Abstract
Rothia species are understudied members of the phylum Actinobacteria and prevalent colonizers of the human and animal upper respiratory tract and oral cavity. The oral cavity, including the palatine tonsils, is colonized by a complex microbial community, which compete for resources, actively suppress competitors and influence host physiology. We analysed genomic data from 43 new porcine Rothia isolates, together with 112 publicly available draft genome sequences of Rothia isolates from humans, animals and the environment. In all Rothia genomes, we identified biosynthetic gene clusters predicted to produce antibiotic non-ribosomal peptides, iron scavenging siderophores and other secondary metabolites that modulate microbe–microbe and potentially microbe–host interactions. In vitro overlay inhibition assays corroborated the hypothesis that specific strains produce natural antibiotics. Rothia genomes encode a large number of carbohydrate-active enzymes (CAZy), with varying CAZy activities among the species found in different hosts, host niches and environments. These findings reveal competition mechanisms and metabolic specializations linked to ecological adaptation of Rothia species in different hosts.
Keywords: antimicrobials, carbohydrate-active enzymes, microbiome, NRPS, Rothia
Data Summary
All datasets generated for this study are included in the manuscript and/or in the Supplementary Files. The whole-genome sequencing data is available at the European Nucleotide Archive (ENA) under BioProject ID PRJEB49523.
Impact Statement.
Rothia species are abundant members of the upper respiratory tract microbiota in humans and animals but are relatively understudied. Like other notable members of the Actinobacteria phylum, Rothia encode novel bioactive compounds in their biosynthetic gene clusters (BGCs) and are an untapped potential source of new antimicrobials. Additionally, our work advances knowledge about the genetics and physiology of Rothia species revealing correlations between BGCs, carbohydrate metabolic activities and the colonization of specific ecological niches.
Introduction
The oropharyngeal cavity including the palatine tonsils is colonized by a complex microbial community influencing pig health and physiology [1]. Rothia bacteria (phylum Actinobacteria) are prevalent members of the upper respiratory tract microbiota of humans [2], pigs [1], rats [3] and birds [4] and early colonizers of the oropharyngeal cavity including the tonsils, tongue and teeth of healthy humans and piglets [5–7]. It is therefore possible that specific Rothia species contribute to the establishment of the microbial communities colonizing healthy animals in early life. Although several different Rothia species have been described, there is little knowledge about genomics in relation to species physiology and their ecological niches.
Rothia bacteria are all Gram-positive rods and to date 14 species have been identified (https://www.bacterio.net/genus/rothia): Rothia aeria [8], Rothia aerolata [9], Rothia amarae [10], Rothia arfidiae [11], Rothia dentocariosa [ 12 ], Rothia endophytica [13], Rothia halotolerans [14], Rothia koreensis [14], Rothia kristinae [14], Rothia marina [15], Rothia mucilaginosa [16], Rothia nasimurium [16], Rothia nasisuis [17] and Rothia terrae [18]. Furthermore, unclassified and uncultured Rothia genomes have been reconstructed from metagenomic data [19]. Members of the Rothia genus, have been isolated from different ecosystems, including environmental samples such as air and water, and host-associated niches including the skin, gut and oral cavity. Microbiota association studies have shown that the genus Rothia mainly correlates with healthy individuals [19–21] whereas specific strains from the species R. aeria , R. dentocariosa and R. mucilaginosa are associated with caries and dental pits, fissures and plaque [22, 23].
Recently, we showed that Rothia nasisuis colonizing the palatine tonsil epithelium of piglets produces the antimicrobial, antiviral and antiparasitic ionophore valinomycin in vivo, via a large multimodule non-ribosomal peptide synthetase (NRPS) enzyme complex encoded by genes tandemly arranged in a biosynthetic gene cluster (BGC) [24]. Biosynthetic gene clusters (BGCs) produce diverse metabolites including natural antimicrobial products that fall into three major metabolite groups: ribosomally synthesized and post-translationally modified peptides (RiPPs), non-ribosomal peptides (NRPs) and polyketides (PKs) [25]. RiPPs form a large heterogeneous group of peptides that are usually classified into post-translationally modified peptides and unmodified peptides [26]. NRPs are relatively small peptides (2–30 amino acids) with hallmarks of nonproteinogenic amino acids and extensive ‘post-translational modifications’. Substrates and intermediates are covalently bound during the assembly pathway and the order of the catalytic domains in the NRPS often parallels the order of their biosynthetic pathway [27, 28]. Finally, PKs are categorized into different classes based on their biochemical mechanisms and enzyme architecture [29]. The presence of specific BGCs in Rothia species can be used as markers to group and distinguish between species, and BGC annotations help to predict ecological traits.
Carbohydrate-active enzymes (CAZy) are widely present in all organisms and play important roles in biological processes and ecological adaptations. CAZy are classified into several families and their annotation can be used to predict the ability of organisms to assemble and break down complex carbohydrates [30], important ecological traits for niche exploitation and colonization of novel environments. Bacteria colonizing different environments, display a range of CAZy metabolic capacities linked to the diversity, structure and composition of carbohydrates available in the habitat [31]. Rothia species in the oral microbiota have been reported as nitrate-reducers [32, 33]. The conversion of nitrate to nitrite and nitric oxide can promote oral health by reducing the acidification of the saliva and inhibiting species contributing to periodontal disease through the antimicrobial activities of nitric oxide. Furthermore, nitrite is swallowed and taken up into the blood circulation where it is converted into nitric oxide, a signalling molecule, which is reported to improve cardiovascular and metabolic health [32, 34].
Here, we report on genomic analyses of 43 novel Rothia strains that we isolated and cultured from the tonsillar microbiota of piglets between 1 and 3 weeks of age. To place the genomic and phylogenetic analysis of these novel porcine strains in a broader ecological context, we analysed these Rothia strains together with publicly available draft genome sequences from human, animal and environmental Rothia strains. We inferred phylogenetic trees based on genomic data that we annotated with origin (niche) and presence/absence of genes of interest including biosynthetic gene clusters, carbohydrate-active enzymes, and antibiotic resistance and nitrate–nitrite metabolism genes, in order to discover ecological adaptations of Rothia species to different mammalian hosts (pigs and human) and other environments.
Methods
Samples
We sampled the palatine tonsil of six random piglets at two high-health status farms in Catalunya, Spain, 1 week before weaning (timepoint −1) and 3 weeks after weaning (timepoint +3). Tonsil biofilms were collected using Puritan HydraFlock Swabs and stored at −80 °C in transport medium [buffered peptone +15 % (v/v) glycerol].
Rothia spp isolation, identification and culture conditions
Dilutions of the tonsil samples between 10−1 and 10−3 were plated onto Sheep Blood Agar [35] (Becton Dickinson, Heidelberg, Germany) and Brain Heart Infusion (BHI) Agar [36] (Becton Dickinson) media. Single isolates were selected based on the morphology (size, shape and colour) of the colonies and transferred to BHI liquid medium (Fig. S1, available in the online version of this article), incubated at 37 °C in the presence of 5 % CO2. Isolates were purified using Gram staining and the quadrant streak plate method [37] and then stored at −80 °C into cryotubes containing BHI and 15 % (v/v) glycerol.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) was used for screening and identification of pure colonies. The samples were spotted in the MALDI-TOF MS target plate and covered with 1 µl of matrix solution (saturated α-cyano 4-hydroxycinnamic acid in 50 % acetonitrile and 2.5 % trifluoroacetic acid) [38]. The samples were analysed using a MICROFLEX spectrometer (Bruker Daltonics) according to the manufacturer’s recommendations. For each spectrum, 90–100 peaks were compared with reference databases at the Department of Medical Microbiology, University Medical Centre Groningen, the Netherlands. An isolate was considered correctly identified at the species level when at least one MALDI-TOF MS spectrum score was ≥1.9 and at the genus level with a score ≥1.7 [38].
Bacterial indicator strains and culture conditions
The following culture media and bacteria were used in overlay inhibition assays (Table 1) to identify Rothia isolates with antagonistic activity.
Table 1.
Indicator bacteria and culture conditions
|
Indicator micro-organism |
Source/strain |
Culture medium/temp (°C) |
|---|---|---|
|
P1/7 |
THB/37 °C |
|
|
S10 |
THB/37 °C |
|
|
J28 |
THB/37 °C |
|
|
ATCC 6538P |
BHI/37 °C |
|
|
L 4242 |
LB/37 °C |
|
|
DSM 23759 |
THY/37 °C |
|
|
DSM 29126 |
THY/37 °C |
|
|
DSM 20725 |
THY/37 °C |
DSM, Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures; ATCC, American Type Culture Collection; L prefix are from the NAICONS pathogens library, Italy. The strains of S. suis strain P1/7 [36], S. suis strain S10 [37] are zoonotic pathogen and the S. suis strain J28 [38] is less pathogenic, an unencapsulated mutant. Escherichia coli L4242, a ΔtolC mutant derivative from MG1061 [39], THB, Todd Hewitt broth (Becton Dickinson); BHI, Brain Heart Infusion (Becton Dickinson); LB, Luria–Bertani (Becton Dickinson); THY; THB enriched with 0.2% (w/w) yeast extract.
Overlay inhibition assays
Rothia isolates were cultured in BHI broth medium and incubated overnight at 37 °C. Using a 96-well replicator (Boekel Scientific), overnight cultures of Rothia were spotted onto BHI agar media in Petri dishes (Fig. S1, available in the online version of this article) and grown for 18 h. Bacterial colonies were inactivated by exposure to UV light for 20 min and overlayed with 20 ml soft agar (0.75 % w/v agar) containing approximately 1×105 c.f.u. ml−1 of the indicator bacteria (Table 2). Antimicrobial activity was determined by the presence of visible zones of growth inhibition (‘halos’) around the Rothia colonies after overnight incubation [39].
Table 2.
In vitro inhibitory activity of the 43 Rothia sp. obtained from porcine palatine tonsil
|
Indicator bacteria |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
ID |
Species |
Streptococcus suis J28 |
Streptococcus suis S10 |
Streptococcus suis P1/7 |
Streptococcus porci DSM 23759 |
Streptococcus porcinus DSM 20725 |
Streptocuccus parasuis DSM 29126 |
Sthaphylococcus aureus ATCC 6538P |
Escherichia coli L4242 |
BGCs from WGS (AntiSMASH 6.0) |
|
|
159RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
56QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing/RiPPs /terpene |
||
|
63RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing/ |
||
|
67.3RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/NRPS |
||
|
68RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/NRPS |
||
|
110RC1 |
+/- |
− |
− |
+/- |
− |
− |
− |
− |
NRPS |
||
|
136RC1 |
− |
− |
− |
+/- |
− |
− |
− |
− |
NRPS |
||
|
207RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
NRPS |
||
|
8QC2CO |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
15QC2CO |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
22QC2CO |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
28QC2CO |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
38QC2CO |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone /RRE-containing |
||
|
48QC2CO |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
54QC2CO |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing /RiPP-like |
||
|
64QC2CO |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing /NRPS |
||
|
2QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
3QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/NRPS |
||
|
4QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing /RiPP-like |
||
|
6QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing /RiPP-like |
||
|
7QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
19QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
34QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone /NRPS |
||
|
36QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing/RiPP-like |
||
|
37QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing/RiPP-like |
||
|
46QC2O2 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing/RiPP-like |
||
|
63QC2CO |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
65RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/NRPS |
||
|
65.2RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/NRPS |
||
|
66RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
NRPS |
||
|
67RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/NRPS |
||
|
123RC1 |
+/- |
− |
− |
+/- |
− |
− |
− |
− |
NRPS |
||
|
124RC1 |
+/- |
− |
− |
+/- |
− |
− |
− |
− |
NRPS |
||
|
213RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing |
||
|
206RC1 |
− |
− |
− |
− |
− |
− |
− |
− |
NRPS |
||
|
15QC4O2 |
− |
− |
− |
− |
− |
− |
− |
+ |
RRE-containing |
||
|
18QC4O2 |
− |
− |
− |
− |
− |
− |
− |
+ |
RiPP-like/RRE-containing |
||
|
31RC1 |
Rothia nasisuis |
+ |
+/- |
+ |
+ |
+ |
+ |
− |
− |
Betalactone/RRE-containing/NRPS (valinomicin) |
|
|
69RC1 |
Rothia nasisuis |
− |
− |
+ |
+ |
+ |
+ |
− |
− |
Betalactone/RRE-containing/NRPS (valinomicin) |
|
|
141RC1 |
Rothia nasisuis |
+ |
+/- |
+ |
+ |
+ |
+ |
− |
− |
Betalactone/RRE-containing/NRPS (valinomicin) |
|
|
152RC1 |
Rothia nasisuis |
+ |
+/- |
+ |
+ |
+ |
+ |
− |
− |
Betalactone/RRE-containing/NRPS (valinomicin) |
|
|
196RC1 |
Rothia nasisuis |
+ |
+/- |
+ |
+ |
+ |
+ |
− |
− |
Betalactone/RRE-containing/NRPS (valinomicin) |
|
|
107RC1 |
Rothia nasisuis |
+/- |
− |
− |
− |
− |
− |
− |
− |
Betalactone/RRE-containing/NRPS (valinomicin) |
|
Inhibition zones were scored after overnight incubation in at least two assays. (−) absence of inhibitory activity; (+/-) inhibition zone between 3 and 6 mm diameter (weak); (+) inhibition zone ≥7 mm diameter [14]. NRPS: non-ribosomal peptide synthetase; RRE-containing: recognition element containing NRPs and/or PKs clusters; RiPPs-like: category of ribosomally synthesized and post-translationally modified peptides.
Genomic DNA extraction and sequencing
Rothia spp. strains were grown overnight in BHI broth at 37 °C. The next day, cells were pelleted by centrifugation, and genomic DNA was extracted using the PowerSoil Genomic Purification Kit (Qiagen) according to the manufacturer’s recommended protocol for Gram-positive bacteria. Recovery of high molecular weight DNA was assessed on a 0.8 % agarose gel (Sigma-Aldrich) in 1× TAE buffer [Tris-HCl 40 mM, 20 mM acetic acid and 1 mM EDTA (pH 8)], stained with 25 µg ml−1 of SYBR Safe and quantified using the Qubit dsDNA Broad-Range (BR) assay and Invitrogen’s Qubit Fluorometer.
Genome sequencing, assembly, annotation and phylogenetic analysis
We obtained genome sequences from 43 tonsil isolates of Rothia from five piglets, randomly selected from different litters on two farms with a high-health status. Genome sequencing was performed on an Illumina HiSeq 2000 platform (Illumina) at MicrobesNG, Birmingham, UK. However, due to lower sequencing depth and shorter read lengths, a few genome sequence assemblies had a high number of contigs (Fig. S3, available in the online version of this article). Reads were trimmed using Trimmomatic 0.30 software with a sliding window set at Q15 [40]. Genome assembly was performed using SKESA 2.4.0 [41]. In total, 112 genome sequences of publicly available Rothia species were downloaded from NCBI’s Reference Sequence database (Table. S1, available in the online version of this article) (download 27 April 2021). All genomes were annotated using Prokka 1.14.6 [42]. The ‘core’ genes were determined based on 90 % protein similarity using Roary 3.13.0 [43] (Table S2, available in the online version of this article). A maximum-likelihood tree was inferred using IQ-TREE 2.1.4 on default parameters from the core gene alignment obtained from Roary 3.13.0. Tree visualization was done in R 4.0.5 using ggtree 2.4.2 [44].
Carbohydrate-active enzyme analysis
Carbohydrate-active enzymes (CAZy) present in the genomes were determined using dbCAN2 2.0.11 using default parameters [45]. CAZy were annotated as auxiliary activities (AA), carbohydrate-binding modules (CBM), carbohydrate esterase (CE), glycoside hydrolases (GH), polysaccharide lyases (PL) or glycosyltransferases (GT). To limit false positives, a minimum of two hits was considered reliable. Hierarchical clustering based on Bray–Curtis dissimilarity was used to cluster the genomes. Five clusters were determined to be the optimal number based on gap statistics. Principal component analysis (PCA) was used to visualize the distribution of the CAZy across the collection genomes in our dataset.
Antimicrobial resistance genes (ARGs) and nitrate–nitrite metabolism genes
Antimicrobial resistance genes (ARGs) were identified using Abricate 1.0.1 (https://github.com/tseemann/abricate) with the ResFinder database [36] at 80 % DNA identity and coverage cut-off for both. The sequences taxonomically related to nitrate/nitrite metabolism in Rothia [32] were compared by similarity using the programme blastx [46] against all the genomes used in this study to identify the potential hits with filtering of e-value <10−5 and as threshold for sequence identity and coverage ≥50 %.
Mining of antimicrobial peptide encoding genes
For identification and annotation of biosynthetic gene clusters (BGCs), draft genomes of all Rothia strains were analysed using antiSMASH 6.0.1 using default detection criteria for the analysis [47]. The BGCs from the strains were sorted into groups based on their predicted activity. In addition, the Biosynthetic Genes Similarity Clustering and Prospecting Engine (BiG-SCAPE) software was used to define a distance metric between gene clusters using a combination of three indices: Jaccard Index of domain types, Domain Sequence Similarity, and the Adjacency Index [48]. Networks were visualized using Cytoscape 3.8.2 [49]. Clinker 0.0.21 was used for the alignment and comparison of BGCs [50].
Results
Rothia spp. isolation and identification corroborated and complemented previous Rothia distribution reports
To obtain upper respiratory isolates of the Rothia genus from pigs we cultured tonsil swabs from five piglets on sheep blood agar plates and purified over 500 isolates by repeated steaking of single colonies on fresh plates. Ninety-three isolates were identified as Rothia spp. by MALDI-TOF MS analysis (data not shown). Forty-six Rothia isolates were then selected for genome sequencing based on differences in colony morphology, growth characteristics and inhibitory activity against different target bacteria in the same niche. The assembled genomes ranged between 2.31–2.79 Mb in size, with a GC-content between 56.7–60.3 % (Table S3, available in the online version of this article). We identified 43 Rothia isolates to the species level using full-length sequences of 16S small subunit ribosomal RNA (rRNA); R. nasimurium (n=36), R. nasisuis (n=5), R. aerolata (n=1) and R. endophytica (n=1).
Genomic analyses and screening the genomic dataset to discover candidate genes involved in colonization and adaptation of the different species in the environment
To identify accessory genes involved in antibiotic resistance, secondary metabolite biosynthesis, and carbohydrate utilization genes, which might play crucial roles in niche adaptation, competition, and persistence, we analysed the genomes of our 43 isolates and 112 publicly available draft genomes of Rothia species and strains (as of April 2021). In total, 155 Rothia genomes were included along with the metadata, sample origin and disease-associated or commensal species. (Fig. 1). The relatedness of the 155 genomes was investigated using core-genome phylogenies and mined for the presence of (i) BCGs, (ii) antimicrobial resistance genes, and (iii) carbohydrate-active enzymes (CAZy) encoded in the genome.
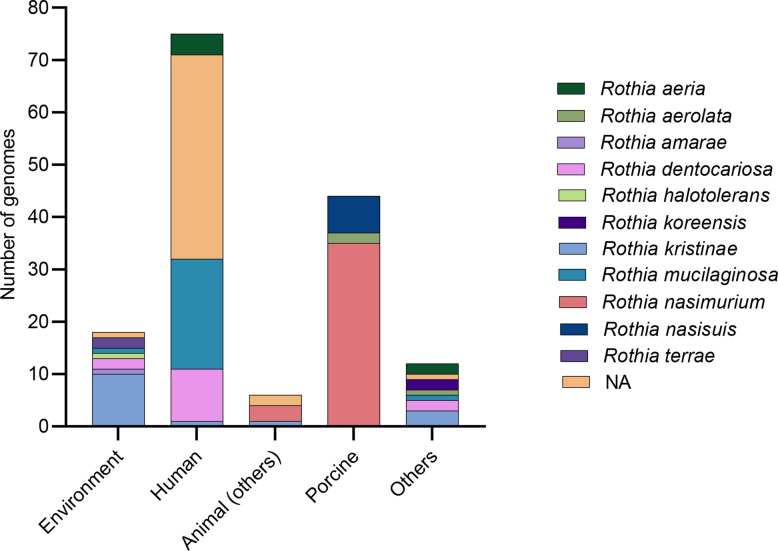
Fig. 1.
Rothia genomes including the species R. aerolata, R. aeria, R. amarae, R. kristinae, R. dentocariosa, R. halotolerans, R. mucilaginosa, R. terrae, R. nasimurium, R. nasisuis, R. koreensis unculturedRothia sp identified in metagenomic data from mainly human clinical samples in the NCBI Reference Sequence Database. The bar chart shows the number of genomes analysed for each Rothia species per sampling site (i.e. origin of isolates); ‘others’ correspond to isolates from food and unspecified samples. Animal (others) corresponds to diverse animal groups (sponge, Mus musculus, duck and marmot) but not pigs. NA no assigned species (Rothia sp).
We constructed phylogenetic trees based on 90 % core protein similarities (encoded by 28 core genes that were shared by the Rothia strains (Table S2, available in the online version of this article) and observed three major clades that overall corresponded to their origin; human, environmental/food and, porcine/other animals (sponge, Mus musculus, duck and marmot). We annotated the phylogeny with the presence/absence of different classes of BGCs and antibiotic resistance genes (Fig. 2).
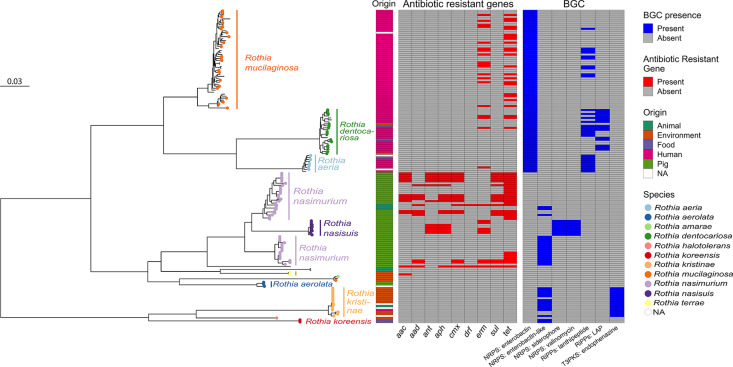
Fig. 2.
Overview of the biosynthetic gene clusters (BGCs) and antimicrobial resistance genes (ARG) in the species of Rothia analysed in this study from different sampling sites. ARG represents multiple variants of the gene ‘families’. The compilation follows: the sampling-sites are represented by the colours shown in the inner colourful column. The rooted tree was constructed based on the core genes (n=28), the species are named and highlighted with the same colour. The antibiotic resistance genes are displayed as presence (red) and absence (grey), which could lead to the selection of potential producers of bioactive compounds. The outermost columns display the presence/absence of biosynthetic gene clusters; distinct clusters were annotated according to antiSMASH in grey (absence) and dark blue (presence). The scale bar indicates nucleotide substitution per site.
Genome mining for antibiotic resistance, nitrate/nitrite reduction genes and BGCs
On average Rothia genomes contain between two to eight antibiotic resistance genes (ARGs) including variants of the gene ‘families’ aac, aad, ant, aph, cmx, dfr, erm, sul and tet. ARGs were identified mainly in Rothia isolates belonging to the species R. nasimurium and R. mucilaginosa sampled from pigs and humans (Fig. 2). Most of the Rothia genomes encode genes involving nitrate/nitrite metabolism as previously reported [51]. The predicted gene functions and number of genes associated with nitrate/nitrite reduction vary across Rothia species and their ecological niche (Table S5, available in the online version of this article). Human-associated Rothia species carry the largest number of genes involved in nitrate and nitrite reduction metabolism, followed by animal-associated Rothia species.
Overall, 386 BGCs were predicted in the 155 genomes used in this study. In 80 % of the Rothia genomes, at least one BGC was annotated as non-ribosomal peptide synthetase (NRPS) or hybrid non-ribosomal peptide-polyketide (NRP/PK) synthase, which together account for 146 of all the predicted clusters (Fig. 2). Other common BGC types were gene clusters annotated as producing betalactone; multidomain polyketide synthases (PKSs), and ribosomally synthesized and post-translationally modified peptides (RiPPs) with a broad range of biological functions including antimicrobial acticvity (Table S3, available in the online version of this article).
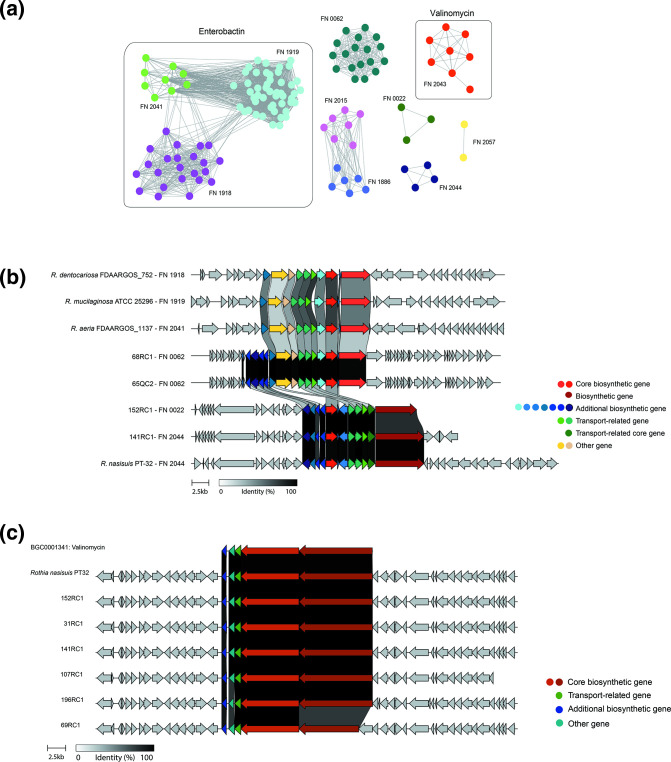
In the 43 porcine Rothia isolates, antiSMASH mainly predicted the presence of NRPs, RiPPs and NRPs (Table 2). NRPS gene cluster (family FN2043), found only in R. nasisuis is predicted to produce the natural antibiotic valinomycin (Fig. 3c). Genomes of R. nasimurium, possess NRPs clusters (family FN0062); members of this family are predicted to produce secondary metabolites (Fig. 3a). These 18 BGCs had low similarity showing potential novelty based on the BigScape analyses.
Fig. 3.
Overview of NRPs-type BGCs from Rothia species. (a) Similarity network of known and putative BGCs annotated as NRPs based on antiSMASH results. The small groups of NPRS families (FN) with unknown clusters containing fewer common genes are drawn outside the boxes: FN2015, FN0022, FN2044 and FN1086. Highlighted groups include widely distributed clusters in many different species. The network containing red nodes represents the valinomycin (FN2043) cluster that was present in all porcine R. nasisuis isolates. The network containing light blue, green and purple nodes represent BGCs corresponding to Enterobactin cluster families FN1918, FN1919, and FN2041 that were present in the species of R. mucilaginosa , R dentocariosa and R. aeria that were mainly sampled from the upper respiratory tract of humans. The network containing dark green nodes represents an NRP-like cluster annotated as family FN0062 that was prevalent in R. nasimurium; this cluster contained conserved genes from the Enterobactin cluster. The families represent the group of BGCs that encode biosynthesis of highly similar or identical metabolites classified in arbitrary numbers based on the outcome of the analyses. (b) Overview of representative species that contain conserved Enterobactin BGCs, or NRP-like cluster annotated as family FN0062 ( R. nasimurium ) that shares genes with Enterobactin BGC as well as the families FN0022 and FN2044. (c) Overview of the conserved valinomycin cluster (BIG-SCAPE family FN2043) in the species R. nasisuis. BIGSCAPE output is visualized as a network using Cytoscape and the cluster organization visualized using Clinker (https://github.com/gamcil/clinker).
Overlay inhibition assays
Inhibition assays were performed using a panel of eight indicator bacteria with all the Rothia strains from this study (n=43) (Table 2). The indicator bacteria included strains of three zoonotic pathogen S. suis , which colonizes the oropharyngeal cavity and tonsil epithelium of pigs, as well as related streptococci S. porci , S. porcinus , S. parasuis, S. aureus and E. coli as a representative Gram-negative bacterium (Table 1). Fourteen Rothia strains were able to inhibit the growth of at least one species of Gram-positive bacteria tested (Table 2). All R. nasisuis and six strains of R. nasimurium showed growth inhibitory activity against streptococci but had no activity against S. aureus and two strains of R. nasimurium had low inhibitory activity against E. coli .
Carbohydrate active enZymes (CAZy)
We discovered more than 5 040 diverse carbohydrate active enZyme (CAZy) genes, which were classified in different families and subfamilies (Table. S4, available in the online version of this article), suggesting there may be a link between CAZy functionality (‘toolbox’) and the ability to colonize and persist in novel niches under varying environmental conditions. Using the gene annotations of Rothia species, our analyses revealed that Rothia species contain five major families of genes predicted to participate in carbohydrate metabolism and energy conversion systems, including genes predicted to encode glycosyl hydrolases (GHs), glycosyl transferases (GT), carbohydrate-binding modules (CBM), carbohydrate esterases (CE), auxiliary activities [36], and other carbohydrate metabolic pathway components (Fig. 4).
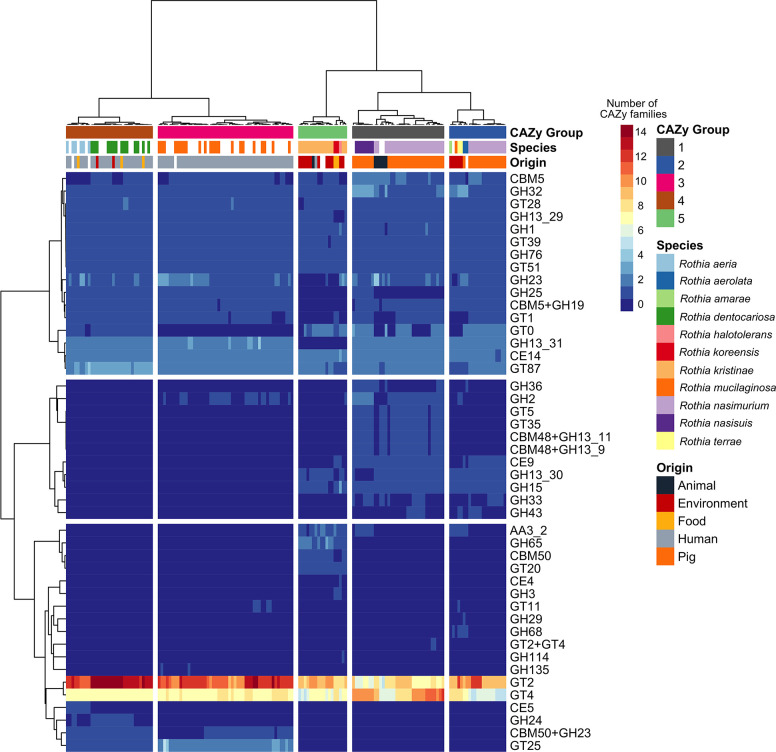
Fig. 4.
Heatmap displaying presence/absence of CAZy families across Rothia genomes. The heatmap includes data from 41 CAZy families (rows); the sampling site and distribution of CAZy families across Rothia species are displayed in bars above the heatmap. ‘CAZy group’ (columns) represents the enzymes present in a specific group of species clustered according to our analysis. All displayed CAZy gene data and annotations were retrieved from the CAZy enzyme database (accessed October 2021). Glycosyl hydrolase (GH), glycosyl transferases (GT), carbohydrate-binding modules (CBM), carbohydrate esterases (CE).
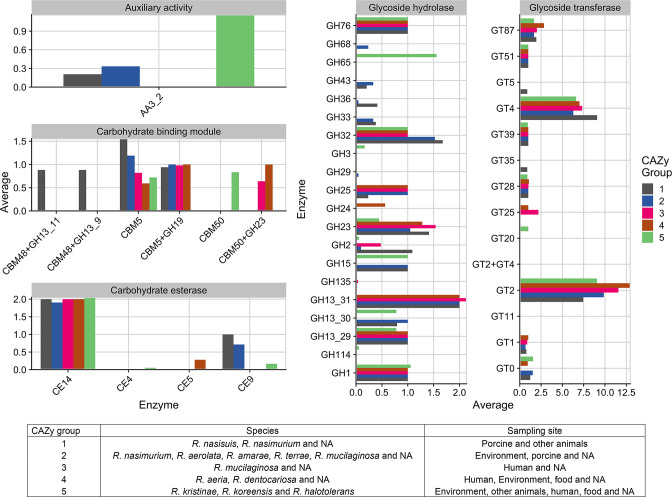
Overall, 41 CAZy families were annotated among the Rothia genomes (21 GHs, 14 GTs, 2 CBMs, 3 CE and one AA) at different abundances per species and niche (Fig. 4). The GHs had highest gene sequence diversity and abundance followed by the GTs. Rothia species have large numbers of GTs representing >65 % of the total CAZy enzymes toolbox. GT families are highly diverse in the CAZy database but in the Rothia genomes just two families, GT2 and GT4, represent a large portion of the total number of GT genes (n=3 372) identified all strains and niches. The first horizontal blocks of the heatmap (Fig. 4) shows clustering between the families CBM5-GT87, which contains enzymes prevalent in several species of Rothia in all niches, suggesting the presence of essential enzymes responsible for carbohydrate metabolism. The CAZy families GH33, GH36, GT5, GT35, CBM48 +GH13-11 and CBM48 +GH13-9 (CAZy Group 1) were mostly discovered in the animal-associated species R. nasimurium and R. nasisuis. The families GT25 and CBM50 +GH23 were more often identified in human-associated Rothia species; these enzymes clustered in CAZy groups 2 and 3. Specifically, the family GT25 was mainly discovered in the genomes of R. dentocariosa and the family complex CBM50 +GH23 was present in 85 % of the Rothia species isolated from human microbiota. On the other hand, CAZy enzymes GH65, CBM50 and GT20 were only discovered in Rothia species sampled from the environment (Fig. 5).
Fig. 5.
Overview of CAZy enzyme families prevalent in the Rothia genus. Based on the distribution of species and sampling origins, CAZy enzymes were clustered in five groups displayed in the first column of the table displayed at the bottom. Glycosyl hydrolases (GHs), glycosyl transferases (GT), carbohydrate-binding modules (CBM), carbohydrate esterases (CE). The bottom table is the overall of the ‘CAZy group’ that represents the enzymes present in a specific group of species clustered according to our analysis (Fig. 4).
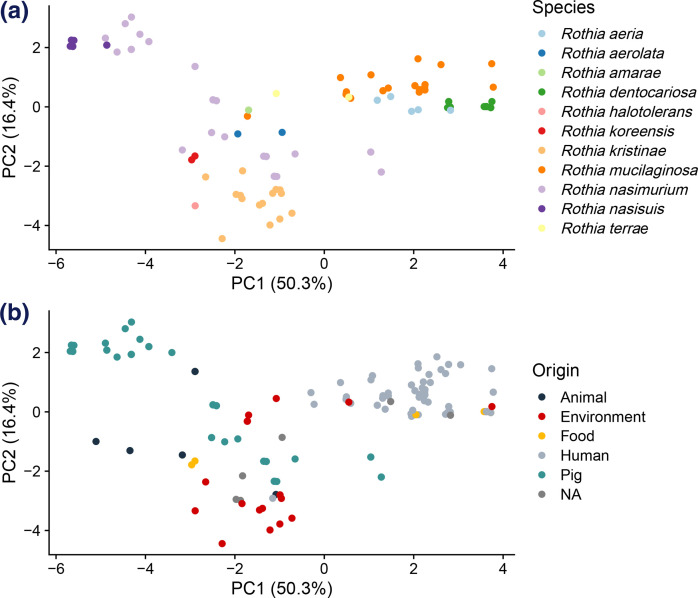
To investigate CAZy distribution across the 155 Rothia genomes representing 12 species originating from different niches, a principal component analysis (PCA) was conducted (Fig. 6). The first two principal components explained 66.7% (50.3 and 16.4 % for PC1 and PC2, respectively) of the total variance.
Fig. 6.
Principal component analysis (PCA) score plot of carbohydrate active enZyme (CAZy) distribution. The first two axes explain 50.3 and 16.4 % of the total variance, respectively. The ecological traits are indicated by (a) taxonomic groups and (b) sampling sites.
The CAZy-based PCA plot clustered species largely by taxonomy; isolates and genomes from the species (Fig. 6a). R. mucilaginosa and Rothia sp. (unclassified Rothia’s) mainly clustered in the upper right area. Most isolates and genomes of the species R. kristinae, R. halotolerans and R. koreensis clustered in the lower-left area. Most isolates and genomes of the species R. aeria and R. aerolata clustered in the middle. Finally, most isolates and genomes of the species R. nasimurium and R. nasisuis clustered in the upper left area of the plot. Those Rothia species clustering together in the PCA also clustered closely in the phylogenetic analysis or colonized very similar niches (Fig. 6b).
Discussion
Recently, Rothia was listed as no. 17 in the top 20 most abundant and prevalent bacterial genera discovered in the human oral cavity [2]. Our recent work on porcine tonsil microbiota revealed Rothia as one of the most prevalent genera with at least 0.98 % mean abundance across individuals; the genus was detected in more than 98 % of the piglets from a data set with over 160 samples, across 11 farms sampled in Europe (unpublished).
The habitat specificity of Rothia species suggest adaptation to certain oral niches such as the teeth surface and the oral mucosa [2]. Here we characterized the genomes of several species of the poorly characterized genus Rothia to gain a better understanding of the interactions and colonization in the upper respiratory tract and the potential ecological role of specialized metabolites and CAZy enzymes. Genome mining of the sequences from the databases of over a hundred Rothia genomes and the recent sequenced porcine isolates identified several BGCs with potential to produce natural antimicrobial compounds. The metabolites predicted to be produced by these gene clusters include compounds that function in competitive interactions between microbes such as antibiotics and siderophores, but also compounds that may function in the host, including immunosuppressants and anticancer drugs. Potentially, the NRPS, PKS and hybrid NRPS-PKS gene clusters could produce groups of bioactive compounds that form a source of antimicrobial products with broad applications [52]. NRPs, PKs as well as other compounds produced by BGCs are released into the surrounding environment upon their biosynthesis. The synthesis is regulated by proteins that are activated by physical and environmental factors, including carbon and nitrogen source availability [52]. Once released, the molecules produced by NRPSs and PKSs or their hybrid gene clusters play roles in niche colonization, including optimization of bacterial proliferation, quorum sensing activity, and providing higher substrate affinity or faster growth rates [53].
Valinomycin, identified only in Rothia nasisuis species, is an NRP ionophore, forming ion channels in membranes that allow free movement K+ ions thereby, altering membrane potential that may lead to disruption of the normal K+ ion membrane gradient [24, 54]. All isolates of R. nasisuis containing the valinomycin-producing NRPs showed inhibitory activity in vitro against Gram-positive target bacteria including different species of Streptococcus sp. and closely related Rothia species as previous reported [55] (Table 2). The presence of the valinomycin clusters in R. nasisuis likely provide this species with a competitive advantage to prevail in the porcine tonsil microbiota and influence the composition of the microbiota in the first weeks of piglet life [7, 54]. We identified 18 novel NRPS gene clusters in the genomes of R. nasimurium species using BigScape. These clustered as family FN0062 and contain genes also present in the FN1919, FN1918 and FN2041 families that have been predicted to produce enterobactins, with strong binding affinity for ferric iron [56] (Fig. 3b). Iron availability is scarce at mucosal surfaces due to host iron-sequestering proteins [57]. NRPS-like clusters from family FN0062 are therefore likely involved in the acquisition of iron from the environment and facilitate colonisation of the human oral cavity by R. mucilaginosa and R. dentocariosa [58]. Six other NRPSs were identified that might produce antimicrobials as judged by growth inhibition of Gram-positive and Gram-negative bacteria by R. nasimurium in vitro. However further research would be needed to verify this conclusion. We also report the first endophytic Rothia isolate originating from pig tonsil microbiota. R. endophytica has been found within healthy plant tissues [13] thus it is possible that the R. endophytica strain we isolated from the piglet tonsil swab originated from ingested plant material. Genomes of Rothia species (i.e. R. nasimurium , R. nasisuis and R. amarae ) found in pigs encoded multidrug-resistance gene elements (Fig. 2), likely due to the strong selection pressure from the use of antibiotics for treatment of bacterial infections or use as growth promoters in livestock animals prior to the European ban in 2006 [Regulation (EC) No. 1831/2003]. Specific tetracycline resistance genes were found in genomes of Rothia strains isolated from human [tet(W)] and porcine strains [tet(M)]. In the past, the presence/absence of tet genes were mainly determined by sampling niche and application of tetracyclines, however, at present, tet genes have evolved and occurred more widely dispersed in different environments being present in commensal and pathogenic strains [59]. Erythromycin is an antibiotic used to treat several respiratory infections and resistance genes [erm(X)], were present in human (n=27) and some porcine strains (n=7). However, a small number of antibiotic resistant genes were found in the Rothia genomes as a reflection of the continuous use in humans and animals on a large scale [60].
As different host organisms and host regions (i.e. tonsil or saliva) can contain different carbohydrates, it is of interest to characterize the carbon-converting enzymatic ‘toolbox’ of Rothia strains sampled from different host organisms and environmental niches. The GH enzymes generally participate in carbohydrate catabolism, suggesting that the enzyme orthologues present in the Rothia genomes have the potential to degrade complex carbohydrates originating from mammals, plants, insects, and fungi including mucins, cellulose, membrane glycoproteins, and chitin in acidic environments [61]. The high number of GTs present in Rothia genomes are likely involved in carbohydrate biosynthesis, although some GTs have been reported to enzymatically attach carbohydrate moieties to precursors of some naturally occurring antibiotics [62, 63].
GTs identified in Rothia genomes can be responsible for the synthesis and assembly of the repetitive units of exopolysaccharides (EPS), which contributes for the biofilm formation, tolerance to desiccation and host antimicrobial peptides, as well as modulation the host immune response by the association with other molecules as glycolipids [64]. The CAZy families GT2 and GT4 are present in all Rothia genomes and are involved in the biosynthesis of various (exo)polysaccharides, which are often involved in biofilm formation, and biosynthesis of natural products [65]. Most Rothia isolates have a mucoid morphology (Table. S1, available in the online version of this article) and aggregate in liquid media, which might be directly associated with the EPS production although the morphology of the colonies varies at the species level depending on the in vitro culture conditions. Exopolysaccharides could also provide alternative carbon sources to support the establishment of a pioneer microbial community in the upper respiratory tract [66]. The genomes of strains from the species R. dentocariosa and R. mucilaginosa, which colonize the human oral cavity, contained the largest numbers of predicted GT2 genes (Fig. 4). Considering exopolysaccharides are one of the main structural components of bacterial extracellular matrix and biofilm [67], the high amount of GTs may favour the formation of structurally organized biofilm consortia composed by a single taxon or a cluster of taxa interacting with Rothia on the oral mucosa and on teeth surfaces [2].
The CAZy families GH33, GH36, GT5, GT35, CBM48 +GH13-11 and CBM48 +GH13-9 (CAZy Group 1) were mostly present in the animal-associated species ( R. nasimurium and R. nasisuis). The GH33 and GH36 families, are mainly present in the porcine isolates and are known to be involved in the utilization of human milk oligosaccharides (HMOs) and O-glycosylated mucins. HMO’s shape the microbiota and have beneficial effects in early life [68]. The capacity to catabolize HMOs and glycosylated mucins may be an advantage to pioneer species and drive microbiota diversity [69]. Several CAZy families contain enzymes involved in carbohydrate digestion and hydrolysation. CAZy family GT35 includes glycogen or starch phosphorylase enzymes, family CBM48 includes enzymes participating in carbohydrate binding and glycogen-binding, and GH13 includes enzymes with glycosyl hydrolase activity. Little information is available about the utilization of complex carbohydrates in the oral cavity, but it is well described that the enzymes that release energy from the breakdown of branched substrates and complex carbohydrates can support proliferation and survival of certain species when there other carbon sources are limiting, and by maintaining microbiota diversity, have a positive impact on health [70].
The families GT25 and CBM50 +GH23 contain enzymes with glycosyltransferase activity and are mostly present in the human-associated Rothia species. The CAZy family GT25 plays important roles in catalysing utilization of monosaccharides, assembly of glycoconjugates and complex carbohydrates by transferring sugar moieties onto growing liposaccharide chains (PF01755). GT25 enzymes may play a role in utilization of carbohydrates from different sources, and in biofilm formation, while the family complex CBM50 +GH23 is associated with lytic transglycosylase activity related to the peptidoglycan metabolic process. Proteins with this annotation are usually enzymes active in the breakdown of chitin or peptidoglycan into molecules that trigger innate immune responses through recognition by host pattern receptors [71].
The CAZy enzymes GH65, CBM50, and GT20 were only identified in Rothia species isolated from environmental niches. The GT20 family contains enzymes annotated with trehalose-phosphatase activity (PF00982) while the GH65 family contains acid trehalases and some phosphorylases that catalyse conversion of trehalose to glucose and glucose-1-phosphate. Trehalose is a common disaccharide used by bacteria, archaea, fungi, invertebrates and others as a carbon source but trehalose also plays an important role in protecting bacteria against stress including desiccation, osmotic stress, oxidation and temperature changes [72]. The CBM50 or LysM domain family members contain a domain of approximately 40–50 amino acid residues that can occur in various carbohydrate-modifying enzymes, for instance, glycoside hydrolase enzymes that play roles in the digestion of complex carbohydrates [73]. The LysM domain is found in carbohydrate-active enzymes, peptidoglycan binding proteins and plant cell surface receptors for fungal chitin oligosaccharides [73, 74]. The LysM domain is also found in proteins correlated with several other biological functions including stress response in plants [75], signalling for specific plant and bacteria interactions [76] and bacterial spore surface development [77, 78]. As these modules are involved in interactions between different organisms, they may play roles during symbiosis and quorum sensing in soil- or plant-associated bacteria during microbiota development. Many of the CAZy enzymes identified in Rothia correlate with the environmental or host niche, emphasizing that these enzymes have important roles in microbial ecology and are relevant determinants of which habitats can be colonized and exploited by microbes. Rothia species encode CAZy genes for breakdown of chitin in fungal and insect cell walls, mucins and milk oligosaccharides into simple carbohydrates that can be used as nutrients and carbon sources for fermentation by other microbiota members [79]. Such cross-feeding might enable symbiotic interactions between Rothia and other microbes and could benefit microbial homeostasis and symbiosis from early life onward.
The presence of genes related to the transport, assimilation and conversion of nitrate to nitrite, denitrification, nitric oxide detoxification and bacterial nitrate reduction enzymes cofactors were identified in several species and highly correlated with the source of isolation (Table S5, available in the online version of this article) . We noted that the human associated species R. dentocariosa, R. mucilaginosa, R. aeria and Rothia sp. contain most of the genes involved in nitrate and nitrite metabolic pathways as reported previously in ten human-associated oral Rothia species isolates [79]. Porcine and other animal associated species as R. nasimurium and R. nasisuis also carry some of the genes involved in nitrate and nitrite metabolism, suggesting that those species might have similar function(s) in the oral cavity of animals, potentially with consequences for health and disease [32]. We noted that environmental species as R. kristinae and R. koreensis appeared to lack most of nitrate/nitrite metabolic genes. Nitrate/nitrite metabolic genes await functional characterization. This topic warrants further studies using physiological measurements of nitrate reduction for different Rothia species.
In summary, we showed that the NRPS producing the antimicrobial valinomycin was specific to R. nasisuis isolates from pig upper respiratory tract and identified novel BGCs encoding NRPSs, PKSs and RiPPs in other Rothia species that may produce antimicrobial compounds. According to our analyses, Rothia species have an extensive number of CAZy (5040 genes annotated in total), many of which are associated with EPS production and catabolism of carbohydrate sources (possibly including milk oligosaccharides) in different hosts and environments. Members of the Rothia genus have the potential to produce novel antimicrobials and may be used as probiotics to shape the microbiota of humans and mammals in early life and provide colonization resistance against pathogens and pathobionts.
Supplementary Data
Funding information
This project was partly funded by the European Research Council (ERC) under the European Union's Horizon 2020 (H2020-EU.1.3.1) and MSCA-ITN-ETN - European Training Networks (grant agreement ID: 765147).
Author contributions
I.M.F.O., Y.K.N., P.v.B. and J.M.W. conceptualized the study. I.M.F.O. and Y.K.N performed the data analysis and interpretation of results under the supervision of P.v.B., M.S., P.S.A and J.M.W. The experiments and strains isolation were performed by I.M.F.O. The manuscript was written by I.M.F.O. and Y.K.N with input from all co-authors. All authors contributed to the article and approved the final manuscript.
Conflicts of interest
The authors declare that they do not have a conflict of interest.
Ethical statement
This study uses samples obtained for diagnostic procedures performed according to the ethical principles and guidelines covered by EU Directive 2010/63/EU.
Footnotes
Abbreviations: ARGs, antimicrobial resistance genes; BGC, biosynthetic gene cluster; CAZy, carbohydrate-active enzymes; NRPs, non-ribosomal peptides; NRPS, non-ribosomal peptide synthetase; PKs, polyketides; RiPPs, ribosomally synthesized and post-translationally modified peptides.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure and five supplementary table are available with the online version of this article.
References
- 1.Lowe BA, Marsh TL, Isaacs-Cosgrove N, Kirkwood RN, Kiupel M, et al. Microbial communities in the tonsils of healthy pigs. Vet Microbiol. 2011;147:346–357. doi: 10.1016/j.vetmic.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 2.Wilbert SA, Mark Welch JL, Borisy GG. Spatial ecology of the human tongue dorsum microbiome. Cell Rep. 2020;30:4003–4015. doi: 10.1016/j.celrep.2020.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manrique P, Freire MO, Chen C, Zadeh HH, Young M, et al. Perturbation of the indigenous rat oral microbiome by ciprofloxacin dosing. Mol Oral Microbiol. 2013;28:404–414. doi: 10.1111/omi.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herder EA, Spence AR, Tingley MW, Hird SM. Elevation correlates with significant changes in relative abundance in hummingbird fecal microbiota, but composition changes little. Front Ecol Evol. 2021;8:534. doi: 10.3389/fevo.2020.597756. [DOI] [Google Scholar]
- 5.Palmer RJ Jr, Shah N, Valm A, Paster B, Dewhirst F, et al. Interbacterial adhesion networks within early oral biofilms of single Human hosts. Appl Environ Microbiol. 2017;83:e00407-17. doi: 10.1128/AEM.00407-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sulyanto RM, Thompson ZA, Beall CJ, Leys EJ, Griffen AL. The predominant oral microbiota is acquired early in an organized pattern. Sci Rep. 2019;9:1–8. doi: 10.1038/s41598-019-46923-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pena Cortes LC, LeVeque RM, Funk J, Marsh TL, Mulks MH. Development of the tonsillar microbiome in pigs from newborn through weaning. BMC Microbiol. 2018;18:35. doi: 10.1186/s12866-018-1176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Kawamura Y, Fujiwara N, Naka T, Liu H, et al. Rothia aeria sp. nov., Rhodococcus baikonurensis sp. nov. and Arthrobacter russicus sp. nov., isolated from air in the Russian space laboratory Mir. Int J Syst Evol Microbiol. 2004;54:827–835. doi: 10.1099/ijs.0.02828-0. [DOI] [PubMed] [Google Scholar]
- 9.Kämpfer P, Kleinhagauer T, Busse H-J, Klug K, Jäckel U, et al. Rothia aerolata sp. nov., isolated from exhaust air of a pig barn. Int J Syst Evol Microbiol. 2016;66:3102–3107. doi: 10.1099/ijsem.0.001153. [DOI] [PubMed] [Google Scholar]
- 10.Fan Y, Jin Z, Tong J, Li W, Pasciak M, et al. Rothia amarae sp. nov., from sludge of a foul water sewer. Int J Syst Evol Microbiol. 2002;52:2257–2260. doi: 10.1099/00207713-52-6-2257. [DOI] [PubMed] [Google Scholar]
- 11.Ko KS, Lee MY, Park YK, Peck KR, Song J-H. Molecular identification of clinical Rothia isolates from Human patients: proposal of a Novel Rothia species, Rothia arfidiae sp. nov. J Bacteriol Virol. 2009;39:159. doi: 10.4167/jbv.2009.39.3.159. [DOI] [Google Scholar]
- 12.Georg LK, Brown JM. Rothia, gen. nov. an aerobic genus of the family Actinomycetaceae. International Journal of Systematic Bacteriology. 1967;17:79–88. doi: 10.1099/00207713-17-1-79. [DOI] [Google Scholar]
- 13.Xiong Z-J, Zhang J-L, Zhang D-F, Zhou Z-L, Liu M-J, et al. Rothia endophytica sp. nov., an actinobacterium isolated from Dysophylla stellata (Lour.) Benth. Int J Syst Evol Microbiol. 2013;63:3964–3969. doi: 10.1099/ijs.0.052522-0. [DOI] [PubMed] [Google Scholar]
- 14.Nouioui I, Carro L, García-López M, Meier-Kolthoff JP, Woyke T, et al. Genome-based taxonomic classification of the Phylum Actinobacteria . Front Microbiol. 2018;9:2007. doi: 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z-X, Yang L-L, Huang Y, Zhao H, Liu H, et al. Rothia marina sp. nov., isolated from an intertidal sediment of the South China Sea. Antonie Van Leeuwenhoek. 2013;104:331–337. doi: 10.1007/s10482-013-9955-8. [DOI] [PubMed] [Google Scholar]
- 16.Collins MD, Hutson RA, Båverud V, Falsen E. Characterization of a Rothia-like organism from a mouse: description of Rothia nasimurium sp. nov. and reclassification of Stomatococcus mucilaginosus as Rothia mucilaginosa comb. nov. Int J Syst Evol Microbiol. 2000;50 Pt 3:1247–1251. doi: 10.1099/00207713-50-3-1247. [DOI] [PubMed] [Google Scholar]
- 17.Schlattmann A, von Lützau K, Kaspar U, Becker K. ‘Rothia nasisuis’ sp. nov., ‘Dermabacter porcinasus’ sp. nov., ‘Propionibacterium westphaliense’ sp. nov. and ‘Tessaracoccus nasisuum’ sp. nov., isolated from porcine nasal swabs in the Münster region, Germany. New Microbes New Infect. 2018;26:114–117. doi: 10.1016/j.nmni.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou Y-J, Chou J-H, Lin K-Y, Lin M-C, Wei Y-H, et al. Rothia terrae sp. nov. isolated from soil in Taiwan. Int J Syst Evol Microbiol. 2008;58:84–88. doi: 10.1099/ijs.0.65172-0. [DOI] [PubMed] [Google Scholar]
- 19.Baker JL, Morton JT, Dinis M, Alvarez R, Tran NC, et al. Deep metagenomics examines the oral microbiome during dental caries, revealing novel taxa and co-occurrences with host molecules. Genome Res. 2021;31:64–74. doi: 10.1101/gr.265645.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agnello M, Marques J, Cen L, Mittermuller B, Huang A, et al. Microbiome associated with severe caries in Canadian First Nations children. J Dent Res. 2017;96:1378–1385. doi: 10.1177/0022034517718819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez A, Espinoza JL, Harkins DM, Leong P, Saffery R, et al. Host Genetic Control of the Oral Microbiome in Health and Disease. Cell Host Microbe. 2017;22:269–278. doi: 10.1016/j.chom.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ihara Y, Takeshita T, Kageyama S, Matsumi R, Asakawa M, et al. Identification of initial colonizing bacteria in dental plaques from young adults using full-length 16S rRNA Gene Sequencing. mSystems. 2019;4:e00360-19. doi: 10.1128/mSystems.00360-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan ST, Ahamed M, Musarrat J, Al-Khedhairy AA. Anti-biofilm and antibacterial activities of zinc oxide nanoparticles against the oral opportunistic pathogens Rothia dentocariosa and Rothia mucilaginosa . Eur J Oral Sci. 2014;122:397–403. doi: 10.1111/eos.12152. [DOI] [PubMed] [Google Scholar]
- 24.Gaiser RA. Antimicrobial Peptides and the Interplay Between Microbes and Host: Towards Preventing Porcine Infections with Streptococcus suis. Wageningen University and Research; 2016. [Google Scholar]
- 25.Wang L, Ravichandran V, Yin Y, Yin J, Zhang Y. Natural products from mammalian gut microbiota. Trends Biotechnol. 2019;37:492–504. doi: 10.1016/j.tibtech.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Letzel A-C, Pidot SJ, Hertweck C. Genome mining for ribosomally synthesized and post-translationally modified peptides (RiPPs) in anaerobic bacteria. BMC Genomics. 2014;15:983. doi: 10.1186/1471-2164-15-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller BR, Gulick AM. Structural biology of nonribosomal peptide synthetases. Nonribosomal Peptide and Polyketide Biosynthesis: Springer. 2016:3–29. doi: 10.1007/978-1-4939-3375-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulick AM. Nonribosomal peptide synthetase biosynthetic clusters of ESKAPE pathogens. Nat Prod Rep. 2017;34:981–1009. doi: 10.1039/c7np00029d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 30.Drula E, Garron M-L, Dogan S, Lombard V, Henrissat B, et al. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022;50:D571–D577. doi: 10.1093/nar/gkab1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onyango SO, Juma J, De Paepe K, Van de Wiele T. Oral and gut microbial carbohydrate-active enzymes landscape in health and disease. Front Microbiol. 2021;12:653448. doi: 10.3389/fmicb.2021.653448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosier BT, Takahashi N, Zaura E, Krom BP, MartÍnez-Espinosa RM, et al. The importance of nitrate reduction for oral health. J Dent Res. 2022;2022:220345221080982. doi: 10.1177/00220345221080982. [DOI] [PubMed] [Google Scholar]
- 33.Sato-Suzuki Y, Washio J, Wicaksono DP, Sato T, Fukumoto S, et al. Nitrite-producing oral microbiome in adults and children. Sci Rep. 2020;10:16652. doi: 10.1038/s41598-020-73479-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundberg JO, Carlström M, Weitzberg E. Metabolic effects of dietary nitrate in health and disease. Cell Metab. 2018;28:9–22. doi: 10.1016/j.cmet.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira AC, Cunha MV. An effective culturomics approach to study the gut microbiota of mammals. Res Microbiol. 2020;171:290–300. doi: 10.1016/j.resmic.2020.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Lagier J-C, Dubourg G, Million M, Cadoret F, Bilen M, et al. Culturing the human microbiota and culturomics. Nat Rev Microbiol. 2018;16:540–550. doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 39.Hockett KL, Baltrus DA. Use of the soft-agar overlay technique to screen for bacterially produced inhibitory compounds. J Vis Exp. 2017;2017:119. doi: 10.3791/55064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Souvorov A, Agarwala R, Lipman DJ. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018;19:153. doi: 10.1186/s13059-018-1540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 43.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. Ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. [Google Scholar]
- 45.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarro-Muñoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, et al. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. 2020;16:60–68. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, et al. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilchrist CLM, Chooi Y-H. Clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics. 2021;37:2473–2475. doi: 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 51.Rosier BT, Moya-Gonzalvez EM, Corell-Escuin P, Mira A. Isolationand characterization of nitrate-reducing bacteria as potential probiotics for oral and systemic health. Front Microbiol. 2020;11:555465. doi: 10.3389/fmicb.2020.555465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esmaeel Q, Pupin M, Jacques P, Leclère V. Nonribosomal peptides and polyketides of Burkholderia: new compounds potentially implicated in biocontrol and pharmaceuticals. Environ Sci Pollut Res Int. 2018;25:29794–29807. doi: 10.1007/s11356-017-9166-3. [DOI] [PubMed] [Google Scholar]
- 53.Ishaque NM, Burgsdorf I, Limlingan Malit JJ, Saha S, Teta R, et al. Isolation, genomic and metabolomic characterization of Streptomyces tendae VITAKN with quorum sensing inhibitory activity from Southern India. Microorganisms. 2020;8:121. doi: 10.3390/microorganisms8010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang S, Liu Y, Liu W-Q, Neubauer P, Li J. The nonribosomal peptide valinomycin: from discovery to bioactivity and biosynthesis. Microorganisms. 2021;9:780. doi: 10.3390/microorganisms9040780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaiser RA, Medema MH, Kleerebezem M, van Baarlen P, Wells JM. Draft genome sequence of a porcine commensal, Rothia nasimurium, encoding a nonribosomal peptide synthetase predicted to produce the ionophore antibiotic valinomycin. Genome Announc. 2017;5:17. doi: 10.1128/genomeA.00453-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uranga CC, Arroyo Jr P, Duggan BM, Gerwick WH, Edlund A, et al. Commensal oral rothia mucilaginosa produces enterobactin, a metal-chelating siderophore. mSystems. 2020;5:e00161-20. doi: 10.1128/mSystems.00161-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganz T. Iron and infection. Int J Hematol. 2018;107:7–15. doi: 10.1007/s12185-017-2366-2. [DOI] [PubMed] [Google Scholar]
- 58.Uranga CC, Arroyo P, Duggan BM, Gerwick WH, Edlund A. Commensal oral Rothia mucilaginosa produces enterobactin, a metal-chelating siderophore. mSystems. 2020;5:e00161-20. doi: 10.1128/mSystems.00161-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gasparrini AJ, Markley JL, Kumar H, Wang B, Fang L, et al. Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance. Commun Biol. 2020;3:241. doi: 10.1038/s42003-020-0966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zainab SM, Junaid M, Xu N, Malik RN. Antibiotics and antibiotic resistant genes (ARGs) in groundwater: a global review on dissemination, sources, interactions, environmental and human health risks. Water Res. 2020;187:116455. doi: 10.1016/j.watres.2020.116455. [DOI] [PubMed] [Google Scholar]
- 61.Weijers C, Franssen MCR, Visser GM. Glycosyltransferase-catalyzed synthesis of bioactive oligosaccharides. Biotechnol Adv. 2008;26:436–456. doi: 10.1016/j.biotechadv.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 62.Losey HC, Peczuh MW, Chen Z, Eggert US, Dong SD, et al. Tandem action of glycosyltransferases in the maturation of vancomycin and teicoplanin aglycones: novel glycopeptides. Biochemistry. 2001;40:4745–4755. doi: 10.1021/bi010050w. [DOI] [PubMed] [Google Scholar]
- 63.Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS One. 2012;7:e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cruz-Aldaco K, Govea-Salas M, Gomes-Araujo R, Dávila-Medina MD, Loredo-Trevino A. Bbioactivities of bacterial polysaccharidesbioactivities of bacterial polysaccharides. 2021.
- 65.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 66.Lin F, Li C, Chen Z. Exopolysaccharide-derived carbon dots for microbial viability assessment. Front Microbiol. 2018;9:2697. doi: 10.3389/fmicb.2018.02697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marvasi M, Visscher PT, Casillas Martinez L. Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett. 2010;313:1–9. doi: 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- 68.Heine RG, AlRefaee F, Bachina P, De Leon JC, Geng L, et al. Lactose intolerance and gastrointestinal cow’s milk allergy in infants and children - common misconceptions revisited. World Allergy Organ J. 2017;10:41. doi: 10.1186/s40413-017-0173-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Herreweghen F, De Paepe K, Roume H, Kerckhof F-M, Van de Wiele T. Mucin degradation niche as a driver of microbiome composition and Akkermansia muciniphila abundance in a dynamic gut model is donor independent. FEMS Microbiol Ecol. 2018;94:12. doi: 10.1093/femsec/fiy186. [DOI] [PubMed] [Google Scholar]
- 70.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Humann J, Lenz LL. Bacterial peptidoglycan degrading enzymes and their impact on host muropeptide detection. J Innate Immun. 2009;1:88–97. doi: 10.1159/000181181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakaguchi M. Diverse and common features of trehalases and their contributions to microbial trehalose metabolism. Appl Microbiol Biotechnol. 2020;104:1837–1847. doi: 10.1007/s00253-019-10339-7. [DOI] [PubMed] [Google Scholar]
- 73.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pham M-L, Tran A-M, Kittibunchakul S, Nguyen T-T, Mathiesen G, et al. Immobilization of β-Galactosidases on the Lactobacillus cell surface using the peptidoglycan-binding Motif LysM. Catalysts. 2019;9:443. doi: 10.3390/catal9050443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tzelepis G, Karlsson M. Killer toxin-like chitinases in filamentous fungi: structure, regulation and potential roles in fungal biology. Fungal Biol Rev. 2019;33:123–132. doi: 10.1016/j.fbr.2018.11.001. [DOI] [Google Scholar]
- 76.Singh K, Upadhyay SK, Shumayla. Madhu LysM domain-containing proteins modulate stress response and signalling in Triticum aestivum L. Environ Exp Bot. 2021;189:104558. doi: 10.1016/j.envexpbot.2021.104558. [DOI] [Google Scholar]
- 77.Spaink HP. Specific recognition of bacteria by plant LysM domain receptor kinases. Trends Microbiol. 2004;12:201–204. doi: 10.1016/j.tim.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 78.Pereira FC, Nunes F, Cruz F, Fernandes C, Isidro AL, et al. A LysM domain intervenes in sequential protein-protein and protein-peptidoglycan interactions important for spore coat assembly in Bacillus subtilis . J Bacteriol. 2019;201:e00642-18. doi: 10.1128/JB.00642-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao B, Gallagher T, Zhang Y, Elbadawi-Sidhu M, Lai Z, et al. Tracking polymicrobial metabolism in cystic fibrosis airways: Pseudomonas aeruginosa metabolism and physiology are influenced by Rothia mucilaginosa-derived metabolites. mSphere. 2018;3:e00151-18. doi: 10.1128/mSphere.00151-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.