One of the most basic tasks of every cell is the synthesis of ribosomes, yet the process is so complex that we are only beginning to comprehend it. In human cells, the final product contains four RNAs and 79 proteins forming a large ribonucleoprotein, the ribosome. About twice as many factors are involved in assembly of these protein synthesizing machines. Additionally, some 200 small nucleolar ribonucleoproteins (snoRNPs) function in the modification of preribosomal RNA (pre-rRNA) at a similar number of ribonucleotides. Modification is essential for proper function and biogenesis of ribosomes (Decatur and Fournier 2002; Sloan et al. 2016). While abrogation of individual modifications has little to no measurable effects, removal of a combination of modifications or of all causes severe to lethal effects. Modifications mostly cluster around the functional centers of the ribosome, and some modifications are important for ribosome assembly. The subject of this perspective is to raise awareness to how modification at so many sites can be achieved while the pre-rRNA is being processed, folded and refolded, and assembled with ribosomal proteins to yield the preribosomal subunits in the nucleolus (Baßler and Hurt 2019). This is not a comprehensive review but a simplification of complex issues providing access to the general reader.
This perspective exclusively concentrates on mammalian snoRNP-mediated RNA modification, which accounts for 95% of all 228 modified residues in human rRNA (Taoka et al. 2018). There are two kinds of snoRNPs: box H/ACA for pseudouridylation and box C/D for 2′-O-methylation (Fig. 1A). Each snoRNP contains a short 60–150 nt-long H/ACA or C/D RNA, which base-pairs with 8 to 20 nt flanking (H/ACA) or overlapping with (C/D) their target nucleotides. In addition to the pseudouridine synthase and methyltransferase for the respective box H/ACA and box C/D snoRNPs, each snoRNP has three other core proteins that comprise the ribonucleoprotein. The snoRNAs guide the enzymes of the RNPs to their specific nucleotide for modification by site-directed base-pairing (Fig. 1A). As most snoRNAs can target two sites in target RNAs (Fig. 1A), they potentially guide modification at two nucleotides (Kiss 2001). Additionally, the occasional snoRNP is not involved in modification, but functions in chaperoning pre-rRNA folding, as most prominently documented by the U3 C/D snoRNP and the U17/snR30 H/ACA snoRNP (Dragon et al. 2002; Fayet-Lebaron et al. 2009). If and how many modification-guide snoRNPs function in similar roles is not known.
FIGURE 1.
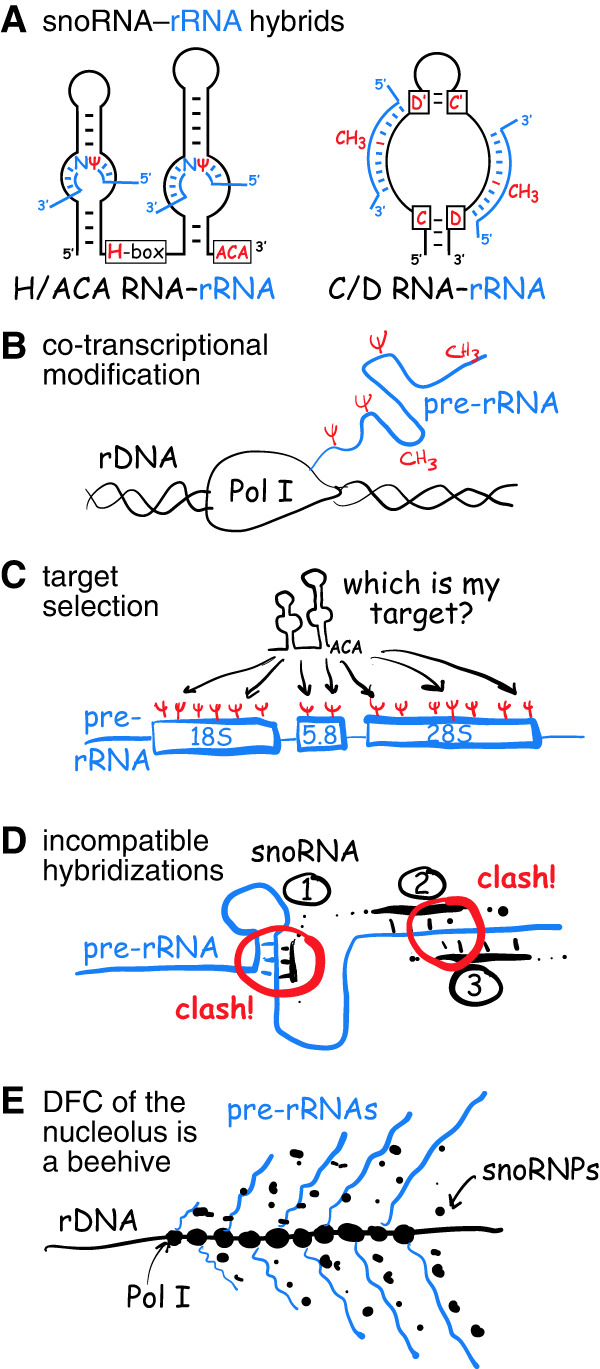
(A) Pre-rRNA (blue) hybridizes with H/ACA snoRNAs to isomerize uridine (U) to pseudouridine (Ψ, red) and with C/D snoRNAs to methylate ribose at the 2′-position (CH3, red). (B) Pre-rRNA is modified while transcribed by RNA polymerase I (Pol I). (C) Only segments incorporated into mature ribosomes are modified (sites are represented by Ψs, red). But how does each snoRNA find its target? (D) Some snoRNA-guided modifications occur in double-stranded regions of mature rRNA (1) and next to each other (2 and 3) presenting potential clashes of two hybrids (circled red). (E) Transcribing Pol I forms trains on rDNA genes, from which nascent pre-rRNAs emerge in a Christmas tree–like fashion on electron microscope grids. This occurs in the dense fibrillar component (DFC) of the nucleolus enveloped in a cloud of snoRNPs looking for target nucleotides to modify and rendering the DFC akin to a beehive.
Pre-rRNAs are modified while in their nascent state when they emerge from RNA polymerase I, that is, cotranscriptionally (Fig. 1B). Using pulse-chase labeling with 14C-methionine to monitor rRNA methylation in HeLa cells, the incorporation into 45S pre-rRNA was originally documented (Greenberg and Penman 1966). Using more precise pulse labeling of yeast pre-rRNA combined with mathematical modeling (Koš and Tollervey 2010), or by RiboMeth-seq analysis of chromatin-associated RNA (Birkedal et al. 2015), cotranscriptional methylation of pre-rRNA was confirmed. Importantly, modification-guide snoRNPs are rarely detected in the cryo-EM structures of preribosomes, suggesting that they have already left after completing their task and before assembly of the earliest defined pre-60S and pre-40S intermediates, for example, in nucleolar pre-60S particles (Kater et al. 2017; Sanghai et al. 2018; Zhou et al. 2019). Consequently, each nascent pre-rRNA needs to be sampled by some 100–200 snoRNPs for base-pairing to complementary sites.
Modification of human RNA must be efficient and fast. Human rRNA is transcribed as a 13 kb (47S) precursor, from which the 18S, 5.8S, and 28S mature rRNAs are processed. Although modification occurs cotranscriptionally, only the segments destined for mature ribosomes are modified (Fig. 1C). RNA polymerase I transcribes rDNA at a rate of about 95 nt per second (Dundr et al. 2002). Consequently, it takes about 2 min and 20 sec to transcribe the entire 47S pre-rRNA, but only 1 min and 15 sec to cover the 18S, 5.8S, and 28S rRNA segments. Given that these segments together incorporate about 200 modifications, each modification must occur on average in a fraction of a second, that is, in 0.37 sec. During that fraction of a second, each one of the 100–200 snoRNPs must sample the entire pre-rRNA, base-pair with it, modify the target nucleotide, and release the pre-rRNA. As rRNA is modified to near completion at most sites (Birkedal et al. 2015; Taoka et al. 2018), modification needs to be continuously monitored and validated or sampling and catalysis must be incredibly efficient.
The problem is complicated by the fact that there appears to be an order to modification (Fig. 1D). Although the cognate guide RNAs for their target nucleotide often lack experimental validation, regions of snoRNA–pre-rRNA hybridization overlap between individual snoRNAs in some 40% of instances (Fig. 1D, #2 and #3; Lestrade and Weber 2006), necessitating sequential modification by those guide RNAs. However, the order of modification is anybody's guess. Additionally, modified rRNA nucleotides often reside in double-stranded regions of the folded product (Fig. 1D, #1), requiring the modifications to occur before folding pre-rRNA in this region to the mature rRNA (Smith and Steitz 1997).
Further increasing the complexity, some 100 RNA polymerase I enzymes form trains on rDNA genes and produce an equal number of nascent pre-rRNA transcripts side-by-side (Fig. 1E). These transcripts have been famously visualized as “Christmas trees” on electron microscopy grids (Miller and Beatty 1969). Every one of these neighboring pre-rRNAs needs to be modified by 100–200 individual snoRNPs, that is, minimally 100–200, but potentially up to 20,000 snoRNPs per rDNA gene locus. Considering that at least 100 rDNA genes are being transcribed simultaneously, the required snoRNP number could increase to 2,000,000, equating to an average of 10,000–20,000 molecules for each individual snoRNA, which is close to that estimated (Yu et al. 1999; Fafard-Couture et al. 2021). Alternatively, if only 100–200 individual snoRNPs service all 100 pre-rRNAs at a given locus, how is it assured that they all get modified to the same extent? Is there competition for snoRNPs between neighboring pre-rRNA transcripts?
The modification of pre-rRNA to near completion at most sites (Birkedal et al. 2015; Taoka et al. 2018; Marchand et al. 2020) is further confounded by the problem of snoRNA abundance. The abundance of human snoRNAs that modify rRNA varies by orders of magnitude, and all are encoded in introns of genes. Let us take the example of the DKC1 gene encoding the pseudouridine synthase of H/ACA RNPs, NAP57/dyskerin (Meier and Blobel 1994; Heiss et al. 1998). DKC1 harbors SNORA36A and SNORA56 in its introns 8 and 13, respectively. The predicted target sites of those two H/ACA RNAs in 18S and 28S rRNA are fully modified in 100% and 97% of rRNAs, respectively (Taoka et al. 2018). Yet, the abundance of SNORA36A and SNORA56 differs by an order of magnitude when averaged over seven human tissues (Fafard-Couture et al. 2021).
Considering these issues and despite the ubiquitous importance of ribosome synthesis, pre-rRNA modification poses a severe temporal and spatial problem to the cell. For example, imagine the topological constraints posed by the “Christmas trees” in three dimensions and when highly compacted (Fig. 1E). All these processes and molecules congregate in the confines of the dense fibrillar component (DFC) of the nucleolus. Recent evidence suggests that the nucleolus, like other nuclear and cytoplasmic membrane-less organelles, is formed through phase separation (Feric et al. 2016; Yao et al. 2019; Lafontaine et al. 2021; Wu et al. 2021). Perhaps it is this specialized phase that allows modification (including snoRNP movement and pre-rRNA sampling) to occur within a fraction of a second. In other words, intrinsically disordered regions of snoRNP proteins may allow for their concentration while permitting rapid movement.
Some explanation may be found in the interaction of bacterial sRNAs with their chaperone Hfq (Roca et al. 2022). In that case, as perhaps in that of snoRNPs and their targets, regulation occurs through on-rates and competition with overlapping hybridization sites. Moreover, different catalytic prowess between individual snoRNPs could explain the discrepancy between enzyme abundance and degree of modification. Such differences in catalysis have been observed between H/ACA snoRNPs and between the two hairpins of the same RNP (Caton et al. 2017; Trucks et al. 2021). Finally, human disease attests to the importance of these “housekeeping” processes when even minor genetic or spontaneous mutations that alter modification cause life-threatening conditions, ribosomopathies (Ruggero and Shimamura 2014). Obviously, we have our work cut out to untangle the daunting task of rRNA modification.
ACKNOWLEDGMENTS
I thank Susan Smith, Eric Phizicky, John Woolford and two other reviewers for improving the content and clarity of this perspective. The work in my laboratory is supported by a grant from the National Institutes of Health (HL136662).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079391.122.
Freely available online through the RNA Open Access option.
REFERENCES
- Baßler J, Hurt E. 2019. Eukaryotic ribosome assembly. Annu Rev Biochem 88: 281–306. [DOI] [PubMed] [Google Scholar]
- Birkedal U, Christensen-Dalsgaard M, Krogh N, Sabarinathan R, Gorodkin J, Nielsen H. 2015. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew Chem Int Ed Engl 54: 451–455. [DOI] [PubMed] [Google Scholar]
- Caton EA, Kelly EK, Kamalampeta R, Kothe U. 2017. Efficient RNA pseudouridylation by eukaryotic H/ACA ribonucleoproteins requires high affinity binding and correct positioning of guide RNA. Nucleic Acids Res 46: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. 2002. rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351. [DOI] [PubMed] [Google Scholar]
- Dragon F, Gallagher JEG, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298: 1623–1626. [DOI] [PubMed] [Google Scholar]
- Fafard-Couture É, Bergeron D, Couture S, Abou-Elela S, Scott MS. 2021. Annotation of snoRNA abundance across human tissues reveals complex snoRNA-host gene relationships. Genome Biol 22: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet-Lebaron E, Atzorn V, Henry Y, Kiss T. 2009. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J 28: 1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165: 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg H, Penman S. 1966. Methylation and processing of ribosomal RNA in HeLa cells. J Mol Biol 21: 527–535. [DOI] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. 1998. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 19: 32–38. [DOI] [PubMed] [Google Scholar]
- Kater L, Thoms M, Barrio-Garcia C, Cheng J, Ismail S, Ahmed YL, Bange G, Kressler D, Berninghausen O, Sinning I, et al. 2017. Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell 171: 1599–1610.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. Embo J 20: 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koš M, Tollervey D. 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. 2021. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Bio 22: 165–182. [DOI] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ. 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34: D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Pichot F, Neybecker P, Ayadi L, Bourguignon-Igel V, Wacheul L, Lafontaine DLJ, Pinzano A, Helm M, Motorin Y. 2020. HydraPsiSeq: a method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic Acids Res 48: gkaa769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT, Blobel G. 1994. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol 127: 1505–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL, Beatty BR. 1969. Visualization of nucleolar genes. Science (New York, NY) 164: 955–957. [DOI] [PubMed] [Google Scholar]
- Roca J, Santiago-Frangos A, Woodson SA. 2022. Diversity of bacterial small RNAs drives competitive strategies for a mutual chaperone. Nat Commun 13: 2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Shimamura A. 2014. Marrow failure: a window into ribosome biology. Blood 124: 2784–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghai ZA, Miller L, Molloy KR, Barandun J, Hunziker M, Chaker-Margot M, Wang J, Chait BT, Klinge S. 2018. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 556: 126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Warda AS, Sharma S, Entian K-D, Lafontaine DLJ, Bohnsack MT. 2016. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol 14: 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89: 669–672. [DOI] [PubMed] [Google Scholar]
- Taoka M, Nobe Y, Yamaki Y, Sato K, Ishikawa H, Izumikawa K, Yamauchi Y, Hirota K, Nakayama H, Takahashi N, et al. 2018. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res 46: 9289–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucks S, Hanspach G, Hengesbach M. 2021. Eukaryote specific RNA and protein features facilitate assembly and catalysis of H/ACA snoRNPs. Nucleic Acids Res 49: 4629–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Xu G, Han C, Luan P-F, Xing Y-H, Nan F, Yang L-Z, Huang Y, Yang Z-H, Shan L, et al. 2021. lncRNA SLERT controls phase separation of FC/DFCs to facilitate Pol I transcription. Science 373: 547–555. [DOI] [PubMed] [Google Scholar]
- Yao R-W, Xu G, Wang Y, Shan L, Luan P-F, Wang Y, Wu M, Yang L-Z, Xing Y-H, Yang L, et al. 2019. Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol Cell 76: 767–783.e11. [DOI] [PubMed] [Google Scholar]
- Yu YT, Scharl EC, Smith CM, Steitz JA. 1999. The growing world of small nuclear ribonucleoproteins. In The RNA world (ed. Gesteland RF, Cech TR, Atkins JF), pp. 487–524. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Zhou D, Zhu X, Zheng S, Tan D, Dong M-Q, Ye K. 2019. Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell 10: 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]


