Abstract
Alzheimer’s disease biomarkers are becoming increasingly important for characterizing the longitudinal course of disease, predicting the timing of clinical and cognitive symptoms, and for recruitment and treatment monitoring in clinical trials. In this work, we develop and evaluate three methods for modelling the longitudinal course of amyloid accumulation in three cohorts using amyloid PET imaging. We then use these novel approaches to investigate factors that influence the timing of amyloid onset and the timing from amyloid onset to impairment onset in the Alzheimer's disease continuum.
Data were acquired from the Alzheimer's Disease Neuroimaging Initiative (ADNI), the Baltimore Longitudinal Study of Aging (BLSA) and the Wisconsin Registry for Alzheimer's Prevention (WRAP). Amyloid PET was used to assess global amyloid burden. Three methods were evaluated for modelling amyloid accumulation using 10-fold cross-validation and holdout validation where applicable. Estimated amyloid onset age was compared across all three modelling methods and cohorts. Cox regression and accelerated failure time models were used to investigate whether sex, apolipoprotein E genotype and e4 carriage were associated with amyloid onset age in all cohorts. Cox regression was used to investigate whether apolipoprotein E (e4 carriage and e3e3, e3e4, e4e4 genotypes), sex or age of amyloid onset were associated with the time from amyloid onset to impairment onset (global clinical dementia rating ≥1) in a subset of 595 ADNI participants that were not impaired before amyloid onset.
Model prediction and estimated amyloid onset age were similar across all three amyloid modelling methods. Sex and apolipoprotein E e4 carriage were not associated with PET-measured amyloid accumulation rates. Apolipoprotein E genotype and e4 carriage, but not sex, were associated with amyloid onset age such that e4 carriers became amyloid positive at an earlier age compared to non-carriers, and greater e4 dosage was associated with an earlier amyloid onset age. In the ADNI, e4 carriage, being female and a later amyloid onset age were all associated with a shorter time from amyloid onset to impairment onset. The risk of impairment onset due to age of amyloid onset was non-linear and accelerated for amyloid onset age >65. These findings demonstrate the feasibility of modelling longitudinal amyloid accumulation to enable individualized estimates of amyloid onset age from amyloid PET imaging. These estimates provide a more direct way to investigate the role of amyloid and other factors that influence the timing of clinical impairment in Alzheimer's disease.
Keywords: amyloid, Alzheimer's, PET, dementia, amyloid onset age
Betthauser et al. validate three methods for modelling amyloid (A) accumulation trajectories using amyloid PET imaging. They show that APOE genotype but not sex is associated with A+ onset age, and that higher APOEe4 dosage, being female and higher A+ onset age decrease the time from A+ onset to dementia.
Introduction
Alzheimer's disease is characterized by the aggregation of beta-amyloid plaques and neurofibrillary tau tangles, followed by subsequent neurodegeneration and progressive cognitive decline.1–4 The disease course consists of an extended ‘preclinical’ phase in which Alzheimer's disease pathology is accumulating before the onset of clinical symptoms.5 The preclinical phase may last 20 years or more, although the empirical basis for this is largely at the group level6–9 where studies have observed predictable change in amyloid levels.7,9–11 Robust methods for estimating amyloid onset and thereby demarcating the onset of preclinical Alzheimer's disease in individuals are needed and appear to be feasible.11–15 Although conceptually promising, existing methods require further validation in additional cohorts to elucidate Alzheimer's disease timing and dementia risk. Additionally, autonomous methods could broaden accessibility to other cohorts and applications such as spatiotemporal modelling. Here, we report two new autonomous methods for estimating individualized amyloid onset age and evaluate these methods with our previously published approach.13 We then use these methods to investigate factors that influence amyloid onset age, amyloid accumulation rates and time from amyloid onset to impairment.
Apolipoprotein E (APOE) genotype, specifically APOE-e4 allele carriage and gene dosage, is the most established risk factor for beta-amyloid accumulation and cognitive impairment in sporadic Alzheimer's disease.16 Dementia prevalence studies demonstrate a gene–dose-dependent relationship wherein dementia risk increases and dementia onset age decreases with lower APOE-e2 dosage and higher e4 dosage.17–19 Some studies have observed an interaction between APOE and sex such that females with one or more APOE-e4 alleles experience earlier dementia onset relative to e4 non-carriers, but a similar result was not observed for male APOE-e4 carriers.20 Biomarker studies indicate amyloid positivity (A+) prevalence increases with increasing APOE-e4 dosage, and decreases with increasing e2 gene dosage (except e2e4 carriers) compared to e3 homozygotes.8,21 Neuroimaging studies have also shown e4 carriers become A+ earlier than non-carriers.8,12,22,23 Thus, an explanation for e4 carriers becoming impaired earlier in life is that they start accumulating beta-amyloid earlier in life. To test this hypothesis, we need to determine whether the relationship between APOE genotype and A+ prevalence is due to an earlier amyloid onset age, a difference in amyloid accumulation rates or a combination of both. There is also a need to determine whether factors such as APOE genotype, sex or age of amyloid onset shorten/lengthen the preclinical timeframe.
This work builds on two previous papers where we demonstrated methods that provide individualized estimated A+ onset age (EAOA) based on one or more 11C-Pittsburgh Compound B (PiB) PET scans. These methods were developed separately in the Baltimore Longitudinal Study on Aging (BLSA)12 and the Wisconsin Registry for Alzheimer's Prevention (WRAP).13 The BLSA study used non-linear mixed-effects modelling with random effects to derive EAOA and showed asymptomatic APOE-e4 carriers exhibited earlier EAOA than non-carriers. The WRAP study used group-based trajectory modelling (GBTM) and Bayes’ theorem to obtain EAOA and showed EAOA could align observations by A+ duration (age at observation minus EAOA; termed amyloid chronicity). Higher amyloid chronicity was associated with faster rates of cognitive decline, increased cognitive impairment risk and was more strongly associated with MK-6240 tau PET than age in initially unimpaired WRAP participants.
In this work, we present a simplification to the GBTM approach and introduce two new autonomous methods for modelling longitudinal amyloid PET trajectories. All algorithms were evaluated in three cohorts: the Alzheimer's Disease Neuroimaging Initiative (ADNI) using 18F-florbetapir, the BLSA using 11C-PiB and the WRAP using 11C-PiB. Our aims were to: (i) cross-validate three methods to model amyloid accumulation, impute EAOA and predict amyloid PET levels; (ii) characterize associations between APOE genotype and sex with EAOA and amyloid accumulation rates; and (iii) characterize dementia risk and time from EAOA to dementia associated with APOE genotype, EAOA and sex.
Materials and methods
Study participants
Participants were included from ADNI, BLSA and WRAP studies if they had at least one amyloid PET scan and a cognitive diagnosis available in addition to each cohort's inclusion/exclusion criteria (see Johnson et al.24 Ferruci,25 Weiner et al.26 and Resnick et al.27 references for cohort information; participant summary in Table 1). Participants’ written consent was obtained in each source study according to the Declaration of Helsinki and under local Institutional Review Board approvals.
Table 1.
Demographics and cohort summaries
| Modelling sets | All participants | |||||
|---|---|---|---|---|---|---|
| ADNIa | BLSAa | WRAPa | ADNI | BLSA | WRAP | |
| N | 739 | 142 | 179 | 1215 | 207 | 272 |
| Female, n (%) | 345 (46.7) | 71 (50.0) | 120 (67.0) | 570 (46.9) | 104 (50.2) | 185 (68.0) |
| Race, n (%) | ||||||
| American Indian or Alaskan Native | 2 (0.3) | 0 | 1 (0.6) | 2 (0.2) | 0 | 4 (1.5) |
| Asian | 11 (1.5) | 2 (1.4) | 1 (0.6) | 19 (1.6) | 4 (1.9) | 1 (0.4) |
| Native Hawaiian or Other Pacific Islander | 1 (0.1) | 2 (1.4) | 0 | 2 (0.2) | 3 (1.4) | 0 |
| Black or African American | 24 (3.2) | 25 (17.6) | 7 (3.9) | 46 (3.8) | 36 (17.4) | 10 (3.7) |
| White | 689 (93.2) | 112 (78.9) | 169 (94.4) | 1127 (92.8) | 162 (78.3) | 256 (94.1) |
| Other or multiple | 11 (1.5) | 1 (0.7) | 1 (0.6) | 16 (1.3) | 2 (1.0) | 1 (0.4) |
| Unavailable/unknown | 1 (0.1) | 0 () | 0 | 3 (0.2) | 0 | 0 |
| APOE genotype, n (%) | ||||||
| e2e2 | 1 (0.1) | 1 (0.7) | 0 | 2 (0.2) | 2 (1.0) | 0 |
| e2e3 | 67 (9.1) | 15 (10.6) | 16 (8.9) | 99 (8.1) | 25 (12.1) | 30 (11.0) |
| e2e4 | 15 (2.0) | 6 (4.2) | 5 (2.8) | 24 (2.0) | 8 (3.9) | 10 (3.7) |
| e3e3 | 371 (50.2) | 84 (59.2) | 88 (49.2) | 589 (48.5) | 118 (57.0) | 132 (48.5) |
| e3e4 | 233 (31.5) | 33 (23.2) | 63 (35.2) | 394 (32.4) | 49 (23.7) | 88 (32.4) |
| e4e4 | 52 (7.0) | 2 (1.4) | 7 (3.9) | 107 (8.8) | 3 (1.4) | 10 (3.7) |
| Unknown | 0 | 1 (0.7) | 0 | 0 | 2 (1.0) | 2 (0.7) |
| Baseline diagnosis, n (%) | ||||||
| Unimpaired | 303 (41.0) | 142 (100) | 177 (98.9) | 461 (37.9) | 207 (100) | 261 (96.0) |
| MCI | 386 (52.2) | 0 | 2 (1.1) | 559 (46.0) | 0 | 9 (3.3) |
| Dementia | 50 (6.8) | 0 | 0 | 195 (16.0) | 0 | 2 (0.7) |
| Baseline age, mean (SD) | 73.4 (7.4) | 75.9 (7.9) | 61.0 (6.2) | 73.7 (7.6) | 75.8 (8.2) | 62.4 (6.7) |
| PET follow-up duration years, mean (SD) | 4.14 (1.93) | 5.13 (3.18) | 6.25 (2.11) | 2.52 (2.52) | 3.52 (3.6) | 4.11 (3.43) |
| Number of amyloid PET scans, n (%) | ||||||
| 1 | — | — | — | 476 (39.2) | 65 (31.4) | 93 (34.2) |
| 2 | 307 (41.5) | 39 (18.8) | 56 (20.6) | 307 (25.3) | 39 (18.8) | 56 (20.6) |
| 3 | 235 (31.8) | 38 (18.4) | 96 (35.3) | 235 (19.3) | 38 (18.4) | 96 (35.3) |
| 4 | 163 (22.1) | 25 (12.1) | 25 (9.2) | 163 (13.4) | 25 (12.1) | 25 (9.2) |
| 5 | 33 (4.5) | 14 (6.8) | 2 (0.7) | 33 (2.7) | 14 (6.8) | 2 (0.7) |
| 6 | 1 (0.1) | 13 (6.3) | 0 | 1 (0.1) | 13 (6.3) | 0 |
| 7 | 0 | 3 (1.4) | 0 | 0 | 3 (1.4) | 0 |
| 8 | 0 | 5 (2.4) | 0 | 0 | 5 (2.4) | 0 |
| 9 | 0 | 3 (1.4) | 0 | 0 | 3 (1.4) | 0 |
| 10 | 0 | 1 (0.5) | 0 | 0 | 1 (0.5) | 0 |
| 11 | 0 | 1 (0.5) | 0 | 0 | 1 (0.5) | 0 |
SD = standard deviation.
Subsets from each cohort with two or more amyloid PET scans used for longitudinal modelling.
Cohort descriptions
ADNI
Data used in this article were obtained from the ADNI database (adni.loni.usc.edu). ADNI was launched in 2003 as a public–private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease. For up-to-date information, see www.adni-info.org. Diagnostic cognitive status was established from the ADNI diagnosis table.
BLSA
At enrolment into the BLSA PET neuroimaging substudy,27 participants were free of CNS disease, severe cardiac disease, severe pulmonary disease and metastatic cancer. Participants with a clinical dementia rating (CDR)28 of zero and ≤3 errors on the Blessed Information-Memory-Concentration test29 were categorized as cognitively normal; otherwise, cognitive status was determined by consensus case conference on review of clinical and neuropsychological data. Dementia and MCI diagnoses were based on Diagnostic and Statistical Manual of Mental Disorders30 (DSM-IIIR) and Petersen criteria,31 respectively.
WRAP
WRAP participants are unimpaired at enrolment and complete comprehensive neuropsychological assessments roughly biennially.24 Cognitive status (unimpaired, MCI, dementia) was established by consensus conference as previously reported for WRAP participants.32
Diagnostic status (unimpaired, MCI, dementia) was established across cohorts for descriptive purposes using similar criteria. Clinical progression was assessed using the CDR scale when applicable.28
Amyloid PET processing, quantification and positivity
Cortical amyloid burden was assessed using either dynamic 11C-PiB (BLSA and WRAP) or late-frame 18F-florbetapir (ADNI) PET imaging using separate processing and quantification methods optimized within each cohort to provide the highest quantitative accuracy and lowest binding estimate variance (see Supplementary material for details).33–37 ADNI cortical Florbetapir standard uptake value ratios (SUVRs; eroded white matter reference region) processed by the Banner Alzheimer's Institute were downloaded from the 22 November, 2019 data freeze. Mean cortical PiB distribution volume ratios (DVRs; cerebellar grey matter reference region) were estimated using different reference tissue methods for BLSA37 (DVRRTM3P) and WRAP (DVRLGA).36 A+ thresholds were established based on separate studies for each cohort. Florbetapir SUVRWM > 0.8 (33.1 Centiloids) was used for ADNI based on receiver operating characteristic analyses (AUC 0.91, 91% negative agreement, 82% positive agreement) comparing SUVRWM with a published threshold from Berkeley processed data.38 BLSA used DVRRTM3P > 1.066 (20.6 Centiloids) derived from Gaussian mixture modelling.12 DVRLGA > 1.19 (21.6 Centiloids) was used for WRAP based on receiver operating characteristic analysis with visual assessment.39 DVR and SUVRWM values were mapped to Centiloids40 using linear regression for reporting and to estimate the time difference between A+ thresholds (Supplementary material).
Amyloid trajectory modelling and estimated amyloid onset age
Three methods were used to model longitudinal amyloid PET trajectories and estimate amyloid onset age (EAOA). Descriptions of each method are provided next with additional details in the supplement. Models were trained separately for each cohort using subsets of participants with ≥2 amyloid PET scans (Table 1, left three columns). EAOA and A+ duration (A+ duration = age at observation—EAOA) were estimated from trained models for all participants.
Group-based trajectory modelling
As previously reported,13 the GBTM EAOA method applies GBTM41,42 to identify the optimal number and shape of age-related DVR/SUVR group trajectory equations (up to cubic polynomial). EAOA is calculated for each group function by solving for age at the A+ threshold. Life expectancy at the last scan is used13 to estimate EAOA for an intercept-only (representing amyloid non-accumulators) group function. Each participant's EAOA is calculated as the weighted sum of group function EAOAs with weights derived from residuals for each function (Supplementary material). DVR/SUVR prediction is accomplished by modelling the DVR/SUVR versus A+ duration curve using piecewise regression, and solving this modelled curve for DVR/SUVR at the time difference between the reference scan and the scan of interest for prediction.
Ordinary differential equation–Gaussian process
The ordinary differential equation–Gaussian process (ODE–GP) algorithm is a gradient matching method43 that fits a non-parametric ODE to data. The first step of ODE–GP estimates DVR/SUVR gradients for each participant using linear regression. The second step solves a non-standard GP regression problem using data from all participants in the sample, accounting for both the noise in measurements and in the estimated gradients. An approximate solution is provided using a second-order Taylor expansion of the Gaussian process using a Gaussian kernel. The kernel radius is estimated by maximizing the marginal likelihood of the data. The fitted model allows for prediction of past and future DVR/SUVR for a participant using a single DVR/SUVR value at a given age as an initial condition for the ODE. A prediction is made by integrating numerically the ODE using the Euler method. Since the fitted model is autonomous (time-invariant), we summarize the fitted data with a growth curve defining the DVR/SUVR as function of the time since the A+ threshold.
Sampled iterative local approximation
The sampled iterative local approximation (SILA) algorithm uses discrete sampling of DVR/SUVR versus age data to establish the relationship between DVR/SUVR rate and DVR/SUVR. Numerical smoothing (robust LOESS) and Euler's method are used to numerically integrate these data to generate a non-parametric DVR/SUVR versus A+ duration curve with zero time corresponding to the A+ threshold. EAOA is calculated for each person by first solving this curve for time using a person's observed DVR/SUVR, and subtracting the estimated A+ duration from their age at that scan. SUVR/DVR is estimated for antecedent or prospective scans by solving the DVR/SUVR versus time curve for DVR/SUVR at the time corresponding to the difference between reference and target scans.
Statistical analyses
Aim 1: evaluating and comparing amyloid modelling methods
Aim 1 compared EAOA and A+ duration outcomes across the three methods and evaluated the predictive performance of each method within each cohort. The composition of the model training and testing sets varied depending on the analysis goals described next. Computational speed was evaluated for autonomous methods ODE–GP and SILA (Supplementary material).
Inter-method comparisons of EAOA and amyloid accumulation curves
EAOA and SUVR/DVR versus A+ duration curves were compared across methods in each cohort. EAOA and A+ duration were estimated for all participants using each participant's observed DVR/SUVR and age at their last available amyloid PET scan. Pearson correlations compared EAOA between methods separately for A+ and A− groups. Amyloid accumulation curves (i.e. DVR/SUVR versus estimated A+ duration) were compared by plotting observed DVR/SUVR values at the first scan as a function of estimated A+ duration referencing the last scan for participants with ≥2 scans. EAOA accuracy was evaluated in A− to A+ converters (n = 72 ADNI; n = 6 BLSA; n = 22 WRAP) by calculating the proportion of times age last A− ≤ EAOA ≤ age first A+ and calculating the conversion midpoint error (years between EAOA and the midpoint age between A− A+ observations).
Method predictive validity
Predictive validity to estimate future and past DVR/SUVR and elevated or non-elevated amyloid PET (A±) status was evaluated using holdout validation (all methods; ADNI) and 10-fold cross-validation (ODE–GP and SILA; all cohorts). Holdout validation was limited to ADNI because the larger sample enabled separate training and testing partitions with similar amyloid distributions. GBTM was omitted from 10-fold cross-validation because it would have required ∼90 h of human time. Stratification methods for 10-fold and holdout validation samples are provided in the Supplementary Material. For each method and cohort, models were trained on subsets of longitudinal data, and then DVR/SUVR and age at a single reference scan (last or first) were used to predict DVR/SUVR at an earlier or later PET scan for participants omitted from model training. Forwards prediction was evaluated by predicting DVR/SUVR at each person's last observation inputting each person's first observation, whereas backwards prediction used the last observation to predict DVR/SUVR at the first observation. DVR/SUVR prediction residuals were used to investigate associations with factors that might influence model prediction (reference DVR/SUVR, age and time from reference scan) and to generate model summary statistics. A± prediction performance was evaluated using balanced accuracy (adjusted for imbalanced A+ and A− frequency), sensitivity and specificity. We also compared EAOA estimates derived from in-sample versus out-of-sample prediction schemes graphically and using Pearson correlations. After observing similar model performance for ODE–GP and SILA methods, analyses for aims 2 and 3 used the average of ODE–GP and SILA EAOA.
Aim 2: APOE and sex associations with EAOA and amyloid accumulation rate
The impact of sex and APOE-e4 carriage on amyloid accumulation rates were evaluated by investigating the relationship between DVR/SUVR rate and DVR/SUVR. Smoothed DVR/SUVR rate versus DVR/SUVR curves were generated for each cohort, and separately for comparison groups (APOE-e4 carriers versus non-carriers; males versus females) by applying local regression with weighted linear least squares to the mean within-person DVR/SUVR rate versus DVR/SUVR data. Ninety-five percent confidence intervals were estimated using 1000 bootstrapped samples with replacement.
The second part of Aim 2 addressed three questions: (i) what age does A+ onset occur; (ii) what is the A+ risk conferred by APOE; and (iii) does A+ onset differ between APOE groups? We assessed whether and how much EAOA differs across APOE genotypes using survival analyses, as previously described.44 The event time was EAOA for A+ individuals and age at last PET observation (right censored) for individuals that were A− but could become A+. Kaplan–Meier plots by APOE-e4 carriage (excluding e2e4), and separately by APOE genotype (e2e3, e2e4, e3e3, e3e4, and e4e4), were compared in each dataset. Survival curves were compared using log-rank tests with pairwise comparisons conducted for APOE groups. Median survival times with 95% confidence intervals for APOE groups (e4 carriers and non-carriers; APOE genotypes) were reported for groups that reached 50% probability of remaining A−. APOE-e2e2 carriers were excluded from genotype analyses due to low frequency. We used Cox proportional hazards models to quantify A+ risk by APOE-e4 carriage, and by APOE genotype in separate models. Accelerated failure time (AFT) models quantified relative differences in EAOA by APOE-e4 carriage and genotype. Cox proportional hazards and the AFT models covaried for sex.
Aim 3: Effects of APOE, sex and EAOA on time from A+ onset to dementia
WRAP and BLSA observed few clinical conversions compared to ADNI. Therefore, we used a subset of 595 ADNI participants to investigate the effects of EAOA, sex and APOE on time from EAOA to impairment. ADNI participants were included if they were not impaired before EAOA, with CDR ≥1 as the criterion for impairment.28APOE-e2e4 carriers were excluded due to the combination of risk and protective effects and low frequency (n = 15). We used Cox survival models to estimate hazard and survival functions of impairment from EAOA and their relationship with APOE-e4 carriage, sex and EAOA. The Cox model outcome was time from EAOA to impairment for those who became impaired, and time from EAOA to age at last CDR (right censored) for those that remained unimpaired. Predictors included education, sex, APOE-e4 carriage, EAOA, EAOA2, sex*EAOA, sex*EAOA2, APOE*EAOA, APOE*EAOA2. EAOA2 was included to allow for accelerated log hazard of impairment with increasing EAOA. Non-significant interactions were removed from the model. Analyses were repeated using APOE genotypes (e3e3, e3e4, e4e4) instead of APOE-e4 carriage. Results are reported as hazard ratios, median unimpaired survival time and the ten-year unimpaired rate with confidence intervals estimated using 1000 bootstrapped samples with replacement.
Data availability
Data from the WRAP (https://wrap.wisc.edu) and BLSA (https://blsa.nih.gov) can be requested through an online submission process. ADNI data can be obtained via the ADNI and LONI websites. ODE–GP and SILA algorithms are available at https://gitlab.com/bilgelm/amyloid-ode-gp and https://github.com/Betthauser-Neuro-Lab/SILA-AD-Biomarker, respectively.
Results
Study sample descriptive statistics and demographics
Table 1 shows demographic and summary statistics for the three cohorts and model training subsets. ADNI and BLSA cohorts were 11.3 and 13.4 years older, respectively, than WRAP, which had a mean age of 62.4 years at baseline amyloid PET scan. BLSA and WRAP samples were primarily unimpaired at baseline PET whereas ADNI had 62% impaired participants (MCI or dementia). Study participants primarily identified as white (ADNI: 93%, BLSA: 78%, WRAP: 94%). Demographic variables were similar for the subsets used for model training and testing. The mean (SD) PET follow-up for these subsets was 4.14 (1.93), 5.13 (3.18) and 6.25 (2.11) years for ADNI, BLSA and WRAP, respectively.
Aim 1: amyloid modelling methods comparison and evaluation
Inter-method comparisons
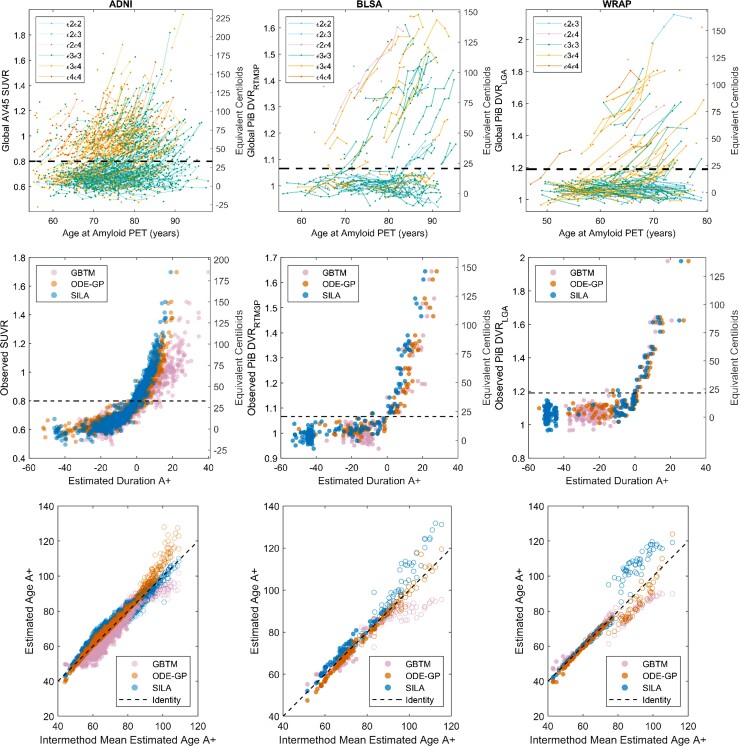
Amyloid versus age data and A+ thresholds are shown for each cohort in Fig. 1 (top row). Each cohort had a visually apparent group of amyloid non-accumulators below the A+ threshold and a group of amyloid accumulators crossing and above the threshold. Overall, the three methods produced similar amyloid accumulation patterns (middle row, Fig. 1), with the largest differences observed in ADNI. EAOA was highly linearly correlated between method pairs in A+ participants (rPearson = 0.88–1.00) with lower correlations observed in A− participants (rPearson = 0.76–0.95; Fig. 1 bottom row; Supplementary Tables 2 and 3). The largest inter-method EAOA differences were observed in A− participants and at high DVR/SUVR where longitudinal observations were sparse (Supplementary Fig. 1). In A± convertors, EAOA accuracy ranged from 16.7% (1/6, GBTM in BLSA) to 77% (17/22, SILA in WRAP), and the mean error ranged from −3.65 (GBTM in ADNI) to 1.96 years (GBTM in BLSA) across all cohorts and methods (Supplementary Fig. 2 and Table 4). GBTM, ODE–GP and SILA had similar EAOA accuracy and errors in BLSA and WRAP cohorts, whereas EAOA accuracy was lower and errors were higher for GBTM in ADNI compared to ODE–GP and SILA.
Figure 1.
Observed and modelled amyloid trajectories and EAOA. Observed amyloid PET by age (top row), modelled amyloid accumulation patterns (middle row) and EAOA (bottom row) for each method and cohort (each column represents a cohort). APOE genotype is colour coded for plots in the top row. Equivalent Centiloid values corresponding DVR and SUVR from each cohort are represented along the right side of the plots in the middle and top row for reporting purposes. A+ thresholds in all cohorts were slightly above the visually apparent group of longitudinal amyloid non-accumulators suggesting those above A+ thresholds had a high likelihood of future amyloid accumulation (see also Supplementary Fig. 8). The plots in the middle row represent backwards prediction of the duration A+ (estimated duration A+ at first scan using last scan for a within-person reference). Open circles in the bottom row indicate participants who were A− at their last observation and demonstrate the instability of estimating the onset age of A+ in cases with low amyloid levels. This suggests that interpretation of EAOA in A− cases is probably limited.
Forwards and backwards predictive validity
The time for prediction evaluation ranged from 0.9 to 13.1 years. Using holdout cross-validation in ADNI, all three methods had high balanced accuracy (94–98%) for predicting future or antecedent A± status (Supplementary Table 5). Sum of squared residuals were ODE–GP < SILA < GBTM for backwards SUVR prediction, and ODE–GP < GBTM < SILA for forwards SUVR prediction with all methods except GBTM reporting higher residual sum of squares for forwards compared to backwards SUVR prediction. Using 10-fold cross-validation in all cohorts, ODE–GP and SILA had similar SUVR/DVR predictive performance with balanced accuracy ranging from 94 to 99% for backwards A± prediction and 82–96% for forwards A± prediction. Prediction error was lower for backwards prediction compared to forwards prediction for ODE–GP and SILA in all three cohorts. Model residuals were weakly associated with age at reference scan (|Spearman’s rho| ≤ 0.26) and time to/from reference scan (|Spearman’s rho| ≤ 0.28) for all methods and cohorts (Supplementary Tables 6–9 and Supplementary Figs 3–6). ODE–GP and SILA had weak to moderate associations between prediction residuals and reference SUVR/DVR (|rSpearman| ≤ 0.41), whereas GBTM had a moderate association in the ADNI dataset (rSpearman = 0.45 and 0.61 for backwards and forwards prediction, respectively). EAOA was highly linearly correlated (rPearson > 0.99) for all methods and cohorts when using out-of-sample (10-fold or holdout cross-validation) versus in-sample training/prediction paradigms (Supplementary Fig. 7). No convergence issues were observed during model fitting.
Aim 2: EAOA, sex and APOE genotypes
APOE, sex and rate of amyloid accumulation
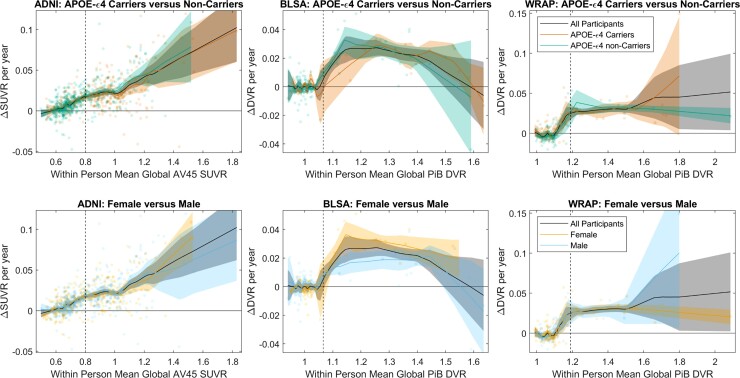
No differences were observed in SUVR/DVR rates versus SUVR/DVR between males and females or between APOE-ε4 carriers and non-carriers (all cohorts) or between impaired versus unimpaired in ADNI (Fig. 2 and Supplementary Figs 8–10).
Figure 2.
Amyloid accumulation rates by APOE Carriage and Sex. Comparisons of amyloid accumulation rates as a function of amyloid level between APOE-e4 carriers and non-carriers (top row) and between males and females (bottom row) for each of the three cohorts (columns). Scatter plots show the within-person amyloid rate as a function of their mean DVR or SUVR value. Local regression with weighted linear least squares lines show the average relationship between the rate of amyloid change and the amyloid level for the entire sample, and for subsets of APOE-e4 carriers and non-carriers and for females and males. Shaded regions represent the 95% confidence intervals derived from 1000 bootstrapped samples. The relationship between the amyloid accumulation rate and the level of amyloid burden was similar between APOE-e4 carriers versus non-carriers, and between males versus females across all cohorts. This suggests the rate of amyloid accumulation does not differ by APOE-e4 carriage or sex.
Effect of APOE and sex on EAOA and A+ risk
The mean (SD) estimated time between A+ thresholds across ODE–GP and SILA methods was 0.42 (0.11) years from BLSA to WRAP and 4.53 (1.11) years from BLSA to ADNI.
At what age does A+ onset occur?
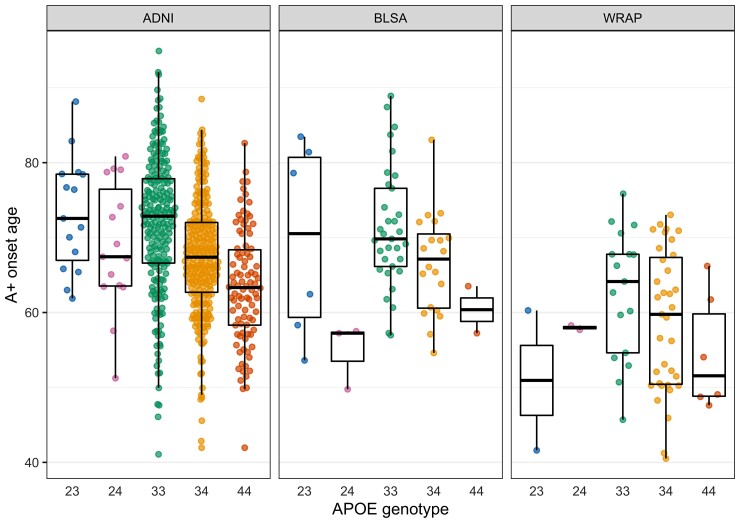
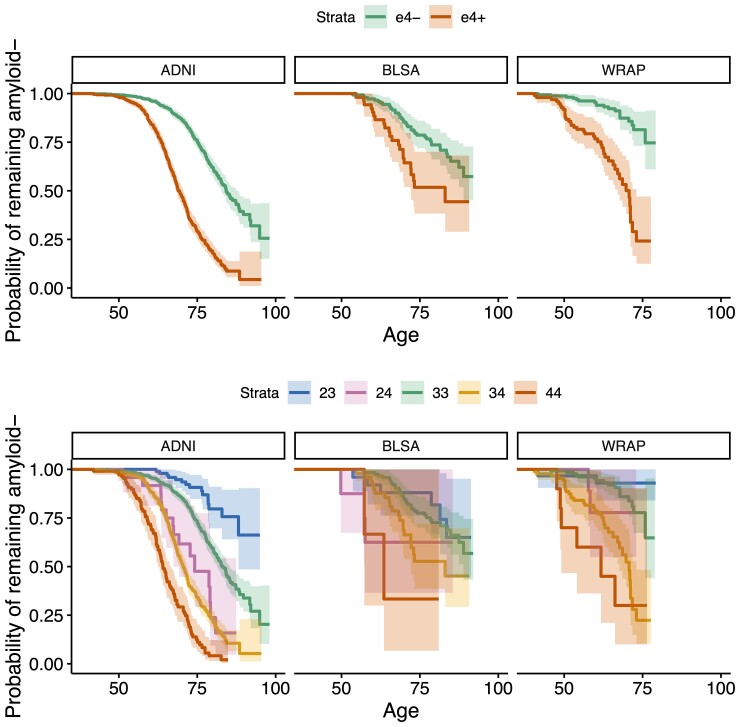
EAOA by APOE genotype is shown for each cohort in Fig. 3 for observed A+ participants. Kaplan–Meier curves by APOE-e4 status and genotype are presented in Fig. 4. The difference in Kaplan–Meier curves between APOE-e4− and e4+ (excluding e2e4) was statistically significant in each dataset (ADNI P < 10−15, BLSA P = 0.003, WRAP P = 10−11). In ADNI, the median A− survival age was 84.2 (95% confidence limits: 82.7, 88.1) for e4− and 69.1 (68.2, 70.5) for e4 + . Median survival times for e4− could not be estimated in BLSA and WRAP because this group did not attain 50% A+ probability. Median A− survival time for e4+ was 83.0 (lower 95% confidence limit = 72.1) in BLSA and 70.7 (67.0, 73.0) in WRAP. In ADNI, all pairwise survival curve comparisons among e2e3, e2e4, e3e3, e3e4 and e4e4 were statistically significant (P < 0.0008) except for differences between e2e4 and e3e4 (P = 0.33). In WRAP, e2e3 versus e3e4, e2e3 versus e4e4, e3e3 versus e3e4, and e3e3 versus e4e4 were significant (P < 0.0002). No significant pairwise comparisons were found in BLSA, but the e3e3 versus e3e4 and e3e3 versus e4e4 comparisons were near significance (P = 0.059). In ADNI, the median A− survival time was 83.1 (81.3, 85.4) for the e3e3, 74.2 (lower 95% confidence limit = 67.4) for e2e4, 70.5 (69.4, 71.8) for e3e4, and 64.0 (62.5, 66.4) for e4e4. In BLSA, the median A− survival time was 83.0 (lower 95% confidence limit = 72.1) for e3e4 and 63.5 (lower 95% confidence limit = 57.2) for e4e4, and in WRAP, 70.7 (67.7, 73.0) and 61.7 (lower 95% confidence limit = 49.1), respectively.
Figure 3.
EAOA by APOE genotype. Estimates are shown only for individuals whose observed A+ onset preceded their last PET visit. Data shown are the average of the estimates computed by the SILA and ODE–GP methods. To assess whether A+ onset age differs by APOE genotype, we conducted pairwise comparisons using AFT models, which allow inclusion of A− participants in the analyses via right censoring. The e3e4 and e4e4 groups had an earlier A+ onset compared to e3e3 in all datasets. The e3e4 A+ onset age was 13, 12 and 20% earlier in the ADNI, BLSA and WRAP, respectively, and the e4e4 A+ onset age was 21, 24 and 30% earlier. The e2e4 group had an earlier amyloid onset than the e3e3 in the ADNI (10% earlier) and BLSA (19% earlier). e2e3 had a later amyloid onset compared to e3e3 in the ADNI only (13% later). The remaining comparisons with the e3e3 group were not statistically significant.
Figure 4.
A− survival as a function of age by APOE-e4 carriage and APOE genotype. Kaplan–Meier curves showing A− survival for APOE e4 carriers versus non-carriers (top row) and for individual APOE genotypes (bottom row) across all cohorts. Individualized estimates of age A+ (average of the SILA and ODE–GP estimates) were used for analyses. Individuals that were A− at their last available PET scan (i.e. those where Estimate Age A+ < age at last scan) are right censored. Cox proportional hazard models indicated significant differences between e4+ and e4− groups. The hazards for e3e4 and e4e4 groups were higher relative to the e3e3 group in all cohorts, and additionally for the e2e4 group in ADNI. e2e3 carriers had a lower hazard in ADNI.
What is the risk of A+ conferred by APOE?
Cox proportional hazards models adjusted for sex showed e4 + was associated with a greater risk of becoming A+ in each dataset [hazard ratio (HR) (95% CI) ADNI: HR = 3.97 (3.35, 4.70), P < 10−15; BLSA: HR = 2.29 (1.35, 3.90), P = 0.002; WRAP: HR = 5.66 (3.24, 9.86), P = 10−9]. Sex and interaction of e4+ by sex were not significant for explaining A+ risk. Results using APOE genotype instead of e4± for A+ risk are shown in Supplementary Table 10. In ADNI, the e2e3 genotype was associated with a lower A+ risk compared to e3e3 [HR = 0.37 (0.22, 0.62), P = 0.0002], whereas e2e4 was associated with higher risk [HR = 2.37 (1.40, 4.00), P = 0.0013]. In each cohort, e3e4 and e4e4 were associated with higher A+ risk, with the point HR estimate for e4e4 being higher. The difference in the risk conferred by e4e4 versus e3e4 was statistically significant in ADNI only [HR = 2.25 (1.78, 2.85), P < 10−10].
Does EAOA differ between APOE groups?
AFT models adjusted for sex indicated e4+ individuals had earlier EAOA [ADNI: β = −0.18 (−0.20, −0.16), P < 10−15; BLSA: β = −0.14 (−0.23, −0.053), P = 0.0017; WRAP: β = −0.26 (−0.35, −0.17), P < 10−8]. Sex differences in EAOA were not significant nor was the interaction between APOE-e4 status and sex in any cohort. In ADNI, compared to e3e3, the e2e3 group had a later EAOA [β = 0.12 (0.064, 0.168), P = 0.00001], whereas e2e4 [β = −0.11 (−0.18, −0.036), P = 0.0034], e3e4 [β = −0.14 (−0.16, −0.12), P < 10−15] and e4e4 [β = −0.24 (−0.28, −0.21), P < 10−15] groups had earlier EAOA (Supplementary Table 11). Additionally, the e4e4 group had earlier EAOA relative to e3e4 [β = −0.10 (−0.14, −0.067), P < 10−7]. In BLSA, compared to e3e3, the e2e4 [β = −0.21 (−0.41, −0.013), P = 0.037], e3e4 [β = −0.13 (−0.23, −0.035), P = 0.0076] and e4e4 [β = −0.28 (−0.57, −0.011), P = 0.059] groups had earlier EAOA. In WRAP, compared to e3e3, the e3e4 [β = −0.23 (−0.33, −0.14), P = 0.000001] and e4e4 [β = −0.35 (−0.53, −0.17), P = 0.00015] groups had earlier EAOA. Kaplan–Meier curves and AFT model results were similar when using left, interval and right censoring with only observed data, but confidence intervals were larger compared to primary analyses using EAOA (Supplementary material).
Aim 3: EAOA, sex, APOE and time to impairment
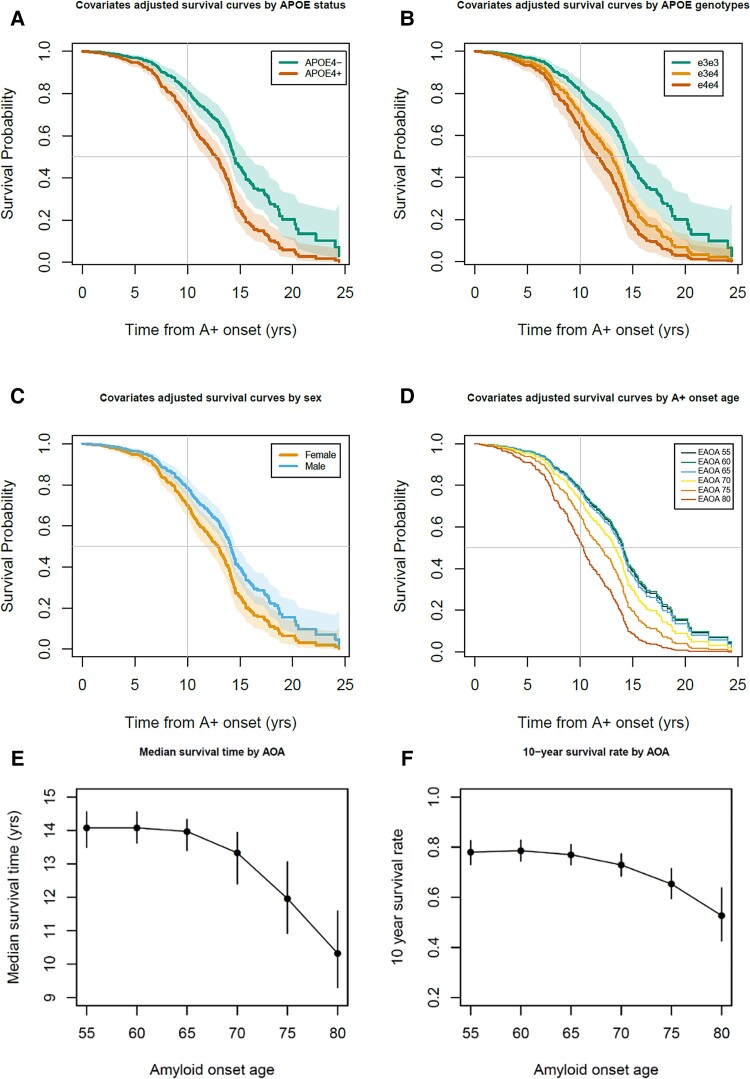
Participant demographics for this ADNI subset are provided in Table 2. Interactions between sex or APOE-e4+/− with linear and quadratic EAOA terms were not significant (global test P = 0.44 and P = 0.17, respectively) in Cox proportional hazard models investigating time from EAOA to impairment onset. Results for HR, median unimpaired survival times and 10-year unimpaired survival rates are reported as estimate (95% confidence interval). In the adjusted Cox hazard model, APOE-e4+ had a 77% greater risk of impairment after EAOA compared to e4− [HR = 1.77 (1.33, 2.36), P < 0.0001] and males had a 32% lower risk compared to females [HR = 0.68, (0.53, 0.88), P = 0.003]. Both EAOA and EAOA2 terms were significant (P < 0.0001). Due to this non-linearity, we report HRs at multiple EAOA with each HR corresponding to a decade difference in EAOA. Impairment risk was small and not significant for EAOA of 65 versus 55 [HR = 1.06 (0.89, 1.26)], but was significant for EAOA of 75 versus 65 [63% greater risk; HR = 1.63 (1.33, 2.00)] and EAOA of 85 versus 75 [HR = 2.51 (1.71, 3.68)]. Groupwise unimpaired survival curves are presented in Fig. 5A, C and D with median survival times and 10-year unimpaired rates included in Supplementary Table 12. The median survival time from EAOA to CDR ≥1 was 13.57 (12.98–14.08) years with a 10-year post-EAOA survival rate of 0.744 (0.703–0.786) across this ADNI subset. Median survival time from EAOA to impairment was shorter for females [12.89 (11.87, 13.66) years] compared to males [14.08 (13.57, 14.55) years], and shorter for APOE-e4+ [12.73 (11.71, 13.42) years] compared to e4− [14.42 (13.97, 15.54) years]. The 10-year post-EAOA unimpaired survival rate was lower for females [0.699 (0.644–0.755) years] compared to males [0.784 (0.736–0.826) years], and lower for APOE-e4+ [0.691 (0.637, 0.741) years] versus e4− [0.812 (0.766, 0.857) years]. Non-linear median survival times and 10-year unimpaired survival rates as a function of EAOA are depicted in Fig. 5E and F and Supplementary Table 13. Analyses using APOE genotype (e3e3, e3e4, e4e4) instead of e4+ versus e4− indicated similar results with significant differences between e3e3 and other genotypes, but not between e3e4 and e4e4 genotypes (Fig. 5B and Supplementary material). Results did not differ when accounting for competing risk of death.
Table 2.
ADNI participant demographics for Aim 3 analyses
| Analysis sample | Cognitively normal during follow-up | Impaired after A+ onset | P-values | |
|---|---|---|---|---|
| n subjects | 595 | 332 (55.8%) | 263 (44.2%) | |
| Males | 327 (55.0%) | 175 (52.7%) | 152 (57.8%) | 0.22 |
| APOE e4+ | 364 (61.2%) | 175 (52.7%) | 189 (71.9%) | <0.0001 |
| White | 566 (95.1%) | 316 (95.2%) | 250 (95.1%) | 0.94 |
| Education | 16.0 (2.8) [6–20] | 16.1 (2.8) [6–20] | 15.8 (2.7) [8–20] | 0.18 |
| A+ onset age | 68.5 (8.7) [41.1–94.9] | 70.8 (8.0) [49.0–94.9] | 65.7 (8.7) [41.1–88.6] | <0.0001 |
| Time (years) to censor/event | 9.4 (4.5) [0.2–24.4] | 8.5 (4.5) [0.2–20.8] | 10.4 (4.4) [0.4–24.4] | <0.0001 |
| AV45 SUVRWM last visit | 1.03 (0.17) [0.80–1.96] | 0.98 (0.14) [0.80–1.59] | 1.09 (0.18) [0.80–1.96] | <0.0001 |
| Age at first CDR | 74.1 (6.9) [55.0–90.3] | 74.6 (6.5) [55.0–90.1] | 73.5 (7.3) [55.1–90.3] | 0.041 |
| Age at last CDR | 78.6 (7.7) [55.7–97.4] | 79.3 (7.5) [59.0–97.4] | 77.7 (7.9) [55.7–96.0] | 0.014 |
| Cognitive follow-up (years) | 4.5 (3.4) [0–15.1] | 4.7 (3.3) [0–15.1] | 4.3 (3.5) [0–14.8] | 0.15 |
| AV45 last age | 77.4 (7.6) [55.6–96.5] | 78.2 (7.3) [59.0–96.5] | 76.5 (7.9) [55.6–96.0] | 0.0061 |
Continuous variables are represented as mean (SD) [range]. Participants were included in aim three analyses if they were not impaired (i.e. CDR < 1) before the estimated age of A+ onset. Fifteen APOE e2e4 carriers were excluded from the analyses due to small sample size and the mixture of risk and protective alleles. Group comparisons (impaired during follow-up versus not impaired during follow-up) for continuous and categorical variables used t-tests and chi-square tests, respectively.
Figure 5.
Time from amyloid onset to impairment in ADNI. (A–D) Covariate adjusted survival curves indicating the probability of remaining unimpaired as a function of the time from A+ onset demonstrated that APOE-e4 carriage, being female and becoming A+ later in life were all associated with a shorter time from A+ onset to impairment onset (i.e. first CDR ≥1). Dotted lines in A–C represent 95% confidence intervals of the covariate adjusted survival functions. (E and F) Median time to impairment and the 10-year unimpaired rate following A+ onset were nonlinearly associated with the EAOA such that people who became A+ at an older age had greater risk of becoming impaired in a shorter time from A+ onset.
Discussion
We evaluated three methods to model amyloid accumulation and estimate A+ onset age from amyloid PET data in three cohorts with different sample characteristics, PET tracers and quantification methods. These novel methods were then applied to further explain the risk and timing of amyloidosis and dementia onset associated with age, sex and APOE genotype. Using individualized EAOA, we found APOE genotype, but not sex, significantly affects EAOA with e4 homozygotes becoming positive ∼6 to 10 years earlier than e3e4 heterozygotes (all cohorts), and approximately two decades earlier than e3 homozygotes (ADNI). Consistent with previous observations, amyloid accumulation rates are predictable and neither APOE genotype, nor sex affected the rate of amyloid accumulation.45,46 In an ADNI subset, time from A+ onset to eventual cognitive impairment was shorter for APOE-e4 carriers, females and people with higher EAOA. Together, these results provide a major advance in characterizing the temporal course of Alzheimer's disease on an individual basis and increase understanding of how common risk factors affect disease proteinopathy and clinical progression.
Method comparisons and amyloid PET inference
Comparisons of the three modelling methods provide insights into model performance, amyloid PET binding estimate interpretation and predictability of amyloid aggregation. All methods had similar DVR/SUVR predictive validity and produced similar EAOA, except GBTM EAOA in ADNI, but differ in efficiency and autonomy. SILA and ODE–GP are autonomous requiring ∼1 min and ∼2.5 h computational time to train and estimate model outputs, respectively, whereas GBTM requires ∼3–4 h of human time. Because of this human resource burden and comparable or better performance of ODE–GP and SILA methods, we forewent additional SUVR/DVR prediction evaluation for GBTM. The autonomy of ODE–GP and SILA enables translation to additional cohorts or cohort subsets, and adaptation to other applications such as spatiotemporal modelling. GBTM was harder to fit in the ADNI dataset, which might suggest ODE–GP and SILA are more robust against differences in PET quantification methods, radiotracers and/or study composition. ODE–GP and SILA also produce non-parametric SUVR/DVR versus A+ time curves that may better depict the observed nature of amyloid accumulation over parametric models that require assumptions of this geometric relationship.
Amyloid accumulation patterns were similar between methods in the A+ range with inter-method differences increasing dramatically below the A+ threshold. This observation, coupled with similarities in DVR/SUVR rates, suggests that amyloid accumulation rates are ostensibly consistent between individuals and predictable once underlying amyloid burden sufficiently exceeds amyloid PET detection limits. This detection limit and binding estimate accuracy and variance are influenced by several factors including tracer affinity and non-specific white matter binding, acquisition length, image reconstruction, scanner resolution and image processing and quantification methodology, which varied across cohorts in this study.47,48 Below the A+ threshold, DVR/SUVR interpretation gradually transitions from being due to amyloid to being due to stochastic measurement error, which is consistent with PET detection physics,47 the concept of subthreshold amyloid49 and supports previous work proposing two thresholds50 to define three interpretation zones: undetectable amyloid, transition and confidently A+. EAOA derived from DVR/SUVR in these zones therefore inherit a similar interpretation, in which EAOA is interpretable in the A+ range and becomes less interpretable as reference DVR/SUVR approaches PET detection limits. Similarly, the accuracy and variability of reference DVR/SUVR affect the accuracy and variability of imputed DVR/SUVR and EAOA. Lower variability of DVR quantification compared to SUVR, along with previous work showing Florbetapir has higher variance compared to PiB after converting to Centiloids,51 might explain why ADNI had a higher Centiloid threshold despite A+ thresholds delineating longitudinal amyloid non-accumulators from accumulators. For this reason and because direct comparisons of factors that affect PET detection limits and variability were not available across cohorts, we used previously validated methods and A+ thresholds from each cohort that minimize PET binding estimate variance and used a study design focused on replicating findings across cohorts and methods rather than attempting pooled analyses across cohorts. As in our previous work13 and a recently published similar work14 estimating individualized EAOA in people who are A+ enables estimation of A+ disease duration from a single amyloid PET scan. This ability to define disease time from amyloid PET provides new insights into how known and unknown factors affect dementia risk and timing in Alzheimer's disease, which has clinical trial implications.
APOE, EAOA and dementia timing and risk
Our results support literature showing higher APOE-e4 dosage associates with earlier amyloid and dementia onset.8,12,17–19,21–23,46 Additionally, we observed significant EAOA differences between almost all compared APOE genotypes in ADNI, with similar patterns observed in BLSA and WRAP (lack of significance potentially due to power limitations and/or cohort makeup). Individualized EAOA allowed us to show e4 carriage also shortens the preclinical time from A+ onset to dementia onset (in initially unimpaired ADNI participants, even after controlling for age and sex). These findings suggest APOE-e4 carriers experience a double-hit wherein they accumulate amyloid earlier in life and have a shorter preclinical disease phase. Effect sizes for APOE genotype differences in EAOA far exceeded differences in time from A+ onset to dementia suggesting that when amyloid accumulation begins is a key factor explaining APOE associated lifetime dementia risk and resilience.
Clinical implications
Consistent with our previous reports,12,13 this work suggests individualized EAOA and A+ duration are relevant to clinical prognosis and treatment planning. Figures 1 and 3 demonstrate EAOA varies widely, even within APOE genotypes, spanning ages 40 to 90+ years across cohorts. Recruitment differences probably explain some cohort EAOA differences. Nevertheless, the broad range of EAOA highlights the heterogeneity of when amyloid pathology begins to accrue. These results and our previous findings13 also underscore the need to consider the magnitude/duration of amyloid exposure to better inform prognosis compared to binary A±. Comparatively less heterogeneity was observed in the time from A+ onset to dementia onset (range 0–25 years post-EAOA, Fig. 6) in initially unimpaired A+ ADNI participants. Combining the amyloid chronicity framework13 and survival analyses we showed APOE-e4 carriers, females, and people with older EAOA have shorter time from A+ onset to dementia, and that the effect of EAOA on this timing accelerated rapidly after age 65. These methods can also establish EAOA, sex and APOE stratified dementia risk probabilities (e.g. 1-, 5-, 10-year impairment probability) that could inform individualized patient prognoses anchored to EAOA, which might also help identify optimal treatment windows for anti-amyloid therapies. More work is needed to better understand additional factors that mediate and moderate both the heterogeneity of EAOA and the timing from EAOA to impairment, and to validate risk probabilities in larger and representative samples.
Figure 6.
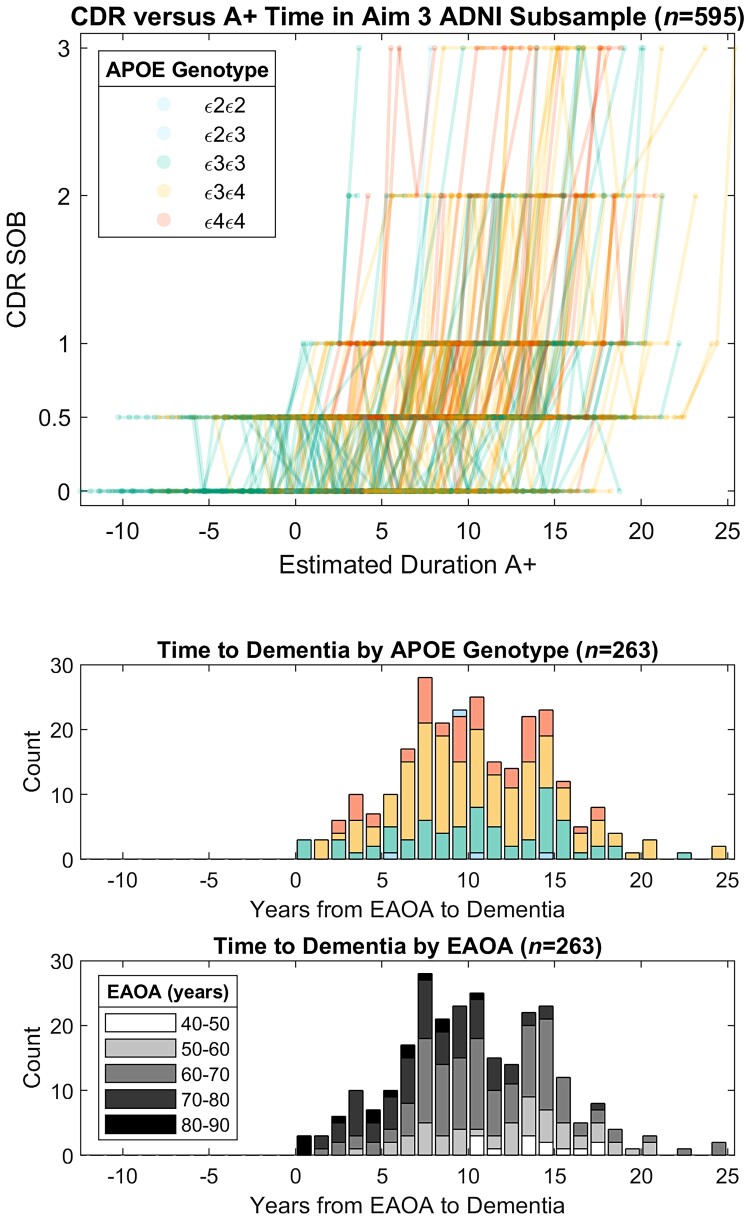
Global CDR scores as a function of the time A+ in ADNI subsample from Aim 3 analyses. Observed longitudinal global CDR scores are plotted as a function of estimated duration A+ for the ADNI subset used in Aim 3 analyses (top). Histograms depict the time from estimated A+ onset age (EAOA) to global CDR = 1 colour coded by APOE genotype (middle) and by EAOA (bottom) for the subset of participants that became impaired (i.e. global CDR ≥1) after A+ onset. Time from A+ onset to global CDR ≥1 in this ADNI subsample ranged from 0 to nearly 25 years demonstrating the heterogeneity in the timing of dementia onset relative to A+ onset.
The effects of EAOA, sex and APOE on the timing between EAOA and dementia have several potential explanations. Similar findings regarding age, but not APOE or sex were recently reported using a similar approach.14 EAOA shortening the preclinical timeframe may be explained by age-associated increases in comorbidity with other age-related diseases such as vascular disease and other neurodegenerative proteinopathies that co-occur in Alzheimer's disease.52APOE-e4 carriage shortening the preclinical timeframe could represent a pleiotropic susceptibility effect of APOE on brain health since APOE is also implicated in cerebrovascular health.53 Females having shorter preclinical disease is consistent with but does not fully explain some epidemiologic reports of sex differences in Alzheimer's disease risk. These results coupled with the observations that sex did not affect EAOA or amyloid accumulation rates is consistent with recent findings that observed no sex differences in Alzheimer's disease biomarker prevalence but did observe differences in MCI prevalence.54 More work is needed to understand the basis for the observed greater vulnerability to dementia for these risk factors. Anchoring such investigations to EAOA may facilitate greater understanding of the relative influence of these and other factors that are associated with increased Alzheimer's disease risk.
Strengths and limitations
This work and other recent publications,3,7,11–15,44,55 demonstrate the robustness and use of the conceptual framework of defining Alzheimer's disease time using amyloid PET (i.e. an ‘Amyloid Clock’) to study disease progression. Strengths of the current study include comparisons and consistency of results across three different methods and cohorts, prediction validation in subsets of observed A− to A+ convertors and a methodological framework for applying individualized EAOA to characterize biomarker and dementia timing and risk in Alzheimer's disease. Further work is needed to better understand the impact of PET quantification and processing methods, radiotracers, different PET and MRI scanners, and cohort composition on temporal amyloid models, which affect EAOA accuracy and harmonization. Additionally, this study used convenience samples that were skewed towards highly educated, non-Hispanic white people. More work is needed to collect biomarkers and validate these approaches in diverse populations.
Conclusions
This work demonstrates multiple methods for modelling amyloid accumulation and estimating the onset age and duration of A+ from PET imaging. These novel approaches and framework can be used to more directly investigate the timing of cognitive and pathological events in Alzheimer's disease on an individual basis, and understand factors that modify this timing.
Supplementary Material
Acknowledgements
We would like to thank all participants, their families and the study teams that make this work possible. This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health. Data collection and sharing for this project was funded by the ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Data used in preparation of this article were obtained from the ADNI database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Abbreviations
- A±
elevated or non-elevated amyloid PET
- ADNI
Alzheimer's Disease Neuroimaging Initiative
- AFT
accelerated failure time
- BLSA
Baltimore Longitudinal Study on Aging
- CDR
clinical dementia rating
- DVR
distribution volume ratio
- EAOA
estimated amyloid onset age
- GBTM
group-based trajectory modelling
- MCI
mild cognitive impairment
- ODE–GP
ordinary differential equation–Gaussian process
- PiB
11C-Pittsburgh compound B
- SILA
sampled iterative local approximation
- SUVR
standard uptake value ratio
- WRAP
Wisconsin Registry for Alzheimer's Prevention
Contributor Information
Tobey J Betthauser, Wisconsin Alzheimer’s Disease Research Center, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Department of Medicine, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, USA.
Murat Bilgel, Laboratory of Behavioral Neuroscience, National Institute on Aging, National Institutes of Health, Baltimore, MD, USA.
Rebecca L Koscik, Wisconsin Alzheimer’s Disease Research Center, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Department of Medicine, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, USA; Wisconsin Alzheimer’s Institute, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA.
Bruno M Jedynak, Department of Mathematics and Statistics, Portland State University, Portland, OR, USA.
Yang An, Laboratory of Behavioral Neuroscience, National Institute on Aging, National Institutes of Health, Baltimore, MD, USA.
Kristina A Kellett, Wisconsin Alzheimer’s Disease Research Center, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Department of Medicine, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, USA.
Abhay Moghekar, Laboratory of Behavioral Neuroscience, National Institute on Aging, National Institutes of Health, Baltimore, MD, USA.
Erin M Jonaitis, Wisconsin Alzheimer’s Disease Research Center, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Department of Medicine, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, USA; Wisconsin Alzheimer’s Institute, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA.
Charles K Stone, Department of Medicine, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, USA.
Corinne D Engelman, Wisconsin Alzheimer’s Disease Research Center, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Wisconsin Alzheimer’s Institute, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Department of Population Health Sciences, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, USA.
Sanjay Asthana, Wisconsin Alzheimer’s Disease Research Center, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Department of Medicine, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, USA; Wisconsin Alzheimer’s Institute, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Geriatric Research Education and Clinical Center, William S. Middleton Veterans Hospital, Madison, WI, USA.
Bradley T Christian, Wisconsin Alzheimer’s Disease Research Center, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Waisman Laboratory for Brain Imaging and Behavior, University of Wisconsin-Madison, Madison, WI, USA; Department of Medical Physics, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, USA.
Dean F Wong, Department of Radiology, Mallinckrodt Institute of Radiology, Neurology, Psychiatry and Neuroscience, Washington University School of Medicine, St. Louis, MO, USA.
Marilyn Albert, Department of Neurology, Division of Cognitive Neuroscience, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
Susan M Resnick, Laboratory of Behavioral Neuroscience, National Institute on Aging, National Institutes of Health, Baltimore, MD, USA.
Sterling C Johnson, Wisconsin Alzheimer’s Disease Research Center, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Department of Medicine, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, USA; Wisconsin Alzheimer’s Institute, University of Wisconsin School of Medicine and Public Health, Madison, WI, USA; Geriatric Research Education and Clinical Center, William S. Middleton Veterans Hospital, Madison, WI, USA.
Funding
The following funding sources contributed to this work: NIH R01 AG021155, R01 AG027161, P50 AG033514, U54 HD090256, S10 OD025245, R01 AG054047, RF1 AG059869; Alzheimer’s Association AARF-19-614533; and NSF 2019216.
Competing interests
S.C.J. has served on a Roche Diagnostics advisory board and receives research support from Cerveau Technologies. D.F.W. receives research support via WUSTL from Roche Neuroscience, Intracellular Technologies. All remaining authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Thal DR, Rüb U, Orantes M, Braak H. Phases of Aβ-deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58:1791–1800. [DOI] [PubMed] [Google Scholar]
- 2. Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. [DOI] [PubMed] [Google Scholar]
- 3. Jack CR, Knopman DS, Jagust WJ, et al. . Tracking pathophysiological processes in Alzheimer’s disease: An updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nelson PT, Alafuzoff I, Bigio EH, et al. . Correlation of Alzheimer disease neuropathologic changes with cognitive status: A review of the literature. J Neuropathol Exp Neurol. 2012;71:362–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sperling RA, Aisen PS, Beckett LA, et al. . Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the national institute on aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bateman RJ, Xiong C, Benzinger TL, et al. . Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Villemagne VL, Burnham S, Bourgeat P, et al. . Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: A prospective cohort study. Lancet Neurol. 2013;12:357–367. [DOI] [PubMed] [Google Scholar]
- 8. Jansen WJ, Ossenkoppele R, Knol DL, et al. . Prevalence of cerebral amyloid pathology in persons without dementia: A meta-analysis. JAMA. 2015;313:1924–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jagust WJ, Landau SM, Alzheimer’s Disease Neuroimaging Initiative . Temporal dynamics of β-amyloid accumulation in aging and Alzheimer disease. Neurology. 2021;96:e1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jack CR, Knopman DS, Jagust WJ, et al. . Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010;9:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Insel PS, Donohue MC, Berron D, Hansson O, Mattsson-Carlgren N. Time between milestone events in the Alzheimer’s disease amyloid cascade. Neuroimage. 2021;227:117676. [DOI] [PubMed] [Google Scholar]
- 12. Bilgel M, An Y, Zhou Y, et al. . Individual estimates of age at detectable amyloid onset for risk factor assessment. Alzheimers Dement. 2016;12:373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koscik RL, Betthauser TJ, Jonaitis EM, et al. . Amyloid duration is associated with preclinical cognitive decline and tau PET. Alzheimers Dement. 2020;12:e12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schindler S, Li Y, Buckles VD, et al. . Predicting symptom onset in sporadic Alzheimer disease with amyloid PET. Neurology. 2021;97:e1823–e1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Therneau TM, Knopman DS, Lowe VJ, et al. . Relationships between β-amyloid and tau in an elderly population: An accelerated failure time model. Neuroimage. 2021;242:118440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Belloy ME, Napolioni V, Greicius MD. A quarter century of APOE and Alzheimer’s disease: Progress to date and the path forward. Neuron. 2019;101:820–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Corder EH, Saunders AM, Strittmatter WJ, et al. . Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. [DOI] [PubMed] [Google Scholar]
- 18. Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer disease. Proc Natl Acad Sci U S A. 1995;92:4725–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van der Lee SJ, Wolters FJ, Ikram MK, et al. . The effect of APOE and other common genetic variants on the onset of Alzheimer’s disease and dementia: A community-based cohort study. Lancet Neurol. 2018;17:434–444. [DOI] [PubMed] [Google Scholar]
- 20. Payami H. Alzheimer’s disease, apolipoprotein E4, and gender. JAMA. 1994;271:1316–1317. [PubMed] [Google Scholar]
- 21. Morris JC, Roe CM, Xiong C, et al. . APOE Predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol. 2010;67:122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Resnick SM, Bilgel M, Moghekar A, et al. . Changes in Aβ biomarkers and associations with APOE genotype in 2 longitudinal cohorts. Neurobiol Aging. 2015;36:2333–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fleisher AS, Chen K, Liu X, et al. . Apolipoprotein E ε4 and age effects on florbetapir positron emission tomography in healthy aging and Alzheimer disease. Neurobiol Aging. 2013;34:1–12. [DOI] [PubMed] [Google Scholar]
- 24. Johnson SC, Koscik RL, Jonaitis EM, et al. . The Wisconsin Registry for Alzheimer’s Prevention: A review of findings and current directions. Alzheimers Dement. 2018;10:130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ferrucci L. The Baltimore longitudinal study of aging (BLSA): A 50-year-long journey and plans for the future. J Gerontol A Biol Sci Med Sci. 2008;63:1416–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weiner MW, Veitch DP, Aisen PS, et al. . Recent publications from the Alzheimer’s disease neuroimaging initiative: Reviewing progress toward improved AD clinical trials. Alzheimers Dement. 2017;13:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Resnick SM, Sojkova J, Zhou Y, et al. . Longitudinal cognitive decline is associated with fibrillar amyloid-beta measured by [11C]PiB. Neurology. 2010;74:807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morris JC. The clinical dementia rating (CDR): current version and scoring rules. Neurology. 1993;43:241.2–241-a. [DOI] [PubMed] [Google Scholar]
- 29. Fuld P. Psychological testing in the differential diagnosis of the dementias. In: Katzman RTerry RD and Bick KL, eds. Alzheimer’s disease: senile dementia and related disorders, Aging, Vol. 7. Ravens Press; 1978: 185–193. [Google Scholar]
- 30. American Psychiatric Association . Diagnostic and statistical manual of mental disorders. 3rd edn.American Psychiatric Association. 1987. [Google Scholar]
- 31. Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183–194. [DOI] [PubMed] [Google Scholar]
- 32. Koscik RL H, Allison BP, Clark S, et al. . Validity evidence for the research category, “cognitively unimpaired—declining,” as a risk marker for mild cognitive impairment and Alzheimer’s disease. Front Aging Neurosci. 2021;13:688478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jagust WJ, Landau SM, Koeppe RA, et al. . The Alzheimer’s disease neuroimaging initiative 2 PET core: 2015. Alzheimer’s Dement. 2015;11:757–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fleisher AS, Chen K, Liu X, et al. . Using positron emission tomography and florbetapir F18 to image cortical amyloid in patients with mild cognitive impairment or dementia due to Alzheimer disease. Arch Neurol. 2011;68:1404–1411. [DOI] [PubMed] [Google Scholar]
- 35. Chen K, Roontiva A, Thiyyagura P, et al. . Improved power for characterizing longitudinal amyloid-β PET changes and evaluating amyloid-modifying treatments with a cerebral white matter reference region. J Nucl Med. 2015;56:560–566. [DOI] [PubMed] [Google Scholar]
- 36. Johnson SC, Christian BT, Okonkwo OC, et al. . Amyloid burden and neural function in people at risk for Alzheimer’s disease. Neurobiol Aging. 2014;35:576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou Y, Resnick SM, Ye W, et al. . Using a reference tissue model with spatial constraint to quantify [11C]Pittsburgh compound B PET for early diagnosis of Alzheimer’s disease. Neuroimage. 2007;36:298–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Landau SM, Fero A, Baker SL, et al. . Measurement of longitudinal β-amyloid change with 18F-florbetapir PET and standardized uptake value ratios. J Nucl Med. 2015;56:567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Racine AM, Clark LR, Berman SE, et al. . Associations between performance on an abbreviated CogState battery, other measures of cognitive function, and biomarkers in people at risk for Alzheimer’s disease. J Alzheimers Dis. 2016;54:1395–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klunk WE, Koeppe RA, Price JC, et al. . The centiloid project: Standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 2015;11:1–15 e1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jones BL, Nagin DS, Roeder K. A SAS procedure based on mixture models for estimating developmental trajectories. Sociol Methods Res. 2001;29:374–393. [Google Scholar]
- 42. Nagin DS. Analyzing developmental trajectories: A semiparametric, group-based approach. Psychol Methods. 1999;4:139–157. [DOI] [PubMed] [Google Scholar]
- 43. Long ZL Y, Ma X, Dong B. PDE-net: learning PDES from data. In: Proceedings of the 35th International Conference on Machine Learning, 2018; vol. 80: pp. 3208–3216. [Google Scholar]
- 44. Bilgel M, Prince JL, Wong DF, Resnick SM, Jedynak BM. A multivariate nonlinear mixed effects model for longitudinal image analysis: application to amyloid imaging. Neuroimage. 2016;134:658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lim YY, Mormino EC, Alzheimer’s disease neuroimaging Initiative . APOE genotype and early β-amyloid accumulation in older adults without dementia. Neurology. 2017;89:1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burnham SC, Laws SM, Budgeon CA, et al. . Impact of APOE-ε4 carriage on the onset and rates of neocortical Aβ-amyloid deposition. Neurobiol Aging. 2020;95:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cherry SR, Sorenson JA, Phelps ME. Physics in nuclear medicine e-Book. Elsevier Health Sciences. 2012. [Google Scholar]
- 48. Logan J, Fowler JS, Volkow ND, Wang G-J, Ding Y-S, Alexoff DL. Distribution volume ratios without blood sampling from graphical analysis of PET data. J Cereb Blood Flow Metab. 1996;16:834–840. [DOI] [PubMed] [Google Scholar]
- 49. Landau SM, Horng A, Jagust WJ, Alzheimer’s disease neuroimaging Initiative . Memory decline accompanies subthreshold amyloid accumulation. Neurology. 2018;90:e1452–e1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bullich S, Roe-Vellve N, Marquie M, et al. . Early detection of amyloid load using (18)F-florbetaben PET. Alzheimers Res Ther. 2021;13:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Su Y, Flores S, Wang G, et al. . Comparison of Pittsburgh compound B and florbetapir in cross-sectional and longitudinal studies. Alzheimers Dement. 2019;11:180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schneider JA, Arvanitakis Z, Bang W, Bennett DA. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology. 2007;69:2197–2204. [DOI] [PubMed] [Google Scholar]
- 53. Zlokovic BV. Cerebrovascular effects of apolipoprotein E: Implications for Alzheimer disease. JAMA Neurol. 2013;70:440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jack CR Jr, Therneau TM, Weigand SD, et al. . Prevalence of biologically vs clinically defined Alzheimer Spectrum entities using the national institute on aging-Alzheimer’s association research framework. JAMA Neurol. 2019;76:1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Whittington A, Sharp DJ, Gunn RN, Alzheimer’s disease neuroimaging Initiative . Spatiotemporal distribution of β-Amyloid in Alzheimer disease is the result of heterogeneous regional carrying capacities. J Nucl Med. 2018;59:822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the WRAP (https://wrap.wisc.edu) and BLSA (https://blsa.nih.gov) can be requested through an online submission process. ADNI data can be obtained via the ADNI and LONI websites. ODE–GP and SILA algorithms are available at https://gitlab.com/bilgelm/amyloid-ode-gp and https://github.com/Betthauser-Neuro-Lab/SILA-AD-Biomarker, respectively.