Abstract
Nucleic acid nanoparticles, or NANPs, rationally designed to communicate with the human immune system, can offer innovative therapeutic strategies to overcome the limitations of traditional nucleic acid therapies. Each set of NANPs is unique in their architectural parameters and physicochemical properties, which together with the type of delivery vehicles determine the kind and the magnitude of their immune response. Currently, there are no predictive tools that would reliably guide the design of NANPs to desired immunological outcome, a step crucial for the success of personalized therapies. Through a systematic approach investigating physicochemical and immunological profiles of a comprehensive panel of various NANPs, our research team has developed and experimentally validated a computational model based on the transformer architecture able to predict the immune activities of NANPs. We anticipate that the freely accessible computational tool that we call an “artificial immune cell,” or AI-cell, will aid in addressing in a timely manner the current critical public health challenges related to safety criteria of nucleic acid therapies and promote the development of novel biomedical tools.
Keywords: NANPs, RNA nanotechnology, immunology, immunorecognition, artificial intelligence, machine learning
Graphical Abstract
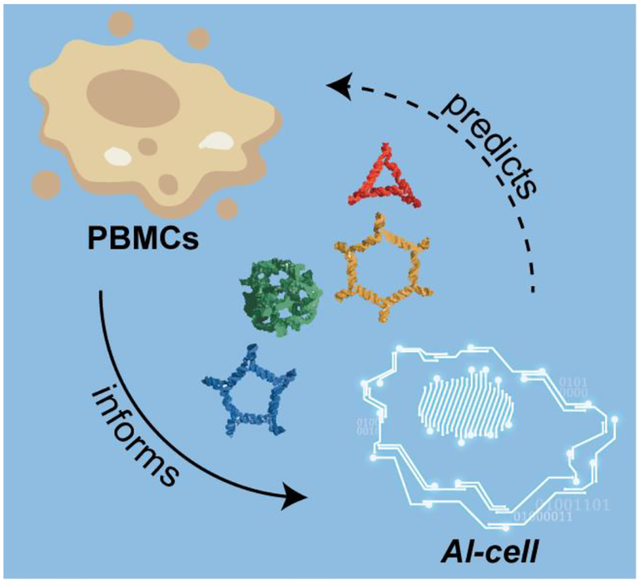
Nucleic acid nanoparticles, or NANPs, can be utilized to communicate with the human immune system, but their immunostimulatory effects can only be evaluated after their design and assembly. AI-cell is a computational model serving as an “artificial immune cell” to drive NANP designing principles based on predictions built from a comprehensive panel of NANP physicochemical and immunological profiles.
INTRODUCTION
Therapeutic nucleic acids (TNAs) have enriched and diversified the landscape of nanomedicine1, and their clinical success brought about the development of a new biomolecular platform, based on nucleic acid nanoparticles, called NANPs2–3. NANP technologies aim to advance the programmability of TNAs, tune their physicochemical and biological properties, and optimize their formulation, storage, and handling processes. The bottom-up assembly of NANPs takes advantage of nucleic acids’ folding pathways along with several computational tools available for precise coordination of sequence design and an expanded repertoire of structural and interacting motifs4–9. A large number of NANPs has been engineered to vary in chemical composition, sizes, and shapes that range from three-dimensional assemblies down to linear nanoscaffolds. Individual oligonucleotides in NANP compositions may be additionally defined in certain lengths and GC content, while also incorporating various TNAs (e.g., siRNAs, aptamers, and CpG DNAs), proteins, small molecules, and imaging agents suitable for biomedical applications10–14. Consequently, a growing library of functional NANPs has been shown to operate in response to other classes of biomolecules, or stimuli while executing therapeutic decisions based on environmental inputs15–17.
While the practicality of NANPs offers new ways to treat a broad spectrum of malignancies that span from cancers to infectious and cardiovascular diseases18, the intended clinical applications and routes of administration prioritize NANPs’ interactions with the human immune system to be carefully considered and understood for further translation of this technology into the clinic19–20. The immune recognition of these novel nanomaterials is inherent to the natural line of immune defense evolved for the detection of nucleic acids associated with pathogen invasion and cellular damage4, 21–24. However, NANPs’ unique architectural parameters and chemical compositions define their immunorecognition which cannot be extrapolated from the immune responses to pathogen- or damaged self-associated nucleic acids and conventional TNAs21. The ability to predict how NANPs interact with the human immune system would allow for tailoring their formulations to the specific biomedical task with maximized therapeutic effects and controlled immunological activity, which collectively are required to achieve desired therapeutic efficacy and safety. In addition, as was revealed by numerous studies10–12, 21, 25–30, NANPs can function not merely as nanoscaffolds for TNAs but also as independent immunostimulatory therapeutics with conditional intracellular activation of intended functions beneficial for vaccines and immunotherapies. Over the last years, our team created a comprehensive library of NANPs, designed by our group and others, and subjected them to detailed physicochemical characterization, sterility and endotoxin assessments, and immunological assays carried out in model cell lines and in primary human peripheral blood mononuclear cells (PBMCs)26. PBMCs were chosen as the most accurate pre-clinical model that produced the most predictive results for cytokine storm toxicity in humans31.
Translating NANP materials from bench to the clinic requires quick coordination of design principles. The incorporation of a particular level of immunostimulation and matching it to the desired application requires feedback from the experimental analysis to the computational design phase, which in turn entails complete recharacterization of NANPs and can delay their production. To improve this pipeline, several design parameters based around a representative set of NANPs have been previously correlated with cytokine production in model cell lines to determine trends of the immune response29.
Deep learning has contributed to major advancements in several research fields ranging from computer vision to natural-language processing. It is also widely applied in biomedical research areas such as drug discovery and genomics32. In genomics, sequence-based deep learning models outperformed classical machine learning33 and also enabled efficient prediction of the function, origin, and properties of DNA and RNA sequences by training neural networks on large datasets34–39. A robust model that can predict immune responses will have an enormous benefit in the design of NANPs. Our earlier quantitative structure-activity relationships (QSAR) modeling utilized a dataset collected for 16 NANPs which were assessed in model cell lines, and demonstrated that computational prediction of experimentally observed immunomodulatory properties is feasible29.
Despite this progress, there is currently no reliable bioinformatics tool to computationally identify optimal NANP structures matched to the desired immunological outcome. Such a tool would tremendously accelerate NANPs design and selection for personalized immunotherapeutic approaches or immunologically safe nanoscaffolds for other indications in which the stimulation of the innate immune responses is not wanted. Therefore, our present study was conducted to improve the communication between biotechnology, immunology, and bioinformatics and to create a new tool which would enable the prediction of NANPs structure-activity relationships in order to better guide the overall designing principles (Figure 1A). In particular, we employed random forest (RF) and two different neural network architectures (a recurrent neural network and a transformer neural network) to develop models that predict immunomodulatory activity for a much larger set of 58 representative NANPs that had been uniformly characterized using previously established, clinically relevant models26. Long-short-term memory (LSTM) architecture was used as the recurrent neural network. While the RF models use physicochemical properties derived from the constructed NANPs, the neural networks learn directly from the NANPs’ sequences. The neural network architectures investigated in this study facilitate discovery of hidden patterns via non-linear transformation of raw sequence data. These methods may also be applied to designing new NANPs (Figure 1B).
Figure 1.
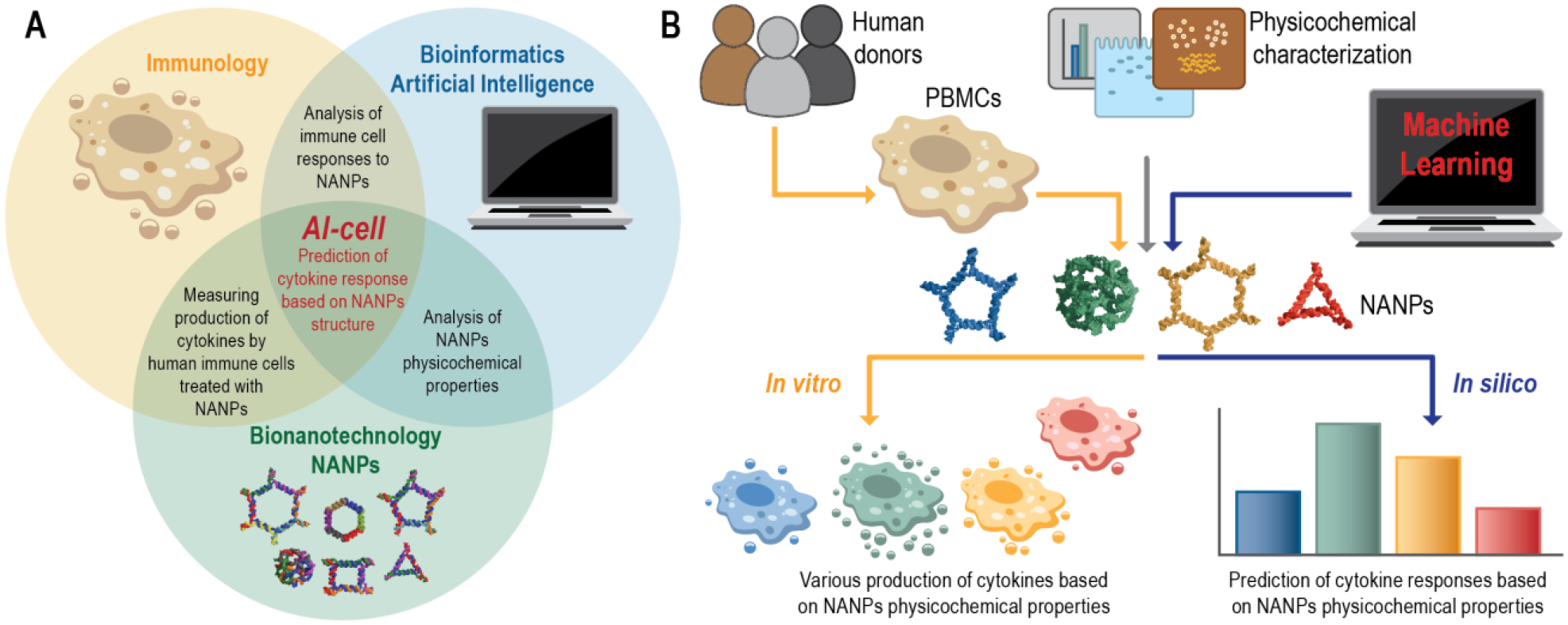
Conceptual representation of artificial immune cell (or AI-cell) tool. (A) The initial design and synthesis of nucleic acid nanoparticles (NANPs) is followed by their physicochemical characterization and assessment of immunostimulatory potential to then be applied for predictive computational analysis of the NANPs immune responses. (B) The experimental workflow used for the development of AI-cell.
The top performing models resulted from this study are freely accessible to the research community via the online tool that was named an “Artificial Immune cell”, or AI-cell, and that now can be applied to predict the immunological responses of any novel nucleic acid architecture (https://aicell.ncats.io/).
METHODS
Preparation of NANP Training Set.
All sequences of tested NANPs are available in SI Tables S1C–D. A database was compiled from previously published NANPs adhering to standard methods of characterization as described below. For each NANP, the sequences of all strands included in the assembly along with the composition (DNA or RNA), quantity, and length (nts) of each strand were recorded. For each fully assembled NANP, the overall composition (DNA, RNA, or hybrid of the two), mass (g/mol), GC content (%), total number of strands in the assembly, number of helices in the structure, number of single-stranded bases, number of RNA bases, number of DNA bases, dimensionality (1D, 2D, or 3D), connectivity (origami or tectoRNA1), diameter (nm), melting temperature (°C), and production of IFN-α, IFN-β, IFN-ω, and IFN-λ (pg/mL) were denoted.
NANP Preparation.
All DNA sequences were purchased from Integrated DNA Technologies, Inc. All RNA sequences were purchased as DNA templates and primers which were PCR-amplified via MyTaq™ Mix, 2x (Bioline) and purified using DNA Clean & Concentrator® (Zymo Research) for the preparation of double-stranded DNA templates containing a T7 RNA polymerase promotor. Templates underwent in vitro transcription with T7 RNA polymerase in 80 mM HEPES-KOH (pH 7.5), 2.5 mM spermidine, 50 mM DTT, 25 mM MgCl2, and 5 mM each rNTP for 3.5 hours at 37 °C and was stopped with the addition of RQ1 RNase-Free DNase (Promega, 3u/50 μL) for 30 minutes at 37 °C. Strands were purified via denaturing polyacrylamide gel electrophoresis (PAGE, 8%) in 8 M urea in 89 mM tris-borate, 2 mM EDTA (TBE, pH 8.2) run at 85 mA for 1.5 hours. Bands in the gel were visualized by UV shadowing, cut, and eluted overnight in 300 mM NaCl, TBE at 4 °C. Precipitation was performed in 2.5 volumes of 100% EtOH at −20 °C for 3 hours, followed by centrifugation at 10.0 G for 30 minutes with two 90% EtOH washes between 10 minute centrifugations at 10.0 G. The pelleted samples were dried in a CentriVap micro IR vacuum concentrator (Labconco) at 55 °C. Pellets were dissolved in HyClone™ Water, Molecular Biology Grade (Cytiva) and concentrations were determined by measuring the A260 on a NanoDrop 2000 (ThermoFisher). NANPs were assembled in HyClone™ Water, Molecular Biology Grade (Cytiva), by adding strands in an equimolar ratio. Each NANP assembly followed previously published respective steps2–5.
Assessment of Immunostimulation in Human PBMCs.
Human whole blood was obtained from healthy donor volunteers under Institutional Review Board-approved NCI-Frederick protocol OH9-C-N046. Each donor was assigned a random number. Vacutainers containing Li-heparin as an anticoagulant were used for blood collection. Research donor blood was processed to isolate PBMC within 2 hours after donation according to the protocol described earlier6. All NANPs were complexed with Lipofectamine 2000 (L2K) before addition to the cells as described earlier7. Culture supernatants were collected 24 hours after addition of NANPs-L2K, and stored at −80 °C before analysis for the presence of type I and type II I interferons using multiplex ELISA. The procedure for the interferon detection along with materials’ sources has been described earlier7. Some NANPs have been previously characterized and reported, whereas others were synthesized and tested de novo to support the computational modeling of the present study (e.g., SI Figures S1–4). More details about new and previously studied NANPs are available in SI Tables S1A–D.
Dataset for Modeling.
In this study, NANP sequences were used to construct computational models that predict their immune responses. Based on the levels of IFN- α, IFN-β, IFN- ω and IFN- λ, four types of immune responses were identified and were used as target variables in the development of models. The complete list of the associated IFN activities and their physicochemical properties are provided in SI Tables S1A–D. We employed 58 NANPs to train the models; evaluated by five-fold cross-validation procedure repeated 10 times.
Tokenization.
Tokenization is considered the first step that processes the input sequence data when building a sequence-to-sequence model. It involves transformation of text input into a sequence of tokens that generally correspond to ‘words’. The nanoparticle sequences were tokenized using the K-mer representation (K=3). The K-mer representation incorporates rich contextual information for each nucleotide base by loosely encoding triplets as codons i.e., all possible combinations of 3-mers of the individual nucleotide bases (A, T, G, C, U). This resulted in a total of 125 codon combinations or tokens, which were then used to create a vocabulary. Each input sequence in the training dataset was then tokenized and passed through an embedding layer, which maps the 3-mers to vector representations.
Generating All Possible Combination of NANPs.
It has been widely acknowledged that different NANP compositions be engineered to produce desired immune responses5, 8. When translating biological activity (in this case an immune response) to sequence-based learning, it is impossible to be certain about the order in which individual strands of each nanoparticle should be connected to achieve a particular immune response. Thus, to address this limitation, we generated all possible configurations using the individual strands for each nanoparticle. For a nanoparticle with ‘n’ strands, a total of ‘n!’ different combinations can be generated. IFN activity values for each combination were assigned as observed for the respective nanoparticle. This process resulted in a significantly larger training dataset. The augmented dataset was used for the training the final models. The models generated using this approach are referred to as Transformer_M1. When partitioning the augmented dataset during the 5-fold external cross-validation, to prevent the leakage of information from the training dataset to the validation dataset, all generated combination of a particular NANP were either present in the training set or the validation set during. Thus, the individual cross-validation runs had no overlap of NANP between the two set. In this scenario, the model statistics were calculated based on the ‘mean prediction values’ across each NANP.
Combining Physicochemical Properties with Sequence-Based Models.
Physicochemical descriptors derived from the constructures nanoparticles were previously reported to improved model performance due to their importance and relevance to the IFN activity (i.e., immune response) of nanoparticles2. Therefore, the physicochemical properties were used together with sequence data in development of sequence-based neural network models. For this purpose, the numerical descriptors were transformed into categories or bins (i.e., each bin encodes a value range) and added as tokens to the vocabulary previously described in the tokenization section. SI Table S3 provides the complete list of categories for each of the eight physicochemical descriptors. Further, when generating a numerical vector for the transformer model, the tokens related to the physicochemical properties were added to the original vector after converting the input sequence to a numerical vector. The models generated using this approach are referred to as Transformer_M2.
Modeling Approaches.
Two different modeling approaches were employed for the development of prediction models. In the first approach, the physicochemical properties of constructed nanoparticles were used as descriptors for creating a regression model using Random Forest (RF). RF is an ensemble of decision trees9 and is widely used in both classification and regression tasks. The number of trees was arbitrarily set to 100, and due to the robustness of RF10, no parameter optimization was performed. In the second approach, two different neural network architectures: LSTM and transformers; were employed to build prediction models that use nanoparticle sequences as input data. LSTM (Long Short-Term Memory) networks are specialized recurrent neural networks that are designed to avoid long-term dependency problem by remembering information for an extended period of time using a gating mechanism11. The readers are encourage to refer to the literature for further reading on LSTM networks12. Transformer networks have been recently introduced in the field of natural language processing13 and were reported to outperform recurrent neural networks architectures such as LSTM and Gated Recurrent Unit (GRU) in several NLP benchmarks on automatic speech recognition, speech translation and text-to-speech14. Transformers use attention mechanism and positional embeddings and facilitate encoding of multiple relationships within a sentence and process complete sentences by learning relationships between the words. The neural network architecture and the parameters used for training each of these models is provided in SI Tables S4A–B.
Evaluation of model statistics.
To evaluate the predictive performance of the developed models, a 5-fold external cross-validation procedure (5-CV)15 was employed. In this procedure, the initial data set was randomly divided into five parts. In each fold, four parts of the data were used as training set for model building and the fifth part was used as test set for assessment of external predictive performance. To be more robust and ensure that the performance obtained is not due to chance correlations, the 5-CV procedure was repeated for a total of 10 times. The performance of each model was assessed on the basis of root mean squared error (RMSE) (Eq. 1), and determination coefficient R2 (Eq. 2),
| (1) |
| (2) |
Yi(cap) is the predicted value for each particular sequence; Yi is the observed value for each particular sequence; Y(bar) is the mean activity value from all the sequences; n is the number of sequences.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism software version 9.0.0 for Windows, GraphPad Software, San Diego, California, USA (www.graphpad.com). The difference between the model performances was evaluated using a nonparametric statistical test (Friedman) to compare pairwise if one approach significantly outperforms the other. All data were presented as mean of several repeats with the sample size (n) specified for each dataset and error bars denoted mean ± SD; p values of less than 0.05 were considered statistically significant.
RESULTS
Representative NANP Database.
We designed a library of representative NANPs to study key structure-activity relationships that define NANP interactions with the cells of the human immune system (Figure 2 and SI Tables 1A–D). Our dataset included different functional and non-functional NANPs made of either DNA or RNA, having planar, globular, or fibrous structures, different sizes, flexibilities, thermodynamic stabilities, and connectivity rules. Some of these datasets have already been published10–11, 21, 30, 40–41, whereas others were newly generated to support the development of the current AI algorithm (all NANPs are itemized in the SI Tables S1A–D).
Figure 2.
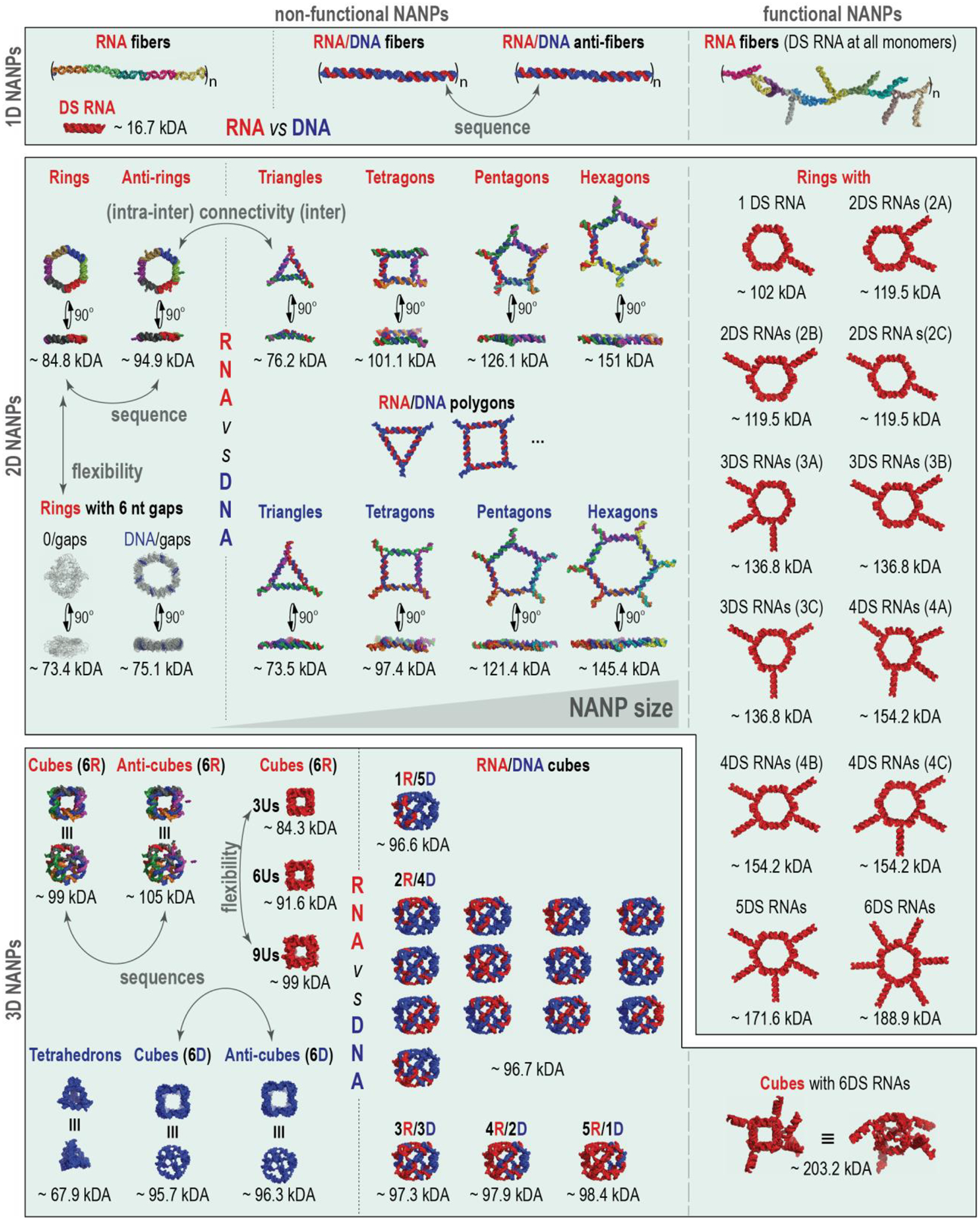
Representative NANPs chosen to collectively address the influence of their physicochemical properties and architectural parameters on their immunorecognition in human PBMC to further the development of AI-cell.
To study the influence of architectural parameters, the immune responses to 1D fibers were compared to 2D planar and to 3D globular NANPs, designed by two different approaches that define the connectivity of NANPs. The first approach, represented by RNA/DNA fibers and all polygons and cubes, relies solely on intermolecular canonical Watson-Crick interactions with all NANP sequences designed to avoid any intramolecular structures42–43. These design principles are characteristic for DNA nanotechnology and DNA origami44–45 and allow for any RNA strand in the NANP’s composition to be substituted with DNA analog. The second approach, called tectoRNA46–47, is exemplified by RNA rings and fibers that employ naturally occurring structural and long-range interacting motifs (e.g., kissing loops) that are rationally combined, similarly to Lego® bricks, to achieve topological control in the bottom-up assembly of NANPs43.
To study the role of chemical composition, origami-like RNA NANPs were compared to their DNA and RNA/DNA analogs. This compositional blend allowed for changes in NANPs’ physicochemical and biological properties in a highly predictable and controlled manner. For example, the responses of individual NANPs to heating become different (Tm~36°C of the DNA cubes vs Tm~55.5°C of the RNA cubes21) and a new version of Hyperfold48 can accurately predict the experimental results10. The chemical makeup also influenced the relative chemical stabilities of NANPs in blood serum and towards degradation by different nucleases10, 21, 25, 49.
To assess the effect of structural flexibility, we included gapped ring structures50 and cubes with different numbers of single-stranded uracils at their corners21, 51, all designed to control the dynamic behavior of 2D and 3D NANPs, respectively. Both experimental results and MD simulations supported the notion that the stability and dynamicity of NANPs can be modulated by changing the number of single-stranded regions in their structures30, 51.
The effect of functionalization was assessed via the addition of Dicer substrate (DS) RNAs52 to the 1D, 2D, and 3D NANPs and for the size contributions, different polygons29 were compared. DS RNAs are widely used for Dicer-assisted intracellular release of siRNAs. The sequence effects were studied through the inclusion of several reverse complement structures (denoted as “anti-“) for 1D40, 2D10, and 3D10 NANPs. All physicochemical properties of NANPs have been assessed under the equivalent conditions and their relative immunostimulation was measured in PBMCs isolated from fresh blood drawn from healthy donors with at least three donors per each NANP. All data have been combined in a single dataset shown in SI Tables S1A–D with all sources for experimental results cited.
IFN Modeling Results.
With a diverse library of NANPs in the dataset, three different methods were employed to build models that predict the immunological activities of NANPs in PBMCs (Figure 3). First, an RF method was applied using the physicochemical descriptors derived from the input sequences. The physicochemical properties of the studied NANPs along with their immune responses are provided in SI Table S1A. Next, the neural network architectures LSTM and transformers were applied that directly learn on the NANP sequences. In both neural network models, the first step involves tokenization of the input sequences. Tokenization was performed using the ‘K-mer’ representation (K=3), which is usually employed for nucleotide sequences and the encoded sequences can be loosely considered as codons. Generating all possible combinations of 3-mers of the individual nucleotide bases (A, T, G, C, U) resulted in a total of 125 codon combinations or tokens. Each model was evaluated using five-fold cross-validation (5-CV) that was repeated 10 times. Figures 4A and 4B provide a comparison of the average performance (R2, RMSE) for different models generated in this study. According to the 5-CV results presented in Figure 4, the models developed using the transformer architecture significantly outperformed other models. The detailed model performance statistics can be found in SI Tables S2A–D.
Figure 3.
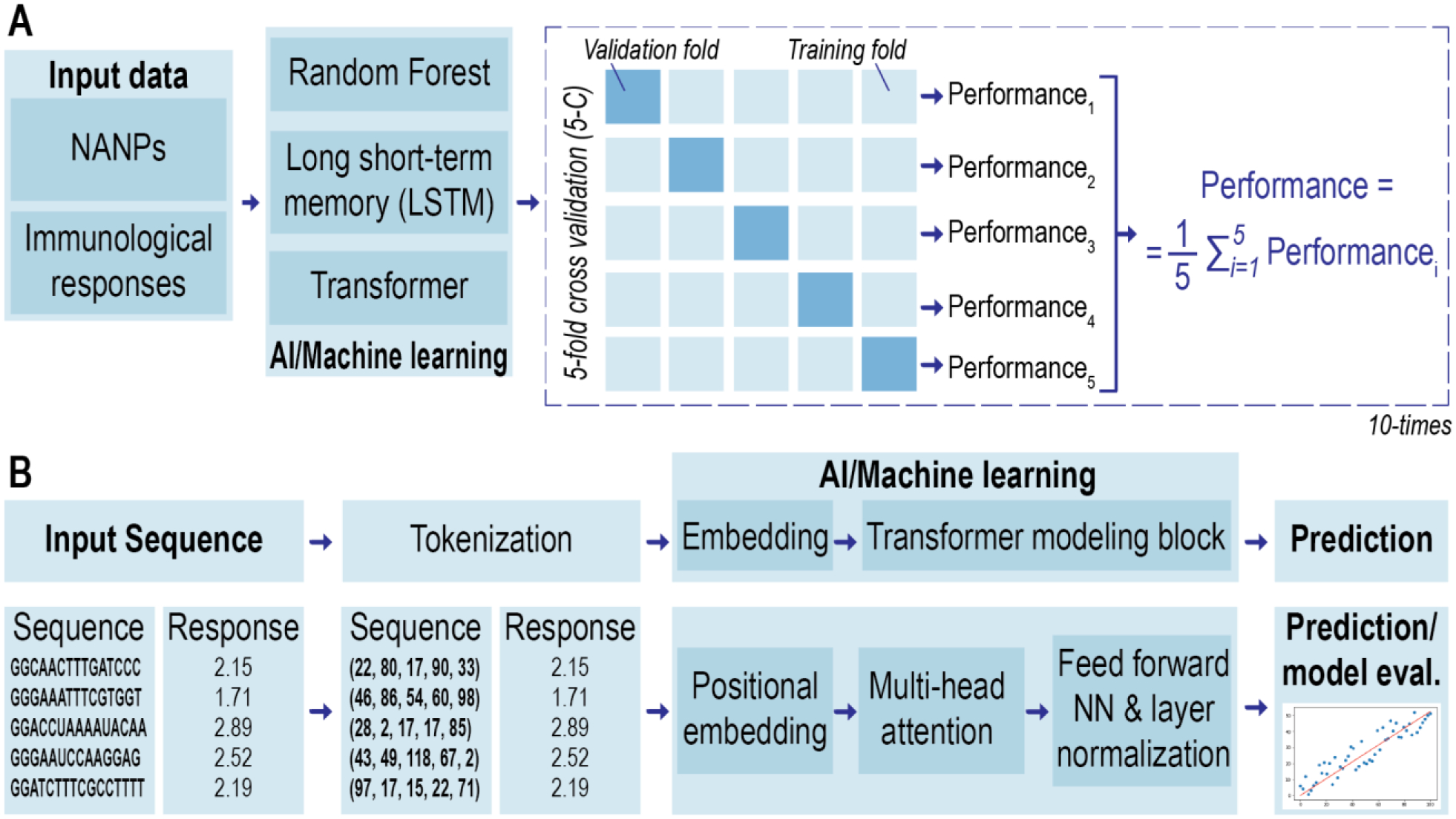
Schematic representation of the quantitative structure–activity relationship (QSAR) methodology used in this project. (A) Modeling workflow: three machine learning approaches are evaluated using five-fold cross-validation (5-CV) repeated 10 times. (B) Overall workflow and the training procedure for prediction of nanoparticle sequence using transformer-based approach: tokenization, embedding followed by transformer modeling and prediction.
Figure 4.
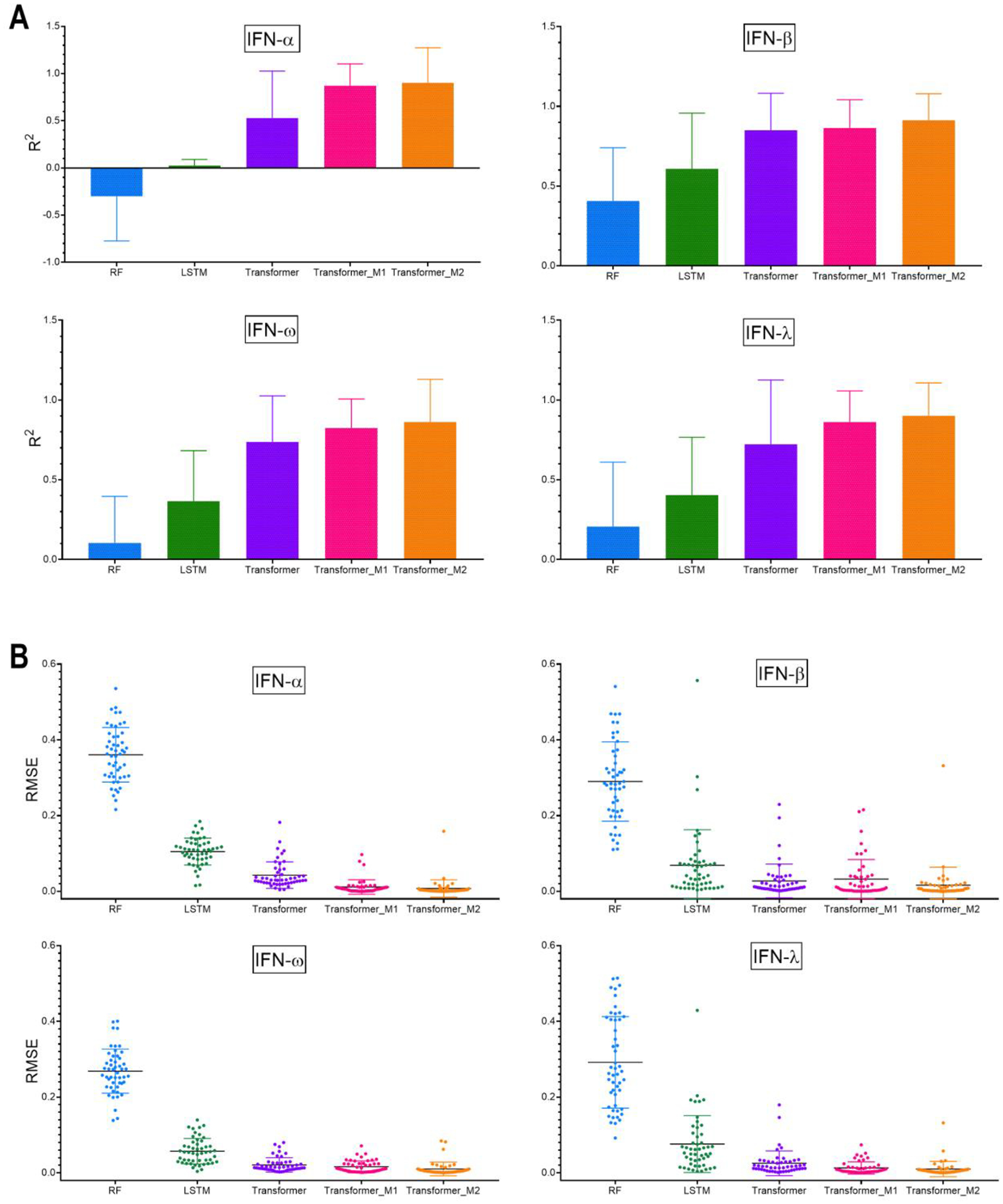
Average performance (A) R2 and (B) RMSE for different modeling approaches over 5-fold cross-validation and repeated 10-times. The error bar represents the standard deviation of the average performance over 5-folds cross-validation repeated 10-times (n=50). Detailed statistical analysis is shown in SI Table S5.
Each NANP is represented by multiple sequences or strands: for example, dA, dB, dC, dD, dE, and dF are the individual sequences that form DNA cube. By default, these sequences were joined in a sequential manner for the purpose of learning. However, it is uncertain if this is the only possible configuration (i.e., arrangement of sequences) for DNA cube. To avoid any bias during learning, the best performing models based on the transformer architecture were evaluated on all possible combinations based on the different strands present in each NANP. This data augmentation step led to a significant increase in the training dataset size. The models generated using this approach are referred to as Transformer_M1. The model performance improved, particularly in the case of IFN-α, where the R2 improved from 0.53 to 0.87. Using this approach (Transformer_M1), all models provided an R2 > 0.80 and RMSE < 0.03 (Figure 4).). The difference between the model performance (R2 and RMSE) was evaluated using the Friedman nonparametric statistical test. SI Tables S5A–D provide the pairwise comparison between the different machine learning methods to compare if one approach significantly outperforms the other.
It is also known that the physicochemical properties of NANPs are important for their immune response29. Therefore, to evaluate the contribution of the physicochemical properties to model performance, we pursued a third approach in which the physicochemical properties were combined with the sequence data to build sequence-based models. To achieve this, the numerical descriptors were converted into categories and added as tokens to the vocabulary. Tokenization was performed in the same manner on triplets. The models generated using this approach are referred to as Transformer_M2. As shown in Figure 4A–B, inclusion of the descriptors to the sequence-based models further improved the model performance (R2 > 0.85). The best prediction performance was obtained using a model that combined physicochemical properties together with sequence-based models. As seen from Figure 4A–B, the sequence information alone demonstrated high predictivity using transformer models (Transformer_M1), especially for IFN-β responses, and thus might play a significant role in predicting the behavior of polygons with more diverse shapes and structures.
Model Interpretability and Implementation.
Since the best-performing model developed in this study involves generating all possible combinations of the input NANP strands and using the K-mer representation (K=3) for tokenization (cf. methods section for details), it is not feasible to determine the importance of individual tokens (K-mer) and thus to obtain the interpretability of our best performing model (Transformer_M1). However, to help the research community to better guide the overall designing principles for the NANPs, and to overcome this limitation, we provide online tool with implementation of the best developed model (https://aicell.ncats.io/). This implementation helps the user to predict the immunological responses of novel nucleic acid architectures while enabling alteration of nucleic acid. Depending on the number of input strands, their sequences and lengths, it takes on average between two and four seconds to predict the immune responses (induction of IFN-α, IFN-β, IFN-ω, and IFN-λ). In contrast, evaluation of the IFN responses using human immune cells takes at the minimum three days; this further emphasizes the convenience and cost-benefit for researchers to use the newly developed on-line model. In addition, our implementation also provides an uncertainty of prediction in terms of the standard deviation calculated from the prediction of all possible combinations of the input nanoparticles strands (cf. the methods section for details). Thus, the online tool can be used for NANPs design to achieve the desired immunological outcome.
DISCUSSION
Machine learning and artificial intelligence (AI) have been increasingly applied in various domain such as computer vision53–54, natural language processing55–57, drug discovery58–59, QSAR60–62, and genomics63–65. AI methods such as convolutional neural networks (CNNs)66 and recurrent neural networks (RNNs)67 that are extensively used in computer vision and natural language processing have been investigated for identifying protein binding sites in DNA and RNA sequences, and achieved state-of-the-art performance68–70. More recently, transformer neural networks were reported to provide superior performance in the field of drug discovery and QSAR71–73 and demonstrated state-of-the-art results on neural machine translation task74–75 including direct and single-step retrosynthesis of chemical compounds76. The Transformer architecture incorporates the mechanism of self-attention together with positional embedding77, which makes them heavily successful in the field of natural language processing (NLP) tasks78–79. Transformer-based models have also been effective in predicting novel drug–target interactions from sequence data and significantly outperformed existing methods like DeepConvDTI80, DeepDTA81, and DeepDTI82 on their test data set for drug–target interaction (DTI)83. Another attractive task that remarkably benefits from the transformer architecture is generative molecular design. It was recently shown that transformer-based generative models demonstrated state-of-the-art performance when compared to previous approaches based on recurrent neural networks84. Additionally, a recent study demonstrated the application of a transformer architecture is development of a SMILES canonicalization approach that extracts information-rich embedding and exposes them for further use in QSAR studies76; however the applicability of this approach to therapeutic nucleic acids and NANPs is unknown. Given the importance of nanoparticles in the field of drug delivery and the ability of NANPs to act as active pharmaceutical ingredients; offering innovative therapeutic strategies and overcoming the limitations of traditional nucleic acid therapies and the lack of predictive tools that would reliably guide NANPs design to the desired immunological outcome, we adopted transformer-based models to predict the immunological activities of the nanoparticles. An earlier study by Johnson et al.29 reports the use of random forest (RF) based QSAR models; however, considering that the nature of input data is sequence/text, transformer neural networks are able better learn the patterns within the data in comparison to other methods used in this study such as random forests.
To the best of our knowledge this is the first study to evaluate and implement the use of state-of-the-art transformer neural networks to predict immunological activity and thus advance the current understanding of the NANP properties that contribute to the observed immunomodulatory activity and establish corresponding designing principles. Our results demonstrate the benefit (significant improvement in prediction statistics; R2 and RMSE) of using a transformer framework that is solely based on sequence data vs. RF models (Figure 4, SI Tables S2A–D). The data augmentation (Transformer_M1) led to a further increase of the model performance. In the case of QSAR modeling, the importance of data augmentation has been shown to be critical for the Transformer models to achieve their high performance85–86. Transformer models extract semantic information in NLP tasks by jointly conditioning on both left and right contexts in all layers75. This is particularly an essential feature in context to biological sequences, which are multidirectional in nature. The inclusion of robust sequence embeddings facilitated the proposed models to score well with the performance metrics (Figure 4). We expect this hybrid architecture will be continually explored for the purpose of studying NANPs.
In summary, we applied a systematic approach to connect physicochemical and immunological properties of a comprehensive panel of various NANPs and developed a computational model based on the transformer architecture. The resulting AI-cell tool predicts the immune responses of NANPs based on the input of their physicochemical properties. This model overcomes the limitations of the previous QSAR model and is imperative for responding timely to critical public health challenges related to drug overdose and the safety of nucleic acid therapies by streamlining the selection of optimal NANP designs for personalized therapies.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would also like to thank Karan Sood for his help with the web-based implementation of the prediction models. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers R01GM120487 and R35GM139587 (to K.A.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The study was also funded in part by federal funds from the National Cancer Institute, National Institutes of Health, under contract 75N91019D00024 (M.A.D. and E.H.). This research was supported in part by the Intramural/Extramural research program of the NCATS, NIH. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
SUPPLEMENTARY MATERIALS
Supplementary Tables S1 – S5 and Figures S1 – S4,
DATA AVAILABILITY
The dataset used for modeling and the scripts generated are available at our GitHub repository (https://github.com/ncats/AI-cell_nano_pred). Furthermore, the top prediction models have also been made publicly available to the research community through a web-based implementation that can be accessed via https://aicell.ncats.io/
REFERENCES
- 1.Kulkarni JA; Witzigmann D; Thomson SB; Chen S; Leavitt BR; Cullis PR; van der Meel R, The current landscape of nucleic acid therapeutics. Nat Nanotechnol 2021, 16 (6), 630–643. [DOI] [PubMed] [Google Scholar]
- 2.Afonin KA; Dobrovolskaia MA; Ke W; Grodzinski P; Bathe M, Critical review of nucleic acid nanotechnology to identify gaps and inform a strategy for accelerated clinical translation. Adv Drug Deliv Rev 2022, 181, 114081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandler M; Johnson B; Khisamutdinov E; Dobrovolskaia MA; Sztuba-Solinska J; Salem AK; Breyne K; Chammas R; Walter NG; Contreras LM; Guo P; Afonin KA, The International Society of RNA Nanotechnology and Nanomedicine (ISRNN): The Present and Future of the Burgeoning Field. ACS Nano 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson MB; Chandler M; Afonin KA, Nucleic acid nanoparticles (NANPs) as molecular tools to direct desirable and avoid undesirable immunological effects. Advanced Drug Delivery Reviews 2021, 173, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Afonin KA; Kasprzak WK; Bindewald E; Kireeva M; Viard M; Kashlev M; Shapiro BA, In silico design and enzymatic synthesis of functional RNA nanoparticles. Acc Chem Res 2014, 47 (6), 1731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veneziano R; Ratanalert S; Zhang K; Zhang F; Yan H; Chiu W; Bathe M, Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352 (6293), 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parlea L; Bindewald E; Sharan R; Bartlett N; Moriarty D; Oliver J; Afonin KA; Shapiro BA, Ring Catalog: A resource for designing self-assembling RNA nanostructures. Methods 2016, 103, 128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afonin KA; Viard M; Tedbury P; Bindewald E; Parlea L; Howington M; Valdman M; Johns-Boehme A; Brainerd C; Freed EO; Shapiro BA, The Use of Minimal RNA Toeholds to Trigger the Activation of Multiple Functionalities. Nano letters 2016, 16 (3), 1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bindewald E; Afonin KA; Viard M; Zakrevsky P; Kim T; Shapiro BA, Multistrand Structure Prediction of Nucleic Acid Assemblies and Design of RNA Switches. Nano letters 2016, 16 (3), 1726–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halman JR; Satterwhite E; Roark B; Chandler M; Viard M; Ivanina A; Bindewald E; Kasprzak WK; Panigaj M; Bui MN; Lu JS; Miller J; Khisamutdinov EF; Shapiro BA; Dobrovolskaia MA; Afonin KA, Functionally-Interdependent Shape-Switching Nanoparticles with Controllable Properties. Nucleic Acids Research 2017, 45 (4), 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke W; Hong E; Saito RF; Rangel MC; Wang J; Viard M; Richardson M; Khisamutdinov EF; Panigaj M; Dokholyan NV; Chammas R; Dobrovolskaia MA; Afonin KA, RNA-DNA Fibers and Polygons with Controlled Immunorecognition Activate RNAi, FRET and Transcriptional Regulation of NF-κB in Human Cells. Nucleic Acids Research 2019, 47 (3), 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson MB; Halman JR; Miller DK; Cooper JS; Khisamutdinov EF; Marriott I; Afonin K. A. J. N. a. r., The immunorecognition, subcellular compartmentalization, and physicochemical properties of nucleic acid nanoparticles can be controlled by composition modification. 2020, 48 (20), 11785–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afonin KA; Viard M; Koyfman AY; Martins AN; Kasprzak WK; Panigaj M; Desai R; Santhanam A; Grabow WW; Jaeger L; Heldman E; Reiser J; Chiu W; Freed EO; Shapiro BA, Multifunctional RNA nanoparticles. Nano letters 2014, 14 (10), 5662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito RF; Rangel MC; Halman JR; Chandler M; de Sousa Andrade LN; Odete-Bustos S; Furuya TK; Carrasco AGM; Chaves-Filho AB; Yoshinaga MY; Miyamoto S; Afonin KA; Chammas R, Simultaneous silencing of lysophosphatidylcholine acyltransferases 1–4 by nucleic acid nanoparticles (NANPs) improves radiation response of melanoma cells. Nanomedicine 2021, 36, 102418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandler M; Afonin KA, Smart-Responsive Nucleic Acid Nanoparticles (NANPs) with the Potential to Modulate Immune Behavior. Nanomaterials (Basel) 2019, 9 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang SX; Ge ZL; Mou S; Yan H; Fan CH, Designer DNA nanostructures for therapeutics. Chem-Us 2021, 7 (5), 1156–1179. [Google Scholar]
- 17.Panigaj M; Johnson MB; Ke W; McMillan J; Goncharova EA; Chandler M; Afonin KA, Aptamers as Modular Components of Therapeutic Nucleic Acid Nanotechnology. ACS Nano 2019, 13 (11), 12301–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binzel DW; Li X; Burns N; Khan E; Lee WJ; Chen LC; Ellipilli S; Miles W; Ho YS; Guo P, Thermostability, Tunability, and Tenacity of RNA as Rubbery Anionic Polymeric Materials in Nanotechnology and Nanomedicine-Specific Cancer Targeting with Undetectable Toxicity. Chem Rev 2021, 121 (13), 7398–7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afonin KA; Dobrovolskaia MA; Church G; Bathe M, Opportunities, Barriers, and a Strategy for Overcoming Translational Challenges to Therapeutic Nucleic Acid Nanotechnology. ACS Nano 2020, 14 (8), 9221–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Surana S; Shenoy AR; Krishnan Y, Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat Nanotechnol 2015, 10 (9), 741–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong E; Halman JR; Shah AB; Khisamutdinov EF; Dobrovolskaia MA; Afonin KA, Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles. Nano letters 2018, 18 (7), 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobrovolskaia MA; Bathe M, Opportunities and challenges for the clinical translation of structured DNA assemblies as gene therapeutic delivery and vaccine vectors. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2021, 13 (1), e1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandler M; Johnson MB; Panigaj M; Afonin KA, Innate immune responses triggered by nucleic acids inspire the design of immunomodulatory nucleic acid nanoparticles (NANPs). Current Opinion in Biotechnology 2020, 63, 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartmann G, Nucleic Acid Immunity. Adv Immunol 2017, 133, 121–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avila YI; Chandler M; Cedrone E; Newton HS; Richardson M; Xu J; Clogston JD; Liptrott NJ; Afonin KA; Dobrovolskaia MA, Induction of Cytokines by Nucleic Acid Nanoparticles (NANPs) Depends on the Type of Delivery Carrier. Molecules 2021, 26 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobrovolskaia MA; Afonin KA, Use of human peripheral blood mononuclear cells to define immunological properties of nucleic acid nanoparticles. Nature Protocols 2020, 15 (11), 3678–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halman JR; Kim KT; Gwak SJ; Pace R; Johnson MB; Chandler MR; Rackley L; Viard M; Marriott I; Lee JS; Afonin KA, A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (Nanps) for controlled gene silencing, immunostimulation, and biodistribution. Nanomedicine 2020, 23, 102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson MB; Halman JR; Burmeister AR; Currin S; Khisamutdinov EF; Afonin KA; Marriott I, Retinoic acid inducible gene-I mediated detection of bacterial nucleic acids in human microglial cells. J Neuroinflammation 2020, 17 (1), 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson MB; Halman JR; Satterwhite E; Zakharov AV; Bui MN; Benkato K; Goldsworthy V; Kim T; Hong E; Dobrovolskaia MA; Khisamutdinov EF; Marriott I; Afonin KA, Programmable Nucleic Acid Based Polygons with Controlled Neuroimmunomodulatory Properties for Predictive QSAR Modeling. Small (Weinheim an der Bergstrasse, Germany) 2017, 13 (42). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sajja S; Chandler M; Fedorov D; Kasprzak WK; Lushnikov A; Viard M; Shah A; Dang D; Dahl J; Worku B; Dobrovolskaia MA; Krasnoslobodtsev A; Shapiro BA; Afonin KA, Dynamic Behavior of RNA Nanoparticles Analyzed by AFM on a Mica/Air Interface. Langmuir 2018, 34 (49), 15099–15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vessillier S; Eastwood D; Fox B; Sathish J; Sethu S; Dougall T; Thorpe SJ; Thorpe R; Stebbings R, Cytokine release assays for the prediction of therapeutic mAb safety in first-in man trials--Whole blood cytokine release assays are poorly predictive for TGN1412 cytokine storm. J Immunol Methods 2015, 424, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wainberg M; Merico D; Delong A; Frey BJ, Deep learning in biomedicine. Nature biotechnology 2018, 36 (9), 829–838. [DOI] [PubMed] [Google Scholar]
- 33.Zou J; Huss M; Abid A; Mohammadi P; Torkamani A; Telenti A, A primer on deep learning in genomics. Nature genetics 2019, 51 (1), 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quang D; Xie X, DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic acids research 2016, 44 (11), e107–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen AA; Voigt CA, Deep learning to predict the lab-of-origin of engineered DNA. Nature communications 2018, 9 (1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trabelsi A; Chaabane M; Ben-Hur A, Comprehensive evaluation of deep learning architectures for prediction of DNA/RNA sequence binding specificities. Bioinformatics 2019, 35 (14), i269–i277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angenent-Mari NM; Garruss AS; Soenksen LR; Church G; Collins JJ, A deep learning approach to programmable RNA switches. Nature communications 2020, 11 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam JH; Li Y; Zhu L; Umarov R; Jiang H; Héliou A; Sheong FK; Liu T; Long Y; Li Y, A deep learning framework to predict binding preference of RNA constituents on protein surface. Nature communications 2019, 10 (1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amin N; McGrath A; Chen Y-PP, Evaluation of deep learning in non-coding RNA classification. Nature Machine Intelligence 2019, 1 (5), 246–256. [Google Scholar]
- 40.Ke W; Hong E; Saito RF; Rangel MC; Wang J; Viard M; Richardson M; Khisamutdinov EF; Panigaj M; Dokholyan NV; Chammas R; Dobrovolskaia MA; Afonin KA, RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-kappaB in human cells. Nucleic Acids Res 2019, 47 (3), 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rackley L; Stewart JM; Salotti J; Krokhotin A; Shah A; Halman JR; Juneja R; Smollett J; Lee L; Roark K; Viard M; Tarannum M; Vivero-Escoto J; Johnson PF; Dobrovolskaia MA; Dokholyan NV; Franco E; Afonin KA, RNA Fibers as Optimized Nanoscaffolds for siRNA Coordination and Reduced Immunological Recognition. Adv Funct Mater 2018, 28 (48). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonin KA; Bindewald E; Yaghoubian AJ; Voss N; Jacovetty E; Shapiro BA; Jaeger L, In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol 2010, 5 (9), 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afonin KA; Grabow WW; Walker FM; Bindewald E; Dobrovolskaia MA; Shapiro BA; Jaeger L, Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc 2011, 6 (12), 2022–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chidchob P; Sleiman HF, Recent advances in DNA nanotechnology. Curr Opin Chem Biol 2018, 46, 63–70. [DOI] [PubMed] [Google Scholar]
- 45.Pinheiro AV; Han D; Shih WM; Yan H, Challenges and opportunities for structural DNA nanotechnology. Nat Nanotechnol 2011, 6 (12), 763–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaeger L; Leontis NB, Tecto-RNA: One-dimensional self-assembly through tertiary interactions. Angew Chem Int Edit 2000, 39 (14), 2521–+. [DOI] [PubMed] [Google Scholar]
- 47.Jaeger L; Westhof E; Leontis NB, TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic acids research 2001, 29 (2), 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bindewald E; Afonin KA; Viard M; Zakrevsky P; Kim T; Shapiro BA, Multistrand Structure Prediction of Nucleic Acid Assemblies and Design of RNA Switches. Nano Lett 2016, 16 (3), 1726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong E; Halman JR; Shah A; Cedrone E; Truong N; Afonin KA; Dobrovolskaia MA, Toll-Like Receptor-Mediated Recognition of Nucleic Acid Nanoparticles (NANPs) in Human Primary Blood Cells. Molecules 2019, 24 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sajja S; Chandler M; Fedorov D; Kasprzak WK; Lushnikov A; Viard M; Shah A; Dang D; Dahl J; Worku B; Dobrovolskaia MA; Krasnoslobodtsev A; Shapiro BA; Afonin KA, Dynamic Behavior of RNA Nanoparticles Analyzed by AFM on a Mica/Air Interface. Langmuir 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afonin KA; Kasprzak W; Bindewald E; Puppala PS; Diehl AR; Hall KT; Kim TJ; Zimmermann MT; Jernigan RL; Jaeger L; Shapiro BA, Computational and experimental characterization of RNA cubic nanoscaffolds. Methods 2014, 67 (2), 256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose SD; Kim DH; Amarzguioui M; Heidel JD; Collingwood MA; Davis ME; Rossi JJ; Behlke MA, Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res 2005, 33 (13), 4140–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chai J; Zeng H; Li A; Ngai EWT, Deep learning in computer vision: A critical review of emerging techniques and application scenarios. Mach Learn Appl 2021, 6, 100134. [Google Scholar]
- 54.Sarraf A; Azhdari M; Sarraf S, A Comprehensive Review of Deep Learning Architectures for Computer Vision Applications. Am Acad Sci Res J Eng Technol Sci 2021, 77, 1–29. [Google Scholar]
- 55.Buchlak QD; Esmaili N; Bennett C; Farrokhi F, Natural Language Processing Applications in the Clinical Neurosciences: A Machine Learning Augmented Systematic Review. Acta Neurochir Suppl 2022, 134, 277–289. [DOI] [PubMed] [Google Scholar]
- 56.Kalyan KS; Rajasekharan A; Sangeetha S, MMU: A survey of transformer-based biomedical pretrained language models. J Biomed Inform 2022, 126, 103982. [DOI] [PubMed] [Google Scholar]
- 57.Yang X; Bian J; Hogan WR; Wu YH, Clinical concept extraction using transformers. J Am Med Inform Assn 2020, 27 (12), 1935–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dara S; Dhamercherla S; Jadav SS; Babu CHM; Ahsan MJ, Machine Learning in Drug Discovery: A Review. Artif Intell Rev 2022, 55 (3), 1947–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vamathevan J; Clark D; Czodrowski P; Dunham I; Ferran E; Lee G; Li B; Madabhushi A; Shah P; Spitzer M; Zhao SR, Applications of machine learning in drug discovery and development. Nat Rev Drug Discov 2019, 18 (6), 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keyvanpour MR; Shirzad MB, An Analysis of QSAR Research Based on Machine Learning Concepts. Curr Drug Discov Technol 2021, 18 (1), 17–30. [DOI] [PubMed] [Google Scholar]
- 61.Muratov EN; Bajorath J; Sheridan RP; Tetko IV; Filimonov D; Poroikov V; Oprea TI; Baskin II; Varnek A; Roitberg A; Isayev O; Curtarolo S; Fourches D; Cohen Y; Aspuru-Guzik A; Winkler DA; Agrafiotis D; Cherkasov A; Tropsha A, QSAR without borders (vol 10, pg 531, 2020). Chem Soc Rev 2020, 49 (11), 3716–3716. [DOI] [PubMed] [Google Scholar]
- 62.Wu Z; Zhu M; Kang Y; Lai-Han Leung E; Lei T; Shen C; Jiang D; Wang Z; Cao D; Hou T, Do we need different machine learning algorithms for QSAR modeling? A comprehensive assessment of 16 machine learning algorithms on 14 QSAR data sets. Brief Bioinform 2021, 22 (4). [DOI] [PubMed] [Google Scholar]
- 63.Angenent-Mari NM; Garruss AS; Soenksen LR; Church G; Collins JJ, A deep learning approach to programmable RNA switches. Nat Commun 2020, 11 (1), 5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watson DS, Interpretable machine learning for genomics. Hum Genet 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yue T; Wang H, Deep Learning for Genomics: A Concise Overview. ArXiv preprint arXiv:1808.03314 2018, 1802.00810. [Google Scholar]
- 66.Lecun Y; Bottou L; Bengio Y; Haffner P, Gradient-based learning applied to document recognition. P Ieee 1998, 86 (11), 2278–2324. [Google Scholar]
- 67.Hochreiter S; Schmidhuber J, Long short-term memory. Neural Comput 1997, 9 (8), 1735–1780. [DOI] [PubMed] [Google Scholar]
- 68.Alipanahi B; Delong A; Weirauch MT; Frey BJ, Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nature Biotechnology 2015, 33 (8), 831–+. [DOI] [PubMed] [Google Scholar]
- 69.Quang D; Xie X, DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res 2016, 44 (11), e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen Z; Bao WZ; Huang DS, Recurrent Neural Network for Predicting Transcription Factor Binding Sites. Sci Rep-Uk 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grechishnikova D, Transformer neural network for protein-specific de novo drug generation as a machine translation problem. Sci Rep-Uk 2021, 11 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Honorio KM; Garratt RC; Polikatpov I; Andricopulo AD, 3D QSAR comparative molecular field analysis on nonsteroidal farnesoid X receptor activators. J Mol Graph Model 2007, 25 (6), 921–927. [DOI] [PubMed] [Google Scholar]
- 73.Zheng SJ; Lei ZR; Ai HT; Chen HM; Deng DG; Yang YD, Deep scaffold hopping with multimodal transformer neural networks. J Cheminformatics 2021, 13 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vaswani A; Shazeer N; Parmar N; Uszkoreit J; Jones L; Gomez AN; Kaiser Ł; Polosukhin I, Attention is all you need. Advances in neural information processing systems 2017, 30. [Google Scholar]
- 75.Devlin J; Chang M-W; Lee K; Toutanova K, BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. ArXiv 2019. [Google Scholar]
- 76.Tetko IV; Karpov P; Van Deursen R; Godin G, State-of-the-art augmented NLP transformer models for direct and single-step retrosynthesis. Nature Communications 2020, 11 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vaswani A; Shazeer N; Parmar N; Uszkoreit J; Jones L; Gomez AN; Kaiser L; Polosukhin I, Attention Is All You Need. Adv Neur In 2017, 30. [Google Scholar]
- 78.Khan S; Naseer M; Hayat M; Zamir SW; Khan FS; Shah M, Transformers in Vision: A Survey. ArXiv 2021. [Google Scholar]
- 79.Yang S; Wang Y; Chu X, A Survey of Deep Learning Techniques for Neural Machine Translation. ArXiv 2020. [Google Scholar]
- 80.Lee I; Keum J; Nam H, DeepConv-DTI: Prediction of drug-target interactions via deep learning with convolution on protein sequences. Plos Comput Biol 2019, 15 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ozturk H; Ozgur A; Ozkirimli E, DeepDTA: deep drug-target binding affinity prediction. Bioinformatics 2018, 34 (17), 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wen M; Zhang ZM; Niu SY; Sha HZ; Yang RH; Yun YH; Lu HM, Deep-Learning-Based Drug-Target Interaction Prediction. J Proteome Res 2017, 16 (4), 1401–1409. [DOI] [PubMed] [Google Scholar]
- 83.Kalakoti Y; Yadav S; Sundar D, TransDTI: Transformer-Based Language Models for Estimating DTIs and Building a Drug Recommendation Workflow. Acs Omega 2022, 7 (3), 2706–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bagal V; Aggarwal R; Vinod PK; Priyakumar UD, MolGPT: Molecular Generation Using a Transformer-Decoder Model. J Chem Inf Model 2022, 62, 2064–2076. [DOI] [PubMed] [Google Scholar]
- 85.Karpov P; Godin G; Tetko IV, Transformer-CNN: Swiss knife for QSAR modeling and interpretation. J Cheminform 2020, 12 (1), 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tetko IV; Karpov P; Bruno E; Kimber TB; Godin G, Augmentation Is What You Need! Lect Notes Comput Sc 2019, 11731, 831–835. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset used for modeling and the scripts generated are available at our GitHub repository (https://github.com/ncats/AI-cell_nano_pred). Furthermore, the top prediction models have also been made publicly available to the research community through a web-based implementation that can be accessed via https://aicell.ncats.io/


