ABSTRACT
Lyssaviruses cause rabies, which is an acute neurological disease responsible for more than 59,000 human deaths annually and has no available effective treatments. The phosphoprotein (P) of lyssaviruses (lyssavirus-P) plays multiple roles in virus replication and immune evasion. Lyssavirus-P has been identified as the major type I interferon (IFN-I) antagonist, while the precise site and precise molecular mechanism remain unclear. Herein, we found that substitution of site 179 of lyssavirus-P from serine (Ser) to proline (Pro) impairs its antagonism function of IFN-I by sequence alignment and site mutations. Subsequent studies demonstrated that lyssavirus-P containing S179 specifically interacted with I-kappa B kinase ε (IKKε). Specifically, lyssavirus-P containing S179 interacted simultaneously with the kinase domain (KD) and scaffold dimerization domain (SDD) of IKKε, competing with TNF receptor-associated factor 3 (TRAF3) and IFN regulatory factor 3 (IRF3) for binding with IKKε, leading to the inhibition of IFN production. Furthermore, S179 was involved in the viral pathogenicity of the typical lyssavirus rabies virus in a mouse model. Interestingly, we found that S179 is conserved among most lyssavirus-P and functional for IFN antagonism. Collectively, we identified S179 of lyssavirus-P is essential for IFN-I inhibition, which provides deep insight into the immune evasion strategies of lyssaviruses.
IMPORTANCE Interferon (IFN) and the IFN-induced cellular antiviral response constitute the first line of defense against viral invasion. Evading host innate immunity, especially IFN signaling, is the key step required for lyssaviruses to establish infection. In this study, S179 of lyssavirus phosphoprotein (lyssavirus-P) was identified as the key site for antagonizing IFN-I production. Mechanistically, lyssavirus-P containing S179 specifically targets the key kinase IKKε and disrupts its interaction with TRAF3 and IRF3. S179P mutation in the P protein of the typical lyssavirus rabies virus (RABV) attenuated its pathogenicity in a mouse model. Our findings provide deep insight into the immune evasion strategies of lyssaviruses, which is helpful for the development of effective antiviral therapeutics.
KEYWORDS: lyssavirus, phosphoprotein, type I interferon, IKKε, immune evasion
INTRODUCTION
The genus Lyssavirus is a diverse group of viruses capable of causing rabies and includes rabies virus (RABV), the classical lyssavirus (genotype 1), and other rabies-related viruses, such as Lagos Bat virus (LBV, genotype 2), Mokola virus (MOKV, genotype 3), Duvenhage virus (DUVV, genotype 4), European Bat Lyssaviruses 1 and 2 (EBLV-1 and EBLV-2, genotypes 5 and 6, respectively), and Australian Bat Lyssavirus (ABLV, genotype 7) (1–8). As a typical lyssavirus, RABV causes fatal encephalitis that leads to more than 59,000 human deaths worldwide every year, which results in heavy economic and social losses (9). RABV is an enveloped, nonsegmented, and single-stranded RNA virus belonging to the genus Lyssavirus within the family Rhabdoviridae (10). Its genome encodes five proteins in the following order: a nucleoprotein (N), a phosphoprotein (P), a matrix protein (M), a single external surface glycoprotein (G), and an virus RNA polymerase (L) (11, 12). In most cases, RABV transmits through bites by rabid dogs. From the entry site, RABV secreted into saliva moves along the peripheral nervous system and eventually reaches the central nervous system (CNS) via retrograde axonal transport (13). Rabies is almost always fatal after the onset of symptoms, and no effective treatment is currently available.
The innate immune response acts as the first line of defense against viral invasion. Specific pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and NOD-like receptors (NLRs), recognize conserved molecular structures known as pathogen-associated molecular patterns (PAMPs). Activation of these molecules leads to the production of chemokines, cytokines, and IFNs (14). Previous studies have shown that RABV infection is recognized by RIG-I (15, 16). Upon initiation, RIG-I induces downstream signaling by binding to an adaptor molecule, mitochondrial antiviral signaling (MAVS) protein, which recruits TANK-binding kinase 1 (TBK1) and IKKε (17). Activated TBK1 and IKKε catalyze the phosphorylation and dimerization of IRF3 in the cytoplasm. Subsequently, IRF3 translocates into the nucleus and activates the transcription of IFNs (18). Secreted IFN-I binds to a common receptor, leading to the activation of the Janus kinase (JAK) signal transducers and activators of transcription (STAT) signaling pathway (19). STATs heterodimerize and bind to the DNA binding protein IFN regulatory factor 9 (IRF9) to form a complex called IFN-stimulated growth factor 3 (ISGF3). The heterotrimer complexes translocate into the nucleus and bind to the IFN-stimulated response element (ISRE), subsequently inducing the production of hundreds of interferon-stimulated genes (ISGs) (20, 21). IFNs act on target cells to confer resistance to viral infection. However, lyssaviruses have evolved multiple strategies of immune evasion. Previous studies have shown that lyssavirus-P functions as the major IFN antagonist, counteracting not only IFN production but also IFN signaling pathways (22).
RABV-P is a multifunctional modular protein with diverse essential tasks during the viral life cycle. First, RABV-P is an essential noncatalytic cofactor of virus RNA polymerase L (23). The 19 N-terminal residues of RABV-P bind to the L protein, promoting polymerase function (24). RABV-P contains two N-binding domains: one is located in the C-terminal part of the protein and another between amino acids 69 and 177 (25, 26). In addition, RABV-P is indispensable for the formation of RABV Negri bodies, where viral transcription and replication take place (27–29). In addition to transcription and replication, RABV-P also plays a central role in the network of viral and host protein interactions. RABV-P interacts with ribosomal protein L9, focal adhesion kinase, and nucleolin to regulate virus replication and infection (30, 31). In addition, RABV-P interacts with mitochondrial complex I and induces mitochondrial dysfunction and oxidative stress (32–34). Importantly, RABV-P is widely recognized as an IFN antagonist that prevents both transcriptional inductions of IFN and IFN-mediated JAK/STAT signaling. The pioneering studies showed that RABV-P impairs TBK1- and IKKε-mediated IRF3 activation and the region of 176 to 186 amino acid (aa) is indispensable for IRF3 inhibition (35–37). RABV-P protein also prevents JAK/STAT signaling through several mechanisms, such as interacting with STAT1 and inhibiting IFN signal transduction pathways (38). Finally, RABV-P interacts with promyelocytic leukemia protein (PML), which is an ISG that has antiviral effects on RABV, causing its mislocalization to the cytoplasm and leading to the disruption of PML (39, 40). However, the detailed mechanism underlying how RABV-P inhibits IFN production and affects viral pathogenicity is not completely understood.
In this study, we compare the ability of the P protein from the laboratory-attenuated RABV strain CVS-B2c (challenge virus standard [CVS]) and the wild-type RABV strain DRV-Mexico (DRV) to inhibit IFN-I production. We find that aa S179 plays an important role in preventing IRF3 activation. Specifically, S179 of RABV-P targets the key kinase IKKε to disrupt its interaction with both TRAF3 and IRF3, thereby blocking the production of IFN-I. Furthermore, we demonstrate that the mutation of S179 in RABV-P affects viral pathogenicity in a mouse model. More importantly, we find that S179 is highly conserved among most lyssavirus P proteins, which play an important role in antagonizing IFN signaling.
RESULTS
DRV-P possesses a stronger ability to inhibit IRF3 activation than CVS-P.
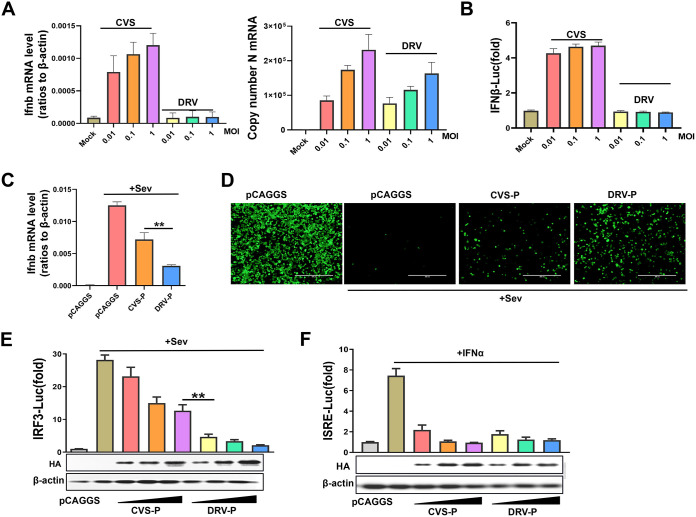
Our previous study suggested that the lab-attenuated RABV strain CVS induces more cytokine and IFN production than the wild-type RABV strain DRV in astrocytes (41). To determine the difference in IFN-β production induced by CVS and DRV, BV2 (mouse microglioma) cells were infected with an indicated multiplicity of infection (MOI) of CVS and DRV for 36 h, and a quantitative PCR (qPCR) assay was performed to evaluate the Ifnb mRNA level and viral replication. As shown in Fig. 1A, CVS, but not DRV, significantly induced the expression of Ifnb mRNA. HEK-293T (human embryonic kidney, 293T) cells were transfected with the firefly luciferase reporter plasmid IFN-β-Luc and Renilla luciferase reporter plasmid pRL-TK (as an internal control) for 12 h, followed by infection with CVS or DRV at the indicated MOI for 36 h. The cell lysates were collected and subjected to a dual-luciferase reporter assay. The results showed that CVS, but not DRV, significantly upregulated the transcription of Ifnb mRNA (Fig. 1B).
FIG 1.
Differential abilities of CVS-P and DRV-P to inhibit IRF3 activity. (A) BV2 cells were infected with CVS or DRV at an MOI of 0.01, 0.1, or 1 for 36 h, and the Ifnb mRNA and viral replication levels were evaluated by qPCR. (B) 293T cells were transfected with IFN-β-Luc and pRL-TK for 12 h and infected with CVS or DRV at an MOI of 0.01, 0.1, or 1 for 36 h. A dual-luciferase reporter assay was performed to investigate the activation of IFN-β-Luc. (C and D) 293T cells were transfected with CVS-P, DRV-P, or the empty vector (pCAGGS) and then maintained as uninfected or infected with SeV (50 hemagglutination units per mL) for 12 h. The cells were collected for qPCR (C), and the supernatants were subjected to UV irradiation treatment, followed by addition to a monolayer of 293T cells in a 24-well plate for 24 h. The cells were infected with VSV-GFP for 12 h, and viral replication in the cells was evaluated under a fluorescence microscope (D). (E) 293T cells were cotransfected with increasing amounts of CVS-P or DRV-P along with IRF3-Luc and pRL-TK for 24 h and maintained as uninfected or infected with SeV for 12 h. (F) HEK-293T cells were cotransfected with increasing amounts of CVS-P or DRV-P along with ISRE-Luc and pRL-TK for 24 h and untreated or treated with IFN-α for 12 h. A dual-luciferase reporter assay was performed to evaluate the activation of IRF3-Luc or ISRE-Luc. Statistical analyses of grouped comparisons were carried out by one-way ANOVA (**, P < 0.01). The error bars represent the standard deviations (SDs); n = 3. Results are representative of two independent assays.
RABV-P is generally considered to be an antagonist of IFN production; thus, we hypothesized that the P proteins of the two RABV strains are responsible for this difference. Next, 293T cells were cotransfected with CVS-P, DRV-P, or the empty vector and treated with Sedai virus (SeV) for 12 h. The mRNA levels of Ifnb were quantified by qPCR. Both CVS-P and DRV-P inhibited SeV-induced Ifnb mRNA transcription, but DRV-P showed a stronger ability to inhibit Ifnb mRNA levels than CVS-P (Fig. 1C). Additionally, an IFN bioassay was performed using an IFN-sensitive vesicular stomatitis virus encoding a green fluorescent protein reporter (VSV-GFP) in 293T cells. The cellular supernatants from SeV-infected cells notably restricted the replication of VSV-GFP. Cells with supernatants expressing DRV-P, rather than CVS-P, were more vulnerable to VSV-GFP infection, which indicated that DRV-P antagonizes IFN production more than CVS-P (Fig. 1D).
RABV-P has been found to inhibit the JAK-STAT pathway, which is downstream of the IFN signaling pathway (38). To investigate whether CVS-P and DRV-P differ in inhibition of JAK-STAT pathway, 293T cells were cotransfected with increasing amounts of CVS-P or DRV-P along with IRF3-Luc or ISRE-Luc and pRL-TK for 24 h, followed by treatment with SeV or IFN-α for 12 h. According to reporter assays, DRV-P more significantly inhibited IRF3-Luc activation than CVS-P (Fig. 1E) but inhibited ISRE-Luc activation to the same extent (Fig. 1F). Together, these results demonstrate that DRV-P possesses a stronger ability to inhibit IRF3 activation than CVS-P.
S179 of RABV-P is essential for the inhibition of IRF3 activation.
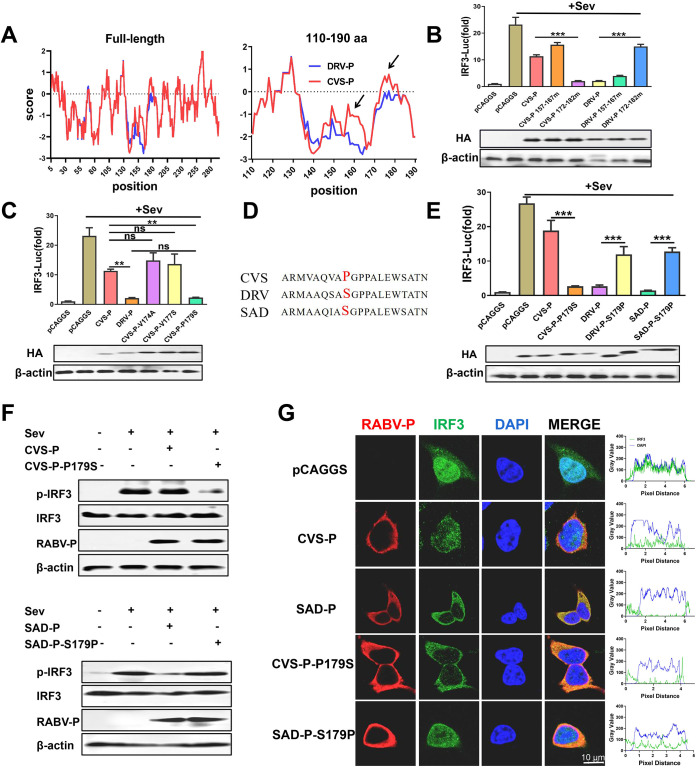
A previous study has shown that the region 176 to 186 of P protein is required for efficient inhibition of IFN induction (36). To clarify how CVS-P and DRV-P differ in the disruption of IRF3 activation, we decided to start by analyzing the protein structure of these two proteins. Considering that neither CVS-P nor DRV-P has the crystal structure data, hydrophobicity is considered to be the most important factor affecting protein structure (42, 43). Therefore, we analyzed and compared the hydrophobicity of the CVS-P and DRV-P protein sequences. The results showed that two regions (157 to 167 aa and 172 to 182 aa) had obvious differences in hydrophobicity (Fig. 2A). To explore which region caused the observed differences, we interchanged the two regions between CVS-P and DRV-P and investigated the inhibitory effect on IRF3 activation by a dual-luciferase reporter assay. The results suggested that the replacement of 172 to 182 aa, but not 157 to 167 aa, was responsible for the differences in IRF3 inhibition (Fig. 2B). There are three different amino acids in the 172- to 182-aa sequences of CVS-P and DRV-P (a valine [Val, V] at 174, a valine [Val, V] at 177, and a proline [Pro, P] at 179 in CVS-P and an alanine [Ala, A] at 174, a Ser at 177, and a Ser at 179 in DRV-P). Next, three mutants, CVS-P-V174A (Val to Ala substitution at CVS-P-179), CVS-P-V177S (Val to Ser substitution at CVS-P-179), and CVS-P-P179S (Pro to Ser substitution at CVS-P-179), were constructed and subjected to a dual-luciferase reporter assay. The results showed that the mutation of Pro to Ser at site 179 in P played a significant role in determining the differences in IRF3 inhibition (Fig. 2C).
FIG 2.
S179 of RABV-P determines its inhibitory effect on IRF3 activation. (A) Hydrophobicity comparison of the protein sequences of CVS-P and DRV-P (left, full-length comparison; right, 110- to 190-aa comparison). (B) 293T cells were cotransfected with IRF3-Luc and pRL-TK along with CVS-P, CVS-P 157 to 167 mutant (mut), CVS-P 172 to 182 mut, DRV-P, DRV-P 157 to 167 mut, DRV-P 172 to 182 mut, or pCAGGS for 24 h and maintained as uninfected or infected with SeV for 12 h. A dual-luciferase reporter assay was performed to investigate the activation of the IRF3 promoter (above). The expression of P and β-actin was detected by a Western blot analysis with antibodies against HA and β-actin, respectively (below). (C) Dual-luciferase reporter assay (above) and Western blot assay (below) (as described in panel B) of 293T cells transfected with CVS-P-V174A, CVS-P-V174A, CVS-P-P179S, or pCAGGS. (D) Sequence alignment of 179 aa of P proteins from CVS, DRV, and SAD. (E) Dual-luciferase reporter assay (above) and Western blot assay (below) (as described in panel B) of 293T cells transfected with CVS-P, CVS-P-P179S, DRV-P, DRV-P S179P, SAD-P, SAD-P-S179P, or pCAGGS. (F) 293T cells were transfected with CVS-P, CVS-P-P179S, SAD-P, SAD-P-S179P, or pCAGGS for 24 h and maintained as uninfected or infected with SeV. At 12 h after infection, the cells were lysed and subjected to the Western blot analysis with antibodies against phosphorylated IRF3 (p-IRF3 Ser386), total IRF3, RABV-P, or β-actin. (G) 293T cells were treated as described in panel F. The cells were fixed and subjected to an immunofluorescence assay with rabbit anti-IRF3 and mouse anti-RABV-P, followed by treatment with Alexa 594-conjugated anti-mouse IgG (red) or Alexa 488-conjugated anti-rabbit IgG (green) secondary antibodies. The nuclei were stained with DAPI (blue). Scale bar, 10 μm. Statistical analyses of grouped comparisons were carried out by one-way ANOVA (**, P < 0.01; ***, P < 0.001; ns, no significant difference). The error bars represent the SDs; n = 3. Results are representative of two independent assays.
RABV-P of the vaccine strain SAD-L16 (SAD-P) has been reported to effectively block IRF3 activation and has a Ser at 179 aa (35). Therefore, the mutants DRV-P-S179P (Ser to Pro at DRV-P-179) and SAD-P-S179P (Ser to Pro at SAD-P-179) were generated, and a dual-luciferase reporter assay was conducted (Fig. 2D and E). CVS-P-P179S, DRV-P, and SAD-P significantly blocked IRF3 activation, among which SAD-P had the most inhibitory effect. However, DRV-P-S179P and SAD-P-S179P showed the same inhibitory effect as CVS-P (Fig. 2E). IRF3 is a key transcription factor for the production of IFN-I and is activated via phosphorylation and nuclear translocation (44). Thus, the effect of P on IRF3 phosphorylation and nuclear translocation was further evaluated by Western blotting and indirect immunofluorescence assays (IFAs). The introduction of CVS-P and SAD-P-S179P had a slight influence on SeV-induced IRF3 phosphorylation, whereas CVS-P-P179S and SAD-P markedly downregulated the level of IRF3 phosphorylation (Fig. 2F). Consistent with the Western blot results, CVS-P-P179S and SAD-P showed stronger inhibition of IRF3 nuclear translocation than CVS-P and SAD-P-S179P in the IFAs (Fig. 2G). Together, the results support that S179 of RABV-P is essential for P blockage of IRF3 activation.
S179 of RABV-P is essential for the interaction between RABV-P and IKKε.
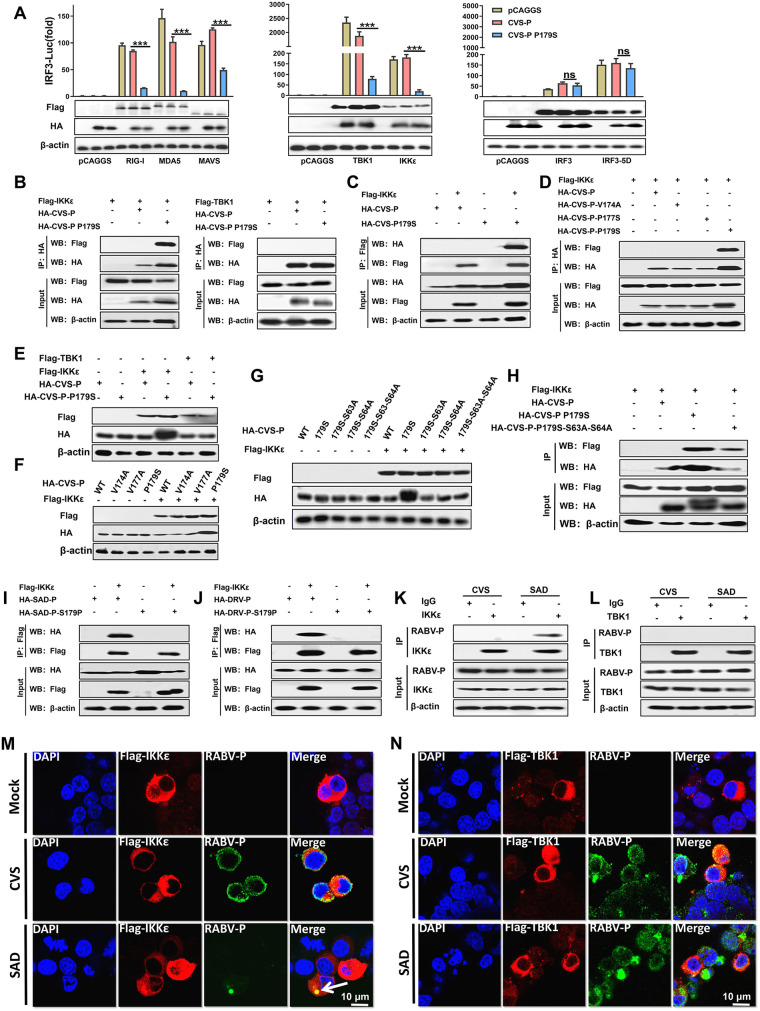
To investigate whether RABV-P containing S179 targeted IRF3 itself or its upstream activation steps, 293T cells were cotransfected with CVS-P or CVS-P-P179S and key molecules of the RLR signaling pathway, including RIG-I, MDA-5, MAVS, TBK1, IKKε, IRF3, or the constitutively active phosphorylation mimetic IRF3-5D (45), for dual-luciferase reporter assays. The results suggested that RIG-I-, MDA-5-, MAVS-, TBK1-, and IKKε-mediated IRF3-Luc activation was blocked by CVS-P and CVS-P-P179S to different degrees, whereas IRF3- and IRF3-5D-mediated IRF3-Luc activation was not affected by CVS-P or CVS-P-P179S, which indicated that S179-RABV-P exerted its function at the TBK1/IKKε level (Fig. 3A). The results of coimmunoprecipitation (co-IP) revealed that IKKε, rather than TBK1, was specifically detected in isolated immune complexes with CVS-P-P179S (Fig. 3B). In the reverse co-IP experiment, CVS-P-P179S was efficiently coimmunoprecipitated by IKKε, (Fig. 3C). In addition, CVS-P-V174A and CVS-P-V177S did not interact with IKKε (Fig. 3D).
FIG 3.
S179 of RABV-P is critical for its interaction with IKKε. (A) 293T cells were cotransfected with HA-CVS-P or HA-CVS-P-P179S and Flag-RIG-I, -MDA-5, -MAVS, -TBK1, -IKKε, -IRF3, the constitutively active phosphorylation mimetic -IRF3-5D, or pCAGGS. Dual-luciferase reporter assay (above) and Western blot (WB; below) assays were performed (as described in Fig. 2B). (B) 293T cells were cotransfected with Flag-TBK1 or Flag-IKKε and HA-CVS-P, HA-CVS-P-P179S, or pCAGGS. At 48 h after transfection, the cell lysates were harvested for co-IP assays with antibodies against HA (IP: HA). Immunoprecipitation (IP, above) and whole-cell lysate (input, below) complexes were analyzed by Western blotting using antibodies against Flag, HA, or β-actin. (C) 293T cells were cotransfected with HA-CVS-P or HA-CVS-P-P179S and Flag-IKKε or pCAGGS for 48 h, and the cell lysates were prepared and subjected to immunoprecipitation analysis with antibodies against Flag (IP: Flag). The input and IP complexes were analyzed as described in panel B. (D) 293T cells were cotransfected with Flag-IKKε and HA-CVS-P, HA-CVS-P-V174A, CVS-P-V174A, HA-CVS-P-P179S, or pCAGGS for 48 h. (E to G) A co-IP assay with antibodies against HA (IP: HA) was performed as described in panel B. 293T cells were cotransfected with HA-CVS-P or HA-CVS-P-P179S and Flag-TBK1, Flag-IKKε, or pCAGGS (E); HA-CVS-P, HA-CVS-P-V174A, HA-CVS-P-V174A, or HA-CVS-P-P179S and Flag-IKKε or pCAGGS (F); and HA-CVS-P, HA-CVS-P-P179S, HA-CVS-P179S-S63A (an Ala to Ser substitution at 63 based on CVS-P-P179S), HA-CVS-P179S-S64A (an Ala to Ser substitution at 64 based on CVS-P-P179S), HA-CVS-P179S-S63A-S64A (an Ala to Ser substitution at 63 and 64 based on CVS-P-P179S), and Flag-IKKε and pCAGGS (G), and Western blot assays were performed at 48 h posttransfection. (H) 293T cells were cotransfected with Flag-IKKε and HA-CVS-P, HA-CVS-P-P179S, HA-CVS-P179S-S63-S64A, or pCAGGS for 48 h. A co-IP assay with antibodies against HA (IP: HA) was performed as described above. (I and J) 293T cells were cotransfected with HA-SAD-P or HA-SAD-P-S179P, HA-DRV-P or HA-DRV-P S179P, and Flag-IKKε or pCAGGS for 48 h, and a co-IP assay with anti-Flag (IP: Flag) was performed as described in panel B. (K and L) N2a cells were mock infected or infected with CVS or SAD at an MOI of 1 for 24 h. A co-IP assay with antibodies against IKKε, TBK1, or IgG (control) was performed as described in panel B. (M and N) N2a cells were transfected with Flag-IKKε and Flag-TBK1 for 12 h and mock infected or infected with CVS or SAD at an MOI of 1 for 24 h. The cells were fixed and subjected to confocal microscopy with anti-Flag (IKKε, red), anti-RABV-P (P, green), and DAPI (nucleus, blue). Scale bar = 10 μm. Statistical analyses of grouped comparisons were carried out by one-way ANOVA (***, P < 0.001; ns, no significant difference). The error bars represent the SDs; n = 3. Results are representative of two independent assays.
Notably, we found that the expression level of CVS-P containing S179 was sharply increased when coexpressed with IKKε (Fig. 3B to D). To confirm this observation, HEK-293T cells were transfected with CVS-P or CVS-P-P179S and IKKε, TBK1, or pCAGGS for Western blot analysis. The results suggested that the expression of CVS-P-P179S increased significantly in the presence of IKKε but not in the presence of TBK1 (Fig. 3E). In addition, HEK-293T cells were transfected with IKKε or pCAGGS and CVS-P, CVS-P-V174A, CVS-P-V177S, or CVS-P-P179S. As shown in Fig. 3F, only the expression of CVS-P-P179S increased in the presence of IKKε, while the parent and the other two mutants did not. A previous study showed that the phosphorylation at S63 and S64 of CSV-P protein increased the expression level of CVS-P (46, 47). Consequently, we generated three mutants based on CVS-P-P179S: CVS-P179S-S63A (Ala to Ser substitution at 63), CVS-P179S-S64A (Ala to Ser substitution at 64), and CVS-P179S-S63A-S64A (Ala to Ser substitution at 63 and 64). Interestingly, the results suggested S63 and S64 mutants alone and in combination abolished this alteration in P protein (Fig. 3G). Importantly, the co-IP assay results implied that both CVS-P-P179S and CVS-P179S-S63A-S64A could interact with IKKε, whereas CVS-P-P179S could pull down more IKKε (Fig. 3H).
SAD-P, SAD-P-S179P, DRV-P, and DRV-P-S179P were also subjected to parallel co-IP experiments. As expected, interactions of IKKε with SAD-P and DRV-P were observed, but the mutation of Pro to Ser at P-179 (SAD-P-S179P and DRV-P-S179P) led to dissociation between RABV-P and IKKε (Fig. 3I and J).
To further investigate whether P was able to interact with IKKε and TBK1 in RABV-infected cells, N2a (mouse neuroblastoma) cells were mock infected or infected with CVS or SAD for a co-IP assay. The results suggested that P interacted with IKKε in the case of SAD infection but not CVS infection (Fig. 3K), but no interaction between RABV-P and TBK1 was observed in either SAD- or CVS-infected cells (Fig. 3L). Furthermore, N2a cells were transfected with Flag-IKKε and Flag-TBK1 for 12 h and mock infected or infected with CVS or SAD for confocal microscopy assays. The results also demonstrated that P from SAD, but not CVS, colocalized with IKKε (Fig. 3M). However, no colocalization was observed in either SAD- or CVS-infected cells (Fig. 3N). Taken together, these results indicated that S179 of RABV-P is essential for the interaction between P and IKKε.
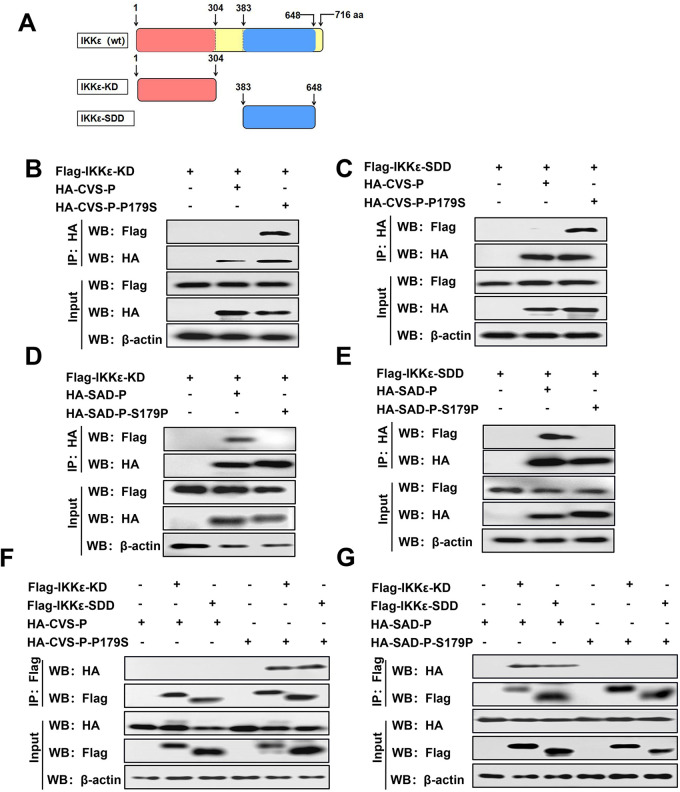
RABV-P containing S179 interacts with the KD and SDD domains of IKKε.
IKKε contains two key domains, KD (1 to 304 aa) and SDD (384 to 648 aa) (Fig. 4A) (48). To map the domains of IKKε that are necessary for its interaction with RABV-P containing S179, co-IP experiments were performed by cotransfection with Flag-IKKε-KD or Flag-IKKε-SDD and HA-CVS-P, HA-CVS-P-P179S, or pCAGGS. CVS-P-P179S interacted with both IKKε-KD and IKKε-SDD, while CVS-P did not interact with either (Fig. 4B and C). Likewise, we executed the same experiment with HA-SAD-P and HA-SAD-P-S179P. SAD-P interacted with both IKKε-KD and IKKε-SDD, but no association between SAD-P-S179P and either domain was observed (Fig. 4D and E). In reverse co-IP experiments, IKKε-KD and IKKε-SDD also coimmunoprecipitated with CVS-P-P179S and SAD-P but not CVS-P or SAD-P-S179P (Fig. 4F and G). These results illustrate that RABV-P containing S179 interacts with the KD and SDD domains of IKKε, while RABV-P containing P179 does not.
FIG 4.
RABV-P containing S179 interacts with the KD and SDD domains of IKKε. (A) Schematic of full-length IKKε and its truncated mutants. (B to E) 293T cells were cotransfected with Flag-IKKε-KD and HA-CVS-P, HA-CVS-P-P179S, or pCAGGS (B); Flag-IKKε-SDD and HA-CVS-P, HA-CVS-P-P179S, or pCAGGS (C); Flag-IKKε-KD and HA-SAD-P, HA-SAD-P-S179P, or pCAGGS (D); and Flag-IKKε-SDD and HA-SAD-P, HA-SAD-P-S179P, or pCAGGS (E); and co-IP assays with antibodies against HA (IP: HA) were performed as described in Fig. 3B. (F and G) 293T cells were cotransfected with HA-CVS-P or HA-CVS-P-P179S and Flag-IKKε-KD, Flag-IKKε-SDD, or pCAGGS (F) and with HA-SAD-P or HA-SAD-P-S179P and Flag-IKKε-KD, Flag-IKKε-SDD, or pCAGGS (G), and a co-IP assay with antibodies against Flag (IP: Flag) was performed as described in Fig. 3B. Results are representative of two independent assays.
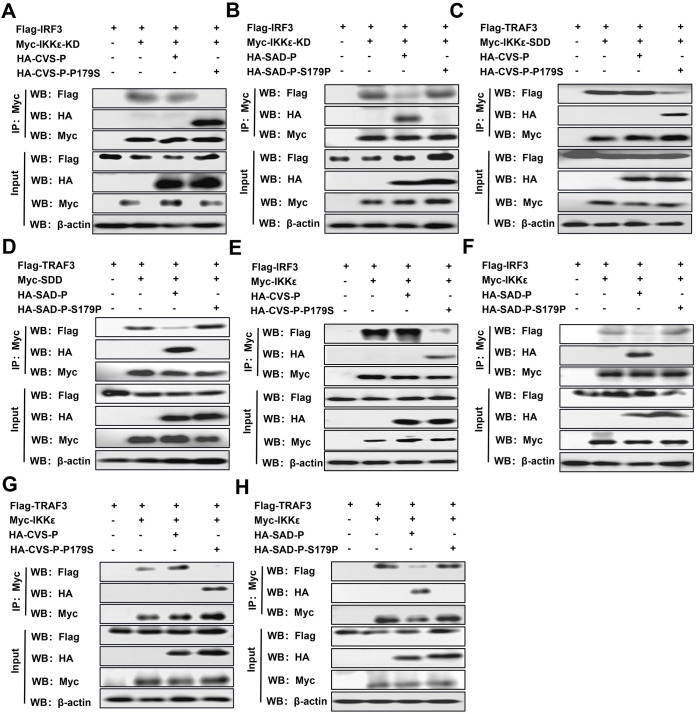
RABV-P containing S179 competes with IRF3 and TRAF3 to bind IKKε.
KD and SDD, as the two functional domains of IKKε, interact with the downstream component IRF3 and the upstream molecule TRAF3, respectively (48). Because RABV-P containing S179 simultaneously binds to the KD and SDD domains of IKKε, RABV-P containing S179 may compete with IRF3 and TRAF3 for binding to IKKε. To confirm this hypothesis, competitive co-IP experiments were performed by cotransfecting IRF3 with IKKε-KD (Fig. 5A and B) or full-length IKKε (Fig. 5E and F) and TRAF3 with IKKε-SDD (Fig. 5C and D) or full-length IKKε (Fig. 5G and H), along with the indicated P. The results demonstrated that IRF3 and TRAF3 were coimmunoprecipitated by KD, SDD, and full-length IKKε. However, the levels of TRAF3 and IRF3 in isolated immune complexes were significantly reduced in the presence of CVS-P-P179S or SAD-P compared to CVS-P and SAD-P-S179P. Taken together, these results indicate that RABV-P with S179 partially functions to disrupt the binding of IKKε with IRF3 and TRAF3.
FIG 5.
(A to H) RABV-P containing S179 interferes with the interaction of IKKε with IRF3 and TRAF3. 293T cells were cotransfected with Flag-IRF3 and Myc-IKKε-KD or pCAGGS along with HA-CVS-P, HA-CVS-P-P179S, HA-SAD-P, HA-SAD-P-S179P, or pCAGGS (A and B); Flag-TRAF3 and Myc-IKKε-SDD or pCAGGS along with P as described above (C and D); Flag-IRF3 and Myc-IKKε or pCAGGS along with P as described above (E and F); and Flag-TRAF3 and Myc-IKKε or pCAGGS along with P described above (G and H); and a co-IP assay with antibodies against HA (IP: Myc) was performed as described in Fig. 3B. Results are representative of two independent assays.
S179 of RABV-P is essential for the inhibition of IFN production.
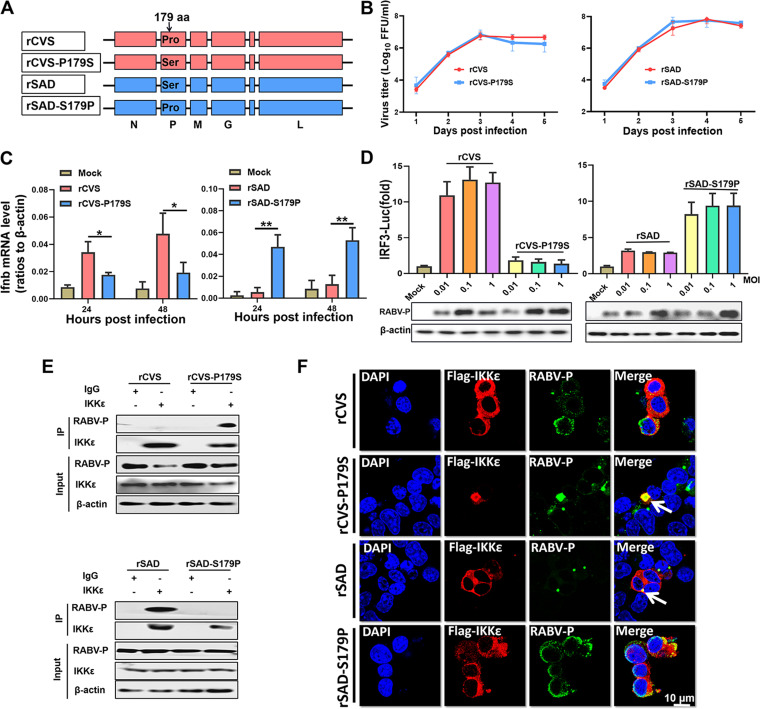
To further investigate whether S179 of RABV-P also plays a role in RABV infection, we constructed the two mutant viruses rCVS-P179S (Pro to Ser substitution at P-179) and rSAD-S179P (Ser to Pro substitution at P-179) based on the recombinant RABV strains CVS-B2c and SAD-L16, respectively (Fig. 6A). Viral growth kinetics showed that the mutant virus replication processes in cells were similar (Fig. 6B). BV2 and 293T cells were infected with the viruses, and qPCR and dual-luciferase reporter assays were performed to evaluate the mRNA level of Ifnb and the activation of IRF3, respectively. Compared with rCVS, rCVS-P179S could not induce Ifnb expression (Fig. 6C) and IRF3 activation (Fig. 6D), while rSAD-S179P could activate the IRF3 pathway compared with rSAD.
FIG 6.
S179 of P is critical for RABV inhibition of IRF3 activation. (A) Four parent and mutant viruses, rCVS (recombinant CVS), rCVS-P179S (A Pro to Ser substitution at P-179), rSAD (recombinant SAD, and rSAD-S179P (A Ser to Pro substitution at P-179), were constructed and rescued. (B) Growth kinetics in BSR cells (MOI = 0.01). (C) BV2 cells were infected with rCVS, rCVS-P179S, rSAD, or rSAD-S179P at an MOI of 1 for 24 h or 48 h, and the Ifnb mRNA levels were quantified by qPCR. (D) 293T cells were infected with the four viruses at an MOI of 0.01, 0.1, or 1 for 36 h. A dual-luciferase reporter assay (above) and Western blot assay (below) were performed as described in Fig. 2B. (E) N2a cells were mock infected or infected with the four viruses at an MOI of 1 for 24 h. A co-IP assay with antibodies against IKKε or IgG (control) was performed. (F) N2a cells were transfected with Flag-IKKε for 12 h and infected with the four viruses at an MOI of 1 for 24 h. A confocal microscopy assay was performed as described in Fig. 3H. Statistical analyses of grouped comparisons were carried out by one-way ANOVA (*, P < 0.05; **, P < 0.01). The error bars represent the SDs; n = 3. Results are representative of two independent assays.
Furthermore, N2a cells were mock infected or infected with four viruses, respectively, for co-IP assays with an IKKε antibody. The results suggested that P interacted with endogenous IKKε in the case of rCVS-P179S and rSAD infection but not rCVS and rSAD-S179P infection (Fig. 6E). In addition, N2a cells transfected with Flag-IKKε and then infected with four viruses, respectively, were examined by confocal microscopy. The results also suggested that P colocalized with IKKε in rCVS-P179S- and rSAD-infected cells but not in rCVS- and rSAD-S179P-infected cells (Fig. 6F). Collectively, these results establish that S179 of RABV-P is required for RABV to interfere with IKKε and inhibit IRF3 activation.
S179 of RABV-P affects viral pathogenicity in a mouse model.
To investigate the contribution of S179 of lyssavirus-P to pathogenicity in vivo, we compared the pathogenicity of parent and mutant viruses in a mouse model. C57BL/6 mice were intradermally (i.d.) infected in the ear with 2.5 × 104 focus-forming unit (FFU) of rCVS or rCVS-P179S or intracerebrally (i.c.) infected with 2.5 × 103 FFU of rSAD or rSAD-S179P, and body weight changes, clinical scores, and survival ratios were monitored daily. The results showed that the weight loss (Fig. 7A) and clinical symptoms (Fig. 7B) of rCVS-P179S-infected mice were more severe than those of rCVS-infected mice. Correspondingly, rCVS-P179S-infected mice developed an earlier onset and a higher mortality than rCVS-infected mice (no significance, P = 0.0723) (Fig. 7C). In contrast, the weight loss (Fig. 7D) and clinical symptoms (Fig. 7E) of rSAD-S179P-infected mice were significantly milder than those of SAD-infected mice. Correspondingly, the survival ratio of rSAD-S179P-infected mice was significantly increased compared with that of SAD-infected mice. Among all mice, 20% of rSAD-S179P-infected mice succumbed to rabies, compared with the 100% mortality observed within 11 days of rSAD infection (Fig. 7F).
FIG 7.
(A to F) S179 of RABV-P affects viral pathogenicity in mice. C57BL/6 mice (n = 10) were intradermally (i.d.) infected with 2.5 × 104 FFU in the ear of rCVS or rCVS-P179S or intracerebrally (i.c.) infected with 2.5 × 103 FFU of rSAD or rSAD-S179P, and body weight changes (A and D), clinical scores (B and E), and survival ratios (C and F) were monitored daily. The mice in panels A to C and in panels D to F are the same bunch. Body weight changes and clinical scores were analyzed by two-way ANOVA. Survival ratios were analyzed by the log-rank (Mantel-Cox) test. The error bars represent the SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (G and H) The mRNA levels of Ifnb and RABV-N in the mouse brain were analyzed by qPCR at 4, 6, and 8 dpi (n = 5). (I and J) The mRNA levels of RABV-N in the cortex, cerebellum, brain stem, and olfactory bulb were analyzed by qPCR at 3, 6, and 9 dpi (n = 5). Statistical analyses of grouped comparisons were carried out by one-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001). The error bars represent the SEMs; n = 5. Results are representative of two independent assays.
Since S179 of RABV-P is critical for lyssavirus inhibition of IRF3 activation, we analyzed the Ifnb mRNA and viral RNA levels in mouse brains by qPCR at 4, 6, and 8 days postinfection (dpi). The results suggested that rCVS (compared with rCVS-P179S) and rSAD-S179P (compared with rSAD) induced higher Ifnb mRNA levels and possessed lower RABV-N mRNA levels in infected mouse brains at 6 and 8 dpi (Fig. 7G and H). Furthermore, the RABV-N mRNA levels in different encephalic regions were also investigated by qPCR. At 6 and/or 8 dpi, we observed dramatically increased RABV N mRNA levels in rCVS-P179S (compared with rCVS)- and rSAD (compared with rSAD-S179P)-infected mice, and these increments were observed in the cortex, cerebellum, brain stem, and olfactory bulb regions on 6 or 8 dpi (Fig. 7I and J).
S179 is conserved among most of the P proteins of lyssaviruses, and the proteins share the same inhibitory mechanism of IRF3 activation.
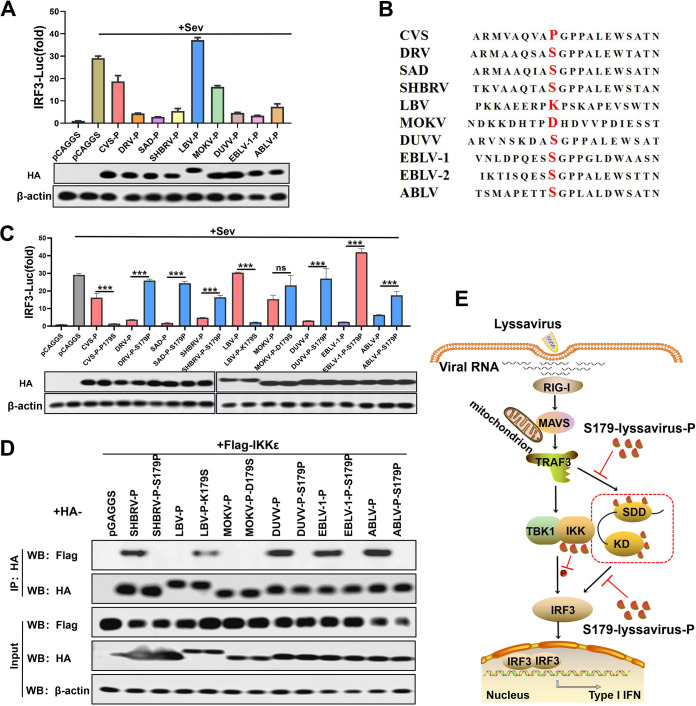
To identify the IRF3-inhibition function of P in other lyssaviruses, a dual-luciferase reporter assay was performed with diversified P from lyssaviruses. The results showed that P from SAD, DRV, silver-haired bat rabies virus (SHBRV), DUVV, EBLV-1, and ABLV blocked IRF3 activation better than P from CVS, LBV, and MOKV (Fig. 8A). Sequence alignment results showed the following identities of 179 aa in diversified P proteins: Pro in CVS; Ser in SAD, DRV, SHBRV, DUVV, EBLV-1, and ABLV; lysine (Lys, K) in LBV; and aspartic acid (Asp, D) in MOKV (Fig. 8B). In the subsequent experiments, 179 aa of lyssavirus-P was substituted with Pro or Ser. Dual-luciferase reporter and co-IP assays were performed on diversified parent and mutant lyssavirus-P. The results of the dual-luciferase reporter assay confirmed that lyssavirus-P containing S179 exhibited dramatic inhibition of IRF3 (except for MOKV-P), while lyssavirus-P containing P179 had no or a weak inhibitory effect on IRF3 (Fig. 8C). We observed an interaction of IKKε with lyssavirus-P containing S179 but not lyssavirus-P containing P179 (except for MOKV-P) (Fig. 8D). These results demonstrated the conserved function of the interaction of lyssavirus-P containing S179 with IKKε to inhibit IRF3 activation.
FIG 8.
Inhibitory effects of P and mutants from different lyssaviruses on IRF3 activity. (A) Dual-luciferase reporter assay (above) and Western blot assay (below) of diverse lyssavirus-P (as described in Fig. 2B). (B) Sequence alignment of diverse lyssavirus P proteins. (C) Dual-luciferase reporter assay (above) and Western blot assay (below) of diverse lyssavirus-P and mutant proteins (as described in Fig. 2B). (D) co-IP assay of IKKε and diverse lyssavirus parent and mutant P proteins (as described in Fig. 3B). (E) Schematic diagram of lyssavirus-P-inhibited IRF3 activation by competing with TRAF3 and IRF3 for binding to IKKε. Statistical analyses of grouped comparisons were carried out by one-way ANOVA (***, P < 0.001; ns, no significant difference). The error bars represent the SDs; n = 3. Results are representative of two independent assays.
DISCUSSION
Lyssavirus-P is known as an interferon antagonist. It was first reported that the P protein of the vaccine RABV strain SAD (SAD-P) inhibited TBK1-induced IRF3 phosphorylation, and a 176- to 186-aa deletion in SAD-P strongly impaired its ability to inhibit IRF3 activation (35, 36). However, the precise site required for SAD-P suppression of IRF3 activation remains to be resolved. A recent study suggested that P from street RABV interacted with IKKε, thus inhibiting the activation of IKKε-mediated IRF3 activity (37). However, P from SAD also inhibits IKKε-mediated IRF3, which is unexplained. In addition to antagonizing IFN production, lyssavirus-P blocks IFN signaling through several mechanisms, such as the sequestration of STAT in the cytoplasm, the inhibition of pSTAT1 and ISGF3 binding to DNA promoters, and the antagonism of cytokine-activated STAT3-STAT1 heterodimers (38, 49–56). In the present study, we found that 179 aa of lyssavirus-P is essential for inhibiting IRF3 activation, which is consistent with the previously reported region of 176 to 186 aa. Mechanistically, lyssavirus-P containing S179 interacts with IKKε and competes with IRF3 and TRAF3 for binding to IKKε. Importantly, S179 of RABV-P correlates with viral pathogenesis, and its antagonism is conserved among most lyssavirus-P.
IKKε and TBK1 are two key components with similar structures and functions in the antiviral immunity signaling pathway. Thus, many viruses have evolved various mechanisms to antagonize the IFN system by targeting these two indispensable kinases. For instance, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nonstructural protein 6 (nsp6) binds TBK1 to suppress IRF3 phosphorylation, and nsp13 binding blocks TBK1 phosphorylation (57). Porcine epidemic diarrhea virus (PEDV) nsp15 directly degrades the RNA levels of TBK1 and IRF3 to suppress IFN production and constrain ISG induction (58). There are also many reports suggesting that viral proteins inhibit interactions between TBK1 or IKKε and their upstream or downstream molecules. Herpes simplex virus 1 (HSV-1) γ134.5 protein (59), PEDV nucleocapsid (N) protein (60), and heartland virus (HRTV) nonstructural proteins (NSs) (61) that interact with TBK1 and block its association with downstream IRF3 have been identified. Other viral proteins, such as Ebola virus (EBOV) protein VP35 and swine acute diarrhea syndrome coronavirus (SADS-CoV) N protein, interact with both TBK1 and IKKε (62, 63). EBOV VP35 antagonizes the early antiviral response by disrupting IKKε interactions with IRF3 and IRF7, whereas SADS-CoV N protein impairs IFN production by blocking the interaction between TRAF3 and TBK1. In this study, we screened the key molecules in the RLR signaling pathway by a co-IP assay and found that P specifically targets IKKε rather than the closely related TBK1. Only three similar conclusions have been reported thus far involving arenavirus lymphocytic choriomeningitis virus (LCMV) nucleoprotein (NP), dengue virus (DENV) NS2B/3, and porcine delta coronavirus (PDCoV) accessory protein NS7a, which specifically target IKKε but not TBK1 to inhibit IFN production (64–66).
Initial studies suggest a more important role of TBK1, rather than IKKε, in the induction of type IFN-I in response to virus infection (67, 68). However, evidence indicated that TBK1 was completely dispensable for IFN responses to virus infection in mouse bone marrow-derived macrophages, where IKKε function is predominant (69). Follow-up studies demonstrated that both TBK1 and IKKε were required for the optimal induction of IFN upon SeV infection (64). Importantly, a previous report showed that mice lacking IKKε are hypersusceptible to viral infection, which indicates the critical role of IKKε in the IFN-inducible antiviral transcriptional response (70). In our study, RABV-P containing S179 inhibited TBK1-induced IFN production, although there was no interaction between RABV-P and TBK1. Previous studies have shown that knockdown of either IKKε or TBK-1 leads to significant impairment of SeV-induced IFN production in 293T cells (64). We infer that the interaction of RABV-P containing S179 with IKKε may affect the function of TBK1 by interfering with the formation of TBK1-IKKε dimers.
The RABV-P protein is reported to be phosphorylated by protein kinase C (PKC) and rabies virus protein kinase (RVPK) (46, 47, 71, 72). The phosphorylation at S63 and S64 of CSV-P protein (37 kDa) by RVPK alters its mobility in gel to migrate at 40 kDa and increases its expression (46). In the present study, we found CVS-P containing S179 was sharply increased that appeared to be accompanied by an alteration from 37 kDa to 40 kDa in the presence of IKKε. Interestingly, the S63 and S64 mutants alone and in combination abolished this increase in CVS-P protein. Accordingly, we suspected that CVS-P containing S179 enhanced its own phosphorylation by IKKε to bind more IKKε. Similar observations have been reported in other members of Mononegavirales, including EBOV, Borna disease virus (BDV), and Paramyxoviruses (62, 73, 74). The BDV P protein was first reported to be phosphorylated by TBK1 and to inhibit the kinase activity of TBK1 (73). Subsequently, the V proteins of several Paramyxoviruses and the VP35 protein of EBOV were also found to serve as substrates of TBK1 and/or IKKε, thus preventing IRF3 phosphorylation (62, 74). Of note, the V protein of Paramyxoviruses is an accessory protein encoded by the P gene and shares a common amino-terminal domain with the P protein. Additionally, the EBOV VP35 protein is functionally equivalent to the P proteins of other members of Mononegavirales. Consequently, the ability to act as an alternative substrate of TBK1 and/or IKKε and inhibit IRF3 phosphorylation appears to be a function common to the P proteins of these viruses. It is worth noting that there are two bands in the input of CVS-P-P179S, while there is only one band in the immunoprecipitation, which imply that only the nonphosphorylated CVS-P-P179S interacts with IKKε. A limitation of the present study is that we have not determined whether IKKε-induced phosphorylation of RABV-P protein occurs during normal infection.
IKKε contains two key functional domains, SDD and KD, which have been confirmed to mediate the formation of TRAF3-IKKε and IKKε-IRF3 complexes, respectively. Previous studies indicated that certain viral proteins, such as LCMV NP and DENV NS2B/3 (64, 65), interact with IKKε KD to impair the IFN response. In our study, RABV-P containing S179 bound both KD and SDD, which is similar to the NS7a protein of PDCoV (66). The interaction between RABV-P containing S179 and IKKε presents two possible mechanisms by which RABV-P containing S179 disrupts the association of IKKε with other molecules. One mechanism is, as observed at the SARS-CoV M protein, to block the complex formation of TBK1/IKKε and the upstream adaptor molecule TRAF3 (75). The other is to prevent the recruitment of IKKε to its downstream transcription factor IRF3, as observed for EBOV VP35 (62). Our results show that both the TRAF3-IKKε/IKKε-KD and IRF3-IKKε/IKKε-SDD interactions were interrupted or decreased in the presence of RABV-P containing S179, which suggests that RABV-P containing S179 acts as a competitor of both TRAF3 and IRF3 for IKKε binding.
Although mutation of 179 aa strongly affected the interaction of P with IKKε, it is uncertain whether S179 is directly involved in the binding with IKKε. Some evidence strongly suggests that mutation of 179 aa affects specific protein folding, which is required for IKKε binding and IFN inhibition. First, similar strong defects in IRF3 inhibition have also been reported with the deletion of 181 to 186 aa in P. Second, the combined deletion of 176 to 181 aa and 181 to 186 aa resulted in less noticeable inhibition of IRF3 than a deletion of either 176 to 181 aa or 181 to 186 aa alone (36). Finally, in this study, RABV-P containing S179 independently binds KD and SDD domains of IKKε and the same S179 mutation abolishes both interactions. Notably, it seems that the CVS strain is unique among RABVs, in having a quite profound Pro substitution at 179 aa. Unfortunately, the crystal structure of RABV-P containing 179 aa has not been resolved yet (76), impeding the further elaboration of a detailed mechanism.
IFNs play an essential role in the onset of rabies disease. Early experiments demonstrated that animal treatment with IFN, IFN-inducing poly(I·C), or IFN-inducing viruses (such as Newcastle disease virus) could control RABV infection (77–79). In contrast, mice that underwent IFN neutralization experienced a significantly shorter morbidity period than controls during RABV infection (80). IFN-I produced in the CNS is efficient enough to reduce viral pathogenicity during RABV infection and partially protect the host from fatal infection (81). A mutant virus with a 176 to 181 aa deletion in RABV-P lost most of its inhibitory activity in preventing IRF3 activation, and this construct was shown to be completely avirulent in adult mice after i.c. administration. Consistently, in our study, the replacement of Pro with Ser at P-179 of SAD significantly limited the proliferation of SAD in the CNS and weakened its viral pathogenicity. In contrast, the substitution of Pro with Ser at P-179 of CVS exacerbates the onset of rabies. Therefore, the capacity of RABV-P containing S179 to interact with IKKε and inhibit IFN production correlates with its viral pathogenicity. The evidence also suggests that in the case of inhibition of JAK/STAT signaling, lyssavirus-induced IRF3 activation is sufficient to trigger a protective response in the host.
In addition to limiting RABV infection, the activation of the IFN pathway exerts a distinct impact on the induction of adaptive immunity. It has been reported that Ifnar−/− mice developed lower levels of VNAs than wild mice (82, 83), which implies the importance of IFN in RABV antibody production. In another study, IFN-β served as a molecular adjuvant in a RABV-based HIV-1 vaccine vector, and recombinant IFN-α1 was also expressed using a recombinant RABV to enhance the immune response (84, 85). Evidence of the high immunogenicity of viruses lacking key IFN antagonists, such as influenza virus, suggests that virus-induced early IFN functions as a powerful adjuvant in vivo (86). In the present study, we find that the P-179 mutant of SAD-P abolishes the ability to efficiently prevent IFN production and does not affect viral replication and proliferation in BSR cells (IFN incompetent). These results suggest the possibility of applying rSAD-S179P as a potential live attenuated vaccine. Immunization studies in foxes and skunks have been undertaken to investigate whether deletion mutants would be more efficacious than the parental SAD strain, but the results were not promising. This is probably due to the reduced viral growth in antigen-presenting cells when the deletion mutants induce increased IFN production (87). Overall, the genetic manipulation of 179 aa in lyssaviruses sheds new light on the development of novel live-attenuated vaccines, while the delicate balance between safety and efficacy has yet to be considered.
In summary, our data demonstrate that S179 site of lyssavirus P is essential for its inhibition of IFN-I signaling. Mechanistically, RABV-P containing S179 specifically targets the key kinase IKKε. In addition, RABV-P containing S179 interacts with both the kinase KD and SDD domains of IKKε, disrupting their interaction with TRAF3 and IRF3. Importantly, S179 of RABV-P is involved in viral pathogenicity, and its function is conserved among most lyssavirus-P. This finding will contribute to fully understanding the immune evasion strategies of lyssaviruses, thereby providing important insights for the development of antiviral drugs and attenuated vaccines.
MATERIALS AND METHODS
Cells, plasmids, viruses, antibodies, and animals.
HEK-293T (human embryonic kidney, 293T), N2a (mouse neuroblastoma), BV2 (mouse microglioma), and BSR (a cloned cell line derived from BHK-21 cells) cells were cultured in Dulbecco’s modified DMEM or RPMI 1640 supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY). The P genes of several RABV strains were amplified and cloned into the pCAGGS-HA vector. The mutations of the P gene were constructed from wt-P. Flag-tagged pCAGGS expression constructs encoding human RIG-I, RIG-IN, MDA5, MAVS, TRAF3, TBK1, IKKε, IRF3, and IRF3-5D were kindly provided by Shengbo Cao (Huazhong Agricultural University, Wuhan, China). pCAGGS-Flag-IKKε-KD, -IKKε-SDD, pCAGGS-Myc-IKKε, -IKKε-KD, and -IKKε-SDD were constructed from full-length IKKε. All expression constructs were validated by DNA sequencing. The RABV strain challenge virus standard (CVS)-B2c, which is a laboratory-adapted virus, was produced from the CVS-24 virus by passaging in BHK-21 (88). DRV-Mexico, which is a dog-derived wild-type RABV strain, was isolated from a human patient (89). The recombinant virus SAD L16 (SAD) was rescued according to the consensus sequence of the attenuated SAD B19 vaccine strain (90). VSV-GFP was propagated in BHK-21 cells and stored in our lab. Monoclonal antibodies (MAb) against RABV-N protein and RABV-P protein were prepared in our laboratory. Other antibodies were purchased from the indicated companies, including antibodies against Flag tag (MBL; M185-3 L; 1:10,000), HA tag (MBL; M180-3; 1:10,000), Myc tag (MBL; M180-3; 1:5,000), β-actin (MBL; M192-3; 1:5,000), IRF3 (Abclonal; A11118; 1:1,000), p-IRF3 (Abclonal; AP0995; 1:1,000), TBK1 (BOSTER; BM4038; 1:2,000), p-TBK1 (Abclonal; S172; AP1026; 1:1,000), and IKKε (Abcam,; ab7891; 1:1,000). Secondary antibodies included goat anti-mouse (BOSTER; BA1051; 1:5,000), goat anti-rabbit (BOSTER; BA1055; 1:5,000), and goat anti-mouse IgG light-chain (Abbkine; A25012; 1:5,000). Female C57BL/6 mice aged 6 to 8 weeks were purchased from the Centers for Disease Control and Prevention of Hubei Province, China, and housed in the Animal Facility at Huazhong Agricultural University.
Viral infection and titration.
293T, N2a, BSR, and BV2 cells were infected with the viruses at the indicated MOIs (diluted in DMEM) for 1 h at 37°C. The cells were washed with phosphate-buffered saline (PBS) three times, fresh medium was added, and the cells were cultured in an incubator at 37°C. The cell culture supernatants were collected at the indicated time points for viral titration. Viral titers were determined by a direct immunofluorescence assay as described previously (91, 92).
Dual-luciferase reporter assay.
The luciferase reporter plasmids IRF3-Luc, NF-κB-Luc, ISRE-Luc, and pRL-TK were kindly provided by Rui Luo (Huazhong Agricultural University, Wuhan, China). For the luciferase reporter assay, cells cultured in 48-well plates were cotransfected with a luciferase reporter plasmid and pRL-TK along with the indicated plasmids or empty vector (pCAGGS) for 24 h and were then infected with SeV (50 hemagglutination units per mL). At 12 h postinfection, the cells were lysed, and firefly luciferase and Renilla luciferase activities were measured using a dual-luciferase reporter assay kit according to the manufacturer’s protocol (Vazyme; DL101-01). Representative data from three independently conducted experiments are shown as the relative firefly luciferase activities normalized to the Renilla luciferase activities.
Western blot.
The cell samples were lysed, collected, and centrifuged. The supernatants were boiled for 5 min with SDS loading buffer. The prepared samples were separated on SDS-PAGE gels and transferred to PVDF membranes (Bio-Rad). The membranes were blocked with TBST containing 5% (wt/vol) nonfat dry milk for 3 h and probed with primary antibodies overnight at 4°C. The proteins were visualized using the BeyoECL Star kit (Beyotime; P0018A), and images were captured with an Amersham Imager 600 (GE Healthcare) imaging system.
RNA extraction and quantitative real-time PCR.
Total RNA was isolated by TRIzol Reagent (Invitrogen, Karlsruhe, Germany) and treated with a TURBO DNA-free kit (Invitrogen; AM1907) according to the manufacturer’s instructions. RNA was quantified using a NanoDrop and Agilent 2100 bioanalyzer (Thermo Fisher Scientific, MA, USA) and converted to cDNA by reverse transcription using FSQ-201 ReverTra Ace (TOYOBO, Osaka, Japan). qPCR was performed using SYBR green Supermix (Bio-Rad; 172-5124) on an Applied Biosystems 7300 real-time PCR system (Applied Biosystems, CA, USA). The primer sets used in this study are listed in Table 1.
TABLE 1.
Primer pairs used for qPCR in this studya
| Primer | Sequence (5′ to 3′) |
|---|---|
| Ifnb-F | CACAGCCCTCTCCATCAAC |
| Ifnb-R | GCATCTTCTCCGTCATCTCC |
| β-Actin-F | AGGTGACAGCATTGCTTCTG |
| β-Actin-R | GCTGCCTCAACACCTCAAC |
| RABV-N-F | CTCTGACAGGAGGCATGGAA |
| RABV-N-R | GGTATAGTACTCCAATTAGCACACAT |
F, forward; R, reverse.
Sequence alignment.
The sequences of various lyssavirus-P were obtained from NCBI (http://www.ncbi.nlm.nih.gov), and the protein IDs are shown in Table 2. Multiple sequence alignment was performed with CLUSTALW (https://www.genome.jp/tools-bin/clustalw).
TABLE 2.
Protein IDs of various lyssavirus-P used in this study
| Protein | Protein ID |
|---|---|
| CVS-P | ABF82951.1 |
| DRV-P | ADU55579.1 |
| SAD-P | ABN11302.1 |
| SHBRV-P | AAU11516.1 |
| LBV-P | ABU87604.1 |
| DUVV | ACF32421.1 |
| MOKV-P | YP_142351.1 |
| EBLV−1-P | SMD52091.1 |
| EBLV-2-P | AAC04591.1 |
| ABLV-P | AIN50170.1 |
Confocal microscopy.
Confocal microscopy assays were performed as previously reported (93, 94). 293T and N2a cells were seeded onto coverslips and infected with RABV or transfected with plasmids by using Jetprime according to the manufacturer’s protocol. At different time points postinfection and posttransfection, the cells were fixed with 4% (vol/vol) paraformaldehyde for 20 min at room temperature, permeabilized with 0.5% Triton X-100 for 10 min at room temperature, blocked in 10% goat serum (diluted with PBS) for 2 h at room temperature, and then probed with primary antibodies against HA tag, Flag tag, RABV-P, or 4′,6-diamidino-2-phenylindole (DAPI) overnight at 4°C. After being washed, the samples were stained with Alexa 488-conjugated or 594-conjugated secondary antibodies for 1 h at room temperature and then stained with DAPI for 10 min. The results were then observed and imaged with a ZEISS confocal microscope under an oil objective.
Coimmunoprecipitation.
293T cells were cotransfected with the specified expression constructs encoding Flag, HA, or Myc-tagged proteins for 48 h. The cells were washed with cold PBS and lysed with NP-40 lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, and 0.5% NP-40) containing an anti-protease cocktail (Roche) and phenylmethylsulfonyl fluoride (1 mM) for 30 min at 4°C. A portion of each supernatant from the lysed cells was used in the whole-cell extract assay. The remaining portions of the supernatants were immunoprecipitated with the indicated antibodies overnight at 4°C, followed by the addition of protein A+G agarose beads for 3 h at 4°C. The agarose beads containing immunocomplexes were washed three to five times with 1 mL of lysis buffer. Whole-cell lysates (input) and immunoprecipitation complexes were analyzed by Western blotting as described above.
Construction and rescue of the recombinant RABVs.
The recombinant viruses were constructed and rescued as described previously (95). To rescue each mutant virus, the plasmid containing the full-length viral genome and helper plasmids expressing N, P, G, and L proteins were cotransfected into N2a cells using Jetprime (Polyplus; number 23Y2307M4). At 6 days posttransfection, the supernatants were harvested and examined for the presence of the rescued viruses using FITC-conjugated anti-RABV-P antibody (prepared in our laboratory). The rescued viruses were propagated and titrated in BSR cells.
Animal experiments.
Six-week-old female C57BL/6 mice were randomly divided into the indicated groups and injected i.d. in the ear (rCVS and rCVS-P179S) with 25 μL of virus at 2.5 × 104 FFU or i.c. (rSAD-wt and rSAD-S179P) with 25 μL of virus at 2.5 × 103 FFU. Body weight changes, clinical scores, and mortality were monitored daily. The clinical scores were assessed as previously described (96). Clinical signs were scored using a scale of 0 to 5: 0, no clinical signs; 1, disordered movement; 2, ruffled fur, hunched back; 3, trembling and shaking; 4, complete loss of motion (complete paralysis); 5, death. Mice with body weight losses of more than 25% were euthanized with CO2. At 4, 6, and 8 dpi, the mice were euthanized with CO2, and the brains were collected for qPCR analysis.
Statistical analysis.
Statistical analysis was performed with GraphPad Prism 8 (GraphPad, La Jolla, CA, USA). For the survival percentage tests, survival curves were analyzed using the log-rank (Mantel-Cox) test. For other data, one-way ANOVA or two-way ANOVA was used to determine statistical significance. The error bars represent the standard deviations (SDs) or standard errors of the mean (SEMs). Asterisks in figures indicate statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ns, no significant difference).
Ethics statement.
All animals used in this study were maintained in the animal facility of Huazhong Agricultural University in compliance with the Regulations for the Administration of Affairs Concerning Experimental Animals presented by the Ministry of Science and Technology of China. The experiments were carried out with the protocol approved by the Scientific Ethics Committee of Huazhong Agricultural University (permit number: HZAUMO-2018-040).
ACKNOWLEDGMENTS
This work was partially supported by Guangdong Major Project of Basic and Applied Basic Research (2020B0301030007 to Z.F.F.) and the National Natural Science Foundation of China (grant number 31872451 to L.Z.; 31720103917 to Z.F.F.).
Contributor Information
Ling Zhao, Email: zling604@outlook.com.
Bryan R.G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Morimoto K, Patel M, Corisdeo S, Hooper DC, Fu ZF, Rupprecht CE, Koprowski H, Dietzschold B. 1996. Characterization of a unique variant of bat rabies virus responsible for newly emerging human cases in North America. Proc Natl Acad Sci USA 93:5653–5658. 10.1073/pnas.93.11.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Familusi JB, Osunkoya BO, Moore DL, Kemp GE, Fabiyi A. 1972. A fatal human infection with Mokola virus. Am J Trop Med Hyg 21:959–963. 10.4269/ajtmh.1972.21.959. [DOI] [PubMed] [Google Scholar]
- 3.Crick J, Tignor GH, Moreno K. 1982. A new isolate of Lagos bat virus from the Republic of South Africa. Trans R Soc Trop Med Hyg 76:211–213. 10.1016/0035-9203(82)90277-2. [DOI] [PubMed] [Google Scholar]
- 4.Foggin CM. 1983. Mokola virus infection in cats and a dog in Zimbabwe. Vet Rec 113:115. 10.1136/vr.113.5.115. [DOI] [PubMed] [Google Scholar]
- 5.Shope RE. 1982. Rabies-related viruses. Yale J Biol Med 55:271–275. [PMC free article] [PubMed] [Google Scholar]
- 6.Bourhy H, Kissi B, Tordo N. 1993. Molecular diversity of the Lyssavirus genus. Virology 194:70–81. 10.1006/viro.1993.1236. [DOI] [PubMed] [Google Scholar]
- 7.Gould AR, Hyatt AD, Lunt R, Kattenbelt JA, Hengstberger S, Blacksell SD. 1998. Characterisation of a novel lyssavirus isolated from Pteropid bats in Australia. Virus Res 54:165–187. 10.1016/s0168-1702(98)00025-2. [DOI] [PubMed] [Google Scholar]
- 8.Regnault B, Evrard B, Plu I, Dacheux L, Troadec E, Cozette P, Chrétien D, Duchesne M, Vallat J-M, Jamet A, Leruez M, Pérot P, Bourhy H, Eloit M, Seilhean D. 2022. First case of lethal encephalitis in Western Europe due to european bat lyssavirus type 1. Clin Infect Dis 74:461–466. 10.1093/cid/ciab443. [DOI] [PubMed] [Google Scholar]
- 9.Fooks AR, Banyard AC, Horton DL, Johnson N, McElhinney LM, Jackson AC. 2014. Current status of rabies and prospects for elimination. Lancet 384:1389–1399. 10.1016/S0140-6736(13)62707-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher CR, Streicker DG, Schnell MJ. 2018. The spread and evolution of rabies virus: conquering new frontiers. Nat Rev Microbiol 16:241–255. 10.1038/nrmicro.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anilionis A, Wunner WH, Curtis PJ. 1981. Structure of the glycoprotein gene in rabies virus. Nature 294:275–278. 10.1038/294275a0. [DOI] [PubMed] [Google Scholar]
- 12.Hidaka Y, Lim C-K, Takayama-Ito M, Park C-H, Kimitsuki K, Shiwa N, Inoue K-I, Itou T. 2018. Segmentation of the rabies virus genome. Virus Res 252:68–75. 10.1016/j.virusres.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Schnell MJ, McGettigan JP, Wirblich C, Papaneri A. 2010. The cell biology of rabies virus: using stealth to reach the brain. Nat Rev Microbiol 8:51–61. 10.1038/nrmicro2260. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 15.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M, Endres S, Hartmann G. 2006. 5′-triphosphate RNA is the ligand for RIG-I. Science 314:994–997. 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 16.Faul EJ, Wanjalla CN, Suthar MS, Gale M, Wirblich C, Schnell MJ. 2010. Rabies virus infection induces type I interferon production in an IPS-1 dependent manner while dendritic cell activation relies on IFNAR signaling. PLoS Pathog 6:e1001016. 10.1371/journal.ppat.1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seth RB, Sun LJ, Ea CK, Chen ZJJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappa B and IRF3. Cell 122:669–682. 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao S-M, Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4:491–496. 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 19.Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu Rev Biochem 67:227–264. 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 20.van Boxel-Dezaire AHH, Rani MRS, Stark GR. 2006. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 25:361–372. 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Blaszczyk K, Olejnik A, Nowicka H, Ozgyin L, Chen Y-L, Chmielewski S, Kostyrko K, Wesoly J, Balint BL, Lee C-K, Bluyssen HAR. 2015. STAT2/IRF9 directs a prolonged ISGF3-like transcriptional response and antiviral activity in the absence of STAT1. Biochem J 466:511–524. 10.1042/BJ20140644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blondel D, Maarifi G, Nisole S, Chelbi-Alix MK. 2015. Resistance to rhabdoviridae infection and subversion of antiviral responses. Viruses 7:3675–3702. 10.3390/v7072794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chenik M, Schnell M, Conzelmann KK, Blondel D. 1998. Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J Virol 72:1925–1930. 10.1128/JVI.72.3.1925-1930.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albertini AAV, Wernimont AK, Muziol T, Ravelli RBG, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RWH. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360–363. 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 25.Chenik M, Chebli K, Gaudin Y, Blondel D. 1994. In-vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N)–existence of 2 N-binding sites on P-protein. J Gen Virol 75:2889–2896. 10.1099/0022-1317-75-11-2889. [DOI] [PubMed] [Google Scholar]
- 26.Mavrakis M, Iseni F, Mazza C, Schoehn G, Ebel C, Gentzel M, Franz T, Ruigrok RWH. 2003. Isolation and characterisation of the rabies virus N degrees-P complex produced in insect cells. Virology 305:406–414. 10.1006/viro.2002.1748. [DOI] [PubMed] [Google Scholar]
- 27.Lahaye X, Vidy A, Pomier C, Obiang L, Harper F, Gaudin Y, Blondel D. 2009. Functional characterization of Negri bodies (NBs) in rabies virus-infected cells: evidence that NBs are sites of viral transcription and replication. J Virol 83:7948–7958. 10.1128/JVI.00554-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ménager P, Roux P, Mégret F, Bourgeois J-P, Le Sourd A-M, Danckaert A, Lafage M, Préhaud C, Lafon M. 2009. Toll-like receptor 3 (TLR3) plays a major role in the formation of rabies virus Negri Bodies. PLoS Pathog 5:e1000315. 10.1371/journal.ppat.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolic J, Le Bars R, Lama Z, Scrima N, Lagaudrière-Gesbert C, Gaudin Y, Blondel D. 2017. Negri bodies are viral factories with properties of liquid organelles. Nat Commun 8:58. 10.1038/s41467-017-00102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Dong W, Shi Y, Deng F, Chen X, Wan C, Zhou M, Zhao L, Fu ZF, Peng G. 2016. Rabies virus phosphoprotein interacts with ribosomal protein L9 and affects rabies virus replication. Virology 488:216–224. 10.1016/j.virol.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 31.Fouquet B, Nikolic J, Larrous F, Bourhy H, Wirblich C, Lagaudrière-Gesbert C, Blondel D. 2015. Focal adhesion kinase is involved in rabies virus infection through its interaction with viral phosphoprotein P J Virol 89:1640–1651. 10.1128/JVI.02602-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kammouni W, Wood H, Jackson AC. 2017. Lyssavirus phosphoproteins increase mitochondrial complex I activity and levels of reactive oxygen species. J Neurovirol 23:756–762. 10.1007/s13365-017-0550-z. [DOI] [PubMed] [Google Scholar]
- 33.Kammouni W, Wood H, Saleh A, Appolinario CM, Fernyhough P, Jackson AC. 2015. Rabies virus phosphoprotein interacts with mitochondrial complex I and induces mitochondrial dysfunction and oxidative stress. J Neurovirol 21:370–382. 10.1007/s13365-015-0320-8. [DOI] [PubMed] [Google Scholar]
- 34.Kammouni W, Wood H, Jackson AC. 2017. Serine residues at positions 162 and 166 of the rabies virus phosphoprotein are critical for the induction of oxidative stress in rabies virus infection. J Neurovirol 23:358–368. 10.1007/s13365-016-0506-8. [DOI] [PubMed] [Google Scholar]
- 35.Brzozka K, Finke S, Conzelmann KK. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J Virol 79:7673–7681. 10.1128/JVI.79.12.7673-7681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rieder M, Brzozka K, Pfaller CK, Cox JH, Stitz L, Conzelmann KK. 2012. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprotein: inhibition of interferon regulatory factor 3 activation is important for pathogenicity (vol 85, pg 842, 2011). J Virol 86:4720–4720. 10.1128/JVI.00338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masatani T, Ozawa M, Yamada K, Ito N, Horie M, Matsuu A, Okuya K, Tsukiyama-Kohara K, Sugiyama M, Nishizono A. 2016. Contribution of the interaction between the rabies virus P protein and I-kappa B kinase to the inhibition of type I IFN induction signalling. J Gen Virol 97:316–326. 10.1099/jgv.0.000362. [DOI] [PubMed] [Google Scholar]
- 38.Vidy A, Chelbi-Alix M, Blondel D. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J Virol 79:14411–14420. 10.1128/JVI.79.22.14411-14420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blondel D, Regad T, Poisson N, Pavie B, Harper F, Pandolfi PP, De Thé H, Chelbi-Alix MK. 2002. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21:7957–7970. 10.1038/sj.onc.1205931. [DOI] [PubMed] [Google Scholar]
- 40.Blondel D, Kheddache S, Lahaye X, Dianoux L, Chelbi-Alix MK. 2010. Resistance to rabies virus infection conferred by the PMLIV isoform. J Virol 84:10719–10726. 10.1128/JVI.01286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian B, Zhou M, Yang Y, Yu L, Luo Z, Tian D, Wang K, Cui M, Chen H, Fu ZF, Zhao L. 2017. Lab-attenuated rabies virus causes abortive infection and induces cytokine expression in astrocytes by activating mitochondrial antiviral-signaling protein signaling pathway. Front Immunol 8:2011. 10.3389/fimmu.2017.02011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kauzmann W. 1959. Some factors in the interpretation of protein denaturation. Adv Protein Chem 14:1–63. 10.1016/S0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 43.Tanford C. 1978. The hydrophobic effect and the organization of living matter. Science 200:1012–1018. 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- 44.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. 1998. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett 425:112–116. 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 45.Lin RT, Heylbroeck C, Pitha PM, Hiscott J. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol 18:2986–2996. 10.1128/MCB.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta AK, Blondel D, Choudhary S, Banerjee AK. 2000. The phosphoprotein of rabies virus is phosphorylated by a unique cellular protein kinase and specific isomers of protein kinase C. J Virol 74:91–98. 10.1128/jvi.74.1.91-98.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toriumi H, Eriguchi Y, Takamatsu F, Kawai A. 2004. Further studies on the hyperphosphorylated form (p40) of the rabies virus nominal phosphoprotein (P). Microbiol Immunol 48:865–874. 10.1111/j.1348-0421.2004.tb03618.x. [DOI] [PubMed] [Google Scholar]
- 48.Fang R, Jiang Q, Zhou X, Wang C, Guan Y, Tao J, Xi J, Feng J-M, Jiang Z. 2017. MAVS activates TBK1 and IKK epsilon through TRAFs in NEMO dependent and independent manner. PLoS Pathog 13:e1006720. 10.1371/journal.ppat.1006720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brzozka K, Finke S, Conzelmann KK. 2006. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent bindinig of STAT1 and STAT2. J Virol 80:2675–2683. 10.1128/JVI.80.6.2675-2683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vidy A, El Bougrini J, Chelbi-Alix MK, Blondel D. 2007. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STATI1. J Virol 81:4255–4263. 10.1128/JVI.01930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito N, Moseley GW, Blondel D, Shimizu K, Rowe CL, Ito Y, Masatani T, Nakagawa K, Jans DA, Sugiyama M. 2010. Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J Virol 84:6699–6710. 10.1128/JVI.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiltzer L, Larrous F, Oksayan S, Ito N, Marsh GA, Wang LF, Blondel D, Bourhy H, Jans DA, Moseley GW. 2012. Conservation of a unique mechanism of immune evasion across the Lyssavirus genus. J Virol 86:10194–10199. 10.1128/JVI.01249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lieu KG, Brice A, Wiltzer L, Hirst B, Jans DA, Blondel D, Moseley GW. 2013. The rabies virus interferon antagonist P protein interacts with activated STAT3 and inhibits Gp130 receptor signaling. J Virol 87:8261–8265. 10.1128/JVI.00989-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiltzer L, Okada K, Yamaoka S, Larrous F, Kuusisto HV, Sugiyama M, Blondel D, Bourhy H, Jans DA, Ito N, Moseley GW. 2014. Interaction of rabies virus P-protein with STAT proteins is critical to lethal rabies disease. J Infect Dis 209:1744–1753. 10.1093/infdis/jit829. [DOI] [PubMed] [Google Scholar]
- 55.Harrison AR, Lieu KG, Larrous F, Ito N, Bourhy H, Moseley GW. 2020. Lyssavirus P-protein selectively targets STAT3-STAT1 heterodimers to modulate cytokine signalling. PLoS Pathog 16:e1008767. 10.1371/journal.ppat.1008767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhan J, Harrison AR, Portelli S, Nguyen TB, Kojima I, Zheng S, Yan F, Masatani T, Rawlinson SM, Sethi A, Ito N, Ascher DB, Moseley GW, Gooley PR. 2021. Definition of the immune evasion-replication interface of rabies virus P protein. PLoS Pathog 17:e1009729. 10.1371/journal.ppat.1009729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia H, Cao Z, Xie X, Zhang X, Chen JY-C, Wang H, Menachery VD, Rajsbaum R, Shi P-Y. 2020. Evasion of type I interferon by SARS-CoV-2. Cell Rep 33:108234. 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Y, Zhang H, Shi Z, Chen J, Li M, Shi H, Shi D, Guo L, Feng L. 2020. Porcine epidemic diarrhea virus nsp15 antagonizes interferon signaling by RNA degradation of TBK1 and IRF3. Viruses 12:599. 10.3390/v12060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verpooten D, Ma YJ, Hou SW, Yan ZP, He B. 2009. Control of TANK-binding kinase 1-mediated signaling by the gamma(1)34.5 protein of herpes simplex virus 1. J Biol Chem 284:1097–1105. 10.1074/jbc.M805905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. 2014. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol 88:8936–8945. 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ning YJ, Feng K, Min YQ, Deng F, Hu ZH, Wang HL. 2017. Heartland virus NSs protein disrupts host defenses by blocking the TBK1 kinase-IRF3 transcription factor interaction and signaling required for interferon induction. J Biol Chem 292:16722–16733. 10.1074/jbc.M117.805127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prins KC, Cardenas WB, Basler CF. 2009. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J Virol 83:3069–3077. 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Z, Sun Y, Xu J, Tang X, Zhou L, Li Q, Lan T, Ma J. 2021. Swine acute diarrhea syndrome coronavirus nucleocapsid protein antagonizes interferon-beta production via blocking the interaction between TRAF3 and TBK1. Front Immunol 12:573078. 10.3389/fimmu.2021.573078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pythoud C, Rodrigo WWSI, Pasqual G, Rothenberger S, Martínez-Sobrido L, de la Torre JC, Kunz S. 2012. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKepsilon. J Virol 86:7728–7738. 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anglero-Rodriguez YI, Pantoja P, Sariol CA. 2014. Dengue virus subverts the interferon induction pathway via NS2B/3 protease-IkappaB kinase epsilon interaction. Clin Vaccine Immunol 21:29–38. 10.1128/CVI.00500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fang P, Fang L, Xia S, Ren J, Zhang J, Bai D, Zhou Y, Peng G, Zhao S, Xiao S. 2020. Porcine deltacoronavirus accessory protein NS7a antagonizes IFN-beta production by competing with TRAF3 and IRF3 for binding to IKKepsilon. Front Cell Infect Microbiol 10:257. 10.3389/fcimb.2020.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S. 2004. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med 199:1641–1650. 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci USA 101:233–238. 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perry AK, Chow EK, Goodnough JB, Yeh WC, Cheng G. 2004. Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J Exp Med 199:1651–1658. 10.1084/jem.20040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tenoever BR, Ng SL, Chua MA, McWhirter SM, Garcia-Sastre A, Maniatis T. 2007. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science 315:1274–1278. 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 71.Tuffereau C, Fischer S, Flamand A. 1985. Phosphorylation of the N and M1 proteins of rabies virus. J Gen Virol 66:2285–2289. 10.1099/0022-1317-66-10-2285. [DOI] [PubMed] [Google Scholar]
- 72.Takamatsu F, Asakawa N, Morimoto K, Takeuchi K, Eriguchi Y, Toriumi H, Kawai A. 1998. Studies on the rabies virus RNA polymerase: 2. Possible relationships between the two forms of the non-catalytic subunit (P protein). Microbiol Immunol 42:761–771. 10.1111/j.1348-0421.1998.tb02350.x. [DOI] [PubMed] [Google Scholar]
- 73.Unterstab G, Ludwig S, Anton A, Planz O, Dauber B, Krappmann D, Heins G, Ehrhardt C, Wolff T. 2005. Viral targeting of the interferon-beta-inducing Traf family member-associated NF-kappa B activator (TANK)-binding kinase-1. Proc Natl Acad Sci USA 102:13640–13645. 10.1073/pnas.0502883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu LL, Puri M, Horvath CM, Sen GC. 2008. Select paramyxoviral v proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kappa B kinase is an element of (IKKe)/TBK1. J Biol Chem 283:14269–14276. 10.1074/jbc.M710089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siu K-L, Kok K-H, Ng M-HJ, Poon VKM, Yuen K-Y, Zheng B-J, Jin D-Y. 2009. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem 284:16202–16209. 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gerard FC, Ribeiro EdeA Jr, Leyrat C, Ivanov I, Blondel D, Longhi S, Ruigrok RW, Jamin M. 2009. Modular organization of rabies virus phosphoprotein. J Mol Biol 388:978–996. 10.1016/j.jmb.2009.03.061. [DOI] [PubMed] [Google Scholar]
- 77.Postic BO, Fenje P. 1971. Effect of administered interferon on rabies in rabbits. Appl Microbiol 22:428–431. 10.1128/am.22.3.428-431.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weinmann E, Majer M, Hilfenhaus J. 1979. Intramuscular and/or intralumbar postexposure treatment of rabies virus-infected cynomolgus monkeys with human interferon. Infect Immun 24:24–31. 10.1128/iai.24.1.24-31.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marcovistz R, Germano PM, Riviere Y, Tsiang H, Hovanessian AG. 1987. The effect of interferon treatment in rabies prophylaxis in immunocompetent, immunosuppressed, and immunodeficient mice. J Interferon Res 7:17–27. 10.1089/jir.1987.7.17. [DOI] [PubMed] [Google Scholar]
- 80.Marcovistz R, Galabru J, Tsiang H, Hovanessian AG. 1986. Neutralization of interferon produced early during rabies virus infection in mice. J Gen Virol 67:387–390. 10.1099/0022-1317-67-2-387. [DOI] [PubMed] [Google Scholar]
- 81.Chopy D, Detje CN, Lafage M, Kalinke U, Lafon M. 2011. The type I interferon response bridles rabies virus infection and reduces pathogenicity. J Neurovirol 17:353–367. 10.1007/s13365-011-0041-6. [DOI] [PubMed] [Google Scholar]
- 82.Li J, Faber M, Dietzschold B, Hooper DC. 2011. The role of toll-like receptors in the induction of immune responses during rabies virus infection. Adv Virus Res 79:115–126. 10.1016/B978-0-12-387040-7.00007-X. [DOI] [PubMed] [Google Scholar]
- 83.Barkhouse DA, Garcia SA, Bongiorno EK, Lebrun A, Faber M, Hooper DC. 2015. Expression of interferon gamma by a recombinant rabies virus strongly attenuates the pathogenicity of the virus via induction of type I interferon. J Virol 89:312–322. 10.1128/JVI.01572-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Faul EJ, Wanjalla CN, McGettigan JP, Schnell MJ. 2008. Interferon-beta expressed by a rabies virus-based HIV-1 vaccine vector serves as a molecular adjuvant and decreases pathogenicity. Virology 382:226–238. 10.1016/j.virol.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y, Tian Q, Xu X, Yang X, Luo J, Mo W, Peng J, Niu X, Luo Y, Guo X. 2014. Recombinant rabies virus expressing IFNalpha1 enhanced immune responses resulting in its attenuation and stronger immunogenicity. Virology 468–470:621–630. 10.1016/j.virol.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 86.García-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 87.Vos A, Conzelmann K-K, Finke S, Müller T, Teifke J, Fooks AR, Neubert A. 2011. Immunogenicity studies in carnivores using a rabies virus construct with a site-directed deletion in the phosphoprotein. Adv Prev Med 2011:898171. 10.4061/2011/898171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang ZW, Sarmento L, Wang Y, Li X-q, Dhingra V, Tseggai T, Jiang B, Fu ZF. 2005. Attenuated rabies virus activates, while pathogenic rabies virus evades, the host innate immune responses in the central nervous system. J Virol 79:12554–12565. 10.1128/JVI.79.19.12554-12565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu F, Zhang G, Zhong X, Han N, Song Y, Zhao L, Cui M, Rayner S, Fu ZF. 2014. Comparison of complete genome sequences of dog rabies viruses isolated from China and Mexico reveals key amino acid changes that may be associated with virus replication and virulence. Arch Virol 159:1593–1601. 10.1007/s00705-013-1966-2. [DOI] [PubMed] [Google Scholar]
- 90.Schnell MJ, Mebatsion T, Conzelmann KK. 1994. Infectious rabies viruses from cloned cDNA. EMBO J 13:4195–4203. 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yuan Y, Wang Z, Tian B, Zhou M, Fu ZF, Zhao L. 2019. Cholesterol 25-hydroxylase suppresses rabies virus infection by inhibiting viral entry. Arch Virol 164:2963–2974. 10.1007/s00705-019-04415-6. [DOI] [PubMed] [Google Scholar]
- 92.Wang Z, Yuan Y, Chen C, Zhang C, Huang F, Zhou M, Chen H, Fu ZF, Zhao L. 2021. Colloidal manganese salt improves the efficacy of rabies vaccines in mice, cats, and dogs. J Virol 95: e0141421. 10.1128/JVI.01414-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tian B, Yuan Y, Yang Y, Luo Z, Sui B, Zhou M, Fu ZF, Zhao L. 2020. Interferon-inducible GTPase 1 impedes the dimerization of rabies virus phosphoprotein and restricts viral replication. J Virol 94: e01203-20. 10.1128/JVI.01203-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sui B, Chen D, Liu W, Wu Q, Tian B, Li Y, Hou J, Liu S, Xie J, Jiang H, Luo Z, Lv L, Huang F, Li R, Zhang C, Tian Y, Cui M, Zhou M, Chen H, Fu ZF, Zhang Y, Zhao L. 2020. A novel antiviral lncRNA, EDAL, shields a T309 O-GlcNAcylation site to promote EZH2 lysosomal degradation. Genome Biol 21:228. 10.1186/s13059-020-02150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tian D, Luo Z, Zhou M, Li M, Yu L, Wang C, Yuan J, Li F, Tian B, Sui B, Chen H, Fu ZF, Zhao L. 2016. Critical role of K1685 and K1829 in the large protein of rabies virus in viral pathogenicity and immune evasion. J Virol 90:232–244. 10.1128/JVI.02050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Faber M, Faber ML, Li J, Preuss MA, Schnell MJ, Dietzschold B. 2007. Dominance of a nonpathogenic glycoprotein gene over a pathogenic glycoprotein gene in rabies virus. J Virol 81:7041–7047. 10.1128/JVI.00357-07. [DOI] [PMC free article] [PubMed] [Google Scholar]