Abstract
Objective
There are altered mucosal functions in irritable bowel syndrome with diarrhoea (IBS-D); ~30% of patients with IBS-D have abnormal bile acid (BA) metabolism (ABAM) and diarrhoea (summarised as BAD).
Aim
To compare biochemical parameters, gastrointestinal and colonic transit, rectal sensation and pathobiological mechanisms in IBS-D without ABAM and in BAD (serum 7C4>52 ng/mL).
Design
In patients with Rome III criteria of IBS-D, we compared biochemical features, colonic transit, rectal sensation, deep genotype of five BA-related genes, ileal and colonic mucosal mRNA (differential expression (DE) analysis) and stool dysbiosis (including functional analysis of microbiome). Results in BAD were compared with IBS-D without ABAM.
Results
Compared with 161 patients with IBS-D without ABAM, 44 patients with BAD had significantly faster colonic transit, lower microbial alpha diversity, different compositional profile (beta diversity) and higher Firmicutes to Bacteroidetes ratio with evidence of decreased expression of bile acid thiol ligase (involved in transformation of primary to secondary BAs) and decreased sulfatases. In BAD (compared with IBS-D without ABAM), terminal ileal biopsies showed downregulation of SLC44A5 (a BA transporter), and ascending colon biopsies showed upregulation in barrier-weakening genes (CLDN2), serine protease inhibitors, immune activation, cellular differentiation and a cellular transporter (FABP6; BA binding). No DE of genes was documented in descending colon biopsies. The two groups had similar rectal sensation.
Conclusion
Though sharing clinical symptoms with IBS-D, BAD is associated with biological differences and mechanisms that have potential to enhance diagnosis and treatment targeting barrier dysfunction, inflammatory and microbial changes.
INTRODUCTION
Previous studies in patients with irritable bowel syndrome (IBS) reported alterations in barrier function, immunological factors and serine protease activity in jejunal and colorectal mucosa.1 About 30% of patients with IBS with diarrhoea (IBS-D) or functional diarrhoea have markers of abnormal bile acid (BA) metabolism (ABAM) and are diagnosed as bile acid diarrhoea (BAD).2 Cholecystectomy may result in BAD; however, it is unclear whether the biochemical parameters related to BA synthesis and excretion or colonic transit differ between patients with BAD, with or without prior cholecystectomy.
Primary BAD has been associated with reduced functional expression of fibroblast growth factor 19 (FGF-19) in ileal biopsies in response to stimulation by BAs3 or by protein expression on immunohistochemistry.4 There are five pivotal genes that may play a role in the absorption (SLC10A2) and feedback regulation of BA synthesis (NR1H4 (gene for the nuclear farnesoid X receptor of ileal enterocyte that determines FGF-19 levels in portal blood), klotho b (KLB) and FGF-4 (FGF-R4) which together determine the FGF-19 feedback regulation of hepatocyte synthesis of bile acids). The fifth gene, TGR5 (also called GPBAR1), is the gene for the receptor mediating effects of BAs in target organs, for example, colonic motility and transit.5 At least 50% of intracolonic BAs are absorbed by passive diffusion in the colon.6
Faecal BAs may be impacted by colonic transit and microbiota. The microbiota impacts the deconjugation, dehydroxylation and sulfation of conjugated BAs in the colon and their biological effects. Increased proportions of chenodeoxycholic acid (CDCA) and cholic acid (CA) were demonstrated in patients with IBS-D with elevated total faecal BA excretion over 48 hours7; this is functionally relevant because of the effects of CDCA on secretion and mucosal permeability.
BAs in the colon enable the survival of gram negative bacteria (eg, Escherichia coli and Campylobacter), which are resistant to BAs compared with gram positive bacteria.8 The bi-directional relationship between bile acids and microbiota is introduced in greater detail in the online supplemental materials.
The aim of our study was to quantify intraluminal BAs and short chain fatty acids (SCFAs), colonic transit and sensation, genetics of the five pivotal BA genes, ileal and colonic mucosal expression of genes potentially impacting mucosal function, as well as stool dysbiosis in BAD compared with patients with IBS-D without ABAM.
METHODS
Regulatory
Data will be available, consistent with data sharing NIH policy for studies supported by NIH (in this case, R01-DK115950); in addition, all relevant data are included in the paper and/or in the online online supplemental materials.
Patient and public involvement
The public is involved in the discussion of the approval of the protocol by the Mayo Clinic’s institutional review board (IRB) since, by law, there must be public representation on the IRB.
In addition, in accordance with the requirements of the NIH for sharing information acquired through NIH funding, the anonymised information will be submitted in accordance with the guidance on the NIH Genomic Data Sharing Policy. Since the information from this research study does not have immediate clinical application, the information is not included in the patients’ medical records or communicated to the patients.
Participants and design
We screened 1744 primary or secondary referral patients with IBS-D (based on Rome III criteria,9 which were standard at the time of commencement of the study) for eligibility to participate in the studies. The same cohort of patients participating in the ‘Aim 1 Transit’ study also underwent studies of intestinal–colonic permeability in BAD compared with IBS-D without ABAM; results are reported elsewhere.10
Participants, who resided within 100 miles of the single centre, where the study was performed (Mayo Clinic, Rochester, Minnesota, USA), were invited to prospectively enrol in the two components of the study reported here (two aims of NIH R01-DK115950). The participants’ enrolment and allocations are summarised in figure 1 as ‘transit’ and ‘colonoscopy’ cohorts, including screen failures. These cohorts are further described in the online supplemental materials.
Figure 1.
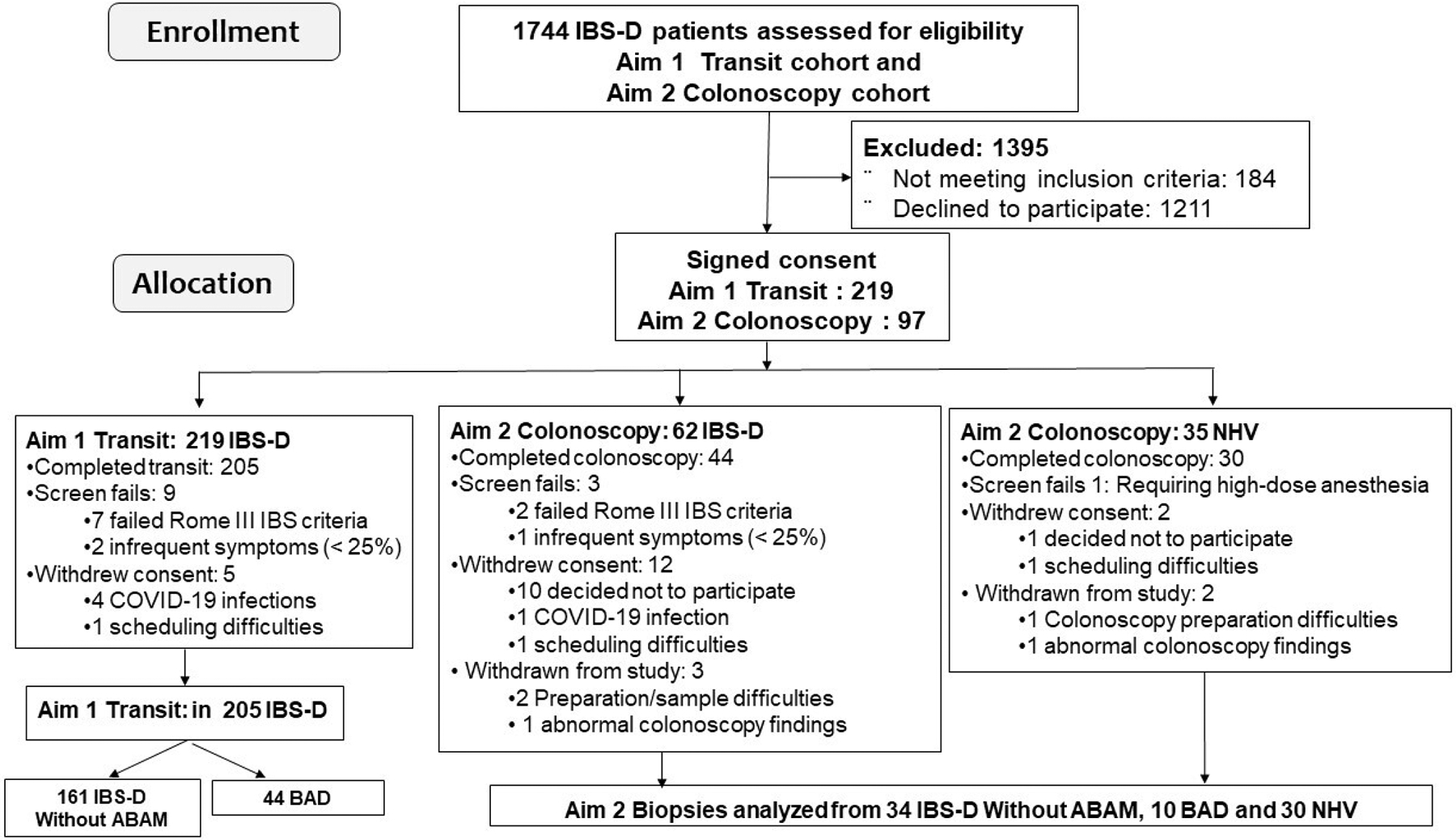
Enrolment and allocation of patients and healthy controls in the two study cohorts reported. Note: some patients with Rome III-positive IBS-D participated in both ‘transit’ and ‘colonoscopy’ aims. The numbers of participants with BAD or IBS-D without ABAM are indicated in the two aims of the study. ABAM, abnormal bile acid metabolism; BAD, bile acid diarrhoea; IBS-D, irritable bowel syndrome with diarrhoea; NHV, normal healthy volunteers.
Other methods in supplemental materials
The online supplemental materials includes information regarding measurements of biological parameters, including biochemical characterisation of bile synthesis and excretion, gastrointestinal and colonic transit by scintigraphy, and rectal sensation and compliance, as in prior studies from our laboratory, as well as faecal SCFAs.11 Additional studies examined venous blood DNA targeted sequencing of five pivotal BA-related genes (as in our prior studies, and FGF-R4 meta-analysis12 with bioinformatics analysis13–16), faecal microbiota,17–20 ileocolonoscopy with mucosal biopsies and RNA sequencing21–28 and statistical analysis.14–28 A power estimate for the colonic transit and expression data is given in online supplemental table 1. In our prior study,29 average SD of fold changes of all the genes of interest in colonic mucosal gene expression were 0.67 for irritable bowel syndrome with constipation (IBS-C) and 2.09 for IBS-D.
RESULTS
Participants, BA parameters and SCFAs
Among 205 patients with Rome III criteria for IBS-D, 44 had BAD; online supplemental table 2 shows the number of patients in the two groups who underwent venous blood DNA assay, serum 7α-hydroxy-4-cholesten-3-one (7αC4), faecal BA and SCFA, colonic transit, rectal sensation and colonoscopy with mucosal biopsies.
Biochemical features differentiating the two groups (BAD or IBS-D without ABAM) are listed in table 1. As previously reported in the same cohort,30 there was a significant difference in significant diarrhoea between the two groups (BAD (n=44) and IBS-D without ABAM (n=160)). The report of loose or watery stools was significantly more prevalent in BAD compared with IBS-D without ABAM (p=0.002), and specifically loose or watery stools more than 75% of the time were reported by 61% of BAD and 30.9% of patients with IBS-D without ABAM. In addition, there was a greater degree of faecal urgency, frequency of bowel movements, fear of faecal incontinence and the need to be closer to bathrooms to defecate in patients with BAD compared with IBS-D without BAD.
Table 1.
Demographics and biochemical features of patients with IBS-D without ABAM and patients with BAD. Data shown are median (IQR)
| IBS-D without ABAM (N=161) | Bad (N=44) | P value | |
|---|---|---|---|
| Demographics | |||
| Age (years) | 37 (26, 50) | 46 (35, 58) | 0.01 |
| BMI (kg/m2) | 28.6 (23.5, 33.9) | 33.9 (29.9, 38.5) | <0.001 |
| % female | 77.6% | 81.4% | |
| Bile acid indices | |||
| Serum FGF-19 (pg/mL) | 103.4 (63.9, 170.5) | 59.1 (30.2, 102.2) | <0.001 |
| Serum 7αC4 (ng/mL) | 19.8 (9.9, 30.9) | 74.3 (61.1, 93.7) | <0.001 |
| Total faecal bile acids (μmol/g stool) | 2.1 (1.2, 3.5) | 3.8 (2.5, 5.1) | <0.001 |
| % faecal 1° BAs (CDCA+CA) | 1.3 (0.7, 4.3) | 15.0 (1.2, 39.8) | <0.001 |
| SCFAs in stool | |||
| Faecal acetic acid (nmol/mg) | 35.2 (25.6, 48.4) | 36.0 (27.6, 49.6) | 0.85 |
| Faecal propionic acid (nmol/mg) | 6.8 (4.7, 10.1) | 7.2 (5.4, 11.6) | 0.39 |
| Faecal isobutyric acid (nmol/mg) | 2.2 (1.6, 3.1) | 1.9 (1.1, 3.0) | 0.15 |
| Faecal isovaleric acid (nmol/mg) | 1.8 (1.2, 2.6) | 1.6 (0.8, 2.4) | 0.37 |
| Faecal valeric acid (nmol/mg) | 0.7 (0.5, 1.0) | 0.6 (0.2, 1.0) | 0.32 |
| Faecal isocaproic acid (nmol/mg) | 0.05 (0.03, 0.07) | 0.04 (0.02, 0.09) | 0.75 |
| Faecal hexanoic acid (nmol/mg) | 0.08 (0.04, 0.24) | 0.06 (0.03, 0.10) | 0.014 |
Isobutyric acid is similar to butyric acid ((CH3)2-CH-COOH).
P values are based on Wilcoxon rank sum test.
ABAM, abnormal bile acid metabolism; BAD, bile acid diarrhoea; BAs, bile acids; BMI, body mass index; CA, cholic acid; CDCA, chenodeoxycholic acid; FGF-19, fibroblast growth factor-19; IBS-D, irritable bowel syndrome with diarrhoea; SCFAs, short chain fatty acids; 7αC4, 7α-hydroxy-4-cholesten-3-one.
Analysis for total faecal BAs and primary BAs was available in 189 patients, and for SCFAs in 194 patients. Fasting serum 7αC4 (available in all 205 patients) was significantly correlated with total faecal BAs (Rs=0.391; p<0.001) and with % primary BAs (Rs=0.293; p<0.001). Stool SCFA concentrations were not statistically different in the two groups.
Clinical and biochemical features of patients with bad with or without cholecystectomy
Among 43 patients with BAD (data inconclusive for 1 patient), 35% had a history of cholecystectomy. Results are summarised in online supplemental table 3. Those with cholecystectomy who reported that the chronic diarrhoea preceded cholecystectomy were significantly older and had numerically higher body mass index. There were no differences in serum FGF-19, serum 7αC4, total faecal BAs and per cent primary BAs (CDCA+CA) in stool in those with or without cholecystectomy. Therefore, studies of mucosal expression and microbiome were grouped for all patients with BAD.
Colonic transit measurements
Among the two groups (totalling 205 patients), there were no differences in gastric or small bowel transit; however, significant acceleration of colonic transit at 24 hours and ascending colon emptying T1/2 were documented in BAD compared with IBS-D without ABAM (online supplemental table 4).
Among patients with BAD, there was a significant Spearman correlation (Rs=0.349; p=0.0275) between fasting serum 7αC4 (reflecting severity of BAD) and colonic transit at 24 hours (figure 2, left panel), and a borderline correlation with ascending colon emptying T1/2 (Rs=−0.288, p=0.0677). Among patients with IBS-D without ABAM, there was also significant Spearman correlation (Rs=0.256; p=0.0013) between fasting serum 7αC4 and colonic transit at 24 hours (figure 2, right panel).
Figure 2.
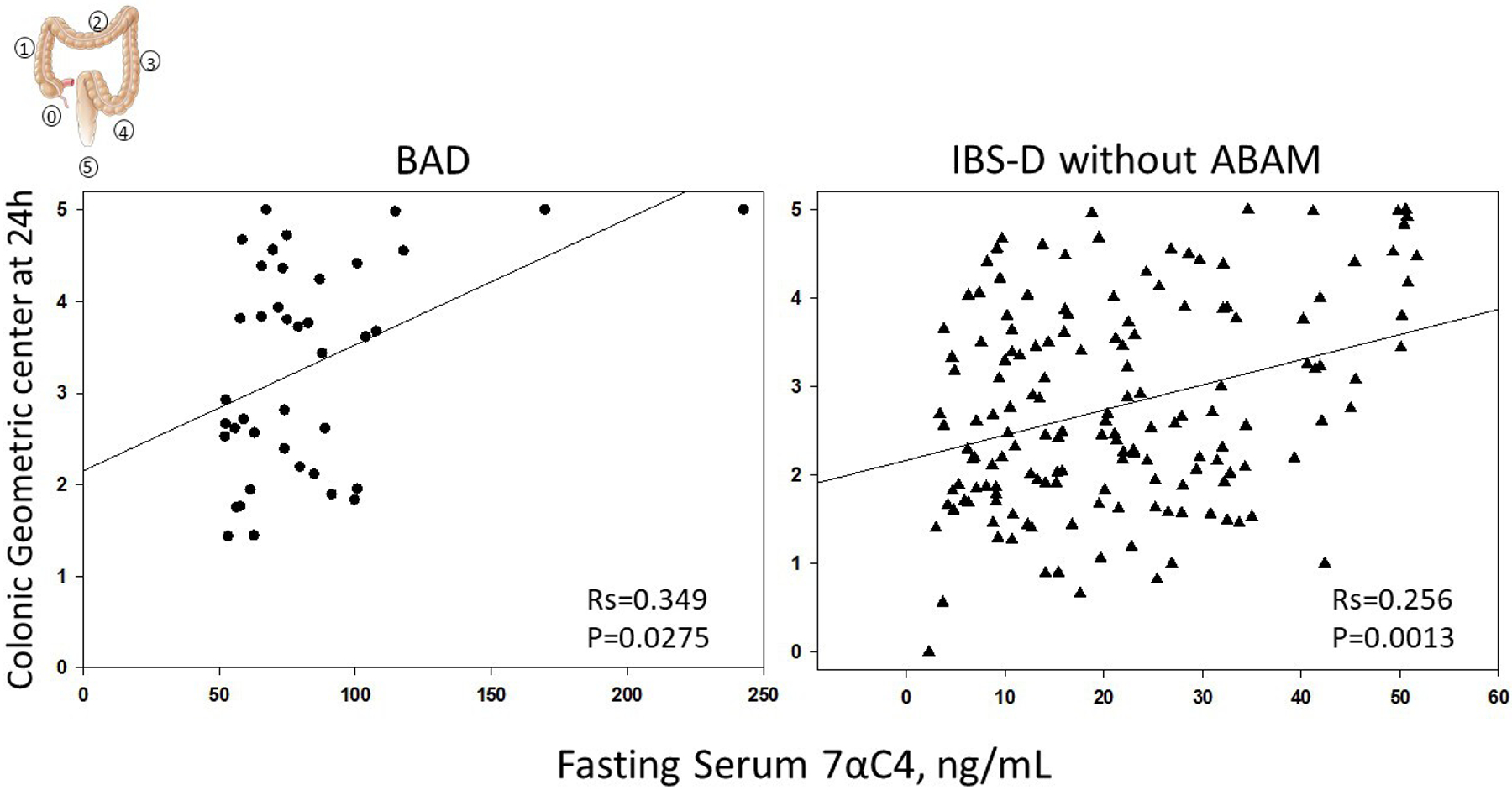
Correlation between fasting serum 7αC4 and colonic transit at 24 hours; geometric centre (GC) 1, all isotope in ascending colon; GC5, all isotope in stool. 7αC4, 7α-hydroxy-4-cholesten-3-one; ABAM, abnormal bile acid metabolism; BAD, bile acid diarrhoea; IBS-D, irritable bowel syndrome with diarrhoea.
Rectal compliance and sensation
Sixty patients underwent rectal sensation (method shown in online supplemental figure 1) and compliance studies: 19 with BAD and 41 with IBS-D without ABAM. Online supplemental table 5 shows sensation thresholds and ratings were not significantly different. There were no differences in rectal compliance between the two groups.
Targeted DNA sequencing of five pivotal genes related to synthesis and effects of bile acids
Candidate variations in exons in the BA genes were based on previous studies. The detailed information appears in online supplemental table 6. There were no significant associations between the previously noted variations in exons and the group (BAD compared with IBS-D without ABAM).
Targeted capture of the DNA sequencing of the 5 pivotal genes identified 8 gene variants that showed different prevalence (unadjusted p<0.05) in allelic distribution in BAD compared with IBS-D without ABAM. These included five single nucleotide polymorphisms with previously identified rs numbers (rs116274139, rs79532857, rs61966074, rs3135918 and rs1966265). Among the eight gene variants, three are associated with synonymous amino acids and all variants except one were associated with extremely low minor allele frequency or unknown functions. The exception was rs1966265 (in FGF-R4); its minor allele frequency was 22% and function was previously demonstrated in relation to colonic transit in patients with IBS-D.31 We identified no significant associations between any of the gene variants identified by targeted capture DNA sequencing and ileal FGF-19 or FXR mRNA expression.
For the DIET1 rs12256835 SNP, in the BAD group, the proportions with T and G allele were respectively 0.857 and 0.142, and in the IBS-D without ABAM, the proportions were 0.805 and 0.195 respectively (Chi-square p=0.275).
mRNA expression study
Table 2 shows demographics, BA-related findings and numbers of biopsies performed in the colonoscopy study in patients with Rome III criteria of IBS-D and healthy controls. There was significant upregulation of FGF-19 (log2 fold change=4.485; adjusted p (padj)=9.13E-05) in ileal mucosa of IBS-D with BAD compared with healthy controls.
Table 2.
In the mRNA expression studies, the table shows participant demographics, measurements of bile acid parameters and # of biopsies at 3 locations in 44 patients with IBS-D and 30 healthy controls
| Healthy (N=30) | IBS-D without ABAM (N=34) | BAD (N=10) | |
|---|---|---|---|
| Female: male | 14:16 | 24:10 | 7:3 |
| Age, years (SD) | 45.6 (13.3) | 38.6 (12.9) | 44.7 (11.8) |
| Serum 7αC4 mean (SD) Median (10th–90th percentile) |
19.1 (29.8) 13.9 (4.1–25.5) |
19.2 (13.4) 14.6 (5.6–37.8) |
84.9 (33.2)* 76.4 (55.3–135.5) |
| Serum FGF-19 mean (SD) Median (10th–90th percentile) |
125.1 (142.9) 74.6 (29.5–210.9) |
103.4 (77.0) 80.2 (29.3–215.9) |
67.3 (46.1) 52.6 (21.9–137.3) |
| % faecal CA+CDCA mean (SD) Median (10th–90th percentile) |
NA | 4.2 (2.3) 1.0 (0.34–10.98) |
23.4 (27.0)* 23.7 (0.08–64.52) |
| TI biopsies (# of participants) | 21 | 27 | 7 |
| RC biopsies (# of participants) | 21 | 34 | 10 |
| LC biopsies (# of participants) | 30 | 34 | 10 |
P<0.05 versus IBS-D without ABAM.
ABAM, abnormal bile acid metabolism; BAD, bile acid diarrhoea; CA, cholic acid; CDCA, chenodeoxycholic acid; FGF-19, fibroblast growth factor 19 ; IBS-D, irritable bowel syndrome with diarrhoea; LC, left colon; RC, right colon; TI, terminal ileum; 7αC4, 7α-hydroxy-4-cholesten-3-one.
Table 3 compares terminal ileum biopsies in BAD compared with IBS-D without ABAM; NUTM2D and SLC44A5 (BA transporter) were downregulated and C6 upregulated. No differences in ileal expressions of genes for FGF-19, FXR and apical sodium-coupled BA transporter were noted in these two groups.
Table 3.
Comparison of expressions of genes in terminal ileal mucosa for patients with BAD compared with those with IBS-D without ABAM
| Gene name | Gene function | Log2 fold change | Padj value | DE |
|---|---|---|---|---|
| Complement component, C6 | Immune activation | 2.117 | 0.007 | Up |
| Solute carrier family 44, member 5 (SLC44A5) | Transport of glucose, other sugars, bile salts, organic acids, metal ions and amines | −4.398 | 0.015 | Down |
| NUTM2D/FAM22D | Unclear function | −1.726 | 0.032 | |
| FGF-19 | Negative feedback of bile acid synthesis | 1.150 | 0.898 | None |
| NR1H4 (FXR gene) | Epithelial cell nuclear receptor leading to FGF-19 synthesis | −0.314 | 0.926 | |
| SLC10A2 (ASBT gene) | Ileal bile acid active transporter | −0.174 | 0.991 |
ABAM, abnormal bile acid metabolism; ASBT, apical sodium-coupled bile acid transporter; BAD, bile acid diarrhoea; DE, differential expression; FGF-19, fibroblast growth factor 19; FXR, farnesoid X receptor; IBS-D, irritable bowel syndrome with diarrhoea.
Differential expression (DE) of several genes was noted in ascending colon biopsies (table 4 and figures 3 and 4) between BAD and IBS-D without ABAM, with upregulation in barrier genes (CLDN2), serine protease inhibitor activity (SPINK4 and SERPINB5), immune activation (C4BPB, CCL25, CXCL5 and IL1RN), cellular differentiation (REG4) and cellular transporters ((FABP6) fatty acid uptake and bile acid binding) and SLC2A2 (Na+-glucose co-transporter)). It is noteworthy that two of the upregulated immune genes (CCL25 and CXCL5) would be expected to reflect immune activation, whereas upregulation of two other genes (C4BPB and IL1RN) reduced complement C4 activation or antagonised the receptor of the cytokine IL-1 and would, therefore, be expected to reduce inflammatory responses. There was downregulation in mRNA expression of barrier function (GALNT15), proton transport (OTOP2) and other genes (NUTM2D and BEX5), whose functional significance is unclear.
Table 4.
Differential gene expressions in ascending colon biopsies of BAD and IBS-D without ABAM
| Gene symbol | Full name of gene | Gene function | Log2 fold change | Padj value | DE |
|---|---|---|---|---|---|
| Permeability/barrier | |||||
| CLDN2 | Claudin 2 | Exclusively in tight junction: Barrier function | 1.737 | 0.007 | Up |
| GALNT15 | Polypeptide N-Acetylgalactos-aminyltransferase 15 | Mucin type O-glycan biosynthesis in goblet cells | −1.019 | 0.025 | Down |
| Serine/cysteine protease activity | |||||
| SPINK4 | Serine Protease Inhibitor, Kazal Type 4 | Serine-type endopeptidase inhibitor | 1.607 | 0.002 | Up |
| SERPINB5 | Serpin Family B Member 5 | Serine protease inhibitor | 1.725 | 0.007 | Up |
| CAPN6 | Calpain 6 | Ca++-dependent, cysteine proteases | 1.185 | 0.031 | Up |
| Inflammation or immune function | |||||
| C4BPB | Complement component 4- binding protein beta chain | Control of complement activation, eg, degradation of C3 convertase | 1.208 | 0.002 | Up |
| CCL25 | C-C Motif Chemokine Ligand 25 | Chemokine in T cell development | 2.451 | 0.014 | Up |
| CXCL5 | C-X-C motif chemokine ligand 5 | Chemokine | 3.173 | 0.014 | Up |
| IL1RN | Interleukin 1 receptor antagonist | IL-1 receptor antagonist | 1.341 | 0.014 | Up |
| Cellular transport/differentiation/proliferation | |||||
| REG4 | Regenerating family member 4 | Cellular differentiation and proliferation | 1.476 | 2.97E-06 | Up |
| FABP6 | Fatty acid binding protein 6 | Fatty acid uptake, transport and metabolism and BA binding | 2.207 | 0.003 | Up |
| SLC2A2 | Solute carrier family 2 member 2 | Na+/glucose co-transporter | 2.998 | 0.0002 | Up |
| OTOP2 | Otopetrin-2 | Proton-selective channel | −1.221 | 0.003 | Down |
| Miscellaneous | |||||
| CP | Ceruloplasmin | Copper-binding glycoprotein | −1.024 | 0.014 | Down |
| NUTM2D | NUT family member 2D | Unclear | −1.015 | 0.017 | Down |
| MYEOV | Myeloma overexpressed | Unclear | 1.323 | 0.008 | Up |
| BEX5 | Brain expressed X-linked 5 | Unclear | −1.328 | 0.003 | Down |
ABAM, abnormal bile acid metabolism; BA, bile acid; BAD, bile acid diarrhoea; IBS-D, irritable bowel syndrome with diarrhoea.
Figure 3.
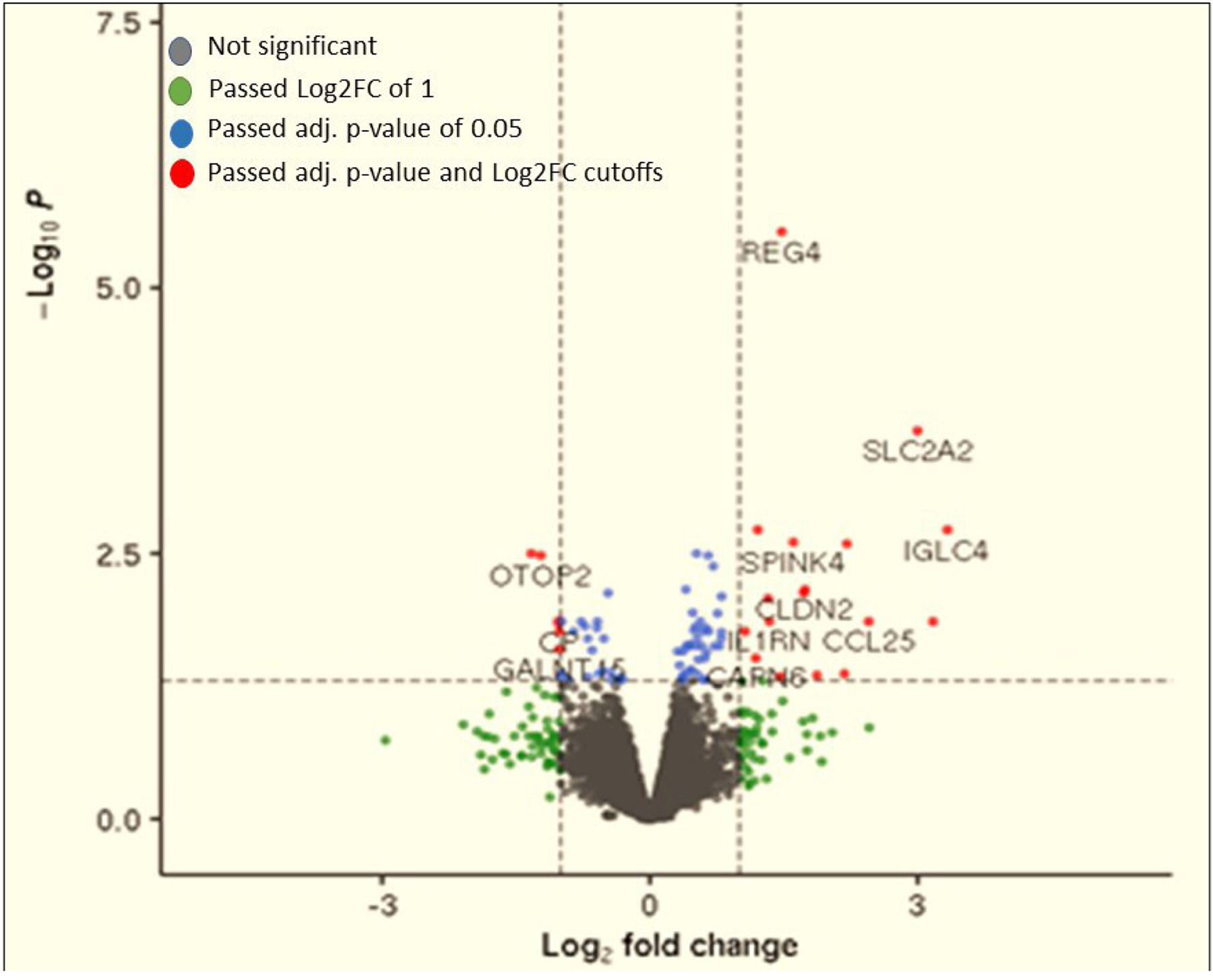
A volcano plot of differential mRNA expression in the right colon mucosal biopsies in patients with BAD compared with IBS-D without ABAM: genes with increased mRNA expression in BAD were CLDN2, SPINK4, SERPINB5, C4BPB, CCL25, CXCL5, IL1RN, REG4 and FABP6 and the gene with decreased mRNA expression in BAD was GALNT15. ABAM, abnormal bile acid metabolism; BAD, bile acid diarrhoea; IBS-D, irritable bowel syndrome with diarrhoea; log2FC, log2 fold change.
Figure 4.
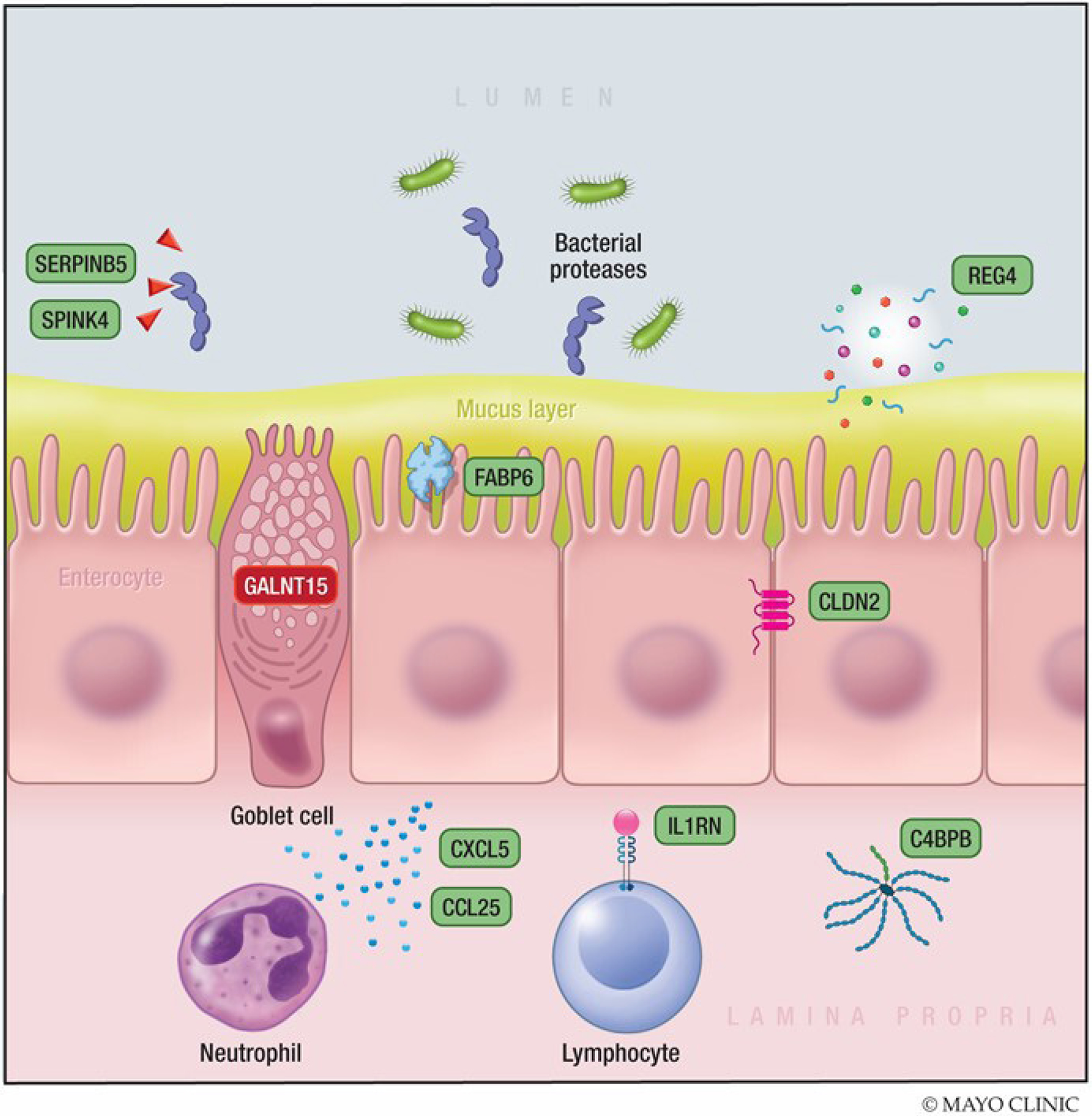
Differential gene expression in ascending colon biopsies of BAD and IBS-D without ABAM. Genes with increased mRNA expression in BAD are shown in green. The gene with decreased mRNA expression in BAD is shown in red. This summary of the detailed information in figure 3 and table 4 suggests that there is upregulation of markers of increased mucosal permeability, immune activation and inhibition of serine proteases. These three factors increase likelihood of mucosal damage and inflammation. ABAM, abnormal bile acid metabolism; BAD, bile acid diarrhoea; IBS-D, irritable bowel syndrome with diarrhoea
There were no differentially expressed genes found in BAD versus IBS-D without ABAM in the left colon biopsies.
Pathway Enrichment Analysis using EnrichR (https://maayanlab.cloud/Enrichr/)
All reported pathways differentiated BAD from IBS-D without ABAM and were confirmed on Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis or gene ontology biological process (http://geneontology.org/) as listed in table 5. The pathway analysis of right colon biopsies suggests reduced mucin O-glycan biosynthesis in BAD, documented with downregulation of GALNT15.
Table 5.
Pathway enrichment analysis using EnrichR identified as differentially expressed in ileal or right colon mucosa in BAD compared with IBS-D without ABAM
| Term | Overlap | Padj value | OR | Combined score | Genes | DE expression | Confirmation of category |
|---|---|---|---|---|---|---|---|
| Terminal ileum mucosa | |||||||
| Complement and coagulation cascades | 1/79 | 0.0118 | 255.4 | 1236.8 | C6 | Up | KEGG_2019_Human |
| Right colon mucosa | |||||||
| Mucin type O-glycan biosynthesis | 1/31 | 0.0125 | 133.1 | 623.1 | GALNT15 | Down | KEGG_2019_Human |
| O-glycan processing | 1/65 | 0.0363 | 62.3 | 245.7 | GALNT15 | Down | GO_Biological _Process |
| Cytokine-cytokine receptor interaction | 3/294 | 0.0224 | 14.5 | 91.4 |
CCL25
CXCL5 IL1RN |
Up | KEGG_2019_Human |
ABAM, abnormal bile acid metabolism; BAD, bile acid diarrhoea; DE, differential expression; IBS-D, irritable bowel syndrome with diarrhoea; Padj, adjusted p.
A second pathway identified in the right colon mucosal biopsies pertains to immune activation with upregulation of chemokines CCL25 and CXCL5, whereas the upregulation of IL-1RN would antagonise the activation of interleukin-1 receptors, which may reflect adaptation to the inflammatory effects of the BAs. There was also upregulation of complement C6 in terminal ileal biopsies in BAD compared with IBS-D without ABAM. However, it is noted that, in each case, the pathway analysis identified only 1.0%–3.2% of the pathway and, in three cases resulting from only 1 gene (table 5). Importantly, these mechanisms had been identified in the analysis of the regulated genes (as detailed in table 4).
Microbiota
For all 194 participants with Rome III-positive IBS-D who provided a stool sample, we appraised the antibiotic, probiotic and fibre use listed in the medical record. Sixteen patients were receiving probiotics, and all were in the group with IBS-D without ABAM. Six patients were receiving antibiotics, one in the BAD group and five in the IBS-D without ABAM group. The prevalence of the use of antibiotics was not significant between the two groups (Fisher’s exact test statistic value=1, p>0.05). Twelve patients were receiving fibre supplements, 1 in the BAD group and 11 in the IBS-D without ABAM group. The prevalence of the use of fibre was not significant between the two groups (Fisher’s exact test statistic value=0.4704, p>0.05).
Patients with BAD had significantly lower alpha diversity (Shannon and Inverse Simpson indices, p≤0.001) and a different compositional profile based on beta diversity (Bray-Curtis and Jaccard distances, p≤0.001) compared with patients with IBS-D without ABAM (figure 5, upper panel). Patients with BAD and patients with IBS-D without ABAM had a different microbiome composition at the phylum, genus and species levels (figure 5, lower panel). Patients with BAD had a higher Firmicutes to Bacteroidetes ratio (p=0.0003, figure 5, upper panel) compared with patients with IBS-D without ABAM.
Figure 5.
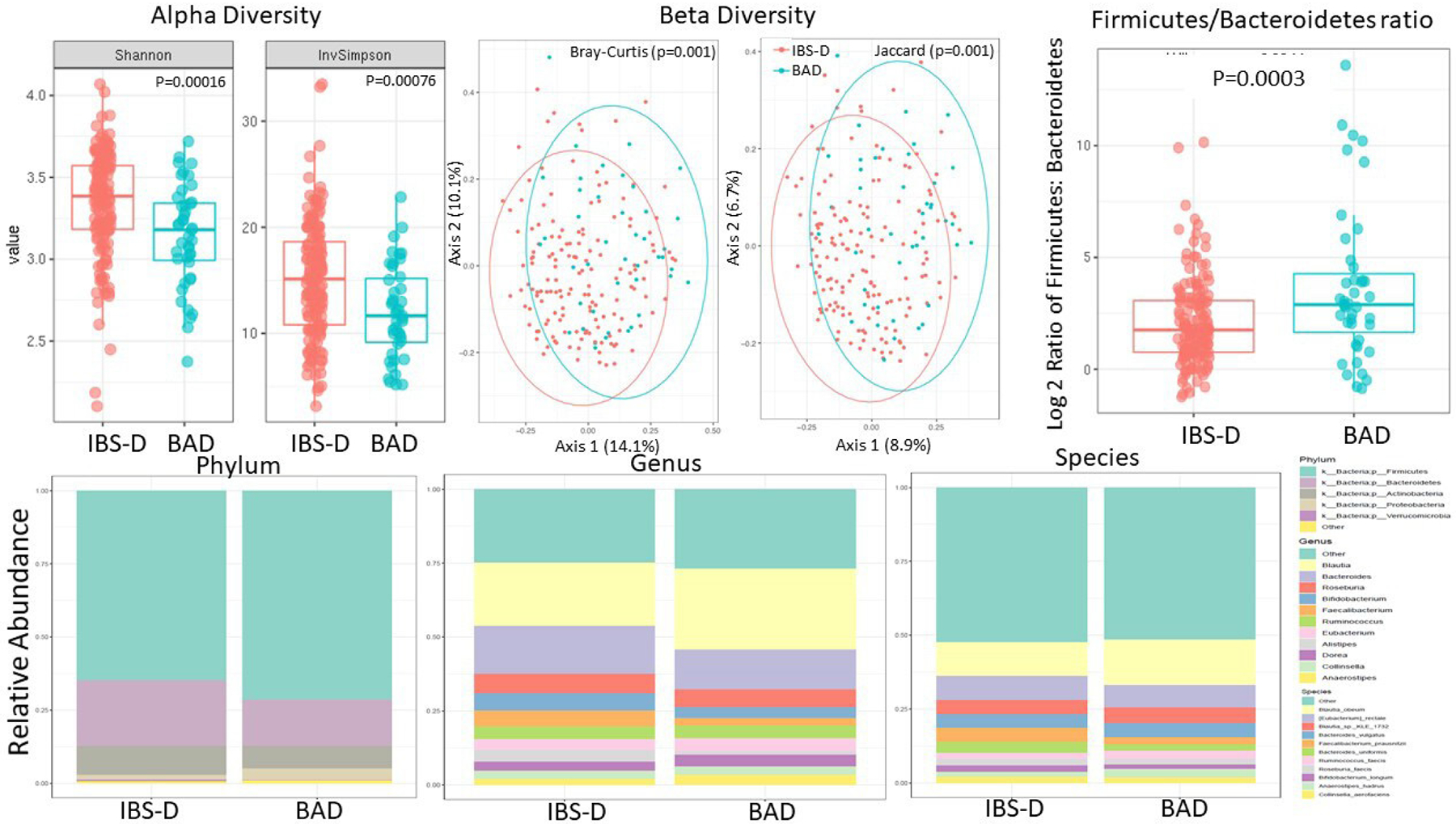
Upper panel: alpha diversity and beta diversity and Firmicutes to Bacteroidetes ratio are significantly different in patients with IBS-D without ABAM compared with patients with BAD. Lower panel: differences in microbiome composition at the phylum, genus and species levels in patients with IBS-D without ABAM compared with patients with BAD. Data based on SHOTGUN metagenomics. Note the reduced alpha diversity in patients with BAD and the different compositional profile based on beta diversity among the two groups. ABAM, abnormal bile acid metabolism; BAD, bile acid diarrhoea; IBS-D, irritable bowel syndrome with diarrhoea.
There are 29 differentially abundant genera of bacteria between patients with BAD and those with IBS-D without ABAM; 26 of the genera were decreased in BAD, including Alistipes, Clostridium and Bacteroides (online supplemental figure 2).
There were 70 differentially abundant species between the two groups, 61 of which were decreased in BAD. These are listed in online supplemental table 8 and in online supplemental figure 3. Among the bacterial species with decreased abundance, there were several Clostridia such as phoceensis, polynesiense and leptum; Faecalibacterium prausnitzii; Alistipes obesi and Alistipes finegoldii. The species with increased abundance included Erysipelatoclostridium ramosum.
Association of microbiota diversity with biochemical indicators of BAD
In addition to the overall association of diagnosed BAD with greater alpha and beta diversity of the microbiota and a higher Firmicutes to Bacteroidetes ratio, the level of fasting serum 7αC4 (more clearly demonstrated with log2 transformation (figure 6, upper panel)) and of the % primary faecal BAs in stool (figure 6, lower panel) were significantly associated with alpha and beta diversity. Moreover, differential taxa diversity was demonstrated with both these biochemical indices associated with BAD.
Figure 6.

Alpha and beta diversity of microbiota associated with log2 serum 7αC4 (upper panel) and per cent primary BAs in stool (lower panel). Patients with higher serum 7αC4 and higher per cent primary BAs had significantly lower alpha diversity. There was a significantly different microbial compositional profile based on beta diversity between participants with higher and lower markers of ABAM based on both parameters of ABAM. ABAM, abnormal bile acid metabolism; BAs, bile acids.
No significant association was found between colonic transit at 24 hours (GC24) and ascending colon emptying time (t50%) with alpha or beta diversity (online supplemental figure 4).
Microbial functional analysis
Functional analysis showed important differences in bacterial-encoded enzymes in microbial species associated with BAD, specifically decreased expression of BA thiol ligase (log2 fold change=−0.292; padj=0.042) by KEGG pathway analysis (online supplemental figure 5, left panel). These are involved in the transformation of primary BAs into secondary BAs and are associated with observed higher per cent primary BAs detected in stool of patients with BAD (1.3% vs 15%). KEGG pathway analysis showed there was also a reduced expression of sulfuric ester hydrolases or sulfatases (log2 fold change=−1.699; padj=0.0088).
By KEGG module analysis (online supplemental figure 5, right panel), patients with BAD also had evidence of reduced methanogenesis (adjusted all p<10−3) from diverse substrates (carbon dioxide, methanol and diverse methylamines). On the other hand, they also had increased tryptophan metabolism to kynurenine (log2 fold change=0.895; padj=0.0035), gamma-aminobutyrate synthesis from putrescine (log2 fold change=1.406; padj=0.041) and tyrosine synthesis (log2 fold change=1.673; padj=0.0002). The key to the modules >1 or <−1 log2 fold change is included in online supplemental table 7.
DISCUSSION
Our study has investigated the multidimensional clinical, biochemical, physiological, genomics, transcriptomics and microbiota of BAD in comparison with the appropriate disease comparator, that is, IBS-D without ABAM.
Clinical, biochemical, motor and sensory characteristics
We used well-established biochemical serum markers to differentiate BAD from IBS-D without ABAM and showed that biochemical indices in serum and stool of patients with BAD, with or without cholecystectomy, are similar. This justifies using the entire BAD cohort for subsequent mechanistic studies.
Our current study confirmed that BAD is associated with acceleration of colonic transit, and we had previously documented (in the same cohorts) the increased intestinal–colonic permeability in BAD compared with IBS-D without ABAM.10 We discuss our findings in perspective to the published literature under three main mechanistic evaluations: targeted genomics, mRNA sequencing in ileal and colonic mucosa, and microbiota.
Targeted DNA sequencing
The findings from the targeted DNA sequencing study of the known five pivotal BA genes confirmed rs1966265 variant in FGF-R4 as potentially relevant.31 FGF-R4 is the receptor protein on the hepatocyte membrane that transduces the signal from portal venous FGF-19 to intracellular synthesis of BAs. The protein KLB is also important for the interaction of FGF-19 with FGF-R4; indeed, prior studies showed that rs1966265 variant in FGF-R4 and rs17618244 variant in KLB are both associated with acceleration of colonic transit in IBS-D.32 Lee et al identified a DIET1 coding variant (rs12256835) that had skewed prevalence between 22 patients with BAD (with 75SeHCAT retention <10%) and 78 healthy controls, or 22 with chronic diarrhoea with 75SeHCAT retention >10%). DIET1 modulates intestinal production of the hormone, FGF-15, which is the murine analogue of the human FGF-19.33
RNA sequencing of mucosal biopsies
These measurements revealed important findings in the ileal and ascending colon biopsies. Thus, BAD was associated with increased ileal FGF-19 expression relative to healthy controls, and this would be consistent with increased BA absorption in BAD, possibly resulting from increased small intestinal BAs (as reflected in the high serum 7αC4). Prior studies documenting a role of FGF-19 in BAD were predominantly based on mucosal FGF-19 expression in response to incubation with CDCA or glyco-CDCA in vitro3; however, the fold changes in FGF-19 expression in established BAD patients were highly variable despite the overall significant correlation.3
The findings in mucosal mRNA expression in the ascending colon mucosal biopsies provide important mechanistic insights on the detergent and proinflammatory effects of BAs, particularly the primary di-α-OH-bile acid, CDCA, which was found at higher concentrations in the stool of patients with BAD compared with IBS-D without ABAM. Thus, upregulation of barrier, immune and inflammatory markers in the ascending colon biopsies in BAD reflects the known effects of BAs on colonic inflammation and increased colonic permeability, as documented in the same cohort of patients.10 The increased expression of the CLDN2 gene identified in the current study conforms with documentation that claudin-2 is a mediator of leaky gut barrier during intestinal inflammation.34 Another effect on barrier function is suggested by the KEGG pathway analysis showing reduced mucin O-glycan biosynthesis in BAD, documented with downregulation of GALNT15 in ascending colon biopsies. Mucins are highly O-glycosylated glycoproteins. This loss of mucin is consistent with the depletion of surface mucus and goblet cell mucus in experimental in vivo and in vitro models of effects of CDCA on colonic mucosa or cell lines.
The upregulation of complement and cytokine mRNA is consistent with the prior report of association of increased secondary BAs with a significant increase in expression of inflammatory cytokines in colonic mucosa in patients with alcoholic cirrhosis.35 Our data also suggest mucosal adaptations to the immune activation, as demonstrated by the upregulation of a gene antagonising the synthesis of the IL-1 receptor. Overall, pathway analysis (confirmed by KEGG pathway analysis or gene oncology biological process) confirmed mechanisms (mucin production and immune activation) identified in the DE of individual genes.
The upregulation of inhibitors of serine proteases may potentially protect the mucosa from such proteases. In a mouse model of IBS exposed to high protease activity documented in supernatant of faeces from patients with post-infectious IBS or IBS-C,36 there was evidence of barrier dysfunction. The increased expression of protease inhibitors may therefore be protective in BAD.
An earlier study had documented DNA sequence variations in FABP6, but overall frequencies were similar in patients with primary BAD and healthy controls.37 Our current study shows upregulated expression of FAPB6 in ascending colon mucosa in BAD. FAPB6 is associated with BA binding and could conceivably impact the biological effects of the higher intraluminal BAs in BAD. The finding of marked downregulation in terminal ileal mucosa of SLC44A5 is intriguing, as this is involved in transport of glucose, other sugars, bile salts, organic acids, metal ions and amines. This requires further research.
The level of TGR5 immunoreactivity in rectosigmoid mucosal biopsies was significantly higher in patients with IBS-D than in healthy controls38; however, we did not find differences in mRNA expression of TGR5 in ascending or descending colon biopsies in BAD compared with IBS-D without ABAM. The lack of differences may reflect that 50%–75% of BAs are absorbed in the mammalian colon6 and that TGR5 is activated in colonic mucosa by several BA species, including the secondary BAs, lithocholic acid (LCA) and deoxycholic acid (DCA), which predominate in IBS-D. We recently showed that, in BAD, the BA sequestrant, colesevelam, decreased expression in sigmoid mucosa of NR1H4 and P2RY4 (relative to baseline) and increased expression of TGR5 compared with placebo.39
Alterations in microbiota and functional analysis
The studies of the faecal microbiota and the associated functional effects provide interesting insights on their potential role in BAD. Thus, patients with BAD have a higher Firmicutes to Bacteroidetes ratio in stool. Increased colonic BAs reduce microbial diversity, decrease Bacteroidetes and increase Firmicutes, which in turn increases secondary BA production, stimulates colonic secretion by DCA and accelerates colonic transit (eg, by stimulation of TGR5 receptors) by both LCA and DCA.40 41 Our observations in patients are consistent with studies in rodents fed BAs or high fat diets that displayed decreased Bacteroidetes and expansion of Firmicutes.42 Conjugated BAs have a lower ability to inhibit intestinal aerobic and anaerobic bacteria compared with deconjugated CA and DCA.43 Bile salt hydrolases (BSH), which deconjugate BAs, are encoded by diverse microorganisms, including Clostridium, Bacteroides, Lactobacillaceae, Bifidobacterium, Enterococcus and Archaea. Deconjugated BAs may inhibit certain bacteria, whereas other bacteria are more sensitive to conjugated BAs and, thus, BSH encoded by the previously mentioned bacteria may help render the colonic environment less toxic.44 It is notable that we observed the patients with BAD had changes in several of these microbial species. Alterations in gut microbiota in relation to BA metabolism in IBS have been summarised.45 Clostridia were associated with enhanced BA excretion in IBS-D with weak correlation (Rs=0.2) with Clostridium scindens and serum 7αC4 level.46
The other major actions of the colonic microbiota on BAs are dehydroxylation and sulfation. Dehydroxylation may result in a balanced effect on the overall detergent properties of BAs, since CDCA is converted to LCA (which has no detergent effects, but it can certainly stimulate colonic motility via TGR5 receptors40 47) and CA is converted to the detergent molecule, DCA. Our functional analysis of the microbiota in BAD revealed decreased BA thiol ligases which are involved in the transformation of primary to secondary BAs. Accordingly, we also observed higher per cent primary BAs in stool of patients with BAD (15%) compared with IBS-D without ABAM (1.3%).
Sulfation could also neutralise the secretory effects of BAs. There are intestinal mucosal41 as well as bacterial sulfotransferase enzymes that may sulfate the BAs. Sulfation reduces the secretory effects of DCA48 and was associated with functional constipation in childhood.49 Efficient sulfation results in rapid faecal excretion of bile acids, so that the total lithocholate pool remains small,50 and reduces stimulation of colonic motility through TGR5 receptors. One of the functional effects of the microbiota in BAD was reduced sulfatase, which would increase levels of sulfated primary BAs and potentially reduce the toxic effects by the di-α-hydroxy BA, CDCA, potentially an adaptation in the microbiota in response to chronically elevated colonic BA levels.
Dysbiosis and altered BA metabolism may also conceivably impact gut inflammation, as demonstrated in patients with inflammatory bowel diseases, with decrease in Firmicutes in remission phylum and a more profound decrease in Firmicutes, and an increase in Lactobacillus and Enterobacteria (E. coli at a species level) during flares.51 The potential association between BA exposure in BAD and its proinflammatory effects (complement and chemokines) in the ascending colon in our study deserves further study in larger cohorts, especially given the potential role of BAs in microscopic colitis.
The functional analysis of the microbiota revealed reduced methanogenesis in BAD. This is consistent with the converse, or increased methanogenesis, previously reported with IBS-C or constipation.52
Short chain fatty acids
The microbiota is involved in metabolism of the complex carbohydrates that reach the colon and in the production of SCFAs53 and, to a lesser extent, organic acids and amino acids. Bacteroidetes mainly produce acetate and propionate, while Firmicutes (in particular, Faecalibacterium prausnitzii and Clostridium leptum of the family Ruminococcaceae, and Eubacterium rectale and Roseburia spp of the family Lachnospiraceae54) mostly produce butyrate in the human gut.
Our observation of no significant difference in SCFAs between BAD and IBS-D without ABAM was somewhat surprising, given the observed Firmicutes to Bacteroidetes ratio.
Proposed integration and interpretation of molecular mechanisms observed in BAD
The observations in patients with BAD are consistent with the known association between increased serum 7αC4, decreased serum FGF-19 and increased percentage of faecal primary BAs in association with induction of diarrhoea. In addition, the functional analyses of the altered faecal microbial composition in patients with BAD shows effects on dehydroxylation and sulfation, which, as explained above, could also be contributing to the development of diarrhoea. In the absence of intervention directed either at the microbiota or at the BAs (eg, with BA sequestrant) which may influence the microbial population, it is not possible to unequivocally determine the biological effects responsible for the diarrhoea between the microbiota and the BAs.
Acceleration of colonic transit was correlated with elevated serum 7C4 in the patients with BAD; it is not possible to be certain that the acceleration of transit in BAD is exclusively caused by the increased synthesis (and presumably excretion) of BAs rather than another mechanism such as increased colonic motility. However, it is worth noting that rectal infusion of CDCA is associated with induction of high amplitude propagated contractions,55 55 and other BAs, including LCA, stimulate the TGR5 receptor resulting in stimulation of colonic motility.47
The mucosal biopsies showed evidence of immune activation or inflammation and increased permeability, which could conceivably be contributing to the diarrhoea in BAD, as shown in figures 3 and 4 and table 4. Importantly, the serine protease inhibition was increased, arguing against a role of serine protease in the induction of diarrhoea in BAD.
Conversely, faecal SCFAs do not appear to be specifically contributing to the development of the diarrhoea because the quantitation is similar in BAD and in IBS-D without ABAM. Similarly, we did not identify a relationship between variations in five genes of interest in the synthesis or function of BAs and the diagnosis of BAD.
Limitations
Only 60 of 205 participants consented to perform the rectal sensation studies. There were >90% of all the patients who participated in the other measurements. While the statistical power calculations for fold change expressions had anticipated 40 patients with IBS-D without ABAM and 20 patients with BAD (online supplemental table 1), the final count (impacted by closure of research unit due to COVID-19 pandemic) was, respectively, patients 34 and 10 patients as well as 30 healthy controls. Nevertheless, several important fold differences, well above the 1.693 predicted in the power calculation, were noted (see table 4) indicating sufficient power.
CONCLUSIONS
Important differences in pathobiological mechanisms between BAD and IBS-D without ABAM have the potential to enhance diagnosis and treatment of BAD by targeting barrier dysfunction and inflammatory and microbial changes.
Supplementary Material
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT?
There are altered mucosal functions in irritable bowel syndrome with diarrhoea (IBS-D); ~30% of patients with IBS-D have bile acid (BA) diarrhoea (BAD). We need to understand mechanisms that mediate BAD to improve diagnosis and treatment.
WHAT ARE THE NEW FINDINGS?
Patients with BAD had significantly faster colonic transit, higher Firmicutes to Bacteroidetes ratio with decreased dehydroxylation and sulfatases, and their ascending colon biopsies showed upregulation in barrier-weakening genes (CLDN2) and immune activation.
HOW MIGHT IT IMPACT ON CLINICAL PRACTICE IN THE FORESEEABLE FUTURE?
Understanding the pathobiology of BAD opens opportunities for novel treatments, including modification of microbiota, facilitating conversion to secondary BAs and possibly anti-inflammatory approaches.
Acknowledgements
The authors wish to thank Michael Ryks and Lisa Tebay, Registered Nurse for technical assistance, Cindy Stanislav for secretarial assistance, and the Immunochemistry Core Lab of the Mayo CCaTS for measurements of stool short chain fatty acids (Mai Petterson, PhD).
Funding
MC is supported from National Institutes of Health (NIH) (grant number: R01-DK115950). The study was facilitated by the CCaTS Clinical Research Trials Unit at Mayo Clinic (grant number: UL1-TR002377) from NIH.
Footnotes
Competing interests None declared.
Patient and public involvement Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Ethics approval This study involves human participants and was approved by Mayo Clinic’s institutional review board (IRB #16–001445). Participants gave informed consent to participate in the study before taking part and for their electronic medical records to be used for research purposes.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. Data transparency statement: data will be available, consistent with data sharing National Institutes of Health (NIH) policy for studies supported by NIH (in this case, R01-DK115950); in addition, all relevant data are included in the paper and/or in the online supplemental materials.
REFERENCES
- 1.Camilleri M, Carlson P, Acosta A, et al. RNA sequencing shows transcriptomic changes in rectosigmoid mucosa in patients with irritable bowel syndrome-diarrhea: a pilot case-control study. Am J Physiol Gastrointest Liver Physiol 2014;306:G1089–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valentin N, Camilleri M, Altayar O, et al. Biomarkers for bile acid diarrhoea in functional bowel disorder with diarrhoea: a systematic review and meta-analysis. Gut 2016;65:1951–9. [DOI] [PubMed] [Google Scholar]
- 3.Johnston IM, Nolan JD, Pattni SS, et al. Characterizing factors associated with differences in FGF19 blood levels and synthesis in patients with primary bile acid diarrhea. Am J Gastroenterol 2016;111:423–32. [DOI] [PubMed] [Google Scholar]
- 4.Chang C, Jiang J, Sun R, et al. Downregulation of serum and distal ileum fibroblast growth factor19 in bile acid diarrhoea patients. Dig Dis Sci 2022;67:872–9. [DOI] [PubMed] [Google Scholar]
- 5.Duboc H, Taché Y, Hofmann AF. The bile acid TGR5 membrane receptor: from basic research to clinical application. Dig Liver Dis 2014;46:302–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mekhjian HS, Phillips SF, Hofmann AF. Colonic absorption of unconjugated bile acids: perfusion studies in man. Dig Dis Sci 1979;24:545–50. [DOI] [PubMed] [Google Scholar]
- 7.Camilleri M, Busciglio I, Acosta A, et al. Effect of increased bile acid synthesis or fecal excretion in irritable bowel syndrome-diarrhea. Am J Gastroenterol 2014;109:1621–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev 2005;29:625–51. [DOI] [PubMed] [Google Scholar]
- 9.Longstreth GF, Thompson WG, Chey WD, et al. Functional bowel disorders. Gastroenterology 2006;130:1480–91. [DOI] [PubMed] [Google Scholar]
- 10.Magnus Y, BouSaba J, Sannaa W, et al. Bile acid diarrhea is associated with increased intestinal permeability compared with irritable bowel Syndrome-Diarrhea. Gastroenterology 2022;162:1343–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mars RAT, Yang Y, Ward T, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell 2020;182:1460–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moazeni-Roodi A, Sarabandi S, Karami S, et al. An updated meta-analysis of the association between fibroblast growth factor receptor 4 polymorphisms and susceptibility to cancer. Biosci Rep 2020;40:BSR20192051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asmann YW, Middha S, Hossain A, et al. TREAT: a bioinformatics tool for variant annotations and visualizations in targeted and exome sequencing data. Bioinformatics 2012;28:277–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kocher J-PA, Quest DJ, Duffy P, et al. The biological reference Repository (BioR): a rapid and flexible system for genomics annotation. Bioinformatics 2014;30:1920–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, Evans JM, Bhagwate AV, et al. PatternCNV: a versatile tool for detecting copy number changes from exome sequencing data. Bioinformatics 2014;30:2678–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bushnell B BBTools software package, 2014. Available: http://sourceforge.net/projects/bbmap
- 18.Chu J, Sadeghi S, Raymond A, et al. BioBloom tools: fast, accurate and memory-efficient host species sequence screening using bloom filters. Bioinformatics 2014;30:3402–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Ghalith GA, Hillmann B, Ang K, et al. SHI7 is a Self-Learning pipeline for multipurpose short-read DNA quality control. mSystems 2018;3:e00202–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H, He K, Chen J. LinDA: linear models for differential abundance analysis of microbiome compositional data. arXiv:2104 2021;00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalari KR, Nair AA, Bhavsar JD, et al. MAP-RSeq: Mayo analysis pipeline for RNA sequencing. BMC Bioinformatics 2014;15:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013;41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z, Bailey A, Kuleshov MV, et al. Gene set knowledge discovery with Enrichr. Curr Protoc 2021;1:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyoto encyclopedia of genes and genomes. Available: https://www.genome.jp/kegg/#
- 27.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon A, Birger C, Thorvaldsdóttir H, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camilleri M, Carlson P, Acosta A, et al. Colonic mucosal gene expression and genotype in irritable bowel syndrome patients with normal or elevated fecal bile acid excretion. Am J Physiol Gastrointest Liver Physiol 2015;309:G10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.BouSaba J, Sannaa W, McKinzie S, et al. Impact of bile acid diarrhea in patients with diarrhea-predominant irritable bowel syndrome on symptoms and quality of life. Clin Gastroenterol Hepatol 2021;S1542–3565:01273–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camilleri M, Klee EW, Shin A, et al. Irritable bowel syndrome-diarrhea: characterization of genotype by exome sequencing, and phenotypes of bile acid synthesis and colonic transit. Am J Physiol Gastrointest Liver Physiol 2014;306:G13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong BS, Camilleri M, Carlson PJ, et al. A Klothoβ variant mediates protein stability and associates with colon transit in irritable bowel syndrome with diarrhea. Gastroenterology 2011;140:1934–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JM, Ong JR, Vergnes L, et al. Diet 1, bile acid diarrhea, and FGF15/19: mouse model and human genetic variants. J Lipid Res 2018;59:429–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luettig J, Rosenthal R, Barmeyer C, et al. Claudin-2 as a mediator of leaky gut barrier during intestinal inflammation. Tissue Barriers 2015;3:e977176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kakiyama G, Hylemon PB, Zhou H, et al. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastrointest Liver Physiol 2014;306:G929–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edogawa S, Edwinson AL, Peters SA, et al. Serine proteases as luminal mediators of intestinal barrier dysfunction and symptom severity in IBS. Gut 2020;69:62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balesaria S, Pell RJ, Abbott LJ, et al. Exploring possible mechanisms for primary bile acid malabsorption: evidence for different regulation of ileal bile acid transporter transcripts in chronic diarrhoea. Eur J Gastroenterol Hepatol 2008;20:413–22. [DOI] [PubMed] [Google Scholar]
- 38.Wei W, Wang H, Zhang Y, et al. Faecal bile acids and colonic bile acid membrane receptor correlate with symptom severity of diarrhoea-predominant irritable bowel syndrome: a pilot study. Dig Liver Dis 2021;53:1120–7. [DOI] [PubMed] [Google Scholar]
- 39.Vijayvargiya P, Camilleri M, Carlson P, et al. Effects of colesevelam on bowel symptoms, biomarkers, and colonic mucosal gene expression in patients with bile acid diarrhea in a randomized trial. Clin Gastroenterol Hepatol 2020;18:2962–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li N, Koester ST, Lachance DM, et al. Microbiome-encoded bile acid metabolism modulates colonic transit times. iScience 2021;24:102508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dew MJ, Hawker PC, Nutter S, et al. Human intestinal sulphation of lithocholate: a new site for bile acid metabolism. Life Sci 1980;27:317–23. [DOI] [PubMed] [Google Scholar]
- 42.Ridlon JM, Harris SC, Bhowmik S, et al. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016;7:22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taranto MP, Perez-Martinez G, Font de Valdez G. Effect of bile acid on the cell membrane functionality of lactic acid bacteria for oral administration. Res Microbiol 2006;157:720–5. [DOI] [PubMed] [Google Scholar]
- 44.Doden HL, Ridlon JM. Microbial hydroxysteroid dehydrogenases: from alpha to omega. Microorganisms 2021;9:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhan K, Zheng H, Li J, et al. Gut microbiota-bile acid crosstalk in diarrheairritable bowel syndrome. Biomed Res Int 2020;2020:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao L, Yang W, Chen Y, et al. A Clostridia-rich microbiota enhances bile acid excretion in diarrhea-predominant irritable bowel syndrome. J Clin Invest 2020;130:438–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alemi F, Poole DP, Chiu J, et al. The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology 2013;144:145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breuer NF, Rampton DS, Tammar A, et al. Effect of colonic perfusion with sulfated and nonsulfated bile acids on mucosal structure and function in the rat. Gastroenterology 1983;84:969–77. [PubMed] [Google Scholar]
- 49.Hofmann AF, Loening-Baucke V, Lavine JE, et al. Altered bile acid metabolism in childhood functional constipation: inactivation of secretory bile acids by sulfation in a subset of patients. J Pediatr Gastroenterol Nutr 2008;47:598–606. [DOI] [PubMed] [Google Scholar]
- 50.Cowen AE, Korman MG, Hofmann AF, et al. Metabolism of lithocholate in healthy man. II. enterohepatic circulation. Gastroenterology 1975;69:67–76. [PubMed] [Google Scholar]
- 51.Duboc H, Rajca S, Rainteau D, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013;62:531–9. [DOI] [PubMed] [Google Scholar]
- 52.Kim G, Deepinder F, Morales W, et al. Methanobrevibacter smithii is the predominant methanogen in patients with constipation-predominant IBS and methane on breath. Dig Dis Sci 2012;57:589–95. [DOI] [PubMed] [Google Scholar]
- 53.Stephen AM, Haddad AC, Phillips SF. Passage of carbohydrate into the colon. direct measurements in humans. Gastroenterology 1983;85:589–95. [PubMed] [Google Scholar]
- 54.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 2017;19:29–41. [DOI] [PubMed] [Google Scholar]
- 55.Bampton PA, Dinning PG, Kennedy ML, et al. The proximal colonic motor response to rectal mechanical and chemical stimulation. Am J Physiol Gastrointest Liver Physiol 2002;282:G443–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information. Data transparency statement: data will be available, consistent with data sharing National Institutes of Health (NIH) policy for studies supported by NIH (in this case, R01-DK115950); in addition, all relevant data are included in the paper and/or in the online supplemental materials.


