Significance
Immune-cell infiltration and cytotoxicity to pathogens and diseased cells are ubiquitous in health and disease. To better understand immune-cell behavior in a 3D environment, we developed an automated high-throughput microfluidic platform that enables real-time imaging of immune-cell infiltration dynamics and killing of the target cancer cells. We trained a deep learning algorithm using clinical data and integrated the algorithm with our microfluidic platform to effectively identify epigenetic drugs that promote T cell tumor infiltration and enhance cancer immunotherapy efficacy both in vitro and in vivo. Our platform provides a unique method to investigate immune-tissue interactions, which can be widely applied to oncology, immunology, neurology, microbiology, tissue engineering, regenerative medicine, translational medicine, and so on.
Keywords: cancer immunotherapy, deep learning, drug screening, immune infiltration, microfluidics
Abstract
Immunocyte infiltration and cytotoxicity play critical roles in both inflammation and immunotherapy. However, current cancer immunotherapy screening methods overlook the capacity of the T cells to penetrate the tumor stroma, thereby significantly limiting the development of effective treatments for solid tumors. Here, we present an automated high-throughput microfluidic platform for simultaneous tracking of the dynamics of T cell infiltration and cytotoxicity within the 3D tumor cultures with a tunable stromal makeup. By recourse to a clinical tumor-infiltrating lymphocyte (TIL) score analyzer, which is based on a clinical data-driven deep learning method, our platform can evaluate the efficacy of each treatment based on the scoring of T cell infiltration patterns. By screening a drug library using this technology, we identified an epigenetic drug (lysine-specific histone demethylase 1 inhibitor, LSD1i) that effectively promoted T cell tumor infiltration and enhanced treatment efficacy in combination with an immune checkpoint inhibitor (anti-PD1) in vivo. We demonstrated an automated system and strategy for screening immunocyte-solid tumor interactions, enabling the discovery of immuno- and combination therapies.
An effective immune system is critically important for fighting against cancer. However, immune evasion is one of the key hallmarks of cancer. During tumor development, lymphocytes infiltrate the tumor and inhibit its progression. Meanwhile, tumors evolve to evade such immune surveillance by downregulating antigen presentation, secreting extracellular matrix (ECM) to physically limit lymphocyte infiltration as well as chemokine/cytokines to repel lymphocytes. (1) High densities of tumor-infiltrating lymphocytes (TILs) correlate with improved prognosis in many cancer types, including breast, colorectal, ovarian, skin, and pancreatic cancers (2–4). In addition to TIL density, clinical evidence shows that the type, infiltration depth, clustering index, and activation status of TILs inside solid tumors are closely associated with disease progression and treatment outcomes (5). Recent advances in cancer immunotherapy, such as immune checkpoint inhibitors (ICIs) that remove the “brakes” on T cell–mediated antitumor immunity (6), point to potentially better outcomes in solid tumor treatment. However, the responses of cancer patients to treatment protocols that entail ICI drugs vary greatly, with overall response rates ranging from <10% to ∼40% for solid tumors (7). The ineffectiveness of ICI therapy can be largely attributed to limited TIL infiltration (8). However, TIL infiltration behavior has not been thoroughly investigated and exploited for cancer immunotherapy discovery. Thus, methods based on TIL behavior may become important tools for the discovery of effective immunotherapies and/or combination therapies that not only improve T cell cytotoxicity but also simultaneously promote T cell tumor infiltration.
To date, efforts to discover cancer immunotherapies have been performed mainly using in vitro two-dimensional (2D) cell cultures and/or in vivo animal models. However, these models have some distinct limitations. The 2D cultures fail to recapitulate key physiological aspects of tumors, such as the tumors’ stromal physical barrier and hypoxic cores (9). Animal models are limited by their scalability and are time-consuming and labor intensive (10). The 2D culture-based screening assays often rely on easily obtainable readouts, such as cytotoxicity, activation status, and cytokine profiles of T cells, while ignoring the more complex dynamics of T cell tumor infiltration behavior. In contrast, 3D tumor cultures, such as organoids/spheroids, can be constructed to include key physiological aspects of an in vivo tumor, including tumor genotypes and phenotypes, cell–cell contacts, ECM barriers, and hypoxic cores (11, 12), thereby benefitting from their potential for elucidating dynamic immunocyte–tumor interactions (13). For example, a large number of colon tumor organoids cultured in a thick basement membrane matrix such as Matrigel were used to isolate tumor-reactive T cells based on the cytotoxicity and proliferation of T cells (14). However, challenges in 3D tumor culturing, such as difficulties in generating scalable, uniformly sized, and standardized organoids/spheroids, as well as obstruction of free immunocyte infiltration and interaction by thick ECM embedding, have limited utility for high-throughput T cell behavior–based phenotypic screening (15). Furthermore, manual acquisition and analysis of the vast amount of data generated from 3D cultures pose barriers to establishing efficient and scalable high-throughput screens. Recently, automated data analysis approaches based on deep learning methods have improved the consistency and accuracy of analyzing TIL scores in pathological slides (16–18). Despite their promise, deep learning methods have not yet been used for TIL pattern analysis in in vitro models. Thus, the development of high-throughput, standardized, and automated technologies incorporating deep learning for automated TIL behavior analysis would pave the way for further advances in immunotherapy screening.
Here, we report a high-throughput, automated system for T cell behavior (e.g., tumor infiltration)–based immunotherapy discovery. This constitutes a unique demonstration of cancer immunotherapy screening that can simultaneously interrogate both T cell infiltration behavior and cytotoxicity. Through a relatively simple yet effective pillar-lattice-array design, our platform provides scalable, standardized, and tunable “core/tumor-shell/stroma” spheroids for robust drug discovery applications. In addition, we demonstrate a strategy using a deep learning algorithm to instruct drug prioritization from real-world features and clinical and pathological datasets. As a proof of concept, we used our intelligent microfluidic system to screen a library of 141 small-molecule epigenetic modulators as potential drug candidates. Among these epigenetic modulators, we have identified a lead compound, GSK-LSD1, which effectively promotes T cell infiltration, and consequently promotes deep tumor cytotoxicity once combined with anti-programmed cell death protein 1 (PD1) treatment. Furthermore, we validated the efficacy of this compound in a B16F10 mouse model in vivo.
Results
Integration of Automated Screening System.
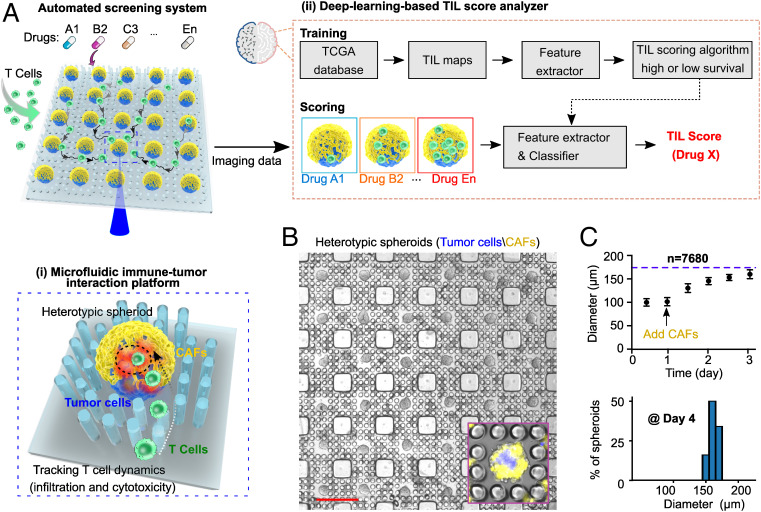
Our intelligent microfluidic screening system incorporates two components (Fig. 1A). (1) A microfluidic immune-tumor interaction platform: Through the introduction of pillar-lattice-arrays into well plates, our platform comprises 7,680 uniformly sized heterotypic tumor spheroids per well plate. Each spheroid consists of a uniform tumor core and stromal outer layer, representing the essential features of a primary tumor. Both the size of the core and the thickness of the outer layer are tunable. With the Matrigel-free culture and pillar immobilization of spheroids, our platform enables perfusion, free interaction, and time-lapse tracking of immunocyte dynamic behaviors within a large number of spheroids under different treatment conditions (e.g., 96, 384, or 1,536 conditions per run, depending on the choice of well plates), mitigating technical inconsistencies of long-term coculturing and imaging, such as fusion, size variation, and motility of spheroids. (2) A deep learning–based TIL score analyzer: To process the vast amount of T cell behavior data from our microfluidic platform, we incorporated automated image acquisition and image-based screening. The deep learning TIL scoring algorithm is trained using clinical data, including digital pathology images and survival data from 411 tumors from 397 melanoma patients in The Cancer Genome Atlas (TCGA) database, and our TIL score analyzer then automatically generates the T cell infiltration scores that correspond to TIL distribution patterns learned from the images of the high-survival (>3 y) or the low-survival patient group, respectively. We integrated our microfluidic platform with the deep learning–based TIL score analyzer to produce an automated screening system to determine immunocyte behaviors and enable screening for immunotherapeutic drugs in a high-throughput and automated manner.
Fig. 1.
Integration of automated screening platform. (A) Schematics of the automated screening platform for cancer immunotherapy screening, consisting of (i) a microfluidic immune-tumor interaction platform for high-throughput tracking of T cell dynamic behaviors and (ii) a deep learning–based TIL score analyzer for image processing and scoring T cell infiltration patterns that correspond to TIL patterns found in the images of high-survival (>3 y) or low-survival patient groups. (B) Scalable microfluidic fabrication of core/tumor-shell/stroma spheroids (tumor core: blue, CAF; shell: yellow). (C) Spheroid growth within pillar-lattice-arrays (arrow indicating CAF seeding after tumor core formation), and size distribution of tumor-stroma spheroids at day 4 of spheroid formation. (Scale bar in B: 500 µm.)
Formation of Scalable Tumor-Stroma Spheroid Array.
We fabricated scalable uniformly sized 3D core/tumor-shell/stroma spheroids (Fig. 1B and SI Appendix, Fig. S1A) in the pillar-lattice-arrays within a well plate using a two-step cell-seeding procedure. Initially, tumor cell suspensions (labeled with membrane DiO green dye, pseudocolored as blue) were seeded into the pillar-lattice-arrays to self-assemble into tumor spheroid “cores” within 12 h. The spheroid formation was driven by the geometry of the hydrophobic pillar lattices, which repel the cells. After 1 d of culture, cancer-associated fibroblasts (CAFs, labeled with membrane Dil red dye) isolated from orthotopic primary tumors were seeded on top of the tumor “cores” to form heterotypic spheroids. A large number of spheroids (7,680) per plate can be formed within pillar-lattice-arrays within one well; they reached their maximum diameter on day 3 and were arrested at uniform sizes (160 ± 9.3 µm) by the pillar-lattice-array design (Fig. 1C). These immobilized uniform-sized spheroids maintained high viability over prolonged culture (SI Appendix, Fig. S1B). This method avoids the risks of spheroids merging or moving in the well during medium changes, which is a common problem with other spheroid fabrication and culture methods, thus highlighting the advantage of our design for high-throughput screening applications. Notably, the 3D core/tumor-shell/stroma spheroids were fabricated to recapitulate tumor immunosuppressive architectures and microenvironments in that CAFs secrete ECM to physically restrict T cell infiltration and secrete chemokines to repel T cells (19). We also demonstrated the fine-tuning of dimension and composition of heterotypic spheroids by controlling the ratio and initial seeding density of tumor cells and CAFs (SI Appendix, Fig. S1C). In addition, we confirmed that tumor-stroma spheroids proliferate faster compared to regular tumor spheroids (SI Appendix, Fig. S1D), indicating that the CAFs retained their in vivo function to promote tumor cell proliferation.
Tracking Dynamic T Cell Behavior within 3D Cultures.
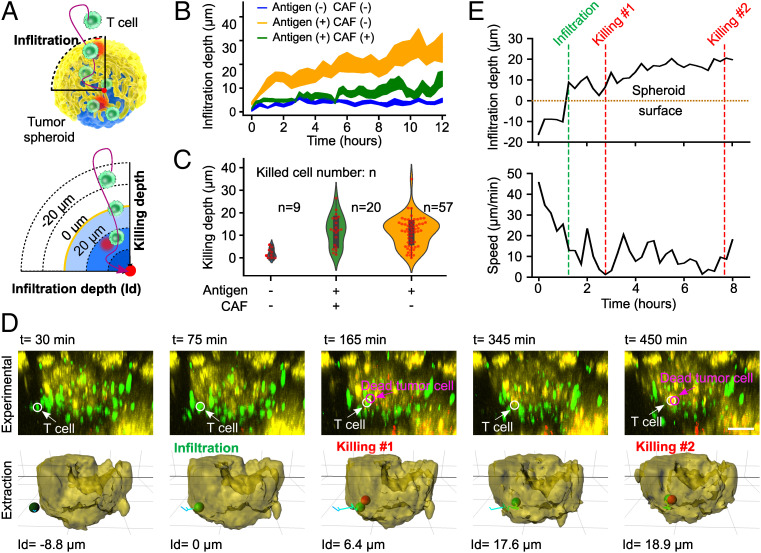
Through time-lapse imaging, our microfluidic platform tracks the dynamic interactions of T cells with tumor-stroma spheroids at a single-cell resolution while facilitating high throughput. To visualize tumor antigen-specific T cell interactions with the tumors, we subjected the tumor core consisting of ovalbumin (OVA257–264) presenting tumor cells with a shell of CAFs to OVA antigen-specific T cells (OT-I cells). Using our microfluidic platform and fluorescence-labeled cells (T cells labeled with blue CMAC cell tracker dye, tumor cells labeled with DiO green dye, CAFs labeled with DiL red dye, and dead cells detected via the SYTOX deep-red dye), we observed and quantified the dynamic migration and killing behaviors of T cells within the tumor spheroids. We defined the infiltration depth (or killing depth) of a T cell as the radial distance between the T cell and the tumor spheroid surface (Fig. 2A). We investigated the impact of antigen (OVA) presentation on tumor cores consisting of UN-KC6141 cells, as well as the stromal layer of shells/CAFs, on the dynamics of immune-tumor interactions (Fig. 2 B and C and SI Appendix, Discussion S1). We found that the CAF shells inhibited both T cell infiltration and cytotoxicity (details in SI Appendix, Fig. S2), and that the presentation of OVA antigen of tumor cores enhanced T cell cytotoxicity (details in SI Appendix, Fig. S3). By using this engineering platform and cell system, we simultaneously tracked the dynamic processes of the tumor infiltration and cytotoxicity of an individual T cell. We further observed that a T cell swarmed toward a tumor spheroid at high speed, infiltrated the spheroid with a slower speed, performed killing of a tumor cell, and then continued to move at elevated speed to locate the next target tumor cell (Fig. 2 D and E and Movie S1). Moreover, by comparing the behaviors of cytotoxic and noncytotoxic T cells (n = 30), we found that the cytotoxic T cells had lower median speeds as well as fewer straight tracks compared to noncytotoxic T cells (SI Appendix, Fig. S1E), which are consistent with other reports in in vivo models (20).
Fig. 2.
Tracking dynamics of T cell tumor infiltration and cytotoxicity. (A) Schematics of dynamic interactions between T cells and a tumor-stroma spheroid. T cell infiltration depth is defined as the radial distance from the cell to the spheroid surface, and infiltration (or killing) depth is positive (or negative) once the T cell has infiltrated into (or is outside) the spheroid. (B) Dependence of T cell infiltration on the antigen presentation of the tumor core (Antigen (+)) and the presence of a stroma/CAF shell (CAF(+)). All of the lines are plotted with a 68% confidence interval (CI). (C) Dependence of T cell killing depth and capability (n: killed cell number) on the antigen presentation of tumor core (Antigen (+)) and the stroma/CAF shell (CAF(+)). (D) Time-lapse images and their extractions of the simultaneous infiltration and killing dynamics of an individual T cell within a tumor-stroma spheroid (from Movie S1). The white (or magenta) arrow and circle indicate the T cell (or dead tumor cells), negative values of infiltration depth (Id) indicate T cells outside of spheroids, and positive values indicate T cells inside of spheroids. (E) Quantification of infiltration depth and speed of the same T cell (in D) over time. The green and red dashed lines indicate the infiltration and killing events of this T cell. (Scale bar in D: 50 µm.)
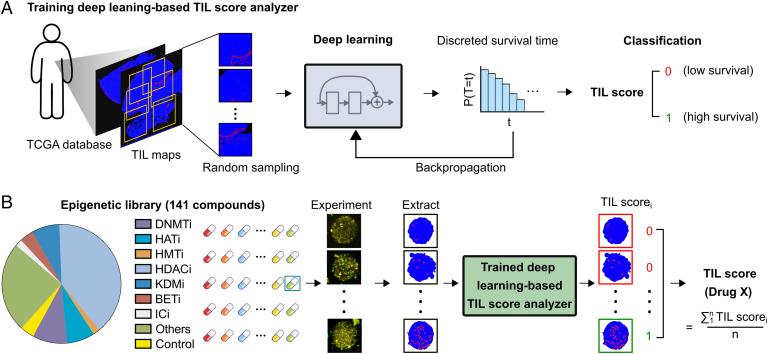
Training the Deep Learning–Based TIL Score Analyzer Using a Clinical Database.
To process the data from very large numbers of images tracking T cell behaviors on many spheroids immobilized in our microfluidic platform, we developed a deep learning–based TIL score analyzer (Fig. 3A and SI Appendix, Fig. S4 and Discussion S2) that consists of an image processer, a deep learning algorithm, and a classifier. We integrated an image processer that extracted TIL maps by digitalizing hematoxylin and eosin (H&E) images of solid tumors (411 images from TCGA database) into a bicolor map in red (lymphocytes) and blue (tumors) (21). Next, we trained the deep learning–based TIL score analyzer to score TIL infiltration based on clinical patient survival data (Fig. 3A). The analyzer used a deep convolutional neural network to extract the features of TILs from training bicolor maps and associated them with discrete patient survival time. Then, the analyzer could assign a TIL score of “0” or “1” by using a 3-y patient survival classifier with an area under the receiver operating characteristic curve of 0.8051 (SI Appendix, Fig. S4C). The TIL score correlates with the TIL number, infiltration depth, and clustering index such as the Ball-Hall or Banfield-Raftery indexes (21), and shows a better correlation with patient survival as compared with single TIL metrics (SI Appendix, Fig. S5). Using the well-trained analyzer, we evaluated the TIL scores of a particular drug X as TIL score (drug X). We defined the TIL score (drug X) by dividing TIL images into high- and low-infiltration groups by binning the cases based on predicted TIL score quartiles. Each screening result of “drug X” was given a TIL scorei of “0” or “1.” To generate the final TIL score (drug X), 10 T cell tumor infiltration images were randomly selected, and the final TIL score (drug X) was calculated by averaging these scorei (Fig. 3B). This automated strategy enables us to evaluate the efficacy of each drug in a large library in an automated, objective, and scalable manner.
Fig. 3.
Scoring based on deep learning and clinical data. (A) Schematics of training a deep learning–based TIL score analyzer using 411 patient tumor H&E images with correlated patient survival information from the TCGA database. (B) Schematics show the screening of a library of 141 small-molecule epigenetic modulators. Our automated system tracks the T cell tumor infiltration images for each drug, extracts the T cell map, assigns TIL scorei (e.g., 0, 1) for each image, and generates a final TIL score for drug X by averaging multiple TIL scorei from the drug treatment images.
Screening of Epigenetic Drugs.
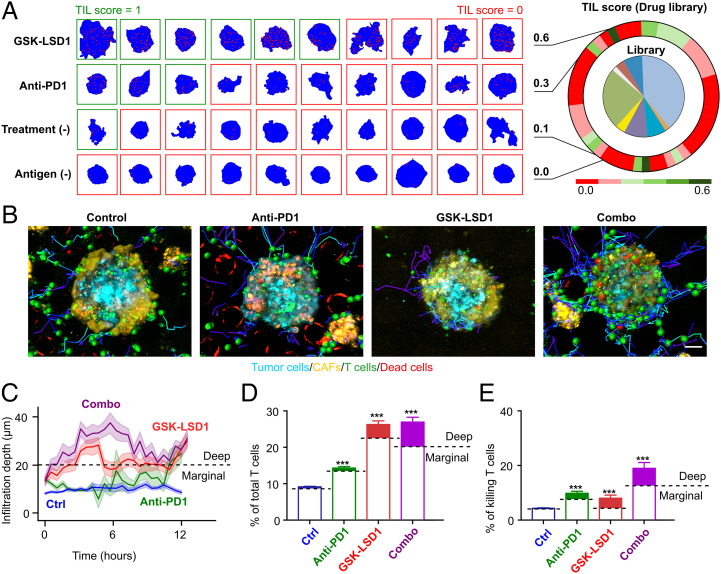
After the establishment of the method, we test-ran our intelligent microfluidic screening system in a proof-of-concept application by screening a library of drugs with epigenetic activities. It is known that epigenetic drugs can promote antigen presentation, reverse T cell exhaustion, or augment inflammation-related genes through the activation of endogenous retrovirus–mediated pathways (22). We chose a commercially available library of 141 epigenetic modulators against common targets, such as histone deacetylase, DNA methyltransferase, histone methyltransferase, and bromodomain and extra-terminal repeat. Since these small molecules may have cytotoxic activities (e.g., anti-proliferation, direct killing) and/or immunomodulating effects (e.g., altering T cell cytotoxicity, T cell infiltration), we performed the screening using our screening system in two steps: (1) drug cytotoxicity screening: Using our automated system, a total of 135,360 tumor-stroma spheroids (from melanoma B16F10 or pancreatic UN-KC6141 cell lines) were formed in our microfluidic platform in 48 h (480 spheroids/6 replicate wells per condition). After treating each epigenetic modulator at a screening concentration of 5 μM, we excluded any single agent that causes >20% cytotoxicity/antiproliferative activity, and finally chose a total number of 50 noncytotoxic agents for B16F10 spheroids and 48 noncytotoxic agents for UN-KC6141 spheroids for further immunotherapeutic screening (SI Appendix, Fig. S6). (2) Immunotherapy screening: Initially, using our engineering platform, 23,520 tumor-stroma spheroids (melanoma: B16F10-CAF; pancreatic UN-KC6141-CAF) were formed in 48 h and treated with the noncytotoxic epigenetic agents at a screening concentration of 5 μM for 24 h. After removal of the epigenetic agents, the tumor spheroids were subjected to fluorescence-labeled T cells (OT-I cells), following tracking of T cell infiltration and cytotoxicity within the tumor spheroids for 12 h. A total of 980 sets of z stack time-lapse images were collected after screening the full epigenetic drug library. Our screening system generated TIL scores for all of the drugs (Fig. 4A and SI Appendix, Fig. S7). The drug GSK-LSD1, a small-molecule inhibitor of LSD1, reached a TIL score of 0.6, which is significantly higher than that of positive control anti-PD1 treatment (0.3), untreated blank control (0.1), and antigen negative control (0.0) (Fig. 4A and SI Appendix, Fig. S4D). This finding echoes previous studies that used animal models with genetic knockout of LSD1, in which the recruitment and activation of T cells are promoted (23).
Fig. 4.
Deep learning–guided screening of an epigenetic drug library. (A) Screening results from the library. The heatmap shows TIL scores of control conditions and drug conditions. Representative images of the distribution of T cells (red dots) inside the spheroids (blue background) under different drug treatments. (B) Extraction of cell position and trajectory from time-lapse images of T cell infiltration and cytotoxicity within heterotypic tumor spheroids under four treatment conditions (untreated control, anti-PD1, GSK-LSD1, and combination treatment [combo]) (from Movie S2). (C) T cell infiltration depth over time under four treatment conditions. All of the lines are plotted with a 68% CI. (D) The average percentage of infiltrated T cells of total T cells. T cells deep within tumor spheroids (infiltration depth >20 µm) are considered to be deep infiltrating T cells and illustrated with filled colors. (E) Average killing T cell percentage of the total T cell population. Killing events located deep within tumor spheroids are labeled with filled colors. Statistical analysis: ANOVA. (Scale bar in B, 30 µm.)
In Vitro Validation of the Top Drug Candidate and Development of Combination Therapy.
To further investigate the efficacy of GSK-LSD1 alone and in combination therapy, we simultaneously tracked the infiltration and cytotoxicity of T cells. We analyzed a total of 19,032 T cell tracks from 40 tumor spheroids treated with GSK-LSD1, anti-PD1, or a combination of GSK-LSD1 and anti-PD1 (Fig. 4B and Movie S2). Analysis of a total of 1,261 infiltrated T cells showed that (1) GSK-LSD1 alone and the combination treatments resulted in significantly enhanced infiltration depths and higher numbers of infiltrating T cells compared to the untreated control and the anti-PD1 treatment (Fig. 4 C and D), and (2) The anti-PD1 treatment increased the ratio of cytotoxic T cells compared to the untreated control and the GSK-LSD1 treatment (Fig. 4E. The combination treatment resulted in the highest proportion of cytotoxic T cells. In conclusion, the combination treatment provided the greatest infiltration depth, number of infiltrating T cells, and number of cytotoxic T cells. Thus, the GSK-LSD1 treatment enhanced T cell infiltration and promoted both deep infiltration and cytotoxicity in combination with anti-PD1 treatment.
In Vivo Validation of the Top Drug Candidates.
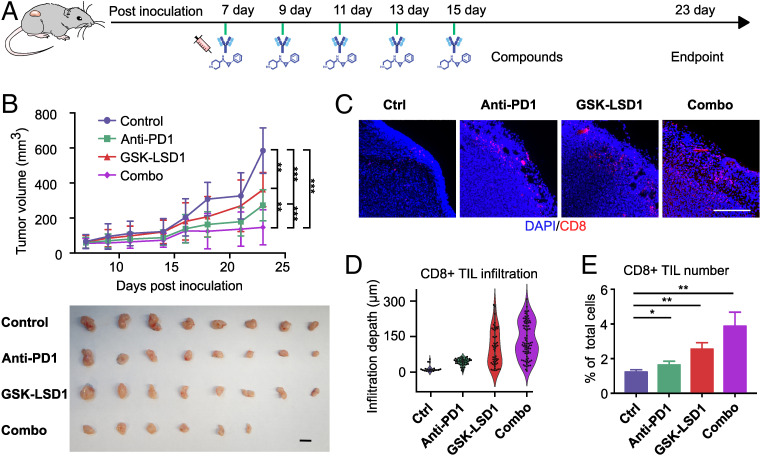
To validate our in vitro results, we tested our top candidate drug in vivo in the B16F10 syngeneic tumor model (Fig. 5A). We examined the antitumor effect of GSK-LSD1 treatment alone and in combination with anti-PD1. A moderate tumor growth inhibition was seen in the GSK-LSD1 treatment group. Moreover, when GSK-LSD1 treatment was applied in combination with anti-PD1 treatment, a greater tumor growth inhibition was observed, with two of eight mice having tumors completely regressed (Fig. 5B). Upon closer examination, a significantly higher number of CD8+ T cell infiltrated into GSK-LSD1-treated tumors compared to tumors of control or anti-PD1-treated mice, as analyzed by immunofluorescence staining (Fig. 5C) and flow cytometry (Fig. 5 D and E and SI Appendix, Fig. S8). Our results are consistent with previous reports using LSD1 knockout mice or LSD1 inhibitor treatment in melanoma and breast cancer models (23, 24). Our in vivo test further validated our in vitro screening results. Thus, we demonstrated that our automated screening system can identify valid T cell tumor infiltration–promoting agents that translate into in vivo results.
Fig. 5.
In vivo validation. (A) Tumor inoculation and treatment timeline. Anti-PD1 and GSK-LSD1 were administered intraperitoneally on days 7, 9, 11, 13, and 15. All of the animals were euthanized on day 24 to harvest tumors. (B) Growth curves and final sizes of tumors in animals of four treatment groups. (Scale bar, 10 mm.) (C) Representative images of infiltrated CD8+ T cells in the tumor slides of the four treatment groups. (Scale bar, 200 µm.) (D) T cell infiltration depth distribution in the tumor slides (n = 5). (E) CD8+ T cell percentage of all tumor cells analyzed by flow cytometry (n = 3).
Discussion
Immune infiltration and cytotoxicity in solid tissues are essential for immune surveillance, inflammation, autoimmune disease, and immunotherapy. However, current in vitro models and methods are largely lacking in high-throughput tracking and analysis of dynamic behaviors of immunocytes within 3D tissues. To address these shortcomings, we developed an automated screening system that generates scalable uniform-sized core/tumor-shell/stroma spheroid arrays, enables free perfusion of immune cells, and achieves automated tracking and analysis of immune cell–tissue interaction dynamics. As a proof-of-concept application for cancer immunotherapy drug screening, our system was used to screen a library of epigenetic modulators in a high-throughput and automated manner. We identified and in vivo validated an epigenetic drug (GSK-LSD1) that effectively promotes T cell tumor infiltration and enhances T cell infiltration and cytotoxicity in combination with anti-PD1 treatment.
Scalable, Standardized 3D Cultures That Represent Key Physiological Features.
Compared with 2D in vitro cultures, 3D cultures can recapitulate key physiological and pathological aspects of primary tumors, including tumor architecture, microenvironment, and drug response vulnerabilities (25). Recently, patient-derived organotypic cultures and tumor spheroids/organoids/clusters were established to model cancer immunity and test treatments, highlighting the uniqueness of these 3D models in preserving the genomic and/or phenotypic features of clinical tumors (26–28). However, current patient-derived 3D cultures suffer from a high heterogeneity in cell composition, cell architecture, and size and shape of the cell mass, limiting their applications for high-throughput screening and robust testing. Attempts have been made to develop methods for the standardization of organoid fabrication and minimization of interorganoid heterogeneity (29). Thus, it is critical to generate scalable standardized 3D cultures that represent key physiological features. In this work, considering tumor stroma as one of the main barriers to immune invasion, we developed a pillar-lattice-array–based microfluidic platform to fabricate uniformly sized core/tumor-shell/stroma spheroids on the scale of 105 spheroids with tunable size, structure, and composition. We believe that our microfluidic platform and core-shell spheroid fabrication strategy can also be used for scalable, standardized 3D cultures with other key physiological features, such as endothelium and immune components.
Automated Culturing and Imaging of 3D Cultures.
High-throughput screening platforms based on 2D culture with automated robotic handling for culturing cells and with imaging or readout systems for analysis have been developed. An automated system has also been published for culturing and maintaining 3D organoid cultures (29). However, challenges remain for tracking the dynamics of drug response or immune interactions of 3D cultures, partly caused by changes that occur during long-term cultures, such as uneven growth, random fusion, movement, rotation, and necrotic cores. To overcome these problems, we integrated a transparent thin polydimethylsiloxane (PDMS) layer of engineered pillar lattices into commonly used well plates for the generation and immobilization of a large number of uniform-sized spheroids. After the incorporation of automated in situ time-lapse imaging and image processing approaches, our platform allows the real-time tracking of 100 spheroids as well as dynamic infiltration and cytotoxic behaviors of T cells from over 7,000 immobilized tumor spheroids under 30 treatment conditions (SI Appendix, Discussion S3), resulting in 1,040 sets of T cell–tumor interaction images. Further efforts will be needed to integrate robotic medium/drug-handling approaches with our automated microfluidic platform for a fully automated high-throughput screening system.
Machine Learning–Based Analysis of the Vast Amount of Screening Data.
Recently, TIL infiltration patterns in patient tumor slides were shown to predict patient survival (5). However, the current evaluation of TIL infiltration mainly relies on a visual inspection and manual qualitative or quantitative analysis by experienced pathologists. Machine learning methods have been used to address the challenges in imaging processing and feature extraction of TIL maps from a vast amount of clinical data, avoiding the inconsistency and bias stemming from human interpretations (16–18). Moreover, simple metrics such as TIL number, infiltration depth, and clustering have been explored individually as predictors of patient survival or immunotherapy outcome, but have not been unified as a comprehensive single “TIL score.” Here, using such clinical TIL map and survival data from the TCGA database, we demonstrated that a machine learning method generated an integrated TIL score that better correlates with patient survival time based on the feature pattern of the TIL map, achieving an excellent median c index of 0.674 and a higher correlation (a coefficient of 0.231) than current methods based on any selected individual parameter, such as TIL clustering index or density (SI Appendix, Fig. S8). Since the distribution of CD8+ T cells highly correlated to that of TILs (21, 30), we used this algorithm and an effect threshold to score for the effect of drugs based on the T cell tumor infiltration images. We believe that the incorporation of machine learning methods is crucial for developing automated, nonbiased, high-throughput screening of TIL infiltration. In addition, TIL infiltration could vary based on disease indication, metastatic sites, genetic background, and prior treatments. Thus, we envision this strategy and algorithm could be adapted, fine-tuned, and trained using unique datasets that are relevant to the disease indication/demographics that the screened therapeutics are intended to treat.
We developed a prototype of an automated screening system and demonstrated a proof-of-concept application for screening immunotherapeutic compounds for treating solid tumors, based on the characteristics of T cell tumor infiltration identified through deep learning. This system may find extensive applications in basic research and translational medicine to treat autoimmune disorders, neuroinflammatory diseases, compromised immune responses, and the like.
Materials and Methods
Device Fabrication.
The pillar-lattice-array devices were fabricated by using the standard soft lithography and PDMS fabrication procedure as described in the SI Appendix, SI Methods and Materials.
Cell Cultures.
The mouse melanoma cell line B16F10 was purchased from the American Type Culture Collection. The mouse pancreatic tumor cell line UN-KC6141 was a kind gift from Dr. Surinder K. Batra (University of Nebraska). CAFs were isolated from orthotopic tumors. The OT-I CD8+ T cells were isolated from OT-I mice spleen. All of the procedures were approved by the Indiana University Institutional Animal Care and Use Committee. The cell isolation and culture followed standard procedures as detailed in the SI Appendix, SI Materials and Methods.
On-Chip Investigation.
The hybrid tumor-CAF spheroids were formed using the fabricated devices. The on-chip T cell infiltration of tumor spheroids was tracked and recorded by a Leica SP8 confocal microscope or Olympus OSR spinning disk confocal microscope, and raw images captured were exported as TIFF image stacks in ImageJ. The detailed on-chip investigation of T cell tumor infiltration as described in the SI Appendix, SI Materials and Methods.
TIL Score Analyzer.
The TIL scoring deep learning system was developed to score TIL infiltration based on clinical pathology data and patient survival data (https://gdc.cancer.gov/). The network architecture, training, prediction, and validation of this system were detailed in the SI Appendix, SI Materials and Methods.
Screening of Epigenetic Drugs.
An epigenetic drug library (Cayman Chemicals, Epigenetics Screening Library, cat. no. 11076) was used to test the intelligent screening system. The TIL score for each drug was determined by using the system. The details of the drug screening process were described in the SI Appendix, SI Materials and Methods.
Validation of Candidate Drug.
The antitumor activity of screened top drug candidates was tested in vivo using a syngeneic mouse tumor model. The details of in vivo validation were described in the SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We gratefully acknowledge the insightful guidance and detailed input from Prof. Subra Suresh at Nanyang Technological University. F.G. acknowledges the departmental startup funds of Indiana University Bloomington, and NIH awards (R03EB030331, DP2AI160242, R01DK133864, and U01DA056242). M.D. acknowledges support from NIH (R01HL154150). Xiongbin Lu, Xin Lu, and F.G. also acknowledge support from the Indiana Clinical and Translational Sciences Institute (funded, in part by grant no. UL1TR002529 from the NIH National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH). The authors thank the Indiana University Imaging Center (NIH1S10OD024988) and Indiana University Flow Cytometry core facility and the Indiana University Nanoscale Characterization Facility for use of their instruments.
Footnotes
Competing interest statement: Z.A., H.C., and F.G. have filed a patent based on the work presented in this paper.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120393119/-/DCSupplemental.
Data, Materials, and Software Availability
All of the study data are included in the article and/or supporting information.
References
- 1.Coussens L. M., Werb Z., Inflammation and cancer. Nature 420, 860–867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denkert C., et al. , Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 28, 105–113 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Naito Y., et al. , CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 58, 3491–3494 (1998). [PubMed] [Google Scholar]
- 4.Clemente C. G., et al. , Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer 77, 1303–1310 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Galon J., et al. , Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Robert C., et al. , Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: A randomised dose-comparison cohort of a phase 1 trial. Lancet 384, 1109–1117 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Yarchoan M., Hopkins A., Jaffee E. M., Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fridman W. H., Pagès F., Sautès-Fridman C., Galon J., The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 12, 298–306 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Ben-David U., Beroukhim R., Golub T. R., Genomic evolution of cancer models: Perils and opportunities. Nat. Rev. Cancer 19, 97–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winters I. P., Murray C. W., Winslow M. M., Towards quantitative and multiplexed in vivo functional cancer genomics. Nat. Rev. Genet. 19, 741–755 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Nath S., Devi G. R., Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 163, 94–108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labernadie A., et al. , A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat. Cell Biol. 19, 224–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman H., Gitschier H. J., Rossi A. E., A novel three-dimensional immune oncology model for high-throughput testing of tumoricidal activity. Front. Immunol. 9, 857 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dijkstra K. K., et al. , Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 174, 1586–1598.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drost J., Clevers H., Organoids in cancer research. Nat. Rev. Cancer 18, 407–418 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Saltz J., et al. , Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 23, 181–193.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X., et al. , Infiltration of CD8+ T cells into tumor cell clusters in triple-negative breast cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 3678–3687 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mobadersany P., et al. , Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. U.S.A. 115, E2970–E2979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monteran L., Erez N., The dark side of fibroblasts: Cancer-associated fibroblasts as mediators of immunosuppression in the tumor microenvironment. Front. Immunol. 10, 1835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris T. H., et al. , Generalized Lévy walks and the role of chemokines in migration of effector CD8+ T cells. Nature 486, 545–548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saltz J., et al. ; Cancer Genome Atlas Research Network, Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 23, 181–193.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Topper M. J., et al. , Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 171, 1284–1300.e21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng W., et al. , LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin Y., et al. , Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene 38, 390–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han K., et al. , CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature 580, 136–141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Ootani A., Kuo C., An air-liquid interface culture system for 3D organoid culture of diverse primary gastrointestinal tissues. Methods Mol. Biol. 1422, 33–40 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Ao Z., et al. , Rapid profiling of tumor-immune interaction using acoustically assembled patient-derived cell clusters. Adv. Sci. (Weinh.) 9, e2201478 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ao Z., et al. , Evaluation of cancer immunotherapy using mini-tumor chips. Theranostics 12, 3628–3636 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandenberg N., et al. , High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat. Biomed. Eng. 4, 863–874 (2020). [DOI] [PubMed] [Google Scholar]
- 30.König L., et al. , Dissimilar patterns of tumor-infiltrating immune cells at the invasive tumor front and tumor center are associated with response to neoadjuvant chemotherapy in primary breast cancer. BMC Cancer 19, 120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the study data are included in the article and/or supporting information.