57, or K58 decreased the antiporter's transport activity. Thus, the hydrophilic N‐terminal part of the protein appears to allosterically auto‐inhibit cation transport of HsNHA2. Our data also show this in vivo approach to be useful for a rapid screening of SNP's effect on HsNHA2 activity.
Keywords: human NHA2, Na+/H+ antiporter, N‐terminal auto‐inhibition, phloretin, yeast
1. INTRODUCTION
The maintenance of ion homeostasis is crucial for any living cell, as it influences various physiological parameters such as cell size, intracellular pH, and membrane potential. Cation/H+ antiporters (CPAs, SLC9 family) are tightly regulated transporters that ensure appropriate intracellular concentrations of monovalent cations in organisms of all kingdoms, from bacteria to mammals. 1 They mediate the exchange of monovalent cations, mainly Na+ and K+, for one or two protons across the membrane. Human cation/H+ antiporters account for 13 isoforms from three subfamilies—NHE (SLC9A), which includes 9 proteins (NHE1‐9), NHA (SLC9B) with two members, NHA1 and NHA2, and the SLC9C subfamily with two proteins (SLC9C1 and SLC9C2) of unknown functions. 2 , 3 Deficiencies in the functions of CPAs are related to a growing list of pathologies, ranging from hypertension to autism spectrum disorder and cancer, which demonstrates their importance for human health. 3
In this work we focus on a member of the SLC9B subfamily encoded by the NHA2 gene. It is a Na+(Li+)/H+ antiporter, which is expressed in multiple tissues, 4 but its physiological functions are relatively poorly defined. Within individual cells, it was shown to be localized to the plasma membrane or intracellularly to endosomes or mitochondria. 4 , 5 It generally contributes to the regulation of intracellular pH, sodium homeostasis, and cellular volume, and it was found to play crucial roles in various physiological processes depending on the specific tissue where it resides.
All knowledge obtained so far indicates that NHA2 performs multiple functions in the human body and its malfunctioning leads to various pathologies ranging from metabolic to fertility disorders. Moreover, the NHA2 gene is located in a human chromosomal region (4q24) which has been associated with hypertension in numerous linkage studies. 6 So far, particular physiological roles of NHA2 have been studied in a range of organisms and tissues. Single RNAi knockdowns of NHA2 or its homolog NHA1 in Drosophila melanogaster reduced survival, and their combination was lethal, suggesting their essentiality for life. 7 In pancreatic β‐cells of mice and humans, NHA2 resides in endosomes and is critical for insulin secretion and clathrin‐mediated endocytosis. 8 The loss of NHA2 also exacerbates obesity‐ and aging‐induced glucose intolerance in mice, 9 and a single‐nucleotide polymorphism of the NHA2 locus was recently associated with type 2 diabetes in humans. 10 In the kidney, NHA2 localizes to distal convoluted tubules, where it is critical for electrolyte (sodium reabsorption) and blood pressure homeostasis. 4 , 11 In 2021, Anderegg et al. corroborated the NHA2 role in blood pressure regulation in mice, and identified it as critical for the maintenance of serine–threonine with‐no‐lysine kinase 4 (WNK4) levels, and thus ultimately for the activity of the Na+/Cl− co‐transporter NCC in the distal convoluted tubules of the kidney. 12 Increased NHA2 expression also promotes cyst development in an in vitro model of polycystic kidney disease. 13 In the testis, NHA2 is relevant for sperm motility and male fertility, since the deletion of NHA2 significantly reduced the percentage of motile sperm (of about 30%) and resulted in a decrease in pregnancy rate (of about 25%) in mice. 14 In addition, several studies showed NHA2 to be expressed in osteoclasts, where NHA2 depletion significantly inhibited osteoclast differentiation in vitro 5 , 15 , 16 ; however, its physiological role in vivo in this type of cell remains to be elucidated. 17 , 18
As for its structure, human NHA2 is a protein 537 amino acids in length and 58 kDa in size. It consists of 14 transmembrane segments (TMS), a hydrophilic N‐terminus approximately 82 amino acids long, and a relatively short (27 aa) hydrophilic C‐terminus. A two‐dimensional representation of HsNHA2 topology is schematically shown in Figure 1a, and in detail in Figure S1. Recently, 3D‐structures of human NHA2 and its bison homolog were determined using cryo‐electron microscopy (Protein Data Bank 7B4M and 7B4L, 23 respectively). However, the hydrophilic N‐terminal and C‐terminal parts of NHA2 were not resolved in both structures. The N‐ and C‐termini are predicted to be unstructured, for example by the recently published AI‐based protein structure prediction algorithm AlphaFold2. 24 The importance of the highly unstructured hydrophilic N‐terminal part for the functioning of NHA2 has not been studied yet. In general, methods such as x‐ray crystallography and cryo‐electron microscopy are limited when it comes to extremely mobile and unstructured regions of proteins. In the case of CPAs, many mammalian antiporters possess extramembrane regions mostly in their N‐ or C‐termini, which are predicted to be unstructured. These are often involved in regulatory mechanisms and interaction with other proteins. 25 , 26 , 27 , 28 Indeed, despite the growing number of CPAs structures our understanding of the function of these regulatory regions remains limited.
FIGURE 1.
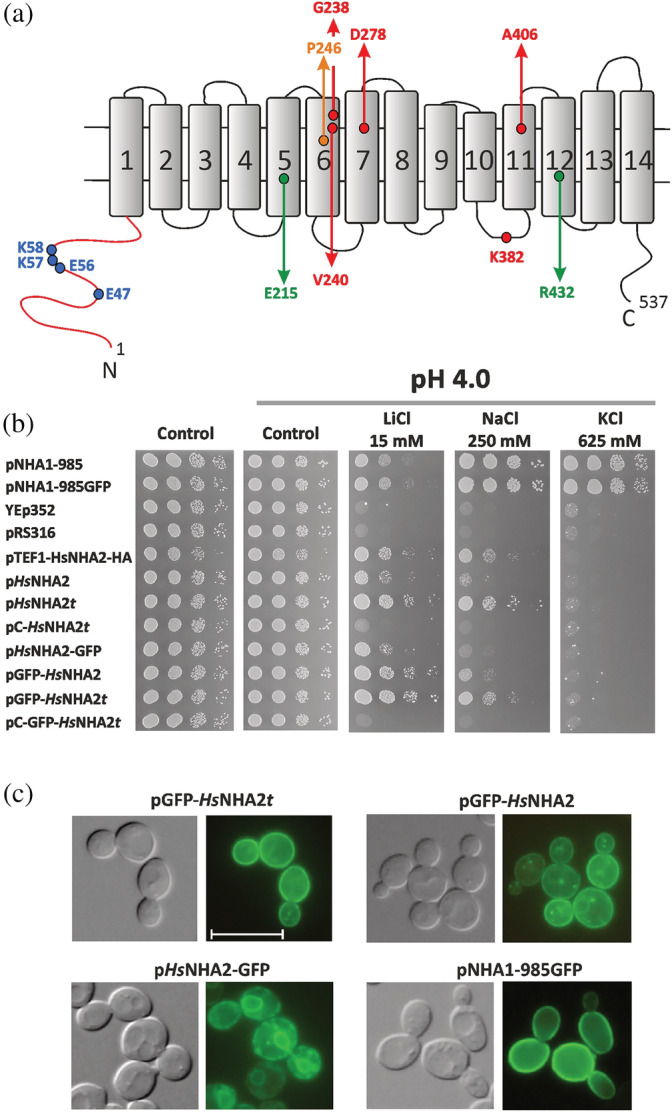
Optimization of conditions for heterologous expression of HsNHA2 in S. cerevisiae. (a) Schematic representation of transmembrane topology of HsNHA2. The positions of residues studied in this work are highlighted. (b) Growth of alkali‐metal cation‐sensitive BW31 strain containing empty vectors (YEp352, pRS316) or expressing ScNha1 or HsNHA2 (non‐tagged or N‐ or C‐terminally tagged with GFP) from various types of plasmids (Table 1) was compared on YNB‐Pro (pH approx. 4.8) or non‐buffered YNB‐Pro plates (pH 4.0) supplemented with LiCl, NaCl, or KCl as indicated (t = TPS1 terminator, pC = centromeric plasmid). Cells were grown at 30°C for 2 (control), 6 (LiCl) or 4 (NaCl, KCl) days. (c) Nomarski (left) and fluorescence (right) micrographs of BW31 cells expressing either N‐terminally GFP‐tagged HsNHA2, C‐terminally GFP‐tagged HsNHA2 or ScNha1‐GFP (expressed from pGFP‐HsNHA2t, pGFP‐HsNHA2, pHsNHA2‐GFP or pNHA1‐985GFP, respectively). Cells were grown in YNB‐Pro (4% glucose) to the early exponential phase. The scale bar corresponds to 10 μm
HsNHA2 mediates an electroneutral efflux of sodium or lithium cations in exchange for external protons across the membrane. 7 , 29 Some studies indicate that, depending on the proton or cation concentrations, it is able to work in both directions and also provides Na+/Li+ counter transport in kidney‐derived MDCK cells. 11 , 30 Pharmacologically, NHA2 is resistant to amiloride, a typical inhibitor of members of the NHE subfamily, but it is a phloretin‐sensitive transport system. 6 , 30
So far, several functional and mutagenesis studies of HsNHA2 have been done upon its expression in the model yeast S. cerevisiae, in a strain that lacked the three Na+ transporters—the Na+/H+ antiporter Nha1 and Na+‐ATPases Ena from the plasma membrane and the endosomal Na+/H+ antiporter Nhx1. 4 , 6 , 23 , 31 This strain is highly sensitive to all alkali‐metal‐cations salts and the expression of HsNHA2 improved the growth of these cells in the presence of sodium or lithium in a pH dependent manner. 4 , 6 Examination of the growth of yeast cells expressing mutated variants of HsNHA2 in the presence of salts was also used to identify some residues that are important for the activity of HsNHA2. 23 , 31 However, the transport activity of the native (or mutated) HsNHA2 antiporter(s) has never been measured directly in yeast cells.
In this work, we first optimized the functional expression of HsNHA2 in S. cerevisiae cells and obtained a highly valuable experimental model that we next used for in vivo determination of effects of HsNHA2 mutations that alter its substrate specificity, transport activity, stability, or are among the single‐nucleotide polymorphisms (SNP) in humans. In addition, for the first time, we demonstrate that the hydrophilic N‐terminus (and negatively charged residues in this part of the protein) has a regulatory (inhibition) effect on HsNHA2 activity. Altogether, our data suggest an allosteric link between the hydrophilic N‐terminus and transmembrane core of the protein.
2. RESULTS
2.1. Optimization of HsNHA2 expression in yeast cells
A eukaryotic cell model organism, the yeast S. cerevisiae, has been used for functional characterization of human proteins for a long time. Several mammalian Na+/H+ antiporters were also studied via their expression in S. cerevisiae, 32 , 33 including Homo sapiens NHA2. 4 , 6 , 23 , 31 In these studies, HsNHA2 was expressed under the control of a strong promoter (PGK1 or TEF1) from a multi‐copy plasmid in a salt‐sensitive strain (AB11c; nha1Δ ena1‐4Δ nhx1Δ). HsNHA2 activity was then observed as a pH‐dependent increase in tolerance to sodium or lithium, estimated by the growth of cells either in liquid media or on plates containing the corresponding salts. 4 , 6 , 23 , 31 However, in our first experiment, we realized that using one of these previously made constructs (pTEF1‐HsNHA2‐HA), in which HsNHA2 is expressed under the control of the TEF1 promoter (Table 1, 4 ) is partially toxic for AB11c cells as well as for another strain, BW31, which only lacks the two plasma‐membrane Na+ exporters (nha1Δ ena1‐4Δ). In both cases, cells expressing HsNHA2 grew under non‐stress conditions significantly slower than cells containing the empty vector (shown for the BW31 strain in Figure 1, control plates). Therefore, we next carefully examined and optimized the conditions of the expression of HsNHA2 in S. cerevisiae (non‐toxic for yeast cells) to obtain a valuable experimental model useful for the in vivo determination of HsNHA2 transport activity. We constructed seven new plasmids (centromeric or multi‐copy; Table 1), in which the HsNHA2 was expressed under the control of a weak and constitutive S. cerevisiae NHA1 promoter. 34 Four plasmids contained a GFP sequence added in frame either at the 5′‐ or 3′‐ends of HsNHA2 to confirm a plasma‐membrane localization of HsNHA2 in yeast cells (Table 1, 6 , 31 ). Since the presence of a terminator behind any gene is important for mRNA export from the nucleus to the cytoplasm, as well as its stability and translation efficiency, 35 an efficient S. cerevisiae terminator TPS1 36 was added behind the HsNHA2 cDNA in four versions of plasmids (Table 1). BW31 cells were transformed with all new plasmids, and we compared the level of cell salt tolerance provided by HsNHA2 expressed from particular plasmids (Figure 1b). Salt tolerance was tested on YNB‐Pro plates with the pH adjusted to 4.0 to ensure the necessary gradient of H+ for the transport activity of HsNHA2. 4 Adjustment of the media to lower pH from approximately 4.8 to 4.0 with HCl accentuated the phenotypes on plates with salts described below (results obtained on non‐adjusted YNB‐Pro plates are not shown). Cells expressing empty vectors (YEp352, pRS316) or S. cerevisiae Nha1 antiporter (non‐tagged or tagged with GFP) were used as negative or positive controls, respectively. BW31 cells containing an empty vector (YEp352 or pRS316; Table 1) were highly sensitive to all three salts tested (LiCl, NaCl, KCl), while cells expressing the ScNha1 antiporter were able to grow in the presence of LiCl, NaCl, and KCl (Figure 1b), as ScNha1 is able to export all three cations from cells. 20 None of the constructs with HsNHA2 improved the cell growth on KCl plates, which confirmed the previous results that HsNHA2 is not transporting K+. 4 , 30
TABLE 1.
Plasmids used in this study
| Plasmid | Description a | Source/reference |
|---|---|---|
| YEp352 | Multi‐copy empty vector | 19 |
| pHsNHA2 | H. sapiens NHA2 in YEp352 | This work |
| pGRU1 | Multi‐copy empty vector with GFP | B. Daignan‐Fornier, NCBI Acc. No. AJ249649 |
| pHsNHA2‐GFP | H. sapiens NHA2 in pGRU1 | This work |
| pGFP‐HsNHA2 | N‐terminally GFP tagged HsNHA2 in YEp352 | This work |
| pHsNHA2t | H. sapiens NHA2 with TPS1 terminator in YEp352 | This work |
| pGFP‐HsNHA2t | N‐terminally GFP tagged HsNHA2 with TPS1 terminator in YEp352 | This work |
| pNHA1‐985 | S. cerevisiae NHA1‐985 in YEp352 | 20 |
| pNHA1‐985GFP | S. cerevisiae NHA1‐985 in pGRU1 | 20 |
| pTEF1‐HsNHA2‐HA | H. sapiens NHA2 in p426TEF | 4 |
| pRS316 | Centromeric empty vector | 21 |
| pC‐HsNHA2t | H. sapiens NHA2 with TPS1 terminator in pRS316 | This work |
| pC‐GFP‐HsNHA2t | N‐terminally GFP tagged HsNHA2 with TPS1 terminator in pRS316 | This work |
| YEp352‐ZrSTL1‐TPS1t‐kanMX | Plasmid for the expression of Zygosaccharomyces rouxii STL1 gene downstream with the TPS1 terminator | 22 |
ScNHA1 or HsNHA2 were expressed under the control of S. cerevisiae NHA1 promoter, except in pTEF1‐HsNHA2‐HA, in which TEF1 promoter was used.
The expression of HsNHA2 from new plasmids was not toxic for cells, as all transformants grew as well as cells containing empty vectors or expressing the yeast Nha1 antiporter on control plates (and better than cells transformed with pTEF1‐HsNHA2‐HA) (Figure 1b). The level of HsNHA2 expression from centromeric plasmids (pC‐HsNHA2t, pC‐GFP‐HsNHA2t) was too low to observe its activity, since cells with these plasmids grew similarly to cells with empty vectors (Figure 1b). On the other hand, the expression of HsNHA2 under the control of the ScNHA1 promoter from new multi‐copy plasmids increased the cell tolerance to LiCl and NaCl, although they differed in the level of tolerance provided (Figure 1b). Cells expressing HsNHA2 from pHsNHA2t or pHsNHA2‐GFPt plasmids were more tolerant to salts than cells with the same constructs without the terminator (Figure 1b). Thus, the use of multi‐copy plasmids with a weak ScNHA1 promoter and TPS1 terminator was the best for studying the transport properties of human NHA2 expressed in yeast.
To consider various types of expression for the correct targeting of HsNHA2 to the plasma membrane, we first compared the localization of GFP‐tagged versions of HsNHA2 in AB11c and BW31 cells, and realized that the lack of intracellular Nhx1 antiporter in the AB11c strain resulted in a higher intracellular stacking of GFP‐HsNHA2 than in BW31 cells (Figure S1). Thus, we subsequently only used the BW31 strain. As shown in Figure 1c, the GFP‐tagging of HsNHA2 at the C‐terminus also resulted in a partial stacking of the antiporter in the endoplasmic reticulum (Figure 1c), and correspondingly, the tolerance of cells expressing HsNHA2‐GFP was lower than that of cells with non‐tagged HsNHA2 or HsNHA2 tagged with GFP at the N‐terminus (Figure 1b). HsNHA2 with a GFP tag at the N‐terminus exhibited similar plasma‐membrane localization to ScNha1‐GFP (Figure 1c). The expression of GFP‐HsNHA2 from a plasmid with a TPS1 terminator (pGFP‐HsNHA2t) resulted in a diminished level of intracellular fluorescent spots observed in cells containing pGFP‐HsNHA2 (Figure 1c, compare the upper two images).
Thus, from all these tested conditions, for the optimal functional expression of HsNHA2 in yeast cells and further experiments, we selected the BW31 strain transformed either with the pHsNHA2t or pGFP‐HsNHA2t plasmids.
2.2. Point mutations of Pro246 modify the ion selectivity of the antiporter for Na+ and/or Li+ and its transport activity at pH 7.0
The membrane parts of Na+/H+ antiporters are highly conserved and are sufficient for ion exchange. 20 , 25 In the last phylogenetic analysis, a minimal set of eight highly conserved positions in the protein core that are present in all CPAs was identified. 2 In HsNHA2, this motif contains, among others, proline 246 in TMS 6 and arginine 432 in TMS 12 (Figures 1a and S1). The replacement of an equivalent proline with a polar amino‐acid residue (serine or threonine) in the homologous Na+, Li+/H+ antiporter Sod2‐22 from Zygosaccharomyces rouxii resulted in an ability to recognize and transport K+ cations. 37 Thus, in the next part of this work, we tested how a substitution of P246 in HsNHA2 can change its substrate specificity and transport activity (Figure 2). Proline 246 was replaced by site‐directed mutagenesis either with glycine, a small hydrophobic amino acid (alanine) or polar residues (serine or threonine), and mutated versions were expressed in the BW31 strain from the corresponding pHsNHA2t or pGFP‐HsNHA2t plasmids. Transformants were tested for their tolerance to LiCl, NaCl or KCl at pH 4.0 or 7.0 in comparison with cells transformed with the empty vector or expressing the native HsNHA2 antiporter (Figure 2a). None of the mutated versions improved the tolerance of cells to KCl (not shown), but we observed that their ability to provide cells with tolerance to LiCl or NaCl within the pH range 4.0–7.4 was different from the native antiporter (Figure 2a,e). The GFP‐tagging had no effect on the substrate specificity of mutated versions (cf. below), but it slightly enhanced the phenotypes observed as cell salt tolerance on plates (compare Figures 2a and S3A). All four mutated versions were localized to the plasma membrane (Figure 2c), like the native GFP‐HsNHA2 (Figure 1c), and similar amounts of proteins in cells were detected by immunoblot for all of them (Figure 2d).
FIGURE 2.
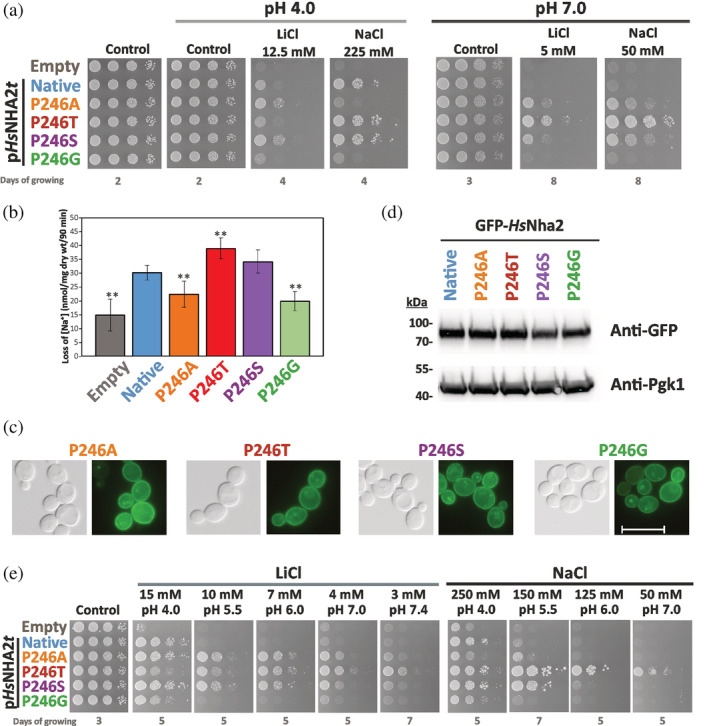
Characterization of HsNHA2 with point mutations at proline 246. (a) Salt tolerance of S. cerevisiae BW31 cells containing empty vector or expressing native HsNHA2 or one of four HsNHA2 versions with single point mutations P246A, T, S or G from pHsNHA2t plasmid. Cells were grown on YNB‐Pro or non‐buffered YNB‐Pro plates with pH adjusted to 4.0 or 7.0 and supplemented with LiCl or NaCl as indicated. Plates were incubated at 30°C and photographed on the indicated day. (b) Loss of Na+ from same cells as described in (a). Cells were grown in YNB‐Pro media, preloaded with Na+, transferred to Na+‐free incubation buffer at pH 4.0 and changes in the intracellular Na+ content were estimated as described in Section 5. Columns show the amount of Na+ lost from cells within 90 min. Data represent the mean of at least 5 replicate values ± SD. Significant differences compared to cells expressing the native HsNHA2 are indicated with asterisks (**p < .01). Localization (c) and immunodetection (d) of N‐terminal GFP‐tagged HsNHA2 with single point mutations at Pro246. BW31 cells expressing variants of GFP‐HsNHA2 from pGFP‐HsNHA2t were grown in YNB‐Pro (4% glucose) to the exponential phase and observed under a fluorescence microscope (c, right). A Nomarski prism was used for whole‐cell imaging (c, left). In (d), protein extracts of cells were prepared as described in Section 5, subjected to SDS‐PAGE (10% gel) and transferred to a nitrocellulose membrane. Native GFP‐HsNHA2 and its variants were detected with an anti‐GFP antibody. The membrane was reprobed and incubated with an anti‐Pgk1 antibody to verify the amount of loaded proteins. (e) Activity of mutated HsNHA2 antiporters at various extracellular pH was determined by growth of cells on YNB‐Pro plates buffered to various pH levels (4.0–7.4) and supplemented with LiCl or NaCl as indicated. Images were taken on the indicated day of incubation at 30°C
As expected, the presence of the native antiporter only increased cell tolerance to sodium and lithium at pH 4.0 (when there is a high H+ gradient across the membrane), but not at pH 7.0 (Figure 2a). The P246G substitution resulted in an almost inactive antiporter, as cells with this mutated version grew only slightly better in the presence of salts than cells containing the empty vector (Figure 2a). The P246A mutation resulted in an antiporter which was able to transport Li+ better than the native antiporter, but its ability to improve the NaCl tolerance was much lower than that of the native HsNHA2 (Figure 2a). In contrast, Pro246 replaced with threonine resulted in an antiporter with the opposite substrate preferences to the P246A version, that is, transporting lithium worse and sodium better than the native HsNHA2 (Figure 2a). Finally, HsNHA2 with the P246S mutation provided a better tolerance of cells to both cations compared to the native HsNHA2 (Figure 2a).
Interestingly, and in contrast to the native antiporter, HsNHA2 harboring the point mutations P246A/S/T also exhibited activity at a higher external pH than 4.0. They improved the tolerance of BW31 cells to both cations on non‐buffered plates with the pH adjusted to 7.0 (Figure 2a), where the P246T version provided the most robust growth (Figure 2a). In the growth test shown in Figure 2e, we verified this observation on plates with buffered media (cf. Section 5). On these plates, extracellular pH should not change with the growth of cells, as in Figure 2a. It is evident that under these conditions, each of the three P246A/T/S versions was able to improve the tolerance of cells to LiCl within the pH range 4.0–7.4, and the P246T/S versions also to NaCl (Figure 2e). Note that in contrast to P246A/S, the ability of the P246T version to improve the tolerance to LiCl increased with increasing extracellular pH (Figure 2e). This drop test also confirmed the decreased activity of the P246G version in comparison with the native HsNHA2, in which Li+ and Na+ transport activity was only observable on plates with low pH 4.0 (Figure 2e or a).
Next, the differences in capacity to transport Na+ cations observed in drop tests (Figure 2a,e) were verified by sodium loss measurements at pH 4.0 (Figure 2b) from sodium‐preloaded cells (cf. Section 5). During the experiment, the intracellular concentration of Na+ also slightly decreased in control cells without any antiporter (Figure 2b, empty vector), but significantly higher efflux was observed in cells with the native HsNHA2 (Figure 2b). In accordance with the drop tests, the amounts of sodium lost from cells expressing the HsNHA2 antiporter with the P246A or P246G mutation were lower than from cells with the native HsNHA2, but higher than from cells with the empty vector (Figure 2b). On the other hand, the P246S, and especially the P246T mutation increased HsNHA2 transport activity for Na+. The amount of sodium lost from these cells was significantly higher than from cells with the native antiporter (Figure 2b). Since the localization and amounts of proteins did not change with the P246 substitutions (Figure 2c,d), sodium loss measurements fully reflected the sodium transport capacity of the particular mutated versions observed as differences in growth on plates with NaCl (Figure 2a,e).
All these data identified proline 246 as a crucial residue involved in the ability to recognize and transport particular substrates (ion selectivity), most likely also influencing the capacity to transport protons, as the pH profile of HsNHA2 transport activity changed, especially when the polar‐branched amino acid threonine was at this position.
2.3. HsNHA2 with substitutions at Pro246 are less sensitive to phloretin inhibition
The activity of HsNHA2 is insensitive to amiloride (a hallmark of plasma‐membrane NHE antiporters), and sensitive to phloretin, 30 even when it is expressed in yeast cells. 6 Phloretin (a flavonoid abundant in apples) is a highly flexible molecule with the ability to bind to biological macromolecules. It has interesting pharmacological and pharmaceutical potential due to its antimicrobial, antioxidant, anti‐inflammatory, and anticancer activities. 38 The mechanism of HsNHA2 inhibition by phloretin is unclear. To determine whether mutations of P246 change the sensitivity of the antiporter to phloretin, we tested the growth of cells expressing the native, or either of the four antiporters mutated at proline P246, on plates supplemented with 15 mM LiCl or 250 mM NaCl and increasing concentrations of phloretin (Figure 3).
FIGURE 3.
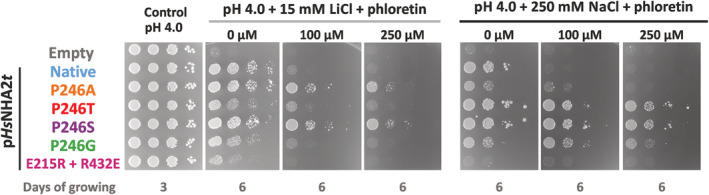
HsNHA2 variants of proline 246 are less sensitive to phloretin inhibition. S. cerevisiae BW31 cells containing empty vector or expressing native HsNHA2 or HsNHA2 versions with point mutations P246A, T, S or G or the double mutation E215R + R432E from pHsNHA2t plasmid, as indicated, were used in this experiment. The inhibition of mutated HsNHA2 versions by phloretin was determined by the growth of cells on non‐buffered YNB‐Pro plates with the pH adjusted to 4.0 and supplemented with LiCl or NaCl, and with or without phloretin at the indicated concentrations. Plates were incubated at 30°C for 3 (controls) or 6 days (phloretin)
In accordance with previous results 6 , the ability of the native HsNHA2 to improve LiCl tolerance at pH 4.0 of yeast cells highly decreased in the presence of phloretin (Figure 3). Here we also demonstrate the same effect of phloretin on Na+ transport activity (Figure 3). Remarkably, the inhibition effect of phloretin decreased when proline 246 was replaced with alanine/glycine or even disappeared when serine or threonine was at this position (Figure 3). BW31 cells transformed with these versions of HsNHA2 grew on plates with LiCl or NaCl and phloretin very similarly to when the inhibitor was not present (Figure 3).
These results reinforced the importance of proline 246 for the proper activity of HsNHA2. They also indicate that this residue could take part in a mechanism of inhibition mediated by phloretin.
2.4. Swapping E215 and R432 results in a Li+‐specific antiporter that is also active at higher pH
As was mentioned above, R432 is one of the eight highly conserved amino acids of the whole CPA family, and it was predicted to potentially form a salt bridge with E215 in TMS 5 (Figure 1a). 2 To experimentally prove the importance and function of these two titratable residues, we prepared three mutated versions of HsNHA2 in which we either introduced the opposite charge at these positions (E215R or R432E versions) or we swapped these two residues (E215R + R432E double mutant). The effect of mutation(s) on the transport properties of HsNHA2 was tested similarly as above.
The single‐mutated versions of the antiporter, E215R or R432E, were non‐functional (Figure 4a). Cells transformed with pHsNHA2t (E215R) or pHsNHA2t (R432E) plasmids did not grow in the presence of NaCl or LiCl (at both pHs, 4.0 or 7.0), just like cells containing the empty vector (Figure 4a). On the other hand, swapping these two residues led to a functional antiporter, however, with substrate specificity limited to Li+ cations. At both of pH levels, cells expressing HsNHA2 (E215R + R432E) were only able to grow on plates with LiCl, but not with NaCl (Figure 4a). Similar results were observed in a drop test with cells expressing the same mutated versions, but tagged with GFP at the N‐terminus (Figure S3B).
FIGURE 4.
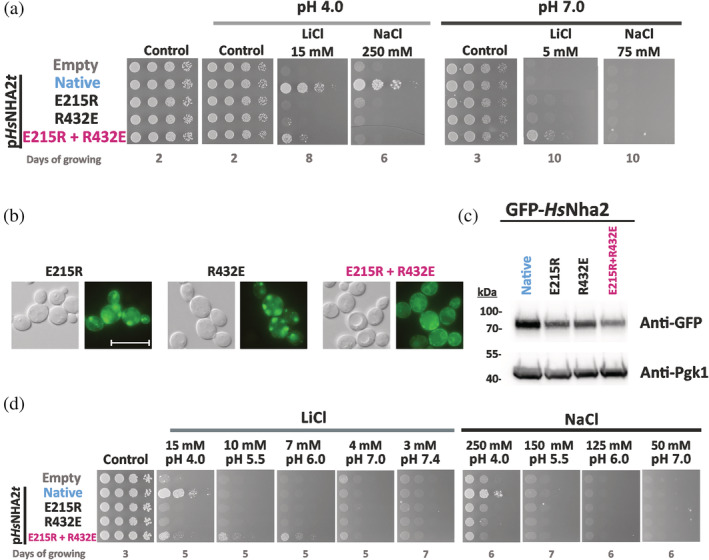
Characterization of HsNHA2 with mutations of residues forming a putative salt bridge. (a) Salt tolerance of S. cerevisiae BW31 cells containing empty vector or expressing native HsNHA2 or its mutated versions (E215R, R432E or E215R + R432E) from pHsNHA2t plasmid. Cells were grown on YNB‐Pro (pH approx. 4.8) or non‐buffered YNB‐Pro plates with the pH adjusted to 4.0 or 7.0 and supplemented with LiCl or NaCl as indicated. Plates were incubated at 30°C and photographed on the indicated day. Localization (b) and immunodetection (c) of N‐terminal GFP‐tagged HsNHA2 mutated versions E215R, R432E or E215R + R432E. Cells were grown in YNB‐Pro (4% glucose) to the exponential phase and observed under a fluorescence microscope (b, right). A Nomarski prism was used for whole‐cell imaging (b, left). The scale bar corresponds to 10 μm. In (c), protein extracts from the same cells as in (b) (with cells expressing native GFP‐HsNHA2 as a control) were prepared as described in Section 5, subjected to SDS‐PAGE (10% gel) and transferred to a nitrocellulose membrane. GFP‐HsNHA2 was detected with an anti‐GFP antibody. The membrane was reprobed and incubated with an anti‐Pgk1 antibody to verify the amount of loaded proteins. (d) Activity of mutated HsNHA2 antiporters at various extracellular pH levels determined by growth of cells on YNB‐Pro plates buffered to various pH levels (4.0–7.4) and supplemented with LiCl or NaCl as indicated. Images were taken on the indicated day of incubation at 30°C
The loss of function of the two single‐mutated versions most likely resulted from their mislocalization in intracellular compartments, observed as fluorescent spots inside cells (Figure 4b). In contrast, the double‐mutated E215R + R432E version tagged with GFP was partially targeted to the plasma membrane and partially observed in the perinuclear ER (Figure 4b). The level of proteins expressed in cells checked by immunoblotting revealed that all three mutated variants were present in cells in lower amounts than the native antiporter (Figure 4c). Since all proteins are expressed under the same conditions (plasmid, promoter, terminator), it indicates that these mutations affected the structure, stability, and plasma‐membrane targeting of HsNHA2.
The drop test on plates with salts and pH buffered to various values (Figure 4d) confirmed the lithium substrate specificity of the E215R + R432E version. In contrast to the native antiporter, it was able to improve the LiCl tolerance of cells not only at pH 4.0, but also at higher pH, up to 6.0 (Figure 4d). Nevertheless, in contrast to P246 variants, phloretin inhibited the Li+ transport ability of the double mutant E215R + R432E, as observed for the native HsNHA2 (Figure 3).
All these results confirmed E215 and R432 to be crucial residues for HsNHA2 functionality. The transport activity of the antiporter with the E215R mutation in TMS 5 could be restored by introducing a second mutation R432E in TMS 12 (Figures 1, S1, and 8). Thus, we experimentally confirmed that in the full version of HsNHA2 (containing the hydrophilic N‐terminus), these two residues are spatially close to each other with a salt bridge between them. The interaction between these two residues is important for the structural integrity of the protein.
FIGURE 8.
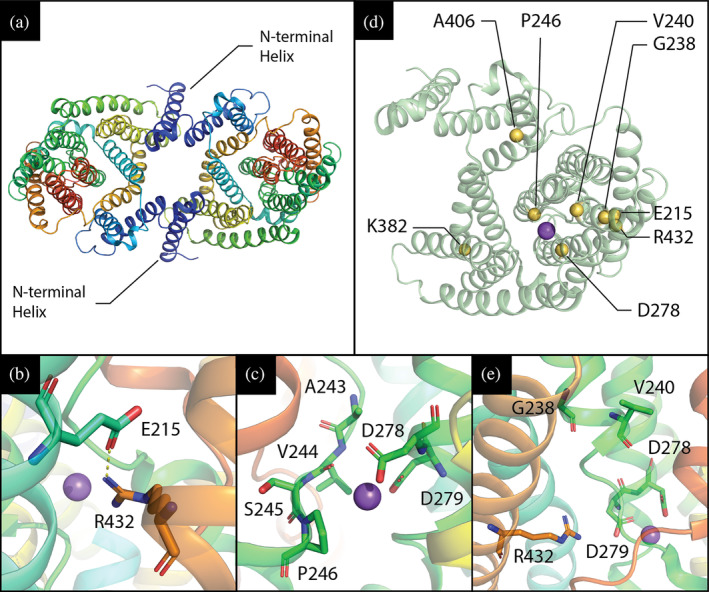
HsNHA2 3D‐structure and location of various point mutations. (a) Structure of HsNHA2 dimer, residues 79–521 (PDB entry 7B4L) in cartoon representation with rainbow coloring corresponding to N‐ through C‐terminus. The additional N‐terminal helix that is missing in other mammalian CPA members with known structures (dark blue) is highlighted. (b, c, e) Close‐up view of studied HsNHA2's point mutations in vicinity of ion‐binding site. (b) Previously predicted E215‐R432 salt bridge was recently confirmed by newly published structures of both human and bison NHA2, and is likely to play a structural role. (c) P246 is located at unwound section of TMS 6. Its surroundings are shown as sticks, and the bound sodium ion is shown as a purple sphere. Notably, the addition of a polar side‐chain in the vicinity of D278 and D279, which mediate sodium and proton binding, could alter the pH‐dependent activity of the antiporter. (e) G238, V240, D278 and R432, though they do not coordinate the sodium ion directly, are all located on the mobile and highly conserved core domain of the antiporter. (d) HsNHA2 monomer, shown in cartoon representation, with position of various point mutations shown as yellow spheres. A sodium ion, observed in the experimental structure, is shown as a purple sphere and denotes the position of NHA2's binding site
2.5. Validation of the system for studying the effect of SNPs in HsNHA2
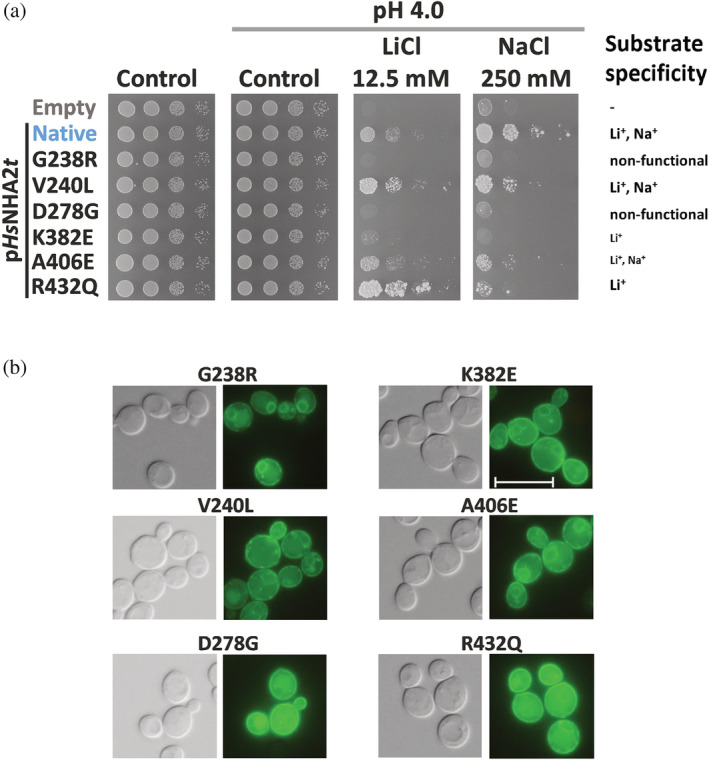
As was mentioned above, HsNHA2 is associated with a growing list of various pathologies. For this reason, we think that our optimized expression system of HsNHA2 in yeast cells could be useful in the future for the characterizing the effects of a broad range of mutations that are present in human genomes. To test this possibility, we selected six point mutations (G238R, V240L, D278G, R432Q, K382E, and A406E) that belong to known human SNPs from patients. 39 The positions of these residues in HsNHA2 are shown in Figures 1a, S1, and 8b,e. Mutated versions were prepared by site‐directed mutagenesis and expressed from pHsNHA2t or pGFP‐HsNHA2t plasmids in BW31 cells, and we tested the tolerance of cells to LiCl and NaCl at pH 4.0 (Figures 5a and S4). The localization of the corresponding GFP‐tagged versions was observed by fluorescence microscopy (Figure 5b).
FIGURE 5.
Phenotypes of SNPs of HsNHA2. (a) Salt tolerance of S. cerevisiae BW31 cells containing empty vector or expressing native HsNHA2 or one of six HsNHA2 mutated versions—G238R, V240L, D278G, K382E, A406E, and R432Q from the pHsNHA2t plasmid. Cells were grown on YNB‐Pro or non‐buffered YNB‐Pro plates with pH adjusted to 4.0 and supplemented with LiCl or NaCl as indicated. Growth was monitored for 2 (controls) or 7 (LiCl or NaCl) days at 30°C. (b) Localization of N‐terminal GFP‐tagged HsNHA2 versions. BW31 cells expressing GFP‐HsNHA2 with single point mutations as in (a) from pGFP‐HsNHA2t were grown in YNB‐Pro (4% glucose) to the exponential phase and observed under fluorescence microscope (b, right). A Nomarski prism was used for whole‐cell imaging (b, left). The scale bar corresponds to 10 μm
All six mutations influenced the transport properties of HsNHA2, but to various extents. The mutation V240L resulted in an antiporter with a preference for transporting Li+ over Na+, while the R432Q or K382E versions narrowed the substrate specificity of the antiporter only for Li+ (Figure 5a; for K382E cf. also Figure S4). The point mutation A406E resulted in a less active antiporter for both cations, as cells with this mutated antiporter grew worse in the presence of both salts than cells with the native one (Figure 5a). Neither of the mutated versions G238R or D278G were functional, because in the presence of salts, cells with these antiporters grew very similarly to cells with the empty vector (Figure 5a). However, the non‐functionality of G238R resulted from misfolding of the antiporter (stacking in the ER; Figure 5b), while for D278G, which was localized in the plasma membrane (Figure 5b), it was instead due to the effect of the mutation on activity (the mechanism of transport). The fluorescence in cells expressing GFP‐HsNHA2 with V240L, K382E, A406E, or R432Q was also observed predominantly in the plasma membrane (Figure 5b).
These results confirmed that in the future our expression system may provide an efficient screening of the effects of mutations present in HsNHA2 versions associated with pathologies or for the large‐scale testing of various compounds that activate/inhibit the HsNHA2 activity.
2.6. Amino acids 1–40 of the hydrophilic N‐terminus inhibit the transport activity of HsNHA2
HsNHA2 and its homologs from other mammals possess an unusually long hydrophilic N‐terminus (about 82 aa, Figure S1), which is not present in the NHE clade. 2 , 23 The structure of the N‐terminus is predicted to be disordered (Alpha‐fold prediction Q86UD5). 24 In the above drop test experiments, we always noticed that N‐terminally GFP‐tagged versions of HsNHA2 provided BW31 cells with a higher tolerance to LiCl and NaCl (Figures S3–S5) than the corresponding variants without the tag (Figures 2, 4, and 5), even though the expression conditions were the same. This observation indicated that GFP‐tagging somehow activated HsNHA2, and that the hydrophilic N‐terminal part of the protein may play a role in the regulation of the transport activity of HsNHA2. In addition, the cryo‐EM structure of bison NHA2 truncated by 69 amino acids from the N‐terminus indicated the presence of an unusual short helical segment (corresponding to amino acids 70–79) prior to TMS 1. 23
To understand the role of the N‐terminus in HsNHA2's function, we constructed a series of step‐by‐step truncated HsNHA2 versions, ranging from the complete antiporter down to the shortest version lacking the first 90 amino acids (Figure S1). In eight newly prepared constructs of HsNHA2, codons for amino acids at positions 20, 40, 60, 70, 75, 80, and 90 were replaced by the start codon ATG for methionine. Truncated HsNHA2 versions were expressed in BW31 cells, and tested for their properties as above (Figure 6). For each version, a corresponding N‐terminal GFP‐tagged construct was used to test the localization and level of expression of the encoded protein (Figure 6c,d).
FIGURE 6.
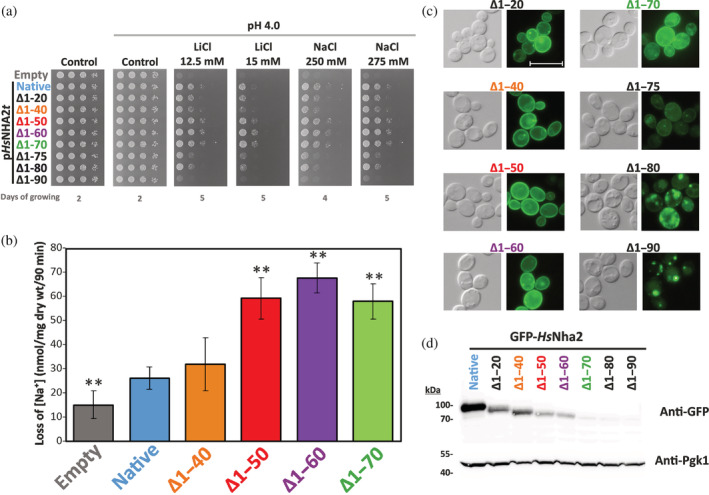
Hydrophilic N‐terminus of HsNHA2 plays inhibitory role. (a) Salt tolerance of S. cerevisiae BW31 cells containing empty vector or expressing native or N‐terminally truncated HsNHA2 versions from plasmids derived from pHsNHA2t. Cells were grown on YNB‐Pro (pH aprox. 4.8) or non‐buffered YNB‐Pro plates with pH adjusted to 4.0 and supplemented with LiCl or NaCl as indicated. Plates were incubated at 30°C and photographed on the indicated day. (b) Loss of Na+ from BW31 cells mediated by N‐terminal truncated HsNHA2 versions. Cells were grown in YNB‐Pro media, preloaded with Na+, transferred to Na+‐free incubation buffer at pH 4.0, and changes in the intracellular Na+ content were estimated as described in Section 5. Columns show the amount of Na+ lost from cells within 90 min. Data represent the mean of at least 3 replicate values ± SD. Significant differences compared to cells expressing the native HsNHA2 are indicated with asterisks (**p < .01). Localization (c) and immunodetection (d) of N‐terminal GFP‐tagged truncated HsNHA2 versions. BW31 cells expressing variants of GFP‐HsNHA2 with truncated N‐terminus from plasmids derived from pGFP‐HsNHA2t were grown in YNB‐Pro (4% glucose) to the exponential phase and observed under a fluorescence microscope (c, right). A Nomarski prism was used for whole‐cell imaging (c, left). The scale bar corresponds to 10 μm. In (d), protein extracts from the same cells as in (c) (with cells expressing native GFP‐HsNHA2 as a control) were prepared as described in Section 5, subjected to SDS‐PAGE (10% gel) and transferred to a nitrocellulose membrane. GFP‐HsNHA2 variants were detected with an anti‐GFP antibody. The membrane was reprobed and incubated with an anti‐Pgk1 antibody to verify the amount of loaded proteins
The lack of the first 40 amino acids had no effect on HsNHA2 transport properties. Cells with Δ1–20 and Δ1–40 truncated versions exhibited the same salt tolerance as the native HsNHA2 (Figure 6a). The lack of the first 40 amino acids also did not change the Na+‐efflux activity of the antiporter (Figure 6b). On the other hand, cells expressing Δ1–50, Δ1–60, or Δ1–70 versions grew better on LiCl and NaCl plates than cells expressing the native antiporter (Figure 6a). This suggested that truncation of the first 50–70 aa increased the activity of the antiporter, which we confirmed by measurements of Na+‐efflux activity (Figure 6b). Notably, the amount of Na+ lost from cells expressing the Δ1–50, Δ1–60, or Δ1–70 HsNHA2 variants was double that of the native antiporter or Δ1–40 version (Figure 6b). Truncations longer than 75 amino acids resulted in less efficient transporters, as they provided cells with lower LiCl and NaCl tolerance than the native antiporter, and the removal of 90 amino acids resulted in a non‐functional protein (Figure 6a). Truncations up to 70 amino acids did not influence the plasma‐membrane localization of the antiporter (Figure 6b). In contrast, the fluorescence signal in cells with the Δ1–75, Δ1–80, or Δ1–90 GFP‐HsNHA2 antiporters was predominantly observed in intracellular compartments (Figure 6c). Thus, these versions were probably not folded properly, which led to their mislocalization. Neither of these truncated versions was able to improve the salt tolerance of BW31 cells on plates with pH 7.0 (results not shown), which means that the dependency on the H+ gradient of HsNHA2 did not change with N‐terminal truncations.
The immunodetection of truncated versions (Figure 6d) showed that the signal of the GFP‐HsNHA2 proteins gradually decreased with larger truncations of the N‐terminus, being the weakest for the last three (Δ1–70, Δ1–80, or Δ1–90). This indicates that the N‐terminus is not only important for the regulation of transport activity, but also for the stability of the protein and/or its trafficking to the plasma membrane. Interestingly, in contrast to the point mutations studied above (localized within the TMS), the GFP‐tagging decreased the ability of truncated HsNHA2 to improve the tolerance of cells to LiCl, but the provided tolerance to NaCl was enhanced with a truncation longer than 40 aa, as with non‐tagged antiporters (compare Figure 6a and S5).
Although the protein signals detected by immunoblotting for the Δ1–50, Δ1–60, or Δ1–70 versions were much lower than that for the native antiporter (Figure 6d), these GFP‐non‐tagged versions exhibited at least double the Na+ efflux activity (Figure 6b). Moreover, truncation of the N‐terminus of 50 aa in GFP‐HsNHA2 also resulted in an increased Na+ efflux activity in comparison with the native version of GFP‐HsNHA2 (57 ± 5 nmol Na+/mg dry wt/90 min vs. 35 ± 6 nmol Na+/mg dry wt/90 min, respectively). Since we observed this phenomenon in yeast cells, which may lack some regulatory mechanisms or protein interactions that regulate the activity of HsNHA2 in mammalian cells, 12 the first 40–50 amino acids of the protein appears to allosterically auto‐inhibit HsNHA2 activity.
2.7. Conserved charged amino acids in the hydrophilic N‐terminus of HsNHA2 are important for its activity
It was found that in the hydrophilic N‐terminus of the Na+/HCO3 − co‐transporter, the presence of charged (positively as well as negatively) residues is essential to fulfilling the auto‐inhibitory function of the transporter. 40 To corroborate the role of the N‐terminus in the regulation of HsNHA2 activity, we compared the sequences of unique homologs of NHA2 from various metazoan species (Figure 7a). In the important region revealed above (amino acids 40–70), we identified four charged residues (3 of them highly conserved; Figure 7a): negatively charged glutamates 47 and 56, and positively charged lysines at positions 57 and 58, and we estimated their possible role in HsNHA2 activity. We replaced these amino acids with neutral alanine or with an amino acid with the opposite charge. The resulting six mutated versions of HsNHA2 (E47A/K, E56A/K, K57A + K58A, K57E + K58E) were tested for their ability to improve the salt tolerance of BW31 cells at pH 4.0 in comparison with the native antiporter (Figure 7a). Fluorescence microscopy of GFP‐tagged versions revealed that none of these mutations changed the plasma‐membrane localization of the antiporter (Figure 7c), or the amount of proteins in cells (Figure 7d).
FIGURE 7.
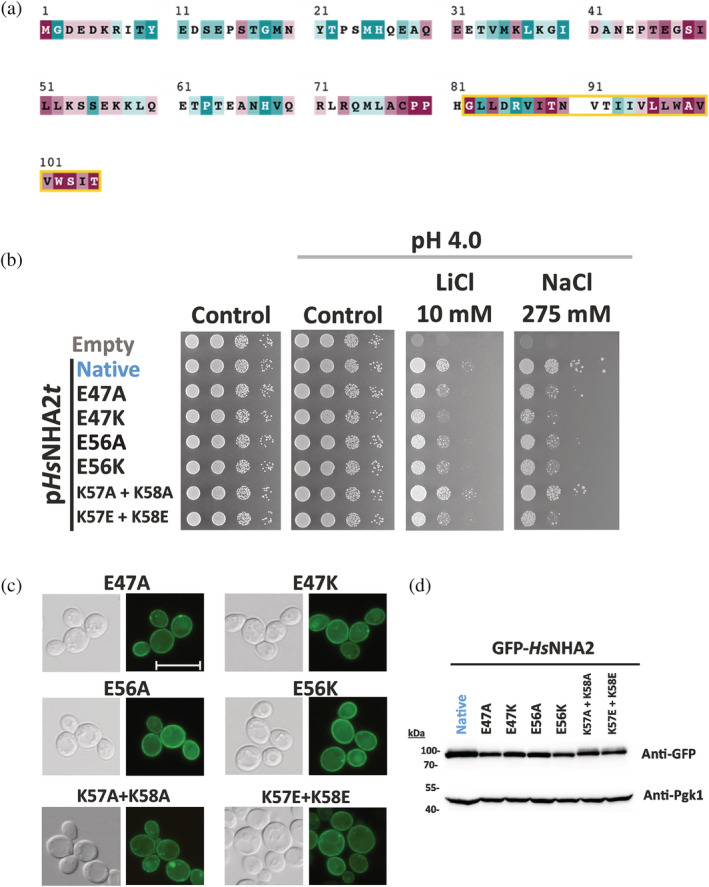
Charged residues in HsNHA2 hydrophilic N‐terminus are important for transport activity. (a) Evolutionary conservation analysis of HsNHA2 N‐terminus. HsNHA2 N‐terminal residues 1‐to‐105 colored based on ConSurf server evolutionary conservation scale, with cyan to maroon representing variable to conserved amino acids, respectively. The position of the unique N‐terminal TMS is highlighted with a yellow box. The conservation analysis was conducted using HsNHA2 N‐terminal amino acids. A total of 99 unique sequences were collected by HMMER from the UniRef90 database with sequence identity ranging from 35 to 95% and an E‐value of at least 0.00001. (b) Salt tolerance of S. cerevisiae BW31 cells containing empty vector or expressing native HsNHA2 or one of six HsNHA2 mutated versions E47A, E47K, E56A, E56K, K57A + K58A, or K57E + K58E from pHsNHA2t. Cells were grown on YNB‐Pro or non‐buffered YNB‐Pro plates with the pH adjusted to 4.0 and supplemented with LiCl or NaCl as indicated. Plates were incubated at 30°C for 2 (controls) or 5 days (salts). Localization (c) and immunodetection (d) of N‐terminal GFP‐tagged HsNHA2 mutated versions as in (b). BW31 cells expressing variants of GFP‐HsNHA2 from the pGFP‐HsNHA2t were grown in YNB‐Pro (4% glucose) to the exponential phase and observed under a fluorescence microscope (c, right). A Nomarski prism was used for whole‐cell imaging (c, left). The scale bar corresponds to 10 μm. In (d), protein extracts from the same cells as in (c) were prepared as described in Section 5, subjected to SDS‐PAGE (10% gel) and transferred to a nitrocellulose membrane. GFP‐HsNHA2 variants were detected with an anti‐GFP antibody. The membrane was reprobed and incubated with an anti‐Pgk1 antibody to verify the amount of loaded proteins
The drop test (Figure 7b) showed that both glutamate residues E47 and E56 are important for the antiporter's activity. Their replacement with alanine resulted in antiporters with a lower ability to improve the lithium and sodium tolerance of cells, and their replacement with an amino‐acid with the opposite charge (lysine) emphasized this phenotype (Figure 7b). The replacement of K57 and K58 with alanines did not affect the antiporter's function, but negatively charged glutamates at these two positions also resulted in an antiporter with decreased activity (Figure 7b).
In summary, these experiments revealed a new role of the N‐terminus in the regulation of HsNHA2 activity and identified four charged amino‐acid residues in the N‐terminus as crucial for determining HsNHA2 transport activity.
3. DISCUSSION
As a unicellular eukaryotic organism, the yeast S. cerevisiae plays a key role in fundamental scientific research in exploring molecular aspects of cellular processes. Thanks to the identification of human orthologs to yeast proteins, including proteins related to human pathologies, yeast is widely used as a simplified cellular model of human diseases. 41 The heterologous expression of human proteins in yeast cells also provides valuable information about the structure of human proteins and their functional domains. 42 We optimized the expression of the human Na+/H+ antiporter NHA2 in yeast cells to be stable, functional, at a non‐toxic level for cells, and with proper targeting to the plasma membrane (Figure 1). For the first time, we set the conditions and methodology for in vivo measurements of its Na+ transport activity (Figures 2 and 6). Using this system enabled us to identify several as‐yet unknown residues crucial for HsNHA2 substrate specificity and transport activity (Figures 2, 4, 5, and 7), and to discover a possible auto‐inhibitory role of the hydrophilic N‐terminal segment (Figures 6 and 7), which appears to allosterically regulate transmembrane ion transport. Our experimental model is also a key tool for better understanding the molecular causes of pathologies related to single‐nucleotide polymorphism of HsNHA2, and to screen for putative modifiers, including drugs, which could alter HsNHA2 activity (similarly as is shown in Figures 5 and 3, respectively).
All Na+/H+ antiporters seem to share a similar transmembrane topology, known as the NhaA fold, organized into two functional domains—a dimerization domain and a conserved core domain encapsulating the ion‐binding site. 43 , 44 According to recently resolved N‐terminally truncated cryoEM structures of two NHA2 proteins (human [PDB 7B4L; Figure 8a], bison 23 ), the antiporter consists of 14 TMS, instead of the 13 TMS observed in mammalian NHE Na+/H+ exchangers 45 , 46 and the prokaryotic NhaP1, NapA and NhaP antiporters 47 , 48 , 49 , 50 or the 12 TMS in Escherichia coli antiporter NhaA. 43 HsNHA2 possesses a long hydrophilic N‐terminus that resides in the cytosol (Figures 1a and S1), which is followed by the first transmembrane helix corresponding to amino acids 82–105 (Figures 7a and 8a). Predicted to be unstructured, the hydrophilic N‐terminal part preceding TMS1 is not present in both resolved NHA2 structures that were recently released (Protein Data Bank 7B4M or 7B4L 23 ). Specifically, in structural studies of bison NHA2, the protein was shortened of 69 amino acids at the N‐terminus to reduce predicted disorder. 23 We have found that this region is important for the regulation of HsNHA2 activity (Figure 6). Our data show that, in HsNHA2, molecular dissection of this region (aa 1–70) neither affected protein localization (Figure 6c) nor substrate specificity (Figure 6a). However, truncation of the first 50–70 aa from the N‐terminus provided cells with higher tolerance to LiCl and NaCl (Figure 6a) and resulted in increased Na+‐transport activity (Figure 6b). The activity of various Na+/H+ antiporters, members of the CPA1 clade, was found to be mainly regulated via their large C‐terminal hydrophilic domains. A number of phosphorylation sites and binding sites that interact with regulatory proteins were found in these C‐terminal segments. 3 , 20 , 25 , 51 Nevertheless, our results revealed a novel and unique auto‐inhibitory function of the N‐terminus among human Na+/H+ antiporters. So far, an auto‐inhibitory role of the hydrophilic N‐terminus has been described in some other transporters, for example, the human Ca2+ and Mn2+ transporter TMEM165 42 or Na+/HCO3 − co‐transporter NBCe1‐B, 40 plant Ca2+/H+ antiporters 52 or yeast vacuolar Na+, K+/H+ antiporter Vnx1. 53
The N‐terminus of HsNHA2 is poorly conserved except for the region between amino acids 45 and 60 (Figure 7a). We identified two glutamates (E47, E56) and two lysines (K57, K58) within this region as important residues for HsNHA2 activity (Figure 7b), but not essential for the protein's stability (Figure 7d) or its localization in the plasma membrane (Figure 7c). In particular, the replacement of these residues with an amino acid with the opposite charge resulted in HsNHA2 with a decreased capacity to transport both its substrates, Li+ and Na+ (Figure 7b).
In bison NHA2ΔN1–69, the TMS1 (aa 86–106) formed a unique homodimer interface with a large intracellular gap between the protomers, which closes in the presence of phosphoinositol lipids, and the lipid‐compacted form was more active. 23 Our results confirm that truncations larger than 75 amino acids in HsNHA2 in the immediate proximity of TMS1 (aa 82–105; Figure 7a) led to the loss of function of the antiporter (Figure 6a), most likely due to protein misfolding, and consequently mislocalization (Figure 6c). The N‐terminus seems to be also important for the stability of the protein, because with truncations larger than 70 amino acids we observed a decrease in the protein amount in cells (Figure 6d). Nevertheless, our results clearly show that the N‐terminal hydrophilic extension preceding TMS1 is an important part of the protein involved in the regulation of HsNHA2's function. Whether the structure of the conserved region between amino acids 45–60 or some posttranslational modification in this region are crucial for the regulation of HsNHA2 is currently studied in our lab. Charged residues present in this segment could determine the flexibility of the N‐terminus or they could be involved in intramolecular interactions and/or substrate attraction.
The core domain of Na+/H+ antiporters encapsulating the ion binding site includes two unwound transmembrane helices that cross each other in the middle of the membrane near the ion‐binding site, creating an X‐shaped structure, characteristic of the NhaA fold. 44 As for the membrane domain of HsNHA2, a comprehensive evolutionary analysis of 6,537 representative CPAs revealed a sequence motif of 8 amino acids residing on different transmembrane segments, but spatially close to each other around the substrate binding site. 2 In HsNHA2, this motif A2431V2442S2453P2464…‐5…D2786D2797….R4328 lacks the glutamate at position 5 present in other electrogenic CPA2 members. It was suggested that the electroneutrality of mammalian NHA2 is instead ensured by another glutamate (E215 in HsNHA2) in TMS 5 (Figures S1 and 8b) which forms a salt bridge with R432 at position 8 of the motif in TMS 12. 2 The glutamate residue located in TMS 5 (Figures 1 and S1) is unique in the HsNHA2 and its homologs and cannot be found in other CPA2 members. 2 Cryo‐EM structures of N‐terminally truncated human (PDB 7B4M or 7B4L) or bison NHA2 confirmed the proximity of these two residues (Figure 8b; Protein Data Bank 7B4M or 7B4L 23 ). Our mutagenesis studies of full version of HsNHA2 corroborate the results obtained for bison NHA2ΔN that, in contrast to non‐functional single‐mutated versions (E215R or R432E), swapping up of E215 and R432 results in a Li+‐selective transporter that transports Li+ within the pH range 4.0–6.0 (Figure 4a,d). In addition, we show that the interaction between these two residues is also important for its structural integrity, and hence for the trafficking of the protein (Figure 4b,c).
Another residue that our study found to be highly important for the determination of HsNHA2 transport properties is Pro246, located at position 4 in the conserved CPA motif close to the cation binding site D279 (Figure 8c). The presence of proline in any TMS perturbs the α‐helical structure due to the flexibility of the ring structure of its side chain and the elimination of helix backbone hydrogen bonds for the carbonyls at positions i‐3 and i‐4. 54 In HsNHA2, Pro246 is located in TMS 6 (Figures 1 and S1), which is one of the two unwound transmembrane segments that create the X‐shaped structure in CPAs' core domain (Figure 8c). Depending on the nature of the amino acid substituted at this position, we observed changes in the substrate specificity (ability to recognize and transport Li+ or Na+) of the antiporter (Figure 2a). Mutations of the corresponding proline residue to polar amino acids (Ser, Thr) in yeast Na+/H+ antiporters also resulted in altered substrate specificity toward larger cations. 37 In human NHE1, the replacement of this residue with alanine or glycine resulted in the loss of transport activity. 55 , 56 , 57 In Schizosacchromyces pombe sod2 antiporter, mutations of corresponding proline affected its structure and localization. 58 According to our results, the replacement of P246 with G, A, S or T did not change the targeting of HsNHA2 to the plasma membrane or its stability (Figure 2b,c). Nevertheless, in particular the presence of the polar‐branched threonine resulted in an antiporter that transports better Na+ than Li+.
The cytosolic pH in growing S. cerevisiae cells is kept around a neutral value 7.0–7.2 59 and the activity of the native HsNHA2 antiporter could only be detected at pH 4.0–5.5 (Figures 2a,e and 4a,d), that is, when there is a high gradient of protons across the plasma membrane. Interestingly, mutations P246S/T altered the pH dependent transport activity of HsNHA2, as it was highly active within the external pH range 4.0–7.4 (Figure 2a,e). Remarkably, measurements of Li+ binding by the purified E. coli NhaA showed that a variant mutated at the corresponding residue at position 4 of the conserved CPA motif to threonine (I134T) also exhibited a decrease in pH dependence, but the affinity of this mutated version to Li+ increased. 60 Similar to what was observed in EcNhaA, the altered pH‐dependent activity of the P246 mutants could arise from the fact that this position is spatially close to the two aspartic acids in the antiporter's binding site (D278 and D279). Specifically, the position equivalent to D279 was shown repeatedly to facilitate sodium binding and proton transport, apparently by alternating between a protonated and deprotonated state. 2 , 31 , 61 , 62 , 63 It follows that the intrinsic pKa of this residue affects the antiporter's pH‐dependent ion transport. D279's pKa shift in the P246 mutants can arise from two reasons. First, replacing the conformational‐rigid proline with any other amino acid could alter the geometry of the highly conserved binding site. This could modify, for example, D279 hydration, thereby altering its pKa. Second, the most significant effect on NHA2's pH‐dependent activity was observed for the P246T mutant. This implies that the addition of a polar residue like threonine in a position that is spatially close to D279 could alter its pKa. Here we show in vivo that indeed mutations in position 246 can modulate cation and proton transport in HsNHA2.
Mutations of proline 246 to G, A, S or T provided the antiporter with highly increased resistance to the HsNHA2‐specific inhibitor phloretin (Figure 3). It was found to efficiently inhibit for example, the enzymatic activity of tyrosinase 64 or glucose transporters GLUT, 65 , 66 and thus it influences various physiological processes. 67 , 68 , 69 Molecular docking showed that hydrophobic interaction and hydrogen bonding are the main non‐covalent interaction forces between phloretin and tyrosinase. 64 So far, the molecular mechanism of the phloretin inhibition of HsNHA2 has not been studied. Here we show that the inhibition effect of phloretin did not change with mutations E215R + R432E in HsNHA2; however, it decreased significantly with alterations at the position of P246 (Figure 3). Thus, our results indicate that phloretin acts directly in the core domain of HsNHA2 and mutations of P246 possibly cause inhibitor resistance by altering the binding site's geometry near the substrate/inhibitor‐binding pocket, resulting from the substitution of the conformational‐rigid proline with other amino acids. Figure 9 depicts a model of the bound phloretin to the native HsNHA2 produced with Glid. 70 , 71 Phloretin is predicted to directly interact with D279 in HsNHA2's binding site.
FIGURE 9.
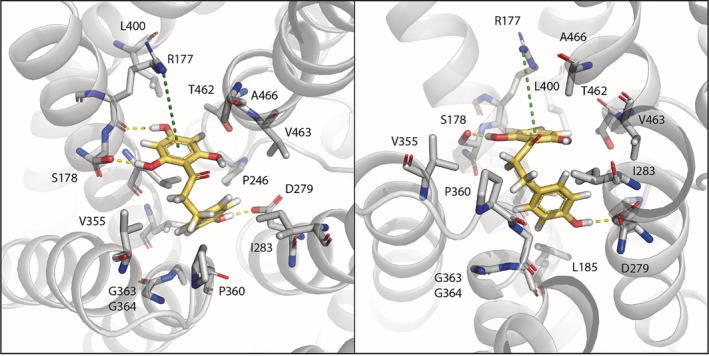
A model of phloretin binding to HsNHA2. Phloretin, shown in yellow, was docked to the crystal structure of human NHA2 (PDB entry 7B4L) as described in Material and Methods and is shown in two different views. Residues within a radius of 5 Å from phloretin are shown as sticks. Phloretin is stabilized by three amino acids. The 2,4,6‐trihydroxyphenyl ring interacts via hydrogen bonds with the side chain hydroxyl of S178 and the backbone carbonyl of R177. It also forms cation‐pi interaction with the side chain guanidinium of R177. The 4‐hydroxyphenyl ring hydrogen bonds with the highly conserved D279 in HsNHA2 binding site, that facilitates both proton and cation transport
There are significant differences between the genomes of human individuals due to single‐nucleotide variants (polymorphisms, SNPs). In this study, we characterized the effect of six mutations existing as SNPs in human NHA2 39 on the antiporter's transport activity, substrate specificity and localization (Figure 5). The positions of mutated residues within the HsNHA2 monomer in relation to the cation‐binding site are shown in Figure 8d. Residues G238, V240, D278 and R432, though they do not coordinate the transported cation (Na+ or Li+) directly, are all located on the mobile and highly conserved core domain of the antiporter (Figure 8e). Our data show that this in vivo experimental approach can be highly useful for a rapid screening of SNP's effect on HsNHA2 activity. Here, it enabled us to distinguish the various effects of mutations on HsNHA2 properties at the molecular level (e.g., effect on substrate specificity—R432Q; transport mechanism—D278G or protein folding and trafficking—G238R; Figure 5). A more detail quantification of the total amount of proteins in cells with the estimation of their amount in the plasma membrane would bring even more knowledge on the effect of particular SNP(s) on the structure, stability and plasma‐membrane targeting of HsNHA2.
Remarkably, the K382E mutation was found to be associated with hypertension. 39 Positively charged residues are more abundant in the cytoplasmic side of many membrane proteins (“positive‐inside rule”) and the substitution of such residue with an opposite charge could potentially affect the proper insertion and orientation of the protein to the membrane. 72 , 73 , 74 Notably, the positively charged K382 is located far from the cation‐binding site, facing the cytoplasm (Figure 8d), its replacement with negatively charged glutamate did not change the plasma‐membrane localization of the antiporter (Figure 5b), but it resulted in restricted substrate specificity for Li+ alone and very low transport activity (ability to improve the LiCl tolerance of cells) (Figure 5a). On the other hand, the V240L mutation in the antiporter's core domain (Figure 8d,e) had only minor effect on HsNHA2's functionality (Figure 5), probably, due to its conservative nature. Furthermore, these results are in line with V240 positioning roughly 12 Å away from where the sodium ion binds (based on the Na+‐bound structure of the HsNHA2; PDB entry 7B4L; Figure 8d). Thus, the combination of our experimental expression model for HsNHA2 with molecular modeling can be used to characterize various pathology‐related SNPs that exist in the HsNHA2 gene in the future.
4. CONCLUSION
In general, our work provides new and important knowledge on using S. cerevisiae as a host for studying human membrane transporters. Specifically, it provides new valuable knowledge on the structure and function of the human Na+/H+ antiporter NHA2 that has broad physiological functions in the human body, and whose mal‐functioning results in a growing list of pathologies. We identified several amino‐acid residues important for HsNHA2 substrate specificity, transport activity, stability and trafficking that had not been characterized previously. Furthermore, our optimized expression system of HsNHA2 in yeast cells may be a powerful tool for the efficient screening of compounds that can activate/inhibit HsNHA2 activity, for the selection of other inhibitor‐resistant mutants to elucidate structure–activity relationships, or for discovering the molecular mechanism of inhibitors' actions in HsNHA2. Newly identified mutated versions of HsNHA2 with higher activity and higher resistance to phloretin than the native antiporter may also serve as controls in future HsNHA2 studies. Moreover, our findings concerning a unique regulatory/autoinhibitory role of the hydrophilic N‐terminal part of the protein will definitely be useful for future studies of NHA2 regulation in human cells.
5. MATERIALS AND METHODS
5.1. Yeast strains and growth media
Alkali‐metal‐cation‐sensitive S. cerevisiae W303‐1A derivatives—BW31 (ena1Δ::HIS3::ena4Δ nha1Δ::LEU2) 37 or AB11c (ena1Δ::HIS3::ena4Δ nha1Δ::LEU2 nhx1Δ::TRP1) 75 lacking genes encoding plasma‐membrane Ena ATPases, plasma‐membrane antiporter Nha1 or intracellular antiporter Nhx1 were used to characterize HsNHA2 activity in vivo. Yeast cultures were routinely grown in YPD (Formedium, Hunstanton, UK) or YNB media (Difco, Sparks, MD) at 30°C. YNB‐based media (referred in this work as YNB‐Pro) contained 0.17% YNB without amino acids and ammonium sulfate, 0.1% proline, and 2 or 4% glucose. Proline was used instead of ammonium sulfate as a nitrogen source. A mixture of appropriate auxotrophic supplements (adenine, tryptophan, leucine, and histidine—each at a final concentration of 20 or 40 μg/ml) was added after autoclaving. YPD media were supplemented with extra adenine (final concentration 20 μg/ml) to avoid additional spontaneous mutagenesis caused by the ade2 mutation present in both strains. Solid media were prepared by adding 2 or 3% (w/v) agar.
5.2. Plasmids
The plasmids used in this study are listed in Table 1. All new plasmids were constructed by homologous recombination in the S. cerevisiae BW31 strain by using a series of overlapping PCR products and a linearized plasmid (pNHA1‐985, pENA1, or pENA1‐GFP; Table 1), in which the original gene was replaced by the corresponding HsNHA2 cDNA or its appropriately truncated versions. The oligonucleotides used for PCR and the construction of particular plasmids are listed in Table S1. HsNHA2 cDNA encoding full or N‐terminally truncated versions of the antiporter were amplified from pTEF1‐HsNHA2‐HA (Table 1). The sequence corresponding to GFP or the ScTPS1 terminator were amplified from pGRU1 or YEp352‐ZrSTL1‐TPS1t‐kanMX, respectively (Table 1). In all new plasmids, the expression of HsNHA2 variants was under the control of the weak and constitutive S. cerevisiae NHA1 promoter. 20 The resulting plasmids were based on multicopy vectors YEp352 or pGRU1 (Table 1). Plasmids containing N‐terminally GFP‐tagged versions of HsNHA2 (native or truncated) were constructed by inserting a GFP‐encoding sequence in frame at the 5′‐end. In pHsNHA2‐GFP, the GFP was added in frame at the 3′‐end in front of the STOP codon. In pHsNHA2t and pGFP‐HsNHA2t plasmids, the ScTPS1 terminator was inserted behind the HsNHA2 cDNA. To generate centromeric versions of pHsNHA2t and pGFP‐HsNHA2t, the 2μ sequence was replaced by homologous recombination with the sequence corresponding to a centromere amplified from pRS316, 21 and the resulting plasmids were named pC‐HsNHA2t and pC‐GFP‐HsNHA2t (Table 1). Escherichia coli XL1‐Blue (Agilent Technologies, Santa Clara, CA) was used for pDNA amplification. Successful cloning was verified by restriction analysis and sequencing.
Point mutations were introduced into pHsNHA2t or pGFP‐pHsNHA2t using a QuikChange XL Site‐Directed Mutagenesis kit (Agilent Technologies) and the corresponding oligonucleotides (Table S1). The accuracy of mutations was confirmed by sequencing.
5.3. Growth assays
To estimate the cell tolerance to different alkali‐metal cations, YNB‐Pro media were supplemented with LiCl, NaCl or KCl at the indicated concentration. HCl or TEA (triethylamine) were used to adjust the pH. Media with buffered pH were supplemented either with 20 mM MES (pH 5.5 and pH 6.0) or 20 mM MOPS (pH 7.0 and pH 7.4), and the required pH was adjusted as above. To test the activity inhibition of HsNHA2 and its variants, media were supplemented with a solution of phloretin (Merck, Darmstadt, Germany; cat. no. P7912) dissolved in DMSO (100 mM stock solution). Tenfold serial dilutions of fresh cell suspensions (OD600 = 2, Eppendorf BioPhotometer, Hamburg, Germany) were spotted on plates as indicated, and growth was monitored for 2–10 days. Representative results of at least three independent experiments are shown.
5.4. Fluorescence microscopy
Microscopic images of yeast cells were acquired with an Olympus BX53 microscope with an Olympus DP73 camera (Olympus, Tokio, Japan). A Cool LED light source with 460 nm excitation and 515 nm emission was used to visualize HsNHA2 and its variants or ScNha1 tagged with GFP (expressed from plasmids pGFP‐HsNHA2t, pHsNHA2‐GFP or pNHA1‐985GFP, respectively, Table 1). Cells containing the corresponding multi‐copy plasmids were grown overnight in YNB‐Pro with 4% glucose and supplemented with adenine and tryptophan (final concentration 40 μg/ml), and leucine and histidine (final concentration 20 μg/ml). Cells were observed when they reached the exponential phase (OD600 ≈ 0.3–0.5). A Nomarski prism was used for whole‐cell images.
5.5. Cation loss determination
Sodium efflux was determined similarly as previously described. 37 Cells were grown in YNB‐Pro medium to the early exponential phase (OD600 ≈ 0.2–0.3, Spekol, Carl Zeiss, Oberkochen, Germany), harvested, resuspended, and incubated in YNB‐Pro supplemented with 100 mM NaCl at pH 7.0 (to preload them with sodium) for 60 min at 30°C. Cells were collected by centrifugation, washed with deionized water, and resuspended in an incubation buffer (10 mM Tris, 0.1 mM MgCl2, 2% glucose). The pH of the buffer was first brought down to 3.9 with citric acid and then adjusted to 4.0 with Ca(OH)2. The buffer was supplemented with 10 mM KCl to prevent Na+ reuptake. Samples of cell suspensions were withdrawn at intervals over 90 min. Cells were harvested by filtration (Millipore membrane filters, Merck‐Millipore, Cork, Ireland), washed with 20 mM MgCl2, acid extracted, and the cation content in samples was determined by atomic absorption spectrometry. 37 Data obtained in at least three independent experiments are shown as the average amount of sodium lost from cells over 90 min ±SD.
5.6. Protein extraction and immunoblotting
Yeast cells containing the pGFP‐HsNHA2t plasmid or its variants were grown in liquid YNB‐Pro media to the exponential phase (OD600 ≈ 0.6–0.8, Spekol, Carl Zeiss), collected by centrifugation, and washed once with deionized cold water. Cell pellets were kept at −80°C. Total extracts of proteins from cells were then prepared as described previously. 76 The RCDC protein assay (Bio‐Rad, Hercules, CA) was used for the quantification of proteins. Samples of total protein extracts (120 μg per sample) were separated by 10% SDS‐PAGE and transferred to nitrocellulose membranes (Trans‐Blot Turbo 0.2 μm nitrocellulose, Bio‐Rad) using a Trans‐Blot Turbo Transfer System (Bio‐Rad). Membranes were incubated overnight at 4°C with an anti‐GFP monoclonal antibody (Roche, Basel, Switzerland; dilution 1:500), washed and then incubated with a secondary anti‐mouse IgG antibody tagged with a horseradish peroxidase (GE Healthcare, Chicago, IL; dilution 1:10,000). Immunoreactive proteins were visualized with a Clarity Max Western ECL substrate kit (Bio‐Rad) in ChemiDoc visualizer (Bio‐Rad). To detect Pgk1 protein (as a protein loading control), the same membranes used for GFP‐HsNHA2 were reprobed after being incubated with an anti‐Pgk1 mouse monoclonal antibody (Abcam, Cambridge, UK; dilution 1:20,000) for 1 hr at room temperature, incubated with the secondary antibody, and detected as above.
5.7. HsNHA2 evolutionary conservation analysis
The conservation analysis for HsNHA2's N‐terminus was conducted using the ConSurf web server. 77 A total of 99 unique sequences were collected by HMMER from the UniRef90 database with sequence identity ranging between 35 and 95% and an E‐value lower than 10−5.
5.8. Modeling of phloretin binding to HsNHA2
A molecule of phloretin was docked to the crystal structure of human NHA2 (PDB entry 7B4L) using Glide. 70 The crystal structure was initially prepared for docking using the Protein Preparation Wizard. 78 The protonation states of all residues were determined with PROPKA3, 79 with an exception of D278 and D279, which were modeled in all combinations of protonation states. In Figure 9, we arbitrarily show both D278 and D279 in their deprotonated state. The docking poses were scored and selected according to the extra precision scoring function (XP). 71 The docking score for the HsNHA2‐phloretin complex ranges from −4.9 to −6.4 kcal/mol depending on the different protonation states of D278 and D279. It should be noted that despite their energy units, docking scores are not equivalent to real world ΔG values, as they are calculated based on a single conformation of the ligand‐protein complex and neglect dynamic and entropic contributions associated with the small molecule binding. The figure was produced using PyMOL (The PyMOL Molecular Graphics System, Version 2.5, Schrödinger).
5.9. Statistics
Data presented are either from a representative experiment or from the mean of replicative values plus or minus standard deviation. Statistically significant differences were analysed by unpaired Student t‐test using Microsoft Office Excel 2016 (**p < .01).
AUTHOR CONTRIBUTIONS
Diego Velázquez: Conceptualization (equal); data curation (equal); investigation (lead); visualization (equal); writing – original draft (equal); writing – review and editing (supporting). Vojtěch Průša: Investigation (supporting). Gal Masrati: Data curation (supporting); investigation (supporting); visualization (supporting); writing – review and editing (supporting). Elon Yariv: Investigation (supporting). Hana Sychrova: Supervision (supporting); writing – review and editing (supporting). Nir Ben‐Tal: Data curation (supporting); funding acquisition (supporting); supervision (equal); writing – review and editing (equal). Olga Zimmermannova: Conceptualization (lead); data curation (equal); formal analysis (lead); funding acquisition (lead); investigation (supporting); methodology (lead); project administration (lead); supervision (lead); validation (lead); writing – original draft (lead); writing – review and editing (lead).
FUNDING INFORMATION
The work of Olga Zimmermannova group was supported by a GAČR grant 21‐08985S. The work of Diego Velázquez was supported by IPHYS Mobility II, CZ.02.2.69/0.0/0.0/18_053/0016977. The work of Nir Ben‐Tal group was supported by Grant 2017293 of the USA–Israel Binational Science Foundation and the Abraham E. Kazan Chair in Structural Biology, Tel Aviv University.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Table S1. Oligonucleotides used in this study.
Figure S1. The membrane topology of HsNHA2.
Figure S2. Comparison of localization of GFP‐HsNHA2 in two yeast backgrounds.
Figure S3. N‐terminal GFP‐tagging does not change substrate specificity, but increases activity of HsNHA2 mutated versions.
Figure S4. N‐terminal GFP‐tagging does not change substrate specificity of HsNHA2 versions with mutations that belong to known human SNPs.
Figure S5. N‐terminal GFP‐tagging altered LiCl tolerance provided by HsNHA2 versions truncated at N‐terminus.
ACKNOWLEDGMENTS
The technical assistance of Pavla Herynková is highly acknowledged. We thank Daniel Fuster, MD, for providing us the plasmid pTEF1‐HsNHA2‐HA. We wish to thank Dr. Raghu Metpally and Prof. Rajini Rao for providing information about mutations identified as SNPs in HsNHA2 that we characterized in this study. Prof. Rajini Rao and Prof. Pierre Falson are highly acknowledged also for a stimulating and fruitful discussion.
Velázquez D, Průša V, Masrati G, Yariv E, Sychrova H, Ben‐Tal N, et al. Allosteric links between the hydrophilic N‐terminus and transmembrane core of human Na+/H+ antiporter NHA2 . Protein Science. 2022;31(12):e4460. 10.1002/pro.4460
Review Editor: John Kuriyan
Funding information Grantová Agentura České Republiky, Grant/Award Number: 21‐08985S; IPHYS Mobility II, Grant/Award Number: CZ.02.2.69/0.0/0.0/18_053/0016977; USA‐Israel Binational Science Foundation and the Abraham E. Kazan Chair in Structural Biology, Tel Aviv University, Grant/Award Number: 2017293
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Padan E, Landau M. Sodium‐proton Na+/H+ antiporters: Properties and roles in health and disease. Metal Ions Life Sci. 2016;16:391–458. 10.1007/978-3-319-21756-7_12. [DOI] [PubMed] [Google Scholar]
- 2. Masrati G, Dwivedi M, Rimon A, et al. Broad phylogenetic analysis of cation/proton antiporters reveals transport determinants. Nat Commun. 2018;9:4205. 10.1038/s41467-018-06770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pedersen SF, Counillon L. The SLC9A‐C mammalian Na+/H+ exchanger family: Molecules, mechanisms, and physiology. Physiol Rev. 2019;99:2015–2113. 10.1152/physrev.00028.2018. [DOI] [PubMed] [Google Scholar]
- 4. Fuster DG, Zhang J, Shi M, Bobulescu IA, Andersson S, Moe OW. Characterization of the sodium/hydrogen exchanger NHA2. J Am Soc Nephrol. 2008;19:1547–1556. 10.1681/ASN.2007111245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Battaglino RA, Pham L, Morse LR, et al. NHA‐oc/NHA2: A mitochondrial cation‐proton antiporter selectively expressed in osteoclasts. Bone. 2008;42:180–192. 10.1016/j.bone.2007.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiang M, Feng M, Muend S, Rao R. A human Na+/H+ antiporter sharing evolutionary origins with bacterial NhaA may be a candidate gene for essential hypertension. Proc Natl Acad Sci U S A. 2007;104:18677–18681. 10.1073/pnas.0707120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chintapalli VR, Kato A, Henderson L, et al. Transport proteins NHA1 and NHA2 are essential for survival, but have distinct transport modalities. Proc Natl Acad Sci U S A. 2015;112:11720–11725. 10.1073/pnas.1508031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deisl C, Simonin A, Anderegg M, et al. Sodium/hydrogen exchanger NHA2 is critical for insulin secretion in beta‐cells. Proc Natl Acad Sci U S A. 2013;110:10004–10009. 10.1073/pnas.1220009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deisl C, Anderegg M, Albano G, et al. Loss of sodium/hydrogen exchanger NHA2 exacerbates obesity‐ and aging‐induced glucose intolerance in mice. PLoS One. 2016;11:e0163568. 10.1371/journal.pone.0163568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu HM, He JY, Zhang Q, et al. Improved detection of genetic loci in estimated glomerular filtration rate and type 2 diabetes using a pleiotropic cFDR method. Mol Genet Genomics. 2018;293:225–235. 10.1007/s00438-017-1381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kondapalli KC, Todd Alexander R, Pluznick JL, Rao R. NHA2 is expressed in distal nephron and regulated by dietary sodium. J Physiol Biochem. 2017;73:199–205. 10.1007/s13105-016-0539-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderegg MA, Albano G, Hanke D, et al. The sodium/proton exchanger NHA2 regulates blood pressure through a WNK4‐NCC dependent pathway in the kidney. Kidney Int. 2021;99:350–363. 10.1016/j.kint.2020.08.023. [DOI] [PubMed] [Google Scholar]
- 13. Prasad H, Dang DK, Kondapalli KC, Natarajan N, Cebotaru V, Rao R. NHA2 promotes cyst development in an in vitro model of polycystic kidney disease. J Physiol. 2019;597:499–519. 10.1113/JP276796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen SR, Chen M, Deng SL, Hao XX, Wang XX, Liu YX. Sodium‐hydrogen exchanger NHA1 and NHA2 control sperm motility and male fertility. Cell Death Dis. 2016;7:e2152. 10.1038/cddis.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ha BG, Hong JM, Park JY, et al. Proteomic profile of osteoclast membrane proteins: Identification of Na+/H+ exchanger domain containing 2 and its role in osteoclast fusion. Proteomics. 2008;8:2625–2639. 10.1002/pmic.200701192. [DOI] [PubMed] [Google Scholar]
- 16. Lee SH, Kim T, Park ES, et al. NHE10, an osteoclast‐specific member of the Na+/H+ exchanger family, regulates osteoclast differentiation and survival [corrected]. Biochem Biophys Res Commun. 2008;369:320–326. 10.1016/j.bbrc.2008.01.168. [DOI] [PubMed] [Google Scholar]
- 17. Hofstetter W, Siegrist M, Simonin A, Bonny O, Fuster DG. Sodium/hydrogen exchanger NHA2 in osteoclasts: Subcellular localization and role in vitro and in vivo. Bone. 2010;47:331–340. 10.1016/j.bone.2010.04.605. [DOI] [PubMed] [Google Scholar]
- 18. Charles JF, Coury F, Sulyanto R, et al. The collection of NFATc1‐dependent transcripts in the osteoclast includes numerous genes non‐essential to physiologic bone resorption. Bone. 2012;51:902–912. 10.1016/j.bone.2012.08.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 20. Kinclova O, Ramos J, Potier S, Sychrova H. Functional study of the Saccharomyces cerevisiae Nha1p C‐terminus. Mol Microbiol. 2001;40:656–668. 10.1046/j.1365-2958.2001.02412.x. [DOI] [PubMed] [Google Scholar]
- 21. Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duskova M, Ferreira C, Lucas C, Sychrova H. Two glycerol uptake systems contribute to the high osmotolerance of Zygosaccharomyces rouxii . Mol Microbiol. 2015;97:541–559. 10.1111/mmi.13048. [DOI] [PubMed] [Google Scholar]
- 23. Matsuoka R, Fudim R, Jung S, et al. Structure, mechanism and lipid‐mediated remodeling of the mammalian Na+/H+ exchanger NHA2. Nat Struct Mol Biol. 2022;29:108–120. 10.1038/s41594-022-00738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Varadi M, Anyango S, Deshpande M, et al. Alphafold protein structure database: Massively expanding the structural coverage of protein‐sequence space with high‐accuracy models. Nucleic Acids Res. 2022;50:D439–D444. 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hendus‐Altenburger R, Kragelund BB, Pedersen SF. Structural dynamics and regulation of the mammalian SLC9A family of Na+/H+ exchangers. Curr Top Membr. 2014;73:69–148. 10.1016/B978-0-12-800223-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 26. Chow CW, Woodside M, Demaurex N, et al. Proline‐rich motifs of the Na+/H+ exchanger 2 isoform‐binding of Src homology domain 3 and role in apical targeting in epithelia. J Biol Chem. 1999;274:10481–10488. 10.1074/jbc.274.15.10481. [DOI] [PubMed] [Google Scholar]
- 27. Onishi I, Lin PJ, Diering GH, Williams WP, Numata M. Rack1 associates with NHE5 in focal adhesions and positively regulates the transporter activity. Cell Signal. 2007;19:194–203. 10.1016/j.cellsig.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 28. Fukura N, Ohgaki R, Matsushita M, Nakamura N, Mitsui K, Kanazawa H. A membrane‐proximal region in the C‐terminal tail of NHE7 is required for its distribution in the trans‐Golgi network, distinct from NHE6 localization at endosomes. J Membr Biol. 2010;234:149–158. 10.1007/s00232-010-9242-9. [DOI] [PubMed] [Google Scholar]
- 29. Uzdavinys P, Coinçon M, Nji E, et al. Dissecting the proton transport pathway in electrogenic Na+/H+ antiporters. Proc Natl Acad Sci U S A. 2017;114:E1101–E1110. 10.1073/pnas.1614521114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kondapalli KC, Kallay LM, Muszelik M, Rao R. Unconventional chemiosmotic coupling of NHA2, a mammalian Na+/H+ antiporter, to a plasma membrane H+ gradient. J Biol Chem. 2012;287:36239–36250. 10.1074/jbc.M112.403550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schushan M, Xiang M, Bogomiakov P, Padan E, Rao R, Ben‐Tal N. Model‐guided mutagenesis drives functional studies of human NHA2, implicated in hypertension. J Mol Biol. 2010;396:1181–1196. 10.1016/j.jmb.2009.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Flegelova H, Haguenauer‐Tsapis R, Sychrova H. Heterologous expression of mammalian Na+/H+ antiporters in Saccharomyces cerevisiae . Biochim Biophys Acta. 2006;1760:504–516. 10.1016/j.bbagen.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 33. Flegelova H, Sychrova H. Mammalian NHE2 Na+/H+ exchanger mediates efflux of potassium upon heterologous expression in yeast. FEBS Lett. 2005;579:4733–4738. 10.1016/j.febslet.2005.07.046. [DOI] [PubMed] [Google Scholar]
- 34. Banuelos MA, Ruiz MC, Jiménez A, Souciet JL, Potier S, Ramos J. Role of the Nha1 antiporter in regulating K+ influx in Saccharomyces cerevisiae . Yeast. 2002;19:9–15. 10.1002/yea.799. [DOI] [PubMed] [Google Scholar]
- 35. Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. 10.1128/MMBR.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamanishi M, Katahira S, Matsuyama T. TPS1 terminator increases mRNA and protein yield in a Saccharomyces cerevisiae expression system. Biosci Biotechnol Biochem. 2011;75:2234–2236. 10.1271/bbb.110246. [DOI] [PubMed] [Google Scholar]
- 37. Kinclova‐Zimmermannova O, Zavrel M, Sychrova H. Identification of conserved prolyl residue important for transport activity and the substrate specificity range of yeast plasma membrane Na+/H+ antiporters. J Biol Chem. 2005;280:30638–30647. 10.1074/jbc.M506341200. [DOI] [PubMed] [Google Scholar]
- 38. Behzad S, Sureda A, Barreca D, Nabavi SF, Rastrelli L, Nabavi SM. Health effects of phloretin: From chemistry to medicine. Phytochem Rev. 2017;16:527–533. 10.1007/s11101-017-9500-x. [DOI] [Google Scholar]
- 39. Dewey FE, Murray MF, Overton JD, et al. Distribution and clinical impact of functional variants in 50,726 whole‐exome sequences from the discover study. Science. 2016;354:aaf6814. 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 40. Su P, Wu H, Wang M, Cai L, Liu Y, Chen LM. Irbit activates NBCe1‐B by releasing the auto‐inhibition module from the transmembrane domain. J Physiol. 2021;599:1151–1172. 10.1113/JP280578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thines L, Deschamps A, Stribny J, Morsomme P. Yeast as a tool for deeper understanding of human manganese‐related diseases. Genes (Basel). 2019;10:545. 10.3390/genes10070545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stribny J, Thines L, Deschamps A, Goffin P, Morsomme P. The human Golgi protein TMEM165 transports calcium and manganese in yeast and bacterial cells. J Biol Chem. 2020;295:3865–3874. 10.1074/jbc.RA119.012249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 44. Padan E. Functional and structural dynamics of NhaA, a prototype for Na+ and H+ antiporters, which are responsible for Na+ and H+ homeostasis in cells. Biochim Biophys Acta. 2014;1837:1047–1062. 10.1016/j.bbabio.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 45. Dong Y, Gao Y, Ilie A, et al. Structure and mechanism of the human NHE1‐CHP1 complex. Nat Commun. 2021;12:3474. 10.1038/s41467-021-23496-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winklemann I, Matsuoka R, Meier PF, et al. Structure and elevator mechanism of the mammalian sodium/proton exchanger NHE9. EMBO J. 2020;39:e105908. 10.15252/embj.2020105908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee C, Kang HJ, Von Ballmoos C, et al. A two‐domain elevator mechanism for sodium/proton antiport. Nature. 2013;501:573–577. 10.1038/nature12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paulino C, Wohlert D, Kapotova E, Yildiz O, Kuhlbrandt W. Structure and transport mechanism of the sodium/proton antiporter MjNhaP1. eLife. 2014;3:e03583. 10.7554/eLife.03583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wohlert D, Kuhlbrandt W, Yildiz O. Structure and substrate ion binding in the sodium/proton antiporter PaNhaP. eLife. 2014;3:e03579. 10.7554/eLife.03579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goswami P, Paulino C, Hizlan D, Vonck J, Yildiz Ö, Kühlbrandt W. Structure of the archaeal Na+/H+ antiporter NhaP1 and functional role of transmembrane helix 1. EMBO J. 2011;30:439–449. 10.1038/emboj.2010.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smidova A, Stankova K, Petrvalska O, et al. The activity of Saccharomyces cerevisiae Na+, K+/H+ antiporter Nha1 is negatively regulated by 14‐3‐3 protein binding at serine 481. Biochim Biophys Acta Mol Cell Res. 2019;1866:118534. 10.1016/j.bbamcr.2019.118534. [DOI] [PubMed] [Google Scholar]
- 52. Pittman JK, Sreevidya CS, Shigaki T, Ueoka‐Nakanishi H, Hirschi KD. Distinct N‐terminal regulatory domains of Ca2+/H+ antiporters. Plant Physiol. 2002;130:1054–1062. 10.1104/pp.008193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cagnac O, Baghour M, Jaime‐Pérez N, et al. Deletion of the N‐terminal domain of the yeast vacuolar Na+, K+/H+ antiporter Vnx1p improves salt tolerance in yeast and transgenic arabidopsis. Yeast. 2020;37:173–185. 10.1002/yea.3450. [DOI] [PubMed] [Google Scholar]
- 54. Visiers I, Braunheim BB, Weinstein H. Prokink: A protocol for numerical evaluation of helix distortions by proline. Protein Eng. 2000;13:603–606. 10.1093/protein/13.9.603. [DOI] [PubMed] [Google Scholar]
- 55. Counillon L, Noel J, Reithmeier RA, Pouyssegur J. Random mutagenesis reveals a novel site involved in inhibitor interaction within the fourth transmembrane segment of the Na+/H+ exchanger‐1. Biochemistry. 1997;36:2951–2959. 10.1021/bi9615405. [DOI] [PubMed] [Google Scholar]
- 56. Slepkov ER, Rainey JK, Li X, et al. Structural and functional characterization of transmembrane segment IV of the NHE1 isoform of the Na+/H+ exchanger. J Biol Chem. 2005;280:17863–17872. 10.1074/jbc.M409608200. [DOI] [PubMed] [Google Scholar]
- 57. Slepkov ER, Chow S, Lemieux MJ, Fliegel L. Proline residues in transmembrane segment IV are critical for activity, expression and targeting of the Na+/H+ exchanger isoform 1. Biochem J. 2004;379:31–38. 10.1042/BJ20030884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ndayizeye M, Touret N, Fliegel L. Proline 146 is critical to the structure, function and targeting of sod2, the Na+/H+ exchanger of Schizosaccharomyces pombe . Biochim Biophys Acta. 2009;1788:983–992. 10.1016/j.bbamem.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 59. Zimmermannova O, Felcmanová K, Rosas‐Santiago P, Papoušková K, Pantoja O, Sychrová H. Erv14 cargo receptor participates in regulation of plasma‐membrane potential, intracellular pH and potassium homeostasis via its interaction with K+‐specific transporters Trk1 and Tok1. BBA‐Mol Cell Res. 2019;1866:1376–1388. 10.1016/j.bbamcr.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 60. Mondal R, Rimon A, Masrati G, Ben‐Tal N, Friedler A, Padan E. Towards molecular understanding of the pH dependence characterizing NhaA of which structural fold is shared by other transporters. J Mol Biol. 2021;433:167156. 10.1016/j.jmb.2021.167156. [DOI] [PubMed] [Google Scholar]
- 61. Furrer EM, Ronchetti MF, Verrey F, Pos KM. Functional characterization of a NapA Na+/H+ antiporter from Thermus thermophilus . FEBS Lett. 2007;581:572–578. 10.1016/j.febslet.2006.12.059. [DOI] [PubMed] [Google Scholar]
- 62. Inoue H, Noumi T, Tsuchiya T, Kanazawa H. Essential aspartic acid residues, Asp‐133, Asp‐163 and Asp‐164, in the transmembrane helices of a Na+/H+ antiporter (NhaA) from Escherichia coli . FEBS Lett. 1995;363:264–268. 10.1016/0014-5793(95)00331-3. [DOI] [PubMed] [Google Scholar]
- 63. Kuwabara N, Inoue H, Tsuboi Y, Mitsui K, Matsushita M, Kanazawa H. Structure‐function relationship of the fifth transmembrane domain in the Na+/H+ antiporter of Helicobacter pylori: Topology and function of the residues, including two consecutive essential aspartate residues. Biochemistry. 2006;45:14834–14842. 10.1021/bi061048d. [DOI] [PubMed] [Google Scholar]
- 64. Chen J, Li Q, Ye Y, Huang Z, Ruan Z, Jin N. Phloretin as both a substrate and inhibitor of tyrosinase: Inhibitory activity and mechanism. Spectrochim Acta A Mol Biomol Spectrosc. 2020;226:117642. 10.1016/j.saa.2019.117642. [DOI] [PubMed] [Google Scholar]
- 65. Lin ST, Tu SH, Yang PS, et al. Apple polyphenol phloretin inhibits colorectal cancer cell growth via inhibition of the type 2 glucose transporter and activation of p53‐mediated signaling. J Agric Food Chem. 2016;64:6826–6837. 10.1021/acs.jafc.6b02861. [DOI] [PubMed] [Google Scholar]
- 66. Liu L, Xie H, Zhao S, Huang X. The GLUT1‐mtORC1 axis affects odontogenic differentiation of human dental pulp stem cells. Tissue Cell. 2022;76:101766. 10.1016/j.tice.2022.101766. [DOI] [PubMed] [Google Scholar]
- 67. Casado‐Diaz A, Rodríguez‐Ramos Á, Torrecillas‐Baena B, Dorado G, Quesada‐Gómez JM, Gálvez‐Moreno MÁ. Flavonoid phloretin inhibits adipogenesis and increases OPG expression in adipocytes derived from human bone‐marrow mesenchymal stromal‐cells. Nutrients. 2021;13:4185. 10.3390/nu13114185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wu KH, Ho CT, Chen ZF, et al. The apple polyphenol phloretin inhibits breast cancer cell migration and proliferation via inhibition of signals by type 2 glucose transporter. J Food Drug Anal. 2018;26:221–231. 10.1016/j.jfda.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vadavanath Prabhakaran V, Kozhiparambil Gopalan R. Phloretin alleviates arsenic trioxide‐induced apoptosis of H9c2 cardiomyoblasts via downregulation in Ca2+/calcineurin/NFATc pathway and inflammatory cytokine release. Cardiovasc Toxicol. 2021;21:642–654. 10.1007/s12012-021-09655-0. [DOI] [PubMed] [Google Scholar]
- 70. Friesner RA, Banks JL, Murphy RB, et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 71. Friesner RA, Murphy RB, Repasky MP, et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein‐ligand complexes. J Med Chem. 2006;49:6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 72. Sipos L, von Heijne G. Predicting the topology of eukaryotic membrane proteins. Eur J Biochem. 1993;213:1333–1340. 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- 73. von Heijne G. Membrane‐protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 74. Baker JA, Wong WC, Eisenhaber B, Warwicker J, Eisenhaber F. Erratum to: Charged residues next to transmembrane regions revisited: “positive‐inside rule” is complemented by the “negative inside depletion/outside enrichment rule”. BMC Biol. 2017;15:72. 10.1186/s12915-017-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Maresova L, Sychrova H. Physiological characterization of Saccharomyces cerevisiae kha1 deletion mutants. Mol Microbiol. 2005;55:588–600. 10.1111/j.1365-2958.2004.04410.x. [DOI] [PubMed] [Google Scholar]
- 76. Horak J, Wolf DH. Glucose‐induced monoubiquitination of the Saccharomyces cerevisiae galactose transporter is sufficient to signal its internalization. J Bacteriol. 2001;183:3083–3088. 10.1128/JB.183.10.3083-3088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ashkenazy H, Abadi S, Martz E, et al. Consurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 79. Olsson MH, Sondergaard CR, Rostkowski M, Jensen JH. Propka3: Consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput. 2011;7:525–537. 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Oligonucleotides used in this study.
Figure S1. The membrane topology of HsNHA2.
Figure S2. Comparison of localization of GFP‐HsNHA2 in two yeast backgrounds.
Figure S3. N‐terminal GFP‐tagging does not change substrate specificity, but increases activity of HsNHA2 mutated versions.
Figure S4. N‐terminal GFP‐tagging does not change substrate specificity of HsNHA2 versions with mutations that belong to known human SNPs.
Figure S5. N‐terminal GFP‐tagging altered LiCl tolerance provided by HsNHA2 versions truncated at N‐terminus.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


