Abstract
The mammalian liver must cope with various metabolic and physiological changes that normally recur every day and primarily stem from daily cycles of rest-activity and fasting-feeding. While large body of evidence supports the reciprocal regulation of circadian rhythms and liver function, the research on the hepatic ultradian rhythms have largely been lagging behind. However, with the advent of more cost-effective high-throughput omics technologies, high-resolution time-lapse imaging, and more robust and powerful mathematical tools, several recent studies have shed new light on the presence and functions of hepatic ultradian rhythms. In this review, we will first very briefly discuss the basic principles of circadian rhythms, and then cover in greater details the recent literature related to ultradian rhythms. Specifically, we will highlight the prevalence and mechanisms of hepatic 12-hour rhythms, and 8-hour rhythms, which cycle at the second and third harmonics of circadian frequency. Finally, we also refer to ultradian rhythms with other frequencies and examine the limitations of the current approaches as well as the challenges related to identifying ultradian rhythm and addressing their molecular underpinnings.
Graphical Abstract
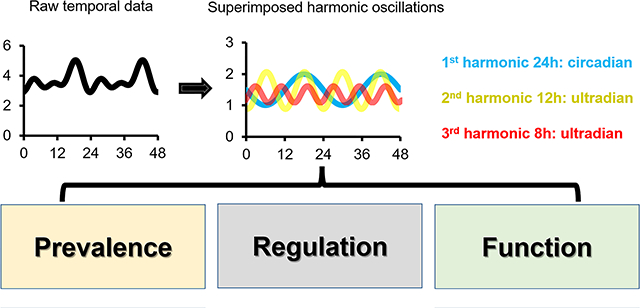
Introduction on biological rhythms
It is all about rhythms…
Rhythms are present in almost every aspect of biology from molecular circuits to whole organism physiology and behavior. Biological rhythms can be categorized into exogenously and endogenously driven. The former is activated in response to rhythmic external / environmental cues, while the latter is driven by internal mechanisms. These two distinct mechanisms can interact with each other to reinforce rhythmicity and align internal rhythms with external cycles. Endogenous rhythms are driven by oscillators which are classified as ultradian, circadian and infradian based on their frequency. There are no clear guidelines but generally speaking If the period is between 20 and 30 hours they are regarded as circadian; ultradian when the period is shorter than 20 hours and can even range between milliseconds to several hours, and infradian when the period is longer than 30 hours, namely from days to years.
Unlike Hourglass mechanisms that determine the timing of biological events that occur only once in life (e.g., sexual maturity, menopause), biological oscillators rely on a fundamental design principle whereby the process repeats itself in the absence of external signals (1, 2). Biological oscillators, per definition, maintain a relative constant frequency yet their amplitude can change. A ‘self-sustained oscillator’ is an oscillator which preserves a relatively constant amplitude, whereas if the amplitude progressively decreases with each consecutive cycle the underlying oscillator is considered as a ‘damped oscillator’. Both self-sustained and damped oscillators play important roles in temporal control of various biological processes (3).
Basic principles of circadian rhythms
The most prevalent, and widely studied rhythms in mammals are circadian rhythms. Rhythms are classified as circadian (from the Latin circa, about and diem, a day) when they persist in an environment devoid of external time-cues, with a ‘free-running’ period that is about 24 hours. A wide variety of biological processes oscillate with a 24-hour rhythmicity, corresponding to the cyclic environment (i.e., daily light-dark cycles) generated by the earth’s rotation on its axis. These rhythms are prevalent in virtually all aspects of mammalian biology, from gene expression, protein modifications and enzymatic activity, through complex metabolic and physiological processes, to behavior (4). Circadian rhythms are not a mere response to environmental changes but are rather endogenously driven by a circadian clock. These clocks are believed to confer an evolutionary advantage by enabling the organism to anticipate, and act a priori to challenges imposed by the cyclic environment.
The circadian clock in mammals is cell-autonomous and relies on autoregulatory transcription-translation feedback loops (TTFL). Clock-genes were among the first genes to be identified as controlling behavior, and their oscillations drive the rhythmicity of different gene networks. The positive arm of the core clock circuitry includes the transcription factors BMAL1 and CLOCK that bind as heterodimers to E-box motifs and drive the expression of rhythmic genes, among others Per1,2,3 and Cry1,2. Subsequently, PERs:CRYs complexes accumulate in the nucleus and repress their own transcription by binding to BMAL1:CLOCK. As repression progresses, Pers and Crys gene transcription declines and the existing PERs:CRYs complexes are degraded. An additional integrated regulatory loop includes members of the nuclear receptor families of REV-ERB and ROR which regulate Bmal1 expression (Figure 1A). These intricate feedback loops generate about 24 hours rhythms in gene expression of clock as well as various output genes (5, 6).
Figure 1. Transcriptional network of circadian and 12-hour oscillator.
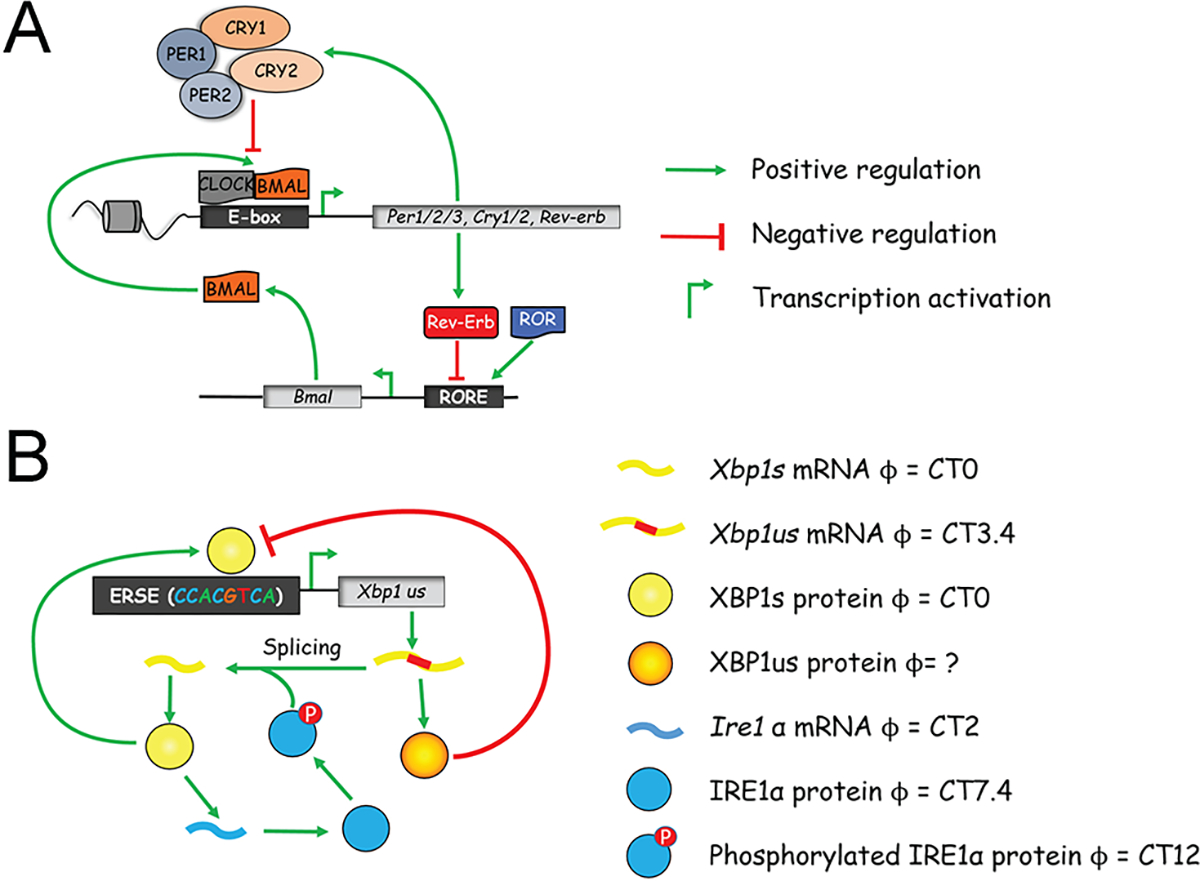
(A) A simplified model of core circadian clock transcriptional network. (B) Proposed transcriptional network of 12-hour oscillator. Please note while Ire1α mRNA exhibits an XBP1s-dependent 12-hour rhythms, XBP1s likely transcriptionally regulates the 12-hour rhythmic Ire1α expression via a yet-to-be-identified intermediate transcriptional factor, as ChIP-seq revealed no XBP1s recruitment to Ire1α gene promoter.
The mammalian circadian timing system is structured in a hierarchical manner and consists of a central pacemaker in the suprachiasmatic nucleus (SCN) region of the brain and peripheral clocks in almost every cell of the body. The master clock in the SCN is entrained by daily light-dark cycles and synchronizes subsidiary oscillators in virtually all tissues and cells of the body. Given that circadian clocks function in a self-sustaining and cell-autonomous manner, at least in culture, it raises the question how billions of individual oscillators in the body tick in synchrony. Since these cellular oscillators anticipate environmental changes and function together in a proactive manner, their temporal orchestration is vital. It is believed that wide variety of endocrine, neuronal and metabolic signals such as body temperature (7), oxygen (8) and nutrient levels (9) participate in communication between clocks in different organs throughout the body to maintain their phase coherence (10, 11). Several recent reviews have addressed in detail the molecular mechanisms underlying the circadian clockwork as well as the molecular, metabolic and physiologic circadian regulation of different organs (12–16) and their potential medical implications (17), in particular in liver (18).
Ultradian rhythms predominantly cycle at the harmonic frequencies of the 24-hour.
The term “harmonic” arises from the musical instrument theory. A harmonic is a sound wave that has a frequency that is an integer multiple of a fundamental tone. The lowest frequency sound that can be produced on string or wind instruments is the fundamental tone frequency. The frequency twice that of the fundamental tone is the second harmonic, and so on and on. It has been well-known that music played with harmonic notes is the most pleasant to the ears. Like music, biological ultradian rhythms also tend to cycle at the harmonic frequencies of the 24-hour, namely, at the ultradian periods of 12-hour, 8-hour, 6-hour, etc., ranging from the transcriptome, proteome, metabolome to behavior levels (19–32). Interestingly, the harmonic periods usually vary concordantly with the exact period of the circadian rhythm under various physiological conditions. For example, in the MMH-D3 hepatocyte cell line, the free-running first, second and third harmonic periods of gene expression are 21.6 hour, 10.8 hour and 7.3 hour, respectively (27). In the fungus Neurospora crassa, after 11 hour/11 hour light-dark entrainment, rhythmic genes cycle predominantly at 7.33-hour, 11-hour, or 22-hour periods (28). In the malaria-causing pathogen Plasmodium falciparum, the periods of second harmonic of cyclic genes are half period of the first harmonic, which is usually between 30~60 hours (29). One plausible reason for the dominant features of harmonic oscillations in different species is the evolutionary advantage of ensuring that all harmonic oscillations are always in-phase with each other during each diurnal cycle (the first harmonic period). After all, if two oscillations cycle at the periods of 17-hour and 25-hour, then it will take 25 and 17 days, respectively, for them to be in-phase again, which would be a major disadvantage for animals that live in a world with regular environmental cycles. Due to the harmonic nature of most ultradian rhythms, early studies posit that these rhythms are likely “derivative” of the circadian rhythms and propose different mechanisms of how circadian clocks can generate ultradian rhythms cycling at the harmonic frequencies (22, 33, 34). However, emerging evidence begin to unveil the tantalizing possibility of the existence of independent ultradian oscillators that are responsible for establishing and maintaining many of these ultradian rhythms (31, 35, 36). In the following sections, we will review the prevalence and proposed mechanisms of ultradian rhythms alongside the challenges related to uncovering them and addressing their molecular makeup, with a focus on the hepatic harmonic oscillations.
Hepatic 12-hour ultradian rhythms
The presence of hepatic harmonic oscillation in mice was first comprehensively revealed using high-resolution microarray (at 1 hour resolution for a total of 48 hours) in a landmark study by Hughes and colleagues in 2009 (23). In the original study, using both Fisher’s G and COSOPT methods, the authors identified ~200 genes cycling with a robust 12-hour period in mouse liver in constant darkness condition (23). Although this number was later found to be a significant underestimation of the true prevalence of 12-hour rhythms due to the limit of statistical tools available at the time; this study, nevertheless, made several key findings about 12-hour rhythms that have been validated by later studies (23). First, the authors found that compared to the more evenly distributed acrophases of circadian genes, the peaks of these 12-hour gene expression are heavily biased toward the two ‘rush hours’ in a day: just after sunrise (CT0) and sunset (CT12) (23). Secondly, these ~200 genes are significantly enriched in unfolded protein response (UPR) pathway. Finally, day-time restricted feeding to an 8-hour window abolishes many of these 12-hour rhythms (23). While the detailed origin, regulation, and function of these hepatic 12-hour ultradian rhythms were not clear back then, these initial findings did imply a role of them in the regulation of proteostasis at the two key transitional periods between fasting-feeding and sleep-wake at dawn and dusk. In the following paragraphs, we will first discuss the prevalence and regulation of the 12-hour rhythms and then elaborate in greater details on our newly gained knowledge of the reciprocal regulation of 12-hour rhythms and hepatic metabolism. In the end, we will touch upon the evolutionary origin of the 12-hour rhythms and speculate on how the metabolic shifts may play a role in the evolvement of 12-hours rhythms in terrestrial mammals.
12-hour rhythms of gene expression are prevalent
While the number of 12-hour cycling hepatic mRNA in mice was originally thought to be relatively small (~200) (23), re-analyzing the same hepatic microarray dataset with a recently developed eigenvalue/pencil method (37) uncovered a much larger repertoire of 3,652 genes (~20% of total hepatic mRNA) (21). The eigenvalue/pencil method is a nonparametric signal processing algorithm that assumes that most oscillations can be approximated by a linear combination of exponentials (standard sine waves with a decay factor) plus noise and therefore can be applied to any situations where such assumptions are valid. As a result, the biggest distinction between the eigenvalue/pencil method and other commonly-used cycle-identification methods [such as JTK_CYCLE (38), Fisher’s G (23), COSOPT (23), ARSER (39) and RAIN (40)] lies in the former’s ability to identify all superimposed oscillation in an unbiased manner simultaneously, while the latter ones require the users to define a narrow period range and thus can only reveal one single dominant oscillation at one time (21, 41). However, a limitation of the eigenvalue/pencil method is the lack of statistical power: the p value for each identified oscillation is not available and only one fixed FDR can be obtained for all identified oscillations through permutation-based method (27). Overall, the eigenvalue/pencil method is more sensitive in detecting weaker ultradian oscillations but at the same time has a higher type I error (higher false positive rate) (27). Therefore, it is recommended that the eigenvalue/pencil method be first used as an exploratory tool to identify oscillations with all possible periods. Oscillations with specific periods of interest can subsequently be further validated by a statistical method like JTK_CYCLE, RAIN, etc (27).
This significant increase of 12-hour genes compared to older studies can be largely attributed to the unique ability of the eigenvalue/pencil to uncover both dominant and other “masked” oscillations with smaller amplitudes. In fact, only 760 genes were found to have dominant 12-hour rhythms (whose 12-hour amplitudes are the greatest among all identified oscillations) in this re-analysis, while the rest were often superimposed with circadian rhythms with larger amplitude, therefore evading detection by traditional methods such as JTK_CYCLE (21). This much wider prevalence of 12-hour hepatic genes was later validated by two high resolution RNA-Seq datasets (27, 42), with the number of dominant 12-hour hepatic genes further updated to be close to 1,700 (27). It is important to note that while the prevalence of hepatic 12-hour transcriptome matches that of hepatic circadian rhythm (43), their amplitudes, on average, are much smaller, often in the range of 1.3 to 4-fold changes (27). Consistent with the 2009 study, the acrophases of 12-hour rhythms identified in these two RNA-seq datasets were also enriched at the two short time windows at sunrise and sunset (27, 42). Besides transcriptome, 12-hour hepatic oscillations are also found abundantly at the proteome level (also in the range of 20%~30% of all hepatic proteome), although a limited overlap with ~12-hour transcriptome (~35%) and a more diverse phase distribution was observed (21, 44, 45). As a final note, besides the liver, prevalent 12-hour rhythms were identified in multiple peripheral tissues as well as the brain (19, 35, 46). Superimposed 12-hour rhythms of locomotive activity were also reported in both mice and rats under standard laboratory 12-h light/12-h dark conditions (25, 47). Since this is a liver-centric review, we will not elaborate on the topic of 12-hour rhythms in extrahepatic tissues, the topic of which was extensively discussed elsewhere (35, 46).
Evidence supporting the existence of an XBP1s-dependent 12-hour oscillator
Several theories have been put forward over the past decade to account for the regulation of hepatic 12-hour rhythms. Early studies favor the hypothesis that the mammalian 12-hour rhythms are not cell-autonomous and instead established by the combined effects of circadian clock and fasting-feeding cues (22, 23, 34). This claim was supported by the observation that many of these hepatic 12-hour rhythms were impaired in Cry1/Cry2 double knockout mice (34). Another study observed a conversion of 12-hour to an apparent 24-hour hepatic gene expression in a mouse model of brain-specific rescue of Clock function (22), suggesting possibly that the central circadian clock in the brain controls peripheral ultradian rhythms. Alternatively, it was suggested that two circadian transcription factors with anti-phasic transcriptional activity and binding cooperativity are theoretically capable of establishing 12-hour rhythms of gene expression in a cell-autonomous manner (33). However, emergent evidence challenges the old paradigm, and instead supports the existence of a cell-autonomous 12-hour oscillator, separate from both the circadian clock and the cell cycle. This 12-hour oscillator is proposed to be responsible for the establishment and maintenance of many 12-hour ultradian rhythms (21, 27, 35, 37, 42, 44, 45). For instance, widespread 12-hour rhythms of gene expression were observed in both serum-synchronized hepatocyte line MMH-D3 in vitro (24, 27) and dexamethasone-synchronized liver slices cultured ex vivo (35, 48). More importantly, many of these 12-hour rhythms of gene expression remain intact in the liver of circadian clock deficient mice (including both BMAL1 knockout and CLOCK Δ19 mice) (21, 49–51).
In two recent studies, XBP1s (the spliced form of XBP1) was identified to be a major transcriptional regulator of the hepatic 12-hour rhythms (27, 42). XBP1s was classically studied as one of the three transcription factors regulating the UPR (along with ATF4 and ATF6). It is unorthodoxly spliced by ER-localized endoribonuclease/kinase phospho-IRE1α in response to ER stress (52). XBP1s, total and phospho-IRE1α exhibited robust 12-hour rhythms of expression in mouse liver (21, 27, 34). Due to the enrichment of hepatic 12-hour transcriptome in UPR pathways, it is not entirely surprising that the same transcription factor regulating canonical UPR also governs the 12-hour rhythms of gene expression. In XBP1 liver-specific knockout mice, ~55% of the 12-hour hepatic transcriptome was abolished, and another ~32% was dampened compared to wild-type mice, while the core circadian clock genes and the vast majority of circadian clock-controlled output genes remained oscillatory with a ~24-hour period (27). These results not only place XBP1s as a central transcriptional regulator of hepatic 12-hour rhythms, but also further support the existence of a dedicated 12-hour oscillator separate from the circadian clock.
XBP1s physical recruitment to the promoter region was only observed for ~550 12-hour genes (27), indicating the existence of additional transcriptional factors for hepatic 12-hour rhythm control. Via bioinformatic and computational analysis, a tripartite regulatory network comprising of E26 transformation-specific (ETS) (including GABPA/GABPB1), Basic Leucine Zipper Domain (bZIP) (including XBP1s/ATF6/CRELD2) and Nuclear transcription factor Y (NFY) (including NFYA/NFYB) family of transcription factors was recently proposed to be the foundation of the mammalian hepatic 12-hour oscillator (46), although the detailed molecular circuits have yet to be elucidated. Within the network, XBP1us (unspliced form of XBP1) was proposed to provide the negative feedback required for sustaining 12-hour oscillations by antagonizing XBP1s-mediated transcription activation (46, 53), while XBP1s positively transcriptionally regulates its own 12-hour expression as well as the 12-hour expression of Ire1α, thereby completing the positive feed forward loops (45) (Figure 1B). In a recent study, Steroid Receptor Coactivator-3 (SRC-3) was further discovered to be a co-activator for XBP1s and transcriptionally regulates the hepatic 12-hour transcriptome and metabolome (54). Future experimental studies with new gain or loss-of-function genetic models combined with mathematical modeling are needed to reveal the complete gene regulatory network underlying the hepatic 12-hour oscillator.
12-hour hepatic mRNA and protein metabolism
Gene Ontology (GO) analysis of the mouse hepatic 12-hour transcriptome revealed top-enriched pathways implicated in the entire central dogma information flow (CEDIF) process, ranging from transcription initiation and elongation, mRNA processing and export, ribosome biogenesis, translation initiation, to protein folding, processing and sorting in the endoplasmic reticulum (ER) and Golgi, and include both anabolic and catabolic processes (27). Intriguingly, detailed phase analysis further unveiled an ~1 hour phase delay from genes involved in mRNA processing (peaking at CT1 and CT13) to those participating in the protein secretory pathway (peaking at CT2 and CT14), consistent with the unidirectional genetic information flow (27). In addition, 12-hour genes involved in metabolic pathways auxiliary to CEDIF were further uncovered (41), including those involved in purine and pyrimidine de novo synthesis and hexosamine biosynthesis pathway. The former provide precursors for mRNA synthesis, while the latter is a branch of glycolysis responsible for the production of the key substrate UDP-GlcNAc for protein glycosylation in the ER and Golgi (55). Importantly, the 12-hour hepatic metabolic enzyme expression closely matches those of 12-hour hepatic metabolome, including various nucleosides, nucleotides and UDP-N-Acetylamino sugars (21, 27, 44, 56). The 12-hour rhythms of metabolites are also temporally coupled with the CEDIF. One prominent example is the observation that the levels of various ribonucleosides/ribonucleotides peak at CT10 and CT22, ~3 hours preceding the peaking of transcription and mRNA processing at CT13 and CT1 (46).
While the regulation of proteostasis by XBP1s is well-established, the control of mRNA metabolism by XBP1s and the mechanistic link between mRNA and protein homeostasis remain poorly characterized. In a recent study, Dion et al identified a previously unappreciated hepatic XBP1/SON axis that underlies the coordinated 12-hour rhythms of mRNA and protein metabolism (45). SON is the scaffold protein of nuclear speckle, a membraneless organelle in the nucleus that plays important roles in nearly all aspects of mRNA metabolism (57–59). As a direct transcriptional target of XBP1s, hepatic SON exhibits a robust 12-hour rhythm at both the mRNA and protein level. As the scaffold protein of liquid condensates, the 12-hour oscillation of SON expression in turn generates a 12-hour rhythm of nuclear speckle liquid-liquid phase separation (LLPS) dynamics that alternates between a round/stagnant (at CT2 and CT14) and a more diffuse/fluid (at CT8 and CT20) state. The 12-hour nuclear speckle LLPS further drives a global 12-hour rhythm of nuclear speckle-chromatin interactions, uncoupled from the transcriptional state of individual genes. In XBP1 liver-specific knockout mice, the oscillations of SON expression, nuclear speckle LLPS dynamics and nuclear speckle-chromatin interactions were all dampened and very interestingly, shortened to ~10 hour. Causally, increasing SON expression (by 1.5~2-fold) is sufficient to give rise to more diffuse and fluid nuclear speckles, increases their interactions with chromatin, preferentially amplifies UPR at the transcriptional level and reduces protein aggregates formation in the ER, while the converse biological effects are observed following reduced SON expression. The fact that Xbp1s expression itself is also augmented in response to SON overexpression indicates the existence of a positive feed-forward loop connecting temporal nuclear speckle LLPS dynamics, mRNA metabolism and proteostasis transcriptional control (45).
To explain the salient feature that the acrophases of 12-hour cycling mRNA and protein metabolism genes are strongly enriched around sunrise and sunset, a vehicle-cargo hypothesis was proposed (27). It was argued that the 12-hour rhythm accommodates increased demands for gene expression and processing at the two biological ‘rush hours’ via elevating the global traffic capacity of the central dogma information flow. This connects and tunes rates of mRNA, protein and lipid metabolism (see later) to the 12-hour cycle of metabolic stress (thus acting as the vehicle). The circadian clock, on the other hand, likely dictates the particular genes/gene products processed at each rush hour (thus acting as the cargo) (27). It was further conjectured that having increased nuclear speckle fluidity at early morning and early afternoon enables the mammals to anticipate, and subsequently to rapidly turn on UPR genes to cope with heightened metabolic stress associated with transition periods later at sunrise and sunset. The hypersensitivity of proteostasis gene expression to nuclear speckle LLPS dynamics would therefore ensure tightly coupled mRNA and protein metabolic processes, which in turn can entail a highly efficient genetic information transfer across multiple compartments within the cell (45).
12-hour hepatic lipid metabolism
Besides regulating mRNA metabolism and proteostasis, XBP1s (and UPR in general) also modulates hepatic lipid metabolism (60). Over the past years, many studies have demonstrated that activating XBP1s and other UPR pathways protect against hepatic steatosis (61–64). One such mechanism is through the regulation of membrane fluidity by controlling the fatty acyl composition of phospholipids. For example, Lysophosphatidylcholine Acyltransferase 3 (Lpcat3) gene expression, which promotes the preferential incorporation of polyunsaturated fatty acids into phosphatidylcholines (PC) in the ER membrane, is increased during high fat diet-induced ER stress (21, 65). Elevated level of polyunsaturated PC in turn increases the ER membrane fluidity and ameliorates hepatic inflammation to attenuate the original ER stress (65). Interestingly, robust 12-hour rhythms of Lpcat3 gene expression and levels of various 2-Lyso-PC species (LPCAT3 conjugates 2-Lyso-PC and unsaturated Acyl-CoA to form PC) were identified in mouse liver (21, 42, 44). In addition, many fatty acid desaturases and elongases such as Scd1 and Elovl6 also exhibit hepatic 12-hour rhythms of gene expression (21, 42). Since reduced fluidity of membrane limits lateral diffusion of molecules within the lipid bilayer, many of which are rate-limiting enzymes involved in diverse metabolic processes in ER, mitochondria, and Golgi apparatus (66, 67), the 12-hour rhythm of lipid composition change is expected to have profound influences on systemic metabolism, and much of the biological significance still awaits to be discovered.
In contrast to wild-type mice, liver specific ablation of XBP1 dampened the hepatic 12-hour oscillation of Lpcat3 gene expression, reduced the hepatic polyunsaturated PC level, leading to a drastic reduction of membrane fluidity and impaired lipid metabolism. As a result, XBP1 liver-specific knockout mice exhibited a much accelerated non-alcoholic fatty liver disease (NAFLD) and liver aging phenotype, concomitant with impaired glucose tolerance and hyperinsulinemia (42). Since the impairment of 12-hour transcriptome precedes the manifestation of the NAFLD phenotype in XBP1 liver-specific knockout mice, it suggests that the loss of 12-hour oscillator is likely a cause, not just a consequence of NAFLD.
One common feature shared among the 12-hour regulation of hepatic mRNA, protein and lipid metabolism is the impact on phase behaviors. 12-hour Son and Lpcat3 expression dictates LLPS dynamics of nuclear speckle condensates and phase behaviors of lipid bilayers, respectively. The mobility (or fluidity) change of these diverse molecules in turn can sense the metabolic stress, transduce the signal and modulate the rates of genetic information transfer: viscous ER membrane rich in saturated fatty acid can activate and transduce UPR (68), while fluid nuclear speckle speeds up transcription and mRNA processing rates of proteostasis genes (45). While it is too early to generalize the concept that the 12-hour oscillator controls genetic information transfer via temporally coordinating phase behaviors across diverse organelles and cellular compartments (45), it remains a tantalizing hypothesis awaiting to be explored further.
Compared to mice, little is known about the role of 12-hour rhythms in regulating hepatic metabolism in humans. It was previously reported that the downregulation of the average 12-hour gene expression is associated with the human progression to hepatic steatosis and nonalcoholic steatohepatitis (NASH), although whether the 12-hour rhythms are also altered in human NAFLD subjects remains to be determined (21, 69). Although the field of 12 ultradian rhythm is still in its infancy, these data have nonetheless unveiled an emerging role of the mammalian hepatic 12-hour oscillator in maintaining metabolic homeostasis and protecting against metabolic syndromes.
Metabolic (dys)-regulation of hepatic 12-hour rhythm: lessons from feeding studies
Most studies on biorhythms were performed on mice fed ad libitum. Since it is well-established that peripheral circadian rhythms can be entrained by feeding cues (70), several studies were undertaken to investigate whether different feeding regimens can also affect 12-hour rhythms of gene expression and metabolism. Under food deprivation condition (after a complete night of fasting), both 12-hour rhythms of hepatic Xbp1s mRNA and phospho-IRE1α were maintained with similar phases (34). Similarly, hepatic 12-hour rhythmic of XBP1s and its co-activator SRC-3 chromatin occupancy were also maintained under 16-hour food deprivation condition (54). On the contrary, restricted feeding to 8-hour during the day when mice are normally asleep and not eating abolished the hepatic 12-hour rhythms of four UPR genes tested (they were actually converted to circadian rhythms) (23). Consistent with this observation, measuring the real-time respiratory exchange ratio (RER) of mice fed either ad libitum or with an 8-hour daytime restricted feeding regimen revealed drastic differences (21). Under ad libitum condition, robust superimposed 12-hour RER oscillation were revealed by the eigenvalue/pencil method as the second harmonic after the dominant circadian rhythm. By contrast, restricted feeding significantly dampened the 12-hour RER rhythmic while maintaining their phases (21). Although the exact mechanisms by which miscued feeding impairs 12-hour rhythms remains elusive, it is likely through altering the ER stress and UPR. As a matter of fact, both tunicamycin and glucose deprivation can reset and synchronize the 12-hour rhythms (but not circadian rhythms) in mouse embryonic fibroblasts (MEF) in vitro by activating XBP1s (21). Together, these results suggest that 12-hour rhythms are cell-autonomous and can persist in the absence of environmental cues like food and light, but in the meantime, they are sensitive to wrongful cues that are misaligned with the normal diel physiological cycle of animals.
Evolutionary origin of the mammalian 12-hour oscillator: co-option of the circatidal clock to adapt to terrestrial life?
The mammalian 12-hour rhythm is reminiscent of the ~12-hour circatidal rhythms of coastal and estuarine animals that modulate their behavior in tune to the ~12.4-hour ebb and flow of the tides (71, 72). Although the mechanisms underlying the marine circatidal rhythms remain an open field of research, more and more studies favor the independent circatidal clock hypothesis (73). In the crustacean E. pulchra, the 12-hour circatidal clock is dissociated from the circadian timekeeping system (74). In circatidal mangrove crickets, 12-hour rhythms persist in constant darkness, even after the removal of the optic lobe. Furthermore, these 12-hour rhythms are intact even when expression of components of the circadian TTFL such as Clock or per are reduced through siRNA-mediated knockdown (75–77).
Comparing 12-hour transcriptomes of circatidal animals and mouse liver revealed stunning overlap. In E. pulchra, robust circatidal rhythms for 10 mitochondrial DNA (mtDNA)-encoded protein-coding genes (Mt-Nd1~6, Mt-Cox1~3 and Cytb) were previously described (78). Remarkably, 12-hour rhythms of global mtDNA-encoded gene transcription is conserved in mouse liver (21). In another study, Pan et al. compared the murine hepatic 12-hour transcriptome with circatidal transcriptomes of two marine species: aposymbiotic sea anemone Aiptasia diaphaha (79) and limpet Cellana rota (80), and found substantial convergence that are enriched in CEDIF pathways, including Xbp1, Gabpa, and Son (27, 45). 12-hour rhythms of gene expression involved in CEDIF were also recently identified in the circatidal mangrove cricket (81). Collectively, these data suggest that the mammalian 12-hour oscillator evolved from the circatidal clock of coastal and estuarine animals (21, 27, 44). This hypothesis is further supported by recent reports of a higher conservation of chromosomal location (20) and an older average evolutionary age for 12-hour over circadian genes (82).
How the 12-hour oscillator provides an evolutionary advantage for terrestrial mammals that no long live around the coast is an important question. One plausible explanation is that the 12-hour oscillator in terrestrial mammals is co-opted to adapt to the two main metabolic shifts at the two transition periods (44). At the subjective dawn (ZT12 in mice), the prolonged absence of energy intake combined with significant energy expenditure during sleep gives rise to a peak of energy ‘shortage’ and thus a greater demand for catabolic pathways to produce more energy; while at the subjective dusk (ZT0 in mice), excessive energy intake during the day concomitant with decreasing energy expenditure (diurnal animals in the wild-type start to rest after sunset) culminates in a peak of energy ‘excess’ and anabolic activity to store energy. Both energy ‘shortage’ (such as glucose deprivation) and energy ‘excess’ (such as high carbohydrate availability) can activate IRE1α-XBP1s signaling to elicit UPR (46, 83–85) (Figure 2). The mammalian 12-hour oscillator, therefore, may provide increased metabolic capacity at a 12-hour interval to anticipate and respond to the shifts in metabolic demands and availability at dawn and dusk.
Figure 2. A diagram summarizing the origin, regulation, function and evolutionary conservation of mammalian hepatic 12-hour rhythms.
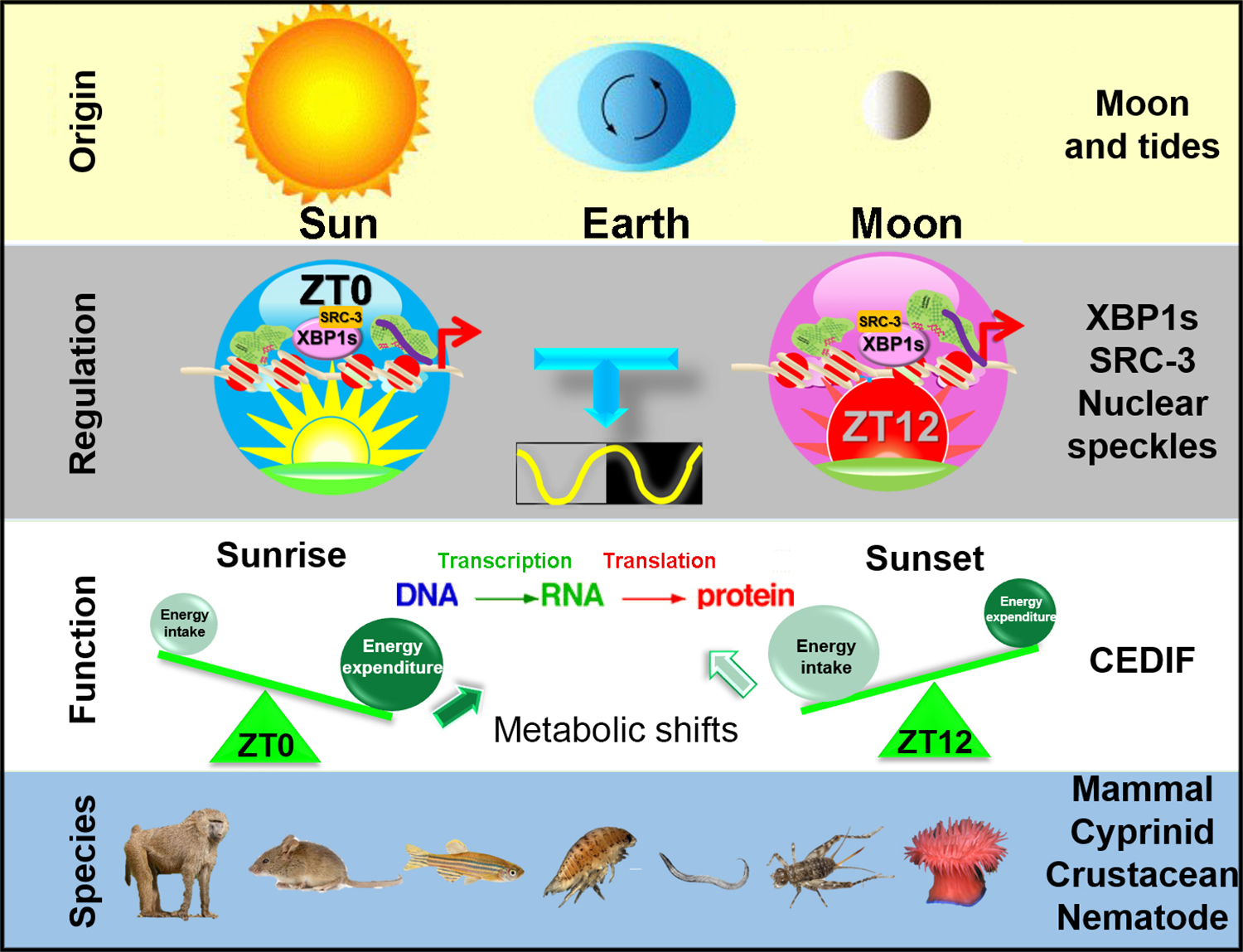
Please refer to the main text for a detailed description of each section
Hepatic 8-hour and other harmonic oscillations with shorter periods
As previously mentioned, hepatic transcriptome and metabolome oscillations predominantly cycle at the harmonic frequency of the circadian rhythm. Besides the 12-hour rhythms, oscillations with periods close to 8-hour (3rd harmonic), 6-hour (4th harmonic) and even 4~5-hour (5th and 6th harmonic) were also revealed (21, 23, 44, 86). Generally, oscillations with shorter periods tend to have smaller amplitudes on average (21). It is important to note that ~4-hour oscillations are near the Nyquist limit of sampling frequency of most temporal transcriptome studies reported so far and therefore the possibility that many of them may arise from technical artifacts or noise cannot be precluded (44). In this section, we will briefly discuss the few literatures on these shorter harmonic oscillations. However, since these shorter oscillations are rarely studied and the regulation and functions of them are still poorly understood, many of the discussions herein may be more speculative than definitive.
8-hour rhythms of gene expression are the third most prevalent populations in mouse liver after circadian and 12-hour rhythms (21, 23). Although more than 3,000 hepatic genes were found to exhibit superimposed 8-hour components, only ~ 1,000 (ranging from ~910 to 1,403 depending on the dataset) hepatic genes exhibit dominant 8-hour oscillations as revealed by the eigenvalue/pencil method (21, 27) (Figure 3A). In the 1-hour high resolution hepatic transcriptome dataset (23), post-hoc GO analysis of 1,403 genes with dominant 8-hour oscillations uncovered in (21) revealed top-enriched pathways in proteosome, ribosome and oxidative phosphorylation (21) (Figure 3B). In addition, post-hoc eigenvalue/pencil analysis of the 1-hour resolution hepatic metabolome dataset (56) revealed 16 metabolites with dominant 8-hour oscillations (Figure 3C). Very intriguingly, many of them are amino acids/amino acid analogs (tyrosine, homocysteic acid, thiamine, aminobenzoates and glutamate) or intermediates involved in oxidative phosphorylation and TCA cycle (citrate, NADH−, oxaloacetate and coenzyme A). 5-methylthioadenosine is further involved in the methionine salvage pathway (87). The high concordance of the enriched biological pathways between hepatic 8-hour genes and metabolites imply that these are real oscillations with potential high biological importance, rather than technical artifact or noises.
Figure 3. Mammalian 8-hour transcriptome and metabolome are prevalent.
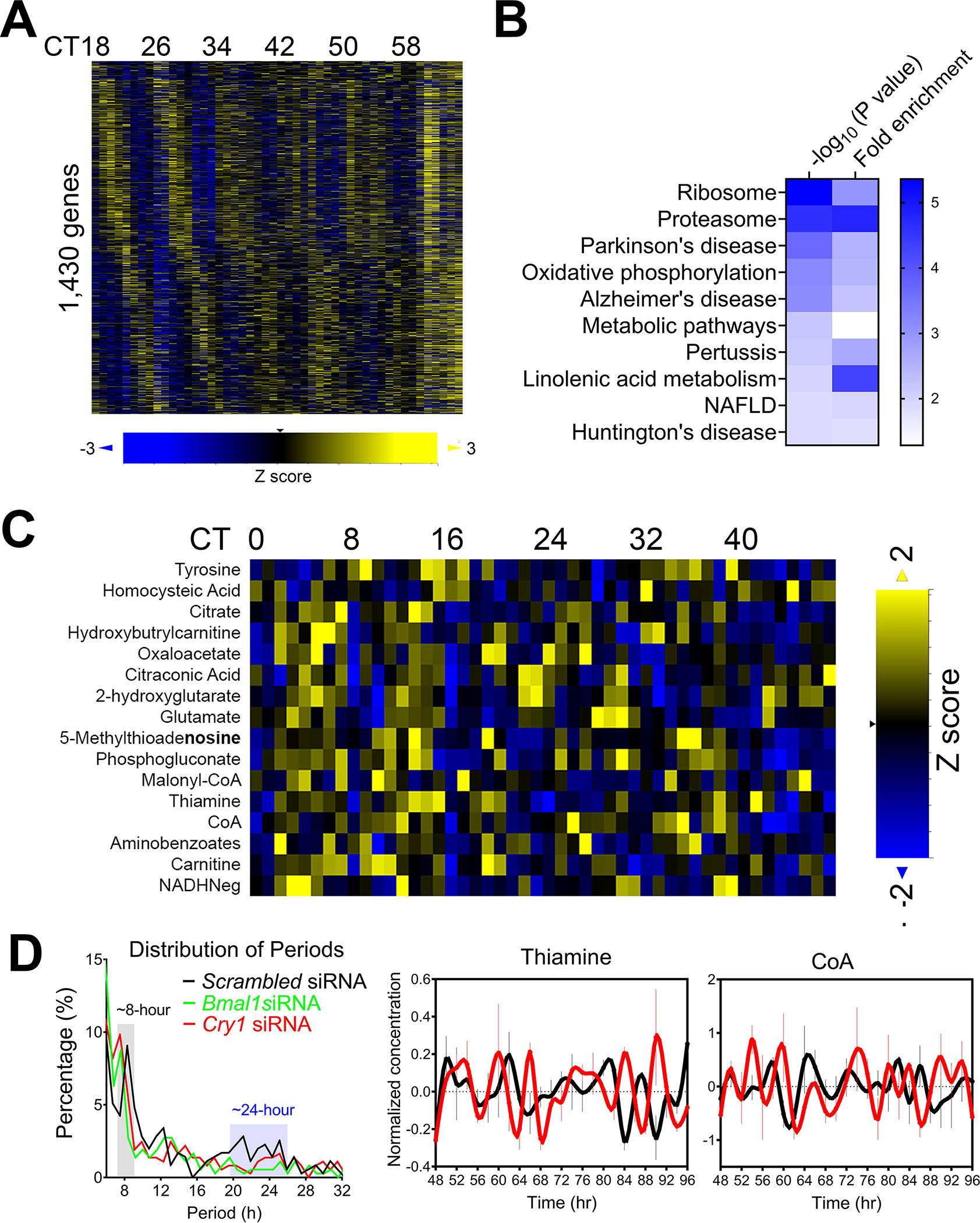
(A) Heat map of hepatic mRNA exhibiting dominant 8-hour oscillation ranked by phase. (B) GO analysis of top enriched KEGG pathways with p value and fold enrichment revealed by David analysis. (C) Heat map of hepatic metabolite exhibiting dominant 8-hour oscillation ranked by phase. (D) U2OS cells are transfected with different siRNAs and synchronized by dexamethasone. Distribution of periods of oscillations revealed by eigenvalue/pencil method under different condition (left) and representative level of thiamine and coenzyme A in cells with scrambled (black) or Cry1 (red) siRNAs.
8-hour rhythms of gene expression are also present in serum-synchronized MMH-D3 hepatocytes in vitro (27), indicating that they are not solely driven by physiological or environmental cues but likely regulated by a cell-autonomous oscillator. So, are 8-hour rhythms established by an independent oscillator or are they under circadian clock regulation? Due to the low resolution of publicly available hepatic transcriptome dataset from circadian clock deficient mice, it is technically challenging to systemically examine whether hepatic 8-hour rhythms of gene expression persist in the absence of a functional circadian clock in vivo. Alternatively, a high resolution (1-hour) metabolome dataset in control and circadian clock genes-knocked down U2OS cells may provide some answers (56). In dexamethasone-synchronized control U2OS cells, eigenvalue/pencil method revealed prevalent circadian, ~12-hour and ~8-hours metabolites as previously reported (Figure 3D) (27). In the presence of circadian clock regulator BMAL1 or CRY1 knocking-down, circadian metabolites are largely abolished as expected; while both ~12-hour and ~8-hour metabolites remain, with the latter including thiamine and coenzyme A that also oscillate with ~8-hour period in mouse liver (Figure 3D) (27, 56). Although irrelevant to hepatic biology, a recent study also reported that incremental growth lines in molar dentin exhibited 8-hour ultradian rhythm in both wild-type and BMAL1 knockout mice (88). Similarly, in the fungus Neurospora crass, many ~7-hour third harmonic transcripts (the circadian rhythm is ~21-hour in Neurospora) were kept after the knock-out of a circadian output regulator (28). Although much more future studies are needed to reveal the exact mechanism by which 8-hour rhythms are regulated, emerging evidence does support a tantalizing possibility that there is a cell-autonomous 8-hour oscillator as well (44).
Literatures on cell autonomous harmonic oscillations with periods shorter than 8-hour are scarcer. In one recent endeavor, Ghenim et al. uncovered a cell-autonomous ~4-hour ultradian rhythm of protein recycling in diverse mammalian cell types, independent from the circadian clock (89). Using lens-free microscopy to make measurements of dry mass (constituted principally of proteins) in diverse types of non-fluorescent and non-synchronized mammalian cells, the authors uncovered that this 4-hour rhythm of dry mass kinetics occurs during the interphase of the cell cycle. In addition, this 4-hour rhythm is temperature-compensated, a characteristic of biological clock. Since this rhythm can be suppressed by proteasome inhibitors and only observed in proliferating cells, it suggests a 4-hour cycle of protein degradation and re-synthesis in growing cells (89).
Combined with the observed 8-hour hepatic rhythms of gene expression enriched in proteasome and ribosome, and metabolite oscillations of various amino acids, these data together suggest a possible wider role of shorter ultradian rhythms in protein and amino acid recycling. If this is indeed the case, then for what reason? One possibility is that besides the de novo synthesis of proteins necessary for cell growth, the much smaller ultradian rhythms of protein recycling may enable animals to adapt to acute micro-environmental changes at a low cost in energy. Alternatively, these ultradian rhythms of protein recycling may reflect part of a defense system against continuous pathogen exposure (cycles of pathogens degradation and antigen generation). Or it could be a cell-intrinsic mechanism against accumulation of misfolded protein and protein aggregates. A high frequency ‘background’ protein recycling cycle would ensure fresher protein pools are always present, which would significantly reduce the possibility of protein misfolding and maintain long-term protein homeostasis.
If independent oscillators exist that can regulate harmonic oscillations, then how do the periods of different harmonic oscillations always seem to be locked to that of the circadian rhythm? One possibility is that different harmonic oscillators can adjust their periods concordantly with that of the circadian period (the 1st harmonic period) in the presence of varying environmental changes. As previous studies showed that the circadian period can fluctuate in response to pH, oxygen, redox potential and temperature alterations (8, 90–95), it is possible that the periods of faster oscillators of higher harmonics can also change in the same direction proportional to the circadian period change. Alternatively, it is equally possible that there are yet-to-be identified cell-intrinsic mechanisms to “period-lock” the different oscillators. Future studies are needed to further investigate these and other possibilities.
Non-harmonic ultradian rhythms – case studies
Recently, Per1/2 null mice were reported to exhibit ~16-hour ultradian oscillations in AKT phosphorylation and related gene expression. Interestingly, these genes were enriched for mitochondria related functions, prominent among them were genes related to mitochondrial gene expression (ribosomal subunits) and mitochondrial respiration, with representatives from all electron transport chain protein complexes (51). Notably, their phases were aligned, i.e., their expression coordinated, which supports functional significance. Interestingly, ~16-hour rhythms in locomotor activity were reported in Cry1, 2 −/− mice (96) as well as Per1,2 −/− animals (97) under specific experimental settings.
It is possible that these rhythms emerge in the absence of a functional circadian clock, yet experiments performed with Bmal1 null mice under the same conditions, namely constant darkness, do not show such rhythms (51). Hence, the emergence of ultradian rhythms in the absence of Pers might be dependent on the presence of Bmal1. Future studies with triple knockout of Pers1/2 and Bmal1 are expected to clarify this point.
The scarcity of information regarding short rhythms specifically in mouse liver and in general in animals raises the questions whether this stems from technical or biological reasons. In this regard, experiments performed in cultured cells can serve as food for thought regarding the presence and potential mechanism underlying such rhythms. The best documented examples in cultured cells relate to the tumor suppressor p53 and the inflammatory response regulator, NF-kB. Although currently there is no evidence that these short oscillations are prevalent in animals, their presence in cell culture are intriguing. Interestingly, DNA damage-dependent cell autonomous and sustained p53-mdm oscillations were reported (98). These p53 oscillations were highly variable between isogenic cells following DNA-damage, with undamped oscillations for at least 3 days in some cells (more than 10 peaks). The amplitude of the oscillations was much more variable than the period, which was on average ~5.5 hour (99). These repeated p53 pulses in response to DNA-damage were suggested to control cell fate (100).
In the case of NF-kB, it displayed intrinsic oscillatory behavior in cultured cells, with about 1 hour period that is entrainable and translates into functionally related patterns of gene expression (101). While non-oscillatory NF-kB was suggested to alter chromatin accessibility, the function of the oscillatory NF-kB was proposed to maintain the epigenomic state while exploiting existing poised enhancers for inflammatory gene activation (102). Whether p53 or NF-kB oscillations persist in animals’ models or convert to oscillation with longer or shorter period remains an open question. Yet the presence of high frequency oscillation of these type in cultured cells supports the presence of high frequency cell autonomous oscillators.
Limitations and perspective.
As aforementioned, the lion’s share of the literature is centered on 24-hour rhythms and the molecular circadian clock. Recent work supports the presence of 12 and even 8-hour rhythms and their potential molecular underpinning. While much less is known about ultradian rhythms, in particular, in animal models with other frequencies. It is conceivable that ultradian rhythms that are not harmonics of circadian oscillations are less prevalent as there was no evolutionary pressure, namely 24-hour environmental rhythms to preserve them. Having said that, it does not exclude the concomitant presence of ultradian rhythm that hitherto were uncovered due to either biological or technical reasons as discussed below.
First, it might be that the circadian clock overrides or masks shorter rhythms and that the latter emerge only in the absence of the clock. An example for that, is the presence of ~16-hour rhythms in AKT phosphorylation and related gene expression, in liver of circadian arrhythmic animals, i.e., the Per1/2 null mice.
There might be several technical reasons that might explain our limited knowledge on ultradian rhythms, many of which stem from the fact that, to date, most studies were designed to detect circadian rhythms and consequently were less informative regarding shorter rhythms. This refers to limitations such as, the temporal sampling resolution as well as the readout that was examined.
First, the issue of sampling; around the clock sampling are often done at 4- or 2-hour intervals, and on rare occasions at 1-hour interval. To detect short rhythms, the sampling time should be performed at shorter intervals or preferably by using continuous measurements, especially in the case of shallow rhythms. Along this line, statistical simulations reveal that the conventional 4 hour sampling frequency is reaching the limit of detecting 12-hour oscillations and below the limit of uncovering 8-hour cycling genes (37). Obviously, continues measurements require prior knowledge on the nature of the rhythms and cannot currently be performed in a high throughput manner.
Second, given the nature of molecular circadian clock as a transcription-translation oscillator and that transcriptomic analyses are readily available, they became the benchmark for circadian rhythmicity. The majority of the ‘around the clock’ collections are gene expression datasets. Since gene expression by itself is a time-demanding, it is conceivable that short rhythm might involve rapid processes such as post translation modifications (e.g., protein phosphorylation, acetylation), or changes in metabolite abundance due changes in enzymatic reactions. Indeed, in recent years around the clock acetylome and phospho proteome were performed as well as metabolomics and lipidomics yet the time resolution was mostly 2–4 hour and hence their value is limited in respect to ultradian rhythms. However, we have reasons to believe that with the ongoing technological burst in the field of imaging, new tools that will allow investigators to monitor dynamics of biochemical reactions and kinetics of metabolites at high temporal and spatial resolution using time-lapse imaging will reveal the “hidden reality” of ultradian rhythms in the near future.
Last but not the least, unlike circadian rhythms for which translational aspects in general and specifically in respect to liver functions have been intensively studied over several decades now, very little is known regarding the functional roles and the translational implications of disrupted harmonic or non-harmonic ultradian rhythms. For the former, one can speculate that given the tight interactions among harmonic oscillations, it is possible that some translational manifestations of disrupted circadian rhythms could stem from alternation in related specific harmonic oscillations. In addition, in light of the strong implications of ~4, ~8 and ~12-hour ultradian rhythms in the regulation of proteostasis, it is imperative for future studies to determine whether diseases with dysregulated proteostasis have altered ultradian rhythms, and more importantly, to establish the potential causality between ultradian rhythm dysregulation and disease progressions.
Acknowledgement:
We apologize for any potential omission of relevant works and citations due to space constraints. B.Z. is supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number DP2GM140924. G.A. is supported by a grant from the European Research Council (ERC-2017 CIRCOMMUNICATION 770869), Abisch Frenkel Foundation for the Promotion of Life Sciences, Adelis Foundation, Susan and Michael Stern. B.Z and G.A have no conflict of interest to declare.
Reference:
- 1.Rensing L, Meyer-Grahle U, Ruoff P. Biological timing and the clock metaphor: oscillatory and hourglass mechanisms. Chronobiol Int 2001;18:329–369. [DOI] [PubMed] [Google Scholar]
- 2.Gliech CR, Holland AJ. Keeping track of time: The fundamentals of cellular clocks. Journal of Cell Biology 2020;219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schibler U The daily rhythms of genes, cells and organs. Biological clocks and circadian timing in cells. EMBO Rep 2005;6 Spec No:S9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aviram R, Adamovich Y, Asher G. Circadian Organelles: Rhythms at All Scales. Cells 2021;10:2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 2017;18:164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol 2014;24:90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol 2002;12:1574–1583. [DOI] [PubMed] [Google Scholar]
- 8.Adamovich Y, Ladeuix B, Golik M, Koeners MP, Asher G. Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1α. Cell Metabolism 2017;25:93–101. [DOI] [PubMed] [Google Scholar]
- 9.Astiz M, Heyde I, Oster H. Mechanisms of Communication in the Mammalian Circadian Timing System. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowalska E, Brown SA. Peripheral clocks: keeping up with the master clock. Cold Spring Harb Symp Quant Biol 2007;72:301–305. [DOI] [PubMed] [Google Scholar]
- 11.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 2010;72:517–549. [DOI] [PubMed] [Google Scholar]
- 12.Reinke H, Asher G. Crosstalk between metabolism and circadian clocks. Nature Reviews Molecular Cell Biology 2019;20:227–241. [DOI] [PubMed] [Google Scholar]
- 13.Guan D, Lazar MA. Interconnections between circadian clocks and metabolism. The Journal of Clinical Investigation 2021;131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asher G, Sassone-Corsi P. Time for Food: The Intimate Interplay between Nutrition, Metabolism, and the Circadian Clock. Cell 2015;161:84–92. [DOI] [PubMed] [Google Scholar]
- 15.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science 2016;354:994–999. [DOI] [PubMed] [Google Scholar]
- 16.Panda S. Circadian physiology of metabolism. Science 2016;354:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cederroth CR, Albrecht U, Bass J, Brown SA, Dyhrfjeld-Johnsen J, Gachon F, Green CB, et al. Medicine in the Fourth Dimension. Cell Metabolism 2019;30:238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinke H, Asher G. Circadian Clock Control of Liver Metabolic Functions. Gastroenterology 2016;150:574–580. [DOI] [PubMed] [Google Scholar]
- 19.El-Athman R, Knezevic D, Fuhr L, Relogio A. A Computational Analysis of Alternative Splicing across Mammalian Tissues Reveals Circadian and Ultradian Rhythms in Splicing Events. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genov N, Castellana S, Scholkmann F, Capocefalo D, Truglio M, Rosati J, Turco EM, et al. A Multi-Layered Study on Harmonic Oscillations in Mammalian Genomics and Proteomics. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu B, Zhang Q, Pan Y, Mace EM, York B, Antoulas AC, Dacso CC, et al. A Cell-Autonomous Mammalian 12 hr Clock Coordinates Metabolic and Stress Rhythms. Cell Metab 2017;25:1305–1319 e1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes ME, Hong HK, Chong JL, Indacochea AA, Lee SS, Han M, Takahashi JS, et al. Brain-specific rescue of Clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet 2012;8:e1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet 2009;5:e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atwood A, DeConde R, Wang SS, Mockler TC, Sabir JS, Ideker T, Kay SA. Cell-autonomous circadian clock of hepatocytes drives rhythms in transcription and polyamine synthesis. Proc Natl Acad Sci U S A 2011;108:18560–18565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasanpour M, Mitricheva E, Logothetis N, Noori HR. Intensive longitudinal characterization of multidimensional biobehavioral dynamics in laboratory rats. Cell Reports 2021;35:108987. [DOI] [PubMed] [Google Scholar]
- 26.Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, Venkataraman A, et al. Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metab 2017;25:1206. [DOI] [PubMed] [Google Scholar]
- 27.Pan Y, Ballance H, Meng H, Gonzalez N, Kim S-M, Abdurehman L, York B, et al. 12-h clock regulation of genetic information flow by XBP1s. PLOS Biology 2020;18:e3000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ananthasubramaniam B, Diernfellner A, Brunner M, Herzel H. Ultradian Rhythms in the Transcriptome of Neurospora crassa. iScience 2018;9:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith LM, Motta FC, Chopra G, Moch JK, Nerem RR, Cummins B, Roche KE, et al. An intrinsic oscillator drives the blood stage cycle of the malaria parasite Plasmodium falciparum. Science 2020;368:754–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Veen DR, Gerkema MP. Unmasking ultradian rhythms in gene expression. FASEB J 2017;31:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blum ID, Zhu L, Moquin L, Kokoeva MV, Gratton A, Giros B, Storch K-F. A highly tunable dopaminergic oscillator generates ultradian rhythms of behavioral arousal. eLife 2014;3:e05105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das B, de Bekker C. Time-course RNASeq of Camponotus floridanus forager and nurse ant brains indicate links between plasticity in the biological clock and behavioral division of labor. BMC Genomics 2022;23:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westermark PO, Herzel H. Mechanism for 12 hr rhythm generation by the circadian clock. Cell Rep 2013;3:1228–1238. [DOI] [PubMed] [Google Scholar]
- 34.Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab 2010;11:47–57. [DOI] [PubMed] [Google Scholar]
- 35.Ballance H, Zhu B. Revealing the hidden reality of the mammalian 12-h ultradian rhythms. Cellular and Molecular Life Sciences 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mofatteh M, Echegaray-Iturra F, Alamban A, Dalla Ricca F, Bakshi A, Aydogan MG. Autonomous clocks that regulate organelle biogenesis, cytoskeletal organization, and intracellular dynamics. eLife 2021;10:e72104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antoulas AC, Zhu B, Zhang Q, York B, O’Malley BW, Dacso CC. A novel mathematical method for disclosing oscillations in gene transcription: A comparative study. PLoS One 2018;13:e0198503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 2010;25:372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang R, Su Z. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics 2010;26:i168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. J Biol Rhythms 2014;29:391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan Y, Ballance H, Schnytzer Y, Chen X, Levy O, Coarfa C, Zhu B. 12h-clock control of central dogma information flow by XBP1s. bioRxiv 2019:559039. [Google Scholar]
- 42.Meng H, Gonzales NM, Lonard DM, Putluri N, Zhu B, Dacso CC, York B, et al. XBP1 links the 12-hour clock to NAFLD and regulation of membrane fluidity and lipid homeostasis. Nature Communications 2020;11:6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012;338:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu B, Dacso CC, O’Malley BW. Unveiling “Musica Universalis” of the Cell: A Brief History of Biological 12-Hour Rhythms. J Endocr Soc 2018;2:727–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dion W, Ballance H, Lee J, Pan Y, Irfan S, Edwards C, Sun M, et al. Four-dimensional nuclear speckle phase separation dynamics regulate proteostasis. Science Advances 2022;8:eabl4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu B. Decoding the function and regulation of the mammalian 12h-clock. Journal of Molecular Cell Biology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dowse H, Umemori J, Koide T. Ultradian components in the locomotor activity rhythms of the genetically normal mouse, Mus musculus. J Exp Biol 2010;213:1788–1795. [DOI] [PubMed] [Google Scholar]
- 48.Ray S, Valekunja UK, Stangherlin A, Howell SA, Snijders AP, Damodaran G, Reddy AB. Circadian rhythms in the absence of the clock gene Bmal1. Science 2020;367:800–806. [DOI] [PubMed] [Google Scholar]
- 49.Yang G, Chen L, Grant GR, Paschos G, Song WL, Musiek ES, Lee V, et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med 2016;8:324ra316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A 2007;104:3342–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aviram R, Dandavate V, Manella G, Golik M, Asher G. Ultradian rhythms of AKT phosphorylation and gene expression emerge in the absence of the circadian clock components Per1 and Per2. PLOS Biology 2022;19:e3001492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hetz C, Zhang K, Kaufman RJ. Mechanisms, regulation and functions of the unfolded protein response. Nature Reviews Molecular Cell Biology 2020;21:421–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol 2006;172:565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meng H, Gonzales NM, Jung SY, Lu Y, Putluri N, Zhu B, Dacso CC, et al. Defining the mammalian coactivation of hepatic 12-h clock and lipid metabolism. Cell Reports 2022;38:110491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horn M, Denzel SI, Srinivasan B, Allmeroth K, Schiffer I, Karthikaisamy V, Miethe S, et al. Hexosamine Pathway Activation Improves Protein Homeostasis through the Integrated Stress Response. iScience 2020;23:100887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, Venkataraman A, et al. Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metabolism 2017;25:961–974.e964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA. Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell 2010;21:650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ilik İA, Malszycki M, Lübke AK, Schade C, Meierhofer D, Aktaş T. SON and SRRM2 are essential for nuclear speckle formation. eLife 2020;9:e60579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fei J, Jadaliha M, Harmon TS, Li ITS, Hua B, Hao Q, Holehouse AS, et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci 2017;130:4180–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moncan M, Mnich K, Blomme A, Almanza A, Samali A, Gorman AM. Regulation of lipid metabolism by the unfolded protein response. J Cell Mol Med 2021;25:1359–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell 2010;21:2975–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell 2008;15:829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, et al. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J 2011;30:1357–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herrema H, Zhou Y, Zhang D, Lee J, Salazar Hernandez MA, Shulman GI, Ozcan U. XBP1s Is an Anti-lipogenic Protein. J Biol Chem 2016;291:17394–17404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rong X, Albert CJ, Hong C, Duerr MA, Chamberlain BT, Tarling EJ, Ito A, et al. LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition. Cell Metab 2013;18:685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanders CR, Hutchison JM. Membrane properties that shape the evolution of membrane enzymes. Current Opinion in Structural Biology 2018;51:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edidin M Lipids on the frontier: a century of cell-membrane bilayers. Nature Reviews Molecular Cell Biology 2003;4:414–418. [DOI] [PubMed] [Google Scholar]
- 68.Volmer R, van der Ploeg K, Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proceedings of the National Academy of Sciences 2013;110:4628–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahrens M, Ammerpohl O, von Schonfels W, Kolarova J, Bens S, Itzel T, Teufel A, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab 2013;18:296–302. [DOI] [PubMed] [Google Scholar]
- 70.Manella G, Sabath E, Aviram R, Dandavate V, Ezagouri S, Golik M, Adamovich Y, et al. The liver-clock coordinates rhythmicity of peripheral tissues in response to feeding. Nature Metabolism 2021;3:829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilcockson D, Zhang L. Circatidal clocks. Curr Biol 2008;18:R753–R755. [DOI] [PubMed] [Google Scholar]
- 72.Andreatta G, Tessmar-Raible K. The Still Dark Side of the Moon: Molecular Mechanisms of Lunar-Controlled Rhythms and Clocks. J Mol Biol 2020;432:3525–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barford E Biological clocks defy circadian rhythms. Nature 2013. [Google Scholar]
- 74.Zhang L, Hastings MH, Green EW, Tauber E, Sladek M, Webster SG, Kyriacou CP, et al. Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr Biol 2013;23:1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takekata H, Matsuura Y, Goto SG, Satoh A, Numata H. RNAi of the circadian clock gene period disrupts the circadian rhythm but not the circatidal rhythm in the mangrove cricket. Biol Lett 2012;8:488–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takekata H, Numata H, Shiga S, Goto SG. Silencing the circadian clock gene Clock using RNAi reveals dissociation of the circatidal clock from the circadian clock in the mangrove cricket. J Insect Physiol 2014;68:16–22. [DOI] [PubMed] [Google Scholar]
- 77.Takekata H, Numata H, Shiga S. The circatidal rhythm persists without the optic lobe in the mangrove cricket Apteronemobius asahinai. J Biol Rhythms 2014;29:28–37. [DOI] [PubMed] [Google Scholar]
- 78.O’Neill JS, Lee KD, Zhang L, Feeney K, Webster SG, Blades MJ, Kyriacou CP, et al. Metabolic molecular markers of the tidal clock in the marine crustacean Eurydice pulchra. Curr Biol 2015;25:R326–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sorek M, Schnytzer Y, Ben-Asher HW, Caspi VC, Chen CS, Miller DJ, Levy O. Setting the pace: host rhythmic behaviour and gene expression patterns in the facultatively symbiotic cnidarian Aiptasia are determined largely by Symbiodinium. Microbiome 2018;6:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schnytzer Y, Simon-Blecher N, Li J, Ben-Asher HW, Salmon-Divon M, Achituv Y, Hughes ME, et al. Tidal and diel orchestration of behaviour and gene expression in an intertidal mollusc. Scientific Reports 2018;8:4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Satoh A, Terai Y. Circatidal gene expression in the mangrove cricket Apteronemobius asahinai. Sci Rep 2019;9:3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Castellana S, Mazza T, Capocefalo D, Genov N, Biagini T, Fusilli C, Scholkmann F, et al. Systematic Analysis of Mouse Genome Reveals Distinct Evolutionary and Functional Properties Among Circadian and Ultradian Genes. Front Physiol 2018;9:1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deng Y, Wang ZV, Tao C, Gao N, Holland WL, Ferdous A, Repa JJ, et al. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J Clin Invest 2013;123:455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shao M, Shan B, Liu Y, Deng Y, Yan C, Wu Y, Mao T, et al. Hepatic IRE1alpha regulates fasting-induced metabolic adaptive programs through the XBP1s-PPARalpha axis signalling. Nat Commun 2014;5:3528. [DOI] [PubMed] [Google Scholar]
- 85.Lipson KL, Fonseca SG, Ishigaki S, Nguyen LX, Foss E, Bortell R, Rossini AA, et al. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab 2006;4:245–254. [DOI] [PubMed] [Google Scholar]
- 86.van der Veen DR, Gerkema MP. Unmasking ultradian rhythms in gene expression. The FASEB Journal 2017;31:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Albers E Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5′-methylthioadenosine. IUBMB Life 2009;61:1132–1142. [DOI] [PubMed] [Google Scholar]
- 88.Ono R, Koike N, Inokawa H, Tsuchiya Y, Umemura Y, Yamamoto T, Kanamura N, et al. Incremental Growth Lines in Mouse Molar Dentin Represent 8-hr Ultradian Rhythm. Acta Histochem Cytochem 2019;52:93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghenim L, Allier C, Obeid P, Herve L, Fortin JY, Balakirev M, Gidrol X. A new ultradian rhythm in mammalian cell dry mass observed by holography. Sci Rep 2021;11:1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 2010;330:379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee SK, Achieng E, Maddox C, Chen SC, Iuvone PM, Fukuhara C. Extracellular low pH affects circadian rhythm expression in human primary fibroblasts. Biochem Biophys Res Commun 2011;416:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu Y, Tang D, Liu N, Xiong W, Huang H, Li Y, Ma Z, et al. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab 2017;25:73–85. [DOI] [PubMed] [Google Scholar]
- 93.Peek CB, Levine DC, Cedernaes J, Taguchi A, Kobayashi Y, Tsai SJ, Bonar NA, et al. Circadian Clock Interaction with HIF1alpha Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab 2017;25:86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pei J-F, Li X-K, Li W-Q, Gao Q, Zhang Y, Wang X-M, Fu J-Q, et al. Diurnal oscillations of endogenous H2O2 sustained by p66Shc regulate circadian clocks. Nature Cell Biology 2019;21:1553–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu Y, Tang D, Liu N, Xiong W, Huang H, Li Y, Ma Z, et al. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metabolism 2017;25:73–85. [DOI] [PubMed] [Google Scholar]
- 96.Putker M, Wong DCS, Seinkmane E, Rzechorzek NM, Zeng A, Hoyle NP, Chesham JE, et al. CRYPTOCHROMES confer robustness, not rhythmicity, to circadian timekeeping. The EMBO Journal 2021;40:e106745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bae K, Weaver DR. Transient, Light-Induced Rhythmicity in mPER-Deficient Mice. Journal of Biological Rhythms 2007;22:85–88. [DOI] [PubMed] [Google Scholar]
- 98.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet 2004;36:147–150. [DOI] [PubMed] [Google Scholar]
- 99.Geva-Zatorsky N, Rosenfeld N, Itzkovitz S, Milo R, Sigal A, Dekel E, Yarnitzky T, et al. Oscillations and variability in the p53 system. Mol Syst Biol 2006;2:2006.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 Dynamics Control Cell Fate. Science 2012;336:1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zambrano S, De Toma I, Piffer A, Bianchi ME, Agresti A. NF-κB oscillations translate into functionally related patterns of gene expression. Elife 2016;5:e09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng QJ, Ohta S, Sheu KM, Spreafico R, Adelaja A, Taylor B, Hoffmann A. NF-κB dynamics determine the stimulus specificity of epigenomic reprogramming in macrophages. Science 2021;372:1349–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]


