Abstract
Many proteins harboring low complexity or intrinsically disordered sequences (IDRs) are capable of undergoing liquid–liquid phase separation to form mesoscale condensates that function as biochemical niches with the ability to concentrate or sequester macromolecules and regulate cellular activity. Engineered disordered proteins have been used to generate programmable synthetic membraneless organelles in cells. Phase separation is governed by the strength of interactions among polypeptides with multivalency enhancing phase separation at lower concentrations. Previously, we and others demonstrated enzymatic control of IDR valency from multivalent precursors to dissolve condensed phases. Here, we develop noncovalent strategies to multimerize an individual IDR, the RGG domain of LAF-1, using protein interaction domains to regulate condensate formation in vitro and in living cells. First, we characterize modular dimerization of RGG domains at either terminus using cognate high-affinity coiled-coil pairs to form stable condensates in vitro. Second, we demonstrate temporal control over phase separation of RGG domains fused to FRB and FKBP in the presence of dimerizer. Further, using a photocaged dimerizer, we achieve optically induced condensation both in cell-sized emulsions and within live cells. Collectively, these modular tools allow multiple strategies to promote phase separation of a common core IDR for tunable control of condensate assembly.
Condensation of proteins and RNAs into micron size “membraneless organelles” is a ubiquitous feature of all cell types facilitating subcellular compartmentalization in the absence of a lipid bilayer.1,2 Polymers can demix from solution via liquid–liquid phase separation (LLPS), forming condensates via multiple weak interactions, often spreading across the polypeptide chain of intrinsically disordered proteins (IDPs).3,4 Many biomolecular condensates are dynamic and exhibit liquid-like behavior including fission and fusion of individual condensates as well as rapid recovery from photobleaching both in vitro and in live cells.5−8 These proteinaceous subcellular compartments serve a wide array of functions, concentrating reactants to accelerate reaction rates or sequestering client factors to shut down pathway activity.1,9 Over the past decade, a number of activities regulated by biomolecular condensates have come into focus. These include those involved in ribosomal biogenesis (nucleoli), RNA processing (Cajal bodies), transcriptional regulation (enhancers and super enhancers), early development (P bodies), and stress response (stress granules).5,10−17 Due to their diverse functionality and feasibility of forming compartments from a single type of disordered polypeptide, condensates are an attractive target for protein engineering, synthetic biology, and construction of new biologically inspired materials.
Unlike typical interactions in a protein complex among folded proteins, disordered polypeptides can self-assemble via multiple, relatively weak interactions between amino acids in the protein chain.18 Because individual interactions are weak, valency is a fundamental strategy of increasing the propensity for LLPS.19−21 If a short peptide sequence is repeated in a larger multimer, the longer polymer has a higher probability of homotypic and heterotypic interactions, thus lowering the required free energy for phase separation.19,22−24 In short, the multivalency of side chain interactions profoundly effects the saturation concentration (Csat) for protein condensation, the liquid versus gel-like properties of condensates, and the kinetics of coarsening.25−28 The extent of multivalency can be hard coded in the amino acid sequence or through post-translational modifications.20,23,29−33 Both strategies have been explored to fine tune the extent of protein condensation under physiological conditions.20,25,31,34−36 In cells, the viscoelastic properties of condensates can be altered by the presence of other multivalent polymers such as RNA.37,38 For example, the C. elegans P granule protein LAF-1 is capable of LLPS independent of RNA in vitro, but the presence of RNA helps fluidize LAF-1 condensates.6 In other cases, multivalency was used to lower the concentration of polypeptide required for self-assembly of elastin and resilin-like polymers.39−41
A challenge in protein engineering is how to regulate the valency of sequence-defined polymers in real time using genetically encoded components under physiological conditions. Simply encoding a longer, higher repeat polypeptide sequence removes the flexibility to alter polymer valency and for temporal control of phase separation. Several strategies exist to increase the chain length of existing polymers. One strategy uses chemical cross-linking such as intermolecular chain cross-linking via cysteine disulfide bonds.42 Such a strategy was used to condense oleosin in a redox-responsive manner.42 Short, elastin-like polypeptides can assemble into multivalent chains via cross-linking of non-native amino acids to control gelation.43 However, due to their length, these sequences are not compatible with tagging with larger folded helices or domains, which in the case of IDRs would increase Csat to ranges of expression that cannot be achieved in cells. Native chemical ligation strategies can be used to multimerize polypeptides, although the chemical conditions may not be ideal for living systems.44 For example, the Spycatcher–spytag system results in a covalent and irreversible conjugation of the two target peptides.45 Alternatively, noncovalent interactions offer an attractive strategy to rapidly multimerize polypeptides using protein interaction domains.46−48 Examples include coiled coils, small-molecule-induced dimerization of FRB-FKBP, optogenetic dimerization of DHFR, Halo, and optogenetic dimerization or multimerization domains.34,47,49 Given the established utility of LAF-1 RGG IDR for engineering synthetic condensates6,20,29,50,51 and the well-characterized biophysical features of its condensates, we chose to use this sequence for testing multimerization strategies in the present study.
The structure, affinity, and design of helical coiled-coil interaction domains are well characterized, inspiring a route for noncovalent control of IDR multivalency compatible via genetic encoding in living cells. Characterized by heptad sequence repeats, alpha-helical coils are common in endogenous proteins, and the rules of their interactions are well established for protein–protein binding.52 They have been engineered to generate orthogonal synthetic heterodimeric helical peptides that have low nanomolar affinities and high sequence selectivity.53−55 A growing body of literature has implemented helical coiled coils to create new connectivities and localizations in cells to regulate cellular functions. We previously used coiled-coil motifs, attached to an IDR and client proteins, to recruit clients to preformed IDR coacervates in vitro and later to insulate endogenous client enzymes within synthetic condensates to control the cell behavior.20,29 Although heterotypic coiled coils have been successful in promoting scaffold–client interactions, their utility for stitching together disordered polypeptides to control protein phase separation has not been explored. Two well-validated examples that we focus on in this work include SYNZIPs and Parallel Peptide Pairs.55−57 In both cases, a variety of short, parallel coiled-coil pairs have been constructed and shown to be orthogonal, interacting with only its designated, cognate coil and no others in the set.56,57 As such, these tools and others like them are ideal for tagging exogenously or endogenously expressed proteins without disrupting their functionality to promote specific, noncovalent binding or dimerization.29
Additional strategies for inducible multimerization or clustering of IDRs leveraged light-responsive protein interaction domains, which have been instrumental in studying the LLPS activity of a number of disordered sequences in cells under illumination conditions.50,58,59 Such tools have been used to enhance or control biochemical reactions in cell culture using the presence and extent of illumination.60,61 Whereas optogenetics require sustained illumination and may be challenging to implement biochemically in vitro, chemogenic and optochemical approaches utilizing small molecular dimerizers and Halo-DHFR or FRB-FKBP domains have been used both in vitro and in cells.49,62 For example, anchoring native factors away from substrates either by relocalization to other native organelles or by insulation within synthetic condensates has been shown to be successful in controlling the cell polarity and proliferation in a predicable manner.29,63,64
In this work, we characterized sets of short, heterodimeric coiled-coil modules to promote the multivalency of a model IDR, the Arg/Gly-rich RGG domain of C. elegans P granule protein LAF-1 and regulate protein condensation in vitro and in live cells. We chose the LAF-1 RGG IDR which has been used to engineer synthetic condensates and whose biophysical determinants for phase separation are understood.6,20,29,50,51 We first identified optimal strategies for tagging the RGG IDP with coiled coils to promote dimerization and condensate formation in vitro. To achieve temporal chemogenic control over condensation, we constructed RGG domains fused to FRB and FKBP domains in vitro and in cells. We also show optical control of condensate formation using a photocaged dimerizer within cell-like compartments in vitro and in live cells, a strategy that requires only a short pulse of light to stably form condensed phases. Collectively, this study reveals new strategies for real-time control of IDP valency providing a regulatory toolkit for protein condensate assembly with applications in protocell engineering and synthetic biology.
Materials and Methods
Protein Expression and Purification
Plasmids encoding N-terminally 6xHis-tagged RGG constructs with coiled-coil tags and 6xHis mCherry-FRB were transformed into BL21(DE3) E. coli cells (Thermo Fisher Scientific; Waltham, MA). Cultures were grown in Luria Broth (LB) containing kanamycin at 37 °C to an OD600 of 0.6–0.8, and expression was induced by 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 16 °C overnight. Cell pellets were collected and stored at −80 °C.
RGG-FKBP and RGG-FRB Purification
Bacterial cell pellets were thawed, resuspended in lysis buffer (50 mM HEPES, pH 6.8, 1 M NaCl, 20 mM imidazole, 1 mM β-mercaptoethanol) containing complete EDTA-free protease inhibitor cocktail (Roche; Mannheim, Germany), and lysed for a total of 3 min of sonication at 50% power using a Branson Sonifier. Lysates were clarified by centrifugation at 13 000 rpm (20,064g) for 20 min in an F21S-8x50y rotor (Thermo Fisher Scientific) at 37 °C and incubated with 0.5 mL of Ni–NTA beads (Thermo Fisher Scientific) at room temperature for 1 h. Beads were then washed three times with 10 mL of lysis buffer. Proteins were eluted by addition of lysis buffer containing 500 mM imidazole and 1 mM DTT. Elutions were diluted to 3 mg/mL in lysis buffer containing 1 mM DTT and dialyzed overnight into single RGG storage buffer (500 mM NaCl, 20 mM HEPES, pH 6.8, 1 mM DTT) using 10 kDa cutoff Slide-A-Lyzer membrane cassettes (Thermo Fisher Scientific). Proteins were concentrated by centrifugation in 4 mL of Amicon filter concentrators with a 10 kDa cutoff (Millipore Sigma; Burlington, MA). TCEP (1 mM) was added prior to snap freezing and storage at −80 °C
Purification of Tandem, RGG-RGG, Fluorescent Tracer (RGG-GFP-RGG), 6xHis-RGG Domains Tagged with Coiled-Coil Dimerizers (P3-6, SZ1/2), and 6xHis-mCherry-FRB
Pellets were thawed and resuspended in lysis buffer. RGG polypeptides that included P3-6 or tags and mCherry-FRB were resuspended in lysis buffer containing 50 mM Tris-HCl, pH 8.5, 1 M NaCl, 20 mM Imidazole, 1 mM β-mercaptoethanol, and a dissolved tablet of protease inhibitor cocktail (Roche). RGG constructs with SZ1/2 tags were treated similarly, but lysis buffer contained 20 mM HEPES pH 6.8. Cells were lysed, cleared by centrifugation, and incubated with Ni–NTA beads as above. Beads were then washed in three column volumes of lysis buffer and eluted with lysis buffer containing 500 mM imidazole and 1 mM DTT. Elutions were diluted to 3 mg/mL in lysis buffer containing 1 mM DTT as above and dialyzed overnight into storage buffer. Constructs with P3–P6 and mCherry-FRB: 1 M NaCl, 20 mM Tris-HCl, pH 8.5, 1 mM DTT. Constructs with SZ1 and SZ2: 1 M NaCl, 20 mM Tris-HCl, pH 6.8, 1 mM DTT. Proteins were concentrated by centrifugation in Amicon filter concentrators with a 10 kDa cutoff before addition of 1 mM TCEP and storage at −80 °C. In all cases, protein concentrations were determined by A280 and Bradford assay (BioRad).
Turbidity Measurements
Protein aliquots were thawed and solubilized at 50 °C and then diluted to 6 μM in buffer to adjust to a final salt concentration of 150 mM in 20 mM, Tris-HCl, pH 8.5. SYNZIP-tagged constructs were similarly adjusted to 10 μM in 150 mM NaCl, 20 mM HEPES, pH 6.8. The protein mixture, 60 μL in volume, was added to quartz microcuvettes (10 mm path length) (Starna Cells, Inc. Atascadero, CA). Cuvettes were inserted into a Cary 3500 UV–vis spectrophotometer controlled by an Agilent multizone Peltier temperature controller (Agilent Technologies; Santa Clara, CA). To test kinetics of rapamycin-induced dimerization and phase separation of FRB and FKBP-tagged RGG constructs, rapamycin (Sigma-Aldrich; St. Louis, MO) was spiked into the protein mixtures to a final concentration of 10 μM and absorbance at 600 nm was measured over time. For mapping the temperature-dependent phase separation, protein mixtures were applied to quartz cuvettes preincubated at 50 °C. Cuvettes were then inserted into the preheated spectrophotometer, set to 50 °C, and samples were cooled to 5 °C at a rate of 1 °C/min while measuring the absorbance at 600 nm.
Imaging of Protein Condensation In Vitro
Fluorescence microscopy imaging of protein droplet formation was performed at ambient temperature (approximately 22 °C) on an Olympus IX81 inverted confocal microscope (Olympus Life Science; Tokyo, Japan) equipped with a Yokogawa CSU-X1 spinning disk, mercury lamp, 488 and 561 nm laser launches, iLas-targeted laser system for photobleaching and an iXon3 EMCCD camera (Andor; Belfast, UK). Multidimensional acquisition was controlled by MetaMorph software (Molecular Devices; Downingtown, PA). Samples were illuminated using a 488 nm laser and imaged through a 100×/1.4 NA oil-immersion objective. To image in vitro droplet formation, proteins were thawed at 50 °C, diluted to 4 μM in a buffer containing 150 mM NaCl and 20 mM Tris-HCl (pH 8.5 unless otherwise specified), and placed on custom-fabricated acrylic gasket chambers adhered to glass coverslips.
Chemical Dimerization
The fluorescent tracer, RGG-GFP-RGG, was present in the protein mixture at 0.1–0.2 μM to track condensation in the 488 nm channel. Chamber wells had been previously passivated overnight in a solution of 10 mg/mL BSA (Thermo Fisher Scientific) at room temperature and then rinsed with sterile ddH2O immediately prior to use. Condensate formation of FRB and FKBP-tagged proteins was induced by addition of rapamycin (Sigma-Aldrich) at a final concentration of 10 μM per reaction. Tandem RGG constructs in the same buffer conditions noted above formed condensates in the absence of additional components. Condensate formation was monitored by time-lapse imaging with bright-field transmitted light and via 488 nm fluorescence. FRAP experiments were conducted on the same microscope using 405 nm light from an iLas targeted laser system. For photobleaching of internal regions of droplets, ROIs of similar sizes were selected and bleached. For photobleaching of whole droplets, a circular ROI encompassing an entire droplet was selected and photobleached as above.
Optical Uncaging of dRap for Light-Induced Condensation
Proteins mixtures were assembled in a dark room. Proteins were diluted to 10 μM in a reaction in a buffer containing 150 mM NaCl and 20 mM HEPES, pH 6.8, and supplemented with 5 μM dRap. Each molecule of dRap, upon uncaging, liberates two molecules of Rap. The protein mixture was then encapsulated inside cell-size water-in-oil emulsions by repeated pipetting of 1 μL of aqueous phase within 50 μL of a 5% (w/v) mixture of Cithrol DPHS (Croda, Inc. Edison, NJ) dissolved in mineral oil (Sigma-Aldrich). This emulsion mixture was then applied to wells in custom imaging chambers that had been pretreated overnight with mineral oil. Emulsions were allowed to settle for 20 min in the dark. To induce condensation, emulsions were subjected to 30 s total of continuous 405 nm light from a mercury lamp applied in steps through the Z planes. Droplet formation inside emulsions was then monitored via time-lapse microscopy. Occasionally, emulsions would drift, and the field of view was recentered manually between imaging intervals.
Yeast Procedures
Standard methodologies were followed for all experiments involving S. cerevisiae.65,66 For Rap and dRap-mediated droplet assembly, a tor1–1 fpr1Δ::KANMX6 strain in the BY4741 genetic background was used. All other yeast strains were of the YEF473 genetic background. RGG constructs with a coiled coil or FRB and FKBP tags were cloned downstream of a galactose-inducible GAL1 promoter and integrated into the yeast URA3 locus using the Yiplac211 integrating vector or LEU2 locus using the Yiplac128 integrating vector by linearizing plasmid with EcoRV just before transformation. All yeast transformations were performed by the standard lithium acetate method.
To induce expression of RGG constructs in yeast, cells were first grown to saturation overnight in liquid YPD media in a 25 °C shaking incubator. Cells were then washed three times in sterile water, diluted in YP + 2% Raffinose, and incubated in a 25 °C shaking incubator for 6–8 h. Finally, yeast cells were diluted to an OD600 of 0.3 in YP + 2% galactose. Induction was allowed to proceed overnight in the same shaking incubator. The final OD600 of cultures used for experiments was between 0.4 and 0.8.
Image and Data Analysis
Quantitation of In Vitro Protein Condensate Formation
We used image segmentation in ImageJ. Two-dimensional maximum intensity projections in the 488 nm channel (RGG-GFP-RGG tracer) were generated for each time point and converted to 8-bit images. A binary mask was generated by automated thresholding with the Intermodes algorithm, objects were cleared from the boundary, and a watershed function was used to split objects. The particle analysis function in ImageJ was used to segment condensates. This provided the droplet number and areas from maximum intensity projections at each time point. Conversion of the droplet areas to volume was performed assuming a spherical shape.
FRAP experiments were analyzed by placing an appropriately sized ROI over the photobleached area of each droplet. The fluorescence profile over time for each ROI over time was recorded, and the maximum value prior to photobleaching was set to 1. Results from FRAP experiments for each type of droplet were then pooled, and the average recovery is shown with standard deviation.
Analysis of condensate formation in cells was performed in a similar manner in ImageJ. Time-lapse images were converted to maximum intensity projections. Individual objects (cells) were first cropped to facilitate condensate segmentation. Images were corrected for bleaching using an exponential fit. Condensate masks were generated from 8-bit images thresholded by the intermodes algorithm. In a small number of cases, despeckling was required prior to thresholding. Cells that could not be reliably thresholded were excluded from analysis. After generating a mask, particle analysis was performed in ImageJ as above to generate the object number and size. The number of droplets is reported as generated by this analysis, and volumes were calculated from the 2D areas assuming objects are spherical.
Results and Discussion
Modular, short protein helical coiled-coil bundles have been used to dimerize and target components in cells for nanoscale origami of protein structures and to generate switches via protein engineering.67−69 Previously, we demonstrated the feasibility of recruiting exogenous proteins to an IDP condensate by tagging scaffold and client proteins with cognate SYNZIP (SZ1 and SZ2) coiled coils.29 We wondered whether we could also use these coiled coils to stitch together higher order assemblies of the model IDR from LAF-1, the RGG domain. We decided to test a handful of coiled coils including the 48 aa SYNZIPs56 and 33 aa Parallel Peptide Pairs.57 We cloned these coiled coils on the N or C terminus of the LAF-1 RGG domain and characterized the phase separation behavior of individual and paired sets of proteins using microscopy and spectrophotometric turbidity assays (Figure 1). The LAF-1 RGG domain displays the upper upper critical solution temperature (UCST) behavior; it is miscible at high temperatures and as the temperature is lowered will condense at its phase boundary or “cloud point”, resulting in turbid solutions.70 Using microscopy, we could image condensate assembly at ambient temperature either in bright field or with fluorescence using a small amount of fluorescent RGG tracer (RGG-GFP-RGG) significantly below its Csat, such that it would not condense alone. As a validation of our biochemical conditions, we show that RGG-RGG (constitutive dimer) formed robust condensates that could be visualized by fluorescence microscopy. (Figure S1A). Under these same conditions, the single RGG domain will not condense because of its much higher Csat.20
Figure 1.
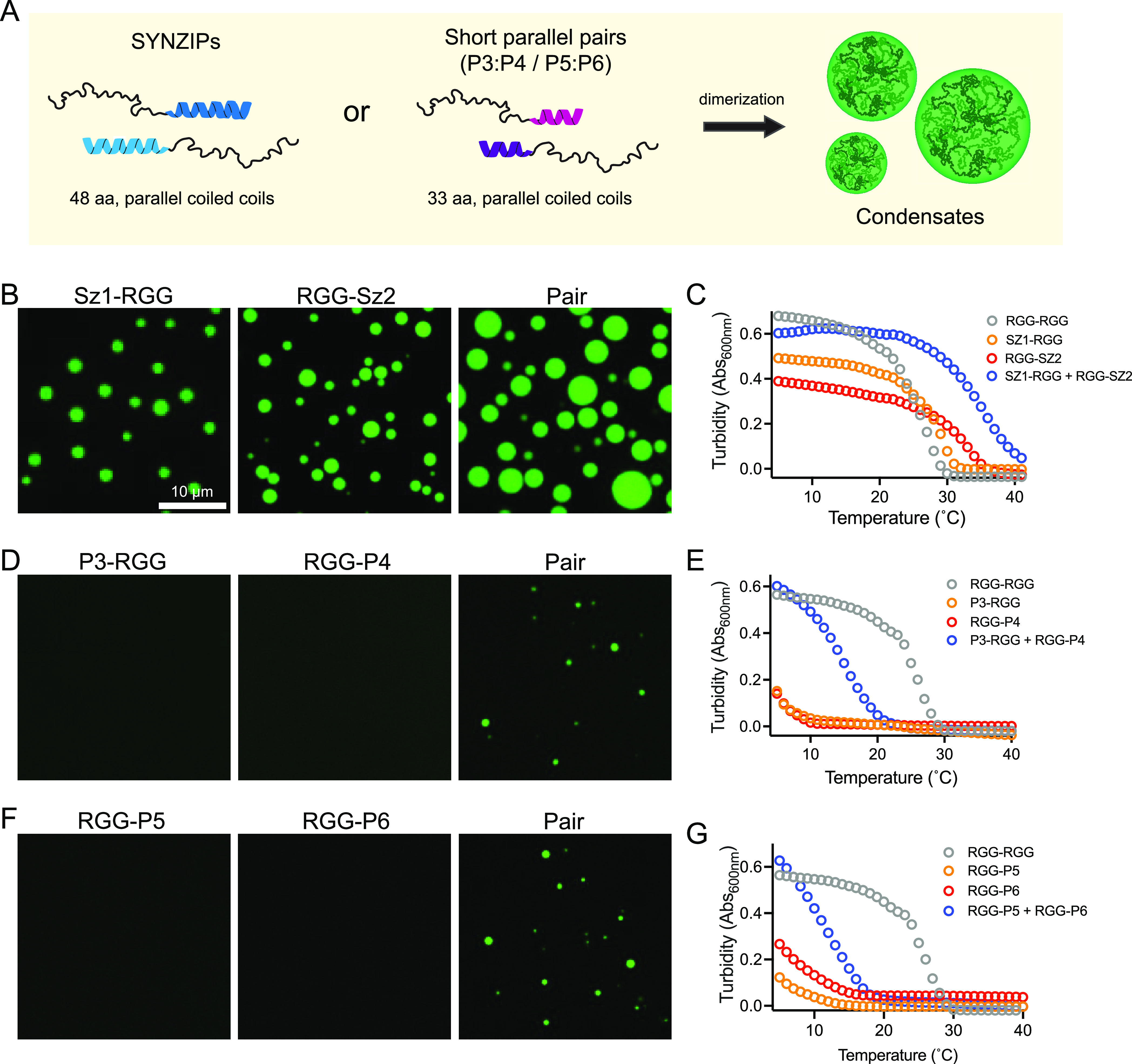
Tuning Csat for condensation using noncovalent RGG multimerization via coiled-coil pairs. (A) Schematic of assembly of higher order RGG disordered polypeptides by genetic fusion to cognate pairs of helical coiled coils. In OFF (monomer) state, monomer concentrations should be below their Csat, and in ON (dimer) state, higher polymer valency (length) lowers Csat such that the dimer condenses into liquid-like droplets. (B, D, and F) Representative fluorescence microscopy images of condensates formed using 4 μM RGG monomer concentrations, 150 mM salt, pH 8.5. (B) images of SZ1-RGG, RGG-SZ2, and a mixture of SZ1-RGG and RGG-SZ2. (C) Turbidity measurements at A600 for 6 μM concentration of the control RGG-RGG dimer, indicated monomers, or mixtures of monomers, over a range of temperatures. (D) Images of P3-RGG, RGG-P4, and combination of P3-RGG and RGG-P4. (E) Turbidity measurement using a 6 μM concentration of the indicated control, monomer, and mixed monomer over a range of temperatures. (F) Representative images of RGG-P5, RGG-P6, and a mixture of RGG-P5 and RGG-P5. (G) Turbidity measurement using a 6 μM concentration of the indicated control dimer, monomer, and mixed monomer over a range of temperatures.
Using these assay conditions, we set out to test whether tagged RGG remains miscible (OFF) in the monomer form but could condense (ON) as a mixed heterodimer. We first tested tagging a single RGG domain tagged with a folded SZ1 or SZ2 coiled coil under physiological conditions at 4 μM protein concentration. Somewhat unexpectedly, SZ1-RGG and RGG-SZ2 both formed condensates on their own (Figure 1B), suggesting a lower Csat than that of the untagged RGG alone.20 By mixing this cognate pair we expected the RGG domain to dimerize and for condensates to grow larger. Indeed, we observe this behavior and a shift in critical temperature in the turbidity assay (Figure 1C). The lowered Csat of SYNZIP-tagged single RGG constructs may be explained by chain collapse due to reciprocal charges between the RGG domains (positively charged) and the coiled-coil tag (negatively charged). The RGG domain is enriched with positively charged arginine residues across its sequences, which are involved in π–π contacts and contribute to its phase separation.51 Electrostatic interactions between the SYNZIPs and the RGG domain could result in partial chain collapse, and several studies have demonstrated that chain collapse is an important feature in the phase separation of disordered proteins; single-chain collapse theory has been used to predict phase boundaries of disordered proteins.3,71,72 It is plausible that by introducing a motif with reciprocal charge, we promoted increased chain collapse, leading to a lower overall Csat and greater propensity to phase separate in the conditions used in the assay.
Because individual RGG domains tagged to SZ1 with SZ2 formed condensates on their own in the OFF state of our assay conditions, we chose to characterize additional coiled-coil pairs. LAF-1 RGG domains tagged with different parallel coiled coils (P3 or P4) at either terminus do not condense at physiological conditions at a 4 μM protein concentration (Figure 1D, Figure S1B). Yet, when paired, they form micron scale condensates within minutes of mixing protein solutions. By increasing the protein concentration, we identified the phase boundary for RGG-P4 alone, yet P3-RGG remained miscible, suggesting the monomers indeed have a higher Csat than the dimer (Figure S1B). By varying the position of the P3 and P4 tags on the RGG termini, we observed no coacervation of any of the monomer variants under these experimental conditions. Of the mixed pairs at the same molar concentrations, P3-RGG + RGG-P4, RGG-P3 + P4-RGG, and RGG-P3 + RGG-P4 displayed condensation (Figure S1C). Importantly, this condensation is specific to heterodimerization as mixtures of noncognate pairs of tagged RGG did not generate condensates (Figure S1D). The lowest critical concentration appeared to be for P3-RGG + RGG-P4 as it formed the largest condensates. The mixed pair of P3-RGG + P4-RGG did not form condensates, suggesting that N-terminal tagging may have an effect on either dimerization or RGG condensation. We note that RGG contains an important 10 aa patch, residues 21–30, which promote condensation.51 Turbidity measurements on P3-RGG and RGG-P4 support the microscopy results and suggest that they are an excellent pair to control RGG valency and Csat (Figure 1E). We also tested the phase separation of RGG tagged with a different set of parallel coiled coils, P5 and P6 (Figure 1F and 1G). At a 4 μM protein concentration, pH 8.5, and physiological salt, the P5-RGG and RGG-P6 monomers showed no visible condensation in the OFF state (Figure 1F). The monomers also showed very little turbidity, even at lower temperatures, and appeared to heterodimerize nicely, creating a large increase in cloud point for the mixed pair, features that suggest they might be an ideal pair for controlling RGG valency. However, at pH 6.8, the same concentration of individual monomers condenses on their own (Figure S1E). Notably, the P3- and P4-tagged RGG monomers do not exhibit such pH sensitivity. Taken together, this data suggested that it is feasible to increase valency via protein interaction tags and that the type and position of the coiled coil influences the level of miscibility in the OFF (monomer) state and efficacy of condensates in the ON state (pair, heterodimer). In addition, these data suggest that the P3 and P4 heterodimers are quite compatible with dimerization and condensation of the LAF-1 RGG IDP and may be broadly useful for generating higher order disordered polymers.
Having validated LAF-1 RGG heterodimerization as a means to increase polymer valency and self-assembly into condensates, we sought to induce coacervation with temporal precision by adding a small molecule. We fused chemically responsive FRB and FKBP tags to the RGG domains (Figure 2). FRB and FKBP domains form a ternary complex with rapamycin (Rap), and thus, when linked to RGG domains, Rap induces formation of a divalent RGG polymer (Figure 2A). Because folded FRB and FKBP domains are larger than the coiled coils, they more significantly alter the order-to-disorder ratio of the polypeptide and increase Csat. As a result, a slightly higher protein concentration was needed to visualize condensation of the dimerized state. A covalently linked RGG-RGG control dimer robustly phase separates into liquid droplets at ambient temperature and 10 μM concentration, consistent with previous observations. Monomeric RGG domains fused to FRB and FKBP tags are miscible in the same aqueous solution (Figure 2B). Addition of Rap triggers rapid and robust phase separation to form micrometer-scale condensates that enrich with the RGG-GFP-RGG tracer (Figure 2B–D, Movie S1). To quantify the kinetics of the heterodimer assembly, we measured turbidity over time in spectrophotometric turbidity assays after addition of Rap (Figure 2E). Reactions lacking Rap do not become turbid, consistent with in vitro microscopy. Addition of Rap results in rapid solution turbidity with a t1/2 of ∼10 s (Figure 2E), demonstrating efficient system responsiveness resulting from multimerization of RGG domains. We also used turbidity assays to compare the UCST behavior of Rap-induced RGG dimers to our control RGG-RGG (Figure 2F). Monomeric RGG domains fused to FRB or FKBP in the absence of Rap have critical temperatures similar to single RGG domains,20 and dimerization in the presence of Rap results in turbidity curves similar to the RGG-RGG control. Finally, we assayed the liquidity of these condensates using fluorescence recovery after photobleaching (FRAP) to measure the dynamics of a fluorescent tracer localized to the condensed phase. Fluorescence of the RGG-GFP-RGG tracer in control condensates and those formed by Rap-mediated dimers rapidly recovers after photobleaching and shows very similar recovery kinetics both via internal diffusion of fluorescent molecules as well as via diffusion from outside condensates (Figure 2G and 2H and Figures S2A and S2B). Taken together, these data demonstrate a strategy for chemogenic heterodimerization of IDP scaffolds to increase the valency and induce formation of liquid-like condensates.
Figure 2.
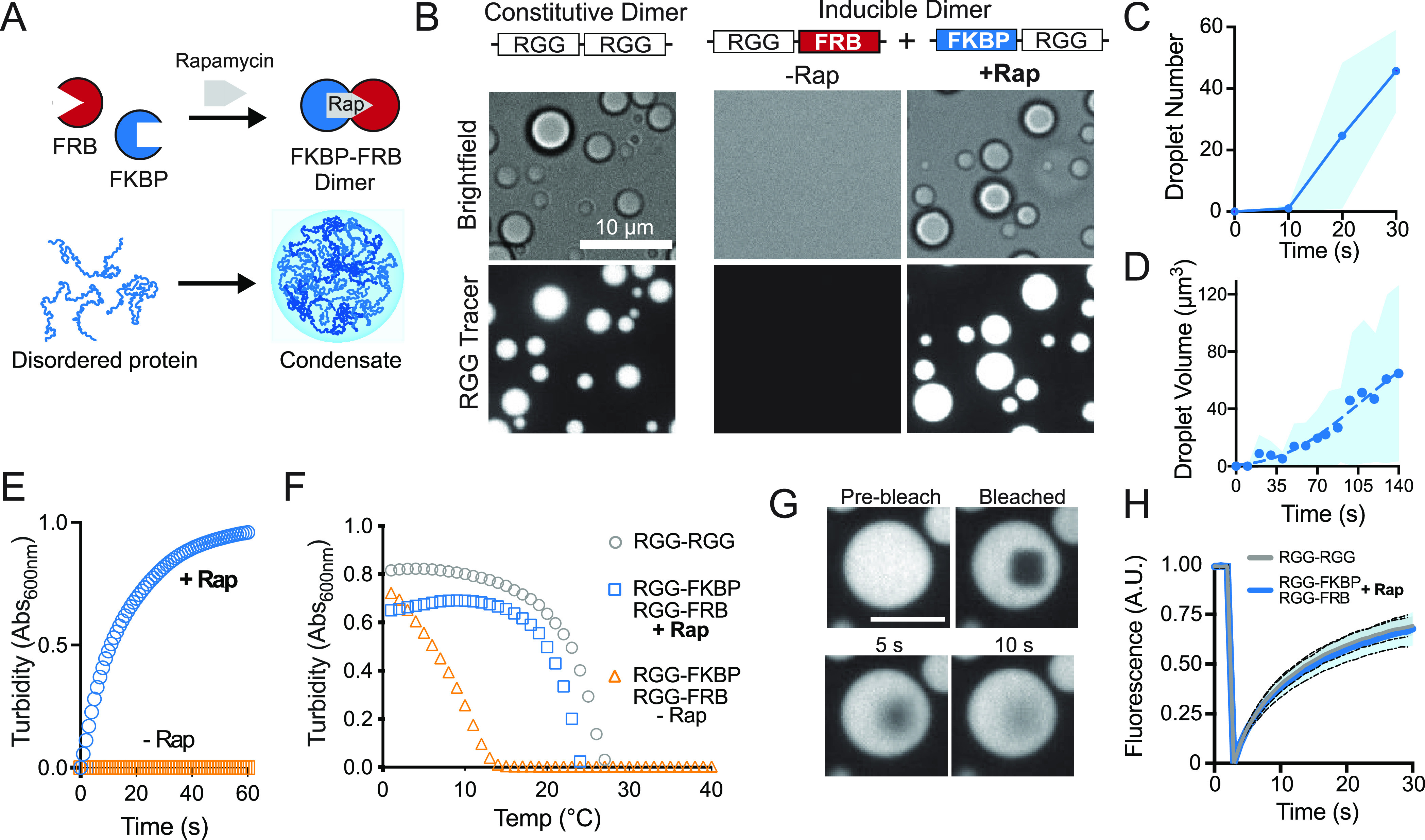
Temporal control of IDR multimerization and phase separation in vitro. (A) Schematic representation for chemogenic dimerization of RGG polypeptides to form mesoscale liquid-like protein condensates. (B) Representative images of liquid droplet formation through increased domain valency upon addition of dimerizer, Rap. Recombinant RGG-FKBP and RGG-FRB proteins, in the absence of Rap, do not form condensates in a buffer containing 10 μM protein and 0.2 μM tracer (RGG-GFP-RGG). Scale bar, 10 μm. Addition of Rap to the reaction rapidly induces dimerization, causing condensate formation similar to the RGG-RGG constitutive dimer. (C and D) Quantitation of the kinetics of droplet formation upon equimolar addition of Rap. Average of three independent trials. Shaded area, StDev. (E) Kinetics of solution clouding after addition of dimerizer in spectrophotometric turbidity assays; average of three experiments area shown. (F) Phase transition temperature measured by turbidity assay shows induced dimerization of RGG-FKBP and RGG-FRB with Rap shifts the cloud point to higher temperatures, similar to constitutive RGG-RGG dimer; average of three experiments. (G) Representative images from photobleaching and recovery of condensates composed of Rap-mediated RGG-FKBP/RGG-FRB dimers marked by 0.2 μM RGG-GFP-RGG tracer. Scale bar 5 μm. (H) Quantification of FRAP indicating similar recovery kinetics of Rap-induced vs constitutive RGG-RGG dimers. n = 30 condensates from two independent trials.
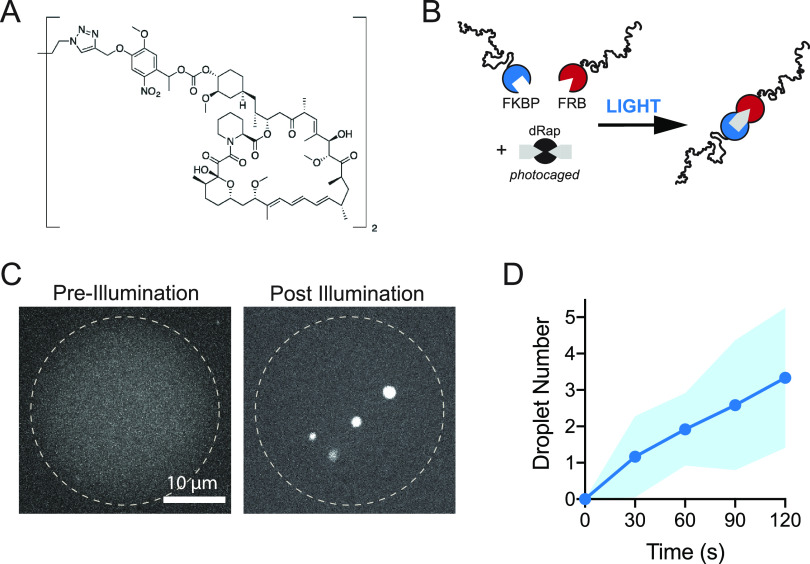
A useful feature of the FRB and FKBP domains is that their dimerization can also be optically regulated.73 A challenge of optogenetic dimerization domains is that their association requires sustained illumination, and many of these systems are difficult to reconstitute in vitro. We sought to trigger irreversible condensation in vitro using only seconds of illumination. To achieve this, we utilized a photocaged version of rapamycin (dRap)47,73 in which a cage occludes the FRB binding sites, thereby preventing FRB-FKBP dimerization in the dark state (Figure 3A and 3 B). To achieve sufficient levels of illumination and mimic a cellular context, we encapsulated RGG-FRB and RGG-FKBP proteins with dRap in cell-size water-in-oil emulsions. This approach confines the reaction to picoliter volumes which can be entirely illuminated on a microscope. Prior to illumination, we observed an evenly dispersed signal from our RGG-GFP-RGG tracer, indicating that no LLPS had occurred (Figure 3C). Upon exposure to 405 nm light, we observed rapid formation of protein droplets, visible within tens of seconds (Figure 3C and 3D, Movie S2). These data demonstrate a robust method for control of RGG domain valency in real time to drive stable protein condensation, responsive to both small-molecule and optical triggers.
Figure 3.
Optochemical regulation of IDR condensation. (A) Chemical structure of photocaged rapamycin, dRap. (B) Schematic of approach: RGG domains fused to FKBP or FRB tags do not dimerize in the presence of dRap. Upon illumination, dRap is uncaged to Rap, resulting in dimerization of RGG-FKBP and RGG-FRB. (C) Pre- and postillumination images of water-in-oil emulsions, stabilized by Cithrol DPHS surfactant, encapsulating 10 μM each of RGG-FKBP and RGG-FRB, 0.2 μM RGG-GFP-RGG as a fluorescent tracer, and 5 μM dRap. Dotted line is the emulsion boundary. Prior to illumination, GFP signal is diffuse, indicating no condensation. After 30 s of illumination and uncaging of Rap, RGG condensates appear, indicating dRap uncaging and protein dimerization. Scale bar 10 μm. (D) Kinetics measured from images of optically induced droplet formation inside emulsions, as in C. n = 10 emulsions. Shaded area, StDev.
To demonstrate that our inducible RGG dimerization and condensation system can be extended to living systems, we encoded the monomer scaffold in a model single-cell organism. Budding yeast, S. cerevisiae, is a well-established system for studying aggregate formation and LLPS in vivo (Figure 4). We genomically integrated sequences encoding single RGG domains fused to FRB and FKBP under the control of an inducible yeast GAL1 promoter. We included a GFP tag on one of the constructs (pGAL1-RGG-GFP-FKBP) in order to visualize condensate formation (Figure 4A). We induced expression of these single RGG constructs via galactose and observed only a diffuse GFP signal, indicating that without dimerizer they do not undergo LLPS (Figure 4B). Upon addition of Rap, fluorescent puncta appeared. Condensates could be initially observed within tens of seconds and formed micron-size structures on the order of minutes (Movie S3), increasing in both number and size over time (Figure 4C and Figure S3A). We then tested whether we could induce the assembly of stable membraneless organelles using a single, short period of light exposure. Using the same strain and encoded constructs as above, we added the photocaged dRap to the cells in media. Upon a 10 s pulse of 405 nm light, round puncta appeared and grew in number and size over time (Figure 4D and 4E), Rap addition or dRap optical uncaging, condensates also grew in size (Figure 4E and Figure S3B). Further, the kinetics of droplet formation is very similar in both strategies above (Figure S3C). In contrast to the available optical LLPS systems in vitro, this strategy requires only a single short pulse of light to induce dimerization of a disordered sequence and cause protein coacervation.
Figure 4.
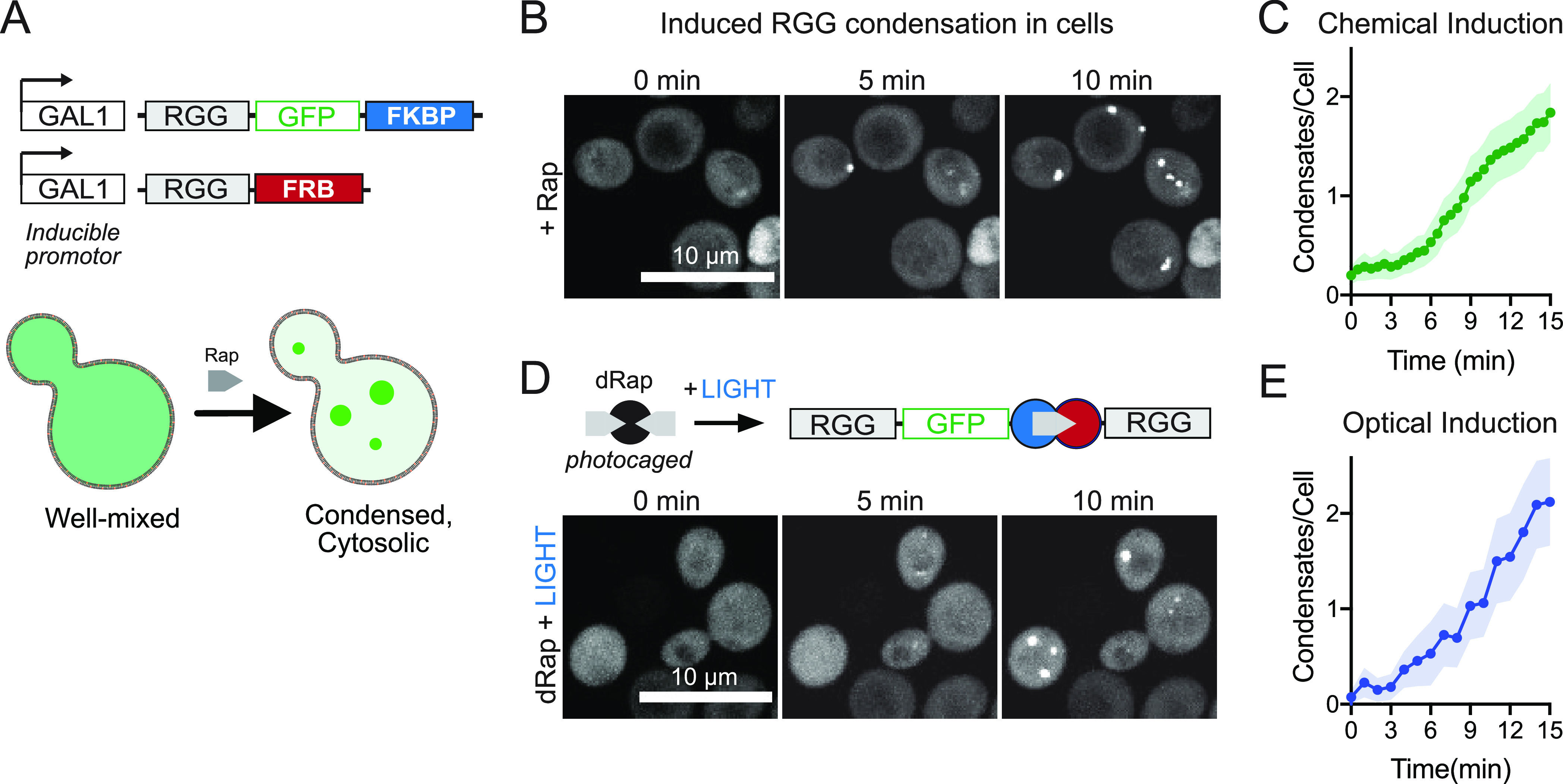
Leveraging induced dimerization to trigger synthetic condensate formation in living cells. (A) Scheme for encoding and expression of RGG polypeptides in cells, sensitive to small-molecule-induced dimerization and condensation. RGG-GFP-FKBP and RGG-FRB constructs are integrated into the yeast genome controlled by an inducible GAL1 promoter. (B) Representative images for strains described in A, showing addition of 20 μM Rap triggers condensate formation within minutes in live cells. (C) Average number of droplets formed per yeast cell in the n = 105 cells. Shaded area, 95% CI. (D) Optical regulation of condensation in cells following illumination to photouncage dRap. (E) Average number of droplets formed per cell following 10 s of 405 nm laser illumination (n = 66 cells). Shaded area, 95% CI.
We also wanted to test whether chemogenic dimerization could be utilized to control recruitment of clients or cargo to the condensed phase, mimicking enzyme partitioning to membraneless organelles. To test the specificity and temporal kinetics of client recruitment, we used a model cargo, mCherry-FRB, and imaged its enrichment in condensates formed from either RGG-RGG or heterodimers of RGG-FKBP + RGG-FRB in the presence of Rap (Figure 5A and 5B). The cargo does not enrich in control RGG-RGG condensates, whereas it is recruited to and enriched in condensates of dimerized RGG-FKBP + RGG-FRB (Figure 5C). This suggests selective client recruitment to the condensed phase dependent on dimerizing to RGG-FKBP. Overall, these data demonstrate a proof-of-concept for rapid and stable multimerization to form condensates both in vitro and in cell culture and provide new avenues for temporally regulating condensates assembly and composition, useful features for cellular engineering.
Figure 5.
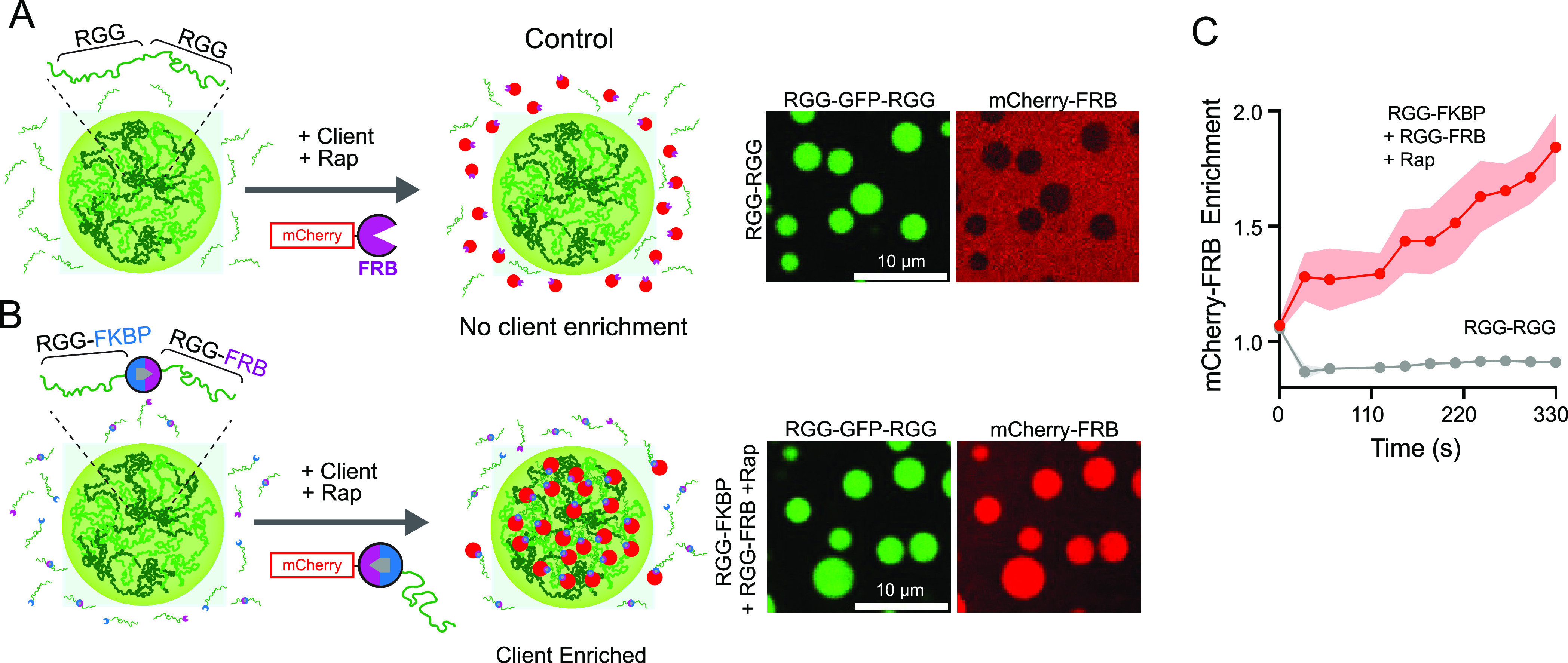
Selective cargo recruitment to condensates via chemogenic dimerization. (A) Schematic of RGG-RGG condensates, which cannot selectively recruit FRB-tagged client protein (mCherry-FRB). Client does not enrich in condensates because it cannot interact with free RGG polypeptides. (Right) Representative fluorescent images of condensates from 10 μM RGG-RGG marked with 0.2 μM RGG-GFP-RGG tracer and 5 μM of client mCherry-FRB. Client is excluded from RGG-RGG condensates. (B) Scheme for cargo recruitment via rapamycin through dimerization with RGG-FKBP which partitions to condensed phase. (Right) Representative images of condensates from 10 μM each of RGG-FKBP and RGG-FRB in the presence of 10 μM Rap and 0.2 μM RGG-GFP-RGG tracer. Addition of 5 μM mCherry-FRB (client) results in robust and selective enrichment to condensed phase. (C) Kinetics of client enrichment after addition of mCherry FRB as in A and B from three independent experiments. Shaded area, 95% CI.
Conclusions
Characterizing the behavior of disordered protein sequences that self-assemble into condensed phases is important for understanding the biology of membraneless organelles and in bioengineering to generate gel-like and other materials for a range of therapeutic and industrial applications.40,61,74−76 Similar to previous work with multivalent folded domains,36,77 we demonstrated that the valency of an IDR determines critical concentrations for phase separation in vitro.20 Although proteolytic cleavage has been used to reduce IDR valency,20,78 we were interested in the converse, building up multivalency of an IDR sequence to regulate protein condensation in a predictable manner. Here, we systematically engineered noncovalent dimerization of the LAF-1 RGG domain via coiled-coil motifs and further demonstrated real-time control of IDR condensation and client recruitment using folded optochemical dimerization domains. On the basis of our initial findings, one can also imagine increasing valency to trimeric and tetramer IDRs to alter Csat or promote temperature-resistant LLPS in cells and also to fine tune the desired physicochemical properties of the condensed phase.
We note there may be several reasons why the terminal position of the coiled coil can influence the LLPS of the monomer IDR. First, the LAF-1 RGG domain harbors a well-conserved and critical motif at its N-terminal end, aa 21–30;51 and thus, tagging may interfere with clustering of this motif. Also, the RGG domain has well-dispersed charge along the polypeptide chain, but it also has slightly more positive charge at its C-terminal end, which may interact with the net negative charge of the helical coils whose isoelectric points are between 4 and 5 and thus affect phase separation by altering the kinetics of chain collapse. Future study using positively charged coiled coils would be of interest. Despite the size and structured nature of FKBP and FRB tags, these tagged constructs formed condensates that behaved similarly to dimeric RGG controls, suggesting that tagging with folded domains did not significantly alter condensation behavior. We note that this is likely due to use of a 168 aa IDR, and the ratio of IDR to folded protein molecular mass can certainly affect LLPS.
Multivalency has been used as a useful strategy for driving self-assembly and coacervation for both structured and disordered proteins.20,23,33 Our study provides paradigms for sequential multimerization of IDRs for applications in materials science, synthetic biology, and cellular engineering. The short coiled coils are readily knocked in using CRISPR to native gene loci without disrupting endogenous protein function.29 Further, more complex assemblies using multiple sets of coiled coils in protein origami could be used to build novel architectures that potentially nucleate condensation.
Acknowledgments
We thank Andrea Stout and the Penn CDB Microscopy Core for imaging and support as well as Erfei Bi and Anuj Kumar for yeast strains and technical expertise for yeast work.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.2c00250.
Additional experiments including varying concentrations and pH of mixtures of RGG tagged with coiled coils, FRAP experiments, quantification of yeast data, and legends describing supplemental movies (PDF)
Bright-field and 488 nm channel images of condensate formation from 10 μM RGGFKBP and RGG-FRB (AVI)
Condensate formation in cell-sized emulsions (AVI)
Rapamycin induced condensate formation in live yeast cells (AVI)
Author Contributions
M.V.G., Z.S., D.A.H., and M.C.G. conceptualized the project and designed experiments. A.D. synthesized the small molecules. M.V.G., Z.S., J.D., and M.T. performed the experiments. M.V.G. and Z.S. analyzed data. M.V.G., Z.S., D.A.H., and M.C.G. wrote the manuscript.
Author Contributions
§ M.V.G. and Z.S.: These authors contributed equally to this work.
This study was supported by a National Institute of Biomedical Imaging and Bioengineering R01 grant, EB028320 (M.C.G. and D.A.H.). Biochemical characterization of disordered proteins was partly funded by National Science Foundation (NSF) MRSEC Seed grant DMR1720530 (M.C.G. and D.A.H.). Synthesis of optochemical dimerizers was supported by NSF grant CHE-1404836 (A.D.). Conceptual development of IDR multimerization and investigator salary support was supported in part by Department of Energy BES Biomolecular Materials grant DE-SC0007063 (M.C.G. and D.A.H.).
The authors declare no competing financial interest.
Supplementary Material
References
- Alberti S. The wisdom of crowds: regulating cell function through condensed states of living matter. J. Cell Sci. 2017, 130 (17), 2789–2796. 10.1242/jcs.200295. [DOI] [PubMed] [Google Scholar]
- Hyman A. A.; Weber C. A.; Julicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev Biol. 2014, 30, 39–58. 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Martin E. W.; Mittag T. Relationship of Sequence and Phase Separation in Protein Low-Complexity Regions. Biochemistry 2018, 57 (17), 2478–2487. 10.1021/acs.biochem.8b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E.; Shorter J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294 (18), 7115–7127. 10.1074/jbc.TM118.001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C. P.; Eckmann C. R.; Courson D. S.; Rybarska A.; Hoege C.; Gharakhani J.; Julicher F.; Hyman A. A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324 (5935), 1729–32. 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S.; Kim Y.; Szczepaniak K.; Chen C. C.; Eckmann C. R.; Myong S.; Brangwynne C. P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (23), 7189–94. 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M. T.; Elbaum-Garfinkle S.; Holehouse A. S.; Chen C. C.; Feric M.; Arnold C. B.; Priestley R. D.; Pappu R. V.; Brangwynne C. P. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 2017, 9 (11), 1118–1125. 10.1038/nchem.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S.; Gladfelter A.; Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176 (3), 419–434. 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeples W.; Rosen M. K. Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat. Chem. Biol. 2021, 17 (6), 693–702. 10.1038/s41589-021-00801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L.; Richardson T. M.; Wacheul L.; Wei M. T.; Feric M.; Whitney G.; Lafontaine D. L. J.; Brangwynne C. P. Controlling the material properties and rRNA processing function of the nucleolus using light. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (35), 17330–17335. 10.1073/pnas.1903870116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M.; Vaidya N.; Harmon T. S.; Mitrea D. M.; Zhu L.; Richardson T. M.; Kriwacki R. W.; Pappu R. V.; Brangwynne C. P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165 (7), 1686–1697. 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R. D.; Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature 2000, 404 (6778), 604–9. 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Nott T. J.; Petsalaki E.; Farber P.; Jervis D.; Fussner E.; Plochowietz A.; Craggs T. D.; Bazett-Jones D. P.; Pawson T.; Forman-Kay J. D.; Baldwin A. J. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 2015, 57 (5), 936–947. 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari B. R.; Dall’Agnese A.; Boija A.; Klein I. A.; Coffey E. L.; Shrinivas K.; Abraham B. J.; Hannett N. M.; Zamudio A. V.; Manteiga J. C.; Li C. H.; Guo Y. E.; Day D. S.; Schuijers J.; Vasile E.; Malik S.; Hnisz D.; Lee T. I.; Cisse I. I.; Roeder R. G.; Sharp P. A.; Chakraborty A. K.; Young R. A. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361 (6400), eaar3958. 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrinivas K.; Sabari B. R.; Coffey E. L.; Klein I. A.; Boija A.; Zamudio A. V.; Schuijers J.; Hannett N. M.; Sharp P. A.; Young R. A.; Chakraborty A. K. Enhancer Features that Drive Formation of Transcriptional Condensates. Mol. Cell 2019, 75 (3), 549–561. 10.1016/j.molcel.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich F.; Bodenmiller B.; Trajkovska M. G.; Wanka S.; Aebersold R.; Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013, 152 (4), 791–805. 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Banani S. F.; Lee H. O.; Hyman A. A.; Rosen M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18 (5), 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C. P.; Tompa P.; Pappu R. V. Polymer physics of intracellular phase transitions. Nat. Phys. 2015, 11 (11), 899–904. 10.1038/nphys3532. [DOI] [Google Scholar]
- Zumbro E.; Alexander-Katz A. Multivalent polymers can control phase boundary, dynamics, and organization of liquid-liquid phase separation. PLoS One 2021, 16 (11), e0245405 10.1371/journal.pone.0245405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster B. S.; Reed E. H.; Parthasarathy R.; Jahnke C. N.; Caldwell R. M.; Bermudez J. G.; Ramage H.; Good M. C.; Hammer D. A. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 2018, 9 (1), 2985. 10.1038/s41467-018-05403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaizar A.; Sanchez-Burgos I.; Collepardo-Guevara R.; Espinosa J. R. Expansion of Intrinsically Disordered Proteins Increases the Range of Stability of Liquid-Liquid Phase Separation. Molecules 2020, 25 (20), 4705. 10.3390/molecules25204705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory P. J.Principles of Polymer Chemistry; Cornell University Pres:, 1953; Vol. 1. [Google Scholar]
- Banani S. F.; Rice A. M.; Peeples W. B.; Lin Y.; Jain S.; Parker R.; Rosen M. K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166 (3), 651–663. 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak C. W.; Kosno M.; Holehouse A. S.; Padrick S. B.; Mittal A.; Ali R.; Yunus A. A.; Liu D. R.; Pappu R. V.; Rosen M. K. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol. Cell 2016, 63 (1), 72–85. 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz F. G.; Chilkoti A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater. 2015, 14 (11), 1164–71. 10.1038/nmat4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Elbaum-Garfinkle S.; Langdon E. M.; Taylor N.; Occhipinti P.; Bridges A. A.; Brangwynne C. P.; Gladfelter A. S. RNA Controls PolyQ Protein Phase Transitions. Mol. Cell 2015, 60 (2), 220–30. 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle T. K.; Sing C. E. Tuning chain interaction entropy in complex coacervation using polymer stiffness, architecture, and salt valency. Molecular Systems Design & Engineering 2018, 3 (1), 183–196. 10.1039/C7ME00108H. [DOI] [Google Scholar]
- Garaizar A.; Espinosa J. R.; Joseph J. A.; Collepardo-Guevara R. Kinetic interplay between droplet maturation and coalescence modulates shape of aged protein condensates. Sci. Rep 2022, 12 (1), 4390. 10.1038/s41598-022-08130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian M. V.; Wang W.; Dabdoub J. B.; Tong M.; Caldwell R. M.; Benman W.; Schuster B. S.; Deiters A.; Good M. C. Designer membraneless organelles sequester native factors for control of cell behavior. Nat. Chem. Biol. 2021, 17 (9), 998–1007. 10.1038/s41589-021-00840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdikari T. M.; Jovic N.; Dignon G. L.; Kim Y. C.; Fawzi N. L.; Mittal J. A predictive coarse-grained model for position-specific effects of post-translational modifications. Biophys. J. 2021, 120 (7), 1187–1197. 10.1016/j.bpj.2021.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen I.; Shewmaker F. The Role of Post-Translational Modifications in the Phase Transitions of Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2019, 20 (21), 5501. 10.3390/ijms20215501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J.; Zwicker D.; Sohrabi-Jahromi S.; Boehning M.; Kirschbaum J. Mechanisms for Active Regulation of Biomolecular Condensates. Trends Cell Biol. 2020, 30 (1), 4–14. 10.1016/j.tcb.2019.10.006. [DOI] [PubMed] [Google Scholar]
- Song D.; Jo Y.; Choi J. M.; Jung Y. Client proximity enhancement inside cellular membrane-less compartments governed by client-compartment interactions. Nat. Commun. 2020, 11 (1), 5642. 10.1038/s41467-020-19476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Zhao R.; Tones J.; Liu M.; Dilley R. L.; Chenoweth D. M.; Greenberg R. A.; Lampson M. A. Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol. Biol. Cell 2020, 31 (18), 2048–2056. 10.1091/mbc.E19-10-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.; Conicella A. E.; Schmidt H. B.; Martin E. W.; Rhoads S. N.; Reeb A. N.; Nourse A.; Ramirez Montero D.; Ryan V. H.; Rohatgi R.; Shewmaker F.; Naik M. T.; Mittag T.; Ayala Y. M.; Fawzi N. L. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. Embo j 2018, 37 (5), e97452 10.15252/embj.201797452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.; Banjade S.; Cheng H. C.; Kim S.; Chen B.; Guo L.; Llaguno M.; Hollingsworth J. V.; King D. S.; Banani S. F.; Russo P. S.; Jiang Q. X.; Nixon B. T.; Rosen M. K. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483 (7389), 336–40. 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden C.; Gladfelter A. S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021, 22 (3), 183–195. 10.1038/s41580-020-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejedor A. R.; Garaizar A.; Ramírez J.; Espinosa J. R. RNA modulation of transport properties and stability in phase-separated condensates. Biophys. J. 2021, 120 (23), 5169–5186. 10.1016/j.bpj.2021.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N. K.; Roberts S.; Quiroz F. G.; Chilkoti A.; Yingling Y. G. Sequence Directionality Dramatically Affects LCST Behavior of Elastin-Like Polypeptides. Biomacromolecules 2018, 19 (7), 2496–2505. 10.1021/acs.biomac.8b00099. [DOI] [PubMed] [Google Scholar]
- Dzuricky M.; Rogers B. A.; Shahid A.; Cremer P. S.; Chilkoti A. De novo engineering of intracellular condensates using artificial disordered proteins. Nat. Chem. 2020, 12 (9), 814–825. 10.1038/s41557-020-0511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basheer A.; Shahid S.; Kang M. J.; Lee J. H.; Lee J. S.; Lim D. W. Switchable Self-Assembly of Elastin- and Resilin-Based Block Copolypeptides with Converse Phase Transition Behaviors. ACS Appl. Mater. Interfaces 2021, 13 (21), 24385–24400. 10.1021/acsami.1c00676. [DOI] [PubMed] [Google Scholar]
- Reed E. H.; Hammer D. A. Redox sensitive protein droplets from recombinant oleosin. Soft Matter 2018, 14 (31), 6506–6513. 10.1039/C8SM01047A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S. A.; Simon J. R.; Amiram M.; Tang L.; Zauscher S.; Brustad E. M.; Isaacs F. J.; Chilkoti A. Photo-Crosslinkable Unnatural Amino Acids Enable Facile Synthesis of Thermoresponsive Nano- to Microgels of Intrinsically Disordered Polypeptides. Adv. Mater. 2018, 30 (5), 1704878. 10.1002/adma.201704878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agouridas V.; El Mahdi O.; Diemer V.; Cargoët M.; Monbaliu J. M.; Melnyk O. Native Chemical Ligation and Extended Methods: Mechanisms, Catalysis, Scope, and Limitations. Chem. Rev. 2019, 119 (12), 7328–7443. 10.1021/acs.chemrev.8b00712. [DOI] [PubMed] [Google Scholar]
- Keeble A. H.; Howarth M. Power to the protein: enhancing and combining activities using the Spy toolbox. Chem. Sci. 2020, 11 (28), 7281–7291. 10.1039/D0SC01878C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L. A.; Liu C. W.; Wandless T. J. Characterization of the FKBP.rapamycin.FRB ternary complex. J. Am. Chem. Soc. 2005, 127 (13), 4715–21. 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- Caldwell R. M.; Bermudez J. G.; Thai D.; Aonbangkhen C.; Schuster B. S.; Courtney T.; Deiters A.; Hammer D. A.; Chenoweth D. M.; Good M. C. Optochemical Control of Protein Localization and Activity within Cell-like Compartments. Biochemistry 2018, 57 (18), 2590–2596. 10.1021/acs.biochem.8b00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister E. R.; Aonbangkhen C.; Mayo A. M.; Lampson M. A.; Chenoweth D. M. Localized light-induced protein dimerization in living cells using a photocaged dimerizer. Nat. Commun. 2014, 5, 5475. 10.1038/ncomms6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inobe T.; Nukina N. Rapamycin-induced oligomer formation system of FRB-FKBP fusion proteins. J. Biosci Bioeng 2016, 122 (1), 40–6. 10.1016/j.jbiosc.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Reed E. H.; Schuster B. S.; Good M. C.; Hammer D. A. SPLIT: Stable Protein Coacervation Using a Light Induced Transition. ACS Synth. Biol. 2020, 9 (3), 500–507. 10.1021/acssynbio.9b00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster B. S.; Dignon G. L.; Tang W. S.; Kelley F. M.; Ranganath A. K.; Jahnke C. N.; Simpkins A. G.; Regy R. M.; Hammer D. A.; Good M. C.; Mittal J. Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (21), 11421–11431. 10.1073/pnas.2000223117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolovic B.; Danial M.; Klok H. A. Coiled coils: attractive protein folding motifs for the fabrication of self-assembled, responsive and bioactive materials. Chem. Soc. Rev. 2010, 39 (9), 3541–75. 10.1039/b914339b. [DOI] [PubMed] [Google Scholar]
- Fletcher J. M.; Boyle A. L.; Bruning M.; Bartlett G. J.; Vincent T. L.; Zaccai N. R.; Armstrong C. T.; Bromley E. H.; Booth P. J.; Brady R. L.; Thomson A. R.; Woolfson D. N. A basis set of de novo coiled-coil peptide oligomers for rational protein design and synthetic biology. ACS Synth. Biol. 2012, 1 (6), 240–50. 10.1021/sb300028q. [DOI] [PubMed] [Google Scholar]
- Majerle A.; Hadži S.; Aupič J.; Satler T.; Lapenta F.; Strmšek Ž.; Lah J.; Loris R.; Jerala R. A nanobody toolbox targeting dimeric coiled-coil modules for functionalization of designed protein origami structures. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (17), e2021899118 10.1073/pnas.2021899118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgoulia P. S.; Bjelic S. Prediction of Protein-Protein Binding Interactions in Dimeric Coiled Coils by Information Contained in Folding Energy Landscapes. Int. J. Mol. Sci. 2021, 22 (3), 1368. 10.3390/ijms22031368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson K. E.; Bashor C. J.; Lim W. A.; Keating A. E. SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains. ACS Synth. Biol. 2012, 1 (4), 118–29. 10.1021/sb200015u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebar T.; Lainšček D.; Merljak E.; Aupič J.; Jerala R. A tunable orthogonal coiled-coil interaction toolbox for engineering mammalian cells. Nat. Chem. Biol. 2020, 16 (5), 513–519. 10.1038/s41589-019-0443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y.; Berry J.; Pannucci N.; Haataja M. P.; Toettcher J. E.; Brangwynne C. P. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell 2017, 168 (1–2), 159–171. 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha D.; Walls M. T.; Wei M. T.; Zhu L.; Kurian M.; Avalos J. L.; Toettcher J. E.; Brangwynne C. P. Mapping Local and Global Liquid Phase Behavior in Living Cells Using Photo-Oligomerizable Seeds. Cell 2018, 175 (6), 1467–1480. 10.1016/j.cell.2018.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S. P.; Qian Z. G.; Hu C. F.; Pan F.; Chen M. T.; Lee S. Y.; Xia X. X. Formation and functionalization of membraneless compartments in Escherichia coli. Nat. Chem. Biol. 2020, 16 (10), 1143–1148. 10.1038/s41589-020-0579-9. [DOI] [PubMed] [Google Scholar]
- Zhao E. M.; Suek N.; Wilson M. Z.; Dine E.; Pannucci N. L.; Gitai Z.; Avalos J. L.; Toettcher J. E. Light-based control of metabolic flux through assembly of synthetic organelles. Nat. Chem. Biol. 2019, 15 (6), 589–597. 10.1038/s41589-019-0284-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aonbangkhen C.; Zhang H.; Wu D. Z.; Lampson M. A.; Chenoweth D. M. Reversible Control of Protein Localization in Living Cells Using a Photocaged-Photocleavable Chemical Dimerizer. J. Am. Chem. Soc. 2018, 140 (38), 11926–11930. 10.1021/jacs.8b07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H.; Nishikawa J.; Laemmli U. K. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 2008, 31 (6), 925–32. 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Woods B.; Kuo C. C.; Wu C. F.; Zyla T. R.; Lew D. J. Polarity establishment requires localized activation of Cdc42. J. Cell Biol. 2015, 211 (1), 19–26. 10.1083/jcb.201506108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J.; Fritsch E. F.; Maniatis T.. MolCecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 1989. [Google Scholar]
- Guthrie C.; Fink G. R.. Guide to yeast genetics and molecular biology. Methods in Enzymogy; Elsevier; , 1991; Vol. 194, pp 1–863. [PubMed] [Google Scholar]
- Langan R. A.; Boyken S. E.; Ng A. H.; Samson J. A.; Dods G.; Westbrook A. M.; Nguyen T. H.; Lajoie M. J.; Chen Z.; Berger S.; Mulligan V. K.; Dueber J. E.; Novak W. R. P.; El-Samad H.; Baker D. De novo design of bioactive protein switches. Nature 2019, 572 (7768), 205–210. 10.1038/s41586-019-1432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng A. H.; Nguyen T. H.; Gómez-Schiavon M.; Dods G.; Langan R. A.; Boyken S. E.; Samson J. A.; Waldburger L. M.; Dueber J. E.; Baker D.; El-Samad H. Modular and tunable biological feedback control using a de novo protein switch. Nature 2019, 572 (7768), 265–269. 10.1038/s41586-019-1425-7. [DOI] [PubMed] [Google Scholar]
- Ljubetič A.; Lapenta F.; Gradišar H.; Drobnak I.; Aupič J.; Strmšek Ž.; Lainšček D.; Hafner-Bratkovič I.; Majerle A.; Krivec N.; Benčina M.; Pisanski T.; Veličković T.; Round A.; Carazo J. M.; Melero R.; Jerala R. Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo. Nat. Biotechnol. 2017, 35 (11), 1094–1101. 10.1038/nbt.3994. [DOI] [PubMed] [Google Scholar]
- Seuring J.; Agarwal S. Polymers with upper critical solution temperature in aqueous solution. Macromol. Rapid Commun. 2012, 33 (22), 1898–920. 10.1002/marc.201200433. [DOI] [PubMed] [Google Scholar]
- Dignon G. L.; Zheng W.; Mittal J. Simulation methods for liquid-liquid phase separation of disordered proteins. Curr. Opin Chem. Eng. 2019, 23, 92–98. 10.1016/j.coche.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H. Y.; Aksimentiev A. Single-Protein Collapse Determines Phase Equilibria of a Biological Condensate. J. Phys. Chem. Lett. 2020, 11 (12), 4923–4929. 10.1021/acs.jpclett.0c01222. [DOI] [PubMed] [Google Scholar]
- Karginov A. V.; Zou Y.; Shirvanyants D.; Kota P.; Dokholyan N. V.; Young D. D.; Hahn K. M.; Deiters A. Light regulation of protein dimerization and kinase activity in living cells using photocaged rapamycin and engineered FKBP. J. Am. Chem. Soc. 2011, 133 (3), 420–3. 10.1021/ja109630v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich M.; Georgeson J. M.; Locatelli E.; Rovigatti L.; Nandi S. K.; Steinberg A.; Nadav Y.; Shimoni E.; Safran S. A.; Doye J. P. K.; Levy E. D. Designer protein assemblies with tunable phase diagrams in living cells. Nat. Chem. Biol. 2020, 16 (9), 939–945. 10.1038/s41589-020-0576-z. [DOI] [PubMed] [Google Scholar]
- Roberts S.; Harmon T. S.; Schaal J. L.; Miao V.; Li K. J.; Hunt A.; Wen Y.; Oas T. G.; Collier J. H.; Pappu R. V.; Chilkoti A. Injectable tissue integrating networks from recombinant polypeptides with tunable order. Nat. Mater. 2018, 17 (12), 1154–1163. 10.1038/s41563-018-0182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings R. L.; Boeynaems S. Designer Condensates: A Toolkit for the Biomolecular Architect. J. Mol. Biol. 2021, 433 (12), 166837. 10.1016/j.jmb.2021.166837. [DOI] [PubMed] [Google Scholar]
- Petka W. A.; Harden J. L.; McGrath K. P.; Wirtz D.; Tirrell D. A. Reversible hydrogels from self-assembling artificial proteins. Science 1998, 281 (5375), 389–92. 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- Kelley F. M.; Favetta B.; Regy R. M.; Mittal J.; Schuster B. S. Amphiphilic proteins coassemble into multiphasic condensates and act as biomolecular surfactants. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (51), e2109967118 10.1073/pnas.2109967118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.