Abstract
Objectives:
Extracellular vesicles (EVs) are lipid bound vesicles secreted by cells into the extracellular environment. Studies have implicated EVs in cell proliferation, epithelial-mesenchymal transition, metastasis, angiogenesis and mediating the interaction of tumor cells and microenvironment. A systematic characterization of EVs from pancreatic cancer cells and cancer-associated fibroblasts (CAFs) would be valuable for studying the roles of EV proteins in pancreatic tumorigenesis.
Methods:
Proteomic and functional analyses were applied to characterize the proteomes of EVs released from five pancreatic cancer lines, two CAF cell lines and a normal pancreatic epithelial cell line (HPDE).
Results:
More than 1400 non-redundant proteins were identified in each EV derived from the cell lines. The majority of the proteins identified in the EVs from the cancer cells, CAFs and HPDE were detected in all three groups, highly enriched in the biological processes of vesicle-mediated transport and exocytosis. Protein networks relevant to pancreatic tumorigenesis, including EMT, complement and coagulation components, were significantly enriched in the EVs from cancer cells or CAFs.
Conclusions:
These findings support the roles of EVs as a potential mediator in transmitting EMT signals and complement response in the tumor microenvironment and possibly contributing to coagulation defects related to cancer development.
Keywords: extracellular vesicles, pancreatic cancer, proteomics, cancer-associated fibroblasts, exosomes
Introduction
Extracellular vesicles (EVs) broadly represent lipid bound vesicles secreted by cells into the extracellular milieu, playing various roles in cell communication. While apoptotic bodies are known to be released by damaged cells, all cells release EVs to maintain cellular homeostasis. These EVs include exosomes (30–100 nm in diameter) released from the intracellular compartment and ectosomes (100–1000 nm in diameter) directly shed from the plasma membrane via outward budding. Depending on their cell of origin, EVs can contain various cell constituents, including DNA, RNA, proteins, lipids and cellular metabolites. Recent studies support a mechanism-driven functional role of EVs in mediating intercellular communication both locally and distantly.1–8 Such cell-cell communication mediated by EVs may induce a biological response of recipient cells to stimulate complex intracellular signaling that can significantly affect cellular functions. Due to their clinical relevance, EVs have become an area of great interest in biomedical research. They can be detected in cancer cell lines, tumor tissues and many biofluids including plasma or serum, making these small vesicles an attractive target for diagnostic and therapeutic applications.
In pancreatic cancer, EVs are involved in cell proliferation, epithelial-mesenchymal transition (EMT), metastasis, angiogenesis and mediating the interaction of tumor cells and tumor microenvironment.9–25 Mutations of common proto-oncogenes and tumor suppressor genes, such as mutant KRAS and TP53, have been detected in circulating EVs from patients with pancreatic ductal adenocarcinoma (PDAC),26–29 even in the early stages of the disease.30 Studies have also found that EVs play roles in immunosuppression and chemoresistance of PDAC.13,20,31–36 Proteins are a major constituent in EVs and play important roles in EV delivery and interactions with recipient cells, which may be involved in pancreatic cancer promotion and metastasis. Studies11,12,15,16,21,27,37–53 have suggested the potential role of EV proteins in diagnosing PDAC54–58 and in understanding tumor biology and therapeutic treatment of PDAC. However, the mechanism of how the EV protein cargo participates in communication with the tumor microenvironment and contributes to carcinogenesis of PDAC remains poorly understood.
A recent study reported a comprehensive proteomic analysis of EVs derived from tumor tissue explants and bodily fluids from multiple cancers, and identified a pan-cancer EV marker panel to distinguish tumor from normal tissues.59 Several other proteomic studies have emphasized the characterization of circulating EVs (eg, exosomes) from PDAC patients for biomarker discovery.60–62 However, EVs derived from bodily fluid or tumor tissue could be a mixture of various cell types. To focus on pancreatic cancer cells, two recent studies investigated the proteome of exosomes derived from several pancreatic cancer cell lines and, and identified cancer related signal pathways enriched in the exosomes from pancreatic cancer cell lines, in comparison to normal pancreatic ductal cells.63,64 Interestingly, about 35% of cellular proteins were sorted into exosomes, and these EV proteins presented significant enrichment of pathways relevant to PDAC, including pathways regulating cell death and survival, cellular movement, and cell-to-cell signaling and interaction, as well as inflammatory response and FXR/RXR activation in biosynthetic pathways.63 Due to the heterogeneity of PDAC, an essential understanding of the proteomes of EVs derived from PDAC cells from different patients would be important to facilitate the studies in EVs for translational and clinical applications. Furthermore, a systematic characterization of the EV proteomes from pancreatic cancer-associated fibroblast (CAF) has not been reported.
We herein report proteomic analysis to characterize the protein composition of EVs enriched from five pancreatic cancer lines, including MIA PaCa-2, PANC-1, BxPC-3, AsPC-1 and CFPAC-1. Three of these cell lines, MIA PaCa-2, PANC-1, and BxPC-3 originated from primary tumors.65 The other two cell lines, AsPC-1 and CFPAC-1, originated from metastasis. These PDAC cell lines originated from different patients and their genomic mutations and related tumorigenic and metastatic properties have been previously characterized (ATCC.org). Two human primary pancreatic CAF cell lines (CAF11–50066 and WR76) were also included in our study, as CAF exosomes may regulate survival and proliferation of pancreatic cancer cells.67 A normal pancreatic epithelial cell line (HPDE)68 was used as the normal control for the comparison. The data obtained in this study provides an in-depth description of EV proteomes from pancreatic CAF cell lines and in comparison to five PDAC cell lines and normal pancreatic ductal cells.
MATERIAL AND METHODS
Cell Lines
MIA PaCa-2, PANC-1, and BxPC-3 cell lines were obtained from ATCC (Manassas, Va). AsPC-1 and CFPAC-1 cell lines were obtained from Dr. Tatjana Crnogorac-Jurcevic (Cancer Research UK Barts Centre, London, UK). The human pancreatic ductal epithelium (HPDE) cell line was obtained from Dr. Ming-Sound Tsao (University of Toronto, Toronto, Canada). CAF11–500 (UM27) cells were obtained from Dr. Diane Simeone (NYU, New York, NY) and WR76 cells were a gift from Dr. David Dawson (UCLA, Los Angeles, Calif). Cells were grown in DMEM or RPMI supplemented with 10% fetal bovine serum and penicillin/streptomycin and maintained in a humidified 37oC incubator with 5% CO2.
EV Enrichment
Cells were grown to 50–60% confluence and switched to serum free media for 48 hours. 40 ml of conditioned media was filtered using a 0.22 μm Steriflip vacuum filter (Millipore #SCGP00525, Burlington, Mass) and concentrated 20-fold using Amicon Ultra-15 3K centrifugal filters (Millipore #UFC900308) as per manufacturer’s instructions and multiple consecutive spins. The filtrate was transferred to a new tube. Twenty percent volume of ExoQuick TC (System Biosciences #EXOTC10A-1, Palo Alto, Calif) was added to each filtrate per manufacturer’s instruction and previous studies.69–71 Samples were mixed by inversion and stored at 4°C overnight. ExoQuick/media mixtures were centrifuged for 30 minutes at 1500 × g at 4°C. The supernatant was aspirated. Tubes were spun for an additional 5 minutes at 1500 × g and residual ExoQuick-TC solution was removed. The EV pellet was resuspended in RIPA buffer plus protease inhibitors. Pellets were incubated on ice for 10 minutes and tubes were spun for 15 min at 13,000 × g at 4°C. Supernatant was transferred to a new tube and protein concentration measured with bicinchoninic acid assay (BCA assay). Protein yields for these cell lines varied, and ranged from 0.13 to 15.91 mg/ml. Enriched EV samples were visualized by transmission electron microscopy (TEM) to confirm the size of EVs. In brief, following ExoQuick-TC precipitation, EV pellets were fixed overnight in half strength Karnovsky’s fixative containing 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer at 4°C and incubated on mesh grids and embedded in 2% methylcellulose. Extracellular vesicles samples were visualized with a FEI Tecnai Spirit BioTwin TEM using AMT software at 80 kV with magnifications of 49,000× and 68,000× (AMT V602.580.06 by AMT Imaging Systems, Woburn, Mass). Images were analyzed for EV size using ImageJ (version 1.53e, National Institutes of Health, Bethesda, Md).
Proteomics Sample Preparation
Fifty μg of EV preparations were transferred to new tubes and brought to 100 μl volume with 1X phosphate buffered saline. Invertase glycoprotein (Sigma I0408, St. Louis, Mo) was added to 0.5% from 2 mg/ml stock solution. Dithiothreitol was added to 10 mM final concentration and tubes incubated at 50oC for 1 hour. Iodoacetamide was added to 25 mM final concentration and tubes incubated at RT in the dark for 30 min. For acetone precipitation, 5X volume of cold acetone (−20°C) was added to each tube. Tubes were vortexed briefly and incubated at −20°C for 1 hour. Then tubes were centrifuged at 13,000 × g for 10 min at 4°C. The supernatant was carefully removed, and tubes were incubated at room temperature uncapped for 5 minutes to allow residual acetone to evaporate. Pellets were then gently resuspended in 50 mM ammonium bicarbonate buffer. The samples were digested with sequencing-grade trypsin (Promega #V5111, Madison, Wis) using 1:50 enzyme to protein weight ratio in a two-step process. In the first step, half of the trypsin was added, and the sample was incubated for 2 h at 37°C and vortexed every 30 min. Then, the remaining trypsin was added, and the mixture was incubated for an additional 16 h at 37°C. Trypsinized samples were purified with C18 columns (Nest Group, Ipswich, Mass), resuspended to 1 μg/μl in 0.1% formic acid for mass spectrometric analysis.
Liquid Chromatography Tandem Mass Spectrometry Analysis
The samples were analyzed by liquid chromatography (LC) tandem mass spectrometry (MS/MS) using an Orbitrap Fusion™ Tribrid™ mass spectrometer (Thermo Fisher Scientific, Waltham, Mass) interfaced with a nanoACQUITY UPLC system (Waters, Milford, Mass). Buffer A and buffer B for the HPLC were water and acetonitrile with 0.1 % formic acid, respectively. One microgram of each sample was loaded from the autosampler onto a Integrafrit trap column (100 μm ID, NewObjective, Littleton, Mass) packed with Reprosil-Pur C18-AQ 120 Å 5 μm material (Dr. Maisch, Ammerbuch-Entringen, Germany) to a bed length of 3 cm. After loading and desalting for 10 min with 2% buffer B at a flow rate of 2 μL/min, the sample was brought in-line with a fused-silica capillary column (75 μm ID) with a pulled tip packed with 25 cm of Reprosil-Pur C18-AQ 120 Å 5 μm (Dr. Maisch). Peptides were then resolved with a 90-minute gradient of 5 to 30% B followed by flushing at 80% B for 10 minutes and column equilibration with 2% B for 20 minutes. The analytical flow rate was 0.3 μL/min and entire data acquisition lasted for 120 minutes. The peptides were analyzed using the data-dependent acquisition (DDA) mode. The survey scan was performed with 120K resolution at 400 m/z from 400 to 1600 m/z with AGC (automatic gain control) target of 4e5 and max injection time of 50 msec. The DDA cycle was limited to 3 seconds. Precursors exhibiting a charge state from 2 to 4 of greater than 5e3 intensity were then selected for further fragmentation within a dynamic exclusion range of 30 seconds. Fragmentation priority was given to the most intense ions. Monoisotopic precursor selection was enabled. Precursor ions were isolated using the quadrupole with an isolation window of 1.6 m/z. Higher-energy collisional dissociation (HCD) was applied with a normalized collision energy of 28% and resulting fragments were detected using the rapid scan rate in the ion trap. The AGC (automatic gain control) target for MS/MS was set to 1e4 and the maximum injection time was limited to 50 msec.
Data Processing and Analysis
The MS raw data files were converted to mzML open format and searched against the UniProt Homo sapiens database for peptide/protein identification using the Comet algorithm72 embedded in the Trans-Proteomic Pipeline.73 The database search was restricted with the following parameters: trypsin was set as the enzyme with maximum missed cleavages set to 2; the precursor ion tolerance was set to 20 ppm, and the fragment ion tolerance to 1.0 Da. Variable modifications were set to methionine oxidation, and carbamidomethylation on cysteine was set as a static modification. The peptide assignment was validated with PeptideProphet,74 and a probability score in correspondence with an error rate of 1% was applied to filter the peptides. The Skyline software75 (version 4.2.0, University of Washington, Seattle, Wash) was used for quantitative analysis of the DDA data. The composite spectral library was built using all of the DDA data collected from the samples analyzed with a PeptideProphet threshold score corresponding to 1% error rate. Quantification was made at MS1 level using the sum of the first 3 monoisotopic peaks. The abundance of each peptide was normalized to total ion current (TIC) and presented as ion per million (IPM) using the following formula: Normalized IPM = Peptide Intensity / TIC * 1,000,000. Protein quantification was achieved by summation of the normalized intensities of the corresponding peptides.
Statistical and Gene Set Enrichment Analysis
The protein network analysis and Gene Ontology (GO) annotation were conducted using STRING.76 The STRING implements well-known classification systems such as GO and KEGG (Kyoto Encyclopedia of Genes and Genomes), as well as new classification systems based on high-throughput text-mining and clustering of the association network itself.76 The relative abundance data was log2 transformed to stabilize the variance. To avoid the issue of log2 zero, we manually added one to the value before the data transformation. The data was further subject to median standardization and the mean of each protein within each sample was calculated. We used the normal control group (HPDE) as the reference and log2 fold change of each protein was calculated. Gene Set Enrichment Analysis (GSEA) pre-ranked analysis was performed and the annotated gene sets of “hallmark”, “gene ontology” and “KEGG” were used in this study. Adjusted P value less than 0.05 was considered to be statistically significant.
RESULTS
EV Characterization
The morphology and sizing of the enriched EVs were examined by TEM. As presented in Figure 1A, the size of vesicles ranged between 50 nm to 350 nm, with an average size of 118 (standard deviation [SD], 56) nm. A recent published MISEV2018 guidelines77 recommended using at least three positive protein markers of EVs, including at least one transmembrane/lipid-bound protein and one cytosolic protein in all bulk EV preparations to demonstrate the presence of EVs. Based on the mass spectrometric data, multiple common EV markers were detected in the enriched EV preparations from all the cell lines (Fig. 1C).
FIGURE 1.
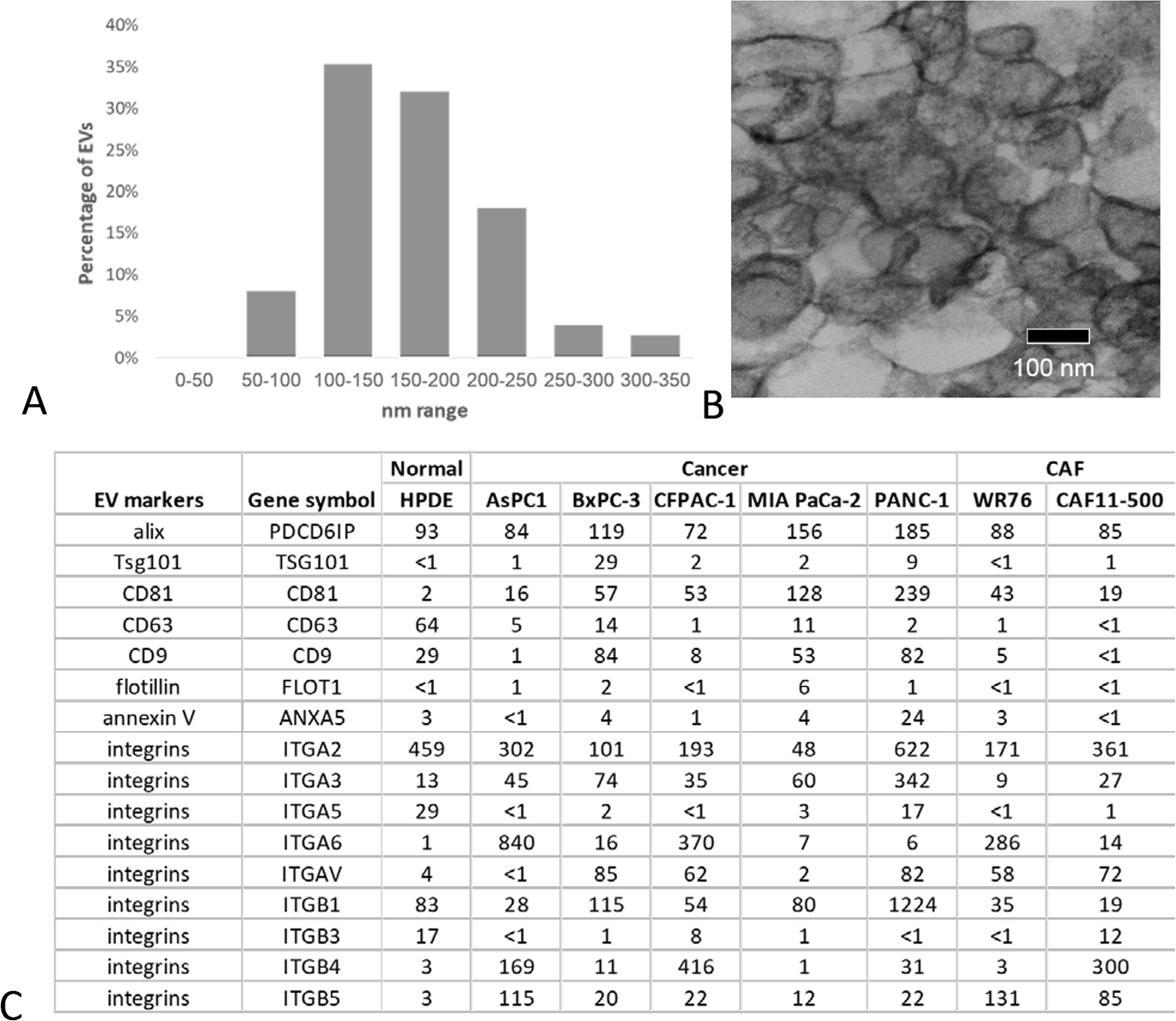
Characterization of enriched EVs. A, Histogram of the size distribution of EVs using analysis of images taken with electron microscopy (n = 150). B, Representative image from electron microscopy of EV enriched sample. The bar indicates 100 nm. C, Expression of common EV markers in the enriched EV samples based on proteomic data. The number presented in the table were the normalized intensity of the protein (IPM).
EV Protein Profiles in PDAC Cells, CAFs, and HPDE
The investigation of EV proteomes from five PDAC cell lines, two pancreatic cancer associated CAF cell lines, as well as a normal pancreas epithelial cell line, HPDE, yielded more than 1400 unique proteins in the EV proteome from each cell line using stringent criteria for peptide and protein identification (1% error rate) (Fig. 2A). In a comparison of the EV proteomes identified in cancer cells, CAFs and HPDE, more proteins were identified in the EVs derived from cancer cells, while a majority of the proteins (75%) were found in all three groups (Fig. 2B). The comparison of the two CAF lines indicates that about 86% of EV proteins identified from each CAF line overlapped with each other (Fig. 2C). Similar observations were made in the comparison of the EV proteomes for the five PDAC cell lines (Fig. 2D). Although each PDAC cell line demonstrated some differences and overlap with others, likely due to inter-tumoral heterogeneity, the majority of the EV proteins were commonly identified among the PDAC cells.
FIGURE 2.
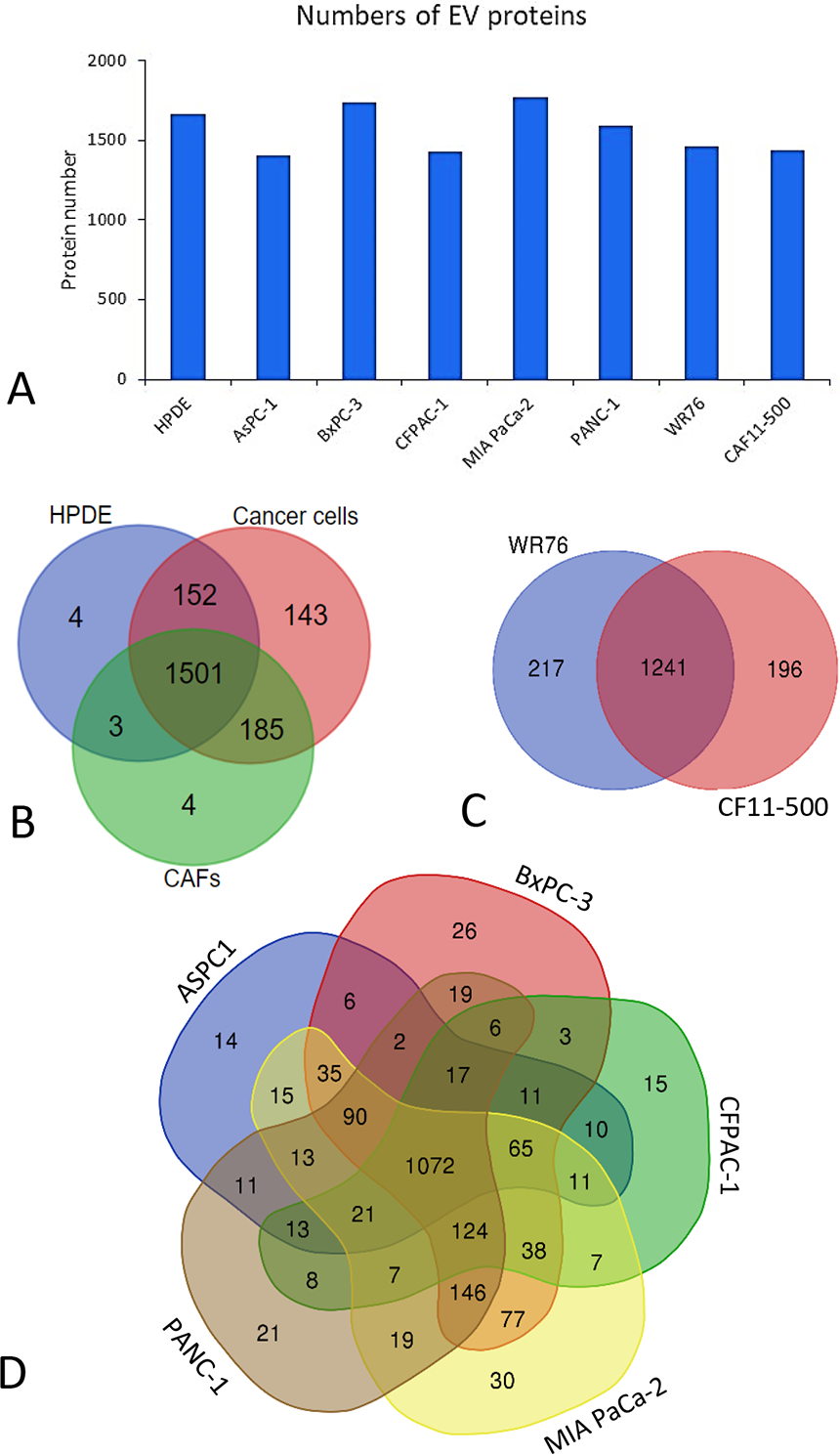
Summary of EV proteins identified from the eight cell lines, including 5 PDAC cell lines, 2 CAF cell lines, and HPDE. A, The number of proteins identified in each of the cell lines with 1% false discovery rate (FDR). B, Overlap of protein identification between the PDAC cell group, CAF group and HPDE. C, Overlap of protein identification between the two CAF cell lines. D) Overlap of protein identification among the five PDAC cell lines.
Composition of EV Proteomes
Among the EVs derived from all eight cell lines (5 PDAC, 2 CAF, and HPDE), 948 proteins were commonly identified. Protein network analysis indicated that these EV proteins represented a highly interactive and complex functional network (Fig. 3A). The composition of these proteins included 30% and 85% of extracellular proteins and intracellular proteins, respectively (a small portion of the proteins presented as located in both the extracellular and intracellular compartments). The extracellular proteins included the proteins that were present in the cell membrane, secretory vesicles, and extracellular matrix (Fig. 3B). The intracellular proteins were comprised of proteins derived from various intracellular components, such as the cytosol, secretory granules, nucleolus, endoplasmic reticulum, Golgi apparatus, and mitochondrion (Fig. 3C).
FIGURE 3.
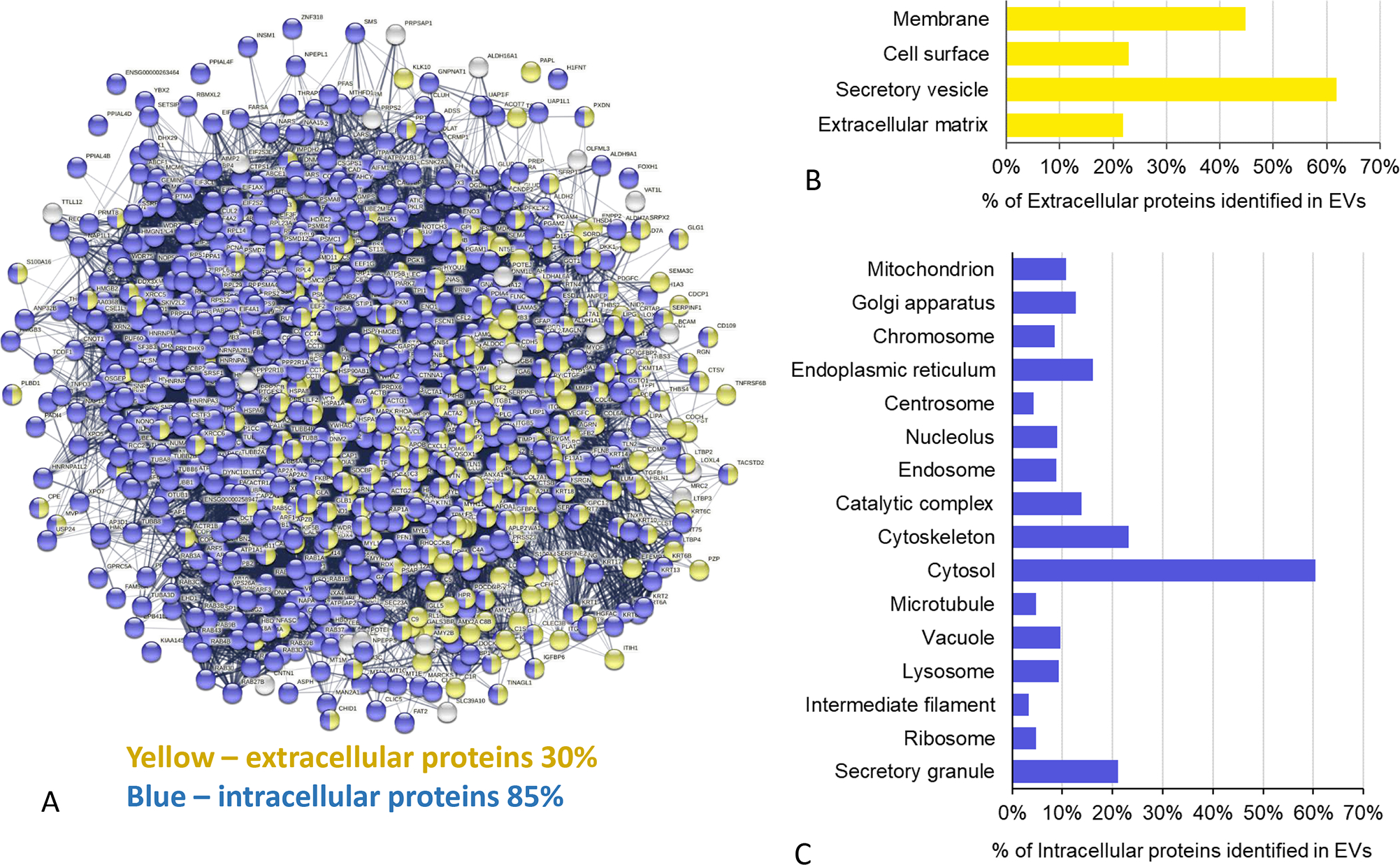
Protein network analysis of common EV proteins. A, The protein network of the common 948 EV proteins which were present in all eight cell lines. The yellow and blue nodes represent the extracellular and intracellular proteins, respectively. The nodes with two colors represent proteins may present in both extracellular and intracellular regions. B, The distributions of the extracellular proteins in the different extracellular regions. C, The distributions of the intracellular proteins among the intracellular components.
Protein Functions and Pathways Commonly Enriched in the EV Proteomes
The functional clustering and interaction analysis indicated that the common EV proteins identified in all eight cell lines were highly enriched in the biological processes of vesicle-mediated transport and regulated exocytosis (Fig. 4A), and molecular functions of bindings, structural molecule and hydrolase activity (Fig. 4B). The top enriched KEGG pathway was focal adhesion, with 55 EV proteins participating in the pathway (false discovery rate [FDR] = 1.55E-21) (Fig. 4C). Other enriched pathways include pathogenic infection and glycolysis. For protein-protein interaction, the most significantly enriched reactome was metabolism, involving 286 EV proteins (FDR = 7.99E-68) (Fig. 4D). Other top enriched reactomes also included the axon guidance and immune system.
FIGURE 4.
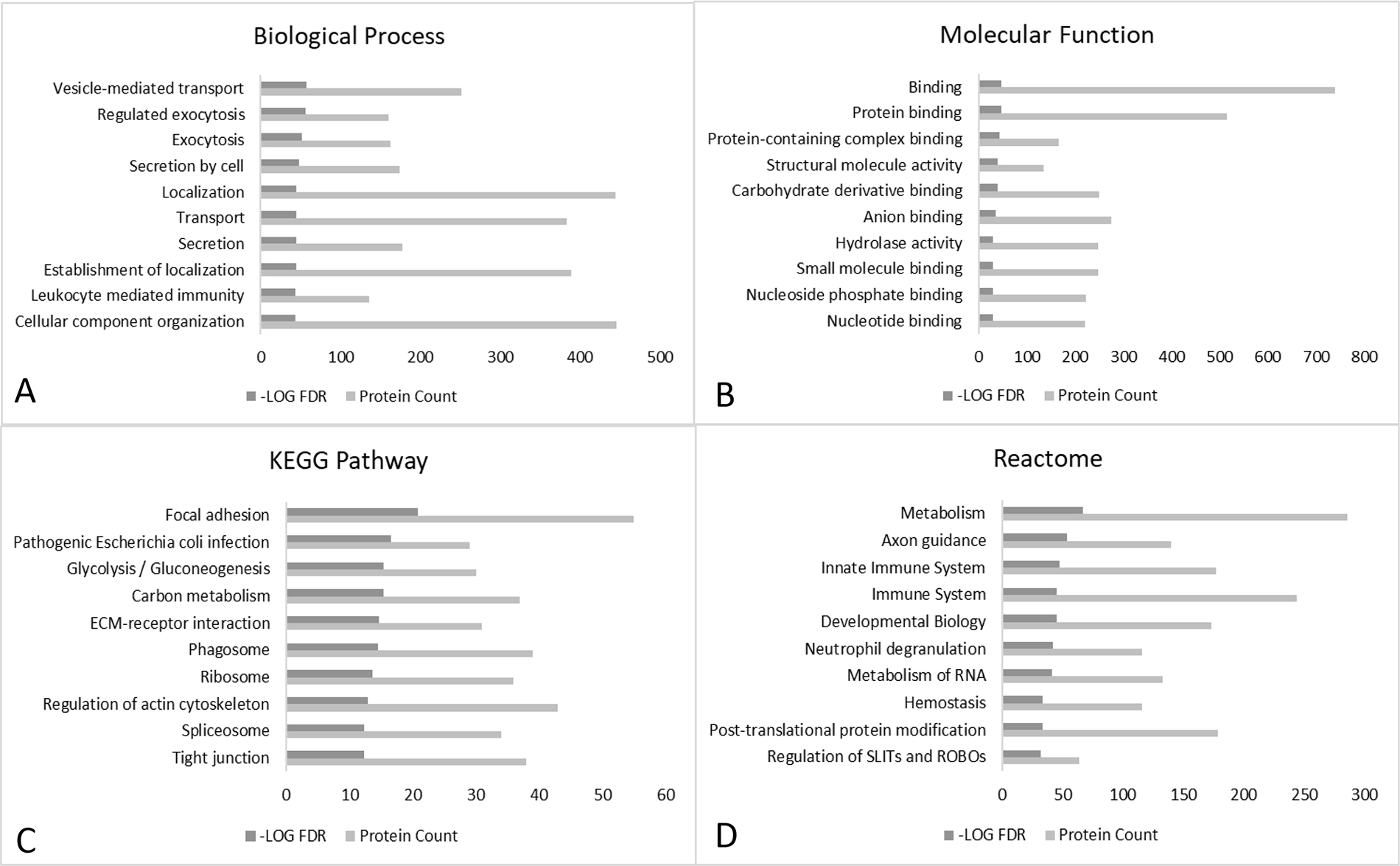
STRING analysis of common EV proteins. The common 948 EV proteins which were present in all eight cell lines were imported to STRING for enrichment analysis based on biological process (A), molecular function (B), KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway (C), and reactome (D). Only the top ten enrichment terms based on false discovery rate were presented.
Alterations of EV Proteomes in PDAC Cells and CAFs
Based on the protein intensity, quantitative assessments were further performed to identify the significant functional networks that were altered in PDAC cells and CAFs in comparison to the normal HPDE cell line. Using GO as the gene sets database, 58 and 56 dysregulated gene sets were identified in cancer group and CAF group, respectively (Supplemental Figs. 1A, B, note that only the top 50 sets were plotted). Using hallmark gene sets as the database, nine gene sets were identified with significant difference in the cancer group, including four gene sets that were highly up-regulated in the EVs from cancer cell lines (Fig. 5A). The GSEA plots of epithelial-mesenchymal transition (EMT), complement components and coagulation were illustrated in Figures 6A–C. For the CAFs, three gene sets were enriched in CAF group with statistical significance (Fig. 5B), including EMT, coagulation, and angiogenesis (Figs. 6D–F).
FIGURE 5.
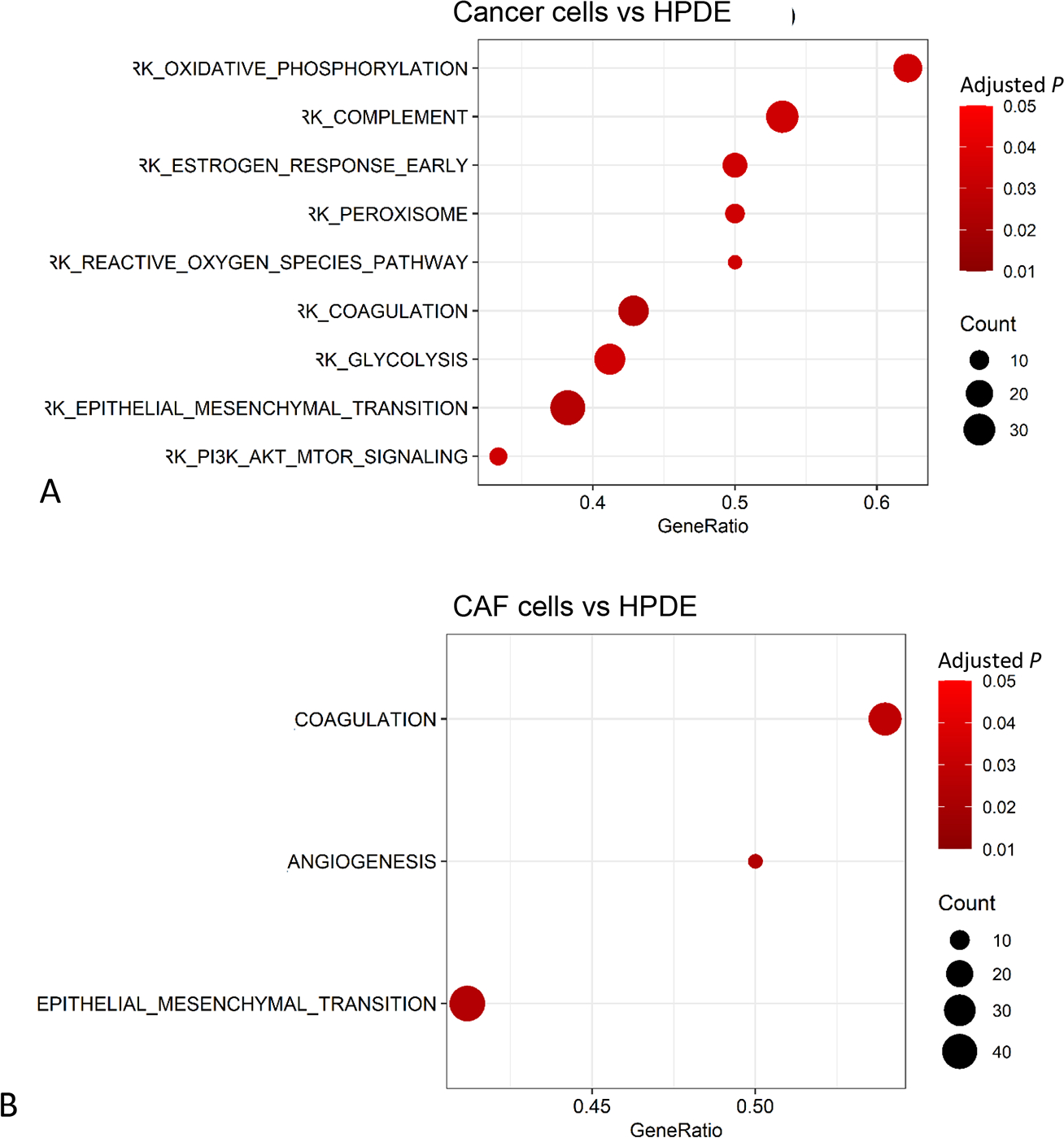
Enrichment analysis of differential proteins using GSEA (Gene Set Enrichment Analysis) with hallmark gene sets. A, PDAC cells versus HPDE. B), CAF cells versus HPDE.
FIGURE 6.
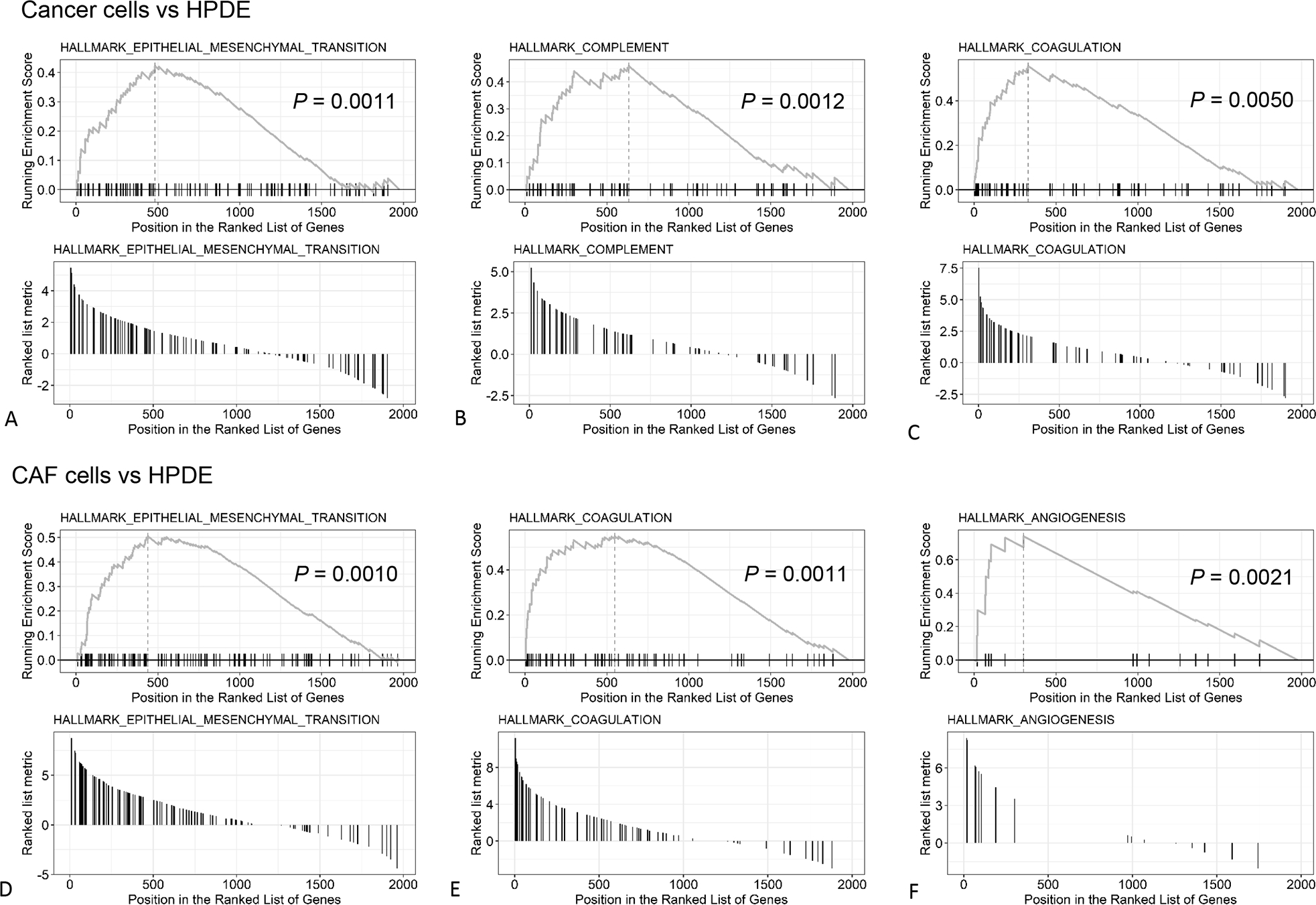
The GSEA (Gene Set Enrichment Analysis) plots of the top 3 up-regulated hallmark gene sets based on the p-value. A-C, PDAC cells versus HPDE. D-F, CAF cells versus HPDE.
Assessment of PDAC Associated Proteins in EV Proteome
In addition to the network analysis, we examined the individual EV proteins that may be associated with pancreatic cancer. Using 50 IPM as an intensity threshold, we focused on proteins that were relatively abundant (≥50 IPM) in the EVs derived from all the PDAC cell lines with >1.5-fold increase in expression compared to HPDE cells. Among the 36 proteins identified, a set of eight proteins (alpha-2-HS-glycoprotein (AHSG),78 apolipoprotein E (APOE),79,80 fibulin-1 (FBLN1),81 aminopeptidase N (ANPEP or CD13),82 plasminogen (PLG)83, gelsolin (GSN),84 S100-A4 (S100A4),79 and pigment epithelium-derived factor (SERPINF1),84 that were previously found to be associated with PDAC, are shown in Figure 7. Except for ANPEP (CD13), the remaining proteins showed increased expression in the EVs secreted from PDAC and CAF cell lines relative to HPDE. ANPEP (CD13) had an increased expression in the EVs from all PDAC cell lines, but not the two CAF cell lines. It is also notable that proteins from 14–3-3 protein family, which are implicated in pancreatic carcinogenesis and chemoresistance, were found significantly decreased in the EVs from all PDAC cell lines and CAF cell lines, including 14–3-3 protein epsilon, gamma, eta, zeta/delta.
FIGURE 7.
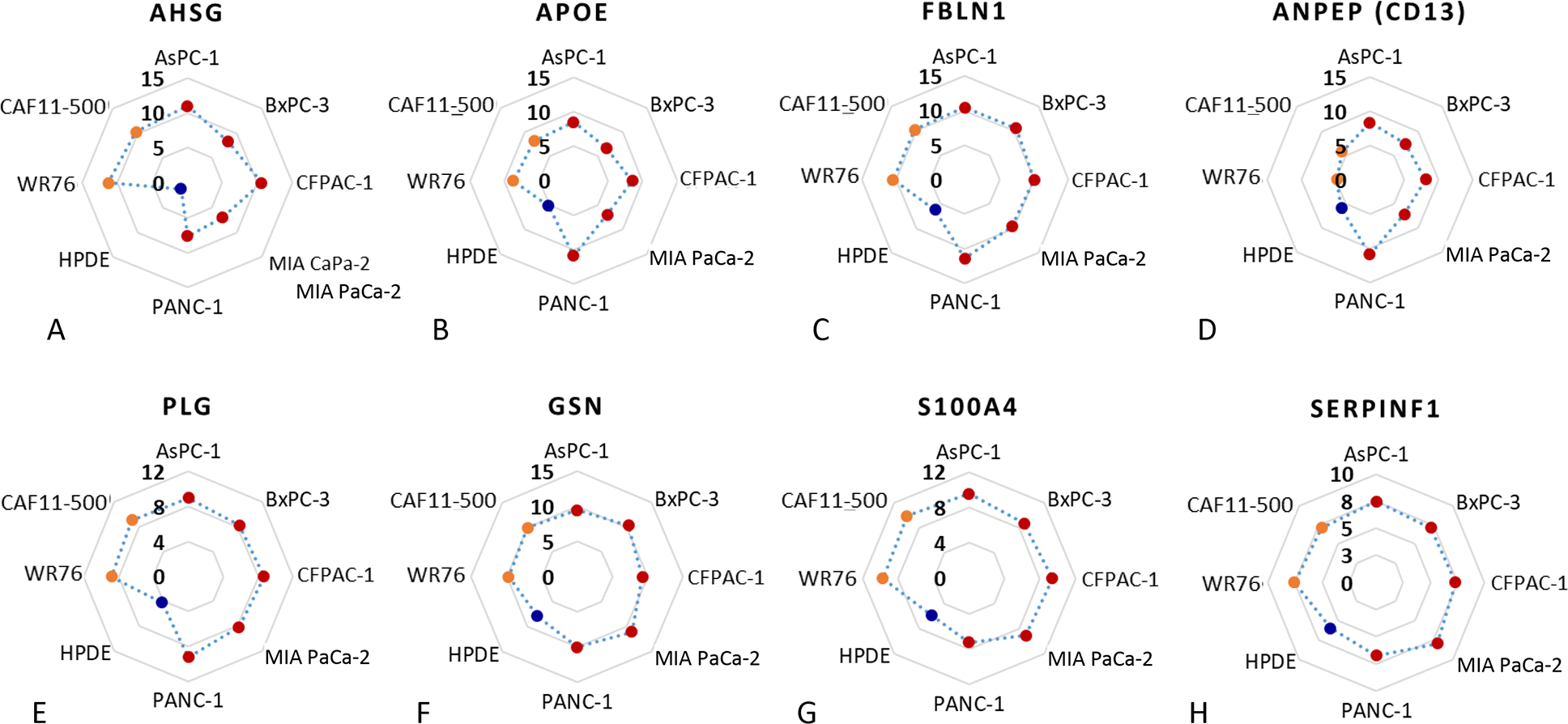
The abundance (represented by normalized intensity in log2 scale) of selected proteins in the EVs secreted from PDAC cells (red dots), CAFs (orange dots), and HPDE (blue dot). A, AHSG, (B) APOE, (C) FBLN1, (D) ANPEP (CD13), (E) PLG, (F) GSN, (G) S100A4, (H) SERPINF1.
DISCUSSION
In this study, we characterized the EV proteomes enriched from eight cell lines, including PDAC cells, CAFs, and normal ductal epithelial cells. As expected, most of the commonly used EV markers were detected in our cell line datasets, including alix, Tsg101, CD81, CD63, CD9, flotillin, integrins, and annexin V. Some EV markers, such as alix, CD81, and integrins, were present across all eight cell lines analyzed, and could thus be considered as pan-EV markers for these cell lines. In addition to these common EV markers, our study identified nearly 1000 proteins that were detected in all eight cell lines, which could be considered as common EV proteins. It is not surprising that 85% of these common EV proteins have been annotated as intracellular proteins, and they were highly enriched in the biological processes related to vesicle-mediated transport and regulated exocytosis.
In the comparison of the cancer cells, CAFs and HPDE, several interesting observations emerged. First, there were more hallmark gene sets enriched in the EVs from cancer cells than the non-cancer cells. Among these, EMT was the most significantly enriched gene set in cancer cells and surprisingly, also in mesenchymal CAFs. EMT is a process that epithelial cancer cells gain migratory and invasive mesenchymal-like properties, which drives cell stemness, invasiveness, and metastases. EVs could thus act as mediators to orchestrate intercellular communication between tumor cells, CAFs, and tumor-associated macrophages. In fact, EMT signals could be transferred between cancer cells and neighboring cells, such as CAFs and immune cells.85,86 Our findings of an enriched EMT gene set in the EVs of cancer cells and CAFs support and reinforce the role of EVs as a mediator in transmitting EMT signals in the tumor microenvironment.
Our study further highlighted the coagulation pathway as the second most significantly enriched gene set in the EVs from pancreatic cancer cells. For pancreatic cancer, studies have shown that EVs released from pancreatic cancer cells can promote thrombin generation.87,88 Interestingly, our data indicated that the EVs from CAFs were also enriched in the coagulation gene set. This confirms the notion that CAFs are critically involved in orchestrating the diverse crosstalk between pancreatic cancer cells and immune cells; together these cells contribute to a prominent PDAC tumor microenvironment that promotes cancer proliferation, metastasis, coagulation defects, and drug resistance.
GPC1 was previously explored as an exosome biomarker for pancreatic cancer detection.54,55,57,89 However, several recent studies have reported discrepant results on its accuracy in early detection of PDAC.89–92 In this study, while GPC1 was identified in the EVs from all cell lines, it showed an increased expression in the EVs from BxPC-3 and PANC-1 cancer cell lines and the two CAF cell lines (WR76, CAF11–500) relative to HPDE, but not in the EVs from the other three PDAC cell lines (AsPC-1, CFPAC-1, and MIA PaCa-2). More efforts may be needed to further evaluate exosomal GPC1 for its clinical value in pancreatic cancer detection. In summary, while a significant overlap was observed between EV proteomes of PDAC cells, CAFs and HPDE cells, there were notable differences between the malignant cells and normal cells. Protein networks relevant to pancreatic tumorigenesis, including EMT, complement response and coagulation processes, were significantly enriched in the EVs from cancer cells or CAFs. These findings support the roles of EVs as a potential mediator in transmitting malignant signals in tumor microenvironment and possibly contributing to coagulation defects related to cancer development. Further study is warranted to investigate potential functional changes or biological relevance of the EVs in relation to the protein abundance changes.
Supplementary Material
Funding
This work was supported in part with funding from Swim Across America and the National Institutes of Health under grant R01CA180949, as well as the support from the Walters Foundation for early detection of pancreatic cancer.
Abbreviations
- EV
Extracellular vesicles
- EMT
epithelial-mesenchymal transition
- CAF
cancer-associated fibroblasts
- HPDE
pancreatic epithelial cell line
- PDAC
pancreatic ductal adenocarcinoma
- GSEA
Gene Set Enrichment Analysis
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
Footnotes
The authors declare no conflict of interest.
REFERENCES
- 1.Choi DS, Kim DK, Kim YK, et al. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554–1571. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. [DOI] [PubMed] [Google Scholar]
- 4.Milane L, Singh A, Mattheolabakis G, et al. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–294. [DOI] [PubMed] [Google Scholar]
- 5.Raimondo F, Morosi L, Chinello C, et al. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. 2011;11:709–720. [DOI] [PubMed] [Google Scholar]
- 6.Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan A, De La Pena H, Seifalian AM. The application of exosomes as a nanoscale cancer vaccine. Int J Nanomedicine. 2010;5:889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villarroya-Beltri C, Baixauli F, Gutierrez-Vazquez C, et al. Sorting it out: regulation of exosome loading. Semin Cancer Biol. 2014;28:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiba M, Kubota S, Sato K, et al. Exosomes released from pancreatic cancer cells enhance angiogenic activities via dynamin-dependent endocytosis in endothelial cells in vitro. Sci Rep. 2018;8:11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman A, Hao W. The role of exosomes in pancreatic cancer microenvironment. Bull Math Biol. 2018;80:1111–1133. [DOI] [PubMed] [Google Scholar]
- 12.Heiler S, Wang Z, Zoller M. Pancreatic cancer stem cell markers and exosomes - the incentive push. World J Gastroenterol. 2016;22:5971–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Javeed N, Gustafson MP, Dutta SK, et al. Immunosuppressive CD14(+)HLA-DR(lo/neg) monocytes are elevated in pancreatic cancer and “primed” by tumor-derived exosomes. Oncoimmunology. 2017;6:e1252013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Javeed N, Sagar G, Dutta SK, et al. Pancreatic cancer-derived exosomes cause paraneoplastic beta-cell dysfunction. Clin Cancer Res. 2015;21:1722–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin H, Wu Y, Tan X. The role of pancreatic cancer-derived exosomes in cancer progress and their potential application as biomarkers. Clin Transl Oncol. 2017;19:921–930. [DOI] [PubMed] [Google Scholar]
- 16.Lan B, Zeng S, Grutzmann R, et al. The role of exosomes in pancreatic cancer. Int J Mol Sci. 2019;20:4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masamune A, Yoshida N, Hamada S, et al. Exosomes derived from pancreatic cancer cells induce activation and profibrogenic activities in pancreatic stellate cells. Biochem Biophys Res Commun. 2018;495:71–77. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa K, Lin Q, Li L, et al. Prometastatic secretome trafficking via exosomes initiates pancreatic cancer pulmonary metastasis. Cancer Lett. 2020;481:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray K Pancreatic cancer: Pancreatic cancer exosomes prime the liver for metastasis. Nat Rev Gastroenterol Hepatol. 2015;12:371. [DOI] [PubMed] [Google Scholar]
- 20.Shen T, Huang Z, Shi C, et al. Pancreatic cancer-derived exosomes induce apoptosis of T lymphocytes through the p38 MAPK-mediated endoplasmic reticulum stress. FASEB J. 2020;34:8442–8458. [DOI] [PubMed] [Google Scholar]
- 21.Sun W, Ren Y, Lu Z, et al. The potential roles of exosomes in pancreatic cancer initiation and metastasis. Mol Cancer. 2020;19:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Sun H, Provaznik J, et al. Pancreatic cancer-initiating cell exosome message transfer into noncancer-initiating cells: the importance of CD44v6 in reprogramming. J Exp Clin Cancer Res. 2019;38:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YF, Zhou YZ, Zhang B, et al. Pancreatic cancer-derived exosomes promoted pancreatic stellate cells recruitment by pancreatic cancer. J Cancer. 2019;10:4397–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng J, Hernandez JM, Doussot A, et al. Extracellular matrix proteins and carcinoembryonic antigen-related cell adhesion molecules characterize pancreatic duct fluid exosomes in patients with pancreatic cancer. HPB (Oxford). 2018;20:597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou W, Zhou Y, Chen X, et al. Pancreatic cancer-targeting exosomes for enhancing immunotherapy and reprogramming tumor microenvironment. Biomaterials. 2021;268:120546. [DOI] [PubMed] [Google Scholar]
- 26.Buscail L Pancreatic cancer: Exosomes for targeting KRAS in the treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2017;14:636–638. [DOI] [PubMed] [Google Scholar]
- 27.Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whiteside TL. Therapeutic targeting of oncogenic KRAS in pancreatic cancer by engineered exosomes. Transl Cancer Res. 2017;6(Suppl 9):S1406–S1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S, Che SP, Kurywchak P, et al. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol Ther. 2017;18:158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan J, Wei Q, Koay EJ, et al. Chemoresistance transmission via exosome-mediated EphA2 transfer in pancreatic cancer. Theranostics. 2018;8:5986–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel GK, Khan MA, Bhardwaj A, et al. Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. Br J Cancer. 2017;116:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Que RS, Lin C, Ding GP, et al. Increasing the immune activity of exosomes: the effect of miRNA-depleted exosome proteins on activating dendritic cell/cytokine-induced killer cells against pancreatic cancer. J Zhejiang Univ Sci B. 2016;17:352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su MJ, Aldawsari H, Amiji M. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of microRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci Rep. 2016;6:30110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y, Liu N, Wei Y, et al. Anticancer effects of miR-124 delivered by BM-MSC derived exosomes on cell proliferation, epithelial mesenchymal transition, and chemotherapy sensitivity of pancreatic cancer cells. Aging (Albany NY). 2020;12:19660–19676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Zhao N, Cui J, et al. Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210. Cell Oncol (Dordr) 2020;43:123–136. [DOI] [PubMed] [Google Scholar]
- 37.Ariston Gabriel AN, Wang F, Jiao Q, et al. The involvement of exosomes in the diagnosis and treatment of pancreatic cancer. Mol Cancer. 2020;19:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armstrong EA, Beal EW, Chakedis J, et al. Exosomes in pancreatic cancer: from early detection to treatment. J Gastrointest Surg. 2018;22:737–750. [DOI] [PubMed] [Google Scholar]
- 39.Batista IA, Melo SA. Exosomes and the Future of Immunotherapy in Pancreatic Cancer. Int J Mol Sci. 2019;20:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erb U, Zoller M. Progress and potential of exosome analysis for early pancreatic cancer detection. Expert Rev Mol Diagn. 2016;16:757–767. [DOI] [PubMed] [Google Scholar]
- 41.Farran B, Nagaraju GP. Exosomes as therapeutic solutions for pancreatic cancer. Drug Discov Today. 2020;25:2245–2256. [DOI] [PubMed] [Google Scholar]
- 42.Lau C, Kim Y, Chia D, et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013;288:26888–26897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu L, Risch HA. Exosomes: potential for early detection in pancreatic cancer. Future Oncol. 2016;12:1081–1090. [DOI] [PubMed] [Google Scholar]
- 44.Massoumi RL, Hines OJ, Eibl G, et al. Emerging evidence for the clinical relevance of pancreatic cancer exosomes. Pancreas. 2019;48:1–8. [DOI] [PubMed] [Google Scholar]
- 45.Mendt M, Kamerkar S, Sugimoto H, et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3:e99263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliveira C, Calmeiro J, Carrascal MA, et al. Exosomes as new therapeutic vectors for pancreatic cancer treatment. Eur J Pharm Biopharm. 2021;161:4–14. [DOI] [PubMed] [Google Scholar]
- 47.Qian L, Yu S, Chen Z, et al. Functions and clinical implications of exosomes in pancreatic cancer. Biochim Biophys Acta Rev Cancer. 2019;1871:75–84. [DOI] [PubMed] [Google Scholar]
- 48.Srinivas L, S D, Kgk D, et al. Current perspectives of exosomes as therapeutic targets and drug delivery vehicles for pancreatic cancer. Crit Rev Oncog. 2019;24:179–190. [DOI] [PubMed] [Google Scholar]
- 49.Takeda Y, Kobayashi S, Kitakaze M, et al. Immuno-surgical management of pancreatic cancer with analysis of cancer exosomes. Cells. 2020;9:1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang CC, Zhao YM, Wang HY, et al. New insight into the role of exosomes in pancreatic cancer. Ann Clin Lab Sci. 2019;49:385–392. [PubMed] [Google Scholar]
- 51.Wu H, Chen X, Ji J, et al. Progress of exosomes in the diagnosis and treatment of pancreatic cancer. Genet Test Mol Biomarkers. 2019;23:215–222. [DOI] [PubMed] [Google Scholar]
- 52.Yan Y, Fu G, Ming L. Role of exosomes in pancreatic cancer. Oncol Lett. 2018;15:7479–7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao C, Gao F, Weng S, et al. Pancreatic cancer and associated exosomes. Cancer Biomark. 2017;20:357–367. [DOI] [PubMed] [Google Scholar]
- 54.Frampton AE, Prado MM, Lopez-Jimenez E, et al. Glypican-1 is enriched in circulating-exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget. 2018;9:19006–19013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herreros-Villanueva M, Bujanda L. Glypican-1 in exosomes as biomarker for early detection of pancreatic cancer. Ann Transl Med. 2016;4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorenzon L, Blandino G. Glypican-1 exosomes: do they initiate a new era for early pancreatic cancer diagnosis? Transl Gastroenterol Hepatol. 2016;1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Huang S, Li P, et al. Pancreatic cancer-derived exosomes suppress the production of GIP and GLP-1 from STC-1cells in vitro by down-regulating the PCSK1/3. Cancer Lett. 2018;431:190–200. [DOI] [PubMed] [Google Scholar]
- 59.Hoshino A, Kim HS, Bojmar L, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–1061 e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.An M, Lohse I, Tan Z, et al. Quantitative proteomic analysis of serum exosomes from patients with locally advanced pancreatic cancer undergoing chemoradiotherapy. J Proteome Res. 2017;16:1763–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiao YJ, Jin DD, Jiang F, et al. Characterization and proteomic profiling of pancreatic cancer-derived serum exosomes. J Cell Biochem. 2019;120:988–999. [DOI] [PubMed] [Google Scholar]
- 62.Tang P, Tao L, Yuan C, et al. Serum derived exosomes from pancreatic cancer patients promoted metastasis: An iTRAQ-based proteomic analysis. Onco Targets Ther. 2019;12:9329–9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han S, Huo Z, Nguyen K, et al. The proteome of pancreatic cancer-derived exosomes reveals signatures rich in key signaling pathways. Proteomics. 2019;19:e1800394. [DOI] [PubMed] [Google Scholar]
- 64.Servage KA, Stefanius K, Gray HF, et al. Proteomic Profiling of small extracellular vesicles secreted by human pancreatic cancer cells implicated in cellular transformation. Sci Rep. 2020;10:7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deer EL, Gonzalez-Hernandez J, Coursen JD, et al. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waghray M, Yalamanchili M, Dziubinski M, et al. GM-CSF mediates mesenchymal-epithelial cross-talk in pancreatic cancer. Cancer Discov. 2016;6:886–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richards KE, Zeleniak AE, Fishel ML, et al. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36:1770–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qian J, Niu J, Li M, et al. In vitro modeling of human pancreatic duct epithelial cell transformation defines gene expression changes induced by K-ras oncogenic activation in pancreatic carcinogenesis. Cancer Res. 2005;65:5045–5053. [DOI] [PubMed] [Google Scholar]
- 69.Chugh PE, Sin SH, Ozgur S, et al. Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog. 2013;9:e1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alvarez ML. Isolation of urinary exosomes for RNA biomarker discovery using a simple, fast, and highly scalable method. Methods Mol Biol. 2014;1182:145–170. [DOI] [PubMed] [Google Scholar]
- 71.Sohel MM, Hoelker M, Noferesti SS, et al. Exosomal and non-exosomal transport of extra-cellular micrornas in follicular fluid: Implications for bovine oocyte developmental competence. PLoS One. 2013;8:e78505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. [DOI] [PubMed] [Google Scholar]
- 73.Deutsch EW, Mendoza L, Shteynberg D, et al. Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics Clin Appl. 2015;9:745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keller A, Nesvizhskii AI, Kolker E, et al. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. [DOI] [PubMed] [Google Scholar]
- 75.MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ochieng J, Nangami G, Sakwe A, et al. Impact of Fetuin-A (AHSG) on Tumor Progression and Type 2 Diabetes. Int J Mol Sci. 2018;19:2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan S, Brentnall TA, Chen R. Proteome alterations in pancreatic ductal adenocarcinoma. Cancer Lett. 2020;469:429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kemp SB, Carpenter ES, Steele NG, et al. Apolipoprotein E promotes immune suppression in pancreatic cancer through NF-kappaB-mediated production of CXCL1. Cancer Res. 2021;81:4305–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mahajan D, Kancharla S, Kolli P, et al. Role of fibulins in embryonic stage development and their involvement in various diseases. Biomolecules. 2021;11:685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pang L, Zhang N, Xia Y, et al. Serum APN/CD13 as a novel diagnostic and prognostic biomarker of pancreatic cancer. Oncotarget. 2016;7:77854–77864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Resovi A, Bani MR, Porcu L, et al. Soluble stroma-related biomarkers of pancreatic cancer. EMBO Mol Med. 2018;10: e8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peng H, Pan S, Yan Y, et al. Systemic Proteome alterations linked to early stage pancreatic cancer in diabetic patients. Cancers (Basel). 2020;12:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim H, Lee S, Shin E, et al. The emerging roles of exosomes as EMT regulators in cancer. Cells. 2020;9:861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang X, Luo G, Zhang K, et al. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kgamma to promote pancreatic cancer metastasis. Cancer Res. 2018;78:4586–4598. [DOI] [PubMed] [Google Scholar]
- 87.Becker A, Thakur BK, Weiss JM, et al. Extracellular vesicles in cancer: Cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rousseau A, Van Dreden P, Khaterchi A, et al. Procoagulant microparticles derived from cancer cells have determinant role in the hypercoagulable state associated with cancer. Int J Oncol. 2017;51:1793–1800. [DOI] [PubMed] [Google Scholar]
- 89.Lai X, Wang M, McElyea SD, et al. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017;393:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lewis JM, Vyas AD, Qiu Y, et al. Integrated analysis of exosomal protein biomarkers on alternating current electrokinetic chips enables rapid detection of pancreatic cancer in patient blood. ACS Nano. 2018;12:3311–3320. [DOI] [PubMed] [Google Scholar]
- 91.Xiao D, Dong Z, Zhen L, et al. Combined exosomal GPC1, CD82, and serum CA19–9 as multiplex targets: a specific, sensitive, and reproducible detection panel for the diagnosis of pancreatic cancer. Mol Cancer Res. 2020;18:300–310. [DOI] [PubMed] [Google Scholar]
- 92.Yang KS, Im H, Hong S, et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci Transl Med. 2017;9:eaal3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


