Abstract
We present a fluorescence fluctuation image correlation analysis method that can rapidly and simultaneously measure the diffusion coefficient, photoblinking rates, and fraction of diffusing particles of fluorescent molecules in cells. Unlike other image correlation techniques, we demonstrated that our method could be applied irrespective of a nonuniformly distributed, immobile blinking fluorophore population. This allows us to measure blinking and transport dynamics in complex cell morphologies, a benefit for a range of super-resolution fluorescence imaging approaches that rely on probe emission blinking. Furthermore, we showed that our technique could be applied without directly accounting for photobleaching. We successfully employed our technique on several simulations with realistic EMCCD noise and photobleaching models, as well as on Dronpa-C12-labeled β-actin in living NIH/3T3 and HeLa cells. We found that the diffusion coefficients measured using our method were consistent with previous literature values. We further found that photoblinking rates measured in the live HeLa cells varied as expected with changing excitation power.
Why it matters
We developed an image correlation fluorescence fluctuation analysis technique that can analyze microscopy images of cells containing nonhomogeneous distributions of immobile biomolecules labeled with fluorophores that are subject to photoblinking and photobleaching. As far as we know, this is the first image correlation method that does this. This enables the study of intricate cellular systems using fluorescent probes with complex photophysical properties. Our method rapidly and simultaneously measures diffusion and photoblinking rates as well as the fraction of diffusing particles. We tested our technique on simulations and Dronpa-C12-labeled β-actin in live NIH/3T3 and HeLa cells. Our measured diffusion coefficients from the live cell experiments were consistent with previously reported values in the literature.
Introduction
The past decade has seen a revolution in optical microscopy, with the advent of far-field super-resolution approaches. Fluorescence super-resolution microscopy has become an invaluable tool for furthering our understanding of biological systems by allowing one to circumvent the diffraction-limited resolution of traditional fluorescence microscopy techniques, leading to important insights in cell biology, neuroscience, and cellular biophysics (1, 2, 3, 4, 5). Among the super-resolution imaging techniques, single-molecule localization microscopy methods are one of the most commonly used. These methods rely on photophysical processes to localize the position of single fluorophores with a spatial uncertainty much lower than the wavelength diffraction limit of light. Some popular examples of such techniques are stochastic optical reconstruction microscopy (6) and photoactivated localization microscopy (PALM) (7). Super-resolution optical fluctuation imaging (SOFI) (8) also relies on the stochastic photoswitching nature of fluorophores but builds super-resolution images using the cumulants of the fluorescence fluctuations. Many applications of super-resolution, however, have so far been limited to studying immobile components in cells or static molecules in chemically fixed cells without examining their dynamic counterparts. Whereas super-resolution has also been coupled with quantitative methods to investigate these dynamics, there are currently only a limited number of approaches that combine super-resolution imaging and measurement of dynamics. Notably, stimulated emission depletion microscopy was combined with fluorescence correlation spectroscopy (FCS) in stimulated emission depletion-FCS (9), which was shown to better characterize heterogeneous diffusive behavior of membrane biomolecules than traditional FCS by reducing the beam spot size. Scanning versions of this technique exist as well (10,11). Single-particle tracking (SPT) and PALM were also combined in sptPALM (12). Minimal photon fluxes (13) is also capable of super-resolved SPT. Another example is fcsSOFI, which combines FCS and SOFI techniques to form super-resolved diffusion maps (14).
Many methods currently exist for measuring the dynamics in biological systems, and a significant subset rely on measurement of fluorescence fluctuations to compute autocorrelations that are then fitted with specific models to measure transport parameters, such as biomolecule diffusion coefficients and directed flow rates. Conventional FCS (15, 16, 17) is a widely used example that analyzes fluorescence fluctuation time series collected from a single fixed laser focal spot. Imaging FCS (18) was developed to allow for multiplexed FCS analysis of pixels forming a fluorescence image. Similarly, image correlation spectroscopy (ICS) methods (19, 20, 21, 22) also analyze fluorescence fluctuations in images but utilize both spatial and temporal information when computing the autocorrelation. One advantage of ICS techniques is that the use of spatial information increases statistical sampling; however, the inherent spatiotemporal heterogeneity in biological systems makes it difficult to simply average over different pixels and frames. ICS techniques usually handle spatiotemporal heterogeneity by analyzing many smaller local regions of interest (ROIs) and then correlating over a chosen time of interest (TOI). This is the approach for generating flow maps when using spatiotemporal ICS (21). The most challenging example of heterogeneity in time in fluorescence microscopy is the issue of photobleaching, which has been addressed using both pre- (23, 24, 25) and postprocessing (26, 27, 28, 29, 30) methods. Another example is anomalous diffusion, which has been investigated in multiple FCS studies (31, 32, 33, 34, 35, 36).
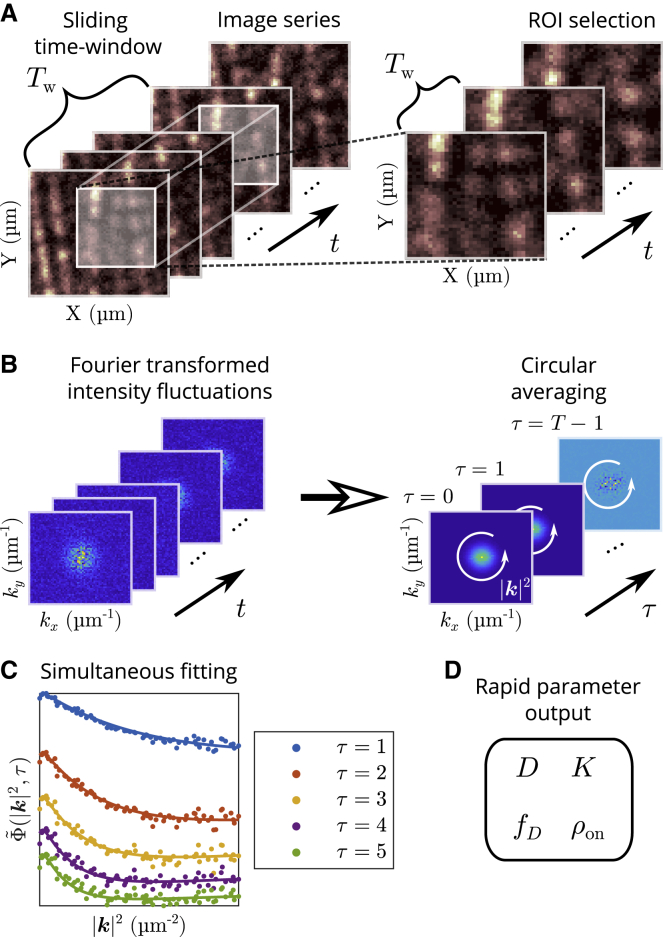
Because super-resolution aims to better resolve complex cell processes in space and time, a technique that can analyze dynamics in the presence of spatially and temporally heterogeneous structures is required. Here, we present an image correlation method that can successfully and rapidly analyze heterogeneous systems for accurate measurement of biomolecule diffusion coefficients, probe photoblinking rates, and the fraction of particles undergoing diffusion (see Fig. 1). Along with measuring dynamic parameters in a complex cell environment, we anticipate that the measurement of probe photoblinking rates will be useful for optimal fluorescent probe development and optimization for methods such as stochastic optical reconstruction microscopy and SOFI. The ability of our method to analyze spatially heterogeneous systems further allows us to significantly increase the spatial sampling used in our autocorrelation computation.
Figure 1.
Schematic illustrating extended kICS method application. (A) An ROI and sliding time window are first chosen from an image series. Intensity fluctuations are computed locally in time, according to the chosen time-window size, to mitigate the effects of photobleaching. (B) The 2D spatial Fourier transforms of the intensity fluctuations are first computed in each frame, then this k-space ROI frame stack is autocorrelated in time. A circular averaging of the autocorrelation is also calculated when the dynamics of the system are isotropic. Nonuniformly distributed, immobile blinking fluorophore populations do not systematically affect the fluctuation-defined autocorrelation. (C) Computed ACF (points) and simultaneous fits (lines) over five time lags. (D) The process of computing the ACF and fitting is rapid (order of seconds for 64 × 64 pixel image series with 2048 frames) and outputs; the diffusion coefficient (D), the sum of the photoblinking rates (K), the fraction of time spent in the on-state (ρon), and the fraction of diffusing particles relative to all particles (fD) are shown.
Our method is based on k-space ICS (kICS) (19), which was originally developed for measuring transport parameters independently from the fluorophore photophysics. Similar to other ICS techniques, one of the underlying assumptions behind kICS is uniformity in space and time in the data being analyzed (20). Nonuniformity in space caused by the presence of immobile structures was commonly filtered out by either subtracting the time average, or equivalently, applying a Fourier filter. This solution does not work, however, when the fluorophores labeling these structures are undergoing photophysical processes (e.g., photoblinking and photobleaching on timescales comparable to the imaging) thus making them difficult to analyze with traditional ICS methods. This is especially relevant in the context of single-molecule localization microscopy and SOFI methods. Therefore, this significant extension of kICS is important for dealing with real heterogeneities in space and time in the complex cellular environment.
We begin by showing that our definition of the autocorrelation is approximately independent of immobile particle positions. We then derive the autocorrelation for a mixed system of photoblinking-immobile and -diffusing particles that is independent of any direct imaging parameters in two dimensions (2D), such as the point spread function (PSF). We proceed to demonstrate the accurate measurement of diffusion and photophysical parameters when applying our technique on simulations. We then employ our method on Dronpa-labeled β-actin imaged in live 3T3 and HeLa cells. Our measured diffusion coefficients for β-actin from these experiments are consistent with previously reported values. Furthermore, we observed that the photoblinking follows the expected dependence on excitation power in our HeLa cell experiments.
Materials and methods
Theory
In the development of the original kICS technique by Kolin et al. (19), it was shown that diffusion and flow dynamics could be recovered regardless of the photophysical properties of the fluorophores (e.g., photoblinking and photobleaching). However, the original method did not consider the presence of immobile particle populations that were also undergoing photophysical processes. We show here that, with the presence of such populations, one cannot simply apply the original kICS analysis technique. We further derive an expression for the autocorrelation function (ACF) that we simultaneously fit for transport and photophysical parameters. This expression is independent of any direct imaging parameters in 2D (e.g., PSF size, amplitude, etc.) as well as the immobile particle positions, allowing us to probe systems with complex immobile particle spatial arrangements and nonuniform cell morphologies. Furthermore, this enables us to maximize statistical spatial sampling when computing the ACF. This is in contrast with other image correlation techniques, which require spatially uniform regions for analysis (20). Consequently, previous image correlation techniques would not be able to include cell boundaries and narrow projections, such as dendritic spines and narrow cellular lamellae, such as dendritic projections, when choosing ROIs for analysis. The 2D model presented here can be used to model membrane dynamics sampled using total internal reflection fluorescence microscopy, for example. We later extend this to the three-dimensional (3D) case when analyzing the live 3T3 and HeLa cell data. We will refer to the developed technique as an extended kICS method.
We begin with the definition of a fluorescence microscopy image series of intensities in 2D, i(r,t):
| (1) |
where is the fluorescence intensity from the labeled particles at position r and time t belonging to population α with common transport parameters; I(r) is the PSF, #r is a spatial convolution, ό(r,t) is an additive noise term (which is assumed to be independent from itself for any (r,t) (r′,t′)), and ρ(α)(r, t) is the apparent particle density of population α (i.e., the density of particles in population α that are emitting and detectable). Note that we only consider dependence of population α on particle position in this work. The apparent density of particles belonging to population α is given by:
| (2) |
In this last equation, δ(⋅) is the 2D Dirac delta function, m is an index denoting the fluorophores (which we also refer to as “particles”), qm,t is the instantaneous rate of detector counts for the mth fluorophore at time t (which depends on several factors, including the photon budget, quantum efficiency of the detector, and camera detector gain), is the position of the mth fluorophore at time t (belonging to population α), and Θm,t is its photo-emissive state, expressed as
| (3) |
Note that we do not consider photophysical transitions between singlet states (i.e., absorption/emission and quenching) because these are far beyond the time-resolution capabilities of electron multiplying charge-coupled device (EMCCD) camera detectors.
Because we developed this approach for widefield fluorescence microscopy, we account for faster processes by also considering the effects of camera detector integration time by substituting (30)
| (4) |
with τi being the integration time (in this work, we consider τi = 1); however, we leave the integration out of the notation because it is straightforward to redo the derivation with it.
Defining the spatial Fourier transform as , we have the Fourier counterparts of Eqs. (1), (2), respectively,
| (5) |
and
| (6) |
In this work, we calculate the autocorrelation as
| (7) |
Practically, the above autocorrelation is realized by computing a time average; therefore, we use the notation to denote an expectation value that only considers random variables that depend on time to be random. We also define δt as a fluctuation with respect to the time average as follows:
| (8) |
Note that in the original kICS work (19), the autocorrelation was defined without the fluctuations. In the Supporting materials and methods, we show that the original definition leads to noisy autocorrelations affected by the immobile blinking particle positions (see Supporting materials and methods, Comparison with original kICS method). Using the Fourier transform of an image series in Eq. 5, we can express the autocorrelation in Eq. 7 as follows:
| (9) |
The terms in the curly brackets are zero, assuming noise and particle positions are independent. We show in Supporting materials and methods, Noise autocorrelation, that the autocorrelation of the noise (i.e., the second term) only affects τ = 0 by a constant offset, which we will denote by . Note that |k|2 = 0 is also affected by the noise but is omitted from the analysis.
Using Eq. 6 for immobile populations we have
| (10) |
where we have assumed that all fluorophores have equal quantal brightness (i.e., qm,t). Furthermore, note that the time indexing is dropped for immobile particle positions so that they are unaffected by the time averaging. Conversely, for mobile populations
| (11) |
because for |k| 0, assuming the mobile particle positions are uniformly distributed in space within the chosen ROI.
Using Eqs. (10), (11), Eq. 9 for one mobile and one immobile population becomes
| (12) |
where we have assumed nonidentical particles are mutually independent so that we can sum their individual autocorrelations and δτ,0 is the Kronecker delta function. Note that the fluctuations are not necessarily expected to vanish in the second term in the curly brackets because, in practice, they are computed by subtracting the sample time average, which does not converge to the ensemble average in nonergodic systems. Furthermore, subtraction by the spatial average only affects the autocorrelation at |k| = 0. Notice, also, that this term is affected by the immobile particle positions, which is why we need to define conditions when it is approximately zero. It is clear that without photobleaching (or any other nonstationary photophysical process) the photophysical fluctuations are indeed zero on average; however, in the presence of bleaching, the fluctuations need to be properly defined to make sure this term is approximately zero. For this reason, we use local time averaging over a subset time window to compute Eq. 8 in practice (see Autocorrelation computation) as follows:
| (13) |
where TW is the window size chosen for the local time average. If the photobleaching is slow and TW is large enough, then Eq. 8 is a good approximation of Eq. 13. On the other hand, if the photobleaching is significant, one must account for TW in the autocorrelation (see Supporting materials and methods, Time-windowed correction, where we provide an expression for this correction). Eq. 8 will also not hold when the dynamics in the system are slow and resemble immobility; in this case, the effects of time windowing should again be accounted for (for example, see Fig. 2 C). Note that if the second term in the curly brackets in Eq. 12 is zero, then the autocorrelation is independent of any assumptions on immobile particle positions, making it a powerful tool to study previously inaccessible systems using image correlation. Time-windowed or moving-average subtraction has been previously used in raster ICS as a way to filter out slow-moving fluorescent objects (22,37).
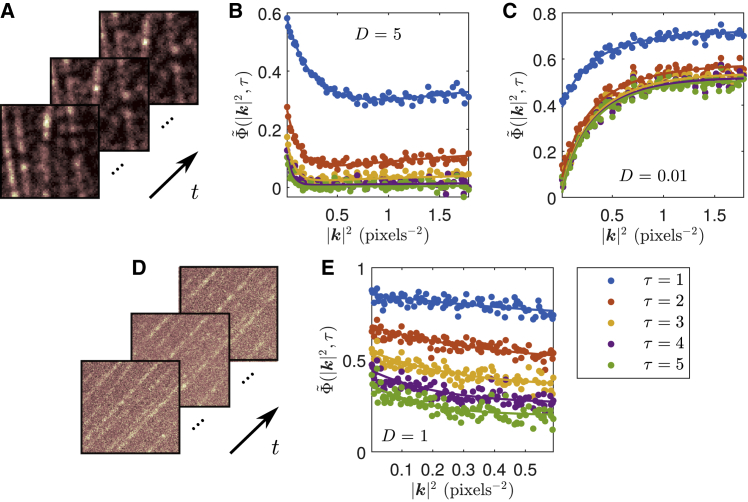
Figure 2.
Example ACFs and fits computed from simulations of filamentous structures composed of static blinking particles, with a second population of freely diffusing particles. Both populations are set to have the same photophysical properties. Each simulation also contained simulated EMCCD noise. The ACF fits were all done over the first 10 time lags; only the first five are shown. Time windowing was done with TW = 200 frames in each case. Simulation and fit details are shown in Table 1. (A) Example of simulated intensity images in time. (B and C) Computed ACFs (points) and corresponding simultaneous fits (lines) for simulations with D = 5 and 0.01 pixels2/frame, respectively. In (C), we utilize a fit model that corrects for the chosen sliding time window. (D) Sample simulation with higher simulated autofluorescence background and lower photon budget than simulation shown in (A). (E) Corresponding computed ACF and fit of simulation shown in (D) with D = 1 pixels2/frame.
To obtain a quantity that is independent of PSF, we define the ACF as follows:
| (14) |
The second term in the denominator removes dependence of the ACF on the noise; practically, it is computed as the large |k|2 offset in the autocorrelation. Furthermore, if the mobile components of the system being analyzed are isotropic, one can circularly average the autocorrelations for statistical sampling purposes (19,20,38). Because the focus of this work will be on such systems, the dependence of the ACF in this last equation is left to be on |k|2. It should also be mentioned that this definition of the ACF leads to division by zero after sufficient decay of the PSF, so that the ACF must be appropriately trimmed in |k|2 before fitting. For a 2D mixture of diffusing and immobile particles, Eq. 14 can be written explicitly as (19,30,39):
| (15) |
where D is the diffusion coefficient, K ≡ kon + koff is the sum of the photoblinking rates, (where kon and koff are the on- and off-blinking rates, respectively), ρon is the fraction of time spent in the fluorescent on-state, and fD is the fraction of particles diffusing. Note that the blinking in this last expression is assumed to follow a two-state on-off model without photobleaching. In the Supporting materials and methods, we include a derivation considering the effects of bleaching and detector time integration as well as a derivation for the 3D effects on the ACF (see Supporting materials and methods, Autocorrelation function derivation).
Live cell imaging
The NIH/3T3 fibroblast and HeLa cells were transfected with plasmids containing either the Dronpa-C12 β-actin fusion protein or with Lipofectamine 2000 using the standard protocol. Before imaging, cell culture media was gently replaced into 1× PBS buffer warmed to 37°C. The cells with prominent actin stress fibers were empirically identified for imaging. The NIH/3T3 fibroblast cell data were imaged with acquisition time of 50 ms per frame, and we observed slow detaching of its focal adhesion sites during the imaging time course. The same data appeared in previous work (40). The HeLa cells were imaged at 10 ms per frame under excitation powers of ∼14, ∼19, and ∼24 mW.
Imaging was performed with an inverted fluorescence microscope using widefield imaging mode (Nikon Eclipse Ti, Tokyo, Japan) equipped with an EMCCD camera (model number DU-897E-CSO-#BV, Andor iXon; Andor Technology, Belfast, Northern Ireland) and a standard EGFP filter cube (460/60 nm band pass excitation filter, 495 nm long pass dichroic, and 520/40 nm band pass emission filter). Excitation of 485/25 nm was used (cyan option, AURA light engine; Lumencor, Beaverton, OR). A 60× oil immersion objective (NA = 1.4) was combined with an extra 1.5× magnification module integrated into the microscope body.
Computer simulations
Simulations were created and analyzed using MATLAB R2020a (The MathWorks, Natick, MA) on a Dell XPS 9530 (Intel(R) Core i7 @ 2.3 GHz, 16 GB RAM) running Windows 10. Simulations were also created using MATLAB R2020a on a dedicated research server (Intel(R) Core i7 @ 3.2 GHz, 64 GB RAM) running Ubuntu version 18.04.
To simulate fluorophores on filaments, we first drew angles from a normal distribution with specified mean and SD. This determined the direction of each filament in the synthetic image series. Starting from a filament’s endpoint (randomly distributed along simulated image edges), particles were then iteratively placed along the filaments with incremental distance (in pixels) drawn from a uniform distribution on (0,1). This process was repeated until the edge of the synthetic image was reached. Each time a simulated emitter was placed, a predefined probability determined whether the position was occupied by an aggregate. Each simulated aggregate had an assigned mean number of fluorophores following a Poisson distribution as well as a random distance from the aggregate center (mean position) following a normal distribution with a chosen standard deviation. Simulated diffusing particles were initialized randomly within the pixel grid and allowed to diffuse with periodic boundaries.
Synthetic emitters were subject to stochastic switching between on- and off-states at specified rates to simulate photoblinking. Photobleaching was assumed to be equal from the on/off-states in all simulations. Both populations of immobile and diffusing particles were simulated to have the same photoblinking and photobleaching rates.
We then convolved the simulated image series with a 2D Gaussian function (integrated over pixel dimensions) to simulate the optical PSF. To emulate the effect of the detector integration time, we split each frame into 50 “subframes,” so that a single frame comprised a sum of its constituent “subframes.” For more simulation details, we refer the reader to Supporting materials and methods, Simulation details.
Synthetic pixel intensity values were assigned using the EMCCD model presented by Hirsch et al. (41), as was previously described in Sehayek et al. (30).
The ACF was fitted to Eq. S7, unless otherwise stated. The fitting model assumed one diffusing and one immobile population. The global fitting of the ACF was done using the built-in MATLAB object GlobalSearch with fmincon as a local solver. The fitted parameters were chosen according to the least-squares method across the specified domain of the ACF. All fitted parameters were constrained to be greater than zero, with the added conditions ρon, PD ≤ 1. We used uniformly drawn random numbers in the interval (0,1) as an initial guess for all fitted parameters to demonstrate the robustness of our method. In the case for which the fit did not visually match the data, we repeated the fitting process until reasonable agreement was achieved. The fits always excluded the points |k|2 = 0 because they are affected by the noise in the system. The τ = 0 curve was also excluded from our fits because it does not contain any useful information when using the definition in Eq. 14. Our analyses were performed on several ROIs and/or TOIs. The reported fitted parameters and their errors were then taken to be the mean and the standard error (SE) of the mean from these analyses.
Autocorrelation computation
The autocorrelation was calculated as follows:
| (16) |
where Ft is the fast Fourier transform in time, and denotes the time-windowed fluctuation, as in Eq. 13; that is, at pixel (x, y) and frame t, we subtract the mean intensity of TW subsequent frames, including frame t (we used the MATLAB movmean function to do this). Defining the fluctuations in this way diminishes the oscillations caused by photobleaching (see Supporting materials and methods, Comparison with original kICS method). The choice of TW should ultimately depend on the photobleaching rate. Note that the Wiener-Khinchin theorem is used in Eq. 16 to minimize autocorrelation computation time via Fourier (reciprocal) space calculations.
The autocorrelation in Eq. 16 was then circularly averaged. This was done by averaging all autocorrelation points with the same value of |k|2. Finally, the ACF was computed as shown in Eq. 14. To determine the “large” |k|2 offset in the denominator of the equation, a range of |k|2-values were chosen after the τ = 0 autocorrelation had sufficiently decayed and averaged over.
Data availability
Data are available from the kICS GitHub repository (https://github.com/ssehayek/kics-project.git) and the SOFI 2.0 GitHub repository (https://github.com/xiyuyi-at-LLNL/pysofi.git).
Results and discussion
Computer simulations
We first apply our extended kICS method on 2D computer simulations of immobile filamentous structures composed of fixed emitting particles with a second simulated population of freely diffusing particles, as shown in Fig. 2. The fit results from this figure are tabulated in Table 1. The fitted parameters demonstrate the wide range of diffusion coefficients that are measurable in heterogeneous morphologies using our extended technique.
Table 1.
Comparison of fitted and simulated parameters
| Fig. 2B | Fig. 2C | Fig. 2E | ||||
| Fit | Simulation | Fit | Simulation | Fit | Simulation | |
| D (pixels2 frame−1) | 4.66 ± 0.06 | 5 | 0.0113 ± 0.0001 | 0.01 | 1.12 ± 0.07 | 1 |
| K (frame−1) | 1.02 ± 0.02 | 1 | 1.68 ± 0.01 | 1.7 | 0.284 ± 0.006 | 0.3 |
| ρon | 0.095 ± 0.006 | 0.1 | 0.59 ± 0.01 | 0.59 | 0.36 ± 0.03 | 0.33 |
| fD | 0.68 ± 0.03 | 0.7 | 0.59 ± 0.02 | 0.65 | 0.36 ± 0.06 | 0.35 |
The parameters are for fits shown in Fig. 2. Each simulation had photobleaching rate kp = 10−4 frame−1 over T =2048 frames. Simulations (Fig. 2B) and (Fig. 2C) were generated on 128 × 128 pixel grids, and (Fig. 2E) was on a 256 × 256 pixel grid. The simulations were assigned 8130, 8411, and 9048 total particles, respectively. In each case, fitted parameters and errors were obtained by splitting the simulation spatially into four equally sized and independent ROIs and then calculating the mean and its SE from their analyses.
The simulations assumed both immobile and diffusing particle populations to have the same photoblinking and photobleaching rates. Each simulation also contained simulated EMCCD noise (see Sehayek et al. (30) for noise model details). We note that our method is not limited to analyzing EMCCD data and could also be applied to data acquired by other camera detectors used in widefield setups. This is because we considered the exposure time of each frame when deriving the ACF (see Eq. 4). Examples of other such detectors are scientific complementary metal-oxide-semiconductors (sCMOS) cameras and single-photon avalanche diode arrays, both of which have been used in multiplexed FCS studies (42,43). An extension of our method to sCMOS data would most likely require characterization of spurious correlations from hot pixels characteristic of such cameras, but this should be possible using a camera-specific masking operation. As well, recent work by Mandracchia et al. (44) presents an adaptive algorithm approach to correct for noise characteristic of sCMOS cameras, and such an approach may be useful before performing kICS.
Note that the nonuniformity in the immobile particle positions placed along the simulated filaments in Fig. 2, which confirms that the technique can be successfully applied in nonhomogeneous systems. Conversely, previous image correlation techniques required ROIs to be selected for which the spatial distribution of particles was homogeneous (e.g., avoiding cell boundaries; see Supporting materials and methods, Comparison with original kICS method, in which we compare our extended kICS technique to the original one) (20). It would be impossible to select such an ROI with both diffusing and immobile particles in the simulations shown in Fig. 2. The ability to analyze larger ROIs further enables us to increase the spatial sampling in our analyses; however, we assume that all the particles within this ROI have transport and photophysical parameters that are drawn from common distributions (i.e., we assume a single diffusing population and a single photophysical population). Although the extension to multiple populations with different dynamic parameters is possible, one must be mindful of the possibility of overfitting. Thus, our analysis offers a coarse-grained approach for quickly measuring these parameters within regions. SPT is beneficial when a common distribution cannot be assumed for such parameters. However, the fluorescence correlation approach can be applied to cell expression systems for which high-density labeling might not permit SPT. SPT is also limited by factors such as photoblinking in transport populations.
In Fig. 2 B, the shape of the ACF is characterized by a decay at low |k|2 and a convergence to a nonzero value at higher |k|2. The former is attributed to the diffusion coefficient, whereas the latter is due to the presence of an immobile particle population, as can be seen from Eq. 15 (note that these fits use the time-integrated version of this equation; see Supporting materials and methods, Autocorrelation function derivation for details). The decrease in the ACF amplitude and offset with increasing time lag is due to the photophysical processes in the system (i.e., photoblinking and photobleaching).
When the ACF is characterized by an initial increase along |k|2, as is the case in Fig. 2 C, one needs to account for the effect of the sliding time window (see Supporting materials and methods, Time-windowed correction). This type of behavior occurs when the diffusion is relatively slow or when the chosen time window is relatively short.
In Fig. 2 D, we show sample images from a simulation with more noise. Specifically, we increased the simulated autofluorescence background while decreasing the photon budget of the simulated fluorophores. To accurately analyze such an image series (Fig. 2 E), we required larger ROIs than the ones used in the previous analyses. Consequently, we generated this simulation on a 256 × 256 pixel grid.
Note that photobleaching was not considered in the fits shown in Table 1. A derivation considering the effects of bleaching on the ACF is included in Supporting materials and methods, Time-windowed correction. Although one can consider these effects, we have demonstrated that we can still obtain accurate parameters in the presence of photobleaching without having to incorporate it into our fit model (given appropriate choice of time window). This is beneficial because bleaching pathways of a fluorescent label are not often known.
We also note that the fraction of time spent in the on-state ρon cannot be measured for a purely immobile population (i.e., fD = 0). This can be seen by the lack of dependence on ρon in Eq. 15 in this limit. Previous techniques have described how to measure the photoblinking rates for immobile emitters (30,45, 46, 47, 48).
To perform our analysis, it is essential to choose a time window, an ROI, a range of time lags to fit simultaneously, and cutoff values for |k|2. As discussed in Theory above, the choice of time window will mainly depend on the photobleaching rate. A value of TW that is too small results in loss of information but a smoother ACF. A value of TW that is too large will result in a noisier ACF that is more influenced by the photobleaching, which may lead to poorer fits. With a simulated bleaching rate of kp = 10−4 frames−1 (with a frame acquisition time of 50 ms, this corresponds to a characteristic bleaching time of 500 s), we find a suitable choice of time window to be TW = 200 frames. We will use this value consistently throughout this work for the same value of kp. The same value of TW can be used for smaller bleaching rates. In general, we found from our simulations that TW can be chosen to be ∼2–5% of the characteristic bleaching time.
As mentioned above, when choosing an ROI, it is necessary to select a relatively large region to avoid aliasing of the ACF along |k|2. This is especially important when dealing with larger diffusion coefficients because the decay will appear in a short range of small |k|2.
Similarly, when choosing a range of time lags to fit, it is best to choose a wide range to capture slower dynamics. If the range is too large, the simultaneous fitting will be visibly biased, and a smaller range should then be used. Trying to fit a larger time-lag range can be complicated by the presence of photobleaching, for example (we show how to account for these effects in Supporting materials and methods, Time-windowed correction). We also remark that the ACF is noisier for higher time lags, so it is informative to compare them with their fits to gauge whether the fitted time-lag range is too wide. We showed that we can achieve reasonable fits for our simulated parameters by fitting the 10 first time lags in our analyses in Table 1. Systematic errors were reduced when including more time lags in the fit or when choosing larger ROIs, but in nonsimulated data, it may be difficult to make such adjustments.
Finally, we discuss choosing cutoff values for |k|2. We discard the value of the ACF at |k|2 = 0 because it is affected by the noise in the system, as was mentioned in the Theory. Excluding a few small |k|2 is also beneficial for avoiding the autocorrelation from the time windowing when not using the time-window correction (see Supporting materials and methods, Time-windowed correction for details). The maximal value for |k|2 can be selected by examining where the ACF begins to diverge because of the normalization in Eq. 14. It is optimal to choose the largest possible range of |k|2 to fit while avoiding points that are too noisy because of the normalization. Choice of the maximal cutoff will depend on PSF size as well as the noise in the system. As was previously mentioned, this is to avoid division by zero that occurs because of our definition of the ACF in Eq. 14. This occurs because the noise is subtracted from the denominator, which is then effectively zero after the PSF has sufficiently decayed. In Fig. 2 E, because of the higher noise in the simulation, the fit has a smaller chosen maximal cutoff of |k|2 because higher values result in a divergence of the ACF.
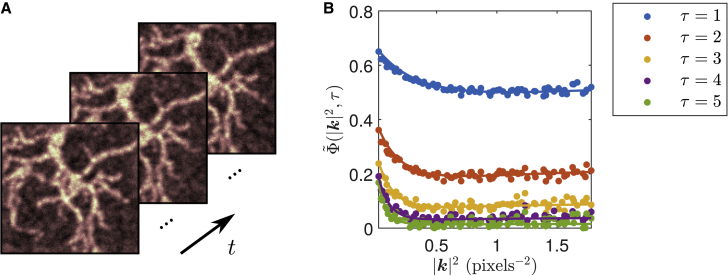
In Fig. 3, we perform the same analysis on a simulated dendritic morphology. A comparison of the simulated and fitted parameters recovered using our analysis is shown in Table 2. This further demonstrates the ability of the technique to be applied independently of immobile particle distribution. In this case, it would again be impossible to select an ROI containing a uniform distribution of both diffusing and immobile populations for analysis using previous image correlation techniques. As mentioned previously, with the extended kICS technique developed in this work, we are no longer restricted by this requirement and can further benefit from large ROIs to increase the spatial sampling in our analysis.
Figure 3.
Example ACF computed from simulation of dendritic structures and fit. (A) Sample-simulated intensity images in time. Simulations contain immobile particles on the dendrites, along with a background of freely diffusing particles. Both populations are assumed to have equal photophysical properties. (B) Computed ACF (points) and corresponding simultaneous fit (lines). The fit is done over the first 10 time lags; only first five are shown. Time windowing was done with TW = 200 frames. Fit details are shown in Table 2.
Table 2.
Comparison of fitted and simulated parameters
| Fit | Simulation | |
| D (pixels2 frame−1) | 2.80 ± 0.08 | 3 |
| K (frame−1) | 0.86 ± 0.01 | 0.9 |
| ρon | 0.38 ± 0.03 | 0.44 |
| fD | 0.26 ± 0.02 | 0.3 |
The parameters are for fits shown Fig. 3. Simulation had photobleaching rate kp = 10−4 frame−1 and was generated on 128 × 128 pixel grid with 10,744 total particles and T =2048 frames. Fitted parameters and errors were obtained by splitting the simulation spatially into five 64 × 64 ROIs, with some overlap between different parts, and then calculating the mean and its SE from their analyses. The ROIs were chosen so that a significant portion of the simulated dendritic structure was encompassed overall.
Live NIH/3T3 cell data
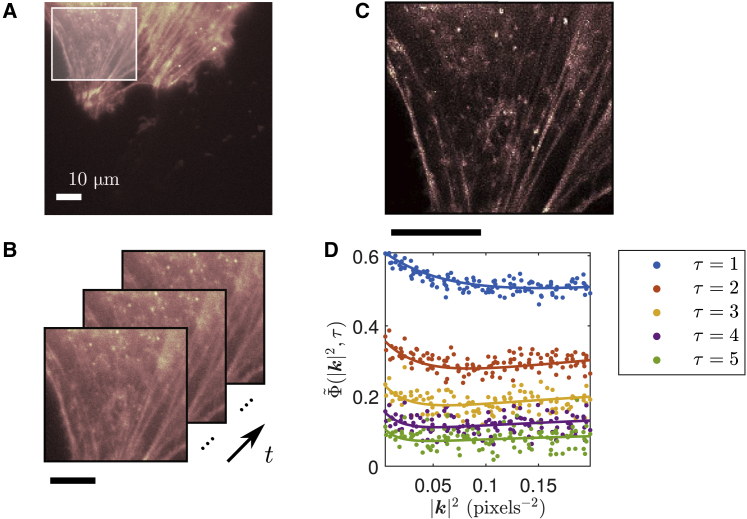
Using a widefield fluorescence microscope equipped with an EMCCD camera detector, we imaged β-actin labeled with Dronpa-C12 in an NIH/3T3 fibroblast cell line expression system. The Dronpa-C12 exhibited blinking and photobleaching during image acquisition, and the β-actin pool was both diffusively mobile within the cell and immobile in actin filaments. A sample of our analysis from the data is shown in Fig. 4.
Figure 4.
ACF computed from Dronpa-C12-labeled actin in a live NIH/3T3 cell and fit. (A) Fluorescence image of Dronpa-C12-labeled actin in a live NIH/3T3 cell, with ROI used in analysis highlighted. (B) Sample fluorescence images in time of ROI shown in (A). (C) SOFI image generated from immobile blinking fluorophores (see GitHub code for SOFI 2.0). (D) Computed ACF from ROI shown in (A) (points) and corresponding simultaneous fit (lines). The fit was done over the first five time lags. Time windowing was done with TW = 100 frames. Fitted parameters are D = 9.2 ± 0.4μm2 s−1, K = 7.6 ± 0.4 s−1, and fD = 0.58 ± 0.03. Fit for ρon is omitted because of inconsistent values between different ROIs and TOIs. Pixel size is 177.78 nm. Frame time is 50 ms. Analysis details are as follows: two spatially independent ROIs (each about 30 × 30 μm2) over two temporally independent TOIs (each ∼50 s in length) were considered in the analysis. The reported fitted parameters and errors are given as the mean and its SE from these analyses. Scale bars: 10 μm.
In Fig. 4 C, we reconfirm that the blinking of immobile fluorophores can be used to obtain a SOFI (8) image using SOFI 2.0 (40,49) (see GitHub code for SOFI 2.0). Along with our dynamic analysis of this data, we demonstrate that we can extract both static and dynamic information from our system with careful selection of correlation analysis approaches.
Because the data were acquired using a widefield fluorescence microscope with actin monomers diffusing in the cytoplasm, we needed to employ a 3D model for the ACF fit (see Supporting materials and methods, Diffusing and immobile populations (3D) for more details). The extension to a 3D model (without considering integration time effects) is achieved through a scaling factor that depends on τ and, consequently, does not affect the behavior of the ACF along |k|2. Using the 2D fit model for the ACF yielded visibly inconsistent fits to the data. Notably, our reported value for the apparent diffusion coefficient (9.2 ± 0.4 μm2 s−1) is within range of the simulated and experimentally verified diffusion coefficient of globular actin (G-actin) in the cytoplasm of ∼3–30 μm2 s−1 (50, 51, 52, 53, 54). This is also in reasonable agreement with the diffusion coefficient of Dronpa-labeled actin in an MCF-7 cell of 13.7 μm2 s−1, reported by Kiuchi et al. (53) Our measurement of a relatively rapid diffusion coefficient is further confirmed by the behavior of the ACF, which exhibits a characteristic initial decay in |k|2, as in Fig. 2 B. This is in contrast to systems with lower diffusion coefficients, which exhibit an initial increase in |k|2, as in Fig. 2 C. As can also be seen from Fig. 4, our assumption of a single diffusing population provides a reasonable fit to the data. Therefore, we argue that it would not be advantageous to include more diffusing components in our fit model because it would risk overfitting our data.
To account for 3D effects, we needed to first estimate the e−2 radius along the axial direction from the data. To this end, we used the Abbe resolution criterion to determine the full width at half maximum of the PSF in z (i.e., 2λ/NA2), which was then converted to an e−2 radius.
McGrath et al. (50) demonstrated that they can simultaneously measure the actin filament turnover rate, fraction of actin in filaments, and actin diffusion using either fluorescence recovery after photobleaching or photoactivation of fluorescence. In their model, the filamentous actin is not necessarily immobile, but it is not diffusing. We show that on the time and spatial scales we considered in our analysis, the actin flow is negligible. Furthermore, filament turnover rate is an important parameter at filament ends, but we chose ROIs away from these ends so that we would not have to consider such effects.
Because flow appears as an imaginary component in the autocorrelation (19), we compared the magnitude of the imaginary part to that of the modulus of the autocorrelation as follows:
| (17) |
This quantity was determined to be very small (close to machine precision), confirming that the flow is negligible relative to other dynamics over the time and spatial scales examined.
From our analysis of the data shown in Fig. 4, we also found a diffusing fraction of fD = 0.58 ± 0.03. Gasilina et al. (55) reported a percentage of filamentous actin of 48 ± 4% from their immunoblotting-based analysis of wild-type NIH/3T3 fibroblast cells. If we assume filamentous actin to be immobile and G-actin to be diffusing, then this value corresponds to fD = 0.52 ± 0.04.
We further tested whether the G-actin was undergoing anomalous subdiffusion in the cell. This can be done approximately by replacing the dependence of the ACF from τ→τα (ignoring detector time integration), where α is the degree of subdiffusion (34). A fit including α as a free parameter yielded a fitted value of α ∼ 1, indicating that the diffusion of the G-actin within this nonmigrating cell was mainly free diffusion.
Live HeLa cell data
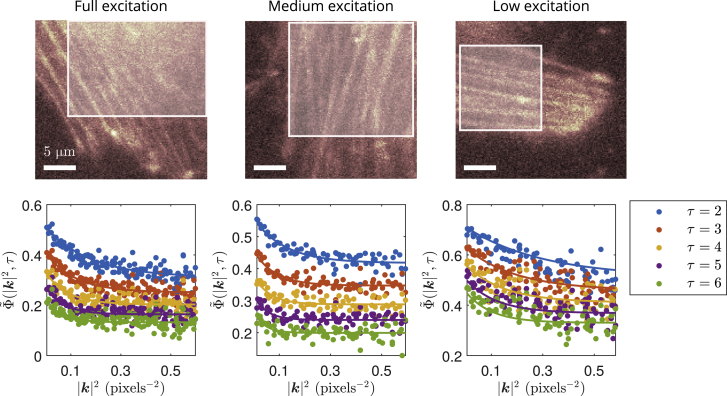
We proceeded to analyze Dronpa-C12-labeled β-actin in live HeLa cells imaged under different excitation intensities. Our analyses are shown in Fig. 5, with results given in Table 3.
Figure 5.
Example ACF analyses of independent HeLa cells irradiated at different excitation powers. Fluorescence images of Dronpa-C12-labeled actin in live HeLa cells, with ROI used in analyses highlighted, are shown in top part of figure. Corresponding computed ACFs from ROIs (points) and respective simultaneous fits (lines) are shown in bottom part of figure. The time-lag ranges for the fits varied from τ = 2 to either τ = 10 or τ = 20. Time windowing was done with TW = 100 frames. Pixel size is 177.78 nm. Frame time is 10 ms. Image series length is 50 s. Scale bars: 5 μm.
Table 3.
Fit parameters measured from independent HeLa cells at different excitation powers
| Excitation Power | D (μm2 s−1) | K (s−1) | ρon | fD |
| Full (∼24 mW) | 10.9 | 23.6 | 0.490 | 0.252 |
| 8.89 | 21.4 | 0.451 | 0.376 | |
| 12.8 | 22.8 | 0.416 | 0.322 | |
| Medium (∼1⩽ mW) | 12.8 | 18.5 | 0.545 | 0.230 |
| 10.9 | 16.6 | 0.495 | 0.323 | |
| 7.39 | 17.2 | 0.501 | 0.290 | |
| Low (∼14 mW) | 5.31 | 10.9 | — | — |
Different rows are ROI analyses of independent cells. Fits for ρon and fD at low power were omitted because of inconsistent fitted values when using different τ and |k|2 fitting ranges.
Measurements of the apparent diffusion coefficient from the HeLa cells are again within the range of ∼3–30 μm2 s−1 for G-actin diffusion in cytoplasm. Furthermore, the diffusion coefficients are about the same as we measured in the 3T3 fibroblast cell. The diffusion coefficient measured from the low-power data set, however, is lower than the ones measured at higher powers. One possible explanation for this observation is the lower excitation power would lead to lower excitation probabilities, especially outside of the focal plane. As such, the signal/noise ratio may not be sufficient to detect as many of the fluorescent proteins diffusing in 3D away from the focal plane, thus reducing the measured apparent diffusion coefficient (for instance, these events might be characterized by our analysis as photoblinking).
We also observed that the sum of the photoblinking rates K and on-time fraction ρon consistently increased with decreasing excitation intensity. Computing the mean on-time residency from Table 3,we obtained ton = 81 ± 3 ms at full excitation power and ton = 118 ± 1 ms at medium excitation power (note that we use the convention koff ≡ 1/ton). We also calculated the mean off-time to be toff = 99 ± 6 ms at full excitation and toff = 113 ± 7 ms at medium excitation. The increasing on-time, with decreasing excitation intensity, as well as the roughly constant off-time is characteristic behavior of any fluorophore because of the long-lived triplet state (or any similar dark state that depletes the ground singlet state) (56). This effect on the photoblinking rates as a function of excitation power was also observed in wild-type Dronpa (57).
We point out that Dronpa is expected to have a nonemissive state with a longer off-time (57), whereas we found . In fact, Habuchi et al. (57) found that wild-type Dronpa has three distinct dark states, of which one is significantly longer than the others. The toff values we measured can therefore depend on the residency times of multiple off-states. We make a simplifying argument to illustrate why our measurement may not be able to detect a much longer off-time. In our technique, we explicitly have ρon ≡ kon/K. If we now assume that kon is a sum of two rates, say and , such that , then we have. In other words, the rate corresponding to the longer time does not contribute to ρon and is thus not detected by our analysis.
At low power, the measured K is consistent with a longer on-time, assuming ton is the only characteristic time that is a function of excitation intensity. Note that we could not properly measure ρon and fD for the low-power data because different fitting ranges of τ and |k|2 would significantly affect these fitted parameters. This is, once more, likely due to the lower signal/noise ratio, causing the ACF to be noisier. In general, ρon and fD varied most among the fitted parameters when fitting the ACF over different ranges.
We also found a mean value for the diffusing fraction of fD = 0.30 ± 0.02. Blikstad and Carlsson (58) have previously reported values of unpolymerized actin measured from HeLa cell homogenates between 35 and 45%.
Note that the time-integrated 3D diffusion model did not fit our HeLa cell data well. This is possibly due to the non-negligible dead time (∼ ms) of the EMCCD camera detector relative to the shorter frame times used for imaging this data (10 ms). Instead of accounting for this effect in our model, we found that simply excluding the first time lag from our analyses gave reasonable fits.
Conclusions
We have presented an extended kICS fluorescence fluctuation rapid analysis method that simultaneously fits for the diffusion coefficient, photoblinking rates, and fraction of diffusing particles from a fluorescence image series. This is done independently from any other parameters. Unlike other image correlation techniques, our current approach can be applied to regions with nonuniform fluorophore distributions, including complex cellular morphologies. This enables us to increase spatial sampling across areas of the cell, which improves the statistical precision of the ACF and extends the dynamic range for transport coefficient measurement. Furthermore, we have shown through physically realistic simulations that we can obtain accurate fit results in the presence of photobleaching without having to consider its effects. We also demonstrated that our method can measure an apparent diffusion coefficient of Dronpa-C12-labeled actin in live NIH/3T3 and HeLa cell data that is consistent with previous literature values. We further observed that the fitted photoblinking parameters, measured from several independent HeLa cells, gave the expected trend as a function of excitation power. Last, our reported values for the diffusing fractions in both 3T3 and HeLa cells agree well with literature values. We anticipate that our technique will be useful in the study of dynamics in super-resolution because of its ability to analyze more intricate systems than previous image correlation methods.
In the future, we plan to apply our method to measure biomolecular binding kinetics because photoblinking and mean-field binding/unbinding are virtually analogous processes mathematically (under certain assumptions). Another potential application could be to use the measured photoblinking rates as probes for sensing changes in a cellular environment.
Author contributions
S.S. and P.W.W. designed research and wrote the manuscript. S.S. developed theory for method, analyzed data, and generated simulations. X.Y. and S.W. provided experimental data, provided super-resolution optical fluctuation imaging analysis, and contributed to method development.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We give special thanks to Paul De Konick (Laval University, Quebec City, Canada) for providing us with an image of a branched neuron that was used for generating simulations.
P.W.W. kindly acknowledges support of a Natural Sciences and Engineering Research Council of Canada Discovery Grant. S.W. was funded by the STROBE National Science Foundation Science and Technology Center (grant DMR-1548924) and by the National Science Foundation (grant CMI-1808766). The work from X.Y. was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344, release number: LLNL-JRNL-816054.
Editor: Andrey Klymchenko.
Footnotes
Supplemental material can be found online at https://doi.org/10.1016/j.bpr.2021.100015.
Supporting material
References
- 1.Huang B., Babcock H., Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sezgin E. Super-resolution optical microscopy for studying membrane structure and dynamics. J. Phys. Condens. Matter. 2017;29:273001. doi: 10.1088/1361-648X/aa7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone M.B., Shelby S.A., Veatch S.L. Super-resolution microscopy: shedding light on the cellular plasma membrane. Chem. Rev. 2017;117:7457–7477. doi: 10.1021/acs.chemrev.6b00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung B.O., Chou K.C. Review of super-resolution fluorescence microscopy for biology. Appl. Spectrosc. 2011;65:967–980. doi: 10.1366/11-06398. [DOI] [PubMed] [Google Scholar]
- 5.Gahlmann A., Moerner W.E. Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nat. Rev. Microbiol. 2014;12:9–22. doi: 10.1038/nrmicro3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betzig E., Patterson G.H., et al. Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 8.Dertinger T., Colyer R., et al. Enderlein J. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI) Proc. Natl. Acad. Sci. USA. 2009;106:22287–22292. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggeling C., Ringemann C., et al. Hell S.W. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 10.Honigmann A., Mueller V., et al. Eggeling C. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat. Commun. 2014;5:5412. doi: 10.1038/ncomms6412. [DOI] [PubMed] [Google Scholar]
- 11.Hedde P.N., Dörlich R.M., et al. Nienhaus G.U. Stimulated emission depletion-based raster image correlation spectroscopy reveals biomolecular dynamics in live cells. Nat. Commun. 2013;4:2093. doi: 10.1038/ncomms3093. [DOI] [PubMed] [Google Scholar]
- 12.Manley S., Gillette J.M., et al. Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 13.Balzarotti F., Eilers Y., et al. Hell S.W. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science. 2017;355:606–612. doi: 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]
- 14.Kisley L., Brunetti R., et al. Landes C.F. Characterization of porous materials by fluorescence correlation spectroscopy super-resolution optical fluctuation imaging. ACS Nano. 2015;9:9158–9166. doi: 10.1021/acsnano.5b03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magde D., Elson E., Webb W.W. Thermodynamic fluctuations in a reacting system-measurement by fluorescence correlation spectroscopy. Phys. Rev. Lett. 1972;29:705–708. [Google Scholar]
- 16.Elson E.L., Magde D. Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers. 1974;13:1–27. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 17.Magde D., Elson E.L., Webb W.W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 18.Krieger J.W., Singh A.P., et al. Wohland T. Imaging fluorescence (cross-) correlation spectroscopy in live cells and organisms. Nat. Protoc. 2015;10:1948–1974. doi: 10.1038/nprot.2015.100. [DOI] [PubMed] [Google Scholar]
- 19.Kolin D.L., Ronis D., Wiseman P.W. k-Space image correlation spectroscopy: a method for accurate transport measurements independent of fluorophore photophysics. Biophys. J. 2006;91:3061–3075. doi: 10.1529/biophysj.106.082768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolin D.L., Wiseman P.W. Advances in image correlation spectroscopy: measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochem. Biophys. 2007;49:141–164. doi: 10.1007/s12013-007-9000-5. [DOI] [PubMed] [Google Scholar]
- 21.Hebert B., Costantino S., Wiseman P.W. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Digman M.A., Brown C.M., et al. Gratton E. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys. J. 2005;89:1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoebe R.A., Van Oven C.H., et al. Manders E.M. Controlled light-exposure microscopy reduces photobleaching and phototoxicity in fluorescence live-cell imaging. Nat. Biotechnol. 2007;25:249–253. doi: 10.1038/nbt1278. [DOI] [PubMed] [Google Scholar]
- 24.Donnert G., Eggeling C., Hell S.W. Major signal increase in fluorescence microscopy through dark-state relaxation. Nat. Methods. 2007;4:81–86. doi: 10.1038/nmeth986. [DOI] [PubMed] [Google Scholar]
- 25.Mondal P.P., Gilbert R.J., So P.T.C. Photobleaching reduced fluorescence correlation spectroscopy. Appl. Phys. Lett. 2010;97:103704. [Google Scholar]
- 26.Song L., Hennink E.J., et al. Tanke H.J. Photobleaching kinetics of fluorescein in quantitative fluorescence microscopy. Biophys. J. 1995;68:2588–2600. doi: 10.1016/S0006-3495(95)80442-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Widengren J., Rigler R. Mechanisms of photobleaching investigated by fluorescence correlation spectroscopy. Bioimaging. 1996;4:149–157. [Google Scholar]
- 28.Kolin D.L., Costantino S., Wiseman P.W. Sampling effects, noise, and photobleaching in temporal image correlation spectroscopy. Biophys. J. 2006;90:628–639. doi: 10.1529/biophysj.105.072322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Digman M.A., Dalal R., et al. Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sehayek S., Gidi Y., et al. Wiseman P.W. A high-throughput image correlation method for rapid analysis of fluorophore photoblinking and photobleaching rates. ACS Nano. 2019;13:11955–11966. doi: 10.1021/acsnano.9b06033. [DOI] [PubMed] [Google Scholar]
- 31.Wachsmuth M., Waldeck W., Langowski J. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol. 2000;298:677–689. doi: 10.1006/jmbi.2000.3692. [DOI] [PubMed] [Google Scholar]
- 32.Sengupta P., Garai K., et al. Maiti S. Measuring size distribution in highly heterogeneous systems with fluorescence correlation spectroscopy. Biophys. J. 2003;84:1977–1984. doi: 10.1016/S0006-3495(03)75006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss M., Hashimoto H., Nilsson T. Anomalous protein diffusion in living cells as seen by fluorescence correlation spectroscopy. Biophys. J. 2003;84:4043–4052. doi: 10.1016/S0006-3495(03)75130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss M., Elsner M., et al. Nilsson T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 2004;87:3518–3524. doi: 10.1529/biophysj.104.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banks D.S., Fradin C. Anomalous diffusion of proteins due to molecular crowding. Biophys. J. 2005;89:2960–2971. doi: 10.1529/biophysj.104.051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsekouras K., Siegel A.P., et al. Pressé S. Inferring diffusion dynamics from FCS in heterogeneous nuclear environments. Biophys. J. 2015;109:7–17. doi: 10.1016/j.bpj.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown C.M., Dalal R.B., et al. Gratton E. Raster image correlation spectroscopy (RICS) for measuring fast protein dynamics and concentrations with a commercial laser scanning confocal microscope. J. Microsc. 2008;229:78–91. doi: 10.1111/j.1365-2818.2007.01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandão H.B., Sangji H., et al. Wiseman P.W. Measuring ligand-receptor binding kinetics and dynamics using k-space image correlation spectroscopy. Methods. 2014;66:273–282. doi: 10.1016/j.ymeth.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 39.Berne B.J., Pecora R. Dynamic Light Scattering: With Applications to Chemistry, Biology, and Physics. Dover Publications; Mineola, NY: 2000. Heterodyne Correlation Function for Particle Diffusion; pp. 57–62. [Google Scholar]
- 40.Yi X. UCLA; 2017. Super resolution of optical fluctuation imaging 2.0 (SOFI-2.0): towards fast super resolved imaging of live cells. PhD thesis. [Google Scholar]
- 41.Hirsch M., Wareham R.J., et al. Rolfe D.J. A stochastic model for electron multiplication charge-coupled devices – from theory to practice. PLoS One. 2013;8:e53671. doi: 10.1371/journal.pone.0053671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh A.P., Krieger J.W., et al. Wohland T. The performance of 2D array detectors for light sheet based fluorescence correlation spectroscopy. Opt. Express. 2013;21:8652–8668. doi: 10.1364/OE.21.008652. [DOI] [PubMed] [Google Scholar]
- 43.Colyer R.A., Scalia G., et al. Michalet X. High-throughput FCS using an LCOS spatial light modulator and an 8 × 1 SPAD array. Biomed. Opt. Express. 2010;1:1408–1431. doi: 10.1364/BOE.1.001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandracchia B., Hua X., et al. Jia S. Fast and accurate sCMOS noise correction for fluorescence microscopy. Nat. Commun. 2020;11:94. doi: 10.1038/s41467-019-13841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yip W.-T., Hu D., et al. Barbara P.F. Classifying the photophysical dynamics of single- and multiple-chromophoric molecules by single molecule spectroscopy. J. Phys. Chem. A. 1998;102:7564–7575. [Google Scholar]
- 46.Yeow E.K.L., Melnikov S.M., et al. Hofkens J. Characterizing the fluorescence intermittency and photobleaching kinetics of dye molecules immobilized on a glass surface. J. Phys. Chem. A. 2006;110:1726–1734. doi: 10.1021/jp055496r. [DOI] [PubMed] [Google Scholar]
- 47.Widengren J., Mets U., Rigler R. Fluorescence correlation spectroscopy of triplet states in solution: a theoretical and experimental study. J. Phys. Chem. 1995;99:13368–13379. [Google Scholar]
- 48.Geissbuehler S., Bocchio N.L., et al. Lasser T. Mapping molecular statistics with balanced super-resolution optical fluctuation imaging (bSOFI) Opt. Nanoscopy. 2012;1:4. [Google Scholar]
- 49.Yi X., Son S., et al. Weiss S. Moments reconstruction and local dynamic range compression of high order superresolution optical fluctuation imaging. Biomed. Opt. Express. 2019;10:2430–2445. doi: 10.1364/BOE.10.002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGrath J.L., Tardy Y., et al. Hartwig J.H. Simultaneous measurements of actin filament turnover, filament fraction, and monomer diffusion in endothelial cells. Biophys. J. 1998;75:2070–2078. doi: 10.1016/S0006-3495(98)77649-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zicha D., Dobbie I.M., et al. Dunn G.A. Rapid actin transport during cell protrusion. Science. 2003;300:142–145. doi: 10.1126/science.1082026. [DOI] [PubMed] [Google Scholar]
- 52.McDonald D., Carrero G., et al. Hendzel M.J. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J. Cell Biol. 2006;172:541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiuchi T., Nagai T., et al. Mizuno K. Measurements of spatiotemporal changes in G-actin concentration reveal its effect on stimulus-induced actin assembly and lamellipodium extension. J. Cell Biol. 2011;193:365–380. doi: 10.1083/jcb.201101035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novak I.L., Slepchenko B.M., Mogilner A. Quantitative analysis of G-actin transport in motile cells. Biophys. J. 2008;95:1627–1638. doi: 10.1529/biophysj.108.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gasilina A., Vitali T., et al. Randazzo P.A. The ArfGAP ASAP1 controls actin stress fiber organization via its N-BAR domain. iScience. 2019;22:166–180. doi: 10.1016/j.isci.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bretschneider S., Eggeling C., Hell S.W. Breaking the diffraction barrier in fluorescence microscopy by optical shelving. Phys. Rev. Lett. 2007;98:218103. doi: 10.1103/PhysRevLett.98.218103. [DOI] [PubMed] [Google Scholar]
- 57.Habuchi S., Ando R., et al. Hofkens J. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc. Natl. Acad. Sci. USA. 2005;102:9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blikstad I., Carlsson L. On the dynamics of the microfilament system in HeLa cells. J. Cell Biol. 1982;93:122–128. doi: 10.1083/jcb.93.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the kICS GitHub repository (https://github.com/ssehayek/kics-project.git) and the SOFI 2.0 GitHub repository (https://github.com/xiyuyi-at-LLNL/pysofi.git).