ABSTRACT
Transcription factors (TFs) bind to the promoters of target genes to regulate gene expression in response to different stimuli. The functions and regulatory mechanisms of transcription factors (TFs) in Verticillium dahliae are, however, still largely unclear. This study showed that a C2H2-type zinc finger TF, VdCf2 (V. dahliae chorion transcription factor 2), plays key roles in V. dahliae growth, melanin production, and virulence. Transcriptome sequencing analysis showed that VdCf2 was involved in the regulation of expression of genes encoding secreted proteins, pathogen-host interaction (PHI) homologs, TFs, and G protein-coupled receptors (GPCRs). Furthermore, VdCf2 positively regulated the expression of VdPevD1 (VDAG_02735), a previously reported virulence factor. VdCf2 thus regulates the expression of several pathogenicity-related genes that also contribute to virulence in V. dahliae. VdCf2 also inhibited the transcription of the Vd276-280 gene cluster and interacted with two members encoding proteins (VDAG_07276 and VDAG_07278) in the gene cluster.
IMPORTANCE Verticillium dahliae is an important soilborne phytopathogen which can ruinously attack numerous host plants and cause significant economic losses. Transcription factors (TFs) were reported to be involved in various biological processes, such as hyphal growth and virulence of pathogenic fungi. However, the functions and regulatory mechanisms of TFs in V. dahliae remain largely unclear. In this study, we identified a new transcription factor, VdCf2 (V. dahliae chorion transcription factor 2), based on previous transcriptome data, which participates in growth, melanin production, and virulence of V. dahliae. We provide evidence that VdCf2 regulates the expression of the pathogenicity-related gene VdPevD1 (VDAG_02735) and Vd276-280 gene cluster. VdCf2 also interacts with VDAG_07276 and VDAG_07278 in this gene cluster based on a yeast two-hybrid and bimolecular fluorescence complementation assay. These results revealed the regulatory mechanisms of a pivotal pathogenicity-related transcription factor, VdCf2 in V. dahliae.
KEYWORDS: Verticillium dahliae, transcription factor, VdCf2, pathogenicity, transcriptome, gene cluster
INTRODUCTION
Verticillium dahliae is a soilborne phytopathogen that causes vascular wilt disease in more than 200 plant species (1–5). The pathogen can survive in the soil by means of microsclerotia for more than 14 years (6). In response to root exudates released in the plant rhizosphere, microsclerotia germinate and produce hyphae, which then directly penetrate host roots to colonize the xylem vessels (2, 7, 8). With the progressive colonization of the xylem vessels, the host begins to show symptoms such as foliar wilting, chlorosis, plant stunting, and vascular browning (9, 10). With further disease progression, infected plants begin to senesce and wilt, and microsclerotia are formed in the necrotic tissue to serve as the primary inoculum for subsequent crops (9). The wilt disease caused by V. dahliae is a difficult disease to manage in commercial agriculture. A greater understanding of the mechanisms of pathogenesis and virulence in V. dahliae may provide knowledge for developing strategies for the management of Verticillium wilt.
Mechanisms of V. dahliae pathogenesis and virulence have been widely studied. Many genes that play important roles in pathogenicity and virulence, including those encoding secreted proteins, such as PevD1 (an Alt a 1-like protein) (11, 12), GH12 (glycoside hydrolase 12) (13), VdSCP41 (effector) (14), and VdPEL1 (pectate lyase) (15), and genes encoding nonsecreted proteins, such as VGB (G protein β subunit) (16), VdMyo5 (myosin V protein) (17), VdNoxB/VdPls1 (NADPH oxidases/tetraspanin) (18), Vst1 (APSES-type transcription factor [TF]) (19), and VdAtf1 (bZIP-type TF) (20), have been identified in V. dahliae. TFs not only play critical roles in pathogenicity and virulence in V. dahliae, but also could interact with other TFs (AtfA, AtfB, AtfC, AtfD) or molecular chaperones (Hsp70, Hdj1) (21, 22).
Based on the differences in the conserved DNA-binding domains, the 530 predicted TFs in the reference genome of V. dahliae strain VdLs.17 were classified into 42 types (23). Of these, the C2H2-type TF VdCrz1 and the MADS-box-type TF VdMcm1 are involved in fungal growth, melanin and microsclerotium formation, and virulence (24, 25). Furthermore, the Zn2Cys6-type TF VdFTF1, APSES-type TF Vst1, and bZIP-type TF VdAtf1 also play roles in virulence (19, 20, 26). When functions of specific TFs in other fungal pathogens were altered, their development and virulence to hosts were also significantly affected (27–29). In Botrytis cinerea, disruption of MADS-box TF Bcmads1 caused defects in growth and sclerotium production and full virulence on apple fruit (30). In Magnaporthe oryzae, deletion of bZIP TF MoAP1 led to defects in aerial hyphal growth and pathogenicity (31). Therefore, the disruption of several types of key TFs influences fungal development, pathogenicity, and virulence.
A total of 93 annotated C2H2-type TFs have been identified in the genome of V. dahliae strain VdLs.17 (23). However, only a few of such TFs, such as Vta2, VdCrz1, and VdMsn2, have been functionally characterized (24, 32, 33). The role of most C2H2-type TFs in virulence is largely unknown. According to our previous transcriptome data (unpublished), VdCf2 was significantly upregulated in the dormant microsclerotia, which encodes a C2H2-type chorion transcription factor in V. dahliae. And VdCf2 was found to be associated with virulence. Cf2 was first discovered in Drosophila melanogaster due to its ability to target various DNA sequences caused by its alternative splicing (34). Previous studies showed that the Drosophila TF Cf2 plays an important role in ensuring an appropriate myofibril size by regulating the transcription level of several muscle structural genes (35–37). Functions of chorion transcription factor Cf2 in phytopathogenic fungi have, however, not been studied yet. The present study showed that the VdCf2 deletion mutant impaired growth and virulence but increased melanin accumulation compared to that of the wild-type strain. Furthermore, transcriptomic analysis indicated that VdCf2 regulated the expression of several pathogenicity-related genes, some of which also contributed to virulence in V. dahliae.
RESULTS
VdCf2 is localized to the nucleus.
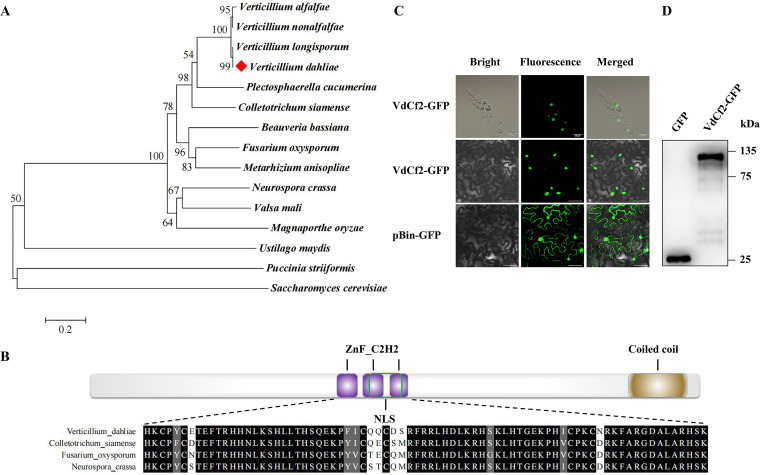
VdCf2 (VDAG_08721) in V. dahliae was annotated as a chorion transcription factor Cf2, and shared 34.33% identity with the Cf2 in D. melanogaster (see Fig. S1 in the supplemental material). Phylogenetic analysis showed high sequence identity with Cf2 in Verticillium homologs (Fig. 1A). It also revealed that VdCf2 was not a specifically conserved transcription factor in phytopathogenic fungi (Fig. 1A). The VdCf2 protein was predicted to possess three C2H2-type zinc finger domains in the middle and one coiled-coil region in the C terminus based on the SMART online tool (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1), as well as a nuclear localization signal based on cNLS Mapper (https://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) (Fig. 1B). Multiple sequence alignments showed that C2H2-type zinc finger domains of Cf2 orthologs from V. dahliae, Colletotrichum siamense, Fusarium oxysporum, and Neurospora crassa were highly conserved (Fig. 1B).
FIG 1.
Sequence analysis and subcellular localization of VdCf2. (A) Dendrogram of VdCf2 and its homologues from Verticillium longisporum, Verticillium nonalfalfae, Verticillium alfalfae, Colletotrichum siamense, Plectosphaerella cucumerina, M. oryzae, Fusarium oxysporum, Valsa mali, Puccinia striiformis, Ustilago maydis, N. crassa, Beauveria bassiana, Metarhizium anisopliae, and Saccharomyces cerevisiae. The phylogenetic tree was constructed using MEGA 7.0 software based on the neighbor-joining method. The number of bootstrap replications was set as 1,000. (B) Schematic representation of the VdCf2 structure containing conserved C2H2 zinc finger domain and coiled-coil region according to the prediction from SMART as well as the nuclear localization signal predicted by cNLS Mapper. Multiple sequence alignment of the C2H2-type zinc finger domain of VdCf2 orthologs from V. dahliae, Colletotrichum siamense, F. oxysporum, and N. crassa was completed using MEGA 7.0 software and displayed using the multiple sequence comparison display online website. (C) The subcellular localization was performed in V. dahliae (first line, bars = 20 μm) and tobacco (N. benthamiana) cells (last two lines, bars = 40 μm). pBin-GFP was used as the control. (D) Western blot analysis of GFP and VdCf2-GFP fusion proteins in tobacco (N. benthamiana) cells.
To investigate the subcellular localization of VdCf2 protein, green fluorescent protein (GFP)-tagged VdCf2 protein was expressed in both V. dahliae and Nicotiana benthamiana cells, and fluorescent signals were detected in the nucleus in both (Fig. 1C, Fig. S2). In contrast, florescent signals were diffused across the plant cell when free GFP was expressed (Fig. 1C). Western blots further verified that GFP and the VdCf2-GFP fusion proteins were expressed successfully in N. benthamiana cells (Fig. 1D). The conservation of C2H2-type zinc finger domain sequences of VdCf2 coupled with the nuclear localization of VdCf2-GFP in fungi and plant cells further suggested that VdCf2 encodes a C2H2-type zinc finger TF because of its common function in nuclear compartmentalization.
VdCf2 is involved in growth but not in osmotic stress response.
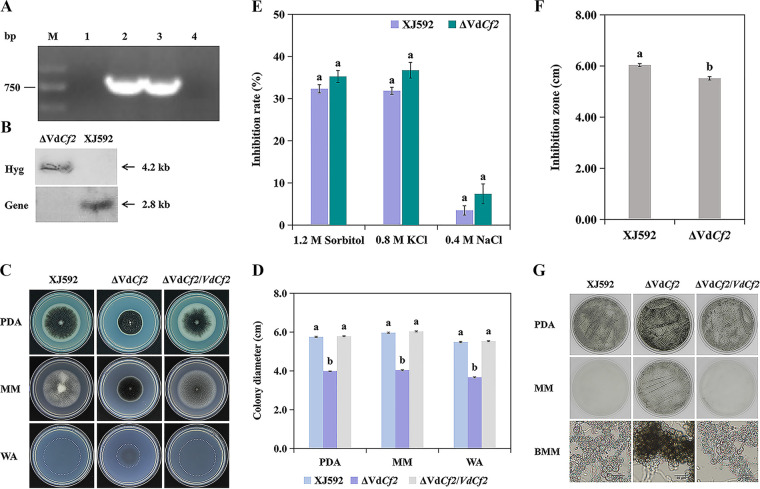
To determine the importance of VdCf2 in V. dahliae, the ΔVdCf2 strain was generated by homologous recombination and verified by PCR and Southern blot assays (Fig. 2A and B). To explore the role of VdCf2 on growth, the wild-type strain XJ592 and the ΔVdCf2 and ΔVdCf2/VdCf2 strains were cultured on potato dextrose agar (PDA), minimal medium (MM), and water agar (WA) and incubated for 14 days. Compared to strains XJ592 and ΔVdCf2/VdCf2, the growth rate of strain ΔVdCf2 was reduced by more than 30% on all three media (Fig. 2C and D), indicating that VdCf2 is critical for the growth of V. dahliae.
FIG 2.
VdCf2 plays key roles in V. dahliae growth and melanin production. (A) The identification electrophoretogram of the ΔVdCf2 and ΔVdCf2/VdCf2 strains. M, 2,000 bp marker; 1, ΔVdCf2 strain; 2, ΔVdCf2/VdCf2 strain; 3, wild-type strain; 4, negative control (sterile water). (B) Confirmation of VdCf2 deletion by Southern blot analysis. Total genomic DNA of the wild-type and ΔVdCf2 strains was digested with BamHI and subjected to Southern blot analysis. The probe in the VdCf2 gene region amplified with primer pair NF/NR was used to confirm the presence or absence of the VdCf2 gene in the wild-type and ΔVdCf2 strains. The probe in the hygromycin B resistance gene region amplified by primer pair HYG-F/HYG-R was used to verify the copy number of the hygromycin B resistance gene in the wild-type and ΔVdCf2 strains. (C) Colony morphology of the XJ592, ΔVdCf2, and ΔVdCf2/VdCf2 strains on PDA, MM, and WA plates. Pictures were taken from the upper side of 14-day-old cultures. (D) Colony diameter of the XJ592, ΔVdCf2, and ΔVdCf2/VdCf2 strains on three media. Error bars are standard errors calculated from three replicates, and different letters represent statistically significant differences (P = 0.05). The SNK test was executed among each strain in each medium. (E) VdCf2 was not required for osmotic stress response. The inhibition rate of colony growth was quantified. Strains XJ592 and ΔVdCf2 were cultured on CM plates containing osmotic stress reagents (1.2 M sorbitol, 0.8 M KCl, or 0.4 M NaCl) for 14 days. Error bars indicate standard errors calculated from three replicates, and different letters represent the statistical significance (P = 0.05). Student’s t test was executed between strains in each osmotic stress reagent. (F) VdCf2 participated in oxidative stress response. The inhibition zones of the XJ592 and ΔVdCf2 strains were quantified. Error bars indicate standard errors calculated from three replicates (Student’s t test, P = 0.05). (G) VdCf2 curtails melanin production. The conidial suspensions of strains XJ592, ΔVdCf2, and ΔVdCf2/VdCf2 were cultured on PDA and MM plates for 3 days and BMM plates for 4 days. The mycelia on the BMM plates were observed under a light microscope (bars = 20 μm).
To explore whether VdCf2 plays a role in the osmotic stress response, strains XJ592 and ΔVdCf2 were cultured on complete medium (CM) containing 1.2 M sorbitol, 0.8 M KCl, or 0.4 M NaCl and incubated for 14 days. The growth inhibition of strain ΔVdCf2 was indistinguishable from that of XJ592 (Fig. 2E), indicating that VdCf2 did not respond to the osmotic stress induced by sorbitol, KCl, or NaCl. To evaluate the role of VdCf2 in oxidative stress, the inhibition zone was measured on the CM containing H2O2. The result showed that strain ΔVdCf2 exhibited smaller inhibition zones than the wild-type strain (Fig. 2F), suggesting that VdCf2 is negatively involved in regulation of the oxidative stress response.
VdCf2 regulates melanin production.
In general, melanin protects pathogens from environmental stresses and is beneficial to the long-term survival of fungi under unfavorable environmental conditions (38–41). Compared to the wild-type XJ592 and ΔVdCf2/VdCf2 strains, strain ΔVdCf2 accumulated more melanin on PDA, MM, and WA media, suggesting that VdCf2 regulates melanin production (Fig. 2C and G). In addition, strain ΔVdCf2 cultured on modified basal agar medium (BMM) produced melanized microsclerotia 4 days after plating, whereas the wild-type XJ592 and ΔVdCf2/VdCf2 strains produced comparable microsclerotia without melanin deposition following a similar incubation time (Fig. 2G). These results suggest that VdCf2 is involved in melanin homeostasis but not microsclerotium formation.
VdCf2 participated in pathogenicity of V. dahliae on cotton.
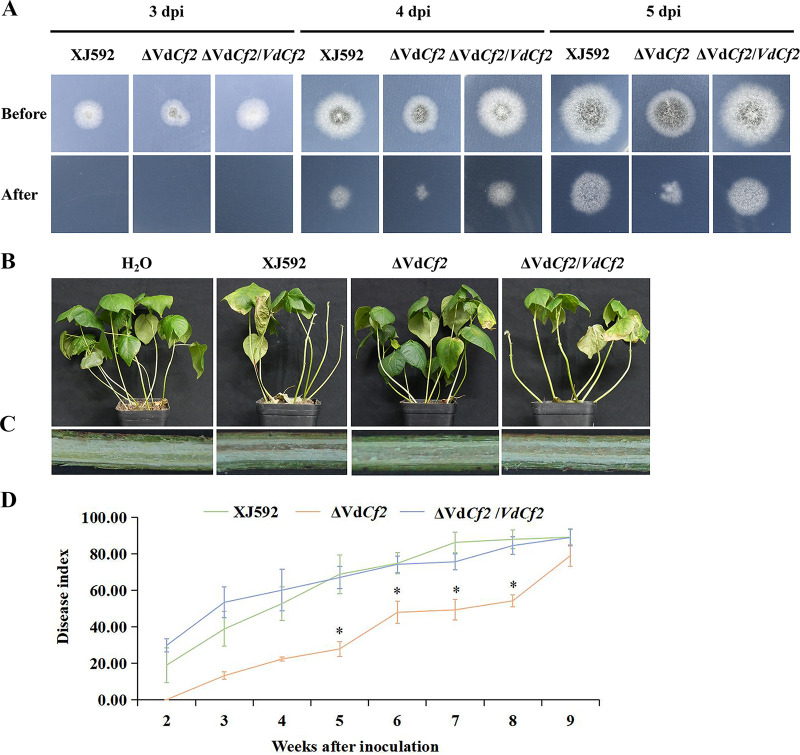
To assess the potential roles of VdCf2 during initial colonization of V. dahliae, the penetration ability of strains XJ592, ΔVdCf2, and ΔVdCf2/VdCf2 were examined by inoculation on MM plates covered with a cellophane membrane. When the cellophane membrane was removed at 4 or 5 days postinoculation, hyphae of strains XJ592 and ΔVdCf2/VdCf2 had penetrated the cellophane membrane and had grown into the medium (Fig. 3A). Similarly, strain ΔVdCf2 maintained the ability to penetrate cellophane membrane (Fig. 3A). This result indicates that VdCf2 is not required for penetration by V. dahliae.
FIG 3.
VdCf2 was required for full virulence on cotton. (A) VdCf2 was not involved in cellophane membrane penetration of V. dahliae. Strains XJ592, ΔVdCf2, and ΔVdCf2/VdCf2 were cultured on top of cellophane membrane overlaid onto MM plates for 3, 4, or 5 days. The cellophane membranes were removed, and all strains were cultured for additional 3 days. (B) Verticillium wilt symptoms on susceptible cotton plants (JM11) inoculated with the wild-type, ΔVdCf2, and ΔVdCf2/VdCf2 strains at 35 days postinoculation. (C) The vascular discoloration in cotton plants inoculated with the wild-type, ΔVdCf2, and ΔVdCf2/VdCf2 strains. (D) Disease index of cotton plants inoculated with the wild-type, ΔVdCf2, and ΔVdCf2/VdCf2 strains. Error bars represent standard errors calculated from three replicates, and asterisks represent the statistical significance (SNK test, P = 0.05).
To evaluate the role of VdCf2 in virulence, seedlings of susceptible cotton cultivar JM11 were inoculated with conidial suspensions of strain XJ592, ΔVdCf2, and ΔVdCf2/VdCf2 and were assessed. The plants inoculated with strains XJ592 and ΔVdCf2/VdCf2 developed chlorosis and defoliation at 35 days postinoculation (Fig. 3B). In contrast, the plants inoculated with strain ΔVdCf2 showed only mild foliar chlorosis and defoliation (Fig. 3B). Furthermore, the xylem in the plants inoculated with strain XJ592 or ΔVdCf2/VdCf2 were brown, whereas the vascular discoloration was incipient in the plants inoculated with strain ΔVdCf2 (Fig. 3C). In addition, the disease index was significantly reduced in the plants inoculated with strain ΔVdCf2 compared with those caused by the XJ592 and ΔVdCf2/VdCf2 strains at 5, 6, 7, and 8 weeks after inoculation (Fig. 3D). These results suggest that VdCf2 plays a role in V. dahliae pathogenicity and virulence.
VdCf2 regulates the transcription of potential pathogenicity-related genes.
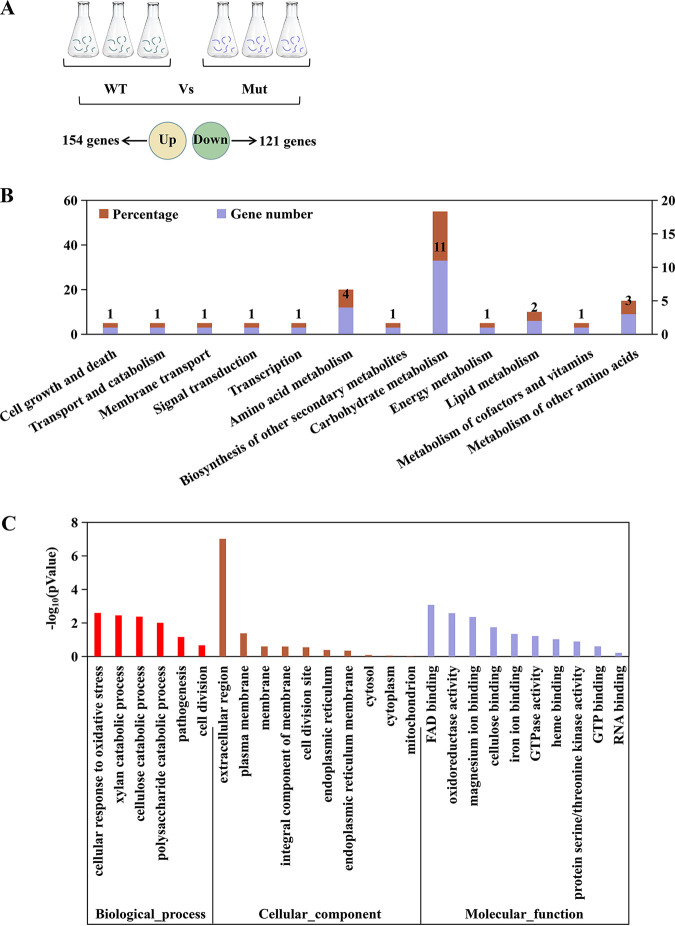
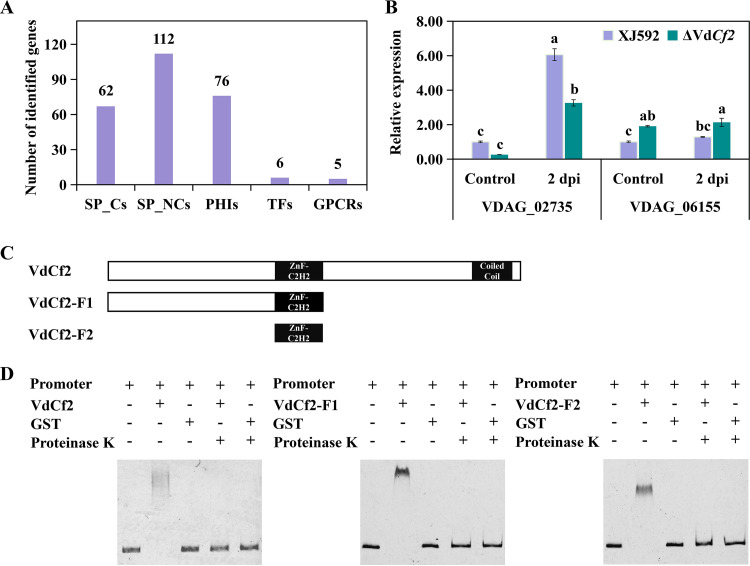
To study the regulatory functions of VdCf2, the transcriptomes between strains XJ592 and ΔVdCf2 were compared. Mycelia of strains XJ592 and ΔVdCf2 cultured in CDM were used for RNA sequencing (RNA-seq). A total of 275 differentially expressed genes (DEGs) were identified: 154 upregulated and 121 downregulated genes (Fig. 4A, Table S1; P < 0.05 with a log2 fold change of ≥ 1.0) in the wild-type compared to the ΔVdCf2 strain. KEGG pathway enrichment analysis showed that the upregulated genes were mainly enriched in carbohydrate and amino acid metabolism (Fig. 4B). Gene Ontology (GO) enrichment analysis revealed that the majority of DEGs were related to the genes encoding extracellular proteins and integral components of the membrane (Fig. 4C, Table S2). Among DEGs, there were 174 secreted proteins, comprising 62 classical secreted proteins and 112 nonclassical secreted proteins, 76 pathogen-host interaction (PHI) homolog proteins, including 33 verified to be associated with pathogenicity (reduced virulence or loss of pathogenicity in their knockout mutants), 6 TFs, and 5 G protein-coupled receptors (GPCRs) (Fig. 5A, Table S1). Seven DEGs (VDAG_02735, VDAG_06200, VDAG_02289, VDAG_10037, VDAG_09577, VDAG_07255, VDAG_09018) were randomly chosen to quantify the gene expression level with a reverse transcriptase quantitative (RT-qPCR) assay to verify the reliability of this transcriptome data. Results showed that their expressions were all upregulated in the XJ592 strain, consistent with the RNA-seq analysis (Fig. S3).
FIG 4.
KEGG and GO enrichment analysis of differentially expressed genes in the transcriptome analysis. (A) Sample preparation of RNA-seq and the number of differentially expressed genes between the wild-type strain and ΔVdCf2 strain. Three fungus blocks (6-mm diameter) were inoculated into the Czapek-Dox medium, and then the mycelia were collected at 8 days postinoculation. WT represents the XJ592 strain and Mut represents the ΔVdCf2 strain. The gene expression was analyzed in the mycelia of the wild-type XJ592 strain compared to that of strain ΔVdCf2. Differentially expressed genes were defined by a log2 fold change of ≥1.0 and P value of <0.05. Three repeats were performed in this transcriptome sequencing assay. (B) KEGG pathway enrichment analysis of upregulated differentially expressed genes in the transcriptome data. The percentage of genes represents the proportion of upregulated differentially expressed genes in this pathway. (C) GO enrichment analysis of all differentially expressed genes in RNA-seq analysis in the mycelia of strain XJ592 compared to that of strain ΔVdCf2.
FIG 5.
VdCf2 regulated the expression of potential pathogenicity-related genes. (A) Number of SP_Cs, SP_NCs, PHIs, TFs, and GPCRs in differentially expressed genes. SP_Cs, classical secretory proteins; SP_NCs, nonclassical secretory proteins; PHIs, pathogen-host interaction-related proteins; TFs, transcription factors; GPCRs, G protein-coupled receptors. (B) The expression level of VDAG_02735 and VDAG_06155 was determined by an RT-qPCR experiment. Conidial suspensions (107 conidia/mL) of the wild-type and ΔVdCf2 strains were cultured on the Czapek-Dox medium for 5 days before being treated with cotton root extracts for 2 days. The β-tubulin gene (VDAG_10074) was used as an internal reference. The expression level of each gene in the wild-type strain in the control was standardized as 1. Error bars represent standard errors calculated from three replicates, and different letters indicate the statistical significance of Tukey’s test at a P value of 0.05. (C) Schematic diagrams of VdCf2-F1 and VdCf2-F2 used in the EMSA. (D) VdCf2, VdCf2-F1, and VdCf2-F2 proteins bound to the promoter of the VDAG_02735 gene according to the EMSA.
VdCf2 regulated the expression of many genes encoding secreted proteins based on the comparative transcriptome analysis, including two previously reported secreted proteins, VdPevD1 (VDAG_02735) and VdPEL1 (VDAG_06155), which were proved to both be positive regulators in the pathogenicity of V. dahliae (15, 42) (Table S1). RNA-seq analysis showed that VdCf2 positively and negatively regulated the expression levels of VdPevD1 and VdPEL1, respectively (Table S1). However, after the fungal sample was treated with cotton root extracts for 2 days, the expression of VdPevD1 was upregulated in the wild type, but the expression of VdPEL1 did not differ significantly (Fig. 5B). To investigate whether VdCf2 regulates the expression of VdPevD1 by directly binding to its promoter, electrophoretic mobility shift assay (EMSA) was performed with purified glutathione S-transferase (GST)-tagged VdCf2 protein and the promoter of VdPevD1. When VdCf2 protein was coincubated with the VdPevD1 promoter, a shift band was observed (Fig. 5C and D), indicating an interaction between them. These results suggest that VdCf2 directly regulates the expression of VdPevD1 in mycelia and the sample treated with cotton root extracts.
To further investigate whether the C2H2-type zinc finger domains of VdCf2 directly bind the promoter region of VdPevD1, EMSA was performed with the truncated VdCf2 protein (VdCf2-F1 or VdCf2-F2) and the promoter of VdPevD1. Both VdCf2-F1 and VdCf2-F2 proteins could bind to the promoter region of VdPevD1 (Fig. 5C and D), suggesting that VdCf2 has a function in binding to the promoter region of VdPevD1 via its C2H2-type zinc finger domain.
VdCf2 regulates the expression of several PHI genes.
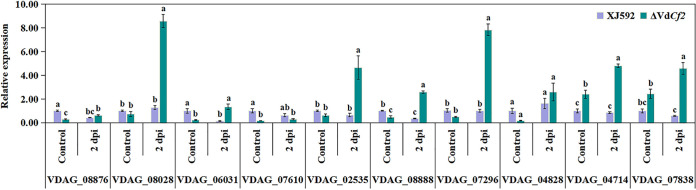
In this transcriptome analysis between the wild-type and ΔVdCf2 strains, VdCf2 regulates 76 PHI genes, including 33 associated with pathogenicity (Table 1). To identify whether these PHI genes associated with pathogenicity respond to cotton root extracts, 10 genes with the highest expression level in all PHI genes which positively regulated pathogenicity were selected for quantitative analysis in fungal samples treated with cotton root extracts for 2 days. These genes include eight upregulated (VDAG_08876, VDAG_08028, VDAG_06031, VDAG_07610, VDAG_02535, VDAG_08888, VDAG_07296, VDAG_04828) and two downregulated (VDAG_04714, VDAG_07838) differential expression genes. Quantitative results showed that the expression level of most PHI genes was significantly induced in strain ΔVdCf2 but not significantly different in strain XJ592 when the fungal sample was treated with cotton root extracts (Fig. 6). These results indicate that VdCf2 inhibits the expression of several PHI genes in fungal samples treated with cotton root extracts for 2 days. However, VdCf2 promoted the expression of a few PHI genes in the control, indicating that VdCf2 played different regulatory roles in fungal samples treated without and with cotton root extracts.
TABLE 1.
The 33 DEGs of PHI reduced virulence or loss of pathogenicity in the transcriptome data
| Gene ID | Phenotype of mutant in other pathogens | Pathogen species | Reference |
|---|---|---|---|
| VDAG_08721 | Reduced virulence | Fusarium graminearum | 72 |
| VDAG_08876 | Reduced virulence | Claviceps purpurea | 73 |
| VDAG_08028 | Reduced virulence | Zymoseptoria tritici | 74 |
| VDAG_06031 | Reduced virulence | M. oryzae | 75 |
| VDAG_07610 | Reduced virulence | M. oryzae | 76 |
| VDAG_02535 | Reduced virulence | Nectria haematococca | No data |
| VDAG_08888 | Reduced virulence | M. oryzae | 76 |
| VDAG_07296 | Reduced virulence | B. bassiana | 77 |
| VDAG_04828 | Reduced virulence | Candida albicans | 78 |
| VDAG_03487 | Reduced virulence | C. albicans | 78 |
| VDAG_10408 | Reduced virulence | Aspergillus fumigatus | 79 |
| VDAG_09443 | Loss of pathogenicity | M. oryzae | 80 |
| VDAG_06032 | Reduced virulence | B. cinerea | 81 |
| VDAG_02241 | Reduced virulence | M. oryzae | 82 |
| VDAG_02899 | Reduced virulence | M. oryzae | 75 |
| VDAG_08498 | Reduced virulence | M. oryzae | 83 |
| VDAG_07251 | Reduced virulence | Ustilago maydis | 84 |
| VDAG_07393 | Reduced virulence | M. oryzae | 82 |
| VDAG_08741 | Reduced virulence | Trichoderma virens | 85 |
| VDAG_09710 | Reduced virulence | M. oryzae | 76 |
| VDAG_04416 | Reduced virulence | T. virens | 85 |
| VDAG_07018 | Reduced virulence | Candida glabrata | 86 |
| VDAG_07929 | Reduced virulence | M. oryzae | 75 |
| VDAG_09581 | Reduced virulence | Grosmannia clavigera | 87 |
| VDAG_09343 | Reduced virulence | C. albicans | 78 |
| VDAG_10092 | Loss of pathogenicity | M. oryzae | 88 |
| VDAG_04729 | Loss of pathogenicity | Erwinia amylovora | 89 |
| VDAG_07983 | Reduced virulence | Histoplasma capsulatum | 90 |
| VDAG_04718 | Reduced virulence | Colletotrichum coccodes | 91 |
| VDAG_06155 | Reduced virulence | N. haematococca | 92 |
| VDAG_07195 | Reduced virulence | F. graminearum | 93 |
| VDAG_04714 | Reduced virulence | Cryptococcus neoformans | 94 |
| VDAG_07838 | Reduced virulence | M. oryzae | 95 |
FIG 6.
VdCf2 regulated the expression of several PHI genes. Sample preparation and data analysis are consistent with Fig. 5B. Error bars represent standard errors calculated from three replicates, and different letters indicate the statistical significance by Tukey’s test at a P value of 0.05.
VdCf2 influences the expression of other TFs.
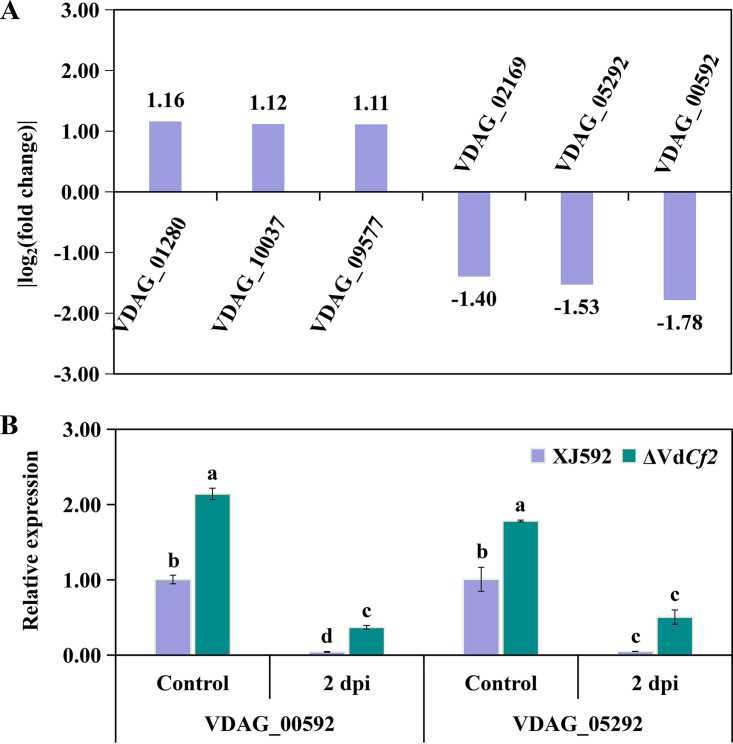
RNA-seq analysis indicated that VdCf2 affected expression of six TFs, including three upregulated (VDAG_01280, VDAG_10037, VDAG_09577) and three downregulated (VDAG_02169, VDAG_00592, VDAG_05292) genes (Fig. 7A, Table S1). VDAG_00592 and VDAG_05292, with the highest expression in all differentially expressed transcription factor genes, encoded meiosis-specific transcription factor NDT80-like protein and transcription factor vib-1, respectively. MoNdt80 played functions in the pathogenesis of M. oryzae (43), and transcription factor vib-1 in N. crassa participated in plant cell wall degradation (44). To further identify whether these differentially expressed TFs respond to cotton root extracts, the expression level of two TFs (VDAG_00592 and VDAG_05292) was quantified in fungal samples treated with cotton root extracts for 2 days. The expression level of VDAG_00592 and VDAG_05292 genes was significantly inhibited in both strains XJ592 and ΔVdCf2 when the fungal sample was treated with cotton root extracts (Fig. 7B). The extent of inhibition was, however, much greater in strain XJ592 than in the ΔVdCf2 strain (Fig. 7B). In the mycelia and the fungal sample treated with cotton root extracts for 2 days, the expression level of the VDAG_00592 gene was significantly inhibited in strain XJ592 compared to the ΔVdCf2 strain, indicating that VdCf2 curtails the expression of the VDAG_00592 gene in both states (Fig. 7B).
FIG 7.
VdCf2 regulated the expression of other TFs. (A) The value of the log2 fold change of genes encoding TFs regulated by VdCf2 according to transcriptome data. (B) The relative expression level of VDAG_00592 and VDAG_05292 genes in the wild-type and ΔVdCf2 strains. Conidial suspensions (107 conidia/mL) were cultured in the Czapek-Dox medium for 5 days before being treated with cotton root extracts for 2 days. The β-tubulin gene (VDAG_10074) was used as the internal reference. The expression level of each gene in the wild-type strain in the control was standardized as 1. Error bars represent standard errors calculated from three replicates, and different letters indicate the statistical significance of Tukey’s test at a P value of 0.05.
VdCf2 regulates the expression of the Vd276-280 gene cluster.
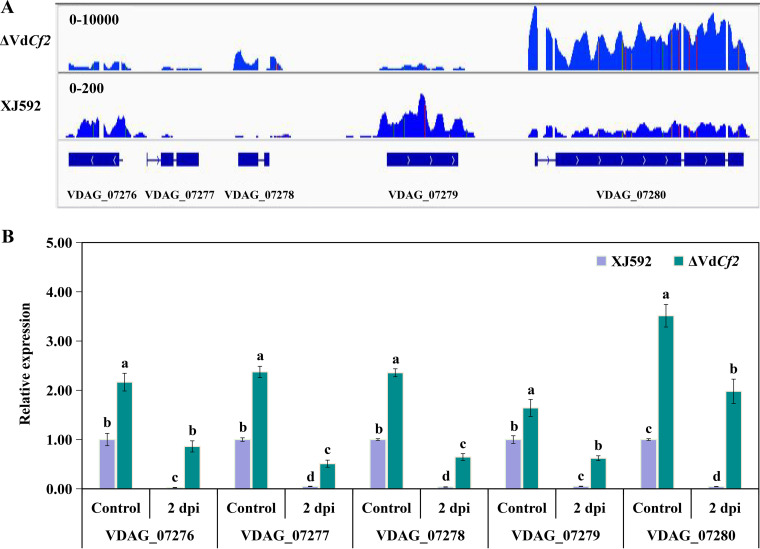
A previous study showed that the GATA-type transcription factor Csm1 inhibited the expression of the putative STC1 (sesquiterpene cyclase) gene cluster in Fusarium fujikuroi (45). This transcriptomic data showed that VdCf2 inhibited the expression of the Vd276-280 gene cluster comprising VDAG_07276 (cystinosin-like protein), VDAG_07277 (hypothetical protein), VDAG_07278 (hypothetical protein), VDAG_07279 (indoleamine 2,3-dioxygenase 1), and VDAG_07280 (alpha-galactosidase B like protein) (Fig. 8A, Fig. S4, Table 2). This gene cluster belongs to a portion of VDAG_07259 to VDAG_07280, predicted to be a secondary metabolism gene cluster (25, 46). To further identify whether the Vd276-280 gene cluster responds to cotton root extracts, the expression level of each gene in the Vd276-280 gene cluster was quantified in fungal samples treated with cotton root extracts for 2 days. The expression level of all genes in the Vd276-280 gene cluster was significantly inhibited in wild-type XJ592 and ΔVdCf2 strains in fungal samples treated with cotton root extracts for 2 days (Fig. 8B). In addition, the expression level of all genes in this gene cluster was also significantly inhibited in the wild-type XJ592 strain compared to those in the ΔVdCf2 strain (Fig. 8B). This suggests that this gene cluster responds to cotton root extracts and that VdCf2 regulates the expression of this gene cluster when the fungal sample was treated with cotton root extracts.
FIG 8.
VdCf2 regulated the expression of a putative secondary metabolism gene cluster. (A) Visualization of RNA-seq coverage of this putative secondary metabolism gene cluster in the wild-type and ΔVdCf2 strains. The blue lines indicate read coverage. (B) Gene expression level of the Vd276-280 gene cluster. The conidial suspensions (107 conidia/mL) were cultured in the Czapek-Dox medium for 5 days before being treated with cotton root extracts for 2 days. The β-tubulin gene (VDAG_10074) was used as the internal reference. The expression level of each gene in the wild-type strain in the control was standardized as 1. Error bars represent standard errors calculated from three replicates, and different letters indicate the statistical significance of Tukey’s test at a P value of 0.05.
TABLE 2.
The top 20 upregulated and 20 downregulated differentially expressed genes in the transcriptome data
| Gene ID | Regulation | Log2 fold change | Annotated gene function |
|---|---|---|---|
| VDAG_07673 | Up | Infinite | Gst19 |
| VDAG_04512 | Up | 5.27 | Uncharacterized protein |
| VDAG_05264 | Up | 4.36 | Hypothetical protein |
| VDAG_09533 | Up | 3.29 | Uncharacterized protein |
| VDAG_08876 | Up | 2.88 | Uncharacterized protein |
| VDAG_08028 | Up | 2.64 | NADH-cytochrome b5 reductase |
| VDAG_09486 | Up | 2.61 | Uncharacterized protein |
| VDAG_03485 | Up | 2.55 | Aflatoxin biosynthesis ketoreductase nor-1 |
| VDAG_03386 | Up | 2.49 | Uncharacterized protein |
| VDAG_09527 | Up | 2.41 | Hydrolase |
| VDAG_09526 | Up | 2.41 | FAD binding domain-containing protein |
| VDAG_06200 | Up | 2.39 | Uncharacterized protein |
| VDAG_03676 | Up | 2.36 | Uncharacterized protein |
| VDAG_06031 | Up | 2.35 | Uncharacterized protein |
| VDAG_00722 | Up | 2.29 | Uncharacterized protein |
| VDAG_09251 | Up | 2.26 | Oxalate decarboxylase oxdC |
| VDAG_09524 | Up | 2.23 | Hypothetical protein |
| VDAG_00625 | Up | 2.21 | Sphingoid long-chain base transporter RSB1 |
| VDAG_09018 | Up | 2.20 | Elongation factor G2 |
| VDAG_01819 | Up | 2.16 | Glutamate decarboxylase |
| VDAG_03806 | Down | –2.59 | Uncharacterized protein |
| VDAG_00794 | Down | –2.65 | Uncharacterized protein |
| VDAG_08603 | Down | –2.67 | Uncharacterized protein |
| VDAG_04714 | Down | –2.73 | Histidine kinase G7 |
| VDAG_07279 | Down | –3.08 | Indoleamine 2,3-dioxygenase 1 |
| VDAG_05839 | Down | –3.09 | Uncharacterized protein |
| VDAG_01292 | Down | –3.25 | Uncharacterized protein |
| VDAG_02862 | Down | –3.25 | Interferon-induced GTP-binding protein Mx |
| VDAG_06156 | Down | –3.35 | Uncharacterized protein |
| VDAG_05535 | Down | –3.35 | Uncharacterized protein |
| VDAG_06272 | Down | –3.70 | 6-Hydroxy-d-nicotine oxidase |
| VDAG_05104 | Down | –3.85 | Uncharacterized protein |
| VDAG_07595 | Down | –4.11 | Alpha-l-fucosidase |
| VDAG_08513 | Down | –4.26 | Epoxide hydrolase |
| VDAG_08707 | Down | –4.31 | Uncharacterized protein |
| VDAG_07838 | Down | –4.33 | Cerato-ulmin |
| VDAG_07277 | Down | –6.29 | Hypothetical protein |
| VDAG_07280 | Down | –7.58 | Alpha-galactosidase B like protein |
| VDAG_07276 | Down | –8.23 | Cystinosin-like protein |
| VDAG_07278 | Down | –8.83 | Hypothetical protein |
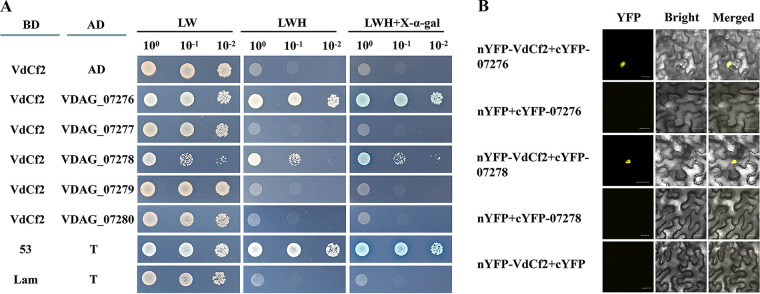
VdCf2 directly interacted with VDAG_07276 and VDAG_07278.
To identify proteins whose functions are related to VdCf2, the interaction targets of VdCf2 were screened based on a yeast two-hybrid (Y2H) cDNA library. We identified several proteins that potentially interact with VdCf2. The interaction was verified with a one-to-one Y2H assay. The yeast cells containing BD-VdCf2 and AD-VDAG_07276 constructs were able to grow on an synthetic dropout (SD)-Trp-Leu-His plate supplemented with 40 mg/L X-α-Gal (5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside) and turned blue (Fig. 9A). These results indicate that VdCf2 directly interacts with VDAG_07276 in the yeast two-hybrid system. To further identify whether other genes in this gene cluster interact with VdCf2, AD-VDAG_07277, AD-VDAG_07278, AD-VDAG_07279, and AD-VDAG_07280 constructs were transformed together with BD-VdCf2 into Y2H yeast competent cells, respectively. The yeast cells containing BD-VdCf2 and AD-VDAG_07278 constructs were able to grow on an SD-Trp-Leu-His plate, suggesting an interaction between VdCf2 and VDAG_07278 (Fig. 9A).
FIG 9.
VdCf2 interacted with VDAG_07276 and VDAG_07278. (A) VdCf2 interacted with VDAG_07276 and VDAG_07278 in a yeast two-hybrid system. For analysis of interaction, the full-length cDNA of VdCf2 was inserted into the pGBKT7 vector to generate the bait construct, and the full-length CDS of VDAG_07276 and VDAG_07278 was inserted into the pGADT7 vector to generate the prey construct. The Y2HGold yeast cells containing the bait construct and pGADT7 vector were used to detect self-activation. The Y2HGold yeast cells containing the bait and prey constructs were used to detect interaction. The Y2HGold yeast cells with 10-fold serial dilutions (100, 10−1, and 10−2) were simultaneously cultured on SD plates –LW (lacking Leu and Trp), –LWH (lacking Leu, Trp, and His), and –LWH (lacking Leu, Trp, and His) supplemented with 40 mg/L X-α-Gal. The Y2HGold yeast cells containing the pGBKT7-53 and pGADT7-T vectors were regarded as the positive control, and the Y2HGold yeast cells containing the pGBKT7-Lam and pGADT7-T vectors were regarded as the negative control. (B) VdCf2 interacted with VDAG_07276 and VDAG_07278 as determined by a bimolecular fluorescence complementation assay. The epidermal cells of N. benthamiana leaves coinfiltrated with nYFP and cYFP-07276, nYFP and cYFP-07278, and nYFP-VdCf2 and cYFP were negative controls. Scale bars = 20 μm.
To further determine the interaction relationship between VdCf2 and VDAG_07276 or VDAG_07278, nYFP-VdCf2 and cYFP-07276, nYFP-VdCf2 and cYFP-07278 were coexpressed in the epidermal cells of N. benthamiana leaves for observation of yellow fluorescent protein (YFP) signals. The tobacco leaves coinfiltrated with nYFP and cYFP-07276, nYFP and cYFP-07278, and nYFP-VdCf2 and cYFP were used as negative controls. The result of bimolecular fluorescence complementation (BiFC) showed that YFP signals were observed in the cell nucleus of N. benthamiana coinfiltrated with nYFP-VdCf2 and cYFP-07276 and with nYFP-VdCf2 and cYFP-07278, whereas no yellow fluorescence signals were observed in negative controls, indicating that there is an interaction relationship between VdCf2 and VDAG_07276, as well as between VdCf2 and VDAG_07278 (Fig. 9B).
DISCUSSION
VdCf2 belongs to TFs of the C2H2 type, which is the second largest type of TFs, following the Zn2Cys6 type in V. dahliae (23). Only a few TFs of the C2H2 type have been characterized, and the mechanisms of most C2H2-type TFs involved in pathogenicity remain undetermined (24, 32, 33). In this study, VdCf2 was found to promote pathogenicity to cotton probably by influencing fungus growth and the expression level of secreted protein-encoding genes.
In this study, the growth of strain ΔVdCf2 was reduced on PDA, MM, and WA media. Transcriptomic analysis showed that five GPCRs were significantly downregulated in the ΔVdCf2 strain—carbon/amino acid receptor (VDAG_05120), RGS_domain GPCR (VDAG_08194), and 3 PTH11-related GPCRs (VDAG_07929, VDAG_02899 and VDAG_06031). GPCRs are mainly responsible for transmitting extracellular signals to the intracellular region to trigger several signaling cascade reactions that are important for fungal development, including the cyclic AMP and mitogen-activated protein kinase pathways (47, 48). The decreased expression of genes encoding GPCRs in strain ΔVdCf2 may thus be responsible for the reduced mycelial growth observed on the different media.
The interplay between virulence and melanin production is complex (32, 49). Strains with reduced melanin production often show reduced virulence (33, 49, 50). In this study, however, even though melanin production increased in the ΔVdCf2 strain, its virulence on cotton seedlings was reduced. The role of VdCf2 in melanin production and virulence was consistent with that of VTA2, which encodes a transcription activator of adhesion (32). It is possible that VdCf2 and VTA2 act as intermediaries to couple virulence and melanin production. Transcription factor VdCmr1, located in a secondary metabolism cluster of genes, is a key regulator of the melanin biosynthesis pathway (51). The influence of VdCf2 and VTA2 on melanin formation may be related to VdCmr1.
Secreted proteins are vital for successful colonization of host plants by fungal pathogens (14, 52, 53). A fungal_trans domain-containing TF (VdFTF1) affects pathogenicity mainly by regulating the expression of those genes encoding secreted proteins (26). Many secreted proteins were differentially expressed in the wild-type strain from the ΔVdCf2 strain, including two virulence-related proteins, VdPevD1 and VdPEL1 (15, 42). Further RT-qPCR analysis verified that VdCf2 promoted the expression of VdPevD1 in the sample treated with cotton root extracts. VdPevD1 promotes pathogenicity by inhibiting antifungal activities of pathogenesis-related protein 5 (PR5)-like GhPR5 (42). VdCf2 probably promotes pathogenicity by enhancing the transcription of VdPevD1 to overcome antifungal activities of host plants via GhPR5 (42). VdPEL1 belongs to the pectate lyase family, which also facilitates pathogenicity in V. dahliae (15). However, the expression of VdPEL1 in the wild-type strain was not induced in the sample treated with cotton root extracts, indicating that VdPEL1 did not participate in this process.
VdFTF1 regulates the expression of genes encoding TFs during V. dahliae infection of cotton (26). Som1 and Vta3 controlled the expression of numerous TFs encoding genes when V. dahliae was cultured on a modified simulated xylem medium (SXM) (54). The present study indicated that VdCf2 regulated the expression of six putative TF-encoding genes, one of which was VDAG_00592. VDAG_00592 belonging to NDT80/PhoG TF was downregulated in the wild-type strain. In Candida parapsilosis, NDT80/PhoG acts as a repressor of virulence attributes (55). However, an Ndt80/PhoG domain-containing TF MoNDT80 in M. oryzae positively participated in pathogenicity to host rice (43). Whether NDT80/PhoG in V. dahliae also has a role in pathogenicity needs further investigation.
In the present study, VdCf2 repressed the expression of a putative secondary metabolite gene cluster. Similarly, the APSES TF Vst1 regulates the expression of seven secondary metabolism gene clusters at the early stages of microsclerotium biogenesis in V. dahliae (19). In this study, VdCf2 inhibits the expression of the Vd276-280 gene cluster, and in yeast two-hybrid and BiFC assays, VdCf2 interacted physically with VDAG_07276 and VDAG_07278, two members of Vd276-280 gene cluster. Thus, it is possible that VdCf2 regulates the expression of VDAG_07276 and VDAG_07278 by interacting with them.
In summary, VdCf2 is an important regulator of growth, melanin production, and virulence in V. dahliae. Transcriptomic analysis showed that VdCf2 curtailed the expression of virulence-related VdPevD1 (VDAG_02735) by directly binding its promoter region, which might explain the reduced virulence of strain ΔVdCf2. VdCf2 also regulates the expression of the Vd276-280 gene cluster and directly interacts with its two members. Further study is under way to research the function of this gene cluster.
MATERIALS AND METHODS
Fungal strains and culture conditions.
A V. dahliae strain (XJ592) isolated from cotton in XinJiang province, China, was used as the wild-type strain in this study. V. dahliae conidial suspensions (1 μL) of approximately 107 conidia/mL were placed on potato dextrose agar (PDA) medium, minimal medium (MM) (2 g/L of dextrose, 0.5 g/L of MgSO4·7 H2O, 0.5 g/L of KCl, 2 g/L of NaNO3, 0.01 g/L of FeSO4·7H2O, 2.6 mg/L of NH4Fe(SO4)2·12 H2O, 0.5 mg/L of CuSO4·5 H2O, 1 g/L of KH2PO4, 0.01 g/L of ZnSO4·7 H2O, 0.1 mg/L of Na2MoO4·2 H2O, 0.01 g/L of citrate, 0.1 mg/L of MnSO4·H2O, 0.1 g/L of H3BO3, 20 g/L of agar) and water agar (WA) and incubated at 25°C for 2 weeks to evaluate colony growth by measuring the diameter. For conidial production and collection, the wild-type, ΔVdCf2, and ΔVdCf2/VdCf2 strains were cultured in liquid Czapek-Dox medium (CDM) for 5 days before the culture was harvested and filtered through Miracloth (Millipore, Billerica, MA, USA). For the osmotic stress response assay, conidial suspensions of 1 μL (107 conidia/mL) of each strain were placed in the center of plates containing complete medium (CM) with the addition of 1.2 M sorbitol, 0.8 M KCl, or 0.4 M NaCl and incubated at 25°C for 2 weeks. For the oxidative stress response assay, conidial suspensions (107 conidia/mL) of the wild-type and ΔVdCf2 strains were coated on CM plates with a piece of circular filter paper (6-mm diameter) containing 1 μL 30% H2O2 and cultured for 48 h. Conidial suspensions from each strain were spread onto the BMM medium (56) overlaid on a piece of cellulose membrane and cultured for 4 days before microsclerotia were assessed and photographed. Conidial suspensions of 1 μL (107 conidia/mL) were cultured on MM covered with a piece of cellulose membrane for 3, 4, or 5 days before the cellulose membrane was removed to analyze the penetration ability. The colonies were photographed 3 days after the cellulose membranes’ removal. Cotton plants with two true leaves (Gossypium hirsutum cv. JIMIAN 11) were used for the virulence assay. Details of this assay are provided below.
Generation of gene deletion and complementation mutants.
The upstream and downstream flanking sequences of VdCf2 were amplified from XJ592 genomic DNA with primer pairs Cf2-AF/Cf2-AR and Cf2-BF/Cf2-BR, respectively. The hygromycin B resistance fragment was amplified from pA-HYG OSCAR vector with primer pair HYG-QF/HYG-QR. These three fragments were fused with the primer pair Cf2-AF/Cf2-BR via fusion PCR to acquire a gene deletion cassette. The fusion fragment was cloned into the pOSCAR vector digested with the restriction enzymes HindIII and EcoRI to generate the knockout plasmid. The resulting vector was transformed into the wild-type strain to generate the ΔVdCf2 strain. The ΔVdCf2 strain was confirmed by PCR with the primer pair Cf2-NF/Cf2-NR. The genomic sequence of VdCf2 was amplified with the primer pair Cf2-CF/Cf2-CR from XJ592 genomic DNA and then cloned into the pOSCAR-C vector digested with the restriction endonuclease XhoI. The resulting construct was then transformed into the ΔVdCf2 strain to generate the ΔVdCf2/VdCf2 strain.
Southern blotting was performed with DIG High Prime DNA labeling and detection starter kit I according to the manufacturer’s protocol (Roche, Switzerland). The genomic DNA of the XJ592 and ΔVdCf2 strains was digested by the restriction endonuclease BamHI. The probe for VdCf2 gene detection was PCR amplified with the primer pair NF/NR, and the probe for hygromycin B resistance gene detection was PCR amplified with the primer pair HYG-F/HYG-R. The primers used in this study are listed in Table 3.
TABLE 3.
List of the primers used in this study
| Primer name | Primer sequence (5′–3′)a | Application |
|---|---|---|
| Cf2-AFa | GATCCAAGCTCAAGCTAAGCTTCAAGCGAGGTAGAGGAGAGG | Amplification of upstream flanking sequence of VdCf2 |
| Cf2-AR | GATGCCGACCGGGAACCAGTTACAGGCACAGAGGTTCAGTT | |
| Cf2-BF | CAATATCAGTTGGGCTGCAGGGCAGCATCAAGGAAGGAGGA | Amplification of downstream flanking sequence of VdCf2 |
| Cf2-BR | GAATTAACGCCGAATTGAATTCGTGGAACATATTCGCCGTCAG | |
| HYG-QF | AACTGGTTCCCGGTCGGCATCTACTC | Amplification of hygromycin-resistant gene |
| HYG-QR | CCTGCAGCCCAACTGATATTGAAGGAGC | |
| Cf2-NF | GAGCCACCTTCTGACACATAGC | Detection of deletion mutant |
| Cf2-NR | GAAGCCTGCGTGACTCGTAAG | |
| Cf2-CF | TGTCTACTGCTGGCCTCGAGGTCCGTTGATGGATGTGGTGTT | Amplification of VdCf2 gene with native promoter |
| Cf2-CR | ATTAACGCCGAATTGAATTCTTGCCGTCCTCCTTCCTTGAT | |
| Cf2pBinF | ACCCCCGGGGTCGACGGATCCATGGCCTCGGAAAAGGGTTC | Vector construction for subcellular localization in tobacco |
| Cf2pBinR | TCTAGTTCATCTAGAGGATCCTGCCTTGGGCGTGAGGGCCT | |
| Cf2pF | AACCTCTAGAGGATCCGCCACCATGGCCTCGGAAAAGGGTTC | Vector construction for subcellular localization in V. dahliae |
| Cf2pR | GCAGCTTCTGCGAATTCTGCCTTGGGCGTGAGGGCCT | |
| Cf2-NF | GAGCCACCTTCTGACACATAGC | Amplification of VdCf2 gene probe in Southern blot |
| Cf2-NR | GAAGCCTGCGTGACTCGTAAG | |
| HYG-F | GCAGACAGGAACGAGGACAT | Amplification of hygromycin-resistant gene probe in Southern blot |
| HYG-R | GCTCCATACAAGCCAACCAC | |
| Cf2pGexF | CCGCGTGGATCCCCGGAATTCATGGCCTCGGAAAAGGGTTC | Vector construction for protein purification |
| Cf2pGexR | GATGCGGCCGCTCGAGTCGACTCATGCCTTGGGCGTGAGGG | |
| VdCf2-F1 | CCGCGTGGATCCCCGGAATTCATGAAGCACAAGTGCCCGTA | |
| VdCf2-R1 | GATGCGGCCGCTCGAGTCGACTCAACCCATGCTCGATCGCC | |
| 02735FamF | CTCGATGACAATGACGAC | Amplification of VDAG_02735 promoter probe in EMSA |
| 02735proR | TTTGATGTTGGGTGAGGTTG | |
| 02735-qF | CATTGCCGACTTCAACGTCC | Validation of the transcriptome data in RT-qPCR |
| 02735-qR | CAGGCTCAGAGCGAACTCAT | |
| 06200-qF | TCCAATGCGTGTCAAACAGG | |
| 06200-qR | TCGCATTTTCCCTCTGCGTA | |
| 02289-qF | ACATGTACGCAGACGAAGCA | |
| 02289-qR | GGGGTACCGTGACTTCTTGG | |
| 10037-qF | TCGTCGTTGTCTTCACTGGG | |
| 10037-qR | TGTGTACCGCTCATGGCTTT | |
| 09577-qF | TATCCGACGCCAGCAAACTT | |
| 09577-qR | ACGACCACCATCAGTCAACC | |
| 07255-qF | ATGGCCGACATCCTCATCAC | |
| 07255-qR | GAGGCTCGACTACGTCTGTG | |
| 09018-qF | GATGGACGCCATCATCGACT | |
| 09018-qR | CCCCCTTTTCTCCGAATGCT | |
| 06155qF | ATCAAGGGTGTCTTCGACGG | Validation of the expression level of secreted protein-encoding genes in RT-qPCR |
| 06155qR | CCAAACGTTCTCGATGGTGC | |
| 08876qF | ACCATATTGCTCGTAGGCGA | Validation of the expression level of PHI protein-encoding genes in RT-qPCR |
| 08876qR | ATGTCGCGGAGCATAGTCAC | |
| 08028qF | TCCCCACGTCACACTGATTG | |
| 08028qR | ACCGACCGGGAAAATCCTTC | |
| 06031qF | CAAGATCTCGGTCCTCGCTC | |
| 06031qR | GCACTGGAAAATGCAAGCCA | |
| 07610qF | CAGCAATTCAGTTCCGCCAC | |
| 07610qR | TGGATTCGGTGGATGGCAAA | |
| 02535qF | GCAAATACGACGCTCCGTTC | |
| 02535qR | GCCCGACTTGCTAGTGACTT | |
| 08888qF | CACTCTCACCTCCAGCAAGG | |
| 08888qR | CGTAAAACGCAGATGAGCCG | |
| 07296qF | GTGTCCCCGGAATGAGGAAA | |
| 07296qR | GCTCAAGCTCCAGATCCCTC | |
| 04828qF | CCGTACGGAGAGAATCTGGC | |
| 04828qR | CTGGCGCATCCGATCTTTTG | |
| 04714qF | AGCAAGAACTCAGCTCGCAT | |
| 04714qR | GGAAGGCGTCCAACTGTGTA | |
| 07838qF | GACCGCCAAGTCTACATCCC | |
| 07838qR | TTGTTGCAGAGGACACCCTG | |
| 00592qF | ATCGACGTGTTTCACCCACA | Validation of the expression level of transcription factors in RT-qPCR |
| 00592qR | TACGTGTTACAGCGGTTGCT | |
| 05292qF | ACACTGGTGGATGCTCTTCG | |
| 05292qR | TGTCGAGACTGGGTAGCTGA | |
| 07276qF | CAGGAAGGAAGAAGCCCGAT | Validation of the expression level of gene clusters in RT-qPCR |
| 07276qR | CTTGGCCTCTTCAGGCTTGG | |
| 07277qF | GACCACGTTATAGGCCCCTG | |
| 07277qR | ACATAACGGTCAACAGGCGT | |
| 07278qF | TCGTCGCGAAGCAGAGATAC | |
| 07278qR | TTGCGCTTCGACTTGGTCTT | |
| 07279qF | AGAGAACCCGACGATCCAGA | |
| 07279qR | TCCCATTCGAGTACGTTGCC | |
| 07280qF | CGTATCTGGAACGACTCGGG | |
| 07280qR | ACTGGACAACAGCATCAGGG | |
| β-tubulin-F | TTTCCAGATCACCCACTCC | Internal control in RT-qPCR |
| β-tubulin-R | ACGACCGAGAAGGTAGCC | |
| CF2-BKF | CCATGGAGGCCGAATTCACCATGGCCTCGGAAAAGGGTTC | Vector construction for yeast two-hybrid assay |
| CF2-BKR | CGCTGCAGGTCGACGGATCCTCATGCCTTGGGCGTGAGGG | |
| 07276-ADF | GCCATGGAGGCCAGTGAATTCACCATGGACACCAAG | |
| 07276-ADR | ATGCCCACCCGGGTGGAATTCTCACAGCAAAGCAAGAGTCA | |
| 07277-ADF | GCCATGGAGGCCAGTGAATTCACCATGGACATCATCGCGCTCCT | |
| 07277-ADR | ATGCCCACCCGGGTGGAATTCTCAGTCCCTGACATACTGGC | |
| 07278-ADF | GCCATGGAGGCCAGTGAATTCACCATGTCGTCGCGAAGCAGAGA | |
| 07278-ADR | ATGCCCACCCGGGTGGAATTCTTACAGGAGAGCAAGCTCCA | |
| 07279-ADF | GCCATGGAGGCCAGTGAATTCACCATGCAGACACCCGCTCCTGC | |
| 07279-ADR | ATGCCCACCCGGGTGGAATTCTCACGAACTCGTACCAATAT | |
| 07280-ADF | GCCATGGAGGCCAGTGAATTCACCATGGCCGACACTACTACGAA | |
| 07280-ADR | ATGCCCACCCGGGTGGAATTCTTATTTGTTCTTTCCCGTGA | |
| Cf2-NeYFP-F | TCTGAGGAGGATCTTCCTAGGATGGCCTCGGAAAAGGGTTC | Vector construction for bimolecular fluorescence complementation (BiFC) |
| Cf2-NeYFP-R | GGGAGGCCTGGATCGACTAGTTGCCTTGGGCGTGAGGGCCT | |
| 07276-CeYFP-F | CTAGTCGACTCTAGCCTCGAGATGGACACCAAGGACCAGCAAC | |
| 07276-CeYFP-R | ATCGTATGGGTACATCCTAGGCAGCAAAGCAAGAGTCAGAT | |
| 07278-CeYFP-F | CTAGTCGACTCTAGCCTCGAGATGTCGTCGCGAAGCAGAGA | |
| 07278-CeYFP-R | ATCGTATGGGTACATCCTAGGCAGGAGAGCAAGCTCCAGAT |
Bold letters in the primer sequences represent the recombination sequence.
RNA isolation and RT-qPCR assays.
For quantitative analysis of potential pathogenicity-related genes, the wild-type and ΔVdCf2 strains were cultured in the CDM for 5 days before treatment with cotton root extracts for another 2 days. Total RNA was extracted using the SV total RNA isolation system (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The cDNA was synthesized with the PrimeScript RT reaction system (TaKaRa, Tokyo, Japan). RT-qPCR was performed with UltraSYBR mixture (CWBIO, Beijing, China) using a LightCycler 480 II device (Roche, Basel, Switzerland). The β-tubulin gene (VDAG_10074) was used as an internal reference (57). The relative expression level of all genes was calculated with the 2–ΔΔCT method (58). Three replicates were used in all experiments. The primers used are listed in Table 3.
Virulence assays.
Susceptible cotton cultivar JM11 plants with two true leaves were used in virulence assays with the quantitative root dip inoculation method (59). Conidia of the XJ592, ΔVdCf2, and ΔVdCf2/VdCf2 strains were collected by filtration and centrifugation and then resuspended in sterile water to a concentration of approximately 107 conidia/mL. Five to eight cotton plants were planted in a pot, which was placed within a petri dish cover and cultured at 25°C under a 16/8-h light-dark photoperiod in a greenhouse. Three pots of cotton plants were prepared for each strain. Conidial suspensions of 35 mL were added into the petri dish cover to inoculate the cotton roots. The control group was dipped in sterile water. Each pot of all treatments was randomly assigned to specific locations in the greenhouse. Wilt development of cotton plants was assessed based on a five-grade infection scale: 0, no symptoms on leaves; 1, one or two cotyledon leaves showing symptoms; 2, a single true leaf showing symptoms; 3, more than two leaves showing symptoms; and 4, plant dead (50). The stems of the cotton plants were longitudinally cut and photographed.
Subcellular localization.
The full-length coding DNA sequence (CDS) of VdCf2 without the termination codon, amplified with primer pair Cf2pF/Cf2pR, was inserted into pNEO binary vector digested with the restriction endonuclease BamHI and EcoRI. The successful construct pNEO-VdCf2-GFP was transformed into Agrobacterium tumefaciens EHA105. The agrobacteria containing pNEO-VdCf2-GFP were cocultured with the conidial suspensions of strain ΔVdCf2 to generate the VdCf2-GFP strain. The GFP signal was visualized with a confocal laser scanning microscope (Olympus Corporation, Tokyo, Japan).
The full-length CDS of VdCf2 without the termination codon was amplified with the primer pair Cf2pBinF/Cf2pBinR and then cloned into the pBin vector digested with the restriction endonuclease BamHI to generate the construct pBin-VdCf2-GFP. The resulting construct was transformed into A. tumefaciens GV3101. The agrobacteria containing pBin-VdCf2-GFP or pBin-GFP were injected into the epidermal cells of 4-week-old Nicotiana benthamiana leaves. Infiltrated tobacco seedlings were grown at 25°C under a 16/8-h light-dark photoperiod in a greenhouse for another 2 to 3 days before assessment. The GFP signals in tobacco leaves were observed and photographed under a laser scanning confocal microscope (Olympus Corporation, Tokyo, Japan).
Protein extraction and Western blot analysis.
Total protein was extracted from N. benthamiana leaves using the plant protein extraction kit (Solarbio, Beijing, China) according to the manufacturer’s instructions. GFP and VdCf2-GFP fusion proteins were detected with the anti-GFP antibody (Beyotime, Shanghai, China).
Bimolecular fluorescence complementation (BiFC) assay.
The full-length CDS of VDAG_07276 and VDAG_07278 was cloned into the pER8-CeYFP vector digested with the restriction endonucleases XhoI and AvrII, respectively. The full-length cDNA of VdCf2 was inserted into the pER8-NeYFP vector digested with AvrII and SpeI. The resulting constructs were transformed into A. tumefaciens GV3101 (Tsingke, China). The agrobacteria harboring the combinations nYFP-VdCf2 and cYFP-07276, nYFP-VdCf2 and cYFP-07278, nYFP and cYFP-07276, nYFP and cYFP-07278, and nYFP-VdCf2 and cYFP together with P19 were injected into the epidermal cells of N. benthamiana leaves. After 2 to 3 days of infiltration, injected leaves were sampled and observed for YFP signals under a laser scanning confocal microscope (Olympus Corporation, Tokyo, Japan).
Electrophoretic mobility shift assay (EMSA).
The full coding region of VdCf2 amplified with primer pair Cf2pGexF/Cf2pGexR was inserted into the pGEX-4T-1 vector digested with the restriction endonucleases EcoRI and SalI to generate the prokaryotic expression construct pGEX-VdCf2. The successful construct was transformed into the Escherichia coli BL21(DE3) strain. VdCf2 protein was purified via the GST-tag protein purification kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. The EMSA experiment was carried out according to the EMSA binding buffer kit (Beyotime) instructions. The 6-carboxyfluorescein (FAM)-labeled probe was amplified with the primer pair 02735FamF/02735proR.
Yeast two-hybrid (Y2H) assay.
The full coding region of VdCf2 amplified with the primer pair CF2-BKF/CF2-BKR was inserted into the pGBKT7 vector digested with the restriction endonucleases EcoRI and SalI to generate the bait construct. The full-length cDNA of VDAG_07276 amplified with primer pair 07276-ADF/07276-ADR was inserted into the pGADT7 vector digested with the restriction endonuclease EcoRI to generate the prey construct. Generation of prey constructs for other candidate proteins was similar to that for VDAG_07276. The bait and prey constructs were cotransformed into Y2HGold yeast cells. The yeast cells containing bait and prey constructs were cultured on SD base medium without Leu, Trp, and His supplemented with 40 mg/L X-α-Gal to determine their interaction. The yeast cells harboring the bait construct and pGADT7 vector were cultured on SD base medium without Leu, Trp, and His to test self-activation. The yeast cells containing the pGBKT7-53 and pGADT7-T vectors and pGBKT7-Lam and pGADT7-T vectors were regarded as the positive and negative controls, respectively.
RNA-seq analysis.
Three fungal mycelial plugs (6-mm diameter) of the wild-type and ΔVdCf2 strains were inoculated into 90 mL liquid CDM and cultured at 150 rpm for 8 days. Fungal mycelia were collected with Miracloth and then rapidly quick-frozen in liquid nitrogen before being delivered to OE Biotech Co., Ltd. (Shanghai, China). Total RNA was extracted with the mirVana microRNA (miRNA) isolation kit (Ambion, Austin, TX, USA) in accordance with the manufacturer’s protocol. RNA-seq was performed on the Illumina HiSeq X Ten sequencing platform, and 150-bp paired-end reads were generated.
Raw data were trimmed and filtered using Trimmomatic to remove the low-quality reads and ploy-N (unreliable sequences) (60). Clean reads were aligned to the reference genome of V. dahliae strain VdLs.17 with HISAT2, and the fragments per kilobase per million (FPKM) of each gene was estimated with Cufflinks (61–63). The DESeq (2012) R package functions estimateSizeFactors and nbinomTest identified differentially expressed genes (DEGs) in the mycelia between the wild-type and ΔVdCf2 strains (64). A P value of <0.05 and fold change of >2 were set as the threshold for DEG identification. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses, which were used to annotate gene functions, were conducted by using R based on the hypergeometric distribution (65). Secreted proteins were predicted based on SignalP 5.0, TMHMM 2.0, and SecretomeP 2.0 (66–68). The prediction of PHI homologs was performed based on the PHI database (69). TFs were identified based on the Fungal Transcription Factor Database (FTFD) and HMMER online website (23). All G protein-coupled receptors (GPCRs) were identified based on a previous analysis (70). The Vd276-280 gene cluster (VDAG_07276, VDAG_07277, VDAG_07278, VDAG_07279, and VDAG_07280) is part of a secondary metabolism gene cluster from VDAG_07259 to VDAG_07280, which was identified based on Secondary Metabolite Unknown Region Finder (25, 46). Visualization of RNA-seq coverage of the Vd276-280 gene cluster in the wild-type and ΔVdCf2 strains was completed using Integrative Genomics Viewer (71).
Statistical analyses.
Statistical analyses were performed using SPSS Statistics 22.0. One-way analysis of variance (ANOVA) was applied, and then the Student-Newman-Keuls (SNK) test with a P value of 0.05 was conducted to determine significant differences among three treatments. When more groups were compared, Tukey’s test with a P value of 0.05 was used to analyze significant differences. The pairwise comparison was conducted using Student’s t test with a P value of 0.05 based on RStudio 1.1.463.
Data availability.
The RNA-seq raw reads were submitted to the NCBI SRA database under BioProject number PRJNA797877 and BioSample numbers SAMN25039392 (XJ592) and SAMN25039393 (ΔVdCf2).
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2018YFE0112500), the Natural Science Basic Research Program of Shaanxi (2021JQ-146), and the Chinese Universities Scientific Fund (2452021136).
We thank Hui-Shan Guo from the Institute of Microbiology, Chinese Academy of Sciences, for providing the pNEO binary vector. We also thank Si-Wei Zhang from Northwest A&F University for providing the pER8-CeYFP and pER8-NeYFP vectors.
X.H. and W.S. designed the research. T.L. and Y.C. mainly contributed to the all experiments. J.C., K.V.S., and X.X. assisted with specific experiments. T.L. and J.Q. prepared the manuscript, and X.H., M.K.M., J.C., K.V.S., and X.X. revised the manuscript. None of the authors have conflicts of interest with this manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Xiaoping Hu, Email: xphu@nwsuaf.edu.cn.
Ning-Yi Zhou, Shanghai Jiao Tong University.
REFERENCES
- 1.Agrios GN. 1997. Plant pathology, 4th ed. Academic Press, San Diego, CA. [Google Scholar]
- 2.Fradin EF, Thomma BPHJ. 2006. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol 7:71–86. 10.1111/j.1364-3703.2006.00323.x. [DOI] [PubMed] [Google Scholar]
- 3.Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV. 2009. Diversity, pathogenicity, and management of Verticillium species. Annu Rev Phytopathol 47:39–62. 10.1146/annurev-phyto-080508-081748. [DOI] [PubMed] [Google Scholar]
- 4.Lu WJ, Liu YJ, Zhu HQ, Shang WJ, Yang JR, Hu XP. 2013. Verticillium wilt of redbud in China caused by Verticillium dahliae. Plant Dis 97:1513. 10.1094/PDIS-08-12-0804-PDN. [DOI] [PubMed] [Google Scholar]
- 5.Inderbitzin P, Subbarao KV. 2014. Verticillium systematics and evolution: how confusion impedes Verticillium wilt management and how to resolve it. Phytopathology 104:564–574. 10.1094/PHYTO-11-13-0315-IA. [DOI] [PubMed] [Google Scholar]
- 6.Wilhelm S. 1955. Longevity of the Verticillium wilt fungus in the laboratory and field. Phytopathology 45:180–181. [Google Scholar]
- 7.Vallad GE, Subbarao KV. 2008. Colonization of resistant and susceptible lettuce cultivars by a green fluorescent protein-tagged isolate of Verticillium dahliae. Phytopathology 98:871–885. 10.1094/PHYTO-98-8-0871. [DOI] [PubMed] [Google Scholar]
- 8.Fitzell R, Evans G, Fahy PC. 1980. Studies on the colonization of plant roots by Verticillium dahliae Klebahn with use of immunofluorescent staining. Aust J Bot 28:357–368. 10.1071/BT9800357. [DOI] [Google Scholar]
- 9.Klimes A, Dobinson KF, Thomma BPHJ, Klosterman SJ. 2015. Genomics spurs rapid advances in our understanding of the biology of vascular wilt pathogens in the genus Verticillium. Annu Rev Phytopathol 53:181–198. 10.1146/annurev-phyto-080614-120224. [DOI] [PubMed] [Google Scholar]
- 10.Yadeta KA, Thomma BPHJ. 2013. The xylem as battleground for plant hosts and vascular wilt pathogens. Front Plant Sci 4:97. 10.3389/fpls.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bu BW, Qiu DW, Zeng HM, Guo LH, Yuan JJ, Yang XF. 2014. A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Rep 33:461–470. 10.1007/s00299-013-1546-7. [DOI] [PubMed] [Google Scholar]
- 12.Liang YB, Li Z, Zhang Y, Meng FL, Qiu DW, Zeng HM, Li GY, Yang XF. 2021. Nbnrp1 mediates Verticillium dahliae effector PevD1-triggered defense responses by regulating sesquiterpenoid phytoalexins biosynthesis pathway in Nicotiana benthamiana. Gene 768:145280. 10.1016/j.gene.2020.145280. [DOI] [PubMed] [Google Scholar]
- 13.Gui YJ, Chen JY, Zhang DD, Li NY, Li TG, Zhang WQ, Wang XY, Short DPG, Li L, Guo W, Kong ZQ, Bao YM, Subbarao KV, Dai XF. 2017. Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ Microbiol 19:1914–1932. 10.1111/1462-2920.13695. [DOI] [PubMed] [Google Scholar]
- 14.Qin J, Wang KL, Sun LF, Xing HY, Wang S, Li L, Chen S, Guo HS, Zhang J. 2018. The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. Elife 14:e34902. 10.7554/eLife.34902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang YK, Zhang Y, Li BB, Yang XF, Dong YJ, Qiu DW. 2018. A Verticillium dahliae pectate lyase induces plant immune responses and contributes to virulence. Front Plant Sci 9:1271. 10.3389/fpls.2018.01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzima AK, Paplomatas EJ, Tsitsigiannis DI, Kang S. 2012. The G protein β subunit controls virulence and multiple growth- and development-related traits in Verticillium dahliae. Fungal Genet Biol 49:271–283. 10.1016/j.fgb.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Feng ZD, Tian J, Han LB, Geng Y, Sun J, Kong ZS. 2018. The Myosin5-mediated actomyosin motility system is required for Verticillium pathogenesis of cotton. Environ Microbiol 20:1607–1621. 10.1111/1462-2920.14101. [DOI] [PubMed] [Google Scholar]
- 18.Zhao YL, Zhou TT, Guo HS. 2016. Hyphopodium-specific VdNoxB/VdPls1-dependent ROS-Ca2+ signaling is required for plant infection by Verticillium dahliae. PLoS Pathog 12:e1005793. 10.1371/journal.ppat.1005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarmiento-Villamil JL, Garcia-Pedrajas NE, Baeza-Montanez L, Garcia-Pedrajas MD. 2018. The APSES transcription factor Vst1 is a key regulator of development in microsclerotium- and resting mycelium-producing Verticillium species. Mol Plant Pathol 19:59–76. 10.1111/mpp.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang C, Li TY, Klosterman SJ, Tian CM, Wang YL. 2020. The bZIP transcription factor VdAtf1 regulates virulence by mediating nitrogen metabolism in Verticillium dahliae. New Phytol 226:1461–1479. 10.1111/nph.16481. [DOI] [PubMed] [Google Scholar]
- 21.Shi YH, Mosser DD, Morimoto RI. 1998. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev 12:654–666. 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva LP, Horta MAC, Goldman GH. 2021. Genetic interactions between Aspergillus fumigatus basic leucine zipper (bZIP) transcription factors AtfA, AtfB, AtfC, and AtfD. Front Fungal Biol 2. 10.3389/ffunb.2021.632048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Park J, Jang S, Kim S, Kong S, Choi J, Ahn K, Kim J, Lee S, Kim S, Park B, Jung K, Kim S, Kang S, Lee YH. 2008. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 24:1024–1025. 10.1093/bioinformatics/btn058. [DOI] [PubMed] [Google Scholar]
- 24.Xiong DG, Wang YL, Tang C, Fang YL, Zou JY, Tian CM. 2015. VdCrz1 is involved in microsclerotia formation and required for full virulence in Verticillium dahliae. Fungal Genet Biol 82:201–212. 10.1016/j.fgb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 25.Xiong DG, Wang YL, Tian LY, Tian CM. 2016. MADS-Box transcription factor VdMcm1 regulates conidiation, microsclerotia formation, pathogenicity, and secondary metabolism of Verticillium dahliae. Front Microbiol 7. 10.3389/fmicb.2016.01192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang WQ, Gui YJ, Short DPG, Li TG, Zhang DD, Zhou L, Liu C, Bao YM, Subbarao KV, Chen JY, Dai XF. 2018. Verticillium dahliae transcription factor VdFTF1 regulates the expression of multiple secreted virulence factors and is required for full virulence in cotton. Mol Plant Pathol 19:841–857. 10.1111/mpp.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi ZQ, Wang Q, Dou XY, Wang W, Zhao Q, Lv RL, Zhang HF, Zheng XB, Wang P, Zhang ZG. 2012. MoSwi6, an APSES family transcription factor, interacts with MoMps1 and is required for hyphal and conidial morphogenesis, appressorial function and pathogenicity of Magnaporthe oryzae. Mol Plant Pathol 13:677–689. 10.1111/j.1364-3703.2011.00779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu YX, Yin ZY, Xu LS, Feng H, Huang LL. 2018. VmPacC is required for acidification and virulence in Valsa mali. Front Microbiol 9:1981. 10.3389/fmicb.2018.01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding ZJ, Xu TW, Zhu WJ, Li LJ, Fu QY. 2020. A MADS-box transcription factor FoRlm1 regulates aerial hyphal growth, oxidative stress, cell wall biosynthesis and virulence in Fusarium oxysporum f. sp. cubense. Fungal Biol 124:183–193. 10.1016/j.funbio.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Zhang ZQ, Li H, Qin GZ, He C, Li BQ, Tian SP. 2016. The MADS-Box transcription factor Bcmads1 is required for growth, sclerotia production and pathogenicity of Botrytis cinerea. Sci Rep 6:33901. 10.1038/srep33901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo M, Chen Y, Du Y, Dong YH, Guo W, Zhai S, Zhang HF, Dong SM, Zhang ZG, Wang YC, Wang P, Zheng XB. 2011. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog 7:e1001302. 10.1371/journal.ppat.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran VT, Braus-Stromeyer SA, Kusch H, Reusche M, Kaever A, Kühn A, Valerius O, Landesfeind M, Aßhauer K, Tech M, Hoff K, Pena-Centeno T, Stanke M, Lipka V, Braus GH. 2014. Verticillium transcription activator of adhesion Vta2 suppresses microsclerotia formation and is required for systemic infection of plant roots. New Phytol 202:565–581. 10.1111/nph.12671. [DOI] [PubMed] [Google Scholar]
- 33.Tian LY, Yu J, Wang YL, Tian CM. 2017. The C2H2 transcription factor VdMsn2 controls hyphal growth, microsclerotia formation, and virulence of Verticillium dahliae. Fungal Biol 121:1001–1010. 10.1016/j.funbio.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Hsu T, Gogos JA, Kirsh SA, Kafatos FC. 1992. Multiple zinc finger forms resulting from developmentally regulated alternative splicing of a transcription factor gene. Science 257:1946–1950. 10.1126/science.1411512. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka KK, Bryantsev AL, Cripps RM. 2008. Myocyte enhancer factor 2 and chorion factor 2 collaborate in activation of the myogenic program in Drosophila. Mol Cell Biol 28:1616–1629. 10.1128/MCB.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gajewski KM, Schulz RA. 2010. CF2 represses actin 88F gene expression and maintains filament balance during indirect flight muscle development in Drosophila. PLoS One 5:e10713. 10.1371/journal.pone.0010713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arredondo JJ, Vivar J, Laine-Menéndez S, Martínez-Morentin L, Cervera M. 2017. CF2 transcription factor is involved in the regulation of Mef2 RNA levels, nuclei number and muscle fiber size. PLoS One 12:e0179194. 10.1371/journal.pone.0179194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawke MA, Lazarovits G. 1995. The role of melanin in the survival of microsclerotia of Verticillium dahliae. Phytoparasitica 23:54. [Google Scholar]
- 39.Butler MJ, Day AW. 1998. Fungal melanins: a review. Can J Microbiol 44:1115–1136. 10.1139/w98-119. [DOI] [Google Scholar]
- 40.Henson JM, Butler MJ, Day AW. 1999. The dark side of the mycelium: melanins of phytopathogenic fungi. Annu Rev Phytopathol 37:447–471. 10.1146/annurev.phyto.37.1.447. [DOI] [PubMed] [Google Scholar]
- 41.Fang YL, Klosterman SJ, Tian CM, Wang YL. 2019. Insights into VdCmr1-mediated protection against high temperature stress and UV irradiation in Verticillium dahliae. Environ Microbiol 21:2977–2996. 10.1111/1462-2920.14695. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Gao YH, Liang YB, Dong YJ, Yang XF, Qiu DW. 2019. Verticillium dahliae PevD1, an Alt a 1-like protein, targets cotton PR5-like protein and promotes fungal infection. J Exp Bot 70:613–626. 10.1093/jxb/ery351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhatt DN, Ansari S, Kumar A, Ghosh S, Narula A, Datta A. 2020. Magnaporthe oryzae MoNdt80 is a transcriptional regulator of GlcNAc catabolic pathway involved in pathogenesis. Microbiol Res 239:126550. 10.1016/j.micres.2020.126550. [DOI] [PubMed] [Google Scholar]
- 44.Xiong Y, Sun JP, Glass NL. 2014. VIB1, a link between glucose signaling and carbon catabolite repression, is essential for plant cell wall degradation by Neurospora crassa. PLoS Genet 10:e1004500. 10.1371/journal.pgen.1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niehaus EM, Schumacher J, Burkhardt I, Rabe P, Spitzer E, Münsterkötter M, Güldener U, Sieber CMK, Dickschat JS, Tudzynski B. 2017. The GATA-Type transcription factor Csm1 regulates conidiation and secondary metabolism in Fusarium fujikuroi. Front Microbiol 8:1175. 10.3389/fmicb.2017.01175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. 2010. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47:736–741. 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maller JL. 2003. Signal transduction. Fishing at the cell surface. Science 300:594–595. 10.1126/science.1083725. [DOI] [PubMed] [Google Scholar]
- 48.Servin JA, Campbell AJ, Borkovich KA. 2012. G protein signaling components in filamentous fungal genomes, p 21–38. In Witzany G (ed), Biocommunication of fungi. Springer, Amsterdam, the Netherlands: ). [Google Scholar]
- 49.Wang YL, Tian LY, Xiong DG, Klosterman SJ, Xiao SX, Tian CM. 2016. The mitogen-activated protein kinase gene, VdHog1, regulates osmotic stress response, microsclerotia formation and virulence in Verticillium dahliae. Fungal Genet Biol 88:13–23. 10.1016/j.fgb.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Fan R, Klosterman SJ, Wang CH, Subbarao KV, Xu XM, Shang WJ, Hu XP. 2017. Vayg1 is required for microsclerotium formation and melanin production in Verticillium dahliae. Fungal Genet Biol 98:1–11. 10.1016/j.fgb.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Wang YL, Hu XP, Fang YL, Anchieta A, Goldman PH, Hernandez G, Klosterman SJ. 2018. Transcription factor VdCmr1 is required for pigment production, protection from UV irradiation, and regulates expression of melanin biosynthetic genes in Verticillium dahliae. Microbiology (Reading) 164:685–696. 10.1099/mic.0.000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rep M. 2005. Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiol Lett 253:19–27. 10.1016/j.femsle.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 53.de Sain M, Rep M. 2015. The role of pathogen-secreted proteins in fungal vascular wilt diseases. Int J Mol Sci 16:23970–23993. 10.3390/ijms161023970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bui TT, Harting R, Braus-Stromeyer SA, Tran VT, Leonard M, Höfer A, Abelmann A, Bakti F, Valerius O, Schlüter R, Stanley CE, Ambrósio A, Braus GH. 2019. Verticillium dahliae transcription factors Som1 and Vta3 control microsclerotia formation and sequential steps of plant root penetration and colonisation to induce disease. New Phytol 221:2138–2159. 10.1111/nph.15514. [DOI] [PubMed] [Google Scholar]
- 55.Branco J, Martins-Cruz C, Rodrigues L, Silva RM, Araújo-Gomes N, Gonçalves T, Miranda IM, Rodrigues AG. 2021. The transcription factor Ndt80 is a repressor of Candida parapsilosis virulence attributes. Virulence 12:601–614. 10.1080/21505594.2021.1878743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu XP, Bai YW, Chen T, Hu DF, Yang JR, Xu XM. 2013. An optimized method for in vitro production of Verticillium dahliae microsclerotia. Eur J Plant Pathol 136:225–229. 10.1007/s10658-013-0170-2. [DOI] [Google Scholar]
- 57.Hu DF, Wang CS, Tao F, Cui Q, Xu XM, Shang WJ, Hu XP. 2014. Whole genome wide expression profiles on germination of Verticillium dahliae microsclerotia. PLoS One 9:e100046. 10.1371/journal.pone.0100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 59.Zhu HQ, Fen ZL, Li ZF, Zhao LH, Shi YQ. 2010. Vermiculite sand bottomless paper port quantitative dip in microbial method appraisal cotton varieties (strains) anti-Verticillium wilt. China Cotton 37:15–17. [Google Scholar]
- 60.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. 2011. Improving RNA-seq expression estimates by correcting for fragment bias. Genome Biol 12:R22. 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S, Huber W. 2012. Differential expression of RNA-Seq data at the gene level - the DESeq package. Heidelberg: European Molecular Biology Laboratory. https://www.semanticscholar.org/paper/Differential-expression-of-RNA-Seq-data-at-the-gene-Anders-Huber/81c42bdf29edd0035b237375e2270f5a31762147. [Google Scholar]
- 65.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. 2007. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484. 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 67.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. 2004. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 17:349–356. 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 68.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 69.Winnenburg R, Urban M, Beacham A, Baldwin TK, Holland S, Lindeberg M, Hansen H, Rawlings C, Hammond-Kosack KE, Köhler J. 2007. PHI-base update: additions to the pathogen-host interaction database. Nucleic Acids Res 36:D572–D576. 10.1093/nar/gkm858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng HX, Zhou L, Dou TH, Han XT, Cai YY, Zhan XY, Tang C, Huang J, Wu QH. 2010. Genome-wide prediction of G protein-coupled receptors in Verticillium spp. Fungal Biol 114:359–368. 10.1016/j.funbio.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Son H, Seo YS, Min K, Park AR, Lee J, Jin JM, Lin Y, Cao PJ, Hong SY, Kim EK, Lee SH, Cho A, Lee S, Kim MG, Kim Y, Kim JE, Kim JC, Choi GJ, Yun SH, Lim JY, Kim M, Lee YH, Choi YD, Lee YW. 2011. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog 7:e1002310. 10.1371/journal.ppat.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheffer J, Chen C, Heidrich P, Dickman MB, Tudzynski PT. 2005. A CDC42 homologue in Claviceps purpurea is involved in vegetative differentiation and is essential for pathogenicity. Eukaryot Cell 4:1228–1238. 10.1128/EC.4.7.1228-1238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Derbyshire MC, Michaelson L, Parker J, Kelly S, Thacker U, Powers SJ, Bailey A, Hammond-Kosack K, Courbot M, Rudd J. 2015. Analysis of cytochrome b(5) reductase-mediated metabolism in the phytopathogenic fungus Zymoseptoria tritici reveals novel functionalities implicated in virulence. Fungal Genet Biol 82:69–84. 10.1016/j.fgb.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tucker SL, Besi MI, Galhano R, Franceschetti M, Goetz S, Lenhert S, Osbourn A, Sesma A. 2010. Common genetic pathways regulate organ-specific infection-related development in the rice blast fungus. Plant Cell 22:953–972. 10.1105/tpc.109.066340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeon J, Park SY, Chi MH, Choi J, Park J, Rho HS, Kim S, Goh J, Yoo S, Choi J, Park JY, Yi M, Yang S, Kwon MJ, Han SS, Kim BR, Khang CH, Park B, Lim SE, Jung K, Kong S, Karunakaran M, Oh HS, Kim H, Kim S, Park J, Kang S, Choi WB, Kang S, Lee YH. 2007. Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat Genet 39:561–565. 10.1038/ng2002. [DOI] [PubMed] [Google Scholar]
- 77.Zhang LB, Tang L, Ying SH, Feng MG. 2016. Distinct roles of two cytoplasmic thioredoxin reductases (Trr1/2) in the redox system involving cysteine synthesis and host infection of Beauveria bassiana. Appl Microbiol Biotechnol 100:10363–10374. 10.1007/s00253-016-7688-0. [DOI] [PubMed] [Google Scholar]
- 78.Braun BR, Head WS, Wang MX, Johnson AD. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31–44. 10.1093/genetics/156.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amich J, Vicentefranqueira R, Mellado E, Ruiz-Carmuega A, Leal F, Calera JA. 2014. The ZrfC alkaline zinc transporter is required for Aspergillus fumigatus virulence and its growth in the presence of the Zn/Mn-chelating protein calprotectin. Cell Microbiol 16:548–564. 10.1111/cmi.12238. [DOI] [PubMed] [Google Scholar]
- 80.Du Y, Zhang HF, Hong L, Wang JM, Zheng XB, Zhang ZG. 2013. Acetolactate synthases MoIlv2 and MoIlv6 are required for infection-related morphogenesis in Magnaporthe oryzae. Mol Plant Pathol 14:870–884. 10.1111/mpp.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Siewers V, Viaud M, Jimenez-Teja D, Collado IG, Gronover CS, Pradier JM, Tudzynski B, Tudzynski P. 2005. Functional analysis of the cytochrome P450 monooxygenase gene bcbot1 of Botrytis cinerea indicates that botrydial is a strain-specific virulence factor. Mol Plant Microbe Interact 18:602–612. 10.1094/MPMI-18-0602. [DOI] [PubMed] [Google Scholar]
- 82.Mir AA, Park SY, Abu Sadat M, Kim S, Choi J, Jeon J, Lee YH. 2015. Systematic characterization of the peroxidase gene family provides new insights into fungal pathogenicity in Magnaporthe oryzae. Sci Rep 5:11831. 10.1038/srep11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang K, Czymmek KJ, Caplan JL, Sweigard JA, Donofrio NM. 2011. Suppression of plant-generated reactive oxygen species is required for successful infection by the rice blast fungus. Virulence 2:559–562. 10.4161/viru.2.6.18007. [DOI] [PubMed] [Google Scholar]
- 84.Valdés-Santiago L, Guzmán-de-Peña D, Ruiz-Herrera J. 2010. Life without putrescine: disruption of the gene-encoding polyamine oxidase in Ustilago maydis odc mutants. FEMS Yeast Res 10:928–940. 10.1111/j.1567-1364.2010.00675.x. [DOI] [PubMed] [Google Scholar]
- 85.Baek JM, Howell CR, Kenerley CM. 1999. The role of an extracellular chitinase from Trichoderma virens Gv29-8 in the biocontrol of Rhizoctonia solani. Curr Genet 35:41–50. 10.1007/s002940050431. [DOI] [PubMed] [Google Scholar]
- 86.Ueno K, Matsumoto Y, Uno J, Sasamoto K, Sekimizu K, Kinjo Y, Chibana H. 2011. Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS One 6:e24759. 10.1371/journal.pone.0024759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y, Lim L, DiGuistini S, Robertson G, Bohlmann J, Breuil C. 2013. A specialized ABC efflux transporter GcABC-G1 confers monoterpene resistance to Grosmannia clavigera, a bark beetle-associated fungal pathogen of pine trees. New Phytol 197:886–898. 10.1111/nph.12063. [DOI] [PubMed] [Google Scholar]
- 88.Wang ZY, Soanes DM, Kershaw MJ, Talbot NJ. 2007. Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta-oxidation during appressorium-mediated plant infection. Mol Plant Microbe Interact 20:475–491. 10.1094/MPMI-20-5-0475. [DOI] [PubMed] [Google Scholar]
- 89.Ramos LS, Lehman BL, Peter KA, McNellis TW. 2014. Mutation of the Erwinia amylovora argD gene causes arginine auxotrophy, nonpathogenicity in apples, and reduced virulence in pears. Appl Environ Microbiol 80:6739–6749. 10.1128/AEM.02404-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garfoot AL, Shen Q, Wüthrich M, Klein BS, Rappleye CA. 2016. The Eng1 β-glucanase enhances Histoplasma virulence by reducing β-glucan exposure. mBio 7:e01388-15. 10.1128/mBio.01388-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ben-Daniel BH, Bar-Zvi D, Lahkim LT. 2012. Pectate lyase affects pathogenicity in natural isolates of Colletotrichum coccodes and in pelA gene-disrupted and gene-overexpressing mutant lines. Mol Plant Pathol 13:187–197. 10.1111/j.1364-3703.2011.00740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rogers LM, Kim YK, Guo W, González-Candelas L, Li D, Kolattukudy PE. 2000. Requirement for either a host- or pectin-induced pectate lyase for infection of Pisum sativum by Nectria hematococca. Proc Natl Acad Sci USA 97:9813–9818. 10.1073/pnas.160271497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dufresne M, van der Lee T, Ben M’barek S, Xu XD, Zhang X, Liu TG, Waalwijk C, Zhang WW, Kema GHJ, Daboussi MJ. 2008. Transposon-tagging identifies novel pathogenicity genes in Fusarium graminearum. Fungal Genet Biol 45:1552–1561. 10.1016/j.fgb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 94.Bahn YS, Kojima K, Cox GM, Heitman J. 2006. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell 17:3122–3135. 10.1091/mbc.e06-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim S, Ahn IP, Rho HS, Lee YH. 2005. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol Microbiol 57:1224–1237. 10.1111/j.1365-2958.2005.04750.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download aem.01385-22-s0001.pdf, PDF file, 1.2 MB (1.2MB, pdf)
Table S1. Download aem.01385-22-s0002.xls, XLS file, 0.1 MB (153.5KB, xls)
Table S2. Download aem.01385-22-s0003.xls, XLS file, 0.03 MB (28KB, xls)
Data Availability Statement
The RNA-seq raw reads were submitted to the NCBI SRA database under BioProject number PRJNA797877 and BioSample numbers SAMN25039392 (XJ592) and SAMN25039393 (ΔVdCf2).