Abstract
Uropathogenic Escherichia coli (UPEC) is the most prevalent cause of urinary tract infections (UTIs). Biofilm formation and antibiotic resistance could be high among the causative agent. The purpose of this study was to determine antibiotic resistance, biofilm production, and biofilm-associated genes, bcsA and csgD, and sub-inhibitory hydrogen peroxide (H2O2) stimulation in UPEC for biofilm formation. A total of 71 UPEC were collected from a tertiary care hospital in Kathmandu and subjected to identify antibiotic susceptibility using Kirby-Bauer disk diffusion. The biofilm formation was assessed using microtiter culture plate method while pellicle formation was tested by a tube method. In representative 15 isolates based on biofilm-forming ability, bcsA and csgD were screened by conventional polymerase chain reaction, and treated with sub-lethal H2O2. The UPEC were found the most susceptible to meropenem (90.2%), and the least to ampicillin (11.3%) in vitro and 90.1% of them were multi-drug resistant (MDR). Most UPEC harbored biofilm-producing ability (97.2%), and could form pellicle at 37°C. Among representative 15 isolates, csgD was detected only among 10 isolates (66.67%) while bcsA gene was present in 13 isolates (86.67%). This study revealed that level of biofilm production elevated after sub-lethal H2O2 treatment (P = .041). These findings suggested that the pathogens are emerging as MDR. The biofilm production is high and the majority of selected strains contained bcsA and csgD genes. Pellicle formation test was suggestive to be an alternative qualitative method to screen biofilm production in UPEC. The sub-inhibitory concentration of H2O2 may contribute in increasing biofilm formation in UPEC.
Keywords: Uropathogenic E. coli, biofilm, antibiotic resistance, bcsA, csgD
Introduction
Uropathogenic E. coli (UPEC) strains are responsible for the majority of urinary tract infections.1-3 UPEC infect the host via cell surface hydrophobicity (CSH), fimbriae, curli fibers, and the colanic capsule, facilitating the bacterial biofilm lifestyle, enhancing persistence and resistance to host innate immune factors, and antibiotic resistance.4-6 Intracellular bacterial communities (IBCs) are formed when intracellular bacteria encase themselves on the bladder surface in a polysaccharide-rich matrix.7 The development of IBCs in bladder epithelial cells involves several phases, including reversible to irreversible attachment, microcolony formation, and maturation.8-10 Environmental factors such as immunological response, oxidative stress, predation, and other environmental pressures influence the production of the extracellular matrix, which is regulated by transcription factors.11 The presence of terminal electron receptors in the urine, together with reduced oxygen stress in the bladder, supports the preferred development of E. coli biofilms.12 The pellicle, which forms at the air-liquid interface and enables adhesion between bacteria and assembles to construct multicellular architectures, is a type of biofilm.13 Polysaccharides are often involved in the establishment of productive cell-to-cell contacts that contribute to the formation of pellicles at liquid and solid interfaces such as clumping of cell aggregates in liquid cultures. This signifies UPEC can form pellicles in vitro in an air-water interface, indicating curli is important for the formation of this kind of biofilm. The switching between pellicle and biofilm during infection or survival in the natural environment is still unfamiliar.14 Curli are amyloid fibers that participate in the generation of biofilms and aid in the adherence of bacteria to the human bladder.1,15 In E. coli, the genes involved in curli production are arranged into 2 operons: csgAB and csgDEFG. The csgAB encodes 2 curli components (cgsA and csgB), while csgDEFG is in charge of control, assembly, and transportation.16 In the bacteria, the master regulatory gene csgD stimulates the production of curli and extracellular matrix.17 The biofilm generated by Enterobacteriaceae contains cellulose as a major component.18 In bacterial biofilms, cellulose acts as a structural component that contributes as a scaffold for biofilm formation.19 The bcsABZC operon contains structural genes for cellulose expression.20 The cellulose synthase enzyme is transcribed by the bcsA gene, and the transcriptional regulator csgD is connected to the regulation of cellulose production.18,21
Increasing antibiotic resistance against UTIs in recent years is emerging to be troublesome, which implies a serious threat to human health.2 Antibiotic-resistant bacteria and their propagation in various settings have evidently become a major concern around the world.22 The biofilm-forming isolates are resistant to antibiotic therapy, posing a major clinical concern in the case of biofilm-related infections.23,24 Biofilm acts as a protective layer around bacteria, preventing antibiotics, immune cells, and host proteins from proliferating.25 Various investigations among UPEC have shown that the production of biofilm is closely linked to antibiotic resistance and MDR.26-28 However, some reports have demonstrated that resistance is not dependent on the production of biofilm.29-32
Reactive oxygen intermediates such as hydrogen peroxide (H2O2) are toxic molecules produced by immune cells in response to bacterial invasion into the host. Bacteria try to protect themselves against the immune system through specific properties such as biofilm formation. This phenomenon occurs also during urinary tract infections.33 The bacterial biofilm is integral to many infections by promoting persistence, protecting from host innate immune factors, and resisting to antibiotics.5,6
Despite the fact that UPEC has been widely reported from clinical samples in Nepal, no study has yet reported the presence of bcsA and csgD genes in the UPEC for biofilm formation across the country. It is important to determine anti-microbial resistance for evaluating the effectiveness of the drugs. Thus, this study attempts to demonstrate antibiotic, and multi-drug resistance status in the pathogens. This research was also hypothesized upon, H2O2 under sub-inhibitory concentration stimulates biofilm production among UPEC and presence of bcsA and cgsD genes is associated with biofilm formation. The findings of this research are anticipated to provide new insights associated with the pathogenicity of the biofilm-producing UPEC.
Materials and Methods
Bacterial strains collection
In Bharosa Hospital, the urine samples were cultured on MacConkey agar (Hi-Media Laboratories Pvt. Ltd., India) and Blood agar (Hi-Media Laboratories Pvt. Ltd., India). The study period ranged from February 2019 to February 2020. The ethical approval from the Institutional Review Committee, Institute of Science and Technology was obtained for the research (IRC/IOST-Regd. No. 1).
A loopful of urine was streaked on the plates and then incubated at 37°C overnight. Colony count was performed to calculate the number of CFU per mL of urine and the bacterial count was reported as insignificant growth for 104 CFU/mL of organisms, 104-105 CFU/mL of organisms as doubtful, and significant bacteriuria was defined when the bacterial colony is more than 105 CFU/mL organisms.34 The identification of E. coli was done by standard laboratory procedures. Gram staining was performed. Identification was carried out by various tests such as positive catalase test, negative oxidase test, motile, indole positive, citrate negative, urea hydrolysis test positive, fermentative in Hugh’s and Leifson’s medium, and TSI (triple sugar iron) test is with A/A with gas production.35
Antibiotics susceptibility testing
The confirmed isolates recovered from urine samples were subjected to antibiotic susceptibility testing (Kirby-Bauer disk diffusion) using Mueller Hinton Agar (Hi-Media Laboratories Pvt. Ltd., India). Altogether, 10 antibiotics (recommended by CLSI guideline 2020) were used which included ampicillin (10 µg), ciprofloxacin (5 µg), cefalexin (30 µg), cefepime (30 µg), ceftriaxone (30 µg), amoxyclav (30 µg), co-trimoxazole (25 µg), nitrofurantoin (300 µg), gentamicin (10 µg), and meropenem (10 µg) (Hi-Media Laboratories Pvt. Ltd., India). The pathogens were categorized as resistant and sensitive. Those bacteria were considered as MDR strains when they were found non-susceptible to at least one agent in 3 or more antimicrobial categories.36
Pellicle test
The isolates were grown without shaking which included overnight incubation in 5 mL Luria broth (LB) at 37ºC and transferred into 4 mL LB in 15 mL glass tubes. After 48 hours at 37ºC, the formation of the pellicle at the air-liquid interface was visually observed.37
Biofilm assay
In this study, 71 isolates were employed for the quantitative test of biofilm as described by Christensen et al.38 A loopful of test organisms isolated from fresh agar plates were inoculated in 1 mL of tryptone soya broth (TSB) (Hi-Media Laboratories Pvt. Ltd., India) with 1% glucose. Broths were incubated at 37ºC for 24 hours which were then diluted at 1:100 with fresh TSB at 100 rpm. Then, 96 well microtiter plate was filled with 200 µL of diluted culture broth in each well and incubated for 48 hours. TSB with 1% glucose was used as the negative control in 1 lane of the microtiter plate and E. coli (ATCC 25922) as a positive control in another 3 wells. After the incubation, the contents of each well were removed by gentle tapping. The wells were then washed with 0.2 mL phosphate-buffered saline (pH 7.3) 4 times to remove the free-floating bacteria. The biofilm formed by the bacteria adherent to the wells were fixed by 2% sodium acetate and then stained by 100 µL of 0.1% crystal violet for 15 minutes at room temperature. Excess stain was removed with deionized water and the biofilm was quantified by measuring the absorbance at 630 nm against a blank in Multiskan Sky/Microtiter spectrophotometer (Thermo Fisher Scientific, USA) equipped with SkanIt software version 5.0. following solubilization of attached biofilm in 95% ethanol.39 The experiment was performed in triplicate and repeated 3 times. The interpretation of biofilm production was done according to the criteria of Stepanović et al.40 The cut-off optical density (ODc) is defined as 3 standard deviations above the mean OD of the negative control.
Detection of bcsA and csgD
Only 15 isolates were subjected to a polymerase chain reaction, which includes 3 strong producers, 5 moderate producers, 5 weak producers, and 2 non-biofilm producers regarding biofilm formation. The primers used for amplifying bcsA (base pair 826 bp) were F: GCTTCTCGGCGCTAATGTTG and R: GAGGTATAGCCACGACGGTG41 and for csgD (base pair 97 bp) were F: CCGCTTGTGTCCGGTTTT and R: GAGATCGCTCGTTCGTTGTTC.42 PCR for bcsA gene was done in a DNA thermal cycler (Applied biosystems, USA) with the setting: initial denaturation for 10 minutes at 95°C, followed by 30 cycles of denaturation for 1 minute at 94°C, annealing for 1 minute 30 seconds at 55°C and extension for 1 minute at 72°C, and a final extension for 10 minutes at 72°C.41 PCR for csgD gene was done in the cycler with the setting: initial denaturation for 5 minutes at 95°C, followed by 35 cycles of denaturation for 1 minute at 94°C, annealing for 1 minute at 57°C and extension for 1 minute at 72°C and a final extension for 10 minutes at 72°C. Electrophoresis was performed in 2.5% gel. Bromphenol blue was employed for loading DNA samples into agarose gel wells as well as tracking migration during electrophoresis.42
Treatment of bacterial strains with H2O2
Those selected 15 E. coli strains were cultured (106 CFU/mL) in Luria Bertani broth at 37°C for 24 hours along 0.625 mM H2O2, sub-inhibitory concentration for bacterial growth and crude catalase was added to stop the reaction after 15 minutes treatment with H2O2.33 The source of catalase was Solanum tuberosum.43 Then, determination of absorbance for biofilm for the treated strains was performed as described in the microtiter plate culture method.
Statistical analysis
All data obtained were analyzed using the statistical program statistical package for social science (SPSS v. 22.0) and OriginPro v. 8.5 for descriptive statistics. Different percentages, chi-square test (antibiotics, biofilm, pellicle, AST, MDR), and t-test (biofilm formation and biofilm-forming genes), chi-square test (association of bcsA and csgD with biofilm) were used to compare groups, and P-values < .05 were considered statistically significant.
Results
UPEC show the highest susceptibility to meropenem and the least susceptibility to ampicillin in vitro
Through the Kirby-Bauer test susceptibility method, the sensitivity of UPEC (n = 71) toward antibiotics was determined. The susceptibility pattern of UPEC isolates to different antimicrobial agents is shown in Table 1. Among the antibiotics, the bacterial resistance was extensively high toward ampicillin (88.7%) followed by cotrimoxazole (73.2%) and ciprofloxacin (40.8%). The isolates showed the least resistance toward meropenem (9.8%) followed by nitrofurantoin (18.3%) and gentamicin (21.1%). Altogether, 64 (90.1%) isolates were multidrug-resistant.
Table 1.
Comparison of antibiotic susceptibility to biofilm producers in the non-biofilm forming environment (n = 71).
| Antibiotics | Sensitive | Resistant | Total resistant-UPEC (%) | P-value | ||
|---|---|---|---|---|---|---|
| Biofilm producers (%) | Biofilm non-producers (%) | Biofilm producers (%) | Biofilm non-producers (%) | |||
| Ampicillin (AMP) | 8 (11.3) | - | 61 (85.9) | 2 (2.8) | 63 (88.7) | .6 |
| Amoxyclav (CAC) | 48 (67.6) | 1 (1.4) | 21 (29.6) | 1 (1.4) | 22 (31) | .56 |
| Ciprofloxacin (CIP) | 41 (57.8) | 1 (1.4) | 28 (39.4) | 1 (1.4) | 29 (40.8) | .79 |
| Cefalexin (CN) | 48 (67.6) | - | 21 (29.7) | 2 (2.8) | 23 (32.4) | .038 |
| Co-Trimoxazole (COT) | 18 (25.4) | 1 (1.4) | 51 (71.8) | 1 (1.4) | 52 (73.2) | .46 |
| Cefepime (CPM) | 46 (64.8) | 1 (1.4) | 23 (32.4) | 1 (1.4) | 24 (33.8) | .62 |
| Ceftriaxone (CTR) | 48 (67.5) | 2 (2.8) | 21 (29.7) | - | 21 (29.7) | .56 |
| Gentamicin (GEN) | 54 (76.1) | 2 (2.8) | 15 (21.1) | - | 15 (21.1) | .46 |
| Meropenem (MRP) | 62 (87.4) | 2 (2.8) | 7 (9.8) | - | 7 (9.8) | .6 |
| Nitrofurantoin (NIT) | 55 (78.9) | 2 (2.8) | 14 (18.3) | - | 14 (18.3) | .042 |
Biofilm-formation is non-significant (P > .05) with AMP, CAC, CIP, COT, CPM, CTR, GEN, and MRP and biofilm-formation is significant (P < .05) with CN and NIT on MHA.
UPEC demonstrate pellicle formation in air-liquid interface and corresponds biofilm formation ability at 37°C
In the test tubes with Luria Bertani broth, 67 (94.34%) isolates were capable of forming pellicles in the air-liquid interface. Through microtiter plate culture assay based on the optical density of negative control, biofilm formation was categorized into 4 groups (Figure 1). The cut-off value of optical density for biofilm production was 0.062 obtained by adding 3 Standard Deviations to the value of negative control. The biofilm formation was observed in 69 (97.1%) isolates. Strong biofilm formation was observed among 3 (4.2%) isolates, weak biofilm formation among 32 (45.1%) isolates, moderate biofilm formation among 34 (47.9%) isolates, and 2 (2.8%) isolates were unable to form biofilm. Furthermore, there was a significant relationship (P = .002) between pellicle formation inside the tube and biofilm formation in the microtiter plate.
Figure 1.
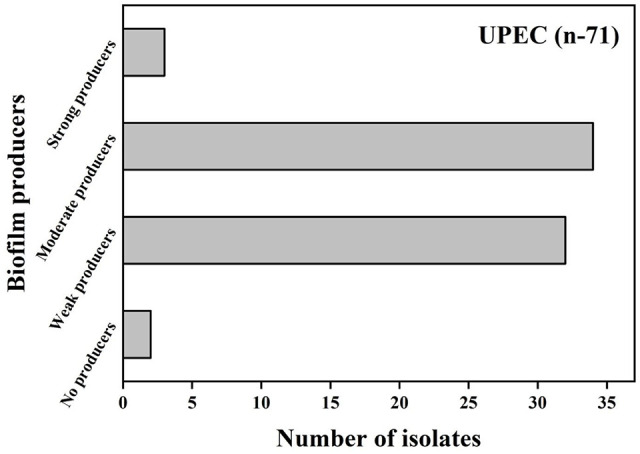
Biofilm production in UPEC via microtiter plate assay (n = 71).
Cephalexin and nitrofurantoin are effective against biofilm-forming UPEC in a non-biofilm forming environment
Comparing antibiotic susceptibility (AST test on MHA) and biofilm formation (Table 1), most of those selected antibiotics showed an insignificant relationship. Against ampicillin, amoxyclav, ciprofloxacin, co-trimoxazole, cefepime, ceftriaxone, gentamicin, and meropenem, the P-value was ⩾.05 which signifies biofilm-formation may not have a relation with these antibiotics used in vitro in non-biofilm-forming environment. Biofilm-forming bacteria in a non-biofilm-forming conditions appeared to be inhibited by administration of cephalexin (P = .038) and nitrofurantoin (P = .042) in vitro.
Co-occurrence of bcsA and csgD in biofilm production is significant
Detection of genes was carried out through conventional PCR, agarose gel electrophoresis, and visualization (Photograph 1) in a UV chamber. Among the 71 isolates, only 15 selected isolates were selected based on biofilm-forming ability. The gene, csgD was detected only among 10 isolates (66.67%) while bcsA gene was detected among 13 isolates (86.67%). The co-occurrence of bcsA and csgD in the isolates was significant (P = .032). Out of the total 13 producers; 12 (92.3%) producers harbored bcsA while 10 producers (76.92%) contained csgD. However, the association of bcsA with biofilm (P = .116) and csgD with biofilm (P = .257) were insignificant for both genes (Table 2). Detection of targeted genotypes (bcsA and csgD) among those UPEC indicates insignificance in biofilm (P = .24). Among 2 non-biofilm-producers, one isolate harbored both bcsA and csgD while the other isolate lacked both genes.
Photograph 1(A).
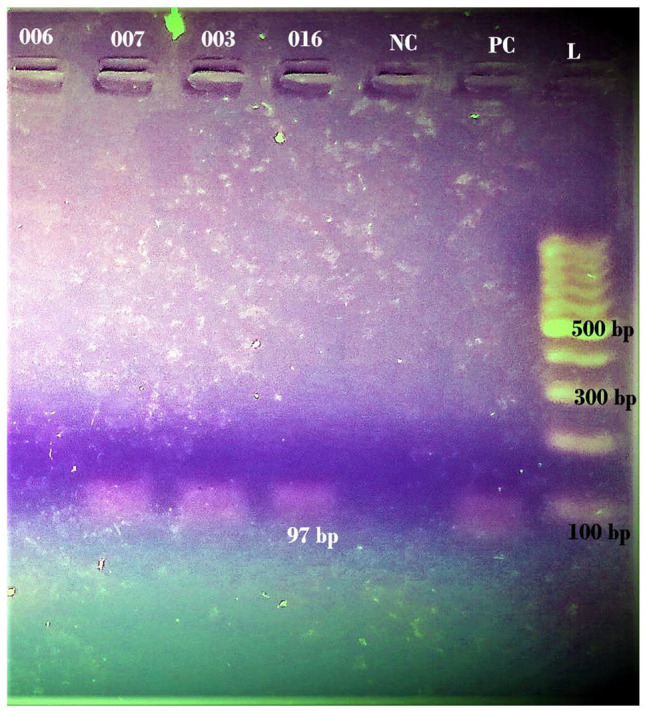
2.5% agarose gel electrophoresis for detection of csgD (97 bp) in UPEC isolates (L = Thermo Scientific GeneRuler 100 bp plus DNA ladder).
Table 2.
Association of bcsA gene and csgD gene with biofilm.
| Genotype | Biofilm | Total | Chi-square test (P-value) | |
|---|---|---|---|---|
| Producers | Non-producers | |||
| bcsA − csgD − | 1 | 1 | 2 | .236 |
| bcsA + csgD + | 9 | 1 | 10 | |
| bcsA + csgD − | 3 | 0 | 3 | |
| Total | 13 | 2 | 15 | |
bcsA – (bcsA absent), bcsA+ (bcsA present), csgD – (csgD absent), csgD + (csgD present).
Photograph 1(B).
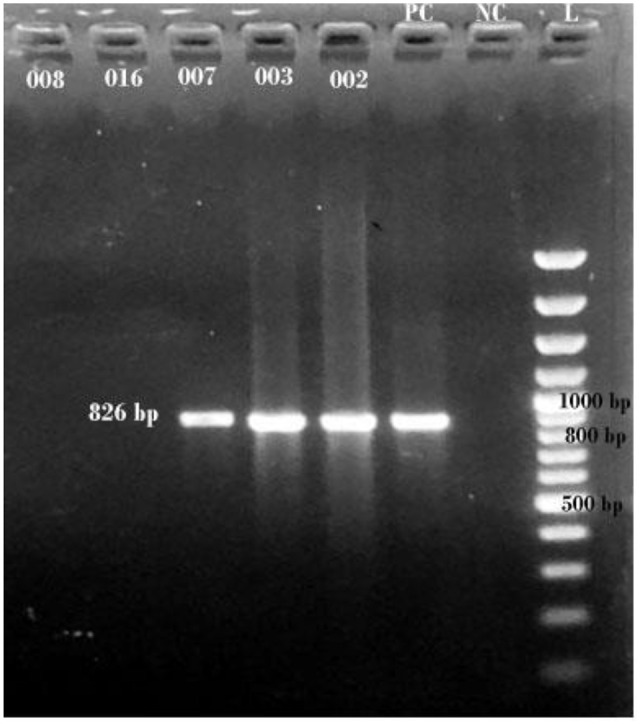
1% agarose gel electrophoresis for detection of bcsA (826 bp) in UPEC isolates (L = Thermo Scientific GeneRuler 100 bp plus DNA ladder).
Sub-inhibitory H2O2 treatment elevated biofilm formation
The level of biofilm formation was measured based on optical density. The level of biofilm production increased after the treatment with sub-inhibitory H2O2, and catalase in non-producers, weak producers, and moderate producers (Photograph 2). In contrast, among the strong producers, the amount of biofilm formed was decreased (Table 3). The relationship between before treatment and after treatment is significant that is, P-value ⩽0.05 which implies hydrogen peroxide can stimulate biofilm formation in the UPEC.
Photograph 2.

Hydrogen peroxide and catalase treatment of UPEC (Luria Bertani broths with hydrogen peroxide + crude catalase).
Table 3.
Effect of hydrogen peroxide (H2O2) treatment on biofilm production.
| Biofilm production level (Sample code) | Optical density before H2O2 | Optical density H2O2 | P-value | |
|---|---|---|---|---|
| Only H2O2 | H2O2 + catalase (stimulation) | |||
| Non-producer | ||||
| 008 | 0.062 | 0.03 | 0.17 | |
| 016 | 0.049 | 0.04 | 0.22 | |
| Weak | ||||
| 003 | 0.09 | 0.06 | 0.61 | |
| 006 | 0.09 | 0.06 | 0.28 | |
| 007 | 0.07 | 0.05 | 0.20 | |
| 009 | 0.07 | 0.05 | 0.29 | |
| 015 | 0.08 | 0.04 | 0.29 | |
| Moderate | .041 | |||
| 001 | 0.17 | 0.10 | 0.41 | |
| 004 | 0.14 | 0.09 | 0.45 | |
| 010 | 0.15 | 0.08 | 0.29 | |
| 011 | 0.19 | 0.10 | 0.37 | |
| 012 | 0.14 | 0.07 | 0.29 | |
| Strong | ||||
| 002 | 0.63 | 0.12 | 0.27 | |
| 005 | 0.33 | 0.10 | 0.22 | |
| 014 | 0.62 | 0.12 | 0.39 | |
Discussion
Different studies display variability in the spectrum and frequency of antibiotic resistance among UPEC.44-49 Our study showed the highest sensitivity toward meropenem since only 9.8% of UPEC are resistant to the antibiotic, which was similar to some studies.49-51 Carbapenems are highly active against E. coli isolates and represent the best treatment option.2,52 The highest resistance was observed with ampicillin (88.7%). Similar bacterial resistance to ampicillin was demonstrated in various studies.48,49,53 Ampicillin was used as empirical therapy for a long time, and resistance may have emerged as a result of self-medication, increased antibiotic intake, and the emergence of resistant isolates.47,54,55 In a research conducted by Yadav and Prakash in Southern Terai of Nepal, it was found that 91.86% of the isolates were MDR.56 The results were similar regarding MDR rates.
The majority of isolates in our study showed pellicle formation. Nascimento et al conducted an investigation in which pellicle production in clinical isolates of atypical enteropathogenic E. coli was demonstrated (aEPEC).37 Pellicles are also known as air-liquid (A-L) biofilms because they form at the air-liquid interface.57 Pellicle development begins with bacteria adhering to the culture device’s wall at the air-liquid contact, followed by the development of a monolayer by attached cells, and finally the formation of the distinctive three-dimensional architecture.58,59 The pellicle is considered a special structure of biofilm.60-62
The formation of pellicle at the air-liquid interface was successfully observed among the isolates. At 37°C, the pathogen pellicles correspond to the ability to produce biofilm. A meta-analysis showed more than 84% of UPEC have the ability to form a biofilm.28 Variation in the level of biofilm production was observed in different researches.27,48,63,64 The study data depends upon the biofilm formation ability of the isolates determined by specific factors such as hydrophobicity, and cellular surface electrical discharge and varies among strains.65 Likewise, the organisms tend to produce more biofilm to establish successful infection, biofilms are formed on urinary catheters or on/within bladder epithelial cells protecting them from the host immune system, antimicrobial therapy, and various dynamic environmental conditions.12 According to our findings, pellicle formation can be used to screen for biofilm formation. Exopolysaccharides are believed to be associated with the production of productive cell-to-cell interactions that contribute to the formation of biofilm communities at liquid and solid interfaces, such as clumping of cell aggregates in liquid cultures, according to a study.66 The primary components for producing the pellicle matrix include oxygen, flagellar motility, and cellulose.67-70
Biofilm-forming bacteria are a prevalent cause of recurring and complex UTIs.6 Biofilm-associated microorganisms are considered to be more resistant to antimicrobial treatments.71 In this study, biofilm producers are more susceptible to cefalexin and nitrofurantoin. The susceptibility test showed cefalexin to be the most effective antibiotic against biofilm producers in non-biofilm forming conditions which was peculiar. Cefalexin displayed low sensitivity against bacteria in another finding.56 However, based on geographical and regional location, antimicrobial sensitivity can vary.72 A meta-analysis showed nitrofurantoin is the best antibiotic for invading UPEC strains.28 According to a study by Makled et al, nitrofurantoin could be considered as selective antibiotics against biofilm structures.73 Also, our finding suggests that nitrofurantoin was also effective against most of the biofilm producers. The findings could aid in the treatment of initial infection when biofilm is not formed in the isolates. Cefalexin and nitrofurantoin are frequently administered in the context of Nepal currently. Individual associations with resistance in E. coli to gentamicin and ceftazidime were seen in a research.32 Antibiotic resistance can develop as a result of the synthesis of the ß-lactamase enzyme, the efflux pump, and decreased antibiotic uptake due to alterations in the outer membrane porin protein.47 Furthermore, antibiotic tolerance is mediated through genetic changes at the bacterial chromosomal level.74 Despite the fact that various integers suggest a link between antibiotic resistance and biofilm, this study found the contrary. As a result, more advanced research on uroepithelial organoids is needed to investigate the molecular links between antibiotic resistance and biofilm.
Our research revealed that bcsA was detected in more isolates than csgD. The expression of the bcsA gene, which codes for cellulose, had previously been linked to the csg operon, which also codes for curli fimbriae.75 However, when comparing biofilm formation in vitro, our investigation found that bcsA was present in more isolates than csgD. The bcsA gene is not necessarily needed for biofilm formation in Enterobacteriaceae because other genes, such as csgD, adrA, and other factors, can also be involved in cellulose expression and regulation.76,77 There could be csgD independent pathway for cellulose formation.77 In UPEC, cyclic AMP (cAMP) is also responsible for regulating curli and cellulose.11 According to reports, the pgaABCD locus found in E. coli is required for biofilm formation.48,78,79 In another investigation, the virulence genes fimH, pap, afa, and sfa were found to be strongly associated with biofilm formation.80 However, Davari Abad et al could not ascertain the connection of biofilm formation with sfa and afa genes.81 A significant correlation was established between biofilm production and the sdiA, rcsA, and rpoS genes.82 These findings, combined with our own, reveal that biofilm formation is a complicated process that will require more research to understand the genetic makeup of biofilm formation.
Our finding suggests that hydrogen peroxide can enhance the biofilm-forming ability among non- producers, weak, and moderate biofilm producers despite the presence or absence bcsA and csgD. The level of biofilm production among the strong producers has been decreased. It may be due to exogenous quorum sensing inhibitor when binds with QS receptor inhibit the signaling and fail to produce further biofilm or the catabolite repression by glucose.83
So far, no investigations have been published in Nepal reporting the detection of bcsA and csgD genes in UPEC isolated from clinical settings. The limitations of the study were the antibiotic susceptibility test (AST) was not performed in the biofilm-forming environment and gene expression was not carried out. All samples were not included in the molecular study since the resources were limited. Only 15 isolates were screened for the target genes and sub-inhibitory H2O2 treatment. There are significant drawbacks to this study, such as it was limited to a single hospital, the short period of the study, and the use of crude catalase extract. To expand about epidemiological or virulence aspects of UPEC, the presence of bcsA and csgD genes could be checked among MDR and XDR biofilm forming strains by in-silico analysis of genomes in further studies. Our study may help researchers in accounting the virulence factor, and multi-drug resistance of the bacteria for developing further treatment strategies.
Conclusion
The effectiveness of meropenem against the isolates was demonstrated to be the highest. About 90% of the pathogens were MDR which indicates alarming threat to public health. The biofilm production was observed in more than 95% isolates. The pellicle formation test appeared to be a potentially viable qualitative approach for detecting biofilm formation as the ability of UPEC to form pellicles at 37°C correlates to biofilm formation capability. Cefalexin and nitrofurantoin were screened to be selective against UPEC capable of forming biofilm in non-biofilm forming conditions. The bcsA and csgD genes were found in the majority of the chosen strains. The sub-lethal dosage of H2O2 may contribute in elevating biofilm forming capacity in UPEC except in strong producers. Further researches must be warranted to encounter the research gaps in biofilm, antibiotic susceptibility, and response toward environmental stress in UPEC.
Acknowledgments
We would like to acknowledge Bharosa Hospital, Kathmandu and Central Department of Microbiology, Tribhuvan University, Kirtipur, Kathmandu, Nepal. Also, we would also like to thank Dr. Rajdeep Bomjan (Washington University, the United States of America) for his constructive suggestions during the research period.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was partially supported by the University Grants Commission, Nepal (UGC Masters Research Support award no.: MRS-75/76-S&T-52). Prabin Dawadi received the grant.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: PD conceived the study design. PD and DRJ contributed to the design. PD planned the study and carried out the experiments and data analysis. AD guided in collecting and processing the samples in the hospital. BLM assisted in the laboratory for conducting the experiments. SanK, SudK, RT, AD, and DRJ guided in the interpretation, and manuscript writing. PD drafted the original manuscript. The manuscript was revised and edited by TPJ, DRJ and PD. All the authors contributed to the article and approved the submitted version.
Availability of Data and Material: All data collected in the study have been presented in the manuscript.
Code Availability: Not applicable
Consent to Participate: Written consent was obtained from the patients before the collection of samples and data.
Consent for Publication: Not applicable
Ethical Approval: Ethical approval was obtained from Institutional Review Committee of Institute of Science and Technology, Tribhuvan University, Kirtipur, Kathmandu (IRC/IOST-Regd. No. 1).
References
- 1. Nhu NTK, Phan MD, Peters KM, et al. Discovery of new genes involved in curli production by a uropathogenic Escherichia coli strain from the highly virulent O45:K1:H7 lineage. mBio. 2018;9:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kot B. Antibiotic resistance among uropathogenic Escherichia coli. Pol J Microbiol. 2019;68:403-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abejew AA, Denboba AA, Mekonnen AG. Prevalence and antibiotic resistance pattern of urinary tract bacterial infections in Dessie area, north-East Ethiopia. BMC Res Notes. 2014;7:687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Honarmand Jahromy S, Zare Karizi S. Evaluation of biofilm formation and frequency of genes encoding curli fiber, colanic acid capsule and f1c fimberia among uropathogenic Escherichia coli isolates with strong cell surface hydrophobicity. Avicenna J Clin Microb Infec. 2019;6:2-8. [Google Scholar]
- 5. Foxman B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am. 2014;28:1-13. [DOI] [PubMed] [Google Scholar]
- 6. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13:269-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105-107. [DOI] [PubMed] [Google Scholar]
- 8. Justice SS, Hung C, Theriot JA, et al. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci USA. 2004;101:1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hadjifrangiskou M, Gu AP, Pinkner JS, et al. Transposon mutagenesis identifies uropathogenic. Escherichia coli biofilm factors. J Bacteriol. 2012;194:6195-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med. 2013;3:a010306-a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hufnagel DA, Evans ML, Greene SE, Pinkner JS, Hultgren SJ, Chapman MR. The catabolite repressor protein-cyclic AMP complex regulates csgD and biofilm formation in uropathogenic Escherichia coli. J Bacteriol. 2016;198:3329-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eberly A, Floyd K, Beebout C, et al. Biofilm formation by uropathogenic Escherichia coli iIs fFavored under oxygen conditions that mimic the bladder environment. Int J Mol Sci. 2017;18:2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20-26. [DOI] [PubMed] [Google Scholar]
- 14. Paytubi S, Cansado C, Madrid C, Balsalobre C. Nutrient composition promotes switching between pellicle and bottom biofilm in Salmonella. Front Microbiol. 2017;8:2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luna-Pineda VM, Moreno-Fierros L, Cázares-Domínguez V, et al. Curli of uropathogenic Escherichia coli enhance urinary tract colonization as a fitness factor. Front Microbiol. 2019;10:2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator csgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol. 2011;193:2587-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma Q, Wood TK. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ Microbiol. 2009;11:2735-2746. [DOI] [PubMed] [Google Scholar]
- 19. York A. Biofilms: naturally modified cellulose in bacterial biofilms. Nat Rev Microbiol. 2018;16:123. [DOI] [PubMed] [Google Scholar]
- 20. Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452-1463. [DOI] [PubMed] [Google Scholar]
- 21. Liu Z, Niu H, Wu S, Huang R. CsgD regulatory network in a bacterial trait-altering biofilm formation. Emerg Microbes Infect. 2014;3:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. PT. 2015;40:277. [PMC free article] [PubMed] [Google Scholar]
- 23. Ciofu O, Rojo-Molinero E, Macià MD, Oliver A. Antibiotic treatment of biofilm infections. APMIS. 2017;125:304-319. [DOI] [PubMed] [Google Scholar]
- 24. Hall MR, McGillicuddy E, Kaplan LJ. Biofilm: basic principles, pathophysiology, and implications for clinicians. Surg Infect. 2014;15:1-7. [DOI] [PubMed] [Google Scholar]
- 25. Kokare CR, Chakraborty S, Khopade AN, et al. Biofilm: importance and applications. Indian J Biotechnol. 2009;8:159-168. [Google Scholar]
- 26. Mittal S, Sharma M, Chaudhary U. Biofilm and multidrug resistance in uropathogenic Escherichia coli. Pathog Glob Health. 2015;109:26-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ponnusamy P, Natarajan V, Sevanan M. In vitro biofilm formation by uropathogenic Escherichia coli and their antimicrobial susceptibility pattern. Asian Pac J Trop Med. 2012;5:210-213. [DOI] [PubMed] [Google Scholar]
- 28. Zhao F, Yang H, Bi D, Khaledi A, Qiao M. A systematic review and meta-analysis of antibiotic resistance patterns, and the correlation between biofilm formation with virulence factors in uropathogenic E. coli isolated from urinary tract infections. Microb Pathog. 2020;144:104196. [DOI] [PubMed] [Google Scholar]
- 29. Shrestha B, Shrestha B, Poudel A, Lekhak B, Upreti MK. In-vitro biofilm detection among uropathogens and their antibiogram profile. Tribhuvan Univ J Microbiol. 2018;5:57-62. [Google Scholar]
- 30. Ochoa SA, Cruz-Córdova A, Luna-Pineda VM, et al. Multidrug- and extensively drug-resistant uropathogenic Escherichia coli clinical strains: phylogenetic groups widely associated with integrons maintain high genetic diversity. Front Microbiol. 2016;7:2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Behzadi P, Urbán E, Gajdács M. Association between biofilm-production and antibiotic resistance in uropathogenic Escherichia coli (UPEC): an in vitro study. Diseases. 2020;8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cepas V, López Y, Muñoz E, et al. Relationship between biofilm formation and antimicrobial resistance in Gram-negative bacteria. Microb Drug Resist. 2019;25:72-79. [DOI] [PubMed] [Google Scholar]
- 33. Adamus-Białek W, Vollmerhausen TL, Janik K. Hydrogen peroxide stimulates uropathogenic Escherichia coli strains to cellulose production. Microb Pathog. 2019;126:287-291. [DOI] [PubMed] [Google Scholar]
- 34. Kass EH. Pyelonephritis and bacteriuria. A major problem in preventive medicine. Ann Intern Med. 1962;56:46-53. [DOI] [PubMed] [Google Scholar]
- 35. Stamm WE. Quantitative urine cultures revisited. Eur J Clin Microbiol. 1984;3:279-281. [DOI] [PubMed] [Google Scholar]
- 36. CLSI. Performance Standards for Antimicrobial Suceptibility Testing. Clinical and Laboratory Standards Institute; 2019. [Google Scholar]
- 37. Nascimento HH, Silva LE, Souza RT, Silva NP, Scaletsky IC. Phenotypic and genotypic characteristics associated with biofilm formation in clinical isolates of atypical enteropathogenic Escherichia coli (aEPEC) strains. BMC Microbiol. 2014;14:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Christensen GD, Simpson WA, Bisno AL, Beachey EH. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Imumun. 1982;37:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanchez CJ, Jr, Mende K, Beckius ML, et al. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis. 2013;13:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of Staphylococcal biofilm formation. J Microbiol Methods. 2000;40:175-179. [DOI] [PubMed] [Google Scholar]
- 41. Schiebel J, Böhm A, Nitschke J, et al. Genotypic and phenotypic characteristics associated with biofilm formation by human clinical Escherichia coli isolates of different pathotypes. Appl Environ Microbiol. 2017;83:e01660-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen C, Liao X, Jiang H, et al. Characteristics of Escherichia coli biofilm production, genetic typing, drug resistance pattern and gene expression under aminoglycoside pressures. Environ Toxicol Pharmacol. 2010;30:5-10. [DOI] [PubMed] [Google Scholar]
- 43. Beaumont F, Jouvec H-M, Gagnon J, Gaillard J, Pelmont J. Purification and properties of a catalase from potato tubers (Solanum tuberosum). Plant Sci. 1990;72:19-26. [Google Scholar]
- 44. Sedighi I, Arabestani MR, Rahimbakhsh A, Karimitabar Z, Alikhani MY. Dissemination of extended-spectrum β-lactamases and quinolone resistance genes among clinical isolates of uropathogenic Escherichia coli in children. Jundishapur J Microbiol. 2015;8:e19184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pourzare M, Derakhshan S, Roshani D. Distribution of uropathogenic virulence genes in Escherichia coli isolated from children with urinary tract infection in Sanandaj, Iran. Arch Pediatr Infect Dis. 2017;5:e41995. [Google Scholar]
- 46. Harwalkar A, Gupta S, Rao A, Srinivasa H. Lower prevalence of hlyD, papC and cnf-1 genes in ciprofloxacin-resistant uropathogenic Escherichia coli than their susceptible counterparts isolated from southern India. J Infect Public Health. 2014;7:413-419. [DOI] [PubMed] [Google Scholar]
- 47. Karam MRA, Habibi M, Bouzari S. Relationships between virulence factors and antimicrobial resistance among Escherichia coli isolated from urinary tract infections and commensal isolates in Tehran, Iran. Osong Public Health Res Perspect. 2018;9:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shrestha R, Khanal S, Poudel P, et al. Extended spectrum β-lactamase producing uropathogenic Escherichia coli and the correlation of biofilm with antibiotics resistance in Nepal. Ann Clin Microbiol Antimicrob. 2019;18:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gurung S, Kafle S, Dhungel B, et al. Detection of OXA-48 gene in carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from urine samples. Infect Drug Resist. 2020;13:2311-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dehbanipour R, Rastaghi S, Sedighi M, Maleki N, Faghri J. High prevalence of multidrug-resistance uropathogenic Escherichia coli strains, Isfahan, Iran. J Nat Sci Biol Med. 2016;7:22-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ali I, Rafaque Z, Ahmed S, Malik S, Dasti JI. Prevalence of multi-drug resistant uropathogenic Escherichia coli in Potohar region of Pakistan. Asian Pac J Trop Biomed. 2016;6:60-66. [Google Scholar]
- 52. Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, Pascual A. Treatment of infections caused by Extended-Spectrum-Beta-Lactamase-, ampc-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev. 2018;31:e00079-NaN17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Malekzadegan Y, Khashei R, Sedigh Ebrahim-Saraie H, Jahanabadi Z. Distribution of virulence genes and their association with antimicrobial resistance among uropathogenic Escherichia coli isolates from Iranian patients. BMC Infect Dis. 2018;18:572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Davies J. Origins and evolution of antibiotic resistance. Microbiología. 1996;12:9-16. [PubMed] [Google Scholar]
- 55. Leekha S, Terrell CL, Edson RS. General principles of antimicrobial therapy. Mayo Clin Proc. 2011;86:156-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yadav K, Prakash S. Screening of ESBL producing multidrug resistant E. coli from urinary tract infection suspected cases in southern terai of Nepal. J Infect Dis Diagn. 2017;2:100116. [Google Scholar]
- 57. Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69-72. [DOI] [PubMed] [Google Scholar]
- 58. O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49-79. [DOI] [PubMed] [Google Scholar]
- 59. Lemon KP, Earl AM, Vlamakis HC, Aguilar C, Kolter R. Biofilm development with an emphasis on Bacillus subtilis. Curr Top Microbiol Immunol. 2008;322:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Joshua GWP, Guthrie-Irons C, Karlyshev AV, Wren BW. Biofilm formation in Campylobacter jejuni. Microbiology. 2006;152:387-396. [DOI] [PubMed] [Google Scholar]
- 61. Goller CC, Romeo T. Environmental influences on biofilm development. Curr Top Microbiol Immunol. 2008;322:37-66. [DOI] [PubMed] [Google Scholar]
- 62. Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73:310-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Suman E, Jose J, Varghese S, Kotian MS. Study of biofilm production in Escherichia coli causing urinary tract infection. Indian J Med Microbiol. 2007;25:305-306. [DOI] [PubMed] [Google Scholar]
- 64. Zamani H, Salehzadeh A. Biofilm formation in uropathogenic Escherichia coli: association with adhesion factor genes. Turk J Med Sci. 2018;48:162-167. [DOI] [PubMed] [Google Scholar]
- 65. Krasowska A, Sigler K. How microorganisms use hydrophobicity and what does this mean for human needs? Front Cell Infect Microbiol. 2014;4:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Simm R, Fetherston JD, Kader A, Römling U, Perry RD. Phenotypic convergence mediated by GGDEF-domain-containing proteins. J Bacteriol. 2005;187:6816-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ude S, Arnold DL, Moon CD, Timms-Wilson T, Spiers AJ. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ Microbiol. 2006;8:1997-2011. [DOI] [PubMed] [Google Scholar]
- 68. Spiers AJ, Bohannon J, Gehrig SM, Rainey PB. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol Microbiol. 2003;50:15-27. [DOI] [PubMed] [Google Scholar]
- 69. Barken KB, Pamp SJ, Yang L, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10:2331-2343. [DOI] [PubMed] [Google Scholar]
- 70. Hölscher T, Bartels B, Lin YC, et al. Motility, chemotaxis and aerotaxis contribute to competitiveness during bacterial pellicle biofilm development. J Mol Biol. 2015;427:3695-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sannes MR, Kuskowski MA, Johnson JR. Geographical distribution of antimicrobial resistance among Escherichia coli causing acute uncomplicated pyelonephritis in the United States. FEMS Immunol Med Microbiol. 2004;42:213-218. [DOI] [PubMed] [Google Scholar]
- 73. Makled AF, Salem EH, Elbrolosy AM. Biofilm formation and antimicrobial resistance pattern of uropathogenic E. Coli : comparison of phenotypic and molecular methods. Egypt J Med Microbiol. 2017;26:37-45. [Google Scholar]
- 74. Coculescu BI. Antimicrobial resistance induced by genetic changes. J Med Life. 2009;2:114-123. [PMC free article] [PubMed] [Google Scholar]
- 75. Saldaña Z, Xicohtencatl-Cortes J, Avelino F, et al. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of fis as a negative regulator of curli. Environ Microbiol. 2009;11:992-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Grimm M, Stephan R, Iversen C, et al. Cellulose as an extracellular matrix component present in Enterobacter sakazakii biofilms. J Food Prot. 2008;71:13-18. [DOI] [PubMed] [Google Scholar]
- 77. Da Re S, Ghigo JM. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J Bacteriol. 2006;188:3073-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chen S, Feng Z, Sun H, Zhang R, Qin T, Peng D. Biofilm-formation-related genes csgD and bcsA promote the vertical transmission of Salmonella enteritidis in chicken. Front Vet Sci. 2020;7:625049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang X, Preston Jf, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tajbakhsh E, Ahmadi P, Abedpour-Dehkordi E, Arbab-Soleimani N, Khamesipour F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob Resist Infect Control. 2016;5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Davari Abad E, Khameneh A, Vahedi L. Identification phenotypic and genotypic characterization of biofilm formation in Escherichia coli isolated from urinary tract infections and their antibiotics resistance. BMC Res Notes. 2019;12:796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Noie Oskouie A, Hasani A, Ahangarzadeh Rezaee M, Soroush Bar Haghi MH, Hasani A, Soltani E. A relationship between o-serotype, antibiotic susceptibility and biofilm formation in uropathogenic Escherichia coli. Microb Drug Resist. 2019;25:951-958. [DOI] [PubMed] [Google Scholar]
- 83. Macedo AJ, Abraham W-R. Can infectious biofilm be controlled by blocking bacterial communication? Med Chem. 2009;5:517-528. [DOI] [PubMed] [Google Scholar]


