Summary
Plants have evolved complex signalling networks to regulate growth and defence responses under an ever‐changing environment. However, the molecular mechanisms underlying the growth‐defence tradeoff are largely unclear. We previously reported that rice CALCIUM‐DEPENDENT PROTEIN KINASE 18 (OsCPK18) and MITOGEN‐ACTIVATED PROTEIN KINASE 5 (OsMPK5) mutually phosphorylate each other and that OsCPK18 phosphorylates and positively regulates OsMPK5 to suppress rice immunity. In this study, we found that OsCPK18 and its paralog OsCPK4 positively regulate plant height and yield‐related traits. Further analysis reveals that OsCPK18 and OsMPK5 synergistically regulate defence‐related genes but differentially regulate development‐related genes. In vitro and in vivo kinase assays demonstrated that OsMPK5 phosphorylates C‐terminal threonine (T505) and serine (S512) residues of OsCPK18 and OsCPK4, respectively. The kinase activity of OsCPK18T505D, in which T505 was replaced by aspartic acid to mimic T505 phosphorylation, displayed less calcium sensitivity than that of wild‐type OsCPK18. Interestingly, editing the MAPK phosphorylation motif in OsCPK18 and its paralog OsCPK4, which deprives OsMPK5‐mediated phosphorylation but retains calcium‐dependent activation of kinase activity, simultaneously increases rice yields and immunity. This editing event also changed the last seven amino acid residues of OsCPK18 and attenuated its binding with OsMPK5. This study presents a new regulatory circuit that fine tunes the growth‐defence tradeoff by modulating OsCPK18/4 activity and suggests that CRISPR/Cas9‐mediated engineering phosphorylation pathways could simultaneously improve crop yield and immunity.
Keywords: rice, CDPK, MAPK, genome editing, immunity, growth
Introduction
Plants have evolved sophisticated immune systems to defend against pathogens, although the immune response is generally activated at the expense of growth and development (Ning et al., 2017; Zust and Agrawal, 2017). Phytohormones and their crosstalk are considered central players in balancing defence responses and growth in plants (De Bruyne et al., 2014; Huot et al., 2014; Yang et al., 2013). For example, Yang et al. (2012) revealed that jasmonic acid prioritizes defence over growth by interfering with gibberellin (GA) signalling. Engineer the expression of NONEXPRESSER OF PR GENES1 (NPR1), a master immune regulator in the salicylic acid (SA) pathway, enhanced disease resistance without fitness cost (Xu et al., 2017). Recent reports suggested that engineering the expression of key regulators of plant development, such as IDEAL PLANT ARCHITECTURE 1 (Liu et al., 2019; Wang et al., 2018c) and miR168 (Wang et al., 2021), could enhance immunity without yield penalties in rice. However, the molecular mechanism through which plants maintain the balance of growth and defence under an ever‐changing environment remains largely unclear.
The mitogen‐activated protein kinase (MAPK or MPK) cascade and calcium‐dependent protein kinase (CDPK or CPK) are pivotal signalling hubs of plant immune responses, abiotic stress tolerance, phytohormone biogenesis and signalling, and development (Boudsocq and Sheen, 2013; Meng and Zhang, 2013; Zhang et al., 2018; Zhou and Zhang, 2020). Rapid activation of the classical MAPK cascade is one of the earliest signalling events after plants sense pathogen invasion. The orthologs of Arabidopsis AtMPK3/4/6, which belong to the subgroup A MAPKs, are predominant regulators of immune signal transduction and have been extensively studied in Arabidopsis (Meng and Zhang, 2013). In rice, OsMPK5 (also referred to as OsMPK3), which is the ortholog of AtMPK3, negatively regulates immunity but positively regulates abiotic stress tolerance (Xiong and Yang, 2003). OsMPK5 is involved in ABA‐induced susceptibility or resistance through suppression of ethylene production (Bailey et al., 2009; De Vleesschauwer et al., 2010; Hu et al., 2011). Interestingly, OsMPK5 also regulates rice tolerance to high salinity and flooding via ethylene signalling (Li et al., 2014; Singh and Sinha, 2016). These reports suggest that OsMPK5 plays a pivotal role in the crosstalk between rice immunity and abiotic stress tolerance.
Elevation of Ca2+ influx and subsequent CDPK activation is another early signalling process in the plant immune response (Couto and Zipfel, 2016; Tian et al., 2019). CDPK consists of four domains: an N‐terminal variable domain (V), a protein kinase domain (K), an autoinhibitory junction (J) peptide, and a calmodulin‐like domain (CaM) for calcium (Ca2+) binding (Harper et al., 2004). The kinase activity of CDPK is activated by the binding of Ca2+; therefore, CDPK functions as a sensor and transducer of Ca2+ signals (Harper et al., 2004). Plant CDPKs are phylogenetically classified into four subgroups (subgroups I–IV; Hamel et al., 2014). Recent studies revealed that the subgroup IV CDPKs, including Arabidopsis AtCPK16/18/28 and rice OsCPK4/18, are important regulators of plant immunity. AtCPK28 negatively regulates immune signalling by promoting BOTRITIS‐INDUCED KINASE 1 (BIK1) degradation (Monaghan et al., 2014; Wang et al., 2018b). AtCPK28 also regulates abiotic stress tolerance and development (Gao et al., 2014; Jin et al., 2017; Matschi et al., 2013, 2015). In rice, OsCPK18 and OsCPK4 are characterized as negative regulators of defence (Wang et al., 2018a; Xie et al., 2014). OsCPK4 phosphorylates and promotes the degradation of RECEPTOR‐LIKE CYTOPLASMIC KINASE 176 (OsRLCK176), which is a rice ortholog of BIK1 (Wang et al., 2018a). OsCPK4 was also shown to act as a positive regulator of salt and drought stress responses (Campo et al., 2014).
We previously found that OsMPK5 and OsCPK18 mutually phosphorylated each other and demonstrated that OsCPK18 phosphorylated and positively regulated OsMPK5 to suppress defence (Xie et al., 2014). Here, we show that OsCPK18 and its paralog OsCPK4 suppress rice disease resistance but promote yield‐related traits. OsMPK5, which is activated by pathogens and abiotic stresses (Xiong and Yang, 2003), phosphorylates OsCPK18 and alters its calcium sensitivity. Interestingly, targeted mutagenesis of the MAPK phosphorylation motif in OsCPK18 and OsCPK4 enhanced disease resistance and yield in rice. Our results reveal that the mutual phosphorylation between OsMPK5 and OsCPK18 constitutes a regulatory circuit to orchestrate growth and defence signalling pathways. Precision manipulation of the OsCPK18–OsMPK5 phosphorylation pathway could simultaneously improve both yield and immunity.
Results
OsCPK18 and OsCPK4 regulate yield‐related traits and defence
We analysed two OsCPK18 knockdown lines (OsCPK18‐RI #10 and #13) that were previously generated using dsRNA‐mediated interference (Xie et al., 2014). In addition to increased resistance against Magnaporthe oryzae, which causes rice blast disease, OsCPK18‐RI plants displayed pleiotropic phenotypes (Figure 1a–e). The shoot length, effective tiller number, and 1000‐grain weight of OsCPK18‐RI plants were significantly reduced (Figure 1a–e). As a result, the yield of the two OsCPK18‐RI lines was reduced by 50.6% and 67.9% in the field compared to wild‐type (WT) plants (Figure 1e, Student's t‐test P = 1.5 × 10−24 and 1.1 × 10−29). We then examined whether OsCPK18 overexpression could increase rice yield. To this end, a truncated OsCPK18 protein, which is activated constitutively (referred to as OsCPK18AC hereafter) by removing the J and CaM domains, was overexpressed in rice with the cauliflower mosaic virus 35S promoter (Figure S1a–c). OsCPK18‐AC plants were taller than WT plants (Figure 1a–e). The yield per plant of two independent OsCPK18‐AC lines increased by 16.5% and 18.1% (Figure 1e, Student's t‐test P = 1.4 × 10−7 and 1.8 × 10−9). However, M. oryzae inoculation experiments showed that OsCPK18‐AC plants had more lesions and fungal growth than WT plants (Figure 1f,g). The expression of defence genes, including PATHOGENESIS‐RELATED 5 (PR5), CHINTINASE11, and HARPIN‐INDUCED 1 (HIN1), was also reduced by approximately 50% in OsCPK18‐AC plants (Figure S1d), suggesting that OsCPK18‐AC plants are more susceptible to disease. Opposite changes in the yield and defence of OsCPK18‐RI and OsCPK18‐AC plants indicate that OsCPK18 regulates the growth‐defence tradeoff in rice.
Figure 1.
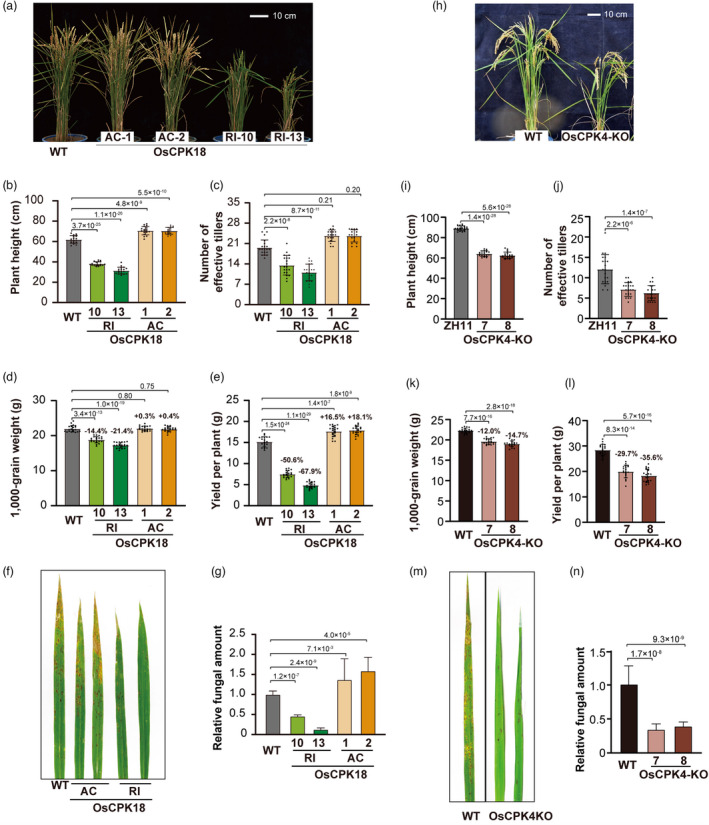
Rice growth and defence were inversely regulated by OsCPK18 and OsCPK4. (a–g) Comparisons of yield‐related traits and disease resistance of the WT, OsCPK18 knockdown (OsCPK18‐RI), and OsCPK18 constitutively activated (OsCPK18‐AC, see also Figure S1) plants. (h–n) Comparisons of yield‐related traits and disease resistance of WT and OsCPK4 knockout plants (OsCPK4‐KO, see also Figure S2). Two independent lines of OsCPK18‐RI (10 and 13), OsCPK18‐AC lines (1 and 2), and OsCPK4‐KO (7 and 8) were analysed. a and h, Morphology of OsCPK18‐RI, OsCPK18‐AC, OsCPK4‐KO, and WT plants at the grain filling stage. (b–e and i–l) Comparisons of yield‐related traits between OsCPK18‐RI, OsCPK18‐AC, OsCPK4‐KO, and WT plants. For each genotype, 20 individual plants were analysed. The traits include plant height (b, i), effective tiller number (c, j), 1000‐grain weight (d, k), and yield per plant (e, l). (f, m) Photos of rice leaves inoculated with M. oryzae. (g, n) The relative fungal amount in rice leaves inoculated with M. oryzae. The data are presented as the mean ± s.d. (n = 3 technical replicates). The P values of Student's t‐tests are shown in the plots (b–l, g, n).
We sought to determine whether OsCPK4 functions similarly to OsCPK18 since their protein sequence alignment showed 84.7% identity. The T‐DNA insertion mutant oscpk4 is semidwarf and shows increased disease resistance (Wang et al., 2018a), which was also observed in OsCPK18‐RI plants. However, another report found that overexpressing OsCPK4 also enhanced rice defence (Bundo and Coca, 2016). To ascertain the function of OsCPK4, we used CRISPR/Cas9 gene editing to knock out OsCPK4 in rice (Figure S2a,b). Similar to OsCPK18‐RI plants, OsCPK4 knockout (OsCPK4‐KO) plants displayed a reduction in yield‐related traits but enhanced disease resistance (Figures 1h–n, S2c,d). In comparison with the WT, the yields of the two OsCPK4‐KO lines were reduced by 29.7% and 35.6% (Figure 1l, Student's t‐test P = 8.3 × 10−14 and 5.7 × 10−16) but their resistance against rice blast was significantly increased (Figure 1m,n). The expression of PR5 and PR10 was also increased approximately 20‐fold in homozygous knockout lines of OsCPK4‐KO (Figure S2e,f). These data suggest that OsCPK4 and OsCPK18 have overlapped functions in regulating growth and defence.
OsCPK18 and OsMPK5 negatively regulate defence genes but differentially regulate growth‐related genes
Previous studies indicate that OsCPK18 and OsMPK5 mutually phosphorylate each other and that OsCPK18‐mediated phosphorylation of OsMPK5 suppresses the basal defence of rice (Xie et al., 2014). We therefore sought to determine whether OsMPK5 regulates yield‐related traits as OsCPK18. In comparison with WT plants, the yield‐related traits including plant height and effective tiller number were not significantly changed in OsMPK5 silencing and overexpressing plants (OsMPK5‐RI and ‐OX, Figure 2a–c).
Figure 2.
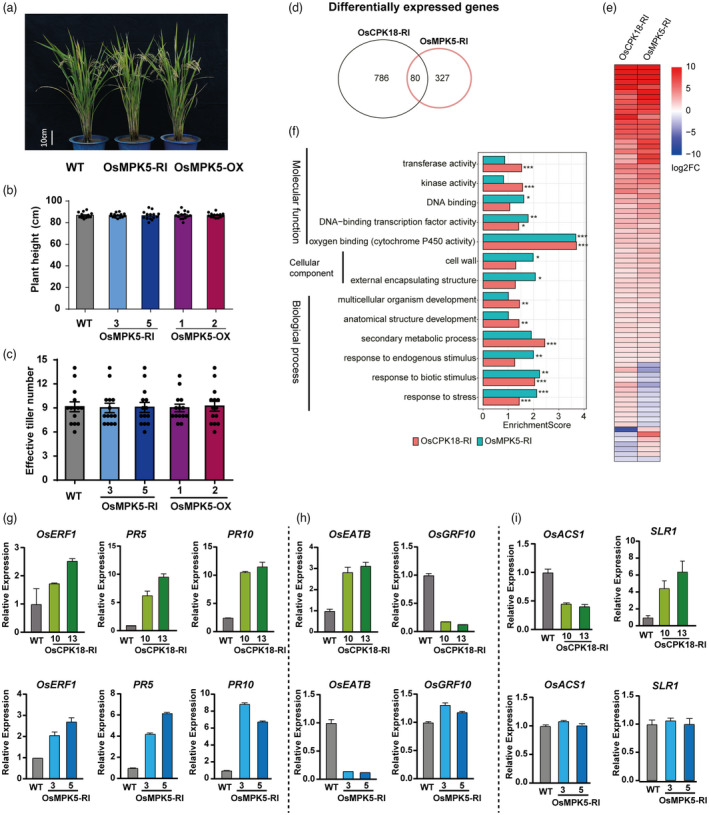
OsCPK18 and OsMPK5 synergistically regulate defence‐related genes but differentially regulated growth‐related genes. (a–c) Morphology, plant height, and effective tiller number of OsMPK5‐RI, OsMPK5‐OX, and WT plants. (d) Venn diagram shows the number of DEGs (fold‐change >2, adjusted P value <0.05) in OsCPK18‐RI and OsMPK5‐RI seedlings. (e) Heatmap shows DEGs shared by OsCPK18‐RI and OsMPK5‐RI. Log2FC, log2 transformed fold‐change. (f) Enriched GO terms of OsCPK18‐RI and OsMPK5‐RI DEGs. *, **, *** indicate adjusted P value <0.05, 0.01, 0.001, respectively. (g–i) Relative expression of defence and growth‐related genes in OsCPK18‐RI, OsMPK5‐RI, and WT plants. Data are presented as the mean ± s.d. (n = 3 technical replicates). [Correction added on 28 October 2022, after first online publication: The gene symbol is changed from OsMPK3 to OsMPK5 in this version].
We then compared the transcriptome of OsMPK5‐RI and OsCPK18‐RI seedlings using RNA‐seq. In comparison with WT, 866 and 407 genes were changed by more than twofold in OsCPK18‐RI and OsMPK5‐RI plants, respectively (Figure 2d; Data S1 and S2, FDR adjusted P < 0.05). A total of 80 differentially expressed genes (DEGs) were regulated by both OsCPK18 and OsMPK5. Among these shared DEGs, 61 DEGs displayed similar foldchange, while 19 DEGs have opposite foldchange, between OsCPK18‐RI and OsMPK5‐RI datasets (Figure 2d,e; Data S3). Gene Ontology (GO) enrichment analysis was used to compare the function of all DEGs in OsCPK18‐RI and OsMPK5‐RI plants. Among GO terms under Biological Process, genes related to the development and secondary metabolic processes were significantly enriched in OsCPK18‐RI DEGs (FDR adjusted P < 0.01) but was not in OsMPK5‐RI DEGs (Figure 2f). By contrast, genes associated with GO term of response to endogenous stimulus were only enriched in OsMPK5‐RI DEGs (FDR adjusted P < 0.001). Furthermore, stress‐responsive genes, including genes response to biotic stimulus, were significantly enriched in both OsCPK18‐RI and OsMPK5‐RI DEGs. In molecular function catalogue, two GO terms (DNA binding transcription factor activity and cytochrome P450 activity) were enriched in both datasets. The expression of defence and growth‐related marker genes were further examined at tillering stage using quantitative reverse transcription PCR (RT‐qPCR). Consistent with these phenotypic changes and RNA‐seq data, the defence‐related genes (PR5, PR10) and ethylene‐responsive genes (Ethylene Response Factor 1, OsERF1), were increased in both OsCPK18‐RI and OsMPK5‐RI plants (Figure 2g). These results confirm that OsMPK5 and OsCPK18 synergistically regulate rice basal defence. However, two growth‐related transcription factors, OsGRF10 (GROWTH‐REGULATING FACTOR 10) (Kuijt et al., 2014) and OsEATB (ERF protein associated with tillering and panicle branching; Qi et al., 2011), were oppositely regulated by OsCPK18 and OsMPK5 (Figure 2h). Furthermore, we also found that 1‐aminocyclopropane‐1‐carboxylate synthase (OsACS1) and SLENDER RICE 1 (SLR1; Ikeda et al., 2001), which are involved in ethylene biogenesis and GA response, respectively, were only changed in OsCPK18‐RI but not in OsMPK5‐RI plants (Figure 2i). These data imply that OsCPK18 and OsMPK5 synergistically regulate a specific set of defence responsive genes but differentially regulate rice growth‐related genes.
OsMPK5 phosphorylates a C‐terminal threonine of OsCPK18
Given the mutual phosphorylation between these two protein kinases, we sought to determine whether OsMPK5‐mediated retrophosphorylation of OsCPK18 was involved in growth‐defence tradeoff. To biochemically characterize OsMPK5‐mediated regulation of OsCPK18, we first mapped the OsMPK5 phosphorylation site in OsCPK18. According to the consensus MAPK phosphorylation motif, which is a threonine (T) or serine (S) before a proline (P; Davis, 1993), only T15, T282, and T505 in OsCPK18 are candidate sites for OsMPK5 phosphorylation (Figure 3a). These three sites were replaced with aspartic acid (D) in OsCPK18DA, which carries the D178A mutation to eliminate autophosphorylation. We performed in vitro kinase assays using polyhistidine‐tagged (His‐) OsMPK5 as a kinase and four mutated His‐OsCPK18 proteins (OsCPK18DA, OsCPK18DA‐T15D, OsCPK18DA‐T282D, and OsCPK18DA‐T505D) as substrates. The kinase reaction products were analysed with Phos‐tag sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), which can detect protein phosphorylation according to electrophoretic mobility shifts (Kinoshita‐Kikuta et al., 2007; Xie et al., 2014). As shown in Figure 3b, single phosphorylated protein bands were detected for all OsCPK18DA‐derived proteins except OsCPK18DA‐T505D in the Phos‐tag gel, suggesting that OsCPK18 T505 is the sole phosphorylation site of OsMPK5. The phosphorylation site was further verified using radioactive in vitro kinase assays using OsCPK18DA‐T505A as substrates (Figure S3). Phosphorylation at OsCPK18 T505 was also validated in vivo. Because the D178A mutation drastically impairs OsCPK18 stability in rice (Figure S4a), a sensitive nanoluciferase (NanoLuc) tag and in‐gel detection method were used to detect OsCPK18DA‐derived proteins in vivo (Li et al., 2021). To activate OsMPK5 in protoplasts, GREEN FLUORESCENCE PROTEIN (GFP)‐tagged OsMKK4DD (carrying T238D and S244D substitutions) with constitutive catalytic activity was coexpressed with FLAG‐tagged OsMPK5 and OsCPK18DA. Except for OsCPK18DA‐T505D, a single phosphorylated protein band of OsCPK18DA‐derived proteins was detected in the presence of an active MAPK cascade consisting of OsMKK4DD‐OsMPK5 (Figure 3c). Together, the in vitro and in vivo experiments indicated that the active OsMKK4‐OsMPK5 cascade phosphorylates OsCPK18 at T505.
Figure 3.
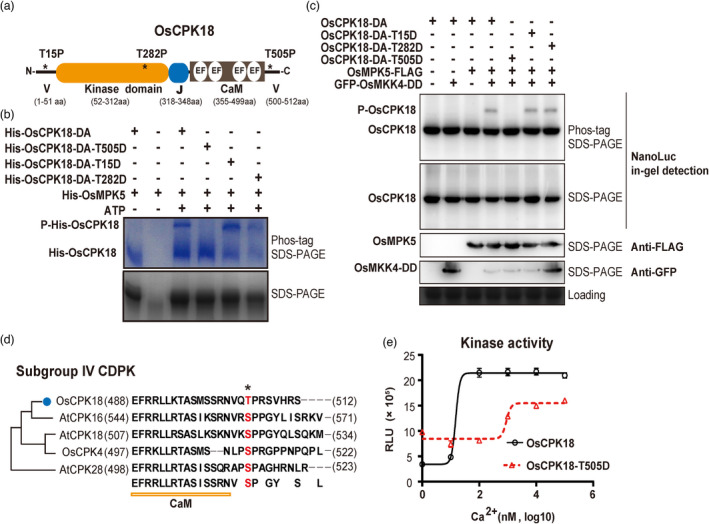
OsMPK5 phosphorylates OsCPK18 T505 and modulates its kinase activity. (a) Schematic illustration of OsCPK18 protein domains. Asterisks (*) denote three MAPK phosphorylation motifs as S/T–P. V, N‐, and C‐terminal variable domains; EF, EF‐hand motif; J, junction peptide; CaM, calmodulin‐like domain for Ca2+ binding. The number at the bottom indicates the position of different protein domains. aa, amino acids. (b) In vitro kinase assays show that His‐OsMPK5 phosphorylated His‐OsCPK18DA, His‐OsCPK18DA‐T15D, and His‐OsCPK18DA‐T282D but not His‐OsCPK18DA‐T505D. The kinase reaction products were analysed using Phos‐tag SDS‐PAGE and regular SDS‐PAGE. The proteins were visualized by Coomassie Brilliant Blue staining. Two phosphorylation reactions without ATP were used as controls to show the mobilities of unphosphorylated proteins. His, polyhistidine tag; P‐His‐OsCPK18, a phosphorylated form of OsCPK18DA recombinant proteins. (c) Activated OsMPK5 phosphorylated OsCPK18 in rice protoplasts. GFP‐tagged OsMKK4DD, FLAG‐tagged OsMPK5, and NanoLuc‐tagged OsCPK18DA with different mutations were coexpressed in rice protoplasts. The rice total proteins were separated by Phos‐tag SDS‐PAGE and regular SDS‐PAGE. The NanoLuc‐tagged proteins were analysed by in‐gel detection (see the Methods), while FLAG‐ and GFP‐tagged proteins were analysed by standard Western blotting. Coomassie Brilliant Blue‐stained gel shows the loading of total protein. P‐OsCPK18, a phosphorylated form of OsCPK18DA‐derived proteins whose mobility was shifted in Phos‐tag SDS–PAGE. (d) Sequence alignment of OsCPK18 and other subgroup IV CDPKs of Arabidopsis and rice. The OsMPK5 phosphorylation site is highlighted in red. The numbers in brackets indicate the positions of the first and last amino acid residues of each protein. (e) Ca2+ sensitivity of OsCPK18 and OsCPK18T505D. The kinase activities were compared using the ADP‐Glo method, which measures the relative amount of ADP produced from phosphorylation reactions. The Histone III‐S protein was used as the substrate in this experiment. RLU, relative luminescent unit. The recombinant protein purification and in vitro kinase reaction were repeated three times with similar results. The data are presented as the mean ± s.d. (n = 3 technical replicates).
OsCPK18T505D displayed less Ca2+ dependency than OsCPK18
Next, we examined the effect of OsCPK18 T505 phosphorylation. T505 is located in the C‐terminal variable region of CDPK and conserved only in subgroup IV CDPKs (Figure 3d). We used the T505D mutation to mimic OsCPK18 T505 phosphorylation. The WT OsCPK18 and OsCPK18T505D proteins displayed similar abundance and subcellular locations after transient expression in rice protoplasts and tobacco leaves (Figure S4b), suggesting that T505 phosphorylation likely did not affect protein stability and subcellular localization. We then hypothesized that T505 phosphorylation may regulate calcium sensitivity since it is located adjacent to the CaM domain (Figure 3d). To explore this possibility, a luminescent adenosine diphosphate (ADP) detection assay (Zegzouti et al., 2009), quantifies the amount of ADP produced in kinase reactions, was used to compare the activity of recombinant OsCPK18 and OsCPK18T505D. To avoid the interference of residual Ca2+ from recombinant protein purification, polyhistidine‐tagged OsCPK18 and OsCPK18T505D proteins were dialyzed against buffer containing ethylenediaminetetraacetic acid (EDTA) to remove free Ca2+. The kinase activity of CDPKs was measured using the generic substrate histone III protein with 0–100 μm CaCl2 in kinase reactions. As shown in Figure 3e, the Ca2+‐dependent activation kinetics of OsCPK18T505D differ drastically from those of OsCPK18. The kinase activity of OsCPK18T505D was higher than that of OsCPK18 when the Ca2+ concentration was less than 10 nm. OsCPK18 displayed typical Ca2+‐dependent activation and reached maximal activity (approximately fivefold activation) when the Ca2+ concentration was 100 nm or higher. In contrast, OsCPK18T505D displayed reduced Ca2+ sensitivity and reached maximal activity (approximately 1.25‐fold activation) when the Ca2+ concentration was higher than 1000 nm. As a result, the maximal kinase activity of OsCPK18 was higher than that of OsCPK18T505D. We concluded that OsMPK5 regulates the Ca2+ dependency of OsCPK18 by phosphorylating T505.
Editing OsMPK5 phosphorylation motif of subgroup IV CDPKs improves rice yield and immunity
To determine whether disconnecting the reciprocal regulation between OsMPK5‐OsCPK18 could disrupt the growth‐defence tradeoff, we used CRISPR/Cas9 gene editing to modify the MAPK phosphorylation motif of OsCPK18. In addition to OsCPK18, OsMPK5 phosphorylated OsCPK4 at S512 (Figures 3d and S5), which functions similarly to OsCPK18; therefore, we also edited the MAPK phosphorylation motif of OsCPK4. Due to the constraints of protospacer‐adjacent‐motif (PAM) requirements and the editing window of CRISPR/Cas9 (Zhu et al., 2020), we could not find appropriate guide RNAs (gRNAs) that can direct Cas9 nuclease or base editor to edit codons encoding the MAPK phosphorylation residues of OsCPK18 or OsCPK4. Instead, gRNAs were available to precisely introduce frameshift mutations immediately after T505 and S512 of OsCPK18 and OsCPK4 (Figure 4a), respectively, which would prevent OsMPK5 phosphorylation by disrupting the S/T–P motif. These frameshift mutations would only change the last 7 and 10 amino acid residues of OsCPK18 and OsCPK4, respectively, and these alterations were expected to not impair their kinase properties. After transforming the gRNA/Cas9 constructs to rice cultivar Zhonghua 11 (ZH11), we obtained a total of 20 and 14 phosphorylation motif edited lines for OsCPK18 and OsCPK4 (referred to as OsCPK18‐GE and OsCPK4‐GE), respectively. Homozygous edited lines with two different frameshift mutations were used for phenotypic analyses: OsCPK18‐GE3 (−2 bp), OsCPK18‐GE8 (−1 bp), OsCPK4‐GE1 (−2 bp), and OsCPK4‐GE3 (−4 bp). As shown in Figure 4a, only the C‐terminal variable peptide after T505 and S512 of these alleles was changed. Except for editing at the desired targeting site, we did not observe Cas9 off‐target editing in these plants (Table S1). Furthermore, the transgene‐free lines were selected in the T2 generation, and their progenies were used to assess yield‐related traits and disease resistance.
Figure 4.
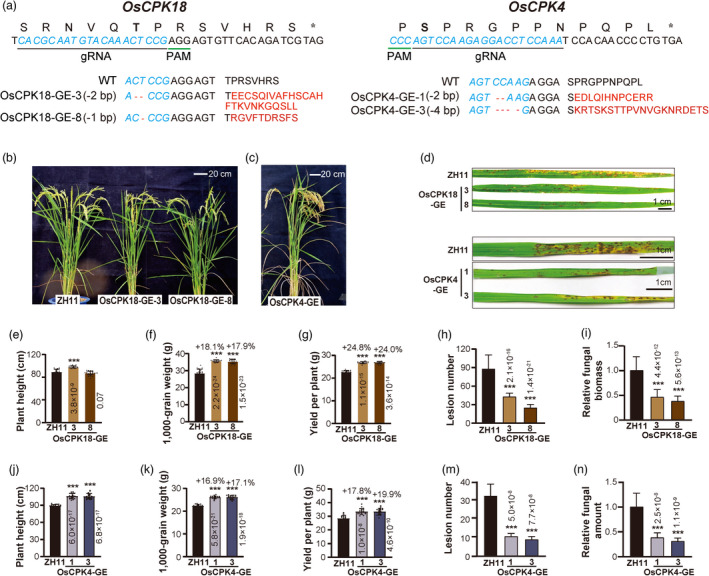
Precise editing of the OsCPK18 T505‐P506 or OsCPK4 S512‐P513 phosphorylation motif increases yield and disease resistance. (a) Schematics showing CRISPR/Cas9‐mediated editing of the MAPK phosphorylation motifs in OsCPK18 and OsCPK4. The 20 bp targeting sequences (blue letter) and corresponding protein sequences are shown on the top. Alignments of the WT sequence and gene‐edited alleles of OsCPK18 and OsCPK4 are shown at the bottom. Red letters indicate the amino acid sequences generated by frameshift mutations in gene‐edited alleles. *, stop codon; −, deletions. (b, c) Morphology of the OsCPK18‐GE, OsCPK4‐GE, and WT (cv ZH11) plants at the grain filling stage. (d) Photos representing the rice blast symptoms of OsCPK18‐GE, OsCPK4‐GE, and WT. Photos were taken 7 days after M. oryzae inoculation. (e–n) Yield‐related traits and disease resistance of OsCPK18‐GE, OsCPK4‐GE, and WT plants. The plant height (e, j), 1000‐grain weight (f, k), and yield per plant (g, l) were measured. The lesion number (h, m) and relative fungal amount (i, n) were measured at 7 days postinoculation of M. oryzae spores. The data are presented as the mean ± s.d. n = 20 for e–h and j–m; n = 3 technical replicates for i and n. The P values (Student's t‐test) are shown in the plots. ***, Statistically significant difference with P < 0.001.
It is remarkable that all phosphorylation motif‐edited plants displayed higher yields and enhanced disease resistance compared with the WT plants (Figures 4b–n and S6). The plant heights were significantly increased in both OsCPK4‐GE and OsCPK18‐GE3. However, the height of OsCPK18‐GE8 was not changed, implying that the function of two OsCPK18 alleles slightly diversified between OsCPK18‐GE3 and OsCPK18‐GE8 plants. As shown in Figure 4, the 1000‐grain weight and yield of OsCPK18‐GE and OsCPK4‐GE plants were increased by 16.9%–24.8% (Figure 4f,g,k,l, Student's t‐test P < 0.001), which resembled the phenotype of the OsCPK18‐AC plants. In these two gene‐edited lines, the panicle size was also slightly increased (Figure S6). Importantly, rice blast resistance of these phosphorylation motif‐edited plants was also significantly improved (Figure 4d,h,i,m,n), which resembles the phenotypes of the OsCPK18‐RI and OsCPK4‐KO mutants (Figures 1 and S2). These results suggest that editing MAPK phosphorylation motifs generated valuable gain‐of‐function alleles of OsCPK18/4 that simultaneously improved rice yield and immunity.
Dual effects of OsCPK18 phosphorylation‐motif editing
To gain mechanistic insights into the OsCPK18‐GE alleles, we analysed the expression and kinase activities of OsCPK18‐GE3 and OsCPK18‐GE8. The mRNA splicing, protein stability and subcellular location of OsCPK18‐GE3 and OsCPK18‐GE8 were the same as those of the WT OsCPK18 (Figure S7). Furthermore, the recombinant OsCPK18‐GE3/8 proteins displayed similar Ca2+ sensitivity as the WT OsCPK18, although the maximal kinase activities of the two OsCPK18‐GE proteins were slightly reduced (Figure 5a). Importantly, neither OsCPK18‐GE3 nor OsCPK18‐GE8 was phosphorylated by the active OsMKK4‐OsMPK5 cascade in rice protoplasts (Figure 5b), indicating that OsMPK5‐mediated regulation of OsCPK18 was precisely abolished in these phosphorylation motif‐edited plants. We also tested the in vivo interactions between OsCPK18‐GE and OsMPK5 proteins using split luciferase complemental assays. Compared to WT OsCPK18, the complemental luciferase activity between OsCPK18‐GE3/GE8 and OsMPK5 was reduced by more than 70% (Figure 5c–f), implying that these two editing events drastically attenuated OsCPK18‐OsMPK5 interaction. The reduced interaction between OsCPK18‐GE and OsMPK5 was further confirmed by coimmunoprecipitation assays (Figure S8). However, replacing T505 with alanine OsCPK18 did not impair its interaction with OsMPK5 (Figure 5e). Together, OsCPK18‐GE alleles escape OsMPK5‐mediated suppression but retain their ability to sense and transduce Ca2+ signals. On the other hand, the weak interaction between OsCPK18‐GE and OsMPK5 also reduced the CDPK‐mediated regulation of OsMPK5. Such dual effects of phosphorylation motif editing thereby result in improved yield and immunity (Figure 5g).
Figure 5.
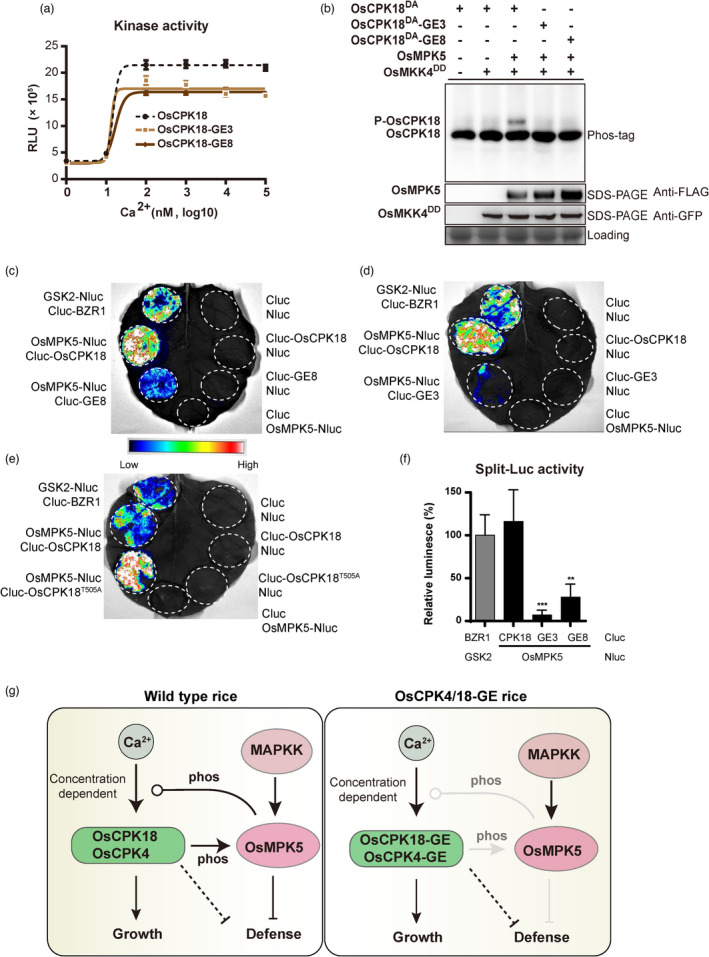
Dual effects of OsCPK18 phosphorylation motif editing. (a) Ca2+ sensitivity of OsCPK18 and OsCPK18‐GE alleles. The kinase activity was analysed with the ADP‐Glo assay using Histone III‐S protein substrate (see the Methods). RLU, relative luminescent unit. The data are presented as the mean ± s.d. (n = 3 technical replicates). (b) OsCPK18‐GE alleles were not phosphorylated by OsMKK4DD‐OsMPK5 in rice protoplasts. NanoLuc‐tagged OsCPK18DA and OsCPK18DA‐GE were analysed by in‐gel detection after Phos‐tag SDS–PAGE. P‐OsCPK18, phosphorylated forms of OsCPK18DA. (c–f) Comparison of protein interactions between OsMPK5 and OsCPK18‐derived proteins using split luciferase (LUC) complementation assay. OsMPK5 is fused with N‐terminal LUC fragments (Nluc) and OsCPK18‐GE3/8 and OsCPK18T505A were fused with C‐terminal LUC fragments (Cluc). These Nluc and Cluc fused proteins were co‐expressed in Nicotiana benthamiana leaves by agroinfiltration. Rice GSK2 and BZR1 were used as positive controls. Representative photos from three independent experiments are shown in (c–e). The relative luminescence was calculated from three independent experiments and presented as mean ± s.d. in (f). ***P < 0.001; **P < 0.01 (paired Student's t‐test). (g) Schematic illustration of the OsCPK18/4‐OsMPK5 regulatory circuit in rice. OsCPK18 and OsCPK4, whose activities are dependent on cellular Ca2+ concentration, positively regulate growth but negatively regulate immunity. OsCPK18/4 suppresses rice immunity through phosphorylating OsMPK5 and other MAPK‐independent pathways. OsMPK5 retrophosphorylates OsCPK18 and attenuates its Ca2+ sensitivity. Cellular Ca2+ level and MAPK phosphorylation together fine tunes OsCPK18/4 activity to balance growth and defence. In OsCPK4/18‐GE plants, OsMPK5‐mediated suppression of OsCPK4/18 is abolished while CDPK‐mediate regulation of OsMPK5 was also attenuated. As a result, OsCPK4/18‐GE plants improve rice yield and defence. The dashed line indicates the uncharacterized signalling pathway. Arrow and blunt lines indicate positive and negative regulation, respectively. Round end lines indicate concentration/dose‐dependent regulation.
Discussion
Cellular signals are often transduced by linear phosphorylation and activation of protein kinases. Mutual phosphorylation of two protein kinases has been observed in animals (Matsuda et al., 1993; Xu et al., 1999), yeast (Jimenez‐Sanchez et al., 2007) and plants (Wang et al., 2018a; Xie et al., 2014), but their biological implications are unclear. In this study, we found that the mutual phosphorylation between OsMPK5 and OsCPK18 constitutes a regulatory circuit to fine tune rice growth and defence. Figure 5g shows a proposed model of OsCPK4/18‐OsMPK5 signalling pathways that orchestrate rice growth and defence. As a positive regulator of growth, OsCPK18 phosphorylates and positively regulates OsMPK5 to suppress rice immunity. OsMPK5, which is activated by disease and abiotic stresses (Xie et al., 2014), retrophosphorylates OsCPK18 to regulate its Ca2+ dependency, likely impairing rice growth under stress conditions (Figure 5g). However, the RNA‐seq data only detected 61 genes that were synergistically regulated by OsCPK18 and OsMPK5, implying that an OsMPK5‐independent pathway should exist for OsCPK18‐mediate suppression of defence (Figure 5g). Interestingly, OsCPK18‐GE and OsCPK4‐GE plants displayed improved disease resistance and growth, suggesting that MAPK‐mediated phosphorylation of OsCPK18/4 plays an important role to coordinate defence and growth signalling. Mutual phosphorylation also occurred between the other subgroup IV CDPKs and subgroup A MAPKs. The reciprocal regulation between these CDPKs and MAPKs, such as in the case of OsMPK5 and OsCPK18, is likely a general mechanism to regulate the crosstalk between different cellular signalling pathways.
Recent studies have revealed that CDPKs undergo multiple layers of regulation during the immune response, including alternative splicing (Dressano et al., 2020), subcellular relocalization (Medina‐Puche et al., 2020), and autophosphorylation (Bredow et al., 2021). Previous biochemical analysis revealed that different CDPKs displayed highly variable Ca2+ dependencies (Boudsocq et al., 2012) to decode different Ca2+ signals. To the best of our knowledge, little is known about whether and how Ca2+ sensitivities of CDPKs are regulated in plants. Our study shows that changing one amino acid at a specific phosphorylation site can drastically affect the Ca2+ sensitivities of recombinant OsCPK18 (Figure 3e). We also noticed that the C‐terminal regions of CDPKs are variable between different subgroups. It will be interesting to investigate whether the amino acids adjacent to the C‐terminus of the CaM domain affect the Ca2+ sensitivity of CDPKs.
It is intriguing that genome editing of the MAPK phosphorylation motif of OsCPK18 and OsCPK4 simultaneously improves disease resistance and growth (Figure 4). Recently, Bredow et al. (2021) found that S318 of AtCPK28, which was phosphorylated by itself and BIK1, was differentially required for its function in immune homeostasis and stem elongation in Arabidopsis. Our data imply that MAPK‐mediated phosphorylation of OsCPK18 T505 and OsCPK4 S512 is important to orchestrate defence and growth signalling pathways. Because OsCPK18‐GE and OsCPK4‐GE plants displayed similar phenotypes despite the difference in the amino acid sequences after T505 and S512, we speculated that improvements in both traits were caused by the dual effects of phosphorylation motif editing on OsCPK18 and OsCPK4 (Figure 5g). First, MAPK‐mediate regulation of Ca2+ dependency of OsCPK18/4 was abolished in OsCPK18/4‐GE plants, which permits full activation of these two CDPKs by Ca2+ and promotes rice growth. Second, the interaction of OsCPK18/4‐GE and OsMPK5 was weaker than that of WT OsCPK18/4 (Figure 5c,d). As a result, CDPK‐mediate phosphorylation and regulation of OsMPK5, which negatively regulates defence (Xie et al., 2014), was also abolished or attenuated. Thus, OsCPK18/4‐GE plants also displayed enhanced disease resistance. Such gain‐of‐function mutations by phosphorylation motif editing create an ideal balance between growth and defence with improvements on both traits in rice (Figure 5g). Full understanding of the mechanism of rebalancing the growth and defence tradeoff in the OsCPK18/4‐GE plant requires further work to investigate how OsCPK18/4 differentially regulates downstream response according to cellular Ca2+ signal and MAPK cascade.
Gene editing technology has become a powerful tool for improving plant disease resistance. A few examples demonstrated that editing susceptibility (S) genes could confer crop disease resistance against various pathogens (Langner et al., 2018). However, targeted mutation of S genes may impact yield‐related traits because they often play important roles in plant growth and development. Based on our understanding of the OsCPK18‐OsMPK5 regulatory circuit, we successfully exploited CRISPR/Cas9 technology to edit the phosphorylation site and selectively block OsMPK5‐mediated phosphorylation of OsCPK18. Remarkably, the such precise engineering of a phosphorylation signalling loop improves rice yield and immunity simultaneously. Since protein phosphorylation is a predominant biochemical mechanism to transduce cellular signals, phosphorylation sites could serve as potential targets for editing and generating valuable gain‐of‐function gene‐edited crops. As shown in this study, genome‐editing‐mediated engineering of plant signalling pathways is broadly applicable in various crops and may simultaneously improve yield, disease resistance, and other agronomic traits.
Experimental procedures
Plant materials and growth conditions
Rice (Oryza sativa L. ssp. Geng) cv Kitaake, Nipponbare (Nip), and Zhonghua 11 (ZH11) were used in this study. OsCPK18‐RI and OsCPK18‐AC were generated in cv Kitaake. OsMPK5‐RI and OsMPK5‐OX were generated in cv Nip (Xiong and Yang, 2003). OsCPK18‐GE, OsCPK4‐GE, and OsCPK4‐KO were generated in cv ZH11 in this study. For the yield‐related trait measurements, the rice plants were grown in a rice field on the campus of Huazhong Agricultural University, Wuhan, China. For disease resistance evaluation, the rice plants were grown in a greenhouse or growth chamber under the following conditions: 28 °C, 14 h day/23 °C, and 10 h night.
DNA vector construction
To overexpress OsCPK18‐AC in rice, pGWB11‐OsCPK18AC was used to transform rice. Briefly, the DNA fragment encoding the N‐terminal V and protein kinase domains of OsCPK18 was amplified with primers CPK18‐ENTR‐F157 and CPK18‐ENRT‐R1121 (Figure S1a). The PCR products were then cloned into the pENTR‐D/TOPO vector and subsequently inserted into pGWB11 (Nakagawa et al., 2007) using the LR reaction (Thermo Fisher Scientific). pGWB11‐OsCPK18AC was transformed into rice by Agrobacterium tumefaciens‐mediated transformation.
To knock out OsCPK4, two gene‐specific gRNAs were designed and used in the CRISPR/Cas9‐mediated targeted mutagenesis (Figure S2a). To edit the MAPK phosphorylation motif, gRNAs were designed based on the available PAM sequences close to the OsCPK4 S512 codon and OsCPK18 T505 codon (Figure 4a). The gRNAs were constructed as tRNA‐gRNA fusions and inserted into the pRGEB32 vector as we described previously (Xie et al., 2015). See Table S2 for the primers used in the CRISPR/Cas9 vector construction.
For protoplast transient expression, pUGW11‐OsMPK5 was used to express FLAG‐tagged OsMPK5 (Xie et al., 2014). For NanoLuc tagging, WT OsCPK18 and its mutant forms were amplified with CPK18‐KpnI‐F and CPK18‐EcoRI‐R. The PCR products were then ligated with the Kpn I‐ and EcoR I‐digested pENTR11 using the ClonExpressII One Step Cloning Kit (Vazyme). Finally, the DNA fragments in pENTR11 were cloned into p34NN (Li et al., 2021) via the LR reaction to obtain plasmid constructs for the expression of NanoLuc‐tagged proteins (Figure S7b). The OsMKK4 gene was amplified from cDNA and cloned into pENTR‐D/TOPO (Thermo Fisher Scientific). The OsMKK4 fragment was subsequently cloned into pUGW5 via LR reaction. See Table S2 for primer sequences.
To express enhanced cyan fluorescent protein (eCFP)‐tagged protein in tobacco, OsCPK18, OsCPK18 T505D , OsCPK18‐GE3, and OsCPK18‐GE8 in pENTR11 were cloned into pEarlyGate102 (Earley et al., 2006) via LR reaction. The resulting binary vector constructs were introduced into A. tumefaciens strain GV3101 for agroinfiltration.
For split luciferase complementation assays, the coding sequence of OsCPK18, OsCPK18‐GE3, OsCPK18‐GE8, OsCPK18 T505A , and OsMPK5 were PCR amplified and then inserted into pCAMBIA‐Cluc and pCAMBIA‐Nluc (Chen et al., 2008) using the ClonExpressII One Step Cloning Kit (Vazyme), respectively. These plasmid constructs were subsequently introduced into A. tumefaciens strain EHA105 for agroinfiltration. See Table S2 for primer sequences.
Targeted gene editing and genotyping
The CRISPR/Cas9 constructs were transformed into rice via A. tumefaciens‐mediated transformation. For genotyping of target genes, rice genomic DNA was isolated from rice leaves using the cetyltrimethylammonium bromide method as we described previously (Xie and Yang, 2013). The target regions were amplified from genomic DNA using gene‐specific primers and analysed by Sanger sequencing. To analyse the potential off‐target editing in OsCPK4‐GE and OsCPK18‐GE plants, the PCR products containing the predicted off‐targets, which were predicted using CRISPR‐P 2.0 (Liu et al., 2017), were analysed by Sanger sequencing. See Table S1 for predicted off‐target sites and Table S2 for PCR primer sequences.
RNA extraction and RT‐qPCR
Total RNA was extracted from rice leaves using TRIzol Reagent (Thermo Fisher Scientific) and treated with DNase I before reverse transcription. cDNA was synthesized from total RNA using reverse transcriptase M‐MLV (RNase H−; Takara). Quantitative PCR was performed with QuantStudio 3 (Applied Biosystems) with a One Step TB Green PrimeScript RT‐PCR Kit (Takara). The relative expression of analysed genes was calculated based on the 2 −ΔΔCt method using rice UBIQUITIN10 as the internal reference gene. The sequences of gene‐specific primers are listed in Table S2.
RNA‐seq analysis
The total RNA was extracted and purified from rice leaf samples of OsCPK18‐RI, OsMPK5‐RI and corresponding WT plants. The RNA‐seq libraries were prepared using NEBNext Ultra RNA Library Prep Kit (NEB) and sequenced using paired‐end sequencing (Novogene). The clean reads were mapped to the rice reference genome using HISAT2 (Kim et al., 2015). The abundance of rice transcripts was summarized using featureCounts (Liao et al., 2014). DESeq2 (Love et al., 2014) was used to identify DEGs (FDR adjust P value <0.05 and ¦log2FC¦ > =1). The rice reference genome sequence and gene annotation were downloaded from Rice Genome Annotation Project (http://rice.uga.edu/index.shtml). The GO enrichment analysis was performed using TBtools (Chen et al., 2020).
Recombinant protein purification
The plasmids for recombinant protein expression were constructed as follows. Briefly, the coding regions of OsCPK18 (and its derived mutants) and OsCPK4 were amplified by PCR using primer pairs CPK18‐PET‐F/CPK18‐PET‐R and pET‐CPK4‐F/pET‐CPK4‐R, respectively. The PCR fragments were cloned into pET28b using a ClonExpress II One Step Cloning Kit (Vazyme). pDEST17‐OsMPK5 was constructed previously (Xie et al., 2014) and used to express His‐OsMPK5 in Escherichia coli. OsCPK18DA, OsCPK18DA‐T15D, OsCPK18DA‐T282D, and OsCPK18DA‐T505D were generated using a MultiSite‐Directed Mutagenesis Kit (Agilent Technologies).
The recombinant proteins were expressed in E. coli BL21(DE3) pLysS. Polyhistidine‐tagged (His‐) OsCPK18 and its mutants were induced by 1 mm IPTG at 37 °C for 4 h. His‐OsMPK5 protein was induced by 0.01 mm IPTG at 12 °C for 12 h. His‐tagged recombinant proteins were purified using a His GraviTrap Column (GE Healthcare) according to the manufacturer's instructions. OsMPK5 protein was dialyzed against a storage buffer containing 20 mm Tris–HCl (pH 7.5), 0.1 mm DTT, and 10% glycerol. The OsCPK18, OsCPK18T505D, OsCPK18‐GE3, and OsCPK18‐GE8 proteins, which were used in Ca2+ sensitivity assays, were dialyzed against storage buffer with 2 mm EDTA to remove residual Ca2+. All proteins were stored at −80 °C in aliquots until use.
In vitro kinase assays
For the in vitro MAPK assays, 5 μg of recombinant His‐OsMPK5 and 20 μg of the substrate (His‐OsCPK18DA, His‐OsCPK18DA‐T15D, His‐OsCPK18DA‐T282D, and His‐OsCPK18DA‐T505D) were mixed in 50 μL reaction buffer containing 25 mm Tris–HCl (pH 7.5), 10 mm MgCl2, 1 mm DTT, 0.2 mm ATP, and 5 mm MnCl2. The reaction was incubated at room temperature (25 °C) for 4 h. Then, the kinase products were desalted by chloroform/methanol precipitation and dissolved in 2 × SDS sample buffer for SDS–PAGE w/o Phos‐tag. Phos‐tag SDS–PAGE was performed as described previously (Xie et al., 2014) using a 7% resolving gel containing 15 μm Phos‐tag (Wako Chemicals) and 60 μm MnCl2. The radioactive in vitro kinase assay was performed as described previously (Xie et al., 2014).
Analysis of protein kinase activity with ADP‐Glo assay
The kinase activity of OsCPK18 and OsCPK18T505D was analysed using an ADP‐Glo Kinase Assay kit (Promega). Briefly, kinase assays were carried out in 96‐well plates in 25 μL reactions containing 2.5 μg His‐OsCPK18 or His‐OsCPK18T505D, 20 μg Histone III‐S protein (Sigma‐Aldrich), 10 μm ATP, 40 mm Tris–HCl, pH 7.5, 20 mm MgCl2, and 0 to 1 × 105 nm CaCl2. After incubation at room temperature for 0.5 h, 25 μL of ADP‐Glo Reagent was added to each well to terminate the kinase reaction and deplete the remaining ATP for 40 min. Finally, 50 μL of Kinase Detection Reagent was added to each well and incubated at room temperature for 1 h. Then, the luminescence of each reaction was read using a SPARK Multimode Microplate Reader (Tecan). For each reaction, three technical replicates were performed. The experiments were repeated three times using recombinant proteins purified from three different batches.
Protoplast transient expression and agroinfiltration
The plasmid constructs were prepared using a Qiagen Plasmid Maxi Kit (Qiagen). Protoplast preparation and transfection were performed as we described previously (Xie and Yang, 2013). To examine OsCPK18DA phosphorylation, 10 μg pUGW5‐OsMKK4DD, 10 μg pUGW11‐OsMPK5, and 20 μg p34NN‐OsCPK18DA/OsCPK18DA‐T15D/OsCPK18DA‐T282D/OsCPK18DA‐T505D plasmids were used to cotransfect 2.0 × 106 protoplasts. The transformed protoplasts were incubated in WI solution (4 mm MES, pH 5.7, 0.6 M mannitol, 4 mm KCl) for 24 h in the dark and then used for protein extraction.
To express eCFP‐tagged proteins in tobacco leaves, pEarlyGate102‐derived constructs were transformed into A. tumefaciens strain GV3101. An orange fluorescent protein fused CBL1n protein was used as a plasma membrane marker (Batistic et al., 2010). For agroinfiltration, overnight bacterial culture was diluted in an LB medium containing kanamycin (50 μg/mL) and rifampicin (50 μg/mL) and continuously grown until the OD600 reached 0.6. Bacterial cells were harvested by centrifugation and then resuspended in an infiltration buffer containing 10 mm MES pH 5.6, 10 mm MgCl2, and 200 μm acetosyringone. Bacterial suspensions were infiltrated into the leaves of 4‐ to 5‐week‐old Nicotiana benthamiana plants. The infiltrated leaves were examined 2–3 days postinoculation using spinning‐disc confocal microscopy (Nikon Eclipse TE2000‐U).
The split luciferase complementation assay was performed as described by Chen et al. (2008). Briefly, Agrobacteria containing Nluc and Cluc‐fusion constructs were equally mixed to a final OD600 = 0.5. Bacterial suspensions were infiltrated into young leaves of N. benthamiana as described above. The split luciferase constructs of GSK2 and BZR1 from Prof. Yibo Li′s laboratory (Huazhong Agricultural University) were used as a positive control in split luciferase assays. The luciferase activities were measured using a Tanon 5200 chemiluminescent imager (Tanon).
Plant protein extraction, Western blotting, and NanoLuc in‐gel detection
Total proteins were extracted from protoplasts using modified RIPA buffer containing 50 mm Tris–HCl, pH 7.5, 150 mm NaCl, 1% Triton X‐100, 0.1% SDS, 1 mm EDTA, 1 mm dithiothreitol, and 1% protease inhibitor cocktail (EDTA‐free, Sigma–Aldrich). The total protein samples were separated in a 10%–12% SDS–PAGE gel and transferred to a polyvinylidene difluoride membrane. Immunodetection of FLAG‐ and GFP‐tagged proteins was performed using anti‐FLAG (1 : 2000 dilution, Sigma–Aldrich) and anti‐GFP (1 : 1000 dilution, ABclonal) primary antibodies, respectively. After hybridization with goat anti‐mouse IgG (whole molecule)‐peroxidase antibody (1 : 10 000 dilution, Sigma–Aldrich), the tagged protein was detected with the Pierce Fast Western blot Kit and SuperSignal West Pico Substrate (Thermo Fisher Scientific) and visualized using a Tanon‐5200 chemiluminescent imager (Tanon).
For coimmunoprecipitation assays, GFP tagged OsCPK18 or OsCPK18‐GE were respectively co‐expressed with FLAG‐tagged OsMPK5 in N. benthamiana leaves. Then, total proteins were extracted from infiltrated tobacco leaves using extraction buffer containing 50 mm Tris–HCl (pH 7.5), 100 mm NaCl, 1 mm EDTA, 10 mm NaF, 5 mm Na3VO4, 0.25% Triton X‐100, 0.25% NP‐40, 1 mm PMSF, and 1% protease inhibitor cocktail (Sigma–Aldrich). For immunoprecipitation, total proteins were incubated with 20 μL of anti‐FLAG Magnetic beads (Thermo Scientific) at 4 °C for 2 h. The beads were washed five times with washing buffer containing 50 mm Tris–HCl (pH 7.5), 100 mm NaCl, 1 mm EDTA, 1 mm PMSF, and 1× protease inhibitor cocktail. Finally, the precipitated proteins were eluted with 2× SDS loading buffer by incubating at 95 °C for 3 min.
The in‐gel detection of NanoLuc‐tagged proteins was performed as previously described (Li et al., 2021). After SDS–PAGE or Phos‐tag SDS–PAGE, the gel was washed at room temperature in SDS‐Removing Buffer (50 mm Tris pH 7.5, 1 mm dithiothreitol, and 0.1 mm EDTA) for approximately 1 h with three changes of buffer. Then, the gel was incubated in Renaturation Buffer (50 mm Tris pH 7.5, 5 mm dithiothreitol, 0.1 mm EDTA, 100 mm NaCl, 5 mm MgCl2, 0.04% Tween‐20) at 4 °C for 4 h. Finally, the gel was rinsed with distilled water and then incubated in Substrate Solution [1× PBS, 0.1% BSA, 1× Nano‐Glo Luciferase Assay Substrate (Promega)]. After 5 min, the NanoLuc luminescence in the gel was imaged using a Tanon 5200 chemiluminescent imager (Tanon).
Rice blast inoculation
M. oryzae isolates 99–20 (Zhou et al., 2018), which are virulent on cv ZH11, cv Nip, and cv Kitaake, were used in this study. Rice seedlings (3 weeks old) were spray‐inoculated with 3 × 105 spores/mL. The lesion number was counted at 7 days postinoculation. The relative growth of fungi in the inoculated leaves was analysed by qPCR as described previously (Qi and Yang, 2002; Xie et al., 2014).
Statistical analysis
Two‐tailed Student's t‐test was performed using GraphPad Prism 8 software to compare the differences between the two groups. For multiple comparisons, a one‐way analysis of variance followed by Tukey's test was performed using the R package (version 4.1.0).
Accession numbers
The RNA‐seq data have been deposited at the National Genomics Data Center of China under accession number PRJCA008323. The accession number of genes described in this study are OsCPK18 (Os07g0409900), OsCPK4 (Os02g0126400), OsMKK4 (Os02g0787300), and OsMPK5 (Os03g0285800).
Funding
This work was supported by funding from the National Natural Science Foundation of China (31622047 and 31821005), the National Transgenic Science and Technology Program (2019ZX08010003 and 2016ZX08010002), the Collaborative Fund of Huazhong Agricultural University and Agricultural Genomics Institute at Shenzhen (SZYJY2021007), the Fundamental Research Funds for the Central Universities (2021ZKPY002), and the Higher Education Discipline Innovation Project (B20051). YY is supported by the USDA National Institute of Food and Agriculture and Hatch Appropriations under Project #PEN04659 and Accession #1016432.
Conflict of interest
The authors declare no competing interests.
Author contributions
K.X. and Y.Y. conceived the projects. H.L., K.X., X.H., L.X., and Y.Y. designed the experiments. H.L., Y.Z., C.W., J.B., M.C., Y.C., and K.X. performed the experiments. H.L., C. J., Y.Y., L.X., Y.M., and K.X. analysed the data. K.X. wrote the manuscript with input from all authors.
Supporting information
Figure S1 Overexpression of truncated OsCPK18 with constitutive kinase activity (OsCPK18‐AC) in rice.
Figure S2 Targeted mutation of OsCPK4 in rice using CRISPR/Cas9 gene editing.
Figure S3 Radioactive in vitro kinase assay shows that OsMPK5 phosphorylated OsCPK18 at T505.
Figure S4 Comparison of OsCPK18, OsCPK18DA, and OsCPK18T505D protein expression and localization in rice protoplasts and tobacco leaves.
Figure S5 OsMPK5 phosphorylated OsCPK4 at S512.
Figure S6 Comparison of panicle size of OsCPK18‐GE, OsCPK4‐GE, and ZH11.
Figure S7 OsCPK18‐GE protein stability and subcellular localization.
Figure S8 Comparisons of OsCPK18, OsCPK18‐GE, and OsMPK5 interactions using Co‐immunoprecipitation assay.
Table S1 Analysis of off‐target editing in OsCPK18‐GE and OsCPK4‐GE plants.
Table S2 DNA oligos used in this study.
Data S1 Differentially expressed genes in OsCPK18‐RI plants.
Data S2 Differentially expressed genes in OsMPK5‐RI plants.
Data S3 Differentially expressed genes shared by OsCPK18‐RI and OsMPK5‐RI plants.
Acknowledgements
We thank Prof. Xuewei Chen at Sichuan Agricultural University for providing the Magnaporthe oryzae isolates 99‐20, Prof. Yibo Li at Huazhong Agricultural University for assistance in split luciferase complementation assays.
Contributor Information
Yinong Yang, Email: yuy3@psu.edu.
Kabin Xie, Email: kabinxie@mail.hzau.edu.cn.
References
- Bailey, T. , Zhou, X. , Chen, J. and Yang, Y. (2009) Role of ethylene, abscisic acid and MAP kinase pathways in rice blast resistance. In Advances in Genetics, Genomics and Control of Rice Blast Disease( Wang, G.‐L. and Valent, B. , eds), pp. 185–190. Netherlands: Springer. [Google Scholar]
- Batistic, O. , Waadt, R. , Steinhorst, L. , Held, K. and Kudla, J. (2010) CBL‐mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61, 211–222. [DOI] [PubMed] [Google Scholar]
- Boudsocq, M. , Droillard, M.‐J. , Regad, L. and Laurière, C. (2012) Characterization of Arabidopsis calcium‐dependent protein kinases: activated or not by calcium? Biochem. J. 447, 291–299. [DOI] [PubMed] [Google Scholar]
- Boudsocq, M. and Sheen, J. (2013) CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredow, M. , Bender, K.W. , Johnson Dingee, A. , Holmes, D.R. , Thomson, A. , Ciren, D. , Tanney, C.A.S. et al. (2021) Phosphorylation‐dependent subfunctionalization of the calcium‐dependent protein kinase CPK28. Proc. Natl. Acad. Sci. U.S.A. 118, e2024272118. 10.1073/pnas.2024272118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundo, M. and Coca, M. (2016) Enhancing blast disease resistance by overexpression of the calcium‐dependent protein kinase OsCPK4 in rice. Plant Biotechnol. J. 14, 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo, S. , Baldrich, P. , Messeguer, J. , Lalanne, E. , Coca, M. and San Segundo, B. (2014) Overexpression of a calcium‐dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Chen, H. , Zhang, Y. , Thomas, H.R. , Frank, M.H. , He, Y. and Xia, R. (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Zou, Y. , Shang, Y. , Lin, H. , Wang, Y. , Cai, R. , Tang, X. et al. (2008) Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol. 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto, D. and Zipfel, C. (2016) Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Davis, R.J. (1993) The mitogen‐activated protein kinase signal transduction pathway. J. Biol. Chem. 268, 14553–14556. [PubMed] [Google Scholar]
- De Bruyne, L. , Hofte, M. and De Vleesschauwer, D. (2014) Connecting growth and defense: the emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol. Plant 7, 943–959. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer, D. , Yang, Y. , Cruz, C.V. and Hofte, M. (2010) Abscisic acid‐induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase‐mediated repression of ethylene signaling. Plant Physiol. 152, 2036–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressano, K. , Weckwerth, P.R. , Poretsky, E. , Takahashi, Y. , Villarreal, C. , Shen, Z. , Schroeder, J.I. et al. (2020) Dynamic regulation of Pep‐induced immunity through post‐translational control of defence transcript splicing. Nat. Plants 6, 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. and Pikaard, C.S. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Gao, A.L. , Wu, Q.Y. , Zhang, Y. , Miao, Y.C. and Song, C.P. (2014) Arabidopsis calcium‐dependent protein kinase CPK28 is potentially involved in the response to osmotic stress. Chin. Sci. Bull. 59, 1113–1122. [Google Scholar]
- Hamel, L.P. , Sheen, J. and Seguin, A. (2014) Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci. 19, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.F. , Breton, G. and Harmon, A. (2004) Decoding Ca(2+) signals through plant protein kinases. Annu. Rev. Plant Biol. 55, 263–288. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Zhou, J. , Peng, X. , Xu, H. , Liu, C. , Du, B. , Yuan, H. et al. (2011) The Bphi008a gene interacts with the ethylene pathway and transcriptionally regulates MAPK genes in the response of rice to brown planthopper feeding. Plant Physiol. 156, 856–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot, B. , Yao, J. , Montgomery, B.L. and He, S.Y. (2014) Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, A. , Ueguchi‐Tanaka, M. , Sonoda, Y. , Kitano, H. , Koshioka, M. , Futsuhara, Y. , Matsuoka, M. et al. (2001) Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height‐regulating gene GAI/RGA/RHT/D8 . Plant Cell 13, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez‐Sanchez, M. , Cid, V.J. and Molina, M. (2007) Retrophosphorylation of Mkk1 and Mkk2 MAPKKs by the Slt2 MAPK in the yeast cell integrity pathway. J. Biol. Chem. 282, 31174–31185. [DOI] [PubMed] [Google Scholar]
- Jin, Y. , Ye, N. , Zhu, F. , Li, H. , Wang, J. , Jiang, L. and Zhang, J. (2017) Calcium‐dependent protein kinase CPK28 targets the methionine adenosyltransferases for degradation by the 26S proteasome and affects ethylene biosynthesis and lignin deposition in Arabidopsis . Plant J. 90, 304–318. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. and Salzberg, S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita‐Kikuta, E. , Aoki, Y. , Kinoshita, E. and Koike, T. (2007) Label‐free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol. Cell. Proteomics 6, 356–366. [DOI] [PubMed] [Google Scholar]
- Kuijt, S.J. , Greco, R. , Agalou, A. , Shao, J. , t Hoen, C.C. , Overnas, E. , Osnato, M. et al. (2014) Interaction between the GROWTH‐REGULATING FACTOR and KNOTTED1‐LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164, 1952–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner, T. , Kamoun, S. and Belhaj, K. (2018) CRISPR crops: plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56, 479–512. [DOI] [PubMed] [Google Scholar]
- Li, C.H. , Wang, G. , Zhao, J.L. , Zhang, L.Q. , Ai, L.F. , Han, Y.F. , Sun, D.Y. et al. (2014) The receptor‐like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26, 2538–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Wu, C. , Du, M. , Chen, Y. , Hou, X. , Yang, Y. and Xie, K. (2021) A versatile nanoluciferase toolkit and optimized in‐gel detection method for protein analysis in plants. Mol. Breed. 41, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G.K. and Shi, W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Ding, Y. , Zhou, Y. , Jin, W. , Xie, K. and Chen, L.L. (2017) CRISPR‐P 2.0: an improved CRISPR‐Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. [DOI] [PubMed] [Google Scholar]
- Liu, M. , Shi, Z. , Zhang, X. , Wang, M. , Zhang, L. , Zheng, K. , Liu, J. et al. (2019) Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice. Nat. Plants 5, 389–400. [DOI] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschi, S. , Hake, K. , Herde, M. , Hause, B. and Romeis, T. (2015) The calcium‐dependent protein kinase CPK28 regulates development by inducing growth phase‐specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in Arabidopsis. Plant Cell 27, 591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschi, S. , Werner, S. , Schulze, W.X. , Legen, J. , Hilger, H.H. and Romeis, T. (2013) Function of calcium‐dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 73, 883–896. [DOI] [PubMed] [Google Scholar]
- Matsuda, S. , Gotoh, Y. and Nishida, E. (1993) Phosphorylation of Xenopus mitogen‐activated protein (MAP) kinase kinase by MAP kinase kinase kinase and MAP kinase. J. Biol. Chem. 268, 3277–3281. [PubMed] [Google Scholar]
- Medina‐Puche, L. , Tan, H. , Dogra, V. , Wu, M. , Rosas‐Diaz, T. , Wang, L. , Ding, X. et al. (2020) A defense pathway linking plasma membrane and chloroplasts and co‐opted by pathogens. Cell 182, 1109–1124 e1125, 1109, 1124.e25. [DOI] [PubMed] [Google Scholar]
- Meng, X. and Zhang, S. (2013) MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. [DOI] [PubMed] [Google Scholar]
- Monaghan, J. , Matschi, S. , Shorinola, O. , Rovenich, H. , Matei, A. , Segonzac, C. , Malinovsky, F.G. et al. (2014) The calcium‐dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16, 605–615. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. , Nakamura, S. , Hino, T. , Maeo, K. , Tabata, R. et al. (2007) Improved gateway binary vectors: high‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Ning, Y. , Liu, W. and Wang, G.L. (2017) Balancing immunity and yield in crop plants. Trends Plant Sci. 22, 1069–1079. [DOI] [PubMed] [Google Scholar]
- Qi, W. , Sun, F. , Wang, Q. , Chen, M. , Huang, Y. , Feng, Y.Q. , Luo, X. et al. (2011) Rice ethylene‐response AP2/ERF factor OsEATB restricts internode elongation by down‐regulating a gibberellin biosynthetic gene. Plant Physiol. 157, 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, M. and Yang, Y. (2002) Quantification of Magnaporthe grisea during infection of rice plants using real‐time polymerase chain reaction and Northern blot/Phosphoimaging analyses. Phytopathology 92, 870–876. [DOI] [PubMed] [Google Scholar]
- Singh, P. and Sinha, A.K. (2016) A positive feedback loop governed by SUB1A1 interaction with MITOGEN‐ACTIVATED PROTEIN KINASE3 imparts submergence tolerance in rice. Plant Cell 28, 1127–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, W. , Hou, C. , Ren, Z. , Wang, C. , Zhao, F. , Dahlbeck, D. , Hu, S. et al. (2019) A calmodulin‐gated calcium channel links pathogen patterns to plant immunity. Nature 572, 131–135. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Grubb, L.E. , Wang, J. , Liang, X. , Li, L. , Gao, C. , Ma, M. et al. (2018b) A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 69, 493–504 e496, 493, 504.e6. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Li, Y. , Chern, M. , Zhu, Y. , Zhang, L.L. , Lu, J.H. , Li, X.P. et al. (2021) Suppression of rice miR168 improves yield, flowering time and immunity. Nat. Plants 7, 129–136. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Wang, S. , Hu, K. , Yang, J. , Xin, X. , Zhou, W. , Fan, J. et al. (2018a) The kinase OsCPK4 regulates a buffering mechanism that fine‐tunes innate immunity. Plant Physiol. 176, 1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Zhou, L. , Shi, H. , Chern, M. , Yu, H. , Yi, H. , He, M. et al. (2018c) A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Xie, K. , Chen, J. , Wang, Q. and Yang, Y. (2014) Direct phosphorylation and activation of a mitogen‐activated protein kinase by a calcium‐dependent protein kinase in rice. Plant Cell 26, 3077–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. , Minkenberg, B. and Yang, Y. (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA‐processing system. Proc. Natl. Acad. Sci. U.S.A. 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. and Yang, Y. (2013) RNA‐guided genome editing in plants using a CRISPR‐Cas system. Mol. Plant 6, 1975–1983. [DOI] [PubMed] [Google Scholar]
- Xiong, L. and Yang, Y. (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid‐inducible mitogen‐activated protein kinase. Plant Cell 15, 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, B. , Wilsbacher, J.L. , Collisson, T. and Cobb, M.H. (1999) The N‐terminal ERK‐binding site of MEK1 is required for efficient feedback phosphorylation by ERK2 in vitro and ERK activation in vivo . J. Biol. Chem. 274, 34029–34035. [DOI] [PubMed] [Google Scholar]
- Xu, G. , Yuan, M. , Ai, C. , Liu, L. , Zhuang, E. , Karapetyan, S. , Wang, S. et al. (2017) uORF‐mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D.L. , Yang, Y. and He, Z. (2013) Roles of plant hormones and their interplay in rice immunity. Mol. Plant 6, 675–685. [DOI] [PubMed] [Google Scholar]
- Yang, D.L. , Yao, J. , Mei, C.S. , Tong, X.H. , Zeng, L.J. , Li, Q. , Xiao, L.T. et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109, E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti, H. , Zdanovskaia, M. , Hsiao, K. and Goueli, S.A. (2009) ADP‐Glo: a bioluminescent and homogeneous ADP monitoring assay for kinases. Assay Drug Dev. Technol. 7, 560–572. [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Su, J. , Zhang, Y. , Xu, J. and Zhang, S. (2018) Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 45, 1–10. [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Liao, H. , Chern, M. , Yin, J. , Chen, Y. , Wang, J. , Zhu, X. et al. (2018) Loss of function of a rice TPR‐domain RNA‐binding protein confers broad‐spectrum disease resistance. Proc. Natl. Acad. Sci. U.S.A. 115, 3174–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M. and Zhang, Y. (2020) Plant immunity: danger perception and signaling. Cell 181, 978–989. [DOI] [PubMed] [Google Scholar]
- Zhu, H. , Li, C. and Gao, C. (2020) Applications of CRISPR‐Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–677. [DOI] [PubMed] [Google Scholar]
- Zust, T. and Agrawal, A.A. (2017) Trade‐offs between plant growth and defense against insect herbivory: an emerging mechanistic synthesis. Annu. Rev. Plant Biol. 68, 513–534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Overexpression of truncated OsCPK18 with constitutive kinase activity (OsCPK18‐AC) in rice.
Figure S2 Targeted mutation of OsCPK4 in rice using CRISPR/Cas9 gene editing.
Figure S3 Radioactive in vitro kinase assay shows that OsMPK5 phosphorylated OsCPK18 at T505.
Figure S4 Comparison of OsCPK18, OsCPK18DA, and OsCPK18T505D protein expression and localization in rice protoplasts and tobacco leaves.
Figure S5 OsMPK5 phosphorylated OsCPK4 at S512.
Figure S6 Comparison of panicle size of OsCPK18‐GE, OsCPK4‐GE, and ZH11.
Figure S7 OsCPK18‐GE protein stability and subcellular localization.
Figure S8 Comparisons of OsCPK18, OsCPK18‐GE, and OsMPK5 interactions using Co‐immunoprecipitation assay.
Table S1 Analysis of off‐target editing in OsCPK18‐GE and OsCPK4‐GE plants.
Table S2 DNA oligos used in this study.
Data S1 Differentially expressed genes in OsCPK18‐RI plants.
Data S2 Differentially expressed genes in OsMPK5‐RI plants.
Data S3 Differentially expressed genes shared by OsCPK18‐RI and OsMPK5‐RI plants.


