Abstract
Allergic asthma is a chronic inflammatory disease in which oxidative stress serves a pivotal role. In clinical practice, dexmedetomidine (DEX), an α-2-adrenergic receptor agonist, is used as a sedative. DEX exhibits antioxidative and organ-protective properties. In a murine model of asthma, DEX has a therapeutic effect via the toll like receptor 4/NF-кB signaling pathway; however, whether DEX can exert an antioxidative effect on asthma has yet to be elucidated. In the present study, a T helper (Th)2-dominant murine asthma model was established. DEX treatment significantly reduced eosinophilic airway inflammation, mucus overproduction and airway hyperresponsiveness, as well as the concentrations of Th2 cytokines. The lung tissues of mice with asthma were characterized by redox imbalance (increased oxidative stress and impaired antioxidant capacity). DEX treatment alleviated this imbalance by decreasing the levels of malondialdehyde and reactive oxygen species, and increasing the levels of glutathione. Furthermore, the nuclear factor erythroid 2-related factor 2 (Nrf2) signaling pathway was inhibited in the lung tissues of asthmatic mice; these effects were noted in its downstream genes, heme oxygenase 1 and glutathione peroxidase 4. In mice with asthma, DEX treatment induced the expression of these antioxidant genes and the activation of Nrf2, whereas ML385 (an inhibitor of Nrf2) partially abrogated the antioxidative and therapeutic effects of DEX. To the best of our knowledge, the present study is the first to demonstrate the protective effect of DEX on Th2-dominant asthma through the activation of the Nrf2 signaling pathway. The results suggested that the antioxidative properties of DEX could be beneficial in clinical application of DEX for the relief of asthmatic symptoms.
Keywords: dexmedetomidine, oxidative stress, allergic asthma, airway inflammation, nuclear factor erythroid 2-related factor 2
Introduction
Asthma has long been a global health issue, affecting approximately 262 million people in 2019 (1) and causing 455,000 mortalities (2). The established asthma risk factors (3) includes air pollutants, allergens, microbial products and obesity. According to the updated National Asthma Education Prevention Program guidelines (4), the combination of inhaled corticosteroids and long-acting β-agonist is the current mainstream asthma treatment. Asthma is a heterogeneous disease with various phenotypes or endotypes, including eosinophilic asthma and non-eosinophilic asthma (5). Eosinophilic airway inflammation is a hallmark of allergic asthma; this type of inflammation may explain the symptoms of asthma, including airway hyperresponsiveness (AHR) (5), airway remodeling (5) and mucus overproduction (6).
Oxidative stress is a cellular condition in which the production of reactive oxygen species (ROS) exceeds the antioxidant content of the cell (7). This process is known to serve a major role in the pathogenesis and progression of asthma, and although it is considered to be a significant part of the inflammatory response, it can also stimulate inflammation in asthma (8). The inflammatory cells, notably eosinophils, are an important source of ROS following antigenic challenge; excessive production of ROS can cause damage to the lipids, proteins and DNA in cells (9). Increased ROS production is closely related to a decrease in the forced expiratory volume in 1 sec. It has also been reported that pulmonary function is negatively related to oxidative stress markers and positively related to serum antioxidant levels (10). The levels of antioxidants, such as malondialdehyde (MDA), and oxidative stress markers, such as reduced glutathione (GSH), have also been reported to be linked to the severity of asthma (11).
The cellular antioxidant system, which maintains redox homeostasis, consists of both enzymatic and non-enzymatic antioxidants. The transcription factor nuclear factor erythroid 2-related factor (Nrf2) is crucial in ROS scavenging (12). As a result of oxidative stress, Nrf2 is dissociated from kelch-like ECH-associated protein-1 (Keap1) and translocated to the nucleus where it can activate >200 antioxidant genes. These gene products are crucial for the antioxidant, anti-inflammatory and cytoprotective functions of the cells (13). Superoxide dismutase (SOD), heme oxygenase-1 (HO-1), quinone oxidoreductase-1 and glutathione peroxidase (GPx) are the main antioxidant molecules. Accumulating evidence has demonstrated the importance of the Nrf2 pathway in asthma, as eosinophilic inflammation; AHR and inflammatory cytokine production have been reported in Nrf2 knockout mice (14–16).
Dexmedetomidine (DEX) is a selective α2-adrenergic receptor agonist, which is commonly used as a sedative in clinical practice (17). Several studies have reported organ-protective effects and anti-inflammatory effects of DEX. For example, it has been reported that DEX attenuates neuroinflammation (18), protects the lung from injury induced by limb ischemia/reperfusion (19), protects hepatic cells from oxygen-glucose deprivation/reperfusion injury (20) and reduces the levels of proinflammatory mediators (IL-6, TNF-α, IL-10 and IL-1) in an acute lung injury model in vivo (21). Oxidative stress serves a major role in inducing organ injury and inflammation, and the antioxidative effects of DEX have also been reported by a number of studies (22–24). Our previous study demonstrated that administration of DEX could reduce airway eosinophilic inflammation, AHR and mucus production in an ovalbumin (OVA)-induced murine asthma model (25). In light of the potential role of oxidative stress in the pathogenesis of asthma, it was hypothesized that DEX could reduce the symptoms of allergic asthma by decreasing oxidative stress. To evaluate this hypothesis, the present study assessed the antioxidative effect of DEX on an allergic murine asthma model.
Materials and methods
Animals
A total of 25 female Balb/c mice (age, 6–7 weeks; weight, 15–20 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The mice were kept in the animal experimental center of the Plastic Surgery Hospital Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China) under a controlled temperature (25°C) and humidity (45–55%), with a 12-h light-dark cycle. The mice were fed with sterilized food and water ad libitum and were acclimated for 1 week prior to the experiment. The protocol of the animal experiments was approved by the Institutional Animal Care and Use Committee of Plastic Surgery Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College [approval no. 2022(201)].
Animal experiments
The mice were divided into five groups (n=5) as follows: i) Control group; ii) OVA group; iii) DEX group; iv) OVA + DEX group; and v) OVA + DEX + ML385 group. The protocol used for the OVA-induced murine asthma model was described in our previous work (25). In brief, the mice in the control group received an intraperitoneal (i.p.) injection of 0.2 ml saline on days 0, 7 and 14 for sensitization, and were administered 30 µl saline intranasally for the challenge on days 21–28. The mice in the DEX group were sensitized with i.p. injection of 0.2 ml saline on days 0, 7 and 14, and received an i.p. injection of 30 µg/kg DEX 1 h prior to intranasal administration of 30 µl saline on days 21–28. The mice in the OVA, OVA + DEX and OVA + DEX + ML385 groups received an i.p. injection of 0.2 ml saline containing 100 µg OVA (cat. no. A5503; Sigma-Aldrich; Merck KGaA) and 10 mg aluminum hydroxide (cat. no. 239186; Sigma-Aldrich; Merck KGaA) on days 0, 7 and 14 for sensitization. From day 21 to 28, the mice in the OVA group were administered 30 µl saline containing 200 µg OVA intranasally for the challenge. The mice in the OVA + DEX group were injected i.p. with 30 µg/kg DEX and were challenged with 30 µl saline containing 200 µg OVA intranasally 1 h after DEX injection; the mice in the OVA + DEX+ML385 group were injected i.p. with 30 mg/kg ML385 working solution (cat. no. 846557-71-9; MedChemExpress) 1 h prior to the i.p. injection of 30 µg/kg DEX and were challenged with 30 µl saline containing 200 µg OVA intranasally 1 h following DEX injection. The preparation method for the ML385 working solution was as follows: ML385 was diluted with DMSO (cat. no. K91281; Beijing Kerhui Technology Co., Ltd.) and corn oil to a concentration of 2 mg/ml; the final concentration of DMSO was 1%. Following the assessment of AHR, bronchoalveolar lavage fluid (BALF) was collected on day 29 for subsequent analysis.
Assessment of AHR
The respiratory resistance (Rrs) (cm H2O.s/ml) was assessed for the mice in each group using a flexiVent lung function system (SCIREQ) 24 h following the last challenge. Prior to the tracheotomy and intubation, the mice were i.p. anesthetized with 2% pentobarbital sodium (50 mg/kg). Following intubation, the mice were fixed to a platform and allowed to inhale aerosolized methacholine (Mch) at different concentrations (0, 6, 12, 24 and 48 mg/ml) through the tube for 6 min. The results presented for the different concentrations of Mch inhaled by each mouse comprised the mean absolute value of the data collected during the 6 min period.
Analysis of BALF
Immediately following AHR assessment, the mice were euthanized by i.p. injection of 2% pentobarbital sodium (100 mg/kg); the mice were considered to be at a terminally-anesthetized state when they stopped breathing, the heart stopped beating and they did not response to stimuli (26). Subsequently, a tube was inserted into the trachea of each mouse for lung lavage. The lungs were lavaged three times using 0.8 ml Dulbecco's phosphate-buffered saline (DPBS) and ~90% of the lavage volume was collected. Following centrifugation at 187 × g at 4°C for 10 min, the supernatant of the BALF was collected and stored at −80°C for use in ELISA and the cells were resuspended in 500 µl DPBS for cell counting. The total cell count in the BALF samples was assessed using a chamber slide and Wright-Giemsa-stained BALF smears were made for eosinophil and lymphocyte counts. The assay was performed using a Wright-Giemsa staining kit (cat. no. G1020; Beijing Solarbio Science & Technology Co., Ltd.) according to the manufacturer's protocol (staining at room temperature for 1.5 min). The smears were assessed under ×200 magnification using a BX53 upright light microscope (Olympus Corporation). The eosinophil and lymphocyte counts were assessed using ImageJ software (version 1.8.0; National Institutes of Health).
Measurement of cytokines in BALF
The levels of Th2 cytokines (including IL-4, IL-5, and IL-13) were detected using commercial ELISA kits purchased from Boster Biological Technology (IL-4: cat. no. EK0405; IL-5: cat. no. EK0408; IL-13: cat. no. EK0425). Detections were performed according to the manufacturer's instructions. The concentrations of cytokines were determined by measuring absorbance at 450 nm using a multimode plate reader (cat. no. HH34000000; PerkinElmer).
Hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS) staining
The right lobes of the lung were collected 24 h after the last challenge, following euthanasia, and fixed using 4% paraformaldehyde (cat. no. P1110; Beijing Solarbio Science & Technology Co., Ltd.) for 48 h at room temperature. The following procedures were all performed at room temperature. H&E and PAS staining were performed on different lung sections. For H&E staining, the lung tissues were embedded in paraffin and subsequently cut into 5 µm thick sections. Deparaffinization of the sections was performed using xylene and hydration was performed by passing the sections through changes of ethanol with different concentrations (100% ethanol for 2 min; 100% ethanol for 2 min; 95% ethanol for 2 min). The sections were subsequently stained with hematoxylin for 1 min at room temperature. Following rinsing in tap water for 5 min, nuclear staining was finished following incubation in an alkaline solution (Scott's Bluing Solution; cat. no. G1865; Beijing Solarbio Science & Technology Co., Ltd.) for 20 sec. Following differentiation of the sections for 3 sec, they were stained using alcoholic-eosin for 1 min and subsequently dehydrated using an ascending alcohol series.
PAS staining was performed using the Periodic Acid-Schiff (PAS) stain kit (cat. no. G1280; Beijing Solarbio Science & Technology Co., Ltd.) according to the manufacturer's protocol. In brief, after being fixed using 4% paraformaldehyde (cat. no. P1110; Beijing Solarbio Science & Technology Co., Ltd.) at room temperature for 48 h and then placed in 30% sucrose (1X PBS) until lung tissues sank, the lung tissues were subsequently embedded in OTC and frozen at −20°C. Then, 5 µm-thick frozen sections were washed with tap water for 2 min and rinsed with double-distilled H2O once. The sections were incubated with the oxidizing agent contained in the PAS stain kit for 6.5 min and rinsed with tap water for 10 min. The slides were incubated with Schiff reagent for 15 min and subsequently washed with tap water for 15 min. The nuclei were stained using hematoxylin for 50 sec. Following washing with tap water for 2 min, the slides were re-stained with an alkaline solution (Scott's Bluing Solution; cat. no. G1865; Beijing Solarbio Science & Technology Co., Ltd.) for 20 sec and differentiated for 3 sec. Five fields of view were assessed by two experienced pathologists in a blinded study design at ×200 magnification using a BX53 upright light microscope (Olympus Corporation). The color imbalance was corrected as described by Marty (27). In brief, auto exposure was used to capture a tissue image and a blank image which were saved and processed with Adobe Photoshop CS6 (Adobe, Inc.) as follows: i) The blank image was inverted and the layer duplicated to the original tissue image; ii) in the window with the tissue image, Color Dodge in the Layers Palette was to achieve a uniform white background; iii) the image was flattened and then saved as the final version.
The scoring systems for H&E and PAS staining were described in our previous work (25). In brief, the score for H&E staining represented the infiltration of inflammatory cells around the airways and was presented as follows: 0, normal; 1, a low number of cells present around the airways; 2, one cell layer was present around the airways; 3, 2–4 cell layers were present around the airways; 4, 5–7 cell layers were present around the airways; 5, 8–10 cell layers were present around the airways; and 6, >10 cell layers were present around the airways. The percentage of PAS-positive cells in each slide was assessed using ImageJ software (version 1.8.0; National Institutes of Health) and scored as follows: 0, <2%; 1, ≥2 to <20%; 2, ≥20 to <40%; 3, ≥40 to <60%; 4, ≥60 to <80%; and 5, ≥80% PAS-positive cells.
Dihydroethidium (DHE) staining
DHE (cat. no. S0063) was purchased from Beyotime Institute of Biotechnology and the staining process was performed according to the manufacturer's protocols. Briefly, frozen lung sections were prepared as those used for PAS staining and were treated with DHE working reagent (5 µM) at 37°C in the dark for 30 min. The slides were sealed with a sealing reagent containing DAPI (cat. no. P0131; Beyotime Institute of Biotechnology) at room temperature for 1 min. The slides were assessed using a BX53 fluorescence microscope (Olympus Corporation) at 535 nm. Unprocessed frozen sections of lung tissues were used as an endogenous control to exclude autofluorescence and no autofluorescence was observed in the unprocessed frozen sections at 488 nm (data not shown).
Measurement of MDA, SOD and GSH levels
Specific commercial kits (purchased from Beyotime Institute of Biotechnology) were used to determine the MDA content (cat. no. S0131S), the activity levels of SOD (cat. no. S0103) and the GSH content (cat. no. S0053) in the supernatant of the homogenized lung tissues. Lung tissues (30 mg) were homogenized in the sample preparation solution which was contained in the kits using a Multi-sample tissue grinder (TissueLyser-24L; Shanghai Jingxin). The assays were performed according to the manufacturer's protocols. Total protein concentration was determined using the BCA method (cat. no. P0012S; Beyotime Institute of Biotechnology) according to the manufacturer's protocols.
Reverse transcription-quantitative PCR (RT-qPCR)
A total of 50 mg lung tissue was homogenized in TRIzol® (cat. no. 15596-026; Invitrogen; Thermo Fisher Scientific, Inc.), which was used for total RNA extraction according to the manufacturer's protocol. The total RNA was resuspended in DNase/RNase-free water (cat. no. R1600; Beijing Solarbio Science & Technology Co., Ltd.), and its concentration was determined using a NanoDrop® 2000 ultraviolet spectrophotometer (Thermo Fisher Scientific, Inc.). RNA samples with a 260/280 absorbance ratio between 1.8 and 2.0 were used. A total of 5 µg RNA was revers transcribed using the TransScript® First-Strand cDNA Synthesis SuperMix (cat. no. AT301-02; Beijing Transgen Biotech Co., Ltd.) in 20 µl reaction volumes according to the manufacturer's protocol. The qPCR primer sequences (Invitrogen; Thermo Fisher Scientific, Inc.) used were as follows: Nrf2 forward (F), 5′-AGATGACCATGAGTCGCTTGC-3′ and reverse (R), 5′-CCTGATGAGGGGCAGTGAAG-3′; Keap1 F, 5′-GCCCCGGGACTCTTATTGTG-3′ and R, 5′-TTAGGGGCCCCGCCAT-3′; HO-1 F, 5′-GCTAGCCTGGTGCAAGATACT-3′ and R, 5′-AAGCTGAGAGTGAGGACCCA-3′; GPx4 F, 5′-CCTCCCCAGTACTGCAACAG-3′ and R, 5′-GCACACGAAACCCCTGTACT-3′; and β-actin F, 5′-CTCTTTTCCAGCCTTCCTTCTT-3′ and R, 5′-AGGTCTTTACGGATGTCAACGT-3′. The relative mRNA expression levels of Nrf2, Keap1, HO-1 and GPx4 were normalized to those of β-actin. The LightCycler® 480 SYBR Green I Master mix (cat. no. 04707516001) and the LightCycler® 96 Instrument for qPCR were purchased from Roche Diagnostics GmbH and all the experimental procedures were performed according to the manufacturer's protocol. The thermocycling conditions for qPCR were as follows: Initial denaturation at 95°C for 180 sec; a two-step amplification of 40 cycles of denaturation at 95°C for 10 sec and extension at 60°C for 30 sec. The data were quantified using the 2−ΔΔCq method (28) and LightCycler® 96 software version 1.1 (Roche Diagnostics GmbH).
Western blotting
The lung tissues were washed with cold PBS and lysed using RIPA lysis buffer (cat. no. C1053; Applygen Technologies, Inc.) in the presence of Protease Phosphatase Inhibitor Cocktail (cat. no. P1045; Beyotime Institute of Biotechnology). The extraction of nuclear proteins was performed using a Nuclear and Cytoplasmic Protein Extraction Kit (cat. no. P0028; Beyotime Institute of Technology). The total and nuclear protein concentrations were determined using the BCA Protein Assay Kit (cat. no. P0012S; Beyotime Institute of Biotechnology). The proteins were dissolved using SDS-PAGE Protein Sample Loading Buffer (cat. no. P0286-15 ml; Beyotime Institute of Biotechnology) and subsequently denatured. Protein samples (20 µg) were separated using SDS-PAGE (4–20%). The protein bands were subsequently transferred to nitrocellulose membranes and blocked using 5% skimmed milk for 2 h at room temperature. The membranes were incubated with primary antibodies overnight at 4°C. The membranes were washed and incubated with the appropriate secondary antibodies for 1 h at room temperature. Finally, the protein bands were visualized using BeyoECL Plus detection reagent (cat. no. P0018S; Beyotime Biotechnology Institute of Biotechnology). Band images were obtained using Chemi-Doc Imaging System (cat. no. 12003153; Bio-Rad Laboratories, Inc.). The semi-quantification of the bands was performed using ImageJ software (version 1.8.0) according to a publicly available protocol (29). The primary antibodies used in the experiments were as follows: anti-Nrf2 (1:1,000; cat. no. 16396-1-AP; ProteinTech Group, Inc.), anti-Keap1 (1:1,000; cat. no. 10503-2-AP; ProteinTech Group, Inc.), anti-HO-1 (1:500; cat. no. sc-390991; Santa Cruz Biotechnology, Inc.), anti-GPx4 (1:1,000; cat. no. 67763-1-Ig; ProteinTech Group, Inc.), anti-β-actin (1:1,000; cat. no. 66009-1-Ig; ProteinTech Group, Inc.), anti-β-tubulin (1:2,000; cat. no. 10094-1-AP; ProteinTech Group, Inc.) and anti-PCNA (1:2,000; cat. no. 10205-2-AP; ProteinTech Group, Inc.). The secondary antibodies used for the experiments were as follows: HRP-conjugated Affinipure goat anti-mouse IgG (H+L). (1:2,000; cat. no. SA00001-1; ProteinTech Group, Inc.;), HRP-conjugated Affinipure goat anti-rabbit IgG (H+L) (1:2,000; cat. no. SA00001-2; ProteinTech Group, Inc.) and anti-mouse-IgGκ binding protein-HRP (1:2,000; cat. no. sc-516102; Santa Cruz Biotechnology, Inc.).
Statistical analysis
The data were analyzed using GraphPad Prism 9 (GraphPad Software, Inc.). Quantitative and semi-quantitative data are presented as mean ± SD, and one-way ANOVA followed by Bonferroni's multiple comparisons test was performed to assess the differences between the groups. The histopathological scores are presented as median + interquartile range, and the Kruskal-Wallis test followed by Dunn's multiple comparisons test was performed to assess the differences between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
DEX attenuates airway inflammation and mucus overproduction
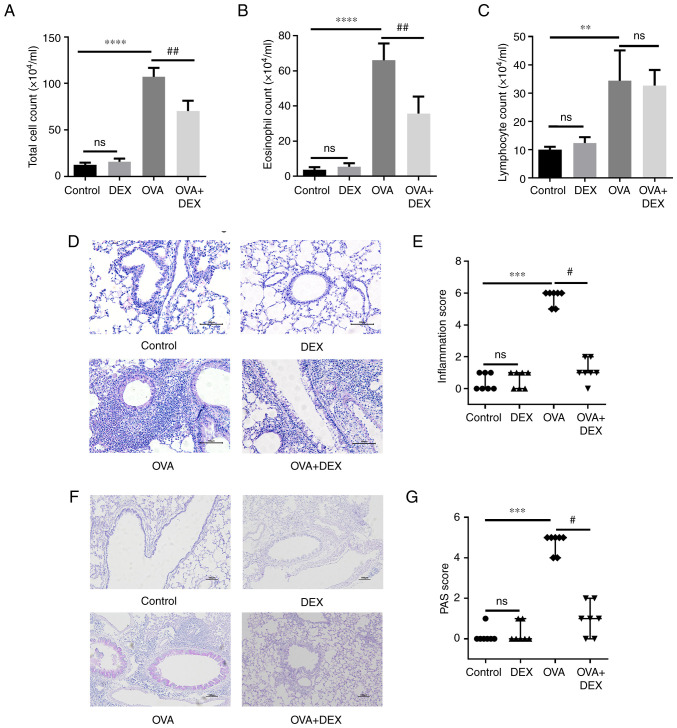
Allergic asthma is characterized by eosinophilic airway inflammation and mucus overproduction (30). To assess airway inflammation and mucus production in the lung tissues, BALF was collected for total cell counting and Wright-Giemsa-stained BALF smears were used for differential cell counts. Significant increases were demonstrated in the total cell counts (P<0.0001), eosinophil counts (P<0.0001) and lymphocyte counts (P<0.01) in the OVA group compared with the control group (Figs. 1A-C and S1). In comparison with the OVA group, DEX treatment significantly reduced the total cell and eosinophil counts (both P<0.01) in the BALF; however no significant difference was demonstrated for the lymphocyte count. Total cell, eosinophil and lymphocyte counts in the DEX group were not significantly different to that of the control group.
Figure 1.
DEX attenuates eosinophilic airway inflammation and mucus overproduction. (A) Total cell, (B) eosinophil and (C) lymphocyte counts in bronchoalveolar lavage fluid samples. (D) H&E and (F) PAS staining were performed to evaluate inflammatory cell infiltration and mucus production in lung tissues, respectively. Magnification for H&E staining, ×200; magnification for PAS staining, ×100; scale bar, 100 µm. The color imbalance was corrected as described by Marty (27). Histological scoring of (E) inflammatory cell infiltration and (G) mucus production were based on the layers of cells around the airways and the percentage of PAS-positive cells, respectively. Quantitative data are presented as the mean ± SD (n=3). Ordinal data are presented as median + interquartile range (n=7). **P<0.01, ***P<0.001 and ****P<0.0001 vs. control; #P<0.05 and ##P<0.01 vs. OVA. DEX, dexmedetomidine; H&E, hematoxylin and eosin; ns, not significant; OVA, ovalbumin; PAS, periodic acid-Schiff.
Lung sections were used for H&E and PAS staining to evaluate the histopathological characteristics. The number of the rings of the inflammatory cells infiltrating around the airways, which were assessed as inflammation scores, was significantly higher in the OVA group compared with the control group (Fig. 1D and E; P<0.001); DEX was able to significantly inhibit inflammatory cell infiltration (P<0.05) compared with the OVA group. A significant increase in the PAS-positive cells was demonstrated in the OVA group compared with the control (Fig. 1F and G; P<0.001), and a significant reduction in mucus overproduction was demonstrated in the OVA + DEX group compared with the OVA group (P<0.05). DEX treatment alone failed to induce histopathological changes in the lung tissues.
DEX reduces T helper (Th)2 cytokine production and AHR
A Th2-dominant asthma model was established and the content of the Th2 cytokines IL-4, IL-5 and IL-13 was assessed using ELISA. The levels of these cytokines in BALF were significantly elevated in the OVA group compared with those in the control group (P<0.01 for IL-4, P<0.001 for IL-5, P<0.0001 for IL-13; Fig. 2A). The levels of IL-4, IL-5 and IL-13 in the BALF were significantly decreased following DEX administration compared with the OVA group (P<0.05 for IL-4, P<0.01 for IL-5, P<0.05 for IL-13; Fig. 2A). DEX administration alone did not significantly affect the number of Th2 cytokines.
Figure 2.
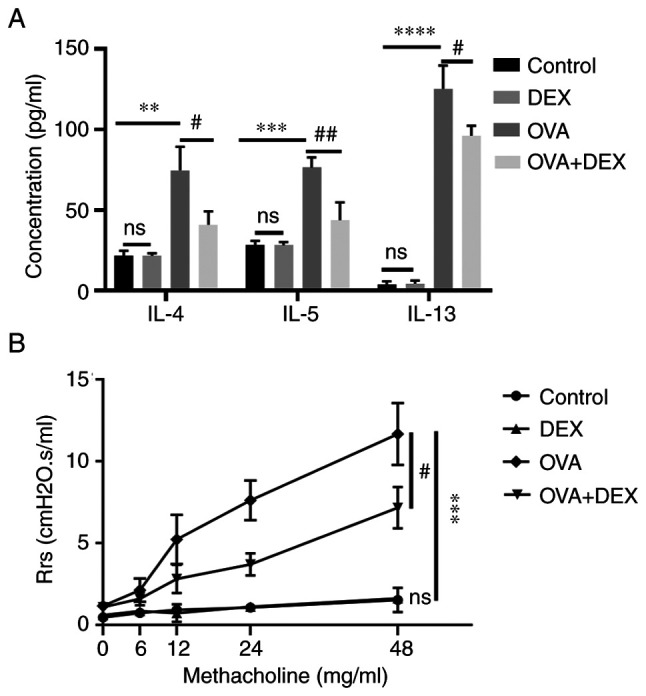
DEX reduces T helper 2 cytokine production and airway hyperresponsiveness. (A) Commercial ELISA kits were used to assess the concentration of IL-4, IL-5 and IL-13 in bronchoalveolar lavage fluid. (B) Airway resistance of mice in response to nebulized methacholine of different concentrations (0, 6, 12, 24 and 48 mg/ml). Quantitative data are presented as the mean ± SD (n=3). **P<0.01, ***P<0.001 and ****P<0.0001 vs. control; #P<0.05 and ##P<0.01 vs. OVA. DEX, dexmedetomidine; OVA, ovalbumin; Rrs, respiratory resistance.
AHR is a crucial feature of asthma; both the airway inflammation and Th2 cytokines (particularly IL-13) contribute to its pathogenesis (31). Compared with the control group, the respiratory resistance to inhaled Mch (48 mg/ml; the Rrs for each concentration of Mch inhaled by each animal was collected every minute for 6 min and the results were presented as the mean absolute value of the data collected during the 6 min period.) was significantly increased in the OVA group (P<0.001; Fig. 2B). DEX treatment significantly reduced the airway resistance compared with the OVA group (P<0.05). DEX administration alone did not significantly affect airway responsiveness compared with the control (Fig. 2).
DEX reduces oxidative stress and restores antioxidant capacity in the murine model of asthma
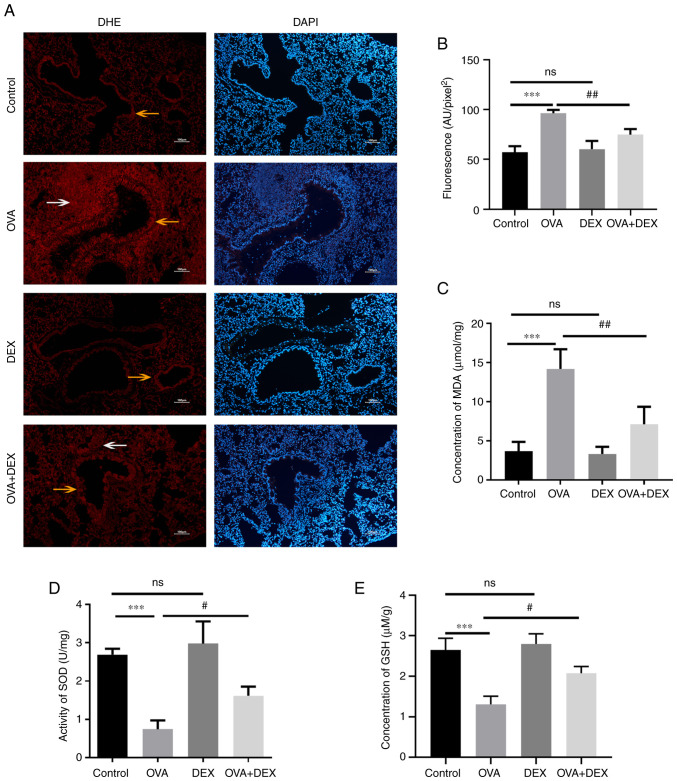
It has been reported that redox imbalance is associated with the development of asthma (32). To evaluate the redox state in the present study, the levels of oxidative stress markers (ROS and MDA) and the antioxidant capacity of the cells (GSH level and SOD activity) were assessed. Lung sections were stained with DHE to evaluate ROS. The data indicated that ROS were mainly generated in airway epithelial cells (yellow arrows) and inflammatory cells (white arrows) around the airways (Fig. 3A). Subsequently, the average fluorescence intensity was calculated (Fig. 3B). ROS levels were significantly increased in the OVA group compared with the control (P<0.001), and DEX treatment significantly decreased ROS levels in the lung compared with the OVA group (P<0.01). The MDA levels in the lung tissue were assessed to evaluate lipid peroxidation (Fig. 3C). In agreement with the ROS results, these data demonstrated that the MDA levels were significantly elevated in the OVA group compared with the control (P<0.001), whereas DEX treatment significantly reduced MDA compared with the OVA group (P<0.01). The activity of SOD and level of GSH were reduced in asthmatic mice compared with the control (both P<0.001), and DEX treatment restored their content compared with the OVA group (both P<0.05; Fig. 3D and E). In comparison with the control group, administration of DEX alone demonstrated no significant effect on the redox state of the lung tissues compared with the control group.
Figure 3.
DEX reduces oxidative stress and restores antioxidant capacity in a murine model of asthma. (A) Representative images of DHE-stained (red fluorescence) and DAPI-stained (blue fluorescence) lung tissue sections. The airway epithelial cells are indicated by yellow arrows and the inflammatory cells around the airways are indicated by white arrows. Magnification, ×100; scale bar, 100 µm. (B) The mean fluorescence density of the DHE-stained lung tissue sections represented ROS generation in the lung tissues. (C) A commercial MDA assay kit was used to assess the MDA content per mg protein. (D) The activity levels of SOD per mg of protein in the lung tissues were assessed using SOD assay kits. (E) GSH levels in the lung tissues were assessed using GSH assay kits. Quantitative data are presented as the mean ± SD (n=3). ***P<0.001 vs. control; #P<0.05 and ##P<0.01 vs. OVA. DEX, dexmedetomidine; DHE, dihydroethidium; GSH, reduced glutathione; MDA, malondialdehyde; OVA, ovalbumin; ROS, reactive oxygen species; SOD, superoxide dismutase.
Nrf2 signaling pathway and its downstream antioxidant genes are activated by DEX treatment
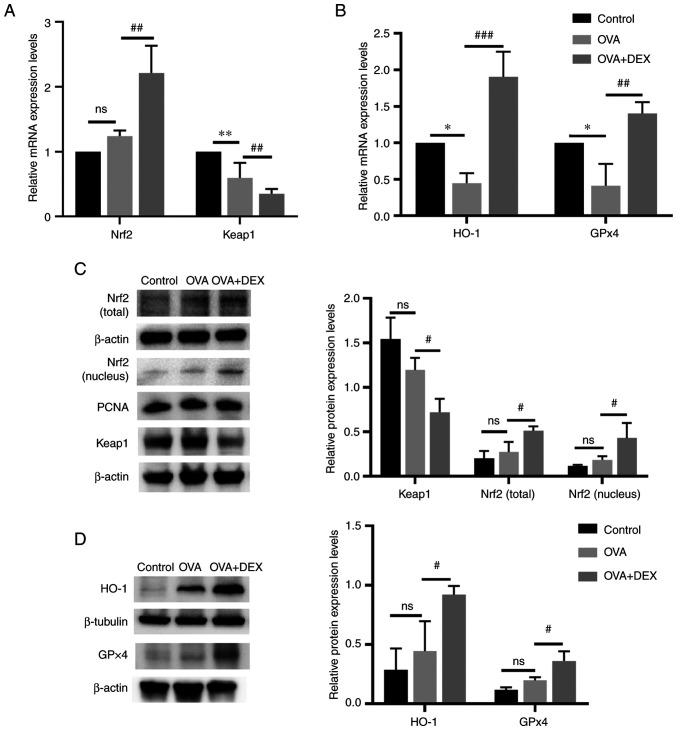
Nrf2 signaling and its downstream genes (HO-1 and GPx4) serve an important role in maintaining redox balance in asthma (33). The mRNA expression levels of Nrf2 in the OVA + DEX group were significantly higher compared with those demonstrated in the OVA group (P<0.01; Fig. 4A), whereas no significant differences were demonstrated between the OVA and the control group. As the activation of Nrf2 is regulated by modification of Keap1 (34), the expression level of Keap1 was detected. The mRNA expression levels of Keap1 were significantly decreased in the OVA group compared with the control (P<0.01), and DEX treatment significantly reduced its levels compared with the OVA group (P<0.01). The mRNA expression levels of HO-1 and GPx4 were significantly reduced in the OVA group compared with the control (P<0.05; Fig. 4B), and DEX administration led to a significant increase in HO-1 (P<0.001) and GPx4 (P<0.01) mRNA expression levels compared with the OVA group. Western blotting demonstrated that although Nrf2 (nuclear and total) and antioxidant (HO-1 and GPX4) protein expression levels were increased and Keap1 protein expression level was reduced in the OVA group, the data were not statistically significant compared with those of the control group (Fig. 4C and D), which suggested that Nrf2 might be activated to a limited extent in asthmatic mice. The presence of DEX further activated Nrf2 and demonstrated significantly increased nuclear and total Nrf2 protein expression levels and significantly decreased Keap1 protein expression levels compared with the OVA group (P<0.05; Fig. 4C). In addition, the protein expression levels of HO-1 and GPx4 were significantly elevated in the OVA + DEX group compared with the OVA group (P<0.05; Fig. 4D).
Figure 4.
Nrf2 signaling and its downstream antioxidant genes are activated by DEX. mRNA expression levels of (A) Nrf2 and Keap1, and (B) HO-1 and GPx4 in lung tissues were assessed using reverse transcription-quantitative PCR. Protein expression levels of (C) Nrf2 (cytoplasm and nucleus), Keap1, (D) HO-1 and GPx4 were assessed by western blotting. Data are presented as the mean ± SD (n=3). #P<0.05, ##P<0.01 and ###P<0.001 vs. OVA; *P<0.05 and **P<0.01 vs. control. DEX, dexmedetomidine; GPx, glutathione peroxidase; HO-1, heme oxygenase 1; Keap1, kelch-like ECH-associated protein-1; Nrf2, nuclear factor erythroid 2-related factor 2; OVA, ovalbumin.
Therapeutic and antioxidative effects of DEX in asthma are partially diminished by ML385
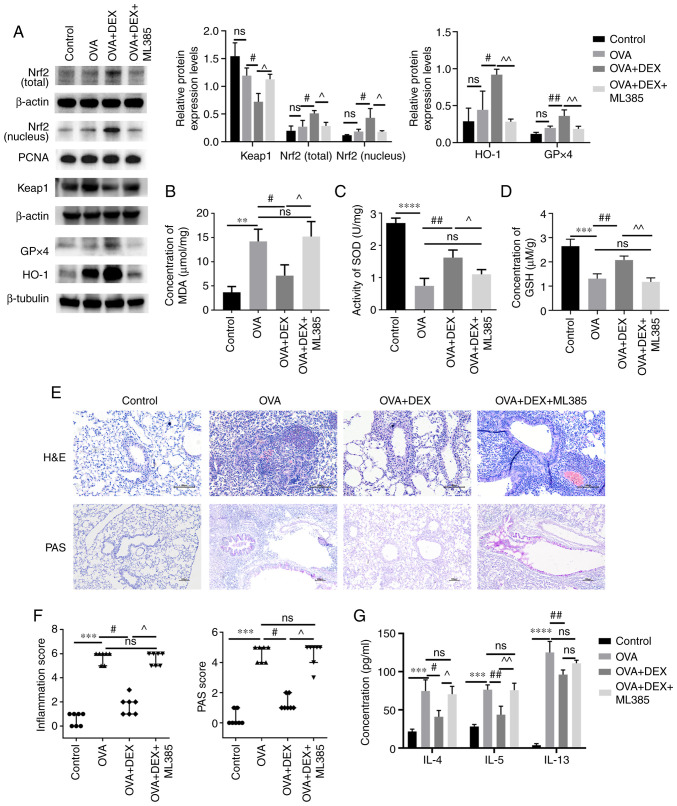
The presence of Nrf2 activation was further explored in the current model by the administration of ML385 (an Nrf2 inhibitor) prior to DEX administration. As demonstrated using western blotting (Fig. 5A), ML385 significantly reduced the total and nuclear Nrf2 protein expression levels (P<0.05), significantly upregulated the protein expression level of Keap1 (P<0.05) and significantly downregulated the protein expression levels of HO-1 and GPx4 (P<0.01) compared with the OVA + DEX group. The MDA content was significantly increased by ML385 treatment compared with the OVA + DEX group (Fig. 5B). Moreover, the increased GSH content and the increased activity levels of SOD caused by DEX treatment were significantly suppressed following administration of ML385 compared with the OVA + DEX group (SOD, P<0.05; GSH, P<0.01; Fig. 5C and D). As shown in Fig. 5E and F, administration of OVA significantly increased inflammatory cells infiltration around airways and mucus production (both P<0.001, vs. the control group), and DEX treatment effectively decreased airway inflammation and mucus overproduction (both P<0.05; vs. the OVA group). Subsequently, ML385 treatment reversed these effects of DEX (P<0.05; Fig. 5E and F). However, administration of ML385 did not significantly increase the airway resistance, which was reduced by DEX treatment (Fig. S2). The concentration of Th2 cytokines in the BALF was also assessed (Fig. 5G); the concentrations of IL-4 (P<0.05) and IL-5 (P<0.01) were significantly elevated following the administration of ML385 compared with the OVA + DEX group. However, the concentration of IL-13 in the OVA + DEX + ML385 group was not significantly different compared with those of the OVA + DEX group.
Figure 5.
Therapeutic and antioxidative effects of DEX in asthma are partly diminished by ML385. (A) The protein expression levels of Nrf2 (cytoplasmic and nuclear), Keap1, HO-1 and GPx4 were assessed using western blotting. (B) MDA content (C) activity levels of SOD and (D) GSH content in lung tissues were assessed using commercial kits. (E) H&E and PAS staining were performed to evaluate inflammatory cell infiltration and mucus production in lung tissues, respectively. Magnification for H&E staining, ×200; magnification for PAS staining, ×100; scale bar, 100 µm. Color imbalance was corrected as described by Marty (27). (F) Histological scoring of inflammatory cell infiltration and mucus production were based on the layers of the cells around the airways and the percentage of PAS-positive cells, respectively. (G) The concentration of IL-4, IL-5 and IL-13 in bronchoalveolar lavage fluid were assessed using commercial ELISA kits. Quantitative data are presented as the mean ± SD (n=3). Ordinal data are presented as the median + interquartile range (n=7). **P<0.01, ***P<0.001 and ****P<0.0001; #P<0.05 and ##P<0.01; ^P<0.05 and ^^P<0.01. DEX, dexmedetomidine; GPx, glutathione peroxidase; GSH, reduced glutathione; H&E, hematoxylin and eosin; HO-1, heme oxygenase 1; Keap1, kelch-like ECH-associated protein-1; MDA, malondialdehyde; Nrf2, nuclear factor erythroid 2-related factor 2; ns, not significant OVA, ovalbumin; PAS, periodic acid-Schiff; SOD, superoxide dismutase.
Discussion
In our previous study, it was demonstrated that 30 µg/kg DEX exerted the optimal therapeutic effect on a murine model of asthma induced by OVA (25). DEX has been reported to have exerted anti-inflammatory and antioxidative effects on brain injury (22), neuroinflammation (24) and hepatic ischemia/reperfusion injury (35) via activation of the Nrf2 signaling pathway. DEX was also reported to lessen oxidative stress-induced alveolar epithelial cell apoptosis in the lung (36). Since oxidative stress serves an important role in the pathogenesis of asthma (10), the present study evaluated the antioxidative effect of DEX on a murine model of allergic asthma. The data demonstrated that DEX significantly reduced the levels of oxidative stress in lung tissues and inhibited the symptoms of asthma via activation of the Nrf2 signaling pathway and significantly promoted the expression of antioxidant factors. To verify that the Nrf2 signaling pathway served a vital role in the antioxidative effect of DEX, the Nrf2 inhibitor ML385 was used. These data demonstrated that the therapeutic effect of DEX was partially reduced following treatment with ML385. However, a limitation of the present study was that a ML385 + OVA group was not included as a control. In general, the present study indicated that DEX may suppress inflammation in the lung through activation of the antioxidant signaling pathway.
Allergic asthma is characterized by eosinophilic airway inflammation, AHR and excessive mucus production (30). Th2 cells and Th2 cytokines serve key roles in allergic asthma development, and the OVA-induced murine model of asthma used in the present study represented a classical allergic asthma model in which Th2 cells were the dominant cell type (37). Cytokines production is one of the hallmarks of Th2 cell activation (38). The secretion of cytokines from activated Th2 cells contributed to the symptoms of asthma by recruiting eosinophils (IL-5), inducing excessive mucus production (IL-4) and leading to airway smooth muscle contraction (IL-13) (30). The results of the present study suggested that DEX treatment lowered the levels of Th2-secreted cytokines prior to each challenge, which in turn attenuated the infiltration of eosinophils in BALF samples and lung tissues, and alleviated mucus overproduction. It was a limitation of the present study that the levels of Th2 cells were not assessed and this should be incorporated in future studies. In addition, no significant differences were observed in the levels of the inflammatory markers between the DEX and the control groups.
The critical role of oxidative stress in the development of asthma has been reported by numerous studies. Oxidative stress has been reported to regulate T cell differentiation (39) and as being closely related to inflammation (40). Eosinophils, which participate in the allergic airway inflammation, are one of the main sources of oxidative stress in asthma; eosinophils produce ROS by releasing eosinophil peroxidase, which can induce bromotyrosine and nitrotyrosine. Both of these two compounds are oxidation products of eosinophils which have been reported to indicate a high risk of asthma exacerbation (41). In addition to excessive production of oxidants, asthmatic lungs possess lower activity levels of antioxidant enzymes (such as SOD and catalase) compared with those levels in normal lungs (32). Certain factors involved in oxidative stress, such as ROS and H2O2, can serve a major role in signal transduction (42). However, the overproduction of oxidants that exceeds the capacity of the cellular antioxidant system can contribute to the symptoms of asthma (43). It has been reported that ROS induces the activation of EGFR, which leads to goblet cell metaplasia (44). ROS have also been reported to result in AHR by induction of airway remodeling (airway stenosis) (45), increased intracellular Ca2+ concentration (46) and vagal tone (47). Furthermore, ROS have been reported to decrease β-adrenergic function in the lungs, which may result in a poor response to traditional bronchodilators (15). Certain oxidative stress biomarkers are reported to be highly related to asthma, including MDA, GSH (11) and SOD (48). MDA is a common indicator of lipid peroxidation and its levels are considerably higher in patients with asthma compared with those in healthy subjects during both acute attacks and symptom-free periods (49). The levels of MDA have also been reported to be positively correlated with the infiltration of eosinophils (8). GSH is the most abundant antioxidant in the airway epithelial lining fluid and is involved in various important functions relevant to asthma, such as detoxification, scavenging of free radicals and modulation of apoptosis and immune function (50). SOD levels are positively correlated with certain lung functions (48), and its inactivation was reported in asthmatic patients (51). In the present study, the levels of these oxidative biomarkers were evaluated, and the data indicated that DEX administration improved the maintenance of the redox balance by significantly reducing the levels of MDA and ROS, and significantly increasing the activity of SOD and the levels of GSH.
Nrf2 is a transcription factor that regulates the majority of antioxidant genes, including HO-1, GPx4 and NAD(P)H quinone dehydrogenase 1 (52). Under oxidative stress conditions, Nrf2 can be activated by phosphorylation (53) or Keap1 modification (54). Subsequently, it translocates to the nucleus by dissociating from Keap1 (55). Park et al (53) reported the involvement of protein kinase C (PKC) in the antioxidant effect of DEX in a rat model of ischemia. PKC phosphorylated Nrf2 and activated its detachment from Keap1. ERK1/2 has also been reported to have mediated the antioxidative effect of DEX by participating in Nrf2 activation and translocation into the nucleus (56). In addition to the two aforementioned kinases, the MAPK signaling pathway (57) and certain microRNAs (miRNAs), such as miRNA (miR)-27a, miR-153, miR142-5p and miR-144 (58), have also been reported to have mediated the regulation of Nrf2 expression and its activation. However, their potential to participate in the regulation of Nrf2 by DEX remains to be elucidated. The modification of Keap1 is used to regulate the activation of the Nrf2 pathway. Several mechanisms have been reported that explain the regulation of the concentration and activity levels of Keap1, such as transcriptional regulation (STAT6 and hypoxia-inducible factor) (59), miRNAs (miR-7 and miR-24-3p) (54), post-translational regulation, such as ubiquitination (60) and S-nitrosylation (61), as well as degradation of the Keap1 protein (p62-dependent autophagy) (62). Liu et al (63) reported the impact of DEX on the regulation of Keap1; however, the underlying mechanism of this effect remains unknown. The possible mechanism of DEX activation of the Nrf2/Keap1 signaling pathway in a murine model of asthma needs further evaluation.
The anti-inflammatory effects of Nrf2 have been reported in numerous disease models, including asthma (64). Rangasamy et al (15) reported that Nrf2 knockout mice were more susceptible to asthma, and Sussan et al (16) reported the role of Nrf2 in the cytoprotective effect on the airway epithelia of subjects with asthma. To assess whether the antioxidant effects of DEX on asthma were mediated by Nrf2, the mRNA and protein expression levels of Nrf2 and its downstream genes were evaluated. Owing to the cytoprotective effect of HO-1 and GPx4 on the airway epithelia (33) and their crucial role as in the cellular antioxidant system (65), the mRNA and protein expression levels of HO-1 and GPx4 were assessed. The results of the present study indicated that DEX treatment significantly increased the expression levels of HO-1 and GPx4 compared with those of the untreated OVA group. Administration of ML385, which significantly inhibited the activation of Nrf2, partly abrogated the antioxidant and anti-inflammatory effects of DEX. The results of the present study indicated that the therapeutic effect of DEX on asthma partly depended on the activation of the Nrf2 signaling pathway.
In conclusion, results from the present study indicated that DEX attenuated airway inflammation, AHR and mucus overproduction in the lung by the reduction of oxidative stress. This effect was at least partially mediated by the Nrf2 signaling pathway. The results suggested the potential protective effects of DEX in asthma. However, this was just a preliminary study on the antioxidant effect of DEX in a murine asthma model with Th2 dominance; further investigation of the mechanisms involved and the evaluation of other endotypes of asthma are required.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by The Plastic Surgery Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China; grant no. YS202006).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SX designed the study and drafted the manuscript. SX and DY critically revised it for important intellectual content. HG and YZ performed the animal experiments and were major contributors to writing the manuscript. SX, HG and YZ performed the experiments and analyzed the data. YZ and DY made substantial contributions to the study conception. SX and DY confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Experimental animals were handled under a protocol approved by the Institutional Animal Care and Use Committee of Plastic Surgery Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College [Beijing, China; approval no. 2022(201)].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ribeiro A, Aguiar R, Morais-Almeida M. Biological therapies, asthma and coronavirus disease 2019. Curr Opin Allergy Clin Immunol. 2021;21:597–601. doi: 10.1097/ACI.0000000000000783. [DOI] [PubMed] [Google Scholar]
- 2.O'Byrne P, Fabbri LM, Pavord ID, Papi A, Petruzzelli S, Lange P. Asthma progression and mortality: The role of inhaled corticosteroids. Eur Respir J. 2019;54:1900491. doi: 10.1183/13993003.00491-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller RL, Grayson MH, Strothman K. Advances in asthma: New understandings of asthma's natural history, risk factors, underlying mechanisms, and clinical management. J Allergy Clin Immunol. 2021;148:1430–1441. doi: 10.1016/j.jaci.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Chipps BE, Murphy KR, Oppenheimer J. 2020 NAEPP guidelines update and GINA 2021-asthma care differences, overlap, and challenges. J Allergy Clin Immunol Pract. 2022;10((1S)):S19–S30. doi: 10.1016/j.jaip.2021.10.032. [DOI] [PubMed] [Google Scholar]
- 5.Nakagome K, Nagata M. Involvement and possible role of eosinophils in asthma exacerbation. Front Immunol. 2018;9:2220. doi: 10.3389/fimmu.2018.02220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djukanovic R. Airway inflammation in asthma and its consequences: Implications for treatment in children and adults. J Allergy Clin Immunol. 2002;109((6 Suppl)):S539–S548. doi: 10.1067/mai.2002.124568. [DOI] [PubMed] [Google Scholar]
- 7.Pizzino G, Irrera N, Cucinotta M, Pallio G, Mannino F, Arcoraci V, Squadrito F, Altavilla D, Bitto A. Oxidative stress: Harms and benefits for human health. Oxid Med Cell Longev. 2017;2017:8416763. doi: 10.1155/2017/8416763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Groot LES, Sabogal Piñeros YS, Bal SM, van de Pol MA, Hamann J, Sterk PJ, Kulik W, Lutter R. Do eosinophils contribute to oxidative stress in mild asthma? Clin Exp Allergy. 2019;49:929–931. doi: 10.1111/cea.13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Checa J, Aran JM. Airway redox homeostasis and inflammation gone awry: From molecular pathogenesis to emerging therapeutics in respiratory pathology. Int J Mol Sci. 2020;21:9317. doi: 10.3390/ijms21239317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riedl MA, Nel AE. Importance of oxidative stress in the pathogenesis and treatment of asthma. Curr Opin Allergy Clin Immunol. 2008;8:49–56. doi: 10.1097/ACI.0b013e3282f3d913. [DOI] [PubMed] [Google Scholar]
- 11.Fatani SH. Biomarkers of oxidative stress in acute and chronic bronchial asthma. J Asthma. 2014;51:578–584. doi: 10.3109/02770903.2014.892965. [DOI] [PubMed] [Google Scholar]
- 12.Wang P, Geng J, Gao J, Zhao H, Li J, Shi Y, Yang B, Xiao C, Linghu Y, Sun X, et al. Macrophage achieves self-protection against oxidative stress-induced ageing through the Mst-Nrf2 axis. Nat Commun. 2019;10:755. doi: 10.1038/s41467-019-08680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang X, He B, Ning Q, Liu Y, Guo J, Niu G, Chen M. Alantolactone suppresses inflammation, apoptosis and oxidative stress in cigarette smoke-induced human bronchial epithelial cells through activation of Nrf2/HO-1 and inhibition of the NF-κB pathways. Respir Res. 2020;21:95. doi: 10.1186/s12931-020-01358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Xu Y, Yan M, Yu Y, Guo Y. 18β-Glycyrrhetinic acid suppresses allergic airway inflammation through NF-κB and Nrf2/HO-1 signaling pathways in asthma mice. Sci Rep. 2022;12:3121. doi: 10.1038/s41598-022-06455-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, Biswal S. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sussan TE, Gajghate S, Chatterjee S, Mandke P, McCormick S, Sudini K, Kumar S, Breysse PN, Diette GB, Sidhaye VK, Biswal S. Nrf2 reduces allergic asthma in mice through enhanced airway epithelial cytoprotective function. Am J Physiol Lung Cell Mol Physiol. 2015;309:L27–L36. doi: 10.1152/ajplung.00398.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gertler R, Brown HC, Mitchell DH, Silvius EN. Dexmedetomidine: A novel sedative-analgesic agent. Proc (Bayl Univ Med Cent) 2001;14:13–21. doi: 10.1080/08998280.2001.11927725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao Y, Zhu Y, He G, Ni H, Liu C, Ma L, Zhang L, Shi D. Dexmedetomidine attenuates neuroinflammation In LPS-stimulated BV2 microglia cells through upregulation of miR-340. Drug Des Devel Ther. 2019;13:3465–3475. doi: 10.2147/DDDT.S210511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue BB, Chen BH, Tang YN, Weng CW, Lin LN. Dexmedetomidine protects against lung injury induced by limb ischemia-reperfusion via the TLR4/MyD88/NF-κB pathway. Kaohsiung J Med Sci. 2019;35:672–678. doi: 10.1002/kjm2.12115. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z, Chen Q, Wan L, Zheng D, Li Z, Wu Z. Dexmedetomidine protects hepatic cells against oxygen-glucose deprivation/reperfusion injury via lncRNA CCAT1. Cell Biol Int. 2018;42:1250–1258. doi: 10.1002/cbin.10996. [DOI] [PubMed] [Google Scholar]
- 21.Geng Y, Li R, He SX, Yang HH, Deng QT, Shao XY, Wu YS, Xu WW, Ma Q. Dexmedetomidine attenuates acute lung injury induced by heatstroke and improve outcome. Shock. 2019;52:532–539. doi: 10.1097/SHK.0000000000001289. [DOI] [PubMed] [Google Scholar]
- 22.Feng X, Ma W, Zhu J, Jiao W, Wang Y. Dexmedetomidine alleviates early brain injury following traumatic brain injury by inhibiting autophagy and neuroinflammation through the ROS/Nrf2 signaling pathway. Mol Med Rep. 2021;24:661. doi: 10.3892/mmr.2021.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Kong GY, Pei WM, Zhou B, Zhang QQ, Pan BB. Dexmedetomidine alleviates hepatic injury via the inhibition of oxidative stress and activation of the Nrf2/HO-1 signaling pathway. Eur Cytokine Netw. 2019;30:88–97. doi: 10.1684/ecn.2019.0431. [DOI] [PubMed] [Google Scholar]
- 24.Li F, Wang X, Zhang Z, Zhang X, Gao P. Dexmedetomidine attenuates neuroinflammatory-induced apoptosis after traumatic brain injury via Nrf2 signaling pathway. Ann Clin Transl Neurol. 2019;6:1825–1835. doi: 10.1002/acn3.50878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao S, Wang Q, Gao H, Zhao X, Zhi J, Yang D. Dexmedetomidine alleviates airway hyperresponsiveness and allergic airway inflammation through the TLR4/NF-κB signaling pathway in mice. Mol Med Rep. 2022;25:74. doi: 10.3892/mmr.2022.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Hoecke L, Job ER, Saelens X, Roose K. Bronchoalveolar lavage of murine lungs to analyze inflammatory cell infiltration. J Vis Exp. 2017;4:55398. doi: 10.3791/55398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marty GD. Blank-field correction for achieving a uniform white background in brightfield digital photomicrographs. Biotechniques. 2007;42:716, 718, 720. doi: 10.2144/000112488. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Davarinejad H. Quantifications of western blots with imageJ. http://www.yorku.ca/yisheng/Internal/Protocols/ImageJ.pdf [Google Scholar]
- 30.Lambrecht BN, Hammad H, Fahy JV. The cytokines of asthma. Immunity. 2019;50:975–991. doi: 10.1016/j.immuni.2019.03.018. [DOI] [PubMed] [Google Scholar]
- 31.Townley RG, Horiba M. Airway hyperresponsiveness: A story of mice and men and cytokines. Clin Rev Allergy Immunol. 2003;24:85–110. doi: 10.1385/CRIAI:24:1:85. [DOI] [PubMed] [Google Scholar]
- 32.Comhair SA, Erzurum SC. Redox control of asthma: Molecular mechanisms and therapeutic opportunities. Antioxid Redox Signal. 2010;12:93–124. doi: 10.1089/ars.2008.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagasaki T, Schuyler AJ, Zhao J, Samovich SN, Yamada K, Deng Y, Ginebaugh SP, Christenson SA, Woodruff PG, Fahy JV, et al. 15LO1 dictates glutathione redox changes in asthmatic airway epithelium to worsen type 2 inflammation. J Clin Invest. 2022;132:e151685. doi: 10.1172/JCI151685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baird L, Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol Cell Biol. 2020;40:e00099–20. doi: 10.1128/MCB.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y, Qiu G, Zhang H, Zhu L, Cheng G, Wang Y, Li Y, Wu W. Dexmedetomidine alleviates hepatic ischaemia-reperfusion injury via the PI3K/AKT/Nrf2-NLRP3 pathway. J Cell Mol Med. 2021;25:9983–9994. doi: 10.1111/jcmm.16871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui J, Zhao H, Wang C, Sun JJ, Lu K, Ma D. Dexmedetomidine attenuates oxidative stress induced lung alveolar epithelial cell apoptosis in vitro. Oxid Med Cell Longev. 2015;2015:358396. doi: 10.1155/2015/358396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casaro M, Souza VR, Oliveira FA, Ferreira CM. OVA-induced allergic airway inflammation mouse model. Methods Mol Biol. 2019;1916:297–301. doi: 10.1007/978-1-4939-8994-2_28. [DOI] [PubMed] [Google Scholar]
- 38.Bosnjak B, Stelzmueller B, Erb KJ, Epstein MM. Treatment of allergic asthma: Modulation of Th2 cells and their responses. Respir Res. 2011;12:114. doi: 10.1186/1465-9921-12-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King MR, Ismail AS, Davis LS, Karp DR. Oxidative stress promotes polarization of human T cell differentiation toward a T helper 2 phenotype. J Immunol. 2006;176:2765–2772. doi: 10.4049/jimmunol.176.5.2765. [DOI] [PubMed] [Google Scholar]
- 40.Mishra V, Banga J, Silveyra P. Oxidative stress and cellular pathways of asthma and inflammation: Therapeutic strategies and pharmacological targets. Pharmacol Ther. 2018;181:169–182. doi: 10.1016/j.pharmthera.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu W, Samoszuk MK, Comhair SA, Thomassen MJ, Farver CF, Dweik RA, Kavuru MS, Erzurum SC, Hazen SL. Eosinophils generate brominating oxidants in allergen-induced asthma. J Clin Invest. 2000;105:1455–1463. doi: 10.1172/JCI9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reczek CR, Chandel NS. ROS-dependent signal transduction. Curr Opin Cell Biol. 2015;33:8–13. doi: 10.1016/j.ceb.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho YS, Moon HB. The role of oxidative stress in the pathogenesis of asthma. Allergy Asthma Immunol Res. 2010;2:183–187. doi: 10.4168/aair.2010.2.3.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casalino-Matsuda SM, Monzón ME, Forteza RM. Epidermal growth factor receptor activation by epidermal growth factor mediates oxidant-induced goblet cell metaplasia in human airway epithelium. Am J Respir Cell Mol Biol. 2006;34:581–591. doi: 10.1165/rcmb.2005-0386OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiegman CH, Michaeloudes C, Haji G, Narang P, Clarke CJ, Russell KE, Bao W, Pavlidis S, Barnes PJ, Kanerva J, et al. Oxidative stress-induced mitochondrial dysfunction drives inflammation and airway smooth muscle remodeling in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2015;136:769–780. doi: 10.1016/j.jaci.2015.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Q, Zhou Y, Zhou L, Fu Z, Yang C, Zhao L, Li S, Chen Y, Wu Y, Ling Z, et al. TRPC6-dependent Ca2+ signaling mediates airway inflammation in response to oxidative stress via ERK pathway. Cell Death Dis. 2020;11:170. doi: 10.1038/s41419-020-2360-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park CS, Kim TB, Lee KY, Moon KA, Bae YJ, Jang MK, Cho YS, Moon HB. Increased oxidative stress in the airway and development of allergic inflammation in a mouse model of asthma. Ann Allergy Asthma Immunol. 2009;103:238–247. doi: 10.1016/S1081-1206(10)60188-3. [DOI] [PubMed] [Google Scholar]
- 48.Comhair SA, Ricci KS, Arroliga M, Lara AR, Dweik RA, Song W, Hazen SL, Bleecker ER, Busse WW, Chung KF, et al. Correlation of systemic superoxide dismutase deficiency to airflow obstruction in asthma. Am J Respir Crit Care Med. 2005;172:306–313. doi: 10.1164/rccm.200502-180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma A, Bansal S, Nagpal RK. Lipid peroxidation in bronchial asthma. Indian J Pediatr. 2003;70:715–717. doi: 10.1007/BF02724313. [DOI] [PubMed] [Google Scholar]
- 50.Fitzpatrick AM, Jones DP, Brown LA. Glutathione redox control of asthma: From molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2012;17:375–408. doi: 10.1089/ars.2011.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Comhair SA, Bhathena PR, Dweik RA, Kavuru M, Erzurum SC. Rapid loss of superoxide dismutase activity during antigen-induced asthmatic response. Lancet. 2000;355:624. doi: 10.1016/S0140-6736(99)04736-4. [DOI] [PubMed] [Google Scholar]
- 52.Liu Q, Gao Y, Ci X. Role of Nrf2 and its activators in respiratory diseases. Oxid Med Cell Longev. 2019;2019:7090534. doi: 10.1155/2019/7090534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park YH, Park HP, Kim E, Lee H, Hwang JW, Jeon YT, Lim YJ. The antioxidant effect of preischemic dexmedetomidine in a rat model: Increased expression of Nrf2/HO-1 via the PKC pathway. Braz J Anesthesiol. 2021 doi: 10.1016/j.bjane.2021.08.005. S0104-0014(21)00331-6. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kopacz A, Kloska D, Forman HJ, Jozkowicz A, Grochot-Przeczek A. Beyond repression of Nrf2: An update on Keap1. Free Radic Biol Med. 2020;157:63–74. doi: 10.1016/j.freeradbiomed.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu W, Du Z, Wu L. Dexmedetomidine attenuates hypoxia-induced cardiomyocyte injury by promoting telomere/telomerase activity: Possible involvement of ERK1/2-Nrf2 signaling pathway. Cell Biol Int. 2022;46:1036–1046. doi: 10.1002/cbin.11799. [DOI] [PubMed] [Google Scholar]
- 57.Sun Z, Huang Z, Zhang DD. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS One. 2009;4:e6588. doi: 10.1371/journal.pone.0006588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng D, Wu R, Guo Y, Kong AN. Regulation of Keap1-Nrf2 signaling: The role of epigenetics. Curr Opin Toxicol. 2016;1:134–138. doi: 10.1016/j.cotox.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee OH, Jain AK, Papusha V, Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J Biol Chem. 2007;282:36412–36420. doi: 10.1074/jbc.M706517200. [DOI] [PubMed] [Google Scholar]
- 60.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kopacz A, Klóska D, Proniewski B, Cysewski D, Personnic N, Piechota-Polańczyk A, Kaczara P, Zakrzewska A, Forman HJ, Dulak J, et al. Keap1 controls protein S-nitrosation and apoptosis-senescence switch in endothelial cells. Redox Biol. 2020;28:101304. doi: 10.1016/j.redox.2019.101304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci USA. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Liu W, Wang XQ, Wan ZH, Liu YQ, Zhang MJ. Dexmedetomidine relieves neuropathic pain in rats with chronic constriction injury via the Keap1-Nrf2 pathway. Front Cell Dev Biol. 2021;9:714996. doi: 10.3389/fcell.2021.714996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rockwell CE, Jin Y, Boss AP, Kaiser LM, Awali S. The complicated role of nuclear factor erythroid-derived 2-like 2 in allergy and asthma. Drug Metab Dispos. 2022;50:500–507. doi: 10.1124/dmd.121.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ammar M, Bahloul N, Amri O, Omri R, Ghozzi H, Kammoun S, Zeghal K, Ben Mahmoud L. Oxidative stress in patients with asthma and its relation to uncontrolled asthma. J Clin Lab Anal. 2022;36:e24345. doi: 10.1002/jcla.24345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.