Abstract
Phenotypic switching in Candida albicans spontaneously generates different cellular morphologies and is manifested in strain WO-1 by the reversible switching between the white and opaque phenotypes. We present evidence that phenotypic switching is regulated by the Efg1 protein, which is known as an essential element of hyphal development (dimorphism). Firstly, EFG1 is expressed specifically in cells of the white but not the opaque phenotype. During mass conversion from the opaque to the white phenotype, the EFG1 transcript level correlates with competence of switching of opaque cells to the white form. Secondly, overexpression of EFG1 by a PCK1p-EFG1 fusion forces opaque-phase cells to switch to the white form with a high level of efficiency. Thirdly, low-level expression of EFG1 in strain CAI-8 generates a cellular phenotype similar to that of opaque cells in that cells bud as short rods, which cannot be induced to form hyphae in standard conditions; such cells (unlike authentic opaque cells) lack typical surface “pimples.” Importantly, the opaque-specific OP4 transcript is induced in the opaque-like cells generated by strain CAI8 as a response to low-level expression of EFG1. The results suggest that high EFG1 expression levels induce and maintain the white cell form while low EFG1 expression levels induce and maintain the opaque cell form. It is proposed that changes in EFG1 expression determine or contribute to phenotypic switching events in C. albicans.
Phenotypic switching spontaneously and reversibly generates different morphological and physiological states in the human pathogen Candida albicans (23, 24, 27–30). The characteristic forms that arise by phenotypic switching differ among C. albicans isolates. Switching in strain WO-1 and its derivatives has been studied extensively and consists of a change between a regular yeast form that grows as smooth and white colonies on solid media (white phase) and an elongated, rod-like form that grows as flattened grey colonies (opaque phase) (1, 23, 28). At a low frequency, both forms spontaneously convert to the alternative form; also, by a temperature upshift, the opaque form can be induced experimentally to grow as the white form (28). Both forms differ not only in their cytologies but also with regard to different physiological characteristics, their interaction with host cells, and their virulence (29). It has been proposed that phenotypic switching in C. albicans is equivalent to phase variation in other organisms, such as Salmonella typhimurium, Neisseria gonorrhoeae, or Trypanosoma brucei and functions to evade host immune systems and change adhesive properties (6, 8, 12, 36). Although genes that are specifically expressed in the white phase (WH11) or in the opaque phase (OP4, SAP1, and CDR3) have been described and some regulatory regions within their promoters have been characterized (3, 19, 20, 31–33), nothing is known about the molecular mechanisms that govern phenotypic switching.
Besides phenotypic switching, which occurs spontaneously, C. albicans is known to change its growth form between a regular budding yeast form and a multicellular filamentous form (hypha or pseudohypha), depending on environmental conditions (dimorphism) (22). Evidence indicates that phenotypic switching and dimorphism are essentially different but nevertheless related processes. Both the white and the opaque forms of strain WO-1 are capable of forming hyphae, although hyphae formation of the opaque form occurs only under special conditions (1). Although some antigens not present in white cells are shared by the opaque form and hyphal forms, other antigens occur only in hyphae generated from both forms; a surface structure seen only on opaque cells (a “pimple”) is absent from hyphae of this form (2). Likewise, although the WH11 gene is expressed only in the white form and not in the opaque and hyphal forms (32), other genes (SAP4-6) are expressed only in hyphae (10).
In recent years, some major signal transduction pathways required for the yeast-to-hypha transition have been discovered. Efg1p is a key regulator of dimorphism, whose presence in C. albicans is required to allow induction of hyphae by serum (18, 35). Mutants lacking EFG1 grow as a yeast, and serum induces only short pseudohyphae rather than true septated hyphae, as in wild-type strains. A second pathway comprising members of a conserved mitogen-activated protein (MAP) kinase pathway (Hst7p and Cek1p) and the transcription factor Cph1p is also involved in hyphal morphogenesis (11, 15, 17). Mutants in this pathway are unable to form hyphae in certain media but still form hyphae in the presence of serum. Mutants lacking Cph1p, as well as Efg1p, grow only in the yeast form even in the presence of serum and are almost completely avirulent (18).
Here we present evidence suggesting that Efg1p not only is an essential element for dimorphism but also is involved in the regulation of the white and opaque phenotypic switching event. We show that the opaque state of strain WO-1 is very similar to the phenotype of cells lacking EFG1, a conclusion that is based on their cytological appearances, the expression of an opaque-state-specific gene, and the reduced ability to form hyphae. Furthermore, overexpression of EFG1 in the opaque form forces rapid conversion to the white growth form. Because the absence and presence of Efg1p determines the opaque and white phenotypes, respectively, Efg1p may be a key regulator of phenotypic switching.
MATERIALS AND METHODS
C. albicans strains and growth conditions.
C. albicans Red3/6 is an ade2/ade2-derivative of strain WO-1, which retains the white and opaque switching phenotype (33). Strain SC5314 and its derivatives CAI4 (ura3/ura3) and CAI8 (ura3/ura3 ade2/ade2) have been described (7). In strain SS4 (Δura3::imm434/Δura3::imm434 ade2::hisG/ade2::hisG efg1Δ::ADE2/[URA3-PCK1p]::EFG1), which is a derivative of CAI8, the only intact remaining EFG1 allele is under transcriptional control of the PCK1 promoter (35). HLC52 has the genotype Δura3::imm434/Δura3::imm434 efg1::hisG/efg1::hisG-URA3-hisG (18). Cells were grown in YPD,SCAA medium (0.67% yeast nitrogen base [Difco]–2% Casamino Acids), in SD medium, or in S4D medium (SD medium containing 4% glucose) (16, 26, 35). Solid media contained 2% agar. Colonies of white-and opaque-phase cells were visualized on medium containing 5 μg of phloxine B/ml (1).
Plasmids.
To construct a convenient ADE2-containing vector, the CaARS2 region contained in pRC2312 (4) was first amplified by PCR using the primers CaARS1 (5′-ATTGACGTCCGGGGTAGCGATGAG) and CaARS2 (5′-TTAGACGTCCAGGACCGGCCAGAC) (AatII site underlined). The 660-bp PCR fragment containing CaARS2 was subcloned into the AatII site of pUC19, resulting in plasmid pARS/AatII. Into the SmaI site of pARS/AatII, the 2.4-kb EcoRV fragment of pMK16 containing ADE2 (13) was inserted to generate the vector pBT-4. An EFG1 expression vector was constructed by ligating the 3.75-kb BamHI-HindIII fragment of pRC2312P-H (fusion of the PCK1 promoter to the coding region of EFG1) (35) to the large BamHI-HindIII fragment of pARS/AatII; the resulting vector, pUC-APE2, was modified further by insertion of the 2.4-kb EcoRV ADE2 fragment into its EheI site, resulting in expression vector pAPE(2)/ADE.
Northern analyses.
RNA from C. albicans strains was isolated and analyzed by Northern analysis as previously described (35). poly(A) RNA was prepared by using a commercial protocol (Oligotex; Qiagen, Hilden, Germany). The probes used were (i) the 1.5-kb NheI fragment of pUC19/EFG1, containing the coding region of EFG1 (35); (ii) the 220-bp BamHI fragment of pWH11, containing a segment of the WH11 coding region (kindly provided by K. Schroeppel); and (iii) the 1-kb NcoI fragment of pOP4/3, containing part of the OP4 coding region (kindly provided by K. Schroeppel). As a loading control standard, the 1.5-kb ClaI fragment of p1595/3, containing ACT1 (5), was used as a probe; alternatively, RNA gels were stained with ethidium bromide before blotting and the migration and intensity of 25S (3.44-kb) and 18S (1.8-kb) rRNA was recorded.
Scanning electron microscopy.
Cells were prepared for microscopy essentially as previously described (1), with some modifications. Cells were washed three times in buffer (1× phosphate-buffered saline) and fixed overnight in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2). After washing three times in 0.1 M cacodylate buffer, cells were allowed to attach to polylysine-coated coverslips by standing for at least 4 h. Samples were dehydrated stepwise by acetone washings (30 to 100%; 15 min per washing) followed by drying in a critical-point dryer (Balzers). Samples were coated with gold palladium and inspected in a scanning electron microscope (Cambridge Stereoscan 200).
RESULTS
EFG1 expression is specific for the white growth phase.
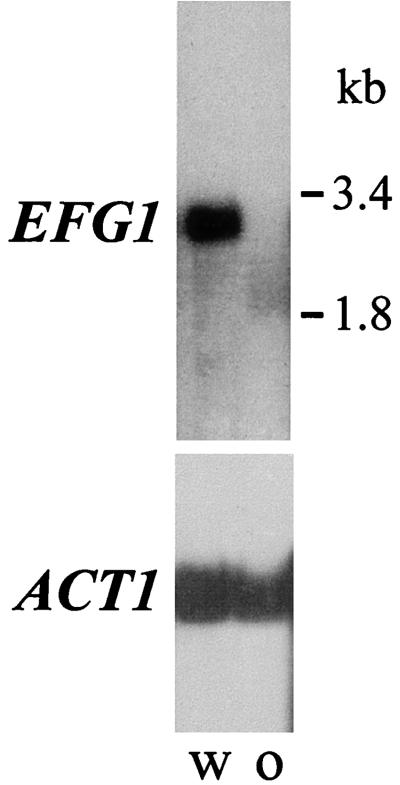
We had shown previously that following induction of hyphal morphogenesis, EFG1 transcript levels decline rapidly (35). Because a similar transcript regulation has been described for the WH11 transcript, which is known to be expressed only in the white phase of strain WO-1 (34), we sought to determine if EFG1 is regulated by the white and opaque switching phenotype of this strain. Both phenotypic forms of strain WO-1 were grown in YPD medium at 25°C, and total RNA and the poly(A) RNA fraction was isolated, followed by Northern blotting. Figure 1 demonstrates that the EFG1 transcript is detectable only in the white phase and not in the opaque-phase cells, while the ACT1 transcript level used as a control is not regulated differently in white and opaque cells. Thus, EFG1 expression is specific for the white phase of strain WO-1.
FIG. 1.
EFG1 expression is specific for the white phase of strain WO-1. poly(A) RNA of the white (w) and opaque (o) phases of strain WO-1 grown at 25°C in YPD medium was analyzed by Northern blotting using EFG1 and ACT1 probes. The migration of rRNA is as indicated.
EFG1 transcript level reflects switching competence.
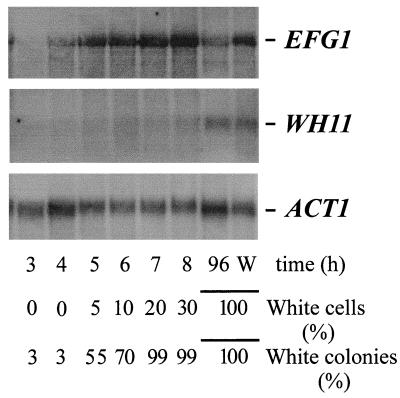
To monitor the time course of EFG1 induction during phenotypic switching, we induced a mass conversion from the opaque to the white phase by temperature shift as described previously (28) and analyzed EFG1 transcript levels (Fig. 2). The opaque phase of strain WO-1 was grown at 25°C in S4D-medium and at time (t) = 0, cells were transferred to 42°C and incubated further at this temperature. The optical densities at 600 nm (OD600) during the incubation at t = 0, 2, 3, 4, 5, 6, 7, and 8 h were 0.6, 1.2, 1.4, 1.6, 2.0, 2.4, 3.3, and 3.6, respectively. At these time points, the percentages of white and opaque cells were determined microscopically. The first white-phase cells appeared after 5 h, i.e., after two to three cell doublings, as reported previously (31). Because after temperature induction cells may still show the opaque phenotype while having acquired the competence to develop white cells in subsequent doublings, we also plated cells on solid SD medium and incubated these plates at 25°C (where the white and opaque forms are stably maintained). Cells in the culture 5 h after the temperature shift developed colonies consisting of a significant percentage of white-phase cells (55%), which is greater than the percentage determined microscopically (5%), indicating that most opaque-form cells in the culture had already been triggered to develop white-phase cells after 5 h, i.e., were switching competent.
FIG. 2.
Transcript levels during a temperature-induced shift from the opaque to the white phase. The opaque phase of strain WO-1 was pregrown at 25°C in S4D medium. At t = 0 h, cells were transferred to 42°C and incubated further at this temperature. Total RNA of cells was isolated at the indicated times and analyzed by using EFG1, WH11, or ACT1 probes. At each time point, the number of white- and opaque-phase cells was determined microscopically. In addition, a sample of the culture was plated on solid SD medium containing phloxine B, followed by incubation at 25°C and determination of white and opaque colonies after 3 days of growth. W, white-phase cells.
By Northern blotting, significant levels of the EFG1 transcript were detectable in cells induced for 5 h; maximum levels were present in cells induced for 7 to 8 h (Fig. 2). In contrast, the WH11 transcript appeared later, with the first signals appearing after 7 to 8 h and reaching maximum levels only after continued incubation. Thus, levels of the WH11 transcript mirrored the percentage of actual white-phase cells present in the culture (as determined microscopically), while EFG1 transcript levels appeared to reflect not only the actual cellular form but also the competence of opaque cells to develop white cells in subsequent divisions (as determined by colony phenotypes).
EFG1 overexpression induces the opaque-to-white switch.
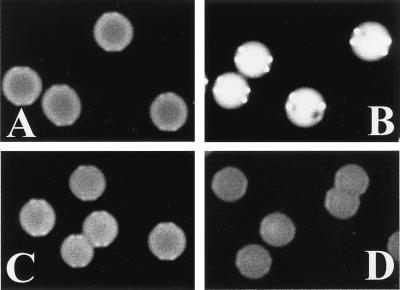
The above experiments suggested that high EFG1 expression was not simply the consequence of the white-phase cell form but also a precondition for the switch from opaque- to white-phase cells during temperature induction. To test if EFG1 expression alone, without temperature induction, was sufficient for switching, we transformed the opaque form of strain Red3/6, an ade2/ade2 derivative of strain WO-1 (28, 33), with plasmid pAPE(2)/ADE containing EFG1 under transcriptional control of the PCK1 promoter (16). Transformants grown in S4D high-glucose medium grown at 25°C remained stably in the opaque form and formed characteristic flattened red colonies on S4D-phloxine B plates (Fig. 3). In contrast, if such cells were plated and incubated on SCAA-phloxine B medium lacking glucose (at 25°C) and EFG1 expression was induced, 100% of colonies showed the white colony phenotype (white, smooth) and contained yeast cells. In some transformants, up to 20% of white colonies also contained one or more red dots indicative of remaining opaque-phase cells, which were verified microscopically (visible as dark spots in colony photographs; Fig. 3B). Colony appearance was independent of the medium used, because control transformants containing an empty vector (pBT-4) stably formed red colonies containing opaque-phase cells. If, following the EFG1-induced opaque-to-white shift, cells were replated on repressing S4D medium, they remained in the white-phase form. These experiments indicate that elevated EFG1 expression suffices to induce the switch from opaque to white cells, but that the return of EFG1 overexpression to wild-type expression levels (by the chromosomal EFG1 alleles) does not trigger a white-to-opaque switch.
FIG. 3.
EFG1 overexpression forces the opaque and white shift. Transformants of the opaque phase of strain Red3/6 contained plasmid pAPE(2)/ADE (PCK1p-EFG1) (A and B) or empty vector pBT-4 (C and D). Cells were pregrown at 25°C in S4D medium and then plated on SCAA medium (EFG1 expression) (B and D) or on S4D medium (no EFG1 expression) (A and C) and incubated at 25°C. Media contained phloxine B, which stains colonies of opaque cells red (which appear grey on figure) but not colonies of white-phase cells.
Similar results were obtained if transformants were grown in liquid S4D or SCAA media. Growth at 25°C in S4D medium led to growth of the Red3/6[pAPE(2)/ADE] transformant in the opaque form, while growth in SCAA medium resulted in only white-phase cells. It should be pointed out that overexpression of EFG1 in the Red3/6 genetic background did not lead to elongated pseudohyphae, as has been reported for strain CAI8 (35); instead, transformants grew only as yeasts. Northern analysis of three transformants grown in liquid media was performed to verify overexpression of the EFG1 transcript (Fig. 4). Clearly, EFG1 transcript levels were elevated in SCAA versus S4D medium, as expected; the degree of overexpression varied between transformants, presumably reflecting different plasmid copy numbers. S4D-grown opaque cells contained high levels of the opaque-specific OP4 transcript (19), while this transcript was missing in SCAA-grown cells. The level of the WH11 transcript, which has been described as a white-specific transcript (31), was lower in S4D- than in SCAA-grown cells. However, we found that the WH11 transcript level is strongly influenced by media composition, in that glucose reduces and the absence of glucose (SCAA or medium containing a nonfermentable carbon source) elevates WH11 transcript levels independent of the cell form (see below). Thus, WH11 transcript levels may be determined by the cell phenotype and/or the type of growth medium.
FIG. 4.
Transcripts in transformants of strain Red3/6. Three transformants of the opaque phase of strain Red3/6 that contained plasmid pAPE(2)/ADE (PCK1p-EFG1) (1 to 3) were diluted into SCAA medium (EFG1 expression) (C) and S4D medium (no EFG1 expression) (S). Cultures were grown at 25°C to an OD600 of 1, and total RNA of cells was prepared. Twenty micrograms of RNA was analyzed by Northern blotting using the indicated probes. Ethidium bromide-stained rRNA was used as loading control. w, white; o, opaque.
Low-level EFG1 expression induces an opaque-like phenotype.
The above experiments indicated that elevated EFG1 expression is associated with and directs the white cell form. These findings led to the question of whether lowered EFG1 expression is in turn correlated with the opaque phase. We could not directly address this question by using strain WO-1 or its derivative Red3/6, because gene disruption using the “URA-blaster” protocol (7) is not possible (WO-1 is prototrophic and Red3/6 has the genotype ade2/ade2). Despite extensive efforts, attempts to delete both EFG1 alleles in strain Red3/6 by mitotic recombination (25) failed, although single-allele disruptions could be obtained. The latter result suggested that strain Red3/6 (similar to strain CAI8 [35]) is more sensitive to the loss of EFG1 than other nonisogenic strains, such as CAI4 (18). Therefore, we tested whether strain SS4 (derivative of CAI8), in which the only remaining EFG1 allele is under control of the glucose-repressible PCK1 promoter (35), would react to the lowering of EFG1 expression on glucose medium by forming the opaque-phase cell form. This experiment was of special interest because strain CAI8 (and other derivatives of SC5314) is not known to undergo the white and opaque phenotypic switching.
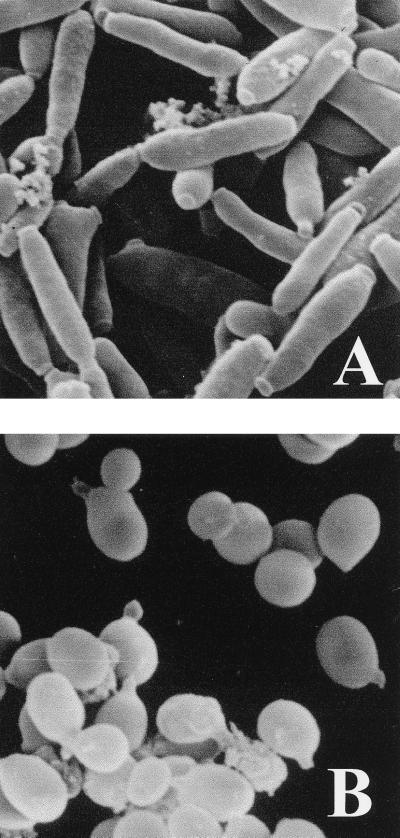
As expected, a EFG1/EFG1 isogenic control strain grew in the regular yeast cell form in S4D high-glucose medium (Fig. 5B). However, strain SS4 grew in S4D medium (low-level EFG1 expression) by unipolar budding in the form of regularly sized, rod-shaped cells. Photographs of cells observed by light microscopy (35) and by scanning electron microscopy (Fig. 5A) revealed that these cells were unlike typical pseudohyphae, which arise in some conditions in laboratory strains (22) or in strains overexpressing EFG1 (35), because of their uniform size and their shortness (length/width ratio = 5 to 6). Instead, they most closely resembled cells of the opaque phase of strains WO-1 or Red3/6. On S4D medium containing phloxine B, strain SS4 grew as a pink colony, resembling the opaque form of strain WO-1, which forms a red colony on this medium (see above). Furthermore, serum did not induce hyphae in strain SS4 at low EFG1 expression levels or in the complete efg1 knockout strain (18, 35) resembling the opaque form of strain WO-1 that is not able to form hyphae in standard induction conditions (1). However, unlike “authentic” opaque cells, the SS4-derived opaque-like cells did not show typical pimples (2) by scanning electron microscopy, even at high magnification (Fig. 5A).
FIG. 5.
Low EFG1 expression levels induce an opaque-phase-like cellular phenotype. Strain SS4 (efg1/PCK1p-EFG1) and strain CAI8 (EFG1/EFG1) containing empty control vectors (pBT-4 and pBI) as a control were grown in S4D medium, in which the PCK1 promoter is repressed (low-level EFG1 expression in strain SS4). Cells were visualized by scanning electron microscopy. Photographs represent images of 20 by 20 μm.
To confirm that the opaque-like cellular phenotype caused by low EFG1 expression in strain SS4 was indeed related to the opaque phenotype of strain WO-1, we tested whether the expression of a gene specifically expressed in opaque cells, OP4, was correlated to EFG1 expression and the cell form in strain SS4. Northern analyses demonstrated a strong transcript in S4D-grown cells, while this transcript was missing in the absence of glucose (Fig. 6A). The presence of this transcript was not due to the S4D medium, since an OP4 transcript was not detected in a control strain. Thus, the upregulation of OP4 in strain SS4 by low-level expression of EFG1 indicates that the opaque-like phenotype that is generated is more than a superficial similarity compared to authentic opaque-phase cells and represents a comparable physiological state of cells. We also tested whether the WH11 transcript, which is specifically expressed in white-phase cells of strain WO-1, is repressed dependent on EFG1 expression. However, these experiments were inconclusive since WH11 transcript levels were almost undetectable in S4D high-glucose medium (Fig. 6B). Further analyses revealed that WH11 is regulated in a strongly glucose-dependent manner, with strong expression occurring only in various media lacking glucose (data not shown). However, because both strain SS4 and the CAI8 control strain contained similar WH11 transcript levels if grown in SCAA medium, there is no evidence that elevated EFG1 transcript levels are correlated with high WH11 transcript levels.
FIG. 6.
Phase-specific transcripts in strains with low-level EFG1 expression. Strains SS4 (efg1/PCK1p-EFG1) and CAI8[pBT-4, pBI] were grown in SCAA medium (EFG1 overexpression in strain SS4) (C) or S4D (low-level EFG1 expression in strain SS4) (S). Total RNA was analyzed by Northern blotting using the indicated probes. 18S rRNA was used as a loading control for Northern gels.
Surprisingly, and at variance with the above results, we verified that complete deletion of both EFG1 alleles in strain CAI4 (strain HLC52 [18]) did not yield any alteration in cellular shape and did not lead to an opaque-like colony phenotype; furthermore, the OP4 transcript was not altered in this strain (data not shown). On the other hand, strain HLC52 is known to be defective in hyphae formation (18).
DISCUSSION
Some evidence indicates that common regulatory circuits govern phenotypic switching and dimorphism of C. albicans. The WH11 gene, which encodes an abundant cytoplasmic protein, is induced both by the opaque-to-white transition and the hypha and yeast morphogenesis of strain WO-1; identical promoter segments mediate both repression events (34). Opaque-phase cells of strain WO-1 contain some specific antigens not present in white-phase cells that also occur in the hyphal form (2). Furthermore, opaque-phase cells are elongated, resembling pseudohyphae. Here we present evidence strongly suggesting that the Efg1p transcription factor, whose presence is essential for hyphal induction (18, 35), is a key element of the white-to-opaque phenotypic switching phenotype in C. albicans WO-1.
EFG1 is expressed specifically in cells of the white phase but not the opaque phase of strain WO-1. During mass conversion from the opaque to the white phase induced by temperature upshift, the EFG1 transcript appears early and reaches maximum levels even before cells in the culture are quantitatively growing in the white phase. Thus, EFG1 transcript levels are not simply the consequence of the white-phase (yeast) cell form, such as the WH11 transcript, whose level closely parallels the percentage of white-phase cells. Instead, plating of cells after induction of switching revealed that the ability to form a white-phase colony, as well as the actual cell morphology, is related to the EFG1 transcript level. It appears that some cells microscopically display a typically opaque cell morphology, while already having acquired the competence to switch in subsequent divisions and to form a white-phase colony; in such cells, EFG1 expression is turned on. We conclude that EFG1 transcript levels reflect the state of competence of opaque cells to switch to the white phenotype as well as the white cell form.
To explore whether Efg1p would be a determining factor of phenotypic switching, we forced expression of EFG1 in opaque cells by use of a PCK1p-EFG1 fusion gene (35). The growth of such transformants in PCK1 promoter repressing medium led to stable growth in the opaque form; in contrast, on inducing medium, EFG1 was expressed and cells switched to the white phenotype (yeast form) at high percentages. Opaque-to-white switching also has been described for the forced expression of the WH11 gene in opaque-phase cells (14). The efficiency of the EFG1-induced switching is greater (close to 100% in different transformants) than that of the reported WH11-induced switching (up to 20%). Because we demonstrated by Northern analysis that WH11 expression is not upregulated by EFG1, it is unlikely that the observed EFG1 expression indirectly leads to switching via WH11 expression.
If Efg1p were indeed a competence factor for phenotypic switching from opaque- to white-phase cells, it conversely was possible that the lowering of EFG1 expression in white-phase cells below wild-type levels would favor switching from the white to the opaque cell form. However, using strain WO-1, such experiments at present cannot be carried out, since a ura3 derivative of this strain in which the URA blaster method (7) can be used for gene disruption does not exist. Therefore, for this analysis, we used strain SS4, a derivative of strain CAI8 in which the only remaining EFG1 allele is under the control of the PCK1 promoter (35). During growth on high-glucose medium, a high percentage of cells switched to an elongated rod-like phenotype strongly resembling the opaque phenotype of strain WO-1. However, surface pimples were not detected on the opaque-like cells of SS-4 but have been observed on authentic opaque cells of strain WO-1 (2); the different genetic backgrounds in both strains may account for this difference. Importantly, the opaque-specific OP4 transcript (19) was strongly induced in SS4 if EFG1 expression was lowered. Thus, opaque-like cells of strain SS4 and opaque cells of strain WO-1 are not only phenotypically related, but appear to represent similar genetic and/or physiological cellular states. Possibly, growth in the yeast form prevents activation of OP4 expression (19), because the OP4 transcript is detected not only in strain SS4 at low EFG1 expression levels and the opaque form of strain WO-1 but also in all variant phenotypes of strain 3153A (21).
A puzzling result of this study was the finding that deletion of EFG1 in strain CAI4 (mutant HLC52 [18]) did not lead to an opaque-like phenotype, such as was observed in the CAI8-derivative SS4. However, although strains CAI4 and CAI8 are derived from a common parental strain, they are not isogenic. Also, it cannot be excluded that during construction of HLC52 mutations or alterations in chromosomal configurations occurred that render cells less responsive to alterations in Efg1p levels. Compensatory mechanisms in strains CAI4 and/or HLC52 could include the expression of genes homologous to EFG1. It should be pointed out that a different response to EFG1 expression between strains CAI8 and Red3/6 was also observed; overexpression of EFG1 in the latter strain did not induce elongated pseudohyphae as has been reported for strain CAI8 (35). Another open question is whether Efg1p, besides regulating the white and opaque phenotypic switching, also determines the switching between other colony phenotypes (27). In summary, we propose a model in which Efg1p at a critical level of concentration or activity is required for switching from the opaque- to the white-phase cell form. After switching, to maintain the white phenotype, EFG1 expression is also needed. It is possible that spontaneous fluctuations in EFG1 expression by epigenetic mechanisms, as has been observed for genes situated in subtelomeric region in Saccharomyces cerevisiae (9), determine or contribute to phenotypic switching.
ACKNOWLEDGMENTS
We thank R. Riehl for his help in scanning electron microscopy, D. Soll for strains, and K. Schroeppel for plasmids. We acknowledge the expert technical assistance of M. Gerads.
This work was supported by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Anderson J, Cundiff L, Schnars B, Gao M, Mackenzie I, Soll D R. Hypha formation in the white-opaque transition of Candida albicans. Infect Immun. 1989;57:458–467. doi: 10.1128/iai.57.2.458-467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson J, Mihalik R, Soll D R. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J Bacteriol. 1990;172:224–235. doi: 10.1128/jb.172.1.224-235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balan I, Alarco A-M, Raymond M. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J Bacteriol. 1997;179:7210–7218. doi: 10.1128/jb.179.23.7210-7218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannon R D, Jenkinson H F, Shepherd M G. Cloning and expression of Candida albicans ADE2 and proteinase genes on a replicative plasmid in C. albicans and in Saccharomyces cerevisiae. Mol Gen Genet. 1992;235:453–457. doi: 10.1007/BF00279393. [DOI] [PubMed] [Google Scholar]
- 5.Delbrück S, Ernst J F. Morphogenesis-independent regulation of actin transcript levels in the pathogenic yeast Candida albicans. Mol Microbiol. 1993;10:859–856. doi: 10.1111/j.1365-2958.1993.tb00956.x. [DOI] [PubMed] [Google Scholar]
- 6.Donelson J E. DNA rearrangements and antigenic variation in African trypanosomes. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 763–782. [Google Scholar]
- 7.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glasgow A C, Hughes K T, Simon M I. Bacterial DNA inversion systems. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 637–660. [Google Scholar]
- 9.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 10.Hube B. Candida albicans secreted aspartyl proteinases. Curr Top Med Mycol. 1996;7:55–69. [PubMed] [Google Scholar]
- 11.Köhler J R, Fink G R. Candida albicans strains heterozygous and homozygous in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kupsch E M, Knepper B, Kuroki T, Heuer I, Meyer T F. Variable opacity (Opa) outer membrane proteins account for the cell tropisms displayed by Neisseria gonorrhoeae for human leukocytes and epithelial cells. EMBO J. 1993;12:641–650. doi: 10.1002/j.1460-2075.1993.tb05697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurtz M B, Cortelyou M W, Miller S M, Lai M, Kirsch D R. Development of autonomously replicating plasmids for Candida albicans. Mol Cell Biol. 1987;7:209–217. doi: 10.1128/mcb.7.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kvaal C A, Srikantha T, Soll D R. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect Immun. 1997;65:4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A R, Brown A J P, Thomas D Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leuker C E, Sonneborn A, Delbrück S, Ernst J F. Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene. 1997;192:235–240. doi: 10.1016/s0378-1119(97)00069-3. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Köhler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1725. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 18.Lo H-J, Köhler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 19.Lockhart S R, Nguyen M, Srikantha T, Soll D R. A MADS box consensus binding site is necessary and sufficient for activation of the opaque-phase-specific gene OP4 of Candida albicans. J Bacteriol. 1998;180:6607–6616. doi: 10.1128/jb.180.24.6607-6616.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrow B, Srikantha T, Soll D R. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrow B, Ramsey H, Soll D R. Regulation of phase-specific genes in the more general switching system of Candida albicans strain 3153A. J Med Vet Mycol. 1994;32:287–294. doi: 10.1080/02681219480000361. [DOI] [PubMed] [Google Scholar]
- 22.Odds F C. Candida and candidosis. London, United Kingdom: Baillière Tindall; 1988. [Google Scholar]
- 23.Rikkerink E H A, Magee B B, Magee P T. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rustchenko E P, Howard D H, Sherman F. Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J Bacteriol. 1994;176:3231–3241. doi: 10.1128/jb.176.11.3231-3241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadhu C, Hoekstra D, McEachern M J, Reed S I, Hicks J B. A G-protein alpha subunit from asexual Candida albicans functions in the mating signal transduction pathway of Saccharomyces cerevisiae and is regulated by the a1-alpha2 repressor. Mol Biol Cell. 1992;12:1977–1978. doi: 10.1128/mcb.12.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman F, Fink G, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 27.Slutsky B, Buffo J, Soll D R. High frequency switching of colony morphology in Candida albicans. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- 28.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll D R. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soll D R, Morrow B, Srikantha T. High-frequency phenotypic switching in Candida albicans. Trends Genet. 1993;9:61–65. doi: 10.1016/0168-9525(93)90189-O. [DOI] [PubMed] [Google Scholar]
- 30.Soll D R. Gene regulation during high-frequency switching in Candida albicans. Microbiology. 1997;143:279–288. doi: 10.1099/00221287-143-2-279. [DOI] [PubMed] [Google Scholar]
- 31.Srikantha T, Soll D R. A white-specific gene in the white-opaque switching system of Candida albicans. Gene. 1993;131:53–60. doi: 10.1016/0378-1119(93)90668-s. [DOI] [PubMed] [Google Scholar]
- 32.Srikantha T, Chandrasekhar A, Soll D R. Functional analysis of the promoter of the phase-specific WH11 gene of Candida albicans. Mol Cell Biol. 1995;15:1797–1805. doi: 10.1128/mcb.15.3.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srikantha T, Morrow B, Schröppel K, Soll D R. The frequency of integrative transformation at phase-specific genes of Candida albicans correlates with their transcriptional state. Mol Gen Genet. 1995;246:342–352. doi: 10.1007/BF00288607. [DOI] [PubMed] [Google Scholar]
- 34.Srikantha T, Tsai L K, Soll D R. The WH11 gene of Candida albicans is regulated in two distinct developmental programs through the same transcription activation sequences. J Bacteriol. 1997;179:3837–3844. doi: 10.1128/jb.179.12.3837-3844.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoldt V R, Sonneborn A, Leuker C, Ernst J F. Efg1, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson J, Koomey J M. Mechanisms for variation of pili and outer membrane protein II in Neisseria gonorrhoea. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 743–762. [Google Scholar]