Abstract
Cancers are often defined by the dysregulation of specific transcriptional programs; however, the importance of global transcriptional changes is less understood. Hypertranscription is the genome-wide increase in RNA output. Hypertranscription’s prevalence, underlying drivers, and prognostic significance are undefined in primary human cancer. This is due, in part, to limitations of expression profiling methods, which assume equal RNA output between samples. Here, we developed a computational method to directly measure hypertranscription in 7494 human tumors, spanning 31 cancer types. Hypertranscription is ubiquitous across cancer, especially in aggressive disease. It defines patient subgroups with worse survival, even within well-established subtypes. Our data suggest that loss of transcriptional suppression underpins the hypertranscriptional phenotype. Single-cell analysis reveals hypertranscriptional clones, which dominate transcript production regardless of their size. Last, patients with hypertranscribed mutations have improved response to immune checkpoint therapy. Our results provide fundamental insights into gene dysregulation across human cancers and may prove useful in identifying patients who would benefit from novel therapies.
Global RNA output increase is a fundamental feature of aggressive tumors, defining the subgroups with worse prognosis across cancer.
INTRODUCTION
Transcriptional misregulation is a defining feature of cancer. However, even consistently misregulated genes often fail to predict prognosis or therapeutic response. The number of genes misregulated, as well as their individual expression levels, is thought to be tightly controlled in cancer. This control helps to maintain cell identity and promote tumor-specific oncogenic signaling. In contrast, tumor DNA can undergo chromosomal doubling (1), massive rearrangements (2), and localized (3) or genome-wide hypermutation (4). Because most mutations are passengers, even global shifts in DNA are tempered by modest changes in RNA expression.
Hypertranscription, also called RNA amplification, refers to the global increase in RNA across all genes. This phenomenon, which is a distinct form of transcriptional misregulation, has been best described in cell lines and model systems (5, 6), not primary human cancers. The prevalence of hypertranscription within and between tumor types is therefore unknown.
Historical observations have associated variable RNA levels with proliferation rates in different cell types (7, 8). For example, early work in a mouse model of leukemia demonstrated that the RNA content of rapidly proliferating transplanted cells is greater than either normal cells or of that of slower growing spontaneous leukemias (4.2-fold change versus 1.6-fold change in transcription above normal cells, respectively) (8). Therefore, the limited available data from cell line studies suggest that cancer cells that globally increase transcription have a growth advantage. Whether hypertranscription occurs in human tumors, and how it may correlate with patient phenotypes and treatment response, remains to be determined.
MYC has been implicated as a driver of hypertranscription in cell lines [acting directly (5) or indirectly (9) via its targets]. We previously observed a correlation between RNA output and expression of estrogen receptor in breast cancer (BRCA), suggesting that it is also a driver of hypertranscription (10). Another open question is whether there are additional drivers and if, collectively, these drivers provide insight into the mechanisms underpinning oncogenic hypertranscription across human cancer.
Here, we use a novel method, called RNAmp, to answer fundamental questions on the prevalence, causes, and consequences of hypertranscription in human cancer. The transcriptional output of 7494 cancer samples from 31 cancer types is measured. We find hypertranscription in most primary human tumors. Specific cancer subtypes exhibit >4-fold higher transcriptional output. Among these previously unidentified subtypes, which are otherwise missed by conventional genomic analyses, hypertranscription confers a worse prognosis, independent of somatic mutation burden, tumor ploidy, tumor stage, patient gender, age, or tumor subtype. Using single-cell analysis of multiple tumor regions, we identity specific clones that consistently produce copious amounts of RNA, irrespective of their clone size. We find that ETS family members are notable drivers of hypertranscription and then validate this in ETS-fused prostate cancer and Ewing sarcoma. In contrast to MYC-driven models, the most prevalent mechanism driving hypertranscription in primary cancer is through loss of transcriptional suppression. Having seen hypertranscription’s ubiquity, prognostic impact, and drivers, we explore whether it led to more expressed neoantigens. Using four cohorts of melanoma treated with immune checkpoint inhibitor, we find that patients with hypertranscription have higher expression of mutations, which predicts improved response to immunotherapy.
RESULTS
Measuring hypertranscription in vivo in human cancers
Gene expression profiling is typically performed by introducing similar amounts of RNA from different sources onto an experimental platform and then normalizing overall signal across samples. Inherent to expression profiling, including RNA sequencing (RNA-seq), is the assumption that each sample’s RNA has come from a similar number of cells. Without accounting for the number of cells the RNA derived from, it is currently not possible to measure hypertranscription (11). To overcome the challenges of analyzing hypertranscription in human tumors, we developed a new computational method. This method distinguishes mRNA transcripts originating from either the cancer or normal cell population within a primary tumor and then statistically models the change in cancer versus normal cell transcript abundance (expressed as a fold change). A key advantage of this approach, called RNAmp, is the ability to analyze already-sequenced human tumors—usually genetically heterogeneous and often nondiploid—whose RNA was derived from bulk tissue composed of an unknown number of cells.
To distinguish cancer cell from normal cell transcription, we used expressed somatic single-nucleotide substitutions (Subs) and germline single-nucleotide polymorphisms (SNPs) contained within regions of loss of heterozygosity (LOH) (Fig. 1A). A typical adult cancer contains ~17,000 somatic substitutions, of which ~134 are coding (12). LOH is also a common feature of cancer cells (13). Heterozygous SNPs in LOH regions will be monoallelically expressed in the tumor, whereas the intermixed normal cells with retained heterozygosity express both alleles. Considered together, expressed Subs and LOH-SNPs form hundreds to thousands of individual markers from which a tumor’s cancer cell–specific RNA output can be detected.
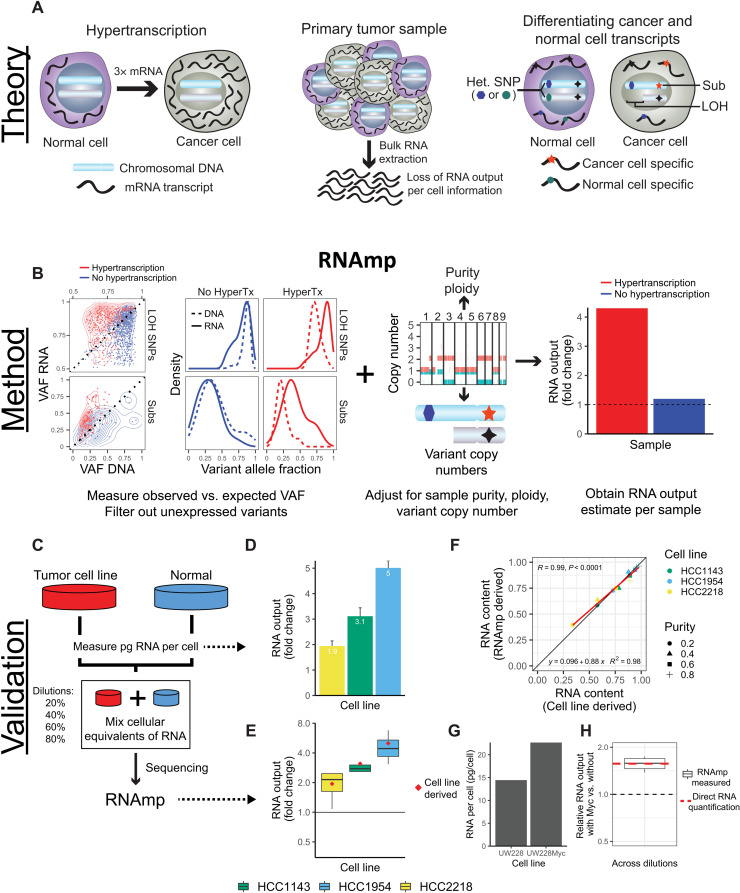
Fig. 1. Overview of RNA output analysis with RNAmp.
(A) Hypertranscription occurs when cancer cells elevate their RNA output above normal cell levels (left). Upon RNA extraction from primary tumor tissue, RNA output per cell information is lost (middle). Cancer cell– and normal cell–specific transcripts can be identified using tumor-specific marker variants, such as somatic substitutions (Subs) and LOH-SNPs (right). (B) DNA and RNA VAF distributions in samples with and without hypertranscription (HyperTX). Positive shifts in the RNA VAF of tumor-specific variants indicate that RNA output has increased. To estimate the overall fold change in RNA output of cancer versus normal cells, RNAmp incorporates these VAF shifts with tumor purity, ploidy, and local copy number data. (C) Cell number–normalized RNA-seq was performed on tumor and normal cell mixtures to validate RNAmp’s accuracy. RNA output per cell was measured before cell mixing. These mixtures were then sequenced and processed by RNAmp. (D) Fold change in RNA output levels of cancer cell lines measured by direct RNA quantification. Error bars correspond to SD. (E) RNAmp-derived RNA output measures (boxplots) compared to direct RNA quantification measures (red diamonds). Boxplot center line corresponds to the median, box limits are upper and lower quartiles, and whiskers represent 1.5 × interquartile range. (F) Pearson correlation of RNAmp-derived tumor RNA content estimates compared to direct RNA content quantification (R = 0.99, P < 0.0001). (G) RNA output per cell measured in medulloblastoma cells with and without MYC induction. (H) RNAmp-derived fold change in RNA output between UW228 Myc and UW228 wild-type cells (boxplot) compared to direct RNA quantification (red line). Boxplots are defined in (E).
RNAmp compares the variant allele fraction (VAF) of these markers in the RNA relative to DNA to quantify cancer cell–specific changes in RNA output (see Materials and Methods and Fig. 1B). When there is no elevation in the cancer’s global transcription, the fraction of reads supporting cancer variants in the RNA would be consistent with that of the DNA (i.e., similar VAFs). In cases of elevated RNA production, an increase in the fraction of RNA reads supporting cancer variants relative to the DNA is expected. To accurately quantify RNA output levels, we removed loci in imprinted regions and unexpressed variants and then corrected for tumor purity and regional DNA copy number (see Materials and Methods). Thus, RNAmp measures the relative fold increase in cancer cell transcription per DNA copy.
To assess the accuracy of RNAmp, we performed experimental analyses on three tumor-derived cell lines, mixed in different proportions with matched normal cells (Fig. 1C). Each of the cancer cell lines showed increased RNA output relative to their matched normal control (Fig. 1D and fig. S1A). The RNA from the mixed dilution samples also displayed increased RNA expression of tumor-specific markers (LOH-SNPs and Subs), relative to the nontumor-specific copy-neutral SNPs (fig. S1B). This was also true for silent mutations, demonstrating that selective pressure on coding mutations did not explain the increased expression of tumor-specific mutations (fig. S1C). We then applied the RNAmp algorithm, which accurately detected the level of hypertranscription in every mixed sample (Fig. 1E). Across all cell lines and at all purity levels, there was a high concordance between the observed and expected tumor RNA content (r = 0.99, P < 0.0001; Fig. 1F). In silico downsampling experiments verified the accuracy of RNAmp even when variant counts are low (down to 10 to 15 variants per sample) (fig. S1, D and E). Regardless, a minimum of 25 somatic substitutions or LOH SNPs and maximum tumor purity of 90% were used in all subsequent analysis.
To further validate RNAmp, we stably overexpressed MYC, a known driver of hypertranscription, in a cell line model of medulloblastoma. As expected, this led to increased RNA output (Fig. 1G and fig. S1F), with a transcriptional increase of ~57% in the Myc-expressing cells (Fig. 1H).
Hypertranscription is a hallmark of human cancer
Having validated that RNAmp could accurately measure cancer cell–specific hypertranscription, we set out to characterize hypertranscription across a spectrum of human cancers. We analyzed 141,167 Subs and 3,906,502 LOH-SNPS in 7494 tumors from 31 cancer types (see Materials and Methods and table S1). We initially measured differences between RNA and DNA VAFs across the whole cohort. A shift in VAF, toward RNA, was seen for both (somatic substitutions and LOH SNPs), suggestive of generally increased RNA output in human cancers (fig. S2A). As expected, no such change in VAF RNA was seen with diploid SNPs. Further, as was the case in our validation experiments, we saw a consistent increase in both missense and silent mutations in transcribed VAF RNA (fig. S2B).
Copy number and tumor purity were integrated, and then RNAmp was applied to the full cohort. Measures of RNA output compared between LOH-SNPs and somatic substitutions were moderately correlated (R = 0.51, P < 2.2 × 10−16; fig. S2C). However, most of RNAmp’s signal was derived from the far more frequent LOH-SNPs, as expected (fig. S2D). Across tumor types, cancer cells were more transcriptionally active than their normal counterparts, with a mean 2.22-fold increase in RNA output (Fig. 2A and table S2). Increased transcription was nearly universal in human cancer (80% of tumor with >1-fold increase), with a 2-fold or greater increase observed in 41% of tumors. RNA output correlated significantly with higher tumor mutation burden (TMB) (Fig. 2B) and ploidy (fig. S2E); particularly in genome-doubled tumors (2.6-fold versus 1.9-fold; P < 2.2 × 10−16; Fig. 2C). Notably, as RNAmp’s measures are normalized per tumor DNA copy, the increased transcription observed in genome-doubled tumors is “above and beyond” what would be expected given their increased DNA copy number.
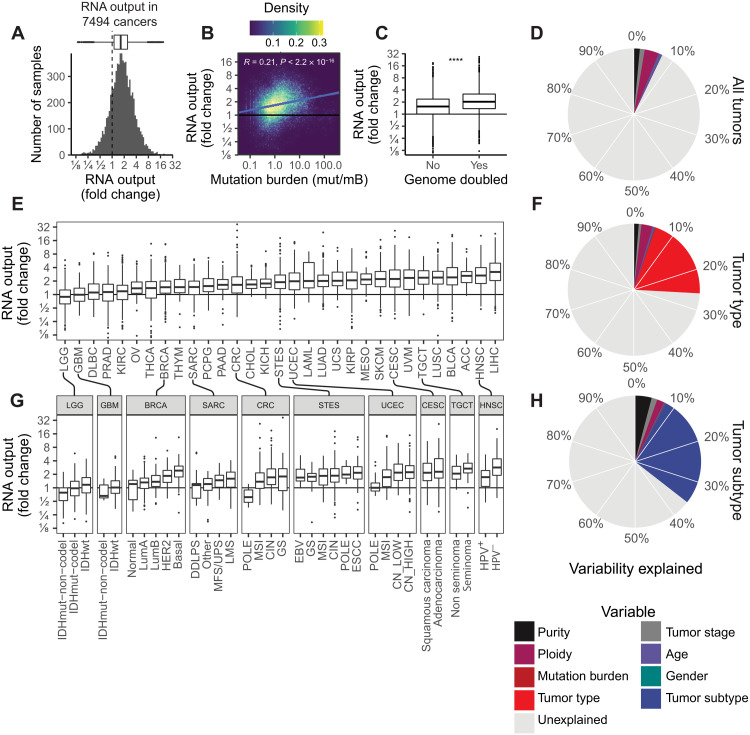
Fig. 2. The landscape of hypertranscription in primary human cancer.
(A) Histogram showing RNA output, expressed as a fold change, across 7494 primary tumor samples. Dashed line indicates onefold, meaning no change in RNA output level. (B) Pearson correlation between RNA output and TMB (P < 0.0001, R = 0.21). (C) Boxplot of RNA output levels in genome-doubled tumors versus nondoubled tumors (****P < 0.0001, Student’s two-sided t test). (D) Pie chart depicting the proportion of variability in RNA output that is explained by clinical features (purity, ploidy, tumor stage, age, mutation burden, and gender). The overall variability (7.1%) is explained by these features. (E) Boxplots of RNA output levels in tumor types. (F) Pie chart depicting the proportion of variability in RNA output explained including tumor type information. Nineteen percent more variance is explained by this model, for a total of 26%. (G) RNA output levels in tumor subtypes. (H) Pie chart depicting the proportion of variability in RNA output explained including tumor-type information. Nine percent more variance is explained by this model, for a total of 35%. Boxplots are defined in Fig. 1E. ESCC, esophageal squamous cell carcinoma; GS, genomically stable; LMS, leiomyosarcoma; SKCM, skin cutaneous melanoma; KIRC, kidney renal clear cell carcinoma; OV, ovarian; PAAD, pancreatic adenocarcinoma; CHOL, cholangiocarcinoma; UCS, uterine carcinosarcoma; KIRP, kidney renal papillary cell carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; UVM, uveal melanoma; LIHC, liver hepatocellular carcinoma. Tumor-type abbreviations can be found in table S1.
We wondered whether variability in RNA output was explained by differences in the tumors’ intrinsic features, such as the cell type they derived from, or somatically acquired changes. We used linear regression modeling to decompose the proportion of variability in RNA output that may be explained by common clinical and molecular features, including tumor stage, ploidy, mutation burden, and patient age. Only 7.1% of the global variability in transcriptional levels could be explained by these factors alone (Fig. 2D and table S3). Notably, tumor purity or the number of genes with zero counts per sample was not considered significant confounders to RNAmp’s measures (fig. S2, F and G).
We therefore further explored differences in RNA output between individual tumor types. Considerable variability in RNA output was seen across tumor types with median levels ranging from 0.9 to 3.2 (Fig. 2E) Some tumor types—such as skin cutaneous melanoma (SKCM), squamous lung cancers (LUSC), and head and neck squamous cell carcinoma (HNSCs)—displayed consistently high levels of hypertranscription (>25% above threefold). In contrast, others—such as brain, prostate, sarcoma and ovarian—had a much lower frequency of hypertranscription (<10% above threefold). Overall, however, individual tumor types accounted for an additional 19% of the variability of RNA output across cancer (total variance explained: 26%; Fig. 2F).
In many cancers, we observed several orders of magnitude separation between the least transcriptionally active samples from the highest. Examining the cohort based on established clinical subtypes resolved a significant amount of heterogeneity within cancer types (5 to 20%; Fig. 2G and fig. S2H), with canonically aggressive subtypes having the highest levels of hypertranscription. For example, in BRCAs, the more clinically aggressive basal-like subtype had the highest levels of hypertranscription (2.55-fold), followed by Her2 (2.13-fold), and then the less aggressive luminal B and A (1.60-fold and 1.38-fold) and normal (1.15-fold) subtypes. Similarly, across all gliomas (low and high grades), the clinically aggressive IDH–wild type samples had notably increased transcription (34% higher than IDH-mutated tumors). In HNSCs, higher-risk human papillomavirus–negative (HPV−) tumors had 80% higher RNA output compared to HPV+ tumors (3.5-fold versus 1.95-fold). In addition to demarcating aggressive subtypes, hypertranscription also correlated with distinct mutational subtypes. For instance, in colorectal cancer (CRC) and uterine corpus endometrial carcinoma (UCEC) types, subgroups that are driven by microsatellite instability (MSI) had more than doubled RNA output compared to the DNA polymerase epsilon, catalytic subunit (POLE)-mutated subtypes (2.5-fold versus 1.2-fold). Overall, tumor subtypes explained an additional 10% of the global variability in hypertranscription, bringing the total variability explained to ~36% (Fig. 2H).
Hypertranscription in single cells reveals transcriptionally dominant subclones
Having seen a high variability in RNA output between cancers (even of the same type), we wondered how much transcriptional heterogeneity exists within a single tumor. The RNA output of individual cells can be measured by incorporating unique molecular identifiers (UMIs), which tag each transcript per cell in standard single-cell RNA-sequencing assays. We obtained UMI-tagged single-cell RNA-seq (scRNA-seq) data from five patients with non–small cell lung cancer [representing The Cancer Genome Atlas (TCGA) types lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC)] (14). Each tumor contained three spatially distinct biopsies, enabling the analysis of transcriptional output differences between cells and tumor regions.
By comparing the overall proportion of transcripts derived from tumor and non-neoplastic cell populations in each tumor region, we can estimate each population’s RNA output fold change, similar to RNAmp (Fig. 3, A and B). Overall, tumor cells had increased RNA output for all patients and tumor regions (Fig. 3C). Furthermore, the RNA output fold change calculated from individual lung cancer cells was highly consistent with values derived from RNAmp applied to bulk-sequenced lung tumors (mean fold changes of 2.57 and 2.59, respectively; P = 0.95; Fig. 3B).
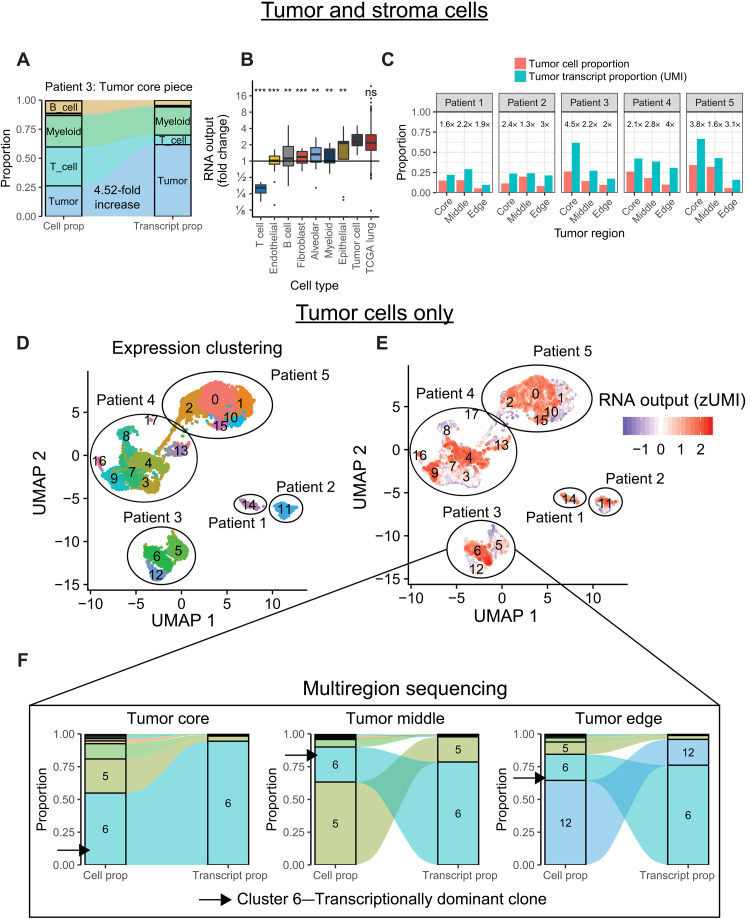
Fig. 3. Hypertranscription in single cells.
(A) Flow diagram depicting the proportional cell counts and transcript counts for different cell types from a primary lung cancer sample. Fold changes in RNA output between tumor and normal cell populations can be estimated from these data, similar to RNAmp. (B) Boxplot summarizing the relative fold change values in RNA output for various cell populations identified from scRNA-seq. Tumor cells have consistently elevated RNA output levels, equivalent to values derived from the bulk TCGA lung dataset (***P < 0.001 and **P < 0.01, Student’s two-sided t test). ns, not significant. (C) Bar charts of tumor cell proportion and tumor transcript proportion from five patients with multiregion scRNA-sequenced lung cancer. Cancer cells consistently increase their relative transcript proportion regardless of tumor region or tumor cellularity. Numbers above each set of bar plots indicate relative fold change in RNA output of tumor cells. (D) Uniform Manifold Approximation and Projection (UMAP) distance plot showing scRNA-seq expression clustering results for tumor cell populations. Subclusters were identified in patients 3 to 5. (E) RNA output of single cells overlaid onto the UMAP expression clusters reveals distinct subclusters of tumor cells within each sample undergoing hypertranscription. (F) Flow diagram depicting the proportional cell counts and transcript counts for different tumor subclusters across spatially distinct tumor regions from patient 3. Subcluster 6 maintains transcriptional dominance across tumor regions, even when it becomes a minority population by cell proportion. Boxplots are defined in Fig. 1E.
Comparing tumor regions to one another, we observed significant variability in hypertranscription between different sites of the same cancer (Fig. 3C). To see whether these spatial differences in RNA output were due to the existence of specific cell populations, we performed gene expression clustering of individual tumor cells (Fig. 3D) and then measured the RNA output of each subcluster (Fig. 3E). Tumor cells from the same patient tended to cluster together, regardless of tumor region, with distinct subclusters identified in three patients. Hypertranscribing tumor cells were primarily localized to only one or two clusters per patient (of three or more clusters). These hypertranscriptional cells were found in each tumor region. They retained their transcriptional dominance even in regions where they represented only a minority of cells (Fig. 3F and fig. S3, A and B). Ultimately, tumor regions with the highest concentration of hypertranscriptional cells were those with the largest fold increase in their RNA output. Together, these data show that specific tumor cell subpopulations are responsible for the majority of transcriptional activity within a tumor. These populations can be unevenly distributed across spatially distinct tumor regions yet still maintain transcriptional dominance irrespective of their clone size.
Consistent signaling pathways underpin oncogenic hypertranscription
Beyond MYC, which contributes to increased transcriptional output in cell lines (5), the drivers of oncogenic hypertranscription are unknown. Much in the same way that cancer genes can be oncogenic or tumor suppressive, we hypothesized that drivers of hypertranscription could do so via their expression being increased (such as MYC) or decreased (Fig. 4A). Because RNAmp uses standard RNA-seq data, it allows for the analysis of focal and global gene expression changes in tandem. We leveraged this to explore genes and pathways differentially expressed in tumors with hypertranscription.
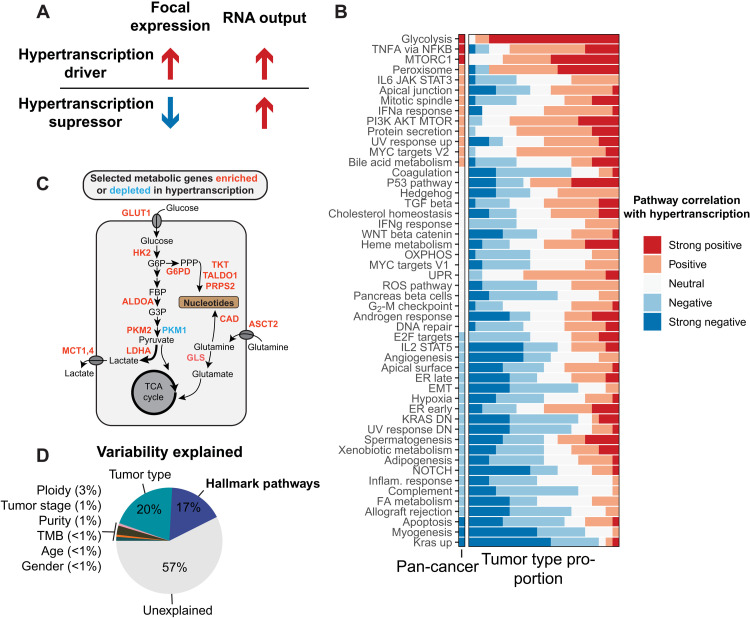
Fig. 4. Integrating focal and global gene expression data reveals pathways of oncogenic hypertranscription.
(A) Hypertranscription can be driven by specific genes and expression pathways either through their focal expression gain (drivers) or through their focal expression loss (suppressors). (B) Correlations between 50 hallmark signaling pathways and RNA output across the pan-cancer cohort and across individual tumor types (displayed as the proportion of tumor types with a given correlation) KRAS DN, KRAS down; DN, down. (C) Diagram depicting selected metabolic genes either enriched (red) or depleted (blue) in hypertranscribing samples. Genes involved in shunting glucose and glutamine toward nucleosynthetic pathways are all elevated in the hypertranscriptional state. TCA, tricarboxylic acid. (D) The proportion of variability explained in the pan-cancer cohort when including hallmark pathway expression. IL6, interleukin-6; JAK, Janus kinase; STAT3, signal transducer and activator of transcription 3; IFNa, interferon-a; PI3K, phosphatidylinositol 3-kinase; UV, ultraviolet; TGF, transforming growth factor; OXPHOS, oxidative phosphorylation; UPR, unfolded protein response; ROS, reactive oxygen species; ER, endoplasmic reticulum; EMT, epithelial-mesenchymal transition; FA, fatty acid.
Using ridge regression, we modeled the associations between hypertranscription and 50 hallmark signaling pathways (Fig. 4B, fig. S4, and table S4) (15). Master signaling pathways including tumor necrosis factor–a (TNFa)/nuclear factor kB (NFkB), mammalian target of rapamycin complex 1 (MTORC1), and peroxisome pathways were associated with hypertranscription. These pathways have been implicated as transcriptional activators across many cancers (16–18). The association between MYC and hypertranscription was confirmed in vivo (fig. S5, A and B). This was particularly evident in CRC and HNSC—tumor types characterized by frequent MYC amplification and elevated expression (fig. S5C) (19). The association in other tumor types was less evident (fig. S5, D and E). Beyond the hallmark pathways, we found that cancers harboring stem-like features display higher levels of RNA output (fig. S6, A and B), which is consistent with hypertranscription in rapidly proliferating stem cells (6, 20).
In general, hallmark pathways were as likely to activate as suppress hypertranscription, with the direction of association depending on tumor type (Fig. 4B). The major exception was glycolysis; in more than 80% of the tumor types analyzed, increased glycolysis was associated with increased hypertranscription. We wondered whether increased glycolysis helped tumors meet the elevated nucleosynthetic demands put upon a cell by hypertranscription itself. This could occur through pathways that shunt glycolytic carbon into nucleotide production. To explore this possibility, we measured the expression of key metabolic genes implicated in generating nucleotide precursors, including the provision of nitrogen and carbon for nucleotide synthesis, and found that nearly every gene was up-regulated in hypertranscribing tumors (Fig. 4C and fig. S6C). This included genes required for glucose and glutamine uptake (GLUT1 and ASCT2) and genes essential in the pentose-phosphate pathway (PPP), responsible for shunting either glycolytic carbon molecules (G6PD, TKT, TALDO1, and PRPS2) or glutamine-derived nitrogen (CAD) toward nucleotide synthesis. We further validated these findings by measuring expression of Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways, confirming that simple sugar metabolism and purine and pyrimidine metabolism are among the most active pathways in hypertranscriptional samples (fig. S6D). This was also validated in our single cell dataset—the same expression pathways that defined hypertranscribing tumors also defined intratumoral heterogeneity in RNA output (fig. S7).
Overall, the expression of hallmark signaling pathways explained a large amount of variability in tumor hypertranscription—an additional 17% of the pan-cancer variability in its RNA output (Fig. 4D). In more than two-thirds of cancer types, most of the variability in RNA output could be explained by the differential expression of these core signaling pathways (fig. S4B).
Oncogenic hypertranscription occurs by loss of transcriptional inhibition
To gain deeper insight into how hypertranscription occurs, we systematically identified and characterized transcription factors (TFs) modulating RNA output. Candidate TFs were identified using a stepwise approach (fig. S8A). First, all genes (including non-TFs) were given a score based on their enrichment in high– or low–RNA output tumors using Fisher’s test. Notably, this distribution was significantly enriched for genes involved in proteasomal degradation, ribosome biogenesis, splicing, and nucleocytoplasmic transport (fig. S8B). We then used this distribution to perform gene set enrichment analysis (GSEA) on TFs (482 total tested), filtering for those where both the TF and its targets showed significant enrichment in either hyper- or hypotranscriptional samples [false discovery rate (FDR) < 0.05]. In this way, we found 202 transcriptional modulators, predicted to regulate global transcriptional levels in one or more cancer types (table S5).
Consistent with our finding of the association between oncogenic signaling pathways and hypertranscription, the TFs identified were significantly enriched in cancer pathways (fig. S9A). Eighteen tumor types contained at least one TF modulator (range 1 to 79 per type, 481 total; fig. S9B) with 22 TFs found in ≥5 cancer types (fig. S9C). Twenty-eight genes were identified as putative drivers of hypertranscription in more than one tumor type (fig. S9D). MYC was among these genes, along with other known cancer drivers DROSHA, HMGA1, ETV4, and HIF1A.
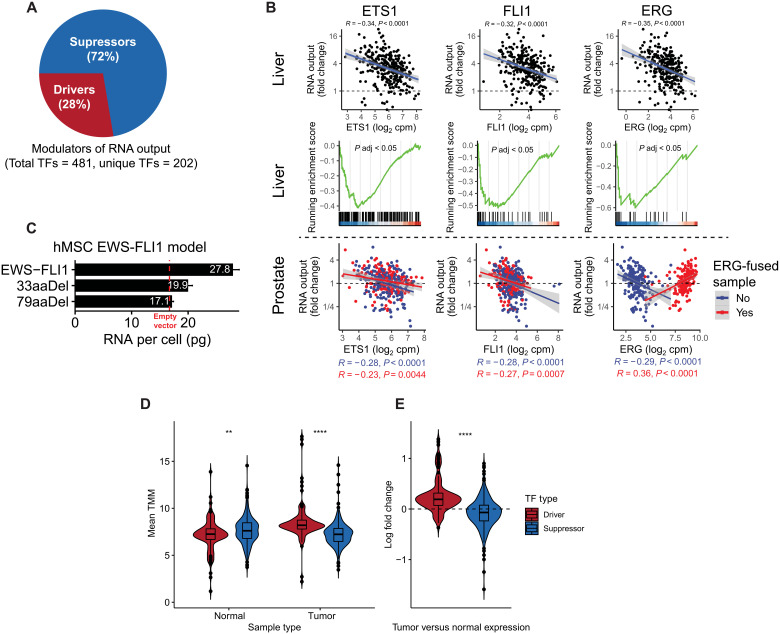
Most of the TF modulators of RNA output displayed a suppressive relationship with RNA output (72%)—that is, their increased expression led to decreased RNA output (Fig. 5A). For example, the expression of ETS family members ERG, FLI1, and ETS1 was significantly diminished in cancers with hypertranscription (Fig. 5B). ETS family members commonly form cancer driving fusions, which lead to their increased expression. Nearly half of prostate cancers harbor TMPRSS2-ERG fusions (21). Consistent with this, in TMPRSS2-ERG prostate cancers, the relationship between ERG expression and RNA output flipped—RNA output increased with elevated ERG expression, in contrast to prostate cancers with wild-type ERG. The EWSR1-FLI1 fusion is pathognomonic for Ewing sarcoma. To validate FLI1’s role as a modulator of hypertranscription, we stably expressed both the full length and a truncated version of the fusion in mesenchymal stem cells, the likely cell of origin of Ewing sarcoma, and then measured RNA output directly (see Materials and Methods). Consistent with our in silico analysis, full-length EWS-FLI1 led to a significant increase in RNA output compared to the empty vector control, while RNA output was restored to near control levels by introducing a C-terminal EWS-FLI1 deletion (Fig. 5C).
Fig. 5. Evidence of transcriptional derepression as a mechanism driving oncogenic hypertranscription.
(A) Pie chart depicting the proportion of TF drivers and suppressors of hypertranscription. (B) Top: Pearson correlation between ETS1, FLI1, and ERG and RNA output in liver cancer. Middle: ETS1, FLI1, and ERG target genes are enriched in hypotranscribing liver cancers and depleted in hypertranscriptional samples. Bottom: Pearson correlation between ETS1, FLI1, and ERG and RNA output in prostate cancers with or without ERG fusions. (C) RNA per cell measurements from a human mesenchymal cell model expression either full-length EWS-FLI1, empty vector, or C-terminal truncating mutations in FLI1 of either 33 or 79 amino acids in length. Error bars correspond to SD. hMSC, human mesenchymal stem cell. (D) Mean expression values of TF drivers and suppressors of transcriptional output in GTEx normal and TCGA tumor samples. TMM, trimmed mean of M values (E) Summarized log fold change in expression of TF driver and suppressor expression between tissue-matched tumor and normal samples. **P < 0.01; ****P < 0.0001.
Last, we compared the expression of TF modulators in tumors and tissue-matched normal samples from Genotype-Tissue Expression (GTEx) (table S6). In normal tissues, transcriptional suppressors were more highly expressed compared with transcriptional drivers, while the opposite trend was observed in tumor samples as expected (Fig. 5D). We then measured the log fold change in tumor versus normal expression for each gene-tumor-tissue–type pair, finding that transcriptional drivers become overexpressed in tumors, whereas transcriptional suppressors become underexpressed compared to their matched normals (Fig. 5E). Overall, these data suggest that loss of transcriptional suppression is critical to development of the hypertranscription phenotype during malignant transformation.
Hypertranscription predicts worse overall survival in multiple cancer types
The association between hypertranscription and aggressive cancer (e.g., basal-like BRCAs and IDH wild-type gliomas) led to the question: Does RNA output add prognostic information beyond what is already known from the tumor’s molecular subtype? Patients were grouped into hyper- and hypotranscription groups using an automated threshold finding approach, and survival analysis was performed (in cancers with sufficient numbers of events; see Materials and Methods). We performed Cox regression analysis, including several clinical and molecular covariates in our models such as tumor type, tumor stage, mutation burden, gender, and age at diagnosis. Hypertranscription predicted worse overall survival across cancer (50% versus 59% Cox-adjusted 5-year survival; fig. S10A). Patients with elevated RNA output had a 42% increased risk of mortality within the first 5 years of diagnosis, even when accounting for tumor type, mutation burden, tumor stage, and gender [fig. S10B; hazard ratio (HR), 1.42; 95% confidence interval (CI), 1.28 to 1.58; P < 0.0001).
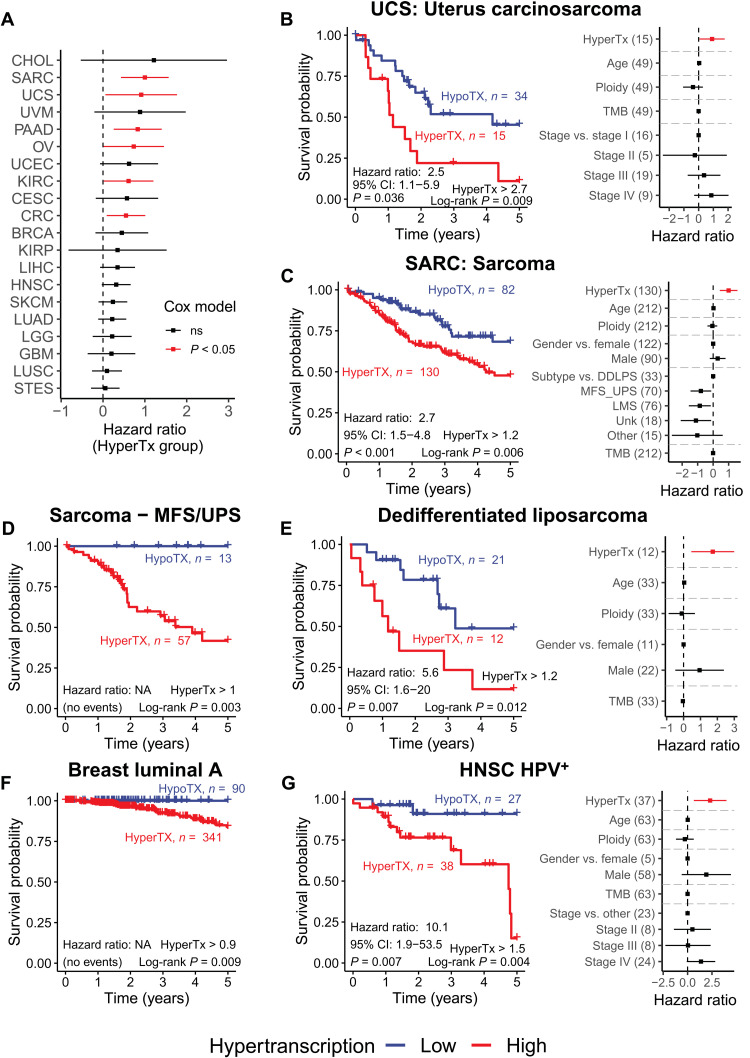
Hypertranscription was an independent prognostic factor in six cancer types (Cox-HR, P < 0.05), defining patient groups with significantly worse survival (even while accounting for somatic mutation burden, tumor ploidy, tumor stage, patient gender, age, or tumor subtype) (Fig. 6, A to C and fig. S10, C to F). Critically, hypertranscription’s prognostic utility across these types was also independent of expression of commonly used proliferative markers, KI67, proliferating cell nuclear antigen (PCNA), and minichromosome maintenance 2 (MCM2), or due to expression of MYC (expect for ovarian cancer; P = 0.068 with MYC included) (fig. S11, A and B). In uterine carcinosarcoma, a heterogeneous tumor of mixed epithelial and mesenchymal origin, the average 5-year survival for the hypertranscriptional group was 11% compared to 45% for the hypotranscriptional group (HR, 2.5; 95% CI, 1.1 to 5.9; P = 0.036; Fig. 5B). Notably, a previous study of this uterine carcinosarcoma cohort did not report significant associations between survival and several clinical and molecular features (22). Bone sarcomas were another tumor type in which hypertranscription had significant prognostic power and correlated with a ~21% decrease in 5-year overall survival (HR, 2.4; 95% CI, 1.4 to 4.2; P = 0.002; Fig. 6C).
Fig. 6. Hypertranscription defines patient subgroups with worse overall survival.
(A) Cox regression HRs for hypertranscriptional patients across 20 tumor types. Hypertranscriptional patients have consistently worse overall survival. In six tumor types, hypertranscription acts as an independent prognostic factor (red bars indicate Cox-HR, P < 0.05). (B to G) Kaplan-Meier survival plots (left) and Cox regression model HRs (right) for (B) uterus carcinosarcoma, (C) sarcoma, (D) myxofibrosarcoma and undifferentiated pleomorphic sarcoma (MFS/UPS), (E) dedifferentiated liposarcoma (DDLPS), (F) luminal A BRCA, and (G) HPV+ HNSC. Only Kaplan-Meier plots are shown for patients with MFS/UPS sarcoma and luminal A BRCA, as all hypotranscriptional patients survive preventing analysis by Cox regression. Error bars on all HR coefficients represent the 95% CI. NA, not applicable.
Hypertranscriptional thresholds were recalculated within each subtype to account for differences in subtypes’ RNA output levels, and survival analyses were reperformed. We again saw a consistent trend of worsened survival corresponding with increased RNA output in nearly every subtype analyzed (fig. S12A). In nine subtypes, hypertranscription correlated with a statistically significant decrease in survival by either the log-rank test or by Cox-adjusted survival (Fig. 6, D to G, and fig. S12, B to F).
For instance, in dedifferentiated liposarcomas (DDLPSs) and myxofibrosarcoma and undifferentiated pleomorphic sarcomas (MFS/UPS), hypertranscriptional patient subgroups had a 37 and 58% decrease in 5-year overall survival, respectively (DDLPS: HR, 3.7; 95% CI, 1.4 to 12.8; P = 0.04; MFS/UPS: log-rank P = 0.003; Fig. 6, D and E). In MFS/UPS, all 13 patients in the hypotranscriptional group survived compared to a 42% survival rate for patients with hypertranscription. Similarly, in luminal A BRCAs, all 90 patients in the hypotranscription group survived compared to the 84% 5-year survival rate in the hypertranscription group (Fig. 6F). In HPV+ and HPV− subtypes of HNSC, hypertranscriptional subgroups had a 76 and 17% decrease in 5-year overall survival, respectively (HPV+: HR, 10.1; 95% CI, 1.9 to 53.5; P = 0.007; HPV−: HR, 1.4; 95% CI, 1.0 to 2.0; P = 0.105; log-rank P = 0.048; Fig. 6G and fig. S12B). Overall, hypertranscription was a significant independent prognostic indicator in six subtypes, highlighting the ability for hypertranscription to uncover “hidden” tumor subtypes (Fig. 6, D to G, and fig. S12, C and D).
Transcriptional mutant abundance predicts immunotherapy response in nonhypermutant patients
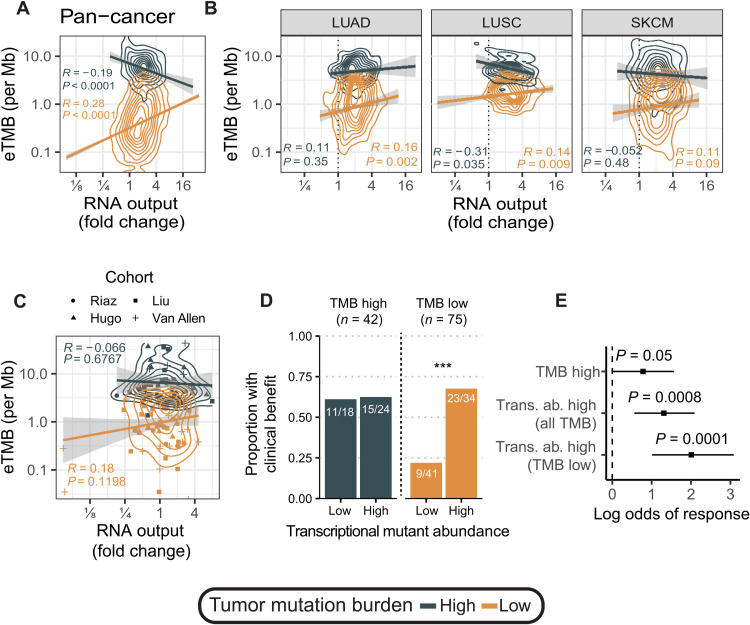
The success of immune checkpoint inhibition (ICI) therapy hinges on the immune system’s ability to recognize tumor cells as foreign. For this reason, high genomic TMB, yielding increased neoepitopes, is associated with ICI responsiveness (23). However, TMB alone is an imperfect predictor of ICI therapeutic response: Low-TMB (nonhypermutant) tumors can respond, while many high-TMB (hypermutant) tumors do not (24). We hypothesized that hypertranscriptional tumors, which, in effect, express more tumor-specific transcripts, including somatic mutations, would invoke a stronger immune response (10). To test this, we first quantified expressed TMB (eTMB) in the TCGA cohort by defining a mutation as expressed if it had ≥3 supporting reads and dividing by exome capture size (~30 Mb) to get expressed mutations per megabase. We then searched for correlations with hypertranscription. In low-TMB cancers (<10 coding mutations per megabase), eTMB increased with RNA output, while the opposite occurred in high-TMB tumors [>10 mutations (mut)/Mb] (Fig. 7A). Within lung and skin cancers, we found significant overlap in eTMB in tumors with low- and high-TMB tumors (Fig. 7B). This suggested that expressed mutation burden due to hypertranscription may better identify patients who would respond to ICI therapy. Low-TMB tumors can effectively “look like” high-TMB tumors in the setting of hypertranscription.
Fig. 7. Transcriptional mutant abundance as a biomarker for ICI response.
(A) Pan-cancer correlation between eTMB and hypertranscription for hypermutant (>10 mut/Mb) and nonhypermutant tumors (<10 mut/Mb). (B) Correlation between eTMB and hypertranscription for hypermutant (>10 mut/Mb) and nonhypermutant tumors (<10 mut/Mb) in lung cancers (LUAD and LUSC), and SKCM. (C) Correlation between eTMB and hypertranscription for hypermutant (>10 mut/Mb) and nonhypermutant tumors (<10 mut/Mb) in four melanoma ICI cohorts. (D) Proportion of patients with clinical benefit from ICI therapy in high- and low-TMB groups split by transcriptional mutant abundance levels. ***P < 0.001. (E) Log odds of response to ICI therapy for different TMB markers. Transcriptional mutant abundance is an overall better predictor of ICI response compared to genomic TMB. Error bars on log odds coefficients represent the 95% CI.
To see whether transcriptional mutant abundance was relevant in the context of ICI treatment, we investigated four clinical melanoma ICI cohorts for which both DNA sequencing and RNA-seq were conducted (25–28). Again, overlap in eTMB was observed for high- and low-TMB tumors (Fig. 7C). Overall, a greater proportion of patients with high TMB had clinical benefit compared to patients with low TMB (62% of hypermutant patients and 43% of nonhypermutant patients; fig. S13A). Because eTMB is simply a count of expressed mutations, it does not effectively capture how abundantly these mutations are expressed in the transcriptome. To measure true transcriptional mutant abundance, we integrated RNA output from RNAmp, VAFs, gene expression count data, and sample purity (see Materials and Methods). We observed no significant difference in transcriptional mutant abundance between low- and high-TMB tumors (fig. S13B). However, transcriptional mutant abundance was significantly elevated in clinically benefitting patients (fig. S13C). Upon closer inspection, we found that expressed mutation abundance was significantly elevated in patients with low TMB with clinical benefit (fig. S13D). Patients with low TMB but high transcriptional mutant abundance were as likely to benefit from ICI therapy as patients with high TMB (68% versus 62%; Fig. 7D). Overall, transcriptional mutant abundance had more predictive value for patients treated with ICI therapy, particularly able to identify nonhypermutant patients for whom ICI therapy was effective (Fig. 7E).
DISCUSSION
This study has shown elevated RNA output across human cancer. The pervasiveness of this phenomenon, seen in nearly every cancer type and frequently predictive of poor survival, strongly suggests that hypertranscription is an essential feature of cancer.
Multiple lines of evidence implicate hypertranscription with tumor aggressiveness. It is especially prevalent in tumors with high mutation load, doubled genomes, or markers of oncogenic stemness. Hypertranscription is not merely a general (nonspecific) phenotype; RNA output levels delineated new cancer subgroups and were independent prognostic factors, even after accounting for established molecular or histopathological markers of prognosis.
In this study, hypertranscription was defined as a relative measure, and hence, our method (RNAmp) was designed to estimate the transcriptional output of cancer cells versus all noncancer cells that are intermixed within a bulk tumor sample, as a relative fold change. We do not differentiate between different types of noncancer cells, which is a current limitation of our method. A recent manuscript, published while ours was under review, used a different approach to report that tumor-specific expression has prognostic and phenotypic importance (29).
What leads to hypertranscription in human cancer? We provide direct in vivo confirmation of MYC’s association with this phenotype. By measuring precise levels of hypertranscription in primary human tumors, we also reveal multiple additional pathways, many tumor type specific. Elevated glycolysis was associated with hypertranscription in almost every cancer type. This suggests that increased glycolytic flux supplies the nucleotides needed for the sustained growth of hypertranscriptional tumors.
In total, 202 putative drivers of hypertranscription were found, many of which are established cancer genes. While exploring how these genes regulated transcriptional output, a notable pattern emerged. Rather than hypertranscription being driven by a positive feedback loop, in which the activation of a key gene contributes to the elevated global expression (as is the case with MYC), we found that inactivation of transcriptional suppressors was a far more common route to achieving hypertranscription. It is likely more efficient to remove a barrier that keeps already poised transcripts from accumulating and then to turn on transcript production genome-wide. In general, studying hypertranscription may shed light on the fundamental nature of gene dysregulation in cancer, in which the balance between activating and suppressive signals is poorly understood.
Analysis of single cells revealed hypertranscriptional subclones responsible for producing the bulk of a tumor’s RNA, irrespective of the clone’s size. By cell proportion, these distinct populations were often minor clones yet still produced most of the tumor’s transcripts. These cells may represent the actively growing component of a tumor. Whether these cells maintain a consistent dependence on this high level of transcription for their survival is unclear. Future studies are warranted to understand the fluctuations in transcriptional output both between cells and across time, in relapsed cancers after therapy, as well as the contribution of epigenetic dysregulation to global transcriptional levels. It may be the case that hypertranscription represents a dynamic phenotype; activated when nutrients are available then turned off, or even reversed toward a “hypotranscriptional” survival state, when the tumor is challenged by therapeutics (30).
No matter how it is initiated, the clinical consequences of hypertranscription are important, suggesting novel drug strategies. Recently, therapies targeting the transcriptional machinery have emerged (31, 32), yet it is not always clear to whom these should be given. Notably, we found that many of the cancer types with reported sensitivity to transcriptional inhibition were those for which hypertranscription identified prognostically significant subgroups. This included HNSC (33) (Fig. 5G and figs. S8B and S9B), CRC (34) (figs. S7C and S9D), kidney cancer (35) (fig. S7E), and other cancers (36–43) (Fig. 5F and figs. S7, D and F; S8C; and S9, E and F). Whether hypertranscription can identify novel subtypes, or individual patients, sensitive to transcriptional inhibition will require future validation. A compelling example in this regard is provided by tumors with ETS fusions, including TMPRSS2-ERG prostate cancer and EWSR1-ETS Ewing sarcoma. In the setting of these fusions, RNA output was elevated. Consistent with this, Ewing sarcomas have been found to be particularly sensitive to transcriptional inhibition (44).
Last, hypertranscription identified patients with melanoma with improved response to immune checkpoint inhibitors, particularly in low-TMB tumors. Intriguingly, the burden of expressed mutations increased with RNA output specifically in patients with low TMB. This was not observed in high-TMB tumors, suggesting a threshold or protective mechanism that avoids excessive mutant overexpression. With accurate measures of hypertranscription, we quantified the abundance of expressed mutations, a powerful predictor of response in patients with low TMB.
Looking more broadly, the combined results reveal a new mechanism to subvert normal transcription used by human tumor cells in vivo. In addition to maintaining aberrant levels of specific genes belonging to select pathways, it is clear that tumors can also sustain increased gene levels across the genome, to their advantage. The relationship between local aberrant gene expression and global hypertranscription is akin to the balance between focal DNA copy number changes, restricted to key loci, and overall ploidy changes, involving the complete set of chromosomes (1). Future research will be needed to understand the relative importance of and balance between local versus global gene expression changes. From these data, it is likely that local and global transcription, when considered together, will explain heterogeneity in clinical presentation and patient survival.
Together, this study has shown that transcription differs in both type and amount across cancer. Hypertranscription represents an unappreciated dimension of oncogenic signaling. While it is often thought of as having carefully balanced levels, gene expression can undergo marked global shifts, with consequences for tumor subtyping, patient prognostication, and response to novel therapies.
MATERIALS AND METHODS
Overview of the RNAmp method
Solid tumors are typically preserved as bulk tissue, which is composed of an unknown number of cells. Without knowing the number of cells from which the nucleic acid was extracted, it is not possible to measure RNA output per cell. Likewise, many tumor specimens are made up of multiple genetically distinct cell populations, which also includes an unknown amount of stromal (normal cell) contamination. Once processed, the tumor cells’ contribution to the total RNA pool becomes unknown. To measure cancer cell–specific transcriptional output, one would need to perform cell sorting (to account for normal cell contamination), then normalize for the number of cells (11), and use RNA spike-in controls mixed into the sequencing run itself (45). Even if these additional steps were technically feasible for ongoing specimens (without destroying the RNA), they have not been used by most publicly available RNA-seq datasets, which includes the nearly 10,000 tumor samples from TCGA.
To overcome these challenges, RNAmp uses somatic substitutions (Subs) and LOH-SNPs as markers of tumor-specific transcription. By quantifying the relative proportion of sequencing reads supporting these marker variants in both the DNA and RNA and integration of tumor copy number and purity, one can assess relative fold change in transcriptional output between cancer and normal cells within a primary tumor sample. The calculations for measuring transcriptional output from Subs and LOH-SNPs are derived separately below. These metrics are then summarized to derive a final fold change estimate for transcriptional output levels.
Measuring transcriptional output using somatic substitutions
The RNA fraction (VAFRNA) of a given mutation (i) at locus (l) can be predicted by dividing the number of mutant RNA transcripts produced per tumor cell at locus (l) by the total number of RNA transcripts (both mutant and nonmutant) produced from that locus by both cancer and normal cells
| (1) |
For a mutation with copy number, CM, in a tumor of a purity, p, local tumor total copy number, CT, and with normal copy number, CN, the RNA fraction can be approximated if the level of hypertranscription (amp) at locus l is known
| (2) |
where CM * amp represents the number of RNA copies produced from chromosomes harboring the mutated allele per cancer cell, CT * amp represents the number of RNA copies produced from both mutant and normal chromosomal alleles per cancer cell, and represents the number of RNA copies produced per contaminating normal cell. The mutation copy number (number of chromosomal alleles harboring the mutation per cancer cell) is given by (46)
| (3) |
Substituting Eq. 3 into Eq. 2 rearranging to solve for amp gives us
| (4) |
Measuring transcriptional output using LOH-SNPs
The RNA fraction (VAFRNA) of a given LOH SNP (i) at locus l is predicted by dividing the number of RNA transcripts with the variant allele produced per tumor and normal cell at a given locus by the total number of RNA transcripts produced from that locus.
| (5) |
For an SNP with copy number, CS (see Eq. 13), in a tumor of a purity, p, local tumor total copy number, CT, with normal copy number, CN, and normal minor copy number CNm, the RNA fraction can be approximated if the level of hypertranscription (amp) at locus l is known
| (6) |
where CS(i,l) * amp(l) represents the number of alternate allele RNA copies produced from the tumor, CT(i,l) * amp(l) represents the total number of RNA copies produced from the tumor, and and represent the number of variant allele and total copies produced per contaminating normal cell, respectively. Substituting 1 and 2 for the minor and total normal copy number (as is expected on normal autosomal chromosomes) and then rearranging to solve for amp give
| (7) |
RNAmp variant filtering and final calculation
To be included in RNAmp’s analysis, variants were filtered for only missense or silent changes in loci with sufficient read depth (>8 reads in the DNA and >30 reads in the RNA) and located in autosomal regions. Somatic variants were filtered to include only clonal mutations as identified using ASCAT (allele-specific copy number analysis of tumors) copy number calls and the ABSOLUTE method (46). These filters ensured that we only considered high-quality variants, in regions that were expressed, and variants that were not affected by strong selection pressures (such as stop-gain or stop-loss mutations).
Our measure of transcriptional output was focused on changes in transcription of both alleles (normal and mutated) across the entire transcriptome. To arrive at a final estimate of global transcriptional output fold change, the VAF DNA and RNA, as well as copy numbers for Subs and LOH-SNPs, are summarized across all variants passing depth filters before applying the RNAmp algorithm outlined above. Samples that do not contain at least 25 Subs or LOH-SNPs are excluded from analysis. For samples with only 25 or more variants of either Sub or LOH-SNPs, the RNAmp estimate derived from that variant type is used as the final RNAmp estimate. For samples that contain 25 or more of both Subs and LOH-SNPs, the fold change estimates are mean-weighted together on the basis of the number of each variant type present, giving the final fold change estimate for transcriptional output. Last, samples with purity above 90% or below 10% are removed from final analysis, as these samples contained insufficient normal cells to estimate RNAmp. This yielded a final dataset of 7494 TCGA tumors for analysis.
Tumor RNA content measurement
The theoretical tumor RNA content per sample—that is, the proportion of all RNA in a tumor sample that is cancer cell–derived—is given by
| (8) |
where p is purity, RNAt is RNA output per tumor cell, and RNAn is RNA output per normal cell. Given that
| (9) |
We then substitute for RNAn in the denominator and simplify to give
| (10) |
Thus, given the relative fold change in transcriptional output of tumor cells versus normal cells and tumor purity and ploidy, we can estimate the proportion of tumor-derived RNA in a mixed sample.
Validation of the RNAmp method
The cell lines HCC1954, HCC1143, HCC2218, HCC1954BL, HCC1143BL, and HCC2218BL were obtained from American Type Culture Collection and cultured in RPMI 1640 with 10% fetal bovine serum (FBS). UW228 cells were obtained from J. R. Silber (University of Washington) and cultured in α–minimum essential medium with 10% FBS. UW228 cells were made to stably express c-Myc by infection with pMN–GFP–c-Myc as previously described (47). Cells were harvested and counted using the Vi-CELL XR Cell Viability Analyzer (Beckman Coulter) before DNA and RNA extraction using the AllPrep DNA/RNA Mini Kit (QIAGEN) and RNA quantification using NanoDrop 1000 (Thermo Fisher Scientific) to generate per cell estimates of RNA output and fold change RNA output values. RNA from tumor and normal cell lines was then mixed in RNA cellular equivalents to create dilutions of 0, 20, 40, 60, 80, and 100% purity. External RNA Controls Consortium (ERCC) RNA spike-ins were added to RNA samples normalized to cell number before sequencing. UW228 does not have a matched normal; therefore, an unmatched peripheral blood cell line was used (HCC1954BL). These mixtures underwent library preparation using NEBNext and RNA-sequenced to at least 100× depth (average per base coverage across each transcript, averaged across all transcripts) using the Illumina HiSeq 2500. All RNA-seq libraries generated were paired-end 2× 126–base pair read length, each with >100 million mapped reads. DNA was extracted from the pure cell lines and underwent whole-exome sequencing (WES) using Agilent’s exome enrichment kit (Agilent SureSelect V5) as previously described (4). All sequencing was performed at The Centre for Applied Genomics (TCAG) at the Hospital for Sick Children. DNA from UW228 and HCC2218 cells was also used for Affymetrix CytoScan HD SNP array analysis. Affymetrix SNP6 array data were downloaded for HCC1954 and HCC1143 cell lines (sample Gene Expression Omnibus accessions: GSM888116 and GSM847319). Mutation calling was performed using MuTect2 (v3.5-0), and DNA copy number was derived using the Tumor Aberration Prediction Suite (TAPs v2.0) (48). For the UW228 cell line, LOH-SNPs were identified by finding the union between heterozygous SNPs in the HCC1954BL normal cell line and matching alleles in LOH regions of the UW228 cell line. DNA VAFs in the impure samples were corrected on the basis of purity and mutation copy number using the following equations for germline and somatic variants, respectively [adapted from (46)]
| (11) |
| (12) |
Samples were then processed using the RNAmp method using parameters identical to those described above.
Downsampling experiment
To test RNAmp’s stability when variant counts are low, we used our validation dataset of three BRCA cell lines (HCC1143, HCC1954, and HCC2218) and took 1000 bootstrapped subsamples of either LOH-SNP or somatic variants at different variant counts (2, 5, 10, 15, 20, 25, 50, 100, 250, 500 or 1000 depending on the total variants in a sample). We then recomputed RNA output for each of these subsamples and compared the resulting value to RNAmp’s original estimate (using the full set of variants).
TCGA dataset
Matched exome (tumor and normal) and RNA-seq (tumor-only) were downloaded from the Genomic Data Commons Portal (https://portal.gdc.cancer.gov/) for 9727 TCGA tumors. Affymetrix SNP6 CEL files (tumor and normal) were downloaded for 9211 tumors. Somatic mutation data in the mutation annotation format (MAF) produced by MuTect were downloaded from the GDC portal (v1.0.1). Clinical and tumor subtype information were obtained from the TCGA Pan-Cancer Atlas (49).
TCGA germline variant calling
Germline SNPs were identified from matched normal exome sequence data using GATK’s best practices (GATK v3.7). Briefly, each sample was first processed using HaplotypeCaller in single-sample genotype discovery mode. Joint genotyping was subsequently performed across the entire cohort. Variants were filtered using GATK’s Variant Quality Score Recalibration using known polymorphic sites from HapMap (v3.3) and Illumina’s Omni 2.5 M SNP chip array for 1000 Genomes samples as true sites and training resources, 1000 Genomes high-confidence SNPs as nontrue training resource, and dbSNP (v138) for known sites but not training. The truth sensitivity filter level was set to 99.5%. Germline SNPs were filtered to select only biallelic heterozygous SNPs with a genotype quality score above 30.
TCGA allele-specific copy number analysis
Raw SNP6 CEL files were first preprocessed using the PennCNV-Affy pipeline (http://penncnv.openbioinformatics.org/en/latest/user-guide/affy/) to generate LogR and BAF values for each sample. Briefly, Affymetrix Power Tools software was used to generate genotype clusters (apt-genotype) and to perform quantile normalization and median polish to produce signal intensities for A and B alleles of SNPs (apt-summarize). PennCNV was then used to convert the signal intensities into LogR and BAF values (normalize_affy_geno_cluster.pl). LogR and BAF files were then processed in R using the ASCAT R package (v2.4) to generate allele-specific copy number calls and purity and ploidy estimates for each sample.
The copy number status of MYC was defined using ASCAT and defined parameters (https://cancer.sanger.ac.uk/cosmic/help/cnv/overview). Briefly, a total copy number greater than or equal to 5 in a sample with ploidy less than 2.7 or a total copy number greater than or equal to 9 in a sample with ploidy greater than 2.7 is defined as copy gain events.
TCGA variant processing and allele counting
Somatic and germline single-base variants were merged into a single VCF file for each sample and annotated using vcf2maf v1.6.12 (https://github.com/mskcc/vcf2maf) and the Ensembl Variant Effect Predictor (v86) to produce annotated MAF files for each sample. Allele counting was performed on variant sites for each sample using GATK’s ASEReadCounter on matched exome and RNA-seq data. Minimum read mapping quality and minimum base quality were set to 10 and 2, respectively. Depth downsampling was turned off.
The copy numbers of each SNP, CS, were determined from tumor exome read count data using the following equation [adapted from (46)]
| (13) |
These values were used to determine whether the reference or alternate allele at a given loci was lost in regions of LOH. SNPs where the exome-derived SNP copy number did not match the copy number status as given by ASCAT were removed before analysis. To harmonize all LOH-SNPs, we inverted the reference and alternate allele counts for SNPs in regions where the alternate allele was lost before analysis.
Variability-explained analysis
To determine the variance explained in transcriptional output levels by predictor variables, we used the relaimpo R package (v2.2-3) setting method = “lmg” and rela = TRUE (50). We assessed the proportion of additional variability explained by tumor types and tumor subtypes by adding each in turn and comparing the differences in variability explained between each model.
Gene expression analysis
Duplicate reads were removed from RNA-seq data using Picard (v2.7.1) MarkDuplicates before gene- and exon-level expression counting. Gene expression counts were generated using HTSeq (v0.6.0). Exon expression counts were created using the dexseq_count.py script (v1.21.1). GENCODE V25 gene annotations were used for both genes and exons. Counts were normalized using the counts per million method for correlation analysis (51).
Gene lists for the 50 hallmark expression pathways were obtained from the Molecular Signatures Database (v6.2). To measure expression of the 50 hallmark expression pathways, we used gene set variation analysis (GSVA; v1.32.0) (52) on Reads per kilobase million (RPKM)–normalized gene expression counts. We trained a ridge regression model using a leave-one-out cross-validation approach. Our model included transcriptional output levels as the outcome variable and hallmark pathway expression data (50 pathways), purity, ploidy, tumor type, mutation burden, tumor stage, gender, and age at diagnosis as predictors. Sixty-four patients had missing values for one of TMB, tumor stage, gender, or age and were removed before ridge regression analysis. We repeated this procedure within tumor types in which at least 80 samples contained information for all included predictors and plotted the resulting normalized coefficients as a heatmap. To assess the variability explained by hallmark pathway expression, we performed analysis of variance (ANOVA) with all 50 pathways included alongside all covariates used in the original variability-explained model and assessed, in aggregate, how much additional variability in each model was explained by inclusion of all hallmark pathway expression levels. This analysis was performed both across the pan-cancer cohort and within individual tumor types. Pathway correlations were summarized into groups on the basis of the strength of the correlation coefficient from the ridge regression as follows: strongly positive > 1, positive > 0.25, neutral between 0.25 and −0.25, negative < −0.25, and strongly negative < −1.
Metabolic gene analysis
A list of relevant metabolic genes involved in either the Warburg effect or rate limiting for nucleotide synthesis in cancer were manually curated from review papers (53, 54). KEGG metabolic pathways were curated from the Molecular Signatures Database and processed by GSVA to produce pathway-level expression values. Pearson correlations between each of these gene or pathway expression values and hypertranscription were determined. P values were adjusted using the FDR method.
Stemness analysis
mRNA expression–based stemness index values were obtained from (55). These values, which scale between 0 and 1, were derived from a one-class logistic regression machine learning algorithm trained on stem cell classes, differentiated ecto-, endo-, and mesoderm progenitors, and then applied to TCGA RNA expression data. Stemness gene sets were curated by literature review and reflect signatures meant to capture stem-like or dedifferentiated cancer cell states (56–60). Pathway activity levels were determined using GSVA on RPKM-normalized gene expression counts. Correlations to hypertranscription levels were determined using Pearson correlation, and adjusted P values were produced using the FDR method.
scRNA-seq analysis
Raw scRNA-seq data were obtained from Lambrechts et al. (14) and reprocessed using the R package Seurat v3.0.1 using the SCTransform R function to perform normalization before plotting by Uniform Manifold Approximation and Projection (UMAP). This dataset contained 15 scRNA-seq experiments representing three spatially distinct tumor regions from five patients with lung cancer. To compare transcriptional output across these samples, UMI counts from each scRNA-seq run were z-scale–normalized. UMIs tag each unique transcript from each cell and therefore represent global transcriptional output in single cells. Fold changes in transcriptional output were estimated by taking the average zUMI score for a given cell population or subcluster compared with all other cells from a given sample, a calculation that is comparable to measurements made by the RNAmp method. We directly compared cell proportions in each tumor piece for each tumor subcluster to the overall proportion of transcripts derived from each subcluster to infer transcriptionally dominant clones. To measure expression of glycolysis, MYC targets, MTORC1, and embryonic stem cell (ESC) pathways, GSVA was performed on single-cell count data using the respective pathways (52).
Hypertranscriptional driver analysis
To determine genes responsible for driving changes in transcriptional output, we reasoned that a putative driver would meet certain criteria. First, we restricted our analysis to TFs. These factors should be themselves correlated with transcriptional output, and expression of their target genes should be enriched in either high– or low–RNA output samples.
TFs and their targets were curated from several public databases (61–65). For the ENCODE database (64), targets were selected on the basis of chromatin immunoprecipitation sequencing peaks with a score of 1000 or more. TFs were filtered to include only those with between 5 and 500 targets. In total, 482 TFs were selected for further analysis on the basis of these filtering criteria.
To create a transcriptional output dataset amenable to GSEA with the TFs and target lists, we scored 16,793 genes for their association with hypertranscription using Fisher’s test after median splitting expression values for each gene. For each gene, this analysis returned an odds ratio related to the enrichment or depletion of a given gene in high–transcriptional output samples. By log-transforming the resulting distribution of 16,793 genes, we obtained a normally distributed log score allowing for GSEA using the TF target gene lists. Fisher’s test and Pearson correlation P values for individual gene correlations were adjusted using FDR within each tumor type. Final TFs were filtered for those with a significant target enrichment in addition to a significant Pearson correlation and Fisher’s test P value, leading to the final list of 202 unique TFs (482 total hits) across 18 tumor types. TFs whose expression was positively correlated with RNA output were considered drivers, and those where their expression was negatively correlated with RNA output were considered suppressors.
For each putative transcriptional driver and suppressor gene, we computed the mean expression of each gene in each tumor type and in a cohort of GTEx tissue-matched normal samples (table S6). We then took the fold change between each genes’ tumor expression level and normal expression level and compared the transcriptional amplifier and suppressor genes distributions. TCGA prostate cancer samples with ERG fusions were identified from (66).
Human mesenchymal stem cell EWS-FLI1 RNA output analysis
Human mesenchymal stem cells (hMSCs) were made to stably express either full-length EWS-FLI1 or EWS-FLI1 C-terminal truncal deletion mutants distal to the DNA binding domain of FLI1 (either 33 or 79 amino acids in length), before cell counting and RNA quantification. Briefly, the p53 and retinoblastoma tumor suppressor pathways were inactivated by introducing the HPV-16 containing E6 and E7. Human telomerase reverse transcriptase was used to then immortalize the hMSCs. Cells were grown in triplicate, and each cell count was also performed in triplicate before RNA quantification using NanoDrop.
Survival analysis
To accommodate the variable follow-up times in each tumor cohort, we focused our analysis on 5-year overall survival. To determine prognostically relevant hypertranscription thresholds in individual tumor types and subtypes, we used the R package OptimalCutpoints (v1.1-4) and maximized Youden’s index (67). Each tumor type or subtype was assigned an independently defined RNA output threshold, above which we considered samples to have hypertranscription.
We filtered out tumor types with 10 or fewer events (which excluded DLBC, KICH, PCPG, PRAD, TGCT, THCA, and THYM), 10 or fewer survivors (which excluded LAML). For the subtype-specific analysis, those without at least five or more events were removed before analysis (which excluded BRCA normal, CRC MSI CIMP, CRC invasive GBM IDHmut-non-codel, SARC other, STES POLE, UCEC CN low, UCEC UCEC MSI, UCEC POLE subtypes).
Instances in which the high- or low-hypertranscription groups made up more than 90% of a tumor type’s samples were removed (which excluded types ACC, BLCA, and MESO). For subtypes, this cutoff was set at 95% [which excluded BRCA basal, HNSC HPV−, LGG IDHmut-codel, LGG IDHwt, STES Epstein-Barr virus (EBV), and STES chromosomal instable (CIN)]. The remaining tumor types (n = 20) and subtypes (n = 15) were used for Kaplan-Meier survival analysis and Cox regression. Tumor type, subtype, stage, age at diagnosis, TMB, purity, ploidy, race, and gender were included in Cox regression models when available. Patients with missing values for one of TMB, tumor stage, gender, or age were removed before survival analysis. To assess MYC, MCM2, KI67, and PCNA expression and survival, we median split each group based on each genes’ expression and included it as a covariate in the Cox regression.
ICI dataset and expressed mutation burden
Whole-exome and RNA-seq data were downloaded for ICI-treated patients with melanoma (25–28). Only ICI-naïve, pretreatment samples were selected for analysis. WES sequence data were aligned as previously described (4). RNA-seq data were aligned using STAR (v2.4.2a) in two-pass mode (68). Somatic mutation data were downloaded from supplementary tables from the original publications. Allele-specific copy number calling and LOH-SNP identification was performed using FACETS (v0.6.1) on the matched tumor-normal WES data (69). Samples were then processed using RNAmp using default parameters, except for the Riaz cohort (27) in which duplicate reads were included in the allele-counting step for the RNA-sequenced data.
Only missense, nonsense, and nonstop mutations were considered for the eTMB analysis. To be considered expressed, a mutation required at least three alternate reads support it in the RNA. To estimate mutation burden per megabase, we only included mutations located within coding exons that were common across multiple exome capture kits (including the TCGA), which totaled 28.7 Mb. Clinical benefit was defined as patients with complete or partial response or those with stable disease after 1 year.
Transcriptional mutant abundance
Transcriptional mutant abundance refers to the average expression level of each mutation in a sample. Gene expression counts from each sample were normalized using GeTMM (70). For each mutation, we estimate the transcriptional mutant abundance by first multiplying the normalized counts for the gene containing the mutation by the VAF of that mutation in the RNA. Then, a correction factor is applied that accounts for tumor purity, hypertranscription, and tumor copy number–related impact on expected mutation counts as follows
| (14) |
| (15) |
where is the tumor ploidy–corrected hypertranscription level and is the normal:tumor cell ratio.
Acknowledgments
We thank all the patients and their families who contributed to this study. We thank TCAG NGS and Biobanking facility for cell line culturing and sequencing services. We thank researchers involved in dbGap study phs000452, L. Garraway, E. Lander, and S. Gabriel and NHGRI (grant #U54 HG003067) for providing melanoma ICI sequencing data used in this study. We also thank T. A. Chan, A. Ribas, and R. S. Lo for access to ICI datasets. We thank B. Thienpont, B. Boeckx, and D. Lambrechts for providing scRNA-seq data. We thank M. Mahendralingam and M. Ramalho-Santos for providing feedback on the manuscript. The results published here are based on data generated by the TCGA Research Network: www.cancer.gov/tcga.
Funding: M.Z. was personally supported by a SickKids Restracomp award. T.S. was supported by a Canadian Institutes of Health Research Canada Graduate Scholarship. A.S. is partially supported by an Early Researcher Award from the Ontario Ministry of Research and Innovation, the Canada Research Chair in Childhood Cancer Genomics, funding from the V Foundation, and the Robert J. Arceci Innovation Award from the St. Baldrick’s Foundation. The Children’s Cancer Foundation Inc. (to J.A.T.) and NIH grant R01CA233619-01A1 (to A.S. and J.A.T.). A.H. received funding from the Canadian Institutes for Health Research (CIHR; grant #178414) and is the Tier 1 Canada Research Chair in Rare Childhood Brain Tumors.
Author contributions: A.S. designed the study. M.Z., F.F., R.R., F.C., and L.-M.E. performed the data analysis. T.S. and S.P.S. performed the experiments and data collection. R.D., G.H.J., F.N., S.G., and M.D.H. provided the sequencing data. F.N., S.G., J.A.T., M.D.H., U.T., and A.H. provided the technical support and conceptual advice. M.Z. and A.S. wrote the manuscript.
Competing interests: M.D.H. reports grants from BMS; personal fees from Achilles, Arcus, AstraZeneca, Blueprint, BMS, Eli Lilly, Genentech/Roche, Genzyme/Sanofi, Janssen, Immunai, Instil Bio, Mana Therapeutics, Merck, Mirati, Natera, Pact Pharma, Shattuck Labs, and Regeneron; and equity options from Factorial, Immunai, Shattuck Labs, and Arcus. A.S. and M.Z. report a filed patent application related to the use of tumor-specific transcription to predict patient prognosis and response to immunotherapy (WO 2022/094720 A1). M.D.H. reports a patent filed by Memorial Sloan Kettering related to the use of tumor mutational burden to predict response to immunotherapy (PCT/US2015/062208) that is pending and licensed by PGDx. J.A.T. is a founder and consultant with Oncternal Therapeutics Inc. No licensed agents were used in these investigations. All authors declare that they have no other competing interests.
Data and materials availability: Source code for RNAmp can be found at GitHub (https://github.com/shlienlab/rnamp) and Zenodo (https://doi.org/10.5281/zenodo.6807299). NGS data generated for the validation experiment have been deposited in EGA (EGAS00001006365). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Other Supplementary Material for this manuscript includes the following:
Data S1 to S6
REFERENCES AND NOTES
- 1.Bielski C. M., Zehir A., Penson A. V., Donoghue M. T. A., Chatila W., Armenia J., Chang M. T., Schram A. M., Jonsson P., Bandlamudi C., Razavi P., Iyer G., Robson M. E., Stadler Z. K., Schultz N., Baselga J., Solit D. B., Hyman D. M., Berger M. F., Taylor B. S., Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet. 50, 1189–1195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y., Roberts N. D., Wala J. A., Shapira O., Schumacher S. E., Kumar K., Khurana E., Waszak S., Korbel J. O., Haber J. E., Imielinski M.; PCAWG Structural Variation Working Group, Weischenfeldt J., Beroukhim R., Campbell P. J.; PCAWG Consortium , Patterns of somatic structural variation in human cancer genomes. Nature 578, 112–121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nik-Zainal S., Alexandrov L. B., Wedge D. C., Van Loo P., Greenman C. D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L. A., Menzies A., Martin S., Leung K., Chen L., Leroy C., Ramakrishna M., Rance R., Lau K. W., Mudie L. J., Varela I., McBride D. J., Bignell G. R., Cooke S. L., Shlien A., Gamble J., Whitmore I., Maddison M., Tarpey P. S., Davies H. R., Papaemmanuil E., Stephens P. J., McLaren S., Butler A. P., Teague J. W., Jönsson G., Garber J. E., Silver D., Miron P., Fatima A., Boyault S., Langerod A., Tutt A., Martens J. W. M., Aparicio S. A. J. R., Borg Å., Salomon A. V., Thomas G., Borresen-Dale A. L., Richardson A. L., Neuberger M. S., Futreal P. A., Campbell P. J., Stratton M. R., Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell B. B., Light N., Fabrizio D., Zatzman M., Fuligni F., de Borja R., Davidson S., Edwards M., Elvin J. A., Hodel K. P., Zahurancik W. J., Suo Z., Lipman T., Wimmer K., Kratz C. P., Bowers D. C., Laetsch T. W., Dunn G. P., Johanns T. M., Grimmer M. R., Smirnov I. V., Larouche V., Samuel D., Bronsema A., Osborn M., Stearns D., Raman P., Cole K. A., Storm P. B., Yalon M., Opocher E., Mason G., Thomas G. A., Sabel M., George B., Ziegler D. S., Lindhorst S., Issai V. M., Constantini S., Toledano H., Elhasid R., Farah R., Dvir R., Dirks P., Huang A., Galati M. A., Chung J., Ramaswamy V., Irwin M. S., Aronson M., Durno C., Taylor M. D., Rechavi G., Maris J. M., Bouffet E., Hawkins C., Costello J. F., Meyn M. S., Pursell Z. F., Malkin D., Tabori U., Shlien A., Comprehensive analysis of hypermutation in human cancer. Cell 171, 1042–1056.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin C. Y., Lovén J., Rahl P. B., Paranal R. M., Burge C. B., Bradner J. E., Lee T. I., Young R. A., Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Percharde M., Bulut-Karslioglu A., Ramalho-Santos M., Hypertranscription in development, stem cells, and regeneration. Dev. Cell 40, 9–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caspersson T., Schultz J., Pentose nucleotides in the cytoplasm of growing tissues. Nature 143, 602–603 (1939). [Google Scholar]
- 8.Petermann M. L., Schneider R. M., Nuclei from normal and leukemic mouse spleen. II. The nucleic acid content of normal and leukemic nuclei. Cancer Res. 11, 485–489 (1951). [PubMed] [Google Scholar]
- 9.Sabò A., Kress T. R., Pelizzola M., de Pretis S., Gorski M. M., Tesi A., Morelli M. J., Bora P., Doni M., Verrecchia A., Tonelli C., Fagà G., Bianchi V., Ronchi A., Low D., Müller H., Guccione E., Campaner S., Amati B., Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shlien A., Raine K., Fuligni F., Arnold R., Nik-Zainal S., Dronov S., Mamanova L., Rosic A., Ju Y. S., Cooke S. L., Ramakrishna M., Papaemmanuil E., Davies H. R., Tarpey P. S., Van Loo P., Wedge D. C., Jones D. R., Martin S., Marshall J., Anderson E., Hardy C., Barbashina V., Aparicio S. A. J. R., Sauer T., Garred Ø., Vincent-Salomon A., Mariani O., Boyault S., Fatima A., Langerød A., Borg Å., Thomas G., Richardson A. L., Børresen-Dale A.-L., Polyak K., Stratton M. R., Campbell P. J., Direct transcriptional consequences of somatic mutation in breast cancer. Cell Rep. 16, 2032–2046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovén J., Orlando D. A., Sigova A. A., Lin C. Y., Rahl P. B., Burge C. B., Levens D. L., Lee T. I., Young R. A., Revisiting global gene expression analysis. Cell 151, 476–482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium , Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zack T. I., Schumacher S. E., Carter S. L., Cherniack A. D., Saksena G., Tabak B., Lawrence M. S., Zhang C.-Z., Wala J., Mermel C. H., Sougnez C., Gabriel S. B., Hernandez B., Shen H., Laird P. W., Getz G., Meyerson M., Beroukhim R., Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwé H., Pircher A., Van den Eynde K., Weynand B., Verbeken E., De Leyn P., Liston A., Vansteenkiste J., Carmeliet P., Aerts S., Thienpont B., Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P., The molecular signatures database hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mossmann D., Park S., Hall M. N., mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 18, 744–757 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Taniguchi K., Karin M., NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 18, 309–324 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Peters J. M., Shah Y. M., Gonzalez F. J., The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat. Rev. Cancer 12, 181–195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaub F. X., Dhankani V., Berger A. C., Trivedi M., Richardson A. B., Shaw R., Zhao W., Zhang X., Ventura A., Liu Y., Ayer D. E., Hurlin P. J., Cherniack A. D., Eisenman R. N., Bernard B., Grandori C.; Cancer Genome Atlas Network , Pan-cancer alterations of the MYC oncogene and its proximal network across the Cancer Genome Atlas. Cell Syst. 6, 282–300.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Percharde M., Wong P., Ramalho-Santos M., Global hypertranscription in the mouse embryonic germline. Cell Rep. 19, 1987–1996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng F. Y., Brenner J. C., Hussain M., Chinnaiyan A. M., Molecular pathways: Targeting ETS gene fusions in cancer. Clin. Cancer Res. 20, 4442–4448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherniack A. D., Shen H., Walter V., Stewart C., Murray B. A., Bowlby R., Hu X., Ling S., Soslow R. A., Broaddus R. R., Zuna R. E., Robertson G., Laird P. W., Kucherlapati R., Mills G. B.; Cancer Genome Atlas Research Network, Weinstein J. N., Zhang J., Akbani R., Levine D. A., Integrated molecular characterization of uterine carcinosarcoma. Cancer Cell 31, 411–423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yarchoan M., Hopkins A., Jaffee E. M., Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman A. M., Kato S., Bazhenova L., Patel S. P., Frampton G. M., Miller V., Stephens P. J., Daniels G. A., Kurzrock R., Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 16, 2598–2608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Allen E. M., Miao D., Schilling B., Shukla S. A., Blank C., Zimmer L., Sucker A., Hillen U., Foppen M. H. G., Goldinger S. M., Utikal J., Hassel J. C., Weide B., Kaehler K. C., Loquai C., Mohr P., Gutzmer R., Dummer R., Gabriel S., Wu C. J., Schadendorf D., Garraway L. A., Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugo W., Zaretsky J. M., Sun L., Song C., Moreno B. H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., Seja E., Lomeli S., Kong X., Kelley M. C., Sosman J. A., Johnson D. B., Ribas A., Lo R. S., Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riaz N., Havel J. J., Makarov V., Desrichard A., Urba W. J., Sims J. S., Hodi F. S., Martín-Algarra S., Mandal R., Sharfman W. H., Bhatia S., Hwu W.-J., Gajewski T. F., Slingluff C. L., Chowell D., Kendall S. M., Chang H., Shah R., Kuo F., Morris L. G. T., Sidhom J.-W., Schneck J. P., Horak C. E., Weinhold N., Chan T. A., Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D., Schilling B., Liu D., Sucker A., Livingstone E., Jerby-Arnon L., Zimmer L., Gutzmer R., Satzger I., Loquai C., Grabbe S., Vokes N., Margolis C. A., Conway J., He M. X., Elmarakeby H., Dietlein F., Miao D., Tracy A., Gogas H., Goldinger S. M., Utikal J., Blank C. U., Rauschenberg R., von Bubnoff D., Krackhardt A., Weide B., Haferkamp S., Kiecker F., Izar B., Garraway L., Regev A., Flaherty K., Paschen A., Van Allen E. M., Schadendorf D., Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao S., Wang J. R., Ji S., Yang P., Dai Y., Guo S., Montierth M. D., Shen J. P., Zhao X., Chen J., Lee J. J., Guerrero P. A., Spetsieris N., Engedal N., Taavitsainen S., Yu K., Livingstone J., Bhandari V., Hubert S. M., Daw N. C., Futreal P. A., Efstathiou E., Lim B., Viale A., Zhang J., Nykter M., Czerniak B. A., Brown P. H., Swanton C., Msaouel P., Maitra A., Kopetz S., Campbell P., Speed T. P., Boutros P. C., Zhu H., Urbanucci A., Demeulemeester J., Van Loo P., Wang W., Estimation of tumor cell total mRNA expression in 15 cancer types predicts disease progression. Nat. Biotechnol. 40, 1624–1633 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rehman S. K., Haynes J., Collignon E., Brown K. R., Wang Y., Nixon A. M. L., Bruce J. P., Wintersinger J. A., Singh Mer A., Lo E. B. L., Leung C., Lima-Fernandes E., Pedley N. M., Soares F., McGibbon S., He H. H., Pollet A., Pugh T. J., Haibe-Kains B., Morris Q., Ramalho-Santos M., Goyal S., Moffat J., O’Brien C. A., Colorectal cancer cells enter a diapause-like DTP state to survive chemotherapy. Cell 184, 226–242.e21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonda T. J., Ramsay R. G., Directly targeting transcriptional dysregulation in cancer. Nat. Rev. Cancer 15, 686–694 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Kwiatkowski N., Zhang T., Rahl P. B., Abraham B. J., Reddy J., Ficarro S. B., Dastur A., Amzallag A., Ramaswamy S., Tesar B., Jenkins C. E., Hannett N. M., McMillin D., Sanda T., Sim T., Kim N. D., Look T., Mitsiades C. S., Weng A. P., Brown J. R., Benes C. H., Marto J. A., Young R. A., Gray N. S., Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W., Ge H., Jiang Y., Huang R., Wu Y., Wang D., Guo S., Li S., Wang Y., Jiang H., Cheng J., Combinational therapeutic targeting of BRD4 and CDK7 synergistically induces anticancer effects in head and neck squamous cell carcinoma. Cancer Lett. 469, 510–523 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Wang J., Li Z., Mei H., Zhang D., Wu G., Zhang T., Lin Z., Antitumor effects of a covalent cyclin-dependent kinase 7 inhibitor in colorectal cancer. Anticancer Drugs 30, 466–474 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Chow P. M., Liu S. H., Chang Y. W., Kuo K. L., Lin W. C., Huang K. H., The covalent CDK7 inhibitor THZ1 enhances temsirolimus-induced cytotoxicity via autophagy suppression in human renal cell carcinoma. Cancer Lett. 471, 27–37 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z., Peng H., Wang X., Yin X., Ma P., Jing Y., Cai M.-C. C., Liu J., Zhang M., Zhang S., Shi K., Gao W.-Q. Q., Di W., Zhuang G., Preclinical efficacy and molecular mechanism of targeting CDK7-dependent transcriptional addiction in ovarian cancer. Mol. Cancer Ther. 16, 1739–1750 (2017). [DOI] [PubMed] [Google Scholar]
- 37.McDermott M. S. J. J., Sharko A. C., Munie J., Kassler S., Melendez T., Lim C. U., Broude E. V., CDK7 inhibition is effective in all the subtypes of breast cancer: Determinants of response and synergy with EGFR inhibition. Cell 9, 638 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong L., Yang S., Jia Y., Lei K., Inhibition of cyclin-dependent kinase 7 suppresses human hepatocellular carcinoma by inducing apoptosis. J. Cell. Biochem. 119, 9742–9751 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Wang C., Jin H., Gao D., Wang L., Evers B., Xue Z., Jin G., Lieftink C., Beijersbergen R. L., Qin W., Bernards R., A CRISPR screen identifies CDK7 as a therapeutic target in hepatocellular carcinoma. Cell Res. 28, 690–692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng W., Wang J., Wang B., Liu F., Li M., Zhao Y., Zhang C., Li Q., Chen J., Zhang L., Tang Y., Ma J., CDK7 inhibition is a novel therapeutic strategy against GBM both in vitro and in vivo. Cancer Manag. Res. 10, 5747–5758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu P., Geng J., Zhang L., Wang Y., Niu N., Fang Y., Liu F., Shi J., Zhang Z. G., Sun Y. W., Wang L. W., Tang Y., Xue J., THZ1 reveals CDK7-dependent transcriptional addictions in pancreatic cancer. Oncogene 38, 3932–3945 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Greenall S. A., Lim Y. C., Mitchell C. B., Ensbey K. S., Stringer B. W., Wilding A. L., O’Neill G. M., McDonald K. L., Gough D. J., Day B. W., Johns T. G., Cyclin-dependent kinase 7 is a therapeutic target in high-grade glioma. Oncogenesis 6, e336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong S., Zhang Y., Yin X., Di W., CDK7 inhibitor suppresses tumor progression through blocking the cell cycle at the G2/M phase and inhibiting transcriptional activity in cervical cancer. Onco. Targets. Ther. 12, 2137–2147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iniguez A. B., Stolte B., Wang E. J., Conway A. S., Alexe G., Dharia N. V., Kwiatkowski N., Zhang T., Abraham B. J., Mora J., Kalev P., Leggett A., Chowdhury D., Benes C. H., Young R. A., Gray N. S., Stegmaier K., EWS/FLI confers tumor cell synthetic lethality to CDK12 inhibition in Ewing sarcoma. Cancer Cell 33, 202–216.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang L., Schlesinger F., Davis C. A., Zhang Y., Li R., Salit M., Gingeras T. R., Oliver B., Synthetic spike-in standards for RNA-seq experiments. Genome Res. 21, 1543–1551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter S. L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P. W., Onofrio R. C., Winckler W., Weir B. A., Beroukhim R., Pellman D., Levine D. A., Lander E. S., Meyerson M., Getz G., Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang A., Ho C. S. W., Ponzielli R., Barsyte-Lovejoy D., Bouffet E., Picard D., Hawkins C. E., Penn L. Z., Identification of a novel c-Myc protein interactor, JPO2, with transforming activity in medulloblastoma cells. Cancer Res. 65, 5607–5619 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Rasmussen M., Sundström M., Kultima H. G., Botling J., Micke P., Birgisson H., Glimelius B., Isaksson A., Allele-specific copy number analysis of tumor samples with aneuploidy and tumor heterogeneity. Genome Biol. 12, R108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J., Lichtenberg T., Hoadley K. A., Poisson L. M., Lazar A. J., Cherniack A. D., Kovatich A. J., Benz C. C., Levine D. A., Lee A. V., Omberg L., Wolf D. M., Shriver C. D., Thorsson V.; Cancer Genome Atlas Research Network, Hu H., An integrated TCGA Pan-Cancer Clinical Data Resource to drive high-quality survival outcome analytics. Cell 173, 400–416.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grömping U., R package relaimpo: Relative importance for linear regression. J. Stat. Softw. 17, 139–147 (2006). [Google Scholar]
- 51.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hänzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pavlova N. N., Thompson C. B., The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hay N., Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 16, 635–649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malta T. M., Sokolov A., Gentles A. J., Burzykowski T., Poisson L., Weinstein J. N., Kamińska B., Huelsken J., Omberg L., Gevaert O., Colaprico A., Czerwińska P., Mazurek S., Mishra L., Heyn H., Krasnitz A., Godwin A. K., Lazar A. J.; Cancer Genome Atlas Research Network, Stuart J. M., Hoadley K. A., Laird P. W., Noushmehr H., Wiznerowicz M., Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173, 338–354.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ben-Porath I., Thomson M. W., Carey V. J., Ge R., Bell G. W., Regev A., Weinberg R. A., An embryonic stem cell–like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong D. J., Liu H., Ridky T. W., Cassarino D., Segal E., Chang H. Y., Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2, 333–344 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer N. P., Schmid P. R., Berger B., Kohane I. S., A gene expression profile of stem cell pluripotentiality and differentiation is conserved across diverse solid and hematopoietic cancers. Genome Biol. 13, R71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim J., Woo A. J., Chu J., Snow J. W., Fujiwara Y., Kim C. G., Cantor A. B., Orkin S. H., A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143, 313–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan X., Ma L., Yi D., Yoon J.-g., Diercks A., Foltz G., Price N. D., Hood L. E., Tian Q., A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc. Natl. Acad. Sci. U.S.A. 108, 1591–1596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han H., Shim H., Shin D., Shim J. E., Ko Y., Shin J., Kim H., Cho A., Kim E., Lee T., Kim H., Kim K., Yang S., Bae D., Yun A., Kim S., Kim C. Y., Cho H. J., Kang B., Shin S., Lee I., TRRUST: A reference database of human transcriptional regulatory interactions. Sci. Rep. 5, 11432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lachmann A., Xu H., Krishnan J., Berger S. I., Mazloom A. R., Ma’ayan A., ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26, 2438–2444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang C., Xuan Z., Zhao F., Zhang M. Q., TRED: A transcriptional regulatory element database, new entries and other development. Nucleic Acids Res. 35, D137–D140 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.The ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., Wingender E., TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao Q., Liang W.-W., Foltz S. M., Mutharasu G., Jayasinghe R. G., Cao S., Liao W.-W., Reynolds S. M., Wyczalkowski M. A., Yao L., Yu L., Sun S. Q.; Fusion Analysis Working Group; Cancer Genome Atlas Research Network, Chen K., Lazar A. J., Fields R. C., Wendl M. C., Van Tine B. A., Vij R., Chen F., Nykter M., Shmulevich I., Ding L., Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 23, 227–238.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.López-Ratón M., Rodríguez-Álvarez M. X., Suárez C. C., Sampedro F. G., OptimalCutpoints: An R package for selecting optimal cutpoints in diagnostic tests. J. Stat. Softw. 61, 1–36 (2014). [Google Scholar]
- 68.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen R., Seshan V. E., FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smid M., Coebergh van den Braak R. R. J., van de Werken H. J. G., van Riet J., van Galen A., de Weerd V., van der Vlugt-Daane M., Bril S. I., Lalmahomed Z. S., Kloosterman W. P., Wilting S. M., Foekens J. A., IJzermans J. N. M.; MATCH study group, Martens J. W. M., Sieuwerts A. M., Gene length corrected trimmed mean of M-values (GeTMM) processing of RNA-seq data performs similarly in intersample analyses while improving intrasample comparisons. BMC Bioinformatics 19, 236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Data S1 to S6